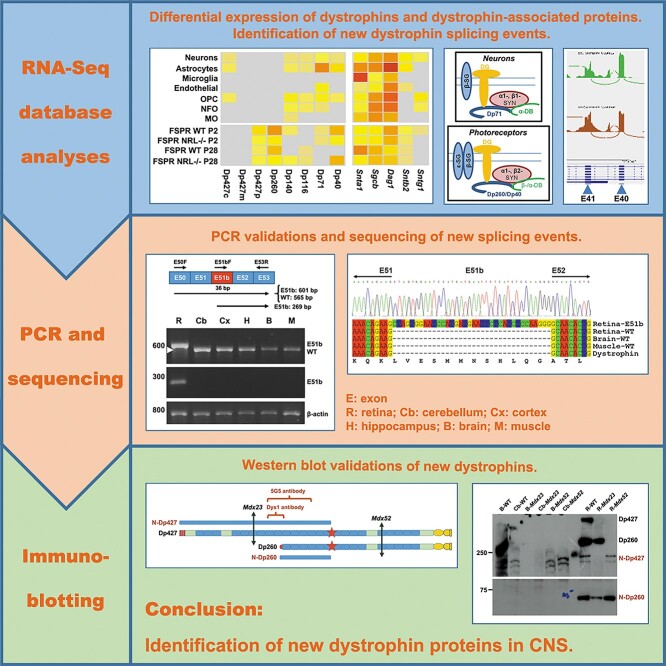
Abstract
The large DMD gene encodes a group of dystrophin proteins in brain and retina, produced from multiple promoters and alternative splicing events. Dystrophins are core components of different scaffolding complexes in distinct cell types. Their absence may thus alter several cellular pathways, which might explain the heterogeneous genotype–phenotype relationships underlying central comorbidities in Duchenne muscular dystrophy (DMD). However, the cell-specific expression of dystrophins and associated proteins (DAPs) is still largely unknown. The present study provides a first RNA-Seq-based reference showing tissue- and cell-specific differential expression of dystrophins, splice variants and DAPs in mouse brain and retina. We report that a cell type may express several dystrophin complexes, perhaps due to expression in separate cell subdomains and/or subpopulations, some of which with differential expression at different maturation stages. We also identified new splicing events in addition to the common exon-skipping events. These include a new exon within intron 51 (E51b) in frame with the flanking exons in retina, as well as inclusions of intronic sequences with stop codons leading to the presence of transcripts with elongated exons 40 and/or 41 (E40e, E41e) in both retina and brain. PCR validations revealed that the new exons may affect several dystrophins. Moreover, immunoblot experiments using a combination of specific antibodies and dystrophin-deficient mice unveiled that the transcripts with stop codons are translated into truncated proteins lacking their C-terminus, which we called N-Dp427 and N-Dp260. This study thus uncovers a range of new findings underlying the complex neurobiology of DMD.
Graphical Abstract
Graphical Abstract.
Introduction
The X-linked Duchenne muscular dystrophy (DMD) gene is among the largest genes reported to date from which a full transcript of about 14 kb can be produced from three independent promoters, driving the expression of full-length dystrophin proteins of 427 kDa (Dp427) in both skeletal muscle and central nervous system (CNS). The Dp427 mRNA is composed of 79 exons and the protein contains four domains: an N-terminal actin-binding domain, a central rod domain, a cysteine rich region and a C-terminal domain that can interact with proteins of the dystrophins-associated glycoprotein complex (DGC) such as the transmembraneous dystroglycans, and cytosolic dystrobrevins and syntrophins (1). Four other internal promoters produce N-terminal truncated dystrophins named Dp260, Dp140, Dp116 and Dp71 according to their molecular weight, which hold in common the C-terminal domains allowing interaction with DGC. Activation of these distinct promoters is regulated in a tissue- and cell-specific manner (see (2) for a review).
The distinct DMD/Dmd-gene products have been detected in the CNS, including brain and retina; however, their precise localization in various cells is still unclear. It is critical to better understand their function and how their loss or altered expression in Duchenne and Becker muscular dystrophies (DMD/BMD), respectively, leads to cognitive, behavioural and sensory deficits (3,4). In the CNS, the full-length Dp427 dystrophins are primarily expressed in cortical/cerebral structures (Dp427c) and in cerebellar Purkinje cells (Dp427p). Expression of the N-terminal truncated dystrophins is also finely regulated in the CNS. Dp260 is expressed in retina from a promoter located in intron 29 and its first unique exon is fused with exon 30. The transcriptional start site (TSS) of Dp140 is in intron 44, while its translation start site is found in exon 51. Dp140 is expressed in CNS and kidney but is absent in muscle. Dp116 is transcribed from a promoter in intron 55 in peripheral-nerve Schwann cells, yet it has also been observed in glioblastoma cells (5). The last promoter located in intron 62 encodes the shortest dystrophins, Dp71 and Dp40. Dp71 is the most widely expressed dystrophin-gene product in non-muscle tissues. Dp40 is expressed from the same promoter but ends at exon 70 due to an alternative polyadenylation signal that is present at intron 70 (6); it has been detected in central neuronal synapses (7) and has an important role in the neuronal differentiation of PC12 cells (8). Among all described dystrophin proteins, Dp40 is the only known isoform truncated at the C-terminal part.
Dystrophins thus constitute a family of proteins with multiple products of various molecular weights. Additionally, splice variants have also been characterized, leading to the existence of subfamilies of dystrophins including several isoforms at the carboxy terminus (9–12). Moreover, the diversity of dystrophin proteins and isoforms promote the formation of different DGCs in distinct CNS cell types (1,13). Similarly, distinct DGCs are also likely present in the retina in association with Dp71 in Müller glial cells, or with other dystrophins (Dp427, Dp260, Dp140) in rod/cone photoreceptor terminals, bipolar and amacrine cells, yet their composition and regulation remain to be determined (14).
In the present study, we performed an in-depth meta-analysis of existing transcriptomic data published by others (15,16) as well as recently generated data by our groups (17) to better detail the expression of dystrophin isoforms and DGC components in both brain and retinal tissues as well as in purified cells. As expected, we found differential expression of dystrophins and associated proteins in distinct brain and retinal tissues and cell types. Recent advances in RNA sequencing have unveiled a range of naturally skipped exons within the DMD gene, as well as pseudoexons or cryptic exons encoding stop codons within large intronic regions (18–20). We therefore sought for non-annotated splicing events to reveal new isoforms. As such, we unveiled the presence in the retina of an in-frame putative exonic sequence within intron 51, between exon 51 and 52, and premature stop codons followed by putative 3′ untranslated regions (UTRs) in intron 40 and 41 that result in truncated dystrophins. These newly identified isoforms were validated by a combination of PCR primers and anti-dystrophin antibodies, which revealed that several dystrophins are affected by these new splicing events, some of which being responsible for the presence of new dystrophin isoforms truncated at the C-terminus.
Results
Dmd-gene mRNAs in CNS tissues and cells
The cell-specific expression of all dystrophin-gene products from mouse brain was identified using a previously published brain RNA-Seq database (16), which includes the transcriptome and splicing database of neuron, astrocyte, microglia, endothelium, oligodendrocyte precursor cells (OPC), newly formed oligodendrocytes (NFO) and myelinating oligodendrocytes (MO) of the cerebral cortex. The DMD gene undergoes complex regulations leading to the specific expression of distinct dystrophins produced from independent internal promoters (Fig. 1A). A basic gene search for expression of Dmd-gene transcript in this database was insufficient to account for this complexity and only revealed that the Dmd gene is active in all cell types, with highest levels in astrocytes (Supplementary Material, Fig. S1). Therefore, we undertook a detailed analysis of this brain database, including manual screening from 5′ to 3′ end seeking for new features, and we also used our unpublished database generated from hippocampal and cerebellar tissue samples of wild-type (WT) mice. To compare with expression of dystrophins in retina, we further analysed other databases (15,17) to determine expression of dystrophins in the retina from P30 WT and rd10 mice, carrying a hypomorphic mutation in Pde6b, a gene involved in rod phototransduction. In this model with autosomal recessive retinitis pigmentosa, retinal degeneration starts around P15. Secondary cone cell death occurs after death of primary rod cells, which have practically all disappeared around P30. Therefore, the use of rd10 at P30 allows removing a vast majority of the genes expressed in rod photoreceptors, which outnumber other retinal cell types. To complete the retinal expression analysis, we added a database from flow-sorted photoreceptor (FSPR) cells at postnatal day 2 (P2) and 28 (P28) from WT mice and Nrl−/− mice. The retina of Nrl−/− mice only contains functional cone-like photoreceptors at the expense of rods, due to a mutation in the rod-specific neural retina leucine zipper transcription factor (21,22).
Figure 1.

Dystrophin-gene transcripts expression in retina and brain tissue and cells. (A) Schematic representation of the Dmd gene and its products. The top drawing shows the position (E: exon; I: intron) of the transcription start of each dystrophin (arrows) within the 79 exons/introns of the gene. The related dystrophin proteins are depicted below, showing the spectrin-like repeats (blue rectangles), hinge domains (green rectangles) and specific N-terminus due to their first unique exons (red symbols). For Dp140, which translation start site is located within exon 51, specific expression in the RNA-Seq database was determined by presence of its specific 5′ UTR. (B) Heatmap of the transcriptional expression levels of the Dmd gene, from top to bottom in purified brain cell types, brain tissues, retinal tissues and purified retinal photoreceptors, as indicated on the right. Each column of the heatmap represents one exon of the Dmd gene (exon numbers indicated at top of heatmap) or specific introns as indicated at bottom to point to promoters and new identified features (*). Relative expression in FPKM is represented by colors from grey to dark red (highest number of reads) as indicated on the color-key scale (bottom). Specific first exons are indicated by arrowheads, for Dp427 in brain tissues (C), muscle (M) and Purkinje cells (P), Dp260, Dp140, Dp116 and Dp71. The specific 3′ UTR of Dp40 is shown by an arrow. Newly identified splicing events, E40e, E41e and E51b, respectively detected in retina samples within introns 40, 41 and 51 are shown by asterisks. WT: wild-type mouse; Rd10: Retinal degeneration 10 mice; OPC: oligodendrocyte precursor cell, NFO: Newly-formed oligodendrocyte; MO: myelinated oligodendrocyte; FSPR: flow-sorted photoreceptors; NRL: mice null for neural retina leucine zipper transcription factor (NRL−/−) which retina contains no rod; P2: postnatal day 2; P28: postnatal day 28.
Using these different datasets, we generated a heatmap representing expression values in log2 (FPKM) for each annotated exons, as well as for the first specific exon of each dystrophin (Fig. 1B). Overall, the exons comprised from E63 to E79, corresponding to the C-terminal part of dystrophins, had the highest brain expression levels. This could be largely attributed to the well-known major expression of Dp71 in brain, as confirmed by the high expression level of the specific first exon of Dp71 in the astrocyte samples. In contrast, but as expected, the exons with highest expression in retina comprised E30 to E79, reflecting main expression of the retina-specific Dp260. As expected, expression levels of the full-length dystrophin mRNAs were lower than the major brain product, Dp71 or the retina-specific Dp260 in retina. The specific 5′ UTR of muscle Dp427m (M in Fig. 1B) was virtually absent or expressed below the detection threshold, in all brain and retina samples. In contrast, Dp427c mRNA (C in Fig. 1B) was expressed in all brain tissue samples (cortex, hippocampus, cerebellum), while Dp427p mRNA (P in Fig. 1B) was only detected above threshold in the cerebellum. In all, the general expression pattern of the distinct CNS dystrophins, determined by presence of detectable amounts of reads in their specific first exons or 3′ UTR (see also Fig. 2A), was in line with current knowledge (2) and with quantification of the 79 Dmd individual exons, thus supporting the reliability of our approach.
