Introduction
In his 2015 State of the Union Address, former President Obama announced the Precision Medicine Initiative signaling continued U.S. government support toward individualized disease detection, prevention, and management strategies.1 Pharmacogenomics is one component of precision medicine and promises to optimize drug therapy through the incorporation of an individual’s genetic information in drug prescribing decisions. Genotype specifically influences pharmacokinetics and pharmacodynamics and allows for prediction of risk for adverse drug effects and likelihood of drug effectiveness.
Following decades of research into genetic determinants of drug response, pharmacogenomics is entering clinical practice at institutions across the U.S. and Europe.2-11 Guidelines by the Clinical Pharmacogenetics Implementation Consortium (CPIC) and Dutch Pharmacogenetics Working Group (DPWG) have facilitated the adoption of genotype-guided therapy by informing implementation priorities and strategies.12,13 These guidelines specifically provide recommendations for how to translate genotype results to prescribing decisions for gene-drug pairs with evidence supporting their incorporation in clinical practice.
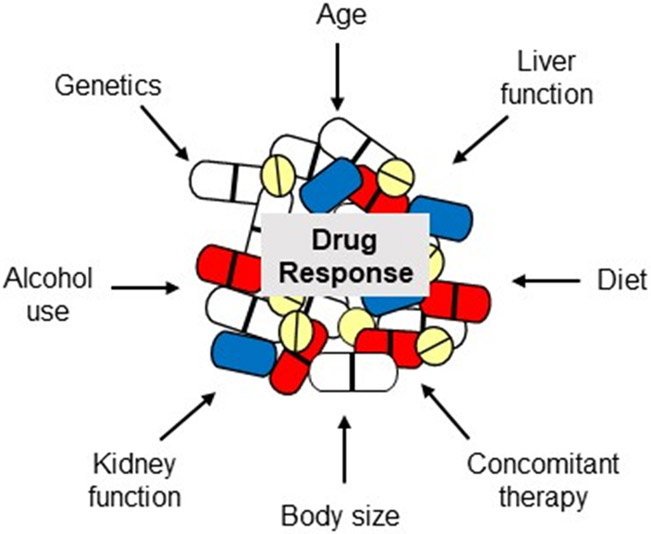
Clinical validity is well established for gene-drug pairs addressed by the CPIC and DPGW. However, evidence supporting the clinical utility of testing is much more limited. While randomized controlled trials (RCTs) are considered the gold standard for establishing clinical utility and informing treatment guidelines, few have been done in the field of pharmacogenomics. Some in fact argue that RCTs should not be the level of evidentiary support required for pharmacogenomic implementation.14-16 Rather, genotype may be viewed as one of a number of patient-specific factors that influence drug response, as shown in Figure 1. Laboratory tests such as serum creatinine, hemoglobin, and serum potassium are routinely ordered prior to initiating drug therapy to guide drug and dose selection in the absence of RCT evidence to support this approach. Genotype is essentially another laboratory test that can be considered in the context of other patient-specific factors to enable individualized drug therapy. Nonetheless, demonstrating value with genotype-guided therapy is needed to influence policy makers and key stakeholders in genomic medicine, including providers and third party payers. Herein, we discuss existing data and on-going efforts to generate evidence in support of genotype-guided therapy approaches and reimbursement for pharmacogenomic testing.
Figure 1.
Patient-Specific Factors commonly influencing drug response
Building Evidence to Support Pharmacogenomic Implementation
Randomized Controlled Trial Evidence
Table 1 summarizes gene-drug pairs investigated in RCTs, which include HLA-B*57:01-abacavir, TPMT-thiopurines, and CYP2C9/VKORC1-warfarin. In the case of abacavir, the PREDICT-1 trial of nearly 2,000 patients showed that prospective screening for the HLA-B*57:01, with avoidance of abacavir in patients testing positive for the allele, resulted in a significant reduction in the risk for immunologically confirmed hypersensitivity reactions, with a 100% negative predictive value.17 Based on these data, the Food and Drug Administration (FDA), European Medicines Agency, CPIC, DPWG, and Human Immunodeficiency Virus treatment guidelines recommend HLA-B*57:01 screening prior to initiation of abacavir-containing regimens.18,19 Similarly, TPMT-guided thiopurine dosing was shown to reduce the risk for adverse hematologic reactions in variant allele carriers, with CPIC and DPWG guidelines available and TPMT genotyping routinely incorporated into thiopurine dosing decisions at many institutions.3,7,8,20,21
Table 1.
