Combined anti-spike TCR repertoire and antibody titers reveal differential T- and B-cell responses in a large cohort of patients with B-cell malignancies after 2 and 3 doses of SARS-CoV-2 mRNA vaccines.
Abstract
The anti-spike T-cell and antibody responses to SARS-CoV-2 mRNA vaccines in patients with B-cell malignancies were examined in a real-world setting. A next-generation sequencing (NGS)–based molecular assay was used to assess SARS-CoV-2–specific T-cell responses. After the second dose, 58% (166/284) of seropositive and 45% (99/221) of seronegative patients display anti-spike T cells. The percentage of patients who displayed T-cell response was higher among patients receiving mRNA-1273 vaccines compared with those receiving BNT162b2 vaccines. After the third vaccination, 40% (137/342) of patients seroconverted, although only 22% displayed sufficient antibody levels associated with the production of neutralizing antibodies. 97% (717/738) of patients who were seropositive before the third dose had markedly elevated anti-spike antibody levels. Anti-spike antibody levels, but not T-cell responses, were depressed by B cell–directed therapies. Vaccinated patients with B-cell malignancies with a poor response to SARS-CoV-2 vaccines may remain vulnerable to COVID-19 infections.
Significance:
This study represents the first investigation of SARS-CoV-2–specific immune responses to vaccination in a patient registry using an NGS-based method for T-cell receptor repertoire–based analysis combined with anti-spike antibody assessments. Vaccinated patients with B cell–derived hematologic malignancies are likely at higher risk of infection or severe COVID-19.
This article is highlighted in the In This Issue feature, p. 476
INTRODUCTION
Neutralizing anti-spike protein (anti-S) antibodies to the SARS-CoV-2 virus in response to SARS-CoV-2 vaccination with mRNA vaccines (mRNA-1273 and BNT162b2) correlate with protection against SARS-CoV-2 infection (1, 2). Emerging evidence suggests that viral antigen-specific T cells can also contribute to protection from SARS-CoV-2 and SARS-CoV-2 variants of concern (reviewed in ref. 3). Moreover, unique methods that facilitate the analysis of T-cell responses in larger patient populations may provide actionable insights for public health (3).
Although immune responses to SARS-CoV-2 mRNA vaccines in healthy individuals are robust, they are reduced in patients with hematologic malignancies (4). However, the correlation between humoral and cellular responses with COVID-19 vaccine efficacy in patients with hematologic malignancies is not well studied (4). We and others reported that serological responses to mRNA vaccinations are impaired in patients with B cell–derived malignancies due to the disease itself and the use of B cell–suppressive therapies (5, 6). Recent data suggest that breakthrough SARS-CoV-2 infections are more common in patients with hematologic malignancies than in healthy individuals (7, 8) and may be more likely among patients with low levels of anti-S antibodies (9).
In this report, we use sequencing of the T-cell receptor (TCR) repertoire from peripheral blood and the enrichment of SARS-CoV-2–specific TCRs (10) to characterize T-cell responses after two doses of mRNA vaccines in patients with B cell–derived malignancies. T-cell responses in this patient population have mostly been characterized by IFNγ production (11–14). Our second effort focused on the humoral response to a third mRNA vaccination across a broad range of hematologic malignancies, which builds upon our previous reports from us and others (5, 15).
RESULTS
Patients with hematologic malignancies who previously were examined for the anti-S antibody response (5) provided additional blood samples for the analysis of SARS-CoV-2–specific T-cell responses after two doses of mRNA vaccines (Supplementary Fig. S1). Samples from nine patients with confirmed SARS-CoV-2 infection or anti-nucleocapsid (anti-N) antibodies were excluded from the analysis. The remaining 505 patients in the analyzed cohort predominantly had chronic lymphocytic leukemia (CLL; n = 285) and non-Hodgkin lymphoma (NHL; n = 154). Most patients (n = 338) were 65 years of age or older. Samples were roughly evenly split between the two mRNA vaccines (mRNA-1273 or BNT162b2; Supplementary Table S1). Immunosequencing of the CDR3 regions of human TCRβ chains on genomic DNA from peripheral blood samples and identification of SARS-CoV-2–specific TCRs were performed as described in Methods. Samples were classified as positive or negative for enrichment of SARS-CoV-2 T cells using Adaptive's T DETECT technology, which underlies the FDA-approved T-Detect assay (16). Briefly, the T-MAP COVID classifier was built using infected and convalescent subjects compared with prepandemic control samples and has a 99.8 target specificity with a range of 75% to 100% sensitivity depending on time since infection (10). As shown in Table 1, of 284 seropositive patients, 58% had a positive call, whereas 34% had a negative call, and 8% had an insufficient number of T-cell rearrangements to make a definitive negative call with the classifier (labeled “No call” in Table 1). All samples including the “No call” samples passed all standard sequencing QC metrics, but “No call” samples had a significantly lower T-cell fraction compared with other samples, likely due to their hematologic disease or prior therapy (P < 1e−15, Wilcox rank sum; Supplementary Table S2). Therefore, we grouped “No call” samples with other seronegative results to capture the entire patient population likely at an elevated risk of infection or severe disease due to reduced vaccine immunogenicity. Among the 221 seronegative patients, 45% had a positive T-cell call, 37% had a negative T-cell call, and 18% had “No call” (Table 1). Thus, the percentage of patients with a positive T-cell call was higher among seropositive patients (58%) than among seronegative patients (45%; P = 0.003, Fisher exact test), suggesting a positive relationship between positive T-cell calls and anti-S antibody response. Notably, the percentage of seropositive hematologic malignancy patients with a positive T-cell call was lower compared with healthy populations using a range of T-cell assessments (17–19). The percentage of patients with a positive T-cell call was higher among patients receiving mRNA-1273 vaccines than among those receiving BNT162b2 vaccines (P = 0.001 Fisher exact test; Table 1). A trend suggestive of differences between the types of mRNA vaccines persisted when subgroups of patients diagnosed with CLL or NHL were examined (Supplementary Table S3).
