Abstract
Resveratrol is an antiaging, antioxidant, and anti-inflammatory natural polyphenolic compound. Growing evidence indicates that resveratrol has potential therapeutic effects for improving aging ovarian function. However, the mechanisms underlying prolonged reproductive longevity remain elusive. We found that resveratrol ameliorates ovarian aging transcriptome, some of which are associated with specific changes in methylome. In addition to known aging transcriptome of oocytes and granulosa cells such as decline in oxidoreductase activity, metabolism and mitochondria function, and elevated DNA damage and apoptosis, actin cytoskeleton are notably downregulated with age, and these defects are mostly rescued by resveratrol. Moreover, the aging-associated hypermethylation of actin cytoskeleton is decreased by resveratrol. In contrast, deletion of Tet2, involved in DNA demethylation, abrogates resveratrol-reprogrammed ovarian aging transcriptome. Consistently, Tet2 deficiency results in additional altered pathways as shown by increased mTOR and Wnt signaling, as well as reduced DNA repair and actin cytoskeleton with mouse age. Moreover, genes associated with oxidoreductase activity and oxidation–reduction process were hypermethylated in Tet2-deficient oocytes from middle-age mice treated with resveratrol, indicating that loss of Tet2 abolishes the antioxidant effect of resveratrol. Taking together, our finding provides a comprehensive landscape of transcriptome and epigenetic changes associated with ovarian aging that can be reprogrammed by resveratrol administration, and suggests that aberrantly increased DNA methylation by Tet2 deficiency promotes additional aging epigenome that cannot be effectively restored to younger state by resveratrol.
Keywords: ovarian aging, oocytes, cumulus cells, transcriptome, methylome
Significance Statement.
Resveratrol has been investigated to effectively delay aging, including reproductive aging and increases fertility, and the effectiveness is dosage and time-dependent. However, the underlying epigenetic mechanisms of ovarian aging, ameliorated by resveratrol, remain elusive. Our study using a model of natural aging mice by long-term-oral administration of resveratrol revealed that resveratrol reverses aging transcriptome and shifts the methylome to a younger profile. However, Tet2 deficiency promotes additional aging epigenome that cannot be effectively restored by resveratrol. Hence, the active DNA demethylation is critical for the beneficial effects of resveratrol on female fertility during reproductive aging. These epigenome data revealing the functions and mechanisms of resveratrol may provide further supports for its use in fertility treatment and postponing reproductive aging.
Introduction
The number and quality of oocytes and follicles decrease significantly with age (1, 2), leading to ovarian aging. After birth, most oocytes become apoptotic because of follicular atresia, and only a small number of oocytes become mature and are ovulated. Moreover, the ability of the oocytes to fertilize and further develop is reduced after birth (1, 3). DNA methylation (DNAm) changes have become a hallmark of aging (4, 5). Epigenetic clocks, built upon age-related differentially methylated positions, can estimate chronological age with remarkable accuracy and across a broad spectrum of the lifespan, from prenatal mammals to the oldest living mammals (6–8). The maintenance of methylome established during development is key to keeping youthful epigenetic states, but is altered with age (9). Consequently, failure in DNAm maintenance results in aging and age-associated disease (10). The de novo DNAm and its dynamics during oocyte growth have been extensively studied (11–13). DNAm dynamics of female germ cell aging also has been explored (14–17). For instance, ribosomal DNAm in human and mouse oocytes increased with age (15), and the rate of epigenetic aging was found to be slower in oocytes compared to that of blood from a bovine model of reproductive aging (16).
Accumulated alterations to the methylome can be alleviated or reversed by longevity-promoting interventions, such as calorie restriction (18, 19), longevity drug [such as rapamycin (20, 21), nicotinamide mononucleotide (22, 23), and α-Ketoglutarate (24, 25)] and epigenetic rejuvenation strategies (26). Resveratrol (3,5,4′-trihydroxystilbene) is a polyphenolic compound found in the skin of red grapes, red wine, and other botanical extracts. Resveratrol has various beneficial effects on health, including antiaging, antioxidant, anti-inflammatory, cardioprotective, and antineoplastic properties (27). Moreover, resveratrol can protect against age-associated infertility in female mice (28–30). Though resveratrol has potential therapeutic effects for prolonging ovarian lifespan (31), the underlying mechanisms have not been well understood. Also, it remains unclear whether resveratrol improves the aging clocks in oocytes to prolong fertility.
DNAm is mediated by DNA methyltransferases. Ten-eleven translocation (TET1, TET2, and TET3) enzymes promote the specific demethylation of 5mC residues in DNA (32). Tet1 or Tet2 single knockout leads to telomere shortening and chromosome fusion (33, 34). Telomere shortening is closely related to aging and age-related diseases (35, 36). Age-related telomere shortening in oocytes and granulosa cells (GCs) is considered one of indicators of reduced oocyte quality (37, 38). Knockout of Tet2 leads to premature ovarian aging and significantly reduced fertility (39). Moreover, maternal resveratrol supplementation in the senescence-accelerated mouse strain produced changes in global 5-mC and 5-hmC as well as changes in their enzymatic machinery such as TET2 and DNMT3A/B in offspring (40, 41). It remains elusive whether resveratrol-induced reprogramming in oocytes requires DNA demethylation activity.
Here, by single-cell RNA sequencing and low-input post-bisulfite adaptor tagging (PBAT) DNAm sequencing, we uncovered that oocyte transcriptome of middle-age or old mice treated with resveratrol maintained younger profile. Moreover, the long-term-oral administration of resveratrol could shift the methylome of aged oocytes to a younger profile. Active DNA demethylation is necessary for the beneficial effects of resveratrol on female fertility during reproductive aging.
Results
Resveratrol improves the fertility of reproductively aged female mice
Although reproductive aging in female mammals is an irreversible process associated with declining oocyte quantity and quality, long-term oral administration of resveratrol or nicotinamide adenine dinucleotide (NAD+) repletion has been shown to rescue female mice fertility during reproductive aging (28, 42).
Our goal was to explore the potential role of resveratrol in counteracting age-related fertility decline associated with aberrant methylome. To this end, wild-type (WT, naturally aging mice) and Tet2−/− female mice (aberrant methylation-induced premature reproductive aging) were treated by providing resveratrol (30 mg/L) in drinking water from 2 to 10 months old (resveratrol-group), whereas the mice in the control group had dimethyl sulfoxide (DMSO) in drinking water (control-group) (Fig. 1A). The mice were tested for their fertility at 6 and 10 months old. Metaphase II (MII) oocytes were harvested from the young (2-month-old), middle age (6-month-old), or old (10-month-old) mice after superovulation treatment. The oocytes were then subjected to single-cell RNA sequencing and low-input PBAT DNAm sequencing.
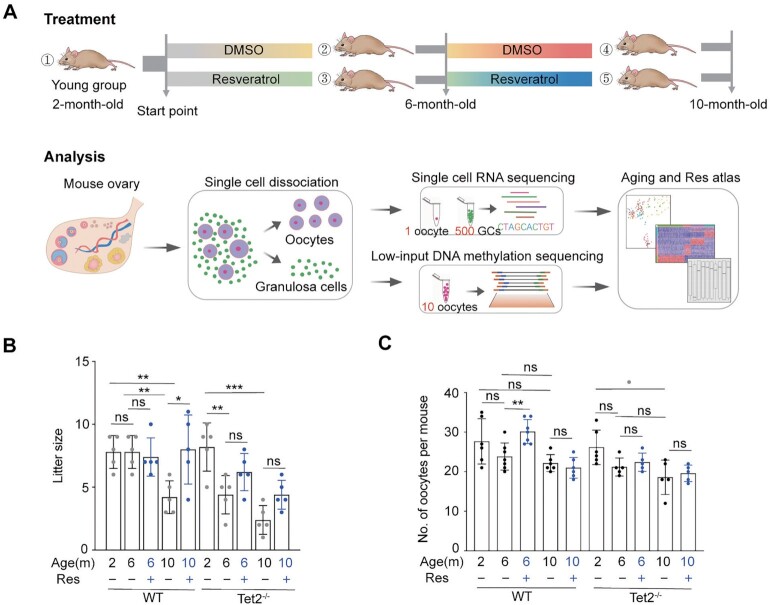
Fig. 1.
Effect of resveratrol on fertility of female mice. (A) Schematic of mice subjected to resveratrol treatment for an 8-month period starting at 2 months old and flowchart overview of the scRNA-Seq and low-input methylation sequencing methodology. (1) 2-month-old mice; (2) 6-month-old mice fed DMSO served as the vehicle control; (3) 6-month-old mice fed resveratrol; (4) 10-month-old mice fed DMSO for 8 months; and (5) 10-month-old mice fed resveratrol for 8 months. MII oocytes were obtained from mice after superovulation. Granulosa cells (GCs) were mural GCs released by puncturing the large follicles after stimulation with pregnant mare serum gonadotrophin (PMSG) (Materials and methods section). (B) Litter size of pregnant mice based on the number of pregnant females (n = 5). Note, knockouts and WT were mated separately. (C) Number of MII oocytes collected following ovulation induction from each mouse treated with (+) or without (–) resveratrol (Res) (n = 5 to 7). *P < 0.05, **P < 0.01, ns: not significant. One-way ANOVA and Tukey’s multiple comparison test were performed to analyze the effect of age on female fertility, and two-way ANOVA performed to analyze the effect of both age and resveratrol treatment on female fertility (B and C).
As expected, mouse fertility was reduced with age. In WT mice without resveratrol administration, the average litter size of old mice (four pups per litter) decreased compared to that of young mice (eight pups per litter) (P = 0.0011, by a one-way ANOVA), while old females who received resveratrol delivered eight pups per litter (Fig. 1B). There was a statistically significant interaction between the effects of age and resveratrol administration (P = 0.019) by two-way ANOVA analysis. Tet2−/− mice exhibited more aggravated infertility associated with aging (P = 0.0003, by one-way ANOVA analysis). The average litter size for both middle-aged and old Tet2−/− females treated with resveratrol was less than that of WT females treated with resveratrol (Fig. 1B).
Moreover, resveratrol treatment significantly enhanced oocyte quantity in middle-aged WT mice. Old WT mice without resveratrol administration had fewer oocytes than did young mice (Fig. 1C). After oral intake of resveratrol, the middle age mice had more oocytes than did the age-matched controls without resveratrol administration (Fig. 1C, P = 0.0026, Tukey’s multiple comparisons). These data showed that resveratrol noticeably improved the fertility of old WT mice. However, Tet2-deficienct old females had significantly fewer oocytes than that of the young mice (Fig. 1C, P = 0.016, by a one-way ANOVA). Tet2−/− mice at middle age or old age after treatment with resveratrol showed no significant difference in the number of ovulated MII oocytes compared with that of the age-matched controls (Fig. 1C).