Examples of randomized controlled trials of genotype-guided therapies
| Gene-Drug Pair | Trial Acronym | Patient Population | Summary of Results |
|---|---|---|---|
| HLA-B*57:01-Abacavir | PREDICT-117 | 1956 patients with HIV randomized to prospective genetic screening with avoidance of abacavir in patients screening positive or to abacavir use without screening | Immunologically confirmed hypersensitivity occurred in 0% of patients in the prospective-screening group vs. 2.7% of controls, p<0.001 |
| TPMT-Thiopurines | TOPIC20 | 783 patients with inflammatory bowel disease randomized to pretreatment screening with thiopurine dose reduction in carriers of a variant allele according to DPWG guidelines or to usual thiopurine dosing without screening | Adverse hematologic reactions did not differ between genotype versus control patients in the population overall, but were less common in TPMT variant allele carriers in the genotype versus control group (2.6% vs 22.9%, RR, 0.11, 95% CI 0.01-0.85) |
| CYP2C9/VKORC1-warfarin | COAG59 | 1015 patients randomized to genotype-guided dosing or clinically-guided dosing | Mean percent of time in therapeutic range in the first 4 weeks was similar between the genotype- and clinically-guided groups (45.2% and 45.4%) for the population overall. Among Blacks, the mean time in range was lower with genotype- vs. clinically-guided dosing (35.2% vs 43.5%, p=0.01) |
| EU-PACT23 | 455 patients randomized to genotype-guided dosing or fixed dosing (5-10 mg day 1, then 5 mg/day days 2-3, then adjustment based on INR) | Mean percent of time in therapeutic range in the initial 12 weeks was lower in the genotype group vs the control group (60.3% vs 67.4%, p<0.001) | |
| GIFT24 | 1597 older patients (≥65 years) undergoing elective hip or knee arthroplasty randomized to genotype-guided dosing or clinically-guided dosing | The rate of the composite outcome of death, venous thromboembolism, major bleeding, or INR ≥4 during the initial 4-6 weeks was 10.8% in the genotype-guided arm and 14.7% in the clinically-guided arm, representing a 27% reduction in the primary endpoint | |
| CYP3A5-Tacrolimus | Not available | 280 renal transplant recipients randomized to CYP3A5-guided tacrolimus dosing or standard dosing60 | A higher proportion of patients in the genotype-guided group achieved therapeutic tacrolimus levels at day 10 post-transplant compared to those in the standard dosing group (43.2% vs 29.1%, p=0.03). |
| CYP2C19-clopidogrel | TAILOR-PCI (ClinicalTrials.gov ID NCT01742117) | 5270 patients undergoing PCI for an acute coronary syndrome or stable coronary disease randomized to CYP2C19 genotype-guided antiplatelet therapy, with clopidogrel avoided in carriers of a nonfunctional allele, or to routine clopidogrel | Estimated to be completed in 2020. The primary outcome is the occurrence of a major adverse cardiovascular event, defined as nonfatal myocardial infarction, non-fatal stroke, cardiovascular mortality, severe recurrent ischemia, and stent thrombosis at one year |
| POPular Genetics (ClinicalTrials.gov ID NCT01761786)44 | 2700 patients with ST-segment elevation myocardial infarction undergoing PCI randomized to CYP2C19-genotype guided antiplatelet therapy, with clopidogrel avoided in carriers of a nonfunctional allele, or to routine ticagrelor or prasugrel | Estimated to be completed in 2019. The primary endpoint is the composite of death, myocardial infarction, stent thrombosis, stroke, and major bleeding at one year. |
CI, confidence interval; DPWG, Dutch Pharmacogenetics Working Group; INR, international normalized ratio; HIV, human immunodeficiency virus; PCI, percutaneous coronary intervention; RR, relative risk
Genotype-guided warfarin dosing is probably the most extensively studied pharmacogenomic intervention, with three large RCTs investigating the efficacy of genotype-guided warfarin dosing.22-24 Only the most recent trial, GIFT, had sufficient power to examine clinical outcomes with warfarin pharmacogenomics, whereas previous trials focused on the endpoint of time in therapeutic range. Among patients who underwent elective hip or knee arthroplasty, GIFT demonstrated a 27% relative risk reduction in the composite outcome of death, venous thromboembolism, major bleeding, or an international normalized ratio (INR) ≥ 4 with a genotype-guided dosing approach versus dosing based on clinical factors.24 These findings are consistent with data from the EU-PACT trial, which demonstrated greater time in the therapeutic INR range with genotype-guided dosing compared to a traditional dosing approach.23 In contrast, the COAG trial, which included a more racially diverse population than either the EU-PACT trial or GIFT, found no difference in time in therapeutic INR range between genotype-guided dosing versus clinically-guided dosing.22 In the subset of African Americans, genotype-guided dosing led to lower time in therapeutic range compared to clinical dosing, which is probably because the study did not genotype for many of the variants influencing warfarin dose requirements in African Americans.25 Other factors that may have contributed to disparate findings across studies are summarized elsewhere.26,27
