Abstract
Up to half of individuals who contract SARS-CoV-2 develop symptoms of long-COVID approximately three months after initial infection. These symptoms are highly variable, and the mechanisms inducing them are yet to be understood. We compared plasma cytokine levels from individuals with long-COVID to healthy individuals and found that those with long-COVID had 100% reductions in circulating levels of Interferon Gamma (IFNγ) and Interleukin-8 (IL-8). Additionally, we found significant reductions in levels of IL-6, IL-2, IL-17, IL-13, and IL-4 in individuals with long-COVID. We propose immune exhaustion as the driver of long-COVID, with the complete absence of IFNγ and IL-8preventing the lungs and other organs from healing after acute infection, and reducing the ability to fight off subsequent infections, both contributing to the myriad of symptoms suffered by those with long-COVID.
Keywords: Long-COVID, PASC, IL-8, IFNγ, Immune exhaustion, SARS-CoV-2
INTRODUCTION
COVID-19, caused by a novel coronavirus known as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), was declared a pandemic by the World Health Organization March 11th, 2020[1]. COVID-19 is responsible for 538.6 million infections and over 6.3 million deaths worldwide as of June 18th, 2022[2]. For up to half of individuals who contract the virus, acute SARS-CoV-2 infection is followed by persistent health issues [3]. These individuals suffer a myriad of symptoms that affect their daily lives, including fatigue and post-exertional malaise, respiratory and cardiac symptoms, neurological symptoms, digestive symptoms, and more (Table 1) [4,5].
Table 1:
Long-COVID symptoms.
| Categories | Neurocognitive | Respiratory | Psychological | Other |
|---|---|---|---|---|
| Brain fog | General fatigue | Post-traumatic stress disorder | Ageusia | |
| Dizziness | Dyspnea | Anxiety | Anosmia | |
| Loss of attention | Cough | Depression | Parosmia | |
| confusion | Throat pain | Insomnia | Skin rash | |
| Symptoms | Autonomic | Gastrointestinal | Musculoskeletal | |
| Chest pain | Diarrhea | Myalgia’s | 23 (38.3) | |
| Tachycardia | Abdominal pain | Arthralgia’s | 23 (38.3) | |
| Palpitations | Vomiting | 23 (38.3) | 23 (38.3) | |
Note: Most commonly reported symptoms associated with long-COVID.
Several names are in use to describe this post-viral syndrome, including long-haul COVID, Post-Acute Sequalae of SARS-CoV-2 (PASC), and long-COVID. The mechanisms driving long-COVID are still poorly understood. We defined long-COVID syndrome patients as those who fulfilled one of the following criteria: (a) Individuals whose symptoms never resolved following acute infection; (b) Individuals whoseCOVID-19 symptoms resolved but subsequently returned; or (c) Individuals who developed new symptoms approximately three months after initial infection [6,7]. Severity of symptoms during acute infection does not appear to predispose to development of long-COVID. Both asymptomatic individuals and those hospitalized due to severe complications develop long-COVID at similar rates [3,8,9].
Post-viral sequelae of Human Coronavirus (HCOV) infections have been well documented [10–12]. Both the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) outbreak in 2003 [13–17] and the Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV) outbreak starting in 2012 [18–20] caused post-viral syndromes with similar symptom profiles to those experienced by individuals with long-COVID [21]. Additionally, other human coronaviruses; especially HCOV 229E and HCOV NL63; that did not reach pandemic status, have been implicated as the etiology of Kawasaki syndrome [10,11]. Chikungunya virus also induces a post-viral syndrome, which presents with symptoms reminiscent of rheumatoid arthritis [22–25].
Long-COVID symptomatology bears similarities to Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). ME/CFS is characterized by 6 months or more of constant or relapsing bouts of excessive fatigue, cognitive impairment, post-exertional malaise, unrefreshing sleep, headaches, and neuroendocrine and immune alterations [26–30]. The number of people affected by ME/CFS is growing each year, currently affecting 0.3%–2.5% of the population globally, depending on the diagnostic criteria used [31,32]. The heterogeneous symptoms of ME/CFS are linked to dysregulation of multiple biological systems including the immune system and inflammation [33–37], cytokines [27,32,38], metabolism [34,39–42], mitochondrial function [34,39,43], oxidative stress [34,36], apoptosis [34,6], and circadian rhythm [34,44]. Additionally, research into cytokine levels as biomarkers for ME/CFS diagnosis or severity metrics has yielded conflicting reports, for a thorough review please see Blundell et al. [32]. There are also disparate theories regarding the origin of ME/CSF including dysbiosis of one’s microbiome [41,45–47], and as a post-viral syndrome following infection with Epstein Barr Virus (EBV) [48–51].Based on the pro-inflammatory basis of other post viral syndromes, we hypothesized that long-COVID is caused by abnormal, sustained, elevated levels of pro-inflammatory cytokines present in the blood after acute SARS-CoV-2 infection has abated. To test this, we assayed plasma from 15 healthy individuals and compared it to plasma from 12 patients at the University of Utah’s long-COVID Clinic.
MATERIALS AND METHODS
Study subjects
We obtained healthy donor blood samples from individuals who were recruited under University of Utah Institutional Review Board (IRB) protocol 131664. These individual were recruited from Salt Lake City, UT and the surrounding metro area between May of 2020 and December of 2021. For the purposes of this study, we define “healthy” as individuals who were uninfected or had been infected but recovered without the sequalae of long-COVID.
In the fall of 2021, the University of Utah opened a long-COVID registry (IRB 140978). Individuals attending University of Utah Comprehensive COVID clinic or self-identified with long-COVID can enroll in the registry, which includes a detailed symptom and health survey and blood draw for biobanking of plasma and PBMCs at the Cellular Translational Research Core (CTRC) at the University of Utah.
