circATP2B4 is found to be associated with the development and progression of epithelial ovarian cancer. Extracellular vesicle-packaged circATP2B4 induces macrophage M2 polarization and cancer cell metastasis via SREBF1/PI3Kα/AKT signaling, suggesting a role as a diagnostic and therapeutic target.
Abstract
Ovarian cancer is one of the most common gynecologic malignancies with a highly immunosuppressive tumor microenvironment (TME) and poor prognosis. Circular RNA (circRNA) is a type of noncoding RNA with high stability, which has been shown to play an important role in biological processes and TME reprogramming in a variety of tumors. The biological function of a novel circRNA, circATP2B4, in epithelial ovarian cancer (EOC) was detected and evaluated. Transmission electron microscopy, differential ultracentrifugation and qRT-PCR were used to verify the existence of extracellular vesicles (EV)-packaged circATP2B4. Macrophage uptake of circATP2B4 was determined by EVs tracing. Dual luciferase reporter, FISH, Western blotting, and flow cytometry assays were used to investigate the interactions between circATP2B4 and miR-532-3p as well as sterol regulatory element-binding factor 1 (SREBF1) expression in macrophages. CircATP2B4 was upregulated in EOC tissues and positively correlated with ovarian cancer progression. Functionally, circATP2B4 promoted carcinogenic progression and metastasis of EOC both in vitro and in vivo. Mechanistically, EV-packaged circATP2B4 in EOC could be transmitted to infiltrated macrophages and acted as competing endogenous RNA of miR-532-3p to relieve the repressive effect of miR-532-3p on its target SREBF1. Furthermore, circATP2B4 induced macrophage M2 polarization by regulating the miR-532-3p/SREBF1/PI3Kα/AKT axis, thereby leading to immunosuppression and ovarian cancer metastasis. Collectively, these data indicate that circATP2B4-containing EVs generated by EOC cells promoted M2 macrophages polarization and malignant behaviors of EOC cells. Thus, targeting EVs-packaged circATP2B4 may provide a potential diagnosis and treatment strategy for ovarian cancer.
Introduction
Ovarian cancer is one of the most common gynecologic malignancies with high mortality worldwide (1). The main histologic type is epithelial ovarian cancer (EOC). Despite the rapid progress of chemotherapy and radiotherapy for ovarian cancer, the anatomic characteristics of ovarian cancer make its early diagnosis difficult (2, 3). Ovarian cancer often develops peritoneal metastasis in the process of progression, which aggravates the outcomes of patients (4, 5). Therefore, the discovery of novel diagnostic biomarkers, therapeutic targets and a better understanding of molecular mechanisms underlying the metastasis of EOC are crucial.
Circular RNAs (circRNA) are a class of regulatory noncoding RNA (ncRNA) molecules which form covalently closed continuous circular structures without 5′ caps and 3′ poly-A tails (6–8). CircRNAs are mainly produced by exon skipping, intron pairing-driven circularization, and RNA-binding proteins (9). Because circRNAs form covalent loop structures, they are more stable than linear RNAs and are resistant to exonucleolytic RNA decay (10, 11). CircRNAs are stable in expression and evolutionarily conserved in organisms, making circRNAs abundant in blood, exosomes, and body fluids (12). Therefore, circRNAs are considered to play key roles in the development of human cancers, including ovarian cancer.
Tumor-associated macrophages (TAM) derived from peripheral blood monocytes are recruited to microenvironment and polarized into M1 or M2 macrophages in response to secreted factors from cancer cells or microenvironmental cells (13). M1 macrophages express high amounts of inducible nitric oxide synthase (iNOS) and TNFα, playing a proinflammatory role and promoting immune responses that prevent oncogenic effects, whereas M2 macrophages express Arginase-1 (ARG1) and produce cytokines, growth factors and protease that are crucial for protumorigenic processes (13, 14). Furthermore, M2 macrophages stimulate cancer cell migration, invasion, tumor angiogenesis, and immunosuppression (13–15). It has been found that the sterol regulatory element-binding factors (SREBF or SREBP) are activated in TAMs during the development of tumors (16). SREBFs activate fatty acid synthesis in macrophages and facilitate macrophages towards M2 polarization, which in turn promotes tumor proliferation and metastasis (17).
In this study, based on publicly available EOC RNA sequencing (RNA-seq) data sets, a novel circRNA termed circATP2B4 derived from exons 20 and 21 of the ATP2B4 (circBase ID of hsa_circ_0000173) was identified. Subsequently, it was found that the expression of circATP2B4 in tumor was associated with the development and progression of EOC. Mechanistically, extracellular vesicles (EV)–packaged circATP2B4 were transmitted into infiltrated macrophages and acted as competing endogenous RNA (ceRNA) of miR-532-3p to relieve the repressive effect of miR-532-3p on its target SREBF1, and activated the SREBF1/PI3Kα/AKT pathway, leading to M2 polarization. Finally, circATP2B4 promoted the metastatic potential of EOC cells in vitro and in vivo. Taken together, our study reveals a novel function of circATP2B4 in tumor immunosuppression and metastasis of EOC.
Materials and Methods
EOC tissues RNA-seq analysis
First, circRNA transcripts were characterized via sequencing analysis of ribosomal RNA and linear RNA. The original fastq files selected for transcriptome sequencing (PRJNA396544) were derived from 6 EOC tissues and 3 healthy ovarian tissues, and 3 fallopian tube epithelium tissues serves as normal tissue controls (18). Each sample was sequenced on Illumina HiSeq and mapped to the human reference genome (GRCh37/hg19) by HISAT2. Qualified reads were sent to the FIND-CIRC v1.2 pipeline to identify and quantify circRNA counts with default parameters (19). A circRNA was recognized with the support of a minimum of two unique back-spliced reads. The data analysis was performed with R software (version 4.0.1) using the differentially expressed genes (DEG) package DEseq2. The threshold set for significant differences was log2|fold change| ≥ 1 and P value < 0.05.
Cell lines and culture
Human monocyte cell line THP-1 were purchased from the ATCC (Manassas, VA) in 2014. Human ovarian cancer cell lines SKOV3, ES-2, Ovcar3, mouse ovarian cancer cells ID8 and human epithelial kidney cell HEK293 were purchased from the Culture Collection of the Chinese Academy of Sciences (Shanghai, China) in 2016, EOC cell lines A2780 were purchased from iCell Bioscience Inc. (Shanghai, China) in 2018, normal ovarian epithelial cell line IOSE80 was purchased from the BeNa Culture Collection (Hebei, China) in 2019. Primary mouse bone marrow–derived macrophages (BMDM) were prepared from C57BL/6 mice by following a standard procedure as previously described (20). The immortalized mouse bone marrow–derived macrophage (iBMDM) cell line was a gift from Prof. Feng Shao (National Institute of Biological Sciences, Beijing, China) in 2015 (21). SKOV3 cells were cultured with MacCoy'5A medium supplemented with 10% FBS. ES-2, ID8 and HEK293 cells were cultured in DMEM supplemented with 10% FBS. A2780, THP1, and iBMDM cells were cultured with RPMI1640 medium supplemented with 10% FBS. Ovcar3 cells were cultured with RPMI1640 medium supplemented with 20% FBS and 0.01 mg/mL bovine insulin. The above media contained 100 U/mL penicillin-100 μg/mL streptomycin. All cell lines were frozen at passages 2 to 5 after purchase. Experiments were performed using passages 3 to 20 after removal from liquid nitrogen. Mycoplasma was tested for routinely using Myco-Lumi Mycoplasma Detector Kit (Beyotime). The most recent cell line authentication was in November 2019 by short tandem repeat analysis in Forensic Science Center, Zhejiang University, China. In a coculture model of macrophages and ovarian cancer cells, THP-1+PMA or BMDMs (1×106) were seeded in the lower chambers (0.4 μmol/L, Corning 3460). Ovarian cancer cells (1×105) were seeded in the upper chambers. Cells were cocultured at 37°C, 5% CO2 condition.
qRT-PCR
Total RNA was extracted from all cells using TRIzol reagent (Life Technologies, Carlsbad, CA). Reverse transcription was performed using 1,000 ng of total RNA and the High Capacity cDNA Reverse Transcription Kit (TaKaRa, Japan) according to the manufacturer's protocol. CFX-96 Quantitative PCR System (Bio-Rad, California) was used for qRT-PCR. The expression of β-actin (ACTB) was used as a control to calibrate the original mRNA concentrations in tissues and cells. Target gene expression was calculated using the 2 −ΔΔCT method. The primer sequences are detailed in the additional file Supplementary Table S3.
RNase R treatment
Total RNA (2 μg) was incubated for 15 min at 37°C with 5 U/μg RNase R (Epicentre Technologies, Madison, WI) and then analysed by RT-PCR.
Lentivirus, siRNA, plasmid construction, and cell transfection
Human full-length circATP2B4 (hsa_circ_0000173) and sh-circATP2B4 were selected and inserted into the pLCDH-ciR-Puro vector Genechem (Shanghai, China) for stable overexpression and knockdown, respectively, with empty plasmid used as a control. And si-circATP2B4, si-SREBF1 were purchased from Genechem (Shanghai, China). For the transfection of siRNAs and plasmids, cells were transfected using the Lipofectamine 3000 kit (Invitrogen, Carlsbad) according to the manufacturer's instructions. All sequences are listed in additional file Supplementary Table S3.
Immunofluorescent and IHC
Paraffin-embedded samples from mouse were sectioned at 5-μm thickness. Antigen retrieval was performed in 0.01 mol/L citrate buffer (pH 6.0) for 20 minutes using a pressure cooker. Cells for immunofluorescence (IF) were fixed with 4% paraformaldehyde for 20 minutes at room temperature and then permeabilized with 0.05% Triton X-100 in PBS for 5 minutes on ice. Samples were blocked in PBS containing 2% BSA for 1 hour at room temperature and incubated with CD68, CD163, CD9, N-Cadherin, E-Cadherin, CA125, and LDHA primary antibodies overnight at 4°C. Fluorescent probes labeled or HRP-conjugated secondary antibodies were incubated for 1 hour at room temperature. Multiplex IF was conducted using a multiplex fluorescence IHC staining kit (abs50013, ABSIN). Nuclei were then counterstained with DAPI. And images were acquired by laser scanning confocal (FV1200, Olympus) or fluorescence microscope (BX63, Olympus).
According to previous reports, CD68, CD163, CD206 staining scores were determined on the basis of tumor area coverage and percentage of positive staining for the detected proteins (22, 23). 3,3N-Diaminobenzidine Tetrahydrochloride (DAB) was used for immunodetection and nuclei were counterstained with hematoxylin. CD9, LDHA, N-Cadherin, E-Cadherin staining scores were determined according to the intensity and proportion of positive cells in 5 random fields under ×40 objective. The proportion of positively stained tumor cells in sections was graded as follows: 0, no positive cells; 1, < 25%; 2, 26% to 50%; 3, 51% to 75%; 4, > 76%. Cells were recorded for each staining intensity on a scale of 0 (no staining), 1 (light brown), 2 (brown), and 3 (dark brown). The staining index (SI) was calculated as follows: SI = staining intensity × proportion of positively stained cells. The CA125 color score was determined on the basis of the percentage of positive staining for CA125 in tumor cell membranes in the tumor area (24). Protein expression signal was excited and recorded with an Olympus BX61 Microscope (BX61, Olympus). Information on all antibodies used is listed in additional file Supplementary Table S4.
