Abstract
Porphyromonas gingivalis, a gram-negative anaerobe, is a major etiological agent in the initiation and progression of severe forms of periodontal disease. An opportunistic pathogen, P. gingivalis can also exist in commensal harmony with the host, with disease episodes ensuing from a shift in the ecological balance within the complex periodontal microenvironment. Colonization of the subgingival region is facilitated by the ability to adhere to available substrates such as adsorbed salivary molecules, matrix proteins, epithelial cells, and bacteria that are already established as a biofilm on tooth and epithelial surfaces. Binding to all of these substrates may be mediated by various regions of P. gingivalis fimbrillin, the structural subunit of the major fimbriae. P. gingivalis is an asaccharolytic organism, with a requirement for hemin (as a source of iron) and peptides for growth. At least three hemagglutinins and five proteinases are produced to satisfy these requirements. The hemagglutinin and proteinase genes contain extensive regions of highly conserved sequences, with posttranslational processing of proteinase gene products contributing to the formation of multimeric surface protein-adhesin complexes. Many of the virulence properties of P. gingivalis appear to be consequent to its adaptations to obtain hemin and peptides. Thus, hemagglutinins participate in adherence interactions with host cells, while proteinases contribute to inactivation of the effector molecules of the immune response and to tissue destruction. In addition to direct assault on the periodontal tissues, P. gingivalis can modulate eucaryotic cell signal transduction pathways, directing its uptake by gingival epithelial cells. Within this privileged site, P. gingivalis can replicate and impinge upon components of the innate host defense. Although a variety of surface molecules stimulate production of cytokines and other participants in the immune response, P. gingivalis may also undertake a stealth role whereby pivotal immune mediators are selectively inactivated. In keeping with its strict metabolic requirements, regulation of gene expression in P. gingivalis can be controlled at the transcriptional level. Finally, although periodontal disease is localized to the tissues surrounding the tooth, evidence is accumulating that infection with P. gingivalis may predispose to more serious systemic conditions such as cardiovascular disease and to delivery of preterm infants.
Periodontal diseases comprise a group of infections involving the supporting tissues of the teeth. These range in severity from mild and reversible inflammation of the gingiva (gum) to chronic destruction of periodontal tissues (gingiva, periodontal ligament, and alveolar bone) with eventual exfoliation of teeth. From a microbiological standpoint, several features of these diseases are of interest. The bacterial etiology is complex, with a variety of organisms responsible for the initiation and progression of disease. Many, if not all, of these organisms may also be present in periodontally healthy individuals and can exist in commensal harmony with the host. Thus, disease episodes may ensue from a shift in the ecological balance between bacterial and host factors, as a result of, for example, alteration in the absolute or relative numbers of certain organisms, changes in pathogenic potential, or modulation of particular host factors. The local environment imposes a variety of unique constraints upon the constituent microbiota of the supragingival tooth surface and the subgingival crevice (the channel between the tooth root and the gingiva that deepens into a periodontal pocket as disease progresses). Both the calcified hard tissues of the tooth and the epithelial cells of the gingiva are available for colonization. These tissues are exposed to host salivary secretions and gingival crevicular fluid (a serum exudate), both of which contain molecules that interact directly with bacteria and alter prevailing environmental conditions. In addition, successful colonizers of the teeth and subgingival area must coexist with many (over 300) other species of bacteria that inhabit these regions. Study of the pathogenesis of periodontal diseases is thus complicated by the ecological intricacy of the microenvironment.
The classification of the various manifestations of periodontal diseases is continually changing, and it will suffice to mention that diseases range in severity, rate of progression, and number of teeth affected and that different age groups can be susceptible following the eruption of primary teeth. The nature of the pathogenic agents varies among these disease entities, as well as among patients and even between different disease sites within a patient. In general, however, severe forms of the disease in adults are associated with a number of gram-negative anaerobic bacteria. Of this group, most evidence points to a pathogenic role for Porphyromonas (formerly Bacteroides) gingivalis. The presence of this organism, acting either alone or as a mixed infection with other bacteria, and possibly in concert with the absence of beneficial species and certain immunological deficiencies in the host, appears to be essential for disease activity (83, 251). Intensive study of P. gingivalis has revealed a vast array of potential virulence factors that are now being defined at the molecular level. In this article, we review the pathogenic mechanisms of P. gingivalis.
INITIAL COLONIZATION OF THE ORAL ENVIRONMENT
Colonization of the oral cavity requires that the bacteria first enter the mouth and then localize at and attach to the available surfaces. Host factors that tend to resist bacterial colonization include the mechanical shearing forces of tongue movement along with saliva and gingival crevicular fluid flow. Furthermore, specific salivary molecules can aggregate organisms and promote their clearance via the processes of expectoration or swallowing (262). Successful oral colonizers therefore possess a variety of attributes to overcome host protective mechanisms. The sessile plaque biofilm that subsequently accumulates on the hard and soft tissues of the mouth is a dynamic system composed of diverse microbial species. P. gingivalis is usually among the late or secondary colonizers of the oral cavity, requiring antecedent organisms to create the necessary environmental conditions. The means by which antecedent bacteria may facilitate colonization by P. gingivalis include provision of attachment sites for interspecies adherence, supply of growth substrates, and reduction of oxygen tension to the low levels required for growth and survival of this obligate anaerobe (262).
Entry into the Oral Cavity
Initial entry of P. gingivalis into the oral cavity is thought to occur by transmission from infected individuals (77). Saliva is considered an important vector for transmission; however, the spouses and children of individuals with P. gingivalis do not always harbor the same genotype (203, 269). Other vectors would therefore also appear to be operational. The clonal diversity of P. gingivalis isolates has been examined by a variety of techniques including ribotyping, restriction endonuclease analysis, restriction fragment length polymorphism, multilocus enzyme electrophoresis, and arbitrary PCR amplification (145, 146, 156, 261). These studies indicate that individuals are colonized by a single (or at least a predominant) genotype, regardless of site of colonization or clinical status. Strains of many different clonal origins, in contrast, are present in different individuals. This supports the concept that P. gingivalis is essentially an opportunistic pathogen, with virulence not being restricted to a particular clonal type.
Adherence to Oral Surfaces
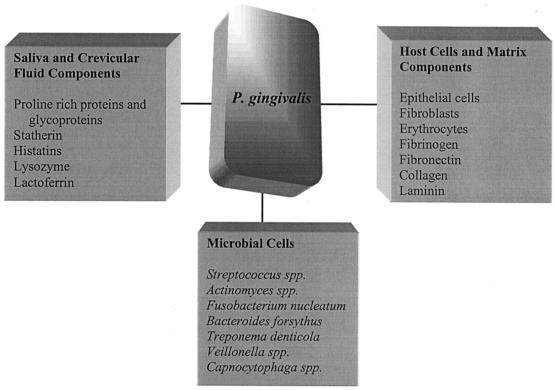
The oral cavity provides a variety of surfaces to which P. gingivalis can adhere (262) (Fig. 1). There are the mineralized hard tissues of the teeth, along with mucosal surfaces including those of the gingiva, cheek, and tongue. Colonization of surfaces remote from the gingival crevice and periodontal pocket can still have significance for the disease process since, once established, pathogenic organisms may reach the subgingival area by spreading proliferation or by translocation of dislodged progeny (244, 259). Oral surfaces generally are coated with a pellicle composed predominantly of salivary molecules but also potentially including serum-derived molecules from gingival crevicular fluid along with products related to host nutrition and epithelial cell turnover (262). It is the components of these pellicles, in particular various salivary molecules adsorbed on the tooth surface, that commonly function as receptors for bacterial adherence. Furthermore, oral surfaces rapidly become colonized with the early commensal microbiota of the mouth, as a consequence of which organisms such as P. gingivalis usually encounter surfaces rich in antecedent bacteria and their products. P. gingivalis can adhere to many of these early plaque organisms such as oral streptococci (Streptococcus gordonii, S. sanguis, S. oralis, S. mitis, and S. crista) and Actinomyces naeslundii (76, 108, 130, 131, 141, 166). These coadherence interactions tend not to result in the formation of large aggregates in suspension (109, 131, 229), a mechanism that may facilitate colonization by diminishing the probability that large coaggregates of organisms would be lost from the mouth by expectoration or swallowing before reaching a solid support. In addition, P. gingivalis can bind to other, later colonizers such as Fusobacterium nucleatum, Treponema denticola, and Bacteroides forsythus (79, 120, 279). These kinds of interspecies binding interactions may not only favor colonization but also promote nutritional interrelationships and intercellular signaling mechanisms (122). Other parameters of relevance to oral adherence include the presence, in the fluid phase, of saliva- and crevicular fluid-derived molecules that have the potential to bind to P. gingivalis and modulate its adhesion. The importance of such interactions is difficult to assess, however, since the degree of inhibition and enhancement varies according to the salivary source, the strains under investigation, and even the assay system used (131, 163, 166, 279).
FIG. 1.
Multiple adhesive interactions of P. gingivalis. Adherence mechanisms may vary according to substrate, and adherence may be multimodal. P. gingivalis releases membrane vesicles containing functional adhesins that may also bind to the substrates shown. Compiled from references 57, 76, 79, 108, 120, 130, 131, 163, 166, and 279.
Perhaps as an evolutionary adaptation to the diversity of available substrates, P. gingivalis displays a variety of distinct adhesive interactions that are associated with both fimbriae and outer membrane proteins (98, 132, 153). Such multimodal adherence mechanisms may not only improve the likelihood of attachment occurring but also increase the avidity of binding to individual substrates. In addition, in some cases, adherence to host cells can be a prelude to the modulation of eucaryotic intracellular signal transduction pathways. Thus, the specificity or strength of the adherence interaction or the sequence in which different adhesins engage their receptors may be important in the manipulation of the biological activity of the host cell by P. gingivalis.
Adhesin Molecules
Fimbriae.
Ultrastructural examination has revealed the presence of peritrichious fimbriae, 0.3 to 3.0 μm long and ca. 5 nm wide, on most strains of P. gingivalis (191, 244). The major class of fimbriae are composed of a fimbrillin monomer subunit that varies in size between 41 and 49 kDa depending on the strain (135). Circular dichroism analysis indicates that the secondary structure contains a high percentage of β-sheet and random coil with no detectable α-helix (253, 284). The gene encoding fimbrillin (fimA) is present in a single copy in the chromosome and is monocistronic (53). Protein sequence analysis reveals no significant homology to fimbrial proteins from other bacteria, indicating that P. gingivalis fimbriae may represent a unique class of gram-negative fimbriae (53).
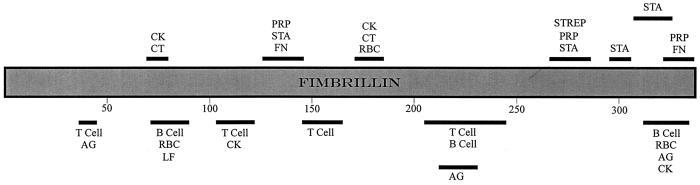
Fimbriae appear to be a major adherence-mediating determinant of P. gingivalis. Investigations variously involving purified fimbriae, recombinant fimbrillin, isogenic mutants deficient in fimbriae production, and antibodies to fimbriae have revealed that the fimbriae per se mediate adherence to a variety of oral substrates and molecules. These include salivary molecules such as proline-rich proteins (PRPs), proline-rich glycoproteins, and statherin; oral epithelial cells; antecedent bacteria such as oral streptococci, and A. naeslundii; and fibrinogen, fibronectin, and lactoferrin (7, 9, 76, 88, 105, 128, 136, 162, 252, 255, 274). Thus, fimbriae have the potential to be involved in most, if not all, of the adherence properties displayed by the organism. The domain structure of fimbrillin has recently received much attention, and studies with synthetic and truncated peptides have contributed greatly to our understanding in this area. Fimbrillin binding to salivary PRP1 and statherin involves separate and multiple binding domains (7, 167). The interactive regions are localized mainly at the C-terminal end of the protein between amino acids (aa) 266 and 337, although a less active domain appears N-terminally located between aa 124 and 146 (Fig. 2). Common sequences comprising VLVxxN and xYDxxxT (where x is any residue) were noted within the PRP1 and statherin binding regions respectively. On the basis of these observations, Amano et al. (7) proposed that the collective activity of all of these domains would be important in the stable association of fimbrillin with its salivary receptor, thus ultimately establishing bacterial adherence to saliva-coated surfaces in the oral cavity. The region from aa 124 to 146 and, to a lesser extent, the region from aa 318 to 337 are also involved in binding fibronectin (255). Amino acid sequences VQxxA or VxxxA were identified as potential interactive domains (255). The fimbrillin C-terminal region is involved in coadherence to S. oralis, with functional domains located in the regions from aa 266 to 286 and aa 287 to 337 (5). A domain map of fimbrillin with respect to a number of binding and immunogenic properties is provided in Fig. 2. Note that the extent to which linearly separated domains are brought into closer proximity upon protein folding is at present unknown.
FIG. 2.
Functional domains of P. gingivalis fimbrillin. Numbers refer to amino acid residues. Abbreviations: AG, immunodominant (IgG binding); B Cell, stimulation of B cells; CK, stimulation of cytokines; CT, chemotaxis; FN, binding to fibronectin; LF, binding to lactoferrin; PRP, binding to salivary PRPs; RBC, binding to erythrocytes; STA, binding to salivary statherin; STREP, binding to Streptococcus oralis; T Cell, stimulation of T cells. The boundaries of the domains are not known precisely. Compiled from references 5, 7, 52, 104, 167, 179–184, 187, 252, and 255.
The functionality of some of the receptor molecules for fimbrillin is also now partially understood. Statherin binds with fimbrillin only after deposition on solid surfaces such as hydroxyapatite (6). It has been theorized that in the aqueous phase statherin forms a dimer with N-terminal negatively charged groups exposed. After adsorption, however, conformational changes occur in the flexible C-terminal region, exposing cryptitopes to which fimbrillin binds (6, 209). Residues 29 and 30 (LY) and 41 to 43 (YTF) appear to constitute the binding domains for fimbrillin (6). Salivary acidic PRPs are also thought to require exposure of cryptitopes for bacterial binding (74). The fimbria binding site of PRP1 has been localized to a PQGPPQ segment that recurs four times within the region spanning aa 75 to 145 (114). Kataoka et al. (114) have proposed that the specificity of this binding interaction is determined by the hydrophobicity of proline and the hydrogen bond provided by glutamine. Less is known about other fimbrial receptors. However, a 48-kDa surface protein on gingival epithelial cells was found to interact with fimbriated but not with nonfimbriated strains of P. gingivalis and thus may function as a cognate receptor for fimbriae (274).
Although most of the evidence to date points to a direct role for the fimbrillin subunit in the various binding interactions, it cannot be stated with certainty that minor components of the fimbriae are not involved in adherence. In this context, it is interesting that the region downstream of fimA contains four open reading frames. These open reading frames, designated 1 to 4, produce proteins of 15, 50, 80, and 19 kDa, respectively, when expressed in Escherichia coli (273, 282). The 50- and 80-kDa products were also detected in P. gingivalis; hence, although the precise relationship is uncertain, these proteins may be associated with the fimbriae and are thus potential candidates for fimbria-associated adhesins (282).
The importance of fimbriae in P. gingivalis infection has been documented in a number of studies. Immunization with purified fimbriae confers protection against periodontal destruction in a gnotobiotic rat model (58). Moreover, insertional inactivation of the fimA gene, with concomitant loss of fimbria production, results in a phenotype significantly less able to cause periodontal bone loss in the gnotobiotic rat model (150). Although individually such studies are not conclusive, collectively they provide strong support for a central role for fimbriae in periodontal disease. The precise nature of this role, however, still remains to be determined. In addition to mediating adherence, fimbriae have a variety of other properties (such as chemotactic properties and cytokine induction), as discussed throughout this article, that may also be involved in pathogenicity. The molecular interplay between P. gingivalis fimbriae and host factors may be a major determinant of the disease process.
The presence of more than one type of fimbria on P. gingivalis has recently become apparent. Electron microscopy of fimA-inactivated strains has revealed that, in addition to the major fimbriae, P. gingivalis possesses shorter fimbriae (87). These structures, designated minor fimbriae, are composed of a protein of 67 kDa that is antigenically distinct from the fimbrillin product of fimA. Another distinct fimbrial structure has been detected by immunoelectron microscopy (188). A 72-kDa protein is the constituent subunit of these fimbriae, which are designated Pg-II. The relationship and functionality of these minor fimbriae has yet to be investigated.
Hemagglutinins.
Hemagglutinin proteins are established virulence factors for a number of bacterial species, and P. gingivalis produces at least five hemagglutinating molecules. When expressed on the bacterial cell surface, hemagglutinins may promote colonization by mediating the binding of bacteria to receptors (usually oligosaccharides) on human cells. Since P. gingivalis utilizes heme for growth, binding of bacterial cells to erythrocytes may also serve a nutritional function (138). Early studies suggested that hemagglutination by P. gingivalis might be fimbria-mediated (191). This concept was subsequently confuted when purified fimbriae were shown not to exhibit hemagglutinating activity (283) and neither fimbriae nor antibodies to fimbriae (105) blocked hemagglutination by P. gingivalis cells. More recent evidence, however, does implicate a role for fimbrillin in hemagglutination, since synthetic peptides based on the sequence of fimbrillin (Fig. 2) clearly possess hemagglutinating activities (180). One current view then is that fimbriae do have hemagglutinating activity (54) but that P. gingivalis cells produce a number of hemagglutinins that are distinct from fimbrial protein synthesis, assembly, and structure.
Hemagglutinin activities expressed by P. gingivalis include those complexed with lipopolysaccharide (LPS) and lipid on the cell surface (21) and a released 40-kDa form of activity designated exohemagglutinin (101). This exohemagglutinin activity was inhibited by arginine (192), while salivary histatins (histidine-rich polypeptides) bind to P. gingivalis 381 cells and inhibit their hemagglutinating activity (164) via an arginine-dependent mechanism (165). Band 3 protein isolated from erythrocyte membranes is a putative receptor for this hemagglutinin (95). Another cell-bound nonfimbrial hemagglutinating adhesin denoted HA-Ag2 was purified from P. gingivalis ATCC 33277 and shown to consist of two polypeptide bands with molecular masses of 43 and 49 kDa (159).
