Abstract
Metabolic reprogramming is required for tumors to meet the bioenergetic and biosynthetic demands of malignant progression. Numerous studies have established a causal relationship between oncogenic drivers and altered metabolism, most prominently aerobic glycolysis, which supports rapid growth and affects the tumor microenvironment. Less is known about how the microenvironment modulates cancer metabolism. In the present study, we found that low extracellular pH, a common feature of solid tumors, provoked PDAC cells to decrease glycolysis and become resistant to glucose starvation. This was accompanied by increased dependency on mitochondrial metabolism, in which long-chain fatty acids became a primary fuel source. Consistent with previous reports, low pH enhanced tumor cell invasive capacity. A novel finding was that limiting PDAC metabolic flexibility by either depletion of oxidative phosphorylation capacity or the pharmacological inhibition of fatty-acid oxidation prevented invasion induced by low extracellular pH. Altogether, our results suggest for the first time that targeting fatty-acid oxidation may be a viable adjunct strategy for preventing metastatic progression of pancreatic cancer mediated by the acidic tumor compartment.
Keywords: pancreatic cancer, acidic microenvironment, aerobic glycolysis, fatty-acid oxidation, EMT, invasion
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with the highest mortality rate among all major cancers [1]. PDAC features an unusually intense desmoplastic reaction that fosters a nutrient-deficient, hypoxic and acidic tumor microenvironment [2]. Recognized as a hallmark of most solid tumors, hypoxia profoundly influences multiple facets of cancer biology through HIF-mediated induction of metabolic reprogramming, neovascularization, epithelial-to-mesenchymal transition (EMT) and metastasis [3, 4]. Importantly, the observed alterations in glucose metabolism has led to improvements in diagnostic imaging and the development of pharmacological inhibitors that target glycolytic pathways [5]. Further understanding of metabolic adaptations that support tumor growth and progression in the PDAC tumor microenvironment will likely uncover additional vulnerabilities that may be therapeutically exploited.
Often colluding with hypoxia, though with little spatial correlation [6], is an acidic tumor microenvironment caused by a combination of increased metabolism, low perfusion, and inflammation. While the extracellular pH (pHe) of most normal tissues range between 7.2 to 7.4, many solid tumors exhibit an acidic pHe ranging from 6.5 to 6.9 [7]. Independent of hypoxia, extracellular acidosis facilitates malignant features including EMT, chemoresistance, and immune evasion [8, 9]. While metabolic reprogramming under hypoxia has been extensively characterized, relatively little is understood regarding the metabolic adaptations that occur in response to acidic conditions and how they support cancer progression. In the present study, we investigated metabolic changes and dependencies of PDAC cells at low pHe and offer novel insight into the influence of pHe on tumor metabolism and potential therapeutic avenues.
2. Materials and Methods
2.1. Cell Culture
S2–013 is a subclone of a human pancreatic tumor cell line (SUIT-2) derived from liver metastasis [10]. T3M4 is a human carcinoembryonic antigen (CEA)-producing cell line established from a primary PDAC transplanted into nude mice [11]. Both S2–013 and T3M4 cells were validated through STR profiling by the Molecular Diagnostics Laboratory at UNMC. Capan-2 were purchased directly from American Type Culture Collection (ATCC). All parental lines were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing high glucose and L-glutamine, supplemented with 10% fetal bovine serum (FBS), at 37°C and 5% CO2. To generate ρ° cells, which are depleted of mitochondrial DNA, parental lines were cultured in 50ng/mL ethidium bromide, 100mM sodium pyruvate, and 5μg/mL uridine for at least one month. For establishing the experimental pH conditions, bicarbonate concentration in DMEM was manipulated according to calculations based on the Henderson-Hasselbalch equation for maintaining a pH of 7.4 (3.7g/L), 6.8 (0.93g/L), or 6.4 (0.37g/L) at 10% CO2; final pH was confirmed using a pH meter. For glucose-starvation, DMEM without glucose was supplemented with 1x glutaMAX and 10% dialyzed FBS.
2.2. In-vitro Growth Assay
Cell growth was assayed using methylene blue as previously described [12]. Briefly, viable cells were seeded and cultured in 96-well plates in replicates of four. After each time point, plates were fixed with 10% formalin and stained with methylene blue (1% w:v in 0.01M borate buffer pH 8.4). Excess dye was completely removed through extensive washing with borate buffer. Dye was solubilized using 0.1M HCl/Ethanol (1:1) and absorbance was measured at 650nm using a microplate reader. Relative growth was calculated based on the absorbance measurements of the first time point.
