Abstract
Objectives: Serine metabolism is essential for tumor cells. Endogenous serine arises from de novo synthesis pathways. As the rate-limiting enzyme of this pathway, PHGDH is highly expressed in a variety of tumors including colon cancer. Therefore, targeted inhibition of PHGDH is an important strategy for anti-tumor therapy research. However, the specific gene expression and metabolic pathways regulated by PHGDH in colon cancer are still unclear. Our study was aimed to clarified the role of PHGDH in serine metabolism in colon cancer to provide new knowledge for in-depth understanding of serine metabolism and PHGDH function in colon cancer. Methods: In this study, we analyzed the gene expression and metabolic remodeling process of colon cancer cells (SW620) after targeted inhibition of PHGDH by gene transcriptomics and metabolomics. LC-MS analysis was performed in 293T cells to PHGDH gene transcription and protein post-translational modification under depriving exogenous serine. Results: We found that amino acid transporters, amino acid metabolism, lipid synthesis related pathways compensation and other processes are involved in the response process after PHGDH inhibition. And ATF4 mediated the transcriptional expression of PHGDH under exogenous serine deficiency conditions. While LC-MS analysis of post-translational modification revealed that PHGDH produced changes in acetylation sites after serine deprivation that the K289 site was lost, and a new acetylation site K21was produced. Conclusion: Our study performed transcriptomic and metabolomic analysis by inhibiting PHGDH, thus clarifying the role of PHGDH in gene transcription and metabolism in colon cancer cells. The mechanism of high PHGDH expression in colon cancer cells and the acetylation modification that occurs in PHGDH protein were also clarified by serine deprivation. In our study, the role of PHGDH in serine metabolism in colon cancer was clarified by multi-omics analysis to provide new knowledge for in-depth understanding of serine metabolism and PHGDH function in colon cancer.
Keywords: colon cancer, PHGDH, transcriptomics, metabolomics, acetylation modification, Abbreviations PHGDH, 3-phosphoglycerate dehydrogenase; PSAT, phosphoserine aminotransferase; PSPH, phosphoserine phosphatase; OPLS-DA, orthogonal partial least-squares discriminant analysis; TIC: total ion chromatograms.
Introduction
Colon cancer is one of the major digestive system cancers with high incidence and mortality.1 The occurrence of colon cancer is related to a variety of factors, such as genetic factors, dietary factors and so on. It should not be ignored that abnormal cellular metabolism, such as glucose metabolism, amino acid metabolism, and lipid metabolism also play an important role in the development of colon cancer.
Serine, as a nonessential amino acid, is an important one-carbon unit raw material. Its main roles include as a raw material for the synthesis of amino acids such as glycine and cysteine, phospholipids, proteins, and nucleic acids; as a donor of methyl groups, participating in the methylation modification of biological macromolecules; and producing NADPH for antioxidant defense. Cells adopt two sets of mechanisms: exogenous uptake and endogenous synthesis to ensure the supply of serine. Exogenous uptake refers to the uptake of serine by cells from the microenvironment by active transport, while endogenous synthesis modes mainly include de novo synthesis, proteolytic recovery, and conversion of glycine. Under fasting conditions, approximately 70% of the intracellular serine is derived from the de novo synthesis pathway (de novo serine synthesis pathway, SSP). The de novo pathway of serine synthesis results from the intermediate metabolite of glycolysis, 3-phosphoglycerate, which is converted to 3-phosphohydroxypyruvate by 3-phosphoglycerate dehydrogenase (PHGDH), then 3-phosphoserine by phosphoserine aminotransferase (PSAT), and then serine by phosphoserine phosphatase (PSPH).2–6
PHGDH, as the rate-limiting enzyme of the endogenous serine de novo synthesis pathway, is highly expressed in a variety of tumors including colon cancer and is significantly negatively correlated with the prognosis of tumors.7–9 However, the changes of transcriptomic and metabolomic involved in the regulation of serine metabolism by PHGDH in colon cancer cells remain unclear. The regulatory mechanisms involved in PHGDH expression and activity are also unclear.
In the present study, we analyzed the pathways involved in gene transcription to metabolism in colon cancer cells after treatment with PHGDH-specific inhibitors by high-throughput techniques such as transcriptomics and metabolomics. Transcriptomic assays revealed a significant enrichment of signaling pathways constituted by amino acid transporters and metabolism-related genes. Corresponding metabolomics tests revealed changes in multiple amino acid metabolic pathways, but also compensatory changes in sphingolipid metabolism. We also found that elevated expression levels of PHGDH were first mediated by an increase in transcript levels by ATF4 under serine-deficient conditions. While LC-MS analysis of post-translational modification revealed that PHGDH produced changes in acetylation sites after serine deprivation that the K289 site was lost, and a new acetylation site K21was produced. Our study reveals the upstream expression regulation and downstream mechanism of PHGDH in response to serine deficiency conditions, providing a new perspective for multiomics combined analysis of amino acid metabolism in tumor cells.
Materials and Methods
Experimental Design
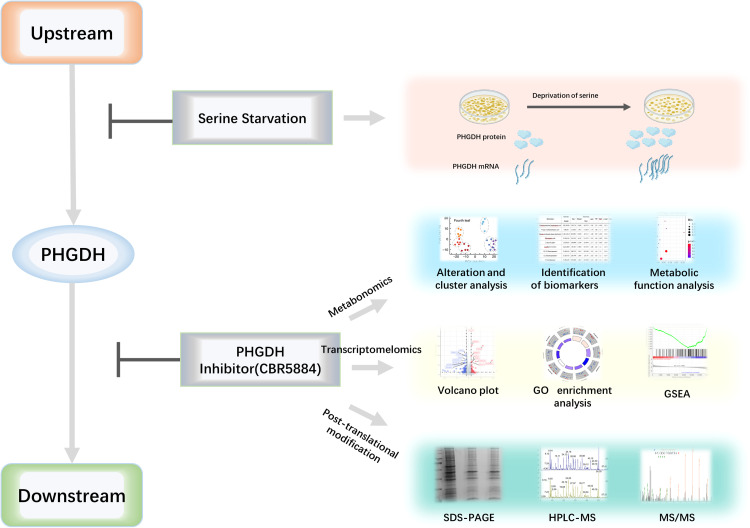
As shown in Figure 1, we drew a picture of the experimental design in order to quickly understand the entire experiment.
