Abstract
Cholera caused by toxigenic Vibrio cholerae is a major public health problem confronting developing countries, where outbreaks occur in a regular seasonal pattern and are particularly associated with poverty and poor sanitation. The disease is characterized by a devastating watery diarrhea which leads to rapid dehydration, and death occurs in 50 to 70% of untreated patients. Cholera is a waterborne disease, and the importance of water ecology is suggested by the close association of V. cholerae with surface water and the population interacting with the water. Cholera toxin (CT), which is responsible for the profuse diarrhea, is encoded by a lysogenic bacteriophage designated CTXΦ. Although the mechanism by which CT causes diarrhea is known, it is not clear why V. cholerae should infect and elaborate the lethal toxin in the host. Molecular epidemiological surveillance has revealed clonal diversity among toxigenic V. cholerae strains and a continual emergence of new epidemic clones. In view of lysogenic conversion by CTXΦ as a possible mechanism of origination of new toxigenic clones of V. cholerae, it appears that the continual emergence of new toxigenic strains and their selective enrichment during cholera outbreaks constitute an essential component of the natural ecosystem for the evolution of epidemic V. cholerae strains and genetic elements that mediate the transfer of virulence genes. The ecosystem comprising V. cholerae, CTXΦ, the aquatic environment, and the mammalian host offers an understanding of the complex relationship between pathogenesis and the natural selection of a pathogen.
Cholera is characterized by a severe watery diarrhea caused by toxigenic Vibrio cholerae, which colonizes the small intestine and produces an enterotoxin, cholera toxin (CT) (93, 108, 149). Cholera is endemic in southern Asia and parts of Africa and Latin America, where seasonal outbreaks occur widely and are particularly associated with poverty and poor sanitation. V. cholerae is a well-defined species on the basis of biochemical tests and DNA homology studies (11), but the species is not homogeneous with regard to pathogenic potential. In assessing the public health significance, two critical properties of V. cholerae are taken into account. These include the production of CT, which is responsible for the severe diarrhea, and the possession of the O1 or O139 antigen, which acts as a marker of epidemic potential, since the actual determinant of such potential is not clearly known (67). However, molecular analysis has revealed that in addition to genes encoding CT, all strains capable of causing cholera invariably carry genes for a colonization factor known as toxin-coregulated pilus (TCP) and a regulatory protein, ToxR, which coregulates the expression of CT and TCP (52). Thus, cholera pathogenesis relies on the synergistic effect of a number of pathogenic factors produced by toxigenic V. cholerae.
A distinctive epidemiological feature of cholera is its appearance in a regular seasonal pattern in areas of endemic infection and in explosive outbreaks often starting in several distinct foci simultaneously (45, 67), indicating a possible role of environmental factors in triggering the epidemic process. However, many aspects of the disease and the pathogen remain unknown, particularly its ecology, the interepidemic reservoirs, and the role of CT and other virulence factors in the natural selection and possible ecological association of the organism with the aquatic environment as well as with the human host.
In V. cholerae, the major virulence-associated genes which encode colonization factors and CT are part of larger genetic elements composed of clusters of genes (73, 104). Although the major subunit of TCP is encoded by the tcpA gene, the formation and function of the pilus assembly require the products of a number of other genes located on a large DNA region referred to as the TCP pathogenicity island, which includes the tcp and acf gene clusters (73). The ctxAB operon, which encodes the A and B subunits of CT, is part of a larger genetic element originally termed the CTX genetic element (104). Recent studies have shown that the CTX genetic element corresponds to the genome of CTXΦ, a lysogenic filamentous bacteriophage (144), and that propagation of CTXΦ may be associated with the origination of novel toxigenic V. cholerae strains from nontoxigenic progenitors (32). The V. cholerae genome has also been found to contain a distinctive class of integrons, which are gene expression elements that acquire open reading frames (ORFs) and convert them to functional genes (80). This permits the bacteria to entrap genes from other microorganisms and thus constitutes a mechanism for the clustering and spread of pathogenic genes as well as genes for other biochemical functions. In agreement with these recent findings from genetic analysis of V. cholerae, molecular epidemiological surveillance of cholera in areas of endemic infection has also revealed temporal changes in the properties of toxigenic V. cholerae and a continual emergence of new epidemic clones which often replace existing clones (33–37, 89, 121). However, the ecology of toxigenic V. cholerae, which maintains the seasonal pattern of epidemics and supports the emergence of new epidemic clones, has not been adequately explained. The purpose of this review is to summarize available information on the epidemiology, genetics, and ecology of toxigenic V. cholerae and to propose a model for the seasonal pattern of cholera outbreaks and the emergence of new epidemic strains. Special emphasis is placed on compiling scientific data obtained from the different aspects of cholera studies and providing an insight into the possible evolutionary significance of V. cholerae virulence factors being encoded by accessory genetic elements.
NOMENCLATURE OF V. CHOLERAE O1 AND NON-O1
In the past, a wide variety of gram-negative, rod-shaped bacteria with polar flagella were classified as belonging to the genus Vibrio. During the mid-1960s, however, some criteria for the taxonomy of the genus Vibrio had been established and the International Subcommittee on Taxonomy of Vibrios recommended a provisional definition, in which the majority of species previously classified as Vibrio were excluded from the genus. On the other hand, taxonomic studies on the related organisms have indicated a close relationship among the three genera Vibrio, Aeromonas, and Plesiomonas (31). On the basis of biochemical characteristics, it is possible to differentiate members of the genus Vibrio from those of allied genera. The current classification has been reviewed by West and Colwell (147). The genus Vibrio contains several species, of which V. cholerae, V. parahaemolyticus, and V. vulnificus are the most important pathogens of humans. Whereas V. parahaemolyticus is an important cause of diarrheal illness, V. vulnificus infections can range from self-limiting gastroenteritis and wound infections to severe necrotizing infections of soft tissues and fatal septicemia (148). V. mimicus has also been implicated in diarrheal disease, and some strains of V. mimicus may produce CT or a heat-stable (ST)-like toxin (90, 110). Besides, many unknown vibrios have been isolated from seawater and sea fish, and these are sometimes confused with vibrios that infect humans. V. cholerae is classified on the basis of its somatic antigens (O antigens) into serovars or serogroups, and there are at least 155 known serogroups (11, 122–124). Until recently, serogroup O:1 was supposed to include all the strains responsible for epidemic and endemic cholera; it has two major serotypes, Ogawa and Inaba, and the Hikojima serotype has been reported rarely. These serotypes can be further distinguished into two biotypes, classical and El Tor, based on some biochemical properties and susceptibility to bacteriophages (114).
There are 154 known serogroups of non-O1 vibrios (124). These vibrios possess biochemical and morphological characteristics very similar to those of the cholera vibrio but are nonagglutinable with polyvalent O:1 antiserum. Such vibrios are agglutinable by their own antisera. Non-O1 serogroups of V. cholerae had been associated mostly with sporadic cases of diarrhea and extraintestinal infections (59) until 1993, when a large cholera-like outbreak in Bangladesh and India was found to be caused by a V. cholerae non-O1 strain (1). This organism did not belong to any of the 138 known O serogroups of V. cholerae but to a new serogroup, which was later designated O139 (123). Since then, V. cholerae O139 has been persisting as a second etiologic agent of cholera. Hence, there are now two serogroups, O1 and O139, that have been associated with epidemic disease, but there are also strains of these serogroups which do not produce CT and are not involved in epidemics.
EPIDEMIOLOGY OF CHOLERA
Outbreaks of cholera cause deaths estimated at 120,000 annually worldwide and many more cases each year, of which the vast majority occur in children (150). Hallmarks of the epidemiology of cholera include (i) a high degree of clustering of cases by location and season, (ii) highest rates of infection in children 1 to 5 years of age in areas of endemic infection, (iii) antibiotic resistance patterns that frequently change from year to year, (iv) clonal diversity of epidemic strains, and (v) protection against the disease by improved sanitation and hygiene and preexisting immunity. Cholera has been categorized as one of the “emerging and reemerging infections” (117) threatening many developing countries. Several recent events that mark the epidemiological importance of the disease include the reemergence of cholera in Latin America in 1991 (77, 101, 113); the explosive outbreak of cholera among Rwandan refugees in Goma, Zaire, which resulted in about 70,000 cases and 12,000 deaths in 1994 (126); and the emergence of V. cholerae O139 in the Indian subcontinent during 1992 to 1993, possibly marking the beginning of the eighth pandemic of cholera (19, 109, 135).
Early Pandemics
It is generally accepted that seven distinct pandemics of cholera have occurred since the onset of the first pandemic in 1817 (106). Except for the seventh pandemic, which originated on the island of Sulawesi in Indonesia (65), the pandemics arose in the Indian subcontinent, usually the Ganges delta, and spread to other continents, affecting many countries and extending over many years (15, 75, 106, 133). The second pandemic of cholera reached the British Isles in the early 1830s, and fundamental epidemiological observations by John Snow on the waterborne transmission of cholera were made in London between 1847 and 1854 during the late second and the third pandemics (133). The second pandemic also reached Canada via ships from Ireland carrying infected immigrants (15). During the third pandemic (1852 to 1859), cholera was rampant in the United States, and toward the end of the fourth pandemic (during the 1870s), cities and towns along the Mississippi, Missouri, and Ohio rivers experienced cholera (12). The fifth pandemic extensively affected South America; it caused large epidemics in many countries and was characterized by high mortality in Argentina, Chile, and Peru (75). During the fifth pandemic, Robert Koch isolated the causative organism of cholera, referred to as “comma bacilli” (72), from rice water stools of patients in Egypt in 1883 and in India in 1884. The sixth pandemic (1899 to 1923) extensively involved populations in the near and middle East and the Balkan peninsula (106). Except for a large epidemic in Egypt in 1947 (125), cholera remained virtually confined to south and southeast Asia from the mid-1920s until the onset of the seventh pandemic in 1961. The sixth pandemic and presumably the fifth pandemic were caused by V. cholerae of the classical biotype.
Seventh Pandemic
The seventh pandemic is the most extensive of the pandemics in geographic spread and in duration, and the causative agent is V. cholerae O1 of the El Tor biotype. The pandemic, which began in 1961 on the island of Sulawesi in Indonesia, spread to other islands, including Java, Sarawak, and Borneo, and then to the Philippines, Sabah, and Taiwan, thereby affecting the entire Southeast Asian archipelago by the end of 1962 (65). During 1963 to 1969, the pandemic spread to the Asian mainland and affected Malaysia, Thailand, Burma, Cambodia, Vietnam, India, Bangladesh, and Pakistan. Soon after El Tor cholera reached Pakistan, outbreaks were reported in Afghanistan, Iran, Iraq, and nearby republics within the Soviet Union (65). By 1970, El Tor cholera had invaded the Arabian Peninsula, Syria, and Jordan, and a limited outbreak occurred in Israel (21). The seventh pandemic reached sub-Saharan West Africa in the early 1970s and caused explosive outbreaks, resulting in more than 400,000 cases with a high mortality rate, due mainly to a lack of background immunity in the population and inadequacies in the health care infrastructure (49). In this epidemic, cholera spread along the coast and into the interior through waterways and was further disseminated into the interior of the Sahelian states by land travel by nomadic tribes. Of the 36 countries that reported cholera in 1970, 28 were newly affected countries and 16 were in Africa (67).
