Abstract
Export of RNA from the nucleus is essential for all eukaryotic cells, with at least three major classes exported, mRNA, tRNA and rRNA. RNA export has emerged as a major step in the control of gene expression, with mRNA molecules required to complete a complex series of processing events and pass a quality control system to protect the cytoplasm from the expression of aberrant proteins. Many of these events are highly conserved across eukaryotes, reflecting their ancient origin, but significant deviation from a canonical pathway as described from animals and fungi has emerged in the trypanosomatids. With significant implications for the mechanisms that control gene expression and hence differentiation, responses to altered environments and fitness as a parasite, these deviations may also reveal additional, previously unsuspected, mRNA export pathways.
Keywords: mRNA export, nuclear pore complex, trypanosomes, parasites, evolution, eukaryogenesis, polycistronic transcription, trans-splicing
Context
All cellular life relies on RNA, and consequently RNA-related processes are highly conserved, including the basic features of transcription and translation. With the major innovation of eukaryogenesis being evolution of the nuclear envelope, which separates transcription from translation, both new challenges and opportunities for RNA metabolism emerged. These include the potential for extensive post-transcriptional processing events, which for mRNA includes splicing, polyadenylation and base modifications throughout, together with a requirement for an export pathway. mRNA export and processing are coupled in modern eukaryotes as a multistep process that essentially safeguards the translational apparatus from aberrant mRNAs encoding potentially toxic products.
Trypanosomes are obligatory parasites of invertebrates, vertebrates and/or vascular plants and cause major public health and economic impact. Their lineage arose from very early separation from the main eukaryotic line and likely shortly following radiation from the Last Eukaryotic Common Ancestor (LECA) [1–3]. Trypanosomes deviate from canonical mechanisms for many aspects of their biology and gene expression especially, with perhaps the headline features being polycistronic transcription together with trans-splicing and the absence of cis-splicing [4, 5]. Early studies intimated that control of individual genes through promoter activity is lacking in trypanosomes due to polycistronic transcription, arguing for control mechanisms focused on mRNA turnover and elements within the 3’ untranslated region. This is however, likely an oversimplification as there are multiple steps between transcription and translation, most of which are shared between essentially all eukaryotes (Figure 1).
Figure 1: Quick start guide to control of eukaryotic gene expression.
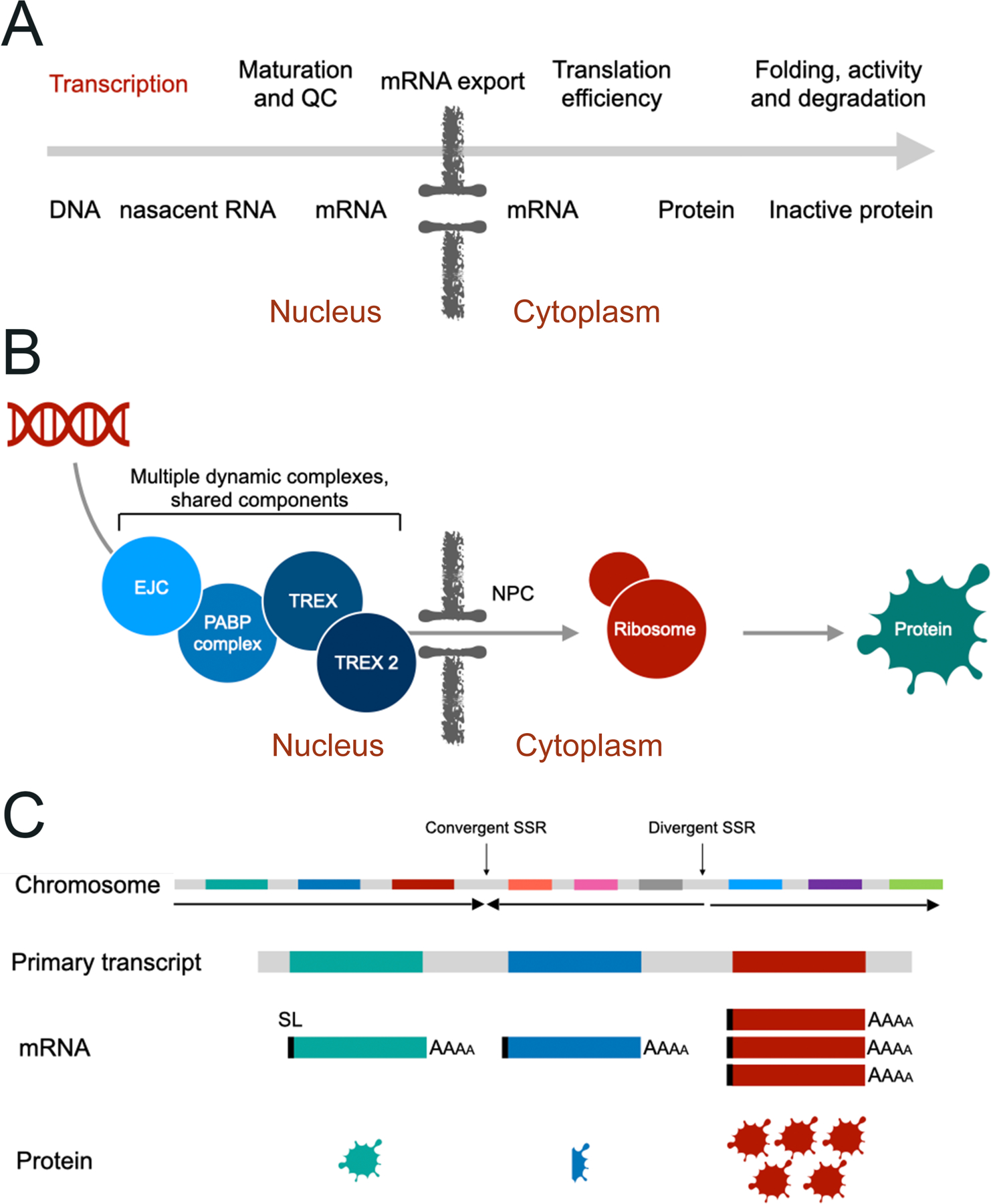
Panel A: Steps in the standard model of transcription and translation. Above the arrow are the processes taking place and which may be regulated by one or more mechanism. Below the arrow are the molecules that encode biological information. Critically, all of these steps, with the exception of control of translation (in red), are common between trypanosomes and other eukaryotes. Panel B: Highly simplified view of mRNA maturation and nuclear export pathways. Multiple complexes are shown in shades of blue that are responsible for the splicing, folding and processing of mRNAs, and which are associated with the nuclear pore complex (NPC). Precise distinctions between these complexes is difficult, as the composition of complexes varies dynamically, with many proteins being shared and/or acting to link complexes. Association with the NPC, by the TREX complexes, is a critical aspect of the export process. Panel C: Simplified scheme for trypanosome polycistronic transcription. Top is an example chromosomal region containing several protein coding sequences, and illustrating convergent and divergent strand with region. Arrow illustrates the direction of RNA transcription. The leftmost cistron is transcribed as a single RNA, and which is resolved into mRNAs following splicing and polyadenylation. Turnover and other processes regulate the copy number of the mRNA, and additional mechanisms, including translational efficiency also contribute to differential protein levels. All of these processes are discussed in detail in the text.
mRNA processing and export, as mapped in animals and fungi, is supported by multiple complexes, amongst which are the EJC (exon-junction complex), CPSF (cleavage and polyadenylation specificity factor), TREX (transcription and export) and TREX-2, as well as the NPC (nuclear pore complex) (Figure 1). Components of each of these complexes are present across the eukaryotic lineage, but several complexes are overall poorly conserved. Hence, despite the core aspects of RNA metabolism, there has been at least one billion years since the eukaryotic lineage arose and expanded, offering considerable scope for diversity to have evolved between these processes in different lineages. Here we will discuss divergence within the NPC and mRNA processing factors that lead to highly distinct mechanisms for sending mRNA to the cytoplasm in trypanosomes, and speculate as to their origins and purpose (Figure 2, Table 1).