The cloning of the first hemagglutinin gene (hagA) from P. gingivalis 381 by Progulske-Fox et al. (207) has led to major advances in our understanding of the genetic and functional complexities of the hemagglutination process. The hagA gene encodes a protein with a predicted molecular mass of 283.3 kDa (2,628 aa) and contains four contiguous direct 440- to 456-aa residue repeat blocks (89). It is likely that each repeat block contains a functional hemagglutinin domain since the polypeptide product of a single repeat clone demonstrated hemagglutinating activity (89). Each block begins with a P-N repeat sequence, which for three of the blocks is PNGTPNPNPNPNPGT. Four additional hag genes have now been cloned and sequenced. The hagB and hagC genes are at distinct chromosomal loci and encode 350-aa polypeptides with inferred molecular masses of 39 kDa (migrating as 49-kDa bands in gel electrophoresis) that are 98.6% identical (138, 208). These polypeptides have no sequence similarities to HagA, and no significant homologies to them can be seen in the protein sequence databases. On the other hand, the HagD polypeptide is 73.8% identical to HagA (139), while HagE, also cloned from strain 381, contains a 523-aa region with 93% homology to HagA (89). These hagA-like sequences are also found at other sites within the P. gingivalis chromosome (discussed below). The precise genetic, structural, and functional relationships between the hag gene products and the aforementioned HA-Ag2 or exohemagglutinin activities are not yet understood.
Important questions thus arising include to what extent these individual gene products contribute both to the hemagglutinating and adhesive activities of P. gingivalis cells and to the infection and disease processes. To begin to answer these questions, Lepine et al. (139) constructed isogenic hag mutants of P. gingivalis 381 and examined their in vitro adhesion phenotypes. Disruption of the hagA, hagB, or hagC gene in each case resulted in reduced hemagglutinating activity of cells, suggesting that all three genes were involved in determining the hemagglutination phenotype (139). The relative expression of hag genes under different environmental conditions, both in vitro and in vivo, was recently considered by Lee et al. (137). A novel vector-reporter construct was used to monitor in vivo expression of the hagB and hagC genes in a mouse subcutaneous abscess model of P. gingivalis infection. Both genes were shown to be expressed in vivo, with the hagB promoter being more active than the hagC promoter. In vitro, the relative expression of the hagB and hagC genes depends upon the phase of bacterial growth and levels of hemin (138). A genetic screen of E. coli clones expressing P. gingivalis genes mediating epithelial-cell attachment identified hagA- and hagD-like sequences (55), thus confirming independently a likely role for these sequences in adhesion to host cells. Epitopes within the HagA and HagD sequences are also recognized by antibodies in sera from periodontitis patients (43), but this immune recognition may not be sufficient to influence adhesion and colonization (118). A monoclonal antibody, which reacts with an epitope within the linear sequence GVSPK VCKDV TVEGS NEFAP VQNLT present within each of the HagA amino acid repeat units (89), inhibits P. gingivalis hemagglutination (43). This sequence might be a potential target for the development of immunization strategies against P. gingivalis.
There have been many reported associations between hemagglutinating and proteolytic activities in P. gingivalis (80, 99, 173, 174). The genetic basis for these is beginning to be explained following the recent activity in sequencing of P. gingivalis genes. Emerging sequence data suggest that hemagglutinin-related sequences not only are present as independently expressed genes, e.g., hagA, but also are coexpressed with genes encoding proteolytic activities. Hence, hemagglutinating and proteinase activities are found in complexes on the cell surface (233). Although it is not yet possible to correlate nucleotide sequences with the genes encoding HA-Ag2, exohemagglutinin, and other hemagglutinins with reported molecular masses in the range of 29 to 49 kDa, it seems plausible to suggest that these may all turn out to be products of two or more conserved hemagglutinin-encoding sequences distributed at multiple sites within the P. gingivalis genome. The genetic complexity should be resolved upon analysis of the complete genomic sequence. This in turn should open the way for more precise functional analyses of the role of hemagglutinin-related sequences in P. gingivalis adhesion and pathogenesis. As considered in more detail below (see “Proteinases”), it is already clear that the genetic and functional determinants of adhesion, hemagglutination, proteolysis, and fimbriation are inextricably linked (33).
INTERACTIONS WITH EPITHELIAL CELLS
The cells of the junctional epithelium provide an early barrier to tissue penetration by periodontal pathogens. Recently, it has become apparent that epithelial cells function both as a mechanical barrier to the ingress of organisms and as sensors of microbial infection (111). Epithelial cells thus comprise an interactive interface with subgingival bacteria, generating and transmitting signals between bacteria and the adjacent and underlying immune cells in the periodontal tissues. Many bacterial species that initiate infections at epithelial cell interfaces can internalize within the epithelial cells and subvert the intracellular information-processing events that control this signaling network. The molecular dialogue that occurs between bacteria and epithelial cells (for which the term “cellular microbiology” has recently been coined [40]) is thus important in the ultimate outcome of the encounter between pathogen and host. Although it has been known for some time that P. gingivalis is capable of penetrating gingival tissues (64, 202, 218–220, 258), recognition of the ability of this organism to subvert host intracellular events and localize intracellularly is more recent.
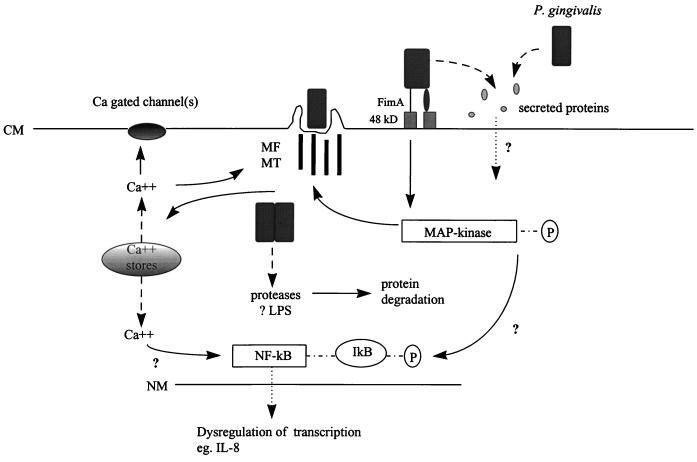
P. gingivalis has invasive potential, actively internalizing within epithelial cells by mechanisms similar to those used by various enteric pathogens (Fig. 3). Invasion of P. gingivalis has been studied in primary cultures of gingival epithelial cells (129, 133), oral epithelial cell lines (56, 175, 223), and cultures of multilayered pocket epithelium (224). Invasion proceeds more efficiently in primary cell cultures than in transformed cells (56, 129), indicating that transformation-induced changes in cell surface receptors or in intracellular signaling pathways may be detrimental to the invasion process. Attachment of P. gingivalis to primary gingival epithelial cells induces the formation of membrane invaginations that surround and engulf the bacteria, effecting their internalization (129, 133). The bacteria rapidly become located in the cytoplasm without, seemingly, being first constrained within a membrane-bound vacuole (129), although such cytoplasmic vacuoles do appear to be present after invasion of KB cells (175). Rearrangement of the host cell cytoskeleton to accommodate these membrane invaginations involves both microfilament and microtubule activity, a characteristic shared with, among others, Neisseria gonorrhoeae, Haemophilus influenzae, and Citrobacter freundii (129, 177, 212, 256). The individual component of the multimodal adherence of P. gingivalis to epithelial cells that is most relevant to subsequent invasion is fimbria mediated. A fimbria-deficient mutant of P. gingivalis exhibited a significantly greater reduction in invasion compared with adherence (274). Fimbriae binding to their cognate receptor(s) may therefore trigger the activation of eucaryotic proteins involved in signal transduction required for internalization (274). The epithelial cell receptor for fimbriae has not been characterized; however, a 48-kDa surface molecule has been found to bind to fimbriae and is thus a potential candidate (274). P. gingivalis is also capable of secreting a novel set of proteins when in contact with epithelial cells (196). Such secretion is generally indicative of the presence of a type III protein secretion pathway which allows bacteria to translocate proteins directly into host cell cytoplasm. These translocated proteins can then impinge upon eucaryotic signaling pathways. The extent to which the P. gingivalis secreted proteins possess intracellular effector functions remains to be determined. Although the ultimate fate of internal P. gingivalis is uncertain, the bacteria can persist for extended periods within host cells and can replicate intracellularly (129, 148). Intercellular spread of the organisms has not been observed. The environment within epithelial cells may thus represent a nutritionally rich, immunologically privileged site for P. gingivalis, which can then serve as a source for recrudescence of active disease episodes. Alternatively, uptake of P. gingivalis by epithelial cells may represent a mechanism by which the host sequesters this pathogenic organism.
FIG. 3.
Model of the currently understood P.
gingivalis interactions with gingival epithelial cells.
Abbreviations: CM, cytoplasmic membrane; MF, actin microfilaments; MT,
tubulin microtubules; NM, nuclear membrane; P, phosphate;
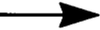 ; pathway with potential
intermediate steps;
; pathway with potential
intermediate steps; 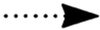 ,
translocation;
,
translocation; 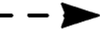 , release;
, release;
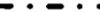 , reversible association.
Compiled from references 47, 55, 106, 129, 175, 196,
223, and 274.
, reversible association.
Compiled from references 47, 55, 106, 129, 175, 196,
223, and 274.
The events that occur within epithelial cells following contact with invasive P. gingivalis are only beginning to be defined (Fig. 3). P. gingivalis induces a transient increase in epithelial cell cytosolic [Ca2+], a characteristic shared with Salmonella typhimurium and enteropathogenic E. coli (12, 106, 195). Elevated intracellular [Ca2+] results from release of Ca2+ from thapsigargin-sensitive intracellular stores; influx of extracellular Ca2+ does not appear to be involved (106). Such Ca2+ ion fluxes are likely to be important in many signaling events and may converge on calcium-gated ion channels in the cytoplasmic membrane, cytoskeletal remodeling, or nuclear transcription factors (17). Eucaryotic cell-signaling pathways also depend on the pattern of protein phosphorylation (40). P. gingivalis invasion is correlated with the tyrosine phosphorylation of a 43-kDa intracellular eucaryotic protein (223), a size consistent with that of mitogen-activated protein (MAP) kinase, which is a target of phosphorylation by invasive S. typhimurium and Listeria monocytogenes (215, 260). Exploitation of MAP kinase-dependent signaling events may also funnel through cytoskeletal remodeling or nuclear transcription factors (63). Furthermore, recent studies in eucaryotic signaling indicate that the cellular skeleton per se may play a direct role in transmitting regulatory information through the cell rather than simply acting as a passive support structure (151, 152). The usurpation of microfilament and microtubule functions by invasive P. gingivalis may therefore be another means by which normal intracellular information flow is compromised. The collective action of these perturbations of epithelial cell intracellular pathways can have phenotypic effects with immediate relevance to the disease process. Regulation of matrix metalloproteinase (MMP) production by gingival epithelial cells is disrupted following contact with the organism (66). Impaired regulation of MMP activation and production will affect extracellular proteolysis in general and, with particular consequence for tissue integrity, interfere with extracellular matrix repair and reorganization. Invasion of P. gingivalis also has implications for innate host immunity. Secretion of interleukin-8 (IL-8) by gingival epithelial cells is inhibited following P. gingivalis invasion (47). P. gingivalis is also able to antagonize IL-8 secretion following stimulation of epithelial cells by common plaque constituents such as Fusobacterium nucleatum (47). Inhibition of IL-8 accumulation by P. gingivalis at sites of bacterial invasion could have a debilitating effect on innate host defense in the periodontium, where bacterial exposure is constant. The host would no longer be able to detect the presence of bacteria and direct leukocytes for their removal. The ensuing overgrowth of bacteria would then contribute to a burst of disease activity.
NUTRIENT ACQUISITION
Peptides
Following entry and attachment, oral bacteria must be able to utilize available nutrients in order to thrive in the oral cavity. P. gingivalis is an asaccharolytic organism, dependent on nitrogenous substrates for energy (230). Although sugars such as glucose can be utilized by the organism, these compounds are not converted to metabolic end products but, rather, are used for the biosynthesis of intracellular macromolecules (230, 231, 234). Among the potential nitrogenous substrates available in the mouth, P. gingivalis has only a limited ability to ferment free amino acids, with the possible exception of aspartic acid and asparagine, which can be metabolized through oxaloacetate, malate, and fumarate to yield succinate (231). In contrast, peptides are efficiently utilized for growth (230). Thus, the action of proteolytic enzymes produced by P. gingivalis, along with other bacterially and host-derived proteases, in the protein-rich subgingival milieu would appear to be pivotal to nutrient acquisition by P. gingivalis. As discussed in considerable detail below, P. gingivalis produces multiple proteases that can degrade a number of potentially important substrates in the gingival crevice, including collagen, fibronectin, fibrinogen, laminin, and keratin (98, 153, 265, 266). Although these proteolytic activities are well established, the identity of the enzyme(s) responsible in each case is less clear. Nonetheless, the high level of proteolytic activity and the wide range of appropriate substrate specificities provide P. gingivalis with the necessary molecular tools to compete metabolically in the protein-rich subgingival environment.
Hemin
P. gingivalis has an obligate iron requirement for growth. However, it appears to lack a siderophore system (22) and utilizes hemin (iron protoporphyrin IX) to satisfy this iron requirement (14, 23, 98, 250). A number of hemin-containing compounds, such as hemoglobin, haptoglobin, myoglobin, hemopexin, methemoglobin, oxyhemoglobin, albumin, lactoperoxidase, catalase, and cytochrome c, can provide hemin following proteolytic processing (14, 23, 67, 98, 250). Although nonhemin iron sources such as ferric, ferrous, and nitrogenous inorganic iron, along with transferrin and lactoferrin, can also support P. gingivalis growth (23, 102), all the iron demands of P. gingivalis can be fulfilled by hemin. Hemin is stored on the cell surface, a feature considered to give rise to the characteristic black-pigmented appearance of P. gingivalis colonies (70). The intact hemin molecule can be transported into the cell in an energy-dependent process regulated by the levels of available hemin (71). Specific interactions between the protoporphyrin IX ring and outer membrane proteins appear to initiate the uptake process (23, 71). A number of potential hemin binding proteins have been described. The work of Bramanti and Holt (24–26) has identified a 26-kDa outer membrane protein that may be involved in both binding and transport of hemin. Fujimura et al. (67, 68) reported a 19-kDa hemoglobulin binding protein, while Smalley et al. (247, 249) and Kim et al. (119) discovered 32 and 30-kDa hemin binding proteins respectively. Karunakaran et al. (113) identified a 48-kDa protein (HemR) with significant homology within the N-terminal region to iron-regulated, TonB-dependent outer membrane receptor proteins IrgA from Vibrio cholerae and BtuB and CirA from E. coli. (In many bacteria, TonB-linked receptors are involved in periplasmic transport of compounds such as colicins, iron, and vitamin B12.) The C-terminal two-thirds of HemR (aa 173 to 419) contains 99% identical residues to a region (aa 575 to 821) of the P. gingivalis PrtT protease, encoded by a gene upstream of hemR. An additional putative TonB-linked and protease-associated protein (Tla) has also been found in P. gingivalis. The tla gene (3) encodes a predicted protein with structural similarity within the N-terminal region to TonB-linked receptors and also contains an internal region of 98% identity to the β domain of RI protease (see below). Mutants inactivated in tla are unable to grow in medium containing low concentrations of hemin and produce significantly lower proteinase activities. The relationship (if any) among the various hemin-binding proteins, the role of TonB in periplasmic translocation of hemin, and the involvement of other hemin-regulated proteins that have not yet been demonstrated to bind hemin (22, 27, 247) remain to be definitively established. Kinetic analyses indicate the existence of more than one hemin-binding mechanism (248, 249) which would allow the participation both of multiple outer membrane proteins and of LPS, another hemin binding molecule (78).
The levels of hemin in the oral cavity are likely to be variable. Bleeding as a result of gingival inflammation will elevate subgingival hemin concentrations and may be one factor that predisposes a site to P. gingivalis accumulation. P. gingivalis has also developed mechanisms that will increase the availability of hemin. Proteolytic activity, perhaps in direct association with hemin-binding proteins, e.g., HemR and Tla (3, 113), will degrade the host hemin-sequestering plasma proteins such as haptoglobulin and albumin (249). Moreover, P. gingivalis produces (at least) two genetically distinct cell- and vesicle-associated hemolysins which will liberate hemoglobulin from erythrocytes (37, 112). Hemin not only is vital for growth but also regulates various virulence-associated activities of P. gingivalis (as discussed below). Furthermore, membrane-bound hemin can scavenge oxygen and help maintain an anaerobic environment (250). More complete elucidation of the complex binding, uptake, and regulatory pathways relating to hemin will provide significant insights into the fundamental nature of the organism.
TOXINS AND TOXIC PRODUCTS
P. gingivalis does not elaborate a single potent exotoxin, but an almost bewildering array of enzymes along with toxic metabolites and cellular constituents are produced by the bacterial cells. These compounds have the potential to impinge upon host tissue integrity and cause loss of the alveolar bone and other supporting periodontal tissues, an important feature of advanced periodontal disease. Furthermore, the host innate and acquired immune defense mechanisms can be adversely affected, prolonging the microbial onslaught on the periodontium and contributing to immunopathogenic mechanisms of tissue destruction.
PROTEINASES
Although the primary function of proteases secreted by asaccharolytic bacteria such as P. gingivalis is to provide peptides for growth, proteases are also involved directly in tissue invasion and destruction by bacteria and in evasion and modulation of host immune defenses. Specific examples of tissue degradation and attenuation of host defense mechanisms include the degradation of extracellular matrix proteins, activation of MMPs, inactivation of plasma proteinase inhibitors, cleavage of cell surface receptors, activation or inactivation of complement factors and cytokines, and activation of the kallikrein-kinin cascade (266) (Table 1). P. gingivalis cells elaborate a number of proteolytic activities that accomplish all these activities. In the realization that periodontitis is a destructive and inflammatory condition, there naturally has been intense research effort directed toward identifying and characterizing the proteinases produced by periodontal pathogens. The current status of over 15 years of work on P. gingivalis proteinases is a somewhat confusing and still emerging story involving multiple genes and multiply processed products of those genes. The confusion is exacerbated by the plethora of strains used, a lack of consensus with respect to genetic nomenclature, incomplete sequence information, and only partial biochemical characterizations of enzymatic activities. Although it is anticipated that the P. gingivalis genomic sequence will begin to clarify matters, functional studies are needed to resolve the complexities of proteinase production and regulation, since these involve extensive posttranslational processing events and probably also translational controls in addition to transcriptional control. Indeed, major advances in understanding of P. gingivalis proteinases have come about recently through close integration of biochemical and genetic studies (18, 200, 210).
