Abstract
The cell wall envelope of gram-positive bacteria is a macromolecular, exoskeletal organelle that is assembled and turned over at designated sites. The cell wall also functions as a surface organelle that allows gram-positive pathogens to interact with their environment, in particular the tissues of the infected host. All of these functions require that surface proteins and enzymes be properly targeted to the cell wall envelope. Two basic mechanisms, cell wall sorting and targeting, have been identified. Cell well sorting is the covalent attachment of surface proteins to the peptidoglycan via a C-terminal sorting signal that contains a consensus LPXTG sequence. More than 100 proteins that possess cell wall-sorting signals, including the M proteins of Streptococcus pyogenes, protein A of Staphylococcus aureus, and several internalins of Listeria monocytogenes, have been identified. Cell wall targeting involves the noncovalent attachment of proteins to the cell surface via specialized binding domains. Several of these wall-binding domains appear to interact with secondary wall polymers that are associated with the peptidoglycan, for example teichoic acids and polysaccharides. Proteins that are targeted to the cell surface include muralytic enzymes such as autolysins, lysostaphin, and phage lytic enzymes. Other examples for targeted proteins are the surface S-layer proteins of bacilli and clostridia, as well as virulence factors required for the pathogenesis of L. monocytogenes (internalin B) and Streptococcus pneumoniae (PspA) infections. In this review we describe the mechanisms for both sorting and targeting of proteins to the envelope of gram-positive bacteria and review the functions of known surface proteins.
The cell wall of gram-positive bacteria is host to a wide variety of molecules and serves a multitude of functions, most of which are critical to the viability of the cell. Although the primary function of the cell wall is to provide a rigid exoskeleton for protection against both mechanical and osmotic lysis (694, 695) the cell wall of gram-positive bacteria also serves as an attachment site for proteins that interact with the bacterial environment. Over the past decade, it has become apparent that the gram-positive bacteria have evolved a number of unique mechanisms by which they can immobilize proteins on their surface. These mechanisms involve either the covalent attachment of protein to the peptidoglycan or the noncovalent binding of protein to either the peptidoglycan or secondary wall polymers such as teichoic acids.
This review describes our current knowledge about surface proteins of gram-positive bacteria and the mechanisms of their anchoring to the cell wall. Functions performed by the various wall proteins are incredibly diverse. For example, many covalently linked surface proteins of gram-positive pathogens are thought to be important for survival within an infected host (713). Other wall-targeted proteins are responsible for the controlled synthesis and turnover of the peptidoglycan at specific sites (division septa) during cell growth and division (348). It is believed that these enzymes are targeted to the division sites through a noncovalent interaction with specifically localized septal receptors. Still other surface proteins of gram-positive bacteria, including the internalin B molecule of Listeria spp., lysostaphin, and S-layer proteins, are immobilized to the cell surface by binding to secondary wall polymers present throughout the cell wall.
To facilitate this discussion of the mechanisms of protein attachment, we briefly discuss the mechanisms of protein secretion in these bacteria. We also summarize what is known about the structure, assembly, and turnover of the cell wall of gram-positive bacteria. For more detailed treatises on these subjects, we refer the reader to other excellent reviews (252, 707).
ARCHITECTURE OF GRAM-POSITIVE BACTERIA
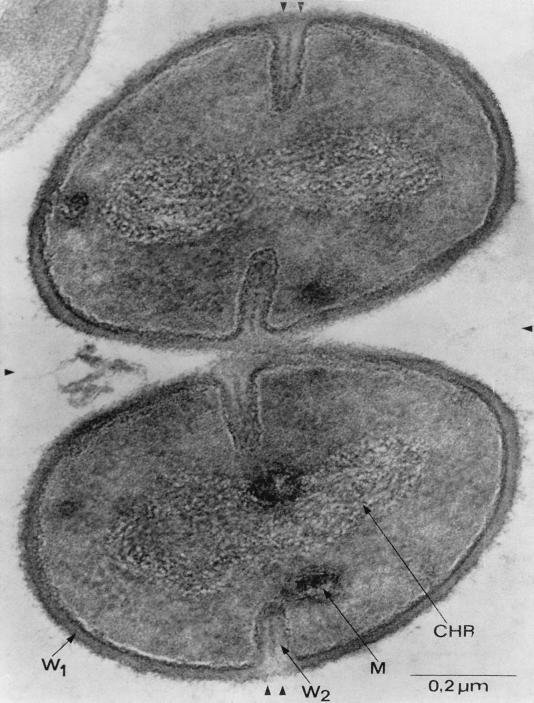
Gram-positive bacteria are simple cells. On the basis of morphological criteria three distinct cellular compartments can be distinguished: the cytosol, a single cytoplasmic membrane, and the surrounding cell wall (261). Some gram-positive bacteria synthesize a large polysaccharide capsule, whereas others elaborate a crystalline layer of surface proteins (739); both structures may envelope the entire cell. Spore-forming gram-positive bacteria, such as Bacillus subtilis, generate morphologically distinct daughter cells by a developmental program of asymmetric cell division (501). The cell wall of spores differs from that of mother cells and contains specific sets of proteins (645, 852). Some gram-positive bacteria divide without separating their cell walls and thus continue to grow as strings of cells (streptococci) or as clusters (staphylococci). Figure 1 is a transmission electron micrograph of thin-sectioned Staphylococcus aureus cells, revealing the characteristic morphology of the subcellular compartments of gram-positive bacteria.
FIG. 1.
Transmission electron micrograph of S. aureus cells undergoing cell division. Completed division septa are indicated by a single arrowhead, and new division sites are marked by double arrowheads. Newly formed cell wall (W2) is distinguished from preexisting cell wall (W1). Also visible are the bacterial chromosome (Chr) and cytoplasmic membranous bodies (M). This electron micrograph was generated by P. Giesbrecht. Reprinted from reference 260 with permission of the publisher.
SYNTHESIS AND STRUCTURE OF THE CELL WALL OF GRAM-POSITIVE BACTERIA
The cell wall of gram-positive bacteria is a peptidoglycan macromolecule with attached accessory molecules such as teichoic acids, teichuronic acids, polyphosphates, or carbohydrates (302, 694). The glycan strands of the cell wall consist of the repeating disaccharide N-acetylmuramic acid-(β1-4)-N-acetylglucosamine (MurNAc-GlcNAc) (254, 255). Glycan strands vary in length and are estimated to contain 5 to 30 subunits, depending on the bacterial species investigated (270, 331, 749). In most cases, the d-lactyl moiety of each MurNAc is amide linked to the short peptide component of peptidoglycan (256, 564, 792). Wall peptides are cross-linked with other peptides that are attached to a neighboring glycan strand (787–789, 791), thereby generating a three-dimensional molecular network that surrounds the cell and provides the desired exoskeletal function (463, 760) (Fig. 2).
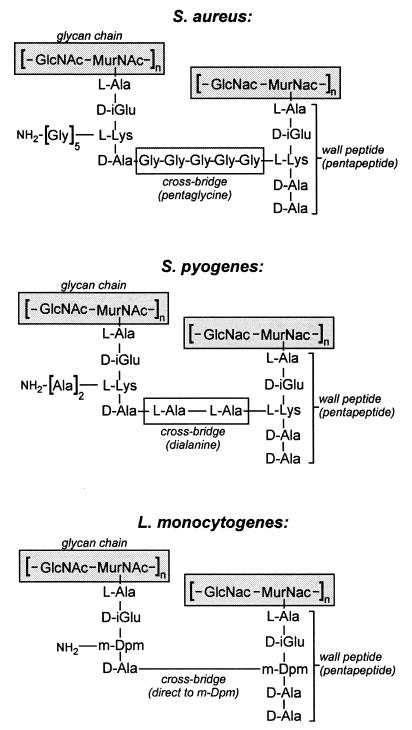
FIG. 2.
Diagram of peptidoglycan structures from S. aureus, S. pyogenes, and L. monocytogenes. Glycan chains composed of a repeating disaccharide, GlcNAc and MurNAc, are linked to short wall peptides through the lactyl moeity of MurNAc. Adjacent wall peptides can be linked through crossbridge peptides (pentaglycine in S. aureus or dialanine in S. pyogenes). In some cases, such as L. monocytogenes, adjacent wall peptides are not cross-linked via peptide crossbridges. Instead, wall peptides are linked via an amide bond between the ɛ-amino group of a meso-diaminopimelic acid (m-Dpm) residue at position 3 of one wall peptide and the carboxy terminus of the d-alanyl residue at position 4 of the adjacent wall peptide.
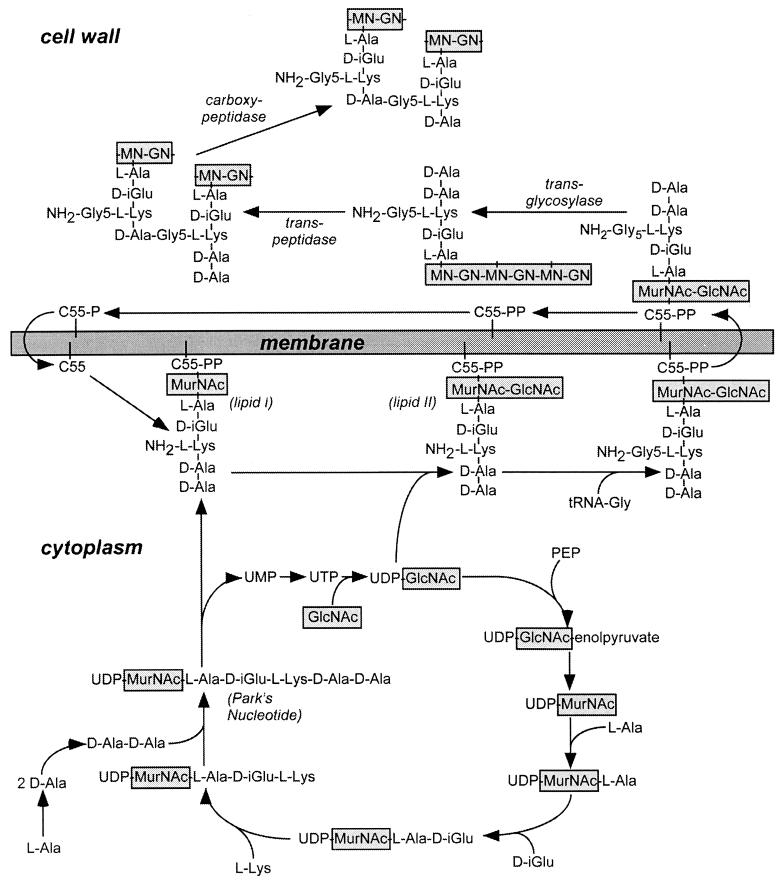
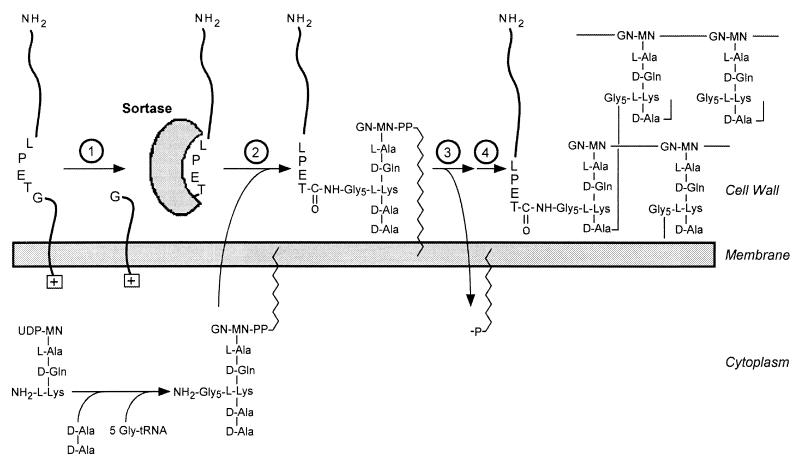
Cell wall synthesis in gram-positive bacteria can be divided into three separate stages that occur in distinct subcellular compartments, the cytoplasm, the membrane, and, finally, the cell wall itself (759, 761) (Fig. 3). Although we discuss here peptidoglycan synthesis in S. aureus, all bacteria use a nearly identical synthesis pathway. Peptidoglycan synthesis begins by generating UDP-MurNAc from UDP-GlcNAc and phosphoenolpyruvate (287, 758). Five amino acids are linked to UDP-MurNAc in four consecutive steps, l-Ala, d-isoGlu, l-Lys, and, finally, the d-Ala–d-Ala dipeptide (370, 554, 572). Synthesis of d-Ala–d-Ala requires two enzymes. The first, d-Ala racemase, converts l-Ala to d-Ala, while the second, d-Ala ligase, creates a peptide bond between two d-Ala residues (113, 578, 823). Synthesis of each peptide (amide) bond within the wall peptide consumes 1 high-energy phosphate (ATP) (812). The end product of the cytoplasmic segment of the cell wall synthesis pathway, UDP-MurNAc–l-Ala–d-isoGlu–l-Lys–d-Ala–d-Ala (UDP-MurNAc-pentapeptide, Park’s nucleotide), has been solubilized by acid extraction of gram-positive cells and purified (616, 617). Radiolabeled Park’s nucleotide was used as substrate for the in vitro synthesis of peptidoglycan, which provided a powerful assay for the elucidation of this biochemical pathway (116). UDP-MurNAc-pentapeptide is phosphodiester linked to an undecaprenyl-pyrophosphate carrier molecule at the expense of UDP (C55–PP-MurNAc–l-Ala–d-isoGlu–l-Lys–d-Ala–d-Ala, or lipid I) (13, 371). UDP-GlcNAc is linked to the muramoyl moiety to generate the disaccharide lipid II precursor [C55–PP-MurNAc(-l-Ala–d-isoGlu–l-Lys(Gly5)–d-Ala–d-Ala)-β1-4-GlcNAc] (335–337). Lipid II is further modified by the addition of amino acids to the ɛ-amino of lysine (12, 417). Figure 3 shows these reactions for the biosynthesis of staphylococcal peptidoglycans, which contain five glycine residues linked to l-Lys. Three glycyl-tRNA species are thought to be dedicated to this biosynthetic pathway (281, 672, 673, 755). Finally, the modified lipid II precursor is translocated across the cytoplasmic membrane and serves as the substrate for the assembly of peptidoglycan (Fig. 3).
FIG. 3.
Diagram of the biosynthetic pathway of cell wall assembly. As discussed in the text, the generation of cell wall precursors begins in the cytoplasm, resulting in the synthesis of UDP-MurNAc–l-Ala–d-iGlu–l-Lys–d-Ala–d-Ala (Park’s nucleotide). The peptidoglycan precursor subunit is transferred to a lipid carrier in the membrane to generate lipid I. After further modification, the lipid-anchored peptidoglycan precursor is translocated to the extracellular face of the cytoplasmic membrane. The peptidoglycan precursor is subsequently incorporated into the cell wall by transpeptidation and transglycosylation reactions with the concomitant displacement of the lipid carrier. The terminal d-alanine of the wall pentapeptide is subject to substitution by cross-linking to other wall subunits or can be removed by the action of a d-alanyl–d-alanine carboxypeptidase. PEP, phosphoenolpyruvate; MN-GN, MurNAc-GlcNAc.
Cell wall assembly is catalyzed by penicillin binding proteins (PBPs) (251). The high-molecular-weight PBPs are bifunctional enzymes that have recently been categorized into one of two classes based on sequence similarity (251, 272). Class A PBPs promote both the polymerization of glycan from its disaccharide precursor, i.e., the successive addition of MurNAc(-l-Ala–d-isoGlu–l-Lys–d-Ala–d-Ala)-GlcNAc to C55–PP-MurNAc(-l-Ala–d-isoGlu–l-Lys–d-Ala–d-Ala)-GlcNAc, and the transpeptidation (cross-linking) of wall peptides (570). The latter reaction results in the proteolytic removal of the d-Ala at the C-terminal end of the pentapeptide and the formation of a new amide bond between the amino group of the crossbridge and the carbonyl group of d-Ala at position 4 (791). The transpeptidation reaction of staphylococcal peptidoglycan is depicted in Fig. 3. This reaction is the target of penicillin and other β-lactam antibiotics which mimic the structure of d-alanyl–d-alanine (791). After cleavage, the β-lactam ring continues to occupy the active site serine residue of PBPs, thereby inhibiting PBPs (871, 872). Less is known about the class B PBPs; however, they are speculated to be involved in morphogenetic networks (272).
Not all cell wall peptides are cross-linked to their neighboring peptidoglycan strands. Unsubstituted (free) peptides may be preserved by trimming the terminal d-Ala off the pentapeptides (839). This reaction is catalyzed by other, low-molecular-weight PBPs that function as carboxypeptidases (372, 805). The reactions carried out by transpeptidases and carboxypeptidases are similar in nature. Transpeptidases use an amino group as a nucleophile to resolve the acyl-enzyme intermediate between the active-site hydroxyl of serine and the carbonyl of d-alanine, whereas carboxypeptidases use water as a nucleophile (251). Consequently, there are sequence and structural similarities between these enzymes (251). The ratio between transpeptidation and carboxypeptidation is thought to be important in generating a three-dimensional structure of the cell wall (463). For example, different degrees of cross-linking could allow wall synthesis at distinct angles and the generation of curves in otherwise cylindrical cells. However, this assumption has never been tested for the distinctly shaped gram-positive bacteria. Peptidoglycan with a low degree of cross-linking is much more sensitive to degradation by cell wall hydrolases (760). It is conceivable that the degree of cross-linking plays a role in specifying sites on the cell wall that are more prone to either degradation or additional synthesis.
While the repeating disaccharide [MurNAc-(β1-4)-GlcNAc] is found in all bacterial peptidoglycans, the wall peptides differ in composition between bacterial species (710). Some organisms, such as Listeria, replace l-Lys with another diamino acid, in this case m-diaminopimelic acid (710). Others add amino acids to the side chain ɛ-amino of l-Lys to synthesize extended peptidoglycan cross-bridges (710). Figure 2 provides an example for the peptidoglycan structures of three different gram-positive bacteria. The wall peptides can either be cross-linked with those of another glycan strand such that the ɛ-amino of l-Lys or any other amino acid added at this position will be amide linked to the carbonyl of d-Ala in the wall peptide or be trimmed to generate an un-cross-linked wall tetrapeptide (710).
The cell wall of many gram-positive bacteria is modified by O-acetylation of MurNAc at C-6 (790). Although the enzymatic mechanism for this decoration remains unknown, it does confer resistance to cell wall degradation by animal lysozymes (97). Other chemical modifications include additions of teichoic acids, phosphorylation, and attachment of carbohydrates (see below). Recent advances in cell wall structure have been made by analyzing peptidoglycan breakdown products by reverse-phase high-pressure liquid chromatography combined with mass spectrometric analysis (144, 244, 269, 801). These techniques allow the characterization of different degrees of cross-linking as well as the identification of new cross-linking reactions within the cell wall (145).
CELL WALL-ASSOCIATED STRUCTURES OF GRAM-POSITIVE BACTERIA
Gram-positive bacteria synthesize several compounds that decorate their peptidoglycan exoskeleton (694). Based on structural differences, one can distinguish teichoic acids, teichuronic acids, lipoteichoic acids, lipoglycans, and polysaccharide modifications (694). Over the past 30 years, the complete structures of some of these compounds as well as the genes required for their synthesis have been characterized. This research has been reviewed in detail elsewhere, and we provide here a brief summary of these elements, since they may be important for protein targeting mechanisms (17, 205, 643).
Teichoic Acids
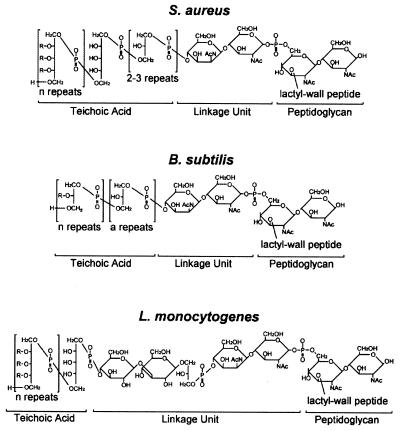
All gram-positive bacteria are thought to synthesize anionic polymers that are covalently attached to the peptidoglycan or tethered to a lipid anchor moiety (18, 23). Typically these polymers consists of polyglycerol phosphate (Gro-P), poly-ribitol phosphate (Rit-P), or poly-glucosyl phosphate (Glc-P) all of which may be glucosylated and/or amino acid esterified (205, 643). Figure 4 compares the structure of staphylococcal cell wall teichoic acid with that of two other bacteria. A polymer of 30 to 50 Rit-P subunits is phosphodiester linked to 2 or 3 Gro-P residues which are linked to the disaccharide ManNac(β1-4)GlcNAc (16, 183, 184, 700, 701). The GlcNAc moiety is (1–6) phosphodiester linked to MurNAc within the peptidoglycan repeating disaccharide (MurNAc-GluNac) (128, 318, 438). The (Gro-P)n-ManNac-GlcNAc tether between wall teichoic acids and the peptidoglycan is referred to as the linkage unit (15). Although many gram-positive bacteria are known to synthesize identical linkage units, some species modify its structure with other carbohydrates whereas a few others use entirely different compounds to tether their wall teichoic acids (15).
FIG. 4.
Structure of cell wall teichoic acids from S. aureus, B. subtilis, and L. monocytogenes. Wall teichoic acids are anionic polymers of glycerol-phosphate, ribitol-phosphate, or glucosyl-phosphate and are tethered to the peptidoglycan. The cell wall linkage unit consists of ManNac(β1-4)GlcNAc, which is 1–6 1-6phosphodiester linked to MurNAc within the glycan strands of the cell wall envelope. One to three glycerol-phosphate moieties serve as a bridge between the linkage unit and the anionic polymer, which is composed of 15 to 60 subunits. R indicates covalent modification of the hydroxyl side chains of either glycerol-phosphate or ribitol-phosphate and can be either GlcNAc or esterified d-alanine.
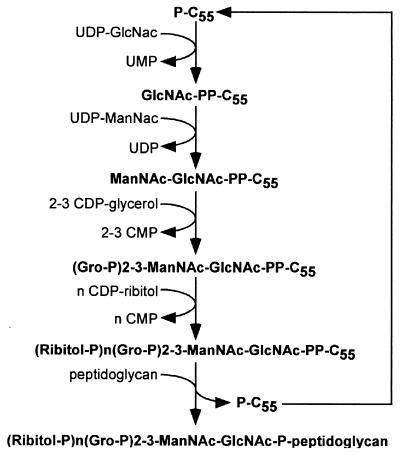
Modifications of the repeating subunits differ from one bacterial species to another (17). Both Gro-P and Rit-P repeating units can be decorated with a variety of different sugars and can also be esterified with d-alanine (78–80). Synthesis of the backbone structure is thought to occur in the bacterial cytoplasm (78, 643). Synthesis begins by fusing UDP-GlcNAc to prenol-phosphate (862, 863) (Fig. 5). The resulting GlcNAc-PP-prenol is then mannosylated with UDP-ManNac to yield ManNac-GlcNAc-PP-prenol (873). The lipid-anchored cell wall linkage unit serves as an attachment site for the addition of two or three Gro-P units and is extended by Rib-P, which is added from CDP-ribitol substrates (267, 268, 521, 642). The end product of this synthetic pathway is presumably translocated across the cytoplasmic membrane via an ATP binding cassette transporter system (472). Attachment of wall teichoic acid to MurNAc proceeds during cell wall assembly; however, the enzymatic machinery required for this process is still unknown.
FIG. 5.
Diagram of the biosynthesis of staphylococcal cell wall teichoic acid. Synthesis begins in the cytoplasm via the addition of UDP-GlcNAc to an undecaprenol carrier molecule (P-C55). UDP-ManNac and 2,3-CDP-glycerol are added to complete the cell wall linkage unit, which is then extended by the polymerization of ribitol-phosphate. This precursor molecule is thought to be translocated across the cytoplasmic membrane and linked to MurNAc within the glycan chains.
Esterification of Gro-P or Rit-P with d-Ala occurs on the surface of the cytoplasmic membrane, i.e., after teichoic acids have been translocated across the membrane (580). Four gene products are required for this process, during which d-Ala is first linked to a diacyl carrier protein and then linked to an undecaprenol phosphate lipid carrier (139, 316, 317, 580, 592, 641). Lipid-linked d-Ala is translocated across the cytoplasmic membrane and serves as a substrate for esterification. This reaction is presumably used for the d-Ala ester modification of cell wall teichoic acids and lipoteichoic acids (579). Cell wall teichoic acids are thought to be uniformly distributed over the entire peptidoglycan exoskeleton (806). Some teichoic acid decorations, for example the esterified d-Ala, are unstable, and S. aureus continuously reesterifies d-Ala residues (431, 433).
Although the structures of cell wall teichoic acids are largely known and some of the genes involved in their synthesis appear to be essential for the growth of gram-positive bacteria, the physiological role of these molecules is still not completely understood (642). It is conceivable that the negatively charged teichoic acids function to capture divalent cations or provide a biophysical barrier to prevent the diffusion of substances (205, 207). However, these claims have been largely speculative and experiments that directly prove or disprove them are difficult to design. Cell wall teichoic acids appear to be the binding sites for some enzymes that cleave the bacterial peptidoglycan (333). For example, the LytA amidase of S. pneumoniae binds to the choline moiety of the cell wall teichoic acids of this organism (351, 352). Conceivably, the affinity for teichoic acids directs murein hydrolases to the cell walls of specific species (discussed below). Thus, teichoic acids may serve as species-specific decorations which allow gram-positive bacteria to synthesize an envelope structure that is chemically distinct from the envelope other organisms that display an otherwise identical peptidoglycan exoskeleton.
Lipoteichoic Acids
Lipoteichoic acids are polyanionic polymers inserted in the outer leaflet of the cytoplasmic membrane via a lipid moiety (205). The polymer extends through the cell wall peptidoglycan onto the surface of gram-positive cells. The precise function of lipoteichoic acids is unknown (205). The cell wall teichoic acids and lipoteichoic acids possess different chemical structures in all gram-positive bacteria except Streptococcus pneumoniae. For example, S. aureus synthesizes poly-Rit-P cell wall teichoic acids, which are modified at the 2′ and 4′ hydroxyl with GlcNAc and d-Ala (701). Staphylococcal lipoteichoic acids are composed of poly-Gro-P, which can be modified at the 2′ hydroxyl of Gro-P with either GlcNAc or esterified d-Ala (165, 208–210). The lipoteichoic acid moiety of S. aureus is attached to Glc(1-4)Glc-(1-3)diacylglycerol (433).
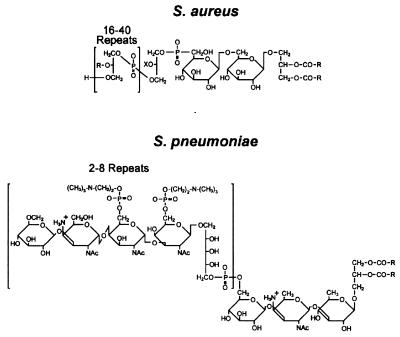
Lipoteichoic acids of other bacteria have different repeating subunits and/or different lipid anchors. Figure 6 compares the structure of staphylococcal lipoteichoic acid with that from S. pneumoniae (205). Lipoglycans are related structures that can be distinguished from lipoteichoic acids by the absence of phosphate from the repeating subunits (205). Synthesis of the lipoteichoic acids is thought to occur on the surface of the cytoplasmic membrane. Thus, the substrates of each lipoteichoic acid constituent must be translocated across the cytoplasmic membrane prior to assembly (205). Indeed, each of the precursor molecules is tethered to a lipophilic moiety: either glycolipid, phosphatidylglycerol, hexosyl-1-phosphoundecaprenol, or undecaprenyl-d-Ala (106, 241) (Fig. 7).
FIG. 6.
Structure of lipoteichoic acid from S. aureus and S. pneumoniae. Staphylococcal lipoteichoic acid is composed of polymerized glycerol-phosphate and is linked to a membrane anchor moiety [Glc(β1–4)Glc(β1–3)diacylglycerol]. The fatty acid side chains of diacylglycerol are indicated by R. The hydroxyl side chains of polymerized glycerolphosphate are modified with GlcNAc or esterified d-alanine. In S. pneumoniae, the repeat unit of lipoteichoic acid, [Glc(β1–3)AATGal(α14)GalNac(6-Cho-P)(α1–3)GalNac(6-Cho-P)(β1-1)ribitol-P], is identical to that of cell wall teichoic acids. The choline-modified repeat unit serves as a targeting receptor for several murein hydrolases and surface protein. Modified from reference 205 with permission of the publisher.
FIG. 7.
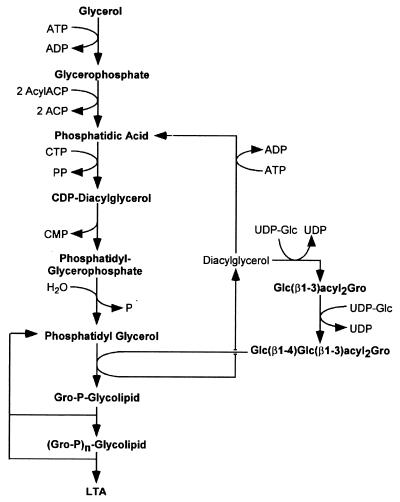
Diagram of the biosynthesis of staphylococcal lipoteichoic acid (LTA). Glycerol-phosphate is acylated with two fatty acids to yield phosphatidic acid. This compound, as well as all other biosynthetic components of lipoteichoic acid, are thought to be located in the outer leaftlet of the cytoplasmic membrane, the presumed site of lipoteichoic acid synthesis. The glycolipid anchor, Glc (β1–4)Glc(β1–3)diacylglycerol, is linked to the first glycerol-phosphate moiety and then further extended by polymerization with phosphatidyl-glycerol as a substrate. Modified from reference 205 with permission of the publisher.
Lipoteichoic acid synthesis begins in the cytoplasm with the synthesis of phosphatidic acid from glycerol, ATP, and two acyl side chains (Fig. 7). Phosphatidic acid is converted to phosphatidyl-glycerophosphate and then to phosphatidylglycerol, which serves as a substrate for the polymerization of Gro-P (266). This synthetic scheme requires large amounts of diacylglycerol, which are recycled to phospatidic acid or used for the biosynthesis of other membrane lipids (433). Glycosylation of lipoteichoic acid requires hexose linked to undecaprenyl, which is presumably synthesized in the cytoplasm and translocated across the membrane (206, 432, 482). It is conceivable that lipoteichoic acids, similarly to the cell wall teichoic acids of S. pneumoniae, also serve as species-specific decorations of the peptidoglycan exoskeleton.
PROTEIN SECRETION IN GRAM-POSITIVE BACTERIA
Protein secretion has been studied extensively in Escherichia coli, and the paradigms established through this work are thought to be true for all bacterial and eukaryotic cells (171, 640, 654). Proteins destined for translocation across the cytoplasmic membrane are marked by a signal (leader) peptide (65) that is generally composed of a core of 15 to 20 hydrophobic residues flanked at the N-terminal end by positively charged residues (182, 728). For secreted proteins, signal peptides are proteolytically removed by signal (leader) peptidases upon translocation across the cytoplasmic membrane (136, 153). Signal peptides are necessary and sufficient for protein translocation across membranes if the fused polypeptide substrate can be maintained in an export competent state, a function that can be achieved by one of two separate pathways (142, 808). In one pathway, a signal recognition particle (SRP) can bind to the signal peptides of nascent chains and temporally arrest their ribosomal translation (824–826). The SRP of E. coli is thought to be a ribonucleoprotein particle consisting of Ffh (also known as P48) and 4.5S RNA (46, 648, 678, 762). Translation resumes after the SRP-ribosome complex docks onto its membrane receptor (FtsY), thereby delivering the nascent polypeptide to the Sec translocation channel (551, 808). Alternatively, signal peptide-bearing precursors may be translocated after their synthesis has been completed, i.e., by a posttranslational translocation process. Binding of a secretion chaperone, SecB, can maintain these precursors in an unfolded, translocation-competent state (452, 661). The SecB protein binds to the mature part of signal peptide-bearing precursors but may also interact with the SecA protein, a component of the secretion machinery itself (90, 151, 341, 662). Once initiated into the secretory pathway by the signal peptide, the SecB chaperone dissociates from the precursor, allowing the translocation of the full-length polypeptide across the membrane (197). The pathways converge at the translocation channel, and the choice of pathway that is used may be a function of the hydrophobicity of the signal peptide (807, 808). Recent observations suggest that the SRP pathway may target proteins destined for the inner membrane to the Sec translocase whereas the SecB pathway seems to be used by proteins that are secreted across the membrane (721, 804).
The gene products necessary for the initiation of leader peptide-containing precursors into the secretory pathway were identified as mutants that restored the β-galactosidase activity of LacZ fusions to the C terminus of signal peptide bearing proteins (600). These LacZ fusion proteins are substrates for export but arrest at an undefined step of the secretion pathway and block the export of other precursors. The results of the sec screens generally matched those of a suppressor screen (prl, for “protein localization”) that scored for the export of a mutant polypeptide harboring a defective signal peptide (181). The functionality of the Sec proteins in membrane translocation of precursor proteins has been elegantly demonstrated in vitro (309, 563). SecYEG is the preprotein translocase channel and requires the SecA ATPase to push polypeptides through a hydrophilic channel (172, 174, 179, 811). The SecDF and YajC proteins function at a later step, perhaps in regulating the activity of SecA (173).
Homologs of SecA, SecD, SecE, SecF, SecY, and YajC have been identified in the sequenced genome of the gram-positive organism B. subtilis; however, no homologs of SecB and SecG were found (454). It is not clear whether SecB and SecG are dispensable for secretion in B. subtilis or whether they have been replaced with other polypeptides that do not display homology to the E. coli gene products. Ffh, 4.5S RNA, and FtsY are conserved between E. coli and B. subtilis (102, 456, 774). Another striking difference between E. coli and B. subtilis is the number of signal peptidases. The gram-positive organism appears to encode seven signal peptidases, whereas E. coli contains only two. Two of the Bacillus genes specify class 2 signal peptidases for the maturation of lipoproteins, whereas the other five signal peptidases are similar to the E. coli lepB gene. Signal peptides of gram-positive bacteria differ from those of their gram-negative counterparts by usually being longer and more hydrophobic and possessing more charge in their N-terminal ends (818, 819). Although these signal peptides of gram-positive organisms generally function in gram-negative bacteria, the same is not always true when signal peptides of gram-negative bacteria are expressed in gram-positive organisms (716). Could it be that gram-positive bacteria require additional components to recognize signal peptides? This has been tested biochemically for both B. subtilis and S. aureus by purifying membranes with bound ribosomes and comparing their protein content with the content of membranes those that did not contain ribosomes (2, 3). Although several different polypeptides could be identified as a secretory (S) complex, analysis of the cloned sequences revealed pyruvate dehydrogenase, an enzyme of intermediary metabolism that is presumably not directly involved in protein secretion (329, 330).
A “PERIPLASMIC SPACE” IN GRAM-POSITIVE BACTERIA?
The existence of a periplasmic space in gram-positive bacteria that is analogous to the one found in gram-negative bacteria has been a matter of speculation for many years (276). Morphological inspection has at times revealed a narrow space between the cytoplasmic membrane and the cell wall peptidoglycan (276). Visualization of this space depended on the mode of sample preparation (275). Others have emphasized an operational definition for the periplasm of gram-positive bacteria as a subcellular compartment (544); i.e., it should contain a subset of secreted proteins that are specifically targeted to this compartment (644).
Several proteins that are periplasmic in gram-negative bacteria were observed to be lipid modified in gram-positive bacteria (264). These lipoproteins function to capture specific import substrates, such as carbohydrates, and deliver them to transport machinery embedded within the cytoplasmic membrane. The homologous proteins in gram-negative cells are generally soluble in a periplasmic space that is bounded by both the bacterial inner and outer membranes. Hence, the lipoyl moiety might be a targeting device to retain polypeptides on the membrane surface of gram-positive bacteria (264, 769, 770). For example, β-lactamase (Bla) is a soluble periplasmic enzyme that confers resistance to β-lactam antibiotics in gram-negative bacteria (444) while the β-lactamases of S. aureus and B. licheniformis are lipoproteins (584, 585). β-Lactamase precursors in gram-positive bacteria appear to be substrates for both type I and type II signal peptidase, resulting in mixed populations of secreted, soluble Bla as well as glyceride-modified, membrane-anchored Bla (573). Mutant Bla exported solely by a type I signal peptide is secreted into the extracellular medium but fails to protect staphylococci from β-lactam antibiotics (573). Thus, tethering of such enzymes to the cytoplasmic membranes is an important factor in both lowering the local concentration of inhibitors such as penicillin and raising the local concentration of necessary nutritional resources.
HISTORY OF SURFACE PROTEIN ANCHORING IN GRAM-POSITIVE BACTERIA
The high incidence of streptococcal and staphylococcal human diseases at the beginning of this century sparked the interest of many medical investigators in these microbes. Initial efforts concentrated on the characterization of the predominant antigens during infection by gram-positive bacteria with the rationale of developing protective vaccines (285, 466). Although vaccines for Streptococcus pyogenes and Staphylococcus aureus are still not commercially available, this research rapidly identified and characterized protein and carbohydrate structures on bacterial surfaces. Lancefield and colleagues classified streptococci on the basis of carbohydrate antigens. The group A streptococcus (GAS), S. pyogenes, is the causative agent of pharyngitis, purulent skin lesions, and postinfectious sequelae. GAS strains display M protein on their surface and can be typed with antisera raised against the N-terminal portion of this α-helical coiled-coil molecule (467). More than 100 types are known, and it was recognized early that these type-specific antibodies conferred immunity to infection of strains carrying one specific M type by promoting the phagocytic killing of GAS (461, 467). Hence, much effort was directed at characterizing the M protein molecule as an antiphagocytic antigen (212).
In 1958, Jensen observed that several S. aureus strains could precipitate immunoglobulins (Ig) from both human and preimmune animal serum in gel precipitation assays (380). The nonimmune precipitation of antibodies is due to the binding of protein A on the staphylococcal surface to the Fc portion of Ig (222). However, antibodies directed specifically at protein A are not protective during animal infections, and strains deficient in protein A do not display a significant reduction in their pathogenic potential in an animal experimental system (403). Although we do not know the precise role of protein A during S. aureus infections, this molecule has served as a model system for immunological, structural, and microbiological studies (588).
It is widely assumed that surface proteins of Gram-positive bacteria might interact with eukaryotic proteins as a means of establishing residence at unique locations or evading the immune system. Although these assumptions have not always been rigorously tested, they have increased research interest in this field. The advent of molecular cloning techniques yielded a rapid accumulation of DNA sequence and allowed structural comparison of surface proteins. The current completion of several microbial genome sequences will soon provide us with a comprehensive view of all sequences. Therefore, we assume that future work will concentrate on the physiological and biochemical characterization of surface proteins in gram-positive bacteria.
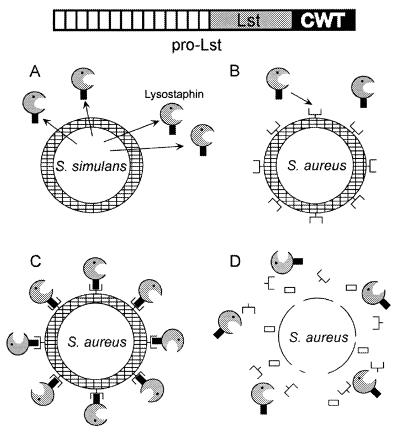
Initial experiments to characterize surface proteins focused on solubilizing and purifying streptococcal M protein and staphylococcal protein A from the bacteria. Lancefield used acid extraction of streptococci to release M proteins from the cell surface (467). This treatment caused partial hydrolysis of polypeptides but not of the bacterial cell wall, and released peptide fragments were recovered in the supernatant after centrifugation of acid extracts. Treatment with detergents, bases, or proteases or by boiling also released some M protein; however, all of these preparations yielded either only peptide fragments or minute amounts of material, indicating that the solubilization technique was insufficient (212). This problem was overcome by the use of cell wall-hydrolytic enzymes, which enabled the efficient solubilization of full-length surface proteins (212, 340). The biochemical analysis of protein A was facilitated by its ability to be purified by affinity chromatography on matrices containing linked immunoglobulins after its enzymatic solubilization from the staphylococcal cell wall (340). When purified S. aureus cell walls were digested with lysostaphin, a bacteriolytic enzyme secreted by Staphylococcus simulans bv. staphylolyticus, protein A was released as a homogeneous population (737). Egg white lysozyme, an N-acetylmuramidase, does not effectively degrade the staphylococcal cell wall. Nevertheless, Sjöquist et al. were able to purify small amounts of lysozyme-released protein A and to show that it migrated more slowly on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) than did the lysostaphin-released counterpart, suggesting an increase in molecular mass (738). Acid hydrolysis of the lysozyme-released species yielded the amino sugars GlcNAc and MurNAc, which were not observed for lysostaphin-released protein A (738). On the basis of these observations, Sjöquist et al. concluded that protein A must be linked to the staphylococcal cell wall (738).
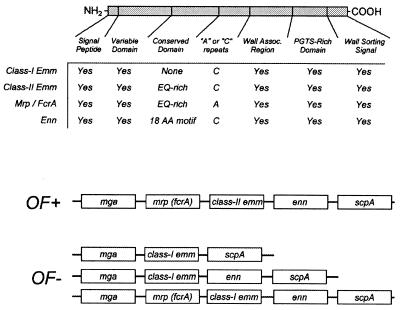
Cloning and sequencing of the emm (streptococcal M protein) and spa (staphylococcal protein A) genes revealed open reading frames that specified polypeptides harboring an N-terminal signal peptide and a C-terminal hydrophobic domain (346, 803). Because hydrophobicity is a universal signal for membrane anchoring of polypeptides (138), it seemed plausible that surface proteins could be tethered to the cytoplasmic membrane of gram-positive bacteria rather than linked to the cell wall. Comparison of several additional surface protein gene sequences allowed a more detailed analysis of their primary structures and revealed a striking C-terminal homology, namely, the presence of an LPXTG sequence motif, where X is any amino acid, followed by a C-terminal hydrophobic domain and a tail of mostly positively charged residues (216). The remarkable conservation of the LPXTG motif in surface proteins of gram-positive bacteria suggested that this element must be involved in the anchor mechanism of surface proteins in gram-positive bacteria.
To characterize the C-terminal part of M proteins, Pancholi and Fischetti digested the surface exposed portion of this molecule with trypsin (613). Streptococci were washed to remove trypsin as well as cleaved peptide fragments and subsequently digested with phage lysin amidase, which resulted in a spectrum of released M-protein fragments with increasing mass when analyzed by SDS-PAGE (613). The fastest-migrating species was further purified and characterized by Edman degradation as well as amino acid composition. The data indicated that the C-terminal end of M protein, beginning at residue 302, is uniformly protease protected (613). Amino acid analysis revealed that the C-terminal peptide did not contain phenylalanine and suggested that the phenylalanine-containing hydrophobic domain might be absent from mature M protein (612). In a subsequent study of the possible membrane anchor cleavage activity, this data was interpreted as indicating a cleavage between the proline and the serine of the LPSTG motif (612), an estimate that was close to the later-identified cleavage site between the threonine and the glycine of the LPXTG motif within protein A (see below). Phage lysin released anchor fragments do not contain amino sugars (396). This can be explained by the amidase activity of this enzyme, which removes the glycan component of the cell wall from its crosslinked peptide backbone (613). Thus, although this has never been demonstrated, the data corroborate a model in which the C-terminal end of M proteins may also be covalently linked to the peptidoglycan.
Chemical analysis of the peptidoglycan of several other bacterial species including S. pneumoniae (476), S. mutans, S. sanguis (64, 680), Lactobacillus fermenti (822), and Mycobacterium tuberculosis (853) revealed the presence of significant amounts of nonpeptidoglycan amino acids. Analysis of the cell walls of S. mutans led to the conclusion that it contained covalently attached proteins, and it was suggested that this may be a feature common to the cell walls of all gram-positive species (577, 689).
SORTING: COVALENT LINKAGE OF SURFACE PROTEINS TO THE CELL WALL
Cell Wall Anchoring of Staphylococcal Protein A
Staphylococcal protein A has been used as a model system to study the anchoring of surface proteins in gram-positive bacteria (716). The cytoplasmic protein A precursor is exported and processed to generate the mature anchored species within 1 min of its synthesis. As described above, the anchored mature species is accessible to protease on the bacterial surface and requires enzymatic release from the staphylococcal cell wall for solubility. Lysostaphin cleaves at the pentaglycine crossbridge of the staphylococcal cell wall and solubilizes protein A as a uniform species on SDS-PAGE (716). In contrast, muramidase cleaves the glycan strands of the cell wall and muramidase-released protein A appears as a spectrum of bands on SDS-PAGE, all of which migrate more slowly than the lysostaphin-released counterpart (715). The notion that these mass differences must be the result of linked peptidoglycan was confirmed by an experiment in which muramidase-released protein A is digested with lysostaphin, which converts all material to the same uniform mobility as that observed for protein A directly solubilized by lysostaphin (715). Because of the uniform migration of lysostaphin-released fragments on SDS-PAGE, the anchoring point of surface proteins must be more proximal to the pentaglycine crossbridge, i.e., the cleavage site of lysostaphin, than to the glycan chains where muramidase cuts.
Mutant Protein A Phenotypes
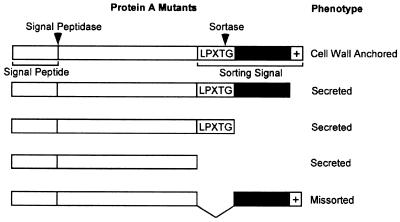
Protein A lacking a C-terminal sorting signal, i.e., the LPXTG motif, hydrophobic domain, and charged tail, is secreted into the medium and thus does not require the enzymatic digestion of the cell wall in order to be soluble when cells are boiled in SDS (716) (Fig. 8). Mutant protein A with the charged tail truncated behaves similarly. Lysostaphin-solubilized protein A migrates faster on SDS-PAGE than the secreted, unanchored species does, suggesting that the polypeptide chain is proteolytically cleaved during the anchoring process (716). To confirm that the secreted species comprised the complete (unprocessed) polypeptide chain, a protein A mutant that contains a single cysteine at the C terminus has been generated. Metabolic labeling of this cysteine revealed that the secreted protein A mutant is uncleaved and that some form of processing must be required for the anchoring of surface proteins (716).
FIG. 8.
Phenotypes of protein A cell wall sorting signal deletion mutants. Full-length protein A is found to be quantitatively anchored to the cell wall. Deletions of the charged tail, charged tail and hydrophobic domain, or the entire sorting signal result in the complete secretion of the polypeptide into the extracellular milieu. Deletion of the LPXTG motif of the C terminal abolishes the anchoring of the polypeptide to the cell wall. In the case of the protein A sorting signal, these mutant proteins are loosely associated with the cytoplasmic membrane.
Mutants with mutations within the LPXTG motif have a different phenotype. These polypeptides are not secreted into the medium but remain cell associated. Compared to the wild type, the LPXTG mutants do not reveal the characteristic size differences of lysostaphin- and muramidase-released species, indicating that these substances are not covalently linked to the cell wall (715). Furthermore, the mutant polypeptides also migrate more slowly on SDS-PAGE than their anchored counterparts do, suggesting that processing, i.e., proteolytic cleavage and cell wall linkage, is disrupted. Thus, the two different mutant phenotypes reveal that surface protein anchoring is arrested at distinct steps. The hydrophobic domain and charged tail cause protein A to be retained from the secretory pathway, whereas the LPXTG motif is needed for cleavage and linkage to the cell wall.
Cell Wall Sorting Signal
To determine the signal sufficient for cell wall anchoring of proteins normally secreted into the medium or lipid anchored to the cytoplasmic membrane, C-terminal protein A sequences have been fused to staphylococcal enterotoxin B (Seb), β-lactamase (BlaZ), or E. coli alkaline phosphatase (PhoA) exported by the protein A signal peptide (715, 716). The C-terminal 35 residues of protein A, comprising the LPXTG motif, hydrophobic domain, and charged tail, are sufficient for cell wall anchoring of each of these fusion proteins. Homologous sequence elements of other surface proteins also function to signal cell wall anchoring (715). When fused to the C terminus of secreted reporter proteins, some of these signals function in a manner indistinguishable from that of the protein A signal. Others fail to signal anchoring but can be mutated to gain this function. In all cases examined, these mutations alter the spacing between the LPXTG motif and the charged tail. The absolute number of residues between the LPXTG motif and the charged tail does not appear to be critical. It has been proposed that the folding of the hydrophobic domain determines the correct spacing between the flanking elements required for proper recognition of the sorting signal (715).
Retention is signaled by the positively charged residues within the charged tail of the sorting signal. Although a single arginine is most effective in signaling retention, this residue can be replaced by lysine when other positively charged residues are present. Increasing the spacing between the LPXTG motif and the charged tail further does not interfere with cell wall sorting but proves to be toxic for staphylococci. The reason for this phenomenon is not known (715).