CPIC guidelines for genotype-guided warfarin dosing were originally published in 2011 prior to the release of data from large RCTs.28 Even so, the guidelines strongly recommended using genotype data to dose warfarin when such data are available based on the strong and consistent evidence that genotype influences dose requirements. The guidelines were updated in 2018 to emphasize the importance of genotyping persons of African ancestry for variants important in this population.29 However, based on the disparate results of the EU-PACT and COAG trials and controversy this generated, there are few examples of genotype-guided warfarin dosing in practice, and most of these involve pharmacogenomic panel-based testing where warfarin-related genotypes are included in among numerous other genotypes with implications for other drug responses.2,6,8,30
Alternative Methods of Evidence Generation
With the rapidly growing number of discoveries in the field of pharmacogenomics, conducting a RCT for each gene-drug pair is impractical from a time and cost perspective and a more efficient approach is needed to generate evidence to support translation into patient care. Pharmacogenomic clinical trials are especially challenging since for any given gene-drug pair only a small portion of the population carries a variant allele(s) associated with drug toxicity or ineffectiveness. Otherwise the drug would never have reached the market. In this regard, pharmacogenomics is a study of outliers, and very large study populations may be needed to detect significant effects. A RCT may be also unethical when the consequences of drug exposure in a genetically predisposed patient can be life-threatening. Such is the case with the HLA-B*15:02 allele, which is found most often in persons of southeastern Asian ancestry and significantly increases the risk for carbamazepine-induced Stevens-Johnson syndrome and toxic epidermal necrolysis. In lieu of a RCT, a prospective cohort study of 4877 carbamazepine candidates was conducted and showed that, compared to historical incidence rates, genetic screening with avoidance of carbamazepine in HLA*15:02 positive patients significantly reduced the occurrence of severe cutaneous reactions.31 The FDA-approved carbamazepine labeling includes a boxed warning about the risk for severe cutaneous reactions in individuals with the HLA-B*15:02 allele and states that genotyping should be done prior to carbamazepine use in at-risk populations (i.e. southeast Asian populations). Similar data exist with HLA-B*58:01 screening to predict risk for several cutaneous adverse reactions to allopurinol.32 Guidelines by both CPIC and the Canadian Pharmacogenomics Network for Drug Safety address risk associated with the HLA-B genotype for carbamazepine, and CPIC provides additional guidelines for allopurinol.33-35
One alternative approach to an RCT is to gather evidence as part of continuing clinical care in a learning health system model through practice-based pragmatic studies.36,37 Unlike RCTs, pragmatic studies are conducted in the context of clinical practice and reflect the effectiveness of an intervention in a real-world setting.38 As such, an advantage of pragmatic study results is that they are more generalizable than those from RCTs, which have strict eligibility criteria and are conducted in controlled settings to minimize selection bias and confounding and maximize internal validity.39 Pragmatic studies are also less rigorous by nature and thus more efficient to conduct. On the other hand, the major limitation with pragmatic studies is that there is less control for sources of bias creating greater uncertainty and necessitating statistical techniques such as propensity score matching to account for differences between treatment groups.39
Pragmatic and Observational Studies of CYP2C19-Guided Antiplatelet Prescribing
CYP2C19-clopidogrel is an example of a gene-drug pair with evidence of benefit from pragmatic studies. Clopidogrel is a prodrug, and the CYP2C19 enzyme has a critical role in clopidogrel bioactivation. Approximately 30% of Whites and African Americans and up to 60% of Asians carry a nonfunctional CYP2C19 allele leading to impaired clopidogrel bioactivation and lesser clopidogrel-mediated effects, the consequences of which are greatest for patients undergoing percutaneous coronary intervention (PCI).40-42 Specifically, numerous studies have demonstrated an increased risk for adverse cardiovascular events after PCI in clopidogrel-treated patients with a nonfunctional allele compared to similarly treated patients with normal or increased function alleles.40,43 Two RCTs investigating the clinical utility of CYP2C19-guided clopidogrel prescribing after PCI are going but not expected to be completed until 2019 to 2020 (ClinicalTrials.gov Identifier: NCT01742117 and NCT01761786).44