Table 1.
T-cell responses by mRNA vaccine type
| T-cell nonpositive | |||||
|---|---|---|---|---|---|
| Vaccine | Anti-spike antibody (AU/mL) | T-cell positive | T-cell negative | T-cell no call | Subtotal |
| Total | <0.8 (221) | 99 (45%) | 82 (37%) | 40 (18%) | 122 (55%) |
| N = 505 | ≥0.8 (284) | 166 (58%)a | 96 (34%) | 22 (8%) | 118 (42%) |
| mRNA-1273 | <0.8 (87) | 45 (52%)b | 27 (31%) | 15 (17%) | 42 (48%) |
| N = 236 | ≥0.8 (149) | 97 (65%) | 37 (25%) | 15 (10%) | 52 (35%) |
| BNT162b | <0.8 (134) | 54 (40%) | 55 (41%) | 25 (19%) | 80 (60%) |
| N = 269 | ≥0.8 (135) | 69 (51%) | 59 (44%) | 7 (5%) | 66 (49%) |
NOTE: SARS-CoV-2–specific T-cell responses were categorized by anti-S antibody levels and mRNA vaccine type. Anti-S antibody levels were analyzed by the semiquantitative Elecsys anti-SARS-CoV-2 S enzyme immunoassay using patient sera. Immunosequencing of the CDR3 regions of human T-cell receptor (TCR) beta chains on genomic DNA from peripheral blood samples and identification of SARS-CoV-2–specific TCRs were performed as described in Methods. Peripheral blood samples for TCR sequencing were collected a median of 147 days after the second dose of vaccination [interquartile range (IQR): 129–164 days]). A classifier trained to diagnose SARS-CoV-2 infection (10) was used to categorize SARS-CoV-2-specific T-cell response calls. “No call” resulting from the insufficient number of T cells in the patient sample for a definitive negative call were grouped together with negative call as “Nonpositive” to reflect the patient population with reduced T-cell activation after two doses of vaccination.
aSignificantly different compared with <0.8 AU/mL, T cell–positive value for the total cohort (P = 0.003, Fisher exact t test).
bSignificantly different compared with <0.8 AU/mL, T cell–positive value for the BTN162b2 cohort (P = 0.001 Fisher exact t test.
To further assess patients likely to be at high risk of infection or severe COVID disease due to insufficient antibody and T-cell responses, we evaluated the data set grouping samples with anti-S antibody levels below 100 AU/mL together with seronegative and “No call” samples. We chose this value based on recent reports indicating that only anti-S antibody levels above 100 AU/mL were associated with protective anti-S antibody titers in primates and conferred 50% protective neutralization in a clinical cohort (20). Supporting this hypothesis, little or no neutralizing antibody activity was detected in several studies in SARS-CoV-2–infected patients or vaccinated individuals with anti-S antibody levels of approximately 30–100 AU/mL (21–23). In this modified analysis, we obtained a similar result finding that 56% (168/301) of patients with no or low anti-S antibodies are likely to lack protective T-cell responses.
Patient age did not significantly affect the T-cell response (Supplementary Table S4). In addition, we examined if the T-cell response correlated with time since the second vaccination. Most patient samples for T-cell analysis were collected 129 to 164 days (25th–75th percentiles) after the second vaccination; this time should reflect more persistent T-cell responses after the initial peak of an antigen-elicited T-cell response (10). We found little change in T-cell positivity in relation to the sampling time for the mRNA-1273 vaccine (Supplementary Table S5). T-cell positivity trended lower beyond 164 days in patients with BNT162b2, but this difference did not reach statistical significance (Supplementary Table S5).
We did not observe an effect of treatment with BTK inhibitors, anti-CD20 antibodies, or venetoclax on T-cell positivity (Supplementary Table S6), consistent with a recent report (11). Very few patients in our study reported active chemotherapy or steroid treatment, the two types of therapies that have been reported to impact T-cell function (11, 13). Therefore, the potential impact of these therapies could not be analyzed.
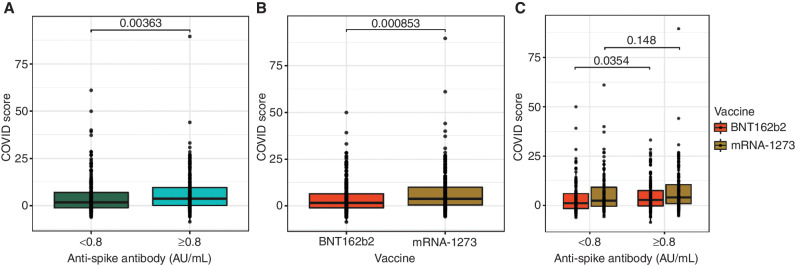
To further the understanding of T-cell responses, we analyzed the COVID score, which is a quantitative value derived from the logistic regression classifier and describes the strength of the overall enrichment of SARS-CoV-2–specific T cells in an antigen-independent manner (16). COVID scores were higher among seropositive compared with seronegative patients after two doses of SARS-CoV-2 mRNA vaccines (P = 0.0036, Wilcoxon rank-sum test; Fig. 1A). Interestingly, COVID scores were higher in patients who received mRNA-1273 than in those who received BNT162b2 vaccine doses (P = 0.0008, Wilcoxon rank-sum test; Fig. 1B and C).
Figure 1.
Association between T-cell COVID scores and anti-S antibody levels (A), type of mRNA vaccine (B), and both anti-S antibody levels and type of mRNA vaccine (C). Anti-S antibody levels were assessed by the semiquantitative Elecsys anti-SARS-CoV-2 S enzyme immunoassay using patient sera. COVID scores were obtained as defined in Methods. The horizontal lines represent median, boxes represent interquartile range, and symbols represent each patient. P values were determined using the Wilcox rank-sum test.