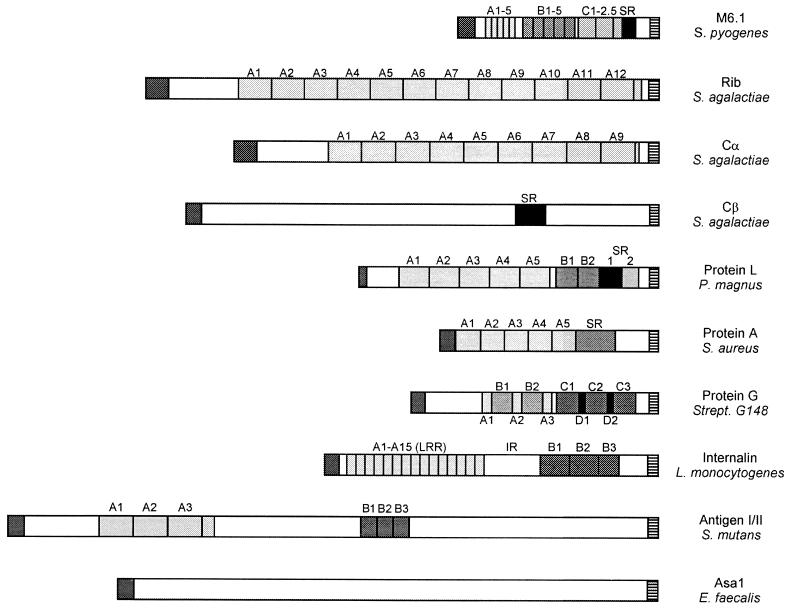
The LPXTG motif is conserved within the sorting signals of all known wall-anchored surface proteins of gram-positive bacteria (Table 1). The threonine (T) displays some variation in that either alanine or serine can be found at this position. A threonine-to-alanine substitution was tested in the sorting signal of staphylococcal protein A, and this mutation does not affect anchoring, which suggests that the identified sequences are indeed functional sorting signals. Other mutations in the LPXTG motif, for example a proline (P)-to-asparagine (N) mutation, do abolish sorting. Nevertheless, a rigorous mutagenesis of this sequence element has never been performed, and the basis for the sequence conservation is thus unknown. The simplest explanation for the conservation of the LPXTG motif may be the preservation of a proteolytic cleavage site. Sortase, the presumed enzymatic activity that cleaves at this point and catalyzes the transpeptidation reaction, may require a specific sequence or fold for substrate recognition. It is surprising that other transpeptidation mechanisms, for example that of glycosylphosphatidylinositol (GPI) anchoring, seemingly have much less stringent substrate requirements (see below).
TABLE 1.
Proteins containing cell wall sorting signals
| Source | Sequence of C-terminal sorting signala | Reference(s) |
|---|---|---|
| Actinomyces naeslundii | ||
| Fim P: type 1 fimbrial subunit (T14V) | LPLTGANGVIFLTIAGALLVAGGAVVAYANKRRHVAKH | 868 |
| Fim A: type 1 fimbrial subunit (T14V) | LPLTGANGMLILTASGASLLMIAVGSVLVARYRERKQNANLAL | 869 |
| Type 2 fimbrial subunit (WVU45) | LPLTGANGMLILTASGAALLMIAVGSVLVARYRERKRNRDLAA | 867 |
| Enterococcus faecalis | ||
| Asa1: aggregation substance | LPQTGEKQNVLLTVAGSLAAMLGLAGLGFKRRKETK | 237 |
| Asc10: aggregation substance | LPKTGEKQNVLLTVVGSLAAMLGLAGLGFKRRKETK | 411 |
| Asp1: aggregation substance | LPHTGEKENTLLSVLGAGMLAGLAWFGLKRKETEK | 236 |
| Sea1: surface exclusion protein | LPHTGEQKSIWLTIFGLFMAVGAISFKNKRRKNS | 840 |
| Sec10: surface exclusion protein | LPQTGEQQSIWLTIIGLLMAAGTINFKNKKRKKNS | 411 |
| PrgC: phermone-responsive surface protein | LPHTGEKFTLLFSVLGSFFVLISGFFFFKKNKKKA | 411, 571 |
| Unknown ORFb (hypothetical 30.5 kDa) | LPKTGETENIALSVLGSLMVLGSAFIFKKRI | 771 |
| Enterococcus faecium | ||
| Ash701: aggregation substance | LPQTGEKQNILLTVAGSLVTMLGLAGVGLKRRKETK | 315 |
| Lactococcus lactis | ||
| CluA: sex factor aggregation protein | LPKTGEGKAALAISIFGAALLGLAAYLKRNWIVSTYRKTVRKIRK | 271 |
| NisP: nisin serine protease | LPVTGDGEDFLPALGIVCISILGILKRKTKN | 810 |
| PrtP: casein serine protease | LPKTGETTERPAFGFLGVIVVSLMGVLGLKRKQREE | 820 |
| Lactobacillus paracasei | ||
| PrtP: casein serine protease | LPKTAETTERPAFGFLGVIVSLMGVLGLKRKQREE | 343 |
| Listeria monocytogenes | ||
| Internalin A | LPTTGDSDNALYLLLGLLAVGTAMALTKKARASK | 235 |
| Internalin C2 | LPTAGDENTMLPIFIGVFLLGTATLILRKTIKVK | 162 |
| Internalin D | LPTAGDENTMLPIFIGVFLLGTATLILRKTIKVK | 162 |
| Internalin E | LPITGDELNVLPIFVGAVLIGIGLVLFRKKRQTK | 162 |
| Internalin F | LPKTGDNAPWKTLFAGILLSSSAFYIWRKKA | 162 |
| Peptostreptococcus magnus | ||
| Protein L | LPKAGSEAEILTLAAAALSTAAGAYVSLKKRK | 412, 567 |
| PAB | LPEAGRRKAEILTLAAASLSSVAGAFISLKKRK | 141 |
| Streptococcus pyogenes (GAS) | ||
| C5A peptidase | LPTTNDKDTNRLHLLKLVMTTFFFGLVAHIFKTKRQKETKK | 118 |
| Trypsin-resistant surface protein (T6) | LPSTGSIGTYLFKAIGSAAMIGAIGIYIVKRRKA | 714 |
| SfbI/protein F | LPATGDIENVLAFLGILILSVLSIFSLLKNKQSNKKV | 720, 778 |
| SfbII/OF | LPASGDKREASFTIVALTIIGAAGLLSKKRRDTEEN | 448, 658 |
| M protein | ||
| Class I Emm (M6) | LPSTGETANPFFTAAALTVMATAGVAAVVKRKEEN | Table 2 |
| Class II Emm (Emm22) | LPSTGEAANPFFTAAAATVMVSAGMLALKRKEEN | Table 2 |
| Mrp/FcrA (Mrp8) | LPSTGEETTNPFFTAAALTVIASAGVLALKRKEEN | Table 2 |
| Enn (Enn49) | LPSTGEAANPFFTAAAATVMVSAGMLALKRKEEN | Table 2 |
| Unknown ORF | LPSTGEKAQPLFLATMTLMSLLGSLLVTKRQKETKK | 639 |
| Streptococcus agalactiae (GBS) | ||
| Alpha antigen | LPATGENATPFFNVAALTIISSVGLLSVSKKKED | 550 |
| Rib protein | LPATGENATPFFNVVALTIMSSVGLLSVSKKKED | 837 |
| Beta antigen (IgA Fc receptor) | LPYTGVASNLVLEIMGLLGLIGTSFIAMKRRKS | 319, 382 |
| C5a peptidase | LPTTNDKDTNRLHLLKLVMTTFFLGLVAHIFKTKRQKETKK | 119 |
| Streptococcus sp. (GCS and GGS) | ||
| Protein G (IgG binding) | LPTTGEGSNPFFTAAALAVMAGAGALAVASKRKED | 194, 290 |
| M protein (Group C) | LPSTGEATNPFFTAAALAVMATAGVAAVVKRKEEN | 776 |
| MIG (strain SC1) | LPTTGEGSNPFFTAAALAVMAGAGALAVASKRKED | 401 |
| MAG (strain 8215) | LPTTGEGSNPFFTAAALAVMAGAGALAVASKRKED | 399 |
| FnBA: fibronectin binding protein | LPQTGDTNKLETFFTITALTVIGAAGLLGKKRRNNQTD | 490 |
| FnBB: fibronectin binding protein | LPAAGEAEHVLSTIVGAMILFLVSLWGLLKRKASKA | 490 |
| GfbA: fibronectin binding protein | LPATGDIENVLAFLGILILSVLPIFSLLKNKQNNKV | 430 |
| Streptococcus gordonii | ||
| SspA/SSP-5 (antigen I/II) | LPKTGTNDSSYMPYLGLAALVGVLGLGQLKRKEDESK | 149 |
| SspB (antigen I/II) | LPKTGTNDATYMPYLGLAALVGFLGLGLAKRKED | 148, 149 |
| CshA | LPRTGSQTSDQTASGLLAAIASLTFFGLANRKKKSKED | 538, 539 |
| Streptococcus mutans | ||
| Antigen I/II (P1, B, PAc) | LPNTGVTNNAYMPLLGIIGLVTSFSLLGLKAKKD | 422, 423, 598 |
| β-d-Fructosidase | LPDTGDHKTDLSQLGVLAMIGSFLVEIASYFKKSKD | 103 |
| Dextranase | LPQTGDNNETRSNLLKVIGAGALLIGAAGLLSLIKGRKND | 363 |
| Glucan binding protein C | LPHTGAAKQNGLATLGAISTAFAAATLIAARKKEN | 704 |
| WapA | LPSTGEQAGLLLTTVGLVIVAVAGVYFYRTRR | 201 |
| Streptococcus pneumoniae | ||
| β-N-Acetylhexoseaminidase | LPETGTHDSAGLVVAGLMSTLAAYGLTKRKED | 122 |
| Neuraminidase | LPETGNKESDLLASLGLTAFFLGLFTLGKKREQ | 107 |
| Streptococcus sobrinus/downeii | ||
| SPAA (antigen I/II; PAg) | LPATGDSSNAYLPLLGLVSLTAGFSLLGLRRKQD | 794 |
| Dextranase | LPKTGDHKTVVLIIVLGLVFVGMTGLLARHEKK | 827 |
| Streptococcus equi | ||
| ZAG | LPTTGEKANPFFTAAALAIMAGAGALAVTSKRQQD | 400 |
| Fibronectin binding protein | LPQTSDMKQLTLSIIGAMSMLLVLCLSLFKRPSKKD | 491, 492 |
| SeM | LPSTGESANPFFTIAALTVIAGAGMAVVSPKRKEN | 784 |
| SzPW60 | LPSTGEATNPFFTAAALAVMAGAGVAAVSTRRKEN | 785 |
| SzPSe | LPSTGEATNPFFTAAALAVMAGAGVAAVSTRRKEN | 784 |
| Streptococcus suis | ||
| 136-kDa muramidase released surface protein | LPNTGEASSVAGALGTAMLVATLAFARKRRRNED | 747 |
| Staphylococcus aureus | ||
| Collagen adhesin | LPKTGMKIITSWITWVFIGILGLYLILRKRFNS | 626 |
| Fibrinogen binding protein (clumping factor) | LPDTGSEDEANTSLIWGLLASIGSLLLFRRKKENKDKK | 531 |
| FNBP: fibronectin binding protein | LPETGGEESTNKGMLFGGLFSILGLALLRRNKKNHKA | 727 |
| Protein A: IgG binding protein | LPETGEENPFIGTTVFGGLSLALGAALLAGRRREL | 726, 803 |
| Staphylococcus epidermidis | ||
| Fibrinogen binding protein | LPDTGANEDYGSKGTLLGTLFAGLGALLLGKRRKNRKNKN | 590 |
The LPXTG motif is shown in boldface type.
ORF, open reading frame.
The hydrophobic domain and charged tail together are thought to retain the polypeptide from the secretory pathway and provide an opportunity for the proteolytic cleavage of surface proteins. One way in which this might be achieved is to anchor the polypeptide in the cytoplasmic membrane. If so, fusion of the C-terminal hydrophobic domain to other secreted proteins such as PhoA or Seb should insert the hybrids in the membrane by acting as a stable transmembrane domain. This, however, is not the case, and the mutant proteins are instead found to be peripherally associated with the membrane (715). Furthermore, mutation or truncations of the charged tail, even at positions that cannot play a role in membrane insertion, lead to the secretion of the polypeptide into the surrounding medium. Perhaps the C-terminal hydrophobic domain and charged tail function to arrest translocation of the polypeptide within the secretion channel but without permitting the actual insertion into the membrane. This arrest in translocation could suffice to allow cleavage of the polypeptide and concomitant cell wall anchoring. A simple test of this hypothesis might be to engineer sorting signals with increased hydrophobicity to allow membrane anchoring. Once inserted into the membrane, the surface protein could diffuse away from the site of cell wall sorting without permitting a transpeptidation reaction at the LPXTG motif.
Proteolytic Cleavage at the LPXTG Motif
The proteolytic cleavage site during the anchoring of surface proteins has been determined by purifying and sequencing the C-terminal cleavage fragment (574). This has been done by using a hybrid protein in which the mature part of maltose binding protein is fused to the C-terminal end of the sorting signal. The hybrid protein is exported by its N-terminal signal peptide and cleaved, and the N-terminal portion is linked to the bacterial cell wall. In contrast, the C-terminal cleavage fragment with the fused maltose binding protein domain remains within the cytoplasm and can be purified by affinity chromatography (574). Edman degradation reveals that the cleavage site is located between the threonine and the glycine of the LPXTG motif (Fig. 9). Deletion of the LPXTG motif abrogates cleavage as well as anchoring, indicating that the two events are linked. Proteolytic cleavage at the LPXTG motif absolutely requires protein export by the Sec machinery. When cells are poisoned with sodium azide, which at low concentrations is an inhibitor of the SecA motor of preprotein translocase (223, 601), no secretion or cleavage of the fusion protein occurs. Similarly, protein A mutants harboring a defective signal peptide are not cleaved at the LPXTG motif. This result suggests that the site of proteolytic cleavage may be either in the membrane, i.e., during secretion of protein A, or on its extracytoplasmic side, which coincides with the locus for cell wall assembly.
FIG. 9.
Proposed model for the cell wall sorting reaction. The model can be divided into four distinct steps. 1, the full-length precursor is exported from the cytoplasm via an N-terminal leader peptide. 2, the protein is prevented from release into the extracellular milieu by the charged tail and hydrophobic domain. 3, the protein is cleaved between the threonyl and glycyl residues of the LPXTG motif. 4, the newly liberated carboxy terminus of threonine is transferred via an amide bond exchange to an amino group found at the end of the wall crossbridge. In this proposed model, steps 3 and 4 are coupled in a two-step reaction. Overall, the reaction bears similarity to the amide bond exchange mechanisms by which proteins are attached to GPI moieties and by which transpeptidation occurs during cell wall synthesis.
Cell Wall Anchor Structure of Protein A
To date, the only cell wall anchor structure to be solved in molecular detail is that of staphylococcal protein A (575, 713, 796, 797). The structure was determined through the use of specifically designed hybrid proteins that enabled the purification of large amounts of surface protein after solubilization from the cell wall with muralytic enzymes. The C-terminal anchor structure was removed and isolated from the full-length hybrid surface protein by proteolysis or cyanogen bromide cleavage at sites specifically engineered into the hybrid protein. The resulting C-terminal anchor peptides allowed detailed studies of associated cell wall structures by chemical analysis, Edman degradation, and mass spectrometry.
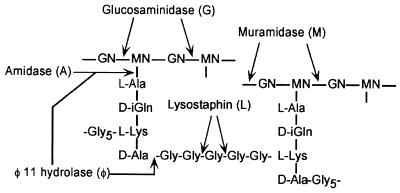
In an early study, a maltose binding protein harboring the C-terminal sorting signal of protein A was solubilized with lysostaphin, purified, and subjected to limited proteolysis to identify C-terminal anchor peptides (713). The C-terminal threonine of the LPXTG motif was found to be amide linked to a triglycine peptide. Because the pentaglycine crossbridge of the staphylococcal cell wall is also the site of lysostaphin cleavage, it was proposed that surface proteins are amide linked to the free amino group of the wall crossbridges. Because both the LPXTG motif and the free amino group of wall crossbridges are conserved features in gram-positive bacteria, this mechanism might be universal for all organisms expressing surface proteins via a C-terminal sorting signal (574, 713).
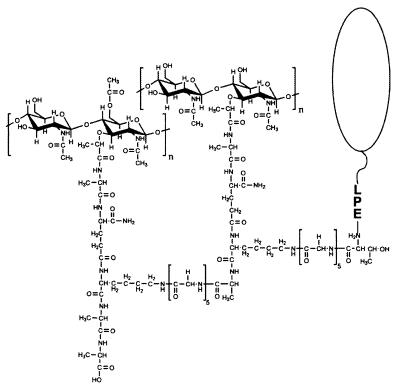
In later studies, a hybrid protein containing the C-terminal sorting signal of protein A fused to Seb was used in similar experiments to analyze the C-terminal anchor structure of surface protein solubilized with several other muralytic enzymes (575, 796, 797). In these studies, six histidyl residues placed at the fusion junction between Seb and the sorting signal facilitated the purification of the full-length protein by chromatography on nickel resin. C-terminal anchor peptides were obtained as follows. The purified surface protein was first cleaved with cyanogen bromide at a methionyl immediately adjacent to the six histidyl. The anchor peptides were then purified by another round of affinity chromatography on nickel resin. As mentioned above, surface protein solubilized from staphylococcal peptidoglycan with muramidases migrate as a ladder of bands on SDS-PAGE (Fig. 10). Muralytic amidases, i.e., enzymes that cut at the amide linkage between the d-lactyl of MurNAc and the l-Ala of the wall peptide, also solubilize surface protein as a ladder of bands (575). Mass-spectrometric analysis of C-terminal anchor peptides from the hybrid surface protein solubilized with these enzymes confirmed that the distinct pattern observed on SDS-PAGE is due to the attachment of a variable number of linked peptidoglycan subunits (575). Amidase-solubilized surface proteins were found to be linked to one or more peptidoglycan subunits of the structure [NH2–l-Ala–d-iGlu–l-Lys(Gly5)–d-Ala–d-Ala], whereas the muramidase-solubilized surface proteins were linked to subunits of the structure MurNAc[-l-Ala–d-iGlu–l-Lys(Gly5)–d-Ala–d-Ala]-GlcNAc. The attached peptidoglycan subunits were found to be linked to one another through their pentaglycine crossbridges. As would be expected, redigestion of amidase- and muramidase-solubilized surface protein with lysostaphin removed the linked peptidoglycan subunits (575). The structure of a surface protein linked to a peptidoglycan dimer is shown in Fig. 11.
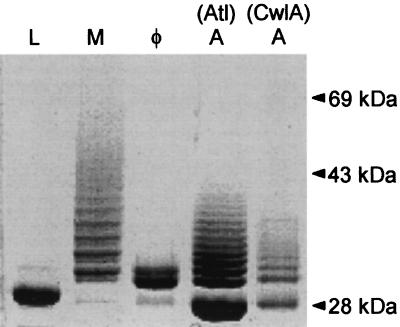
FIG. 10.
SDS-PAGE of surface protein solubilized with five different muralytic enzymes. Shown is a Coomassie blue-stained SDS-PAGE gel of a hybrid surface protein containing the cell wall sorting signal of protein A after solubilization from the staphylococcal peptidoglycan with lysostaphin (L), mutanolysin (M), the staphylococcal phage φ11 murein hydrolase (φ), the amidase portion of the major staphylococcal autolysin (A, Atl), or the lytic amidase from B. subtilis (A, CwlA). Proteins solubilized with these enzymes display distinct mobilities on SDS-PAGE due to the presence of covalently linked cell wall subunits (see the text). See Fig. 17 for a diagram of the enzymatic activities of these enzymes. A species of 28 kDa is apparent in the amidase-, phage hydrolase-, and mutanolysin-digested samples, which migrates faster than the lysostaphin-solubilized protein. The generation of this species is apparently due to degradation during the prolonged incubation necessary for complete solubilization of the cell wall with these enzymes. Reprinted from reference 575 with permission of the publisher.
FIG. 11.
Structure of a surface protein linked to the peptidoglycan of S. aureus. As shown here, the protein is linked to a wall subunit that has been incorporated into the peptidoglycan network by both transpeptidation and transglycosylation. The neighboring wall subunit exists as a pentapeptide and is therefore not substituted. Adapted from reference 575 with permission of the publisher.
On the basis of sequence homology and limited biochemical analysis, the murein hydrolase of staphylococcal bacteriophage φ11 was predicted to be an amidase (835). Hybrid surface proteins solubilized from the staphylococcal cell wall with the φ11 hydrolase migrate as a doublet of bands on SDS-PAGE, in marked contrast to the ladder pattern observed when the protein is solubilized with other well-characterized amidases (796) (Fig. 10). Analysis of φ11 hydrolase-solubilized C-terminal anchor peptides revealed that surface proteins were linked to a single peptidoglycan subunit, of which approximately 50% lacked the GlcNAc-MurNAc disaccharide, indicating that cross-linked peptidoglycan subunits had been removed (796). Recent results reveal that the φ11 hydrolase displays two activities, the expected amidase as well as a d-Ala-Gly peptidase activity (575, 576).
Peptidoglycan Substrate of the Sorting Reaction
Genetic analysis of staphylococcal methicillin resistance has provided new insights into the synthesis of the peptidoglycan crossbridge (43). Staphylococcal strains expressing PBP2a (PBP2′) are resistant to most β-lactam antibiotics including methicillin (310, 520). Genetic screens designed to identify other elements necessary for methicillin resistance yielded mutations in at least 10 different genes (44, 45, 146). Some of these genes are involved in the synthesis of the pentaglycine crossbridge (145, 331, 513, 756) or the amidation of d-isoglutamyl within the wall peptide (292, 394). Staphylococcal strains harboring mutations in the femA, femB, or femX gene synthesize altered cell wall crossbridges with either three glycyl (femB), one glycyl (femA), or a combination of no or one glycyl. The last phenotype has been reported for a mutant with a combination of a femA mutation and a second one leading to a partially nonfunctional FemX protein (442). Although this has not yet been demonstrated directly, it seems likely that FemA, FemB, and presumably also FemX catalyze the addition of glycyl to the ɛ-amino group of l-lysyl in the wall peptide portion of lipid II. Ton-That et al. tested whether the sorting reaction could proceed in staphylococcal strains carrying mutations in any of the three genes femA, femB, and femX (797). Although sorting was slowed in all fem mutant strains, surface protein was found anchored to peptidoglycan-bearing crossbridges with only three or one glycyl residues as well as crossbridges containing seryl moieties. However, species anchored directly to the ɛ-amino of l-lysyl in the wall peptide were not observed, suggesting that the sorting reaction has restricted substrate specificity for the amino group acceptor of this amide bond exchange mechanism (797).
Although it is now clear that protein A and other staphylococcal surface proteins are linked to the pentaglycine cell wall crossbridge, the specific peptidoglycan substrate of the sorting reaction has thus far not been identified. If the sorting reaction requires mature, assembled peptidoglycan as a substrate, staphylococcal protoplasts in which the peptidoglycan had been removed enzymatically should be unable to catalyze the cleavage of sorting precursors. Movitz studied the synthesis of protein A in staphylococcal protoplasts and reported that most of this polypeptide was released into the extracellular medium (561). A similar result was observed for the secretion of M protein by streptococcal protoplasts (809). Staphylococcal protoplasts generated by lysostaphin digestion and osmotically stabilized with sucrose cleaved sorting precursors similarly to intact cells, suggesting that the mature, assembled cell wall is not required for the sorting reaction (797a). Protein A molecules that were cleaved at the LPXTG motif were released into the extracellular medium. Nevertheless, it is not clear whether these released protein A molecules contain linked peptidoglycan fragments.
On the other hand, if the sorting reaction uses the peptidoglycan synthesis precursor, lipid II, as a substrate, inhibition of peptidoglycan synthesis with known antibiotics might also interfere with the sorting reaction. This question has also been addressed by Movitz (562). Although the anchoring mechanism was unknown at that time, the author suspected that protein A was linked to peptidoglycan and measured the amount of pulse-labeled protein A in the cell walls of staphylococci that had been treated with vancomycin. The results showed that vancomycin inhibited cell wall synthesis but did not interfere with the anchoring of protein A. Movitz concluded that protein A may be linked to mature, assembled cell wall. Vancomycin binds to d-alanyl–d-alanine within the wall peptide, and treatment of staphylococci with this antibiotic leads to the accumulation of lipid II molecules (789). If lipid II served as substrate for the sorting reaction, Movitz’s experiments could not have distinguished between protein A molecules that were linked to lipid II and others that were tethered to the assembled cell wall. We think that the identification of the substrate for the sorting reaction will require biochemical studies with purified sortase enzyme as well as the rigorous characterization of presumed sorting intermediates.
Cell Wall Sorting Mechanism: a Model
The sorting of surface proteins to the cell wall of gram-positive bacteria begins in the cytoplasm with the initiation of precursor molecules into the protein export (Sec) pathway via their N-terminal leader peptides (Fig. 9). By default, this pathway leads to the secretion of polypeptides into the surrounding medium of staphylococci; however, the C-terminal hydrophobic domain and charged tail function to retain protein A along the pathway, perhaps within the preprotein translocase. This retention allows recognition of the LPXTG motif and a proteolytic cleavage between the threonine and the glycine of the LPXTG motif. The carbonyl of threonine may be acylated to an active-site thiol or hydroxyl of sortase, and a subsequent nucleophilic attack of the free amino at the end of the pentaglycine cell wall crossbridge results in the transpeptidation of surface protein to peptidoglycan precursor. The sorting intermediate may subsequently be incorporated into the cell wall. Proteolytic degradation of anchored proteins as well as the physiological turnover of peptidoglycan will finally remove the anchored polypeptides.
Murein Lipoprotein of Gram-Negative Bacteria: a Comparison
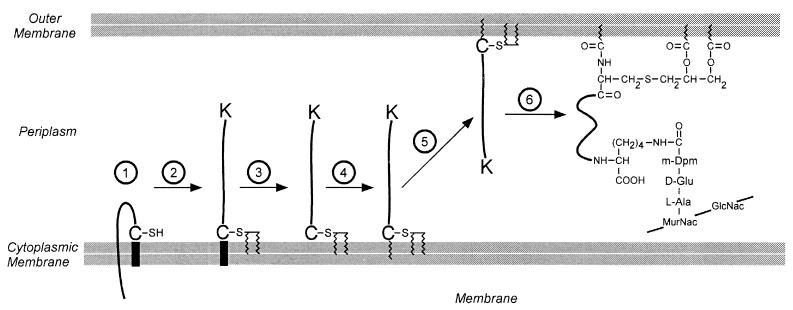
Braun’s murein lipoprotein (LPP) serves as a paradigm for the synthesis and structure of cell wall-linked proteins in gram-negative bacteria (85) (Fig. 12). The N-terminal signal peptide of LPP directs the polypeptide into the secretory pathway (367). A cysteine residue within the signal peptide is glyceride modified via a thioester linkage prior to proteolytic cleavage at its amino group (305). The amino group of the N-terminal cysteine is acylated prior to the insertion of LPP into the outer membrane (485). The 70-residue polypeptide of mature LPP trimerizes in the periplasmic space (84, 87). About one-third of all lipoprotein is covalently linked to the peptidoglycan layer, thereby tethering the outer membrane to the peptidoglycan skeleton of E. coli (86). Lipid modification of murein lipoprotein first requires membrane-embedded glyceryl transferase and O-acyl transferase activities (240, 289, 783). The glycerol-modified LPP precursor is then cleaved by signal peptidase (type II), and the liberated amino group of cysteine is modified by an N-acyl transferase (365). The ɛ-amino of the C-terminal lysine is amide linked to the carboxyl group at the D center of meso-diaminopimelic acid within the E. coli cell wall peptides (88, 89). Neither the chemical reaction nor the enzymes responsible for this linkage have been characterized. It is conceivable that each trimeric murein lipoprotein unit is covalently linked to the cell wall. It also seems plausible that the amide bond between LPP and cell wall is generated at the expense of another amide bond, namely, that between meso-diaminopimelic acid and d-Ala in cell wall tetrapeptides, similar to penicillin-sensitive transpeptidation and the sorting mechanism of surface proteins anchored to the cell wall of gram-positive bacteria.
FIG. 12.
Lipid and peptidoglycan attachment of the murein (Braun’s) lipoprotein of E. coli. The complete processing of the lipoprotein occurs in several discrete steps. 1, The protein is exported from the cytosol by virtue of a type II N-terminal leader peptide. 2, The cysteine residue of the leader peptide is modified by the addition of diacylglycerol. 3, The leader peptide is removed by the type II leader peptidase. 4, The newly liberated amino terminus is acylated. 5, The protein is translocated to the periplasmic side of the outer membrane. 6, The C-terminal lysine residue of the lipoprotein is amide linked to a carboxyl group of the diaminopimelic acid at position 3 of a wall peptide. Like the surface proteins of gram-positive bacteria, the lipoprotein is amide linked to the peptidoglycan. However, in this case the cell wall donates the carboxyl group rather than the amino group to this bond.
Amide Linkage of Eukaryotic Surface Proteins to GPI Anchors: a Comparison
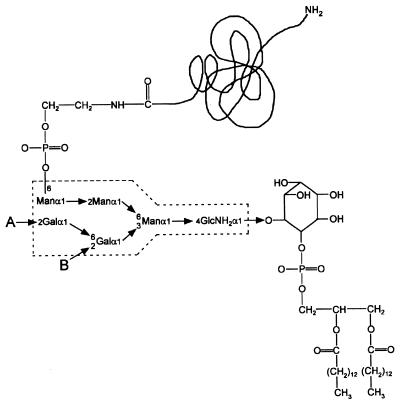
Some proteins displayed on the apical surface of polarized eukaryotic cells are modified by the GPI anchor (187, 198, 527) (Fig. 13). These polypeptides are synthesized with an N-terminal signal peptide and a C-terminal hydrophobic domain that functions to transiently retain these polypeptides in the endoplasmic reticulum (ER) membrane. Precursors are recognized in the ER as substrates by a GPI-modifying enzyme which proteolytically cleaves the peptide backbone upstream of the hydrophobic domain and amide links the N-terminal cleavage fragment to the free amino group of ethanolamine within the GPI moiety (69). GPI-decorated surface protein species are thought to be sorted at the Golgi and trans-Golgi network to lipid vesicles containing ceramide rafts that are marked for fusion with the apical portion of the cytoplasmic membrane (96, 495, 674). The exact mechanism of vesicle targeting is not known; however, the apical membrane differs in lipid composition from the basolateral membrane, which does not contain sphingomyelin and cerebrosides (203, 874). GPI-anchored surface proteins can be internalized by the budding of surface lipids into a structure referred to as caveoli (421). These vesicles are destined for fusion with lysosomes, which results in the degradation of both the GPI-anchored proteins and their bound ligands (522).
FIG. 13.
Core structure of a GPI anchor. This diagram shows the core GPI anchor structure found to be common among almost all known GPI anchors from several species. Species-specific variation of the anchor structure can occur through the addition of glycan moieties that can be attached at the points labeled A and B.
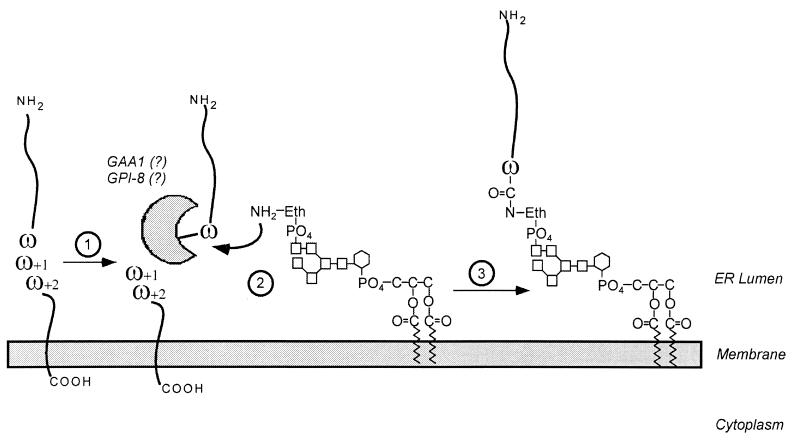
Synthesis of the GPI moiety occurs in the ER and begins on the cytoplasmic leaflet of the membrane (523, 524, 526, 813). After linkage of GlcNAc to phosphatidylinositol, a three-mannose core glycan is added and capped with phosphoethanolamine. The GPI moiety can translocate to the luminal side of the ER membrane and then serve as a substrate for transamidation (187, 775) (Fig. 14). The requirements for substrate recognition of precursor proteins have been thoroughly investigated (248, 434, 435, 548, 549, 556, 557). No conserved amino acids are present at or near the cleavage site, a situation more reminiscent of N-terminal leader peptides than of C-terminal cell wall sorting signals. Nevertheless, extensive mutagenesis of this region revealed that the residue at the cleavage site (ω) could be glycine, alanine, cysteine, serine, or asparagine but was none of the other 15 amino acids found in proteins. The ω+1 position, i.e., the residue at the N terminus of the C-terminal cleavage fragment, could tolerate almost all substitutions except proline. However, ω+2 appears to require the presence of either glycine, serine, threonine, or alanine, i.e., small-chain and uncharged amino acids (435). Furthermore, although the hydrophobic domains of GPI-modified proteins suffice to transiently insert precursors into the membrane, these sequences are also not characteristic membrane anchors and often lack the terminal charged residues as well as the degree of hydrophobicity found in peptide membrane anchors. Their function may therefore be similar to that of the C-terminal hydrophobic domain and charged tails of gram-positive sorting signals, i.e., to temporally retain the polypeptide chains from the secretory pathway and to permit a C-terminal transpeptidation reaction. Single point mutations within the hydrophobic domain suffice to stabilize the peptide membrane anchor and allow insertion into the basolateral portion of the cytoplasmic membrane (828).
FIG. 14.
Attachment of a GPI anchor to the C-terminal end of a polypeptide chain. 1, polypeptides entering the secretory pathway are transiently anchored to the membrane of the ER via the hydrophobic portion of the C-terminal GPI-anchoring signal. 2, cleavage occurs between the ω and ω+1 residues. 3, the newly liberated carboxy-terminus of the ω residue is amide linked to the amino group of the ethanolamine of the GPI anchor. This mechanism is similar to the mechanism by which cell wall sorting of surface proteins is proposed to occur in gram-positive bacteria.
Genes encoding the enzymes involved in synthesis of the GPI anchor were identified by screening for yeast mutants defective in the incorporation of [3H]inositol into the GPI-anchored protein GAS1 (604). Mutants with mutations in three genes, gpi-1, gpi-2, and gpi-3, are defective in GPI anchoring and temperature sensitive for growth, suggesting that these genes may be essential. Strains carrying mutations in any of three genes were found to be defective in the first step of GPI anchoring, the transfer of UDP-GlcNAc to phosphatidylinositol (478–480, 838). A second approach screened for yeast mutants defective in the surface presentation of GPI-anchored agglutinin, and the identified mutants could be grouped into six complementation classes, gpi-4 to gpi-9 (33). One of those strains, carrying a mutant gpi-8 allele, synthesizes a complete GPI moiety but does not transfer it onto precursor proteins, whereas all other mutants are defective in GPI assembly (32). GPI8 is a type I transmembrane ER protein with homology to cysteine proteases (32). Another transmembrane ER protein, GAA1, is also required for the attachment of GPI anchors to polypeptides (300, 338). Furthermore, it has been reported that soluble ER components are also necessary for the anchoring process (814). The role of these factors in catalyzing the transpeptidation reaction of GPI anchoring will have to be determined in a biochemical assay.
In vitro GPI anchoring of precursor protein has been demonstrated independently by several different laboratories (196, 525, 802). Mayor et al. established an assay that allows measurements of GPI anchoring without protein translocation into microsomal membranes (525). This is by far the simplest system and should allow characterization of the required enzymatic activities. The in vitro reaction requires precursor protein and GPI but no ATP or GTP, consistent with a transpeptidation (transamidation) mechanism. It is conceivable that the GPI8 enzyme catalyzes this reaction via an active-site sulfhydryl; a definitive result should be forthcoming.
Sorting at a Distinct Site?
Cole and Hahn examined the sites of cell wall and surface protein synthesis in Streptococcus pyogenes (127). This gram-positive organism forms chains of coccal cells due to the incomplete separation of its peptidoglycan. Thus, although the individual cells are round, the chain appearance permits an easy orientation of cell division sites at the contact points between cells. New cell wall appeared to be synthesized only at the contact points between cells. This experiment was extended to the synthesis of surface proteins by first proteolytically removing all surface (M) proteins and detecting the newly synthesized polypeptide by immunofluorescent staining of type-specific antiserum. All newly synthesized M protein first appeared at the contact sites between streptococcal cells and then extended slowly over the entire surface of the gram-positive cell. This result suggests that streptococci incorporate their surface proteins together with peptidoglycan at a defined site. Such localization may be required to coordinate protein sorting and cell wall synthesis, but these sites could also represent defined secretion sites in the cell wall peptidoglycan of gram-positive bacteria.
Peptidoglycan Turnover or Catalyzed Release of Cell Wall-Anchored Surface Proteins?
Pancholi and Fischetti observed that streptococci pretreated with phage lysin amidase would release large amounts of M protein (612). Phage lysin treatment is performed at pH 5.5 for optimal enzymatic activity, and during this treatment relatively little surface protein is released. However, once the pH of the buffer containing predigested streptococci was adjusted to pH 7.0 or higher, large amounts of M protein were released into the medium. The authors investigated the enzymatic activity that releases M proteins into the medium, assuming that it would cleave a membrane anchor of M proteins. The releasing activity is inhibited by pHMB, zinc, or other divalent cations. In view of the sorting hypothesis described above, it is conceivable that the measured solubilization is that of cell wall-linked M proteins. A change in the pH of intact streptococci does not lead to the release of M proteins.
The release of surface protein upon binding of specific antibody has been investigated for S. mutans P1 protein (476a–476c). Incubation at pH 6 caused the release of the surface protein antigen with its bound antibody from the streptococci (476a, 476b). The biological relevance of this was tested in an experiment that measured the detachment of S. mutans cells from a biofilm, which occurred only under conditions that also permitted the release of surface protein (476c). Shedding of P1 antigen required live cells, suggesting a pathogenic mechanism conceptually similar to another established paradigm, the shedding of variable surface antigen by trypanosome species (227). These organisms anchor their surface antigen via a GPI moiety to the cytoplasmic membrane, and cleavage with a phospholipase C allows the release of these peptides into the surrounding medium upon incubation with cross-linking antibodies. This phenomenon is thought to be one essential aspect of a complex mechanism by which trypanosomes establish antigenic variation of their surface protein antigen.
Cell Wall-Spanning Segments of Surface Proteins
The domain of protein A that is protected from added protease by the staphylococcal cell wall (region X) has a remarkable repeat structure and is about 175 residues long (291). The fact that thermolysin digestion generated a uniformly protected C-terminal peptide suggests that the distance between the cell wall-anchoring point and the surface-displayed portion is similar for all protein A molecules. Hence, protein A molecules that are linked to the cell wall do not “migrate” toward the cell surface, as would be implied by a model in which the cell wall is synthesized from the inside out, like the bark of an aging tree. Rather, all protein A molecules are linked to the cell wall and remain at that distance as the cell wall grows longitudinally over the growing cytoplasmic membrane. If this is so, the distance of the wall-spanning segment between the anchoring and the folded portions of surface protein should be critical in determining the functionality of these molecules. This was tested for S. aureus fibronectin-binding protein (FnBPB) expressed in Staphylococcus carnosus, and it was found that decreasing amounts of wall-spanning region caused reduced surface display of a fused lipase or β-lactamase domain (757). Furthermore, if the wall-spanning segment of FnBPB was shorter than 90 amino acids, the activity of the recombinant enzymes was reduced or abolished, presumably because the peptidoglycan exoskeleton interfered with the folding of the fused lipase or β-lactamase domains.
Other surface proteins harbor wall-spanning segments that also display a repeat structure, albeit with differences in sequence. Protease protection experiments of M proteins have yielded similar results to those described for protein A above (613). The wall-spanning domain of clumping factor (ClfA) of S. aureus consists of tandem repeats of an aspartyl-serine dipeptide. This domain functions to present the fibrinogen binding domain on the staphylococcal surface, and successive truncations of the repeat domain cause reductions in surface display (308).
ENGINEERED DISPLAY OF PROTEINS ON THE SURFACE OF GRAM-POSITIVE BACTERIA
Many gram-positive species colonize the mucosal surfaces of humans or animals without ever causing disease. These species constantly interact with the host immune system, which prevents their penetration into deeper tissues and establishment of disease. Although the cellular immune system and the phagocytosis of bacteria are critical in defending the host mucosal surfaces, it is thought that the humoral immune system also contributes to this defense via secretory IgA molecules (581). Other, more pathogenic microorganisms have the ability to penetrate mucosal surfaces and multiply within the host. On a first encounter with such microbes, the host may be defenseless, and the outcome of this interaction might result in disease. Pozzi and coworkers developed the gram-positive commensal organism Streptococcus gordonii as a delivery system to generate a mucosal immune response against such pathogens (215, 649, 650). Surface display of epitopes or entire proteins requires an N-terminal signal peptide and a C-terminal sorting signal for cell wall anchoring (716). For increased stability and surface display, several vectors that allow fusions with the scaffold of surface proteins of gram-positive bacteria are available (593). Inoculation of these engineered S. gordonii strains into naive animals led to colonization of mucosal surfaces and to the generation of a mucosal immune response (158, 540–542). Similar strategies for the immunization of mucosal surfaces have been established for Staphylococcus xylosus and Lactococcus lactis, two other nonpathogenic commensal species (583, 631, 752).
Surface display of engineered proteins might also be useful for biotechnological purposes. For example, recombinant antibodies are needed for diagnostic and therapeutic approaches. To facilitate their selection, these antibodies are often displayed on the surface of filamentous phages or on the surface of E. coli (518). Certain limitations apply, because these antibodies have to fit within the scaffold of phage proteins that are incorporated into an infectious particle. Several laboratories have used staphylococci as hosts for the expression of recombinant surface proteins. Engineered S. carnosus and S. xylosus were shown to display IgG as well as several other polypeptides on the cell surface (288, 582, 583, 697, 757). Surface-displayed antibodies may be manipulated by genetic or chemical means, and this technology may provide some advantage over phage display.
CELL WALL-ANCHORED SURFACE PROTEINS IN GRAM-POSITIVE BACTERIA
Surface proteins are displayed on the cell surface in order to interact with a substrate located in the surrounding environment. These proteins can have a wide range of functions including binding of host tissues, binding to specific immune system components, protein processing, nutrient acquisition, and interbacterial aggregation for the conjugal transfer of DNA. This section briefly reviews the various proteins identified thus far that possess C-terminal cell wall sorting signals. It is intended to provide a reference for those who are interested in the functions of various surface proteins and to highlight the incredible functional diversity found in this class of proteins. It is important to note that covalent attachment of proteins to the peptidoglycan has been conclusively demonstrated only for a few of the proteins reviewed below. Future studies should determine conclusively whether cell wall sorting is truly a universal process among gram-positive bacteria. It should be noted, however, that no proteins containing cell wall-sorting signals have been identified in the spore-forming gram-positive genera such as Bacillus and Clostridium.
Common Features of Anchored Surface Proteins
Anchored surface proteins must span the thick cell wall of gram-positive bacteria in order to display their functional domains to the surrounding environment. Despite their diverse range of functions, several common themes in the design of cell wall-anchored proteins of gram-positive bacteria are apparent (Fig. 15). N-terminal domains that contain the binding or catalytic activities are frequently followed by a set of repeat domains that may or may not possess activity. Often there is also a proline-rich stretch of amino acids immediately preceding the LPXTG motif that is thought to introduce random coils in the structure to assist in traversing the peptidoglycan network (212). It is possible, however, that such a domain has another function, since the proline-rich domains of several proteins are found to be more N-terminal than are regions predicted to span the cell wall.
FIG. 15.
Diagram of the structures of several anchored surface proteins from various gram-positive bacteria. All surface proteins possess an N-terminal leader peptide (N) and a C-terminal cell wall sorting signal. Many surface proteins possess several repeating domains. Particularly striking are the Cα and Rib proteins of S. agalactiae, which are composed of 9 or 12 nearly identical repeat domains, respectively (see the text). References for these proteins are given in Table 1.
Many cell wall-sorted surface proteins contain a number of tandem repeat domains that can vary in size from a few to several hundred amino acids. Striking examples are the alpha and Rib proteins of group B streptococci, which possess 9 or 13 repeats identical in both amino acid and nucleotide sequence (837). Strain-to-strain variation in the exact number of tandem repeat domains has been observed for many surface proteins, indicating that these domains are targets for recombination and/or duplication. Several mechanisms are likely to be involved in the generation of these repeat domains (139, 419, 420).
Functions of Cell Wall-Anchored Surface Proteins
Binding of immunoglobulins.
Much is known about the Ig binding surface proteins of gram-positive bacteria. Since the discovery of staphylococcal protein A, several other Ig binding proteins have been characterized and many of their genes have been cloned and sequenced. In 1977, Myhre and Kronvall proposed three different classes of Fc receptors based on the ability of certain bacteria to bind several different Ig subtypes from various mammalian species (569). Types I and II were the designations given to staphylococcal protein A and the streptococcal Fc receptors (M proteins) respectively. Type III was the designation given the Fc binding protein of group C and G streptococci (protein G). These designations were established before the genes encoding any of the Ig binding proteins had been determined; however, this nomenclature is still widely used today. For proteins A, G, and L, the three-dimensional structures of the antibody binding domains have been determined (143, 286, 854). With the exception of protein L, all surface protein Ig receptors bind to the heavy-chain portion of the antibody at the interface between the Fc CH2 and CH3 domains (231). Remarkably, despite their similar binding specificity, proteins A and G and the streptococcal Fc receptors (M proteins) have no structural similarities and appear to be the result of convergent evolution (231). Although several gram-positive bacterial species possess IgG binding activity, only two, the group A and B streptococci, have demonstrated the ability to bind IgA. Similar to the situation for the IgG receptors, the IgA receptors of GBS and GAS are unrelated to each other and are also most probably the result of convergent evolution.
Binding of serum and ECM proteins (MSCRAMMs).
Adherence to host tissues is the first step in the establishment of an infection. Many pathogenic gram-positive bacteria do not express structures such as pili and instead utilize cell wall-anchored surface proteins to adhere either to the extracellular matrix (ECM) or to other components present in host tissues. Höök and colleagues have given the name MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) to surface proteins that bind to components of the ECM (621, 624). MSCRAMMs have been found in almost all pathogenic gram-positive species, and their modular design and common binding domains suggests that they have arisen from a series of recombinational events and horizontal gene transfer. Many MSCRAMMs are capable of binding to more than one ECM component, and a single strain often possesses several different proteins that bind the same host component. For example, S. pyogenes has at least three proteins capable of binding fibronectin while Streptococcus dysgalactiae and some S. aureus strains have two (see below). MSCRAMMs and other adhesive surface proteins are the subject of a number of reviews (621, 624, 844).
Other surface proteins bind a wide variety of serum proteins including albumin, complement regulatory factors, soluble forms of fibronectin and fibrinogen, and the proinflammatory molecules plasmin(ogen) and kininogen. In addition, several bacterial surface proteins bind molecules present on the surface of host cells, a function that may help the bacteria either adhere or invade. Surface proteins that bind serum molecules or molecules found on the cell surface are not technically defined as MSCRAMMs even though these molecules may play a role in adherence to host tissues (624).
Wall-anchored enzymes.
Several examples have emerged where a metabolic enzyme is surface attached, particularly in cases where the function of the enzyme is to break down a large, nontransportable nutrient polymer into smaller subunits that can subsequently be taken up into the cell by a permease system. This is the case for both the dextranases and fructosidases of the mutans streptococci and the casein peptidase of lactococci. Not all surface-localized enzymes perform a nutrient acquisition function for the cell, however. For example, the nisin leader peptidase is involved in the processing of the nisin precursor to an active form by the removal of its leader peptide. Another surface protease, the streptococcal C5a peptidase, is involved in the degradation of this chemoattractant, presumably to prevent the recruitment of polymorphonuclear neutrophils (PMNs) and macrophages to the site of infection (see below).
Bacterial aggregation.
Both the conjugal transfer of DNA and the formation of dental plaque require that bacterial cells be able to adhere to one another through the specific interactions of surface proteins (160, 168, 441). In a majority of cases, the gram-positive bacteria employ cell wall-anchored surface proteins for this purpose. A notable exception is the fimbriae of the actinomycetes, whose subunits surprisingly contain C-terminal cell wall sorting signals (see below).
Streptococcus pyogenes (Group A Streptococci)
The work of Rebecca Lancefield, which began over 70 years ago, demonstrated that streptococcal surface proteins are mediators of virulence as well as targets for acquired immunity (467). Early studies determined that the surface proteins of GAS displayed a high degree of antigenic diversity. These findings led to the currently used typing system for GAS that is based on antisera raised against a set of variable surface antigens designated M, T, and R (212, 468). Of these, the M antigens are the most serologically diverse, with over 80 different serotypes identified thus far (see below). Considerable effort has been put into determining the relationship between the antigenic profile of a strain of GAS and its ability to cause disease (213, 420).
