Abstract
Host-oriented antiviral therapeutics are promising treatment options to combat COVID-19 and its emerging variants. However, relatively little is known about the cellular proteins hijacked by SARS-CoV-2 for its replication. Here we show that SARS-CoV-2 induces expression and cytoplasmic translocation of the nucleolar protein, nucleolin (NCL). NCL interacts with SARS-CoV-2 viral proteins and co-localizes with N-protein in the nucleolus and in stress granules. Knockdown of NCL decreases the stress granule component G3BP1, viral replication and improved survival of infected host cells. NCL mediates viral-induced apoptosis and stress response via p53. SARS-CoV-2 increases NCL expression and nucleolar size and number in lungs of infected hamsters. Inhibition of NCL with the aptamer AS-1411 decreases viral replication and apoptosis of infected cells. These results suggest nucleolin as a suitable target for anti-COVID therapies.
Keywords: Nucleolin, Nucleoprotein, SARS-CoV-2, Aptamer, COVID
Graphical abstract
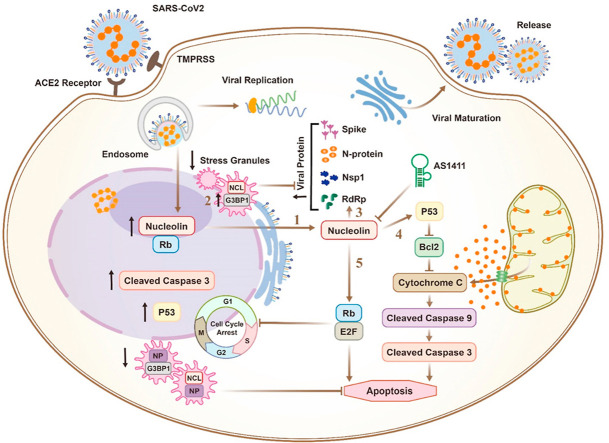
Mechanisms of nucleolin-mediated SARS-CoV-2 infection: 1. SARS-CoV-2 induces nucleolar to cytoplasmic translocation of nucleolin; In the cytoplasm, nucleolin interacts with the viral nucleoprotein and host G3BP1 in stress granules and increases viral proliferation via a direct increase in translation of replication proteins such as G3BP1 (2), nucleoprotein and RdRp (3); Nucleolin mediates viral-induction of apoptosis (4) and cell cycle arrest (5) via stimulation of p53 and Rb.
Mechanisms of nucleolin-mediated SARS-CoV-2 infection: 1. SARS-CoV-2 induces nucleolar to cytoplasmic translocation of nucleolin; In the cytoplasm, nucleolin interacts with the viral nucleoprotein and host G3BP1 in stress granules and increases viral proliferation via a direct increase in translation of replication proteins such as G3BP1 (2), nucleoprotein and RdRp (3); Nucleolin mediates viral-induction of apoptosis (4) and cell cycle arrest (5) via stimulation of p53 and Rb.
1. Introduction
SARS-CoV-2 is a positive-strand RNA virus closely related to SARS-CoV and several members of the beta coronavirus family (Lam et al., 2020; Zhou et al., 2020). Although the mechanisms of SARS-CoV-2 cellular entry via angiotensin-converting enzyme II (ACE2) have been extensively described (Zhou et al., 2020), less is known about SARS-CoV-2 replication (V'Kovski et al., 2021). SARS-CoV-2 must hijack a large series of host factors to enable its replication (V'Kovski et al., 2021). Host-directed drugs are promising therapeutic options to combat emerging viral variants as host genes possess a lower propensity to mutate compared to viral genes (Reuschl et al., 2021). Despite the wide variety of cellular mechanisms co-opted by the virus, a common feature of viral families is their interaction with the nucleolus (Salvetti and Greco, 2014). During viral infection numerous viral components localize within nucleoli, while various host nucleolar proteins are redistributed to other cellular compartments or are modified, and non-nucleolar cellular proteins reach the nucleolus (Salvetti and Greco, 2014).
Nucleolin (NCL) is a multifunctional nucleolar shuttling protein implicated in a variety of cellular functions such as ribosome biogenesis, DNA and RNA metabolism, cell proliferation, apoptosis, stress response and microRNA processing (Jia et al., 2017). NCL has been implicated in entry and replication of several viruses (Jia et al., 2017), including entry of influenza A viruses (Chan et al., 2016) and replication of the coronavirus infectious bronchitis virus (IBV) (Emmott et al., 2013). It is as a key interacting factor for viral replication in certain cases and is involved in nucleo-cytoplasmic export of the viral genome or viral egress (Jia et al., 2017). Recently, NCL was predicted to bind tightly to the 5′-region of the SARS-CoV-2 genome (Vandelli et al., 2020). Here we investigated the role of NCL in SARS-CoV-2 replication and provided mechanistic rationale for the use of a NCL-specific aptamer AS-1411 (Reyes-Reyes et al., 2010) to inhibit SARS-CoV-2 infection.
2. Results
2.1. SARS-CoV-2 proteins interact with nucleolin from host cells
To identify the role of NCL on SARS-CoV-2 replication we first determined whether NCL interacts with SARS-CoV-2 proteins. NCL is a protein localized primarily in the nucleolus, but is also found in the nucleus, cytoplasm and cell membrane of cancer and virally infected cells (Jia et al., 2017). Confocal immunofluorescence analysis of Vero E6 cells transfected with plasmids expressing viral proteins revealed that the spike-receptor binding domain (RBD) and N-protein of SARS-CoV-2 are present with NCL in the cytoplasm (Fig. 1 A, Figs. S1A and B and Video S1A-I). Untransfected cells which do not express the viral proteins showed exclusively nucleolar localization of NCL (Fig. 1A and Figs. S1A and B). The N-protein functions in packaging and transcription of the viral genome and virion replication (Cong et al., 2020). SARS-CoV-2 also uses the RNA-dependent RNA polymerase (RdRp)/nonstructural protein 12 (Nsp12) for replication of its genome and the transcription of its genes (V'Kovski et al., 2021).
Fig. 1.
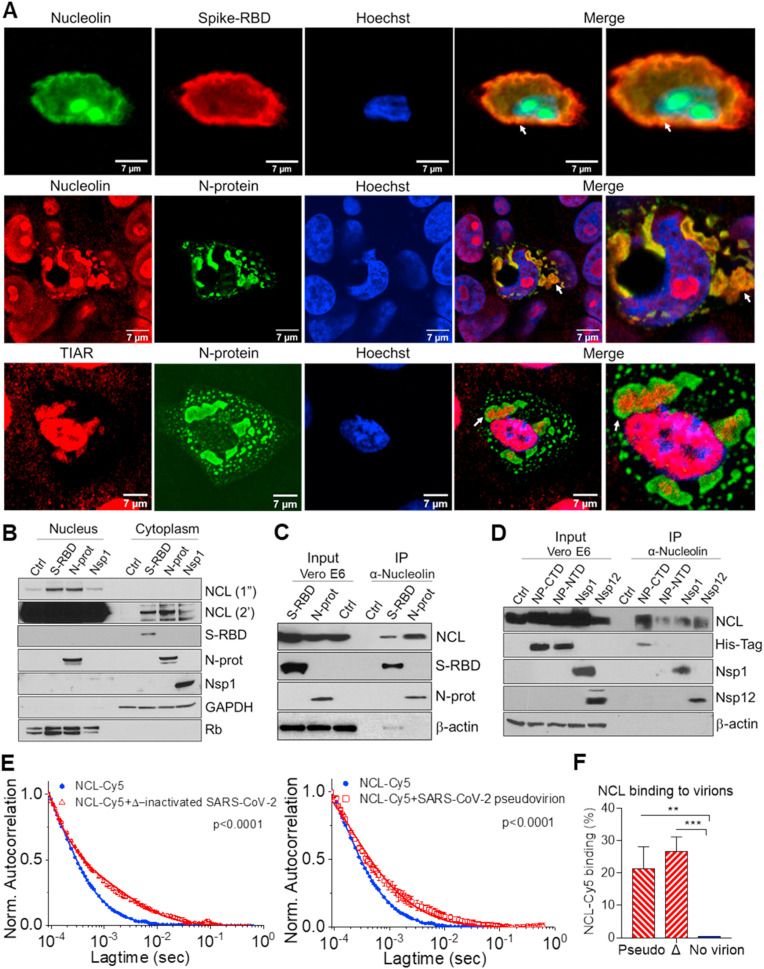
SARS-CoV-2 viral proteins interact with nucleolin and induce its cytoplasmic translocation. A. Immunofluorescence staining and confocal images of Vero E6 cells transfected with spike-RBD and N-protein for 48h, with anti-nucleolin (NCL, green or red), anti-spike-RBD (red), N-protein (green), TIAR (red), and Hoechst (blue). The merge of the fluorescent channels, including higher magnification, are shown (right). White arrows indicate co-localization. B. Western blot determination of NCL and SARS-CoV-2 proteins in the nuclear and cytoplasmic fractions of Vero E6 cells transfected with control (ctrl) empty plasmid (pcDNA3.1+), spike-RBD, N-protein and Nsp1 for 48h. Rb and GAPDH were used as nuclear and cytoplasmic markers, respectively. Exposure time for NCL development were 1 s (1″) and 2 min (2′). Lysates from Vero E6 cells transfected with spike-RBD, total N-protein, and control (pcDNA3.1+) (C) and C- and N-terminal domains (conjugated to a histidine-tag), Nsp1 and Nsp12/RdRp (D) were co-immunoprecipitated with NCL. Western blot determination of NCL and SARS-CoV-2 proteins, using total protein lysate (input) or immunoprecipitated complexes (IP). β-actin: loading control. Fluorescence correlation spectroscopy (FCS) auto-correlation plot for Cy5 labeled NCL alone or in complex with SARS-CoV-2 heat-inactivated virion (E) and percentage of NCL-Cy5 binding to virion (F). Average values from triplicate measurements are shown. Error bars indicate standard deviations. *P < 0.05, **P < 0.01 and ***P < 0.001.
