SF3B1-mutant MDS-RS is associated with buildup of erythroid progenitor cells upregulating EIF2AK1-associated heme deficiency response and autophagy genes. Targeting EIF2AK1 relieves the dysregulated progenitor phenotype.
Abstract
SF3B1 mutations, which occur in 20% of patients with myelodysplastic syndromes (MDS), are the hallmarks of a specific MDS subtype, MDS with ringed sideroblasts (MDS-RS), which is characterized by the accumulation of erythroid precursors in the bone marrow and primarily affects the elderly population. Here, using single-cell technologies and functional validation studies of primary SF3B1-mutant MDS-RS samples, we show that SF3B1 mutations lead to the activation of the EIF2AK1 pathway in response to heme deficiency and that targeting this pathway rescues aberrant erythroid differentiation and enables the red blood cell maturation of MDS-RS erythroblasts. These data support the development of EIF2AK1 inhibitors to overcome transfusion dependency in patients with SF3B1-mutant MDS-RS with impaired red blood cell production.
Significance:
MDS-RS are characterized by significant anemia. Patients with MDS-RS die from a shortage of red blood cells and the side effects of iron overload due to their constant need for transfusions. Our study has implications for the development of therapies to achieve long-lasting hematologic responses.
This article is highlighted in the In This Issue feature, p. 476
INTRODUCTION
Cancer-causing mutations can dysregulate transcriptional programs and cause mutant cells to become dependent on certain molecular regulators or signaling pathways. The regulators or pathways that cancer cells become reliant upon vary based on the cell type and the dynamics of the cell states affected by the mutations (1).
Some of the best examples of hematologic alterations that can induce specific cell identity–based morphologic and clinical phenotypes are hotspot mutations in the splicing factor SF3B1 (SF3B1MT). SF3B1MT are highly recurrent initiating events in myelodysplastic syndromes (MDS), arising in the most primitive hematopoietic stem cells (HSC; ref. 2). Although found in different hematopoietic cell types (3), SF3B1MT preferentially deregulate erythroid progenitors and are hallmarks of a subtype of MDS characterized by the accumulation of ringed sideroblasts (RS) in the bone marrow (BM), MDS-RS (4–7).
Several studies have dissected the molecular mechanisms by which SF3B1MT modulate RNA splicing in MDS-RS. The SF3B1MT-induced aberrant splicing of genes involved in heme biosynthesis and mitochondrial iron transport leads to the abnormal deposition of iron in erythroid cells, resulting in dysfunctional hemoglobin synthesis and RS formation (8–12). However, given that few studies have elucidated how SF3B1MT affect distinct transcriptional states in erythroid progenitors and precursors, the dysregulated transcriptional programs induced by iron accumulation and the ways by which these programs arrest erythroid differentiation are largely unknown.
Here, using single-cell technologies, we sought to elucidate how SF3B1MT disrupt the transcriptional states of MDS-RS hematopoietic stem and progenitor cells (HSPC) and downstream differentiating erythroblasts to find disease dependencies that are not predicted by SF3B1MT-induced aberrant splicing.
RESULTS
SF3B1 MT Do Not Impair Erythropoiesis at the Level of HSPCs
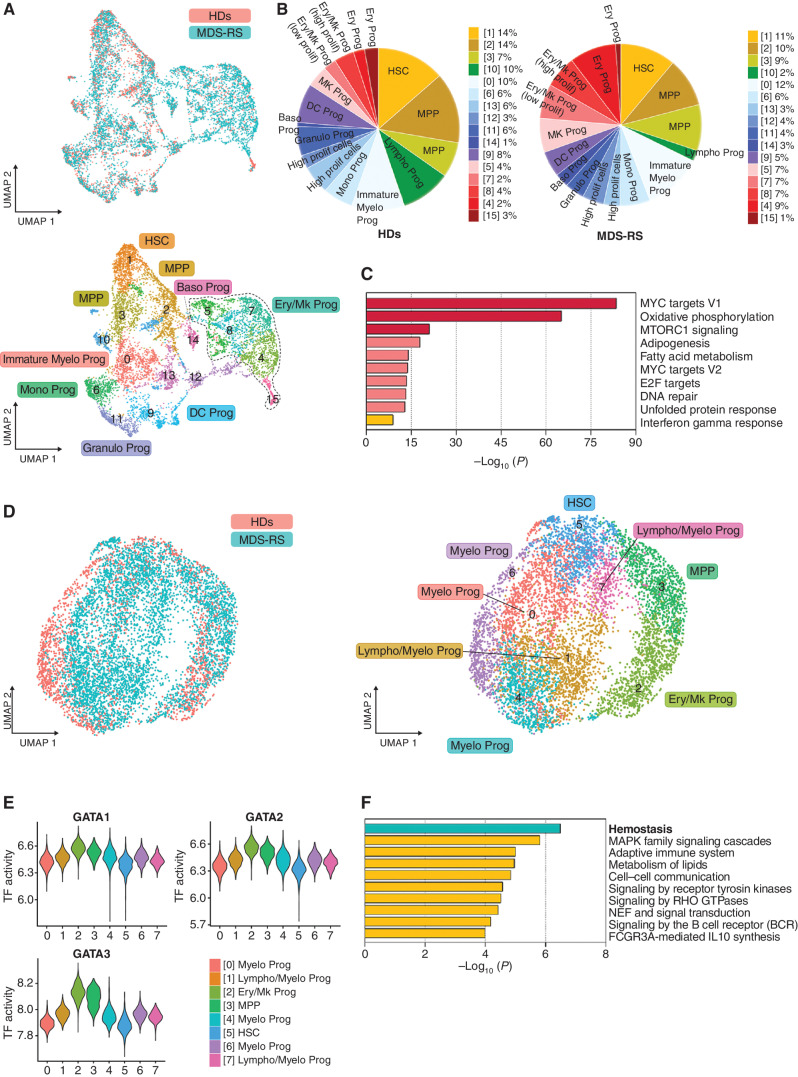
To dissect at the single-cell level the cellular and transcriptomic changes induced by SF3B1MT in cells undergoing erythroid differentiation, we performed single-cell RNA-sequencing (scRNA-seq) analysis of lineage-negative (Lin−) CD34+ HSPCs isolated from the BM of five untreated SF3B1-mutant MDS-RS patients and two age-matched healthy donors (HD; Supplementary Table S1). Our analyses identified 16 cellular clusters driven by cellular differentiation profiles (Fig. 1A) that we defined based on the differential expression of validated lineage-specific transcriptional factors (TF) and cellular markers (refs. 13–16; Supplementary Fig. S1A; Supplementary Table S2). To dissect the changes in lineage specification induced by SF3B1MT, we compared the distribution of HD and SF3B1-mutant HSPCs in the distinct hematopoietic clusters and found that HSPCs from MDS-RS patients had a predominant erythroid/megakaryocytic (Ery/Mk) differentiation (Fig. 1B). Importantly, MDS-RS Ery/Mk cells had higher expression of KLF1 (P = 0) and lower expression of FLI1 (P = 4.68−36), the two major regulators of Ery and Mk differentiation, respectively (17), which suggests that these cells were more erythroid-biased than those from HDs were (Supplementary Fig. S1B). Differential expression analyses revealed that SF3B1-mutant HSPCs undergoing Ery/Mk differentiation, compared with their healthy counterparts, exhibited upregulation of genes in the MYC signaling and oxidative phosphorylation pathways (Fig. 1C; Supplementary Fig. S1C and S1D), which underscores the state of metabolic activation in SF3B1-mutant cells. Similar results were observed when we analyzed the expression profile of most immature hematopoietic cells (cluster 1; Supplementary Fig. S1E and S1F).
Figure 1.
SF3B1 MT do not impair erythropoiesis at the level of HSPCs. A, UMAP plots of scRNA-seq data displaying pooled single Lin−CD34+ cells isolated from two HD (n = 2,324) and five SF3B1-mutant MDS-RS (n = 5,544) samples. Each dot represents one cell. Different colors represent the sample origin (top) and cluster identity (bottom). B, Distribution of HD (left) and MDS-RS (right) Lin−CD34+ cells among the clusters shown in A defined by distinct lineage differentiation profiles. Prolif, proliferative. C, Pathway enrichment analysis of the genes that were significantly upregulated in the MDS-RS Ery/Mk clusters as compared with the HD Ery/Mk clusters shown in A (adjusted P ≤ 0.05). The top 10 Hallmark gene sets are shown. D, UMAP plots of scATAC-seq data for pooled Lin−CD34+ cells isolated from 2 HD (n = 1,844) and 3 SF3B1-mutant MDS-RS (n = 5,203) samples. Each dot represents one cell. Different colors represent the sample origin (left) and cluster identity (right). E, Violin plots showing the activities of the TFs GATA1, GATA2, and GATA3 across the 8 clusters shown in D. F, Pathway enrichment analysis of the genes whose distal elements were enriched in open chromatin regions in HD cells from cluster 2 shown in D as compared with those of MDS-RS cells (adjusted P ≤ 0.05). The top 10 Reactome gene sets are shown. Baso, basophilic; DC, dendritic cell; Ery/Mk, erythroid/megakaryocytic; Granulo, granulocytic; HSC, hematopoietic stem cells; Mono, monocytic; MPP, multipotent progenitors; Myelo, myeloid; Prog, progenitors; UMAP, uniform manifold approximation and projection.
