Abstract
Lately, nitrogenous heterocyclic antivirals, such as nucleoside-like compounds, oxadiazoles, thiadiazoles, triazoles, quinolines, and isoquinolines, topped the therapeutic scene as promising agents of choice for the treatment of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections and their accompanying ailment, the coronavirus disease 2019 (COVID-19). At the same time, the continuous emergence of new strains of SARS-CoV-2, like the Omicron variant and its multiple sublineages, resulted in a new defiance in the enduring COVID-19 battle. Ensitrelvir (S-217622) is a newly discovered orally active noncovalent nonpeptidic agent with potential strong broad-spectrum anticoronaviral activities, exhibiting promising nanomolar potencies against the different SARS-CoV-2 variants. S-217622 effectively and nonspecifically hits the main protease (Mpro) enzyme of a broad scope of coronaviruses. Herein, in the present computational/biological study, we tried to extend these previous findings to prove the universal activities of this investigational agent against any coronavirus, irrespective of its type, through synchronously acting on most of its main unchanged replication enzymes/proteins, including (in addition to the Mpro), e.g., the highly conserved RNA-dependent RNA polymerase (RdRp) and 3′-to-5′ exoribonuclease (ExoN). Biochemical evaluation proved, using the in vitro anti-RdRp/ExoN bioassay, that S-217622 can potently inhibit the replication of coronaviruses, including the new virulent strains of SARS-CoV-2, with extremely minute in vitro anti-RdRp and anti-RdRp/ExoN half-maximal effective concentration (EC50) values of 0.17 and 0.27 μM, respectively, transcending the anti-COVID-19 drug molnupiravir. The preliminary in silico results greatly supported these biochemical results, proposing that the S-217622 molecule strongly and stabilizingly strikes the key catalytic pockets of the SARS-CoV-2 RdRp’s and ExoN’s principal active sites predictably via the nucleoside analogism mode of anti-RNA action (since the S-217622 molecule can be considered as a uridine analog). Moreover, the idealistic druglikeness and pharmacokinetic characteristics of S-217622 make it ready for pharmaceutical formulation with the expected very good clinical behavior as a drug for the infections caused by coronaviruses, e.g., COVID-19. To cut it short, the current critical findings of this extension work significantly potentiate and extend the S-217622’s previous in vitro/in vivo (preclinical) results since they showed that the striking inhibitory activities of this novel anti-SARS-CoV-2 agent on the Mpro could be extended to other replication enzymes like RdRp and ExoN, unveiling the possible universal use of the compound against the next versions of the virus (i.e., disclosing the nonspecific anticoronaviral properties of this compound against almost any coronavirus strain), e.g., SARS-CoV-3, and encouraging us to rapidly start the compound’s vast clinical anti-COVID-19 evaluations.
1. Introduction
For the successive 4th year since 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections remain blazing across the planet, causing more and more coronavirus disease 2019 (COVID-19) cases around the globe. Searching for very potent antiviral agents that effectively stop or, at least, disrupt SARS-CoV-2 transmission, cellular entry, progressive replication, and pathogenicity is the major concern shared by all relevant scientists worldwide to combat this resistant COVID-19 disease.1−7 Agents that primarily act to fulfill the above-mentioned antireplication requirement are comparatively very few to date. Principally, compounds having nitrogenous heterocyclic aromatic cores in their scaffolds, e.g., nucleoside-like compounds (nucleoside analogs), oxadiazoles/thiadiazoles, triazoles, quinolines/isoquinolines, and some polyphenolics, have presented significant successful lead and superiority as SARS-CoV-2 inhibitors and coronavirustatic/coronavirucidal agents, with the two FDA-approved medications molnupiravir and nirmatrelvir on top.2,5,6,8−24
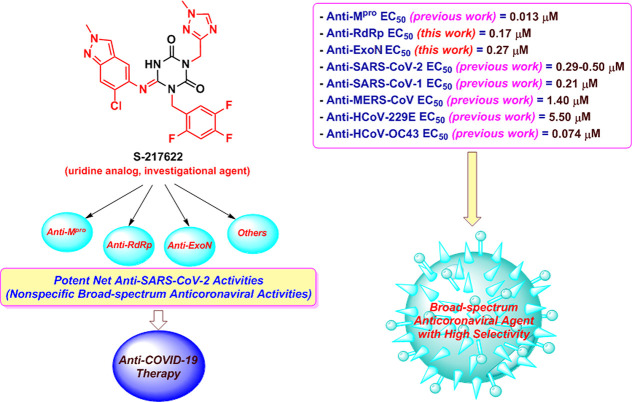
Ensitrelvir (S-217622) (Figure 1A) is an anticoronaviral uridine/pyrimidine analog (as it can be seen as a derivative of the nucleoside uridine, containing a slightly modified uracil base which is one of the four RNA nucleobases) with a nonpeptidic nature. It was recently discovered by Unoh and colleagues and reported in the Journal of Medicinal Chemistry as a very potent orally active noncovalent anti-SARS-CoV-2 agent.16 This chemical was primarily proposed through computational drug design/optimization strategies followed by its successful experimental synthesis and biological evaluation.16 Chemically, the S-217622 molecule (its IUPAC name is (6E)-6-[(6-chloro-2-methyl-2H-indazol-5-yl)imino]-3-[(1-methyl-1H-1,2,4-triazol-3-yl)methyl]-1-(2,4,5-trifluorobenzyl)-1,3,5-triazinane-2,4-dione) is of relatively smaller molecular weight (about 531.88 Da) and volume when compared to its corresponding larger peptidic analogs, and it is characterized by being rich in heterocyclic rings and nitrogen atoms (along with fluorine atoms), which are the reasons for the optimized anticoronaviral activities as compared to its parent molecules.16 According to Unoh-and-colleagues’ paper, S-217622 is effective against almost all coronaviral strains known to date.16 The in vitro biochemical assays revealed the extraordinary nanomolar potencies of this compound against the diverse strains of coronaviruses, displaying anticoronaviral half-maximal effective concentration (EC50) values of 13, 290–500, 210, 1400, 5500, and 74 nM against the main protease (Mpro, also called 3-chymotrypsin-like protease “3CLpro” or nonstructural protein 5 “nsp5”), SARS-CoV-2, severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1), Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus 229E (HCoV-229E, which is a member of the viral genus Alphacoronavirus), and human coronavirus OC43 (HCoV-OC43, which is a member of the viral genus Betacoronavirus), respectively.16 Surprisingly, S-217622 also exhibited very potent broad-spectrum efficacy against all of the different strains of SARS-CoV-2 itself, e.g., the wild/Wuhan-type or WT strain (WK-521; EC50 = 370 nM), Alpha strain (QK002; EC50 = 330 nM), Beta strain (TY8-612; EC50 = 400 nM), Gamma strain (TY7-501; EC50 = 500 nM), Delta strain (TY11-927-P1; EC50 = 410 nM), and Omicron strain (TY38-873; EC50 = 290 nM).16 To express the inhibitory effects of S-217622 on all of these diverse strains of coronaviruses, Unoh-and-colleagues’ paper used mainly the 50% cytopathic effects (CPE) effective concentration (CPEEC50), which is the lowest concentration of the tested compound S-217622 that resulted in 50% reduction of the CPE caused by the coronavirus.16 Encouragingly, S-217622 has a very important clinically strategic advantage of not noticeably affecting the human host cells since it showed no inhibitory activities on the human cell proteases at all (even in very high concentrations of more than 100 000 nM, i.e., the S-217622 molecule has very high favorable selectivity for coronaviral proteases).16 S-217622 is also characterized by having a nonpeptidic scaffold, which supports and boosts the clinical success of the compound with improved oral bioavailability/bioactivity, membrane permeability, target selectivity, nanomolar potency, dose-dependent anticoronaviral effectiveness, metabolic stability, safety margin, and pharmacokinetic profiles, along with optimized molecular size/weight and adjusted intrinsic nature of the molecular reactivity, to overcome the numerous traditional disadvantages of the classical covalent peptide-like coronaviral Mpro inhibitors (which usually have larger molecular size).25,26 Although the S-217622 molecule is moderately similar to nucleos(t)ide analogs (i.e., atypical nucleoside analog) from the structural point of view, it can be mainly considered as an anti-SARS-CoV-2 agent of the nucleosidic type.
Figure 1.