FISH and in situ hybridization
RNA FISH was performed using the Fluorescent In Situ Hybridization Kit (C10910, RiboBio) according to the manufacturer's instructions. Cy3-labeled probes targeting circATP2B4 and FITC-labeled SREBF1 probes (see Supplementary Table S3) were synthesized by RiboBio Technology Co. Ltd. Fluorescence was excited and recorded with a Olympus BX63 Microscope (BX63, Olympus). Fifteen EOC patient FFPE samples were used in the in situ hybridization (ISH) assay, which were diagnosed and tumor tissue surgically removed at Zhejiang University Women's Hospital in 2019 after informed consent by patients and included 10 patients without metastasis and 5 patients with peritoneal metastasis. The protocol of ISH performed on paraffin sections of ovarian cancer tissues or animal tumors was taken from previous literature (25). Briefly, after dewaxing and rehydration, the sections were digested with pepsin, and then hybridized with the digoxin-labelled nucleotide sequence (Synbio-Tech, China) overnight at 37°C. Next, the sections were incubated with anti-digoxin overnight at 4°C. The sections were stained with DAB and counterstained with hematoxylin. RNA expression excited and recorded with a Olympus BX61 Microscope (BX61, Olympus).
EV experiments
EVs from tumor culture medium were isolated as described by Chen and colleagues, purified and collected from ovarian cancer cell culture supernatants by differential ultracentrifugation (26). Briefly, cells were cultured in serum-free medium for 48 hours, then cell culture supernatants were collected and centrifuged at 3,000 rpm for 10 minutes at 4°C to remove cells and 1× 104g for 30 minutes at 4°C to remove cells and cell debris. The resulting supernatant was filtered through a 0.22-μm filter (Millipore) and then ultracentrifuged at 1×105g for 1 hour at 4°C. The pellet was resuspend in PBS and ultracentrifuge again at 1×105g for 1 hour at 4°C. ExoQuick Exosome Precipitation Solution (SBI) was used to isolate exosomes from the supernatant, and EVs RNA was extracted using the miRNeasy Mini Kit (Qiagen, CA).
Western blotting
Protein extracts were isolated from cells involved in this study using RIPA lysate kit (P0013B, Beyotime), quantified BCA Protein Assay Kit (P0012, Beyotime), and then electrophoresed through SDS-PAGE gels and transferred onto nitrocellulose filter membranes (Millipore) in equal amount, blocked with protein blocking buffer (C530040, Sangon) and incubated with primary antibodies at 4°C overnight. Membranes were then incubated with HRP-conjugated secondary antibodies, illuminated using chemiluminescence regents and visualized under a molecular imaging system (CLINX).
Northern blot analysis
CircATP2B4-specific probes (Supplementary Table S3) were synthesized by Fenghbio (China). circRNA was purified and enriched through RNase R treatment followed by polyadenylation and poly (A)+ RNA depletion, electrophoresed in a 2% agarose gel with 4% formaldehyde and transferred to charged nitrocellulose membrane (Millipore), UV cross-linked and hybridized with Cy3-labeled probes and radioactivated on an X-ray film. Analysis of circATP2B4 was performed in reference to a GeneRuler DNA ladder (SM0332, ThermoFisher) that was Cy3-coupled via a customized protocol (Telenbiotech).
Luciferase reporter assay
Similar luciferase reporter assay protocols were described in our previous report (27). Wild-type (WT) and binding site-specific mutated circATP2B4 and SREBF1-3′ UTR sequences were synthesized and integrated into psiCHECK 2.0 vectors (GenePharma, China) and co-transfected into HEK293 cells with miR-532-3p mimics or miR-532-3p inhibitors, respectively, using Lipofectamine 3000 kit (Invitrogen, Carlsbad). Cells were lysed and luciferase intensity was measured 48 hours after transfection. The ratio of Renilla luciferase over Firefly luciferase absorbance was calculated with the DualLuciferase Reporter Assay System (Beyotime, China). To quantitatively assess the interaction between circATP2B4 and miR-532-3p.
Enzyme-linked immunosorbent assay
The supernatant was collected for enzyme-linked immunosorbent assay with IL1β (MLbio, China), TNFα (MultiSciences, China), Arginase-1 (MultiSciences, China) and IL10 (MLbio, China) according to the manufacturer's instructions, and assessment of the levels of extracellular macrophage polarization markers. Results are expressed in pg/mL.
Flow cytometry analysis
Anti–CD11b-FITC and anti–CD206-PE were used to stain cells to detect CD11b + CD206+ macrophages. Anti–CD11b-FITC and anti–iNOS-PE were used to stain cells to detect CD11b + iNOS+ macrophages. Isotype controls were run in parallel. Flow cytometry (FCM) was performed using a CytoFLEX flow cytometer (Beckman Coulter). Antibody information is listed in Supplementary Data file Table S4.
Animal experiments
All mice (female 5-week-old BALB/c nude mice and female 5-week-old C57BL/6 mice) used in this study were housed and manipulated in the Laboratory Animal Center of Zhejiang University in obedience with the institutional guidelines for animal use and care. Mice were accommodated for adaption for 1 week before being injected with cells either subcutaneously or into intraperitoneal injection. GW4869 (inhibitor of extracellular vesicles release, TargetMol) was initially dissolved in DMSO into a stock solution of 100 mmol/L before dilution in PBS to 20 μmol/L for final injection and final DMSO concentration of 0.01%. The impact of vehicle (0.01% DMSO) on EVs secretion was also tested in C57BL/6 mice as a control. Anesthesia was induced and maintained by 10% isoflurane during exposure. Subcutaneous and intraperitoneal injection was performed with 100 μL normal saline suspension of 106 cells using a 1 mL insulin syringe. Weight of BALB/c nude mice and C57BL/6 mice was monitored weekly following injection. Euthanasia was initiated by CO2 suffocation and finished by cervical dislocation. Xenograft tumors and peritoneal tissues were collected following euthanasia.
Single-cell RNA-seq data analyses
Ovarian Cancer Metastasis Samples single-cell RNA-seq (scRNA-seq) data set were downloaded from Sequence Read Archive: PRJNA609094 and PRJNA612966 (28, 29). The CellRanger software (version 2.1.0) was used to process single-cell data to demultiplex, align reads against the GRCh38 human reference genome, and generate feature-barcode matrices. Raw matrix was preprocessed using computational methods deposited in the Seurat R package. To remove empty and low-quality droplets, containing dying cells, barcodes were removed from the analysis if they met one of the following conditions: genes per barcode < 500 or > 6,000, unique molecular identifiers (UMI) per barcode < 500, ratio of number of genes per number of UMI > 0.8 and a ratio of mitochondrial genes compared with all genes > 0.25. To cluster single cells by their expression profiles, we chose 20 principal components and used an unsupervised graph-based clustering algorithm Louvain (resolutions: 1.5). Normalization, clustering, differential gene expression analyses, and visualization were performed using the R package Seurat 4.1. Clustering of cells was performed using the implemented community identification method in the 'FindClusters' function. The specific marker genes of cell clusters were identified using the Seurat 'FindMarkers' function. Specifically, DEGs packages for a specific cluster were identified by comparing cells from that cluster to all other cells using the Wilcoxon Rank-Sum test.
Analysis of immune infiltrating cells from external cohorts
Identifying Ovarian cancer tissue infiltration immune cells in CIBERSORT software base on locally in R software 4.0.1 (30). TCGA-OV cohort inclusion criteria: human ovarian cancer cohort, RNAseq or microarray data from ovarian cancer tissues. Exclusion criteria: fewer than 40 patients in a cohort, no prognostic information. The RNA-seq (Counts or RPKM format) of TCGA-OV, AOCS(GSE9899) and GSE30161 was analyzed to obtain the abundance ratio matrix of CIBERSORT 22 Immune Cells. Then, the correlation analysis was conducted among the contents of the Macrophages M2 type score in the 3 data sets samples. Next, the Kaplan–Meier analysis for overall survival was calculated on the basis of Macrophages M2 score cut-off value. Log-rank test was used to detect the difference in survival, and log-rank P < 0.05 was considered to be statistically significant. We also used the Tumor Immune Estimation Resource (TIMER2.0) database (http://timer.cistrome.org/) to analyze the correlation of ovarian cancer-infiltrating macrophages with gene expression.
Statistical analysis
Bioinformatic prediction and circRNA-miRNA network was generated in miRDB, miRarBase, miRwalk, and TargetScan. Ordinary one-way ANOVA analysis (multiple comparisons were included when necessary) was performed in GraphPad Prism 8.0 (GraphPad Software) to compare protein expression differences among groups in Western blots and IF assays. Means Embedded Image SD were presented in quantification bar graphs. Kaplan–Meier and Cox regression analyses were conducted in SPSS (V22.0, IBM). Changes in mouse body weight among groups were compared by t test and one-way ANOVA analysis (multiple comparisons were included when necessary). A value of P < 0.05 was considered statistically significant.
Ethics approval and consent to participate
The collection and use of patient samples complied with the Declaration of Helsinki and were approved by the Ethics Committee of Zhejiang University Women's Hospital (IRB020200135), and informed written consent was obtained from patients. Animals experiment ethics approval was issued by the Ethics Committee of Laboratory Animal Center of Zhejiang University (ZJU20210181).
Data availability
All data obtained in this work can be acquired from the National Center for Biotechnology Information: RNA-seq data set (PRJNA396544), scRNA-seq data sets (PRJNA609094 and PRJNA612966). TCGA-OV database (https://xenabrowser.net/datapages/), AOCS and GSE30161 database (CuratedOvarianData for R package), and the analysis results of circRNA sequencing and mRNA sequencing data were released when the paper was published. The data generated in this study are available upon request from the corresponding author.
