Abstract
The suprachiasmatic nuclei (SCN) of the anterior hypothalamus host the circadian pacemaker that synchronizes mammalian rhythms with the day–night cycle. SCN neurons are intrinsically rhythmic, thanks to a conserved cell-autonomous clock mechanism. In addition, circuit-level emergent properties confer a unique degree of precision and robustness to SCN neuronal rhythmicity. However, the multicellular functional organization of the SCN is not yet fully understood. Indeed, although SCN neurons are well-coordinated, experimental evidences indicate that some neurons oscillate out of phase in SCN explants, and possibly to a larger extent in vivo. Here, to tackle this issue we used microendoscopic Ca2+i imaging and investigated SCN rhythmicity at a single cell resolution in free-behaving mice. We found that SCN neurons in vivo exhibited fast Ca2+i spikes superimposed upon slow changes in baseline Ca2+i levels. Both spikes and baseline followed a time-of-day modulation in many neurons, but independently from each other. Daily rhythms in basal Ca2+i were highly coordinated, while spike activity from the same neurons peaked at multiple times of the light cycle, and unveiled clock-independent coactivity in neuron subsets. Hence, fast Ca2+i spikes and slow changes in baseline Ca2+i levels highlighted how multiple individual activity patterns could articulate within the temporal unity of the SCN cell network in vivo, and provided support for a multiplex neuronal code in the circadian pacemaker.
Keywords: circadian rhythms, in vivo imaging, calcium signaling
Significance Statement.
Living organisms have evolved exquisite mechanisms to anticipate and cope with changes imposed by the day–night cycle in their environment. The molecular machinery that sets the physiology of individual cells on a 24-hour period has now been deciphered. However, it is still unclear how multiple and diverse clock cells tick all together in the suprachiasmatic nuclei (SCN) of the hypothalamus, where the mammalian circadian pacemaker resides. In this study, we performed deep-brain calcium imaging to investigate daily rhythmicity at a single-cell resolution in free-behaving mice. We found a combination of two independent calcium dynamics in individual neurons of the SCN, which provides support for encoding both temporal unity and functional diversity in this composite neural system.
Introduction
Circadian clocks rely on a conserved molecular mechanism expressed in virtually all body cells (1). In the suprachiasmatic nuclei (SCN) of the anterior hypothalamus, circuit-level interactions confer a unique degree of precision and robustness to circadian neuronal rhythms, which turns cell-autonomous oscillators into the central pacemaker that synchronizes mammalian physiology and behaviors with the day–night cycle (2–6). Deciphering the multicellular functioning of the SCN will, thus constitute a milestone on the way from clock genes to complex, overt rhythms.
Despite a high degree of coordination, SCN neurons are not always perfectly in phase. Stereotyped circadian waves in clock gene expression, intracellular calcium concentration (Ca2+i), and spontaneous firing rate of action potentials outline an approximate 6-hour phase gradient throughout SCN slices (3, 7–10). However, recent evidences challenged the rhythmic unity in the SCN, with some neurons exhibiting extreme phase lag. Phaseoids were defined as groups of neurons expressing the circadian clock protein PERIOD2 stably out of phase with their neighbors (11). Similarly, some SCN neurons appear electrophysiologically more active during nighttime, in total opposition of phase relative to the ensemble rhythm (12, 13). Last but not least, the circadian amplitude in multiunit electrophysiological activity is dramatically downsized in vivo as compared to in vitro preparations (14), suggesting even reduced neuronal synchronicity in living animals. How such a diversity in neuronal activity in vivo articulates within a coherent SCN network remains unclear.
Typically, fast Ca2+i spikes driven by action potentials provide a fairly reliable estimate of neuronal activity. To date, variations in Ca2+i have been extensively described at the circadian timescale in individual SCN neurons ex vivo (7, 8, 15–17) and at a neuronal population level in vivo (18, 19), but the spatiotemporal organization of SCN Ca2+i spikes has been barely investigated, possibly because of technical issues with BAPTA-based Ca2+i probes (20, 21). Here, we performed microendoscopic imaging of the Ca2+i indicator GCamp6f at a single-cell resolution through a graded-index (GRIN) lens aimed at the SCN of freely behaving mice. Fast Ca2+i spikes revealed unexpected diversity in daily neuronal activity patterns, and circuit-level signaling all around the light–dark cycle. Yet, we found that this diversity builds upon a highly coordinated wave in baseline Ca2+i levels in the same cells. Together, Ca2+i spikes and baseline Ca2+i levels were independently modulated with the time of day in vivo, and reconciled the apparent discrepancy between functional cell diversity and network unity in the SCN.
Results
After surgical preparation, mice were trained to the imaging setup for at least 3 weeks. They exhibited low levels of circulating corticosterone during their resting phase, which increased at the beginning of their active phase (Figure S1, Supplementary Material). Moreover, longitudinal monitoring of the global GCamp6f signal from the SCN (n = 12 mice) revealed daily changes, with peak and nadir in the middle of the light and dark phase (Fig. 1a and b), respectively, resembling the typical Ca2+i rhythm recorded at the neuron population level using in vivo fiber photometry (18, 19). Hence, GRIN lens-implanted mice were free of chronic stress and maintained normal daily physiology under our experimental conditions.