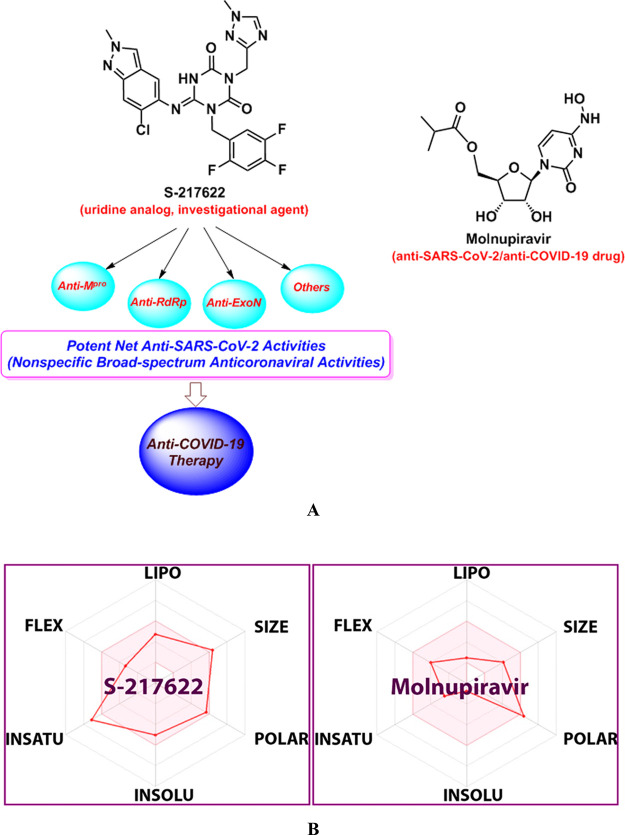
(A) Chemical structures of the investigational anticoronaviral agent S-217622 (along with its expected comprehensive extended and broad-spectrum anticoronaviral activities against the diverse nonspecific proteins of coronaviruses) and the reference anti-SARS-CoV-2 drug molnupiravir, respectively. (B) Comparative physicochemical radar (preliminary in silico pharmacokinetics/druglikeness assessment; LIPO: Lipophilicity, INSOLU: Insolubility, INSATU: Insaturation, and FLEX: Flexibility) of S-217622 versus molnupiravir, generated using the SwissADME webserver.
When we are planning to obtain nonspecific and broad universal targeting of any coronavirus irrespective of its type or mutated version (i.e., if we are seeking to get broad-spectrum anticoronaviral agents effective against all or most coronaviral strains), hitting the changeable spike (S) proteins would not be that efficient or attractive approach. On the other hand, targeting the comparatively universal or conserved (fixed) coronaviral proteins which have no significant mutations near the principal catalytic centers, e.g., proteolysis Mpro, replication RNA-dependent RNA polymerase (RdRp), and proofreading 3′-to-5′ exoribonuclease (ExoN) enzymes, is much more efficient and time-saving tactic in these frequent battles, even against the prospectively coming resistant coronaviral strains. Moreover, drugs targeting the rapidly mutated S protein have only one chance (or very few chances) to hinder the coronaviral-2 infection since after the passage of any viral particles inside the host body (or if these therapies were taken after the occurrence of the infection), there will not be any further abilities of these therapies to stop virus propagation and infection, unlike medications targeting the proteolysis, replication, and proofreading proteins, which have an unlimited number of continuous chances to stop the virus and its successors (its offspring) and prevent their further multiplication throughout the entire human body (even if these therapies were taken after the occurrence of the infection).
Mpro (along with other minor proteases like papain-like protease “PLpro”, which is the protease domain of nonstructural protein 3 “nsp3”) plays a crucial role in the coronaviral replication through generating a functional replicase complex to enable viral particles spread in the infected cells, and its inhibition will eventually have corresponding inhibitory effects on the formation of essential replication enzymes like RdRp (which needs, prior to becoming fully functional, to be proteolytically released, which is mainly induced and regulated by Mpro);16 however, synchronously inhibiting several coronaviral replication enzymes (not only Mpro) have beneficial striking synergistic antimultiplication effects on the virus due to many reasons. Of these several reasons, there are three major important reasons. First, before administering the Mpro inhibitor into the infected patient’s biological system, there are already-formed replication enzymes, like RdRp and ExoN, that will remain present and effectively act to reproduce the viral particles, even after impairing Mpro activity. Second, inhibition and disruption of Mpro by its reversible noncovalent chemical ligands (inhibitors) will never reach a 100% blockade with the safe therapeutic doses taken of the inhibitor, thus relatively considerable amounts of the replication-essential enzymes like RdRp will be adequately released and will be fully functional. Third, integrative disruption of the complicated replication system of coronavirus guarantees and ensures to a far extent that the virus will never be adequately replicated in the infected patient’s biological system soon. Based on all of the previous facts, this condensed anti-SARS-CoV-2-replication strategy has been suggested in the current research work.
As clearly seen in Figure 1A, one of the prominent major differences between the structure of S-217622 molecule and that of molnupiravir molecule is the excessive or substantial aromatic fluorine substitution (three fluorine atoms) in the 217622 molecule. Many investigational anti-COVID-19 agents/medicines of the anti-RdRp category (e.g., favipiravir, 4′-fluorouridine, AT-527, and sofosbuvir) are characterized by their fluorinated skeletons.27 Although fluorine atoms/ions are absent in almost all biological systems, they were recently used/benefited to tailor the pharmacological behaviors of some designed/repurposed compounds for an improvement of their anti-SARS-CoV-2/anti-COVID-19 therapeutic efficacies.27 This therapeutic improvement may be specifically related to the fruitful effects fluorosubstitution exerts on the structural conformation, membrane permeability, metabolic directions, target protein binding, and overall pharmacokinetic properties of these compounds or drugs.27
In the therapeutic antiviral nucleoside mimicry strategy, we mainly draw on the similarity degree of the designed nucleoside-like compound with the normal endogenous human RNA nucleosides/nucleotides to mislead and deceive the SARS-CoV-2 RdRp (the nonstructural protein complex 12/7/8 or nsp12–nsp7–nsp8) and ExoN (the nonstructural protein complex 14/10 or nsp14–nsp10) enzymes.28 In the current research work, we have tried to investigate the possible combined inhibitory activities of this new promising anti-COVID-19 agent, S-217622, on other SARS-CoV-2 proteins (i.e., other than the already-confirmed blocking activities on Mpro), like RdRp and ExoN enzymes,29 as a novel effective approach to close all avenues for the coronavirus to reproduce with a view to synergistically and thoroughly battle COVID-19, SARS, MERS, and potentially all coming coronaviral infections and diseases of the next versions of the virus, hence also proving the universal broad-spectrum antiviral nature of this investigational agent on coronaviruses and their viral successors of the RNA type (Figure 1A). The extensive dual computational–biochemical evaluation approach was used for this striking mission, employing molnupiravir as a potent positive control (reference) anticoronaviral drug (Figure 1A). Physicochemically, the S-217622 molecule exhibits excellent and temperate in silico pharmacokinetic properties (e.g., relatively moderate molecular size, balanced high molecular flexibilities, balanced lipophilic/hydrophilic characteristics, excellent gastrointestinal (GI) absorption, and favorable solubilities; as illustrated in the physicochemical radar of Figure 1B)30 that significantly uphold its druglikeness, pharmaceutical formulation, and clinical use potentials (even much more than the corresponding properties of molnupiravir; see Figure 1B).
2. Materials and Methods
2.1. In Silico Computational Evaluation Methodology
2.1.1. Preparation of the Selected Target Coronaviral-2 Proteins
The 3D structures of the target SARS-CoV-2 RdRp and ExoN proteins were obtained from the RCSB Protein Data Bank (PDB) with PDB identification codes 7BV2 and 7MC6, respectively. Both enzymatic proteins were obtained in the complex forms with their protein cofactors (i.e., they were obtained cocrystallized in the nsp12–nsp7–nsp8 and nsp14–nsp10 complex forms, respectively) to increase nature simulation. The PDB files of the two proteins were properly downloaded. Proteins were viewed through Pymol Molecular Graphic Visualizer software 2.4, and their predetected active site residues (with their closest neighboring residues) were then checked for complete presence and correctness. The catalytic active site residues highlighted through Pymol software were noted for the next in silico studies.
