Abstract
Taxadiene is the first dedicated intermediate in the biosynthetic pathway of the anticancer compound Taxol. Recent studies have taken advantage of heterologous hosts to produce taxadiene and other isoprenoid compounds, and such ventures now offer research opportunities that take advantage of the engineering tools associated with the surrogate host. In this study, metabolic engineering was applied in the context of over-expression targets predicted to improve taxadiene production. Identified targets included genes both within and outside of the isoprenoid precursor pathway. These targets were then tested for experimental over-expression in a heterologous Escherichia coli host designed to support isoprenoid biosynthesis. Results confirmed the computationally predicted improvements and indicated a synergy between targets within the expected isoprenoid precursor pathway and those outside this pathway. The presented algorithm is broadly applicable to other host systems and/or product choices.
Keywords: Taxol, Taxadiene, Taxadiene synthase, Over-expression, E. coli, Heterologous biosynthesis, Metabolic engineering
Introduction
Taxol and other isoprenoids possess a wide range of therapeutic potential (Ajikumar et al. 2008). Many of these compounds derive from plant sources, and many production methods have relied on adapting the plant hosts for process development of the desired isoprenoid compound (Lewis and Ausubel 2006; Wink 2010). Taxol has been a prominent example with both semi-synthetic and cell culture methods reliant upon plants for eventual production of the final compound (Cragg et al. 1993). More recently, heterologous biosynthesis of plant-derived natural products has emerged as an alternative to native plant production systems (Kirby and Keasling 2009; Leonard et al. 2009). In this approach, the genetic material required for eventual product biosynthesis is transferred to a surrogate host. The new host generally possesses some advantageous properties when compared to the original host, and these often include faster growth kinetics, more advanced engineering tools, and a wider genetic and physiologic knowledge base (Zhang et al. 2011). Recent high-profile examples of heterologous biosynthesis include the production of artemisinic acid through Saccharomyces cerevisiae (Ro et al. 2006) and the production of taxadien-5α-ol through Escherichia coli (Ajikumar et al. 2010). These and numerous other examples support continued research into the prospect of using heterologous biosynthesis as an economically viable route to the commercial production of natural products (Kirby and Keasling 2009; Leonard et al. 2009).
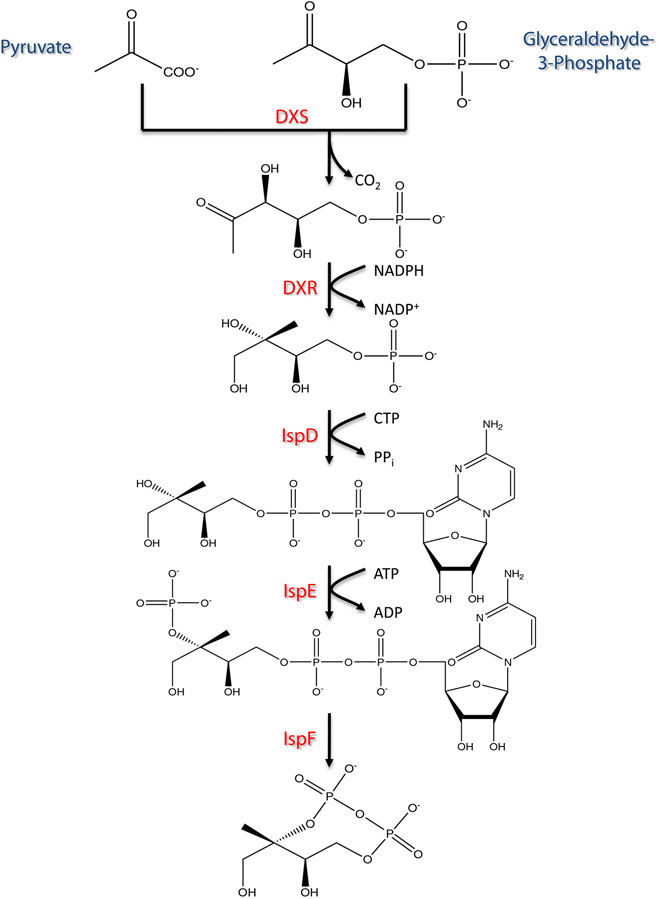
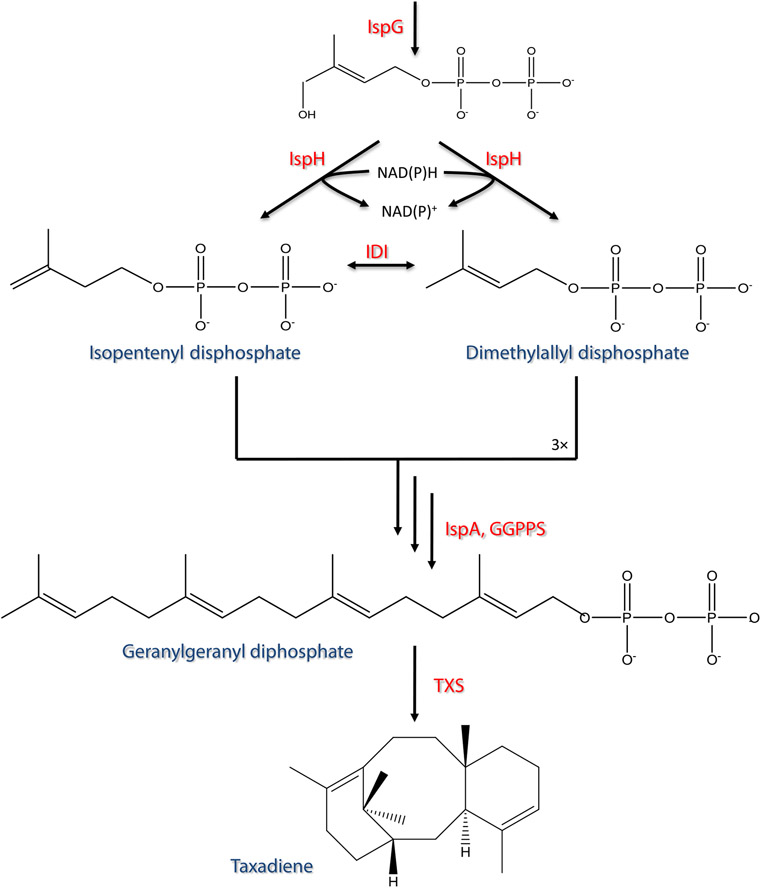
As mentioned, the choice of heterologous biosynthesis offers new engineering tools to aid in process development. This is especially important to improve the often low titers, yields, and specific productivities associated with heterologous production attempts (Boghigian and Pfeifer 2008). One of the most common approaches towards doing so is the application of metabolic engineering in the form of cellular-based modeling of metabolism (Boghigian et al. 2010b; Kim et al. 2008; Park et al. 2009). The underlying goal is to improve carbon flow to a metabolite of interest. In the case of the heterologous Taxol system, a key milestone is to improve carbon flow to early intermediates thus far produced through E. coli. As seen in Fig. 1, the production of taxadiene (the first dedicated intermediate of the Taxol pathway) is produced by the combination of an upstream native precursor pathway, termed the methylerythritol phosphate (MEP) pathway (or the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway), and a downstream heterologous biosynthetic pathway, which converts the universal isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) to taxadiene. The Fig. 1 depiction of the upstream and downstream pathways is a simplification of taxadiene biosynthesis within the complex, surrounding E. coli reaction network. The holistic view of the cell must be considered when optimizing the production of heterologous taxadiene through metabolic engineering.