Blood and tissue samples
15 mL of total blood was collected by phlebotomy-certified research staff into two BD Vacutainer EDTA Additive Blood Collection Tubes. Tubes were gently inverted 8-times to mix the blood and EDTA and were then centrifuged at 150 g for 20 minutes at room temperature. Blood plasma was collected following centrifugation and cryopreserved in sterile cryovials at −80°C. Peripheral Blood Mononuclear Cells (PBMCs) were isolated by Ficoll density gradient (Histopaque-1077, Sigma), and were cryopreserved in 1 ml aliquots in 80% complete culture media (endothelial cell media), 10% Fetal Bovine Serum (FBS), and 10% Di-Methyl Sulf-Oxide (DMSO) in sterile cryovials at −80°C.
Determination of cytokine concentration in plasma
The Luminex based (Luminex Corp, TX) multiplexed cytokine assay was performed using a modified version of our previously published method [52,53]. Briefly, monoclonal antibodies to human IL-2, sIL-2r, IL-4, IL-6, IL-8, IL-10, TNFα (BD Biosciences, Franklin Lakes, NJ), IL-13, IL-17, IFNγ (eBioscience-ThermoFisher Scientific, Waltham, MA), and IL-1β, IL-5 (R and D Systems Minneapolis, MN), IL-12 p35/p70 (Cell Sciences, Newburyport MA), were covalently coupled to MagPlex microsphere particles (Luminex Corporation) using a 2-step carbodiimide reaction, as previously described (Staros, Wright et al.). A standard curve was generated by mixing known concentrations of recombinant human cytokine receptor IL-2r (R and D Systems), and recombinant human cytokines IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IL-13, IL-17, TNFα and IFNγ (R and D Systems). Biotinylated secondary antibodies were purchased from the following sources: eBioscience-ThermoFisher Scientific (IL-1β, IL-4, IL-6, TNFα, IL-12p70, IFNγ, IL-13, IL-17) and BD Biosciences (IL-2, IL-5, IL-8, IL-10, IL-2r). Performance parameters including specimen dilution/recovery, detection capability, precision, interference due to hemolysis, specimen stability, and linearity were validated following Clinical and Laboratory Standards Institute (CLSI) guidelines.
Statistical analysis
The individual cytokine values for the healthy and long-COVID cohorts were analyzed for statistical significance using unpaired, two-tailed, nonparametric Mann-Whitney tests with Prism 9.0 (GraphPad Software, San Diego, CA, USA).
RESULTS
Our long-COVID cohort included 12 individuals, who we compared to 15 matched, healthy controls (Table 2). The cytokines assayed included IL-1β, IL-2, sIL-2R, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, IL-13, IL-17, IFNγ, and TNFα. Figure 1 shows the plasma concentration in pg/ml of each cytokine. Individuals in the long-COVID cohort have decreased levels in most cytokines tested. Most notably, individuals with long-COVID have a 100% reduction in plasma levels of Interferon Gamma (IFNγ) and IL-8, yielding p-values of <0.0001 and 0.0011, respectively (Figure 1).
Table 2:
Cohort demographics.
| All Participants | Sex | Age ranges | |||
|---|---|---|---|---|---|
| Male | Female | ||||
| n | 27 | 7 | 20 | 23–70 | |
| Healthy | 15 | 7 | 8 | 27–65 | |
| Long- COVID | 12 | -- | 12 | 23–70 | |
Note: Demographics of our healthy and long-COVID cohorts.
Figure 1:
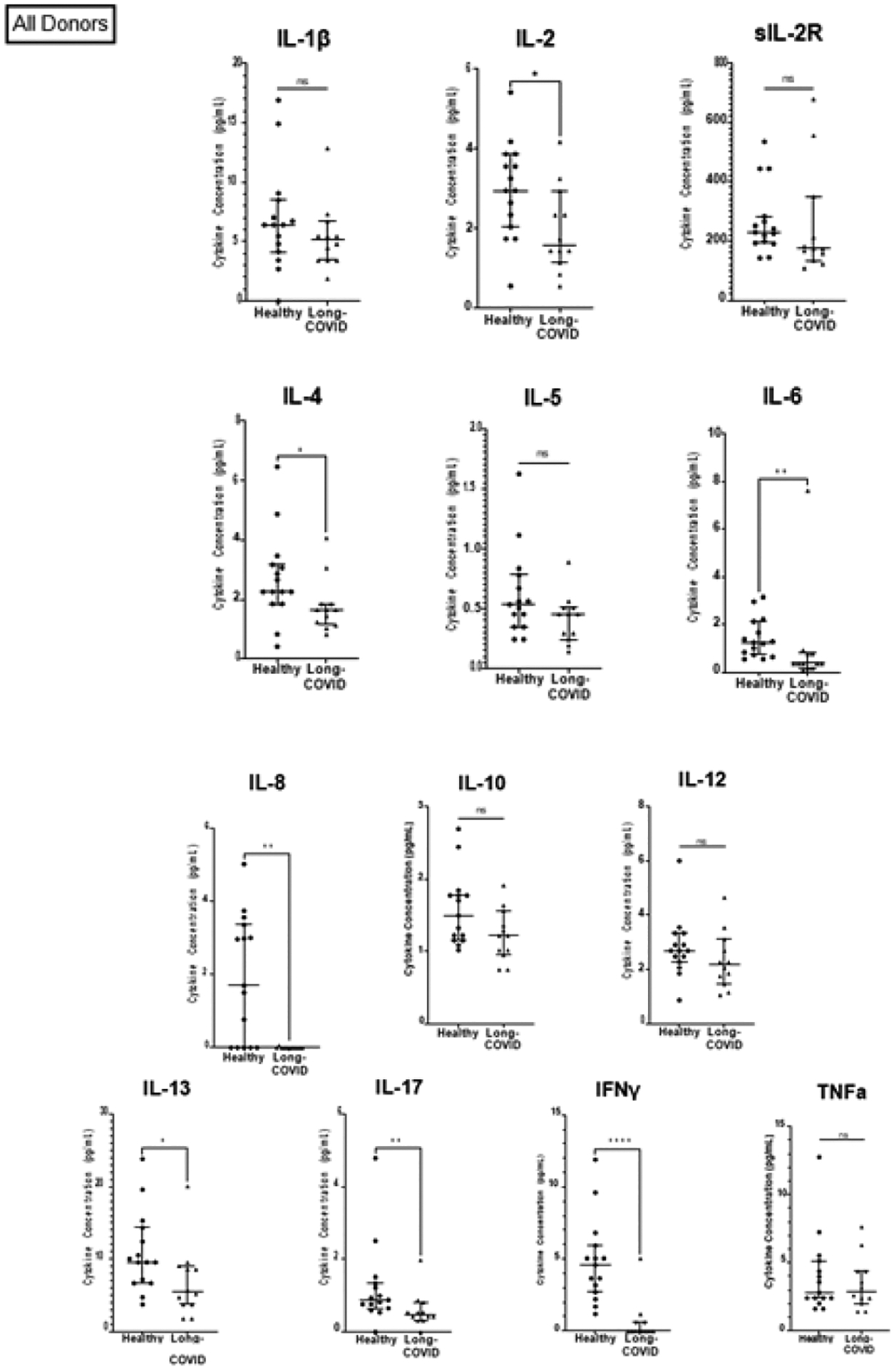
Comparison of pro-inflammatory cytokines in healthy individuals and those with long-COVID. Note: Comparison of cytokines between the healthy cohort (n=15) and the long-COVID cohort (n=12). (*) p ≤ 0.05; (**) p ≤ 0.01; (***) p ≤ 0.001; (****) p ≤ 0.0001.