TABLE 1.
Functions of P. gingivalis proteinasesa
| Impairment of tissue integrity | Perturbation of host defenses | Bacterial function |
|---|---|---|
| Degradation of extracellular matrix proteins (fibronectin, laminin) | Degradation of immunoglobulins | Release of hemin and iron from host proteins |
| Hydrolysis of collagens I, III, IV, and V | Inactivation or activation of complement components | Exposure of host and bacterial cryptitopes |
| Degradation of fibrinogen | Destruction of cytokines and chemokines | Posttranslational processing of proteases, fimbrillin, and outer membrane proteins |
| Inactivation of tissue and plasma proteinase inhibitors | Cleavage of leukocyte surface receptors | Involvement in intracellular invasion |
| Activation of matrix metalloproteinases | Degradation of antimicrobial peptides | |
| Activation of the kallikrein/kinin cascade |
A number of proteases produced by P. gingivalis are thiol-dependent enzymes that cleave C-terminal to arginine (Arg-Xaa) or lysine (Lys-Xaa) within protein or peptide substrates and thus were designated, on the basis of substrate specificity, “trypsin-like” enzymes. Biochemically, these enzymes are members of the cysteine-proteinase family, which includes the papains, calpains, streptopains (from Streptococcus), clostripain (Clostridium), various endopeptidases, and the type IV prepilin leader peptidase. In addition to the cysteine proteinase enzymes, P. gingivalis produces serine proteinase activities (81, 96). Enzymes that degrade immunoglobulins, collagen and/or gelatin, complement factors, fibrinogen, and fibronectin can almost always be ascribed to one of these two families.
Arg-X- and Lys-X-specific proteinases.
Before the genes encoding cysteine proteinases of P. gingivalis were cloned and sequenced, the proteolytic enzymatic activities that were purified or semipurified from cell surface extracts or culture fluid were given a variety of names. Thus, gingivain (232), gingipain (34), and argingipain (110) all describe Arg-X-specific proteinase activities which are probably identical. It has been proposed (158, 205) that the genes encoding Arg-X-specific enzymes be designated rgp and those encoding Lys-X-specific enzymes be designated kgp, a classification scheme that would certainly simplify the nomenclature.
A number of genes encoding Arg-X- and Lys-X-specific proteases from different strains of P. gingivalis have now been isolated and sequenced (Table 2). The genes within the two Arg-X protease families exhibit some differences in sequence and in organization of the coding regions within the genes; however, virtually all these differences are possibly simply strain related and/or related to sequencing or cloning artifacts (205). The major difference between the two Arg-X families is the absence of the hemagglutinin/adhesin domain in the second grouping (rgp-2, etc). The N-terminal catalytic domain sequences are, however, structurally and kinetically almost identical (210). Overall, their extensive sequence and structural similarities lead to the conclusion that all these genes were derived from a single ancestral genetic locus. Inactivation of both genes in strain W50 is necessary for abrogation of Arg-X proteinase activity. This indicates that both genes are expressed in culture and that both their products contribute to extracellular Arg-X protease activity.
TABLE 2.
P. gingivalis Proteinases: Substrate Specificity and Genes
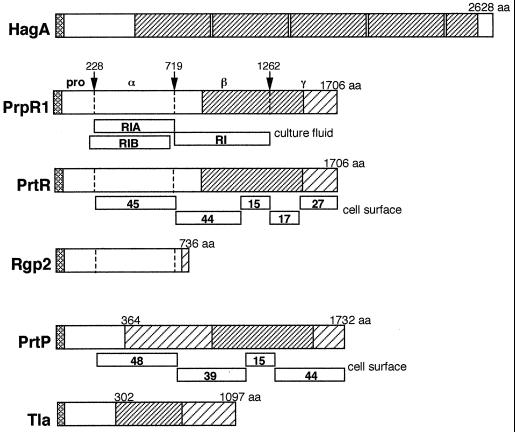
Multiple forms of Arg-X proteases are recovered from the cell surface of P. gingivalis and from the culture fluid. These forms are believed to result mainly from posttranslational processing of larger polyprotein precursor products. For example, the prpR1 gene has a primary transcript of approximately 6 kb and the inferred polyprotein comprises 1,706 aa. Three forms of culture supernatant Arg-X proteinase activity, all products of the prpR1 gene, have been designated RI, RIA, and RIB (210). RI is a heterodimer composed of a catalytic (α) chain with a molecular mass of approximately 53 kDa, derived from the N-terminal region of the polyprotein, held in noncovalent association with a second (β) chain of about the same mass, derived from the central region of the polyprotein (Fig. 4). The RIA and RIB proteins are monomeric forms, RIA being the α chain only and RIB being a lipid-modified form of α that may be associated with extracellular vesicles. The α and β components of RI are contiguous on the initial translation product and are flanked by an N-terminal propeptide (preceded by a typical leader peptide) and a C-terminal extension (termed γ) (Fig. 4). The putative processing sites within the precursor each carry an Arg residue, and therefore it is proposed that the polyprotein is processed by an Arg-X protease.
FIG. 4.
P. gingivalis gene products encoding cysteine proteinase activity and/or containing HagA (hemagglutinin)-related sequences. The HagA (GenBank accession no. U41807) precursor comprises 2628 aa and contains four contiguous repeat blocks of 440 to 456 aa each, with >98% identical residues (▨). Sequences within the other proteins shown that are 90% or more identical to the HagA sequence are also indicated by this pattern. All polypeptides carry a putative leader peptide (▩) at the NH2 terminus. PrpR1 (accession no. X82680) Arg-X-specific proteinase precursor is proteolytically cleaved at the amino acid residues indicated (arrows) to generate α and β species, as described by Rangarajan et al. (210). Culture supernatant proteinase activities are associated with three forms designated RI (αβ), RIA (α), and RIB (*α), shown below PrpR1 and discussed in the text. Homologous proteins to PrpR1, from a number of different P. gingivalis strains, include PrtR (L26341), Rgp-1 (U15282), RgpA (A55426), Agp (D26470), and CpgR (X85186). The P. gingivalis strains examined all contain a second gene encoding an Arg-X proteinase designated Rgp-2 (U85038), RgpB (D64081), and PrtRII (AF007124). Rgp-2 consists of a discrete N-terminal catalytic domain without the associated C-terminal adhesin region (158). Cell surface-associated Arg-X-specific proteinase activity is found as a multimeric complex (18) consisting of five polypeptides derived from cleavage of the polyprotein (shown below PrtR) and four proteolytic products of the Lys-X-specific proteinase shown below PrtP (U42210), also designated PrtK (U75366) or Kgp (1,723 aa) (D83258 and U54691). The HagD (hemagglutinin) sequence begins at aa 364 within PrtP. The Tla protein (Y07618) is a putative outer membrane-associated receptor that, over 795 aa, has 99% identity to the COOH-terminal half of PrtK (PrtP, Kgp). Sequences with 90% or greater identity to HagD are indicated (▨). The patterns of regional sequence similarities amongst the various gene products are far more complex than depicted in this figure, which has been simplified for clarity, and the reader is referred to reference 13 for a detailed comparison of sequence identities between HagA, Rgp-1, and PrtP.
Cell surface-associated Arg-X protease activity in P. gingivalis W50 is found within a complex that contains at least nine polypeptides (18). These are believed to be derived from proteolytic processing of PrpR1 polyprotein [designated PrtR by Bhogal et al. (18)] and from processing of the polyprotein product of the prtK gene that encodes the proteinase with Lys-X specificity (Table 2). The PrtK (Kgp) precursor may be processed in much the same way as the PrtR (PrpR1) precursors are processed (189, 206). Enzymatic cleavage at specific Arg-X or Lys-X sites within the polyprotein generates a 48-kDa Lys-X specific protease, a 39-kDa adhesin (corresponding to the β domain in PrpR1), and two other adhesins of 15 and 44 kDa (Fig. 4). Likewise, processing of PrtR generates a 45-kDa Arg-X-specific proteinase, a 44-kDa adhesin, and 15-, 17-, and 27-kDa adhesins corresponding to the β and γ domains (Fig. 4). The adhesin molecules all exhibit considerable cross-homology and the 15-kDa proteins are identical (18). The formation of such a cell surface complex may be the consequence, at least in part, of the mechanisms involved in secretion and activation of the proteases. There is good evidence that processing of the PrtK (Kgp) precursor polyprotein requires Arg-X-specific protease activity, since Lys-X protease activity is greatly decreased in rgpA rgpB double mutants that are abrogated in Arg-X-specific protease activity (189). Thus, it might be expected that the polyproteins encoding both Arg-X-specific and Lys-X-specific proteolytic activities would be coprocessed. Furthermore, although the monomeric RIA form of PrpR1 is proteolytically active and Lys-X activity of the 48-kDa Kgp protein is not dependent upon the adhesin (β and γ) domains for activity (200), there is some evidence that the adhesin domains are necessary for secretion of the polyproteins (190).
Relationship between hemagglutinin and proteinase.
Historically, the hemagglutinating activities and adherence of P. gingivalis cells have both been associated with protease activity (80, 99, 173, 204). Inactivation of a cysteine protease gene in P. gingivalis 381 had a direct effect on hemagglutinin activity (280) as well as on the ability of P. gingivalis to bind gram-positive bacteria, oral epithelial cells, and extracellular matrix proteins (263). While some of these effects could be the result of a pleiotropic mutation (as discussed below), the sequences of proteinase genes reveal a possible molecular basis for the association of protease and adhesive activities. It is now evident that the C-terminal coding regions of the genes encoding the RgpA family of cysteine proteinases (equivalent to the β and γ regions within PrpR1) encode extensive amino acid blocks that have up to 90% identity to sequences that are also found within the hagA, hagD and hagE genes encoding hemagglutinins (13) (Fig. 4). For example, Rgp1, Agp, PrpR1, and PrtR Arg-X-specific proteinases each contain a 522-aa residue region with 93% identical residues to HagA (89). This region carries a hemoglobin receptor domain (171), localized within the 15-kDa segment of PrtR and PrtP, as shown in Fig. 4. It has not yet been determined if these protease-associated sequences are functional hemagglutinins, in addition to the hemagglutinating activities that have been assigned to HagA, HagB, and HagC. Moreover, it now appears that hagD specifically comprises the sequence encoding the C-terminal region of the Lys-X proteinase, encoded by a single gene in P. gingivalis and variously designated prtK, kpg, and prtP (38) (Fig. 4). As discussed above, the Lys-X proteinase is proposed to be generated by processing of a polyprotein (PrtP is 1,732 aa long) into an active protease (comprising aa 229 to 738 of PrtP) and C-terminally derived hemagglutinin sequences. However, it is not firmly established that posttranslational modifications of the polyproteins account exclusively for the various forms of proteinase and hemagglutinins identified. It is formally possible that translational initiation occurs internally in the primary mRNA transcripts, effectively generating an additional level of control over hemagglutinin production.
hagA-like sequences have now been found within at least five separate genetic loci in P. gingivalis. As well as being a component of the Arg-X (RgpA family) and Lys-X proteinase gene sequences, the HagA sequence is present within Tla (1,097 aa) (Fig. 4), an outer membrane-associated polypeptide thought to be involved in hemin recognition (3), as discussed above. Given that there are estimated to be in excess of 20 sites within the P. gingivalis chromosome that hybridize with a hagA repeat probe (13), it would be prudent to suggest that the genetic complexity of the protease/hemagglutinin system has been underestimated. The acquisition of a conserved sequence encoding a surface protein at multiple genomic sites has been characteristically associated with evasion, subversion, or deflection of host immune defenses in a number of pathogens.
The proteinase gene designated prtH was cloned and sequenced from strain W83 (60), and although prtH contained regions of considerable homology to the other Arg-X proteinase genes rgp-1, prpR1, prtR, and agp, there was some question whether it was a true homolog of these. The product of prtH, a 97-kDa proteinase found in membrane vesicles and with complement C3-degrading activity, contains a region of 270 aa with 95% homology to the hagA gene product (89), and it now appears that prtH is indeed homologous to rgp-1, etc. (140).
Other nonrelated proteinases.
Three additional genes (prtT, prtC, and tpr) encoding proteases have been cloned and sequenced from P. gingivalis. These genes are clearly distinguished from the previous protease genes, and they do not contain sequence homologies to hagA. Nevertheless, prtT (from strain ATCC 53977) does possess a C-terminal coding region for hemagglutinin activity (194) that is distinct from the hagA, hagB, hagC, and hagD sequences. The prtT gene is transcribed as a 3.3-kb mRNA (147) and encodes a protein of approximately 99 kDa. This polypeptide contains 31% identical residues (over 401 aa within the N-terminal region) to streptococcal pyrogenic exotoxin B, which is itself related to streptococcal cysteine proteinase (streptopain). The prtT gene is immediately downstream of prtC, which encodes a putative collagenase comprising 417 aa (115). It is not known how this relates, if at all, to the reports of 55-kDa (16, 124) and 44-kDa (110) collagenolytic activities isolated from P. gingivalis. Furthermore, sequences with significant amino acid identities (up to 55%) to PrtC are found within the inferred amino acid sequences of putative proteins from a range of bacteria including Methanobacterium, Methanococcus, Bacillus subtilis, H. influenzae, and E. coli. Lastly, the tpr gene, which appears to be present as a single copy in strains W83, W50, and ATCC 33277 (198, 199), is expressed as a 1.7-kb mRNA (197). The gene encodes a thiol-dependent, papain-related, protease with a molecular mass of approximately 55 kDa that has no significant sequence homology to any of the other proteases (20). Evidence suggests that this proteinase, although not acting on native collagen, may be involved in the later stages of collagen degradation, since it is active in hydrolyzing gelatin and the bacterial collagenase substrate Pz-peptide (197). Another Pz-peptidase has been isolated from P. gingivalis ATCC 33277 (254) and may be distinct from Tpr, based on N-terminal amino acid sequence information.
Concerted activities.
To recapitulate, P. gingivalis contains two homologous genes encoding Arg-X-specific proteinases, one gene encoding a Lys-X specific proteinase (Table 2), and three genes encoding a putative collagenase (PrtC), a streptopain-related protease (PrtT), and a Pz-peptidase (Tpr). The activity of these cloned gene products accounts for most of the wide variety of substrates degraded by P. gingivalis (Table 1). However, because the enzyme activities derived from a single gene product are present as several molecular species and are complexed with the products of both homologous genes and heterologous genes, it has been immensely difficult if not impossible to ascertain unequivocally the substrate specificities of individual proteinases “purified” from P. gingivalis (50). For example, a protease activity purified from spent culture medium of P. gingivalis was shown to run as a single 55-kDa band upon gel electrophoresis and was found to hydrolyze collagens type I, III, IV, and V, complement factor C3, fibrinogen, fibronectin, α1-antitrypsin, α2-macroglobulin, apotransferrin, and human serum albumin (16). Likewise, the 44-kDa Rgp proteinase purified from P. gingivalis culture fluid was shown to degrade collagens type I and IV and immunoglobulin G (110). However, it is reported that the purified recombinant gingipains (RgpA, RgpB, and Kgp) do not degrade native collagen (205), and so a broad range of substrates degraded by an apparently single enzyme species from P. gingivalis culture fluid may in fact be due to the combined activities of two or more copurified enzyme species. Binding studies with the Arg-X- and Lys-X-specific proteinases Rgp and Kgp, respectively, show that they both bind to and degrade fibrinogen, fibronectin, and laminin (204) and that purified PrtP hydrolyses fibrinogen (38). The combined effects of these activities, in conjunction with the ability of P. gingivalis proteases to activate MMPs (51), would therefore lead to extensive host tissue disruption. In addition, evidence suggests that Arg-X proteinases activate the kallikrein/kinin and complement pathways, disrupt polymorphonuclear leukocyte (PMN) functions (107, 170), inactivate tumor necrosis factor alpha (TNF-α) (30), and modify neutrophil elastase (1). These activities, together with the evidence that Kgp (Lys-X) proteinase is not inhibited by plasma α2-macroglobulin (82), suggest that P. gingivalis proteinases are major virulence factors in the development of periodontal disease (265).
Role in virulence.
Biochemical and genetic evidence points to an essential but highly complex role for proteinases in P. gingivalis metabolism and virulence. While molecular genetics has demonstrated that P. gingivalis cells express multiple proteinases, the relative roles that these enzymes play in adhesion, nutrition, and virulence are hard to determine because these processes are all proteolytically interrelated and interdependent. A current molecular picture of the P. gingivalis cell surface would include major macromolecular complexes of adhesins (hemagglutinins), proteinases, outer membrane-associated protein receptors, and LPS. The complexing of adhesin domains and proteinases might increase the repertoire of targets for the proteinases. At the same time, the hemagglutinin sequences are highly antigenic and might act to shield essential proteolytic functions from the host immune system. Given this potential complexity of proteinase-associated phenotypes and the number of distinct proteinase genes identified, it is perhaps not surprising that the analysis of gene knockout mutants has in the main simply confirmed that proteinases impinge on multiple metabolic and virulence-related processes. A novel erythromycin resistance determinant cassette, developed to insertionally inactivate the prtH gene in P. gingivalis W83, resulted in a strain deficient in Arg-X proteinase activity, with decreased virulence in a mouse model of infection (61). In addition, the isogenic mutant was less able to degrade complement factor C3, and opsonized mutant cells were taken up in much larger numbers by human PMN than were wild-type cells (226). In separate experiments, a double-knockout rgpA rgpB mutant of P. gingivalis ATCC 33277 that was abrogated in Arg-X proteinase activity showed much less inhibition of leukocyte bactericidal function than did the wild-type strain, as well as having markedly reduced hemagglutinin activity (170). Another isogenic mutant with a mutation in the rgp-1 (Arg-X) protease gene of strain 381 exhibited decreased binding to epithelial cells, gram-positive bacteria, extracellular matrix proteins, and type 1 collagen (263). These experiments all confirm the notion that proteinases are obligatory for P. gingivalis growth and survival in the host. However, since it has been shown that Arg-X-specific proteinase mutants are also deficient in Lys-X-specific proteinase activity, the data do not point to a specific role that any one proteinase might play in pathogenesis. This information will be important if inhibition of proteinase function is to be pursued as a viable method of controlling the growth of and tissue destruction by periodontal pathogens such as P. gingivalis.