Figure 1.
The experimental design of this study. This experiment is mainly divided into the upstream part of PHGDH expression regulation under the condition of exogenous serine deficiency and the downstream part of downstream genome and metabolism regulated by PHGDH. Serine deprivation increased the expression of PHGDH in colon cancer. Then, we analyzed the gene expression and metabolic remodeling process of colon cancer cells after targeted inhibition of PHGDH by gene transcriptomics and metabolomics. LC-MS analysis was performed in 293T cells to PHGDH gene transcription and protein post-translational modification under depriving exogenous serine.
Cell Culture
Human colon cancer cells (SW620, TCHu101; SW480, TCHu172; HT29, TCHu103; HCT116, TCHu 99) were cultured in RPMI-1640 medium containing 10% (v/v) fetal bovine serum (Every Green, China) and 1% penicillin (100 U/mL) and 1% streptomycin (100 μg/mL) (Thermo Scientific, USA) and maintained at 37°C in a humidified 5% CO2 incubator (Thermo Scientific, USA). All cell lines were purchased from the Shanghai Institute of Cell Research, Chinese Academy of Sciences (Shanghai, China).
Cell Proliferation Assay CCK-8 Assay
Human colon cancer cells were seeded in 96-well plates (1 × 104 cells/well). 100 μL RPMI-1640 complete medium was added to each well and incubated for 24 h in an incubator containing 5% CO2 at 37°C. After the cells adhered and recovered their morphology, the medium was replaced with serine-depleted medium and cultured for 24 h. CCK-8 testing was then performed. Briefly, the medium in the culture wells was discarded and 100 μL of medium containing 10% CCK-8 reagent (Beyotime, China) was added and incubated for 1 h at 37°C in the dark. Finally, the optical density (OD) value at 450 nm was detected by a microplate reader (Synergy HT ZX-22; Bio-Tek Instruments, VT, USA).10
Colony Formation Assay
The methods for colony formation have been published in our previous study.10
Western Blot Assay
The protein was resolved on 10% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, followed by immunoblot detection and visualization. Immunoblotting was performed using anti-PHGDH (66350, dilution: 1/1000; cell signaling technology), anti-ATF4 (11815, dilution: 1/1000; cell signaling technology), anti-β-Actin (3700, dilution: 1/1000; cell signaling technology) at 37°C for 2 h, followed by incubation with goat antirabbit horseradish peroxidase-conjugated immunoglobulin G secondary antibodies (A0408, dilution: 1/1000; Beyotime) and goat antimouse horseradish peroxidase-conjugated immunoglobulin G secondary antibodies (A0412, dilution: 1/1000; Beyotime) for 1 h at room temperature. Primary antibody dilution buffer (P0256, Beyotime) and second antibody dilution buffer (P0023D, Beyotime) were purchased from Beyotime Biotechnology, Shanghai, China. ChemiDoc image analysis system (Bio-Rad Laboratories, Inc.) analyzed and quantified the relative protein levels.
RNA-Sequencing Analysis of SW620 Cells
TRIzol reagent (Carlsbad, CA, USA) was used to extract total RNA from SW620 cells and SW620 cells treated with PHGDH inhibitor (CBR5884) (MCE, Shanghai, China). Three independent replicates were analyzed. The methods for RNA-sequencing (RNA-seq) have been published in our previous study.10
Quantitative Real-Time PCR (qRT-PCR)
The methods for qRT-PCR have been published in our previous study.10 The primers of PHGDH, ATF4 and β-actin were synthesized by TSINGKE Biological Technology (Beijing, China). β-actin was served as an internal reference of RNA integrity. The sequences are as follows:
PHGDH-F-5’-AACCGCAGCTTCTTGGCTTA-3’
PHGDH-R-5’-TAAGGCCTTCACAGTCCTGC-3’
ATF4-F-5’-TCAAACCTCATGGG-TTCTCC-3’
ATF4-R-5’-GTGTCATCCAACGTGGTCAG-3’
β-actin-F 5’-ACTCTTCCAGCCTTCCTTCC-3’
β-actin-R 5’-CGTCATACTCCTGCTTGCTG-3’
LC-MS Analysis of Post-Translational Modification
Sample Preparation of 293T Cells for Post-Translational Modification
SDT buffer was added to 293T cells transfected with flag-PHGDH plasmid (PHGDH cDNA ORF Clone, Sino Biological, China). The lysate was sonicated and then boiled for 15 min. After centrifuged at 14000 g for 40 min, the supernatant was quantified with the BCA Protein Assay Kit (Beyotime, China). 20 µg protein from each sample was mixed with 5 × loading buffer and boiled in a metal bath for 5 min. Boiled proteins were then individually loaded onto 12% SDS-PAGE gels and run at a constant current of 14 mA for 90 min to separate proteins. The proteins were separated and purified in gel to obtain purified PHGDH protein. Protein bands were visualized by Coomassie Blue R-250 staining. The gel containing PHGDH protein (∼57 kDa) was dissected for digestion.
In Gel Digestion of Proteins
The gel containing PHGDH protein was excised from SDS PAGE. Gel pieces were treated with 30% ACN (100 mM NH4HCO3) until destaining. Then, the destained gels were dried in a vacuum centrifuge. The protein was then reduced with dithiothreitol (10 mM DTT, 100 mM NH4HCO3) for 30 min at 56°C. Iodoacetamide (200 mM IAA, 100 mM NH4HCO3) was then alkylated for 30 min at room temperature in the dark. After rinsing the gel with 100 mM NH4HCO3 and ACN, it was then digested overnight with trypsin (12.5 ng/μL, 100 mM NH4HCO3). Peptides were extracted with 60% ACN (0.1% TFA) in triplicate. The extracts were combined and dried completely in a vacuum centrifuge.