Another indicator of T-cell activation is the breadth of spike-specific TCRs, defined as the number of spike-specific TCRs divided by the number of TCRs in the repertoire as a whole. We compared the published mapping of TCRs selectively identified in SARS-CoV-2–infected individuals versus noninfected controls (10, 16) to the TCRs enriched in our cohort. As expected, increased T-cell breadth was observed for TCRs associated with responses to SARS-CoV-2 spike protein and not for TCRs associated with responses to other viral ORFs, whereas healthy controls who developed COVID-19 infections demonstrated T-cell responses to other SARS-CoV-2 antigenic proteins as well (Supplementary Fig. S2). We observed a greater breadth of spike protein–associated T-cell responses in seropositive patients compared with seronegative patients (Supplementary Fig. S3A; P = 0.00873, Wilcoxon rank-sum test). This correlation was preserved when we further analyzed the breadth of spike protein–associated CD4 and CD8-specific T-cell responses (Supplementary Fig. S3A–S3C). The median CD4 signal was higher than the median CD8 signal (3.02e−5 vs. 0). This finding is likely based on both T-cell biology (more HLA degeneracy and greater CD4 response) and the used technology (Adaptive's CD4 panel has longer peptides compared with the CD8 panel) and is consistent with published literature that SARS-CoV-2 vaccines elicit a stronger CD4 response (24, 25) that is associated with a lower incidence of prolonged COVID-19 infections (26). We also observed a trend for increased breadth of spike-specific T cells in response to vaccination with mRNA-1273 compared with the BNT162b2 vaccine. This trend was preserved in CD4 T-cell responses but not in CD8 T-cell responses (Supplementary Fig. S3D–S3F), largely due to the overall smaller CD8 signal with the used method. Non-spike protein–associated T-cell responses were not affected by anti-S antibody levels nor types of mRNA vaccines (Supplementary Fig. S3G and S3H).
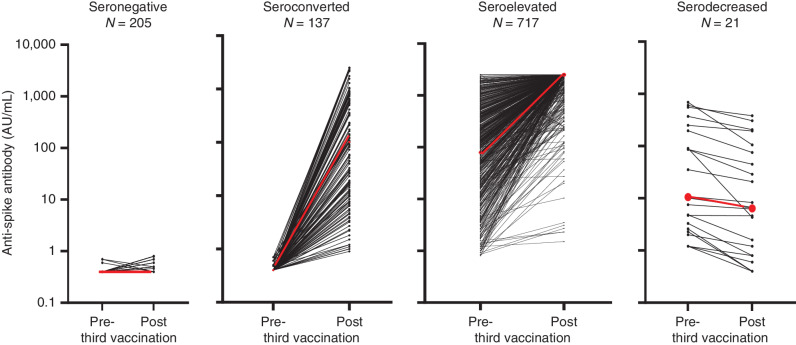
To address our second question, we compared anti-S antibody responses of 1,080, anti–N-negative patients after the second and the third dose of SARS-CoV-2 mRNA vaccine (Supplementary Fig. S1). Ninety-nine percent of these patients received SARS-CoV-2 mRNA vaccines as the initial vaccination, whereas all of these patients received SARS-CoV-2 mRNA vaccines as the third vaccination (Supplementary Table S7). The cohort was predominantly composed of patients with B-cell malignancies (Supplementary Table S8). Anti-S antibody values were obtained at a median of 112 AU/mL [interquartile range (IQR), 52–144] and 29 AU/mL (IQR, 8–34) days after the second and third vaccinations, respectively. Of the 1,080 patients who had third vaccinations, 32% were seronegative (i.e., <0.8 AU/mL), whereas 68% were seropositive for anti-S antibodies prior to the third vaccination (Supplementary Fig. S1). Four main types of serological responses were observed after the third SARS-CoV-2 mRNA vaccine dose (Fig. 2; Supplementary Fig. S1). Among patients who were seronegative after the second vaccination, 40% achieved a vaccine-induced antibody response (seroconverted) after the third vaccination with a wide range of anti-S antibody levels (median, 101 AU/mL: IQR, 9.95–826 AU/mL). Forty-five percent of these patients had anti-S antibody levels below 100 AU/mL. When we grouped these patients with low antibodies together with seronegative patients, only 22% of patients who had no detectable anti-S antibodies after the second vaccination produced antibody levels >100 AU/mL after the third SARS-CoV-2 mRNA vaccine dose.
Figure 2.
The serological response to second and third vaccinations in individual patients with hematologic malignancies was analyzed for anti-S antibodies as described in the Methods. Individual lines represent each patient. Four types of responses were observed. Those patients who were seronegative prior to the third vaccination either remained persistent seronegative or seroconverted. Those patients who were seropositive prior to the third vaccination either had an increase in anti-S antibody levels (seroelevated) or had a small decrease in anti-S antibody levels (serodecreased). The red line in each graph represents the median anti-S antibody level.
Two types of responses were observed in patient who were seropositive after the second vaccination (Fig. 2; Supplementary Fig. S1). Ninety-seven percent of these patients had sustained high or markedly elevated levels of anti-S antibodies after the third vaccination (seroelevated). The median anti-S antibody levels before and after the third vaccination were 231 AU/mL and 2,500 AU/mL (IQRs, 46–638 AU/mL and 2,500–2,500 AU/mL, respectively). Within the seroelevated group, only 2.4% had anti-S antibody levels <100 AU/mL after the third vaccination. In the last group, a few patients experienced a serodecreased response, but these patients typically had low anti-S antibody levels after the second vaccine.
