VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic) syndrome is a recently described entity that is associated with severe autoinflammatory manifestations.1 This syndrome is due to an acquired somatic mutation in the gene coding the ubiquitin-activating enzyme 1, UBA1, leading to the expression of a catalytically impaired cytoplasmic isoform that has been linked to inflammation.1 Most mutations affect methionine 41, but alternative mutations have been described affecting the splice acceptor site of exon 3 or serine 56.2,3 Although definitive diagnosis of VEXAS syndrome requires UBA1 molecular testing, this remains expensive and can be difficult to obtain in low-income countries. There is therefore a need to identify screening criteria to filter out patients with autoinflammatory manifestations who should undergo testing for the UBA1 mutation.
One of the defining features of VEXAS syndrome is the presence of vacuoles in neutrophilic and erythroid precursors. However, vacuolization of hematopoietic cells has also been described in various contexts, including inflammatory conditions, alcohol abuse, copper deficiency, and malnutrition.4 Based on a retrospective study of 6 VEXAS syndrome patients, Lacombe et al5 recently reported that a threshold of 10% of neutrophilic precursors with at least 1 vacuole provided high sensitivity and specificity for identifying VEXAS syndrome. However, the control population was mostly composed of patients with myelodysplastic syndrome and not patients suspected of having VEXAS syndrome, and in the absence of an external validation cohort, the generalizability of the results has not been assessed.
Herein, a quantitative assessment of myeloid precursor vacuolization was performed in bone marrow smears from patients referred for UBA1 mutation testing between December 2020 and March 2022 to the Lyon university hospital (exploratory cohort, n = 32). The demographic characteristics of the 17 UBA1-mutated and 15 UBA1-WT patients were similar. However, UBA1-mutated patients had more frequently fever, chondritis, and ocular involvement; there was also a trend toward more frequent skin lesions, arthralgia and arthritis, vasculitis, and lung involvement. UBA1-mutated patients also had higher of C-reactive protein levels, and higher mean erythrocyte corpuscular volume values. Patients without VEXAS more often had a definite diagnosis of myelodysplastic syndrome (Suppl. Table).
We first assessed interobserver variability in the quantitative measurement of vacuolization. Four expert cytologists (AR, CD, LJ, and FM) from 3 different hospitals reviewed the same 28 bone marrow smears from a fraction of the exploratory cohort, including follow-up bone marrow smears (20 from UBA1-mutated samples, and 8 from UBA1-WT samples). The experts were asked to determine the proportion of cells with vacuoles as well as the number of vacuoles per cell in early neutrophilic precursors (myeloblasts and promyelocytes), late neutrophilic precursors (myelocytes and metamyelocytes), early erythroid precursors (proerythroblasts and basophilic erythroblasts), and late erythroid precursors (polychromatophilic and acidophilic erythroblasts). All smears were analyzed in a blinded fashion, cytologists having no information about the clinical characteristics of the patients or the results of the UBA1 test. There was a satisfactory interobserver correlation (mean standard deviations of 7% for early neutrophilic precursors and 8% for early erythroid precursors), with a higher standard deviation in cases with the highest proportion of vacuolized cells (Suppl. Figure S1). We thus concluded that the quantitative assessment of vacuolization was sufficiently robust to be used as a diagnostic tool, so we analyzed a larger series of patients to decipher if a diagnostic threshold could reliably identify UBA1-mutated patients.