Figure 2.
Relative expression of mRNA from genes coding dystrophin and associated proteins. (A) Heatmaps of the mRNA expression levels for dystrophin (left panel) and associated proteins (right panel) in brain (cortex) and retinal purified cell types are indicated between the two heatmaps. Expression of dystrophins based on reads corresponding to specific first exons or 3′ UTR. WT: flow-sorted rod/cone photoreceptors; NRL: flow-sorted cells from cone-only retinas of NRL−/− mice. P2/P28: postnatal days 2 and 28. Gene/protein names are as follows: Utrn/UTR: utrophin, snt/SYN: syntrophin, Dtn/DB: dystrobrevin, Sgc/SG: sarcoglycan, Dag1/DG: dystroglycan. (B) Schematic representation of presumed dystrophin-associated complexes in brain cell types based on relative expression in heatmaps. Protein names below each diagram correspond to detected transcripts with lower relative expression level in heatmaps compared with isoforms of the same DAP family included in the diagram. (C) Schematic representation of presumed dystrophin-associated complexes in retinal cell types based on relative expression in heatmaps. Estimation of expression in rod photoreceptors is based on a larger expression in flow-sorted cone/rod cells (WT) compared with cone-only cells (NRL). Immature (P2); mature (P28). Protein names below each diagram correspond to detected transcripts with lower relative expression level in heatmaps compared with isoforms of the same DAP family included in the diagram.
Our in-depth analysis of the heatmap also unveiled several new findings. Importantly, we first found that dystrophins are virtually absent in microglial cells. Secondly, we found that Dp116, usually described as a specific dystrophin of the peripheral nerve, was transcribed in whole cortex samples from WT mice, but not in hippocampus and cerebellum. In purified cortical cell samples, Dp116 mRNA was detected in neuron, astrocyte, OPC and NFO. This suggests expression of Dp116 in specific subdomains or subpopulation of brain cells. Dp116 mRNA was also detected in mature photoreceptors in the retina, which further support the presence of this dystrophin in CNS. Thirdly, we found that several dystrophins can be expressed in the same tissues and/or cell types (see also Fig. 2A). In the brain, Dp427c mRNA was detected in cerebellar samples, indicating that Dp427p is not the unique full-length dystrophin transcript in cerebellum. Reads of sequences from Dp427c mRNA were not only found in neurons, which was expected, but also in astrocyte and OPC. Dp140 mRNA was detected in neuron, astrocyte and in all oligodendrocyte-related cells (OPC, NFO, MO). Dp71 mRNA showed expression in all cell types except microglia and MO; it was the only Dmd product found in endothelial cells. Dp71’s strongest expression was in astrocytes, and lower levels were detected in neuron, endothelial cell, OPC and NFO. In contrast, its truncated isoform transcript, Dp40, was detected in brain cortex tissue samples from WT mice, and at the cell level in neuron, as reported (7), as well as in astrocyte and OPC. In all, oligodendrocyte-related cells had different profiles of dystrophins expression, with highest levels and number of dystrophins in OPC compared with NFO and MO.
Several Dmd-gene products were also detected in retina. While full-length Dp427 has been previously detected in retina (14), here we surprisingly found that Dp427p was the only Dp427 transcript expressed in all cell and tissue retinal samples. The Dp427p has two developmentally regulated isoforms, Dp427p1 and Dp427p2, but only Dp427p1 translates into a protein (GenBank NM_004009 and NM_004010). We confirmed that only Dp427p1 was detected in all our samples. The transcript of Dp260, known to be selectively expressed in retina, was not found in the brain tissues and only detected in retina samples. Two isoforms of Dp260 have been reported, Dp260–1 and Dp260–2 (GenBank NM 004011 and NM 004012), but only Dp260–2 was detected and analysed. As expected, detection of E30 to E79 reflecting Dp260 was drastically reduced due to retinal degeneration in the rd10 mice, confirming Dp260 is a major dystrophin of the photoreceptors. Dp140 expression was also absent in rd10 mice, confirming it is also expressed in photoreceptors (14). Several dystrophin mRNAs were also detected in FSPR of the retina. Dp260 mRNA was highly expressed in FSPR; its expression was lower in cone-only FSPR of Nrl−/− mice, suggesting it is more expressed in rods than cones.
We also found that expression of retinal dystrophins is regulated during photoreceptor maturation. Thus, Dp116 mRNA was only detected in mature photoreceptors (P28) but not at P2. Dp140 mRNA was not detected in cone-only (Nrl−/−) retina at P2, suggesting it is likely expressed in rods at this age, but not in cones. However, the Dp140 transcript was detected in mature cones at P28. Dp71 mRNA was only expressed in immature photoreceptors (P2) but not in mature photoreceptors (P28), in line with its selective expression in Müller glial cells in the mature retina (23). The Dp40 isoform was found at P2 and was selectively detected in cone-like cells from Nrl−/− mice at P28.
Distribution of distinct dystrophin-associated complexes in CNS
A search in the databases for dystrophin-associated proteins (DAPs) revealed a complex cell-dependent distribution of a range of DAP isoforms (Fig. 2A). Dystroglycan (Dag1 gene), the key transmembrane component of DAP complexes, was found in all brain and retina cell types, as well as the cytosolic α1-syntrophin (α1-SYN, Snta1 gene), α-dystrobrevin (α-DB, Dtna gene) and β-dystrobrevin (β-DB, Dtnb gene), and the transmembrane β- and ε-sarcoglycans (SG) (Sgcb and Sgce genes, respectively). The β2-syntrophin (β2-SYN) isoform (Sntb2 gene) was found in all cells except MO. The mRNA of the dystrophin paralogue, utrophin (UTR, Utrn gene), was found in all cell types except in mature (P28) flow-sorted photoreceptors of WT mice, suggesting a main expression in mature cone-like cells in the retina. The other DAPs detected in subsets of cell types included β1-, γ1- and γ2-SYN (Sntb1, Sntg1 and Sntg2 genes), and α-, δ- and γ-SG (Sgca, Sgcd and Sgcg genes). A detailed review of DAPs expression allowed us to represent the putative DAP complexes expressed in each brain and retina cell type (Fig. 2B and C, respectively), based on the DAPs showing higher transcription levels compared with other isoforms of the same family. Figure 2C shows the main components of DAP complexes in neurons (Dp71, α1/β1-SYN, α-DB, β-SG), astrocytes (Dp71/Dp40, α1-SYN, α-DB, β-SG), endothelial cells (UTR, β2-SYN, β/ε-SG), microglia (UTR, α1-SYN, ε-SG), OPC (Dp140/Dp71, α1/γ1-SYN, α-DB, β-SG), NFO (Dp140/Dp116, α1-SYN, α-DB, β-SG), MO (Dp140, α1-SYN, α-DB, β-SG), immature cones (Dp260/Dp40, α1/β2-SYN, ε/β-SG), mature cones (Dp40, α1/γ2-SYN, β-DB, β-SG), immature rods (Dp140/Dp260/Dp40/Dp427p, UTR, γ1/β1/β2-SYN) and mature rods (Dp260, α1/γ2-SYN, β-DB). Many other Dmd-gene products and DAPs also detected at an apparent lower level in these cells are listed below each diagram. This highlights the diversity of the DAP complexes that could be detected among and within brain and retina cell types.
Alternative splicing affecting dystrophins in CNS
Alternative splicing of specific exons, exon junctions and inclusions of intronic sequences were analysed using Sashimi plots in the IGV tool (Supplementary Material, Fig. S2A). We first quantified the number of reads corresponding to junctions between E77 and E79, reflecting skipping of E78 (ΔE78), compared with reads reflecting mRNA containing E78 (+E78). This known alternative splicing of the penultimate exon of the DMD gene may likely affect several if not all dystrophin-gene products, and it is commonly used to define the two main families of Dp71 proteins, the Dp71d family that contains E78 and the Dp71f family that lacks E78 (24). The presence of E78 was observed in the majority of transcripts in mature/adult cells and tissues (Supplementary Material, Fig. S2B and C). However, E78 exclusion (ΔE78) was also detected in substantial quantities in both brain and retinal adult tissues (Supplementary Material, Fig. S2B), yet its calculated proportion (100 × reads with ΔE78/total number of reads) remained <30% as compared with mRNA containing E78 (+E78) in all tissues. Likewise, ΔE78 events were observed in about 18% of transcripts in purified cortical cells (169 reads in 12 samples) (Supplementary Material, Fig. S2C), in line with our previous study showing that E78 exclusion in Dp71, the most abundant dystrophin in the brain, is found in about 20% of Dp71 isoforms in the adult brain (24). However, the relative proportion of ΔE78 transcripts differed among cell types: There was a ratio of 20/80% for ΔE78/E78 in neurons and OPC, 15/85% in astrocyte and NFO, while it was of 55/45% in endothelial cells and absent in microglia where dystrophin expression was barely undetectable. In retinal photoreceptors (Supplementary Material, Fig. S2D), whether they were purified from WT or Nrl−/− mice, the majority of mRNA contained E78 at the mature stage (P28), while an opposite distribution was observed in immature photoreceptors at P2, with a major occurrence of ΔE78 events, indicating that loss of exon 78 may be regulated during postnatal development. In summary, all mature brain and retinal tissues and cells expressed both mRNAs containing and lacking E78, while only retinal immature (P2) FSPR expressed a majority of mRNA with splicing out of E78.
Other exon skipping events were also detected in brain and retinal tissue and cell samples. Events were detected as few reads (<5 reads per sample) leading to exclusion of exons 34, 38, 39, 38–39, 41, 72–74, 73–74 and 71–78. As expected, however, skipping of annotated exons were mostly located in the distal part of the genomic sequence between exons 68 and 79 (Supplementary Material, Fig. S3A). As expected from previous reports, ΔE71 was frequently detected in the brain, yet with some differences depending on the brain structure (60% in cerebellum, 48% in hippocampus, 21% in cortex). It was predominantly found in neurons (33%), then in OPC (20%) and NFO (21%). Its higher frequency in retina from rd10 mice (44%) compared with WT retina (16%) suggests that it rather occurs in non-photoreceptoral cells. Skipping of E71 to E74 (ΔE71–74) was mostly found in hippocampal tissue (19%), but it was not frequent in purified cortical cell types. In all, loss of E78 (ΔE78) was the main splicing-out event in the distal part of the Dmd gene in CNS purified cells, except for cortical neurons (only 48%), followed by ΔE71, while ΔE71–74 was mainly found in hippocampus and WT retina.