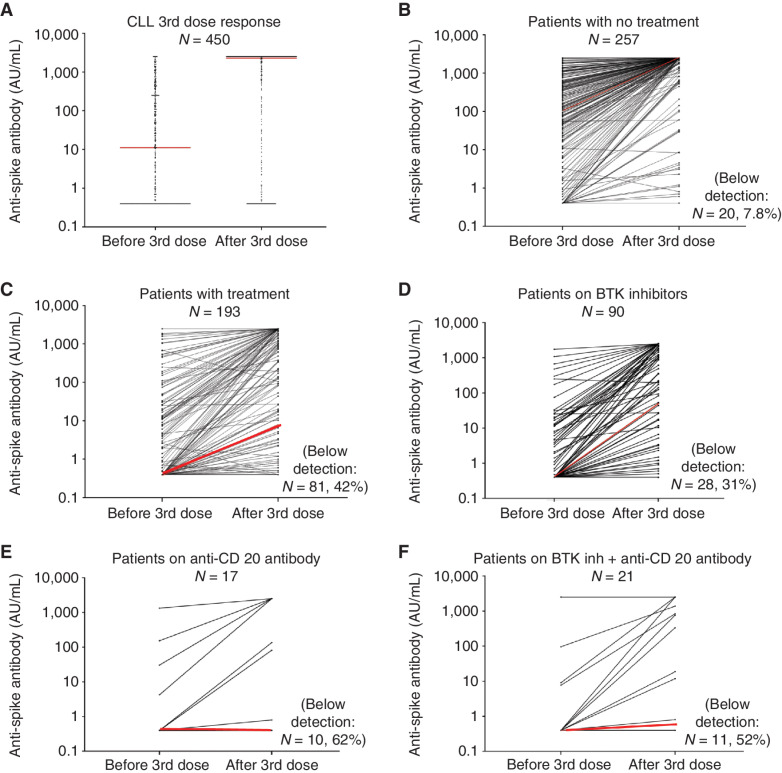
The distribution of the four response groups by disease type is shown in Supplementary Fig. S4. In general, in our cohort impaired responses to the third vaccination were greatest in patients with B cell–derived malignancies, except for patients with multiple myeloma or Hodgkin lymphoma, who responded well. The data for a large population of patients with CLL (N = 450) allowed us to examine response rates by treatment types when examining individual patients (Fig. 3) or as a group (Supplementary Table S9). Patients were typically treated with therapies that included anti-CD20 antibodies or BTK inhibitors. Although the magnitude of the third dose response was high at the population level (Fig. 3A), responses for individual CLL patients varied depending on the type of treatment (Fig. 3B–F). Most patients who had no treatment demonstrated a dramatic increase in anti-S antibodies [median, 112 AU/mL (IQR, 2–472 AU/mL) to 2,500 AU/mL (IQR, 1,826–2,500 AU/mL), before and after the third SARS-CoV-2 mRNA vaccine dose, respectively; Fig. 3B]. In contrast, patients on treatment had a smaller increase in anti-S antibodies [median, 0.4 AU/mL (IQR, 0.4–8.5 AU/mL) to 10 AU/mL (IQR, 10–1,428 AU/mL)] before and after the third SARS-CoV-2 mRNA vaccine dose, respectively; Fig. 3C). The anti-S antibody response was reduced among individual patients treated with B cell–suppressive or depleting therapies, including BTK inhibitors (Fig. 3D), anti-CD20 antibodies (Fig. 3E), or both (Fig. 3F). The response was highly variable in patients treated with BTK inhibitors as a monotherapy [median, 0.4 AU/mL (IQR, 0.4–7 AU/mL) to 49 AU/mL (IQR, 0.4–1,392 AU/mL)], before and after the third SARS-CoV-2 mRNA vaccine dose, respectively; Fig. 3D). In contrast, two distinct types of responses were observed in patients treated with anti-CD20 antibodies. Many patients had a weak anti-S antibody response [median, 0.4 AU/mL (IQR, 0.4–4.2 AU/mL) to 0.4 AU/mL (IQR, 0.4–1,384 AU/mL)] before and after the third SARS-CoV-2 mRNA vaccine dose, respectively; Fig. 3E. In contrast, 5 of 7 patients who had very good antibody responses reported that they had been treated with anti-CD20 antibodies 1 to 2 years prior to the third vaccination. A similar response profile was found in patients with Waldenstrom's macroglobulenima (Supplementary Fig. S5A and S5B) who were either not treated or treated with similar therapies as CLL patients (Supplementary Fig. S5C and S5D; Supplementary Table S9). The third largest subset of NHL patients was diagnosed with follicular lymphoma. Treatment within anti-CD20 antibodies was also associated with an impaired serological response to vaccines in these patients (Supplementary Table S9). 75% (12/16) who remained seronegative after third vaccination had been treated with an anti-CD20 antibody with a median of 185 days prior to the third vaccine, whereas 9 patients who had a marked increase in the anti-S antibody levels had been treated with an anti-CD20 antibody with a median of 393 days prior to the third vaccination.
Figure 3.
The anti-S antibody level in CLL patients was assessed as described in the Methods. Overall response in patients (A); individual dots represent each patient. Response of individual patients before and after the third vaccination for patients with no treatment (B) or treatment (C). Patients treated with BTK inhibitors (D), anti-CD20 antibodies (E), or a combination (F) are shown. Individual lines represent each patient (B–E). Red lines indicate the median response.
DISCUSSION
Here we report data on SARS-CoV-2 mRNA vaccination–induced immune responses obtained from our registry of patients with hematologic malignancies. We demonstrate the utility of a molecular assay for T-cell analysis, based on sequencing of TCR beta chains (10), in a real-world setting. More than half of the patients in this cohort with no detectable anti-S antibodies (55%, 122/221) also did not exhibit spike protein-directed T-cell responses to two full doses of the SARS-CoV-2 mRNA vaccines. These findings are consistent with other reports in patients with cancer (4, 11–14). Our work and published studies also identify patients who have limited or no anti-S antibody response due to anti-CD20 treatments but are able to mount T-cell responses to SARS-CoV-2 vaccination (11, 12, 26–29). Such patients might experience protection against poor outcomes after SARS-CoV-2 infections based on their T-cell responses. For a subset of 279 patients, we examined a potential relationship between T-cell positivity after the second dose and antibody responses after the third dose of mRNA vaccine. However, the rates of seroconversion after the third dose were similar for patients with positive and negative T-cell responses. Similarly, others have reported that only 5% (2/42) of immunocompromised patients developed higher cellular responses after the third vaccination compared with the primary vaccination (30). Sample size for the analysis of the T-cell response to a third dose of vaccine was too small for a thorough analysis.
