Abstract
RNA viruses exhibit a great tendency to mutate. Mutations occur in the parts of the genome that encode the spike glycoprotein and less often in the rest of the genome. This is why Gibbs energy of binding changes more than that of biosynthesis. Starting from 2019, the wild type that was labeled Hu-1 has during the last 3 years evolved to produce several dozen new variants, as a consequence of mutations. Mutations cause changes in empirical formulas of new virus strains, which lead to change in thermodynamic properties of biosynthesis and binding. These changes cause changes in the rate of reactions of binding of virus antigen to the host cell receptor and the rate of virus multiplication in the host cell. Changes in thermodynamic and kinetic parameters lead to changes in biological parameters of infectivity and pathogenicity. Since the beginning of the COVID-19 pandemic, SARS-CoV-2 has been evolving towards increase in infectivity and maintaining constant pathogenicity, or for some variants a slight decrease in pathogenicity. In the case of Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants pathogenicity is identical as in the Omicron BA.2.75 variant. On the other hand, infectivity of the Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants is greater than those of previous variants. This will most likely result in the phenomenon of asymmetric coinfection, that is circulation of several variants in the population, some being dominant.
Keywords: COVID-19, Elemental composition, Thermodynamic property, Variant of concern (VOC), Virus-host interaction, Binding affinity
Graphical abstract
1. Introduction
Viruses represent open thermodynamic systems (von Bertalanffy, 1950, 1971; Popovic and Minceva, 2020a; Şimşek et al., 2021). Viruses have characteristic empirical formulas (Popovic and Minceva, 2020b; Degueldre, 2021), as well as thermodynamic properties of biosynthesis (Popovic, 2022a, 2022b, 2022c) and binding (Gale, 2022, 2020, 2019, 2018; Popovic and Popovic, 2022; Popovic, 2022d, 2022e, 2022f, 2022g). Antigen-receptor binding represents a chemical reaction similar to protein-ligand interaction (Du et al., 2016; Popovic and Popovic, 2022). Life processes - replication, transcription and translation represent chemical processes – polymerization of nucleotides into nucleic acid and amino acids into proteins (Pinheiro et al., 2008; Johansson and Dixon, 2013; Dodd et al., 2020; Lee et al., 2020). Driving force for all chemical processes is Gibbs energy (Demirel, 2014; Balmer, 2010; Atkins and de Paula, 2011, 2014; von Stockar, 2013a, 2013b). To understand biological processes, which have their chemical and biothermodynamic side, it is necessary to know the driving forces for these processes (Popovic, 2022i, 2022j).
RNA viruses, including SARS-CoV-2, exhibit a great tendency to mutate (Duffy, 2018). All variants of SARS-CoV-2, from Wild type Hu-1 to Omicron BA.2.75, have been chemically and thermodynamically characterized (Popovic and Popovic, 2022; Popovic, 2022a, 2022b, 2022c, 2022d, 2022e, 2022f, 2022g, Popovic and Minceva, 2021a, 2022b). Thermodynamics has been used to study various aspects of SARS-CoV-2. These include the binding of SARS-CoV-2 particles to host cells (Gale, 2022; Rombel-Bryzek et al., 2023; Garcia-Iriepa et al., 2020), immune response to SARS-CoV-2 (Ngo et al., 2021), energy content of SARS-CoV-2 particles (Şimşek et al., 2021), energetic cost of infection for the host organism (Özilgen and Yilmaz, 2021; Yilmaz et al., 2020), changes in host cell metabolism (Lucia et al., 2021, 2020a; Head et al., 2022), epidemiology of COVID-19 (Lucia et al., 2020b; Kaniadakis et al., 2020) and effects of COVID-19 on the society (Nadi and Özilgen, 2021). Change in thermodynamic properties of binding and biosynthesis during the time evolution of SARS-CoV-2 has been reported in (Popovic, 2022f).
During the last several months, the Omicron variant has developed new mutations, giving rise to new variants, including BQ.1, BQ.1.1 and XBB. These were labeled variants of concern (VOC) by WHO (2022). The BA.2.75 and BQ.1 have been dominating lately (Zappa et al., 2022). The question is raised whether they are competitive (Zappa et al., 2022). Various viruses compete “for soil” (Popovic and Minceva, 2021a). The result of this competition can be coinfection or interference (Popovic and Minceva, 2021a). Experience learns us that newer variants have caused successive pandemic waves, suppressing the older variants (Popovic, 2022f).
The aim of this paper is to chemically and thermodynamically characterize the new Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants and to compare their thermodynamic properties of biosynthesis and binding with other SARS-CoV-2 variants. The physicochemical properties of interest include empirical formulas, molar masses, Gibbs energies of antigen-receptor binding, as well as enthalpies, entropies and Gibbs energies of formation and biosynthesis of nucleocapsid live matter. These properties will then be used to shed more light on the time evolution of SARS-CoV-2.
2. Methods
2.1. Data sources
The genetic sequences of the BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2 were taken from GISAID, the global data science initiative (Khare et al., 2021; Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017). The BQ.1 genetic sequence can be found under the accession ID: EPI_ISL_15921176. It is labeled as hCoV-19/Israel/ICH-741192169/2022. It has been collected on November 16, 2022 by a lab in Tel Aviv, Israel. The BQ.1.1 genetic sequence can be found under the accession ID: EPI_ISL_15938021. It is labeled hCoV-19/USA/MT-MTPHL-4070970/2022. It was isolated on November 23, 2022 by a lab in Helena, Montana (USA). The XBB genetic sequence can be found under the accession ID: EPI_ISL_15936982. It is labeled hCoV-19/Austria/LB-R00113-S325/2022. It was isolated on November 23, 2022 by a lab in Vienna, Austria. The XBB.1 genetic sequence can be found under the accession ID: EPI_ISL_15927855. It is labeled hCoV-19/Turkey/KSS-UEAH4881829747/2022. It was isolated on November 7, 2022 by a lab in Istanbul, Turkey. The findings of this study are based on metadata associated with 4 sequences available on GISAID up to December 2, 2022, and accessible at 10.55876/gis8.221202be.
The sequence of the nucleocapsid phosphoprotein of SARS-CoV-2 was obtained from the NCBI database (Sayers et al., 2022; National Center for Biotechnology Information, 2022), under the accession ID: UKQ14424.1. The number of copies of the nucleocapsid phosphoprotein in virus particle was taken from (Neuman and Buchmeier, 2016; Neuman et al., 2011; Neuman et al., 2006).