Identification of new splicing events in Dmd gene
Numerous peaks of reads within intronic regions (introns 40, 41 and 51) of dystrophin mRNA were detected in retinal samples and in FSPR cells (asterisks in Fig. 1B and table in Supplementary Material, Fig. S3B). The peak of reads within intron 51 was the most frequent posttranscriptional modification, relatively more expressed in mature (P28) compared with immature (P2) photoreceptors. Importantly, these reads did not correspond to a UTR and were connected to neighbouring exons according to Sashimi plots constructed with the IGV software (Supplementary Material, Fig. S4A). This suggested that it could be considered, at least in the retina, as a putative exonic sequence that we called E51b. Reads within intron 40 were also connected to neighbouring exons, while reads of intron 41 were connected to exon 41 but not exon 42 (Supplementary Material, Fig. S4B). These observations strongly suggested the expression of new Dmd isoforms, produced by the addition of a new unannotated exon E51b or by partial inclusion of intronic elements fused with known annotated exons, which we named E40e and E41e as they may lead to elongation of exons 40 and 41. Based on the position of these clusters of reads, we deduced that they potentially belong to several dystrophin mRNAs (Supplementary Material, Fig. S4C).
Validation of an alternative unannotated exon 51 (E51b)
Sequence analysis revealed that E51b corresponds to an in-frame sequence of 36 bp of intron 51 inserted between E51 and E52. The mouse intron 51 has 57 507 bp and E51b corresponded to 52 544–52 579 bp within intron 51 and ChrX: 84526910 to 84 526 945 at the genomic level (ChrX: 31733081-31 733 116 for human genomic position). To evaluate the presence and strength of alternative splice sites, we performed in silico analyses using splicing prediction tools. The NetGene2–2.42 server (https://services.healthtech.dtu.dk/service.php?NetGene2-2.42) (25) and the Splice Site Prediction by Neural Network–BDGP (https://www.fruitfly.org/seq_tools/splice.html) (26) predicted that E51b could have a cryptic consensus sequence adjacent to the canonical dinucleotide GT-AG splice sites (Supplementary Material, Fig. S5A and B). Prediction scores of these putative acceptor and donor sequences were close to those of the WT splice sites (Supplementary Material, Fig. S5C and D), indicating that they could function as donor and acceptor splice sites. Similar scores were obtained for the human and rat sequences (data not shown).
To validate its presence specifically in retinal samples at the mRNA level, we first designed two sets of primers (Supplementary Material, Table S1). One set was flanking this region (E50F and E53R) and the other set included one internal primer of E51b (E51bF) and E53R (Fig. 3A). PCR products from RT-PCR using primers E50F and E53R revealed a band corresponding to the mRNAs that included E51b, which was selectively detected in retina at the expected size (601 bp) (Fig. 3A, upper panel). A faint band corresponding to the WT sequence was also observed (white arrowhead), thus showing that this inclusion of E51b is more frequent in retina. In contrast, cerebellum, cortex, hippocampus, brain and skeletal muscle only expressed the WT mRNA (565 bp, arrowhead). To confirm that the E51b is only present in retina, we used E51bF and E53R primers. As expected, the band corresponding to cDNAs with the E51b sequence (269 bp) was only found in the retina sample (Fig. 3A, middle panel). Moreover, we found that the band corresponding to this mRNA was only present in WT mice, not in exon 52-deleted mdx52 mice, which further confirmed that it belongs to dystrophin transcripts (Fig. 3B).
Figure 3.
Validation of a small new exon within intron 51 (E51b). (A–D) Total RNA obtained from retina (R), cerebellum (Cb), cortex (Cx), hippocampus (H), whole brain (B) and skeletal muscle (M) was used to perform RT-PCR assays using specific primers to detect selective dystrophin exonic and intronic regions, as shown in the associated diagrams where the size of intron 51, E51b, position of primers and expected size of PCR products is indicated. Molecular weight (bp) is indicated on the left for each gel figure. (A) Upper panel: PCR products obtained with E50F and E53R primers, revealing selective amplification of mRNA containing E51b (601 bp) in retina, and presence of the WT band (565 bp) in all tissues. Note presence of a faint WT band in retina (arrowhead). Middle panel: PCR product detected using primers E51bF and E53R showing E51b (269 bp) mRNA amplification only in retina. Lower panel: Control amplification of β-actin mRNA. (B) PCR product obtained with E51bF and E53R primers, revealing selective amplification of mRNA containing E51b (269 bp) in retina of WT mice and its absence in the exon 52-deleted mdx52 mouse. (C) Analysis of transformant colonies with pGEM-T Easy-WT/E51b. Left panel: PCR products from retina (as shown in A, upper panel) were gel-purified, ligated into the pGEM-T Easy vector and used to transform Escherichia coli DH5α cells. Transformant colonies were screened by PCR with primers E50F and E53R. Lane 1 and 3: Colonies with E51b sequence. Lane 2: Colony with WT sequence. Right panel: DNA sequencing of the WT and E51b-containing plasmids, aligned with the sequenced WT PCR product from brain and muscle, and with the reference dystrophin sequence from Genebank (dystrophin). Sequencing was carried out with primers E50F and E53R. Putative amino acid sequence of E51b is shown at the bottom. (D) PCR products corresponding to Dp427, Dp260 and Dp140 mRNA containing E51b (E51b-Dp427/Dp260/Dp140) in mouse retina.
The PCR products (E50–E53) from retina shown in Fig. 3A were subcloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) followed by Escherichia coli DH5α cell transformation. PCR screening on the colonies was performed using E50F and E53R primers. Among seven colonies containing the insert, six were identified with E51b and only one contained the WT product (Fig. 3C). This agrees with the band intensity of the PCR results in Fig. 3A. Plasmid sequencing confirmed that both E51b-containing and WT cDNA sequences were expressed in the retina, while the sequencing of PCR products from brain and muscle tissues only corresponded to the WT sequence (Fig. 3C). DNA sequencing demonstrated that the presence of E51b preserves the open reading frame, and thus introduces 12 additional amino acids (LVESMMNSHLQG) between E51 and E52 (Fig. 3C).
Finally, using forward primers corresponding to dystrophins’ specific first exons to Dp260 and Dp140, or primer E29F to exon 29 for Dp427, associated with the reverse primer E51bR (Supplementary Material, Table S1), we detected intense bands for E51b-containing mRNAs with expected sizes corresponding to Dp427, Dp260 and Dp140 mRNAs (Fig. 3D). Additional faint bands of lower molecular weights might correspond to mRNA with additional splicing out of exons, as it was occasionally detected between E29 and E53 in the heatmap’s analysis.
Validation of intron 3’UTR insertions
Our RNA-Seq analyses revealed a junction of a part of I40 with E40 or I41 with E41, which may lead to an elongated exon 40 that we called E40e, and an elongated E41 that we called E41e (asterisks in Fig. 1B). We hypothesized that the contigs formed by the sequences in intron 40, E41 and sequences in intron 41 formed putative 3′UTRs of the Dmd transcripts. For validation by PCR from cDNA, we designed primers for exons 39 and 42 flanking these regions (E39F and E42R), as well as internal primers corresponding to insertions of introns 40 (E40eR) and 41 (E41eR) sequences (Supplementary Material, Table S1). The E39F and E42R primers amplified an mRNA at the expected size of a WT sequence (450 bp) (Fig. 4A, top gel panel). This WT sequence was detected in all tissue samples (retina, cerebellum, cortex, hippocampus, brain and skeletal muscle). The E39F-E42R primers also revealed an additional band of 267 bp (not shown), likely corresponding to the occasional skipping of exon 41 (27). The E39F and E40eR primers amplified two bands (Fig. 4A, middle gel panel). The first band had an expected size of 512 bp corresponding to the cDNA containing E40e and was detected in retina, cerebellum, cortex and whole brain, but not in hippocampus and skeletal muscle. This however suggested that E40e is expressed at low levels in some brain structure, in addition to its main expression in retina. The second band was detected at 214 bp. DNA sequencing of PCR products from retina and cerebellum, and in silico analyses (Fig. 4B), revealed that the upper band corresponded to the sequence containing E39 and E40 fused with I40 (E40e), while the lower band corresponded to a loss of the first 298 bp of the 5′ intronic region (ΔE40e). Both E40e and ΔE40e were predicted to generate truncated proteins due to the presence of a stop codon (Fig. 4B).
Figure 4.
Validation of elongated exons, E40e and E41e. (A and C–E) Total RNA was obtained from retina (R), cerebellum (Cb), cortex (Cx), hippocampus (H), brain (B) and skeletal muscle (M) tissues, and RT-PCR assays were carried out using specific primers as depicted. Molecular weight (bp) is indicated on the left for each gel figure. (A) Detection of mRNAs including E40e. The diagram shows the size of intron 40, position of primers and expected sizes of PCR products. Gels show detected PCR products of WT mRNA (450 bp, top panel), E40e-containing mRNA (512 bp) and additional lower band corresponding to splicing leading to partial deletion of the E40e (214 bp, middle panel), and β-actin mRNA as loading control (bottom). (B) Sequencing and in silico alignment with reference dystrophin sequence from GenBank, showing stop codons in E40e and ΔE40e. Corresponding amino acid sequence is shown at bottom. Sequencing was carried out with primers E39F and E42R or E40eR for wild-type or E40e transcripts. (C) Diagram on the left shows position of primers and expected sizes of retinal PCR products detected on gels (right panel) as WT and Dp427 and Dp260-related mRNA containing E40e or the partially deleted ΔE40e (two bands). (D) Detection of mRNAs including E41e. The diagram shows the size of intron 41, position of primers and expected sizes of PCR products (top) and gels showing presence of three bands corresponding to mRNA containing E41e, E41e + ΔE40e, E41e + E40e (from lower to upper band). (E) Diagram showing position of primers and expected sizes of PCR products (top) and gels showing Dp427 or Dp260-related PCR products containing E41e, as two bands corresponding to absence or additional presence of E40e, as indicated on the right.
Thus, the RT-PCR analyses revealed two main transcripts, one containing 328 bp of intron 40 (E40e, 512 bp) and another containing the spliced-out form lacking 298 bp of the 5′ sequence of intron 40 (ΔE40e, 214 bp). Interestingly, retention of full-length intron 40 or alternatively spliced intron 40 has been previously reported in human tissues and cells (28–30), in line with our present observations. Here, Sashimi plots (Supplementary Material, Fig. S4B) only detected transcripts containing ΔE40e, which show overt connection with E40 and E41. However, our RT-PCR assays (Fig. 4) revealed that both the unspliced form (E40e) corresponding to the full-length intron 40 (805 bp) and the spliced form (ΔE40e) that contains the last 507 bp of intron 40 may be included in the mature transcripts. As for E51b above, splicing prediction tools revealed that ΔE40e may have new putative acceptor sequence with prediction score higher than those of the WT splice sites (Supplementary Material, Fig. S5C and D), indicating that it could function as acceptor splice site. Due to the presence of premature stop codons, both E40e and ΔE40e-containing transcripts were predicted to translate as truncated proteins.
To determine if the cDNAs containing E40e take part in Dp427 and/or Dp260 transcripts in retina, we then used specific forward primers for E29 of Dp427 (E29F) and specific exon 1 of Dp260 (I29F) combined with the E40eR primer, or with E42R primer to amplify the WT sequences (Fig. 4C). Both E29F and I29F primers combined with E40eR revealed a doublet of bands suggesting that E40e may be present in these two dystrophins, either complete or partially deleted due to the splicing of E40e described in Fig. 4A.