In addition, individuals with long-COVID have a 70% reduction in levels of IL-6. Levels of IL-2, IL-17, and IL-13 were reduced more than 40% in individuals with long-COVID (p-values 0.0285, 0.0082, and 0.0176, respectively; Figure 1 and Table 3). Individuals with long-COVID also had a reduction in levels of IL-4 (26%; p=0.0266). Differences in plasma levels of soluble IL-2 receptor (sIL-2R), IL-1β, IL-12, IL-10, IL-5, and TNFα between the long-COVID and healthy groups were not statistically significant. Given that all the participants in the long-COVID group (12 out of 12) were female (Table 2), we sought to investigate whether the observed cytokine deficits in the long-COVID group were perhaps linked to biological sex. To do this, we performed two comparisons. First, we compared cytokine levels between males and females in the healthy group. These results showed that in healthy individuals, 12 out of the 13 cytokines assayed were not significantly different between healthy males and females. One cytokine, IL-2, was 42% lower in healthy males than in healthy females (p=0.0367; Figure 3 and Table 4). Secondly, we compared cytokine levels between the long-COVID group (all females) and the female participants in the healthy group (n=8). We continue to observe a 100% reduction in IFNγ and IL-8 levels with p-values of <0.0001 and 0.0144, respectively (Figure 2). We also observed a 72% reduction in IL-6 (p=0.0062), 55% lower levels of IL-2 (p=0.0028), a 59% decrease in IL-13 levels (p=0.0189), and IL-4 levels are reduced by 44% (p=0.0362) in females with long-COVID (Figure 2 and Table 3). One notable difference in the results from this female-female analysis is that the observed decrease in IL-5 levels (26%) in long-COVID females becomes statistically significant with a p-value of 0.0323, whereas the decrease between the healthy cohort when it contains both males and females and the long- COVID cohort is only 14% and is not statistically significant (Figure 1 and Table 3). The changes observed in sIL-2R, IL-1β, IL-12, IL-10, and TNFα levels remain not statistically significant whether the healthy cohort includes the males or not (Figure 2 and Table 3).
Table 3:
Statistical summary of cytokine comparison.