Dissociation equilibrium constants of the BQ.1, BQ.1.1, XBB, XBB.1, BA.2 and BA.4/5 variants were taken from Wang et al. (2022). They were measured using surface plasmon resonance at 25°C (Wang et al., 2022; Rusnati et al., 2015).
Data on standard Gibbs energies of binding of the Wild type Hu-1, Alpha B.1.1.7, Beta B.1.351, Gamma P.1, Delta B.1.617 and Omicron BA.1 variants of SARS-CoV-2 were taken from (Popovic, 2022b). All the data are at 25°C.
Data on standard Gibbs energies of biosynthesis of biosynthesis of the Hu-1, Delta B.1.617.2, Omicron BA.1.1.429, Omicron BA.2 and Omicron BA.2.75 variants of SARS-CoV-2 were taken from (Popovic, 2022a, 2022b, 2022c).
2.2. Empirical formulas and biosynthesis reactions
The genetic and protein sequences were used to find empirical formulas of nucleocapsids of the BQ.1, BQ.1.1 and XBB variants of SARS-CoV-2. This was done using the atom counting method (Popovic, 2022h). The atom counting method is implemented using a computer program (Popovic, 2022h). The input are genetic and protein sequences of the virus of interest, as well as the number of copies of proteins in the virus particle and the virus particle size (Popovic, 2022h). The program goes along the nucleic acid and protein sequences and adds atoms coming from each residue in the sequence, to find the number of atoms contributed by that macromolecule to the virus particle (Popovic, 2022h). The contributions of viral proteins are multiplied by their copy numbers, since proteins are present in multiple copies in virus particles (Popovic, 2022h). The output of the program is elemental composition of virus particles, in the form of empirical formulas, and molar masses of virus particles (Popovic, 2022h). The advantage of the atom counting method is that it can provide the empirical formulas of virus particles, based on widely available data on genetic and protein sequences (Popovic, 2022h). The atom counting method was shown to give results in good agreement with experimental results (Popovic, 2022h).
The empirical formulas of virus particles were used to construct biosynthesis reactions, summarizing conversion of nutrients into new live matter (von Stockar, 2013a, 2013b; Battley, 1998). The biosynthesis reaction for virus particles has the general form
| (Amino acid) + O2 + HPO42− + HCO3−→ (Bio) + SO42− + H2O + H2CO3 | (1) |
where (Bio) represents new live matter, described by an empirical formula given by the atom counting method (Popovic, 2022a, 2022b). (Amino acid) represents a mixture of amino acids with the empirical formula CH1.798O0.4831N0.2247S0.022472 (expressed per mole of carbon), representing the source of energy, carbon, nitrogen and sulfur (Popovic, 2022a, 2022b). O2 is the electron acceptor (Popovic, 2022a, 2022b). HPO4 2− is the source of phosphorus (Popovic, 2022a, 2022b). HCO3 − is a part of the bicarbonate buffer that takes excess H+ ions that are generated during biosynthesis (Popovic, 2022a, 2022b). SO4 2− is an additional metabolic product that takes excess sulfur atoms (Popovic, 2022a, 2022b). H2CO3 takes the oxidized carbon atoms and is also a part of the bicarbonate buffer (Popovic, 2022a, 2022b).
2.3. Thermodynamic properties of live matter and biosynthesis
Empirical formulas of virus nucleocapsids were used to find standard thermodynamic properties of their live matter, using predictive biothermodynamic models: the Patel-Erickson and Battley equations (Patel and Erickson, 1981; Battley, 1999, 1998, 1992). The Patel-Erickson equation was used to find enthalpy of live matter, based on its elemental composition. The Patel-Erickson equation gives standard enthalpy of combustion, ΔCH⁰, of live matter
| (2) |
where E is number of electrons transferred to oxygen during combustion (Patel and Erickson, 1981; Battley, 1998, 1992; Popovic, 2019). E can be calculated from the empirical formula of live matter
| (3) |
where nC, nH, nO, nN, nP and nS represent the number of C, H, O, N, P and S atoms in the live matter empirical formula, respectively (Patel and Erickson, 1981; Battley, 1998, 1992; Popovic, 2019). Once calculated using the Patel-Erickson equation, ΔCH⁰ can be converted into standard entropy of formation, ΔfH⁰, of live matter. ΔCH⁰ is the enthalpy change of the reaction of complete combustion of live matter.
| CnCHnHOnONnNPnPSnS + (nC + ¼ nH + 1¼ nP + 1½ nS - ½ nO) O2 → nC CO2 + ½ nH H2O + ½ nN N2 + ¼ nP P4O10 + nS SO3 | (4) |
This means that ΔCH⁰ can be used to find ΔfH⁰ of live matter using the equation, using the equation (Popovic, 2022b; Atkins and de Paula, 2011, 2014; Popovic, 2019)
| (5) |
2.4. The Battley equation gives standard molar entropy of live matter, S⁰m, based on its empirical formula
| (6) |
where nJ is the number of atoms of element J in the empirical formula of live matter (Battley, 1999; Battley and Stone, 2000; Popovic, 2019). S⁰m and aJ are standard molar entropy and number of atoms per formula unit of element J in its standard state elemental form (Battley, 1999; Battley and Stone, 2000; Popovic, 2019). The Battley equation can be modified to give standard entropy of formation, ΔfS⁰, of live matter (Battley, 1999; Battley and Stone, 2000; Popovic, 2019)
| (7) |
Finally, ΔfH⁰ and ΔfS⁰ are combined to give standard Gibbs energy of formation of live matter, ΔfG⁰.
| (8) |
Once live matter is characterized by finding its ΔfH⁰, S⁰m and ΔfG⁰, these properties can be combined with biosynthesis reactions to find standard thermodynamic properties of biosynthesis. Standard thermodynamic properties of biosynthesis include standard enthalpy of biosynthesis, ΔbsH⁰, standard entropy of biosynthesis, ΔbsS⁰, and standard Gibbs energy of biosynthesis, ΔbsG⁰. These properties are found by applying the Hess's law to biosynthesis reactions
| (9) |
| (10) |
| (11) |
where ν represents a stoichiometric coefficient (Popovic, 2022b; Atkins and de Paula, 2011, 2014; von Stockar, 2013a, 2013b; Battley, 1998). The most important of these three properties is standard Gibbs energy of biosynthesis, which represents the thermodynamic driving force for growth of all organisms (von Stockar, 2013a, 2013b; von Stockar and Liu, 1999), including viruses (Popovic, 2022b).