Insertions of sequences from intron 41 (E41e) were first revealed by Sashimi plots showing reads for E41 fused with reads of E42 (WT mRNA) as well as reads spanning intron 41, fused with E41 but not with exon 42 in both mouse (Supplementary Material, Fig. S4B) and human tissues (Fig. 5B). The size of intron 41 is 25 977 bp for mouse and 31 823 for human, while the size of intron 41 retention ranged from 4000 to 6000 bp. Three different primers within I41 were used for positive amplification of this region, at position 1122, 5024 and 6186 bp. The primer at position 1122 bp was the only one in which amplification was observed, likely because the lengths of PCR products obtained with the other primers were excessive (>5000 bp). No canonical AATAAA polyadenylation signals were found by direct exploration of the sequence until 1122 bp of intron 41 (positive amplification with the E41eR primer) and by alignment (Supplementary Material, Fig. S6A) with the human sequence in which a polyadenylation site was recently identified (31). However, several putative polyadenylation sites were found by direct screening from 2383 to 6212 bp of intron 41 (Supplementary Material, Fig. S6B) that could function as polyadenylation sites.
Figure 5.
Detection of the new splicing events E51b, E40e and E41e in human retina. (A, B) Example of Sashimi plots with reads corresponding to new splicing events responsible for presence of new unannotated exon and elongated exons in human whole-retina samples (each row is a distinct sample); database from Ratnapriya et al. (67). (A) Inclusion of the new exon E51b between E51 and E52. (B) The left image shows the intronic sequences within intron 40 responsible for presence of E40e (red window) and contigs within intron 41 connected to E41 (black accolade) responsible for E41e. The right panel is a zoomed view of the region that includes sequences in intron 40 connected to adjacent exons. Exon junctions and intron inclusion are indicated.
RT-PCR assays validated the presence of E41e in retina, and to a lower extent in brain and muscle tissues. This E39F-E41eR primer pair identified three main bands (arrowheads in Fig. 4D) with expected sizes likely corresponding to mRNAs containing E41e (1489 bp), E41e and the spliced out of I40 (E41e + ΔE40e, 1996 bp), E41e and the full-length I40 (E41e + E40e, 2294 bp) (28,29). We then used specific primers to amplify mRNAs corresponding to Dp427 (E29F) and Dp260 (I29F) paired with primer E41eR (Fig. 4E). Detected bands had expected sizes corresponding to inclusion of E41e in both Dp427 and Dp260-related mRNAs. There were doublets of bands for each, likely corresponding to absence or additional presence of E40e.
Overall, we identified new Dmd isoforms which may include insertion of a new unannotated exon (E51b) in Dp427, Dp260 and Dp140, as well as elongated exons, E40e and E41e, introducing stop codons in both Dp427 and Dp260 mRNAs. Importantly, we also found that these new alternative events are all expressed in the human whole retina, as shown in Sashimi plots derived from a human RNA-Seq database (Fig. 5).
Translation of new truncated dystrophins in CNS
The putative expression of new truncated dystrophins due to the translation of mRNAs containing E40e or E41e was tested by means of a combination of specific antibodies and mouse models of DMD lacking distinct dystrophins. As shown in Fig. 6A, we used the monoclonal antibody DYS1, expected to selectively detect Dp427 and the truncated Dp427 proteins, and the monoclonal 5G5 antibody expected to recognize both Dp427 and Dp260 full-length and truncated dystrophins. The in silico analyses revealed that truncated Dp427 should have a molecular weight of 220–230 kDa while truncated Dp260 should have a molecular weight of 66–74 kDa, depending on presence of E40e or E41e and on their occurrence in specific Dp427/Dp260 known isoforms (Fig. 6B). Moreover, all Dp427 isoforms (WT and truncated) should be absent in tissues from mdx23 mice, due to their mutation generating a stop codon in exon 23, while Dp260 isoforms (WT and truncated) should be unaffected. In the exon52-deleted mdx52 mice, the full-length Dp427 and Dp260 proteins should be absent while their truncated forms could theoretically be expressed.
Figure 6.
Translation of dystrophin transcripts containing E40e and/or E41e. (A) Diagram depicting the experimental design with epitopes for DYS1 and 5G5 antibodies on the dystrophin proteins Dp427 and Dp260, the positions of the intronic insertions (red star), the expected size of the truncated dystrophins (shown in blue) and the position of the mutations leading to interruption of translation in mdx23 and mdx52 mice (green double-head arrows). (B) Expected molecular weight of dystrophin isoforms estimated by in silico analyses. Truncated Dp427 proteins should have a molecular weight of 220–230 kDa, while truncated Dp260 should have a molecular weight of 66–74 kDa, depending on inclusion of the 3′ UTR of intron 40 or 41, and depending on occurrence in specific Dp427/Dp260 known Dmd-gene products (Dp427c, Dp427m, Dp427p, Dp260p-2) that differ in size. (C) Comparative expression of dystrophin isoforms in western blots performed using the DYS1 antibody with whole brain (Br), cerebellar (Cb) and retinal (Ret) extracts from WT, mdx23 and mdx52 mice, as indicated. (D) Western blots performed using the 5G5 antibody with the same tissue samples. Molecular weight markers are indicated on the left of the gels, name of dystrophin isoforms on the right. Isoforms with truncated C-terminus are called N-Dp427 and N-Dp260.
Indeed, the DYS1 antibody detected the full-length Dp427 in both cerebral and retinal tissues from WT mice, but not in mdx23 and mdx52 mice (Fig. 6C). The DYS1 antibody also revealed a doublet band of about 220–230 kDa, which likely corresponded to the truncated Dp427 isoforms generated from two distinct mRNAs containing different intron 40 and/or 41 insertions. As expected, the doublet band was absent in tissues from mdx23 mice, whereas it was readily detectable in both cerebral and retinal tissues from mdx52 mice. The bands above 250 kDa in WT brain (Br-wt) and cerebellum (Cb-wt) cannot correspond to the retina-specific Dp260 as they were not detected in the WT retina and were also absent in samples from mdx23 mice that still express Dp260. They might reflect presence of some degradation truncated products from Dp427. No product of 66–75 kDa was detected with DYS1. As the truncated Dp427 lacks its C-terminal part, we called it N-Dp427.
The 5G5 antibody (Fig. 6D) readily detected both Dp427 and Dp260 full-length dystrophins in tissues of WT mice, but not in mdx23 and mdx52 mice. In retina, there was a very strong immunoreactive signal for Dp260 in both WT and mdx23 mice. In contrast, there was no immunoreactive signal in mdx52 mice, thus confirming that deletion of exon 52 prevents expression of both Dp427 and Dp260. There was an apparent doublet band in retina of WT and mdx23 mice. In a separate experiment (Fig. 6D, bottom panel), we changed the migration conditions to better separate these bands, which unveiled presence of Dp260 in both WT and mdx23 mice, not in mdx52 mice. Strikingly, both WT and mdx52 mice, but not mdx23 mice, expressed a doublet band of about 220–230 kDa likely reflecting presence of the truncated N-Dp427, as concluded from the immunoblot performed with the DYS1 antibody. Importantly, the 5G5 antibody also reacted with protein products of about 66–70 kDa, only in retina samples, suggesting detection of the short-truncated forms of Dp260 that we called N-Dp260. Visualization of a doublet of bands for N-Dp260 was not possible preventing to firmly conclude that this protein product was translated from transcripts containing E40e and/or only E41e. In agreement with our initial hypotheses (Fig. 6A), this short retinal product was detected in WT, mdx23 and mdx52 mice.
Discussion
The CNS (brain and retina) expresses most of the annotated dystrophin-gene products and splice variants, which may explain that distinct DMD gene mutations lead to heterogeneous brain and retinal phenotypes. The present study provides a first RNA-Seq-based reference of the distribution of Dmd-gene products, dystrophin variants and DAPs in mouse brain cells and retinal photoreceptors, including a first description of differential expression in rod and cone photoreceptors. It highlights that multiple dystrophins and associated scaffolding complexes are differentially regulated in CNS in distinct cell types and during cell development, suggesting that a given cell type may express several dystrophins and DAP complexes in distinct cell functional domains and/or cell subpopulations. Another main finding is the identification of new splicing events leading to alternative transcripts, including a new exon within intron 51 in retina (E51b) and elongated exons 40 and 41 (E40e, E41e). Importantly, elongated exons included in these new transcripts contain stop codons, leading to the translation of dystrophins that are truncated at their C-terminus in both retina and brain tissues.
Differential expression of dystrophins and DAPs in brain cell types
The reliability of our in-depth meta-analysis of RNA-Seq data is supported by the identification of well-recognized expression patterns of the dystrophins in CNS. We indeed confirm that Dp71 is the main dystrophin expressed in brain tissues, particularly in astrocytes, while Dp260 is the major product in retina but is absent in brain. As expected, the full-length Dp427c is detected in brain and neurons, and Dp427p selectively in the cerebellum, while muscle Dp427m is undetectable or expressed below the detection threshold in CNS samples.
Our analysis is in line with a recent RNA-Seq and co-expression study suggesting functional association of dystrophins with distinct cellular pathways in the human brain (32). In this latter study, it was reported that Dp427p expression was minimal in the adult human cerebellum as compared with Dp427c, contrary to the expression pattern found in the mouse brain. Our present analysis reveals that both Dp427p and Dp427c mRNA show substantial expression levels in the mouse cerebellum, which does not support a predominant expression of Dp427p in cerebellum. Surprisingly, we found detectable amounts of reads specific of Dp116 in mouse cortical neurons, astrocytes and oligodendrocytes. This suggests that Dp116 is not selectively expressed in Schwann cells of peripheral nerves.
One major conclusion of the present study is that each dystrophin can be expressed in several cell types, and that most CNS cell types may express several dystrophins. Several dystrophins have been recently detected in oligodendrocyte cell types (33), but here we show that this type of expression pattern is not restricted to oligodendrocytes. For instance, we found expression of all brain dystrophins in neurons. In principal neurons, it is well admitted that Dp427 is expressed in GABAergic inhibitory synapses (2), while distinct Dp71 isoforms may be expressed in neuronal soma and glutamatergic excitatory synpases (34,35), or in axon terminals for the Dp40 variant (7). However, we also found presence of Dp140 and Dp116 in neurons. This had never been reported, yet it was recently suggested for Dp140 in studies of neural stem/progenitor cell differentiation (36). In vivo, Dp140 was mostly detected during the foetal stage in mice, and at lower level compared with other brain dystrophins in the adult brain (37,38), notably in oligodendrocytes (33). Here, we confirm that Dp140 is more expressed than Dp427 in this cell lineage. Two studies reported detection of the Dp140 protein along the walls of blood vessels (39,40), but here, we found Dp140 in astrocytes, not endothelial cells, suggesting expression in perivascular astrocyte endfeet. Our analysis also confirms that Dp71 is the main dystrophin expressed in astrocytes, where it is known to have a role in the clustering of AQP4 and Kir4.1 channels (1,41). Interestingly, its shortest isoform, Dp40, is also expressed at relatively important levels in these cells, suggesting that it is not solely expressed in neuronal domains. Moreover, we also detected mRNA of Dp427c, Dp427p, Dp140 and Dp116 in astrocytes. Importantly, we provide the first evidence that Dp71 mRNA is expressed in endothelial cells, while this cell type was believed to selectively express utrophin paralogues (42). Important as well, adult microglia do not express any dystrophin at detectable levels. In contrast, the utrophin paralogue was clearly detected in microglial cells.