The importance of T-cell immune responses as contributors to resolution of SARS-CoV-2 infections is supported by the observation that patients with X-linked agammaglobulinemia, who are genetically incapable of B-cell production but mount T-cell responses, can clear SARS-CoV-2 infection (31). The preservation of T-cell responses may be particularly critical in the context of SARS-CoV-2 variants of concern (4). T-cell epitopes demonstrate a higher level of conservation between viral variants than B-cell epitopes, and cross-reactive T cells may provide protection from SARS-CoV-2 variants of concern (32). Our data suggest that the mRNA-1273 vaccine elicits a stronger T-cell and antibody response than the BNT162b2 mRNA vaccine, a difference we and others have observed previously in the production of anti-S antibodies (4, 5). These differences may be attributed to the larger amount of mRNA within the dose of mRNA-1273 (100 μg) versus BNT162b2 (30 μg), other components of these products, or dose scheduling differences (33). Additional factors contributing to different T-cell responses in patients with hematologic malignancies remain under active investigation.
The immunosequencing assay used here provides a practical method to measure vaccine-induced T-cell responses in a real-world setting. This method is based on the Multiplex Identification of Antigen-Specific TCR Assay (34) applied to SARS-CoV2 antigens (10, 16). It was also recently used to characterize T-cell responses in patients with cancer with delayed clearance of SARS-CoV-2 infection (26). Classic assessments of T-cell responses using functional assays such as ELISpot, intracellular cytokine staining, or activation-induced marker assays typically require extensive handling of live cells, immediate processing, temperature control of samples, restimulation and incubation, and are generally not amenable to implementation in population-level studies (discussed in ref. 3). Although the use of multimeric reagents for the detection of antigen-specific T cells does not require restimulation of cells, the HLA specificity of these reagents limits their wider scale use. Moreover, none of these assays capture all aspects of relevant T-cell function related to the breadth of cytokine production and cell killing ability. In contrast, the immunosequencing-based T-cell assay used here is based on DNA, which is extremely stable, has demonstrated high sensitivity, and is easily amenable to standardization. The method is robust and easily scalable. It can, therefore, be applied in real-world settings to provide actionable insights to inform public health strategies. Although this assay does not provide a direct functional assessment of the T-cell response, it captures the clonal breadth of SARS-CoV-2–specific T cells, which is a measure of T-cell activation and proliferation.
Our data support the utility of a third vaccination for patients with B-cell malignancies. Seroconversion (values >0.8 AU/mL) occurs in approximately 40% of patients who failed to produce anti-S antibodies after a second SARS-CoV-2 mRNA vaccine dose. However, when a minimum anti-S antibody level of 100 AU/mL was used to define a meaningful serologic response associated with detection of neutralizing antibodies based on previous work (21–23), only 22% of patients acquired meaningful serological responses after the third dose (T-cell response positivity rate was not altered). In contrast, 97% of patients who were seropositive after the second mRNA vaccine dose experienced persistently high or increased anti-S antibodies following the third dose. The production of anti-S antibodies after the third dose I is likely mediated, in part, by the activation of memory B cells or long-lived plasma cells in the bone marrow (35, 36). The reconstitution of normal B cells after prolonged B-cell suppression may also contribute to the third dose responses in patients who had completed treatment with anti-CD20 antibodies therapy approximately one year prior to vaccination, as has been described by others (27–29). In our study, the response to vaccination in patients on BTK inhibitors was variable. This may be related to the ability of BTK inhibitors to decrease naïve B-cell counts (37), yet promote helper T cells, which could lead to an overall enhanced immune response (38). Although others have reported lower seroconversion rates (20%–24%) in patients with CLL (12, 39) compared with our results (34%; Supplementary Table S9), higher rates of seroconversion (30%–60%) have been reported in patients with lymphoid malignancies or solid organ transplants who are immunosuppressed (19, 40). The basis for the differences needs further investigation, but may include differences in vaccine type, time of analysis after vaccination, the specific population studied, percentage of patients on active therapies, and the use of anti-S assays with different performance characteristics.
This study has limitations. First, the diagnoses and treatments were self-reported by patients. However, our conclusions are consistent with the T-cell and anti-S antibody response profiles reported by others under more controlled conditions (6). Second, our data were obtained before January 2022, and therefore, mostly prior to the emergence of the SARS-CoV-2 the initial Omicron variant (December 2021). The neutralizing capacity of anti-S antibodies induced by SARS-CoV-2 vaccination is reduced for infections with Omicron and subvariants compared with the prior SARS-CoV-2 variants for both healthy individuals (41–43) and patients with hematologic malignancies (44). This suggests an amplified risk of Omicron or future variant infections in this patient population. Third, our analysis did not examine neutralizing antibody levels. Although the titer of neutralizing antibodies tends to be correlated with anti-S antibody levels in patients with blood cancer, this correlation is only observed for anti-S antibody levels above a threshold of above approximately 30 to 100 AU/mL (21–23). The correlation between neutralizing antibody levels and protection against SARS-CoV-2 infection, particularly against new SARS-COV-2 variants, is not well defined. Finally, the impact of deficient anti-S antibody and T-cell responses on the likelihood of breakthrough infections in hematologic patients needs further study.
In summary, we identify patients with hematologic malignancies with no or low spike-directed T-cell and antibody responses after two SARS-CoV-2 mRNA vaccinations, and those with a persistent lack of serological responses after three vaccinations. Such patients may be at the greatest risk of infection or severe COVID-19. Alternative preventive and therapeutic approaches, including the use of monoclonal antibodies or antiviral drugs, may be justified in patients with hematologic malignancies.