2.3. Immunoblotting
Whole cell lysates following 24 hour incubation in pH manipulated media were prepared using RIPA buffer (1% sodium deoxycholate, 0.15 M NaCl, 0.1% SDS, 1% (v/v) Triton X-100, 0.05 M Tris HCl, pH 7.4) supplemented with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA). Protein concentrations was determined by Pierce BCA protein assay (Thermo Fisher Scientific). Equal amounts of total protein were resolved on 4–20% SDS-PAGE gels (Bio-Rad, Hercules, CA, USA) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Burlington, MA, USA), which were blocked with 5% bovine serum albumen and incubated with primary antibodies (S1 Table) overnight at 4 °C. Following incubation with HRP-conjugated secondary antibodies, the antigen-antibody complex was developed using Bio-Rad enhanced chemiluminescence (ECL) Prime Western Blotting detection reagent (GE Life Sciences, Pittsburgh, PA, USA).
2.4. Invasion Assay
Relative invasion was determined using 24-well Matrigel invasion chambers with 8 μm PET membranes (Corning, Corning, NY,USA). Inserts were rehydrated in serum-free medium per manufacturer’s instructions. Complete medium (pH 7.4, 6.8, or 6.4) was used as chemoattractant. Suspensions of 5 × 104 cells in 500 μL of serum-free medium (pH 7.4, 6.8, or 6.4) were added into the chambers and incubated for 22 hours at 37°C in 10% CO2. For FAO inhibition,100μM etomoxir was added into the chambers. Non-invading cells were removed with a cotton swab while invaded cells were fixed in formalin and stained with methylene blue. Images were captured using an EVOS Cell Imaging System (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at 10x magnification. Quantification was performed by solubilizing the dye with 0.1M HCl/Ethanol (1:1) and measuring absorbance at 650nm.
2.5. Metabolic Flux Assays
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) of live cells were measured in real-time using the Seahorse XFe96 Analyzer (Agilent, Santa Clara, CA, USA). Cells were pre-incubated at pH 7.4, 6.8, or 6.4 for 24 hours and then seeded at a concentration of 2 ×105 (S2–013), 3 ×105 (Capan-2), or 3 ×105 (T3M4) cells per well in 96-well assay plates and allowed to attach overnight. For glycolysis stress test, culture medium was replaced with Seahorse XF DMEM (Agilent) supplemented with 2mM glutamine, pH 7.4 one hour prior to the assay. Measurements were taken before and after each sequential treatment with glucose (10mM), oligomycin (1μM), and 2-DG (50 mM). For mitochondrial stress test, culture medium was replaced with Seahorse XF DMEM supplemented with 10mM glucose, 1mM sodium pyruvate, and 2mM glutamine, pH 7.4 one hour prior to the assay. Cells were sequentially treated with oligomycin (1μM), FCCP (1μM), and rotenone (0.5μM). For determining FAO and glutamine-dependent OCR, culture medium was replaced with unbuffered DMEM, which was pH adjusted with 1N HCL or 1N NaOH to 7.4, 6.8, or 6.4 and supplemented with 2% FBS and 2mM glutamine, one hour prior to the assay. Following basal measurements, cells were treated with either etomoxir (50μM) or CB-839 (2μM). FAO and glutamine-dependent OCR were determined as the loss of OCR following etomoxir or CB-839 injection. All measurements were normalized to total protein as determined by BCA assay.
2.6. Glucose Uptake Measurements
Cells were seeded in a 96-well plate and incubated at pH 7.4, 6.8, or 6.4 for 24 hours. Uptake of glucose was determined using Glucose Uptake-Glo Assay (Promega, Madison, WI, USA) per manufacturer’s instructions.
2.7. Cell Viability Assays
Cells were pre-incubated at pH 7.4, 6.8, or 6.4 for 24 hours and then starved of glucose for the indicated time points or treated with the following metabolic inhibitors: etomoxir (50μM) (Sigma-Aldrich, St Louis, MO, USA), CB-839 (2μM) (Selleckchem, Houston, TX, USA), oligomycin (1μM) (Sigma-Aldrich), or 2-deoxy-D-glucose (2-DG) (20mM) (Sigma-Aldrich) for 48 hours. Cell viability was determined by adding alamarBlue (Bio-Rad) in an amount equal to 10% of the volume in the well and incubating for 2 hours. Fluorescence (560ex/590em) was measured using a microplate reader. Relative viability was calculated based on the fluorescence intensity of the untreated or control wells.
2.8. PCR Determination of mtDNA Copy Number
Mitochondrial DNA content, relative to nuclear DNA, was quantified using real-time PCR analysis as previously described [13]. Briefly, total DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RT-PCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). The primer sets for smtDNA and nucDNA are listed in S2 Table. Mitochondrial DNA copy number was determined by the following equation: 2 × 2ΔCT where ΔCT = (nucDNA CT – mtDNA CT).