| Cytokine | Healthy | Long-COVID | % Change in long-COVID compared to healthy (M+F) | % Change in long-COVID compared to healthy (F only) | |||
|---|---|---|---|---|---|---|---|
| (M+F) | (F only) | ||||||
| n= | 15 | 8 | 12 | ||||
| IFNγ | 4.586 | 5.263 | 0.0000 | −100% | <0.0001**** | −100% | <0.0001**** |
| IL-8 | 1.700 | 0.3848 | 0.0000 | −100% | 0.0011** | −100% | 0.0144* |
| IL-6 | 1.248 | 1.317 | 0.3685 | −70.46% | 0.0016** | −72.05% | 0.0062** |
| IL-2 | 2.953 | 3.567 | 1.588 | −46.19% | 0.0285* | −55.45% | 0.0028** |
| IL-17 | 0.8802 | 1.024 | 0.4846 | −44.94% | 0.0082** | −52.68% | 0.0191* |
| IL-13 | 9.564 | 13.40 | 5.462 | −42.89% | 0.0176* | −59.23% | 0.0189* |
| IL-4 | 2.262 | 2.969 | 1.653 | −26.92% | 0.0266* | −44.32% | 0.0362* |
| sIL-2R | 229.2 | 205.6 | 174.2 | −24.03% | 0.1138 | −15.32% | 0.4269 |
| IL-1α | 6.425 | 7.469 | 5.153 | −19.8% | 0.2701 | −31.01% | 0.0779 |
| IL-12 | 2.687 | 2.900 | 2.162 | −19.53% | 0.1320 | −25.43% | 0.0788 |
| IL-10 | 1.494 | 1.598 | 1.219 | −18.38% | 0.0717 | −23.69% | 0.1506 |
| IL-5 | 0.5346 | 0.6157 | 0.4542 | −14.71% | 0.0884 | −26.23% | 0.0323* |
| TNFα | 2.812 | 3.397 | 2.909 | 3.47% | 0.7815 | −14.35% | 0.7201 |
Figure 3:
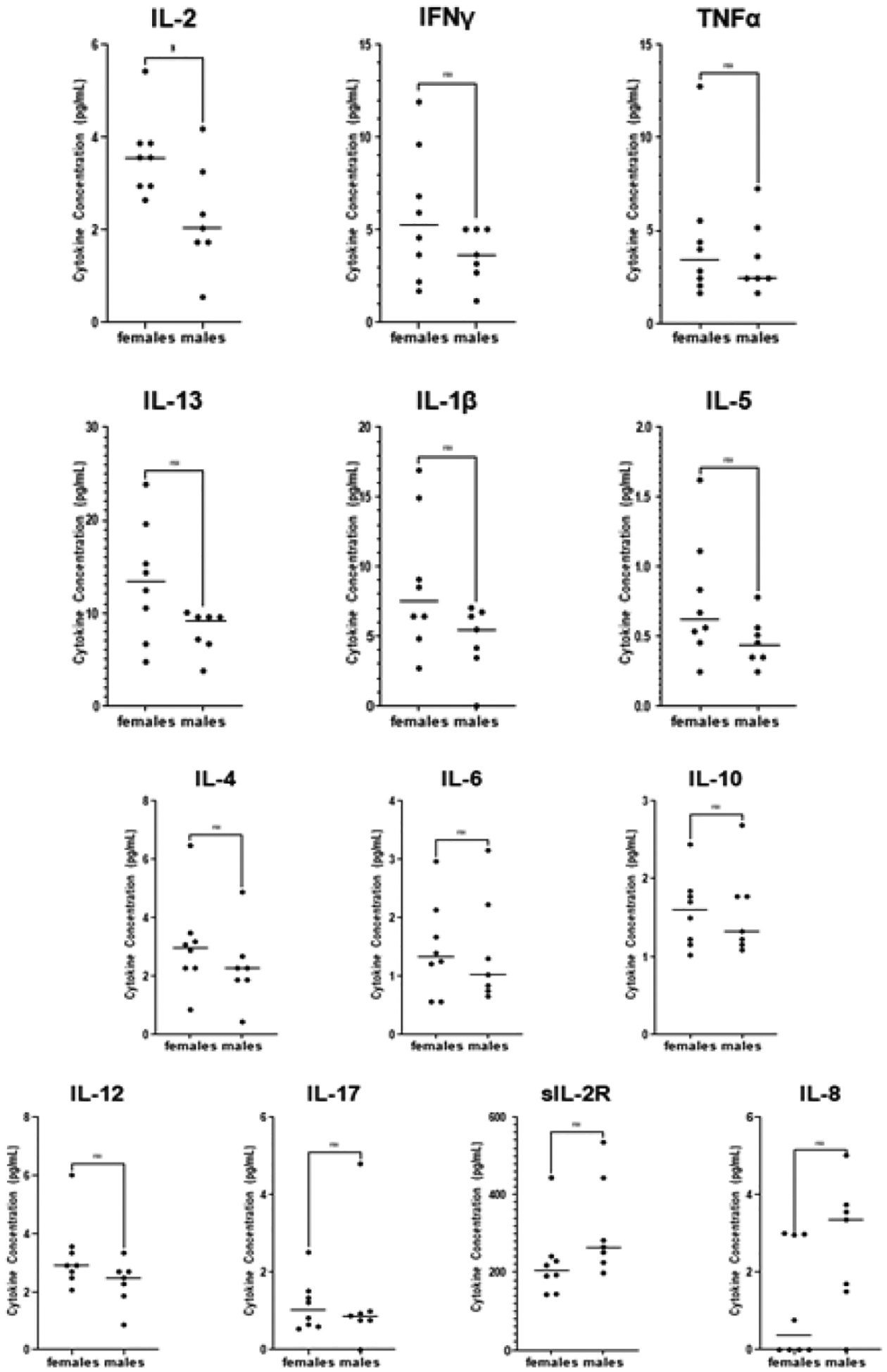
Comparison between healthy males and healthy females.
Note: Comparison of cytokines between the healthy females (n=8) healthy males (n=7). (*) p ≤ 0.05.
Table 4:
Statistical summary of cytokine comparison.
| Cytokine | Healthy | 1.219 | 1.219 | ||
|---|---|---|---|---|---|
| Females | Males | ||||
| n= | 8 | 7 | |||
| 1.219 | Median | Median | % | p= | |
| Change | |||||
| IL-2 | 3.567 | 2.04 | −42.81% | 0.0367* | |
| IFNα | 5.263 | 3.657 | −30.51% | 0.2793 | |
| TNFα | 3.397 | 2.419 | −28.79% | 0.6929 | |
| IL-13 | 13.4 | 9.564 | −28.62% | 0.0533 | |
| IL-1α | 7.469 | 5.479 | −26.63% | 0.1786 | |
| IL-5 | 0.6157 | 0.4542 | −26.23% | 0.1304 | |
| IL-4 | 2.969 | 2.262 | −23.82% | 0.2678 | |
| IL-6 | 1.317 | 1.016 | −22.87% | 0.9259 | |
| IL-10 | 1.598 | 1.322 | −17.27% | 0.8872 | |
| IL-12 | 2.9 | 2.476 | −14.62% | 0.1131 | |
| IL-17 | 1.024 | 0.8802 | −14.04% | 0.8424 | |
| sIL-2R | 205.6 | 264.9 | 22.37% | 0.0541 | |
| IL-8 | 0.3848 | 3.356 | 88.52% | 0.0667 | |
Note: Percent change and p-values for comparisons between males and females within the healthy cohort.
