Abstract
During the first 2 years of life, the infant gut microbiome is rapidly developing, and gut bacteria may impact host health through the production of metabolites that can have systemic effects. Thus, the fecal metabolome represents a functional readout of gut bacteria. Despite the important role that fecal metabolites may play in infant health, the development of the infant fecal metabolome has not yet been thoroughly characterized using frequent, repeated sampling during the first 2 years of life. Here, we described the development of the fecal metabolome in a cohort of 101 Latino infants with data collected at 1-, 6-, 12-, 18-, and 24-months of age. We showed that the fecal metabolome is highly conserved across time and highly personalized, with metabolic profiles being largely driven by intra-individual variability. Finally, we also identified several novel metabolites and metabolic pathways that changed significantly with infant age, such as valerobetaine and amino acid metabolism, among others.
Subject terms: Metabolism, Metabolomics
Introduction
For infants, the first 1000 days represent a critical developmental window, during which metabolic, endocrine, neural, and immune systems are rapidly maturing. In tandem, the infant gut microbiome is also rapidly developing, until 2–3 years of age, when it reaches a more adult-like configuration1. This early colonization of the infant gut has far-reaching health implications, affecting immune system development2, asthma and allergy3–5, obesity and rapid growth6–9, and cognitive development10. While the exact mechanisms by which the gut microbiome impacts human health are still being characterized, gut bacteria can impact host physiological systems through the production of metabolites that can have systemic health effects11–13. High resolution metabolomics is an emerging, analytical, omics-based technology which offers global detection and characterization of the human metabolome, affording insights into how the metabolome interacts with both exogenous and endogenous exposures, and downstream health implications.
To date, several studies have examined the impact of mode of delivery, antibiotic usage, and early life feeding practices on the infant fecal metabolome. For example, 6-week old infants born vaginally compared to caesarian section (CS) were shown to have enriched metabolic pathways related to carbohydrate metabolism including glycolysis/gluconeogenesis and glyoxylate and dicarboxylate metabolism14. Amino acid metabolism has also been shown to be pronounced in infants exposed to antibiotics during the first weeks of life16,17 as well as among formula-fed infants, compared to breastfed infants14. Further, breastfed infants have different metabolic profiles compared to their formula-fed counterparts at 3-, 6-, and 9-months of age, including differences in butyric acid, d-sphingosine, betaine, and kynurenic acid18. However, few studies have characterized the fecal metabolome using frequent repeated sampling during the first 2 years of life. This information is critical to understanding immune system development as infants increase food diversity and gain mobility.
While prior research offers important insight into the development of the fecal metabolome, previous longitudinal studies followed infants only through the first year of life15–20, and none have been conducted in a Latino cohort. The aim of this study was to contribute to the existing literature by systematically describing the development of the infant fecal metabolome during the first 2 years of life. Specifically, we examined the fecal metabolome at 1-, 6-, 12-, 18-, and 24-months of age in 101 Latino infants from the Southern California Mother’s Milk Study – a well-established cohort that was recruited based on an intention to breastfeed for at least three months. As a secondary aim, we sought to assess which factors were important predictors of overall fecal metabolites and fecal metabolomic pathways, including mode of delivery, antibiotic exposure, and infant age. Future work in this cohort will expand on this initial analysis to examine how breastfeeding, formula feeding, solid food introduction, and environmental exposures impact fecal metabolome throughout the first 2 years of life.
Results
Study population characteristics
General population characteristics are shown in Table 1. At the 1-month postpartum visit, mothers were 29 ± 6 years old (18–45), and most had a body mass index (BMI) in the overweight (33%) or obese (40%) category. The average infant age at fecal metabolome assessment was 1.1, 6.2, 12.3, 18.3, and 25.1 months, respectively, at planned 1-, 6-, 12-, 18-, and 24-month collection21. Half of the infants were female, 74% were born vaginally, and 10.9% received antibiotics in the first 2 years of life. Most (97%) were breastfed at 1-month, and 41% were breastfed at 24-months of age.
Table 1.
Characteristics of 101 mother-infant dyads from the Southern California Mother’s Milk Study, 2016–2019. Data reported are mean and standard deviation (SD) unless otherwise noted.
| Mean ± SD or N, % N = 101 |
|
|---|---|
| Maternal characteristics | |
| Age (years) at 1-month postpartum visit | 29 ± 6 |
| Socioeconomic status (SES)a | 27 ± 12 |
| Pre-pregnancy BMI (kg/m2) | 28.7 ± 6.0 |
| Infant characteristics | |
| Age (days) at 1-month postpartum visit | 32.8 ± 3.1 |
| Sex (Female, Male, %Female) | 51, 50, 50.5% |
| Age of solid foods (months)b | 5.9 ± 1.7 |
| Antibiotics (Yes, No, %Yes) | 11, 90, 10.9% |
| Birth mode (Vaginal, C-section, %Vaginal) | 75, 26, 74.3% |
| Birth weight (kg) | 3.4 ± 0.4 |
| Birth length (cm) | 50.4 ± 2.5 |
| Gestational age | |
| Early (< 38 weeks gestation) | 26 (25.7%) |
| On time (38–42 weeks gestation) | 54 (53.5%) |
| Late (> 42 weeks gestation) | 21 (20.8%) |
a Median replacement was performed for missing values (N = 2).
b N = 100.
SES, socioeconomic status; BMI, body mass index.
