Abstract
Background:
Pediatric-onset multiple sclerosis (POMS) represents the earliest stage of disease pathogenesis. Investigating the cerebrospinal fluid (CSF) proteome in POMS may provide novel insights into early MS processes.
Objective:
To analyze CSF obtained from children at time of initial central nervous system (CNS) acquired demyelinating syndrome (ADS), to compare CSF proteome of those subsequently ascertained as having POMS versus monophasic acquired demyelinating syndrome (mADS).
Methods:
Patients were selected from two prospective pediatric ADS studies. Liquid chromatography–mass spectrometry (LC-MS) was performed in a Dutch discovery cohort (POMS n = 28; mADS n = 39). Parallel reaction monitoring–mass spectrometry (PRM-MS) was performed on selected proteins more abundant in POMS in a combined Dutch and Canadian validation cohort (POMS n = 48; mADS n = 106).
Results:
Discovery identified 5580 peptides belonging to 576 proteins; 58 proteins were differentially abundant with ⩾2 peptides between POMS and mADS, of which 28 more abundant in POMS. Fourteen had increased abundance in POMS with ⩾8 unique peptides. Five selected proteins were all confirmed within validation. Adjusted for age, 2 out of 5 proteins remained more abundant in POMS, that is, Carboxypeptidase E (CPE) and Semaphorin-7A (SEMA7A).
Conclusion:
This exploratory study identified several CSF proteins associated with POMS and not mADS, potentially reflecting neurodegeneration, compensatory neuroprotection, and humoral response in POMS. The proteins associated with POMS highly correlated with age at CSF sampling.
Keywords: Cerebrospinal fluid, proteins, proteomics, mass spectrometry, neurons, immune system, pediatrics, multiple sclerosis
Introduction
Despite extensive research, the exact pathogenesis of MS remains to be elucidated. Substantial (indirect) evidence indicates that biological start of disease likely substantially precedes first clinical symptoms, potentially with onset during childhood even when disease does not manifest until years into adulthood.1,2 However, in 3%–5% of all MS patients, first clinical symptoms occur in childhood.3 Therefore, pediatric-onset multiple sclerosis (POMS) provides a unique opportunity to gain key insights into an early stage of the disease. As cerebrospinal fluid (CSF) potentially reflects processes occurring within the central nervous system (CNS),4 proteins in CSF of POMS patients might reveal crucial information about very early MS disease biology.
Previously, one Dutch and one Canadian study have focused on CSF proteins in POMS.5,6 Due to the rarity of POMS, these studies included a limited number of patients. The aim of current collaborative study was to explore and validate differences in the CSF proteome in a larger, combined Dutch and Canadian cohort of children with an initial CNS acquired demyelinating syndrome (ADS), comparing those subsequently ascertained as having either POMS or monophasic ADS (mADS) .
Materials and methods
Patients and samples
Patients were selected from two independent, prospective observational cohort studies including ADS patients <18 years: the Dutch PROUD-kids study (PRedicting the OUtcome of a first Demyelinating event in childhood)7 and Canadian Pediatric Demyelinating Disease Study.8 Patients with available CSF samples collected at incident demyelinating attack were included if, at final follow-up, patients were diagnosed with (1) POMS or (2) mADS, according to International Pediatric MS Study Group criteria.9 Demographic, clinical, and laboratory data collected in both prospective cohort studies were used.
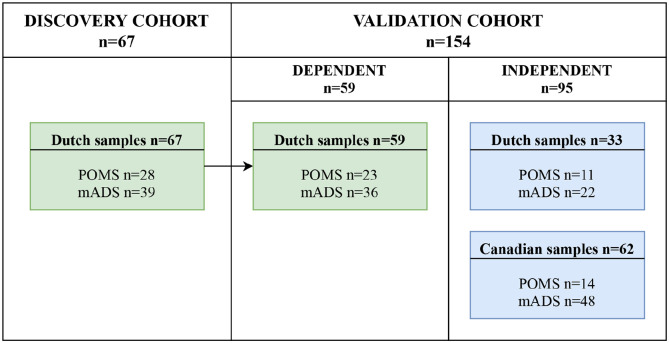
For this study, we used samples from the previously performed Dutch5 and Canadian6 proteomic studies, supplemented by new samples. Figure 1 shows the sample selection for this study. For the untargeted discovery analysis, only Dutch samples were selected (discovery cohort). For the following targeted validation analysis, both dependent (from discovery analysis) and independent (newly acquired) samples were selected (validation cohort). The dependent samples were used for technical validation regarding the different techniques used for discovery and validation. The independent samples consisted of Canadian and newly acquired Dutch samples. CSF samples of adult symptomatic controls (n = 16), having neurological symptoms but no objective (para)clinical findings to define a specific neurological disease,10 were pooled for technical quality control (QC) in validation analysis.
Figure 1.
Sample selection of discovery and validation cohort.
mADS: monophasic acquired demyelinating syndrome; POMS: pediatric-onset multiple sclerosis.
Ethical approval and patients consents
Protocols of the Dutch and Canadian prospective cohort study were approved by the local Medical Ethical Committee, and all patients and/or their legal representatives gave written informed consent.
Discovery analysis
Liquid chromatography–mass spectrometry (LC-MS)
For discovery analysis, CSF proteins were typically digested and prepared.11 Subsequently, digested samples were measured on a nano-liquid chromatography–Orbitrap Fusion mass spectrometer liquid chromatography-mass spectrometry, and the data were subsequently processed in a fragment mass spectrometer database search and label-free quantitative analysis.12 A detailed description and adjustments with regard to cited references are provided in Supplemental Material S1. Liquid chromatography-mass spectrometry (LC-MS) data have been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with dataset identifier PXD031004 and 10.6019/PXD031004.
