Abstract
Amyotrophic lateral sclerosis is a neurodegenerative disease characterized by the degeneration of motor neurons. There is no treatment for this disease that affects the ability to move, eat, speak and finally breathe, causing death. In an Italian family, a heterozygous pathogenic missense variant has been previously discovered in Exon 6 of the gene TARDBP encoding the TAR DNA-binding protein 43 protein. Here, we developed a potential therapeutic tool based on allele-specific small interfering RNAs for familial amyotrophic lateral sclerosis with the heterozygous missense mutation c.1127G>A. We designed a small interfering RNA that was able to diminish specifically the expression of the exogenous Green Fluorescent Protein (TAR DNA-binding protein 43G376D mutant protein) in HEK-293T cells but not that of the Green Fluorescent Protein (TAR DNA-binding protein 43 wild-type). Similarly, this small interfering RNA silenced the mutated allele in fibroblasts derived from patients with amyotrophic lateral sclerosis but did not silence the wild-type gene in control fibroblasts. In addition, we established that silencing the mutated allele was able to strongly reduce the pathological cellular phenotypes induced by TAR DNA-binding protein 43G376D expression, such as the presence of cytoplasmic aggregates. Thus, we have identified a small interfering RNA that could be used to silence specifically the mutated allele to try a targeted therapy for patients carrying the p.G376D TAR DNA-binding protein 43 mutation.
Keywords: ALS, TDP-43, siRNA therapy, antisense oligonucleotides, allele-specific silencing
Romano et al. identified a small interfering RNA able to specifically silence the allele of the TARDBP gene mutated in a familial form of amyotrophic lateral sclerosis due to the heterozygous missense mutation c.1127G>A. The treatment with this siRNA reverted the pathological phenotype in cellular models, representing a potential therapeutic tool.
Graphical Abstract
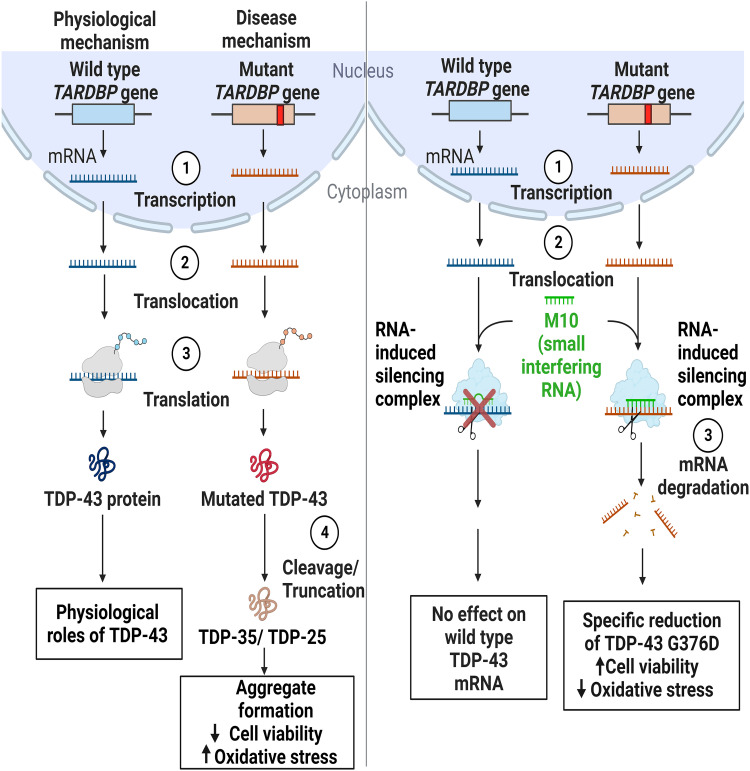
Graphical abstract.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disease characterized by the loss of motor neurons caused by axonal degeneration and thus, the destruction of nerve terminals. The progressive loss of motor neurons leads to muscle weakness and wasting, affecting gradually the ability to speak, move, eat and eventually breathe, causing death.1 TAR DNA-binding protein 43 (TDP-43) is a major disease-related protein in sporadic and familial ALS.2 TDP-43 is a highly conserved, ubiquitously expressed nuclear protein involved in several steps of RNA metabolism, such as transcription, translation, mRNA transport, mRNA stabilization, microRNA and long non-coding RNA processing.3 TDP-43 is mainly in the nucleus, but it can shuttle between the nucleus and cytoplasm through active or passive transport.4 Regarding its structure, an N-terminal domain, two RNA recognition motifs (RRM1 and RRM2), a nuclear export signal and a C-terminal domain composed of TDP-43 protein.2 In the N-terminal domain is localized the nuclear localization sequence, mutations in which cause cytoplasmic aggregation of TDP-43.5 This protein plays an important role in transcriptional repression, pre-mRNA alternative splicing and translational regulation by binding UG/TG-rich single-stranded or double-stranded DNA/RNA through RRM domains.6,7 The C-terminal domain is rich in glycine, glutamine and asparagine, representing the prion-like domain and it is important for the solubility and cellular localization of TDP-43.8,9
Most ALS-related mutations are localized in the C-terminal domain.10,11 Indeed, it was demonstrated that G294V, A315T, M337V, A382T and G376D mutations, all lying in the C-terminal region, promote cytoplasmic mislocalization and aggregation. However, the mechanisms that underlie this phenotype are not yet known.2 ALS aggregates are characterized by the presence of truncated forms of TDP-43. In particular, the most frequent forms are the C-terminal fragments CTF35 (35 kDa) and CTF25 (25 kDa), which can be produced after Caspase 3, 4, and 7 cleavages.12 Therefore, the pathological hallmarks of TDP-43 include nucleus-to-cytoplasm mislocalization, protein truncation leading to the formation of toxic C-terminal TDP-43 fragments and protein aggregation and the deposition of ubiquitinated and hyperphosphorylated forms of TDP-43.3,13–15 At present, there is no treatment available that can prevent or reverse the neurodegenerative condition. Thus, downregulation of the mutant allele without suppression of the corresponding wild-type allele could be a promising therapeutic strategy. One way to target the mutant allele in cases of ALS is by using allele-specific small interfering RNAs (siRNAs), an approach that has been already described in several autosomal dominant diseases such as Parkinson’s disease,16 Alzheimer’s disease17 and Huntington’s disease.18,19 Silencing of a superoxide dismutase 1 (SOD1) mutant allele has been successfully achieved using siRNAs and short hairpin RNA in cells and animal models of ALS.20–22
A heterozygous missense mutation, 1127G>A in the Exon 6 of the TARDBP gene has been discovered in an Italian family with several members affected by ALS.23 To try to put the bases for treatment, we identified a siRNA complementary to the region comprising the point mutation c.1127G>A that specifically targets the TDP-43 mutant allele and that can counteract the cellular phenotypes induced by mutant allele expression.
Materials and methods
Antibodies
Primary antibodies used in this study were: rabbit polyclonal anti-TDP-43 (1:3000 for Western blot analysis, 10782-2-AP) and rabbit polyclonal anti-TDP-43 C-terminal (1:100 for immunofluorescence analysis, 12892-1-AP), both from Proteintech (Rosemont, IL, USA); rabbit polyclonal anti-histone H3 (1:10 000, ab1791) from Abcam (Cambridge, UK); rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:2000, sc-25778) from Santa Cruz Biotechnology (Dallas, TX, USA).
Cells and reagents
We used dermal fibroblasts derived from two healthy individuals (control fibroblasts) and two ALS patients belonging to an Apulian family from Lecce carrying the p.G376D TDP-43 mutation. Fibroblasts were previously obtained, after informed consent, by skin biopsies and then cultured under protocols approved by the Università Cattolica del Sacro Cuore Ethics Committee (Protocol P/740/CE/2012) and the Palermo 1 Ethics Committee (Protocols 7/2017 and 4/2019). HEK-293T and Neuro2A cells were from the ATCC. Fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% foetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin and 10 mg/ml streptomycin, while HEK-293T and Neuro2a cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and 10 mg/ml streptomycin in a 5% CO2 incubator at 37°C. We periodically checked that the cell lines used were free from mycoplasma infections. The reagents for tissue culture were from Sigma-Aldrich (St. Louis, MO, USA) or Gibco (Grand Island, NY, USA).