2.9. Lipid Uptake Assay
Fluorescently labeled palmitate, BODIPY C16 (Invitrogen, Carlsbad, CA, USA), was prepared as a 5mM solution in DMSO. Lipids were diluted in serum-free media without glucose and incubated at a 6:1 molar ratio with defatted bovine serum albumin (BSA) (GoldBio, St Louis, MO, USA) for 1 hour at 37°C to allow for conjugation. Cells were seeded in a 96-well plate and incubated at pH 7.4, 6.8, or 6.4 for 24 hours. One hour prior to lipid exposure, culture media was replaced with serum-free media without glucose. BODIPY C16:BSA solution was added to each well and uptake was allowed to proceed for 15 minutes. Cells were washed with PBS and fluorescence (485ex/520em) was measured using a microplate reader. Normalization was performed with BCA total protein quantification.
2.10. Statistical Analysis
Statistical significance of differences between mean values was assessed using an unpaired, two-sided t-test. Differences were considered significant at P < 0.05. Unless indicated otherwise, data are shown as mean ± SD. Statistical analyses are reported for biological replicates.
3. Results
3.1. Acidic conditions drive an EMT phenotype associated with reduced glucose metabolism
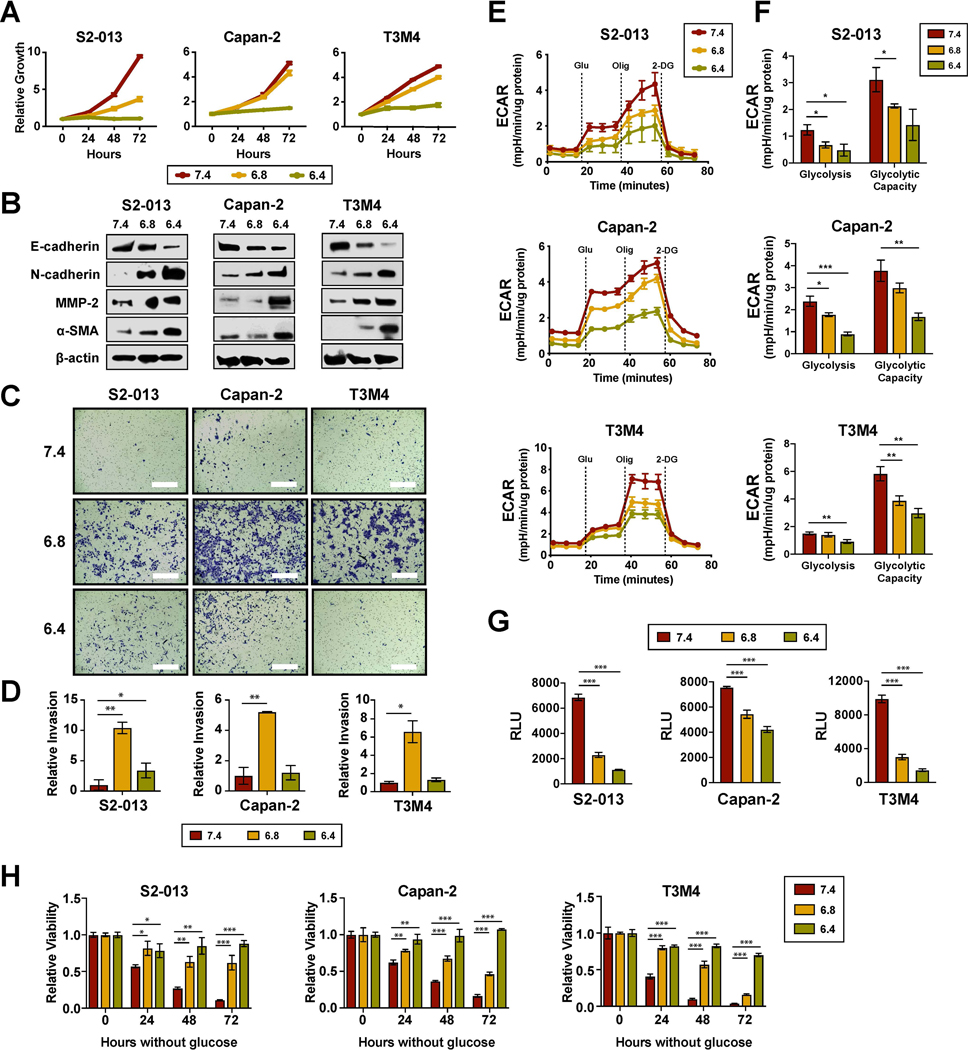
To investigate the influence of acidosis on proliferation, PDAC cells were exposed to three different pH conditions − 7.4, 6.8, or 6.4. Compared to normal conditions, low extracellular pH negatively affected the growth rates in vitro (Fig. 1A). While cells maintained the ability to proliferate at pH 6.8, little to no measurable growth was observed at pH 6.4. Accompanying the impaired growth were morphological changes, specifically from epithelial-like to mesenchymal. Previous studies have demonstrated that low extracellular pH can promote EMT-associated phenotypes, such as invasion and metastasis [14]. Consistent with those findings, we observed the induction of EMT by extracellular acidification as evidenced by increased expression of N-cadherin and decreased expression of E-cadherin (Fig. 1B). Likewise, invasion was significantly increased at pH 6.8 in all three PDAC cell lines (Fig. 1C and 1D). Interestingly, this phenotype was abrogated upon further media acidification to pH 6.4.