2.1.2. S-217622 and Molnupiravir Ligands Preparation
Prior to beginning the different procedures of the comprehensive computational study of the target compound S-217622 (versus the reference drug molnupiravir) against SARS-CoV-2 RdRp and ExoN enzymes using the Molecular Operating Environment (MOE) platform (Chemical Computing Group), the chemical structures of S-217622 and molnupiravir were appropriately prepared and validated using ChemDraw Professional 16.0 software (authorized new version) to be eligible for the following in silico investigations.
2.1.3. Molecular Docking Protocol
Blind docking of the selected two molecules, S-217622 and molnupiravir, in SARS-CoV-2 RdRp and ExoN proteins was performed via MOE. Molnupiravir was used as a positive control anti-SARS-CoV-2 reference, having proven potent RdRp/ExoN inhibitory activities. Prior to starting these docking procedures, some important preparations (mainly additions and corrections) are required. All of the missed atoms/residues in the SARS-CoV-2 RdRp and ExoN structures were added via MOE structure modeling. The aforementioned two proteins were precisely prepared to be valid and ready for the different molecular docking processes by the proper addition of hydrogen atoms to their structures using the 3D-protonation module of the used MOE software; any partial charges were also corrected for both proteins. RdRp and ExoN were energy minimized in their complex forms via the Amber-99 force field, which is available in MOE. Similarly, the structures of S-217622 and molnupiravir were also adequately energy minimized in MOE. For docking of the target/reference ligands with the two proteins, the known London-dG scoring functions were utilized for binding energy calculations. For the docked target/reference molecule, the MOE software produced about 20 different poses with each docked SARS-CoV-2 protein. Of all of the docking poses for each molecule with each protein, the one with the highest number of best molecular interactions, i.e., the top-ranked pose of the best interactions, was recorded and saved. Please note that MOE gives a numerical value for the interaction of any candidate ligand with any certain protein in the form of a docking S-score (docking scores are expressed in kcal/mol). This docking binding energy or S-score represents the net energy of the formed protein–ligand complex, and it also primarily reflects the degree of its expected stability (i.e., it provides a primary idea about the predicted stability of this formed complex prior to performing the more detailed robust computations via the molecular dynamics “MD” simulations). It should also be noted that the results of the computational molecular docking are the primary starting point for the present research. MOE software shows all of the possible molecular interactions (of all types) made during the docking process; these include, e.g., hydrogen bonding (H-bonds), hydrophobic interactions, ionic interactions/bonds, and salt bridges. For the two ligands, S-217622 and molnupiravir, respectively, the 2D and 3D output images of all of the produced protein–ligand complexes (showing almost all of the possible interactions, especially in the 3D images) were saved for reporting and further investigative analysis.
2.1.4. Molecular Dynamics (MD) Simulation Protocol
After organizing and storing the top computational docking results of S-217622 and molnupiravir, e.g., the best molecular interactions, lowest docking score (S-score), and lowest root-mean-square deviation (RMSD), computed through MOE and the apo-enzyme against both proteins, the two ligands were then employed for further in silico studies, mainly the MD simulation studies, using Schrodinger’s Desmond module MD-Simulation software. For MD simulation of the target compound, S-217622, and the reference drug, molnupiravir, the stored best docking poses of these ligands in complexes with the SARS-CoV-2 RdRp and ExoN enzymes were kept in PDB format in MOE to be used for further virtual stability studies in Schrodinger’s Desmond module. The in-built Desmond System Builder tool was used in this current protocol to create the solvated water-soaked MD-Simulation system. The TIP3P model was utilized as the solvating model in the present experiment. With periodic boundary conditions, an orthorhombic box was accurately simulated with a good boundary distance of at least 10 Å from the outer surface of each of the two coronaviral-2 proteins. The simulation systems were neutralized of complex charges by the addition of a reasonably sufficient amount of counter ions. The isosmotic state was maintained by adding 0.10 mol/L sodium and chloride ions, i.e., 0.10 M NaCl, into the simulation panel. Prior to beginning the simulation process, a predefined equilibration procedure was done. The system of the MD simulation was equilibrated by employing the standard Desmond protocol at a constant pressure of about 1.0 bar and a constant temperature of 300 K (NPT ensemble; considering the viral nature of the two target enzymatic proteins) and also by employing the known Berendsen coupling protocol with one temperature group. H-bond length was properly constrained using the validated SHAKE algorithm. The particle Mesh Ewald (PME) summation method was used to specifically model long-range electrostatic interactions. On the other hand, an exact cutoff of 10 Å was specifically assigned for van der Waals and short-range electrostatic interactions. As previously mentioned, the MD simulation was run at ambient pressure conditions of a value of 1.013 bar while the used temperature was exactly set to 300 K for each 100 ns period of this MD simulation, and 1000 frames were saved into the simulation trajectory file. The simulation run time for each complex system and apo system was fixed to 100 ns as a total. After simulations, the trajectory file of the simulated system was used for the calculation of the various structural parameters required, e.g., RMSD (Å), root-mean-square fluctuation (RMSF; Å), the radius of gyration (rGyr; Å), number of protein–ligand contacts (# of total contacts), interactions fractions (%), intermolecular H-bonds (from all aspects), molecular surface area (MolSA; Å2), solvent-accessible surface area (SASA; Å2), and polar surface area (PSA; Å2), to extensively perform the stability studies of the complex and apo systems. The results of the promising target compound S-217622 along with those of the reference drug molnupiravir were saved to be reported, discussed, and compared in the present paper.
2.2. In Vitro Biological Evaluation Methodology
2.2.1. Specifications of the Bioassayed Compounds
A free small sample of the target compound S-217622 (for scientific research purposes only) was obtained from the Shionogi Pharmaceutical Research Center, Osaka 561-0825, Japan (purity: ≥96%).16 While the reference anti-COVID-19 drug molnupiravir (EIDD-2801, CAS Registry Number: 2349386-89-4) was purchased from Biosynth Carbosynth (Carbosynth Ltd., Berkshire, U.K.) (Product Code: AE176721, purity: ≥98%). The ultrapure solvent dimethylsulfoxide (DMSO, CAS Registry Number: 67-68-5) was purchased from a local distributor, El-Gomhouria Company For Drugs (El-Gomhouria Co. For Trading Drugs, Chemicals & Medical Supplies, Mansoura Branch, Egypt) (purity: ≥99.9% “anhydrous”).
2.2.2. In Vitro Anti-RdRp/Anti-ExoN Assay (SARS-CoV-2-RdRp-Gluc Reporter Assay) of the Target Compound S-217622
It is very important to know that RdRp and ExoN proteins are almost the same or very similar in most coronaviruses (e.g., HCoVs, MERS-CoV, SARS-CoV-1, and nearly all SARS-CoV-2 strains) due to the high degree of conservation and universality of these coronaviral proteins among all coronavirus species, and that is why the results of this assay are very reflective/representative (expressive) in all coronaviruses (i.e., this biochemical assay is very suitable for testing compounds as prospective broad-spectrum anticoronaviral agents).31 The first step of this assay involves the careful keeping of the used cells, 293T cells (ATCC CRL-3216), in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 10% (v/v) fetal bovine serum (FBS; Gibco). In the next step, these cells were cultured at 37 °C in a humidified atmosphere of CO2 (5%). After that, HEK293T cells were transfected using Vigofect transfection reagents (Vigorous) according to the strict instructions of the manufacturer. The required plasmid DNAs, antibodies, and reagents were purchased and treated exactly as in the literature procedures.32,33 The tested investigational agent S-217622, the positive control molnupiravir, and the negative control DMSO are as described in Subsection 2.2.1. Also, western blotting (for the collected transfected HEK293T cells), real-time RT-PCR (for the extracted total RNA of transfected HEK293T cells), and cell viability test (using Cell Counting Kit-8 (CCK8), Beyotime) were exactly performed as in the typical procedures of the literature.32,33 The steps of the well-designed in vitro SARS-CoV-2-RdRp-Gluc Reporter Assay were accurately carried out according to the same original method of literature but with almost all of the proteins modified and made relevant to the SARS-CoV-2 Omicron variant “B.1.1.529/BA.1 sublineage” (HEK293T cells were transfected in this biochemical assay with CoV-Gluc, nsp12, nsp7, and nsp8 plasmid DNAs at the ratio of 1:10:30:30, and with CoV-Gluc, nsp12, nsp7, nsp8, nsp10, and nsp14 plasmid DNAs at the ratio of 1:10:30:30:10:90).32,33 Exactly as instructed in the original assay, a stock of coelenterazine-h was dissolved in absolute ethanol (of very pure analytical grade) to a concentration of 1.022 mM.32,33 Directly before each assay, the stock was diluted in phosphate-buffered saline (PBS) to a concentration of 16.7 μM and incubated in the dark for 30 min at room temperature.32,33 For the proper measurement in the luminescence assay, 10 μL of the supernatant was added to each well of a white and opaque 96-well plate, then 60 μL of 16.7 μM coelenterazine-h was injected, and luminescence was measured for 0.5 s using the Berthold Centro XS3 LB 960 microplate luminometer.32,33 Final results were statistically represented as the mean (μ) ± standard deviation (SD) from at least three independent experiments (i.e., triplicates). Statistical analysis was performed using SkanIt 4.0 Research Edition software (Thermo Fisher Scientific) and Prism V5 software (GraphPad). All resultant data were considered statistically significant at p < 0.05.
