Han and Deng et al. show the relationship between follicular lymphoma tumor B-cell mutations and expression patterns, and associations with characteristics of tumor-infiltrating T-cell subsets.
Abstract
Follicular lymphoma (FL) is a B-cell malignancy with a complex tumor microenvironment that is rich in nonmalignant immune cells. We applied single-cell RNA sequencing to characterize the diverse tumor and immune cell populations of FL and identified major phenotypic subsets of FL T cells, including a cytotoxic CD4 T-cell population. We characterized four major FL subtypes with differential representation or relative depletion of distinct T-cell subsets. By integrating exome sequencing, we observed that somatic mutations are associated with, but not definitive for, reduced MHC expression on FL cells. In turn, expression of MHCII genes by FL cells was associated with significant differences in the proportions and targetable immunophenotypic characteristics of T cells. This provides a classification framework of the FL microenvironment in association with FL genotypes and MHC expression, and informs different potential immunotherapeutic strategies based upon tumor cell MHCII expression.
Significance:
We have characterized the FL-infiltrating T cells, identified cytotoxic CD4 T cells as an important component that is associated with tumor cell–intrinsic characteristics, and identified sets of targetable immune checkpoints on T cells that differed from FLs with normal versus low MHC expression.
See related commentary by Melnick, p. 374.
This article is highlighted in the In This Issue feature, p. 369
INTRODUCTION
Follicular lymphoma (FL) is an indolent lymphoma of germinal center B cells that maintain follicle-like architecture and interact closely with T cells and other immune cells. These immune interactions are critical to FL etiology (1) and can be perturbed by somatic mutations that are frequent in FLs (2–4). Understanding the lymphoma microenvironment (LME) of FL and the interplay between perturbed immune interactions and distinct LME T-cell populations will be important for designing rational immunotherapeutic strategies, but these concepts have yet to be comprehensively addressed using high-throughput approaches. Single-cell RNA sequencing (scRNA-seq) is a powerful and high-throughput approach that has revealed the deregulation of normal B-cell developmental programs and allowed for the characterization of targetable immune checkpoints and receptor–ligand pairs on LME T cells in FL (5, 6). However, these studies have been limited to a few patients and have not yet been used to investigate broader LME profiles or the relationship between somatic mutations, tumor B-cell expression profiles, and changes in the LME. Using scRNA-seq of FL lymph node biopsies, we characterized phenotypically distinct subsets of LME T cells, including a cytotoxic CD4 T-cell population, and validated in a large series that the composition of these T-cell subsets defines four distinct subtypes of LME in FLs. By integrating exome-sequencing and scRNA-seq data, we identified chromatin-modifying gene mutation-associated and -independent perturbation of immune interaction genes encoding proteins such as major histocompatibility complex (MHC) class I and II on tumor cells, which is in turn associated with changes in the frequencies and targetable immune profiles of T-cell subsets in FL tumors.
RESULTS
scRNA-seq of follicular lymphoma
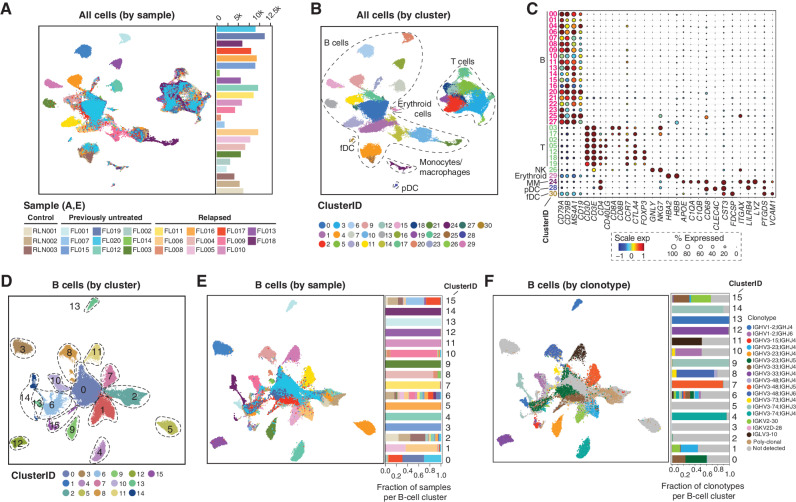
We performed scRNA-seq of 20 FL lymph node biopsies and three reactive lymph nodes (RLN) using the 10X Chromium platform to profile the transcriptome, T-cell receptor (TCR), and immunoglobulin (Ig) repertoires (Supplementary Table S1). RLNs provide a control nonmalignant lymph node with active germinal centers for comparison with malignant FL lymph node tissue. Each biopsy was analyzed fresh to retain cell types that are sensitive to cryopreservation such as plasma cells. This study included 11 previously untreated and nine relapsed FLs (median 1 line of prior therapy; range, 1–6) that were grades 1 to 2 (n = 14) or 3A (n = 6) and 3 control RLN (n = 3) samples. We sequenced a median of 6,138 (range, 635–11,070) cells per sample to a median of 57,933 (range, 49,833–324,873) reads per cell and detected a median of 1,115 (range, 447–2,979) genes per cell. After rigorous quality filtering (Supplementary Fig. S1A), 137,147 cells were retained for subsequent analyses (Fig. 1A; Supplementary Fig. S1B–S1C). We opted to use stringent doublet filtering criteria including the removal of B cells expressing canonical T-lineage markers or productive TCR rearrangements and vice versa. This may remove a minor subset of singlets with coexpression of B/T-lineage markers or antigen receptors, but ensures that the analyses on retained cells are not confounded by potential doublet contamination. Unsupervised clustering analysis following batch-effect correction identified six major cell lineages: B-cell, T-cell, monocyte/macrophage, follicular dendritic cell (fDC), plasmacytoid dendritic cell (pDC), and erythroid cell clusters, as determined by cluster marker genes (Fig. 1B and C; Supplementary Table S2). Cells from RLNs fell into T-cell, monocyte/macrophage, fDC, pDC, and B-cell clusters corresponding to quiescent, proliferating, and plasma cells (Supplementary Fig. S2A–S2D).
Figure 1.
Overview of major cell types and clusters from scRNA-seq of 20 FL tumors. A and B, Uniform Manifold Approximation and Projection (UMAP) plots show 137,147 cells from 20 FL tumors and 3 RLN controls by sample ID (A) and cluster ID (B). Major cell types are annotated in B. C, Bubble plot of cell lineage marker genes are shown for B-cell, T-cell, NK-cell, erythroid, monocyte/macrophage (MM), plasmacytoid dendritic cell (pDC), and follicular dendritic cell (fDC) clusters. D–F, UMAP plots show reclustering of 99,610 B cells by cluster ID (D), sample ID (E), and immunoglobulin clonotype (F). Among B-cell clusters, we identified those corresponding to nonmalignant B cells (C2), proliferating cells (C6), and plasma cells (C15). A malignant B-cell cluster bearing cells from multiple samples was identified (C0). The contribution of each sample to each cluster is shown in the bar graph in E, with many clusters consisting of tumor B cells from a single sample as determined by immunoglobulin clonotype (F) or patterns of inferred copy-number variation (Supplementary Fig. S1).
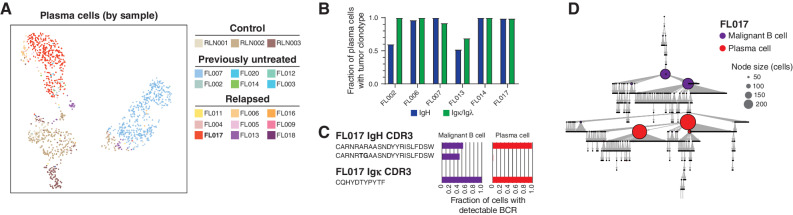
B cells were reclustered (Fig. 1D and E; Supplementary Fig. S3A), and cells were defined as either tumor or nonmalignant by the presence/absence of a clonal immunoglobulin sequence (Fig. 1F; Supplementary Fig. S3B) or arm-level DNA copy-number alterations (Supplementary Fig. S3C). Tumor immunoglobulin clonotypes were not detected for three tumors, presumably due to somatic hypermutation that interfered with primer binding. Clusters of nonmalignant B cells (C2), plasma cells (C15), and proliferating B cells (C6) included cells from both FL and RLN samples (Fig. 1D and E). Notably, plasma cells included those originating from the malignant clone, as determined by the presence of the same immunoglobulin clonotype as that observed in malignant B cells (Fig. 2A and B; Supplementary Table S3), indicating that a subset of malignant clones have the ability to terminally differentiate rather than being developmentally “stalled.” In one of these cases, we observed differences in the hierarchies of immunoglobulin somatic hypermutation between malignant B cells and tumor clonotype-bearing plasma cells (Fig. 2C and D), suggesting that there may be clonal differences in the potential to terminally differentiate. A central cluster (C0) was also found to contain cells from multiple samples but consisted exclusively of clonal malignant B cells from FLs, suggesting that tumor cells from a subset of cases have shared transcriptional characteristics. The FL tumors in the shared C0 cluster were not enriched for any given mutation (Supplementary Fig. S3A) and consisted of both grades 1 to 2 and grade 3A tumors, but tended to have relatively less extensive inferred copy-number variations (Supplementary Fig. S3C) and more frequently originated from treatment-naïve tumors (Supplementary Table S1), suggesting that tumor B cells from relapsed FLs have a greater intersample divergence in transcriptional profiles compared with treatment-naïve FLs that may be associated with chromosomal DNA copy-number changes.
Figure 2.
Plasma cells linked to malignant FL clones. A, UMAP plot showing reclustering of plasma cells (n = 1,275), colored by sample ID. B, Bar plot showing fractions of plasma cells carrying the same clonal expanded immunoglobulin clonotypes, heavy (IgH) and light (Igk/l) chain, as observed in malignant B cells in 6 FL tumors. C, the IgH and IgK CDR3 sequences for the representative case FL-17 and fractions of malignant B and plasma cells with detectable BCR. D, Reconstruction of the immunoglobin variable region gene lineage tree (igTree) based on somatic hypermutation of immunoglobulin genes using scBCR-seq data. Each tree node represents a single mutation (point mutations, deletions, or insertions) separating the CDR3 sequences and the size of the node corresponds to the number of cells carrying a specific CDR3 sequence.
