Significance
In Drosophila, long-term memory (LTM) of learned odors by aversive conditioning usually requires strong spaced training to induce CREB-dependent gene regulation. Here, we report a spaced training-induced CREBB repression-dependent gating mechanism for memory regulation. Weak learning induces 5-HT1A protein synthesis in the early α/β subset of mushroom body neurons that inhibits LTM formation. Alternatively, strong spaced learning induces CREBB synthesis, which then suppresses this inhibitory effect and enables LTM formation. Our deconvolution of a cellular mechanism in identified neurons presages the level of assessment needed to understand the neural basis of memory in any species.
Keywords: memory regulation, memory consolidation, memory inhibition, CREBB, 5-HT1A
Abstract
Learned experiences are not necessarily consolidated into long-term memory (LTM) unless they are periodic and meaningful. LTM depends on de novo protein synthesis mediated by cyclic AMP response element-binding protein (CREB) activity. In Drosophila, two creb genes (crebA, crebB) and multiple CREB isoforms have reported influences on aversive olfactory LTM in response to multiple cycles of spaced conditioning. How CREB isoforms regulate LTM effector genes in various neural elements of the memory circuit is unclear, especially in the mushroom body (MB), a prominent associative center in the fly brain that has been shown to participate in LTM formation. Here, we report that i) spaced training induces crebB expression in MB α-lobe neurons and ii) elevating specific CREBB isoform levels in the early α/β subpopulation of MB neurons enhances LTM formation. By contrast, learning from weak training iii) induces 5-HT1A serotonin receptor synthesis, iv) activates 5-HT1A in early α/β neurons, and v) inhibits LTM formation. vi) LTM is enhanced when this inhibitory effect is relieved by down-regulating 5-HT1A or overexpressing CREBB. Our findings show that spaced training-induced CREBB antagonizes learning-induced 5-HT1A in early α/β MB neurons to modulate LTM consolidation.
Only certain learned episodic events are consolidated into long-term memory (LTM). These tend to be meaningful, recurrent, and their predictability is adaptive (1). Whereas the biochemistry of LTM consolidation is known to involve both positive and negative gene regulation in response to salient periodic experiences (2), the details of this process and the specific neural circuit elements involved remain poorly understood.
Studies of aversive olfactory associative memory in Drosophila have found several distinct genetically defined phases. Most are labile and decay in a short time. However, one type – LTM – is protein synthesis dependent and can persist from several days to a week (3). Weaker learning and labile memory can be generated with a single cycle of training (1×), three spaced cycles with 15-min rest intervals (3×S), or ten massed cycles of training without rest intervals (10×M). Strong learning and persistent LTM consolidation require five or more spaced cycles of training (5×S, 10×S) (3, 4).
Studies of aversive olfactory associative memory in Drosophila also continue to reveal neural components of an olfactory memory circuit. Early investigators noted a correlation between MB size and the complexity of insect social behavior (5). MBs were later found to support odor-shock associative learning and cAMP signaling-mediated subsequent memory formation (6–9). Each paired MB consists of about 2,500 intrinsic neurons belonging to three classes (γ, α’/β’ and α/β) distinguished by their morphology, spatial organization, and birth sequences (10). α/β neurons are subdivided further into three types: pioneer α/β, early α/β and late α/β neurons defined by similar criteria (10). Among these subpopulations of MB neurons, late and pioneer α/βneurons together with various MB extrinsic neurons participate in spaced training-induced LTM formation and retrieval, respectively (11–13). Some claim that learning-induced protein synthesis in intrinsic MB neurons is required for LTM formation (14). Inhibition of protein synthesis in MB after strong spaced training, however, did not reduce LTM (12). Instead, several studies have shown that de novo protein synthesis is only required in a small number of extrinsic MB output neurons to promote LTM consolidation (3, 12, 13). A resolution of these contradictory results is long overdue.
LTM consolidation has also been shown to require cyclic AMP response element-binding protein (CREB) in MB (15). However, protein synthesis and CREB involvement in the biochemistry of LTM gating remain poorly understood. Moreover, the specific regulators and effectors of this biochemistry have not been characterized within identified neurons. CREB proteins are conserved transcription factors shown to regulate LTM formation in mollusks, nematodes, insects, and rodents (2–4, 12, 14–20). Two creb genes are described in Drosophila. One CREBA isoform is encoded by crebA, whereas several distinct CREBB isoforms are generated from crebB (16, 17). CREBB-a is a synthetic (chimeric) protein, whereas isoforms −b, −c, and −d can be readily detected in fly brains (17, 18). Among these, CREBB-c was the first isoform described in several independent studies (16–18). Interpreting the putative roles of certain CREB isoforms in LTM formation has been challenging, due to a diversity of expression patterns, methodologies, and apparently conflicting behavioral outcomes published since 1995 (4).
Broad CREBB-a and CREBB-b expression is reported to enhance, inhibit, or have no effect on LTM formation (3, 4, 12, 14, 18, 19). Interestingly, CREBB in MB was shown to be necessary for LTM formation. Activation of a signaling pathway upstream of CREBB was also found to elevate LTM (15), whereas inducible MB-targeted overexpression of CREBB-b did not impair LTM (12). By comparison, crebA expression levels in MB are much lower than crebB (20), and not surprisingly CREBA in MB does not influence LTM (15). CREBB does regulate transcription in MB during LTM formation, but it is unclear which isoforms regulate which effector genes in which MB compartments in response to spaced training. Elsewhere in the olfactory memory circuit of the fly, LTM was shown to depend on protein synthesis in a pair of dorsal anterior lateral (DAL) neurons (12). CREBA and CREBB in these neurons were found to activate and block gene expression, and to enhance and inhibit LTM formation, respectively (20).