HPLC-MS for Post-Translational Modification
Each fraction was injected for nano LC-MS/MS analysis. Samples were tested by Applied Protein Technology Co., Ltd., and the test method was based on a previous study.11
Data Analysis for Post-Translational Modification
The MS data were analyzed using MaxQuant software version 1.5.3.17 (Max Planck Institute of Biochemistry in Martinsried, Germany).12 MS data were searched against the UniProtKB Escherichia coli database (2,585,998 total entries, downloaded 06/07/12). The precursor mass search range was 6 ppm. The search tolerates a maximum of two missed cleavage sites and the process follows the enzymatic cleavage rules for trypsin/P with a mass error tolerance range of 20 ppm for fragment ions. Enzyme = trypsin, missed cleavage = 2, Fixed modification: carbamidomethyl (C), Variable modification: oxidation (M), phospho (ST), phosoho (Y), GlyGly (K), acetyl (K), acetyl(protein N-term), Decoy database pattern = Reverse. The cutoff of global false discovery rate (FDR) for peptide and protein identification was set to 0.01.13
Metabolomic Analyses
Sample Preparation and HPLC-MS of SW620 Cells for Metabolomic Analyses
The methods for sample preparation and HPLC-MS of SW620 cells for metabolomic analyses have been published in our previous study.10,14
Data Preprocessing and Metabolite Identification
The method of data preprocessing has been published in our previous study.10,15 Briefly, Raw HPLC–MS data was extracted as matrix data and imported into SIMCA14.1 for multivariate statistical analysis. Orthogonal partial least-squares discriminant analysis (OPLS-DA) was performed to visualize the metabolic difference between SW620-Con group and SW620-CBR5884 group. The screened differential metabolites need to meet three conditions simultaneously: (1) VIP >1; (2) p|corr| value >0.58; (3) confidence interval crossed zero in jack-knifed loading plot. The potential biomarkers were identified by MS2 combined with LIPIDMAPS database and HMDB database. Pathway analysis was performed using MetaboAnalyst 5.0 (http://www.MetaboAnalyst.ca/).
Statistical Analysis
The methods for statistical analysis have been published in our previous study.10 All experiments have three repetitions or three parallel samples.
Results
Transcriptomic Analysis of PHGDH Inhibition in Colon Cancer Cells
In the de novo serine synthesis pathway, PHGDH is present as a rate-limiting enzyme (Figure 2A). It is known that PHGDH is rapidly activated and expressed when the serine concentration is insufficient in the environment, and the PHGDH expression level is significantly higher in a variety of tumor tissues including colon cancer than in normal tissues. Therefore, targeted inhibition of PHGDH is considered to be an important strategy for antitumor therapy research. Currently, a variety of inhibitors targeting PHGDH have been reported. Among them, CBR5884 can effectively block the de novo serine synthesis pathway and is selectively toxic to tumor cells.16 To investigate the effects of PHGDH inhibition by CBR5884 on the proliferation and gene expression of colon cancer cells, we performed colony formation assay and transcriptomic analysis after treating SW620 cells with CBR5884.The results of colony formation assay showed that CBR5884 could significantly inhibit SW620 cell proliferation (Figure 2B, C).The reference transcriptomic analysis revealed that 249 genes were altered in expression after CBR5884 treatment, including 88 up-regulated genes and 161 down-regulated genes (Figure 2D,2E, Table S1). Significant enrichment of signaling pathways such as amino acid transporters, amino acid metabolism, and cell proliferation was found by GO (Figure 2F) and KEGG (Figure 2G) analysis. Ribosomal, oxidative phosphorylation, proteasome and pyrimidine signaling pathways were further found to be significantly upregulated. Meanwhile, cell adhesion and signal transduction pathways were significantly downregulated by GSEA analysis (Figure 2H). These results suggest that alterations in cellular transcriptomics after inhibition of PHGDH involve processes such as amino acid metabolism, cell proliferation, and apoptosis.
Figure 2.
Transcriptomic analysis of PHGDH inhibition in colon cancer cells. (A) L-serine synthesis pathway. (B) and (C) The cell proliferations of SW620 treated with CBR5884 (500 nM) were measured by colony formation assay at the indicated days (n = 3 per group). (D) Volcano plot illustrating the differently expressed genes in SW620 cells (n = 3) and SW620- CBR5884 cells (n = 3). (E) Heat map showed 30 genes that were most significantly different between SW620 cells with SW620-CBR5884 cells. (F) GO annotations of differentially expressed mRNAs with the top 10 enrichment scores of biological processes. (G) KEGG annotations and enrichment of differentially expressed genes (Q-value) in SW620-CBR5884 cells compared to SW620 cells. Nodes in red indicate significance (Q < 0.05), and the size of the nodes indicates gene number. (H) Gene set enrichment analysis (GSEA) of RNA-seq data in SW620-CBR5884 relative to SW620 cells.
Metabolomic Analysis of PHGDH Inhibition in Colon Cancer Cells
Genomic assays suggest changes that may occur in cells, while metabolomics suggests changes that are occurring in cells, therefore, we further investigated the changes in the metabolic status of colon cancer cells by inhibiting PHGDH and used metabolomics techniques to detect metabolites after CBR5884 treatment of SW620. A total of 7240 peaks (ESI + ) and 6744 peaks (ESI−) were detected in SW620-Con group and SW620-CBR5884 group. Good repeatability was also demonstrated by overlapping total ion chromatograms (TIC) of six QC samples (Figure 3A). Meantime, six selected extracted ion chromatograms in QC samples were used to assess the system repeatability and stability. Taking peaks in positive mode as an example, the retention times, peak areas, and mass accuracies of six selected peaks showed acceptable RSDs. RSDs of these six selected peaks were 0.01% to 0.10% for retention time, 1.41% to 7.28% for peak area, 5.95 × 10−5% to 14.80 × 10−5% for mass accuracies (Table S2). It indicates that the instrument and samples were stabilized.