The seventh pandemic reached South America in the form of an explosive epidemic that began in Peru in January 1991 and thus caused the return of cholera to the continent after more than a century (77, 101, 113). Subsequently, epidemic cholera was reported in neighboring Ecuador and then in Colombia. In each of these countries, populations at a low socioeconomic level, lacking proper drinking water and sanitation facilities, were the most severely affected (101). By April 1991, a small outbreak occurred in Santiago, the capital of Chile (77), and cholera began to travel along the Pacific coast of South America to progressively enter more countries in South and Central America. The Pan American Health Organization estimates that during 1991 and 1992 there were 750,000 cases of cholera and 6,500 deaths in the Americas (101).
In recent times, one of the worst cholera outbreaks occurred in Goma, Eastern Zaire, in July 1994 (126). Conflicts between tribes in neighboring Rwanda had displaced nearly a million people to Zaire, and they were sheltered in refugee camps. Outbreak of cholera in the poverty-stricken refugee camps led to the death of an estimated 12,000 Rwandan refugees during a 3-week period (126). The seventh pandemic is ongoing, and it continues to cause seasonal outbreaks in many developing countries, especially Bangladesh and India. However, in 1992, V. cholerae belonging to a non-O1 serogroup (now referred to as O139) caused large epidemics of cholera in India and Bangladesh and spread to some other countries; this may represent the beginning of the eighth pandemic.
Eighth Pandemic
In late 1992, epidemic cholera was reported in Madras and other places in India and in Southern Bangladesh (19, 109). Although the clinical syndrome was typical of cholera, the causative agent was a V. cholerae non-O1 strain, which was later serogrouped as O139 (123). The epidemic continued through 1993, and V. cholerae O139 spread throughout Bangladesh and India and neighboring countries. Outbreaks or cases due to V. cholerae O139 have since been reported in Pakistan, Nepal, China, Thailand, Kazakhastan, Afghanistan, and Malaysia (14, 20, 107, 135). Imported cases have been reported in the United Kingdom and the United States (14, 107). Recent surveillance during 1996 and 1997 has shown that V. cholerae O139 continues to cause cholera outbreaks in India and Bangladesh and coexists with the El Tor vibrios (35, 89, 91). If outbreaks of cholera due to this new serogroup continue to occur and to affect more countries, this may represent the eighth pandemic (135).
Epidemiology of Cholera in Bangladesh
Surveillance of cholera in Bangladesh, has provided important information about the epidemiology of the disease and about temporal changes in the properties of V. cholerae strains isolated from different epidemics. Bangladesh is situated in the Ganges delta, the region where all the cholera pandemics except the seventh started. Because of the low-lying deltaic environment, the water volumes in ponds and rivers in Bangladesh change between the dry and wet seasons. Besides raising the river levels, the monsoons wash the sewage from the villages into the rivers. In rural areas, the people come into direct contact with the surface water for drinking, bathing, cooking, and irrigation. The growth of the population has far outstripped the capacity for adequate housing and sanitary facilities. Cholera is endemic in Bangladesh, and outbreaks occur in a regular seasonal pattern.
Systematic surveillance of cholera has been carried out in Bangladesh by the International Centre for Diarrhoeal Disease Research (ICDDR,B) and the former Pakistan Seato Cholera Research Laboratory for more than 35 years (78, 81, 116, 128, 129). A number of studies have shown that epidemic outbreaks in Bangladesh usually occur twice during a year, with the largest number of cases occurring during September to December, just after the monsoon (7, 45, 78, 81, 116, 129, 134). A somewhat smaller peak of cholera cases is also observed in the spring between March and May. Until 1970, more than 90% of cholera in Bangladesh was caused by the classical Inaba serotype; by 1972, 85% of all cases were due to the classical Ogawa serotype (70). The El Tor biotype of V. cholerae O1 appeared in Bangladesh in 1969/1973, and since this biotype had completely replaced the classical biotype. However, in 1982, the classical biotype reemerged as the predominant epidemic biotype in Bangladesh (70, 115) and coexisted with the El Tor vibrios until 1992. Data obtained from studies of diarrhea epidemics in nearly 400 rural subdistricts by ICDDR,B medical teams between 1985 and 1991 showed that V. cholerae O1 was the most frequently (40%) isolated enteropathogen during the epidemics (129). The 1991 epidemic was estimated to have caused between 210,000 and 235,000 cases and over 8,000 deaths (129).
In December 1992, an epidemic of severe acute watery diarrhea, clinically resembling cholera and affecting mainly adults, occurred in southern Bangladesh and later spread to other parts of the country including the capital city, Dhaka (1, 19). Similar cholera-like outbreaks were also reported from several places in neighboring India (109). The bacterium responsible for the outbreaks in Bangladesh and India resembled V. cholerae O1 in its culture and biochemical characteristics but did not agglutinate with V. cholerae O1 antisera. The new epidemic strain of V. cholerae was later serogrouped as O139 and given the synonym name “Bengal” (123). V. cholerae O139 Bengal, which caused explosive epidemics throughout Bangladesh, India, and neighboring countries, was recognized as the second causative agent of cholera after the O1 serogroup. In the beginning, the new strain totally displaced the existing V. cholerae O1 strains, including both classical and El Tor biotypes, which coexisted only in Bangladesh. However, during 1994 and until the middle of 1995, in most northern and central areas of Bangladesh, including Dhaka, the O139 vibrios were replaced by a new clone of V. cholerae O1 of the El Tor biotype, whereas in the southern coastal regions the O139 vibrios continued to exist (36, 127). During the second half of 1995 and in 1996, nearly 4 years after the initial detection of O139 vibrios, cases due to both V. cholerae O1 and O139 were detected in various regions of Bangladesh. Recent surveillance under the epidemic control preparedness program of ICDDR,B revealed the coexistence of V. cholerae O1 and O139 in Dhaka as well as in at least 4 of 10 rural districts of the northern, central, and southern regions of the country affected by outbreaks of cholera between June and December 1996 (35). In keeping with the observation in Bangladesh, a resurgence of V. cholerae O139 infection for the first time since its initial predominance in 1993 has been reported in Calcutta, India, (89). Analysis of V. cholerae O139 strains isolated between 1993 and 1996 in Bangladesh and India has revealed temporal changes in genetic and phenotypic properties of strains belonging to this serogroup (35, 91). Cholera surveillance in Bangladesh showed that until 1992 V. cholerae O1 belonging to both biotypes caused regular epidemics and since then both V. cholerae O1 and O139 have been significant causes of infection and morbidity, although the frequency of infection varies from year to year in different regions of the country (7, 127, 129, 134).
Antibiotic Resistance among Toxigenic V. cholerae Strains
In 1979, an outbreak of cholera due to multiple-drug-resistant V. cholerae O1 occurred in Matlab, a rural subdistrict of Bangladesh (43, 44). Screening of isolates from the outbreak showed that 16.7% of the isolates were resistant to the five antibiotics tetracycline, ampicillin, kanamycin, streptomycin, and trimethoprim-sulfamethoxazole and that another 10% of the isolates were resistant to any four of these antibiotics including tetracycline. An antibiotic resistance plasmid was identified in these isolates and was transferable to an Escherichia coli K-12 recipient by conjugation. Epidemiological assessment of the outbreak suggested that the outbreak began with the introduction into the area of a single multiple-drug-resistant strain of V. cholerae O1 (44). By 1986, the drug resistance pattern had changed, and screening of V. cholerae O1 isolated from cholera patients in January 1986 in Dhaka showed that none of these isolates was resistant to tetracycline, streptomycin, chloramphenicol, amoxicillin, or nalidixic acid (92). However, during 1988 and 1989, nearly all classical V. cholerae strains isolated in Bangladesh were resistant to tetracycline whereas strains belonging to the El Tor biotype were sensitive to the drug (130). After almost a decade, reemergence of tetracycline resistant El Tor strains was observed during the 1991 epidemic in Bangladesh (129). In October 1995, the emergence of nalidixic acid-resistant V. cholerae O1 was observed in southern India (60). The susceptibility of V. cholerae O1 strains to certain antibiotics changed depending on the isolation time and geographical location. The reemergence of tetracycline resistance was observed during the 1991 epidemic in Bangladesh, and 70% of strains isolated were resistant to tetracycline, often in addition to other antibiotics (129). Between March 1994 and December 1996, 80 to 100% of V. cholerae O1 isolates in Kenya and south Sudan and 65 to 90% of isolates in Somalia were sensitive to tetracycline (79) whereas all isolates in Tanzania and Rwanda were resistant. In Kenya and Somalia, the percentage of isolates resistant to chloramphenicol and co-trimoxazole markedly increased from 15% in 1994 to more than 90% in 1996 (79). The O139 serogroup of V. cholerae which emerged during 1992 to 1993 was sensitive to tetracycline (19). Although the new serogroup showed a trend of increased resistance to trimethoprim-sulfamethoxazole, it was more susceptible to ampicillin and tetracycline than the O1 serogroup (119). Waldor et al. (146) reported the presence of a 62-kb self-transmissible transposon-like element (SXT element) encoding resistance to sulfamethoxazole, trimethoprim, and streptomycin in V. cholerae O139. The SXT element could be conjugally transferred from V. cholerae O139 to V. cholerae O1 and E. coli strains, where it integrated into recipient chromosomes in a site-specific recA-independent manner (146).
Comparison of the antibiotic resistance patterns in the O139 strains isolated during 1992 and 1993 and those isolated in 1996 and 1997 in India showed that the latter strains were susceptible to co-trimoxazole, unlike the former. Recent studies have shown that O139 strains are becoming increasingly resistant to ampicillin and neomycin but increasingly susceptible to chloramphenicol and streptomycin (91). Considering the rapidly changing pattern of antibiotic resistance observed among V. cholerae strains, it appears that there is substantial mobility in genetic elements encoding antibiotic resistance in V. cholerae.