NCL associates with the N-protein in stress granule (SG)-like structures, positive for T cell internal antigen 1 receptor (TIAR) (Fig. 1A and Figs. S1B and C). Cytoplasmic SGs are generally triggered by stress-induced translation arrest for storing mRNAs (Miller, 2011). Viruses manipulate SGs to promote synthesis of viral proteins during replication (Miller, 2011). Nsp1 from both SARS-CoV (Kamitani et al., 2006) and SARS-CoV-2 (Thoms et al., 2020) viruses are key factors in viral-induced downregulation of host gene expression and disruption of the cell cycle.
NCL is a nuclear-cytoplasmic shuttling protein (Jia et al., 2017). Post-translational modification of NCL, including N- and O-glycosylation, promotes its localization and function on the plasma membrane (Losfeld et al., 2009). The cytoplasmic redistribution of NCL, by several viral proteins, is closely related to viral replication (Jia et al., 2017). NCL is mainly expressed in the nucleolus and nucleus of Vero E6 cells, however upon transfection with SARS-CoV-2 spike-RBD, N-protein and Nsp1, part of NCL translocates to the cytoplasm (Fig. 1A, B and Figs. S1A, B, D). Similarly, SARS-CoV Nsp1 has been shown to alter the nuclear–cytoplasmic distribution of NCL (Gomez et al., 2019).
To confirm the interaction of SARS-CoV-2 with NCL, we evaluated the interaction of viral proteins from Vero E6 transfected cells with NCL by immunoprecipitation using an anti-NCL antibody. Immunoblotting analysis of NCL-associated proteins confirmed interaction of NCL with spike-RBD and N-protein (Fig. 1C). NCL also associates with SARS-CoV-2 Nsp1 and RdRP/Nsp12 (Fig. 1D). NCL did not immunoprecipitate in control lysate from cells transfected with empty vector (Fig. 1C and D). Accordingly, nucleolar proteins are not isolated using a low-salt lysis buffer. These data support viral-induced mobilization of NCL to the cytoplasm and co-immunoprecipitation in cells transfected with viral proteins (Fig. 1C and D). We used domain mapping and observed that all the RNA binding domains (RBD) from NCL appear to have a strong negative region on one side and a more positive region on the other side (Figs. S1E and F). The binding domains of the N-protein appear to have only a strongly positive region (Figs. S1E and F). Docking analysis showed that the positive region of the N-protein binding domains complex to the most negative regions on the NCL RBDs (Fig. S1G). Detailed analysis showed that NCL binds preferentially to the C-terminal domain (CTD) of the N-protein (Fig. 1D). The CTD of the N-protein forms a compact dimer that has been proposed to bind to RNA, regulate viral transcription, assembly, N-protein oligomerization and SG localization (McBride et al., 2014).
We evaluated the binding of recombinant NCL to pseudovirus containing SARS-CoV-2 spike protein and heat inactivated virus by fluorescence correlation spectroscopy (FCS) (Hess et al., 2002). The diffusion coefficient of NCL-Cy5 alone was 105 ± 2 μm2/s. Upon binding to ∼100 nm virion particles the diffusion coefficient was 6 ± 2 μm2/s (Fig. 1E). We observed on average 21 and 27% of total recombinant NCL bound to pseudovirus and heat inactivated virus, respectively (Fig. 1E and F). Collectively, these data showed that NCL interacts with viral proteins and is recruited to the cytoplasm in the presence of SARS-CoV-2 proteins.
2.2. SARS-CoV-2 induces nucleolin expression and cytoplasmic translocation
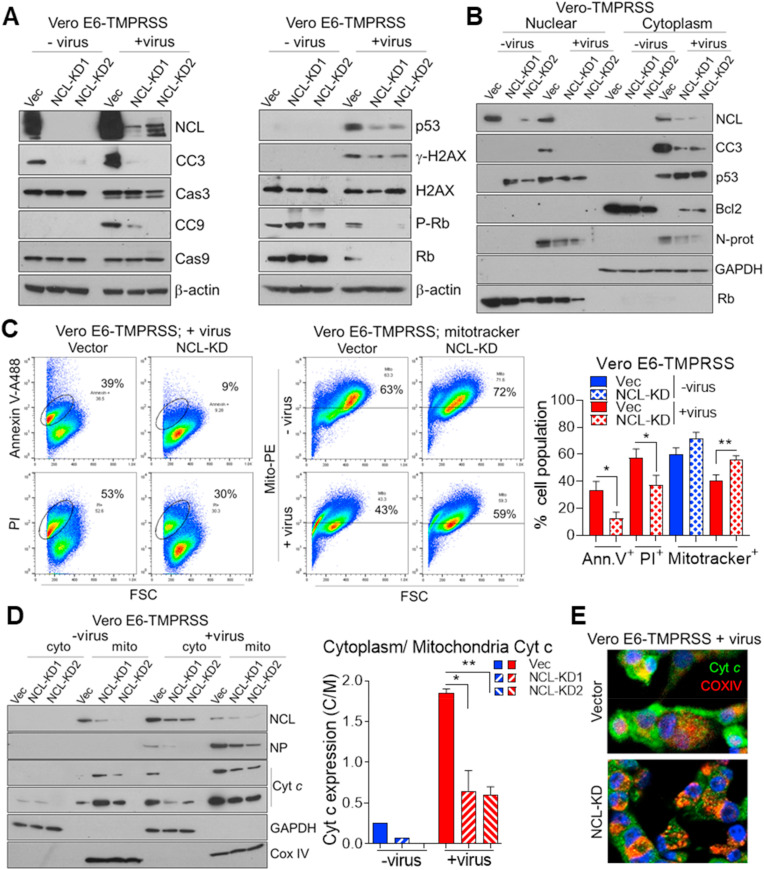
Cells expressing a subgenomic replicon of SARS-CoV have previously shown increased levels of NCL (Zhang et al., 2010). Infection with SARS-CoV-2 resulted on average in 3.6- and 2.3-fold higher levels of NCL in Vero E6-TMPRSS and human Caco-2 cells, respectively (Fig. 2 A). To address if NCL is involved in SARS-CoV-2 infection, and avoid off-target effects, we used two independent NCL knockdown (KD) cells (Fig. 2A and Figs. S2A and B).
Fig. 2.