Fig. 1.
The pathway from central metabolites pyruvate and glyceraldehyde-3-phosphate to taxadiene
While most previous metabolic engineering efforts have applied computational methods to model cellular metabolism and predict gene deletions to improve specified metabolite levels, this study presents the development and application of an algorithm for identifying over-expression targets to improve product titer. Specifically, a genome-scale metabolic network was assessed with over-expression of genetic targets to improve taxadiene biosynthetic flux. Those targets were then tested experimentally through the heterologous system currently available. The results support the use of this approach to identify over-expression targets capable of improving the titer of a desired product. Such an approach emphasizes the expanded repertoire of engineering options available with well-characterized heterologous systems and will be used together with accompanying genetic and process engineering tools to continually improve titers of isoprenoid and other medicinally relevant natural products.
Materials and methods
Model construction
The E. coli genome-scale metabolic model iAF1260 was used as the base model in this study. This model contains 2,077 reactions, 1,039 metabolites, and 1,261 genes (Feist et al. 2007). Reactions then had to be added to account for steps catalyzed by two heterologous enzymes introduced to produce taxadiene through E. coli. These reactions are: 1) a geranylgeranyl diphosphate synthase (GGPPS; to catalyze: farnesyl diphosphate + isopentenyl diphosphate → geranylgeranyl diphosphate + diphosphate), 2) a cyclizing taxadiene synthase (TXS; to catalyze: geranylgeranyl diphosphate → taxa-4,11-diene + diphosphate), and 3) a taxadiene transport reaction (taxa-4,11-diene → [extracellular]) (Ajikumar et al. 2008, 2010).
Calculations were conducted in MATLAB® 7.4 (Math-works Inc.; Natick, MA) utilizing the SBMLToolbox (version 2.0.2, http://sbml.org/software/sbmltoolbox/) (Keating et al. 2006; Schmidt and Jirstrand 2006) and the COBRA Toolbox (version 1.3.3, http://gcrg.ucsd.edu/) (Becker et al. 2007). Optimization was undertaken using the CPLEX (version 11.0) algorithm of the TOMLAB™ Optimization Environment (TOMLAB™/CPLEX) interfaced with the COBRA Toolbox and MATLAB® 7.4.
Over-expression target identification algorithm
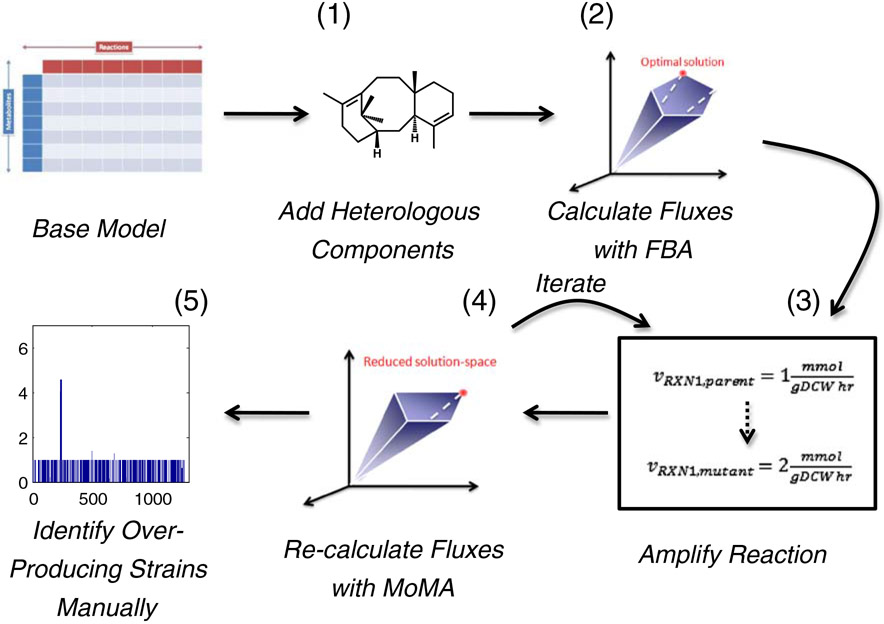
The over-expression algorithm involves: 1) imposing a taxadiene production flux (as determined experimentally), 2) solving a Flux Balance Analysis (FBA) problem (Edwards et al. 2002, 2001; Edwards and Palsson 2000), 3) imposing an amplification in individual reaction fluxes (to simulate the effect of gene over-expression), 4) solving a Minimization of Metabolic Adjustment (MoMA) problem (Segre et al. 2002), and 5) identifying over-expressions that led to a phenotype fraction value, fPH (the product of weighted and dimensionless biomass and taxadiene flux values; Boghigian et al. 2010a), greater than unity (an overview of this algorithm can be seen in Fig. 2). Steps 3 and 4 were iterated for every reaction within the network.