Molecular Epidemiology
Epidemiological surveillance of cholera was limited before the 1970s by the lack of suitable typing systems. However, recent developments in DNA analysis techniques have introduced several new typing methods and have permitted studies of the epidemiology of V. cholerae on a larger global perspective (17, 24, 33, 34, 37, 38, 66, 141–143). These techniques include the analysis of restriction fragment length polymorphisms (RFLPs) in different genes. The use of gene probes to study RFLPs in the ctxAB genes and their flanking DNA sequences, which are part of a larger genetic element (CTX element), indicated that the U.S. Gulf Coast isolates of toxigenic V. cholerae are clonal and that they are different from other seventh-pandemic isolates (66). RFLPs in conserved rRNA genes have also been used to differentiate V. cholerae strains into different ribotypes. Analysis of isolates from the Latin American epidemic in 1991 showed that they were related to the seventh-pandemic isolates from other parts of the world and that the Latin American cholera epidemic was an extension of the seventh pandemic (38, 141, 142). Analysis of toxigenic El Tor strains by multilocus enzyme electrophoresis has also been used to group the El Tor strains into major clonal groups. The clones seem to reflect broad geographical and epidemiological associations. The clonal diversity and epidemiological associations of toxigenic V. cholerae have been reviewed by Wachsmuth et al. (143). Comparative analysis of the El Tor strains of V. cholerae O1 and the epidemic O139 strains suggested that the O139 strains are related to El Tor strains and were derived from them by possible genetic changes in the serotype-specific gene clusters (34, 143). Numerical analysis of ribotype patterns (37) has also revealed that V. cholerae strains belonging to the non-O1 non-O139 serogroups diverge widely from the O1 and O139 V. cholerae strains.
Molecular analysis of V. cholerae strains isolated during epidemics between 1961 and 1996 in Bangladesh revealed clonal diversity among strains isolated during different epidemics (33–37). These studies demonstrated the transient appearance and disappearance of more than six ribotypes of classical vibrios, at least five ribotypes of El Tor vibrios, and three different ribotypes of V. cholerae O139. Different ribotypes often showed different CTX genotypes resulting from differences in the copy number of the CTX element and variations in the integration site of the CTX element in the chromosome (35, 36). These studies indicated that there had been a continual emergence of new clones of toxigenic V. cholerae which replaced existing clones, possibly through natural selection involving unidentified environmental factors and immunity of the host population.
VIRULENCE-ASSOCIATED GENES IN V. CHOLERAE
The pathogenesis of cholera is a complex process and involves a number of factors which help the pathogen to reach and colonize the epithelium of the small intestine and produce the enterotoxin that disrupts ion transport by intestinal epithelial cells. Although production of CT, encoded by the ctxAB genes, is directly responsible for the manifestation of diarrhea, cholera pathogenesis relies on the synergistic action of a number of other genes, including the genes for one or more colonization factors (67). In V. cholerae, the major virulence genes appear to exist in clusters, and there are at least two regions of the V. cholerae chromosome in which genes encoding virulence factors are clustered (29, 50, 98, 104, 139). These include the CTX element, which has now been shown to be the genome of a filamentous bacteriophage (144), and the TCP-accessory colonization factor (ACF) gene cluster, referred to as the TCP pathogenicity island (73). The pathogenicity island shares several characteristics with those of other species of pathogenic bacteria. These include the presence of groups of virulence genes, a regulator of virulence genes, a transposase gene, specific (att-like) attachment sites flanking each end of the island, and an integrase with homology to a phage integrase gene (69, 73). Thus, the TCP pathogenicity island appears to have a phage origin but may now be defective (73).
Since colonization is a prerequisite to establishing a productive infection by V. cholerae, the existence of other possible factors responsible for colonization has been investigated. The mannose-fucose-resistant cell-associated hemagglutinin (MFRHA) has been implicated as a virulence determinant (41), but its exact role in colonization is not clear. An isogenic strain mutated in the gene encoding MFRHA activity was markedly attenuated when tested for virulence in infant mice. A 6.3-kb BamHI fragment of the chromosome of V. cholerae 569B which includes the sequence encoding MFRHA has been sequenced. Ten ORFs were apparent within the 6.3-kb region, and two of these, designated mrhA and mrhB (encoding 7- and 27-kDa proteins, respectively), were associated with MRHA activity (41, 140). A striking feature of the sequenced region is the occurrence of nine copies of a 124-bp direct repeat, all of which are located outside predicted ORFs. It has been suggested recently that the organization of the ORFs and the 124-bp repeats is similar to that of an “integron-gene cassette” structure, which is formed when gene expression elements known as integrons acquire ORFs and convert them to functional genes (5, 80).
The mannose-sensitive hemagglutinin (MSHA), which is expressed mostly by strains of the El Tor biotype, is a flexible pilus composed of subunits (61). The gene encoding the structural subunit of MSHA pili was cloned and sequenced (62). The mshA gene was localized on a 2.6-kb SalI-EcoRI fragment, of the chromosome of an El Tor strain, Phil 6973, and sequencing of the entire fragment revealed a total of six ORFs. The mshA gene encodes an 18,094-Da prepilin protein, which in its mature form has a size of 17,436 Da (62). Protection studies in animal models suggested that MSHA pili may be important in the pathogenesis of El Tor cholera (99, 100). However, a more recent study (3) with El Tor strains of V. cholerae with deletions in the mshA gene did not show any significant attenuation or loss of colonization potential in an infant-mouse model. Antibodies to the pilin subunit were also not sufficient to mediate significant protection against a challenge with the wild-type El Tor strain (3).
Antibodies to some purified outer membrane proteins (OMPs) of V. cholerae have been shown to inhibit intestinal colonization in the infant mouse model (120). A 77-kDa OMP designated IrgA, whose expression is regulated by iron concentration, has been shown to be important for virulence in infant mice (47). The role of other OMPs in virulence has not been established in either animal or human studies. Other factors that have been examined for possible roles in virulence include the core-encoded pilus, which is encoded by the cep gene located within the CTX genetic element (104), and several possible adhesins such as the lipopolysaccharide (18) and a slime agglutinin present on the flagellum (4). Although some of these factors including MFRHA, MSHA, and OMPs are suspected to play a role in enhancing adhesion and colonization, possibly in association with other factors, when tested in animal models, their exact role in the virulence of V. cholerae in humans is still uncertain. Studies to date have shown that the major virulence genes of V. cholerae required for pathogenesis in humans as well as in animal models are the genes involved in the production of TCP and CT. The structures of the TCP pathogenicity island and the CTX genetic element are suggestive of horizontal transfer of these gene clusters as a possible mechanism for the origination of new pathogenic clones of V. cholerae. It seems possible that the acquisition of the TCP pathogenicity island and the CTX element has allowed specific strains of V. cholerae to become adapted to the human intestinal environment.
TCP Pathogenicity Island
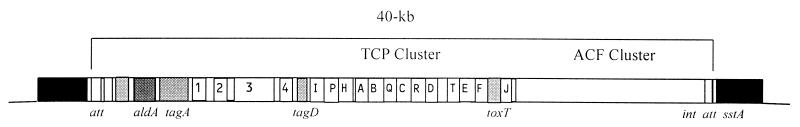
Colonization of brush borders in the small intestine, a crucial component of the infection strategy, is assumed to be mediated by a rigid pilus colonization factor, TCP, since it is under the same genetic control as CT (138). Early studies established that the genes encoding TCP were clustered and present in clinical isolates of O1 El Tor and classical vibrios but not in environmental isolates of V. cholerae O1, with the exception of a few strains from the Gulf Coast of the United States (137). Expression of CT and TCP are coregulated by the ToxR regulatory system, which includes the ToxT protein (27). The genes encoding ToxT and TCP are located in the same chromosomal region (13), together with other ToxR-regulated genes including those for ACF (30, 56, 74). Molecular analysis has revealed that although the major subunit of TCP is encoded by the tcpA gene, the formation and function of the pilus assembly require the products of a number of other genes located on the chromosome adjacent to the tcpA gene, and that these constitute the tcp gene cluster (98). At least 15 ORFs are found in the tcp cluster, which is located immediately downstream of the tagD gene. The tcpH and tcpI genes are two ToxR-regulated genes that influence TcpA synthesis. Inactivation of tcpH results in decreased pilin synthesis, whereas inactivation of tcpI leads to increased synthesis of TcpA. It has been suggested that regulators such as TcpI, which acts downstream of ToxR and ToxT, may function to fine-tune the expression of the TCP virulent determinant throughout the pathogenic cycle of V. cholerae (50). More recently, Hase and Mekalanos presented evidence that TcpP and TcpH constitute homologues of ToxR and ToxS and cooperate with ToxR and ToxS in the transcriptional activation of the ToxT promoter (51). Immediately adjacent to and downstream of the tcp cluster is located the acf gene cluster. The exact nature of the colonization factor is not clear, but acfD, one of the four ORFs (acfABCD), encodes a lipoprotein (102). Further analysis revealed the presence of a putative integrase gene (int) and a putative att-like 20-bp attachment site adjacent to the TCP-ACF gene cluster (73). Sequencing of the 13-kb region upstream of the tcp gene cluster showed the aldA and tagA genes, four additional ORFs of unknown function, a sequence with significant homology to the bfpM gene of enteropathogenic Escherichia coli, a gene for a transposase found in V. parahaemolyticus, and a second copy of an att-like sequence (69). The entire region of nearly 40 kb (Fig. 1) flanked by the att-like sequences and including the TCP-ACF gene clusters, the integrase, and the transposase genes appears to constitute a pathogenicity island. It has been suggested that the 9-kb tagA-tagD region (Fig. 1) is associated with epidemic and pandemic strains and is absent from nontoxigenic environmental strains (69, 73). It appears that the TCP pathogenicity island is the initial genetic element required for the origination of epidemic strains, since CTXΦ uses TCP as its receptor (144). Also, the role of TCP as an essential colonization factor inside the host intestine is well recognized (2, 52).
FIG. 1.
Genetic organization of the TCP pathogenicity island of V. cholerae. Dark boxes show flanking DNA, att sites are shown at both ends, and the putative integrase gene is shown near the right end of the island. The transposase sequence is indicated by a light grey box near the left end of the island. See the text for details.
CTX Genetic Element
Toxigenic V. cholerae carries one or more copies of CT genes (ctxAB). The A and B subunits of CT are encoded by two separate but overlapping ORFs. V. cholerae also produces a putative toxin known as zonula occludens toxin (Zot), which increases the permeability of the small intestinal mucosa by affecting the structure of the intercellular tight junction, or zonula occludens (10, 39). The zot gene consists of a 1.3-kb ORF, which could potentially encode a 44.8-kDa polypeptide and is located immediately upstream of the ctxA gene (10). A third toxin that has been described is accessory cholera enterotoxin (Ace) which is capable of inducing fluid accumulation in rabbit ligated ileal loops (139).
The genes encoding the toxins (ace, zot, and ctxAB) and a core-encoded pilin (cep) and an ORF of unknown function (orfU) are located on a 4.5-kb “core region,” flanked by one or more copies of a repetitive sequence called RS1 (104). Together, these DNA units comprise the CTX element, which had been perceived as a transposon-like genetic element. Further analysis showed divergence between repetitive sequence and revealed the presence of two nearly identical sequences, designated RS1 (2.7 kb) and RS2 (2.4 kb), which were generically referred to as the RS sequence (104). Naturally occurring isolates of V. cholerae which do not produce CT lack sequences homologous to ctxAB and the rest of the core region (68). However, these nontoxigenic strains contain an 18-bp sequence called attRS1. In toxigenic strains, the RS sequence of the CTX genetic element encodes a site-specific recombination system, which allows integration of the CTX element into the 18-bp attRS1 site in a recA-independent manner (104). It has recently been demonstrated by Waldor and Mekalanos (144) that the CTX genetic element corresponds to the genome of a lysogenic filamentous bacteriophage designated CTXΦ and that genes in the core region of the CTX element, particularly zot and orfU, are crucial for the morphogenesis of the phage.