Results
Identification of differentially expressed circATP2B4 in EOC
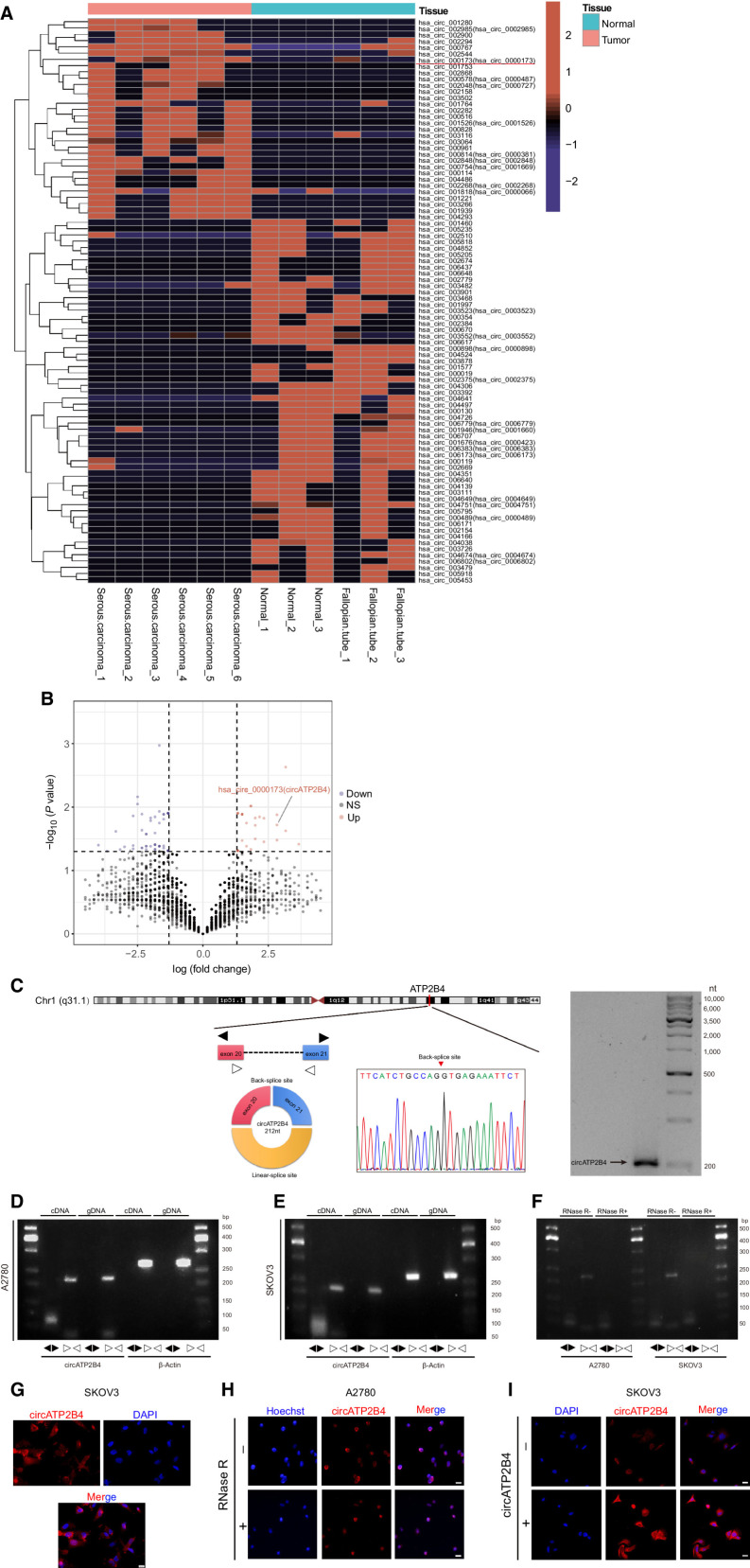
CircRNA expression profiles in six EOCs and paired six normal control tissues were analyzed to screen for differentially expressed circRNAs in ovarian cancer. In all samples, a total of 6,959 distinct circRNA candidates were found. Among them, 32 circRNAs were upregulated and 58 circRNAs were downregulated in EOC tissues compared with control tissues (Fig. 1A and B, Supplementary Fig. S1A and S1B, log2|fold change| ≥ 1 and P value < 0.05). The circBase database (http://circrna.org/) describes circATP2B4 (hsa_circ_0000173, Alias_000173) as back-spliced between exon 20 and exon 21 of the ATP2B4 gene with a length of 212 nt. Northern blot analysis identified the expression of circATP2B4 in ovarian cancer cells, and Sanger sequencing further confirmed the sequence of the head-to-tail junction site (Fig. 1C).
Figure 1.
CircRNA expression profiles in EOC and characterization of circATP2B4. A, Hierarchical clustering of circRNA differential expression profiles between 6 EOC samples and 6 normal samples. The heat map was generated from differentially expressed circRNAs. B, Volcano map of deferentially expressed circRNAs. C, Schematic illustration of the genomic location and back splicing of circATP2B4 with the splicing site validated by Sanger sequencing and Northern blotting detection results of circATP2B4. D, Combining PCR with an electrophoresis assay indicated the presence of circATP2B4 using divergent and convergent primers from cDNA or genomic DNA (gDNA) in A2780 cells. E, Combining PCR with an electrophoresis assay indicated the presence of circATP2B4 using divergent and convergent primers from cDNA or gDNA in SKOV3 cells. F, The expression of circATP2B4 and ATP2B4 mRNA in both A2780 and SKOV3 cell lines was detected by PCR assay followed by nucleic acid electrophoresis in the presence or absence of RNase R. G, FISH analysis showed that circATP2B4 was mainly localized in the cytoplasm and a small part in the nucleus. Nucleus were stained blue with DAPI and cytoplasmic circATP2B4 was stained Cy3 (red). Scale bars, 20 μmol/L. H, RNA FISH analysis revealed the cellular localization of circATP2B4 in EOC cells when cells were treated with RNase R. Scale bars, 20 μmol/L. I, RNA FISH analysis revealed the expression of circATP2B4 in SKOV3 cells when cells were transfection of circATP2B4 plasmid. Scale bars, 20 μmol/L. The symbols * and ** show P < 0.05 and 0.01, respectively.
Next, the expression of circATP2B4 was checked in the A2780, ES-2, OVCAR3, SKOV3 cell lines and ovarian epithelial cell line IOSE80. Among these cell lines, A2780 and SKOV3 showed the highest expression and the lowest expression of circATP2B4, respectively (Supplementary Fig. S1C). Thus, we selected A2780 and SKOV3 cell lines to investigate the expression regulatory models of circATP2B4. PCR and agarose gel electrophoresis were performed with divergent and convergent primers, in which we found back-spliced or canonical forms of circATP2B4, rather than linear ATP2B4 (Fig. 1D and E). RNase R digestion resistance substantiated the existence of the circular form of circATP2B4 (Fig. 1F; Supplementary Fig. S1D–S1F).
Moreover, we conducted RNA-FISH, which revealed that circATP2B4 was mainly located in the cytoplasm, with expression also observed in the nucleus (Fig. 1G). RNase R digestion did not reduce the expression of circATP2B4, and upregulating the expression of circATP2B4 increased its FISH signal (Fig. 1H and I; Supplementary Fig. S1G–S1J).
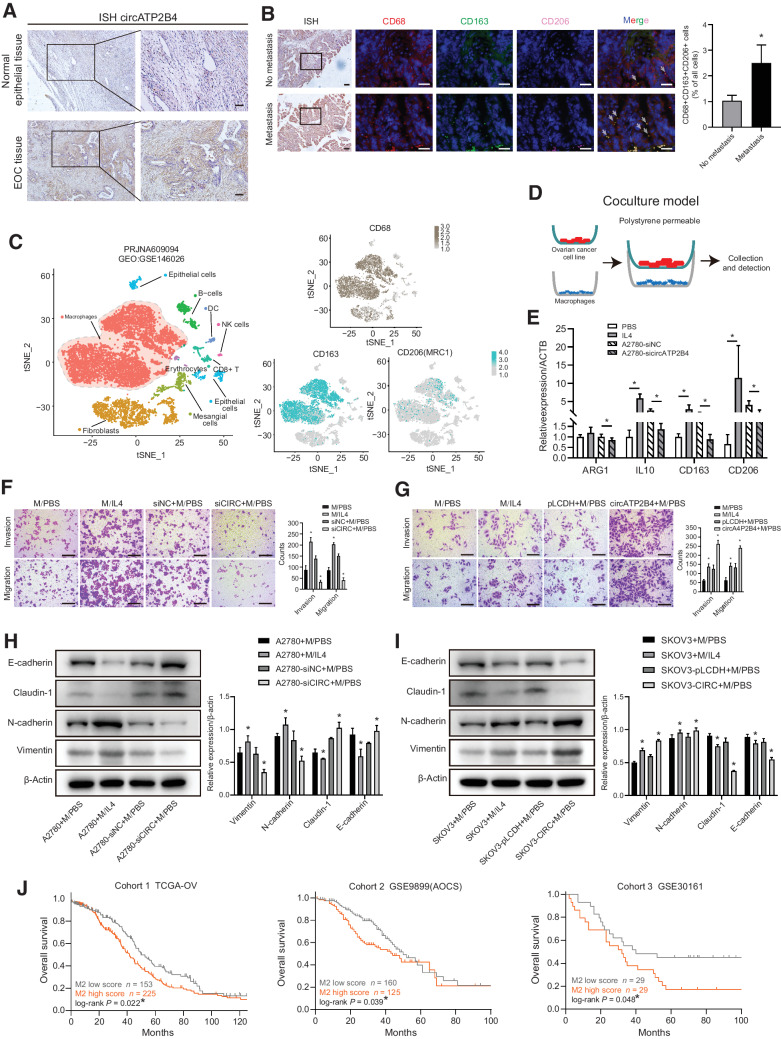
CircATP2B4 overexpression is associated with M2 macrophages and promotes peritoneal metastasis of ovarian cancer cells
To investigate the potential biological role of circATP2B4 in the progression of ovarian cancer, we performed ISH of excised EOCs tissue sections, which indicated that the expression of circATP2B4 in EOC tissues was higher than that in the normal epithelial tissue (Fig 2A; Supplementary Fig. S1G). To further explore the correlation between infiltration of TAMs and metastasis of ovarian cancer, we performed immunostaining to evaluate CD68+ TAMs and the markers of M2 macrophages (CD163+, CD206+). We observed high abundance of circATP2B4 expression in metastatic ovarian cancer tissues, accompanied by high M2 macrophage infiltration in ovarian cancer tissues (Fig 2B; Supplementary Fig. S2A). To investigate the cellularity of the microenvironment of ovarian cancer peritoneal metastasis, and to understand the distribution and types of macrophages in peritoneal metastasis, we analyzed the ovarian cancer peritoneal metastasis data set PRJNA609094 (GEO:GSE146026). The cellular composition was explored with unbiased clustering across all cells by Principal Component Analysis and visualized by t-distributed stochastic neighbor embedding (TSNE). There were a large number of macrophages infiltrating the microenvironment of ovarian cancer peritoneal metastasis, and the high expression of CD163+ and CD206+ suggested M2 polarization (Fig. 2C). Similar results were found in the analysis of the scRNA data set PRJNA612966 (GEO:GSE147082) for ovarian cancer (Supplementary Fig. S2B).
Figure 2.
The correlation of circATP2B4 and M2 macrophages affects ovarian cancer progression. A, Expression of circATP2B4 was examined by ISH in normal ovarian epithelial tissues and EOC tissues. Scale bars, 50 μm. B, ISH and multiplex fluorescent IHC of EOC nonmetastatic and metastatic tissue sections, DAB-circATP2B4, blue-DAPI, red-CD68, green-CD163, pink-CD206, representative images and IF with quantification. Scale bars, 20 μm. C, TSNE plot showed the clusters of scRNA-seq in the ovarian cancer peritoneal metastasis samples from PRJNA609094 (GEO:GSE146026). Each dot is a cell colored by its analyzed cell types. D, Schematic illustration of the in vitro indirect coculture system. E, Macrophages (THP-1 with PMA) were cocultured with A2780-siNC or A2780-sicircATP2B4, or treated with control (PBS and IL4). After 48 hours, qRT-PCR was applied using primers for M2 markers (ARG1, IL10, CD163, CD206). The group treated with IL4 was used as positive control. F, Migration and invasion capacity of A2780 cocultured with macrophages (M, THP-1 with PMA) cocultured with A2780-siNC or A2780-sicircATP2B4, or treated with control (PBS and IL4) by the in vitro transwell coculture system. Scale bars, 300 μm. G, Migration and invasion capacity of SKOV3 cocultured with macrophages (M, THP-1 with PMA) cocultured with SKOV3-pLCDH or SKOV3-circATP2B4, or treated with control (PBS and IL4) by the in vitro transwell coculture system. Representative photographs of migratory or invaded cells on the membrane coated with or without Matrigel (magnification, ×100) are shown. Scale bars, 300 μm. H, The effect of the supernatants of macrophages cocultured with A2780-siNC or A2780-sicircATP2B4, or treated with control (PBS and IL4) on the epithelial-mesenchymal transition (EMT) of A2780 was analyzed by Western blot analysis (*, P < 0.05). I, The effect of the supernatants of macrophages cocultured with SKOV3-pLCDH or SKOV3-circATP2B4, or treated with control (PBS and IL4) on the EMT of SKOV3 was analyzed by Western blot analysis. Densitometry shows relative protein expression normalized for β-actin. Data are representative of three independent experiments (*, P < 0.05). J, Kaplan–Meier survival curves in human ovarian cancer according to M2 macrophages infiltration abundance in TCGA-OV, AOCS, and GSE30161 data sets. *, log-rank P < 0.05.