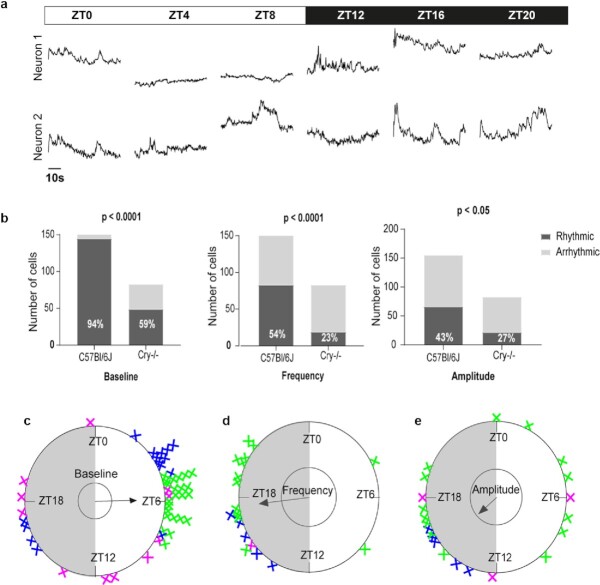
Fig. 1.
Daily diversity and unity of two Ca2+i dynamics in individual SCN neurons in vivo. (a) Time-lapse projection of GCamp6f fluorescence monitored every 2 hours from zeitgeber time ZT = 0 to ZT = 22, through a GRIN lens aimed at the SCN of a freely moving mouse. Scale bar, 100 µm. (b) Daily variations in global GCamp6f signal intensity from one representative SCN field of view. The solid line represents the best cosine fit of experimental data points, measured every 30 minutes over four consecutive day–night cycles. (c) Longitudinal 1-minute recordings of GCamp6f fluorescence over 1 day–night cycle, from two SCN neurons within the same field of view. (d)–(i) Daily variation in GCamp6f baseline (d)–(f) and spike frequency (g)–(i), in SCN neurons. Example from one representative neuron (d) and (g), heat maps obtained from 52 neurons recorded in the same SCN field of view (e) and (h), and Rayleigh plots of significant individual daily peak phases measured from four different mice (f) and (i). Each color represents neurons from one mouse, examples were selected from the mouse color-coded in green. The inner circle represents the statistical threshold (P < 0.05) for the mean vector of the circular distribution of the aggregate data. The white and black boxes depict the light and dark phase, respectively. (j) Phase–lag relationship between spike frequency and the GCamp6f baseline.
At the individual cell level, two dynamics in GCamp6f intensity emerged from SCN neurons recorded in vivo, consisting in short-lived spikes (Movie 1) superimposed upon slow-evolving changes in basal fluorescence (Fig. 1c). To assess how these slow and fast dynamics evolved with the time of day, we monitored GCamp6f fluctuations during 1-minute episodes, every 30 minutes. We were able to follow 155 individual SCN neurons longitudinally over at least 24 hours, from four different mice. The baseline GCamp6f level displayed a significant daily pattern in up to 145 neurons (70% to 100%), as assessed by nonparametric JTK-Cycle analysis (22). In every rhythmic neuron, the baseline level reached its maximum during daytime (Fig. 1d–f; Figure S2, Supplementary Material), around zeitgeber time ZT = 6 (with ZT = 0 at lights on, 95% CI [ZT = 03:28, ZT = 07:11], P < 0.001 Moore’s modified Rayleigh test). The interindividual variability between all four mouse SCN (P < 10–7, F(3, 141), Watson–Williams F-test) suggested subtle topographical heterogeneity that was reminiscent of the phase gradient in circadian Ca2+i rhythm observed in SCN slices (7, 8). Hence, the daily variations in baseline Ca2+i levels underscored the unity of individual neuronal oscillators in the SCN in vivo.
By contrast, fast GCamp6f spikes from the same neurons could occur at all times of day, and revealed the diversity in daily Ca2+i activity patterns in the SCN. A total of 38% to 69% of the recorded neurons exhibited a significant daily organization in spike frequency (Fig. 1g–i; Figure S3, Supplementary Material). In two out of four mice, most neurons had their peak activity phase at the light–dark transition or in the early night. In the other two mice, the peak phase was more widely distributed. This resulted in a globally large phase dispersion of fast spikes (P > 0.1 Moore’s modified Rayleigh test, Fig. 1i), and a stratified phase–lag relationship with the daily rhythm in baseline levels (Fig. 1j), which indicated that fast Ca2+i spike activity and basal Ca2+i levels in SCN neurons were independently modulated with the time of day.
To gain insight into the contribution of spikes to the daily Ca2+i rhythm in SCN neurons, relatively to baseline changes, we measured the amplitude of changes in GCamp6f fluorescence in each 1-minute recording (Fig. 2a). This parameter followed a daily organization in about half (33% to 52%) of the recorded neurons (Fig. 2b–d; Figure S4, Supplementary Material), roughly phase-locked to spike frequency (Spearman r = 0.94, P < 0.0001; Fig. 2e; Figure S5, Supplementary Material). Spiking activity was generally one-to-two orders of magnitude smaller than the variation in basal GCamp6f fluorescence measured over 24 hours in the same neuron (Fig. 2f). Therefore, spikes contributed moderately to the daily rhythmicity in fluorescence, regardless of whether they occurred at high or low baseline levels (Fig. 2g and h). Together, our data indicated that slow changes in basal Ca2+i levels and fast Ca2+i spikes were nested dynamics that respectively supported the daily rhythm unity and functional diversity in the SCN cell network in vivo.