Proteinases and fimbriae.
A further complicating factor in the study of proteinase function in P. gingivalis is the discovery that the production and activity of the major fimbriae are modulated by proteolytic activity. This control appears to occur on at least three levels: posttranslational modification, transcriptional modulation, and substrate activation. The work of Onoe et al. (193) suggested a role for trypsin-like proteases in the processing of the leader peptide from the fimbrillin precursor. The subsequent observations of Nakayama et al. (172) that the ATCC 33277 rgpA rgpB double-mutant cells possessed very few fimbriae on their surface compared with rgpA or rgpB single-mutant or wild-type cells demonstrated that fimbrial production required the expression of Arg-X and/or Lys-X-specific proteinases. These data suggest that proteinase activity was necessary for processing and maturation of fimbrillin, in addition to its role in modifying other cell surface-associated proteins. Control at the transcriptional level is inferred from the finding that inactivation of the rgp-1 (Arg-X proteinase) gene in P. gingivalis 381 resulted in reduced production of the 43-kDa fimbrillin subunit as well as reduced transcription of fimA (263). Since fimbriae are implicated in a large number of adhesive interactions of P. gingivalis (see above), most notably binding to host and bacterial cells, it would not be possible through simply generating proteinase gene knockout mutants to delineate precisely which adhesive functions are related directly to protease/hemagglutinin activity and which are the result of secondary effects on fimbriation. The third control level, substrate activation, is facilitated by the ability of P. gingivalis to bind to and degrade (perhaps progressively) human plasma fibronectin, along with other host proteins such as laminin, fibrinogen, and collagen. Both the fimbriae and the Arg-X-specific (204) and Lys-X-specific (134) proteinases are associated with these binding and degradative processes. Hydrolysis of fibronectin or other matrix proteins such as collagen by P. gingivalis Arg-X proteinase enhances the binding of fimbriae to these substrates (124). Specifically, it seems that the Arg-X proteinase is able to expose sequences, within host matrix protein molecules, that carry C-terminal Arg residues, thus promoting adhesion of the organism through a fimbria-Arg interaction (123). Proteolytic exposure of cryptic binding domains on host molecules and potentially on epithelial cells and other bacteria (35, 97, 124, 130) could involve host-derived as well as bacterial enzymes. It has been conjectured (35, 97) that this may represent one mechanism by which initial gingivitis progresses to more severe periodontitis. A buildup of plaque bacteria in the gingival sulcus may irritate the tissues and cause increased proteolytic activity in the gingival milieu. The resulting exposure of previously hidden receptors would then enhance colonization by P. gingivalis. Subsequently, P. gingivalis proteases themselves could contribute to further exposure of adherence receptors.
P. gingivalis cells are geared to an asaccharolytic life-style, and this underpins the intimate relationships that have evolved between fimbriation, adhesion, hemagglutination, and proteinase activities, in particular centered around the recognition of arginine and lysine. Since these bacterial proteinases have unusual primary sequences, they are attractive targets for the development of novel and specific inhibitors. Critical to the development of new inhibitory compounds will be an understanding of the controls operating on proteinase gene expression and proteinase production in vivo. Evidence exists that proteinase production might be transcriptionally regulated in response to environmental conditions such as availability of hemin and nutrients (3, 27, 72, 197). Trp proteinase production, for example, is enhanced in less nutritious growth medium (197). This raises the possibility that there are as yet undiscovered P. gingivalis proteolytic activities that are expressed poorly, if at all, under conventional culture conditions and might be expressed only under in vivo environmental conditions.
Nonproteolytic, Potentially Destructive Compounds
In addition to proteolytic activity, P. gingivalis can degrade the glycosaminoglycans hyaluronate, chondroitin sulfate, and heparin (98). The repertoire of enzymes and metabolites that could be detrimental to the host also includes phospholipase A, which can provide the prostaglandin precursors that could stimulate prostaglandin-mediated bone resorption (29, 75); alkaline and acid phosphatases, which may contribute to alveolar bone breakdown (65, 242); DNase and RNase (217); sialidase (98); volatile sulfur compounds such as hydrogen sulfide, methylmercaptan, and dimethyl disulfide, which are cytotoxic and can inhibit protein synthesis (243, 264); butyrate and propionate, which are cytotoxic for epithelial cells, fibroblasts, and lymphocytes (125, 238, 243); and indole and ammonia, which also exhibit cytotoxicity (243, 270).
Impact on Bone Metabolism
By a variety of intricate and interconnected mechanisms, P. gingivalis can contribute to alveolar bone loss by stimulating bone resorption, inducing bone destruction, and inhibiting bone formation (98, 144). P. gingivalis LPS can activate osteoclasts directly and causes the release of prostaglandin E2 and of the cytokines IL-1β and TNF-α from macrophages, monocytes, and fibroblasts (28, 142, 214, 239, 276, 278). These compounds are potent local mediators of bone resorption and, moreover, can inhibit collagen synthesis by osteoblasts and induce the production of host metalloproteases that destroy connective tissue and bone (94, 98). In addition to LPS, other cellular constituents may be involved in bone loss. Heat-stable polysaccharide antigens can stimulate the release of IL-1β from monocytes (168), and saline-extracted proteins can stimulate bone resorption in in vitro models (275). The identities of some of the active P. gingivalis proteins have been revealed. A 24-kDa outer membrane protein has bone-resorptive and -destructive activity in vitro (157), while an outer membrane protein of 75 kDa and a 12-kDa lectin-like protein can stimulate macrophages to secrete IL-1β (221, 272). P. gingivalis fimbriae (fimA gene product) stimulate bone resorption in vitro (116), probably by inducing the expression and production of IL-1β and TNF-α from monocytes, macrophages, and gingival fibroblasts (90, 92, 181, 187) and by stimulating the expression of IL-1β and granulocyte-macrophage colony-stimulating factor in bone cells themselves (116). The amino acid sequence LTxxLTxxN, which occurs within fimbrillin aa 61 to 80 and 171 to 185 (Fig. 2), appears to be involved in cytokine stimulation in monocytes (187). Within this domain, the xLTxx sequence may be the minimum essential unit (184). Fimbrial binding, and subsequent stimulation of cytokines and bone resorption can be inhibited by N-acetyl-d-galactosamine (161) or by interaction with fibronectin through the heparin binding and cell attachment domains in the fibronectin structure (117). Fibronectin domains and other receptor analogs therefore have the potential to act as antagonists to neutralize fimbria-mediated bone resorption. The intracellular signalling events that are the target of fimbriae appear to involve eucaryotic cell kinase activity. Tyrosine phosphorylation of several proteins in osteoclasts and macrophages is induced by fimbriae, and inhibition of tyrosine kinase prevents cytokine stimulation and fimbria-mediated bone resorption in vitro (91, 221). The 12-kDa lectin-like surface component also acts via activation of tyrosine kinases within macrophages (222).
The list of P. gingivalis components and products with the capability of compromising tissue integrity is long. Undoubtedly, many of these activities, alone or in combination, are important in vivo. Definitive evidence, however, about the ones that are operational in the disease process is, unfortunately, still lacking.
ENCOUNTER WITH HOST DEFENSE MECHANISMS
Bacterial colonizers of the periodontal pocket will encounter the cells and extracellular effector molecules of the host immune system. Bacterial modulation of host immune processes is an important determinant in their long-term survival and pathogenic potential. Both innate and acquired defense mechanisms are operational in the periodontal pocket. The relative contributions of each component, both to protection and to possible immune system-mediated tissue destruction, have not been precisely delineated.
Of the various immune mechanisms in the periodontal pocket, PMNs appear to play a major role in controlling the overgrowth of periodontal bacteria (49). P. gingivalis impinges on almost all aspects of PMN recruitment and activity. Although there are numerous phenomenological reports of the organism adversely affecting PMNs, this section will concentrate on processes for which functionality has been associated with specific molecules. Neutrophil chemotaxis is inhibited by low-molecular-weight fatty acids, such as succinic acid, that are produced by the organism (19, 216). Succinate may act by reducing the intracellular pH of neutrophils (216). P. gingivalis can also immobilize PMN responses to chemotactic peptides by depolarizing PMN membranes (176). This activity is associated with an outer membrane protein of 31.5 kDa, which may be a porin. Novak and Cohen (176) proposed that depolarization could be caused by translocation of this porin into the PMN membrane, thus producing an ion channel that the PMN would be unable to regulate. Package of the porin in extracellular vesicles could then be a means by which P. gingivalis could act at remote sites to inhibit PMN influx in the periodontal pocket.
As is frequently the case with investigation of bacterial interactions with the host innate immune response, different bacterial components can demonstrate diametrically opposing affects. In the case of P. gingivalis, LPS strongly activates complement, generating the chemotactic product C5a. However, the activity of cell-associated LPS may be tempered by capsular polysaccharide that can physically mask LPS on the cell surface (228). The major fimbriae induce the expression of neutrophil chemotactic factor KC in macrophages (93). Upregulation of KC gene expression appears to be mediated via protein kinase C-dependent phosphorylation of a 68-kDa protein (pp68) in the macrophages (160). Protein kinase C also mediates LPS-engendered production of IL-1, TNF-α, and nitric oxide from macrophages (235). Both LPS and fimbriae are directly chemotactic for monocytes (180, 182). The fimbrial consensus sequence LTxxLTxxN (also involved in cytokine stimulation [see above and Fig. 2]) is involved in binding to monocytes and in the induction of migration-stimulating activity (180, 182).
In addition to direct effects on the professional phagocytes, P. gingivalis and its components can induce the expression of a variety of cytokines and chemokines (as introduced above). The cytokine response of PMNs, monocytes/macrophages, fibroblasts, and epithelial cells is summarized in Table 3. Increases in the levels of proinflammatory cytokines such as IL-1β, TNF-α, IL-6, and IL-8 are likely to not only promote inflammation but also stimulate bone and tissue destruction in the periodontal area. However, there is more than one facet to the nature of P. gingivalis-cytokine interactions. P. gingivalis can induce anti-inflammatory cytokines, degrade existing cytokines, and antagonize IL-8 production by epithelial cells (47, 62, 276, 281). In addition, P. gingivalis LPS does not stimulate E-selectin expression in endothelial cell layers and can inhibit E-selectin expression by LPS from other periodontal bacteria (48). Moreover, P. gingivalis LPS appears to be a poor (compared with E. coli LPS) activator of monocyte production of IL-1β and TNF-α (185, 186), cytokines that are indirect activators of selectin expression in humans. Again, P. gingivalis LPS is able to antagonize the stimulatory action of LPS from other bacteria (185). Since E-selectin is required for neutrophil adhesion and subsequent diapedesis, a reduction in expression would tend to decrease the local inflammatory response. Consistent with this stealth-like role for the organism, Reife et al. (211) found that in a mouse model of early inflammation, P. gingivalis LPS, in contrast to E. coli LPS, was a poor inducer of the chemokines monocyte chemoattractant protein 1 (MCP-1) and fibroblast-induced cytokine and of E- and P-selectins.
TABLE 3.
Cytokine induction by P. gingivalis and its cellular constituentsa
| Inducer | Cell(s) of origin | Cytokine | Pro- or anti-inflammatory actionb |
|---|---|---|---|
| P. gingivalis LPS | Fibroblasts | IL-1α | + |
| IL-1β | + | ||
| IL-6 | + | ||
| IL-8 | + | ||
| MCP-1 | + | ||
| Monocytes/macrophages | IL-1β | + | |
| TNF-α | + | ||
| IL-1rac | − | ||
| IL-6 | + | ||
| IL-8 | + | ||
| GM-CSFc | + | ||
| IFN-γ | + | ||
| PMNs | TNFα | + | |
| IL-8 | + | ||
| IL-1ra | − | ||
| P. gingivalis lipid A-associated proteins | Fibroblasts | IL-6 | + |
| P. gingivalis fimbriae | Fibroblasts | IL-1β | + |
| Monocytes/macrophages | IL-1α | + | |
| IL-1β | + | ||
| TNF-α | + | ||
| IL-6 | + | ||
| IL-8 | + | ||
| KCc | + | ||
| Calvarial bone cells | IL-1β | + | |
| GM-CSF | + | ||
| PMNs | IL-6 | + | |
| T cells | IL-2 | + | |
| IL-4 | − | ||
| TNF-α | + | ||
| IFN-γc | + | ||
| P. gingivalis 75-kDa surface protein | Monocytes/macrophages | IL-1 | + |
| P. gingivalis 12-kDa antigen | Monocytes/macrophages | IL-1β | + |
| P. gingivalis PS | Monocytes/macrophages | IL-1β | + |
| P. gingivalis whole cells | PMNs | IL-8 | + |
| MCP-1 | + |
Compiled from references 90, 92, 93, 116, 142, 168, 181, 184, 186, 187, 221, 239, 272, 275, 276, 278, and 281.
+, proinflammatory action; −, anti-inflammatory action.
IL-1ra, IL-1 receptor antagonist; GM-CSF, granulocyte-macrophage colony-stimulating factor; KC, neutrophil chemoaltractant killer cells; IFN-γ, gamma interferon.
Assessment of the role of P. gingivalis in cytokine induction and suppression is complicated by the lack of consistency among the results of in vitro studies. These discrepancies can be partially explained by differences in assay conditions related to factors such as inclusion of human serum (as a source of LPS binding protein [LPB] and soluble CD14), concentration and purity of the test bacterial molecules, source and activation state of the test host cells, and P. gingivalis strain variability (46, 213, 236). Nonetheless, it would appear that a somewhat paradoxical situation exists for P. gingivalis, in that the organism is capable of both activating and suppressing components of the host innate immune response. The question then arises as to real interactive nature of the organism—an overt aggressive invader or a covert stealth raider. Both of these personas may be true, depending on the circumstances. It could be postulated that in the early stages of infection P. gingivalis inhibits the host response to both itself and other bacteria in the periodontal pocket. This would facilitate overgrowth of previously constrained subgingival bacteria, which may contribute to disease (49). Furthermore, a delay in PMN recruitment from the vasculature could cause premature release of lytic enzymes and contribute to tissue destruction (268). Increasing evidence is implicating LPS as a mediator of initial arrest of the host defense. P. gingivalis LPS differs from E. coli LPS by, among other things, being Schwartzman negative; lacking heptose, 2-keto-3-deoxyoctulosonic acid, and 3-hydroxymyristic acid; and exhibiting a unique pattern of fatty acid acylations and monophosphorylation (69, 186). Possibly due to these structural differences, P. gingivalis LPS binds poorly to LPB and consequently is poorly transferred to soluble CD14 (42). Furthermore, although site-specific mutagenesis has revealed that CD14 aa E47 is associated with P. gingivalis LPS binding (237), the specificity of ligand recognition appears to occur downstream of CD14 binding (41). Since CD14 serves as a pattern recognition molecule that orchestrates activation pathways in myeloid and nonmyeloid cells involved in innate host defense, P. gingivalis interactions with a number of aspects of CD14-dependent defense systems may result in tolerance to the presence of the organism. Additionally, penetration of the gingival tissues by released LPS or LPS-containing outer membrane vesicles will allow immune system-modulating “action at a distance.” Outer membrane vesicles will also saturate antibodies before they can reach the bacterial cells. Nonetheless, host PMNs and other defense mechanisms do eventually become mobilized, as evidenced by the pyogenic nature of most periodontal diseases. The overgrowth of subgingival plaque bacteria, or of P. gingivalis itself, that ensues after initial immune suppression may trigger reactivation of the immune response. Alternatively or concomitantly, the encounter with different host cells as the infection progresses may result in a more vigorous immune response.
Even after the host immune response has been mobilized, P. gingivalis is recalcitrant to elimination by the immune system. Proteolytic enzymes have a role to play here, as with most aspects of P. gingivalis infection. Proteinases can degrade complement components, cytokines, antibodies, as well as PMN receptors for antibodies, complement, and the FMLP receptor protein (62, 107, 127, 266). In addition, proteolytic activation of C3 in the serum (by a process that mimics the activity of complement factor D) prevents C3 accumulation on the cell surface (225). Another important factor is the presence of polysaccharide capsular material on most strains of P. gingivalis, in either an amorphous or a fibrous layer (153, 227, 245). This polysaccharide can impart resistance to phagocytosis (45, 257). The composition of the polysaccharide varies among strains but usually contains a high proportion of amino sugars along with glucose and/or galactose (227). The hydrophilic and anionic properties of the polysaccharide contribute to a decrease both in adherence to neutrophils and in deposition of complement components (227). The end products of metabolism may also affect the immune responses. Butyric acid, a volatile fatty acid produced by P. gingivalis, can induce B- and T-cell death through apoptosis (125, 126). Penetration of the periodontal tissue by butyric acid could thus suppress immune system function. Apoptosis of T and B cells has also been recorded in vivo following administration of P. gingivalis LPS (103). Within professional phagocytes, the production of superoxide dismutase (SOD) may protect the organisms from oxygen-dependent killing. P. gingivalis elaborates a single SOD of 21.5 kDa (10, 11). Amino acid and nucleotide sequencing indicate that the P. gingivalis enzyme belongs to the cambialistic class of SODs, showing activity with either Mn2+ or Fe2+ incorporated into the same active site (11, 169). The sod gene is downstream of the prtC (collagenase) gene in P. gingivalis (36), raising the possibility of coordinate regulation of these potential virulence factors. Protection from oxidative damage may also be provided by surface accumulations of hemin, which will form μ-oxo dimers in the presence of reactive oxygen compounds (250).
It is also pertinent that P. gingivalis does not exist as a monoculture or exclusively as planktonic cells in the periodontal pocket. The organism is a constituent of a complex biofilm and, in this context, may be phenotypically distinct from laboratory-grown cultures. The ability of P. gingivalis to adhere to other oral bacteria could also have implications for resistance to phagocytosis, since ingestion of several aggregated bacteria may be less efficient than ingestion of single cells. Another consideration is that P. gingivalis may be sequestered from the immune response by internalizing within epithelial cells, as noted above.