Statistical analysis of discovery proteomics
The normalized abundances on all individual peptides were compared between POMS and mADS by performing a Wilcoxon rank-sum test. Proteins were deemed to be significantly differentially abundant between the groups if they were identified by two or more peptides and passed a set of three separate stringent criteria: (1) At least 25% of the peptides of the protein had a very low p value (p < 0.01); (2) at least 50% of the peptides of the protein had a low p value (p < 0.05); and (3) at least 75% of the peptides of the protein were altered in the same direction between groups (i.e. increased or decreased abundance in POMS), with a slight modification to criteria published previously.14 False discovery rate (FDR) of the detection of significant proteins was determined by performing the statistical analysis 50 times on permutated datasets, whereby for each permutation, the samples were assigned randomly to a category.
Validation analysis
Protein selection
After discovery analysis, proteins identified more abundant in POMS with eight or more peptides belonging to one protein were considered for validation. Five proteins were selected for validation based on (1) their potential function (CNS or immune related) and (2) their established significant fold change in the discovery analysis. Proteins with both low- and high-fold changes were selected to cover the complete fold-change range. Selected proteins were validated in the validation cohort (Figure 1), using parallel reaction monitoring-mass spectrometry (PRM-MS).
Parallel reaction monitoring-mass spectrometry (PRM-MS)
Validation analysis was conducted as previously described,12 with small alterations. Details of sample preparations and data acquisition are reported in Supplemental Material S1 and information on the set of target peptides in Supplemental Table S2. PRM-MS data have been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with dataset identifier PXD031022 and 10.6019/PXD031022. The PRM-MS signals were integrated using Skyline software.15
Statistical analysis of patient characteristic and validation proteomics
SPSS software version 25.0 was used, with a significance level of p < 0.05. Demographic data were compared between POMS and mADS groups. Chi-square (or Fisher’s exact) and Student’s t-test (or Wilcoxon rank sum test) were used when appropriate for categorical and continuous data, respectively. Because protein concentrations expressed by PRM-MS ratios were neither normally distributed, nor after log-transformation, non-parametric tests were used. Comparison of PRM-MS ratios between groups was performed with Wilcoxon rank sum test. Correlations were studied using Spearman’s rank correlation coefficient. Effects of covariates on PRM-MS ratios were assessed by linear regression.
Results
Patient characteristics
The discovery cohort consisted of 67 Dutch patients (POMS n = 28, mADS n = 39; Figure 1). In 59 patients, remaining CSF samples were available for further analyses. Subsequently, these 59 dependent samples were used in the validation cohort to enable technical validation. In addition, 95 independent newly acquired CSF samples (33 Dutch and 62 Canadian) were selected for the validation cohort, resulting in a total validation cohort of 154 patients (POMS n = 48, mADS n = 106; Figure 1).
Table 1 shows patient characteristics of the discovery and validation cohort, the latter divided into dependent and independent samples. In the dependent and independent validation samples, the same differences were observed; patients in the POMS group compared with the mADS group were older (14.6 vs. 6.4 and 14.5 vs. 8.4 years for the dependent and independent validation samples, respectively, both p < 0.001), more often had unique oligoclonal bands (OCBs) in CSF (100% vs. 13% and 96% vs. 12%, respectively, both p < 0.001), and had longer time between onset of disease and lumbar puncture (22.0 vs. 12.0 and 28.0 vs. 4.5 days, respectively, both p < 0.001). The mADS group compared with POMS group more often included patients with acute disseminated encephalomyelitis (ADEM; 61% vs. 0% and 40% vs. 0%, respectively, both p < 0.001) and more often included myelin oligodendrocyte glycoprotein antibody (MOG-ab) seropositive patients (27% vs. 0% (p = 0.018) and 33% vs. 0% (p = 0.001), respectively), tested by cell-based assay. Seven Dutch patients included in the POMS group had an unknown or inconclusive MOG-ab status; however, none of these patients was qualified as typical MOG-ab associated disorder; six patients had a typical clinical and radiological MS disease course, and one patient was diagnosed with tumefactive MS. This latter patient had negative aquaporin-4 antibody testing and an inconclusive MOG-ab test (both by cell-based assay). CSF examination in this patient showed a high IgG index and presence of OCB. Besides OCBs, other CSF parameters available and follow-up duration were not different between groups. Finally, comparing POMS and mADS separated for Dutch and Canadian validation samples showed the same results (Supplemental Table S3).
Table 1.
Patient characteristics of the samples used in the discovery and validation cohort.