2.5. Thermodynamic properties of antigen-receptor binding
In order to multiply inside the cytoplasm, a virus must first enter its host cell. The first step in this process is binding of the virus antigen to the host cell receptor. The antigen of SARS-CoV-2 is the spike glycoprotein trimer (SGP) (Duan et al., 2020), while the host cell receptor is angiotensin-converting enzyme 2 (ACE2) (Scialo et al., 2020). The process of antigen-receptor binding is, in its essence, a chemical reaction, similar to protein-ligand interactions (Du et al., 2016; Popovic and Popovic, 2022). Thus, the binding of SGP to ACE2 can be described through the chemical reaction
| (An) + (Re) = (An-Re) | (12) |
Where (An) represents the virus antigen (SGP in the case of SARS-CoV-2), (Re) represents the host cell receptor (ACE2 for SARS-CoV-2), while (An-Re) represents the antigen-receptor complex (Du et al., 2016; Popovic and Popovic, 2022).
Like for all other chemical reactions, laws of chemical thermodynamics apply and the process of antigen-receptor binding can be described through several thermodynamic parameters. The dissociation equilibrium constant, Kd, is defined as
| (13) |
where [An] is the concentration of the virus antigen, [Re] the concentration of the host receptor and [An-Re] the concentration of the antigen-receptor complex (Du et al., 2016; Popovic and Popovic, 2022). The reciprocal of Kd is the binding equilibrium constant, KB, (Du et al., 2016; Popovic and Popovic, 2022)
| (14) |
The binding equilibrium constant can be used to find standard Gibbs energy of binding, ΔBG⁰, through the equation
| (15) |
Where T is temperature and R is the universal gas constant (Du et al., 2016; Popovic and Popovic, 2022).
3. Results
In this research, for the first time the empirical formula of BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2 were calculated. They are given in Table 1 . The empirical formula of the nucleocapsid of the BQ.1 variant is CH1.574O0.3427N0.3124P0.006027S0.003358. The molar mass of the BQ.1 nucleocapsid empirical formula is 23.749 g/C-mol, while the molar mass of the entire BQ.1 nucleocapsid is 117.22 MDa. The empirical formula of nucleocapsid of the BQ.1.1 variant is CH1.574O0.3426N0.3124P0.006006S0.003359. The molar mass of the BQ.1.1 nucleocapsid empirical formula is 23.747 g/C-mol, while the molar mass of the entire BQ.1.1 nucleocapsid is 117.28 MDa. The empirical formula of the nucleocapsid of the XBB variant is CH1.574O0.3427N0.3124P0.006028S0.003358. The molar mass of the XBB nucleocapsid empirical formula is 23.749 g/C-mol, while the molar mass of the entire XBB nucleocapsid is 117.22 MDa. The empirical formula of the nucleocapsid of the XBB.1 variant is CH1.574O0.3426N0.3124P0.006012S0.003359. The molar mass of the XBB.1 nucleocapsid empirical formula is 23.748 g/C-mol, while the molar mass of the entire XBB.1 nucleocapsid is 117.19 MDa.
Table 1.
Empirical formulas of nucleocapsids of the BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2. The empirical formulas have the general form CnCHnHOnONnNPnPSnS. All formulas are expressed per mole of carbon. Molar masses are provided in two forms. Mr denotes the molar mass of the empirical formula and is expressed in g/C-mol (Daltons). Mr(nc) denotes the molar mass of the entire nucleocapsid (entire viral genome and all nucleoprotein copies) and is expressed in MDa.
| Variant | C | H | O | N | P | S | Mr (g/C-mol) | Mr(nc) (MDa) |
|---|---|---|---|---|---|---|---|---|
| BQ.1 nucleocapsid | 1 | 1.574 | 0.3427 | 0.3124 | 0.006027 | 0.003358 | 23.749 | 117.22 |
| BQ.1.1 nucleocapsid | 1 | 1.574 | 0.3426 | 0.3124 | 0.006006 | 0.003359 | 23.747 | 117.18 |
| XBB nucleocapsid | 1 | 1.574 | 0.3427 | 0.3124 | 0.006028 | 0.003358 | 23.749 | 117.22 |
| XBB.1 nucleocapsid | 1 | 1.574 | 0.3426 | 0.3124 | 0.006012 | 0.003359 | 23.748 | 117.19 |
Based on the empirical formulas of the SARS-CoV-2 variants, biosynthesis reactions were formulated. They are presented in Table 2 . The biosynthesis reaction of the BQ.1 nucleocapsid is
| 1.3901 CH1.798O0.4831N0.2247S0.022472 + 0.4913 O2 + 0.0060 HPO42− + 0.0437 HCO3−→ CH1.574O0.3427N0.3124P0.006027S0.003358 + 0.0279 SO42− + 0.0538 H2O + 0.4338 H2CO3 | (16) |
Table 2.
Biosynthesis reactions of the BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2.
| Variant | Reactants | Products | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Amino acid | O2 | HPO42− | HCO3− | → | Bio | SO42− | H2O | H2CO3 | |
| BQ.1 nucleocapsid | 1.3901 | 0.4913 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| BQ.1.1 nucleocapsid | 1.3900 | 0.4911 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4337 |
| XBB nucleocapsid | 1.3901 | 0.4913 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4338 |
| XBB.1 nucleocapsid | 1.3900 | 0.4912 | 0.0060 | 0.0437 | → | 1 | 0.0279 | 0.0538 | 0.4337 |
The biosynthesis reaction of the BQ.1.1 nucleocapsid is
| 1.3900 CH1.798O0.4831N0.2247S0.022472 + 0.4911 O2 + 0.0060 HPO42− + 0.0437 HCO3−→ CH1.574O0.3426N0.3124P0.006006S0.003359 + 0.0279 SO42− + 0.0538 H2O + 0.4337 H2CO3 | (17) |
The biosynthesis reaction of the XBB nucleocapsid is
| 1.3901 CH1.798O0.4831N0.2247S0.022472 + 0.4913 O2 + 0.0060 HPO42− + 0.0437 HCO3−→ CH1.574O0.3427N0.3124P0.006028S0.003358 + 0.0279 SO42− + 0.0538 H2O + 0.4338 H2CO3 | (18) |
The biosynthesis reaction of the XBB.1 nucleocapsid is
| 1.3900 CH1.798O0.4831N0.2247S0.022472 + 0.4912 O2 + 0.0060 HPO42− + 0.0437 HCO3−→ CH1.574O0.3426N0.3124P0.006012S0.003359 + 0.0279 SO42− + 0.0538 H2O + 0.4337 H2CO3 | (19) |
Standard thermodynamic properties of live matter of nucleocapsids of the BQ.1, BQ.1.1, XBB and XBB.1 variants were calculated and are shown in Table 3 . Standard enthalpy of formation of the BQ.1 nucleocapsid is -75.40 kJ/C-mol, its standard molar entropy is 32.49 J/C-mol K, while its standard Gibbs energy of formation is -33.28 kJ/C-mol. Standard enthalpy of formation of the BQ.1.1 nucleocapsid is -75.38 kJ/C-mol, its standard molar entropy is 32.49 J/C-mol K, while its standard Gibbs energy of formation is -33.26 kJ/C-mol. Standard enthalpy of formation of the XBB nucleocapsid is -75.40 kJ/C-mol, its standard molar entropy is 32.49 J/C-mol K, while its standard Gibbs energy of formation is -33.28 kJ/C-mol. Finally, standard enthalpy of formation of the XBB.1 nucleocapsid is -75.38 kJ/C-mol, its standard molar entropy is 32.49 J/C-mol K, while its standard Gibbs energy of formation is -33.26 kJ/C-mol.