REGULATION OF GENE EXPRESSION
Bacteria in the environment are subject to continually changing conditions. In the subgingival area, bacteria experience dramatic environmental changes as a consequence of host eating and oral hygiene patterns, gingival crevicular flow rate variability, and degree of bleeding. In response to these kinds of dynamic processes, bacteria often regulate gene expression to maintain optimal phenotypic properties. Expression of virulence factors in a wide range of bacteria is tightly regulated in response to environmental cues (155). Although the study of gene regulation in P. gingivalis is in its infancy, it is likely that an organism with such fastidious requirements with regard to oxygen and iron availability will regulate gene expression accordingly.
The temperature in the subgingival region can vary in relation to tooth location and level of inflammation (59, 84–86). In general, subgingival temperatures vary from around 34 to 37°C at healthy sites to around 39°C in inflamed periodontal pockets. By using a fimA promoter-lacZ reporter chromosomal fusion, Xie et al. (277) found that expression of the P. gingivalis fimA gene decreased about 10-fold as the temperature increased from 34 to 39°C. Thus, P. gingivalis appears to downregulate fimbrial expression in response to conditions that develop as disease progresses. It can be hypothesized, therefore, that fimbriae are more important for the organism during the early stages of infection, facilitating adherence and invasion of epithelial cells. Subsequently, fimbrial production is repressed to reduce the expression of undesirable fimbrial properties such as chemotaxis, chemokine induction, and immunogenicity. A reduction in fimbrial expression may not even adversely affect preestablished adherence, since P. gingivalis possesses a variety of nonfimbrial adhesins that may be operational after the initial fimbria-mediated adherence. Elevated temperatures also influence the levels of SOD. In contrast to the situation with fimbriae, an increase in SOD mRNA levels at 39°C has been reported (8). Since host PMN mobilization will be contemporaneous with an increase in subgingival temperature, elevated SOD activity may be beneficial for the organisms in resisting PMN intracellular killing. Whether fimA and sod form part of a regulon remains to be determined. The presence of a GroEL-like stress-related protein in P. gingivalis (149, 271), however, indicates, by analogy to other bacteria, that coordinate control of gene expression in response to heat or other environmental stresses may occur. In E. coli, for example, the induction of stress proteins by heat shock is under the control of two alternate sigma factors, ς32 and ςE (285). In P. gingivalis, the groESL operon itself appears to be regulated at the transcriptional level through a ς32-like consensus sequence and by a CIRCE (controlling inverted repeat of chaperone expression) (100).
Genetic regulation mediated by the iron concentration is well documented in a variety of gram-negative bacteria (143). As described above, acquisition of hemin by P. gingivalis is important for survival; however, little information is available on the role that hemin/iron may play in gene regulation. Certainly, the presence or absence of hemin affects the expression of outer membrane proteins. Furthermore, LPS from P. gingivalis grown under hemin-limited conditions has a reduced capacity for neutrophil priming (32). Growth in hemin-replete media, conversely, results in the expression of a 26-kDa LPS-associated moiety that is associated with hemin binding (44). By using an in vivo mouse model, McKee et al. (154) found that P. gingivalis cells grown under hemin excess were more virulent than organisms grown under hemin limitation. Hemin-limited (relatively avirulent) cells possessed few fimbriae per cell but produced large numbers of extracellular vesicles. In accord with these studies, production of trypsin-like proteinases has been shown to increase upon culture in hemin-rich media (31, 246). More recently, reverse transcription-PCR analysis of mRNA has demonstrated that the steady-state levels of the hemagglutinin genes hagB and hagC decrease under hemin limitation (138). Data such as these have led to the proposal (31, 154) that the arrival of hemin (in the form of erythrocytes) as a result of inflammation converts previously benign P. gingivalis to a virulent phenotype as a result of hemin-controlled coordinate regulation of virulence genes. Not all studies, however, support this attractive hypothesis. Enhanced virulence and expression of virulence factors such as hemolysins and proteinases under conditions of hemin limitation has been reported (27, 72, 267). These results tend to suggest that P. gingivalis is initially more proactive in inducing conditions of hemin excess, which then switch the organism to a less virulent phenotype, possibly avoiding host defense mechanisms. More complete genetic analyses may help clarify the situation. In this respect, Xie et al. (277) investigated the transcriptional activity of the fimA gene and reported that hemin limitation downregulated promoter activity by about 50%, implying that colonization of areas with low hemin levels may be disfavored. Genco et al. (72) reported that a transposon Tn4351 mutant of P. gingivalis that was impaired in hemin uptake had increased hemolytic, hemagglutinating, and trypsin-like protease activities. Additionally, compared with the parent strain, this mutant was more pathogenic in a mouse subcutaneous chamber model. Elucidation of the genetic regulatory system that responds to the hemin concentration and to other environmental cues, such as cell density and serum and saliva components that the organism may be able to sense (277), promises to be an important area for future study. The recent development of an in vivo expression technology vector for P. gingivalis (137) should prove extremely useful in this regard.
SYSTEMIC MANIFESTATIONS OF P. GINGIVALIS INFECTION
At first glance, it seems that infections caused by P. gingivalis are exquisitely site specific. It now appears, however, that periodontal diseases might be associated with an increased incidence of preterm delivery of low-birth-weight infants and with cardiovascular disease (15, 178). Local persistent infections could exert systemic manifestations in a number of ways. For example, the release of antigens or the modulation of systemic cytokine levels could predispose to infection at remote sites. P. gingivalis LPS, which can be released from the cell in membrane vesicles and could impinge upon the cytokine network, is therefore a potential effector molecule. Indeed, intravenous exposure to P. gingivalis LPS in pregnant hamsters produces significant fetal morbidity and mortality (39). The ability of P. gingivalis molecules to induce apoptosis and modulate eucaryotic cell-signaling pathways may also have a bearing on systemic disease. Improved understanding of the interaction between periodontal bacteria and host cells at the molecular and cellular level, may therefore ultimately have relevance to the overall well-being of the host.
ACKNOWLEDGMENTS
We thank Mike Curtis for critical review of the manuscript and Don Demuth, Rich Darveau, Yoonsuk Park, Frank Roberts, Maria Tran, and Hua Xie for helpful discussions.
Work in our laboratories is supported by the National Institute of Dental Research (R.J.L.) and the Health Research Council of New Zealand and British Heart Foundation (H.F.J.).
REFERENCES
- 1.Abrahamson M, Kikstrom M, Potempa J, Renvert S, Hall A. Modification of cystatin C activity by bacterial proteinases and neutrophil elastase in periodontitis. Mol Pathol. 1997;50:291–297. doi: 10.1136/mp.50.6.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aduse-Opoku J, Muir J, Slaney J M, Rangarajan M, Curtis M A. Characterization, genetic analysis, and expression of a protease antigen (PrpRI) of Porphyromonas gingivalisW50. Infect Immun. 1995;63:4744–4754. doi: 10.1128/iai.63.12.4744-4754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aduse-Opoku J, Slaney J M, Rangarajan M, Muir J, Young K A, Curtis M A. The Tla protein of Porphyromonas gingivalisW50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J Bacteriol. 1997;179:4778–4788. doi: 10.1128/jb.179.15.4778-4788.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allaker R P, Aduse-Opoku J, Batten J E, Curtis M A. Natural variation within the principal arginine-specific protease gene, prpR1, of Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:298–302. doi: 10.1111/j.1399-302x.1997.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 5.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Shamra A, Sojar H T, Genco R J, Hamada S, Shizukuishi S. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralisthrough specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 6.Amano A, Katoka K, Raj P A, Genco R J, Shizukuishi S. Binding sites of salivary statherin for Porphyromonas gingivalis. Infect Immun. 1996;64:4249–4254. doi: 10.1128/iai.64.10.4249-4254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amano A, Sharma A, Lee J-Y, Sojar H T, Raj P A, Genco R J. Structural domains of Porphyromonas gingivalisrecombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64:1631–1637. doi: 10.1128/iai.64.5.1631-1637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amano A, Sharma A, Sojar H T, Kuramitsu H K, Genco R J. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–4685. doi: 10.1128/iai.62.10.4682-4685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalisfimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amano A, Shizukuishi S, Tamagawa H, Iwakura K, Tsunasawa S, Tsunemitsu A. Characterization of superoxide dismutases purified from either anaerobically maintained or aerated Bacteroides gingivalis. J Bacteriol. 1990;172:1457–1463. doi: 10.1128/jb.172.3.1457-1463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amano A, Shizukuishi S, Tsunemitsu A, Maekawa K, Tsunasawa S. The primary structure of superoxide dismutase purified from anaerobically maintained Bacteroides gingivalis. FEBS Lett. 1990;272:217–220. doi: 10.1016/0014-5793(90)80488-5. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin T J, Ward W, Aitken A, Knutton S, Williams P H. Elevation of intracellular free calcium levels in HEp-2 cells infected with enteropathogenic Escherichia coli. Infect Immun. 1991;59:1599–1604. doi: 10.1128/iai.59.5.1599-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkocy-Gallagher G A, Han N, Patti J M, Whitlock J, Progulske-Fox A, Lantz M S. Analysis of the prtP gene encoding porphypain, a cysteine proteinase of Porphyromonas gingivalis. J Bacteriol. 1996;178:2734–2741. doi: 10.1128/jb.178.10.2734-2741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barua P K, Dyer D W, Neiders M E. Effect of iron limitation on Bacteroides gingivalis. Oral Microbiol Immunol. 1990;5:263–268. doi: 10.1111/j.1399-302x.1990.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 15.Beck J, Garcia R, Heiss G, Vokonas P S, Offenbacher S. Periodontal disease and cardiovascular disease. J Periodontol. 1996;67:1123–1137. doi: 10.1902/jop.1996.67.10s.1123. [DOI] [PubMed] [Google Scholar]
- 16.Bedi G S, Williams T. Purification and characterization of a collagen-degrading protease from Porphyromonas gingivalis. J Biol Chem. 1994;269:599–606. [PubMed] [Google Scholar]
- 17.Berridge M J. Calcium signalling and cell proliferation. Bioessays. 1995;17:491–500. doi: 10.1002/bies.950170605. [DOI] [PubMed] [Google Scholar]
- 18.Bhogal P S, Slakeski N, Reynolds E C. A cell-associated protein complex of Porphyromonas gingivalisW50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology. 1997;143:2485–2495. doi: 10.1099/00221287-143-7-2485. [DOI] [PubMed] [Google Scholar]
- 19.Botta G A, Eftimiadi C, Costa A, Tonetti M, van Steenbergen T J M, de Graaff J. Influence of volatile fatty acids on human granulocyte chemotaxis. FEMS Microbiol Lett. 1985;27:69–72. [Google Scholar]
- 20.Bourgeau G, Lapointe H, Peloquin P, Mayrand D. Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992;60:3186–3192. doi: 10.1128/iai.60.8.3186-3192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd J, McBride B C. Fractionation of hemagglutinating and bacterial binding adhesins of Bacteroides gingivalis. Infect Immun. 1984;45:403–409. doi: 10.1128/iai.45.2.403-409.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bramanti T E, Holt S C. Iron-regulated outer membrane proteins in the periodontopathic bacterium, Bacteroides gingivalis. Biochem Biophys Res Commun. 1990;166:1146–1154. doi: 10.1016/0006-291x(90)90986-w. [DOI] [PubMed] [Google Scholar]
- 23.Bramanti T E, Holt S C. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalisW50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bramanti T E, Holt S C. Effect of porphyrins and host iron transport proteins on outer membrane protein expression in Porphyromonas (Bacteroides) gingivalis: identification of a novel 26 kDa hemin-repressible surface protein. Microb Pathog. 1992;13:61–73. doi: 10.1016/0882-4010(92)90032-j. [DOI] [PubMed] [Google Scholar]
- 25.Bramanti T E, Holt S C. Localization of a Porphyromonas gingivalis26-kilodalton heat-modifiable, hemin-regulated surface protein which translocates across the outer membrane. J Bacteriol. 1992;174:5827–5839. doi: 10.1128/jb.174.18.5827-5839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bramanti T E, Holt S C, Ebersole J L, Van Dyke T. Regulation of Porphyromonas gingivalisvirulence: hemin limitation effects on the outer membrane protein (OMP) expression and biological activity. J Periodontal Res. 1993;28:464–466. doi: 10.1111/j.1600-0765.1993.tb02103.x. [DOI] [PubMed] [Google Scholar]
- 28.Bramanti T E, Wong G G, Weintraub S T, Holt S C. Chemical characterization and biologic properties of lipopolysaccharide from Bacteroides gingivalisstrains W50, W83, and ATCC 33277. Oral Microbiol Immunol. 1989;4:183–192. doi: 10.1111/j.1399-302x.1989.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 29.Bulkacz J, Erbland J F, MacGregor J. Phospholipase A activity in supernatants from cultures of Bacteroides melaninogenicus. Biochim Biophys Acta. 1981;664:148–155. doi: 10.1016/0005-2760(81)90037-0. [DOI] [PubMed] [Google Scholar]
- 30.Calkins C C, Platt K, Potempa J, Tavis J. Inactivation of tumor necrosis factor α by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 31.Carman R J, Ramakrishnan M D, Harper F H. Hemin levels in culture medium of Porphyromonas (Bacteroides) gingivalisregulate both hemin binding and trypsinlike protease production. Infect Immun. 1990;58:4016–4019. doi: 10.1128/iai.58.12.4016-4019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champagne C M, Holt S C, van Dyke T E, Gordon B J, Shapira L. Lipopolysaccharide isolated from Porphyromonas gingivalisgrown in hemin-limited chemostat conditions has a reduced capacity for human neutrophil priming. Oral Microbiol Immunol. 1996;5:319–325. doi: 10.1111/j.1399-302x.1996.tb00188.x. [DOI] [PubMed] [Google Scholar]
- 33.Chandad F, Mayrand D, Grenier D, Hinode D, Mouton C. Selection and phenotypic characterization of nonhemagglutinating mutants of Porphyromonas gingivalis. Infect Immun. 1996;64:952–958. doi: 10.1128/iai.64.3.952-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Potempa J, Polanowski A, Wikstom M, Travis J. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J Biol Chem. 1992;267:18896–18901. [PubMed] [Google Scholar]
- 35.Childs W C, Gibbons R J. Selective modulation of bacterial attachment to oral epithelial cells by enzyme activities associated with poor oral hygiene. J Periodontol Res. 1990;25:172–178. doi: 10.1111/j.1600-0765.1990.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 36.Choi J-I, Takahashi N, Kato T, Kuramitsu H K. Isolation, expression, and nucleotide sequence of the sod gene from Porphyromonas gingivalis. Infect Immun. 1991;59:1564–1566. doi: 10.1128/iai.59.4.1564-1566.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu L, Bramanti T E, Ebersole J L, Holt S C. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect Immun. 1991;59:1932–1940. doi: 10.1128/iai.59.6.1932-1940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciborowski P, Nishikata M, Allen R D, Lantz M S. Purification and characterization of two forms of a high-molecular-weight cysteine proteinase (porphypain) from Porphyromonas gingivalis. J Bacteriol. 1994;176:4549–4557. doi: 10.1128/jb.176.15.4549-4557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins J G, Smith M A, Arnold R R, Offenbacher S. Effects of Escherichia coli and Porphyromonas gingivalislipopolysaccharide on pregnancy outcome in the golden hamster. Infect Immun. 1994;62:4652–4655. doi: 10.1128/iai.62.10.4652-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 41.Cunningham, M. D., J. Bajorath, J. E. Sommerville, and R. P. Darveau. Escherichia coli and Porphyromonas gingivalis interactions with CD14: implications for myeloid and non-myeloid cell activation. J. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 42.Cunningham M D, Seachord C, Radcliffe K, Bainbridge B, Aruffo A, Darveau R P. Helicobacter pylori and Porphyromonas gingivalislipopolysaccharides are poorly transferred to recombinant soluble CD14. Infect Immun. 1996;64:3601–3608. doi: 10.1128/iai.64.9.3601-3608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis M A, Aduse-Opoku J, Slaney J M, Rangarajan M, Booth V, Cridland J, Shepherd P. Characterization of an adherence and antigenic determinant of the ArgI protease of Porphyromonas gingivaliswhich is present on multiple gene products. Infect Immun. 1996;64:2532–2539. doi: 10.1128/iai.64.7.2532-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutler C W, Eke P I, Genco C A, van Dyke T E, Arnold R R. Hemin-induced modifications of the antigenicity and hemin-binding capacity of Porphyromonas gingivalislipopolysaccharide. Infect Immun. 1996;64:2282–2287. doi: 10.1128/iai.64.6.2282-2287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutler C W, Kalmar J R, Genco C A. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- 46.Darveau, R. P. Personal communication.
