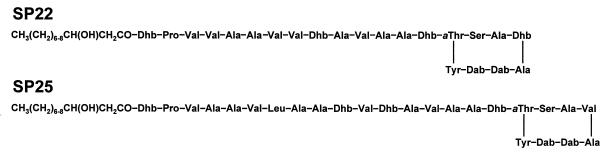Abstract
Coronatine, syringomycin, syringopeptin, tabtoxin, and phaseolotoxin are the most intensively studied phytotoxins of Pseudomonas syringae, and each contributes significantly to bacterial virulence in plants. Coronatine functions partly as a mimic of methyl jasmonate, a hormone synthesized by plants undergoing biological stress. Syringomycin and syringopeptin form pores in plasma membranes, a process that leads to electrolyte leakage. Tabtoxin and phaseolotoxin are strongly antimicrobial and function by inhibiting glutamine synthetase and ornithine carbamoyltransferase, respectively. Genetic analysis has revealed the mechanisms responsible for toxin biosynthesis. Coronatine biosynthesis requires the cooperation of polyketide and peptide synthetases for the assembly of the coronafacic and coronamic acid moieties, respectively. Tabtoxin is derived from the lysine biosynthetic pathway, whereas syringomycin, syringopeptin, and phaseolotoxin biosynthesis requires peptide synthetases. Activation of phytotoxin synthesis is controlled by diverse environmental factors including plant signal molecules and temperature. Genes involved in the regulation of phytotoxin synthesis have been located within the coronatine and syringomycin gene clusters; however, additional regulatory genes are required for the synthesis of these and other phytotoxins. Global regulatory genes such as gacS modulate phytotoxin production in certain pathovars, indicating the complexity of the regulatory circuits controlling phytotoxin synthesis. The coronatine and syringomycin gene clusters have been intensively characterized and show potential for constructing modified polyketides and peptides. Genetic reprogramming of peptide and polyketide synthetases has been successful, and portions of the coronatine and syringomycin gene clusters could be valuable resources in developing new antimicrobial agents.
Pseudomonas spp. produce a wide spectrum of phytotoxic compounds (Table 1). Among the most well-characterized bacterial phytotoxins are those produced by the plant pathogen Pseudomonas syringae. This review summarizes our current understanding of the mechanism of action, biosynthesis, and regulation of four distinct classes of phytotoxins, including the lipodepsipeptides (syringomycins, syringopeptins), coronatines, phaseolotoxin, and tabtoxin.
TABLE 1.
Phytotoxins produced by Pseudomonas spp.
| Toxin | Producing organism | Chemical class or biosynthetic origin | Reference(s) |
|---|---|---|---|
| Coronatine | P. syringae pv. atropurpurea, glycinea, maculicola, morsprunorum, tomato | Polyketide | 108 |
| Corpeptin | P. corrugata | Lipodepsipeptide | 68 |
| Fuscopeptin | P. fuscovaginae | Lipodepsipeptide | 10 |
| Persicomycins | P. syringae pv. persicae | Fatty acid | 17 |
| Phaseolotoxin | P. syringae pv. actinidiae, phaseolicola | Sulfodiaminophosphinyl peptide | 157 |
| Rhizobitoxine | P. andropogonis | Vinylglycine | 167 |
| Syringomycinsa | P. syringae pv. syringae, aptata, atrofaciens | Lipodepsinonapeptide | 12, 13, 77, 275 |
| P. fuscovaginae | 19 | ||
| Syringopeptins | P. syringae pv. syringae | Lipodepsipeptide | 9 |
| Tabtoxin | P. syringae pv. tabaci, coronafaciens, garcae | β-Lactam | 255 |
| Tagetitoxin | P. syringae pv. tagetis | Unknown | 217 |
| Tolaasin | P. tolaasii | Lipodepsipeptide | 211 |
| Viscosin | P. marginalis (P. fluorescens) | Lipodepsipeptide | 134 |
Includes the related toxins syringotoxin, syringostatin, and pseudomycin.
Biology and Pathogenicity of P. syringae
P. syringae is reported to induce a wide variety of symptoms on plants, including blights (rapid death of tissue), leaf spots, and galls. The species is divided into pathogenic variants (pathovars), which vary in host range. Two distinct reactions are possible when P. syringae cells are infiltrated into plant tissue. One potential outcome is a compatible, susceptible interaction which is characterized by a symptom called water soaking, a reaction which is followed by pathogen proliferation and advanced symptom development. In contrast, resistant host cells undergo a reaction known as the hypersensitive response and become necrotic 12 to 24 h after inoculation. A cluster of genes termed the hrp region (for “hypersensitive response and pathogenicity”) is conserved in phytopathogenic prokaryotes and affects the ability of a bacterium to induce a hypersensitive response in nonhost plants, pathogenicity in host plants, and the ability to grow within or on the surface of plants (83). It is important to note that the hrp gene cluster is required for pathogenicity of P. syringae on plant hosts. The hrp genes are known to encode genes for the regulation and biosynthesis of a type III secretion pathway that is similar in both plant and animal pathogens and is used to secrete virulence proteins (228). It is becoming increasingly evident that mechanisms which function in clinical pathogens of animals, such as the type III secretion systems in Salmonella, Shigella, and Yersinia, are similar to those in phytopathogenic species (206, 221). However, in addition to the hrp genes, phytopathogenic pseudomonads encode gene products that significantly enhance pathogen virulence, including extracellular polysaccharides, phytotoxins, cell wall-degrading enzymes, and phytohormones (3, 49, 56, 92).
Phytotoxins Produced by P. syringae
Phytotoxins are products of plant pathogens or of the host-pathogen interaction that directly injure plant cells and influence the course of disease development or symptoms. Both fungal and bacterial pathogens produce a number of secondary metabolites that are toxic to plant cells; however, these metabolites may not be important in plant disease. Consequently, phytopathologists have developed criteria for assessing the involvement of toxins in plant disease. These include (i) reproduction of disease symptoms with the purified toxin, (ii) a correlation between toxin yield and pathogenicity, (iii) production of the toxin during active growth of the pathogen in planta, and (iv) reduced virulence or lack of virulence in nontoxigenic strains. Phytotoxins may be host specific and exhibit the same specificity as the producing pathogen, or they may lack specificity and exhibit a wider host range of activity than the producing pathogen. Most toxins produced by P. syringae lack host specificity and cause symptoms on many plants which cannot be infected by the toxin-producing pathogen.
Visual assessment of phytotoxin production in planta can be somewhat subjective. The phytotoxins produced by P. syringae generally induce chlorosis (coronatine, phaseolotoxin, and tabtoxin) or necrosis (syringomycin and syringopeptin). However, studies of particular phytotoxins are probably influenced by the visible evidence of their activity. Some phytotoxins may instead act by changing metabolic processes in the host in such a way that the deleterious activity might be manifested only at the biochemical level.
Although phytotoxins are not required for pathogenicity in P. syringae, they generally function as virulence factors for this pathogen, and their production results in increased disease severity. For example, P. syringae phytotoxins can contribute to systemic movement of bacteria in planta (198), lesion size (24, 287), and multiplication of the pathogen in the host (24, 75, 172). The phytotoxins produced by P. syringae can substantially enhance the virulence of producing pathogens, even though some disease can occur in their absence.
BIOSYNTHESIS OF PHYTOTOXINS BY NONRIBOSOMAL ENZYME SYSTEMS
The toxins produced by P. syringae are varied in origin and include monocyclic β-lactam (tabtoxin), sulfodiaminophosphinyl peptide (phaseolotoxin), lipodepsinonapeptide (syringomycin), and polyketide (coronatine) structures (163). Knowledge of phytotoxin structure is extremely important since structural information may provide important clues about the biosynthetic processes involved. Fortunately, several P. syringae phytotoxins have structural analogies to antibiotics that are produced via nonribosomal mechanisms in Streptomyces and Bacillus spp. These pathways have served as predictive models for the synthesis of selected phytotoxins.
It has been difficult to obtain information on intermediates in the biosynthetic pathways to various phytotoxins. One reason for the lack of characterized intermediates in these diverse pathways is that nonribosomal synthesis is generally catalyzed by multifunctional proteins or polypeptide complexes and intermediates are transferred between enzymatic domains and not released into the cytoplasm. Furthermore, conversion of intermediates to the final product may occur very rapidly and impede detection and characterization.
Peptide Synthetases
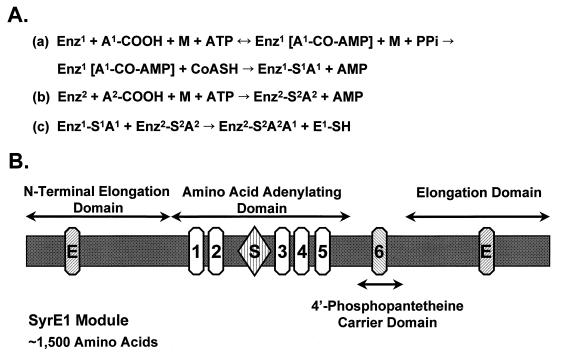
The biosynthesis of nonribosomal peptides has been intensively investigated for a number of years, and there are several excellent reviews on this subject (126, 147, 247, 278). According to the current model, these peptides are synthesized via a thiotemplate mechanism by large multisubunit enzymes ranging from 100 to 1,600 kDa (247). All thiotemplate multienzymatic systems are composed of amino acid-activating domains that catalyze the adenylation of the constituent amino acids and the formation of thioesters. The general sequence of reactions includes (i) carboxyl activation of the substrate amino acid by adenylate formation, (ii) acylation of enzyme-attached pantothenoyl-thiols, and (iii) directed transfer to the next acyl intermediate with condensation (Fig. 1A). The completed peptide is released from the enzyme complex by cyclization, amidation, or hydrolysis (126).
FIG. 1.
(A) Reaction sequence catalyzed by multifunctional peptide synthetases. (a) Carboxyl activation of the first amino acid (A1) and formation of the aminoacyl adenylate; (b) activation of the second amino acid (A2) and formation of the aminoacyl adenylate; and (c) the condensation reaction. Abbreviations: Enz, enzyme; A, amino acid, M, divalent metal ion (Mg2+, Mn2+, Ca2+). Numbering indicates specific domains within an individual multienzyme; for example, Enz1 and Enz2 are two distinct domains within the same enzyme. Amino acids (A1 and A2) are two individual amino acids. For additional information, see reference 278. (B) Domain structure of the amino acid-activating module SyrE1. The SyrE1 module contains approximately 1,500 amino acids organized into four domains as defined by Stein and Vater (252). The relative positions of conserved core sequences are shown for each domain. The two elongation domains contain a characteristic HHxxxDG motif (E) (54). The five core sequences described by Stachelhaus and Marahiel (247) are located within the amino acid-adenylating domain. Core 2 has a sequence (SGTTGxPKGV) resembling the Walker type A motif involved in ATP binding, and the core 4 sequence (TGD) carries a motif associated with ATPase activity. A role in catalyzing aminoacyl adenylate formation is suggested for cores 3 and 5 (247). A region between cores 2 and 3 is associated with substrate recognition (S) (47), and SyrE1 exhibits substrate specificity for l-Ser (95). Core 6, with a characteristic LGGHSL motif, is located in the 4′-phosphopantetheine carrier domain. The motif contains a conserved serine to which the 4′-phosphopantetheine cofactor is covalently attached, which in turn is the site of thioester formation. The next module, SyrE2, carrying three domains (i.e., amino acid adenylating, 4′-phosphopantetheine carrier, and elongation domains), follows the SyrE1 module.
The isolation, sequencing, and characterization of genes encoding multifunctional peptide synthetases has indicated a multidomain arrangement in which an adenylation domain consisting of 600 amino acid residues is highly conserved and repeated. Biochemical studies with specific domains have confirmed the multidomain structure predicted by sequence data (147). An elongation domain of about 500 amino acids separates successive adenylation domains (54). The organization of the multifunctional peptide synthetase is colinear to the amino acid sequence of the corresponding peptide product (247). Turgay et al. (266) described a superfamily of adenylate-forming enzymes which includes all peptide synthetases and several adenylating enzymes. A typical adenylation-thiolation module of a peptide synthetase contains a series of conserved sequences with the same order and spacing (252). Five regions originally designated as core motifs (266) are conserved in the adenylation domain of peptide synthetases (Fig. 1B). Although the function of core 1 remains unknown, cores 2 to 5 are presumed to be involved in ATP binding and hydrolysis (Fig. 1B). Core 2 has a glycine-rich sequence that contains a potential phosphate-binding loop, whereas core 4 shows relatedness to ATPases. Conti et al. (47) identified a substrate-binding pocket between cores 2 and 3 of a gramicidin S synthetase module by analysis of the crystal structure of the adenylation domain. Substrate specificity in various peptide synthetases is presumably mediated by the amino acids lining the substrate-binding pocket. Core 6, which is located in the 4-phosphopantetheine carrier domain adjacent to the adenylation module (Fig. 1B), is associated with covalent binding of the substrate amino acid and contains a serine residue to which the cofactor 4′-phosphopantetheine is covalently attached (147).
The involvement of peptide synthetases and adenylate-forming enzymes in the biosynthesis of coronatine and syringomycin has been established and is discussed in further detail below.
Polyketide Synthases
Polyketides constitute a huge family of structurally diverse natural products including those with antibiotic, chemotherapeutic, and antiparasitic activities. Most of the research on polyketide synthesis in bacteria has focused on compounds synthesized by Streptomyces or other actinomycetes, and several excellent reviews have been recently published (99, 116, 123). However, in addition to coronatine, it is important to note that Pseudomonas produces a variety of antimicrobial compounds from the polyketide pathway, including mupirocin (pseudomonic acid) (73), pyoluteorin (52), and 2,4-diacetylphloroglucinol (14).
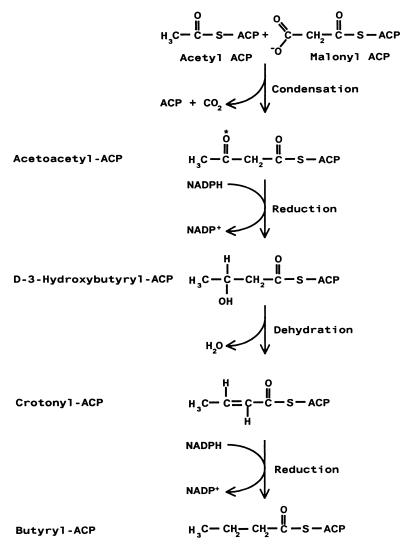
Polyketide synthesis is related to fatty acid biosynthesis; the latter begins with the condensation of acetyl coenzyme A (acetyl-CoA) (starter unit) and malonyl-CoA (chain extender unit) and then proceeds with a cycle of modifications on the carbonyl group of malonyl-CoA (i.e., reduction to a hydroxyl, dehydration to produce a double bond, and further reduction to form a saturated fatty acid) (Fig. 2). Unlike fatty acid synthases, a polyketide synthase (PKS) can accept additional substrates in the starter and extender groups and the products vary in the extent of reduction. PKSs are generally classified as type I or II systems and consist of protein complexes that act on covalently bound substrates that are attached as thioesters to an acyl carrier protein (ACP) (117). Polyketides synthesized by type I PKSs usually have a fairly reduced structure and are synthesized by large multifunctional proteins that consist of individual domains which catalyze specific and discrete reactions in a nonreiterative fashion (Fig. 3) (102). The functional activities catalyzed by domains within the type I PKS are often apparent in the structure of the growing polyketide chain (Fig. 3C), and nucleotide sequencing has become an important tool in predicting the biosynthetic route to polyketides synthesized by a type I PKS. Conversely, type II PKSs are most often associated with synthesis of aromatic polyketides, and biosynthesis occurs on monofunctional proteins that associate in a complex. Unlike the type I system, the type II PKS may utilize one or more enzymes in a reiterative fashion.
FIG. 2.
Reaction sequence in the synthesis of fatty acids. The starting units for the fatty acid synthase are acetyl-CoA and malonyl-CoA; these are converted into acetyl-ACP and malonyl-ACP by acetyl and malonyl transacylase, respectively. The fatty acid synthase proceeds with the condensation of these two precursors and then continues with a cycle of reduction, dehydration, and further reduction of the keto group (asterisk). Two key differences between polyketide synthases (PKS) and fatty acid synthase include the choice of starter units and the extent of reduction.
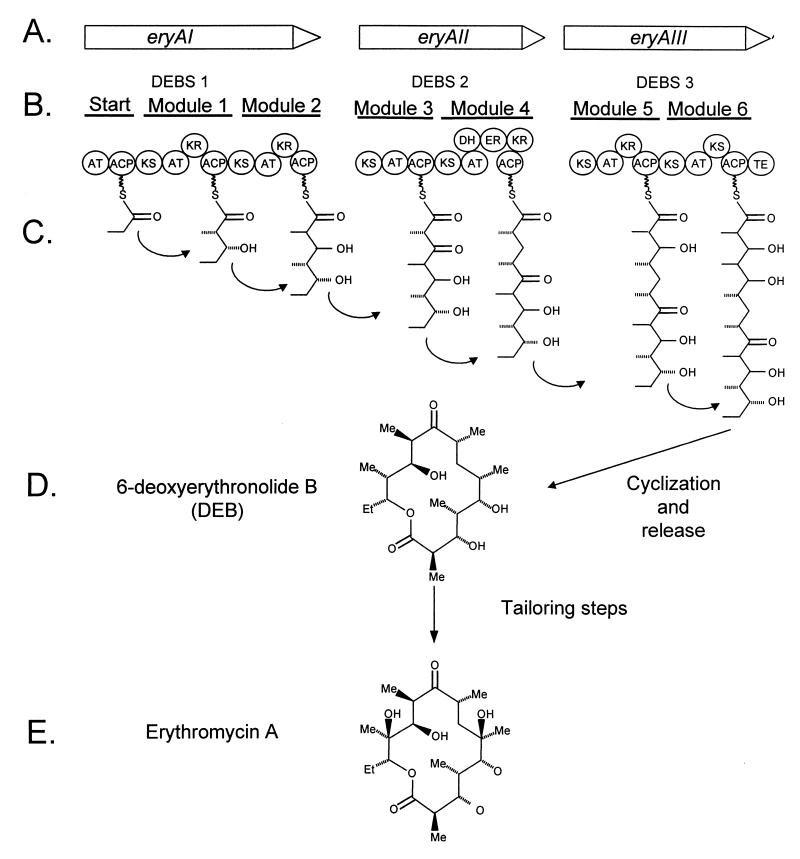
FIG. 3.
Biosynthesis of erythromycin A. (A) The nucleotide sequence of the eryA region contains three ORFs designated eryAI, eryAII, and eryAIII (29, 60). (B) These genes encode three proteins which constitute erythromycin B synthetase (DEBS); these are designated DEBS 1, DEBS 2, and DEBS 3 (39). (C) Each DEBS protein contains two modules, each with domains for acetyltransferase (AT), ACP, and β-keto synthase (KS) activity. Some modules contain additional domains for dehydratase (DH), enoyl reductase (ER), ketoreductase (KR), and TE activity. (D) Cyclization and release of the DEBS 3 product results in the formation of 6-deoxyerythronolide B (DEB), and additional tailoring steps result in the production of erythromycin A (E). Modified from reference 99.
CORONATINE
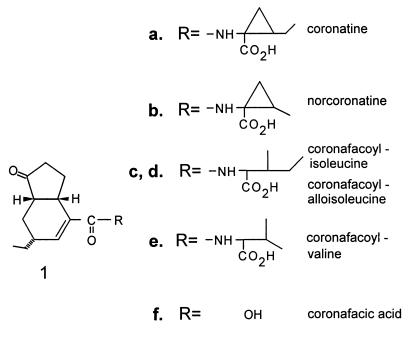
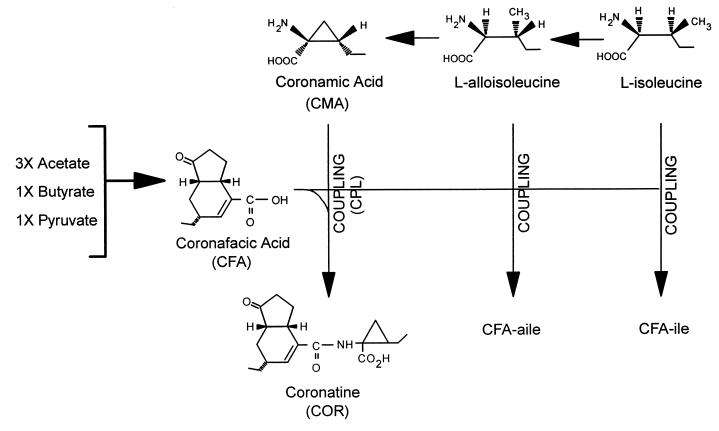
The structure of coronatine (COR) (Fig. 4a) is unusual and has two distinct components: the polyketide coronafacic acid (CFA) (Fig. 4f) and coronamic acid (CMA), an ethylcyclopropyl amino acid derived from isoleucine (108, 160, 196). COR is generally the predominant coronafacoyl compound synthesized by COR producers and also the most toxic; however, other coronafacoyl compounds which contain various amino acid substituents conjugated to CFA via an amide linkage may be synthesized (Fig. 4b to e) (161, 165, 166, 169).
FIG. 4.
Structures of COR and coronafacoyl compounds.
Generalized Biosynthetic Route
Precursor feeding studies with 13C-labeled substrates demonstrated that CFA is a novel polyketide synthesized from one unit of pyruvate, one unit of butyrate, and three acetate residues (196) (Fig. 5). Recent studies have suggested that the pyruvate used for CFA biosynthesis is converted into α-ketoglutarate before incorporation to CFA and that α-ketoglutarate may serve as the starter unit for CFA assembly (194). Little information is available about potential intermediates in the biosynthetic route to CFA, probably because such intermediates remain enzyme bound. However, Mitchell et al. (171) have identified a cyclopentenone compound, 2-(1-oxo-2-cyclopenten-2-ylmethyl)butanoic acid, which may function as an intermediate or shunt product of the CFA biosynthetic pathway.
FIG. 5.
Biochemical pathways involved in the synthesis of COR and coronafacoyl compounds in P. syringae pv. glycinea PG4180. COR consists of a polyketide component, CFA, coupled (CPL) via amide-bond formation to an amino acid component, CMA. CFA is synthesized as a branched polyketide from three acetate units, one pyruvate unit, and one butyrate unit via an unknown sequence of events (196). CMA is derived from isoleucine via alloisoleucine and cyclized by an unknown mechanism (160, 195). CMA functions as an intermediate in the COR biosynthetic pathway, which indicates that cyclization of l-alloisoleucine to form CMA occurs before CFA and CMA are coupled (170). The coronafacoyl analogues, CFA-Ile and CFA-aIle result from amide bond formation between CFA and isoleucine and alloisoleucine, respectively, and are not utilized further in the synthesis of COR.