Resveratrol reverses aging-related gene expression changes in oocytes
To investigate the effects of aging and resveratrol on global gene expression of oocytes, we harvested oocytes from young, middle-age and old female mice in both control- and resveratrol-groups. SMART-seq2 (scRNA-seq) was performed for RNA transcript profiling (Fig. 1A). First, we used principal component analysis (PCA) to explore the effect of resveratrol on the transcriptome of aging oocytes. The PCA plot showed the clustering of oocytes based on gene expression patterns exhibited by PC1 and PC2 (Fig. 2A). Young oocytes were easily separated from middle-aged or old oocytes along the PC1 axis. The variable distributions of the two groups along the PC2 axis indicated increased heterogeneity in gene expression in old oocytes; a fraction of aged oocytes expressed a middle-age-like transcriptome, which suggested that an unknown contributor associated with age drove a greater extent of heterogeneity in the aging oocytes. Several previous studies have demonstrated that chromatin condensation configuration contributes to oocyte aging (43–45). Notably, the oocytes from middle-age mice fed resveratrol were clustered closely to that of young mice, indicating that these middle-aged oocytes have a younger transcriptome. Moreover, the oocytes from old mice fed resveratrol were remarkably similar to those of middle age mice. Therefore, our data revealed that resveratrol promotes maintenance of youthful transcriptome in oocytes during aging.
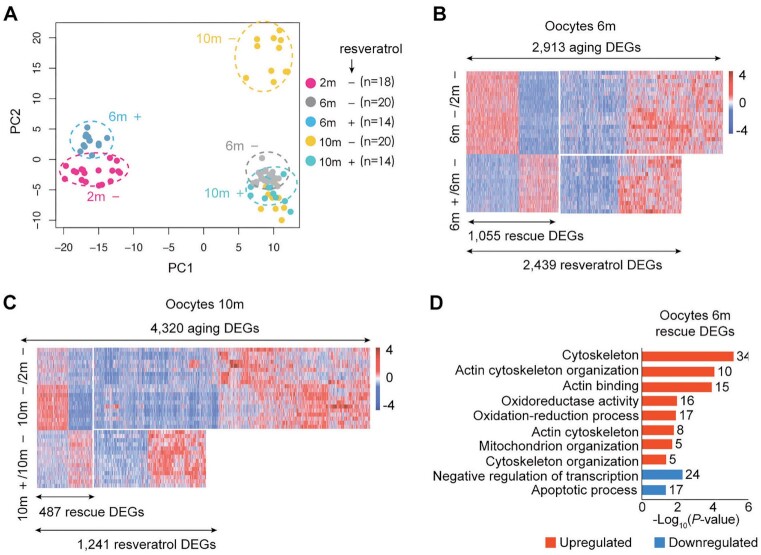
Fig. 2.
Aging transcriptome of oocytes from natural aging mice and intervention with resveratrol. (A) PCA based on RNA-Seq data. (+): mice treated with resveratrol, (–): control without exposure to resveratrol, (n): number of oocytes. (B) Heatmaps displaying the distribution of oocyte DEGs for 6-month (6 m). Each row represents one oocyte and each column represents one gene. Red, upregulated [log2FoldChange > log2(1.5) and padj < 0.05]; blue, downregulated [log2FoldChange < −log2(1.5) and padj < 0.05]. “6 m –/2 m –” represents oocytes of 6-month-old mice compared to those of 2-month-old mice. “6 m +/6 m –” represents oocytes of 6-month-old mice treated with or without (DMSO only) resveratrol. (C) Heatmaps displaying the distribution of oocyte DEGs for 10-month-old mice (10 m). Each row represents one oocyte and each column represents one gene. Red, upregulated [log2FoldChange > log2(1.5) and padj < 0.05]; blue, downregulated [log2FoldChange < −log2(1.5) and padj < 0.05]. “10 m –/2 m –” represents oocytes of 10-month-old mice compared to those of 2-month-old mice. “10 m +/10 m –” represents oocytes of 10-month-old mice treated with or without (DMSO only) resveratrol. (D) KEGG and GO enrichment of oocyte DEGs rescued by resveratrol in 6-month-old. Red represents upregulation and blue represents downregulation. The number on the right side of the bar indicates the number of genes enriched in the signaling pathway.
To unveil the molecular events associated with aging and resveratrol, thousands of differentially expressed genes (DEGs) were identified among young, middle-aged and aged oocytes and between control-group and Resveratrol-group and referred to as “aging DEGs” and “resveratrol DEGs,” respectively. Integrative comparative analysis of these DEGs further identified aging DEGs that were partially rescued to the young-like by resveratrol and thereby referred to as “rescue DEGs.” Compared to young oocytes, middle-aged oocytes exhibited 2,913 aging DEGs and old oocytes showed 4,320 aging DEGs (Fig. 2B and C). That is a greater variability in gene expression was observed in old oocytes. A total of 2,439 resveratrol DEGs were identified in middle-aged oocytes from mice fed resveratrol compared to age-matched oocytes without resveratrol. Remarkably, 1,055 rescue DEGs (43%) of 2,439 resveratrol DEGs were identified (Fig. 2B). Moreover, 1,241 resveratrol DEGs were observed in old oocytes from mice with resveratrol treatment, among which, 487 (39%) were identified as rescue DEGs (Fig. 2C). Consequently, resveratrol protects against transcriptional aging of mouse oocytes.
Next, we performed the biological connotations of aging-, resveratrol-, and rescue-DEGs using Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis. In the oocytes from both middle age and old mice in control-group, the downregulated aging DEGs were enriched in cytoskeleton, actin cytoskeleton organization, actin binding, oxidoreductase activity, oxidation-reduction process, metabolic process, regulation of actin cytoskeleton organization, regulation of actin cytoskeleton, metabolic pathways, and protein autophosphorylation (Fig. S1A and B), and their downregulation was antagonized by resveratrol (Fig. S1C and D). Aging also led to a synchronous enhancement of multiple processes involved in the apoptotic process, response to oxidative stress, response to reactive oxygen species and cellular response to DNA damage stimulus (Fig. S1A and B), and this enhancement was weakened by resveratrol (Fig. S1C and D). The upregulated rescue-DEGs in oocytes from the middle-age mice were enriched in GO terms associated with cytoskeleton, actin cytoskeleton organization, actin binding, actin cytoskeleton, cytoskeleton organization, oxidoreductase activity, oxidation-reduction process, and mitochondrion organization (Fig. 2D). Additionally, the upregulated rescue-DEGs in oocytes from old mice were enriched in GO terms associated with ATP binding, protein autophosphorylation, and enzyme binding (Fig. S1E). Collectively, all of these biological processes are highly related to actin cytoskeleton organization and redox process. Given resveratrol has been established to inhibit oxidative stress, we then focus on actin skeleton organization as novel resveratrol effectors in aged oocytes.
To verify the effect of resveratrol on actin skeleton organization in aged oocytes, we visualized the transcription changes of the set of actin-related DEGs during aging and resveratrol treatment by creating heatmap. We observed that Myo1b, Myh11, Shroom3, Synpo, Fmnl3, Nf2, Fgd1, Abl2, Arpc3, Arhgef5, Ablim1, Klhl20, Pak3, Ablim3, Park2, Cap1, Sntg2, Arhgef18, Myo1a, and Syne1 were downregulated during aging and were mostly restored by resveratrol in the oocytes from 6-month old mice (Fig. S2A). Moreover, we confirmed that actin was reduced in aged oocytes by performing immunofluorescence staining for actin filament protein (Fig. S2B). We also observed that genes related to oxidoreductase activity or the oxidative reduction process were downregulated in middle-aged or aged oocytes, and their expression was reversed to young state by resveratrol in oocytes from middle-age mice (Fig. S2C). We treated oocytes with H2O2 or combined with resveratrol during in vitro maturation (IVM) of oocytes, and the ROS-sensitive probe H2DCFDA was used to detect intracellular ROS levels. Resveratrol abolished ROS induced by H2O2 treatment (Fig. S2D and E). Finally, we sought to determine the effect of ROS and resveratrol on actin protein expression level. F-actin decreased in oocytes following H2O2 treatment, which was rescued by the addition of resveratrol (Fig. S2F). Altogether, these observations suggest that aging-induced actin downregulation in oocytes could be improved with resveratrol administration.
Resveratrol ameliorates aging methylome in middle-aged oocytes
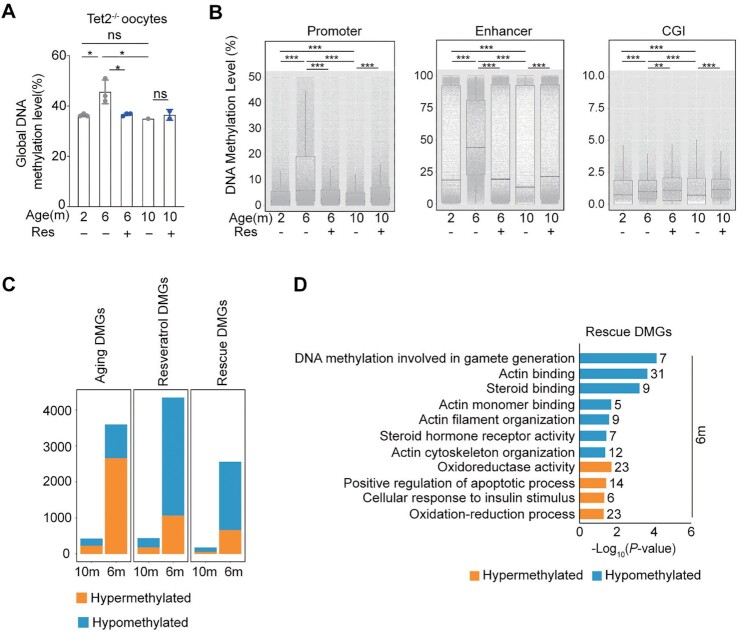
To test potential effects on the epigenome of resveratrol following long-term-oral administration, we performed low-input methylome sequencing analysis to generate methylome profiles of oocytes collected from young, middle-age and old mice fed with or without resveratrol (Fig. 1A, 10 oocytes per sample). The global DNAm level of middle-aged oocytes was lower than that of young oocytes, whereas the global methylation level of old oocytes was comparable to that of young oocytes (Fig. 3A). Moreover, lower methylation levels were also displayed in the promoter, enhancer, and CpG islands (CGIs) of middle-aged oocytes, while comparatively higher levels were detected in old oocytes (Fig. 3B and Fig. S3A). It appeared that resveratrol had no significant effect on the alteration of whole genome methylation levels in middle-aged or aged oocytes (Fig. 3A). However, resveratrol treatment increased the average methylation levels of the promoter, enhancer, and CGIs regions in middle-aged oocytes and reduced their average methylation levels in old oocytes (Fig. 3B and Fig. S3A).