3. Results and Discussion
3.1. Computational Molecular Modeling of S-217622 as a Potential Anti-RdRp-ExoN-System (Anti-CoV/Anticoronaviral) Drug
General preliminary and preparatory computational screening of the new synthetic compound S-217622 using many in silico techniques revealed its potential ideal pharmacodynamic/pharmacokinetic characteristics with respect to the predicted anti-SARS-CoV-2 activities (e.g., see Figure 1B). In the next step, further molecular docking, specifically against SARS-CoV-2 RdRp and ExoN, unveiled that the target compound S-217622 has very good low inhibitory binding energies (ranging from about −6.9 kcal/mol against RdRp to −8.7 kcal/mol against ExoN), which are significantly comparable to the reference anti-RdRp/anti-ExoN drug molnupiravir (which has binding energies ranging from about −6.7 kcal/mol against RdRp to −7.1 kcal/mol against ExoN), as shown in Table 1. The catalytic pockets (i.e., active sites) of the two coronaviral-2 enzymes, RdRp (which is the key enzyme responsible for replication and transcription of the coronaviral RNA genome) and ExoN (it is important to recall that nsp14 or the proofreading exoribonuclease of coronavirus has two active sites; the exoribonuclease active site, the principal one that we are concerned with in the current study, and the methyltransferase active site), were nearly explored and fully detected through many preceding computational, crystallographic, and biochemical experiments in the literature.34−37 Investigating and analyzing the resultant in silico interactions of the S-217622 molecule with the residues of SARS-CoV-2 RdRp and ExoN enzymes showed that this molecule significantly strikes most of the active amino acid residues of the catalytic pockets of both enzymes with strong interactions (of gradual forces), including, mainly, H-bonds, hydrophobic interactions, ionic bonds, and water bridges, of comparatively short bond distances and low binding energies.
Table 1. Binding Affinity Energy Values (Docking S-Scores) Estimated During Molecular Docking of the Screened S-217622 and Molnupiravir (the Positive Control Drug), Respectively, Against the Two SARS-CoV-2 Proteins, RdRp (PDB Code: 7BV2) and ExoN (PDB Code: 7MC6) Enzymes.
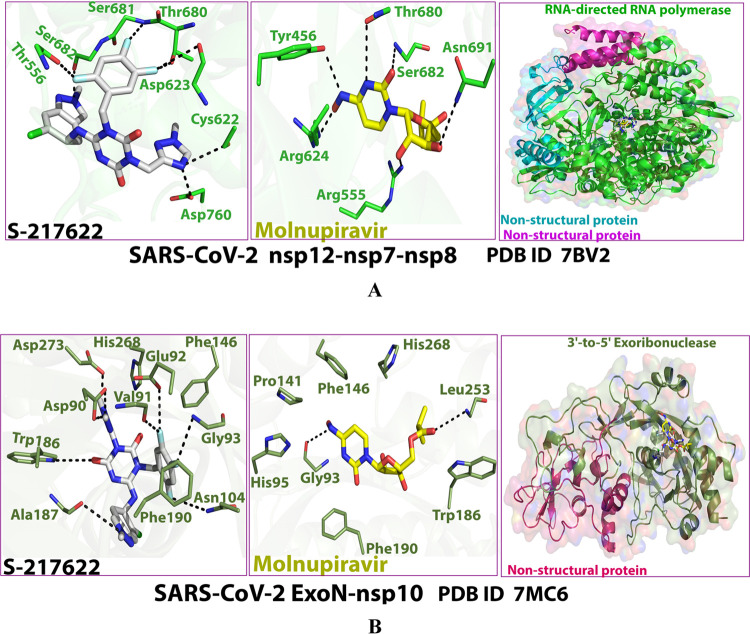
For the itemized protein–compound interactions, Figures 2A,B and 3A,B show the detailed 2D and 3D representations of the most intelligible intermolecular interactions between each ligand of the two ones (including the reference RdRp-ExoN system ligand, molnupiravir) with each of the two coronaviral-2 enzymes, respectively. The 2D representations generally display the surrounding and interacting pocket(s) of amino acids of each protein with each ligand, while the 3D representations specifically focus mainly on displaying the shortest/strongest bonds. Interestingly, the S-217622 molecule strongly hits most of the neighboring active residues of the major catalytic pocket of SARS-CoV-2 RdRp (in chain A, i.e., 7BV2-A receptor), e.g., Asp452, Arg553, Arg555, Thr556, Val557, Ala558, Tyr619, Lys621, Cys622, Asp623, Arg624, Thr680, Ser681, Ser682, Thr687, Ala688, Asn691, Ser759, and Asp760 (interactions with the two catalytic amino acids Arg553 and Ala558 obviously appear in the MD simulation outcomes later). Surprisingly, the S-217622 molecule also powerfully interacts with most of the adjacent active residues of the major catalytic pocket (exoribonuclease active site) of SARS-CoV-2 ExoN (in chain A; QHD43415_13 receptor), e.g., Asp90, Val91, Glu92, Gly93, His95, Asn104, Pro141, Phe146, Trp186, Ala187, His188, Gly189, Phe190, Gln191, Asn252, Leu253, Gln254, His268, and Asp273 (interactions with the three catalytic amino acids Asn252, Leu253, and Gln254 clearly appear in the MD simulation outcomes later). All of the above-mentioned in silico interactions are very encouraging and very comparable to, or even in some cases considerably better than, those of the potent control drug molnupiravir with the same two enzymes.
Figure 2.
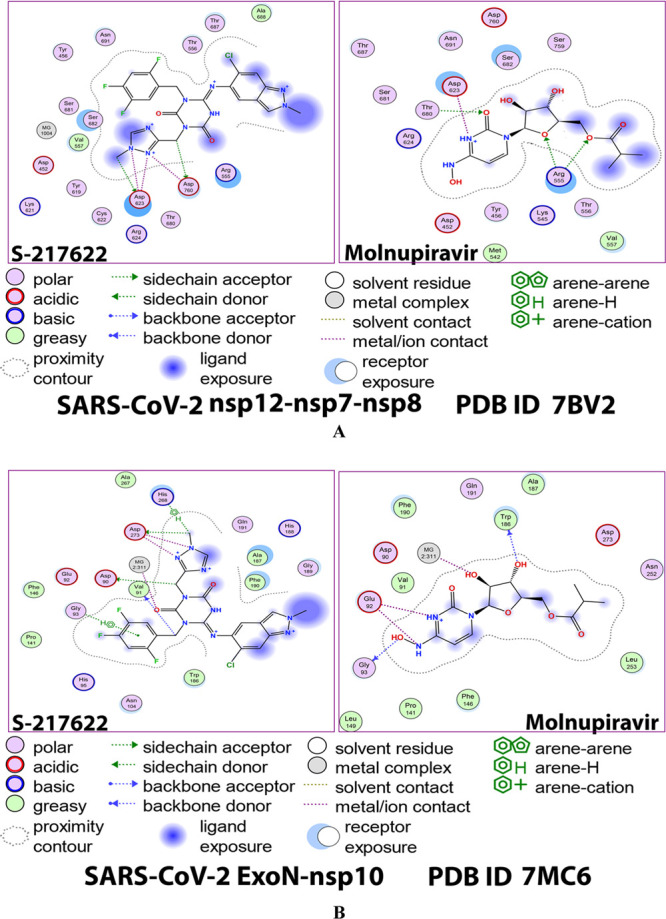
2D images of the postdocking interactions of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
Figure 3.