Parry et al. (195) provided an important clue about the route to CMA by demonstrating that l-alloisoleucine was a more immediate precursor to CMA than was isoleucine. Initially, two possible pathways to COR from CFA were proposed; one route involved the direct coupling of isoleucine (or alloisoleucine) to CFA to form the coronafacoyl conjugates coronafacoylisoleucine (CFA-Ile), and coronafacoylalloisoleucine (CFA-aIle), followed by an oxidative cyclization on the amino acid moiety of the conjugate to form COR (289). This scheme was proposed based on the natural occurrence of CFA-Ile and CFA-aIle in a variety of COR producers (169). An alternative route involved the isomerization of isoleucine to form alloisoleucine, cyclization of alloisoleucine to form CMA, and conjugation of CFA and CMA via amide bond formation (Fig. 5). Support for the latter route developed from the demonstration of CMA as a defined intermediate in the COR pathway (170). The biosynthetic block to COR in several mutants was eliminated when CMA was exogenously supplied, and other mutants were found to excrete CMA when CFA synthesis was blocked (22, 170). Furthermore, CFA-negative mutants could produce COR when supplied with exogenous CFA but not with CFA-Ile or CFA-aIle, indicating that the latter compounds were not operative in coronatine synthesis (170). Our current understanding of the COR biosynthetic pathway is summarized in Fig. 5.
The final step in the pathway to COR is presumed to be the ligation or coupling of CFA and CMA by an amide linkage. The enzyme(s) catalyzing this reaction is thought to lack rigid specificity for the amino acid substrate since a variety of coronafacoyl-amino acid conjugates have been isolated, including CFA-Ile, CFA-aIle, coronafacoylvaline, norcoronatine, and CFA conjugated to serine and threonine (161, 165, 166, 169).
Biological Effects and Mode of Action
The primary symptom elicited by COR is a diffuse chlorosis that can be induced on a wide variety of plant species (82). Interestingly, the reaction of Arabidopsis thaliana to exogenously applied COR is atypical; instead of chlorosis, anthocyanins accumulate at the site of inoculation and the tissue develops a strong purple hue (27). COR is also known to induce hypertrophy, inhibit root elongation, and stimulate ethylene production (74, 122, 226, 277). Several research groups have noted the remarkable structural and functional homologies between COR and methyl jasmonate (MeJA), a plant growth regulator derived from the octadecanoid signaling pathway which is elicited by biological stress (241, 280). COR and MeJA induce analogous biological responses in Arabidopsis seedlings, Eschscholtzia californica cell cultures, and potato tissue; these results have led researchers to suggest that COR functions as a molecular mimic of the octadecanoid signaling molecules produced by higher plants (75, 85, 129, 276, 283). Furthermore, Feys et al. (75) generated a coronatine-insensitive (coi1) mutant of Arabidopsis that was insensitive to the effects of both COR and MeJA, suggesting a similar mode of action.
Light microscopy was used to compare the effects of COR, CFA, and MeJA on tomato tissue (190). Several changes were induced in tomato tissue exposed to the phytotoxin; for example, the epidermal wall was significantly thicker in COR-treated tissue and the chloroplasts stained more intensively and were smaller (190). One of the most pronounced differences was the appearance of spherical and cubical proteinaceous structures in the vacuole of COR-treated tomato tissue. These structures were markedly similar to the proteinase inhibitors which had been previously found in plant tissues exposed to various biological stresses (2, 243, 244). The presence of proteinase inhibitors in the COR-treated tissue was confirmed by demonstrating that this tissue significantly inhibited the activity of both chymopapain and chymotrypsin (190). Recently, polyclonal antibodies to both chymopapain (34) and chymotrypsin (225) inhibitors were used to confirm the identity of the proteinaceous structures in COR-treated tomato tissue. When COR-treated tissue was incubated with antisera to chymopapain inhibitor and then with a secondary antibody conjugated to gold, the cubical crystals were densely labeled with gold particles, indicating that these structures were chymopapain inhibitor (188). A similar experiment indicated that the spherical crystals were chymotrypsin inhibitor (188); thus, we concluded that both chymopapain and chymotrypsin inhibitors are specifically induced in response to COR in tomato tissue.
Although COR, CFA, and MeJA induced the production of proteinase inhibitors, only COR caused cell wall thickening, changes in chloroplast structure, and chlorosis; CFA and MeJA did not induce these changes in tomato tissue (190). Consequently, the CMA moiety, or perhaps the amide linkage between CFA and CMA, may impart additional biological activities to COR in tomato. Further differentiation of COR and MeJA was demonstrated by Krumm et al. (131), who showed that jasmonic acid and COR induce the production of distinctly different volatile compounds in Phaseolus lunatus. Therefore, COR does not function solely as a molecular mimic of MeJA in some plant species, and the mechanism of action of COR may remain unclear until putative receptors for the toxin are localized in various plant species.
Genetic Studies and Involvement of Plasmids in Production
Production of the phytotoxin COR has been demonstrated in five pathovars of P. syringae, i.e., pv. atropurpurea, glycinea, maculicola, morsprunorum, and tomato, which infect ryegrass, soybean, crucifers, Prunus spp., and tomato, respectively (159, 168, 282). Although production of COR outside the species P. syringae is thought to be rare, Xanthomonas campestris pv. phormiicola, a pathogen of New Zealand flax, also produces several coronafacoyl compounds (162, 261).
Tn5 mutagenesis has been used to obtain COR-defective (COR−) mutants of P. syringae pv. atropurpurea, glycinea, morsprunorum, and tomato (23, 25, 176, 289). In several studies, COR was shown to play a distinct role in virulence (24, 172, 229); however, it is important to note that strains of P. syringae pv. glycinea, maculicola, morsprunorum, and tomato that do not produce COR have been isolated (159, 168, 271). Several reports have shown that the COR biosynthetic cluster occurs on indigenous plasmids (23, 25, 138, 229, 297); consequently, the potential instability of plasmid-located COR genes might explain the variability in COR production among strains of P. syringae (51, 271). Although the COR gene cluster has been frequently associated with large (80- to 110-kb) plasmids, these genes can also be chromosomal (51).
Biosynthesis in P. syringae pv. glycinea PG4180
COR biosynthesis has been intensively studied in P. syringae pv. glycinea PG4180 because this strain is easy to manipulate genetically, consistently synthesizes large amounts of COR in vitro (20 to 40 mg/liter), and infects soybean, a host which is easy to cultivate (26). Transposon mutagenesis indicated that the COR biosynthesis genes in P. syringae pv. glycinea PG4180 are located on a 90-kb plasmid designated p4180A (25). The involvement of p4180A in COR production was demonstrated by transforming this plasmid into two nonproducers of COR, P. syringae pv. syringae PS51 and PS61 (22). Organic acids were then extracted from PS51 and PS61 transformants containing p4180A and analyzed by high-pressure liquid chromatography (HPLC) and combined gas chromatography-mass spectrometry. PS51 and PS61 transformants containing p4180A produced both CFA and COR, indicating that p4180A encodes all genes necessary for the biosynthesis of coronafacoyl compounds in P. syringae (22).
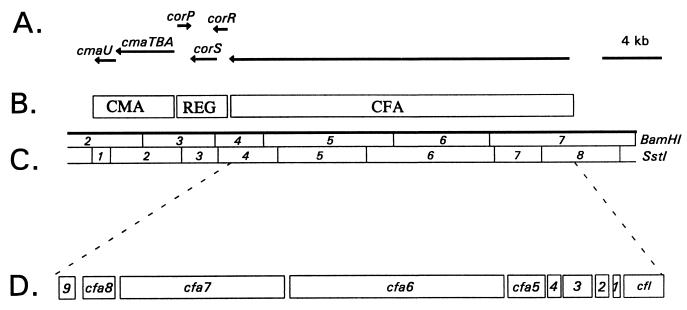
A variety of approaches have been used to characterize the COR biosynthetic cluster encoded by p4180A: (i) saturation Tn5 mutagenesis, (ii) feeding studies using exogenously supplied CFA and CMA, (iii) complementation of selected mutants with cloned DNA, (iv) expression of selected regions of the COR gene cluster in COR nonproducers, and (v) nucleotide sequence analysis (22, 143, 203, 213, 270, 272, 273, 289). Saturation Tn5 mutagenesis indicated that a 32-kb contiguous region was absolutely required for COR biosynthesis, and a physical map was developed by using the restriction enzymes BamHI and SstI (Fig. 6C) (22, 272, 289). Two regions in the COR biosynthetic cluster contained structural genes for CMA and CFA biosynthesis; these were separated by a 3.4-kb regulatory region (REG; Fig. 6B). Transcripts in the COR biosynthetic gene cluster were identified by a combination of the following approaches: (i) complementation of selected mutants with subcloned DNA from the COR biosynthetic cluster, (ii) expression of functional regions of the COR gene cluster in COR nonproducers, (iii) nucleotide sequence and primer extension analyses, and (iv) transcriptional fusions to a promoterless glucuronidase gene (142, 213, 270, 272, 273). These approaches indicated that the COR gene cluster in PG4180 consists of six transcripts (Fig. 6A).
FIG. 6.
Functional and physical map of the COR biosynthetic gene cluster. (A) Horizontal lines with arrowheads indicate the transcriptional organization of the COR gene cluster. (B) Functional regions of the COR biosynthetic cluster: CMA, CMA biosynthetic gene cluster; REG, regulatory region; CFA, CFA biosynthetic gene cluster. (C) Physical map of the COR gene cluster; the enzymes used for restriction mapping were BamHI and SstI. (D) Expanded view of SstI fragments 4 to 8, which contain the CFA biosynthetic gene cluster. Abbreviations: 1, cfa1; 2, cfa2; 3, cfa3; 4, cfa4; 9, cfa9.
The nucleotide sequence of the 6.9-kb region containing the CMA biosynthetic gene cluster revealed the presence of four genes designated cmaA, cmaB, cmaT, and cmaU (37, 197, 270) (Fig. 6A). The CMA biosynthetic gene cluster was shown to encode two transcripts; one transcript was monocistronic and contained cmaU, and the second was polycistronic and contained three cotranscribed genes designated cmaA, cmaB, and cmaT (Fig. 6A). Start sites for both transcripts were determined by primer extension (270). The deduced amino acid sequence of cmaA indicated that the enzyme contains an amino acid-activating domain, whereas cmaB showed extensive homology to syrB2, a gene encoding an enzyme required for syringomycin synthesis (94, 95, 292). The deduced amino acid sequence of cmaT suggested that it functions as a thioesterase (TE), providing further support to the role of a thiotemplate mechanism for CMA biosynthesis (270). CmaT has now been overproduced in E. coli, and assays with a variety of esters and thiolesters indicated that the overproduced protein was a functional esterase in vitro (197). However, sequence analysis of cmaU has failed to reveal anything useful about its potential function in CMA biosynthesis.
A region required for the coupling of CFA and CMA via amide bond formation was sequenced, and a 1.4-kb gene designated cfl (coronafacate ligase) was identified (Fig. 6D) (22, 143). Cfl is most closely related to enzymes that activate carboxylic acids by adenylation; consequently, this enzyme may catalyze the adenylation of CFA and the ligation of the CFA-adenylate to CMA. Coronafacate ligase has been overproduced in Escherichia coli and P. syringae in soluble form (214). Although the precise function of the enzyme remains unclear, construction of a nonpolar mutation in cfl suggested that the enzyme may also function in CFA biosynthesis (214).
Complementation experiments with CFA-defective mutants and an extensive series of subclones demonstrated that the cfl-CFA region was contained in a single 19-kb transcriptional unit (Fig. 6A) (142). Transcriptional fusions to a promoterless glucuronidase gene and primer extension analysis indicated that transcription initiated upstream of cfl in SstI fragment 8 and proceeded through SstI fragment 4 (142, 143) (Fig. 6). The 5′ end of the transcript contains six discrete ORFs including cfl and cfa1 to cfa5 (143, 203) (Fig. 6D). Sequence analysis of cfa1, cfa2, and cfa3 revealed relatedness to ACP, fatty acid dehydratase, and β-ketoacyl synthetase, respectively (203). The function of cfa4 could not be predicted from database searches, whereas the translation product of cfa5 showed relatedness to acyl-CoA ligases (203). Both cfa1 and cfa3 were overexpressed in E. coli, and protein products close to the predicted size were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (203).
The continued sequencing of SstI fragments 5 and 6 indicated the presence of two large open reading frames (ORFs) that were designated cfa6 (8.0 kb) and cfa7 (6.2 kb) (Fig. 6D) (212). Both proteins exhibit a high degree of similarity to 6-deoxyerythronolide B synthase (Fig. 3), suggesting that a type I PKS participates in CFA synthesis. Both Cfa6 and Cfa7 were overproduced in E. coli and shown to encode multifunctional PKSs with antigenic similarity to DEBS 2 (212). Two additional genes, cfa8 and cfa9, mapped downstream of cfa7 (Fig. 6D); cfa8 was required for the biosynthesis of both CFA and COR, and the predicted translational product showed similarity to crotonyl-CoA reductases from Streptomyces spp. (213). Crotonyl-CoA reductase catalyzes one step in the conversion of acetoacetyl-CoA to butyryl-CoA, and the latter product is used as a 4C extender in polyketide synthesis (80). Consequently, the recruitment of a ccr gene into the CFA gene cluster may reflect the requirement of butyryl-CoA as a precursor for CFA synthesis. cfa9 showed relatedness to thioesterases (TEs) involved in the synthesis of gramicidin, tyrocidine, and tylosin (130, 155, 178). Furthermore, Cfa9 contained the GxSxG and GxH motifs characteristic of diisopropyl fluorophosphate-sensitive animal and avian TEs (43). Analysis of a cfa9 mutant indicated that this gene was dispensable for CFA and COR production but may increase the release of enzyme-bound products from the COR pathway (213). The complete nucleotide sequence of the CFA biosynthetic gene cluster has facilitated the development of a model which incorporates the activities of both the mono- and multifunctional proteins (212).
Regulation of Production
A variety of nutritional and environmental factors have been examined for their effect on COR production in P. syringae pv. glycinea PG4180 (189). Temperature had a highly significant effect on COR biosynthesis in P. syringae pv. glycinea PG4180, with maximal production at 18°C and negligible yields at 30°C (189). Interestingly, growth of PG4180 was relatively unaffected over a range of temperatures tested (14 to 30°C). This response to temperature is consistent with symptom development in the field, since P. syringae pv. glycinea is predominantly a cool-weather pathogen. CFA and CMA were also subject to the same pattern of temperature control, with optimal production at 18°C (187, 270). Recently, Rohde et al. (220) showed that COR production was thermoregulated in selected strains of P. syringae pv. atropurpurea, maculicola, morsprunorum, and tomato, which may indicate that temperature is a common regulatory control for COR biosynthesis in other pathovars of P. syringae.
The production of both CFA and CMA in PG4180 is regulated at the transcriptional level by temperature. Transcriptional fusions of the CFA and CMA promoter regions to a promoterless glucuronidase gene indicated that transcriptional activity in both biosynthetic gene clusters was maximal at 18°C and significantly lower at 28°C (142, 191, 214, 270). The higher level of transcriptional activity for the CMA and CFA biosynthetic promoters at 18°C helps explain why COR production is optimal at this temperature.
A regulatory region was isolated which controls both CFA and CMA production; the nucleotide sequence of this region revealed the presence of three genes, corP, corS, and corR (Fig. 6A) (273). The deduced amino acid sequences of corP and corR indicated relatedness to response regulators that function as members of two-component regulatory systems, and the translational product of corS showed sequence similarity to histidine protein kinases which function as environmental sensors (273). Response regulators control the adaptive response in two-component regulatory systems and are characterized by an N-terminal receiver domain which functions as the phosphorylation site and a C-terminal effector domain with a DNA-binding, helix-turn-helix (H-T-H) motif (192, 193). Both domains are strongly conserved in CorR; CorP, however, contains the highly conserved receiver domain (at least two aspartate residues and a conserved lysine) but lacks the H-T-H motif. The N-terminal receiver domains of CorR and CorP are almost identical when aligned, suggesting a shared specificity for the same phosphodonor protein(s). The COR regulatory system is modified from the two-component paradigm since it contains two response regulator proteins together with a single sensor protein.
Both CorR and CorP showed relatedness to response regulators in the ROIII group, which includes NarL, BvgA, and FixJ (193). Several of these response regulators function as positive activators of transcription and bind to specific target sequences upstream of the promoters they regulate (1, 36). Complementation analysis of a corR mutant, PG4180.P2, and transcriptional fusions to a promoterless glucuronidase gene (uidA) indicated that CorR functions as a positive regulator of COR gene expression (202). Deletion analysis of the cfl upstream region was used to define the minimal amount of DNA required for full transcriptional activity of a cfl::uidA fusion (202). A fragment located upstream from the cfl transcriptional start site was used in gel retardation and DNase I footprinting assays to define the specific bases bound by CorR. This region was also conserved in the promoter region for cmaA, suggesting that both the CFA and CMA structural genes are controlled by CorR, a positive activator of COR gene expression (202).
Gene fusions indicated that expression of corR and corP was not significantly different at 18 and 28°C. In contrast, expression of corS was regulated by temperature, and a corS::uidA fusion showed maximal transcriptional activity at 18°C and 15-fold less activity at 28°C (273). The use of transcriptional fusions and complementation analyses indicated that each regulatory gene was independently transcribed (Fig. 6A). Furthermore, experiments with the corS::uidA fusion indicated that transcription of corS was autoregulated and required functional copies of corR, corS, and corP (273).
COR production in PG4180 was significantly affected by the carbon source, glucose levels, amino acid supplements, complex carbon and nitrogen sources, and osmolarity (189). Transcriptional fusions were used to determine if any of these factors impacted transcriptional activity in the COR gene cluster (191). Glucose levels and selected carbon and amino acid sources significantly affected the expression of the cfl and cmaA operons. In general, gene expression increased with increasing amounts of glucose but was strongly repressed when selected carbon sources (xylose and fructose) and amino acids (isoleucine and valine) were added to the medium. Interestingly, changes in osmolarity and the addition of complex C and N sources did not significantly affect COR gene expression. In contrast, several researchers have shown that transcription of the hrp and avirulence genes (avr) genes in P. syringae is repressed by complex C and N sources and by increased osmolarity (106, 109, 144, 210, 227). Furthermore, the hrp and avr genes were shown to be transcriptionally activated in response to fructose (106, 144, 227). Obviously, some of the signals for activation of the hrp and COR gene clusters are different.
Several approaches have been used to investigate the potential stimulation of COR synthesis by host plants. Palmer and Bender amended the growth medium for PG4180 with extracts from soybean tissue or with plant-derived secondary metabolites but found no evidence that these substances substantially increased COR production in vitro (189). In a subsequent study, the activities of cmaA::uidA and cfl::uidA transcriptional fusions were compared in vitro and in soybean leaves; however, there was no evidence that COR gene expression in PG4180 was higher in plant tissue (188). In contrast, Ma et al. (145) showed that COR biosynthesis in P. syringae pv. tomato DC3000 is plant inducible. Gene fusions indicated that a single transcriptional unit designated CorII was expressed at a higher level in planta than in vitro. Other results indicate that shikimic and quinic acids may be signals for COR gene induction in DC3000 (137). These observations suggest that the signals for induction of COR synthesis differ in PG4180 and DC3000.
SYRINGOMYCIN AND RELATED LIPODEPSINONAPEPTIDES
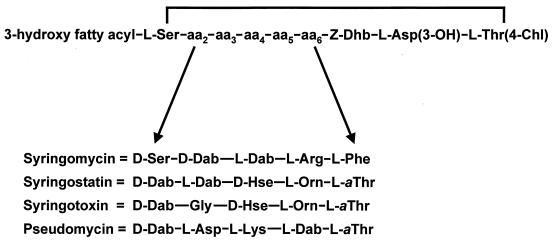
Syringomycin is representative of the cyclic lipodepsinonapeptide class of phytotoxins, which are composed of a polar peptide head and a hydrophobic 3-hydroxy fatty acid tail (77, 240) (Fig. 7). Characteristically, three forms of syringomycin are produced, differing only by the length of the 3-hydroxy fatty acid moiety, which is either decanoic (SRA1), dodecanoic (SRE), or tetradecanoic (SRG) acid. An amide bond attaches the 3-hydroxy fatty acid to an N-terminal serine residue, which in turn is linked to 4-chlorothreonine at the C terminus by an ester linkage to form a macrocyclic lactone ring. Other distinctive structural features are a trio of uncommon amino acids (2,3-dehydroaminobutyric acid, 3-hydroxyaspartic acid, and 4-chlorothreonine) at the C terminus and the presence of d-isomers of serine and 2,4-diaminobutyric acid (231). Chlorination of the syringomycin molecule is important for biological activity (86). Syringomycin is produced by most strains of P. syringae pv. syringae isolated from a wide range of host plants. Syringotoxin and syringostatin are related lipodepsinonapeptides produced by strains isolated from citrus and lilac hosts, respectively (13, 76). The saprophytic strain MSU 16H from barley produces a fourth type of lipodepsinonapeptide called pseudomycin (12). All the lipodepsinonapeptides differ in amino acid sequence between positions 2 and 6, as depicted in Fig. 7. Despite these structural differences, the various types of lipodepsinonapeptides exhibit similar degrees of biological activity (245).
FIG. 7.
Structures of syringomycin, syringostatin, syringotoxin, and pseudomycin. The four lipodepsinonapeptides differ in the amino acid sequence between positions 2 and 6. The 3-hydroxy fatty acyl group is a derivative of either decanoic acid (syringomycin), dodecanoic acid (syringomycin and syringostatin), tetradecanoic acid (all four lipodepsinonapeptides), or hexadecanoic acid (pseudomycin); some forms of pseudomycin are acylated by 3,4-dihydroxytetradecanoate or 3,4-dihydroxyhexadecanoate. Abbreviations of nonstandard amino acids: Asp(3-OH), 3-hydroxyaspartic acid; Dab, 2,4-diaminobutyric acid; Dhb, 2,3-dehydroaminobutyric acid; Hse, homoserine; Orn, ornithine; Thr(4-Chl), 4-chlorothreonine; aThr, allothreonine.