The macrophage polarization and induction of ovarian cancer cell migration, invasion, and epithelial-mesenchymal transition (EMT) of ovarian cancer cells were subsequently evaluated using the in vitro indirect coculture system (Fig. 2D). Compared with A2780 cells treated by negative control siRNA (si-NC), the expression of M2 markers, including Arginase-1 (ARG1), IL10 (IL10), CD163, CD206 were significantly decreased in macrophages cocultured with A2780 cells treated with si-circATP2B4 (Fig. 2E). Macrophages treated with IL4 or PBS were taken as positive and NCs for M2 polarization, respectively. M2-polarized macrophages activated by IL4 significantly enhanced the invasion and migration ability of A2780 cells, which could be significantly suppressed by silencing circATP2B4 (Fig. 2F). The invasion and migration ability of SKOV3 was significantly enhanced with IL4-activated M2 macrophages in comparison to that of PBS-treated macrophages and further by ectopic expression of circATP2B4 (Fig. 2G).
In addition, we further assessed whether activated M2 macrophages could regulate EMT of ovarian cancer cells. After coculturing with macrophages induced by treatment of EOC cell lines culture supernatant for 24 hours, the expression of epithelial cell markers (E-cadherin, Claudin-1) decreased and the expression of mesenchymal cell markers (N-cadherin, Vimentin) increased. Correspondingly, knockdown of circATP2B4 suppressed EMT in A2780 cells (Fig. 2H). Similar results were obtained in SKOV3 cells (Fig. 2I). To evaluate the effect of M2 macrophage infiltration on the development of ovarian cancer, we selected 3 public ovarian cancer cohorts (TCGA, AOCS, GSE30161). Using Cibersort to analyze bulk RNA-seq data of ovarian cancer tissues (Supplementary Fig. S2C and S2D), we found that the prognosis of patients with high abundance of M2 macrophages was significantly worse than that of patients with low abundance of M2 macrophages (log-rank P < 0.05; Fig. 2J).
Taken together, these data suggest that ovarian cancer cell circATP2B4 induced M2 macrophage activation and promoted ovarian cancer cell migration, invasion, and EMT.
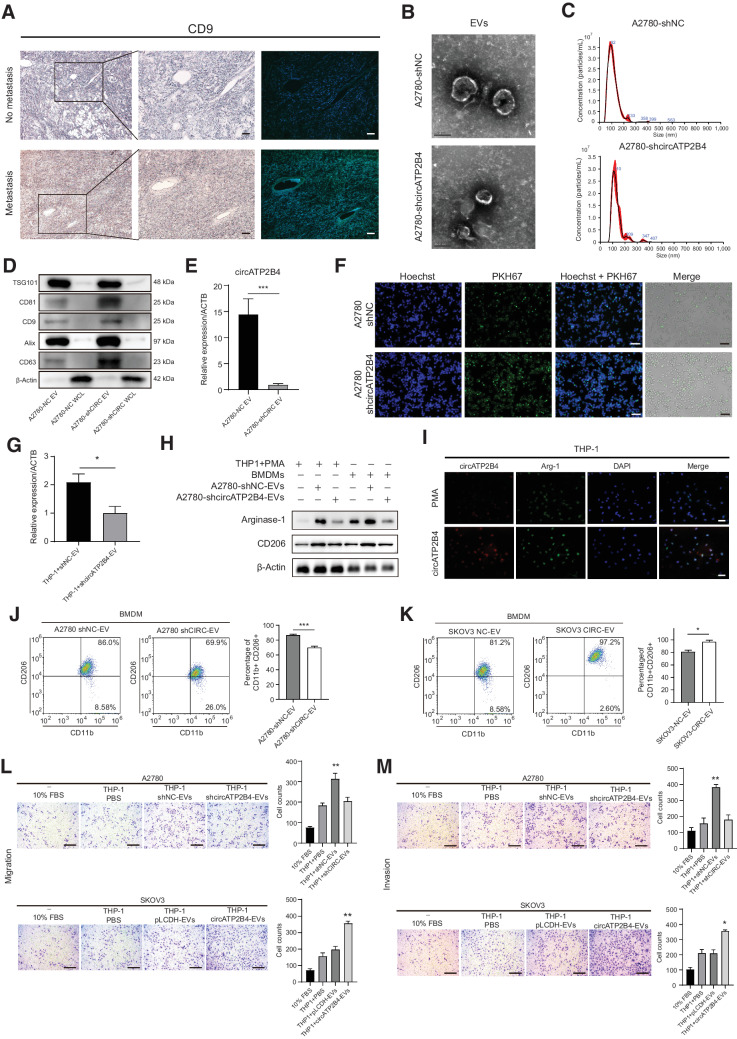
CircATP2B4 is highly expressed in EVs from EOC cells and can be transferred into macrophages
To investigate the mechanism by which EVs in EOC cells induce M2 polarization of macrophages in the tumor microenvironment, the expression of CD9 (a marker for EVs) was examined by IHC and IF in metastatic samples and was significantly higher compared with that in ovarian cancer tissues without metastasis (Fig. 3A). Next, EVs were isolated from conditioned media of A2780 cells after 24 hours and quantified by electron microscopy and nanoparticle tracking analysis (NTA). Electron microscopy revealed typical rounded particles ranging from 50 to 150 nm in diameter (Fig. 3B), and NTA revealed a similar size distribution of EVs (Fig. 3C). Western blot analysis of the protein extract from EVs confirmed the presence of markers of the exosomal proteins CD63, ALIX, CD9, CD81, and TSG101, where the whole cell lysate served as the control (Fig. 3D). Moreover, the expression of circATP2B4 was lower in EVs of A2780-shcircATP2B4 compared with that of circATP2B4 in EVs of A2780-shNC cells (P < 0.01; Fig. 3E).
Figure 3.
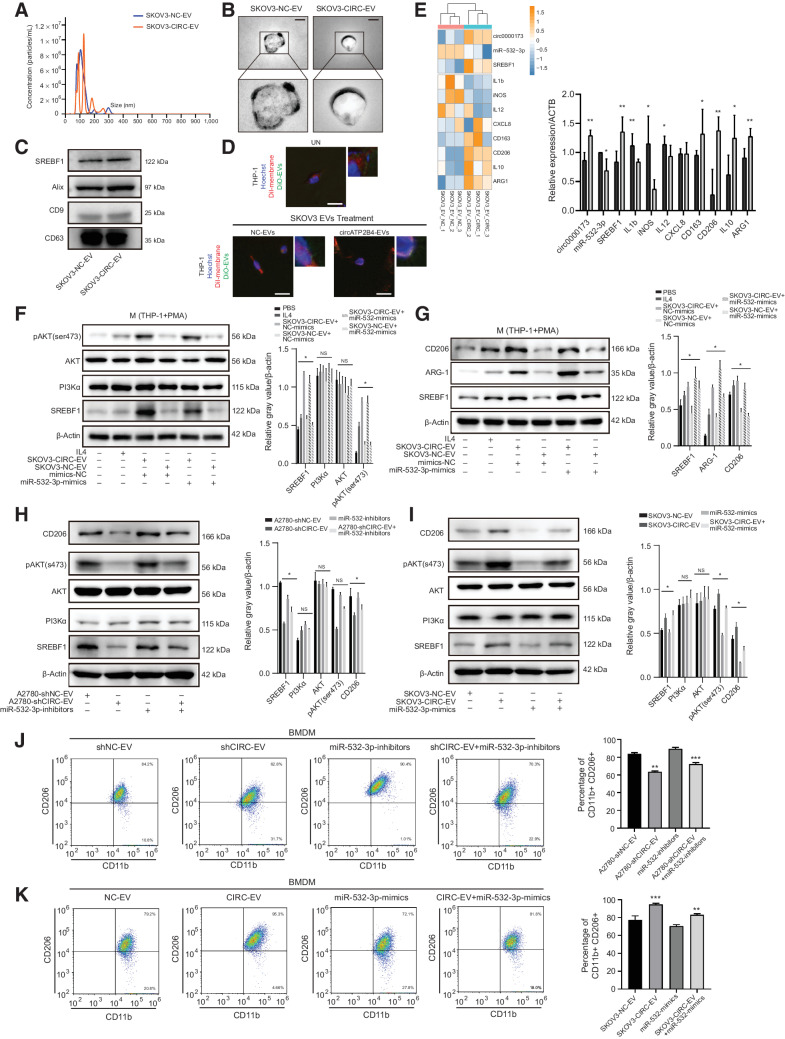
EVs-packaged circATP2B4 induces the polarization of M2 macrophages to promote the migration, invasion, and epithelial-mesenchymal transition (EMT) of ovarian cancer cells. A, Expression of CD9 was examined by IHC and IF in EOC non-metastatic and metastatic tissue section (magnification, ×100 and ×200). Scale bars, 50 μm. B, Electron microscopy images of EVs isolated from conditioned medium of A2780-shNC and A2780-shcircATP2B4. Scale bars, 100 nm. C, NTA analysis of EVs isolated from A2780-shNC and A2780-shcircATP2B4 cell culture supernatants. D, Western blot analysis for EVs marker proteins CD63, ALIX, CD9, CD81, and TSG101, whole cell lysate protein as control. E, The expression of circATP2B4 in EVs extracted from A2780-shNC and A2780-shcircATP2B4 were detected by qRT-PCR. F, Representative IF image shows the internalization of PKH67-labeled A2780-derived EVs (green) by macrophages. Scale bars, 100 μm. G, qRT-PCR detection of A2780-shNC and A2780-shcircATP2B4 EVs added to macrophages for 24 hours, the expression of circATP2B4 in macrophages. H, Western blotting analysis was used to detect M2 marker Arginase-1 and CD206 of Macrophages (THP-1+PMA and BMDMs), expression was normalized to β-actin. I, FISH-IF detected the RNA and arginase-1 protein expression of circATP2B4 in macrophages (THP-1+PMA) after adding A2780-EVs. Scale bars, 20 μm. J and K, FCM analysis macrophages (BMDMs) treated with EVs from A2780 (A2780-shNC and A2780-shcircATP2B4) or SKOV3 (SKOV3-NC and SKOV3-circATP2B4), M2 macrophages (CD11b+ and CD206+). L, Migration capacity of A2780 (10%FBS, THP-1+PBS, THP-1+shNC-EVs, THP-1+shcircATP2B4-EVs) and SKOV3 (10%FBS, THP-1+PBS, THP-1+pLCDH-EVs, THP-1+circATP2B4-EVs) after coculture with macrophages, by the in vitro transwell coculture system without Matrigel are shown. Scale bars, 300 μm. M, Invasion capacity of A2780 (10%FBS, THP-1+PBS, THP-1+shNC-EVs, THP-1+shcircATP2B4-EVs) and SKOV3 (10%FBS, THP-1+PBS, THP-1+pLCDH-EVs, THP-1+circATP2B4-EVs) after coculture with macrophages, by the in vitro transwell coculture system with Matrigel are shown. Scale bars, 300 μm. Data are shown as mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. not significant.