Cholera Toxin-Converting Bacteriophage
It has been demonstrated that under appropriate conditions, toxigenic V. cholerae strains can be induced to produce extracellular CTXΦ particles (32, 144). The phage can be propagated in recipient V. cholerae strains in which the CTXΦ genome either integrates chromosomally at a specific site, forming stable lysogens, or is maintained extrachromosomally as a replicative form (RF) of the phage DNA (144). Cultures of V. cholerae harboring the RF of CTXΦ produce high titers of the phage in their supernatants. The bacteriophage uses the TCP as a receptor, and hence expression of TCP by the bacterium is a prerequisite for its susceptibility to the phage. Thus, a virulence factor of the bacterium in humans also serves as a receptor for CTXΦ, demonstrating a coevolution of genetic elements mediating the transfer of virulence genes with the pathogenic bacterial species they infect.
The CTXΦ genome has two regions, the core and RS2 (145). Genes with related functions are clustered in the genome of CTXΦ similarly to those of other filamentous phages. Analysis of phage morphogenesis revealed that most of the genes of the core region are essential for the formation of the CTXΦ particles and hence for its propagation as an infectious phage. The ORFs in RS2 were designated rstR, rstA2, and rstB2 and were found to encode products required for the integration, replication, and regulatory functions of CTXΦ (145). The deletion of a portion of the genes encoding CT by marker exchange, however, did not affect the morphogenesis of the bacteriophage. It thus seems that the ctxAB genes do not participate directly in the formation of phage particles but are important for the phage to provide a survival advantage to its host bacteria in the gastrointestinal environment (144).
Reexamination of Ace and Zot as Proposed Toxins
In previous reports, the CTX genetic element was suggested to encode at least two additional toxins (Ace and Zot), which were thought to play a role in the pathogenesis of V. cholerae (10, 139). Since the CTX element has been shown to be the genome of a filamentous phage, the structure and role of these previously proposed virulence-associated genes were reexamined. The deduced amino acid sequence of the gene products encoded by the core of the CTX element was compared with those of other filamentous phages including the male (F+)-specific coliphage M13 (144). Comparison of the relative positions and sizes of the genes involved in the morphogenesis of CTXΦ and coliphage M13 showed that the cep, orfU, ace, and zot genes were comparable to genes VIII, III, VI, and I, respectively, of coliphage M13. The zot gene product was found to be homologous to a family of proteins which includes the gene I product of M13 and the corresponding gene I products of several other filamentous bacteriophages of E. coli, Pseudomonas, and Xanthomonas. The gene I product is known to be an inner membrane protein required for the assembly of filamentous phages. In view of the observation that mutation in the zot gene impairs morphogenesis of CTXΦ and in view of the homology between the zot gene product and gene I product of other filamentous phages, Waldor and Mekalanos proposed that the previously described zonula occludens toxin was probably not directly associated with the zot gene product, unless its product has dual functions (144). Similarly, the ace gene corresponds to gene VI of coliphage M13. Alignment of the ace gene product with the corresponding gene VI homolog of pseudomonas filamentous phage Pf1 (ORF141) revealed 61% similarity and 27% identity. The gene VI product of coliphage M13 and other filamentous phages is a small, hydrophobic protein that assembles into the virion particles. Thus, the previously proposed role of the ace gene product as an accessory cholera enterotoxin may not be appropriate. Kaper and coworkers described a volunteer study that tested a V. cholerae strain with specific deletions of sequences encoding Zot, Ace, and the A subunit of CT. This strain, CVD110, still caused mild to moderate diarrhea in volunteers (85, 136). This study clearly indicated that residual diarrhea caused by CT-negative mutants of V. cholerae may also be due to factors other than the zot and ace gene products.
Integron System for Acquisition of Heterologous Genes by V. cholerae
Recent studies have shown the presence of a versatile system in V. cholerae for acquisition of genes from other organisms. This consists of a distinctive class of integrons, which are gene expression elements that may capture ORFs and convert them to functional genes (80). Insertion of an ORF or a gene cassette into an integron takes place by site-specific recombination between the circularized cassette and the recipient integron, which carries an integrase gene and a specific attachment site. All integron-inserted gene cassettes possess an imperfect inverted-repeat sequence located at the 3′ end of the gene that functions as a recognition site for the site-specific integrase. The repeat sequence is referred to as the 59-base element; it is a diverse family of sequences that are recognized by the integrase. Until recently, all known integrons were associated only with genes conferring antibiotic resistance (111, 112). Recently, a gene designated intl4, which encodes a previously unknown integrase, has been identified in V. cholerae (80). This integrase recognizes a family of V. cholerae repeated sequences (VCRs) and is associated with a “gene-VCR” organization, similar to that of the well-characterized antibiotic resistance integrons (80, 112). The VCRs are a family of 123- to 126-bp sequences that are highly repeated (60 to 100 copies) and situated in a region corresponding to about 10% of the V. cholerae genome (6). Two such sequences flank a heat-stable toxin gene in both O1 and non-O1 V. cholerae isolates (97), and nine other VCRs have been found in a 6-kb DNA fragment containing the genes for MFRHA, a lipoprotein gene, and eight other unidentified ORFs (5). The variation observed in codon usage of the gene-VCR cassettes, as well as their G+C content (between 33 and 45%, compared with 47% for the V. cholerae genome), suggested that the VCR-associated genes were probably recruited by V. cholerae from other microbial sources (80). Thus, the demonstration of the distinctive integron system in V. cholerae suggested that integrons may also play a role in the acquisition of pathogenic genes as well as genes for different biological functions of V. cholerae.
REGULATION OF VIRULENCE
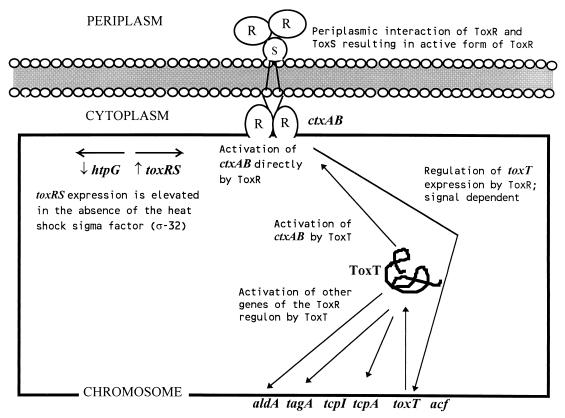
There are multiple systems involved in the regulation of virulence-associated genes in V. cholerae. Expression of several critical virulence genes in V. cholerae is coordinately regulated so that multiple genes respond in a similar fashion to environmental conditions (27, 132). Coordinate expression of virulence genes results from the activity of a cascading system of regulatory factors. ToxR, a 32-kDa transmembrane protein, is the master regulator and is itself regulated by environmental signals. The ToxR protein binds to a tandemly repeated 7-bp DNA sequence found upstream of the ctxAB structural gene and increases the transcription of ctxAB, resulting in higher levels of CT expression (27, 87, 88). The activity of ToxR is enhanced by another 19-kDa transmembrane protein, ToxS, which interacts with ToxR (Fig. 2). ToxS serves to assemble or stabilize ToxR monomers into the dimeric form (26). ToxR regulates not only the expression of ctxAB but also that of at least 17 distinct genes, which constitute the ToxR regulon. These include the TCP colonization factor (138), the accessory colonization factor (105), the OMPs OmpT and OmpU (86), and three other lipoproteins (102). Except for the ctxAB genes, other genes in the ToxR regulon are controlled through another regulatory factor called ToxT, a 32-kDa protein. ToxR controls the transcription of the toxT gene, which encodes a member of the AraC family of bacterial transcription activators (53). The resulting increased expression of the ToxT protein then leads to activation of other genes in the ToxR regulon. Thus, ToxR is at the top of the regulatory cascade that controls the expression of CT and other important virulence factors in V. cholerae, while the expression of ToxR itself remains under the control of environmental factors (132). It has also been recognized that V. cholerae has ToxT-dependent and ToxT-independent branches of the ToxR regulon (16). The regulatory cascade (Fig. 2) has been reviewed by DiRita (25). Interestingly, it has been reported recently that expression of CT from the RF of CTXΦ is independent of ToxR. This indicates that phage induction may provide another mechanism for the regulation of CT production (76).
FIG. 2.
Model for the ToxR/ToxT regulatory cascade of V. cholerae. See the text for details.
The coordinate regulation of virulence genes through the ToxR regulon demonstrates that the organism has developed a mechanism of sampling and responding to its environment. Parsot and Mekalanos (103) have proposed a mechanism for the regulation of virulence genes by V. cholerae in response to temperature variation. Immediately upstream of the toxR gene is the htpG gene, which encodes a heat shock protein (103). The toxR and htpG genes, which are transcribed in opposite directions, have their promoters so close that only one RNA polymerase can bind in the intergenic region. The proposed mechanism is that the normal ς70 RNA polymerase binds to the toxR promoter and transcribes the toxR gene only at low temperatures. At elevated temperatures, ς32, the RNA polymerase sigma subunit involved in the transcription of heat shock genes, binds to the htpG promoter, thus repressing the toxR promoter. This is consistent with the observation that the CT and TCP gene products are expressed in vitro at 30°C but repressed at 37°C in classical V. cholerae strains. However, the decreased expression at 37°C in vitro of virulence factors which are adequately expressed at 37°C in the intestinal environment needs to be explained. It has been suggested that the in vitro temperature effect may be due to the lack of other signals in vitro that are present at 37°C in vivo (25). Expression of CT, TCP, and other virulence factors differs between the classical and El Tor biotypes of V. cholerae (63). It has been suggested that the differential expression of the ToxR regulon in classical and El Tor vibrios is due to biotype-specific control over toxT expression (28).
Expression of certain genes in response to low iron concentration is another distinct regulatory system in V. cholerae, that controls additional putative virulence genes including hemolysins and several OMP that are not expressed when cells are grown in iron-rich media (131). It has been suggested that the intestinal site for growth of V. cholerae is a low-iron environment which triggers the expression of these iron-regulated genes in vivo (118). Low-iron-induced expression of genes in V. cholerae involves a protein known as Fur, which has considerable homology to the E. coli Fur protein (46). The Fur protein binds in the presence of iron to a 21-bp operator sequence found in the promoter of iron-regulated genes, thereby repressing transcription (46). In V. cholerae, Fur acts as a repressor for the irgA and viuA genes. IrgA, a 77-kDa OMP, is important for virulence in infant mice (47), whereas ViuA, a 74-kDa OMP, is the receptor for vibriobactin, which is produced under low-iron conditions and acts to bind iron extracellularly and transport it into the cell. Regulation of irgA also requires another protein, IrgB, which acts as a positive activator of transcription (48). The irgB gene is located immediately upstream of the irgA gene, and the transcription of irgB itself is repressed by Fur in the presence of iron. In addition to the genes whose expression is regulated by ToxR or iron, there are genes that are expressed in vivo but not in vitro (64). Many but not all of these proteins also appear to be induced under iron-limiting conditions. Additional genes that are expressed in vitro but are down regulated during in vivo growth have also been described. Different regulatory systems in V. cholerae apparently allow the bacterium to vary the expression of its genes to optimize survival in different environments, which include the human intestine and the estuarine environment.