Syringomycin Activity Is Centered Around Transmembrane Pore Formation
Syringomycin induces necrosis in plant tissues, and early studies of its mode of action established that the plasma membrane of host cells is the primary target (8, 199). The amphipathic lipopeptide structure of syringomycin promotes its insertion into the lipid bilayers of membranes to form pores that are freely permeable to cations (105). The toxin causes an increase in transmembrane fluxes of K+, H+, and Ca2+ that are deadly to cells (32, 182, 258). Pore formation in lipid bilayers is a highly efficient process based on evidence that only nanomolar amounts of syringomycin are required for measurable activity. This is especially apparent in assays of tobacco protoplasts, where 45Ca2+ influx and membrane lysis occur at a threshold syringomycin concentration of 50 ng/ml (103). Furthermore, Hutchison et al. (105) demonstrated pore-forming activity for SRE in a pure black-lipid membrane, thereby showing that the activity does not arise from opening of native ion channels found in membranes patched from cells. Many medically important bacteria produce pore-forming proteins or peptides that cause cytolysis as a result of massive ion fluxes (31). Syringomycin represents the first example of a virulence factor from a plant-pathogenic bacterium that targets host plasma membranes to form ion channels in lipid bilayers and causes cytolysis (103, 105).
From biophysical analysis of channel formation in planar lipid bilayer membranes, a picture is emerging of how syringomycin pores function and are formed (118). Shortly after insertion into the lipid bilayer, monomers of syringomycin aggregate into pore complexes. Based on the voltage-dependent behavior of ion channels, Feigin et al. (71) concluded that the channel was formed by at least six molecules of syringomycin. The lipophilic portion of each toxin subunit resides in the core of the bilayer, and the hydrophilic peptide head resides close to the surface of the membrane. Individual channels can become aggregated into clusters that exhibit synchronous opening and closing (118). The channel radius is approximately 1 nm for a syringomycin pore (105, 118). This is comparable in size to the channel dimensions of transmembrane pores formed by tolaasin, a lipodepsipeptide produced by the mushroom pathogen Pseudomonas tolaasii (211), and alpha-hemolysin, a cytolytic protein produced by E. coli (30).
It appears that sterols influence channel formation by syringomycin but are not components of the channel structure (70). Julmanop et al. (112) and Taguchi et al. (256) reported that sterols, particularly ergosterol, promoted the binding of syringomycin to cells. Because sterols can play a significant role in channel formation and are known to be essential for the cytotoxic activity of many pore-forming cytotoxins, such as streptolysin O (62), and lipopeptides, such as iturin A (132), an analogous role for sterols was proposed in channel formation by syringomycin. However, Feigin et al. (71) showed that the toxin readily formed ion channels in artificial membrane bilayers that lacked sterols. Subsequently, Feigin et al. (70) demonstrated that the addition of 50 mol% of ergosterol, stigmasterol, or cholesterol (sterols abundant in fungal, plant, and animal cells, respectively) to bilayers failed to alter the channel conductance properties of syringomycin.
The syringomycin pores are freely permeable to a series of monovalent and divalent cations (105). Based on studies by Takemoto and associates (215, 216, 293), an influx of H+ appears to be accompanied by an efflux of K+ across the syringomycin channel. This K+-H+ exchange generates an electrochemical gradient and collapses the pH gradient of the plasma membrane, resulting in acidification of the cytoplasm. The most conspicuous effect of channel formation by syringomycin is a rapid and sustained influx of Ca2+ ions that activates a cascade of events associated with cellular signaling in plants (120, 259, 260). For example, cytoplasmic influxes of Ca2+ caused by low concentrations of syringomycin lead to induction of kinase-mediated phosphorylation of membrane proteins (32) and the incorporation of 1,3-β-callose into plant cell walls (120). Consequently, the phosphorylation of a proton pump ATPase, as observed by Bidwai and Takemoto (32), appears to result from activation of protein kinases as modulated by a free Ca2+ signal (119). A short-term stimulation of the plasma membrane ATPase occurs (33). The resultant increase in ATP hydrolysis promotes the pumping of H+ and Ca2+ back out into the extracellular space, which in the long-term is ineffective against the collapse of the cation gradients of the cell. The ultimate benefit to the bacterium from pore formation is the systematic release of nutrients into the intercellular spaces of host tissues (105) and the alkalization of intercellular fluids, resulting in a more favorable environment for bacterial growth (42).
Sphingolipids, which are major lipid components of eukaryotic plasma membranes, have been associated with cell sensitivity to syringomycin (45, 90). A SYR2 mutant of the fungus Saccharomyces cerevisiae is highly resistant to syringomycin due to a failure to produce 4-hydroxylated sphingolipids such as phytoceramide (90). The total sphingolipid content of a SYR2 mutant is unchanged from the wild-type strain, and the mutant exhibited an abundance of ceramide moieties containing only dihydrosphingosine but not phytosphingosine. The significance of these observations on the biological activity of syringomycin is unclear, but sphingolipids play important roles in signal transduction as mediators of growth suppression and programmed cell death (185).
Another feature of the amphipathic syringomycin molecule is that it exhibits potent biosurfactant activity capable of lowering the interfacial tension of water to 31 mN/m, compared to a value of 73 mN/m for HPLC-grade water (105). Although pure preparations of syringomycin have a critical micelle concentration of 1.2 mg/ml, the surfactant properties are apparent at much lower concentrations. The surface-active properties of syringomycin are similar to those of other biosurfactants produced by fluorescent pseudomonads, including viscosin (183) and tolaasin (104). Biosurfactant activity appears to play an important role in spread of the bacterium across plant surfaces by reducing the surface tension of water and by concentrating relatively sparse nutrients at solvent interfaces (105).
Besides being phytotoxic, syringomycin and related lipodepsinonapeptides exhibit fungicidal activity toward a broad spectrum of filamentous fungi, such as Geotrichum candidum, and yeasts, such as Rhodotorula pilimanae (133, 245). Accordingly, assays for antifungal activity are conveniently used in bioassays of the toxins, which are active at concentrations as low as 0.8 μg/ml against yeasts (245). The related lipodepsinonapeptides syringotoxin and syringostatin have nearly equivalent broad-spectrum antifungal activity. Syringomycin also lyses erythrocytes, but 10-fold-higher concentrations of the toxin (i.e., 0.75 μg/ml) are required for comparable activity, as observed in assays of tobacco protoplasts (103, 105). Recently, efforts have been made to capitalize on the potent antifungal activities of syringomycin to control clinically important fungi, such as Candida spp. (245), and postharvest fungal pathogens of citrus, such as Penicillium digitatum (38). However, the strong hemolytic activity of syringomycin and related lipodepsinonapeptides remains a serious obstacle to commercial development.
Biosynthesis of Syringomycin Occurs by a Nonribosomal Mechanism of Peptide Synthesis
Syringomycin biosynthesis occurs on a multifunctional complex of enzymes by a thiotemplate mechanism as originally described for peptide antibiotics produced by Bacillus, Streptomyces, and filamentous fungi (126). The first evidence for a nonribosomal mechanism of toxin synthesis originated with the association of large proteins, 470 kDa or larger, with lipodepsinonapeptide production (179, 288). Nontoxigenic (Tox−) mutants were identified that were altered in the formation of these large proteins, which were speculated to function as peptide synthetases. Another important milestone was the resolution of the cyclic lipodepsipeptide structure of syringomycin containing nonproteinogenic amino acids and d-amino acids (77, 240). The occurrence of these unusual amino acids in the syringomycin peptide chain is indicative of nonribosomal multifunctional synthetases that catalyze the formation of peptides that contain modified amino acids (126, 252). Subsequently, Grgurina and Mariotti (89) used 14C-labeled amino acids to determine that l-threonine is the precursor of both 2,3-dehydroaminobutyric acid and 4-chlorothreonine and that aspartic acid was incorporated into 2,4-diaminobutyric and 3-hydroxyaspartic acids. The 3-hydroxy fatty acid tail of syringomycin appears to be derived from 3-hydroxyalkanoates that are accumulated in fluorescent pseudomonads and utilized as carbon and energy reserves (67).
Molecular genetic evidence for a thiotemplate mechanism of syringomycin synthesis came from analysis of the syrB1 gene that encodes an amino acid activation module characteristic of peptide synthetases (291). The SyrB1 amino acid sequence revealed six core sequences typical of the amino acid-activating modules described previously (247) (Fig. 1B). The core sequences of SyrB1 exhibited the characteristic order and spacing of thioester-forming modules involved in the synthesis of gramicidin S and other peptide antibiotics (291). Recent biochemical analysis demonstrated that the amino acid-activating module of SyrB1 catalyzes the recognition and activation of l-threonine (95). Thus, the syringomycin multienzyme system represents one of the first described for Pseudomonas, although it appears that all fluorescent pseudomonads synthesize peptide-containing metabolites, such as pyoverdin (154), by the thiotemplate mechanism.
Organization of the Syringomycin Gene Cluster Encoding Peptide Synthetases
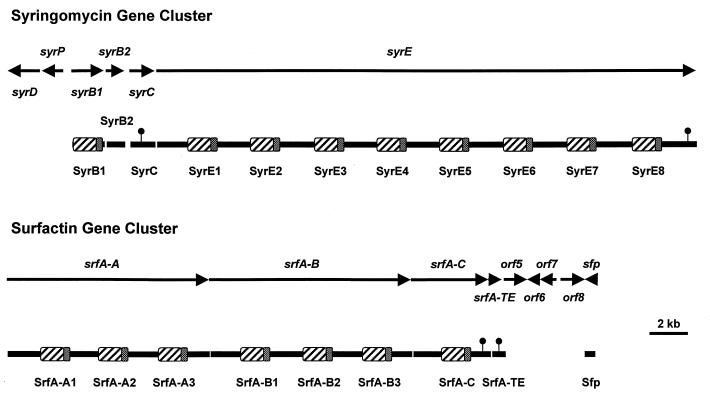
The syringomycin (syr) gene cluster encompasses a DNA region of approximately 37 kb on the chromosome of P. syringae pv. syringae B301D (Fig. 8). Six ORFs in the syr gene cluster are predicted to encode proteins involved in the synthesis (syrB1, syrB2, syrC, and syrE) (95, 291), secretion (syrD) (209), and regulation (syrP) (292) of syringomycin. The most striking feature of the syr gene cluster is the presence of an enormous (28.4-kb) ORF, designated syrE (95). Sequence analysis indicated that syrE encodes a 1,039-kDa synthetase containing eight amino acid activation modules (95). Thus, SyrE represents the largest protein reported for a prokaryote to date. Precedence for the formation of large synthetases exists in the fungus Tolypocladium niveum, where the simA gene encodes a 1,689-kDa synthetase involved in cyclosporin A synthesis (281).
FIG. 8.
Physical map of the syringomycin gene cluster of P. syringae pv. syringae compared to that of the surfactin gene cluster of B. subtilis. The amino acid-adenylating domains and 4′-phosphopantetheine carrier domains are indicated by cross-hatched and stippled regions, respectively, for each module. The small circles attached above the proteins indicate the regions carrying motifs characteristic of thioesterases. The 37-kb syr gene cluster encodes four proteins (SyrB1, SyrB2, SyrC, and SyrE) involved in biosynthesis. The syrD and syrP genes encode proteins predicted to be involved in secretion and regulation, respectively. The 31-kb srf gene cluster encodes five proteins (SrfA-A, SrfA-B, SrfA-C, SrfA-TE, and Sfp) involved in surfactin synthesis. The sfp gene product has been proposed to function as a 4′-phosphopantetheine transferase (147). The functions of the products of orf5 through orf8 are unknown.
The domain organization of the predicted SyrE synthetase is illustrated in Fig. 8; each amino acid activation module contains domains for elongation, adenylation, and thiolation. The various SyrE adenylation domains exhibit a high degree of identity ranging from 45 to over 90%. Because the adenylation domain harbors the regions responsible for amino acid recognition and activation (247), the E1 adenylation domain of syrE was amplified, cloned, overexpressed, and used in biochemical assays (95). The E1 adenylation domain recognized l-serine as a substrate in ATP-pyrophosphate (PPi) exchange reactions, indicating that syringomycin synthesis is initiated by activating the N-terminal amino acid, l-serine. The adenylation domain of E1 is preceded by an elongation domain, which is presumed to catalyze the acyl transfer of the 3-hydroxy fatty acid from SyrC to l-Ser bound to E1 as a thioester (54, 252). Elongation domains were previously shown to precede the first amino acid activation domain in SrfA-A which initiates the synthesis of the lipopeptide surfactin (252).
A distinctive structural feature of the SyrE synthetase is the fusion of a TE domain to the last amino acid activation module, E8 (Fig. 8) (95). This domain contains the GxSxG motif characteristic of TEs involved in the synthesis of fatty acids, polyketides, and peptide antibiotics (252). All bacterial peptide synthetases exhibit a TE domain at the C-terminal end of the synthetase that carries the last amino acid activation module; the TE domain presumably catalyzes a termination reaction in which the thioesterified peptide is released by hydrolytic cleavage from the synthetase. The importance of the TE domain in peptide antibiotic production in bacteria was demonstrated by Schneider and Marahiel (237); these researchers deleted the TE region of srfA to srfC, which encodes the last synthetase in surfactin synthesis, and reported a large reduction in surfactin yield. Unique to SyrE is the positioning of the TE domain relative to the E8 module, which are separated by intervening elongation and thiolation domains (95). In effect, the C terminus of SyrE contains elements of a ninth module that lacks an adenylation domain.
The ninth adenylation domain appears to be encoded by syrB (291). It is now known that syrB represents an operon that expresses two proteins (290): the 68-kDa SyrB1 protein carrying adenylation and thiolation domains, and the 35-kDa SyrB2 protein homologous to the CmaB protein in the COR gene cluster (37). The substrate specificity of the overproduced SyrB1 adenylation domain was analyzed in ATP-PPi exchange reactions (95). SyrB1 activated l-threonine in exchange reactions, whereas a series of amino acids including l-serine and 4-chlorothreonine were not recognized as substrates. Therefore, SyrB1 presumably activates and binds l-threonine as a thioester, which may be subsequently modified to yield 4-chlorothreonine prior to transfer to the SyrE synthetase. Although the function of SyrB2 in syringomycin synthesis is unknown, one can speculate that it functions in the modification of l-threonine bound to the SyrB1 thiolation domain or in the cyclization of the mature peptide.
The syrC gene is located between the syrB operon and syrE and encodes an enzyme with TE activity (Fig. 8) (291). SyrC is similar to several proteins containing TE motifs, including CmaT, which is presumed to function as a coronamic acid thioesterase (270). The 48-kDa SyrC protein contains a GxCxG motif with a Cys substituted for Ser as the active site, a change that does not significantly affect the catalytic activity of TEs (285). The TE activity of SyrC was demonstrated by Grgurina et al. (87), who overproduced SyrC as an N-terminal His-Tag fusion protein in E. coli. SyrC catalyzed the hydrolysis of CoA from a 3-hydroxydodecanoyl-CoA substrate, supporting the hypothesis that SyrC functions as a thioesterase in syringomycin biosynthesis with a potential role in acyltransfer of a 3-hydroxy fatty acid. SyrC recognized a series of linear long chain acyl-CoA derivatives as substrates but did not utilize medium- or short-chain fatty acid derivatives. Site-directed mutagenesis of syrC was used to generate Cys-to-Gly mutations within the TE motif of SyrC, resulting in a mutant SyrC which lacked TE activity (290). Thus, it appears that SyrC catalyzes the transfer of the 3-hydroxydodecanoyl moiety from the corresponding CoA derivative to the amino group of serine bound to the E1 module of SyrE (Fig. 8).
In summary, syringomycin synthesis is catalyzed by four proteins, namely, SyrB1, SyrB2, SyrC, and SyrE (Fig. 8). In comparison, the surfactin gene cluster contains synthetases carrying seven amino acid activation modules (Fig. 8) (247). The adenylation of the N-terminal serine and covalent attachment of the amino acid to the synthetase by a carboxyl thioester initiate the biosynthetic process on the E1 module of SyrE (Fig. 1B) (95). Within the adenylation domain of E1, a serine-specific pocket located between core 2 and core 4 (47, 291) recognizes l-serine. ATP is bound to core 2 and subsequently cleaved by an ATPase located at core 4, leading to the formation of serine adenylate. The activated serine is then transferred to the thiolation domain and covalently linked by a thioester bond to the cofactor, 4′-phosphopantetheine, at a conserved serine residue within core 6 (247, 252). The TE activity of SyrC catalyzes the hydrolysis of 3-hydroxydodecanoyl-CoA and may transfer the 3-hydroxy fatty acid to the amino group of serine bound to the E1 module, thus forming a 3-hydroxydodecanoyl-l-serine conjugate. The intermediate is transferred to the E2 module, where the second amino acid, d-serine, is covalently bound as a thioester to 4′-phosphopantetheine. Each SyrE module carries a molecule of 4′-phosphopantetheine covalently bound to the thiolation domain, and this mediates the sequential transfer of carboxyl thioester-activated amino acids between aligned modules (253). Synthesis continues with a series of elongation cycles until an octapeptide is synthesized and bound at the thiolation domain of the E8 module of SyrE. At this point, the adenylation domain of SyrB1 interacts with the E8 module to incorporate the last amino acid, l-threonine, which is ultimately modified as 4-chlorothreonine. The TE domain at the C terminus of SyrE is presumed to release the mature syringomycin product from the synthetase. SyrB2 is speculated to function either in modifying l-threonine bound to SyrB2 or in cyclization of the mature peptide to form a lactone ring. Once cyclized, syringomycin is exported across the cytoplasmic membrane by the ATP-binding cassette (ABC) transporter protein, SyrD (209).
Regulation of Syringomycin Production
The regulation of syringomycin production is complex, based on evidence that both nutritional factors and plant signal molecules modulate toxin production by P. syringae pv. syringae (92). Iron exerts a positive regulatory effect on syringomycin production based on evidence that Fe3+ concentrations of 2 μM or higher are required for expression of a syrB-lacZ transcriptional fusion (174) and for maximum yields of toxin by strain B301D (91). In contrast, syringomycin production is repressed by inorganic phosphate concentrations of 1 mM or higher (91), and this resembles the phosphate-mediated down-regulation of antibiotic biosynthesis genes in many bacteria (139). The discovery that specific plant signal molecules also play a significant role in activating syrB gene expression and syringomycin production (discussed below) demonstrates that diverse environmental factors control the expression of genes dedicated to toxigenesis in P. syringae pv. syringae. Unfortunately, little is known about the complex genetic network responsible for the perception and transduction of these signals to the syr transcriptional apparatus.
Toxin gene clusters encoding a multienzyme system of peptide synthesis commonly carry regulatory elements that directly control the expression of biosynthesis genes (126). Surprisingly, the 37-kb syr cluster contains only one gene, syrP, that exerts regulatory effects on syringomycin production (292). A syrP mutant has an unusual pleiotropic phenotype with respect to syringomycin production and is substantially reduced in virulence on immature cherry fruits. In particular, syringomycin production by a syrP mutant is relatively insensitive to high inorganic phosphate concentrations in agar media. The syrP gene is located between the syrB and syrD genes (Fig. 8) and encodes a 40-kDa protein that may function in a phosphorelay signal transduction pathway. The SyrP protein exhibits similarity to the phosphoacceptor/transfer regions of histidine kinases such as CheA (296) and KinA (204), which are regulatory elements in phosphorelay pathways (7). Thus, SyrP may function in a phosphorelay system as an intermediate phosphate transmitter between a sensor protein and a response regulator (292). Phosphorelays govern major developmental commitments in microorganisms, and an important advantage of phosphorelays is that multiple signals can be integrated at intermediate steps in the regulatory network (7). Nevertheless, a role for SyrP in a phosphorelay mechanism of toxin production remains speculative until other members of such a regulatory system are identified and characterized. It is unclear whether additional regulatory genes are located within the right border region of the syr cluster (i.e., downstream of syrE) or are linked to the syringopeptin gene cluster (Fig. 8).
Global regulators of syringomycin production which are not physically linked to the syr cluster have been identified (101, 219). The gacS (lemA) and gacA genes encode members of a two-component sensory transduction system in P. syringae pv. syringae that regulates toxigenesis and the ability to cause necrotic lesions in plants. Sequence analysis indicated that GacS is a transmembrane protein which presumably functions as a histidine protein kinase that undergoes phosphorylation in response to environmental stimuli (101). GacA is a response regulator protein that is presumably phosphorylated by GacS (219). Like other members of the FixJ subclass of response regulators (41), GacA carries the phosphorylation site at the N terminus and a H-T-H motif at the C terminus (219). Nevertheless, it has not been demonstrated that GacS interacts directly with GacA to facilitate phosphate transfer in a two-component signal transduction system. An unusual feature of the gacS-gacA two-component system is that the two genes are not physically linked. Furthermore, the GacS-GacA homologs are conserved in fluorescent pseudomonads and control the expression of a diversity of cellular functions including production of protease, pectate lyase, pigments, and various antifungal metabolites (78, 125).
The GacS-GacA protein pair appears to be at the top of the regulatory hierarchy controlling syringomycin production, and little is known about the intermediary regulators. Kitten et al. (125) identified salA as a member of the gacS-gacA regulon that can restore syringomycin production to a gacS mutant if salA is overexpressed. A salA mutant is phenotypically distinguished from gacS or gacA mutants by a lack of suppression of protease production. Correspondingly, expression of a syrB-lacZ reporter was reduced to less than 3% in a salA mutant. The predicted SalA protein sequence exhibits an H-T-H DNA-binding motif with similarity to response regulators such as FixJ (5). It remains to be determined if SalA, as a putative response regulator, binds directly to the promoter region of the syrB operon or, rather, activates the syrB operon indirectly through one or more intermediary regulators such as SyrP. Thus, it appears that salA, as an element in the gacS-gacA regulon, controls the expression of the pathway that leads to syringomycin production and formation of necrotic lesions in plants (125). The gacS-gacA regulon also controls the expression of the ahlIPss gene specifying the synthesis of N-acylhomoserine lactone by P. syringae pv. syringae (61). However, syringomycin production is not controlled by the N-acylhomoserine lactone quorum-sensing signal. The environmental signals that activate the gacS-gacA regulon have not been identified, although they do not appear to be phenolic plant signal molecules such as arbutin (175, 219).
The syrA gene identified by Xu and Gross (288) appears to encode a regulatory protein required for syringomycin production and pathogenicity. The syrA gene, which lies outside the syr gene cluster, has not been sequenced, and its position in the regulatory network controlling toxigenesis remains to be determined.
