Abstract
The Canadian Sentinel Practitioner Surveillance Network estimated vaccine effectiveness (VE) during the unusually early 2022/23 influenza A(H3N2) epidemic. Like vaccine, circulating viruses were clade 3C.2a1b.2a.2, but with genetic diversity affecting haemagglutinin positions 135 and 156, and reassortment such that H156 viruses acquired neuraminidase from clade 3C.2a1b.1a. Vaccine provided substantial protection with A(H3N2) VE of 54% (95% CI: 38 to 66) overall. VE was similar against H156 and vaccine-like S156 viruses, but with potential variation based on diversity at position 135.
Keywords: Influenza, A(H3N2), vaccine effectiveness, clade, test-negative design, observational study, reassortment, imprinting
During the 2021/22 season, Canada experienced a delayed influenza A(H3N2) epidemic caused by clade 3C.2a1b.2a.2 (‘2a.2’) viruses that peaked in May 2022 (week 19) [1,2]. Thereafter, Canada [3,4], like other countries of Europe and the United States (US) [5,6], experienced an early 2022/23 A(H3N2) epidemic, also caused by 2a.2 viruses, that began in late October 2022 (week 43), peaked in November (weeks 47–48) and subsided by January 2023 (week 1) [3,4]. The Canadian Sentinel Practitioner Surveillance Network (SPSN) aimed to assess 2022/23 vaccine effectiveness (VE) against influenza A(H3N2), including genetic characterisation of contributing viruses for context- and variant-specific comparison.
Vaccine effectiveness evaluation
Influenza VE was estimated by test-negative design. Eligible patients aged ≥ 1 year presented within 7 days of influenza-like-illness onset to community-based sentinel practitioners in Alberta, British Columbia (BC), Ontario and Quebec [1,7,8]. Analyses included nasal or nasopharyngeal specimens collected from eligible patients between 1 November 2022 (week 44) and 6 January 2023 (week 1), tested for influenza by real-time RT-PCR and/or multiplex assays. Influenza vaccination status was based on self-report. In SPSN provinces, > 99% of publicly funded vaccines were egg-based inactivated. Elderly adults ≥ 65 years were administered MF59-adjuvanted or high-dose vaccines [9], the latter including facility residents in BC and Quebec but all elderly adults in Ontario and Alberta.
The 2022/23 influenza A(H3N2) vaccine was updated from the 2021/22 clade 3C.2a1b.2a.1 to a 2a.2 strain [10]. The representative 2a.2 anchor strain was A/Bangladesh/4005/2020 but vaccines used A/Darwin/6/2021 (cell-passaged) and A/Darwin/9/2021 (egg-passaged) 2a.2 reference viruses [10-12]. Like virtually all A(H3N2) viruses since 2002, A/Bangladesh has H156 in the haemagglutinin (HA) whereas A/Darwin reflects recent 2a.2 viruses in 2021 and 2022 with an H156S substitution (i.e. A/Darwin vaccines are S156) [10-14].
Virological evaluation
Sanger sequencing of the HA gene was undertaken on a convenience sample of original patient specimens. Whole genome sequencing (WGS), adapted from published protocols [15-18], was applied to a further subset to characterise the neuraminidase (NA). Viruses were classified per recent nomenclature from the European Centre for Disease Prevention and Control (ECDC) as genetic subgroups i–iv; for cross reference, current Nextstrain terminology is as follows for corresponding ECDC subgroups i (‘2b’), ii (‘2a.3a.1’), iii (‘2a.1’) and iv (‘2a.1b’) [12,14]. We indicate HA amino acid substitutions in relation to A/Bangladesh, including antigenic sites in parentheses, and annotate involvement of the receptor-binding site (RBS) and potential gain or loss of N-linked glycosylation (+/− CHO) [19,20]. In Supplementary Table S1 we provide detailed sequencing findings, including additional HA substitutions and NA characterisation. For the 387 viruses with sufficient sequence quality for upload, data are available in the Global Initiative on Sharing All Influenza Data (GISAID) database with GISAID identification numbers: EPI_ISL_16755416 to EPI_ISL_16755802 [21].