To further evaluate whether the fate of SF3B1-mutant Ery/Mk cells was biased toward erythroid differentiation at the expense of megakaryopoiesis, we performed single-cell assays for transposase-accessible chromatin with high-throughput sequencing (scATAC-seq) to profile the chromatin accessibility landscape in Lin− CD34+ HSPCs isolated from three SF3B1-mutant MDS-RS patients and two age- and gender-matched HDs (Supplementary Table S1). We identified 8 cellular clusters with distinct TF binding motif enrichment in open chromatin regions (Fig. 1D; Supplementary Table S3). Cells in cluster 5 were in the earliest differentiation state and were characterized by the highest activities of TFs involved in retinoid and nuclear factor family signaling (Supplementary Fig. S1G). Conversely, cells in clusters 2 and 3 showed the highest activities of TFs involved in Ery/Mk specification, including many members of the GATA family of TFs (Fig. 1E). Cells in clusters 1 and 4 were myeloid-biased and had the highest activities of the TFs SPI1 (PU.1) and CEBPA (Supplementary Fig. S1H). Differential analysis of chromatin accessibility among the samples showed that MDS-RS HSPCs at the latest stage of Ery/Mk differentiation before lineage specification (cluster 2) had significantly decreased open chromatin peaks at the distal elements of genes involved in platelet production (hemostasis; Fig. 1F; Supplementary Table S4). Because distal elements are better than promotors in defining how cells are poised for further differentiation (18), these data demonstrate that SF3B1MT do not arrest erythropoiesis at the HSPC level but instead induce HSC metabolic activation and promote differentiation toward the erythroid lineage at the expense of megakaryopoiesis.
SF3B1 MT Arrest Erythroblasts’ Terminal Differentiation and Activate the EIF2AK1-Induced Response Pathway to Heme Deficiency
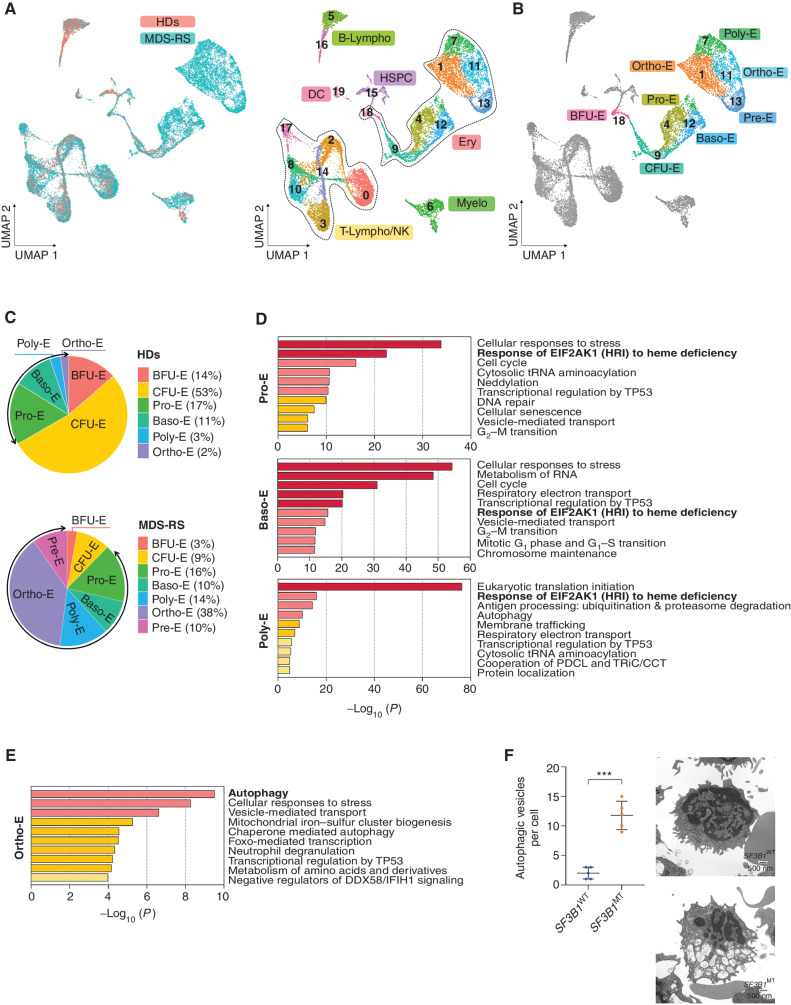
To evaluate whether SF3B1MT affects terminal erythroid differentiation, we performed scRNA-seq analysis of mononuclear cells (MNC) isolated from the BM of the same five SF3B1-mutant MDS-RS patients and two HDs whose HSPC compartments we analyzed previously. Our analysis identified 20 cellular clusters inclusive of all major BM cell types, which we defined according to known cellular surface markers and lineage potential (ref. 19; Fig. 2A; Supplementary Fig. S2A; Supplementary Table S5). As expected, differential analysis of BM cell type composition revealed that SF3B1-mutant MNCs, compared with HD MNCs, were characterized by an erythroid predominance (Supplementary Fig. S2B), which is consistent with the accumulation of RS in the BM of these patients (Supplementary Table S1). To determine the mechanistic basis by which SF3B1MT affect erythroid maturation and impair the release of red blood cells into the peripheral blood (PB), we analyzed the frequency and expression profile of each erythroid cluster within the erythroblastic population. We identified 8 cellular clusters representing 7 different stages of erythroblast differentiation (Fig. 2B), which we defined based on the expression of previously validated surface markers, TFs, and effector molecules involved in erythropoiesis (20, 21). Cells in the early stages of erythropoiesis, including phenotypic burst-forming unit-erythroid cells (BFU-E; cluster 18) and colony-forming unit-erythroid cells (CFU-E; cluster 9), were characterized by variable expression of the TF GATA2 and the surface markers CD34, CD36, CD117 (cKit), and CD105 (ENG), whereas cells undergoing terminal differentiation, including pro-erythroblasts (Pro-E; cluster 4), basophilic erythroblasts (Baso-E; cluster 12), polychromatophilic erythroblasts (Poly-E; cluster 7), and orthochromatic erythroblasts (Ortho-E; clusters 1 and 11), were characterized by progressively increased expression of the TFs GATA1 and KLF1, the erythropoietin and transferrin receptors (EPOR and TFRC, i.e., CD71), the membrane protein glycophorin A (GYPA, i.e., CD235a), and hemoglobin subunits. Pre-erythrocytes (Pre-E; cluster 13)—erythroblasts in the reticulocyte maturation phase, which occurs before the release of erythrocytes into the PB—expressed genes involved in hemoglobin synthesis most highly and surface markers that characterize the upstream stages of erythroblast differentiation most lowly (Supplementary Fig. S2C). Differential analysis of the cluster distribution between HD and SF3B1-mutant samples showed that SF3B1-mutant erythroblasts had an increased frequency of cells at the latest stages of erythroid differentiation, mainly at the Ortho-E stage (Fig. 2C). To exclude the possibility that the arrest of the SF3B1-mutant erythroblasts in the late stages of differentiation was the result of an increase in EPO in the BM of MDS-RS patients, we analyzed the changes induced by EPO administration in the BM MNCs of one MDS-RS patient carrying the SF3B1K700E mutation before and after treatment with EPO as supportive therapy (Supplementary Fig. S2D; Supplementary Table S6). EPO administration did not induce any changes in either the frequencies of the erythroid populations (Supplementary Fig. S2E) or these cells’ transcriptional profile (Supplementary Table S7).
Figure 2.