Activation of Syringomycin Production by Plant Signal Molecules
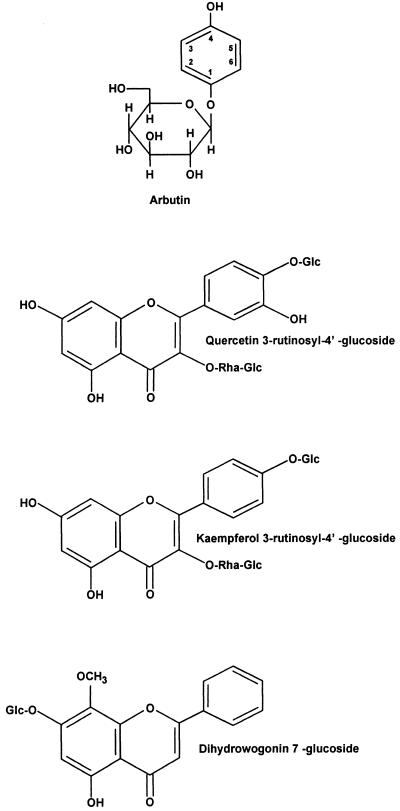
Phytotoxin production by P. syringae pv. syringae is modulated by the perception of signals in the plant environment. The primary signals are specific phenolic glycosides that are abundant in the leaves, bark, and flowers of many plant species parasitized by P. syringae pv. syringae (175). For example, arbutin (Fig. 9) is a phenolic β-glucoside distributed to more than 10 dicot families; in pear (Pyrus communis), arbutin constitutes 3 to 5% of the leaf material (156). In cherry (Prunus avium) leaves, two flavonol glycosides (quercetin 3-rutinosyl-4′-glucoside and kaempferol 3-rutinosyl-4′-glucoside) and one flavanone (dihydrowogonin 7-glucoside) glycoside (Fig. 9) have been identified as signal molecules based on the induction of a syrB-lacZ fusion as a reporter of gene activity (173). The plant signal molecules that activate toxin production by P. syringae pv. syringae are chemically distinct from those that activate the vir genes of Agrobacterium tumefaciens and the nod genes of rhizobia (152, 205). An intact glucosidic linkage is a structural feature of all syrB-inducing phenolic signal molecules; plants accumulate and store phenolic compounds as glycosides, which are more water soluble and less chemically reactive (100). The flavonoid signal molecules from cherry are equivalent in syrB-inducing activity and are abundant, as evidenced by the recovery of more than 11 mg (dry weight) of dihydrowogonin 7-glucoside per g from phloem (264).
FIG. 9.
Structures of phenolic plant signal molecules known to activate the syrB gene involved in syringomycin biosynthesis by P. syringae pv. syringae. Arbutin is found in several plant species including pear (Pyrus communis L.). The flavonol glycosides (quercetin 3-rutinosyl-4′-glucoside and kaempferol 3-rutinosyl-4′-glucoside) and the flavanone glucoside (dihydrowogonin 7-glucoside) are abundant in the leaves of sweet cherry (Prunus avium L.).
Phenolic signal activity is markedly enhanced in the presence of sugars that occur in large quantities in leaf tissue (173, 175). Sucrose and d-fructose are the most active sugars, causing a 10-fold stimulation of signal activity when phenolic signals occur at low concentrations. These two sugars also exhibit intrinsic low-level syrB-inducing signal activity in the absence of the phenolic inducer. Cherry contains an abundance of sucrose and fructose based on estimates of more than 3% of the dry weight of various tissues (121, 236). Consequently, the concentrations of both sugars exceed the threshold of 10 ppm required for significant induction of the syrB operon (175). The mechanism by which sugars augment the sensitivity of P. syringae pv. syringae to the phenolic signal is unknown. In A. tumefaciens, the ChvE sensory system mediates enhanced induction of vir genes (40). However, the two plant-microbe systems differ in sugar specificity, with the most conspicuous distinction being the inability of sucrose and d-fructose to enhance vir gene induction in the presence of the phenolic signal acetosyringone (4, 242).
In addition to the specific activation of syrB, the entire syringomycin biosynthetic apparatus is stimulated by plant signals in nearly all strains of the bacterium. The stimulatory effect of plant signal molecules on syringomycin was quite evident for some strains of P. syringae pv. syringae based on the recovery of up to 10-fold-higher toxin yields in a defined medium supplemented with arbutin and d-fructose (208). Furthermore, some strains required plant signal molecules for the production of syringomycin. In strains producing syringotoxin or syringostatin instead of syringomycin, plant signal molecules also stimulated toxigenesis. Such a network of communication between the plant and the bacterium that controls toxigenesis reflects the ability of P. syringae pv. syringae to adapt to a dynamic plant environment. The sensory mechanism favors the bacterium by detecting specific phenolic glycosides that signal the bacterium to rapidly activate virulence genes. P. syringae pv. syringae aggressively attacks a wide range of plants, and it would not be surprising to find that all host plants contain phenolics with the fundamental chemical structures responsible for signal activity.
SYRINGOPEPTIN
Syringopeptins represent a second class of lipodepsipeptide phytotoxins produced by strains of P. syringae pv. syringae (9). In contrast to lipodepsinonapeptides, syringopeptins contain either 22 or 25 amino acids depending on the specific bacterial strain (Fig. 10). The N-terminal amino acid, 2,3-dehydro-2-aminobutyric acid, is acylated by either 3-hydroxydecanoic or 3-hydroxydodecanoic acid. An ester bond between allothreonine and the C-terminal tyrosine residue forms a lactone ring. A high percentage of hydrophobic amino acids are found in syringopeptin, and Ballio et al. (11) determined that d-amino acids compose most of the syringopeptin peptide chain. The octapeptide cationic loop formed by a lactone ring, together with the hydrophobic tail, is predicted to function as a membrane-permeabilizing motif critical to biological activity (11). Analysis of syringopeptins from several strains of P. syringae pv. syringae demonstrated diversity in the peptide sequences of syringopeptins (9, 110). For example, a strain of P. syringae pv. syringae isolated from laurel produced Phe25-syringopeptin 25A with phenylalanine as the C-terminal amino acid instead of tyrosine (232).
FIG. 10.
Structures of syringopeptin forms SP22 and SP25. The fatty acid can be either 3-hydroxydecanoic or 3-hydroxydodecanoic acid. Abbreviations of nonstandard amino acids: Dab, 2,4-diaminobutyric acid; Dhb, 2,3-dehydroaminobutyric acid; aThr, allothreonine. d-Amino acids are common in both SP22 (13 of 22 residues) and SP25 (15 of 25 residues). A P. syringae pv. syringae strain from laurel produces a form of SP25 that differs from the above structure by the replacement of Phe with Tyr at the C terminus (232). Strain SC1 from sugarcane produces a form of SP22 that differs from the above structure by the replacement of Leu at amino acid positions 4 and 7 and 2-aminodehydropropionic acid (dehydroalanine) at position 9 (110).
Cytotoxic Pore-Forming Activity
Syringopeptin exhibits extraordinary similarity to syringomycin in phytotoxic activity. Iacobellis et al. (107) first described the ability of syringopeptin to cause electrolyte leakage of plant cells, which leads to the development of necrotic symptoms. Subsequently, syringopeptin SP22A was shown to alter the distribution of H+ across the plasma membrane of maize (57) and promote stomatal closure in detached leaves of Xanthium strumarium due to a rapid K+ efflux (58). Thus, syringopeptin appears to induce an H+-K+ exchange response in plant cells analogous to that induced by syringomycin (182).
The phytotoxic activity of syringopeptin is centered on an ability to form pores in plant plasma membranes, thereby promoting transmembrane ion flux and cell death. As a pore-forming lipodepsipeptide, syringopeptin is able to form ion channels in black-lipid membranes, to lyse both tobacco protoplasts and erythrocytes, and to generate a rapid and sustained influx of 45Ca2+ across the plasma membrane of tobacco protoplasts (103). For example, the extreme sensitivity of tobacco protoplasts was observed at syringopeptin concentrations as low as 100 ng/ml, at which about half of the protoplasts lysed after a 30-min incubation. Pores formed by syringopeptin in protoplasts are highly permeable to cations such as Ca2+, with an uptake rate between 0.6 to 0.8 nCi of 45Ca2+ min/ml at a toxin concentration of 500 ng/ml. Both SP22A and SP22B exhibited equivalent pore-forming activities despite differences in the length of the hydrophobic 3-hydroxy fatty acid tail. In direct comparisons of the cytotoxic activities of syringopeptin and syringomycin, both caused lysis of tobacco protoplasts and 45Ca2+ uptake at threshold concentrations of 50 ng/ml; erythrocytes were lysed at threshold toxin concentrations between 0.75 to 1 μg/ml. Thus, on a molar basis, syringopeptin is more active in cytotoxic assays than syringomycin, reflecting the much larger size of the syringopeptin molecule and a concomitant increase in pore formation. Other studies reported syringopeptin to be several times more active than syringomycin in causing necrosis and electrolyte leakage in plant tissues (107, 133) and in affecting stomatal conductance in X. strumarium leaves (58). Although Hutchison and Gross (103) found both toxins roughly equivalent in toxicity to tobacco protoplasts at 0.04 μM, whole tissues treated with toxin concentrations of 0.4 μM or higher showed syringopeptin to be more phytotoxic than syringopeptin (133).
The biophysical characteristics of syringopeptin pores have not been studied, although it is predicted that aggregates of syringopeptin monomers are required for pore formation in lipid bilayers (103). Fewer molecules of syringopeptin may be required to form a functional pore, because it has a larger charged head than syringomycin. The two lipopeptide toxins may interact synergistically in the plant-pathogen interaction (107), but there is no evidence that pores composed of a chimera of syringopeptin and syringomycin are formed (103).
Syringopeptin displays strong biosurfactant activity based on the ability of SP22A and SP22B to lower the interfacial tension of water to approximately 35 mN/m at concentrations of ≥10 μg/ml (103). Syringopeptin has a lower critical micelle concentration than syringomycin does (i.e., 0.8 mg/ml for SP22B and 1.25 mg/ml for syringomycin form SRE). By reducing the contact angle of water, syringopeptin together with syringomycin may facilitate the spread of P. syringae on plant surfaces. The amount of toxin produced in the plant environment is unknown, but a typical strain of P. syringae pv. syringae has the capacity to produce abundant quantities of the two lipodepsipeptides in vitro. For example, strain B301D produces 5 to 15 μg of both toxins per ml in vitro (103, 105) and plant signal molecules stimulate toxin production in most strains of the bacterium (173, 208).
Syringopeptin has antimicrobial activity against certain gram-positive bacteria and fungi. Interestingly, syringopeptin has an antimicrobial spectrum of activity distinct from that of syringomycin and related lipodepsinonapeptides (133). For example, some strains of Botrytis cinerea are highly sensitive to syringopeptin but resistant to syringomycin. In contrast, Geotrichum candidum is sensitive to syringomycin and resistant to syringopeptin. Bacillus megaterium was the most sensitive to syringopeptin of a wide spectrum of microorganisms assayed by Lavermicocca et al. (133). Because B. megaterium is resistant to syringomycin, this bacterium is used in routine bioassays of syringopeptin activity (88, 103). The distinct antimicrobial activities of the two classes of lipodepsipeptides are in sharp contrast to the relatively similar pore-forming activities for the toxins in assays of tobacco protoplasts, erythrocytes, and artificial membrane bilayers (103). The biological basis for these differences is unknown.
Biosynthesis and Genetic Organization
Syringopeptin is synthesized by multienzymatic peptide synthetases distinct from those involved in syringomycin synthesis (88). Because the isoform of syringopeptin produced by strain B301D contains 22 amino acids (9), the syp gene cluster is predicted to exceed 60 kb based on the size of known synthetase domains (247, 252). Mapping and sequence analysis have indicated that the syp gene cluster is located adjacent to the syr gene cluster (95, 238). Although a comprehensive map of the syp cluster is unavailable, a syp region encoding one or more peptide synthetases lies downstream of syrD. For example, the sypA ORF begins 138 bp from the 3′ end of syrD and is transcribed in the same direction as syrD (Fig. 8). The first amino acid activation module encoded by sypA has the same domain organization as the SyrE1 module (Fig. 1B). Strains carrying a transposon insertion in sypA failed to produce syringopeptin and were unaffected in syringomycin production.
Regulation of Production
Syringopeptin is produced under the same set of culture conditions as syringomycin (88), suggesting that synthesis of both classes of toxins is activated by a common regulatory system. Nevertheless, it is unknown if the gacS-gacA regulon (219) and the syrP gene (293) that control syringomycin production play a corresponding role in controlling syringopeptin production. A more thorough analysis of the syr and syp gene clusters is needed to identify other regulatory elements that control the production of one or both lipopeptide toxins.
Common Mechanism of Secretion for Syringopeptin and Syringomycin
The syrD gene (Fig. 8) encodes a member of the ABC transporter family which functions in the export of cytotoxic or proteolytic proteins that enhance virulence in prokaryotes (209). ABC transporter proteins have been associated with the secretion of both nonribosomally (79, 178) and ribosomally encoded peptide antibiotics (44), but SyrD is the first ABC transporter protein to be implicated in secretion of a lipopeptide antibiotic. SyrD exhibits high similarity to PvdE, an ABC transporter required for pyoverdin secretion in P. aeruginosa (151). An ATP-binding pocket of SyrD is located in the hydrophilic C terminus and is cytoplasmic, whereas the largely hydrophobic N terminus is predicted to reside in the inner membrane. Consequently, SyrD functions as an ATP-driven efflux pump for syringomycin secretion across the cytoplasmic membrane. Grgurina et al. (88) showed that syrD mutants of B301D fail to produce both syringomycin and syringopeptin, suggesting that secretion of both lipodepsipeptides is linked in P. syringae pv. syringae.
The highly efficient SyrD-mediated export system may explain the resistance of P. syringae pv. syringae to lipodepsipeptide toxins. Syringomycin is toxic to P. syringae at high concentrations (93), and so a deficiency in toxin export would be lethal unless toxin biosynthesis is reduced. Accordingly, strains carrying mutations in the syrD gene do not accumulate the toxins, and expression of a plasmid-borne syrB-lacZ reporter gene fusion is reduced substantially (88, 209). An active efflux by ABC transporter proteins is frequently the basis for autoresistance to antibiotics produced by actinomycetes (153).
Relative Contribution of Syringopeptin and Syringomycin to Virulence
All strains of P. syringae pv. syringae that have been analyzed produce both classes of the lipodepsipeptide phytotoxins, and this suggests interrelated roles for the toxins in the plant-pathogen interaction. It is now recognized that both syringomycin and syringopeptin are pore-forming cytotoxins that cause necrosis in plants by similar mechanisms (103). The advantages of producing two phytotoxic cytotoxins during pathogenesis are unclear, but their contribution to virulence is significant based on assays of mutants defective in the synthesis of one or both types of lipopeptide toxins. For example, strains carrying mutations in either the syrB1 gene, encoding a syringomycin synthetase, or the sypA gene, encoding a syringopeptin synthetase, exhibit a 35 to 50% reduction in virulence compared to the wild-type strain in assays of immature cherry fruits (209, 239). Strains carrying mutations in both syrB1 and sypA showed the greatest reduction in virulence based on measurements of lesion size in cherry fruits. The virulence of a syrB1-sypA double mutant was roughly equivalent to that of the syrD mutant BR105 (209) defective in secretion of both classes of lipodepsipeptides (88). In contrast, a mutation in the syrB operon of strain B728a pathogenic to bean was reported to have no effect on virulence in bean pathogenicity tests (125). Thus, the contribution of toxin production to virulence may vary with the plant host parasitized by P. syringae pv. syringae.
TABTOXIN
Tabtoxin is a monocyclic β-lactam produced by P. syringae pv. tabaci, coronafaciens, and garcae (163). The dipeptide toxin contains tabtoxinine-β-lactam (TβL) linked by a peptide bond to threonine (Fig. 11). Although tabtoxin is the primary intracellular metabolite produced (64), the chlorosis-inducing activity occurs only after hydrolysis of the peptide bond by aminopeptidases of plant or bacterial origin (136, 269). Cleavage of the peptide bond in tabtoxin releases TβL, the toxic moiety (136, 269).
FIG. 11.
Structure of tabtoxin, which consists of the toxic moiety, TβL, linked via an amide bond to threonine. The arrow shows the site of aminopeptidase cleavage, which releases TβL.
Mode of Action
TβL irreversibly inhibits glutamine synthetase (263). The inhibition of glutamine synthetase has at least two major effects; first, the enzyme is presumably the major route for glutamine synthesis in plants, and second, the enzyme is the only way to efficiently detoxify ammonia. A variety of harmful effects have been attributed to ammonia in plants, including disruption of the thylakoid membrane of the chloroplast and uncoupling of photophosphorylation (66, 267). Protection of the bacteria from the toxin has been associated with the adenylation of glutamine synthetase, which renders the target enzyme less susceptible to inactivation by TβL (127). A second potential detoxification mechanism involves the production of β-lactamases which hydrolyze the β-lactam ring of TβL to liberate the nontoxic metabolite, tabtoxinine (46, 128).
Genetic Aspects of Production
TβL is associated with the symptoms of wildfire disease on tobacco and halo blight of oats but is considered to be a virulence factor rather than an essential component of these diseases (63). For example, P. syringae pv. angulata, which induces necrotic spots on tobacco leaves without producing chlorotic halos, is considered to be a spontaneous toxin-deficient derivative of P. syringae pv. tabaci (124). Similarly, P. syringae pv. striafaciens is thought to be a nontoxigenic derivative of P. syringae pv. coronafaciens, the causal agent of halo blight of oats (284). The natural occurrence of tabtoxin-defective strains is caused by the instability of the tabtoxin biosynthetic gene cluster, which excises from the chromosome at fairly high frequencies (124, 268). Kinscherf et al. (124) showed that a clone containing a 31-kb DNA fragment conferred tabtoxin production to P. syringae BR2 and Cit7, suggesting that all the genes necessary for tabtoxin synthesis in P. syringae were clustered in this 31-kb region.
Biosynthesis and Regulation
The biosynthetic precursors of tabtoxin were identified by the incorporation of 13C-labeled compounds and shown to consist of l-threonine and l-aspartate for the side chain and pyruvic acid and the methyl group of l-methionine for the β-lactam moiety (222, 274). A biosynthetic model for the formation of TβL resembles that of lysine, where the first dedicated step is the DapA-catalyzed condensation of aspartic acid semialdehyde with pyruvate to form l-2,3-dihydropicolinate (DHDPA) (Fig. 12). The next step, the pyridine nucleotide-linked reduction of DHDPA to l-2,3,4,5-tetrahydropicolinate (THDPA), is catalyzed by DapB (Fig. 12). Previous studies have suggested that tabtoxin biosynthesis branches off from the lysine biosynthetic pathway before the formation of diaminopimelate (DAP) (222, 274).
FIG. 12.
Biochemical pathways involved in the synthesis of lysine and TβL. In lysine biosynthesis, the first committed step is the DapA-catalyzed condensation of aspartic acid semialdehyde with pyruvate to form DHDPA. DHDPA is then reduced to THDPA, a reaction catalyzed by DapB. Tabtoxin biosynthesis is thought to diverge from the lysine biosynthetic pathway after THDPA synthesis and before DAP formation (vertical arrow). TabB is related to DapD, a gene encoding THDPA succinyl-CoA succinyltransferase. TabB is thought to function as an acetyltransferase that converts an unknown compound (xTHDPA) to an acetyl derivative, which is further metabolized to TβL.
Engst and Shaw (69) identified a gene, tabA, which is essential for tabtoxin production. The discovery of this gene provided the first experimental data to support the hypotheses that the precursors for tabtoxin originate from the lysine biosynthetic pathway. The deduced amino acid sequence of tabA showed significant relatedness to lysA, which encodes DAP decarboxylase in bacteria. Although tabA was not required for lysine biosynthesis, Engst and Shaw (69) proposed that the tabA translational product is an enzyme which recognizes a substrate analogue of a compound in the biosynthetic route to lysine. In a subsequent study, Liu and Shaw (141) showed that dapB was required for both DAP and TβL synthesis. Their results indicated that the most likely branch point of the lysine and TβL pathways occurred after THDPA synthesis and before DAP formation (Fig. 12).
The deduced product of a tabB, also located in the TβL biosynthetic region (140), showed relatedness to dapD (Fig. 12), a gene encoding THDPA succinyl-CoA succinyltransferase (THDPA-ST). Complementation studies and enzymatic assays indicated that tabB encodes a product with THDPA-ST activity. Liu and Shaw (140) proposed that TabB is an acetyltransferase that converts an unknown compound to an acetyl derivative, which is further metabolized to TβL (Fig. 12). This is consistent with the proposal of Feistner et al. (72) that acetylated intermediates are involved in tabtoxin biosynthesis.
In summary, DapB is essential for both lysine and tabtoxin biosynthesis and THDPA may be an intermediate in both pathways. Three genes have been characterized in the 31-kb region which contains all genes necessary for TβL synthesis and tabtoxin resistance: tabA, tabB, and tblA (15, 69, 140). Although there is no obvious relationship between TblA and known polypeptides, TabA has significant sequence homology to LysA from E. coli and P. aeruginosa whereas TabB shows relatedness to DapD.
Some progress has been made on elucidating factors that regulate tabtoxin biosynthesis in P. syringae. Durbin and Uchytil (65) showed that production of TβL by P. syringae pv. tabaci required the addition of zinc to the growth media. In a subsequent study, zinc was shown to be required for the aminopeptidase activity which hydrolyzes tabtoxin to release TβL (136). Barta et al. (16) showed that tblA, a gene required for tabtoxin synthesis, is regulated by the gacS (lemA) gene in P. syringae pv. coronafaciens. As mentioned above, the gacS locus was conserved among P. syringae pathovars and contained domains that are characteristic of histidine protein kinases which function as environmental sensors (101, 218).
PHASEOLOTOXIN
Phaseolotoxin is produced by P. syringae pv. phaseolicola and actinidiae, which cause halo blight on legumes and bacterial canker on kiwifruit, respectively (157, 230). The structure of phaseolotoxin was elucidated by Mitchell (157), with minor revision by Moore et al. (177), and consists of a sulfodiaminophosphinyl moiety linked to a tripeptide consisting of ornithine, alanine, and homoarginine (Fig. 13A).
FIG. 13.
Structure of phaseolotoxin (A) and octicidine (B). Plant peptidases cleave phaseolotoxin (arrow) to release the alanine and homoarginine residues, a reaction which results in octicidine formation.