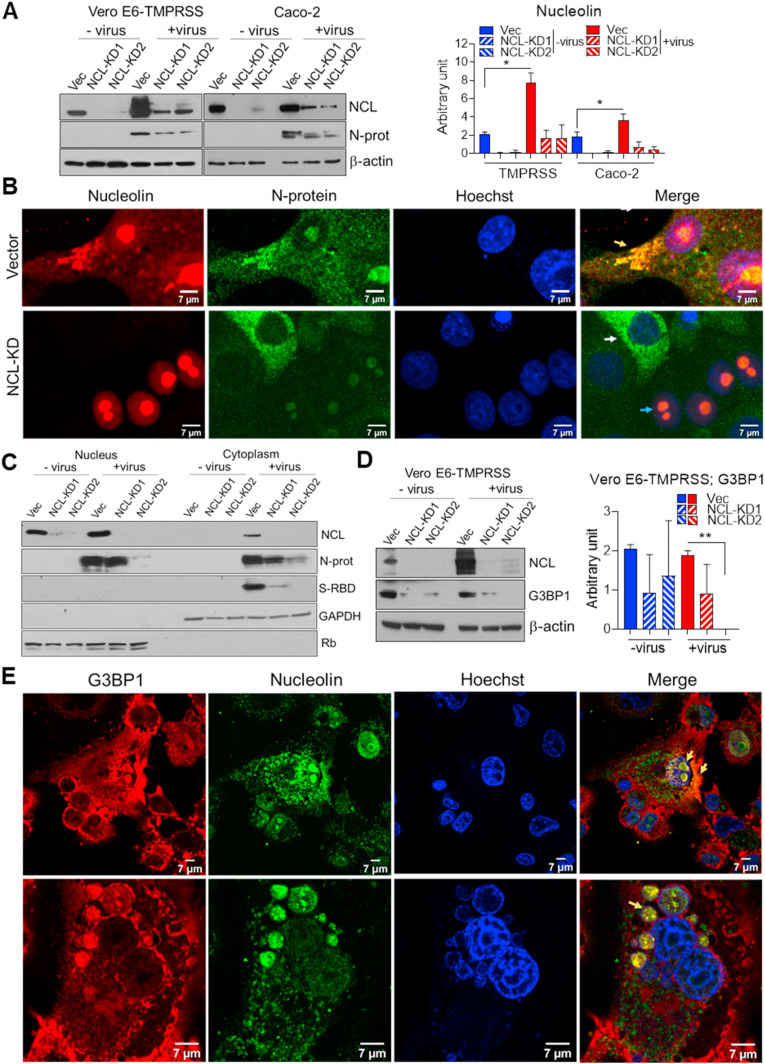
SARS-CoV-2 increases nucleolin levels and translocation to cytoplasm. A. Western blot determination, in triplicate, and ImageJ quantification of NCL levels in Vero E6-TMPRSS and Caco-2 cells, transduced with empty Lenticrispr V2 containing CAS9 vector control and 2 different NCL-knockdown lines (KD1 and KD2), and infected with SARS-CoV-2 for 48h (MOI 0.01). N-protein was used to detect the presence of virus. β-actin: loading control. B. Confocal immunofluorescence of NCL (red) and N-protein (green) in Vero E6-TMPRSS vector and NCL-KD following viral infection (as above). Hoechst (blue) and the fluorescence merge are shown. Yellow, blue and white arrows indicate the SG-like structures, NCL-proficient and -deficient cells, respectively. Western blot determination of NCL, N-protein and spike-RBD in nuclear and cytoplasmic fractions (C) and G3BP1 in total lysate (D) of Vero E6-TMPRSS control and NCL-KD cells infected for 48 h with mock and SARS-CoV-2 (MOI 0.01). Rb and GAPDH were used as nuclear and cytoplasmic markers, respectively. The student t-test was performed. E. Confocal immunofluorescence of G3BP1 (red) and NCL (green) in Vero E6-TMPRSS cells following viral infection (as above). Hoechst stain (blue) and the fluorescence merge are shown. Yellow arrow indicates G3BP1 and NCL co-localization. *P < 0.05, **P < 0.01 and ***P < 0.001.
Coronavirus N-proteins localized to both cytoplasm and nucleolus in virus-infected cells and may disrupt host cell division (Ariumi, 2022; Savastano et al., 2020; Timani et al., 2005). Computational modeling of SARS-CoV-2 viral RNA localization predicts that the SARS-CoV-2 RNA genome and all sgRNAs are enriched in the host mitochondrial matrix and nucleolus (Wu et al., 2020). Similarly to SARS-CoV (Timani et al., 2005), we observed that SARS-CoV-2 N-protein co-localized with NCL mainly in the cytoplasm but also in the nucleolus of Vero E6-TMPRSS infected cells (Fig. 2B and Figs. S2C, D, E). N-protein was absent from the nucleoli of cells depleted of NCL (Fig. 2B and Fig. S2D). Similar to fluorescence images, N-protein was detected in both nuclear and cytoplasmic fractions of infected cells (Fig. 2C). SARS-CoV-2, SARS-CoV and MERS-CoV N-proteins present nuclear localization signals (NLSs) and nuclear export signal (NES), which appears to be associated with high case fatality rate of these coronaviruses (Gussow et al., 2020). NCL could facilitate N-protein nucleolar localization.
SARS-CoV-2 was recently shown to subvert SG machinery and exploit Ras GTPase-activating protein-binding protein 1 (G3BP1) for its replication (Ciccosanti et al., 2021). We observed decreased levels of G3BP1 in Vero E6-TMPRSS NCL-KD cells (Fig. 2D), suggesting NCL up-regulation of G3BP1 in both mock and SARS-CoV-2 infected cells. NCL and G3BP1 co-localized in the nucleoli and SG of Vero E6-TMPRSS infected cells (Fig. 2E and Figs. S2F and G). There was no association of those proteins in mock-infected cells (Fig. S2F). NCL-regulation of G3BP1 and its interaction with N-protein, which are proteins involved in viral replication, prompted us to address the role of NCL in this process.
2.3. Nucleolin regulates replication of SARS-CoV-2
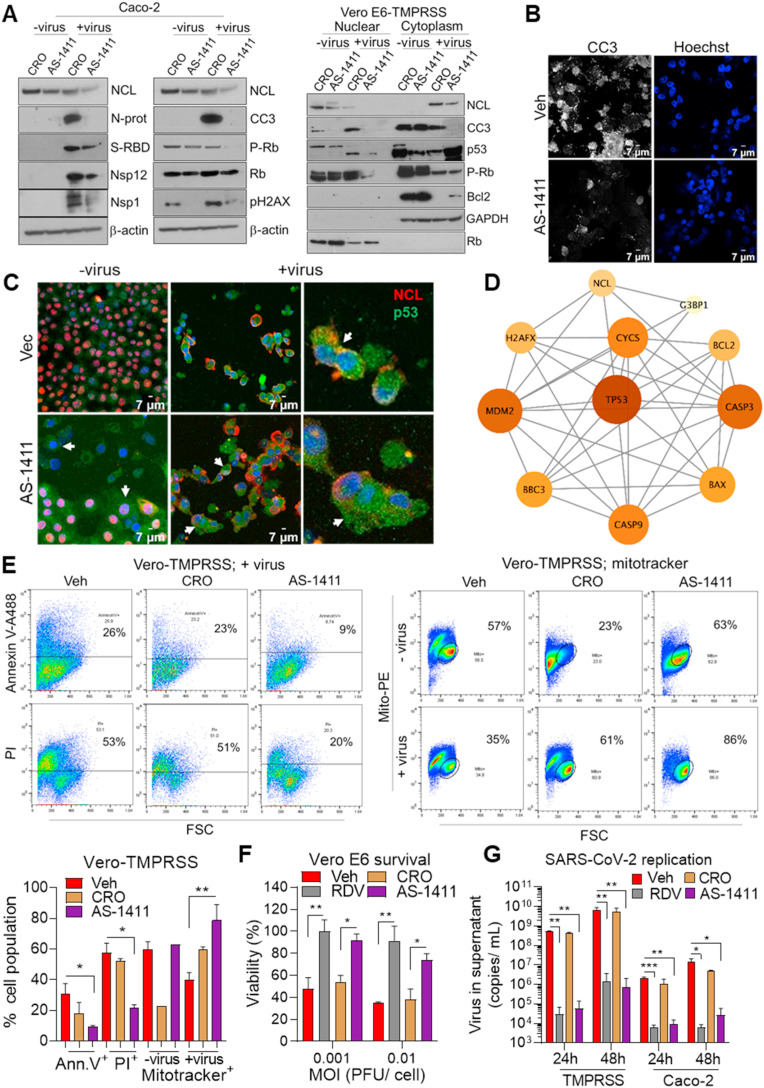
NCL participates extensively in RNA regulatory mechanisms, including transcription, ribosome assembly, mRNA stability and translation, and microRNA processing (Jia et al., 2017). Accordingly, knockdown of NCL in Vero-TMPRSS (Figs. S3A and B) and Caco-2 (Fig. 3 A and Fig. S3B) cells resulted in significantly lower levels of viral proteins related to replication, including, N-protein, Nsp1, and Nsp12/RdRp and spike-RBD. There was a lower effect of NCL-knockdown on the expression of the envelope E-protein (Fig. 3A and Figs. S3A and B). These data suggest that NCL may regulate translation of viral proteins involved in replication. In addition, the lower levels of viral proteins in NCL-KD cells may also be associated to decreased viral titer.
Fig. 3.