Here, we report a compelling biochemical gating mechanism in MB that explains how spaced training removes the antagonizing effects of learning on LTM. Our data show that 1) weak training induces 5-HT1A in early α/β neurons to inhibit LTM, and 2) strong spaced training induces CREBB in early α/β neurons to repress 5-HT1A and relieve this inhibitory effect on LTM.
Results
Spaced Training Induces crebB Transcription.
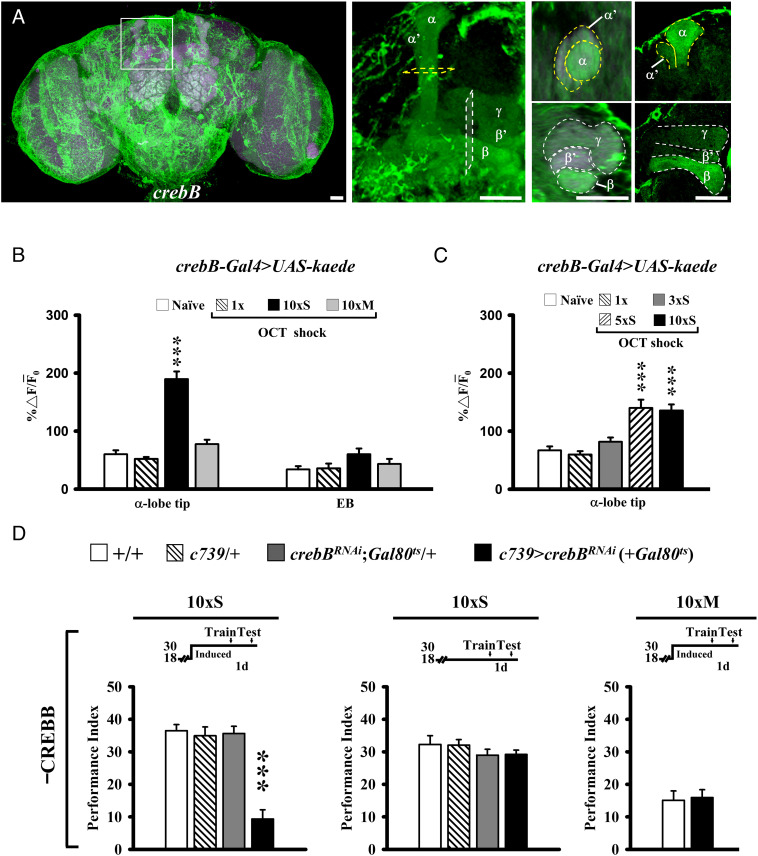
To address whether spaced training is required to induce crebB expression, we generated a crebB promoter-driven Gal4 transgene containing an 11-kb 5′ genomic sequence just upstream of the first open reading frame (see Methods) (17). This crebB-Gal4 drives GFP expression in most glia cells and brain neurons, including most MB neurons, though higher levels of expression can be seen in α/β compared with α’/β’ or γ neurons (Fig. 1A). By photo converting preexisting green KAEDE to red prior to training (see Methods) (12), we measured significantly more newly synthesized green KAEDE driven by the activated crebB-Gal4 in the MB α-lobe during 24-h intervals after 5×S or 10×S training, but not after 1×, 3×S, or 10×M training in comparison with naïve control flies (Fig. 1 B and C and SI Appendix, Fig. S1). This training-induced increase in crebB KAEDE appeared specific to the MB neurons because spaced training did not significantly change the levels of new crebB KAEDE in the ellipsoid body (EB) (control) (Fig. 1 B, Right). Moreover, using adult stage-specific Gal4/UAS system-targeted RNA interference (RNAi) to knock down CREBB in MB α/β neurons under the temporal control of a temperature-sensitive tub-Gal80ts protein (the inhibitory effect of Gal80ts protein on GAL4 expression at 18 °C is inactivated at 30 °C), we observed impairment of 1-d memory after 10×S (Fig. 1 D, Left). Control flies kept at 18 °C after 10×S training or at 30 °C after 10×M training were unaffected (Fig. 1 D, Center and Right). These results suggest that multiple sessions of spaced training induce CREBB-dependent modulation of LTM.
Fig. 1.
Spaced training activates crebB transcription. (A) CREBB expression visualized in dissected brains with crebB-Gal4-driven UAS-mCD8::GFP (green), counterstained with DLG-antibody immunostaining (magenta), viewed under a confocal microscope. crebB-Gal4 labels all MB lobes (Center). Optical slices of vertical and horizontal MB lobes (Right) show more prominent expression in α/β neurons than in α’/β’ and γ neurons. (Scale bar, 10 μm.) (B and C) Promotor activation of crebB 24 h after training reported by de novo KAEDE synthesis, estimated by the ratio of new (green, 488 nm) and preexisting (red, 561 nm) proteins (% ∆ F/F¯0). For each brain, single optical slices through the MB α-lobe tip or EB were imaged under identical conditions. (B) Spaced training stimulates crebB activity preferentially in the α-lobe, in comparison with EB controls. (C) A minimum of 5×S training cycles are necessary to observe KAEDE synthesis reflecting crebB activity. Bars represent mean ± SE, n ≥ 8. (D) adult-stage-specific RNAi downregulation of CREBB proteins (–CREBB) in MB α/β neurons impairs 1-d memory after 10×S training (Left). Gal4-targeted RNAi downregulation is induced at the restrictive temperature for tub-Gal80ts (30 °C) from 7 d before training until testing. Memory is unaffected in these flies held at the permissive temperature for tub-Gal80ts (18 °C) after 10×S (Center) and at 30 °C after 10×M (Right). In the same figure, all experiments were done within the same experimental group. Memory performance indices are calculated as the normalized percent avoidance of shock-paired odor. Bars represent mean ± SE, n = 8/bar unless stated otherwise. *P < 0.05; **P < 0.01, ***P < 0.001. All genotypes are listed in SI Appendix, Table S2.