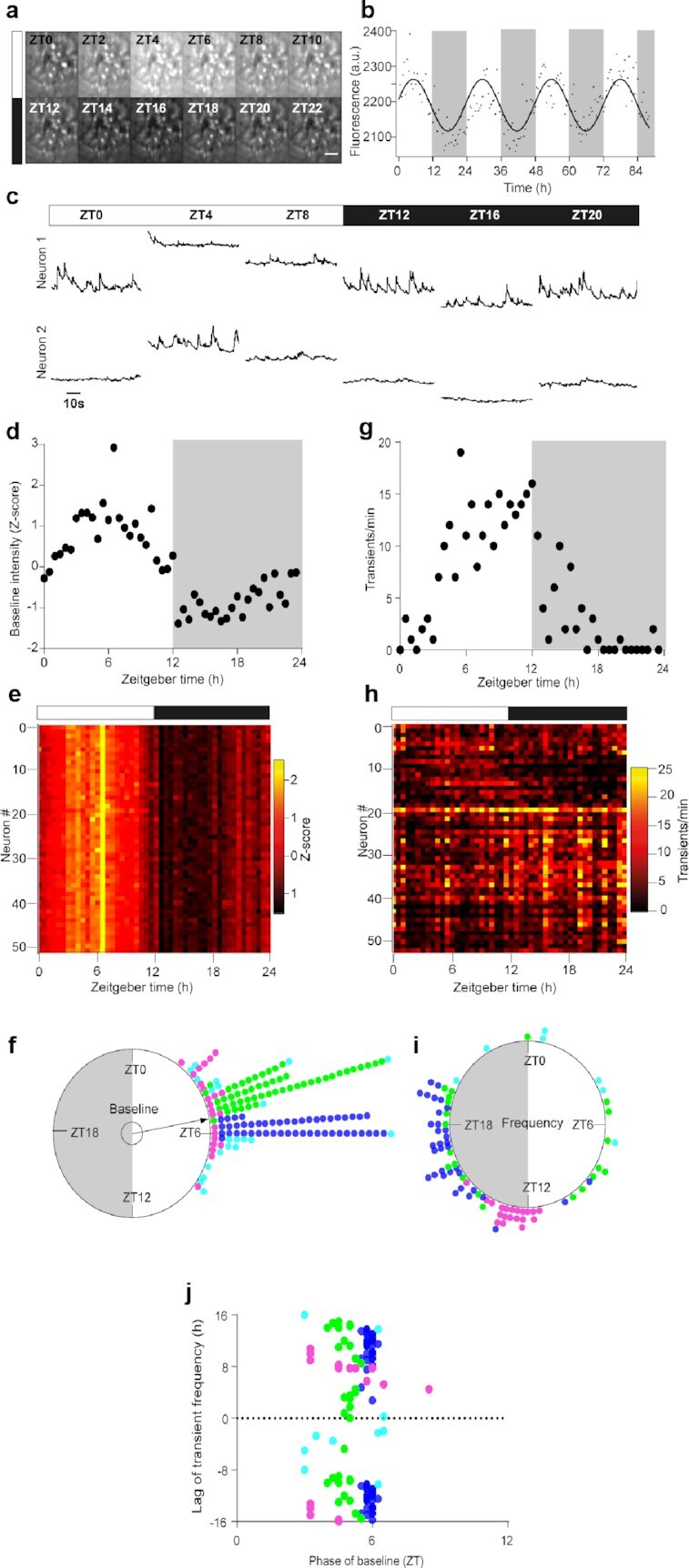
Fig. 2.
Small Ca2+i spikes built upon large daily changes in basal Ca2+i levels. (a) 1-minute recording of GCamp6f fluorescence, with thresholds for the 5th and 95th percentiles (baseline and topline, black and red triangles, respectively) used to calculate the signal amplitude. (b)–(d) Daily variations in GCamp6f signal amplitude in the same neuron (b) and same SCN field (c) as in Fig. 1. (d) Rayleigh plot of significant individual daily peak phases in signal amplitude for the aggregate data. (e) Phase relationship between spike frequency and signal amplitude. (f) Distribution of the ratio values calculated, for each 1-minute recording, between the daily amplitude in baseline GCamp6f (ΔF24h) and the signal amplitude over 1 minute (ΔF1min), for each mouse SCN (colored lines) and the aggregate data (black bars). (g) and (h) Daily profiles in GCamp6f baseline and topline (as depicted in a) from two representative SCN neurons, with larger signal amplitude during the day (g) or night (h). The solid lines represent the best cosine wave for each dataset.
How much is the circadian clock responsible for the daily organization of each of these Ca2+i dynamics? To address this issue, we conducted an independent set of GCamp6f measurements in the SCN of globally clockless Cry1-/- Cry2-/- mutant mice (23). Longitudinal recordings of 83 individual neurons from three mice revealed conserved fast spikes and slow changes in GCamp6f baseline (Fig. 3a), but with a disturbed daily distribution. Cry1-/- Cry2-/- mice are known to maintain a marked alternation of diurnal rest and nocturnal locomotion under normal light–dark conditions such as those used in our study (23, 24). However, the proportion of neurons exhibiting a significant daily organization in GCamp6f signals was reduced by about half in absence of a functional circadian clock as compared to control mice, for both the basal level and the frequency or amplitude of fast spikes (Fig. 3b–e; Figure S2–S4, Supplementary Material). Moreover, although a majority of Cry1-/- Cry2-/- SCN neurons still reached their maximal baseline level during daytime, the overall peak phase distribution was wider than for control mice, with a number of SCN neurons peaking at night in two out of three mice (P > 0.1 Moore’s modified Rayleigh test, Fig. 3e; Figure S2, Supplementary Material). These results indicated that the daily organization of both slow basal Ca2+i changes and fast Ca2+i spikes depended on a functional circadian clock.