Figure 2:
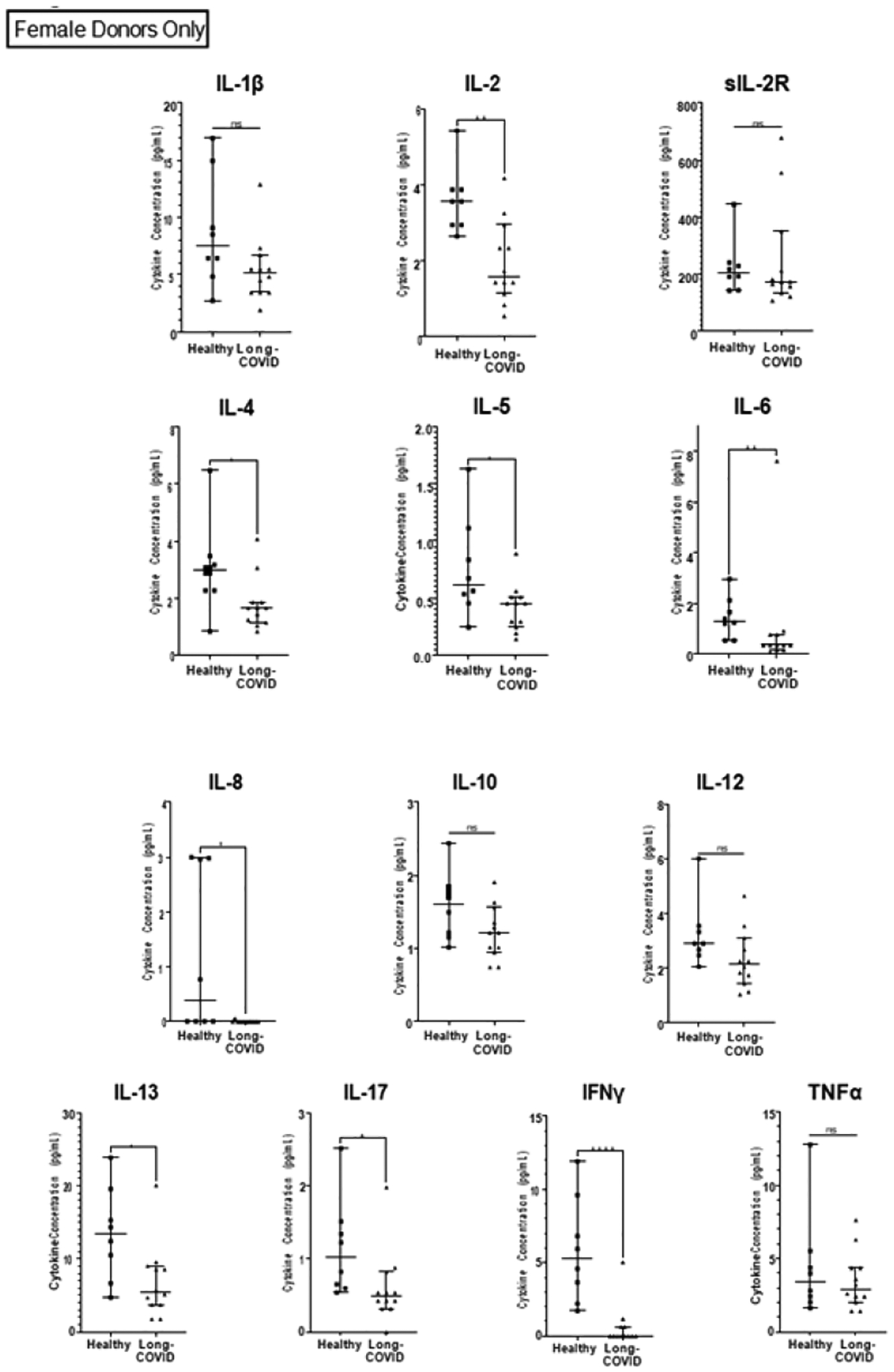
Comparison of pro-inflammatory cytokines present in healthy females and those with long-COVID.
DISCUSSION
Upon infection with SARS-CoV-2 the innate immune system recognizes both Pathogen- and Damage- Associated Molecular Patterns (PAMPs and DAMPs, respectively) and responds by activating the NLRP3 (NOD-, LRR-and pyrin domain-containing protein 3) inflammasome [54,55]. Monocytes and macrophages respond to PAMPs and DAMPs by secreting type I IFN and the pro-inflammatory cytokines IL-1, IL-2, IL-6, IL-12, and TNFα [54,56]. To evaluate the possibility that dysregulated secretion of pro-inflammatory cytokines can be observed in the context of long COVID, we measured levels of 13 plasma cytokines via Luminex assay in samples from 12 donors diagnosed with long- COVID and compared them to 15 healthy controls (Table 2). All the statistically significant differences between the long-COVID cohort and healthy controls represented reductions in cytokine levels rather than the expected increases based on previous studies of other post-viral syndromes (Figure 1 and Table 3) [32,57]. 34 Pro-inflammatory cytokines have been implicated in multiple aspects of acute COVID- 19 pathogenesis. For example, increased levels of IL-1β are linked to lymphopenia in COVID-19 patients, presumably due to ongoing inflammation-induced pyroptosis[54,58]. Macrophages express Angiotensin-Converting Enzyme 2 (ACE2) receptors, making it possible for SARS-CoV-2 to directly infect them, and, as COVID-19 severity increases, activated macrophages congregate in the lungs, where even if not productively infected, an abortive infection of macrophages by SARS-CoV-2 is sufficient to induce cytokine storm [54,59,60]. Lastly, supporting the importance of Th17 in the pathogenesis of COVID-19, is research showing that there are increased numbers of Th17 cells present in blood samples of COVID-19 patients [54,61]. 45 The two most drastically decreased cytokines in our study were IFNγ and IL-8, each reduced by 100% in our long-COVID cohort. IL-8 is produced by many cell types, including epithelial cells, fibroblasts, endothelial cells, macrophages, lymphocytes and mast cells [62]. The secretion of IL-8 is induced in part by levels of IL-1β. However we found that there is no significant difference in IL-1β levels between individuals with long- COVID and healthy controls. Also referred to as the neutrophil chemotactic factor, IL-8 recruit’s neutrophils and NK-cells to sites of inflammation where they can clear infected cells and promote wound healing. It is possible that the apparent lack of IL-8 in long-COVID patients may be responsible for at least some of the debilitating symptoms including post-exertional malaise, fatigue, and persistent cough, shortness of breath and chest pain. In this scenario, the acute SARS-CoV-2 infection damages the lungs, the cytokine milieu unfolds as described above, recruiting cells to the site of damage where the cells can either (a) help control the infection and induce a wound healing environment and the individual recovers normally; or (b) the infection causes abundant cellular infiltration leading to a high concentration of immune cells in a relatively small physical space, ultimately causing more tissue damage, which is not efficiently repaired in the absence of IL-8. Predictably, under scenario ‘b’ the individual remains having difficulty with oxygen transfer from the lungs into the blood stream. Therefore, if the macrophages and other cells that secrete IL-8 become exhausted or are otherwise incapable of secreting IL-8, neutrophils will not be recruited to assist in the wound healing process in the lung once the infection has been cleared [63]. Scenario ‘b’ therefore emerges as a potential model to explain certain long-COVID complications based on lack of IL-8. IFNγ is secreted by the innate immune Natural Killer cells (NK) and Natural Killer T cells (NKT) as well as the adaptive immune CD4+ Th1 and CD8+ Cytotoxic T Lymphocytes (CTL) after the development of antigen-specific immunity [64]. Together with IL-12, IFNγhelps drive the differentiation of Th1 cells, which in turn can secrete IL-2, TNFα, and IFNγ [65]. The observed lack of circulating IFNγ (Figure 1 and Table 2) in the plasma of patients suggests either severe immune dysfunction or exhaustion. We observed no significant difference in the levels of IL-12 (−19%), or TNFα (3%), in individuals with long-COVID. Levels of IL-2 and IL-4 were decreased by 46% and 26%, respectively, in individuals long-COVID (Figure 1 and Table 2). It is possible that fewer T cells