Abstract
Proteins that perform their activity within the cytoplasmic membrane or outside this cell boundary must be targeted to the translocation site prior to their insertion and/or translocation. In bacteria, several targeting routes are known; the SecB- and the signal recognition particle-dependent pathways are the best characterized. Recently, evidence for the existence of a third major route, the twin-Arg pathway, was gathered. Proteins that use either one of these three different pathways possess special features that enable their specific interaction with the components of the targeting routes. Such targeting information is often contained in an N-terminal extension, the signal sequence, but can also be found within the mature domain of the targeted protein. Once the nascent chain starts to emerge from the ribosome, competition for the protein between different targeting factors begins. After recognition and binding, the targeting factor delivers the protein to the translocation sites at the cytoplasmic membrane. Only by means of a specific interaction between the targeting component and its receptor is the cargo released for further processing and translocation. This mechanism ensures the high-fidelity targeting of premembrane and membrane proteins to the translocation site.
The ultrastructure of a bacterial cell such as Escherichia coli may appear simple compared to that of eukaryotic cells, but nevertheless the cell contains several compartments. Starting from the inside, there is the cytosol, the cytoplasmic membrane, the periplasm, the outer membrane, and, finally, the exterior. Proteins synthesized in the cytosol at ribosomes must be targeted to the right compartment in order to fulfil their specific function. This review deals with the first step in the process of the delivering proteins to their final destination: the targeting of presecretory proteins from the cytosol to the translocation site in the cytoplasmic membrane. The targeting to their final destination following their translocation across the inner membrane is not discussed. Several targeting routes for presecretory proteins have been identified, of which the signal recognition particle (SRP)- and SecB-dependent targeting routes are the best studied. Recently, the twin-arginine route has been proposed as a new pathway.
TRANSLOCATION MACHINERY
The E. coli translocation machinery is a well-studied enzyme complex, which consists of several integral membrane proteins and a peripheral bound ATPase (reviewed in references 46, 48, and 50). The core of this multisubunit enzyme is formed by the integral membrane proteins SecY, SecE, and SecG, together with the peripherally bound SecA (22, 45, 65) (see Fig. 3). Collectively, they are termed translocase. Additionally, there are two membrane proteins, SecD and SecF, that are not essential for protein translocation per se but that stabilize SecA in its active conformation (49). The Sec system is involved in the translocation of precursor proteins across the cytoplasmic membrane, and recent data suggests that it is also needed for the insertion of integral membrane proteins.
FIG. 3.
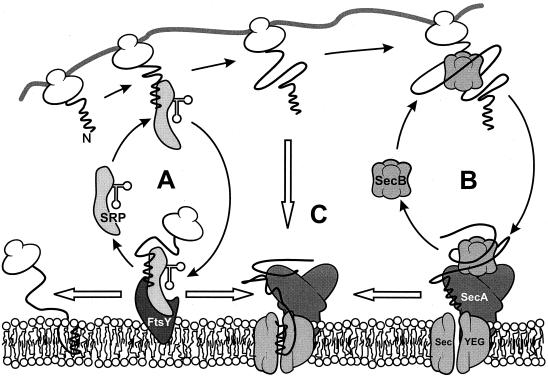
Coexisting SRP- and SecB-dependent targeting routes. When a preprotein or integral membrane protein emerges from the ribosome, the signal sequence domain is exposed first. (A) When this domain is highly hydrophobic, it will bind SRP and the nascent preprotein will be targeted to the membrane-bound FtsY at the membrane. At the membrane, SRP is released from the preprotein in a GTP-dependent manner. The preprotein or integral membrane protein is subsequently transferred to the translocase, which consists of the integral membrane proteins SecY, SecE, and SecG and the peripherally bound ATPase SecA, or inserts into the membrane via a different pathway. (B) When the signal sequence escapes SRP binding and the mature domain possesses the right features, SecB will bind the mature domain and targets the preprotein to the membrane-bound SecA. At the membrane, SecB donates the preprotein to SecA. Upon the binding of ATP by SecA, translocation is initiated and SecB is released from the ternary complex. (C) When the preprotein is not recognized by SecB, the signal sequence may target it directly to the membrane-bound SecA.
Preprotein Translocation Is Governed by the ATPase SecA
The key player in the translocation of preproteins across the cytoplasmic membrane is the ATPase SecA. SecA supplies the energy for the preprotein translocation reaction by mediating repeated cycles of ATP binding and hydrolysis (159). SecA performs a dazzling array of activities: it binds to almost all the components of the translocation process, exhibits ATPase activity, and regulates its own expression by binding to its own mRNA (reviewed in reference 47). When bound to the membrane at the SecYEG complex, SecA is “activated” for the high-affinity recognition of the SecB export chaperone (67), for the leader (signal) region of preproteins, and for the mature domain of precursor proteins. Binding of the precursor protein activates SecA for the exchange of bound ADP for ATP at one of its two ATP-binding sites (105). The energy of ATP binding is thought to drive the insertion of SecA domains (51) plus a loop of the signal sequence and the amino-terminal region of the preprotein across the membrane (159). After insertion, release of the inserted preprotein requires hydrolysis of ATP. Retraction of SecA from its membrane-inserted state occurs upon binding and hydrolysis of a second ATP at a distinct ATP-binding site on the SecA molecule (52). Repeated cycles of SecA preprotein association, ATP binding and hydrolysis, and SecA-preprotein dissociation result in the stepwise translocation of the entire preprotein across the membrane.
TARGETING SIGNALS
Precursor proteins are equipped with signals that are recognized by targeting factors to direct them to the translocation site. For Sec-dependent translocation, these signals are located both in the mature part of the preprotein and in a short amino-terminal extension of the protein, the signal sequence. Not all proteins that are targeted to the membrane have to cross this barrier. Cytoplasmic membrane proteins are anchored in the inner membrane with hydrophobic α-helical stretches that function as internal degenerative signal sequences, sometimes referred to as stop-transfer signals. For these proteins, only the periplasmic loops have to be translocated across the membrane.
Signal Sequence
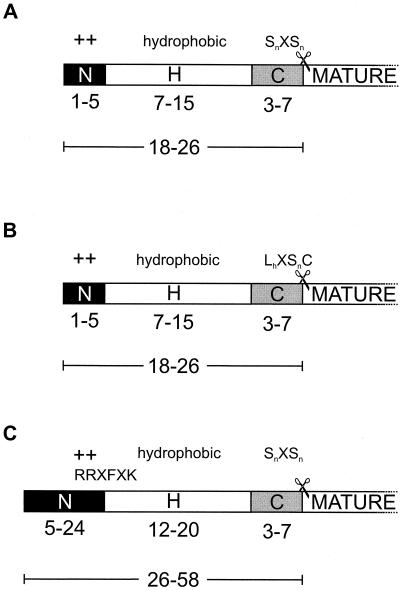
The signal sequence of secretory proteins functions as both the targeting and recognition signal and ranges in length from 18 to about 30 amino acid residues. It is composed of three domains: the positively charged amino terminus (N region); the nonpolar, hydrophobic core region (H region); and the more polar cleavage region (C region) (Fig. 1) (183). The amino acid sequences of these domains are not well conserved among the many signal sequences, but their physicochemical properties are (78).
FIG. 1.
Domain structure of the signal sequence of precursor proteins. (A) Signal sequences of SRP- or SecB-dependent preproteins have a net positive charge in the N region (indicated by +), a hydrophobic H region, and a C region with the signal peptidase cleavage site (✂) preceded by the motif SnXSn, in which Sn stands for an amino acid with a small neutral side chain and X stands for any amino acyl residue. (B) Type II signal sequences LhXSnC, in which Lh stands for an amino acid with a large hydrophobic side chain and C stands for cysteine. The cleavage site is located between Sn and C. (C) Signal sequences of precursor proteins that are dependent on the twin-Arg route resemble normal signal sequences but have an extended N region and possess the RRXFXK motif, which straddles the H and C domains (9). For both types of signal sequences, the variation in length of the different regions and of the total signal sequence is indicated.
N domain.
The presence of a net positive charge in the N region, introduced by lysine or arginine residues, enhances the processing and translocation rates of a precursor protein but is not essential. Preproteins with signal sequences that carry a neutral or even negatively charged N region can be processed, albeit at reduced rates (64). With increasing positive charge at the N region of the signal sequence, the SecA requirement for translocation is reduced while the interaction of the preprotein with SecA is enhanced (1). This suggests that the N region is involved in the targeting of the preprotein to the translocase. Interestingly, a secretion deficiency caused by a reduction in the number of positive charges can be restored by mutations in secA (140, 141). However, the same mutations in secA also suppress mutations in the hydrophobic core of the signal sequence (57, 76), indicating that the suppressing mutation may not simply restore the recognition of the defective signal sequence by SecA. The N region has been suggested to bind the negatively charged surface of the lipid bilayer of the membrane (42). A reduction in the number of positive charges in the N region results in inefficient interaction with the membrane; this phenomenon can be compensated for by an increased hydrophobicity of the H region (134). Strikingly, this is accompanied by a restoration of the translocation defect, suggesting that the interaction of the signal sequence domain with the membrane is an important step in targeting and/or translocation. The positive charges in the N region are thought to orient the signal sequence of secretory proteins or the stop-transfer signal of membrane proteins correctly within the lipid bilayer. The transmembrane electrical potential (Δψ, inside negative) prevents the translocation of positively charged residues and facilitates that of negatively charged residues (5). In this manner, the Δψ would contribute to the realization of the correct topology of integral membrane proteins, which obey the “positive-inside rule” (184). On the other hand, in acidophilic bacteria and archaea, the Δψ has a reversed polarity, i.e., inside positive versus outside. The topology of the inner membrane proteins of these bacteria also obeys the positive-inside rule (179), whereas the signal sequences of the secretory proteins identified thus far are undistinguishable from those of neutro- or alkalophiles. This challenges the electrophoretic mechanism and suggests that Δψ may affect protein translocation and membrane insertion by another mechanism that would involve the translocation apparatus in a more direct manner.
H domain.