Next, the effects of EVs on macrophage polarization were examined. When cocultured with macrophages, EVs labeled with fluorescent PKH67 from A2780 cells without or with circATP2B4 knockdown were internalized by unstained macrophages over time (Fig. 3F). The expression of circATP2B4 in macrophages treated with circATP2B4 knockdown EVs was significantly lower than that in macrophages treated with circATP2B4-NC EVs (P < 0.01; Fig. 3G). The expression of M2 markers (ARG-1 and CD206) increased in macrophages incubated with EVs from A2780 cells without circATP2B4 knockdown. In contrast, incubation of EVs from A2780 cells with circATP2B4 knockdown EVs resulted in decreased expression of M2 markers, which was validated in both THP1 and BMDMs (Fig. 3H). FISH and IF assays showed that the RNA abundance of circATP2B4 was upregulated in THP-1 cells, and the Arginase-1 protein expression was also increased after adding A2780-EVs (Fig. 3I). FCM results further displayed that circATP2B4 significantly upregulated the M2 macrophage markers CD11b and CD206. EVs from A2780 transfected with shcircATP2B4 and incubated with BMDM reduced the proportion of M2 macrophages (Fig. 3J). Consistent with this, incubation of BMDM with SKOV3-circATP2B4 EVs can increase the proportion of M2 macrophages (Fig. 3K). Furthermore, validation experiments of downregulation and upregulation of circATP2B4 were also performed on SKOV3 and A2780, respectively (Supplementary Fig. S3A and Supplementary Fig. S3B). EVs from SKOV3 transfected with shcircATP2B4 and incubated with BMDM reduced the proportion of M2 macrophages (Supplementary Fig. S3C), and incubation of BMDM with SKOV3-circATP2B4 EVs can increase the proportion of M2 macrophages (Supplementary Fig. S3D).
Moreover, both the migration and invasion abilities of A2780 cells cocultured with macrophages transfected with shcircATP2B4 were decreased (Fig. 3L and M). Similarly, the migration and invasion abilities of SKOV3 cells cocultured with macrophages transfected with circATP2B4 were increased (Fig. 3L and M). Together, these in vitro data indicate that ovarian cancer cells could transport circATP2B4 into macrophages through EVs, thereby promoting M2 polarization of macrophages.
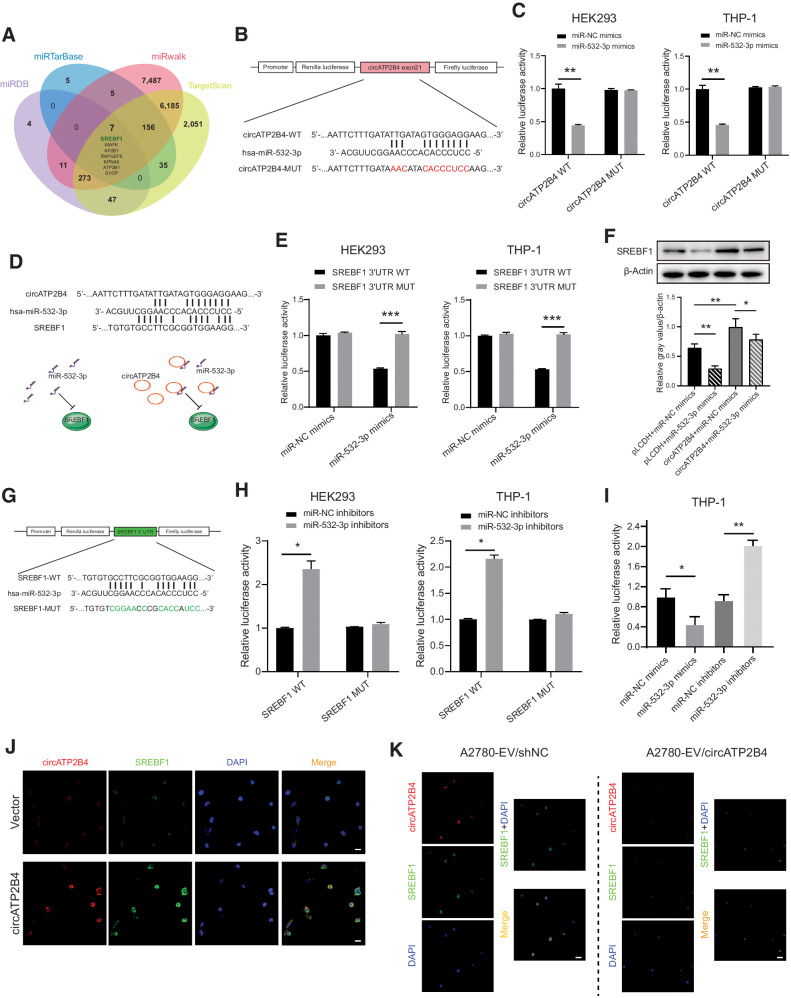
CircATP2B4 directly sponges miR-532-3p with SREBF1 as a downstream target
According to the ceRNA theory, circRNAs are able to function as miRNA sponges and subsequently regulate mRNA expression (31). The potential targets of circATP2B4 were predicted using the database circBASE and CSCD (Cancer-specific CircRNA Database), which indicated that miR-532-3p was a downstream target of circATP2B4. Seven candidate genes were screened for further validation according to the combination with the results predicted by the software miRDB, miRarBase, miRwalk, and TargetScan (Fig. 4A; Supplementary Fig. S4A and Supplementary Fig. S4B). TIMER software was used to evaluate the correlation of candidate genes with the expression of ovarian cancer-infiltrating macrophages, and SREBF1 was identified as the target mRNA for exploration (Supplementary Fig. S4C). To determine the complementarily base-pairing interaction between circATP2B4 and miR-532-3p, the binding site on circATP2B4 was mutated (Fig. 4B). As a result, dual-luciferase reports indicated that by loss of binding site, circATP2B4 failed to sequester miR-532-3p (Fig. 4C; Supplementary Fig. S4D).
Figure 4.
SREBF1 was increased in M2 macrophages regulated by circATP2B4 directly sponges miR-532-3p. A, Schematic illustration exhibiting overlapping of the target of circATP2B4 predicted by miRDB, miRarBase, miRwalk and TargetScan. B and C, Illustration of the WT and mutant (MUT) binding site sequences in circATP2B4 for miR-532-3p. Relative luciferase intensity was quantified as ratio of Renilla (R) over Firefly (F) luciferase activities in HEK293 and macrophages (THP-1+PMA). D and E, Schematic illustration of the WT and mutant (MUT) binding site sequences in miR-532-3p for circATP2B4 and SREBF1. Relative luciferase intensity was quantified as ratio of Renilla (R) over Firefly (F) luciferase activities in HEK293 and macrophages (THP-1+PMA). F, Expression of SREBF1 protein after treatment with circATP2B4, mimics of miR-532-3p and NCs were measured by western blot analysis. G and H, Schematic illustration of the WT and mutant (MUT) binding site sequences in SREBF1 for miR-532-3p in HEK293 and macrophages (THP-1+PMA). Relative luciferase intensity was quantified as ratio of Renilla (R) over Firefly (F) luciferase activities. I, Relative luciferase intensity in mimics or inhibitors of miR-532-3p was quantified as ratio of Renilla (R) over Firefly (F) luciferase activities in macrophages (THP-1+PMA). J, Expression of circATP2B4 and SREBF1 detected by IF-FISH in macrophages transfected with circATP2B4. Scale bars, 20 μm. K, Macrophages were treated to shNC-EV and shCIRC-EV of A2780 cells, and the expression of circATP2B4 and SREBF1 were detected by IF-FISH. Scale bars, 20 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. not significant.
We then performed a luciferase reporter assay with SREBF1 3′ UTR luciferase reporter plasmids (SREBF1 3′UTR-MUT) that containing mutated sequences of these recognition sites (Fig. 4D). The results showed that the luciferase activity was dramatically decreased after co-transfection of miR-532-3p mimics and SREBF1 3′ UTR-WT compared with that of SREBF1 3′UTR-MUT in both HEK293 and THP-1 cells (Fig. 4E). The result indicates that miR-532-3p interacted with SREBF1 3′UTR through recognition of these sequences. Moreover, the results of Western blot analysis further showed that overexpression of circATP2B4 can increase the expression of SREBF1, and overexpression of miR-532-3p can reduce the expression of SREBF1 (Fig. 4F; Supplementary Fig S4E–S4H).
Next, to verify the results of bioinformatics prediction analysis, dual-luciferase reporter assays were performed in THP-1 and HEK293 cell lines. SREBF1 and its mutant version without miR-532-3p binding sites were subcloned into reporter plasmids (Fig. 4G). The results showed that the luciferase activity of SREBF1-WT group significantly decreased in the miR-532-3p mimic group and increased in the miR-532-3p inhibitor group compared with the NC group in HEK293 and THP-1 cell line model (Fig. 4H; Supplementary Fig S4E). And there was no difference among miR-532-3p mimics, inhibitors group and NC group in the luciferase activity of SREBF1-MUT group (Supplementary Fig. S4I). Similar results were observed in THP-1 cells transfected with either miR-532-3p mimics or miR-532-3p inhibitors (Fig. 4I). These results suggest that direct interaction might exists between SREBF1 and miR-532-3p. Considering that circRNAs may serve as miRNA sponges in the cytoplasm, FISH assays were performed in THP-1 cells to observe the subcellular localization of circATP2B4 and SREBF1. We noticed that most of circATP2B4 (red) and SREBF1 (green) were colocalized in the cytoplasm (Fig. 4J; Supplementary Fig. S4J). After collecting and analyzing EVs from ovarian cancer cells and then incubating with THP-1 cells, we found that the expression of circATP2B4 and SREBF1 was relatively reduced in A2780 cells and macrophages incubated with shcircATP2B4 EVs, compared with the NC group (Fig. 4K). Collectively, these data demonstrate that circATP2B4 serves as a ceRNA for miR-532-3p to regulate SREBF1 expression, thus leading to M2 polarization of macrophages.