3D images of the postdocking interactions of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
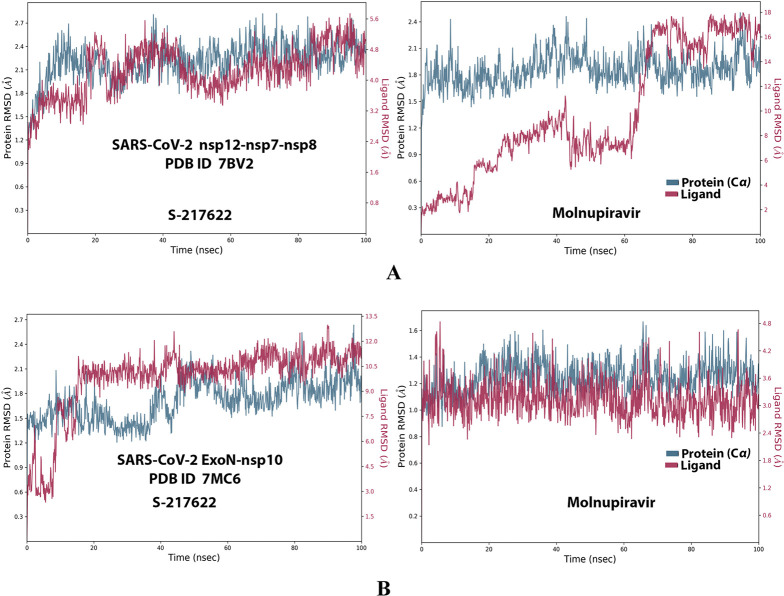
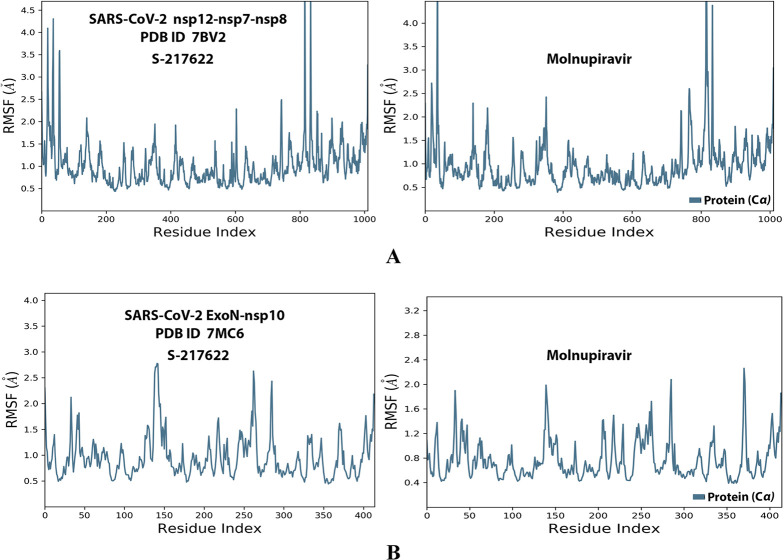
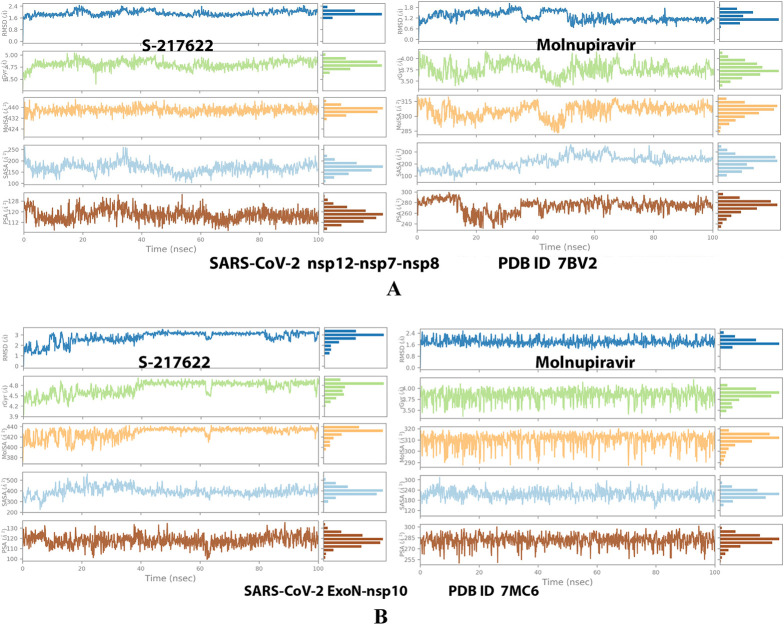
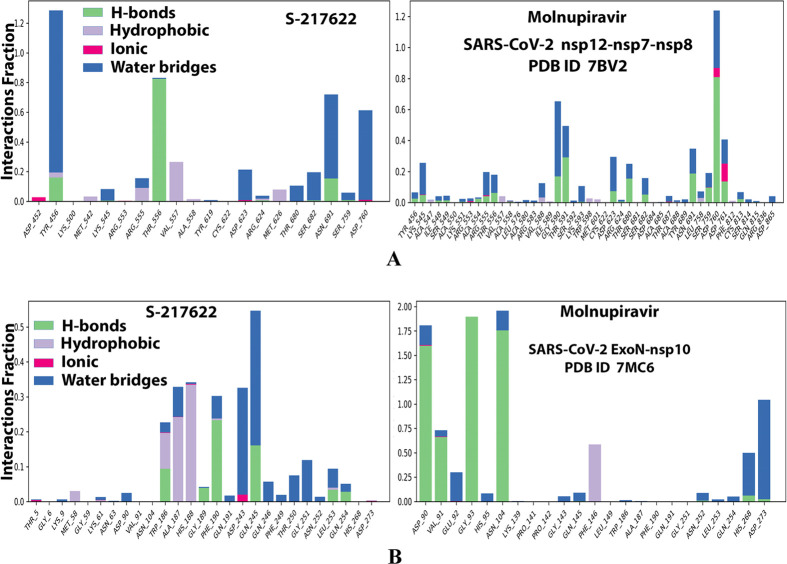
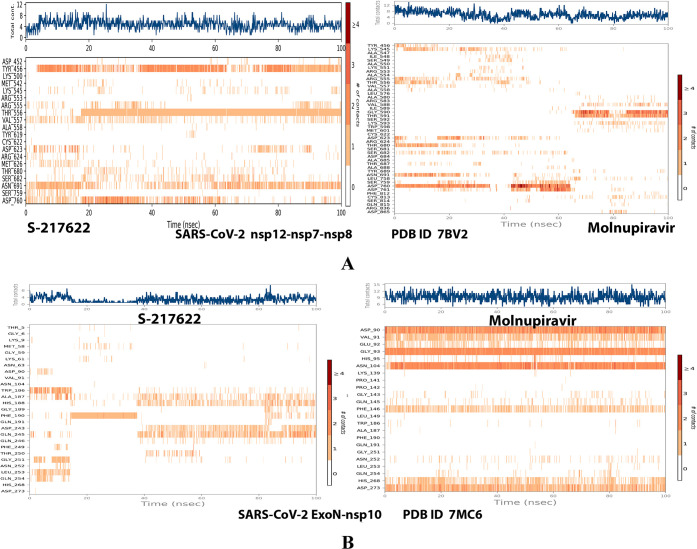
Analysis of the MD simulation results revealed the relative considerable stabilities of the formed RdRp–S-217622 and ExoN–S-217622 complexes. These stabilities are very comparable to the stabilities of the reference RdRp–molnupiravir and ExoN–molnupiravir complexes, respectively. The RdRp–S-217622 complex is more stable than the ExoN–S-217622 complex, and with comparatively less number/intensities of fluctuations than those of the ExoN–S-217622 complex. The RdRp–S-217622 complex is also more stable than the corresponding RdRp–molnupiravir complex, and as well with apparently less number/intensities of fluctuations and comparable RMSD and RMSF values (Å) than/to those of the RdRp–molnupiravir complex. Interestingly, S-217622 displayed encouraging superiority over molnupiravir in some of the compared MD items during the simulation. Comprehensively, the current in silico estimations showed that the RdRp–S-217622 and ExoN–S-217622 complexes are expected to be reasonably stable (i.e., stable with relatively acceptable degrees) in vivo (favorable clinical expectations). The few early fluctuations (which were very temperate) in RMSF and RMSD trajectories may be an indication of some forced conformational changes inside the enzymatic complex systems as a result of the appropriate redirecting and compatible repositioning of S-217622 molecules to harmonically fit within the catalytic binding sites, which take some nanotime till the formation of very interesting strong molecular binding interactions. Possible unrevealed allosteric modulations, especially in the case of the larger protein complex SARS-CoV-2 nsp12–nsp7–nsp8, should also be taken into account. The ExoN–S-217622 and RdRp–S-217622 complexes have close rGyr values (mostly around 4.8 Å), indicating similar degrees of compaction and stability in their produced protein–ligand systems. In addition, from the in silico point of view, the formed S-217622 complexes with both enzymes have very ideal and balanced MolSA, SASA, and PSA values (considerably lower than many of the corresponding values of the formed molnupiravir complexes with the same two enzymes). Interestingly, S-217622 displayed a large interactions fraction of H-bond(s) (of about 0.9% of the total binding interactions predicted) with the hit SARS-CoV-2 RdRp protein. This significantly strong binding interaction occurs specifically with the catalytic amino acid residue Thr556 of the large coronaviral-2 protein nsp12–nsp7–nsp8 (in the stable RdRp–S-217622 complex), indicating the worthy potentials of S-217622 to afford strongly inhibited/blocked statuses of the RdRp and ExoN enzymes. MD simulation results also confirmed nearly all of the primary molecular docking data in respect to, for example, the interacting amino acids along with the numbers/types/strengths of the formed bonds. Figures 4A,B, 5A,B, 6A,B, 7A,B, and 8A,B present the detailed results of MD simulation of the interactions between S-217622 and each of the two SARS-CoV-2 enzymes, RdRp and ExoN, respectively (in comparison with the reference FDA-approved anti-SARS-CoV-2 RdRp drug, molnupiravir). The previous computational findings were very motivating to encourage us to transfer to the experimental/biological assessment part of the current study.