The H domain is the hydrophobic core of a signal sequence and varies in length from 7 to 15 amino acids. It is the most important part of the signal sequence; this is best illustrated by the observation that an increase in the total hydrophobicity of this domain can overcome mutations in the other regions of the signal sequence (see, e.g., references 74, 111, and 134). To some extent, the total hydrophobicity of the H region determines the efficiency of translocation: the translocation efficiency increases with the length and hydrophobicity of the H region (26). This relation is sigmoidal, and a minimum hydrophobicity is required for translocation (44). The residues in the H region are responsible for the α-helical conformation which extends from the N region. Frequently, a so-called helix breaker, i.e., a glycyl or prolyl residue, is found in the middle of the H region. This may allow the signal sequence to form a hairpin-like structure that can insert into the lipid bilayer. According to the unlooping model, the signal sequence inserts into the membrane by extension through unlooping of this hairpin (42, 165). Indeed, nuclear magnetic resonance spectroscopy studies have shown that in a membranous environment, signal sequences may adopt a two-domain conformation consisting of an amino-terminal α-helix and a more flexible carboxy-terminal domain (27, 151, 188). Also, when two cysteines are introduced into the signal sequence, translocation under oxidized conditions is hampered (128), indicating that the formation of the loop prevents translocation. Unlooping would be facilitated by the Δψ to orient the signal sequence within the electrical field. In this respect, some replacements of the α-helix breakers with α-helix-forming residues render preprotein translocation less dependent on the proton motive force (129), although the effects are rather subtle. In our opinion, it seems that the formation of the loop slows translocation rather than being an essential element of the translocation reaction. The unlooping model does not explain how the signal sequence inserts into the membrane while bound to SecA. It is more likely that both SecA and the signal sequence insert simultaneously at the translocation site upon the binding of ATP.
C domain.
The leader peptidase cleavage site (C domain) is the only part of the signal sequence that demands some primary sequence specificity. Two types of leader peptidases are known, type I, serving ordinary preproteins, and type II, cleaving the leaders of lipoproteins. For the signal sequences that rely on type I leader peptidases, the constraints are on the residues located at positions −1 and −3 relative to the start of the mature part (182). This domain interacts with the leader peptidase which cleaves off the signal sequence (for a review, see reference 34). Usually, these residues have small neutral side chains, such as alanine, glycine, serine, and threonine, with a preference for alanine (182). Lipoproteins rely on type II leader peptidases, and the demands differ from those for type I peptidases only at the −3 and +1 positions (154). Precursors of lipoproteins contain larger hydrophobic amino acid residues at the −3 position, with a preference for leucine. At the +1 position, a cysteine is always present and has to undergo modification prior to processing. For both prelipoproteins and prenonlipoproteins, the position relative to the H region is also important, since the active site of the leader peptidase is located near the surface of the membrane (36). However, processing is not essential for the actual translocation process, and the cleavage site can be omitted, resulting in a protein that remains anchored to the membrane by an uncleaved signal sequence. After the signal sequence has been removed, it is degraded completely by a number of peptidases (184).
Interaction between the Signal Sequence Domain and the Translocase
Although the signal sequence can be divided into three domains, it functions as a single entity. This is illustrated by the observation that deleterious alterations in one domain can be restored by mutations in another (64, 78). Also, extragenic mutations that relieve the translocation blockade caused by the signal sequence modifications have been found. These are termed prl mutations, for “protein localization.” Such mutations are found in secA, secY, secE (for reviews, see references 11 and 158), and secG (16), suggesting a direct interaction of the signal sequence with all of the components of the translocase. Biochemically, however, only SecA has been shown to interact directly with signal peptides, since they can alter the ATPase activity of SecA (33, 105) and can be chemically cross-linked to it (116). It is not clear whether the signal sequence first interacts with the integral membrane components of the translocation machinery and then inserts into the membrane, or reverse.
Role of the Mature Domain
Targeting information may also be located within the mature part of precursor proteins. This is underscored by the observation that proteins can be translocated even when they lack a signal sequence (41, 59, 138). In these situations, the preprotein relies totally on targeting factors such as SecB. Also, not all proteins that are artificially equipped with a signal sequence are indeed translocated (101). Translocation is often prevented by the presence of positively charged residues at the beginning of the mature domain (101, 104, 197). This is also true for signal sequence-less precursor proteins (138), suggesting that even in the absence of a signal sequence, the interaction of the mature domain of secretory proteins with SecA is similar to that of native precursors. When the positively charged residues are removed, the translocation of the preprotein is restored. Also, an increase in the length of the H domain of the signal sequence can restore normal translocation (112). It could well be that a positive charge juxtaposed to the signal sequence interferes with the simultaneous insertion of SecA and the preprotein, because it will oppose the positive-inside rule (see above).
SECB-DEPENDENT TARGETING
The first indication that preproteins in E. coli rely on a factor early in the translocation route came from the discovery of the secB gene (87, 88). Deletion of this gene resulted in a mild translocation defect of only a subset of precursor proteins (88). SecB is a highly acidic protein, with a predicted molecular mass of about 17 kDa (90), that exists as a tetramer (166, 189). It plays a dedicated role in guiding precursor proteins to the cytoplasmic membrane (86). Unlike many other chaperones, SecB is not an ATPase. So far, homologues of SecB have been found only in gram-negative bacteria, and completed sequences are available for Buchnera aphidicola (95) and Haemophilus influenzae (58).
Heat Shock Proteins Cannot Substitute for the SecB Function
SecB is not an essential protein, and only under certain conditions, i.e., growth on rich medium, was the absence of SecB believed to be lethal (88). In assays based on the restoration of growth on rich medium, it was found that the heat shock proteins GroEL and DnaK could restore the growth defect of a secB-null strain (4, 195). However, the inability to grow on rich medium can be accounted for by the lack of gpsA expression in the secB-null strain (164). This gene codes for sn-glycerol-3-phosphate dehydrogenase, which is involved in phospholipid synthesis. The coding region of gpsA begins within the termination codon of secB (168), and deletion or disruption of the secB gene leads to inactivation of gpsA expression, which causes the growth defect on rich medium. Although the chaperones GroEL and trigger factor are capable of maintaining a preprotein in a translocation-competent state in vitro, they fail to stimulate the translocation (99), suggesting that they can substitute for SecB in the translocation reaction with regard to the interaction with the preprotein, but cannot properly target the preprotein to the translocase. Large amounts of trigger factor even inhibit translocation.
Lowering the level of the general heat shock proteins GroE, DnaK, DnaJ, and GrpE results in increased SecB production, while disturbance of translocation caused by mutations in other sec genes has no effect on secB expression (123). Conversely, accumulation of preproteins induces the production of the heat shock proteins but not that of SecB (94, 195). This indicates that the secB promoter is not sensitive for a translocation feedback mechanism but responds to a lack of other chaperones. The secB promoter is also sensitive to the carbon source, since SecB production in the presence of acetate, glycerol, and maltose is higher than in the presence of glucose (161). Addition of cyclic AMP and the presence of the cyclic AMP receptor can partially restore the lower SecB levels in glucose medium, indicating that the secB gene is under the control of catabolite repression (161).
The processing of the SecB-independent precursor of alkaline phosphatase (AP) is disturbed in cells lacking DnaK or DnaJ (194), and it has been suggested that these chaperones play a more general role in the targeting of SecB-independent precursor proteins. Overexpression of DnaK can even protect the cells from the toxic effect of jamming of the translocation site by precursor proteins fused to β-galactosidase (132). Taken together, these data point to an early but marginal role for DnaK and DnaJ in the targeting of precursors to the translocase.
Selective Binding of Preproteins by SecB
In vivo, SecB binds highly selectively to only a subset of precursor proteins (85, 91), but in vitro, it interacts with all kinds of proteins provided that they are in a nonnative conformation (54, 100). Although conflicting opinions exist (190, 191), it is generally believed that SecB binds the preprotein at its mature domain (145). The signal sequence provides no positive contribution to the binding energy or binding affinity of the interaction of the preprotein with SecB (147), but the signal sequence slows the folding of the mature domain (66, 130). It has been suggested that this slowing process allows SecB to discriminate between the precursor proteins and other proteins in the cell, as formulated in the kinetic partitioning model (66). In this model, the cytosolic proteins would escape stable interaction by folding more rapidly than precursor proteins do. The final distribution of the precursor protein among the different pathways in the cell is then determined by partitioning that is dependent on the rate of folding or aggregation relative to the rate of binding to the chaperone (145). Indeed, when the folding rate of the SecB-dependent precursors of maltose-binding protein (MBP) and galactose-binding protein (GBP) was increased, the amount of unbound ligand increased as well (174). However, several other observations are against the folding rate as the determining factor for SecB selectivity. First, in vivo, the folding rate is determined by the rate of chain elongation, and SecB is known to bind nascent chains (91) regardless of the presence of a signal sequence (146). Second, SecB is capable of binding fully folded proteins, albeit with low affinity, and can even unfold them and guide them back to the transport pathway (199). Third, when the folding rate of barnase is changed, the affinity for SecB is not. Moreover, barnase folds into its native conformation while bound to SecB (169). Fourth, the binding occurs at rates that are much higher than the folding rates (54). Finally, precursor proteins that lack a signal sequence and are not slowed in their folding become totally dependent on SecB for their translocation (41, 56, 59, 138, 180).
SecB maintains the precursor protein in a loosely folded conformation, which is competent for translocation (30, 99, 100, 144, 193). However, not all proteins that are SecB dependent for in vivo translocation are maintained in vitro in a translocation-competent state by SecB (37, 60). This clearly points to an additional and presumably a more important role for SecB as targeting factor. This was already suggested by Kumamoto and Gannon (89), who demonstrated that the deletion of the secB gene caused a defect in cotranslational translocation. In agreement with this, the interaction between SecB and preproteins had already occurred when the preprotein was still nascent and emerged from the ribosome (91, 146). For this interaction, the presence of a signal sequence is not a prerequisite (146). In fact, when the signal sequence is omitted, targeting and translocation become entirely SecB dependent (41, 56, 59, 138, 180).
