Abstract
Enveloped viruses mature by budding at cellular membranes. It has been generally thought that this process is driven by interactions between the viral transmembrane proteins and the internal virion components (core, capsid, or nucleocapsid). This model was particularly applicable to alphaviruses, which require both spike proteins and a nucleocapsid for budding. However, genetic studies have clearly shown that the retrovirus core protein, i.e., the Gag protein, is able to form enveloped particles by itself. Also, budding of negative-strand RNA viruses (rhabdoviruses, orthomyxoviruses, and paramyxoviruses) seems to be accomplished mainly by internal components, most probably the matrix protein, since the spike proteins are not absolutely required for budding of these viruses either. In contrast, budding of coronavirus particles can occur in the absence of the nucleocapsid and appears to require two membrane proteins only. Biochemical and structural data suggest that the proteins, which play a key role in budding, drive this process by forming a three-dimensional (cage-like) protein lattice at the surface of or within the membrane. Similarly, recent electron microscopic studies revealed that the alphavirus spike proteins are also engaged in extensive lateral interactions, forming a dense protein shell at the outer surface of the viral envelope. On the basis of these data, we propose that the budding of enveloped viruses in general is governed by lateral interactions between peripheral or integral membrane proteins. This new concept also provides answers to the question of how viral and cellular membrane proteins are sorted during budding. In addition, it has implications for the mechanism by which the virion is uncoated during virus entry.
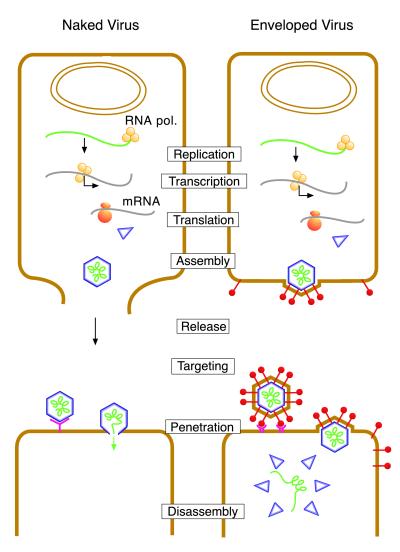
Enveloped viruses possess a membrane that surrounds the nucleocapsid (NC) or core. The membrane is acquired at a late stage of virus assembly, which is referred to as budding (55). In this process, the NC becomes progressively wrapped in a cellular membrane that is modified by virus-specific envelope proteins. Budding ultimately results in pinching off of the virion from the host membrane, thereby releasing the virus into the extracellular space. Some viruses bud at the plasma membrane (PM), whereas others are assembled at intracellular membranes along the secretory pathway. Envelope proteins serve to target the virion to specific receptors at the surface of uninfected cells (291). They also direct virus penetration into cells by inducing fusion between the viral and the host cell membrane, either at the PM or in endosomes (284). The budding and entry processes of an enveloped virus are schematically shown in Fig. 1. For comparison, the release and entry processes of naked viruses are illustrated as well. These viruses are released from the infected cell by the disruption of the PM. Some naked viruses, e.g., adenoviruses, penetrate the cytoplasm of an uninfected cell through lysis of the endosomal membrane (96, 198, 242) whereas others, e.g., picornaviruses, may form a genome-conducting channel in the membrane (205, 211). Thus, the budding and fusion reactions offer enveloped viruses the advantage to exit and enter cells without disrupting the cellular membrane barriers.
FIG. 1.
Replication of naked and enveloped viruses in eucaryotic cells.
In this review, we discuss the mechanisms underlying virus budding. Important questions include (i) the viral proteins involved in budding, (ii) the kind of protein-protein and protein-lipid interactions that drive budding, (iii) the nature of these interactions, (iv) how these interactions are controlled in virus budding and penetration, (v) whether host proteins play a role in budding, and (vi) how viral and host proteins are sorted in the formation of the envelope. All these questions are dealt with in this review. A concise overview of novel technology that has been applied to address these questions is presented as well. We discuss neither the synthesis of the viral proteins nor their intracellular transport to the site of assembly in the cell. Encapsidation of the genome is also outside the scope of this review. In addition, we do not deal with the budding of large and complicated enveloped DNA viruses, e.g., vaccinia virus and herpesvirus. For these and other aspects of virus assembly, the reader is referred to other reviews (17, 29, 55, 87, 99, 113, 127, 207, 235, 257, 285, 289).
GENERAL FEATURES OF ENVELOPED VIRUSES
Most of the new and interesting data about the budding of simple animal viruses have been obtained with alphaviruses (e.g., Semliki Forest virus [SFV], Sindbis virus [SIN], and Ross River virus [RRV]), coronaviruses (e.g., mouse hepatitis virus [MHV]), retroviruses (e.g., human immunodeficiency virus [HIV], simian immunodeficiency virus [SIV], Rous sarcoma virus [RSV], Mason-Pfizer monkey virus [MPMV], and murine leukemia virus [MLV]), rhabdoviruses (e.g., rabies virus and vesicular stomatitis virus [VSV]), paramyxoviruses (e.g., Newcastle disease virus and Sendai virus), orthomyxoviruses (e.g., influenza A virus), and hepadnaviruses (e.g., hepatitis B virus [HBV] and duck hepatitis virus). Before discussing these data, we introduce some basic features of the listed viruses.
Alphavirus particles (diameter, ∼70 nm) contain an NC that consists of a positive, single-stranded RNA genome complexed with several copies of capsid (C) protein (257). The membrane of the virion is covered with spikes. These consist of clusters of three heterodimers that in turn are composed of two transmembrane proteins, E1 and E2. Virus maturation occurs by budding at the PM.
Coronavirus particles (diameter, 80 to 120 nm) contain a long, positive, single-stranded RNA genome that is complexed with NC protein into an NC (55, 251). The membrane typically contains large spikes that consist of homo-oligomers of the transmembrane S protein. In addition, the envelope harbors an abundant membrane protein (M) and a few copies of the small envelope protein (E). The M protein traverses the membrane three times and is predominantly embedded in the envelope. Virus budding occurs intracellularly at membranes of the intermediate compartment between the endoplasmic reticulum (ER) and the Golgi complex. Newly assembled virions are transported by vesicular transport to the cell surface, where they are released via exocytosis.
Retrovirus particles (diameter, 100 to 130 nm) contain an internal core that is made of Gag and Gag-Pol protein enclosing two copies of a positive, single-stranded RNA genome (113, 289). The viral membrane accommodates trimeric clusters of the Env protein (34). Virus budding occurs at the PM. Characteristically, the cores of HIV, SIV, RSV, and MLV are made concomitantly with budding (type C morphogenesis) whereas the core of MPMV is formed in the cytoplasm before this process (type D morphogenesis). Gag and Gag-Pol molecules are proteolytically processed during and/or shortly after budding. The Gag cleavage products include a matrix (MA) protein associated with the viral membrane, a capsid (CA) protein forming the shell of the core, and an NC protein complexed with the genome inside the core. The Gag-Pol cleavage products also include three enzymes that are required for proteolytic cleavage, DNA synthesis, and integration (145).
Rhabdoviruses, orthomyxoviruses, and paramyxoviruses have a negative, single-stranded RNA genome which is either linear (rhabdoviruses and paramyxoviruses) or distributed over eight separate segments (orthomyxovirus) (55, 125, 130, 274). Rhabdovirus particles are bullet shaped (180 nm long by 75 nm wide), whereas orthomyxovirus and paramyxovirus particles are spherical (diameter, 80 to 120 and 150 to 350 nm, respectively). The genomes are packaged with nucleoprotein (NP) into ribonucleoprotein cores (RNPs, sometimes called NCs). Associated with the RNP are the polymerase proteins, which are necessary to initiate replication and transcription early in infection. A common feature of these viruses is the presence of a so-called matrix protein (M or M1) lining the inner face of the viral membrane. The transmembrane proteins of rhabdoviruses (G protein), orthomyxoviruses (hemagglutinin protein [HA] and neuraminidase protein [NA]), and paramyxoviruses (hemagglutinin-neuraminidase protein [HN] and fusion protein [F]) all form homo-oligomeric complexes or spikes. RNA replication and transcription take place in the cytoplasm (paramyxoviruses and rhabdoviruses) or in the nucleus (orthomyxoviruses). All of these negative-strand RNA viruses are assembled at the PM.
Hepadnavirus virions (also called Dane particles in HBV) measure about 42 nm in diameter and contain a circular, partially double-stranded DNA genome (80, 81). Replication starts in the nucleus and involves transcription of the genome into an RNA molecule. This so-called pregenome is encapsidated by NC protein into an NC. Within this structure, the RNA is reverse transcribed and polymerized into partially double-stranded DNA. The envelope accommodates three different but related membrane proteins: the small (S), middle (M), and large (L) proteins. These are synthesized from a common coding unit by using different sites for initiation of translation. Hepadnavirus assembly takes place intracellularly by budding at membranes of the ER. Like coronavirions, hepadnavirus particles use the constitutive secretory pathway to exit the cell.
Note that the description of subviral structures differs between viruses. For instance, surface protein is called spike or Env protein and the internal structure of the virion is referred to as NC, RNP, or core.
NEW TECHNIQUES IN RESEARCH ON BUDDING
Progress in research on the mechanisms of virus budding has been dependent on the development of advanced technology. One important breakthrough has been the construction of cDNA clones of viral RNA genomes from which infectious virus can be expressed. This has opened the possibility of using reverse genetics for testing several hypotheses about virus assembly and entry mechanisms. The first infectious cDNA clones were developed for alphaviruses (46, 131, 149, 220). The complete genomes of these positive-strand RNA viruses have been reverse transcribed and polymerized into double-stranded DNA and subcloned into transcription vectors. From these vectors, replication-competent RNA can be transcribed in vitro. When cells are transfected with the RNA, infectious virus is generated. The system can be used to study the assembly phenotypes of both viable and nonviable mutants of alphaviruses.
The generation of negative-strand RNA viruses from cloned cDNA is more complicated; it requires both the viral transcriptase and a structural framework in the form of an RNP (42). The first important step towards an expression system for these viruses was made by Palese and colleagues (160). They showed that a recombinant influenza virus RNA segment that had been transcribed in vitro from cDNA and complexed with polymerase and NP proteins into RNP was able to replicate when introduced by transfection into cells that were infected with helper virus. Moreover, the recombinant RNP was found to be incorporated into progeny virus (the so-called transfectant), demonstrating the functionality of the method. However, a major problem with the influenza virus expression system is that the eight genomic RNA segments have to be coexpressed for virus production. Therefore, this system has been limited to the generation of helper virus-recombinant RNP reassortants that can be selected with, for example, antibodies or neuraminadase. Virus with lethal mutations in the structural protein genes cannot be generated.
In contrast, for rhabdoviruses, which contain a nonsegmented negative-sense RNA genome, it is possible to recover infectious virus from recombinant DNA in the absence of helper virus. This requires the transcription of full-length genomic cDNA into positive-sense RNA that is complementary to the genome. When coexpressed in cells with the NP and polymerase proteins, the RNA transcripts are packaged into RNPs that can start an infectious cycle resulting in the generation of recombinant virus. In principle, nonviable virus with mutations in the structural genes can be produced if the corresponding wild-type (wt) genes are expressed in trans. This system was first developed by Conzelmann and colleagues for rabies virus (238) and is now also available for VSV (137, 283). Recently, similar expression systems have been developed for several paramyxoviruses including measles virus (212), Sendai virus (82, 122), human parainfluenza virus (58, 109), rinderpest virus (9), and simian virus 5 (103).
It has been straightforward to recover retroviruses and hepadnavirus from cloned DNA, since both viruses use DNA templates in the replication of their genomes. Transfection of cloned genomic DNA into susceptible cells results in the production of infectious virus. However, the expression level is low compared to that of alphaviruses and rhabdoviruses. Therefore, efficient heterologous expression systems, like those based on SFV, baculovirus, and vaccinia virus, have been used to increase the production of these viruses in tissue culture cells (101, 146, 294).
Another important new technique is cryoelectron microscopy (cryo-EM) in combination with computer-aided image processing to determine the structure of a virion (5, 54, 74). Among enveloped viruses, alphaviruses are particularly appropriate for this kind of analysis (36, 73, 195, 272). Alphavirus particles are easily purified and have a homogeneous protein composition, and the envelope and NC both display an icosahedral symmetry. With conventional electron microscopes, the resolution obtained is about 20 Å. This can be improved by using instruments with highly coherent electron beams (200- to 300-kV field emission gun electron microscope). Furthermore, it is possible to determine the high-resolution structure of the complete virion if the cryo-EM analysis data are combined with data from the X-ray crystallographic or nuclear magnetic resonance spectroscopy solution structure determination of the individual viral proteins (36, 37). In the latter approach, the atomic structures of the viral proteins are fitted into the EM density map of the virion.
PROTEIN COMPONENTS OF THE BUDDING APPARATUS
Early Model for Budding
An attractive model for budding was suggested in 1974 (84); it was based on studies with SFV showing that the spikes are composed of transmembrane proteins. It was assumed that the cytoplasmic domain (tail) of the spike protein undergoes interactions with the NC. Accordingly, budding is initiated by the association of the NC with a few spikes in the membrane. Due to its reduced mobility, the initial NC-spike complex stimulates the binding of more spikes to the NC. The NC-spike association subsequently induces the membrane to curve and to eventually enwrap the NC completely.
This model is very illustrative; it explains how an NC becomes surrounded by a membrane and how it chooses the budding site by selective binding to the spikes. Furthermore, a budding mechanism based on specific interactions between the NC and the spikes explains how it is possible to form a virus particle with a homogeneous protein composition. Because the spikes of all enveloped viruses were later shown to consist of transmembrane proteins, the NC-spike interaction was widely believed to drive virus budding in general. The model reveals a fundamental prediction that both the NC and spike proteins are required for budding and particle release.
Budding of Retrovirus Particles Requires Core Proteins Only
In 1989, the “early” budding model was clearly shown not to apply to HIV-1 and SIV, because it was found that the Env protein is dispensable for budding of these viruses (48, 90). This was demonstrated by independent expression of the gag gene in insect cells with a baculovirus vector. The expression resulted in the production of enveloped “Gag” particles which strongly resembled immature virions. By using various heterologous and homologous expression systems, it was subsequently shown that the Gag protein constitutes the budding apparatus of all retroviruses (178, 217, 290). This finding emphasizes the importance of earlier studies showing that certain defective retroviruses direct the formation of particles that do not contain Env (124, 208, 233, 244).