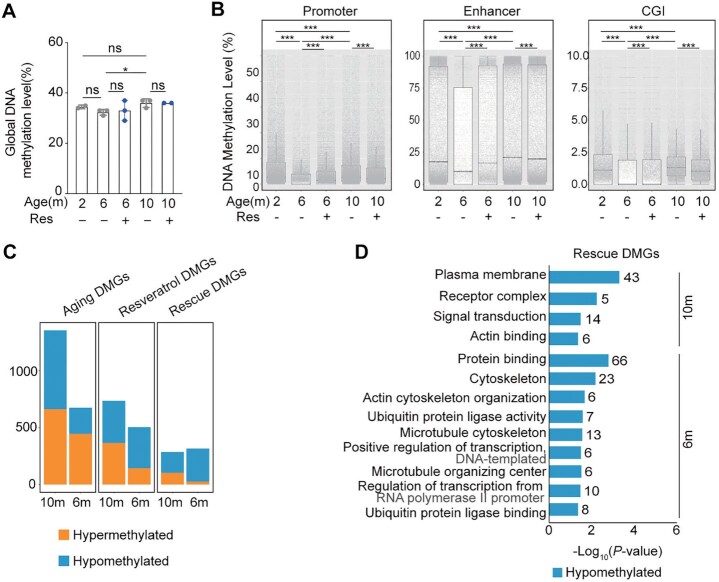
Fig. 3.
Aging methylome of oocytes from natural aging mice and intervention with resveratrol. (A) Global DNAm levels of oocytes from natural aging mice and intervention with resveratrol (Res). (B) Boxplot of the mean DNAm levels in different genomic elements determined by low-input methylation sequencing (promoter is defined as ± 1000 bp of TSS). The bottom and top of the boxes indicate the first and third quartiles, respectively, and the lines inside the boxes indicate the medians of the data. The bars show the mean ± SEM. (C) Histogram showing the number of aging differentially methylated genes (DMGs), resveratrol DMGs, and rescue DMGs in the three groups. “Aging DEGs” represents 10-month-old or 6-month-old (old or middle-age mice treated with DMSO) compared to 2-month-old (young) mice. “Resveratrol DEGs” represents mice treated with resveratrol compared to mice treated with DMSO as control. “Rescue DEGs” represents genes that were upregulated (or downregulated) in old mice but reversed after resveratrol treatment. (D) KEGG and GO enrichment of rescued DMGs by resveratrol. Orange represents hypermethylation and blue represents hypomethylation. The number on the right side of the bar indicates the number of genes enriched in the signaling pathway. *P < 0.05; ***P < 0.001; ns: not significant. One-way ANOVA and Tukey’s multiple comparison test for A (Age), and two-way ANOVA and Tukey's multiple comparison test for A (Age and Res). Two-way ANOVA and Tukey’s multiple comparison test for B.
Furthermore, we assessed the effect of age and resveratrol on the methylation according to the differentially methylated regions (DMRs). DMRs between young and middle-aged oocytes consisted of 2,144 hypermethylated regions and 873 hypomethylated regions (Fig. S3B). DMRs between oocytes from middle-age resveratrol-group and oocytes from the age-matched Control-group consisted of 609 hypermethylated regions and 256 hypomethylated regions (Fig. S3C). The number of age-induced DMRs is much more than that of resveratrol-induced. Therefore, this result indicates that age has a greater impact on methylation profiles than does resveratrol.
To further unveil the methylation variation implications associated with aging and resveratrol, thousands of DMGs were identified according to the methylation changes in the promoter region among young, middle-aged and aged oocytes and between control- and resveratrol-group and referred as “aging DMGs” and “resveratrol DMGs,” respectively. Integrative comparative analysis of these DMGs further identified aging DMGs that were partially rescued to the younger state by resveratrol and thereby referred to as “rescue DMGs.” In line with the DMRs, the number of aging DMGs is much more than that of resveratrol DMGs in oocytes from 6-month old mice or 10-month old mice. Moreover, most of rescue DMGs are hypomethylated (Fig. 3C).
Then, we explored the biological connotations of aging-, resveratrol-, and rescue- DMGs using GO and KEGG pathway analysis. The promoter regions of microtubules and actin cytoskeleton-related genes were hypermethylated, and hypomethylated aging-DMGs were enriched in GO terms associated with aging and Wnt signaling pathway (Fig. S3D). Interestingly, most of hypomethylated resveratrol-DMGs in oocytes from middle-age or old mice were enriched in GO terms of cytoskeleton, microtubule cytoskeleton, actin cytoskeleton organization, microtubule cytoskeleton organization, autophagy, ubiquitin protein ligase activity, transcription, and ATP binding (Fig. S3E). Enrichment analysis further showed the hypomethylated rescue-DMGs were enriched in GO terms associated with protein binding, cytoskeleton, actin cytoskeleton organization, plasma membrane, actin binding, and microtubule cytoskeleton (Fig. 3D). Collectively, some of these rescued biological processes are also highly related to actin skeleton organization, which prompted us to further assess promoter methylation status of actin-related genes and their transcription associated with aging and resveratrol. We generated Venn diagrams of rescue DEGs with the gene set of actin related genes and showed that 53 rescue DEGs were related to actin organization and regulation (Fig. S3F). Then, we performed an association analysis on the methylation and transcriptome of these 53 genes. The results showed that most of these genes increased their promoter methylation level with age, while their expression downregulated (Fig. S3G). In contrast, the methylation level of these genes was reduced and their expression was upregulated in oocytes from middle age mice after resveratrol treatment (Fig. S3H). Together, our data demonstrate that resveratrol ameliorates the transcription of actin-related genes by reprogramming their aging methylome in oocytes with age.
Tet2-deficiency abrogates the effect of resveratrol on reprogramming the aging transcriptome
Our data showed that most of rescue DMGs in oocytes from middle-age WT mice with resveratrol administration are hypomethylated. We asked whether the active DNA demethylation is necessary for the beneficial effects of resveratrol on female fertility during reproductive aging.
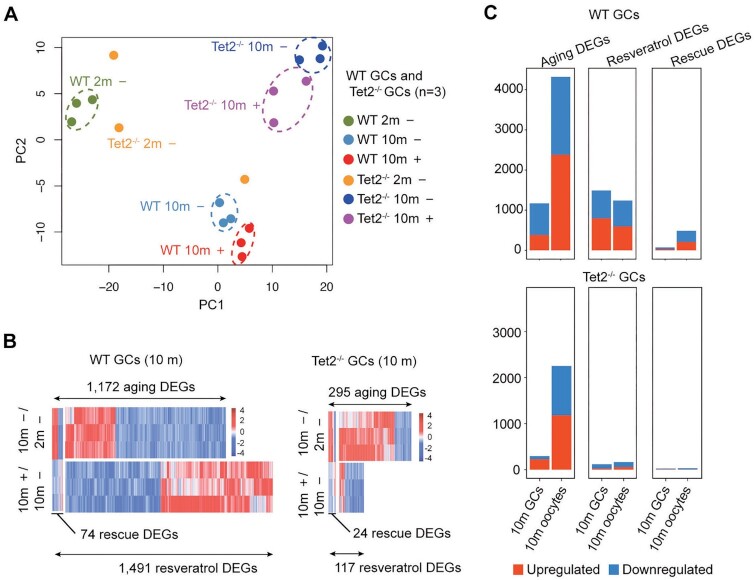
We were curious to know the single-cell transcriptome profiles of oocytes collected from Tet2−/− mice fed with or without resveratrol. The PCA plot demonstrated that young WT oocytes were separated apparently from young Tet2−/− oocytes, middle-aged oocytes, old oocytes. This means that both age and Tet2 greatly impacted the transcriptome dynamics. Moreover, after administration of resveratrol, only the oocytes from middle-age WT mice fed with resveratrol were clustered closely to that of young WT mice, indicating that resveratrol could effectively reverse aging-related gene expression changes in WT middle-aged oocytes (Fig. 4A). These data demonstrated that the loss of Tet2 led to a more stochastic global pattern of gene expression within both middle-aged and old oocytes. In the oocytes from Tet2−/− mice treated with resveratrol, resveratrol had a minimal protective effect on the transcriptome with age. Next, we explored the effects of aging and resveratrol on Tet2−/− oocytes according to DEGs. Compared to the young Tet2−/− oocytes from mice without resveratrol, middle-aged Tet2−/− oocytes exhibited 3,769 aging DEGs and old Tet2−/− oocytes showed 2,253 aging DEGs (Fig. 4B and C). Yet, fewer resveratrol DEGs were detected in both middle-aged Tet2−/− oocytes (124 resveratrol DEGs) and old Tet2−/− oocytes (165 resveratrol DEGs). Moreover, only 78 rescue DEGs were identified in middle-aged Tet2−/− oocytes and just 31 rescue DEGs were observed in old Tet2−/− oocytes (Fig. 4B and C). These rescue DEGs accounted for only approximately 2% or 1% of the aging DEGs in middle-aged oocytes or old oocytes respectively, suggesting that resveratrol has minimal effects on the global transcriptomic profile in Tet2−/− oocytes during aging. The stack bar graph revealed that the counts of resveratrol DEGs and rescue DEGs in old WT oocytes are larger than those in old Tet2−/− oocytes (Fig. 4D), indicating that Tet2-deficiency abrogates the effect of resveratrol on reprogramming the aging transcriptome.
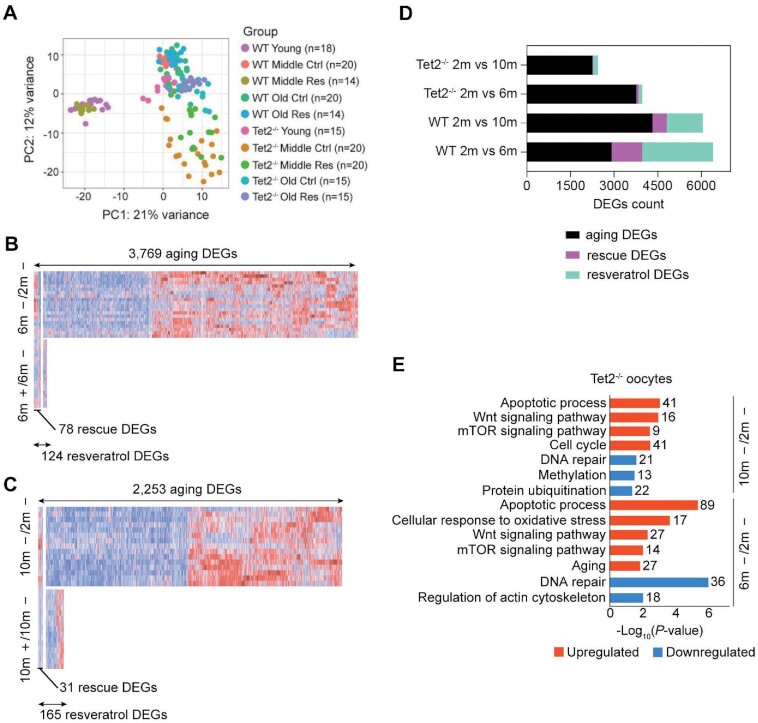
Fig. 4.