Figure 2: An illustration of the differences between protein and mRNA export in opisthokonts versus trypanosomes.
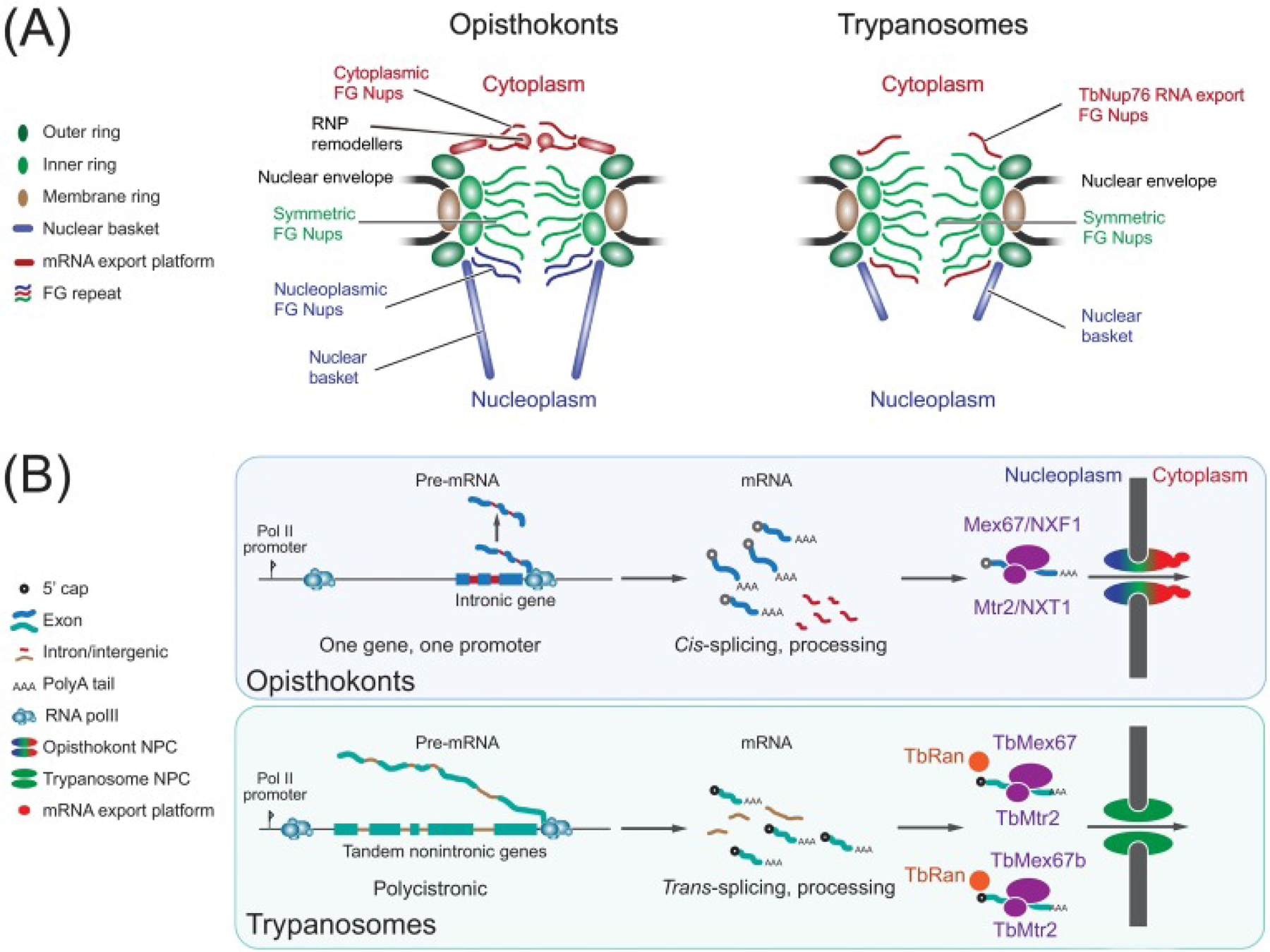
(A) A comparison of the nuclear pore complex (NPC) structure in opisthokonts versus trypanosomes. The arrangement of the major subcomplexes of the NPC are highlighted. The membrane, inner, and outer rings are structural components that act as a scaffold for the nuclear basket and phenylalanine glycine (FG) repeat containing nucleoporins (Nups) that maintain the permeability barrier of the NPC. The NPC in opisthokonts differs from that in trypanosomes in having a well-recognized mRNA export platform on the cytoplasmic side of the NPC which is evolutionarily divergent in trypanosomes. In addition, the nuclear basket is smaller in trypanosomes. and the mechanism of anchoring the NPC to the nuclear envelope is also divergent. (B) A comparison of transcription and export between opisthokonts and trypanosomes. Opisthokonts have individual RNA Pol II promoters for each gene while trypanosomes have single RNA Pol II-like promoter elements at the beginning of each polycistronic transcription unit comprised of several nonfunctionally related tandem genes. Individual mRNAs from each polycistronic transcription unit are resolved by trans-splicing. mRNA export in opisthokonts relies on an ATP-dependent DEAD box helicase [ribonucleoprotein (RNP) modelers] to drive directionality of mRNA transport from the nucleus into the cytoplasm. In trypanosomes, it has been postulated that mRNA export is dependent on the GTPase Ran, a radical departure from opisthokonts, concomitant with a lack of an obvious dedicated cytoplasmic mRNA export platform.
Table 1: The nuclear pore complex components involved in mRNA export.