Temporal trends in fecal metabolites
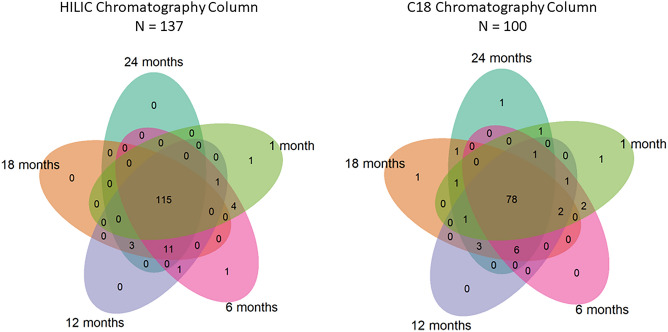
Overall, there were 11,345 metabolic features extracted from the HILIC and 8,609 in the C18 chromatography after removal of features that were present in less than 10% of samples. From this, we confirmed the chemical identities of 143 unique metabolites from HILIC chromatography and 104 metabolites from C18 chromatography with Level 1 evidence (i.e., features whose m/z and retention time could be matched to authentic standards with MS/MS under identical conditions). Among the confirmed metabolites, many were highly conserved over time. For instance, 115/143 metabolites in the HILIC column were observed in at least 50% of samples at each timepoint, as were 78/104 metabolites in the C18 column (Fig. 1). As a sensitivity analysis, we additionally explored metabolites that were present in 25% and 75% of samples (Supplemental Fig. 1 and Supplemental Fig. 2). We found that there were 135 and 97 confirmed metabolites that were present in at least 25% of samples at each timepoint in the HILIC and C18 column, respectively. There were 72 and 50 metabolites present in 75% or more of samples at every timepoint in the HILIC and C18 columns, respectively.
Figure 1.
Confirmed metabolites observed longitudinally in at least 50% of samples during the first 2 years of life in the HILIC (left) and C18 (right) chromatography columns.
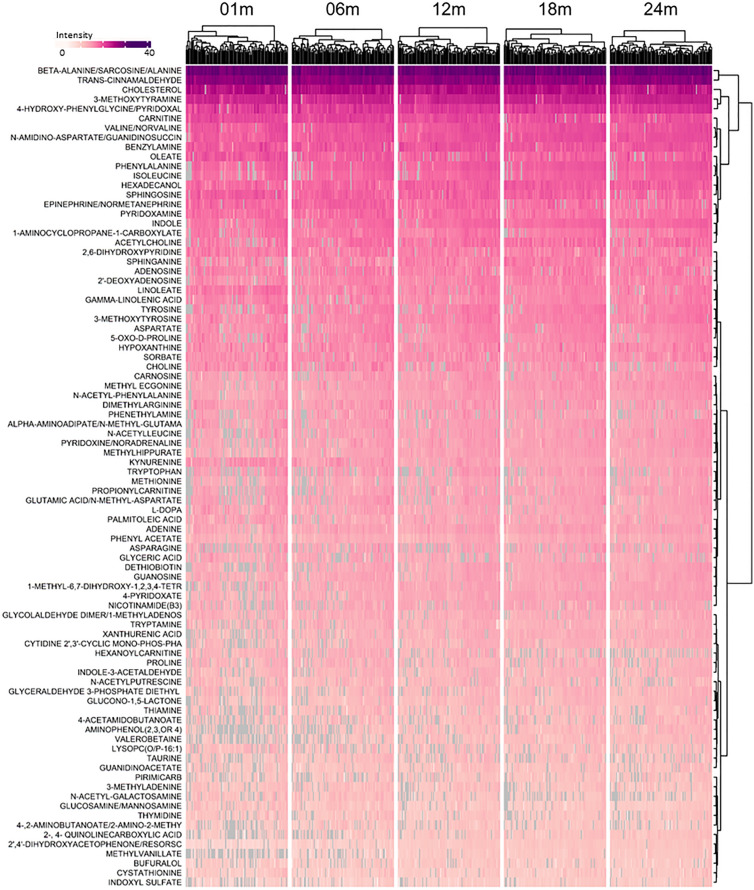
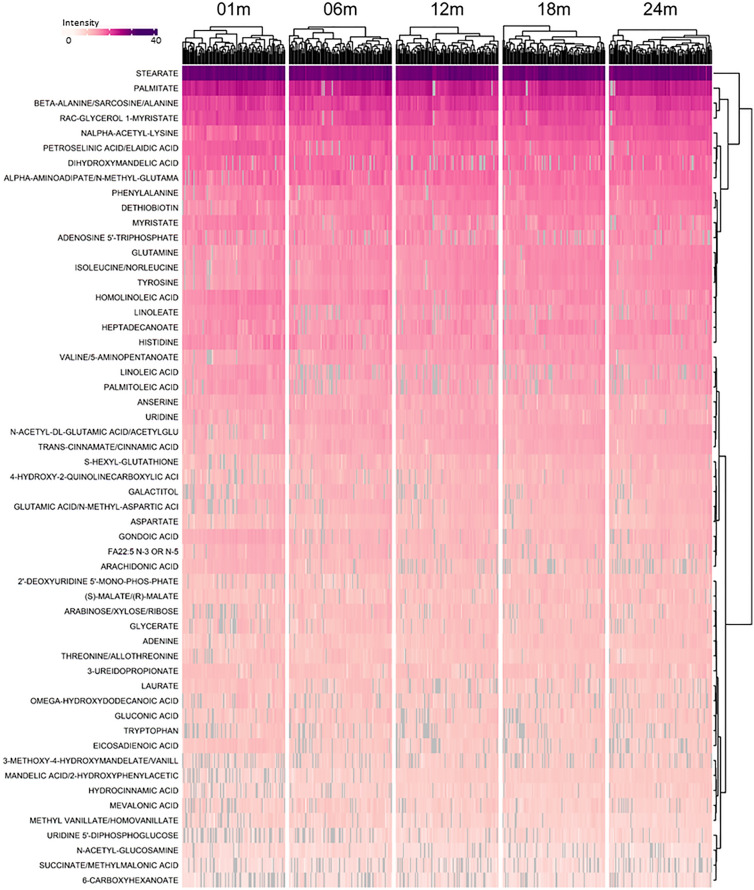
Observed metabolites included (Figs. 2 and 3) amino acids (e.g., tyrosine, tryptophan, methionine, isoleucine, aspartate, proline), amino acid metabolites (e.g., phenylacetate, oxoproline, taurine, cystathionine), purines (adenine, guanine, hypoxanthine), pyrimidine (thymine) and vitamin metabolites [thiamine (B1), nicotinamide (B3), pyridoxine (B6), pyridoxate, alpha-tocopherol (E), dethiobiotin). Lipid metabolites included sphingosine, sphinganine, choline, carnitine, cholesterol, free fatty acids (e.g., linoleate, oleate) and lysophosphatidyl choline. Microbial metabolites with known adverse activities (indoxyl sulphate, valerobetaine) were present, and dietary (cinnamaldehyde) and environmental chemicals (pirimicarb) were also detected.