A number of institutions have clinically implemented CYP2C19 testing to guide post-PCI antiplatelet therapy ahead of clinical trial results.45 In fact, CYP2C19-clopidogrel is one of the most common gene-drug pairs implemented in practice.8 As part of the National Institutes of Health-funded Implementing GeNomics In pracTicE (IGNITE) Network Pharmacogenomics Working Group, seven U.S. institutions pooled data for 1,850 patients who were genotyped for CYP2C19 variants at the time of emergent or elective PCI, with genotype results placed in the electronic health record.8,46,47 Consistent with a pragmatic study design, alternative antiplatelet therapy (e.g. prasugrel or ticagrelor) was recommended in patients with one or two nonfunctional alleles (i.e. intermediate or poor metabolizers), but the ultimate prescribing decision was left to the discretion of the physician. Approximately 31% of patients had a nonfunctional allele, and alternative therapy was prescribed in 61% of these patients, while the remainder were treated with clopidogrel. In contrast, only 15.5% of patients without a nonfunctional allele were prescribed prasugrel or ticagrelor. After propensity scoring to account for differences between groups, the risk for major adverse cardiovascular events (defined as the composite outcome of death, myocardial infarction, or ischemic stroke) over a median follow-up of approximately 5 months, was significantly higher in carriers of a nonfunctional allele prescribed clopidogrel versus alternative therapy (adjusted hazard ratio 2.26, 95% CI 1.18-4.32). There was no difference in risk for cardiovascular events between carriers of a nonfunctional allele prescribed alternative therapy and those without a non-functional allele. The group is currently conducting a cost effectiveness study based on these data.
These data are consistent with findings from a Dutch study of patients genotyped at the time of elective PCI.9 In contrast to the U.S. study, alternative antiplatelet therapy was only recommended in poor metabolizers (with two nonfunctional alleles). Over an 18-month follow-up period, there were significantly more adverse cardiovascular events in poor metabolizers treated with clopidogrel versus alternative antiplatelet therapy. An additional study, conducted in Spain, compared outcomes between patients who received genotype-guided antiplatelet therapy after PCI and historical controls who were mostly treated with clopidogrel.10 Both CYP2C19 and ABCB1 genotypes were determined, and alternative therapy was prescribed to patients with either a CYP2C19 non-functional allele or the ABCB1 rs1045642 TT genotype. Compared to controls who underwent PCI prior to genotype implementation, there were significantly fewer cardiovascular events in the genotype group.
Pragmatic Trial of Pharmacogenomic-Guided Treatment of Depression and Anxiety
Both the CYP2D6 and CYP2C19 genes influence the pharmacokinetics and response to multiple antidepressants, and there are other genes in the serotonergic pathway with potential effects on antidepressant drug response.48,49 The clinical effectiveness of genotype-guided management of depression and anxiety has been the subject of several pragmatic clinical trials and observational studies.50-52 In a recent multi-center study, 685 patients with depression and/or anxiety who were either new to treatment or had inadequately controlled symptoms were randomized to a genotype-guided approach to therapy or usual care.50 Patients were blinded to treatment assignment. Patients in the genotype group were tested using a commercially available panel of 10 genes, including CYP2D6 and CYP2C19. Genotype results were provided to the physician to consider when making drug prescribing decisions. Patients in the genotype arm reported greater response rates for both depression and anxiety at 8 and 12 weeks compared to those in the control group. A smaller pragmatic trial52and observation study51 also showed favorable effects with a genotype-guided strategy of antidepressant prescribing.
Pragmatic Trials of Panel-Based Pharmacogenomic Testing
The examples discussed above focus on individual drugs or drug classes. As of early 2018, CPIC and DPWG guidelines are available for at least 19 gene-drug pairs. Thus, a panel-based approach to testing whereby multiple variants with implications for multiple drugs are tested at once has been proposed as a more practical approach to pharmacogenomic testing than testing genes one at a time. Over 90% of the population is estimated to have at least one variant associated with reduced drug response or increased risk for toxicity, supporting a panel-based approach done preemptively so that genotype data are readily available to inform prescribing decisions across a person’s lifetime.53
Initial efforts have established the feasibility of incorporating multiple genotypes from panel-based testing into clinical care, and pragmatic trials are examining the clinical utility of this strategy.2,3,5,6,11 The INdiana GENomics Implementation: an Opportunity for the UnderServed (INGENIOUS) trial, funded by NHGRI, is examining the economic impact of genotyping for 43 variants in 14 pharmacogenes on the incidence of adverse events and healthcare cost.6 The Ubiquitous Pharmacogenomics Consortium, funded by the European Commission’s Horizon-2929 program, is examining similar outcomes with panel-based pharmacogenomic testing in the PREemptive Pharmacogenomics testing for prevention of Adverse drug REactions (PREPARE) trial.11 The trial is being conducted in seven European countries using a block-randomized design. Participating countries are randomized to an 18-month block of pharmacogenomics-guided prescribing or standard of care, after which they switch to the opposite strategy for a second 18-month block. The trial is targeting enrollment of 4,050 total patients during each block. The pharmacogenomics panel includes 50 variants in 13 genes, and drug therapy recommendations are provided according to DPWG guidelines.