A comparison of the components of the mRNA export platform between trypanosomes, opisthokonts (yeast and vertebrates) and plants. Trypanosomes have a Nup82 ortholog, but then lacks several known components of the mRNA export platform in the NPC, instead having species specific proteins that may reflect their unusual mode of gene regulation. Additionally, trypanosomes have seemingly different nuclear basket components, that are half the size of those in opisthokonts and plants.
| Major secondary structures | Trypanosomes | Yeast | Vertebrates | Plants | NPC subcomplex |
|---|---|---|---|---|---|
| β-propeller, coiled coil | TbNup76 | Nup82 | Nup88 | Nup88 | Cytoplasmic (opisthokonts), Possibly Cytoplasmic and Nucleoplasmic (Trypanosomes) |
| FG repeats, putative ZnFs | TbNup149 | x | x | x | Possibly Cytoplasmic and Nucleoplasmic - part of the TbNup76 complex |
| FG repeats | TbNup140 | x | x | x | |
| α-solenoid (tryps only), FG repeats | TbNup158# | Nup116 and Nup100* | Nup98 | Nup98 | Cytoplasmic and Nucleoplasmic, and also on Nup82 complex (Yeast) |
| Coiled coil, FG repeats | TbNup62 | Nsp1 | Nup62 | Nup62 | Inner ring and Cytoplasmic Nup82/88 complex (Inner ring only in trypanosomes) |
| β-propeller, FG repeats, coiled coil | x | Nup159 | Nup214 | Nup214 | Cytoplasmic - Nup82/88 complex |
| FG repeats | x | Nup42 | Nlp1 | CG1 | |
| Auxiliary Factors | x | Dbp5 | DDX19 | RH38 | Dock on the Cytoplasmic - Nup82/88 complex (Unknown in trypanosomes) |
| x | Gle1 | Gle1 | Gle1 | ||
| TbGle2 | Gle2 | Rae1 | Rae1 | ||
| IP6 | IP6 | IP6 | IP6 | ||
| FG repeats, ZnFs (Nup153 only) | x | Nup60* and Nup1* | Nup153 | Nup153 | Nucleoplasmic |
| FG repeats | x | Nup2 | Nup50 | Nup50 | |
| Coiled Coil | TbNup110 (110 kDa) | x | x | x | Nuclear basket |
| TbNup92 (92 kDa) | x | x | x | ||
| x | Mlp1 (218 kDa) | TPR (267 kDa) | NUA (237 kDa) | ||
| x | Mlp2 (195 kDa) | x | x |
KEY:
IP6 is inositol hexakisphosphate and ZnFs is Zinc Fingers
represents yeast specific gene duplications found at the NPC for which there's a single ortholog in other eukarya
TbNup158 exists as two separate polypeptides in other eukarya (a N-terminal FG repeat protein and a C-terminal α-solenoid which autocleave to form two distinct proteins). TbNup158 is a single protein containing both domains
The pore is the core
After transcription, processing and maturation, most RNAs are transported from the nucleus through NPCs, macromolecular protein assemblies embedded within the nuclear envelope (NE) and which facilitate selective transport between the nucleoplasm and cytoplasm [6]. This process is highly complex with evidence for lineage-specific mechanisms, of which kinetoplastids and optisthokonts (animals and yeast) provide exemplars (Figure 2, Table 1). Amongst these are structural and compositional changes within the NPC and other RNA processing complexes [7].
Nuclear pore complexes are octagonal structures composed of ~30 different proteins termed nucleoporins (Nups), present in multiple copies to comprise approximately 500 total proteins per NPC (Figure 2) [8–10]. Substructures within the NPC, such as a proteinaceous membrane ring anchoring the NPC to the NE and a core structural scaffold attest to a modular evolution and functionality (Figure 2). The scaffold anchors a class of nucleoporins that contain disordered regions of phenylalanine glycine (FG) and related dipeptide repeats. These proteins are primarily responsible for the selective permeability barrier of the NPC. Transport factors, variously called karyopherins, importins, exportins (and other synonyms ad nauseam) facilitate transport by virtue of specific interactions with FG-repeats, while these same regions exclude non-karyopherin-bound proteins [6, 11, 12]. The scaffold also anchors a subset of asymmetrically positioned peripheral Nups (Figure 2) [13], which include nucleoplasm oriented FG-Nups, as well as the nuclear basket, which interacts with nuclear mRNA processing complexes which constitute an RNA export platform (Table 1) [14–17]. The NPC thus acts as an interacting platform, especially the peripheral substructures, to provide a hub for multiple steps in gene expression.
Trypanosomes as models to study evolution of RNA processing and export
Significantly, the overview above is fully applicable to trypanosomes. Their NPC appears to be of similar overall architecture and complexity to animals and fungi, retains a nuclear basket and FG-Nups that mediate gating [18–20], together with a cohort of karyopherins, other transport factors and the small GTPase Ran, an essential mediator of transport direction and fidelity [6]. Trypanosomes offer an excellent system through which to compare various mRNA processing steps, and many of the divergent features place greater reliance on post-transcriptional mechanisms than in animals and fungi. More recently roles for mRNA-binding proteins (RBPs) are being uncovered, a significant number of which are lineage-specific [21]. Some RBPs mediate expression of entire cohorts of mRNAs, acting as master regulators [22–24]. It is formally possible that trypanosome transcription and mRNA processing mechanisms are simply reduced compared with animals and fungi and reflect an absence of control over the environment, negating any need for complex responses to improve fitness [25]. However, several other trypanosome cellular systems exhibit incomplete retention of metazoan machinery, but have emerged as possessing alternate components, rather than simple reduction [26]. We suggest this is also the case for gene expression.
From transcription to quality control and export
In metazoa and fungi, protein coding RNAs are transcribed exclusively by RNA Pol II, with mRNA export being initiated co-transcriptionally (Figure 1). Several protein factors associate with the nascent message to form messenger ribonucleoprotein complexes (mRNPs). RNA export is integrated with mRNA biogenesis and processing, amongst these factors are several transcription – export (TREX) complexes. TREX is comprised of two different cohorts; THO components Tho2, Hpr1, Mft1 and Thp2 [27], together with two TREX-specific components Sub2 and Yra1, which act as adaptors for mRNA export proteins [28]. The THO/TREX complex mediates transcription elongation in yeast, splicing of mRNAs in vertebrates and co-transcriptional recruitment of the mRNA export machinery (Figure 1) [27, 29]. Beyond animals, fungi and plants, the evolutionary conservation of THO/TREX complex proteins, their functions and pathways becomes more difficult to decipher.
The DEAD box helicase Sub2 (UAP56 in vertebrates) is the only evolutionarily conserved TREX-complex protein and has been characterized in diverse protists, including Plasmodium, Toxoplasma and trypanosomes [30, 31], suggesting a very high level of conservation [30–33]. Silencing of Sub2 results in the accumulation of polyA mRNA in the nucleus of trypanosomes and decreased translation [31]. Toxoplasma Sub2 is also heavily involved in export and disruption using CRISPR blocks mRNA export [30]. Additional TREX complex proteins are either so diverged as to be undetectable or absent in trypanosome and Apicomplexa genomes [32]. Thus, our appreciation of the players and processes underlying mRNA processing, from the point of transcription through to export to the cytoplasm, remains incomplete in protists, albeit with the clear indication that Sub2 at least has conserved functions, and potentially with backfilling replacing the absent TREX subunits [32]. What remains unclear is if this represents a secondary loss or later evolution of Sub2-interacting proteins, as recently demonstrated in a proteomic study in the American trypanosome T. cruzi, where several kinetoplastid-specific proteins were found to be involved in RNA processing and splicing in addition to more evolutionarily conserved factors [34]. Although these newly identified kinetoplastid-specific factors may perform analogous functions to THO/TREX components, they are yet to be functionally interrogated.