Figure 2.
Longitudinal patterns of metabolite intensity of confirmed fecal metabolites detected in the HILIC chromatography column during the first 2 years of life. Intensity presented is standardized by dividing by intensity standard deviation, and ranges from 3.1 to 32.4, with dark purple representing the highest intensity and white representing the lowest intensity. Grey indicates a missing value. Metabolites detected in at least 80% of samples were included. Observations are grouped by visit, with the age in days corresponding with each visit as follows: 01 m (25–46 days), 06 m (164–219 days), 12 m (351–429 days), 18 m (517–582 days), and 24 m (709–916 days).
Figure 3.
Longitudinal patterns of metabolite intensity of confirmed fecal metabolites detected in the C18 chromatography column during the first 2 years of life. Intensity presented is standardized by dividing by intensity standard deviation, and ranges from 4.7 to 37.6, with dark purple representing the highest intensity and white representing the lowest intensity. Grey indicates a missing value. Metabolites detected in at least 80% of samples were included. Observations are grouped by visit, with the age in days corresponding with each visit as follows: 01 m (25–46 days), 06 m (164–219 days), 12 m (351–429 days), 18 m (517–582 days), and 24 m (709–916 days).
Changes in fecal metabolites were largely driven by age and intra-infant variability
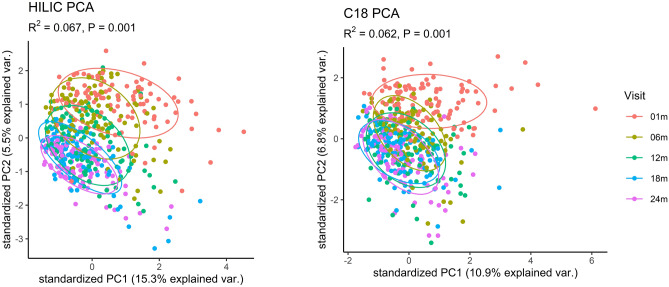
Given that the intensities of many confirmed metabolites changed during the first 24-months of life (Fig. 2 and 3), we performed non-parametric univariate permutational multivariate analysis of variance tests (PERMANOVA) to assess which variables explained the most variance in overall metabolite composition. We found that metabolic variation was largely driven by intra-individual variability such that 29.2% (P = 0.001) of the variability in confirmed metabolites in the HILIC column, and 30.0% (P = 0.001) of the variability in confirmed metabolites in the C18 column could be attributed to the infant providing the sample.
In addition to individual variability, we also examined the impact of infant sex, antibiotic exposure (i.e., having received any antibiotics since birth), and infant age in days on overall profiles of fecal metabolites. Infant sex and antibiotic exposure were not important predictors of metabolites in the HILIC column, with sex explaining just 0.3% of variability (P = 0.1) and antibiotic exposure explaining 0.4% of variability (P = 0.02). Similarly, infant sex explained 0.2% of variability in the metabolites in the C18 column (P = 0.3) and antibiotic exposure explained 0.3% of variability (P = 0.1). Despite this, infant age explained much more of the variability in fecal metabolites where 6.7% (P = 0.001) and 6.2% (P = 0.001) of the variability in confirmed metabolites in the HILIC and C18 columns were explained by infant age (Fig. 4). Indeed, after adjustment for multiple testing using the Benjamini-Hochberg (BH) procedure, we found there were 87 confirmed metabolites in the HILIC chromatography column and 77 in the C18 chromatography column which were significantly associated with infant age in days (PBH < 0.05). Table 2 summarizes the top 10 most significant results for each chromatography column. These showed increases in some diet-derived metabolites (methyl vanillate, hydrocinnamic acid) and decreases in free fatty acids (arachidonate, laurate, homolinoleic acid). A complete list of statistically significant results can be found in Supplemental Table 2.
Figure 4.
Ordination plot of principal components 1 and 2 for confirmed metabolites detected in HILIC (left) and C18 (right) by infant age in days. R2- and P- values calculated using Permutational Multivariate Analysis of Variance, with age in days as the explanatory variable. Points are colored by visit, with the age in days corresponding with each visit as follows: 01 m (25–46 days), 06 m (164–219 days), 12 m (351–429 days), 18 m (517–582 days), and 24 m (709–916 days).
Table 2.
Top ten confirmed metabolites detected by HILIC and C18 chromatography columns that were most significantly associated with infant age, based on the results of linear mixed effects models with random intercepts for individual to account for repeated measures. Results from these models were adjusted using the Benjamini–Hochberg (BH) procedure. Overall, there were 87 metabolites significantly associated with infant age in days after adjustment for multiple testing in the HILIC chromatography column and 77 in the C18 chromatography column.