While previous work already demonstrated that distinct dystrophin-associated complexes may be found in neurons and astrocytes (13), our data further suggest that several complexes may co-exist in a given cell type. Dystrophins and the transmembrane dystroglycan represent the core of DAP complexes in most cells and are complemented by various isoforms of syntrophins, dystrobrevins and sarcoglycan subunits. This likely reflects that interchangeable scaffolding components of the complex may link the core to distinct functional units, i.e. signalling proteins and ion channels in specific cells and functional cell subdomains. We confirm that α-SYN and α-DB are components of the DAP complex in astrocytes, in association with Dp71 and/or Dp40, but we further uncover that these DAPs are present in other cell types including neurons, microglia and oligodendrocytes. Interestingly, γ1-SYN was only detected in oligodendrocyte precursor cells, not in myelinated oligodendrocytes, suggesting that it is only present during early development of oligodendrocytes, while α1-SYN was present at all stages. The β-SG subunit was detected at higher level than other sarcoglycans in most brain cells expressing dystrophins, except for endothelial and microglial cells in which ε-SG showed a relatively important level of expression.
Overall, the observed patterns of expression strongly suggest that several dystrophin complexes may play distinct roles in a given cell type. This might reflect expression of distinct dystrophins in several subtypes or subpopulations of cells, e.g. in neuronal subtypes with distinct neurotransmitter signatures. Alternatively, distinct dystrophins could be expressed in different subcellular domains. As an example, distinct Dp71 isoforms have been selectively detected in nucleus, postsynaptic or presynaptic neuronal domains (43).
Finally, distinct dystrophins and associated complexes may be expressed at different stages of cell maturation (44–46). The transcriptomic brain database used in the present study (16) included neurons, astrocytes and endothelial cells purified at P7, an age at which differentiated cells are present and can be purified with minimal activation and with gene expression profiles close to mature cells, while P17 was chosen for oligodendrocytes to allow purification of all associated lineages. In line with a previous study (33), we show differential expression of several dystrophins in oligodendrocyte precursor cells, newly formed oligodendrocytes and myelinated oligodendrocytes.
Differential expression of dystrophins and DAPs in retinal photoreceptors
This study provides the first transcriptomic-based characterization of dystrophins and DAPs expression in immature and mature retina. We confirm that both Dp260 and Dp427 are expressed in photoreceptors, with Dp260 being more expressed than Dp427 in mature rods (P28). Surprisingly, the full-length form detected in the whole retina and purified photoreceptors is the Purkinje form, Dp427p. This further supports that the P promoter can be regulated in various tissues (47) and may display a broader and more complex range of expression than initially reported. Alike in brain, we also found detectable amounts of Dp116, selectively in mature flow-sorted cone photoreceptors of the retina. Our data show that photoreceptors contain mRNAs from all dystrophins, but their expression appears to be regulated during cell maturation, Dp40 being relatively more expressed in mature cones and Dp260 in mature rods. Dp71 is known to be mainly expressed in Müller glial cells and astrocytes (23,48), but our data comparing WT and cone-only purified photoreceptors indicate it may also be expressed in immature cones at P2. Moreover, we found that Dp40, a synaptic N-terminal isoform of Dp71, is the main dystrophin transcript found in mature cones.
The major differences between immature (P2) and mature (P28) photoreceptor cells suggest an orchestrated involvement of multiple dystrophins during photoreceptor formation, with specific dystrophins predominating in mature cones (Dp40) and rods (Dp260). Analysis of DAPs expression revealed that α1/γ2-SYNs and β-DB are the main isoforms in mature photoreceptors. Regarding sarcoglycans, the β and ε subunits are clearly expressed in flow-sorted photoreceptors from cone-only retina. However, there is no overt difference in their expression levels compared in flow-sorted cone/rod cells (WT) and cone-only cells (NRL), suggesting they are absent or expressed at very low levels in rods. It is noteworthy that α-SG, δ-SG and γ-SG are not detected in photoreceptors, suggesting that some of them may be expressed in other cell types of the retina, as suggested by others (23,49).
Skipping of Dmd-gene exons in CNS
We report the relative expression of exon-junction reads reflecting exon skipping, which was a common form of splicing event in our CNS databases, as previously shown for human muscle (20). Skipping of the penultimate exon 78 (ΔE78), then of exon 71 (ΔE71), is the most frequent alternative splicing events detected in all cell samples, suggesting a ubiquitous role in cell function. Skipping of exon 78 has been described as a main splicing-out event affecting Dp71 (11,24), but it may also affect other dystrophins (10,12,28,50). We show that all cell types contain both +E78 and ΔE78, indicating that E78 skipping is not selective of a given one. Inclusion of E78 (+E78) is more frequent than ΔE78, but we cannot rule out that ΔE78 selectively affects distinct dystrophins. We have previously shown that the adult retina mainly express Dp71 with ΔE78 (24). In the present analyses, we however show that ΔE78 predominates in immature retina (P2), not in mature retina (P28). This suggests that ΔE78 mainly affects Dp71, while the majority of other dystrophins expressed in retina may preferentially include E78. The data also indicate that ΔE78 is developmentally regulated. This is in agreement with previous studies reporting that ΔE78 occurs in early embryonic tissues (9), while inclusion of E78 would rather be required to maintain cell membrane integrity in adult tissues (27,50).
The impact of ΔE71, ΔE74 and more strickingly of ΔE71–74 may also be critical for cell function and integrity, as these splicing-out events are expected to suppress the syntrophin-binding site and affect interaction with dystrobrevin (51). We also detected ΔE69 in neurons, which is expected to alter the C-terminal domain (52).
We suggest that the skipping of these specific exons at the 3′ end of the gene likely supports their appropriate expression in specific cellular environments, at distinct stages of development and/or during activity-dependent processes, to remodel protein–protein interaction networks, at least with components of the dystrophin–glycoprotein complexes.
New alternative splicing events and truncated dystrophins in CNS
In line with the large intron coverage of the DMD gene, we have identified a range of exon-skipping events associated with usage of alternative splice sites, leading to the inclusion of intronic sequences such as cryptic exons or pseudoexons into the processed mRNA, some of which encoding premature stop codons (see (53,54) for related reviews). Importantly, two main identified events, E51b and ΔE40e, were predicted by in silico analyses to have cryptic consensus sequences corresponding to alternative acceptor and donor splice sites with scores close to WT splicing sites.
We first characterized a new exon, E51b, as a cluster of reads between E51 and E52 (within the annotated intron 51), selectively included in mature dystrophin transcripts in retina, not in brain. E51b expression was found in all retinal samples from WT mice, which excludes any relation with pathogenesis conditions (19). According to our RT-PCR assays, E51b-containing transcripts appears to be expressed at much higher levels than the WT mRNAs (without E51b), and the 36 bp of E51b is in frame within Dp427, Dp260 and Dp140 mRNAs, adding 12 amino acids between E51 and E52. E51b thus shows a strong exon-like expression profile and we suggest classifying it as a canonical exon in the retina, rather than a pseudoexon or cryptic exon. A recent RNA-Seq study reports the in-frame inclusion of a pseudoexon located in intron 51 in muscle biopsies of healthy human subjects (20). However, this pseudoexon, located from 21 435 to 21 518 bp within intron 51 in human muscle, does not show any overlap with the retina-specific E51b exon described in the present study, which is located closer to E52 (from 40 862 to 40 897 bp within I51 in human retina) and has overt connections with E51 and E52.
E51b expression is higher in immature photoreceptors (at P2) from WT retina compared with cone-only retina from NRL−/− mice, suggesting a major expression in immature rod photoreceptors. In contrast, E51b expression is relatively comparable in mature cones and rods at P28, indicating that expression E51b-containing transcripts is likely regulated during photoreceptor development. A putative translation of E51b-containing transcripts could not be addressed experimentally due to the very small size of this insertion. Therefore, its role in cell function and/or development remains to be determined. If some dystrophin proteins do include the E51b insertion, this would likely alter their secondary structure and function, as well as a spectrin-like repeat involved in the binding to microtubules (55).
Importantly, we also found long contiguous series of reads spanning E40 and I40, and/or E41 and I41. We demonstrate that these intronic insertions in both Dp427 and Dp260 mRNAs produce dystrophin proteins with truncated C-terminus. We initially detected these splicing-out events in mouse and human retinal samples in our RNA-Seq analyses. However, RT-PCR assays revealed that they are also expressed at lower levels in brain, i.e. in cortex and cerebellum for E40e, and in cortex, cerebellum and hippocampus for E41e. The detection of several amplicons in our PCR experiments suggests that E40e and E41e also occur in combination with additional alternative splicing events, highlighting a complex regulation of these intronic insertions.
We further demonstrate that these alternative transcripts are translated into proteins. This is an important finding, as previous studies suggested that most of the intronic insertions with premature stop codons may generate non-coding mRNAs, eventually part of a posttranscriptional switch system controlling gene expression, or aberrant splicing products degraded by nonsense-mediated mRNA decay (18,56,57). Here, we show that these splicing-out events induce translation of at least two main N-terminal dystrophins, i.e. truncated of their C-terminus, which may correspond either to Dp427 or Dp260, and that we called N-Dp427 and N-Dp260. A previous study reported a tissue-specific insertion of the complete intron 40 between E40 and E41 in DMD transcripts, at high level in kidney (36%) and very low levels in brain (<5%) and other tissues (29). Full or partial insertion of I40 combined with E78 skipping was also reported in CRL-2061 rhabdomyosarcoma cells (30), and partial insertion of I40 was found in the Dp427b transcript in the SH-SY5Y neuronal cell line (28). A recent study also identified alternative polyadenylation of I41 in the Dp427m transcript in a human-derived cell line, responsible for the generation of an N-terminal dystrophin of about 230–235 kDa, selectively expressed in cardiomyocytes derived from induced pluripotent stem cells (31). This likely does not correspond to the E41e-containing transcripts that we found in both CNS and muscle tissues, because this polyadenylation site (AATAAA) in I41 (position 406–411 bp of I41) is not present in mouse I41, but it is present in rat I41 (Supplementary Material, Fig. S6A). Notably, however, we manually found several AATAAA sequences from 2383 to 6212 bp of I41 (Supplementary Material, Fig. S6B), associated with many reads, which may function as polyadenylation sites. We show that Dp427c, Dp427p and Dp260 transcripts may include E40e or E41e, or parts of both intronic insertions. We demonstrate expression of N-Dp427 and N-Dp260 at the protein level in the mouse retina, and that of N-Dp427 in forebrain and cerebellum. The doublets of bands revealed in our immunoblots with the antibody directed against Dp427 suggest that both E40e and E41e-containing mRNAs induce expression of two distinct isoforms of N-Dp427 proteins in retina and brain. For Dp260, a large single band was detected, suggesting that only one type of transcript was translated.