As a prelude to export in metazoa and fungi, Sub2 is displaced by the mRNA export factors Mex67 and Mtr2. These remodeled complexes are now export competent, but pause at the nuclear basket and engage a quality control checkpoint. This is facilitated by the TREX2 complex, which is tethered to the nuclear periphery via the nuclear basket nucleoporins Mlp1/Mlp2 (Figure 2, Table 1) [35, 36]. In this context it is relevant that Tpr, the vertebrate nuclear basket nucleoporin, is not simply a passive interaction platform for TREX2, but rather an integral part of the complex itself whose disruption leads to abnormal transcription and export [37]. The TREX2 component Sac3 provides a scaffold for Thp1, Sem1, Cdc31 and Sus1 [38]. Metazoa Sac3 differs from the yeast ortholog in that metazoan Sac3 shuttles between sites of active transcription and the NPC while binding directly to Mex67, thus facilitating intranuclear translocation of mRNPs from transcription sites to the NPC in preparation for export [35, 39].
TREX2 functions as a staging post for both mRNA processing and for export proteins to interact and facilitate association and repositioning of actively transcribed genes to NPCs in conjunction with the transcription coactivator SAGA (Spt-Ada-Gcn5 acetyltransferase) [40]. The SAGA complex is comprised of ~20 subunits and, due to the presence of Gcn5 was initially considered as a histone acetyltransferase. However, SAGA also contains a histone de-ubiquitinase and subunits interacting with transcriptional activators and the general transcription machinery, indicating coordination of a broad range of functions [41]. Just as most THO/TREX components are either cryptic or absent from trypanosomes, there is scant evidence for SAGA components and the recent T. cruzi immuno-isolation proteomic study amongst others supports the apparent absence of several canonical orthologs of SAGA components [34].
Thus, it is possible that trypanosomes have a wholly divergent system for processing mRNA, supported by a considerable cohort of kinetoplastid-specific proteins interacting with an evolutionarily preserved Sub2 and other conserved components of the mRNA processing system. Indeed, evidence suggests a tightly coupled system stretching from transcription to translation, supported by evidence that trypanosomes can initiate mRNA export cotranscriptionally [42]. Surprisingly, there is no quality control checkpoint at the trypanosome NPC prior to export of mRNA through the NPC in trypanosomes [37]. Blocking trans-splicing, and thus faithful resolution of individual mRNAs from polycistronic mRNAs, does not initiate a “pause” at the NPC, instead allowing export of non-spliced, non-resolved mRNAs [42].
Notably trypanosomes have significant species-specific differences in NPC architecture, and which are mainly focused around the mRNA export machinery, especially the nuclear basket and cytoplasmic mRNA export platforms [18, 20, 43]. The nuclear basket proteins Tpr and Mlp1/Mlp2 in animals and fungi respectively range from 200–270 kDa [44–46], but in trypanosomes these are represented by two proteins of 92kDa and 110kDa in size, suggesting significant evolutionary divergence of this important NPC subcomplex (Table 1) [18, 20, 43]. Unlike other nuclear basket Nups, TbNup92 uniquely possesses a C-terminal BRCT domain; however, it intimately associates with the mitotic spindle and spindle organizer at mitosis, and is a functional analog of yeast Mlp2 [47], which similarly relocates to spindle organizers in a cell cycle dependent manner [43, 47]. TbNup110 is essential for cellular growth in bloodstream form of T. brucei [48] and, analogous to Mlp1, extends circa 40nm into the nucleoplasm from the NPC [20]. However, the function of TbNup110 in quality control of RNA export is unexplored.
mRNA transport factors in canonical organisms
After pausing at the nuclear basket, mRNA export through the NPC in animals and fungi occurs in a matter of milliseconds. This transport is mediated by non-karyopherin transport factors Mex67 and Mtr2 [49, 50]. Mex67 is a multi-domain protein with a cargo-binding domain consisting of an RNA recognition motif, a leucine rich repeat (LRR) which mediate interactions with RNA and auxiliary RNA processing proteins, an NPC-binding domain consisting of an NTF2 domain required to form a heterodimer with Mtr2 and a C-terminal ubiquitin-associated (UBA) domain, mediating interactions with FG-Nups [51, 52]. The final steps of mRNA export and remodelling in animals and fungi are performed by Nups located on the cytoplasmic face of the NPC [14, 16, 53] (Figure 2, Table 1).
The main component of the cytoplasmic mRNA export platform in yeast is the Nup82 complex, a tetrameric assembly comprised of Nup82, Nup159, Nsp1 and Dyn2 (dynein light chain) [14]. Nup82 and Dyn2 are purely structural proteins, whilst Nsp1 and Nup159 also carry FG repeats. The DEAD box RNA helicase Dbp5 and an RNA export mediator Gle1 associate with Nup159 and together they remodel mRNPs exiting the NPC in an ATP-dependent manner [15, 17, 53–58]. This allows the Mex67:Mtr2 complex to disengage from export cargo and recycle into the nucleus, providing both directionality and energy to drive mRNA export [49, 50, 59, 60].
Trypanosomes lack the canonical NPC mRNA export platform
Trypanosomes have orthologs of most of the major transport factors present in animals and fungi [18, 61], suggesting at some level a high degree of evolutionary conservation. As such it is presumed (albeit unproven) that most of these homologs function as in higher eukaryotes. Indeed, the main mRNA export factor Mex67 and its partner Mtr2 are conserved [62–64]. Given this, it is significant to find major differences in mRNA transport mechanisms associated with the trypanosome NPC [20]. Remarkably, orthologs of yeast Nup159, Gle1 and Dbp5 are absent from the trypanosome NPC, the key components of the animal and fungal cytoplasmic RNA export platform [14–17, 53, 54, 56, 65].
In yeast the Nup82 complex is anchored over the central NPC channel by the outer ring complex [14, 66], an asymmetric position crucial for driving unidirectional ATP-dependent mRNA export [67, 68]. Nup159 can be distinguished from all other FG-Nups due to the presence of a unique N-terminal β-propeller, that acts as an interaction platform for Dbp5 (Table 1) [17]. TbNup76 appears to be the trypanosome ortholog of Nup82 and forms a complex with two large FG-Nups, TbNup140 and TbNup149 (Figure 3). The genes encoding the two proteins are adjacent in kinetoplastid genomes and separated by an unusually small (122bp) intergenic region [18]. Neither TbNup140 nor TbNup149 possess a β-propeller, consistent with the absence of Dbp5 from trpypanosomes [17]. TbNup140 contains ~100 FG dipeptides spanning ~120 kDa, with an N-terminal 20kDa coiled-coil motif acting as the NPC anchor. TbNup149 has considerably fewer FG-Nups but is built from three repetitive segments in T. brucei (Figure 3). The ortholog is larger in T. cruzi and Leishmania major (170 kDa and 382 kDa respectively). The standout feature of TbNup149 are three zinc finger-like motifs that are well conserved between the kinetoplastids [20] (Figure 3). Hence, the entire architecture of this NPC subcomplex is remodelled to a remarkable degree and precludes the presence of a canonical mRNA export mechanism. Significantly, this configuration is restricted to trypanosomes as Euglena gracilis, which possesses a Dbp5 ortholog, appears more conventional [69]. The absence of the Nup159 – Dbp5 system removes the ATP-mediated steps from mRNA export, which asks how this process is powered and mRNPs appropriately remodelled upon entry to the cytoplasm in trypanosomes.