Fig. 2.
An overview of the proposed algorithm for identifying over-expression targets to improve product titer
Calculations were made under conditions to simulate complex medium. Medium composition can be approximated by setting uptake rates of specific chemical components known to exist in the medium of interest. The “computational complex medium” contained all twenty naturally occurring amino acids (l-isomers). The lower bounds of the amino acid transport reactions were set to −0.1 mmol/g DCW/h (a negative sign indicates metabolite uptake into the cell) and were chosen based upon previous literature values and because they satisfied the relative biomass differences experimentally observed between media (Oh et al. 2007; Selvarasu et al. 2009). Glycerol transport rates were set to −3.0 mmol/g DCW/h, as described previously (Boghigian et al. 2010a).
Strains and plasmids
Strain YW22(pTrcHis2B-TXS-GGPPS) was used as the base strain for taxadiene production in this study (Boghigian et al. 2011b). This strain is a derivative of JM109(DE3) containing a T7prom-dxs-T7term-T7prom-idi-T7term-T7prom-ispDF-T7term construct in the araA chromosomal locus. Plasmid pTrcHis2B-TXS-GGPPS is a carbenicillin-resistant plasmid with a pBR322 origin of replication, containing synthetic txs and ggpps genes under the control of an IPTG-inducible Trc promoter; the ggpps gene encodes for a geranylgeranyl diphosphate synthase (Boghigian et al. 2011b).
Plasmids containing over-expression targets as identified by the algorithm were obtained from the ASKA library of the National Institute of Genetics in Japan (Kitagawa et al. 2005). The genes are cloned into a derivative of pQE31 (called pCA24N) under the control of an IPTG-inducible T5 promoter with an N-terminal 6× histidine tag (Kitagawa et al. 2005). These plasmids are chloramphenicol-resistant and contain a ColE1 origin of replication (and are therefore compatible with pTrcHis2B-TXS-GGPPS).
The idi gene was PCR-amplified from pASKA-idi using primers 5′-GGGGAATTC ATGCAAACGGAACACGTCAT-3′ and 5′-GGGAAGCTTTTATTTAAGCTGGGTAAATG CAGA-3′. The gene was placed into the pCOLADuet-1 (which has a ColA origin of replication) expression vector using EcoRI and HindIII (restriction sites italicized in the previous sentence). The resulting plasmid, pCOLADuet-idi, was transformed with pTrcHis2B-TXS-GGPPS and certain pASKA plasmids, as indicated, to test the cumulative effects of the over-expression idi isoprenoid precursor pathway target and targets outside the pathway.
Small-scale production cultures
A stab of glycerol stock was inoculated into 2 ml LB medium with appropriate antibiotics and grown overnight at 37 °C and 250 rpm. For production cultures, 3 ml production medium (5 g/l yeast extract, 10 g/l tryptone, 10 g/l sodium chloride, 15 g/l glycerol, 3 ml/l 50% (v/v) Antifoam B, 100 mM HEPES; adjusted to pH 7.60 with 5 M sodium hydroxide) was inoculated into 16 × 100 -mm culture tubes with the pre-cultures to an OD600 nm=0.1. These production cultures were grown for 120 h at 22 °C and 250 rpm. At the end of the culture period, cell-density was measured spectrophotometrically at 600 nm and a single, 1-ml aliquot was stored at −20 °C for subsequent analysis. When required, antibiotics were supplemented at concentrations of 100 mg/l for carbenicillin, 50 mg/l for kanamycin, and 34 mg/l for chloramphenicol, and IPTG was added at a concentration of 100 μM.
Taxadiene quantification
For taxadiene quantification, a culture aliquot was supplemented with (−)-trans-caryophyllene (TC) at a final concentration of 1 μg/l to serve as an internal standard. The samples were then extracted with an equal volume of hexane, followed by 20 s of vortexing and centrifugation for 10 min at 10,000 × g. The hexane layer (150 μl) was removed and stored in glass vials at −20 °C until analysis by gas chromatography–mass spectrometry (GC–MS).
Samples were analyzed on a Shimadzu QP5050A GC–MS using split-less injection. Gas chromatography was run on a non-polar Rxi®-XLB column (30 m × 0.25 mm ID, 0.25 μm). The inlet pressure for the column was set at 120 kPa, and column flow velocity was 1.6 ml/min. The flow rate of the ultra-high-purity helium carrier gas was 20 ml/min. Temperature of the column was initially set and maintained at 100 °C for 2 min and was then increased to 235 °C at a rate of 15.0 °C/min. The column was then maintained at this temperature for 1 min. Mass spectrometry was performed in Single Ion Monitoring (SIM) mode scanning for mass-to-charge ratios of 107 m/z, 122 m/z, and 272 m/z, corresponding to the principle daughter ions and parent ion of taxadiene, respectively (Koepp et al. 1995). Under these conditions, TC and taxadiene eluted at approximately 6.8 and 11.4 min, respectively. Quantification of taxadiene was accomplished based on a calibration curve of taxadiene (kindly provided by Drs. Ajikumar Parayil and Gregory Stephanopoulos) concentration versus the peak area ratio of TC to taxadiene.