Both Nucleocapsid and Spikes Are Prerequisites for Budding of Alphavirus and Hepadnavirus
The results for retrovirus budding cast doubt on whether the “early” budding model would apply to any virus. However, studies with SFV unambiguously demonstrated the applicability of this model to alphaviruses (262). This was shown by expression of SFV genome variants that lacked either the spike or the C coding region. It was observed that the NCs, assembled in the cytoplasm, were not able to associate with membranes in the absence of spikes. However, when the spikes were coexpressed, the NCs were found to form spike-containing particles at the PM by budding. Furthermore, independent expression of the genome variant lacking C resulted in the production of spike protein that was unable to drive particle formation by itself. Thus, these results clearly showed that alphavirus budding is critically dependent on both internal (C) and envelope proteins (Fig. 2). Similar interpretations were made previously on the basis of studies of alphaviruses with temperature-sensitive (ts) mutations in their spike proteins (20, 232, 249).
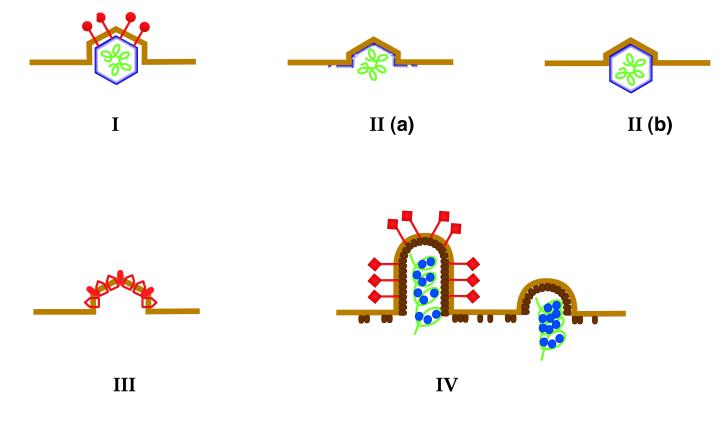
FIG. 2.
Viral proteins that drive budding. (I) Spike (red)- and NC (blue)-dependent budding of alphaviruses. (IIa) Gag protein-driven budding of a type C retrovirus. The membrane is shown in yellow, and the submembrane layer of Gag protein is depicted in blue. An RNA molecule (green) is also indicated, but it is unclear whether this is necessary for budding. (IIb) Budding of a type D retrovirus “Gag” particle. In this case, Gag molecules are assembled into a complete core in the cytoplasm prior to membrane attachment. (III) M (red, unfilled) and E (red, filled) membrane protein-driven budding of coronavirus. (IV) Rhabdovirus budding is depicted as an efficient (left) or inefficient (right) process depending on the presence of the spike proteins (red). The M protein layer below the membrane is shown in brown, and the RNP is depicted as a green helix with proteins (blue).
Expression of HBV genome variants in hepatoma cells revealed that both the NC and the membrane proteins are also required for the budding of hepadnavirus. In particular, it has been shown that NCs are not released from cells in the absence of the envelope proteins L and S (M is dispensable, however) (22). Furthermore, it has been found that the envelope proteins also exhibit an independent budding activity because they can efficiently form small (diameter, 22 nm) capsid-free membrane particles by themselves (200).
Nucleocapsid-Independent Budding of Coronavirus Envelopes
A novel budding principle was recently identified by Rottier and colleagues in a study of the assembly of coronaviruses (270). They found that coexpression of the MHV envelope protein genes (S, M, and E) from transfected cDNA resulted in the release of membrane particles morphologically indistinguishable from authentic virions. This clearly shows that the membrane proteins can assemble into virtually bona fide envelopes without the NC. Like wt virions, the membrane particles contained much M protein and very little E protein. The S protein was dispensable for particle formation but was incorporated when present. The apparent lack of a function for S in budding confirms the results of earlier studies showing that spikeless and hence noninfectious particles are released from infected cells treated with tunicamycin (110, 225). Under these conditions, the S protein is not available for assembly because the drug, which inhibits N-linked glycosylation, induces its aggregation and retention in the ER (270). Also, various ts mutants of coronaviruses carrying defects in the S gene have been reported to form particles with a spikeless phenotype (159, 219). Together, these results point to a critical role of the M and E proteins in coronavirus budding.
Matrix Protein Plays a Key Role in Rhabdovirus Budding
Studies dealing with rhabdoviruses, in particular, have given new insight into the budding of negative-strand RNA viruses. Conzelmann and colleagues have made recombinant rabies virus lacking the gene encoding the G protein by use of the cDNA expression system that they had developed previously (170, 238). They found that this mutant virus was able to form spikeless virus-like particles; however, the efficiency of particle release was about 30-fold lower than that for wt virus. This indicates that the RNP and the M protein together can mediate budding in the absence of the G protein but that the latter promotes this process remarkably. The critical role of M protein in budding is supported by earlier studies with ts mutants of VSV. For instance, cells infected with VSV tsG31, tsG33 or tsM301-303 and incubated at the nonpermissive temperature either do not produce particles or release noninfectious particles that contain reduced amounts of NP (10% of the amount in wt virus) and that have a lower density than wt VSV (240). VSV carrying ts mutations in the G gene has also been isolated (68). The best-characterized one is VSV tsO45. The G protein of this mutant virus is unstable and degraded at the restrictive temperature, resulting in the production of spikeless particles (239). For a long time, this result was taken as evidence that VSV does not require its spike protein for budding. However, this relation became unclear when it was shown later that the spikeless VSV particles did contain a full complement of fragments corresponding to the membrane-spanning and cytoplasmic parts of the G protein (175). Apparently, the tsO45 G protein is engaged in budding before degradation of its ectodomain, or, alternatively, the remnant membrane-spanning fragment is assembly competent as such.
In more recent studies, Wagner and colleagues used baculovirus and vaccinia virus vectors to express the VSV M protein in insect and mammalian cells, respectively (120, 147). The analyses showed that the M protein binds to the PM and induces slow but efficient release of M-protein-containing vesicles from cells. In the same studies, the G protein was also coexpressed with the M protein. Surprisingly, the G protein was not included in the M-protein-containing vesicles, suggesting that the RNP is required to organize the M and G proteins in such a way that they can collaborate in forming a complete particle. Collectively, the data suggest that the M protein plays a key role in the budding of rhabdoviruses but that the RNP and the G protein are actively engaged in this process as well.
Role of Matrix and Spike Proteins in Orthomyxovirus and Paramyxovirus Budding
The fact that orthomyxoviruses, paramyxoviruses, and rhabdoviruses have a layer of M protein underneath the envelope suggests that they all possess a similar mechanism for budding. The role of the spike proteins in orthomyxovirus and paramyxovirus budding, however, has been a subject of controversy. Earlier work indicated that one of the spike proteins, either HA or NA, is not required for budding of orthomyxoviruses. For instance, cells infected with influenza virus expressing a ts transport-defective HA protein release particles at the nonpermissive temperature (199). The particles were devoid of HA but contained more than normal amounts of NA. Other studies showed that an influenza virus variant lacking the NA gene was able to mature at the PM, although the released virions were found to aggregate heavily (152, 153). Nevertheless, recent discoveries point to a crucial role of the cytoplasmic portions of these spike proteins in virus maturation. By using reverse genetics, it has been possible to obtain influenza virus transfectants containing either NA or HA protein lacking their cytoplasmic tails (116, 176). Deletion of the tail of HA slightly affected its incorporation into the envelope and modestly lowered the efficiency of budding but had no effect on virion morphology. However, virus lacking the tail of NA exhibited a tendency to form long, filamentous particles rather than the spherical particles seen among wt virions, and the incorporation of the NA protein into the envelope was impaired. Even more pronounced effects were observed with an influenza virus reassortant lacking the tails of both proteins (117). The double deletion rendered the virus 10-fold less infectious, and its efficiency of budding, compared to that of control virus possessing intact membrane proteins, was decreased to the same extent. Most strikingly, the mutant virions formed a population of very irregularly shaped particles with a greatly increased length, clearly distinguishable from wt virions and from the ones lacking only one of the spike protein tails. Thus, these data suggest that the cytoplasmic portions of the spike proteins are required for efficient budding and that they are necessary for the formation of uniform, spherical particles. Nevertheless, it seems that the spikes are not absolutely required for budding. Therefore, it is likely that orthomyxovirus budding is, like that of rhabdovirus, directed predominantly by the M1 protein.
The exact role of the paramyxovirus envelope proteins in budding also remains to be determined. Studies of Sendai virus and Newcastle disease virus with conditional defects in the HN and F protein, respectively, indicated that each of these proteins is dispensable for budding (166, 258). Similarly, a cold-passaged respiratory syncytial virus variant which grows efficiently in tissue culture although it lacks most of its attachment protein gene has been isolated (121). In contrast, experiments with mutant Sendai virus possessing a ts M protein have clearly shown that M plays an important role in paramyxovirus budding (300, 301).
General Conclusions about Budding Strategies
From the above data, it is evident that enveloped viruses use different kinds of protein for budding. As a synopsis of our survey of the structural components that play a key role in this process, we have classified the types of budding strategies as follows: type I, budding dependent on both capsid and spike proteins (alphavirus and hepadnavirus); type II, budding mediated by capsid or core protein only (retrovirus); type III, budding accomplished by membrane proteins only (coronavirus); and type IV, budding driven by matrix protein with the assistance of spikes and RNP (rhabdovirus and possibly paramyxovirus and orthomyxovirus). This is schematically depicted in Fig. 2.
Notably, it has been shown that recombinant SFV-driven expression of the gene encoding the VSV G protein or a truncated form of the MLV Env protein can lead to the formation of enveloped, infectious particles (138, 223, 224). These particles, also called minimal viruses, contain the respective proteins as well as the SFV replicon encoding these proteins. This phenomenon has sometimes been put forward to support a theory assuming that these viral membrane proteins themselves possess a membrane-bending potential (29, 170). However, because such minimal viruses are heterogeneous in size and are formed very inefficiently, we find it impossible to distinguish the process of their formation from that of PM-derived vesicles. The latter has also been referred to as membrane vesiculation, blebbing, or ectocytosis and seems to serve important cellular functions (141, 172, 194, 214). Thus, minimal viruses may arise accidentally if self-replicating RNA, coding for a viral membrane protein that can mediate receptor binding and membrane fusion, and the encoded protein are coincorporated into PM-derived vesicles.
MOLECULAR INTERACTIONS INVOLVED IN BUDDING
The series of events that lead to budding is governed by complex molecular interactions between the structural components. These interactions not only underlie the association of the participating molecules but also control the timing and location of the budding reaction. Moreover, they may provide the forces that drive budding. An understanding of the mechanism of budding thus requires the characterization of all intermolecular interactions that are essential in this process. In this section, we describe the features of the interactions which have been identified so far and discuss models that explain how they operate in the budding reaction.
Spike-Nucleocapsid Budding Apparatus of Alphaviruses
Organization of the membrane protein heterodimers and C protein in the virion.
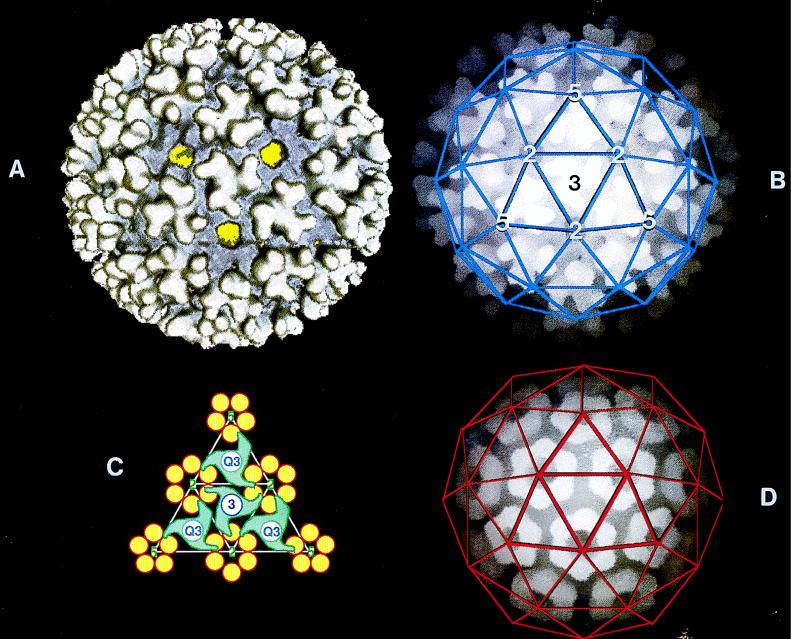
Recently, cryo-EM analysis and image processing of alphavirus particles revealed the first detailed structure of a complete enveloped virion (Fig. 3) (36). The results showed unambiguously that the envelope as well as the NC displays an icosahedral surface symmetry with a triangulation number (T) of 4. This means that both structures are composed of 240 building blocks, which in turn consist of an E1-E2 heterodimer and a C molecule, respectively. Surprisingly, the data showed that the C protein is organized in 12 pentameric and 30 hexameric capsomers whereas the E1-E2 heterodimers appear as 80 trimeric projections or spikes. In the virion, the spikes are located at the threefold and quasi-threefold symmetry axes whereas the pentameric and hexameric capsomers are situated around the fivefold and twofold axes, respectively (Fig. 3B and D).
FIG. 3.
Reconstruction of an alphavirus (RRV) particle from cryo-EM analysis and image processing. (A) Three-dimensional surface structure of the virion (diameter, ∼70 nm) viewed along an icosahedral threefold axis. The spikes ([E1-E2]3), located at the threefold and quasi-threefold axes, have a flower-like head with three bilobal petals. The spikes are engaged in extensive lateral interactions close to the lipid bilayer via their skirts. These parts of the spikes are colored bluish. The lipid bilayer (yellow) is seen through openings in the spike-skirt protein layer at the twofold and fivefold symmetry axes. (B) Depth-cued representation of the structure in panel A. Blue lines indicating the T = 4 lattice are superimposed. (C) Schematic representation of the interactions between the spikes (green) at the threefold (circled 3) or quasi-threefold (circled Q3) axes and the C molecules (yellow) of the capsomers in the underlying NC. (D) Depth-cued representation of the three-dimensional structure of the NC viewed along an icosahedral threefold axis. The COOH-terminal protease domains of the C molecules form hexameric and pentameric capsomers at the twofold and fivefold axes. Red lines indicating the T = 4 lattice are superimposed. Modified from reference 36 with permission of the publisher.