ECOLOGY OF V. CHOLERAE
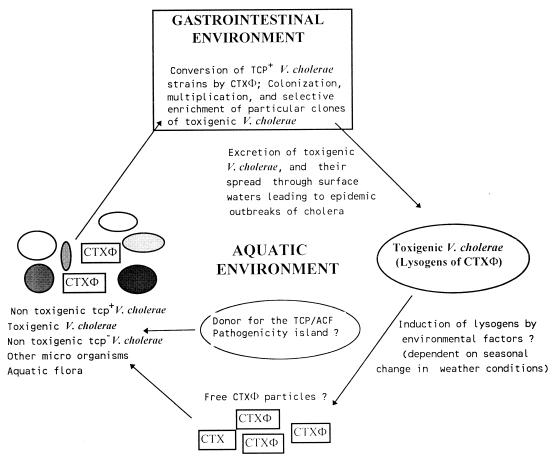
V. cholerae has been regarded as a member of a group of organisms whose major habitats are aquatic ecosystems (22). Although, V. cholerae is part of the normal, free-living bacterial flora in riverine and estuarine areas, non-O1 and non-O139 strains are more commonly isolated from the environment than are O1 and O139 strains. Moreover, outside of areas of epidemic infection and away from areas that may have been contaminated by cholera patients, environmental isolates of V. cholerae O1 have been found to be mostly CT negative. It seems logical that the natural habitat for toxigenic V. cholerae O1 and O139, which in most cases produce various factors necessary for colonization of the mammalian intestine, is likely to be the gastrointestinal tract. The major pathogenic genes in V. cholerae are clustered in several regions of the V. cholerae chromosome, and the structure of these pathogenic gene clusters indicates that these are capable of being propagated horizontally (69, 73, 104, 144). This suggests that environmental strains of V. cholerae may develop the ability to adapt to the intestinal environment through acquisition of the virulence genes. In view of the available information on the epidemiology of cholera, the lysogenic conversion by a bacteriophage encoding CT, and the survival and enrichment of V. cholerae under in vivo and in vitro conditions, it is apparent that the ecosystem for V. cholerae should have a number of components. These include the bacterium, the aquatic environment, CTXΦ and other unidentified genetic elements involved in the transfer of virulence genes, and the intestinal environment of the host population. In this section, we summarize information from studies of the survival of toxigenic V. cholerae in the aquatic environment and in the host intestine and propose a model for the ecology and evolution of V. cholerae (Fig. 3) and the possible role of accessory genetic elements that mediate the horizontal transfer of virulence genes.
FIG. 3.
Proposed model for the emergence of new strains of toxigenic V. cholerae: possible role of CTXΦ, environmental factors, and the human host in the ecology of toxigenic V. cholerae. See the text for details.
Environmental Survival and Persistence
The physicochemical conditions for the survival of V. cholerae O1 have been investigated, and the possibility of survival of the organism in an estuarine environment and other brackish waters is widely accepted (22, 23). However, the nature of the survival and persistence of toxigenic V. cholerae O1 or O139 in aquatic ecosystems and the factors involved in the conservation of the CTX element (the lysogenic form of CTXΦ) and other pathogenic genes in the aquatic environment are not clear. The survival may be dependent on several factors, such as the occurrence of particular physicochemical conditions, a specific association of the bacteria with aquatic plants or animals, and/or the existence of specific ecological associations involving several components of the aquatic environment. It has been postulated that under stress conditions the vibrios are converted to a viable but nonculturable (VNC) form that cannot be recovered by standard culture techniques and that such VNC forms are able to cause infection and can revert to the culturable form (23). In contrast to this proposition, studies of a marine vibrio strain, ANT-300, by Novitsky and Morita (94–96) showed that the organism responded to starvation by reducing its metabolic activities and undergoing morphological changes, e.g., changing from a rod shape to a coccoid shape and producing progeny cells significantly decreased in volume, but that it still remained culturable. It has, however, been argued that in this work the investigators had studied only the cells which remained culturable, not recognizing cells that were possibly VNC forms (23). The public health and ecological importance of the possible survival forms such as VNC depends on whether these forms can be converted back to live infectious bacteria. There is, however, very little evidence to conclusively establish that the possible nonculturable phenomenon is reversible. Hence there is considerable scope to further investigate the role of postulated VNC forms of V. cholerae through carefully controlled studies.
In areas of endemic infection, cholera epidemics occur in a regular seasonal pattern. It is not clear what determines the seasonal appearance of epidemic V. cholerae strains and outbreaks of cholera, although it has been suggested that during interepidemic periods toxigenic V. cholerae exists in an unexplained ecological association with aquatic organisms, possibly in the VNC form, until the next epidemic season, when environmental factors trigger the dormant bacteria to multiply and lead to cholera outbreaks (57, 58). However, differences in genetic or phenotypic properties have been often noticed among V. cholerae O1 and O139 strains isolated during different epidemics (91, 92, 130). Analysis of rRNA gene restriction patterns of V. cholerae strains has also shown clonal diversity among epidemic strains (33–37, 121). These events have raised questions about whether seasonal epidemics are caused by periodic appearances of the same strains of V. cholerae or are due to a continual emergence of new toxigenic clones from nontoxigenic progenitors.
The concept of an aquatic reservoir of V. cholerae O1 or O139 implies not only that the vibrios survive, in whatever form, but also that they form an essential component of the ecosystem. Laboratory studies of microcosms have illustrated the ability of V. cholerae O1 to associate with a variety of zooplankton, phytoplankton, and algae (58). The associations prolong survival, and presumably the vibrios gain nutrients from the host. However, these studies did not explain whether such association is specific for the epidemic serogroups of V. cholerae or is a general phenomenon for all other V. cholerae serogroups and thus does not seem to be a mechanism for selective enrichment of toxigenic V. cholerae O1 or O139. Moreover, the benefit imparted to the pathogen by possessing and maintaining the virulence-associated genes with respect to its survival and persistence in the environment is not clear. The role of extracellular enzymes including CT in the environmental microecology of V. cholerae O1 is uncertain, although it has been suggested that in freshwater systems, local ionic microenvironment can be controlled by V. cholerae O1 by use of toxin acting on other living cells. However, such hypothesis has yet to be proven with specific experimental data, and hence further studies are required to understand the more definitive roles of the virulence-associated factors and environmental selection pressures for toxigenic V. cholerae.
Enrichment of Toxigenic Strains in the Intestinal Environment
Although we now have some understanding of the mechanism by which CT causes diarrhea, we do not yet understand clearly why V. cholerae should infect and elaborate the lethal toxin in the host system. It seems worth speculating whether the role of the toxin is to simply cause diarrhea and thus disseminate the organism to its next victim or whether the toxin is providing a more crucial function for the enrichment and continued existence of the bacteria.
Studies directed toward the development of attenuated V. cholerae mutants altered in toxin production for use as live oral cholera vaccines provided a means of investigating the role of CT in the intestine. In 1971, Howard (55) reported the isolation of nontoxigenic mutants of the classical strain 569B by mutagenesis with nitrosoguanidine. The mutants were unable to induce a secretory response in the rabbit intestinal loop model and did not survive or multiply in the intestinal environment. In 1974 and 1975, Finkelstein et al. (40) and Holmes et al. (54) observed that the ability of various mutants to grow in the intestinal environment correlates with the ability of the mutants to induce a residual secretory response in the infant rabbit model. Several hypotoxigenic mutants were noted to be unstable in the rabbit intestinal loop model; during passage, they produced toxigenic revertants which eventually displaced the mutant strains in vivo (9, 84). A variety of different toxin-deficient mutants of V. cholerae tested in rabbit and infant-mouse models also suggested that the mutants showed enhanced killing and mechanical clearance in the intestinal environment compared to the toxigenic parental strain (8, 9). It seems possible that many of these early mutants were altered in TCP as well as CT expression, perhaps by carrying mutations in either toxR, toxS, toxT, tcpP, or tcpH. The selection for their reversion in vivo may therefore have been driven by the need to up regulate the expression of TCP more than that of CT.
Mekalanos demonstrated in 1983 that significant amplification of the genes encoding CT occurred in hypertoxigenic variants of V. cholerae selected during intestinal passage in rabbits (83). The nature of the selective pressure causing this enrichment of hypertoxigenic variants in vivo is unknown but is probably related to the in vivo selection process that was involved in the reversion of hypotoxigenic mutants observed in earlier animal studies (54, 84). In agreement with these observations, many clinical isolates of V. cholerae O1 or O139 carry multiple copies of the CTX genetic element and hence the ctxAB operon (34, 83). In the light of the recent observation by Lazar and Waldor (76), it seems possible that spontaneous induction of the CTXΦ in vivo plays a role in the amplification of the CTX element (83). The characterization in vivo of site-specific ctxAB mutants constructed by in vitro recombinant DNA methods provided the most convincing evidence that the toxin is beneficial to growth in the intestinal environment. It was demonstrated that the ctx mutants colonized rabbit intestines about 10- to 100-fold less efficiently than the parental strain (82). At least two possible mechanisms have been proposed to explain how CT might enhance intestinal colonization by V. cholerae. These are by relief of nutritional deprivation (42) and by inhibition of bactericidal activity produced by epithelial cells (8, 42, 71). The details of these mechanisms have been reviewed by Mekalanos (82).
Studies so far suggest that causation of cholera in humans is also linked to a natural process of enrichment of toxigenic V. cholerae and partly explains the benefit imparted to the pathogen during the disease in humans. However, to understand the general epidemiological behavior of V. cholerae, which includes mechanisms leading to seasonal pattern of epidemics, transient appearance and disappearance of different clones, and emergence of new epidemic clones, it is important to study the interactions among the bacteria, genetic elements mediating the transfer of virulence genes, the human host, and possible environmental factors.