Plasmid construction
The TDP-43wt full-length construct used in this study has been described previously.22 We used the green fluorescent protein (GFP)-TDP-43wt plasmid as a template to generate the GFP-TDP-43G376D construct through the PfuTurbo DNA polymerase (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions and primers containing the desired mutation. The oligonucleotides used to generate the TDP43 mutant cDNA were 5′-GGAAATAACTCTTATATAGTGACTCTAATTCTGGTGCAGC-3′ and 5′-GCTGCACCAGAATTAGAGTCACTATAAGAGTTATTTCC-3′.
Transfection and silencing
HEK-293T cells were transfected and silenced using oligofectamine transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction and processed after 72 h, as previously described.24 Fibroblasts were silenced using Metafectene SI (Biontex, Martinsried, Germany) or Amaxa Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) according to the protocols provided by the manufacturers and then analysed after 6 days or 48 h, respectively. Neuro2A cells were transfected using Amaxa Cell Line Nucleofector Kit V, as previously described,25 or with Metafectene SI (Biontex, München, Germany) and processed 72 h after transfection. We designed siRNAs complementary to the region comprising the point mutation c.1127G>A in the mRNA of the human TARDBP gene. TDP-43 siRNAs are listed in Table 1. The siRNAs were produced by Eurofins Genomics (Ebersberg, Germany).
Table 1.
siRNAs sequences
| Sequences | |
|---|---|
| siM9 | 5′-UUAUAGUGACUCUAAUUCUdTdT-3′ 3′-dTdT AAUAUCACUGAGAUUAAGA-5′ |
| siM10 | 5′-CUUAUAGUGACUCUAAUUCdTdT-3′ 3′-dTdTGAAUAUCACUGAGAUUAAG-5′ |
| siM9/10 | 5′-UUAUAGUGAGUCUAAUUCUdTdT-3′ 3′-dTdT AAUAUCACUCAGAUUAAGA-5′ |
| siM10/9 | 5′-CUUAUAGUCACUCUAAUUC-dTdxT3′ 3′-dTdTGAAUAUCAGUGAGAUUAAG-5′ |
| TDP-43i | 5′-AAGCAAAGCCAAGAUGAGCCU-dTdT-3′ 3′-dTdTAGGCUCAUCUUGGCUUUGCUU-5′ |
Western blotting
HEK-293T cells and fibroblasts were lysed in Laemmli buffer [100 mM Tris–HCl, pH 6.8, 4% (w/v) sodium dodecyl sulphate (SDS), 0.2% (w/v) bromophenol blue, 20% glycerol and 200 mM dithiothreitol (dithiothreitol)] and subjected to Western blot analysis as previously described.25–27 Signals were visualized using Bio-Rad ChemiDoc MP Imaging Systems (Bio-Rad, Hercules, CA, USA) and densitometric analysis of band intensities was performed with Image Lab software (Bio-Rad).
Confocal immunofluorescence
Fibroblasts and Neuro2A cells were treated as previously described.28 Briefly, cells were fixed with paraformaldehyde at 4% for 30 min at 37°C and then permeabilized using a solution of 10% Triton X-100 diluted 1:5000 in PBS. After 1 h of blocking, cells were incubated with the primary antibody for 40 min at room temperature and, after washes, with the secondary antibody for 20 min at room temperature. Coverslips were mounted using Mowiol solution. Images were acquired using Zeiss LSM700 confocal laser scanning microscopy (Zeiss, Oberkochen, Germany) or the EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific, Waltham, MA, USA) and cells with aggregates were manually counted.
Quantitative real-time polymerase chain reaction
RNA from HEK-293T and fibroblasts was extracted using the RNeasy Micro Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). cDNA was prepared using SuperScript IV reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. The polymerase chain reaction (PCR) reaction was prepared with SYBR™ Green PCR Master Mix (Applied Biosystem, Watham, MA, USA) and run in C1000 Touch Thermal Cycler (Bio-Rad), using the following thermal protocol: 94°C 5 min, 40 cycles of 94°C 30 s, 60°C 30 s, 72°C. The expression level was calculated with the 2−ΔCT method as previously described.26 The normalization was performed using Rplp0 (ribosomal protein large P0).
The primers used were:
Rplp0: forward: 5′-TCGACAATGGCAGCATCTAC-3′
reverse: 5′-ATCCGTCTCCACAGACAAGG-3′
GFP: forward: 5′-CTGCTGCCCGACAACCAC-3′
reverse: 5′-TCACGAACTCCAGCAGGAC-3′
TDP-43: forward: 5′-GTATGATGGGCATGTTAGC-3′
wild-type reverse: 5′-CTGCACCAGAATTAGAGC-3′
G376D reverse: 5′-CTGCACCAGAATTAGAGT-3′
Nuclear and cytoplasmic fractionation
Cells were lysed in lysis buffer (50 mM Tris pH 7.5, 0.1% Triton X-100, 137.5 mM NaCl, 10% glycerol, 5 mM EDTA) supplemented with Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland), and then incubated on ice for 15 min. After centrifugation at 7000 rpm for 2 min at 4°C, the supernatant, representing the cytosolic fraction, was collected. The pellet, which contained nuclei, was washed three times with lysis buffer. After the last centrifugation, the pellet containing the nuclear fraction was re-suspended in lysis buffer supplemented with 0.5% SDS. The fractions were then analysed by Western blot analysis.
Cell viability assay
The sulforhodamine B (SRB) assay was performed to determine cell viability. Briefly, Neuro2A cells were seeded at the density of 2 × 104 in a 24-well plate. After 24 h, cells were transfected with plasmids coding for GFP, GFP-TDP-43wt and GFP-TDP-43G376D and treated with control RNA or M10. After 72 h the medium was changed, and the cells were fixed with 125 µL of 50% cold (trichloroacetic acid) for 1 h at 4°C. The samples were washed five times with water and then let dry overnight at room temperature. 100 µL of SRB solution (0.4% SRB in 1% acetic acid) were added per well. After 30 min, the SRB solution was removed, and the cells were washed four times with 200 µL of 1% acetic acid. The retained SRB was solubilized by adding 800 µL of Tris 10 mM pH 10.5 per well for 10 min. Absorbance was read in a microplate reader (Victor X5, Perkin Elmer) at 565 nm.
2′,7′-Dichlorodihydrofluorescein diacetate staining
2′,7′-Dichlorodihydrofluorescein diacetate (DCFH-DA) staining was performed to determine total cellular reactive oxygen species (ROS) that underlies oxidative stress. Fibroblasts were seeded (2 × 104 per well) in a 24-well plate, treated with control RNA or M10 for six days and then total ROS were detected by the DCFH-DA (Thermo Fisher Scientific) assay. The drug was added to each well at a concentration of 10 µM and incubated at 37°C for 30 min. Then, cells were washed three times with PBS, lysed in 100 µL of RIPA (radioimmunoprecipitation assay) buffer for 5 min on ice and centrifuged at 10 000 rpm for 10 min at 4°C. 90 µL of the supernatant were transferred to a 96-well plate and the fluorescence was read at 490 nm in a microplate reader (Victor X5, Perkin Elmer). The remaining part was used to measure protein concentration by Bradford assay and normalize fluorescence intensities with the protein concentration of each sample.
Statistical analysis
The experiments were performed at least in triplicate. Data were statistically analysed using one-way ANOVA followed by Dunnett’s, Sidak’s or Tukey’s multiple comparison tests (*P < 0.05, **P < 0.01 and ***P < 0.001). Graphs represent mean value ± standard error mean (SEM).
