Abstract
RNA interference (RNAi) is a well-established research tool and is also maturing as a novel therapeutic approach. For the latter, microRNA-like off-target activity of short interfering RNAs (siRNAs) remains as one of the main problems limiting RNAi drug development. In this Communication, we report that replacement of a single internucleoside phosphodiester in the seed region (nucleotides 2 to 7) of the guide strand with an amide linkage suppressed the undesired microRNA-like off-target activity by at least an order of magnitude. For the specific siRNA targeting the PIK3CB gene, an amide modification between the third and fourth nucleotide of the guide strand showed the strongest enhancement of specificity (completely eliminated off-target silencing) while maintaining high on-target activity. These results are important because off-target activity is one of the main remaining roadblocks for RNA based drug development.
Graphical Abstract
RNA interference (RNAi) has become a powerful and broadly applicable tool for functional genetics and basic biology research. RNAi has also become a novel therapeutic approach.1,2 Since the approval of the first RNAi drug, patisiran in 2018, Alnylam has introduced four other short interfering RNAs (siRNAs) for clinical use to treat liver associated diseases. Gene silencing through the RNAi mechanism is mediated by two classes of small RNAs: 1) microRNAs (miRNAs) that are endogenous regulators of gene expression, and 2) short interfering RNAs (siRNAs) that in mammalian cells are exogenous regulatory molecules. Because they share the same gene silencing machinery, siRNAs can inherently act as miRNAs resulting in a sequence-dependent off-target activity that may cause false-positives in RNAi assays and toxicity in clinical trials.3,4 While careful selection of siRNA sequences may reduce some of the off-target activity, it cannot be completely eliminated and remains a significant concern for RNAi-based assays and a limitation in therapeutic development of siRNAs.
Chemical modifications of siRNAs such as 2′-OMe, 2′-F, locked (LNA) and unlocked (UNA) nucleic acids, and phosphorothioate linkages (Figure 1) are widely used to increase metabolic stability, decrease immune stimulation, and modulate binding affinity of siRNAs.5,6 Chemical modifications of siRNA can also be used to optimize specificity of siRNA by decreasing the miRNA-like off-target activity. Jackson and co-workers showed that miRNA-like off-target activity of siRNAs was widespread7 but could be at least partially mitigated by a single 2′-OMe modification at the second nucleotide of the guide strand.8
Figure 1.
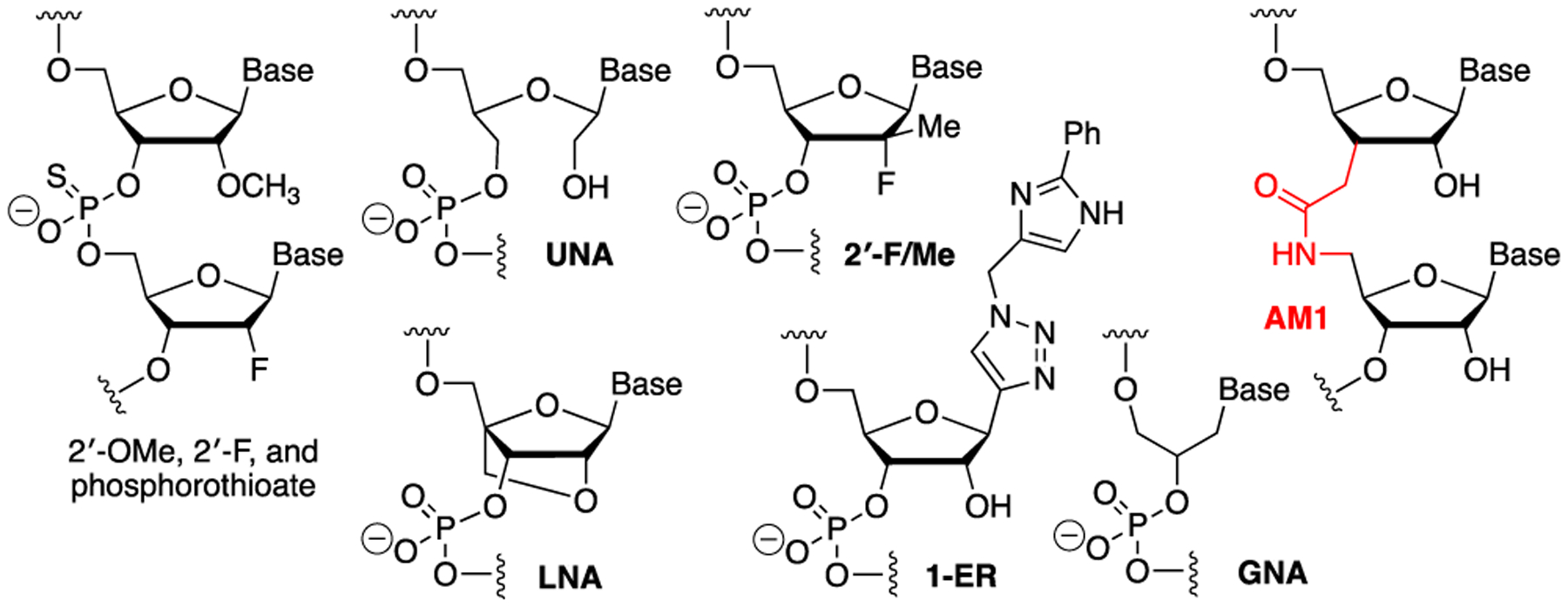
Structures of RNA modifications to improve metabolic stability and on-target activity of siRNAs.