Reimbursement for Pharmacogenetic Testing
Reimbursement for pharmacogenomics testing involves many constantly moving parts. First there must be evidence (i.e. clinical validity) to obtain a Current Procedural Terminology® (CPT) code. Once a CPT® code is obtained, like a catalog number, a value must be assigned to it. Then payers whether governmental or private decide whether or not to reimburse the pharmacogenomics test (or panel). This section will provide an overview of evidence needed for reimbursement for pharmacogenomics.
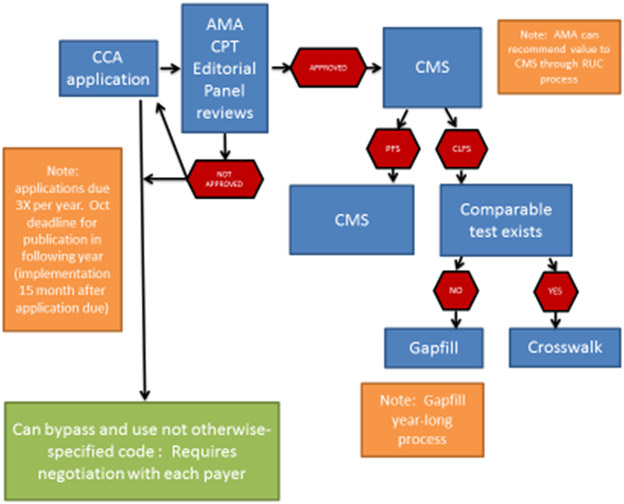
Billing and reimbursement for medical services including pharmacogenetics testing are codified by a code set that is maintained and administered by the American Medical Association (AMA). CPT® codes are considered level 1 Healthcare Common Procedure Coding System (HCPCS, pronounce as “hickspicks”) which is overseen by the Centers for Medicare and Medicaid Services (CMS). To obtain a CPT® code, the medical service must demonstrate clinical validity (Figure 2). CPT® codes are assigned categories. Category I are procedures that are consistent with contemporary medical practice and are widely performed. Category III are temporary codes for emerging technology, services and procedures. For molecular pathology procedures in which pharmacogenomics falls, the AMA created additional criteria to help guide category I status as follows:54
Figure 2.
The process map for requesting a new CPT® from the American Medical Association (AMA) with how the US governmental payer determines a reimbursement value. The CCA = coding change application, CMS = Centers for Medicare and Medicaid Services, PFS = Physician fee schedule, CLFS = Clinical Lab fee schedule, RUC = Specialty Society Relative Value Scale Update Committee or Relative Value Update Committee (pronounced "ruck“)
For Mendelian and somatic disorders, there is a demonstrated relationship between biomarker and phenotype (ie, clinical validity).
Biomarkers (eg, SNPs) that have an association but not a proven causative effect to a known clinical phenotype(s) should have demonstrated clinical usefulness (eg, high positive predictive value, high negative predictive value, directing therapy/management).
The US government, through programs like CMS, pays for approximately half of the country's health care.55 CMS uses Medicare Administrative Contractors (MACs), who are private health care insurers, to administer the Medicare program. The MACs are responsible for various geographic jurisdictions to process Medicare Part A and Part B (A/B) medical claims such as pharmacogenomic testing for Medicare fee-for-service beneficiaries. Medicare Part A is hospital insurance which includes, but not limited to, inpatient care in hospitals, nursing homes, skilled nursing facilities, and critical access hospitals. Medicare Part B covers two types of services: 1) medically necessary services that are needed to diagnose or treat a medical condition and that meet accepted standards of medical practice, and 2) preventive services. Pharmacogenomics testing can occur in both inpatient and outpatient setting and would be covered in Part A/B. Title XVIII of the Social Security Act, Section 1862(a)(1)(A) prohibits Medicare payment "…for items or services which are not reasonable and necessary (often equated to clinical validity) for the diagnosis and treatment of illness or injury…" with certain exceptions. The MACs use a process called Local Coverage Determinations (LCDs) to determine whether they will cover a particular service on a MAC-wide, basis. National Coverage Decisions (NCDs) are issued by CMS for the entire country.