differentiated into Th2 cells due to lower levels of IL-2 and IL-4, which could potentially lead to lower levels of the cytokines that Th2 cells secrete (IL-4, IL-5, IL-6, IL-9, and IL-13). This scenario may be supported by our data as we observed significantly lower levels of IL-4, IL-6, and IL-13 in individuals with long-COVID (Figure 1 and Table 2). Additionally, when we compare the long-COVID cohort, which includes only females, to only the females from the healthy cohort, the decrease in IL-5 between females with long-COVID and healthy becomes statistically significant (p=0.0323; Figure 3 and Table 2). IL-6 is involvedin the differentiation of Th17 cells. It is possible that the lower levels of IL-6 we observed in long COVID patients hindered the ability of the Th17 cells to properly differentiate. Supporting this possibility, we see significantly lower levels of IL-17 in individuals with long-COVID (p=0.0082), the main cytokine secreted by Th17 cells.
To ensure that none of the reported differences were due to inherent sex differences we cytokine levels between males and females within the healthy cohort. From this, we only observed one significant difference, a 42% reduction of IL-2 in healthy males compared to healthy females (Figure 3 and Table 3). This sex-associated difference IL-2 levels between healthy males and females informed us that the most accurate way to analyze IL-2 levels in our long-COVID cohort was to only consider healthy female IL-2 values. Analyzed in this way, long-COVID females show a 55% reduction in IL-2 levels (p=0.0028; Figure 2 and Table 2). The comparison including both males and females in the healthy cohort is also significant, although the inherently lower values in males complicate the interpretation. Additionally, having re-analyzed the data to only include healthy females, it became clear that the differences that we observe between the long-COVID females, and the entire healthy population are not due solely to sex-specific differences in cytokine levels. In fact, the only cytokine that differed in having statistical significance between the healthy female to long-COVID female comparison, and the healthy male+female to long-COVID female comparison was IL-5. The removal of the male values from the analysis caused the percent change between the healthy males+females and the long-COVID females to increase from 14% to 26% (between the healthy females and long-COVID females) with a p-value of 0.0323 (Table 2 and Figures 2 and 3). Earlier we described the heterogeneous nature of the symptoms and cytokines associated with ME/CFS. Even though the symptoms of long-COVID are thought to be similar or overlapping to those of ME/CFS, when we compare our long-COVID cytokine to those from ME/CFS patients there are glaring differences. Specifically, the pro-inflammatory cytokines IL-1β, TNFα, and IL-6 tend to be reported as being elevated in patients with ME/CFS [32], whereas we observed significant decreases in IL-6 levels in individuals with long-COVID, and no differences in levels of IL-1β and TNFα. A detailed comparison of cytokines levels published in the context of ME/CFS [32] and our long-COVID levels is provided in Supplementary Table 1.
CONCLUSION
Based on our results we propose that immune exhaustion perpetuates long-COVID due to the seemingly complete reduction of IFNγ and IL-8, as well as significant decreases in IL-2, IL-4, IL-6, IL-13, and IL-17. Identifying these and other deficiencies will provide clues towards methods to intervene and possibly restore immune function in the context long-COVID. Although functional assays that test the ability of immune cells from individuals with long-COVID to respond to pathogenic stimuli will be required to support this theory.
Supplementary Material
FUNDING
This work was supported by the University of Utah Inflammation, Immunology, and Infectious Diseases (3i) Initiative’s Emerging Infectious Diseases Fellowship (recipient, ESCPW) and by a seed grant from the University of Utah Vice President for Research and the Immunology, Inflammation, and Infectious Disease Initiative (recipient, VP). VP and MC were supported by NIH GRANT 5R01-AI143567.
Footnotes
ETHICAL STATEMENT
Blood samples from donors in both our healthy and long-COVID cohorts were collected from individuals who were recruited in compliance with University of Utah Institutional Review Board (IRB) protocols 131664 and 140978, respectively.
REFERENCES
- 1.CDC Museum COVID-19 Timeline. https://www.cdc.gov/museum/timeline/covid19.htm.
- 2.COVID-19 Dashboard https://coronavirus.jhu.edu/map.html. (Accessed June 18th 2022).