- 47.Darveau R P, Belton C M, Reife R A, Lamont R J. Local chemokine paralysis: a novel pathogenic mechanism of Porphyromonas gingivalis. Infect Immun. 1997;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darveau R P, Cunningham M D, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, Page R C, Aruffo A. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun. 1995;63:1311–1317. doi: 10.1128/iai.63.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darveau R P, Tanner A, Page R C. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 50.DeCarlo A A, Harber G J. Hemagglutinin activity and heterogeneity of related Porphyromonas gingivalisproteinases. Oral Microbiol Immunol. 1997;12:47–56. doi: 10.1111/j.1399-302x.1997.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 51.DeCarlo A A, Windsor L J, Bodden M K, Harber G J, Birkedal-Hansen B, Birkedal-Hansen H. Activation and novel processing of matrix metalloproteinases by thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J Dent Res. 1997;76:1260–1270. doi: 10.1177/00220345970760060501. [DOI] [PubMed] [Google Scholar]
- 52.Deslauriers M, Haque S, Flood P M. Identification of murine protective epitopes on the Porphyromonas gingivalisfimbrillin molecule. Infect Immun. 1996;64:434–440. doi: 10.1128/iai.64.2.434-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickinson D P, Kubiniec M A, Yoshimura F, Genco R J. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–1665. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du L, Pellen-Mussi P, Chandad F, Mouton C, Bonnaure-Mallet M. Fimbriae and the hemagglutinating adhesin HA-Ag2 mediate adhesion of Porphyromonas gingivalisto epithelial cells. Infect Immun. 1997;65:3875–3881. doi: 10.1128/iai.65.9.3875-3881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duncan M J, Emory S A, Almira E C. Porphyromonas gingivalisgenes isolated by screening for epithelial cell attachment. Infect Immun. 1996;64:3624–3631. doi: 10.1128/iai.64.9.3624-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivaliswith epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eke P I, Rotimi V O, Laughon B E. Coaggregation of black-pigmented Bacteroideswith other oral bacteria. J Med Microbiol. 1989;28:1–4. doi: 10.1099/00222615-28-1-1. [DOI] [PubMed] [Google Scholar]
- 58.Evans R T, Klausen B, Sojar H T, Bedi G S, Sfintescu C, Ramamurthy N S, Golub L M, Genco R J. Immunization with Porphyromonas (Bacteroides) gingivalisfimbriae protects against periodontal destruction. Infect Immun. 1992;60:2926–2935. doi: 10.1128/iai.60.7.2926-2935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedi P F, Killoy W J. Temperature differences at periodontal sites in health and disease. J Periodontol. 1992;63:24–27. doi: 10.1902/jop.1992.63.1.24. [DOI] [PubMed] [Google Scholar]
- 60.Fletcher H M, Schenkein H A, Macrina F L. Cloning and characterization of a new protease gene (prtH) from Porphyromonas gingivalis. Infect Immun. 1994;62:4279–4286. doi: 10.1128/iai.62.10.4279-4286.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtHgene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fletcher J, Nair S, Poole S, Henderson B, Wilson M. Cytokine degradation by biofilms of Porphyromonas gingivalis. Curr Microbiol. 1998;36:216–219. doi: 10.1007/s002849900297. [DOI] [PubMed] [Google Scholar]
- 63.Force T, Pombo C M, Avruch J A, Boventre J V, Kyriakis J M. Stress-activated protein kinases in cardiovascular disease. Circ Res. 1996;78:947–953. doi: 10.1161/01.res.78.6.947. [DOI] [PubMed] [Google Scholar]
- 64.Frank R M. Bacterial penetration in the apical pocket wall of advanced human periodontitis. J Periodontal Res. 1980;15:563–573. doi: 10.1111/j.1600-0765.1980.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 65.Frank R M, Voegel J C. Bacterial bone resorption in advanced cases of human periodontitis. J Periodontal Res. 1978;13:251–261. doi: 10.1111/j.1600-0765.1978.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 66.Fravalo P, Menard C, Bonnaure-Mallet M. Effect of Porphyromonas gingivalison epithelial cell MMP-9 type IV collagenase production. Infect Immun. 1996;64:4940–4945. doi: 10.1128/iai.64.12.4940-4945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujimura S, Shibata Y, Hirai K, Nakamura T. Some binding properties of the envelope of Porphyromonas gingivalisto hemoglobin. FEMS Immunol Med Microbiol. 1995;10:109–114. doi: 10.1111/j.1574-695X.1995.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 68.Fujimura S, Shibata Y, Hirai K, Nakamura T. Binding of hemoglobin to the envelope of Porphyromonas gingivalisand isolation of the hemoglobin-binding protein. Infect Immun. 1996;64:2339–2342. doi: 10.1128/iai.64.6.2339-2342.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujiwara T, Ogawa T, Sobue S, Hamada S. Chemical, immunobiological and antigenic characterizations of lipopolysaccharides from Bacteroides gingivalisstrains. J Gen Microbiol. 1990;136:319–326. doi: 10.1099/00221287-136-2-319. [DOI] [PubMed] [Google Scholar]
- 70.Genco C A. Regulation of hemin and iron transport in of Porphyromonas gingivalis. Adv Dent Res. 1995;9:41–47. doi: 10.1177/08959374950090010801. [DOI] [PubMed] [Google Scholar]
- 71.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalisare induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Genco C A, Simpson W, Forng R Y, Egal M, Odusanya B M. Characterization of a Tn4351-generated hemin uptake mutant of Porphyromonas gingivalis: evidence for coordinate regulation of virulence factors by hemin. Infect Immun. 1995;63:2459–2466. doi: 10.1128/iai.63.7.2459-2466.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gharbia, S. E., and H. N. Shah. 1995. Molecular analysis of surface-associated enzymes of Porphyromonas gingivalis. Clin. Infect. Dis. 20(Suppl. 2):S160–S166. [DOI] [PubMed]
- 74.Gibbons R J, Hay D I, Childs W C, Davis G. Role of cryptic receptors (cryptitopes) in bacterial adhesion to oral surfaces. Arch Oral Biol. 1990;35:107–114. doi: 10.1016/0003-9969(90)90139-2. [DOI] [PubMed] [Google Scholar]
- 75.Goodson J M, McClatchy K, Revell C. Prostaglandin induced bone resorption of the adult rat calvarium. J Dent Res. 1974;53:670–677. [Google Scholar]
- 76.Goulbourne P A, Ellen R P. Evidence that Porphyromanas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. 1991;173:5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greenstein G, Lamster I. Bacterial transmission in periodontal diseases: a critical review. J Periodontol. 1997;68:421–431. doi: 10.1902/jop.1997.68.5.421. [DOI] [PubMed] [Google Scholar]
- 78.Grenier D. Hemin-binding property of Porphyromonas gingivalisouter membranes. FEMS Microbiol Lett. 1991;77:45–50. doi: 10.1016/0378-1097(91)90011-x. [DOI] [PubMed] [Google Scholar]
- 79.Grenier D. Demonstration of a bimodal coaggregation reaction between Porphyromonas gingivalis and Treponema denticola. Oral Microbiol Immunol. 1992;7:280–284. doi: 10.1111/j.1399-302x.1992.tb00589.x. [DOI] [PubMed] [Google Scholar]
- 80.Grenier D. Further evidence for a possible role of trypsin-like activity in the adherence of Porphyromonas gingivalis. Can J Microbiol. 1992;38:1189–1192. doi: 10.1139/m92-195. [DOI] [PubMed] [Google Scholar]
- 81.Grenier D, McBride B C. Isolation of a membrane-associated Bacteroides gingivalisglycylprolyl protease. Infect Immun. 1987;55:3131–3136. doi: 10.1128/iai.55.12.3131-3136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gron H, Pike R, Potempa J, Travis J, Thogersen I B, Enghild J J, Pizzo S V. The potential role of α2-macroglobulin in the control of cysteine proteinases (gingipains) from Porphyromonas gingivalis. J Periodontal Res. 1997;32:61–68. doi: 10.1111/j.1600-0765.1997.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 83.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 84.Haffajee A D, Socransky S S, Goodson J M. Subgingival temperature. I. Relation to baseline clinical parameters. J Clin Periodontol. 1992;19:401–408. doi: 10.1111/j.1600-051x.1992.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 85.Haffajee A D, Socransky S S, Goodson J M. Subgingival temperature. II. Relation to future periodontal attachment loss. J Clin Periodontol. 1992;19:409–416. doi: 10.1111/j.1600-051x.1992.tb00671.x. [DOI] [PubMed] [Google Scholar]
- 86.Haffajee A D, Socransky S S, Dibart S S, Goodson J M. Subgingival temperature. III. Relation to microbial counts. J Clin Periodontol. 1992;19:417–422. doi: 10.1111/j.1600-051x.1992.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 87.Hamada N, Sojar H T, Cho M-I, Genco R J. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect Immun. 1996;64:4788–4794. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hamada N, Watanabe K, Sasakawa C, Yoshikawa M, Yoshimura F, Umemoto T. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect Immun. 1994;62:1696–1704. doi: 10.1128/iai.62.5.1696-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han N, Whitlock J, Progulske-Fox A. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis381 contains four large, contiguous, direct repeats. Infect Immun. 1996;64:4000–4007. doi: 10.1128/iai.64.10.4000-4007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanazawa S, Hirose K, Ohmori Y, Amano S, Kitano S. Bacteroides gingivalisfimbriae stimulate production of thymocyte-activating factor by human gingival fibroblasts. Infect Immun. 1988;56:272–274. doi: 10.1128/iai.56.1.272-274.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanazawa S, Kawata Y, Murakami Y, Naganuma K, Amano S, Miyata Y, Kitano S. Porphyromonas gingivalisfimbria-stimulated bone resorption in vitro is inhibited by a tyrosine kinase inhibitor. Infect Immun. 1995;63:2374–2377. doi: 10.1128/iai.63.6.2374-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanazawa S, Murakami Y, Hirose K, Amano S, Ohmori Y, Higuchi H, Kitano S. Bacteroides (Porphyromonas) gingivalisfimbriae activate mouse peritoneal macrophages and induce gene expression and production of interleukin-1. Infect Immun. 1991;59:1972–1977. doi: 10.1128/iai.59.6.1972-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanazawa S, Murakami Y, Takeshita A, Kitami H, Ohta K, Amano S, Kitano S. Porphyromonas gingivalisfimbriae induce expression of the neutrophil chemotactic factor KC gene of mouse peritoneal macrophages: role of protein kinase C. Infect Immun. 1992;60:1544–1549. doi: 10.1128/iai.60.4.1544-1549.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Havemose-Poulsen A, Holmstrup P. Factors affecting IL-1-mediated collagen metabolism by fibroblasts and the pathogenesis of periodontal disease: a review of the literature. Crit Rev Oral Biol Med. 1997;8:217–236. doi: 10.1177/10454411970080020801. [DOI] [PubMed] [Google Scholar]
- 95.Hayashi H, Nagata A, Hinode D, Sato M, Nakamura R. Survey of a receptor protein in human erythrocytes for hemagglutinin of Porphyromonas gingivalis. Oral Microbiol Immunol. 1992;7:204–211. doi: 10.1111/j.1399-302x.1992.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 96.Hinode D, Hayashi H, Nakamura R. Purification and characterization of three types of proteases from culture supernatants of Porphyromonas gingivalis. Infect Immun. 1991;59:3060–3068. doi: 10.1128/iai.59.9.3060-3068.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hirose K, Isogai E, Mizugai H, Ueda I. Adhesion of Porphyromonas gingivalisfimbriae to human gingival cell line Ca9-22. Oral Microbiol Immunol. 1996;11:402–406. doi: 10.1111/j.1399-302x.1996.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 98.Holt S C, Bramanti T E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 99.Hoover C I, Ng C Y, Felton J R. Correlation of haemagglutination activity with trypsin-like protease activity of Porphyromonas gingivalis. Arch Oral Biol. 1992;7:515–520. doi: 10.1016/0003-9969(92)90133-s. [DOI] [PubMed] [Google Scholar]
- 100.Hotokezaka H, Ohara N, Hayashida H, Matsumoto S, Matsuo T, Naito M, Kobayashi K, Yamada T. Transcriptional analysis of the groESL operon from Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:236–239. doi: 10.1111/j.1399-302x.1997.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 101.Inoshita E, Amano A, Hanioka T, Tamagawa H, Shizukuishi S, Tsunemitsu A. Isolation and some properties of exohemagglutinin from the culture medium of Bacteroides gingivalis381. Infect Immun. 1986;52:421–427. doi: 10.1128/iai.52.2.421-427.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Inoshita E, Iwakura K, Amano A, Tamagawa H, Shizukuishi S. Effect of transferrin on the growth of Porphyromonas gingivalis. J Dent Res. 1991;70:1258–1261. doi: 10.1177/00220345910700090501. [DOI] [PubMed] [Google Scholar]
- 103.Isogai E, Isogai H, Kimura K, Fujii N, Takagi S, Hirose K, Hayashi M. In vivo induction of apoptosis and immune responses in mice by administration of lipopolysaccharide from Porphyromonas gingivalis. Infect Immun. 1996;64:1461–1466. doi: 10.1128/iai.64.4.1461-1466.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Isogai E, Isogai H, Takagi S, Ishii N, Fujii N, Kimura K, Hayashi M, Yoshimura F. Fimbriae-specific immune response in various inbred mice inoculated with Porphyromonas gingivalis381. Oral Microbiol Immunol. 1994;9:118–122. doi: 10.1111/j.1399-302x.1994.tb00045.x. [DOI] [PubMed] [Google Scholar]
- 105.Isogai H, Isogai E, Yoshimura F, Suzuki T, Kagota W, Takano K. Specific inhibition of adherence of an oral strain of Bacteroides gingivalis381 to epithelial cells by monoclonal antibodies against the bacterial fimbriae. Arch Oral Biol. 1988;33:479–485. doi: 10.1016/0003-9969(88)90028-3. [DOI] [PubMed] [Google Scholar]
- 106.Izutsu K T, Belton C M, Chan A, Fatherazi S, Kanter J P, Park Y, Lamont R J. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;144:145–150. doi: 10.1111/j.1574-6968.1996.tb08521.x. [DOI] [PubMed] [Google Scholar]
- 107.Jagels M A, Travis J, Potempa J, Pike R, Hugli T E. Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect Immun. 1996;64:1984–1991. doi: 10.1128/iai.64.6.1984-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jenkinson H F, Demuth D R. Structure, function, and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 109.Jenkinson H F, Lamont R J. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 110.Kadowaki T, Yoneda M, Okamoto K, Maeda K, Yamamoto K. Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J Biol Chem. 1994;269:21371–21378. [PubMed] [Google Scholar]
- 111.Kagnoff M F, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:S51–S55. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karunakaran T, Holt S C. Cloning of two distinct hemolysin genes from Porphyromonas (Bacteroides) gingivalis in Escherichia coli. Microb Pathog. 1993;15:37–49. doi: 10.1006/mpat.1993.1055. [DOI] [PubMed] [Google Scholar]
- 113.Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kataoka K, Amano A, Kuboniwa M, Horie H, Nagata H, Shizukuishi S. Active sites of salivary proline-rich protein for binding to Porphyromonas gingivalisfimbriae. Infect Immun. 1997;65:3159–3164. doi: 10.1128/iai.65.8.3159-3164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kato T, Takahashi N, Kuramitsu H K. Sequence analysis and characterization of the Porphyromonas gingivalis prtCgene, which expresses a novel collagenase activity. J Bacteriol. 1992;174:3889–3895. doi: 10.1128/jb.174.12.3889-3895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kawata Y, Hanazawa S, Amano S, Murakami Y, Matsumoto T, Nishida K, Kitano S. Porphyromonas gingivalisfimbriae stimulate bone resorption in vitro. Infect Immun. 1994;62:3012–3016. doi: 10.1128/iai.62.7.3012-3016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawata Y, Iwasaka H, Kitano S, Hanazawa S. Porphyromonas gingivalisfimbria-stimulated bone resorption is inhibited through binding of the fimbriae to fibronectin. Infect Immun. 1997;65:815–817. doi: 10.1128/iai.65.2.815-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kelly C G, Booth V, Kendal H, Slaney J M, Curtis M A, Lehner T. The relationship between colonization and haemagglutination inhibiting and B cell epitopes of Porphyromonas gingivalis. Clin Exp Immunol. 1997;110:285–291. doi: 10.1111/j.1365-2249.1997.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim S-J, Chu L, Holt S C. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 120.Kinder S A, Holt S C. Characterization of coaggregation between Bacteroides gingivalis T22 and Fusobacterium nucleatumT18. Infect Immun. 1989;57:3425–3433. doi: 10.1128/iai.57.11.3425-3433.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kirszbaum L, Sotiropoulos C, Jackson C, Cleal S, Slakeski N, Reynolds E C. Complete nucleotide sequence of a gene prtR of Porphyromonas gingivalisW50 encoding a 132 kDa protein that contains an arginine-specific thiol endopeptidase domain and a haemagglutinin domain. Biochem Biophys Res Commun. 1995;207:424–431. doi: 10.1006/bbrc.1995.1205. [DOI] [PubMed] [Google Scholar]
- 122.Kolenbrander P E, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalisto matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 124.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kurita-Ochiai T, Fukushima K, Ochiai K. Butyric acid-induced apoptosis of murine thymocytes, splenic T cells, and human Jurkat T cells. Infect Immun. 1997;65:35–41. doi: 10.1128/iai.65.1.35-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kurita-Ochiai T, Ochiai K, Fukushima K. Volatile fatty acid, metabolic by-product of periodontopathic bacteria, induces apoptosis in WEHI 231 and RAJI B lymphoma cells and splenic B cells. Infect Immun. 1998;66:2587–2594. doi: 10.1128/iai.66.6.2587-2594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lala A, Amano A, Sojar H T, Radel S J, Denardin E. Porphyromonas gingivalistrypsin-like protease: a possible natural ligand for the neutrophil formyl receptor. Biochem Biophys Res Commun. 1994;199:1489–1496. doi: 10.1006/bbrc.1994.1399. [DOI] [PubMed] [Google Scholar]
- 128.Lamont R J, Bevan C A, Gil S, Persson R E, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 129.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalisinvasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lamont R J, Gil S, Demuth D R, Malamud D, Rosan B. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology. 1994;140:867–872. doi: 10.1099/00221287-140-4-867. [DOI] [PubMed] [Google Scholar]
- 131.Lamont R J, Hersey S G, Rosan B. Characterization of the adherence of Porphyromonas (Bacteroides) gingivalisto oral streptococci. Oral Microbiol Immunol. 1992;7:193–197. doi: 10.1111/j.1399-302x.1992.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 132.Lamont R J, Hsiao G W, Gil S. Identification of a molecule of Porphyromonas gingivalis that binds to Streptococcus gordonii. Microb Pathog. 1994;17:355–360. doi: 10.1006/mpat.1994.1081. [DOI] [PubMed] [Google Scholar]