| Discovery cohort | Validation cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dependent samples | Independent samples | ||||||||
| POMS n = 28 |
mADS n = 39 |
p value* | POMS n = 23 |
mADS n = 36 |
p value* | POMS n = 25 |
mADS n = 70 |
p value* | |
| Sex, no of females (%) | 16 (57) | 24 (62) | NS | 13 (57) | 21 (58) | NS | 15 (60) | 28 (40) | NS |
| Age at onset, y, median (IQR) | 14.9 (13.5–16.0) | 5.8 (3.2–10.8) | <0.001 | 14.6 (13.3–15.9) | 6.4 (3.0–10.9) | <0.001 | 14.5 (13.6–15.3) | 8.4 (4.5–12.2) | <0.001 |
| ADEM, n (%) | 0 | 25 (64) | <0.001 | 0 | 22 (61) | <0.001 | 0 | 28 (40) | <0.001 |
| Blood analyses | |||||||||
| MOG-ab seropositive, n (%) | 0/22 | 9/35 (26) | 0.009 | 0/19 | 9/33 (27) | 0.018 | 0/24 | 23/69 (33) | 0.001 |
| AQP4-ab seropositive, n (%) | 0/11 | 0/18 | na | 0/8 | 0/17 | na | 0/23 | 0/66 | na |
| CSF analyses | |||||||||
| RBC, median (IQR) | 0.0 (0.0–207.0) | 0.0 (0.0–12.0) | NS | 0.5 (0.0–53.0) | 0.0 (0.0–9.0) | NS | 0.0 (0.0-–2.0) | 0.0 (0.0–8.0) | NS |
| WBC, median (IQR) | 8.0 (5.0–28.0) | 8.0 (3.0–41.3) | NS | 11.5 (5.0–28.3) | 7.0 (3.0–37.0) | NS | 8.0 (4.0–20.5) | 9.0 (2.1–32.0) | NS |
| Total protein, median (IQR) | 0.32 (0.25–0.37) | 0.29 (0.22–0.43) | NS | 0.33 (0.25–0.38) | 0.28 (0.21–0.42) | NS | 0.34 (0.27–0.41) | 0.27 (0.22–0.40) | NS |
| Unique OCBs, n (%) | 25/26 (96) | 4/32 (13) | <0.001 | 22/22 (100) | 4/30 (13) | <0.001 | 24/25 (96) | 6/62 (12) | <0.001 |
| Time between disease onset and LP, d, median (IQR) | 23.0 (9.25–68.5) | 11.0 (5.0–20.0) | 0.007 | 22.0 (9.0–71.0) | 12.0 (5.0–21.5) | 0.026 | 28.0 (5.0–43.5) | 4.5 (3.0–12.8) | <0.001 |
| DMT use at time of LP (%) | 0 | 0 | na | 0 | 0 | na | 0 | 0 | na |
| FU, m, median (IQR) | 67.5 (46.3–98.0) | 87.0 (40.0–107.0) | NS | 62.0 (45.0–98.0) | 75.5 (40.3–109.3) | NS | 36.0 (25.5–109.0) | 56.5 (17.3–96.0) | NS |
POMS vs. mADS using chi-square (Fisher’s exact) or Wilcoxon rank-sum test.
ADEM: acute disseminated encephalomyelitis; AQP4-ab: aquaporin-4 antibody; CSF: cerebrospinal fluid; DMT: disease modifying therapy; FU: follow-up; IQR: interquartile range; LP: lumbar puncture; mADS: monophasic acquired demyelinating syndrome; MOG-ab: myelin oligodendrocyte glycoprotein antibody; na: not applicable; OCBs: oligoclonal bands; POMS: pediatric-onset multiple sclerosis; RBC: red blood cell count; WBC: white blood cell count.
Discovery of proteins with LC-MS
Using LC-MS in CSF samples of the discovery cohort (n = 67), a total of 5580 peptides was identified, belonging to 576 proteins; 58 proteins were found with two or more peptides significantly different between POMS and mADS (FDR 4.8%; Table 2), of which 28 with increased abundance in POMS.
Table 2.
Discovery analysis—Identification of CSF proteins differentially abundant POMS (n = 28) and pediatric mADS (n = 39).
| Direction of difference in POMS | # | Accession | Gene | Protein description | Fold changea | # of peptidesb |
|---|---|---|---|---|---|---|
| Increased abundance | 1 | P01599 | IGKV1-17 | Immunoglobulin kappa variable 1-17 | 5.168 | 7 |
| 2 | P06331 | IGHV4-34 | Immunoglobulin heavy variable 4-34 | 4.768 | 3 | |
| 3 | P01611 | IGKV1D-12 | Immunoglobulin kappa variable 1D-12 | 4.362 | 4 | |
| 4 | P06312 | IGKV4-1 | Immunoglobulin kappa variable 4-1 | 4.267 | 10 | |
| 5 | P06310 | IGKV2-30 | Immunoglobulin kappa variable 2-30 | 4.14 | 4 | |
| 6 | P01615 | IGKV2D-28 | Immunoglobulin kappa variable 2D-28 | 3.342 | 6 | |
| 7 | P01593 | IGKV1D-33 | Immunoglobulin kappa variable 1D-33 | 3.284 | 14 | |
| 8 | P01824 | IGHV4-39 | Immunoglobulin heavy variable 4-39 | 2.656 | 2 | |
| 9 | P01594 | IGKV1-33 | Immunoglobulin kappa variable 1-33 | 2.368 | 4 | |
| 10 | P01834 | IGKC | Immunoglobulin kappa constant | 2.092 | 12 | |
| 11 | P01857 | IGHG1 | Immunoglobulin heavy constant gamma 1 | 2.06 | 25 | |
| 12 | P01859 | IGHG2 | Immunoglobulin heavy constant gamma 2 | 1.834 | 25 | |
| 13 | P01763 | IGHV3-48 | Immunoglobulin heavy variable 3-48 | 1.76 | 4 | |
| 14 | O75326 | SEMA7A | Semaphorin-7A | 1.736 | 8 | |
| 15 | P04430 | IGKV1-16 | Immunoglobulin kappa variable 1-16 | 1.637 | 4 | |
| 16 | Q96KN2 | CNDP1 | Beta-Ala-His dipeptidase | 1.553 | 25 | |
| 17 | P20933 | AGA | N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase | 1.516 | 3 | |
| 18 | P34096 | RNASE4 | Ribonuclease 4 | 1.509 | 2 | |
| 19 | P16870 | CPE | Carboxypeptidase E | 1.463 | 13 | |
| 20 | Q7Z7M0 | MEGF8 | Multiple epidermal growth factor-like domains protein 8 | 1.438 | 13 | |