Results
Over-expression target identification
For identifying knockout targets, the binary nature of the problem (the gene and therefore reaction either exists or does not exist) simplifies its mathematical abstraction. For identifying over-expression targets, the problem is now non-binary, meaning that describing gene over-expression in this context is more difficult. Over-expressing a gene on a five-copy plasmid does not necessarily correlate to five times the amount of transcript relative to a one-copy plasmid, nor does it necessarily correlate to five times the amount of soluble protein, nor five times the amount of metabolic flux through that reaction. Nonetheless, this study aimed to formulate a mathematical abstraction for modeling gene over-expression, as well as to simulate metabolic fluxes and identify over-expression targets for improving heterologous isoprenoid titer.
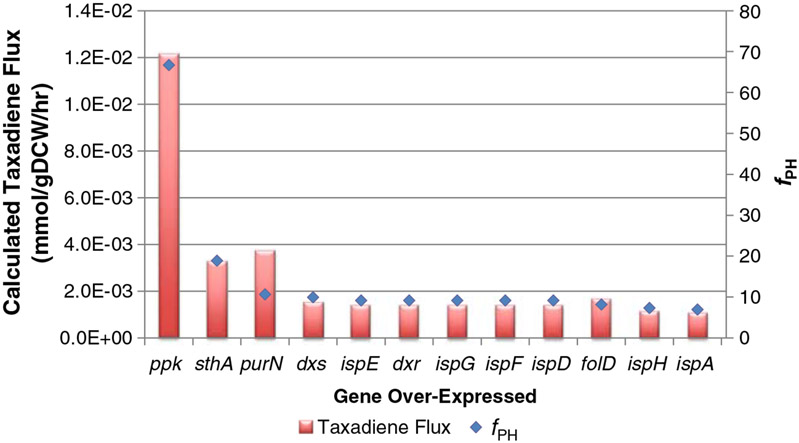
The first demonstration of the over-expression algorithm was undertaken in glycerol-based complex medium (to mimic the medium used in previous studies (Boghigian et al. 2011a; Boghigian et al. 2011b). The experimental specific production rate of YW22 (pTrcHis2B-TXS-GGPPS) was determined to be 1.53×10−4 mmol/g DCW/h and was set as the lower bound for taxadiene transport flux. The specific uptake rate of glycerol was 3.0 mmol/g DCW/h while the specific uptake rates of all twenty l-amino acids were set to 0.1 mmol/g DCW/h. Under these carbon-limited conditions, the specific growth rate was determined by FBA to be 0.2671 h−1. Next, all reactions that had a non-zero flux value in the FBA simulations were over-expressed computationally. Once all reactions had been cycled, reactions that produced an fPH value of greater than 1 were chosen as potential over-expression targets. Figure 3 shows all of these genes and their corresponding fPH values.
Fig. 3.
Calculated taxadiene production flux (left-hand y-axis) and fPH (right-hand y-axis) as a function of gene over-expressed
Of the 12 targets identified, four (ppk, sthA, purN, and folD) were outside of the native isoprenoid precursor pathway. The other eight (dxs, ispE, dxr, ispG, ispF, ispD, ispH, and ispA) were within the MEP pathway (Fig. 1). While the MEP targets each produced an fPH greater than 1, it was expected that amplifying a reaction within the linear isoprenoid precursor pathway would improve taxadiene flux and therefore provided an internal control for verification of the algorithm itself. As a result, the four targets chosen for experimental implementation were ppk, sthA, purN, and folD, as summarized in Table 1.
Table 1.
Genetic targets identified by the over-expression algorithm for experimental implementation
| Gene | Reaction name | Reaction stoichiometry | Predicted fPH |
|---|---|---|---|
| ppk | Polyphosphate kinase | ATP + Pi ↔ ADP + PPi | 66.7 |
| sthA | Pyridine nucleotide transhydrogenase | NAD+ + NADPH ↔ NADH + NADP+ | 18.9 |
| purN | Phosphoribosylglycinamide formyltransferase | 5-Phospho-ribosyl-glycineamide + 10-formyl-tetrahydrofolate ↔ 5′-phosphoribosyl-N-formylglycineamide + tetrahydrofolate + 3 H+ | 10.6 |
| folD | 5,10-Methylene-tetrahydrofolate dehydrogenase/cyclohydrolasea | NADP+ + 5,10-methylene-THF + H2O ↔ NADPH + 10-formyl-tetrahydrofolate + H+ | 8.1 |
The folD gene product is a bi-functional enzyme. The net reaction is shown
As a means of implementing a control, the same algorithm was run with glucose as the principle carbon source instead of glycerol. New targets identified using glucose (determined to be fumA, fumB, fumC, and mdh) were also included in the subsequent experimental analysis. Experimentally, these targets should not be able to improve titer because the primary carbon source used is glycerol.