Specific Isoforms of CREBB in Early α/β Neurons Enhance LTM.
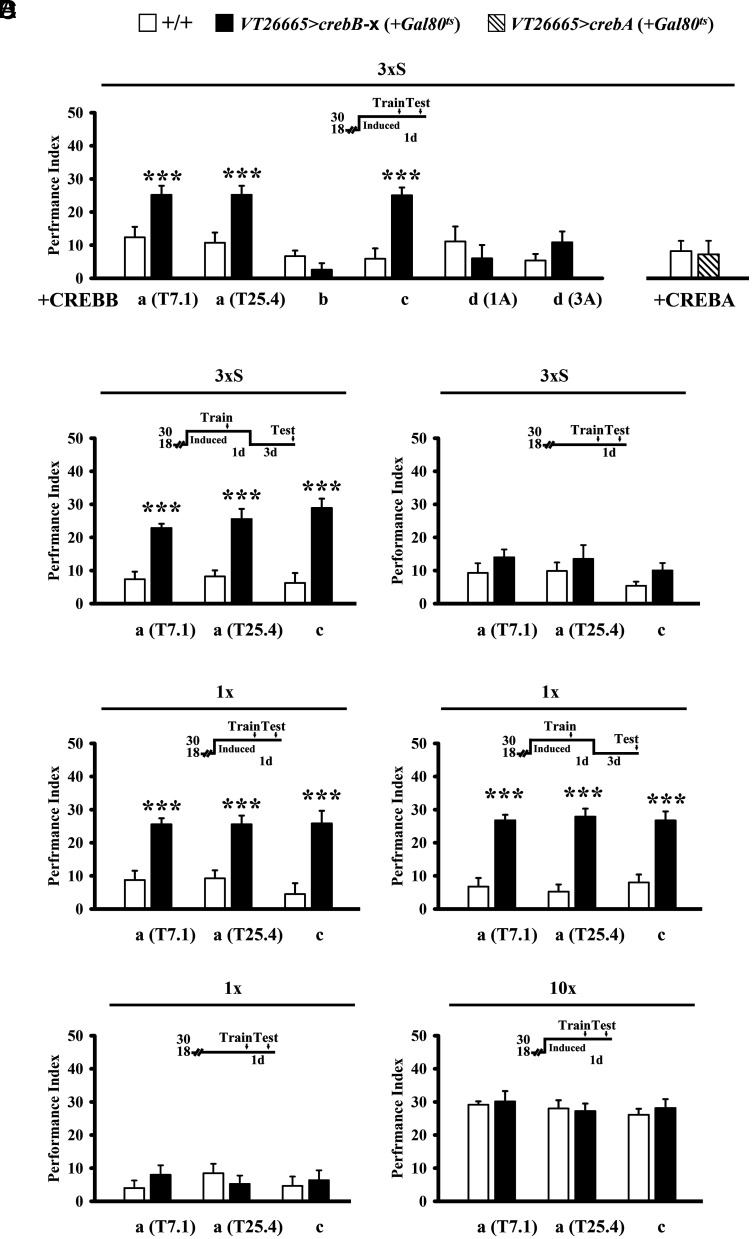
Next, we wanted to verify whether spaced training-induced CREBB expression in MB enhances LTM formation. The crebB gene produces several isoforms (17). Among these, acute expression of a chimeric crebB-a transgene throughout the fly was first reported by Yin et al. (4) to function as a transcriptional activator and to enhance LTM formation. Interestingly, a replication of this study by Perazzona et al. (18) did not support this finding. These authors also identified a frame-shift mutation that led to a premature stop codon in the original transgene. Further tests using a corrected crebB-a transgene also failed to enhance LTM. These conflicting early studies using transgene manipulations in whole flies predated the refinement of Gal4 targeting reagents that can now deconvolute cellular processes to subsets of identified neurons. Here we have reinvestigated the involvement of CREBB in MB during LTM formation. We first repeated the original experiments and confirmed that heat-shock-induced CREBB-a (corrected) transgene overexpression neither enhanced LTM after 3×S or 1× nor impaired LTM after 10×S (SI Appendix, Fig. S2A). Surprisingly, we found that this hs-Gal4 protocol drives strong GFP expression in glia cells and variable expression in brain neurons, including especially weak expression in the MB (SI Appendix, Fig. S2B). Independent of any possible CREBB-a influence in MB during LTM formation, these observations suggest that low levels of expression by hs-Gal4 induction may contribute to the observed inconsistent results. We next targeted crebB-a transgene expression (two independent lines) to all intrinsic MB neurons (OK107) or to α/β neurons (c739) and observed enhanced 1-d memory after 3×S to levels normally seen after 10×S. By comparison, crebB-a transgene expression in γ neurons (VT44966) or α’/β’ neurons (VT30604) did not affect LTM (SI Appendix, Fig. S3A). Moreover, among these α/β neurons, only transgene expression in early α/β (VT26665) neurons contributed to crebB-a-enhanced LTM but not in pioneer (VT9843) or late α/β (VT0110) neurons (Fig. 2A and SI Appendix, Fig. S4A). These observations verify that expression of chimeric crebB-a in Drosophila can enhance LTM. Notably however, full length crebB-a cDNA or mRNA have not been detected or isolated in the fly brain, and our results probably involve biochemical processes that do not occur naturally (17, 18).
Fig. 2.