Tumor-Infiltrating T Cells within the follicular lymphoma microenvironment
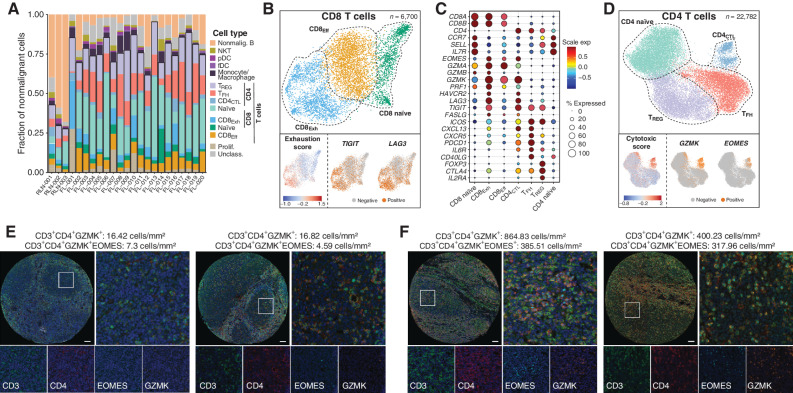
T cells comprised a median of 87.6% (range, 73.8%–98.9%) of the nonmalignant cells within the LME (Fig. 3A). We further characterized phenotypically distinct subsets of CD4 and CD8 T cells by partitioning T cells into major subsets (CD4, CD8, proliferating T, NKT; Supplementary Fig. S4A–S4B), followed by subclustering analysis (Fig. 3B-D; Supplementary Table S4). We did not observe any significant difference in the frequencies of T-cell clusters between grades 1 to 2 and 3A or between previously untreated and relapsed tumors (Supplementary Table S5). Clusters of CD8 T cells included naïve (CCR7, SELL, and IL7R), effector (CD8Eff; granzymes A/B/K and PRF1), and exhausted (CD8Exh; high expression of inhibitory immune-checkpoint genes such as TIGIT and LAG3, and a high exhaustion score) subsets (Fig. 3B and C). Trajectory analysis showed that these represent a functional continuum from naïve through to exhausted states (Supplementary Fig. S5A). Subclustering analysis of CD4 T cells identified four transcriptome states, each with a unique expression of specific marker genes (Fig. 3D; Supplementary Fig. S5B), including naïve (CCR7, SELL, and IL7R), T-regulatory (Treg; FOXP3, CTLA4, and IL2RA), T follicular helper (TFH; PDCD1, TOX, TOX2, CXCR5, and CD40LG), and cytotoxic CD4 T cells (CD4CTL; GZMA/K, NKG7, and EOMES), all of which were detected in both FL and RLN samples. Although naïve, Treg and TFH cells are well-described components of FL (1, 7, 8), there are no prior reports of CD4CTL cells in the LME of FL or any other germinal center-derived lymphoma. CD4CTL cells express CD4 but not CD8A/B, expressed a high cytotoxicity gene signature score that is comparable with CD8 T cells (Supplementary Fig. S6), and with GZMK being the top marker gene having expression detectable in 89.6% of cells. The EOMES transcription factor, which has been implicated in CD4CTL development (9, 10), was also expressed within a subset of cells. In addition, CD4CTL cells bear some similarities to TFH cells, including high expression of CXCL13 and PDCD1, and are transcriptomically most closely related to TFH cells by trajectory analysis (Supplementary Fig. S5B). Although cytotoxic CD4 T cells were not reported by prior scRNA-seq studies of FL, reanalysis of an independent data set from Roider and colleagues (ref. 8; Supplementary Fig. S7A–S7C) identified a population showing similar expression profiles (Supplementary Fig. S7D). We further validated the presence of CD4CTL cells within the neoplastic follicles of an independent series of previously untreated FL tumors (n = 17) using multispectral immunofluorescence (mIF) imaging. Coexpression of the CD3, CD4, and GZMK genes that were characteristic of CD4CTL was detected <2% of other quiescent T-cell and natural killer (NK)/NKT cell subsets and was, therefore, used together (CD3+CD4+GZMK+) as the most comprehensive phenotype to capture CD4CTL by mIF. The EOMES transcription factor was additionally analyzed to define EOMES-negative and EOMES-positive subsets of CD4CTL. As expected from the scRNA-seq data, mIF analysis of FL tumors identified both EOMES-positive and -negative CD4CTL cells, consistent with the scRNA-seq data, with an average density of 346 (range, 64–856) and 192 (range, 41–420) cells/mm2, or 1.94% (range, 0.36%–5.03%) and 1.08% (range, 0.24%–2.2%) of all cells, inside the neoplastic follicle for CD3+CD4+GZMK+ and CD3+CD4+GZMK+EOMES+ cells, respectively (Fig. 3E and F; Supplementary Fig. S8). There was no apparent association between CD4CTL frequency and tumor grade. Thus, our scRNA-seq analysis revealed a cytotoxic CD4 T-cell component within nonmalignant lymph nodes and the FL microenvironment. Further investigation is required to determine the functional characteristics of CD4CTL.
Figure 3.
Tumor-infiltrating T-cell populations in FL. A, A bar graph shows the composition of nonmalignant immune cell populations within FL, with the majority of cells belonging to the T-cell lineages. B, UMAP plots from reclustering of 6,700 CD8 T cells show 3 major populations aligning with naïve, effector (eff), and exhausted (exh) states. Single-cell GSVA of a CD8 T-cell exhaustion signature shows the highest expression in the CD8Exh cluster, which is also characterized by high expression of TIGIT and LAG3. C, A bubble plot shows the proportion of cells of CD8 and CD4 T-cell clusters expressing known phenotypic marker genes (size of circles) and their average expression levels (color of circles). D, UMAP plots from reclustering of 22,782 CD4 T cells shows 4 major subpopulations aligning with naïve, Treg, TFH, and CD4CTL states. Single-cell GSVA of a cytotoxic score including immune effector molecules shows high expression in the CD4CTL cluster, which is also characterized by high expression of GZMK and EOMES. E and F, Exemplar multispectral immunofluorescence images show the abundance of cytotoxic CD4 T cells within tumors with low (E) or high (F) abundance within the neoplastic follicle. Scale bar, 50 μm.
Hierarchical Clustering of scRNA-seq–Defined T-cell Subset Signatures Identifies Four Major follicular lymphoma microenvironment Subtypes
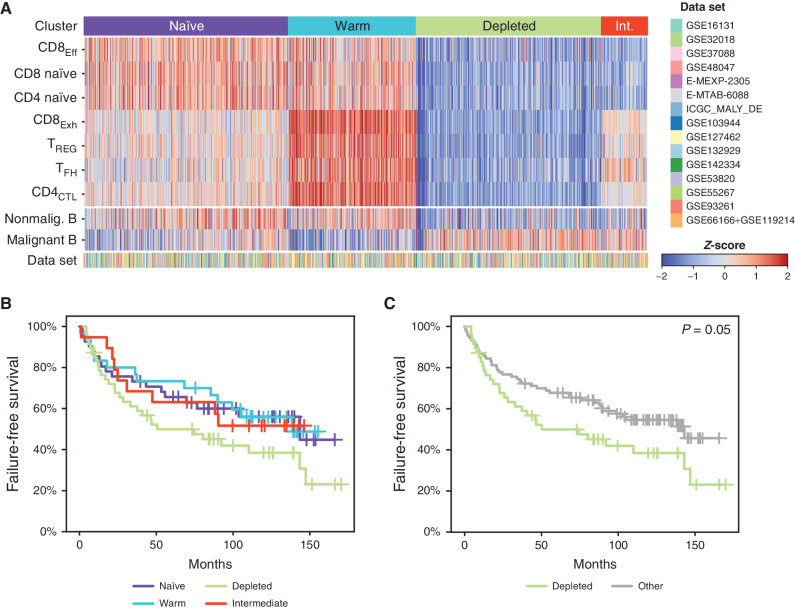
The abundance of phenotypically distinct LME T-cell populations that we characterized by scRNA-seq was highly variable across patients (Fig. 3A). We therefore assessed their representation across a large external validation series of bulk gene-expression profiling (GEP) from 1,269 FLs compiled from 15 data sets (11–24) using hierarchical clustering of the scRNA-seq–derived signatures. Specifically, marker genes from each CD4 and CD8 T-cell cluster were used to infer the abundance and frequencies of each cell type using a microenvironment classification approach (25, 26) to define sets of tumors with similar LME T-cell profiles (Fig. 4A). Signatures from malignant and nonmalignant B cells were also analyzed but were not utilized for the T cell–based clustering. Signatures from CD4CTL and CD8Exh populations had multiple overlapping genes and may therefore be partially confounded during analysis due to some genes contributing to the single sample gene set enrichment analysis (ssGSEA) scores of both CD4CTL and CD8Exh signatures. However, these signatures have a sufficient number of unique genes (89 genes for CD8Exh, 179 genes for CD4CTL) to make them amenable for discrimination using the approach that we used for clustering. Based on the median scaled ssGSEA scores of T-cell populations, samples were clustered into four groups using hierarchical unsupervised clustering. After clustering, using a similar approach, the values for B-cell signatures were calculated and projected onto the heat map. Analysis revealed four distinct subtypes of LME in primary human FL based on the relative abundance of these T-cell subsets: (naïve, n = 461) high in CD8Eff, CD8 naïve, and CD4 naïve; (warm, n = 288) high in CD8Exh, Treg, TFH, and CD4CTL; (depleted, n = 418) high in malignant B cells and depleted of T-cell subsets; (intermediate, n = 102) high in malignant B cells and depletion of CD8Eff, CD8 naïve, and CD4 naïve. Failure-free survival (FFS) data were available for a subset of 137 advanced-stage FL patients treated with first-line R-CHOP, from Pastore and colleagues (14). Patients with naïve, warm, and intermediate LME subsets showed similar trends for FFS (Fig. 4B). Compared with these collective subsets, patients with a T cell–depleted LME had a significantly inferior FFS (log-rank P = 0.05, Fig. 4C). We did not observe any difference in grade or stage between LME subsets (Supplementary Fig. S9A–S9D). The T-cell landscape as defined by scRNA-seq cell composition and measured in bulk GEP data, therefore, defines four distinct LME subsets in primary human FL, including a T cell–depleted subset with an inferior clinical outcome following R-CHOP chemotherapy.
Figure 4.
Deconvolution of T-cell signatures in a large independent series of FL tumors. A, A heat map shows the relative proportions of CD8 and CD4 LME T-cell populations inferred by deconvolution of publicly available bulk gene-expression microarray or RNA-sequencing data sets (n = 1,269 FL tumors from 15 data sets). Unsupervised clustering identified 4 characteristic patterns (naïve, warm, depleted, intermediate (Int.)) with a different relative abundance of tumor-infiltrating T-cell populations. Relative B-cell proportion is shown for reference using the same method but was not used for clustering. B and C, Kaplan–Meier curves of FFS for 137 R-CHOP-treated FL patients are shown according to all LME subtypes (B) or comparing the depleted subtype to others (C). P values were calculated using a log-rank test.