Table 3.
Standard thermodynamic properties of live matter for the nucleocapsids of Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2. This table presents standard enthalpies of formation, ΔfH⁰, standard molar entropies, S⁰m, and standard Gibbs energies of formation, ΔfG⁰, at 25°C (298.15 K).
| Variant | ΔfH⁰ (kJ/C-mol) | S⁰m (J/C-mol K) | ΔfG⁰ (kJ/C-mol) |
|---|---|---|---|
| BQ.1 nucleocapsid | -75.40 | 32.49 | -33.28 |
| BQ.1.1 nucleocapsid | -75.38 | 32.49 | -33.26 |
| XBB nucleocapsid | -75.40 | 32.49 | -33.28 |
| XBB.1 nucleocapsid | -75.38 | 32.49 | -33.26 |
In this research, for the first time the Gibbs energy of biosynthesis is calculated for BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2. They are presented in Table 4 . Standard enthalpy of biosynthesis of the BQ.1 nucleocapsid is -232.33 kJ/C-mol, its standard entropy of biosynthesis is -37.34 J/C-mol K, while its standard Gibbs energy of biosynthesis is -221.24 kJ/C-mol. Standard enthalpy of biosynthesis of the BQ.1.1 nucleocapsid is -232.27 kJ/C-mol, its standard entropy of biosynthesis is -37.33 J/C-mol K, while its standard Gibbs energy of biosynthesis is -221.18 kJ/C-mol. Standard enthalpy of biosynthesis of the XBB nucleocapsid is -232.34 kJ/C-mol, its standard entropy of biosynthesis is -37.34 J/C-mol K, while its standard Gibbs energy of biosynthesis is -221.25 kJ/C-mol. Finally, standard enthalpy of biosynthesis of the XBB.1 nucleocapsid is -232.28 kJ/C-mol, its standard entropy of biosynthesis is -37.33 J/C-mol K, while its standard Gibbs energy of biosynthesis is -221.19 kJ/C-mol.
Table 4.
Standard thermodynamic properties of biosynthesis for the nucleocapsids of BQ.1, BQ.1.1, XBB and XBB.1 variants of SARS-CoV-2. This table gives data on standard enthalpy of biosynthesis, ΔbsH⁰, standard entropy of biosynthesis, ΔbsS⁰, and standard Gibbs energy of biosynthesis, ΔbsG⁰, at 25°C (298.15 K).
| Variant | ΔbsH⁰ (kJ/C-mol) | ΔbsS⁰ (J/C-mol K) | ΔbsG⁰ (kJ/C-mol) |
|---|---|---|---|
| BQ.1 nucleocapsid | -232.33 | -37.34 | -221.24 |
| BQ.1.1 nucleocapsid | -232.27 | -37.33 | -221.18 |
| XBB nucleocapsid | -232.34 | -37.34 | -221.25 |
| XBB.1 nucleocapsid | -232.28 | -37.33 | -221.19 |
Based on Kd data reported in Wang et al. (2022), standard thermodynamic properties of antigen-receptor binding have been determined for BQ.1, BQ.1.1, XBB, XBB.1, BA.2 and BA.4/5 variants of SARS-CoV-2. They are given in Table 5 and include the binding equilibrium constant, KB, and standard Gibbs energy of binding, ΔBG⁰. For the BQ.1 variant, the binding equilibrium constant is KB = 1.613 × 109 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -52.55 kJ/mol. For the BQ.1.1 variant, the binding equilibrium constant is KB = 1.786 × 109 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -52.81 kJ/mol. For the XBB variant, the binding equilibrium constant is KB = 5.000 × 108 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -49.65 kJ/mol. For the XBB.1 variant, the binding equilibrium constant is KB = 4.854 × 108 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -49.58 kJ/mol. For the BA.2 variant, the binding equilibrium constant is KB = 1.053 × 109 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -51.50 kJ/mol. For the BA.4/5 variants, the binding equilibrium constant is KB = 1.639 × 109 M−1, while the standard Gibbs energy of antigen-receptor binding is ΔBG⁰ = -52.59 kJ/mol.
Table 5.
Standard thermodynamic properties of antigen-receptor binding of SARS-CoV-2 variants. This table shows data on dissociation equilibrium constants, Kd, binding equilibrium constants, KB, and standard Gibbs energies of binding, ΔBG⁰. The data are for binding of the spike glycoprotein trimer (SGP) of SARS-CoV-2 to the human ACE2 receptor. All the data are at 25°C (298.15 K). The Kd values were taken from Wang et al. (2022).