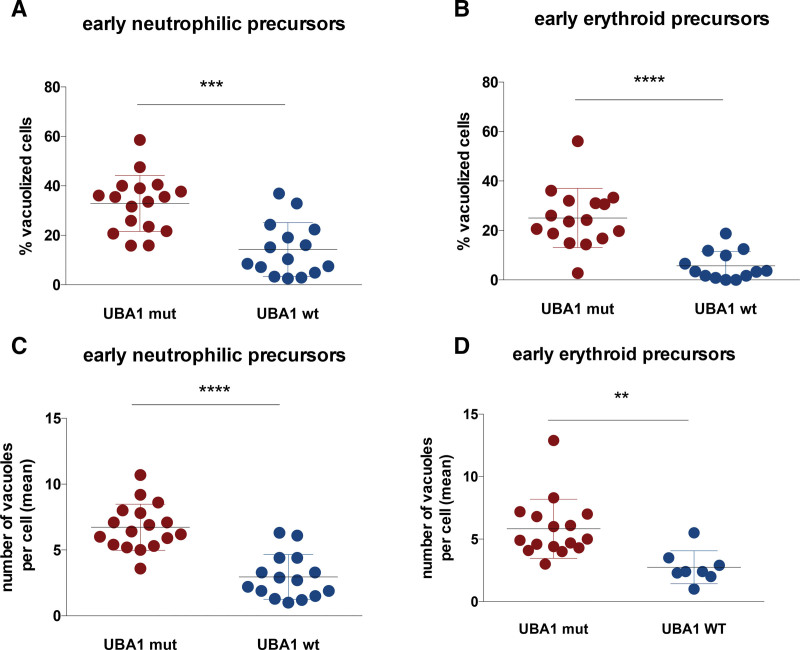
Analysis of smears from the first bone marrow aspiration of 17 UBA1-mutated patients and 15 UBA1-WT patients found a greater proportion of vacuolized cells in early neutrophil precursors (mean, 35% versus 14%, P = 0.001) and early erythroid precursors (mean, 25% versus 6%, P < 0.001) for UBA1-mutated patients (Figure 1A and B). There were very few vacuolized cells in the more mature precursors (<3%), and there was no significant difference between UBA1-mutated or WT patients (data not shown). Of note, there was no correlation between the percentage of vacuolized cells and the variant allele frequency (VAF) of UBA1 mutation among the 7 patients with available data (VAF ranging from 21 to 81%; Suppl. Figure S2). In addition, UBA1-mutated patients also had a higher number of vacuoles per cell in both early neutrophilic precursors (mean, 6.7 vacuoles per cell in UBA1-mutated patients versus 2.9 in WT patients, P < 0.001) and in early erythroid precursors (mean, 5.8 vacuoles per cell in UBA1-mutated patients versus 2.7 in WT patients, P = 0.001; Figure 1C and D). However, considering the high variability of the number of vacuoles per cell and the time required for this procedure, this parameter was not retained in the diagnostic strategy described below.
Figure 1.
Quantitative assessment of vacuolization of myeloid precursors. Relative proportions of vacuolized cells and mean number of vacuoles/cell in early neutrophil precursors (A and C), early erythroid precursors (B and D) in the exploratory cohort of 17 UBA1-mutated patients and 15 UBA1-WT. ***P < 0.001.
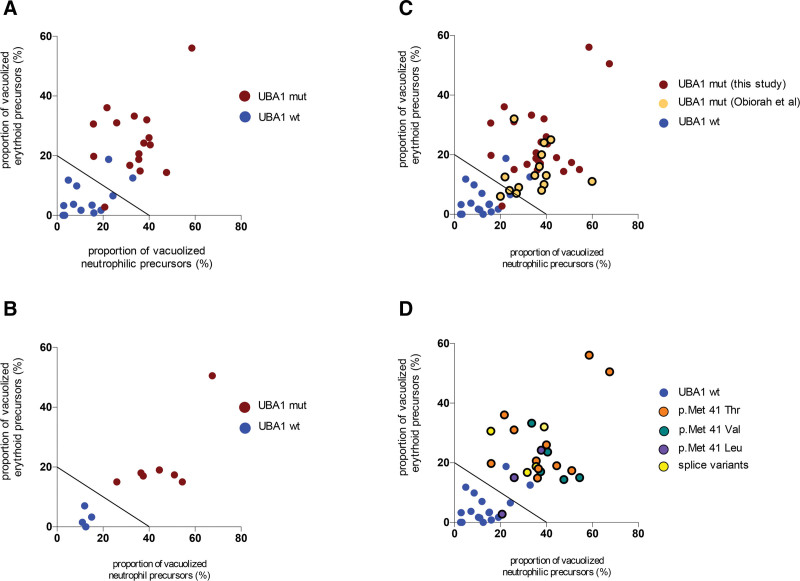
Applying the proposed threshold of 10% vacuolized neutrophilic precursors5 to the 32 patients included herein had an excellent sensitivity (100%) but a low specificity of only 47%, highlighting the need to refine the performance of vacuole evaluation in VEXAS diagnosis. To optimize the diagnostic performance of quantitative vacuolization assessment, the proportion of vacuolized cells in early erythroid precursors was plotted versus the proportion of vacuolized cells in early neutrophilic precursors. On the resulting graph, UBA1 mutant versus WT patients could clearly be discriminated (Figure 2A). The following formula achieved a sensitivity of 93.7% and a specificity of 83% to identify UBA1-mutated patients:
Figure 2.