Figure 3:
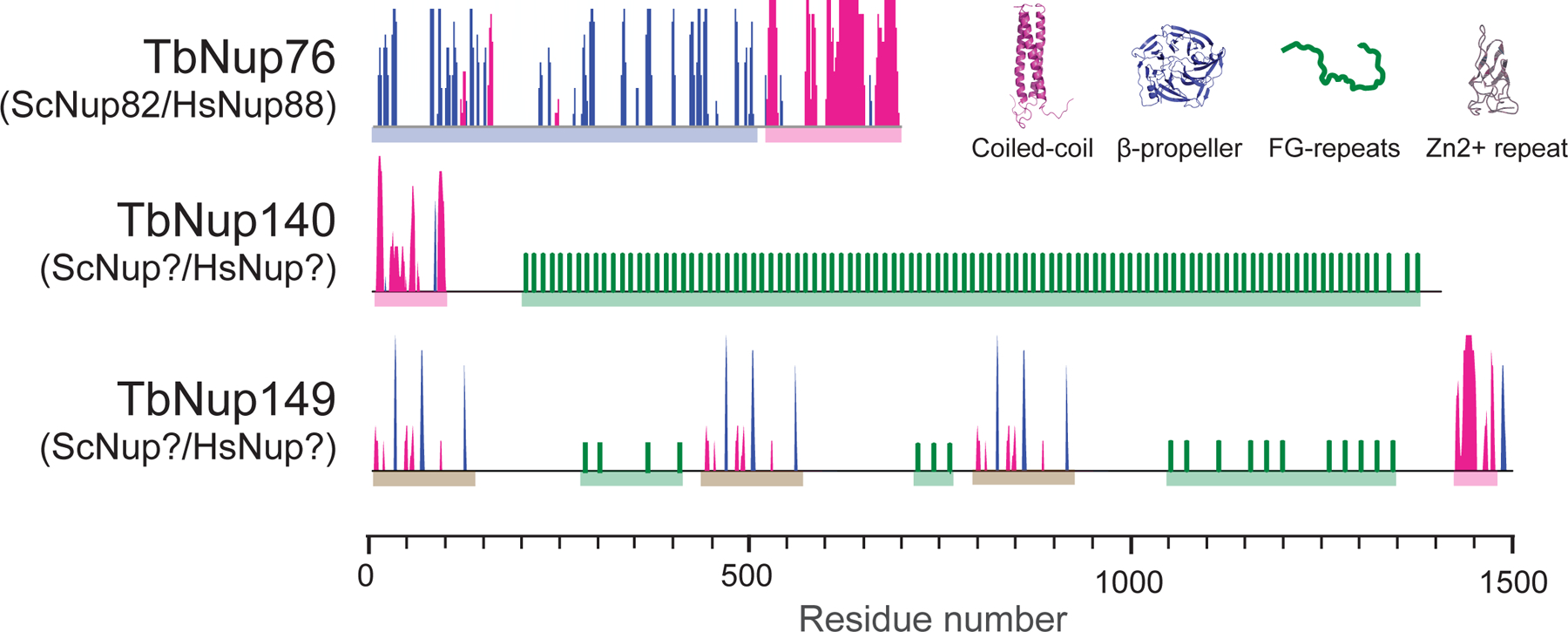
Components of the TbNup76 complex, the trypanosome mRNA export platform. TbNup76 complex comprises of the β-propeller, coiled-coil protein, Nup76, the structural ortholog of Nup82/88 in yeast and vertebrates, respectively. TbNup76 forms a complex with two large FG-Nups with no obvious orthologs in opisthokonts. TbNup 149 appears to have a number of putative zinc fingers (Zn2+) on three repeats on the protein.
Typically, nucleocytoplasmic transport is mediated by two classes of soluble protein: karyopherins which (in)export proteins and non-coding RNAs (rRNA, miRNA, tRNA and snoRNA) and non-karyopherin (nuclear transport factor 2 (NTF2)) type transport factors, which export mRNA [70]. A RanGTP/GDP gradient represents the vectoral driver of nucleocytoplasmic transport [70]. RanGTP is involved in the export of proteins from the nucleus through cooperative interactions with exportins (Figure 2). Once in the cytoplasm, Ran-bound GTP is rapidly hydrolyzed to RanGDP through interaction with Ran GTPase activating protein (RanGAP) and the cofactor Ran binding protein 1 (RanBP1). A conformational change allows Ran to be released from the exportin complex and to bind NTF2, whose major purpose is to actively import RanGDP back into the nucleus to be reactivated into RanGTP, thus maintaining the gradient. By contrast, directionality of bulk mRNA export in animals and fungi is independent of Ran, relying instead on the ATP-dependent Dbp5 path (Figure 2).
Remarkably, immunoisolation of trypanosome Mex67 recovers stoichiometric quantities of Ran, RanBP1 and a putative Ran GTPase activating protein, even though it is well established that neither yeast nor vertebrate Mex67 or Mtr2 can bind Ran [20, 71, 72]. Thus, the interaction of trypanosome Mex67:Mtr2 with Ran is highly atypical. Moreover, once on the cytoplasmic side of the NPC, unknown factors tether the mRNA in granules, to the vicinity of the NPC [42]. This, coupled with the absence of discernible orthologs of TREX2 complex components suggests that the trypanosome nuclear basket cannot function analogously to animals and fungi. It is however interesting that unspliced mRNA is tethered to RNA granules peripheral to the cytoplasmic side of the NPC, hinting at a mechanism for quality control of trypanosome mRNA processing [34].
Increased complexity of mRNA export factors in metazoa and trypanosomes.
The frequency of alternative splicing increased with diversification of cell types in multicellular organisms [73]. Multicellular organisms also have additional Mex67 variants (nuclear exchange factor or NXF in metazoa), some of which are themselves generated as splice variants and exhibit tissue specificity [74–78]. NXF1 is highly expressed in all mammalian tissues, whereas the other paralogs in humans, mice, fruit flies and nematodes tend to be expressed at lower levels, are tissue specific and/or developmentally regulated [76]. Humans and mice have at least four NXF gene products; NXF1,2,3, and 5 in humans and NXF1,2,3 and 7 in mice. NXF1 and NXF2 predominantly localize to the nucleoplasm and display mRNA export activities, whilst NXF3, NXF5 and NXF7 are mainly cytoplasmic, highlighting potential functional differences [76, 79]. NXF2 is expressed in testes and neurons [80, 81], whilst NXF3 is expressed mainly in testes [74]. NXF3 lacks the C-terminal UBA domain required for direct interactions with the NPC [51, 52], instead having a novel XPO1-dependent nuclear export signal that compensates in cis for loss of the canonical NPC targeting domain [74] [82]. Lastly, NXF5 and NXF7 localize to neurons and associate with translating ribosomes, stress granules and P-bodies [78, 80, 83–85]. Fruit flies also have four NXFs: NXF1,2,3 and 4, of which only NXF1 is essential and responsible for mRNA export [86]. This suggests that NXF1 is the global mRNA exporter in metazoa whilst NXF2, NXF3, NXF5 and NXF7 have tissue specific functions, some of which remain cryptic. Importantly, NXF1, 2 and 3 form heterodimers with NXT1 (nuclear transport factor 2-like export factor 1), the metazoan ortholog of Mtr2, which facilitates NPC localization and translocation [49, 52, 77, 87–89].