The way the spikes and capsomers communicate with each other is most interesting. The analyses showed that the three heterodimers of each spike diverge above the external surface of the viral membrane, traverse the lipid bilayer individually, and interact with three underlying C molecules that belong to three separate capsomers (Fig. 3C). Thus, the spikes at the quasi-threefold axes interact with C molecules of one pentameric capsomer and two neighboring hexameric capsomers whereas the spikes at the threefold axes interact with C molecules of three neighboring hexameric capsomers. This creates a complex network of molecular interactions in which the spike-C interactions not only mediate the binding of the NC to the spikes but also stabilize the connections between the capsomers.
Furthermore, the cryo-EM analyses revealed that the skirts of the heterodimeric subunits of neighboring spikes entertain extensive lateral interactions with each other around the icosahedral two- and fivefold axes (Fig. 3A). Thus, each membrane protein heterodimer is involved in a trimeric structure (i.e., the projecting part of the spike) as well as in a hexameric or pentameric structure located between the projections, around the two- or fivefold axes. The existence of spike-spike interactions had been proposed much earlier on the basis of EM analysis of NC-free viral envelopes (273). These envelopes, which were prepared by treating virus particles with small amounts of a mild detergent, displayed a symmetrical spike lattice similar to that of intact virus.
Structural features of the E2 tail-C binding.
The molecular domains engaged in the spike-C interaction have been mapped by a combination of different techniques. The C protein of SIN has been crystallized, and the structure of its COOH-terminal protease domain has been resolved at high resolution by X-ray diffraction (38). Because the amino acid sequence of this domain is highly conserved among various alphaviruses, it was possible to fit the protease structure of the SIN C protein into the cryo-EM-derived electron density map of the capsomer monomers in the RRV NC (36). This has revealed the part of the C protein surface which is exposed toward the tail domains of the spike. Molecular modelling studies have been performed to identify the amino acid residues involved in spike-C interactions. For this purpose, a structure model of a portion of the 31 amino acid residues of the SFV E2 protein tail has been used to find a binding site in the C protein (247). A region around Tyr residue 399 of the E2 tail was chosen for modelling because earlier data, especially genetic data, suggested that the aromatic side chain of this amino acid residue represents a central element in the spike-NC interaction (11, 77, 114, 157, 174, 307). The structure model for this peptide was obtained by analyzing similar peptides that were present in proteins whose structures had previously been determined. Subsequent searching for a binding site at the surface of the C protein resulted in the identification of a cavity that could accommodate Tyr 399 and its flanking regions. The side chain of Tyr 399 is predicted to interact with those of Tyr 184 and Trp 251 of the C protein in this cavity. Therefore, it has been postulated that these aromatic interactions underlie the binding between the NC and the spikes (247).
It is also noteworthy that an anti-idiotype antibody approach has been used to reconstruct the C-E2 interaction in SFV (269). In this study, an anti-idiotype antibody was raised against an E2 tail-specific (idiotype) antibody. The anti-idiotype antibody was claimed to react with the C protein, suggesting that the E2 tail is indeed involved in interactions with the C protein. However, this conclusion appeared unjustified, since it was discovered later that this antibody reacted with the C protein nonspecifically and that it was instead directed against another antigen present in viral RNA replication centers inside the infected cell (261).
Interactions between E1-E2 heterodimers and their role in budding.
Although the E1-E2 heterodimers are obviously involved in multiple kinds of lateral interactions to form spike, hexameric, or pentameric structures, no information about the nature of these interactions is available. The solution to this problem awaits, above all, determination of the three-dimensional structure of the E1-E2 heterodimer. It is also unclear whether E1-E2 heterodimers bind to the NC during budding individually or whether they form multimeric complexes (e.g., trimers or hexamers) beforehand. This question has remained open mainly because the higher-order forms of the heterodimers are unstable under mild-detergent conditions (221, 311, 312). Therefore, it has not been possible to investigate biochemically the stage during virus assembly at which the complexes of the heterodimers are formed. Nevertheless, membrane protein complexes larger than heterodimers have been detected by chemical cross-linking in SIN-infected cells (180).
The lateral interactions between the heterodimers in the viral membrane probably facilitate budding in two ways. First, they result in the formation of larger membrane protein units displaying a polyvalent NC-binding site. Second, these interactions most probably assist in the formation of the icosahedral lattice of membrane protein heterodimers and may thereby provide a force for membrane bending. The role of a polyvalent NC-binding site of a heterodimer cluster in budding is supported by the results of several genetic studies. In one study, a wt-like, budding-competent virus was expressed in cells together with a budding-incompetent variant in which the NC-binding site in the cytoplasmic tail of E2 had been knocked out (65). It was found that the E1-E2 heterodimers with a defective NC-binding site prevented the wt-like heterodimers from participating in virus budding in a concentration-dependent manner. This suggests that wt and mutant heterodimers were preassembled into clusters that were mostly incompetent for NC binding. Presumably, only clusters displaying a (nearly) full complement of intact binding sites can stably associate with the NC. In another study, the phenotype of an SFV variant lacking the E1 gene has been characterized (10). It was observed that E2 is able to reach the cell surface efficiently by itself; however, the NCs failed to bind to the E2 protein at the PM under these conditions. A possible explanation for this result is that separately expressed E2 does not oligomerize into structures exposing a multivalent binding site for the NC.
There is still no direct experimental evidence for a membrane-bending effect of the lateral interactions between the membrane protein heterodimers. However, there are several SIN heterodimer ectodomain mutants and a SIN chimera containing E1 from RRV which have defects in the budding process, although apparently normal heterodimers are expressed at the PM. These phenotypes can be explained by the mutations (or chimeric heterodimer) affecting interactions in the spike projection or in the skirt region, with the result that they cannot support the bending of the membrane (20, 102, 150, 256, 295, 296).
Preassembled NCs are not required for alphavirus budding.
The earliest EM examinations of alphavirus-infected cells revealed the abundant presence of cytoplasmic NCs as free particles, groups of particles, and part of the budding structures at the PM (1). Therefore, it has been postulated that the preassembled NC represents a pivotal element of the budding mechanism of this virus (19, 181, 246, 257). This theory was strengthened by biochemical analyses showing that the majority of the cell-associated C protein is part of fully assembled NCs (250). Surprisingly, it has recently been found that preassembled NCs are not required for the budding of SFV (69). A small deletion in the C gene, which did not affect the efficiency of budding, completely abolished the preformation of cytoplasmic NCs. Apparently, assembly of the mutant NC took place concomitantly with budding at the PM. This phenotype can be explained if one assumes that the deletion in the C protein causes a weakening of the C-C interactions so that stable NCs cannot be formed in the cytoplasm. As a result, the assembly of the mutant NC requires spike-C interactions, which decrease the mobility of the C molecules (or complexes), thereby stabilizing the C-C interactions.
Refined model for alphavirus budding.
We propose that alphavirus budding is initiated by binding of the NC to a cluster of E1-E2 heterodimers at the PM. The cluster could represent a spike-like trimeric complex or a multimer, e.g., a hexamer, that is maintained through interactions found in the skirt region of the spikes. The multivalent nature of this initial membrane protein-NC interaction is strong enough to maintain the resulting complex until it is consolidated by the binding of additional (clusters of) E1-E2 heterodimers. The latter are recruited into the complex by cooperative heterodimer-heterodimer and heterodimer-C interactions. This process continues until envelopment is completed.
It is likely that the establishment of the icosahedral lattice of E1-E2 heterodimers at the surface of the virion directly promotes membrane bending. However, because the membrane proteins are unable to support budding on their own, it is evident that lateral (sideward) heterodimer-heterodimer interactions and heterodimer-NC interactions cooperate in the formation of the surface lattice. At present, we do not know how much of the membrane-bending force is derived from heterodimer-heterodimer interactions and how much is derived from C-E2 interactions. It is conceivable that the former interactions provide the main force for budding. The amount of energy released from these interactions is unknown. It has been calculated that covering the hydrophobic surfaces in the cavity of the C protein and the E2 tail would correspond to 7.6 kcal mol−1 (140).
Gag Budding Apparatus of Retroviruses
Submembrane Gag lattice.
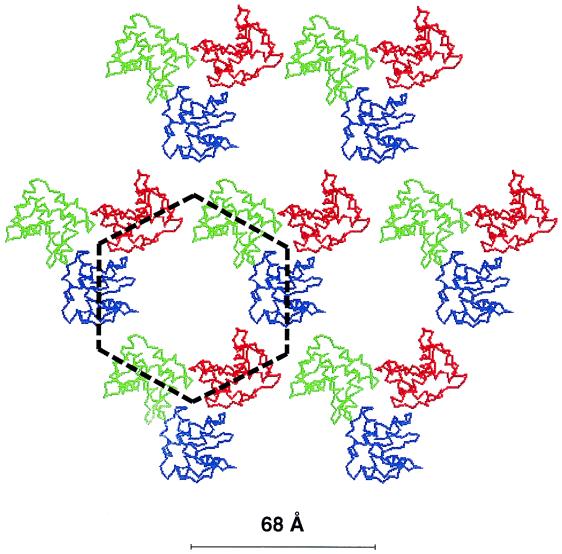
EM data of HIV-1-like “Gag” particles, produced in insect cells by using a baculovirus expression vector, suggest that the Gag molecules are arranged in an icosahedral network below the lipid bilayer (183, 184). The network seems to consist of a lattice of hexameric and pentameric Gag-rings with a T of 63. This organization of Gag molecules was also observed beneath the PM of cells producing the Gag protein (184). The structure of the Gag lattice is compatible with the crystal structures of the HIV-1 and SIV MA proteins. These demonstrated that the MA molecules formed trimers whose dimensions were such that they fit into the Gag lattice observed by EM (108, 213). The MA lattice can thus be depicted as a hexameric array in which each MA trimer donates its monomers to three hexameric rings (Fig. 4). This reconstruction model does not reveal any evident interactions between the trimers. However, it is possible that these interactions are primarily mediated by the CA domain of the Gag protein and not by the MA domain. The latter has been shown to have homo-oligomerization activities in its NH2- and COOH-terminal parts (8, 32, 49, 51, 63, 79, 112, 118, 177, 254).
FIG. 4.
MA protein trimers organized in a hexameric lattice. The MA protein monomers are shown as red, blue, and green polypeptide chain drawings. They form trimeric clusters in a hexameric lattice. One hexameric ring is indicated by the dashed line. The center-to-center distance between neighboring hexameric rings is 68 Å. Modified from reference 213 with permission.
Gag-membrane interactions.
In early studies, it was clearly demonstrated that the Gag precursor and the MA protein bind to membranes. In particular, it was shown that lipids in the envelope of avian and murine retroviruses can be coupled preferentially to the MA protein with a cross-linking reagent (204). Peptide mapping also revealed that it is the NH2-terminal part of the MA protein that associates with the lipids (203). In addition, it has been possible to extract MA protein from a murine retrovirus and to reconstitute it into artificial membranes (6).
The nature of the membrane affinity of the Gag molecule has been studied more recently by using many different systems. These include (i) the expression of Gag precursors and precursor variants in cells and the analyses of their membrane-binding and particle-forming capabilities (90, 118, 215, 271, 310), (ii) in vitro synthesis of Gag related peptides and analysis of their interaction with liposomes (309), and (iii) purification of Gag precursors (and precursor variants) and analysis of their interaction with liposomes (64). The results of these studies suggest that the Gag protein of most retroviruses possesses a bipartite membrane-binding structure in its MA domain. This consists of a myristate modification of the conserved NH2-terminal Gly residue (106) and a cluster of basic amino acid residues in the NH2-terminal part of the molecule. However, in the RSV Gag protein, which is not myristoylated, other features of its NH2-terminal part are responsible for membrane binding instead (271). The bipartite membrane-binding structure is supposed to mediate interactions with the hydrophobic interior of the lipid bilayer and with the negatively charged phospholipid molecules in the cytoplasmic leaflet of the membrane. These conclusions have been supported by the structural data on the MA protein of HIV and SIV (41, 108, 165, 167, 213). Both the X-ray crystal and nuclear magnetic resonance spectroscopy solution structures of the MA proteins revealed a globular molecule with a striking clustering of basic residues at one part of the surface. Although these studies focused on nonmyristoylated MA protein, a model has been proposed in which an MA trimer is oriented below the lipid bilayer in such a way that all three myristoyl groups and the surface patches of basic amino acid residues interact with the membrane, as suggested above (108).
Model for retrovirus budding.
In type C retroviruses, the Gag molecules most probably form complexes at the PM through cooperative Gag-membrane and Gag-Gag interactions. Such cooperation is expected on the basis of theoretical considerations (202) and is also supported by studies showing that particle formation through Gag-Gag interactions is dependent on the myristate-mediated membrane binding of the Gag protein (215, 226). Furthermore, the results of in vitro studies suggest that the Gag protein increases its membrane affinity through homo-oligomerization mediated by its CA and NC domains (60, 210). After binding to the membrane, the Gag molecules probably arrange themselves into a hexagonal lattice. Budding could then, for example, be initiated and possibly driven by the introduction of Gag pentamers in such a lattice.
Formation of the core of type D retroviruses is thought to be completed before it binds to the membrane (217). In this case, budding is supposedly driven by Gag-membrane interactions only. The driving force for budding would thus be provided by the energy released from these interactions. This corresponds to the sum of the energies obtained by burying the myristoyl groups into the hydrophobic interior of the lipid bilayer (8 kcal mol−1) and by neutralization of the basic amino acid residues by the negatively charged head groups of phospholipids (∼1.4 kcal mol−1 for each positive charge) (309).
Most interestingly, Rhee and Hunter (218) have shown that the morphogenetic pathway of MPMV, a type D retrovirus, can be switched to that of a type C retrovirus by changing a single amino acid residue in the MA domain of the Gag protein. The authors suggested that the mutation had inactivated a putative cytoplasmic retention signal in the MPMV Gag protein and, as a result, the mutant Gag molecules were directly targeted to the PM before they engaged in core assembly. Analogous to the explanation for the NC assembly mutant of SFV (69), we would like to offer the hypothesis that the MA point mutation had caused a weakening of the Gag-Gag interactions, thereby preventing the formation of stable cores in the cytoplasm, but that core assembly can still take place at the cell surface, where the Gag-Gag contacts are stabilized through Gag-membrane interactions.