Virological findings
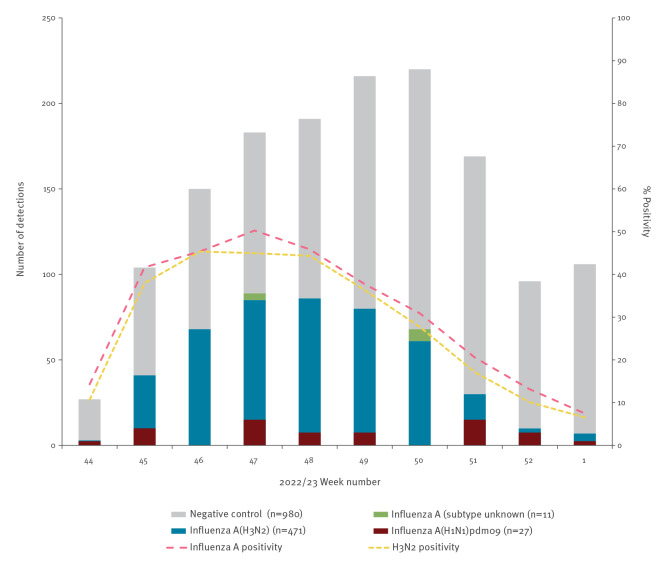
Among 1,490 eligible specimens, 510 (34%) tested influenza-positive, including 509 influenza A and one influenza B (Figure 1). Of the 498 (98%) subtyped influenza A viruses, 471 (95%) were A(H3N2). The HA of 391 (83%) A(H3N2) viruses was genetically characterised, including 89 of 97 (92%), 62 of 77 (81%), 163 of 213 (77%) and 77 of 84 (92%) from BC, Alberta, Ontario and Quebec, respectively.
Figure 1.
Influenza detections among eligible patients presenting with influenza-like illness, by week of specimen collection, SPSN, Canada, 1 November 2022–6 January 2023 (weeks 44–1) (n = 1,490)
SPSN: Sentinel Practitioner Surveillance Network.
Among eligible patients presenting with influenza-like illness, influenza test-positive cases and test-negative controls are displayed by week of specimen collection in 2022/23. Tallies correspond with participant profiles shown below but with influenza A(H1N1)pdm09 and unsubtyped influenza A detections additionally displayed. Not displayed is a single influenza B detection in week 49. The figure excludes cases and controls vaccinated within 2 weeks of illness onset and those with missing information for covariates used in primary vaccine effectiveness analyses. As per usual SPSN approach, missing specimen collection dates were imputed as the date the specimen was received and processed at the laboratory minus 2 days.
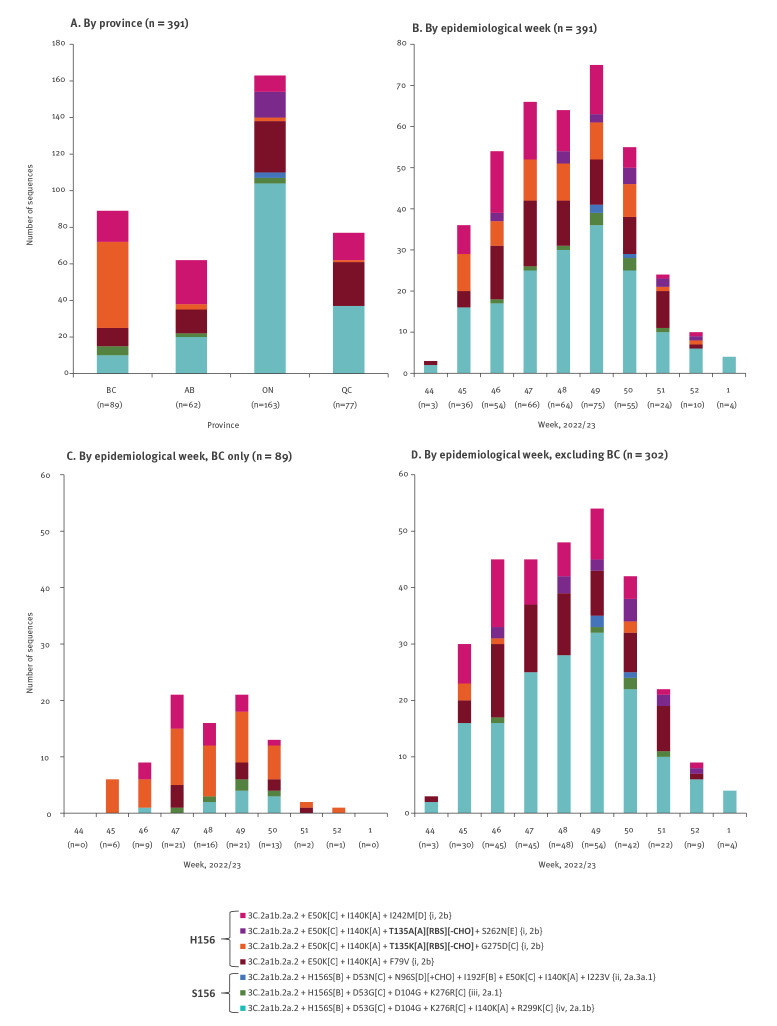
Supplementary Table S1 provides detailed gene sequencing findings. All genetically characterised viruses were clade 2a.2, with 207 of 391 (53%) belonging to ECDC subgroup i, defined by H156 with E50K(C) (Figure 2). In BC, most characterised viruses (83%; 74/89) belonged to the ECDC subgroup i, with most of these (64%; 47/74) bearing additional T135K(A)(RBS)(−CHO) substitution. In other SPSN provinces, less than half (44%; 133/302) of characterised viruses belonged to subgroup i, of which few (9%; 6/133) bore the additional T135K substitution. Of note, position 135 is an accessory site previously involved in the major antigenic cluster transition to A/Sydney/5/97-like viruses in 1997 [13]. Prior to that, K135 viruses had briefly predominated between 1993 and 1996, but with K135T(A)(RBS)(+CHO) substitution accompanying A/Sydney emergence, T135 viruses have predominated since [21,22]. During the 2022/23 epidemic, H156 viruses with T135K substitution comprised more than half (53%; 47/89) of characterised viruses in BC, but < 5% in other SPSN provinces (2%; 6/302). In Ontario, 14 of 163 (9%) characterised viruses were instead H156 with T135A(A)(RBS)(−CHO), a substitution not found in other SPSN provinces.