| HILIC chromatography column | C18 chromatography column | ||||
|---|---|---|---|---|---|
| Metabolite | Direction | PBH | Metabolite | Direction | PBH |
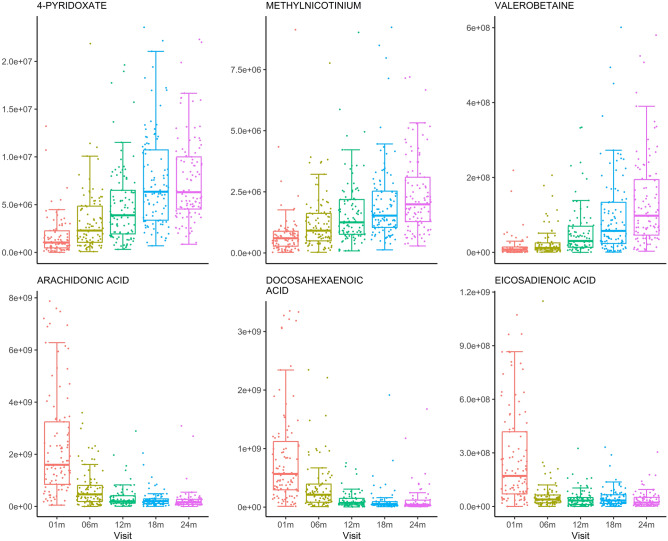
| 4-Pyridoxate | ↑ | 1.2 × 10–47 | Arachidonic Acid | ↓ | 2.3 × 10–34 |
|
Dihydroxyacetophenone/ Resorscinol monoacetate/ Methylparaben |
↑ | 5.7 × 10–55 | Docosahexaenoic acid | ↓ | 1.8 × 10–34 |
| Hexadecanol | ↑ | 4.2 × 10–18 | Eicosadienoic acid | ↓ | 3.5 × 10–20 |
| Kynurenine | ↓ | 1.3 × 10–26 | Gondoic acid | ↓ | 3.1 × 10–17 |
| Methylnicotinium* | ↑ | 8.3 × 10–27 | Homolinoleic acid | ↓ | 2.3 × 10–18 |
| Methylhippurate | ↑ | 1.0 × 10–17 | Hydrocinnamic acid | ↑ | 1.3 × 10–18 |
| Methyl vanillate | ↑ | 2.5 × 10–21 | Hypoxanthine | ↓ | 1.0 × 10–15 |
| N-Acetyl-phenylalanine | ↑ | 3.3 × 10–19 | Laurate | ↓ | 1.6 × 10–16 |
| Valerobetaine | ↑ | 2.7 × 10–35 | Mandelic acid/2-,3-Hydroxyphenylacetic acid | ↑ | 3.0 × 10–41 |
| Xanthurenic acid | ↑ | 3.6 × 10–22 | N-alpha-acetyl-L-lysine | ↑ | 3.0 × 10–27 |
*This compound was unidentified but is an accurate mass match for methylnicotinium.
As shown in Fig. 5, we examined selected metabolites that were most significantly associated with infant age in days in the HILIC and C18 chromatography columns. Mean intensity of valerobetaine, a microbiome-derived metabolite associated with increased adiposity22, increased by 588% between 1- and 24-months of age. Other confirmed metabolites in the HILIC column that decreased with infant age included kynurenine, a metabolite associated with immune cell function, whose mean intensity decreased by 79.1%. In the C18 column, mean intensity of the polyunsaturated fatty acids arachidonic acid and eicosadienoic acid decreased with infant age (an 88.5% and 85.5% decrease, respectively). In addition to examining the level of intensity over time, we also visualized the prevalence of each metabolite that was most significantly associated with infant age (Supplemental Fig. 3). While most of these metabolites were detected with high prevalence across all timepoints, valerobetaine prevalence increased dramatically over time and prevalence of arachidonic and docosahexaenoic acid decreased precipitously between 1- and 24-months.
Figure 5.
Visual representation of the intensity of selected confirmed metabolites detected in the HILIC (top) and C18 (bottom) chromatography columns which varied most significantly with infant age. Observations are grouped by visit, with the age in days corresponding with each visit as follows: 01 m (25–46), 06 m (164–219), 12 m (351–429), 18 m (517–582), and 24 m (709–916). The metabolite labeled methylnicotinium was unidentified but was an accurate mass match for methylnicotinium.
Amino acid, bile acid, and carbohydrate metabolism pathways were enriched with infant age
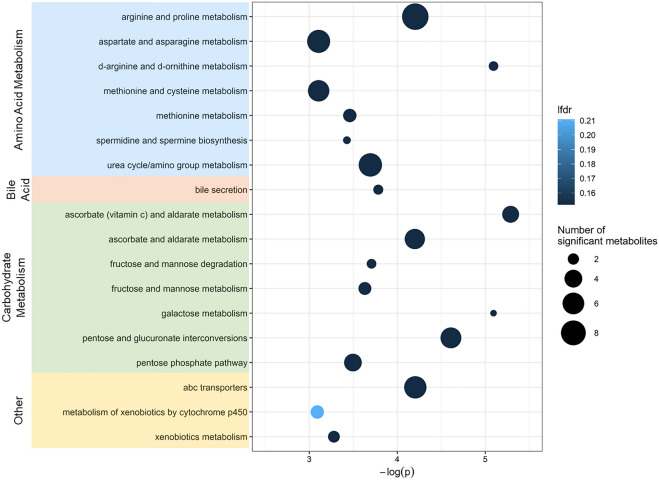
Results from the linear mixed effects models were used to perform pathway enrichment analyses using Metapone23. There were 18 metabolic pathways significantly associated with infant age (P ≤ 0.05) and with weighted number of significant metabolites ≥ 1.5. Pathways associated with infant age are summarized in Fig. 6. Of the pathways associated with infant age, 7 pathways were related to amino acid metabolism, including both non-essential and essential amino acid pathways and the urea cycle. These pathways are linked to many of the metabolites in Table 2 (e.g., pyridoxine is the vitamin supporting amino acid nitrogen elimination through the urea cycle). Two pathways containing methionine are connected to individual metabolites described above (e.g., taurine), and to the bile acid pathway and the spermidine/spermine pathway. Seven pathways were related to carbohydrate metabolism, and one was related to bile acid metabolism. Other significant pathways included metabolism of xenobiotics and membrane transport (ATP-binding cassette transporters).
Figure 6.
Overview of enriched pathways by confirmed HILIC and C18 chromatography column metabolites that were significantly associated with infant age, identified using Metapone. Pathways with P ≥ 0.05 and weighted number of significant metabolites ≥ 1.5 are included. Point size indicates the number of significant metabolites identified within each pathway and point color indicates significance after adjustment for multiple testing via conditional local false discovery rate (lfdr). Pathways are grouped by super pathway (amino acid metabolism, bile acid, carbohydrate metabolism, lipid, and other), indicated by color.