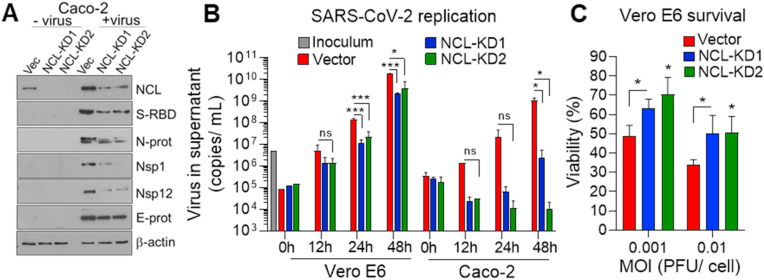
Nucleolin increases viral protein and replication. A. Western blot determination, in triplicate, of NCL and viral proteins in Caco-2 cells control and NCL-KD infected with mock and SARS-CoV-2 (MOI 0.01) for 48 h. β-actin: loading control. B. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), using primers against N-protein, to determine the viral titer in the supernatant of Vero E6 and Caco-2 control and NCL-KD cells at the indicated time points, following SARS-CoV-2 infection (MOI 0.001). Four independent infection experiments and qRT-PCR were performed. C. Cell titer glow assay, in triplicate, to determine cell viability at 48h of Vero E6 control and NCL-KD cells (1 × 104 cells/well in 96 well plate) following viral infection (MOI 0.001 and 0.01). The viral inoculums were removed after 1 h of incubation in both replication and survival assays. Cell viability is represented as percentage relative to non-infected cells. The student t-test was performed. NS: not significant. *P < 0.05, **P < 0.01 and ***P < 0.001.
We addressed the role of NCL in viral replication. The supernatant of Vero E6 and Caco-2 cells, deficient in NCL, contained significantly lower levels of virus in comparison to vector control cells (Fig. 3B). Accordingly, decrease of NCL in Vero E6 cells improved survival of infected cells in comparison to control cells (Fig. 3C). These data indicate a role for NCL in SARS-CoV-2 replication and survival of host cells.
2.4. Nucleolin mediates SARS-CoV-2 induction of stress response
The induction of apoptosis is a hallmark of SARS-CoV-2 infection (Donia and Bokhari, 2021). Since NCL knockdown decreased respiratory syncytial virus (RSV)-induced apoptosis of mouse neuronal-2a (N2a) cells (Hu et al., 2019), we sought to investigate if NCL mediates SARS-CoV-2 induction of apoptosis and DNA damage.
SARS-CoV-2 increased both cleaved caspase 9 and 3 (CC9 and 3) in Vero E6-TMPRSS and Caco-2 cells (Fig. 4 A and Figs. S4A and B). Knockdown of NCL resulted in decreased viral activation of these caspases (Fig. 4A and Figs. S4A and B). As a critical enzyme in promoting apoptosis, caspase-3 becomes activated and then enters the nucleus to exert its function (Kamada et al., 2005). We observed that infection of control cells induced CC3 expression in both cytoplasmic and nuclear fractions (Fig. 4B). No nuclear CC3 expression was detected in NCL-KD cells (Fig. 4B), suggesting decreased apoptosis. Confocal microscopic images of infected cells confirmed increase in CC3 by SARS-CoV-2 and its decrease in NCL-KD cells (Figs. S4C and D).
Fig. 4.
Nucleolin mediates SARS-CoV-2 induction of the stress response and apoptosis. Western blot determination of NCL, apoptotic, stress response and DNA damage proteins in total cell lysate (A) and nuclear and cytoplasmic fractions (B) of Vero E6-TMPRSS cells, control and NCL-KD infected with mock and SARS-CoV-2 (MOI 0.01) for 48 h. β-actin: loading control. Rb and GAPDH were used as nuclear and cytoplasmic markers, respectively. C. Flow cytometric determination, in triplicate, and ImageJ quantification of apoptosis by staining of Vero E6-TMPRSS control and NCL-KD cells with annexin V-Alexa 488, propidium iodide (PI) and mitotracker conjugated with phycoerythrin (PE). The number of positive cells is shown (%). D. Western blot determination in triplicate and ImageJ quantification of NCL, N-protein and cytochrome C (Cyt c) in cytoplasmic and mitochondrial fractions of Vero E6-TMPRSS cells, control and NCL-KD, infected with mock and SARS-CoV-2 (as above). The ratio of Cyt c expression in cytoplasm (C) by mitochondria (M) is shown. E. Merged confocal immunofluorescence image of cytochrome c (Cyt c, green), mitochondrial marker Cox IV (red) and Hoechst (blue) in Vero E6-TMPRSS cells infected with SARS-CoV-2 (MOI 0.01) for 48 h. The student t-test was performed. *P < 0.05, **P < 0.01 and ***P < 0.001.
The p53 is a well-known tumor suppressor protein that is activated by varied stress signals (Levine and Oren, 2009). In order to spread, some viruses cause p53-mediated cell death of the host cell by different mechanisms such as cell lysis and apoptosis (Aloni-Grinstein et al., 2018). SARS-CoV-2 infection of Vero E6-TMPRSS cells increased p53 and the DNA damage marker phospho-H2AX, which were decreased by knockdown of NCL (Fig. 4A and Fig. S4A). Cell fractionation of infected cells showed nuclear activation of p53 and decreased cytoplasmic p53 in Vero E6-TMPRSS control cells (Fig. 4B). On the contrary, in NCL-KD cells p53 was lower in the nucleus and enriched in the cytoplasm (Fig. 4B), the site of p53 degradation (O'Brate and Giannakakou, 2003), supporting the lower levels of p53 detected in the absence of NCL (Fig. 4A). The expression of p53, as well as the downstream apoptotic effectors PUMA and Bax were lower in Caco-2 NCL-KD cells in comparison to control cells (Fig. S4B). Expression of the pro-survival protein Bcl2 was high in mock-infected cells, silenced in virus-infected control cells, and increased in NCL-KD cells (Fig. 4B).
SARS-CoV-2 infection has been shown to induce cell cycle arrest (Bouhaddou et al., 2020). The retinoblastoma tumor suppressor protein (Rb) is a negative regulator of cell proliferation. Rb is targeted by viruses to modulate the host cell cycle under growth limiting conditions (Fan et al., 2018). We observed that Rb levels in viral-infected cells were much lower than in mock-infected cells (Fig. 4A and Figs. S4A and B). However, in infected control cells, which express NCL, Rb levels were higher than NCL-KD cells (Fig. 4A and Figs. S4A and B). Rb expression could contribute to virally induced cell cycle arrest. These data indicate that targeting NCL could revert Rb-induced arrest of host cells. In addition to induction of apoptosis, p53 induces cell cycle arrest upon DNA damage and viral infection (Fan et al., 2018). Accordingly, increase of both Rb and p53 by NCL may mediate SARS-CoV-2-induced cell cycle arrest. Notably, NCL has been shown to interact with Rb (Grinstein et al., 2006) and to regulate the stability of p53 (Takagi et al., 2005) and Bcl2 (Sengupta et al., 2004).
Collectively, these data suggest that NCL mediates SARS-CoV-2-induction of proteins with a role in survival and apoptosis of host cells. In addition, the decrease of viral titer in NCL-KD cells (Fig. 3B) may also contribute to decreased apoptosis.
2.5. Nucleolin mediates SARS-CoV-2-induction of host cell apoptosis
We next sought to investigate if NCL plays a role in viral induction of apoptosis. For that, we stained infected cells with annexin V and a mitochondrial viability marker, mitotracker, and evaluated the translocation of cytochrome c (Cyt c).
Knockdown of NCL significantly decreased the number of apoptotic cells (annexin V and propidium iodide (PI)-positive) and increased the number of live cells (mitotracker-positive) in virally infected Vero E6-TMPRSS cells (Fig. 4C). Pro-apoptotic stimuli promote the mobilization of Cyt c from the mitochondria to cytosol. In the cytosol, Cyt c mediates activation of caspase-9 and caspase-3 (Garrido et al., 2006). Cellular localization of Cyt c was determined by immunofluorescence co-localization with the mitochondrial marker Cox IV and by immunoblotting of fractionated cells. Viral infection resulted in increased cytoplasmic release of Cyt c (Fig. 4D, E and Figs. S4E and F). Knockdown of NCL decreased the ratio of cytoplasmic to mitochondrial Cyt c expression in comparison to infected control cells (Fig. 4D, E and Figs. S4E and F). The SARS-CoV-2 RNA genome was predicted to be enriched in host mitochondrial matrix (Wu et al., 2020). Mitochondria might be closely associated with SARS-CoV2 replication and host cell defense (Shang et al., 2021). Accordingly, we observed expression of viral N-protein as well as NCL in the mitochondrial cell fraction (Fig. 4D). These results suggest that NCL may mediate viral induction of apoptosis.