Multiple trypanosomes paralogs of Mex67 have also recently been identified and characterized (BioRxiv, in preparation). Unlike metazoa, these paralogs are not relatively minor splice variants, but encoded by separate genes, are structurally diverse and have discrete functions. Immunoisolation of TbMtr2 demonstrated an interaction with TbMex67, which has been well characterized; TbMex67 has a non-canonical N-terminal CCCH zinc finger that is essential and appears, thus far, unique to trypanosomes [64]. Additionally, TbMtr2 interacts with TbMex67b and TbMex67-like or TbMex67L. All three possess a NTF2-like domain in addition to the typical LRR domains found in Mex67, while TbMex67 also has a C-terminal UBA. Significantly, neither TbMex67b nor TbMex67L retain the UBA domain, and while not unique to trypanosomes (mammalian also NXF3 lacks this domain), this indicates a distinct modality separating TbMex67 from TbMex67b/TbMex67L [51, 52]. Despite this, Mex67b still interacts with the trypanosome splicing machinery [34]. TbMex67L is considerably larger than TbMex67 and TbMex67b due to an extended N-terminal domain.
The genes encoding TbMex67 and TbMex67b are close on chromosome 11, indicative of a gene duplication event. Moreover, this chromosomal region, including the syntenic arrangement of TbMex67 and TbMex67b genes is conserved throughout the kinetoplastids, and phylogenetic reconstruction indicates that TbMex67 and TbMex67b are more closely related than they are to Mex67L. Orthologs of TbMex67 and TbMex67b are recovered from all kinetoplastids, including the free-living bodonid, Bodo saltans, but TbMex67L is not, albeit retaining a presence within all other kinetoplastids, indicating a more recent addition to the repertoire than diversification of TbMex67/TbMex67b. TbMex67 and TbMex67b localize to the nucleolus as well as to NPCs at the NE periphery, consistent with roles in RNA export, whilst TbMex67L localizes exclusively to the perinucleolar foci in a manner reminiscent of Pol I [91], suggestive of a role specific to rRNA processing. Affinity capture of TbMex67 and TbMex67b co-isolates NPC components while TbMex67L does not, instead co-isolating with ribosome biogenesis proteins and ribosomal proteins (BioRXiv, in preparation). Thus, trypanosomes are the first unicellular organism to have multiple orthologs of Mex67identified, two of which appear to be involved in RNA export and one with a specialized role at the nucleolus and ribosomal biosynthesis.
Mex67 is also involved in the transport of certain non-coding RNAs
In opisthokonts, Mex67/Mtr2 function with XPO1, the most abundant export factor that mediates rRNA export, with involvement in 60S and 40S ribosomal subunits and 5S rRNA [92, 93]. These additional Mex67 activities appear conserved in trypanosomes, with export of 60S and 40S subunits partially dependant on TbMex67 and TbMtr2. Defects in processing 60S rRNA and aberrations in ribosome assembly occur after silencing [94], while TbMex67/TbMtr2 interacts with protein components of the 5S RNP [95]. TbMex67 and TbMtr2 are also involved in tRNA export [96], a role fulfilled by exportin-T (XPOT) in animals and fungi [70]. Silencing XPOT does not perturb tRNA export in trypanosomes [96], but rather knockdown of TbMex67 (partially) and TbMtr2 (fully) blocks tRNA export [96]. As only TbMtr2 fully blocks tRNA translocation, this suggests roles for at least one of the additional TbMex67 paralogs in this pathway. It was also recently shown that silencing of two inner ring FG-Nups TbNup62 (Table 1) and TbNup53a directly affected tRNA export, suggesting that these two specific Nups are part of the tRNA export pathway in trypanosomes [19]
Evolutionary divergence in mRNA export mechanisms
Despite making strides in deciphering trypanosome RNA export pathways, we have neither a clear mechanistic paradigm nor an understanding of how the various complexes coordinate. eIF4AIII is a conserved nucleocytoplasmic shuttling RNA DEAD-box helicase and in trypanosomes depends on TbMex67b for function in RNA export [97]. Although eIF4AIII locates at the cytoplasmic side of trypanosome NPCs, it is also present in the nucleoplasm and cytoplasm and knockdown of TbMex67b leads to nuclear accumulation of eIF4AIII, indicating a functional interaction [97]. Further, DRBD18, an essential and abundant T. brucei RNA-binding protein associates with TbMex67 and TbMtr2 in vivo, probably through interactions with TbMtr2 [98]. RNAi knockdown of DRBD18 leads to partial nuclear retention of mRNA and an export block of a subset of mRNAs, but has no effect on the export of tRNA [98].
Additional complexity in trypanosomes involves XPO1, which exports only some mRNAs in trypanosomes [99]. Further, leptomycin B treatment of T. cruzi, an XPO1 inhibitor, leads to a partial accumulation of a subset of mRNAs, specifically those encoding HSP70, the RNA-binding proteins TcUBP1/TcUBP2 and polyA-binding protein PABP1 [99]. XPO1 is involved in the export of some mRNA in vertebrates, as well as those viral RNAs bypassing the surveillance system that prevents normal export of unspliced RNAs. Lacking an RNA-binding domain itself, XPO1 relies on interactions with additional proteins to export different classes of mRNA, e.g. human antigen R (HuR) and eIF4E- and NXF3-dependent mRNA export [100–104]. If such adaptors are present in trypanosomes with similar functions remains to be established, but the observation of only partial blockade to mRNA export following XPO1 silencing in trypanosomes suggests that this is highly likely.
In fungi XPO1 also mediates export of the large ribosomal subunit, a pathway that depends on Nmd3, an adaptor protein that recruits Crm1 to the 60S subunit in the nucleus in preparation for transport [105, 106]. Nmd3 is extremely well conserved and present in both eukaryotes and archea [107], and TbNmd3 regulates both mRNA and rRNA nuclear export via an XPO1-linked pathway [108]. Silencing TbNmd3 leads to upregulation of procyclin-associated gene transcripts [108] which are transcribed by RNA Pol I [109]. Interestingly, silencing or inhibiting TbXPO1 with leptomycin B or silencing TbMex67 has a similar phenotype. Considering the evolutionarily conserved relationship between Nmd3 and XPO1, this provides further support for divergence as procyclin-associated genes are mRNAs and not rRNAs, albeit Pol I transcripts, suggesting crossover in Nmd3 function between mRNA and rRNA metabolism in trypanosomes. Presently, we have snapshots of several processes, but lack a holistic view.