Figure 2.
Distribution of clade 3C.2a1b.2a2 variants among genetically characterised influenza A(H3N2) viruses contributing to influenza vaccine effectiveness analyses, SPSN, Canada, 1 November–28 December 2022 (weeks 44–1) (n = 391)
BC: British Columbia; CHO: carbohydrate; ECDC: European Centre for Disease Prevention and Control; RBS: receptor-binding site; SPSN: Sentinel Practitioner Surveillance Network.
−CHO and +CHO refer to mutations resulting in the loss or gain of a potential glycosylation site, respectively. Genetically characterised influenza A(H3N2) variants contributing to influenza vaccine effectiveness analyses with associated substitutions and ECDC subgroup designations, followed by Nextstrain nomenclature, noted in curly brackets. Colour coding corresponds with subgroups displayed in Supplementary Table S1, the latter including further substitutions and neuraminidase characterisation.
The remaining 184 of 391 (47%) SPSN viruses not belonging to subgroup i were H156S(B) (i.e. S156, like the A/Darwin vaccines) with additional substitutions placing them within ECDC subgroups ii (< 1%; 3/391), iii (3%; 10/391) or iv (44%; 171/391), the latter S156 viruses including two also bearing T135K substitution (Figure 2).
Among SPSN viruses subjected to WGS (33%; 154/471), we additionally identified a reassortment combining the HA segment from clade 3C.2a1b.2a.2 with the NA segment from clade 3C.2a1b.1a. This reassortment affected 90% (95/106) of H156 subgroup i viruses, including all with T135K substitution, whereas all S156 subgroup iii and iv viruses retained 2a.2 NA. See Supplementary Table S1 for NA characterisation by HA subgroup.
Epidemiological findings
Participants 1–19 years-old contributed disproportionately to cases (46%; 216/471) vs controls (32%; 312/980) (p < 0.001) and had the highest influenza A(H3N2) per cent positivity (41%; 216/528) (Table 1). We further explored age-related risk by genetic subgroup, re-categorising unvaccinated participants as ≤ 25 vs > 25 years to differentiate those born since 1997 with potential T135 priming, from those born before 1997 with potential K135 or other priming history, as outlined in Supplementary Table S2. Among 619 test-negative controls, 272 (44%) were ≤ 25 years-old. Relative to controls, individuals ≤ 25-year-olds were disproportionately represented among A(H3N2) cases overall (57%; 226/396; p < 0.001) but this was more pronounced for H156 viruses with K135 (80%; 32/40; p < 0.001) than H156 with T135 (49%; 58/118; p = 0.30), or S156 with T135 (56%; 88/156; p = 0.006). Recognising their lower contribution overall, individuals 26–35 years of age, with potential K135 priming history, were comparably represented among influenza A(H3N2) cases (15%; 58/396) and controls (13%; 81/619; p = 0.48) but contributed none of the H156 cases with K135 (0/40; p=0.09). Similar age patterns were observed with restriction to BC and, acknowledging smaller sample size, for A135 viruses in Ontario.
Table 1. Participant profile, influenza A(H3N2) test-positive cases and influenza test-negative controls, 2022/23 influenza vaccine effectiveness evaluation, SPSN, Canada, 1 November 2022–6 January 2023 (n = 1,451).