Figure 1. Extracellular acidification induces EMT phenotype with a concomitant decrease in glycolysis.
(A) In vitro proliferation of PDAC cells cultured at pHe 7.4, 6.8, or 6.4 using a methylene blue-based assay. (B) Immunoblot analysis from whole cell lysates following 24-hour incubation at the indicated pHe conditions, with β-actin as loading control. (C) Representative images from Matrigel invasion assays. Bar, 400μm. (D) Quantification of relative invasion (n=2). (E) Extracellular acidification rates (ECAR) measured at baseline conditions and following sequential injections of glucose, oligomycin, and 2-DG using the Seahorse XFe96 Analyzer. (F) Parameters of the glycolytic stress test calculated from the ECAR measurements (n=3). (G) Relative uptake of glucose measured using Promega Glucose Uptake-Glo Assay (n=3). (H) Relative cell viability following glucose starvation compared to unstarved controls (n=3). Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
Although metabolic reprogramming linked to EMT at normal pH is typically characterized by increased aerobic glycolysis [15, 16], we found that acidosis-induced EMT was instead accompanied by a decrease in glycolysis as measured by a Seahorse Extracellular Flux assay (Fig. 1E). The parameters determined from glycolytic stress tests showed significant reduction in both glycolysis and glycolytic capacity (Fig. 1F). To investigate whether the observed decrease in extracellular acidification rate (ECAR) was due to glucose flux being diverted to oxidative phosphorylation or resulted from a reduction in overall glucose utilization, a luminescent glucose uptake assay was performed. Supporting the latter hypothesis, a significant decrease in glucose uptake was observed with exposure to acidic stress (Fig. 1G). Furthermore, consistent with reduced dependency on glucose metabolism, PDAC cells under acidic conditions exhibited increased levels of resistance to glucose starvation (Fig. 1H).
3.2. Increased invasion mediated by extracellular acidification requires oxidative phosphorylation
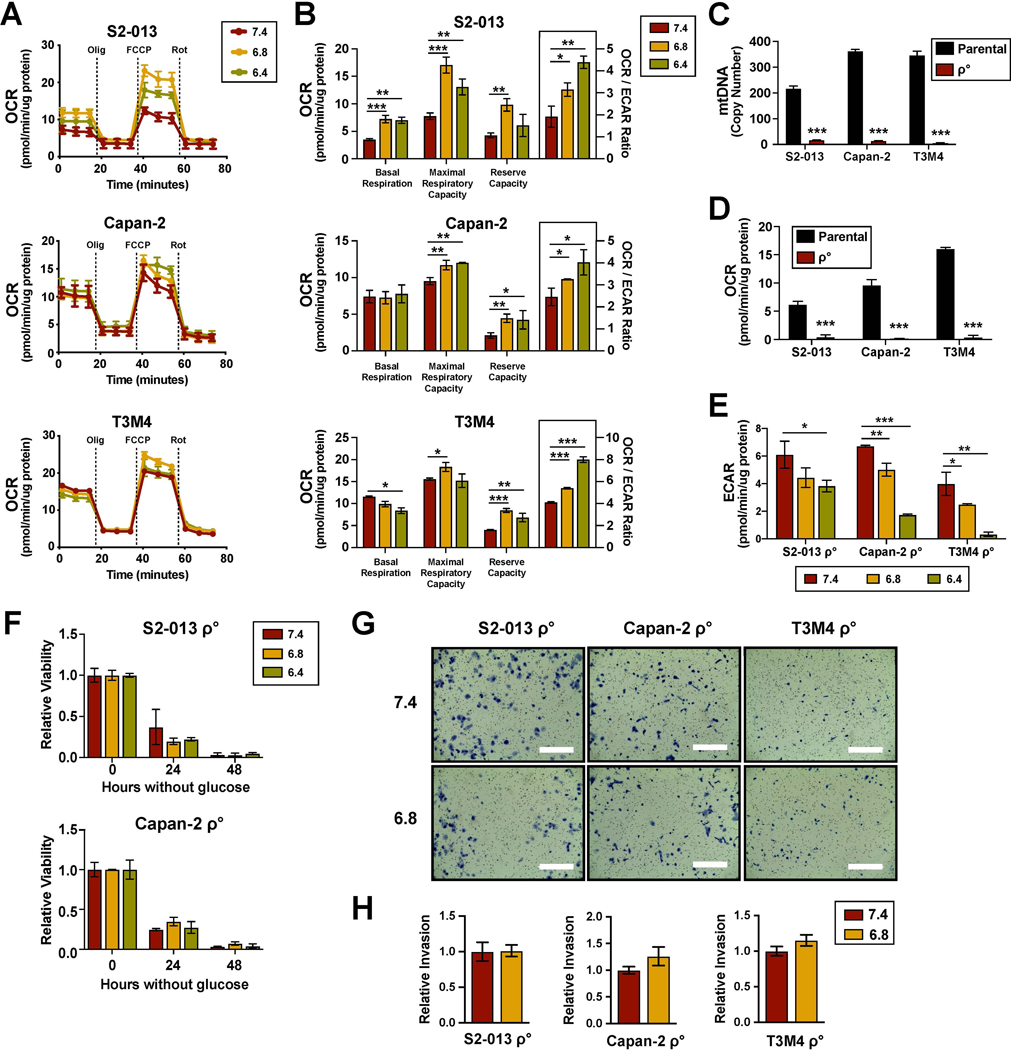
Given the observed decrease in glucose metabolism, we investigated the effects of media acidification on mitochondrial metabolism. Using a Seahorse mitochondrial stress test, we found that exposure to low extracellular pH did not drastically alter overall oxygen consumption rate (OCR) profiles (Fig. 2A). While no conclusive change was determined in basal respiration, the maximal respiration and reserve capacity (Fig. 2B) were elevated. Notably, the OCR to ECAR ratio was significantly increased with media acidification, implying a metabolic shift in dependency from aerobic glycolysis to oxidative phosphorylation (OXPHOS) (Fig. 2B).