Putative roles for the N-Dp427 and N-Dp260 proteins are unclear, since they lack the main cysteine-rich and C-terminal domains required to form DAP complexes. However, N-Dp427 and N-Dp260 still comprise actin-binding sites and spectrin-like repeats that have been shown, at least in muscle cells, to enable non-canonical binding to the cell membrane (55,58). This is further supported by two studies reporting DMD/BMD patients whose specific mutations generate N-terminal dystrophins with preserved ability to bind the muscle cell membrane (59,60). Moreover, our data indicate a higher expression at P2 compared with P28 in retina, suggesting that their involvement in cell function is developmentally regulated. This bundle of information suggests that, structurally, N-Dp427 and N-Dp260 may therefore have a function in brain and retinal cells.
In conclusion, we provide a first RNA-Seq-based reference showing tissue- and cell-specific differential expression of dystrophins, splice variants and DAPs in mouse brain and retina. The new identified splicing events may affect several dystrophin transcripts, and those including stop codons are translated into proteins lacking their C-terminus, which we called N-Dp427 and N-Dp260. Identification of alternative splicing events in the DMD gene is likely relevant to improve diagnosis and identify pathogenic variants and modifiers of disease severity in dystrophinopathies (18,19,61,62). It is worth to note that the loss of the new dystrophins identified here, N-Dp427 and N-Dp260, may contribute to the phenotypes displayed by patients and DMD mouse models. This may also be particularly relevant for the currently developed splice-switching therapeutic strategies, which are also developed to target brain and retina, but may face-specific challenges due to the specificities of Dmd-gene post-transcriptional regulations in the CNS. Finally, few BMD patients may have in-frame mutations around exons 40/41 that would abrogate the insertion of the intronic sequences responsible for the expression of these N-terminal dystrophins in CNS, without affecting the C-terminal domains of the other dystrophins (63). These patients have abnormal retinal physiology (64) and may also display other central comorbidities (65). Our study thus provides an important new piece of knowledge to better understand the basic splicing mechanisms of the DMD gene and the complex etiology of central comorbidities associated with DMD/BMD.
Materials and Methods
Animals and tissue processing
C57BL/6JRj adult (3 months old) male mice were produced in our transgenic mouse facility at Université Paris-Saclay in Orsay (France) and at CINVESTAV (Mexico). Age-matched Dp427-deficient C57BL/10ScSn-Dmdmdx/J (mdx) and exon52-deleted mdx52 mouse models of DMD were produced at Université Paris-Saclay in Orsay (France). The mdx mice were originally purchased from Charles River Laboratories (Saint-Germain sur l’Arbresle). The mdx52 mouse model was originally developed by the group of Dr M. Katsuki (Neurological Institute, Kyushu University, Fukuoka, 812, Japan) (66); mdx52 breeders were generously provided by Dr Jun Tanihata and Dr Shin’ichi Takeda (National Center of Neurology and Psychiatry, Tokyo, Japan). For validation assays, after mouse euthanasia, the eyes were rapidly enucleated to dissect retina, the brain removed from skull to dissect out hippocampus and cerebellum and the skeletal muscle was dissected out for control experiments. Tissue samples were immediately frozen and processed for western blot and/or RT-PCR.
Experiments in Mexico adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were conducted following the federal and local regulations approved by CINVESTAV-UPEAL and Mexican Official Norm (NOM-062-ZOO-1999). Experiments in France adhered to the guidelines of the local mouse facility in France (agreement D91–471-104) in compliance with European Directive 2010/63/EU and French National Committee (87/848).
RNA-Seq libraries
For meta-analysis purposes, we re-analysed the previously published whole transcriptome data from P17 mouse cerebral cortices and from main brain cells purified from the mouse cerebral cortex (16) (GEO accession # GSE52564). We also included hippocampus and cerebellum of 4-month-old male C57BL/6 mice from our RNA-Seq library generated by the I2BC-CNRS platform for high-throughput sequencing (Gif-sur-Yvette, France). In this latter library, RNA was extracted using Nucleospin RNA Plus kit, which includes DNase I treatment (Macherey-Nagel, Düren, Germany). RNA quality and quantity were evaluated using a BioAnalyzer 2100 with RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). The library was constructed from polyadenylated fraction of high-quality RNA of biological triplicates (HiSeq 2500 system Illumina; NextSeq High Output 43 paired-end sequencing cycles).
For mouse retinal expression analysis, we first used our published transcriptomic data (17) (GEO accession # GSE94534) from P30 C57BL6 WT and rd10 mouse retina to estimate distribution of dystrophins’ mRNAs in healthy and degenerative retina displaying photoreceptor degeneration, respectively. Finally, we completed our analysis using rod and cone flow-sorted photoreceptor cell transcriptomes from Nrlp-GFP (WT) and from cone-only retina (Nrl−/−; Nrlp-GFP mice), respectively. These data were generated from postnatal day 2 (P2) and 28 (P28) retina (15) (GEO accession # GSE74660). For analysis of expression in human whole retina, we used a published transcriptomic database ((67); GEO accession # GSE115828).
Analyses of RNA-Seq libraries
Pass-filtered reads from all datasets were mapped using HISAT2 and aligned to mouse reference genome GRCm38.84. Count tables for both genes and exon were obtained using FeatureCounts (68). Normalization and expression analysis were computed using EdgeR (69). To detail the pattern of expression of all dystrophins in these samples (Fig. 1A), we took advantage that several of them have a specific first exon (https://www.dmd.nl/isoforms.html#proms). Because the first exons of dystrophins were mostly reported for human, it was necessary to define them for the mouse. For Dp427 mRNA, no signal for exon 1 was found in mouse genome mm10 (GRCm38.94). However, there were reads in three different positions corresponding to 5′ UTRs of the three Dp427 transcripts that use alternative promoters in brain (Dp427c), muscle (Dp427m) and Purkinje cells (Dp427p). To determine the expression level of the mouse Dp260 first exon, which corresponds to the Dp260–2 isoform in human, the exon had to be annotated in the mouse genome (mm10). Dp140 was identified by its specific 5′ UTR in intron 44, Dp116 by its specific exon in intron 55 and Dp71 by its specific first exon in intron 62. Dp40 ends with a specific 3′ UTR that we have used to identify this important dystrophin (Fig. 1B).
To detect splicing events and new features in the Dmd gene, BAM files were uploaded in the Integrative Genomics Viewer (IGV) software and the whole gene was screened manually from 5′ to 3′ end. Transcripts with spliced-out annotated Dmd exons were identified by the presence of specific splice junctions. We also analysed splicing-out events that did not involve annotated exons solely. We considered as a new feature a pile of reads in the intronic region of the Dmd gene that did not match the 79 known annotated exons. Sashimi plots from IGV were also generated for analysis purposes. We then generated a new gtf files from our curated analysis including all known and new features, as well as promoter positions corresponding to specific first exon for each dystrophin isoform and re-run the counting of the reads in each sample and quantified the expression of each feature (annotated and new). Expression level estimation of each exon and genes (for DGC expression analysis) was reported as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). Heatmaps were generated using gplots, ggplot2 and apcluster packages from R based on the mean of each group.
RT-PCR and sequencing assays
Total RNA was extracted with TRIzol reagent (Invitrogen) and treated with DNAse I according to manufacturer’s protocol. Two micrograms of total RNA was used to synthesize cDNA using random primers, or E51bR in some experiments, a reverse primer specific of the unannotated exon that we identified within intron 51 and called E51b, and the reverse transcriptase SuperScript III First-Strand Synthesis Kit (Invitrogen). The E51bR primer was used to amplify E51b-Dp427, E51b-Dp260 and E51b-Dp140. Canonical sequences and new elements (in-frame putative new exon and 3′ UTRs) of dystrophin mRNAs were validated by PCR with 3–5 μL of cDNA, using Fermentas Taq DNA Polymerase (Thermo Fisher Scientific) for short PCR products (up to 1500 bp) and Phusion High-Fidelity DNA Polymerase (New England Biolabs) for long PCR products.
The primers used to validate the presence of new features in dystrophin RNA (unannotated exon E51b and putative 3′ UTRs in introns 40 and 41 that we called E40e and E41e) are detailed in Supplementary Material, Table S1 and in corresponding diagrams in figures. To determine which dystrophin mRNAs contained new features, we used primers corresponding to exon 29 of Dp427 (E29F), or first specific exons of Dp260 (I29F) and Dp140 (I44F).
PCR was carried out according to each Taq DNA Polymerase manufacturer’s protocol and considering the size of the expected PCR products. For the following primer pairs, E50F-E53R, E51bF-E53R, E39F-E42R, E39F-E40eR and ActF-ActR, the PCR reactions were carried out at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, during 35 cycles. To amplify Dp427, Dp260 and Dp140 mRNAs, their specific forward primers were mixed with reverse primers E51bR, E40eR and E41eR (Supplementary Material, Table S1) and PCR reactions were performed at 98°C for 30 s, 63–65°C for 30 s and 72°C for 2–3 min during 35 cycles. PCR products were electrophoresed in 1.5% agarose gel pre-stained with ethidium bromide. PCR products were gel-purified and sequenced with the Dye DeoxyTerminator Cycle Sequence Kit (Applied Biosystems, Foster City, CA, USA). To sequence E51b, the PCR product obtained with the E50F and E53R primers was gel-purified and ligated in the pGEM-T easy vector which was used to transform E. coli DH5α. Resistant colonies were selected with LB/Amp solid medium. A candidate for each sequence was selected and sequenced.
Western blotting
Total protein extracts were treated with a RIPA-5% sodium dodecyl sulfate (SDS) extraction and lysis buffer containing protease inhibitors (Roche, Inc., Indianapolis, IN, USA). Protein concentration was calculated using the DC™ Protein Assay kit (Bio-Rad, France). Protein extracts (10 μg for retina and 50 μg for brain samples) were resolved in 5% sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred to 5% polyvinylidene fluoride membranes and incubated 1 h at room temperature (RT) with 4% milk blocking buffer, then overnight at 4°C with the monoclonal DYS1 and 5G5 primary antibodies (1/50). The commercial DYS1 antibody (NCL-DYS1; Leica Microsystem, France) is directed against the human R8 repeat of the central rod domain of dystrophin (between amino acids 1181 and 1388) and reacts with Dp427. The 5G5 antibody (D Mornet, Montpellier, France) was generated using a recombinant protein corresponding to mouse amino acids 1173–1728 of the central rod domain and reacts with brain and retinal Dp427 as well as with retinal Dp260 (14). This was followed by incubation with a goat anti-Mouse IgG-horseradish peroxidase secondary antibody (1:10 000; A4416, Sigma-Aldrich) for 1 h at RT. Immunoreactive products were revealed by a chemiluminescent reaction using the SuperSignal West Femto substrate (Thermo Fisher Scientific, France). All protein extracts were obtained by three independent experiments. Tubulin was used as loading control.
Supplementary Material
Acknowledgements
We are grateful to Dr Shin’ichi Takeda and Dr Jun Tanihata (National Center of Neurology and Psychiatry, Tokyo, Japan) for kindly providing mdx52 mouse breeders, to Dr Dominique Mornet (INSERM, Montpellier, France) for providing the 5G5 antibody, and to Dr Alvaro Rendon (Institut de la vision, Paris, France) for fostering France-Mexico collaborations. We also thank the Zootechnic platforms of the Institut des Neurosciences Paris Saclay and CINVESTAV for mouse breeding, care and genotyping, and Mayram González-Reyes for technical support (brain dissections and RNA isolation). This work has benefited from the facilities and expertise of the high throughput sequencing core facility of I2BC (Centre de Recherche de Gif—http://www.i2bc-saclay.fr/). We are grateful to Ecos Nord-CONACyT (grant number M16SO1/276330, 2017) for supporting France-Mexico exchanges that enabled the development of the RNA-Seq techniques during the PhD thesis of CG-C in 2018, and CONACYT (grant number CB-2017-2018-A1-S-24868).