Most if not all the MACs have issued LCDs limiting pharmacogenomics testing reimbursement to few medications and associated gene testing.56 For a medical test, the MACs are using the concept of reasonable and necessary to equate to clinical utility. While there is no standardized definition for clinical utility, in its simplest form, is equated to a change in medical management. Testing for pharmacogenomic variants can determine if a medication is predicted to be effective at current dosage, should be discontinued, or should be a dosing change which should equate to a change in medical management of a patient.
Government reimbursement has an impact on private insurance. Often when the MACs or CMS make LCDs or NCDs to limit coverage on services such as pharmacogenomics testing due to not being reasonable or necessary, private insurance companies often follow these same decisions. This is usually determined by the employer’s contract with the private insurance companies. In some instances, employers have determined that a covering a service may make them a differentiator and save them money in the long term. In addition, because reimbursements from government programs like Medicare and Medicaid are lower than the average cost of serving those patients, laboratories or other providers may charge privately insured patients higher rates in order to recoup their costs. This increase in private sector prices to adjust for government payment levels is called “cost-shifting,” and is a controversial concept.
CPIC evaluates both clinical and research studies of pharmacogenes and administered medications and publishes the drug gene pair clinical validity12. The stance of CPIC is to guide medication dosing or selection if the pharmacogenomic results are already present in the medical record. They do not make statements on whether or not pharmacogenomics testing should be performed prior to or concurrently to medication administration. This is left to the U.S. FDA in the package insert for a medication and to the prescriber to follow. The FDA also can make determinations of a boxed warning, commonly referred to as a “black box” warning for prescribers about serious adverse reactions. It is still up to the medical practice of the prescriber based on their knowledge of the patient to follow (or not) boxed warnings. Even with a boxed warning for pharmacogenomic testing, there is no requirement for testing to be performed. As such, payers would prefer that CPIC support pre-emptive testing in their guidelines.57
Research projects such as the National Institutes of Health-sponsored IGNITE Network and the Ubiquitous Pharmacogenomics Consortium PREPARE trial are using pragmatic trials to build further evidence of clinical utility of pharmacogenomics as well as other genomics. While the IGNITE network as a whole aims to identify and addresses barriers to implementing genomics, the INGenious project when completed will look at the economic value of panel-based pharmacogenomics testing.58 Similar data are expected from the PREPARE trial. Together, these data may demonstrate a cost-savings to the healthcare system with pre-emptive pharmacogenomic testing.
Conclusion
Pharmacogenomic research and implementation continues to be an active area in molecular medicine. In some cases such as with abacavir and HLA-B*57:01, pharmacogenomics can help reduce the frequency of adverse drug reactions (ADRs) by allowing pre-emptive identification of at-risk individuals in whom the drug can be avoided. In other cases, such as with clopidogrel and CYP2C19 testing, pharmacogenomics allows for predicting patients unlikely to respond in whom alternative therapy may be initiated. Together, such examples can have a tremendously positive impact on morbidity and mortality of patients. The trial-and-error approach to prescribing where one-drug-fits-all that is current practice all too often results in a medication being ineffective, thus causing wasted treatment time, high health care and drug costs, and, most importantly, therapeutic failures or alternatively in a medication causing toxicity, also causing significant medical expenditure and threatening patient life and well-being. As additional evidence is gained for the clinical benefit of genotype-guided therapy, it is expected that reimbursement for pharmacogenomic testing will follow of which both are critical to support broader adoption and sustainability of pharmacogenomics in practice.
KEY POINTS.
The evidence gap in outcomes with pharmacogenomic testing and testing reimbursement are major challenges to pharmacogenomic implementation.
While randomized controlled trials (RCTs) are considered the gold standard for establishing clinical utility and informing treatment guidelines, few have been done in the field of pharmacogenomics.
Conducting a RCT for each gene-drug pair is impractical from a time and cost perspective, and more efficient approaches are needed for evidence generation.
Outcomes data with pharmacogenomic testing are emerging from pragmatic and observational studies, and further data are expected from on-going pragmatic clinical trials.
It is expected that reimbursement will follow evidence for benefit with pharmacogenomic testing.
SYNOPSIS.