| Variant | Kd (M) | KB (M−1) | ΔBG⁰ (kJ/mol) |
|---|---|---|---|
| BQ.1 | 6.2E-10 | 1.613E+09 | -52.55 |
| BQ.1.1 | 5.6E-10 | 1.786E+09 | -52.81 |
| XBB | 2E-09 | 5.000E+08 | -49.65 |
| XBB.1 | 2.06E-09 | 4.854E+08 | -49.58 |
| BA.2 | 9.5E-10 | 1.053E+09 | -51.50 |
| BA.4/5 | 6.1E-10 | 1.639E+09 | -52.59 |
4. Discussion
Zappa et al. (2022) have raised an interesting question: will the new Omicron variants compete with the currently dominant BA.2.75 variant. Having in mind that viruses are biological, chemical and thermodynamic entities (Ozilgen and Sorgüven, 2017; Balmer, 2010; von Stockar, 2013a; Wimmer, 2006; Molla et al., 1991), and that during their life cycle they perform chemical reactions (binding, replication, transcription and translation), this means that they compete for resources. The rate of the mentioned chemical reactions, according to the phenomenological equations, which belong to nonequilibrium thermodynamics, directly depends on Gibbs energy of the process (Popovic and Popovic, 2022). Indeed, the rate of binding, rB, is given by the phenomenological equation
| (20) |
where ΔBG is the Gibbs energy of binding, T is temperature, while LB is the binding phenomenological coefficient (Popovic and Popovic, 2022; Popovic, 2022b, 2022g). Furthermore, the rate of biosynthesis, rbs, of viral components is given by the biosynthesis phenomenological equation
| (21) |
where ΔbsG is the Gibbs energy of biosynthesis, while Lbs is the biosynthesis phenomenological coefficient (Popovic, 2022a, 2022b, 2022c). Through analysis of the two phenomenological equations, it is possible to predict the outcome of competition of two viruses, if they appear at the same time in the same place and compete for the same population. If Gibbs energies of binding are approximately same, then the result of competition will be coinfection (Popovic and Minceva, 2021a). Furthermore, if Gibbs energies of biosynthesis are similar, the result of competition will be coinfection (Popovic and Minceva, 2021a). However, if Gibbs energies of binding and biosynthesis are slightly different, the result will be asymmetric coinfection (Popovic and Minceva, 2021a). Asymmetric coinfection means that both variants will appear in the population, but one of them, the one characterized by a more negative Gibbs energy of binding and biosynthesis, will dominate (Popovic and Minceva, 2021a). A large difference in Gibbs energies of binding and/or biosynthesis will lead to the phenomenon of interference (Popovic and Minceva, 2021a).
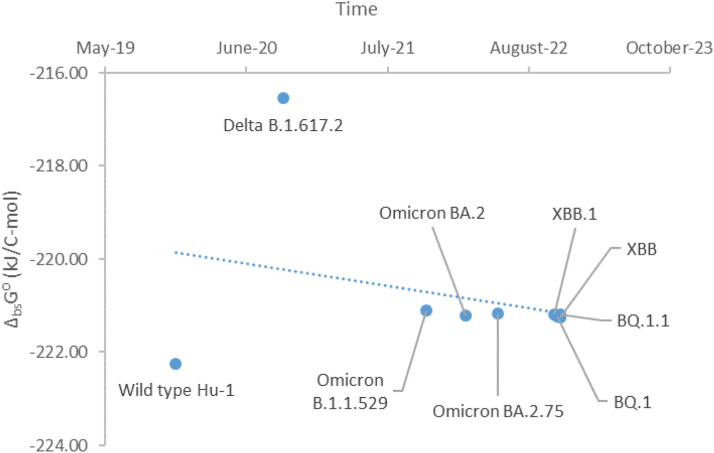
In this paper, standard Gibbs energies of biosynthesis were determined for BQ.1 (-221.24 kJ/C-mol), BQ.1.1 (-221.18 kJ/C-mol), XBB (-221.25 kJ/C-mol) and XBB.1 (-221.19 kJ/C-mol). Gibbs energy of biosynthesis for the BA.2.75 subvariant is -221.18 kJ/C-mol (Popovic, 2022a). From these data, we can conclude that in case of competition there is no winner, that all the variants multiply at the same rate, leading to coinfection, since they all have similar Gibbs energies of biosynthesis.
The next interesting question is related to pathogenicity of the new variants. Pathogenicity is related to the rate of multiplication of viruses. Greater multiplication rate leads to greater damage to host cells (Popovic, 2022f). Greater damage to host cells is a good indicator of virus pathogenicity. Gibbs energies of biosynthesis of viral components for the BA.2.75, BQ.1, BQ.1.1 and XBB variants are approximate the same. Thus, the multiplication rate of the new variants is approximately equal to the multiplication rate of BA.2.75, which leads to the conclusion that the pathogenicity of the new variants is identical to that of BA.2.75. Fig. 1 shows a graphical representation of time evolution of SARS-CoV-2, including the new BQ.1, BQ.1.1, XBB and XBB.1 variants. The graph shows that Gibbs energies of biosynthesis of BA.2.75 and the new variants differ on the second decimal. This difference is negligible and is not sufficient to show the difference in pathogenicity. The Omicron variant has evolved towards maintaining constant multiplication rate and pathogenicity.
Fig. 1.
Time evolution of SARS-CoV-2 variants through standard Gibbs energies of biosynthesis. This time shows standard Gibbs energy of biosynthesis, ΔbsG⁰, of various SARS-CoV-2 variants, which appeared at different times.
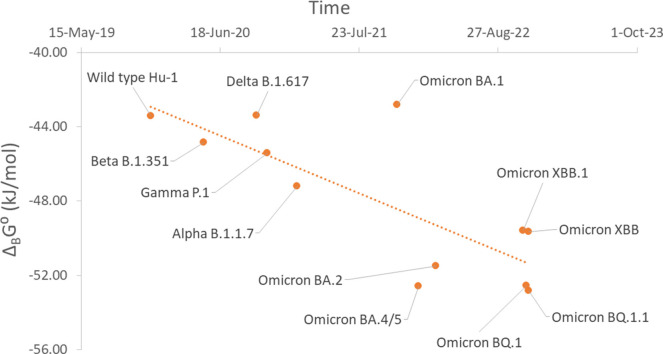
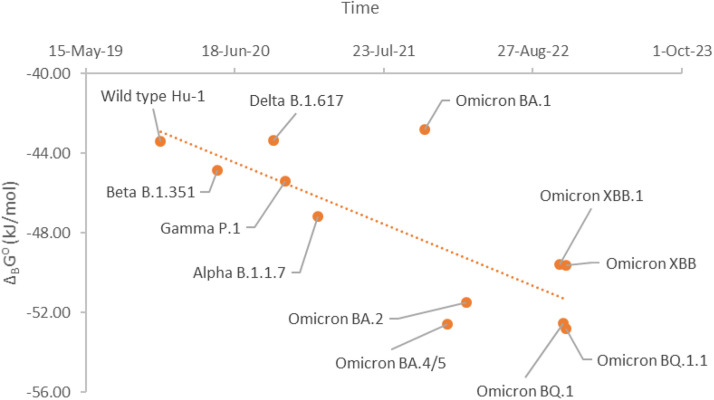
Infectivity depends on the affinity of antigen-receptor binding, and the rate of virus binding and entry into host cells, according to the binding phenomenological Eq. (20). Gibbs energy of binding is the driving force for the reaction of antigen-receptor binding (Popovic and Popovic, 2022; Popovic, 2022b, 2022d). Standard Gibbs energy of binding for the Omicron BA.2.75 variant is −49.41 kJ/mol (Popovic, 2022g). In this research, standard Gibbs energies of binding of the new SARS-CoV-2 variants were found to be: -52.55 for the BQ.1 variant, -52.81 kJ/mol for the BQ.1.1 variant, -49.65 kJ/mol for the XBB variant and -49.58 kJ/mol for the XBB.1 variant. The XBB and XBB.1 variants have very similar standard Gibbs energies of binding to BA.2.75, meaning that they will have an almost identical entry rates. However, BQ.1 and BQ.1.1 have slightly more negative Gibbs energy of binding that BA.2.75. This implies that the entry rates of BQ.1 and BQ.1.1 into host cells will be greater than that of BA.2.75. Having in mind that the difference in Gibbs energies of binding is not great, in case of simultaneous circulation of Omicron BA.2.75, BQ.1 and BQ.1.1 variants, in the population, there will be asymmetric coinfection.