Overexpression of CREB in early α/β neurons enhances LTM formation. (A) Overexpressing CREBB-a (with two independent lines: T7.1 or T25.4) or CREBB-c but not CREBB-b, CREBB-d, or CREBA proteins in early α/β neurons enhances 1-d memory after 3×S. Gal4-targeted transgene overexpression is induced at the restrictive temperature for tub-Gal80ts (30 °C) from 5 d before training until testing. (B) This enhancement lasts at least 4 d (Left). Memory is unaffected in these flies held at the permissive temperature for tub-Gal80ts (18 °C) after 3×S (Right) (C) Overexpressing CREBB-a (2 copies of T7.1 or T25.4 wild-type transgenes) in early α/β neurons enhances 1-d memory after only 1× (Left) and lasts at least 4 d (Right). (D) One-day memory is also unaffected in these flies held 18 °C after 1× (Left) and at 30 °C after 10×S (Right).
Next, we investigated whether overexpressing naturally occurring CREB isoforms in early α/β neurons (VT26665) may also promote or enhance LTM. Interestingly, we found that inducible expression of crebB-c enhanced LTM after 3×S. The effect was not observed when expressing crebB-b, crebB-d, or crebA transgenes (Fig. 2A). crebB-a and crebB-c memory enhancement after 3×S persisted for at least 4 d (Fig. 2 B, Left and SI Appendix, Fig. S3B), while control flies kept at 18 °C after spaced training (Fig. 2 B, Right and SI Appendix, Fig. S3 C, Left) or at 30 °C after massed training were unaffected (SI Appendix, Fig. S3 C, Right and SI Appendix, Fig. S4B). Similarly, crebB-enhanced LTM was also observed after 1× training and the effect persisted for at least 4 d (Fig. 2C), while control flies kept at 18 °C after one session training were unaffected (Fig. 2 D, Left). We observed no enhancement of memory after 10×S training (Fig. 2 D, Right and SI Appendix, Fig. S3D). Together, these findings suggest that specific crebB isoforms (CREBB-a and CREBB-c) in early α/β neurons can promote LTM consolidation.
Activation of 5-HT1A Receptors Inhibits LTM Formation.
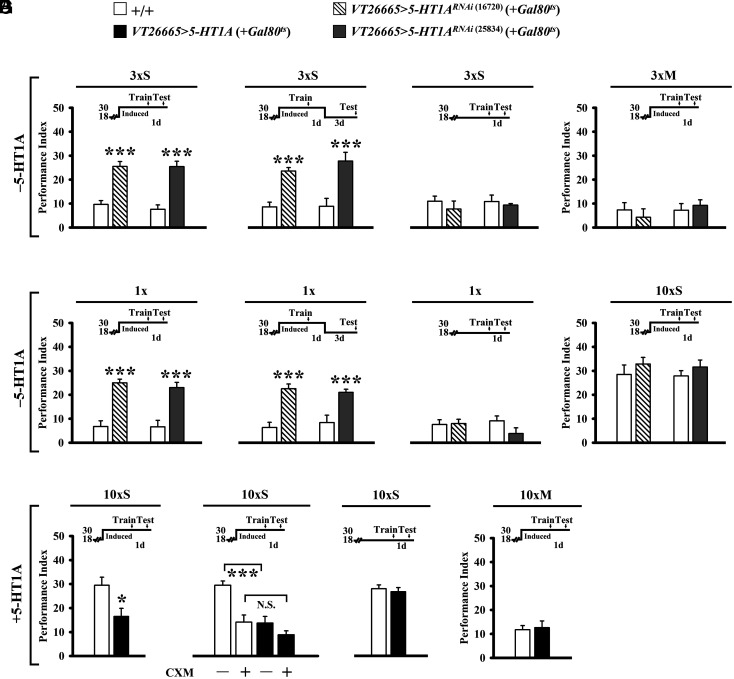
Next, we sought to identify CREBB-regulated gene products that are involved in LTM formation. An intriguing candidate for investigation was the conserved G protein-coupled 5-hydroxytryptamine receptor 1A (5-HT1A) (21, 22). In Drosophila, activation of 5-HT1A which is negatively coupled with adenylyl cyclase decreases cAMP levels (21). cAMP signaling in MB α/β neurons is required for LTM formation (9). Consistent with this observation, studies in mice and humans showed that 5-HT1A agonists inhibit memory formation, whereas 5-HT1A antagonists prevent memory loss (22). To investigate 5-HT1A regulation of LTM formation in flies, we manipulated its expression levels in MB α/β neurons. RNAi knockdown of 5-HT1A in early α/β neurons was sufficient to enhance both 1- and 4-d memory after 3×S or 1× (Fig. 3 A and B). This manipulation had no effect on control flies kept at 18 °C after 3×S, after 1× (Fig. 3 C and D), at 30 °C after massed training or after 10×S training (Fig. 3 E and F). In contrast, overexpression of the UAS-5-HT1A transgene in early α/β neurons impaired 1-d memory after 10×S (Fig. 3 G, Left). We found no further impairment in combination with the protein synthesis inhibitor, cycloheximide (CXM), suggesting that elevated 5-HT1A levels were sufficient to block memory consolidation (Fig. 3 G, Center Left). This manipulation had no effect on control flies kept at 18 °C after 10×S (Fig. 3 G, Center Right) or at 30 °C after massed training (Fig. 3 G, Right). Collectively, these data indicate that 5-HT1A receptors are integral to the molecular mechanism in early α/β neurons that inhibits LTM formation.
Fig. 3.