Does divergence in mRNA metabolism provide therapeutic opportunities?
Common wisdom suggests that a route to specificity for developing therapeutics against trypanosomes can be achieved via targeting divergent mechanisms. Given both the essentiality of mRNA processing and significant evidence for divergence, the process should be a fertile space. However, for existing therapeutics mechanisms targeting both parasite-specific and conserved pathways have been identified. For example, while both pentamidine and suramin require trypanosome-specific factors [110], drugs targeting proteasomal or N-myristoylation are examples of conserved pathways [111, 112]. In the case of acosiborole, a new therapeutic entering the pipeline for treatment of trypanosomiasis, the target is CPSF73, a component of the mRNA maturation machinery. Specificity apparently arises from minor divergence within the acosiborole binding site [113, 114]. CPSF73 arose in archaea, indicating an origin predating eukaryogenesis, and is highly conserved acres the eukaryotes. Clearly then, mechanistic divergence is not a pre-requisite for therapeutic intervention targeting mRNA export and conserved systems or components offer considerable potential. However, small changes between trypanosomes and host, such as in CPSF3, are challenging to predict.
Concluding remarks
Eukaryotic gene expression involves multiple near-ubiquitous processes, many of which have been inherited directly from prokaryotes. Additionally species-specific proteins and pathways are involved that likely arose as adaptations to the specific biology of each organism. For trypanosomes, current understanding has indicated those areas where mRNA processing/export pathways are clearly modified, and which have probably arisen due to polycistronic transcription and the consequences of that mode of gene expression. mRNA export and processing are Ran-dependent in trypanosomes, representing a fundamental distinction to how the pathways are controlled compared with the canonical pathways of animals, fungi, plants and most other lineages. It is presently unclear if Ran-dependent RNA export represents the ancestral state, which would unite all export under a Ran umbrella, or arose during evolution of the Discoba, the trypanosome lineage. Multiple Mex67 paralogs likewise could represent a basal configuration, but the absence of the canonical cytoplasmic mRNA export/QC platform is clearly a secondary loss. Given many additional examples of highly distinct nuclear functions restricted to kinetoplastids, including the lamina, nuclear basket and kinetochores, together with novel proteins interacting with mRNA processing pathways, how these divergent systems integrate will provide significant insights into the origins of the nucleus and eukaryogenesis itself.
Acknowledgements
We are grateful to the Wellcome Trust (Investigator award 204697/Z/16/Z to MCF), the National Institutes of Health (R01 AI140429-01A1 to MPR and SO) and the National Science Foundation (Proposal 1818129 to SO) for financial support.
References
- 1.Kaufer A, et al. (2017) The evolution of trypanosomatid taxonomy. Parasit Vectors 10, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukes J, et al. (2018) Trypanosomatids Are Much More than Just Trypanosomes: Clues from the Expanded Family Tree. Trends Parasitol 34, 466–480 [DOI] [PubMed] [Google Scholar]
- 3.Maslov DA, et al. (2019) Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146, 1–27 [DOI] [PubMed] [Google Scholar]
- 4.Clayton CE (2016) Gene expression in Kinetoplastids. Curr Opin Microbiol 32, 46–51 [DOI] [PubMed] [Google Scholar]
- 5.Kramer S (2012) Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol 181, 61–72 [DOI] [PubMed] [Google Scholar]
- 6.Wente SR and Rout MP (2010) The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer S (2021) Nuclear mRNA maturation and mRNA export control: from trypanosomes to opisthokonts. Parasitology, 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akey CW, et al. (2022) Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 185, 361–378 e325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, et al. (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampoelz B, et al. (2019) Structure and Assembly of the Nuclear Pore Complex. Annual review of biophysics 48, 515–536 [DOI] [PubMed] [Google Scholar]
- 11.Knockenhauer KE and Schwartz TU (2016) The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 164, 1162–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paci G, et al. (2021) Cargo transport through the nuclear pore complex at a glance. J Cell Sci 134 [DOI] [PubMed] [Google Scholar]
- 13.Rout MP, et al. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Martinez J, et al. (2016) Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcazar-Roman AR, et al. (2006) Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8, 711–716 [DOI] [PubMed] [Google Scholar]
- 16.Folkmann AW, et al. (2011) Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus 2, 540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weirich CS, et al. (2004) The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell 16, 749–760 [DOI] [PubMed] [Google Scholar]
- 18.DeGrasse JA, et al. (2009) Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8, 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegedusova E, et al. (2022) Trafficking and/or division: Distinct roles of nucleoporins based on their location within the nuclear pore complex. RNA Biol 19, 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obado SO, et al. (2016) Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol 14, e1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Gaudenzi J, et al. (2005) RNA-binding domain proteins in Kinetoplastids: a comparative analysis. Eukaryot Cell 4, 2106–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Pablos LM, et al. (2017) Characterization of RBP9 and RBP10, two developmentally regulated RNA-binding proteins in Trypanosoma brucei. Open Biol 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mony BM, et al. (2014) Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature 505, 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mugo E and Clayton C (2017) Expression of the RNA-binding protein RBP10 promotes the bloodstream-form differentiation state in Trypanosoma brucei. PLoS Pathog 13, e1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koumandou VL, et al. (2008) The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manna PT, et al. (2017) Lineage-specific proteins essential for endocytosis in trypanosomes. J Cell Sci 130, 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavez S, et al. (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19, 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rondon AG, et al. (2010) The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochim Biophys Acta 1799, 533–538 [DOI] [PubMed] [Google Scholar]
- 29.Custodio N, et al. (2004) In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA 10, 622–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serpeloni M, et al. (2016) UAP56 is a conserved crucial component of a divergent mRNA export pathway in Toxoplasma gondii. Mol Microbiol 102, 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serpeloni M, et al. (2011) An essential nuclear protein in trypanosomes is a component of mRNA transcription/export pathway. PLoS One 6, e20730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serpeloni M, et al. (2011) Comparative genomics of proteins involved in RNA nucleocytoplasmic export. BMC Evol Biol 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adl SM, et al. (2012) The revised classification of eukaryotes. J Eukaryot Microbiol 59, 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue AH, et al. (2022) Proteomics Uncovers Novel Components of an Interactive Protein Network Supporting RNA Export in Trypanosomes. Mol Cell Proteomics 21, 100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jani D, et al. (2012) Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res 40, 4562–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umlauf D, et al. (2013) The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126, 2656–2667 [DOI] [PubMed] [Google Scholar]
- 37.Aksenova V, et al. (2020) Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat Commun 11, 4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart M (2019) Structure and Function of the TREX-2 Complex. Subcell Biochem 93, 461–470 [DOI] [PubMed] [Google Scholar]
- 39.Wickramasinghe VO, et al. (2010) mRNA export from mammalian cell nuclei is dependent on GANP. Curr Biol 20, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Oliver E, et al. (2012) mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta 1819, 555–565 [DOI] [PubMed] [Google Scholar]