Figure 4.
RMSD trajectories (during a simulation period of 100 ns) of the α-carbon of amino acid residues of the protein (blue color) and the ligand (maroon color) in the protein–ligand complexes of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
Figure 5.
RMSF trajectories (along the different residue regions) of the α-carbon of amino acid residues of the protein in the protein–ligand complexes of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
Figure 6.
Collective post-MD simulation analysis of the protein–ligand complexes properties (RMSD, rGyr, MolSA, SASA, and PSA) of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
Figure 7.
Histograms of the protein–ligand interactions fractions throughout the simulative interaction trajectories of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
Figure 8.
Plots of the distribution of the total number of interactions (contacts) in each trajectory framework of the protein–ligand complexes of S-217622 and molnupiravir, respectively, with: (A) SARS-CoV-2 RdRp “nsp12” enzyme cocrystallized with its protein cofactors nsp7 and nsp8 (PDB ID: 7BV2). (B) SARS-CoV-2 ExoN “nsp14” enzyme cocrystallized with its protein cofactor nsp10 (PDB ID: 7MC6).
3.2. Experimental Biological Evaluation of S-217622 as a Potential Anti-RdRp-ExoN-System (Anti-SARS-CoV-2/Anticoronaviral) Drug
The in vitro preclinical evaluation of the current mixed computational–experimental research study depends principally on the validated cell-based test, the in vitro anti-SARS-CoV-2-RdRp/ExoN biochemical assay, which was latterly developed using Gaussia-luciferase (Gluc) as the reporter to assess the anticoronaviral-2 RdRp activity of mainly the nucleoside analogs (the prodrugs of nucleotides and/or the nucleotide analogs) and similar nitrogenous heterocyclic aromatic derivatives.32,33 Additionally, the resultant data of the current work were adequately organized to clearly display the estimated effects of the target anticoronaviral candidate S-217622 on the coronaviral-2 ExoN activity too, making the protocol of this assessment assay very ideal for screening and exploring the potential dual-action SARS-CoV-2 RdRp/ExoN inhibitors (or, generally, the potential dual coronaviral RdRp/ExoN inhibitors, according to the proteins used in the assay).
As formerly mentioned, we chiefly focus in this preclinical research study on exploring the possible abilities of S-217622 to effectively inhibit enzymes that are controlling and activating the coronaviral multiplication, other than Mpro, specifically the two major protein complexes that catalyze and control the SARS-CoV-2 replication/transcription processes, nsp12–nsp7–nsp8 polymerase complex and nsp14–nsp10 exoribonuclease complex. This is why the current assay was designed to be significantly analogical to the corresponding original replication processes that occur for the SARS-CoV-2 genome, as it functionally mimics the RNA bioprocessing steps driven and proofread mainly by the SARS-CoV-2 RdRp and ExoN, respectively.38−40Table 2 displays the detailed values obtained from this in vitro anti-SARS-CoV-2-RdRp/ExoN bioassay. The obtained data showed that S-217622 demonstrated better inhibitory activities than the reference molnupiravir. Experimentally, S-217622 inhibited SARS-CoV-2 RdRp and RdRp/ExoN activities with very good small EC50 values of 0.17 and 0.27 μM, respectively, indicating the strong inhibitory/blocking activities of S-217622 on the SARS-CoV-2 replicating/proofreading (RdRp/ExoN) system. These potent activities, in turn, elucidate that S-217622 might act on SARS-CoV-2 RdRp and ExoN enzymes using the nucleoside analogism mechanism as we previously suggested in the study design and rationale. Mutations in the ExoN protein (i.e., the mutated version of the ExoN enzyme; e.g., mutations of the major active catalytic residues Asp90/Glu92 in nsp14 as in our current case) reinforced the anti-RdRp/ExoN activity of S-217622 to a very interesting EC50 value of 0.22 μM (i.e., slightly lower than that resulted in the presence of the normal wild type of ExoN; this very slight change also reflected, as mentioned above, the potent activities of this investigational anticoronaviral agent against the SARS-CoV-2 ExoN in its unmutated natural type from the beginning prior to implementing any intended mutations for testing and evaluation, and this slight decrease concurrently proves that S-217622 can easily resist and accommodate any mutations of the coronavirus and its principal enzymes, hence the universal broad-scope antiviral nature of this new therapeutic agent). These previous experimental values of the anti-SARS-CoV-2-RdRp/ExoN activities of S-217622 even beat those of the potent reference anti-SARS-CoV-2/anti-COVID-19 agent, molnupiravir, which showed higher inhibitory concentrations (EC50 values of 0.24, 0.46, and 0.35 μM, respectively), pointing out to the expected superiority of S-217622 over molnupiravir in clinical investigations. The findings also proved that molnupiravir comparatively could not resist/inhibit the catalytic activities of the Omicron variant ExoN with the same efficiency and potency S-217622 does against this coronaviral enzyme.
Table 2. Anti-SARS-CoV-2-RdRp/ExoN Activities (along with Respective Ratios) of the Target Synthetic Compound S-217622 (Using Molnupiravir as the Positive Control/Reference Drug and DMSO as the Negative Control/Placebo Drug) in HEK293T Cells, Expressed as EC50 Values in μMa.
| inhibition
of SARS-CoV-2 RdRp in vitro (EC50 in μM)b |
respective
ratios of EC50 |
|||||
|---|---|---|---|---|---|---|
| classification | compound name | Nsp12 | Nsp12 + Nsp14 | Nsp12 + Nsp14mutant | (Nsp12 + Nsp14)/Nsp12 | (Nsp12 + Nsp14mutant)/Nsp12 |
| target agent | S-217622 | 0.17 ± 0.02 | 0.27 ± 0.03 | 0.22 ± 0.03 | 1.59 | 1.29 |
| reference drug | molnupiravir | 0.24 ± 0.04 | 0.46 ± 0.06 | 0.35 ± 0.04 | 1.92 | 1.46 |
| placebo solvent | DMSO | >100 | >100 | >100 | N.A.c | N.A. |
nsp12 refers to the nsp12/7/8 complex, nsp14 refers to the nsp14/10 complex, and nsp14mutant refers to the nsp14mutant/10 complex.
EC50 or 50% effective concentration is the concentration of the tested compound that is required for 50% reduction in the COVID-19 polymerase (SARS-CoV-2 RdRp) activity in vitro. EC50 is expressed in μM.
N.A. means not available or not applicable (i.e., it was not determined).