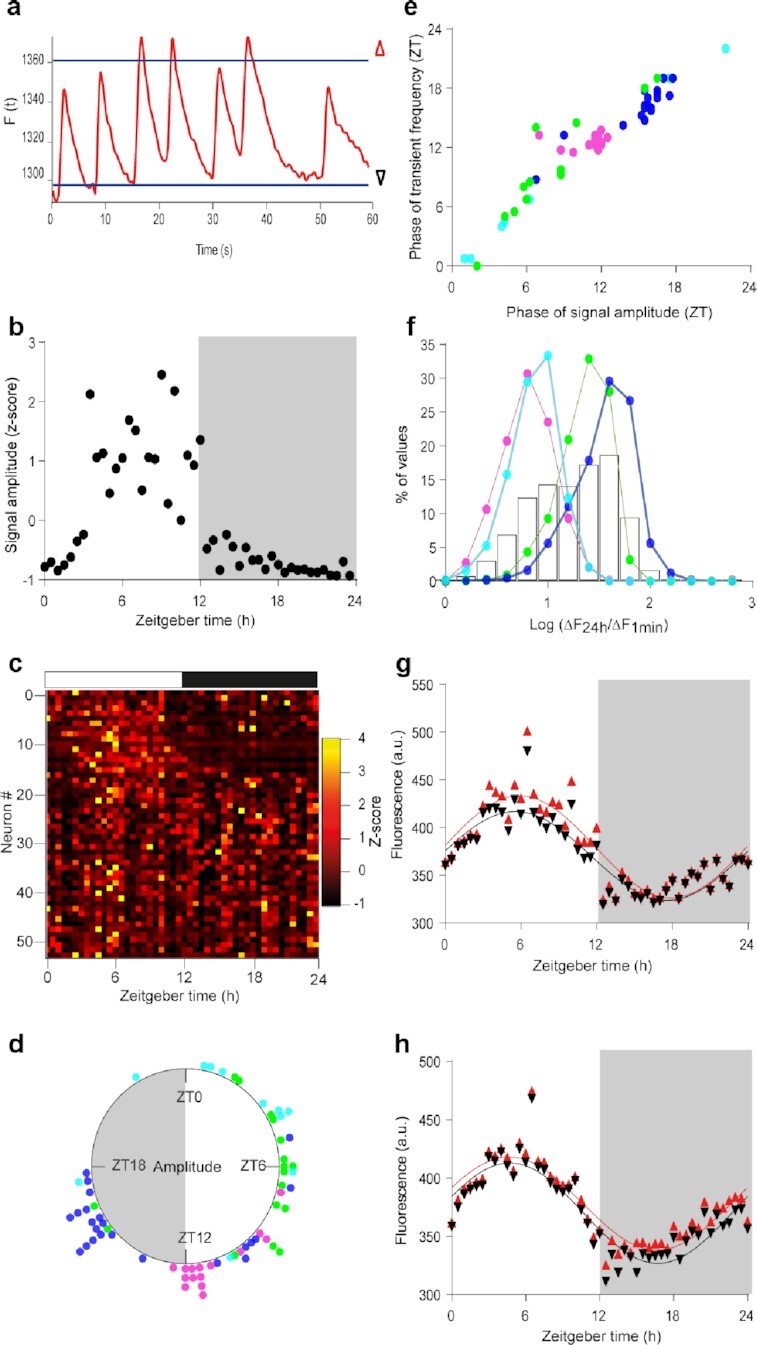
Fig. 3.
Daily variations in slow and fast Ca2+i dynamics in Cry1-/- Cry2-/- mice. (a) Longitudinal 1-minute recordings of GCamp6f fluorescence over 1 day–night cycle, from two SCN neurons of one Cry1-/- Cry2-/- mouse. (b) Quantitation of SCN neurons exhibiting a significant daily rhythm in GCamp6f baseline level (left), spike frequency (middle), and signal amplitude (right), in C57Bl/6 J and Cry1-/- Cry2-/- mice. The P-values indicate a significant genotype-dependence in the proportion of rhythmic neurons for each parameter (Fisher's exact contingency test). (c)–(e) Rayleigh plots of significant individual daily peak phases in baseline level (c), spike frequency (d), and signal amplitude (e) measured from three Cry1-/- Cry2-/- mice. Each color represents neurons from one mouse, examples in (a) were selected from the mouse color-coded in blue.
Next, we explored the spatiotemporal organization of GCamp6f spikes at the multicellular scale to question the circuit-level significance of the fast Ca2+i activity in the SCN. We found that fast GCamp6f spikes could occur within the same time frame in clusters of SCN neurons that partially overlapped each other and involved enough neurons to generate noticeable changes in the global field signal (Fig. 4a and b; Movie 1). The extent of coactivity clusters in the field of view, as expressed in cell.clusters/cell/min, correlated positively to the density of neuronal activity (Spearman r = 0.71, P < 0.0001, Fig. 4c). Since we computed coactivity only when detected above chance threshold (see methods), this correlation between coactivity and activity density could not result solely from random coincidence of Ca2+i spikes in highly active neurons. More likely, the clusters of coactivity depicted circuit-level Ca2+i signaling in the SCN (7).
Fig. 4.