| Characteristics | All participantsa (column %) | Proportion influenza vaccinatedb (row %) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | Influenza A(H3N2) cases | Influenza virus test- negative controls | p valuec | Overall | p valuec | Influenza A(H3N2) casesd | Influenza virus test- negative controlsd | |||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |||
| N (row %) | 1,451 | 100 | 471 | 32 | 980 | 68 | NA | 436 | 30 | NA | 75 | 16 | 361 | 37 |
| Age group (years) | ||||||||||||||
| 1–19 | 528 | 36 | 216 | 46 | 312 | 32 | < 0.001 | 106 | 20 | < 0.001 | 25 | 12 | 81 | 26 |
| 20–64 | 704 | 49 | 212 | 45 | 492 | 50 | 184 | 26 | 29 | 14 | 155 | 32 | ||
| ≥ 65 | 219 | 15 | 43 | 9 | 176 | 18 | 146 | 67 | 21 | 49 | 125 | 71 | ||
| Median (range) | 33 (1–92) | 23 (1–87) | 37 (1–92) | < 0.001 | 50 (1–92) | < 0.001 | 39 (1–84) | 53 (1–92) | ||||||
| Interquartile range | 11–55 | 9–40 | 13–59 | NA | 22–70 | NA | 13–66 | 27–71 | ||||||
| Sex | ||||||||||||||
| Female | 888 | 61 | 278 | 59 | 610 | 62 | 0.19 | 286 | 32 | 0.022 | 47 | 17 | 239 | 39 |
| Male | 551 | 38 | 191 | 41 | 360 | 37 | 146 | 27 | 28 | 15 | 118 | 33 | ||
| Unknown | 12 | 1 | 2 | 0 | 10 | 1 | NA | 4 | 33 | NA | 0 | 0 | 4 | 40 |
| Comorbiditye | ||||||||||||||
| No | 1,068 | 74 | 365 | 77 | 703 | 72 | 0.008 | 280 | 26 | < 0.001 | 45 | 12 | 235 | 33 |
| Yes | 299 | 21 | 78 | 17 | 221 | 23 | 133 | 44 | 22 | 28 | 111 | 50 | ||
| Unknown | 84 | 6 | 28 | 6 | 56 | 6 | NA | 23 | 27 | NA | 8 | 29 | 15 | 27 |
| Province | ||||||||||||||
| Alberta | 275 | 19 | 77 | 16 | 198 | 20 | 0.20 | 89 | 32 | < 0.001 | 12 | 16 | 77 | 39 |
| British Columbia | 293 | 20 | 97 | 21 | 196 | 20 | 115 | 39 | 21 | 22 | 94 | 48 | ||
| Ontario | 656 | 45 | 213 | 45 | 443 | 45 | 183 | 28 | 33 | 16 | 150 | 34 | ||
| Quebec | 227 | 16 | 84 | 18 | 143 | 15 | 49 | 22 | 9 | 11 | 40 | 28 | ||
| Weeks of specimen collection, 2022/23 | ||||||||||||||
| 44–45 | 131 | 9 | 44 | 9 | 87 | 9 | < 0.001 | 30 | 23 | < 0.001 | 6 | 14 | 24 | 28 |
| 46–47 | 329 | 23 | 153 | 32 | 176 | 18 | 56 | 17 | 21 | 14 | 35 | 20 | ||
| 48–49 | 407 | 28 | 166 | 35 | 241 | 25 | 111 | 27 | 25 | 15 | 86 | 36 | ||
| 50–51 | 382 | 26 | 91 | 19 | 291 | 30 | 132 | 35 | 21 | 23 | 111 | 38 | ||
| 52–1 | 202 | 14 | 17 | 4 | 185 | 19 | 107 | 53 | 2 | 12 | 105 | 57 | ||
NA: not applicable; SPSN: Sentinel Practitioner Surveillance Network.
a Presenting within 7 days of influenza-like illness onset defined as acute onset of fever and cough and one other symptom including sore throat, myalgia, arthralgia or prostration. Fever was not a required symptom in adults ≥ 65 years.
b Vaccination status based on patient self-report; defined as receipt of 2022/23 seasonal influenza vaccine at least 2 weeks before symptom onset. Patients vaccinated less than 2 weeks before onset of symptoms or with unknown vaccination status or timing were excluded.
c p values for comparison between cases and controls or for the proportion vaccinated were derived by chi-squared test or Wilcoxon rank-sum test.
d Without regard to time before illness onset, 101 of 497 (20%) cases and 406 of 1,025 (40%) controls were vaccinated (p < 0.001).
e Includes chronic comorbidities that place individuals at higher risk of serious complications from influenza as defined by Canada’s National Advisory Committee on Immunization, including: heart, pulmonary (including asthma), renal, metabolic (such as diabetes), blood, cancer or immunocompromising conditions, conditions that compromise management of respiratory secretions and increase risk of aspiration, or morbid obesity (body mass index ≥ 40) [9].
Unless otherwise specified, values displayed in the columns represent the number of specimens per category and percentages are relative to the total.
Crude and adjusted VE estimates are shown in Table 2. Neither sex nor comorbidity affected estimates (based on investigation provided in Supplementary Table S3) and were not included as covariates. Influenza A(H3N2) VE was 54% (95% confidence interval (CI): 38 to 66) overall. VE was similar against vaccine-like S156 viruses at 53% (95% CI: 25 to 70) and H156 viruses at 50% (95% CI: 24 to 67). Against H156 viruses with T135, the VE was 52% (95% CI: 19 to 71) and against H156 viruses with K135 substitution was 46% (95% CI: −13 to 74), the latter similar at 44% (95% CI: −22 to 75) with restriction to BC where most (47/53; 89%) such variants were detected.