- 3.Chen C, Haupert SR, Zimmermann L, Shi X, Fritsche LG, Mukherjee B. Global Prevalence of Post COVID-19 Condition or Long COVID: A Meta-Analysis and Systematic Review. J. Infect. Dis 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Leon S, Wegman-Ostrosky T, Perelman C, Sepulveda R, Rebolledo PA, Cuapio A, et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Scientific reports 2021;11(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández-de-Las-Peñas C, Palacios-Ceña D, Gómez-Mayordomo V, Cuadrado ML, Florencio LL. Defining post-COVID symptoms (post-acute COVID, long COVID, persistent post-COVID): an integrative classification. Int. J. Environ. Res 2021;18(5):2621.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nabavi N. Long covid: How to define it and how to manage it. [DOI] [PubMed]
- 7.Peluso MJ, Donatelli J, Henrich TJ. Long-term immunologic effects of SARS-CoV-2 infection: leveraging translational research methodology to address emerging questions. Transl Res 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malkova A, Kudryavtsev I, Starshinova A, Kudlay D, Zinchenko Y, Glushkova A, et al. Post COVID-19 Syndrome in Patients with Asymptomatic/Mild Form. Pathogens 2021;10(11):1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peghin M, Palese A, Venturini M, De Martino M, Gerussi V, Graziano E, et al. Post-COVID-19 symptoms 6 months after acute infection among hospitalized and non-hospitalized patients. Clin Microbiol Infect 2021;27(10):1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez SR, Anderson MS, Glodé MP, Robinson CC, Holmes KV. Blinded case-control study of the relationship between human coronavirus NL63 and Kawasaki syndrome. J. Infect. Dis 2006;194(12):1697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirato K, Imada Y, Kawase M, Nakagaki K, Matsuyama S, Taguchi F. Possible involvement of infection with human coronavirus 229E, but not NL63, in Kawasaki disease. J. Med. Virol 2014;86(12):2146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Algahtani H, Subahi A, Shirah B. Neurological complications of Middle East respiratory syndrome coronavirus: a report of two cases and review of the literature. Case reports in neurological medicine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tansey CM, Louie M, Loeb M, Gold WL, Muller MP, de Jager J, et al. One-year outcomes and health care utilization in survivors of severe acute respiratory syndrome. Arch. intern. med. res 2007;167(12):1312–20. [DOI] [PubMed] [Google Scholar]
- 14.Gardner PJ, Moallef P. Psychological impact on SARS survivors: Critical review of the English language literature. Can Psychol 2015; 56(1):123. [Google Scholar]
- 15.Moldofsky H, Patcai J. Chronic widespread musculoskeletal pain, fatigue, depression and disordered sleep in chronic post-SARS syndrome; a case-controlled study. BMC neurology 2011;11(1):1–7.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sim K, Chua HC. The psychological impact of SARS: a matter of heart and mind. Cmaj 2004;170(5):811–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan KS, Zheng JP, Mok YW, Li YM, LIU YN, Chu CM, et al. SARS: prognosis, outcome and sequelae. Respirology 2003;8:S36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung H, Jung SY, Lee MH, Kim MS. Assessing the presence of post-traumatic stress and turnover intention among nurses post–Middle East respiratory syndrome outbreak: the importance of supervisor support. Workplace health & safety 2020;68(7):337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batawi S, Tarazan N, Al-Raddadi R, Al Qasim E, Sindi A, Al Johni S, et al. Quality of life reported by survivors after hospitalization for Middle East respiratory syndrome (MERS). Health Qual. Life Outcomes 2019;17(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Shin HS, Park HY, Kim JL, Lee JJ, Lee H, et al. Depression as a mediator of chronic fatigue and post-traumatic stress symptoms in Middle East respiratory syndrome survivors. Psychiatry Investig 2019;16(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Sullivan O Long-term sequelae following previous coronavirus epidemics. Clin Med 2021; 21(1):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duvignaud A, Fianu A, Bertolotti A, Jaubert J, Michault A, Poubeau P, et al. Rheumatism and chronic fatigue, the two facets of post-chikungunya disease: the TELECHIK cohort study on Reunion island. Epidemiol. Infect 2018;146(5):633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foissac M, Javelle E, Ray S, Guérin B, Simon F. Post-chikungunya rheumatoid arthritis, Saint Martin. Emerg. Infect. Dis;21(3):530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imad HA, Matsee W, Kludkleeb S, Asawapaithulsert P, Phadungsombat J, Nakayama EE, et al. Post–chikungunya virus infection musculoskeletal disorders: Syndromic sequelae after an outbreak. Trop. med. infect 2021;6(2):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Javelle E, Ribera A, Degasne I, Gaüzère BA, Marimoutou C, Simon F. Specific management of post-chikungunya rheumatic disorders: a retrospective study of 159 cases in Reunion Island from 2006–2012. PLoS Negl Trop Dis 2015;9(3):e0003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornig M, Gottschalk CG, Eddy ML, Che X, Ukaigwe JE, Peterson DL, et al. Immune network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome with atypical and classical presentations. Transl psychiatry 2017; 7(4):e1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montoya JG, Holmes TH, Anderson JN, Maecker HT, Rosenberg-Hasson Y, Valencia IJ, et al. Cytokine signature associated with disease severity in chronic fatigue syndrome patients. Proc. Natl. Acad. Sci 2017;114(34):E7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornig M, Montoya JG, Klimas NG, Levine S, Felsenstein D, Bateman L, et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Sci Adv 201; 1(1):e1400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deumer US, Varesi A, Floris V, Savioli G, Mantovani E, López-Carrasco P, et al. Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): an overview. J Clin Med 2021;10(20):4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carruthers BM, van de Sande MI, De Meirleir KL, Klimas NG, Broderick G, Mitchell T, et al. Myalgic encephalomyelitis: international consensus criteria. J Intern Med 2011;270(4):327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prins JB, van der Meer JW, Bleijenberg G. The Impact of Chronic Social Stress on Emotional Behavior in Mice and the Therapeutic Effect of Peripheral Mild-Heat Stimulation. The Lancet 2006;367(9507):346–55. [DOI] [PubMed] [Google Scholar]
- 32.Blundell S, Ray KK, Buckland M, White PD. Chronic fatigue syndrome and circulating cytokines: a systematic review. Brain Behav. Immun 2015;50:186–95. [DOI] [PubMed] [Google Scholar]