Results
Pedigree of the family carrying the p.G376D TDP-43 mutation
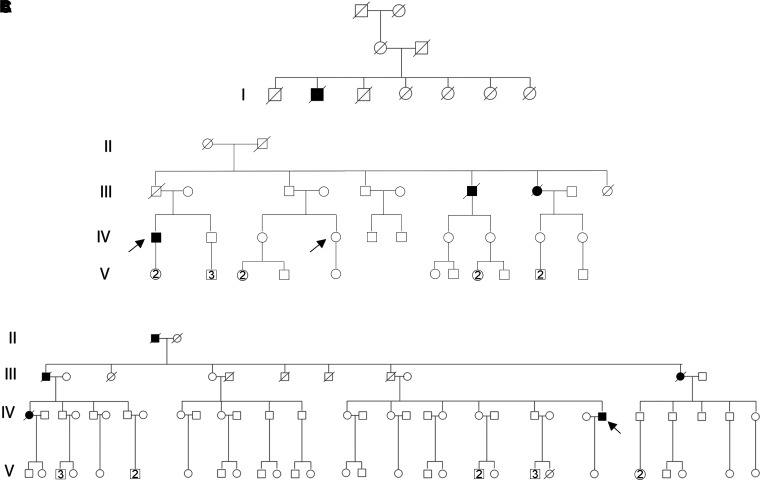
In this study, we performed our experiments using dermal fibroblasts from patients belonging to the same Apulian family from Lecce carrying the p.G376D TDP-43 mutation. The first case of ALS was reported in a man in the first generation of the family tree (Fig. 1A). In the second generation of this pedigree, there are seven branches, in three of which several members affected appear, for a total of 31 cases of ALS in this family, with 19 men and 12 women affected.
Fig. 1.
Pedigree of the family affected by ALS for the TDP-43G376D mutation. Square indicates male; circle female. Arrows indicate patients from which fibroblasts were obtained. Circles and squares crossed by a line indicate deceased female and male, respectively. (A) Family tree showing the first generation in which the first case of ALS was reported. (B) Family tree of Patient 1. (C) Family tree of Patient 2.
In our experiments, we used ALS1 and ALS2 cells: which were obtained from affected individuals belonging to the fourth generation of different branches of the family tree. We obtained dermal fibroblasts from an ALS1 patient when the disease was diagnosed (ALS1O, onset) and four years after diagnosis (ALS1A, advanced; IV.1, Fig. 1B). We also obtained dermal fibroblasts from a healthy woman of the same generation (CTRL2, IV.4, Fig. 1B) not carrying the p.G376D TDP43 mutation. ALS2A patient is indicated in Fig. 1C (IV.26). Clinical characteristics of patients are reported in Table 2.
Table 2.
Clinical and demographic characteristics of the two TARDBP G376D ALS patients in the advanced stage included in the study
| ALS1A | ALS2A | |
|---|---|---|
| Age of onset (years) | 37 | 27 |
| Disease duration from onset to tracheotomy (months) | 16 | 10 |
| Gender | Male | Male |
| Education (years) | 13 | 8 |
| ALSFRS-R at diagnosis (normal value: 48) | 40 | 34 |
| Disease progression (ΔFS) at diagnosis | 1.33 | 1.56 |
| Site of onset | Spinal | Spinal |
Identification of allele-specific siRNA for TDP-43G376D
We designed four allele-specific siRNAs targeting the c.1127G>A mutation that defined M9, M10, M9/10 and M10/9 (Table 1). Early studies on the RNAi pathway showed that a mismatch that occurs at the centre of the siRNA structure abolished the RNAi effect.31 Therefore, the position of the mutated nucleotide in the siRNA is important to discriminate the mutant from the wild-type allele. Allele-specific siRNAs targeting different regions of the same transcript display differences in silencing efficiency; therefore, we designed siRNAs, where the mutated nucleotide varies within the siRNA structure. The allele-specific siRNAs M9 (the mutated nucleotide is at Position 9) and M10 (the mutated nucleotide is at Position 10) are perfectly matched with the mutant allele but have a single mismatch with the wild-type counterpart. However, it is likely that not all single mismatches would create silencing selectivity, and some mismatches might be tolerated by the RNA-induced silencing complex, compromising the allele specificity.32 For this reason, we designed siRNAs containing a double mismatch with the wild-type (siM9/10 and siM10/9), hence creating a single mismatch with the mutant allele.
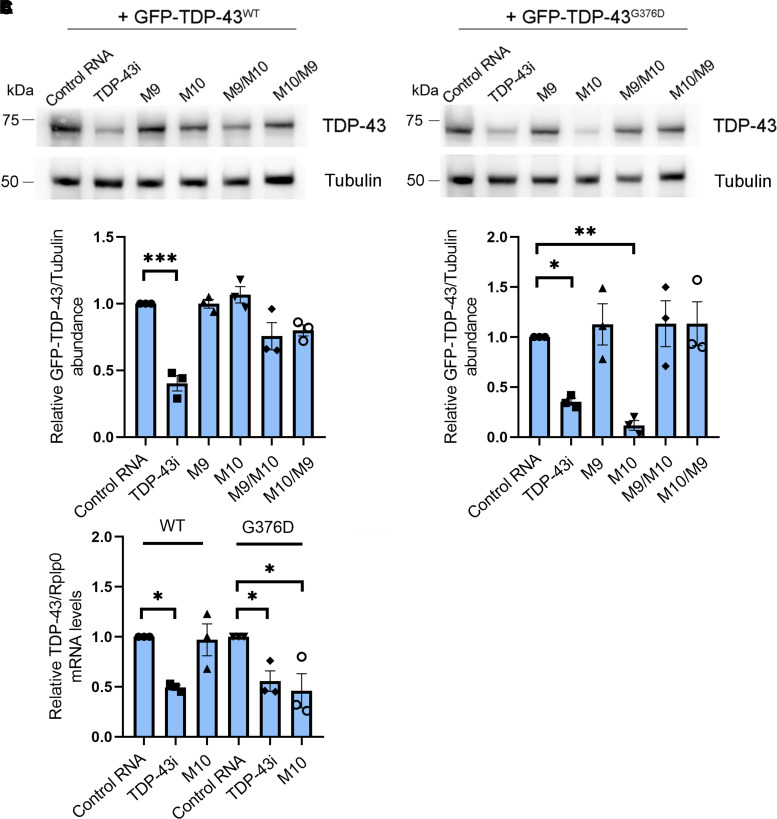
To determine if these allele-specific siRNAs specifically knock down the G376D allele, HEK-293T cells were transiently transfected with full-length GFP-TDP-43wt or GFP-TDP-43G376D in combination with allele-specific siRNAs for 72 h and analysed by Western blot. Non-targeting siRNA control (control RNA) and siRNA targeting the 5′ regions of the TDP-43 transcript (TDP-43i) were used as controls. As expected, TDP-43i was able to silence both GFP-TDP-43 wild-type and GFP-TDP-43G376D (P = 0.0001 and P = 0.0477), while siM10 caused a significant reduction only of GFP-TDP-43G376D protein levels (P = 0.0072), as GFP-TDP-43 wild-type levels remained unchanged by transfection (Fig. 2A and B).
Fig. 2.
An allele-specific siRNA downregulates the GFP-tagged TDP-43G376D mutant allele. (A) HEK-293T cells were transfected with plasmids encoding the GFP-TDP-43WT or GFP-TDP-43G376D proteins and with control RNA, TDP-43 siRNA or putative allele-specific siRNAs (M9, M10, M9/10 and M10/9). Cells were then lysed and analysed by Western blotting using specific anti-TDP-43 and anti-tubulin antibodies, in order to analyse GFP-TDP-43 levels and equal loading, respectively. See Supplementary Fig. 1 for uncropped blots. (B) Quantification of the amount of GFP-TDP-43 wild-type and mutant proteins upon transfection of the different siRNAs. Data are the mean ± SEM of at least three independent experiments. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 17.92 (for GFP-TDP-43WT); F = 8.488 (for GFP-TDP-43G376D). (C) The amount of GFP-TDP-43 transcript was quantified, compared with the Rplp0 transcript as control, using real-time PCR in HEK-293T cells transfected with GFP-TDP-43WT or GFP-TDP-43G376D plasmids and treated with control RNA, TDP-43 siRNA or allele-specific siRNA M10. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Sidak’s multiple comparisons test. *P < 0.05; ** P < 0.01; ***P < 0.001; F = 6.606.
Having determined that an allele-specific siRNA can reduce the protein abundance of the mutant allele specifically, we sought to determine whether this RNAi approach could induce a block of the translation or degradation of TDP-43G376D mRNA; therefore, the expression of the corresponding mRNA was quantified by real-time PCR using primers against GFP. The analysis showed a substantial reduction of GFP-TDP-43 wild-type and GFP-TDP-43G376D transcripts in the presence of TDP-43i (P = 0.0199 and P = 0.0436), while GFP-TDP-43G376D transcript was reduced in the presence of siM10 (P = 0.0131), which did not influence the wild-type transcript abundance (Fig. 2C).