Aging transcriptome of oocytes from Tet2−/− mice and intervention with resveratrol. (A) PCA plot based on RNA-Seq data of both WT and Tet2−/− oocytes. (B) Heatmaps displaying the distribution of Tet2−/− oocyte DEGs for 6-month (6 m). Each row represents one oocyte and each column represents one gene. Red, Upregulated [log2FoldChange > log2(1.5) and padj < 0.05]; blue, downregulated [log2FoldChange < −log2(1.5) and padj < 0.05]. “6 m –/2 m –” represents oocytes of 6-month-old mice compared to those of 2-month-old mice. “6 m +/6 m –” represents oocytes of 6-month-old mice treated with or without (DMSO only) resveratrol. (C) Heatmaps displaying the distribution of oocyte DEGs for 10-month-old mice (10 m). Each row represents one oocyte and each column represents one gene. Red, Upregulated [log2FoldChange > log2(1.5) and padj < 0.05]; blue, downregulated [log2FoldChange < −log2(1.5) and padj < 0.05]. “10 m –/2 m –” represents oocytes of 10-month-old mice compared to those of 2-month-old mice. “10 m +/10 m –” represents oocytes of 10-month-old mice treated with or without (DMSO only) resveratrol. (D) Histogram showing the counts of aging DEGs, resveratrol DEGs, and resveratrol rescued DEGs in WT and Tet2−/− oocytes. (E) KEGG and GO enrichment of aging DEGs from middle-aged and aged Tet2−/− oocyte. Red represents upregulation and blue represents downregulation. The number on the right side of the bar indicates the number of genes enriched in the signaling pathway.
Notably, Tet2-deficient aging oocytes exhibited some unique gene expression signatures. In the middle-aged or old Tet2−/− oocytes, the downregulated aging DEGs were enriched in DNA repair, regulation of actin cytoskeleton, methylation and protein ubiquitination, and the upregulated aging DEGs enhanced multiple processes associated with apoptotic process, cellular response to oxidative stress, Wnt signaling pathway, mTOR signaling pathway, aging and cell cycle (Fig. 4E). Given FOXO3a is a major target of inactivation by mTOR signaling (46) and modulates WNT/beta-catenin signaling (47), we further verified the expression patterns of FoxO3a, which plays an essential role in longevity. Based on RNA-Seq data, the expression of FoxO3a declined significantly in naturally aging oocytes, while in Tet2−/− mice, FoxO3a expression was maintained at a lower level in both the young oocytes and the old oocytes (Fig. S4A). By immunofluorescence, the expression level of FoxO3a in old MII oocytes was significantly lower than that in young oocytes (Fig. S4B). Moreover, immunofluorescence staining for FoxO3a in ovarian sections demonstrated that FoxO3a was significantly decreased in aging oocytes, but was maintained at a low level in Tet2−/− oocytes (Fig. S4C). Together, our data indicates that mTOR signaling pathway and Wnt signaling pathway are enhanced in Tet2-deficient aging oocytes.
Tet2-deficiency perturbs the effect of resveratrol on reprogramming of methylome
Given minimal effects of resveratrol on reversing the global transcriptomic profile in Tet2−/− oocytes during aging, we hypothesized that Tet2 altered the effect of resveratrol in aged oocytes by remodeling the methylome. Interestingly, trends of methylation in the global genome, promoter, enhancer and CGI regions were opposite in WT and Tet2−/− oocytes during aging (Fig. 5A and B; and Fig. S5A). In Tet2−/− oocytes, the middle-aged oocytes showed higher methylation level in global genome, promoter, enhancer and CGI regions, while the methylation level of old oocytes was lowered and was generally similar to that of young oocytes (Fig. 5A and B; and Fig. S5A). Notably, resveratrol administration reduced the genome-wide methylation level in the middle-aged Tet2−/− oocytes, as well as the methylation levels of promoter, enhancer and CGI regions (Fig. 5A and B; and Fig. S5A). Consistent with this, many aging DMGs, resveratrol DMGs and rescue DMGs were observed in middle-aged oocytes, and fewer aging DMGs, resveratrol DMGs, and rescue DMGs were identified in old oocytes (Fig. 5C).
Fig. 5.
Aging methylome of oocytes from Tet2−/− mice and intervention with resveratrol. (A) Global DNAm levels of oocytes from Tet2−/− mice and intervention with resveratrol (Res). (B) Boxplot of the mean DNAm levels in different genomic elements determined by low-input methylation sequencing (promoter is defined as ± 1000 bp of TSS). The bottom and top of the boxes indicate the first and third quartiles, respectively, and the lines inside the boxes indicate the medians of the data. The bars show the mean ± SEM. (C) Histogram showing the number of Aging DEGs, Resveratrol DEGs and Rescue DEGs in middle-age or old Tet2−/− mice. (D) KEGG and GO enrichment of rescued DMGs by resveratrol. Orange represents hypermethylation and blue represents hypomethylation. The number on the right side of the bar indicates the number of genes enriched in the signaling pathway. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant. One-way ANOVA and Tukey’s multiple comparison test for A (Age), and Two-way ANOVA and Tukey’s multiple comparison test for A (Age and Res). Two-way ANOVA and Tukey’s multiple comparison test for B.
Next, we explored the biological connotations of aging-, resveratrol-, and rescue- DMGs in middle-aged Tet2−/− oocytes, revealing the hypermethylated aging DEGs enriched in GO terms, including DNAm involved in gamete generation, steroid hormone receptor activity, steroid binding, proteolysis, oogenesis, and actin filament organization (Fig. S5B). The hypomethylated aging DEGs enriched in negative regulation of transcription from RNA polymerase (Fig. S5B). However, in the oocytes from the middle-age mice treated with resveratrol, the most hypomethylated resveratrol DMGs were enriched in GO terms associated with inflammatory response, actin binding, DNAm involved in gamete generation, actin filament organization, actin cytoskeleton organization, and steroid hormone receptor activity, while the hypermethylated GO terms enriched in cancer and apoptotic pathway for resveratrol DEGs (Fig. S5C). Therefore, the loss of Tet2 might abolish the anti-inflammatory effect of resveratrol on aged oocytes. Based on rescue-DMGs in middle-aged Tet2−/− oocytes, the hypomethylated rescue-DMGs were enriched in GO terms associated with DNAm involved in gamete generation, actin binding, steroid binding, actin monomer binding, and actin cytoskeleton organization. Therefore, resveratrol also can reverse the methylation patterns of genes involved in actin cytoskeleton organization in Tet2−/− oocytes from middle age mice. Unexpectedly, the hypermethylated rescue-DMGs in middle-aged Tet2−/− oocytes were enriched in GO terms associated with oxidoreductase activity, oxidation-reduction process, cellular response to insulin stimulus, and positive regulation of apoptotic process (Fig. 5D), suggesting that the Tet2-deficiency might abrogate the antioxidant effect of resveratrol on aged oocytes.
Oxidative stress accelerates telomere shortening (48). We estimated the telomere length and verified that telomere became shorter in aged oocytes from WT mice, and that resveratrol prevented telomere shortening in oocytes from middle-age or old mice (Fig. S5D). However, resveratrol escalated telomere shortening of Tet2−/− oocytes from middle-aged or old mice (Fig. S5D). The key genes (such as Dkc1, Tert, Terc, Terf1, and Terf2) related to telomeres and telomerase showed no significant changes at the transcriptional level in oocytes with or without resveratrol treatment (Fig. S5E). Therefore, Tet2-deficiency abrogated the effect of resveratrol on methylome reprogramming in aged oocytes, likely abolishing the protection function of resveratrol against oxidative stress.
Resveratrol slightly improves the aging-associated transcriptome of ovarian GCs
We compared the transcription profiles of GCs between young and old mice as well. PCA cluster results indicated that young and old GCs were well distinguished by age and resveratrol in both WT and Tet2−/− mice, while young Tet2−/− GCs clustered loosely (Fig. 6A). Compared to the young WT GCs, old GCs exhibited 1,172 aging DEGs, and old Tet2−/− GCs exhibited 295 aging DEGs. Additionally, 74 rescue DEGs of 1,491 resveratrol-DEGs were identified in old WT GCs and 24 rescue DEGs of 117 resveratrol-DEGs found in old Tet2−/− GCs (Fig. 6B). Also, in both natural aging WT and Tet2−/− mice, the number of DEGs in GCs was less than that in oocytes (Fig. 6C).
Fig. 6.
Aging transcriptome of granulosa cells (GCs) and intervention with resveratrol. (A) PCA plot based on RNA-Seq data of GCs from mice at 2 or 10 months old. (+): Mice treated with resveratrol; (–): control without exposure to resveratrol; and (n): number of mouse samples. (B) Heatmaps showing the distribution of aging DEGs, resveratrol DEGs, and rescue DEGs in GCs from WT or Tet2−/− mice. Each row represents one sample, and each column represents one gene. Red, upregulated [log2FoldChange >log2(1.5) and padj <0.05]; blue, downregulated [log2FoldChange <−log2(1.5) and padj <0.05]. (C) Histogram showing the number of aging DEGs, resveratrol DEGs, and resveratrol rescued DEGs in WT and Tet2−/− GCs or oocytes. “Aging DEGs” represents 10-month-old (old) mice treated with DMSO compared to 2-month-old (young) mice. “Resveratrol DEGs” represents mice treated with resveratrol compared to mice treated with DMSO as control. “Rescue DEGs” represents genes that were upregulated (or downregulated) in old mice but downregulated (or upregulated) after resveratrol treatment.
We performed GO and KEGG enrichment analysis to explore the biological connotations of aging- and resveratrol- DEGs in GCs. In WT GCs, downregulated aging DEGs were enriched in cell cycle, positive regulation of transcription from RNA polymerase II promoter, protein processing in endoplasmic reticulum, DNA replication, cholesterol metabolism, and insulin resistance (Fig. S6A). Aging also enhanced the process involved in response to endoplasmic reticulum stress. While the upregulated resveratrol- DEGs mainly activated the processes involved in the ATP binding, ubiquitin protein ligase binding, metabolic pathways and actin binding. Apoptotic process, response to oxidative stress and cellular response to DNA damage stimulus in old GCs were reduced by resveratrol (Fig. S6B). The rescue DEGs in naturally aging GCs were the upregulated genes related to process of ATP binding and enzyme binding (Fig. S6C). In Tet2−/− GCs, upregulated aging- DEGs were enriched in mitochondrial inner membrane, immune response and inflammatory response. Interestingly, upregulated aging DEGs related to immune response and inflammatory response in old Tet2−/− GCs were antagonized by resveratrol (Fig. S6D, E, and F). To test whether resveratrol could suppress inflammation response in normal GCs, we treated GCs with lipopolysaccharide (LPS) with or without resveratrol. Resveratrol treatment suppressed LPS-induced IFN-γ production in ovarian GCs (Fig. S6G). Apoptotic process-related genes, which were upregulated in old WT GCs, were slightly suppressed by resveratrol (Fig. S6H). Furthermore, the increase in apoptosis in old GCs was confirmed by TdT-mediated dUTP nick end labeling (TUNEL) assay (Fig. S6I). To test whether resveratrol inhibited apoptosis in GCs, we treated GCs with H2O2 and resveratrol. Resveratrol significantly decreased the number of TUNEL-positive GCs induced by H2O2 (Fig. S6J). These data strengthened the notion that resveratrol has antioxidant effects. Taken together, our data indicated that resveratrol slightly improved age-dependent transcriptome of ovarian GCs.