Analysis of the interaction between Bacillus amyloliquefaciens RNase (barnase) and SecB by two-dimensional nuclear magnetic resonance spectroscopy revealed that SecB can bind to native barnase, and the data indicate that SecB could even fully unfold barnase (199). It was concluded that SecB has the potential to act as an unfoldase by using protein-protein binding energy to denature the bound polypeptide chain to its fully unfolded state, allowing misfolded states to be corrected and guided back to the transport pathway. Although SecB can bind to fully unfolded and fully folded barnase, the majority of the bound substrate is in the intermediate folding state (169, 198), suggesting that the unfolding activity is more a backup system than a primary task of SecB.
The structural features of the SecB-binding frame in preproteins have been assessed by determination of the nature of SecB-protected proteolytic preprotein fragments (80, 167, 173) and deletion analysis (2, 3). For the outer membrane protein LamB, a region between amino acids 320 and 380 has been proposed to be the main determinant for SecB binding (2, 3). The SecB-binding frame of the periplasmic proteins MBP (173), GBP (80), and oligopeptide-binding protein (OppA) (167) is centrally located in the primary structure of these proteins. For MBP and GBP, the binding frame is about 160 amino acyl residues long, while that of OppA covers about 460 amino acids. Comparison of these binding frames has not revealed any obvious resemblance. Recently, it was suggested that the SecB-binding site on precursor proteins is composed of a stretch of several basic residues (81). This was based on studies with sequence modules that were inserted into the mature domain of AP, which is normally SecB independent for its targeting. Insertion of several basic residues conferred SecB dependence on the precursor, suggesting that this introduced a SecB-binding site into the protein. However, when the signal sequence of AP is removed, translocation also becomes SecB dependent (41). This implies that a SecB-binding site is already present in AP. Therefore, it could well be that the introduced basic residues negatively affect the targeting information, for instance by interacting with the signal sequence or preventing the proper interaction with SecA. This would make AP dependent on SecB for proper targeting. Alternatively, the SecB-binding site could normally be obscured and become exposed upon the introduction of basic residues into the polypeptide sequence. Overall, it can be concluded that the structural basis of preprotein recognition is not well understood at this juncture.
Sites of Preprotein and SecA Binding on SecB
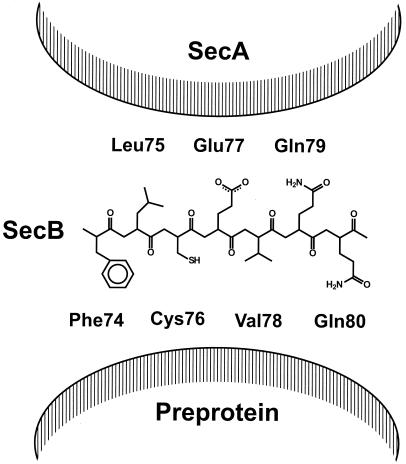
In contrast to the undefined SecB-binding motif in preproteins, specific mutations in SecB that disrupt the interaction between SecB and preproteins have been characterized (63, 82). These mutations are restricted to a short polypeptide stretch that contains mainly hydrophobic residues. These residues are conserved in the known SecB proteins, and their mutation causes a failure in the formation of stable complexes between SecB and preMBP but results in only mild translocation defects (82). Since the mutations that affect preprotein binding occur in an alternating fashion in this restricted region of SecB, it has been suggested that the interaction between SecB and preproteins occurs at a β-β interface and is hydrophobic (Fig. 2) (18, 82). The major substrates for SecB indeed have a high β-sheet content (113), while binding of the preprotein has been shown to involve hydrophobic interactions (54).
FIG. 2.
The SecA- and preprotein-binding sites of SecB are predicted to face in opposite directions in a β-structural conformation. The mainly hydrophobic residues Phe74, Cys76, Val78, and Gln80 are involved in preprotein binding, while the alternating residues Leu75 and Glu77 are involved in SecA binding.
The receptor for SecB at the membrane is SecA (67), but SecA and SecB can already interact in solution (67, 75). This latter interaction is of low affinity (Kd ≈ 1.6 μM) (40), and its physiological relevance is not clear. The SecB-binding site on SecA is located at the carboxy terminus (19) and consists of only the last 22 amino acyl residues (55). This domain, which is functional as a dimer (55), is highly conserved among the bacterial SecA proteins except for those of Streptomyces, Mycobacterium, and Mycoplasma species. It is rich in arginyl and lysyl residues and is therefore positively charged. Due to the high content of glycyl and prolyl residues, this region is probably very flexible. Another remarkable feature is the presence of three cysteinyl residues. Replacement of these residues with serines does not influence the biological function of SecA, as judged from complementation studies with mildly overexpressed SecA (143), but alters the translocation ATPase activity (142). Furthermore, the processing rate of the SecB-dependent precursor proOmpA is decreased whereas that of pre-β-lactamase, which is SecB independent (96), remains unaffected (142).
Based on mutagenesis studies and biochemical characterization, it appears that part of the SecA-binding site on SecB overlaps the preprotein-binding site in an alternating fashion. When the conserved residues Leu75 and Glu77 (Fig. 2) are mutated, SecB still interacts with preproteins but fails to target them to the membrane-bound SecA as a result of the impaired SecA-SecB interaction (56). In a β structural conformation (53), the amphipatic, hydrophobic preprotein-binding site and the polar, negatively charged SecA-binding site are located opposite each other (Fig. 2). Other residues in SecB that are presumably involved in SecA binding are Asp20 and Glu24 (82). These two residues are also conserved among the different SecB proteins. Together with Leu75 and Glu77, they may form a negatively charged surface that can interact electrostatically with the positively charged SecB-binding domain of SecA.
Preprotein Transfer upon SecA-SecB Interaction
In the presence of preprotein, SecB binds SecA with higher affinity (55, 67). Only the signal sequence is needed to stimulate SecA for this high-affinity SecB recognition (56). Upon interaction of SecB with SecA, SecB releases the precursor protein, which is subsequently transferred to SecA (55, 56). This reaction involves binding of the signal sequence of the preprotein to SecA. Upon binding SecB, SecA might lower the affinity of SecB for the preprotein by modulating the conformation of preprotein binding site of SecB. This site is located at the opposite face of the putative β strand that constitutes the SecA-binding site. According to this scenario, the SecA-bound SecB will be unable to interact with a new preprotein, and therefore the binding reaction precludes other proteins from entering the translocation pathway at a site that is already occupied. Only upon initiation of translocation, i.e., after the binding of ATP to SecA, is SecB released from the membrane to bind a new preprotein in the cytosol (55).
SRP-DEPENDENT TARGETING
Until the discovery of an SRP-like particle in E. coli, most of our knowledge on SRP-dependent targeting was based on studies with mammalian cells (reviewed in references 108, 148, and 187). Soluble proteins that have to cross the endoplasmatic reticulum (ER) membrane or membrane proteins that have to integrate into this membrane are targeted to the translocon by SRP and SRP receptor. Targeting occurs cotranslationally with the proteins still attached to the ribosome. Directly after the signal sequence emerges from the ribosome, the nascent chain is bound by SRP. This interaction results in a pause in the translation of the nascent chain, and the ribosome-bound complex is then targeted to the SRP receptor at the ER membrane. The interaction between SRP and its receptor releases SRP from the ribosome and the signal sequence. Subsequently, the translation arrest is relieved. The ribosome binds the membrane-embedded translocon, and the protein is threaded across or into the ER membrane by chain elongation.
Components of the Eukaryotic SRP Route
The eukaryotic SRP is a ribonucleoprotein complex consisting of one 7S RNA molecule as the central core to which six proteins of different sizes from 9 to 72 kDa are attached (185, 186). The 54-kDa protein (SRP54) is the only subunit that interacts with signal sequences (83, 92). SRP54 harbors a GTP-binding motif which is functional in GTP binding and hydrolysis (117). The SRP receptor consists of two GTP-binding proteins, SRα and SRβ (172). SRα is peripherally attached to the membrane via its N-terminal domain (98), while SRβ is a true integral membrane protein (172). Based on the similarity in the GTP-binding boxes, SRP54 and SRα are classified together in a separate subgroup of GTPases (17).
Bacterial SRP
The E. coli homologues of SRP54 and 7S RNA are Ffh (“fifty-four homologue,” also called P48) and 4.5S RNA, respectively (9, 135, 136, 152, 170). These components were identified by sequence comparison and together suffice to constitute a functional SRP. The 4.5S RNA of E. coli SRP is contained in the ffs gene and is much smaller than its eukaryotic and archaeal counterparts and those of most other bacteria (97, 136). 4.5S RNA can functionally be replaced by 7S RNA of human SRP (150). When E. coli cells are depleted of 4.5S RNA or when a dominant mutation in ffs is present, the translocation of preBla is strongly impaired but pre-MBP, pre-RBP and pro-OmpA are processed normally (136, 150). Both SRP54 and Ffh specifically bind to 4.5S RNA in vitro (103, 150), and in turn, Ffh can replace SRP54 in the mammalian SRP for SRP assembly, signal sequence recognition, and translation arrest but not for translocation of preprolactin into microsomal membranes (10). SRP54 fails to restore the defects in an ffs conditional mutant (131). Ffh is essential, since E. coli cells which lack a functional Ffh fail to grow (133). Depletion of Ffh results in an elongated cell phenotype. Furthermore, the translocation of pre-AP, pre-Bla, and pre-RBP in Ffh-depleted cells is strongly hampered while that of pre-MBP, pre-LamB, pre-OmpF, and pro-OmpA is less affected (131, 133).
Based on the primary sequence, Ffh can be divided into three domains: the N domain at the amino terminus; the G domain, which harbors the GTPase activity; and the methionine-rich M domain, which for SRP54 binds both RNA and the signal sequence (153, 203). The M domain of Ffh, however, has been implicated only in RNA binding, while the N and G domains bind signal peptides (201, 202). The crystal structure of the protease-resistant N and G domains (NG domain) of Ffh of Thermus aquaticus has recently been resolved at the atomic level and shows that the GTPase domain resembles that of Ras-GTPases. The two domains are closely associated by a short linker peptide of only 10 residues (61).