Validation of allele-specific siRNA in fibroblasts isolated from ALS patients carrying the p.TDP-43 mutation
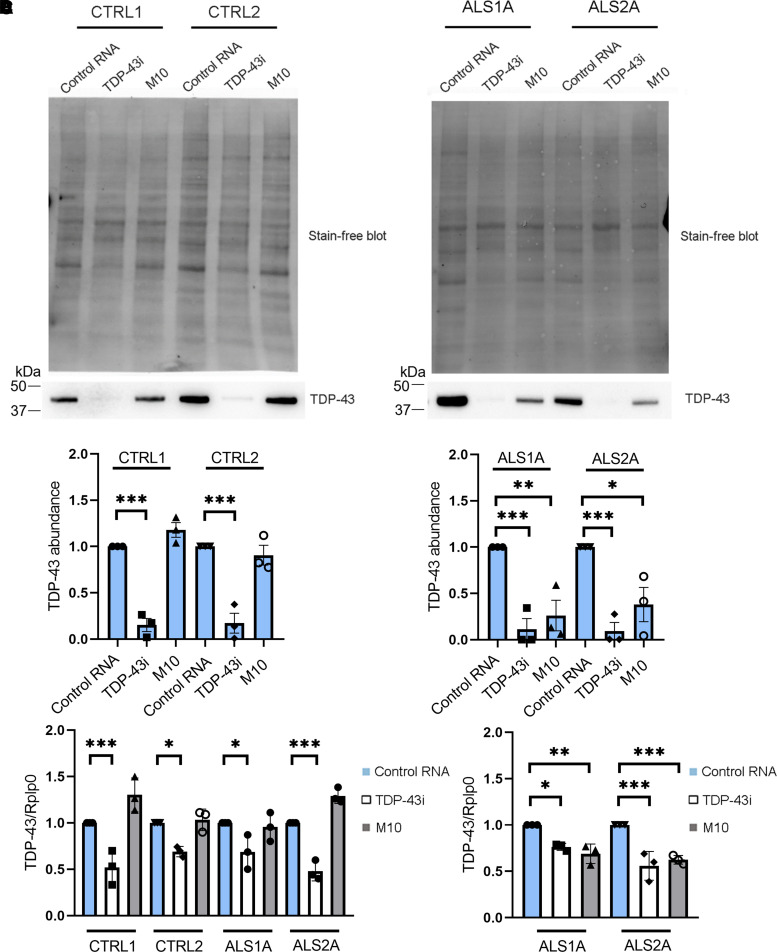
In HEK-293T cells overexpressing GFP-TDP-43G376D, we showed that siM10 could selectively reduce the TDP-43G376D protein amounts. To test whether allele-specific siRNA could decrease endogenous TDP-43G376D levels in cells derived from patients, we transfected dermal fibroblasts previously isolated from ALS patients carrying the p.G376D TDP-43 mutation (ALS fibroblasts), and healthy individuals without the mutation (CTRL = control patients) with siM10, TDP-43i and control RNA. Western blot analysis showed that TDP-43i induced a significant reduction of TDP-43 abundance in control cells (CTRL1 P < 0.0001; CTRL2 P < 0.0001) and ALS fibroblasts (ALS1A P = 0.0007; ALS2A P = 0.0006), while transfection with allele-specific siM10 did not change the protein amount in controls but significantly reduced TDP-43 levels in ALS patient cells (ALS1A P = 0.0032; ALS2A P = 0.0114; Fig. 3A and B). Moreover, we performed a real-time PCR using a primer pair able to recognize the wild-type sequence of TDP-43. The transfection with siM10 did not change the TDP-43 wild-type mRNA in controls and patients while the treatment with TDP-43i significantly reduced TDP-43 transcript in controls (CTRL1 P = 0.0003; CTRL2 P = 0.0248) and patients (ALS1A P = 0.0216; ALS2A P < 0.0001; Fig. 3C). On the contrary, when we used a primer pair that specifically recognizes the mutant sequence, we observed reduced levels of TDP-43 transcript in ALS fibroblasts treated with TDP-43i (ALS1A P = 0.0138; ALS2A P < 0.0001), but mostly, a significant reduction of TDP-43G376D mRNA levels after siM10 transfection (ALS1A P = 0.0018; ALS2A P = 0.0004; Fig. 3D).
Fig. 3.
Allele-specific siM10 decreases TDP-43G376D abundance in ALS fibroblasts. (A) Fibroblasts isolated from two controls and two ALS patients carrying the TDP-43G376D mutation (ALS1, ALS2) were transfected with siM10, TDP-43i and control RNA for 6 days, lysed and subjected to Western blot analysis using anti-TDP-43 antibody and Stain-Free Imaging Technology (Bio-Rad) for loading control. See Supplementary Fig. 2 for uncropped blots. (B) Quantification of the amount of TDP-43 upon transfection with control RNA, TDP43 siRNA and allele-specific siRNA M10. Statistical analysis was performed using one-way ANOVA followed by Sidak’s multiple comparisons test. Data represent the mean ± SEM of at least three experiments. *P < 0.05; ** < 0.01; ***P < 0.001; F = 35.65 (for control cells); F = 12.77 (for ALS cells). (C) The amount of TDP-43 wild-type transcript was quantified, compared with the Rplp0 transcript as control, using real-time PCR in control and ALS fibroblasts treated with control RNA, TDP-43 siRNA or allele-specific siRNA M10. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Sidak’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 16.03. (D) The amount of TDP-43 mutant transcript was quantified, compared with the Rplp0 transcript as control, using real-time PCR in ALS fibroblasts treated with control RNA, TDP-43 siRNA or allele-specific siRNA M10. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Sidak’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 16.68.
Allele-specific siRNA reduced cytoplasmic inclusion in ALS fibroblast carrying the p.G376D TDP-43 mutation and in Neuro2A cells transfected with the mutant plasmid
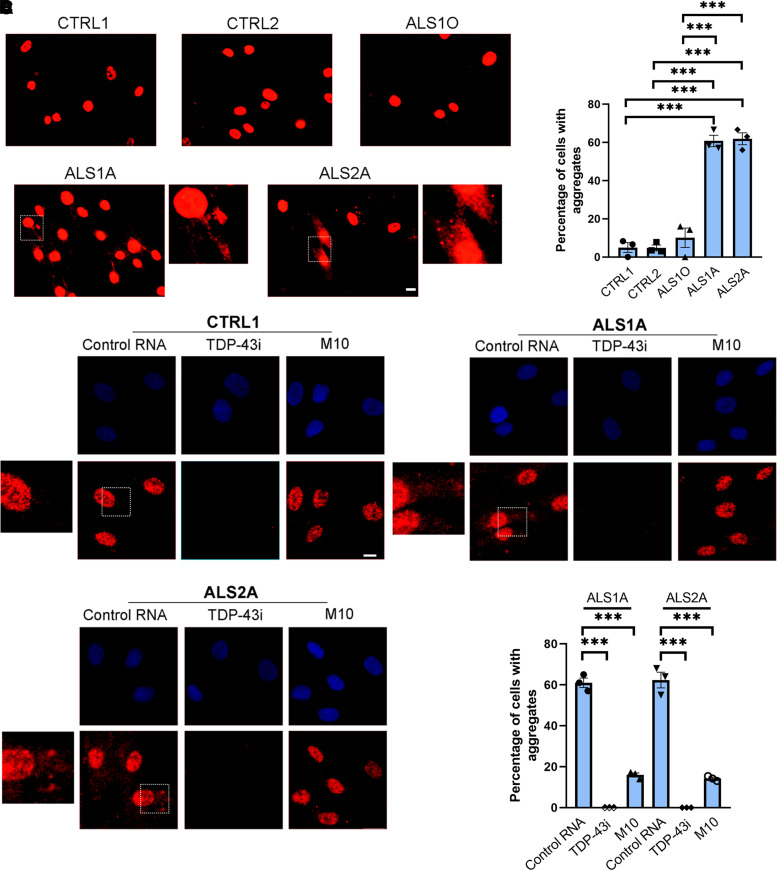
Both familial and sporadic ALS are characterized by TDP-43 cytoplasmic aggregates, which represent a hallmark of the disease.12 We used an antibody able to specifically recognize the C-terminal region of TDP-43 to visualize TDP-43 aggregates in fibroblasts by immunofluorescence experiments. We confirmed that cytoplasmic inclusions are visible in ALS1A and ALS2A patients but not in control cells. Moreover, we highlighted differences in ALS1 patient considering that TDP-43 aggregates are considerably more abundant in the stage of the advanced disease (ALS1A cells) compared with the early stage (ALS1O cells) (Fig. 4A). Indeed, the quantification of TDP-43 inclusions showed that about 60% of the cells of both patients present the aggregates while in ALS1O cells inclusions were not significantly different from control cells (Fig. 4B; P < 0.0001).