Later studies by several groups showed that a single UNA modification at position 7 of the seed region (nucleotides 2–8 of the guide strand) strongly reduced miRNA-like off-target activity while maintaining the desired on-target activity of siRNAs.9,10 The authors proposed that UNA modification reduced the off-target activity by thermodynamic destabilization of the seed-mRNA complex, which at position 7 was more critical for the partially complementary miRNA off-targets than for the fully complementary siRNA targets.9,10 Consistent with this notion, other thermally destabilizing modifications, such as 2′-F/Me at position 7 (Figure 1)11 or substitution of all eight seed ribonucleotides with DNA12 reduced the miRNA-like off-target activity of the modified guide strands. Beal and co-workers showed that the 1-ER triazole nucleobase substitution (Figure 1) at the first position of the guide strand (G1) improved siRNA specificity by reducing the miRNA-like off-target activity while improving the on-target activity.13 The latter study was a rare example of using nucleobase modification to modulate siRNA-protein interactions to improve siRNA specificity.
Recent Alnylam studies provided compelling evidence that miRNA-like off-target activity is a major cause of hepatotoxicity of some siRNA sequences in rodents.14,15 The authors showed that a single glycol nucleic acid (GNA, Figure 1) between nucleotides 5 and 8 of the guide strand significantly reduced the miRNA-like off-target activity and toxicity of these siRNAs in animal models,14,15 allowing Alnylam to reintroduce a previously toxic siRNA in clinical trials after GNA modification at position 6.15 Taken together, previous studies illustrate that chemical modifications of the seed region can have strong impact on specificity of siRNAs.
In our earlier studies, we found that replacement of the internucleoside phosphates with amide linkages had a surprisingly small effect on the structure and thermal stability of double helical RNA.16–18 We19,20 and others21 have studied amide modifications in siRNAs. Isolated amide linkages were well tolerated in siRNA guide strands, except at the first position (G1), and at some positions, even increased the RNAi activity of the modified siRNAs.19,20 When placed at the first position, a single amide linkage completely eliminated the undesired activity of an siRNA’s passenger strand.22 Our results suggested that the latter was due to conformational rigidity of the amide linkage that prevented a backbone twist required for docking of the first nucleotide in the MID domain of Ago2. This result inspired a hypothesis that amide modifications elsewhere in the guide strand might disfavor conformational transitions required to accommodate the partially complementary miRNA off-targets, which would improve the specificity of the modified siRNAs. In the present study, we show that amide modifications at positions G2, G3, G6, and G7 in the seed region improved specificity of an siRNA by reducing the miRNA-like off-target activity while in some cases also slightly improving the on-target activity. Beneficial effects were also observed at positions G12, G18, and G20. Our results suggest that amide linkages at certain positions of the guide strand can be used to optimize siRNA specificity and may be promising modifications to suppress the off-target activity of therapeutic siRNAs.