Emergence of Novel Toxigenic Strains: A Hypothesis
It has been demonstrated that naturally occurring strains of toxigenic V. cholerae O1 and O139 are inducible lysogens of CTXΦ. The phage can be induced in vitro, but the induction is not normally associated with cholera pathogenesis in humans (32). It seems possible that in the natural ecological settings, unidentified environmental factors induce lysogenic CTXΦ in toxigenic V. cholerae, resulting in the release of extracellular CTXΦ particles into the aquatic environment. The cell-free phage particles participate in the emergence of novel toxigenic strains of V. cholerae through interactions with nontoxigenic strains which exist in the environment and in the human population that consumes the environmental waters. CTXΦ uses TCP as its receptor, and hence the phage can infect only V. cholerae cells expressing TCP. The TCP genes, which are part of a greater genetic element referred to as the TCP pathogenicity island, appear to be the initial genetic factors required for the origination of epidemic strains. Analysis of the structure of the pathogenicity island suggests that it could be of phage origin, and it has been speculated that it can be transferred by transducing phages (69, 73). Since genes responsible for the production of TCP are carried mostly by V. cholerae O1 or O139, while other serotypes of V. cholerae usually do not carry genes for TCP, it is obvious that the CTX element is also found mostly in the O1 and O139 vibrios, whereas most non-O1 vibrios are usually nontoxigenic. This further supports the assumption that in natural settings, CTXΦ probably plays an important role in the origination of new toxigenic strains of V. cholerae. Since V. cholerae strains which are TCP positive but CTX negative are not frequently isolated during environmental sampling, it is possible that such strains are normally present in very small numbers in the environment, but following conversion by CTXΦ to toxigenicity, the strains are enriched in the gastrointestinal environment and later become detectable as new strains of toxigenic V. cholerae (Fig. 3). Subsequent increases in the concentration of toxigenic V. cholerae in the aquatic environment may lead to epidemic outbreaks of cholera.
It has been demonstrated that CTXΦ infects recipient V. cholerae strains more efficiently in the intestinal environment, where virulence factors such as TCP are adequately expressed (32, 144). While the conversion of nontoxigenic V. cholerae is favored within the gastrointestinal tract of the mammalian host, the natural selection and persistence of the novel toxigenic strains may involve both intestinal and environment factors, the immune status of the host population, and antigenic properties of the new pathogenic strain. The induction of CTXΦ lysogens is probably controlled by precise environmental signals such as optimum temperature, sunlight, and osmotic conditions (Fig. 3), and this may also account for the observed seasonal outbreaks of cholera in regions of endemic infection. However, to test this hypothesis, it is necessary to carry out further studies to (i) screen environmental water samples in a cholera-endemic region for the presence of cell-free CTXΦ particles during epidemic and interepidemic seasons, (ii) determine the stability of free CTXΦ particles in the aqueous environment and their ability to infect and lysogenize nontoxigenic strains of V. cholerae in animal models and laboratory microcosms, and (iii) characterize toxigenic and nontoxigenic V. cholerae strains isolated from the environment and cholera patients by genetic fingerprinting. Further studies are also required to confirm the origin of the TCP pathogenicity island and its role in the ecology and evolution of epidemic V. cholerae.
Natural Selection of Toxigenic Strains
V. cholerae offers a genetic system to study the relationship between pathogenesis and the natural selection of pathogens so as to ensure their continued existence. It appears that acquisition of pathogenic gene clusters by V. cholerae, which is normally a marine or brackish-water species, has allowed the bacterium to become adapted to the human intestinal environment. Although the donor organism of the TCP pathogenicity island has not been identified, the pathogenicity island seems to have a phage origin, which may now be defective (69, 73). On the other hand the donor for the CTX element has been confirmed to be a filamentous bacteriophage, which has been characterized in some details (144, 145). The acquisition of the CTX genetic element by V. cholerae provides a survival advantage to the bacterium, and hence to the bacteriophage, and leads to enrichment of toxigenic V. cholerae in the intestinal environment. Thus, CTXΦ confers increased evolutionary fitness to its host and hence to its own nucleic acids. With growing immunity in the host population against certain toxigenic clones of V. cholerae, new toxigenic clones emerge and replace existing clones by a process of natural selection. Thus, the continual emergence of new strains of toxigenic V. cholerae and their selective enrichment during cholera outbreaks constitute an essential component of the ecosystem for the survival and evolution of V. cholerae and the genetic elements that mediate the transfer of virulence genes.
CONCLUDING REMARKS
In spite of numerous studies over more than a century, the epidemiology and ecology of cholera remain mysterious and challenging to investigators in the field. Recent studies of the pathogenicity island, the discovery of the bacteriophage encoding CT, and the distinctive system for acquisition of different genes by V. cholerae have provided an impetus for further study of the organism to understand the molecular basis for the emergence of pathogenesis and natural phenomena controlling the conservation of particular genetic traits. V. cholerae provides a natural system to study the coevolution of bacteria and the virulence-associated genetic elements and the mutual benefits imparted in terms of attaining greater evolutionary fitness.
ACKNOWLEDGMENTS
We thank Firdausi Qadri and Tasnim Azim for their invaluable suggestions during the preparation of the manuscript and Manzurul Huq for secretarial assistance.
Research in the ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. A large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Attridge S R, Voss E, Manning P A. The role of toxin co-regulated pili in the pathogenesis of Vibrio cholerae O1 El Tor. Microb Pathog. 1993;15:421–431. doi: 10.1006/mpat.1993.1091. [DOI] [PubMed] [Google Scholar]
- 3.Attridge S R, Manning P A, Holmgren J, Jonson G. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attridge S R, Rowley D. The specificity of V. cholerae adherence and significance of the slime agglutinin as a second mediator of in vitro attachment. J Infect Dis. 1983;147:873–881. doi: 10.1093/infdis/147.5.873. [DOI] [PubMed] [Google Scholar]
- 5.Baker A, Clark C A, Manning P A. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J Bacteriol. 1994;176:5450–5458. doi: 10.1128/jb.176.17.5450-5458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker A, Manning P A. VlpA of Vibrio cholerae O1: the first bacterial member of the alpha 2-microglobulin lipocalin superfamily. Microbiology. 1997;143:1805–1813. doi: 10.1099/00221287-143-6-1805. [DOI] [PubMed] [Google Scholar]
- 7.Baqui A H, Yunus M D, Zaman K, Mitra A K, Hossain K M B. Surveillance of patients attending a rural diarrhoea treatment centre in Bangladesh. Trop Geogr Med. 1991;43:17–22. [PubMed] [Google Scholar]
- 8.Baselski V S, Medina R A, Parker C D. Survival and multiplication of Vibrio cholerae in the upper bowel of infant mice. Infect Immun. 1978;22:435–440. doi: 10.1128/iai.22.2.435-440.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baselski V S, Medina R A, Parker C D. In vivo and in vitro characterization of virulence-deficient mutants of Vibrio cholerae. Infect Immun. 1979;24:111–116. doi: 10.1128/iai.24.1.111-116.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudry B, Fasano A, Ketley J, Kaper J B. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumann P, Furniss A L, Lee J V. Vibrio. In: Kreig N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 518–538. [Google Scholar]
- 12.Billings J S, McClellan E, Peters J C. Document 95, 43rd Congress, 2nd session. U.S. Washington, D.C: Government Printing Office; 1975. The cholera epidemic of 1873 in the United States; pp. 1–1025. [Google Scholar]
- 13.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control. Imported cholera associated with a newly described toxigenic Vibrio cholerae O139 strain—California, 1993. Morbid Mortal Weekly Rep. 1993;42:501–503. [PubMed] [Google Scholar]
- 15.Chambers J S. The conquest of cholera. New York, N.Y: Macmillan; 1938. pp. 24–44. [Google Scholar]
- 16.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterisation of Tox T mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, Evins G M, Cook W L, Almeida R, Bean H N, Wachsmuth I K. Genetic diversity among toxigenic and non-toxigenic Vibrio cholerae O1 isolated from the Western Hemisphere. Epidemiol Infect. 1991;107:225–233. doi: 10.1017/s0950268800048846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chitnis D S, Sharma K D, Kamat R S. Role of somatic antigen of Vibrio cholerae in adhesion to intestinal mucosa. J Med Microbiol. 1982;5:53–61. doi: 10.1099/00222615-15-1-53. [DOI] [PubMed] [Google Scholar]
- 19.Cholera Working Group, International Center for Diarrhoeal Disease Research, Bangladesh. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 20.Chongsa-nguan M, Chaicumpa W, Moolasart P, Kandhasingha P, Shimada T, Kurazono H, Takeda Y. Vibrio cholerae O139 Bengal in Bangkok. Lancet. 1993;342:430–431. doi: 10.1016/0140-6736(93)92841-g. [DOI] [PubMed] [Google Scholar]
- 21.Cohen J, Schwartz T, Klasmer R, Pridan D, Ghalayini H, Davies A M. Epidemiological aspects of cholera El Tor outbreak in a non-endemic area. Lancet. 1971;ii:86–89. doi: 10.1016/s0140-6736(71)92056-3. [DOI] [PubMed] [Google Scholar]
- 22.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Co.; 1992. pp. 107–127. [Google Scholar]
- 23.Colwell R R, Huq A. Vibrios in the environment:viable but non-culturable Vibrio cholerae. In: Wachsmuth I K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C: ASM Press; 1994. pp. 117–133. [Google Scholar]
- 24.Cook W L, Wachsmuth K, Feeley J, Huq I. The question of classical cholera. Lancet. 1983;i:879–880. doi: 10.1016/s0140-6736(83)91424-1. [DOI] [PubMed] [Google Scholar]
- 25.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 26.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 27.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascades controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiRita V J, Neely M, Taylor R K, Bruss P M. Differential expression of ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over ToxT expression. Proc Natl Acad Sci USA. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everiss K D, Hughes K J, Kovach M E, Peterson K M. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect Immun. 1994;62:3289–3298. doi: 10.1128/iai.62.8.3289-3298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everiss K D, Hughes K J, Peterson K M. The accessory colonization factor and toxin-coregulated pilus gene clusters are physically linked on the Vibrio cholerae O395 chromosome. DNA Seq. 1994;5:51–55. doi: 10.3109/10425179409039704. [DOI] [PubMed] [Google Scholar]