Table 2. Vaccine effectiveness estimates against influenza A(H3N2), overall and variant-specific, SPSN, Canada, 1 November 2022–6 January 2023 (weeks 44–1) (n = 1,451).
| Total | Cases | Controls | Unadjusted VEa | Adjustedb VEa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | n vacc | N | % | n vacc | N | % | % | 95% CI | % | 95% CI | ||||
| Primary analysis | 1,451 | 75 | 471 | 16 | 361 | 980 | 37 | 68 | 57 to 75 | 54 | 38 to 66 | |||
| Age-stratified | ||||||||||||||
| 1–19 years | 528 | 25 | 216 | 12 | 81 | 312 | 26 | 63 | 39 to 77 | 47 | 11 to 69 | |||
| 20–64 years | 704 | 29 | 212 | 14 | 155 | 492 | 32 | 66 | 47 to 78 | 58 | 33 to 73 | |||
| ≥ 65 years | 219 | 21 | 43 | 49 | 125 | 176 | 71 | 61 | 23 to 80 | 59 | 15 to 80 | |||
| Variant-specific | ||||||||||||||
| All S156d viruses | 1,164 | 27 | 184 | 15 | 361 | 980 | 37 | 71 | 55 to 81 | 53 | 25 to 70 | |||
| All H156e viruses | 1,187 | 38 | 207 | 18 | 361 | 980 | 37 | 61 | 44 to 74 | 50 | 24 to 67 | |||
| H156 with T135f | 1,120 | 22 | 140 | 16 | 361 | 980 | 37 | 68 | 49 to 80 | 52 | 19 to 71 | |||
| H156 with K135g, all SPSN provinces | 1,033 | 13 | 53 | 25 | 361 | 980 | 37 | 44 | −6 to 71 | 46 | −13 to 74 | |||
| H156 with K135g, British Columbia only | 243 | 13 | 47 | 28 | 94 | 196 | 48 | 59 | 17 to 79 | 44 | −22 to 75 | |||
CI: confidence interval; ECDC: European Centre for Disease Prevention and Control; VE: vaccine effectiveness; OR: odds ratio; PLR: penalised logistic regression; SPSN: Sentinel Practitioner Surveillance Network; vac: vaccinated; VE: vaccine effectiveness.
a OR compared influenza test positivity between vaccinated and unvaccinated participants using logistic regression, adjusting for confounders as specified. VE was calculated as (1 − OR) × 100%.
b Adjusted for age group (1–19, 20–64, ≥ 65 years), province (Alberta, British Columbia, Ontario, Quebec) and calendar time based on bi-weekly categories (44–45, 46–47, 48–49, 50–51, 52–1). Age stratified estimates not further adjusted for finer age sub-categories.
c Vaccination status based on patient self-report; defined as receipt of 2022/23 seasonal influenza vaccine at least 2 weeks before symptom onset. Patients vaccinated less than 2 weeks before onset of symptoms or with unknown vaccination status or timing were excluded.
d 3C.2a1b.2a.2 viruses that are H156S (B) like the A/Darwin/9/2021 (egg-passaged) and A/Darwin/6/2021 (cell-passaged) vaccine strains (i.e. S156 ECDC subgroups ii-iv) [12] (Supplementary Table S1).
e 3C.2a1b.2a.2 viruses that remain H156 (B) and with E50K (C) (as per H156 ECDC subgroup i).
f 3C.2a1b.2a.2 viruses that remain H156 (B) and with E50K (C) (as per H156 ECDC subgroup i) but excluding viruses bearing K or A substitution at position 135 (i.e. T135 viruses, excluding viruses with T135K or T135A (A)(RBS)(−CHO) substitutions).
g 3C.2a1b.2a.2 viruses that remain H156 (B) and with E50K (C) (as per H156 ECDC subgroup i) plus additional substitutions F79V + T135K (A)(RBS)(−CHO) + I140K (A) + G275D(C).
In sensitivity analyses, VE findings were similar using Firth’s method of penalised logistic regression to address small sample size, as shown in Supplementary Table S4 [23,24], and when excluding SARS-CoV-2 test-positive specimens from influenza test-negative controls as shown in Supplementary Table S3 [25].
Discussion
During an unusually early influenza A(H3N2) epidemic caused by 2a.2 viruses, the 2022/23 influenza vaccine reduced the risk of medically attended illness by about 50%. This VE estimate is higher than typically reported for influenza A(H3N2), often < 40% [8], and higher than estimated by the Canadian SPSN (36%) [1], Europe (29%) [26] or the US (36%) [27] during the delayed 2021/22 influenza A(H3N2) epidemic, similarly caused by 2a.2 viruses. Shorter time since vaccination and/or better match with the updated vaccine may have improved VE. The extent to which relative pause in influenza virus circulation during the coronavirus disease (COVID-19) pandemic may have affected the current VE estimates, is uncertain.