Fig. 4.
Allele-specific siM10 decreases TDP-43 cytoplasmic inclusions in ALS fibroblasts. (A) Control and ALS fibroblasts were fixed and immunolabelled with anti-C-terminal TDP-43 antibody followed by Alexa-568-conjugated secondary antibody. Bar 50 µM. (B) Quantification of the percentage of cells showing TDP-43 cytoplasmic aggregates. At least 50 cells per sample were analysed. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 84.83. (C) Control and ALS fibroblasts treated with control RNA, TDP-43 siRNA or allele-specific siRNA M10 were fixed and immunolabelled with anti-C-terminal TDP-43 antibody followed by Alexa-568-conjugated secondary antibody. Nuclei were stained with DAPI. Bar 10 µM. Quantification of the percentage of cells with TDP-43 aggregates is also shown. At least 50 cells per sample were analysed. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 474.4 (for ALS1A); F = 209.6 (for ALS2A).
Having determined that ALS cells showed TDP-43 cytoplasmic aggregates, we wondered if allele-specific siRNA could reverse this phenotype. Confocal microscopy showed that M10 had no effect on control cells while it could strongly reduce the aggregates in both ALS fibroblasts. The quantification showed that the percentage of ALS1A and ALS2A fibroblasts with aggregates drops from about 60 to 15% following M10 treatment (ALS1A: control RNA/TDP-43i P < 0.0001; control RNA/M10 P < 0.0001; ALS2A: control RNA/TDP-43i P < 0.0001; cRNA/M10 P < 0.0001; Fig. 4C).
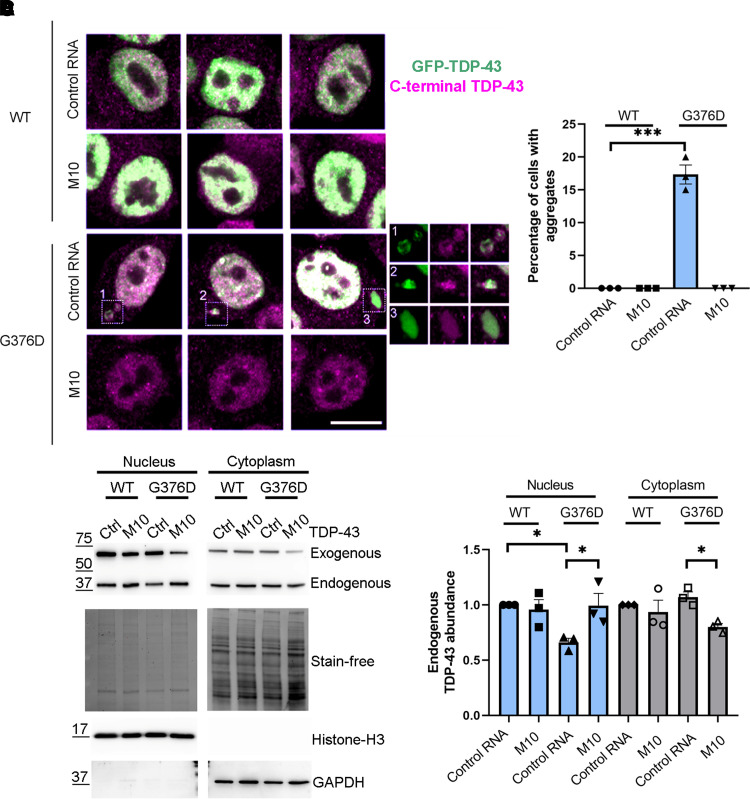
To further confirm that the TDP-43G376D mutation is responsible for cytoplasmic inclusion and that M10 can prevent TDP-43 aggregates, we transfected Neuro2A cells with a plasmid encoding GFP-TDP-43 wild-type or G376D and we treated them with control RNA or M10. Transfection with GFP-TDP-43G376D led to the appearance of large cytoplasmic inclusions, which were no longer visible following M10 treatment, confirming that only the mutant form of TDP-43 forms aggregates and that this cellular phenotype is reverted following the treatment with allele-specific siRNA (Fig. 5A and B).
Fig. 5.
Neuro2A cells transfected with GFP-TDP-43G376D show aggregates which are no longer visible following siM10 treatment. (A) Neuro2A cells were transfected with plasmids encoding the GFP-TDP-43WT or GFP-TDP-43G376D proteins and with control RNA or M10 siRNA. Seventy-two hours after transfection, cells were fixed and immunolabelled with anti-C-terminal TDP-43 antibody followed by Alexa-568-conjugated secondary antibody. Bar 10 µM. (B) Quantification of the percentage of cells showing TDP-43 cytoplasmic aggregates. At least 50 cells per sample were analysed. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 142.3. (C) HEK-293T cells were transfected with plasmids encoding the GFP-TDP-43WT or GFP-TDP-43G376D proteins and with control RNA or M10 siRNA. Seventy-two hours after transfection cells were lysed and cytoplasmic and nuclear fractions were separated. Samples were analysed by Western blotting using anti-TDP-43, anti-GAPDH, anti-histone H3 antibodies. Stain-Free Imaging Technology (Bio-Rad) was used for loading control. GAPDH and histone H3 were used. See Supplementary Figure 3 for uncropped blots. (D) Quantification of endogenous TDP-43 was shown. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 4.945 (for nucleus); F = 3.710 (for cytoplasm).
In cellular models, truncated forms of TDP-43 induced the mislocalization of endogenous TDP-43, which showed an altered nucleocytoplasmic distribution with the depletion of wild-type TDP-43 from the nucleus.33,34
In order to establish if mutant allele-specific silencing results in decreased retention of endogenous TDP-43 in the cytoplasm, we transfected HEK-293T with a plasmid coding for TDP-43 wild-type or the G376D mutant and with control RNA or M10. After 72 h, we lysed cells and separated nuclear and cytoplasmic fractions. Samples were subjected to Western blot analysis to evaluate the localization of endogenous TDP-43. The expression of TDP-43G376D decreased the amount of endogenous TDP-43 in the nucleus and slightly increased its presence in the cytoplasm. Following M10 treatment, TDP-43 localization is similar to control (Fig. 5C and D). This result suggests that M10 treatment could enhance the amount of functional TDP-43 in the nucleus.
Allele-specific siRNA ameliorated cell viability and oxidative stress in Neuro2A cells and ALS fibroblasts
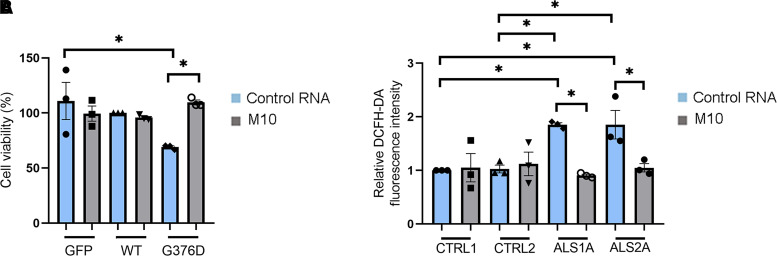
Having demonstrated that M10 treatment could reduce TDP-43 aggregation in the cytoplasm, we wondered if the treatment with this siRNA could help to improve cell viability and reduce oxidative stress in cellular models. Fang and co-workers35 demonstrated that inclusion bodies formed by full-length and truncated TDP-43 were neurotoxic. In order to evaluate if the expression of mutant TDP-43 affected cell viability, we performed an SRB assay. This method is based on SRB, which binds stoichiometrically to proteins under mild acidic conditions and can be extracted using basic conditions, thus, the bound dye reflects the number of cells.36 This assay demonstrated that expression of TDP-43G376D reduced cell viability of Neuro2A cells (P = 0.0189). Importantly, M10 treatment restored cell viability to levels comparable to control cells (P = 0.0234; Fig. 6A).