Discussion
This study provides important new information concerning the longitudinal development of the fecal metabolome in Latino infants from 1 to 24 months, a time period with far-reaching health implications for immune system development2, asthma and allergy3–5, obesity and growth6–9, and cognitive development10. At least 50% prevalence at each timepoint was observed for about 200 confirmed metabolites, indicating that the fecal metabolome was relatively stable. Variability was primarily explained by the individual providing the sample as well as infant age. Metabolites associated with infant age included amino acid metabolites, such as kynurenine, and polyunsaturated fatty acids, arachidonic and docosahexaenoic acid. Of potential importance relative to weight gain and obesity, valerobetaine increased substantially over the first 24 months. Valerobetaine is a metabolite produced by the intestinal microbiome which was recently found to inhibit fatty acid oxidation by mitochondria22. In mice, valerobetaine causes increased adiposity and in humans, valerobetaine is associated with high BMI and fatty liver22. The latter association may be especially relevant to Latino health because adolescent Latino children have increased risk of non-alcoholic fatty liver disease (NAFLD)24,25. Other metabolites whose intensity was significantly associated with infant age were also associated with pathways related to amino acid metabolism, carbohydrate metabolism, bile acid metabolism, and lipid metabolism.
A previous study on preterm infants found that intra-individual variability explained 43% of variation in fecal metabolites during the first months of life; whereas, health outcomes (sepsis and necrotizing enterocolitis), delivery mode, and antibiotics were not significantly associated with metabolomic variation26. Several of the metabolites that we found to be related to infant age have also been identified previously. For example, we found that uracil, aspartate, methionine, tyrosine, phenylalanine, and valine were significantly increased with infant age, while glucose and fucose metabolites were significantly decreased. Interestingly, previous work has found these same metabolites to be associated with either formula or breast feeding14. We also observed that methyl vanillate, which is a compound identified in cow’s milk27 and also produced by Brewer’s yeast (Saccharomyces cerevisiae)28, significantly increased with infant age. Given this, the changes that we observed in metabolite intensity over time may reflect increased formula or cow’s milk consumption, increased consumption of solid foods, and/or decreased breastfeeding or weaning as infants age. Several metabolites that changed significantly over time in our study are found in breast milk and infant formula, including kynurenine, which is present in both breast milk and formula, but is found in higher amounts in formula compared to breast milk29. Arachidonic acid and docosahexaenoic acid (DHA) are both long-chain polyunsaturated fatty acids that are found in breast milk30. Infant formulas are also supplemented with DHA31. These observations further bolster the hypothesis that shifts in the early-life fecal metabolome are driven largely by dietary alterations during the first 2 years of life. Given these observations, future work in this cohort will explore the associations between the fecal metabolome with early life feeding practices, the infant gut microbiota, and adiposity.
In this study, we found that pathways related to amino acid metabolism, carbohydrate metabolism, and bile acid metabolism were associated with increased infant age. Interestingly, several of these pathways have also been linked with infant breast or formula feeding14. For example, we observed that seven amino acid pathways increased with infant age, one of which (arginine and proline metabolism) has been shown to be enriched in the fecal metabolome of formula-fed infants compared to breastfed infants at 6-weeks of age14. We also found seven carbohydrate metabolism pathways enriched with infant age, two of which (galactose metabolism and fructose and mannose metabolism) were enriched in breastfed infants, compared to formula-fed infants14.
Pathways that we identified in the current analysis have also been linked with antibiotic exposure and mode of delivery in other fecal metabolomics studies that investigated infants. For example, amino acid metabolism has previously been shown to be enriched in infants exposed to antibiotics during the first weeks of life16,17. We also found metabolites linked to ABC transporters were enriched with increasing infant age. Previous studies have observed fecal metabolome enrichment of this pathway among infants aged approximately 15-weeks and born cesarean-section, compared to vaginally-born infants15 and among preterm infants with antibiotic exposure during the first 14 days of life, compared to preterm infants who did not receive antibiotics16. Lastly, one carbohydrate metabolism pathway (galactose metabolism), which we found was enriched with infant age, has previously been shown to be enriched in 6-week old infants who were born by cesarean section14.
While this study had several strengths, including repeated sampling and comprehensive metabolic profiling of the infant fecal metabolome in a well-established cohort of infants, some limitations are worth noting. First, stool was collected using OMNIGene GUT kits, which may have decreased the number of identified metabolites33. However, previous studies have found that a much higher proportion of variability in the fecal metabolome is attributable to individual, compared with collection method32. Given the young age of our participants, we were unable to collect blood samples, which limited our ability to examine the circulating metabolome. Therefore, future studies should incorporate fecal metagenomics and serum metabolomics since gut bacteria likely play an important role in shaping the fecal metabolome. Additionally, as this study was focused on metabolites that were identified with Level 1 evidence, we largely characterized patterns in a relatively limited number of endogenous metabolites over the first 2 years of life. This study was also conducted in an exclusively Latino cohort with exclusions such as preterm birth or low birth weight, cigarette smoking or recreational drug use, which may limit the generalizability of these findings to the broader population. In particular, given that our cohort was comprised of Latinos we were unable to assess whether race/ethnicity is an important predictor of the fecal metabolome. Future work is needed to replicate these findings in other populations. Nevertheless, we detected numerous metabolites that were previously observed in other infant populations. Lastly, exploring feeding practices in this cohort is complex because mothers were recruited based on an intention to breastfeed. Furthermore, while weaning may impact the fecal metabolome, we were unable to address weaning in this study due to incomplete information. Therefore, while examining infant feeding was beyond the scope of the current study, future work will be needed to tease apart the relative intake of formula and breast milk over time and characterize how breastfeeding, formula feeding, and solid food introduction impact fecal metabolome throughout the first 2 years of life.