To test our hypothesis that amide modification at selected positions of the guide strand may increase the specificity of siRNAs we chose a short fragment (positions 998 to 1017) of human PIK3CB mRNA (Figure 2) previously used by others to study miRNA-like off target activity.8,13 This siRNA sequence has well-characterized off-target mRNAs, YY1 and FADD that provide convenient assays for the effect of chemical modifications on siRNA specificity.8,13 The siRNAs G0 through G20 were synthesized using AM1-linked dinucleoside phosphoramidites (Figure 2) following our previously reported methodology.19,20,22
Figure 2.

Sequences of PIK3CB mRNA (on-target), AM1-modified siRNA guide strands, YY1 and FADD off-target mRNAs (off-target), and phosphoramidite building blocks to introduce AM1 modification. ‘G’ denotes guide strand targeting PIK3CB and the number denotes the position of AM1 modification; ‘p’ denotes phosphodiester and ‘a’ denotes amide internucleoside linkages; the mismatched nucleotides in YY1 and FADD sequences are highlighted in pink.
The on- and off-target activity of G0-G20 was measured using a dual luciferase assay and three reporter plasmids (psiCHECK-2 vector) with the sequences of PIK3CB (a single copy), YY1 (four copies), and FADD (four copies) inserted into the 3′-UTR of the Renilla luciferase mRNA. In our study, we followed the methodology developed by Beal and co-workers13 that used four copies of YY1 and FADD sequences to facilitate detection of off-target activity. Firefly luciferase, also encoded in these plasmids, served for internal normalization of data. This assay measures the production of protein (luciferase) and, hence, reports on various miRNA-like off target activity mechanisms, such as repression of mRNA translation and destabilization of mRNA.15 Full IC50 curves (Figures S29–S30) were constructed by testing siRNAs at eight or more concentrations; the IC50 values are summarized in Figure 3 and Table S1.
Figure 3.
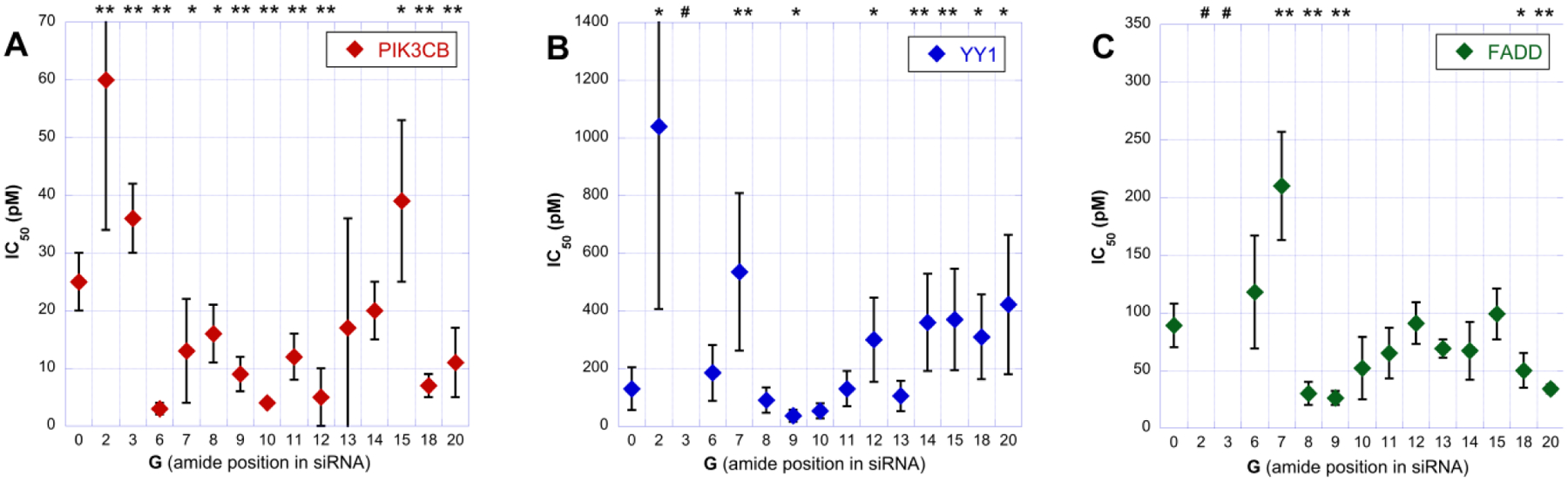
Dual-luciferase assay IC50 values of unmodified (G0) and AM1-modified (G2-G20) siRNAs targeting PIK3CB (A), YY1 (B), and FADD (C). # indicates that IC50 values for G3 targeting YY1 and G2 and G3 targeting FADD could not be calculated because the expression did not go below 60% at the maximum concentration (20 nM); when comparing the data with G0, * and ** indicate two-tailed P values of less than 0.05 and 0.01, respectively, as calculated using Student’s t-test.