Effectiveness of the S156 vaccine did not differ overall with S vs H variation at position 156, notwithstanding reassorted NA additionally distinguishing most H156 viruses. Among H156 viruses we identified a subset with an additional T135K substitution for which the VE point estimate was below 50%, but with wide CI that overlapped those of other estimates. Although involving different vaccine and circulating strains, the SPSN has previously reported lower VE against T135K-bearing variants in 2016/17 (ca 20%) and 2017/18 (ca 10%), as have European investigators in 2018/19 (ca 10%) [7,28]. Such K135 variants, however, have not predominated overall and were ultimately outcompeted by T135 viruses.
Although geographic variation is not unexpected, it is unclear why viruses with T135K substitution established a stronger foothold in BC than elsewhere during the 2022/23 season. Among 118 other viruses collected in BC for non-SPSN clinical purposes between 2 November and 8 December 2022, 81 (69%) were also H156 with T135K substitution (GISAID identifiers in Supplementary Table S5) [21]. Conversely, among > 17,000 global sequences in GISAID (identifiers in Supplementary Table S6) with collection dates between 1 January and 30 December 2022, just 14 bore T135K, namely three from Saskatchewan, nine from the US and two from Denmark, the earliest of which was collected 3 August 2022 (in the US) [21]. As in Ontario, a greater subset in Europe and the US was instead T135A. Among unvaccinated SPSN participants, those ≤ 25 years born during T135 predominance (since 1997), comprised most (ca 80%) of the K135 variant cases, with no K135 cases observed among potentially K135-primed adults. Our exploration and interpretation in relation to potential imprinting effects is hypothesis-generating only [29]. More definitive evaluation, including the implications for VE estimation, requires larger datasets, similarly standardised for testing indication and vaccine status and conducted across the full epidemic to address potential differences in the timing of age-related transmission and risk [30,31].
We report reassortment whereby H156 clade 3C.2a1b.2a.2 viruses, including those with T135K, acquired NA from clade 3C.2a1b.1a. When that occurred is unknown, with analysis of internal gene segments ongoing, but evidence of the same reassortment this season is found among NA phylogenies available in Nextstrain [14]. Inter-clade reassortments involving NA and other gene segments have been shown in other recent seasons, such as between clades 3C.2a2 and 3C.2a1a in 2017/18 [7,11,14,32] and between clades 3C.3a1 and 3C.2a in 2018/19 [14]. The implications of such seasonal reassortments on epidemic severity, as hypothesised for the 2017/18 season [32], or for VE estimates (noting NA inclusion in vaccines), remains uncertain. Strategic use of WGS in sentinel networks may help further elucidate the prevalence, persistence and potential impact of reassortment viruses.
Our study has limitations, including small sample size with wide CI in stratified analyses. Residual bias and confounding cannot be ruled out. Measuring VE while both vaccine coverage and epidemic risk are evolving makes calendar time adjustment especially challenging but important. We genetically characterised most (> 80%) of our contributing viruses but generalisation elsewhere with a different mix of variants (or vaccines) requires caution.
Conclusions
During an unusually early influenza A(H3N2) epidemic in Canada, the 2022/23 influenza vaccine provided substantial protection, reducing the risk of medically attended influenza A(H3N2) illness by about half among vaccinated compared with unvaccinated individuals. The potential impact of unique genetic diversity, including T135K substitution and reassortment, warrants further evaluation elsewhere to inform vaccine update and to refine or refute imprinting considerations.
Ethical statement
Institutional review boards provided approval in Alberta: REB15-0587 and Ontario: 2017-057.06. In British Columbia both the University of British Columbia Clinical and Behavioural Research Ethics Board (REB)s waived review because such evaluations are considered within the core public health mandate of the BC Centre for Disease Control (BCCDC). In Quebec, such evaluations are similarly considered part of core public health surveillance with the Centre Hospitalier Universitaire de Québec REBs providing waiver of review.
Funding statement
Funding was provided by the BCCDC Foundation for Public Health, BC Ministry of Health, Alberta Health and Wellness, Public Health Ontario, the Ministère de la santé et des services sociaux du Québec and the Public Health Agency of Canada. The views expressed herein do not necessarily represent the view of the Public Health Agency of Canada. Funders had no role in data analysis, interpretation or the decision to publish.
Acknowledgements
The authors gratefully acknowledge the contribution of sentinel sites whose regular submission of specimens and data provide the basis of our analyses. We wish to acknowledge the administrative, coordination, data entry and/or management support in participating provinces including: Gabriel Canizares and Yuping Zhan at the British Columbia Centre for Disease Control; Aunshu Goyal and Rhoda Komolafe for TARRANT in Alberta; Sophie Auger and Lauriane Padet for Institut National de Santé Publique du Québec; and Mandy Kwok for Public Health Ontario. We would like to thank Tracy Lee, Kevin Kuchinski and James Zlosnik of the British Columbia Centre for Disease Control Public Health Laboratory for development of whole genome sequencing laboratory and bioinformatics methods and sequence generation. We also wish to thank those who provided additional laboratory and technical support in each province at the British Columbia Centre for Disease Control Public Health Laboratory, the Alberta Provincial Laboratory for Public Health (ProvLab), Public Health Ontario Laboratory and the Laboratoire de santé publique du Québec.