SF3B1 MT arrest erythroid terminal differentiation and activate the EIF2AK1-induced response pathway to heme deficiency. A, UMAP plots of scRNA-seq data for pooled single MNCs isolated from two HD (n = 5,049) and five SF3B1-mutant MDS-RS (n = 16,212) samples. Each dot represents one cell. Different colors represent the sample origin (left) and cluster identity (right). B, UMAP plot of scRNA-seq data from A showing the 7 different stages of erythroid differentiation in the total erythroblast population. C, Distribution of the 7 stages of erythroid differentiation in the HD (top) and MDS-RS (bottom) erythroblast populations shown in A. Arrows indicate the terminal steps of erythroid differentiation. D, Pathway enrichment analyses of the genes that were significantly upregulated in Pro-E (top), Baso-E (middle), Poly-E (bottom) from MDS-RS samples as compared with those from HD samples (P ≤ 0.01) The top 10 Reactome gene sets are shown. E, Pathway enrichment analysis of the genes that were significantly upregulated in Ortho-E from MDS-RS samples as compared with those from HD samples (P ≤ 0.01) The top 10 Reactome gene sets are shown. F, Left, the number of autophagic vesicles per cell in SF3B1WT and SF3B1-mutant erythroblasts from one representative SF3B1WT and one SF3B1-mutant sample. Statistically significant differences were detected using a two-tailed Student t test. Right, representative transmission electron microscopy images of erythroblasts from the BM sections of one SF3B1WT and one SF3B1-mutant MDS sample. Scale bars, 500 nm. Baso-E, basophilic erythroblasts; BFU-E, burst-forming unit-erythroid cells; B-Lympho, B-lymphocytes; CFU-E, colony formation unit-erythroid cells; DC, dendritic cells; Ery, erythroblasts; HSPC, hematopoietic stem and progenitor cells; Myelo, myeloid cells; Ortho-E, orthochromatic erythroblasts; Poly-E, polychromatophilic erythroblasts; Pre-E, pre-erythrocytes; Pro-E, pro-erythroblasts; T-Lympho/NK, T-lymphocytes, and natural killer cells; UMAP, uniform manifold approximation and projection.
Together, these data suggest that SF3B1MT arrest erythropoiesis at the final stage of erythroblast maturation by inhibiting the transition from Ortho-E to Pre-E, which results in the accumulation of terminally differentiated cells in the BM. Consistent with these observations, immunophenotypic analysis showed that, compared with those from HDs, terminally differentiated erythroblasts from SF3B1-mutant MDS-RS patients had a significantly higher frequency of cells in the orthochromatic stage V of differentiation (Supplementary Fig. S2F), as reported previously (22, 23).
To dissect the molecular mechanisms underlying the SF3B1MT-induced arrest of terminal erythroid differentiation, we analyzed the differential expression profiles between HD and SF3B1-mutant cells at each stage of erythroblast differentiation. These analyses revealed that SF3B1-mutant Pro-E, Baso-E, and Poly-E had significantly upregulated genes involved in the response of eukaryotic translation initiation factor 2 alpha kinase 1 (EIF2AK1) to heme deficiency, including EIF2AK1, EIF2S1, ATF4, and DDIT3 (Fig. 2D; Supplementary Fig. S2G). EIF2AK1 is a metabolic stress-sensing kinase that in homeostasis binds to its natural ligand, heme (24). In conditions that limit heme production, EIF2AK1 phosphorylates its main target, EIF2S1, and leads to decreased translation of globin mRNAs and increased expression of the stress response effector ATF4 (25). ATF4 and its downstream effector, DDIT3, are major regulators of autophagy (26) and activate a set of targets involved in autophagy to maintain cell survival in response to different types of stress stimuli, including those affecting mitochondrial functions (27–29). Accordingly, we observed that SF3B1-mutant Ortho-E exhibited a significant upregulation of genes involved in mitochondrial autophagy (Fig. 2E), including PINK1, SQSTM1 (p62), and LAMPTOR5 (Supplementary Fig. S2H). These data are consistent with an attempt by Ortho-E to remove dysfunctional mitochondria, alleviate the iron overload caused by SF3B1MT (8, 30), and maintain their survival.
To functionally validate these findings, we analyzed the morphology of erythroid cells isolated from the BM of SF3B1-mutant and SF3B1-wild-type (SF3B1WT) MDS-RS patients to assess differences in the ultrastructural features of autophagy. Transmission electron microscopy showed that SF3B1-mutant erythroblasts had significantly more autophagic vesicles than SF3B1WT erythroblasts do (Fig. 2F). To determine whether SF3B1MT instigated autophagy activation, we used CRISPR/Cas9 genome editing technology to introduce the SF3B1 missense mutation c.2098A>G (K700E) into the erythroblastic cell line K562. This mutation results in the K700E amino acid substitution, which is recurrently detected in patients with SF3B1-mutant MDS-RS (30). Consistent with our findings in primary samples, SF3B1-mutant cells in physiologic conditions or after hemin-induced erythroid differentiation had EIF2AK1 pathway upregulation (Supplementary Fig. S2I) and had significantly more autophagic vesicles (Supplementary Fig. S2J and S2K) and higher expression of LC3B protein (Supplementary Fig. S2L) than their parental counterparts did. Together, these data demonstrate that SF3B1MT arrest erythroid terminal differentiation and activate the EIF2AK1-induced response pathway to maintain erythroblasts’ survival in response to heme deficiency.
Hypomethylating Agent Therapy Inhibits the EIF2AK1-Induced Response Pathway to Heme Deficiency
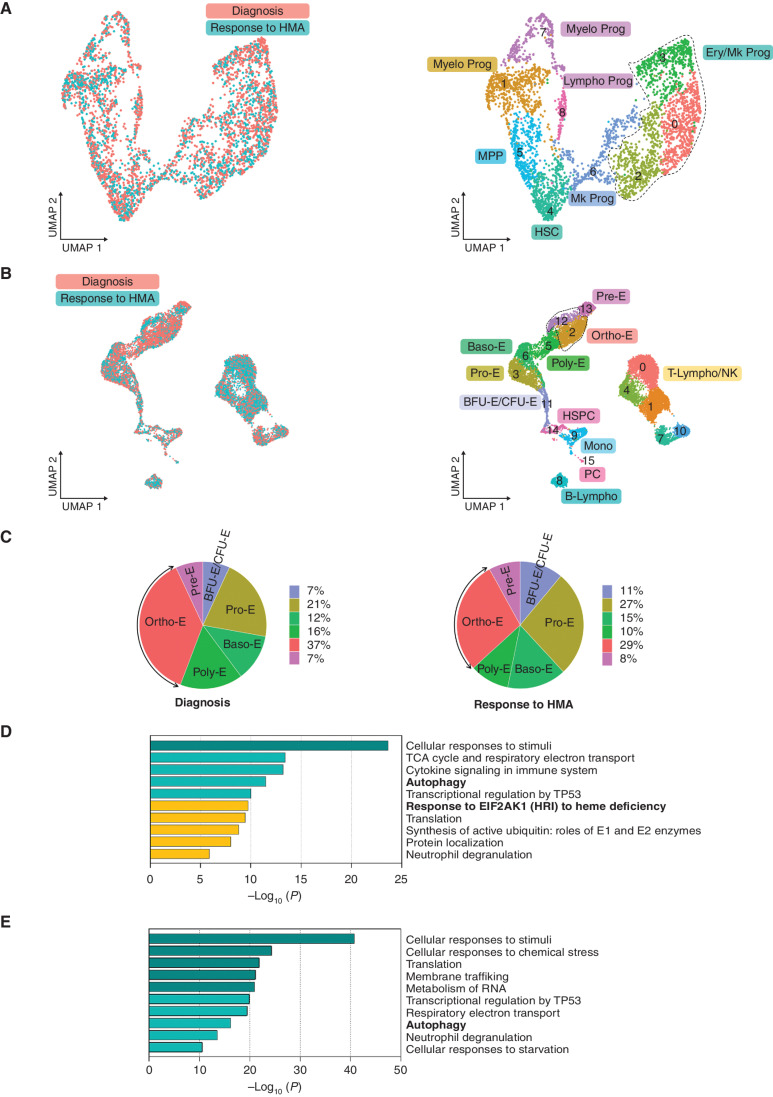
We hypothesized that if EIF2AK1 pathway activation plays a role in arresting the maturation of terminally differentiated SF3B1-mutant erythroblasts, it should progressively decrease once patients achieve hematologic remission and become transfusion-independent following hypomethylating agent (HMA) therapy. To test this hypothesis, we dissected at the single-cell level the cellular and transcriptomic changes induced by HMA therapy in Lin−CD34+ HSPCs and BM MNCs, which were isolated from two SF3B1-mutant MDS-RS patients at the time of diagnosis and at the time of hematologic response following HMA therapy (Supplementary Table S1). Although HMA therapy did not overcome the aberrant differentiation of HSCs toward the erythroid lineage (Fig. 3A; Supplementary Fig. S3A and S3B) or significantly decrease the SF3B1MT burden in BM MNCs (Supplementary Fig. S3C; Supplementary Table S8), it did reduce the accumulation of erythroblasts in the BM (Fig. 3B; Supplementary Fig. S3D and S3E; Supplementary Table S9) by inducing these cells’ differentiation. Indeed, primary cells derived from SF3B1-mutant CD34+ MDS-RS cells cultured in erythroid differentiation media for 8 days and then treated with 5-azacitidine for 3 days had a higher percentage of terminally differentiated CD235a (GYPA)+ erythroblasts than those treated with vehicle (21.9% vs. 7.62%; Supplementary Fig. S3F).