In our dual luciferase assay, the unmodified siRNA (G0) showed IC50 values of 25 pM (PIK3CB, Figure 3A), 130 pM (YY1, Figure 3B) and 90 pM (FADD, Figure 3C). For the on-target PIK3CB, the amide-modified siRNAs showed slightly increased activity with IC50 values ranging from 3 to 20 pM, except for G2, G3 and G15 that showed decreased activity with 60, 36 and 39 pM, respectively. For YY1 and FADD, amide modifications in G7 in the seed region decreased off-target activity while the apparent slight decrease for G6 was not statistically significant. Amide modifications in G8-G20 had relatively small and sequence dependent effect on the silencing of YY1 and FADD off-targets. For YY1, G12 and G14-G20 had slightly decreased activity, while for FADD, G8, G9, G18, and G20 had slightly increased activity. The sequence dependence was not surprising because YY1 and FADD differ significantly in mismatch positions (pink in Figure 2) at the 3′-end of guide strand.
Most interestingly, G2 and G3 caused strong suppression of miRNA-like activity for both YY1 and FADD off-targets. Except for IC50 of ~1000 pM for G2 in YY1, we could not calculate IC50 values because the expression of YY1 and FADD did not go below 60% even at the maximum siRNA treatment concentration (20 nM). While the amide modification in G2 also suppressed on-target activity (IC50 = 60), G3 was unique in causing a relatively small loss of on-target activity (IC50 = 36) combined with strong suppression of off-target activity for both YY1 and FADD (Figure 4).
Figure 4.
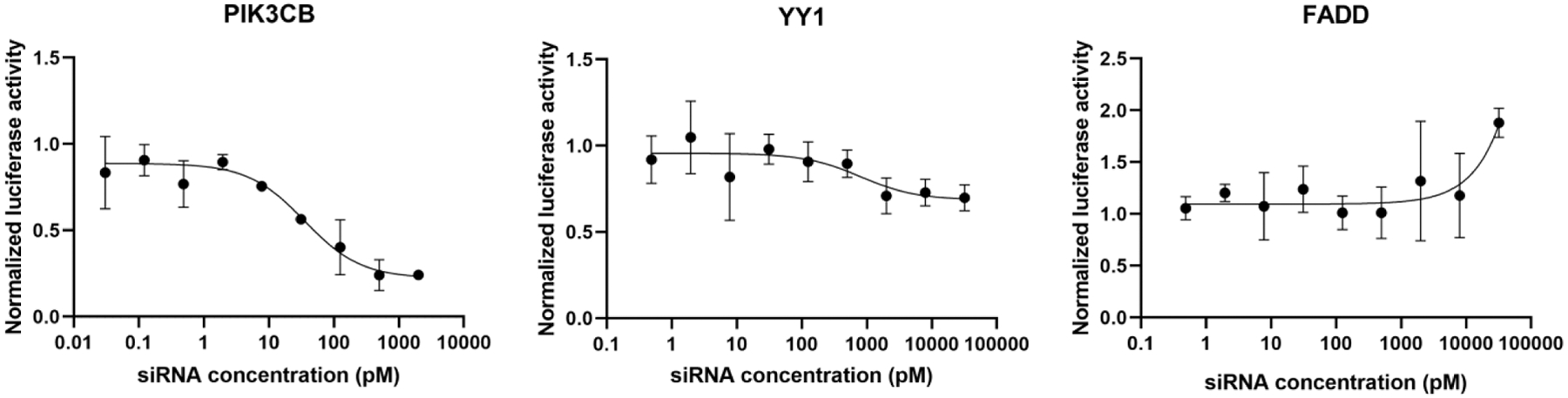
IC50 curves of dual luciferase assay of PIK3CB, YY1, and FADD silencing by G3. Lack of YY1 and FADD silencing below 70% even at 20 nM G3 indicates strong suppression of off-target activity.