Fig. 6.
M10 treatment rescues cell viability and oxidative stress. (A) Neuro2A cells were transfected with plasmids encoding the GFP-TDP-43WT or GFP-TDP-43G376D proteins and with control RNA or M10 siRNA. Seventy-two hours after transfection, cells were subjected to SRB assay to measure cell viability. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 4.060. (B) Fibroblasts isolated from two controls and two ALS patients carrying the TDP-43G376D mutation (ALS1A, ALS2A) were transfected with control RNA and M10 for 6 days, treated with 10 µM DCFH-DA for 30 min and then lysed. Fluorescence intensity was read at 490 nm in a microplate reader. Data represent the mean ± SEM of at least three experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05; ** < 0.01; ***P < 0.001; F = 5.946.
Oxidative stress is elevated in ALS. Indeed, fibroblasts carrying the TDP-43A382T and TDP-43G294V mutations showed enhanced oxidative stress compared with control fibroblasts.37 We wondered if ALS1A and ALS2A patients showed this alteration and, to answer this question, we performed the DCFH-DA staining. DCFH-DA is taken up by cells where the acetyl groups were cleaved off by cellular esterase, resulting in DCFH. Oxidation of DCFH by ROS converts the molecule to dichlorofluorescein, which emits green fluorescence.38 Therefore, fluorescence intensity depends on the amount of ROS in cells. ALS1A and ALS2A patients showed increased oxidative stress compared with control cells (CTRL1/SLA1A P = 0.0268; CTRL1/SLA2A P = 0.0266; CTRL2/SLA1A P = 0.0336; CTRL2/SLA2A P = 0.0333). Notably, treatment of ALS fibroblasts with M10 reduced DCFH-DA fluorescence indicating an improvement of oxidative stress (ALS1A P = 0.0119; ALS2A P = 0.0403; Fig. 6B).
Altogether, these data suggest that the TDP-43G376D mutation reduces cell viability and increases oxidative stress in cellular models and demonstrate that M10 treatment reverts both of these phenotypes.
Discussion
In 1869, Jean–Martin Charcot defined ALS as a pure motor neuron disease, but now it is known that it is a multisystem neurodegenerative disorder characterized by clinical, genetic and neuropathological heterogeneity.39 Considering the genetic heterogeneity, ALS is associated with alterations in >20 genes. The most common genes involved are Chromosome 9 open reading frame 72 (C9orf72), SOD1, TDP-43, fused in sarcoma and TANK-binding kinase 1, which together cause 15% of the ALS cases.40–42
Here, we analysed an Apulian family from Lecce affected by familial ALS caused by the c. 1127G>A pathogenic variant in the TARDBP gene, which determines the aminoacidic substitution G376D in the TDP-43 protein and that it is inherited in autosomal dominant manner.23 This mutation has also been found in an Asian and a Swiss family with an autosomal dominant transmission and a clinical phenotype similar to that of the Italian family.43,44 Noteworthy, the Swiss family is genetically connected to the Apulian family (Association 2HE, personal communication).
Nowadays, ALS does not have an effective treatment. Indeed, the complexity of this disease has been further demonstrated by the failure of many randomized controlled trials, which did not lead to an improvement in disease progression or survival.45 Currently, the antiglutaminergic Riluzole is the most used drug in ALS patients, but the mean patient survival is prolonged by only 3–6 months.46–48 Hence, the need for new therapeutic strategies. One possibility is represented by antisense oligonucleotides (ASOs), short, synthetic, single-stranded oligodeoxynucleotides that bind a target mRNA, bringing it to degradation and reducing the levels of a toxic protein. Recently, this strategy received the approval of the Food and Drug Administration for Duchenne muscular dystrophy and spinal muscular atrophy, and several clinical trials are in progress.49 ASOs have also been considered for ALS.50 In the light of this, we designed allele-specific siRNAs for the c.1127G>A (p.G376D) mutation in TDP-43 and tested their efficacy in HEK-293T cells transfected with GFP-TDP-43 wild-type or G376D. We demonstrated that among the four ASOs designed, M10 reduced GFP-TDP-43G376D mRNA and protein without affecting the wild-type (Fig. 2A–C).
Then, we tested M10 in fibroblasts derived from patients carrying the TDP-43G376D mutation and belonging to the Italian family previously described.23 Also, in these cells, M10 effectively reduced the level of the mutated protein and mRNA (Fig. 3A–D). In particular, using primer pairs to discriminate against the wild-type and the mutated alleles, we demonstrated that only the G376D allele is affected by M10 treatment (Fig. 3C and D).
In 95% of ALS cases, the neuropathological signature of the disease is represented by the appearance of TDP-43 aggregates in the cytoplasm.51 In a previous work, TDP-43 was found in the cytoplasm of primary fibroblasts carrying TARDBP c*4G>A and p.A382T mutations.52 To test whether G376D fibroblasts are also characterized by this phenotype, we performed an immunofluorescence assay using an antibody able to recognize the C-terminal region of the TDP-43 protein. We demonstrated not only that ALS1A and ALS2A fibroblasts are rich in TDP-43 aggregates, which, on the contrary, were not present in control cells, but also that cells taken from Patient 1 at the onset of the disease were similar to controls. In contrast, those taken from the same patient at an advanced stage of the disease showed a cytoplasm rich in aggregates (Fig. 4A and B). This result highlighted that the formation of the aggregates takes place gradually and this phenomenon is associated with the progression of the disease, confirming that the loss of functional TDP-43 in the nucleus and its increased deposition in the cytoplasm is mostly seen in the end stages of the disease.53
We wondered if M10 could revert this phenotype; therefore, we treated fibroblasts with control RNA and M10 and we observed a reduction in the number of ALS cells showing aggregates following M10 treatment, while the cytoplasmic TDP-43 deposits were still visible in the cells of the same patient treated with control RNA (Fig. 4C and D).
Finally, to confirm that the aggregation of TDP-43 is specifically caused by the p.G376D mutation of TDP-43, we transfected Neuro2a cells with GFP-TDP-43 wild-type or mutant, demonstrating the appearance of cytoplasmic aggregates only in cells transfected with the plasmid encoding GFP-TDP-43G376D. Moreover, the treatment of these cells with M10 led to the disappearance of aggregates in this cellular model (Fig. 5A and B). Compared with fibroblasts from patients, in Neuro2a cells expressing the mutant protein, we observed the presence of single aggregates. This could possibly be related to the transient transfection method that we used. Indeed, several studies using this method reported cells with single aggregates.54,55 Larger aggregates are likely to accumulate in the cytoplasm with time.
Increasing evidence suggests that TDP-43 mislocalization determines cellular toxicity in ALS. This could be due to TDP-43 nuclear depletion, which alters several cellular processes.12 Through nuclear cytoplasmic separation, we evaluated the localization of endogenous TDP-43 and we demonstrated that the expression of p.G376D TDP-43 reduced the amount of endogenous TDP-43 in the nucleus, while M10 treatment, in TDP-43G376D-overexpressing cells increased nuclear TDP-43, possibly increasing functional protein levels (Fig. 5C and D).
To test whether the presence of mutated TDP-43 is associated with other phenotypic alterations, we evaluated the viability of Neuro2a overexpressing GFP, GFP-TDP-43 wild-type or GFP-TDP-43G376D and treated the cells with control RNA or M10. Notably, the expression of mutated TDP-43 affected cell viability which was restored following M10 treatment (Fig. 6A). This allele-specific siRNA is also able to reduce the oxidative stress that characterizes ALS fibroblasts (Fig. 6B). Interestingly, recent work showed that mouse and human neurons are more sensitive to oxidative stress when TDP-43 aggregates are present since they sequester microRNAs and proteins, interfering with their function. Many of these proteins are mitochondrial proteins encoded by the nuclear genome and their alterations result in mitochondrial dysfunctions that increase oxidative stress. This gives rise to a vicious circle that could underlie ALS onset and progression.56 Considering our data, allele-specific silencing could put an end to this circle by reducing TDP-43 aggregation and oxidative stress and, maybe, ameliorating mitochondrial function. This point should be investigated in the future.