| 21 | Q06828 | FMOD | Fibromodulin | 1.39 | 3 | |
| 22 | Q92876 | KLK6 | Kallikrein-6 | 1.386 | 11 | |
| 23 | P26992 | CNTFR | Ciliary neurotrophic factor receptor subunit alpha | 1.341 | 3 | |
| 24 | Q96 GW7 | BCAN | Brevican core protein | 1.329 | 14 | |
| 25 | Q7Z3B1 | NEGR1 | Neuronal growth regulator 1 | 1.318 | 8 | |
| 26 | Q02818 | NUCB1 | Nucleobindin-1 | 1.282 | 9 | |
| 27 | Q9UBP4 | DKK3 | Dickkopf-related protein 3 | 1.232 | 18 | |
| 28 | P80748 | IGLV3-21 | Immunoglobulin lambda variable 3-21 | 1.17 | 4 | |
| Decreased abundance | 29 | P18669 | PGAM1 | Phosphoglycerate mutase 1 | 0.847 | 5 |
| 30 | P01023 | A2M | Alpha-2-macroglobulin | 0.823 | 109 | |
| 31 | O43866 | CD5 L | CD5 antigen-like | 0.798 | 8 | |
| 32 | P01024 | C3 | Complement C3 | 0.748 | 147 | |
| 33 | P04217 | A1BG | Alpha-1B-glycoprotein | 0.712 | 22 | |
| 34 | Q96IY4 | CPB2 | Carboxypeptidase B2 | 0.684 | 3 | |
| 35 | P00450 | CP | Ceruloplasmin | 0.682 | 65 | |
| 36 | P10643 | C7 | Complement component C7 | 0.644 | 27 | |
| 37 | Q06033 | ITIH3 | Inter-alpha-trypsin inhibitor heavy chain H3 | 0.617 | 9 | |
| 38 | P23083 | IGHV1-2 | Immunoglobulin heavy variable 1-2 | 0.598 | 2 | |
| 39 | P02788 | LTF | Lactotransferrin | 0.596 | 4 | |
| 40 | P62937 | PPIA | Peptidyl-prolyl cis-trans isomerase A | 0.568 | 8 | |
| 41 | Q86VB7 | CD163 | Scavenger receptor cysteine-rich type 1 protein M130 | 0.559 | 16 | |
| 42 | Q9Y6R7 | FCGBP | IgGFc-binding protein | 0.533 | 55 | |
| 43 | P80723 | BASP1 | Brain acid soluble protein 1 | 0.522 | 10 | |
| 44 | P02763 | ORM1 | Alpha-1-acid glycoprotein 1 | 0.517 | 15 | |
| 45 | P00739 | HPR | Haptoglobin-related protein | 0.482 | 20 | |
| 46 | P59665 | DEFA1 | Neutrophil defensin 1 | 0.445 | 2 | |
| 47 | O75015 | FCGR3B | Low-affinity immunoglobulin gamma Fc region receptor III-B | 0.425 | 2 | |
| Decreased abundance | 48 | P00738 | HP | Haptoglobin | 0.42 | 34 |
| 49 | P62328 | TMSB4X | Thymosin beta-4 | 0.395 | 6 | |
| 50 | P29966 | MARCKS | Myristoylated alanine-rich C-kinase substrate | 0.387 | 2 | |
| 51 | P31946 | YWHAB | 14-3-3 protein beta/alpha | 0.371 | 2 | |
| 52 | P61626 | LYZ | Lysozyme C | 0.346 | 4 | |
| 53 | Q08ET2 | SIGLEC14 | Sialic acid-binding Ig-like lectin 14 | 0.346 | 4 | |
| 54 | P10153 | RNASE2 | Non-secretory ribonuclease | 0.328 | 2 | |
| 55 | P08637 | FCGR3A | Low-affinity immunoglobulin gamma Fc region receptor III-A | 0.305 | 2 | |
| 56 | Q8TEU8 | WFIKKN2 | WAP, Kazal, immunoglobulin, Kunitz, and NTR domain-containing protein 2 | 0.289 | 8 | |
| 57 | Q9Y279 | VSIG4 | V-set and immunoglobulin domain-containing protein 4 | 0.195 | 4 | |
| 58 | P48539 | PCP4 | Calmodulin regulator protein PCP4 | 0.121 | 2 |
Proteins listed in table were identified differentially abundant between POMS and mADS with at least two unique peptides significantly different in discovery analysis. In bold, the proteins with increased abundance in POMS with at least eight unique peptides were significantly different.
Fold change > 1 = increased abundance in POMS. Fold change < 1 = decreased abundance in POMS.
Number of differentially identified peptides for the same protein.
Selection of proteins for validation
Proteins with increased abundance in POMS with eight or more peptides significantly different were considered for validation (Table 2; in bold). Based on function and significant fold change, the five proteins selected for validation included SEMA7A, CPE, Multiple epidermal growth factor-like domains protein 8 (MEGF8), Neuronal growth regulator 1 (NEGR1), and Nucleobindin-1 (NUCB1) (Table 3). Immunoglobulins (Igs) were not selected for validation, as their role in POMS diagnosis is already established.16,17
Table 3.
Proteins selected for validation.
| Direction of difference in POMS | # | Accession | Gene | Protein description | Fold changea | No. of peptidesb |
|---|---|---|---|---|---|---|
| Increased abundance | 1 | O75326 | SEMA7A | Semaphorin-7A | 1.736 | 8 |
| 2 | P16870 | CPE | Carboxypeptidase E | 1.463 | 13 | |
| 3 | Q7Z7M0 | MEGF8 | Multiple epidermal growth factor-like domains protein 8 | 1.438 | 13 | |
| 4 | Q7Z3B1 | NEGR1 | Neuronal growth regulator 1 | 1.318 | 8 | |
| 5 | Q02818 | NUCB1 | Nucleobindin-1 | 1282 | 9 |
Proteins listed in table were proteins with increased abundance in POMS with eight or more peptides significantly different belonging to one protein in discovery analyses, and finally selected for validation based on their function (CNS or immune related) and their fold change (both low and high ranges).