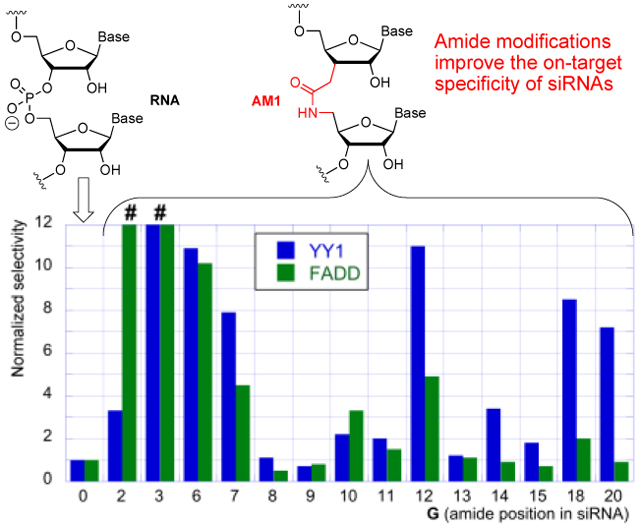
To gauge the improvements in on-target specificity of amide-modified siRNAs, the IC50 values of YY1 and FADD for each guide strand (G0-G20) were divided by the IC50 values of PIK3CB for each guide strand. These ratios were then normalized to unmodified siRNA (G0) and plotted in Figure 5, with values less than 1 indicating a decrease in specificity and values greater than 1 indicating an increase in specificity, relative to unmodified siRNA. The normalized selectivity ratios (Figure 5) showed strong effect of amide modification at the positions tested in the seed region (G2, G3, G6, and G7) as well as notable improvements at positions 12, 18, and 20.
Figure 5.
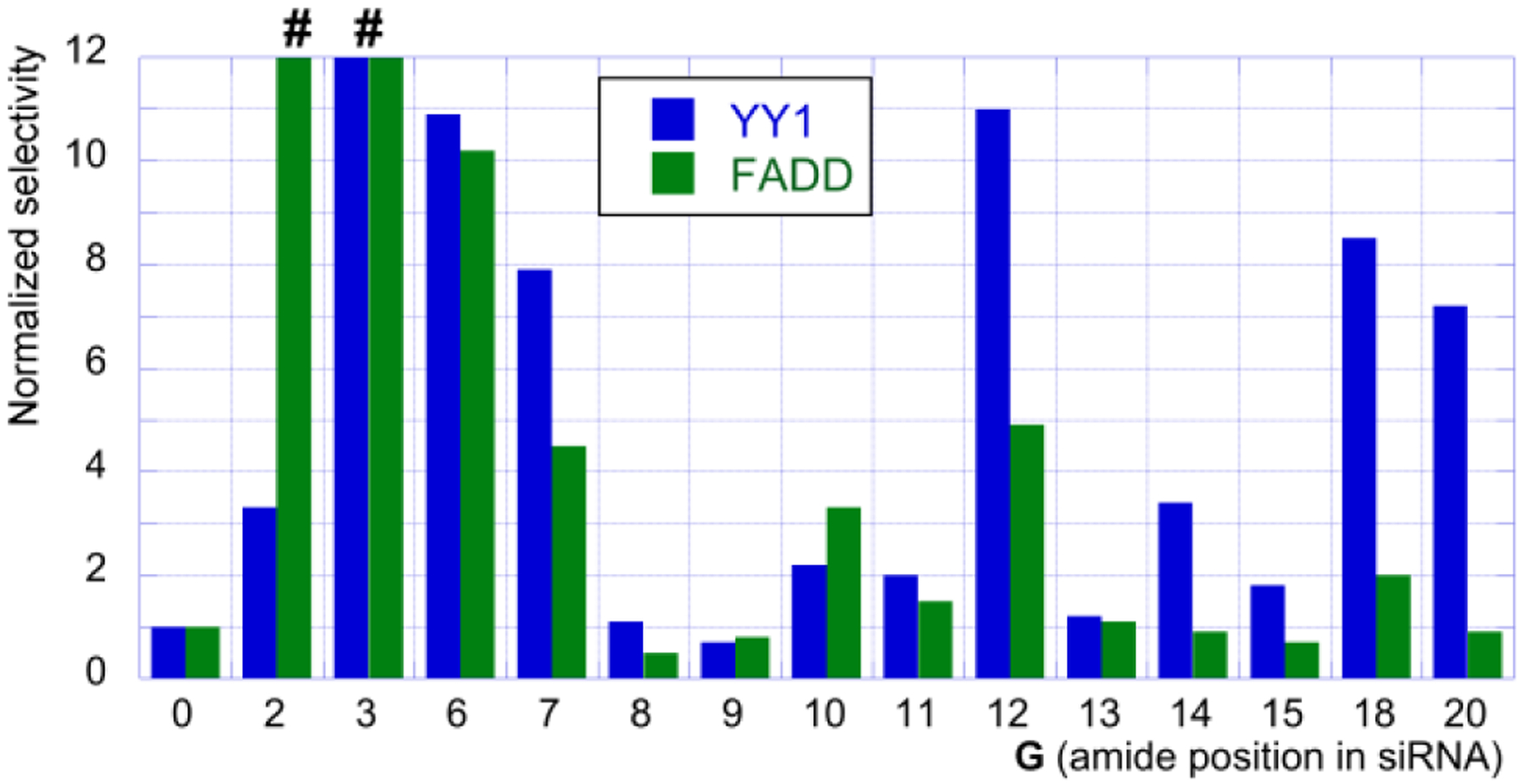
Normalized activity ratios show fold change in siRNA specificity compared to unmodified siRNA. # at G2 and G3 indicates that the high selectivity cannot be precisely calculated because of absence of IC50 values for YY1 and FADD.
The increase in specificity for G2 and G3 was driven entirely by suppression of off-target cleavage of YY1 and FADD; however, this positive result was somewhat offset by concurrent decrease in on-target activity, especially for G2 (Figure 3). In contrast, G6 showed ~10-fold increase in specificity over both off-targets largely due to increased on-target activity for PIK3CB (c.f., Figure 3A with 3B and 3C). G7 showed more balanced increase in specificity by increasing on-target and decreasing off-target activity. The increase in specificity shown by G12, G18, and G20 was also driven by increased on-target activity, enhanced by suppression of YY1 off-target cleavage (Figure 3).
The increase in specificity was not due to destabilization of guide-mRNA complex. UV melting results (Table S2) showed that the single amide linkages caused relatively little change in thermal stability of siRNA duplexes (ΔTm varied from −0.5 to +1.0 °C). This was not unexpected because our previous studies had shown that a single amide modification had relatively little effect on thermal stability of siRNAs.19,20 The improvements of siRNA specificity did not correlate with Tm of amide-modified siRNAs. For example, G2 and G3 had opposite ΔTm of −0.5 to +0.9 °C despite similar activity profiles. The effect of GNA modification was also suggested to be due to conformational flexibility rather than simple thermal destabilization of the seed.15
In the present model system, we systematically replaced the phosphates of the guide strand one by one with amide linkages. We did not test G1 because it was known from our previous studies to strongly inhibit on-target activity,22 and we were unable to test G4 and G5, and G16, G17 and G19 because the required GAM1G and GAM1A dimers have not been synthesized. Synthesis of amide-linked dimers containing guanosine is significantly more difficult than other dimers; so far, we have succeeded with preparation of AAM1G,23 which was also used in the present study. It is conceivable that G4 and G5 may also offer similar improvements in siRNA specificity as G3 and G6.