EVs-packaged circATP2B4 polarize M2 macrophages through miR-532-3p/SREBF1/PI3K/AKT
Next, we explored the mechanism of how EVs circATP2B4 released by EOCs affect macrophages M2 polarization. The concentration and size distribution of EVs derived from SKOV3 cells with stable circATP2B4 overexpression were analyzed by the Nanosight LM10 and TEM, as shown in Fig. 5A and B, respectively. Consistent with the results above, the presence of the exosomal proteins CD63, CD9, ALIX was verified (Fig. 5C). There was no significant difference in the expression of SREBF1 protein in the EVs of SKOV3-NC and SKOV3-circATP2B4. The biomarkers of the EVs were also evaluated by dynamic confocal microscopy, showing that EV enters into macrophage (THP1+PMA) after treatment (Fig. 5D). The detection of circATP2B4 by FISH showed that treated with SKOV3-circATP2B4 EVs increased the expression of circATP2B4 in THP-1, whereas the expression of circATP2B4 was lower in macrophages treated with EVs of SKOV3-NC (Supplementary Fig. S5A).
Figure 5.
EVs-packaged circATP2B4 polarized the M2 macrophage via SREBF1/PI3Kα/AKT signaling pathway. A, NTA analysis of EVs isolated from SKOV3-NC and SKOV3-circATP2B4 cell culture supernatants. B, Electron microscopy images of EVs isolated from conditioned medium of SKOV3-NC and SKOV3-circATP2B4. Scale bars, 50 nm. C, Western blot analysis for EVs marker proteins (CD63, ALIX, and CD9) and SREBF1 protein expression in SKOV3-NC and SKOV3-circATP2B4 EVs. D, Representative confocal microscopy images of the THP-1(PMA) cells assessing the colocalization of EVs (DiO), cell membranes (DiI), and cell nucleus (Hoechst). Boxes indicate the areas shown at higher magnification in the upper right corner. UN, THP-1(PMA) without any treatment. Scale bars, 10 μm. E, RT-PCR for M1 markers (IL1b, iNOS, IL12, CXCL8) and M2 markers (ARG1, IL10, CD163, CD206) on macrophages (THP-1 with PMA) treated with EVs from SKOV3-NC or SKOV3-circATP2B4. Relative expression to β-actin (ACTB). F–G, The effect of the supernatants of macrophages treated with SKOV3-EVs and miR-532-3p mimics, with control (IL4) on SREBF1, PI3Kα/AKT (F) and M2 marker genes (G) analyzed by Western blot analysis. H and I, The effect of macrophages treated with EVs from A2780 (A2780-shNC and A2780-shcircATP2B4) or SKOV3 (SKOV3-NC and SKOV3-circATP2B4) and miR-532-3p inhibitors or miR-532-3p mimics were analyzed by Western blot analysis. J and K, FCM analysis macrophages (BMDM) treated with EVs from A2780 (A2780-shNC and A2780-shcircATP2B4) or SKOV3 (SKOV3-NC and SKOV3-circATP2B4) and miR-532-3p inhibitors or miR-532-3p mimics, M2 macrophages (CD11b+ and CD206+). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, macrophages were treated with EVs from SKOV3 and the expression of M1 and M2 marker genes in macrophages was observed. The expression of M1 markers (IL1b, iNOS, IL12, and CXCL8) decreased, whereas that of M2 markers (CD163, CD206, IL10, and ARG1) increased in macrophages incubated with SKOV3-circATP2B4 compared with SKOV3-NC (Fig 5E). Moreover, overexpression of miR-532-3p significantly inhibited SREBF1 mRNA and protein in macrophages, and inhibition of miR-532-3p revealed the opposite effects (Supplementary Fig. S4E–S4G and Supplementary Fig. S5B).
Activation of the PI3Kα (PI3 Kinase p110α) signaling pathway participates in M2 polarization, which facilitates tumor progression (32). Thus, it was speculated that EOC-derived EVs could promote polarization of macrophages through the PI3Kα signaling pathway. Our results showed that the expression of PI3Kα/AKT in macrophages treated with EOC EVs was higher compared with that in macrophages treated with A2780-shcircATP2B4 EVs (Fig. 5F and G). In addition, the expression of SREBF1 as well as the phosphorylation of AKT increased after treatment with EVs derived from EOC cells (A2780 and SKOV3) (Supplementary Fig. S5C). To investigate whether EVs-packaged circATP2B4 regulates macrophage polarization via the miR-532-3p/SREBF1/PI3Kα signaling pathway, the expression of CD206, PI3Kα, and phosphorylation of AKT in macrophages treated with A2780-shcircATP2B4 EVs or transfected with miR-532-3p inhibitors/mimics was further detected. As shown in Fig. 5H and I, the expression of SREBF1, PI3Kα, and pAKT (ser473) was downregulated in the circATP2B4 knocked down-EVs-treated macrophages groups. Moreover, the expression of both SREBF1 and pAKT (ser473) by macrophages cocultured with A2780 transfected with miR-532-3p inhibitors was increased, and this was reversed in circATP2B4 knockdown cells. The changes in protein expression of SREBF1 and pAKT were also demonstrated in a coculture of SKOV3 cells overexpressing circATP2B4 and after the addition of miR-532-3p mimics (Fig. 5H and I; Supplementary Fig. S4D–S4G).
FCM results further demonstrated that circATP2B4 overexpression significantly upregulated the M2 macrophage markers (CD11+ and CD206+) and downregulated the M1 macrophage markers (CD11+ and iNOS+; Fig. 5J). A2780 EVs transfected with shcircATP2B4 and incubated with BMDM reduced the proportion of M2 macrophages, whereas transfection of macrophages with miR-532-3p inhibitor restored this, increased the proportion of M2 marker-positive macrophages andalso reduced the proportion of M1 marker-positive macrophages (Supplementary Fig. S5G). Consistent with this, incubation of BMDM with SKOV3-circATP2B4 EVs increased the proportion of M2 macrophages, and transfection of macrophages with miR-532-3p mimics reversed the increase, down-regulated the proportion of M2 macrophages (Fig. 5K), and upregulated the proportion of M1 macrophages (Supplementary Fig. S5H). Overall, these results suggested that EOC cell EVs could deliver circATP2B4 to TAMs, thereby enabling them to acquire M2 polarization and immunosuppressive properties.
EOCs releasing circATP2B4 in EVs enhance migration, invasion, and EMT of ovarian cancer cells
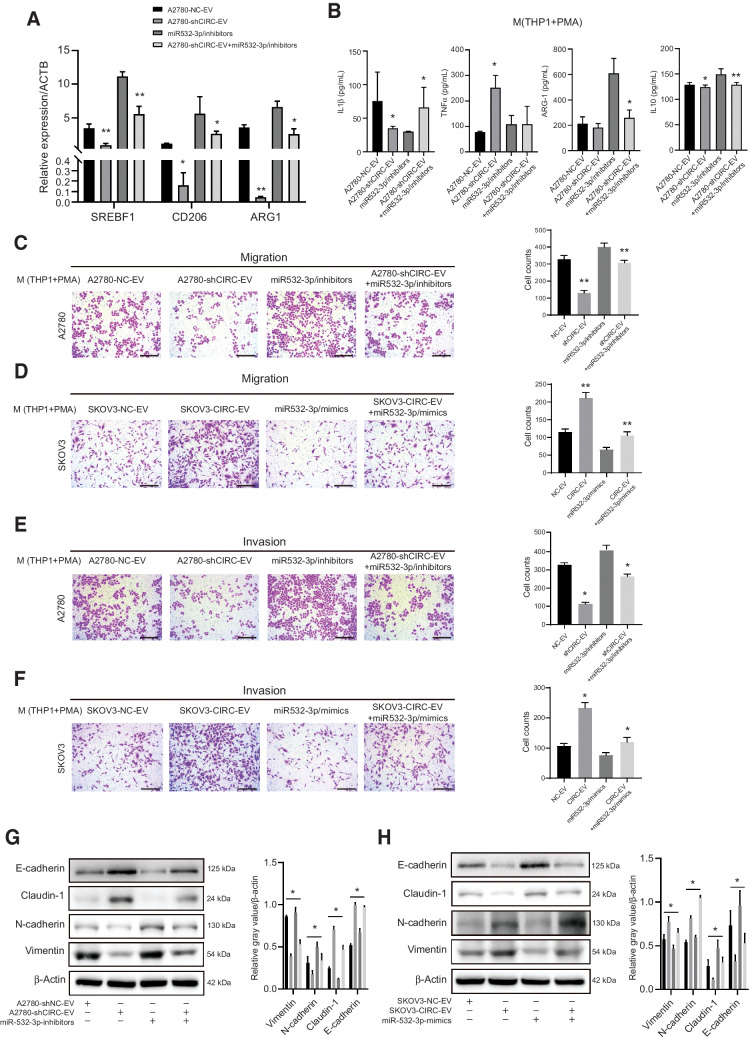
SREBF1 is a downstream target of miR-532-3p and circATP2B4. Therefore, it was further evaluated whether circATP2B4 upregulated EMT through its role as a sponge of miR-532-3p. The expression of SREBF1, CD206, and ARG-1 in EV-treated macrophages was determined by qRT-PCR. Compared with EV-shNC, the expression of SREBF1, CD206, and ARG-1 was significantly decreased in EV-shcircATP2B4-treated macrophages, and miR-532-inhibitors rescued the mRNA expression (Fig. 6A). To further assess the function of circATP2B4 EVs in transforming the M1–M2 phenotype of macrophages, M1 and M2 cytokines was measured. M1 markers IL1β and TNFα were upregulated after knockdown of circATP2B4 but decreased after transfection of miR-532-3p inhibitors. The opposite effect was observed for M2 markers ARG-1 and IL10 (Fig. 6B).
Figure 6.
EVs-packaged circATP2B4 promotes the migration, invasion, and EMT of ovarian cancer cells. A, qRT-PCR assays showing the relative mRNA expression of SREBF1 in macrophages (THP-1+PMA) transfected with A2780-EVs (A2780-NC-EV, A2780-shCIRC-EV, miR-532-3p/inhibitors and A2780-shCIRC-EV+miR-532-3p/inhibitors), n = 3. B, ELISA assays showing the M1 (ILβ, TNFα) and M2 (Arginase-1, IL10) secreted cytokines in macrophages (THP-1+PMA) transfected with A2780-EVs (A2780-NC-EV, A2780-shCIRC-EV, miR-532-3p/inhibitors and A2780-shCIRC-EV) +miR-532-3p/inhibitors), n = 3. C–F, Migration andinvasion capacity of A2780 cocultured with macrophages (A2780-NC-EV, A2780-shCIRC-EV, miR-532-3p/inhibitors and A2780-shCIRC-EV+miR-532-3p/inhibitors) and SKOV3 (SKOV3-NC-EV, SKOV3-CIRC-EV, miR-532-3p/mimics and SKOV3-CIRC-EV+miR-532-3p/mimics) cocultured with macrophages, by an in vitro transwell coculture system without Matrigel are shown. Scale bars, 300 μm. G and H, Western blotting detection of EMT protein (E-cadherin, Claudin-1, N-cadherin, Vimentin) expression after cocultured of A2780 and SKOV3 with macrophages, protein expression relative to β-actin. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Silencing circATP2B4 significantly inhibited the migration abilities of A2780 cells and the effect of circATP2B4 knockdown could be abrogated by miR-532-3p inhibitors (Fig. 6C). Whereas in SKOV3-circATP2B4, the migration ability of SKOV3 was significantly increased, and such an effect could be abrogated by miR-532-3p mimics (Fig. 6D). Similar was observed in the examination of EOC invasion ability. We found that silencing circATP2B4 significantly inhibited the invasion abilities of A2780 cells, and the effect of circATP2B4 knockdown can be abrogated by miR-532-3p inhibitors (Fig. 6E). Whereas in SKOV3-circATP2B4, invasion of SKOV3 was significantly increased, abrogated by miR-532-3p mimics (Fig. 6F).