SRP Interacts with Hydrophobic Signal Sequences
SRP recognizes its substrate by the presence of a hydrophobic signal sequence, hence the name “signal recognition particle.” Photo-cross-linking experiments demonstrate that the signal sequence binds to the mammalian SRP54 (83, 92), and the E. coli Ffh (107). Just like its eukaryotic homologue (163), Ffh binds only when the preprotein is still associated with the ribosome and when it is between 70 and 150 amino acids long (106). Furthermore, in a heterologous system, SRP54 and Ffh compete for signal sequence binding, but in contrast to SRP54, Ffh binds the signal sequence only when it is associated with 4.5S RNA (107). The interaction of Ffh with the signal sequences increases with the hydrophobicity of the signal sequence. Ffh also binds to internal signal anchor domains like the transmembrane segments of integral membrane proteins (176). It has been suggested that the magnitude of the interaction between SRP and the signal sequence correlates with the translocation efficiency (176), since precursors with a more hydrophobic signal sequence are translocated more efficiently (44). Likewise, the interaction between signal sequences and the SRP54 homologues of plant chloroplasts and yeast is dictated by the signal sequence hydrophobicity (73, 125). The binding of Ffh to the signal sequence in a heterologous system does not result in translational arrest, but the Ffh-4.5S RNA complex can target the eukaryotic ribosome-bound preprotein toward microsomal membranes in an FtsY-dependent fashion and thus can support cotranslational translocation (137). This implies a specific interaction between the ribosome and translocase, since in heterologous systems containing wheat germ ribosomes and E. coli SRP, the nascent preproteins are not targeted to E. coli membranes, nor is SRP released by purified or membrane-bound FtsY (177). By making use of a homologous E. coli system, Valent et al. (177, 178) have shown that E. coli SRP indeed interacts with presecretory and membrane proteins synthesized at the E. coli ribosome. As in the eukaryotic system (68), the additional presence of cytoplasmic membranes is a prerequisite for the GTP-dependent release of the signal sequence (178).
SRP Receptor FtsY
Although an E. coli homologue for SRα was found together with that for SRP54 (9, 152), it was some time before a SRP receptor function was biochemically demonstrated for FtsY (109, 118, 137). For a strong interaction between FtsY and the Ffh-4.5S RNA complex, the presence of a nonhydrolyzable GTP homologue, such as GMP-PNP, is required (118). This resembles the interaction between the mammalian SRP and its receptor (31). Ffh alone seems not to interact with FtsY, even in the presence of GMP-PNP (118). Ffh possesses GTPase activity, and although FtsY also contains a GTPase domain (9, 152) and binds GTP (93), it hardly hydrolyzes GTP on its own (93, 118). The interaction between 4.5S RNA, Ffh, and FtsY results in a large enhancement of the overall GTPase activity (118). Both this elevated GTPase activity and the Ffh GTPase activity are inhibited by the presence of a functional signal peptide, suggesting that FtsY stimulates the GTPase activity of Ffh (118). Mutations in the GTP-binding site of FtsY interfere with GTP binding and hydrolysis and, as a consequence, reduce the ability of FtsY to interact with SRP (93). The phenotypic characteristics of the depletion of FtsY resemble those of the depletion of Ffh and cause the accumulation of the precursor forms of Bla, OmpF, and RBP, whereas those of OmpA, OmpC, and MBP are processed normally (109). Interestingly, overexpression of wild-type FtsY also causes a decreased translocation efficiency of pre-Bla and pre-OmpF, probably because of nonproductive interactions between FtsY and SRP (93, 109). In this respect, the translocation of pro-OmpA and pre-OmpC is unaffected. A similar phenomenon is observed with an FtsY mutant bearing a GTPase defect (93).
FtsY contains two distinct domains: a highly charged N-terminal domain (the A domain) and a C-terminal domain (the NG domain). The crystal structure of the NG domain has recently been solved (119). This domain can structurally be divided into three segments: the N domain; the GTPase domain, which displays similarity to the Ras-related GTPases (17); and an α-β-α insertion into the GTPase domain called the I box, which is absent in Ras-related GTPase and unique to all FtsY and Ffh homologues. The N domain is linked to the GTPase domain by an extended arm that reaches along the surface of the GTPase domain. The linker is much longer than the corresponding linker of Ffh (61) and is sensitive to proteolysis in the absence of nucleotides (93). The affinity of FtsY for GTP is rather low (93, 118), which can partially be explained by the open structure of the nucleotide-binding site (119). It was also found that the nucleotide dissociation rate of FtsY is much higher than in Ras but is similar to those seen in GTPases in the presence of an exchange factor. Since the I box constitutes the difference between the GTPase domain of FtsY and Ras, it has been postulated that this domain functions as a built-in exchange factor (121).
Like its mammalian counterpart, SRα, FtsY is peripherally associated with the membrane (39, 109). Both domains of FtsY associate with the membrane, but the nature of the interaction differs since urea removes the A-domain from the membrane under conditions where the NG domain remains tightly associated (39). Also the interaction between wild-type FtsY and membranes can be disrupted by urea (109), suggesting that FtsY is associated with the membrane via the A domain. This suggestion is reinforced by the finding that expression of the A domain alone results in a lowering of the amount of wild-type FtsY associated with inner membrane vesicles (39). This observation indicates the presence of a specific binding site for FtsY at the membrane, but no homologue of SRβ has so far been identified. FtsY lacking a functional A domain exhibits a lower translocation efficiency because it has reduced binding to microsomal membranes. The translocation activity can be restored by the addition of high concentrations of the mutated FtsY (137). When the NG domain of FtsY is fused to a membrane anchor, it operates normally (200). This demonstrates that the main function of the A domain is the targeting of the NG domain to the membrane. Together, the E. coli SRP and FtsY can functionally replace their mammalian counterparts in targeting nascent secretory proteins to microsomal membranes in vitro (137).
A THIRD TYPE OF TARGETING IN BACTERIA?
In chloroplasts of higher plants, both the Sec machinery and the SRP-dependent route are conserved but there is also another translocation mechanism. This route is ΔpH dependent and mediates the Sec-independent translocation of proteins across the thylakoid membrane (21, 29, 32, 70, 122, 126). Characteristic of this route is the so-called Sec avoidance motif found in the signal peptide domain of the preprotein. This motif consists of a twin arginine at the N terminus and a lysine near the C terminus of the signal sequence (Fig. 1) (14, 20). The twin-Arg motif is essential for targeting the preprotein to the ΔpH-dependent pathway (25), and the lysyl residue in the C domain seems to be the only barrier to Sec-dependent translocation (14). It has been shown that deletion of the hcf106 gene in corn disturbs the localization of proteins transported through this pathway (181). The hcf106 gene codes for a protein that is anchored by a single membrane-spanning domain in the thylakoid membrane, exposing a large fraction of the protein to the stromal side (162). Strikingly, this protein has homologues in some bacteria whose chromosomes are completely sequenced (162). E. coli contains two homologues of Hcf106, which are coded for by ybeC (or tatA) and mttA (or tatB [formerly known as yigT]). Recently, both genes have been shown to be part of a system that mediates Sec-independent protein translocation (156, 192). A mutation of mttA (192) or disruption of tatB or tatA (156) prevented the correct localization of several redox enzymes equipped with a twin-Arg signal sequence. In the gram-negative bacterium Azotobacter chroococcum, the homologous gene is located in a cluster of genes required for H2-dependent respiration (162). For this process, which also occurs in E. coli, hydrogenases are secreted to recycle H2 produced by nitrogen fixation and fermentation (102). Hydrogenase consists of several subunits, and E. coli contains at least three hydrogenases (110), of which two are coded for by the hya and hyb operons (114, 115). Interestingly, the precursors of the known β subunits, HyaA and HybA, are equipped with a twin-Arg-bearing signal sequence (114, 115). The complete genome sequence of E. coli (12) also disclosed a putative protein named f372 that has homology to HyaA and contains the twin-Arg motif in the signal sequence. This protein might be the β subunit of the third E. coli hydrogenase. Expression of the hya operon is induced during anaerobic growth (157). Strikingly, under these conditions, processing of β-lactamase with the HyaA signal sequence is enhanced (127), suggesting that the expression of the translocation machinery is also enhanced. Little is known about the influence of oxygen pressure on the expression of components of the Sec machinery, but since the expression of most of the components is decreased at lower growth rates (139), it is highly unlikely that the expression is elevated during anaerobic growth. Mutational analysis of the HyaA signal sequence revealed that the twin-Arg motif is part of the RRXFXK-motif that is essential for efficient translocation (127). The same motif is also found in the signal sequence domain of bacterial periplasmic proteins binding redox cofactors (8). The properties of these translocated proteins are quite different from those of ordinary preproteins in that the latter are kept in a loosely folded conformation to enable passage through the translocase while the former are believed to be translocated in a completely folded state, sometimes even in association with their cofactors (8). The translocation of a periplasmic E. coli molybdoenzyme TorA, which contains a twin-Arg signal sequence, requires the acquisition of the molybdocofactor in the cytoplasm and bypasses the Sec machinery, including SecB. Furthermore, depletion of the SRP function only marginally affects the export, perhaps indirectly, indicating that pre-TorA is targeted and translocated in a unique manner (155). Remarkably, during ΔpH-dependent translocation across the thylakoid membrane, precursors remain competent for translocation even when they are in a tightly folded conformation (28, 32), a feature also characteristic of the translocation of proteins across the peroxisomal membrane (171). No other stromal factors are needed for translocation (77), but this does not rule out the presence of specific chaperones, since SecB is also not obligatory for translocation. In this respect, it is highly unlikely that the chloroplast homologue of SRP54, 54CP, targets preproteins to the ΔpH-dependent system, since it fails to recognize the required signal sequence (73). The E. coli Sec machinery is unable to translocate thylakoid preproteins with a ΔpH-dependent signal sequence, while a chloroplast Sec-dependent signal sequence is recognized and able to support translocation (71). This suggests that the Sec system can discriminate between normal and ΔpH-dependent signal sequences. Conversely, a twin-Arg-bearing signal sequence of E. coli can direct exclusive translocation via the ΔpH-dependent pathway in thylakoids (120), underscoring the genetic link between the two pathways. It is also important to note that the translocation of precursors along the ΔpH-dependent pathway is not sensitive to azide, an inhibitor of SecA. Taking these results together, a unique ΔpH-dependent export system dedicated to the translocation of folded proteins, which require totally different targeting components than SRP or SecB, seems to exist in E. coli and other bacteria.