Multiple Mechanisms of MHCII Loss on follicular lymphoma Tumor B Cells
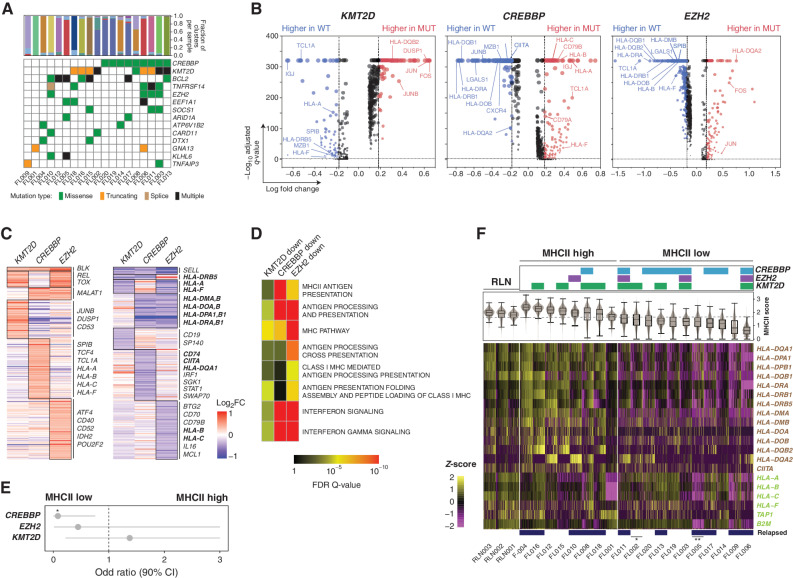
Mutations in chromatin-modifying genes (CMG) are a hallmark of FL (27) and affect the expression of genes in tumor B cells through epigenetic dysregulation. The most frequently mutated CMGs (KMT2D, CREBBP, and EZH2) have each been implicated in deregulating interactions between tumor cells and T cells (3, 4, 28), leading us to hypothesize that these mutations may underlie tumor cell–intrinsic gene-expression changes that drive differential LME T-cell profiles. Using the whole-exome sequencing of tumors with available DNA (n = 19; Supplementary Table S6; Fig. 5A), we applied single-cell differential GEP to identify genes that were significantly altered in association with these mutations (Fig. 5B; Supplementary Tables S7). Only genes with a high variant allele frequency (>20%) were considered, to avoid the confounding effect of subclonal variants that cannot be genotyped at the single-cell level using our scRNA-seq approach. Collectively, the union of genes with significantly reduced expression (FDR q-value <0.05, fold change >1.2; n = 355; Supplementary Table S7) in association with one or more of these mutations was significantly enriched for genes involved in immune cell interactions (P = 1.4 × 10−7) including those with a role in antigen processing and presentation (P = 2.2 × 10−29), confirming that these mutations alter genes involved in immune cell interactions (Fig. 5C). In line with prior reports, CREBBP and EZH2 mutations were both associated with reduced expression of multiple genes involved in antigen presentation through the MHC molecules (refs. 3, 28; Fig. 5D; Supplementary Table S8), which present antigens that are recognized by T-cell receptors and therefore affect T-cell activation. Mutations of CREBBP were detected in nine tumors, cooccurred with EZH2 mutations in three out of four EZH2-mutant tumors, and were predominantly associated with lower MHCII expression (Fig. 5C–E). In contrast, EZH2 mutations were selectively associated with lower MHC class I (MHCI) expression (Fig. 5C and D). KMT2D mutations were also associated with reduced expression of a subset of MHCI genes and cooccurred with EZH2 mutations in three of four EZH2-mutant tumors. Using nonmalignant B cells from RLNs as a reference to define normal expression levels of MHCI and MHCII genes that were found to be deregulated in association with somatic mutations, we observed that loss of MHCI and/or MHCII was not restricted to EZH2 and/or CREBBP-mutant tumors (Fig. 5F). Specifically, MHCII loss was most prevalent and observed in 58% (11/19) of tumors, but 27% (3/11) of MHCII-low tumors had no detectable CREBBP or EZH2 mutations. Further, one CREBBP-mutant tumor showed minimal change in MHCII expression. CMG mutations in FL are therefore associated with perturbed expression of immune interaction genes on tumor B cells, but additional mechanisms exist for MHCI and MHCII loss, such as B2M (29) or CIITA (30) mutations, respectively (Fig. 5F), that are likely to have an equal impact on tumor-infiltrating immune cells via deregulation of immune synapse formation.
Figure 5.
Effect of somatic mutations on tumor B-cell expression profiles. A, An oncoplot shows recurrently mutated genes in the 19 FL tumors with available DNAs. B, Volcano plots displaying differentially expressed genes (DEG) between tumor B cells from KMT2D (left), CREBBP (middle), or EZH2 (right) wild-type and mutant tumors. Examples are annotated and the full list is provided in Supplementary Table S6. C, Heat maps displaying DEGs with increased (left) or decreased (right) expression associated with each mutation, with genes encoding cell-surface proteins annotated. Genes highlighted in bold belong to the MHCI and MHCII pathways defined by the KEGG pathway database. D, Pathway enrichment analysis of downregulated DEGs associated with each mutation showing an association between MHCII and CREBBP mutation, MHCI and EZH2 mutation, and IFN signaling with both CREBBP and EZH2 mutations. A full list of pathways is in Supplementary Table S8. E, Odds ratio (± 95% CI) is shown for the association between individual mutations and MHCII status (two-tailed Fisher exact test P = 0.028). F, The expression of MHCII (brown, above) and MHCI genes (green, below) is shown for individual tumor B cells from each tumor with available mutation data. Mutations of CREBBP, EZH2, and KMT2D are annotated at the top. Sample IDs are labeled at the bottom and tumors with additional mutations in *CIITA and **B2M that may also affect MHCII and MHCI expression, respectively, annotated by asterisks.
Frequencies of lymphoma microenvironment T cells Are Associated with Tumor B-cell MHCII Expression
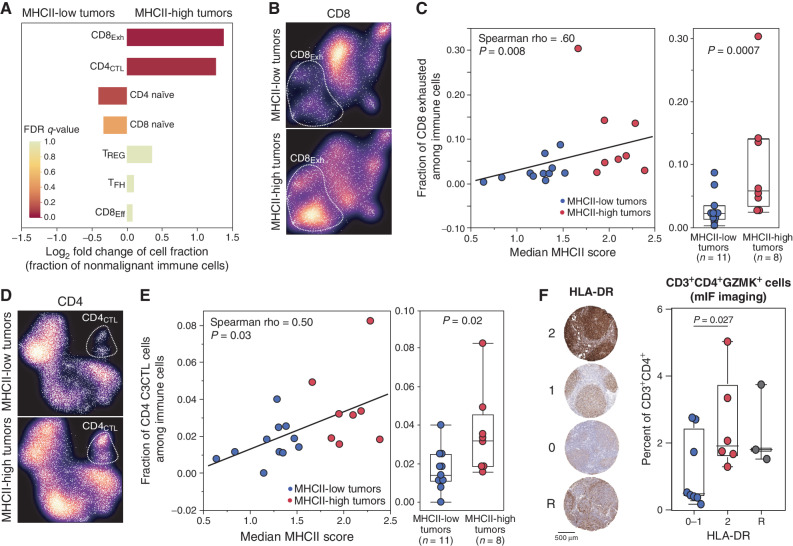
Having observed different patterns of LME T cells in FL, and mutation-associated changes in MHCI and MHCII expression on tumor B cells, we next evaluated whether these features were associated. Tumor MHCII loss was more significantly associated with LME T-cell frequencies than somatic mutations of CREBBP, EZH2, or KMT2D (Supplementary Table S9) and was more frequent than MHCI loss, so we focused on this feature. MHCII-low tumors had significantly reduced levels of CD8Exh (Fig. 6A–C) and CD4CTL subsets (Fig. 6D and E) that are high in the “warm” LME subtype and low in the “depleted” LME subtype. In contrast, MHCII-low tumors had significantly higher frequencies of naïve CD4 and CD8 T cells, features of the “naïve” LME subtype, though to a lesser degree of significance (Fig. 6A). Deconvolution of pseudobulk transcriptomes from our scRNA-seq cohort using the Kassandra algorithm similarly supported these observations (Supplementary Fig. S10A). Analysis of TCR clonotypes did not reveal any significant difference in CD4 or CD8 T-cell clonality between LME subtypes (Supplementary Fig. S10B–S10C). Despite a relatively modest sample size, we observed both a quantitative and qualitative relationship between MHCII expression/status and the frequencies of CD8Exh (Fig. 6C) and CD4CTL (Fig. 6E) subsets. We, therefore, sought to validate the association between MHCII expression and CD4CTL abundance in an independent series by performing IHC staining for HLA-DR in FL tumors for which mIF imaging of CD4CTL had been performed (Fig. 3E and F). Tumors were scored by two trained hematopathologists according to their staining intensity for HLA-DR within the neoplastic follicle (Fig. 6F), highlighting 6 MHCII-high tumors (score 2), 8 MHCII-low tumors (scores 0–1), and 3 tumors with reactive (R) MHCII-high cells that likely correspond to macrophages within a background of MHCII-low cells. CD3+CD4+GZMK+ cells, the phenotype best encompassing the majority of CD4CTL, were significantly higher in frequency within the neoplastic follicles of MHCII-high tumors compared with MHCII-low tumors (Fig. 6F; one-tailed t test P = 0.027). In mantle cell lymphoma, tumor cell immunopeptidome profiling revealed the presentation of tumor idiotype peptides in MHCII that were recognized by CD4CTL in the peripheral blood (31). We, therefore, reasoned that loss of MHCII may be selectively acquired in cells that have accumulated immunogenic mutations in their idiotype sequences, and thus may be restricted to immunogenic clades of the immunoglobulin hierarchy. By evaluating paired single-cell BCR sequencing data, we only found a single anecdotal example of this (Supplementary Fig. S11) with all remaining MHCII-low tumors having uniformly low expression throughout the immunoglobulin hierarchy. Thus, the frequencies of FL LME T-cell subsets, including cytotoxic CD4 T cells, are correlated with tumor B-cell MHCII expression.