Figure 3.
Metabolic analysis of PHGDH inhibition in colon cancer cells. (A) The overlapping typical TICs of six QC samples were obtained from LC-MS in positive mode (ESI + ) and negative mode (ESI−). (B) OPLS-DA score plots of positive ion mode, R2X = 1, R2Y = 1, Q2 = 1. OPLS-DA score plots of positive ion mode and negative ion mode, R2X = 0.414, R2Y = 1, Q2 = 0.993. (C) Heat map showed 313 metabolites that were significantly different between SW620 cells with SW620-CBR5884 cells. (D) S-plot represented the changing trend of all metabolites. Red circles represent significantly changed metabolites, the first quadrant represents down-regulated metabolites, and the third quadrant represents up-regulated metabolites. (E) Disordered pathways in SW620-CBR5884 cells (n = 3) compared with SW620 cells (n = 3). In the bubble plot, Y-axis represents enriched metabolic pathways, X-axis means pathway impacts. The size of each bubble represents the number of metabolites enriched and the color indicates the p value (take the negative natural logarithm). Small p value and big pathway impact factor revealed the pathway is significantly affected. (F) The relative content of biomarkers in SW620-Con (n = 3) and SW620-CBR5884 (n = 3), *p < 0.05.
OPLS-DA plots and heat map indicated that CBR5884 treatment could lead to significant alterations in the metabolic profile of SW620 cells (Figure 3B,3C). Under the analysis of the supervised mode, as shown in the score plots of OPLS-DA in positive mode (Figure 3B), the model described 100% of the variation in X (R2X = 1), 100% of the variation in the response Y (class) (R2Y = 1), and which also predicted 100% of the variation in the model (Q2 = 1). The score plot of OPLS-DA in negative mode was also shown in Figure 3B). Then, based on the characteristic ions and neutral losses, a total of 313 peaks as potential biomarkers were found (Figure 3C) and 27 metabolites were identified. Among the 27 biomarkers, 12 metabolites were upregulated, and 15 metabolites were downregulated (Table 1). The levels of many of these amino acids changed. The levels of methionine, isoleucine and proline are significantly reduced, accompanied by a significant increase in Spermidine. Fluctuations in phospholipids are also prominent. We observed a decrease in a variety of phosphatidylcholines and sphingomyelins, such as LysoPC (16:0), LysoPC (18:1), and phytosphingosine (Figure 3D, F). Pathway analysis was performed for significantly altered metabolites. Pathway analysis revealed that cysteine and methionine metabolism, arginine and proline metabolism, nicotinate and nicotinamide metabolism were the most important metabolic pathways affected. The alterations in glycerophospholipid metabolism and sphingolipid metabolism are also notable. These results indicate that the metabolomic changes after inhibition of PHGDH include processes such as imbalance and compensation of amino acid metabolism and lipid metabolism.
Table 1.
Identification of Biomarkers between SW620-Con and SW620-CBR5884
| NO. | Abbreviation | Molecular formula | Mass | Rt (min) | Theoretical mass | ppm | VIP | Folda | p value | Trenda |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | [Nitrilotris(methylene)] trisphosphonic acid | C3H12NO9P3 | 298.9724 | 14.269 | 298.9725 | −0.34 | 2.33 | >100 | 0.031916 | ↑ |
| 2 | 4-Amino-2- methylenebutanoic acid | C5H9NO2 | 115.0635 | 1.441 | 115.0633 | 1.74 | 1.80 | 1.1 | 0.053669 | ↑ |
| 3 | Deamino nicotinamide adenine dinucleotide | C21H27N6O15P3 | 665.0991 | 19.039 | 665.101 | −2.86 | 9.59 | >100 | 0.001035 | ↑ |
| 4 | LysoPE (24:1) | C29H58NO7P | 563.395 | 13.991 | 563.3951 | −0.18 | 2.24 | 1.2 | 0.012495 | ↑ |
| 5 | N1,N12-Diacetylspermine | C14H30N4O2 | 286.2372 | 1.242 | 286.2369 | 1.05 | 1.14 | 1.2 | 0.036913 | ↑ |
| 6 | n1,n8-diacetylspermidine | C11H23N3O2 | 229.1796 | 1.308 | 229.179 | 2.62 | 4.33 | 1.3 | 0.031669 | ↑ |
| 7 | N1-Acetylspermine | C12H28N4O | 244.2266 | 1.181 | 244.2263 | 1.23 | 1.20 | 1.3 | 0.023241 | ↑ |
| 8 | N8-Acetylspermidine | C9H21N3O | 187.1686 | 1.251 | 187.1685 | 0.53 | 5.06 | 1.3 | 0.002054 | ↑ |
| 9 | PRE | C44H44O24 | 956.2245 | 9.209 | 956.2222 | 2.41 | 1.06 | 3.2 | 0.07348 | ↑ |
| 10 | Propionyl-L-carnitine | C10H20NO4 | 218.138 | 1.776 | 218.1386 | −2.75 | 4.09 | 1.4 | 0.063807 | ↑ |
| 11 | Spermidine | C7H19N3 | 145.1577 | 1.374 | 145.1579 | −1.38 | 3.89 | 5.8 | 0.013145 | ↑ |
| 12 | Tetrahydrofurfuryl acetate | C7H12O3 | 144.0785 | 1.311 | 144.0786 | −0.69 | 1.28 | 1.2 | 0.04509 | ↑ |