After the coculture of EOC cell line A2780 with macrophages treated with EVs from shcircATP2B4 for 24 hours, the expression of epithelial cell markers (E-cadherin, Claudin-1) increased and the expression of mesenchymal cell markers (N-cadherin, vimentin) decreased, which could be abrogated by miR-532-3p inhibitors (Fig. 6G). Consistent with this, treatment of SKOV3-circATP2B4 EVs in macrophages for 24 hours decreased the expression of epithelial cell markers (E-cadherin, Claudin-1) and increased the expression of mesenchymal cell markers (N-cadherin, Vimentin) in SKOV3 cells with circATP2B knockdown. The effect of circATP2B4 was abrogated by miR-532-3p mimics (Fig. 6H).
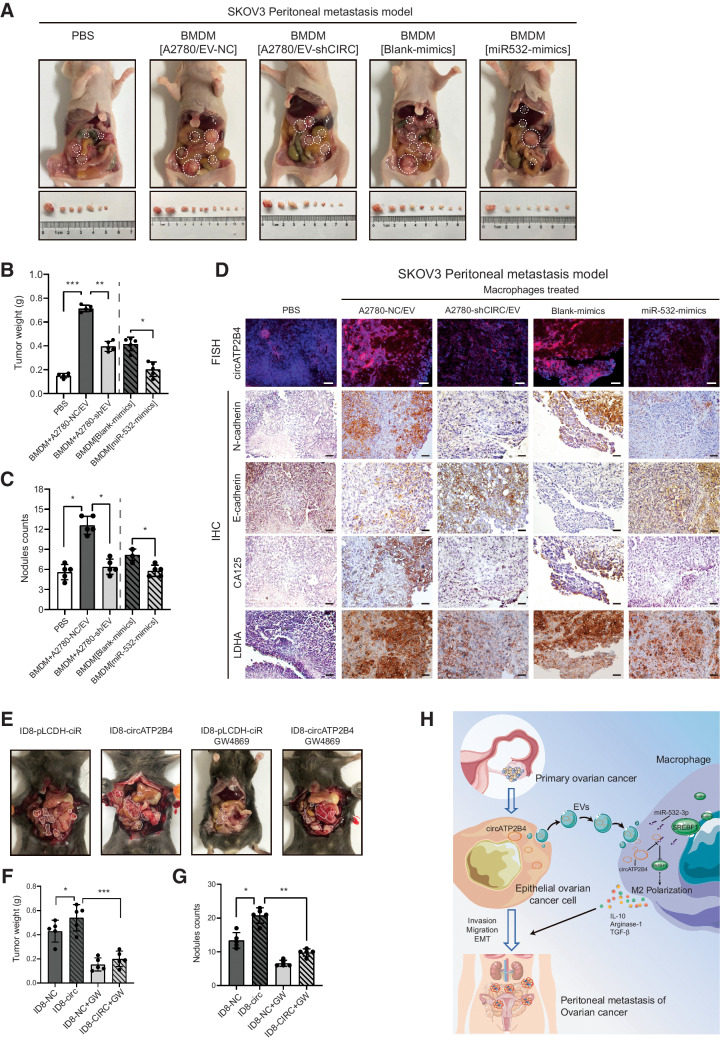
CircATP2B4 in EVs facilitates the metastasis of ovarian cancer cells in vivo
SKOV3 cells mixed with conditioned macrophages stimulated with EVs from A2780 cells transfected with shcircATP2B4 or transfected with miR-532-3p mimics were intraperitoneally injected into mice to further assess the function of circATP2B4 in EVs and miR-532-3p in macrophages with a M2 phenotype, which stimulate the progression of ovarian cancer in vivo. After 5 weeks, SKOV3 cells mixed with macrophages treated with EVs from A2780 transfected with shNC cells generated more peritoneal metastases compared with those generated by SKOV3 cells alone or when mixed with macrophages treated with EVs from A2780 cells transfected with shcircATP2B4. Likewise, significantly fewer visible peritoneal metastases were observed in the SKOV3+ miR-532-3p mimics group compared with the control group (Fig. 7A; Supplementary Fig. S6A). Inoculation of SKOV3 cells and EVs with high expression of circATP2B4 significantly increased tumor weight and nodules counts in the abdominal cavity, whereas EVs with low expression of circATP2B4 and miR-532-3p mimics suppressed this phenotype (Fig. 7B and C). Moreover, markers of tumor peritoneal metastasis were also expressed consistently. FISH detected circATP2B4 in metastatic tissues, and IHC detected EMT (N-cadherin and E-cadherin) of ovarian cancer and malignant markers of EOC (CA125 and LDHA; Fig. 7D; Supplementary Fig. S6B). These results indicated that exosomal circATP2B4 targeted miR-532-3p in macrophage facilitate EOC peritoneal metastasis in vivo.
Figure 7.
EV-packaged circATP2B4 facilitates the peritoneum metastasis of EOCs via inducing M2 macrophage in vivo. A, SKOV3 co-injected with macrophages pelvic peritoneal invasion in nude BALB/C mice. Arrows show tumors in the abdominal cavity, (n = 5 per group). B and C, Tumor weight and nodules counts analysis of nude mice intraperitoneal metastasis model. D, FISH analysis of circATP2B4, IHC analysis of E-cadherin, N-cadherin, CA125 and LDHA proteins. Scale bars, 50 μm. E, ID8 and EVs inhibitor (GW4869) injected pelvic peritoneal invasion in C57BL/6 mice. Arrows show tumors in the abdominal cavity. F and G, Tumor weight and nodules counts analysis of C57BL/6 mice intraperitoneal metastasis model, (n = 5 per group). H, Schematic model of EV-packaged circATP2B4 mediating M2 macrophage polarization via circATP2B4/miR-532-3p/SREBF1, which facilitates the EMT, migration, invasion, and metastatic potential of ovarian cancer cells.*, P < 0.05; **, P < 0.01; ***, P < 0.001.
A mouse model of ovarian cancer cell ID8 peritoneal metastasis was constructed in C57BL/6 mice to study the effect of endogenous macrophages on the peritoneal metastasis of ovarian cancer (Fig. 7E; Supplementary Fig. S6C). Tumor weight and nodule counts in the peritoneum of the extracellular-vesicles inhibitor (GW4869) group were significantly reduced compared with the control (PBS) group after injection of ID8 cells (Fig. 7F and G). Moreover, circATP2B4 overexpressing ID8 cells had more peritoneal metastases compared with the control group (Supplementary Fig. S6D). Furthermore, the increased rumor metastasis induced by circATP2B4 overexpression could be abolished by GW4869 (Supplementary Fig. S6E and S6F). And in our survival analysis of the TCGA-OV cohort, high expression of SREBF1 was associated with worse prognosis (log-rank P = 0.016; Supplementary Fig. S6G).
Discussion
Emerging evidence suggests that circRNAs are a type of noncoding RNAs that play critical roles in various types of tumors (33, 34). Studies strongly support that EOC can originate from the fallopian tube epithelium with evidence from mouse models and genetic evolutionary studies (35). In this study, we first demonstrated that circATP2B4 is upregulated in EOC tissues. CircATP2B4 is known to be circularized in the direction of exons 20 to 21 of the chromosome 1 ATP2B4 host-gene and is stably expressed in ovarian cancer cells. CircATP2B4 aberrant expression correlates to increased TAM infiltration in ovarian cancer tumor microenvironment (TME) and ovarian cancer progression. Functionally, we demonstrated that circATP2B4 promoted EOC tumorigenesis and progression in vitro and in vivo. Mechanistically, circATP2B4 was secreted into macrophages by EVs, through competitive endogenous inhibition of miR-532-3p, targeting the expression of SREBF1. In TAMs, the upregulation of SREBF1 activated the PI3Kα/AKT signaling pathway, and the level of activated phosphorylated AKT promoted the polarization of macrophages to the immunosuppressive M2 type. Finally, M2-polarized macrophages promoted EOC migration, invasion, EMT and peritoneal metastasis through their immunosuppressive properties and secreted cytokines in vivo. The overall function and mechanisms of circATP2B4 are summarized in Fig. 7G. A unique property of our study is that we demonstrate that circATP2B4 could influence EOC progression through cancer cells and their complex network of interactions with macrophages, a key component of the TME.CircATP2B4 acts on TAM polarization in an EV-packaged manner to influence tumorigenic process, highlighting that circATP2B4 and circATP2B4-packed EVs may be potential prognostic biomarkers in ovarian cancer and therapeutic targets.
CircRNAs are considered markers and targets for many malignancies. In a previous ovarian cancer study, circRNA_0049116 (circMUC16) in ovarian cancer was shown to upregulate the expression of ATG13 in ovarian cancer cells by binding to ATG13 protein and enhancing autophagy, thereby promoting the proliferation and metastasis of ovarian cancer cells (36). In another study, circRNA_0132980 (circSLC26A4) was shown to upregulate HOXA7 by competitive binding to miR-1287-5p, which in turn promotes the progression of cervical cancer (37). CircPLEKHM3 acts as a suppressor molecule through regulation of the BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer (38). Accumulating evidence indicates that circRNAs have great potential in the research of female reproductive system tumors. The expression of hsa_circ_0000745 was associated with tumor progression, FIGO stage and ERBB4 level in ovarian cancer (39). In this study, expression of a novel circRNA, circATP2B4, that we discovered and identified was increased in EOC samples, accompanied by enhanced M2 polarization of macrophages. CircATP2B4 acts on TAMs through EVs to promote ovarian cancer invasion, migration, and EMT. Therefore, circATP2B4 is a prospective biomarker for EOC.
TAMs are the critical infiltrating immune cell type in the TME of solid tumors. In ovarian cancer, TAMs have been identified as key cellular components for the tumorigenesis and malignant progression of EOC (40). The activation of M1-like TAMs is induced by pro-inflammatory signals, such as interferon-γ (IFN-γ) and lipopolysaccharide (LPS), and linked to anabolism, including aerobic glycolysis and fatty acid synthesis (41). In contrast, the activation of M2-like TAMs is driven by anti-inflammatory cytokines, such as IL4 and IL10, and by catabolism, including oxidative phosphorylation and fatty acid oxidation (FAO) (42). The tumor-promoting function of preferentially targeting TAMs in the context of cancer immunotherapy has received extensive research attention. SREBF1/SREBP1 is an important regulator of cellular energy absorption and metabolism. In this study, SREBF1 was identified in macrophages as a target molecule that competes endogenously with EOC-released circATP2B4 in EVs for binding to miR-532-3p. Consistent with our results, previous studies have shown that activation of SREBF1-dependent de novo fatty acid synthesis is a key mechanism that tumor cell development and progression utilizes to rapidly divide cells to maintain high-energy-demand intracellular fatty acid pools that contribute to cancer (16). Studies using TME immune cells showed that SREBP1-dependent intracellular fatty acid homeostasis promotes M2-like TAM survival and its protumor phenotype and that M2-like macrophages preferentially utilize lipids during differentiation (43). In addition, it has also been shown that in TME, SREBF1 in TAMs reduces oxidative stress by increasing fatty acid synthesis, promoting TAM polarization to the M2 type and produces immunosuppression through the inhibition of CD8+ T cells by Tregs (44). However, a deeper understanding of the mechanisms underlying the regulation of TAM maturation, differentiation, metabolism, and function may facilitate the development of more targeted and effective immunotherapies. In this study, our data showed that EVs-packaged circATP2B4 from EOCs are translocated into macrophages. In addition, in macrophages circATP2B4 regulates SREBF1 by binding to miR-532-3p. Therefore, the regulation of SREBF1 in macrophages regulates M2 polarization of macrophages through SREBF1/PI3Kα/pAKT and affects the progression of EOC.