While complete understanding of the beneficial effect of amide modifications of the seed region will need additional structural studies, current and previous studies may offer some clues. Our previous molecular modeling and dynamics study showed that amides in G2 and G3 were not able to mimic original phosphate interactions with Ago2 amino acids at these positions.20 After conformational reorientation amide in G2 was able to recover hydrogen bonding to K566 of Ago2, while amide in G3 was only transiently able to re-establish hydrogen bonding with Y790 and R792 side chains of Ago2.20 It is conceivable that the conformational changes required to reorient amides in G2 and G3 were more unfavorable for the partially complementary miRNA off-target complexes than for the fully matched siRNA complex. Beneficial effect of G6 and G7 is most likely related to base pairing dynamics at the junction of seed and central regions. Helix-7 of Ago2 docks into minor groove between nucleotides 6 and 7 of the seed-target duplex probing an A-form duplex and disfavoring mismatches and non-canonical pairing.24 It is likely that amide modifications disfavor conformational transitions of partially complementary miRNA off-target complexes critical for displacement of helix-7 of Ago2 and ultimately the activity of miRNAs.
In conclusion, our study demonstrates that a single amide linkage in the seed region of a guide strand targeting PIK3CB strongly reduced microRNA-like silencing of known off-targets YY1 and FADD. G3 was the best siRNA showing almost complete suppression of off-target silencing while having only slightly reduced on-target activity (IC50 increased from 25 to 36 pM). It is conceivable that for other siRNAs, as well as for other off-targets, an amide modification at a different position may give the best result. Figure 5 illustrates some of the sequence dependency of amide placement. For example, G2 strongly suppressed FADD while having relatively modest effect on YY1, but the benefits of G12, G18, and G20 were reversed. Most likely, amide-modification placement will have to be optimized for any new siRNA sequence and its specific application, as has been previously found for the GNA modifications.15 Brown and co-workers recently demonstrated that amide linkages in DNA, when combined with other modifications such as LNA and phosphorothioates, improved metabolic stability and cellular uptake, and reduced toxicity of antisense oligonucleotides.25,26 Collectively, the present and previous studies,22,25,26 add AM1 amide linkages to the nucleic acid chemist’s toolkit of chemical modifications to modulate the specificity and optimize therapeutic potential of antisense oligonucleotides, siRNAs and crRNAs.
Supplementary Material
ACKNOWLEDGMENT
We thank P. A. Beal for a generous gift of PIK3CB and FADD plasmids and C. Kleeschulte for her help with cloning. This work was supported by the National Institutes of Health (R35 GM130207 to E.R.).
Footnotes
Supporting Information. General experimental procedures, synthesis, purification, and MS characterization of amide-modified dimers and siRNAs; experimental details and results of dual luciferase and UV melting assays; NMR spectra of new compounds.
The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- (1).Setten RL; Rossi JJ; Han S.-p. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov 2019, 18, 421–446. [DOI] [PubMed] [Google Scholar]
- (2).Goga A; Stoffel M Therapeutic RNA-silencing oligonucleotides in metabolic diseases. Nat. Rev. Drug Disc 2022, 21, 417–439. [DOI] [PubMed] [Google Scholar]
- (3).Jackson AL; Linsley PS Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov 2010, 9, 57–67. [DOI] [PubMed] [Google Scholar]
- (4).Doench JG; Petersen CP; Sharp PA siRNAs can function as miRNAs. Genes & Development 2003, 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Khvorova A; Watts JK The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol 2017, 35, 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shen X; Corey DR Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res 2018, 46, 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jackson AL; Burchard J; Schelter J; Chau BN; Cleary M; Lim L; Linsley PS Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 2006, 12, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jackson AL; Burchard J; Leake D; Reynolds A; Schelter J; Guo J; Johnson JM; Lim L; Karpilow J; Nichols K; Marshall W; Khvorova A; Linsley PS Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 2006, 12, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bramsen