Experimental implementation
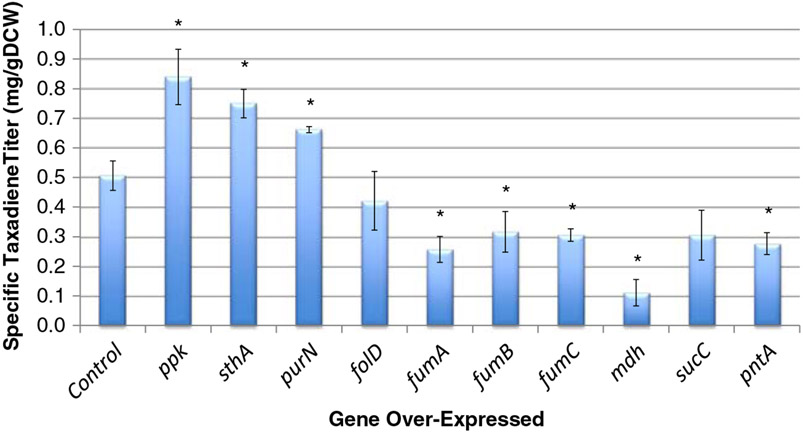
The four positive targets (ppk, sthA, purN, and folD) and four algorithm controls (fumA, fumB, fumC, and mdh) were tested in the context of E. coli taxadiene production. Also tested were two experimental controls encoding for only a subunit of a functional enzyme (sucC and pntA) to ensure that other experimental factors associated with plasmid-based over-expression did not contribute to taxadiene titer. The genes were over-expressed using a T5 promoter from a chloramphenicol-resistant ColE1-based plasmid in YW22 also co-expressing GGPPS and TXS to produce taxadiene. At the end of the culture period (120 h), taxadiene was extracted and quantified using GC–MS. Cell-density was also measured spectrophotometrically at 600 nm. Another aliquot of the culture was stored for SDS–PAGE analysis to verify gene expression of txs and ggpps and the over-expression gene target (data not shown). Raw taxadiene titer was normalized by cell-density (1 OD600 nm=0.52 gDCW/l), creating a specific taxadiene titer (Fig. 4).
Fig. 4.
Experimental specific taxadiene titer data for the computationally identified gene over-expression targets and corresponding controls. The “Control” sample is YW22 (pTrcHis2B-TXS-GGPPS). Error bars represent ± one standard deviation of three independent replicates. *Statistically significant (p<0.05, as determined by paired Student’s t-test) difference from the YW22 (pTrcHis2B-TXS-GGPPS) control
Three (ppk, sthA, purN) of the four experimental targets improved titer (p<0.05 when compared to the YW22 (pTrcHis2B-TXS-GGPPS) control); however, the experimental improvements were below algorithm predictions. The over-expression of ppk, sthA, and purN improved specific production of taxadiene 1.66-fold, 1.48-fold, and 1.31-fold, respectively. The fourth target, folD, appears to decrease titer, but the difference is statistically insignificant from the control (p=0.400). Except for folD, the results showed the same trend of improvement as predicted by the model (ppk > sthA > purN). The three fumarase isozymes (fumA, fumB, and fumC) all decreased titer, while mdh decreased titer even more significantly. For the two experimental controls, over-expression of sucC had no effect on specific taxadiene titer (p=0.119), while over-expression of pntA decreased specific taxadiene titer to roughly half of the control (p=0.010). As a result, all six of the chosen computational and experimental controls were verified as having no positive effect on taxadiene titer. The over-expression of the control genes likely increases metabolic burden on the host, which may partially account for the decrease in taxadiene production (an issue that may also accompany the results of the identified over-expression targets). While this is certainly not an exhaustive analysis of potential control targets, no previous knowledge was used to inherently bias the choice of these targets.
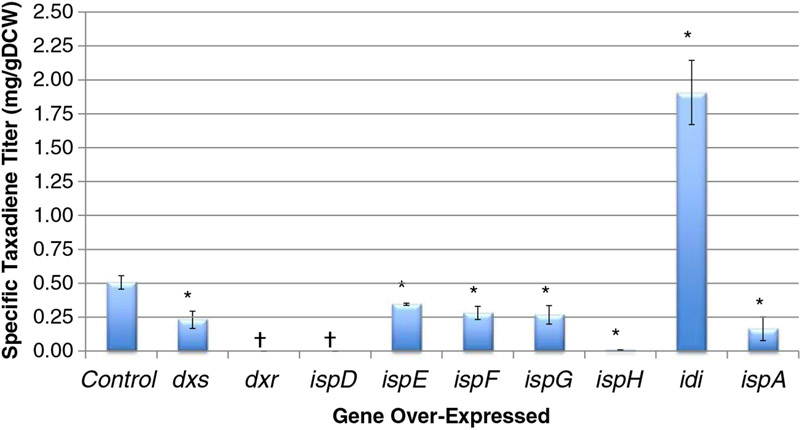
The lack of dramatic improvement in specific taxadiene titer prompted us to investigate the role of a potential bottleneck in the MEP pathway. It has been shown that a complex response (isoprenoid production titer) results when modulating the expression of the upstream and downstream portions of the complete isoprenoid biosynthetic pathway (Ajikumar et al. 2010). Even though YW22 contains chromosomal over-expressed versions of the dxs, idi, ispD, and ispF genes, higher levels of some of these or other precursor pathway genes might be required to fully access improved precursor flux and to debottleneck this pathway. As a result, each upstream pathway gene was individually over-expressed (from the ASKA library plasmids) together with the pTrcHis2B-TXS-GGPPS. Six of these genes produced less taxadiene: dxs, ispE, ispF, ispG, ispH, and ispA (p<0.05). Oddly, the dxr plasmid could not be stably transformed (multiple trials) into YW22(pTrcHis2B-TXS-GGPPS). Over-expression of ispD did not produce any taxadiene, but did produce a viable strain. One target, idi, improved specific production of taxadiene 3.77-fold (p=0.008) (Fig. 5).
Fig. 5.