METHODS
Participation in the study was open to all patients diagnosed with hematologic malignancies. Data were collected using The Leukemia and Lymphoma Society National Registry with written informed consent and sourced by Ciitizen/Invitae (https://www.ciitizen.com/lls/; NCT04794387, NCT04806295, and NCT04898985). Data were collected between May 2021 and December 2021, including demographics, diagnosis, treatments within the past two years, vaccination type, and dates as self-reported by the patients. Anti-S antibody levels were assessed by the semiquantitative Elecsys anti-SARS-CoV-2 S enzyme immunoassay for the detection of IgG anti-S1-RBD-SARS-CoV-2 (range, 0.8–2,500 AU/mL; patients who had <0.8 AU/mL were considered seronegative) as described previously (5). Simultaneously, the detection of high-affinity antibodies to the SARS-CoV-2 nucleocapsid protein (anti-N) was assessed (LabCorp test 164068) to determine if patients might have an infection close to the time of the second or third vaccination. Patients were informed of their serological results, as stated in the IRB consent for this trial. The median time for serological assessment was 64 days (IQR, 61–74 days) before and 29 days (IQR, 26–34 days) after the third dose of the mRNA vaccine. Only 26 patients received the adenoviral-based vaccine AD26COV-2 and the associated data were removed from the analysis.
TCR Variable Beta Chain Sequencing
Immunosequencing of the CDR3 regions of human TCRβ chains was performed on genomic DNA from PBMC samples using the ImmunoSEQ Assay (Adaptive Biotechnologies). ImmunoSEQ is only for research use and not for use in diagnostic procedures. The extracted genomic DNA was amplified using bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered to identify and quantify the absolute abundance of each unique TCRβ CDR3 region for further analysis, as previously described (45–47). Standard sample-level QC was performed (47), and additional pairwise repertoire comparisons were made to confirm that no samples showed evidence of unintended material transfer between different individuals. The fraction of T cells was calculated by normalizing TCR-β template counts to the total amount of DNA usable for TCR sequencing. The amount of usable DNA was determined by PCR amplification and sequencing of several reference genes that are expected to be present in all nucleated cells.
Mapping of SARS-CoV-2 TCRβ Sequences
TCR sequences from peripheral repertoires were mapped against a set of TCR sequences known to react with SARS-CoV-2. Briefly, SARS-CoV-2–specific sequences were identified at Adaptive by multiplex identification of TCR antigen specificity, which identifies TCRs stimulated by particular antigens as well as identifies MHC class presentation (34). TCRs that expanded in response to stimulation were further screened for enrichment in COVID-19–positive repertoires collected as part of immuneCODE (10) compared with COVID-19–negative repertoires to remove TCRs that may be highly public or cross-reactive to common antigens. The filtered list represents a set of TCRs that are both experimentally observed expanding to COVID antigens as well as enriched in COVID-19 subjects (16). Individual response to vaccination can be quantified by the relative number (breadth) and/or sum frequency (depth) of SARS-CoV-2 TCRs observed post-vaccination. TCRs were further stratified for specific viral ORF based on MIRA associations, as well as MHC presentation.
All samples were classified as positive or negative for the detection and enrichment of COVID-specific T cells using Adaptive's T DETECT COVID classifier (ref. 16; https://www.fda.gov/media/146481/download). The classifier uses a simple 2-feature logistic regression, with independent variables E and N, where E is the number of unique TCR-β DNA sequences that encode an enhanced sequence and N is the total number of unique productive TCR-β DNA sequences in that subject. We define the COVID score to be the log-odds of the probability of this logistic regression model. The classifier was trained comparing peripheral repertoires from COVID+ and convalescent subjects with control samples collected pre-pandemic (10). T-cell responses are categorized as negative, positive, and “No call” (representing samples with an insufficient number of T-cell rearrangements to make a definitive negative call). For categorical T-cell response variables, Fisher exact test was used to compare across patient groups and determine statistically significant differences. ImmuneCODE data resources, including the COVID-19 MIRA data and COVID-19 study immunosequencing data, are available for analysis and downloaded from the Adaptive Biotechnologies immuneACCESS site under the immuneACCESS Terms of Use at https://clients.adaptivebiotech.com/pub/covid-2020 (16). ImmuneCODE is used only for research purposes and is not used in diagnostic procedures.
Statistical Analyses
Anti-SARS-CoV-2 antibody levels are reported as a continuous variable in arbitrary units or as a categorical variable using predefined cutoff points. Categorical variables for serostatus are presented as seronegative (<0.8 AU/mL) or seropositive (≥0.8 AU/mL) and categorical variables for the change in anti-SARS-CoV-2 antibody levels between doses two and three of the SARS-CoV-2 mRNA vaccine are presented as persistent seronegative (patient remained seronegative), serodecreased (patient was seropositive and demonstrated a reduced in antibody levels after third vaccine dose), seroconverted (patient was seronegative and subsequently achieved a vaccine-induced increase in antibody levels after third vaccine dose), or seroelevated (patient was seropositive and achieved a vaccine-induced increase in antibody levels after third vaccine dose). For categorical serostatus variables, Fisher exact test was used to compare groups and determine statistically significant differences. For continuous serostatus variables, a two-sample log-rank test with hypergeometric variance was used to compare the groups and determine statistically significant differences.
Descriptive statistics are presented as frequencies and percentages. The reported P values are two-sided. An imputation program was not applied to the data, and differences in sample size due to missing data are reported. No outliers were removed, and all data are shown. Analyses were univariate, and P values are unadjusted. All T-cell statistics were run in R 4.1.x.
Data Availability
T-cell repertoire profiles and antigen annotation data from multiplexed antigen-stimulation experiments are available through the ImmuneCODE resource and can be downloaded from the Adaptive Biotechnologies immuneACCESS site under the immuneACCESS Terms of Use at clients.adaptivebiotech.com/pub/greenberger-2022-bcd (doi: 10.21417/LMG2022BCD).