Figure 6.
Association between tumor MHCII status and tumor-infiltrating T-cell populations. A, A bar graph shows the fold change in cellular fractions of CD8 and CD4 T-cell populations between MHCII-low and MHCII-high tumors, colored by Fisher exact FDR q-value. The CD8Exh (fold change 3.43, Fisher exact q = 0.05) and CD4CTL (fold change 2.11; Fisher exact q = 0.07) populations are significantly higher in MHCII-high tumors compared with MHCII-low tumors. B, UMAP density plots show the relative representation of CD8Exh between MHCII-low (above) or MHCII-high (below) tumors. C, Scatter plot and bar plot show the quantitative and qualitative association between CD8Exh population and either the expression or status of MHCII on tumor B cells, respectively. D, UMAP density plots show the relative representation of CD4CTL, between MHCII-low (above) or MHCII-high (below) tumors. E, Scatter plot and bar plot show the quantitative and qualitative association between CD4CTL population and either the expression or status of MHCII on tumor B cells, respectively. F, IHC staining of HLA-DR was used to assign tumors into high (2), low (0–1), and reactive (R) groups, which were evaluated for the intrafollicular density of CD3+CD4+GZMK+ cytotoxic CD4 T cells. P values between MHCII-high and -low groups were calculated using a one-tailed t test.
Tumor B-cell MHCII Expression Is Associated with Differential Expression of Immunotherapeutic Targets on lymphoma microenvironment T Cells
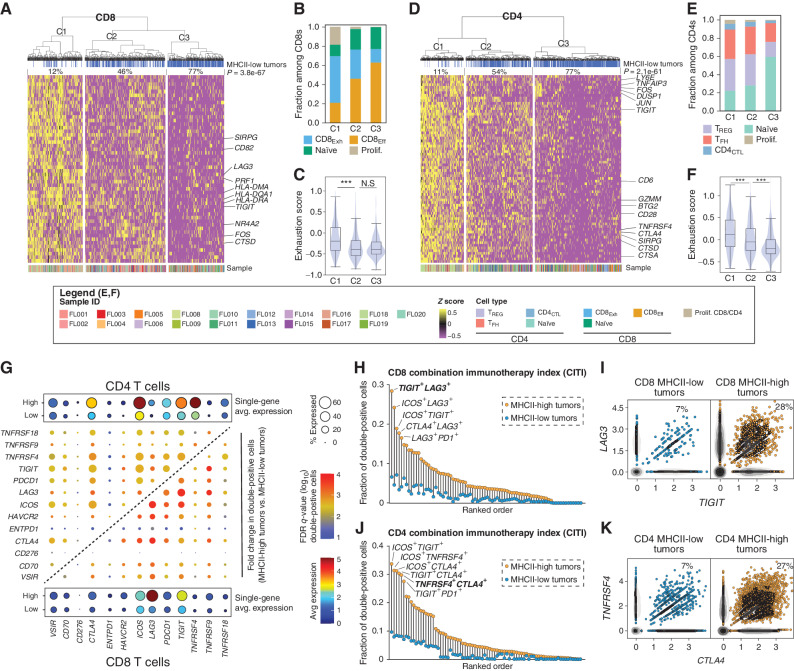
Having observed changes in T-cell frequencies correlated with MHCII expression on tumor B cells, we next explored differences in gene-expression profiles of LME CD4 and CD8 T cells by MHCII status using single-cell differential gene-expression analysis (Supplementary Tables S10–S11; Fig. 7A–F). Cells were clustered within the space of the differentially expressed genes (DEG), which revealed three clusters for both CD4 and CD8 T cells that had a significantly different representations of cells originating from MHCII-high versus MHCII-low tumors (Fig. 7A, CD8, P = 3.8 × 10−67; Fig. 7D, CD4, P = 2.1 × 10−61). The DEGs include markers of T-cell activation, transcription factors, and multiple targetable cell-surface immune-checkpoint molecules (e.g., LAG3, TIGIT, and CTLA4). C1 clusters that have the lowest frequency of cells from MHCII-low tumors expressed the highest level of these genes, in comparison with the C3 clusters that have the greatest frequency of cells from MHCII-low tumors and express low levels of these genes. This is suggestive of higher levels of T-cell activation and exhaustion in tumors that have retained MHCII expression, as supported by the composition of cell types (Fig. 7B and E) and GSVA analysis of a previously described exhaustion score (Fig. 7C and F). Tumors with higher levels of T-cell exhaustion dominate the activated/exhausted C1 cluster (Supplementary Fig. S12A), with the frequency of cells in the CD4 and CD8 activated/exhausted C1 cluster being highly correlated in each tumor (Supplementary Fig. S12B), in line with variable levels of both CD4 and CD8 T-cell activation/exhaustion being associated with high MHCII expression.
Figure 7.
Association between tumor MHCII status and targetable features of LME T cells. A, DEGs between CD8 T cells from MHCII-low vs. MHCII-high tumors were subjected to unsupervised hierarchical clustering which identified 3 clusters with significantly different proportions of cells from MHCII-low/-high tumors (top track, P = 3.8 × 10−67). B, A bar graph of cell state composition in each DEG cluster from A, which shows higher fractions of CD8Exh cells in C1 and a higher fraction of CD8Eff cells in C3. C, GSVA showed higher expression of exhaustion signature genes in cells within C1 compared with either C2 or C3. D, DEGs between CD4 T cells from MHCII-low vs. MHCII-high tumors were subjected to unsupervised hierarchical clustering which identified 3 clusters with significantly different proportions of cells from MHCII-low/high tumors (top track, P = 2.1 × 10−61). E, A bar graph of cell state composition in each DEG cluster from D, which shows relatively higher fractions of CD4CTL, TFH, and Tregs in C1 and a higher fraction of CD4 naïve cell in C3. F, GSVA showed higher expression of exhaustion signature genes in cells within C1 compared with either C2 or C3. G, A bubble plot shows the average expression of immune-modulatory genes on CD4 (top left) or CD8 (below right) T cells in MHCII-low or -high tumors. The center grid shows the fold change and significance of pairs of immune-modulatory genes between MHCII-low and MHCII-high tumors for CD4 and CD8 T cells. H, The CITI shows the dual expression of pairs of ICT targets in CD8 T cells from MHCII-low (blue) and MHCII-high (yellow) tumors, in the same ranked order. I, Scatter plots show the coexpression of the ICT targets LAG3 and TIGIT on CD8 T cells from MHCII-low (blue) and MHCII-high (yellow) tumors. J, The CITI shows the dual expression of pairs of ICT targets in CD4 T cells from MHCII-low (blue) and MHCII-high (yellow) tumors, in the same ranked order. K, Scatter plots show the coexpression of the ICT targets TNFRSF4 (OX40) and CTLA4 on CD4 T cells from MHCII-low (blue) and MHCII-high (yellow) tumors.
We next assessed the expression of pairs of markers that are targetable using current immune-checkpoint therapies (ICT), within the CD4 and CD8 T-cell compartments (Fig. 7G; Supplementary Table S12). FL tumors with low MHCII showed a uniformly lower expression of each individual ICT target on both CD4 and CD8 LME T cells, and a concordantly reduced expression of all ICT target pairs (Fig. 7G). We summarized this as a combination immunotherapy index (CITI), corresponding to the fraction of cells expressing both targets from each ICT pair, and compared CITI between CD8 and CD4 T cells from MHCII-high and MHCII-low tumors (Fig. 7H–K). For all evaluable pairs of ICT targets, the fraction of cells expressing both targets was uniformly lower in MHCII-low tumors compared with MHCII-high tumors. Within the CD8 T-cell compartment, LAG3 and TIGIT showed the highest CITI in MHCII-high tumors and significantly reduced frequency in MHCII-low tumors (fold change = 4.3; FDR q-value = 1.6 × 10−3; Fig. 7H and I). Among ICT pairs with the highest CITI in CD4 T cells from MHCII-high tumors were TNFRSF4 (aka. OX40) and CTLA4, which also showed significantly reduced frequency in MHCII-low tumors (fold change = 3.9; FDR q-value = 0.01; Fig. 7J and K). Thus, tumor cell MHCII expression correlates with the frequency and targetable immune profile of LME T cells in FL, highlighting subsets of FL that are likely to have differential responses to specific immune-checkpoint blockade.
DISCUSSION
FL is an indolent disease, with some patients having equivalent overall survival to age-matched controls (32, 33). Decreasing the use of cytotoxic chemotherapy in the treatment of FL is therefore a priority. The LME of FL is a complex ecosystem that includes large numbers of T cells that provide survival signals that are integral to disease etiology, offering an attractive opportunity for immunotherapeutics that target critical nexuses. However, single-agent checkpoint blockers such as anti–PD-1/PD-L1 are largely ineffective in FL (34). Understanding the characteristics of the FL LME and how it is modulated by tumor-cell-intrinsic characteristics is, therefore, an important step toward the rational design of combination immunotherapeutic strategies that may have increased efficacy.
Prior scRNA-seq studies of FL and DLBCL have provided insight into major immune cell populations and highlighted critical receptor–ligand pairs that likely regulate T-cell activity (6–8). The large number of cells that we sequenced afforded us the power to identify an additional transcriptionally distinct subset of CD4 T cells expressing effector molecules such as granzymes and perforin (CD4CTL). These cells have not been previously appreciated as a component of the FL LME, and have been infrequently described in other cancers such as bladder cancer (35) and mantle cell lymphoma (31). CD4CTL play an important role in antiviral immune responses (36), and their development in this context has been shown to be mediated by the transcription factors T-bet or EOMES (36). Consistent with this, we observed high expression of EOMES in a subset of the CD4CTL that we defined. Interestingly, CD4CTL were also detected within RLN samples, suggesting that these cells may be a normal component of the lymph node microenvironment. In addition, we identified multiple potential therapeutic targets on CD4CTL, including markers of exhaustion phenotype such as CTLA4, LAG3, and HAVCR2 (a.k.a. TIM-3). Khodadoust and colleagues (31) identified cytotoxic CD4 T cells within the peripheral blood of mantle cell lymphoma patients following tumor idiotype vaccination and showed that cytotoxic CD4 T cells recognizing idiotype peptides presented in MHCII are capable of killing cognate tumor cells (31). Furthermore, we have shown that loss of MHCII expression on FL tumor cells is associated with reduced proliferation of CD4 T cells (28). It is therefore plausible that loss of MHCII by FL tumor cells is a mechanism of immune escape from CD4CTL and results in their reduced proliferation and underrepresentation within the FL LME. In line with this, we observed reduced frequencies of CD4CTL in tumors with low MHCII expression. Future studies are needed to characterize the role of CD4CTL in normal and malignant lymphoid tissues, and whether these cells can be targeted to induce antilymphoma immunity.