Discussion
Here, we present a wealth of molecular information on ovarian aging and the effect of resveratrol in ovarian aging. We observed age-associated alterations in telomere, single-cell transcriptome, and methylome, which collectively provide insights into the mechanism underlying ovarian aging. We discover that long-term-oral resveratrol administration yields a younger transcriptome, improving the fertility in old female mice. Moreover, our study reports that resveratrol shifts the methylome to a younger profile by attenuating the age-related DNAm alterations in oocytes from middle-age or old mice. However, resveratrol exerts minimal effects on rescuing the reproductive aging-related phenotypic alterations in Tet2−/− mice. We elucidate unique gene expression signatures of Tet2−/− oocytes during aging. Tet2-deficiency abrogates the effect of resveratrol on reprogramming the aging transcriptome, as well as perturbs the effect of resveratrol on methylome reprogramming in aged oocytes. Together, our results demonstrate that reprogramming of the aging epigenome of oocytes by resveratrol can be found at levels of telomeres, transcriptome and methylome, and the active DNA demethylation is necessary for the reprogramming to occur (Fig. S7).
Several research groups have identified the transcriptomic changes and methylome variations during aging in ovaries at the single-cell level (14, 17, 49–55). These studies detailed transcriptomic changes and methylome variations between young and old oocytes. The molecular signatures of middle-aged oocytes and the molecular dynamics underlying oocytes aging remain to be explored. We delineate comprehensive age-associated gene-expression variation and DNAm changes in young, middle-aged and aged oocytes. Inactivated antioxidative pathways, increased reactive oxygen species and aging-induced actin organization dysfunction were observed in oocytes from middle-age mice. Consistently, oxidative damage has been recognized as a crucial factor in ovarian functional decline with age (49, 53). Moreover, several previous studies in human oocytes demonstrated the analogous observations in mice that genes involved in oxidoreductase activity, oxidation-reduction process, mitochondria function and cytoskeletal structure are all affected by age (56, 57). Thus, our study provided age-associated alterations in single-cell transcriptome and methylome to support the idea that the ovary exhibits early-onset aging-associated dysfunction prior to the middle age (3, 58). In support of our findings, age-associated alterations to the methylome showed that the underlying cellular and molecular changes began much earlier (59, 60). Meanwhile, our data revealed the disconnection between declining reproductive performance with age (breeding trials) and global DNAm level, which is unchanged between 2- and 10-month-old females but dips lower in 6-month-old female mice, whereas 6-month-old WT female mice have normal breeding performance. These data imply that the global DNAm dynamic patterns may not closely parallel with reproductive aging.
Resveratrol possesses various bioactivities, such as antioxidant, anti-inflammatory, cardiovascular protection, anticancer, and antiaging effects. Consistent with previous studies (28, 29), our findings validated that resveratrol can protect against age-associated infertility in female mice. Our results demonstrate that genes related to the oxidoreductase activity, oxidation-reduction process, response to oxidative stress and response to reactive oxygen species were dysregulated in middle-aged and aged oocytes, but recovered following resveratrol administration. A recent study of the mouse liver proteome showed that metformin, resveratrol and rapamycin have reprogramming effects on the fundamental processes such as mitochondrial function, oxidative stress, and RNA splicing (61). Therefore, in terms of mitochondria function, it appears that resveratrol has similar reprogramming effects in ovaries like in liver. Moreover, we showed that the downregulated actin-related genes also are rescued by resveratrol, suggesting that the beneficial effect of resveratrol on aged oocytes might be partially mediated by actin organization function. Actin plays a very extensive role in oocytes meiosis involved in nuclear positioning, germinal vesicle breakdown, spindle migration chromosome segregation, and other processes (62). Consistently, Fmnl3 is involved in actin-mediated spindle migration and cytokinesis in mouse oocytes and its deficiency leads to the failure of polar body extrusion and large polar bodies (63). Abnormal actin cytoskeleton has also been found to involve in oocyte aging (64–66). Interestingly, at methylation level, hypermethylated aging DMGs in aged oocytes related to actin cytoskeleton organization are changed into hypomethylated state by resveratrol.
Moreover, DNA demethylation activity is necessary for this reprogramming to occur by resveratrol in aged oocytes. We find that Tet2-deficient aging oocytes exhibit unique gene expression signatures, such as enhancement of multiple processes associated with mTOR signaling pathway and cellular response to oxidative stress. In line with our findings, a reduction in 5-hmC activates mTOR signaling pathway (67). Even though resveratrol is reported to have a major role in the epigenetic regulation of transcription as well as in influencing translation via mTOR (61, 68), mTOR signaling pathway and cellular response to oxidative stress in Tet2−/− oocytes are not effectively reprogrammed by resveratrol. Furthermore, Tet2−/− oocytes display an upshift in global DNAm levels in middle-aged oocytes, which could be due to the Tet-mediated demethylation. Consistent with this, reduced demethylation by oocyte TET3 insufficiency contributes to hypermethylation at the paternal alleles of several insulin secretion genes (69). Unexpectedly, genes associated with oxidoreductase activity and oxidation-reduction process were hypermethylated in Tet2−/− oocytes from middle-age mice fed with resveratrol, which indicates the Tet2-deficiency abrogates the antioxidant effect of resveratrol on middle-aged oocytes. It remains to be understood regarding the causality of gene expression changes according to the specific DNAm alterations in genes related to oxidoreductase activity and oxidation-reduction process (70). Also, resveratrol administration escalates telomere shortening in Tet2−/− oocytes. Indeed, oxidative stress caused by excess ROS accelerates telomere shortening (48). Interestingly, rapamycin decreases life span in short telomere G3 Terc KO mice, which showed activated mTOR (71). Thus, mTOR is important for survival in the context of short telomeres. Tet2-deficient oocytes seem to have similar phenotypes of premature aging like G3 Terc KO mice as to elevated mTOR signaling pathway, and resveratrol fails in suppressing mTOR in Tet2−/− mice, so these mice continue to undergo premature aging. Together, our data suggest that resveratrol-induced epigenetic reprogramming in oocytes requires active demethylation.
Ovarian microenvironment can affect the quality of oocytes, and even accelerate the oocyte aging and lead to infertility. Ovarian microenvironment is composed of different types of cells within the ovary, and communication between the oocyte and the microenvironment is mediated by direct contact with surrounding cells, the extracellular matrix (ECM), and signaling molecules, including hormones, growth factors, and metabolites (72). Bidirectional communication between the oocyte and its associated somatic cells plays a key role in fertility and embryogenesis. For example, cumulus cells can provide essential nutrients for oocyte maturation, and they are also involved in accelerating oocyte senescence by activating the Fas/FasL pathway (73–77). GCs are in direct contact with the oocyte, playing a fundamental role in its growth and development. Low doses of resveratrol protect human GCs from induced-oxidative stress (78). Our results demonstrate that genes involved in cellular response to DNA damage stimulus, response to oxidative stress and apoptotic process are downregulated in old ovarian GCs by resveratrol.
Altered homeostasis between synthesis and degradation of ECM components affects tissue structure and function, and excessive accumulation of ECM contributes to tissue fibrosis. Fibrosis within the mouse ovarian stroma increases with reproductive age, and it is an early marker of ovarian stroma aging (79, 80). Ovarian fibrosis is reversible, and antifibrosis drugs (pirfenidone and BGP-15) were reported to eliminate fibrotic collagen and restore ovulation in reproductively old and obese mice, in association with dampened M2 macrophage polarization and upregulated MMP13 protease (81). Our data showed that the upregulated rescue-DEGs in normal aging GCs were related to process of ATP binding and enzyme binding. Moreover, resveratrol has been reported to resist cardiac fibrosis, pulmonary fibrosis and lung fibrosis through several cell signaling pathways (82–84). Although activation of inflammatory response has been recognized as indicators of aging (85, 86), our transcriptome data did not show inflammation response in oocytes or GCs of WT mice at 6 or 10 months old. Recent research also pointed that inflammation did not occur until 12 moths in C57BL/6 inbred mice (87, 88). However, inflammatory factor interleukin 6 (IL-6) is highly expressed in GCs of Tet2-deficient mice at 10-months old. In a clinical study, older IVF patients have higher IL-6 levels compared to younger patients and higher IL-6 levels are associated with decreased possibility of clinical pregnancy (89). Our data suggest that resveratrol could improve oocyte quality during aging partially by alleviating the ovarian microenvironment.
Taken together, our findings reveal that resveratrol retains a younger ovarian epigenome in mice, which extends the natural reproductive lifespan by mechanisms of antioxidation and remodeling of methylation. Notably, Tet2 deficiency disrupts the ability of resveratrol to retain the younger epigenome of aging mouse ovaries. Our work provides further insights into the epigenome regulation of ovarian aging and into new functions of resveratrol with Tet2 in retaining a younger epigenome.
Materials and Methods
Tet2 knockout mice
Tet2 knockout mice were generated from a 129 and C57BL/6 J mixed genetic background (90). Most of the WT and Tet2 knockout mice were generated from Tet2 heterozygote mating. All mice in the study were maintained in a mixed 129 and C57BL/6 background. All mouse experiments were approved by and performed in accordance with the guidelines and regulations of the Institutional Animal Care and Use Committee of Nankai University. For fertility analysis, females were crossed with 3- to 4-month-old C57 male mice until they gave birth, and the number of litters was counted.
Genotyping
Postnatal mice at 2 weeks old were genotyped using DNA extracted from their ears. Polymerase chain reaction (PCR) was conducted at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. DNA fragments were visualized by agarose gel electrophoresis.
Resveratrol treatment
Resveratrol has low solubility in water and is easily decomposed when exposed to light. Therefore, 30 mg resveratrol (R5010, Sigma) was dissolved in 500-μl DMSO (D2650, Sigma), and then 1 L 37°C sterile water was added and the solution was incubated in a water bath at 37°C for 2 to 3 h until dissolved (28). The control-group was treated with 500 μl DMSO per 1 L of drinking water. Drinking water was freshly prepared and changed every 5 days. The bottles containing resveratrol were covered with aluminum foil to protect from light. Mice were treated with medication at the age of 2 months (the young group), and the treatment was continued until the mice were 6 or 10 months old.
MII oocytes
Female mice from the different age groups were superovulated by injection of 5 IU pregnant mare’s serum gonadotrophin (PMSG, Millipore, 5 IU per mouse), followed by 5 IU hCG 46 to 48 h later. Females were humanely sacrificed and oocytes were enclosed in cumulus masses collected into potassium simplex optimized medium (KSOM) by releasing from oviduct ampullae 14 h after hCG injection. Cumulus cells were removed by pipetting after brief incubation in 0.03% hyaluronidase prepared in KSOM containing 14 mM HEPES and 4 mM sodium bicarbonate (HKSOM).