SARS-CoV-2 has continued to evolve in the direction it has taken since the beginning of the COVID-19 pandemic, related to slight increase in infectivity with maintaining pathogenicity at a relatively constant level. Thus, it is not possible to give a definite response to the question of the time of arrival of the pandemic phase “game over”.
Fig. 2 shows the time evolution of Gibbs energy of binding of SARS-CoV-2. From the graph, a trend can be observed towards a more negative Gibbs energy of binding of the new variants. As was extrapolated in the paper (Popovic, 2022f), new mutations on the receptor binding domain (RBD) have developed towards increase in binding affinity. The mutations have led to change in elemental composition and empirical formulas, which characterize the new variants. Change in empirical formulas have led to changes in thermodynamic properties of new variants, in the exact direction predicted by the theory of evolution.
Fig. 2.
Time evolution of SARS-CoV-2 variants through standard Gibbs energies of binding. This time shows standard Gibbs energy of binding, ΔBG⁰, of various SARS-CoV-2 variants, which appeared at different times.
5. Conclusions
Empirical formulas and molar masses have been determined for the nucleocapsids of BQ.1, BQ.1.1, XBB and XBB.1 virions. The empirical formula of the BQ.1 nucleocapsid is CH1.574O0.3427N0.3124P0.006027S0.003358. For the BQ.1.1 nucleocapsid the empirical formula is CH1.574O0.3426N0.3124P0.006006S0.003359. For the XBB nucleocapsid, the empirical formula is CH1.574O0.3427N0.3124P0.006028S0.003358. For the XBB.1 nucleocapsid, the empirical formula is CH1.574O0.3426N0.3124P0.006012S0.003359. The molar mass of the BQ.1 nucleocapsid empirical formula is 23.749 g/C-mol, while the molar mass of the entire BQ.1 nucleocapsid is 117.22 MDa. The molar mass of the BQ.1.1 nucleocapsid empirical formula is 23.747 g/C-mol, while the molar mass of the entire BQ.1.1 nucleocapsid is 117.28 MDa. The molar mass of the XBB nucleocapsid empirical formula is 23.749 g/C-mol, while the molar mass of the entire XBB nucleocapsid is 117.22 MDa. The molar mass of the XBB.1 nucleocapsid empirical formula is 23.748 g/C-mol, while the molar mass of the entire XBB.1 nucleocapsid is 117.19 MDa.
Thermodynamic properties of biosynthesis of BQ.1, BQ.1.1, XBB and XBB.1 variants differ very little from those of other Omicron variants. Thus, pathogenicity of new variants has not changed compared to BA.2.75.
The BQ.1 and BQ.1.1 variants have evolved towards more negative standard Gibbs energy of binding, leading to increase in infectivity. This is in agreement with the expectation of the evolution theory and empirical observations.
New variants that compete with BA.2.75 should exhibit the phenomenon of coinfection. Several variants can simultaneously circulate in the population, causing a new wave of the pandemic.
CRediT authorship contribution statement
Marko Popovic: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization.
Declaration of Competing Interest
The author declares no conflict of interest.
Acknowledgements
The author gratefully acknowledges all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
References
- Atkins P.W., de Paula J. 2nd ed. W. H. Freeman and Company; 2011. Physical Chemistry for the Life Sciences. ISBN-13: 978-1429231145. [Google Scholar]
- Atkins P.W., de Paula J. 10th ed. W. H. Freeman and Company; New York: 2014. Physical Chemistry: Thermodynamics, Structure, and Change. ISBN-13: 978-1429290197. [Google Scholar]
- Balmer R.T. Academic Press; Cambridge, MA: 2010. Modern Engineering Thermodynamics. [DOI] [Google Scholar]
- Battley E.H., Stone J.R. A comparison of values for the entropy and the entropy of formation of selected organic substances of biological importance in the solid state, as determined experimentally or calculated empirically. Thermochim. Acta. 2000;349(1-2):153–161. doi: 10.1016/S0040-6031(99)00509-2. [DOI] [Google Scholar]
- Battley E.H. An empirical method for estimating the entropy of formation and the absolute entropy of dried microbial biomass for use in studies on the thermodynamics of microbial growth. Thermochim. Acta. 1999;326(1-2):7–15. doi: 10.1016/S0040-6031(98)00584-X. [DOI] [Google Scholar]
- Battley E.H. The development of direct and indirect methods for the study of the thermodynamics of microbial growth. Thermochim. Acta. 1998;309(1-2):17–37. doi: 10.1016/S0040-6031(97)00357-2. [DOI] [Google Scholar]
- Battley E.H. On the enthalpy of formation of Escherichia coli K-12 cells. Biotechnol. Bioeng. 1992;39:5–12. doi: 10.1002/bit.260390103. [DOI] [PubMed] [Google Scholar]
- Degueldre C. Single virus inductively coupled plasma mass spectroscopy analysis: a comprehensive study. Talanta. 2021;228 doi: 10.1016/j.talanta.2021.122211. [DOI] [PubMed] [Google Scholar]
- Demirel Y. 3rd ed. Elsevier; Amsterdam: 2014. Nonequilibrium Thermodynamics: Transport and Rate Processes in Physical, Chemical and Biological Systems. ISBN: 9780444595812. [Google Scholar]
- Dodd T., Botto M., Paul F., et al. Polymerization and editing modes of a high-fidelity DNA polymerase are linked by a well-defined path. Nat. Commun. 2020;11:5379. doi: 10.1038/s41467-020-19165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Liu S.Q. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. PMCID: 31565258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Using thermodynamic equilibrium models to predict the effect of antiviral agents on infectivity: Theoretical application to SARS-CoV-2 and other viruses. Microb. Risk Anal. 2022;21 doi: 10.1016/j.mran.2021.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. How virus size and attachment parameters affect the temperature sensitivity of virus binding to host cells: predictions of a thermodynamic model for arboviruses and HIV. Microb. Risk Anal. 2020;15 doi: 10.1016/j.mran.2020.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Towards a thermodynamic mechanistic model for the effect of temperature on arthropod vector competence for transmission of arboviruses. Microb. Risk Anal. 2019;12:27–43. doi: 10.1016/j.mran.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale P. Using thermodynamic parameters to calibrate a mechanistic dose-response for infection of a host by a virus. Microb. Risk Anal. 2018;8:1–13. doi: 10.1016/j.mran.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Iriepa C., Hognon C., Francés-Monerris A., Iriepa I., Miclot T., Barone G., Marazzi M. Thermodynamics of the interaction between the spike protein of severe acute respiratory syndrome coronavirus-2 and the receptor of human angiotensin-converting enzyme 2. Effects of possible ligands. J. Phys. Chem. Lett. 2020;11(21):9272–9281. doi: 10.1021/acs.jpclett.0c02203. [DOI] [PubMed] [Google Scholar]