JB; Pakula MM; Hansen TB; Bus C; Langkjaer N; Odadzic D; Smicius R; Wengel SL; Chattopadhyaya J; Engels JW; Herdewijn P; Wengel J; Kjems J A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res 2010, 38, 5761–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Vaish N; Chen F; Seth S; Fosnaugh K; Liu Y; Adami R; Brown T; Chen Y; Harvie P; Johns R; Severson G; Granger B; Charmley P; Houston M; Templin MV; Polisky B Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res 2011, 39, 1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Guenther DC; Mori S; Matsuda S; Gilbert JA; Willoughby JLS; Hyde S; Bisbe A; Jiang Y; Agarwal S; Madaoui M; Janas MM; Charisse K; Maier MA; Egli M; Manoharan M Role of a “Magic” Methyl: 2′-Deoxy-2′-α-F-2′-β-C-methyl Pyrimidine Nucleotides Modulate RNA Interference Activity through Synergy with 5′-Phosphate Mimics and Mitigation of Off-Target Effects. J. Am. Chem. Soc 2022, 144, 14517–14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ui-Tei K; Naito Y; Zenno S; Nishi K; Yamato K; Takahashi F; Juni A; Saigo K Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008, 36, 2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Suter SR; Sheu-Gruttadauria J; Schirle NT; Valenzuela R; Ball-Jones AA; Onizuka K; MacRae IJ; Beal PA Structure-Guided Control of siRNA Off-Target Effects. J. Am. Chem. Soc 2016, 138, 8667–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Janas MM; Schlegel MK; Harbison CE; Yilmaz VO; Jiang Y; Parmar R; Zlatev I; Castoreno A; Xu H; Shulga-Morskaya S; Rajeev KG; Manoharan M; Keirstead ND; Maier MA; Jadhav V Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun 2018, 9, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schlegel MK; Janas MM; Jiang Y; Barry JD; Davis W; Agarwal S; Berman D; Brown CR; Castoreno A; LeBlanc S; Liebow A; Mayo T; Milstein S; Nguyen T; Shulga-Morskaya S; Hyde S; Schofield S; Szeto J; Woods LB; Yilmaz VO; Manoharan M; Egli M; Charissé K; Sepp-Lorenzino L; Haslett P; Fitzgerald K; Jadhav V; Maier MA From bench to bedside: Improving the clinical safety of GalNAc–siRNA conjugates using seed-pairing destabilization. Nucleic Acids Res. 2022, 50, 6656–6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rozners E; Katkevica D; Bizdena E; Strömberg R Synthesis and Properties of RNA Analogs Having Amides as Interuridyl Linkages at Selected Positions. J. Am. Chem. Soc 2003, 125, 12125–12136. [DOI] [PubMed] [Google Scholar]
- (17).Selvam C; Thomas S; Abbott J; Kennedy SD; Rozners E Amides as Excellent Mimics of Phosphate Linkages in RNA. Angew. Chem., Int. Ed 2011, 50, 2068–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kotikam V; Rozners E Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs. Acc. Chem. Res 2020, 53, 1782–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mutisya D; Selvam C; Lunstad BD; Pallan PS; Haas A; Leake D; Egli M; Rozners E Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Mutisya D; Hardcastle T; Cheruiyot SK; Pallan PS; Kennedy SD; Egli M; Kelley ML; Smith Anja van B.; Rozners E Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45, 8142–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gong W; Desaulniers J-P Gene-silencing properties of siRNAs that contain internal amide-bond linkages. Bioorg. Med. Chem. Lett 2012, 22, 6934–6937. [DOI] [PubMed] [Google Scholar]
- (22).Hardcastle T; Novosjolova I; Kotikam V; Cheruiyot SK; Mutisya D; Kennedy SD; Egli M; Kelley ML; van Brabant Smith A; Rozners E A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol 2018, 13, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kotikam V; Gajula PK; Coyle L; Rozners E Amide Internucleoside Linkages Are Well Tolerated in Protospacer Adjacent Motif-Distal Region of CRISPR RNAs. ACS Chem. Biol 2022, 17, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Klum SM; Chandradoss SD; Schirle NT; Joo C; MacRae IJ Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018, 37, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Epple S; Thorpe C; Baker YR; El-Sagheer AH; Brown T Consecutive 5′- and 3′-amide linkages stabilise antisense oligonucleotides and elicit an efficient RNase H response. Chem. Commun 2020, 56, 5496–5499. [DOI] [PubMed] [Google Scholar]
- (26).Baker YR; Thorpe C; Chen J; Poller LM; Cox L; Kumar P; Lim WF; Lie L; McClorey G; Epple S; Singleton D; McDonough MA; Hardwick JS; Christensen KE; Wood MJA; Hall JP; El-Sagheer AH; Brown T An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides. Nat. Commun 2022, 13, 4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