Establishment and validation of a diagnostic threshold. Graphical distribution of UBA1-mutated patients (red dots) and UBA1-WTpatients (blue dots) in the exploratory (A) and validation (B) cohorts according to their proportion of vacuolized cells in early erythroid and neutrophilic precursors; (C) distribution of VEXAS syndrome cases described in Obiorah et al6; (D) distribution of cases according to the nature of the mutation.
where VacEP is the percentage of vacuolized early erythroid precursors and VacEN the percentage of vacuolized eary neutrophilic precursors.
The performance of this threshold was then tested in an external validation cohort of 7 UBA1-mutated and 4 UBA1-WT patients referred for UBA1 mutation testing at the Nantes University Hospital; all patients were correctly classified after quantitative vacuole assessment (Figure 2B).
Data published by Obiorah et al6 reporting the proportion of erythroid precursors (not restricted to early precursors) and immature myeloid precursors in 15 patients with VEXAS syndrome were also analyzed. Patients in this cohort had a similar vacuolization profile, leading to a sensitivity of the proposed threshold of 93.7% (1 false negative in 15 patients; Figure 2C). Finally, given the differential impact of UBA1 mutations on residual UBA1b translation,7 we evaluated whether the nature of UBA1 mutation might impact the magnitude of vacuolization, but we did not observe any clustering according to the nature of the UBA1 mutation (Figure 2D).
The present study found that the quantitative assessment of the proportion of vacuolized precursors on bone marrow smears is reproducible and allows the identification of VEXAS syndrome patients with a high level of sensitivity and specificity.
VEXAS is a recently recognized syndrome that is important to detect to avoid diagnostic wandering and propose specific therapeutic interventions that have proven to be effective, such as ruxolitinib.8 The clinical presentation of VEXAS syndrome is heterogeneous but generally easy to recognize for experienced physicians. However, an increasing number of reports describe unusual clinical presentations of VEXAS syndome and expand the phenotypic spectrum of the disease.9–11 Molecular confirmation is mandatory to definitively confirm or exclude the diagnosis; this is based on UBA1 sequencing on DNA extracted from blood or bone marrow samples, but access to molecular testing can be difficult for patients in low-income countries or in countries without public reimbursement for molecular testing.12 In this context, the diagnostic threshold proposed herein is an easy, free and performing way to specify the pretest probability of UBA1 mutation, which can be used to select patients for molecular testing. Still, the management of VEXAS patients would remain complicated in low-income countries, especially to access to effective treatments such as ruxolitinib8 or azacytidine.13 The results presented here refine a previously proposed threshold.5 One obvious explanation for this difference is the size of the cohort used herein, which is 3 times larger, and probably better captures the diversity of VEXAS syndrome phenotypes. However, the cohort size is still limited and larger cohorts are needed to confirm our results. Another explanation is methodological, as the control population comprised patients with clinical manifestations suggestive of VEXAS syndrome, whereas Lacombe et al5 included mostly patients with myelodysplastic syndromes. The excellent performance of the threshold described herein in a completely independent cohort confirms its diagnostic value. As specificity is not absolute, molecular tests should still be performed to confirm a suspected diagnosis. On the other hand, patients who are predicted to be UBA1 WT after quantitative assessment of vacuolization should still be referred for molecular testing in case of strong clinical suspicion of VEXAS. In this cohort as well as in previously published cohorts,5,6 there was no UBA1-mutated patients with less than 10% vacuolized neutrophilic precursors, which could be a good threshold to rule out the diagnosis. However, it should be recalled that rare cases of VEXAS syndrome without vacuoles have also been described,14 therefore UBA1 sequencing should always be performed in cases of strong clinical suspicion.