2.6. Targeting nucleolin decreased viral replication and increased host cell viability
We next sought to investigate if NCL could be exploited therapeutically to decrease SARS-CoV-2 replication and associated host cell damage. Similar to NCL-KD cells (Fig. 3A), pharmacological inhibition of NCL, with the NCL-specific aptamer AS-1411, decreased significantly the expression of viral proteins, with a role in replication, in comparison to the control aptamer CRO (Fig. 5 A and Fig. S5A). We observed that treatment with the aptamer decreased NCL and Nsp12 proteins in both cytoplasm and nuclear compartments, and Spike and N-protein in the cytoplasm (Fig. S5B). The aptamer mediated decrease of N-protein and Nsp12 replication proteins in the cytoplasm (Fig. S5B) may have a role on viral titer. Treatment with AS-1411 decreased viral-mediated increase of the apoptotic protein CC3 and DNA damage phospho-H2AX and restored the expression of the survival protein Bcl2 in infected cells (Fig. 5A and Fig. S5A). AS-1411 also decreased nuclear phospho-Rb, p53 and CC3 (Fig. 5A). Confocal immunofluorescence confirmed the decrease of CC3 (Fig. 5B) and increase of cytoplasmic p53 (Fig. 5C and Fig. S5C) by AS-1411 in viral-infected Vero E6-TMPRSS cells. The interaction of NCL, G3BP and p53 pathway was visualized by protein interaction network (Fig. 5D).
Fig. 5.
Nucleolin inhibition decreases SARS-CoV-2 replication and apoptosis of host cells. A. Western blot determination of NCL, apoptosis, stress response, DNA damage and viral proteins in total cell lysate from Caco-2 cells and nuclear and cytoplasmic fraction from Vero E6-TMPRSS cells treated with CRO and AS-1411 (10 μM) for 24 h prior to SARS-CoV-2 (MOI 0.01) infection for 48 h. β-actin: loading control. Confocal immunofluorescence of mock- and viral-infected Vero E6-TMPRSS cells stained with anti-cleaved caspase 3 (CC3) (white) and Hoechst (blue) (B) and merged images of NCL (red), p53 (green) and Hoechst (blue) (C). White arrows indicate cytoplasmic p53. D. String interaction network of NCL, G3BP1 and p53 apoptotic pathway. E. Flow cytometry determination, in triplicate, of apoptosis by staining Vero E6-TMPRSS cells treated with vehicle, CRO and AS-1411 (10 μM) for 24 prior to SARS-CoV-2 infection (MOI 0.01) for 48 h. The percentage of annexin V, PI- and mitotracker-positive cells is shown. Cell viability assay of Vero E6 cells (F) and qRT-PCR detection of SARS-CoV-2 E-protein in the supernatant of Vero E6-TMPRSS and Caco-2 cells (G) treated with vehicle, remdesivir (5 μM), CRO and AS1411 (10 μM) 24 h prior to viral infection (MOI 0.001 for replication). Cell viability is represented as percentage relative to non-infected treated cells. Four independent infection experiments and qRT-PCR were performed. *P < 0.05, **P < 0.01 and ***P < 0.001.
According to the decrease of apoptotic and DNA damage markers (Fig. 5A), AS-1411 treatment significantly decreased virally induced apoptotic cells, annexin V- and PI-positive, and increased viable cells, mitotracker-positive (Fig. 5E). Similar to remdesivir, approved by the Food and Drug Administration (FDA) for the treatment of COVID-19 (Beigel et al., 2020), AS-1411 improved survival of Vero E6 infected cells in comparison to treatment with vehicle and CRO controls (Fig. 5F). Notably, the NCL aptamer decreased viral titers in the supernatant of Vero E6-TMPRSS and human Caco-2 cells, with comparable potency to remdesivir (Fig. 5G). Collectively, these results suggest that targeting NCL is a novel approach to control viral replication and improve host cell viability.
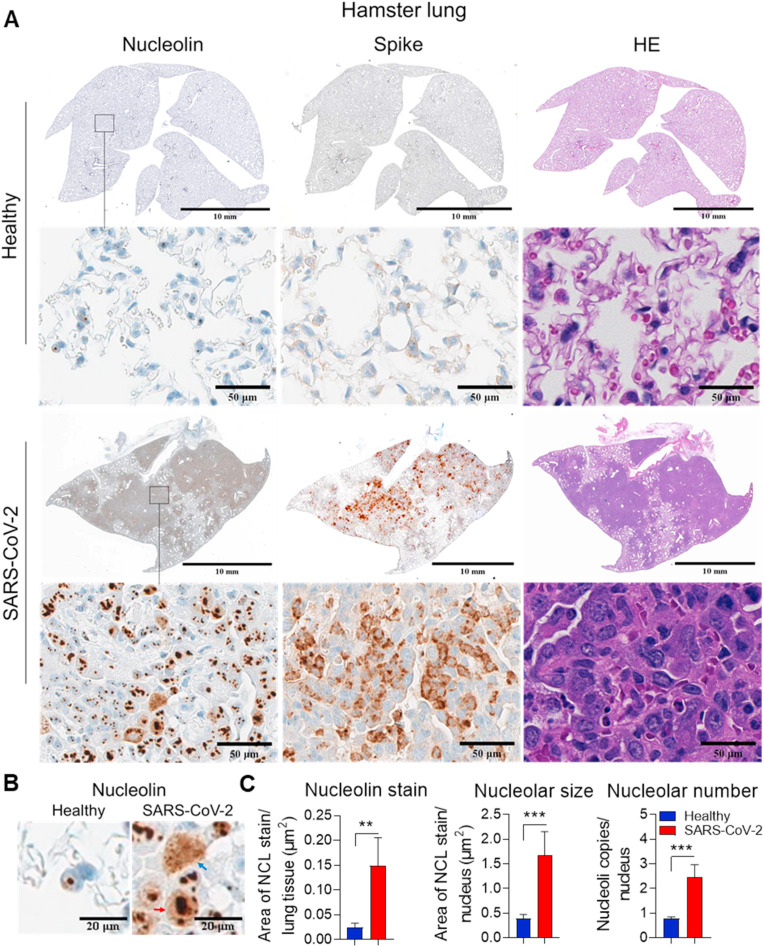
2.7. Higher nucleolin expression and nucleolar size and number in lungs of SARS-CoV-2 infected hamsters
Since we observed that NCL participates in the replication of SARS-CoV-2 (Fig. 3B) and induction of apoptosis of host cells (Fig. 4C), we investigated their interaction in vivo.
Confocal immunofluorescence of lungs of SARS-CoV-2-inoculated hamsters and mice overexpressing human ACE2 showed that NCL co-localized with spike and N-protein (Figs. S6A and B). NCL co-localizes with N-protein in the cytoplasm and nucleolus of infected lungs (Fig. S6B).
SARS-CoV-2 inoculation resulted in significant up-regulation of NCL in specific areas that showed hyperplasia of pneumocytes (Sia et al., 2020) and the presence of virus (Fig. 6 A). NCL is up-regulated in the nucleoli of pneumocytes of hamsters (Fig. 6A–C). In addition, viral infection resulted in increased size and number of nucleoli (Fig. 6A–C), suggesting nucleolar stress. Similar to infected cells (Fig. 2B), we observed that SARS-CoV-2 induced mobilization of NCL to the nucleus and cytoplasm in vivo (Fig. 6B and Fig. S6C). These results suggest that SARS-CoV-2 regulation of NCL in vivo may result in damage of lung tissue.
Fig. 6.
SARS-CoV-2 interacts with nucleolin and regulates its expression in lungs of infected hamsters. Representative consecutive images (A) and higher magnification (B) of immunohistochemistry (IHC) detection of NCL and Spike in lungs of hamsters inoculated with mock and SARS-CoV-2 (n = 5 each). Hematoxylin and eosin (HE) stain of tissue sections is shown. Blue and red arrows indicate NCL cytoplasmic and nuclear stain, respectively. C. Quantification of total NCL stain, nucleolar size and number using Halo software. The student t-test was performed. *P < 0.05, **P < 0.01 and ***P < 0.001.
3. Discussion
In addition to employing cellular machines like the ribosome to translate their proteins and manipulating cellular membranes during RNA synthesis and viral morphogenesis, several coronavirus proteins modify the cellular environment in ways that may influence viral pathogenesis and replication in vivo. These include cell cycle regulation, intracellular protein transport, apoptosis, and others (V'Kovski et al., 2021). Targeting host pathways and cellular proteins that are hijacked by viruses can potentially offer broad-spectrum targets for the development of future antiviral drugs (Tao et al., 2021). Here we showed that SARS-CoV-2 increased nucleolin (NCL) levels and hijacks this multifunctional nucleolar protein (Jia et al., 2017) for its replication and induction of apoptosis of host cells.