The GAS are also subclassified by whether they express the serum opacity factor (OF), an apoproteinase whose expression causes serum to become opaque (see below). Approximately half of streptococcal isolates express OF, and this expression pattern generally correlates with certain M serotypes. For example, M types 2, 4, 8, 9, 13, 22, 49, 59, 62, and 76 are often found to be OF+ whereas M types 1, 3, 5, 6, 12, 14, 18, 19, 24, 55, 57, and 80 are usually OF−. The M41 strain D421 is OF+, whereas another M41 strain, D463, is OF−, demonstrating that exceptions to this general rule exist (658). A table compiling the various M, T, and OF types has been published (389), and streptococcal surface proteins and typing systems have been the subject of a number of excellent past reviews (212, 419, 467). Therefore, this section is focused primarily on findings that have been made in the past decade.
It has recently become possible to analyze the genetic elements responsible for the antigenic diversity displayed by GAS at the nucleotide level. Until a decade ago, it was assumed that the antigenic properties of the various M types were due to the expression of a single variable M antigen. However, it is now evident that many strains of GAS, including almost all OF+ isolates, express more than one member of the M-protein family (295, 607, 632, 753) and that there can be considerable genetic diversity within a single M serotype (152, 627). In addition, there are many cases where M proteins from strains considered to be of a different M serotype have identical N termini (193, 848), suggesting that some type-specific antisera may be directed against more than one of the expressed M proteins (see below). Such findings have led many to reassess the previous assumptions that stemmed from early serological studies.
Mga regulon.
The genes encoding the OF, the C5a peptidase, and M proteins are all regulated at least in part by a multiple gene regulator called Mga (formerly virR or mry [111, 718, 729]) (117, 470, 634, 639). The mga gene encodes a 62-kDa DNA binding protein that activates transcription by binding to a consensus 45-nucleotide binding site found in the promoters of the regulon members (533, 536). Mga also positively regulates the expression of the genes encoding a secreted complement inhibitor (sic) (9, 427), streptococcin A (scnA), a cysteine protease (speB), and an oligopeptide permease system (opp) while down-regulating the expression of streptolysin S (639). There is some evidence, however, that other elements are involved in the regulation of these genes, including transcriptional terminators that serve to down-regulate the expression of the scpA gene and a neighboring open reading frame (71, 637, 639, 652). Mga bears some homology to the effector proteins of bacterial two-component regulatory systems, although a cognate sensor-kinase for this protein has not been identified. The expression of Mga and surface proteins in GAS is regulated by a variety of environmental conditions (534) including elevated levels of carbon dioxide (110, 595, 637) and/or the growth phase (535).
Mga regulons are ubiquitous throughout GAS (633), but their exact organization varies from strain to strain, particularly in the number and types of M-protein genes present (295, 296, 344, 632). In all cases, the mga gene is located immediately upstream of the M-protein genes (mrp, emm, or enn) and the gene encoding the C5a peptidase (scpA).
Streptococcal M-protein family (Emm, Enn, and Mrp).
The M family of streptococcal surface proteins is composed of the related Emm (class I and II), Mrp (FcrA), and Enn proteins. The Mga regulons of OF+ strains of GAS always contain all three M-family proteins in the order mrp, emm, and enn followed by the scpA gene. In contrast, the organization of the Mga regulons in OF− strains is much more variable and may include either one, two, or all three of the M-family genes, although the emm gene is always found (344). In at least one case, an additional open reading frame encoding a surface protein of unknown function (orfX) controlled by Mga has been found downstream of scpA (639).
Members of the M family of proteins are elongated dimeric alpha-helical coiled-coil molecules that can bind a variety of host components including Igs (5, 7, 75, 229, 320), fibrinogen (358, 409, 830, 850), kininogens (35), plasminogen (42), and albumin (229, 669) as well as factors that inhibit the deposition of complement on the bacterial surface (354, 780). A source of some confusion is that different laboratories have assigned different names to similar M-family proteins (e.g., the various class II Emm proteins have been designated Arp, Sir, or Emm). To assist the reader, the different names and ligands of several M-protein family members that have been sequenced are summarized in Table 2.
TABLE 2.
M proteins and their ligands
| M protein | Strain | Reference(s) | M type | Experimentally demonstrated ligands |
|---|---|---|---|---|
| Class I Emm | ||||
| M1 (Emm1) | AP1 | 8 | 1 | Albumin (712), fibrinogen (8, 712), IgG (712), kininogen (33) |
| M1.0 (Emm1.0) | CS130 | 297, 306 | 1 | |
| M1.1 (Emm1.1) | CS190 | 306 | 1 | |
| M1.2 (Emm1.2) | T1/29/58 | 306 | 1 | |
| M3 (Emm3) | 4/55 | 416, 666 | 3 | Albumin (666), fibrinogen (666), fibronectin (666) |
| M5 (Emm5) | Manfredo | 552 | 5 | Fibrinogen (851), factor H (445), FHL-1 (445) |
| M5.8193 | NCTC 8193 | 849 | 5 | Fibrinogen (849) |
| M6 (Emm6.1) | D471 | 346 | 6 | CD46 (596), factor H (354), FHL-1 (445), fibrinogen (851), kininogen (33) |
| M12 (Emm12) | CS24 | 671 | 12 | Albumin (666), IgG (666) |
| M18.1 (Emm18.1) | 87-282 | 137 | 18 | |
| M24 (Emm24) | Vaughn | 560 | 24 | Fibrinogen (850) |
| M41 (Emm41) | 71-710 | 632 | 41 | Plasminogen (112) |
| M52 (Emm52) | 71-719 | 632 | 52 | Plasminogen (112) |
| EmmL55 (Emm55) | A928 | 77 | 55 | Albumin (77), fibrinogen (77), IgG (77) |
| M57 (Emm57) | A995 | 515 | 57 | |
| Class II Emm | ||||
| Emm2 (EmmL2.1) | T2/MR | 51 | 2 | |
| Emm4 (Arp4) | AP4 | 232 | 4 | C4BP (780), IgA (5), IgG (753) |
| Emm9 | 71-683 | 635 | 9 | IgG (635) |
| EmmL15 (Emm15) | EF1949 | 415 | 15 | |
| Emm22 (Sir22) | AL168 | 754 | 22 | C4BP (780), IgA (753), IgG (753) |
| Emm28 (Sir28) | AL368 | 488, 753 | 28 | IgA (753), IgG (753) |
| Emm49 (EmmL49) | CS101 | 295 | 49 | Albumin (229), IgG (229) |
| Emm50 (EmmL50) | B514 | 877 | 50 | IgG (877) |
| Emm53 (PAM) | M53 | 42 | 53 | Plasminogen (42, 112) |
| Emm60 (Arp60) | AW43 | 320 | 60 | C4BP (780), IgA (320), IgG (486) |
| Emm64/14 | 64/14 | 75 | NT | IgG (75) |
| Mrp | ||||
| Mrp4 | AP4 | 607 | 4 | Fibrinogen (607), IgG (607) |
| Mrp8 | 4025 | 447 | 8 | Fibrinogen (447), IgG (447) |
| Mrp9 | 71-683 | 447 | 9 | Fibrinogen (447), IgG (447) |
| Mrp13 | 71-686 | 447 | 13 | Fibrinogen (447), IgG (447) |
| Mrp15 | EF1949 | 415 | 15 | |
| Mrp22 | AL168 | 754 | 22 | Fibrinogen (753), IgG (753) |
| Mrp28 | AL368 | 753 | 28 | Fibrinogen (753), IgG (753) |
| Mrp49 (FcrA49) | CS101 | 636 | 49 | Fibrinogen (447), IgG (636) |
| Mrp50 | B514 | 877 | 50 | IgG (877) |
| Mrp60 | AW43 | 753 | 60 | Fibrinogen (753), IgG (753) |
| Mrp76 (FcrA76) | CS110 | 313 | 76 | Fibrinogen (607), IgG (607) |
| Mrp64/14 (Fcr64/14) | 64/14 | 75 | NT | Fibrinogen (447), IgG (75, 447) |
| Enn (Emm Iib) | ||||
| Enn2 (EmmL2.2) | T2/MR | 51 | 2 | IgA (51) |
| Enn4 | AP4 | 381 | 4 | IgA (391) |
| Enn15 | EF1949 | 415 | 15 | |
| Enn22 | AL168 | 754 | 22 | |
| Enn49 (EnnX) | CS101 | 295 | 49 | |
| Enn50 | B514 | 877 | 50 | IgA (877) |
| Mosaic Enn | ||||
| Protein H | AP1 | 273 | 1 | Albumin (229), C4BP (393), IgG (7), fibronectin (230), MHC-II (8), N-CAM (230) |
| Enn5.8193 | NCTC8193 | 849 | 5 | IgG (849) |
| Enn64/14 | 64/14 | 635 | NT | IgG3 (635) |
The designation “M protein” has traditionally been reserved for members of the M family that are known to possess antiphagocytic properties, whereas all similar proteins with unknown or no antiphagocytic properties are generally designated “M-like.” Recent studies, however, have blurred this distinction, and it has become necessary to reassess the classical definition of an M protein. Separate laboratories have found that both the Mrp and Emm proteins can have antiphagocytic properties in a given strain (638, 781). It is also unclear whether M proteins play an equal role in protection from phagocytosis in every strain (137, 496) or whether a given M protein will have antiphagocytic properties when expressed in all strains (361, 781). Also, recent findings suggest that heparin, added as a blood anticoagulant in most phagocytosis assays, may interfere with the function of some M proteins by competing against the binding of factor H (62, 445, 781) (see below). If this is true, the antiphagocytic function of proteins previously considered to be M-like may have to be reassessed. For these reasons, we choose to adopt the functionally neutral terms Mrp, Emm, and Enn used by other laboratories to identify individual proteins based on their specific structural features while using the term “M protein” to designate all members of the family irrespective of their antiphagocytic function (345, 635, 781, 848). The mechanisms by which M proteins are thought to prevent phagocytosis are discussed below.
Sequencing of the various M proteins has revealed several common features in their molecular design, which are summarized in Fig. 16. The N-terminal hypervariable domains of mature M proteins account for the observed antigenic diversity of these molecules. In contrast, the central and C-terminal domains are almost completely conserved within each specific M subfamily (420). Throughout almost the entire M-protein sequence are heptad repeat motifs whose first and fourth amino acids are apolar. This arrangement is typical of coiled-coil molecules and serves to display the hydrophobic amino acids on the face of an alpha helix serving as a surface for dimerization (212). The heptad repeats are found even in the highly variable N-terminal domains, although not always in an optimal distribution, indicating that there is considerable selective pressure to retain the coiled-coil conformations (212, 217, 515, 586).
FIG. 16.
Domain structure of the M-protein family and the structure of the Mga regulons from OF+ and OF− strains of GAS (see the text).
All members of the M family possess an N-terminal leader peptide and a typical C-terminal cell wall sorting signal. The cell wall localization of M and M-like proteins was determined as early as 1952 (27, 228, 696), and it was demonstrated that purified cell walls could be used as a source for M protein (410, 446). Covalent attachment to the cell wall has been demonstrated by the fact that M protein solubilized from the cell wall with phage-associated lysin migrates as a ladder of bands on SDS-PAGE. More recently, it has been shown that the M6 protein is cell wall anchored when expressed in the gram-positive lactic acid bacteria (631).
Emm proteins (class I and class II).
The class I Emm proteins are found almost exclusively in OF− GAS. They were originally distinguished from the class II Emm proteins on the basis of their reactivity with a set of monoclonal antibodies raised against their C-terminal repeats (49). The binding properties of the class I Emm proteins are remarkably diverse (Table 2), but most have been shown to bind fibrinogen and/or albumin. These binding functions appear to have been divided between the Emm (class II) and Mrp proteins in the OF+ strains of GAS (see below).
The class II Emm proteins are typically found in GAS of the OF+ lineage and are the molecules usually recognized by M-typing antisera. These proteins were initially distinguished from their class I counterparts due to their inability to react with certain monoclonal antibodies directed against the C repeats of class I Emm molecules. None of the class II Emm proteins have been found to bind fibrinogen, a function that seems to be compensated for by the Mrp proteins in the OF+ strains. In addition, all class II Emm proteins studied thus far bind Ig, although their exact binding preferences differ (see Table 2).
Mrp (FcrA).
Mrp proteins are usually found in GAS strains of the OF+ lineage and are always encoded immediately downstream of the mga gene (Fig. 16). All Mrp proteins characterized thus far bind both human IgG (subclasses 1, 2, and 4) and fibrinogen (753). They can be distinguished from the Emm and Enn proteins by the absence of the conserved C-repeat domains. Instead, these proteins possess two or more copies of a different type of central repeat designated “A repeats” (295, 607), in which the IgG binding properties are thought to reside (8, 312). Upstream of the conserved repeats in both the Mrp and class II Emm proteins is a domain of unknown function that is rich in glutamate and glutamine (EQ domains). Unlike in the Emm proteins, the N-terminal domains of the Mrp proteins are not highly variable. Sequencing of the N-terminal regions of Mrp proteins from 37 different M serotypes revealed the existence of only six different types of N-terminal Mrp domains (447). In at least four cases (Mrp2, Mrp22, Mrp49, and Mrp64/14), an Mrp protein has shown evidence of being antiphagocytic (638, 781), and this property may be due to their ability to bind fibrinogen (see below).
Enn.
The genes encoding the Enn proteins are found almost exclusively in OF+ strains as the third open reading frame following mga in the Mga regulon (Fig. 16). Instead of the EQ-rich domains found in the class II Emm and Mrp proteins, the “C-repeat” domain of Enn proteins are usually preceded by a conserved set of amino acids (EDLKTTLAKTTKEN). The Enn proteins, like the Mrp proteins, do not display the same high degree of variability displayed by the Emm proteins (847). Recombinant Enn proteins expressed in E. coli are generally found to have IgA binding activity (51).
Thus far, no members of the Enn family have been demonstrated to have antiphagocytic properties. Under laboratory conditions, it has been demonstrated that the expression of enn genes is consistently more than 30-fold lower than the expression levels found for both mrp and emm (51, 381, 496, 849, 877). The low expression level has led to the hypothesis that the enn genes act primarily as a “sequence reservoir” for recombination with the emm and mrp genes. Indeed some unusual “mosaic” Enn proteins including protein H, Enn 5.8193, and Enn 64/14 all appear to have been generated by recombination between an emm gene and an enn gene (see below). Low expression of Enn has not been demonstrated during in situ, however, and it is possible that the Enn proteins do play a role during an infection.
Binding properties of M proteins: role in antiphagocytosis, adhesion, inflammation, and invasion.
It was demonstrated long ago that GAS strains are resistant to phagocytosis in human blood in the absence of specific antibodies directed against the M proteins (467). In 1979, it was demonstrated that M-protein-deficient strains of GAS could be efficiently opsonized and cleared by the alternative complement pathway (55, 630). The alternative complement pathway ultimately results in the deposition of C3b on the surface of the bacteria, which can subsequently act as a receptor for phagocytic immune system cells. Jacks-Weis and colleagues found that strains of GAS expressing M protein accumulate approximately fourfold less C3b on their surface than did mutant strains (374). Several laboratories have since attempted to elucidate the molecular basis of the antiphagocytic properties of the M proteins and their possible effect on the alternative complement pathway. These studies have variously attributed the antiphagocytic property of the M proteins to their ability to bind fibrinogen (355), factor H (62, 214, 354), FHL-1, C4BP (392, 393, 780), or C1q (443).
Factor H, FHL-1, and C4BP belong to the RCA (for “regulators of complement activation”) family of molecules that are capable of regulating complement activity. The proteins in this family are composed of multiple, highly homologous domains known as short consensus repeats (SCRs) (356). SCRs domains are each composed of approximately 60 amino acids, of which 4 cysteines and a few other residues are conserved and provide the structural framework for the domain while the remaining amino acids are thought to contribute to the binding specificity. SCRs, like many other common protein domains, are modular and are believed to act independently of other domains in a protein (28). Factor H is a soluble molecule composed of 20 SCRs, which compose almost the entire protein (356). FHL-1 (for “factor H-like”) is a truncated variant of factor H generated by alternate mRNA splicing and is composed of the first seven SCR domains of factor H (356, 553). C4BP is a soluble regulator of complement that is composed of seven alpha chains and one beta chain, which contain eight and three SCRs each, respectively (356).
The finding that an M protein could bind a down-regulator of the alternative complement pathway was first demonstrated for factor H (354). The binding of factor H to the class I M6 protein was mapped to the C-repeat region (214); however, this finding has not been without some controversy. For example, the C-repeat domains was shown to bind albumin by another laboratory (8, 229, 230), although the possibility exists that the C repeats can bind multiple ligands. In addition, fibrinogen was found to compete for factor H binding to the M6 protein even though fibrinogen binding is believed to occur in the B repeats of M6, upstream of the C repeats (355). It was suggested in that study, however, that fibrinogen could form a complex with factor H and that this fibrinogen-factor H complex would be responsible for providing protection from phagocytosis (355). This hypothesis may explain the previously observed antiphagocytic properties of fibrinogen. One study of a GAS strain (JRS251) expressing an M6 protein lacking the C repeats indicated that this strain was no longer able to bind factor H even though it was still resistant to phagocytosis (628). A more recent study, however, has demonstrated that these mutant cells can bind factor H, albeit at lower levels (724). Given this data, it appears unlikely that the binding site of factor H lies within the C-repeat domain.
Recent findings have suggested that the physiologically relevant binding partner of the class I Emm proteins is not factor H but, rather, FHL-1 (392, 445). Surprisingly, the binding of FHL-1 occurs through the hypervariable N-terminal domain of the M protein. Three separate laboratories have identified a domain found in both factor H and FHL-1 (SCR7) as being responsible for interacting with the M proteins (62, 445, 724). Given this data, it is difficult to envision how the N-terminal domains would be capable of selectively binding FHL-1 over factor H. It has been noted, however, that the properties of FHL-1 and factor H differ both structurally and functionally (326). In addition, it has been demonstrated that the binding of radiolabeled FHL-1 to the M5 protein can be inhibited only weakly by factor H and not at all by fibrinogen and that FHL-1 bound to bacteria retains its complement inhibitory function (392). Also, the binding of FHL-1 appears to occur under physiologically relevant conditions whereas the binding of factor H is inhibited by fibrinogen at the concentrations present in plasma (355, 392). This data suggests that there are two factor H binding sites on the M5 protein, one of which binds factor H (see above) while the other has a greater affinity for FHL-1 and lies outside of the fibrinogen binding and C-repeat regions. Such a model is in agreement with other observations made by Sharma and Pangburn (724) and would explain why M proteins lacking C repeats retain antiphagocytic activity (628). It should be noted that the characterization of the N-terminal FHL-1 binding domain was carried out by fusing domains onto an M protein without FHL-1 binding activity. This type of “gain-of-function” binding study avoids certain experimental concerns raised by the negative data obtained from binding studies that employ deletion mutants, such as the possibility of deleterious effects caused by conformational changes.
Several Emm proteins, particularly those in the class II subfamily, do not have the ability to bind either factor H or FHL-1. Instead, it has been demonstrated that many of these proteins bind C4BP. Similar to the binding studies of FHL-1, it was determined that the binding of C4BP occurs through the hypervariable N-terminal domain (393). The FHL-1 and C4BP binding data suggest a model whereby the N-terminal hypervariable domains of M proteins have evolved the ability to bind complement regulatory molecules (392). Interestingly, no consensus amino acid motif has been identified in the hypervariable regions demonstrated to bind C4BP (393). This data suggests that the N-terminal domains of the M proteins arise from two opposing selective pressures, the ability to maintain their antiphagocytic function while simultaneously altering their antigenic structure (393). This model may explain why the N-terminal variable domain is so critical to the function of M proteins and has not been lost due to selective pressure. It would also explain why mutant M6 proteins lacking C-repeat domains retain their antiphagocytic function.
Very recently, C1q, another complement component, was suggested to bind to the M proteins FcrA76 (Mrp76) and M5 (443). The binding studies were carried out on crude acid extracts, and the identification of the two M proteins was based solely on their apparent masses on SDS-PAGE. More studies are therefore necessary to demonstrate if this binding actually occurs through the M proteins and is biologically relevant in vivo.
At this point it should be mentioned that there is still some debate about whether M proteins are either necessary or sufficient for protection from phagocytosis in all GAS strains. For example, a mutant strain that lacks a hyaluronic acid capsule but expresses wild-type levels of M protein is sensitive to phagocytosis whereas the parent strain (strain 87-282) is resistant (841). In a later study, it was shown that strain Vaughn (M type 24) required fibrinogen for optimal resistance to opsonization and phagocytosis whereas in surprising contrast, strain 87-282 (M type 18) was opsonized by C3b in either the presence or absence of fibrinogen but in both cases remained resistant to phagocytosis (137). These results suggest that in some strains the deposition of C3b does not necessarily lead to phagocytic killing. It has recently been reported that a mutant of 87-282, in which the M protein was insertionally inactivated, remained resistant to phagocytotic killing in blood (496). The 87-282 mutant strain used in these studies still had an intact enn gene, although the Enn expression levels appeared to be quite low. The Emm18.1 protein from this strain was demonstrated to confer protection from phagocytosis to a strain of a different M type, indicating that the M protein from strain 87-282 is indeed functional (134). Nevertheless, these data suggest that the hyaluronic acid capsule can, by itself, protect the bacteria from phagocytosis in strains like 87-282 whereas other strains also require the expression of an M protein.
The ability of many different M proteins to bind Ig has been clearly demonstrated. The IgA binding site of Arp4 (Emm4) has been mapped by separate laboratories to a short stretch of residues near the N terminus (50, 391). A consensus binding site was identified and found to be present in most known IgA binding M proteins including Arp60, Enn4, Enn2.2, Enn50, and Sir22. Binding to IgG has also been clearly demonstrated, but the situation is much less clear. It is apparent that some M proteins bind human IgG1, IgG2, and IgG4 whereas others bind only IgG3 and a few M proteins have the ability to bind all four subclasses (summarized in reference 75). This leads to the tempting speculation that two or more different binding sites exist on M proteins, one of which binds IgG1, IgG2, and IgG4 while the other binds IgG3. Indeed, Björck and colleagues have found two separate IgG binding sites on protein H and M1. Domains homologous to the protein H IgG binding site were also found in the A repeats of the Mrp proteins, suggesting that such domains may bind IgG1, IgG2, and IgG4 (229). The IgG binding site of M1 is homologous to a region found in Arp4, a protein that can bind only a limited subset of IgG molecules, including IgG3 but not IgG1, IgG2, or IgG4 (8). Despite this correlation, the protein H and M1 IgG binding domains are found only in a subset of the Emm proteins known to have IgG binding activity. Obviously, more experiments must be performed to determine the locations of the various IgG binding domains.
As mentioned above, surface proteins can be released from some gram-positive cells by the action of specific proteases. In GAS, biologically active fragments of protein H, M1, and C5a peptidase can all be released from the cell surface by the action of SCP, a streptococcal cysteine protease (39). It has recently been shown that a soluble protein H-antibody complex can mediate the activation of the classical (antibody-mediated) pathway of complement (40). When immobilized on a surface, however, the protein H-antibody complex was found to instead inhibit the activation of complement. These findings suggest that the role of the Ig binding domains may involve inhibition of the classical complement-mediated pathway at the surface of the bacteria while the soluble forms of these proteins may serve to deplete complement at sites further away from the bacteria (40).
M proteins have been suggested to play a role in the generation of an inflammatory response by the binding of fibrinogen (831), kininogen (34), or plasminogen (76). By doing so, the bacteria may gain the ability to invade deeper tissues by increasing vascular permeability, dissolving clots, and disrupting the tight interactions within the ECM. Björck and colleagues tested the kininogen binding abilities of 49 strains of GAS and found that 41 were able to bind radiolabeled kininogen to their surface (35). This binding ability correlated well with the presence of M protein, since none of six M-protein-deficient strains were capable of binding kininogens. Direct analysis of kininogen binding to three M proteins revealed that the interaction occurs within the N-terminal portion of the M protein at a site that does not overlap the albumin, fibrinogen, or IgG binding sites (35).
Plasminogen is the biologically inactive precursor to plasmin, a fibrinolytic molecule speculated to aid in the invasive properties of cells (76). The role of plasmin(ogen) binding on the surface of GAS is not entirely clear but is apparently of some importance since more than 10 different species of invasive bacterial pathogens have been shown to bind these molecules (76). Most strains of GAS possess a surface-exposed glyceraldehyde-3-dehydrogenase-like molecule (SDH) that binds plasmin and not plasminogen (614). In addition, five GAS strains, of M serotypes 33, 41, 52, 53, and 56, that are usually associated with skin infections possess M proteins with plasminogen binding activity (42, 112). The common plasminogen binding motif in these M proteins was identified and localized to a set of tandem repeats found within the N-terminal domain (112). Surprisingly, Pancholi and Fischetti have recently reported the presence of a surface-localized alpha-enolase (SEN) that binds both plasmin and plasminogen (610). The finding that SEN is expressed ubiquitously throughout all strains of GAS conflicts with the finding that surface binding of plasminogen is a rare occurrence (112, 610). The reason for this discrepancy is unclear. Both SEN and SDH belong to a recently identified group of unusual enzymes that have the ability to bind plasma components. Their role in virulence and even their surface localization are under considerable debate, since they do not possess N-terminal leader peptides. These proteins are discussed further below.
Like plasminogen, kininogen is a biologically inactive precursor molecule. Proteolytic activation of kininogen generates the peptide hormone bradykinin, a vasoactive molecule that may assist in the generation of a localized inflammatory response near the site of infection. The effect of M-protein binding to either plasminogen or kininogen does not interfere with the ability of either molecule to be proteolytically converted into a biologically active form by their respective cognate proteases (34, 112).
It has long been speculated that M proteins may also act as adhesins to host tissue, but the mechanisms by which this may occur remain unclear (180), although adhesion has recently been implicated in the ability of GAS to generate a proinflammatory response in keratinocytes (829). Expression of the Emm5, Emm18, and Emm24 proteins in a deletion mutant of strain Vaughn enables these cells to adhere to HEp-2 cells. The M6 protein has been shown to mediate adherence to both HEp-2 cells (832) and keratinocytes (597). The binding of M6-expressing cells to HEp-2 cells appears to require fucose-containing glycoproteins (833). On the other hand, the ligand on the surface of keratinocytes responsible for binding M proteins was identified as the membrane cofactor protein (MCP or CD46 [596]). Interestingly, MCP is a membrane-bound complement regulator that contains SCRs and belongs to the RCA family of proteins like C4BP, factor H, and FHL-1. The binding of M proteins to the MCP molecule is speculated to occur through the C-repeat domains and can be inhibited by factor H (596). In contrast, the binding of HEp-2 cells appears to require the N-terminal domain of the M6 protein (832). It is clear that other receptors are responsible for mediating streptococcal adherence to many other cell types including fibroblasts (597). Other proteins implicated in the adhesion of GAS to host tissues are the fibronectin binding proteins FBP54 (132) and SfbI/protein F (see below).
Antigenic variation of M proteins: evidence of horizontal gene transfer.
A fundamental question in the biology of M proteins is how they evolve to vary their antigenic epitopes without disturbing the critical contacts necessary to maintain their structural and ligand binding characteristics. This question is particularly valid in light of the findings that the N-terminal hypervariable domains bind complement regulatory proteins. Both sequencing of several M protein genes and PCR analysis have allowed several laboratories to study the mechanisms by which sequence variation can occur. M proteins generate sequence diversity by point mutations, small insertions and deletions, and intragenic recombination between tandem repeat domains (212, 306, 347).
Analysis of several Mga regulons by PCR suggested that the evolution of the Mga regulons involved gene duplication followed by sequence divergence (344). Other studies have shown that the GAS also possess the ability to horizontally transfer gene segments, not only with group A organisms but possibly also with other streptococcal groups (420, 730, 849). Unusual mosaic genes that possess sequence elements from both enn and emm genes have been observed in three GAS strains, indicating that M proteins probably have the ability to recombine with one another (635, 849). Recently an insertion sequence has been identified within the Mga regulon of strain AP1, suggesting at least one possible mechanism by which the transfer of gene segments could occur (41). Another laboratory found another insertion sequence, IS1548, immediately downstream of the scpA gene in several GAS and GBS strains (277). It is possible that the considerable genetic diversity observed in the streptococcal Mga regulons is assisted by the presence of nearby insertion sequences.
The discovery of horizontal transfer of M-protein genes has challenged the previous assumption that the relatedness of GAS strains could be determined through the serologic and genetic properties of their M proteins. A comparison of 79 different M serotypes by multilocus enzyme electrophoresis revealed that the variation of the emm genes and overall genetic relationship between the various GAS strains are not congruent (848). This finding was supported by another study of several clinical M type 1 isolates that found that identity of scpA and emm genes between strains did not correlate with similarity in overall genetic structure (568). Therefore, the fact that two GAS strains possess identical or similar M serotypes does not by necessity indicate that the two strains are closely related. These findings have significant implications for the epidemiological study of GAS.
In summary, M proteins are multifunctional binding proteins that have evolved the ability to bind several of the common building block domains of eukaryotic plasma and matrix proteins including albumins, fibrinogens, fibronectins, Ig, plasmins, and proteins containing SCRs. These binding functions may serve the multiple purposes of protecting the bacteria from the various phagocytic mechanisms that exist within the host, assisting in the adhesion to certain cell types, generating an inflammatory response, and triggering the invasion of deeper tissues. The conserved C repeats may mediate adhesion to keratinocytes by binding MCP and/or factor H, but the binding properties of this domain are still the subject of considerable debate. Indeed, despite recent advances, the role of M proteins in protection from phagocytosis remains something of an enigma.
Fibronectin binding protein (protein F/Sfb I).
Many different pathogenic bacterial species have the ability to bind fibronectin, and the ability of GAS to bind fibronectin was established long before genes encoding the protein receptors responsible for the binding had been cloned. Fibronectin itself is a very large, multifunctional glycoprotein that is found as a disulfide-linked, soluble dimer in most body fluids or in a matrix form that is insoluble and is a major component of both basement membranes and ECM. Fibronectin has multiple binding domains that are capable of binding to a wide variety of compounds such as collagens, fibrin, heparin, and actin. It stands to reason, therefore, that GAS would use this molecule as a target for binding because it is ubiquitous throughout the human body and is especially prevalent at sites, such as the dermis, where an infection would be initiated.
Many factors have been implicated in the binding of fibronectin to the surface of GAS. Although some early studies had suggested that lipoteichoic acid is responsible for the binding of GAS to fibronectin, later studies refuted those claims. There are reports in the literature of at least five streptococcal fibronectin binding factors including Emm3 (666) and protein H (230), SDH (614), OF/SfbII (448, 658), FBP54 (133), and protein F/SfbI (303, 778). Given the rather promiscuous binding abilities of fibronectin itself, the results of any in vitro binding study must be viewed with caution until the biological relevance of such an interaction can be clearly demonstrated.
The best-characterized group of streptococcal fibronectin binding proteins are the SfbI/protein F family members. SfbI and protein F are essentially identical fibronectin binding proteins that were cloned from different strains of GAS. Southern blot analysis of several streptococcal isolates has revealed that SfbI-like proteins are present in approximately 70% of GAS strains. The gene encoding protein F, prtF, was cloned by screening a streptococcal expression library in E. coli for the expression of a protein which could inhibit the binding of radiolabeled fibronectin to the surface of GAS (303). A role for this protein in binding fibronectin has been demonstrated, since mutants generated by insertional inactivation are unable to adhere to either fibronectin or respiratory epithelial cells (303) and the exogenous addition of purified protein F/SfbI is able to inhibit the binding of fibronectin to the surface of GAS. More convincingly, expression of protein F in both nonadherent streptococci and enterococci enables these bacteria to bind fibronectin and respiratory epithelial cells (304).
The SfbI/F proteins contain a rather large N-terminal leader peptide followed by a slightly variable aromatic region of approximately 200 amino acids and a set of conserved proline-rich repeat domains (RD1). The fibronectin binding region begins immediately downstream from RD1 and consists of a stretch of 43 amino acids (UR or UFBD) followed by up to five repeat domains (RD2) which are homologous in primary sequence to other fibronectin binding proteins including that of S. aureus (720, 777, 778). Both the number of repeats and the sequence variation within the N-terminal variable region account for the heterogeneity between strains. Binding studies have demonstrated that the minimal functional unit of the RD2 domains is generated at the junction between two adjacent repeats and that the RD2 and UR domains of protein F bind different domains of the fibronectin molecule (609). A more recent study has indicated that protein F can also bind fibrinogen through the N-terminal variable domain that precedes the RD1 repeat domains (414).
C5a peptidase.
One of the terminal steps of both the alternative and classical complement cascades is the proteolytic conversion of the full-length C5 protein into the biologically active C5a and C5b molecules by the C5 convertase complex. C5b plays a role in the membrane attack complex, which is generally ineffective against gram-positive bacteria. C5a, on the other hand, is a potent chemoattractant that recruits PMNs to the site of infection (360). To inhibit the recruitment of these PMNs, the streptococci produce a protease that proteolytically inactivates C5a (845, 846).
The gene for the C5a peptidase (scpA) encodes an 1,167-residue protein with a molecular mass of approximately 128 kDa (118). A 31-residue N-terminal leader peptide and a very large N-terminal catalytic domain are followed by four short repeats that lie almost immediately upstream of the cell wall sorting signal. The enzymatic domain bears significant homology to the subtilisin family of proteases, but, unlike the broad activities displayed by many members of this class of enzymes, the C5a peptidase shows remarkable specificity by being unable to cleave the full-length C5 molecule and cleaving only a single bond within C5a (125).
Insertional inactivation of the scpA gene demonstrated that the C5a peptidase plays a role in virulence in a mouse model (386). Mutant strains were cleared more efficiently than wild-type strains were. In addition, the mutant strains were trafficked to the lymph nodes whereas the wild-type strain was found predominantly in the spleen. The C5a peptidase has been investigated as a vaccine candidate due to its widespread conservation throughout GAS (385). Genes nearly identical to scpA have also been found in both group B and G streptococci, although the expression of scpA in the group G streptococci appears to be restricted to the specific strains that are capable of infecting humans (119, 123, 124).
Serum opacity factor/SfbII.
OF is an apoproteinase responsible for the ability of some strains of GAS to cause opalescence in serum (703). As mentioned above, the expression of OF is usually restricted to GAS strains that express class II M proteins. The cloning of OF in 1995 by two independent groups showed unequivocally that OF is distinct from M proteins (448, 658).
Whereas Rakonjac et al. cloned the OF gene on the basis of its ability to cause opalescence in serum (658), Kreikmeyer et al. cloned the gene based on its ability to bind fibronectin (448) and hence named the protein SfbII. The OF/SfbII protein itself is very large (1,025 residues) and contains an N-terminal leader peptide and a C-terminal cell wall sorting signal with a seryl residue substituted instead of the threonyl of the LPXTG motif. A domain repeated 2.5 times near the C terminus of the protein is homologous to the RD2 repeats from SfbI/protein F, and purified fusion proteins containing these domains are capable of competitively inhibiting the binding of radiolabeled fibronectin to streptococcal cells. Both groups noted that the gene is present only in a subset of streptococcal isolates, which correlates with what was previously known about OF (see above).
T antigen.
T antigens form a group of approximately 25 serologically distinct surface molecules found on the surface of group A, B, C, and G streptococci that have the common property of being resistant to digestion with trypsin (437, 469). Antibodies against T antigens are not protective, and therefore very few studies on the biological relevance of these molecules have been carried out. Although T antigens are expressed independently of M antigens and more than one T antigen can be expressed simultaneously on the same strain, most M types are associated with a single T type (389). T antigens have been used as a means of subclassifying different strains of GAS and are used in cases where M antigen, due to either a lack of available typing sera or a lack of expression, cannot be used. Although antibodies to T antigen are generated during the course of an infection, these antibodies are not protective against subsequent infections.
Although two T antigens have been purified (390, 502), to date the nucleotide sequence of only one T-antigen gene, tee6 from the M6 serotype strain D471, has been determined (714). The tee6 gene encodes a 55-kDa polypeptide of 537 amino acids (T6) that possesses a C-terminal cell wall sorting signal. The protein is rich in serine, threonine, and lysine residues and is rare among the streptococcal surface proteins in containing no discernible repeat domains. DNA hybridization studies demonstrated that the 5′ end of the tee6 gene hybridized with chromosomal DNA of only 10 of the 25 known T types, and probes constructed from the 3′ end of tee6 hybridized with even fewer isolates (397). Indeed, chromosomal DNA from several T-type isolates failed to hybridize with any tee6 probe. These findings leave open the possibility that the T antigens are actually several different molecules that have in common only their ability to resist trypsin digestion.
Streptococcus agalactiae (Group B Streptococci)
Group B streptococci (GBS or S. agalactiae) are responsible for the majority of cases of neonatal sepsis and meningitis in developed countries (25). GBS are frequently present in the vaginal flora of humans and can be transmitted to the neonate during or before birth. GBS can also cause invasive infections in adults, particularly the immunocompromised and the elderly (195). GBS can be subclassified immunologically according to their polysaccharide capsules, which fall into at least nine distinct types (436, 842). A majority of the invasive GBS infections are due to strains expressing the type III capsular polysaccharide, although strains expressing other capsular types are increasingly gaining in importance (66). The GBS can also be grouped according to the expression of a set of surface proteins designated Cα (alpha), Cβ, and Rib (202). In experiments with animal models, it has been determined that antibodies against either the polysaccharide capsule or the surface proteins are protective against infection with GBS (52, 685, 751, 843). Capsular polysaccharides can be poorly immunogenic, however, and this has led many laboratories to focus on the use of surface proteins as possible vaccines (471, 842). Unfortunately, research on the GBS surface proteins has focused primarily on their vaccine potential and relatively little has been determined about the role of these proteins in virulence.
Cα (alpha, alpha C) and Rib.
Cα and Rib are homologous trypsin-resistant proteins found on the surface of the GBS. Rib is almost always found on the more invasive GBS strains of capsular type III, whereas Cα is usually expressed on strains expressing other capsular types (220, 751). Recently another Cα-like protein has been identified on a type V strain, but the sequence of this protein has not yet been published (464, 465). Rarely, if ever, are Cα and Rib expressed together on the same cell (220). The genes encoding Cα (bca) and Rib (rib) were sequenced from the type Ia GBS strain A909 and the type III strain BM110, respectively (550, 837). The two proteins have a similar organization with unusually long leader peptides followed by N-terminal domains of ≈170 amino acids that are 61% identical to each other. The N-terminal domains are followed by set of 9 (Cα) or 12 (Rib) tandem repeat domains that in both cases are followed by a classical C-terminal cell wall sorting signal. The repeat domains of Cα and Rib contain 82 and 79 amino acids, respectively, and are 47% identical to one another. Strikingly, there is no variation between individual repeats in either protein, even at the nucleotide level. In both cases the repeat domains constitute greater than 70% of the total length of the protein.
Both Cα and Rib proteins are capable of eliciting protective immunity against GBS, but immunity against one does not confer immunity against GBS strains expressing the other even though the two proteins are highly homologous (471, 751). Since almost all strains of GBS express either alpha or Rib, a vaccine composed of both proteins should, in theory, provide broad-range immunity against these bacteria. The number of repeat domains among Cα proteins of different clinical isolates of GBS can vary significantly but averages between 8 and 9 (509). It is apparent that the repeat domains in both Cα and Rib are subject to insertions and deletions due to homologous recombination. There is some evidence that strains of GBS expressing Cα antigens with fewer repeat domains are less susceptible to the host immune response than are those that express Cα antigens containing larger numbers of repeats (280, 510). Conversely, it has been shown that Cα proteins with fewer repeat domains are more immunogenic (278, 279). It would therefore appear that the selective advantage conferred by Cα molecules containing fewer repeats is counterbalanced by their ability to generate a more vigorous immune response.
The role of Rib and Cα in the pathogenesis of GBS is not understood; however, a recent experiment has shown that a mutant GBS strain in which the bca gene was insertionally inactivated is attenuated in its ability to cause disease in mice (483). More studies are necessary to determine the exact role of these proteins in virulence.
Cβ (beta antigen, Bac, IgA binding protein).
The Cβ antigen is found on less than half of GBS strains, and many of these strains appear to express a truncated, secreted form of the protein (81, 512). The role of Cβ in virulence is unclear, but the protein has a high affinity for serum IgA and, under some conditions, secretory IgA (52, 81, 487, 691). Immunization of pregnant mice with purified Cβ can confer resistance against infection with GBS onto neonatal pups (511). The gene encoding Cβ (bac) was cloned from GBS strains SB35 (319) and A909 (382) by two independent laboratories. Both genes possess a 37-residue leader peptide and a classical C-terminal cell wall sorting signal. Approximately 180 residues upstream of the sorting signal is an unusual domain composed almost entirely of a triplet repeat, in which the first residue is a proline, the second residue alternates between a positively and a negatively charged amino acid, and the third residue is uncharged. The two genes differ slightly in the proline-rich repeat region, with that from the SB35 strain containing two short deletions. Analysis of recombinant Cβ expressed in E. coli indicated that the Cβ protein contains two or more separate IgA binding domains within the N-terminal half of the molecule, designated A and B (382). A later study mapped the binding site of the A domain to a region centered around a hexapeptide motif (MLKKIE), but it was not conclusively determined that this motif actually participated in the binding of IgA (383). Neither IgA binding domain on Cβ bears any significant homology to the IgA binding domains found in M proteins (50, 391). The Cβ typing antisera appear to recognize a separate domain that lies C-terminal to the IgA binding domain(s) (319).
Group C and G Streptococci
Streptococci in Lancefield groups C and G (GCS and GGS) comprise a group of closely related species that are capable of causing disease in both humans and a wide variety of animals. The large-colony forms of GCS, including the alpha-hemolytic S. dysgalactiae and beta-hemolytic S. equi (subspp. equi, equisimilis, and zooepidemicus), are common veterinary pathogens. Less often, these strains will infect humans, with S. equi subsp. equisimilis (S. equisimilis) being the most frequently isolated along with the small-colony form, S. anginosus (56). GGS are generally not given a species designation and are also associated with disease in both humans and animals.
M protein.
Clinical isolates of GCS and GGS can possess antiphagocytic M proteins similar to those found in GAS (56, 57, 129, 395, 745). However, several studies have suggested that the expression of such proteins is limited to the specific strains that can infect humans (555, 717, 731). As mentioned previously, the GGS and GAS appear to be able to share their M protein genes by horizontal transfer (730, 750). A recent study of 38 human GGS isolates revealed that each possessed a single class I emm gene that could be divided into one of 14 distinct types based on the N-terminal sequence of its protein (717). The sequences of some of these proteins was similar to known N-terminal sequences from GAS M proteins, whereas others were found to be quite divergent.
SzP and SeM.
S. equi and S. zooepidemicus frequently colonize the nasopharynx and genital mucosa of horses. S. equi is the causative agent of strangles, a highly contagious disease of horses, whereas S. zooepidemicus is a normal mucosal commensal organism that can cause disease in an opportunistic fashion. Recent genetic studies have indicated that S. equi (also known as S. equi subsp. equi) is actually a variant strain of the more genetically diverse S. zooepidemicus. Strains of S. zooepidemicus express an acid-extractable surface antigen (SzP) that, like the GAS M proteins, is antigenically variable among strains. In contrast, S. equi appears to express two invariant surface proteins, one of which is related to SzP while the other appears to be unique to this strain (SeM).
The genes encoding the SzP protein have been sequenced from S. equi (SzPSe) and S. zooepidemicus W60 (SzPW60), as has the gene encoding SeM (784, 785). Both SzP and SeM bind equine fibrinogen, and it appears that these proteins can limit the deposition of equine C3 on the bacterial surface (72, 73, 784). Antibodies against SzP are opsonigenic against S. zooepidemicus (but not against S. equi), whereas S. equi is opsonized by antibodies against SeM (784). For these and other reasons, the SzP and SeM proteins were designated “M-like.” However, despite their functional similarities and the presence of an N-terminal leader peptide and C-terminal cell wall sorting signal, SeM and SzP do not bear significant homology to each other or to the M proteins of GAS. SzPSe and SzPW60 are 85% homologous to one another and do not possess large tandem repeats; however, the proline-rich region of each is composed of several copies of a 4-residue repeat, PEPK. SeM contains two tandem repeats of 21 residues each and several shorter C-terminal tandem repeats. No specific functions have yet been ascribed to any domain of either SeM or SzP.
Protein G and related IgG binding proteins (protein G, MIG, MAG, ZAG).
As mentioned above, Myhre and Kronvall, in 1977, classified the IgG binding activities of the gram-positive bacteria, giving the distinction “type III” to proteins with binding properties similar to those found on the GCS and GGS (569). Shortly thereafter, Myhre and Kronvall also identified an albumin-binding activity on the surface of the GCS and GGS (569a). It has since become apparent that a single protein, protein G, confers both the IgG and albumin binding properties on these bacteria. Although possessing multiple ligand binding properties similar to those found in some GAS M proteins, protein G and related proteins are structurally distinct from the M proteins. Over the past decade, protein G-like molecules have been identified in several group C and G species of human and bovine origin as well as in S. dysgalactiae (MIG and MAG) (398, 399, 401), S. zooepidemicus (ZAG) (400), and the distantly related anaerobe Peptostreptococcus magnus (see below).
Protein G was first isolated based on its IgG binding activity from the group G strain G148 (60) and from the group C strain C26RP66 (667, 668). The albumin binding properties of this molecule were not described until the gene had been isolated and expressed in E. coli (59). Genes encoding protein G have been sequenced from strains G148 (290, 603), GX7805 (204), GX7809 (194), G43, and the group C strain C43 (734), allowing a detailed comparison of their structures. Unlike the GAS M proteins, the binding activities of the protein G family are contained within distinct modular domains, whose number can vary from strain to strain. The globular structures of these domains are more reminiscent of staphylococcal protein A (see below) than the extended alpha-helical coiled-coil structure of the M proteins.
The binding of albumin and that of IgG occur within separate domains of protein G, each encoded by a set of tandem repeats (6, 735). The modular albumin binding domains (GA modules) are contained within the N-terminal region of protein G, whereas the IgG binding domains are encoded in a more C-terminal region of the molecule. The structures of both domains have been determined (286, 387, 388). GA modules are found in a wide variety of streptococcal and peptostreptococcal albumin binding proteins; however, these domains are not present in all protein G molecules from human clinical isolates (388, 733, 734). Protein G also binds the protease inhibitor α2-macroglobulin (α2M), but this binding is inhibited by IgG and it is unclear whether it is physiologically relevant (736).
MIG and MAG are surface proteins cloned from two different strains of S. dysgalactiae, whereas ZAG was isolated from a strain of S. zooepidemicus (399–401). Earlier studies had suggested that S. zooepidemicus possessed a unique type (type V) of IgG binding protein; however, sequence analysis revealed that MIG, MAG, and ZAG are similar to type III protein G. Like protein G, MAG and ZAG bind albumin (MIG does not), and all three of these proteins have been shown to bind α2M. The binding of α2M occurs through an N-terminal domain unique to MIG, MAG, and ZAG that is not found in protein G.
Fibronectin binding proteins.
Fibronectin binding proteins have been cloned from the GCS strains S. dysgalactiae, S. equisimilis, and S. zooepidemicus (490–492). S. dysgalactiae has two adjacent genes, fnbA and fnbB, that encode fibronectin binding proteins (490). Unlike the two fibronectin binding proteins from S. aureus, however, FnbA and FnbB are quite dissimilar and are most probably not the product of gene duplication. In fact, the C-terminal fibronectin binding domains of FnbA and FnbB each have greater homology to protein F than they do to one another. The reasons why this bacteria contains two proteins with this activity are unclear, although under laboratory conditions only FnbB appears to be expressed (490). The C-terminal domain of FnbB is also highly homologous to the fibronectin binding protein of S. equisimilis (491).
The fibronectin binding protein of S. zooepidemicus, FNZ, is structurally similar to protein F, including the presence of nearly identical fibronectin repeat domains and the presence of an UR-like sequence. The UR domain of FNZ, however, lies between the first and second fibronectin binding repeats rather than immediately upstream as is found in protein F. FNZ, at ≈60 kDa, is rather small in comparison to the rather large (>100-kDa) fibronectin binding proteins of S. dysgalactiae and S. equisimilis (492).
Streptococcus pneumoniae
The pneumococci are one of the leading causes of septicemia, meningitis, and lower respiratory tract infections in humans. Despite rapid and aggressive antibiotic therapy, pneumococcal infections can lead to a variety of debilitating complications including hearing loss. The polysaccharide capsule expressed by these bacteria is required for virulence but in itself is not toxic. Although proteins are obviously important, their role in the pathogenicity of these bacteria has not been extensively studied (620). S. pneumoniae produces two hemolysins, one of which, the well-studied pneumolysin, has been clearly demonstrated to contribute to the disease process (38, 109, 620, 663). In addition, the pneumococci produce two adhesins, PspA and PsaA, which may also contribute to the ability of these bacteria to establish an infection within a host (48, 875). Only two cell wall-anchored enzymes, a neuraminidase and a β-N-acetylglycosaminidase, have thus far been cloned in S. pneumoniae. Both of these glycosidases probably play a role in virulence as well.
Neuraminidase.