Fold change > 1 = increased abundance in POMS.
Number of differentially identified peptides for the same protein.
Validation of proteins with PRM-MS
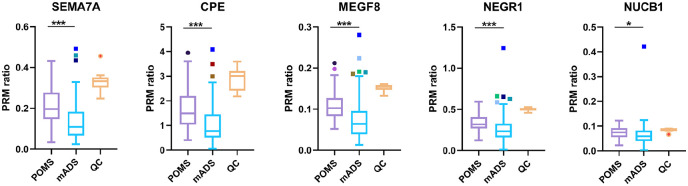
Using PRM-MS, in the total validation cohort (n = 154), all five selected proteins were confirmed more abundant in POMS (p < 0.001 for SEMA7A, CPE, MEGF8, and NEGR1; p = 0.020 for NUCB1; Figure 2). In the dependent validation samples (technical validation), all proteins were confirmed more abundant in POMS. In the independent validation samples, four of five proteins were confirmed more abundant in POMS (NUCB1 not). Analyzing the Dutch (n = 92) and Canadian (n = 62) samples of the total validation cohort separately, again four of five proteins were confirmed more abundant in POMS in both groups (NUCB1 not).
Figure 2.
PRM-MS ratios of selected proteins for validation with PRM-MS measurements in POMS vs. mADS.
CSF samples of adult controls (n = 16) were pooled and included for technical QC.
Using Wilcoxon rank sum test; *p < 0.05, **p < 0.01, and ***p < 0.001.
CPE: Carboxypeptidase E; mADS: monophasic acquired demyelinating syndrome; MEGF8: Multiple epidermal growth factor-like domains protein 8; NEGR1: Neuronal growth regulator 1; NUCB1: Nucleobindin-1; POMS: pediatric-onset multiple sclerosis; SEMA7A: Semaphorin-7A; QC: quality control.
Further sub-analyses of the five selected proteins were performed within the total validation cohort in order to utilize all acquired quantitative PRM-MS data.
Effect of clinical phenotype and age
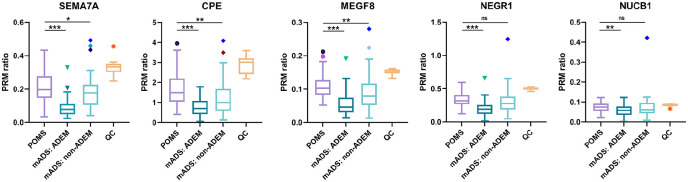
Some overlap was observed in PRM-MS ratios of POMS and mADS groups (Figure 2), but as described above, comparing the groups still resulted in significant differences. Further sub-analyses in the total validation cohort showed a clear distinction within the mADS group based on clinical phenotype, with lower PRM-MS ratios in mADS patients with an ADEM phenotype and higher PRM-MS ratios in mADS patients with a non-ADEM phenotype (Figure 3). However, compared with mADS non-ADEM group, SEMA7A (p = 0.047), CPE (p = 0.004), and MEGF8 (p = 0.006) remained significantly more abundant in POMS.
Figure 3.
PRM-MS ratios of selected proteins for validation with PRM-MS measurements separated for clinical phenotype within mADS group in ADEM and non-ADEM patients.
CSF samples of adult controls (n = 16) were pooled and included for technical QC.
Using Wilcoxon rank sum test: *p < 0.05, **p < 0.01, and ***p < 0.001.
ADEM: acute disseminated encephalomyelitis; CPE: Carboxypeptidase E; mADS: monophasic acquired demyelinating syndrome; MEGF8: Multiple epidermal growth factor-like domains protein 8; NEGR1: Neuronal growth regulator 1; NUCB1: Nucleobindin-1; POMS: pediatric-onset multiple sclerosis; SEMA7A: Semaphorin-7A; QC: quality control.
Because mADS non-ADEM patients were older than mADS ADEM patients (11.1 vs. 4.6 years, p < 0.001), we hypothesized that age could contribute to observed differences in PRM-MS ratios. A significant positive correlation was found between age and PRM-MS ratio in the complete validation cohort, but also within mADS group (Spearman’s ρ ranging between 0.203 and 0.541 for all selected proteins) and mADS non-ADEM group (Spearman’s ρ ranging between 0.284 and 0.404 for all selected proteins except for NUCB1; Supplemental Table S4). Subsequent linear regression analysis showed that all selected proteins were significantly dependent on increasing age when adjusted for diagnosis (POMS vs. mADS). Conversely, proteins SEMA7A and CPE remained significantly more abundant in POMS compared with mADS when adjusted for age (SEMA7A: Canadian samples p = 0.041; CPE: complete validation cohort p = 0.036 and Canadian samples p = 0.018).
Discussion
This exploratory collaborative study discovered and validated potentially interesting CSF proteins that may be involved in POMS and not mADS. Discovery analysis (LC-MS) with use of stringent statistical criteria resulted in a total of 58 identified proteins with at least two unique peptides differentially abundant between POMS and mADS, of which 28 with increased abundance in POMS. Applying even more stringent criteria, selecting only the proteins with at least eight unique peptides different identified 14 proteins with increased abundance in POMS (Table 2; in bold). A selection of five of these proteins, covering the entire fold-change range of discovery analysis, was made for subsequent validation with a high-resolution targeted quantitative proteomic approach (PRM-MS).18 All these five proteins were confirmed in the dependent validation samples and overall validation cohort, including the proteins with lowest fold-change ranges in the discovery analysis (e.g. NUCB1 and NEGR1), indicating that our discovery analysis was already accurate, possibly due to the use of very stringent criteria. Therefore, the complete list of those 14 proteins may include potentially promising proteins involved in POMS.