Conflict of Interest statement. The authors have no relevant financial or non-financial interests to disclose.
Contributor Information
César García-Cruz, Departamento de Genética y Biología Molecular, Centro de Investigación y de Estudios Avanzados del IPN, 07360 Ciudad de México, Mexico; Institut des Neurosciences Paris Saclay, Université Paris-Saclay, CNRS, 91400 Saclay, France.
Jorge Aragón, Departamento de Genética y Biología Molecular, Centro de Investigación y de Estudios Avanzados del IPN, 07360 Ciudad de México, Mexico.
Sophie Lourdel, Institut des Neurosciences Paris Saclay, Université Paris-Saclay, CNRS, 91400 Saclay, France.
Ahrmad Annan, Institut des Neurosciences Paris Saclay, Université Paris-Saclay, CNRS, 91400 Saclay, France.
Jérôme E Roger, Institut des Neurosciences Paris Saclay, Université Paris-Saclay, CNRS, 91400 Saclay, France; CERTO-Retina France, 91400 Saclay, France.
Cecilia Montanez, Departamento de Genética y Biología Molecular, Centro de Investigación y de Estudios Avanzados del IPN, 07360 Ciudad de México, Mexico.
Cyrille Vaillend, Institut des Neurosciences Paris Saclay, Université Paris-Saclay, CNRS, 91400 Saclay, France.
Funding
Centre National de la Recherche Scientifique (CNRS) and Université Paris-Sud (France), by a grant obtained in 2017 from Coopération et Partenariat France—Amérique Latine (ECOS Nord-CONACyT) supporting France-Mexico collaborative research (M16SO1/276330 to C.M. and C.V.), a grant from Consejo Nacional de Ciencia y Tecnología (CONACyT) (CB-2017-2018-A1-S-24868 to C.M.) and a PhD fellowship from CONACyT (394374 to C.G.-C.).
References
- 1. Waite, A., Brown, S.C. and Blake, D.J. (2012) The dystrophin-glycoprotein complex in brain development and disease. Trends Neurosci., 35, 487–496. [DOI] [PubMed] [Google Scholar]
- 2. Perronnet, C. and Vaillend, C. (2010) Dystrophins, utrophins, and associated scaffolding complexes: role in mammalian brain and implications for therapeutic strategies. J. Biomed. Biotechnol., 2010, 849426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ricotti, V., Mandy, W.P.L., Scoto, M., Pane, M., Deconinck, N., Messina, S., Mercuri, E., Skuse, D.H. and Muntoni, F. (2016) Neurodevelopmental, emotional, and behavioural problems in Duchenne muscular dystrophy in relation to underlying dystrophin gene mutations. Dev. Med. Child Neurol., 58, 77–84. [DOI] [PubMed] [Google Scholar]
- 4. Ricotti, V., Jägle, H., Theodorou, M., Moore, A.T., Muntoni, F. and Thompson, D.A. (2016) Ocular and neurodevelopmental features of Duchenne muscular dystrophy: a signature of dystrophin function in the central nervous system. Eur. J. Hum. Genet., 24, 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rani, A.Q.M., Maeta, K., Kawaguchi, T., Awano, H., Nagai, M., Nishio, H. and Matsuo, M. (2019) Schwann cell-specific Dp116 is expressed in glioblastoma cells, revealing two novel DMD gene splicing patterns. Biochem. Biophys. Rep., 20, 100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tinsley, J.M., Blake, D.J. and Davies, K.E. (1993) Apo-dystrophin-3: a 2.2kb transcript from the DMD locus encoding the dystrophin glycoprotein binding site. Hum. Mol. Genet., 2, 521–524. [DOI] [PubMed] [Google Scholar]
- 7. Fujimoto, T., Itoh, K., Yaoi, T. and Fushiki, S. (2014) Somatodendritic and excitatory postsynaptic distribution of neuron-type dystrophin isoform, Dp40, in hippocampal neurons. Biochem. Biophys. Res. Commun., 452, 79–84. [DOI] [PubMed] [Google Scholar]
- 8. García-Cruz, C., Merino-Jiménez, C., Aragón, J., Ceja, V., González-Assad, B., Reyes-Grajeda, J.P. and Montanez, C. (2022) Overexpression of the dystrophins Dp40 and Dp40L170P modifies neurite outgrowth and the protein expression profile of PC12 cells. Sci. Rep., 12, 1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bies, R.D., Phelps, S.F., Cortez, M.D., Roberts, R., Caskey, C.T. and Chamberlain, J.S. (1992) Human and murine dystrophin mRNA transcripts are differentially expressed during skeletal muscle, heart, and brain development. Nucleic Acids Res., 20, 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feener, C.A., Koenig, M. and Kunkel, L.M. (1989) Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature, 338, 509–511. [DOI] [PubMed] [Google Scholar]
- 11. Austin, R., Howard, P.L., D’souza, V.N., Klamut, H.J. and Ray, P.N. (1995) Cloning and characterization of alternatively spliced isoforms of Dp71. Hum. Mol. Genet., 4, 1475–1483. [DOI] [PubMed] [Google Scholar]
- 12. Lidov, H.G.W. and Kunkel, L.M. (1997) Dp140: alternatively spliced isoforms in brain and kidney. Genomics, 45, 132–139. [DOI] [PubMed] [Google Scholar]
- 13. Blake, D.J., Hawkes, R., Benson, M.A. and Beesley, P.W. (1999) Different dystrophin-like complexes are expressed in neurons and glia. J. Cell Biol., 147, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wersinger, E., Bordais, A., Schwab, Y., Sene, A., Bénard, R., Alunni, V., Sahel, J.-A., Rendon, A. and Roux, M.J. (2011) Reevaluation of dystrophin localization in the mouse retina. Invest. Ophthalmol. Vis. Sci., 52, 7901–7908. [DOI] [PubMed] [Google Scholar]
- 15. Kim, J.W., Yang, H.J., Oel, A.P., Brooks, M.J., Jia, L., Plachetzki, D.C., Li, W., Allison, W.T. and Swaroop, A. (2016) Recruitment of rod photoreceptors from short-wavelength-sensitive cones during the evolution of nocturnal vision in mammals. Dev. Cell, 37, 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang, Y., Chen, K., Sloan, S.A., Bennett, M.L., Scholze, A.R., O’Keeffe, S., Phatnani, H.P., Guarnieri, P., Caneda, C., Ruderisch, N.et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci., 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamon, A., Masson, C., Bitard, J., Gieser, L., Roger, J.E. and Perron, M. (2017) Retinal degeneration triggers the activation of YAP/TEAD in reactive Müller cells. Invest. Ophthalmol. Vis. Sci., 58, 1941–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang, Z., Habara, Y., Nishiyama, A., Oyazato, Y., Yagi, M., Takeshima, Y. and Matsuo, M. (2007) Identification of seven novel cryptic exons embedded in the dystrophin gene and characterization of 14 cryptic dystrophin exons. J. Hum. Genet., 52, 607–617. [DOI] [PubMed] [Google Scholar]
- 19. Xie, Z., Tang, L., Xie, Z., Sun, C., Shuai, H., Zhou, C., Liu, Y., Yu, M., Zheng, Y., Meng, L.et al. (2020) Splicing characteristics of dystrophin pseudoexons and identification of a novel pathogenic intronic variant in the DMD gene. Genes (Basel), 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bougé, A.L., Murauer, E., Beyne, E., Miro, J., Varilh, J., Taulan, M., Koenig, M., Claustres, M. and Tuffery-Giraud, S. (2017) Targeted RNA-Seq profiling of splicing pattern in the DMD gene: exons are mostly constitutively spliced in human skeletal muscle. Sci. Rep., 7, 39094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akimoto, M., Cheng, H., Zhu, D., Brzezinski, J.A., Khanna, R., Filippova, E., Oh, E.C.T., Jing, Y., Linares, J.L., Brooks, M.et al. (2006) Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U. S. A., 103, 3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mears, A.J., Kondo, M., Swain, P.K., Takada, Y., Bush, R.A., Saunders, T.L., Sieving, P.A. and Swaroop, A. (2001) Nrl is required for rod photoreceptor development. Nat. Genet., 29, 447–452. [DOI] [PubMed] [Google Scholar]
- 23. Claudepierre, T., Mornet, D., Pannicke, T., Forster, V., Dalloz, C., Bolaños, F., Sahel, J., Reichenbach, A. and Rendon, A. (2000) Expression of Dp71 in Müller glial cells: a comparison with utrophin- and dystrophin-associated proteins. Invest. Ophthalmol. Vis. Sci., 41, 294–304. [PubMed] [Google Scholar]
- 24. Aragón, J., González-Reyes, M., Romo-Yáñez, J., Vacca, O., Aguilar-González, G., Rendón, A., Vaillend, C. and Montañez, C. (2018) Dystrophin Dp71 isoforms are differentially expressed in the mouse brain and retina: report of new alternative splicing and a novel nomenclature for Dp71 isoforms. Mol. Neurobiol., 55, 1376–1386. [DOI] [PubMed] [Google Scholar]
- 25. Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouzé, P. and Brunak, S. (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res., 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reese, M.G., Eeckman, F.H., Kulp, D. and Haussler, D. (1997) Improved splice site detection in Genie. J. Comput. Biol., 4, 311–323. [DOI] [PubMed] [Google Scholar]
- 27. Miro, J., Bougé, A.L., Murauer, E., Beyne, E., Da Cunha, D., Claustres, M., Koenig, M. and Tuffery-Giraud, S. (2020) First identification of RNA-binding proteins that regulate alternative exons in the dystrophin gene. Int. J. Mol. Sci., 21, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishida, A., Minegishi, M., Takeuchi, A., Awano, H., Niba, E.T.E. and Matsuo, M. (2015) Neuronal SH-SY5Y cells use the C-dystrophin promoter coupled with exon 78 skipping and display multiple patterns of alternative splicing including two intronic insertion events. Hum. Genet., 134, 993–1001. [DOI] [PubMed] [Google Scholar]
- 29. Nishida, A., Minegishi, M., Takeuchi, A., Niba, E.T.E., Awano, H., Lee, T., Iijima, K., Takeshima, Y. and Matsuo, M. (2015) Tissue- and case-specific retention of intron 40 in mature dystrophin mRNA. J. Hum. Genet., 60, 327–333. [DOI] [PubMed] [Google Scholar]