Experimental specific taxadiene titer data for the isoprenoid precursor pathway gene over-expression targets. The “Control” sample is YW22 (pTrcHis2B-TXS-GGPPS). Error bars represent ± one standard deviation of three independent replicates. *Statistically significant (p<0.05, as determined by paired Student’s t-test) difference from the YW22 (pTrcHis2B-TXS-GGPPS) control. †Taxadiene was not detected by GC–MS
Combined expression of algorithm targets and idi
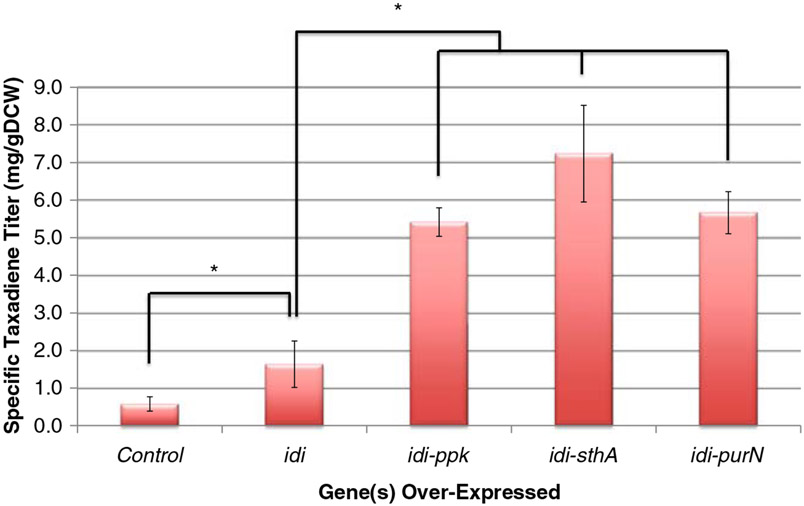
As a result of the positive effect observed for the over-expression of idi, this gene was tested together with those non-pathway targets identified by the algorithm. The expectation was an improvement in taxadiene titer beyond the results from the over-expression of the ppk, sthA, and purN targets alone. Figure 6 presents the cumulative effects of idi and the ppk, sthA, and purN over-expression targets and indicates positive synergy upon taxadiene titer. By over-expressing ppk, sthA, and purN with idi, titers improved to 5.42, 7.23, and 5.66 mg/g DCW, respectively. The final strain in this study has a cumulative improvement in specific taxadiene titer of over 12-fold from an already significantly engineered strain and plasmid system (Boghigian et al. 2011b).
Fig. 6.
Experimental specific taxadiene titer data for the computationally identified targets combined with over-expression of idi. The “Control” sample is YW22 (pTrcHis2B-TXS-GGPPS). Error bars represent ± one standard deviation of three independent replicates. *Statistically significant (p<0.05, as determined by paired Student’s t-test) difference from the YW22 (pTrcHis2B-TXS-GGPPS) control
Discussion
The use of systematic methods for identifying over-expression targets, at the start of this study, had been explored once theoretically (Pharkya and Maranas 2006), but had not been implemented experimentally. The goal of this study was to use a previously developed optimization strategy, modify its use to identify over-expression targets, and implement it experimentally for improving taxadiene production through E. coli. A variation of the MoMA algorithm was used as an extension to indentifying gene over-expression targets to improve a product titer. As has been described previously, MoMA was proposed as an alternative to FBA as a means of quantifying metabolic fluxes in networks that had been perturbed, originally by gene knockouts (Segre et al. 2002). In a similar sense, forced over-expression of a particular gene could also be considered a genetic perturbation, and this computational framework was extended for identifying genetic over-expression targets. Unfortunately, there is no experimental data available on global flux distributions upon over-expression of single genes in E. coli, so there was no means of verifying this algorithm short of testing it in our case study of improving taxadiene production.
The algorithm identified four targets as candidates to implement experimentally. While an in-depth examination of the physiological effects of over-expression of these four targets was outside the scope of this work, it would appear that all four targets improve cofactor availability. The over-expression of ppk allows for the reversible generation of ATP and Pi from ADP and the PPi generated by the IspA and GGPPS reactions in the isoprenoid precursor pathway. The next three targets are involved with improving NADPH supply for isoprenoid biosynthesis. Transhydrogenases are responsible for the reversible NAD+ + NADPH ↔ NADP+ + NADH reaction and can therefore theoretically be used to modulate the level of reduction within the cell. Over-expression of sthA led to improved production of poly (3-hydroxybutyrate) in E. coli (requiring NADPH reducing equivalents) (Sanchez et al. 2006). The same function appears relevant here, as one molecule of taxadiene requires four molecules of NADPH (at the DXR reaction step). The DXP metabolite node itself serves as a branch point between the isoprenoid biosynthetic pathway, the pyridoxal 5′-phosphate pathway for vitamin B6 biosynthesis, and the thiamin biosynthetic pathway. It has been previously shown that high concentrations of NADPH strongly shift the equilibrium of the reversible DXS towards production of MEP (Koppisch et al. 2002), thereby reducing metabolic flux towards the vitamin B6 and thiamin biosynthetic pathways. As a result, it is conceivable that general conditions that favor the production of NADPH may also improve flux towards the isoprenoid pathway relative to alternative destinations.
Last, purN and folD are a two-step linear pathway in tetrahydrofolate biosynthesis. While it appears that tetrahydrofolate has no immediate metabolic relation to the isoprenoid biosynthetic pathway, this step also produces an equivalent of NADPH.
Of the four targets identified, three improved specific production titer while all six controls implemented showed either no change or a decrease in specific taxadiene titer. However, the observed improvements did not exceed 2-fold and did not reproduce the flux improvements as predicted by the algorithm. At this point, the hypothesis was that a step within the isoprenoid precursor pathway was rate-limiting, such that over-expressing other genes potentially affecting reaction steps further upstream would have little or no effect on taxadiene titer. This drove us to individually over-express the genes within the MEP pathway itself, which identified IDI as a rate-limiting step in the current experimental system. Over-expressing idi with the computationally identified targets further increased taxadiene titers and provided better agreement with algorithm outputs.