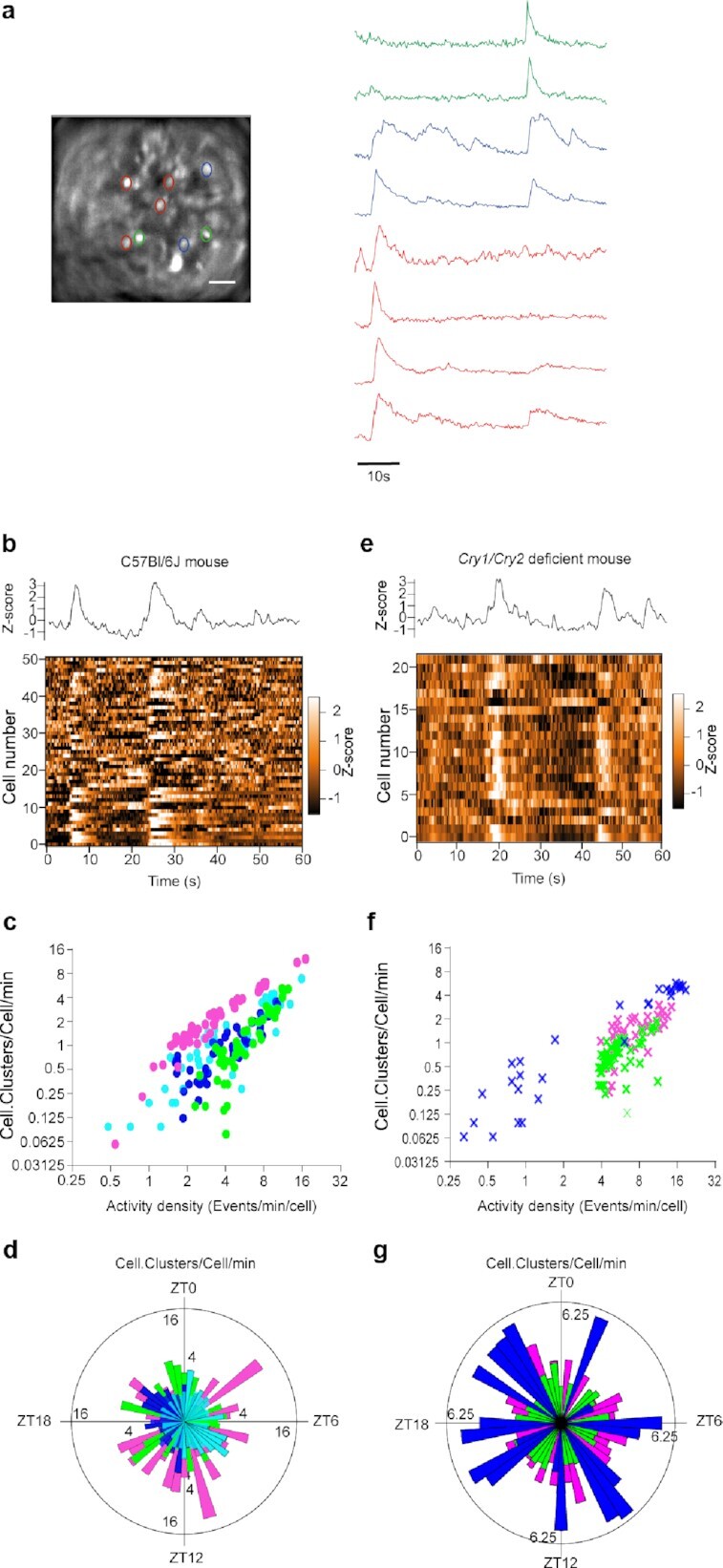
Synchronous fast Ca2+i spikes depicted clock-independent cell–cell signaling in the SCN. (a) 1-minute recordings of GCamp6f fluorescence from SCN neurons in a same field of view. Note the occurrence of synchronous spikes in neuron subsets (delineated by three different colors). Scale bar, 100 µm. (b)–(g) Analysis of clusters of coactivity from the SCN of C57Bl/6 J (b)–(d) and Cry1-/- Cry2-/- (e)–(g) mice. (b) and (e) Global GCamp6f fluorescence (upper panel) and heat map of signal intensity from individual neurons (lower panel), recorded during 1 minute. (c), (d), (f), and (g) Extent of clusters of coactivity as a function of the density in activity (c) and (f) and time of day (d) and (g).
Accordingly, the extent of coactivity was maximum in each mouse when most neurons reached their peak in activity (Figures S3 and S6, Supplementary Material). If one considered data from all four mice globally, coactivity spread evenly around the clock (P > 0.5, Moore’s modified Rayleigh test, Fig. 4d), suggesting that synchronizing cues circulated across the SCN cell network in vivo at all times of the light–dark cycle. Moreover, coactivity persisted under the conditions of an altered circadian clock, both during jetlag, when the global GCamp6f rhythm was progressively resetting to a new light phase (n = 3 mice, Figure S7, Supplementary Material), and in Cry1-/- Cry2-/- mice, in a similar relationship with activity density as in control mice (Spearman r = 0.88, P < 0.0001, Fig. 4e–g). Together, these data revealed extensive cell–cell coordination independent from the circadian clock in the SCN of free-behaving mice.
Discussion
In this study, we used in vivo Ca2+i imaging to investigate spatiotemporal Ca2+i signaling at a single-cell resolution in the SCN of free-behaving mice. We found that daily Ca2+i rhythmicity in SCN neurons is composed of fast Ca2+i spikes superimposed upon slow variations in baseline Ca2+i levels. Similarly, such slow and fast Ca2+i dynamics were recently reported in circadian pacemaker neurons of Drosophila (25). Altogether, these results reveal conserved properties between Invertebrates and Vertebrates in the cell biology of daily Ca2+i rhythms in circadian timekeeping systems.