Supplementary Data
Conflict of interest: DMS is Principal Investigator on grants received to her institution from the Public Health Agency of Canada and the BCCDC Foundation for Public Health in support of this work. She has received grants from the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research for unrelated work, also paid to her institution. JBG is a paid consultant scientific editor for GIDEON Informatics, Inc., which is unrelated to the current work. Other authors have no conflicts of interest to declare.
Authors’ contributions: Principal investigators (epidemiological): DMS (National and British Columbia); JAD (Alberta); and GDS (Québec). Principal investigator (laboratory): ANJ (British Columbia); NZ (Alberta); JBG and RO (Ontario); HC (Québec); NB (National). Data collection oversight: SK. Genetic analyses: SS and SEK. Epidemiological data analysis: ESYC and DMS. Draft revision and approval: all.
References
- 1. Kim S, Chuang ESY, Sabaiduc S, Olsha R, Kaweski SE, Zelyas N, et al. Influenza vaccine effectiveness against A(H3N2) during the delayed 2021/22 epidemic in Canada. Euro Surveill. 2022;27(38):2200720. 10.2807/1560-7917.ES.2022.27.38.2200720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buckrell S, Ben Moussa M, Bui T, Rahal A, Schmidt K, Lee L, et al. National influenza annual report, Canada, 2021-2022: A brief, late influenza epidemic. Can Commun Dis Rep. 2022;48(10):473-83. 10.14745/ccdr.v48i10a07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Public Health Agency of Canada (PHAC). FluWatch report: January 8 to January 14, 2023 (week 2). Ottawa: PHAC; 2023. [Accessed: 31 Jan 2023). Available from: https://www.canada.ca/en/public-health/services/diseases/flu-influenza/influenza-surveillance/weekly-reports-2022-2023-season.html
- 4. Ben Moussa M, Buckrell S, Rahal A, Schmidt K, Lee L, Bastien N, et al. National influenza mid-season report, 2022-2023: A rapid and early epidemic onset. Can Commun Dis Rep. 2023;49(1):10-4. 10.14745/ccdr.v49i01a03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Centre for Disease Prevention and Control (ECDC). Flu New Europe: Joint ECDC-WHO/Europe weekly influenza update. Week 02/2023 (9 January – 15 January 2023). [Accessed: 31 Jan 2023]. Stockholm: ECDC; 2022. Available from: http://flunewseurope.org
- 6.Centers for Disease Control and Prevention (CDC). FluView: Past weekly surveillance reports. 2022-2023, January 14, 2023—Week 2. Atlanta: CDC; 2022. [Accessed: 31 Jan 2023]. Available from: https://www.cdc.gov/flu/weekly/pastreports.htm
- 7. Skowronski DM, Leir S, Sabaiduc S, Chambers C, Zou M, Rose C, et al. Influenza vaccine effectiveness by A(H3N2) phylogenetic subcluster and prior vaccination history: 2016-2017 and 2017-2018 epidemics in Canada. J Infect Dis. 2022;225(8):1387-98. 10.1093/infdis/jiaa138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canadian Sentinel Practitioner Surveillance Network. Canadian Sentinel Practitioner Surveillance Network (SPSN) influenza vaccine effectiveness estimates % (95% CI), 2004-05 to 2021-22 seasons. [Accessed: 16 Jan 2023]. Available from: http://www.bccdc.ca/resource-gallery/Documents/Statistics%20and%20Research/Publications/Epid/Influenza%20and%20Respiratory/SPSN_VE_By_Year_Table.pdf [DOI] [PMC free article] [PubMed]
- 9.National Advisory Committee on Immunization. Canadian immunization guide chapter on influenza and statement on seasonal influenza vaccine for 2022-2023. Ottawa (ON): Public Health Agency of Canada; 2022. [Accessed: 22 Jan 2023]. Available from: https://www.canada.ca/en/public-health/services/publications/vaccines-immunization/canadian-immunization-guide-statement-seasonal-influenza-vaccine-2022-2023.html
- 10.World Health Organization (WHO). Recommended composition of influenza virus vaccines for use in the 2022-2023 northern hemisphere influenza season. Geneva: WHO; 2022. Available from: https://www.who.int/publications/m/item/recommended-composition-of-influenza-virus-vaccines-for-use-in-the-2022-2023-northern-hemisphere-influenza-season
- 11.Francis Crick Institute. Worldwide influenza centre. Annual and interim reports. February 2022 and September 2022 interim reports. London: Francis Crick Institute. [Accessed: 16 Jan 2023]. Available from: https://www.crick.ac.uk/research/platforms-and-facilities/worldwide-influenza-centre/annual-and-interim-reports
- 12.European Centre for Disease Prevention and Control (ECDC). Influenza virus characterization, summary Europe, November 2022. Stockholm: ECDC; 2023. Available from: https://www.ecdc.europa.eu/en/publications-data/influenza-virus-characterization-summary-europe-november-2022