5-HT1A receptors in early α/β neurons inhibits LTM formation. (A and B) Adult-stage-specific RNAi downregulation of proteins in early α/β neurons (with two independent RNAi constructs; two copies of each transgene) enhances 1-d memory after 3×S or only 1× training (Left) and lasts at least 4 d (Right). Gal4-targeted 5-HT1A downregulation is induced at the restrictive temperature for tub-Gal80ts (30 °C) from 5 d before training until testing. (C and D) Memory is unaffected in these flies held at the permissive temperature for tub-Gal80ts (18 °C) after 3×S or 1× training. (E and F) One-day memory is also unaffected in these flies held at 30 °C after 3×M or 10×S training. (G) Overexpressing 5-HT1A proteins in early α/β neurons impaired 1-d memory after 10×S (Left). The impairments were similar to those induced by CXM feeding (Center Left). Memory was unaffected at 18 °C after 10×S (Center Right) and at 30 °C after 10×M (Right).
5-HT1A Transcription Is Induced after Weak Learning.
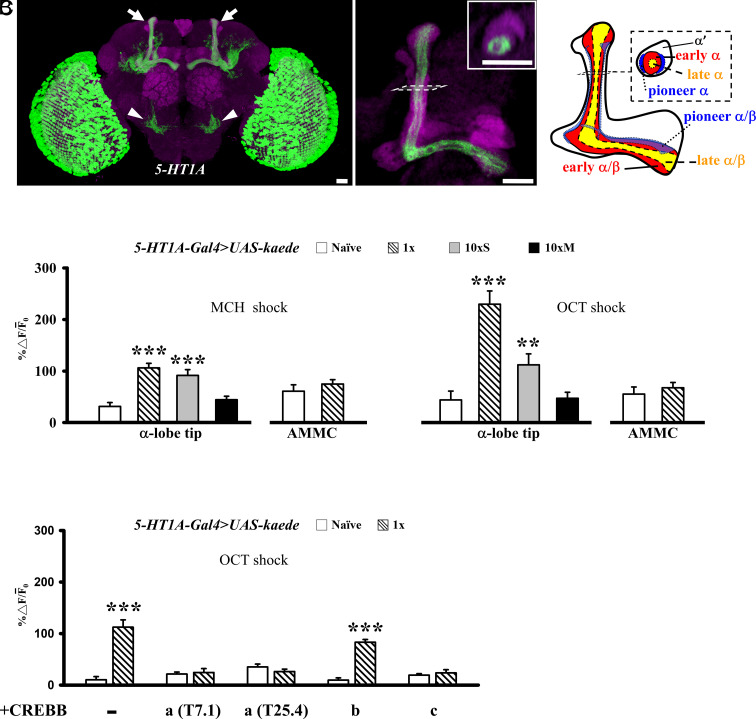
We next analyzed whether transcription of 5-HT1A was activated in trained flies. To visualize transcriptional activity during LTM formation, we generated a 5-HT1A-Gal4 line containing the 5.2-kb 5-HT1A promoter region (see Methods). 5-HT1A-Gal4 was expressed preferentially in ~420 early α/β neurons (SI Appendix, Table S1), in addition to the antennal mechanosensory and motor center (AMMC) and the medulla (Fig. 4A). When driving GFP expression with VT26665-Gal4 alone, 5-HT1A-Gal4 alone, or with both, the total number of labeled MB neurons remained similar (SI Appendix, Table S1), indicating that the two Gal4 lines were expressed in the same population of early α/β neurons.
Fig. 4.
Learning-induced activation of the 5-HT1A gene is suppressed by CREBB. (A) Preferential expression of 5-HT1A-Gal4 >UAS-mCD8::GFP (green) at MB (arrows), AMMC (arrowheads) and medulla. Inset, a cross-section of MB vertical lobes shows specific expression in early α/β neurons. Right, a schematic representation of the spatial distributions of three types of α/β neurons. Inset, a cross-section at the vertical lobes. (Scale bar, 10 μm.) (B) 5-HT1A promoter activity reported by KAEDE fluorescent protein synthesis, determined by the ratio between new (green, 488 nm) to preexisting (red, 561 nm) KAEDE (% ∆ F/F¯0) 24 h after no training (naïve), 1×, and 10×S or 10×M. (C) Effects of CREBB isoform expression on 1×-induced 5-HT1A gene activation. In control 5-HT1A-Gal4>UAS-kaede flies, an elevation of 5-HT1A gene activity was monitored by green KAEDE synthesis measured at the tip of α lobes 24 h after 1×. The 5-HT1A activation was abolished by overexpressing CREBB-a (two copies of T7.1 or T25.4 wild-type transgenes) or CREBB-c, but not CREBB-b. See SI Appendix, Table S1 for details.
To evaluate whether training might induce 5-HT1A expression, we quantified levels of 5-HT1A promoter-driven photoconvertible fluorescent protein KAEDE (see Methods) after 1× training. Surprisingly, we found significantly higher levels of newly synthesized green KAEDE (reflecting 5-HT1A activation) at the MB α-lobe during the 24-h interval after 1× training, but not after 10×M training or in naïve control flies (Fig. 4B). Importantly, 10×S training also increased KAEDE levels. This training-induced increase in 5-HT1A KAEDE appeared specific to the early α/β neurons because 1× training did not significantly change the levels of new 5-HT1A KAEDE in the AMMC (control) (Fig. 4B).
To clarify the mechanism underlying the increased 5-HT1A transcription after learning, we pharmacologically manipulated serotonin levels in 5-HT1A KAEDE flies by administering a serotonin precursor, L-5-hydroxytryptophan, or a tryptophan hydroxylase inhibitor, DL-p-chlorophenyl-alanine, to increase or reduce serotonin levels, respectively (23). We found that increasing serotonin enhanced new KAEDE synthesis at the MB α-lobe during the 24-h interval after 1× training, whereas decreasing serotonin reduced new KAEDE synthesis during the same period (SI Appendix, Fig. S5A). Decreasing serotonin levels also reduced new KAEDE synthesis in the MB α-lobe in naïve 5-HT1A KAEDE flies but not in naïve cryptochrome KAEDE (control) flies (SI Appendix, Fig. S5B) (12), verifying that the KAEDE signal was an accurate and specific reporter of 5-HT1A gene activation. Thus, 5-HT1A activation in early α/β neurons is dependent on both training and 5HT levels.