- 31.Ewing W H, High R, Johnson J G. Studies on the aeromonas group. Atlanta, Ga: Communicable Disease Center, U.S. Department of Health, Education and Welfare; 1961. [Google Scholar]
- 32.Faruque S, Asadulghani M, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic V. cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faruque S M, Alim A R M A, Rahman M M, Siddique A K, Sack R B, Albert M J. Clonal relationships among classical Vibrio cholerae O1 strains isolated between 1961 and 1992 in Bangladesh. J Clin Microbiol. 1993;31:2513–2516. doi: 10.1128/jcm.31.9.2513-2516.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faruque S M, Alim A R M A, Roy S K, Khan F, Nair G B, Sack R B, Albert M J. Molecular analysis of rRNA and cholera toxin genes carried by the new epidemic strain of toxigenic Vibrio cholerae O139 synonym Bengal. J Clin Microbiol. 1994;32:1050–1053. doi: 10.1128/jcm.32.4.1050-1053.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faruque S M, Ahmed K M, Siddique A K, Zaman K, Alim A R M A, Albert M J. Molecular analysis of toxigenic Vibrio cholerae O139 Bengal isolated in Bangladesh between 1993 and 1996: evidence for the emergence of a new clone of the Bengal vibrios. J Clin Microbiol. 1997;35:2299–2306. doi: 10.1128/jcm.35.9.2299-2306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faruque S M, Ahmed K M, Alim A R M A, Qadri F, Siddique A K, Albert M J. Emergence of a new clone of toxigenic Vibrio cholerae O1 biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J Clin Microbiol. 1997;35:624–630. doi: 10.1128/jcm.35.3.624-630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faruque S M, Roy S K, Alim A R M A, Siddique A K, Albert M J. Molecular epidemiology of toxigenic V. cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J Clin Microbiol. 1995;33:2833–2838. doi: 10.1128/jcm.33.11.2833-2838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faruque S M, Albert M J. Genetic relation between Vibrio cholerae O1 strains in Ecuador and Bangladesh. Lancet. 1992;339:740–741. doi: 10.1016/0140-6736(92)90636-h. [DOI] [PubMed] [Google Scholar]
- 39.Fasano A, Baudry B, Pumplin D W, Wasserman S S, Tall B D, Ketley J M, Kaper J B. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc Natl Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finkelstein R A, Vasil M L, Holmes R K. Studies on toxinogenesis in Vibrio cholerae. I. Isolation of mutants with altered toxinogenicity. J Infect Dis. 1974;129:117–123. doi: 10.1093/infdis/129.2.117. [DOI] [PubMed] [Google Scholar]
- 41.Franzon V F, Baker A, Manning P A. Nucleotide sequence and construction of a mutant in the mannose-fucose-resistant hemagglutinin (MFRHA) of Vibrio cholerae O1. Infect Immun. 1993;61:3032–3037. doi: 10.1128/iai.61.7.3032-3037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freter R, Brein O P C M. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: fitness and virulence of nonchemotactic Vibrio cholerae mutants in infant mice. Infect Immun. 1981;34:222–233. doi: 10.1128/iai.34.1.222-233.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glass R I, Huq I, Alim A R M A, Yunus M. Emergence of multiple antibiotic-resistant Vibrio cholerae in Bangladesh. J Infect Dis. 1980;142:939–942. doi: 10.1093/infdis/142.6.939. [DOI] [PubMed] [Google Scholar]
- 44.Glass R I, Huq M I, Lee J V, Threlfall E J, Khan M R, Alim A R M A, Rowe B, Gross R J. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1, biotype El Tor: evidence of a point-source outbreak in Bangladesh. J Infect Dis. 1983;147:204–209. doi: 10.1093/infdis/147.2.204. [DOI] [PubMed] [Google Scholar]
- 45.Glass R I, Becker S, Huq M I, Stoll B J, Khan M U, Merson M H, Lee J V, Black R E. Endemic cholera in rural Bangladesh, 1966–1980. Am J Epidemiol. 1982;116:959–970. doi: 10.1093/oxfordjournals.aje.a113498. [DOI] [PubMed] [Google Scholar]
- 46.Goldberg M B, Boyko S A, Calderwood S B. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli Fur system. J Bacteriol. 1990;172:6863–6870. doi: 10.1128/jb.172.12.6863-6870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg M B, DiRita V J, Calderwood S B. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg M B, Boyko S A, Calderwood S B. Transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodgame R W, Greenough W B., III Cholera in Africa: a message for the West. Ann Intern Med. 1975;82:101–106. doi: 10.7326/0003-4819-82-1-101. [DOI] [PubMed] [Google Scholar]
- 50.Harkey C W, Everiss K D, Peterson K M. The Vibrio cholerae toxin-coregulated pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect Immun. 1994;62:2669–2678. doi: 10.1128/iai.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hase C C, Mekalanos J J. TcpP is a positive regulator of virulence factor gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and ToxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgins D E, DiRita V J. Genetic analysis of the interaction between Vibrio cholerae transcription activator ToxR and ToxT promoter DNA. J Bacteriol. 1996;178:1080–1087. doi: 10.1128/jb.178.4.1080-1087.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmes R K, Vasil M L, Finkelstein R A. Studies on toxino-genesis in Vibrio cholerae. III. Characterization of nontoxigenic mutants in vitro and in experimental animals. J Clin Invest. 1975;55:551–556. doi: 10.1172/JCI107962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howard B D. A prototype live oral cholera vaccine. Nature. 1971;230:97–99. doi: 10.1038/230097a0. [DOI] [PubMed] [Google Scholar]
- 56.Hughes K J, Everiss K D, Harkey C W, Peterson K M. Identification of a Vibrio cholerae ToxR-activated gene (tagD) that is physically linked to the toxin coregulated pilus (tcp) gene cluster. Gene. 1994;148:97–100. doi: 10.1016/0378-1119(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 57.Islam M S, Drasar B S, Sack R B. The aquatic environment as reservoir of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;11:197–206. [PubMed] [Google Scholar]
- 58.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 59.Janda J M, Powers C, Bryant R G, Abbott S L. Current perspective on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jesudason M V, Saaya R. Resistance of Vibrio cholerae O1 to nalidixic acid. Indian J Med Res. 1997;105:153–154. [PubMed] [Google Scholar]
- 61.Jonson G, Holmgren J, Svennerholm A M. Identification of a mannose-binding pilus on V. cholerae El Tor. Microb Pathog. 1991;11:433–441. doi: 10.1016/0882-4010(91)90039-d. [DOI] [PubMed] [Google Scholar]
- 62.Jonson G, Lebens M, Holmgren J. Cloning and sequencing of Vibrio cholerae mannose-sensitive hemagglutinin pilin gene: localization of mshA within a cluster of type 4 pilin genes. Mol Microbiol. 1994;13:109–108. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 63.Jonson G, Svennerholm A-M, Holmgren J. Expression of virulence factors by classical and El Tor Vibrio cholerae in vivo and in vitro. FEMS Microbiol Ecol. 1990;74:221–228. [Google Scholar]
- 64.Jonson G, Svennerholm A-M, Holmgren J. Vibrio cholerae expresses cell surface antigens during intestinal infection which are not expressed during in vitro culture. Infect Immun. 1989;57:1809–1815. doi: 10.1128/iai.57.6.1809-1815.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamal A M. The seventh pandemic of cholera. In: Barua D, Burrows W, editors. Cholera. Philadelphia, Pa: The W. B. Saunders Co.; 1974. pp. 1–14. [Google Scholar]
- 66.Kaper J B, Bradford H B, Roberts N C, Falkow S. Molecular epidemiology of Vibrio cholerae in the U.S. Gulf Coast. J Clin Microbiol. 1982;16:129–134. doi: 10.1128/jcm.16.1.129-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaper J B, Morris J G, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaper J B, Moseley S L, Falkow S. Molecular characterization of environmental and nontoxigenic strains of V. cholerae. Infect Immun. 1981;32:661–667. doi: 10.1128/iai.32.2.661-667.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan M U, Samadi A R, Huq M I, Greenough W B. Reappearance of classical Vibrio cholerae in Bangladesh. In: Kuwahara S, Pierce N F, editors. Advances in research on cholera and related diarrheas. Vol. 3. Tokyo, Japan: KTK Scientific Publishers; 1986. pp. 3–12. [Google Scholar]
- 71.Knop J, Rowley D. Protection against cholera. A bactericidal mechanism on the mucosal surface of the small intestine of mice. Aust J Exp Biol. 1975;53:155–165. [PubMed] [Google Scholar]
- 72.Koch R. An address on cholera and its bacillus. Br Med J. 1884;1884:403–407. doi: 10.1136/bmj.2.1235.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 74.Kovach M E, Hughes K J, Everiss K D, Peterson K M. Identification of a ToxR-activated gene, tagE, that lies within the accessory colonization factor gene cluster of Vibrio cholerae O395. Gene. 1994;148:91–95. doi: 10.1016/0378-1119(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 75.Laval E. El colera en Chile (1886–1888) Rev Chil Infect. 1989;6:96–99. [Google Scholar]
- 76.Lazar S, Waldor M K. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect Immun. 1998;66:394–397. doi: 10.1128/iai.66.1.394-397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levine M M. South America: the return of cholera. Lancet. 1991;338:45–46. [Google Scholar]
- 78.Martin A R, Mosley W H, Sau B B, Ahmed S, Huq I. Epidemiologic analysis of endemic cholera in urban East Pakistan, 1964–1966. Am J Epidemiol. 1969;89:572–582. doi: 10.1093/oxfordjournals.aje.a120970. [DOI] [PubMed] [Google Scholar]
- 79.Materu S F, Lema O E, Mukunza H M, Adhiambo C G, Carter J Y. Antibiotic resistance pattern of Vibrio cholerae and Shigella causing diarrhoea outbreaks in the eastern Africa region: 1994–1996. East Afr Med J. 1997;74:193–197. [PubMed] [Google Scholar]
- 80.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 81.McCormack W M, Mosley W H, Fahimuddin M, Benenson A S. Endemic cholera in rural East Pakistan. Am J Epidemiol. 1969;89:393–404. doi: 10.1093/oxfordjournals.aje.a120953. [DOI] [PubMed] [Google Scholar]
- 82.Mekalanos J J. Cholera toxin: genetic analysis, regulation and role in pathogenesis. Curr Top Microbiol Immunol. 1985;118:97–118. doi: 10.1007/978-3-642-70586-1_6. [DOI] [PubMed] [Google Scholar]
- 83.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 84.Mekalanos J J, Collier R J, Roming W R. Affinity filters, a new approach to the isolation of tox mutants of Vibrio cholerae. Proc Natl Acad Sci USA. 1978;75:941–945. doi: 10.1073/pnas.75.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Michalski J, Galen J E, Fasano A, Kaper J B. V. cholerae CVD110: a live oral attenuated Eltor vaccine strain. Infect Immun. 1993;61:4462–4468. doi: 10.1128/iai.61.10.4462-4468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller V L, Mekalanos J J. A novel suicide vector and is use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 89.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 90.Morris J G, Jr, Black R E. Cholera and other vibrios in the United States. N Eng J Med. 1985;312:343–350. doi: 10.1056/NEJM198502073120604. [DOI] [PubMed] [Google Scholar]