We observed that NCL interacts with the SARS-CoV-2 N-protein, spike-RBD, Nsp1 and Nsp12 (Fig. 1A, C, D). NCL has previously been shown to interact with the N-protein of different viruses (Jia et al., 2017), including the coronavirus IBV (Chen et al., 2002). We observed that SARS-CoV-2 induced NCL translocation from the nucleolus to the cytoplasm (Fig. 2C). SARS-CoV Nsp1 was also shown to alter the nuclear–cytoplasmic distribution of NCL (Gomez et al., 2019). NCL interacts with SARS-CoV-2 N-protein in stress granule (SG) structures (Figs. 1A and 2B, E). SGs are membraneless organelles composed of mRNAs and RNA binding proteins (RBP) which undergo assembly in response to stress-induced inactivation of translation initiation (Miller, 2011). NCL is a stress responsive RBP that plays important roles in gene transcription and RNA metabolism (Jia et al., 2017). NCL was shown to be recruited to perinuclear SGs following unfolded protein response (UPR) activation (Child et al., 2021). N-protein interacts with the SG component G3BP1, which is required for efficient SARS-CoV-2 replication (Ciccosanti et al., 2021). We showed that NCL also interacts with G3BP1 in infected cells (Fig. 2E) and decrease of NCL resulted in lower levels of G3BP1 (Fig. 2D). Collectively these data suggest that regulation of G3BP1 by NCL may contribute to viral replication. In agreement with viral inhibition of SG to favor viral translation, we observed that SARS-CoV-2 induced NCL levels, which in turn induced translation of viral proteins with a role in replication (Fig. 3A). Consistently, NCL knockdown decreased viral levels in the supernatant of infected cells (Fig. 3B). NCL has previously been shown to play an important role in the synthesis of viral protein and replication of different viruses (Jia et al., 2017). NCL induces lyssaviruses P-protein expression and infectious virus production (Oksayan et al., 2015).
Apoptotic cell death triggered by viruses has a complex role in host antiviral immunity and might facilitate viral clearance or act as a mechanism for virally induced tissue injury and disease progression (Zhou et al., 2017). Protein network analysis based on interactions between SARS-CoV-2 and human host proteins identified that the host systems most affected were apoptotic and mitochondrial pathways (Guzzi et al., 2020). SARS-CoV-2 has been shown to induce apoptosis in lung tissue of animals (Mehta et al., 2020) and humans (Liu et al., 2021) via activation of caspase-3, 8, and 9. We observed that NCL increased those pro-apoptotic caspases, p53 and the DNA damage marker phospho-H2AX, and decreased the pro-survival protein Bcl2 in viral infected cells (Fig. 4A). NCL mediates the SARS-CoV-2- induction of p53-dependent apoptosis (Fig. 5D). Similar to our findings, SARS-CoV-induced cell death has been shown to involve downregulation of Bcl-2, release of cytochrome c from mitochondria, and activation of caspase 3 (Tan et al., 2007). The p53 signaling was one of the most enriched pathway in bioinformatics studies upon SARS-CoV-2 infection (Kwan et al., 2021; Rahaman et al., 2021). The human double minute 2 (Hdm2) oncoprotein (with non-human homologs referred to as Mdm2) is a well-studied E3 ubiquitin ligase that targets p53 for proteasomal degradation (Marine and Lozano, 2010). NCL was shown to inhibit Hdm2 leading to p53 stabilization (Saxena et al., 2006; Takagi et al., 2005). Recently it was reported an increase in MDM2 in lungs of hamsters following spike pseudovirus administration (Lei et al., 2021). It is possible that SARS-CoV-2 increases NCL to inhibit the MDM2 effect and stabilize p53. In addition to induction of apoptosis, p53 induces cell cycle arrest upon DNA damage and viral infection (Fan et al., 2018). SARS-CoV-2 infection promoted shutdown of mitotic kinases, resulting in cell cycle arrest (Bouhaddou et al., 2020). SARS-CoV induces cell cycle arrest via the Rb pathway (Yuan et al., 2006). MHV virus with mutations in Rb exhibited reduced viral titer (Bhardwaj et al., 2012), suggesting that Rb may have a role in viral replication. Targeting NCL decreased Rb in infected cells (Figs. 4A and 5A), and could revert Rb- induction of cell cycle arrest.
AS-1411, a G-rich DNA aptamer against NCL (Bates et al., 2009), was evaluated in clinical trials for cancer treatment (Mongelard and Bouvet, 2010; Rosenberg et al., 2014). NCL inhibition with the aptamer AS-1411 decreased apoptosis of infected cells (Fig. 5E and F) and viral replication (Fig. 5G). Accordingly, treatment of cells with AS-1411 results in a significant reduction of viral titers after dengue (Balinsky et al., 2013) and human respiratory syncytial (Mastrangelo et al., 2017) viruses infection. We observed that SARS-CoV-2 up-regulated NCL in the nucleoli of pneumocytes of hamsters (Fig. 6A). In addition, viral infection resulted in increased size and number of nucleoli (Fig. 6B). Accordingly, infection of cells with the coronavirus IBV has been shown to disrupt nucleolar architecture (Dove et al., 2006b), and cause arrest of the cell cycle (Dove et al., 2006a). Infection with SARS-CoV-2 virus resulted in multinucleated cells with prominent nucleoli (Draeger et al., 2022; Xia et al., 2022). RNA viruses interact with the nucleolus to usurp host-cell functions and recruit nucleolar proteins to facilitate viral replication (Hiscox, 2007; Rawlinson and Moseley, 2015). Although the principal site of replication of the majority of RNA viruses is the cytoplasm (Lam et al., 2020; Zhou et al., 2020), one of the most abundant viral proteins produced during infection, N-protein, can localize to the cytoplasm and nucleolus in infected cells (Wurm et al., 2001). We showed that SARS-CoV-2 induced translocation of NCL to cytoplasm in the lungs of infected hamsters (Fig. 6B). SARS-CoV-2 up-regulation of NCL correlated with hyperplasia of the pneumocytes (Sia et al., 2020) and the expression of spike protein (Fig. 6A), suggesting a causal relationship. SARS-CoV-2 infects predominantly alveolar epithelial cells and induces a localized hyperinflammatory cell state that is associated with reactive pneumocyte hyperplasia and diffuse alveolar damage (Bradley et al., 2020). SARS-CoV-2 has evolved strategies to co-opt host NCL to induce host cell apoptosis in order to favor replication. Targeting viral replication and apoptosis with the therapeutics targeting NCL could decrease viral burden and tissue damage of COVID-19 patients.
4. Materials and methods
Cells and viruses. All cells were obtained from the American Type Culture Collection. Normal lung epithelial cell lines HAEo and HBEC were obtained from Millipore Sigma. Cells were authenticated using short tandem repeat (STR) profiling and tested for mycoplasma using MycoAlert PLUS mycoplasma detection kit (Lonza). HCT-8 [HRT-18] and Vero C1008 [Vero 76, clone E6, Vero E6] cells were used for growing virus and determining virus stock titers. Vero E6/TMPRSS2 cells (JCRB #1819) were obtained from the Japanese Collection of Research Bioresources, distributed by Sekisui XenoTech (Matsuyama et al., 2020). Human SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281 was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). The virus stocks were stored at −80 °C and titers were determined by tissue culture infectious dose 50 (TCID50) assay. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and ABSL3 facilities at Johns Hopkins University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Drugs and constructs. Remdesivir was obtained from Cayman Chemical. The nucleolin aptamer AS-1411 (5′-GGTGGTGGTGGTTGTGGTGGTGGTGG) and CRO control (5′-CCTCCTCCTCCTTCTCCTCCTCCTCC) were purchased from Integrated DNA technologies. The following SARS-CoV-2 reagents were obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein Receptor Binding Domain (RBD), NR-52309; Nucleocapsid Protein RNA Binding Domain Gene NR-52429; Nucleocapsid Protein C-Terminal Domain, NR-52434; Non-Structural Protein 12, NR-53503. Expression plasmids for Nucleocapsid ORF (VG40588-UT) and Nsp1-2xStrep-IRES-Puro (141367) were obtained from Sino Biological and Addgene, respectively. The plasmids were transiently transfected into Vero E6 cells using Lipofectamine 2000 transfection reagent (ThermoFisher).