Mechanism of Action
Phaseolotoxin competitively inhibits ornithine carbamoyl transferase (OCTase), a critical enzyme in the urea cycle which converts ornithine and carbamoyl phosphate to citrulline (Fig. 14). Although phaseolotoxin is a reversible inhibitor of OCTase, it is hydrolyzed in planta by peptidases to produce Nδ-(N′sulfodiaminophosphinyl)-l-ornithine, also called octicidine or PSorn (Fig. 13B). Octicidine is an irreversible inhibitor of OCTase and the predominant form of the toxin in infected tissues (164). Inhibition of OCTase causes an accumulation of ornithine and a deficiency in intracellular pools of arginine (Fig. 14), leading to chlorosis. The urea cycle is a critical pathway in both prokaryotes and eukaryotes, and P. syringae pv. phaseolicola deals with the toxic effect of phaseolotoxin by producing two isozymes of OCTase. One isozyme is resistant to the toxin (ROCTase), and one is sensitive (SOCTase). During conditions favorable to phaseolotoxin production, P. syringae pv. phaseolicola synthesizes the ROCTase isoform (201, 251, 262).
FIG. 14.
Mechanism of action of octicidine (PSorn), the toxic moiety of phaseolotoxin. PSorn is an irreversible inhibitor of OCTase, a critical enzyme in the urea cycle which converts ornithine and carbamoyl phosphate to citrulline. Inhibition of OCTase causes an accumulation of ornithine and a deficiency in intracellular pools of arginine, leading to chlorosis. Diagonal parallel lines indicate the block in the urea cycle elicited by PSorn.
Genetic Aspects of Production
The importance of phaseolotoxin in the virulence of P. syringae pv. phaseolicola was demonstrated in an early study by Patil et al. (198), who showed that phaseolotoxin-defective mutants did not move systemically in bean plants. Mutational complementation has since been used to isolate genes involved in phaseolotoxin production (200, 294). Peet et al. (200) reported the first cloning of genes required for phaseolotoxin biosynthesis on a cosmid clone designated pRCP17. In a later study, pRCP17 was shown to contain the argK gene, which encodes ROCTase (201). A 2.6-kb fragment from pRCP17 complemented several Tox− mutants and was subsequently used as a probe for detection of strains which produce phaseolotoxin (233).
In an independent study, Zhang et al. (294) isolated a clone designated pHK120 which contained a 25-kb insert and restored phaseolotoxin production to Tox− mutants. Pairwise complementation analysis indicated that pHK120 contained a minimum of eight transcriptional units designated phtA through phtH. A comparison of the insert from pHK120 with that from pRCP17 revealed that the inserts in these two cosmids overlap; pHK120 lacked the argK gene but included regions missing from pRCP17 (294). Zhang and Patil (295) determined the nucleotide sequence of the phtE locus, a region transcribed as a 6.4-kb operon. Six ORFs were identified in the phtE transcript, and three of these showed significant relatedness to genes or motifs deposited in various databases. The ORF3 amino acid sequence showed significant relatedness to acetylornithine aminotransferase (ACOAT) and ornithine aminotransferase (OAT) from various organisms. ACOAT catalyzes the reversible conversion of N-acetylglutamate-5-semialdehyde and glutamate to N2-acetylornithine and 2-oxoglutarate, whereas OAT catalyzes the transfer of the acetyl group of N2-acetylornithine onto glutamate to yield ornithine and N-acetylglutamate (50). These results suggest that ORF3 is involved in the production of ornithine, a constituent of phaseolotoxin (295). ORF5 contained an H-T-H domain and a putative leucine zipper, motifs which suggest a regulatory role for ORF5 in phaseolotoxin production. The translational product of ORF6 showed relatedness to a number of fatty acid desaturases; these enzymes are known to generate unsaturated fatty acids for the synthesis of phospholipids in the cytoplasmic membrane (279). Independently, Hatziloukas et al. (97) sequenced the 2.6-kb fragment used for detection of P. syringae pv. phaseolicola and showed that it contained a gene (designated ptx) related to fatty acid desaturases. Although several differences were noted between the nucleotide sequence of ORF6 and ptx, they appear to be the same gene. Hatziloukas et al. (97) speculated that the fatty acid desaturase encoded by ptx may facilitate the export of phaseolotoxin across the bacterial membrane at the low temperatures conducive to phaseolotoxin production.
Biosynthesis and Regulation
Biosynthetic precursors for the Nδ-(N′sulfodiaminophosphinyl) moiety of phaseolotoxin have not been identified. Märkisch and Reuter (148) demonstrated that the homoarginine and ornithine residues of phaseolotoxin are synthesized by a transamidination reaction from arginine and lysine. The amindinotransferase had an Mr of about 200,000 and showed high substrate specificities for arginine and lysine in phaseolotoxin-producing strains of P. syringae pv. phaseolicola. Zhang and Patil (295) suggested that the ORF3 product of the phtE may catalyze the formation of the ornithine needed for phaseolotoxin production, but biochemical evidence for this function is lacking.
A nonribosomal, thiotemplate mechanism similar to that used for the synthesis of peptide antibiotics is likely to be required for biosynthesis of phaseolotoxin, since this molecule contains a tripeptide moiety consisting of ornithine, alanine, and homoarginine. As noted above, peptide synthetases contain adenylation and thiolation domains consisting of conserved core sequences with the same order and spacing (Fig. 1B and 8). Turgay et al. (265) synthesized degenerate primers derived from the ATPase and thioester-forming domains (cores 4 and 6, respectively). These degenerate primers were used to amplify and clone a putative peptide synthetase from P. syringae pv. phaseolicola. Although the involvement of this region in phaseolotoxin synthesis has not yet been demonstrated, this approach was previously used to isolate portions of the surfactin synthetase from B. subtilis (35).
Temperature is a factor which regulates phaseolotoxin production in P. syringae pv. phaseolicola. Goss (84) showed that the chlorosis associated with P. syringae pv. phaseolicola infection of bean was induced at lower temperatures (18 to 20°C) and absent at higher temperatures (28 to 32°C). Subsequent studies showed that phaseolotoxin production decreased progressively at temperatures above 18°C (158, 184). Rowley et al. (223) described the production of a repressor when P. syringae pv. phaseolicola was grown at 28°C, a temperature unfavorable for phaseolotoxin synthesis. Production of this repressor at the nonpermissive temperature may explain the thermoregulation of phaseolotoxin biosynthesis.
DETECTION OF PHYTOTOXINS AND TOXIN-PRODUCING BACTERIA
Visual assessment of phytotoxin production in planta can be somewhat subjective since there are a limited number of ways in which plants react visibly to phytotoxins (stunting, chlorosis, necrosis, and wilting). However, it should be emphasized that studies of particular phytotoxins are probably influenced by the visible evidence of their activity. Some phytotoxins may instead act by changing metabolic processes in the host in such a way that the deleterious activity might be manifested only at the biochemical level.
When a toxic metabolite is suspected of contributing to disease, extraction of the component from the producing organism is required. If the toxin is produced in vitro, the initial purification may proceed from aqueous or organic phases of the culture supernatant. Some knowledge of organic chemistry is required at this juncture since the ultimate aim will be to obtain or verify the structure of the toxin by mass spectrometry, nuclear magnetic resonance spectroscopy, or other analytical methods. Reproduction of some aspect of the disease (for example, chlorosis or necrosis) by using the purified compound is essential in proving the role of the phytotoxin in symptom development.
As noted in the previous sections on individual phytotoxins, molecular techniques have been used to construct Tox− derivatives of phytopathogenic bacteria. Mutational cloning has been used successfully to recover genes required for phytotoxin production, and Tox− mutants have been used to assess the contribution of phytotoxins to disease development and symptomology (24, 124, 177, 200, 287, 294). A priori, it is necessary to establish a reliable and simple method for detecting production of the phytotoxin.
Bioassays for Toxins Produced by P. syringae
Some phytotoxins are antimicrobial and can be detected in bioassays with sensitive fungi or bacteria. For example, phaseolotoxin production can be detected at picogram levels by growth inhibition of E. coli K-12 (250). Both Geotrichum candidum and Rhodotorula pilimanae are sensitive to syringomycin and can be used in bioassays for this phytotoxin (93, 293). In contrast, syringopeptin production is detected in bioassays with Bacillus megaterium (133). Tabtoxin also has antimicrobial activity and can be detected in bioassays with toxin-sensitive bacteria or fungi (81). Although COR is not antimicrobial, it can be detected by its ability to induce chlorosis in a variety of plants; however, this assay is qualitative rather than quantitative. Völksch et al. (277) have described a semiquantitative bioassay for COR in which a hypertrophic reaction on potato tissue is used to detect the toxin. Although this assay is sensitive, some variability can occur depending on the potato cultivar used and the age of the potato tissue.
Analytical Methods for Assessing Toxin Production
Quantitative chromatographic methods are available for detecting COR (23), tabtoxin (15), and lipodepsipeptides, syringomycin, and syringopeptin (9). To facilitate genetic studies of COR biosynthesis, a rapid extraction and fractionation method for COR was developed which involves the direct extraction of organic acids from 0.5 ml of culture supernatant, a 9-min fractionation on a reverse-phase C8 column in a gradient of acetonitrile and water, and quantitative detection at 208 nm (189). The extraction can be performed in microcentrifuge tubes and takes approximately 3 min, and HPLC fractionation accurately separates and quantifies CFA, COR, coronafacoylvaline, and CFA-Ile. The availability of quantitative detection methods for COR and the two defined intermediates in the COR pathway (CFA and CMA) has greatly facilitated the analysis of mutant phenotypes (21).
Molecular Detection of Phytotoxins and Toxin Synthesis Genes
One outcome of the mutational cloning of toxin gene clusters has been the development of DNA probes or PCR primers for the identification of phytotoxin-producing strains of P. syringae. For example, a DNA probe containing the ptx gene was used to detect and identify P. syringae pv. phaseolicola from mixed cultures and diseased specimens (233). Oligonucleotide primers derived from the DNA probe were sensitive enough to detect P. syringae pv. phaseolicola in commercial seed lots (234). Recently, PCR primers based on the sequence of the gene encoding ROCTase, argK, were used for specific detection of P. syringae pv. phaseolicola and actinidiae (181, 230). The PCR amplification procedure was applied directly to bacteria present in seed extracts and shown to be extremely sensitive (181). Similarly, DNA probes and PCR primer sets from the COR biosynthetic gene cluster have proven useful for the detection of COR-producing pathovars of P. syringae (28, 53, 257).
The utility of toxin synthesis genes in strain identification was further demonstrated by Quigley and Gross (208), who used DNA probes containing syrB1 and syrD to show the conservation of these sequences among syringomycin-producing strains of P. syringae. Scheck et al. (235) used the syrB1 and syrD genes in combination with bioassays on lilac plantlets to identify strains of P. syringae pv. syringae pathogenic on woody ornamentals. More recently, Sorensen et al. used PCR to amplify a 752-bp fragment of syrB1 and successfully detected P. syringae pv. syringae strains which produced syringotoxin and syringostatin, cyclic lipodepsinonapeptide toxins related to syringomycin (246).
Although DNA probes and PCR primers have been extremely helpful in bacterial detection, they do not indicate whether the phytotoxin is actively synthesized in vitro or in planta. Although serological methods are more attractive for this type of analysis, the phytotoxins discussed in this review are too small to be antigenic and must be conjugated to a carrier protein to form a hapten conjugate that is large enough to generate antibodies. Leary et al. (135) reported on the use of polyclonal antibodies to detect COR in vitro, suggesting that an immunological approach could be used for COR detection. More recently, Jones et al. (111) constructed a COR-ovalalbumin conjugate where COR was linked to ovalbumin at the free carboxyl group present on CMA. This hapten conjugate was used to produce monoclonal antibodies that were subsequently used in a competitive enzyme-linked immunosorbent assay. One monoclonal line recognized COR and coronafacoylvaline equally well and CFA-Ile and CFA-aIle to a lesser extent. Furthermore, the monoclonal antibody did not recognize CFA or CMA, which are the nonphytotoxic intermediates in the pathway to COR. Since COR-producing P. syringae strains often synthesize coronafacoyl analogues (161, 165, 169), this monoclonal antibody should be useful in detecting the entire family of coronafacoyl phytotoxins.
ENGINEERING PLANTS WITH PHYTOTOXIN RESISTANCE
Several novel strategies have been used to develop plants with resistance to phytotoxins. When phytotoxins are broadly antimicrobial, they are also frequently toxic to the producing organism. Consequently, one potential source of toxin resistance is the phytotoxin producer, P. syringae.
Transgenic Plants with Phaseolotoxin and Tabtoxin Resistance
The cloning of the gene encoding ROCTase (argK) from P. syringae pv. phaseolicola and its subsequent use as a transgene have been successful in the development of phaseolotoxin-resistant plants. As mentioned above, phaseolotoxin competitively inhibits OCTase, which converts ornithine and carbamoyl phosphate to citrulline (Fig. 14). The argK gene has been cloned and sequenced (97, 180), and two laboratories have used it as a source of phaseolotoxin resistance in tobacco (55, 96). In plant cells, OCTase is produced in the chloroplast, and so it was necessary to target the product of the argK gene into the chloroplast. This was done by fusing the argK coding region to the transit peptide of the small subunit of ribulose bisphosphate carboxylase (Fig. 15) (55). This construct was cloned into a plant transformation vector, introduced into A. tumefaciens, and then transformed into tobacco leaf disks. Rooted plants were then transplanted to soil and screened for OCTase activity and phaseolotoxin sensitivity. Control tobacco plants which contained SOCTase showed chlorosis in response to phaseolotoxin treatment and had a reduced chlorophyll content. Transgenic plants expressing ROCTase did not turn chlorotic when treated with phaseolotoxin and had no change in chlorophyll content. Control tobacco plants were systemically infected when inoculated with P. syringae pv. phaseolicola, but the transgenic plants expressing ROCTase showed a localized HR in response to infection. Therefore, a level of host resistance to both the pathogen and the toxin was expressed in the transgenic plants (55). The next challenge will be to introduce argK into bean plants, the natural host for P. syringae pv. phaseolicola.
FIG. 15.
Construct used for obtaining transgenic tobacco with resistance to phaseolotoxin. LB and RB indicate the left and right border regions, respectively, of the T-DNA region in the Ti plasmid of Agrobacterium tumefaciens. The argK gene encodes ROCTase, the toxin-resistant form of ornithine carbamoyltransferase. A transit peptide (tp) sequence was inserted at the 5′ end of argK to facilitate targeting of the gene product to the chloroplast. The argK gene was expressed under control of the 35S promoter of CaMV. The neomycin transferase gene (NPTII) was used as a selective marker and expressed by using the nos (nopaline synthetase) promoter. Arrowheads indicate the direction of transcription. See reference 55 for additional details.
As mentioned above, tabtoxin and TβL inhibit glutamine synthetase; consequently, tabtoxin-producing strains of P. syringae must protect themselves from these compounds. Anzai et al. (6) isolated a gene designated ttr (for “tabtoxin resistance”), which conferred resistance to transgenic tobacco harboring this gene. Presumably, ttr functions to acetylate tabtoxin and TβL, and the acetylated forms of these toxins are nontoxic (6). Batchvarova et al. (18) reported that the resistance conferred by ttr was heritable in transgenic tobacco. Other potential sources of resistance to tabtoxin include the β-lactamases synthesized by the producing organism and the enzymes which adenylate glutamate synthetase, rendering it resistant to tabtoxin and TβL (46, 127, 128).
Selection for Coronatine Resistance in A. thaliana
Since the mode of action of COR is unclear, precise modification of the target site to render it resistant to the toxin is not currently possible. Moreover, COR does not show antibiotic activity to prokaryotic cells, so that COR producers do not possess a resistance gene that could be used to develop transgenic plants with insensitivity to the toxin. However, one alternative strategy is to use mutagenesis to select COR-resistant (Corr) plants and then identify the target site in the mutants by map-based cloning. For example, the coi1 mutant of Arabidopsis thaliana was obtained by exposing seeds to mutagenic concentrations of ethyl methanesulfonate (75). The coi1 mutants were insensitive to MeJA and resistant to P. syringae infection (75). The wild-type allele (COI1) was recently localized by mapped-based cloning, sequenced, and shown to contain leucine-rich repeats and an F-box motif, which suggests that the COI1 protein plays a role in protein-protein interactions (286). Ultimately, COI1 and the identification of related genes may facilitate the introduction of Corr into agronomically important host plants, such as tomato and soybean.
ENGINEERING NEW COMPOUNDS BY USING PHYTOTOXIN GENES
It is now possible to take a genetic rather than synthetic approach to biosynthesize novel peptides and polyketides with altered biological properties. Much of the reprogramming or engineering of novel peptides and polyketides has involved actinomycete or fungal gene clusters, but much of this technology could also be applied to compounds synthesized by Pseudomonas.
Peptide Synthetases
As mentioned previously, the nonribosomal synthesis of peptides is catalyzed by using a protein template that contains the correct number and order of activating units (126). This knowledge has been used for the rational design of bioactive peptides with altered biological properties. Marahiel’s group (248, 249) was the first to successfully reprogram a peptide synthetase to produce a novel peptide. These experiments involved the srfA operon (Fig. 8), which encodes the biosynthesis of surfactin in B. subtilis. The recombination was achieved by a combination of gene disruption and replacement. The result was the replacement of the Bacillus subtilis leucine domain with the phenylalanine, ornithine, and leucine domains from the grs operon of Bacillus brevis and the cysteine and valine domains of the ACV synthetase from Penicillium chrysogenum (248, 249). The surfactin derivatives produced by the engineered Bacillus strains were characterized by infrared spectroscopy and mass spectrometry and shown to contain the predicted amino acid substitutions. Furthermore, the genetically modified surfactin derivatives retained biological activity. These results are exciting because they indicate that amino acid-activating domains from different organisms can be exchanged and used to produce novel compounds. Ultimately, it should be possible to conduct similar experiments with Pseudomonas by using the alloisoleucine-activating domain encoded by cmaA or one of the amino acid-activating domains encoded by the syr or syp gene clusters.
Polyketide Synthases
Both type I and II PKSs have been exploited in the construction of novel products via genetic engineering (59, 113–115, 146, 149, 150, 186, 207, 224). One important innovation was the development of a Streptomyces host-vector system which facilitated the efficient construction and expression of recombinant polyketides (150). The shuttle vector pRM5 contained the minimal set of type II genes required for polyketide synthesis but lacked the tailoring genes which diversify the resulting product (150). The first hybrids to be constructed contained various combinations of the actinorhodin, granaticin, and tetracenomycin gene clusters, and the majority of recombinants generated novel polyketide products (150).
The Streptomyces coelicolor CH999(pRM5) expression system proved immensely useful for analyzing the function of type II PKS genes. However, the same system has also been used to generate recombinant polyketides containing the type I eryA genes. For example, overproduction of the DEBS 1 protein in pRM5 resulted in the synthesis of a triketide lactone which was the predicted product of modules 1 and 2 (114). The quantity of this product increased when the TE domain located at the C terminus of DEBS 3 was added to DEBS 1 (48, 115). Oliynyk et al. (186) further utilized the pRM5 expression system in a subsequent experiment where the acetyltransferase (AT) domain from module 1 of DEBS 1 was replaced with the AT from module 2 of the rapamycin PKS. This experiment resulted in the synthesis of two novel triketide lactones with the predicted structures (186).
Although the reprogramming experiments discussed above involve PKSs from gram-positive bacteria, exploitation of these approaches has been extended to fungal PKSs. For example, the fungal polyketide 6-methylsalicylic acid is synthesized by 6-methylsalicylic acid synthase, a multifunctional PKS. Bedford et al. (20) expressed the 6-methylsalicylic acid synthase genes in S. coelicolor CH999 and showed that the recombinant strain produced significant amounts of 6-methylsalicylic acid. These results are especially relevant to the potential expression of Pseudomonas polyketides in heterologous hosts. The expression of selected multifunctional or monofunctional PKS genes from the CFA region in pRM5 could be used to generate new polyketides with altered biological activities. For example, the starter unit for CFA synthesis is presumed to be α-ketoglutarate (194). The monofunctional proteins, Cfa1 to Cfa5, are presumably involved in the conversion of the starter unit into a cyclopentenone product, which is then loaded to Cfa6 (197). Some of these genes could be exploited to engineer the incorporation of unique precursor molecules into existing polyketides. Such changes might change or expand the biological activity of the resulting polyketide product.
PERSPECTIVES
Our understanding of phytotoxin biosynthesis, regulation, and mode of action has expanded largely due to multidisciplinary studies which integrate genetics, biochemistry, and organic chemistry. Additional work is clearly needed to define the structural genes involved in the synthesis of tabtoxin, phaseolotoxin, and syringopeptin. For COR and syringomycin, biochemical studies to examine proposed gene functions based on sequence analyses have been completed or are under way. The results of these experiments will facilitate the development or revision of models for phytotoxin assembly. The mechanisms of polyketide and peptide toxin biosynthesis in Pseudomonas exhibit unique features that expand the boundaries of antimicrobial metabolite synthesis. These systems offer valuable insight into the ecological significance of toxin synthesis and how they may be exploited in the quest to bioengineer new pharmaceutical products.
The regulatory circuits which modulate phytotoxin synthesis in P. syringae are poorly understood. Although some of the signals for phytotoxin production have been defined, the mechanisms used for integration of these signals and the regulatory cascades involved remain unclear. An especially intriguing question is the potential relationship between phytotoxin synthesis and the type III secretion system encoded by the hrp gene cluster. The coordinated regulation of genes involved in pathogenicity (hrp cluster) and virulence (phytotoxin production) seems logical but has not been established.
The evolution of phytotoxin gene clusters remains a mystery. The structural gene clusters for COR, syringomycin, and syringopeptin indicate biosynthetic relatedness to compounds synthesized by nonribosomal, multifunctional enzymes. However, each phytotoxin gene cluster has distinctive features, suggesting that evolutionary divergence from systems defined in gram-positive bacteria has occurred. Furthermore, genes related to various transposases have been identified in the borders of phytotoxin gene clusters (270). Obviously, the potential occurrence of phytotoxin gene clusters within pathogenicity islands warrants investigation.
CONCLUSIONS
This review focuses on the five most intensively studied phytotoxins of P. syringae, all of which contribute significantly to bacterial virulence. COR functions partly as a mimic of MeJA, a potent hormone synthesized by plants undergoing biological stress. Syringomycin and syringopeptin form pores in plasma membranes, a process that leads to electrolyte leakage. Tabtoxin and phaseolotoxin are strongly antimicrobial and function by inhibiting glutamine synthetase and OCTase, respectively.
Genetic analysis has revealed the mechanisms responsible for toxin biosynthesis. COR biosynthesis requires the cooperation of PKSs and peptide synthetases for the CFA and CMA moieties, respectively. Tabtoxin is derived from the lysine biosynthetic pathway, whereas peptide synthetases are required for syringomycin, syringopeptin, and phaseolotoxin assembly.