Supplementary Material
Acknowledgments
This study was supported by The Leukemia and Lymphoma Society. We thank our generous donors and patients who have participated in the LLS National Registry, a project of the Michael J. Garil Data Collaborative. We thank Ciitizen, a division of Invitae, and LabCorp for their collaboration in this effort, as well as Drs. Neil Kay (Mayo Clinic) and Barbara Banbury (Adaptive Biotechnologies) for their contribution to this research effort.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
L.M. Greenberger reports grants from BeiGene, Pharmacyclics an AbbVie Company, Kite a Gilead Company, and Janssen Biotech during the conduct of the study; grants from Bristol Myers Squibb and Pfizer outside the submitted work; and employees from Adaptive Biosciences (Fields and Sanders) are authors on this manuscript, participated in generating data, and providing data analysis. This work at Adaptive was paid for by the Leukemia and Lymphoma Society. Employees from Ciitizen, a division of Invitae, were contracted by LLS to collect data that entered the patient registry. This service was paid for solely by the Leukemia and Lymphoma Society. No Invitae employee is listed as an author. P.A. Fields reports a financial interest in Adaptive Biotechnologies. C. Sanders reports other support from Adaptive Biotechnologies outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
L.M. Greenberger: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing. L.A. Saltzman: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, project administration. L.M. Gruenbaum: Data curation, formal analysis, supervision, writing–original draft, writing–review and editing. J. Xu: Data curation, formal analysis. S.T. Reddy: Data curation, visualization. J.W. Senefeld: Data curation, formal analysis, supervision, writing–review and editing. P.W. Johnson: Formal analysis. P.A. Fields: Data curation, software, formal analysis, visualization, methodology, writing–review and editing. C. Sanders: Data curation, software, visualization, writing–review and editing. L.J. DeGennaro: Writing–review and editing. G.L. Nichols: Supervision, writing–review and editing.
References
- 1. Gilbert PB, Montefiori DC, McDermott AB, Fong Y, Benkeser D, Deng W, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022;375:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021;7:1205–11. [DOI] [PubMed] [Google Scholar]
- 3. Vardhana S, Baldo L, Morice WG II, Wherry EJ. Understanding T cell responses to COVID-19 is essential for informing public health strategies. Sci Immunol 2022;7:eabo1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fendler A, de Vries EGE, GeurtsvanKessel CH, Haanen JB, Wörmann B, Turajlic S, et al. COVID-19 vaccines in patients with cancer: immunogenicity, efficacy and safety. Nat Rev Clin Oncol 2022;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greenberger LM, Saltzman LA, Senefeld JW, Johnson PW, DeGennaro LJ, Nichols GL. Antibody response to SARS-CoV-2 vaccines in patients with hematologic malignancies. Cancer Cell 2021;39:1031–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ribas A, Dhodapkar MV, Campbell KM, Davies FE, Gore SD, Levy R, et al. How to provide the needed protection from COVID-19 to patients with hematologic malignancies. Blood Cancer Discov 2021;2:562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mittleman N, Magen O, Barda N, Dagan N, Oster HS, Leader A, et al. Effectiveness of the BNT162b2mRNA covid-19 vaccine in patients with hematological neoplasms. Blood 2022;139:1439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song Q, Bates B, Shao YR, Hsu FC, Liu F, Madhira V, et al. National COVID Cohort Collaborative Consortium. Risk and outcome of breakthrough COVID-19 infections in vaccinated patients with cancer: real-world evidence from the National COVID Cohort Collaborative. J Clin Oncol 2022;40:1414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pagano L, Salmanton-García J, Marchesi F, Lopez-Garcia A, Lamure S, Itri F, et al. COVID-19 in vaccinated adult patients with hematological malignancies: preliminary results from EPICOVIDEHA. Blood 2022;139:1588–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R, et al. Magnitude and dynamics of the T cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv 2020. doi: 10.1101/2020.07.31.20165647. [Google Scholar]
- 11. Ehmsen S, Asmussen A, Jeppesen SS, Nilsson AC, Osterlev S, Vesteraard H, et al. Antibody and T cell immune responses following mRNA COVID-19 vaccination in patients with cancer. Cancer Cell 2022;39:1034–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parry H, Bruton R, Roberts T, McIlroy G, Damery S, Sylla P, et al. COVID-19 vaccines elicit robust cellular immunity and clinical protection in chronic lymphocytic leukemia. Cancer Cell 2022;40:584–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiménez M, Roldán E, Fernández-Naval C, Villacampa G, Martinez-Gallo M, Medina-Gil D, et al. Cellular and humoral immunogenicity of the mRNA-1273 SARS-CoV-2 vaccine in patients with hematologic malignancies. Blood Adv 2022;6:774–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Re D, Seitz-Polski B, Brglez V, Carles M, Graça D, Benzaken S, et al. Humoral and cellular responses after a third dose of SARS-CoV-2 BNT162b2 vaccine in patients with lymphoid malignancies. Nat Commun 2022;13:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenberger LM, Saltzman LA, Senefeld JW, Johnson PW, DeGennaro LJ, Nichols GL. Anti-S antibody response to SARS-CoV-2 booster vaccination in patients with B cell-derived hematologic malignancies. Cancer Cell 2021;39:1297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nolan S, Vignali M, Klinger M, Dines JN, Kaplan IM, Svejnoha E, et al. A large-scale database of T cell receptor beta (TCRβ) sequences and binding associations from natural and synthetic exposure to SARS-CoV- 2. Res Sq 2020. doi: 10.21203/rs.3.rs-51964/v1. [Google Scholar]
- 17. Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020;586:594–602. [DOI] [PubMed] [Google Scholar]