Conclusions
This study characterizes the development of the infant fecal metabolome during the first 2 years of life. Overall, we found evidence that the metabolome was relatively stable across time and highly personalized, with metabolic profiles being largely driven by intra-individual variability. We also identified several novel metabolites, such as valerobetaine, and pathways, including amino acid biosynthesis, which were significantly associated with infant age.
Methods
Study population
The Southern California Mother’s Milk Study is an ongoing, longitudinal cohort of 219 Latino mother-infant pairs who were recruited between 2016 and 2019 from Los Angeles County maternity clinics, which has been previously described21. Participants were eligible to participate if they were ≥ 18 years old at time of delivery; had a healthy, singleton birth; enrolled in the study by 1-month postpartum; and could read at a 5th grade level in either Spanish or English. Potential participants were excluded if they had any diagnoses known to impact mental/physical health, nutritional status, or metabolism; were currently using tobacco or recreational drugs; had infants who were pre-term or low birth weight; or had infants with clinically diagnosed fetal abnormalities. The Institutional Review Boards of the University of Southern California, Children’s Hospital Los Angeles, and the University of Colorado Boulder approved of the study procedures and all research was performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from participants at time of enrollment.
Study design
Participants were enrolled by 1-month postpartum and attended follow-up visits at 6-, 12-, 18-, and 24-months postpartum. 219 mother-infant dyads were initially enrolled in the Mother’s Milk cohort. As previously reported34, the primary aim of the Mother’s Milk Study was to assess the impact of sugars and human milk oligosaccharides on the infant microbiome and obesity. Briefly, socioeconomic status was estimated using a modified Hollingshead index, as previously described21,35. Maternal self-report was used to classify infants’ gestational age as early (< 38 weeks gestation), on time (38–42 weeks gestation), and late (> 42 weeks gestation). Questionnaires were used to determine birth mode, infant antibiotic exposure, and infant feeding practices. Additional funding supported the analysis of 600 fecal metabolomics samples in this cohort and a subset of 127 participants were selected to undergo fecal metabolomics analysis, to maximize the number of participants with repeated fecal metabolome samples. 101 infants had complete fecal metabolomics samples at all 5 visits and were included in this analysis. Those individuals that were excluded due to missing data did not differ significantly from those who were included in the analysis (Supplemental Table 1).
High-resolution metabolomics
OmniGene GUT kits were used to collect infant stool samples at 1-, 6-, 12-, 18-, and 24- months of age. Untargeted high-resolution metabolomics analysis was carried out by the Emory Clinical Biomarkers Laboratory, as previously described36,37. To precipitate proteins, stool samples were first added to ice-cold acetonitrile. Samples were then kept on ice for 30 min, centrifuged for 10 min at 14,000 g, and kept at 4 °C until analysis. Extractants were examined in triplicate using high-resolution mass spectrometry (LC-HRMS) (Dionex Ultimate 3000, Thermo Scientific Orbitrap Fusion).
Instrumentation and analytical conditions
Hydrophilic interaction liquid chromatography (HILIC) (Waters XBridge BEH Amide XP HILIC column; 2.1 × 50 mm2, 2.6 μm particle size) with positive electrospray ionization (ESI) and reverse phase (C18) chromatography (Higgins Targa C18 2.1 × 50 mm2, 3 μm particle size) with negative ESI were used. HILIC analyte separation was conducted using water, acetonitrile, and 2% formic acid mobile phases following the subsequent gradient elution. The initial 1.5-min period consisted of 22.5% water, 75% acetonitrile, and 2.5% formic acid followed by a linear increase to 75% water, 22.5% acetonitrile, and 2.5% formic acid at 4 min, followed by a final hold for 1 min. Analyte separation for the C18 chromatography column was conducted using water, acetonitrile, and 10 mM ammonium acetate mobile phases under the following gradient elution. The initial 1-min period consisted of 60% water, 35% acetonitrile, and 5% ammonium acetate followed by a linear increase to 0% water, 95% acetonitrile, and 5% ammonium acetate at 3 min with a final hold for the last 2 min. Mobile phase flow rate was 0.35 mL/min for the first minute and was increased to 0.4 mL/min for the last 4 min for both the HILIC and C18 chromatography columns. LC-HRMS was run in full scan mode, with 120 k resolution and had a range of mass-to-charge ratio (m/z) from 85 to 1,275. Tuning parameters for sheath gas were 45 (arbitrary units) for positive ESI and 30 for negative ESI. For positive ESI, auxiliary gas was set to 25 (arbitrary units) and spray voltage was set at 3.5 kV, and for negative ESI, auxiliary gas was set to 5 and spray voltage was set to − 3.0 kV. Internal standards included pooled stool and standard reference materials for human metabolites in stool. These internal standards were added at the beginning and end of each 20-sample batch for quality control and standardization.
Metabolite confidence and identification
Data from positive and negative ion modes were analyzed separately, and raw files were converted to the .mzXML format. Then, metabolomic signals (i.e., metabolic features) were extracted and aligned using apLCMS with modification of xMSanalyzer for quality control and reduction of batch effects following instrument analysis38,39. The coefficients of variation (CV) of metabolites were assessed as part of quality control. Metabolites whose intensity had CV > 30% were removed and intensities of metabolic features were averaged across triplicates. Metabolic features which were detected in < 10% of samples were excluded. Outliers were assessed visually using principal component analysis (PCA) of the log2 transformed metabolite intensities. In a sensitivity analysis, samples with PCA score was > 3 standard deviations for PC 1 or PC 2 were removed (not shown). However, there were no important differences in results, so these observations were not removed. Metabolomic features were then annotated and confirmed using the Metabolomics Standards initiative criteria40. Level 1 confidence was assigned to features whose m/z and retention time matched the authentic standards analyzed with MS/MS under identical conditions (within 10 ppm and 50 s).