Granulosa cells (GCs)
Isolation of GCs was based on the previous method (91). Briefly, PMSG was injected into the abdominal cavity of mice 46 h before isolating GCs. The mice were humanely sacrificed and the ovaries were dissected. Insulin syringes were used to puncture visible follicles on the surface of ovaries under a stereomicroscope to release GCs into culture medium, avoiding isolated GCs from small follicles. In addition, oocytes were filtered out using a 40-μm cell strainer (Falcon). Then, the isolated GCs were washed in phosphate-buffered saline (PBS), dispersed into a single-cell suspension, and then counted to take 50 cells for RNA-Seq library construction.
Oocyte IVM
Female mice were humanely sacrificed after PMSG injection at 46 to 48 h intervals. After ovaries were dissected, fully grown germinal vesicle (GV) oocytes were collected under a microscope by pricking follicles using an insulin syringe in IVM medium [α-MEM (Invitrogen) with 5% fetal bovine serum (FBS), 0.24 mM sodium pyruvate, 1 IU/ml PMSG, and 1.5 IU/ml hCG]. The obtained GV oocytes were divided into two equal parts and then placed in IVM medium or IVM medium containing 1 μM resveratrol. Oocytes were matured in IVM medium for 17 to 18 h at 37°C (92).
Immunofluorescence microscopy of oocytes
Based on a previous method (93), the spindle, actin, and chromatin of oocytes were stained and observed by immunofluorescence microscopy. Oocytes were fixed in fixative (MTSB XF) at 37°C for at least 30 min and then washed four times with washing buffer (PBS, supplemented with 0.02% NaN3, 0.01% Triton X-100, 0.2% nonfat dry milk, 2% goat serum, 2% bovine serum albumin, and 0.1 M glycine). Oocytes were left in washing buffer for 2 h at 37°C for blocking. Oocytes were incubated with FITC-α-tubulin (1:100 F2168, Sigma, Beijing, China) and Phalloidin-iFluor 594 Reagent-cytopainter (1:1000, ab176757, Abcam, Beijing, China) for 2 h at 37°C. Oocytes were washed and stained with Hoechst 33,342 (1:200, H3570, Life Technologies, Shanghai, China) to label DNA. Oocytes were mounted on glass slides, sealed with nail polish, and examined with an Axio-Imager Z2 Fluorescence Microscope (Carl Zeiss, Shanghai, China). For the expression of Foxo3a, fixed oocytes were left in washing buffer for 2 h at 37°C for blocking, and then incubated with anti-Foxo3a antibody (1:200,12829S, Cell Signaling Technology, Shanghai, China) overnight at 4°C. Oocytes were washed and incubated with secondary goat anti-rabbit IgG Alexa Fluor 488 antibody (1:200, A-21,208, Thermo Scientific, Shanghai, China) at 37°C for 2 h and stained with Hoechst 33,342 to label DNA. Oocytes were mounted on glass slides, sealed with nail polish, and imaged using an Axio-Imager Z2 Fluorescence Microscope (Carl Zeiss). ImageJ was used for relative fluorescence quantification.
Immunofluorescence microscopy of ovarian sections
Sections were deparaffinized, rehydrated, and washed with 0.01 M PBS (pH, 7.2), and then incubated with 3% H2O2 for 10 min at room temperature to block endogenous peroxidase. Antigen recovery was achieved with 0.01 M citrate buffer (pH, 6.0) at high pressure for 3 min. Sections were incubated with blocking solution containing 5% goat serum and 0.1% bovine serum albumin (BSA) for 2 h at room temperature, and then with diluted Foxo3a antibody overnight at 4°C. Blocking solution without Foxo3a antibody served as the negative control. Sections were washed with PBS, incubated with goat anti-rabbit IgG Alexa Fluor 488 antibodies, and stained with Hoechst 33,342. Fluorescence was imaged using an Axio-Imager Z2 Fluorescence Microscope.
Detection of ROS levels in oocytes
We used 2′, 7′-dichlorodihydrofluorescein diacetate (H2DCFDA, HY-D0940, MCE), a cell-permeable nonfluorescent (but fluorescein-containing) probe for detecting oxidants, to detect the ROS levels (94). Detection of ROS levels in mouse oocytes has been described previously (95). Briefly, denuded oocytes were incubated in Hepes-buffered KSOM (HKSOM) medium containing 5 μM H2DCFDA in the dark for 30 min at 37°C, before being washed three times in HKSOM medium, mounted on glass slides, and imaged using a fluorescence microscope.
TUNEL assay of ovarian tissues and GCs
Apoptosis detection by TUNEL assay of tissues or cells was performed strictly in accordance with the instructions provided in the DeadEnd™ kit (G3250, Promega). Briefly, for tissues, after deparaffinization, the slide was fixed with 4% paraformaldehyde for 15 min at room temperature, before being incubated with 20 μg/ml proteinase K solution at room temperature for 10 min and washed with PBS. Cells were covered with 100 μl equilibration buffer and equilibrated at room temperature for 10 min, before adding rTdT incubation buffer and incubating at 37°C for 60 min to induce a tailing reaction. Slides were immersed in the 2 × saline sodium citrate (SSC) solution for 15 min at room temperature to stop the tailing reaction, and stained with Hoechst to reveal the nucleus. The slides were mounted and analyzed under a fluorescence microscope. Cells were fixed with 4% paraformaldehyde at 4°C for 25 min and then permeabilized with 0.2% TritonX-100 solution at room temperature for 5 min. The next steps were the same as those described above for tissues.
H2O2 treatment
Appropriate concentrations of H2O2 were used to induce ROS (96). Oocytes were treated with 100 μM, 150 μM H2O2, or both, with or without 1 μM resveratrol in the IVM medium. Oocytes were placed in IVM medium and matured for 17 to 18 h at 37°C. Following treatment of GCs with 5 mM H2O2 for 2 h, the medium was changed to fresh medium added with or without 1 μM resveratrol and cultured for 24 h at 37°C in a 5% CO2 incubator with humidified air (49, 97–102).
Lipopolysaccharide treatment
GCs were treated with 100 ng LPS (L2880, Sigma, Beijing, China) to induce an inflammatory response and 1 μM resveratrol was added for 24 h. After treatment, the cells were washed three times with PBS and then fixed with 3.7% paraformaldehyde for 30 min on ice. Permeabilizing solution (0.1% Triton X-100) was added to cells for 30 min at room temperature, before being carefully washed with PBS. Cells were blocked in blocking solution (3% goat serum plus 0.1% BSA in PBS) for 2 h. Cells were incubated with primary antibody IFN-γ (1:50,505,706, BioLegend, San Diego, CA, USA) overnight at 4°C. Next, the cells were washed three times with blocking solution (15 min per wash), and then incubated with Cy3-labeled Goat anti-Rat IgG (H + L) (1:200, A0507, Beyotime, Shanghai, China) for 2 h. Cells were washed and counterstained with Hoechst 33,342 (H3570, Thermo Scientific, Shanghai, China). Fluorescence was detected and imaged by an Axio-Imager Z2 Fluorescence Microscope (Carl Zeiss, Shanghai, China).
Single-cell telomere length measurement
This experiment was based on a method described previously (103). Three or more oocytes were collected from each mouse for this experiment. The amplification efficiency was measured according to the standard curves of Tel and mB1. The relative telomere length (T/R ratio) was calculated by 2−ΔΔCt.
Single-cell RNA-seq library construction and sequencing
RNA-Seq libraries were prepared using Smart-Seq2 methods as previously described (104). Briefly, the libraries were prepared using a TruePrep DNA Library Prep Kit V2 for Illumina® (TD503-02, Vazyme Biotech, Nanjing, China) according to the instruction manual. Samples were barcoded during library preparation and multiplex sequenced, with a 50-bp single-end sequencing strategy on a HiSeq x10 (Illumina, San Diego, CA, US).
Single-cell RNA-seq data analysis
Reads were mapped to mm10 from the UCSC genome (http://genome.ucsc.edu/) by Hisat2 (v2.1.0) with default parameters (105, 106). Read counts of each gene annotated in Gencode vM17 were calculated using featureCounts with default parameters. Sum factor normalization was applied with deconvolution of size factors within different batch samples using SCnorm. Raw counts were normalized by library size via TPM, and TPM was used to perform PCA for all groups. DEGs between different groups were analyzed using Deseq2. DEGs were defined as fold change of log2 transformed larger than log2(1.5) and an adjusted P-value < 0.05. Adjusted P-values were computed in DESeq2 using the Wald test, and were adjusted for multiple testing using the Benjamini and Hochberg procedure. GO and KEGG pathway enrichment analyses were performed via DAVID (https://david.ncifcrf.gov/home.jsp) based on DEGs.
DNAm library preparation and sequencing
DNAm libraries of mice MII oocytes were constructed according to a previously reported method (107), with minor changes. Ten MII oocytes from three mice (≥ 3 oocytes each) were collected in one PCR tube as a sample, and 2 to 4 samples in each group were used as replicates. The bisulfite conversion was performed on cell lysates with the following modifications: incubation at 98°C for 10 min and 64°C for 120 min. DNA was eluted in 10 mM Tris-HCl (pH 8.5) and combined with 10 mM dNTPs, 5 μM BioPEA_N4_37 (5′-biotin-ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNN N-3′), and 10 × NEBuffer 2 (E7645S, NEB, Ipswich, England) before incubation at 95°C for 5 min followed by a 4°C pause for 2 min. Next, 75 U of Klenow Fragment (M0212M, NEB, Ipswich, England) was added, and the samples were incubated at 4°C for 5 min, +1°C/15 s to 37°C, and 37°C for 30 min. Samples were incubated at 95°C for 1 min and transferred immediately to ice before the addition of fresh 1 mM dNTPs, 10 nM BioPEA_N4_37, 10 × NEBuffer 2, and 75 U Klenow fragment in a 2.75 μl total volume. The samples were incubated at 4°C for 5 min, and + 1°C/15 s to 37°C for 30 min. This random priming and extension were repeated a further three times (five rounds in total). Samples were then incubated with 40 U exonuclease I (M0293V, NEB, Ipswich, England) for 1 h at 37°C before DNA was purified using 1 × Agencourt Ampure XP beads (A63881, Beckman) according to the manufacturer’s guidelines. Samples were eluted in 10 mM Tris-Cl (pH 8.5) and incubated with washed M-280 Streptavidin Dynabeads (65,001, Life Technologies, Shanghai, China) for 30 min with rotation at room temperature. Beads were washed twice with 0.1 N NaOH, twice with 10 mM Tris-Cl (pH 8.5), and resuspended in 48 μl of 10 mM dNTPs, 10 × NEBuffer 2, and 10 μM Primer 2.0 (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNN-3′). Samples were incubated at 95°C for 45 s and transferred immediately to ice before the addition of 100 U Klenow Fragment and incubation at 4°C for 5 min, +1°C/15 s to 37°C, and 37°C for 90 min. Beads were washed with 10 mM Tris-Cl (pH 8.5) and resuspended in 50 μl of 1 U KAPA HiFi HotStart DNA Polymerase (KK2801, KAPA Biosystems), 10 μM Primer1.0 (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′), and 10 μM Index. The libraries were then amplified by PCR as follows: 98°C 45 s, 8 repeats of (98°C 15 s, 65°C 30 s, 72°C 30 s), 72°C 1 min, and 4°C hold. Amplified libraries were purified using 0.8 × Agencourt Ampure XP beads, according to the manufacturer’s guidelines. Samples were eluted in another 27 μl of 1 U KAPA HiFi HotStart DNA Polymerase (KK2801, KAPA Biosystems, Shanghai, China), 10 μM Primer 1.0 forward primer, and 10 μM index, and amplified by PCR for a further eight repeats. The amplified libraries were assessed for quality and quantity using High-Sensitivity DNA chips on the Agilent Bioanalyser. DNAm libraries were sequenced by an Annoroad using a HiSeq 4000 platform.