It has long been appreciated that all clinical isolates of S. pneumoniae possess neuraminidase activity (425, 608). Neuraminidases, found in a wide variety of pathogenic microbes, cleave terminal sialic acid residues from a variety of glycosylated proteins, lipids, and oligosaccharides found both in body fluids and on the surfaces of cells. Neuraminidase activity correlates with virulence in pneumococcal meningitis, and early studies with crudely purified pneumococcal neuraminidase demonstrated its toxicity in mice (426, 608). At least two genes encode neuraminidases in the pneumococci (47, 108). One gene, nanB, encodes a putative secreted protein, while a gene 4.5 kb upstream, nanA, encodes a protein with the typical characteristics of a gram-positive surface proteins (47, 107). NanA and NanB have no significant homology to one another and appear to have optimal activities at different pH values, suggesting that they may act at different sites or stages during an infection (47). NanA is >100 kDa and has four copies of a consensus bacterial neuraminidase sequence motif (SXDXGXTW), suggesting that it belongs to the “large” family of bacterial neuraminidases (107, 675). The surface localization of NanA has been confirmed by immunoelectron microscopy (107).
The exact role(s) that the neuraminidases play in virulence is unclear, although it has been suggested that removal of sialic acid from surface molecules can “unmask” carbohydrate ligands for pneumomcoccal adhesion (449, 489). A recent experiment demonstrated that a strain of S. pneumoniae with an interrupted nanA gene was not defective in its ability to cause cochlear damage in an experimental guinea pig meningitis model whereas strains deficient in pneumolysin were avirulent (856). These strains, however, still possessed an intact nanB gene, although the authors claimed that the activity from this gene was minimal.
β-N-Acetylhexoseaminidase.
The gene for the pneumococcal β-N-acetylglucosaminidase (strH) was cloned from a phage lambda expression library from S. pneumoniae by screening for plaques with enzymatic activity (122). StrH is very large (144 kDa) and possesses both an N-terminal leader peptide and a typical C-terminal cell wall sorting signal. The protein also contains two rather large (335-residue) tandem repeat domains, which account for approximately half of the mature protein. The repeats, separated by approximately 100 residues, each contain a 30-residue sequence motif that is homologous to conserved sequences found in other bacterial hexosaminidases. Host tissues are abundant in surface molecules containing GlcNAcβ1-4-linked residues, a substrate for the enzyme, but the role of the StrH protein in the biology of S. pneumoniae is unclear.
Oral Streptococci (Viridans Group)
The oral cavity is typically host to several species of gram-positive bacteria, most notably the viridans group streptococci. Important among the viridans group are S. mutans and S. sobrinus, which have been implicated as primary causes of dental caries (376, 497). Colonization of the oral cavity requires that the bacteria bind to salivary components, to other bacteria, and to both soluble (dextran) and insoluble (mutan) extracellular polysaccharides that are synthesized by the bacteria themselves. Surface hydrophobicity has been speculated to play a role in the ability of bacteria to adhere to the oral pellicle, and surface proteins have been implicated in contributing to overall surface hydrophobicity.
Surface proteins from these organisms have been widely studied for their role in oral colonization as well as their potential as vaccine targets. At least 20 polypeptides are exposed on the cell surface of S. gordonii and S. sanguis (14, 377). Sequence analysis of several of these surface proteins has revealed that viridans streptococci use similar adhesins to attach to both host tissue components and other bacteria (160).
Antigen I/II (P1 protein, B antigen, PAc, IF, sr, SpaA).
The streptococcal antigen I/II proteins have been found on the surface of almost every oral streptococcal species studied thus far. Antigen I/II (688) of S. mutans serotype c has also previously been designated B antigen (689), IF (359), P1 (221), MSL-1 (150), and PAc (598). Genes homologous to antigen I/II have been cloned and sequenced in S. mutans serotype f (sr protein; [1]), S. gordonii (SspA and SspB [149]), and S. sobrinus (SpaA [94] and pag).
These proteins are almost identical and have both N-terminal leader peptides and C-terminal cell wall sorting signals. They are large (1,500 to 1,566 amino acid residues) and, like many other surface proteins, contain multiple binding activities and repeat domains (378). I/II antigens have been reported to be able to bind salivary glycoproteins as well as other oral microbes. The structure and function of this group of proteins have been the subject of several recent reviews (375, 376, 378).
The surface localization and cell wall attachment of P1 to the streptococcal cell wall has recently been demonstrated (353, 565, 566). A mutation was isolated in S. mutans GS-5 that resulted in the complete loss of surface attachment of P1 (565). Sequencing of the GS-5 variant revealed a frameshift mutation that resulted in the production of a I/II variant which was lacking the C-terminal end. Homonylo-McGavin and Lee found that antigen I/II of S. mutans could be localized to the surface of S. mutans, S. gordonii, and Enterococcus faecalis only if the C-terminal domain was intact (353). In addition, antigen I/II remained associated with the cell after boiling in SDS only if the cell wall sorting signal was present. This was confirmed in a later study in which it was shown that removal of the charged tail led to the complete solubilization of antigen I/II from the cell surface (566).
CshA of S. gordonii.
CshA is a very large (≈290-kDa, 2,508-amino-acid) surface protein which was identified in S. gordonii (538, 539). Cells defective in the expression of this protein have decreased cell surface hydrophobicity and a defect in their ability to coaggregate with oral actinomycetes (538, 539) and other bacteria (537), suggesting that this protein is probably necessary for colonization and adherence to the oral pellicle. Indeed, cshA mutants are defective in their ability to colonize the oral cavity of mice (539).
Like many other surface proteins, much of CshA is composed of repeating domains (539). The N-terminal domain of the mature protein (amino acids 42 to 878) is nonrepetitive, whereas the central portion of the protein (amino acids 879 to 2417) is composed of 16 repeat regions of ≈100 amino acids each. All of the binding regions thus far identified in CshA have been mapped by antibody inhibition studies to the N-terminal nonrepetitive domain (537). The C terminus of CshA is necessary for the retention of the polypeptide on the cell surface, since C-terminal mutations which truncate CshA to a protein of ≈260 kDa result in the complete secretion of CshA into the growth medium and a subsequent loss of surface hydrophobicity (538).
β-d-Fructosidase (FruA) of S. mutans.
Oral streptococci produce polysaccharides that have a short half-life in dental plaque and are thought to be used for short-term sugar storage. These polymers are synthesized by a set of glucosyl-transferases (see below). Unlike the insoluble mutan polysaccharides, which are composed of α1-3 linkages and are unable to be enzymatically degraded, the storage polysaccharides are composed of glycosidic bonds that are susceptible to enzymatic lysis. β-d-Fructosidase (FruA) is a surface-associated enzyme capable of liberating fructose through the degradation of levans as well as sucrose, raffinose, and inulins. The fruA gene was cloned and sequenced from S. mutans, but Northern blot analysis suggests that homologues are likely to exist in all of the mutans streptococci (103). FruA is notable in having almost no proline or glycine residues in the area of the protein predicted to span the cell wall.
Interestingly, a study of S. mutans grown in a chemostat demonstrated that over 95% of the fructosidase activity was located in the supernatant (105) whereas a vast majority of the activity was cell associated in batch culture (103). An investigation into this phenomenon revealed that most of the fructosidase can be found in the supernatant when the bacterium is grown at a pH of 7.0, whereas approximately half is cell surface bound when the organism is grown at a pH of 5.0. The study also demonstrated that the release of fructosidase into the extracellular media can be inhibited by copper, a phenomenon also observed for the release of antigen I/II (104).
Dextranases of S. mutans and S. sobrinus.
Cell surface dextranases cleave glycosidic bonds and are thought to be involved both in the metabolic utilization of sugar and in controlling the amount and content of extracellular polymerized glucans (126, 249). The streptococcal dextranases degrade dextran into isomaltosaccharides (362). Genes encoding extracellular dextranases have been cloned from S. mutans (363), S. sobrinus (827), S. salivarius (594), and S. suis (723), although only the dextranases from S. mutans and S. sobrinus possess C-terminal cell wall sorting signals. The S. mutans and S. sobrinus dextranases are 47% homologous to one another, although the S. sobrinus enzyme is somewhat larger. In addition, the two enzymes have similar enzymatic properties and pH optima, indicating functional similarity (827). Inactivation of the dextranase gene of S. mutans appears to increase the ability of the bacteria to adhere to smooth surfaces but has no effect on sucrose-dependent cell-cell aggregation (126).
Glucan binding protein of S. mutans.
Under conditions of stress, certain strains of S. mutans express a glucan binding protein that enables these cells to aggregate in the presence of dextran (704). The gene for this protein, gbpC, has recently been cloned and has been shown to encode a ≈60-kDa protein that contains both an N-terminal leader peptide and classic C-terminal cell wall sorting signal (704). The glucan binding activity of GbpC has been demonstrated in extracts of E. coli which harbor a plasmid encoding the gene (704). GbpC has limited homology to streptococcal antigen I/II, and the protein cross-reacts with anti-antigen I/II antisera. More work is required to determine why there appear to be several glucan binding proteins in S. mutans. The only other glucan binding protein to be sequenced thus far has no C-terminal cell wall sorting signal (26).
WapA (protein antigen III) of S. mutans.
Wall-associated protein antigen A (WapA [201, 689]) and antigen III (686) were recently shown to be identical (687). WapA is a 45-kDa surface antigen of S. mutans, which has been implicated in helping the bacteria to bind smooth surfaces and to undergo sucrose-dependent aggregation (655), although this is a matter of some dispute (307, 687). Also disputed is its ability to serve as an effective vaccine against dental caries (687).
Fimbriae of Actinomyces
The actinomycetes, including Actinomyces naeslundii and, perhaps, A. odontolyticus, are some of the most common bacteria in the oral flora. Like the oral streptococci, these bacteria are involved in the early stages of plaque development and have the ability to bind both the tooth surface and other oral bacteria (441, 484). A. naeslundii binds to the tooth surface via a fimbrial structure, designated type 1, which has affinity for proline-rich proteins that coat the tooth enamel (121, 257). The adherence of actinomycetes to eukaryotic host cells and other bacteria, including several streptococcal species, is directed by a separate set of fimbriae designated type 2. Type 2 fimbriae appear to utilize a lectin-like activity to bind to the surface of target cells, and this binding can often be disrupted by high concentrations of sugars such as lactose (298, 440).
Strains of A. naeslundii may express either one or both types of fimbriae on their surface. One strain, T14V, expresses both types of fimbriae and has been used as a model system for the study of these structures in detail at the molecular level (120). The gene encoding the type 1 fimbrial subunit (fimP) of strain T14V has been cloned and sequenced, as has the gene encoding the type 2 fimbrial subunits (fimA) of T14V and another strain, WVU45 (867, 868). All three cloned subunit genes encode proteins of ≈54 to 59 kDa that have typical N-terminal leader peptides as well as C-terminal cell wall sorting signals. The type 2 subunit from T14V has a very high degree of homology (≈65% identity) to the type 2 subunit from WVU45 but only moderate homology (≈31% identity) to the type 1 subunit from T14V (868, 869).
The presence of the cell wall sorting signals is somewhat unexpected, and their possible role in fimbrial assembly is unclear. Accessory genes that appear to be involved in the assembly of intact fimbriae from individual subunits have been identified for both the type 1 and 2 fimbriae from strain T14V, but no pathway for assembly has been determined and specific roles for these genes have not yet been determined (869, 870). A clue to the role of the C-terminal cell wall sorting signal may be revealed by the fact that it is not possible to dissociate assembled fimbriae into their individual monomer subunits, suggesting that they may be covalently linked to one another. Additionally, antibodies raised to the C-terminal domain of a recombinant monomer subunit are unable to recognize the intact fimbriae, suggesting that the C-terminal domain may be proteolytically removed (869). This evidence, although far from conclusive, has led Yeung et al. to suggest that perhaps the C-terminal sorting signal directs the covalent multimerization of the fimbrial subunits (869). If this were true, it would represent a novel use of the sorting machinery to synthesize a complex fimbrial organelle. Obviously the verification of this model will require the structural characterization of both free and complex-bound fimbrial subunits.
Protein L and PAB of Peptostreptococcus magnus
The gram-positive anaerobe Peptostreptococcus magnus is a common human commensal organism that can also cause a variety of diseases including vaginosis and urethritis. Approximately 10% of clinical isolates express a unique Ig receptor, protein L (PPL), that has the unusual ability to bind the variable portions of Ig kappa light chains (4, 58, 188, 587, 855). This unusual binding specificity confers upon PPL the ability to bind antibodies of almost any subclass, and therefore this molecule has generated much interest as a tool for purifying and studying Ig. The gene encoding PPL has been cloned and sequenced from P. magnus 312 (413) and 3316 (567). A comparison of their structures reveals that these proteins have a modular design similar to that found for protein G from the GGS. PPL3316 and PPL312 possess four and five tandem repeat domains, respectively, that encode the Ig light-chain binding modules, whose three-dimensional structure has recently been determined (854). The two proteins also possess common tandem repeat domains of unknown function. PPL3316, but not PPL312, possesses four copies of the albumin binding GA modules similar to those found in protein G, and, as expected, strain 3316 binds albumin whereas strain 312 does not (413, 567).
A physiological role for the light-chain binding function of PPL has been proposed. Addition of strain 312 to a culture of human basophils was found to stimulate the release of histamines, whereas no such stimulation was observed upon the addition of a strain that lacks PPL (619). A similar effect was observed upon the addition of purified recombinant PPL to the cultures, presumably due to cross-linking of the membrane-bound IgE receptors present on these types of host cells.
Approximately half of P. magnus strains that do not possess PPL bind albumin through a receptor known as PAB. The gene encoding PAB has been sequenced from strain ALB8 and was found to encode a 43-kDa protein that, like protein G and PPL, is strikingly modular in its design (141). Although PAB lacks the kappa light-chain binding domains found in PPL, it does have many similarities to PPL, including the presence of a “GA module” as well as a sequence homologous to one of the repeats of unknown function from PPL312. The structural similarities between PAB and PPL seem to indicate that these two proteins are actually derived from a common ancestral molecule that evolved through the shuffling of the relevant binding modules (140). Indeed, a recent study has identified short nucleotide sequences within PPL and PAB that appear to facilitate the shuffling of these modular domains in a manner similar to the swapping of exons in eukaryotic cells (140).
Enterococci
Enterococci possess a unique and highly efficient mechanism for the conjugal transfer of plasmid DNA (166, 178). Expression of this transfer system in donor cells is induced by an oligopeptide mating pheromone that is secreted by recipient cells (167). Induction results in the increased expression of surface protein (aggregation substance [AS]) on the donor that acts as an adhesin specific for the surface of recipient cells (239). An additional surface protein (surface exclusion protein) is involved in preventing donor cells from conjugating with identical donors (170). The pheromone-induced expression of aggregation protein appears to be controlled by a novel posttranscriptional mechanism (36, 37). The genetics of this system has recently been reviewed in detail (168, 169). A similar conjugative plasmid transfer system has recently been identified and sequenced in the Enterococcus faecium plasmid pHKK701 (315).
Aggregation substance.
The genes encoding AS from three conjugal plasmids, pCF10 (411), pAD1 (237), and pPD1 (236), have been sequenced, and it is believed that almost all enterococcal conjugal plasmids encode similar aggregation proteins (238, 339, 857). The three known AS proteins are all large (≈145-kDa) proteins that have significant sequence homology to each other and possess a typical C-terminal cell wall sorting signal as well as a proline-rich domain. The domain structure of these proteins is similar to that found in both the antigen I/II proteins of the viridans streptococci and the CluA protein of L. lactis, consistent with their roles as mediators of bacterial aggregation. The ligand for AS, designated enterococcal binding substance (EBS), is unidentified to date. The inactivation of several unlinked genes is required to obtain an EBS-negative phenotype, and it is speculated that lipoteichoic acid may be a component (711). The expression of both AS and EBS has been suggested to play a role in virulence in addition to their role in conjugal transfer of DNA (602, 711).
Surface exclusion protein.
As mentioned above, surface exclusion proteins reduce the probability of mating between enterococci carrying identical plasmids, although the exact mechanism underlying this phenotype is unclear. The surface exclusion proteins of the plasmids of pAD1 (840) and pCF10 (411) have been sequenced. Both genes encode homologous 95-kDa proteins that have both N-terminal leader peptides and C-terminal cell wall sorting signals. The gene encoding the surface exclusion protein of plasmid pPD1 has also been sequenced and has been found to contain an insertion sequence that results in a truncated protein without a C-terminal cell wall sorting signal (339). Cells containing plasmid pPD1 lack the surface exclusion phenotype, strongly suggesting that cell wall anchoring is required for the surface exclusion phenotype (339).
Lactococci
CluA.
Not unlike their close relatives the enterococci, certain strains of lactococci are able to exchange genetic information through conjugal mating. Recently a gene encoding the aggregation factor was cloned from Lactococcus lactis subsp. cremoris (271). The gene encodes a polypeptide of 1,243 residues that has a modular structure similar to other surface proteins. Sequence analysis revealed homology to the aggregation proteins of E. faecalis. Three central domains have homology to both the E. faecalis domains and the antigen I/II proteins from the viridans streptococci.
NisP.
Lantibiotics are ribosomally synthesized antimicrobial oligopeptides that are produced by several gram-positive species. These (β-methyl)lanthionine-containing peptides display activity primarily against other gram-positive bacteria and have been the subject of considerable interest because of their ability to inhibit food spoilage (147). Nisin, produced by L. lactis, is one of the best characterized of the lantibiotics. The genetics and modification of nisin and other lantibiotics have recently been the subject of several reviews (154, 190, 373, 693).
Nisin is synthesized as a 57-amino-acid precursor peptide in the cytoplasm, at which point it undergoes a series of posttranslational modifications before being exported from the cell. The N-terminal 23 amino acids of pre-nisin serve as an atypical leader peptide that assists in the secretion of pre-nisin from the cytoplasm by a putative ATP binding cassette transporter (186, 451). The exported nisin pre-peptide is inactive due to the presence of the N-terminal leader sequence but is converted to an active form upon removal of its leader peptide by a surface-located serine protease (product of the nisP gene). NisP has also been implicated in providing immunity to nisin, although the mechanism by which this would occur is unclear (866).
Sequence analysis of NisP has revealed that it contains both an N-terminal leader sequence and C-terminal cell wall sorting signal (810). Neither the surface localization nor attachment of NisP to the peptidoglycan has been verified experimentally. It is unclear why such an attachment would be necessary, given that functionally homologous lantibiotic-peptidases from other species do not have a cell wall sorting signal in their sequence (154).
PrtP.
During growth in milk, the primary source of amino acids for the lactococci are the milk proteins (caseins), since the supply of free amino acids in milk is relatively limited (405). The caseins are initially broken down by the bacterium into small peptides, which are subsequently taken up by an oligopeptide permease system (453). The imported peptides can then be further degraded into individual amino acids by a series of intracellular proteases. The complete protease system of L. lactis has recently been reviewed (453).
The casein degradation pathway is initiated by a cell wall-bound serine protease called PrtP or CEP (192, 744). The prtP gene has been cloned and sequenced in a number of L. lactis strains (155, 429, 439), and each displays >95% homology to the other prtP genes. Sequence analysis has revealed that the proteins encoded by prtP are approximately 200 kDa and contain both an N-terminal leader peptide and a C-terminal cell wall sorting signal. The substrate specificity of the PrtP proteinase varies from strain to strain (192) but generally falls into one of two classes, designated PI and PIII, depending on the specific casein that the enzyme preferentially degrades (453). It has been demonstrated that PrtP is synthesized as a preproenzyme and, after being exported from the cytosol, is activated by the action of another cell surface protease, PrtM (293, 821), which is a lipoprotein (294). The attachment of PrtP to the cell wall and the role of the C terminus had been suggested several years before the cell wall sorting mechanism had been elucidated for protein A in S. aureus (155, 821).
Interestingly, a distantly related gram-positive bacterium, Lactobacillus paracasei, has a surface protein (PepP) which is homologous to the lactococcal PrtP. The putative C-terminal cell wall sorting sequence is unusual in that the ultimate glycine of the consensus motif has been replaced with an alanine (LPKTA) (343). It remains to be proven whether this sorting signal is functional.
Listeria monocytogenes (InlA, InlC2, InlD to InlF)
Listeria monocytogenes is an invasive intracellular pathogen that can infect animal hosts from within the intestinal lumen. Once inside the host, these bacteria invade eukaryotic cells and replicate intracellularly, thereby escaping clearance by the humoral immune response (131). The ability of L. monocytogenes to enter eukaryotic cells has been traced to a family of surface and secreted proteins, the internalins. Thus far, seven members of the internalin family have been found to exist (InlA to InlC, InlC2, and InlD to InlF) and all have been found to share certain structural features (162, 235). All feature a N-terminal leucine-rich-repeat domain (LRR) followed by a conserved inter-repeat region (IR) (Fig. 15). InlA, InlC2, and InlD to InlF possess C-terminal cell wall sorting sequences at their C termini; InlB is targeted to the cell envelope (see below) (82); and InlC (also called IrpA) is secreted (159, 185, 493). It has been demonstrated that the LRRs and IR of InlA are sufficient to confer an invasive phenotype when expressed on the surface of latex beads or the noninvasive bacteria L. innocua or E. faecalis (475). Similar findings have recently been made for InlB (83, 83a).
InlA binds to E-cadherin, and it has been shown that this interaction is critical for internalin-mediated invasion of epithelial cell lines (543). It has also been demonstrated the LRR and IR are both necessary and sufficient for this interaction (475). No ligand has been identified for any of the other internalin family members. InlA is one of the few proteins for which a covalent linkage to the cell wall has been demonstrated (474).
The purpose of multiple internalin proteins is unclear, but it has been hypothesized that they confer tropism toward different cell types. For example, it has been suggested that InlB is critical for the invasion of Listeria into hepatocytes as well as several other cell types (161, 368). This result is disputed by in vivo experiments in which it was determined that InlB deletion mutants are able to invade hepatocytes (283) but are unable to escape the phagocytic vacuole and replicate intracellularly (284). Indeed, the precise role of any of the internalins in vivo is not yet entirely clear (131).
Staphylococci
Staphylococci are common, highly virulent nosocomial pathogens that are receiving widespread attention due to the appearance of strains that are resistant to all useful antibiotics. Primary to their ability to establish infection in the hospital setting is their ability to bind biomaterial deposited on catheters and other foreign bodies (226). With the exception of protein A, every staphylococcal wall-anchored surface protein thus far characterized is an MSCRAMM. The regulation of surface proteins in S. aureus is widely believed to be under the control of the agr virulence regulon, which induces the expression of surface proteins during growth at low cell density (384, 591, 665). However, a recent study of the collagen adhesion protein suggests that it is under the control of another, overlapping regulatory system, sar (262).
It has been reported that the staphylococcal epidermal cell differentiation inhibitor (EDIN) (763) has a cell wall sorting signal; however, this report appears to be incorrect (215). The C-terminal domain of the protein contains a sequence, LPRGT, which is similar but not identical to the consensus LPXTG motif, and has a tail containing two lysine residues (366). EDIN, however, does not possess a significantly hydrophobic stretch of residues at its C-terminal end. In addition, the protein is found primarily in culture supernatants, and amino acid analysis demonstrates that EDIN is not processed at its C-terminal end, thereby indicating that this protein is not anchored to the cell wall (366, 763).
Protein A.
Staphylococcal protein A was the first nonimmune Ig binding protein identified (222). Since its discovery, it has become the best studied and most widely used surface protein of gram-positive bacteria. Early studies established the localization of protein A to the cell wall and even suggested that the protein was covalently attached to the staphylococcal peptidoglycan. The sequence of protein A was determined in 1983 and 1984; however, it should be noted that the earlier publications included a sequencing error at the 3′ end of the gene that eliminated the proper translation of the coding sequence for the cell wall sorting signal (291, 803). The IgG binding properties of protein A are due to the presence of five (in some strains, four) almost identical globular domains that bind the Fc regions of IgG1, IgG2, and IgG4. The three-dimensional structure of a protein A-antibody complex revealed that the protein A-Ig interaction occurs at the CH2 and CH3 domains of the Fc fragment (143). Despite all that is known about the structure and binding properties of protein A, almost nothing is known about its exact role in virulence. Studies on protein A mutants in animal model systems have yielded conflicting results, suggesting that other proteins may play a greater role in staphylococcal pathogenesis than protein A (226, 618).
Collagen adhesion protein of S. aureus.
The gene encoding the collagen adhesion protein of S. aureus (cna) was cloned from a phage expression library by screening with antibodies raised against a purified collagen adhesion protein (626). Like other MSCRAMMs of other Gram-positive bacteria, the Cna protein has a large unique N-terminal region followed by a set of 187 amino acid repeats, a proline-rich region, and a classic cell wall sorting signal. The collagen-binding domain had been localized to the unique N-terminal domain (622, 625), an observation now verified by the recent determination of the crystal structure (773). The role of the repeat domains, which can vary in number from strain to strain (772), remains unclear.
The fact that cna is both necessary and sufficient for the binding of S. aureus to cartilage has been demonstrated convincingly in vitro (772). In addition, mutants with mutations in the cna gene are less virulent in a rat experimental endocarditis model and in a mouse septic arthritis model (623). However, a recent study of clinical isolates from patients with endocarditis or bone or joint infection found that strains from only half of these patients possessed the cna gene, which indicates that collagen binding is probably not a prerequisite for these types of infection (692). The potential of collagen adhesin as a staphylococcal vaccine is being explored (589).
Fibronectin binding proteins of S. aureus.
S. aureus was the first bacterium in which specific binding to fibronectin could be demonstrated, although the gene product responsible remained elusive for almost a decade (462). A >200-kDa fibronectin binding protein was purified from S. aureus Newman (191, 233), and soon thereafter a gene encoding a similar activity was cloned from strain 8325-4 (218). Sequencing of this gene, fnbA, revealed that it encodes a surface protein, designated FnBPA, with a predicted Mr of 108,000 (727). The gene for another fibronectin binding protein, FnBPB, was discovered only 682 nucleotides downstream of the fnbA gene (402), although not all S. aureus strains possess both genes (226).
Both FnBPA and FnBPB are similar in their general organization to the FnBPs from other gram-positive cocci such as Streptococcus pyogenes (see above). FnBPA is a 982-residue protein composed of a unique N-terminal domain that is interrupted by two copies of a 35-amino-acid repeat, termed B1 and B2 (727). Downstream from the B repeats are 3.5 repeats of 38 amino acids that are homologous to repeat domains found in the other fibronectin binding MSCRAMMs from S. pyogenes and S. dysgalactiae. FnBPB does not possess B repeats and is only 45% similar to FnBPA in the N-terminal domain, but it is essentially identical to FnBPA in all other aspects (402).
A study of S. aureus 8325-4 revealed that binding of S. aureus to fibronectin-coated coverslips was not significantly affected in either an fnbA or fnbB single mutant but was completely abolished in a double mutant. Complementation with either one of the two fnb genes restored the fibronectin binding phenotype to near wild-type levels (282), thereby demonstrating that both genes can be expressed in S. aureus and that both are capable of mediating staphylococcal adherence to fibronectin. Antibodies to this protein are capable of providing protection against infection in several model animal systems, indicating that this protein may be a potential vaccine candidate (514, 683, 684, 708). The role of this protein in pathogenicity is still the subject some debate, since mutants do not display a virulence defect in a rat model of endocarditis (219).
Fibrinogen binding protein of S. aureus (clumping factor).
S. aureus forms clumps in plasma due to its ability to bind fibrinogen, a multisubunit glycoprotein that is proteolytically converted into fibrin, a major component in blood clots. Fibrinogen is deposited in large quantities at wound sites and is quickly deposited on synthetic material such as catheters (226). Fibrinogen binding, therefore, is seen as a major primary virulence determinant in S. aureus, especially in infections due to the presence of foreign bodies.
The fibrinogen binding protein of S. aureus was identified by transposon mutagenesis followed by screening for mutants unable to clump in plasma (531). All four mutants isolated had a defect in a gene (clfA) encoding an 896-amino-acid protein that harbors an N-terminal leader peptide and a C-terminal cell wall sorting signal. The protein has an N-terminal nonrepetitive domain that is responsible for binding fibrinogen. The clumping-factor protein does not possess a typical proline-rich wall-spanning segment but instead has a very unusual stretch of 154 amino acids composed almost entirely of alternating serine and aspartate residues. This domain is not involved in ligand binding and has recently been shown to be required for the proper display of the molecule on the surface of the cell (308). Mutants with mutations in clfA are defective for adherence to fibrinogen-coated surfaces including coated catheter material, suggesting that clumping factor is involved in the establishment of infections due to the presence of foreign bodies (226).
It had been proposed that coagulase, a mostly secreted enzyme found in S. aureus, was responsible for the clumping phenotype, since it has been shown to bind fibrinogen in vitro (67). Coagulase mutants, however, were still able to clump in plasma and were unable to cause clotting, whereas clfA mutants were unable to clump but were still able to clot plasma (532). These results indicate that the fibrinogen binding clumping factor is the product of the clfA gene and not of coa. A detailed kinetic study to address the relative contributions of coagulase and clumping factor to the binding of S. aureus to fibrinogen-coated surfaces was carried out with a radial-flow chamber (157). By using this method, it was possible to determine the binding constants of intact cells under a variable set of shear force conditions. Clumping factor, not coagulase, was found to be primarily responsible for both the initial attachment and the resulting adhesion.
Fibrinogen binding protein of S. epidermidis.
Like S. aureus, the coagulase-negative bacterium Staphylococcus epidermidis is a frequent nosocomial pathogen and has been shown to bind to fibrinogen deposited on synthetic material such as catheters (629). The gene encoding a fibrinogen binding protein from S. epidermidis HB has recently been cloned and has been designated fbe (590). Although they are not identical in primary sequence, the basic structure and organization of the Fbe protein is almost identical to that of the fibrinogen binding protein from S. aureus (see above), including the unusual region of alternating serine and aspartate residues immediately preceding the cell wall sorting signal. The fibrinogen binding site was identified by using a phage display technique, and the domain was mapped to the same region found to be responsible for fibrinogen binding in the S. aureus homologue. Strangely, although this protein does bind fibrinogen and is similar to the clumping factor from S. aureus, S. epidermidis cells do not clump in the presence of plasma (590).
TARGETING: BINDING OF POLYPEPTIDES TO THE CELL SURFACE
The protein secretory pathway (Sec) releases polypeptides into the surrounding medium of gram-positive bacteria. Some of these secreted products bind to the bacterial surface of microorganisms and play critical roles in the synthesis and turnover of the peptidoglycan exoskeleton as well as the pathogenesis of animal infections. We describe here what is known about the mechanisms by which secreted proteins are directed to the cell surface and the physiological role these proteins play.
Murein Hydrolases
The peptidoglycan of gram-positive organisms is hydrolyzed at specific times and sites during physiological growth of the exoskeleton (261). This is accomplished by murein hydrolases. These enzymes have been studied extensively, and an excellent review summarizes what is known about their activities and physiological roles (725). Murein hydrolases can be grouped according to their enzymatic activities; the various peptidoglycan cleavage sites of these enzymes are diagrammed in Fig. 17 (250). N-Acetylmuramidases (muramidase) and N-acetylglucosaminidases (glucosaminidase) cleave MurNAc(β1-4)GlcNAc and GlcNAc(β1-4)MurNAc, respectively (793). Lytic transglycosylases and lysozymes also attack the glycan chains (349); however, these enzymes have not been found in gram-positive bacteria and are not discussed here. Several different classes of enzymes attack the peptide backbone of the cell wall. N-Acetylmuramoyl-l-alanine amidase (amidase) cleaves the amide bond between the d-lactyl group of MurNAc and the amino group of l-Ala. Lysostaphin and Myxobacter AL1 proteases are glycyl-glycine endopeptidases that cleave the pentaglycine crossbridge of staphylococcal peptidoglycans (189, 709). Another class of enzyme cleaves the attachment site of the peptidoglycan crossbridges: d-Ala–X (250). In staphylococci, the d-Ala is linked to pentaglycine, and this bond is cleaved by the murein hydrolase of phage φ11 (LytA) (576). In other bacterial species, the d-Ala can be linked to l-Ala–l-Ala, d-iAsn, or the side chain amino group of m-diaminopimelic acid, d-ornithine, and l-Lys (710). Enzymatic activities that cleave these peptide bonds of d-Ala have been found; however, the primary structures of these enzymes have hitherto not been identified (250). Carboxypeptidases trim the murein pentapeptide subunits by removing the terminal residue of d-Ala–d-Ala; these membrane-anchored PBPs do not hydrolyze the peptidoglycan and are therefore also not considered here (761).
FIG. 17.
Sites of hydrolysis of various muralytic enzymes on the peptidoglycan of S. aureus (see the text). Modified from reference 575 with permission of the publisher.
Sequence alignment of murein hydrolases of gram-positive bacteria revealed that most of these enzymes display a domain structure (404). In general, murein hydrolases harbor an N-terminal signal peptide followed by a second domain containing the enzymatic activity (404). In addition, these proteins harbor repeat structures that flank either the N- or C-terminal side of the enzymatic domain (404). Figure 18 displays the domain structure of murein hydrolases from S. aureus. Although the enzymatic domains are often conserved between enzymes from different species, the repeat domains are not. The functions of these repeat domains have been studied most extensively for Streptococcus pneumoniae LytA amidase, Staphylococcus simulans lysostaphin (Lst), and S. aureus Atl and are discussed in detail below. Ghuysen et al. reported sequence and structural similarity between the noncatalytic regions of muramidases and amidases from Clostridium acetobutylicum and Bacillus spp. and the crystallized structure of Sreptomyces albus G Zn dd-peptidase (253). This region is required for the binding of dd-peptidase to insoluble peptidoglycan, suggesting that murein hydrolases from several other gram-positive bacteria retain this domain to bind to their substrate (253). This substrate binding domain is distinct from targeting domains, which direct some murein hydrolases to specific sites on the bacterial cell surface (see below).
FIG. 18.
Domain structure of the peptidoglycan hydrolases of S. aureus. Autolysin is synthesized as a preproenzyme, and after cleavage of its N-terminal signal peptide (SP), soluble pro-Atl is released into the extracellular milieu. The repeat domains R1 to R3 function to direct pro-Atl to its receptor on the equatorial surface rings of staphylococci. Proteolytic cleavage generates a propeptide of unknown function (PP), mature amidase with C-terminal R1 and R2 repeats (Atl-A), and glucosaminidase with an N-terminal R3 repeat (Atl-G). Lysostaphin is also synthesized as a preproenzyme, and after signal peptide cleavage, soluble pro-Lst is released into the extracellular milieu. Proteolytic cleavage of 15 tandem repeats of a 13-residue peptide generate mature Lst, which functions as a glycyl-glycine endopeptidase. The C-terminal cell wall-targeting domain (CWT) directs Lst to its receptor on the surface of S. aureus. LytM endopeptidase is probably exported by an N-terminal signal peptide. Its C-terminal domain displays some homology to the endopeptidase domain of lysostaphin, whereas the N-terminal domain of LytM is uncharacterized. φ11 murein hydrolase is encoded by a lysogenic phage (φ11) and exported by a lysis mechanism requiring another phage protein (holein). The enzyme is composed of two domains, one of which is thought to be responsible for amidase activity (LytA-A) and the other is responsible for d-Ala-gly endopeptidase activity (LytA-DL). The C-terminal cell wall-targeting signal of LytA displays homology to that of lysostaphin and also functions to direct this enzyme to its receptor in the cell wall envelope of S. aureus.
Staphylococcus simulans Lysostaphin
Staphylococcus simulans bv. staphylocolyticus secretes lysostaphin (709), which hydrolyzes the peptidoglycan of all the staphylococci that synthesize peptidoglycan with pentaglycine crossbridges (95, 878) (Fig. 19). In this way, lysostaphin functions as a bacteriocin to kill competing microorganisms in mixed bacterial populations. Lysostaphin is synthesized as a preproenzyme and initiated into the secretory pathway by an N-terminal leader peptide. The proenzyme is released into the culture medium and contains 15 tandem repeats of a 13-residue peptide at the N-terminal end (324). Prolysostaphin is 4.5-fold less active than mature lysostaphin, and the N-terminal repeats are removed in a growth phase-dependent manner by a secreted cysteine protease (782). Mature lysostaphin cleaves the peptidoglycan of S. aureus cells much more actively than it cleaves the peptidoglycan of its S. simulans host (314). There appear to be two reasons for this. First, S. simulans cells elaborate an immunity factor that causes the incorporation of serine residues into the cell wall crossbridge (782). Although, on balance, only one or two serine residues are found in the otherwise poly-glycyl crossbridge, this modification renders S. simulans resistant (immune) to this bacteriocin (782). The S. simulans gene required for incorporation of the serine residues, lif (lysostaphin immunity factor), displays striking homology to the femA and femB genes (145, 331, 513). The latter genes are thought to specify enzymes which synthesize the cell wall crossbridge by using lipid II peptidoglycan precursor and charged Gly-tRNA as substrates (43). Consistent with the notion that Lif is the functional equivalent of a FemA/FemB enzyme is the observation that Lif can complement femB mutant staphylococci (798). Furthermore, immediately adjacent to the immunity factor gene is a locus encoding seryl-tRNA, suggesting that Lif may also utilize charged tRNAs as substrates (782). Recently, a glycyl-glycine endopeptidase of Staphylococcus capititis was described and characterized (764, 765). The molecular structure, targeting mechanism, and immunity of this enzyme appear to be identical to those described for lysostaphin.
FIG. 19.
Targeting of lysostaphin to the cell wall envelope of S. aureus. (A and B) After secretion by S. simulans bv. staphylolyticus (A), prolysostaphin is converted to the active mature species and targeted to those staphylococci that harbor the appropriate receptor on the bacterial surface (B). (C and D) Mature lysostaphin is a glycyl-glycine endopeptidase that cleaves the peptidoglycan crossbridge of staphylococci (C), thereby hydrolyzing the cell wall envelope and lysing the bacteria (D).
Purified lysostaphin binds much more avidly to S. aureus than to S. simulans cells (21, 519). When added to mixed bacterial populations, purified lysostaphin kills 1,000 S. aureus cells for every S. simulans cell (21). This selectivity requires the C-terminal targeting domain of Lst, which is located in the 92 C-terminal amino acids (21). When deleted from lysostaphin, this mutant enzyme no longer binds to staphylococci and has lost the ability to distinguish between S. aureus and S. simulans cells (21). Furthermore, when the targeting domain is linked to the C terminus of secreted enterotoxin B, the fusion protein is exported but directed to the cell wall compartment of S. aureus cells. Similarly, when fused to the C terminus of reporter proteins such as glutathione S-transferase, the recombinant protein binds to the surface of S. aureus cells but not of S. simulans cells (21). Hence, lysostaphin is specifically directed to the surface of S. aureus by its C-terminal targeting domain (20). This targeting mechanism functions in mixed bacterial populations and therefore must be species specific. In other words, murein hydrolases such as lysostaphin are probably designed such that the targeting domain recognizes receptors found on the surface of specific species of microorganisms. It follows that the targeting domain cannot recognize peptidoglycan alone, because its chemical structure is largely conserved in all gram-positive bacteria (20).
Staphylococcal Autolysin
Not all murein hydrolases function to kill microorganisms. Staphylococci divide in a direction perpendicular to the previous cell division plane (258, 259, 261). These coccal cells require hydrolysis of their thick peptidoglycan layer at the midcell to synthesize new cell wall and separate the dividing daughter cells (261). This depends on the Atl enzyme, because atl mutants show characteristic defects in cell separation and grow as large clusters of staphylococci (606, 766). The atl gene specifies a large polypeptide (preproenzyme) with several domains (225, 605). An N-terminal signal peptide is required for export (22). Secreted Pro-Atl is processed at two sites, residues 198 and 775 (22, 605). These cleavage events generate an N-terminal propeptide of unknown function as well as two domains with enzymatic functions, amidase and glucosaminidase (732, 786). Three repeat domains are located at the center of pro-Atl, such that mature amidase retains two C-terminal repeats and mature glucosaminidase retains one N-terminal repeat domain (605). The repeat domains function to direct Atl to the future cell division site of staphylococci (22), a structure named the equatorial surface ring (766, 767, 864) (Fig. 20). Targeting of pro-Atl occurs before it is processed; however, mature amidase and glucosaminidase remain bound at the surface ring structure (22, 864). The significance of the timing of processing and targeting is thus far not understood. Fusion of the repeat domains to reporter proteins directs the hybrid proteins to the equatorial surface rings, demonstrating that the repeat domains of Atl properly address the enzyme to the cell division site (22). This mechanism implies that a specific receptor for Atl might be located at this site to attract the secreted enzyme for localized hydrolysis (22). Although this hypothesis provides a model for cell separation in gram-positive bacteria, several questions are left unanswered. To define a spatial location on the cell surface, staphylococci presumably synthesize a specific decoration of the peptidoglycan. This decoration must be positioned in a manner that defines both the equatorial surface ring and future cell division sites. The same dilemma exists in the bacterial cytoplasm, where cell division occurs via constriction of an FtsZ ring structure which is assembled at midcell from soluble components and then constricts to fuse the membrane, thereby separating two daughter cells (54, 505). It is conceivable that the definition of the cell division site is provided by the synthesis of a chemical decoration in the rigid exoskeleton, which might provide an anchoring point for cell wall hydrolases and the membrane fusion machinery alike (681, 682).
FIG. 20.
Targeting of Atl to the equatorial surface rings of S. aureus. Autolysin is exported from the bacterial cytoplasm (A), and extracellular pro-Atl is targeted to receptor molecules representing the equatorial surface rings of staphylococci that mark the future cell division site (B). Targeting requires the repeats domains (R1 to R3) or pro-Atl. Proteolytic cleavage of pro-Atl generates mature amidase (Atl-A) and glucosaminidase (Atl-G), each of which retains one or two repeat domains and remains bound to the surface rings. The enzymes hydrolyze the cell wall at the cell division site, an event that is probably synchronized with intracellular contraction of FtsZ rings (C), thereby generating two fully separated daughter cells (D). In staphylococci, division occurs perpendicular to the previous cell division plane and targeting of pro-Atl to the second equatorial surface ring can initiate hydrolysis at the future cell division site.
As mentioned previously, coagulase-negative staphylococci, such as S. epidermidis, are important nosocomial pathogens that adhere to foreign bodies implanted into human tissues (629). Adherence of staphylococci to catheters and other materials is followed by the production of large amounts of biofilm (629). Götz and coworkers screened a collection of Tn917 insertional mutants of S. epidermidis for their inability to produce biofilm and/or adhere to polystyrene surfaces (321). Wild-type staphylococci synthesize a linear carbohydrate polymer composed of β1-6-linked 2-deoxy-2-amino-d-glucopyranosyl, in which almost all moieties are N-acetylated, named polysaccharide intercellular adhesin (PIA) (506, 507). One class of Tn917 mutants is defective in the production of PIA, and the mutation maps to the icaABC operon (323, 508). The ica genes specify the enzymatic machine that synthesizes PIA in both mutant S. epidermidis and S. carnosus, a microorganism otherwise unable to synthesize biofilm (323). A second class of mutants is unable to adhere to polystyrene surfaces, and its Tn917 insertion maps to the autolysin gene of S. epidermidis (atlE) (322). Binding of staphylococci to foreign materials is thought to be aided by human plasma proteins. A search for bound plasma proteins reveals that vitronectin may bind to AtlE (322). The authors observed less vitronectin binding to mature Atl-amidase and Atl-glucosaminidase than to pro-Atl, suggesting that the propeptide may be required for binding (322). The autolysin of S. saprophyticus is thought to be involved in binding to fibronectin proteins (325). This microorganism causes a large number of female urinary tract infections and expresses a surface protein which binds to fibronectin (246). A search for fibronectin binding activity of cloned staphylococcal sequences revealed the aas gene, which is homologous to the atlE and atl genes (325). Mutant S. saprophyticus strains carrying a defective aas gene were unable to bind either fibronectin or erythrocytes, and the fibronectin binding activity of purified Aas protein was mapped to the R1 to R3 repeats (325). Thus, the autolysin of staphylococci appears to be a multifunctional protein, required for cell wall hydrolysis at a designated site as well as for attachment of bacteria to eukaryotic proteins and/or mammalian tissues.
Phage-Encoded Murein Hydrolases
Bacteriophage lambda of E. coli employs the S lysis protein (holin) to disrupt the host cell membrane (11, 63, 245, 664). This mechanism allows phage-encoded intrabacterial murein hydrolases to leave the cytoplasm and degrade the peptidoglycan sacculus, which is located in the periplasmic space of gram-negative bacteria (61, 494). Without murein hydrolysis, phage particles could not be released into the extracellular medium (664). A similar strategy is used by the bacteriophages of gram-positive organisms (679). For example, S. aureus phage φ11 expresses a murein hydrolase (LytA) devoid of an N-terminal signal peptide (834, 835). The LytA protein has a C-terminal targeting signal similar to that of lysostaphin (21), and its N-terminal domains code for both its d-Ala–Gly endopeptidase and amidase activities (576). Adjacent to the lytA gene is an open reading frame specifying a lysis protein with sequence homology to the holins of bacteriophages of gram-negative bacteria (70). A similar genetic arrangement to that of φ11 has been observed for staphylococcal phages 80α and Twort (68, 498). Thus, although this has never been demonstrated directly, it seems likely that phage-encoded murein hydrolases are released from the cytoplasm of gram-positive bacteria only after the holin-induced disruption of the cytoplasmic membrane that occurs during the phage lytic cycle.
Several groups have searched for autolysins, enzymes that degrade the cell wall of gram-positive bacteria at the end of logarithmic growth. Autolysin activity can be measured on agar plates containing heat-killed bacteria (274, 606). Mutant colonies that fail to hydrolyze peptidoglycan can be screened from a population of transposon insertion mutants. Several such mutants were isolated in S. aureus, and mutations in lytA, atl, lytM, and lytRS were found in screen (98, 605, 660, 835). It now appears that two of these genes encode staphylococcal autolysins. The Atl enzyme, specifying amidase as well as glucosaminidase activity, is described above. The lytM gene encodes a protein harboring an N-terminal signal peptide followed by an enzymatic domain that is homologous to that of lysostaphin and probably functions as a glycyl-glycine endopeptidase (660). LytM does not harbor any of the known targeting signals of either lysostaphin or Atl, and its location on the staphylococcal surface is still unknown. The lytA gene is encoded by bacteriophage φ11, which has lysogenized strain 8325 (834). The lytRS genes display homology to a two-component regulatory system involved in signal transduction (98). The lytRS locus controls the activity of two other genes, lrgA and lrgB, encoding a staphylococcal holin lysis protein (LrgA) similar to those described for phages φ11, 80α, and Twort and a protein with unknown function (LrgB) (99).
Murein hydrolases of gram-positive bacteria appear to fall into two classes: the enzymes which are encoded by genes on the bacterial chromosome, exported by an N-terminal signal peptide, and presumably involved in physiological cell wall turnover (class I), and others that are located on the chromosome of bacteriophages, devoid of signal peptides, and exported by a general lysis mechanism (class II). The latter class of genes found in the screen described above were identified due to the induction of lysogenic phages at the end of logarithmic growth.
S. pneumoniae
S. pneumoniae synthesizes a major autolysin which hydrolyzes the cell walls of these bacteria in stationary-phase cultures (795). The enzyme is thought to exist in two states, a cytoplasmic inactive species and a membrane-associated active counterpart (91). The inactive species (E) accumulates in cells that are grown in ethanolamine, whereas the more active enzyme (C) accumulates in cultures grown in media containing choline (91). The inactive enzyme has been purified to homogeneity by affinity chromatography on choline-Sepharose (91) or by DEAE ion-exchange chromatography (702). The pure enzyme can be activated by the addition of choline (352), suggesting that choline is a necessary cofactor (265). Presumably, LytA undergoes a conformational change when encountering choline, resulting in enzymatic activity (350–352, 698). Cloning and sequencing of the lytA gene revealed an open reading frame specifying a 31-kDa polypeptide (242). The open reading frame displays sequence homology to the gene sequence encoding enzymatic domains of many other known amidases (677). However, six repeat domains located at the C-terminal end of LytA (242, 699) are not found in the amidase sequences from other bacterial species but are present in a variety of phage-encoded murein hydrolases of S. pneumoniae (499). When expressed in E. coli and purified to homogeneity, these LytA repeats were shown to bind choline and hence have been named choline binding repeats. Thus, LytA binds to choline-containing teichoic acids, resulting in its enzymatic activation and peptidoglycan hydrolysis on the cell surface (350–352). The common repeat structure of pneumococcal cell wall teichoic acid and lipoteichoic acid (31, 207, 211, 379) [Glc(β1-3)AATGal(α1-4)GalNac(6-Cho-P)(α1-3)GalNac(6-Cho-P)(β1-1)ribitol-P] suggests that LytA can bind to both structures. The lytA open reading frame did not reveal an N-terminal signal peptide (242), and either LytA is not exported under physiological conditions or its export may occur by a pathway other than the known Sec machinery (156). When expressed in E. coli, LytA appears to be translocated across the cytoplasmic membrane and was detected by immunogold labeling on the periplasmic side of the cytoplasmic membrane (156). Immunogold labeling of S. pneumoniae revealed LytA staining on the cell surface, indicating that the enzyme is also translocated and surface displayed (156). Future work is needed to further characterize the export mechanism of LytA.