- 91.Mukhopadhyay A K, Basu A, Garg P, Bag P K, Ghosh A, Bhattacharya S K, Takeda Y, Nair G B. Molecular epidemiology of reemergent Vibrio cholerae O139 Bengal in India. J Clin Microbiol. 1998;36:2149–2152. doi: 10.1128/jcm.36.7.2149-2152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakasone N, Iwanaga M, Eeckels R. Characterization of Vibrio cholerae O1 recently isolated in Bangladesh. Trans R Soc Trop Med Hyg. 1987;81:876–878. doi: 10.1016/0035-9203(87)90059-9. [DOI] [PubMed] [Google Scholar]
- 93.Norris H T. Cholera. In: Barua D, Burrows W, editors. The pathology of cholera. Philadelphia, Pa: The W. B. Saunders Co.; 1974. pp. 160–188. [Google Scholar]
- 94.Novitsky J S, Morita R Y. Morphological characterization of small cells resulting from nutrient starvation of a psychrophylic marine vibrio. Appl Environ Microbiol. 1976;32:617–622. doi: 10.1128/aem.32.4.617-622.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Novitsky J S, Morita R Y. Survival of a psychrophilic marine vibrio under long-term nutrient starvation. Appl Environ Microbiol. 1977;33:635–641. doi: 10.1128/aem.33.3.635-641.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novitsky J S, Morita R Y. Possible strategy for the survival of marine bacteria under starvation conditions. Mar Biol. 1978;48:289–295. [Google Scholar]
- 97.Ogawa A, Takeda T. The gene encoding the heat-stable enterotoxin of Vibrio cholerae is flanked by 123-base pair direct repeats. Microbiol Immunol. 1993;37:607–616. doi: 10.1111/j.1348-0421.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 98.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 99.Osek J, Svennerholm A M, Holmgren J. Protection against Vibrio cholerae El Tor infection by specific antibodies against mannose-binding hemagglutinin pili. Infect Immun. 1992;60:4961–4964. doi: 10.1128/iai.60.11.4961-4964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Osek J, Jonson G, Svennerholm A M, Holmgren J. Role of antibodies against biotype-specific Vibrio cholerae pili in protection against experimental classical and El Tor cholera. Infect Immun. 1994;62:2901–2907. doi: 10.1128/iai.62.7.2901-2907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pan American Health Organization. Cholera situation in the Americas. Epidemiol Bull. 1991;12:1–4. [PubMed] [Google Scholar]
- 102.Parsot C, Taxman E, Mekalanos J J. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1641–1645. doi: 10.1073/pnas.88.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parsot C, Mekalanos J J. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modified by the heat shock response. Proc Natl Acad Sci USA. 1990;87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pollitzer R. History of the disease. In: Pollitzer R, editor. Cholera. Geneva, Switzerland: World Health Organization; 1959. pp. 11–50. [Google Scholar]
- 107.Public Health Laboratory Service. Vibrio cholerae O139 and epidemic cholera. Commun Dis Rep Weekly. 1993;3:173. [PubMed] [Google Scholar]
- 108.Rabbani G H, Greenough W B. Cholera. In: Lebenthal E, Duffy M, editors. Textbook of secretory diarrhea. New York, N.Y: Raven Press; 1990. pp. 233–253. [Google Scholar]
- 109.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of a novel strain of Vibrio cholerae with epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 110.Ramamurthy T, Albert M J, Huq A, Colwell R R, Takeda Y, Takeda T, Shimada T, Mandal B K, Nair G B. Vibrio mimicus with multiple toxin types isolated from human and environmental sources. J Med Microbiol. 1994;40:194–196. doi: 10.1099/00222615-40-3-194. [DOI] [PubMed] [Google Scholar]
- 111.Recchia G D, Hall M R. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 112.Recchia G D, Hall M R. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 113.Ries A A, Vugia D J, Beingolea L, Palacios A M, Vasquez E, Wells J G, Baca N G, Swerdlow D L, Pollack M, Bean N H, Seminario L, Tauxe R V. Cholera in Piura, Peru: a modern urban epidemic. J Infect Dis. 1992;166:1429–1433. doi: 10.1093/infdis/166.6.1429. [DOI] [PubMed] [Google Scholar]
- 114.Sakajaki R. Classification and characteristics of vibrios. Public health papers no. 40. Geneva, Switzerland: World Health Organization; 1970. [PubMed] [Google Scholar]
- 115.Samadi A R, Huq M I, Shahid N, Khan M U, Eusof A, Rahman A S M M, Yunus M M, Faruque A S G. Classical Vibrio cholerae biotype displaces El Tor in Bangladesh. Lancet. 1983;i:805–807. doi: 10.1016/s0140-6736(83)91860-3. [DOI] [PubMed] [Google Scholar]
- 116.Samadi A R, Chowdhury M K, Huq M I, Khan M U. Seasonality of classical and El Tor cholera in Dhaka, Bangladesh: 17 year trends. Trans R Soc Trop Med Hyg. 1983;77:853–856. doi: 10.1016/0035-9203(83)90306-1. [DOI] [PubMed] [Google Scholar]
- 117.Satcher D. Emerging infections: getting ahead of the cure. Emerg Infect Dis. 1995;1:1–6. doi: 10.3201/eid0101.950101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sciortino C V, Finkelstein R A. Vibrio cholerae expresses iron-regulated outer membrane proteins in vivo. Infect Immun. 1983;42:990–996. doi: 10.1128/iai.42.3.990-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sciortino C V, Johnson J A, Hamad A. Vitek system antimicrobial susceptibility testing of O1, O139, and non-O1 Vibrio cholerae. J Clin Microbiol. 1996;34:897–900. doi: 10.1128/jcm.34.4.897-900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sengupta D K, Sengupta T K, Ghose A C. Major outer membrane proteins of Vibrio cholerae and their role in induction of protective immunity through inhibition of intestinal colonization. Infect Immun. 1992;60:4848–4855. doi: 10.1128/iai.60.11.4848-4855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 122.Shimada T, Sakazaki R. Additional serovars and inter-O antigenic relationships of V. cholerae. Jpn J Med Sci Biol. 1977;30:275–277. doi: 10.7883/yoken1952.30.275. [DOI] [PubMed] [Google Scholar]
- 123.Shimada T, Nair G B, Deb B C, Albert M J, Sack R B, Takeda Y. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet. 1993;341:1347. doi: 10.1016/0140-6736(93)90855-b. [DOI] [PubMed] [Google Scholar]
- 124.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakajato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 125.Shousha A T. Cholera epidemic in Egypt (1947). A preliminary report. Bull W H O. 1947;1:353–381. [PMC free article] [PubMed] [Google Scholar]
- 126.Siddique A K, Salam A, Islam M S, Akram K, Majumdar R N, Zaman K, Fronczak N, Laston S. Why treatment centres failed to prevent cholera deaths among Rwandan refugees in Goma, Zaire. Lancet. 1995;345:359–361. doi: 10.1016/s0140-6736(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 127.Siddique A K, Akram K, Zaman K, Mutsuddy P, Eusof A, Sack R B. Vibrio cholerae O139: how great is the threat of a pandemic? Trop Med Int Health. 1996;1:393–398. doi: 10.1046/j.1365-3156.1996.d01-54.x. [DOI] [PubMed] [Google Scholar]
- 128.Siddique A K, Baqui A H, Eusof A, Haider K, Hossain M A, Bashir I, Zaman K. Survival of classical cholera in Bangladesh. Lancet. 1991;337:1125–1127. doi: 10.1016/0140-6736(91)92789-5. [DOI] [PubMed] [Google Scholar]
- 129.Siddique A K, Zaman K, Baqui A H, Akram K, Mutsuddy P, Eusof A, Haider K, Islam S, Sack R B. Cholera epidemics in Bangladesh: 1985–1991. J Diarrhoeal Dis Res. 1992;10:79–86. [PubMed] [Google Scholar]
- 130.Siddique A K, Zaman K, Mazumder Y. Simultaneous outbreaks of contrasting drug resistant classic and El Tor Vibrio cholerae O1 in Bangladesh. Lancet. 1989;i:396. doi: 10.1016/s0140-6736(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 131.Sigel S P, Payne S M. Effect of iron limitation on growth, siderophore production, and expression of outer membrane proteins of Vibrio cholerae. J Bacteriol. 1982;150:148–155. doi: 10.1128/jb.150.1.148-155.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 133.Snow J. On the mode of communication of cholera. 2nd ed. London, England: J. Churchill; 1855. [Google Scholar]
- 134.Stoll B J, Glass R I, Huq M I, Khan M U, Holt J E, Banu H. Surveillance of patients attending a diarrhoeal disease hospital in Bangladesh. Br Med J. 1982;285:1185–1188. doi: 10.1136/bmj.285.6349.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Swerdlow D L, Ries A A. Vibrio cholerae non-O1—the eighth pandemic? Lancet. 1993;342:382–383. doi: 10.1016/0140-6736(93)92806-5. [DOI] [PubMed] [Google Scholar]
- 136.Tacket C O, Losonsky G, Nataro J P, Cryz S J, Edelman R, Kaper J B, Levine M M. Safety, immunogenicity, and transmissibility of live oral cholera vaccine candidate CVD 110, a ΔctxA Δzot Δace derivative of El Tor Ogawa Vibrio cholerae. J Infect Dis. 1993;168:1536–1540. doi: 10.1093/infdis/168.6.1536. [DOI] [PubMed] [Google Scholar]
- 137.Taylor R, Shaw C, Peterson K, Spears P, Mekalanos J. Safe, live Vibrio cholerae vaccines? Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 138.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Trucksis M, Galen J E, Michalski J, Fasano A, Kaper J B. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc Natl Acad Sci USA. 1993;90:5267–5271. doi: 10.1073/pnas.90.11.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Dongen W M A M, DeGraaf F K. Molecular cloning of a gene coding for a Vibrio cholerae hemagglutinin. J Gen Microbiol. 1986;132:2225–2234. doi: 10.1099/00221287-132-8-2225. [DOI] [PubMed] [Google Scholar]
- 141.Wachsmuth I K, Bopp C A, Fields P I, Carrilo C. Difference between toxigenic Vibrio cholerae O1 from South America and US Gulf Coast. Lancet. 1991;337:1097–1098. doi: 10.1016/0140-6736(91)91744-f. [DOI] [PubMed] [Google Scholar]
- 142.Wachsmuth I K, Evins G M, Fields P I, Olsvic O, Popovic T, Bopp C A, Wells J G, Carrillo C, Blake P A. The molecular epidemiology of cholera in Latin America. J Infect Dis. 1993;167:621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 143.Wachsmuth I K, Olsvik O, Evins G M, Popovic T. Molecular epidemiology of cholera. In: Wachsmuth K, Blake P A, Olsvik O, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington, D.C: American Society for Microbiology; 1994. pp. 357–370. [Google Scholar]
- 144.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 145.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXΦ are encoded by regions RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 146.Waldor M K, Tschape H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.West P A, Colwell R R. Identification and classification of Vibrionaceae—an overview. In: Colwell R R, editor. Vibrios in the environment. New York, N.Y: John Wiley & Sons, Inc.; 1984. pp. 285–363. [Google Scholar]
- 148.West P A. The human pathogenic vibrios—a public health update with environmental perspectives. Epidemiol Infect. 1989;103:1–34. doi: 10.1017/s0950268800030326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.WHO Scientific Working Group. Cholera and other Vibrio-associated diarrhoeas. Bull W H O. 1980;58:353–374. [PMC free article] [PubMed] [Google Scholar]
- 150.World Health Organization. Meeting on The Potential Role of New Cholera Vaccines in the Prevention and Control of Cholera Outbreaks during Acute Emergencies. Document CDR/GPV/95.1. Geneva, Switzerland: World Health Organization; 1995. [Google Scholar]