Figure 2. Mitochondrial respiration is required for increased invasion induced by acidic conditions.
(A) Oxygen consumption rates (ECAR) measured at baseline conditions and after sequential injections of oligomycin, FCCP, and rotenone using the Seahorse XFe96 Analyzer. (B) Parameters of mitochondrial respiration calculated from the OCR measurements (n=3). (C) Quantification of mitochondrial DNA copy number through qPCR of total DNA extract using genomic DNA as reference (n=3). (D) OCR measurements of ρ° cells and their parental counterparts (n=3). (E) ECAR measurements ρ° cells pre-cultured under indicated pHe conditions (n=3). (F) Relative viability of ρ° cells following glucose starvation compared to unstarved controls (n=3). (G) Representative images from Matrigel invasion assays with ρ° cells. Bar, 400μm. (H) Quantification of relative invasion (n=2). Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
To further examine OXPHOS dependency under acidic conditions, cancer cells with depleted mitochondrial DNA (ρ°) were generated using low dose ethidium bromide. Loss of mitochondrial DNA copy number was confirmed through qPCR analysis (Fig. 2C). As expected, the ρ° cells had no detectable levels of oxygen consumption (Fig. 2D). Rather, these cells relied heavily on glycolysis and demonstrated relatively high levels of ECAR. However, resembling the parental lines, ρ° cells also exhibited reduced glycolysis in response to acidic stress (Fig. 2E). The exceptionally low levels of ECAR detected at pH 6.4 was partially due to loss of viability, as ρ° cells were less tolerant of media acidification. To test whether resistance to glucose starvation under acidic conditions was dependent on OXPHOS, we measured the viability of ρ° cells in the absence of glucose. We found that irrespective of pH, ρ° PDAC cells no longer survived without glucose (Fig. 2F). Furthermore, unlike their parental counterparts, ρ° cells did not exhibit increased invasion at low extracellular pH (Fig. 2G and 2H). Altogether, these results suggested that under low extracellular pH, PDAC cells require the ability to shift their dependency from aerobic glycolysis in order to support the invasive phenotype.