Briles et al. identified PspA (pneumococcal surface protein A) as a major antigen during animal infections by S. pneumoniae (93). Antibodies raised against purified PspA protect mice from challenge with virulent strains (92) and pneumococcal pspA mutants are less virulent in a mouse model system (530). PspA displays size and antigenic variation; however, the immunodominant antibodies are opsonic (phagocytic) and cross-reactive with PspA species of other strains (528, 529, 659, 779). Thus, PspA might serve as a vaccine candidate in protecting animals from pneumococcal infections. Sequencing of the structural gene revealed that PspA is synthesized with an N-terminal signal peptide (875). Because much of the N-terminal part of the mature polypeptide showed a 7-residue periodicity of hydrophilic and hydrophobic residues, it is likely that this domain of PspA assumes a coiled-coil structure. At the C terminus, PspA contains repeat domains homologous to those of LytA (875). Shortening of the C-terminal domain to five repeats abolished surface display and released the mutant protein into the culture supernatant (876). These results are comparable to those obtained for LytA, which showed that binding to the pneumococcal surface required as many as four repeat domains (243, 699). The LytA repeat domains are not only required but also sufficient for binding to choline-containing teichoic acids. This has been demonstrated both by expressing fusion proteins with LytA repeats in pneumococci (676, 746) and by adding purified recombinant proteins to cultures of S. pneumoniae (135).
Bacillus
Bacillus and Clostridium are spore-forming species. During sporulation, the mother cell undergoes asymmetric cell division to engulf and nurture the spore (501). The cell wall peptidoglycan and associated structures (cortex) of the spore are distinct from that of cells growing under vegetative conditions (646, 836, 852). During germination, the mother and spore peptidoglycan are dissolved and the released spore once again grows as a vegetative cell (101). Sequencing of the B. subtilis genome revealed at least 16 genes with homologies to known murein hydrolases, most of which are amidases (454). To characterize these enzymes, several investigators have used zymography, i.e., the activity measurement of renatured proteins after their separation on SDS-PAGE by overlaying with heat-killed bacteria (748). Murein hydrolase activity can be visualized as a zone of bacteriolysis representing a protein with unique mobility. When this experiment is performed with proteins of B. subtilis cell extracts, at least 10 proteins with bacteriolytic activity can be revealed (224), suggesting that most if not all of the 16 genes specifying murein hydrolases are indeed expressed. Clues to the function of these genes came from the generation of specific knockout mutations. The two most prominent zymographic species, a 90-kDa glucosaminidase (LytD) and a 50-kDa amidase (LytC), may not function as autolysins, because strains carrying knockout mutations in their structural genes revealed either no (lytD) or little (lytC) decreased autolysis of stationary-phase cultures (460, 516, 517). No effects on either cell separation, sporulation, or germination of this organism were observed. The lytC gene is located within the lytRABC operon specifying the regulatory transcription factor LytR, lipoprotein LytA, and LytB, the modifier protein for the LytC amidase (473). LytB binds to LytC, altering its enzymatic activity from a random to a processive amidase removal of peptide substituents along single glycan strands (332, 333). LytB is similar in sequence to SpoIID (458, 473), a factor required for dissolution of the forespore septum with the mother cell (364). Transcription of lytRABC is regulated in part by the flagellar transcription factor sigma D (327), which also controls all lytD expression (459, 473). Because sigD mutants grow as long filaments, it is suspected that this transcription factor may control the expression of murein hydrolases required for cell division (328). There appears to be at least one enzyme other than lytC involved in this, because lytC mutants do not display filamentation (748). With the completion of the B. subtilis genome sequence, it is presumed that all open reading frames specifying murein hydrolases have been identified. Little is known, however, about the targeting of any of these enzymes. The SpoIID protein displays sequence homology to the modifier protein (LytB) of the LytC amidase (458). Whether this homologous domain is involved in directing these proteins to the septum is still unknown. Mother cell lysis depends on the compensatory effect of the LytC (CwlB) and CwlC hydrolases, suggesting that these two enzymes have redundant functions (457). The CwlD hydrolase is thought to be required for spore cortex maturation (19, 645, 719). Germination of spores also requires murein hydrolases, and several enzymes appear to fulfill compensatory functions, for example CwlJ and SleB (369, 558, 559). Whether any of these Bacillus enzymes are directed to their subcellular location by a specific targeting event is also still unknown.
Clostridium
Clostridium difficile, the causative agent of pseudomembranous colitis and antibiotic-associated colitis in humans, releases two toxins, TcdA and TcdB, which are thought to be major virulence factors of this organism (406, 408, 424). Both toxins are targeted into the cytosol of eukaryotic cells, where they function as glucosidases to modify small GTP binding proteins such as Rho, Rac, and Cdc42 by using UDP-Glc as a cosubstrate (10, 815). Glucosidation leads to inactivation of GTPase activity and of the signaling function of these molecules, causing actin rearrangements, fluid loss, and, finally, cell death (407). Injection of glucosylated Rho into eukaryotic cells has the same cytotoxic effect as injection of TcdB, indicating that Rho is the main target of toxin action and is dominant over unmodified Rho (407). C. sordellii and C. novyi toxins also glucosylate small GTP binding proteins, although the alpha-toxin of C. novyi employs UDP-GlcNAc as a substrate for modification (647, 722).
Neither TcdA, TcdB, nor any other member of a family of large clostridial toxins is synthesized with an N-terminal signal peptide (815). Although the mechanism by which these toxins are secreted and/or released from the bacteria is unknown, once in the extracellular milieu, toxins display binding affinity for mammalian cells and hence are taken up into the eukaryotic cytosol (705). Clostridial toxins A and B are very large polypeptides (308 and 270 kDa) (29, 706), composed of an N-terminal domain containing glucosyl transferase activity (342, 651) and a C-terminal domain with tandem repeats of a 30-residue peptide (705). The repeat domains are responsible for binding to carbohydrate compounds, and TcdA recognizes Galα1-3Galβ1-4GlcNAc (450). Such carbohydrate structures are displayed on the surface of animal cells, accounting for the hemagglutination of rabbit erythrocytes by TcdA (800). Although similar in sequence, TcdB does not hemagglutinate the same cells, presumably because its repeats do not recognize the same carbohydrates (800). Antibodies directed against the C-terminal repeats of TcdA prevent toxicity in a mouse model system, as does preincubation of cells with recombinant proteins carrying the C-terminal repeat domains but not the N-terminal domain with enzymatic activity (705). Thus, the C-terminal repeat domains function as a targeting device that directs clostridial toxins to certain eukaryotic cells.
Several authors observed that the repeat elements found in clostridial toxins have some sequence similarity to the repeat domains of streptococcal glucan binding proteins as well as the LytA repeats of pneumococcal proteins (816, 817, 860). This suggested that the ability to bind carbohydrate elements may be a common property of these repeat structures. In keeping with this hypothesis, aromatic amino acid residues (W, Y, and F) are found regularly in the repeat structures of proteins of gram-positive bacteria with targeting elements, and they may serve as stacking devices for the interaction with carbohydrate ring structures, as has been observed for sugar binding proteins in the periplasm of gram-negative bacteria (860).
Listeria
InlB is a member of the internalin family of bacterial invasins and promotes the entry of Listeria into hepatocytes and several other cell lines (161, 163, 368). The broad tropism apparently conferred by InlB might indicate that the InlB receptor is more widespread than E-cadherin, the receptor for InlA. During the infection of mice, Listeria has been observed in the liver, and InlB is thought to be essential for multiplication in this organ (130, 161). InlB is synthesized as a precursor bearing an N-terminal signal peptide (235). The N-terminal domain of mature InlB displays sequence homology to other members of the internalin family whereas the C-terminal 232 amino acids are homologous to the targeting domain of a Listeria amidase (82). When added externally to Listeria, purified InlB binds to the bacterial surface (82). Binding of InlB is dependent on the C-terminal targeting signal, since mutant proteins without this sequence are found in the extracellular medium (82). The addition of purified InlB to other bacteria or to latex beads also promotes uptake of these organisms or particles into hepatocytes, indicating that InlB alone is sufficient to promote this function (83). It is not clear why Listeria chooses to display InlB via a cell wall-targeting mechanism whereas all other internalins are apparently linked (sorted) to the bacterial peptidoglycan. To investigate this further, Cossart and coworkers designed a recombinant InlA protein which was displayed on the listerial surface via the C-terminal targeting signal of InlB (82). This hybrid protein was functional in promoting invasion in a manner indistinguishable from wild-type InlA (162, 164). It is conceivable that Listeria regulates the surface display of its different internalins (162) and that this regulation is, at least in part, carried out by their anchoring and release from the cell surface.
Listeria p60 is required for cell growth and intestinal invasion of this organism (334, 861). p60 homologs have been found in many listerial species (100). The N- and C-terminal domains of p60 species are conserved in all Listeria species; however, the central sequences are variable (100). Cytotoxic T cells that recognize p60 epitopes are protective against Listeria infections in a mouse model system, suggesting that this surface protein presents an immunodominant antigen (74, 247). Purified p60 displays murein hydrolase activity, and overexpression of p60 leads to the appearance of significantly shortened Listeria cells. The C-terminal domain of p60 displays homology to the repeat domains of an autolysin of Enterococcus faecium (234), suggesting that p60 may be targeted to the cell surface and required for peptidoglycan hydrolysis similar to the staphylococcal Atl enzyme.
Streptococci
Pancholi and Fischetti described the purification of several different glycolytic enzymes from S. pyogenes cell extracts (614). SDH (glyceraldehyde-3-phosphate dehydrogenase) and enolase were isolated from cell wall extracts obtained after enzymatic peptidoglycan hydrolysis and removal of the resulting protoplasts by sedimentation (610). Immunoelectron microscopy, reactivity of affinity-purified antibodies, and enzyme assays with intact streptococci have each indicated that these glycolytic enzymes are found in two locations, the bacterial cytoplasm and the streptococcal surface (610). Surface-exposed and cytoplasmic enzyme appear to be one and the same species, synthesized from a single gene and devoid of an N-terminal signal peptide. Clearly, these glycolytic enzymes are not exported, sorted, or targeted to the envelope of this gram-positive bacterium by any of the established pathways. It will be interesting to see by which mechanism streptococci can display enzymes in two different locations. SDH binds to a variety of eukaryotic proteins (fibronectin, lysozyme, myosin, and actin) (615). Furthermore, purified SDH activates eukaryotic protein tyrosine kinase and protein kinase C by ADP-ribosylation (611). Inhibitors of protein kinase activity prevented S. pyogenes invasion of pharyngeal cells, suggesting that host protein phosphorylation plays a role in the uptake of this pathogen (615). Streptococcal enolase binds to plasmin and plasminogen (610). The precise role of this binding in streptococcal invasion and disease is not yet clear. Others have reported that high-affinity plasminogen binding to the surface of S. pyogenes is caused by certain M-protein-like proteins (see above) (858).
S. mutans is one of the principal etiologic agents of dental caries (497). This pathogen synthesizes both water-soluble α(1-6) glucan and insoluble α(1-3) glucan by hydrolyzing dietary sucrose (311) and polymerizing the resulting monomeric glucose residues (455). Both types of glucan are components of dental plaque, and the water-insoluble species mediates the adherence of S. mutans to smooth dental surfaces (299). Synthesis of glucan is stimulated by the GtfA enzyme, which acts as a sucrose phosphorylase to synthesize Glc-1P (200, 690). The Gtf-I (gtfB and gtfC) (301) and Gtf-S (gtfD) (301) enzymes are responsible for synthesizing water-insoluble and water-soluble glucans, respectively, and streptococcal strains carrying mutations in any of these genes display a defect in the pathogenesis of dental caries in a pathogen-free rat model (865). Sequence analysis of the gtf genes and their encoded polypeptides revealed that each of these enzymes is exported by an N-terminal signal peptide. A large N-terminal enzymatic domain is followed by a C-terminal domain with tandem repeats of 20 to 48 amino acids (26, 199, 263, 653, 859). Successive truncations of the C-terminal repeats yielded enzymes that were still active but had impaired binding activity for α1-6-linked glucans, the substrates for their glucosyltransferase activity (199). Thus, the C-terminal repeat domains of Gtf enzymes target these enzymes to their glucan substrates, which are deposited on the dental surface. As mentioned above, the repeat sequences are homologous to those found in large clostridial toxins and pneumococcal choline binding domains (817).
Targeting of S-Layer Subunits
S-layers are two-dimensional proteinaceous crystalline arrays formed by proteinaceous subunits that cover the outer surface of many different kinds of unicellular organisms (53, 741, 742). S-layers have been identified in a range of species from the Archaea, Bacteria, and Eucarya and diverge greatly in their structures and functions. We make no attempt here to cover the many aspects of the hundreds of S-layers that have been identified and will instead focus on what is known about the mechanisms by which these S-layers are targeted to and assembled on the cell surface of gram-positive bacteria. For a more extensive review of what is known about the biology of S-layers, the reader is directed to a series of recent reviews (24, 53, 547, 657, 739).
S-layers are formed by the entropy-driven aggregation of monomer subunits which, in most cases, are exported from the cytoplasm via the general secretory pathway by an N-terminal leader peptide. S-layers are not covalently attached to the cell surface and can usually be extracted either as sheets (30) or as individual subunits in the presence of dissociating agents such as 5 M lithium chloride (500) or EDTA (768) or chaotropic denaturants such as guanidine hydrochloride or urea (357, 740, 743). In many cases the monomeric subunits can spontaneously reassemble into a two-dimensional lattice once the dissociating agent is removed. Lattices formed in vitro usually bear the same hallmark geometric crystal pattern of the original cell-bound S-layer and are often capable of reassociation with the bacterial cell surface. The spontaneous nature by which S-layers assemble means that the primary energetic cost to the cell of manufacturing the S-layer stems from synthesizing the enormous number of subunits necessary to cover the entire cell surface. S-layer proteins are often the most abundant species in cells, comprising up to 15% of the total cellular protein (742). This is not surprising considering that approximately 5 × 105 subunits are required to cover an average-sized cell (741).
S-layers are usually surface exposed, although in at least one strain of Bacillus anthracis the S-layer can coexist with a surface-exposed capsule and it appears that the expression and display of one does not interfere with that of the other (545). Multiple S-layers can be simultaneously expressed on a single cell, and it is believed that in some cases one S-layer can serve as the primary attachment or crystallization surface for other layers (418, 428, 546, 799). The S-layer in Bacillus stearothermophilus is the envelope component responsible for the attachment of a surface-displayed amylase (175, 177), indicating that these layers can serve as targeting ligands for other proteins.
SLH domains.
Only recently has any insight been gained into the manner by which S-layers are attached to the gram-positive cell surface. Recent structural studies have revealed that several S-layer subunits share a region of homology in a domain that has been shown to interact with the cell wall (481, 504, 599). These surface layer homology (SLH) domains have been identified in S-layer subunit protein sequences from several gram-positive species as well as in some other types of surface proteins including the cellulosome of Clostridium thermocellum (477). All SLH domains identified thus far have been found at the very beginning or end of the mature proteins (503). An SLH domain is usually composed of either a single or three repeating SLH motifs of approximately 50 to 60 residues (503); a comparison of these SLH motifs reveals that they are fairly divergent with an average identity of 27% and only one universally conserved residue (504).
SLH domains have been speculated to interact either directly with peptidoglycan (599) or with other wall-associated polymers. Recent studies of three different S-layer proteins from different strains of B. stearothermophilus have identified two apparently unique secondary wall polymers as the surface ligands responsible for the adherence of the S-layer to the cell surface (176, 670). The SLH receptor on the surface of PV72/p2 was isolated by extracting isolated cell walls with hydrofluoric acid (670). Chemical analysis determined that the receptor molecule was a polymer composed of GlcNAc and ManNAc in a molar ratio of 3.5:1 and with an approximate molecular mass of 20,000 Da (670).
An S-layer subunit from Thermus thermophilus containing a single SLH motif seems to bind directly to the peptidoglycan (599). Although T. thermophilus is not taxonomically classified as a gram-positive bacterium, it is known to have a similar subcellular architecture. Chemical analysis of the envelope from this bacteria has indicated that the peptidoglycan does not contain any associated secondary wall polymers (656). It therefore appears likely that SLH domains have evolved the ability to recognize a wide variety of surface ligands, including, in some cases, the peptidoglycan itself. This property is perhaps reflected in the loose sequence conservation found among the various SLH domains.
Other S-layer-targeting domains.
It is important to note that S-layer proteins from several gram-positive bacteria do not have SLH domains, demonstrating that different adherence mechanisms have evolved for different S-layers. The PS2 protein of Corynebacterium glutamicum contains a C-terminal hydrophobic domain of approximately 79 residues that can be removed by protease treatment, resulting in an intact S-layer sheet that is not able to adhere to the Corynebacterium cell surface (114, 115). More recently, a secondary wall polymer ligand has been partially characterized for the S-layer proteins from B. stearothermophilus ATCC 12980 and the PV72 variant, PV72/p6 (176), both of which lack SLH domains. The composition of this wall polymer appears to differ significantly from that of the ligand for the PV72/p2 S-layer protein; the polymer appears to be composed of glucose and glucosamine in roughly equal ratio (176). It is almost certain that other adherence mechanisms will become apparent as S-layers from more gram-positive bacteria are studied in the future.
CONCLUSIONS AND FUTURE DIRECTIONS
We have summarized here the currently known surface proteins of gram-positive bacteria and the mechanisms of their anchoring to the cell wall envelope. The completion of several microbial genome sequences will soon provide us with a more comprehensive view of the number and nature of these molecules. If this information is combined with biochemical analysis, our understanding of surface protein function will probably expand rapidly. Nevertheless, future work must also further investigate the mechanisms of surface protein anchoring to the cell wall envelope. The cell wall sacculus provides rigid exoskeletal functions for microorganisms and requires proper targeting of surface proteins for its concerted assembly and turnover. A detailed knowledge of these mechanisms appears absolutely necessary to our understanding other physiological events such as bacterial cell division and separation. We apologize to all those authors whose work was not mentioned here either because of space constraints or because of our oversight.
ACKNOWLEDGMENTS
We are indebted to Marjorie Russel and Peter Model for their interest and relentless encouragement. We thank Gunnar Lindahl, Maria K. Yeung, Pascale Cossart, Agnès Fouet, and Stéphane Mesnage for their comments that much improved the manuscript. We also thank Tadashi Baba, Hung Ton-That, Sarkis Mazmanian, Vincent Lee, Kumaran Ramamurthi, Debbie Anderson, and Kym F. Faull for their support and contributions to this work.
O.S. acknowledges grant support (AI33985 and AI38897) from the Infectious Disease Branch, NIAID, NIH. W.W.N. was supported by a Cellular and Molecular Biology Training Grant (GM 07185-22) from the Public Health Service to UCLA.
REFERENCES
- 1.Ackermans F, Klein J P, Ogier J, Bazin H, Cormont F, Frank R M. Purification and characterization of a saliva-interacting cell-wall protein from Streptococcus mutansserotype f by using monoclonal-antibody immunoaffinity chromatography. Biochem J. 1985;228:211–217. doi: 10.1042/bj2280211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler L A, Arvidson S. Cloning and expression in Escherichia coli of genes encoding a multiprotein complex involved in secretion of proteins from Staphylococcus aureus. J Bacteriol. 1988;170:5337–5343. doi: 10.1128/jb.170.11.5337-5343.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler L A, Arvidson S. Correlation between the rate of exoprotein synthesis and the amount of the multiprotein complex on membrane-bound ribosomes (MBRP-complex) in Staphylococcus aureus. J Gen Microbiol. 1987;133:803–813. doi: 10.1099/00221287-133-3-803. [DOI] [PubMed] [Google Scholar]
- 4.Åkerström B, Björck L. Protein L: an immunoglobulin light chain-binding bacterial protein. Characterization of binding and physicochemical properties. J Biol Chem. 1989;264:19740–19746. [PubMed] [Google Scholar]
- 5.Åkerström B, Lindqvist A, Lindahl G. Binding properties of protein Arp, a bacterial IgA-receptor. Mol Immunol. 1991;28:349–357. doi: 10.1016/0161-5890(91)90147-c. [DOI] [PubMed] [Google Scholar]
- 6.Åkerström B, Nielsen E, Björck L. Definition of IgG- and albumin-binding regions of streptococcal protein G. J Biol Chem. 1987;262:13388–13391. [PubMed] [Google Scholar]
- 7.Åkesson P, Cooney J, Kishimoto F, Björck L. Protein H—a novel IgG binding bacterial protein. Mol Immunol. 1990;27:523–531. doi: 10.1016/0161-5890(90)90071-7. [DOI] [PubMed] [Google Scholar]
- 8.Åkesson P, Schmidt K H, Cooney J, Björck L. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994;300:877–886. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Åkesson P, Sjöholm A G, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenesinterfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 10.Aktories K. Rho proteins: targets for bacterial toxins. Trends Microbiol. 1997;5:282–288. doi: 10.1016/S0966-842X(97)01067-6. [DOI] [PubMed] [Google Scholar]
- 11.Altman E, Altman R K, Garrett J M, Grimaila R J, Young R. S gene product: identification and membrane localization of a lysis control protein. J Bacteriol. 1983;155:1130–1137. doi: 10.1128/jb.155.3.1130-1137.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson J S, Matsuhashi M, Haskin M A, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. II. Phospholipid carriers in the reaction sequence. J Biol Chem. 1967;242:3180–3190. [PubMed] [Google Scholar]
- 13.Anderson J S, Strominger J L. Isolation and utilization of phospholipid intermediates in cell wall glycopeptide synthesis. Biochem Biophys Res Commun. 1965;21:516–521. doi: 10.1016/0006-291x(65)90414-6. [DOI] [PubMed] [Google Scholar]
- 14.Appelbaum B, Rosan B. Cell surface proteins of oral streptococci. Infect Immun. 1984;46:245–250. doi: 10.1128/iai.46.1.245-250.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki Y, Ito E. Linkage units in cell walls of Gram-positive bacteria. Crit Rev Microbiol. 1989;17:121–135. doi: 10.3109/10408418909105745. [DOI] [PubMed] [Google Scholar]
- 16.Archibald A R, Baddiley J, Heckels J E. Molecular arrangement of teichoic acid in the cell wall of Staphylococcus lactis. Nat New Biol. 1973;241:29–31. doi: 10.1038/newbio241029a0. [DOI] [PubMed] [Google Scholar]
- 17.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 18.Armstrong J, Baddiley J, Buchanan J G, Carss B. Nucleotides and the bacterial cell wall. Nature. 1958;181:1692–1693. doi: 10.1038/1811692a0. [DOI] [PubMed] [Google Scholar]
- 19.Atrih A, Zollner P, Allmaier G, Foster S J. Structural analysis of Bacillus subtilis168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baba T, Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. doi: 10.1016/S0966-842X(97)01196-7. [DOI] [PubMed] [Google Scholar]
- 21.Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–4797. [PMC free article] [PubMed] [Google Scholar]
- 22.Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17:4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baddiley J. Teichoic acids in cell walls and membranes of bacteria. Essays Biochem. 1972;8:35–77. [PubMed] [Google Scholar]
- 24.Bahl H, Scholz H, Bayan N, Chami M, Leblon G, Gulik-Krzywicki T, Shechter E, Fouet A, Mesnage S, Tosi-Couture E, Gounon P, Mock M, Conway de Macario E, Macario A J, Fernandez-Herrero L A, Olabarriá G, Berenguer J, Blaser M J, Kuen B, Lubitz W, Sára M, Pouwels P H, Kolen C P, Boot H J, Resch S. Molecular biology of S-layers. FEMS Microbiol Rev. 1997;20:47–98. doi: 10.1111/j.1574-6976.1997.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 25.Baker C, Edwards M S. Group B streptococcal infections. In: Remmington J, Klein J O, editors. Infectious diseases of the fetus and newborn infants. Philadelphia, Pa: The W. B. Saunders Co.; 1995. pp. 980–1054. [Google Scholar]
- 26.Banas J A, Russell R R, Ferretti J J. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutansIngbritt. Infect Immun. 1990;58:667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkulis S S, Jones M F. Studies of streptococcal cell walls. I. Isolation, chemical composition, and preparation of M protein. J Bacteriol. 1957;74:207–216. doi: 10.1128/jb.74.2.207-216.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barlow P N, Campbell I D. Strategy for studying modular proteins: application to complement modules. Methods Enzymol. 1994;239:464–485. doi: 10.1016/s0076-6879(94)39018-5. [DOI] [PubMed] [Google Scholar]
- 29.Barroso L A, Wang S Z, Phelps C J, Johnson J L, Wilkins T D. Nucleotide sequence of Clostridium difficiletoxin B gene. Nucleic Acids Res. 1990;18:4004. doi: 10.1093/nar/18.13.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumeister W, Karrenberg F, Rachel R, Engel A, ten Heggeler B, Saxton W O. The major cell envelope protein of Micrococcus radiodurans(R1). Structural and chemical characterization. Eur J Biochem. 1982;125:535–544. doi: 10.1111/j.1432-1033.1982.tb06715.x. [DOI] [PubMed] [Google Scholar]
- 31.Behr T, Fischer W, Peter-Katalinic J, Egge H. The structure of pneumococcal lipoteichoic acid. Improved preparation, chemical and mass spectrometric studies. Eur J Biochem. 1992;207:1063–1075. doi: 10.1111/j.1432-1033.1992.tb17143.x. [DOI] [PubMed] [Google Scholar]
- 32.Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- 33.Benghezal M, Lipke P N, Conzelmann A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J Cell Biol. 1995;130:1333–1344. doi: 10.1083/jcb.130.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben Nasr A, Herwald H, Sjöbring U, Renne T, Müller-Esterl W, Björck L. Absorption of kininogen from human plasma by Streptococcus pyogenesis followed by the release of bradykinin. Biochem J. 1997;326:657–660. [PMC free article] [PubMed] [Google Scholar]
- 35.Ben Nasr A B, Herwald H, Müller-Esterl W, Björck L. Human kininogens interact with M protein, a bacterial surface protein and virulence determinant. Biochem J. 1995;305:173–180. doi: 10.1042/bj3050173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensing B A, Dunny G M. Pheromone-inducible expression of an aggregation protein in Enterococcus faecalisrequires interaction of a plasmid-encoded RNA with components of the ribosome. Mol Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3311709.x. [DOI] [PubMed] [Google Scholar]
- 37.Bensing B A, Manias D A, Dunny G M. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 38.Benton K A, Paton J C, Briles D E. Differences in virulence for mice among Streptococcus pneumoniaestrains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect Immun. 1997;65:1237–1244. doi: 10.1128/iai.65.4.1237-1244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 40.Berge A, Kihlberg B M, Sjöholm A G, Björck L. Streptococcal protein H forms soluble complement-activating complexes with IgG, but inhibits complement activation by IgG-coated targets. J Biol Chem. 1997;272:20774–20781. doi: 10.1074/jbc.272.33.20774. [DOI] [PubMed] [Google Scholar]
- 41.Berge A, Rasmussen M, Björck L. Identification of an insertion sequence located in a region encoding virulence factors of Streptococcus pyogenes. Infect Immun. 1998;66:3449–3453. doi: 10.1128/iai.66.7.3449-3453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berge A, Sjöbring U. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J Biol Chem. 1993;268:25417–25424. [PubMed] [Google Scholar]
- 43.Berger-Bachi B. Expression of resistance to methicillin. Trends Microbiol. 1994;2:389–393. doi: 10.1016/0966-842x(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 44.Berger-Bachi B, Barberis-Maino L, Strassle A, Kayser F H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989;219:263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- 45.Berger-Bachi B, Strassle A, Gustafson J E, Kayser F H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 47.Berry A M, Lock R A, Paton J C. Cloning and characterization of nanB, a second Streptococcus pneumoniae neuraminidase gene, and purification of the NanB enzyme from recombinant Escherichia coli. J Bacteriol. 1996;178:4854–4860. doi: 10.1128/jb.178.16.4854-4860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry A M, Paton J C. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64:5255–5262. doi: 10.1128/iai.64.12.5255-5262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bessen D, Jones K F, Fischetti V A. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J Exp Med. 1989;169:269–283. doi: 10.1084/jem.169.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bessen D E. Localization of immunoglobulin A-binding sites within M or M-like proteins of group A streptococci. Infect Immun. 1994;62:1968–1974. doi: 10.1128/iai.62.5.1968-1974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bessen D E, Fischetti V A. Nucleotide sequences of two adjacent M or M-like protein genes of group A streptococci: different RNA transcript levels and identification of a unique immunoglobulin A-binding protein. Infect Immun. 1992;60:124–135. doi: 10.1128/iai.60.1.124-135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bevanger L, Naess A I. Mouse-protective antibodies against the Ibc proteins of group B streptococci. Acta Pathol Microbiol Immunol Scand Sect B. 1985;93:121–124. doi: 10.1111/j.1699-0463.1985.tb02862.x. [DOI] [PubMed] [Google Scholar]
- 53.Beveridge T J, Pouwels P H, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer E M, Schocher I, Sleytr U B, Morelli L, Callegari M L, Nomellini J F, Bingle W H, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Beguin P, Ohayon H, Gounon P, Matuschek M, Koval S F. Functions of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 54.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 55.Bisno A L. Alternate complement pathway activation by group A streptococci: role of M-protein. Infect Immun. 1979;26:1172–1176. doi: 10.1128/iai.26.3.1172-1176.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bisno A L, Collins C M, Turner J C. M proteins of group C streptococci isolated from patients with acute pharyngitis. J Clin Microbiol. 1996;34:2511–2515. doi: 10.1128/jcm.34.10.2511-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bisno A L, Craven D E, McCabe W R. M proteins of group G streptococci isolated from bacteremic human infections. Infect Immun. 1987;55:753–757. doi: 10.1128/iai.55.3.753-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Björck L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988;140:1194–1197. [PubMed] [Google Scholar]
- 59.Björck L, Kastern W, Lindahl G, Wideback K. Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Mol Immunol. 1987;24:1113–1122. doi: 10.1016/0161-5890(87)90080-0. [DOI] [PubMed] [Google Scholar]
- 60.Björck L, Kronvall G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J Immunol. 1984;133:969–974. [PubMed] [Google Scholar]
- 61.Black L W, Hogness D S. The lysozyme of bacteriophage lambda. I. Purification and molecular weight. J Biol Chem. 1969;244:1968–1975. [PubMed] [Google Scholar]
- 62.Blackmore T K, Fischetti V A, Sadlon T A, Ward H M, Gordon D L. M protein of the group A Streptococcusbinds to the seventh short consensus repeat of human complement factor H. Infect Immun. 1998;66:1427–1431. doi: 10.1128/iai.66.4.1427-1431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blasi U, Chang C Y, Zagotta M T, Nam K B, Young R. The lethal lambda S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bleiweis A S, Craig R A, Zinner D D, Jablon J M. Chemical composition of purified cell walls of cariogenic streptococci. Infect Immun. 1971;3:189–191. doi: 10.1128/iai.3.1.189-191.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blumberg H M, Stephens D S, Modansky M, Erwin M, Elliot J, Facklam R R, Schuchat A, Baughman W, Farley M M. Invasive group B streptococcal disease: the emergence of serotype V. J Infect Dis. 1996;173:365–373. doi: 10.1093/infdis/173.2.365. [DOI] [PubMed] [Google Scholar]
- 67.Bodén M K, Flock J I. Fibrinogen-binding protein/clumping factor from Staphylococcus aureus. Infect Immun. 1989;57:2358–2363. doi: 10.1128/iai.57.8.2358-2363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bon J, Mani N, Jayaswal R K. Molecular analysis of lytic genes of bacteriophage 80α of Staphylococcus aureus. Can J Microbiol. 1997;43:612–616. doi: 10.1139/m97-087. [DOI] [PubMed] [Google Scholar]
- 69.Boothroyd J C, Cross G A, Hoeijmakers J H, Borst P. A variant surface glycoprotein of Trypanosoma bruceisynthesized with a C-terminal hydrophobic ‘tail’ absent from purified glycoprotein. Nature. 1980;288:624–626. doi: 10.1038/288624a0. [DOI] [PubMed] [Google Scholar]
- 70.Borchardt S A, Babwah A V, Jayaswal R K. Sequence analysis of the region downstream from a peptidoglycan hydrolase-encoding gene from Staphylococcus aureusNCTC8325. Gene. 1993;137:253–258. doi: 10.1016/0378-1119(93)90016-v. [DOI] [PubMed] [Google Scholar]
- 71.Bormann N E, Cleary P P. Transcriptional analysis of mga, a regulatory gene in Streptococcus pyogenes: identification of monocistronic and bicistronic transcripts that phase vary. Gene. 1997;200:125–134. doi: 10.1016/s0378-1119(97)00392-2. [DOI] [PubMed] [Google Scholar]
- 72.Boschwitz J S, Timoney J F. Characterization of the antiphagocytic activity of equine fibrinogen for Streptococcus equi subsp. equi. Microb Pathog. 1994;17:121–129. doi: 10.1006/mpat.1994.1058. [DOI] [PubMed] [Google Scholar]
- 73.Boschwitz J S, Timoney J F. Inhibition of C3 deposition on Streptococcus equi subsp. equiby M protein: a mechanism for survival in equine blood. Infect Immun. 1994;62:3515–3520. doi: 10.1128/iai.62.8.3515-3520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouwer H G, Hinrichs D J. Cytotoxic-T-lymphocyte responses to epitopes of listeriolysin O and p60 following infection with Listeria monocytogenes. Infect Immun. 1996;64:2515–2522. doi: 10.1128/iai.64.7.2515-2522.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boyle M D, Hawlitzky J, Raeder R, Podbielski A. Analysis of genes encoding two unique type IIa immunoglobulin G-binding proteins expressed by a single group A streptococcal isolate. Infect Immun. 1994;62:1336–1347. doi: 10.1128/iai.62.4.1336-1347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boyle M D, Lottenberg R. Plasminogen activation by invasive human pathogens. Thromb Haemostasis. 1997;77:1–10. [PubMed] [Google Scholar]
- 77.Boyle M D, Weber-Heynemann J, Raeder R, Podbielski A. Characterization of a gene coding for a type IIo bacterial IgG-binding protein. Mol Immunol. 1995;32:669–678. doi: 10.1016/0161-5890(95)00022-7. [DOI] [PubMed] [Google Scholar]
- 78.Bracha R, Chang M, Fiedler F, Glaser L. Biosynthesis of teichoic acids. Methods Enzymol. 1978;50:387–400. doi: 10.1016/0076-6879(78)50046-3. [DOI] [PubMed] [Google Scholar]
- 79.Bracha R, Glaser L. In vitro system for the synthesis of teichoic acid linked to peptidoglycan. J Bacteriol. 1976;125:872–879. doi: 10.1128/jb.125.3.872-879.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bracha R, Glaser L. An intermediate in teichoic acid biosynthesis. Biochem Biophys Res Commun. 1976;72:1091–1098. doi: 10.1016/s0006-291x(76)80244-6. [DOI] [PubMed] [Google Scholar]
- 81.Brady L J, Boyle M D. Identification of non-immunoglobulin A-Fc-binding forms and low-molecular-weight secreted forms of the group B streptococcal beta antigen. Infect Immun. 1989;57:1573–1581. doi: 10.1128/iai.57.5.1573-1581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InIB: an invasion protein of Listeria monocytogeneswith a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 83.Braun L, Ohayon H, Cossart P. The InIB protein of Listeria monocytogenesis sufficient to promote entry into mammalian cells. Mol Microbiol. 1998;27:1077–1087. doi: 10.1046/j.1365-2958.1998.00750.x. [DOI] [PubMed] [Google Scholar]
- 83a.Braun, L., and P. Cossart. Personal communication.