CRISPR gene silencing. Nucleolin guide RNAs were designed using sgRNA online web page from Broad Institute and cloned into Lenticrispr V2 (Addgene). 293T cells were transfected with the lentivirus constructs using Lipofectamine and virus were used to infect the cells (Merino et al., 2016). Cell pool generation were obtained following puromycin selection. High knockout efficiencies (80–90% knockout frequency) were generated in a heterogeneous population of cells, referred to as a knockout cell pool or knockdown.
Cytopathic effect (CPE) inhibition assay. We determined the CPE of SARS-CoV-2 virus at a multiplicity of infection (MOI) of 0.001 (Hoffmann et al., 2020) and 0.01 (Bojkova et al., 2020) infecting Vero E6 control and nucleolin-knockout (NCL-KO) cells (1 × 104 cells/well in 96 well plate) for 48h. The CPE inhibitory effect of remdesivir (5 μM) and CRO and AS-1411 aptamers (10 μM) was determined by treatment with the drugs for 24h prior to viral infection. The viral inoculums were removed after 1 h of incubation. Cell viability was assessed by measuring ATP levels using Promega's Cell Titer Glo reagent (#G7571). Cell viability is represented as percentage relative to uninfected cells. The viability of uninfected host cells after exposure to compounds for 72 h is measured in a counter screen to determine and exclude compounds with cytotoxic effects. Luminescence readouts were obtained in a FLUOstar Omega plate reader (BMG Labtech) (Gorshkov et al., 2021).
Viral titer determination. TRIzol reagent (ThermoFisher) was used to isolate RNA from supernatants of mock- and viral-infected cells according to the manufacturer's protocol. cDNA synthesis was performed using iScript cDNA Synthesis kit (BioRad) following the manufacture's protocol. Real-time quantitative reverse transcription PCR (RT-qPCR) was performed in triplicate using iTaq Universal SYBR Green supermix (BioRad) on a StepOne Plus Real Time PCR machine (Applied Biosystems). Primers against SARS-CoV-2 N- and E−protein were used with supernatant of NCL-knockout and treated cells, respectively (Supplementary Methods).
Hamsters and mice infection. Animal studies were performed according to the guidelines and approval of the Animal Care Committee of the Johns Hopkins School of Medicine. Heterozygous K18-hACE C57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. After induction of anesthesia with ketamine hydrochloride and xylazine, 5–6 weeks old male golden Syrian hamsters (Envigo) and 6–8 weeks old male mice were infected intranasally with 5.2 × 105 and 8.4 × 105 TCID50 of SARS-CoV-2/USA/WI1/2020 (NR-52384), respectively. Uninfected animals received intranasally the same volume of vehicle. Weights were monitored daily, the animals were sacrificed 7 days post-infection by isoflurane overdose, and the tissues were harvested following perfusion with PBS. Tissues were fixed on 10% neutral-buffered formalin.
Western blot, Immunohistochemistry and Immunofluorescence. Western blot, immunohistochemistry (IHC) (Merino et al., 2016) and immunofluorescence (IF) (Foss et al., 2013) were performed as previously described using antibodies against nucleolin (Cell Signaling Technology) and viral proteins (Supplementary Methods). For IF, the slides with transfected and viral-infected cells or formalin-fixed lung tissues were probed with the following primary antibodies: nucleolin (#14574, Cell Signaling Technology), spike (#GTX635692, GeneTex), other viral-specific antibodies (Supplementary Methods), and nuclear staining (Hoechst; Fisher Scientific). Slides were viewed using a LSM 880 Zeiss Confocal microscope. IHC on lung sections was performed using primary antibody rabbit anti-nucleolin and anti-spike-RBD (#GTX135356, GeneTex) (Supplementary Methods). ImageJ and Halo software were used for quantification of Western blot and IHC, respectively.
Flow cytometry and Apoptosis assays. Vero-TMPRSS NCL-knockout and AS-1411 treated cells infected with mock and SARS-CoV-2 (MOI 0.01) for 48h were stained at room temperature for 15 min with AnnexinV-Alexa Fluor 488 conjugate and propidium iodide (PI) in Annexin buffer (10 mm HEPES (pH 7.4), 150 mm NaCl, 2.5 mm CaCl2, pH 7.4). Another set of adherent cells were stained with MitoTracker Red CMXRos (#9082, Cell Signaling Technology) following manufacturer's instructions. Virus was inactivated by cell fixation in 1% formaldehyde. Samples were run on the BD FACSCalibur system (Becton Dickinson). FCS files were analyzed using Flowjo v10.6.2 software.
Statistical Analysis. Two-tailed nonparametric Mann Whitney Test and parametric student's t-test were performed on pairwise combinations of non- normally and normally distributed data, respectively, to determine statistical significance defined as P < 0.05. Data were analyzed using Prism 9.0.1 (GraphPad).
Author contributions
Conception and experimental design: Y.Y., V.M., A.O., and M.P. Performed the experiments: Y.Y., A.O., K.B., K.R., V.M. Acquisition of data: Y.Y., A.O., K.B., K.R., V.M. Analysis and interpretation of data: Y.Y., A.O., K.B., A.L., H.S., K.R., T.H., S.J., M.P., V.M. Writing and/or revision of the manuscript: Y.Y., A.O., K.B., K.R., T.H., S.J., M.P., V.M. Study supervision: A.O., S.J., M.P., V.M.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was funded by the Division of Nuclear Medicine and Molecular Imaging, Center for Infection and Inflammation Imaging, EB024495 and AI153349.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105550.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Aloni-Grinstein R., Charni-Natan M., Solomon H., Rotter V. p53 and the viral connection: back into the future (double dagger) Cancers. 2018;10 doi: 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y. Host cellular RNA helicases regulate SARS-CoV-2 infection. J. Virol. 2022;96 doi: 10.1128/jvi.00002-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Liu P., Leibowitz J.L., Kao C.C. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012;86:4294–4304. doi: 10.1128/JVI.07012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Munch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Huttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Felix E., Shi Y., Goff M., Lim J.K., McBride T., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., De Wit E., Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B.R., Mullins R.D., Fischer E.R., Kochs G., Grosse R., Garcia-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712 e619. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., Yarid N., Marshall D.A. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Chu H., Zhang A.J., Leung L.H., Sze K.H., Kao R.Y., Chik K.K., To K.K., Chan J.F., Chen H., Jin D.Y., Liu L., Yuen K.Y. Hemagglutinin of influenza A virus binds specifically to cell surface nucleolin and plays a role in virus internalization. Virology. 2016;494:78–88. doi: 10.1016/j.virol.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child J.R., Chen Q., Reid D.W., Jagannathan S., Nicchitta C.V. Recruitment of endoplasmic reticulum-targeted and cytosolic mRNAs into membrane-associated stress granules. RNA. 2021;27:1241–1256. doi: 10.1261/rna.078858.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccosanti F., Di Rienzo M., Romagnoli A., Colavita F., Refolo G., Castilletti C., Agrati C., Brai A., Manetti F., Botta L., Capobianchi M.R., Ippolito G., Piacentini M., Fimia G.M. Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection. Antivir. Res. 2021;190 doi: 10.1016/j.antiviral.2021.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V'Kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia A., Bokhari H. Apoptosis induced by SARS-CoV-2: can we target it? Apoptosis : an international journal on programmed cell death. 2021;26:7–8. doi: 10.1007/s10495-021-01656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove B., Brooks G., Bicknell K., Wurm T., Hiscox J.A. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 2006;80:4147–4156. doi: 10.1128/JVI.80.8.4147-4156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove B.K., You J.H., Reed M.L., Emmett S.R., Brooks G., Hiscox J.A. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell Microbiol. 2006;8:1147–1157. doi: 10.1111/j.1462-5822.2006.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draeger T.B., Tedesco S., Andaz S.K., Gibson V.R. Histologic evidence of tracheal stenosis directly resulting from SARS-CoV-2 tissue infiltration, a case series. J. Cardiothorac. Surg. 2022;17:128. doi: 10.1186/s13019-022-01839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Munday D., Bickerton E., Britton P., Rodgers M.A., Whitehouse A., Zhou E.M., Hiscox J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013;87:9486–9500. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Sanyal S., Bruzzone R. Breaking bad: how viruses subvert the cell cycle. Front. Cell. Infect. Microbiol. 2018;8:396. doi: 10.3389/fcimb.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss C.A., Harper J.S., Wang H., Pomper M.G., Jain S.K. Noninvasive molecular imaging of tuberculosis-associated inflammation with radioiodinated DPA-713. J. Infect. Dis. 2013;208:2067–2074. doi: 10.1093/infdis/jit331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C., Galluzzi L., Brunet M., Puig P.E., Didelot C., Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- Gomez G.N., Abrar F., Dodhia M.P., Gonzalez F.G., Nag A. SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2019;97:758–766. doi: 10.1139/bcb-2018-0394. [DOI] [PubMed] [Google Scholar]