CONVERGING TARGETING ROUTES
When a nascent chain emerges from the ribosome, a range of chaperones, folding catalysts, and targeting factors are waiting to bind the new protein (24). At that moment, the different targeting factors will compete for preprotein binding. Most chaperones use ATP and cochaperones as cofactors and bind the emerging polypeptide repeatedly, cycling the nascent chain between free and chaperone-bound forms (23).
Competition between Chaperones for the Nascent Chain
The first chaperone encountered by a nascent chain is probably trigger factor (for a review, see reference 72). This ribosome-bound peptidyl-prolyl cis/trans isomerase does not distinguish between secretory and cytosolic proteins (176, 177), and scans the nascent chain for proline residues. A likely candidate to follow trigger factor is DnaJ, which bind to the N-terminal segment of the nascent protein. DnaJ is accompanied by DnaK, and subsequent association of GrpE leads to ATP-dependent dissociation of the complex (84). When the nascent chain resembles a preprotein bearing a hydrophobic signal sequence, this team of chaperones has to compete with SRP for stable binding (Table 1). In contrast to the “general” chaperones, the factors that cause SRP to dissociate from the nascent chain are specifically localized at the cytoplasmic membrane, i.e., the membrane-bound FtsY (Fig. 3) (178). In our opinion, this enables SRP to outcompete the other chaperones for nascent-chain binding and to stably interact with the signal sequence until it arrives at its destination. The data so far indicate that E. coli SRP fails to cause translational arrest. Moreover, when the nascent chain reaches about 150 amino acids, SRP is released from the ribosome spontaneously (106). This would imply that the time available for functional targeting is limited. When the signal sequence is not hydrophobic enough, it does not interact with SRP (176, 177). This could provide other chaperones with an opportunity to interact with the growing nascent chain. Such a chaperone might be GroEL, but this chaperone binds only after the protein has been released from the ribosome (62, 149). When the proper conditions are met, SecB will bind the nascent chain (Fig. 3; Table 1). SecB cannot be cross-linked to short nascent chains (176), and stable SecB binding depends on the type of precursor. For both pro-OmpA (7) and pre-MBP (146), ∼200 amino acids suffices for the nascent chain, whereas binding to pre-LamB requires very long nascent chains (91). These chain lengths correspond to the assumed location of SecB binding on these proteins. SecB requires the first 229 amino acids of the mature domain of pro-OmpA for proper binding (112), and pre-MBP is bound in the middle of its sequence (30, 173). The SecB-binding site on pre-LamB is located at the carboxy terminus (3). Consistent with this hypothesis is the observation that a fair portion of pre-MBP is translocated cotranslationally whereas pre-LamB is mainly translocated posttranslationally (79). Unlike SRP, SecB can interact with preproteins that already are released from the ribosome. However, we believe that this does not influence the basic mechanism of SecB-dependent targeting, since, similar to SRP, SecB releases its substrate only after interaction with its membrane-localized receptor, SecA (55, 56). SecB thus seems to act as a cochaperone of SecA. This mechanism ensures an efficient targeting of the preprotein from the ribosome to the translocase. Proteins that are targeted to the ΔpH-dependent translocation system demand special requirements, since they appear to be fully folded at the time of arrival at the membrane (Table 1). It is highly unlikely that either SRP or SecB plays a role in the targeting of such presecretory proteins, because SRP fails to bind proteins that are released from the ribosome whereas SecB can interact efficiently only with nonnative proteins. After completion of translation, these proteins may be assisted in their folding by GroEL and targeted to the membrane solely by their specialized signal sequences or by their folded structure.
TABLE 1.
Requirements of signal sequence and mature domain for different targeting routes
| Structure | Requirement for targeting route:
|
||
|---|---|---|---|
| SRP | SecB | Twin Arg | |
| Signal sequence | Hydrophobic | Optional | RRXFXK motif |
| Mature domain | Nascent membrane proteins (?) | Nonnative proteins, SecB-binding domain (?) | Folded redox proteins |
The fact that GroEL, together with DnaK and DnaJ, seems to be part of a backup system for protein targeting toward the membrane is a consequence of their ability to bind unfolded or even misfolded proteins. The targeting is most probably the result of the presence of a signal sequence domain of the preprotein and is not directly related to the backup system, since this system has no affinity for membrane components or for members of the translocation machinery. Recently, it has been suggested that GroEL interacts with the membrane via SecA (13). This is based on observations that GroEL and membrane-bound SecA could be cross-linked to each other and that GroEL facilitates the release of SecA from the membrane. However, these results cannot be interpreted clearly, since the specificity of the reaction has not been demonstrated.
Targeting of Integral Membrane Proteins
Evidence that integral membrane proteins are the main substrate for E. coli SRP is accumulating. When SRP is disrupted by the presence of a dominant lethal 4.5S mutant or by the depletion of Ffh, the amount of functional lactose permease (LacY) inserted into the inner membrane is decreased (111). Also, leader peptidase (Lep) and a mutant with inverted topology, Lep-inv, are dependent on functional SRP for their insertion into the inner membrane (38). Furthermore, mainly polytopic inner membrane proteins have emerged from a genetic screen for substrates of SRP (175). Finally, depletion of FtsY is suggested to cause failure of the biogenesis of the membrane proteins SecY and LacY (160). These observations are consistent with the notion that SRP has a preference for the highly hydrophobic internal signal sequences of membrane proteins (177). SecB has never been found to be associated with membrane proteins, and depletion of SecB affects only the processing of precursor proteins that are independent of SRP. On the other hand, an impaired SRP function affects mainly the processing and translocation of SecB-independent preproteins. Also, although the timing of interaction with the nascent chain is different, this strongly suggests that, by default, SRP and SecB are components of two different targeting routes which converge only at the translocase (Fig. 3).
The requirement for SecY and SecA differs among the SRP-dependent membrane proteins. LacY does not require SecA, but it probably uses SecY (111). Insertion of Lep is dependent on SecA and SecY (196), while that of Lep-inv is independent of both (38). Also, the demand on SecY differs between the insertion of membrane proteins and the translocation of preproteins. This can be concluded from studies of the secY40 allele, which shows defects in the integration of membrane proteins but enables normal translocation (124). So far, no components that mediate the Sec-independent export of N-terminal tails of polytopic membrane proteins are known (35). For the integration of proteins into the inner membrane of mitochondria, a process that resembles the bacterial N-tail export in its energetics and topogenic signal requirements, the Oxa1p protein has been implicated (69). This mitochondrial protein has homologs in bacteria (6, 15), and it may well be that these proteins facilitate the Sec-independent integration of membrane proteins into the cytoplasmic membrane of E. coli. Nascent chains of the membrane protein FtsQ bind very efficiently to SRP and can be transferred from SRP to SecA and SecY (178). Combined with the previous data, this suggests that the requirements for SRP binding are determined by the highly hydrophobic signal sequence, but it is totally unclear what governs the targeting to the translocase. In this respect, it is interesting that SecB targets its substrate specifically to the translocation site, where it is handed over to SecA. SRP targets its substrate merely to the membrane and releases it upon interaction with FtsY. This can be taken as an indication that SRP-targeted proteins do not always rely on the Sec machinery for insertion or translocation.
CONCLUDING REMARKS
Depending on their intrinsic properties, precursor proteins take different targeting routes. When they are very hydrophobic, such as membrane proteins, they have to be recognized early in the route to the membrane to prevent their aggregation. SRP takes care of this by virtue of its high affinity for hydrophobic signal sequences. The proteins are targeted to FtsY at the membrane, where the cargo is released from SRP in a GTP-dependent manner. Unlike the eukaryotic SRP system, no specific FtsY receptor has been identified in bacteria. It might be that, depending on the insertion mechanism, other membrane proteins like SecY function as the receptor.
Preproteins that escape the SRP route but have difficulties in finding the translocase are directed to the membrane by SecB. It is still unclear which structural feature of a preprotein determines the SecB dependence. This will be solved only when a structure of SecB at the atomic level is available. Furthermore, the high on and off rates of the SecB-preprotein interaction give us an opportunity to study this interaction with nuclear magnetic resonance spectroscopy techniques, thereby elucidating its nature. A clear homologue of SecB has not been found in gram-positive bacteria. However, it is to be expected that a protein with a similar function exists. Such a chaperone protein probably has different structural features, since outer membrane proteins rich in β-structure, which are the main substrates for SecB in E. coli, are absent in gram-positive bacteria. It remains to be investigated whether this chaperone uses the conserved C terminus of SecA as a docking site.
Finally, the twin-Arg route functions as a specialized system to target proteins that can adopt a tertiary and even quaternary conformation prior to their translocation. However, the components and mechanisms of this route have yet to be established.
ACKNOWLEDGMENTS
We thank Joen Luirink for sharing unpublished information.
The investigations related to this review were supported by a PIONIER grant of the Netherlands Organization for Scientific Research (N.W.O.).