DNAm sequencing data analysis
Sequencing reads were trimmed with the default setting of Trimmomatic software to remove the adapters and low-quality reads. Trimmed reads were aligned to the mouse reference mm10 using Bismark (v0.16.312.5). PCR duplications were removed with Picard (http://broadinstitute.github.io/picard/), and the overlapped regions in uniquely mapped paired reads were clipped with the clipOverlap function of BamUtil (http://genome.sph. umich.edu/wiki/BamUtil: clipOverlap). CpG and non-CpG site methylation levels were extracted using a custom script (108), with the mpileup function of Samtools (v0.1.19). Strands were merged to calculate the CpG methylation level per site. Promoters are defined as regions 1 kb upstream and 1 kb downstream from the transcriptional start sites (TSSs) of mouse RefSeq transcripts (mm10). Only promoters containing at least 5 CpGs and covered by at least 10 reads were considered for further analysis. The methylation level of each promoter was calculated as the average methylation level of all CpG sites in this promoter.
Identification of DMRs and association analysis of DNA demethylation status and gene expression levels
We first aggregated DNAm data into two groups (e.g., 2 m vs 6 m Ctrl oocytes, 6 m Ctrl vs 6 m Res oocytes). The CpG sites covered by both groups were used for further DMR analyses. Then, the methylation levels of 1000-bp tiles (with ≥3 CpGs) of each group were calculated. DMRs were identified using the methylKit R package based on the following criteria: difference in methylation >20% and adjusted P-value <0.05, which were adjusted using the SLIM method. To correlate epigenetic and transcriptome datasets, DMRs were annotated to the nearest transcription start site using the annotatePeaks.pl function in HOMER v4.11.1. Genes displaying FDR <0.05 and fold change >1.2 for the indicated comparisons were used for association analysis. Gene sets related to actin were download from GO terms, including actin binding (GO:0,003,779) and actin cytoskeleton organization (GO:0,030,036), from Gene Ontology resource.
Statistical analysis
Data were analyzed by Student’s t-test, one-way analysis of variance (ANOVA) (multiple comparisons) and Two-way ANOVA by using GraphPad Prism 8.0 Software or StatView software from the SAS Institute, Inc. Significant differences were defined as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank, Guoliang Xu for support of the study by generously providing the Tet2 knockout mice.
Notes
Competing Interest Statement: The authors declare no competing interests.
Contributor Information
Mo Gou, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Jie Li, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Lizhi Yi, CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, 100101 Beijing, China.
Huiyu Li, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Xiaoying Ye, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Huasong Wang, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Linlin Liu, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Baofa Sun, Department of Zoology, College of Life Science, Nankai University, Tianjin 300071, China.
Song Zhang, Department of Biochemistry and Molecular Biology, College of Life Science, Nankai University, Tianjin 300071, China.
Zhengmao Zhu, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China.
Jiang Liu, CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, 100101 Beijing, China.
Lin Liu, Department of Genetics and Cell Biology, College of Life Science, Nankai University, Tianjin 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300350, China; Institute of Translational Medicine, Nankai Union Medical Center, Nankai University, Tianjin 300000, China.
Funding
The work was funded by the National Natural Science Foundation of China (91749129; 32030033; 31970667) and China National Key R&D Program (2018YFC1003004).
Authors' Contributions
M.G. performed major experiments and data analysis and prepared the manuscript draft. J.L. performed validation experiments and some bioinformatics analysis, and prepared the manuscript. L.Z.Y. performed the bioinformatics analysis of the methylation sequencing samples. H.Y.L., X.Y.Y., and H.S.W. performed the experiments. L.L.L. performed the bioinformatics analysis of the RNA-seq samples. B.F.S. advised on methylation sequencing analysis and revised manuscript. S.Z. advised on the validation experiment of inflammation and provided reagents. J.L. advised experimental design for methylation sequencing, provided reagents, discussed the data, and revised the manuscript. Z.M.Z. discussed, wrote and revised the manuscript. L.L. designed the study, interpreted the data, and wrote and revised the manuscript. The authors declare no competing interests.
Data Availability
All study data are included in the article and/or Supplementary Material. RNA-sequencing and methylation-sequencing data were deposited in the Gene Expression Omnibus (GEO) database (accession number GSE184637). The authors declare no competing interests.
References
- 1. Li Q, et al. 2012. Current understanding of ovarian aging. Sci China Life Sci. 55:659–669. [DOI] [PubMed] [Google Scholar]
- 2. Qiao J, et al. 2014. The root of reduced fertility in aged women and possible therapentic options: current status and future perspects. Mol Aspects Med. 38:54–85. [DOI] [PubMed] [Google Scholar]
- 3. Broekmans FJ, Soules MR, Fauser BC. 2009. Ovarian aging: mechanisms and clinical consequences. Endocr Rev. 30:465–493. [DOI] [PubMed] [Google Scholar]
- 4. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell. 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seale K, Horvath S, Teschendorff A, Eynon N, Voisin S. 2022. Making sense of the ageing methylome. Nat Rev Genet. 23:585–605. [DOI] [PubMed] [Google Scholar]
- 6. Ake Lu VP, Fei Z, Raj K, Horvath S. 2021. Universal DNA methylation age across mammalian tissues. Innov Aging. 5:410–410. [Google Scholar]
- 7. Horvath S 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horvath S, Raj K. 2018. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 19:371–384. [DOI] [PubMed] [Google Scholar]
- 9. Dabin J, Fortuny A, Polo SE. 2016. Epigenome maintenance in response to DNA damage. Mol Cell. 62:712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ermolaeva M, Neri F, Ori A, Rudolph KL. 2018. Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol. 19:594–610. [DOI] [PubMed] [Google Scholar]
- 11. Yan R, et al. 2021. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell. 28:1641–1656. [DOI] [PubMed] [Google Scholar]
- 12. Shirane K, et al. 2013. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLos Genet. 9:e1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stewart KR, et al. 2015. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 29:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang FL, et al. 2022. Parallel bimodal single-cell sequencing of transcriptome and methylome provides molecular and translational insights on oocyte maturation and maternal aging. Genomics. 114:110379. [DOI] [PubMed] [Google Scholar]
- 15. Potabattula R, et al. 2022. Ribosomal DNA methylation in human and mouse oocytes increases with age. Aging (Albany NY). 14:1214–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kordowitzki P, et al. 2021. Epigenetic clock and methylation study of oocytes from a bovine model of reproductive aging. Aging Cell. 20:e13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castillo-Fernandez J, et al. 2020. Increased transcriptome variation and localised DNA methylation changes in oocytes from aged mice revealed by parallel single-cell analysis. Aging Cell. 19:e13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sziraki A, Tyshkovskiy A, Gladyshev VN. 2018. Global remodeling of the mouse DNA methylome during aging and in response to calorie restriction. Aging Cell. 17:e12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maegawa S, et al. 2017. Caloric restriction delays age-related methylation drift. Nat Commun. 8:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harrison DE, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 460:392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shindyapina A, et al. 2022. Rapamycin treatment during development extends lifespan and healthspan. bioRxiv 481092. 10.1101/2022.02.18.481092, preprint: not peer reviewed. [DOI]
- 22. Katsyuba E, Romani M, Hofer D, Auwerx J. 2020. NAD(+) homeostasis in health and disease. Nat Metab. 2:9–31. [DOI] [PubMed] [Google Scholar]
- 23. Partridge L, Fuentealba M, Kennedy BK. 2020. The quest to slow ageing through drug discovery. Nat Rev Drug Discovery. 19:513–532. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, et al. 2020. Alpha-ketoglutarate ameliorates age-related osteoporosis via regulating histone methylations. Nat Commun. 11:5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asadi Shahmirzadi A, et al. 2020. Alpha-Ketoglutarate, an endogenous metabolite, extends lifespan and compresses morbidity in aging mice. Cell Metab. 32:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Y, et al. 2020. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 588:124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou DD, et al. 2021. Effects and mechanisms of resveratrol on aging and age-Related diseases. Oxid Med Cell Long. 2021:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu M, et al. 2013. Resveratrol protects against age-associated infertility in mice. Hum Reprod. 28:707–717. [DOI] [PubMed] [Google Scholar]
- 29. Sun YL, Tang SB, Shen W, Yin S, Sun QY. 2019. Roles of resveratrol in improving the quality of postovulatory aging oocytes in vitro. Cells. 8:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu MJ, et al. 2018. Resveratrol improves in vitro maturation of oocytes in aged mice and humans. Fertil Steril. 109:900–907. [DOI] [PubMed] [Google Scholar]
- 31. Ochiai A, Kuroda K. 2020. Preconception resveratrol intake against infertility: friend or foe?. Reprod Med Biol. 19:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ito S, et al. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 466:1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang J, et al. 2016. Tet enzymes regulate telomere maintenance and chromosomal stability of mouse escs. Cell Rep. 15:1809–1821. [DOI] [PubMed] [Google Scholar]
- 34. Khoueiry R, et al. 2017. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat Genet. 49:1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang S, et al. 2004. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 36:877–882. [DOI] [PubMed] [Google Scholar]
- 36. Sanders JL, Newman AB. 2013. Telomere length in epidemiology: a biomarker of aging, age-related disease, both, or neither?. Epidemiol Rev. 35:112–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng EH, et al. 2013. Evaluation of telomere length in cumulus cells as a potential biomarker of oocyte and embryo quality. Hum Reprod. 28:929–936. [DOI] [PubMed] [Google Scholar]
- 38. Yamada-Fukunaga T, et al. 2013. Age-associated telomere shortening in mouse oocytes. Reprod Biol Endocrinol. 11:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H, et al. 2021. Roles of Tet2 in meiosis, fertility and reproductive aging. Protein Cell. 12:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Izquierdo V, et al. 2019. Maternal resveratrol supplementation prevents cognitive decline in senescent mice offspring. Int J Mol Sci. 20:1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He X, et al. 2022. Punicalagin attenuates neuronal apoptosis by upregulating 5-hydroxymethylcytosine in the diabetic mouse brain. J Agric Food Chem. 70:4995–5004. [DOI] [PubMed] [Google Scholar]