Figure 3.
Hypomethylating agent therapy inhibits the EIF2AK1-induced response pathway to heme deficiency in terminally differentiated cells in patients who became transfusion independent. A, UMAP plots of scRNA-seq data for pooled single Lin-CD34+ cells isolated from two SF3B1-mutant MDS-RS patients at the time of diagnosis (n = 2,372) and at the time of response to HMA therapy (n = 1,551). Each dot represents one cell. Different colors represent the sample origin (left) and cluster identity (right). B, UMAP plots of scRNA-seq data for pooled single MNCs isolated from three SF3B1-mutant MDS-RS patients at the time of diagnosis (n = 6,089) and response to HMA therapy (n = 6,156). Each dot represents one cell. Different colors represent the sample origin (left) and cluster identity (right). C, Distribution of the stages of erythroid differentiation in the total erythroblast population shown in B at the time of diagnosis (left) and at the time of response to HMA therapy (right). Arrows indicate Ortho-E. D, Pathway enrichment analysis of the genes in the MDS-RS Pro-E, Baso-E, and Poly-E clusters shown B that were significantly downregulated at the time of response to HMA therapy as compared with the time of diagnosis (adjusted P ≤ 0.05). The top 10 Reactome gene sets are shown. E, Pathway enrichment analysis of the genes in the MDS-RS Ortho-E shown in B that were significantly downregulated at the time of response to HMA therapy as compared with the time of diagnosis (P ≤ 0.01). The top 10 Reactome gene sets are shown. Baso-E, basophilic erythroblasts; BFU-E, burst-forming unit-erythroid cells; B-Lympho, B-lymphocytes; CFU-E, colony formation unit-erythroid cells; Ery/Mk, erythroid/megakaryocytic; HSC, hematopoietic stem cells; HSPC, hematopoietic stem and progenitor cells; Lympho, lymphoid; Mk, megakaryocytic; Mono, monocytes; MPP, multipotent progenitors; Myelo, myeloid; NK, natural killer cells; Ortho-E, orthochromatic erythroblasts; PC, plasma cells; Poly-E, polychromatophilic erythroblasts; Pre-E, pre-erythrocytes; Pro-E, pro-erythroblasts; Prog, progenitors; T-Lympho, T-lymphocytes; UMAP, uniform manifold approximation and projection.
Differential analysis of the cluster distribution in the erythroid compartment showed that, compared with those from untreated patients, erythroblasts from HMA-treated patients who achieved hematologic remission had a decreased frequency of Ortho-E (Fig. 3C). Differential expression profiles of untreated and HMA-treated cells at different stages of terminal erythroid differentiation revealed that HMA-treated Pro-E, Baso-E, and Poly-E (clusters 3, 6, and 5, respectively) had significant downregulation of the major effectors of the EIF2AK1 response (Fig. 3D; Supplementary Fig. S3G). Accordingly, the expression of autophagy genes was significantly decreased in HMA-treated Ortho-E (clusters 2 and 12; Fig. 3E; Supplementary Fig. S3H). Together, these data suggest that HMA therapy can overcome EIF2AK1 pathway activation in terminally differentiated erythroblasts in patients who have had hematologic remission and have become transfusion-independent.
Inhibition of EIF2AK1 Overcomes the Accumulation of RS and Enables Red Blood Cell Production
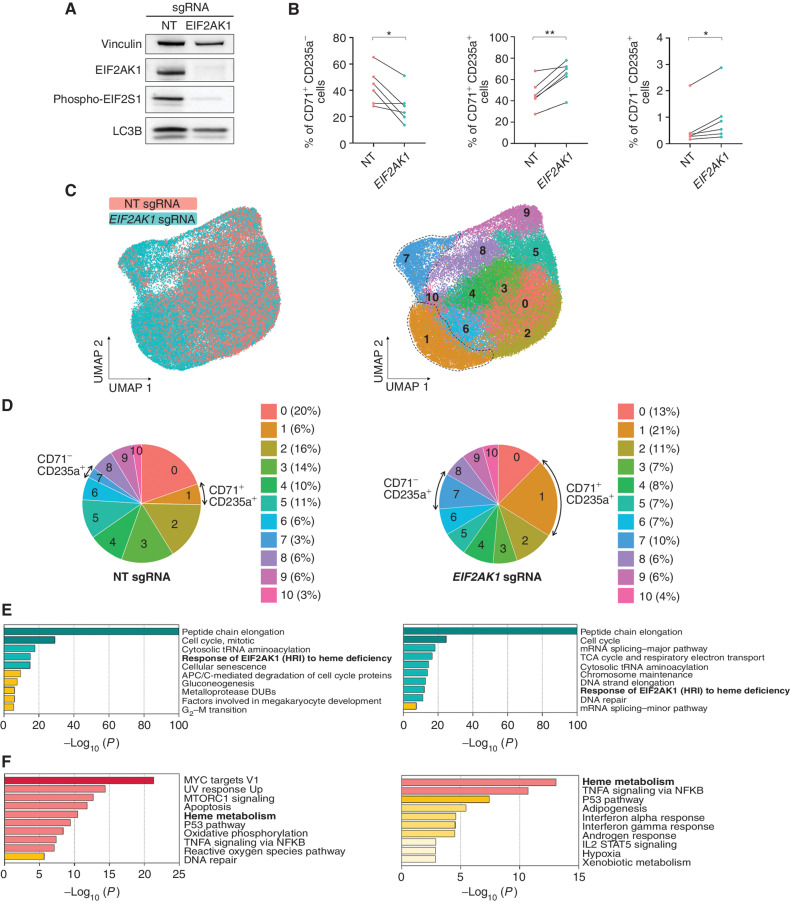
We hypothesized that the inhibition of EIF2AK1 pathway activation induces the maturation of SF3B1-mutant erythroblasts. To test this hypothesis, we used CRISPR/Cas9 genome editing technology to delete EIF2AK1 in primary erythroblasts derived from a three-phase erythroid differentiation culture system of SF3B1-mutant CD34+ cells (31). EIF2AK1 depletion in MDS-RS cells using previously validated single-guide RNAs (sgRNA; ref. 32; Fig. 4A) significantly increased terminally differentiated CD71 (TFRC)+CD235a (GYPA)+ and CD71−CD235a+ erythroblasts at the expense of less-differentiated CD71+CD235a− cells after 13 days of culture (Fig. 4B; Supplementary Fig. S4A). In contrast, EIF2AK1 depletion in HD CD34+ cells had no significant effect on erythroid differentiation (Supplementary Fig. S4B and S4C). To validate these results, we performed an scRNA-seq analysis of three MDS-RS samples treated with nontargeting (NT) or EIF2AK1 sgRNAs at day 13 of erythroid differentiation (Fig. 4C). Differential analysis of the cluster distribution showed that, compared with those treated with the NT sgRNAs, cells treated with EIF2AK1 sgRNAs had a significantly higher frequency of terminally differentiated cells (clusters 1 and 7; Fig. 4D; Supplementary Fig. S4D). These cells were characterized by the highest expression of both CD71 and CD235a (cluster 1) or CD235a (cluster 7; Supplementary Fig. S4E) and genes involved in heme metabolism (Supplementary Fig. S4F; Supplementary Table S10). Differential expression analysis of these clusters revealed that, as expected, EIF2AK1 sgRNA–treated MDS-RS erythroblasts were characterized by the downregulation of genes involved in the response of EIF2AK1 to heme deficiency (Fig. 4E), including the major effectors ATF4 and DDIT3 (Supplementary Fig. S4G). EIF2AK1-mediated pathway downregulation was associated with the significantly increased expression of genes that regulate heme metabolism and are involved in erythroid differentiation (Fig. 4F; Supplementary Fig. S4H). Similar results were obtained from bulk RNA-seq analysis of three other SF3B1-mutant MDS-RS samples treated with NT or EIF2AK1 sgRNAs at 13 days of erythroid differentiation (Supplementary Fig. S4I). Notably, downregulation of EIF2AK1 signaling was associated with an increased expression of genes involved in heme biosynthesis and mitochondrial iron transport, including all regulators of protoporphyrin IX biosynthesis, such as ALAD, ALAS2, HMBS, UROD, ABCB6, CPOX, TMEM14C, PPOX, and FECH (Supplementary Fig. S4J). Given that the missplicing of TMEM14C diminishes porphyrin abundance within the mitochondria, resulting in iron accumulation and heme deficiency, and that the increased expression of TMEM14C partially overcomes RS formation (12), these results may explain why EIF2AK1-depleted erythroblasts undergo terminal differentiation. As the SF3B1MT burden did not differ between NT sgRNA– and EIF2AK1 sgRNA–treated MDS-RS cells (Supplementary Fig. S4K), these data suggest that EIF2AK1 inhibition overcomes the arrest of erythroid differentiation induced by SF3B1MT.