- 84.Braun V, Bosch V. In vivo biosynthesis of murein-lipoprotein of the outer membrane of E. coli. FEBS Lett. 1973;34:302–306. doi: 10.1016/0014-5793(73)80817-8. [DOI] [PubMed] [Google Scholar]
- 85.Braun V, Hantke K. Biochemistry of bacterial cell envelopes. Annu Rev Biochem. 1974;43:89–121. doi: 10.1146/annurev.bi.43.070174.000513. [DOI] [PubMed] [Google Scholar]
- 86.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. colicell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 87.Braun V, Rotering H, Ohms J P, Hagenmaier H. Conformational studies on murein-lipoprotein from the outer membrane of Escherichia coli. Eur J Biochem. 1976;70:601–610. doi: 10.1111/j.1432-1033.1976.tb11051.x. [DOI] [PubMed] [Google Scholar]
- 88.Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia colicell wall. The attachment site of the lipoprotein on the murein. Eur J Biochem. 1970;13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 89.Braun V, Wolff H. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem. 1970;14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 90.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 91.Briese T, Hakenbeck R. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur J Biochem. 1985;146:417–427. doi: 10.1111/j.1432-1033.1985.tb08668.x. [DOI] [PubMed] [Google Scholar]
- 92.Briles D E, King J D, Gray M A, McDaniel L S, Swiatlo E, Benton K A. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine. 1996;14:858–867. doi: 10.1016/0264-410x(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 93.Briles D E, Yother J, McDaniel L S. Role of pneumococcal surface protein A in the virulence of Streptococcus pneumoniae. Rev Infect Dis. 1988;10:S372–S374. doi: 10.1093/cid/10.supplement_2.s372. [DOI] [PubMed] [Google Scholar]
- 94.Brooks W, Burnie J P. Cloning and sequencing the endocarditis immunodominant antigen of Streptococcus sobrinusstrain MUCOB 263. J Med Microbiol. 1994;40:330–337. doi: 10.1099/00222615-40-5-330. [DOI] [PubMed] [Google Scholar]
- 95.Browder H P, Zygmunt W A, Young J R, Tavoramina P A. Lysostaphin: enzymatic mode of action. Biochem Biophys Res Commun. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- 96.Brown D A, Rose J K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 97.Brumfitt W, Wardlaw A C, Park J T. Development of lysozyme-resistance in Micrococcus lysodiekticusand its association with increased O-acetyl content of the cell wall. Nature. 1958;181:1783–1784. doi: 10.1038/1811783a0. [DOI] [PubMed] [Google Scholar]
- 98.Brunskill E W, Bayles K W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brunskill E W, Bayles K W. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bubert A, Kuhn M, Goebel W, Kohler S. Structural and functional properties of the p60 proteins from different Listeriaspecies. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buchanan C E, Henriques A O, Piggot P J. Cell wall changes during bacterial endospore formation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishing, Inc.; 1994. pp. 167–186. [Google Scholar]
- 102.Bunai K, Takamatsu H, Horinaka T, Oguro A, Nakamura K, Yamane K. Bacillus subtilisFfh, a homologue of mammalian SRP54, can intrinsically bind to the precursors of secretory proteins. Biochem Biophys Res Commun. 1996;227:762–767. doi: 10.1006/bbrc.1996.1582. [DOI] [PubMed] [Google Scholar]
- 103.Burne R A, Penders J E. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-beta-d-fructosidase. Infect Immun. 1992;60:4621–4632. doi: 10.1128/iai.60.11.4621-4632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burne R A, Penders J E. Differential localization of the Streptococcus mutansGS-5 fructan hydrolase enzyme, FruA. FEMS Microbiol Lett. 1994;121:243–249. doi: 10.1111/j.1574-6968.1994.tb07105.x. [DOI] [PubMed] [Google Scholar]
- 105.Burne R A, Schilling K, Bowen W H, Yasbin R E. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cabacungan E, Pieringer R A. Mode of elongation of the glycerol phosphate polymer of membrane lipoteichoic acid of Streptococcus faeciumATCC 9790. J Bacteriol. 1981;147:75–79. doi: 10.1128/jb.147.1.75-79.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Camara M, Boulnois G J, Andrew P W, Mitchell T J. A neuraminidase from Streptococcus pneumoniaehas the features of a surface protein. Infect Immun. 1994;62:3688–3695. doi: 10.1128/iai.62.9.3688-3695.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Camara M, Mitchell T J, Andrew P W, Boulnois G J. Streptococcus pneumoniae produces at least two distinct enzymes with neuraminidase activity: cloning and expression of a second neuraminidase gene in Escherichia coli. Infect Immun. 1991;59:2856–2858. doi: 10.1128/iai.59.8.2856-2858.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Canvin J R, Paton J C, Boulnois G J, Andrew P W, Mitchell T J. Streptococcus pneumoniaeproduces a second haemolysin that is distinct from pneumolysin. Microb Pathog. 1997;22:129–132. doi: 10.1006/mpat.1996.0098. [DOI] [PubMed] [Google Scholar]
- 110.Caparon M G, Geist R T, Perez-Casal J, Scott J R. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carlsson Wistedt A, Ringdahl U, Müller-Esterl W, Sjöbring U. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol Microbiol. 1995;18:569–578. doi: 10.1111/j.1365-2958.1995.mmi_18030569.x. [DOI] [PubMed] [Google Scholar]
- 113.Carpenter C V, Neuhaus F C. Enzymatic synthesis of d-alanyl-d-alanine. Two binding modes for product on d-alanine:d-alanine ligase (ADP) Biochemistry. 1972;11:2594–2598. doi: 10.1021/bi00764a007. [DOI] [PubMed] [Google Scholar]
- 114.Chami M, Bayan N, Dedieu J, Leblon G, Shechter E, Gulik-Krzywicki T. Organization of the outer layers of the cell envelope of Corynebacterium glutamicum: a combined freeze-etch electron microscopy and biochemical study. Biol Cell. 1995;83:219–229. doi: 10.1016/0248-4900(96)81311-6. [DOI] [PubMed] [Google Scholar]
- 115.Chami M, Bayan N, Peyret J L, Gulik-Krzywicki T, Leblon G, Shechter E. The S-layer protein of Corynebacterium glutamicumis anchored to the cell wall by its C-terminal hydrophobic domain. Mol Microbiol. 1997;23:483–492. doi: 10.1046/j.1365-2958.1997.d01-1868.x. [DOI] [PubMed] [Google Scholar]
- 116.Chatterjee A N, Park J T. Biosynthesis of cell wall mucopeptide by a particulate fraction from Staphylococcus aureus. Proc Natl Acad Sci USA. 1964;51:9–16. doi: 10.1073/pnas.51.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 118.Chen C C, Cleary P P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1990;265:3161–3167. [PubMed] [Google Scholar]
- 119.Chmouryguina I, Suvorov A, Ferrieri P, Cleary P P. Conservation of the C5a peptidase genes in group A and B streptococci. Infect Immun. 1996;64:2387–2390. doi: 10.1128/iai.64.7.2387-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cisar J O, Vatter A E, Clark W B, Curl S H, Hurst-Calderone S, Sandberg A L. Mutants of Actinomyces viscosusT14V lacking type 1, type 2, or both types of fimbriae. Infect Immun. 1988;56:2984–2989. doi: 10.1128/iai.56.11.2984-2989.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Clark W B, Wheeler T T, Cisar J O. Specific inhibition of adsorption of Actinomyces viscosusT14V to saliva-treated hydroxyapatite by antibody against type 1 fimbriae. Infect Immun. 1984;43:497–501. doi: 10.1128/iai.43.2.497-501.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clarke V A, Platt N, Butters T D. Cloning and expression of the beta-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J Biol Chem. 1995;270:8805–8814. doi: 10.1074/jbc.270.15.8805. [DOI] [PubMed] [Google Scholar]
- 123.Cleary P P, Handley J, Suvorov A N, Podbielski A, Ferrieri P. Similarity between the group B and A streptococcal C5a peptidase genes. Infect Immun. 1992;60:4239–4244. doi: 10.1128/iai.60.10.4239-4244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cleary P P, Peterson J, Chen C, Nelson C. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect Immun. 1991;59:2305–2310. doi: 10.1128/iai.59.7.2305-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cleary P P, Prahbu U, Dale J B, Wexler D E, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60:5219–5223. doi: 10.1128/iai.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Colby S M, Whiting G C, Tao L, Russell R R. Insertional inactivation of the Streptococcus mutans dexA(dextranase) gene results in altered adherence and dextran catabolism. Microbiology. 1995;141:2929–2936. doi: 10.1099/13500872-141-11-2929. [DOI] [PubMed] [Google Scholar]
- 127.Cole R M, Hahn J J. Cell wall replication in Streptococcus pyogenes. Science. 1962;135:722–724. doi: 10.1126/science.135.3505.722. [DOI] [PubMed] [Google Scholar]
- 128.Coley J, Archibald A R, Baddiley J. A linkage unit joining peptidoglycan to teichoic acid in Staphylococcus aureusH. FEBS Lett. 1976;61:240–242. doi: 10.1016/0014-5793(76)81047-2. [DOI] [PubMed] [Google Scholar]
- 129.Collins C M, Kimura A, Bisno A L. Group G streptococcal M protein exhibits structural features analogous to those of class-I M protein of group A streptococci. Infect Immun. 1992;60:3689–3696. doi: 10.1128/iai.60.9.3689-3696.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Conlan J W, North R J. Neutrophil-mediated dissolution of infected host cells as a defense strategy against a facultative intracellular bacterium. J Exp Med. 1991;174:741–744. doi: 10.1084/jem.174.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cossart P, Lecuit M. Interactions of Listeria monocytogeneswith mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Courtney H S, Dale J B, Hasty D I. Differential effects of the streptococcal fibronectin-binding protein, FBP54, on adhesion of group A streptococci to human buccal cells and HEp-2 tissue culture cells. Infect Immun. 1996;64:2415–2419. doi: 10.1128/iai.64.7.2415-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Courtney H S, Li Y, Dale J B, Hasty D L. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect Immun. 1994;62:3937–3946. doi: 10.1128/iai.62.9.3937-3946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Courtney H S, Liu S, Dale J B, Hasty D L. Conversion of M serotype 24 of Streptococcus pyogenesto M serotypes 5 and 18: effect on resistance to phagocytosis and adhesion to host cells. Infect Immun. 1997;65:2472–2474. doi: 10.1128/iai.65.6.2472-2474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Croux C, Ronda C, López R, García J L. Interchange of functional domains switches enzyme specificity: construction of a chimeric pneumococcal-clostridial cell wall lytic enzyme. Mol Microbiol. 1993;9:1019–1025. doi: 10.1111/j.1365-2958.1993.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 136.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dale J B, Washburn R G, Marques M B, Wessels M R. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect Immun. 1996;64:1495–1501. doi: 10.1128/iai.64.5.1495-1501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davis N G, Model P. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell. 1985;41:607–614. doi: 10.1016/s0092-8674(85)80033-7. [DOI] [PubMed] [Google Scholar]
- 139.Debabov D V, Heaton M P, Zhang Q, Stewart K D, Lambalot R H, Neuhaus F C. The D-Alanyl carrier protein in Lactobacillus casei: cloning, sequencing, and expression of dltC. J Bacteriol. 1996;178:3869–3876. doi: 10.1128/jb.178.13.3869-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.de Château M, Björck L. Identification of interdomain sequences promoting the intronless evolution of a bacterial protein family. Proc Natl Acad Sci USA. 1996;93:8490–8495. doi: 10.1073/pnas.93.16.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Château M, Björck L. Protein PAB, a mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J Biol Chem. 1994;269:12147–12151. [PubMed] [Google Scholar]
- 142.de Gier J W, Valent Q A, Von Heijne G, Luirink J. The E. coliSRP: preferences of a targeting factor. FEBS Lett. 1997;408:1–4. doi: 10.1016/s0014-5793(97)00402-x. [DOI] [PubMed] [Google Scholar]
- 143.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureusat 2.9- and 2.8-Å resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 144.de Jonge B L, Chang Y S, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureusstrain. The role of penicillin binding protein 2A. J Biol Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 145.de Jonge B L, Sidow T, Chang Y S, Labischinski H, Berger-Bachi B, Gage D A, Tomasz A. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femAlocus. J Bacteriol. 1993;175:2779–2782. doi: 10.1128/jb.175.9.2779-2782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Delves-Broughton J, Blackburn P, Evans R J, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 148.Demuth D R, Davis C A, Corner A M, Lamont R J, Leboy P S, Malamud D. Cloning and expression of a Streptococcus sanguissurface antigen that interacts with a human salivary agglutinin. Infect Immun. 1988;56:2484–2490. doi: 10.1128/iai.56.9.2484-2490.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Demuth D R, Duan Y, Brooks W, Holmes A R, McNab R, Jenkinson H F. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordoniito human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 150.Demuth D R, Lammey M S, Huck M, Lally E T, Malamud D. Comparison of Streptococcus mutans and Streptococcus sanguisreceptors for human salivary agglutinin. Microb Pathog. 1990;9:199–211. doi: 10.1016/0882-4010(90)90022-i. [DOI] [PubMed] [Google Scholar]
- 151.den Blaauwen T, Terpetschnig E, Lakowicz J R, Driessen A J. Interaction of SecB with soluble SecA. FEBS Lett. 1997;416:35–38. doi: 10.1016/s0014-5793(97)01142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Desai M, Tanna A, Efstratiou A, George R, Clewley J, Stanley J. Extensive genetic diversity among clinical isolates of Streptococcus pyogenesserotype M5. Microbiology. 1998;144:629–637. doi: 10.1099/00221287-144-3-629. [DOI] [PubMed] [Google Scholar]
- 153.Dev I K, Ray P H. Signal peptidases and signal peptide hydrolases. J Bioenerg Biomembr. 1990;22:271–290. doi: 10.1007/BF00763168. [DOI] [PubMed] [Google Scholar]
- 154.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 155.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremorisSK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 156.Díaz E, García E, Ascaso C, Méndez E, López R, García J L. Subcellular localization of the major pneumococcal autolysin: a peculiar mechanism of secretion in Escherichia coli. J Biol Chem. 1989;264:1238–1244. [PubMed] [Google Scholar]
- 157.Dickinson R B, Nagel J A, McDevitt D, Foster T J, Proctor R A, Cooper S L. Quantitative comparison of clumping factor- and coagulase-mediated Staphylococcus aureusadhesion to surface-bound fibrinogen under flow. Infect Immun. 1995;63:3143–3150. doi: 10.1128/iai.63.8.3143-3150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Di Fabio S, Medaglini D, Rush C M, Corrias F, Panzini G L, Pace M, Verani P, Pozzi G, Titti F. Vaginal immunization of Cynomolgus monkeys with Streptococcus gordoniiexpressing HIV-1 and HPV 16 antigens. Vaccine. 1998;16:485–492. doi: 10.1016/s0264-410x(97)80002-3. [DOI] [PubMed] [Google Scholar]
- 159.Domann E, Zechel S, Lingnau A, Hain T, Darji A, Nichterlein T, Wehland J, Chakraborty T. Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogeneswhose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats. Infect Immun. 1997;65:101–109. doi: 10.1128/iai.65.1.101-109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Douglas C W. Bacterial-protein interactions in the oral cavity. Adv Dent Res. 1994;8:254–262. doi: 10.1177/08959374940080021901. [DOI] [PubMed] [Google Scholar]
- 161.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenesinto hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–261. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 162.Dramsi S, Dehoux P, Lebrun M, Goossens P L, Cossart P. Identification of four new members of the internalin multigene family of Listeria monocytogenesEGD. Infect Immun. 1997;65:1615–1625. doi: 10.1128/iai.65.5.1615-1625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dramsi S, Kocks C, Forestier C, Cossart P. Internalin-mediated invasion of epithelial cells by Listeria monocytogenesis regulated by the bacterial growth state, temperature and the pleiotropic activator PrfA. Mol Microbiol. 1993;9:931–941. doi: 10.1111/j.1365-2958.1993.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 164.Dramsi S, Lebrun M, Cossart P. Molecular and genetic determinants involved in invasion of mammalian cells by Listeria monocytogenes. Curr Top Microbiol Immunol. 1996;209:61–77. doi: 10.1007/978-3-642-85216-9_4. [DOI] [PubMed] [Google Scholar]
- 165.Duckworth M, Archibald A R, Baddiley J. Lipoteichoic acid and lipoteichoic acid carrier in Staphylococcus aureusH. FEBS Lett. 1975;53:176–179. doi: 10.1016/0014-5793(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 166.Dunny G M, Brown B L, Clewell D B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dunny G M, Craig R A, Carron R L, Clewell D B. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid. 1979;2:454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 168.Dunny G M, Leonard B A. Cell-cell communication in Gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 169.Dunny G M, Leonard B A, Hedberg P J. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol. 1995;177:871–876. doi: 10.1128/jb.177.4.871-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dunny G M, Zimmerman D L, Tortorello M L. Induction of surface exclusion (entry exclusion) by Streptococcus faecalissex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc Natl Acad Sci USA. 1985;82:8582–8586. doi: 10.1073/pnas.82.24.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the Gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 172.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFYajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Duong F, Wickner W. Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J. 1998;17:696–705. doi: 10.1093/emboj/17.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 175.Egelseer E, Schocher I, Sára M, Sleytr U B. The S-layer from Bacillus stearothermophilusDSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J Bacteriol. 1995;177:1444–1451. doi: 10.1128/jb.177.6.1444-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Egelseer E M, Leitner K, Jarosch M, Hotzy C, Zayni S, Sleytr U B, Sára M. The S-layer proteins of two Bacillus stearothermophiluswild-type strains are bound via their N-terminal region to a secondary cell wall polymer of identical chemical composition. J Bacteriol. 1998;180:1488–1495. doi: 10.1128/jb.180.6.1488-1495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Egelseer E M, Schocher I, Sleytr U B, Sára M. Evidence that an N-terminal S-layer protein fragment triggers the release of a cell-associated high-molecular-weight amylase in Bacillus stearothermophilusATCC 12980. J Bacteriol. 1996;178:5602–5609. doi: 10.1128/jb.178.19.5602-5609.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ehrenfeld E E, Clewell D B. Transfer functions of the Streptococcus faecalisplasmid pAD1: organization of plasmid DNA encoding response to sex pheromone. J Bacteriol. 1987;169:3473–3481. doi: 10.1128/jb.169.8.3473-3481.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Eichler J, Wickner W. Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc Natl Acad Sci USA. 1997;94:5574–5581. doi: 10.1073/pnas.94.11.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ellen R P, Gibbons R J. M protein-associated adherence of Streptococcus pyogenesto epithelial surfaces: prerequisite for virulence. Infect Immun. 1972;5:826–830. doi: 10.1128/iai.5.5.826-830.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Emr S D, Hanley-Way S, Silhavy T J. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 182.Emr S D, Hedgpeth J, Clement J M, Silhavy T J, Hofnung M. Sequence analysis of mutations that prevent export of lambda receptor, an Escherichia coliouter membrane protein. Nature. 1980;285:82–85. doi: 10.1038/285082a0. [DOI] [PubMed] [Google Scholar]
- 183.Endl J, Seidl H P, Fiedler F, Schleifer K H. Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch Microbiol. 1983;135:215–223. doi: 10.1007/BF00414483. [DOI] [PubMed] [Google Scholar]
- 184.Endl J, Seidl P H, Fiedler F, Schleifer K H. Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch Microbiol. 1984;137:272–280. doi: 10.1007/BF00414557. [DOI] [PubMed] [Google Scholar]
- 185.Engelbrecht F, Chun S K, Ochs C, Hess J, Lottspeich F, Goebel W, Sokolovic Z. A new PrfA-regulated gene of Listeria monocytogenesencoding a small, secreted protein which belongs to the family of internalins. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 186.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian K D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992;58:3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Englund P T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 188.Enokizono J, Wikström M, Sjöbring U, Björck L, Forsén S, Arata Y, Kato K, Shimada I. NMR analysis of the interaction between protein L and Ig light chains. J Mol Biol. 1997;270:8–13. doi: 10.1006/jmbi.1997.1090. [DOI] [PubMed] [Google Scholar]
- 189.Ensign J C, Wolfe R S. Characterization of a small proteolytic enzyme which lyses bacterial cells. J Bacteriol. 1966;91:524–534. doi: 10.1128/jb.91.2.524-534.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Entian K D, de Vos W M. Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics. Antonie Leeuwenhoek. 1996;69:109–117. doi: 10.1007/BF00399416. [DOI] [PubMed] [Google Scholar]
- 191.Espersen F, Clemmensen I. Clumping of Staphylococcus aureusby human fibronectin. Acta Pathol Microbiol Scand Sect B. 1981;89:317–321. doi: 10.1111/j.1699-0463.1981.tb00195_89b.x. [DOI] [PubMed] [Google Scholar]
- 192.Exterkate F A, Alting A C, Bruinenberg P G. Diversity of cell envelope proteinase specificity among strains of Lactococcus lactisand its relationship to charge characteristics of the substrate-binding region. Appl Environ Microbiol. 1993;59:3640–3647. doi: 10.1128/aem.59.11.3640-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Facklam R, Beall B. Anomalies in emmtyping of group A streptococci. Adv Exp Med Biol. 1997;418:335–337. doi: 10.1007/978-1-4899-1825-3_80. [DOI] [PubMed] [Google Scholar]
- 194.Fahnestock S R, Alexander P, Nagle J, Filpula D. Gene for an immunoglobulin-binding protein from a group G streptococcus. J Bacteriol. 1986;167:870–880. doi: 10.1128/jb.167.3.870-880.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Farley M M, Harvey R C, Stull T, Smith J D, Schuchat A, Wenger J D, Stephens D S. A population-based assessment of invasive disease due to group B Streptococcusin nonpregnant adults. N Engl J Med. 1993;328:1807–1811. doi: 10.1056/NEJM199306243282503. [DOI] [PubMed] [Google Scholar]
- 196.Fasel N, Rousseaux M, Schaerer E, Medof M E, Tykocinski M L, Bron C. In vitro attachment of glycosyl-inositolphospholipid anchor structures to mouse Thy-1 antigen and human decay-accelerating factor. Proc Natl Acad Sci USA. 1989;86:6858–6862. doi: 10.1073/pnas.86.18.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fekkes P, van der Does C, Driessen A J. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ferguson M A, Homans S W, Dwek R A, Rademacher T W. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma bruceivariant surface glycoprotein to the membrane. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 199.Ferretti J J, Gilpin M L, Russell R R. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinusMFe28. J Bacteriol. 1987;169:4271–4278. doi: 10.1128/jb.169.9.4271-4278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ferretti J J, Huang T T, Russell R R. Sequence analysis of the glucosyltransferase A gene (gtfA) from Streptococcus mutansIngbritt. Infect Immun. 1988;56:1585–1588. doi: 10.1128/iai.56.6.1585-1588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ferretti J J, Russell R R, Dao M L. Sequence analysis of the wall-associated protein precursor of Streptococcus mutansantigen A. Mol Microbiol. 1989;3:469–478. doi: 10.1111/j.1365-2958.1989.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 202.Ferrieri P. Surface-localized protein antigens of group B streptococci. Rev Infect Dis. 1988;10:S363–S366. doi: 10.1093/cid/10.supplement_2.s363. [DOI] [PubMed] [Google Scholar]
- 203.Fiedler K, Kobayashi T, Kurzchalia T V, Simons K. Glycosphingolipid-enriched, detergent-insoluble complexes in protein sorting in epithelial cells. Biochemistry. 1993;32:6365–6373. doi: 10.1021/bi00076a009. [DOI] [PubMed] [Google Scholar]
- 204.Filpula D, Alexander P, Fahnestock S R. Nucleotide sequence of the protein G gene from Streptococcus GX7805, and comparison to previously reported sequences. Nucleic Acids Res. 1987;15:7210. doi: 10.1093/nar/15.17.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fischer W. Lipoteichoic acids and lipoglycans. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishing B.V.; 1994. pp. 199–216. [Google Scholar]
- 206.Fischer W. Molecular analysis of lipid macroamphiphiles by hydrophobic interaction chromatography, exemplified with lipoteichoic acids. Anal Biochem. 1993;208:49–56. doi: 10.1006/abio.1993.1007. [DOI] [PubMed] [Google Scholar]
- 207.Fischer W, Behr T, Hartmann R, Peter-Katalinic J, Egge H. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniaepossess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide) Eur J Biochem. 1993;215:851–857. doi: 10.1111/j.1432-1033.1993.tb18102.x. [DOI] [PubMed] [Google Scholar]
- 208.Fischer W, Koch H U, Haas R. Improved preparation of lipoteichoic acids. Eur J Biochem. 1983;133:523–530. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 209.Fischer W, Koch H U, Rosel P, Fiedler F. Alanine ester-containing native lipoteichoic acids do not act as lipoteichoic acid carriers. Isolation, structural and functional characterization. J Biol Chem. 1980;255:4557–4562. [PubMed] [Google Scholar]
- 210.Fischer W, Koch H U, Rosel P, Fiedler F, Schmuck L. Structural requirements of lipoteichoic acid carrier for recognition by the poly(ribitol phosphate) polymerase from Staphylococcus aureusH. A study of various lipoteichoic acids, derivatives, and related compounds. J Biol Chem. 1980;255:4550–4556. [PubMed] [Google Scholar]
- 211.Fischer W, Markwitz S, Labischinski H. Small-angle X-ray scattering analysis of pneumococcal lipoteichoic acid phase structure. Eur J Biochem. 1997;244:913–917. doi: 10.1111/j.1432-1033.1997.00913.x. [DOI] [PubMed] [Google Scholar]
- 212.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fischetti V A. The streptococcus and the host: present and future challenges. ASM News. 1997;63:541–545. doi: 10.1007/978-1-4899-1825-3_5. [DOI] [PubMed] [Google Scholar]
- 214.Fischetti V A, Horstmann R D, Pancholi V. Location of the complement factor H binding site on streptococcal M6 protein. Infect Immun. 1995;63:149–153. doi: 10.1128/iai.63.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fischetti V A, Medaglini D, Pozzi G. Gram-positive commensal bacteria for mucosal vaccine delivery. Curr Opin Biotechnol. 1996;7:659–666. doi: 10.1016/s0958-1669(96)80079-6. [DOI] [PubMed] [Google Scholar]
- 216.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 217.Fischetti V A, Parry D A, Trus B L, Hollingshead S K, Scott J R, Manjula B N. Conformational characteristics of the complete sequence of group A streptococcal M6 protein. Proteins. 1988;3:60–69. doi: 10.1002/prot.340030106. [DOI] [PubMed] [Google Scholar]
- 218.Flock J I, Froman G, Jonsson K, Guss B, Signäs C, Nilsson B, Raucci G, Höök M, Wadström T, Lindberg M. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 1987;6:2351–2357. doi: 10.1002/j.1460-2075.1987.tb02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Flock J I, Hienz S A, Heimdahl A, Schennings T. Reconsideration of the role of fibronectin binding in endocarditis caused by Staphylococcus aureus. Infect Immun. 1996;64:1876–1878. doi: 10.1128/iai.64.5.1876-1878.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Flores A E, Ferrieri P. Molecular species of R-protein antigens produced by clinical isolates of group B streptococci. J Clin Microbiol. 1989;27:1050–1054. doi: 10.1128/jcm.27.5.1050-1054.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Forester H, Hunter N, Knox K W. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans. J Gen Microbiol. 1983;129:2779–2788. doi: 10.1099/00221287-129-9-2779. [DOI] [PubMed] [Google Scholar]
- 222.Forsgren A, Sjöquist J. “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol. 1966;97:822–827. [PubMed] [Google Scholar]
- 223.Fortin Y, Phoenix P, Drapeau G R. Mutations conferring resistance to azide in Escherichia coli occur primarily in the secAgene. J Bacteriol. 1990;172:6607–6610. doi: 10.1128/jb.172.11.6607-6610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Foster S J. Analysis of the autolysins of Bacillus subtilis168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Foster S J. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus8325/4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Foster T J, McDevitt D. Surface-associated proteins of Staphylococcus aureus: their possible roles in virulence. FEMS Microbiol Lett. 1994;118:199–205. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]
- 227.Fox J A, Duszenko M, Ferguson M A, Low M G, Cross G A. Purification and characterization of a novel glycan-phosphatidylinositol-specific phospholipase C from Trypanosoma brucei. J Biol Chem. 1986;261:15767–15771. [PubMed] [Google Scholar]
- 228.Freimer E H, Krause R M, McCarty M. Studies of L forms and protoplasts of group A streptococci. I. Isolation, growth and bacteriologic characteristics. J Exp Med. 1959;110:853–874. doi: 10.1084/jem.110.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Frick I M, Åkesson P, Cooney J, Sjöbring U, Schmidt K H, Gomi H, Hattori S, Tagawa C, Kishimoto F, Björck L. Protein H—a surface protein of Streptococcus pyogeneswith separate binding sites for IgG and albumin. Mol Microbiol. 1994;12:143–151. doi: 10.1111/j.1365-2958.1994.tb01003.x. [DOI] [PubMed] [Google Scholar]
- 230.Frick I M, Crossin K L, Edelman G M, Björck L. Protein H—a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J. 1995;14:1674–1679. doi: 10.1002/j.1460-2075.1995.tb07156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Frick I M, Wikström M, Forsén S, Drakenberg T, Gomi H, Sjöbring U, Björck L. Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc Natl Acad Sci USA. 1992;89:8532–8536. doi: 10.1073/pnas.89.18.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Frithz E, Hedén L O, Lindahl G. Extensive sequence homology between IgA receptor and M proteins in Streptococcus pyogenes. Mol Microbiol. 1989;3:1111–1119. doi: 10.1111/j.1365-2958.1989.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 233.Froman G, Switalski L M, Speziale P, Höök M. Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J Biol Chem. 1987;262:6564–6571. [PubMed] [Google Scholar]
- 234.Furst P, Mosch H U, Solioz M. A protein of unusual composition from Enterococcus faecium. Nucleic Acids Res. 1989;17:6724. doi: 10.1093/nar/17.16.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gaillard J L, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenesinto cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 236.Galli D, Friesenegger A, Wirth R. Transcriptional control of sex-pheromone-inducible genes on plasmid pAD1 of Enterococcus faecalisand sequence analysis of a third structural gene for (pPD1-encoded) aggregation substance. Mol Microbiol. 1992;6:1297–1308. doi: 10.1111/j.1365-2958.1992.tb00851.x. [DOI] [PubMed] [Google Scholar]
- 237.Galli D, Lottspeich F, Wirth R. Sequence analysis of Enterococcus faecalisaggregation substance encoded by the sex pheromone plasmid pAD1. Mol Microbiol. 1990;4:895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 238.Galli D, Wirth R. Comparative analysis of Enterococcus faecalissex pheromone plasmids identifies a single homologous DNA region which codes for aggregation substance. J Bacteriol. 1991;173:3029–3033. doi: 10.1128/jb.173.9.3029-3033.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Galli D, Wirth R, Wanner G. Identification of aggregation substances of Enterococcus faecaliscells after induction by sex pheromones. An immunological and ultrastructural investigation. Arch Microbiol. 1989;151:486–490. doi: 10.1007/BF00454863. [DOI] [PubMed] [Google Scholar]
- 240.Gan K, Gupta S D, Sankaran K, Schmid M B, Wu H C. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimuriumdefective in prolipoprotein modification. J Biol Chem. 1993;268:16544–16550. [PubMed] [Google Scholar]
- 241.Ganfield M C, Pieringer R A. The biosynthesis of nascent membrane lipoteichoic acid of Streptococcus faecium (S. faecalisATCC 9790) from phosphatidylkojibiosyl diacylglycerol and phosphatidylglycerol. J Biol Chem. 1980;255:5164–5169. [PubMed] [Google Scholar]
- 242.García E, García J L, García P, Arrarás A, Sánchez-Puelles J M, López R. Molecular evolution of lytic enzymes of Streptococcus pneumoniaeand its bacteriophages. Proc Natl Acad Sci USA. 1988;85:914–918. doi: 10.1073/pnas.85.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.García J L, Díaz E, Romero A, Garcia P. Carboxy-terminal deletion analysis of the major pneumococcal autolysin. J Bacteriol. 1994;176:4066–4072. doi: 10.1128/jb.176.13.4066-4072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Garcia-Bustos J F, Chait B T, Tomasz A. Structure of the peptide network of pneumococcal peptidoglycan. J Biol Chem. 1987;262:15400–15405. [PubMed] [Google Scholar]
- 245.Garrett J, Fusselman R, Hise J, Chiou L, Smith-Grillo D, Schulz J, Young R. Cell lysis by induction of cloned lambda lysis genes. Mol Gen Genet. 1981;182:326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- 246.Gatermann S, Meyer H G. Staphylococcus saprophyticushemagglutinin binds fibronectin. Infect Immun. 1994;62:4556–4563. doi: 10.1128/iai.62.10.4556-4563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Geginat G, Lalic M, Kretschmar M, Goebel W, Hof H, Palm D, Bubert A. Th1 cells specific for a secreted protein of Listeria monocytogenesare protective in vivo. J Immunol. 1998;160:6046–6055. [PubMed] [Google Scholar]
- 248.Gerber L D, Kodukula K, Udenfriend S. Phosphatidylinositol glycan (PI-G) anchored membrane proteins. Amino acid requirements adjacent to the site of cleavage and PI-G attachment in the COOH-terminal signal peptide. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- 249.Germaine G R, Harlander S K, Leung W L, Schachtele C F. Streptococcus mutansdextransucrase: functioning of primer dextran and endogenous dextranase in water-soluble and water-insoluble glucan synthesis. Infect Immun. 1977;16:637–648. doi: 10.1128/iai.16.2.637-648.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ghuysen J-M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 251.Ghuysen J M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 252.Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. [Google Scholar]
- 253.Ghuysen J M, Lamotte-Brasseur J, Joris B, Shockman G D. Binding site-shaped repeated sequences of bacterial wall peptidoglycan hydrolases. FEBS Lett. 1994;342:23–28. doi: 10.1016/0014-5793(94)80577-6. [DOI] [PubMed] [Google Scholar]
- 254.Ghuysen J-M, Strominger J L. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. I. Preparation of fragments by enzymatic hydrolysis. Biochemistry. 1963;2:1110–1118. doi: 10.1021/bi00905a035. [DOI] [PubMed] [Google Scholar]
- 255.Ghuysen J-M, Strominger J L. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of the disaccharides. Biochemistry. 1963;2:1119–1125. doi: 10.1021/bi00905a036. [DOI] [PubMed] [Google Scholar]
- 256.Ghuysen J-M, Tipper D J, Birge C H, Strominger J L. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. VI. The soluble glycopeptide and its sequential degradation by peptidases. Biochemistry. 1965;4:2244–2254. doi: 10.1021/bi00879a016. [DOI] [PubMed] [Google Scholar]
- 257.Gibbons R J, Hay D I, Cisar J O, Clark W B. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosusT14V-J1 on apatitic surfaces. Infect Immun. 1988;56:2990–2993. doi: 10.1128/iai.56.11.2990-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Giesbrecht P, Kersten T, Wecke J. Fan-shaped ejections of regularly arranged murosomes involved in penicillin-induced death of staphylococci. J Bacteriol. 1992;174:2241–2252. doi: 10.1128/jb.174.7.2241-2252.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Giesbrecht P, Labischinski H, Wecke J. A special morphogenetic wall defect and the subsequent activity of “murosomes” as the very reason for penicillin-induced bacteriolysis in staphylococci. Arch Microbiol. 1985;141:315–324. doi: 10.1007/BF00428843. [DOI] [PubMed] [Google Scholar]
- 260.Giesbrecht P, Wecke J. Contribution to the morphogenesis of the cell wall of staphylococci. I. The formation of the cross wall and the cell separation. Cytobiologie. 1971;4:349–368. [Google Scholar]
- 261.Giesbrecht P, Wecke J, Reinicke B. On the morphogenesis of the cell wall of staphylococci. Int Rev Cytol. 1976;44:225–318. doi: 10.1016/s0074-7696(08)61651-4. [DOI] [PubMed] [Google Scholar]
- 262.Gillaspy A F, Patti J M, Smeltzer M S. Transcriptional regulation of the Staphylococcus aureus collagen adhesion gene, cna. Infect Immun. 1997;65:1536–1540. doi: 10.1128/iai.65.4.1536-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gilmore K S, Russell R R, Ferretti J J. Analysis of the Streptococcus downei gtfSgene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect Immun. 1990;58:2452–2458. doi: 10.1128/iai.58.8.2452-2458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gilson E, Alloing G, Schmidt T, Claverys J P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in Gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Giudicelli S, Tomasz A. Attachment of pneumococcal autolysin to wall teichoic acids, an essential step in enzymatic wall degradation. J Bacteriol. 1984;158:1188–1190. doi: 10.1128/jb.158.3.1188-1190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Glaser L, Lindsay B. The synthesis of lipoteichoic acid carrier. Biochem Biophys Res Commun. 1974;59:1131–1136. doi: 10.1016/s0006-291x(74)80096-3. [DOI] [PubMed] [Google Scholar]
- 267.Glaser L, Loewy A. Control of teichoic acid synthesis during phosphate limitation. J Bacteriol. 1979;137:327–331. doi: 10.1128/jb.137.1.327-331.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Glaser L, Loewy A. Regulation of teichoic acid synthesis during phosphate limitation. J Biol Chem. 1979;254:2184–2186. [PubMed] [Google Scholar]
- 269.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 270.Glauner B, Höltje J-V, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 271.Godon J J, Jury K, Shearman C A, Gasson M J. The Lactococcus lactis sex-factor aggregation gene cluA. Mol Microbiol. 1994;12:655–663. doi: 10.1111/j.1365-2958.1994.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 272.Goffin C, Ghuysen J M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Gomi H, Hozumi T, Hattori S, Tagawa C, Kishimoto F, Björck L. The gene sequence and some properties of protein H. A novel IgG-binding protein. J Immunol. 1990;144:4046–4052. [PubMed] [Google Scholar]
- 274.Good C M, Tipper D J. Conditional mutants of Staphylococcus aureusdefective in cell wall precursor synthesis. J Bacteriol. 1972;111:231–241. doi: 10.1128/jb.111.1.231-241.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Graham L L, Beveridge T J. Structural differentiation of the Bacillus subtilis168 cell wall. J Bacteriol. 1994;176:1413–1421. doi: 10.1128/jb.176.5.1413-1421.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Graham L L, Beveridge T J, Nanninga N. Periplasmic space and the concept of the periplasm. Trends Biochem Sci. 1991;16:328–329. doi: 10.1016/0968-0004(91)90135-i. [DOI] [PubMed] [Google Scholar]
- 277.Granlund M, Öberg L, Sellin M, Norgren M. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J Infect Dis. 1998;177:967–976. doi: 10.1086/515233. [DOI] [PubMed] [Google Scholar]
- 278.Gravekamp C, Horensky D S, Michel J L, Madoff L C. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect Immun. 1996;64:3576–3583. doi: 10.1128/iai.64.9.3576-3583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Gravekamp C, Kasper D L, Michel J L, Kling D E, Carey V, Madoff L C. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infect Immun. 1997;65:5216–5221. doi: 10.1128/iai.65.12.5216-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Gravekamp C, Rosner B, Madoff L C. Deletion of repeats in the alpha C protein enhances the pathogenicity of group B streptococci in immune mice. Infect Immun. 1998;66:4347–4354. doi: 10.1128/iai.66.9.4347-4354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Green C J, Vold B S. Staphylococcus aureushas clustered tRNA genes. J Bacteriol. 1993;175:5091–5096. doi: 10.1128/jb.175.16.5091-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Greene C, McDevitt D, Francois P, Vaudaux P E, Lew D P, Foster T J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnbgenes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 283.Gregory S H, Sagnimeni A J, Wing E J. Expression of the inlAB operon by Listeria monocytogenesis not required for entry into hepatic cells in vivo. Infect Immun. 1996;64:3983–3986. doi: 10.1128/iai.64.10.3983-3986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Gregory S H, Sagnimeni A J, Wing E J. Internalin B promotes the replication of Listeria monocytogenesin mouse hepatocytes. Infect Immun. 1997;65:5137–5141. doi: 10.1128/iai.65.12.5137-5141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Griffith F. The serological classification of Streptococcus pyogenes. J Hyg. 1934;34:542–584. doi: 10.1017/s0022172400043308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Gronenborn A M, Filpula D R, Essig N Z, Achari A, Whitlow M, Wingfield P T, Clore G M. A novel highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 287.Gunetileke K G, Anwar R A. Biosynthesis of uridine diphospho-N-acetyl muramic acid. J Biol Chem. 1966;241:5740–5743. [PubMed] [Google Scholar]
- 288.Gunneriusson E, Samuelson P, Uhlén M, Nygren P A, Stahl S. Surface display of a functional single-chain Fv antibody on staphylococci. J Bacteriol. 1996;178:1341–1346. doi: 10.1128/jb.178.5.1341-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Gupta S D, Gan K, Schmid M B, Wu H C. Characterization of a temperature-sensitive mutant of Salmonella typhimuriumdefective in apolipoprotein N-acyltransferase. J Biol Chem. 1993;268:16551–16556. [PubMed] [Google Scholar]
- 290.Guss B, Eliasson M, Olsson A, Uhlén M, Frej A K, Jornvall H, Flock J I, Lindberg M. Structure of the IgG-binding regions of streptococcal protein G. EMBO J. 1986;5:1567–1575. doi: 10.1002/j.1460-2075.1986.tb04398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Guss B, Uhlén M, Nilsson B, Lindberg M, Sjöquist J, Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 292.Gustafson J, Strassle A, Hachler H, Kayser F H, Berger-Bachi B. The femC locus of Staphylococcus aureusrequired for methicillin resistance includes the glutamine synthetase operon. J Bacteriol. 1994;176:1460–1467. doi: 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Haandrikman A J, Kok J, Laan H, Soemitro S, Ledeboer A M, Konings W N, Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989;171:2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Haandrikman A J, Meesters R, Laan H, Konings W N, Kok J, Venema G. Processing of the lactococcal extracellular serine proteinase. Appl Environ Microbiol. 1991;57:1899–1904. doi: 10.1128/aem.57.7.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Haanes E J, Cleary P P. Identification of a divergent M protein gene and an M protein-related gene family in Streptococcus pyogenesserotype 49. J Bacteriol. 1989;171:6397–6408. doi: 10.1128/jb.171.12.6397-6408.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Haanes E J, Heath D G, Cleary P P. Architecture of the virregulons of group A streptococci parallels opacity factor phenotype and M protein class. J Bacteriol. 1992;174:4967–4976. doi: 10.1128/jb.174.15.4967-4976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Haanes-Fritz E, Kraus W, Burdett V, Dale J B, Beachey E H, Cleary P. Comparison of the leader sequences of four group A streptococcal M protein genes. Nucleic Acids Res. 1988;16:4667–4677. doi: 10.1093/nar/16.10.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hallberg K, Holm C, Öhman U, Strömberg N. Actinomyces naeslundii displays variant fimP and fimAfimbrial subunit genes corresponding to different types of acidic proline-rich protein and beta-linked galactosamine binding specificity. Infect Immun. 1998;66:4403–4410. doi: 10.1128/iai.66.9.4403-4410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Hamada S, Slade H D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Hamburger D, Egerton M, Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Hanada N, Kuramitsu H K. Isolation and characterization of the Streptococcus mutans gtfDgene, coding for primer-dependent soluble glucan synthesis. Infect Immun. 1989;57:2079–2085. doi: 10.1128/iai.57.7.2079-2085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hancock I C. Bacterial cell surface carbohydrates: structure and assembly. Biochem Soc Trans. 1997;25:183–187. doi: 10.1042/bst0250183. [DOI] [PubMed] [Google Scholar]
- 303.Hanski E, Caparon M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Hanski E, Horwitz P A, Caparon M G. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenesJRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect Immun. 1992;60:5119–5125. doi: 10.1128/iai.60.12.5119-5125.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hantke K, Braun V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coliouter membrane. Eur J Biochem. 1973;34:284–296. doi: 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- 306.Harbaugh M P, Podbielski A, Hügl S, Cleary P P. Nucleotide substitutions and small-scale insertion produce size and antigenic variation in group A streptococcal M1 protein. Mol Microbiol. 1993;8:981–991. doi: 10.1111/j.1365-2958.1993.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 307.Harrington D J, Russell R R. Multiple changes in cell wall antigens of isogenic mutants of Streptococcus mutans. J Bacteriol. 1993;175:5925–5933. doi: 10.1128/jb.175.18.5925-5933.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hartford O, Francois P, Vaudaux P, Foster T J. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureuscell surface. Mol Microbiol. 1997;25:1065–1076. doi: 10.1046/j.1365-2958.1997.5291896.x. [DOI] [PubMed] [Google Scholar]
- 309.Hartl F U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coliplasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 310.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Hayakawa M, Aoki H, Kuramitsu H K. Isolation and characterization of the sucrose 6-phosphate hydrolase gene from Streptococcus mutans. Infect Immun. 1986;53:582–586. doi: 10.1128/iai.53.3.582-586.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Heath D G, Boyle M D, Cleary P P. Isolated DNA repeat region from fcrA76, the Fc-binding protein gene from an M-type 76 strain of group A streptococci, encodes a protein with Fc-binding activity. Mol Microbiol. 1990;4:2071–2079. doi: 10.1111/j.1365-2958.1990.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 313.Heath D G, Cleary P P. Fc-receptor and M-protein genes of group A streptococci are products of gene duplication. Proc Natl Acad Sci USA. 1989;86:4741–4745. doi: 10.1073/pnas.86.12.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Heath H E, Heath L S, Nitterauer J D, Rose K E, Sloan G L. Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem Biophys Res Commun. 1989;160:1106–1109. doi: 10.1016/s0006-291x(89)80117-2. [DOI] [PubMed] [Google Scholar]
- 315.Heaton M P, Discotto L F, Pucci M J, Handwerger S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faeciumsex pheromone-response plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 316.Heaton M P, Neuhaus F C. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J Bacteriol. 1992;174:4707–4717. doi: 10.1128/jb.174.14.4707-4717.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Heaton M P, Neuhaus F C. Role of the d-alanyl carrier protein in the biosynthesis of d-alanyl-lipoteichoic acid. J Bacteriol. 1994;176:681–690. doi: 10.1128/jb.176.3.681-690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Heckels J E, Archibald A R, Baddiley J. Studies on the linkage between teichoic acid and peptidoglycan in a bacteriophage-resistant mutant of Staphylococcus aureusH. Biochem J. 1975;149:637–647. doi: 10.1042/bj1490637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hedén L O, Frithz E, Lindahl G. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur J Immunol. 1991;21:1481–1490. doi: 10.1002/eji.1830210623. [DOI] [PubMed] [Google Scholar]
- 320.Hedén L O, Lindahl G. Conserved and variable regions in protein Arp, the IgA receptor of Streptococcus pyogenes. J Gen Microbiol. 1993;139:2067–2074. doi: 10.1099/00221287-139-9-2067. [DOI] [PubMed] [Google Scholar]
- 321.Heilmann C, Gerke C, Perdreau-Remington F, Götz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidisaffected in biofilm formation. Infect Immun. 1996;64:277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Heilmann C, Gotz F. Further characterization of Staphylococcus epidermidistransposon mutants deficient in primary attachment or intercellular adhesion. Zentbl Bakteriol. 1998;287:69–83. doi: 10.1016/s0934-8840(98)80149-7. [DOI] [PubMed] [Google Scholar]
- 323.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidisto a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 324.Heinrich P, Rosenstein R, Bohmer M, Sonner P, Gotz F. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol Gen Genet. 1987;209:563–569. doi: 10.1007/BF00331163. [DOI] [PubMed] [Google Scholar]
- 325.Hell W, Meyer H G, Gatermann S G. Cloning of aas, a gene encoding a Staphylococcus saprophyticussurface protein with adhesive and autolytic properties. Mol Microbiol. 1998;29:871–881. doi: 10.1046/j.1365-2958.1998.00983.x. [DOI] [PubMed] [Google Scholar]
- 326.Hellwage J, Kühn S, Zipfel P F. The human complement regulatory factor-H-like protein 1, which represents a truncated form of factor H, displays cell-attachment activity. Biochem J. 1997;326:321–327. doi: 10.1042/bj3260321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 328.Helmann J D, Marquez L M, Chamberlin M J. Cloning, sequencing, and disruption of the Bacillus subtilissigma 28 gene. J Bacteriol. 1988;170:1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Hemila H, Palva A, Paulin L, Adler L, Arvidson S, Palva I. The secretory S complex in Bacillus subtilisis identified as pyruvate dehydrogenase. Res Microbiol. 1991;142:779–785. doi: 10.1016/0923-2508(91)90055-f. [DOI] [PubMed] [Google Scholar]
- 330.Hemila H, Palva A, Paulin L, Arvidson S, Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990;172:5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bachi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993;175:1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Herbold D R, Glaser L. Bacillus subtilisN-acetylmuramic acid L-alanine amidase. J Biol Chem. 1975;250:1676–1682. [PubMed] [Google Scholar]
- 333.Herbold D R, Glaser L. Interaction of N-acetylmuramic acid L-alanine amidase with cell wall polymers. J Biol Chem. 1975;250:7231–7238. [PubMed] [Google Scholar]
- 334.Hess J, Dreher A, Gentschev I, Goebel W, Ladel C, Miko D, Kaufmann S H. Protein p60 participates in intestinal host invasion by Listeria monocytogenes. Zentbl Bakteriol. 1996;284:263–272. doi: 10.1016/s0934-8840(96)80102-2. [DOI] [PubMed] [Google Scholar]
- 335.Higashi Y, Siewert G, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XIX. Isoprenoid alcohol phosphokinase. J Biol Chem. 1970;245:3683–3690. [PubMed] [Google Scholar]