- 33.Sweetman E, Noble A, Edgar C, Mackay A, Helliwell A, Vallings R, et al. Current research provides insight into the biological basis and diagnostic potential for myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Diagnostics 2019;9(3):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweetman E, Ryan M, Edgar C, MacKay A, Vallings R, Tate W. Changes in the transcriptome of circulating immune cells of a New Zealand cohort with myalgic encephalomyelitis/chronic fatigue syndrome. Int J immunopathol pharmacol 2019; 33:2058738418820402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenu EW, Ashton KJ, Batovska J, Staines DR, Marshall-Gradisnik SM. High-throughput sequencing of plasma microRNA in chronic fatigue syndrome/myalgic encephalomyelitis. PLoS One 2014;9(9):e102783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gow JW, Hagan S, Herzyk P, Cannon C, Behan PO, Chaudhuri A. A gene signature for post-infectious chronic fatigue syndrome. BMC med genomics 2009;2(1):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen CB, Alsøe L, Lindvall JM, Sulheim D, Fagermoen E, Winger A, et al. Whole blood gene expression in adolescent chronic fatigue syndrome: an exploratory cross-sectional study suggesting altered B cell differentiation and survival. J Transl Med 2017;15(1):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broderick G, Katz BZ, Fernandes H, Fletcher MA, Klimas N, Smith FA, et al. Cytokine expression profiles of immune imbalance in post-mononucleosis chronic fatigue. J Transl Med 2012;10(1):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fluge Ø, Mella O, Bruland O, Risa K, Dyrstad SE, Alme K, et al. Metabolic profiling indicates impaired pyruvate dehydrogenase function in myalgic encephalopathy/chronic fatigue syndrome. JCI insight 2016;1(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naviaux RK, Naviaux JC, Li K, Bright AT, Alaynick WA, Wang L, et al. Metabolic features of chronic fatigue syndrome. Proc Natl Acad Sci 2016;113(37):E5472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proal A, Marshall T. Myalgic encephalomyelitis/chronic fatigue syndrome in the era of the human microbiome: persistent pathogens drive chronic symptoms by interfering with host metabolism, gene expression, and immunity. Front Pediatr 2018;6:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamano E, Watanabe Y, Kataoka Y. Insights into metabolite diagnostic biomarkers for myalgic encephalomyelitis/chronic fatigue syndrome. Int. J. Mol. Sci 2021; 22(7):3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomas C, Brown A, Strassheim V, Elson J, Newton J, Manning P. Cellular bioenergetics is impaired in patients with chronic fatigue syndrome. PLoS One 2017;12(10):e0186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Vega WC, Herrera S, Vernon SD, McGowan PO. Epigenetic modifications and glucocorticoid sensitivity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). BMC med genomics 2017;10(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navaneetharaja N, Griffiths V, Wileman T, Carding SR. A role for the intestinal microbiota and virome in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS)?. J Clin Med 2016. Jun 6;5(6):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giloteaux L, Goodrich JK, Walters WA, Levine SM, Ley RE, Hanson MR. Reduced diversity and altered composition of the gut microbiome in individuals with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 2016;4(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newberry F, Hsieh SY, Wileman T, Carding SR. Does the microbiome and virome contribute to myalgic encephalomyelitis/chronic fatigue syndrome?. Clin Sci 2018;132(5):523–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz-Pablos M, Paiva B, Montero-Mateo R, Garcia N, Zabaleta A. Epstein-Barr Virus and the origin of myalgic encephalomyelitis or chronic fatigue syndrome. Front immunol 2021:4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shikova E, Reshkova V, capital K. Cytomegalovirus, Epstein-Barr virus, and human herpesvirus-6 infections in patients with myalgic small ie, Cyrillicncephalomyelitis/chronic fatigue syndrome. J Med Virol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ariza ME. Myalgic encephalomyelitis/chronic fatigue syndrome: the human herpesviruses are back!. Biomolecules 2021; 11(2):185.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortes Rivera M, Mastronardi C, Silva-Aldana CT, Arcos-Burgos M, Lidbury BA. Myalgic encephalomyelitis/chronic fatigue syndrome: a comprehensive review. Diagnostics 2019;9(3):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martins TB, Pasi BM, Litwin CM, Hill HR. Heterophile antibody interference in a multiplexed fluorescent microsphere immunoassay for quantitation of cytokines in human serum. Clin Diagn Lab Immunol 2004;11(2):325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins TB, Burlingame R, von Mühlen CA, Jaskowski TD, Litwin CM, Hill HR. Evaluation of multiplexed fluorescent microsphere immunoassay for detection of autoantibodies to nuclear antigens. Clin Diagn Lab Immunol 2004;11(6):1054–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel M, Shahjin F, Cohen JD, Hasan M, Machhi J, Chugh H, et al. The immunopathobiology of SARS-CoV-2 infection. FEMS Microbiol Rev 2021; 45(6):fuab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov 2019;5(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang G, Ghosh S. Toll-like receptor–mediated NF-αB activation: a phylogenetically conserved paradigm in innate immunity. J Clin Invest 200;107(1):13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryabkova VA, Churilov LP, Shoenfeld Y. Influenza infection, SARS, MERS and COVID-19: Cytokine storm–The common denominator and the lessons to be learned. Clin Immunol 2021;223:108652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams ES, Martins TB, Hill HR, Coiras M, Shah KS, Planelles V, et al. Plasma cytokine levels reveal deficiencies in IL-8 and gamma interferon in Long-COVID. medRxiv 2022. [Google Scholar]
- 59.Nguyen T, Russell J. The regulation of FasL expression during activation-induced cell death (AICD). Immunology 2001;103(4):426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions. Life sci 2020; 257:118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J Microbiol Immunol Infect 2020;53(3):368–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baggiolini M, Walz A, Kunkel SL. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J. Clin. Investig 1989;84(4):1045–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metzemaekers M, Gouwy M, Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol 2020;17(5):433–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schoenborn JR, Wilson CB. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol 2007; 96:41–101. [DOI] [PubMed] [Google Scholar]
- 65.Gulati K, Guhathakurta S, Joshi J, Rai N, Ray AJ. Cytokines and their role in health and disease: a brief overview. Moj Immunol 2016;4(2):1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