Role of RNA in retrovirus budding.
Although the Gag protein can drive budding in the absence of viral genomic RNA, it may, under these conditions, interact with cellular RNA instead. In keeping with this possibility, the binding of Gag to the viral RNA, which is mediated by the NC domain, involves both specific and nonspecific interactions (45, 228, 263). Moreover, several studies have shown that foreign RNA molecules can be incorporated into retrovirus particles (40, 75, 78, 92, 143, 144, 173). Hence, it cannot be excluded that Gag-RNA interactions are involved in the budding of type C retroviruses. These interactions might, for instance, cooperate with Gag-lipid and Gag-Gag interactions in forming and curving the submembrane Gag lattice. Interestingly, it has been shown that Gag protein lacking the complete NC domain, or parts of it, can also drive the budding of particles. These particles, however, had a significantly lower density than those formed by intact Gag protein (13, 118, 119, 282). The “light” particles presumably lacked RNA, since the mutant Gag protein was most probably crippled in its ability to bind nucleic acid. Nevertheless, the change in the density of the particle is probably not caused by the lack of RNA alone, because the RNA constitutes only ∼1% of the mass of a retrovirus particle (266). Therefore, the particles may have contained a less densely packed Gag lattice due to the absence of Gag-RNA interactions. In vitro studies have also suggested a role for RNA in assembly, since it was found to facilitate the formation of cylindrical structures (diameter, 30 nm) from purified CA-NC fragments of both RSV and HIV-1 Gag proteins. These structures were sensitive to RNase, protease, and nonionic detergent, and their lengths were determined by the size of the RNA (32). Recent assembly studies dealing with p10-CA-NC fragments of the RSV Gag protein have shown the formation of predominantly spherical virus-like particles (31). This was also dependent on the addition of RNA to the reaction mixture.
Role of actin in retrovirus budding.
There are several studies suggesting the participation of the host cytoskeleton, especially actin, in the budding reaction. For instance, the HIV Gag protein is associated with polymerized actin (F actin) in cells and binds F actin in vitro (216). Furthermore, muscle and nonmuscle actin, as well as regulatory and structural actin-binding proteins, have been found in HIV preparations (3, 191). Finally, preparations of mouse mammary tumor virus have been shown to contain actin (44). In the latter study, morphological observations suggesting the involvement of actin filaments in the budding process have been reported as well. One possibility is that the Gag protein has similarities to the cytoplasmic ERM (ezrin-radixin-moesin) proteins, which link the PM to actin filaments (267), and that actin polymerization serves as an additional force for membrane bending during budding.
M-E Budding Apparatus of Coronaviruses
It has been suggested that coronavirus envelope formation is dominated by laterally interacting M molecules that form a two-dimensional lattice in intracellular membranes (187). This model is supported by biochemical analyses of M protein in cells. The MHV M protein has been found to form higher-order complexes when expressed in the absence of other coronavirus proteins (129). Interestingly, the association of M molecules could be detected only under specific detergent conditions; a mixture of nonionic (Nonidet P-40) and ionic (deoxycholic acid) detergents, or 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) alone, was found to be optimal whereas nonionic detergents (Nonidet P-40 or Triton X-100) alone failed to preserve the complexes. Multimeric complexes of the M protein also occur in infected cells and virions (186, 187). However, these complexes also contain the S protein and are maintained by both M-M and M-S interactions.
It is unknown how the M and E proteins cooperate in budding. Because there are no indications that the M protein causes bulging of the membrane by itself, it is believed that the involvement of the E protein in the M lattice is decisive in inducing curving and budding of the membrane (270).
Function of M Protein in Budding of Negative-Strand RNA Viruses
Submembrane layer of M protein.
EM analyses of several negative-strand RNA viruses have demonstrated that the M protein forms a dense layer tightly associated with the inner leaflet of the lipid bilayer (27, 182). Indeed, results of morphological examinations of the influenza virus envelope suggest that the M1 protein replaces the inner lipid leaflet completely (72). These observations fit with the results of biochemical studies of purified virions. In particular, studies with VSV have shown that membrane-reactive photoactivatable probes can be coupled preferentially to the M protein (142). In addition, treatment of rhabdovirus and paramyxovirus particles with bifunctional protein-reacting reagents results in the efficient cross-linking of M protein into homo-oligomers (56, 163).
The association of M with lipids has been directly studied in vitro with purified M protein and liposomes. The results showed that the M protein of all negative-strand viruses binds efficiently to liposomes (25, 67, 97). The interactions are hydrophobic and, at least for VSV, also electrostatic. By using fragments of the M protein, it has been possible to map the regions in the influenza virus and VSV M molecules that mediate the hydrophobic interactions (98, 297, 298). Not surprisingly, these regions were found to be enriched in hydrophobic amino acid residues. They did not, however, contain stretches of amino acid residues that are long enough to span the lipid bilayer as a typical α-helical transmembrane peptide (201). The electrostatic interaction of the VSV M protein with lipids has been shown, as in the case of retrovirus Gag proteins, to be mediated by a cluster of positively charged amino acid residues located at the NH2 terminus of the molecule (39, 298).
Recently, the crystal structure of an NH2-terminal fragment of the influenza virus M1 protein has been solved (243). The fragment, which encompasses residues 2 to 158 of the 252-residue M1 polypeptide, was found to be folded into two domains, an N (NH2-terminal) domain and an M (middle) domain, each consisting of four α-helices. The N domain contains a region with potential lipid-binding properties, and the M domain possesses a positively charged region that might interact with the viral RNA. These regions correspond roughly to the lipid and RNA binding M1 peptides mapped by biochemical experiments (see below).
The crystal structure of the influenza virus M1 protein fragment also provided some clues about how this protein associates with itself. The fragment formed dimers which, in the crystal, were stacked on one another in a way suggesting that M1 can form a polyprotein ribbon along the RNP. There are also several biochemical results suggesting the existence of interactions between M1 molecules. For instance, it has been shown that M1 protein, which had been isolated from virions by using high-salt buffer and detergent, self-associates when the salt concentration is lowered (104, 234). Furthermore, studies dealing with the VSV M protein have demonstrated that polymerization in vitro, in the presence of ZnCl2, is dependent on the formation of a nucleation site consisting of three or four M molecules (85). Finally, expression of influenza virus M1 in BHK-21 cells by using a SFV vector has shown that this protein forms homomultimers (307a). The tendency of M protein to polymerize both in vitro and in vivo might be indicative of a lattice-forming function during budding.
M-spike interactions.
Early reassociation studies with fractionated Sendai virus NCs and M and spike proteins suggested the existence of M-spike interactions. These studies revealed that the M protein is required for spike-NC complex formation (301). Later, specific M-spike interactions were identified biophysically in studies demonstrating that the oligomeric form of the VSV G protein is stabilized by the M protein (161). In addition, M-spike interactions in orthomyxoviruses and paramyxoviruses have been demonstrated by flotation analysis. Studies in which the Sendai virus HN and M genes (231) or the influenza virus HA, NA, and M genes (66) were expressed have shown that the fraction of membrane-bound M molecules clearly increases in the presence of the spikes. It should be noted, however, that the latter results could not be reproduced by other groups (128, 306).
The nature of M-spike interactions has not been solved yet for any of the negative-strand RNA viruses. Genetic data, however, suggest that the cytoplasmic domain of the rhabdovirus G protein is involved in M-spike interactions. For example, it has been shown that the HIV-1 Env protein is incorporated into the rhabdovirus envelope only when its cytoplasmic tail is replaced by that of the G protein (169, 193). The presence of an “incorporation signal” in the G-protein tail was also suggested by transcomplementation studies which revealed that at least the nine membrane-proximal amino acid residues of the tail (with a total length of 29 residues) are required for efficient incorporation of G into VSV tsO45 particles (286).
It has also been shown that the cytoplasmic tail of the G protein is required for efficient virus budding (170). A recombinant rabies virus lacking the G-protein tail was found to produce about fivefold less virus particles than the wt virus did. Intriguingly, recent studies have suggested that it is the length rather than the amino acid sequence of the G protein tail that determines the budding facilitation effect (236). For instance, a chimeric G protein carrying the tail of CD4 seemed to facilitate budding to the same extent as wt G protein did. These data are difficult to interpret and obscure our understanding of the mechanism by which the G and M proteins cooperate in budding. Interactions between G and M are even more puzzling since immuno-EM analysis of isolated VSV skeletons has suggested that the M protein resides predominantly inside the RNP and that it is exposed only at the surface of the RNPs curved extremity (7). Possibly, the interactions between the M and G proteins that facilitate budding are required only at the beginning of virus maturation, i.e., during the formation of the hemispheric part of the bullet-shaped virion.
The tails of the influenza virus HA and NA proteins are not strictly necessary for incorporation of the spikes into the viral envelope (116, 176). However, as with the VSV G protein, they seem to be involved in budding, since it has been shown that influenza virus encoding tail-less HA and NA proteins was significantly impaired in budding whereas influenza virus coding for either a complete HA or NA protein was able to bud efficiently (117). Thus, the presence of either tail appears to be sufficient for efficient budding. Clearly, further studies are also required to solve the question how the influenza virus spikes communicate with M1 during budding.
M-RNP interactions.
Binding of M protein to the RNP has been convincingly shown by treating virus particles with detergents and salt (134, 245). Treatment of virions with a mild detergent alone solubilizes the lipid bilayer and the spikes but leaves the M proteins attached to the RNP. The M proteins can be released by further treatment of the particles with high-salt buffers. For influenza virus, it has been shown that M1-RNP interactions are also sensitive to low pH (26, 308). Additional evidence for M-RNP interactions was obtained in reassembly studies with VSV, which showed that the M protein was required for condensation of the RNP (185). The M-protein extraction studies indicate that M-RNP interactions are electrostatic. The binding is probably to the RNA of the RNP. Indeed, it has been clearly shown that influenza virus M1 binds to RNA in vitro and that this binding is mediated by a positively charged peptide segment (residues 80 to 111) in the M1 molecule (276, 277, 299). In addition, the crystal structure of the dimer of M1 revealed that 10 positively charged amino acid side chains are exposed to a platform surface, formed by α-helices of the M (middle) domain, that can interact with the negatively charged phosphate groups of the viral RNA (243).
Model for the budding of negative-strand RNA viruses.
The structural organization of M in the virion and its involvement in multiple interactions with other viral proteins and RNP support the assumption that budding of negative-strand RNA viruses is orchestrated by this protein. M protein may self-assemble into a membrane-associated lattice with the assistance of both RNP and spikes. The involvement of the latter components in the formation of the M lattice may promote its curving as well. It is also possible that actin plays a role in this process. As in retroviruses, actin has been found in purified paramyxovirus particles (133, 189, 268) and actin filaments have been seen by EM in association with budding virions (16). Furthermore, the formation of complexes between the paramyxovirus M protein and actin has been demonstrated in vitro (100).
General Concept of Budding
For a long time, it was thought that budding in general is driven by interactions between the spikes and the internal components of the virion. It is now becoming clear that various other types of intermolecular interactions are engaged in budding as well. These include lateral (sideward) interactions between core (Gag), matrix, or membrane proteins and interactions between core or matrix proteins and the lipid bilayer. Each virus has evolved a unique fine mechanism of budding directed by one or more of these kinds of interactions (Fig. 2).
Interestingly, the budding processes of the different viruses have in common the fact that they involve a peripheral or integral membrane protein that is engaged in lateral interactions. A clear example is coronavirus budding, which seems to be directed entirely by lateral interactions between the envelope proteins. It is therefore tempting to speculate that the budding of most viruses is mastered by lateral interactions between integral or peripheral membrane proteins. According to this model, establishment of lateral interactions between the membrane proteins results in the formation of a protein lattice within or at the surface of the membrane. Hence, the budding reaction is in essence the result of the association of membrane proteins. Notably, the lateral interactions between the membrane proteins in many viruses is controlled by additional protein-protein or by protein-RNA interactions. For example, retrovirus Gag protein possesses a strong self-assembly capacity yet the density of the Gag lattice and its curvature may be influenced by Gag-RNA interactions. The budding activity of the M proteins of negative-strand RNA viruses seems to be highly dependent on the RNP and the spikes. The latter components interact with the laterally interacting M molecules and may thereby assist in the formation of the M lattice and induce its curving. The formation of the alphavirus membrane protein lattice might be controlled largely by interactions between the NC and the membrane protein heterodimers. The association of the NC with a cluster of E1-E2 heterodimers might offer a nucleation site for the assembly of the membrane protein lattice, and NC-heterodimer interactions could assist in the further recruitment of membrane protein heterodimers into this lattice. It is even possible that the attraction of membrane protein heterodimers to the NC directly results in bending and curving of the membrane. However, by analogy to the proposed budding mechanisms of other viruses, we expect that the establishment of lateral interactions between the membrane protein heterodimers at the outer surface of the membrane also has a pivotal function in the budding of alphaviruses.
PINCHING-OFF REACTION
The process of budding is completed when the virus particle has pinched off from the membrane. One can imagine that the pinching-off reaction includes two dynamic steps: (i) formation of a budding pore and (ii) membrane fission. The budding pore represents the narrow aqueous connection between the interior of the virion and the cytoplasm at the stage at which the viral and host membranes are still continuous. In the fission reaction, the lipids surrounding the pore mix and reorganize into separate membranes. The pinching-off reaction possibly requires special functions mediated by viral proteins. For instance, alphaviruses might use some structurally altered membrane protein heterodimers for pore formation and membrane fission. These processes could be similar to membrane fusion during virus entry; the reactions just occur in the opposite direction. In retroviruses, which do not require spikes for budding, similar pore-forming and fission functions might be carried out by the Gag protein.