Tumor B cells have been studied in FL and other germinal center-derived lymphomas by scRNA-seq and found to have a perturbed expression of genes and signatures that are normally dynamically regulated during B-cell development (5, 8). Deregulation of normal B-cell development programs via somatic mutations is a hallmark of FL and has been thought to result in a “block” in terminal differentiation (27). However, our use of freshly processed tumors in this study allowed us to retain plasma cells, which we observed to bear tumor immunoglobulin rearrangements in multiple cases. To the best of our knowledge, this is the first direct evidence that FL tumor cells can retain the potential to terminally differentiate.
Here we also evaluated for the first time in the context of lymphoma using scRNA-seq the relationship between tumor cell gene-expression changes and somatic mutations. Loss of MHCII is common in FL and diffuse large B-cell lymphoma and has been linked to recurrent mutations in CREBBP and EZH2 (3, 28). We not only confirmed this association at the single-cell level but also identified multiple cases of FL with CREBBP and EZH2 mutation-independent loss of MHC expression. Furthermore, we found that the MHCII expression status is more significantly associated with LME T-cell characteristics than somatic mutations. Responses to therapy in FL are likely to be influenced by the LME, as has previously been indicated by bulk gene-expression microarray (37) and NanoString analysis (38). For example, Tobin and colleagues recently showed that FLs with low immune infiltration were enriched for cases with the progression of disease within 24 months (38). By leveraging our discovery-based derivation of T-cell signatures from scRNA-seq data to deconvolute immune cell frequencies from bulk expression data, we identified four major LME subtypes of FLs characterized by a different abundance of LME T cells, including a “depleted” subtype with relatively lower frequencies of all LME T-cell subsets. A “depleted” LME subtype was associated with reduced FFS in R-CHOP–treated patients, consistent with prior observations from Tobin and colleagues (38) in the setting of FL and by Kotlov and colleagues in the setting of diffuse large B-cell lymphoma (25). However, associations between LME characteristics and outcome in FL were observed within the context of rituximab-chemotherapy (R-chemo) regimens and therefore need to be prospectively evaluated with alternative first-line therapies such as bendamustine and rituximab (BR) or revlamid and rituximab (R2). Furthermore, we expect that FL LME subtypes or other associated LME characteristics are likely to be associated with differential responses in subsequent lines of therapy with chimeric antigen receptor (CAR) T cells, immunotherapies or T-cell engagers. We provided some evidence for this by demonstrating that the expression of immunotherapeutic targets is significantly different in tumors with high or low MHCII expression, including higher frequencies of LAG3+TIGIT+ CD8 T cells and CTLA4+TNFRSF4+ CD4 T cells in tumors with high MHCII. Importantly, a combination of CTLA4-blocking and TNFRSF4-agonist antibodies have shown efficacy in lymphoma preclinical models (39). Our hypothesis that tumor MHCII expression may be associated with differential response to immunotherapy is also consistent with observations in classic Hodgkin lymphoma, in which expression of MHCII on malignant Reed-Sternberg cells is associated with response to PD-1 blockade (40). Moreover, MHCII can be therapeutically modulated using HDAC3-selective inhibitors (41) or EZH2 inhibitors (3), suggesting that MHCII-related LME profiles may be potentially amenable to therapeutic manipulation. We, therefore, posit that it is important to evaluate responses to immune therapies within the context of FL LME subtypes and/or other characteristics such as tumor cell MHCII expression, in the event that high response rates are observed only within patients with specific LMEs and therefore appear underwhelming in an unselected population and/or in order to identify opportunities for rational therapeutic combinations.
Caveats to this study include the lack of mutation data for tumors evaluated by bulk GEP to validate the relationship between tumor microenvironment subtype and mutations of CREBBP or EZH2. However, the relationship between CREBBP mutations and MHCII expression is well established and has been independently validated by multiple groups (28, 41–43). Tumor MHCII status also cannot be predicted from bulk GEP data due to highly variable frequencies of tumor-infiltrating T cells and other antigen-presenting cells that express MHCII, and will therefore require prospective validation using orthogonal approaches. Consistent with our scRNA-seq data, T-cell subsets that express high levels of exhaustion markers (CD4CTL and CD8Exh) were also correlated in their relative representation across these LME subtypes defined by GEP deconvolution. We also chose to restrict our analysis to mutations with a high variant allele frequency so as to avoid the possible confounding effects of subclonal mutations on tumor B-cell expression profiles. Although CREBBP mutations have been found to be predominantly clonal and invariable from site-to-site and during disease evolution of FL (28, 44), Haebe and colleagues recently showed site-to-site variability in expression patterns of malignant B cells including variable MHCII expression in some cases (7). Subclonal patterns of gene expression have also been shown to be important in therapeutic resistance in B-cell lymphoma (8). However, the relative representation of immune cell populations is remarkably consistent between sites, suggesting that tumor site sampling bias may only have a limited impact on LME profiles. Nonetheless, patterns of clonal heterogeneity linked with subclonal mutations, and the spatial relationships between subclones and unique LME components, will be important considerations as single-cell technologies are developed that allow for simultaneous and spatially resolved genotyping and GEP.
In conclusion, the results of this work have important implications for FL biology and therapy. We show that the FL LME is highly variable across patients, influenced by tumor-cell–intrinsic characteristics such as somatic mutations and MHCII expression, and associated with patient outcome. Specifically, we implicate mutation-driven loss of MHCII as a potential mechanism of evasion from a cytotoxic CD4 T-cell population, and highlight differential expression of immunotherapeutic targets in MHCII high and low tumors. These observations provide insight into the potential selective advantage of MHCII loss in FL, and an important context in which to design and evaluate cellular or immune-therapeutic strategies to improve the outcomes of FL patients.
METHODS
Sample Collection and Single-Cell Preparation
FL tumor specimens and reactive lymph node specimens (Supplementary Table S1) were obtained with written informed consent in accordance with protocols approved by the review board of the University of Texas MD Anderson Cancer Center (protocols 2005-0656 and PA19-0420) and the Declaration of Helsinki. Race and ethnicity data were not available. Reactive lymph node biopsies were obtained due to suspicion for lymphoma, but diagnosed as RLN by pathology review of an alternate core taken during the same biopsy. All samples were mechanically dissociated into single-cell suspension. PBS containing 0.04% UltraPure BSA (50 mg/mL) was used for cell washing and resuspension to minimize cell losses and aggregation. Cell viability was assessed by trypan blue examination, and samples with more than 80% viable cells were chosen for Chromium Single-Cell Immune Profiling Solution, according to Chromium Single-Cell 5′ Library and Gel Bead Kits User Guide (v1 Chemistry). Briefly, the final cell concentration was adjusted to ∼1,000 cells/μL, single-cell suspension mixed with reverse transcription (RT) master mixture was loaded on a 10X Genomics single-cell instrument. GEMs were broken and single-strand cDNA was cleaned up with DynaBeads. Amplified cDNA quality and quantity were assessed by High Sensitivity D5000 DNA Screen Tape analysis (Agilent Technologies) and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific).
Single-cell RNA-Library Construction and Sequencing
We used the 10X Genomics Single-Cell 5′ Library Kit (PN-1000020) to construct indexed sequencing libraries following the manufacturer's protocol. Sequencing with indexing the Chromium i7 Samplex index Kit (PN-120262) was conducted on an Illumina NovaSeq sequencer with 2 × 100 bp paired reads to achieve a depth of at least 50,000 read pairs per cell.
Single-cell V(D)J Enrichment Library Construction and Sequencing
The Chromium Single-Cell V(D)J Enrichment Kit (Human B Cell, PN-1000016; Human T-cell, PN-1000005) from 10X Genomics was used to enrich immune repertoire, TCR or B-cell immunoglobulin (Ig) transcripts. Fifty nanograms of enriched BCR or TCR products was used for library construction, according to the manufacturer's protocol. The single-cell V(D)J enriched library was indexed by the Chromium i7 Samplex index Kit (PN-120262) and multiple samples were pooled and sequenced in a lane of an Illumina HiSeq 4000 at 2 × 150 bp with a minimum depth of 5,000 read pairs per cell.
scRNA-seq Bioinformatics
Raw Sequencing Data Processing, Quality Check, Data Filtering, and Normalization.
The raw scRNA-seq data were preprocessed (demultiplex cellular barcodes, reads aligned (genome reference build: hg19), and feature-barcode matrix) using Cell Ranger (10X Genomics, v3.0.2). Detailed QC metrics were generated and evaluated. Genes detected in <3 cells and cells where <200 genes had nonzero counts were filtered out and excluded from subsequent analyses. Low-quality cells where >15% of the read counts derived from the mitochondrial genome were also discarded. In addition, cells with a number of detected genes >6,000 were discarded to remove likely doublet or multiplet captures. The resulting cells were further filtered using the following criteria to clean additional possible doublets: (i) The cells with both productive BCRs and TCRs were removed. (ii) For the T, NK, myeloid, and B cells derived from unsupervised clustering analysis, we further cleaned out cells expressing discrepant canonical markers. For example, in the T and NK cell lineages, the cells or cell clusters that have productive BCRs, or express lineage-specific B or myeloid cell markers were removed. Similarly, in the myeloid cell populations, the cells or cell clusters that have productive TCRs/BCRs, or expressing canonical T or B-cell markers were removed. Finally, cell clusters that have productive TCRs or express T-cell lineage markers were removed from malignant/nonmalignant B-cell lineages. We next performed batch-effect correction on LME cell populations using Harmony (45). The results of principal component analysis, uniform manifold approximation and projection (UMAP; ref. 46) plots, and sample-by-cluster distribution were carefully reviewed. Seurat (version 3.2.0; ref. 47) was applied to the filtered gene–cell matrix to generate the normalized UMI counts using the NormalizeData function.
Unsupervised Cell Clustering and Dimensionality Reduction.
Seurat version 3 was applied to the normalized gene–cell matrix to identify highly variable genes. The elbow plot was generated with the ElbowPlot function of Seurat (48) and, based on which, the number of significant principal components (PC) was determined. Different resolution parameters for unsupervised clustering were then examined in order to determine the optimal number of clusters. For this study, the first 100 PCs calculated using the top 2,000 highly variable genes identified by Seurat were used for unsupervised clustering analysis with the resolution set to 0.5, yielding a total of 31 cell clusters. Dimensionality reduction and 2-D visualization of the single-cell clusters were performed using UMAP3 with Seurat function RunUMAP.