REFERENCES
- 1.Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. [PubMed] [Google Scholar]
- 2.Altman E, Bankaitis V A, Emr S D. Characterization of a region in mature LamB protein that interacts with a component of the export machinery of Escherichia coli. J Biol Chem. 1990;265:18148–18153. [PubMed] [Google Scholar]
- 3.Altman E, Emr S D, Kumamoto C A. The presence of both the signal sequence and a region of mature LamB protein is required for the interaction of LamB with the export factor SecB. J Biol Chem. 1990;265:18154–18160. [PubMed] [Google Scholar]
- 4.Altman E, Kumamoto C A, Emr S D. Heat-shock proteins can substitute for SecB function during protein export in Escherichia coli. EMBO J. 1991;10:239–245. doi: 10.1002/j.1460-2075.1991.tb07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson H, von Heijne G. Membrane protein topology: effects of ΔμH+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J. 1994;13:2267–2272. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer M, Behrens M, Esser K, Michaelis G, Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol Gen Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- 7.Behrmann M, Koch H-G, Hengelage T, Wieseler B, Hoffschulte H K, Müller M. Requirements for the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J Biol Chem. 1998;273:13898–13904. doi: 10.1074/jbc.273.22.13898. [DOI] [PubMed] [Google Scholar]
- 8.Berks B. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein H D, Zopf D, Freymann D M, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci USA. 1993;90:5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieker K L, Phillips G J, Silhavy T J. The sec and prl genes of Escherichia coli. J Bioenerg Biomembr. 1990;22:291–310. doi: 10.1007/BF00763169. [DOI] [PubMed] [Google Scholar]
- 12.Blattner F R, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Bochkareva E S, Solovieva M E, Girshovich A S. Targeting of groEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc Natl Acad Sci USA. 1998;95:478–483. doi: 10.1073/pnas.95.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogsch E, Brink S, Robinson C. Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 16.Bost S, Belin D. prl Mutations in the Escherichia coli secG gene. J Biol Chem. 1997;272:4087–4093. doi: 10.1074/jbc.272.7.4087. [DOI] [PubMed] [Google Scholar]
- 17.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 18.Breukink E, Kusters R, de Kruijff B. In vitro studies on the folding characteristics of the Escherichia coli precursor protein prePhoE: evidence that SecB prevents the precursor from aggregating by forming a functional complex. Eur J Biochem. 1992;208:419–425. doi: 10.1111/j.1432-1033.1992.tb17203.x. [DOI] [PubMed] [Google Scholar]
- 19.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C-terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 20.Brink S, Bogsch E, Mant A, Robinson C. Unusual characteristics of aminoterminal and hydrophobic domains in nuclear-encoded thylakoid signal peptides. Eur J Biochem. 1997;245:340–348. doi: 10.1111/j.1432-1033.1997.00340.x. [DOI] [PubMed] [Google Scholar]
- 21.Brock I W, Mills J D, Robinson D, Robinson C. The ΔpH-dependent, ATP-independent protein translocation mechanism in the chloroplast thylakoid membrane: kinetics and energetics. J Biol Chem. 1995;270:1657–1662. doi: 10.1074/jbc.270.4.1657. [DOI] [PubMed] [Google Scholar]
- 22.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 23.Buchner J. Supervising the fold: functional principles of molecular chaperones. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 24.Bukau B, Hesterkamp T, Luirink J. Growing up in a dangerous environment: a network of multiple targeting and folding pathways for nascent polypeptides in the cytosol. Trends Cell Biol. 1996;6:480–486. doi: 10.1016/0962-8924(96)84946-4. [DOI] [PubMed] [Google Scholar]
- 25.Chaddock A M, Mant A, Karnouchov I, Brink S, Herrmann R G, Klösgen R B, Robinson C. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou M M, Kendall D A. Polymeric sequences reveal a functional interrelationship between hydrophobicity and length of signal peptides. J Biol Chem. 1990;265:2873–2880. [PubMed] [Google Scholar]
- 27.Chupin V, Killian J A, Breg J, de Jongh H H J, Boelens R, Kaptein R, de Kruijff B. PhoE signal peptide inserts into micelles as a dynamic helix-break-helix structure, which is modulated by the environment. Biochemistry. 1995;34:11617–11624. doi: 10.1021/bi00036a038. [DOI] [PubMed] [Google Scholar]
- 28.Clark S A, Theg S M. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell. 1997;8:923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cline K, Ettinger W F, Theg S M. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J Biol Chem. 1992;267:2688–2696. [PubMed] [Google Scholar]
- 30.Collier D N, Bankaitis V A, Weiss J B, Bassford P J., Jr The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 31.Connolly T, Rapiejko P J, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 32.Creighton A M, Hulford A, Mant A, Robinson D, Robinson C. A monomeric, tightly folded stromal intermediate on the ΔpH-dependent thylakoidal protein transport pathway. J Biol Chem. 1995;270:1663–1669. doi: 10.1074/jbc.270.4.1663. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham K, Wickner W. Specific recognition of the leader region of precursor proteins is required for the activation of translocation ATPase of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:8630–8634. doi: 10.1073/pnas.86.22.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalbey R E, von Heijne G. Signal peptidases in prokaryotes and eukaryotes-a new protease family. Trends Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- 35.Dalbey R E, Kuhn A, Von Heijne G. Directionality in protein translocation across membranes: the N-tail phenomenon. Trends Cell Biol. 1995;5:380–383. doi: 10.1016/s0962-8924(00)89079-0. [DOI] [PubMed] [Google Scholar]
- 36.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Cock H, Tommassen J. SecB-binding does not maintain the translocation-competent state of prePhoE. Mol Microbiol. 1992;6:599–604. doi: 10.1111/j.1365-2958.1992.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 38.De Gier J-W L, Mansournia P, Valent Q A, Phillips G J, Luirink J, von Heijne G. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 39.De Leeuw E, Poland D, Mol O, Sinning I, ten Hagen-Jongman C M, Oudega B, Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- 40.Den Blaauwen T, Terpetschnig E, Lakowicz J R, Driessen A J M. Interaction of SecB with soluble SecA. FEBS Lett. 1997;416:35–38. doi: 10.1016/s0014-5793(97)01142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derman A I, Puziss J W, Bassford P J, Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Vrije T, Batenburg A M, Killian J A, de Kruijff B. Lipid involvement in protein translocation in Escherichia coli. Mol Microbiol. 1990;4:143–150. doi: 10.1111/j.1365-2958.1990.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 43.Diamond D L, Randall L L. Kinetic partitioning. Poising SecB to favor association with a rapidly folding ligand. J Biol Chem. 1997;272:28994–28998. doi: 10.1074/jbc.272.46.28994. [DOI] [PubMed] [Google Scholar]
- 44.Doud S K, Chou M M, Kendall D A. Titration of protein transport by incremental changes in signal peptide hydrophobicity. Biochemistry. 1993;32:1251–1256. doi: 10.1021/bi00056a008. [DOI] [PubMed] [Google Scholar]
- 45.Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 46.Driessen A J M. Translocation of proteins across the bacterial cytoplasmic membrane. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biophysics. 2. Transport processes in membranes. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1996. pp. 759–790. [Google Scholar]
- 47.Driessen A J M, de Wit J G, Kuiper W, van der Wolk J P W, Fekkes P, van der Does C, van Wely K, Manting E H, den Blaauwen T. SecA, a novel ATPase that converts chemical energy into a mechanical force to drive precursor protein translocation. Biochem Soc Trans. 1995;23:981–985. doi: 10.1042/bst0230981. [DOI] [PubMed] [Google Scholar]
- 48.Driessen A J M, Fekkes P, van der Wolk J P W. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 49.Duong F, Wickner W. Distinct catalytic role of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the Gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 51.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 52.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 53.Fasman G D, Park K, Randall L L. Chaperone SecB: conformational changes demonstrated by circular dichroism. J Protein Chem. 1995;14:595–600. doi: 10.1007/BF01886885. [DOI] [PubMed] [Google Scholar]
- 54.Fekkes P, den Blaauwen T, Driessen A J M. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry. 1995;34:10078–10085. doi: 10.1021/bi00031a032. [DOI] [PubMed] [Google Scholar]
- 55.Fekkes P, van der Does C, Driessen A J M. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fekkes P, de Wit J G, van der Wolk J P W, Kimsey H H, Kumamoto C A, Driessen A J M. Preprotein transfer to the Escherichia coli translocase requires the cooperative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 57.Fikes J D, Bassford P J., Jr Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J Bacteriol. 1989;171:402–409. doi: 10.1128/jb.171.1.402-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 59.Flower A M, Doebele R C, Silhavy T J. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol. 1994;176:5607–5614. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francetiç O, Kumamoto C A. Escherichia coli SecB stimulates export without maintaining export competence of ribose-binding protein signal sequence mutants. J Bacteriol. 1996;178:5954–5959. doi: 10.1128/jb.178.20.5954-5959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freymann D M, Keenan R J, Stroud R M, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 62.Gaitanaris G A, Vysokanov A, Hung S, Gottesman M E, Gragerov A. Successive action of Escherichia coli chaperones in vivo. Mol Microbiol. 1994;14:861–869. doi: 10.1111/j.1365-2958.1994.tb01322.x. [DOI] [PubMed] [Google Scholar]
- 63.Gannon P M, Kumamoto C A. Mutations of the molecular chaperone protein SecB which alter the interaction between SecB and maltose-binding protein. J Biol Chem. 1993;268:1590–1595. [PubMed] [Google Scholar]
- 64.Gennity J, Goldstein J, Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990;22:233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- 65.Hanada M, Nishiyama K, Mizushima S, Tokuda H. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12) J Biol Chem. 1994;269:23625–23631. [PubMed] [Google Scholar]
- 66.Hardy S J S, Randall L L. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science. 1991;251:439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- 67.Hartl F-U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 68.Hauser S, Bacher G, Dobberstein B, Lütcke H. A complex of the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J. 1995;14:5485–5493. doi: 10.1002/j.1460-2075.1995.tb00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hell K, Herrmann J M, Pratje E, Neupert W, Stuart R A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry R, Kapazoglou A, McCaffery M, Cline K. Differences between lumen targeting domains of chloroplast transit peptides determine pathway specificity for thylakoid transport. J Biol Chem. 1994;269:10189–10192. [PubMed] [Google Scholar]