- Head R.J., Lumbers E.R., Jarrott B., Tretter F., Smith G., Pringle K.G., Islam S., Martin J.H. Systems analysis shows that thermodynamic physiological and pharmacological fundamentals drive COVID-19 and response to treatment. Pharmacol. Res. Perspect. 2022;10(1):e00922. doi: 10.1002/prp2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E., Dixon N. Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013;5(6) doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniadakis G., Baldi M.M., Deisboeck T.S., Grisolia G., Hristopulos D.T., Scarfone A.M., Sparavigna A., Wada T., Lucia U. The κ-statistics approach to epidemiology. Sci. Rep. 2020;10(1):19949. doi: 10.1038/s41598-020-76673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., et al. GISAID's role in pandemic response. China CDC Wkly. 2021;3(49):1049–1051. doi: 10.46234/ccdcw2021.255. PMCID: 8668406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Schwarz K.J., Kim D.S., Moore J.S., Jewett M.C. Ribosome-mediated polymerization of long chain carbon and cyclic amino acids into peptides in vitro. Nat. Commun. 2020;11(1):4304. doi: 10.1038/s41467-020-18001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia U., Grisolia G., Deisboeck T.S. Thermodynamics and SARS-CoV-2: neurological effects in post-Covid 19 syndrome. Atti della Accad. Peloritana dei Pericolanti. 2021;99(2):A3. doi: 10.1478/AAPP.992A3. [DOI] [Google Scholar]
- Lucia U., Grisolia G., Deisboeck T.S. Seebeck-like effect in SARS-CoV-2 bio-thermodynamics. Atti della Accademia Peloritana dei Pericolanti-Classe di Scienze Fisiche. Mat. Nat. 2020;98(2):6. doi: 10.1478/AAPP.982A6. [DOI] [Google Scholar]
- Lucia U., Deisboeck T.S., Grisolia G. Entropy-based pandemics forecasting. Front. Phys. 2020;8:274. doi: 10.3389/fphy.2020.00274. [DOI] [Google Scholar]
- Molla A., Paul A.V., Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254(5038):1647–1651. doi: 10.1126/science.1661029. (New York, N.Y.) [DOI] [PubMed] [Google Scholar]
- Nadi F., Özilgen M. Effects of COVID-19 on energy savings and emission reduction: a case study. Int. J. Glob. Warm. 2021;25(1):38–57. doi: 10.1504/IJGW.2021.117432. [DOI] [Google Scholar]
- National Center for Biotechnology Information (2022). NCBI database [online]. Available at: https://www.ncbi.nlm.nih.gov/(Accessed on December 2, 2022).
- Neuman B.W., Buchmeier M.J. Supramolecular architecture of the coronavirus particle. Adv. Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in Coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80(16):7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo S.T., Nguyen T.H., Pham D.H., Tung N.T., Nam P.C. Thermodynamics and kinetics in antibody resistance of the 501Y.V2 SARS-CoV-2 variant. RSC Adv. 2021;11(53):33438–33446. doi: 10.1039/d1ra04134g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özilgen M., Yilmaz B. COVID-19 disease causes an energy supply deficit in a patient. Int. J. Energy Res. 2021;45(2):1157–1160. doi: 10.1002/er.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozilgen M., Sorgüven E. CRC Press; Boca Raton: 2017. Biothermodynamics: Principles and Applications. [DOI] [Google Scholar]
- Patel S.A., Erickson L.E. Estimation of heats of combustion of biomass from elemental analysis using available electron concepts. Biotechnol. Bioeng. 1981;23:2051–2067. doi: 10.1002/bit.260230910. [DOI] [Google Scholar]
- Pinheiro A.V., Baptista P., Lima J.C. Light activation of transcription: photocaging of nucleotides for control over RNA polymerization. Nucleic Acids Res. 2008;36(14):e90. doi: 10.1093/nar/gkn415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Popovic M. Strain Wars: Competitive interactions between SARS-CoV-2 strains are explained by Gibbs energy of antigen-receptor binding. Microb. Risk Anal. 2022 doi: 10.1016/j.mran.2022.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Thermodynamic properties of microorganisms: determination and analysis of enthalpy, entropy, and Gibbs free energy of biomass, cells and colonies of 32 microorganism species. Heliyon. 2019;5(6) doi: 10.1016/j.heliyon.2019.e01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Omicron BA.2.75 sublineage (centaurus) follows the expectations of the evolution theory: less negative Gibbs energy of biosynthesis indicates decreased pathogenicity. Microbiol. Res. 2022;13(4):937–952. MDPI AG. Retrieved from 10.3390/microbiolres13040066. [Google Scholar]
- Popovic M. Strain wars 3: Differences in infectivity and pathogenicity between Delta and Omicron strains of SARS-CoV-2 can be explained by thermodynamic and kinetic parameters of binding and growth. Microb. Risk Anal. 2022 doi: 10.1016/j.mran.2022.100217. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Strain Wars 4 - Darwinian evolution through Gibbs’ glasses: Gibbs energies of binding and growth explain evolution of SARS-CoV-2 from Hu-1 to BA.2. Virology. 2022 doi: 10.1016/j.virol.2022.08.009. [DOI] [PubMed] [Google Scholar]