CREBB Antagonizes Learning-Induced 5-HT1A.
Our results suggest that learning-induced protein synthesis in early α/β neurons inhibits LTM formation. What spaced training-responsive mechanism might relieve the 5-HT1A-dependent inhibitory effect in these neurons to gate LTM formation? We tested the hypothesis that spaced training-induced CREBB protein suppresses 5-HT1A by monitoring gene activation reported by KAEDE fluorescent protein synthesis after targeting CREBB overexpression to early α/β neurons. Our results showed that 5-HT1A gene activation after 1× training was repressed by overexpressing CREBB-a (T7.1), CREBB-a (T25.4), or CREBB-c but not by CREBB-b, in comparison with naïve control flies (Fig. 4C). In considering whether GAL4 levels may be limiting or subthreshold for driving expression of multiple UAS-transgenes, we noted that KAEDE was coexpressed together with CREBB-b to the same level as in control flies carrying a single UAS-driven KAEDE transgene (Fig. 4C).
Lastly, we tested whether CREBB-c can antagonize 5-HT1A regulation of LTM formation (Fig. 3G). Indeed, we observed normal 1-d LTM after 10×S when both 5-HT1A and CREBB-c were simultaneously overexpressed (SI Appendix, Fig. S6, Left). Interestingly, these manipulations also enhanced 1-d memory after 3×S or 1× (SI Appendix, Fig. S6, Center and Right). In all cases, the enhanced memory was similar to memory in the wild-type control flies after 10×S. In summary, learning-induced 5-HT1A produced an inhibitory effect on LTM formation. This can be relieved by CREBB expression, suggesting that an inhibition/disinhibition mechanism in early α/β MB neurons gates LTM formation.
Discussion
Recurrent spaced learning has been shown to relieve inhibition and gate LTM formation in animal models (2–4). However, gene regulatory mechanisms that act to filter relevant signals of repeated events and override inhibitory constraints in identified circuit elements remain unknown. Here, our data suggest that MB neurons in Drosophila provide a compelling cellular gating mechanism for LTM formation. Weak learning is sufficient to increase 5-HT1A synthesis in early α/β neurons, and these neurons produce a downstream inhibitory effect on LTM formation. After spaced training, CREBB expression represses further 5-HT1A synthesis, thereby relieving the inhibitory effect on LTM formation. These conclusions are supported by several lines of evidence: i) CREBB transcription increased after 5×S or 10×S but not after 1× (Fig. 1); and ii) RNAi-mediated knockdown of CREBB in α/β impaired LTM (Fig. 1), while overexpression of a crebB-a or -c transgene enhanced LTM (Fig. 2). iii) Conversely, RNAi-mediated knockdown of 5-HT1A in early α/β neurons enhanced LTM, while overexpression of a 5-HT1A transgene impaired LTM (Fig. 3); and iv) 1× was sufficient to activate 5-HT1A (Fig. 4B), and this activation was inhibited by expression of CREBB proteins (Fig. 4C). v) Furthermore, overexpression of 5-HT1A-mediated LTM impairment was fully rescued by CREBB overexpression (SI Appendix, Fig. S6). Together, these findings suggest that synthesis of 5-HT1A and CREBB proteins in response to training operate like an opposing molecular switch (3, 4) to inhibit or disinhibit downstream LTM formation, respectively.
Previous reports suggested that expression of a chimeric CREBB-a transcriptional activator and a CREBB-b transcriptional repressor throughout whole fly enhanced and impaired LTM formation, respectively (3, 4, 17, 24). Subsequently, CREBB-a-dependent enhancement of LTM was not observed using a hs-Gal4 driver (18) that we showed has low expression in MB (SI Appendix, Fig. S2). Chronic expression of a CREBB-b in all α/β neurons was shown to impair 1-d memory after spaced training (14). Chen et al. (12) documented, however, that these chronic disruptions of CREBB-b produced developmental abnormalities in MB structure. In contrast, acute induced expression of CREBB-b only in adult α/β neurons did not impair 1-d memory after spaced training (and did not produce structural defects). Using a different inducible system (MB247-Switch) to acutely expresses CREBB-b in γ and α/β neurons, Hirano et al. (19) showed a mild impairment of 1-d memory after spaced training. More interestingly, they used various molecular genetic tools to show that interactions among CREBB, CREB-binding protein, and CREB-regulated transcription coactivator in MB were clearly involved in LTM formation or maintenance, respectively. Using the same inducible gene switch tool, Miyashita et al. (25) showed a positive regulatory loop between Fos and CREBB in MB during LTM formation – but they did not show behavioral data pertaining to manipulation of CREBB per se – nor did they restrict their experiments to early α/β neurons.
Zhang et al. (26) expressed a CRE-luciferase transgene in different subpopulations of MB neurons and then monitored luciferase activity in live flies at various times after spaced training. Immediately after spaced training, some patterns of luciferase expression decreased (OK107 expressing in all MB neurons; c739 expressing in all α/β neurons; 1471 expressing in γ neurons), or increased (c747 and c772 expressing variably in all MB neurons), or showed no detectable change (c320 expressing variably in γ, α’/β’ and α/β subpopulation, 17d expressing primarily in late α/β and in early α/β neurons). Indeed, these authors pointed out that, because the CRE-reporter was expressed in more than one subpopulation of MB neurons, only net effects of CREB function could be quantified. Furthermore, this study did not elucidate which CREBB isoforms might increase or decrease after spaced training. Obviously, this information would be critical if different isoforms have opposing activator and repressor functions in specific MB neuron subpopulations. Our study provides a dramatic example of this point. By restricting our manipulation to early α/β neurons in adult stage animals, we show that enhanced LTM formation after acute CREBB-c overexpression is comparable to the net effect of chimeric CREBB-a overexpression in whole flies (Figs. 1 and 2) (4), and that spaced training serves to increase the expression of CREBB in these early α/β neurons (Fig. 1).