- 133.Lamont R J, Oda D, Persson R E, Persson G R. Interaction of Porphyromonas gingivaliswith gingival epithelial cells maintained in culture. Oral Microbiol Immunol. 1992;7:364–367. doi: 10.1111/j.1399-302x.1992.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 134.Lantz M S, Allen R D, Duck L W, Blume J L, Switalski L M, Hook M. Identification of Porphyromonas gingivaliscomponents that mediate its interactions with fibronectin. J Bacteriol. 1991;173:4263–4270. doi: 10.1128/jb.173.14.4263-4270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee J Y, Sojar H T, Bedi G S, Genco R J. Porphyromonas (Bacteroides) gingivalisfimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun. 1991;59:383–389. doi: 10.1128/iai.59.1.383-389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee J Y, Sojar H T, Bedi G S, Genco R J. Synthetic peptides analogous to the fimbrillin sequence inhibit adherence of Porphyromonas gingivalis. Infect Immun. 1992;60:1662–1670. doi: 10.1128/iai.60.4.1662-1670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee S-W, Hillman J D, Progulske-Fox A. The hemagglutinin genes hagB and hagC of Porphyromonas gingivalisare transcribed in vivo as shown by use of a new expression vector. Infect Immun. 1996;64:4802–4810. doi: 10.1128/iai.64.11.4802-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lepine G, Progulske-Fox A. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:65–78. doi: 10.1111/j.1399-302x.1996.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 139.Lepine G, Ellen R P, Progulske-Fox A. Construction and preliminary characterization of three hemagglutinin mutants of Porphyromonas gingivalis. Infect Immun. 1996;64:1467–1472. doi: 10.1128/iai.64.4.1467-1472.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lewis J P, Macrina F L. IS195, an insertion sequence-like element associated with protease genes in Porphyromonas gingivalis. Infect Immun. 1998;66:3035–3042. doi: 10.1128/iai.66.7.3035-3042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li J, Ellen R P, Hoover C J, Felton J R. Association of proteases of Porphyromonas (Bacteroides) gingivalis with its adhesion to Actinomyces viscosus. J Dent Res. 1991;70:82–86. doi: 10.1177/00220345910700021501. [DOI] [PubMed] [Google Scholar]
- 142.Lindemann R A, Economou J S, Rothermel H. Production of interleukin-1 and tumor necrosis factor by human peripheral monocytes activated by periodontal bacteria and extracted lipopolysaccharides. J Dent Res. 1988;67:1131–1135. doi: 10.1177/00220345880670081401. [DOI] [PubMed] [Google Scholar]
- 143.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Loomer P M, Sigusch B, Sukhu B, Ellen R P, Tenenbaum H C. Direct effects of metabolic products and sonicated extracts of Porphyromonas gingivalis2561 on osteogenesis in vitro. Infect Immun. 1994;62:1289–1297. doi: 10.1128/iai.62.4.1289-1297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Loos B G, Dyer D W, Whittam T S, Selander R K. Genetic structure of populations of Porphyromonas gingivalisassociated with periodontitis and other oral infections. Infect Immun. 1993;61:204–212. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Loos B G, Mayrand D, Genco R J, Dickinson D P. Genetic heterogeneity of Porphyromonas (Bacteroides) gingivalisby genomic fingerprinting. J Dent Res. 1990;69:1488–1493. doi: 10.1177/00220345900690080801. [DOI] [PubMed] [Google Scholar]
- 147.Madden T E, Clark V L, Kuramitsu H K. Revised sequence of the Porphyromonas gingivalisPrtT cysteine protease/hemagglutinin gene: homology with streptococcal pyrogenic exotoxin B/streptococcal proteinase. Infect Immun. 1995;63:238–247. doi: 10.1128/iai.63.1.238-247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Madianos P N, Papapanou P N, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalisFDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maeda H, Miyamoto M, Hongyo H, Nagai A, Kurihara H, Muryama Y. Heat shock protein 60 (GroEL) from Porphyromonas gingivalis: molecular cloning and sequence analysis of its gene and purification of the recombinant protein. FEMS Microbiol Lett. 1994;124:121–122. doi: 10.1111/j.1574-6968.1994.tb07271.x. [DOI] [PubMed] [Google Scholar]
- 150.Malek R, Fisher J G, Caleca A, Stinson M, van Oss C J, Lee J Y, Cho M I, Genco R J, Evans R T, Dyer D W. Inactivation of Porphyromonas gingivalis fimAgene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Maniotis A J, Bojanowski K, Ingber D E. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J Cell Biochem. 1997;65:114–130. [PubMed] [Google Scholar]
- 152.Maniotis A J, Chen C S, Ingber D E. Demonstration of mechanical connections between integrins cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mayrand D, Holt S C. Biology of asaccharolytic black-pigmented Bacteroidesspecies. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McKee A S, McDermid A S, Baskerville A, Dowsett B, Ellwood D C, Marsh P D. Effect of hemin on the physiology and virulence of Bacteroides gingivalisW50. Infect Immun. 1986;52:349–455. doi: 10.1128/iai.52.2.349-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalisassessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mihara J, Yoneda T, Holt S C. Role of Porphyromonas gingivalis-derived fibroblast-activating factor in bone resorption. Infect Immun. 1993;61:3562–3564. doi: 10.1128/iai.61.8.3562-3564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mikolajczyk-Pawlinska J, Kordula T, Pavloff N, Pemberton P A, Chen W C, Travis J, Potempa J. Genetic variation of Porphyromonas gingivalisgenes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem. 1998;379:205–211. doi: 10.1515/bchm.1998.379.2.205. [DOI] [PubMed] [Google Scholar]
- 159.Mouton C, Ni Eidhin D, Deslauriers M, Lamy L. The hemagglutinating adhesin HA-Ag2 of Bacteroides gingivalisis distinct from fimbrillin. Oral Microbiol Immunol. 1991;6:6–11. doi: 10.1111/j.1399-302x.1991.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 160.Murakami Y, Hanazawa S, Watanabe A, Naganuma K, Iwasaka H, Kawakami K, Kitano S. Porphyromonas gingivalisfimbriae induce a 68-kilodalton phosphorylated protein in macrophages. Infect Immun. 1994;62:5242–5246. doi: 10.1128/iai.62.12.5242-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Murakami Y, Hanazawa S, Nishida K, Iwasaka H, Kitano S. N-Acetyl-d-galactosamine inhibits TNFα gene expression induced in mouse peritoneal macrophages by fimbriae of Porphyromonas (Bacteroides) gingivalis, an oral anaerobe. Biochem Biophys Res Commun. 1993;192:826–832. doi: 10.1006/bbrc.1993.1489. [DOI] [PubMed] [Google Scholar]
- 162.Murakami Y, Iwahashi H, Tasuda H, Umemoto T, Namikawa I, Kitano S, Hanazawa S. Porphyromonas gingivalisfimbrillin is one of the fibronectin-binding proteins. Infect Immun. 1996;64:2571–2576. doi: 10.1128/iai.64.7.2571-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Murakami Y, Nagata H, Amano A, Takagaki M, Shizukuishi S, Tsunemitsu A, Aimoto S. Inhibitory effects of human salivary histatins and lysozyme on coaggregation between Porphyromonas gingivalis and Streptococcus mitis. Infect Immun. 1991;59:3284–3286. doi: 10.1128/iai.59.9.3284-3286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Murakami Y, Shizukuishi S, Tsunemitsu A, Nakashima K, Kato Y, Aimoto S. Binding of a histidine-rich peptide to Porphyromonas gingivalis. FEMS Microbiol Lett. 1991;82:253–256. doi: 10.1016/0378-1097(91)90269-g. [DOI] [PubMed] [Google Scholar]
- 165.Murakami Y, Tamagawa H, Shizukuishi S, Tsunemitsu A, Aimoto S. Biological role of an arginine residue present in a histidine-rich peptide which inhibits hemagglutination of Porphyromonas gingivalis. FEMS Microbiol Lett. 1992;98:201–204. doi: 10.1016/0378-1097(92)90156-i. [DOI] [PubMed] [Google Scholar]
- 166.Nagata H, Murakami Y, Inoshita E, Shizukuishi S, Tsunemitsu A. Inhibitory effect of human plasma and saliva on co-aggregation between Bacteroides gingivalis and Streptococcus mitis. J Dent Res. 1990;69:1476–1479. doi: 10.1177/00220345900690080501. [DOI] [PubMed] [Google Scholar]
- 167.Nagata H, Sharma A, Sojar H T, Amano A, Levine M J, Genco R J. Role of the carboxyl-terminal region of Porphyromonas gingivalisfimbrillin in binding to salivary proteins. Infect Immun. 1997;65:422–427. doi: 10.1128/iai.65.2.422-427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Naito Y, Ohno J, Takazoe I, Okuda K. The relationship between polysaccharide antigen and interleukin-1β producing activity in Porphyromonas gingivalis. Bull Tokyo Dent Coll. 1992;33:187–195. [PubMed] [Google Scholar]
- 169.Nakayama K. The superoxide dismutase-encoding gene of the obligately anaerobic bacterium Bacteroides gingivalis. Gene. 1990;96:149–150. doi: 10.1016/0378-1119(90)90357-w. [DOI] [PubMed] [Google Scholar]
- 170.Nakayama K, Kadowaki T, Okamoto K, Yamamoto K. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J Biol Chem. 1995;270:23619–23626. doi: 10.1074/jbc.270.40.23619. [DOI] [PubMed] [Google Scholar]
- 171.Nakayama K, Ratnayake D B, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 172.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nishikata M, Yoshimura F. Characterization of Porphyromonas (Bacteroides) gingivalishemagglutinin as a protease. Biochem Biophys Res Commun. 1991;178:336–342. doi: 10.1016/0006-291x(91)91819-x. [DOI] [PubMed] [Google Scholar]
- 174.Nishikata M, Yoshimura F. Active site structure of a hemagglutinating protease from Porphyromonas gingivalis: similarity to clostripain. Biochem Mol Biol Int. 1995;37:547–553. [PubMed] [Google Scholar]
- 175.Njoroge T, Genco R J, Sojar H T, Genco C A. A role for fimbriae in Porphyromonas gingivalisinvasion of oral epithelial cells. Infect Immun. 1997;65:1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Novak M J, Cohen H J. Depolarization of polymorphonuclear leukocytes by Porphyromonas (Bacteroides) gingivalis381 in the absence of respiratory burst activation. Infect Immun. 1991;59:3134–3142. doi: 10.1128/iai.59.9.3134-3142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oelschlaeger T A, Guerry P, Kopecko D J. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc Natl Acad Sci USA. 1993;90:6884–6888. doi: 10.1073/pnas.90.14.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Offenbacher S, Katz V, Fertik G, Collins J, Boyd D, Maynor G, McKaig R, Beck J. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol. 1996;67:1103–1113. doi: 10.1902/jop.1996.67.10s.1103. [DOI] [PubMed] [Google Scholar]
- 179.Ogawa T. The potential protective immune response to synthetic peptides containing conserved epitopes of Porphyromonas gingivalisfimbrial protein. J Med Microbiol. 1994;41:349–358. doi: 10.1099/00222615-41-5-349. [DOI] [PubMed] [Google Scholar]
- 180.Ogawa T, Hamada S. Hemagglutinating and chemotactic properties of synthetic peptide segments of fimbrial protein from Porphyromonas gingivalis. Infect Immun. 1994;62:3305–3310. doi: 10.1128/iai.62.8.3305-3310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ogawa T, Kusumoto Y, Uchida H, Nagashima S, Ogo H, Hamada S. Immunobiological activities of synthetic peptide segments of fimbrial protein from Porphyromonas gingivalis. Biochem Biophys Res Commun. 1991;180:1335–1341. doi: 10.1016/s0006-291x(05)81342-7. [DOI] [PubMed] [Google Scholar]
- 182.Ogawa T, Ogo H, Hamada S. Chemotaxis of human monocytes by synthetic peptides that mimic segments of Porphyromonas gingivalisfimbrial protein. Oral Microbiol Immunol. 1994;9:257–261. doi: 10.1111/j.1399-302x.1994.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 183.Ogawa T, Ogo H, Uchida H, Hamada S. Humoral and cellular immune responses to the fimbriae of Porphyromonas gingivalisand their synthetic peptides. J Med Microbiol. 1994;40:397–402. doi: 10.1099/00222615-40-6-397. [DOI] [PubMed] [Google Scholar]
- 184.Ogawa T, Uchida H. A peptide, ALTTE, within the fimbrial subunit protein from Porphyromonas gingivalis, induces production of interleukin 6, gene expression and protein phosphorylation in human peripheral blood mononuclear cells. FEMS Immunol Med Microbiol. 1995;11:197–206. doi: 10.1111/j.1574-695X.1995.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 185.Ogawa T, Uchida H. Differential induction of IL-1β and IL-6 production by the nontoxic lipid A from Porphyromonas gingivalis in comparison with synthetic Escherichia colilipid A in human peripheral blood mononuclear cells. FEMS Immunol Med Microbiol. 1996;14:1–13. doi: 10.1111/j.1574-695X.1996.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 186.Ogawa T, Uchida H, Amino K. Immunobiological activities of chemically defined lipid A from lipopolysaccharides of Porphyromonas gingivalis. Microbiology. 1994;140:1209–1216. doi: 10.1099/13500872-140-5-1209. [DOI] [PubMed] [Google Scholar]
- 187.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalisfimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–242. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 188.Ogawa T, Yasuda K, Yamada K, Mori H, Ochiai K, Hasegawa M. Immunochemical characterization and epitope mapping of a novel fimbrial protein (Pg-II fimbria) of Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 1995;11:247–256. doi: 10.1111/j.1574-695X.1995.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 189.Okamoto K, Kadowaki T, Nakayama K, Yamamoto K. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain) J Biochem (Tokyo) 1996;120:398–406. doi: 10.1093/oxfordjournals.jbchem.a021426. [DOI] [PubMed] [Google Scholar]
- 190.Okamoto K, Misumi Y, Kadowaki T, Yoneda M, Yamamoto K, Ikehara Y. Structural characterization of argingipain, a novel arginine-specific cysteine proteinase as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch Biochem Biophys. 1995;316:917–925. doi: 10.1006/abbi.1995.1123. [DOI] [PubMed] [Google Scholar]
- 191.Okuda K, Slots J, Genco R J. Bacteroides gingivalis, Bacteroides asaccharolyticus, and Bacteroides melanogenicussub-species: cell surface morphology and adherence to erythrocytes and human buccal epithelial cells. Curr Microbiol. 1981;6:7–12. [Google Scholar]
- 192.Okuda K, Yamamoto A, Naito Y, Takazoe I, Slots J, Genco R J. Purification and properties of hemagglutinin from culture supernatant of Bacteroides gingivalis. Infect Immun. 1986;54:659–665. doi: 10.1128/iai.54.3.659-665.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Onoe T, Hoover C I, Nakayama K, Ideka T, Nakamura H, Yoshimura F. Identification of Porphyromonas gingivalisprefimbrilin possessing a long leader peptide: possible involvement of trypsin-like protease in fimbrilin maturation. Microb Pathog. 1995;19:351–364. doi: 10.1016/s0882-4010(96)80006-4. [DOI] [PubMed] [Google Scholar]
- 194.Otogoto J-I, Kuramitsu H K. Isolation and characterization of the Porphyromonas gingivalis prtTgene, coding for protease activity. Infect Immun. 1993;61:117–123. doi: 10.1128/iai.61.1.117-123.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pace J, Hayman M J, Galan J E. Signal transduction and invasion of epithelial cells by S. typhimurium. Cell. 1993;72:505–514. doi: 10.1016/0092-8674(93)90070-7. [DOI] [PubMed] [Google Scholar]
- 196.Park Y, Lamont R J. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Park Y, Lu B, Mazur C, McBride B C. Inducible expression of a Porphyromonas gingivalisW83 membrane-associated protease. Infect Immun. 1997;65:1101–1104. doi: 10.1128/iai.65.3.1101-1104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Park Y, McBride B C. Cloning of a Porphyromonas (Bacteroides) gingivalisprotease gene and characterization of its product. FEMS Microbiol Lett. 1992;71:273–278. doi: 10.1016/0378-1097(92)90721-y. [DOI] [PubMed] [Google Scholar]
- 199.Park Y, McBride B C. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalisW83. Infect Immun. 1993;61:4139–4146. doi: 10.1128/iai.61.10.4139-4146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pavloff N, Pemberton P A, Potempa J, Chen W C, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and characterization of Porphyromonas gingivalislysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem. 1997;272:1595–1600. doi: 10.1074/jbc.272.3.1595. [DOI] [PubMed] [Google Scholar]
- 201.Pavloff N, Potempa J, Pike R N, Prochazka V, Kiefer M C, Travis J, Barr P J. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase-adhesin polyprotein. J Biol Chem. 1995;270:1007–1010. doi: 10.1074/jbc.270.3.1007. [DOI] [PubMed] [Google Scholar]
- 202.Pekovic D D, Fillery E D. Identification of bacteria in immunopathological mechanisms of human periodontal diseases. J Periodontal Res. 1984;19:329–351. doi: 10.1111/j.1600-0765.1984.tb01007.x. [DOI] [PubMed] [Google Scholar]
- 203.Petit M D A, van Steenbergen T J M, Timmerman M F, de Graaff J, van der Velden U. Prevalence of periodontitis and suspected periodontal pathogens in families of adult periodontitis patients. J Clin Periodontol. 1994;21:76–85. doi: 10.1111/j.1600-051x.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 204.Pike R N, Potempa J, McGraw W, Coetzer T H T, Travis J. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J Bacteriol. 1996;178:2876–2882. doi: 10.1128/jb.178.10.2876-2882.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Potempa J, Pavloff N, Travis J. Porphyromonas gingivalis: a proteinase/gene accounting audit. Trends Microbiol. 1995;3:430–434. doi: 10.1016/s0966-842x(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 206.Potempa J, Pike R, Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalisare due to the presence of either Arg-gingipain or Lys-gingipain. Infect Immun. 1995;63:1176–1182. doi: 10.1128/iai.63.4.1176-1182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Progulske-Fox A, Tumwasorn S, Holt S C. The expression and function of a Bacteroides gingivalis hemagglutinin gene in Escherichia coli. Oral Microbiol Immunol. 1989;4:121–131. doi: 10.1111/j.1399-302x.1989.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 208.Progulske-Fox A, Tumwasorn S, Lepine G, Whitlock J, Savett D, Ferretti J J, Banas J A. The cloning, expression and sequence analysis of second Porphyromonas gingivalisgene that encodes for a protein involved in hemagglutination. Oral Microbiol Immunol. 1995;10:311–318. doi: 10.1111/j.1399-302x.1995.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 209.Ramasubbu N, Thomas L M, Bhandary K K, Levine M J. Structural characteristics of human salivary statherin: a model for boundary lubrication at the enamel surface. Crit Rev Oral Biol Med. 1993;4:363–370. doi: 10.1177/10454411930040031501. [DOI] [PubMed] [Google Scholar]
- 210.Rangarajan M, Aduse-Opoku J, Slaney J M, Young K A, Curtis M A. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalisW50 produce five biochemically distinct enzymes. Mol Microbiol. 1997;23:955–965. doi: 10.1046/j.1365-2958.1997.2831647.x. [DOI] [PubMed] [Google Scholar]
- 211.Reife R A, Shapiro R A, Bamber B A, Berry K K, Mick G E, Darveau R P. Porphyromonas gingivalislipopolysaccharide is poorly recognized by molecular components of innate host defense in a mouse model of early inflammation. Infect Immun. 1995;63:4686–4694. doi: 10.1128/iai.63.12.4686-4694.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Richardson W P, Sadoff J C. Induced engulfment of Neisseria gonorrhoeaeby tissue culture cells. Infect Immun. 1988;56:2512–2514. doi: 10.1128/iai.56.9.2512-2514.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Roberts, F. A. Personal communication.