3.3. Fatty acid-oxidation specifically supports cell viability and invasion of PDAC cells under low extracellular pH
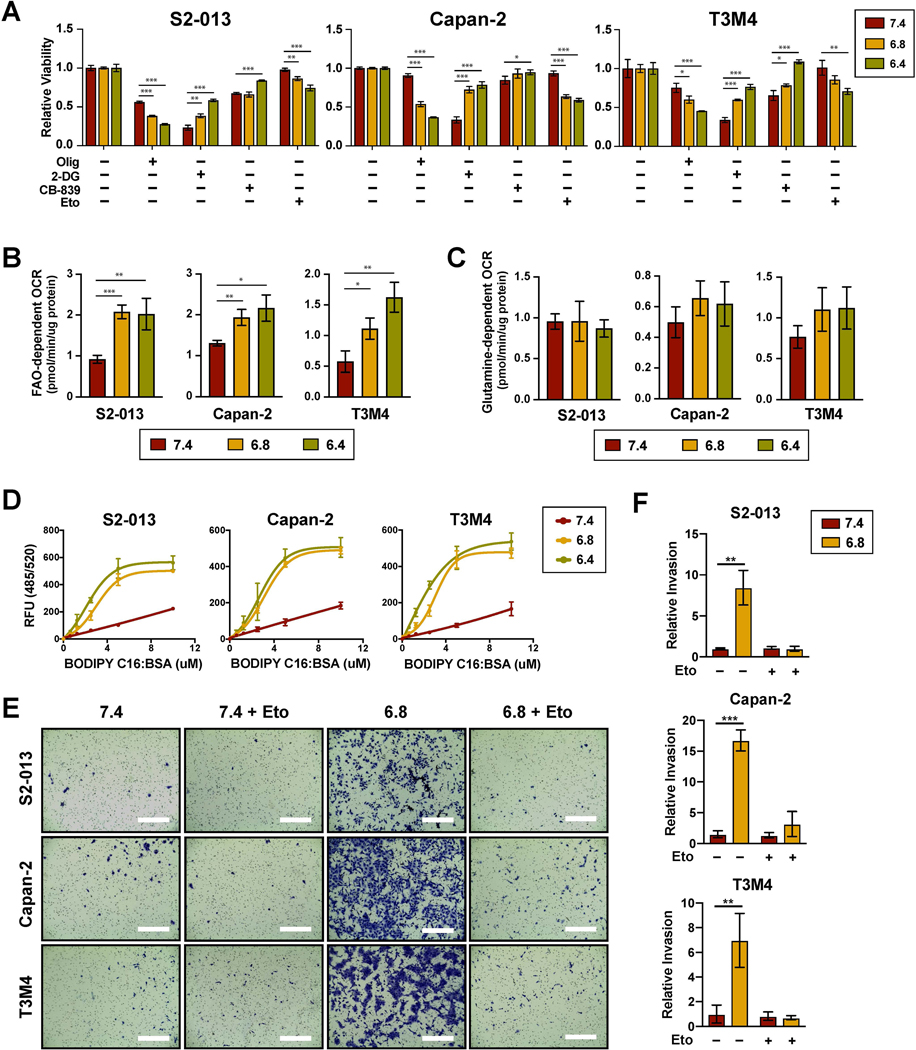
Consistent with increased OXPHOS dependency, extracellular acidification increased the sensitivity of PDAC cells to oligomycin, an ATP synthase inhibitor (Fig. 3A). To determine the major fuel source for mitochondrial metabolism under low extracellular pH, PDAC cells were treated with metabolic inhibitors of glucose, glutamine, or long-chain fatty acid oxidation (FAO) pathways using 2-deoxyglucose (2-DG), CB-839, and etomoxir, respectively. Only treatment with etomoxir mirrored the effects observed with OXPHOS inhibition, suggesting a metabolic shift in dependency from glucose to fatty acids (Fig. 3A). An increase in FAO-dependent OCR was also confirmed with a Seahorse assay (Fig. 3B). This was in contrast to glutamine-dependent OCR which was unaffected by extracellular acidification (Fig. 3C). Furthermore, PDAC cells exhibited increased lipid uptake under acidic culture conditions, as measured by incubating the cells with fluorescently labelled palmitate (BODIPY C16) (Fig. 3D). Finally, we tested whether blocking FAO with etomoxir could inhibit invasion at pH 6.8. Similar to what was observed in respiratory-deficient cells, treatment with etomoxir effectively prevented low pH-induced invasion (Fig. 3E and 3F).
Figure 3. Low pHe increases PDAC dependency on fatty acid oxidation.