Another interesting aspect of the present study is to propose an objective evaluation of the vacuolization phenotype of VEXAS syndrome, which could be useful to further characterize this syndrome. First, emerging data suggest that the phenotype of VEXAS syndrome differs according to the nature of the mutation, possibly due to a different translation rate of the cytoplasmic isoform UBA1b.7 This observation prompted us to analyze whether the vacuolization phenotype was also influenced by the nature of the mutation, but we did not find a clear clustering. Second, sequencing of the entire UBA1 gene revealed atypical mutations such as p.Ser56Phe3 or others of undetermined significance (unpublished data). In these cases, quantitative assessment of vacuolization could be a valuable tool to decide whether the patient should be considered as having VEXAS or not.
Taken together, the present study provides a framework to quantitatively measure myeloid precursor vacuolization in VEXAS syndrome, which could be useful both for diagnosis and for better characterization of this recently described disease.
AUTHOR CONTRIBUTIONS
AR, LJ, CD, and FM did the reproducibility experiment. CR and ME provided the cases of the validation cohort. EB, MH, MGF, and YJ provided the cases of the exploratory cohort. PS designed the study and wrote the manuscript. All authors revised the manuscript.
DISCLOSURES
The authors do not have any conflict of interest to disclose.
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
REFERENCES
- 1.Beck DB, Ferrada MA, Sikora KA, et al. Somatic mutations in UBA1 and severe adult-onset autoinflammatory disease. N Engl J Med. 2020;383:2628–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourbon E, Heiblig M, Gerfaud-Valentin M, et al. Therapeutic options in Vexas syndrome: insights from a retrospective series. Blood. 2021;137:3682–3684. [DOI] [PubMed] [Google Scholar]
- 3.Poulter JA, Collins JC, argo C, et al. Novel somatic mutations in UBA1 as a cause of VEXAS syndrome. Blood. 2021;137:3676–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurnari C, Pagliuca S, Durkin L, et al. Vacuolization of hematopoietic precursors: an enigma with multiple etiologies. Blood. 2021;137:3685–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacombe V, Prevost M, Bouvier A, et al. Vacuoles in neutrophil precursors in VEXAS syndrome: diagnostic performances and threshold. Br J Haematol. 2021;195:286–289. [DOI] [PubMed] [Google Scholar]
- 6.Obiorah IE, Patel BA, Groarke EM, et al. Benign and malignant hematologic manifestations in patients with VEXAS syndrome due to somatic mutations in UBA1. Blood Adv. 2021;5:3203–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrada MA, Savic S, Ospina Cardona D, et al. Translation of cytoplasmic UBA1 contributes to VEXAS syndrome pathogenesis. Blood. 2022;140:1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heiblig M, Ferrada MA, Koster MJ, et al. Ruxolitinib is more effective than other JAK Inhibitors to treat VEXAS Syndrome: a retrospective multi center study. Blood. 2022;140:927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto H, Asano T, Tsuchida N, et al. Behçet’s disease with a somatic UBA1 variant:Expanding spectrum of autoinflammatory phenotypes of VEXAS syndrome. Clin Immunol. 2022;238:108996. [DOI] [PubMed] [Google Scholar]
- 10.Ribereau-Gayon E, Heiblig M, Bourbon E, et al. Atypical extensive lupus tumidus-like eruption as an early presentation of VEXAS syndrome. Int J Dermatol. 2022;61:e89–e91. [DOI] [PubMed] [Google Scholar]
- 11.Georgin-Lavialle S, Terrier B, Guedon AF, et al. Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a multicentre case series of 116 French patients. Br J Dermatol. 2021;186:564–574. [DOI] [PubMed] [Google Scholar]
- 12.Drake TM, Knight SR, Harrison EM, et al. Global inequities in precision medicine and molecular cancer research. Front Oncol. 2018;8:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comont T, Heiblig M, Rivière E, et al. Azacitidine for patients with Vacuoles, E1 Enzyme, X-linked, Autoinflammatory, Somatic syndrome (VEXAS) and myelodysplastic syndrome: data from the French VEXAS registry. Br J Haematol. 2021;196:969–974. [DOI] [PubMed] [Google Scholar]
- 14.Templé M, Duroyon E, Croizier C, et al. Atypical splice-site mutations causing VEXAS syndrome. Rheumatology. 2021;60:e435–e437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.