In mouse SCN neurons as well as in Drosophila clock neurons, fast Ca2+i spikes underscored a wide array in daily activity patterns. This diversity was obvious between the SCN from different mice, but also between neurons from the same individual, suggesting a topographical distribution of activity patterns, like in Drosophila. Overall, a majority of SCN neurons were more active at night. As Ca2+i spikes are typically associated with electrophysiological activity (21, 25), this observation may first appear surprising in view of the many evidences of daytime-biased firing of action potentials in the SCN (3–5, 14, 26). Yet, it is actually in line with the high firing rate measured during nighttime from the SCN of live animals as compared to slice preparations (14), and suggests a widespread occurrence in vivo of the night-active neuron populations reported in SCN slices (12, 13).
Unlike in Drosophila clock neurons (25), SCN rhythms in fast Ca2+i activity and basal Ca2+i levels are not cophasic. Daily variations in basal Ca2+i levels reach their peak phase around midday in all the recorded SCN neurons. This homogeneity contrasts with the diversity in fast Ca2+i activity patterns, and depicts the global unity of individual neuron rhythmicity within the SCN cell network assumed to confer robustness and precision to the mammalian circadian pacemaker (4–6).
Since slow and fast Ca2+i dynamics are independently modulated with the time of day in the mouse SCN, they provide an additional level of combination as compared to Drosophila clock neurons. The occurrence of small and fast Ca2+i spikes at various time points upon the large and slow change in baseline extents the repertoire of Ca2+i signaling in the SCN, from the typical binary alternation of up and down states, toward the possibility of an activity-dependent multiplex code. It is then tempting to speculate that neurons exhibiting fast Ca2+i spikes during e.g. their down Ca2+i state may contribute in different functions than neurons active during their up state. Conversely, neurons using one same Ca2+i code might constitute functional units, gating specific body rhythms (13, 27–30). The composite tuning of Ca2+i-dependent downstream effectors identified in SCN neurons, such as the multisite phosphorylation of CREB (31, 32), affords a possible molecular mechanism to decipher this Ca2+i code.
One limitation of our study is that we could not locate precisely the recorded cells, nor establish whether the diversity in activity patterns corresponds to the diversity in peptidergic content among SCN neurons (33–37). Topographical differences in the timing and origin of Ca2+i signaling have been reported in SCN slices (7, 8, 38). As we implanted GRIN lenses dorsally to the SCN in order not to lesion circadian neurons, we assume that we performed recordings in the shell SCN, enriched in vasopressin-expressing neurons. The slow entrainment of the GCamp6f rhythm during simulated jetlag (Figure S7b, Supplementary Material) is in line with this assumption as retino-recipient VIP-containing neurons are expected to phase-shift rapidly (19). However, the AAV-driven expression of GCamp6f under a nonspecific promoter does not allow the assignment of activity patterns to identified neuronal subtypes. Further experiments targeting either SCN neuronal population (39) will be needed to fill this gap.
At the multicellular level, fast Ca2+i spikes revealed evidence of circuit-level communication. Synchronous events occurred at all times of day and persisted under circadian clock disruption, in line with clock-independent network properties in the SCN (10, 40). Hence, fast Ca2+i spikes are both hints of individual neuron activity, and mediate intercellular signaling, indicating that the mammalian circadian pacemaker ticks also at the timescale of seconds. As individuating part-whole relations in neural circuits has become an important challenge (41, 42), our study illustrates how the combination of slow and fast Ca2+i dynamics in the SCN of free-behaving mice supports the diversity in individual neuron activity patterns within the temporal unity of the global network.
Materials and methods
Animals
All mouse experiments complied with the European Directive 2010/63/UE, and were registered under the reference APAFIS#15,032–2018050918181227 v2. Male C57Bl/6 J mice (5 to 6 weeks old) were purchased from Janvier Labs (Le Genest-Saint-Isle, France). Cry1-/- Cry2-/- mice (23) were produced by breeding double-heterozygous males and females from our colony maintained in a C57Bl6/J background for more than 20 generations (43). All mice were housed in ventilated microisolator cages, under a 12 h:12 h light:dark cycle, with free access to food and water. For simulated jetlag experiments, the light–dark cycle was advanced by 6 hours, starting with the shift of light onset.
Surgery
At the age of 8 weeks, mice were anesthetized with a cocktail of Ketamine (Imalgene 500, 75 mg/kg) and Xylazine (Rompun, 10 mg/kg). A skin incision exposed the skull, and small craniotomies were made dorsal to the injection site with a drill mounted on a stereotaxic apparatus. The virus solution (1,000 nl, titer ≥ 1,013 vg/ml, #100836-AAV9 from Addgene) was injected in the SCN (ML +/−0.2, AP 0, and DV −5.7) at a rate of 50 nl/minute using a microinjector-controlled syringe (micro-4, World Precision Instruments) and needle (Nanofil 33 G beveled needle, World Precision Instruments). The needle was kept in place for 10 minutes following injection to allow suitable diffusion, and reduce backflow during withdraw. Mice were then implanted with a GRIN lens (diameter 0.6 mm, length 7.44 mm, and working distance 150 µm, #AB000436 from GRINTECH), aimed 50 to 100 µm dorsally to the virus injection site. The lens was stabilized with dental cement (Metabond) and Kwik-Silsealant (World Precision Instruments). After 3 to 4 weeks, mice were anesthetized for placement of the microendoscope baseplate (#1050–004638, Inscopix), and a baseplate cover (#1050–004639, Inscopix) was used to protect the lens when not in use.
After surgery, mice were housed in individual cages, and regularly habituated to the imaging setup, with a dummy microendoscope (DMS-2, Inscopix) for at least 3 weeks. To ascertain well-being and normal daily physiology under these conditions, tail-tip blood (6 µl) was collected from four mice at ZT7 and ZT12 to check corticosterone levels (ELISA kit From Assaypro).