| 13 | (3R,7R)-13,7-Octanetriol | C8H18O3 | 162.1259 | 7.630999 | 162.1256 | 1.85 | 1.23 | 0.7 | 0.058409 | ↓ |
| 14 | 26-Methyl nigranoate | C31H48O4 | 484.3557 | 4.823999 | 484.3553 | 0.83 | 1.34 | 0.9 | 0.029497 | ↓ |
| 15 | 2-Phenylethyl octanoate | C16H24O2 | 248.1775 | 15.498 | 248.1776 | −0.40 | 1.19 | 0.4 | 0.001203 | ↓ |
| 16 | Glyceryl lactopalmitate | C20H16N6O2S | 404.1053 | 7.376 | 404.1055 | −0.50 | 1.55 | 0.1 | 0.027572 | ↓ |
| 17 | Guanidinosuccinic Acid | C5H9N3O4 | 175.0592 | 1.674 | 175.0593 | −0.57 | 1.27 | 0.0 | 0.002199 | ↓ |
| 18 | L-isoleucyl-L-proline | C11H20N2O3 | 228.1474 | 1.657 | 228.1474 | 0.00 | 1.02 | 0.7 | 0.003456 | ↓ |
| 19 | LysoPC (16:0) | C24H51NO7P | 495.3323 | 14.939 | 495.3325 | −0.40 | 2.13 | 0.7 | 0.057755 | ↓ |
| 20 | LysoPC (16:1) | C24H49NO7P | 493.3161 | 13.835 | 493.3168 | −1.42 | 1.40 | 0.7 | 0.04405 | ↓ |
| 21 | LysoPC (18:1) | C26H53NO7P | 521.3491 | 15.42 | 521.3481 | 1.92 | 6.11 | 0.5 | 0.001664 | ↓ |
| 22 | LysoPC (18:2) | C26H51NO7P | 519.3311 | 14.366 | 519.3325 | −2.70 | 1.42 | 0.4 | 0.006986 | ↓ |
| 23 | LysoPC (P-16:0) | C24H51NO6P | 479.3373 | 15.465 | 479.3376 | −0.63 | 1.23 | 0.4 | 0.018421 | ↓ |
| 24 | Methionyl-Methionine | C10H20N2O3S2 | 280.0904 | 5.862999 | 280.0915 | −3.93 | 1.37 | 0.6 | 0.041698 | ↓ |
| 25 | Phytosphingosine | C18H39NO3 | 317.2933 | 8.921999 | 317.293 | 0.95 | 1.28 | 0.7 | 0.003959 | ↓ |
| 26 | S-Adenosylmethionine | C15H23N6O5S | 399.1451 | 1.547 | 399.1451 | 0.00 | 1.19 | 0.5 | 0.000483 | ↓ |
| 27 | Tripropylamine | C9H21N | 143.1672 | 4.580001 | 143.1673 | −0.70 | 1.55 | 0.7 | 0.003644 | ↓ |
Serine Deprivation Increased the Expression of PHGDH in Colon Cancer
After investigating the downstream genomic and metabolic regulated by PHGDH, we focused on the regulation of PHGDH expression under exogenous serine deficiency conditions. We treated a variety of colon cancer cells (SW480, SW620, HT29, HCT116) with serine-deprived medium for 24 h. The results showed that both mRNA and protein expression levels of PHGDH were significantly increased in these cells (Figure 4A, B). After treatment with serine-deprived medium for different periods of time in SW620 cells, it was found that the mRNA level of PHGDH was significantly increased at 4 h. While the protein level was started to increase at 12 h (Figure 4C, D). These results suggested that after serine deprivation in the environment, colon cancer cells lead to increased PHGDH protein levels by rapidly activating gene transcription of PHGDH.
Figure 4.
Serine deprivation increased the expression of PHGDH in colon cancer. (A) Quantification of PHGDH mRNA levels in colon cancer cells (SW620, SW480, HT29 and HCT116) cultured in basic and serine-deprived medium for 24 h. Error bars represent mean ± SD (n = 3). *p < 0.05. (B) Immunoblotting analysis of PHGDH protein expression in colon cancer cells (SW620, SW480, HT29 and HCT116) cultured in basic and serine-deprived medium for 24 h. (C) and (d) SW620 was cultured in basic and serine-deprived medium and harvested at the indicated hours; mRNA levels and protein levels (D) of PHGDH. Error bars represent mean ± SD (n = 3), *p < 0.05. (E) Immunoblotting analysis of ATF4 protein expression in SW620 cells cultured in basic and serine-deprived medium for 12 h and harvested at the indicated hours. (F) and (G) SW620 transfected with indicated si-ATF4 (1#, 2#) and cultured in basic and serine-deprived medium; the mRNA expressions of ATF4 and PHGDH, and protein levels of PHGDH. Error bars represent mean ± SD (n = 3), *p < 0.05.
Increased ATF4 Expression upon Serine Deprivation Activates Gene Transcription of PHGDH
ATF4 is a major factor in response to amino acid starvation.17 ATF4 plays an important role in promoting the transcriptional activation process of PHGDH.18 We found that in SW620 the protein expression level of ATF4 was significantly increased after 1 h of serine deprivation, and that the protein expression level of ATF4 continued to increase over time (Figure 4E). Moreover, ATF4 mRNA was also significantly increased after serine deprivation. While treatment with siRNA significantly inhibited ATF4 mRNA expression under serine deprivation conditions. More meaningfully, knockdown of ATF4 expression could inhibit the mRNA and protein expression of PHGDH, allowing the expression level of PHGDH under serine deprivation to return to the resting level (Figure 4F and G). These results illustrate that ATF4 is a major transcription factor that promotes increased PHGDH expression under serine starvation conditions in colon cancer cells.