- 41.Koutelou E, et al. (2010) Multiple faces of the SAGA complex. Curr Opin Cell Biol 22, 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goos C, et al. (2019) Trypanosomes can initiate nuclear export co-transcriptionally. Nucleic Acids Res 47, 266–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holden JM, et al. (2014) Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol Biol Cell 25, 1421–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordes VC, et al. (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. The Journal of cell biology 136, 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu XM, et al. (2007) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. The Plant cell 19, 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strambio-de-Castillia C, et al. (1999) Proteins connecting the nuclear pore complex with the nuclear interior. The Journal of cell biology 144, 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niepel M, et al. (2005) The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. The Journal of cell biology 170, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsford S, et al. (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katahira J, et al. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J 18, 2593–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segref A, et al. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16, 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liker E, et al. (2000) The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J 19, 5587–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suyama M, et al. (2000) Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep 1, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams RL, et al. (2014) Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics 197, 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodge CA, et al. (1999) Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J 18, 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodge CA, et al. (2011) The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev 25, 1052–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montpetit B, et al. (2011) A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noble KN, et al. (2011) The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev 25, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weirich CS, et al. (2006) Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 59.Kang Y and Cullen BR (1999) The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev 13, 1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bayliss R, et al. (2002) Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J 21, 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Reilly AJ, et al. (2011) Evolution of the karyopherin-beta family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PloS one 6, e19308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwede A, et al. (2009) The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res 37, 5511–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kramer S, et al. (2010) Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major. BMC Genomics 11, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dostalova A, et al. (2013) The nuclear mRNA export receptor Mex67-Mtr2 of Trypanosoma brucei contains a unique and essential zinc finger motif. Mol Microbiol 88, 728–739 [DOI] [PubMed] [Google Scholar]
- 65.Clouse KN, et al. (2001) A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol 3, 97–99 [DOI] [PubMed] [Google Scholar]
- 66.Teimer R, et al. (2017) A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun 8, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurwitz ME, et al. (1998) Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proceedings of the National Academy of Sciences of the United States of America 95, 11241–11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt C, et al. (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18, 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebenezer TE, et al. (2019) Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol 17, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohler A and Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 71.Fribourg S, et al. (2001) Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8, 645–656 [DOI] [PubMed] [Google Scholar]
- 72.Oeffinger M, et al. (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 73.Bush SJ, et al. (2017) Alternative splicing and the evolution of phenotypic novelty. Philos Trans R Soc Lond B Biol Sci 372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang J, et al. (2001) Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol Cell 8, 397–406 [DOI] [PubMed] [Google Scholar]
- 75.Kerkow DE, et al. (2012) The structure of the NXF2/NXT1 heterodimeric complex reveals the combined specificity and versatility of the NTF2-like fold. J Mol Biol 415, 649–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herold A, et al. (2000) TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol 20, 8996–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Izaurralde E (2002) A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol 81, 577–584 [DOI] [PubMed] [Google Scholar]
- 78.Jun L, et al. (2001) NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr Biol 11, 1381–1391 [DOI] [PubMed] [Google Scholar]
- 79.Tan W, et al. (2005) Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucleic Acids Res 33, 3855–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai D, et al. (2006) The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. RNA 12, 1446–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alber F, et al. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 82.Kneuss E, et al. (2019) Specialization of the Drosophila nuclear export family protein Nxf3 for piRNA precursor export. Genes Dev 33, 1208–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katahira J, et al. (2008) Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs. Nucleic Acids Res 36, 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vanmarsenille L, et al. (2013) Generation and characterization of an Nxf7 knockout mouse to study NXF5 deficiency in a patient with intellectual disability. PloS one 8, e64144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang M, et al. (2007) Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc Natl Acad Sci U S A 104, 10057–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herold A, et al. (2001) NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7, 1768–1780 [PMC free article] [PubMed] [Google Scholar]
- 87.Levesque L, et al. (2001) RNA export mediated by tap involves NXT1-dependent interactions with the nuclear pore complex. J Biol Chem 276, 44953–44962 [DOI] [PubMed] [Google Scholar]
- 88.Wiegand HL, et al. (2002) Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol Cell Biol 22, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santos-Rosa H, et al. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol 18, 6826–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Braun IC, et al. (2002) Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol Cell Biol 22, 5405–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Landeira D and Navarro M (2007) Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. The Journal of cell biology 176, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao W, et al. (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26, 51–62 [DOI] [PubMed] [Google Scholar]
- 93.Faza MB, et al. (2012) Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet 8, e1002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rink C, et al. (2019) The Nuclear Export Receptors TbMex67 and TbMtr2 Are Required for Ribosome Biogenesis in Trypanosoma brucei. mSphere 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rink C and Williams N (2019) Unique Interactions of the Nuclear Export Receptors TbMex67 and TbMtr2 with Components of the 5S Ribonuclear Particle in Trypanosoma brucei. mSphere 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hegedusova E, et al. (2019) The general mRNA exporters Mex67 and Mtr2 play distinct roles in nuclear export of tRNAs in Trypanosoma brucei. Nucleic Acids Res 47, 8620–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inoue AH, et al. (2014) Identification of a novel nucleocytoplasmic shuttling RNA helicase of trypanosomes. PloS one 9, e109521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mishra A, et al. (2021) Selective nuclear export of mRNAs is promoted by DRBD18 in Trypanosoma brucei. Mol Microbiol 116, 827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cuevas IC, et al. (2005) Insights into a CRM1-mediated RNA-nuclear export pathway in Trypanosoma cruzi. Mol Biochem Parasitol 139, 15–24 [DOI] [PubMed] [Google Scholar]
- 100.Fischer U, et al. (1999) Rev-mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle. Nucleic Acids Res 27, 4128–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fischer U, et al. (1994) Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J 13, 4105–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fridell RA, et al. (1996) Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci U S A 93, 4421–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stade K, et al. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 104.Neville M and Rosbash M (1999) The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18, 3746–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ho JH, et al. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. The Journal of cell biology 151, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gadal O, et al. (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21, 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aravind L and Koonin EV (2000) Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res 10, 1172–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buhlmann M, et al. (2015) NMD3 regulates both mRNA and rRNA nuclear export in African trypanosomes via an XPOI-linked pathway. Nucleic Acids Res 43, 4491–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koenig-Martin E, et al. (1992) A procyclin-associated gene in Trypanosoma brucei encodes a polypeptide related to ESAG 6 and 7 proteins. Mol Biochem Parasitol 55, 135–145 [DOI] [PubMed] [Google Scholar]
- 110.Alsford S, et al. (2012) High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bijlmakers MJ (2020) Ubiquitination and the Proteasome as Drug Targets in Trypanosomatid Diseases. Front Chem 8, 630888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frearson JA, et al. (2010) N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature 464, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Begolo D, et al. (2018) The trypanocidal benzoxaborole AN7973 inhibits trypanosome mRNA processing. PLoS Pathog 14, e1007315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wall RJ, et al. (2018) Clinical and veterinary trypanocidal benzoxaboroles target CPSF3. Proc Natl Acad Sci U S A 115, 9616–9621 [DOI] [PMC free article] [PubMed] [Google Scholar]