Determination of Major Cell Types and Cell States.
To determine the cell types and cell states, cluster marker genes were calculated using FindAllMarkers function in Seurat R package. The significant DEGs (FDR q-value < 0.05; fold change > 1.2) were examined and an integrative approach was used to determine cell types and states. The major cell type (CD4 and CD8) was defined by marker gene expression (CD3D, CD4, CD40LG, CD8A, and CD8B) by 10× transcriptome and CapID sequencing data. The functional state of each cluster (e.g., activated, memory, exhausted, and regulatory) was determined using markers described by Sade-Feldman and colleagues (49) and Zheng and colleagues (50).
Inferring Cell-Cycle Stage, Hierarchical Clustering, DEGs.
The cell-cycle stage was computationally assigned to each individual cell using the R code implemented in Seurat based on expression profiles of the cell-cycle–related signature genes, as previously described (51). DEGs were identified for each cluster using the FindMarkers function of Seurat R package, and the DEG list was filtered with the default criteria to select genes that are expressed in 10% or a greater fraction in the more abundant group with the absolute expression fold change >1.2 and FDR q-value <0.05. Hierarchical clustering was performed for each cell type using the Ward's minimum variance method. Heat map was then generated using the heat map function in pheatmap R package for selected DEGs.
TCR V(D)J Sequence Assembly, Paired Clonotype Calling, TCR Diversity and Clonality Analysis, and Integration with scRNA-seq Data.
Cell Ranger v3.0.2 for V(D)J sequence assembly was applied for TCR reconstruction and paired TCR clonotype calling. The CDR3 motif was located and the productivity was determined. The clonotype landscape was then assessed and the clonal fraction of each identified clonotype was calculated. The TCR clonotype diversity matrix was calculated using the tcR R package (52). TCR clonality was defined as 1-Peilou's evenness and was calculated on productive rearrangements as previously described (53). Clonality values approaching 0 indicate a very even distribution of clone frequencies, whereas values approaching 1 indicate an increasingly asymmetric distribution in which a few clones are present at high frequencies. The TCR clonotype data were then integrated with the T-cell phenotype data inferred from single-cell gene-expression analysis based on their shared unique cell barcodes.
Analysis of Large-Scale Copy-Number Variations.
To quantify the level of aneuploidy, profiles of copy-number variation (CNV) inferred by inferCNV (https://github.com/broadinstitute/inferCNV) using scRNA-seq data were aggregated using a similar strategy adopted from a previous study (54). We first computed arm-level CNV scores as the mean of the squares of CNV values across each chromosomal arm. The arm-level CNV scores were further aggregated across all chromosomes by taking the average.
Whole-Exome Sequencing Data Processing and Genotyping QC
Library Preparation and Hybrid Capture Sequencing.
Genomic DNA (gDNA) was extracted from these remaining cells of prepared 10X single-cell suspension using AllPrep DNA/RNA Mini Kit (QIAGEN). Fifty to 200 ng of gDNA was applied to DNA library preparation using the KAPA HyperPlus Kit (Roche), according to the manufacturer's protocol. TruSeq adapters (Bioo Scientific) were utilized at the recommended ratio to input DNA. Library quality was assessed by High Sensitivity D1000 DNA Screen Tape analysis (Agilent Technologies) and Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Libraries were 6-plexed in equal quantities and a 1 μg of pooled libraries were enriched by hybrid capture using a Nimblegen SeqCap Exome v3 (Roche), according to the manufacturer's protocol. Each capture pool was sequenced on a lane of a HiSeq4000 to generate 2 × 100 bp reads.
Somatic Mutation Calling, Filtering, and Functional Annotation.
Mutation detection and filtering were performed without the use of paired germline control, as previously described (18). Raw FASTQ files were assessed for quality using FASTQC. Samples with high-quality metrics were run through our in-house pipeline. FASTQ files were (i) aligned to the human genome (hg19) using BWA-Mem (55); (ii) deduplicated using Picard MarkDuplicates; (iii) realigned around InDels using GATK (56); and (iv) recalibrated by base score using GATK (56). On-target rate and coverage over the targeted region were calculated by Picard CalculateHSMetrics. All samples achieved our standard minimum threshold of >50× average on-target coverage, with an average of 93.7× (minimum 63×, maximum 171×; Supplementary Table S1). For samples with additional sequencing performed to obtain sufficient coverage, FASTQC was performed on individual sequencing events and bam files from the same sample were merged using BamTools Mappings Merger following alignment. Variants were called by GATK Unified Genotyper and VarScan2 (57). Only variants called by both tools, with a minimum coverage of 30× and ≥3 supporting reads were retained, which we have previously shown to provide a sensitivity of 96.7% and specificity of 92.9% (28). All variants were annotated using SeattleSeq (58). To avoid mapping artifacts in repetitive regions, all variants within RepeatMasker or tandemRepeat annotated regions were filtered from the data set. All variants in dbSNP build 32 or the Genome Aggregation Database (gnomAD) were removed (59) to control for potential germline variants. We have shown previously that this approach effectively filters germline variants (18). Only genes previously shown to be recurrently targeted by somatic mutation in FL, genes with mutations present within ≥3 tumors, and variants with >20% variant allele fraction were considered for analysis.
Validation of the scRNA-seq Signatures with a Public scRNA-seq Data Set
To additionally validate that the identified signatures were expressed in the proposed cell types, we used a public scRNA-seq data set including B-cell lymphoma data (8). The data were downloaded and analyzed according to best practices (60). In brief, cells with high mitochondrial expression (>10% of total UMIs per cell) were excluded from further analysis. After the selection of 2,000 highly variable genes and the removal of expression linked with the total unique molecular identifier (UMI) counts and the percent of mitochondrial UMIs, cells from different patients were integrated using Harmony (45). Using the Leiden algorithm (ref. 61; with resolution = 1.5), based on the community graph constructed on the first 15 Harmony components and with 50 local neighborhoods (Supplementary Fig. S7A–S7C). Gene signature scores were calculated using the scanpy (62) package function score_genes. Next, we used median scaling to calculate the z-score for each of the gene signatures across the defined cell populations and compared the value of the corresponded cell population with others (Supplementary Fig. S7D). The CD4_Naive, Treg, and CD8_Eff signatures had the highest Z-scores in the corresponding cell types. The CD8Exh and CD4CTL groups had comparable Z-scores within the Roider and colleagues’ data set (8) due to lower sequencing depth compared with this study, and the overlap of highly expressed genes between cluster markers.
lymphoma microenvironment Identification with Public Bulk RNA-seq Data Sets Using Unsupervised Clustering
Marker gene signatures from CD4 and CD8 T-cell clusters identified from the scRNA-seq analysis (Supplementary Table S4) were utilized for clustering with 15 bulk FL cohorts (11–24): GSE127462, GSE53820, GSE93261, ICGC_MALY_DE, GSE66166, GSE55267, GSE37088, E-MTAB-6088, E-MEXP-2305, GSE132929, GSE103944, GSE32018, GSE48047, GSE142334, and GSE16131. RNA-seq data sets were processed using Kallisto (63). To estimate the intensity of each of the gene sets in bulk expression data (bulk RNA-seq or microarray data), gene signature scores were quantified using in-house python implementation of ssGSEA (64) as previously described in Kotlov and colleagues (25) and Bagaev and colleagues (26) and below. After exclusion of non-FL samples, expression data were subjected to ssGSEA, which is a method of estimating the expression intensity of custom genesets (in this case obtained from the scRNA-seq). For each gene set, a score within each sample is ascertained, resulting in a score matrix for each expression data set. Gene signature scores were median scaled within each data set and combined to increase the number of samples (n = 1,269). LME subtype identification was performed on the combined score matrix, and a hierarchical clustering approach was utilized (signature scores of the gene sets were used as features, a distance matrix was calculated using the Euclidian metric). We revealed four major clusters that contained 461, 288, 418, and 102 samples each (Fig. 4).
Deconvolution to Determine Tissue Cell Composition from Pseudobulk Gene-Expression Data
A deconvolution algorithm, Kassandra (65), was used for cell deconvolution to further reconstruct the cell composition of each LME from bulk RNA-seq. The Kassandra algorithm uses a two-stage hierarchical learning procedure for gradient boosting a LightGBM model that included training on artificial RNA-seq mixtures of different cell types. This stepwise approach allowed the model to adapt using information from other cell types and allowed all data sets hierarchically to be included for artificial transcriptome creation. Additionally, a transcripts per million (TPM)-based mathematical noise model was developed to take into account technical variability caused by different sequencing methods. The algorithm estimates the RNA proportion of a cell type from bulk RNA-seq from a sample, which is converted into cell percentage if the RNA concentration of a cell type is known (66). Kassandra was trained on over 9,400 tissue and blood-sorted cell RNA profiles for microenvironment reconstruction. Accounting for technical and biological variability, inclusion of aberrant cancer cell expression, and quantification and normalization of transcript expression produced a robust algorithm. Accuracy was confirmed on over 4,000 H&E tissue slides and 1,000 normal and tumor tissues with scRNA-seq, cytometry, or IHC comparisons.
Next, the Kassandra algorithm was applied to our scRNA-seq data. For this application, we converted the original scRNA-seq data to pseudobulk RNA-seq. To create the pseudobulk RNA-seq, all expression vectors (umi counts) of cells belonging to each patient sample were summarized, normalized to the sum of umi counts, and multiplied by 1 million. Therefore, for each patient, we created a pseudobulk expression vector that was approximate to TPM. The pseudobulk expression vector was then analyzed by the Kassandra ML algorithm for deconvolution of this pseudobulk RNA-seq to predict the percentages of major cell populations (Supplementary Fig. S10). Specifically, the Kassandra algorithm via python was applied to first to predict the RNA proportions on different hierarchical levels (main cell populations: B cells, CD4 T cells, CD8 T cells, NK cells, monocytes, macrophages, neutrophils, fibroblasts, endothelium, other; additional T-cell populations: T helpers, Treg, CD8 T cells with high PD1 phenotype, and CD8 T cells with low PD1 phenotype) and second to convert RNA signals into cell proportions by precalculated Kassandra transformation coefficients (that converts the RNA levels to absolute cell numbers for the designated cell populations).