Post-Translational Modification Changes of PHGDH Protein Under Serine Deprivation
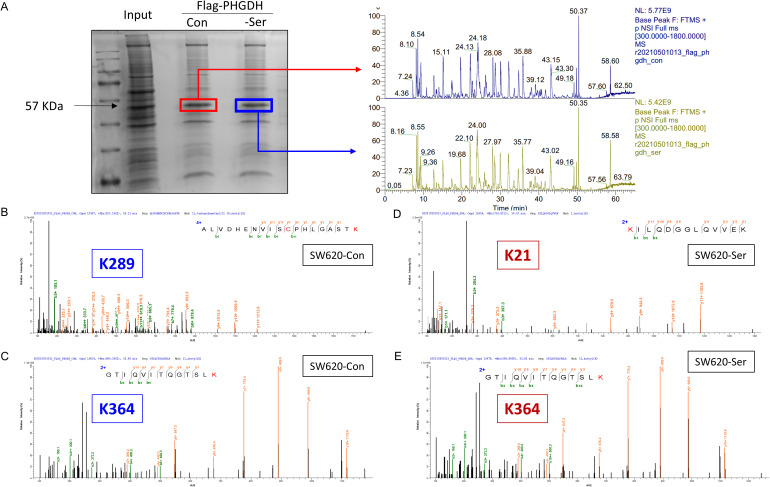
After clarifying that high PHGDH expression upon serine deprivation is mainly driven by transcript levels, we focused on what post-translational modifications occur in PHGDH proteins and how they are altered after serine deprivation. Therefore, protein modificationomics analysis was performed. 293T cells have high transfection efficiency and are usually used for target protein overexpression. We transfected the flag-tagged PHGDH expression plasmid in 293T cells, then serine was deprived, and flag antibody was used for immunoprecipitation, the gel at the molecular weight position where PHGDH is located was used for LC-MS identification (Figure 5A). The results showed that PHGDH could undergo acetylation modification, with specific sites K289 and K364 (Figure 5B, C). Interestingly, under serine starvation condition, the acetylation site at position K364 still existed, while loss of the K289 site, in turn, produced a new acetylation site K21(Figure 5D, E). These results preliminarily suggest that acetylation modification plays an important role in PHGDH response to serine deprivation.
Figure 5.
Post-translational modification changes of PHGDH protein under serine deprivation. (A) 293T cells transfected with Flag-PHGDH plasmid in complete medium and serine withdrawal medium, respectively, and the protein samples collected after Immunoprecipitation were electrophoresed by SDS-PAGE and the protein distribution maps stained with Kaomasbrilliant blue. The target protein (∼57KD) was collected and analyzed by HPLC-MS. The figure on the right shows the primary mass spectrum of the target protein. (B) and (C) Under complete medium (SW620-Con), PHGDH undergone acetylation modification, with specific sites K289 and K364. (D) and (E) Under serine starvation conditions (SW620-Ser), the acetylation site at position K364 and K21.
Discussion
Abnormal amino acid metabolism plays an important role in the occurrence and development of various tumors. Therefore, amino acid metabolic restriction strategies have become one of the hotspots in anti-tumor research.19 Amino acid metabolic restriction strategies can be divided into limiting amino acid intake and targeted inhibition of amino acid metabolic enzymes. Many amino acid metabolizing enzymes are expressed at significantly elevated levels in tumors, such as GLS, MAT2A, PHGDH, PSAT1, etc.20–23 Studies targeting the expression and function of amino acid metabolizing enzymes will help to improve antitumor strategies for amino acid metabolic restriction.
As the rate-limiting enzyme of endogenous serine de novo synthesis pathway, the role of PHGDH in various tumors and the research and development of inhibitors have been widely concerned.24–28 Studies have reported that PHGDH protein is highly expressed in colon cancer and has a significant negative correlation with the prognosis of colon cancer.7,8 Therefore, employing the treatment of colon cancer cells by PHGDH-specific inhibitors and dissecting the alterations occurring intracellularly by multiomics techniques will contribute to the understanding of PHGDH function. Serine is a nonessential amino acid that can be taken up either from the external environment or synthesized via the endogenous pathway. When the balance of extrinsic and intrinsic pathways is broken, cells need to alter the gene transcriptional state to control the metabolic level, thus achieving remodeling of the metabolic state. In this study, we analyzed genomic transcriptional and metabolic changes in colon cancer cells after PHGDH inhibition using reference transcriptomics and metabolomics techniques. Transcriptomic analysis revealed that many amino acid metabolic pathways were significantly enriched when PHGDH was inhibited, such as regulation of transmembrane receptor protein serine/threonine kinase signaling pathway, glycine, serine and threonine metabolism, cysteine and methionine metabolism. Transcriptomics also suggested that survival proliferation-related signaling pathways were highly enriched. Altered expression of these genes facilitated metabolic reprogramming, so we also performed metabolomic analysis. Many amino acids were found to be altered in content, such as methionine, proline, and isoleucine. The enrichment analysis also shows that amino acid metabolic pathway, lipid synthesis related pathways and phospholipid metabolic pathway were highly enriched. Interestingly, inhibition of PHGDH revealed enrichment of sphingolipid metabolic pathways, whereas metabolic alterations of sphingolipids have been reported to occur following serine deprivation, supporting tumor growth.28,29 These studies suggested that either exogenous restriction of serine utilization or endogenous inhibition of serine synthesis can produce partially identical metabolomic alterations. The close link between sphingolipid metabolism and serine metabolism was also demonstrated again. Therefore, remodeling studies on gene expression changes and metabolic status involved in serine metabolism that are conducive to in-depth understanding of the complex interactive link between cellular gene expression and metabolism.
The vast majority of enzymes are essentially proteins. The synthesis of proteins undergoes a process of transcription and translation from genes. Metabolic enzymes actively respond to changes in metabolite content. Therefore, we analyzed the gene and protein expression of PHGDH under serine-deficient conditions in this project. ATF4 is a major transcription factor in response to amino acid restriction. When amino acids in the environment are deficient, free tRNA increases, which activates GCN2, promotes phosphorylation of eIF2, and increases ATF4 mRNA translation into protein.17 We observed a significant increase in protein levels of ATF4 in intestinal cancer cells as soon as 1 h after serine deprivation, verifying that the increase of ATF4 under amino acid deprivation conditions occurred first at the translational level. However, knockdown of ATF4 expression in colon cancer cells restored PHGDH expression levels to resting levels under serine deprivation conditions, which is also consistent with the previous report that ATF4 is a factor in the transcriptional regulation of PHGDH.18 Transcription factor functioning requires accurate localization to the promoter region of a gene, during which histone methylation modifications play an important role. It has been reported that histone methyltransferase G9a can promote the transcriptional expression of PHGDH under serine deprivation conditions.30 These studies illustrate that transcriptional regulation is essential for PHGDH expression during the response to serine deprivation.