It would be advantageous to investigate aspects of the algorithm that might further improve predictability. For better validation of this algorithm, strains with various single over-expressions should be cultured under different carbon sources. Then, 13C-MFA could be used to quantify fluxes in central metabolism and observe how well the predicted fluxes compare with measured fluxes under varying environmental and genetic conditions. A further extension would be to couple the computational approach described here with a genetic algorithm for a computationally tractable means of surveying the effect of product titer when over-expressing multiple genes.
It has been demonstrated and recognized that the first step in the MEP-based isoprenoid precursor pathway is rate-limiting (Begley et al. 1999; Lawhorn et al. 2004; Matthews and Wurtzel 2000). A bacterial d-1-deoxyxylulose 5-phosphate synthase (encoded by dxs), catalyzing the condensation of pyruvate and d-glyceraldehyde-3-phosphate to d-1-deoxyxylulose 5-phosphate (requiring thiamine diphosphate as a cofactor and Mg2+ as a metal adduct), has a turnover rate of 1.9 s −1 (the value for the E. coli enzyme has not been reported) (Eubanks and Poulter 2003). While the rate of the next step in the pathway, the 1-deoxy-d-xylulose-5-phosphate reductoisomerase (encoded by dxr), has been reported to be between 29 s−1 and 38 s−1 for E. coli (Fox and Poulter 2005a, 2005b) (with a kcat/KM value of 2.2 × 107 M−1s−1), providing in vitro evidence that DXS activity may bottleneck the pathway from the beginning. In vivo, it has been shown in numerous cases that over-expression of dxs improves isoprenoid or carotenoid titers in native and heterologous hosts (Alper et al. 2005a; Alper et al. 2005b; Brown et al. 2010; Chiang et al. 2008; Choi et al. 2010; Leonard et al. 2010; Morrone et al. 2010; Yuan et al. 2006). However, the lack of effect in this case could be due to increased expression of one rate-limiting enzyme (DXS) shifting control of pathway flux to another enzyme (perhaps IDI) (Kacser and Burns 1973). While it seems unusual that an isomerization reaction would be limiting, the turnover number for the E. coli IDI has been reported to be 0.33 s−1 (also having a smaller kcat/KM value of 4.2×104 M−1s−1) (Hahn et al. 1999). A highly complex phenotype has been observed recently with respect to the upstream and downstream isoprenoid pathways (Ajikumar et al. 2010). As such, systematic over-expression of the genes in the MEP pathway may lead to further pathway unbalancing and to decreased or no taxadiene production. However, it appears that IDI is the rate-limiting step under the current experimental conditions. The simultaneous over-expression of idi and ppk, sthA, and purN resulted in the highest taxadiene titers within this study.
During the course of this study, an algorithm was developed by another group for identifying over-expression targets and applied towards heterologous lycopene production in E. coli (Choi et al. 2010). This algorithm, called Flux Scanning based on Enforced Objective Flux (FSEOF), is based on simulating cellular metabolism by maximizing biomass formation (utilizing FBA) and then imposing specific production rates of a product of interest. Fluxes that increase through a reaction step as the product flux increases are considered to be over-expression targets. Interestingly, both fumarase (encoded by the homologous fumA, fumB, and fumC genes) and malate dehydrogenase (encoded by mdh) were identified as targets in this method for producing lycopene from glucose; these same genes were identified as targets for producing taxadiene from glucose in this study (as taxadiene and lycopene are generated from the same isoprenoid precursors). Numerous targets were implemented experimentally (pfkA, pgi, fbaA, tpiA, icdA, and mdh); however, each was tested with plasmid-based dxs and idi over-expression. Three of the six targets (pfkA, pgi, and icdA) decreased or did not change lycopene titer, while the other three (fbaA, tpiA, mdh) improved titer between 3-fold and 4-fold. The top over-expression strain (dxs, idi, and mdh) improved titer from 2.52 mg/l to 12.85 mg/l. Upon combining with MoMA deletion targets, titer increased to 26.77 mg/l (with a ΔlacI, ΔgdhA, and ΔgpmB strain).
Acknowledgements
The authors acknowledge the financial support from the NIH (GM085323), the Milheim Foundation (Grant for Cancer Research No. 2006–17), and the Tufts Summer Scholars Program and the technical advice and the GC–MS assistance from Professor Kyongbum Lee and Dr. David Wilbur, respectively, at Tufts University.