Alternatively, host proteins may assist in these reactions. In this respect, the finding by Wills and collaborators about a late assembly domain in the p2b region of the RSV Gag protein is interesting (288, 293). Mutations in this region seem to cause a block in the pinching-off reaction because the mutant virions remain attached to the PM by a thin membrane “stalk.” The domain contains a Pro sequence motif, Pro-Pro-Pro-Pro-Tyr, which is also found in the Gag protein of many other retroviruses although not in HIV-1. In the latter virus, another Pro-rich domain with similar late assembly functions has been identified instead (93, 196). Surprisingly, the common Pro sequence motif is identical to the binding site of the Trp-Trp (WW) motif found in certain cellular signalling and cytoskeleton proteins. It is therefore possible that the Gag precursors interact with such proteins for the purpose of pinching off. Indeed, an interaction between the RSV Gag protein and the WW domain of Yap has been demonstrated in vitro (83). Yap is a signalling molecule that interacts with Yes, which in turn is a PM-associated tyrosine kinase (260).
CONTROL OF ACTIVITIES THAT DRIVE BUDDING
Control Mechanisms in the Infected Cell
Virus assembly occurs at distinct places in the cell and requires the colocalization of the structural components. Newly synthesized proteins are therefore prevented from initiating budding reactions until they have been transported to the site of assembly. Thus, the budding-driving activity of viral proteins is strictly regulated in time and space. When different components cooperate in budding, e.g., the membrane protein heterodimers and the NC of alphaviruses, it is probably enough to control the binding activity of only one of the partners. In fact, it has been shown that the tail of the SIN E2 membrane protein is not exposed to the cytoplasm when the protein is still located at the beginning of the secretory pathway (154). This should effectively prevent premature association between the NC and the heterodimers. Similar conclusions have been drawn about the exposure of the tail of the VSV G protein (162). The question how the corresponding control mechanism operates for retroviruses is intriguing. It is particularly difficult to understand how the membrane-binding activity of the type D retrovirus core is suppressed before it reaches the PM. It might involve shielding of the NH2-terminal myristic acid of the Gag protein (168).
Recent studies have revealed an interesting kind of fine-tuning of the budding process in hepadnaviruses (89). By use of polymerase mutants of HBV and duck hepatitis virus, it has been demonstrated that budding occurs only if the proviral genome has been reverse transcribed inside the NC. This suggests that the latter process primes the NC to interact with the envelope proteins. Accordingly, capsids lacking the genome and/or polymerase cannot be used for budding.
Control Mechanisms during Virus Entry
An important step in virion maturation is that the budding-driving interactions have to be weakened or abrogated at some stage after the virion has formed. This is necessary to enable the NC of the incoming virion to penetrate the cytoplasm of an uninfected cell after fusion. At present, we can only speculate how the strength of the envelope-NC interactions is controlled. By analogy to the well-defined control mechanisms in virus-mediated membrane fusion (28, 33, 105, 229, 275), those of NC uncoating are probably based on specific triggers that change the structure and hence the function and activity of the proteins that were engaged in budding. To understand how the trigger might control the budding-driving activity, it is important to consider two general aspects of budding. First, budding is driven by numerous repetitive protein-protein and/or protein-lipid interactions. Small changes in the strength of these interactions may therefore have a drastic impact on the stability of the particle whose integrity is maintained by the concerted forces of these interactions. Second, a newly formed virion is stabilized through its closed structure. Therefore, the envelope-NC interactions may be abrogated after the virion has pinched off while the envelope remains intact. Below, we describe how the envelope-NC interactions might be controlled in some viruses.
Hepadnaviruses may exploit a novel mechanism to control the strength of envelope protein-NC interactions during virus maturation and entry. Binding of envelope protein to the NC is mediated by the NH2-terminal, pre-S portion of the L protein (21, 24, 59). Translocation of this multispanning membrane protein is initiated by an internal signal sequence which is located downstream of the pre-S region (61, 62). This leaves the pre-S region exposed to the cytoplasm available for interaction with the NC. However, in about half of the L molecules, pre-S is translocated posttranslationally to the luminal side of the membrane (23, 264). Consequently, about half of the pre-S regions is exposed at the surface of virus particles, where they function as ligands for receptors on host cells (126). It is possible that a high concentration of L molecules with a cytoplasmically exposed pre-S region is required to initiate binding to the NC. Translocation of the pre-S region might then occur before or shortly after completion of budding. This should weaken the NC-envelope protein interaction inside the virion and thereby facilitate membrane fusion and subsequent release of the NC into the cytoplasm during virus entry.
Unwrapping the retrovirus envelope is probably facilitated through cleavage of the Gag protein. The enzyme responsible for this cleavage is an aspartyl protease, which resides in the Gag-Pol fusion protein (123). It becomes active upon its dimerization after incorporation of Gag-Pol into the Gag lattice. One result of the cleavage is that the membrane-interacting part of the Gag lattice, i.e., the MA domain, is released from the other Gag products (17, 88). There are reasons to believe that the remaining layer of MA protein underneath the envelope is less stable than the original Gag lattice (108, 184, 213). First, it lacks the CA domain, which is, as already discussed, supposedly engaged in extensive intermolecular interactions in the Gag lattice. Second, the cleavage of Gag possibly induces structural changes in the MA protein that loosen its association with the membrane. This is supported by membrane-binding studies that have been done with the Gag precursor and MA protein of HIV both in cells that express these proteins and in vitro with purified proteins and liposomes (64, 310). The results showed clearly that the MA protein binds less efficiently to membranes than the Gag precursor does. Therefore, the cleavage of the Gag protein most probably represents the trigger controlling the membrane-core interactions in retroviruses. This model predicts that the core of immature retrovirus particles is unable to penetrate the cytoplasm during entry.
The penetration of influenza A virus into new host cells is probably facilitated by a structural change in the submembrane layer of M1 protein. This might be triggered by the low pH that the incoming virion encounters in endosomes. Protons are probably transported across the viral membrane into the interior of the virion through the ion channel formed by the minor envelope protein, M2 (132, 209). This model is supported by indirect evidence. For instance, it has been shown that the activity of the M2 channel is important for the disruption of M1-RNP interactions in incoming virus. If the channel activity is inhibited by the drug amantadine M1-RNP dissociation does not occur and the RNP cannot be routed to the nucleus to start replication (164). In addition, it has been demonstrated that the M1-RNP interaction is pH sensitive (26, 308). However, it is not yet clear whether acidification also affects M1-M1 or M1-membrane interactions. Nevertheless, a strong indication that this might be true for M1-lipid membrane interaction is obtained from X-ray analysis of the M1 protein crystal (243). This revealed that the proposed membrane-binding region of the N domain is buried within the interface of the N and M domains at pH 4.0. It has been proposed that the hydrophobic region is exposed to interactions with the membrane during virus assembly and that the low pH in the endosome triggers a conformational switch, i.e., flipping of the N domain, that leads to the structure found in the crystal (243).
ASSEMBLY OF VIRAL COMPONENTS THAT DO NOT DRIVE BUDDING
Many viruses possess structural components that are not actively engaged in budding but are essential in other processes of the virus life cycle. These include, for retroviruses, the Env and Gag-Pol proteins and the genome and, for coronaviruses, the S protein and the NC. To become part of the virion these components have to interact with the budding driving proteins. Below, we discuss the mechanisms that control the inclusion of the additional components into the virion.
Incorporation of Genomes, Env, and Gag-Pol into Retroviruses
The mechanism for uptake of Env protein into the retroviral membrane has been rather puzzling for a long time, since it was very difficult to identify Env-Gag interactions. This issue was initially addressed by studying the effects of genetic engineering of the tail of the Env protein. The rationale of the approach was that disruption or deletion of a potential Gag-binding site in the Env tail would prevent the incorporation of Env into the virion. Surprisingly, it was found that the tailless form of the RSV, HIV, and SIV Env proteins is assembled into the envelope of the respective viruses with an efficiency at least as high as that for the wt Env protein (76, 206, 287, 313). A glycosylphosphatidylinositol (GPI)-anchored version of the HIV-1 Env protein was also found to enter virus particles (230). This suggested that Env is incorporated into the virion not through specific interactions with the Gag protein but by some other, nonspecific mechanism instead. However, it has also been shown that many irrelevant PM-resident proteins are not excluded from the retrovirus envelope (see below). This means that the incorporation assay with engineered Env proteins would probably fail to reveal any specific Env-Gag interactions. The results of the above-described experiments are therefore inconclusive. However, other studies provided convincing evidence for the existence of Env-Gag interactions.
First, Cosson (43a) has obtained biochemical data for HIV Env-Gag interactions in an in vitro binding assay in which soluble MA–β-galactosidase fusion proteins were bound to glutathione S-transferase–Env tail fusion proteins. It was found that a peptide corresponding to the 67 COOH-terminal amino acid residues of the Env tail (comprising 150 residues) can bind to the MA protein. The reaction was specific, since several mutations in the Env tail and MA protein abolished the interaction.
Second, Compans and coworkers have used polarized epithelial MDCK cells to study Env-Gag interactions in HIV (192). They found that the expression of gag alone results in the release of Gag particles from both apical and basolateral membranes. However, when env was coexpressed budding was restricted to the basolateral membrane. Since the Env protein is intrinsically targeted to the latter membrane, it is likely that the location of budding was determined by specific Env-Gag interactions. This explanation has been supported by analyzing Env tail and Gag-MA mutants in a similar assay (155).
Third, primary dorsal root ganglion cells have been used to show that the MLV and HIV Env proteins can restrict the subcellular localization of the homologous Gag protein to the somatodendritic region (278). When expressed separately, the Env proteins are localized to the somatodendritic regions whereas the Gag protein is found in the axons as well.
Fourth, several reports describe the introduction of point mutations and deletions in the MA coding region of the HIV-1, SIV, and MPMV genomes (52, 71, 139, 218, 303). Expression of many of these mutant genomes resulted in the production of Gag particles containing little or no Env protein, suggesting that an Env-binding site in the MA domain of the Gag protein had been inactivated by the mutations.
Finally, it should be mentioned that some mutant Env proteins of HIV, MPMV, and MLV having point mutations, partial deletions, or linker insertions in the tail domain, respectively, fail to be incorporated into the viral membrane (18, 53, 94, 95, 115, 304). Although these results fit with the existence of an MA-binding site in the Env tail, it should be noted that there are alternative explanations for the lack of incorporation. For instance, similar results would have been obtained if the mutations affected the transport of Env to the budding site (e.g., by causing a change in the protein’s properties of being subjected to exo- or endocytosis).
The mechanism of incorporation of Gag-Pol into retrovirus particles has been studied in cells coexpressing wt and mutant forms of Gag and Gag-Pol. The results showed that the Gag molecules trap Gag-Pol protein into the particles through specific interactions mediated by the CA domain (197, 248, 253). It has also been demonstrated that the NC domain of the Gag protein interacts specifically with an encapsidation signal, ψ, in the genome (2, 12, 14, 45, 47, 57, 173, 179, 228, 235).
Incorporation of S Protein and the Nucleocapsid into Coronavirus Virions
The coronavirus NC presumably needs to interact with the envelope proteins to ensure its uptake into virions. A likely partner engaged in such interactions is the M protein because of its abundance and its prominent role in assembly. Although M-NC interactions have not been characterized in detail, it has been demonstrated that the NC and the M protein of detergent-solubilized virions associate at 37°C (259). In addition, M protein has been found in purified NCs derived from virus particles that had been mildly treated with detergents (222).
The S protein seems to be incorporated into virions through specific interactions with the M protein. Heteromultimeric complexes of M and S proteins have been identified in lysates of infected cells and virions by use of coimmunoprecipitation and sedimentation assays. The two envelope proteins also associate into large complexes when they are coexpressed in the absence of other coronavirus proteins (187). Under these conditions, the S protein is prevented from being transported to the PM, but instead it is retained intracellularly by the M protein (M accumulates within cells by itself). Thus, specific M-S interactions determine the intracellular transport of the S protein and direct its assembly into virus particles. Accordingly, the incorporation of spikes into virions does not depend on the NC. This has been verified in studies showing that S is also assembled into envelopes that are formed in the absence of the NC (270).
PROTEIN SORTING IN BUDDING
The formation of the viral membrane is thought to be a process of stringent protein sorting during which cellular proteins are displaced by viral envelope proteins. Indeed, highly purified preparations of SIN virions are virtually devoid of host contaminants (255). The efficient exclusion of host proteins is probably due to the extensive and specific interactions formed between the membrane protein heterodimers and between the latter and the NC (36, 73). This leaves little space for host membrane proteins.
However, other viruses may be less selective in the assembly of their envelope. For instance, it has been shown in a large number of studies that foreign membrane proteins are frequently taken up into retrovirus particles. Phenotypic mixing observed in cells coinfected with RSV and VSV exemplifies that retroviruses allow the incorporation of foreign spike proteins (280, 281). In an extreme case, foreign spike protein can completely substitute for homologous Env protein, resulting in the generation of so-called pseudotyped virions. This phenomenon has been repeatedly noticed in studies in which Env-defective retroviruses were coexpressed with heterologous env or foreign spike protein genes (50, 135, 136).
Retroviruses tolerate also the incorporation of several host PM proteins. For instance, significant amounts of HLA DR protein (about 15% of the Gag content) have been detected in HIV-1 particles produced in H9 cells (3). Similarly, host cell adhesion receptors, Thy-1, and CD4 molecules have been found in the envelopes of HIV-1, MLV, and avian leukosis virus, respectively (30, 70, 188, 190, 302).
However, serious criticism has recently been raised against the finding of foreign membrane proteins in retrovirus particles, particularly in HIV-1. Studies of HIV-1-producing lymphocytes demonstrated that these cells endogeneously release PM-derived microvesicles which copurify with virus particles (15, 91, 214). Therefore, the amounts of foreign proteins in retrovirus particles might be overestimated and should be reconsidered. Nevertheless, the phenomenon of incorporation of unrelated proteins most probably remains true. This is convincingly illustrated by the fact that retroviruses can be neutralized, both in vitro and in vivo, with antibodies specific for foreign membrane proteins (4, 35, 280, 281). Moreover, foreign proteins have been detected in situ, i.e., at the surface of the virions, by immunogold labelling and EM analysis (86, 107, 171).
Foreign membrane proteins have also been found in the envelopes of rhabdoviruses. For instance, phenotypically mixed VSV particles are formed in cells coinfected with VSV and other enveloped RNA viruses (279, 292). Furthermore, cell surface iodination revealed that some host-specific PM proteins enter the VSV envelope during budding (156). Similarly, CD4 molecules, expressed by a recombinant vaccinia virus, are incorporated into VSV tsO45 particles (241). Finally, Rose and colleagues have studied a recombinant VSV expressing the CD4 and measles virus H and F genes (237). They found that the foreign proteins were expressed at levels corresponding to 25 to 60% of that of the G protein and that all of them entered the VSV envelope to a significant extent (20 to 30% of G) without displacing any of the homologous spikes. Collectively, these results suggest that budding of rhabdoviruses is not accompanied by extensive extrusion of foreign membrane proteins from the viral envelope. The fact that phenotypic mixing occurs in both paramyxoviruses and orthomyxoviruses suggests that these viruses also fail to exclude irrelevant membrane proteins during budding (305).