- 30. Niba, E.T.E., Yamanaka, R., Rani, A.Q.M., Awano, H., Matsumoto, M., Nishio, H. and Matsuo, M. (2017) DMD transcripts in CRL-2061 rhabdomyosarcoma cells show high levels of intron retention by intron-specific PCR amplification. Cancer Cell Int., 17, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rani, A.Q.M., Yamamoto, T., Kawaguchi, T., Maeta, K., Awano, H., Nishio, H. and Matsuo, M. (2020) Intronic alternative polyadenylation in the middle of the DMD gene produces half-size N-terminal dystrophin with a potential implication of ECG abnormalities of DMD patients. Int. J. Mol. Sci., 21, 3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doorenweerd, N., Mahfouz, A., vanPutten, M., Kaliyaperumal, R., T’Hoen, P.A.C., Hendriksen, J.G.M., Aartsma-Rus, A.M., Verschuuren, J.J.G.M., Niks, E.H., Reinders, M.J.T.et al. (2017) Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Sci. Rep., 7, 12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aranmolate, A., Tse, N. and Colognato, H. (2017) Myelination is delayed during postnatal brain development in the mdx mouse model of Duchenne muscular dystrophy. BMC Neurosci., 18, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodríguez-Munoz, R., Cardenas-Aguayo, M.D.C., Aleman, V., Osorio, B., Chavez-Gonzalez, O., Rendon, A., Martínez-Rojas, D. and Meraz-Ríos, M.A. (2015) Novel nuclear protein complexes of dystrophin 71 isoforms in rat cultured hippocampal GABAergic and glutamatergic neurons. PLoS One, 10, e0137328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daoud, F., Candelario-Martínez, A., Billard, J.-M., Avital, A., Khelfaoui, M., Rozenvald, Y., Guegan, M., Mornet, D., Jaillard, D., Nudel, U.et al. (2009) Role of mental retardation-associated dystrophin-gene product Dp71 in excitatory synapse organization, synaptic plasticity and behavioral functions. PLoS One, 4, e6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Romo-Yáñez, J., Rodríguez-Martínez, G., Aragón, J., Siqueiros-Márquez, L., Herrera-Salazar, A., Velasco, I. and Montanez, C. (2020) Characterization of the expression of dystrophins and dystrophin-associated proteins during embryonic neural stem/progenitor cell differentiation. Neurosci. Lett., 736, 135247. [DOI] [PubMed] [Google Scholar]
- 37. Morris, G.E., Simmons, C. and Man, N.T. (1995) Apo-dystrophins (Dp140 and Dp71) and dystrophin splicing isoforms in developing brain. Biochem. Biophys. Res. Commun., 215, 361–367. [DOI] [PubMed] [Google Scholar]
- 38. Schofield, J.N., Blake, D., Simmons, C., Morris, G.E., Tinsley, J.M., Davies, K.E. and Edwards, Y.H. (1994) Apo-dystrophin-1 and apo-dystrophin-2, products of the Duchenne muscular dystrophy locus: expression during mouse embryogenesis and in cultured cell lines. Hum. Mol. Genet., 3, 1309–1316. [DOI] [PubMed] [Google Scholar]
- 39. Caudal, D., François, V., Lafoux, A., Ledevin, M., Anegon, I., leGuiner, C., Larcher, T. and Huchet, C. (2020) Characterization of brain dystrophins absence and impact in dystrophin-deficient Dmdmdx rat model. PLoS One, 15, e0230083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lidov, H.G.W., Selig, S. and Kunkel, L.M. (1995) Dp140: a novel 140 kDa CNS transcript from the dystrophin locus. Hum. Mol. Genet., 4, 329–335. [DOI] [PubMed] [Google Scholar]
- 41. Belmaati Cherkaoui, M., Vacca, O., Izabelle, C., Boulay, A.C., Boulogne, C., Gillet, C., Barnier, J.V., Rendon, A., Cohen-Salmon, M. and Vaillend, C. (2021) Dp71 contribution to the molecular scaffold anchoring aquaporine-4 channels in brain macroglial cells. Glia, 69, 954–970. [DOI] [PubMed] [Google Scholar]
- 42. Perronnet, C., Chagneau, C., leBlanc, P., Samson-Desvignes, N., Mornet, D., Laroche, S. and de la porte, S. and Vaillend, C. (2012) Upregulation of brain utrophin does not rescue behavioral alterations in dystrophin-deficient mice. Hum. Mol. Genet., 21, 2263–2276. [DOI] [PubMed] [Google Scholar]
- 43. Naidoo, M. and Anthony, K. (2020) Dystrophin Dp71 and the neuropathophysiology of Duchenne muscular dystrophy. Mol. Neurobiol., 57, 1748–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paúl-González, S., Aragón, J., Rodríguez-Martínez, G., Romo-Yáñez, J. and Montanez, C. (2021) Differential expression of Dp71 and Dp40 isoforms in proliferating and differentiated neural stem cells: identification of Dp40 splicing variants. Biochem. Biophys. Res. Commun., 560, 152–158. [DOI] [PubMed] [Google Scholar]
- 45. Simon, M.J., Murchison, C. and Iliff, J.J. (2018) A transcriptome-based assessment of the astrocytic dystrophin-associated complex in the developing human brain. J. Neurosci. Res., 96, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lange, J., Gillham, O., Alkharji, R., Eaton, S., Ferrari, G., Madej, M., Flower, M., Tedesco, F.S., Muntoni, F. and Ferretti, P. (2022) Dystrophin deficiency affects human astrocyte properties and response to damage. Glia, 70, 466–490. [DOI] [PubMed] [Google Scholar]
- 47. Holder, E., Maeda, M. and Bies, R.D. (1996) Expression and regulation of the dystrophin Purkinje promoter in human skeletal muscle, heart, and brain. Hum. Genet., 97, 232–239. [DOI] [PubMed] [Google Scholar]
- 48. Giocanti-Auregan, A., Vacca, O., Bénard, R., Cao, S., Siqueiros, L., Montañez, C., Paques, M., Sahel, J.A., Sennlaub, F., Guillonneau, X.et al. (2016) Altered astrocyte morphology and vascular development in dystrophin-Dp71-null mice. Glia, 64, 716–729. [DOI] [PubMed] [Google Scholar]
- 49. Fort, P., Estrada, F.J., Bordais, A., Mornet, D., Sahel, J.A., Picaud, S., Vargas, H.R., Coral-Vázquez, R.M. and Rendon, A. (2005) The sarcoglycan-sarcospan complex localization in mouse retina is independent from dystrophins. Neurosci. Res., 53, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rau, F., Lainé, J., Ramanoudjame, L., Ferry, A., Arandel, L., Delalande, O., Jollet, A., Dingli, F., Lee, K.Y., Peccate, C.et al. (2015) Abnormal splicing switch of DMD’s penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat. Commun., 6, 7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sadoulet-Puccio, H.M., Rajala, M. and Kunkel, L.M. (1997) Dystrobrevin and dystrophin: an interaction through coiled-coil motifs. Proc. Natl. Acad. Sci. U. S. A., 94, 12413–12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ponting, C.P., Blake, D.J., Davies, K.E., Kendrickjones, J. and Winder, S.J. (1996) ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci., 21, 11–13. [PubMed] [Google Scholar]
- 53. Keegan, N.P. (2020) Pseudoexons of the DMD gene. J. Neuromuscul. Dis., 7, 77–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tuffery-Giraud, S., Miro, J., Koenig, M. and Claustres, M. (2017) Normal and altered pre-mRNA processing in the DMD gene. Hum. Genet., 136, 1155–1172. [DOI] [PubMed] [Google Scholar]
- 55. Zhao, J., Kodippili, K., Yue, Y., Hakim, C.H., Wasala, L., Pan, X., Zhang, K., Yang, N.N., Duan, D. and Lai, Y. (2016) Dystrophin contains multiple independent membrane-binding domains. Hum. Mol. Genet., 25, 3647–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Surono, A., Takeshima, Y., Wibawa, T., Pramono, Z.A.D. and Matsuo, M. (1997) Six novel transcripts that remove a huge intron ranging from 250 to 800 kb are produced by alternative splicing of the 5′ region of the dystrophin gene in human skeletal muscle. Biochem. Biophys. Res. Commun., 239, 895–899. [DOI] [PubMed] [Google Scholar]
- 57. Jacob, A.G. and Smith, C.W.J. (2017) Intron retention as a component of regulated gene expression programs. Hum. Genet., 136, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Allen, D.G., Whitehead, N.P. and Froehner, S.C. (2016) Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev., 96, 253–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bulman, D.E., Murphy, E.G., Zubrzycka-Gaarn, E.E., Worton, R.G. and Ray, P.N. (1991) Differentiation of Duchenne and Becker muscular dystrophy phenotypes with amino- and carboxy-terminal antisera specific for dystrophin. Am. J. Hum. Genet., 48, 295–304. [PMC free article] [PubMed] [Google Scholar]
- 60. Helliwell, T.R., Ellis, J.M., Mountford, R.C., Appleton, R.E. and Morris, G.E. (1992) A truncated dystrophin lacking the C-terminal domains is localized at the muscle membrane. Am. J. Hum. Genet., 50, 508–514. [PMC free article] [PubMed] [Google Scholar]
- 61. Kumari, A., Sedehizadeh, S., Brook, J.D., Kozlowski, P. and Wojciechowska, M. (2022) Differential fates of introns in gene expression due to global alternative splicing. Hum. Genet., 141, 31–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jones, H.F., Bryen, S.J., Waddell, L.B., Bournazos, A., Davis, M., Farrar, M.A., McLean, C.A., Mowat, D.R., Sampaio, H., Woodcock, I.R.et al. (2019) Importance of muscle biopsy to establish pathogenicity of DMD missense and splice variants. Neuromuscul. Disord., 29, 913–919. [DOI] [PubMed] [Google Scholar]
- 63. Deburgrave, N., Daoud, F., Llense, S., Barbot, J.C., Récan, D., Peccate, C., Burghes, A.H.M., Béroud, C., Garcia, L., Kaplan, J.C.et al. (2007) Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum. Mutat., 28, 183–195. [DOI] [PubMed] [Google Scholar]
- 64. Pillers, D.A.M., Fitzgerald, K.M., Duncan, N.M., Rash, S.M., White, R.A., Dwinnell, S.J., Powell, B.R., Schnur, R.E., Ray, P.N., Cibis, G.W.et al. (1999) Duchenne/Becker muscular dystrophy: correlation of phenotype by electroretinography with sites of dystrophin mutations. Hum. Genet., 105, 2–9. [DOI] [PubMed] [Google Scholar]
- 65. Young, H.K., Barton, B.A., Waisbren, S., Portales Dale, L., Ryan, M.M., Webster, R.I. and North, K.N. (2008) Cognitive and psychological profile of males with Becker muscular dystrophy. J. Child Neurol., 23, 155–162. [DOI] [PubMed] [Google Scholar]
- 66. Araki, E., Nakamura, K., Nakao, K., Kameya, S., Kobayashi, O., Nonaka, I., Kobayashi, T. and Katsuki, M. (1997) Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem. Biophys. Res. Commun., 238, 492–497. [DOI] [PubMed] [Google Scholar]
- 67. Ratnapriya, R., Sosina, O.A., Starostik, M.R., Kwicklis, M., Kapphahn, R.J., Fritsche, L.G., Walton, A., Arvanitis, M., Gieser, L., Pietraszkiewicz, A.et al. (2019) Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet., 51, 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liao, Y., Smyth, G.K. and Shi, W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- 69. Chen, Y., Lun, A.T.L. and Smyth, G.K. (2014) Differential expression analysis of complex rna-seq experiments using edgeR. In Datta, S. and Nettleton, D. (eds), Statistical Analysis of Next Generation Sequencing Data. Frontiers in Probability and the Statistical Sciences. Springer, Cham, pp. 51–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.