- Popovic M. Strain wars 2: binding constants, enthalpies, entropies, Gibbs energies and rates of binding of SARS-CoV-2 variants. Virology. 2022;570:35–44. doi: 10.1016/j.virol.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Strain Wars 5: Gibbs energies of binding of BA.1 through BA.4 variants of SARS-CoV-2. Microb. Risk Anal. 2022 doi: 10.1016/j.mran.2022.100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Beyond COVID-19: do biothermodynamic properties allow predicting the future evolution of SARS-CoV-2 variants? Microb. Risk Anal. 2022;22 doi: 10.1016/j.mran.2022.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Omicron BA.2.75 subvariant of SARS-CoV-2 is expected to have the greatest infectivity compared with the competing BA.2 and BA.5, due to most negative Gibbs energy of binding. BioTech. 2022;11(4):45. doi: 10.3390/biotech11040045. MDPI AG. Retrieved from 10.3390/biotech11040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M. Atom counting method for determining elemental composition of viruses and its applications in biothermodynamics and environmental science. Comput. Biol. Chem. 2022;96 doi: 10.1016/j.compbiolchem.2022.107621. [DOI] [PubMed] [Google Scholar]
- Popovic M. Formulas for death and life: Chemical composition and biothermodynamic properties of Monkeypox (MPV, MPXV, HMPXV) and Vaccinia (VACV) viruses. Therm. Sci. 2022;26(6A) doi: 10.2298/TSCI220524142P. [DOI] [Google Scholar]
- Popovic M. Vol. 100236. 2022. Why doesn't Ebola virus cause pandemics like SARS-CoV-2? (Microb. Risk Anal.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. Coinfection and interference phenomena are the results of multiple thermodynamic competitive interactions. Microorganisms. 2021;9(10):2060. doi: 10.3390/microorganisms9102060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. A thermodynamic insight into viral infections: do viruses in a lytic cycle hijack cell metabolism due to their low Gibbs energy? Heliyon. 2020;6(5):e03933. doi: 10.1016/j.heliyon.2020.e03933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. Thermodynamic insight into viral infections 2: empirical formulas, molecular compositions and thermodynamic properties of SARS, MERS and SARS-CoV-2 (COVID-19) viruses. Heliyon. 2020;6(9):e04943. doi: 10.1016/j.heliyon.2020.e04943. 10.1016%2Fj.heliyon.2020.e04943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombel-Bryzek A., Miller A., Witkowska D. Thermodynamic analysis of the interactions between human ACE2 and spike RBD of Betacoronaviruses (SARS-CoV-1 and SARS-CoV-2) FEBS Open Bio. 2023;13(1):174–184. doi: 10.1002/2211-5463.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnati M., Chiodelli P., Bugatti A., Urbinati C. Bridging the past and the future of virology: surface plasmon resonance as a powerful tool to investigate virus/host interactions. Crit. Rev. Microbiol. 2015;41(2):238–260. doi: 10.3109/1040841X.2013.826177. [DOI] [PubMed] [Google Scholar]
- Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S., Madej T., Marchler-Bauer A., Lanczycki C., Lathrop S., Lu Z., Thibaud-Nissen F., Murphy T., Phan L., Skripchenko Y., Tse T., Sherry S.T. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50(D1):D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialo F., Daniele A., Amato F., Pastore L., Matera M.G., Cazzola M., Castaldo G., Bianco A. ACE2: the major cell entry receptor for SARS-CoV-2. Lung. 2020;198(6):867–877. doi: 10.1007/s00408-020-00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: from vision to reality. EuroSurveillance. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. PMCID: PMC5388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şimşek B., Özilgen M., Utku F.Ş. How much energy is stored in SARS-CoV-2 and its structural elements? Energy Storage. 2021:e298. doi: 10.1002/est2.298. [DOI] [Google Scholar]
- Von Bertalanffy L. The theory of open systems in physics and biology. Science. 1950;111(2872):23–29. doi: 10.1126/science.111.2872.23. [DOI] [PubMed] [Google Scholar]
- Von Bertalanffy L. George Braziller Inc; New York, NY: 1971. General System Theory: Foundations, Development, Applications. ISBN-13: 978-0807604533. [Google Scholar]
- Von Stockar U. In: Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering. von Stockar U., editor. EPFL Press; Lausanne: 2013. Live cells as open non-equilibrium systems; pp. 475–534. [DOI] [Google Scholar]
- Von Stockar U. In: Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering. von Stockar U., editor. EPFL Press; Lausanne: 2013. Biothermodynamics of live cells: energy dissipation and heat generation in cellular structures; pp. 475–534. [DOI] [Google Scholar]
- von Stockar U., Liu J. Does microbial life always feed on negative entropy? Thermodynamic analysis of microbial growth. Biochim. Biophys. Acta. 1999;1412(3):191–211. doi: 10.1016/s0005-2728(99)00065-1. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Iketani, S., Li, Z., Liu, L., Guo, Y., Huang, Y., & Ho, D. D. (2022). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. bioRxiv, 2022.11.23.517532. 10.1101/2022.11.23.517532. [DOI] [PMC free article] [PubMed]
- WHO . World Health Organization; 2022. TAG-VE Statement on Omicron Sublineages BQ.1 and XBB.https://www.who.int/news/item/27-10-2022-tag-ve-statement-on-omicron-sublineages-bq.1-and-xbb [Online]Available at: (Accessed on November 29, 2022) [Google Scholar]
- Wimmer E. The test-tube synthesis of a chemical called poliovirus. The simple synthesis of a virus has far-reaching societal implications. EMBO Rep. 2006 doi: 10.1038/sj.embor.7400728. 7 Spec No(Spec No), S3–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B., Ercan S., Akduman S., Özilgen M. Energetic and exergetic costs of COVID-19 infection on the body of a patient. Int. J. Exergy. 2020;32(3):314–327. 10.1504/IJEX.2020.10030515. [Google Scholar]
- Zappa M., Verdecchia P., Angeli F. The new phase of pandemic: are BA.2.75 and BQ.1 competitive variants? An in silico evaluation. Eur. J. Intern. Med. 2022 doi: 10.1016/j.ejim.2022.11.006. S0953-6205(22)00389-2. Advance online publication. [DOI] [PubMed] [Google Scholar]