Taken together, these data suggest that envelope formation is not generally accompanied by stringent protein sorting. Instead, the extent of sorting varies between viruses. The efficiency of sorting most probably depends on the specificity of the molecular interactions involved in the formation of the particle. These interactions underlie the specific association of the structural components, thereby displacing nonrelated proteins. In addition, the structural organization of the viral proteins plays an important role in sorting. Basically, envelope assembly can be envisaged as the formation of a lattice by integral or peripheral membrane protein. Such a lattice offers room to the full complement of structural envelope components that are incorporated through specific molecular interactions. If not all positions are occupied by viral proteins or when additional space is available, nonrelated proteins may remain in the membrane. Accordingly, foreign membrane proteins are expected to be passively incorporated into the envelope if they do not sterically hinder the process of envelope formation.
Protein sorting in budding presumably forms a primary constraint on virus evolution. Viruses may often have the opportunity to diverge through mutation and recombination. Mutations in the structural proteins are tolerated if they do not interfere with the formation of viable progeny. It is also possible that viruses assume a nonrelated protein, for instance from another virus during mixed infections or from the host cell, to supplement the virion. Complementation of a virus with a nonrelated structural protein, however, largely depends on whether this protein withstands the pressure of sorting. The rate at which the nature and diversity of structural components evolve is therefore particularly determined by the selection criteria imposed by viral sorting mechanisms.
In this context, it is interesting that some coronavirus variants are complemented with a fourth structural envelope protein, the hemagglutinin-esterase (HE) protein (111, 251). This protein may facilitate infection and has a significant homology to the HE protein of influenza C virus. It has been speculated that coronaviruses acquired the HE-encoding gene by recombination with the latter virus (158) or through recombination with a host mRNA (43). Liao et al. (148) have shown that exogeneously expressed HE protein is also incorporated into the progeny of an HE-defective virus. Apparently, the coronavirus envelope has the flexibility to accommodate a variable number of different proteins. It is perhaps because of this feature that coronaviruses could adopt a nonrelated structural protein during evolution.
CONCLUDING REMARKS AND PROSPECTS
Most significantly, the progress in research on virus budding has changed the idea that the capsid simply recruits a membrane around itself by attracting virus-specific proteins associated with the membrane. Rather, formation of the envelope appears to be a distinct step in virion assembly that requires specific functions of one or a few structural proteins. These proteins can be either spike proteins (alphavirus), matrix proteins (orthomyxoviruses, paramyxoviruses, and rhabdovirus), membrane proteins (coronavirus), or core (Gag) proteins (retrovirus) that engage in lateral interactions. They thereby pack into lattices to form a cage-like construction at the outer surface of, within, or underneath the membrane. The budding reaction as such is fully integrated into this process; the membrane adopts the curvature of the “cage” while it is being assembled.
Clearly, detailed characterization of the proteins and molecular interactions involved in budding, as well as structural analysis of the assembled virion, has proven to be invaluable in providing insights into the budding of the viruses described in this review. Nevertheless, the picture of the budding mechanism of these viruses, even of those that have been very extensively studied, is still incomplete. Thus, it will be of great importance to obtain detailed information about the structure of the entire membrane-associated protein lattice or cage and to unravel the process by which it is assembled during virus maturation and disassembled during virus entry. For instance, determination of the ultrastructure of the alphavirus heterodimeric membrane protein subunit (E1-E2) would probably illuminate the nature of the heterodimer-heterodimer interactions at the external surface of the viral membrane. For retroviruses, it will be necessary to resolve the structure of the Gag lattice to high resolution in order to establish its geometry and to determine how the postulated Gag trimers interact with each other. Structural determination of the membrane-associated protein lattices in other viruses is still in its infancy. It is expected that concerted efforts to solve the structures of these lattices by cryo-EM and by high-resolution analysis of individual proteins or protein domains will be amply rewarded with novel insights in the budding mechanism of these viruses.
It will be a great challenge to develop a system in which budding can be reconstituted in vitro by using purified viral constituents and artificial membranes. This approach might be most feasible for retroviruses. Such a system could, for instance, be used to directly test the involvement of RNA in budding. In vitro-reconstituted budding would also open the possibility of measuring the effects on budding of various factors and conditions, such as protein concentration, lipids, ions, and temperature. It should be noted that some promising attempts in establishing in vitro assembly systems for retroviruses have been reported (31, 151, 227, 252, 265).
Some aspects of budding have remained almost completely mysterious. In particular, the pinching-off reaction and the role of host components (e.g., actin) in budding have not been elucidated. The use of perforated cells in which budding can be controlled by conditioning the cytoplasm from outside could be one approach to studying these subjects.
Finally, it will be of interest to investigate whether and how host proteins and lipids are sorted during envelope formation. For this purpose, the protein and lipid composition of the viral envelope and the donor (host) membrane should be compared quantitatively and qualitatively. One of the major preconditions for these studies is that the viral envelopes and the cellular host membranes can be purified from contaminants. Analysis of the criteria by which proteins and lipids are excluded from the envelope would be informative in identifying the mechanism of sorting.
Overall, we conclude that there are still many important and interesting questions about virus budding. The field of virus budding will therefore remain the subject of intensive study in the future.
ACKNOWLEDGMENTS
We thank Ingrid Sigurdsson and Aila Holappa for typing, Carin Lindgren for making drawings, and Holland Cheng for critical reading of the manuscript.
REFERENCES
- 1.Acheson N H, Tamm I. Replication of Semliki Forest virus: an electron microscopic study. Virology. 1967;32:128–143. doi: 10.1016/0042-6822(67)90261-9. [DOI] [PubMed] [Google Scholar]
- 2.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid cys-his motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur L O, Bess J W, Jr, Sowder II R C, Benveniste R E, Mann D L, Chermann J-C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 4.Arthur L O, Bess J W, Jr, Urban R G, Strominger J L, Morton W R, Mann D L, Henderson L E, Benvesite R E. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J Virol. 1995;69:3117–3124. doi: 10.1128/jvi.69.5.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 6.Barbacid M, Aaronson S A. Membrane properties of the gag gene-coded p15 protein of mouse type-C RNA tumor viruses. J Biol Chem. 1978;253:1408–1414. [PubMed] [Google Scholar]
- 7.Barge A, Gaudin Y, Coulon P, Ruigrok R W H. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J Virol. 1993;67:7246–7253. doi: 10.1128/jvi.67.12.7246-7253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D. Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron M D, Barret T. Rescue of rinderpest virus from cloned cDNA. J Virol. 1997;71:1265–1271. doi: 10.1128/jvi.71.2.1265-1271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barth B U, Garoff H. The nucleocapsid-binding spike subunit E2 of Semliki Forest virus requires complex formation with the E1 subunit for activity. J Virol. 1997;71:7857–7865. doi: 10.1128/jvi.71.10.7857-7865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barth B U, Suomalainen M, Liljeström P, Garoff H. Alphavirus assembly and entry: role of the cytoplasmic tail of the E1 spike subunit. J Virol. 1992;66:7560–7564. doi: 10.1128/jvi.66.12.7560-7564.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkowitz R D, Ohagen Å, Höglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bess J W, Jr, Gorelick R J, Bosche W J, Henderson L E, Arthur L O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 16.Bohn W, Rutter G, Hohenberg H, Mannweiler K, Nobis P. Involvement of actin filaments in budding of measles virus: studies on cytoskeletons of infected cells. Virology. 1986;149:91–106. doi: 10.1016/0042-6822(86)90090-5. [DOI] [PubMed] [Google Scholar]
- 17.Bolognesi D P, Montelaro R C, Frank H, Schäfer W. Assembly of type C oncornaviruses: a model. Science. 1978;199:183–186. doi: 10.1126/science.202022. [DOI] [PubMed] [Google Scholar]
- 18.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown D T. Assembly of alphaviruses. In: Schlesinger R W, editor. The togaviruses. New York, N.Y: Academic Press, Inc.; 1980. pp. 473–501. [Google Scholar]
- 20.Brown D T, Smith J F. Morphology of BHK-21 cells infected with Sindbis virus temperature sensitive mutants in complementation groups D and E. J Virol. 1975;15:1262–1266. doi: 10.1128/jvi.15.5.1262-1266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruss V, Lu X, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bucher D J, Kharitonenkow L G, Zakomiridin J A, Griboriev V B, Klimenko S M, Davis J F. Incorporation of influenza virus M-protein into liposomes. J Virol. 1980;36:586–590. doi: 10.1128/jvi.36.2.586-590.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Büechi M, Bächi T. Microscopy of internal structures of Sendai virus associated with the cytoplasmic surface of host membranes. Virology. 1982;120:349–359. doi: 10.1016/0042-6822(82)90036-8. [DOI] [PubMed] [Google Scholar]
- 28.Bullough P A, Hugson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 29.Cadd T L, Skoging U, Liljeström P. Budding of enveloped viruses from the plasma membrane. Bioessays. 1997;19:993–1000. doi: 10.1002/bies.950191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calafat J, Janssen H, Démant P, Hilgers J, Závada J. Specific selection of host cell glycoproteins during assembly of murine leukaemia virus and vesicular stomatitis virus: presence of Thy-1 glycoprotein and absence of H-2, Pgp-1 and T-200 glycoproteins on the envelopes of these virus particles. J Gen Virol. 1983;64:1241–1253. doi: 10.1099/0022-1317-64-6-1241. [DOI] [PubMed] [Google Scholar]
- 31.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 34.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 35.Chan W L, Rodgers A, Grief C, Almond N, Ellis S, Flanagan B, Silvera P, Bootman J, Stott J, Kent K, Bomford R. Immunization with class I human histocompatibility leukocyte antigen can protect macaques against challenge infection with SIVmac-32H. AIDS. 1995;9:223–228. [PubMed] [Google Scholar]
- 36.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu W, Smith T J. Structural studies of virus-antibody complexes by electron cryomicroscopy and X-ray crystallography. Curr Opin Struct Biol. 1994;4:219–224. [Google Scholar]
- 38.Choi H-K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 39.Chong L D, Rose J K. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coffin J. Structure of the retrovirus genome. In: Weiss R, Teich N, Varmus H E, Coffin J M, editors. RNA tumor viruses. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 261–268. [Google Scholar]
- 41.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conzelmann K-K. Genetic manipulation of non-segmented negative-strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 43.Cornelissen L A H M, Wierda C M H, van der Meer F J, Herrewegh A A P M, Horzinek M C, Egberink H F, de Groot R J. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 44.Damsky C H, Sheffield J B, Tuszynski G P, Warren L. Is there a role for actin in virus budding? J Cell Biol. 1977;75:593–605. doi: 10.1083/jcb.75.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis N L, Willis L V, Smith J F, Johnston R E. In vitro synthesis of infectious venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology. 1989;171:189–204. doi: 10.1016/0042-6822(89)90526-6. [DOI] [PubMed] [Google Scholar]
- 47.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 48.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deminie C A, Emerman M. Incorporation of human immunodeficiency virus type 1 gag proteins into murine leukemia virus virions. J Virol. 1993;67:6499–6506. doi: 10.1128/jvi.67.11.6499-6506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong J, Roth M G, Hunter E. A chimeric avian retrovirus containing the influenza virus hemagglutinin gene has an expanded host range. J Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorfman T, Bukovsky A, Öhagen Å, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 55.Dubois-Dalcq M, Holmes K V, Rentier B, Kingsbury D W, editors. Assembly of enveloped RNA viruses. Vienna, Austria: Springer-Verlag KG; 1984. [Google Scholar]
- 56.Dubovi E, Wagner R R. Spatial relationships of the proteins of vesicular stomatitis virus: induction of reversible oligomers by cleavable protein cross-linking and oxidation. J Virol. 1977;22:500–509. doi: 10.1128/jvi.22.2.500-509.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dupraz P, Spahr P-F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durbin A P, Hall S L, Siew J W, Whitehead S S, Collins P L, Murphy B R. Recovery of infectious human parainfluenza virus type 3 from cDNA. Virology. 1997;235:323–332. doi: 10.1006/viro.1997.8697. [DOI] [PubMed] [Google Scholar]
- 59.Dyson M R, Murray K. Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of hepatitis B virus. Proc Natl Acad Sci USA. 1995;92:2194–2198. doi: 10.1073/pnas.92.6.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 61.Eble B E, Lingappa V R, Ganem D. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol. 1990;64:1414–1419. doi: 10.1128/jvi.64.3.1414-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eble B E, MacRae D R, Lingappa V R, Ganem D. Multiple topogenic sequences determine the transmembrane orientation of hepatitis B surface antigen. Mol Cell Biol. 1987;7:3591–3601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ehrlich L S, Fong S, Scarlata S, Zybarth G, Carter C. Partitioning of HIV-1 gag and gag-related proteins in membranes. Biochemistry. 1996;35:3933–3943. doi: 10.1021/bi952337x. [DOI] [PubMed] [Google Scholar]
- 65.Ekström M, Liljeström P, Garoff H. Membrane protein lateral interactions control Semliki Forest virus budding. EMBO J. 1994;13:1058–1064. doi: 10.1002/j.1460-2075.1994.tb06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faaberg K S, Peeples M E. Association of soluble matrix protein of Newcastle disease virus with liposomes is independent of ionic conditions. Virology. 1988;166:123–132. doi: 10.1016/0042-6822(88)90153-5. [DOI] [PubMed] [Google Scholar]
- 68.Flamand A. Etude génétic du virus de la stomatite vésiculaire: classement de mutants thermosensibles spontanés en groupes de complémentation. J Gen Virol. 1970;8:187–195. doi: 10.1099/0022-1317-8-3-187. [DOI] [PubMed] [Google Scholar]