- 13. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976-9. 10.1126/science.1244730 [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Neher R, Bedford T. Real-time tracking of influenza A/H3N2 evolution. Nextstrain. [Accessed: 30 Jan 2023]. Available from: https://nextstrain.org/flu/seasonal/h3n2/ha/2y
- 15. Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83(19):10309-13. 10.1128/JVI.01109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman R, Nguyen J, Lee TD, Tyson JR, Azana R, Tsang F, et al. Rapid, high-throughput, cost effective whole genome sequencing of SARS-CoV-2 using a condensed one hour library preparation of the Illumina DNA Prep kit. medRxiv. 2022.02.07.22269672. Pre-print. [Accessed: 26 Jan 2023] 10.1101/2022.02.07.22269672. 10.1101/2022.02.07.22269672 [DOI] [PMC free article] [PubMed]
- 17.Illumina. Sequencing the rapidly evolving influenza A virus on the MiSeq system. San Diego: Illumina; 2017. Available from: https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/miseq-nextera-xt-influenza-application-note-770-2015-053.pdf
- 18.Kuchinski K. FluViewer [Source code]. 2022 Mar 17 [Accessed: 26 Jan 2023]. GitHub. Available from: https://github.com/KevinKuchinski/FluViewer
- 19. Eisler D, Fornika D, Tindale LC, Chan T, Sabaiduc S, Hickman R, et al. Influenza Classification Suite: An automated Galaxy workflow for rapid influenza sequence analysis. Influenza Other Respir Viruses. 2020;14(3):358-62. 10.1111/irv.12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ndifon W, Wingreen NS, Levin SA. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci USA. 2009;106(21):8701-6. 10.1073/pnas.0903427106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13):30494. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skowronski DM, Sabaiduc S, Leir S, Rose C, Zou M, Murti M, et al. Paradoxical clade- and age-specific vaccine effectiveness during the 2018/19 influenza A(H3N2) epidemic in Canada: potential imprint-regulated effect of vaccine (I-REV). Euro Surveill. 2019;24(46):1900585. 10.2807/1560-7917.ES.2019.24.46.1900585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80(1):27-38. 10.1093/biomet/80.1.27 [DOI] [Google Scholar]
- 24. Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21(16):2409-19. 10.1002/sim.1047 [DOI] [PubMed] [Google Scholar]
- 25. Doll MK, Pettigrew SM, Ma J, Verma A. Effects of confounding bias in coronavirus disease 2019 (COVID-19) and influenza vaccine effectiveness test-negative designs due to correlated influenza and COVID-19 vaccination behaviors. Clin Infect Dis. 2022;75(1):e564-71. 10.1093/cid/ciac234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kissling E, Pozo F, Martínez-Baz I, Buda S, Vilcu A-M, Domegan L, et al. Influenza vaccine effectiveness against influenza A subtypes in Europe: Results from the 2021-2022 I-MOVE primary care multicentre study. Influenza Other Respir Viruses. 2023;17(1):e13069. 10.1111/irv.13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Price AM, Flannery B, Talbot HK, Grijalva CG, Wernli KJ, Phillips CH, et al. Influenza vaccine effectiveness against influenza A(H3N2)-related illness in the United States during the 2021-2022 influenza season. Clin Infect Dis. 2022;ciac941. 10.1093/cid/ciac941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kissling E, Pozo F, Buda S, Vilcu A-M, Gherasim A, Brytting M, et al. Low 2018/19 vaccine effectiveness against influenza A(H3N2) among 15-64-year-olds in Europe: exploration by birth cohort. Euro Surveill. 2019;24(48):1900604. 10.2807/1560-7917.ES.2019.24.48.1900604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Francis ME, King ML, Kelvin AA. Back to the future for influenza preimmunity-looking back at influenza virus history to infer the outcome of future infections. Viruses. 2019;11(2):E122. 10.3390/v11020122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Griggs EP, Flannery B, Foppa IM, Gaglani M, Murthy K, Jackson ML, et al. Role of age in the spread of influenza, 2011-2019: Data from the US influenza vaccine effectiveness network. Am J Epidemiol. 2022;191(3):465-71. 10.1093/aje/kwab205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schanzer D, Vachon J, Pelletier L. Age-specific differences in influenza A epidemic curves: do children drive the spread of influenza epidemics? Am J Epidemiol. 2011;174(1):109-17. 10.1093/aje/kwr037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potter BI, Kondor R, Hadfield J, Huddleston J, Barnes J, Rowe T, et al. Evolution and rapid spread of a reassortant A(H3N2) virus that predominated the 2017-2018 influenza season. Virus Evol. 2019;5(2):vez046. 10.1093/ve/vez046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.