Yin et al. (17) reported that the CREBB-a isoform functions as a PKA-responsive transcriptional activator and the CREBB-b isoform functions as a repressor of CREBB-a-induced gene activation. Using new KAEDA synthesis as a reporter for temporal gene activation, we have previously shown that CREBB-b in DAL neurons represses CREBA-mediated gene activation to inhibit LTM formation (12, 20). Here, in early α/β MB neurons, our KAEDA experiments indicate that CREBB-a and CREBB-c, but not CREBB-b, both repress 5-HT1A-mediated inhibition to gate LTM formation (Fig. 4). These findings demonstrate a neuron- and training-specific CREBA activation and CREBB repression of effecter genes involved in modulating LTM formation (20, 24). Although crebB promoter-driven Gal4 expression, crebBRNAi downregulation (Fig. 1), and cell-type specific transcriptomes (27) show CREBB expression in early α/β neurons, it remains unclear whether specific naturally occurring CREBB isoforms in these neurons serve to modulate LTM formation.
How is the learning-induced LTM gating mechanism differentially regulated by different (1×, 10×M or 10×S) training protocols? Expression of both 5-HT1A and crebB in early α/β MB neurons was elevated 24 h after 10×S, whereas only 5-HT1A was induced after 1×, and neither gene was induced after 10×M (Figs. 1 and 4B). Why do we still see elevated 5-HT1A after 10×S (Fig. 4B), when constitutive expression of CREBB proteins suppresses 5-HT1A expression (Fig. 4C)? A possible explanation is that 5-HT1A may be normally activated as an early response to 1×, whereas crebB induction by 10×S is not evident for about 3 h (4, 25, 28). Gradual cessation of 5-HT1A transcription by the delayed 10×S-induced CREBB expression may account for lower KAEDE levels observed in one odor/shock pairing experiment (Fig. 4B). Interestingly, our data showed that even with elevated 5-HT1A, CREBB proteins can still enhance 1-d memory (SI Appendix, Fig. S6), suggesting that CREBB-mediated inhibition is rather complex.
Massed training appears not to activate or suppress learning-induced transcriptional activity in early α/β neurons, and 5-HT1A nor crebB is activated after 10×M (Figs. 1 and 4). Nevertheless, massed training may antagonize LTM formation. For instance, in MB neurons, spaced training induces repetitive waves of Ras/mitogen-activated protein kinase (MAPK) activity (28), activates MAPK translocation to the nucleus mediated by importin-7 (29), increases CREBB expression (25) and, in DAL neurons, training induces activity-dependent crebA, CamKII, and per gene expression (12, 20) – all of which are not activated after massed training. These notions above suggest that massed training produces a more upstream general suppression of these 1×- and 10×S-induced genes required for inhibitory/gating mechanisms allocated in MB and DAL neurons, respectively.
An LTM enhancing role associated with CREBB expression and protein synthesis inhibition is a novel aspect of this gating mechanism (Figs. 2, 3, and 4). Our previous study showed that inhibition of protein synthesis in MB after strong spaced training did not reduce LTM. Since it would not be possible to detect enhanced performance in these experiments, we cannot exclude the possibility that this inhibition might eliminate downregulation of LTM effector genes, with a net effect of promoting the formation of LTM rather than impairing it (12). Here, we identified that synthesis of new 5-HT1A proteins in early α/β neurons after weak learning provides negative regulation and produces a downstream inhibitory effect on LTM formation (Figs. 3 and 4). Surprisingly, CREBB protein synthesis in early α/β neurons after strong spaced training provides positive regulation by antagonizing this negative effect of 5-HT1A on LTM (Figs. 1, 2, and 4). Thus, CREBB-mediated repression is equivalent to the net effect of blocking protein synthesis in MB. Both relieve downstream inhibition and enhance rather than impair LTM formation. We propose that CREBB-mediated inhibition operates both directly by repressing gene transcription and indirectly through activating their downstream translational suppression (30).
Together, our experiments uncover a biochemical LTM gating mechanism that requires delicate regulation of protein synthesis and repression after training within identified neurons. More broadly, our observations also highlight the need to confirm the regulatory functions of specific CREB isoforms in identified neuronal subtypes before making conclusions about their roles in LTM formation.
The discovery that molecules in early α/β neurons inhibit LTM formation is relevant to our future studies. Another persistent anesthesia-resistant form of memory (ARM) is also mediated by α/β neurons (23) and has been shown to inhibit LTM formation (31). 5-HT1A appears to be a key protein involved in both ARM (23) and LTM (Fig. 3). Furthermore, the interaction of serotonin released from dorsal paired medial neurons and 5-HT1A in α/β neurons is necessary for sleep (32). CREBB expression in MB is also under circadian regulation (33), which together suggests mechanistic links between ARM, LTM, sleep, and circadian timing in early α/β neurons.