Statistical analysis
Descriptive statistics for key variables were performed on the full analytic data set. We used the VennDiagram package in R to visualize how many metabolites were present in 25%, 50%, and 75% of samples at each visit41. Heatmaps to visualize log2 transformed metabolite intensities over time were generated using the ComplexHeatmap package in R42. Next, we performed PCA on the log2 transformed metabolite intensities to visualize overall metabolite profiles over time. We used permutational multivariate ANOVA (PERMANOVA) tests to explore how overall fecal metabolite intensities changed over time, and in relation to individual, infant sex, and antibiotic exposure using the adonis2 function implemented by the vegan package in R, using Euclidian distance (permutations = 1000)43. Linear mixed effects models were then used to estimate the relationship between the log2 transformed intensity of each confirmed metabolite and infant age in days using the lme4 package in R44. Models included random intercepts to account for repeated measures and were adjusted for multiple testing using the Benjamini–Hochberg procedure45. Boxplots were used to visualize the intensity of 6 selected confirmed metabolites associated with infant age in days in the HILIC and C18 chromatography columns by visit. Finally, we used the Metapone package in R23 to jointly assess which metabolic pathways were associated with infant age in days, with combined linear mixed effects model results generated for the HILIC and C18 chromatography columns, using a weighted gene set enrichment analysis (GSEA) modified to untargeted metabolomics data. Briefly, Metapone employs established online MS databases of metabolites to putatively annotate metabolic features, incorporates a weight to limit the influence of single features that are annotated to multiple metabolites, and finally applies the modified GSEA test to focus on groups of metabolic features playing a role in the same biological pathways23. Metapone uses local false discovery rate (lfdr) to adjust for multiple testing, which is a Bayesian approach with minimal a priori assumptions and which does not rely on the assumption that individual statistical tests are independent46.
Supplementary Information
Acknowledgements
This project is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK110793), The Gerber Foundation (15PN-013), the National Institute of Environmental Health Sciences (R00ES027853), a Health Effects Institute Rosenblith Award, and the National Heart, Lung, and Blood Institute (T32 HL149646). Research described in this article was conducted under contract to the Health Effects Institute (HEI), an organization jointly funded by the United States Environmental Protection Agency (EPA) (Assistance Award No. CR 83,998,101) and certain motor vehicle and engine manufacturers. The contents of this article do not necessarily reflect the views of HEI, or its sponsors, nor do they necessarily reflect the views and policies of the EPA or motor vehicle and engine manufacturers.
Author contributions
E.A.H., B.C., and T.L.A. had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis. E.A.H., N.S., B.C., and Z.L. analyzed the data. V.T. conducted the experiments, H.H.C. provided statistical consultation, J.A.S., D.P.J., M.I.G., and H.H.C. assisted with manuscript preparation and interpretation of results. All authors provided feedback and approved the final manuscript. M.I.G. conceived of the original Mother’s Milk Cohort and D.L. and T.L.A. conceived of the study and designed the experiments.
Data availability
Data cannot be shared publicly because they include potentially identifying information on human subjects. The data that support the findings of this study are available upon reasonable request from the corresponding author, TLA.
Competing interests
MIG receives book royalties and is a scientific advisor for Yumi. The authors declare no additional competing interests to disclose.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-28862-z.
References
- 1.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francino MP. Early development of the gut microbiota and immune health. Pathogens. 2014;3(3):769–790. doi: 10.3390/pathogens3030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Trans. Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 4.Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. 2016;130(23):2199–207. doi: 10.1042/CS20160349. [DOI] [PubMed] [Google Scholar]
- 5.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin. Exp. Allergy. 2015;45(3):632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 6.Korpela K, Zijlmans MAC, Kuitunen M, Kukkonen K, Savilahti E, Salonen A, et al. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome. 2017;5(1):26. doi: 10.1186/s40168-017-0245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalliomäki M, Carmen Collado M, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008;87(3):534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 8.Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first 3 years of life: an observational study. Gut Pathogens. 2011;3(1):8. doi: 10.1186/1757-4749-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alderete TL, Jones RB, Shaffer JP, Holzhausen EA, Patterson WB, Kazemian E, et al. Early life gut microbiota is associated with rapid infant growth in Hispanics from Southern California. Gut Microbes. 2021;13(1):1961203. doi: 10.1080/19490976.2021.1961203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson AL, Xia K, Azcarate-Peril MA, Goldman BD, Ahn M, Styner MA, et al. Infant gut microbiome associated with cognitive development. Biol. Psychiat. 2018;83(2):148–159. doi: 10.1016/j.biopsych.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma N, Ma X. Dietary amino acids and the gut-microbiome-immune axis: physiological metabolism and therapeutic prospects. Comp. Rev. Food Sci. Food Saf. 2019;18(1):221–242. doi: 10.1111/1541-4337.12401. [DOI] [PubMed] [Google Scholar]
- 12.Kriaa A, Bourgin M, Potiron A, Mkaouar H, Jablaoui A, Gérard P, et al. Microbial impact on cholesterol and bile acid metabolism: current status and future prospects. J. Lipid Res. 2019;60(2):323–332. doi: 10.1194/jlr.R088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JAM, et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 2015;117(9):817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoen AG, Coker MO, Madan JC, Pathmasiri W, McRitchie S, Dade EF, et al. Association of cesarean delivery and formula supplementation with the stool metabolome of 6-week-old infants. Metabolites. 2021;11(10):702. doi: 10.3390/metabo11100702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N, Liang S, Chen Q, Zhao L, Li B, Huo G. Distinct gut microbiota and metabolite profiles induced by delivery mode in healthy Chinese infants. J. Proteom. 2021;10(232):104071. doi: 10.1016/j.jprot.2020.104071. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, Huang Q, Wei B, Chen Y. Effects of β-lactam antibiotics on gut microbiota colonization and metabolites in late preterm infants. Curr. Microbiol. 2020;77(12):3888–3896. doi: 10.1007/s00284-020-02198-7. [DOI] [PubMed] [Google Scholar]