Microendoscopic data acquisition and processing
Images were acquired with a head-mounted miniaturized microscope (nVista, Inscopix) at 4 frames per second over 1 minute (240 ms exposure time, 10% to 20% LED illumination, 1.5 to 2.5x gain). The 1-minute movies were spatially filtered (high-pass filter, cutoff at 40 µm), corrected for motion artifacts (Inscopix Data Processing Software), and saved as stacks of frames in TIFF format. Regions of interest (ROI) were defined as nonoverlapping bright regions that displayed fast activity in any stack. They were applied to every stack (ImageJ), and numerical data were saved as text files for further processing in Matlab (MathWorks).
Data analysis and statistics
A Butterworth low-pass filter was applied (cutoff frequency at 1.1 Hz). After detrending, the 5th and 95th values were computed for each 1-minute recording. The 5th percentile value was considered as the baseline level, and the difference between the 5th and 95th values defined the signal amplitude. Fast spikes were detected as fast and large events with a two-threshold procedure. Respectively, threshold 1 selected the fastest events from all recordings (90th percentile of all derivative values from all recordings, from all ROIs), and threshold 2 eliminated the smallest of these events for each recorded neuron (set at 2 SDs of all the raw signals over the course of a 24-hour period for each ROI). For each recording, the spike frequency was calculated as the number of spikes detected during 1 minute. The activity density was computed as the spike frequency divided by the number of recorded cells in the field of view.
The onset of spikes (derivative crossing above 0) was computed as their time of occurrence. Ca2+i spikes occurring in a high-activity frame in a statistically significant number of neurons delineated the clusters of coactivity. The threshold corresponding to a significance level of P < 0.05 was defined as the number of activated cells in a single frame that exceeded only 5% of 500 surrogate datasets obtained by randomly transposing intervals of activity within each cell (44). The extent of clusters of coactivity was computed as the sum of all the cells participating in any of the coactive events during a 1-minute recording, normalized by the total number of cells recorded in the field (expressed in Cell.Clusters/Cell/min).
Nonparametric JTK-Cycle analysis (22) was used to estimate the daily phase of the GCamp6f signal parameters from datasets recorded over one 24-hour period. Circular analysis and statistics were conducted using Oriana (Kovach Computing Services). Other numerical and statistical analyses were conducted with IgorPro (WaveMetrics, Inc), Prism 7 (GraphPad Software, Inc).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ombeline Hoa and Pauline Campos for advices with GRIN lens implantation, and the iExplore and IPAM platforms in Montpellier for technical support. We are grateful to Robert J. Lucas and Marco Brancaccio for reading an earlier draft of the manuscript.
Notes
Competing Interest: The authors declare no competing interest.
Contributor Information
Lama El Cheikh Hussein, Institut de Génomique Fonctionnelle, Université de Montpellier, CNRS, INSERM, IGF, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France; BioCampus Montpellier, Université de Montpellier, CNRS, INSERM, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France.
Pierre Fontanaud, Institut de Génomique Fonctionnelle, Université de Montpellier, CNRS, INSERM, IGF, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France; BioCampus Montpellier, Université de Montpellier, CNRS, INSERM, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France.
Patrice Mollard, Institut de Génomique Fonctionnelle, Université de Montpellier, CNRS, INSERM, IGF, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France; BioCampus Montpellier, Université de Montpellier, CNRS, INSERM, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France.
Xavier Bonnefont, Institut de Génomique Fonctionnelle, Université de Montpellier, CNRS, INSERM, IGF, 141 Rue de la Cardonille, F-34094 Montpellier, Cedex 5, France.
Funding
This work was supported by the Centre National de la Recherche Scientifique (P.M., P.F., and X.B.), the Agence Nationale pour la Recherche (ANR) grant nos. ANR-15-CE14-0012 and ANR-18-CE14-0017-01, and the France BioImaging ANR-10-INSB-04 “Investments for the Future” to P.M., the Fédération pour la Recherche sur le Cerveau (X.B.), and a fellowship from the Secours Populaire Libanais to L.E.C.H.
Authors’ contributions
L.E.C.H. conducted the experiments, analyzed the data, and edited the manuscript. P.F. contributed analytical tools and edited the manuscript. P.M. obtained funding, provided intellectual contribution, and edited the manuscript. X.B. designed the experiments, analyzed and interpreted the data, wrote the manuscript, and obtained funding.
Data availability
All data is included in the manuscript and/or supporting information.