- 214.Roberts F A, Richardson G J, Michalek S M. Effects of Porphyromonas gingivalis and Escherichia colilipopolysaccharide on mononuclear phagocytes. Infect Immun. 1997;65:3248–3254. doi: 10.1128/iai.65.8.3248-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rosenshine I, Ruschkowski S, Foubister V, Finlay B B. Salmonella typhimuriuminvasion of epithelial cells: role of induced host cell tyrosine phosphorylation. Infect Immun. 1994;62:4969–4974. doi: 10.1128/iai.62.11.4969-4974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rotstein O D, Pruett T L, Fiegel V D, Nelson R D, Simmons R L. Succinic acid, a metabolic by-product of Bacteroidesspecies, inhibits polymorphonuclear leukocyte function. Infect Immun. 1985;48:402–408. doi: 10.1128/iai.48.2.402-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rudek W, Haque R U. Extracellular enzymes of the genus Bacteroides. J Clin Microbiol. 1976;4:458–460. doi: 10.1128/jcm.4.5.458-460.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Saglie F R, Carranza F A, Newman M G, Cheng L, Lewin K J. Identification of tissue-invasive bacteria in human periodontal disease. J Periodontal Res. 1982;17:452–455. doi: 10.1111/j.1600-0765.1982.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 219.Saglie F R, Marfany A, Camargo P. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalisin active destructive periodontal lesions. J Periodontol. 1988;59:259–265. doi: 10.1902/jop.1988.59.4.259. [DOI] [PubMed] [Google Scholar]
- 220.Saglie F R, Smith C T, Newman M G, Carranza F A, Pertuiset J H, Cheng L, Auil E, Nisengard R J. The presence of bacteria in oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J Periodontol. 1986;57:492–500. doi: 10.1902/jop.1986.57.8.492. [DOI] [PubMed] [Google Scholar]
- 221.Saito A, Sojar H T, Genco R J. Porphyromonas gingivalissurface components induce interleukin-1 release and tyrosine phosphorylation in macrophages. FEMS Immunol Med Microbiol. 1996;15:51–57. doi: 10.1111/j.1574-695X.1996.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 222.Saito A, Sojar H T, Genco R J. Interleukin-1 gene expression in macrophages induced by surface protein components of Porphyromonas gingivalis: role of tyrosine kinases in signal transduction. Oral Microbiol Immunol. 1997;12:135–140. doi: 10.1111/j.1399-302x.1997.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 223.Sandros J, Madianos P N, Papapanou P N. Cellular events concurrent with Porphyromonas gingivalisinvasion of oral epithelium in vitro. Eur J Oral Sci. 1996;104:363–371. doi: 10.1111/j.1600-0722.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 224.Sandros J, Papapanou P N, Nannmark U, Dahlen G. Porphyromonas gingivalis invades human pocket epithelium in vitro. J Periodontal Res. 1994;29:62–69. doi: 10.1111/j.1600-0765.1994.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 225.Schenkein H A. Complement factor D-like activity of Porphyromonas gingivalisW83. Oral Microbiol Immunol. 1991;6:216–220. doi: 10.1111/j.1399-302x.1991.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 226.Schenkein H A, Fletcher H M, Bodnar M, Macrina F L. Increased opsonization of a prtH-defective mutant of Porphyromonas gingivalisW83 is caused by reduced degradation of complement-derived opsonins. J Immunol. 1995;154:5331–5337. [PubMed] [Google Scholar]
- 227.Schifferle R E, Reddy M S, Zambon J J, Genco R J, Levine M J. Characterization of a polysaccharide antigen from Bacteroides gingivalis. J Immunol. 1989;143:3035–3042. [PubMed] [Google Scholar]
- 228.Schifferle R E, Wilson M E, Levine M J, Genco R J. Activation of serum complement by polysaccharide-containing antigens of Porphyromonas gingivalis. J Periodontal Res. 1993;28:248–254. doi: 10.1111/j.1600-0765.1993.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 229.Schwarz S, Ellen R P, Grove D A. Bacteroides gingivalis-Actinomyces viscosuscohesive interactions as measured by a quantitative binding assay. Infect Immun. 1987;55:2391–2397. doi: 10.1128/iai.55.10.2391-2397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shah H N, Gharbia S E. Ecological events in subgingival dental plaque with reference to Bacteroides and Fusobacteriumspecies. Infection. 1989;17:264–268. doi: 10.1007/BF01639537. [DOI] [PubMed] [Google Scholar]
- 231.Shah H N, Gharbia S E. Studies on the physiology and ecology of black-pigmented gram-negative anaerobes which may be important in disease development. FEMS Immunol Med Microbiol. 1993;6:165–172. doi: 10.1111/j.1574-695X.1993.tb00320.x. [DOI] [PubMed] [Google Scholar]
- 232.Shah H N, Gharbia S E, Kowlessur D, Wilkie E, Brocklehurst K. Gingivain; a cysteine proteinase isolated from Porphyromonas gingivalis. Microb Ecol Health Dis. 1991;4:319–328. [Google Scholar]
- 233.Shah H N, Gharbia S E, Progulske-Fox A, Brocklehurst K. Evidence for independent molecular identity and functional interaction of the haemagglutinin and cysteine proteinase (gingivain) of Porphyromonas gingivalis. J Med Microbiol. 1992;36:239–244. doi: 10.1099/00222615-36-4-239. [DOI] [PubMed] [Google Scholar]
- 234.Shah H N, Williams R A D. Utilization of glucose and amino acids by Bacteroides intermedius and Bacteroides gingivalis. Curr Microbiol. 1987;15:241–246. [Google Scholar]
- 235.Shapira L, Sylvia V L, Halabi A, Soskolne W A, Van Dyke T E, Dean D D, Boyan B D, Schwartz Z. Bacterial lipopolysaccharide induces early and late activation of protein kinase C in inflammatory macrophages by selective activation of PKC-ɛ. Biochem Biophys Res Commun. 1997;240:629–634. doi: 10.1006/bbrc.1997.7717. [DOI] [PubMed] [Google Scholar]
- 236.Shapira L, Champagne C, van Dyke T E, Amar S. Strain-dependent activation of monocytes and inflammatory macrophages by lipopolysaccharide of Porphyromonas gingivalis. Infect Immun. 1998;66:2736–2742. doi: 10.1128/iai.66.6.2736-2742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shapiro R A, Cunningham M D, Ratcliffe K, Seachord C, Blake J, Bajorath J, Aruffo A, Darveau R P. Identification of CD14 residues involved in specific lipopolysaccharide recognition. Infect Immun. 1997;65:293–297. doi: 10.1128/iai.65.1.293-297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Singer R E, Buckner B A. Butyrate and propionate: important components of toxic dental plaque extracts. Infect Immun. 1981;32:458–463. doi: 10.1128/iai.32.2.458-463.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sismey-Durrant H J, Hopps R M. Effect of lipopolysaccharide from Porphyromonas gingivalison prostaglandin E2 and interleukin-1β release from rat periosteal and human gingival fibroblasts in vitro. Oral Microbiol Immunol. 1991;6:378–380. doi: 10.1111/j.1399-302x.1991.tb00510.x. [DOI] [PubMed] [Google Scholar]
- 240.Slakeski N, Bhogal P S, O’Brien-Simpson N M, Reynolds E C. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtKproteinases and adhesins into large complexes. Microbiology. 1998;144:1583–1592. doi: 10.1099/00221287-144-6-1583. [DOI] [PubMed] [Google Scholar]
- 241.Slakeski N, Cleal S M, Reynolds E C. Characterization of a Porphyromonas gingivalis gene prtRthat encodes an arginine-specific thiol proteinase and multiple adhesins. Biochem Biophys Res Commun. 1996;224:605–610. doi: 10.1006/bbrc.1996.1073. [DOI] [PubMed] [Google Scholar]
- 242.Slots J. Characterization of some oral and nonoral gram-negative bacteria with the API ZYM system. J Clin Microbiol. 1981;14:288–294. doi: 10.1128/jcm.14.3.288-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Slots J, Genco R J. Microbial pathogenicity. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitansin human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984;63:412–421. doi: 10.1177/00220345840630031101. [DOI] [PubMed] [Google Scholar]
- 244.Slots J, Gibbons R J. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticusto oral surfaces and its possible role in colonization of the mouth and periodontal pockets. Infect Immun. 1978;19:254–264. doi: 10.1128/iai.19.1.254-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Slots J, Listgarten M A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitansin human periodontal disease. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 246.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-restriction influences haemin-binding, haemagglutination and protease activity of cells and extracellular membrane vesicles of Porphyromonas gingivalisW50. FEMS Microbiol Lett. 1991;61:63–67. doi: 10.1016/0378-1097(91)90647-s. [DOI] [PubMed] [Google Scholar]
- 247.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-binding proteins of Porphyromonas gingivalisW50 grown in a chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 248.Smalley J W, Birss A J, McKee A S, Marsh P D. Changes in the affinity of haemin-binding by Porphyromonas gingivalisW50 under different environmental conditions. Microb Ecol Health Dis. 1994;7:9–15. [Google Scholar]
- 249.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin binding as a factor in the virulence of Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;141:65–70. doi: 10.1111/j.1574-6968.1996.tb08364.x. [DOI] [PubMed] [Google Scholar]
- 250.Smalley J W, Silver J, Marsh P D, Birss A J. The periodontopathogen Porphyromonas gingivalisbinds iron protoporphyrin IX in the μ-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 252.Sojar H T, Hamada N, Genco R J. Structures involved in the interaction of Porphyromonas gingivalisfimbriae and human lactoferrin. FEBS Lett. 1998;422:205–208. doi: 10.1016/s0014-5793(98)00002-7. [DOI] [PubMed] [Google Scholar]
- 253.Sojar H T, Lee J-Y, Bedi G S, Cho M-I, Genco R J. Purification, characterization and immunolocalization of fimbrial protein from Porphyromonas (Bacteroides) gingivalis. Biochem Biophys Res Commun. 1991;175:713–719. doi: 10.1016/0006-291x(91)91624-l. [DOI] [PubMed] [Google Scholar]
- 254.Sojar H T, Lee J-Y, Bedi G S, Genco R J. Purification and characterization of a protease from Porphyromonas gingivaliscapable of degrading salt-solubilized collagen. Infect Immun. 1993;61:2369–2376. doi: 10.1128/iai.61.6.2369-2376.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sojar H T, Lee J-Y, Genco R J. Fibronectin binding domain of P. gingivalisfimbriae. Biochem Biophys Res Commun. 1995;216:785–792. doi: 10.1006/bbrc.1995.2690. [DOI] [PubMed] [Google Scholar]
- 256.St. Geme J W, Falkow S. Haemophilus influenzaeadheres to and enters cultured human epithelial cells. Infect Immun. 1990;58:4036–4044. doi: 10.1128/iai.58.12.4036-4044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sundqvist G, Figdor D, Hanstrom L, Sorlin S, Sandstrom G. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand J Dent Res. 1991;99:117–129. doi: 10.1111/j.1600-0722.1991.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 258.Takarada H, Cattoni M, Sujimoto A, Rose G G. Ultrastructural studies of human gingiva. II. The lower part of the pocket epithelium in chronic periodontitis. J Periodontol. 1974;45:155–159. doi: 10.1902/jop.1974.45.3.155. [DOI] [PubMed] [Google Scholar]
- 259.Takazoe I, Nakamura T, Okuda K. Colonization of the subgingival area by Bacteroides gingivalis. J Dent Res. 1984;63:422–426. doi: 10.1177/00220345840630031201. [DOI] [PubMed] [Google Scholar]
- 260.Tang P, Rosenshine I, Finlay B B. Listeria monocytogenes, an invasive bacterium, stimulates MAP kinase upon attachment to epithelial cells. Mol Cell Biol. 1994;5:455–464. doi: 10.1091/mbc.5.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Teanpaisan R, Douglas C W I, Eley A R, Walsh T F. Clonality of Porphyromonas gingivalis, Prevotella intermedia and Prevotella nigrescensisolated from periodontally diseased and healthy sites. J Periodontal Res. 1996;31:423–432. doi: 10.1111/j.1600-0765.1996.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 262.Theilade E. Factors controlling the microflora of the healthy mouth. In: Hill M J, Marsh P D, editors. Human microbial ecology. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 1–56. [Google Scholar]
- 263.Tokuda M, Duncan M, Cho M I, Kuramitsu H K. Role of Porphyromonas gingivalisprotease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tonzetich J, McBride B C. Characterization of volatile sulphur production by pathogenic and non-pathogenic strains of oral Bacteroides. Arch Oral Biol. 1981;26:963–969. doi: 10.1016/0003-9969(81)90104-7. [DOI] [PubMed] [Google Scholar]
- 265.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalisproteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 266.Travis J, Potempa J, Maeda H. Are bacterial proteinases pathogenic factors? Trends Microbiol. 1995;3:405–407. doi: 10.1016/s0966-842x(00)88988-x. [DOI] [PubMed] [Google Scholar]
- 267.U S, Harper F, Curtis M A. Haemin inhibits the trypsin-like enzyme activity of Porphyromonas gingivalisW83. FEMS Microbiol Lett. 1990;60:169–172. doi: 10.1111/j.1574-6968.1990.tb03883.x. [DOI] [PubMed] [Google Scholar]
- 268.Van Dyke T E. Neutrophil receptor modulation in the pathogenesis of periodontal diseases. J Dent Res. 1984;63:452–454. doi: 10.1177/00220345840630031701. [DOI] [PubMed] [Google Scholar]
- 269.Van Steenbergen T J M, Petit M D A, Scholte L H M, van der Velden U, de Graaff J. Transmission of Porphyromonas gingivalisbetween spouses. J Clin Periodontol. 1993;20:340–345. doi: 10.1111/j.1600-051x.1993.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 270.Van Winkelhoff A J, van Steenbergen T J M, de Graaff J. The role of black-pigmented Bacteroidesin human oral infections. J Clin Periodontol. 1988;15:145–155. doi: 10.1111/j.1600-051x.1988.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 271.Vayssier C, Mayrand D, Grenier D. Detection of stress proteins in Porphyromonas gingivalisand other oral bacteria by western immunoblotting analysis. FEMS Microbiol Lett. 1994;121:303–307. doi: 10.1111/j.1574-6968.1994.tb07117.x. [DOI] [PubMed] [Google Scholar]
- 272.Watanabe K, Kumada H, Yoshimura F, Umemoto T. The induction of polyclonal B-cell activation and interleukin-1 production by the 75-kDa cell surface protein from Porphyromonas gingivalisin mice. Arch Oral Biol. 1996;41:725–731. doi: 10.1016/s0003-9969(96)00074-x. [DOI] [PubMed] [Google Scholar]
- 273.Watanabe K, Onoe T, Ozeki M, Shimizu Y, Sakayori T, Nakamura H, Yoshimura F. Sequence and product analyses of the four genes downstream from the fimbrillin gene (fimA) of the oral anaerobe Porphyromonas gingivalis. Microbiol Immunol. 1996;40:725–734. doi: 10.1111/j.1348-0421.1996.tb01133.x. [DOI] [PubMed] [Google Scholar]
- 274.Weinberg A, Belton C M, Park Y, Lamont R J. Role of fimbriae in Porphyromonas gingivalisinvasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Wilson M, Meghji S, Barber P, Henderson B. Biological activities of surface-associated material from Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 1993;6:147–155. doi: 10.1111/j.1574-695X.1993.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 276.Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodontopathogenic bacteria. J Periodontal Res. 1996;31:393–407. doi: 10.1111/j.1600-0765.1996.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 277.Xie H, Cai S, Lamont R J. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun. 1997;65:2265–2271. doi: 10.1128/iai.65.6.2265-2271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Yamaji Y, Kubota T, Sasaguri K, Sato S, Suzuki Y, Kumada H, Umemoto T. Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect Immun. 1995;63:3576–3581. doi: 10.1128/iai.63.9.3576-3581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yao E S, Lamont R J, Leu S P, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. 1996;11:35–41. doi: 10.1111/j.1399-302x.1996.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 280.Yoneda M, Kuramitsu H K. Genetic evidence for the relationship of Porphyromonas gingivaliscysteine protease and hemagglutinin activities. Oral Microbiol Immunol. 1996;11:129–134. doi: 10.1111/j.1399-302x.1996.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 281.Yoshimura A, Hara Y, Kaneko T, Kato I. Secretion of IL-1β, TNF-α, IL-8 and IL-1ra by human polymorphonuclear leukocytes in response to lipopolysaccharides from periodontopathic bacteria. J Periodontal Res. 1997;32:279–286. doi: 10.1111/j.1600-0765.1997.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 282.Yoshimura F, Takahashi Y, Hibi E, Takasawa T, Kato H, Dickinson D P. Proteins with molecular masses of 50 and 80 kilodaltons encoded by genes downstream from the fimbrillin gene (fimA) are components associated with fimbriae in the oral anaerobe Porphyromonas gingivalis. Infect Immun. 1993;61:5181–5189. doi: 10.1128/iai.61.12.5181-5189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yoshimura F, Takashi K, Nodasaka Y, Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J Bacteriol. 1984;160:949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yoshimura F, Takasawa T, Yoneyama M, Yamaguchi T, Shiokawa H, Suzuki T. Fimbriae from the oral anaerobe Bacteroides gingivalis: physical, chemical, and immunological properties. J Bacteriol. 1985;163:730–734. doi: 10.1128/jb.163.2.730-734.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]