Among these 14 promising proteins are Igs, which are a hallmark of MS disease and have been implemented in most recent MS diagnostic criteria in the form of oligoclonal Ig gamma (IgG) bands.16 Also, the kappa subtype of Ig has been studied as a potential biomarker for MS diagnosis and alternative for OCB.19 Two of five implicated Igs have been previously associated with POMS, namely Ig heavy constant gamma 15 and Ig heavy constant gamma 2.6 These Igs underline the key role of the humoral immune system in POMS. The last implicated protein with an immunological function is SEMA7A, known to be involved in T-cell-mediated inflammation, and associated with more inflammatory lesions and demyelination in MS and experimental autoimmune encephalomyelitis.20
Our two previously conducted exploratory studies in POMS reported an increased abundance of predominantly gray matter5 and axoglial6 proteins in CSF. Similar to findings in these two studies,5,6 the remainder of the 14 promising proteins we implicate here in POMS mainly has neuron-related functions, including Brevican core protein (BCAN), NEGR1, SEMA7A, CPE, and NUCB1. BCAN inhibits neurite outgrowth,21 while NEGR1 promotes neuronal growth.22,23 SEMA7A, in addition to its above-described immune-related function, is also expressed in the CNS by different (injured) neuronal and glial cells.24 CPE is a protease catalyzing a wide range of (neuro)peptides which are involved in neuronal structure and survival,25 and finally, NUCB1 is exclusively found in neurons.26 Both CPE and NUCB1 are reported to be upregulated after (oxidative) stress.25,27
We speculate that the multiple neuron-related proteins we implicate here in POMS versus mADS may reflect a combination of neurodegenerative and potentially compensatory neuroprotective mechanisms manifesting in MS but not in monophasic CNS inflammatory conditions. Neurodegeneration is considered also in other studies to be involved in POMS.28,29 Due to the ongoing CNS damage, compensatory neuroprotective pathways may have been initiated. In contrast, in mADS patients (in whom clinical and biological onset of the single event likely coincide), neurodegeneration may not be an ongoing process (or may not yet have been triggered), and hence, compensatory neuroprotective pathways are not initiated.
Three of these five neuron-related proteins have been reported previously with increased abundance in MS, of which CPE5,6 and NEGR5 only in POMS, and BCAN both in POMS5 and adult-onset multiple sclerosis (AOMS).30 Strikingly, SEMA7A was observed in lower concentrations in adults with clinically isolated syndromes (CIS) subsequently ascertained as having clinically definite MS versus CIS patients who remained monophasic during multiple years of follow-up,31 while in our study, we found an increased abundance in POMS versus mADS. Although our study does not include a direct AOMS CSF proteomic comparison, the fact that currently implicated proteins with potentially neuroprotective functions are reported mainly in POMS and less in AOMS, if confirmed, could indicate that repair and/or compensatory mechanisms might be superior in POMS compared with AOMS. This would be in line with other evidence that POMS patients exhibit less physical disability in the first-decade post-onset and have longer time to disease progression in spite of having increased inflammatory activity.32 Finally, protein NUCB1 has never been described in MS, but interestingly, it has been associated with primary neurodegenerative disorders.33 However, as this protein was not confirmed significantly (only as a trend) in our independent validation samples, further studies are needed to determine whether there is a role of this protein in POMS.
Remaining proteins of the complete list of 14 include Kallikrein-6 (KLK6), Dickkopf-related protein 3 (DKK3), MEGF8, and Beta-Ala-His dipeptidase. Oligodendrocyte-derived KLK-6 has been studied quite extensively in the context of MS (models) and is observed to be elevated in CSF of AOMS34 and POMS.5 Finally, DKK35 and MEGF86 were previously also found more abundant in POMS.
In the quantification analysis, it was possible to study the contribution of factors other than diagnosis on the concentrations of specific proteins. The clinical phenotype was found to be related to protein concentrations. However, age at time of CSF sampling was revealed to be the main contributing factor, as only SEMA7A and CPE remained significantly increased in POMS when corrected for age, substantiating the need for correction for age.
This study has several advantages and limitations. First, CSF remains the closest compartment to the CNS tissue which is still reasonably accessible. As the CSF composition may reflect the processes occurring in the diseased MS CNS,4 the analysis of the CSF proteome may provide unique insights into disease biology. Although it should be recognized that one is only indirectly (and likely only partially) sampling such processes. Second, although our sample size remains relatively limited, the current combination of samples from previously conducted studies5,6 with the addition of new samples increased sample size substantially and enabled both validation of multiple proteins (the overlap has been described above in detail) and extension of previous results in a combined cohort of pediatric ADS patients from separate countries. Third, the use of mADS as representative of other CNS inflammatory disorders serves as an excellent control group to identify CSF proteins specific for MS.10 Although our study has multiple years of follow-up (Table 1), future relapses in mADS patients who are MOG-ab seropositive cannot be ruled out, as a small proportion of these patients might experience new disease activity years after disease onset.35 Studying samples from age-matched healthy children would also be of interest, particularly to determine the extent to which observed differences between POMS and mADS CSF profiles may be driven by age-related changes in pediatric CSF composition. However, obtaining pediatric healthy control CSF samples is extremely difficult for ethical reasons, and CSF collected from healthy adults is not adequate for this purpose. Fourth, we focused on proteins with increased abundance in POMS, while proteins with decreased abundance in POMS could also be interesting in terms of disease biology. We decided to select five proteins more abundant in POMS for validation, which covered the complete fold-change range, in order to explore the accuracy of the LC-MS measurements, and we conclude that the remaining of the 14 proteins found in the discovery analysis are worth validating in an independent cohort as well.