It is apparently observed from the important data in Table 2 that as much as the EC50 values of the investigated compound (potential inhibitor) against the polymerase alone and against the polymerase in the presence of the exoribonuclease are close to each other, the more potent this inhibitor/blocker is (i.e., as more predicted for this investigated compound to be an ideally effective anti-SARS-CoV-2 replication). According to the results demonstrated in Table 2, S-217622 exhibited higher resistance than molnupiravir against the coronaviral-2 nsp14 exoribonuclease activity in HEK293T cells. The very promising capabilities of S-217622 to disrupt the nsp12 polymerase and nsp14 exoribonuclease activities of the coronaviral-2 Omicron variant interestingly boost the therapeutic potentials of S-217622 to succeed in the clinical settings as a potent anti-COVID-19 drug. In addition, the broad-scope activities of S-217622 against the several universal fixed coronaviral enzymes (such as Mpro, RdRp, and ExoN) significantly potentiate and support the potential use of this compound as a universal broad-spectrum inhibitor against most coronaviral infections in general. The present biochemical outcomes concerning the potent inhibitory SARS-CoV-2 RdRp-binding and ExoN-binding properties of S-217622 are in an ideal correspondence with almost all of the computed parameters of the prior in silico part of this comprehensive research, which was discussed in details in Subsection 3.1.
4. Conclusions and Future Therapeutic Applications
In the last weeks, synthetic and/or natural nitrogenous heterocyclic aromatic compounds with potent antiviral activities topped the scene as first and excellent choices as coronavirucidals for COVID-19 therapy. S-217622 is a newly designed synthetic uridine derivative with exceptional strong inhibitory effects against almost all of the species of infectious coronaviruses (e.g., HCoV-OC43, HCoV-229E, MERS-CoV, SARS-CoV-1, and all SARS-CoV-2 strains) and most of their diverse unchangeable proteins (e.g., Mpro, RdRp, and ExoN). This precious compound successfully demonstrated a remarkable unique superiority over molnupiravir, as a potent broad-spectrum anticoronaviral drug, in most of all of the investigated anticoronaviral properties. This assessed superiority is extended to nearly all species, strains, and variants of coronaviruses (as enumerated above). Physicochemically and pharmacokinetically, the S-217622 molecule exhibits very good and balanced drug-like behavior, rendering it ready to enter the in vivo/clinical phases of drug evaluation and development (the compound will already begin its first journey in clinical trials in the few coming days). The current dual in silico/in vitro preclinical research study investigated and revealed the anti-COVID-19 (in specific)/anticoronaviral-infection (in general) potentials of S-217622. Interestingly, the integrative findings of both the current study and the previous work16 disclosed the very promising potent mutagenic capabilities of S-217622 on the SARS-CoV-2 RNA or, at least, the promising inhibitory activities of this compound on the coronaviral replication in general, with selective deteriorating effects targeting only the viral enzymes/genomes, not the human ones.
Physically, the S-217622 molecule has a flexible three-arm scaffold and can easily tolerate chemical reactions in the laboratory (as a parent anticoronaviral compound ready for several types of derivatization based on the drug design modeling and optimization techniques) and chemical modifications in biosystem (e.g., inside the active site pockets and cavities of coronaviral enzymes). It was also clearly proved in the current research study that coronaviral particles are very sensitive to nonpeptidic molecules having a bioactive uracil/uracil analog nucleus (scaffold) along with a considerable number of nitrogen heteroatoms and/or aromatic fluorines (substituents) in their structures. We suggest that S-217622 may effectively stop SARS-CoV-2 spreadability and pathogenicity (and, consequently, end COVID-19 infection as a whole) in the human body, mainly through severely hindering SARS-CoV-2 replication via, at least for now, a synergistic triple noncovalent inhibitory (i.e., multi-inhibitory) mode of action against the three principal SARS-CoV-2 enzymes Mpro, RdRp, and ExoN. These concurrent integrative three selective mechanisms of antiviral action could be extended to four, five, or more mechanisms if other inhibitory effects of this potential anticoronaviral agent against the other diverse coronaviral enzymes/proteins and biochemical processes/pathways are extensively explored and proved in the next studies. Although the S-217622 molecule as a whole, from the medicinal chemical point of view, is not a straightforward typical nucleoside compound, its structure has common dual nucleoside/non-nucleoside amphoteric chemical features (i.e., it contains both nucleoside-like and non-nucleosidic moieties). Based on the current research observations, which are complementary to the previous research findings, we can finally conclude that the novel synthetic nucleoside analog S-217622 has the priority to be pharmacologically and clinically investigated as a candidate oral anti-COVID-19 therapeutic agent (with very promising broad-spectrum anticoronaviral activities of EC50 values of 0.013, 0.17, 0.27, 0.29–0.50, 0.21, 1.40, 5.50, and 0.074 μM against the coronaviral Mpro, RdRp, ExoN, SARS-CoV-2 strains, SARS-CoV-1, MERS-CoV, HCoV-229E, and HCoV-OC43, respectively).
Acknowledgments
This new discovery research did not receive any external funding or specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The authors gratefully thank and deeply acknowledge anyone who helped to make this new discovery and work coming out to light.
The authors declare no competing financial interest.
References
- Chitalia V. C.; Munawar A. H. A painful lesson from the COVID-19 pandemic: the need for broad-spectrum, host-directed antivirals. J. Transl. Med. 2020, 18, 390 10.1186/s12967-020-02476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Cao R.; Zhang H.; Liu J.; Xu M.; Hu H.; Li Y.; Zhao L.; Li W.; Sun X.; Yang X.; Shi Z.; Deng F.; Hu Z.; Zhong W.; Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery 2020, 6, 28 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H.; Sarma P.; Bhattacharyya A.; Sharma S.; Chhimpa N.; Prajapat M.; Prakash A.; Kumar S.; Singh A.; Singh R.; Avti P.; Thota P.; Medhi B. Efficacy and safety of dihydroorotate dehydrogenase (DHODH) inhibitors ″leflunomide″ and ″teriflunomide″ in Covid-19: A narrative review. Eur. J. Pharmacol. 2021, 906, 174233 10.1016/j.ejphar.2021.174233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Teriflunomide: A possible effective drug for the comprehensive treatment of COVID-19. Curr. Res. Pharmacol. Drug Discovery 2021, 2, 100055 10.1016/j.crphar.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Cyanorona-20: The first potent anti-SARS-CoV-2 agent. Int. Immunopharmacol. 2021, 98, 107831 10.1016/j.intimp.2021.107831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ip A.; Ahn J.; Zhou Y.; Goy A. H.; Hansen E.; Pecora A. L.; Sinclaire B. A.; Bednarz U.; Marafelias M.; Sawczuk I. S.; Underwood J. P.; Walker D. M.; Prasad R.; Sweeney R. L.; Ponce M. G.; La Capra S.; Cunningham F. J.; Calise A. G.; Pulver B. L.; Ruocco D.; Mojares G. E.; Eagan M. P.; Ziontz K. L.; Mastrokyriakos P.; Goldberg S. L. Hydroxychloroquine in the treatment of outpatients with mildly symptomatic COVID-19: a multi-center observational study. BMC Infect. Dis. 2021, 21, 72 10.1186/s12879-021-05773-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif J.-C.; Bouabdallaoui N.; L’Allier P. L.; Gaudet D.; Shah B.; Pillinger M. H.; Lopez-Sendon J.; Da Luz P.; Verret L.; Audet S.; Dupuis J.; Denault A.; Pelletier M.; Tessier P. A.; Samson S.; Fortin D.; Tardif J.-D.; Busseuil D.; Goulet E.; Lacoste C.; Dubois A.; Joshi A. Y.; Waters D. D.; Hsue P.; Lepor N. E.; Lesage F.; Sainturet N.; Roy-Clavel E.; Bassevitch Z.; Orfanos A.; Stamatescu G.; Grégoire J. C.; Busque L.; Lavallée C.; Hétu P.-O.; Paquette J.-S.; Deftereos S. G.; Levesque S.; Cossette M.; Nozza A.; Chabot-Blanchet M.; Dubé M.-P.; Guertin M.-C.; Boivin G. for the COLCORONA Investigators. Colchicine for community-treated patients with COVID-19 (COLCORONA): a phase 3, randomised, double-blinded, adaptive, placebo-controlled, multicentre trial. Lancet Respir. Med. 2021, 9, 924–932. 10.1016/S2213-2600(21)00222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- Imran M.; Kumar Arora M.; Asdaq S. M. B.; Khan S. A.; Alaqel S. I.; Alshammari M. K.; Alshehri M. M.; Alshrari A. S.; Mateq Ali A.; Al-shammeri A. M.; Alhazmi B. D.; Harshan A. A.; Alam M. T.; Abida Discovery, Development, and Patent Trends on Molnupiravir: A Prospective Oral Treatment for COVID-19. Molecules 2021, 26, 5795 10.3390/molecules26195795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moirangthem D. S.; Surbala L. Remdesivir (GS-5734) in COVID-19 Therapy: The Fourth Chance. Curr. Drug Targets 2021, 22, 1346–1356. 10.2174/1389450121999201202110303. [DOI] [PubMed] [Google Scholar]