While pharmacogenomic testing has entered clinical practice in some centers, the demand for evidence of clinical benefit with genotype-guided therapy approaches and reimbursement for testing hinder wider adoption. Few randomized controlled trials have been done in the area of pharmacogenomics, and financial, time, and ethical concerns limit the ability to conduct an RCT for each gene-drug pair example. Thus, more practical approaches are needed, such as evidence generation through practice-based pragmatic studies. This review provides a summary of existing outcomes data and on-going efforts to generate evidence in support of genotype-guided therapy approaches and reimbursement for pharmacogenomic testing.
Funding:
This work is supported by NIH/NHGRI grant U01 HG007269 and NIH/NCATS grant UL1 TR001427 to LHC. VMP is supported by the IGNITE project grant (U01HG007762) and the Indiana University Health – Indiana University School of Medicine Strategic Research Initiative. The Indiana University School of Medicine Pharmacogenomics Laboratory is a fee-for-service clinical laboratory that offers clinical pharmacogenetic testing.
Footnotes
DISCLOSURE STATEMENT
Disclosure of any relationship with a commercial company that has a direct financial interest in subject matter or materials discussed in article or with a company making a competing product
Contributor Information
Larisa H. Cavallari, Department of Pharmacotherapy and Translational Research, Director, Center for Pharmacogenomics Associate Director, Personalized Medicine Program, University of Florida, Gainesville, FL USA.
Victoria M. Pratt, Pharmacogenomics and Molecular Genetics Laboratories, Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis IN USA.
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clin Pharmacol Ther 2017;102:859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014;166C:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson JF, Field JR, Unertl KM, et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin Pharmacol Ther 2016;100:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012;92:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eadon MT, Desta Z, Levy KD, et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin Pharmacol Ther 2016;100:63–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks JK, Stowe D, Willner MA, et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy 2016;36:940–8. [DOI] [PubMed] [Google Scholar]
- 8.Cavallari LH, Beitelshees AL, Blake KV, et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci 2017;10:143–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deiman BA, Tonino PA, Kouhestani K, et al. Reduced number of cardiovascular events and increased cost-effectiveness by genotype-guided antiplatelet therapy in patients undergoing percutaneous coronary interventions in the Netherlands. Neth Heart J 2016;24:589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Ramos J, Davila-Fajardo CL, Toledo Frias P, et al. Results of genotype-guided antiplatelet therapy in patients who undergone percutaneous coronary intervention with stent. Int J Cardiol 2016;225:289–95. [DOI] [PubMed] [Google Scholar]
- 11.van der Wouden CH, Cambon-Thomsen A, Cecchin E, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther 2017;101:341–58. [DOI] [PubMed] [Google Scholar]
- 12.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011;89:464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther 2011;89:662–73. [DOI] [PubMed] [Google Scholar]
- 14.Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol 2010;11:507–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khoury MJ. Dealing with the evidence dilemma in genomics and personalized medicine. Clin Pharmacol Ther 2010;87:635–8. [DOI] [PubMed] [Google Scholar]
- 16.van der Wouden CH, Swen JJ, Samwald M, Mitropoulou C, Schwab M, Guchelaar HJ. A brighter future for the implementation of pharmacogenomic testing. Eur J Hum Genet 2016;24:1658–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358:568–79. [DOI] [PubMed] [Google Scholar]
- 18.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents Living with HIV. Department of Health and Human Services. Available at http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. [Google Scholar]
- 19.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 2011;89:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coenen MJ, de Jong DJ, van Marrewijk CJ, et al. Identification of Patients With Variants in TPMT and Dose Reduction Reduces Hematologic Events During Thiopurine Treatment of Inflammatory Bowel Disease. Gastroenterology 2015;149:907–17 e7. [DOI] [PubMed] [Google Scholar]
- 21.Weitzel KW, Smith DM, Elsey AR, et al. Implementation of Standardized Clinical Processes for TPMT Testing in a Diverse Multidisciplinary Population: Challenges and Lessons Learned. Clin Transl Sci 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013;369:2283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013;369:2294–303. [DOI] [PubMed] [Google Scholar]