In conclusion, this collaborative study found several CSF proteins with expected immunological but also neuronal functions to be involved in POMS, representing the earliest stage of MS that can be evaluated. These proteins provide potential insights into the underlying biology of POMS regarding neurodegeneration and possible compensatory neuroprotection. Besides our current confirmation of increased SEMA7A and CPE in POMS CSF, validation of remaining potentially interesting proteins is needed to determine their possible role after adjusting for age, as we note that differences in CSF proteins between pediatric patient cohorts are strongly age-associated. Further international collaborative efforts will be critical to enable both protein validation and additional discovery in this rare but relevant group of pediatric patients. Finally, a direct comparison between the CSF proteome of AOMS and POMS is of interest to confirm potential differences in pathobiology of early versus later-onset MS.
Supplemental Material
Supplemental material, sj-docx-1-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-2-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-3-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-4-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-5-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Acknowledgments
The authors thank the patients and their families for their participation in this study as well as members of the Dutch Pediatric MS and ADEM Study Group and the Canadian Pediatric Demyelinating Disease Network for their contributions. Furthermore, the authors would like to dedicate this manuscript to the memory of Prof. Rogier Q. Hintzen, who was actively involved in the original concept of the study and in the first analyses, but unexpectedly passed away on 15 May 2019.
Footnotes
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: A.L.B., C.S., C.G., M.P.S., Y.Y.M.W., E.D.vP. B.L.B., T.M.L., and R.F.N. declared no conflicts of interest. A.B-O. has participated as a speaker in meetings sponsored by and received consulting fees and/or grant support from Accure, Atara Biotherapeutics, Biogen, BMS/Celgene/Receptos, GlaxoSmithKline, Gossamer, Janssen/Actelion, Medimmune, Merck/EMD Serono, Novartis, Roche/Genentech, and Sanofi-Genzyme.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported in part by the Dutch MS Research Foundation and the Canadian Multiple Sclerosis Scientific Research Foundation. This study was not industry sponsored.
Supplemental material: Supplemental material for this article is available online.
ORCID iDs: Arlette L Bruijstens  https://orcid.org/0000-0002-7990-5894
https://orcid.org/0000-0002-7990-5894
Rinze F Neuteboom  https://orcid.org/0000-0001-6136-4981
https://orcid.org/0000-0001-6136-4981
Contributor Information
Arlette L Bruijstens, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Christoph Stingl, Laboratory of Neuro-Oncology, Clinical and Cancer Proteomics, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Coşkun Güzel, Laboratory of Neuro-Oncology, Clinical and Cancer Proteomics, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Marcel P Stoop, Laboratory of Neuro-Oncology, Clinical and Cancer Proteomics, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Yu Yi M Wong, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
E Daniëlle van Pelt, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Brenda L Banwell, Division of Child Neurology, Department of Neurology, The Children’s Hospital of Philadelphia, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Amit Bar-Or, Center for Neuroinflammation and Experimental Therapeutics and Division of Multiple Sclerosis and Related Disorders, Department of Neurology, Perelman Center for Advanced Medicine (PCAM), University of Pennsylvania.
Theo M Luider, Laboratory of Neuro-Oncology, Clinical and Cancer Proteomics, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
Rinze F Neuteboom, Department of Neurology, Erasmus University Medical Center, Rotterdam, The Netherlands.
References
- 1. Miller DH, Chard DT, Ciccarelli O. Clinically isolated syndromes. Lancet Neurol 2012; 11(2): 157–169. [DOI] [PubMed] [Google Scholar]
- 2. Bjornevik K, Munger KL, Cortese M, et al. Serum neurofilament light chain levels in patients with presymptomatic multiple sclerosis. JAMA Neurol 2020; 77(1): 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chitnis T, Glanz B, Jaffin S, et al. Demographics of pediatric-onset multiple sclerosis in an MS center population from the Northeastern United States. Mult Scler 2009; 15(5): 627–631. [DOI] [PubMed] [Google Scholar]
- 4. Thygesen C, Larsen MR, Finsen B. Proteomic signatures of neuroinflammation in Alzheimer’s disease, multiple sclerosis and ischemic stroke. Expert Rev Proteomics 2019; 16(7): 601–611. [DOI] [PubMed] [Google Scholar]
- 5. Singh V, van Pelt ED, Stoop MP, et al. Gray matter-related proteins are associated with childhood-onset multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2015; 2(5): e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dhaunchak AS, Becker C, Schulman H, et al. Implication of perturbed axoglial apparatus in early pediatric multiple sclerosis. Ann Neurol 2012; 71(5): 601–613. [DOI] [PubMed] [Google Scholar]
- 7. Ketelslegers IA, Catsman-Berrevoets CE, Neuteboom RF, et al. Incidence of acquired demyelinating syndromes of the CNS in Dutch children: A nationwide study. J Neurol 2012; 259(9): 1929–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banwell B, Bar-Or A, Arnold DL, et al. Clinical, environmental, and genetic determinants of multiple sclerosis in children with acute demyelination: A prospective national cohort study. Lancet Neurol 2011; 10(5): 436–445. [DOI] [PubMed] [Google Scholar]
- 9. Krupp LB, Tardieu M, Amato MP, et al. International Pediatric Multiple Sclerosis Study Group criteria for pediatric multiple sclerosis and immune-mediated central nervous system demyelinating disorders: Revisions to the 2007 definitions. Mult Scler 2013; 19(10): 1261–1267. [DOI] [PubMed] [Google Scholar]