References
- Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G (2008) Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm 5:167–190 [DOI] [PubMed] [Google Scholar]
- Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Jin YS, Moxley JF, Stephanopoulos G (2005a) Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7:155–164 [DOI] [PubMed] [Google Scholar]
- Alper H, Miyaoku K, Stephanopoulos G (2005b) Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol 23:612–616 [DOI] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2:727–738 [DOI] [PubMed] [Google Scholar]
- Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon AP, Taylor S, Campobasso N, Chiu HJ, Kinsland C, Reddick JJ, Xi J (1999) Thiamin biosynthesis in prokaryotes. Arch Microbiol 171:293–300 [DOI] [PubMed] [Google Scholar]
- Boghigian BA, Pfeifer BA (2008) Current status, strategies, and potential for the metabolic engineering of heterologous polyketides in Escherichia coli. Biotechnol Lett 30:1323–1330 [DOI] [PubMed] [Google Scholar]
- Boghigian BA, Lee K, Pfeifer BA (2010a) Computational analysis of phenotypic space in heterologous polyketide biosynthesis—applications to Escherichia coli, Bacillus subtilis, and Saccharomyces cerevisiae. J Theor Biol 262:197–207 [DOI] [PubMed] [Google Scholar]
- Boghigian BA, Seth G, Kiss R, Pfeifer BA (2010b) Metabolic flux analysis and pharmaceutical production. Metab Eng 12:81–95 [DOI] [PubMed] [Google Scholar]
- Boghigian BA, Myint M, Wu J, Pfeifer BA (2011a) Simultaneous production and partitioning of heterologous polyketide and isoprenoid natural products in an Escherichia coli two-phase bioprocess. J Ind Microbiol Biotechnol. doi: 10.1007/s10295-011-0969-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boghigian BA, Salas DF, Ajikumar PK, Stephanopoulos G, Pfeifer BA (2011b) Analysis of heterologous taxadiene production in K- and B-derived Escherichia coli. Appl Microbiol Biotechnol. doi: 10.1007/s00253-011-3528-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Eberl M, Crick DC, Jomaa H, Parish T (2010) The nonmevalonate pathway of isoprenoid biosynthesis in Mycobacterium tuberculosis is essential and transcriptionally regulated by Dxs. J Bacteriol 192:2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CJ, Chen PT, Chao YP (2008) Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli. Biotechnol Bioeng 101:985–995 [DOI] [PubMed] [Google Scholar]
- Choi HS, Lee SY, Kim TY, Woo HM (2010) In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol 76:3097–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg GM, Schepartz SA, Suffness M, Grever MR (1993) The taxol supply crisis. new NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J Nat Prod 56:1657–1668 [DOI] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci USA 97:5528–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Ibarra RU, Palsson BO (2001) In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol 19:125–130 [DOI] [PubMed] [Google Scholar]
- Edwards JS, Covert M, Palsson B (2002) Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 4:133–140 [DOI] [PubMed] [Google Scholar]
- Eubanks LM, Poulter CD (2003) Rhodobacter capsulatus 1-deoxy-d-xylulose 5-phosphate synthase: steady-state kinetics and substrate binding. Biochemistry 42:1140–1149 [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Poulter CD (2005a) Mechanistic studies with 2-C-methyl-d-erythritol 4-phosphate synthase from Escherichia coli. Biochemistry 44:8360–8368 [DOI] [PubMed] [Google Scholar]
- Fox DT, Poulter CD (2005b) Synthesis and evaluation of 1-deoxy-d-xylulose 5-phosphoric acid analogues as alternate substrates for methylerythritol phosphate synthase. J Org Chem 70:1978–1985 [DOI] [PubMed] [Google Scholar]
- Hahn FM, Hurlburt AP, Poulter CD (1999) Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J Bacteriol 181:4499–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1973) The control of flux. Symp Soc Exp Biol 27:65–104 [PubMed] [Google Scholar]
- Keating SM, Bornstein BJ, Finney A, Hucka M (2006) SBMLToolbox: an SBML toolbox for MATLAB users. Bioinformatics 22:1275–1277 [DOI] [PubMed] [Google Scholar]
- Kim HU, Kim TY, Lee SY (2008) Metabolic flux analysis and metabolic engineering of microorganisms. Mol Biosyst 4:113–120 [DOI] [PubMed] [Google Scholar]
- Kirby J, Keasling JD (2009) Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol 60:335–355 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299 [DOI] [PubMed] [Google Scholar]
- Koepp AE, Hezari M, Zajicek J, Vogel BS, LaFever RE, Lewis NG, Croteau R (1995) Cyclization of geranylgeranyl diphosphate to taxa-4(5),11(12)-diene is the committed step of taxol biosynthesis in Pacific yew. J Biol Chem 270:8686–8690 [DOI] [PubMed] [Google Scholar]
- Koppisch AT, Fox DT, Blagg BSJ, Poulter CD (2002) E. coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry 41:236–243 [DOI] [PubMed] [Google Scholar]
- Lawhorn BG, Gerdes SY, Begley TP (2004) A genetic screen for the identification of thiamin metabolic genes. J Biol Chem 279:43555–43559 [DOI] [PubMed] [Google Scholar]
- Leonard E, Runguphan W, O'Connor S, Prather KJ (2009) Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol 5:292–300 [DOI] [PubMed] [Google Scholar]
- Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KL (2010) Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci USA 107:13654–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K, Ausubel FM (2006) Prospects for plant-derived antibacterials. Nat Biotechnol 24:1504–1507 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Wurtzel ET (2000) Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl Microbiol Biotechnol 53:396–400 [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol 85:1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 282:28791–28799 [DOI] [PubMed] [Google Scholar]
- Park JM, Kim TY, Lee SY (2009) Constraints-based genome-scale metabolic simulation for systems metabolic engineering. Biotechnol Adv 27:979–988 [DOI] [PubMed] [Google Scholar]
- Pharkya P, Maranas CD (2006) An optimization framework for identifying reaction activation/inhibition or elimination candidates for overproduction in microbial systems. Metab Eng 8:1–13 [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943 [DOI] [PubMed] [Google Scholar]
- Sanchez AM, Andrews J, Hussein I, Bennett GN, San KY (2006) Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol Prog 22:420–425 [DOI] [PubMed] [Google Scholar]
- Schmidt H, Jirstrand M (2006) Systems Biology Toolbox for MATLAB: a computational platform for research in systems biology. Bioinformatics 22:514–515 [DOI] [PubMed] [Google Scholar]
- Segre D, Vitkup D, Church GM (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA 99:15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarasu S, Ow DS, Lee SY, Lee MM, Oh SK, Karimi IA, Lee DY (2009) Characterizing Escherichia coli DH5alpha growth and metabolism in a complex medium using genome-scale flux analysis. Biotechnol Bioeng 102:923–934 [DOI] [PubMed] [Google Scholar]
- Wink M (2010) Biochemistry of plant secondary metabolism, 2nd edn. Wiley, Chichester [Google Scholar]
- Yuan LZ, Rouviere PE, Larossa RA, Suh W (2006) Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng 8:79–90 [DOI] [PubMed] [Google Scholar]
- Zhang H, Boghigian BA, Armando J, Pfeifer BA (2011) Methods and options for the heterologous production of complex natural products. Nat Prod Rep 28:125–151 [DOI] [PMC free article] [PubMed] [Google Scholar]