Construction of Immunoglobulin Hierarchy Tree
The immunoglobulin hierarchy tree was constructed using the Alakazam module of the Immcantation analysis framework (67). BCR data output from Cell Ranger was first reformatted to a compatible file format using the change-10x.sh tool (version 2.7.0). V(D)J gene annotations for these cells were determined with IGBLAST (version 1.14.0; ref. 68). The ParseDb.py function was used to extract productive sequences for downstream clonotype analysis. For simplicity, the heavy-chain information was used to reconstruct the Ig-specific lineage tree. The CreateGermlines.py function was used to reconstruct the germline sequences of BCRs. The IMGT database was used in germline sequence reconstruction. We used the function DefineClones.py with the dist parameter set to 0.15 to cluster BCR sequences and define clonal groups, and then selected cells with expression values from the major expanded clones to construct the hierarchy tree. For a selected expanded clone, the Ig lineage hierarchy tree was reconstructed using the buildPhylipLineage function with default parameters. The buildPhylipLineage builds the lineage tree of a set of unique Ig sequences via maximum parsimony through an external call to the dnapars application of the PHYLIP package (version 3.697; ref. 69). Distance between two sequences is calculated using seqDist. The igraph R package (version: 1.2.6) was used for visualization.
Statistical Analysis
In addition to the bioinformatics approaches described above for scRNA/TCR-seq data analysis, all other statistical analysis was performed using statistical software R v3.5.2. Analysis of differences in immunologic features (continuous variables) between patient groups (MHC II high vs. MHC II low) was determined by the nonparametric Mann–Whitney U test. To control for multiple hypothesis testing, we applied the Benjamini–Hochberg method to correct P values, and the false discovery rates (q-values) were calculated. All statistical significance testing was two-sided and results were considered statistically significant at P < 0.05 and FDR q-value <0.10 unless otherwise stated.
Multiplex Immunofluorescence Staining and Image Analysis
Multiplex immunofluorescence (mIF) staining was performed using a similar method that has been previously described and optimized (70, 71). Briefly, 4-μm-thick formalin-fixed, paraffin-embedded TMA sections were stained using a mIF panel containing antibodies against CD19 (clone LE-CD19, Dako), granzyme K (clone HPA063181, Sigma-Aldrich), EOMES (clone EPR21950-2411, Abcam), CD4 (clone EPR6855, Abcam), HLA-DR (clone EPR3692, Abcam), CD8 (clone C8/144B, Thermo Scientific), and CD3ε (clone D7AA6E, CST). All the markers were stained in sequence using their respective fluorophore containing in the Opal 7 kit (cat. #NEL797001KT; Akoya Biosciences) and the individual tyramide signal amplification fluorophore Opal Polaris 480 (cat. #FP1500001KT, Akoya Biosciences). The staining TMA was imaged using the VectraPolaris spectral imaging system (Akoya Biosciences) using the fluorescence protocol at 10 nm λ from 420 nm to 720 nm. The slide was scanned in low magnification at ×10 and then each individual core in a high magnification at ×20. Each marker was analyzed at single-cell level, and a supervised algorithm (InForm image analysis software, Akoya Biosciences) for phenotyping was built for each marker. Cell density for each marker and possible combinations were consolidated using R studio 3.5.3 (Phenopter 0.2.2 packet, Akoya Biosciences). Data for cores in which CD19 segmentation failed or high background was observed were filtered from the analysis. For tumors that had multiple cores on the TMA, the median value of the independent cores was used for statistical analysis.
IHC for HLA-DR
Lymph node formalin-fixed paraffin-embedded FL tissues were used to build a tissue microarray (TMA) carrying 3 cores with 1 mm diameter for each case (72). The IHC protocol is briefly described: TMA blocks were sectioned at 4 μm thickness and stained using Leica Bond RX automated stainer (Leica Biosystems). The antigen retrieval was performed with Bond ER Solution #1 (AR9961, Leica Biosystems) equivalent to citrate buffer, pH 6.0 for 20 minutes at 100°C. Primary antibody against HLA-DR (clone EPR3692; cat. #ab92511, dilution 1:4,000) was used and incubated for 15 minutes at room temperature. The antibody was detected using the Bond Polymer Refine Detection kit (DS9800, Leica Biosystems) with Diaminobenzidine (DAB) as chromogen. All the slides were counterstained with hematoxylin, dehydrated, and coverslipped. Tonsils were used as an external positive control. All cores were analyzed using standard microscopy by two pathologists, and the intensity was scored based on the predominant pattern (0: negative, 1: weak, 2: strong, R: weak with reactive cells) in the intrafollicular area.
Data Availability
Raw sequencing data are available through the European Genome-Phenome Archive (EGA), Accession EGAS00001006052. Single-cell RNA-sequencing data have been uploaded in a browsable format to the Chan Zuckerburg cellxgene repository (https://cellxgene.cziscience.com/collections/968834a0–1895–40df-8720–666029b3bbac).
Supplementary Material
Acknowledgments
This work was supported by the NIH/NCI R01 CA201380 (MRG), the MD Anderson Cancer Center Support Grant (P30 CA016672), the Jaime Erin Follicular Lymphoma Research Consortium (M.R. Green and S.S. Neelapu), the Futcher Foundation (L.J. Nastoupil and M.R. Green), and an MD Anderson Institutional Research Grant program (L. Wang). M.R. Green is a Scholar of the Leukemia and Lymphoma Society. H. Yang is a Fellow of the Leukemia and Lymphoma Society. P. Strati is supported by a Lymphoma Research Foundation Career Development Award. L. Wang and M.R. Green are supported by Andrew Sabin Family Fellow Awards.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
O. Kudryashova reports personal fees from BostonGene during the conduct of the study; personal fees from BostonGene outside the submitted work; in addition, O. Kudryashova has a patent for BostonGene pending; and BostonGene employee. M. Meerson reports personal fees from BostonGene during the conduct of the study; personal fees from BostonGene outside the submitted work; in addition, M. Meerson has a patent for BostonGene pending; and BostonGene employee. S. Isaev reports personal fees from BostonGene outside the submitted work. N. Kotlov reports other support from BostonGene Corporation during the conduct of the study; in addition, N. Kotlov has a patent for BostonGene Corporation pending and issued. K.J. Nomie reports being an employee of BostonGene. A. Bagaev reports personal fees from BostonGene during the conduct of the study; in addition, A. Bagaev has a patent for BotonGene pending, issued, and licensed. S. Parmar reports grants from Cellenkos Inc outside the submitted work; and scientific founder of Cellenkos Inc with equity interest. S.P. Iyer reports other support from Seattle Genetics, Merck, Rhizen, Trillium, Innate, Crispr, Curio Science, Targeted Oncology, Acrotech, Legend, and Ono outside the submitted work. R. Steiner reports grants from SeaGen, Bristol Myers Squibb, Lymphoma Research Foundation, GSK, and Rafael Pharmaceuticals during the conduct of the study. N.H. Fowler reports other support from BostonGene outside the submitted work. C.R. Flowers reports personal fees from AstraZeneca, Bayer, BeiGene, BioAscend, Bristol Myers Squibb, Celgene, Curio Sciences, Denovo Biopharma, Epizyme/Incyte, Foresight Diagnostics, Genentech/Roche, Genmab, MEI Pharmaceuticals, MorphoSys AG, Pharmacyclics/Janssen, SeaGen and grants from 4D, AbbVie, Acerta, Adaptimmune, Allogene, Amgen, Bayer, Celgene, Cellectis, Emanuel Merck, Darmstadt (EMD), Gilead, Genentech/Roche, Guardant, Iovance, Janssen Pharmaceutical, Kite, Morphosys, Nektar, Novartis, Pfizer, Pharmacyclics, Sanofi, Takeda, TGTherapeutics, Xencor, Ziopharm, Burroughs Wellcome Fund, Eastern Cooperative Oncology Group, NCI, V Foundation, Cancer Prevention and Research Institute of Texas: CPRIT Scholar in Cancer Research outside the submitted work. P. Strati reports other support from AstraZeneca, ALX Oncology and Hutchison, TGT, ADCT, and Roche outside the submitted work. J.R. Westin reports grants and personal fees from Kite/Gilead, Bristol Myers Squibb, Novartis, Genentech, Morphosys/Incyte, ADC Therapeutics, and AstraZeneca outside the submitted work. S.S. Neelapu reports grants from Kite/Gilead, Bristol Myers Squibb, Cellectis, Poseida, Allogene, Unum Therapeutics, Precision Biosciences, Adicet BioKite/Gilead, Bristol Myers Squibb, Cellectis, Poseida, Allogene, Unum Therapeutics, Precision Biosciences, Adicet Bio, personal fees from Kite/Gilead, Merck, Novartis, Sellas Life Sciences, Athenex, Allogene, Incyte, Adicet Bio, Bristol Myers Squibb, Legend Biotech, Bluebird Bio, Sana Biotechnology, Caribou, Astellas Pharma, and other support from Takeda Pharmaceuticals during the conduct of the study; in addition, S.S. Neelapu has a patent for cell therapy pending. L.J. Nastoupil reports personal fees from ADC Therapeutics, Bristol Myers Squibb, Genentech/Roche, Gilead/Kite, Janssen, Morphosys, Novartis, and Takeda outside the submitted work. M.R. Green reports grants from Kite/Gilead, Sanofi, Allogene, Tessa Therapeutics, Monte Rosa Therapeutics, Daiichi Sankyo, and other support from KDAc Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
G. Han: Data curation, formal analysis, writing–review and editing. Q. Deng: Formal analysis, investigation, methodology, writing–review and editing. M.L. Marques-Piubelli: Formal analysis, validation, methodology, writing–review and editing. E. Dai: Data curation, writing–review and editing. M. Dang: Data curation, writing–review and editing. M.C.J. Ma: Data curation, writing–review and editing. X. Li: Data curation. H. Yang: Investigation, writing–review and editing. J. Henderson: Project administration. O. Kudryashova: Formal analysis, writing–review and editing. M. Meerson: Formal analysis, writing–review and editing. S. Isaev: Formal analysis, writing–review and editing. N. Kotlov: Formal analysis, writing–review and editing. K.J. Nomie: Formal analysis, writing–review and editing. A. Bagaev: Formal analysis, writing–review and editing. E.R. Parra: Methodology, writing–review and editing. L.M. Solis Soto: Methodology, writing–review and editing. S. Parmar: Resources, writing–review and editing. F.B. Hagemeister: Resources, writing–review and editing. S. Ahmed: Resources, writing–review and editing. S.P. Iyer: Resources, writing–review and editing. F. Samaniego: Resources, writing–review and editing. R. Steiner: Resources, writing–review and editing. L. Fayad: Resources, writing–review and editing. H. Lee: Resources, writing–review and editing. N.H. Fowler: Resources, writing–review and editing. C.R. Flowers: Resources, writing–review and editing. P. Strati: Resources, writing–review and editing. J.R. Westin: Resources, writing–review and editing. S.S. Neelapu: Resources, writing–review and editing. L.J. Nastoupil: Resources, writing–review and editing. F. Vega: Supervision, writing–review and editing. L. Wang: Supervision, funding acquisition, writing–review and editing. M.R. Green: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest 2012;122:3424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Green MR, Alizadeh AA. Common progenitor cells in mature B-cell malignancies: implications for therapy. Curr Opin Hematol 2014;21:333–40. [DOI] [PubMed] [Google Scholar]