- 17.Patton L, Li N, Garrett TJ, Ruoss JL, Russell JT, de la Cruz D, et al. Antibiotics effects on the fecal metabolome in preterm infants. Metabolites. 2020;10(8):331. doi: 10.3390/metabo10080331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brink LR, Mercer KE, Piccolo BD, Chintapalli SV, Elolimy A, Bowlin AK, et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am. J. Clin. Nutr. 2020;111:1190–1202. doi: 10.1093/ajcn/nqaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Parenti M, Grip T, Lönnerdal B, Timby N, Domellöf M, et al. Fecal microbiome and metabolome of infants fed bovine MFGM supplemented formula or standard formula with breast-fed infants as reference: a randomized controlled trial. Sci. Rep. 2019;9(1):11589. doi: 10.1038/s41598-019-47953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conta G, Del Chierico F, Reddel S, Marini F, Sciubba F, Capuani G, Tomassini A, Cocco MED, Laforgia N, Baldassarre ME, Putignani L, Miccheli A. Longitudinal multi-omics study of a mother-infant dyad from breastfeeding to weaning: an individualized approach to understand the interactions among diet, fecal metabolome and microbiota composition. Front. Mol. Biosci. 2021 doi: 10.3389/fmolb.2021.688440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson WB, Glasson J, Naik N, Jones RB, Berger PK, Plows JF, et al. Prenatal exposure to ambient air pollutants and early infant growth and adiposity in the Southern California Mother’s Milk Study. Environ. Health. 2021;20(1):67. doi: 10.1186/s12940-021-00753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu KH, Owens JA, Saeedi B, Cohen CE, Bellissimo MP, Naudin C, et al. Microbial metabolite delta-valerobetaine is a diet-dependent obesogen. Nat Metab. 2021;3(12):1694–1705. doi: 10.1038/s42255-021-00502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L, Li Z, Ma G, Zhang X, Tang Z, Wang S, Kang J, Liang D, Tianwei Y. Metapone: a bioconductor package for joint pathway testing for untargeted metabolomics data. Bioinformatics. 2022;38(14):3662–3664. doi: 10.1093/bioinformatics/btac364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goran MI, Walker R, Allayee H. Genetic-related and carbohydrate-related factors affecting liver fat accumulation. Curr. Opin. Clin. Nutr. Metab. Care. 2012;15(4):392–396. doi: 10.1097/MCO.0b013e3283544477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider ALC, Lazo M, Selvin E, Clark J M. Racial differences in nonalcoholic fatty liver disease in the U.S. population. Obesity. 2014;22(1):292–299. doi: 10.1002/oby.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K. The microbiome and metabolome of preterm infant stool are personalized and not driven by health outcomes, including necrotizing enterocolitis and late-onset sepsis. mSphere. 2018 doi: 10.1128/mSphere.00104-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brewington CR, Parks OW, Schwartz DP. Conjugated compounds in cow’s milk. J. Agric. Food Chem. 1973;21(1):38–39. doi: 10.1021/jf60185a015. [DOI] [PubMed] [Google Scholar]
- 28.PubChem. Methyl vanillate. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/19844
- 29.Marszalek-Grabska M, Stachniuk A, Iwaniak P, Gawel K, Sumara A, Kocki T, et al. Unexpected content of kynurenine in mother’s milk and infant formulas. Sci. Rep. 2022;12(1):6464. doi: 10.1038/s41598-022-10075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salem N, Van Dael P. Arachidonic acid in human milk. Nutrients. 2020;12(3):626. doi: 10.3390/nu12030626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horrocks LA, Yeo YK. Health benefits of docosahexaenoic acid (DHA) Pharmacol. Res. 1999;40(3):211–225. doi: 10.1006/phrs.1999.0495. [DOI] [PubMed] [Google Scholar]
- 32.Guan H, Pu Y, Liu C, Lou T, Tan S, Kong M, et al. Comparison of fecal collection methods on variation in gut metagenomics and untargeted metabolomics. mSphere. 2021;6(5):e00636. doi: 10.1128/mSphere.00636-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Zolnik CP, Qiu Y, Usyk M, Wang T, Strickler HD, et al. Comparison of fecal collection methods for microbiome and metabolomics studies. Front. Cell Infect. Microbiol. 2018;28(8):301. doi: 10.3389/fcimb.2018.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plows JF, Berger PK, Jones RB, Alderete TL, Yonemitsu C, Najera JA, et al. Longitudinal changes in human milk oligosaccharides (HMOs) over the course of 24 months of lactation. J. Nutr. 2021;151(4):876–882. doi: 10.1093/jn/nxaa427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollingshead, A. B. Four factor index of social status. Yale J. Sociol.8 (2011).
- 36.Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci. 2015;148(2):531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang D, Moutinho JL, Golan R, Yu T, Ladva CN, Niedzwiecki M, et al. Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ. Int. 2018;120:145–154. doi: 10.1016/j.envint.2018.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformat. 2013;14(1):15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu T, Jones DP. Improving peak detection in high-resolution LC/MS metabolomics data using preexisting knowledge and machine learning approach. Bioinformatics. 2014;30(20):2941–2948. doi: 10.1093/bioinformatics/btu430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, H. In VennDiagram: Generate High-Resolution Venn and Euler Plots. Available from: https://CRAN.R-project.org/package=VennDiagram (2022).
- 42.Gu, Z. In: ComplexHeatmap: Make Complex Heatmaps. Bioconductor version: Release (3.15). Available from: https://bioconductor.org/packages/ComplexHeatmap/ (2022).
- 43.Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R. et al. In: Vegan: Community Ecology Package. Available from: https://CRAN.R-project.org/package=vegan (2022).
- 44.Bates, D., Maechler, M., Bolker [aut, B., cre, Walker, S., Christensen, R. H. B. et al. In: lme4: Linear Mixed-Effects Models Using “Eigen” and S4. Available from: https://CRAN.R-project.org/package=lme4 (2022)
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57(1):289–300. [Google Scholar]
- 46.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet. Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data cannot be shared publicly because they include potentially identifying information on human subjects. The data that support the findings of this study are available upon reasonable request from the corresponding author, TLA.