- Gorshkov K., Chen C.Z., Bostwick R., Rasmussen L., Tran B.N., Cheng Y.S., Xu M., Pradhan M., Henderson M., Zhu W., Oh E., Susumu K., Wolak M., Shamim K., Huang W., Hu X., Shen M., Klumpp-Thomas C., Itkin Z., Shinn P., Carlos de la Torre J., Simeonov A., Michael S.G., Hall M.D., Lo D.C., Zheng W. The SARS-CoV-2 cytopathic effect is blocked by lysosome alkalizing small molecules. ACS Infect. Dis. 2021;7:1389–1408. doi: 10.1021/acsinfecdis.0c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstein E., Shan Y., Karawajew L., Snijders P.J., Meijer C.J., Royer H.D., Wernet P. Cell cycle-controlled interaction of nucleolin with the retinoblastoma protein and cancerous cell transformation. J. Biol. Chem. 2006;281:22223–22235. doi: 10.1074/jbc.M513335200. [DOI] [PubMed] [Google Scholar]
- Gussow A.B., Auslander N., Faure G., Wolf Y.I., Zhang F., Koonin E.V. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master regulator analysis of the SARS-CoV-2/human interactome. J. Clin. Med. 2020;9 doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S.T., Huang S., Heikal A.A., Webb W.W. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Yu H., Lu M., Yuan X., Wu X., Qiu H., Chen J., Huang S. TLR4 and nucleolin influence cell injury, apoptosis and inflammatory factor expression in respiratory syncytial virus-infected N2a neuronal cells. J. Cell. Biochem. 2019;120:16206–16218. doi: 10.1002/jcb.28902. [DOI] [PubMed] [Google Scholar]
- Jia W., Yao Z., Zhao J., Guan Q., Gao L. New perspectives of physiological and pathological functions of nucleolin (NCL) Life Sci. 2017;186:1–10. doi: 10.1016/j.lfs.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Kamada S., Kikkawa U., Tsujimoto Y., Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J. Biol. Chem. 2005;280:857–860. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan P.K.W., Cross G.B., Naftalin C.M., Ahidjo B.A., Mok C.K., Fanusi F., Permata Sari I., Chia S.C., Kumar S.K., Alagha R., Tham S.M., Archuleta S., Sessions O.M., Hibberd M.L., Paton N.I. A blood RNA transcriptome signature for COVID-19. BMC Med. Genom. 2021;14:155. doi: 10.1186/s12920-021-01006-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., Leung G.M., Holmes E.C., Hu Y.L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lei Y., Zhang J., Schiavon C.R., He M., Chen L., Shen H., Zhang Y., Yin Q., Cho Y., Andrade L., Shadel G.S., Hepokoski M., Lei T., Wang H., Zhang J., Yuan J.X., Malhotra A., Manor U., Wang S., Yuan Z.Y., Shyy J.Y. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ. Res. 2021;128:1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Garron T.M., Chang Q., Su Z., Zhou C., Qiu Y., Gong E.C., Zheng J., Yin Y.W., Ksiazek T., Brasel T., Jin Y., Boor P., Comer J.E., Gong B. Cell-type apoptosis in lung during SARS-CoV-2 infection. Pathogens. 2021;10 doi: 10.3390/pathogens10050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losfeld M.E., Khoury D.E., Mariot P., Carpentier M., Krust B., Briand J.P., Mazurier J., Hovanessian A.G., Legrand D. The cell surface expressed nucleolin is a glycoprotein that triggers calcium entry into mammalian cells. Exp. Cell Res. 2009;315:357–369. doi: 10.1016/j.yexcr.2008.10.039. [DOI] [PubMed] [Google Scholar]
- Marine J.C., Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Mastrangelo P., Norris M.J., Duan W., Barrett E.G., Moraes T.J., Hegele R.G. 2017. Targeting Host Cell Surface Nucleolin for RSV Therapy: Challenges and Opportunities. Vaccines 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration, U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino V.F., Nguyen N., Jin K., Sadik H., Cho S., Korangath P., Han L., Foster Y.M.N., Zhou X.C., Zhang Z., Connolly R.M., Stearns V., Ali S.Z., Adams C., Chen Q., Pan D., Huso D.L., Ordentlich P., Brodie A., Sukumar S. Combined treatment with epigenetic, differentiating, and chemotherapeutic agents cooperatively targets tumor-initiating cells in triple-negative breast cancer. Cancer Res. 2016;76:2013–2024. doi: 10.1158/0008-5472.CAN-15-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.L. Stress granules and virus replication. Future Virol. 2011;6:1329–1338. doi: 10.2217/fvl.11.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F., Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Therapeut. 2010;12:107–114. [PubMed] [Google Scholar]
- O'Brate A., Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist. Updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2003;6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Oksayan S., Nikolic J., David C.T., Blondel D., Jans D.A., Moseley G.W. Identification of a role for nucleolin in rabies virus infection. J. Virol. 2015;89:1939–1943. doi: 10.1128/JVI.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman M., Komanapalli J., Mukherjee M., Byram P.K., Sahoo S., Chakravorty N. Decrypting the role of predicted SARS-CoV-2 miRNAs in COVID-19 pathogenesis: a bioinformatics approach. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlinson S.M., Moseley G.W. The nucleolar interface of RNA viruses. Cell Microbiol. 2015;17:1108–1120. doi: 10.1111/cmi.12465. [DOI] [PubMed] [Google Scholar]
- Reuschl A.K., Thorne L., Zuliani Alvarez L., Bouhaddou M., Obernier K., Soucheray M., Turner J., Fabius J., Nguyen G.T., Swaney D., Rosales R., White K.M., Aviles P., Kirby I.T., Melnyk J.E., Shi Y., Zhang Z., Shokat K., Garcia-Sastre A., Jolly C., Towers G.J., Krogan N.J. Host-directed therapies against early-lineage SARS-CoV-2 retain efficacy against B.1.1.7 variant. bioRxiv : the preprint server for biology. 2021 [Google Scholar]
- Reyes-Reyes E.M., Teng Y., Bates P.J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N., Jr., Harzstark A.L., Wagle N., Figlin R.A., Smith G.W., Garraway L.A., Choueiri T., Erlandsson F., Laber D.A. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. N. Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A., Greco A. Viruses and the nucleolus: the fatal attraction. Biochim. Biophys. Acta. 2014;1842:840–847. doi: 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A., Rorie C.J., Dimitrova D., Daniely Y., Borowiec J.A. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- Sengupta T.K., Bandyopadhyay S., Fernandes D.J., Spicer E.K. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- Shang C., Liu Z., Zhu Y., Lu J., Ge C., Zhang C., Li N., Jin N., Li Y., Tian M., Li X. SARS-CoV-2 causes mitochondrial dysfunction and mitophagy impairment. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.780768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W. Regulation of cell death during infection by the severe acute respiratory syndrome coronavirus and other coronaviruses. Cell Microbiol. 2007;9:2552–2561. doi: 10.1111/j.1462-5822.2007.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Bonilla H., Jagannathan P., Shafer R.W. Clinical microbiology reviews; 2021. SARS-CoV-2 Antiviral Therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Sturzel C.M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani K.A., Liao Q., Ye L., Zeng Y., Liu J., Zheng Y., Ye L., Yang X., Lingbao K., Gao J., Zhu Y. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelli A., Monti M., Milanetti E., Armaos A., Rupert J., Zacco E., Bechara E., Delli Ponti R., Tartaglia G.G. Structural analysis of SARS-CoV-2 genome and predictions of the human interactome. Nucleic Acids Res. 2020;48:11270–11283. doi: 10.1093/nar/gkaa864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.E., Fazal F.M., Parker K.R., Zou J., Chang H.Y. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell systems. 2020;11:102–108 e103. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Hsu Lin L., Sun W., Moreira A.L., Simsir A., Brandler T.C. Effusion fluid cytology and COVID-19 infection. Cancer cytopathology. 2022;130:183–188. doi: 10.1002/cncy.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wu J., Shan Y., Yao Z., Dong B., Chen B., Zhao Z., Wang S., Chen J., Cong Y. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. 2006;346:74–85. doi: 10.1016/j.virol.2005.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang Z.P., Zhang X.E., Lin F.S., Ge F. Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication. J. Virol. 2010;84:6050–6059. doi: 10.1128/JVI.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jiang W., Liu Z., Liu S., Liang X. Virus infection and death receptor-mediated apoptosis. Viruses. 2017;9 doi: 10.3390/v9110316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.