- Yan V. C.; Muller F. L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med. Chem. Lett. 2020, 11, 1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunotte L.; Zheng S.; Mecate-Zambrano A.; Tang J.; Ludwig S.; Rescher U.; Schloer S. Combination Therapy with Fluoxetine and the Nucleoside Analog GS-441524 Exerts Synergistic Antiviral Effects against Different SARS-CoV-2 Variants In Vitro. Pharmaceutics 2021, 13, 1400 10.3390/pharmaceutics13091400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication. ACS Omega 2022, 7, 2960–2969. 10.1021/acsomega.1c05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Efficacious Preclinical Repurposing of the Nucleoside Analogue Didanosine against COVID-19 Polymerase and Exonuclease. ACS Omega 2022, 7, 21385–21396. 10.1021/acsomega.1c07095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q.; Yang M.; Liu D.; Chen J.; Shu D.; Xia J.; Liao X.; Gu Y.; Cai Q.; Yang Y.; Shen C.; Li X.; Peng L.; Huang D.; Zhang J.; Zhang S.; Wang F.; Liu J.; Chen L.; Chen S.; Wang Z.; Zhang Z.; Cao R.; Zhong W.; Liu Y.; Liu L. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020, 6, 1192–1198. 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoh Y.; Uehara S.; Nakahara K.; Nobori H.; Yamatsu Y.; Yamamoto S.; Maruyama Y.; Taoda Y.; Kasamatsu K.; Suto T.; Kouki K.; Nakahashi A.; Kawashima S.; Sanaki T.; Toba S.; Uemura K.; Mizutare T.; Ando S.; Sasaki M.; Orba Y.; Sawa H.; Sato A.; Sato T.; Kato T.; Tachibana Y. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 2022, 65, 6499–6512. 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Two antioxidant 2,5-disubstituted-1,3,4-oxadiazoles (CoViTris2020 and ChloViD2020): successful repurposing against COVID-19 as the first potent multitarget anti-SARS-CoV-2 drugs. New J. Chem. 2021, 45, 761–771. 10.1039/D0NJ03708G. [DOI] [Google Scholar]
- Rabie A. M. Potent toxic effects of Taroxaz-104 on the replication of SARS-CoV-2 particles. Chem.–Biol. Interact. 2021, 343, 109480 10.1016/j.cbi.2021.109480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. Discovery of Taroxaz-104: The first potent antidote of SARS-CoV-2 VOC-202012/01 strain. J. Mol. Struct. 2021, 1246, 131106 10.1016/j.molstruc.2021.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrou A.; Zagaliotis P.; Theodoroula N. F.; Mystridis G. A.; Vizirianakis I. S.; Walsh T. J.; Geronikaki A. Thiazole/Thiadiazole/Benzothiazole Based Thiazolidin-4-One Derivatives as Potential Inhibitors of Main Protease of SARS-CoV-2. Molecules 2022, 27, 2180 10.3390/molecules27072180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck I.; Nguemo F. Triazole, imidazole, and thiazole-based compounds as potential agents against coronavirus. Results Chem. 2021, 3, 100132 10.1016/j.rechem.2021.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttens A.; Gullberg H.; Abdurakhmanov E.; Vo D. D.; Akaberi D.; Talibov V. O.; Nekhotiaeva N.; Vangeel L.; De Jonghe S.; Jochmans D.; Krambrich J.; Tas A.; Lundgren B.; Gravenfors Y.; Craig A. J.; Atilaw Y.; Sandström A.; Moodie L. W. K.; Lundkvist Å.; van Hemert M. J.; Neyts J.; Lennerstrand J.; Kihlberg J.; Sandberg K.; Danielson U. H.; Carlsson J. Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J. Am. Chem. Soc. 2022, 144, 2905–2920. 10.1021/jacs.1c08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J.; Sedova A.; Galanie S.; Kneller D. W.; Davidson R. B.; Maradzike E.; Del Galdo S.; Labbé A.; Hsu D. J.; Agarwal R.; Bykov D.; Tharrington A.; Parks J. M.; Smith D. M. A.; Daidone I.; Coates L.; Kovalevsky A.; Smith J. C. Hit Expansion of a Noncovalent SARS-CoV-2 Main Protease Inhibitor. ACS Pharmacol. Transl. Sci. 2022, 5, 255–265. 10.1021/acsptsci.2c00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V. A.; Saundane A. R.; Shamrao R.; Meti R. S.; Shinde V. M. Novel indolo [3,2-c]isoquinoline-5-one-6-yl [1,2,4]triazolo [3,4-b] [1,3,4]thiadiazole analogues: Design, synthesis, anticancer activity, docking with SARS-CoV-2 Omicron protease and MESP/TD-DFT approaches. J. Mol. Struct. 2022, 1264, 133153 10.1016/j.molstruc.2022.133153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelly S.; Liotta D. Potent SARS-CoV-2 Direct-Acting Antivirals Provide an Important Complement to COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 396–399. 10.1021/acscentsci.1c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist H. M.; Mekonnen D.; Mohammed A.; Shi R.; Jin T. Potency, Safety, and Pharmacokinetic Profiles of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Pharmacol. 2021, 11, 630500 10.3389/fphar.2020.630500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Fluorine in Medicinal Chemistry: In Perspective to COVID-19. ACS Omega 2022, 7, 18206–18212. 10.1021/acsomega.2c01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M.; Anderson T. K.; Jockusch S.; Tao C.; Li X.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater S.; Kumar P.; Dasgupta N.; Das G.; Ray S.; Prakash A. Combining SARS-CoV-2 Proofreading Exonuclease and RNA-Dependent RNA Polymerase Inhibitors as a Strategy to Combat COVID-19: A High-Throughput in silico Screening. Front. Microbiol. 2021, 12, 647693 10.3389/fmicb.2021.647693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SwissADME Web Tool. Available from SwissADME “Swiss Institute of Bioinformatics”. http://www.swissadme.ch (last accessed May 10, 2022).
- Malone B.; Urakova N.; Snijder E. J.; Campbell E. A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Liu Q.; Yi D.; Li Q.; Guo S.; Ma L.; Zhang Y.; Dong D.; Guo F.; Liu Z.; Wei T.; Li X.; Cen S. 5-Iodotubercidin inhibits SARS-CoV-2 RNA synthesis. Antiviral Res. 2022, 198, 105254 10.1016/j.antiviral.2022.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Guo S.; Yi D.; Li Q.; Ma L.; Zhang Y.; Wang J.; Li X.; Guo F.; Lin R.; Liang C.; Liu Z.; Cen S. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res. 2021, 190, 105078 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doharey P. K.; Singh V.; Gedda M. R.; Sahoo A. K.; Varadwaj P. K.; Sharma B. In silico study indicates antimalarials as direct inhibitors of SARS-CoV-2-RNA dependent RNA polymerase. J. Biomol. Struct. Dyn. 2022, 40, 5588–5605. 10.1080/07391102.2021.1871956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RdRp. Available from DrugDevCovid19. http://clab.labshare.cn/covid/php/database_target.php?target=RdRp&id=P0DTD1 (last accessed May 28, 2022).
- Moeller N. H.; Shi K.; Demir Ö.; Belica C.; Banerjee S.; Yin L.; Durfee C.; Amaro R. E.; Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. U. S. A. 2022, 119, e2106379119 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsp14. Available from DrugDevCovid19. http://clab.labshare.cn/covid/php/database_target.php?target=nsp14&id=P0DTD1 (last accessed May 29, 2022).
- Hillen H. S.; Kokic G.; Farnung L.; Dienemann C.; Tegunov D.; Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Ferron F.; Subissi L.; Silveira De Morais A. T.; Le N.; Sevajol M.; Gluais L.; Decroly E.; Vonrhein C.; Bricogne G.; Canard B.; Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E162–E171. 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C.; Blanc H.; Surdel M. C.; Vignuzzi M.; Denison M. R. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathog. 2013, 9, e1003565 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]