References
- 1. Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 18(3):164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El Cheikh Hussein L, Mollard P, Bonnefont X. 2019. Molecular and cellular networks in the suprachiasmatic nuclei. Int J Mol Sci. 20(8):2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hastings MH, Maywood ES, Brancaccio M. 2018. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci. 19(8):453–469. [DOI] [PubMed] [Google Scholar]
- 4. Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. 2004. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms. 19(1):35–46. [DOI] [PubMed] [Google Scholar]
- 5. Honma S, Nakamura W, Shirakawa T, Honma K. 2004. Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci Lett. 358(3):173–176. [DOI] [PubMed] [Google Scholar]
- 6. Liu AC, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 129(3):605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. 2013. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 78(4):714–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Enoki R, et al. 2017. Synchronous circadian voltage rhythms with asynchronous calcium rhythms in the suprachiasmatic nucleus. Proc Natl Acad Sci. 114(12):E2476–E2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. 2013. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 80(4):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pauls S, et al. 2014. Differential contributions of intra-cellular and inter-cellular mechanisms to the spatial and temporal architecture of the suprachiasmatic nucleus circadian circuitry in wild-type, cryptochrome-null and vasoactive intestinal peptide receptor 2-null mutant mice. Eur J Neurosci. 40(3):2528–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshikawa T, et al. 2021. Phase gradients and anisotropy of the suprachiasmatic network: discovery of phaseoids. eNeuro. 8(5):ENEURO.0078–ENEU21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belle MD, Diekman CO, Forger DB, Piggins HD. 2009. Daily electrical silencing in the mammalian circadian clock. Science. 326(5950):281–284. [DOI] [PubMed] [Google Scholar]
- 13. Collins B, et al. 2020. Circadian VIPergic neurons of the suprachiasmatic nuclei sculpt the sleep-wake cycle. Neuron. 108(3):486–499.e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meijer JH, Schaap J, Watanabe K, Albus H. 1997. Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Res. 753(2):322–327. [DOI] [PubMed] [Google Scholar]
- 15. Colwell CS. 2000. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 12(2):571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda M, et al. 2003. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 38(2):253–263. [DOI] [PubMed] [Google Scholar]
- 17. Noguchi T, et al. 2017. Calcium circadian rhythmicity in the suprachiasmatic nucleus: cell autonomy and network modulation. eNeuro. 4(4):ENEURO.0160–ENEU17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones JR, Simon T, Lones L, Herzog ED. 2018. SCN VIP neurons are essential for normal light-mediated resetting of the circadian system. J Neurosci. 38(37):7986–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mei L, et al. 2018. Long-term in vivo recording of circadian rhythms in brains of freely moving mice. Proc Natl Acad Sci USA. 115(16):4276–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong JH, et al. 2010. Intracellular calcium spikes in rat suprachiasmatic nucleus neurons induced by BAPTA-based calcium dyes. PLoS ONE. 5(3):e9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irwin RP, Allen CN. 2007. Calcium response to retinohypothalamic tract synaptic transmission in suprachiasmatic nucleus neurons. J Neurosci. 27(43):11748–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 25(5):372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Horst GT, et al. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 398(6728):627–630. [DOI] [PubMed] [Google Scholar]
- 24. Mrosovsky N. 2001. Further characterization of the phenotype of mCry1/mCry2-deficient mice. Chronobiol Int. 18(4):613–625. [DOI] [PubMed] [Google Scholar]
- 25. Liang X, Holy TE, Taghert PH. 2022. Circadian pacemaker neurons display cophasic rhythms in basal calcium level and in fast calcium fluctuations. Proc Natl Acad Sci USA. 119(17):e2109969119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Albus H, et al. 2002. Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr Biol. 12(13):1130–1133. [DOI] [PubMed] [Google Scholar]
- 27. Gizowski C, Zaelzer C, Bourque CW. 2016. Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature. 537(7622):685–688. [DOI] [PubMed] [Google Scholar]
- 28. Jones JR, Chaturvedi S, Granados-Fuentes D, Herzog ED. 2021. Circadian neurons in the paraventricular nucleus entrain and sustain daily rhythms in glucocorticoids. Nat Commun. 12(1):5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Todd WD, et al. 2018. A hypothalamic circuit for the circadian control of aggression. Nat Neurosci. 21(5):717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yulyaningsih E, et al. 2014. Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell Metab. 19(1):58–72. [DOI] [PubMed] [Google Scholar]
- 31. Gau D, et al. 2002. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 34(2):245–253. [DOI] [PubMed] [Google Scholar]
- 32. Deisseroth K, Tsien RW. 2002. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron. 34(2):179–182. [DOI] [PubMed] [Google Scholar]
- 33. Abrahamson EE, Moore RY. 2001. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916(1–2):172–191. [DOI] [PubMed] [Google Scholar]
- 34. Lee IT, et al. 2015. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron. 85(5):1086–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morris EL, et al. 2021. Single-cell transcriptomics of suprachiasmatic nuclei reveal a prokineticin-driven circadian network. EMBO J. 40(20):e108614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wen S, et al. 2020. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci. 23(3):456–467. [DOI] [PubMed] [Google Scholar]
- 37. Xu P, et al. 2021. NPAS4 regulates the transcriptional response of the suprachiasmatic nucleus to light and circadian behavior. Neuron. 109(20):3268–3282.e3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Enoki R, Ono D, Kuroda S, Honma S, Honma KI. 2017. Dual origins of the intracellular circadian calcium rhythm in the suprachiasmatic nucleus. Sci Rep. 7:41733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stowie A, et al. 2021. Arginine-vasopressin expressing neurons in the murine suprachiasmatic nucleus exhibit a circadian rhythm in network coherence in vivo. BioRxiv. DOI: 10.1101/2021.12.07.471437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ko CH, et al. 2010. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 8(10):e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carrillo-Reid L, Yuste R. 2020. Playing the piano with the cortex: role of neuronal ensembles and pattern completion in perception and behavior. Curr Opin Neurobiol. 64:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaiser MI. 2018. Individuating part–whole relations in the biological world. In: Bueno O, Chen R-L, Bonnie Fagan M, editors. Individuation, process, and scientific practices. Oxford: Oxford Scholarship Online. [Google Scholar]
- 43. Bur IM, et al. 2009. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J Biol Chem. 284(14):9066–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller JE, Ayzenshtat I, Carrillo-Reid L, Yuste R. 2014. Visual stimuli recruit intrinsically generated cortical ensembles. Proc Natl Acad Sci USA. 111(38):E4053–E4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is included in the manuscript and/or supporting information.