For proteins, post-translational modifications can regulate their stability and activity. Post-translational modifications include ubiquitination, methylation, acetylation, sumo oxidation, etc.31,32 Mass spectrometry is an effective method to detect post-translational modifications. By comparing the post-translational modification of PHGDH before and after serine deprivation, we found that the acetylation site at position K364 still existed after serine deprivation, while the loss of position K289 produced a new acetylation site K21. The findings illustrate that acetylation modification has an important role for PHGDH in response to serine deprivation, but the specific mechanism is not clear. Acetylation modification sites of PHGDH have also been reported to be K21, K33, K58, K146 and K289, and this study concluded that acetylation modification at position K58 is essential for the protein stability of PHGDH.33 Considering that the high expression of PHGDH under serine deprivation is mainly because of increased transcriptional regulation, whether acetylation modification has an important effect on the enzymatic activity of PHGDH is a question worthy of in-depth study.
The disadvantage of this study is that we did not verify the above findings at the clinical level. The research of PHGDH in colon cancer is still in progress. We are now constructing colon cancer organoids, and in the future, we will conduct in-depth studies in patient-derived colon cancer organoids and tissues. We hope our research can contribute to the intervention of tumor nutrition in tumor therapy.
Conclusion
Our study clarified the role of PHGDH in gene transcription and metabolism in colon cancer cells by performing transcriptomic and metabolomic analysis via inhibition of PHGDH. The mechanism of high PHGDH expression in colon cancer cells and the acetylation modification that occurs in PHGDH protein were also clarified by serine deprivation. Our study will provide new knowledge for in-depth understanding of serine metabolism and PHGDH function in colon cancer.
Supplemental Material
Supplemental material, sj-xlsx-1-tct-10.1177_15330338221145994 for Multi-omics Analysis of the Role of PHGDH in Colon Cancer by Zhihui Dai, Lin Chen, KaiLing Pan, XiaoYa Zhao, WenXia Xu, JinLin Du and Chungen Xing in Technology in Cancer Research & Treatment
Supplemental material, sj-xlsx-2-tct-10.1177_15330338221145994 for Multi-omics Analysis of the Role of PHGDH in Colon Cancer by Zhihui Dai, Lin Chen, KaiLing Pan, XiaoYa Zhao, WenXia Xu, JinLin Du and Chungen Xing in Technology in Cancer Research & Treatment
Acknowledgment
Thanks to the platform support of the Central Laboratory, Affiliated Jinhua Hospital, Zhejiang University School of Medicine.
Footnotes
Author Contributions: Zhihui Dai: Writing - Original draft preparation and formal analysis. Lin Chen: Data curation, Methodology. Kailing Pan: Formal analysis, Software. Xiaoya Zhao: Methodology, Data curation. Wenxia Xu: Writing - Review & Editing, Funding acquisition, Resources. Jinlin Du: Funding acquisition, Supervision, Project administration. Chungen Xin: Visualization, Supervision, Funding acquisition.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Our study did not require an ethical board approval because it did not contain human or animal trials. All experimental protocols were approved by the Medical Ethics Committee of Jinhua Hospital of Zhejiang University.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Social Development Key Project of Jinhua Science and Technology Planning Project, CN (2019-3-004); Jinhua Science and Technology Research Program, CN (2020-3-046); Jinhua Science and Technology Research Program, CN (2021-3-036); Jinhua Science and Technology Research Program, CN (2018-3-001e); Program of Zhejiang Province Natural Science Foundation, CN (LY21H160014); Special Research Fund for Basic Research of Jinhua Central Hospital, CN (JY2020-6-08).