Figure 4.
Inhibition of EIF2AK1 overcomes the accumulation of RS and enables red blood cell production. A, Representative western blot analysis of EIF2AK1, phospho-EIF2S1, and LC3B in nontargeting (NT) sgRNA– and EIF2AK1 sgRNA–treated SF3B1-mutant MDS-RS cells at day 13 of culture. Vinculin was used as a loading control. B, Frequencies of CD71+CD235a− (left), CD71+CD235a+ (middle), and CD71−CD235a+ (right) erythroblasts in NT sgRNA– or EIF2AK1 sgRNA–treated MDS-RS samples (n = 6) at day 13 of culture. Each symbol represents one sample; lines connect paired samples. Statistical significance was calculated using paired t tests. C, UMAP plots of scRNA-seq data for single cells from NT sgRNA–treated (n = 30,307) or EIF2AK1 sgRNA–treated (n = 36,825) cells from 3 pooled SF3B1-mutant MDS-RS samples at day 13 of erythroid culture. Each dot represents one cell. Different colors represent the sample origin (left) and cluster identity (right). Dotted lines indicate terminally differentiated erythroblasts. D, Distribution of cells from NT (left) and EIF2AK1 (right) sgRNA–treated SF3B1-mutant MDS-RS samples among the clusters shown in C. Arrows indicate clusters 1 and 7.E, Pathway enrichment analyses of the significantly downregulated genes in EIF2AK1 sgRNA–treated SF3B1-mutant MDS-RS cells from clusters 1 (left) and 7 (right; adjusted P ≤ 0.05). The top 10 Reactome or Hallmark gene sets are shown. F, Pathway enrichment analyses of the significantly upregulated genes in EIF2AK1 sgRNA–treated SF3B1-mutant MDS-RS cells from clusters 1 (left) and 7 (right; adjusted P ≤ 0.05). The top 10 Hallmark gene sets are shown.
DISCUSSION
Despite mechanistic insights into SF3B1MT-induced tumorigenesis, effective therapies targeting SF3B1MT-induced aberrations in MDS have not yet been developed, and therapeutic approaches based on global splicing inhibition have fallen short of expectations. Results from a phase I clinical trial of the oral SF3B1 modulator H3B-8800 in patients with myeloid neoplasms did not recapitulate the results of the preclinical studies performed using mouse models (33). These studies demonstrate that the widespread inhibition of splicing activity does not yield significant clinical improvements (34). Taken together, these data suggest that alternative therapeutic approaches are needed and that targeting specific downstream effectors of SF3B1MT in erythroblasts may represent a novel strategy to overcome ineffective erythropoiesis before other secondary genetic lesions lead to MDS progression.
In our study, we used single-cell technologies to elucidate the biological mechanisms of the SF3B1MT-induced arrest of erythropoiesis. We demonstrated that SF3B1MT significantly inhibit the differentiation of Ortho-E to Pre-E, thus impairing the release of red blood cells into the PB. We analyzed the molecular pathways that were transcriptionally upregulated in Ortho-E independently of splicing alterations and identified the EIF2AK1 response to heme deficiency as a potential driver of the SF3B1MT-induced arrest of erythroid differentiation. EIF2AK1 pathway activation in terminally differentiated SF3B1-mutant erythroblasts resulted in increased ATF4 expression, which in turn upregulated the expression of genes involved in autophagy and RS survival. We would, therefore, hypothesize that EIF2AK1 signaling exacerbates RS formation and anemia in SF3B1-mutant MDS-RS patients by reducing oxidative stress in response to heme deficiency through the EIF2AK1-induced inhibition of globin protein and biosynthetic heme enzyme expression and translation (25, 35). Consistent with this hypothesis, EIF2AK1 inhibition significantly increased the expression of both mitochondrial heme biosynthetic enzymes and iron transporters and improved the terminal differentiation of SF3B1-mutant MDS-RS erythroblasts (Fig. 5). We could not directly assess RS formation in our functional validation studies owing to the limited number of primary cells available for our assays.
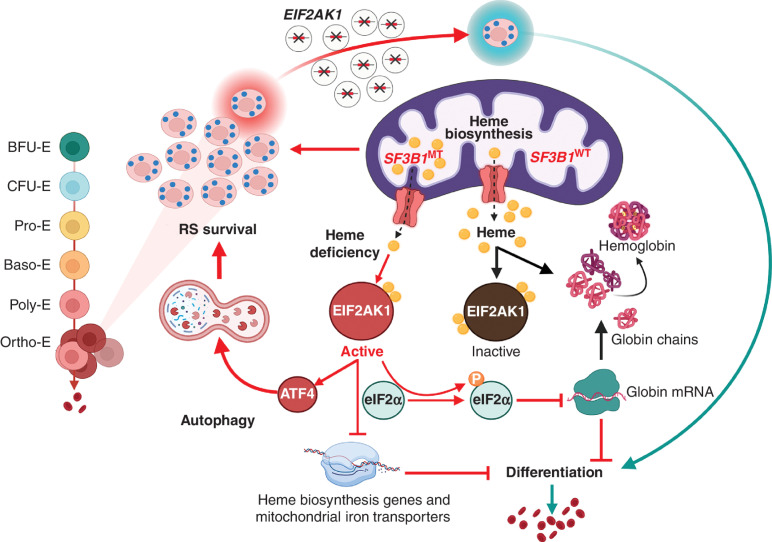
Figure 5.
Proposed model. Under normal physiologic conditions, heme binds to EIF2AK1 and represses its activation. In conditions that limit heme production, such as those induced by SF3B1MT in terminally differentiated erythroblasts, EIF2AK1 is activated. EIF2AK1 pathway activation promotes RS survival and inhibits erythroid maturation by increasing the expression of ATF4, which in turn upregulates the expression of genes involved in autophagy, transcriptionally inhibits genes involved in heme biosynthesis and mitochondrial iron transport, and translationally inhibits globin production. Targeting EIF2AK1 pathway activation by depleting EIF2AK1 rescues erythroid differentiation and red blood cell production.
Growing evidence suggests that the EIF2AK1 pathway affects erythropoiesis in response to several types of insults. The CRISPR/Cas9-mediated deletion of EIF2AK1 in sickle erythroblasts increases fetal hemoglobin production and reduces sickling in cultured erythroid cells (31, 32). EIF2AK1-mediated fetal hemoglobin regulation proceeds via the modulation of the fetal hemoglobin repressor BCL11A (36). A clinical study based on the transplantation of autologous HSCs transduced by lentiviral vectors delivering shRNAs targeting BCL11A in patients with sickle cell disease demonstrated a significant improvement of anemia and other disease manifestations (37), suggesting that pharmacologically inhibiting of EIF2AK1 may lead to similar clinical outcomes. Thus, EIF2AK1 inhibition represents one of the most promising therapeutic approaches for sickle cell anemia to date.
The EIF2AK1 effector DDIT3 is overexpressed in SF3B1WT MDS HSCs (38). DDIT3 overexpression delays the erythroid differentiation of MDS CD34+ cells by affecting the activation of erythroid transcriptional programs. The inhibition of DDIT3 in CD34+ cells isolated from MDS patients with anemia restores proper erythroid differentiation (38). Our study showed that the activation of the EIF2AK1 pathway occurs in the later stages of erythroblast maturation, as SF3B1MT-induced aberrant splicing primarily affected the function of terminally differentiated erythroblasts with the highest mitochondrial content.
Although the EIF2AK1-mediated response to heme deficiency is not completely conserved in mice, mouse models in which the gene Eif2ak1 has been knocked out have normal hematologic indices (36). Moreover, EIF2AK1 depletion is well tolerated in human erythroid cells in steady-state conditions (31, 32), as also confirmed in our study. These data suggest that the EIF2AK1 pathway could be a therapeutic target for not only sickle cell disease but also MDS-RS or other types of disorders with impaired red blood production.