EVs are extracellular lipid bilayer vesicles with a diameter of 30 to 150 nm that are released by almost all cells under physiologic and pathologic conditions (45). From biological fluids, including cell secretions and plasma, it acts on other tissues and cells through the local microenvironment, blood, lymph, and pleural effusion (46). It is well known that EVs, or exosomes, exert their biological functions by carrying various molecules such as mRNA, proteins and noncoding RNAs, which play important roles in promoting the development of cancers through interaction between tumor cells and infiltrating immune cells of TME (47–49). For the role of EVs and tumor development in ovarian cancer, it has been reported that ovarian cancer exosomes secrete miR-21 to promote resistance to paclitaxel by targeting APAF1 (50); ovarian cancer exosomes secrete RNA to promote peritoneal metastasis (51); and exosomes carrying miR-223 promote TAM polarization (52). In this study, we identified circATP2B4 as a novel circRNA which is highly expressed in ovarian cancer and stably expressed in EVs. After being transmitted into macrophages, it specifically sponges miR-532-3p, thereby preventing miR-532-3p from targeting its downstream gene SREBF1. Such biological process promotes M2 polarization of TAMs and ultimately increases the metastatic capacity of ovarian cancer. This provides a solution to antagonize the nucleic acid target of circATP2B4 encapsulated by vesicles and regulates lipid metabolism by inhibiting SREBF1 in TAM through nucleic acid molecular drugs, which plays a potential role in the study of ovarian epithelial tumors.
In conclusion, our results demonstrate that circATP2B4 is expressed in ovarian cancer and its translocation into macrophages induces macrophage M2 polarization via miR-532-3p signaling through activation of the SREBF1/PI3K/AKT pathway, which in turn promotes migration, invasion, and EMT of ovarian cancer cells. These findings reveal a novel function of EV circATP2B4 in EOC progression and show the potential of circATP2B4 as a diagnostic and therapeutic target for ovarian cancer.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figures legends
Supplementary table S1-5
Supplementary table S6-8
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no: 21976155, 81773016), Zhejiang Provincial Natural Science Foundation of China (grant no: LY18C060001), and CAMS Innovation Fund for Medical Sciences (CIFMS; grant no: 2019-I2M-5-044). We thank Professor Paul Héroux (McGill University, QC, Canada) for his linguistic advices. Thanks Dr. Jiajia Wang and Jingyao Chen for their technical support from the Core Facilities, Zhejiang University School of Medicine.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
F. Wang: Conceptualization, resources, data curation, software, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Niu: Conceptualization, resources, data curation, software, formal analysis, methodology, writing–original draft. K. Chen: Resources, data curation, software, formal analysis, funding acquisition, investigation, visualization, methodology. X. Yuan: Data curation, validation, methodology. Y. Qin: Resources, validation, methodology. F. Zheng: Data curation, formal analysis, supervision. Z. Cui: Resources, data curation, investigation, visualization, methodology. W. Lu: Conceptualization, resources, supervision, validation, investigation, visualization, methodology. Y. Wu: Conceptualization, resources, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. D. Xia: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ 2020;371:m3773. [DOI] [PubMed] [Google Scholar]
- 3. Monk BJ, Colombo N, Oza AM, Fujiwara K, Birrer MJ, Randall L, et al. Chemotherapy with or without avelumab followed by avelumab maintenance versus chemotherapy alone in patients with previously untreated epithelial ovarian cancer (JAVELIN Ovarian 100): an open-label, randomized, phase III trial. Lancet Oncol 2021;22:1275–89. [DOI] [PubMed] [Google Scholar]
- 4. Anastassiou D, Rumjantseva V, Cheng W, Huang J, Canoll PD, Yamashiro DJ, et al. Human cancer cells express slug-based epithelial–mesenchymal transition gene expression signature obtained in vivo. BMC Cancer 2011;11:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawakami E, Tabata J, Yanaihara N, Ishikawa T, Koseki K, Iida Y, et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin Cancer Res 2019;25:3006–15. [DOI] [PubMed] [Google Scholar]
- 6. Wilusz JE, Sharp PA. A circuitous route to noncoding RNA. Science 2013;340:440–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie C. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol 2019;58:90–9. [DOI] [PubMed] [Google Scholar]
- 8. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 2022;19:188–206. [DOI] [PubMed] [Google Scholar]
- 9. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475–90. [DOI] [PubMed] [Google Scholar]
- 10. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176:869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 2020;19:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest 2013;123:1580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019 Jul 16;51:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suarez-Lopez L, Sriram G, Kong YW, Morandell S, Merrick KA, Hernandez Y, et al. MK2 contributes to tumor progression by promoting M2 macrophage polarization and tumor angiogenesis. Proc Natl Acad Sci USA 2018;115:E4236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 2012;23:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, et al. Vignali DAA Treg cells promote the SREBP1-dependent metabolic fitness of tumor-promoting macrophages via repression of CD8+ T cell–derived Interferon-γ. Immunity 2019;51:381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ducie J, Dao F, Considine M, Olvera N, Shaw PA, Kurman RJ, et al. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat Commun 2017;8:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 20. Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci USA 2013;110:14408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011;477:596–600. [DOI] [PubMed] [Google Scholar]
- 22. Huang YK, Wang M, Sun Y, Di Costanzo N, Mitchell C, Achuthan A, et al. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun 2019;10:3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raggi C, Correnti M, Sica A, Andersen JB, Cardinale V, Alvaro D, et al. Cholangiocarcinoma stem-like subset shapes tumor-initiating niche by educating associated macrophages. J Hepatol 2017;66:102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosenthal AN, Fraser L, Manchanda R, Badman P, Philpott S, Mozersky J, et al. Results of annual screening in phase I of the United Kingdom familial ovarian cancer screening study highlight the need for strict adherence to screening schedule. J Clin Oncol 2013;31:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu G, Cao L, Zhu J, Tan Z, Tang M, Li Z, et al. Loss of RBMS3 confers platinum resistance in epithelial ovarian cancer via activation of miR-126-5p/β-catenin/CBP signaling. Clin Cancer Res 2019;25:1022–35. [DOI] [PubMed] [Google Scholar]
- 26. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle–packaged HIF-1α-stabilizing lncRNA from tumor-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 2019;21:498–510. [DOI] [PubMed] [Google Scholar]
- 27. Lin H, Xu X, Chen K, Fu Z, Wang S, Chen Y, et al. MiR-23b Cluster and SMAD3 form a novel positive feedback loop to promote epithelial–mesenchymal transition and metastasis in ovarian cancer. Int J Biol Sci 2022 Feb 21;18:1989–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Izar B, Tirosh I, Stover EH, Wakiro I, Cuoco MS, Alter I, et al. A single-cell landscape of high-grade serous ovarian cancer. Nat Med 2020;26:1271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olalekan S, Xie B, Back R, Eckart H, Basu A. Characterizing the tumor microenvironment of metastatic ovarian cancer by single-cell transcriptomics. Cell Rep 2021;35:109165. [DOI] [PubMed] [Google Scholar]
- 30. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. A robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675–91. [DOI] [PubMed] [Google Scholar]
- 32. Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol 2017;198:1006–14. [DOI] [PubMed] [Google Scholar]
- 33. Guarnerio J, Zhang Y, Cheloni G, Panella R, Katon JM, Simpson M, et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res 2019;29:628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Wang H, Li Z, Li F, Liang L, Zou Y, et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol 2022;76:135–47. [DOI] [PubMed] [Google Scholar]
- 35. Hu Z, Artibani M, Alsaadi A, Wietek N, Morotti M, Shi T, et al. Ahmed AA The repertoire of serous ovarian cancer Non-genetic heterogeneity revealed by single-cell sequencing of normal fallopian tube epithelial cells. Cancer Cell 2020 Feb 10;37:226–42. [DOI] [PubMed] [Google Scholar]
- 36. Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer 2020;19:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids 2020;19:413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer 2019;18:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang S, Li Z, Zhu G, Hong L, Hu C, Wang K, et al. RNA-binding protein IGF2BP2 enhances circ_0000745 abundancy and promotes aggressiveness and stemness of ovarian cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J Ovarian Res 2021;14:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Long L, Hu Y, Long T, Lu X, Tuo Y, Li Y, et al. Tumor-associated macrophages induced spheroid formation by CCL18-ZEB1-M-CSF feedback loop to promote transcoelomic metastasis of ovarian cancer. J Immunother Cancer 2021;9:e003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eguchi K, Manabe I, Oishi-Tanaka Y, Ohsugi M, Kono N, Ogata F, et al. Saturated fatty acid and TLR signaling link β cell dysfunction and islet inflammation. Cell Metab 2012;15:518–33. [DOI] [PubMed] [Google Scholar]
- 42. Liu PS, Wang H, Li X, Chao T, Teav T, Christen S, et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017;18:985–94. [DOI] [PubMed] [Google Scholar]
- 43. Oliveira MC, Menezes-Garcia Z, Henriques MC, Soriani FM, Pinho V, Faria AM, et al. Acute and sustained inflammation and metabolic dysfunction induced by high refined carbohydrate-containing diet in mice. obes 2013;21:E396–406. [DOI] [PubMed] [Google Scholar]
- 44. Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, et al. Treg cells promote the SREBP1-dependent metabolic fitness of tumor-promoting macrophages via repression of CD8 T cell–derived interferon-γ. Immunity 2019;51:381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213–28. [DOI] [PubMed] [Google Scholar]
- 46. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016;30:836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arab T, Mallick ER, Huang Y, Dong L, Liao Z, Zhao Z, et al. Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single-particle analysis platforms. J Extracell Vesicles 2021;10:e12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carolyn M, Bartholomew S, Denis W. Extracellular vesicles in immunomodulation and tumor progression. Nat Immunol 2021;22:560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian W, Lei N, Zhou J, Chen M, Guo R, Qin B, et al. Extracellular vesicles in ovarian cancer chemoresistance, metastasis, and immune evasion. Cell Death Dis 2022;13:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yeung CLA, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun 2016;7:11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feng W, Dean DC, Hornicek FJ, Shi H, Duan Z. Exosomes promote premetastatic niche formation in ovarian cancer. Mol Cancer 2019;18:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, et al. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res 2019;38:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figures legends
Supplementary table S1-5
Supplementary table S6-8
Data Availability Statement
All data obtained in this work can be acquired from the National Center for Biotechnology Information: RNA-seq data set (PRJNA396544), scRNA-seq data sets (PRJNA609094 and PRJNA612966). TCGA-OV database (https://xenabrowser.net/datapages/), AOCS and GSE30161 database (CuratedOvarianData for R package), and the analysis results of circRNA sequencing and mRNA sequencing data were released when the paper was published. The data generated in this study are available upon request from the corresponding author.