ORCID iDs: JinLin Du https://orcid.org/0000-0003-0135-3935
Chungen Xing https://orcid.org/0000-0001-7865-1258
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA. Cancer J Clin. 2018;68(6):394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16(10):650-662. 10.1038/nrc.2016.81 [DOI] [PubMed] [Google Scholar]
- 3.Montrose DC, Saha S, Foronda Met al. Exogenous and endogenous sources of serine contribute to colon cancer metabolism, growth, and resistance to 5-fluorouracil. Cancer Res. 2021;81(9):2275-2288. 10.1158/0008-5472.Can-20-1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H, Park YJ. Links between serine biosynthesis pathway and epigenetics in cancer metabolism. Clin Nutr Res. 2018;7(3):153-160. 10.7762/cnr.2018.7.3.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattaini KR, Sullivan MR, Vander Heiden MG. The importance of serine metabolism in cancer. J Cell Biol. 2016;214(3):249-257. 10.1083/jcb.201604085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snell K. Enzymes of serine metabolism in Normal, developing and neoplastic rat tissues. Adv Enzyme Regul. 1984;22:325-400. https://pubmed.ncbi.nlm.nih.gov/6089514. [DOI] [PubMed] [Google Scholar]
- 7.Jia X-Q, Zhang S, Zhu H-Jet al. et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9(3):191-196. 10.1016/j.tranon.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon S, Kim JG, Seo ANet al. Clinical implication of serine metabolism-associated enzymes in colon cancer. Oncology. 2015;89(6):351-359. 10.1159/000439571 [DOI] [PubMed] [Google Scholar]
- 9.Ngo B, Kim E, Osorio-Vasquez Vet al. Limited environmental serine and Glycine confer brain metastasis sensitivity to PHGDH inhibition. Cancer Discov. 2020;10(9):1352-1373. 10.1158/2159-8290.CD-19-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Fu J, Hu Bet al. Serine metabolism regulates YAP activity through USP7 in colon cancer. Front Cell Dev Biol. 2021;9:639111. 10.3389/fcell.2021.639111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S-Z, Wang J, Zhu L-Bet al. et al. Quantitative label-free proteomic analysis reveals differentially expressed proteins in the digestive juice of resistant versus susceptible silkworm strains and their predicted impacts on BmNPV infection. J Proteomics. 2020;210:103527. 10.1016/j.jprot.2019.103527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367-1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Chen G, Huang J. Shot-gun proteome and transcriptome mapping of the jujube floral organ and identification of a pollen-specific S-locus F-box gene. PeerJ. 2017;5:e3588. 10.7717/peerj.3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Dai Z, Ge Cet al. et al. Specific metabolic response of patient-derived organoids to curcumin of colorectal cancer. J Chromatogr B, Anal Technol Biomed Life Sci. 2022;1203:123260. 10.1016/j.jchromb.2022.123260 [DOI] [PubMed] [Google Scholar]
- 15.Shi Z-Q, Wang L-Y, Zheng J-Y, Xin G-Z, Chen L. Lipidomics characterization of the mechanism of Cynomorium songaricum polysaccharide on treating type 2 diabetes. J Chromatogr B, Anal Technol Biomed Life Sci. 2021;1176:122737. 10.1016/j.jchromb.2021.122737 [DOI] [PubMed] [Google Scholar]
- 16.Mullarky E, Lucki NC, Beheshti Zavareh Ret al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci U S A. 2016;113(7):1778-1783. 10.1073/pnas.1521548113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20(9):436-443. 10.1016/j.tem.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282(23):16744-16753. 10.1074/jbc.M610510200 [DOI] [PubMed] [Google Scholar]
- 19.Pathria G, Ronai ZA. Harnessing the co-vulnerabilities of amino acid-restricted cancers. Cell Metab. 2021;33(1):9-20. 10.1016/j.cmet.2020.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Yang X, Zhang Q, Sun L, Yuan S, Xin Y. Targeting GLS1 to cancer therapy through glutamine metabolism. Clin Transl Oncol Off Publ Fed Spanish Oncol Soc Natl Cancer Inst Mex. 2021;23(11):2253-2268. 10.1007/s12094-021-02645-2 [DOI] [PubMed] [Google Scholar]
- 21.Sánchez-Castillo A, Vooijs M, Kampen KR. Linking Serine/Glycine metabolism to radiotherapy resistance. Cancers (Basel). 2021;13(6):1191. 10.3390/cancers13061191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maldonado LY, Arsene D, Mato JM, Lu SC. Methionine adenosyltransferases in cancers: Mechanisms of dysregulation and implications for therapy. Exp Biol Med (Maywood). 2018;243(2):107-117. 10.1177/1535370217740860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Fu J, Du J, Xu W. The role of D-3-phosphoglycerate dehydrogenase in cancer. Int J Biol Sci. 2020;16(9):1495-1506. 10.7150/ijbs.41051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C, Zheng K, Jiang Ket al. et al. The alternative activity of nuclear PHGDH contributes to tumour growth under nutrient stress. Nat Metab. 2021;3:1357-1371. 10.1038/s42255-021-00456-x [DOI] [PubMed] [Google Scholar]
- 25.Itoyama R, Yasuda-Yoshihara N, Kitamura Fet al. Metabolic shift to serine biosynthesis through 3-PG accumulation and PHGDH induction promotes tumor growth in pancreatic cancer. Cancer Lett. 2021;523:29-42. 10.1016/j.canlet.2021.09.007 [DOI] [PubMed] [Google Scholar]
- 26.D’Avola A, Legrave N, Tajan Met al. PHGDH is required for germinal center formation and is a therapeutic target in MYC-driven lymphoma. J Clin Invest. 2022;132(9):e153436. 10.1172/JCI153436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spillier Q, Frédérick R. Phosphoglycerate dehydrogenase (PHGDH) inhibitors: A comprehensive review 2015-2020. Expert Opin Ther Pat. 2021;31(7):597-608. 10.1080/13543776.2021.1890028 [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Lee K, Reid MAet al. et al. Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep. 2018;22(13):3507-3520. 10.1016/j.celrep.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muthusamy T, Cordes T, Handzlik MKet al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature. 2020;586(7831):790-795. 10.1038/s41586-020-2609-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J, Li T, Wang Xet al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18(6):896-907. 10.1016/j.cmet.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Yang L, Liu M, Luo J. Protein post-translational modifications in the regulation of cancer hallmarks. Cancer Gene Ther. 2022. 10.1038/s41417-022-00464-3 [DOI] [PubMed] [Google Scholar]
- 32.Salas-Lloret D, González-Prieto R. Insights in post-translational modifications: Ubiquitin and SUMO. Int J Mol Sci 2022;23(6):3281. 10.3390/ijms23063281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Wan X, Yu Tet al. Acetylation stabilizes phosphoglycerate dehydrogenase by disrupting the interaction of E3 ligase RNF5 to promote breast tumorigenesis. Cell Rep. 2020;32(6):108021. 10.1016/j.celrep.2020.108021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-tct-10.1177_15330338221145994 for Multi-omics Analysis of the Role of PHGDH in Colon Cancer by Zhihui Dai, Lin Chen, KaiLing Pan, XiaoYa Zhao, WenXia Xu, JinLin Du and Chungen Xing in Technology in Cancer Research & Treatment
Supplemental material, sj-xlsx-2-tct-10.1177_15330338221145994 for Multi-omics Analysis of the Role of PHGDH in Colon Cancer by Zhihui Dai, Lin Chen, KaiLing Pan, XiaoYa Zhao, WenXia Xu, JinLin Du and Chungen Xing in Technology in Cancer Research & Treatment