In conclusion, our study highlights the importance of dissecting the conserved molecular mechanisms of the erythroid stress response to develop novel approaches that achieve lasting hematologic remission in patients with different types of ineffective erythropoiesis or aberrant erythroblast functions.
METHODS
Human Primary Samples
BM aspirates from 17 patients with SF3B1-mutant MDS-RS who were referred to the Department of Leukemia at MD Anderson Cancer Center, the Cleveland Clinic Foundation, the University of California San Diego, or the Josep Carreras Leukaemia Research Institute were obtained after the approval of the corresponding Institutional Review Boards and in accordance with the Declaration of Helsinki. Diagnosis of MDS-RS was assigned according to the World Health Organization criteria (39). SF3B1MT were determined by a targeted amplicon-based next-generation sequencing (40). All available samples from patients with MDS-RS and SF3B1MT were included in the study. Baseline BM aspirates were collected from patients before any treatment. For patients for whom baseline samples were available, sequential BM samples were collected during HMA treatment. The clinical characteristics of the individuals with SF3B1-mutant MDS-RS are shown in Supplementary Table S1. BM samples from HDs were obtained from AllCells, the Department of Stem Cell Transplantation at MD Anderson Cancer Center, and the Cleveland Clinic Foundation. Written informed consent was obtained from all donors.
MNCs were collected from each BM sample immediately after BM aspiration using the standard gradient separation approach with Ficoll-Paque PLUS (catalog number #45-001-752, Thermo Fisher Scientific). MNCs were cryopreserved and stored in liquid nitrogen until use. For cell-sorting applications, MNCs were enriched in CD34+ cells using magnetic-activated cell sorting (MACS) with the CD34 Microbead Kit (catalog number #130-046-702, Miltenyi Biotec) and further purified by fluorescence-activated cell sorting (FACS) as described below.
Generation of the SF3B1K700E Cell Line
The K562 cell line was obtained by ATCC in 2017. Mycoplasma testing was routinely performed on the cell line, and cell identity was validated by short tandem repeat DNA fingerprinting before performing any experiment. Cells were cultured in 10% FBS (catalog number #S11150, Atlanta Biologicals) Iscove's modified Dulbecco's medium (catalog number #12440053, Thermo Fisher Scientific). Cells for genome editing were used after 4 passages from reception. The CRISPR/Cas9-based extreme genome editing system developed by Biocytogen (https://biocytogen.com/gene-editing) was used to generate the heterozygous SF3B1K700E cell line. The nonconserved region of the SF3B1 intron 14 (near exon 15) was targeted by Cas9 endonuclease using the following guide RNA sequence: 5′-GGACAGCTGTCCTAAAATTT GGG-3′. The repaired template DNA included at least 1-kb 5′ and 3′ homologous arms, the puromycin gene, and the K700E mutation in exon 15 (the sequence of the repaired DNA template can be provided upon request). CRISPR plasmids and template DNA (Biocytogen) were introduced into the K562 cells by electroporation. Single clones were selected by puromycin (catalog number #194539, Sigma-Aldrich) treatment (1.5 μg/mL). Mutant clones were screened by PCR using the following primers: 5′-GCATCAAGCTTGGTACCGATATTCTTATGGGCTGTGCCATCTTGC-3′ and 5′-ACTTAATCGTGGAGGATGATAGGTAATTGGTGGATTTACCTTTCCTCT3′.
SF3B1 K700E mutation was confirmed by DNA sequencing. To induce erythroid differentiation, parental and SF3B1K700E K562 cells were treated with 30 μmol/L hemin (catalog number # 51280, Sigma-Aldrich) for 3 days, as described previously (23).
Fluorescence-Activated Cell Sorting
FACS of Lin−CD34+ cells was performed using antibodies against CD2 (RPA-2.10), CD3 (SK7), CD14 (MφP9), CD19 (SJ25C1), CD20 (2H7), CD34 (581), CD56 (B159), and CD235a (HIR2; all from BD Biosciences); CD4 (S3.5), CD11b (ICRF44), and CD33 (P67.6; all from Thermo Fisher Scientific); CD7 (6B7; BioLegend); and CD10 (SJ5-1B4; Leinco Technologies), as we described previously. Samples were acquired with a BD Influx Cell Sorter (BD Biosciences).
Cell Culture
MACS-purified CD34+ cells were subjected to a three-phase in vitro culture system as described previously (36). Briefly, for phase I medium, StemSpan SFEM II (catalog number #09605, Stem Cell Technologies) was supplemented with 100 ng/mL of human stem cell factor (catalog number #300-07, PeproTech), 1 ng/mL of IL3 (catalog number #200-03, PeproTech), 3 units per mL of erythropoietin (catalog number #100-64, PeproTech), 200 μg/mL of holotransferrin (catalog number #T4132, Sigma-Aldrich), 10 μg/mL of insulin (catalog number # I9278, Sigma-Aldrich), 5% human male A/B plasma (catalog number #H4522, Sigma-Aldrich), and 10 μg/mL of heparin (catalog number #7980, Stem Cell Technologies). For phase II medium, IL3 was withdrawn after 9 days of culture. For phase III medium, the cells were cultured with StemSpan SFEM II supplemented with 3 units per mL of erythropoietin, 1 mg/mL of holotransferrin, 10 μg/mL of insulin, 5% human A/B plasma, and 10 μg/mL of heparin. Cells were maintained in phase I from the day of collection until day 8, at which time the cells underwent ribonucleoprotein-based gene editing and were transitioned to phase II medium. On day 13 of culture, the cells were transitioned to phase III medium. Samples for western blot and flow cytometry analyses were harvested on day 13 or 15 of culture, respectively.
To evaluate whether HMA therapy induces erythroblast differentiation, erythroblasts derived from SF3B1-mutant CD34+ MDS-RS cells were cultured in phase I differentiation medium for 8 days and then treated with vehicle or 0.1 μmol/L 5-azacitidine (catalog number #A2385, Sigma-Aldrich) for 3 days. Samples for flow cytometry analysis were harvested on day 13 of culture.
Ribonucleoprotein Electroporation in CD34+ Cells
Ribonucleoprotein-based gene editing was performed as described previously (32). Briefly, CD34+ cells were electroporated with the previously validated EIF2AK1-targeting sgRNAs TTGTTGGCTATCACACCGCG and ATAGTCGAGAGAAACAAGCG or the nontargeting sgRNAs GCACTACCAGAGCTAACTCA and GTACGTCGGTATAACTCCTC, together with the Cas9 Nuclease V3 (catalog number #1081060, Integrated DNA Technologies) using the P3 Primary Cell 4D-Nucleofector X Kit S (catalog number # V4XP-3032, Lonza). Chemically modified sgRNAs were purchased from Synthego.
Flow Cytometry
For erythroblast staging analysis, HD and SF3B1-mutant MDS-RS BM MNCs were analyzed as described previously (22). Briefly, MNCs were stained with the following antibodies: PE-CD45 (clone HI30, BD Biosciences), PE-CY7-CD235a (clone HI264, BioLegend), APC-CD49d (clone 9F10, BioLegend), and FITC–band 3 (a generous gift from Dr. Xiuli An, New York Blood Cancer Center).
For erythroid differentiation analyses, cells on day 15 of culture were stained with antibodies against PE-CD45, APC-CD71 (clone mA712, BD Biosciences), and FITC-CD235a (clone HIR2, BD Biosciences). Dead cells were excluded with DAPI staining (catalog number D1306, Thermo Fisher Scientific). Samples were acquired with a BD LSRFortessa Cell Analyzer, and the cell populations were analyzed using FlowJo software (version 10.5.3, FlowJo, LLC).