Materials and Methods
In this report, we used an automated olfactory aversive learning task (3, 4) and assessed LTM after RNAi knockdown or expression of transgenes in these temporally and spatially restricted domains to identify the subsets of neurons critical for this task. We evaluated training-responsive CREBB or 5-HT1A expression with confocal microscopy using a Gal4-targeted UV-sensitive KAEDE reporter system (12). In various experiments, flies were fed CXM to provide a systemic level of protein synthesis inhibition. See SI Appendix, SI Materials and Methods for details of fly strains, reagents, and all procedures.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the Bloomington Drosophila stock center, Vienna Drosophila RNAi Center and Kyoto Drosophila Genomics Resource Centers for fly stocks. We also thank the Developmental Studies Hybridoma Bank for the antibodies. We thank Chia–Ling Wu for preliminary cloning of the 5-HT1A promoter and Yuan-Ruei Huang for preliminary counting of the number of MB neurons. This work was supported by the Brain Research Center under the Higher Education Sprout Project cofunded by the Ministry of Education and the National Science and Technology Council in Taiwan, the Yushan Scholar Program from the Ministry of Education in Taiwan, and Dart NeuroScience LLC.
Author contributions
H.-W.L., C.-C.C., T.T., and A.-S.C. designed research; H.-W.L. and C.-C.C. performed research; R.-Y.J. and L.C. contributed new reagents/analytic tools; H.-W.L., C.-C.C., J.S.d.B., T.T., and A.-S.C. analyzed data; and H.-W.L., C.-C.C., J.S.d.B., T.T., and A.-S.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Chun-Chao Chen, Email: chenchunchao@gapp.nthu.edu.tw.
Ann-Shyn Chiang, Email: aschiang@life.nthu.edu.tw.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Kelley P., Whatson T., Making long-term memories in minutes: A spaced learning pattern from memory research in education. Front. Hum. Neurosci. 7, 589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abel T., Kandel E., Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res. Brain Res. Rev. 26, 360–378 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Yin J. C., Tully T., CREB and the formation of long-term memory. Curr. Opin. Neurobiol. 6, 264–268 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Yin J. C., Del Vecchio M., Zhou H., Tully T., CREB as a memory modulator: Induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81, 107–115 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Heisenberg M., What do the mushroom bodies do for the insect brain? An introduction. Learn Mem. 5, 1–10 (1998). [PMC free article] [PubMed] [Google Scholar]
- 6.Heisenberg M., Borst A., Wagner S., Byers D., Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 2, 1–30 (1985). [DOI] [PubMed] [Google Scholar]
- 7.de Belle J. S., Heisenberg M., Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science 263, 692–695 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Zars T., Fischer M., Schulz R., Heisenberg M., Localization of a short-term memory in Drosophila. Science 288, 672–675 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Blum A. L., Li W., Cressy M., Dubnau J., Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr. Biol. 19, 1341–1350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H. H., Lai J. S., Chin A. L., Chen Y. C., Chiang A. S., A map of olfactory representation in the Drosophila mushroom body. Cell 128, 1205–1217 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Perisse E., Burke C., Huetteroth W., Waddell S., Shocking revelations and saccharin sweetness in the study of Drosophila olfactory memory. Curr. Biol. 23, R752–R763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C. C., et al. , Visualizing long-term memory formation in two neurons of the Drosophila brain. Science 335, 678–685 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Pai T. P., et al. , Drosophila ORB protein in two mushroom body output neurons is necessary for long-term memory formation. Proc. Natl. Acad. Sci. U.S.A. 110, 7898–7903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D., Akalal D. B., Davis R. L., Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron 52, 845–855 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee P. T., et al. , A kinase-dependent feedforward loop affects CREBB stability and long term memory formation. Elife 7, e33007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usui T., Smolik S. M., Goodman R. H., Isolation of Drosophila CREB-B: A novel CRE-binding protein. DNA Cell Biol. 12, 589–595 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Yin J. C., et al. , A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol. Cell Biol. 15, 5123–5130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perazzona B., Isabel G., Preat T., Davis R. L., The role of cAMP response element-binding protein in Drosophila long-term memory. J. Neurosci. 24, 8823–8828 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano Y., et al. , Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science 339, 443–446 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Lin H. W., Chen C. C., de Belle J. S., Tully T., Chiang A. S., CREBA and CREBB in two identified neurons gate long-term memory formation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 118, e2100624118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tierney A. J., Structure and function of invertebrate 5-HT receptors: A review. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 128, 791–804 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Huang Y., Thathiah A., Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett. 589, 1607–1619 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lee P. T., et al. , Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 108, 13794–13799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tubon T. C. Jr., et al. , dCREB2-mediated enhancement of memory formation. J. Neurosci. 33, 7475–7487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyashita T., Kikuchi E., Horiuchi J., Saitoe M., Long-term memory engram cells are established by c-Fos/CREB transcriptional cycling. Cell Rep. 25, 2716–2728.e2713 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Tanenhaus A. K., Davis J. C., Hanlon B. M., Yin J. C., Spatio-temporal in vivo recording of dCREB2 dynamics in Drosophila long-term memory processing. Neurobiol. Learn. Mem. 118, 80–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih M. M., Davis F. P., Henry G. L., Dubnau J., Nuclear transcriptomes of the seven neuronal cell types that constitute the Drosophila mushroom bodies. G3 (Bethesda) 9, 81–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagani M. R., Oishi K., Gelb B. D., Zhong Y., The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell 139, 186–198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., et al. , Importin-7 mediates memory consolidation through regulation of nuclear translocation of training-activated MAPK in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, 3072–3077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubnau J., et al. , The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Isabel G., Pascual A., Preat T., Exclusive consolidated memory phases in Drosophila. Science 304, 1024–1027 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Haynes P. R., Christmann B. L., Griffith L. C., A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4, e03868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fropf R., et al. , Time of day influences memory formation and dCREB2 proteins in Drosophila. Front. Syst. Neurosci. 8, 43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.