Activation of toxin synthesis is controlled by diverse environmental factors including plant signal molecules and temperature. Genes involved in the regulation of phytotoxin synthesis have been located within the COR and syringomycin gene clusters; however, additional regulatory genes are required for the synthesis of these and other phytotoxins. Global regulatory genes such as gacS modulate phytotoxin production in certain pathovars, indicating the complexity of the regulatory circuits controlling phytotoxin synthesis.
The COR and syringomycin gene clusters have been intensively characterized and show potential for constructing modified polyketides and peptides. Genetic reprogramming of peptide and polyketide synthetases is feasible, and portions of the COR and syringomycin gene clusters could be valuable resources in developing new antimicrobial agents. Finally, characterization of phytotoxin gene clusters has led to improved methods of disease detection and diagnosis.
ACKNOWLEDGMENTS
This work was supported by a grant from the Oklahoma Agricultural Experiment Station (C.B.) and by National Science Foundation grants MCB-9603618 and MCB-9807774 (C.B.). D.G. acknowledges support from the National Research Initiative Competitive Grants Program of the USDA (grants 92-37303-7732 and 97-35303-4460).
REFERENCES
- 1.Agron P G, Monson E K, Ditta G S, Helinski D R. Oxygen regulation of expression of nitrogen fixation genes in Rhizobium meliloti. Res Microbiol. 1994;145:454–459. doi: 10.1016/0923-2508(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 2.Akers C P, Hoff J E. Simultaneous formation of chymopapain inhibitor activity and cubical crystals in tomato leaves. Can J Bot. 1980;58:1000–1003. [Google Scholar]
- 3.Alfano J B, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankenbauer R G, Nester E W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990;172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- 6.Anzai H, Yoneyama K, Yamaguchi I. Transgenic tobacco resistant to a bacterial disease by the detoxification of a pathogenic toxin. Mol Gen Genet. 1989;219:492–494. [Google Scholar]
- 7.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 8.Backman P A, DeVay J E. Studies on the mode of action and biogenesis of the phytotoxin syringomycin. Physiol Plant Pathol. 1971;1:215–234. [Google Scholar]
- 9.Ballio A, Barra D, Bossa F, Collina A, Grgurina I, Marino G, Moneti G, Paci M, Pucci P, Segre A, Simmaco M. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 1991;291:109–112. doi: 10.1016/0014-5793(91)81115-o. [DOI] [PubMed] [Google Scholar]
- 10.Ballio A, Bossa F, Camoni L, Di Giorgio D, Flamand M-C, Maraite H, Nitti G, Pucci P, Scaloni A. Structure of fuscopeptins, phytotoxic metabolites of Pseudomonas fuscovaginae. FEBS Lett. 1996;381:213–216. doi: 10.1016/0014-5793(96)00043-9. [DOI] [PubMed] [Google Scholar]
- 11.Ballio A, Bossa F, Di Giorgio D, Di Nola A, Manetti C, Paci M, Scaloni A, Segre A L. Solution conformation of the Pseudomonas syringae pv. syringae phytotoxic lipodepsipeptide syringopeptin 25-A: two-dimensional NMR, distance geometry and molecular dynamics. Eur J Biochem. 1995;234:747–758. doi: 10.1111/j.1432-1033.1995.747_a.x. [DOI] [PubMed] [Google Scholar]
- 12.Ballio A, Bossa F, Di Giorgio D, Ferranti P, Paci M, Pucci P, Scaloni A, Segre A, Strobel G A. Novel bioactive lipodepsipeptides from Pseudomonas syringae: the pseudomycins. FEBS Lett. 1994;355:96–100. doi: 10.1016/0014-5793(94)01179-6. [DOI] [PubMed] [Google Scholar]
- 13.Ballio A, Collina A, Di Nola A, Manetti C, Paci M, Segre A. Determination of structure and conformation in solution of syringotoxin, a lipodepsipeptide from Pseudomonas syringae pv. syringae by 2D NMR and molecular dynamics. Struct Chem. 1994;5:43–50. [Google Scholar]
- 14.Bangera M G, Thomashow L S. Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by the biological control agent Pseudomonas fluorescens Q2-87. Mol Plant-Microbe Interact. 1996;9:83–90. doi: 10.1094/mpmi-9-0083. [DOI] [PubMed] [Google Scholar]
- 15.Barta T M, Kinscherf T G, Uchytil T F, Willis D K. DNA sequence and transcriptional analysis of the tblA gene required for tabtoxin biosynthesis by Pseudomonas syringae. Appl Environ Microbiol. 1993;59:458–466. doi: 10.1128/aem.59.2.458-466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barta T M, Kinscherf T G, Willis D K. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J Bacteriol. 1992;174:3021–3029. doi: 10.1128/jb.174.9.3021-3029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barzic M R, Guittet E. Structure and activity of persicomycins, toxins produced by a Pseudomonas syringae pv. persicae/Prunus persica isolate. Eur J Biochem. 1996;239:702–709. doi: 10.1111/j.1432-1033.1996.0702u.x. [DOI] [PubMed] [Google Scholar]
- 18.Batchvarova R, Nikolaeva V, Slavov S, Bossolova S, Valkov V, Atanassova S, Guelemerov S, Atanassov A, Anzai H. Transgenic tobacco cultivars resistant to Pseudomonas syringae pv. tabaci. Theor Appl Genet. 1998;97:986–989. [Google Scholar]
- 19.Batoko H, de Kerchove d’Exaerde A, Kinet J M, Bouharmont J, Gage R A, Maraite H, Boutry M. Modulation of plant plasma membrane H+-ATPase by phytotoxic lipodepsipeptides produced by the plant pathogen Pseudomonas fuscovaginae. Biochim Biophys Acta. 1998;17:216–226. doi: 10.1016/s0005-2736(98)00060-1. [DOI] [PubMed] [Google Scholar]
- 20.Bedford D J, Schweizer E, Hopwood D A, Khosla C. Expression of a functional fungal polyketide synthase in the bacterium Streptomyces coelicolor A3(2) J Bacteriol. 1995;177:4544–4548. doi: 10.1128/jb.177.15.4544-4548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender C, Palmer D, Peñaloza-Vázquez A, Rangaswamy V, Ullrich M. Biosynthesis of coronatine, a thermoregulated phytotoxin produced by the phytopathogen Pseudomonas syringae. Arch Microbiol. 1996;166:71–75. [Google Scholar]
- 22.Bender C L, Liyanage H, Palmer D, Ullrich M, Young S, Mitchell R. Characterization of the genes controlling biosynthesis of the polyketide phytotoxin coronatine including conjugation between coronafacic and coronamic acid. Gene. 1993;133:31–38. doi: 10.1016/0378-1119(93)90221-n. [DOI] [PubMed] [Google Scholar]
- 23.Bender C L, Malvick D K, Mitchell R E. Plasmid-mediated production of the phytotoxin coronatine in Pseudomonas syringae pv. tomato. J Bacteriol. 1989;171:807–812. doi: 10.1128/jb.171.2.807-812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender C L, Stone H E, Sims J J, Cooksey D A. Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol. 1987;30:272–283. [Google Scholar]
- 25.Bender C L, Young S A, Mitchell R E. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl Environ Microbiol. 1991;57:993–999. doi: 10.1128/aem.57.4.993-999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender C L, Young S A, Mitchell R E. Ecological and genetic studies of coronatine synthesis in Pseudomonas syringae. In: Galli E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 56–63. [Google Scholar]
- 27.Bent A F, Innes R W, Ecker J R, Staskawicz B J. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant-Microbe Interact. 1992;5:372–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- 28.Bereswill S, Bugert P, Völksch B, Ullrich M, Bender C L, Geider K. Identification and relatedness of coronatine-producing Pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl Environ Microbiol. 1994;60:2924–2930. doi: 10.1128/aem.60.8.2924-2930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bevitt D J, Cortes J, Haydock S F, Leadlay P F. Deoxyerythronolide-B synthase from Saccharopolyspora erythraea. Cloning of the structural gene, sequence analysis and inferred domain structure of the multifunctional enzyme. Eur J Biochem. 1992;204:39–49. doi: 10.1111/j.1432-1033.1992.tb16603.x. [DOI] [PubMed] [Google Scholar]
- 30.Bhakdi S, Mackman N, Nicaud J M, Holland I B. Escherichia coli hemolysin damages target cell membranes by generating transmembrane pores. Infect Immun. 1986;52:63–69. doi: 10.1128/iai.52.1.63-69.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhakdi S, Tranum-Jensen J. Damage to mammalian cells by proteins that form transmembrane pores. Rev Physiol Biochem Pharmacol. 1987;107:147–223. doi: 10.1007/BFb0027646. [DOI] [PubMed] [Google Scholar]
- 32.Bidwai A P, Takemoto J Y. Bacterial phytotoxin, syringomycin, induces a protein kinase-mediated phosphorylation of red beet plasma membrane polypeptides. Proc Natl Acad Sci USA. 1987;84:6755–6759. doi: 10.1073/pnas.84.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bidwai A P, Zhang L, Bachmann R C, Takemoto J Y. Mechanism of action of Pseudomonas syringae phytotoxin, syringomycin. Stimulation of red beet plasma membrane ATPase activity. Plant Physiol. 1987;83:39–43. doi: 10.1104/pp.83.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolter C J. Methyl jasmonate induces papain inhibitor(s) in tomato leaves. Plant Physiol. 1993;103:1347–1353. doi: 10.1104/pp.103.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borchert S, Patil S S, Marahiel M A. Identification of putative multifunctional peptide synthetase genes using highly conserved oligonucleotide sequences derived from known synthetases. FEMS Microbiol Lett. 1992;92:175–180. doi: 10.1016/0378-1097(92)90508-l. [DOI] [PubMed] [Google Scholar]
- 36.Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators. Insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J Mol Biol. 1994;241:363–377. doi: 10.1006/jmbi.1994.1513. [DOI] [PubMed] [Google Scholar]
- 37.Budde I P, Rohde B H, Bender C L, Ullrich M S. Growth phase and temperature influence promoter activity, transcript abundance and protein stability during biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1998;180:1360–1367. doi: 10.1128/jb.180.6.1360-1367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bull C T, Wadsworth M L, Sorensen K N, Takemoto J Y, Austin R K, Smilanick J L. Syringomycin E produced by biological control agents controls green mold on citrus. Biol Control. 1998;12:89–95. [Google Scholar]
- 39.Caffrey P, Bevitt D J, Staunton J, Leadlay P F. Identification of DEBS 1, DEBS 2, and DEBS 3, the multienzyme polypeptides of the erythromycin-producing polyketide synthase from Saccharopolyspora erythraea. FEBS Lett. 1992;304:225–228. doi: 10.1016/0014-5793(92)80624-p. [DOI] [PubMed] [Google Scholar]
- 40.Cangelosi G A, Ankenbauer R G, Nester E W. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci USA. 1990;87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charles T C, Jin S, Nester E W. Two-component sensory transduction systems in phytobacteria. Annu Rev Phytopathol. 1992;30:463–484. doi: 10.1146/annurev.py.30.090192.002335. [DOI] [PubMed] [Google Scholar]
- 42.Che F-S, Kasamo K, Fukuchi N, Isogai A, Suzuki A. Bacterial phytotoxins, syringomycin, syringostatin and syringotoxin, exert their effect on the plasma membrane H+-ATPase partly by a detergent action and partly by inhibition of the enzyme. Physiol Plant. 1992;86:518–524. [Google Scholar]
- 43.Cho H, Cronan J E., Jr Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J Biol Chem. 1993;268:9238–9245. [PubMed] [Google Scholar]
- 44.Chung Y J, Steen M T, Hansen J N. The subtilin gene of Bacillus subtilis ATCC 6633 is encoded in an operon that contains a homolog of the hemolysin B transport protein. J Bacteriol. 1992;174:1417–1422. doi: 10.1128/jb.174.4.1417-1422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cliften P, Wang Y, Mochizuki D, Miyakawa T, Wangspa R, Hughes J, Takemoto J Y. SYR2, a gene necessary for syringomycin growth inhibition of Saccharomyces cerevisiae. Microbiology. 1996;142:477–484. doi: 10.1099/13500872-142-3-477. [DOI] [PubMed] [Google Scholar]
- 46.Coleman R H, Shaffer J, True H. Properties of β-lactamase from Pseudomonas syringae. Curr Microbiol. 1996;32:147–150. doi: 10.1007/s002849900026. [DOI] [PubMed] [Google Scholar]
- 47.Conti E, Stachelhaus T, Marahiel M A, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes J, Wiesmann K E H, Roberts G A, Brown M J B, Staunton J, Leadlay P F. Repositioning of a domain in a modular polyketide synthase to promote specific chain cleavage. Science. 1995;268:1487–1489. doi: 10.1126/science.7770773. [DOI] [PubMed] [Google Scholar]
- 49.Costacurta A, Vanderleyden J. Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol. 1995;21:1–18. doi: 10.3109/10408419509113531. [DOI] [PubMed] [Google Scholar]
- 50.Cunin R, Glansdorff N, Piérard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuppels D C, Ainsworth T. Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl Environ Microbiol. 1995;61:3530–3536. doi: 10.1128/aem.61.10.3530-3536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuppels D A, Howell C R, Stipanovic R D, Stoessl A, Stothers J B. Biosynthesis of pyoluteorin: a mixed polyketide-tricarboxylic acid cycle origin demonstrated by [1,2-13C2]acetate incorporation. Z Naturforsch. 1986;41:532–536. [Google Scholar]
- 53.Cuppels D A, Moore R A, Morris V L. Construction and use of a nonradioactive DNA hybridization probe for detection of Pseudomonas syringae pv. tomato on tomato plants. Appl Environ Microbiol. 1990;56:1743–1749. doi: 10.1128/aem.56.6.1743-1749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Crécy-Lagard V, Marlière P, Saurin W. Multienzymatic non ribosomal peptide biosynthesis: identification of the functional domains catalysing peptide elongation and epimerisation. C R Acad Sci Paris. 1995;318:927–936. [PubMed] [Google Scholar]
- 55.De La Fuente-Martinez J, Mosqueda-Cano G, Alvarez-Morales A, Herrera-Estrella L. Expression of a bacterial phaseolotoxin-resistant ornithyl transcarbamylase in transgenic tobacco confers resistance to Pseudomonas syringae pv. phaseolicola. Bio/Technology. 1992;10:905–909. doi: 10.1038/nbt0892-905. [DOI] [PubMed] [Google Scholar]
- 56.Denny T P. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 57.Di Giorgio D, Camoni L, Ballio A. Toxins of Pseudomonas syringae pv. syringae affect H+-transport across the plasma membrane of maize. Physiol Plant. 1994;91:741–746. [Google Scholar]
- 58.Di Giorgio D, Camoni L, Mott K A, Takemoto J Y, Ballio A. Syringopeptins, Pseudomonas syringae pv. syringae phytotoxins, resemble syringomycin in closing stomata. Plant Pathol. 1996;45:564–571. [Google Scholar]
- 59.Donadio S, McAlpine J B, Sheldon P J, Jackson M, Katz L. An erythromycin analog produced by reprogramming of polyketide synthesis. Proc Natl Acad Sci USA. 1993;90:7119–7123. doi: 10.1073/pnas.90.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donadio S, Staver M J, McAlpine J B, Swanson S, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 61.Dumenyo C K, Mukherjee A, Chun W, Chatterjee A K. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur J Plant Pathol. 1998;104:569–582. [Google Scholar]
- 62.Duncan J L, Schlegel R. Effect of streptolysin O on erythrocyte membranes, liposomes, and lipid dispersions: a protein-cholesterol interaction. J Cell Biol. 1975;67:160–173. doi: 10.1083/jcb.67.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durbin R D. Bacterial phytotoxins: mechanism of action. Experientia. 1991;47:776–783. [Google Scholar]
- 64.Durbin R D, Uchytil T F. The role of intercellular fluid and bacterial isolate on the in vivo production of tabtoxin and tabtoxinine-β-lactam. Physiol Plant Pathol. 1984;24:25–31. [Google Scholar]
- 65.Durbin R D, Uchytil T F. The role of zinc in regulating tabtoxin production. Experientia. 1985;41:136–137. [Google Scholar]
- 66.Durbin R D, Uchytil T F. The mechanism for self-protection against bacterial phytotoxins. Annu Rev Phytopathol. 1988;26:313–329. [Google Scholar]
- 67.Eggink G, de Waard P, Huijberts G N M. The role of fatty acid biosynthesis and degradation in the supply of substrates for poly(3-hydroxyalkanoate) formation in Pseudomonas putida. FEMS Microbiol Rev. 1992;103:159–164. [Google Scholar]
- 68.Emanuele M C, Scaloni A, Lavermicocca P, Iacobellis N S, Camoni L, Di Giorgio D, Pucci P, Paci M, Segre A, Ballio A. Corpeptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata. FEBS Lett. 1998;433:317–320. doi: 10.1016/s0014-5793(98)00933-8. [DOI] [PubMed] [Google Scholar]
- 69.Engst K, Shaw P D. Identification of a lysA-like gene required for tabtoxin biosynthesis and pathogenicity in Pseudomonas syringae pv. tabaci strain PTBR2.024. Mol Plant-Microbe Interact. 1992;5:322–329. doi: 10.1094/mpmi-5-322. [DOI] [PubMed] [Google Scholar]
- 70.Feigin A M, Schagina L V, Takemoto J Y, Teeter J H, Brand J G. The effect of sterols on the sensitivity of membranes to the channel-forming antifungal antibiotic, syringomycin E. Biochim Biophys Acta. 1997;1324:102–110. doi: 10.1016/s0005-2736(96)00214-3. [DOI] [PubMed] [Google Scholar]
- 71.Feigin A M, Takemoto J Y, Wangspa R, Teeter J H, Brand J G. Properties of voltage-gated ion channels formed by syringomycin E in planar lipid bilayers. J Membr Biol. 1996;149:41–47. doi: 10.1007/s002329900005. [DOI] [PubMed] [Google Scholar]
- 72.Feistner G J, Uchytil T F, Knoche K K, Durbin R D. A tabtoxinine-related metabolite from Pseudomonas syringae pv. tabaci. J Org Chem. 1991;56:2922–2925. [Google Scholar]
- 73.Feline T C, Jones R B, Mellows G, Phillips L. Pseudomonic acid. 2. Biosynthesis of pseudomonic acid A. J Chem Soc Perkin Trans I. 1977;1977:309–318. [PubMed] [Google Scholar]
- 74.Ferguson I B, Mitchell R E. Stimulation of ethylene production in bean leaf discs by the pseudomonad phytotoxin coronatine. Plant Physiol. 1985;77:969–973. doi: 10.1104/pp.77.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukuchi N, Isogai A, Nakayama J, Takayama S, Yamashita S, Suyama K, Suzuki A. Isolation and structural elucidation of syringostatins, phytotoxins produced by Pseudomonas syringae pv. syringae lilac isolate. J Chem Soc Perkin Trans I. 1992;1992:875–880. [Google Scholar]
- 77.Fukuchi N, Isogai A, Nakayama J, Takayama S, Yamashita S, Suyama K, Takemoto J Y, Suzuki A. Structure and stereochemistry of three phytotoxins, syringomycin, syringotoxin and syringostatin, produced by Pseudomonas syringae pv. syringae. J Chem Soc Perkin Trans I. 1992;1992:1149–1157. [Google Scholar]
- 78.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di Maio J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, Kempf H-J, Becker J O. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 79.Gaisser S, Hughes C. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol Gen Genet. 1997;253:415–427. doi: 10.1007/s004380050339. [DOI] [PubMed] [Google Scholar]
- 80.Gandecha A R, Large S L, Cundliffe E. Analysis of four tylosin biosynthetic genes from tylM region of the Streptomyces fradiae genome. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 81.Gasson M J. Indicator technique for antimetabolic toxin production by phytopathogenic species of Pseudomonas. Appl Environ Microbiol. 1980;39:25–29. doi: 10.1128/aem.39.1.25-29.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gnanamaickam S S, Starratt A N, Ward E W B. Coronatine production in vitro and in vivo and its relation to symptom development in bacterial blight of soybean. Can J Bot. 1982;60:645–650. [Google Scholar]
- 83.Gopalan S, He S Y. Bacterial genes involved in the elicitation of hypersensitive response and pathogenesis. Plant Dis. 1996;80:604–609. [Google Scholar]
- 84.Goss R W. The relation of temperature to common and halo blight of beans. Phytopathology. 1940;30:258–264. [Google Scholar]
- 85.Greulich F, Yoshihara T, Ichihara A. Coronatine, a bacterial phytotoxin, acts as a stereospecific analog of jasmonate type signals in tomato cells and potato tissues. J Plant Physiol. 1995;147:359–366. [Google Scholar]
- 86.Grgurina I, Barca A, Cervigni S, Gallo M, Scaloni A, Pucci P. Relevance of chlorine-substituent for the antifungal activity of syringomycin and syringotoxin, metabolites of the phytopathogenic bacterium Pseudomonas syringae pv. syringae. Experientia. 1993;50:130–133. doi: 10.1007/BF01984950. [DOI] [PubMed] [Google Scholar]
- 87.Grgurina I, Gross D C, Deligiovas I, Zhang J-H. SyrC, an enzyme involved in syringomycin biosynthesis, shows thioesterasic activity. In: Rudolf K, Burr T J, Mansfield J W, Stead D, Vivian A, Von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 192–197. [Google Scholar]
- 88.Grgurina I, Gross D C, Iacobellis N S, Lavermicocca P, Takemoto J Y, Benincasa M. Phytotoxin production by Pseudomonas syringae pv. syringae: syringopeptin production by syr mutants defective in biosynthesis or secretion of syringomycin. FEMS Microbiol Lett. 1996;138:35–39. [Google Scholar]