- 24.Gage BF, Bass AR, Lin H, et al. Effect of Genotype-Guided Warfarin Dosing on Clinical Events and Anticoagulation Control Among Patients Undergoing Hip or Knee Arthroplasty: The GIFT Randomized Clinical Trial. JAMA 2017;318:1115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drozda K, Wong S, Patel SR, et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet Genomics 2015;25:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavallari LH. Time to revisit warfarin pharmacogenetics. Future Cardiol 2017;13:511–3. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med 2015;25:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther 2011;90:625–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther 2017;102:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nutescu EA, Drozda K, Bress AP, et al. Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy 2013;33:1156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Lin JJ, Lu CS, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med 2011;364:1126–33. [DOI] [PubMed] [Google Scholar]
- 32.Ko TM, Tsai CY, Chen SY, et al. Use of HLA-B*58:01 genotyping to prevent allopurinol induced severe cutaneous adverse reactions in Taiwan: national prospective cohort study. BMJ 2015;351:h4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips EJ, Sukasem C, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin Pharmacol Ther 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito Y, Stamp LK, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin Pharmacol Ther 2016;99:36–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amstutz U, Shear NH, Rieder MJ, et al. Recommendations for HLA-B*15:02 and HLA-A*31:01 genetic testing to reduce the risk of carbamazepine-induced hypersensitivity reactions. Epilepsia 2014;55:496–506. [DOI] [PubMed] [Google Scholar]
- 36.Ginsburg G Medical genomics: Gather and use genetic data in health care. Nature 2014;508:451–3. [DOI] [PubMed] [Google Scholar]
- 37.Rosenthal GE. The role of pragmatic clinical trials in the evolution of learning health systems. Trans Am Clin Climatol Assoc 2014;125:204–16; discussion 17-8. [PMC free article] [PubMed] [Google Scholar]
- 38.Patsopoulos NA. A pragmatic view on pragmatic trials. Dialogues Clin Neurosci 2011;13:217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terry SF. An Evidence Framework for Genetic Testing. Genet Test Mol Biomarkers 2017;21:407–8. [DOI] [PubMed] [Google Scholar]
- 40.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 2013;94:317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mega JL, Close SL, Wiviott SD, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation 2009;119:2553–60. [DOI] [PubMed] [Google Scholar]
- 42.Sorich MJ, Rowland A, McKinnon RA, Wiese MD. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circ Cardiovasc Genet 2014;7:895–902. [DOI] [PubMed] [Google Scholar]
- 43.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 2010;304:1821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergmeijer TO, Janssen PW, Schipper JC, et al. CYP2C19 genotype-guided antiplatelet therapy in ST-segment elevation myocardial infarction patients-Rationale and design of the Patient Outcome after primary PCI (POPular) Genetics study. Am Heart J 2014;168:16–22 e1. [DOI] [PubMed] [Google Scholar]
- 45.Empey PE, Stevenson JM, Tuteja S, et al. Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weitzel KW, Alexander M, Bernhardt BA, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 2016;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavallari LH, Lee CR, Beitelshees AL, et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv 2018;11:181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 2015;98:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley P, Shiekh M, Mehra V, et al. Improved efficacy with targeted pharmacogenetic-guided treatment of patients with depression and anxiety: A randomized clinical trial demonstrating clinical utility. J Psychiatr Res 2018;96:100–7. [DOI] [PubMed] [Google Scholar]
- 51.Brennan FX, Gardner KR, Lombard J, et al. A Naturalistic Study of the Effectiveness of Pharmacogenetic Testing to Guide Treatment in Psychiatric Patients With Mood and Anxiety Disorders. Prim Care Companion CNS Disord 2015;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez V, Salavert A, Espadaler J, et al. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double-blind clinical trial. BMC Psychiatry 2017;17:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Driest SL, Shi Y, Bowton EA, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther 2014;95:423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.American Medical Association. CPT Code Application. (Accessed February 25, 2018, at https://www.ama-assn.org/practice-management/applying-cpt-codes.) [Google Scholar]
- 55.Center for Medicare & Medicaid Services. NHE Fact Sheet. (Accessed February 25, 2018, at https://www.cms.gov/research-statistics-data-and-systems/statistics-trends-and-reports/nationalhealthexpenddata/nhe-fact-sheet.html.) [Google Scholar]
- 56.Center for Medicare & Medicaid Services. Medicare Coverage Database. (Accessed February 25, 2018, at https://www.cms.gov/medicare-coverage-database.) [Google Scholar]
- 57.Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenman MB, Decker B, Levy KD, Holmes AM, Pratt VM, Eadon MT. Lessons Learned When Introducing Pharmacogenomic Panel Testing into Clinical Practice. Value Health 2017;20:54–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimmel SE, Christie J, Kealey C, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J 2008;8:53–60. [DOI] [PubMed] [Google Scholar]
- 60.Thervet E, Loriot MA, Barbier S, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther 2010;87:721–6. [DOI] [PubMed] [Google Scholar]