- 69.Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fortin J F, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol. 1997;71:3588–3596. doi: 10.1128/jvi.71.5.3588-3596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freed E O, Martin M A. Virus incorporation of envelope glycoprotein with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujiyoshi Y, Kume N P, Sakata K, Sato S B. Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J. 1994;13:318–326. doi: 10.1002/j.1460-2075.1994.tb06264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 74.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles: the uncommon line. J Struct Biol. 1996;116:55–60. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 75.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 76.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein’s cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 78.Gallis B, Linial M, Eisenman R. An avian oncovirus deficient in genomic RNA: characterization on the packaged RNA as cellular messenger RNA. Virology. 1979;94:146–161. doi: 10.1016/0042-6822(79)90445-8. [DOI] [PubMed] [Google Scholar]
- 79.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 80.Ganem D. Assembly of hepadnaviral virions and subviral particles. Curr Top Microbiol Immunol. 1991;168:61–83. doi: 10.1007/978-3-642-76015-0_4. [DOI] [PubMed] [Google Scholar]
- 81.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 82.Garcin D, Pelet T, Calain P L R, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 84.Garoff H, Simons K. Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc Natl Acad Sci USA. 1974;71:3988–3992. doi: 10.1073/pnas.71.10.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaudin Y, Sturgis J, Doumith M, Barge A, Robert B, Ruigrok R W H. Conformational flexibility and polymerization of vesicular stomatitis virus matrix protein. J Mol Biol. 1997;274:816–825. doi: 10.1006/jmbi.1997.1439. [DOI] [PubMed] [Google Scholar]
- 86.Gelderblom H, Reupke H, Winkel T, Kunze R, Pauli G. MHC-antigens: constituents of the envelopes of human and simian immunodeficiency viruses. Z Naturforsch Sect C. 1987;42:1328–1334. doi: 10.1515/znc-1987-11-1230. [DOI] [PubMed] [Google Scholar]
- 87.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 88.Gelderblom H R, Hausmann E H S, Özel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 89.Gerelsaikhan T, Tavis J E, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 91.Gluschankof P, Mondor I, Gelderblom H R, Sattentau Q J. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 1997;230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 92.Gorelick R J, Hendenson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Granowitz C, Colicelli J, Goff S P. Analysis of mutations in the envelope gene of Moloney murine leukemia virus: separation of infectivity from superinfection resistance. Virology. 1991;183:545–554. doi: 10.1016/0042-6822(91)90983-i. [DOI] [PubMed] [Google Scholar]
- 95.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greber U F, Willets M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 97.Gregoriades A. Interaction of influenza M protein with viral lipids and phosphatidylcholine vesicles. J Virol. 1980;36:470–479. doi: 10.1128/jvi.36.2.470-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gregoriades A, Frangione B. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J Virol. 1981;40:323–328. doi: 10.1128/jvi.40.1.323-328.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guiffre R M, Tovell D R, Kay C M, Tyrrell D L J. Evidence for an interaction between the membrane protein of a paramyxovirus and actin. J Virol. 1982;42:963–968. doi: 10.1128/jvi.42.3.963-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haffar O, Garrigues J, Travis B, Moran P, Zarling J, Hu S-L. Human immunodeficiency virus-like, nonreplicating, gag-env particles assemble in a recombinant vaccinia virus expression system. J Virol. 1990;64:2653–2659. doi: 10.1128/jvi.64.6.2653-2659.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hahn C S, Rice C M, Strauss G, Lenches E M, Strauss J H. Sindbis virus ts103 has a mutation in glycoprotein E2 that leads to defective assembly of virions. J Virol. 1989;63:3459–3465. doi: 10.1128/jvi.63.8.3459-3465.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 104.Heggeness M H, Smith P R, Choppin P W. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc Natl Acad Sci USA. 1982;79:6232–6236. doi: 10.1073/pnas.79.20.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heinz F X, Stiasny K, Püschner-Auer G, Holzmann H, Allison S L, Mandl C W, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 106.Henderson L E, Krutzsch H C, Oroszlan S. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational protein modification. Proc Natl Acad Sci USA. 1983;80:339–343. doi: 10.1073/pnas.80.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Henderson L E, Sowder R, Copeland T D, Oroszlan S, Arthur L O, Robey W G, Fischinger P J. Direct identification of class II histocompatibility DR proteins in preparations of human T-cell lymphotropic virus type III. J Virol. 1987;61:629–632. doi: 10.1128/jvi.61.2.629-632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoffman M A, Banerjee A K. An infectious clone of human parainfluenza virus type 3. J Virol. 1997;71:4272–4277. doi: 10.1128/jvi.71.6.4272-4277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holmes K V, Doller E W, Sturman L S. Tunicamycin resistant glycosylation of a coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holmes K V, Lai M M C. Coronaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1075–1093. [Google Scholar]
- 112.Hong S S, Boulanger P. Assembly-defective point mutants of the human immunodeficiency virus type 1 gag precursor phenotypically expressed in recombinant baculovirus-infected cells. J Virol. 1993;67:2787–2798. doi: 10.1128/jvi.67.5.2787-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 114.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jin H, Leser G P, Lamb R A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jin H, Leser G P, Zhang J, Lamb R A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jones T, Blaug G, Hansen M, Barklis E. Assembly of gag–β-galactosidase proteins into retrovirus particles. J Virol. 1990;64:2265–2279. doi: 10.1128/jvi.64.5.2265-2279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jowett J B M, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 120.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clementsmann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro—clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 123.Katz R A, Skalka A-M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 124.Kawai S, Hanafusa H. Isolation of defective mutant of avian sarcoma virus. Proc Natl Acad Sci USA. 1973;70:3493–3497. doi: 10.1073/pnas.70.12.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. [Google Scholar]
- 126.Klingmüller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kräusslich, H. G. 1998. Morphogenesis and maturation of retroviruses. Curr. Top. Microbiol. Immunol., 214.
- 128.Kretzschmar E, Bui M, Rose J K. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology. 1996;220:37–45. doi: 10.1006/viro.1996.0283. [DOI] [PubMed] [Google Scholar]
- 129.Krijnse Locker J K, Opstelten D-J E, Ericsson M, Horzinek M C, Rottier P J M. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 130.Krug R M. The influenza viruses. New York, N.Y: Plenum Press; 1989. [Google Scholar]
- 131.Kuhn R J, Niesters H G M, Hong Z, Strauss J H. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 132.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–87. [Google Scholar]
- 133.Lamb R A, Mahy B W J, Choppin P W. The synthesis of Sendai virus polypeptides in infected cells. Virology. 1976;69:116–131. doi: 10.1016/0042-6822(76)90199-9. [DOI] [PubMed] [Google Scholar]
- 134.Lamb R A, Zebedee S L, Richardson C D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 135.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lebedeva I, Fujita K, Nihrane A, Silver J. Infectious particles derived from Semliki Forest virus vectors encoding murine leukemia virus envelopes. J Virol. 1997;71:7061–7067. doi: 10.1128/jvi.71.9.7061-7067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee S, Owen K E, Choi H-K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 141.Lee T-L, Lin Y-C, Mochitate K, Grinnell F. Stress-relaxation of fibroblasts in collagen matrices triggers ectocytosis of plasma membrane vesicles containing actin, annexins II and VI, and β1 integrin receptors. J Cell Sci. 1993;105:167–177. doi: 10.1242/jcs.105.1.167. [DOI] [PubMed] [Google Scholar]
- 142.Lenard J, Vanderoef R. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J Virol. 1990;64:3486–3491. doi: 10.1128/jvi.64.7.3486-3491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Levin J G, Seidman J G. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J Virol. 1981;38:403–408. doi: 10.1128/jvi.38.1.403-408.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Levin J G, Seidman J G. Selective packaging of host tRNAs by murine leukemia virus particles does not require genomic RNA. J Virol. 1979;29:328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 146.Li K-J, Garoff H. Production of infectious recombinant Moloney murine leukemia virus particles in BHK cells using Semliki Forest virus-derived RNA expression vectors. Proc Natl Acad Sci USA. 1996;93:11658–11663. doi: 10.1073/pnas.93.21.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Y, Luo L, Schubert M, Wagner R R, Kang C Y. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J Virol. 1993;67:4415–4420. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liao C-L, Zhang X, Lai M M C. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology. 1995;208:319–327. doi: 10.1006/viro.1995.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lindqvist B H, DiSalvo J, Rice C M, Strauss J H, Strauss E G. Sindbis virus mutant ts20 of complementation group E contains a lesion in glycoprotein E2. Virology. 1986;151:10–20. doi: 10.1016/0042-6822(86)90099-1. [DOI] [PubMed] [Google Scholar]
- 151.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu C, Air G M. Selection and characterization of a neuraminidase-minus mutant of influenza virus and its rescue by cloned neuraminidase genes. Virology. 1993;194:403–407. doi: 10.1006/viro.1993.1276. [DOI] [PubMed] [Google Scholar]
- 153.Liu C, Eichelberger M C, Compans R W, Air G M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu N, Brown D T. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J Cell Biol. 1993;120:877–883. doi: 10.1083/jcb.120.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lodge R, Göttlinger H, Gabuzda D, Cohen É, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lodish H F, Porter M. Specific incorporation of host cell surface proteins into budding vesicular stomatitis virus particles. Cell. 1980;19:161–169. doi: 10.1016/0092-8674(80)90397-9. [DOI] [PubMed] [Google Scholar]
- 157.Lopez S, Yao J-S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Luytjes W, Bredenbeek P J, Noten A F H, Horzinek M C, Spaan W J M. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luytjes W, Gerritsma H, Bos E, Spaan W J M. Characterization of two temperature-sensitive mutants of coronavirus mouse hepatitis virus strain A59 with maturation defects in the spike protein. J Virol. 1997;71:949–955. doi: 10.1128/jvi.71.2.949-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Amplification, expression and packaging of a foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 161.Lyles D S, McKenzie M, Parce J W. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J Virol. 1992;66:349–358. doi: 10.1128/jvi.66.1.349-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mack D, Kluxen B, Kruppa J. Accessibility to proteases of the cytoplasmic G protein domain of vesicular stomatitis virus is increased during intracellular transport. J Cell Biol. 1989;109:2057–2065. doi: 10.1083/jcb.109.5.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Markwell M A K, Fox C F. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J Virol. 1980;33:152–166. doi: 10.1128/jvi.33.1.152-166.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 165.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 166.Matsumura H, Futaesaku Y, Kohno S, Sugiura A, Kohase M. A temperature-sensitive mutant of Newcastle disease virus defective in intracellular processing of fusion protein. J Virol. 1990;64:1410–1413. doi: 10.1128/jvi.64.3.1410-1413.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, Kingsman S, Kingsman A, Campbell I. Structural similarity between the p17 matrix protein of HIV-1 and interferon-γ. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 168.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 169.Mebatsion T, Conzelmann K-K. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mebatsion T, König M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 171.Meerloo T, Sheikh M, Bloem A, de Ronde A, Schutten M, van Els C A, Roholl P J, Joling P, Goudsmit J, Schuurman H-J. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. J Gen Virol. 1993;74:129–135. doi: 10.1099/0022-1317-74-1-129. [DOI] [PubMed] [Google Scholar]
- 172.Mehul B, Hughes R C. Plasma membrane targing, vesicular budding and release of galectin 3 from the cytoplasm of mammalian cells during secretion. J Cell Sci. 1997;110:1169–1178. doi: 10.1242/jcs.110.10.1169. [DOI] [PubMed] [Google Scholar]
- 173.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Metsikkö K, Garoff H. Oligomers of the cytoplasmic domain of the p62/E2 membrane protein of Semliki Forest virus bind to the nucleocapsid in vitro. J Virol. 1990;64:4678–4683. doi: 10.1128/jvi.64.10.4678-4683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Metsikkö K, Simons K. The budding mechanism of spikeless vesicular stomatitis virus particles. EMBO J. 1986;5:1913–1920. doi: 10.1002/j.1460-2075.1986.tb04444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mitnaul L J, Castrucci M R, Murti K G, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 178.Morikawa S, Booth T F, Bishop D H L. Analyses of the requirements for the synthesis of virus-like particles by feline immunodeficiency virus gag using baculovirus vectors. Virology. 1991;183:288–297. doi: 10.1016/0042-6822(91)90141-w. [DOI] [PubMed] [Google Scholar]
- 179.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mulvey M, Brown D T. Assembly of the Sindbis virus spike protein complex. Virology. 1996;219:125–132. doi: 10.1006/viro.1996.0229. [DOI] [PubMed] [Google Scholar]
- 181.Murphy F A. Togavirus morphology and morphogenesis. In: Schlesinger R W, editor. The togaviruses: biology, structure, replication. New York, N.Y: Academic Press; 1980. pp. 241–316. [Google Scholar]
- 182.Murti K G, Brown P S, Bean W J, Jr, Webster R G. Composition of the helical internal components of influenza virus as revealed by immunogold labeling/electron microscopy. Virology. 1992;186:294–299. doi: 10.1016/0042-6822(92)90084-3. [DOI] [PubMed] [Google Scholar]
- 183.Nermut M V, Grief C, Hashmi S, Hockley D J. Further evidence of icosahedral symmetry in human and simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:929–938. doi: 10.1089/aid.1993.9.929. [DOI] [PubMed] [Google Scholar]
- 184.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 185.Newcomb W W, Tobin G J, McGowan J J, Brown J C. In vitro reassembly of vesicular stomatitis virus skeletons. J Virol. 1982;41:1055–1062. doi: 10.1128/jvi.41.3.1055-1062.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Opstelten D-J E, Horzinek M C, Rottier P J M. Complex formation between the spike protein and the membrane protein during mouse hepatitis virus assembly. Adv Exp Med Biol. 1994;342:189–195. doi: 10.1007/978-1-4615-2996-5_30. [DOI] [PubMed] [Google Scholar]
- 187.Opstelten D-J E, Raamsman M J B, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Orentas R J, Hildreth J E K. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res Hum Retroviruses. 1993;9:1157–1165. doi: 10.1089/aid.1993.9.1157. [DOI] [PubMed] [Google Scholar]
- 189.Örvell C. Structural polypeptides of mumps virus. J Gen Virol. 1978;41:527–539. doi: 10.1099/0022-1317-41-3-527. [DOI] [PubMed] [Google Scholar]