- 18. Alter G, Yu J, Liu J, Chandrashekar A, Borducchi EN, Tostanoski LH, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 2021;596:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swanson PA 2nd, Padilla M, Hoyland W, McGlinchey K, Fields PA, Bibi S, et al. AstraZeneca/Oxford/VRC Study Group. AZD1222/ChAdOx1 nCoV-19 vaccination induces a polyfunctional spike protein-specific TH1 response with a diverse TCR repertoire. Sci Transl Med 2021;13:eabj7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall VG, Ferreira VH, Ku T, Ierullo M, Majchrzak-Kita B, Chaparro C, et al. Randomized trial of a third dose of mRNA-1273 vaccine in transplant recipients. N Engl J Med 2021;385:1244–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rubio-Acero R, Castelletti N, Fingerle V, Olbrich L, Bakuli A, Wölfel R, et al. KoCo19 study team. In search of the SARS-CoV-2 protection correlate: head-to-head comparison of two quantitative S1 assays in pre-characterized oligo-/asymptomatic patients. Infect Dis Ther 2021;10:1505–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nooka AK, Shanmugasundaram U, Cheedarla N, Verkerke H, Edara VV, Valanparambil R, et al. Determinants of neutralizing antibody response after SARS CoV-2 vaccination in patients with myeloma. J Clin Oncol 2022;40:3057–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chung DJ, Shah GL, Devlin SM, Ramanathan LV, Doddi S, Pessin MS, et al. Disease- and therapy-specific impact on humoral immune responses to COVID-19 vaccination in hematologic malignancies. Blood Cancer Discov 2021;2:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Painter MM, Mathew D, Rishi RR, Apostolidis SA, Pattekar A, Kuthuru O, et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 2021;54:2133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarke A, Sidney J, Methot N, Zhang Y, Dan JM, Goodwin B, et al. Negligible impact of SARS-CoV-2 variants on CD4+ and CD8+ T cell reactivity in COVID-19 exposed donors and vaccinees. BioRxiv preprint 2021. doi.org/10.1101/2021.02.27.433180. [Google Scholar]
- 26. Lyudovyk O, Kim JY, Quails D, Hwee MA, Lin Y-H, Boutemine SRet al. Impaired humoral immunity is associated with prolonged COVID-19 despite robust CD8 T cell responses. Cancer Cell 2022;40:738–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shree T, Shankar V, Lohmeyer JJK, Czerwinski DK, Schroers-Martin JG, Rodriguez GM, et al. CD20-targeted therapy ablates de novo antibody response to vaccination but spares preestablished immunity. Blood Cancer Discov 2022;3:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liebers N, Speer C, Benning L, Bruch PM, Kraemer I, Meissner J, et al. Humoral and cellular responses after COVID-19 vaccination in anti-CD20-treated lymphoma patients. Blood 2022;139:142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim SH, Stuart B, Joseph-Pietras D, Johnson M, Campbell N, Kelly A, et al. Immune responses against SARS-CoV-2 variants after two and three doses of vaccine in B cell malignancies: UK PROSECO study. Nat Cancer 2022;3:552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang LM, Costales C, Ramanathan M, Bulterys PL, Murugesan K, Schroers-Martin J, et al. Cellular and humoral immune response to SARS-CoV-2 vaccination and booster dose in immunosuppressed patients: an observational cohort study. J Clin Virol 2022;153:105217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Foca E, et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatric Allergy Immunol 2020;31:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wherry EJ, Barouch DH. T cell immunity to COVID-19 vaccines. Science 2022;377:821–2. [DOI] [PubMed] [Google Scholar]
- 33. Heinz FX, Stiasny K. Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 2021;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klinger M, Pepin F, Wilkins J, Asbury T, Wittkop T, Zheng J, et al. Multiplex identification of antigen-specific T cell receptors using a combination of immune assays and immune receptor sequencing. PLoS One 2015;10:e0141561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, et al. SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 2021;595:421–5. [DOI] [PubMed] [Google Scholar]
- 36. Gabler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021;591:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schliffke S, Sivina M, Kim E, von Wenserski L, Thiele B, Akyüz N, et al. Dynamic changes of the normal B lymphocyte repertoire in CLL in response to ibrutinib or FCR chemoimmunotherapy. Oncoimmunology 2018;7:e1417720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mundy BL, Gordon A, Lehman AM, Maddocks KJ, Cheney C, Jones JA, et al. Ibrutinib treatment improves T cell number and function in CLL patients. J Clin Invest 2017;127:3052–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herishanu Y, Rahav G, Levi S, Braester A, Itchaki G, Bairey O, et al. Efficacy of a third BNT162b2 mRNA COVID-19 vaccine dose in patients with CLL who failed standard 2-dose vaccination. Blood 2022;139:678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petrelli F, Luciani A, Borgonovo K, Ghilardi M, Parati MC, Petrò D, et al. Third dose of SARS-CoV-2 vaccine: a systematic review of 30 published studies. J Med Virol 2022;94:2837–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hachmann NP, Miller J, Collier AY, Ventura JD, Yu J, Rowe M, et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med 2022;387:86–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022;185:2422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022;608:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fendler A, Shepherd STC, Au L, Wu M, Harvey R, Schmitt AM, et al. Omicron neutralising antibodies after third COVID-19 vaccine dose in patients with cancer. Lancet 2022;399:905–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T cell receptor beta-chain diversity in alphabeta T cells. Blood 2009;114:4099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 47. Elyanow R, Snyder TM, Dalai SC, Gittelman RM, Boonyaratanakornkit J, Wald A, et al. T cell receptor sequencing identifies prior SARS-CoV-2 infection and correlates with neutralizing antibodies and disease severity. JCI Insight 2022;7:e150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
T-cell repertoire profiles and antigen annotation data from multiplexed antigen-stimulation experiments are available through the ImmuneCODE resource and can be downloaded from the Adaptive Biotechnologies immuneACCESS site under the immuneACCESS Terms of Use at clients.adaptivebiotech.com/pub/greenberger-2022-bcd (doi: 10.21417/LMG2022BCD).