Western Blots
Cells were washed with a 10% FBS/phosphate-buffered saline solution (catalog numbers #16-140-07 and #14-190-250, respectively, Thermo Fisher Scientific), and pellets were resuspended in the Mammalian Cell and Tissue Extraction Kit buffer (catalog number #K269-500, BioVision Incorporated) and incubated for 15 minutes with gentle shaking. Lysates were then collected after centrifugation at 12,000 rpm for 20 minutes at 4°C. The amount of protein was quantified using the Qubit Protein Assay Kit (catalog number #Q33212, Thermo Fisher Scientific) and a Qubit Fluorometer (Thermo Fisher Scientific). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blotting were performed following standard protocols. Blotted membranes were incubated with primary polyclonal antibodies against EIF2AK1 (MBS2538144, Mybiosource), phospho-EIF2S1 (ab32157, Abcam), LC3B (2775, Cell Signaling), and vinculin (clone hVIN-1, Sigma-Aldrich), and with secondary anti-mouse and anti-rabbit digital antibodies (R1005 and R1006, respectively, Kindle Biosciences LLP). Membranes were developed using the SuperSignal West Pico PLUS Chemiluminescent Substrate (catalog number #34580, Thermo Fisher Scientific) in a KwikQuant Imager (Kindle Biosciences LLP).
scRNA-seq Analysis
Lin−CD34+ and MNC samples were enriched by FACS. Sample preparation and sequencing were performed at MD Anderson Cancer Center's Advanced Technology Genomics Core. Sample concentration and cell suspension viability were evaluated using a Countess II FL Automated Cell Counter (Thermo Fisher Scientific). Samples were normalized for input onto the Chromium Single-Cell A Chip Kit (catalog number #PN120236, 10X Genomics), and single cells were lysed and barcoded for reverse transcription. Equal amounts of each uniquely indexed sample library were pooled together. Pooled libraries were sequenced using a NovaSeq6000 SP 100-cycle flow cell (Illumina). Sequencing analysis was carried out using 10X Genomics’ Cell Ranger software, version 3.0.2.
The adapter trimming was done automatically using the Cell Ranger software. Alignment was performed using “Star” as a part of the Cell Ranger analysis. Cell Ranger default settings were used for all quality thresholds. The sequencing was run as a paired-end using the following run format: 28 cycles Read 1, 8 cycles Index 1, 0 cycles Index2, and 91 cycles Read2. Barcodes and unique molecular identifiers were sequenced in Read1, and the transcriptional information was sequenced in Read2. The samples were single-indexed.
After sequencing, raw reads were aligned to the human genome (hg38), and the digital expression matrix was generated using cellranger count. Individual samples were merged to generate the digital expression matrix using cellranger aggr. The R package Seurat was used to analyze the digital expression matrix. Cells with fewer than 100 genes and fewer than 500 unique molecular identifiers detected were removed from further analysis. The Seurat function NormalizeData was used to normalize the raw counts. Variable genes were identified using the FindVariableGenes function. The Seurat ScaleData function was used to scale and center expression values in the data set for dimensional reduction. Default parameters were used for the Seurat functions. When needed, samples were integrated using the Seurat functions FindIntegrationAnchors and IntegrateData. Principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) were used to reduce the dimensions of the data, and the first two dimensions were used in plots. To cluster the cells and determine the marker genes for each cluster, we used the FindClusters and FindAllMarkers functions, respectively. Differential expression analysis of the samples was performed using the FindMarkers function and the Wilcoxon rank-sum test. The Benjamini–Hochberg procedure was applied to adjust the false discovery rate.
The normalized expression matrix for the scRNA-seq data was extracted and uploaded to the CytoTRACE web tool (https://cytotrace.stanford.edu/; ref. 41), and the output CytoTRACE score for each cell was then plotted on the UMAP.
Total RNA-seq Analysis
Total RNA from NT sgRNA– or EIF2AK1 sgRNA–treated SF3B1-mutant MDS-RS CD34+ cells at 13 days of erythroid differentiation was purified using the PicoPure RNA Isolation Kit (catalog number #KIT0204, Thermo Fisher Scientific). Quality checks and quantifications were performed with the 2100 Expert Bioanalyzer (Agilent). Illumina low input total RNA libraries were prepared using the SMARTer Stranded Total RNA-seq Kit v2–Pico Input Mammalian (catalog number #634412; Takara Bio). Briefly, 10 ng of total RNA was converted to cDNA using Takara's SMART (Switching Mechanism At 5′ End of RNA Template) technology. Illumina adapters with specific barcodes (Supplementary Table S11) were added to the double-stranded cDNA using 12 cycles of PCR. The PCR products were purified using Ampure Beads (Agencort); then, ribosomal cDNA was specifically depleted using ZapR V2. The remaining cDNA fragments were enriched by 12 cycles of PCR amplification using Illumina universal library primers. The enriched library was purified to yield the final cDNA library. Libraries were combined and sequenced on the Illumina NextSeq500 sequencer using the High Output 150 cycle flow cell with the 75nt PE format. The mapping of the RNA-seq reads to the human reference genome hg19 was performed with Tophat2. The quality of the reads was verified using the FASTQC software (v0.11.8). HTseq software (v0.11.2) was used to summarize the gene-expression counts from the Tophat2 alignment data after sorting the BAM files. Unsupervised analysis was performed using PCA and hierarchical clustering to verify any outliers and the overall similarities or differences among all samples. Differential expression analysis was performed using the DEseq2 package, as we described previously (42).
scATAC-seq Analysis
The scATAC Low Cell Input Nuclei Isolation protocol (10X Genomics) was used to isolate nuclei from FACS-purified cells. The trypan blue exclusion assay was performed to check for intact nuclei. The remaining extracted nuclei were used for the consecutive steps of the scATAC-seq library preparation protocol following 10X Genomics guidelines. Equal molar concentrations of uniquely indexed samples were pooled together. Pooled libraries were sequenced using a NextSeq500 150-cycle flow cell (Illumina). Reads were aligned to the human genome (hg38), and peaks were called using the cellranger-atac count pipeline. Individual samples were merged using the cellranger-atac pipeline to generate the peak-barcode matrix and TF-barcode matrix. To identify specific TF activity for each cell cluster, we used the R package Seurat to analyze the TF-barcode matrix. The raw counts were normalized by the sequencing depth for each cell and scaled for each TF using the NormalizeData and ScaleData functions. PCA and UMAP were applied to reduce the dimensions of the data, and the first two dimensions were plotted. The FindClusters function was used to cluster the cells. The FindAllMarkers function was used to determine the TF markers for each cluster. Differential analysis of TF activity in the samples was performed using the FindMarkers function and the Wilcoxon rank-sum test. Cluster identity was determined based on the activity of master regulators of lineage commitment, as we (43) and others (44, 45) described previously.
Briefly, the HSC cluster was defined based on the lowest activity of lineage-specific markers and highest activity of TFs that regulate stemness, such as the retinoic acid receptor, NRF, and EGR families of TFs. Lymphoid and myeloid bipotent-biased clusters were defined based on the coactivities of major regulators of lymphoid differentiation (e.g., POU4F, TBX families of TFs) and myeloid differentiation (RUNX1 and CEBPA); myeloid clusters were defined based on the highest activities of the TFs SPI1 and CEBPA (master regulators of myelopoiesis); erythroid/megakaryocyte clusters were defined based on the highest activity of the GATA family of TFs.
Cluster-specific peaks were determined using the FindAllMarkers function, and differentially accessible peaks between the samples were determined using the FindMarkers function. Each peak was associated with a specific gene based on its distance to that gene's transcription start site (TSS). Peaks overlapping with a promoter region (−1,000 bp, +100 bp) of any TSS were annotated as peaks in promoters, whereas peaks not in promoter regions but within 200 kb of the closest TSS were annotated as peaks in the distal elements. Peaks not mapped in either the promoters or distal elements were annotated as peaks in intergenic regions.
Targeted Deep Sequencing
Selected samples were sequenced at MD Anderson's Advanced Technology Genomics Core facility. We used a custom SureSelect panel of 200 genes (Agilent Technologies) that are recurrently mutated in hematologic malignancies. DNA from MNCs was fragmented and bait-captured in solution according to the manufacturer's protocols. Captured DNA libraries were then sequenced using a NovaSeq sequencer (Illumina) with 76-bp paired-end reads. Raw sequencing data from the Illumina platform were converted to fastq format and aligned to the hg19 reference genome using the Burrows-Wheeler Aligner. The aligned BAM files were subjected to mark duplication, realignment, and recalibration using Picard and GATK (https://www.broadinstitute.org/gatk/guide/best-practices?bpm=DNAseq). Preprocessed BAM files were then analyzed to detect single-nucleotide variants and small indels using the MuTect and Pindel algorithms, respectively, against a virtual normal sequence developed in-house.
Transmission Electron Microscopy
Fixed MNC samples were washed in 0.1 mol/L sodium cacodylate buffer (catalog number #RT12310, Electron Microscopy Sciences) and treated with 0.1% Millipore-filtered cacodylate buffered tannic acid (catalog number #RT21700, Electron Microscopy Sciences), postfixed with 1% buffered osmium tetroxide (catalog number #RT19172, Electron Microscopy Sciences), and stained with 1% Millipore-filtered uranyl acetate (catalog number #RT22400, Electron Microscopy Sciences). The samples were dehydrated in increasing concentrations of ethanol, and then infiltrated and embedded in LX-112 medium (catalog number #LX112-21212, Ladd Research Industries). The samples were polymerized in a 60°C oven for approximately 3 days. Ultrathin sections were cut in an Ultracut microtome (Leica), stained with uranyl acetate and lead citrate (catalog number #RT17800, Electron Microscopy Sciences), and examined with a JEM 1010 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV. Digital images were obtained using an AMT Imaging System (Advanced Microscopy Techniques Corp).