(A) Relative cell viability following 48-hour treatment with the indicated metabolic inhibitors compared to untreated controls (n=3). FAO and glutamine-dependent OCR as determined by the decrease in basal OCR following injection of etomoxir (B) (n=3) or CB-839 (C) (n=3), respectively. (D) Lipid uptake measured by incubating cells with fluorescently labeled palmitate (BODIPY C16) conjugated to fatty acid-free BSA for 15 min following 24-hour pre-culturing under the indicated pHe conditions (n=3). (C) Representative images from Matrigel invasion assays. Bar, 400μm. (D) Quantification of relative invasion (n=2). Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
4. Discussion
Despite substantial progress in overall cancer treatment, PDAC remains a disease with poor prognosis and limited therapeutic options. While targeted therapy represents a promising strategy, KRAS mutations, the primary driver in up to 90 percent of PDAC patients, is currently undruggable [17]. Alternative approaches have focused on exploiting KRAS-regulated metabolic pathways, such as enhanced aerobic glycolysis [18]. The genetic factors that influence tumor metabolism are relatively well understood; however, much of how extrinsic factors alter metabolic dependencies is unclear. This knowledge gap may partially explain why targeting glucose metabolism has not yet translated to clinical success [19]. Significantly, our results show that acidic conditions can drive KRAS-mutant PDAC cell lines to shift away from aerobic glycolysis and become resistant to glucose starvation. Thus, efforts to inhibit glycolytic metabolism may not be effective without concurrently targeting intratumoral acidity.
Further impetus for targeting the acidic tumor compartment comes from evidence that it drives local invasion and malignant progression [20]. Consistent with previous studies, we observed that extracellular acidification of culture media to pH 6.8 induced expression of EMT markers and increased invasiveness of PDAC cells. Unanticipated, however, was that a further decrease in acidity to pH 6.4 inhibited invasion, despite elevated EMT markers. Although further investigation is necessary, we suspect this could be due to decreased motility from bioenergetic deficiency. As shown from our results, glycolysis is increasingly compromised with media acidification, without an inverse correlation in OXPHOS compensation. Furthermore, our results showed that increased acidic conditions may have a negative effect on cell migration (Supplementary Fig.1A). While unexpected, the uncoupling of migration and invasion during cancer associated EMT has been previously documented [21]. In addition, our studies demonstrated that exogenous ATP supplementation could increase invasion at pH 6.4 (Supplementary Fig. 1B). Thus, there is likely an optimal extracellular pH that supports increased invasion – where an EMT phenotype is induced but the bioenergetic demands of motility are still met.
Finally, we confirmed that PDAC cells under acidic conditions increase their dependency on mitochondrial metabolism. Notably, our study is the first to document that OXPHOS is specifically required for low pH-mediated invasion. This finding is particularly interesting given that several groups have reported OXPHOS deficiency to promote invasion [22–26]. However, our results suggest that the phenotypic response to OXPHOS inhibition is likely pH dependent. While reduced OXPHOS may prompt invasion of cancer cells residing in a non-acidic environment, it significantly dampens the invasiveness of cells under acidic stress. Furthermore, we found that low extracellular pH enhanced the uptake of exogenous lipids in PDAC cells, suggesting that fatty acids are the predominant fuel source for mitochondrial respiration under acidic conditions. This is similar to the results from a previous study that reported an increase in FAO, associated with alterations in mitochondrial and histone acetylation, in cancer cells adapted to chronic acidosis [27]. Our results extend those findings by showing that changes in fatty acid metabolism can occur following acute (24 hours) exposure to low pH conditions, indicating that a rapid stress response mechanism may be involved. Nevertheless, these results are especially pertinent in light of a recent study demonstrating that resident pancreatic stellate cells, once activated into cancer-associated fibroblasts, secrete abundant lipids into the tumor microenvironment [28]. Importantly, we found that inhibiting fatty-acid oxidation with etomoxir effectively suppressed low pH-induced invasion without altering invasiveness at pH 7.4. Based on these results, future studies to investigate fatty-acid oxidation as a potential target for adjunct therapy in metastasis prevention are warranted.
Supplementary Material
Highlights.
Low pHe induces EMT and a metabolic shift away from aerobic glycolysis
Invasive phenotype under acidic conditions require mitochondrial metabolism
Etomoxir, an FAO inhibitor, specifically inhibits growth and invasion under low pHe
Funding
This work was supported in part by funding from the National Institutes of Health (P50CA127297, U01CA210240, P01 CA217798, R01 CA128108) and DHHS/NE-LB506.
Footnotes
Conflicts of Interest
The authors have no competing interests to disclose regarding the publication of this article.