- 89.Grgurina I, Mariotti F. Biosynthesis of bioactive lipodepsipeptides by Pseudomonas syringae pv. syringae. In: Rudolf K, Burr T J, Mansfield J W, Stead D, Vivian A, Von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 182–187. [Google Scholar]
- 90.Grilley M M, Stock S D, Dickson R C, Lester R L, Takemoto J Y. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 91.Gross D C. Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J Appl Bacteriol. 1985;58:167–174. doi: 10.1111/j.1365-2672.1985.tb01444.x. [DOI] [PubMed] [Google Scholar]
- 92.Gross D C. Molecular and genetic analysis of toxin production by pathovars of Pseudomonas syringae. Annu Rev Phytopathol. 1991;29:247–278. [Google Scholar]
- 93.Gross D C, DeVay J E. Population dynamics and pathogenesis of Pseudomonas syringae in maize and cowpea in relation to the in vitro production of syringomycin. Phytopathology. 1977;67:475–483. [Google Scholar]
- 94.Gross D C, Scholz-Schroeder B K, Zhang J-H, Grgurina I, Mariotti F, Della Torre G, Guenzi E, Grandi G. Characterization of the thiotemplate mechanisms of syringomycin and syringopeptin synthesis by Pseudomonas syringae pv. syringae. In: Kohmoto, Yoder O C, editors. Molecular genetics of host-specific toxins in plant disease. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 91–98. [Google Scholar]
- 95.Guenzi E, Galli G, Grgurina I, Gross D C, Grandi G. Characterization of the syringomycin synthetase gene cluster: a link between prokaryotic and eukaryotic peptide synthetases. J Biol Chem. 1998;273:32857–32863. doi: 10.1074/jbc.273.49.32857. [DOI] [PubMed] [Google Scholar]
- 96.Hatziloukas E, Panopoulos N J. Origin, structure, and regulation of argK, encoding the phaseolotoxin-resistant ornithine carbamoyltransferase in Pseudomonas syringae pv. phaseolicola, and functional expression of argK in transgenic tobacco. J Bacteriol. 1992;174:5895–5909. doi: 10.1128/jb.174.18.5895-5909.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hatziloukas E, Panopoulos N J, Delis S, Prosen D E, Schaad N W. An open reading frame in the approximately 28-kb tox-argK gene cluster encodes a polypeptide with homology to fatty acid desaturases. Gene. 1995;166:83–87. doi: 10.1016/0378-1119(95)00569-5. [DOI] [PubMed] [Google Scholar]
- 98.Hoffmann K, Schneider-Scherzer E, Kleinkauf H, Zocher R. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J Biol Chem. 1994;269:12710–12714. [PubMed] [Google Scholar]
- 99.Hopwood D A. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 100.Hösel W. Glycosylation and glycosides. In: Conn E E, editor. The biochemistry of plants. 7. Secondary plant products. New York, N.Y: Academic Press, Inc.; 1981. pp. 725–753. [Google Scholar]
- 101.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hutchinson C R, Fujii I. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu Rev Microbiol. 1995;49:201–238. doi: 10.1146/annurev.mi.49.100195.001221. [DOI] [PubMed] [Google Scholar]
- 103.Hutchison M L, Gross D C. Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol Plant-Microbe Interact. 1997;10:347–354. doi: 10.1094/MPMI.1997.10.3.347. [DOI] [PubMed] [Google Scholar]
- 104.Hutchison M L, Johnstone K. Evidence for the involvement of the surface active properties of the extracellular toxin tolaasin in the manifestation of brown blotch disease symptoms by Pseudomonas tolaasii on Agaricus bisporus. Physiol Mol Plant Pathol. 1993;40:107–116. [Google Scholar]
- 105.Hutchison M L, Tester M A, Gross D C. Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: A model for the mechanism of action in the plant-pathogen interaction. Mol Plant-Microbe Interact. 1995;8:610–620. doi: 10.1094/mpmi-8-0610. [DOI] [PubMed] [Google Scholar]
- 106.Huynh T V, Dahlbeck D, Staskawicz B J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 107.Iacobellis N S, Lavermicocca P, Grgurina I, Simmaco M, Ballio A. Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol. 1992;40:107–116. [Google Scholar]
- 108.Ichihara A, Shiraishi K, Sato H, Sakamura S, Nishiyama K, Sakai R, Furusaki A, Matsumoto T. The structure of coronatine. J Am Chem Soc. 1977;99:636–637. [Google Scholar]
- 109.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Isogai A, Iguchi H, Nakayama J, Kusai A, Takemoto J Y, Suzuki A. Structural analysis of new syringopeptins by tandem mass spectrometry. Biosci Biotechnol Biochem. 1995;59:1374–1376. doi: 10.1271/bbb.59.1374. [DOI] [PubMed] [Google Scholar]
- 111.Jones W T, Harvey D, Mitchell R E, Ryan G B, Bender C L, Reynolds P H S. Competitive ELISA employing monoclonal antibodies specific for coronafacoyl amino acid conjugates. Food Agric Immunol. 1997;9:67–76. [Google Scholar]
- 112.Julmanop C, Takano Y, Takemoto J Y, Miyakawa T. Protection by sterols against the cytotoxicity of syringomycin in yeast Saccharomyces cerevisiae. J Gen Microbiol. 1993;139:2323–2327. doi: 10.1099/00221287-139-10-2323. [DOI] [PubMed] [Google Scholar]
- 113.Kao C M, Katz L, Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 114.Kao C M, Luo G, Katz L, Cane D E, Khosla C. Engineered biosynthesis of a triketide lactone from an incomplete modular polyketide synthase. J Am Chem Soc. 1994;116:11612–11613. [Google Scholar]
- 115.Kao C M, Luo G, Katz L, Cane D E, Khosla C. Manipulation of macrolide ring size by directed mutagenesis of a modular polyketide synthase. J Am Chem Soc. 1995;117:9105–9106. [Google Scholar]
- 116.Katz L. Manipulation of modular polyketide synthetases. Chem Rev. 1997;97:2557–2575. doi: 10.1021/cr960025+. [DOI] [PubMed] [Google Scholar]
- 117.Katz L, Donadio S. Polyketide synthesis: prospects for hybrid antibiotics. Annu Rev Microbiol. 1993;47:875–912. doi: 10.1146/annurev.mi.47.100193.004303. [DOI] [PubMed] [Google Scholar]
- 118.Kaulin Y A, Schagina L V, Bezrukov S M, Malev V V, Feigin A M, Takemoto J Y, Teeter J H, Brand J G. Cluster organization of ion channels formed by the antibiotic syringomycin E in bilayer lipid membranes. Biophys J. 1998;74:2918–2925. doi: 10.1016/S0006-3495(98)77999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kauss H. Some aspects of calcium-dependent regulation in plant metabolism. Annu Rev Plant Physiol. 1987;38:47–72. [Google Scholar]
- 120.Kauss H, Waldmann T, Jeblick W, Takemoto J Y. The phytotoxin syringomycin elicits Ca2+-dependent callose synthesis in suspension-cultured cells of Catharanthus roseus. Physiol Plant. 1991;81:134–138. [Google Scholar]
- 121.Keller J D, Loescher W H. Nonstructural carbohydrate partitioning in perennial parts of sweet cherry. J Am Soc Hortic Sci. 1989;114:969–975. [Google Scholar]
- 122.Kenyon J S, Turner J G. The stimulation of ethylene synthesis in Nicotiana tabacum leaves by the phytotoxin coronatine. Plant Physiol. 1992;100:219–224. doi: 10.1104/pp.100.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khosla C. Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev. 1997;97:2577–2590. doi: 10.1021/cr960027u. [DOI] [PubMed] [Google Scholar]
- 124.Kinscherf T G, Coleman R H, Barta T M, Willis D K. Cloning and expression of the tabtoxin biosynthetic region from Pseudomonas syringae. J Bacteriol. 1991;173:4124–4132. doi: 10.1128/jb.173.13.4124-4132.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 126.Kleinkauf H, von Döhren H. A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 127.Knight T J, Durbin R D, Langston-Unkefer P J. Role of glutamine synthetase adenylation in the self-protection of Pseudomonas syringae subsp. “tabaci” from its toxin, tabtoxinine-β-lactam. J Bacteriol. 1986;166:224–229. doi: 10.1128/jb.166.1.224-229.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Knight T J, Durbin R D, Langston-Unkefer P J. Self-protection of Pseudomonas syringae pv. “tabaci” from its toxin, tabtoxinine-β-lactam. J Bacteriol. 1987;169:1954–1959. doi: 10.1128/jb.169.5.1954-1959.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koda Y, Takahashi K, Kikuta Y, Greulich F, Toshima H, Ichihara A. Similarities of the biological activities of coronatine and coronafacic acid to those of jasmonic acid. Phytochemistry. 1996;41:93–96. [Google Scholar]
- 130.Krätschmar J, Krause M, Marahiel M. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krumm T, Bandemer K, Boland W. Induction of volatile biosynthesis in the Lima bean (Phaseolus lunatus) by leucine and isoleucine conjugates of 1-oxo- and 1-hydroxyindan-4-carboxylic acid: evidence for amino acid conjugates of jasmonic acid as intermediates in the octadecanoid signaling pathway. FEBS Lett. 1995;377:523–529. doi: 10.1016/0014-5793(95)01398-9. [DOI] [PubMed] [Google Scholar]
- 132.Latoud C, Peypoux F, Michel G. Interaction of iturin A, a lipopeptide antibiotic, with Saccharomyces cerevisiae cells: influence of the sterol membrane composition. Can J Microbiol. 1990;36:384–389. doi: 10.1139/m90-067. [DOI] [PubMed] [Google Scholar]
- 133.Lavermicocca P, Iacobellis N S, Simmaco M, Graniti A. Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol. 1997;50:129–140. [Google Scholar]
- 134.Laycock M V, Hildebrand P D, Thibault P, Walter J A, Wright J L C. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. J Agric Food Chem. 1991;39:483–489. [Google Scholar]
- 135.Leary J V, Roberts S, Willis J W. Detection of coronatine toxin of Pseudomonas syringae pv. glycinea with an enzyme-linked immunoabsorbent assay. Phytopathology. 1988;78:1498–1500. [Google Scholar]
- 136.Levi C, Durbin R D. The isolation and properties of a tabtoxin-hydrolyzing aminopeptidase from the periplasm of Pseudomonas syringae pv. tabaci. Physiol Mol Plant Pathol. 1986;28:345–352. [Google Scholar]
- 137.Li X-Z, Staratt A N, Cuppels D A. Identification of tomato leaf factors that activate toxin gene expression in Pseudomonas syringae pv. tomato DC3000. Phytopathology. 1998;88:1094–1100. doi: 10.1094/PHYTO.1998.88.10.1094. [DOI] [PubMed] [Google Scholar]
- 138.Liang L Z, Sobiczewski P, Paterson J M, Jones A L. Variation in virulence, plasmid content, and genes for coronatine synthesis between Pseudomonas syringae pv. morsprunorum and P. s. syringae from Prunus. Plant Dis. 1994;78:389–392. [Google Scholar]
- 139.Liras P, Asturias J A, Martín J F. Phosphate control sequences involved in transcriptional regulation of antibiotic biosynthesis. Trends Biotechnol. 1990;8:184–189. doi: 10.1016/0167-7799(90)90170-3. [DOI] [PubMed] [Google Scholar]
- 140.Liu L, Shaw P D. A possible role for acetylated intermediates in diaminopimelate and tabtoxinine-β-lactam biosynthesis in Pseudomonas syringae pv. tabaci BR2.024. J Bacteriol. 1997;179:5922–5927. doi: 10.1128/jb.179.18.5922-5927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu L, Shaw P D. Characterization of dapB, a gene required by Pseudomonas syringae pv. tabaci BR2.024 for lysine and tabtoxinine-β-lactam biosynthesis. J Bacteriol. 1997;179:507–513. doi: 10.1128/jb.179.2.507-513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liyanage H, Palmer D A, Ullrich M, Bender C L. Characterization and transcriptional analysis of the gene cluster for coronafacic acid, the polyketide component of the phytotoxin coronatine. Appl Environ Microbiol. 1995;61:3843–3848. doi: 10.1128/aem.61.11.3843-3848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liyanage H, Penfold C, Turner J, Bender C L. Sequence, expression and transcriptional analysis of the coronafacate ligase-encoding gene required for coronatine biosynthesis by Pseudomonas syringae. Gene. 1995;153:17–23. doi: 10.1016/0378-1119(94)00661-b. [DOI] [PubMed] [Google Scholar]
- 144.Lorang J M, Keen N T. Characterization of avrE from Pseudomonas syringae pv. tomato: a hrp-linked avirulence locus consisting of at least two transcriptional units. Mol Plant-Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 145.Ma S-W, Morris V L, Cuppels D A. Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol Plant-Microbe Interact. 1991;4:69–74. [Google Scholar]
- 146.Madduri K, Kennedy J, Rivola G, Inventi-Solari A, Filippini S, Zanuso G, Colombo A L, Gewain K M, Occi J L, MacNeil D J, Hutchinson C R. Production of the antitumor drug epirubicin (4′-epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nat Biotechnol. 1998;16:69–74. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]
- 147.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 148.Märkisch U, Reuter G. Biosynthesis of homoarginine and ornithine as precursors of the phytoeffector phaseolotoxin by the amidinotransfer from arginine to lysine catalyzed by an amidinotransferase in Pseudomonas syringae pv. phaseolicola. J Basic Microbiol. 1990;30:425–433. [Google Scholar]
- 149.Marsden A F A, Wilkinson B, Cortes J, Dunster N J, Staunton J, Leadlay P F. Engineering broader specificity into an antibiotic-producing polyketide synthase. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 150.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 151.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterization of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 152.Melchers L S, Regensburg-Tuïnk A J G, Schilperoort R A, Hooykaas P J J. Specificity of signal molecules in the activation of Agrobacterium virulence gene expression. Mol Microbiol. 1989;3:969–977. doi: 10.1111/j.1365-2958.1989.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 153.Mèndez C, Salas J A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol Lett. 1998;158:1–8. doi: 10.1111/j.1574-6968.1998.tb12792.x. [DOI] [PubMed] [Google Scholar]
- 154.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Merson-Davies L A, Cundliffe E. Analysis of five tylosin biosynthetic genes from the tyl1BA region of the Streptomyces fradiae genome. Mol Microbiol. 1994;13:349–355. doi: 10.1111/j.1365-2958.1994.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 156.Miller L P. Glycosides. In: Miller L P, editor. Phytochemistry. 1. The process and products of photosynthesis. New York, N.Y: Van Nostrand Reinhold Co.; 1973. pp. 297–375. [Google Scholar]
- 157.Mitchell R E. Isolation and structure of a chlorosis-inducing toxin of Pseudomonas phaseolicola. Phytochemistry. 1976;15:1941–1947. [Google Scholar]
- 158.Mitchell R E. Halo blight of beans: toxin production by several Pseudomonas phaseolicola isolates. Physiol Plant Pathol. 1978;13:37–49. [Google Scholar]
- 159.Mitchell R E. Coronatine production by some phytopathogenic pseudomonads. Physiol Plant Pathol. 1982;20:83–89. [Google Scholar]
- 160.Mitchell R E. Coronatine biosynthesis: incorporation of l-[U-14C]isoleucine and l-[U-14C]threonine into the 1-amido-1-carboxy-2-ethylcyclopropyl moiety. Phytochemistry. 1985;24:247–249. [Google Scholar]
- 161.Mitchell R E. Norcoronatine and N-coronafacoyl-l-valine, phytotoxin analogues of coronatine produced by a strain of Pseudomonas syringae pv. glycinea. Phytochemistry. 1985;24:1485–1488. [Google Scholar]
- 162.Mitchell R E. Coronatine analogues produced by Xanthomonas campestris pv. phormiicola. Phytochemistry. 1991;30:3917–3920. [Google Scholar]
- 163.Mitchell R E. Implications of toxins in the ecology and evolution of plant pathogenic microorganisms: bacteria. Experientia. 1991;47:791–803. doi: 10.1007/BF01922459. [DOI] [PubMed] [Google Scholar]
- 164.Mitchell R E, Bieleski R L. Involvement of phaseolotoxin in halo blight of beans. Transport and conversion to functional toxin. Plant Physiol. 1977;60:723–729. doi: 10.1104/pp.60.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mitchell R E, Ford K L. Chlorosis-inducing products from Pseudomonas syringae pathovars: new N-coronafacoyl compounds. Phytochemistry. 1998;49:1579–1583. doi: 10.1016/s0031-9422(98)00279-9. [DOI] [PubMed] [Google Scholar]
- 166.Mitchell R E, Frey E J. Production of N-coronafacoyl-L-amino acid analogues of coronatine by Pseudomonas syringae pv. atropurpurea in liquid cultures supplemented with l-amino acids. J Gen Microbiol. 1985;132:1503–1507. [Google Scholar]
- 167.Mitchell R E, Frey E J, Benn M H. Rhizobitoxine and l-threo-hydroxythreonine production by the plant pathogen Pseudomonas andropogonis. Phytochemistry. 1986;25:2711–2715. [Google Scholar]
- 168.Mitchell R E, Hale C N, Shanks J C. Production of different pathogenic symptoms and different toxins by strains of Pseudomonas syringae pv. tomato not distinguishable by gel-immunodiffusion assay. Physiol Plant Pathol. 1983;23:315–322. [Google Scholar]
- 169.Mitchell R E, Young H. N-coronafacoyl-l-isoleucine and N-coronafacoyl-l-alloisoleucine, potential biosynthetic intermediates of the phytotoxin coronatine. Phytochemistry. 1985;24:2716–2717. [Google Scholar]
- 170.Mitchell R E, Young S A, Bender C L. Coronamic acid, an intermediate in coronatine biosynthesis by Pseudomonas syringae. Phytochemistry. 1994;35:343–348. [Google Scholar]
- 171.Mitchell R E, Young H, Liddell M J. Isolation and structural characterization of 2-[1-oxo-2-cyclopenten-2-ylmethyl]-butanoic acid, a polyketide product of coronatine-producing Pseudomonas spp. Tetrahedron Lett. 1995;36:3237–3240. [Google Scholar]
- 172.Mittal S M, Davis K R. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant-Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 173.Mo Y-Y, Geibel M, Bonsall R F, Gross D C. Analysis of sweet cherry (Prunus avium L.) leaves for plant signal molecules that activate the syrB gene required for synthesis of the phytotoxin, syringomycin, by Pseudomonas syringae pv. syringae. Plant Physiol. 1995;107:603–612. doi: 10.1104/pp.107.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mo Y-Y, Gross D C. Expression in vitro and during plant pathogenesis of the syrB gene required for syringomycin production by Pseudomonas syringae pv. syringae. Mol Plant-Microbe Interact. 1991;4:28–36. [Google Scholar]
- 175.Mo Y-Y, Gross D C. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1991;173:5784–5792. doi: 10.1128/jb.173.18.5784-5792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moore R A, Starratt A N, Ma S-W, Morris V L, Cuppels D A. Identification of a chromosomal region required for biosynthesis of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Can J Microbiol. 1989;35:910–917. [Google Scholar]
- 177.Moore R E, Niemczura W P, Kwok O C H, Patil S S. Inhibitors of ornithine carbamoyltransferase from Pseudomonas syringae pv. phaseolicola. Tetrahedron Lett. 1984;25:3931–3934. [Google Scholar]
- 178.Mootz H D, Marahiel M A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Morgan M K, Chatterjee A K. Genetic organization and regulation of proteins associated with production of syringotoxin by Pseudomonas syringae pv. syringae. J Bacteriol. 1988;170:5689–5697. doi: 10.1128/jb.170.12.5689-5697.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mosqueda G, Van den Broeck G, Saucedo O, Bailey A, Herrera-Estrella L. Isolation and characterization of the gene from Pseudomonas syringae pv. phaseolicola encoding the phaseolotoxin-insensitive ornithine carbamoyltransferase. Mol Gen Genet. 1990;222:461–466. doi: 10.1007/BF00633857. [DOI] [PubMed] [Google Scholar]
- 181.Mosqueda-Cano G, Herrera-Estrella L. A simple and efficient PCR method for the specific detection of Pseudomonas syringae pv. phaseolicola in bean seeds. World J Microbiol Biotechnol. 1997;13:463–467. [Google Scholar]
- 182.Mott K A, Takemoto J Y. Syringomycin, a bacterial phytotoxin, closes stomata. Plant Physiol. 1989;90:1435–1439. doi: 10.1104/pp.90.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Neu T R, Hartner T, Poralla K. Surface active properties of viscosin: a peptidolipid antibiotic. Appl Microbiol Biotechnol. 1990;32:518–520. [Google Scholar]
- 184.Nüske J, Fritsche W. Phaseolotoxin production by Pseudomonas syringae pv. phaseolicola: the influence of temperature. J Basic Microbiol. 1989;29:441–447. doi: 10.1002/jobm.3620290713. [DOI] [PubMed] [Google Scholar]
- 185.Obeid L M, Hannun Y A. Ceramide: a stress signal and mediator of growth suppression and apoptosis. J Cell Biochem. 1995;58:191–198. doi: 10.1002/jcb.240580208. [DOI] [PubMed] [Google Scholar]
- 186.Oliynyk M, Brown M J B, Cortés J, Staunton J, Leadlay P F. A hybrid modular polyketide synthase obtained by domain swapping. Chem Biol. 1996;3:833–839. doi: 10.1016/s1074-5521(96)90069-1. [DOI] [PubMed] [Google Scholar]
- 187.Palmer D A. Regulation and mode of action of the polyketide phytotoxin coronatine. Ph.D. dissertation. Stillwater: Oklahoma State University; 1995. [Google Scholar]
- 188.Palmer, D. A., and C. L. Bender. Unpublished data.