Further studies are necessary to confirm the efficacy of M10 in the treatment of ALS caused by the p.G376D mutation. An important point that should be considered is the effect of decreased TDP-43 levels caused by mutated allele silencing, considering that the overexpression but also the downregulation of TDP-43 may lead to neuronal dysfunction.57 However, in heterozygous null TARDPB mouse models the levels of TDP-43 are normal, suggesting tightly controlled compensation mechanisms following the loss of one allele.58,59 Indeed, TDP-43 can regulate the level of its own transcript in vivo.60 Another important point to consider regards the fact that TDP-43 aggregation mimics its knockdown since the expression of several proteins is affected in both conditions in the same way.61 Therefore, on the one hand, allele-specific silencing could reduce TDP-43 aggregation by restoring the functionality of several proteins, on the other hand, TDP-43 could regulate its own expression to compensate for the loss of the mutated allele. Clearly, this aspect should be evaluated further in vivo.
Still, our results are promising, considering the reversion of the phenotypes observed in the cellular models that we used. The results of Phase 1–2 trials based on the use of tofersen, an antisense nucleotide for ALS caused by SOD1 mutations, have been recently published. The treatment has proved to reduce SOD1 levels in the cerebrospinal fluid, but the study was not designed to test the safety and clinical efficacy of the drug.62 However, in October 2021, the results of another study were released. It was a 6-month Phase 3 randomized study, VALOR, which did not reach the primary endpoint represented by change in the ALS Functional Rating Scale-Revised (ALSFRS-R), even though signs of reduced disease progression were observed. Still, recently, at the European Network to cure ALS, Biogen Inc. presented new 12-month data that highlighted how starting tofersen treatment early slowed ALS progression. Indeed, the results showed a slowed decline of respiratory and clinical functions (measured by ALSFRS-R), strength and quality of life as well as a reduction in neurofilament, a marker of neurodegeneration (investors.biogen.com). Moreover, tofersen is being studied in the ATLAS study designed to evaluate the impact of initiating the treatment in the pre-symptomatic carriers of SOD1 variants.63
A further point that should be considered regards the application of siRNA treatment to neuronal diseases, which has limitations due to the lack of an efficient method of siRNA delivery in neurons. However, in recent years, several attempts have been made to chemically modify siRNAs in order to make them more effective.64 For instance, the generation of nanocomplexes or hydrophobically modified siRNAs were able to cause gene silencing in neurons in vitro and in mice.65,66
In the light of all this, the efficacy of M10 should be evaluated in neurons considering the possibility of chemically modifying it to improve its uptake by neurons and, in the future, it could be considered as a therapeutic agent in trials possibly evaluating its effects in the early stages of the disease.
Conclusions
We identified a siRNA able to specifically silence the mutated allele of TDP-43. The treatment of TDP-43G376D expressing cells with this siRNA strongly reduced the presence of TDP-43 aggregates in the cytoplasm. This treatment eased the burden of aggregates and thereby increased cell viability and diminished oxidative stress. This represents a potentially powerful therapeutic tool for the future that should be tested on iPS-derived neurons.
Supplementary Material
Acknowledgement
The authors thank Christopher E. Shaw for the pGFP-TDP43 construct and Association 2HE for constant support. Graphical abstract was created with Biorender.com.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- ASOs
antisense oligonucleotides
- DCFH-DA
2′,7′-dichlorodihydrofluorescein diacetate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- ROS
reactive oxygen species
- RRM
RNA recognition motifs
- SDS
sodium dodecyl sulphate
- SEM
standard error mean
- siRNA
small interfering RNA
- SOD1
superoxide dismutase 1
- TDP-43
TAR DNA-binding protein 43
Contributor Information
Roberta Romano, Department of Biological and Environmental Sciences and Technologies (DiSTeBA), Via Provinciale Lecce-Monteroni n.165, 73100 Lecce, Italy.
Maria De Luca, Department of Biological and Environmental Sciences and Technologies (DiSTeBA), Via Provinciale Lecce-Monteroni n.165, 73100 Lecce, Italy.
Victoria Stefania Del Fiore, Department of Biological and Environmental Sciences and Technologies (DiSTeBA), Via Provinciale Lecce-Monteroni n.165, 73100 Lecce, Italy.
Martina Pecoraro, ALS Clinical Research Center, P Giaccone University Hospital and Department of Biomedicine, Neuroscience and advanced Diagnostic (BIND), University of Palermo, via Gaetano La Loggia n° 1, 90129 Palermo, Italy.
Serena Lattante, Section of Genomic Medicine, Department of Life Sciences and Public Health, Università Cattolica del Sacro Cuore, Rome, Italy; Unit of Medical Genetics, Department of Laboratory and Infectious Disease Sciences, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy.
Mario Sabatelli, Adult NEMO Clinical Center, Unit of Neurology, Department of Aging, Neurological, Orthopedic and Head-Neck Sciences, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy; Section of Neurology, Department of Neuroscience, Faculty of Medicine and Surgery, Università Cattolica del Sacro Cuore, Rome, Italy.
Vincenzo La Bella, ALS Clinical Research Center, P Giaccone University Hospital and Department of Biomedicine, Neuroscience and advanced Diagnostic (BIND), University of Palermo, via Gaetano La Loggia n° 1, 90129 Palermo, Italy.
Cecilia Bucci, Department of Biological and Environmental Sciences and Technologies (DiSTeBA), Via Provinciale Lecce-Monteroni n.165, 73100 Lecce, Italy.
Funding
The work was partially supported by Association 2HE (Center for Human Health and Environment) and by Regione Puglia - Grant Malattie Rare DUP n. 246 of 2019.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
Data availability
All data are available from the first author on request.