- 10. Teunissen C, Menge T, Altintas A, et al. Consensus definitions and application guidelines for control groups in cerebrospinal fluid biomarker studies in multiple sclerosis. Mult Scler 2013; 19(13): 1802–1809. [DOI] [PubMed] [Google Scholar]
- 11. Stoop MP, Singh V, Stingl C, et al. Effects of natalizumab treatment on the cerebrospinal fluid proteome of multiple sclerosis patients. J Proteome Res 2013; 12(3): 1101–1107. [DOI] [PubMed] [Google Scholar]
- 12. Van der Ende EL, Meeter LH, Stingl C, et al. Novel CSF biomarkers in genetic frontotemporal dementia identified by proteomics. Ann Clin Transl Neurol 2019; 6(4): 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perez-Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res 2019; 47(D1): D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van den Berg CB, Duvekot JJ, Guzel C, et al. Elevated levels of protein AMBP in cerebrospinal fluid of women with preeclampsia compared to normotensive pregnant women. Proteomics Clin Appl 2017; 11: 1600082. [DOI] [PubMed] [Google Scholar]
- 15. Pino LK, Searle BC, Bollinger JG, et al. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom Rev 2020; 39(3): 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 2018; 17(2): 162–173. [DOI] [PubMed] [Google Scholar]
- 17. Wong YYM, de Mol CL, van der Vuurst de Vries RM, et al. Real-world validation of the 2017 McDonald criteria for pediatric MS. Neurol Neuroimmunol Neuroinflamm 2019; 6(2): e528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peterson AC, Russell JD, Bailey DJ, et al. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics 2012; 11(11): 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Presslauer S, Milosavljevic D, Huebl W, et al. Validation of kappa free light chains as a diagnostic biomarker in multiple sclerosis and clinically isolated syndrome: A multicenter study. Mult Scler 2016; 22(4): 502–510. [DOI] [PubMed] [Google Scholar]
- 20. Okuno T, Nakatsuji Y, Kumanogoh A. The role of immune semaphorins in multiple sclerosis. FEBS Lett 2011; 585(23): 3829–3835. [DOI] [PubMed] [Google Scholar]
- 21. Yamada H, Fredette B, Shitara K, et al. The brain chondroitin sulfate proteoglycan brevican associates with astrocytes ensheathing cerebellar glomeruli and inhibits neurite outgrowth from granule neurons. J Neurosci 1997; 17(20): 7784–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schäfer M, Bräuer AU, Savaskan NE, et al. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol Cell Neurosci 2005; 29(4): 580–590. [DOI] [PubMed] [Google Scholar]
- 23. Singh K, Loreth D, Pottker B, et al. Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated Negr1 gene. Front Mol Neurosci 2018; 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eixarch H, Gutiérrez-Franco A, Montalban X, et al. Semaphorins 3A and 7A: Potential immune and neuroregenerative targets in multiple sclerosis. Trends Mol Med 2013; 19(3): 157–164. [DOI] [PubMed] [Google Scholar]
- 25. Cheng Y, Cawley NX, Loh YP. Carboxypeptidase E (NF-α1): A new trophic factor in neuroprotection. Neurosci Bull 2014; 30(4): 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tulke S, Williams P, Hellysaz A, et al. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience 2016; 314: 179–188. [DOI] [PubMed] [Google Scholar]
- 27. Ning M, Sarracino DA, Kho AT, et al. Proteomic temporal profile of human brain endothelium after oxidative stress. Stroke 2011; 42(1): 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yeh EA, Weinstock-Guttman B, Ramanathan M, et al. Magnetic resonance imaging characteristics of children and adults with paediatric-onset multiple sclerosis. Brain 2009; 132(Pt. 12): 3392–3400. [DOI] [PubMed] [Google Scholar]
- 29. Bartels F, Nobis K, Cooper G, et al. Childhood multiple sclerosis is associated with reduced brain volumes at first clinical presentation and brain growth failure. Mult Scler 2019; 25(7): 927–936. [DOI] [PubMed] [Google Scholar]
- 30. Schutzer SE, Angel TE, Liu T, et al. Gray matter is targeted in first-attack multiple sclerosis. PLoS ONE 2013; 8(9): 0066117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cantó E, Tintoré M, Villar LM, et al. Validation of semaphorin 7A and ala-beta-his-dipeptidase as biomarkers associated with the conversion from clinically isolated syndrome to multiple sclerosis. J Neuroinflammation 2014; 11: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chitnis T, Aaen G, Belman A, et al. Improved relapse recovery in paediatric compared to adult multiple sclerosis. Brain 2020; 143(9): 2733–2741. [DOI] [PubMed] [Google Scholar]
- 33. Bonito-Oliva A, Barbash S, Sakmar TP, et al. Nucleobindin 1 binds to multiple types of pre-fibrillar amyloid and inhibits fibrillization. Sci Rep 2017; 7: 42880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hebb AL, Bhan V, Wishart AD, et al. Human kallikrein 6 cerebrospinal levels are elevated in multiple sclerosis. Curr Drug Discov Technol 2010; 7(2): 137–140. [DOI] [PubMed] [Google Scholar]
- 35. Hacohen Y, Wong YY, Lechner C, et al. Disease course and treatment responses in children with relapsing myelin oligodendrocyte glycoprotein antibody-associated disease. JAMA Neurol 2018; 75(4): 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-2-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-3-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-4-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal
Supplemental material, sj-docx-5-msj-10.1177_13524585221125369 for Neurodegeneration and humoral response proteins in cerebrospinal fluid associate with pediatric-onset multiple sclerosis and not monophasic demyelinating syndromes in childhood by Arlette L Bruijstens, Christoph Stingl, Coşkun Güzel, Marcel P Stoop, Yu Yi M Wong, E Daniëlle van Pelt, Brenda L Banwell, Amit Bar-Or, Theo M Luider and Rinze F Neuteboom in Multiple Sclerosis Journal