- 71.Henry R, Carrigan M, McCaffery M, Ma X, Cline K. Targeting determinants and proposed evolutionary basis for the Sec and the delta pH protein transport systems in chloroplast thylakoid membranes. J Cell Biol. 1997;136:823–832. doi: 10.1083/jcb.136.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hesterkamp T, Bukau B. The Escherichia coli trigger factor. FEBS Lett. 1996;389:32–34. doi: 10.1016/0014-5793(96)00582-0. [DOI] [PubMed] [Google Scholar]
- 73.High S, Henry R, Mould R M, Valent Q, Meacock S, Cline K, Gray J C, Luirink J. Chloroplast SRP54 interacts with a specific subset of thylakoid precursor proteins. J Biol Chem. 1997;272:11622–11628. doi: 10.1074/jbc.272.17.11622. [DOI] [PubMed] [Google Scholar]
- 74.Hikita C, Mizushima S. The requirement of a positive charge at the amino terminus can be compensated for by a longer central hydrophobic stretch in the functioning of signal peptides. J Biol Chem. 1992;267:12375–12379. [PubMed] [Google Scholar]
- 75.Hoffschulte H K, Drees B, Müller M. Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J Biol Chem. 1994;269:12833–12839. [PubMed] [Google Scholar]
- 76.Huie J L, Silhavy T J. Suppression of signal sequence defects and azide resistance in Escherichia coli commonly result from the same mutations in secA. J Bacteriol. 1995;177:3518–3526. doi: 10.1128/jb.177.12.3518-3526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hulford A, Hazell L, Mould R M, Robinson C. Two distinct mechanisms for the translocation of proteins across the thylakoid membrane, one requiring the presence of a stromal protein factor and nucleotide triphosphates. J Biol Chem. 1994;269:3251–3256. [PubMed] [Google Scholar]
- 78.Izard J W, Kendall D A. Signal peptides: exquisitely designed transport promoters. Mol Microbiol. 1994;13:765–773. doi: 10.1111/j.1365-2958.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 79.Josefsson L G, Randall L L. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell. 1981;25:151–157. doi: 10.1016/0092-8674(81)90239-7. [DOI] [PubMed] [Google Scholar]
- 80.Khisty V J, Munske G, Randall L L. Mapping of the binding frame for the chaperone SecB within a natural ligand, galactose-binding protein. J Biol Chem. 1995;270:25920–25927. doi: 10.1074/jbc.270.43.25920. [DOI] [PubMed] [Google Scholar]
- 81.Kim J, Kendall D A. Identification of a sequence motif that confers SecB dependance on a SecB-independent secretory protein in vivo. J Bacteriol. 1998;180:1396–1401. doi: 10.1128/jb.180.6.1396-1401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kimsey H H, Dagarag M D, Kumamoto C A. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- 83.Krieg U C, Walter P, Johnson A E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kudlicki W, Odom O W, Kramer G, Hardesty B. Binding of an N-terminal rhodanes peptide to DnaJ and to ribosomes. J Biol Chem. 1996;271:31160–31165. doi: 10.1074/jbc.271.49.31160. [DOI] [PubMed] [Google Scholar]
- 85.Kumamoto C A. Escherichia coli SecB protein associates with exported protein precursors in vivo. Proc Natl Acad Sci USA. 1989;86:5320–5324. doi: 10.1073/pnas.86.14.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumamoto C A. Molecular chaperones and protein translocation across the Escherichia coli inner membrane. Mol Microbiol. 1991;5:19–22. doi: 10.1111/j.1365-2958.1991.tb01821.x. [DOI] [PubMed] [Google Scholar]
- 87.Kumamoto C A, Beckwith J. Mutations in a new gene, secB, cause defective protein localization in Escherichia coli. J Bacteriol. 1983;154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumamoto C A, Beckwith J. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumamoto C A, Gannon P M. Effects of Escherichia coli secB mutations on pre-maltose binding protein conformation and export kinetics. J Biol Chem. 1988;263:11554–11558. [PubMed] [Google Scholar]
- 90.Kumamoto C A, Nault A K. Characterization of the Escherichia coli protein-export gene secB. Gene. 1989;75:167–175. doi: 10.1016/0378-1119(89)90393-4. [DOI] [PubMed] [Google Scholar]
- 91.Kumamoto C A, Francetiç O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kurzchalia T V, Wiedmann M, Girshovich A S, Bochkareva E S, Bielka H, Rapoport T A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 93.Kusters R, Lentzen G, Eppens E, van Geel A, van der Weijden C C, Wintermeyer W, Luirink J. The functioning of the SRP receptor FtsY in protein-targeting in E. coli is correlated with its ability to bind and hydrolyse GTP. FEBS Lett. 1995;372:253–258. doi: 10.1016/0014-5793(95)00997-n. [DOI] [PubMed] [Google Scholar]
- 94.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lai C-Y, Baumann P. Sequence analysis of a DNA fragment from Buchnera aphidicola (an endosymbiont of aphids) containing genes homologous to dnaG, rpoD, cysE, and secB. Gene. 1992;119:113–118. doi: 10.1016/0378-1119(92)90074-y. [DOI] [PubMed] [Google Scholar]
- 96.Laminet A A, Kumamoto C A, Plückthun A. Folding in vitro and transport in vivo of pre-β-lactamase are SecB independent. Mol Microbiol. 1991;5:117–122. doi: 10.1111/j.1365-2958.1991.tb01832.x. [DOI] [PubMed] [Google Scholar]
- 97.Larsen N, Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991;19:209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lauffer L, Garcia P D, Harkins R N, Coussens L, Ullrich A, Walter P. Topology of the SRP receptor in the endoplasmic reticulum membrane. Nature. 1985;318:334–338. doi: 10.1038/318334a0. [DOI] [PubMed] [Google Scholar]
- 99.Lecker S H, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford P J, Jr, Kumamoto C A, Wickner W. Three pure chaperone proteins of Escherichia coli—SecB, trigger factor and GroEL—form soluble complexes with precursor proteins in vitro. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lecker S H, Driessen A J M, Wickner W. ProOmpA contains secondary and tertiary structure prior to translocation and is shielded from aggregation by association with SecB protein. EMBO J. 1990;9:2309–2314. doi: 10.1002/j.1460-2075.1990.tb07402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee C, Li P, Inouye H, Brickman E R, Beckwith J. Genetic studies on the inability of β-galactosidase to be translocated across the Escherichia coli cytoplasmic membrane. J Bacteriol. 1989;171:4609–4616. doi: 10.1128/jb.171.9.4609-4616.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee J H, Patel P, Sankar P, Shanmugan K T. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol. 1985;162:344–352. doi: 10.1128/jb.162.1.344-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lentzen G, Dobberstein B, Wintermeyer W. Formation of SRP-like particle induces a conformational change in E. coli 4.5S RNA. FEBS Lett. 1994;348:233–238. doi: 10.1016/0014-5793(94)00599-0. [DOI] [PubMed] [Google Scholar]
- 104.Li P, Beckwith J, Inouye H. Alteration of the amino terminus of the mature sequence of a periplasmic protein can severely affect protein export in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:7685–7689. doi: 10.1073/pnas.85.20.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lill R, Dowan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 106.Luirink, J. Personal communication.
- 107.Luirink J, High S, Wood H, Giner A, Tollervey D, Dobberstein B. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature. 1992;359:741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- 108.Luirink J, Dobberstein B. Mammalian and Escherichia coli signal recognition particles. Mol Microbiol. 1994;11:9–13. doi: 10.1111/j.1365-2958.1994.tb00284.x. [DOI] [PubMed] [Google Scholar]
- 109.Luirink J, ten Hagen-Jongman C M, van der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lutz S, Jacobi A, Schlensog V, Böhm R, Sawers G, Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991;5:123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- 111.MacFarlane J, Müller M. The functional integration of a polytopic membrane protein in Escherichia coli is dependent on the bacterial signal-recognition particle. Eur J Biochem. 1995;233:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 112.MacIntyre S, Eschbach M-L, Mutschler B. Export incompatibility of N-terminal basic residues in a mature polypeptide of Escherichia coli can be alleviated by optimising the signal peptide. Mol Gen Genet. 1990;221:466–474. doi: 10.1007/BF00259413. [DOI] [PubMed] [Google Scholar]
- 113.MacIntyre S, Mutschler B, Henning U. Requirement of the SecB chaperone for export of a non-secretory polypeptide in Escherichia coli. Mol Gen Genet. 1991;227:224–228. doi: 10.1007/BF00259674. [DOI] [PubMed] [Google Scholar]
- 114.Menon N K, Robbins J, Peck H D, Jr, Chatelus C Y, Choi E, Przybyla A E. Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J Bacteriol. 1990;172:1969–1977. doi: 10.1128/jb.172.4.1969-1977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Menon N K, Chatelus C Y, DerVartanian M, Wendt J C, Shanmugan K T, Peck H D, Jr, Przybyla A E. Cloning, sequencing and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miller A, Wang L, Kendall D A. Synthetic signal peptides specifically recognize SecA and stimulate ATPase activity in the absence of preprotein. J Biol Chem. 1998;273:11409–11412. doi: 10.1074/jbc.273.19.11409. [DOI] [PubMed] [Google Scholar]
- 117.Miller J D, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;336:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 118.Miller J D, Bernstein H D, Walter P. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 119.Montoya G, Svensonn C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 120.Mori H, Cline K. A signal peptide that directs non-Sec transport in bacteria also directs efficient and exclusive transport on the thylakoid delta pH pathway. J Biol Chem. 1998;273:11405–11408. doi: 10.1074/jbc.273.19.11405. [DOI] [PubMed] [Google Scholar]
- 121.Moser C, Mol O, Goody R S, Sinning I. The signal recognition particle receptor of Escherichia coli (FtsY) has a nucleotide exchange factor built into the GTPase domain. Proc Natl Acad Sci USA. 1997;94:11339–11344. doi: 10.1073/pnas.94.21.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mould R M, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J Biol Chem. 1991;266:12189–12193. [PubMed] [Google Scholar]
- 123.Müller J P. Influence of impaired chaperone or secretion function on SecB production in Escherichia coli. J Bacteriol. 1996;178:6097–6104. doi: 10.1128/jb.178.21.6097-6104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Newitt J A, Bernstein H D. A mutation in the Escherichia coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
- 125.Ng D T W, Brown J D, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nielsen V S, Mant A, Knoetzel J, Møller B L, Robinson C. Import of barley photosystem I subunit N into the thylakoid lumen is mediated by a bipartite presequence lacking an intermediate processing site. J Biol Chem. 1994;269:3762–3766. [PubMed] [Google Scholar]
- 127.Nivière V, Wong S, Voordouw G. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents the export of a β-lactamase fusion protein. J Gen Microbiol. 1992;138:2173–2183. doi: 10.1099/00221287-138-10-2173. [DOI] [PubMed] [Google Scholar]
- 128.Nouwen N, Tommassen J, de Kruijff B. Requirement for conformational flexibility in the signal sequence of precursor protein. J Biol Chem. 1994;269:16029–16033. [PubMed] [Google Scholar]
- 129.Nouwen N, de Kruijff B, Tommassen J. ΔμH+ dependency of in vitro protein translocation into E. coli inner membrane vesicles varies with the signal sequence core region composition. Mol Microbiol. 1996;19:1205–1214. doi: 10.1111/j.1365-2958.1996.tb02466.x. [DOI] [PubMed] [Google Scholar]
- 130.Park S, Liu G, Topping T B, Cover W H, Randall L L. Modulation of folding pathways of exported proteins by the leader sequence. Science. 1988;239:1033–1035. doi: 10.1126/science.3278378. [DOI] [PubMed] [Google Scholar]