References
- 1.Gentile F, Scarlino S, Falzone YM, et al. The peripheral nervous system in amyotrophic lateral sclerosis: Opportunities for translational research. Front Neurosci. 2019;13:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jo M, Lee S, Jeon YM, Kim S, Kwon Y, Kim HJ. The role of TDP-43 propagation in neurodegenerative diseases: Integrating insights from clinical and experimental studies. Exp Mol Med. 2020;52(10):1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinarbasi ES, Cağatay T, Fung HYJ, Li YC, Chook YM, Thomas PJ. Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci Rep. 2018;8(1):7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30(2):639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acid Res. 2009;37(6):1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukavsky PJ, Daujotyte D, Tollervey JR, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20(12):1443–1449. [DOI] [PubMed] [Google Scholar]
- 8.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11(3):344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191(4):1041–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284(30):20329–20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suk TR, Rousseaux MWC. The role of TDP-43 mislocalization in amyotrophic lateral sclerosis. Mol Neurodegener. 2020;15(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baralle M, Buratti E, Baralle FE. The role of TDP-43 in the pathogenesis of ALS and FTLD. Biochem Soc Trans. 2013;41:1536–1540. [DOI] [PubMed] [Google Scholar]
- 14.Da Cruz S, Cleaveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Wang L, Huntley ML, Perry G, Wang X. Pathomechanisms of TDP-43 in neurodegeneration. J Neurochem. 2018;146:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Ynifo-Mojado L, Martin-Ruiz I, Sutherland JD. Efficient allele-specific targeting of LRRK2 R1441 mutations mediated by RNAi. PLoS One. 2011;6(6):e21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez-Lebrón E, Gouvion CM, Moore SA, Davidson BL, Paulson HL. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer’s disease. Mol Ther. 2009;17:1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardi MS, Jaspers L, Spronkmans C, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217(2):312–319. [DOI] [PubMed] [Google Scholar]
- 19.Fiszer A, Ellison-Klimontowicz ME, Krzyzosiak WJ. Silencing of genes responsible for polyQ diseases using chemically modified single-stranded siRNAs. Acta Biochim Pol. 2016;63(4):759–764. [DOI] [PubMed] [Google Scholar]
- 20.Maxwell MM, Pasinelli P, Kazantsev AG, Brown RHJ. RNA interference-mediated silencing of mutant superoxide dismutase rescues cyclosporin A-induced death in cultured neuroblastoma cells. Proc Natl Acad Sci U S A. 2004;101:3178–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralph GS, Radcliffe PA, Day DM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med. 2005;11(4):429–433. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura AL, Shum C, Scotter EL, et al. Allele-specific knockdown of ALS-associated mutant TDP-43 in neural stem cells derived from induced pluripotent stem cells. PLoS One. 2014;9(3):e91269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conforti FL, Sproviero W, Simone IL, et al. TARDBP gene mutations in south Italian patients with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82(5):587–588. [DOI] [PubMed] [Google Scholar]
- 24.De Luca M, Romano R, Bucci C. Role of the V1G1 subunit of V-ATPase in breast cancer cell migration. Sci Rep. 2021;11(1):4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romano R, Calcagnile M, Margiotta A, et al. RAB7A regulates vimentin phosphorylation through AKT and PAK. Cancers (Basel). 2021;13(9):2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano R, Rivellini C, De Luca M, et al. Alteration of the late endocytic pathway in Charcot-Marie-Tooth type 2B disease. Cell Mol Life Sci. 2021;78(1):351–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano R, Del Fiore VS, Saveri P, et al. Autophagy and lysosomal functionality in CMT2B fibroblasts carrying the RAB7K126R mutation. Cells. 2022;11(3):496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cicardi ME, Cristofani R, Rusmini P, et al. Tdp-25 routing to autophagy and proteasome ameliorates its aggregation in amyotrophic lateral sclerosis target cells. Sci Rep. 2018;8(1):12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS-R: A revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS study group (phase III). J Neurol Sci. 1999;169(1–2):13–21. [DOI] [PubMed] [Google Scholar]
- 30.Kimura F, Fujimura C, Ishida S, et al. Progression rate of ALSFRS-R at time of diagnosis predicts survival time in ALS. Neurology. 2006;66(2):265–267. [DOI] [PubMed] [Google Scholar]
- 31.Ledford H. Gene-silencing technology gets first drug approval after 20-year wait. Nature. 2018;560(7718):291–292. [DOI] [PubMed] [Google Scholar]
- 32.Towne C, Raoul C, Schneider BL, Aebischer P. Systemic AAV6 delivery mediating RNA interference against SOD1: Neuromuscular transduction does not alter disease progression in fALS mice. Mol Ther. 2008;16(6):1018–1025. [DOI] [PubMed] [Google Scholar]
- 33.Che MX, Jiang LL, Li HY, Jiang YJ, Hu HY. TDP-35 sequesters TDP-43 into cytoplasmic inclusions through binding with RNA. FEBS Lett. 2015;589(15):1920–1928. [DOI] [PubMed] [Google Scholar]
- 34.Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem. 2008;283(19):13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang YS, Tsai KJ, Chang YJ, et al. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat Commun. 2014;5:4824. [DOI] [PubMed] [Google Scholar]
- 36.Orellana EA, Kasinski AL. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio Protoc. 2016;6(21):e1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano N, Catalani A, Lattante S, et al. ALS skin fibroblasts reveal oxidative stress and ERK1/2-mediated cytoplasmic localization of TDP-43. Cell Signal. 2020;70:109591. [DOI] [PubMed] [Google Scholar]
- 38.Kim H, Xue XY. Detection of total reactive oxygen species in adherent cells by 2′,7′-dichlorodihydrofluorescein diacetate staining. J Vis Exp. 2020;160:10.3791/60682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masrori P, Van Damme P. Amyotrophic lateral sclerosis: A clinical review. Eur J Neurol. 2020;27(10):1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardiman O, Al-Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3:17085. [DOI] [PubMed] [Google Scholar]
- 41.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377(2):162–172. [DOI] [PubMed] [Google Scholar]
- 42.van Es MA, Hardiman O, Chio A, et al. Amyotrophic lateral sclerosis. Lancet. 2017;390(10107):2084–2098. [DOI] [PubMed] [Google Scholar]
- 43.Mitsuzawa S, Akiyama T, Nishiyama A, et al. TARDBP P.G376D mutation, found in rapid progressive familial ALS, induces mislocalization of TDP-43. eNeurologicalSci. 2018;11:20–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czell D, Andersen PM, Morita M, Neuwirth C, Perren F, Weber M. Phenotypes in Swiss patients with familial ALS carrying TARDBP mutations. Neurodegener Dis. 2013;12(3):150–155. [DOI] [PubMed] [Google Scholar]
- 45.Mitsumoto H, Brooks BR, Silani V. Clinical trials in amyotrophic lateral sclerosis: Why so many negative trials and how can trials be improved? Lancet Neurol. 2014;13(11):1127–1138. [DOI] [PubMed] [Google Scholar]
- 46.Lacomblez L, Bensimon G, Leigh PN, Guillet P, Meininger V. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis/riluzole study group II. Lancet. 1996;347(9013):1425–1431. [DOI] [PubMed] [Google Scholar]
- 47.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/riluzole study group. New Engl J Med. 1994;330(9):585–591. [DOI] [PubMed] [Google Scholar]
- 48.Hinchcliffe M, Smith A. Riluzole: Real-world evidence supports significant extension of median survival times in patients with amyotrophic lateral sclerosis. Degener Neurol Neuromuscul Dis. 2017;7:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinaldi C, Wood MJA. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9–21. [DOI] [PubMed] [Google Scholar]
- 50.Klim JR, Vance C, Scotter EL. Antisense oligonucleotide therapies for amyotrophic lateral sclerosis: Existing and emerging targets. Int J Biochem Cell Biol. 2019;110:149–153. [DOI] [PubMed] [Google Scholar]
- 51.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 52.Sabatelli M, Zollino M, Conte A, et al. Primary fibroblasts cultures reveal TDP-43 abnormalities in amyotrophic lateral sclerosis patients with and without SOD1 mutations. Neurobiol Aging. 2015;36(5):2005.e5–2005.e13. [DOI] [PubMed] [Google Scholar]
- 53.de Boer EMJ, Orie VK, Williams T, et al. TDP-43 proteinopathies: A new wave of neurodegenerative diseases. J Neurol Neurosurg Psychiatry. 2021;92(1):86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y-J, Xu Y-F, Cook C, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106(18):7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyerowitz J, Parker SJ, Vella LJ, et al. C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol Neurodegener. 2011;6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuo X, Zhou J, Li Y, et al. TDP-43 aggregation induced by oxidative stress causes global mitochondrial imbalance in ALS. Nat Struct Mol Biol. 2021;28(2):132–142. [DOI] [PubMed] [Google Scholar]
- 57.Riku Y, Seilhean D, Duyckaerts C, et al. Pathway from TDP-43-related pathology to neuronal dysfunction in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Int J Mol Sci. 2021;22(8):3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kraemer BC, Schuck T, Wheeler JM, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119(4):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sephton CF, Good SK, Atkin S, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285(9):6826–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ayala YM, De Conti L, Avendaño-Vázquez SE, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30(2):277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prpar Mihevc S, Baralle M, Buratti E, Rogelj B. TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci Rep. 2016;6:33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller T, Cudkowicz M, Shaw PJ, et al. Phase 1-2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2020;383(2):109–119. [DOI] [PubMed] [Google Scholar]
- 63.Benatar M, Wuu J, Andersen PM, et al. Design of a randomized, placebo-controlled, phase 3 trial of tofersen initiated in clinically presymptomatic SOD1 variant carriers: The ATLAS study. Neurotherapeutics. 2022;19(4):1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu B, Zhong L, Weng Y, et al. Therapeutic siRNA: State of the art. Signal Transduct Target Ther. 2020;5(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo Q, Zheng X, Yang P, et al. Small interfering RNA delivery to the neurons near the amyloid plaques for improved treatment of Alzheimer’s disease. Acta pharma Sin B. 2019;9(3):590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alterman JF, Hall LM, Coles AH, et al. Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol Ther Nucleic Acids. 2015;4(12):e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the first author on request.