- 336.Higashi Y, Strominger J L, Sweeley C C. Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus. J Biol Chem. 1970;245:3697–3702. [PubMed] [Google Scholar]
- 337.Higashi Y, Strominger J L, Sweeley C C. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol. Proc Natl Acad Sci USA. 1967;57:1878–84. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hiroi Y, Komuro I, Chen R, Hosoda T, Mizuno T, Kudoh S, Georgescu S P, Medof M E, Yazaki Y. Molecular cloning of human homolog of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett. 1998;421:252–258. doi: 10.1016/s0014-5793(97)01576-7. [DOI] [PubMed] [Google Scholar]
- 339.Hirt H, Wirth R, Muscholl A. Comparative analysis of 18 sex pheromone plasmids from Enterococcus faecalis: detection of a new insertion element on pPD1 and implications for the evolution of this plasmid family. Mol Gen Genet. 1996;252:640–647. doi: 10.1007/BF02173969. [DOI] [PubMed] [Google Scholar]
- 340.Hjelm H, Hjelm K, Sjöquist J. Protein A from Staphylococcus aureus. Its isolation by affinity chromatography and its use as an immunosorbent for isolation of immunoglobulins. FEBS Lett. 1972;28:73–76. doi: 10.1016/0014-5793(72)80680-x. [DOI] [PubMed] [Google Scholar]
- 341.Hoffschulte H K, Drees B, Muller M. Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J Biol Chem. 1994;269:12833–12839. [PubMed] [Google Scholar]
- 342.Hofmann F, Busch C, Prepens U, Just I, Aktories K. Localization of the glucosyltransferase activity of Clostridium difficiletoxin B to the N-terminal part of the holotoxin. J Biol Chem. 1997;272:11074–11078. doi: 10.1074/jbc.272.17.11074. [DOI] [PubMed] [Google Scholar]
- 343.Holck A, Naes H. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracaseiNCDO 151. J Gen Microbiol. 1992;138:1353–1364. doi: 10.1099/00221287-138-7-1353. [DOI] [PubMed] [Google Scholar]
- 344.Hollingshead S K, Arnold J, Readdy T L, Bessen D E. Molecular evolution of a multigene family in group A streptococci. Mol Biol Evol. 1994;11:208–219. doi: 10.1093/oxfordjournals.molbev.a040103. [DOI] [PubMed] [Google Scholar]
- 345.Hollingshead S K, Bessen D E. Evolution of the emmgene family: virulence gene clusters in group A streptococci. Dev Biol Stand. 1995;85:163–168. [PubMed] [Google Scholar]
- 346.Hollingshead S K, Fischetti V A, Scott J R. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J Biol Chem. 1986;261:1677–1686. [PubMed] [Google Scholar]
- 347.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 348.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Höltje J-V, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Höltje J-V, Tomasz A. Biological effects of lipoteichoic acids. J Bacteriol. 1975;124:1023–1027. doi: 10.1128/jb.124.2.1023-1027.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Höltje J-V, Tomasz A. Lipoteichoic acid: a specific inhibitor of autolysin activity in Pneumococcus. Proc Natl Acad Sci USA. 1975;72:1690–1694. doi: 10.1073/pnas.72.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Höltje J-V, Tomasz A. Specific recognition of choline residues in the cell wall teichoic acid by the N-acetylmuramyl-l-alanine amidase of Pneumococcus. J Biol Chem. 1975;250:6072–6076. [PubMed] [Google Scholar]
- 353.Homonylo-McGavin M K, Lee S F. Role of the C terminus in antigen P1 surface localization in Streptococcus mutansand two related cocci. J Bacteriol. 1996;178:801–807. doi: 10.1128/jb.178.3.801-807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Horstmann R D, Sievertsen H J, Knobloch J, Fischetti V A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Horstmann R D, Sievertsen H J, Leippe M, Fischetti V A. Role of fibrinogen in complement inhibition by streptococcal M protein. Infect Immun. 1992;60:5036–5041. doi: 10.1128/iai.60.12.5036-5041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Hourcade D, Holers V M, Atkinson J P. The regulators of complement activation (RCA) gene cluster. Adv Immunol. 1989;45:381–416. doi: 10.1016/s0065-2776(08)60697-5. [DOI] [PubMed] [Google Scholar]
- 357.Howard L, Tipper D J. A polypeptide bacteriophage receptor: modified cell wall protein subunits in bacteriophage-resistant mutants of Bacillus sphaericusstrain P-1. J Bacteriol. 1973;113:1491–1504. doi: 10.1128/jb.113.3.1491-1504.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Hryniewicz W, Lipinski B, Jeljaszewicz J. Nature of the interaction between M protein of Streptococcus pyogenesand fibrinogen. J Infect Dis. 1972;125:626–630. doi: 10.1093/infdis/125.6.626. [DOI] [PubMed] [Google Scholar]
- 359.Hughes M, Machardy S M, Sheppard A J, Woods N C. Evidence for an immunological relationship between Streptococcus mutansand human cardiac tissue. Infect Immun. 1980;27:576–588. doi: 10.1128/iai.27.2.576-588.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Hugli T E. Structure and function of the anaphylatoxins. Springer Semin Immunopathol. 1984;7:193–219. doi: 10.1007/BF01893020. [DOI] [PubMed] [Google Scholar]
- 361.Husmann L K, Scott J R, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Igarashi T, Yamamoto A, Goto N. Characterization of the dextranase purified from Streptococcus mutansIngbritt. Microbiol Immunol. 1992;36:969–976. doi: 10.1111/j.1348-0421.1992.tb02100.x. [DOI] [PubMed] [Google Scholar]
- 363.Igarashi T, Yamamoto A, Goto N. Sequence analysis of the Streptococcus mutans Ingbritt dexAgene encoding extracellular dextranase. Microbiol Immunol. 1995;39:853–860. doi: 10.1111/j.1348-0421.1995.tb03282.x. [DOI] [PubMed] [Google Scholar]
- 364.Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: roles of sigma E and sigma F in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Innis M A, Tokunaga M, Williams M E, Loranger J M, Chang S Y, Chang S, Wu H C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci USA. 1984;81:3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Inoue S, Sugai M, Murooka Y, Paik S Y, Hong Y M, Ohgai H, Suginaka H. Molecular cloning and sequencing of the epidermal cell differentiation inhibitor gene from Staphylococcus aureus. Biochem Biophys Res Commun. 1991;174:459–464. doi: 10.1016/0006-291x(91)91438-i. [DOI] [PubMed] [Google Scholar]
- 367.Inouye S, Hsu C P, Itakura K, Inouye M. Requirement for signal peptide cleavage of Escherichia coliprolipoprotein. Science. 1983;221:59–61. doi: 10.1126/science.6344218. [DOI] [PubMed] [Google Scholar]
- 368.Ireton K, Payrastre B, Chap H, Ogawa W, Sakaue H, Kasuga M, Cossart P. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 1996;274:780–782. doi: 10.1126/science.274.5288.780. [DOI] [PubMed] [Google Scholar]
- 369.Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilisspores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Ito E, Strominger J L. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. III. Purification and properties of l-lysine-adding enzyme. J Biol Chem. 1964;239:210–214. [PubMed] [Google Scholar]
- 371.Izaki K, Matsuhashi M, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. 8. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reaction in strains of Escherichia coli. Proc Natl Acad Sci USA. 1966;55:656–663. doi: 10.1073/pnas.55.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Izaki K, Matsuhashi M, Strominger J L. Glycopeptide transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Proc Natl Acad Sci USA. 1966;55:656–663. doi: 10.1073/pnas.55.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Jacks-Weis J, Kim Y, Cleary P P. Restricted deposition of C3 on M+ group A streptococci: correlation with resistance to phagocytosis. J Immunol. 1982;128:1897–1902. [PubMed] [Google Scholar]
- 375.Jenkinson H F. Adherence and accumulation of oral streptococci. Trends Microbiol. 1994;2:209–212. doi: 10.1016/0966-842x(94)90114-k. [DOI] [PubMed] [Google Scholar]
- 376.Jenkinson H F. Cell surface protein receptors in oral streptococci. FEMS Microbiol Lett. 1994;121:133–140. doi: 10.1111/j.1574-6968.1994.tb07089.x. [DOI] [PubMed] [Google Scholar]
- 377.Jenkinson H F. Cell-surface proteins of Streptococcus sanguisassociated with cell hydrophobicity and coaggregation properties. J Gen Microbiol. 1986;132:1575–1589. doi: 10.1099/00221287-132-6-1575. [DOI] [PubMed] [Google Scholar]
- 378.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 379.Jennings H J, Lugowski C, Young N M. Structure of the complex polysaccharide C-substance from Streptococcus pneumoniaetype 1. Biochemistry. 1980;19:4712–4719. doi: 10.1021/bi00561a026. [DOI] [PubMed] [Google Scholar]
- 380.Jensen K. A normally occurring staphylococcus antibody in the human serum. Acta Pathol Microbiol Scand. 1958;44:421–428. [Google Scholar]
- 381.Jeppson H, Frithz E, Hedén L-O. Duplication of a DNA sequence homologous to genes for immunoglobulin receptors and M proteins in Streptococcus pyogenes. FEMS Microbiol Lett. 1992;92:139–146. doi: 10.1016/0378-1097(92)90502-f. [DOI] [PubMed] [Google Scholar]
- 382.Jerlström P G, Chhatwal G S, Timmis K N. The IgA-binding β antigen of the c protein complex of group B streptococci: sequence determination of its gene and detection of two binding regions. Mol Microbiol. 1991;5:843–849. doi: 10.1111/j.1365-2958.1991.tb00757.x. [DOI] [PubMed] [Google Scholar]
- 383.Jerlström P G, Talay S R, Valentin-Weigand P, Timmis K N, Chhatwal G S. Identification of an immunoglobulin A binding motif located in the beta-antigen of the c protein complex of group B streptococci. Infect Immun. 1996;64:2787–2793. doi: 10.1128/iai.64.7.2787-2793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Ji Y, Carlson B, Kondagunta A, Cleary P P. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect Immun. 1997;65:2080–2087. doi: 10.1128/iai.65.6.2080-2087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Johansson M U, de Château M, Björck L, Forsén S, Drakenberg T, Wikström M. The GA module, a mobile albumin-binding bacterial domain, adopts a three-helix-bundle structure. FEBS Lett. 1995;374:257–261. doi: 10.1016/0014-5793(95)01121-t. [DOI] [PubMed] [Google Scholar]
- 388.Johansson M U, de Château M, Wikström M, Forsén S, Drakenberg T, Björck L. Solution structure of the albumin-binding GA module: a versatile bacterial protein domain. J Mol Biol. 1997;266:859–865. doi: 10.1006/jmbi.1996.0856. [DOI] [PubMed] [Google Scholar]
- 389.Johnson D R, Kaplan E L. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J Med Microbiol. 1993;38:311–315. doi: 10.1099/00222615-38-5-311. [DOI] [PubMed] [Google Scholar]
- 390.Johnson R H, Vosti K L. Purification and characterization of group A streptococcal T-1 antigen. Infect Immun. 1977;16:867–875. doi: 10.1128/iai.16.3.867-875.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Johnsson E, Andersson G, Lindahl G, Hedén L O. Identification of the IgA-binding region in streptococcal protein Arp. J Immunol. 1994;153:3557–3564. [PubMed] [Google Scholar]
- 392.Johnsson E, Berggård K, Kotarsky H, Hellwage J, Zipfel P F, Sjöbring U, Lindahl G. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161:4894–4901. [PubMed] [Google Scholar]
- 393.Johnsson E, Thern A, Dahlbäck B, Hedén L O, Wikström M, Lindahl G. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J Immunol. 1996;157:3021–3029. [PubMed] [Google Scholar]
- 394.Jolly L, Wu S, van Heijenoort J, de Lencastre H, Mengin-Lecreulx D, Tomasz A. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J Bacteriol. 1997;179:5321–5325. doi: 10.1128/jb.179.17.5321-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Jones K F, Fischetti V A. Biological and immunochemical identity of M protein on group G streptococci with M protein on group A streptococci. Infect Immun. 1987;55:502–506. doi: 10.1128/iai.55.3.502-506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Jones K F, Khan S A, Erickson B W, Hollingshead S K, Scott J R, Fischetti V A. Immunochemical localization and amino acid sequences of crossreactive epitopes within the group A streptococcal M6 protein. J Exp Med. 1986;164:1226–1238. doi: 10.1084/jem.164.4.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Jones K F, Schneewind O, Koomey J M, Fischetti V A. Genetic diversity among the T-protein genes of group A streptococci. Mol Microbiol. 1991;5:2947–2952. doi: 10.1111/j.1365-2958.1991.tb01854.x. [DOI] [PubMed] [Google Scholar]
- 398.Jonsson H, Burtsoff-Asp C, Guss B. Streptococcal protein MAG—a protein with broad albumin binding specificity. Biochim Biophys Acta. 1995;1249:65–71. doi: 10.1016/0167-4838(95)00065-3. [DOI] [PubMed] [Google Scholar]
- 399.Jonsson H, Frykberg L, Rantamaki L, Guss B. MAG, a novel plasma protein receptor from Streptococcus dysgalactiae. Gene. 1994;143:85–89. doi: 10.1016/0378-1119(94)90609-2. [DOI] [PubMed] [Google Scholar]
- 400.Jonsson H, Lindmark H, Guss B. A protein G-related cell surface protein in Streptococcus zooepidemicus. Infect Immun. 1995;63:2968–2975. doi: 10.1128/iai.63.8.2968-2975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Jonsson H, Muller H P. The type-III Fc receptor from Streptococcus dysgalactiaeis also an alpha 2-macroglobulin receptor. Eur J Biochem. 1994;220:819–826. doi: 10.1111/j.1432-1033.1994.tb18684.x. [DOI] [PubMed] [Google Scholar]
- 402.Jonsson K, Signäs C, Muller H P, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 403.Jonsson P, Lindberg M, Haraldsson I, Wadström T. Virulence of Staphylococcus aureusin a mouse mastitis model: studies of alpha hemolysin, coagulase, and protein A as possible virulence determinants with protoplast fusion and gene cloning. Infect Immun. 1985;49:765–769. doi: 10.1128/iai.49.3.765-769.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Joris B, Englebert S, Chu C P, Kariyama R, Daneo-Moore L, Shockman G D, Ghuysen J M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalisautolysin. FEMS Microbiol Lett. 1992;70:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- 405.Juillard V, Le Bars D, Kunji E R, Konings W N, Gripon J C, Richard J. Oligopeptides are the main source of nitrogen for Lactococcus lactisduring growth in milk. Appl Environ Microbiol. 1995;61:3024–3030. doi: 10.1128/aem.61.8.3024-3030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Just I, Fritz G, Aktories K, Giry M, Popoff M R, Boquet P, Hegenbarth S, von Eichel-Streiber C. Clostridium difficiletoxin B acts on the GTP-binding protein Rho. J Biol Chem. 1994;269:10706–10712. [PubMed] [Google Scholar]
- 407.Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficiletoxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 408.Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. The enterotoxin from Clostridium difficile(ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 409.Kantor F S. Fibrinogen precipitating by streptococcal M protein. I. Identity of the reactants and stoichiometry of the reaction. J Exp Med. 1965;121:849–859. doi: 10.1084/jem.121.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Kantor F S, Cole R M. Preparation and antigenicity of M protein released from group A, type 1, streptococcal cell walls by phage-associated lysin. J Exp Med. 1960;112:77–85. doi: 10.1084/jem.112.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Kao S M, Olmsted S B, Viksnins A S, Gallo J C, Dunny G M. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J Bacteriol. 1991;173:7650–7664. doi: 10.1128/jb.173.23.7650-7664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Kastern W, Holst E, Nielsen E, Sjöbring U, Björck L. Protein L, a bacterial immunoglobulin-binding protein and possible virulence determinant. Infect Immun. 1990;58:1217–1222. [Google Scholar]
- 413.Kastern W, Sjöbring U, Björck L. Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J Biol Chem. 1992;267:12820–12825. [PubMed] [Google Scholar]
- 414.Katerov V, Andreev A, Schalén C, Totolian A A. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology. 1998;144:119–126. doi: 10.1099/00221287-144-1-119. [DOI] [PubMed] [Google Scholar]
- 415.Katerov V, Schalén C, Totolian A A. Sequencing of genes within the vir regulon of Streptococcus pyogenestype M15—an opacity factor-positive serotype with low opacity factor expression. Mol Gen Genet. 1994;245:78–85. doi: 10.1007/BF00279753. [DOI] [PubMed] [Google Scholar]
- 416.Katsukawa C. Cloning and nucleotide sequence of type 3 M protein gene (emm3) consisting of an N-terminal variable portion and C-terminal conserved C repeat regions: relation to other genes of Streptococcus pyogenes. J Jpn Assoc Infect Dis. 1994;68:698–705. doi: 10.11150/kansenshogakuzasshi1970.68.698. [DOI] [PubMed] [Google Scholar]
- 417.Katz W, Matsuhashi M, Dietrich C P, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. IV. Incorporation of glycine in Micrococcus lysodeikticus. J Biol Chem. 1967;242:3207–3217. [PubMed] [Google Scholar]
- 418.Kawata T, Takeoka A, Takumi K, Masuda K. Demonstration and preliminary characterization of a regular array in the cell wall of Clostridium difficile. FEMS Microbiol Lett. 1984;24:323–328. [Google Scholar]
- 419.Kehoe M A. Cell-wall-associated proteins in Gram-positive bacteria. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 217–261. [Google Scholar]
- 420.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 421.Keller G A, Siegel M W, Caras I W. Endocytosis of glycophospholipid-anchored and transmembrane forms of CD4 by different endocytic pathways. EMBO J. 1992;11:863–874. doi: 10.1002/j.1460-2075.1992.tb05124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Kelly C, Evans P, Bergmeier L, Lee S F, Progulske-Fox A, Harris A C, Aitken A, Bleiweis A S, Lehner T. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 423.Kelly C, Evans P, Ma J K, Bergmeier L A, Taylor W, Brady L J, Lee S F, Bleiweis A S, Lehner T. Sequencing and characterization of the 185 kDa cell surface antigen of Streptococcus mutans. Arch Oral Biol. 1990;35:33S–38S. doi: 10.1016/0003-9969(90)90128-w. [DOI] [PubMed] [Google Scholar]
- 424.Kelly C P, Pothoulakis C, LaMont J T. Clostridium difficilecolitis. N Engl J Med. 1994;330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 425.Kelly R T, Farmer S, Greiff D. Neuraminidase activities of clinical isolates of Diplococcus pneumoniae. J Bacteriol. 1967;94:272–273. doi: 10.1128/jb.94.1.272-273.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Kelly R T, Greiff D. Toxicity of pneumococcal neuraminidase. Infect Immun. 1970;2:115–117. doi: 10.1128/iai.2.1.115-117.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Kihlberg B M, Cooney J, Caparon M G, Olsén A, Björck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 428.Kist M L, Murray R G. Components of the regular surface array of Aquaspirillum serpensMW5 and their assembly in vitro. J Bacteriol. 1984;157:599–606. doi: 10.1128/jb.157.2.599-606.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Kiwaki M, Ikemura H, Shimizu-Kadota M, Hirashima A. Molecular characterization of a cell wall-associated proteinase gene from Streptococcus lactisNCDO763. Mol Microbiol. 1989;3:359–369. doi: 10.1111/j.1365-2958.1989.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 430.Kline J B, Xu S, Bisno A L, Collins C M. Identification of a fibronectin-binding protein (GfbA) in pathogenic group G streptococci. Infect Immun. 1996;64:2122–2129. doi: 10.1128/iai.64.6.2122-2129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Koch H U, Doker R, Fischer W. Maintenance of d-alanine ester substitution of lipoteichoic acid by reesterification in Staphylococcus aureus. J Bacteriol. 1985;164:1211–1217. doi: 10.1128/jb.164.3.1211-1217.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Koch H U, Fischer W. Acyldiglucosyldiacylglycerol-containing lipoteichoic acid with a poly(3-O-galabiosyl-2-O-galactosyl-sn-glycero-1-phosphate) chain from Streptococcus lactisKiel 42172. Biochemistry. 1978;17:5275–5281. doi: 10.1021/bi00617a030. [DOI] [PubMed] [Google Scholar]
- 433.Koch H U, Haas R, Fischer W. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur J Biochem. 1984;138:357–363. doi: 10.1111/j.1432-1033.1984.tb07923.x. [DOI] [PubMed] [Google Scholar]
- 434.Kodukula K, Cines D, Amthauer R, Gerber L, Udenfriend S. Biosynthesis of phosphatidylinositol-glycan (PI-G)-anchored membrane proteins in cell-free systems: cleavage of the nascent protein and addition of the PI-G moiety depend on the size of the COOH-terminal signal peptide. Proc Natl Acad Sci USA. 1992;89:1350–1353. doi: 10.1073/pnas.89.4.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Kodukula K, Gerber L D, Amthauer R, Brink L, Udenfriend S. Biosynthesis of glycosylphosphatidylinositol (GPI)-anchored membrane proteins in intact cells: specific amino acid requirements adjacent to the site of cleavage and GPI attachment. J Cell Biol. 1993;120:657–664. doi: 10.1083/jcb.120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Kogan G, Uhrin D, Brisson J R, Paoletti L C, Blodgett A E, Kasper D L, Jennings H J. Structural and immunochemical characterization of the type VIII group B Streptococcuscapsular polysaccharide. J Biol Chem. 1996;271:8786–8790. doi: 10.1074/jbc.271.15.8786. [DOI] [PubMed] [Google Scholar]
- 437.Kohler W. A-Streptokokken-T-antigene bei den Streptokokkengruppen B, C und G. Z Immunitaets-Allergieforsch. 1963;125:141–144. [PubMed] [Google Scholar]
- 438.Kojima N, Araki Y, Ito E. Structure of linkage region between ribitol teichoic acid and peptidoglycan in cell walls of Staphylococcus aureusH. J Biol Chem. 1983;258:9043–9045. [PubMed] [Google Scholar]
- 439.Kok J, Leenhouts K J, Haandrikman A J, Ledeboer A M, Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremorisWg2. Appl Environ Microbiol. 1988;54:231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Kolenbrander P E. Intergeneric coaggregation among human oral bacteria and ecology of denal plaque. Annu Rev Microbiol. 1988;42:627–656. doi: 10.1146/annurev.mi.42.100188.003211. [DOI] [PubMed] [Google Scholar]
- 441.Kolenbrander P E, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Kopp U, Roos M, Wecke J, Labischinski H. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb Drug Resist. 1996;2:29–41. doi: 10.1089/mdr.1996.2.29. [DOI] [PubMed] [Google Scholar]
- 443.Koroleva I V, Sjöholm A G, Schalén C. Binding of complement subcomponent C1q to Streptococcus pyogenes: evidence for interactions with the M5 and FcRA76 proteins. FEMS Immunol Med Microbiol. 1998;20:11–20. doi: 10.1111/j.1574-695X.1998.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 444.Koshland D, Botstein D. Secretion of beta-lactamase requires the carboxy end of the protein. Cell. 1980;20:749–760. doi: 10.1016/0092-8674(80)90321-9. [DOI] [PubMed] [Google Scholar]
- 445.Kotarsky H, Hellwage J, Johnsson E, Skerka C, Svensson H G, Lindahl G, Sjöbring U, Zipfel P F. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J Immunol. 1998;160:3349–3354. [PubMed] [Google Scholar]
- 446.Krause R N. Studies on the bacteriophages of hemolytic streptococci. II. Antigens released from the streptococcal cell wall by a phage-associated lysin. J Exp Med. 1958;108:803–818. doi: 10.1084/jem.108.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Krebs B, Kaufhold A, Boyle M D, Podbielski A. Different alleles of the fcrA/mrp gene of Streptococcus pyogenesencode M-related proteins exhibiting an identical immunoglobulin-binding pattern. Med Microbiol Immunol. 1996;185:39–47. doi: 10.1007/s004300050013. [DOI] [PubMed] [Google Scholar]
- 448.Kreikemeyer B, Talay S R, Chhatwal G S. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol Microbiol. 1995;17:137–145. doi: 10.1111/j.1365-2958.1995.mmi_17010137.x. [DOI] [PubMed] [Google Scholar]
- 449.Krivan H C, Roberts D D, Ginsburg V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc-(β1-4)-Gal found in some glycolipids. Proc Natl Acad Sci USA. 1988;85:6157–6161. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Krivan H C, Wilkins T D. Purification of Clostridium difficiletoxin A by affinity chromatography on immobilized thyroglobulin. Infect Immun. 1987;55:1873–1877. doi: 10.1128/iai.55.8.1873-1877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Kuipers O P, Beerthuyzen M M, Siezen R J, De Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisIgenes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 452.Kumamoto C A, Francetic O. Highly selective binding of nascent polypeptides by an Escherichia colichaperone protein in vivo. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Kunji E R, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 454.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 455.Kuramitsu H K, Wondrack L. Insoluble glucan synthesis by Streptococcus mutansserotype c strains. Infect Immun. 1983;42:763–770. doi: 10.1128/iai.42.2.763-770.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Kurita K, Honda K, Suzuma S, Takamatsu H, Nakamura K, Yamane K. Identification of a region of Bacillus subtilisFfh, a homologue of mammalian SRP54 protein, that is essential for binding to small cytoplasmic RNA. J Biol Chem. 1996;271:13140–13146. doi: 10.1074/jbc.271.22.13140. [DOI] [PubMed] [Google Scholar]
- 457.Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Kuroda A, Rashid M H, Sekiguchi J. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIIDproduct. J Gen Microbiol. 1992;138:1067–1076. doi: 10.1099/00221287-138-6-1067. [DOI] [PubMed] [Google Scholar]
- 459.Kuroda A, Sekiguchi J. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J Bacteriol. 1993;175:795–801. doi: 10.1128/jb.175.3.795-801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Kuroda A, Sekiguchi J. Molecular cloning and sequencing of a major Bacillus subtilisautolysin gene. J Bacteriol. 1991;173:7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Kuttner A G, Lenert T F. The occurrence of bacteriostatic properties in the blood of patients after recovery from streptococcal paryngitis. J Clin Invest. 1944;23:151–158. doi: 10.1172/JCI101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Kuusela P. Fibronectin binds to Staphylococcus aureus. Nature. 1978;276:718–720. doi: 10.1038/276718a0. [DOI] [PubMed] [Google Scholar]
- 463.Labischinski H, Maidhof H. Bacterial peptidoglycan: overview and evolving concepts. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam: Elsevier Science BV; 1994. pp. 23–38. [Google Scholar]
- 464.Lachenauer C S, Madoff L C. Cloning and expression in Escherichia coliof a protective surface protein from type V group B streptococci. Adv Exp Med Biol. 1997;418:615–618. doi: 10.1007/978-1-4899-1825-3_143. [DOI] [PubMed] [Google Scholar]
- 465.Lachenauer C S, Madoff L C. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect Immun. 1996;64:4255–4260. doi: 10.1128/iai.64.10.4255-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Lancefield R C. The antigenic complex of Streptococcus hemolyticus. I. Demonstration of a type-specific substance in extracts of Streptococcus hemolyticus. J Exp Med. 1928;47:91–103. doi: 10.1084/jem.47.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 468.Lancefield R C. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933;57:571–595. doi: 10.1084/jem.57.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Lancefield R C, Dole V P. The properties of T antigens extracted from the group A hemolytic streptococci. J Exp Med. 1946;84:449–471. doi: 10.1084/jem.84.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.La Penta D, Zhang X P, Cleary P P. Streptococcus pyogenestype IIa IgG Fc receptor expression is co-ordinately regulated with M protein and streptococcal C5a peptidase. Mol Microbiol. 1994;12:873–879. doi: 10.1111/j.1365-2958.1994.tb01075.x. [DOI] [PubMed] [Google Scholar]
- 471.Larsson C, Stalhåmmar-Carlemalm M, Lindahl G. Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and alpha. Infect Immun. 1996;64:3518–3523. doi: 10.1128/iai.64.9.3518-3523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 473.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 474.Lebrun M, Mengaud J, Ohayon H, Nato F, Cossart P. Internalin must be on the bacterial surface to mediate entry of Listeria monocytogenesinto epithelial cells. Mol Microbiol. 1996;21:579–592. doi: 10.1111/j.1365-2958.1996.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 475.Lecuit M, Ohayon H, Braun L, Mengaud J, Cossart P. Internalin of Listeria monocytogeneswith an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun. 1997;65:5309–5319. doi: 10.1128/iai.65.12.5309-5319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Lee C J, Liu T Y. The autolytic enzyme activity upon pneumcoccal cell wall. Int J Biochem. 1977;8:573–580. [Google Scholar]
- 476a.Lee S F. Identification and characterization of a surface protein-releasing activity in Streptococcus mutansand other pathogenic streptococci. Infect Immun. 1992;60:4032–4039. doi: 10.1128/iai.60.10.4032-4039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476b.Lee S F. Active release of bound antibody by Streptococcus mutans. Infect Immun. 1995;63:1940–1946. doi: 10.1128/iai.63.5.1940-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476c.Lee S F, Li Y H, Bowden G H. Detachment of Streptococcus mutansbiofilm cells by an endogenous enzymatic activity. Infect Immun. 1996;64:1035–1038. doi: 10.1128/iai.64.3.1035-1038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Leibovitz E, Ohayon H, Gounon P, Beguin P. Characterization and subcellular localization of the Clostridium thermocellumscaffoldin dockerin binding protein SdbA. J Bacteriol. 1997;179:2519–2523. doi: 10.1128/jb.179.8.2519-2523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Leidich S D, Drapp D A, Orlean P. Isolation and characterization of yeast glycosylphosphatidylinositol anchoring mutants. Methods Enzymol. 1995;250:560–571. doi: 10.1016/0076-6879(95)50097-9. [DOI] [PubMed] [Google Scholar]
- 479.Leidich S D, Kostova Z, Latek R R, Costello L C, Drapp D A, Gray W, Fassler J S, Orlean P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the gpi2gene. J Biol Chem. 1995;270:13029–13035. doi: 10.1074/jbc.270.22.13029. [DOI] [PubMed] [Google Scholar]
- 480.Leidich S D, Orlean P. Gpi1, a Saccharomyces cerevisiaeprotein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J Biol Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- 481.Lemaire M, Ohayon H, Gounon P, Fujino T, Beguin P. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177:2451–2459. doi: 10.1128/jb.177.9.2451-2459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Leopold K, Fischer W. Hydrophobic interaction chromatography fractionates lipoteichoic acid according to the size of the hydrophilic chain: a comparative study with anion-exchange and affinity chromatography for suitability in species analysis. Anal Biochem. 1992;201:350–355. doi: 10.1016/0003-2697(92)90350-g. [DOI] [PubMed] [Google Scholar]
- 483.Li J, Kasper D L, Ausubel F M, Rosner B, Michel J L. Inactivation of the α C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proc Natl Acad Sci USA. 1997;94:13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Liljemark W F, Bloomquist C G, Brandt C L, Pihlstrom B L, Hinrichs J E, Wolff L F. Comparison of the distribution of Actinomycesin dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease. Oral Microbiol Immunol. 1993;8:5–15. doi: 10.1111/j.1399-302x.1993.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 485.Lin J J, Kanazawa H, Ozols J, Wu H C. An Escherichia colimutant with an amino acid alteration within the signal sequence of outer membrane prolipoprotein. Proc Natl Acad Sci USA. 1978;75:4891–4895. doi: 10.1073/pnas.75.10.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Lindahl G, Åkerström B. Receptor for IgA in group A streptococci: cloning of the gene and characterization of the protein expressed in Escherichia coli. Mol Microbiol. 1989;3:239–247. doi: 10.1111/j.1365-2958.1989.tb01813.x. [DOI] [PubMed] [Google Scholar]
- 487.Lindahl G, Åkerström B, Vaerman J P, Stenberg L. Characterization of an IgA receptor from group B streptococci: specificity for serum IgA. Eur J Immunol. 1990;20:2241–2247. doi: 10.1002/eji.1830201013. [DOI] [PubMed] [Google Scholar]
- 488.Lindahl G, Stenberg L. Binding of IgA and/or IgG is a common property among clinical isolates of group A streptococci. Epidemiol Infect. 1990;105:87–93. doi: 10.1017/s0950268800047683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Linder T E, Daniels R L, Lim D J, DeMaria T F. Effect of intranasal inoculation of Streptococcus pneumoniaeon the structure of the surface carbohydrates of the chinchilla eustachian tube and middle ear mucosa. Microb Pathog. 1994;16:435–441. doi: 10.1006/mpat.1994.1043. [DOI] [PubMed] [Google Scholar]
- 490.Lindgren P E, McGavin M J, Signäs C, Guss B, Gurusiddappa S, Höök M, Lindberg M. Two different genes coding for fibronectin-binding proteins from Streptococcus dysgalactiae. The complete nucleotide sequences and characterization of the binding domains. Eur J Biochem. 1993;214:819–827. doi: 10.1111/j.1432-1033.1993.tb17985.x. [DOI] [PubMed] [Google Scholar]
- 491.Lindgren P E, Signäs C, Rantamaki L, Lindberg M. A fibronectin-binding protein from Streptococcus equisimilis: characterization of the gene and identification of the binding domain. Vet Microbiol. 1994;41:235–247. doi: 10.1016/0378-1135(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 492.Lindmark H, Jacobsson K, Frykberg L, Guss B. Fibronectin-binding protein of Streptococcus equi subsp. zooepidemicus. Infect Immun. 1996;64:3993–3999. doi: 10.1128/iai.64.10.3993-3999.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Lingnau A, Chakraborty T, Niebuhr K, Domann E, Wehland J. Identification and purification of novel internalin-related proteins in Listeria monocytogenes and Listeria ivanovii. Infect Immun. 1996;64:1002–1006. doi: 10.1128/iai.64.3.1002-1006.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Lis J, Schleif R. Lambda lysozyme synthesis in the absence of N protein. Virology. 1971;45:532–533. doi: 10.1016/0042-6822(71)90356-4. [DOI] [PubMed] [Google Scholar]
- 495.Lisanti M P, Caras I W, Davitz M A, Rodriguez-Boulan E. A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol. 1989;109:2145–2156. doi: 10.1083/jcb.109.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Liu S, Courtney H S, Bessen D E, Hasty D L, Dale J B. M protein expression is not required for resistance to phagocytosis of type 18 group A streptococci. Adv Exp Med Biol. 1997;418:725–727. doi: 10.1007/978-1-4899-1825-3_170. [DOI] [PubMed] [Google Scholar]
- 497.Loesche W J. Role of Streptococcus mutansin human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Loessner M J, Gaeng S, Wendlinger G, Maier S K, Scherer S. The two-component lysis system of Staphylococcus aureusbacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- 499.López R, García E, García P, García J L. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb Drug Resist. 1997;3:199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- 500.Lortal S, van Heijenoort J, Gruber K, Sleytr U B. S-layer of Lactobacillus helveticusATCC 12046: isolation, chemical characterization and re-formation after extraction with lithium chloride. J Gen Microbiol. 1992;138:611–618. [Google Scholar]
- 501.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 502.Ludwicka A, Kloczewiak M. Characterization of group A streptococcal T-12 protein purified by ion-exchange column chromatography. Infect Immun. 1978;21:940–945. doi: 10.1128/iai.21.3.940-945.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Lupas A. A circular permutation event in the evolution of the SLH domain? Mol Microbiol. 1996;20:897–898. doi: 10.1111/j.1365-2958.1996.tb02528.x. [DOI] [PubMed] [Google Scholar]
- 504.Lupas A, Engelhardt H, Peters J, Santarius U, Volker S, Baumeister W. Domain structure of the Acetogenium kivuisurface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Lutkenhaus J. Bacterial cytokinesis: let the light shine in. Curr Biol. 1997;7:R573–575. doi: 10.1016/s0960-9822(06)00285-5. [DOI] [PubMed] [Google Scholar]
- 506.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidisis a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Mack D, Haeder M, Siemssen N, Laufs R. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis. 1996;174:881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- 508.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidisimpaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Madoff L C, Hori S, Michel J L, Baker C J, Kasper D L. Phenotypic diversity in the alpha C protein of group B streptococci. Infect Immun. 1991;59:2638–2644. doi: 10.1128/iai.59.8.2638-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Madoff L C, Michel J L, Gong E W, Rodewald A K, Kasper D L. Protection of neonatal mice from group B streptococcal infection by maternal immunization with beta C protein. Infect Immun. 1992;60:4989–4994. doi: 10.1128/iai.60.12.4989-4994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Maeland J A, Brakstad O G, Bevanger L, Kvam A I. Streptococcus agalactiae β gene and gene product variations. J Med Microbiol. 1997;46:999–1005. doi: 10.1099/00222615-46-12-999. [DOI] [PubMed] [Google Scholar]
- 513.Maidhof H, Reinicke B, Blumel P, Berger-Bachi B, Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureusstrains. J Bacteriol. 1991;173:3507–3513. doi: 10.1128/jb.173.11.3507-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Mamo W, Jonsson P, Flock J I, Lindberg M, Muller H P, Wadström T, Nelson L. Vaccination against Staphylococcus aureus mastitis: immunological response of mice vaccinated with fibronectin-binding protein (FnBP-A) to challenge with S. aureus. Vaccine. 1994;12:988–992. doi: 10.1016/0264-410x(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 515.Manjula B N, Khandke K M, Fairwell T, Relf W A, Sriprakash K S. Heptad motifs within the distal subdomain of the coiled-coil rod region of M protein from rheumatic fever and nephritis associated serotypes of group A streptococci are distinct from each other: nucleotide sequence of the M57 gene and relation of the deduced amino acid sequence to other M proteins. J Protein Chem. 1991;10:369–384. doi: 10.1007/BF01025251. [DOI] [PubMed] [Google Scholar]
- 516.Margot P, Karamata D. Identification of the structural genes for N-acetylmuramoyl-l-alanine amidase and its modifier in Bacillus subtilis168: inactivation of these genes by insertional mutagenesis has no effect on growth or cell separation. Mol Gen Genet. 1992;232:359–366. doi: 10.1007/BF00266238. [DOI] [PubMed] [Google Scholar]
- 517.Margot P, Mauel C, Karamata D. The gene of the N-acetylglucosaminidase, a Bacillus subtilis168 cell wall hydrolase not involved in vegetative cell autolysis. Mol Microbiol. 1994;12:535–545. doi: 10.1111/j.1365-2958.1994.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 518.Marks J D, Hoogenboom H R, Griffiths A D, Winter G. Molecular evolution of proteins on filamentous phage. Mimicking the strategy of the immune system. J Biol Chem. 1992;267:16007–16010. [PubMed] [Google Scholar]
- 519.Marova I, Dadak V. Modified simplified method for isolation of lysostaphin from the culture filtrate of Staphylococcus staphylolyticus. Folia Microbiol. 1993;38:245–252. doi: 10.1007/BF02814386. [DOI] [PubMed] [Google Scholar]
- 520.Matsuhashi M, Song M D, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167:975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Mauel C, Young M, Karamata D. Genes concerned with synthesis of poly(glycerol phosphate), the essential teichoic acid in Bacillus subtilisstrain 168, are organized in two divergent transcription units. J Gen Microbiol. 1991;137:929–941. doi: 10.1099/00221287-137-4-929. [DOI] [PubMed] [Google Scholar]
- 522.Maxfield F R, Mayor S. Cell surface dynamics of GPI-anchored proteins. Adv Exp Med Biol. 1997;419:355–364. doi: 10.1007/978-1-4419-8632-0_47. [DOI] [PubMed] [Google Scholar]
- 523.Mayor S, Menon A K, Cross G A. Galactose-containing glycosylphosphatidylinositols in Trypanosoma brucei. J Biol Chem. 1992;267:754–761. [PubMed] [Google Scholar]
- 524.Mayor S, Menon A K, Cross G A. Glycolipid precursors for the membrane anchor of Trypanosoma bruceivariant surface glycoproteins. II. Lipid structures of phosphatidylinositol-specific phospholipase C sensitive and resistant glycolipids. J Biol Chem. 1990;265:6174–6181. [PubMed] [Google Scholar]
- 525.Mayor S, Menon A K, Cross G A. Transfer of glycosyl-phosphatidylinositol membrane anchors to polypeptide acceptors in a cell-free system. J Cell Biol. 1991;114:61–71. doi: 10.1083/jcb.114.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Mayor S, Menon A K, Cross G A, Ferguson M A, Dwek R A, Rademacher T W. Glycolipid precursors for the membrane anchor of Trypanosoma bruceivariant surface glycoproteins. I. Can structure of the phosphatidylinositol-specific phospholipase C sensitive and resistant glycolipids. J Biol Chem. 1990;265:6164–6173. [PubMed] [Google Scholar]
- 527.McConville M J, Ferguson M A. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.McDaniel L S, Ralph B A, McDaniel D O, Briles D E. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniaebetween amino acid residues 192 and 260. Microb Pathog. 1994;17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 529.McDaniel L S, Sheffield J S, Swiatlo E, Yother J, Crain M J, Briles D E. Molecular localization of variable and conserved regions of pspA and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb Pathog. 1992;13:261–269. doi: 10.1016/0882-4010(92)90036-n. [DOI] [PubMed] [Google Scholar]
- 530.McDaniel L S, Yother J, Vijayakumar M, McGarry L, Guild W R, Briles D E. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA) J Exp Med. 1987;165:381–394. doi: 10.1084/jem.165.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 532.McDevitt D, Vaudaux P, Foster T J. Genetic evidence that bound coagulase of Staphylococcus aureusis not clumping factor. Infect Immun. 1992;60:1514–1523. doi: 10.1128/iai.60.4.1514-1523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.McIver K S, Heath A S, Green B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpAgenes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.McIver K S, Heath A S, Scott J R. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emmtranscription. Infect Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.McIver K S, Scott J R. Role of mgain growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenesM49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol Lett. 1995;128:45–51. doi: 10.1111/j.1574-6968.1995.tb07498.x. [DOI] [PubMed] [Google Scholar]
- 537.McNab R, Holmes A R, Clarke J M, Tannock G W, Jenkinson H F. Cell surface polypeptide CshA mediates binding of Streptococcus gordoniito other oral bacteria and to immobilized fibronectin. Infect Immun. 1996;64:4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.McNab R, Jenkinson H F. Gene disruption identifies a 290 kDa cell-surface polypeptide conferring hydrophobicity and coaggregation properties in Streptococcus gordonii. Mol Microbiol. 1992;6:2939–2949. doi: 10.1111/j.1365-2958.1992.tb01753.x. [DOI] [PubMed] [Google Scholar]
- 539.McNab R, Jenkinson H F, Loach D M, Tannock G W. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol Microbiol. 1994;14:743–754. doi: 10.1111/j.1365-2958.1994.tb01311.x. [DOI] [PubMed] [Google Scholar]
- 540.Medaglini D, Oggioni M R, Pozzi G. Vaginal immunization with recombinant Gram-positive bacteria. Am J Reprod Immunol. 1998;39:199–208. doi: 10.1111/j.1600-0897.1998.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 541.Medaglini D, Pozzi G, King T P, Fischetti V A. Mucosal and systemic immune responses to a recombinant protein expressed on the surface of the oral commensal bacterium Streptococcus gordoniiafter oral colonization. Proc Natl Acad Sci USA. 1995;92:6868–6872. doi: 10.1073/pnas.92.15.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Medaglini D, Rush C M, Sestini P, Pozzi G. Commensal bacteria as vectors for mucosal vaccines against sexually transmitted diseases: vaginal colonization with recombinant streptococci induces local and systemic antibodies in mice. Vaccine. 1997;15:1330–1337. doi: 10.1016/s0264-410x(97)00026-1. [DOI] [PubMed] [Google Scholar]
- 543.Mengaud J, Ohayon H, Gounon P, Mege R M, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenesinto epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 544.Merchante R, Pooley H M, Karamata D. A periplasm in Bacillus subtilis. J Bacteriol. 1995;177:6176–6183. doi: 10.1128/jb.177.21.6176-6183.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Mesnage S, Tosi-Couture E, Gounon P, Mock M, Fouet A. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J Bacteriol. 1998;180:52–58. doi: 10.1128/jb.180.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. Molecular characterization of the Bacillus anthracismain S-layer component: evidence that it is the major cell-associated antigen. Mol Microbiol. 1997;23:1147–1155. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 547.Messner P, Allmaier G, Schaffer C, Wugeditsch T, Lortal S, Konig H, Niemetz R, Dorner M. Biochemistry of S-layers. FEMS Microbiol Rev. 1997;20:25–46. doi: 10.1111/j.1574-6976.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 548.Micanovic R, Gerber L D, Berger J, Kodukula K, Udenfriend S. Selectivity of the cleavage/attachment site of phosphatidylinositol-glycan-anchored membrane proteins determined by site-specific mutagenesis at Asp-484 of placental alkaline phosphatase. Proc Natl Acad Sci USA. 1990;87:157–161. doi: 10.1073/pnas.87.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Micanovic R, Kodukula K, Gerber L D, Udenfriend S. Selectivity at the cleavage/attachment site of phosphatidylinositol-glycan anchored membrane proteins is enzymatically determined. Proc Natl Acad Sci USA. 1990;87:7939–7943. doi: 10.1073/pnas.87.20.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Michel J L, Madoff L C, Olson K, Kling D E, Kasper D L, Ausubel F M. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci USA. 1992;89:10060–10064. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Miller J D, Bernstein H D, Walter P. Interaction of E. coliFfh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 552.Miller L, Burdett V, Poirier T P, Gray L D, Beachey E H, Kehoe M A. Conservation of protective and nonprotective epitopes in M proteins of group A streptococci. Infect Immun. 1988;56:2198–2204. doi: 10.1128/iai.56.8.2198-2204.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Misasi R, Huemer H P, Schwaeble W, Solder E, Larcher C, Dierich M P. Human complement factor H: an additional gene product of 43 kDa isolated from human plasma shows cofactor activity for the cleavage of the third component of complement. Eur J Immunol. 1989;19:1765–1768. doi: 10.1002/eji.1830190936. [DOI] [PubMed] [Google Scholar]
- 554.Mizuno Y, Yaegashi M, Ito E. Purification and properties of uridine diphosphate N-acetylmuramate:l-alanine ligase. J Biochem Tokyo. 1973;74:525–538. doi: 10.1093/oxfordjournals.jbchem.a130273. [DOI] [PubMed] [Google Scholar]
- 555.Mogollon J D, Pijoan C, Murtaugh M P, Cleary P P, Collins J E. Testing meningeal strains of Streptococcus suisto detect M protein genes. Res Vet Sci. 1992;53:244–246. doi: 10.1016/0034-5288(92)90116-j. [DOI] [PubMed] [Google Scholar]
- 556.Moran P, Caras I W. Requirements for glycosylphosphatidylinositol attachment are similar but not identical in mammalian cells and parasitic protozoa. J Cell Biol. 1994;125:333–343. doi: 10.1083/jcb.125.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Moran P, Raab H, Kohr W J, Caras I W. Glycophospholipid membrane anchor attachment. Molecular analysis of the cleavage/attachment site. J Biol Chem. 1991;266:1250–1257. [PubMed] [Google Scholar]
- 558.Moriyama R, Hattori A, Miyata S, Kudoh S, Makino S. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J Bacteriol. 1996;178:6059–6063. doi: 10.1128/jb.178.20.6059-6063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Moriyama R, Kudoh S, Miyata S, Nonobe S, Hattori A, Makino S. A germination-specific spore cortex-lytic enzyme from Bacillus cereusspores: cloning and sequencing of the gene and molecular characterization of the enzyme. J Bacteriol. 1996;178:5330–5332. doi: 10.1128/jb.178.17.5330-5332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Mouw A R, Beachey E H, Burdett V. Molecular evolution of streptococcal M protein: cloning and nucleotide sequence of the type 24 M protein gene and relation to other genes of Streptococcus pyogenes. J Bacteriol. 1988;170:676–684. doi: 10.1128/jb.170.2.676-684.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Movitz J. Formation of extracellular protein A by Staphylococcus aureus. Eur J Biochem. 1976;68:291–299. doi: 10.1111/j.1432-1033.1976.tb10788.x. [DOI] [PubMed] [Google Scholar]
- 562.Movitz J. A study on the biogenesis of protein A in Staphylococcus aureus. Eur J Biochem. 1974;48:131–136. doi: 10.1111/j.1432-1033.1974.tb03750.x. [DOI] [PubMed] [Google Scholar]
- 563.Muller M, Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Munoz E, Ghuysen J-M, Leyh-Bouille M, Petit J F, Heymann H, Bricas E, Lefrancier P. The peptide subunit na-(l-alanyl-d-isoglutaminyl)-l-lysyl-d-alanine in cell wall peptidoglycans of Staphylococcus aureus strain Copenhagen, Micrococcus aureus R27 and Streptococcus pyogenesgroup A, type 14. Biochemistry. 1966;5:3748–3764. [Google Scholar]
- 565.Murakami Y, Nakano Y, Yamashita Y, Koga T. Identification of a frameshift mutation resulting in premature termination and loss of cell wall anchoring of the PAc antigen of Streptococcus mutansGS-5. Infect Immun. 1997;65:794–797. doi: 10.1128/iai.65.2.794-797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Murakami Y, Yamashita Y, Nakano Y, Ito H O, Yu H, Koga T. Role of the charged tail in localization of a surface protein antigen of Streptococcus mutans. Infect Immun. 1997;65:1531–1535. doi: 10.1128/iai.65.4.1531-1535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Murphy J P, Trowern A R, Duggleby C J. Nucleotide sequence of the gene for peptostreptococcal protein L. DNA Seq. 1994;4:259–265. doi: 10.3109/10425179409020850. [DOI] [PubMed] [Google Scholar]
- 568.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationships among Streptococcus pyogenesstrains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Myhre E B, Kronvall G. Heterogeneity of nonimmune immunoglobulin Fc reactivity among gram-positive cocci: description of three major types of receptors for human immunoglobulin G. Infect Immun. 1977;17:475–482. doi: 10.1128/iai.17.3.475-482.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569a.Myhre E B, Kronvall G. Demonstration of specific binding sites for human serum albumin in group C and G streptococci. Infect Immun. 1980;27:6–14. doi: 10.1128/iai.27.1.6-14.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Nakagawa J, Tamaki S, Tomioka S, Matsuhashi M. Functional biosynthesis of cell wall peptidoglycan by polymorphic bifunctional polypeptides. Penicillin-binding protein 1Bs of Escherichia coliwith activities of transglycosylase and transpeptidase. J Biol Chem. 1984;259:13937–13946. [PubMed] [Google Scholar]
- 571.Nakayama J, Ruhfel R E, Dunny G M, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalistetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Nathensen S G, Strominger J L, Ito E. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. IV. Purification and properties of d-glutamic acid-adding enzyme. J Biol Chem. 1964;239:1773–1776. [PubMed] [Google Scholar]
- 573.Navarre W W, Daefler S, Schneewind O. Cell wall sorting of lipoproteins in Staphylococcus aureus. J Bacteriol. 1996;178:441–446. doi: 10.1128/jb.178.2.441-446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 575.Navarre W W, Ton-That H, Faull K F, Schneewind O. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J Biol Chem. 1998;273:29135–29142. doi: 10.1074/jbc.273.44.29135. [DOI] [PubMed] [Google Scholar]
- 576.Navarre, W. W., H. Ton-That, K. F. Faull, and O. Schneewind. Unpublished data.