- 3. Ennishi D, Takata K, Beguelin W, Duns G, Mottok A, Farinha P, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov 2019;9:546–63. [DOI] [PubMed] [Google Scholar]
- 4. Wang G, Chow RD, Zhu L, Bai Z, Ye L, Zhang F, et al. CRISPR-GEMM pooled mutagenic screening identifies KMT2D as a major modulator of immune checkpoint blockade. Cancer Discov 2020;10:1912–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Milpied P, Cervera-Marzal I, Mollichella ML, Tesson B, Brisou G, Traverse-Glehen A, et al. Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat Immunol 2018;19:1013–24. [DOI] [PubMed] [Google Scholar]
- 6. Andor N, Simonds EF, Czerwinski DK, Chen J, Grimes SM, Wood-Bouwens C, et al. Single-cell RNA-seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood 2019;133:1119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haebe S, Shree T, Sathe A, Day G, Czerwinski DK, Grimes SM, et al. Single-cell analysis can define distinct evolution of tumor sites in follicular lymphoma. Blood 2021;137:2869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roider T, Seufert J, Uvarovskii A, Frauhammer F, Bordas M, Abedpour N, et al. Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels. Nat Cell Biol 2020;22:896–906. [DOI] [PubMed] [Google Scholar]
- 9. Curran MA, Geiger TL, Montalvo W, Kim M, Reiner SL, Al-Shamkhani A, et al. Systemic 4-1BB activation induces a novel T cell phenotype driven by high expression of eomesodermin. J Exp Med 2013;210:743–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirschhorn-Cymerman D, Budhu S, Kitano S, Liu C, Zhao F, Zhong H, et al. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med 2012;209:2113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brodtkorb M, Lingjaerde OC, Huse K, Troen G, Hystad M, Hilden VI, et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood 2014;123:1051–4. [DOI] [PubMed] [Google Scholar]
- 13. Huet S, Tesson B, Jais JP, Feldman AL, Magnano L, Thomas E, et al. A gene-expression profiling score for prediction of outcome in patients with follicular lymphoma: a retrospective training and validation analysis in three international cohorts. Lancet Oncol 2018;19:549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pastore A, Jurinovic V, Kridel R, Hoster E, Staiger AM, Szczepanowski M, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015;16:1111–22. [DOI] [PubMed] [Google Scholar]
- 15. Guo S, Chan JK, Iqbal J, McKeithan T, Fu K, Meng B, et al. EZH2 mutations in follicular lymphoma from different ethnic groups and associated gene expression alterations. Clin Cancer Res 2014;20:3078–86. [DOI] [PubMed] [Google Scholar]
- 16. Oricchio E, Ciriello G, Jiang M, Boice MH, Schatz JH, Heguy A, et al. Frequent disruption of the RB pathway in indolent follicular lymphoma suggests a new combination therapy. J Exp Med 2014;211:1379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dreyling M, Santoro A, Mollica L, Leppa S, Follows GA, Lenz G, et al. Phosphatidylinositol 3-kinase inhibition by copanlisib in relapsed or refractory indolent lymphoma. J Clin Oncol 2017;35:3898–905. [DOI] [PubMed] [Google Scholar]
- 18. Ma MCJ, Tadros S, Bouska A, Heavican T, Yang H, Deng Q, et al. Subtype-specific and co-occurring genetic alterations in B-cell non-Hodgkin lymphoma. Haematologica 2021;107:690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horn H, Kohler C, Witzig R, Kreuz M, Leich E, Klapper W, et al. Gene expression profiling reveals a close relationship between follicular lymphoma grade 3A and 3B, but distinct profiles of follicular lymphoma grade 1 and 2. Haematologica 2018;103:1182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomez-Abad C, Pisonero H, Blanco-Aparicio C, Roncador G, Gonzalez-Menchen A, Martinez-Climent JA, et al. PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood 2011;118:5517–27. [DOI] [PubMed] [Google Scholar]
- 21. Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet 2012;44:1316–20. [DOI] [PubMed] [Google Scholar]
- 22. Takata K, Tanino M, Ennishi D, Tari A, Sato Y, Okada H, et al. Duodenal follicular lymphoma: comprehensive gene expression analysis with insights into pathogenesis. Cancer Sci 2014;105:608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parsa S, Ortega-Molina A, Ying HY, Jiang M, Teater M, Wang J, et al. The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nat Cancer 2020;1:653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leich E, Salaverria I, Bea S, Zettl A, Wright G, Moreno V, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood 2009;114:826–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kotlov N, Bagaev A, Revuelta MV, Phillip JM, Cacciapuoti MT, Antysheva Z, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov 2021;11:1468–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65. [DOI] [PubMed] [Google Scholar]
- 27. Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood 2018;131:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Nat Acad Sci U S A 2015;112:E1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Challa-Malladi M, Lieu YK, Califano O, Holmes AB, Bhagat G, Murty VV, et al. Combined genetic inactivation of beta2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 2011;20:728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steidl C, Shah SP, Woolcock BW, Rui L, Kawahara M, Farinha P, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011;471:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khodadoust MS, Olsson N, Wagar LE, Haabeth OA, Chen B, Swaminathan K, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 2017;543:723–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rivas-Delgado A, Magnano L, Moreno-Velazquez M, Garcia O, Nadeu F, Mozas P, et al. Response duration and survival shorten after each relapse in patients with follicular lymphoma treated in the rituximab era. Br J Haematol 2019;184:753–9. [DOI] [PubMed] [Google Scholar]
- 33. Magnano L, Alonso-Alvarez S, Alcoceba M, Rivas-Delgado A, Muntanola A, Nadeu F, et al. Life expectancy of follicular lymphoma patients in complete response at 30 months is similar to that of the Spanish general population. Br J Haematol 2019;185:480–91. [DOI] [PubMed] [Google Scholar]
- 34. Flowers CR, Leonard JP, Nastoupil LJ. Novel immunotherapy approaches to follicular lymphoma. Hematology Am Soc Hematol Educ Program 2018;2018:194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell 2020;181:1612–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. Cytotoxic CD4 T cells-friend or foe during viral infection? Front Immunol 2017;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med 2004;351:2159–69. [DOI] [PubMed] [Google Scholar]
- 38. Tobin JWD, Keane C, Gunawardana J, Mollee P, Birch S, Hoang T, et al. Progression of disease within 24 months in follicular lymphoma is associated with reduced intratumoral immune infiltration. J Clin Oncol 2019;37:3300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marabelle A, Kohrt H, Sagiv-Barfi I, Ajami B, Axtell RC, Zhou G, et al. Depleting tumor-specific tregs at a single site eradicates disseminated tumors. J Clin Invest 2013;123:2447–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roemer MGM, Redd RA, Cader FZ, Pak CJ, Abdelrahman S, Ouyang J, et al. Major histocompatibility complex class ii and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol 2018;36:942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mondello P, Tadros S, Teater M, Fontan L, Chang AY, Jain N, et al. Selective inhibition of HDAC3 targets synthetic vulnerabilities and activates immune surveillance in lymphoma. Cancer Discov 2020;10:440–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hashwah H, Schmid CA, Kasser S, Bertram K, Stelling A, Manz MG, et al. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc Nat Acad Sci U S A 2017;114:9701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov 2017;7:38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kridel R, Chan FC, Mottok A, Boyle M, Farinha P, Tan K, et al. Histological transformation and progression in follicular lymphoma: a clonal evolution study. PLoS Med 2016;13:e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arxiv 2020;1802.0346v3. [Google Scholar]
- 47. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56. [DOI] [PubMed] [Google Scholar]
- 51. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nazarov VI, Pogorelyy MV, Komech EA, Zvyagin IV, Bolotin DA, Shugay M, et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinf 2015;16:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reuben A, Gittelman R, Gao J, Zhang J, Yusko EC, Wu CJ, et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov 2017;7:1088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An Integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWAMEM. arXiv 2013;1303:3997. [Google Scholar]
- 56. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 2019;15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Traag VA, Waltman L, van Eck NJ. From louvain to leiden: guaranteeing well-connected communities. Sci Rep 2019;9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 64. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zaytcev A, Chelushkin M, Nuzhdina K, Bagaev A, Dyykanov D, Zyrin V, et al. Abstract 853: novel machine learning based deconvolution algorithm results in accurate description of tumor microenvironment from bulk RNAseq. Cancer Res 2020;80:853. [Google Scholar]
- 66. Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6:e26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stern JN, Yaari G, Vander Heiden JA, Church G, Donahue WF, Hintzen RQ, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med 2014;6:248ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 2013;41:W34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Felsenstein J. PHYLIP – phylogeny inference package (Version 3.2). Cladistics 1989;5:164–6. [Google Scholar]
- 70. Hernandez S, Rojas F, Laberiano C, Lazcano R, Wistuba I, Parra ER. Multiplex immunofluorescence tyramide signal amplification for immune cell profiling of paraffin-embedded tumor tissues. Front Mol Biosci 2021;8:667067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Parra ER, Jiang M, Solis L, Mino B, Laberiano C, Hernandez S, et al. Procedural requirements and recommendations for multiplex immunofluorescence tyramide signal amplification assays to support translational oncology studies. Cancers (Basel) 2020;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hedvat CV, Hegde A, Chaganti RS, Chen B, Qin J, Filippa DA, et al. Application of tissue microarray technology to the study of non-Hodgkin's and Hodgkin's lymphoma. Hum Pathol 2002;33:968–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available through the European Genome-Phenome Archive (EGA), Accession EGAS00001006052. Single-cell RNA-sequencing data have been uploaded in a browsable format to the Chan Zuckerburg cellxgene repository (https://cellxgene.cziscience.com/collections/968834a0–1895–40df-8720–666029b3bbac).