- 190.Ott D E. Cellular proteins in HIV virions. Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 191.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Owens R J, Rose J K. Cytoplasmic domain requirement for incorporation of a foreign envelope protein into vesicular stomatitis virus. J Virol. 1993;67:360–365. doi: 10.1128/jvi.67.1.360-365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pan B-T, Teng K, Wu C, Adam M, Johnstone R M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bradford Bowzard J, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pastan I, Seth P, Fitzgerald D, Willingham M C. Adenovirus entry into cells: some new observations on an old problem. In: Notkins A L, Oldstone M B A, editors. Concepts in viral pathogenesis. New York, N.Y: Springer-Verlag; 1986. pp. 141–146. [Google Scholar]
- 199.Pattnaik A K, Brown D J, Nayak D P. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J Virol. 1986;60:994–1001. doi: 10.1128/jvi.60.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Patzer E J, Nakamura G R, Simonsen C C, Levinson A D, Brands R. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J Virol. 1986;58:884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Peeples M. Paramyxovirus M protein: pulling it all together and taking it on the road. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 427–479. [Google Scholar]
- 202.Peitzsch R M, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 203.Pepinsky R B. Localization of lipid-protein and protein-protein interactions within the murine retrovirus gag precursor by a novel peptide-mapping technique. J Biol Chem. 1983;258:11229–11235. [PubMed] [Google Scholar]
- 204.Pepinsky R B, Vogt V M. Identification of retrovirus matrix proteins by lipid-protein cross-linking. J Mol Biol. 1979;131:819–837. doi: 10.1016/0022-2836(79)90203-1. [DOI] [PubMed] [Google Scholar]
- 205.Pérez L, Carrasco L. Entry of poliovirus into cells does not require a low-pH step. J Virol. 1993;67:4543–4548. doi: 10.1128/jvi.67.8.4543-4548.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Perez L G, Davis G L, Hunter E. Mutants of Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J Virol. 1987;61:2981–2988. doi: 10.1128/jvi.61.10.2981-2988.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 208.Pinter A, deHarven E. Protein composition of a defective murine sarcoma virus particle possessing the enveloped type-A morphology. Virology. 1979;99:103–110. doi: 10.1016/0042-6822(79)90041-2. [DOI] [PubMed] [Google Scholar]
- 209.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 210.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Prchla E, Plank C, Wagner E, Blaas D, Fuchs R. Virus-mediated release of endosomal content in vitro: different behavior of adenovirus and rhinovirus serotype 2. J Cell Biol. 1995;131:111–123. doi: 10.1083/jcb.131.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 214.Raposo G, Nijman H W, Stoorvogel W, Leijendekker R, Harding C V, Melief C J M, Geuze H J. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rey O, Canon J, Krogstad P. HIV-1 gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 217.Rhee S S, Hui H, Hunter E. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J Virol. 1990;64:3844–3852. doi: 10.1128/jvi.64.8.3844-3852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 219.Ricard C S, Koetzner C A, Sturman L S, Masters P S. A conditional-lethal murine coronavirus mutant that fails to incorporate the spike glycoprotein into assembled virions. Virus Res. 1995;39:261–276. doi: 10.1016/0168-1702(95)00100-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rice C M, Strauss J H. Association of Sindbis virion glycoproteins and their precursors. J Mol Biol. 1982;154:325–438. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- 222.Risco C, Antón I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Rolls M M, Haglund K, Rose J K. Expression of additional genes in a vector derived from a minimal RNA virus. Virology. 1996;218:406–411. doi: 10.1006/viro.1996.0211. [DOI] [PubMed] [Google Scholar]
- 224.Rolls M M, Webster P, Balba N H, Rose J K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 225.Rottier P J M, Horzinek M C, van der Zeijst B A M. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Royer M, Cerutti M, Gay B, Hong S-S, Devauchelle G, Boulanger P. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 227.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sakalian M, Wills J W, Vogt V M. Efficiency and selectivity of RNA packaging by Rous sarcoma virus gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Salzwedel K, Johnston P B, Roberts S J, Dubay J W, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sanderson C M, McQueen N L, Nayak D P. Sendai virus assembly: M protein binds to viral glycoproteins in transit through the secretory pathway. J Virol. 1993;67:651–663. doi: 10.1128/jvi.67.2.651-663.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Saraste J, von Bonsdorff C-H, Hashimoto K, Kääriäinen L, Keränen S. Semliki Forest virus mutants with temperature-sensitive transport defect of envelope proteins. Virology. 1980;100:229–245. doi: 10.1016/0042-6822(80)90516-4. [DOI] [PubMed] [Google Scholar]
- 233.Scheele C M, Hanafusa H. Proteins of helper-dependent RSV. Virology. 1971;45:401–410. doi: 10.1016/0042-6822(71)90341-2. [DOI] [PubMed] [Google Scholar]
- 234.Scheid A, Choppin P W. Isolation and purification of the envelope proteins of Newcastle disease virus. J Virol. 1973;11:263–271. doi: 10.1128/jvi.11.2.263-271.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schlesinger S, Makino S, Linial M L. cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Schnell M J, Mebatsion T, Conzelmann K-K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schnitzer T J, Dickson C, Weiss R A. Morphological and biochemical characterization of viral particles produced by the tsO45 mutant of vesicular stomatitis virus at restrictive temperature. J Virol. 1979;29:185–195. doi: 10.1128/jvi.29.1.185-195.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schnitzer T J, Lodish H F. Noninfectious vesicular stomatitis virus particles deficient in the viral nucleocapsid. J Virol. 1979;29:443–447. doi: 10.1128/jvi.29.2.443-447.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Schubert M, Joshi B, Blondel D, Harmison G G. Insertion of the human immunodeficiency virus CD4 receptor into the envelope of vesicular stomatitis virus particles. J Virol. 1992;66:1579–1589. doi: 10.1128/jvi.66.3.1579-1589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Seth P. Mechanism of adenovirus-mediated endosome lysis: role of the intact adenovirus capsid structure. Biochemistry. 1994;205:1318–1324. doi: 10.1006/bbrc.1994.2809. [DOI] [PubMed] [Google Scholar]
- 243.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 244.Shields A, Witte O N, Rothenberg E, Baltimore D. High frequency of aberrant expression of Moloney murine leukemia virus in clonal infections. Cell. 1978;14:601–609. doi: 10.1016/0092-8674(78)90245-3. [DOI] [PubMed] [Google Scholar]
- 245.Shimizu K, Ishida N. The smallest protein of Sendai virus: its candidate function of binding nucleocapsid to envelope. Virology. 1975;67:427–437. [PubMed] [Google Scholar]
- 246.Simons K, Garoff H. The budding mechanisms of enveloped animal viruses. J Gen Virol. 1980;50:1–21. doi: 10.1099/0022-1317-50-1-1. [DOI] [PubMed] [Google Scholar]
- 247.Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 248.Smith A J, Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Smith J F, Brown D T. Envelopment of Sindbis virus: synthesis and organization of proteins in cells infected with wild type and maturation-defective mutants. J Virol. 1977;22:662–678. doi: 10.1128/jvi.22.3.662-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Söderlund H. Kinetics of formation of the Semliki Forest virus nucleocapsid. Intervirology. 1973;1:354–361. doi: 10.1159/000148864. [DOI] [PubMed] [Google Scholar]
- 251.Spaan W, Cavanagh D, Horzinek M C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 252.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and gag-pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Strambio-de Castillia C, Hunter E. Mutational analyses of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Strauss E G. Mutants of Sindbis virus. III. Host polypeptides present in purified HR and ts103 virus particles. J Virol. 1978;28:466–474. doi: 10.1128/jvi.28.2.466-474.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Strauss E G, Birdwell C R, Lenches E M, Staples S E, Strauss J H. Mutants of Sindbis virus. II. Characterization of a maturation-defective mutant, ts103. Virology. 1977;82:122–149. doi: 10.1016/0042-6822(77)90038-1. [DOI] [PubMed] [Google Scholar]
- 257.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Stricker R, Roux L. The major glycoprotein of Sendai virus is dispensable for efficient virus particle budding. J Gen Virol. 1991;72:1703–1707. doi: 10.1099/0022-1317-72-7-1703. [DOI] [PubMed] [Google Scholar]
- 259.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 261.Suomalainen M, Garoff H. Alphavirus spike-nucleocapsid interaction and network antibodies. J Virol. 1992;66:5106–5109. doi: 10.1128/jvi.66.8.5106-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Surovoy A, Dannull J, Moelling K, Jung G. Conformational and nucleic acid binding studies on the synthetic nucleocapsid protein of HIV-1. J Mol Biol. 1993;229:94–104. doi: 10.1006/jmbi.1993.1011. [DOI] [PubMed] [Google Scholar]
- 264.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tanchou V, Gabus C, Rogemond V, Darlix J-L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 266.Teich N. Taxonomy of retroviruses. In: Weiss R, Teich N, Varmus H E, Coffin J M, editors. RNA tumor viruses. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 25–207. [Google Scholar]
- 267.Tsukita S, Yonemura S, Tsukita S. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- 268.Tyrrell D L J, Norrby E. Structural polypeptides of measles virus. J Gen Virol. 1978;39:219–229. doi: 10.1099/0022-1317-39-2-219. [DOI] [PubMed] [Google Scholar]
- 269.Vaux D J T, Helenius A, Mellman I. Spike-nucleocapsid interaction in Semliki Forest virus reconstructed using network antibodies. Nature. 1988;336:36–42. doi: 10.1038/336036a0. [DOI] [PubMed] [Google Scholar]
- 270.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Vogel R H, Provencher S W, von Bonsdorff C-H, Adrian M, Dubochet J. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature. 1986;320:533–535. doi: 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- 273.von Bonsdorff C-H, Harrison S C. Hexagonal glycoprotein arrays from Sindbis virus membranes. J Virol. 1978;28:578–583. doi: 10.1128/jvi.28.2.578-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wagner, R. R. (ed.). The rhabdoviruses. Plenum Press, New York, N.Y.
- 275.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wakefield L, Brownlee G G. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix proteins. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Weclewicz K, Ekström M, Kristensson K, Garoff H. Specific interactions between retrovirus env and core proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Weiss R A. Rhabdovirus pseudotypes. In: Bishop D H L, editor. Rhabdoviruses. Cleveland, Ohio: CRC Press, Inc.; 1981. pp. 51–65. [Google Scholar]
- 280.Weiss R A, Boettiger D, Murphy H M. Pseudotypes of avian sarcoma viruses with the envelope properties of vesicular stomatitis virus. Virology. 1977;76:808–825. doi: 10.1016/0042-6822(77)90261-6. [DOI] [PubMed] [Google Scholar]
- 281.Weiss R A, Wong A L. Phenotypic mixing between avian and mammalian RNA tumor viruses. I. Envelope pseudotypes of Rous sarcoma virus. Virology. 1977;76:826–834. doi: 10.1016/0042-6822(77)90262-8. [DOI] [PubMed] [Google Scholar]
- 282.Weldon R A, Jr, Wills J W. Characterization of a small (25-Kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Whelan S P J, Ball L A, Barr J N, Wertz G T W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 285.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 286.Whitt M A, Chong L, Rose J K. Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J Virol. 1989;63:3569–3578. doi: 10.1128/jvi.63.9.3569-3578.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 288.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 290.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wimmer E. Cellular receptors for animal viruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 292.Witte O N, Baltimore D. Mechanism of formation of pseudotypes between vesicular stomatitis virus and murine leukemia virus. Cell. 1977;11:505–511. doi: 10.1016/0092-8674(77)90068-x. [DOI] [PubMed] [Google Scholar]
- 293.Xiang Y, Cameron C, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late-assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Yamshchikov G V, Ritter G D, Vey M, Compans R W. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214:50–58. doi: 10.1006/viro.1995.9955. [DOI] [PubMed] [Google Scholar]
- 295.Yao J, Strauss E G, Strauss J H. Molecular genetic study of the interaction of Sindbis virus E2 with Ross river virus E1 for virus budding. J Virol. 1998;72:1418–1423. doi: 10.1128/jvi.72.2.1418-1423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Yao J S, Strauss E G, Strauss J H. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ye Z, Pal R, Fox J W, Wagner R R. Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J Virol. 1987;61:239–246. doi: 10.1128/jvi.61.2.239-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ye Z, Sun W, Suryanarayana K, Justice P, Robinson D, Wagner R R. Membrane-binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J Virol. 1994;68:7386–7396. doi: 10.1128/jvi.68.11.7386-7396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ye Z P, Baylor N W, Wagner R R. Transcription inhibition and RNA-binding domains of influenza A virus matrix protein mapped with anti-idiotypic antibodies and synthetic peptides. J Virol. 1989;63:3586–3594. doi: 10.1128/jvi.63.9.3586-3594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Yoshida T, Nagai Y, Maeno K, Iinuma M, Hamaguchi M, Matsumoto T, Nagayoshi S, Hoshino M. Studies on the role of M protein in virus assembly using a ts mutant of HVJ (Sendai virus) Virology. 1979;92:139–154. doi: 10.1016/0042-6822(79)90220-4. [DOI] [PubMed] [Google Scholar]
- 301.Yoshida T, Nagai Y, Yoshii S, Maeno K, Matsumoto T, Hoshino M. Membrane (M) protein of HVJ (Sendai virus): its role in virus assembly. Virology. 1976;71:143–161. doi: 10.1016/0042-6822(76)90101-x. [DOI] [PubMed] [Google Scholar]
- 302.Young J A T, Bates P, Willert K, Varmus H E. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science. 1990;250:1421–1423. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- 303.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Závada J. The pseudotypic paradox. J Gen Virol. 1982;63:15–24. doi: 10.1099/0022-1317-63-1-15. [DOI] [PubMed] [Google Scholar]
- 306.Zhang J, Lamb R A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
- 307.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C-H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307a.Zhao H, Garoff H. The M1 and NP proteins of influenza A virus form homo- but not heterooligomeric complexes when coexpressed in BHK-21 cells. J Gen Virol. 1998;79:2435–2446. doi: 10.1099/0022-1317-79-10-2435. [DOI] [PubMed] [Google Scholar]
- 308.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]
- 309.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ziemiecki A, Garoff H. Subunit composition of the membrane glycoprotein complex of Semliki Forest virus. J Mol Biol. 1978;122:259–269. doi: 10.1016/0022-2836(78)90189-4. [DOI] [PubMed] [Google Scholar]
- 312.Ziemiecki A, Garoff H, Simons K. Formation of the Semliki Forest virus membrane glycoprotein complexes in the infected cell. J Gen Virol. 1980;50:111–123. doi: 10.1099/0022-1317-50-1-111. [DOI] [PubMed] [Google Scholar]
- 313.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]