References
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2019. CA Cancer J Clin, 2019. 69(1): p. 7–34. [DOI] [PubMed] [Google Scholar]
- 2.Pandol S, Edderkaoui M, Gukovsky I, Lugea A, and Gukovskaya A, Desmoplasia of pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol, 2009. 7(11 Suppl): p. S44–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouyssegur J, Dayan F, and Mazure NM, Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature, 2006. 441(7092): p. 437–43. [DOI] [PubMed] [Google Scholar]
- 4.Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, and Jin WL, Reprogramming of the Tumor in the Hypoxic Niche: The Emerging Concept and Associated Therapeutic Strategies. Trends Pharmacol Sci, 2017. 38(8): p. 669–686. [DOI] [PubMed] [Google Scholar]
- 5.Hay N, Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer, 2016. 16(10): p. 635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helmlinger G, Yuan F, Dellian M, and Jain RK, Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med, 1997. 3(2): p. 177–82. [DOI] [PubMed] [Google Scholar]
- 7.Kimbrough CW, Khanal A, Zeiderman M, Khanal BR, Burton NC, McMasters KM, et al. , Targeting Acidity in Pancreatic Adenocarcinoma: Multispectral Optoacoustic Tomography Detects pH-Low Insertion Peptide Probes In Vivo. Clin Cancer Res, 2015. 21(20): p. 4576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki A, Maeda T, Baba Y, Shimamura K, and Kato Y, Acidic extracellular pH promotes epithelial mesenchymal transition in Lewis lung carcinoma model. Cancer Cell Int, 2014. 14(1): p. 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, et al. , Acidity generated by the tumor microenvironment drives local invasion. Cancer Res, 2013. 73(5): p. 1524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwamura T, Katsuki T, and Ide K, Establishment and characterization of a human pancreatic cancer cell line (SUIT-2) producing carcinoembryonic antigen and carbohydrate antigen 19–9. Jpn J Cancer Res, 1987. 78(1): p. 54–62. [PubMed] [Google Scholar]
- 11.Okabe T, Yamaguchi N, and Ohsawa N, Establishment and characterization of a carcinoembryonic antigen (CEA)-producing cell line from a human carcinoma of the exocrine pancreas. Cancer, 1983. 51(4): p. 662–8. [DOI] [PubMed] [Google Scholar]
- 12.Oliver MH, Harrison NK, Bishop JE, Cole PJ, and Laurent GJ, A rapid and convenient assay for counting cells cultured in microwell plates: application for assessment of growth factors. J Cell Sci, 1989. 92 ( Pt 3): p. 513–8. [DOI] [PubMed] [Google Scholar]
- 13.Rooney JP, Ryde IT, Sanders LH, Howlett EH, Colton MD, Germ KE, et al. , PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol Biol, 2015. 1241: p. 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rofstad EK, Mathiesen B, Kindem K, and Galappathi K, Acidic extracellular pH promotes experimental metastasis of human melanoma cells in athymic nude mice. Cancer Res, 2006. 66(13): p. 6699–707. [DOI] [PubMed] [Google Scholar]
- 15.Liu M, Quek LE, Sultani G, and Turner N, Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab, 2016. 4: p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sciacovelli M. and Frezza C, Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J, 2017. 284(19): p. 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. , Whole genomes redefine the mutational landscape of pancreatic cancer. Nature, 2015. 518(7540): p. 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 2012. 149(3): p. 656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vander Heiden MG and DeBerardinis RJ, Understanding the Intersections between Metabolism and Cancer Biology. Cell, 2017. 168(4): p. 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohani N, Hao L, Alexis MS, Joughin BA, Krismer K, Moufarrej MN, et al. , Acidification of Tumor at Stromal Boundaries Drives Transcriptome Alterations Associated with Aggressive Phenotypes. Cancer Res, 2019. 79(8): p. 1952–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeffer D, Somarelli JA, Hanna G, Palmer GM, and Garcia-Blanco MA, Cellular migration and invasion uncoupled: increased migration is not an inexorable consequence of epithelial-to-mesenchymal transition. Mol Cell Biol, 2014. 34(18): p. 3486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. , ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science, 2008. 320(5876): p. 661–4. [DOI] [PubMed] [Google Scholar]
- 23.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, et al. , mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A, 2005. 102(3): p. 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Waveren C, Sun Y, Cheung HS, and Moraes CT, Oxidative phosphorylation dysfunction modulates expression of extracellular matrix--remodeling genes and invasion. Carcinogenesis, 2006. 27(3): p. 409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, and Avadhani NG, Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene, 2002. 21(51): p. 7839–49. [DOI] [PubMed] [Google Scholar]
- 26.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, and Avadhani NG, Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J, 2001. 20(8): p. 1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, and Feron O, Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab, 2016. 24(2): p. 311–23. [DOI] [PubMed] [Google Scholar]
- 28.Auciello FR, Bulusu V, Oon C, Tait-Mulder J, Berry M, Bhattacharyya S, et al. , A Stromal Lysolipid-Autotaxin Signaling Axis Promotes Pancreatic Tumor Progression. Cancer Discov, 2019. 9(5): p. 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.