Statistical Analysis
Flow cytometry data were analyzed with Prism 8 software (GraphPad). The figure legends indicate the statistical test(s) used in each experiment. Statistical significance was represented as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Figure 5 was made using Biorender.com.
Supplementary Material
Acknowledgments
This work was supported by philanthropic contributions to The University of Texas MD Anderson AML/MDS Moon Shot, by the NIH through MD Anderson's Leukemia SPORE grant (P50 CA100632), by a Ladies Leukemia League grant to R. Kanagal-Shamanna, and by an NIH/NIDDK 5U54DK106857 grant to the Animal Modeling Core of the Yale Cooperative Center of Excellence in Hematology. S. Colla is a Scholar of the Leukemia and Lymphoma Society. N. Thongon was supported by the 2020 ASH restart award. This work used MD Anderson's South Campus Flow Cytometry and Cellular Imaging Core, the Advanced Technology Genomics Core, and the High-Resolution Electron Microscopy Facility, which are supported in part by the NIH (NCI) through MD Anderson's Cancer Center Support Grant (P30 CA16672). The authors thank Joseph Munch and Kelly Soltysiak for assistance with manuscript editing.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Data Availability
data sets generated by scRNA-seq are accessible at GEO under accession number GSE188367. data sets generated by bulk RNA-seq are accessible at GEO under accession number GSE202722.
Authors’ Disclosures
J.P. Maciejewski reports grants from the Leukemia and Lymphoma Society, the U.S. Department of Defense, and NIH during the conduct of the study. R. Bejar reports other support from Aptose Biosciences, personal fees from Gilead and Epizyme outside the submitted work. J.S. Carew reports other support from Majestic Therapeutics, LLC outside the submitted work; in addition, J.S. Carew has a patent 9926326 issued. S. Halene reports grants and other support from NIH and other support from The Frederick A DeLuca Foundation during the conduct of the study; personal fees from FORMA Therapeutics outside the submitted work; in addition, S. Halene has a pending patent. V. Santini reports personal fees from BMS, AbbVie, Geron, Gilead, Takeda, Novartis, and Menarini outside the submitted work. K. Clise-Dwyer reports grants from NCI during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
V. Adema: Data curation, formal analysis, methodology. F. Ma: Data curation, software, methodology. R. Kanagal-Shamanna: Methodology. N. Thongon: Methodology. G. Montalban-Bravo: Data curation. H. Yang: Methodology. S.A. Peslak: Supervision. F. Wang: Software, formal analysis. P. Acha: Methodology. F. Sole: Supervision. P. Lockyer: Methodology. M. Cassari: Methodology. J.P. Maciejewski: Supervision. V. Visconte: Methodology. I. Ganan-Gomez: Methodology. Y. Song: Resources, formal analysis. C. Bueso-Ramos: Supervision. M. Pellegrini: Supervision. T.M. Tan: Methodology. R. Bejar: Supervision. J.S. Carew: Methodology. S. Halene: Supervision. V. Santini: Supervision. G. Al-Atrash: Supervision. K. Clise-Dwyer: Methodology. G. Garcia-Manero: Resources, supervision. G.A. Blobel: Supervision. S. Colla: Conceptualization, resources, supervision, investigation, writing–original draft, writing–review and editing.
References
- 1. Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell 2017;168:629–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mian SA, Rouault-Pierre K, Smith AE, Seidl T, Pizzitola I, Kizilors A, et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat Commun 2015;6:10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Awada H, Kerr CM, Durmaz A, Adema V, Gurnari C, Pagliuca S, et al. Clonal trajectories and cellular dynamics of myeloid neoplasms with SF3B1 mutations. Leukemia 2021;35:3324–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64–9. [DOI] [PubMed] [Google Scholar]
- 5. Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011;365:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011;118:6239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 2012;26:542–5. [DOI] [PubMed] [Google Scholar]
- 8. Visconte V, Avishai N, Mahfouz R, Tabarroki A, Cowen J, Sharghi-Moshtaghin R, et al. Distinct iron architecture in SF3B1-mutant myelodysplastic syndrome patients is linked to an SLC25A37 splice variant with a retained intron. Leukemia 2015;29:188–95. [DOI] [PubMed] [Google Scholar]
- 9. Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015;29:1092–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 2016;30:2322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shiozawa Y, Malcovati L, Galli A, Sato-Otsubo A, Kataoka K, Sato Y, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clough CA, Pangallo J, Sarchi M, Ilagan JO, North K, Bergantinos R, et al. Coordinated mis-splicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 2021;139:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol 2013;14:756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 2015;16:302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017;19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belluschi S, Calderbank EF, Ciaurro V, Pijuan-Sala B, Santoro A, Mende N, et al. Myelo-lymphoid lineage restriction occurs in the human haematopoietic stem cell compartment before lymphoid-primed multipotent progenitors. Nat Commun 2018;9:4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 2015;163:1663–77. [DOI] [PubMed] [Google Scholar]
- 18. Yu VWC, Yusuf RZ, Oki T, Wu J, Saez B, Wang X, et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 2017;168:944–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol 2018;68:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mello FV, Land MGP, Costa ES, Teodosio C, Sanchez ML, Barcena P, et al. Maturation-associated gene expression profiles during normal human bone marrow erythropoiesis. Cell Death Discov 2019;5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang P, Zhao Y, Zhong J, Zhang X, Liu Q, Qiu X, et al. Putative regulators for the continuum of erythroid differentiation revealed by single-cell transcriptome of human BM and UCB cells. Proc Natl Acad Sci U S A 2020;117:12868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 2013;121:3246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lieu YK, Liu Z, Ali AM, Wei X, Penson A, Zhang J, et al. SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc Natl Acad Sci U S A 2022;119:e2111703119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, et al. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 2001;20:6909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen JJ. Translational control by heme-regulated eIF2alpha kinase during erythropoiesis. Curr Opin Hematol 2014;21:172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010;40:280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with bortezomib. Cancer Res 2009;69:4415–23. [DOI] [PubMed] [Google Scholar]
- 28. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010;120:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 2013;41:7683–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015;126:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grevet JD, Lan X, Hamagami N, Edwards CR, Sankaranarayanan L, Ji X, et al. Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 2018;361:285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peslak SA, Khandros E, Huang P, Lan X, Geronimo CL, Grevet JD, et al. HRI depletion cooperates with pharmacologic inducers to elevate fetal hemoglobin and reduce sickle cell formation. Blood Adv 2020;4:4560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med 2016;22:672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steensma DP, Wermke M, Klimek VM, Greenberg PL, Font P, Komrokji RS, et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 2021;35:3542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood 2007;109:2693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang P, Peslak SA, Lan X, Khandros E, Yano JA, Sharma M, et al. The HRI-regulated transcription factor ATF4 activates BCL11A transcription to silence fetal hemoglobin expression. Blood 2020;135:2121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esrick EB, Lehmann LE, Biffi A, Achebe M, Brendel C, Ciuculescu MF, et al. Post-transcriptional genetic silencing of BCL11A to treat sickle cell disease. N Engl J Med 2021;384:205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berastegui N, Ainciburu M, Romero JP, Alfonso-Pierola A, Philippe C, Vilas-Zornoza A, et al. Transcriptional regulation of HSCs in aging and MDS reveals DDIT3 as a potential driver of transformation. bioRxiv 2021: 2021.09.08.459384. [Google Scholar]
- 39. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009;114:937–51. [DOI] [PubMed] [Google Scholar]
- 40. Kanagal-Shamanna R, Singh RR, Routbort MJ, Patel KP, Medeiros LJ, Luthra R. Principles of analytical validation of next-generation sequencing based mutational analysis for hematologic neoplasms in a CLIA-certified laboratory. Expert Rev Mol Diagn 2016;16:461–72. [DOI] [PubMed] [Google Scholar]
- 41. Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020;367:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ganan-Gomez I, Yang H, Ma F, Montalban-Bravo G, Thongon N, Marchica V, et al. Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy. Nat Med 2022;28:557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thongon N, Ma F, Santoni A, Marchesini M, Fiorini E, Rose A, et al. Hematopoiesis under telomere attrition at the single-cell resolution. Nat Commun 2021;12:6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 2018;173:1535–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
data sets generated by scRNA-seq are accessible at GEO under accession number GSE188367. data sets generated by bulk RNA-seq are accessible at GEO under accession number GSE202722.