- 189.Palmer D A, Bender C L. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–1626. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Palmer D A, Bender C L. Ultrastructure of tomato leaf tissue treated with the pseudomonad phytotoxin coronatine and comparison with methyl jasmonate. Mol Plant-Microbe Interact. 1995;8:683–692. [Google Scholar]
- 191.Palmer D A, Bender C L, Sharma S B. Use of Tn5-gusA5 to investigate environmental and nutritional effects on gene expression in the coronatine biosynthetic gene cluster of Pseudomonas syringae pv. glycinea. Can J Microbiol. 1997;43:517–525. doi: 10.1139/m97-074. [DOI] [PubMed] [Google Scholar]
- 192.Pao G M, Tam R, Lipschitz L S, Saier M H. Response regulators: structure, function, and evolution. Res Microbiol. 1994;145:356–362. doi: 10.1016/0923-2508(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 193.Parkinson J S, Kofoid E C. Communication modules in bacterial signalling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 194.Parry R J, Jiralerspong S, Mhaskar S, Alemany L, Willcott R. Investigations of coronatine biosynthesis. Elucidation of the mode of incorporation of pyruvate into coronafacic acid. J Am Chem Soc. 1996;118:703–704. [Google Scholar]
- 195.Parry R J, Lin M T, Walker A E, Mhaskar S. Biosynthesis of coronatine: investigations of the biosynthesis of coronamic acid. J Am Chem Soc. 1991;113:1849–1850. [Google Scholar]
- 196.Parry R J, Mhaskar S V, Lin M-T, Walker A E, Mafoti R. Investigations of the biosynthesis of the phytotoxin coronatine. Can J Chem. 1994;72:86–99. [Google Scholar]
- 197.Patel J, Hoyt J C, Parry R J. Investigations of coronatine biosynthesis. Overexpression and assay of CmaT, a thioesterase involved in coronamic acid biosynthesis. Tetrahedron. 1998;54:15927–15936. [Google Scholar]
- 198.Patil S S, Hayward A C, Emmons R. An ultraviolet-induced nontoxigenic mutant of Pseudomonas phaseolicola of altered pathogenicity. Phytopathology. 1974;64:590–595. [Google Scholar]
- 199.Paynter V A, Alconero R. A specific fluorescent antibody for detection of syringomycin in infected peach tree tissues. Phytopathology. 1979;69:493–496. [Google Scholar]
- 200.Peet R C, Lindgren P B, Willis D K, Panopoulos N J. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. “phaseolicola.”. J Bacteriol. 1986;166:1096–1105. doi: 10.1128/jb.166.3.1096-1105.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Peet R C, Panopoulos N J. Ornithine carbamoyltransferase genes and phaseolotoxin immunity in Pseudomonas syringae pv. phaseolicola. EMBO J. 1987;6:3585–3591. doi: 10.1002/j.1460-2075.1987.tb02689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Peñaloza-Vázquez A, Bender C L. Characterization of CorR, a transcriptional activator which is required for biosynthesis of the phytotoxin coronatine. J Bacteriol. 1998;180:6252–6259. doi: 10.1128/jb.180.23.6252-6259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Penfold C N, Bender C L, Turner J G. Characterisation of genes involved in biosynthesis of coronafacic acid, the polyketide component of the phytotoxin coronatine. Gene. 1996;183:167–173. doi: 10.1016/s0378-1119(96)00550-1. [DOI] [PubMed] [Google Scholar]
- 204.Perego M, Cole S P, Burbulys D, Trach K, Hoch J A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Peters N K, Verma D P S. Phenolic compounds as regulators of gene expression in plant-microbe interactions. Mol Plant-Microbe Interact. 1990;3:4–8. doi: 10.1094/mpmi-3-004. [DOI] [PubMed] [Google Scholar]
- 206.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 207.Pieper R, Luo G, Cane D E, Khosla C. Cell-free synthesis of polyketides by recombinant erythromycin polyketide synthases. Nature. 1995;378:263–266. doi: 10.1038/378263a0. [DOI] [PubMed] [Google Scholar]
- 208.Quigley N B, Gross D C. Syringomycin production among strains of Pseudomonas syringae pv. syringae: conservation of the syrB and syrD genes and activation of phytotoxin production by plant signal molecules. Mol Plant-Microbe Interact. 1994;7:78–90. doi: 10.1094/mpmi-7-0078. [DOI] [PubMed] [Google Scholar]
- 209.Quigley N B, Mo Y-Y, Gross D C. SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol Microbiol. 1993;9:787–801. doi: 10.1111/j.1365-2958.1993.tb01738.x. [DOI] [PubMed] [Google Scholar]
- 210.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rainey P B, Brodey C L, Johnstone K. Biological properties and spectrum of activity of tolaasin, a lipodepsipeptide toxin produced by the mushroom pathogen Pseudomonas tolaasii. Physiol Mol Plant Pathol. 1991;39:57–70. [Google Scholar]
- 212.Rangaswamy V, Jiralerspong S, Parry R, Bender C L. Biosynthesis of the Pseudomonas polyketide coronafacic acid requires monofunctional and multifunctional polyketide synthase proteins. Proc Natl Acad Sci USA. 1998;95:15469–15474. doi: 10.1073/pnas.95.26.15469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rangaswamy V, Mitchell R, Ullrich M, Bender C L. Analysis of genes involved in the synthesis of the polyketide phytotoxin coronatine. J Bacteriol. 1998;180:3330–3338. doi: 10.1128/jb.180.13.3330-3338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rangaswamy V, Ullrich M, Jones W, Mitchell R, Parry R, Reynolds P, Bender C L. Expression and analysis of coronafacate ligase, a thermoregulated gene required for production of the phytotoxin coronatine in P. syringae. FEMS Microbiol Lett. 1997;154:65–72. doi: 10.1111/j.1574-6968.1997.tb12625.x. [DOI] [PubMed] [Google Scholar]
- 215.Reidl H H, Grover T A, Takemoto J Y. 31P-NMR evidence for cytoplasmic acidification and phosphate extrusion in syringomycin-treated cells of Rhodotorula pilimanae. Biochim Biophys Acta. 1989;1010:325–329. doi: 10.1016/0167-4889(89)90056-6. [DOI] [PubMed] [Google Scholar]
- 216.Reidl H H, Takemoto J Y. Mechanism of action of bacterial phytotoxin, syringomycin. Simultaneous measurement of early responses in yeast and maize. Biochim Biophys Acta. 1987;898:59–69. [Google Scholar]
- 217.Rhodehamel N H, Durbin R D. Toxin production by strains of Pseudomonas syringae pv. tagetis. Physiol Mol Plant Pathol. 1989;35:301–311. [Google Scholar]
- 218.Rich J J, Hirano S S, Willis D K. Pathovar-specific requirement for the Pseudomonas syringae lemA gene in disease lesion formation. Appl Environ Microbiol. 1992;58:1440–1446. doi: 10.1128/aem.58.5.1440-1446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rohde B H, Pohlack B, Ullrich M S. Occurrence of thermoregulation of genes involved in coronatine biosynthesis among various Pseudomonas syringae strains. J Basic Microbiol. 1998;38:41–50. [PubMed] [Google Scholar]
- 221.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Roth P, Hädener A, Tamm C. Further studies on the biosynthesis of tabtoxin (wildfire toxin): incorporation of [2,3-13C2]pyruvate into the β-lactam moiety. Helv Chim Acta. 1990;73:476–482. [Google Scholar]
- 223.Rowley K B, Clements D E, Mandel M, Humphreys T, Patil S S. Multiple copies of a DNA sequence from Pseudomonas syringae pathovar phaseolicola abolish thermoregulation of phaseolotoxin production. Mol Microbiol. 1993;8:625–635. doi: 10.1111/j.1365-2958.1993.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 224.Ruan X, Pereda A, Stassi D L, Zeidner D, Summers R G, Jackson M, Shivakumar A, Kakavas S, Staver M J, Donadio S, Katz L. Acyltransferase domain substitutions in erythromycin polyketide synthase yield novel erythromycin derivatives. J Bacteriol. 1997;179:6416–6425. doi: 10.1128/jb.179.20.6416-6425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ryan C. Quantitative determination of soluble cellular proteins by radial diffusion in agar gels containing antibodies. Anal Biochem. 1967;19:434–440. doi: 10.1016/0003-2697(67)90233-3. [DOI] [PubMed] [Google Scholar]
- 226.Sakai R, Nishiyama K, Ichihara A, Shiraishi K, Sakamura S. The relation between bacterial toxic action and plant growth regulation. In: Daly J M, Uritani I, editors. Recognition and specificity in plant host-parasite interactions. Baltimore, Md: University Park Press; 1979. pp. 165–179. [Google Scholar]
- 227.Salmeron J M, Staskawicz B J. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 228.Salmond G P C. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu Rev Phytopathol. 1994;32:181–200. [Google Scholar]
- 229.Sato M, Nishiyama K, Shirata A. Involvement of plasmid DNA in the productivity of coronatine by Pseudomonas syringae pv. atropurpurea. Ann Phytopathol Soc Jpn. 1983;49:522–528. [Google Scholar]
- 230.Sawada H, Takeuchi T, Matsuda I. Comparative analysis of Pseudomonas syringae pv. actinidiae and pv. phaseolicola based on phaseolotoxin-resistant ornithine carbamoyltransferase gene (argK) and 16S-23S rRNA intergenic spacer sequences. Appl Environ Microbiol. 1997;63:282–288. doi: 10.1128/aem.63.1.282-288.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Scaloni A, Bachmann R C, Takemoto J Y, Barra D, Simmaco M, Ballio A. Stereochemical structure of syringomycin, a phytotoxic metabolite of Pseudomonas syringae pv. syringae. Nat Prod Lett. 1994;4:159–164. [Google Scholar]
- 232.Scaloni A, Camoni L, Di Giorgio D, Scortichini M, Cozzolino R, Ballio A. A new syringopeptin produced by a Pseudomonas syringae pv. syringae strain isolated from diseased twigs of laurel. Physiol Mol Plant Pathol. 1997;51:259–264. [Google Scholar]
- 233.Schaad N W, Azad H, Peet R C, Panopoulos N J. Identification of Pseudomonas syringae pv. phaseolicola by a DNA hybridization probe. Phytopathology. 1989;79:903–907. [Google Scholar]
- 234.Schaad N W, Cheong S S, Tamaki S, Hatziloukas E, Panopoulos N J. A combined biological and enzymatic amplification (BIO-PCR) technique to detect Pseudomonas syringae pv. phaseolicola in bean seed extracts. Phytopathology. 1995;85:243–248. [Google Scholar]
- 235.Scheck H J, Canfield M L, Pscheidt J W, Moore L W. Rapid evaluation of pathogenicity in Pseudomonas syringae pv. syringae with a lilac tissue culture bioassay and syringomycin DNA probes. Plant Dis. 1997;81:905–910. doi: 10.1094/PDIS.1997.81.8.905. [DOI] [PubMed] [Google Scholar]
- 236.Schmid P P S, Feucht W. Carbohydrates in the phloem of Prunus avium/Prunus cerasus graftings and of homospecific controls. Angew Bot. 1986;60:201–208. [Google Scholar]
- 237.Schneider A, Marahiel M A. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch Microbiol. 1998;169:404–410. doi: 10.1007/s002030050590. [DOI] [PubMed] [Google Scholar]
- 238.Scholz-Schroeder, B. K., I. Grgurina, and D. C. Gross. Unpublished data.
- 239.Scholz-Schroeder, B. K., M. L. Hutchison, I. Grgurina, and D. C. Gross. Unpublished data.
- 240.Segre A, Bachmann R C, Ballio A, Bosa F, Grgurina I, Iacobellis N S, Marino G, Pucci P, Simmaco M, Takemoto J Y. The structure of syringomycins A1, E and G. FEBS Lett. 1989;255:27–31. doi: 10.1016/0014-5793(89)81054-3. [DOI] [PubMed] [Google Scholar]
- 241.Sembdner G, Parthier B. The biochemistry and the physiological and molecular actions of jasmonates. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:569–580. [Google Scholar]
- 242.Shimoda N, Toyoda-Yamamoto A, Nagamine J, Usami S, Katayama M, Sakagami Y, Machida Y. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci USA. 1990;87:6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Shumway L K, Rancour J M, Ryan C A. Vacuolar protein bodies in tomato leaf cells and their relationship to storage of chymotrypsin inhibitor I protein. Planta. 1970;93:1–14. doi: 10.1007/BF00387647. [DOI] [PubMed] [Google Scholar]
- 244.Shumway L K, Yang V V, Ryan C A. Evidence for the presence of proteinase inhibitor I in vacuolar protein bodies of plant cells. Planta. 1976;129:161–165. doi: 10.1007/BF00390023. [DOI] [PubMed] [Google Scholar]
- 245.Sorensen K N, Kim K-H, Takemoto J Y. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsipeptides produced by Pseudomonas syringae pv. syringae. Antimicrob Agents Chemother. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sorensen K N, Kim K-H, Takemoto J Y. PCR detection of cyclic lipodepsinonapeptide-producing Pseudomonas syringae pv. syringae and similarity of strains. Appl Environ Microbiol. 1998;64:226–230. doi: 10.1128/aem.64.1.226-230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Stachelhaus T, Marahiel M. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 248.Stachelhaus T, Schneider A, Marahiel M. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science. 1995;269:69–73. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 249.Stachelhaus T, Schneider A, Marahiel M. Engineered biosynthesis of peptide antibiotics. Biochem Pharm. 1996;52:177–186. doi: 10.1016/0006-2952(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 250.Staskawicz B J, Panopoulos N J. A rapid and sensitive microbiological assay for phaseolotoxin. Phytopathology. 1979;69:663–666. [Google Scholar]
- 251.Staskawicz B J, Panopoulos N J, Hoogenraad N J. Phaseolotoxin-insensitive ornithine carbamoyltransferase of Pseudomonas syringae pv. phaseolicola: Basis for immunity to phaseolotoxin. J Bacteriol. 1980;142:720–723. doi: 10.1128/jb.142.2.720-723.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Stein T, Vater J. Amino acid activation and polymerization at modular multienzymes in nonribosomal peptide biosynthesis. Amino Acids. 1996;10:201–227. doi: 10.1007/BF00807324. [DOI] [PubMed] [Google Scholar]
- 253.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 254.Stein T, Vater J, Kruft V, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. Detection of 4′-phosphopantetheine at the thioester binding site for l-valine of gramicidin S synthetase 2. FEBS Lett. 1994;340:39–44. doi: 10.1016/0014-5793(94)80169-x. [DOI] [PubMed] [Google Scholar]
- 255.Stewart W W. Isolation and proof of structure of wildfire toxin. Nature. 1971;229:174–178. doi: 10.1038/229174a0. [DOI] [PubMed] [Google Scholar]
- 256.Taguchi N, Takano Y, Julmanop C, Wang Y, Stock S, Takemoto J, Miyakawa T. Identification and analysis of the Saccharomyces cerevisiae SYR1 gene reveals that ergosterol is involved in the action of syringomycin. Microbiology. 1994;140:353–359. doi: 10.1099/13500872-140-2-353. [DOI] [PubMed] [Google Scholar]
- 257.Takahashi Y, Omura T, Hibino H, Sato M. Detection and identification of Pseudomonas syringae pv. atropurpurea by PCR amplification of specific fragments from an indigenous plasmid. Plant Dis. 1996;80:783–788. [Google Scholar]
- 258.Takemoto J Y. Bacterial phytotoxin syringomycin and its interaction with host membranes. In: Verma D P S, editor. Molecular signals in plant-microbe communications. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 247–260. [Google Scholar]
- 259.Takemoto J Y, Giannini J L, Vassey T, Briskin D P. Syringomycin effects on plasma membrane Ca2+ transport. In: Graniti A, Durbin R D, Ballio A, editors. Phytotoxins and plant pathogenesis. Berlin, Germany: Springer-Verlag KG; 1989. pp. 167–175. [Google Scholar]
- 260.Takemoto J Y, Zhang L, Taguchi N, Tachikawa T, Miyakawa T. Mechanism of action of the phytotoxin syringomycin: a resistant mutant of Saccharomyces cerevisiae reveals an involvement of Ca2+ transport. J Gen Microbiol. 1991;137:653–659. [Google Scholar]
- 261.Tamura K, Takikawa Y, Tsuyumu S, Goto M, Watanabe M. Coronatine production by Xanthomonas campestris pv. phormiicola. Ann Phytopathol Soc Jpn. 1992;58:276–281. [Google Scholar]
- 262.Templeton M D, Sullivan P A, Shepherd M G. Phaseolotoxin-insensitive l-ornithine transcarbamoylase from Pseudomonas syringae pv. phaseolicola. Physiol Mol Plant Pathol. 1986;29:393–403. [Google Scholar]
- 263.Thomas M D, Langston-Unkefer P J, Uchytil T F, Durbin R D. Inhibition of glutamine synthetase from pea by tabtoxinine-β-lactam. Plant Physiol. 1983;71:912–915. doi: 10.1104/pp.71.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Treutter D, Galensa R, Feucht W, Schmid P P S. Flavanone glucosides in callus and phloem of Prunus avium: identification and stimulation of their synthesis. Physiol Plant. 1985;65:95–101. [Google Scholar]
- 265.Turgay K, Bachmann A S, Marahiel M, Patil S S. Cloning of a putative peptide synthetase gene involved in the synthesis of phaseolotoxin. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, Von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 248–254. [Google Scholar]
- 266.Turgay K, Krause M, Marahiel M A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 267.Turner J G, Debbage J M. Tabtoxin-induced symptoms are associated with accumulation of ammonia formed during photorespiration. Physiol Plant Pathol. 1982;20:223–233. [Google Scholar]
- 268.Turner J G, Taha R R. Contribution of tabtoxin to the pathogenicity of Pseudomonas syringae pv. tabaci. Physiol Plant Pathol. 1984;25:55–69. [Google Scholar]
- 269.Uchytil T F, Durbin R D. Hydrolysis of tabtoxins by plant and bacterial enzymes. Experientia. 1980;36:301–302. [Google Scholar]
- 270.Ullrich M, Bender C L. The biosynthetic gene cluster for coronamic acid, an ethylcyclopropyl amino acid, contains genes homologous to amino acid-activating enzymes and thioesterases. J Bacteriol. 1994;176:7574–7586. doi: 10.1128/jb.176.24.7574-7586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ullrich M, Bereswill S, Völksch B, Fritsche W, Geider K. Molecular characterization of field isolates of Pseudomonas syringae pv. glycinea differing in coronatine production. J Gen Microbiol. 1993;139:1927–1937. doi: 10.1099/00221287-139-8-1927. [DOI] [PubMed] [Google Scholar]
- 272.Ullrich M, Guenzi A C, Mitchell R E, Bender C L. Cloning and expression of genes required for coronamic acid (2-ethyl-1-aminocyclopropane 1-carboxylic acid), an intermediate in the biosynthesis of the phytotoxin coronatine. Appl Environ Microbiol. 1994;60:2890–2897. doi: 10.1128/aem.60.8.2890-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ullrich M, Peñaloza-Vázquez A, Bailey A M, Bender C L. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1995;177:6160–6169. doi: 10.1128/jb.177.21.6160-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Unkefer C J, London R E, Durbin R D, Uchytil T F, Langston-Unkefer P J. The biosynthesis of tabtoxinine-β-lactam. Use of specifically 13C-labeled glucose and 13C NMR spectroscopy to identify its biosynthetic precursors. J Biol Chem. 1987;262:4994–4999. [PubMed] [Google Scholar]
- 275.Vassilev V, Lavermicocca P, Di Giorgio D, Iacobellis N S. Production of syringomycins and syringopeptins by Pseudomonas syringae pv. atrofaciens. Plant Pathol. 1996;45:316–322. [Google Scholar]
- 276.Vignutelli A, Wasternack C, Apel K, Bohlmann H. Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J. 1998;14:285–295. doi: 10.1046/j.1365-313x.1998.00117.x. [DOI] [PubMed] [Google Scholar]
- 277.Völksch B, Bublitz F, Fritsche W. Coronatine production by Pseudomonas syringae pathovars: screening method and capacity of product formation. J Basic Microbiol. 1989;29:463–468. [Google Scholar]
- 278.Von Döhren H, Keller U, Vater J, Zocher R. Multifunctional peptide synthetases. Chem Rev. 1997;97:2675–2705. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 279.Wada H, Gombos Z, Murata N. Enhancement of chilling tolerance of a cyanobacterium by genetic manipulation of fatty acid desaturation. Nature. 1990;347:200–203. doi: 10.1038/347200a0. [DOI] [PubMed] [Google Scholar]
- 280.Wasternack C, Parthier B. Jasmonate-signaled plant gene expression. Trends Plant Sci. 1997;2:302–307. [Google Scholar]
- 281.Weber G, Schörgendorfer K, Schneider-Scherzer E, Leitner E. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8 kilobase open reading frame. Curr Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- 282.Wiebe W L, Campbell R N. Characterization of Pseudomonas syringae pv. maculicola and comparison with P.s. tomato. Plant Dis. 1993;77:414–419. [Google Scholar]
- 283.Weiler E W, Kutchan T M, Gorba T, Brodschelm W, Neisel U, Bublitz F. The Pseudomonas phytotoxin coronatine mimics octadecanoid signaling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 284.Willis D K, Barta T M, Kinscherf T G. Genetics of toxin production and resistance in phytopathogenic bacteria. Experientia. 1991;47:765–771. [Google Scholar]
- 285.Witkowski A, Naggert J, Witkowska H E, Randhawa Z I, Smith S. Utilization of an active serine 101 → cysteine mutant to demonstrate the proximity of the catalytic serine 101 and histidine 237 residues in thioesterase II. J Biol Chem. 1992;267:18488–18492. [PubMed] [Google Scholar]
- 286.Xie D-X, Feys B F, James S, Nieto-Rostro M, Turner J G. COI1: an Arabidopsis gene required for jasmonate-regulated defence and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 287.Xu G-W, Gross D C. Evaluation of the role of syringomycin in plant pathogenesis by using Tn5 mutants of Pseudomonas syringae pv. syringae defective in syringomycin production. Appl Environ Microbiol. 1988;54:1345–1353. doi: 10.1128/aem.54.6.1345-1353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Xu G-W, Gross D C. Physical and functional analyses of the syrA and syrB genes involved in syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1988;170:5680–5688. doi: 10.1128/jb.170.12.5680-5688.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Young S A, Park S K, Rodgers C, Mitchell R E, Bender C L. Physical and functional characterization of the gene cluster encoding the polyketide phytotoxin coronatine in Pseudomonas syringae pv. glycinea. J Bacteriol. 1992;174:1837–1843. doi: 10.1128/jb.174.6.1837-1843.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhang, J.-H., I. Grgurina, and D. C. Gross. Unpublished data.
- 291.Zhang J-H, Quigley N B, Gross D C. Analysis of the syrB and syrC genes of Pseudomonas syringae pv. syringae indicates that syringomycin is synthesized by a thiotemplate mechanism. J Bacteriol. 1995;177:4009–4020. doi: 10.1128/jb.177.14.4009-4020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhang J-H, Quigley N B, Gross D C. Analysis of the syrP gene, which regulates syringomycin synthesis by Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:2771–2778. doi: 10.1128/aem.63.7.2771-2778.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang L, Takemoto J Y. Effects of Pseudomonas syringae phytotoxin, syringomycin, on plasma membrane functions of Rhodotorula pilimanae. Phytopathology. 1987;77:297–303. [Google Scholar]
- 294.Zhang Y, Rowley K B, Patil S S. Genetic organization of a cluster of genes involved in the production of phaseolotoxin, a toxin produced by Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1993;175:6451–6464. doi: 10.1128/jb.175.20.6451-6458.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhang Y X, Patil S S. The phtE locus in the phaseolotoxin gene cluster has ORFs with homologies to genes encoding amino acid transferases, the AraC family of transcriptional factors, and fatty acid desaturases. Mol Plant-Microbe Interact. 1997;8:947–960. doi: 10.1094/MPMI.1997.10.8.947. [DOI] [PubMed] [Google Scholar]
- 296.Zhou H, McEvoy M M, Lowry D F, Swanson R V, Simon M I, Dahlquist F W. Phosphotransfer and the CheY-binding domains of the histidine autokinase CheA are joined by a flexible linker. Biochemistry. 1996;35:433–443. doi: 10.1021/bi951960e. [DOI] [PubMed] [Google Scholar]
- 297.Zhu Y, Tamura K, Watanabe M, Matsuda I, Sato M. Plasmid-mediated coronatine production in Pseudomonas syringae pv. maculicola. Ann Phytopathol Soc Jpn. 1995;61:569–574. [Google Scholar]