- 42. Bertoldo MJ, et al. 2020. NAD(+) repletion rescues female fertility during reproductive aging. Cell Rep. 30:1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Monti M, et al. 2013. Developmental arrest and mouse antral not-surrounded nucleolus oocytes. Biol Reprod. 88:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma JY, et al. 2013. Maternal factors required for oocyte developmental competence in mice: transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes. Cell Cycle. 12:1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuccotti M, Piccinelli A, Giorgi Rossi P, Garagna S, Redi CA. 1995. Chromatin organization during mouse oocyte growth. Mol Reprod Dev. 41:479–485. [DOI] [PubMed] [Google Scholar]
- 46. Santo EE, et al. 2013. FOXO3a is a major target of inactivation by PI3K/AKT signaling in aggressive neuroblastoma. Cancer Res. 73:2189–2198. [DOI] [PubMed] [Google Scholar]
- 47. Liu H, et al. 2015. FOXO3a modulates WNT/beta-catenin signaling and suppresses epithelial-to-mesenchymal transition in prostate cancer cells. Cell Signal. 27:510–518. [DOI] [PubMed] [Google Scholar]
- 48. Fouquerel E, et al. 2019. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol Cell. 75:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miao Y, Cui Z, Gao Q, Rui R, Xiong B. 2020. Nicotinamide mononucleotide supplementation reverses the declining quality of maternally aged oocytes. Cell Rep. 32:107987. [DOI] [PubMed] [Google Scholar]
- 50. Barone S, et al. 2020. Chromosome missegregation in single human oocytes is related to the age and gene expression profile. Int J Mol Sci. 21:1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mishina T, et al. 2021. Single-oocyte transcriptome analysis reveals aging-associated effects influenced by life stage and calorie restriction. Aging Cell. 20:e13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang JJ, et al. 2020. Advanced maternal age alters expression of maternal effect genes that are essential for human oocyte quality. Aging (Albany NY). 12:3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang S, et al. 2020. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 180:585–600. [DOI] [PubMed] [Google Scholar]
- 54. Lv Y, et al. 2021. Single-oocyte gene expression suggests that curcumin can protect the ovarian reserve by regulating the PTEN-AKT-FOXO3a pathway. Int J Mol Sci. 22:6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Llonch S, et al. 2021. Single human oocyte transcriptome analysis reveals distinct maturation stage-dependent pathways impacted by age. Aging Cell. 20:e13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reyes JM, et al. 2017. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum Reprod. 32:2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Steuerwald NM, Bermudez MG, Wells D, Munne S, Cohen J. 2007. Maternal age-related differential global expression profiles observed in human oocytes. Reprod Biomed Online. 14:700–708. [DOI] [PubMed] [Google Scholar]
- 58. Tilly JL, Sinclair DA. 2013. Germline energetics, aging, and female infertility. Cell Metab. 17:838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rando TA, Wyss-Coray T. 2021. Asynchronous, contagious and digital aging. Nat Aging. 1:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gladyshev VN 2021. The ground zero of organismal life and aging. Trends Mol Med. 27:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Le Couteur DG, et al. 2021. Nutritional reprogramming of mouse liver proteome is dampened by metformin, resveratrol, and rapamycin. Cell Metab. 33:2367–2379. [DOI] [PubMed] [Google Scholar]
- 62. Duan X, Sun SC. 2019. Actin cytoskeleton dynamics in mammalian oocyte meiosis. Biol Reprod. 100:15–24. [DOI] [PubMed] [Google Scholar]
- 63. Pan MH, et al. 2020. FMNL3 regulates FASCIN for actin-mediated spindle migration and cytokinesis in mouse oocytesdagger. Biol Reprod. 102:1203–1212. [DOI] [PubMed] [Google Scholar]
- 64. Brunet S, Verlhac MH. 2011. Positioning to get out of meiosis: the asymmetry of division. Hum Reprod Update. 17:68–75. [DOI] [PubMed] [Google Scholar]
- 65. Zhou Z, et al. 2021. Temporal transcriptomic landscape of postnatal mouse ovaries reveals dynamic gene signatures associated with ovarian aging. Hum Mol Genet. 30:1941–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim MJ, et al. 2020. Association between functional activity of mitochondria and actin cytoskeleton instability in oocytes from advanced age mice. Reprod Sci. 27:1037–1046. [DOI] [PubMed] [Google Scholar]
- 67. Takayama K, et al. 2015. TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat Commun. 6:8219. [DOI] [PubMed] [Google Scholar]
- 68. Fernandes GFS, et al. 2017. Epigenetic regulatory mechanisms induced by resveratrol. Nutrients. 9:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen B, et al. 2022. Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature. 605:761–766. [DOI] [PubMed] [Google Scholar]
- 70. Jones PA 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13:484–492. [DOI] [PubMed] [Google Scholar]
- 71. Ferrara-Romeo I, et al. 2020. The mTOR pathway is necessary for survival of mice with short telomeres. Nat Commun. 11:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ahmed TA, et al. 2020. Oocyte aging: the role of cellular and environmental factors and impact on female fertility. Adv Exp Med Biol. 1247:109–123. [DOI] [PubMed] [Google Scholar]
- 73. May-Panloup P, et al. 2016. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update. 22:725–743. [DOI] [PubMed] [Google Scholar]
- 74. Zhu J, et al. 2016. The signaling pathways by which the Fas/FasL system accelerates oocyte aging. Aging (Albany NY). 8:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhu J, et al. 2015. Cumulus cells accelerate oocyte aging by releasing soluble Fas ligand in mice. Sci Rep. 5:8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kujjo LL, Perez GI. 2012. Ceramide and mitochondrial function in aging oocytes: joggling a new hypothesis and old players. Reproduction. 143:1–10. [DOI] [PubMed] [Google Scholar]
- 77. Perez GI, et al. 2005. A central role for ceramide in the age-related acceleration of apoptosis in the female germline. FASEB J. 19:860–862. [DOI] [PubMed] [Google Scholar]
- 78. Moreira-Pinto B, Costa L, Felgueira E, Fonseca BM, Rebelo I, 2021.Low doses of resveratrol protect human granulosa cells from induced-oxidative stress. Antioxidants (Basel). 10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Briley SM, et al. 2016. Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction. 152:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Amargant F, et al. 2020. Ovarian stiffness increases with age in the mammalian ovary and depends on collagen and hyaluronan matrices. Aging Cell. 19:e13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Umehara T, et al. 2022. Female reproductive life span is extended by targeted removal of fibrotic collagen from the mouse ovary. Sci Adv. 8:eabn4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yu D, et al. 2021. Resveratrol against cardiac fibrosis: research progress in experimental animal models. Molecules. 26:6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang J, et al. 2018. Resveratrol inhibits pulmonary fibrosis by regulating miR-21 through MAPK/AP-1 pathways. Biomed Pharmacother. 105:37–44. [DOI] [PubMed] [Google Scholar]
- 84. Azmoonfar R, et al. 2020. Mitigation of radiation-induced pneumonitis and lung fibrosis using alpha-lipoic acid and resveratrol. Antiinflamm Antiallergy Agents Med Chem. 19:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Santoro A, Bientinesi E, Monti D. 2021. Immunosenescence and inflammaging in the aging process: age-related diseases or longevity?. Ageing Res Rev. 71:101422. [DOI] [PubMed] [Google Scholar]
- 86. Kang C, et al. 2015. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 349:aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lliberos C, et al. 2021. Evaluation of inflammation and follicle depletion during ovarian ageing in mice. Sci Rep. 11:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lliberos C, Liew SH, Mansell A, Hutt KJ. 2020. The inflammasome contributes to depletion of the ovarian reserve during aging in mice. Front Cell Dev Biol. 8:628473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Altun T, Jindal S, Greenseid K, Shu J, Pal L. 2011. Low follicular fluid IL-6 levels in IVF patients are associated with increased likelihood of clinical pregnancy. J Assist Reprod Genet. 28:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hu X, et al. 2014. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 14:512–522. [DOI] [PubMed] [Google Scholar]
- 91. Mao J, Liu L. 2016. Generation of iPS cells from Granulosa cells. Methods Mol Biol. 1357:451–464. [DOI] [PubMed] [Google Scholar]
- 92. Eppig JJ, et al. 2009. Effect of in vitro maturation of mouse oocytes on the health and lifespan of adult offspring. Hum Reprod. 24:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Allworth AE, Albertini DF. 1993. Meiotic maturation in cultured bovine oocytes is accompanied by remodeling of the cumulus cell cytoskeleton. Dev Biol. 158:101–112. [DOI] [PubMed] [Google Scholar]
- 94. Lyublinskaya OG, et al. 2017. Redox environment in stem and differentiated cells: a quantitative approach. Redox Biol. 12:758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu W, et al. 2018. Putrescine delays postovulatory aging of mouse oocytes by upregulating PDK4 expression and improving mitochondrial activity. Aging (Albany NY). 10:4093–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu L, Trimarchi JR, Keefe DL. 2000. Involvement of mitochondria in oxidative stress-induced cell death in mouse zygotes. Biol Reprod. 62:1745–1753. [DOI] [PubMed] [Google Scholar]
- 97. Bogliolo S, Nadalini C, Iacobone AD, Musacchi V, Carus AP. 2013. Vaginal cuff closure with absorbable bidirectional barbed suture during total laparoscopic hysterectomy. Eur J Obstet Gynecol Reprod Biol. 170:219–221. [DOI] [PubMed] [Google Scholar]
- 98. Choi WJ, et al. 2007. Oxidative stress and tumor necrosis factor-alpha-induced alterations in metaphase II mouse oocyte spindle structure. Fertil Steril. 88:1220–1231. [DOI] [PubMed] [Google Scholar]
- 99. Di Emidio G, et al. 2014. SIRT1 signalling protects mouse oocytes against oxidative stress and is deregulated during aging. Hum Reprod. 29:2006–2017. [DOI] [PubMed] [Google Scholar]
- 100. Li L, et al. 2020. Characterization of metabolic patterns in mouse oocytes during meiotic maturation. Mol Cell. 80:525–540.e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Takahashi T, Takahashi E, Igarashi H, Tezuka N, Kurachi H. 2003. Impact of oxidative stress in aged mouse oocytes on calcium oscillations at fertilization. Mol Reprod Dev. 66:143–152. [DOI] [PubMed] [Google Scholar]
- 102. Zeng J, et al. 2018. SIRT4 is essential for metabolic control and meiotic structure during mouse oocyte maturation. Aging Cell. 17:e12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wang F, et al. 2013. Robust measurement of telomere length in single cells. Proc Natl Acad Sci USA. 110:E1906–E1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Picelli S, et al. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 9:171–181. [DOI] [PubMed] [Google Scholar]
- 105. Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Smallwood SA, et al. 2014. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 11:817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang L, et al. 2014. Programming and inheritance of parental DNA methylomes in mammals. Cell. 157:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or Supplementary Material. RNA-sequencing and methylation-sequencing data were deposited in the Gene Expression Omnibus (GEO) database (accession number GSE184637). The authors declare no competing interests.