- 131.Patel S, Austen B M. Substitution of fifty four homologue (Ffh) in Escherichia coli with the mammalian 54-kDa protein of signal-recognition particle. Eur J Biochem. 1996;238:760–768. doi: 10.1111/j.1432-1033.1996.0760w.x. [DOI] [PubMed] [Google Scholar]
- 132.Phillips G J, Silhavy T J. Heat-shock proteins DnaK and GroEL facilitate export of LacZ hybrid proteins in E. coli. Nature. 1990;344:882–884. doi: 10.1038/344882a0. [DOI] [PubMed] [Google Scholar]
- 133.Phillips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 134.Phoenix D A, Kusters R, Hikita C, Mizushima S, de Kruijff B. OmpF-Lpp signal sequence mutants with varying charge hydrophobicity ratios provide evidence for a phosphatidylglycerol-signal sequence interaction during protein translocation across the Escherichia coli inner membrane. J Biol Chem. 1993;268:17069–17073. [PubMed] [Google Scholar]
- 135.Poritz M A, Strub K, Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- 136.Poritz M A, Bernstein H D, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 137.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Prinz W A, Spiess C, Ehrmann M, Schierle C, Beckwith J. Targeting of signal sequenceless proteins for export in Escherichia coli with altered protein translocase. EMBO J. 1996;15:5209–5217. [PMC free article] [PubMed] [Google Scholar]
- 139.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Puziss J W, Fikes J D, Bassford P J., Jr Analysis of mutational alterations in the hydrophilic segment of the maltose-binding protein signal peptide. J Bacteriol. 1989;171:2303–2311. doi: 10.1128/jb.171.5.2303-2311.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Puziss J W, Strobel S M, Bassford P J., Jr Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations. J Bacteriol. 1992;174:92–101. doi: 10.1128/jb.174.1.92-101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rajapandi T, Oliver D. Carboxy-terminal region of Escherichia coli SecA ATPase is important to promote its protein translocation activity in vivo. Biochem Biophys Res Commun. 1994;200:1477–1483. doi: 10.1006/bbrc.1994.1617. [DOI] [PubMed] [Google Scholar]
- 143.Ramamurthy V, Oliver D. Topology of integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 144.Randall L L, Hardy S J S. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 145.Randall L L, Hardy S J S. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem Sci. 1995;20:65–69. doi: 10.1016/s0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 146.Randall L L, Topping T B, Hardy S J S, Pavlov M Y, Freistroffer D V, Ehrenberg M. Binding of SecB to ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc Natl Acad Sci USA. 1997;94:802–807. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Randall L L, Topping T B, Suciu D, Hardy S J S. Calorimetric analysis of the interaction between SecB and its ligands. Protein Sci. 1998;7:1195–1200. doi: 10.1002/pro.5560070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rapoport T A, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmatic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 149.Reid B G, Flynn G C. GroEL binds to and unfolds rhodanese posttranslationally. J Biol Chem. 1996;271:7212–7217. doi: 10.1074/jbc.271.12.7212. [DOI] [PubMed] [Google Scholar]
- 150.Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 151.Rizo J, Blanco F J, Kobe B, Bruch M D, Gierasch L M. Conformational behavior of Escherichia coli OmpA signal peptides in membrane mimetic environments. Biochemistry. 1993;32:4881–4894. doi: 10.1021/bi00069a025. [DOI] [PubMed] [Google Scholar]
- 152.Römisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 153.Römisch K, Webb J, Lingelbach K, Gausepohl H, Dobberstein B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1990;111:1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sankaran K, Wu H C. Signal peptidase II-specific signal peptidase for bacterial lipoproteins. In: von Heijne G, editor. Signal peptidases. Austin, Tex: Landes Co.; 1994. pp. 17–29. [Google Scholar]
- 155.Santini C-L, Ize B, Chanal A, Müller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sawers R G, Ballantine S P, Boxer D H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol. 1985;164:1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schatz P J, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 159.Schiebel E, Driessen A J M, Hartl F-U, Wickner W. ΔμH+ and ATP function at different steps in the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 160.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 161.Seoh H, Tai P C. Carbon source-dependent synthesis of SecB, a cytosolic chaperone involved in protein translocation across Escherichia coli membranes. J Bacteriol. 1997;179:1077–1081. doi: 10.1128/jb.179.4.1077-1081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 163.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–1775. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shimizu H, Nishiyama K, Tokuda H. Expression of gpsA encoding biosynthetic sn-glycerol 3-phosphate dehydrogenase suppresses both the LB− phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol Microbiol. 1997;26:1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x. [DOI] [PubMed] [Google Scholar]
- 165.Shinde U P, Guru Row T N, Mayway Y R. Export of proteins across membranes: the helix reversion theory. Biosci Rep. 1989;9:737–745. doi: 10.1007/BF01114812. [DOI] [PubMed] [Google Scholar]
- 166.Smith V F, Schwartz B L, Randall L L, Smith R D. Electrospray mass spectrometric investigation of the chaperone SecB. Protein Sci. 1996;5:488–494. doi: 10.1002/pro.5560050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith V F, Hardy S J S, Randall L L. Determination of the binding frame of the chaperone SecB within the physiological ligand oligopeptide-binding protein. Protein Sci. 1997;6:1746–1755. doi: 10.1002/pro.5560060815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sofia H J, Burland V, Daniels D L, Plunket III G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 min. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stenberg G, Fersht A R. Folding of barnase in the presence of the molecular chaperone SecB. J Mol Biol. 1997;274:268–275. doi: 10.1006/jmbi.1997.1398. [DOI] [PubMed] [Google Scholar]
- 170.Struck J C R, Toschka H Y, Specht T, Erdmann V A. Common structural features between eukaryotic 7SL RNAs, eubacterial 4.5S RNA and scRNA and archaebacterial 7S RNA. Nucleic Acids Res. 1988;16:7740. doi: 10.1093/nar/16.15.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- 172.Tajima S, Lauffer L, Rath V L, Walter P. The signal recognition particle is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Topping T B, Randall L L. Determination of the binding frame within a physiological ligand for the chaperone SecB. Protein Sci. 1994;3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Topping T B, Randall L L. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligand. J Biol Chem. 1997;272:19314–19318. doi: 10.1074/jbc.272.31.19314. [DOI] [PubMed] [Google Scholar]
- 175.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 176.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Valent Q A, de Gier J-W L, von Heijne G, Kendall D A, ten Hagen-Jongman C M, Oudega B, Luirink J. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 178.Valent Q A, Scotti P A, High S, de Gier J-W L, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The E. coli SRP and Sec targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van de Vossenberg J L C M, van der Does C, Albers S V, Driessen A J M, van Klompenburg W. The positive inside rule is not determined by the polarity of the Δψ. Mol Microbiol. 1998;29:1125–1126. doi: 10.1046/j.1365-2958.1998.01001.x. [DOI] [PubMed] [Google Scholar]
- 180.Van der Wolk J P W, Fekkes P, Boorsma A, Huie J L, Silhavy T J, Driessen A J M. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Von Heijne G. How signal sequences maintain cleavage specific. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 183.Von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 184.Von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 185.Walter P, Blobel G. Purification of membrane-associated protein complex required for protein translocation across the endoplasmatic reticulum. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmatic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 187.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 188.Wang Z, Jones J D, Rizo J, Gierasch L M. Membrane-bound conformation of a signal peptide: a transferred nuclear Overhauser effect analysis. Biochemistry. 1993;32:13991–13999. doi: 10.1021/bi00213a032. [DOI] [PubMed] [Google Scholar]
- 189.Watanabe M, Blobel G. Cytosolic factor purified from Escherichia coli is necessary and sufficient for the export of a preprotein and is a homotetramer of SecB. Proc Natl Acad Sci USA. 1989;86:2728–2732. doi: 10.1073/pnas.86.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Watanabe M, Blobel G. SecB functions as a cytosolic signal recognition factor for protein export in E. coli. Cell. 1989;58:695–705. doi: 10.1016/0092-8674(89)90104-9. [DOI] [PubMed] [Google Scholar]
- 191.Watanabe M, Blobel G. High-affinity binding of Escherichia coli SecB to the signal sequence region of a presecretory protein. Proc Natl Acad Sci USA. 1995;92:10133–10136. doi: 10.1073/pnas.92.22.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 193.Weiss J B, Ray P H, Bassford P J., Jr Purified SecB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc Natl Acad Sci USA. 1988;85:8978–8982. doi: 10.1073/pnas.85.23.8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wild J, Altman E, Yura T, Gross C A. DnaK and DnaJ heat shock proteins participate in protein export in E. coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 195.Wild J, Walter W A, Gross C A, Altman E. Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J Bacteriol. 1993;175:3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wolfe P B, Rice M, Wickner W. Effects of two sec genes on protein assembly into the plasma membrane of Escherichia coli. J Biol Chem. 1985;260:1836–1841. [PubMed] [Google Scholar]
- 197.Yamane K, Mizushima S. Introduction of basic amino acid residues after the signal peptide inhibits protein translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1988;263:19690–19696. [PubMed] [Google Scholar]
- 198.Zahn R, Perrett S, Fersht A R. Conformational states bound by the molecular chaperones GroEL and SecB: a hidden unfolding (annealing) activity. J Mol Biol. 1996;261:43–61. doi: 10.1006/jmbi.1996.0440. [DOI] [PubMed] [Google Scholar]
- 199.Zahn R, Perrett S, Stenberg G, Fersht A R. Catalysis of amide proton exchange by the molecular chaperones GroEL and SecB. Science. 1996;271:642–645. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
- 200.Zelazny A, Seluanov A, Cooper A, Bibi E. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc Natl Acad Sci USA. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zheng N, Gierasch L M. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]
- 202.Zopf D, Bernstein H D, Johnson A E, Walter P. The methionine-rich domain of the 54 kD protein subunit of the signal recognition particle contains a RNA binding site and can be cross-linked to a signal sequence. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zopf D, Bernstein H D, Walter P. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J Cell Biol. 1993;120:1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]