- 577.Nesbitt W E, Staat R H, Rosan B, Taylor K G, Doyle R J. Association of protein with the cell wall of Streptococcus mutans. Infect Immun. 1980;28:118–126. doi: 10.1128/iai.28.1.118-126.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Neuhaus F C. The enzymatic synthesis of d-alanyl-d-alanine. I. Purification and properties of d-alanyl-d-alanine synthetase. J Biol Chem. 1962;237:778–786. [PubMed] [Google Scholar]
- 579.Neuhaus F C, Heaton M P, Debabov D V, Zhang Q. The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb Drug Resist. 1996;2:77–84. doi: 10.1089/mdr.1996.2.77. [DOI] [PubMed] [Google Scholar]
- 580.Neuhaus F C, Linzer R, Reusch V M., Jr Biosynthesis of membrane teichoic acid: role of the d-alanine-activating enzyme and d-alanine: membrane acceptor ligase. Ann N Y Acad Sci. 1974;235:502–518. doi: 10.1111/j.1749-6632.1974.tb43287.x. [DOI] [PubMed] [Google Scholar]
- 581.Neutra M R, Pringault E, Kraehenbuhl J P. Antigen sampling across epithelial barriers and induction of mucosal immune responses. Annu Rev Immunol. 1996;14:275–300. doi: 10.1146/annurev.immunol.14.1.275. [DOI] [PubMed] [Google Scholar]
- 582.Nguyen T N, Gourdon M H, Hansson M, Robert A, Samuelson P, Libon C, Andreoni C, Nygren P A, Binz H, Uhlén M, et al. Hydrophobicity engineering to facilitate surface display of heterologous gene products on Staphylococcus xylosus. J Biotechnol. 1995;42:207–219. doi: 10.1016/0168-1656(95)00081-z. [DOI] [PubMed] [Google Scholar]
- 583.Nguyen T N, Hansson M, Stahl S, Bachi T, Robert A, Domzig W, Binz H, Uhlén M. Cell-surface display of heterologous epitopes on Staphylococcus xylosusas a potential delivery system for oral vaccination. Gene. 1993;128:89–94. doi: 10.1016/0378-1119(93)90158-y. [DOI] [PubMed] [Google Scholar]
- 584.Nielsen J B, Caulfield M P, Lampen J O. Lipoprotein nature of Bacillus licheniformismembrane penicillinase. Proc Natl Acad Sci USA. 1981;78:3511–3515. doi: 10.1073/pnas.78.6.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Nielsen J B, Lampen J O. Glyceride-cysteine lipoproteins and secretion by gram-positive bacteria. J Bacteriol. 1982;152:315–322. doi: 10.1128/jb.152.1.315-322.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Nilson B H, Frick I M, Åkesson P, Forsén S, Björck L, Åkerström B, Wikström M. Structure and stability of protein H and the M1 protein from Streptococcus pyogenes. Implications for other surface proteins of Gram-positive bacteria. Biochemistry. 1995;34:13688–13698. doi: 10.1021/bi00041a051. [DOI] [PubMed] [Google Scholar]
- 587.Nilson B H, Solomon A, Björck L, Åkerström B. Protein L from Peptostreptococcus magnusbinds to the kappa light chain variable domain. J Biol Chem. 1992;267:2234–2239. [PubMed] [Google Scholar]
- 588.Nilsson B, Abrahmsén L. Fusions to staphylococcal protein A. Methods Enzymol. 1990;185:144–161. doi: 10.1016/0076-6879(90)85015-g. [DOI] [PubMed] [Google Scholar]
- 589.Nilsson I M, Patti J M, Bremell T, Höök M, Tarkowski A. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J Clin Invest. 1998;101:2640–2649. doi: 10.1172/JCI1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Nilsson M, Frykberg L, Flock J I, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66:2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 592.Ntamere A S, Taron D J, Neuhaus F C. Assembly of d-alanyl-lipoteichoic acid in Lactobacillus casei: mutants deficient in the d-alanyl ester content of this amphiphile. J Bacteriol. 1987;169:1702–1711. doi: 10.1128/jb.169.4.1702-1711.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Oggioni M R, Pozzi G. A host-vector system for heterologous gene expression in Streptococcus gordonii. Gene. 1996;169:85–90. doi: 10.1016/0378-1119(95)00775-x. [DOI] [PubMed] [Google Scholar]
- 594.Ohnishi Y, Kubo S, Ono Y, Nozaki M, Gonda Y, Okano H, Matsuya T, Matsushiro A, Morita T. Cloning and sequencing of the gene coding for dextranase from Streptococcus salivarius. Gene. 1995;156:93–96. doi: 10.1016/0378-1119(95)00071-d. [DOI] [PubMed] [Google Scholar]
- 595.Okada N, Geist R T, Caparon M G. Positive transcriptional control of mryregulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 596.Okada N, Liszewski M K, Atkinson J P, Caparon M. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc Natl Acad Sci USA. 1995;92:2489–2493. doi: 10.1073/pnas.92.7.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Okada N, Pentland A P, Falk P, Caparon M G. M protein and protein F act as important determinants of cell-specific tropism of Streptococcus pyogenesin skin tissue. J Clin Invest. 1994;94:965–977. doi: 10.1172/JCI117463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 599.Olabarriá G, Carrascosa J L, de Pedro M A, Berenguer J. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J Bacteriol. 1996;178:4765–4772. doi: 10.1128/jb.178.16.4765-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Oliver D B, Beckwith J. E. colimutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 601.Oliver D B, Cabelli R J, Dolan K M, Jarosik G P. Azide-resistant mutants of Escherichia colialter the SecA protein, an azide-sensitive component of the protein export machinery. Proc Natl Acad Sci USA. 1990;87:8227–8231. doi: 10.1073/pnas.87.21.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Olmsted S B, Dunny G M, Erlandsen S L, Wells C L. A plasmid-encoded surface protein on Enterococcus faecalisaugments its internalization by cultured intestinal epithelial cells. J Infect Dis. 1994;170:1549–1556. doi: 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 603.Olsson A, Eliasson M, Guss B, Nilsson B, Hellman U, Lindberg M, Uhlén M. Structure and evolution of the repetitive gene encoding streptococcal protein G. Eur J Biochem. 1987;168:319–324. doi: 10.1111/j.1432-1033.1987.tb13423.x. [DOI] [PubMed] [Google Scholar]
- 604.Orlean P, Leidich S D, Drapp D A, Colussi P. Isolation of temperature-sensitive yeast GPI-anchoring mutants. Braz J Med Biol Res. 1994;27:145–150. [PubMed] [Google Scholar]
- 605.Oshida T, Sugai M, Komatsuzawa H, Hong Y M, Suginaka H, Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci USA. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Oshida T, Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992;174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.O’Toole P, Stenberg L, Rissler M, Lindahl G. Two major classes in the M protein family in group A streptococci. Proc Natl Acad Sci USA. 1992;89:8661–8665. doi: 10.1073/pnas.89.18.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.O’Toole R D, Goode L, Howe C. Neuraminidase activity in bacterial meningitis. J Clin Invest. 1971;50:979–985. doi: 10.1172/JCI106591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Ozeri V, Tovi A, Burstein I, Natanson-Yaron S, Caparon M G, Yamada K M, Akiyama S K, Vlodavsky I, Hanski E. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 1996;15:989–998. [PMC free article] [PubMed] [Google Scholar]
- 610.Pancholi V, Fischetti V A. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 611.Pancholi V, Fischetti V A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci USA. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Pancholi V, Fischetti V A. Identification of an endogenous membrane anchor-cleaving enzyme for group A streptococcal M protein. Its implication for the attachment of surface proteins in Gram-positive bacteria. J Exp Med. 1989;170:2119–2133. doi: 10.1084/jem.170.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Pancholi V, Fischetti V A. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J Bacteriol. 1988;170:2618–2624. doi: 10.1128/jb.170.6.2618-2624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Pancholi V, Fischetti V A. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J Exp Med. 1997;186:1633–1643. doi: 10.1084/jem.186.10.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Park J T. Uridine-5′-pyrophosphate derivatives. I. Isolation from Staphylococcus aureus. J Biol Chem. 1952;194:877–904. [PubMed] [Google Scholar]
- 617.Park J T, Johnson M J. Accumulation of labile phosphate in Staphylococcus aureusgrown in the presence of penicillin. J Biol Chem. 1949;179:585–592. [PubMed] [Google Scholar]
- 618.Patel A H, Nowlan P, Weavers E D, Foster T. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureusisolated by allele replacement. Infect Immun. 1987;55:3103–3110. doi: 10.1128/iai.55.12.3103-3110.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Patella V, Casolaro V, Björck L, Marone G. Protein L. A bacterial Ig-binding protein that activates human basophils and mast cells. J Immunol. 1990;145:3054–3061. [PubMed] [Google Scholar]
- 620.Paton J C, Andrew P W, Boulnois G J, Mitchell T J. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu Rev Microbiol. 1993;47:89–115. doi: 10.1146/annurev.mi.47.100193.000513. [DOI] [PubMed] [Google Scholar]
- 621.Patti J M, Allen B L, McGavin M J, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 622.Patti J M, Boles J O, Höök M. Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry. 1993;32:11428–11435. doi: 10.1021/bi00093a021. [DOI] [PubMed] [Google Scholar]
- 623.Patti J M, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Höök M. The Staphylococcus aureuscollagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Patti J M, Höök M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol. 1994;6:752–758. doi: 10.1016/0955-0674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 625.Patti J M, House-Pompeo K, Boles J O, Garza N, Gurusiddappa S, Höök M. Critical residues in the ligand-binding site of the Staphylococcus aureuscollagen-binding adhesin (MSCRAMM) J Biol Chem. 1995;270:12005–12011. doi: 10.1074/jbc.270.20.12005. [DOI] [PubMed] [Google Scholar]
- 626.Patti J M, Jonsson H, Guss B, Switalski L M, Wiberg K, Lindberg M, Höök M. Molecular characterization and expression of a gene encoding a Staphylococcus aureuscollagen adhesin. J Biol Chem. 1992;267:4766–4772. [PubMed] [Google Scholar]
- 627.Penney T J, Martin D R, Williams L C, de Malmanche S A, Bergquist P L. A single emm gene-specific oligonucleotide probe does not recognize all members of the Streptococcus pyogenesM type 1. FEMS Microbiol Lett. 1995;130:145–149. doi: 10.1111/j.1574-6968.1995.tb07711.x. [DOI] [PubMed] [Google Scholar]
- 628.Perez-Casal J, Okada N, Caparon M G, Scott J R. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol Microbiol. 1995;15:907–916. doi: 10.1111/j.1365-2958.1995.tb02360.x. [DOI] [PubMed] [Google Scholar]
- 629.Peters G, Locci R, Pulverer G. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J Infect Dis. 1982;146:479–482. doi: 10.1093/infdis/146.4.479. [DOI] [PubMed] [Google Scholar]
- 630.Peterson P K, Schmeling D, Cleary P P, Wilkinson B J, Kim Y, Quie P G. Inhibition of alternative complement pathway opsonization by group A streptococcal M protein. J Infect Dis. 1979;139:575–585. doi: 10.1093/infdis/139.5.575. [DOI] [PubMed] [Google Scholar]
- 631.Piard J C, Hautefort I, Fischetti V A, Ehrlich S D, Fons M, Gruss A. Cell wall anchoring of the Streptococcus pyogenesM6 protein in various lactic acid bacteria. J Bacteriol. 1997;179:3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Podbielski A. Three different types of organization of the virregulon in group A streptococci. Mol Gen Genet. 1993;237:287–300. doi: 10.1007/BF00282810. [DOI] [PubMed] [Google Scholar]
- 633.Podbielski A. Ubiquitous occurrence of virR and scpAgenes in group A streptococci. Med Microbiol Immunol. 1992;181:227–240. doi: 10.1007/BF00215768. [DOI] [PubMed] [Google Scholar]
- 634.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural virregulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Podbielski A, Hawlitzky J, Pack T D, Flosdorff A, Boyle M D. A group A streptococcal Enn protein potentially resulting from intergenomic recombination exhibits atypical immunoglobulin-binding characteristics. Mol Microbiol. 1994;12:725–736. doi: 10.1111/j.1365-2958.1994.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 636.Podbielski A, Kaufhold A, Cleary P P. PCR-mediated amplification of group A streptococcal genes encoding immunoglobulin-binding proteins. ImmunoMethods. 1993;2:55–64. [Google Scholar]
- 637.Podbielski A, Peterson J A, Cleary P. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol Microbiol. 1992;6:2253–2265. doi: 10.1111/j.1365-2958.1992.tb01401.x. [DOI] [PubMed] [Google Scholar]
- 638.Podbielski A, Schnitzler N, Beyhs P, Boyle M D. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 639.Podbielski A, Woischnik M, Pohl B, Schmidt K H. What is the size of the group A streptococcal virregulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 640.Pohlschroder M, Prinz W A, Hartmann E, Beckwith J. Protein translocation in the three domains of life: variations on a theme. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 641.Pollack J H, Ntamere A S, Neuhaus F C. d-Alanyl-lipoteichoic acid in Lactobacillus casei: secretion of vesicles in response to benzylpenicillin. J Gen Microbiol. 1992;138:849–859. doi: 10.1099/00221287-138-5-849. [DOI] [PubMed] [Google Scholar]
- 642.Pooley H M, Abellan F X, Karamata D. A conditional-lethal mutant of Bacillus subtilis168 with a thermosensitive glycerol-3-phosphate cytidylyltransferase, an enzyme specific for the synthesis of the major wall teichoic acid. J Gen Microbiol. 1991;137:921–928. doi: 10.1099/00221287-137-4-921. [DOI] [PubMed] [Google Scholar]
- 643.Pooley H M, Karamata D. Teichoic acid synthesis in Bacillus subtilis: genetic organization and biological roles. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 187–196. [Google Scholar]
- 644.Pooley H M, Merchante R, Karamata D. Overall protein content and induced enzyme components of the periplasm of Bacillus subtilis. Microb Drug Resist. 1996;2:9–15. doi: 10.1089/mdr.1996.2.9. [DOI] [PubMed] [Google Scholar]
- 645.Popham D L, Helin J, Costello C E, Setlow P. Analysis of the peptidoglycan structure of Bacillus subtilisendospores. J Bacteriol. 1996;178:6451–6458. doi: 10.1128/jb.178.22.6451-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Popham D L, Helin J, Costello C E, Setlow P. Muramic lactam in peptidoglycan of Bacillus subtilisspores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci USA. 1996;93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Popoff M R, Chaves-Olarte E, Lemichez E, von Eichel-Streiber C, Thelestam M, Chardin P, Cussac D, Antonny B, Chavrier P, Flatau G, Giry M, de Gunzburg J, Boquet P. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordelliilethal toxin glycosylation. J Biol Chem. 1996;271:10217–10224. doi: 10.1074/jbc.271.17.10217. [DOI] [PubMed] [Google Scholar]
- 648.Poritz M A, Strub K, Walter P. Human SRP RNA and E. coli4.5S RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- 649.Pozzi G, Contorni M, Oggioni M R, Manganelli R, Tommasino M, Cavalieri F, Fischetti V A. Delivery and expression of a heterologous antigen on the surface of streptococci. Infect Immun. 1992;60:1902–1907. doi: 10.1128/iai.60.5.1902-1907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Pozzi G, Oggioni M R, Manganelli R, Fischetti V A. Expression of M6 protein gene of Streptococcus pyogenes in Streptococcus gordoniiafter chromosomal integration and transcriptional fusion. Res Microbiol. 1992;143:449–457. doi: 10.1016/0923-2508(92)90090-b. [DOI] [PubMed] [Google Scholar]
- 651.Prepens U, Just I, Hofmann F, Aktories K. ADP-ribosylating and glucosylating toxins as tools to study secretion in RBL cells. Adv Exp Med Biol. 1997;419:349–353. doi: 10.1007/978-1-4419-8632-0_46. [DOI] [PubMed] [Google Scholar]
- 652.Pritchard K H, Cleary P P. Differential expression of genes in the vir regulon of Streptococcus pyogenesis controlled by transcription termination. Mol Gen Genet. 1996;250:207–213. doi: 10.1007/BF02174180. [DOI] [PubMed] [Google Scholar]
- 653.Pucci M J, Macrina F L. Molecular organization and expression of the gtfA gene of Streptococcus mutansLM7. Infect Immun. 1986;54:77–84. doi: 10.1128/iai.54.1.77-84.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Qian H, Dao M L. Inactivation of the Streptococcus mutans wall-associated protein A gene (wapA) results in a decrease in sucrose-dependent adherence and aggregation. Infect Immun. 1993;61:5021–5028. doi: 10.1128/iai.61.12.5021-5028.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Quintela J C, Pittenauer E, Allmaier G, Aran V, de Pedro M A. Structure of peptidoglycan from Thermus thermophilusHB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Rachel R, Pum D, Smarda J, Smajs D, Komrska J, Krzyzanek V, Rieger G, Stetter K O. Fine structure of S-layers. FEMS Microbiol Rev. 1997;20:13–23. [Google Scholar]
- 658.Rakonjac J V, Robbins J C, Fischetti V A. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect Immun. 1995;63:622–631. doi: 10.1128/iai.63.2.622-631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Ralph B A, Briles D E, McDaniel L S. Cross-reactive protection eliciting epitopes of pneumococcal surface protein A. Ann N Y Acad Sci. 1994;730:361–363. doi: 10.1111/j.1749-6632.1994.tb44293.x. [DOI] [PubMed] [Google Scholar]
- 660.Ramadurai L, Jayaswal R K. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol. 1997;179:3625–3631. doi: 10.1128/jb.179.11.3625-3631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Randall L L. Peptide binding by chaperone SecB: implications for recognition of nonnative structure. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]
- 662.Randall L L, Topping T B, Hardy S J. No specific recognition of leader peptide by SecB, a chaperone involved in protein export. Science. 1990;248:860–863. doi: 10.1126/science.2188362. [DOI] [PubMed] [Google Scholar]
- 663.Rayner C F, Jackson A D, Rutman A, Dewar A, Mitchell T J, Andrew P W, Cole P J, Wilson R. Interaction of pneumolysin-sufficient and -deficient isogenic variants of Streptococcus pneumoniaewith human respiratory mucosa. Infect Immun. 1995;63:442–447. doi: 10.1128/iai.63.2.442-447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology. 1971;43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- 665.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 666.Reichardt W, Gubbe K, Schmidt K H. M3-protein with close sequence homology to M12 protein binds fibrinogen, albumin, fibronectin, but not to any subclass of IgG-localization of binding regions. Dev Biol Stand. 1995;85:179–182. [PubMed] [Google Scholar]
- 667.Reis K J, Ayoub E M, Boyle M D. Streptococcal Fc receptors. I. Isolation and partial characterization of the receptor from a group C streptococcus. J Immunol. 1984;132:3091–3097. [PubMed] [Google Scholar]
- 668.Reis K J, Ayoub E M, Boyle M D. Streptococcal Fc receptors. II. Comparison of the reactivity of a receptor from a group C streptococcus with staphylococcal protein A. J Immunol. 1984;132:3098–3102. [PubMed] [Google Scholar]
- 669.Retnoningrum D S, Cleary P P. M12 protein from Streptococcus pyogenesis a receptor for immunoglobulin G3 and human albumin. Infect Immun. 1994;62:2387–2394. doi: 10.1128/iai.62.6.2387-2394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Ries W, Hotzy C, Schocher I, Sleytr U B, Sára M. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilusPV72/p2 recognizes a secondary cell wall polymer. J Bacteriol. 1997;179:3892–3898. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 671.Robbins J C, Spanier J G, Jones S J, Simpson W J, Cleary P P. Streptococcus pyogenestype 12 M protein gene regulation by upstream sequences. J Bacteriol. 1987;169:5633–5640. doi: 10.1128/jb.169.12.5633-5640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672.Roberts R J. Staphylococcal transfer ribonucleic acids. II. Sequence analysis of isoaccepting glycine transfer ribonucleic acids IA and IB from Staphylococcus epidermidisTexas 26. J Biol Chem. 1974;249:4787–4796. [PubMed] [Google Scholar]
- 673.Roberts R J, Lovinger G G, Tamura T, Strominger J L. Staphylococcal transfer ribonucleic acids. I. Isolation and purification of the isoaccepting glycine transfer ribonucleic acids from Staphylococcus epidermidisTexas 26. J Biol Chem. 1974;249:4781–4786. [PubMed] [Google Scholar]
- 674.Rodgers W, Crise B, Rose J K. Signals determining protein tyrosine kinase and glycosyl-phosphatidylinositol-anchored protein targeting to a glycolipid-enriched membrane fraction. Mol Cell Biol. 1994;14:5384–5391. doi: 10.1128/mcb.14.8.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Roggentin P, Schauer R, Hoyer L L, Vimr E R. The sialidase superfamily and its spread by horizontal gene transfer. Mol Microbiol. 1993;9:915–921. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 676.Romero A, López R, Garcia P. Lytic action of cloned pneumococcal phage lysis genes in Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;108:87–92. doi: 10.1016/0378-1097(93)90492-k. [DOI] [PubMed] [Google Scholar]
- 677.Romero A, López R, Garcia P. Sequence of the Streptococcus pneumoniaebacteriophage HB-3 amidase reveals high homology with the major host autolysin. J Bacteriol. 1990;172:5064–5070. doi: 10.1128/jb.172.9.5064-5070.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coliproteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 679.Ronda-Lain C, López R, Tapia A, Tomasz A. Role of the pneumococcal autolysin (murein hydrolase) in the release of progeny bacteriophage and in the bacteriophage-induced lysis of the host cells. J Virol. 1977;21:366–374. doi: 10.1128/jvi.21.1.366-374.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Rosan B. Relationship of the cell wall composition of group H streptococci and Streptococcus sanguisto their serological properties. Infect Immun. 1976;13:1144–1153. doi: 10.1128/iai.13.4.1144-1153.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 682.Rothfield L I, Zhao C R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 683.Rozalska B, Wadström T. Protective opsonic activity of antibodies against fibronectin-binding proteins (FnBPs) of Staphylococcus aureus. Scand J Immunol. 1993;37:575–580. doi: 10.1111/j.1365-3083.1993.tb02574.x. [DOI] [PubMed] [Google Scholar]
- 684.Rozalska B, Wadström T. Role of antibodies against fibronectin-, collagen-binding proteins and alphatoxin in experimental Staphylococcus aureusperitonitis and septicaemia in neutropenic mice. Int J Med Microbiol Virol Parasitol Infect Dis. 1994;281:495–501. doi: 10.1016/s0934-8840(11)80337-3. [DOI] [PubMed] [Google Scholar]
- 685.Rubens C E, Wessels M R, Heggen L M, Kasper D L. Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc Natl Acad Sci USA. 1987;84:7208–7212. doi: 10.1073/pnas.84.20.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Russell M W. Purification and properties of a protein surface antigen of Streptococcus mutants. Microbios. 1979;25:7–18. [PubMed] [Google Scholar]
- 687.Russell M W, Harrington D J, Russell R R. Identity of Streptococcus mutanssurface protein antigen III and wall-associated protein antigen A. Infect Immun. 1995;63:733–735. doi: 10.1128/iai.63.2.733-735.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Russell M W, Lehner T. Characterisation of antigens extracted from cells and culture fluids of Streptococcus mutansserotype c. Arch Oral Biol. 1978;23:7–15. doi: 10.1016/0003-9969(78)90047-x. [DOI] [PubMed] [Google Scholar]
- 689.Russell R R. Wall-associated protein antigens of Streptococcus mutans. J Gen Microbiol. 1979;114:109–115. doi: 10.1099/00221287-114-1-109. [DOI] [PubMed] [Google Scholar]
- 690.Russell R R, Mukasa H, Shimamura A, Ferretti J J. Streptococcus mutans gtfAgene specifies sucrose phosphorylase. Infect Immun. 1988;56:2763–2765. doi: 10.1128/iai.56.10.2763-2765.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Russell-Jones G J, Gotschlich E C, Blake M S. A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J Exp Med. 1984;160:1467–1475. doi: 10.1084/jem.160.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Ryding U, Flock J I, Flock M, Soderquist B, Christensson B. Expression of collagen-binding protein and types 5 and 8 capsular polysaccharide in clinical isolates of Staphylococcus aureus. J Infect Dis. 1997;176:1096–1099. doi: 10.1086/516520. [DOI] [PubMed] [Google Scholar]
- 693.Sahl H G, Jack R W, Bierbaum G. Biosynthesis and biological activities of antibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 694.Salton M R J. The bacterial cell envelope—a historical perspective. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 1–22. [Google Scholar]
- 695.Salton M R J. Cell wall of Micrococcus lysodeikticusas the substrate of lysozyme. Nature. 1952;170:746–747. doi: 10.1038/170746a0. [DOI] [PubMed] [Google Scholar]
- 696.Salton M R J. The nature of the cell walls of some Gram-positive and Gram-negative bacteria. Biochim Biophys Acta. 1952;9:334–335. doi: 10.1016/0006-3002(52)90172-8. [DOI] [PubMed] [Google Scholar]
- 697.Samuelson P, Hansson M, Ahlborg N, Andreoni C, Gotz F, Bachi T, Nguyen T N, Binz H, Uhlén M, Stahl S. Cell surface display of recombinant proteins on Staphylococcus carnosus. J Bacteriol. 1995;177:1470–1476. doi: 10.1128/jb.177.6.1470-1476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 698.Sánchez-Puelles J M, García J L, López R, García E. 3′-End modifications of the Streptococcus pneumoniae lytAgene: role of the carboxy terminus of the pneumococcal autolysin in the process of enzymatic activation (conversion) Gene. 1987;61:13–19. doi: 10.1016/0378-1119(87)90360-x. [DOI] [PubMed] [Google Scholar]
- 699.Sánchez-Puelles J M, Sanz J M, García J L, García E. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene. 1990;89:69–75. doi: 10.1016/0378-1119(90)90207-8. [DOI] [PubMed] [Google Scholar]
- 700.Sanderson A R, Juergens W G, Strominger J L. Chemical and immunochemical structure of teichoic acid from Staphylococcus aureus. Biochem Biophys Res Commun. 1961;5:472–476. [Google Scholar]
- 701.Sanderson A R, Strominger J L, Nathenson S G. Chemical structure of teichoic acid from Staphylococcus aureus, strain Copenhagen. J Biol Chem. 1962;237:3603–3613. [PubMed] [Google Scholar]
- 702.Sanz J M, López R, García J L. Structural requirements of choline derivatives for ‘conversion’ of pneumococcal amidase. A new single-step procedure for purification of this autolysin. FEBS Lett. 1988;232:308–312. doi: 10.1016/0014-5793(88)80759-2. [DOI] [PubMed] [Google Scholar]
- 703.Saravani G A, Martin D R. Opacity factor from group A streptococci is an apoproteinase. FEMS Microbiol Lett. 1990;56:35–39. doi: 10.1111/j.1574-6968.1990.tb04118.x. [DOI] [PubMed] [Google Scholar]
- 704.Sato Y, Yamamoto Y, Kizaki H. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect Immun. 1997;65:668–675. doi: 10.1128/iai.65.2.668-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Sauerborn M, Leukel P, von Eichel-Streiber C. The C-terminal ligand-binding domain of Clostridium difficiletoxin A (TcdA) abrogates TcdA-specific binding to cells and prevents mouse lethality. FEMS Microbiol Lett. 1997;155:45–54. doi: 10.1111/j.1574-6968.1997.tb12684.x. [DOI] [PubMed] [Google Scholar]
- 706.Sauerborn M, von Eichel-Streiber C. Nucleotide sequence of Clostridium difficiletoxin A. Nucleic Acids Res. 1990;18:1629–1630. doi: 10.1093/nar/18.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Schatz P J, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 708.Schennings T, Heimdahl A, Coster K, Flock J I. Immunization with fibronectin binding protein from Staphylococcus aureusprotects against experimental endocarditis in rats. Microb Pathog. 1993;15:227–236. doi: 10.1006/mpat.1993.1073. [DOI] [PubMed] [Google Scholar]
- 709.Schindler C A, Schuhardt V T. Lysostaphin: a new bacteriolytic agent for the staphylococcus. Proc Natl Acad Sci USA. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Schlievert P M, Gahr P J, Assimacopoulos A P, Dinges M M, Stoehr J A, Harmala J W, Hirt H, Dunny G M. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalisendocarditis. Infect Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Schmidt K H, Wadström T. A secreted receptor related to M1 protein of Streptococcus pyogenesbinds to fibrinogen, IgG, and albumin. Int J Med Microbiol. 1990;273:216–228. doi: 10.1016/s0934-8840(11)80252-5. [DOI] [PubMed] [Google Scholar]
- 713.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 714.Schneewind O, Jones K F, Fischetti V A. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J Bacteriol. 1990;172:3310–3317. doi: 10.1128/jb.172.6.3310-3317.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 717.Schnitzler N, Podbielski A, Baumgarten G, Mignon M, Kaufhold A. M or M-like protein gene polymorphisms in human group G streptococci. J Clin Microbiol. 1995;33:356–363. doi: 10.1128/jcm.33.2.356-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Scott J R, Cleary P, Caparon M G, Kehoe M, Hedén L, Musser J M, Hollingshead S, Podbielski A. New name for the positive regulator of the M protein of group A streptococcus. Mol Microbiol. 1995;17:799. doi: 10.1111/j.1365-2958.1995.mmi_17040799.x. [DOI] [PubMed] [Google Scholar]
- 719.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Sela S, Aviv A, Tovi A, Burstein I, Caparon M G, Hanski E. Protein F: an adhesin of Streptococcus pyogenesbinds fibronectin via two distinct domains. Mol Microbiol. 1993;10:1049–1055. doi: 10.1111/j.1365-2958.1993.tb00975.x. [DOI] [PubMed] [Google Scholar]
- 721.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 722.Selzer J, Hofmann F, Rex G, Wilm M, Mann M, Just I, Aktories K. Clostridium novyialpha-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J Biol Chem. 1996;271:25173–25177. doi: 10.1074/jbc.271.41.25173. [DOI] [PubMed] [Google Scholar]
- 723.Serhir B, Dugourd D, Jacques M, Higgins R, Harel J. Cloning and characterization of a dextranase gene (dexS) from Streptococcus suis. Gene. 1997;190:257–261. doi: 10.1016/s0378-1119(97)00004-8. [DOI] [PubMed] [Google Scholar]
- 724.Sharma A K, Pangburn M K. Localization by site directed mutagenesis of the site in human complement factor H that binds to Streptococcus pyogenesM protein. Infect Immun. 1997;65:484–487. doi: 10.1128/iai.65.2.484-487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 131–166. [Google Scholar]
- 726.Shuttleworth H L, Duggleby C J, Jones S A, Atkinson T, Minton N P. Nucleotide sequence analysis of the gene for protein A from Staphylococcus aureus Cowan 1 (NCTC8530) and its enhanced expression in Escherichia coli. Gene. 1987;58:283–295. doi: 10.1016/0378-1119(87)90383-0. [DOI] [PubMed] [Google Scholar]
- 727.Signäs C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Silhavy T J, Benson S A, Emr S D. Mechanisms of protein localization. Microbiol Rev. 1983;47:313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Simpson W J, LaPenta D, Chen C, Cleary P P. Coregulation of type 12 M protein and streptococcal C5a peptidase genes in group A streptococci: evidence for a virulence regulon controlled by the virRlocus. J Bacteriol. 1990;172:696–700. doi: 10.1128/jb.172.2.696-700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Simpson W J, Musser J M, Cleary P P. Evidence consistent with horizontal transfer of the gene (emm12) encoding serotype M12 protein between group A and group G pathogenic streptococci. Infect Immun. 1992;60:1890–1893. doi: 10.1128/iai.60.5.1890-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Simpson W J, Robbins J C, Cleary P P. Evidence for group A-related M protein genes in human but not animal-associated group G streptococcal pathogens. Microb Pathog. 1987;3:339–350. doi: 10.1016/0882-4010(87)90004-0. [DOI] [PubMed] [Google Scholar]
- 732.Singer H J, Wise E M, Jr, Park J T. Properties and purification of N-acetylmuramyl-l-alanine amidase from Staphylococcus aureusH. J Bacteriol. 1972;112:932–939. doi: 10.1128/jb.112.2.932-939.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Sjöbring U, Björck L, Kastern W. Protein G genes: structure and distribution of IgG-binding and albumin-binding domains. Mol Microbiol. 1989;3:319–327. doi: 10.1111/j.1365-2958.1989.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 734.Sjöbring U, Björck L, Kastern W. Streptococcal protein G. Gene structure and protein binding properties. J Biol Chem. 1991;266:399–405. [PubMed] [Google Scholar]
- 735.Sjöbring U, Falkenberg C, Nielsen E, Åkerström B, Björck L. Isolation and characterization of a 14-kDa albumin-binding fragment of streptococcal protein G. J Immunol. 1988;140:1595–1599. [PubMed] [Google Scholar]
- 736.Sjöbring U, Trojnar J, Grubb A, Åkerström B, Björck L. Ig-binding bacterial proteins also bind proteinase inhibitors. J Immunol. 1989;143:2948–2954. [PubMed] [Google Scholar]
- 737.Sjöquist J, Meloun B, Hjelm H. Protein A isolated from Staphylococcus aureusafter digestion with lysostaphin. Eur J Biochem. 1972;29:572–578. doi: 10.1111/j.1432-1033.1972.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 738.Sjöquist J, Movitz J, Johansson I B, Hjelm H. Localization of protein A in the bacteria. Eur J Biochem. 1972;30:190–194. doi: 10.1111/j.1432-1033.1972.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 739.Sleytr U B. Basic and applied S-layer research: an overview. FEMS Microbiol Rev. 1997;20:5–12. [Google Scholar]
- 740.Sleytr U B, Glauert A M. Ultrastructure of the cell walls of two closely related clostridia that possess different regular arrays of surface subunits. J Bacteriol. 1976;126:869–882. doi: 10.1128/jb.126.2.869-882.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Sleytr U B, Messner P. Crystalline surface layers in procaryotes. J Bacteriol. 1988;170:2891–2897. doi: 10.1128/jb.170.7.2891-2897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 742.Sleytr U B, Messner P, Pum D, Sára M. Crystalline bacterial cell surface layers. Mol Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 743.Sleytr U B, Thorne K J. Chemical characterization of the regularly arranged surface layers of Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. J Bacteriol. 1976;126:377–383. doi: 10.1128/jb.126.1.377-383.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Smid E J, Poolman B, Konings W N. Casein utilization by lactococci. Appl Environ Microbiol. 1991;57:2447–2452. doi: 10.1128/aem.57.9.2447-2452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Smirnov O, Denesyuk A I, Zakharov M V, Abramov V M, Zav’yalov V P. Protein V, a novel type-II IgG receptor from Streptococcus sp.: sequence, homologies and putative Fc-binding site. Gene. 1992;120:27–32. doi: 10.1016/0378-1119(92)90005-a. [DOI] [PubMed] [Google Scholar]
- 746.Smith G A, Portnoy D A, Theriot J A. Asymmetric distribution of the Listeria monocytogenesActA protein is required and sufficient to direct actin-based motility. Mol Microbiol. 1995;17:945–951. doi: 10.1111/j.1365-2958.1995.mmi_17050945.x. [DOI] [PubMed] [Google Scholar]
- 747.Smith H E, Vecht U, Gielkens A L, Smits M A. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suistype 2. Infect Immun. 1992;60:2361–2367. doi: 10.1128/iai.60.6.2361-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Smith T J, Blackman S A, Foster S J. Peptidoglycan hydrolases of Bacillus subtilis168. Microb Drug Resist. 1996;2:113–118. doi: 10.1089/mdr.1996.2.113. [DOI] [PubMed] [Google Scholar]
- 749.Snowden M A, Perkins H R. Peptidoglycan cross-linking in Staphylococcus aureus. An apparent random polymerisation process. Eur J Biochem. 1990;191:373–377. doi: 10.1111/j.1432-1033.1990.tb19132.x. [DOI] [PubMed] [Google Scholar]
- 750.Sriprakash K S, Hartas J. Lateral genetic transfers between group A and G streptococci for M-like genes are ongoing. Microb Pathog. 1996;20:275–285. doi: 10.1006/mpat.1996.0026. [DOI] [PubMed] [Google Scholar]
- 751.Stalhåmmar-Carlemalm M, Stenberg L, Lindahl G. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J Exp Med. 1993;177:1593–1603. doi: 10.1084/jem.177.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Steidler L, Viaene J, Fiers W, Remaut E. Functional display of a heterologous protein on the surface of Lactococcus lactis by means of the cell wall anchor of Staphylococcus aureusprotein A. Appl Environ Microbiol. 1998;64:342–345. doi: 10.1128/aem.64.1.342-345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Stenberg L, O’Toole P, Lindahl G. Many group A streptococcal strains express two different immunoglobulin-binding proteins, encoded by closely linked genes: characterization of the proteins expressed by four strains of different M-type. Mol Microbiol. 1992;6:1185–1194. doi: 10.1111/j.1365-2958.1992.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 754.Stenberg L, O’Toole P W, Mestecky J, Lindahl G. Molecular characterization of protein Sir, a streptococcal cell surface protein that binds both immunoglobulin A and immunoglobulin G. J Biol Chem. 1994;269:13458–13464. [PubMed] [Google Scholar]
- 755.Stewart T S, Roberts R J, Strominger J L. Novel species of tRNA. Nature. 1971;230:36–38. doi: 10.1038/230036a0. [DOI] [PubMed] [Google Scholar]
- 756.Stranden A M, Ehlert K, Labischinski H, Berger-Bachi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Strauss A, Gotz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- 758.Strominger J L. Enzymatic transfer of pyruvate to uridine diphosphoacetylglucosamine. Biochim Biophys Acta. 1959;30:645–646. doi: 10.1016/0006-3002(58)90119-7. [DOI] [PubMed] [Google Scholar]
- 759.Strominger J L. Penicillin-sensitive enzymatic reactions in bacterial cell wall synthesis. Harvey Lect. 1968;64:179–213. [PubMed] [Google Scholar]
- 760.Strominger J L, Ghuysen J M. Mechanisms of enzymatic bacteriaolysis. Cell walls of bacteria are solubilized by action of either specific carbohydrases or specific peptidases. Science. 1967;156:213–221. doi: 10.1126/science.156.3772.213. [DOI] [PubMed] [Google Scholar]
- 761.Strominger J L, Izaki K, Matsuhashi M, Tipper D J. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed Proc. 1967;26:9–22. [PubMed] [Google Scholar]
- 762.Struck J C, Toschka H Y, Specht T, Erdmann V A. Common structural features between eukaryotic 7SL RNAs, eubacterial 4.5S RNA and scRNA and archaebacterial 7S RNA. Nucleic Acids Res. 1988;16:7740. doi: 10.1093/nar/16.15.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Sugai M, Enomoto T, Hashimoto K, Matsumoto K, Matsuo Y, Ohgai H, Hong Y M, Inoue S, Yoshikawa K, Suginaka H. A novel epidermal cell differentiation inhibitor (EDIN): purification and characterization from Staphylococcus aureus. Biochem Biophys Res Commun. 1990;173:92–98. doi: 10.1016/s0006-291x(05)81026-5. [DOI] [PubMed] [Google Scholar]
- 764.Sugai M, Fujiwara T, Akiyama T, Ohara M, Komatsuzawa H, Inoue S, Suginaka H. Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitisEPK1. J Bacteriol. 1997;179:1193–1202. doi: 10.1128/jb.179.4.1193-1202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 765.Sugai M, Fujiwara T, Ohta K, Komatsuzawa H, Ohara M, Suginaka H. epr, which encodes glycylglycine endopeptidase resistance, is homologous to femAB and affects serine content of peptidoglycan cross bridges in Staphylococcus capitis and Staphylococcus aureus. J Bacteriol. 1997;179:4311–4318. doi: 10.1128/jb.179.13.4311-4318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Sugai M, Komatsuzawa H, Akiyama T, Hong Y M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 767.Sugai M, Yamada S, Nakashima S, Komatsuzawa H, Matsumoto A, Oshida T, Suginaka H. Localized perforation of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J Bacteriol. 1997;179:2958–2962. doi: 10.1128/jb.179.9.2958-2962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768.Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J Bacteriol. 1990;172:7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Sutcliffe I C, Russell R R. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Sutcliffe I C, Tao L, Ferretti J J, Russell R R. MsmE, a lipoprotein involved in sugar transport in Streptococcus mutans. J Bacteriol. 1993;175:1853–1855. doi: 10.1128/jb.175.6.1853-1855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Swinfield T J, Oultram J D, Thompson D E, Brehm J K, Minton N P. Physical characterisation of the replication region of the Streptococcus faecalisplasmid pAMβ1. Gene. 1990;87:79–90. [PubMed] [Google Scholar]
- 772.Switalski L M, Patti J M, Butcher W, Gristina A G, Speziale P, Höök M. A collagen receptor on Staphylococcus aureusstrains isolated from patients with septic arthritis mediates adhesion to cartilage. Mol Microbiol. 1993;7:99–107. doi: 10.1111/j.1365-2958.1993.tb01101.x. [DOI] [PubMed] [Google Scholar]
- 773.Symersky J, Patti J M, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas L J, Höök M, Narayana S V. Structure of the collagen-binding domain from a Staphylococcus aureusadhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 774.Takamatsu H, Bunai K, Horinaka T, Oguro A, Nakamura K, Watabe K, Yamane K. Identification of a region required for binding to presecretory protein in Bacillus subtilisFfh, a homologue of the 54-kDa subunit of mammalian signal recognition particle. Eur J Biochem. 1997;248:575–582. doi: 10.1111/j.1432-1033.1997.00575.x. [DOI] [PubMed] [Google Scholar]
- 775.Takeda J, Kinoshita T. GPI-anchor biosynthesis. Trends Biochem Sci. 1995;20:367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]
- 776.Talay S R, Grammel M P, Chhatwal G S. Structure of a group C streptococcal protein that binds to fibrinogen, albumin and immunoglobulin G via overlapping modules. Biochem J. 1996;315:577–582. doi: 10.1042/bj3150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Talay S R, Valentin-Weigand P, Jerlström P G, Timmis K N, Chhatwal G S. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect Immun. 1992;60:3837–3844. doi: 10.1128/iai.60.9.3837-3844.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Talay S R, Valentin-Weigand P, Timmis K N, Chhatwal G S. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol Microbiol. 1994;13:531–539. doi: 10.1111/j.1365-2958.1994.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 779.Talkington D F, Voellinger D C, McDaniel L S, Briles D E. Analysis of pneumococcal PspA microheterogeneity in SDS polyacrylamide gels and the association of PspA with the cell membrane. Microb Pathog. 1992;13:343–355. doi: 10.1016/0882-4010(92)90078-3. [DOI] [PubMed] [Google Scholar]
- 780.Thern A, Stenberg L, Dahlbäck B, Lindahl G. Ig-binding surface proteins of Streptococcus pyogenesalso bind human C4b-binding protein (C4BP), a regulatory component of the complement system. J Immunol. 1995;154:375–386. [PubMed] [Google Scholar]
- 781.Thern A, Wästfelt M, Lindahl G. Expression of two different antiphagocytic M proteins by Streptococcus pyogenesof the OF+ lineage. J Immunol. 1998;160:860–869. [PubMed] [Google Scholar]
- 782.Thumm G, Gotz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol. 1997;23:1251–1265. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- 783.Tian G, Wu H C, Ray P H, Tai P C. Temperature-dependent insertion of prolipoprotein into Escherichia colimembrane vesicles and requirements for ATP, soluble factors, and functional Sec Y protein for the overall translocation process. J Bacteriol. 1989;171:1987–1997. doi: 10.1128/jb.171.4.1987-1997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Timoney J F, Artiushin S C, Boschwitz J S. Comparison of the sequences and functions of Streptococcus equiM-like proteins SeM and SzPSe. Infect Immun. 1997;65:3600–3605. doi: 10.1128/iai.65.9.3600-3605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Timoney J F, Walker J, Zhou M, Ding J. Cloning and sequence analysis of a protective M-like protein gene from Streptococcus equi subsp. zooepidemicus. Infect Immun. 1995;63:1440–1445. doi: 10.1128/iai.63.4.1440-1445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Tipper D J. Mechanism of autolysis of isolated cell walls of Staphylococcus aureus. J Bacteriol. 1969;97:837–847. doi: 10.1128/jb.97.2.837-847.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Tipper D J. Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureus Copehagen. II. Structure of neutral and basic peptides from hydrolysis with the MyxobacterAL-1 peptidase. Biochemistry. 1969;8:2192–2202. doi: 10.1021/bi00833a061. [DOI] [PubMed] [Google Scholar]
- 788.Tipper D J, Berman M F. Structures of the cell wall peptidoglycans of Staphylococcus epidermidis Texas 26 and Staphylococcus aureusCopenhagen. I. Chain length and average sequence of cross-bridge peptides. Biochemistry. 1969;8:2183–2192. doi: 10.1021/bi00833a060. [DOI] [PubMed] [Google Scholar]
- 789.Tipper D J, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XII. Inhibition of cross-linking by penicillins and cephalosporins: studies in Staphylococcus aureus in vivo. J Biol Chem. 1968;243:3169–3179. [PubMed] [Google Scholar]
- 790.Tipper D J, Strominger J L. Isolation of 4-O-β-N-acetylmuramyl-N-acetylglucosamine and 4-O-β-N, 6-O-diacetylmuramyl-N-acetylglucosamine and the structure of the cell wall polysaccharide of Staphylococcus aureus. Biochem Biophys Res Commun. 1966;22:48–56. doi: 10.1016/0006-291x(66)90601-2. [DOI] [PubMed] [Google Scholar]
- 791.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Tipper D J, Strominger J L, Ensign J C. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. VII. Mode of action of the bacteriolytic peptidase from Myxobacterand the isolation of intact cell wall polysaccharides. Biochemistry. 1967;6:906–920. doi: 10.1021/bi00855a035. [DOI] [PubMed] [Google Scholar]
- 793.Tipper D J, Strominger J L, Ghuysen J-M. Staphylolytic enzyme from Chalaropsis: mechanism of action. Science. 1964;146:781–782. doi: 10.1126/science.146.3645.781. [DOI] [PubMed] [Google Scholar]
- 794.Tokuda M, Okahashi N, Takahashi I, Nakai M, Nagaoka S, Kawagoe M, Koga T. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infect Immun. 1991;59:3309–3312. doi: 10.1128/iai.59.9.3309-3312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 796.Ton-That H, Faull K F, Schneewind O. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 797.Ton-That H, Labischinski H, Berger-Bachi B, Schneewind O. Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J Biol Chem. 1998;273:29143–29149. doi: 10.1074/jbc.273.44.29143. [DOI] [PubMed] [Google Scholar]
- 797a.Ton-That, H., and O. Schneewind. Unpublished data.
- 798.Tschierske M, Ehlert K, Stranden A M, Berger-Bachi B. Lif, the lysostaphin immunity factor, complements FemB in staphylococcal peptidoglycan interpeptide bridge formation. FEMS Microbiol Lett. 1997;153:261–264. doi: 10.1016/s0378-1097(97)00234-6. [DOI] [PubMed] [Google Scholar]
- 799.Tsuboi A, Uchihi R, Tabata R, Takahashi Y, Hashiba H, Sasaki T, Yamagata H, Tsukagoshi N, Udaka S. Characterization of the genes coding for two major cell wall proteins from protein-producing Bacillus brevis47: complete nucleotide sequence of the outer wall protein gene. J Bacteriol. 1986;168:365–373. doi: 10.1128/jb.168.1.365-373.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Tucker K D, Wilkins T D. Toxin A of Clostridium difficilebinds to the human carbohydrate antigens I, X, and Y. Infect Immun. 1991;59:73–78. doi: 10.1128/iai.59.1.73-78.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 801.Tuomanen E, Schwartz J, Sande S, Light K, Gage D. Unusual composition of peptidoglycan in Bordetella pertussis. J Biol Chem. 1989;264:11093–11098. [PubMed] [Google Scholar]
- 802.Udenfriend S, Kodukula K, Amthauer R. Cell-free processing of nascent proteins designed to be linked to the plasma membrane by a phosphatidylinositol-glycan anchor. Cell Mol Biol. 1992;38:11–16. [PubMed] [Google Scholar]
- 803.Uhlén M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 804.Ulbrandt N D, Newitt J A, Bernstein H D. The E. colisignal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 805.Umbreit J N, Strominger J L. d-Alanine carboxypeptidase from Bacillus subtilismembranes. I. Purification and characterization. J Biol Chem. 1973;248:6759–6766. [PubMed] [Google Scholar]
- 806.Umeda A, Yokoyama S, Arizono T, Amako K. Location of peptidoglycan and teichoic acid on the cell wall surface of Staphylococcus aureusas determined by immunoelectron microscopy. J Electron Microsc. 1992;41:46–52. [PubMed] [Google Scholar]
- 807.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Valent Q A, Scotti P A, High S, de Gier J W, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coliSRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.van de Rijn I, Fischetti V A. Immunochemical analysis of intact M protein secreted from cell wall-less streptococci. Infect Immun. 1981;32:86–91. doi: 10.1128/iai.32.1.86-91.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 810.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, De Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.van der Wolk J P, de Wit J G, Driessen A J. The catalytic cycle of the Escherichia coliSecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.van Heijenoort J. Biosynthesis of the bacterial peptidoglycan unit. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 39–54. [Google Scholar]
- 813.Vidugiriene J, Menon A K. Biosynthesis of glycosylphosphatidylinositol anchors. Methods Enzymol. 1995;250:513–535. doi: 10.1016/0076-6879(95)50094-4. [DOI] [PubMed] [Google Scholar]
- 814.Vidugiriene J, Menon A K. Soluble constituents of the ER lumen are required for GPI anchoring of a model protein. EMBO J. 1995;14:4686–4694. doi: 10.1002/j.1460-2075.1995.tb00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.von Eichel-Streiber C, Boquet P, Sauerborn M, Thelestam M. Large clostridial cytotoxins—a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 1996;4:375–382. doi: 10.1016/0966-842X(96)10061-5. [DOI] [PubMed] [Google Scholar]
- 816.von Eichel-Streiber C, Sauerborn M. Clostridium difficiletoxin A carries a C-terminal repetitive structure homologous to the carbohydrate binding region of streptococcal glycosyltransferases. Gene. 1990;96:107–113. doi: 10.1016/0378-1119(90)90348-u. [DOI] [PubMed] [Google Scholar]
- 817.von Eichel-Streiber C, Sauerborn M, Kuramitsu H K. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutansglycosyltransferases. J Bacteriol. 1992;174:6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 819.von Heijne G, Abrahmsén L. Species-specific variation in signal peptide design. Implications for protein secretion in foreign hosts. FEBS Lett. 1989;244:439–446. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]
- 820.Vos P, Simons G, Siezen R J, de Vos W M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989;264:13579–13585. [PubMed] [Google Scholar]
- 821.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactisSK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Wallinder I B, Neujahr H Y. Cell wall and peptidoglycan from Lactobacillus fermenti. J Bacteriol. 1971;105:918–926. doi: 10.1128/jb.105.3.918-926.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 823.Walsh C T. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 824.Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Wanda S Y, Curtiss R., III Purification and characterization of Streptococcus sobrinus dextranase produced in recombinant Escherichia coliand sequence analysis of the dextranase gene. J Bacteriol. 1994;176:3839–3850. doi: 10.1128/jb.176.13.3839-3850.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Waneck G L, Stein M E, Flavell R A. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1988;241:697–699. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]
- 829.Wang B, Ruiz N, Pentland A, Caparon M. Keratinocyte proinflammatory responses to adherent and nonadherent group A streptococci. Infect Immun. 1997;65:2119–2126. doi: 10.1128/iai.65.6.2119-2126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Wang H, Lottenberg R, Boyle M D. Analysis of the interaction of group A streptococci with fibrinogen, streptokinase and plasminogen. Microb Pathog. 1995;18:153–166. doi: 10.1016/s0882-4010(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 831.Wang H, Lottenberg R, Boyle M D. A role for fibrinogen in the streptokinase-dependent acquisition of plasmin(ogen) by group A streptococci. J Infect Dis. 1995;171:85–92. doi: 10.1093/infdis/171.1.85. [DOI] [PubMed] [Google Scholar]
- 832.Wang J-R, Stinson M W. M protein mediates streptococcal adhesion to HEp-2 cells. Infect Immun. 1994;62:442–448. doi: 10.1128/iai.62.2.442-448.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 833.Wang J-R, Stinson M W. Streptococcal M protein binds to fucose-containing glycoproteins on cultured human epithelial cells. Infect Immun. 1994;62:1268–1274. doi: 10.1128/iai.62.4.1268-1274.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 834.Wang X, Mani N, Pattee P A, Wilkinson B J, Jayaswal R K. Analysis of a peptidoglycan hydrolase gene from Staphylococcus aureusNCTC 8325. J Bacteriol. 1992;174:6303–6306. doi: 10.1128/jb.174.19.6303-6306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 835.Wang X, Wilkinson B J, Jayaswal R K. Sequence analysis of a Staphylococcus aureusgene encoding a peptidoglycan hydrolase activity. Gene. 1991;102:105–109. doi: 10.1016/0378-1119(91)90547-o. [DOI] [PubMed] [Google Scholar]
- 836.Warth A D, Strominger J L. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry. 1972;11:1389–1396. doi: 10.1021/bi00758a010. [DOI] [PubMed] [Google Scholar]
- 837.Wästfelt M, Stalhåmmar-Carlemalm M, Delisse A M, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem. 1996;271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- 838.Watanabe R, Inoue N, Westfall B, Taron C H, Orlean P, Takeda J, Kinoshita T. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 839.Waxman D J, Strominger J L. Penicillin-binding proteins and the mechanism of action of β-lactam antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 840.Weidlich G, Wirth R, Galli D. Sex pheromone plasmid pAD1-encoded surface exclusion protein of Enterococcus faecalis. Mol Gen Genet. 1992;233:161–168. doi: 10.1007/BF00587575. [DOI] [PubMed] [Google Scholar]
- 841.Wessels M R, Moses A E, Goldberg J B, DiCesare T J. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Wessels M R, Paoletti L C, Guttormsen H K, Michon F, D’Ambra A J, Kasper D L. Structural properties of group B streptococcal type III polysaccharide conjugate vaccines that influence immunogenicity and efficacy. Infect Immun. 1998;66:2186–2192. doi: 10.1128/iai.66.5.2186-2192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 843.Wessels M R, Rubens C E, Benedi V J, Kasper D L. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc Natl Acad Sci USA. 1989;86:8983–8987. doi: 10.1073/pnas.86.22.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 845.Wexler D E, Cleary P P. Purification and characteristics of the streptococcal chemotactic factor inactivator. Infect Immun. 1985;50:757–764. doi: 10.1128/iai.50.3.757-764.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 846.Wexler D E, Nelson R D, Cleary P P. Human neutrophil chemotactic response to group A streptococci: bacteria-mediated interference with complement-derived chemotactic factors. Infect Immun. 1983;39:239–246. doi: 10.1128/iai.39.1.239-246.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Whatmore A M, Kapur V, Musser J M, Kehoe M A. Molecular population genetic analysis of the enn subdivision of group A streptococcal emm-like genes: horizontal gene transfer and restricted variation among enngenes. Mol Microbiol. 1995;15:1039–1048. doi: 10.1111/j.1365-2958.1995.tb02279.x. [DOI] [PubMed] [Google Scholar]
- 848.Whatmore A M, Kapur V, Sullivan D J, Musser J M, Kehoe M A. Non-congruent relationships between variation in emmgene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 849.Whatmore A M, Kehoe M A. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 850.Whitnack E, Beachey E H. Biochemical and biological properties of the binding of human fibrinogen to M protein in group A streptococci. J Bacteriol. 1985;164:350–358. doi: 10.1128/jb.164.1.350-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Whitnack E, Dale J B, Beachey E H. Common protective antigens of group A streptococcal M proteins masked by fibrinogen. J Exp Med. 1984;159:1201–1212. doi: 10.1084/jem.159.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Wickus G G, Warth A D, Strominger J L. Appearance of muramic lactam during cortex synthesis in sporulating cultures of Bacillus cereus and Bacillus megaterium. J Bacteriol. 1972;111:625–627. doi: 10.1128/jb.111.2.625-627.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Wietzerbin-Falszpan J, Das B C, Gros C, Petit J F, Lederer E. The amino acids of the cell wall of Mycobacterium tuberculosis var. bovis, strain BCG. Presence of a poly(l-glutamic acid) Eur J Biochem. 1973;32:525–532. doi: 10.1111/j.1432-1033.1973.tb02637.x. [DOI] [PubMed] [Google Scholar]
- 854.Wikström M, Drakenberg T, Forsén S, Sjöbring U, Björck L. Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G. Biochemistry. 1994;33:14011–14017. doi: 10.1021/bi00251a008. [DOI] [PubMed] [Google Scholar]
- 855.Wikström M, Sjöbring U, Drakenberg T, Forsén S, Björck L. Mapping of the immunoglobulin light chain-binding site of protein L. J Mol Biol. 1995;250:128–133. doi: 10.1006/jmbi.1995.0364. [DOI] [PubMed] [Google Scholar]
- 856.Winter A J, Comis S D, Osborne M P, Tarlow M J, Stephen J, Andrew P W, Hill J, Mitchell T J. A role for pneumolysin but not neuraminidase in the hearing loss and cochlear damage induced by experimental pneumococal meningitis in guinea pigs. Infect Immun. 1997;65:4411–4418. doi: 10.1128/iai.65.11.4411-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Wirth R, Friesenegger A, Horaud T. Identification of new sex pheromone plasmids in Enterococcus faecalis. Mol Gen Genet. 1992;233:157–160. doi: 10.1007/BF00587574. [DOI] [PubMed] [Google Scholar]
- 858.Wistedt A C, Ringdahl U, Müller-Esterl W, Sjöbring U. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol Microbiol. 1995;18:569–578. doi: 10.1111/j.1365-2958.1995.mmi_18030569.x. [DOI] [PubMed] [Google Scholar]
- 859.Wong C, Hefta S A, Paxton R J, Shively J E, Mooser G. Size and subdomain architecture of the glucan-binding domain of sucrose:3-alpha-d-glucosyltransferase from Streptococcus sobrinus. Infect Immun. 1990;58:2165–2170. doi: 10.1128/iai.58.7.2165-2170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Wren B W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 861.Wuenscher M D, Kohler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenesis essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Wyke A W, Ward J B. The biosynthesis of muramic acid phosphate in Bacillus licheniformis. FEBS Lett. 1977;73:159–163. doi: 10.1016/0014-5793(77)80971-x. [DOI] [PubMed] [Google Scholar]
- 863.Wyke A W, Ward J B. Biosynthesis of wall polymers in Bacillus subtilis. J Bacteriol. 1977;130:1055–1063. doi: 10.1128/jb.130.3.1055-1063.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.Yamashita Y, Bowen W H, Burne R A, Kuramitsu H K. Role of the Streptococcus mutans gtfgenes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866.Ye S Y, Koponen O, Qiao M, Immonen T, Saris P E. NisP is related to nisin precursor processing and possibly to immunity in Lactococcus lactis. J Tongji Med Univ. 1995;15:193–197. doi: 10.1007/BF02887942. [DOI] [PubMed] [Google Scholar]
- 867.Yeung M K, Cisar J O. Cloning and nucleotide sequence of a gene for Actinomyces naeslundiiWVU45 type 2 fimbriae. J Bacteriol. 1988;170:3803–3809. doi: 10.1128/jb.170.9.3803-3809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Yeung M K, Cisar J O. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomycesspp. J Bacteriol. 1990;172:2462–2468. doi: 10.1128/jb.172.5.2462-2468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Yeung M K, Donkersloot J A, Cisar J O, Ragsdale P A. Identification of a gene involved in assembly of Actinomyces naeslundiiT14V type 2 fimbriae. Infect Immun. 1998;66:1482–1491. doi: 10.1128/iai.66.4.1482-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Yeung M K, Ragsdale P A. Synthesis and function of Actinomyces naeslundiiT14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Yocum R R, Rasmussen J R, Strominger J L. The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilusd-alanine carboxypeptidase. J Biol Chem. 1980;255:3977–3986. [PubMed] [Google Scholar]
- 872.Yocum R R, Waxman D J, Rasmussen J R, Strominger J L. Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial d-alanine carboxypeptidases. Proc Natl Acad Sci USA. 1979;76:2730–2734. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 873.Yokoyama K, Miyashita T, Araki Y, Ito E. Structure and functions of linkage unit intermediates in the biosynthesis of ribitol teichoic acids in Staphylococcus aureus H and Bacillus subtilisW23. Eur J Biochem. 1986;161:479–489. doi: 10.1111/j.1432-1033.1986.tb10469.x. [DOI] [PubMed] [Google Scholar]
- 874.Yoshimori T, Keller P, Roth M G, Simons K. Different biosynthetic transport routes to the plasma membrane in BHK and CHO cells. J Cell Biol. 1996;133:247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875.Yother J, Briles D E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876.Yother J, Handsome G L, Briles D E. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspAgene. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Yung D L, Hollingshead S K. DNA sequencing and gene expression of the emmgene cluster in an M50 group A streptococcus strain virulent for mice. Infect Immun. 1996;64:2193–2200. doi: 10.1128/iai.64.6.2193-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Zygmunt W A, Browder H P, Tavormina P A. Lytic action of lysostaphin on susceptible and resistant strains of Staphylococcus aureus. Can J Microbiol. 1967;13:845–853. doi: 10.1139/m67-111. [DOI] [PubMed] [Google Scholar]