Significance
R-loop plays a role in transcription regulation, but the presence of R-loop structure in our genome also poses a threat to genome stability. Tumor suppressor BRCA1 has been implicated in repairing R-loop-associated DNA damage and suppressing R-loop formation during RNA polymerase II-mediated production of messenger RNA via protein–protein interactions with transcription factors and enzymes that resolve R-loops. In this study, we describe a mechanism by which BRCA1 facilitates antisense ribosomal RNA production and double-stranded ribosomal RNA formation to suppress RNA polymerase I-induced ribosomal R-loops. Through this function, BRCA1 also contributes to regulating ribosomal biogenesis, an essential cellular process known to be altered in Fanconi anemia and linked to the etiology of bone marrow failure.
Keywords: BRCA1, R-loops, ribosomal RNA, antisense RNA
Abstract
R-loops, or RNA:DNA hybrids, can induce DNA damage, which requires DNA repair factors including breast cancer type 1 susceptibility protein (BRCA1) to restore genomic integrity. To date, several pathogenic mutations have been found within the tandem BRCA1 carboxyl-terminal (BRCT) domains that mediate BRCA1 interactions with proteins and DNA in response to DNA damage. Here, we describe a nonrepair role of BRCA1 BRCT in suppressing ribosomal R-loops via two mechanisms. Through its RNA binding and annealing activities, BRCA1 BRCT facilitates the formation of double-stranded RNA between ribosomal RNA (rRNA) and antisense-rRNA (as-rRNA), hereby minimizing rRNA hybridization to ribosomal DNA to form R-loops. BRCA1 BRCT also promotes RNA polymerase I-dependent transcription of as-rRNA to enhance double-stranded rRNA (ds-rRNA) formation. In addition, BRCA1 BRCT-mediated as-rRNA production restricts rRNA maturation in unperturbed cells. Hence, impairing as-rRNA transcription and ds-rRNA formation due to BRCA1 BRCT deficiency deregulates rRNA processing and increases ribosomal R-loops and DNA breaks. Our results link ribosomal biogenesis dysfunction to BRCA1-associated genomic instability.
DNA damage can be generated both by environmental assaults, such as radiation, and during endogenous cellular processes such as transcription (1–5). Although transcription is essential for cell growth, this process creates unusual nucleic acid structures, such as RNA:DNA hybrids, known as R-loops, that form between the DNA template and newly synthesized RNA (1–5). During transcription in unperturbed cells, positively supercoiled DNA accumulates ahead of the active RNA polymerase (RNAP) elongation complex, and negatively supercoiled DNA is generated behind the RNAP. Negatively supercoiled DNA, if not removed by topoisomerases, can facilitate the formation of R-loops. In addition to the action of topoisomerases, R-loop formation can also be avoided by the binding of RNA processing factors to the newly synthesized RNA to prevent nascent RNA from invading the DNA template to generate R-loops (3–5). The formation and accumulation of R-loops can stall DNA replication forks, leading to DNA double-strand breaks, which require DNA repair factors, such as breast cancer type 1 susceptibility protein (BRCA1), to restore genomic integrity (4). The critical role of BRCA1 in repairing R-loop-induced DNA breaks is underscored by the observation that in brca1-deficient cells, R-loop-induced DNA breaks accumulate at the subsets of RNA polymerase II (RNAPII) pausing sites, near promoters and transcription termination regions (6–8).
Mutations in BRCA1 predispose individuals to several types of cancer, including breast, ovarian, prostate, and pancreatic cancers (9–11). In addition to increasing cancer incidence, a subset of BRCA1 mutations also contribute to Fanconi anemia, a rare recessive disorder characterized by bone marrow failure and acute myeloid leukemia (12). BRCA1 is a 220 kDa protein containing a RING domain and a nuclear export signal at the amino (N) terminus; its carboxyl (C) terminus includes a coiled-coil (CC) domain and tandem BRCA1 C terminus (BRCT) domains (10). Cumulative studies reported the involvement of BRCA1 in multiple cellular processes including DNA damage repair, and transcription and translational regulation. For example, BRCA1 participates in the activation of homology-directed repair (HDR) to repair DNA breaks by counteracting p53-binding protein 1 (53BP1), allowing DNA end resection to generate single-stranded DNA (ssDNA) as a substrate for homology searching and pairing by RAD51 to initiate DNA recombination (9). The role of BRCA1 in HDR requires functional RING, CC, and BRCT domains (10). The RING domain of BRCA1 interacts with BRCA1-associated RING domain protein 1 (BARD1) to form an active E3-ubiquitin ligase, which ubiquitinates histone H2A to facilitate the removal of 53BP1 from DNA breaks (13, 14). The CC and BRCT domains of BRCA1 participate in protein–protein interactions with factors such as PALB2, BRCA2, and CtIP to initiate HDR (9, 15–17). In addition to interacting with proteins, the tandem BRCT domains of BRCA1 are capable of binding to DNA (18), and BRCA1 DNA binding affinity is enhanced by the interaction of its RING domain with BARD1 (19).
BRCA1 not only participates in repairing DNA damage induced by transcription but also has a direct role in transcriptional regulation. For example, BRCA1 copurifies with the RNAPII holoenzyme via its BRCT domains (20). Furthermore, BRCA1 interacts with the estrogen receptor through its N terminus to inhibit estrogen-mediated, RNAPII-dependent transcriptional activation, and this regulation is abolished by BRCA1 mutations that are linked to increasing cancer incidence (21). Interestingly, BRCA1 is involved in regulating RNAPII pausing at both promoter and termination pause sites via its interactions with NELFB and senataxin, and this function helps to suppress R-loop accumulation at promoter and termination pause sites (6, 8, 22). In addition, BRCA1 has been implicated in binding to a subset of messenger RNAs (mRNA) to regulate their protein translation (23). However, it remains unclear whether RNA interaction involves the BRCT domains that bind to DNA. In addition to RNAPII-dependent transcription, BRCA1 also facilitates RNA polymerase I (RNAPI)-dependent ribosomal RNA (rRNA) transcription (24). This function depends on a direct interaction with RNAPI, and this interaction is weakened by DNA damage, which causes cells to halt RNAPI transcription in response to cellular stress (24). BRCA1 may also act as a repressor for RNAPIII-dependent tRNA and small nucleolar RNA transcription (25).
In this study, we provide evidence that BRCA1 promotes double-stranded rRNA (ds-rRNA) formation between rRNA and antisense-rRNA (as-rRNA) via its BRCT domain to suppress ribosomal R-loops and restrict rRNA maturation. BRCA1 BRCT binds to both rRNA and as-rRNA in cells. Through its RNA binding, BRCA1 BRCT facilitates the annealing between two complementary RNA strands, and this activity correlates with reduced rRNA hybridization to ribosomal DNA (rDNA) to form ribosomal R-loops. In addition, we show that in unperturbed cells, ds-rRNA formation can be enhanced by the RNAPI-mediated as-rRNA production in a manner also dependent on BRCA1 BRCT. The BRCA1-dependent as-rRNA production reduces rRNA processing efficiency likely due to the formation of ds-rRNA precursors that hinder RNA endo- and exonuclease cleavages. Consequently, BRCA1 deficiency or expression of BRCA1 pathogenic S1655 mutation within BRCT that hinders RNA binding and ds-rRNA formation leads to elevated ribosomal R-loops and altered ribosomal biogenesis.
Results
BRCA1 BRCT Suppresses Ribosomal R-Loops.
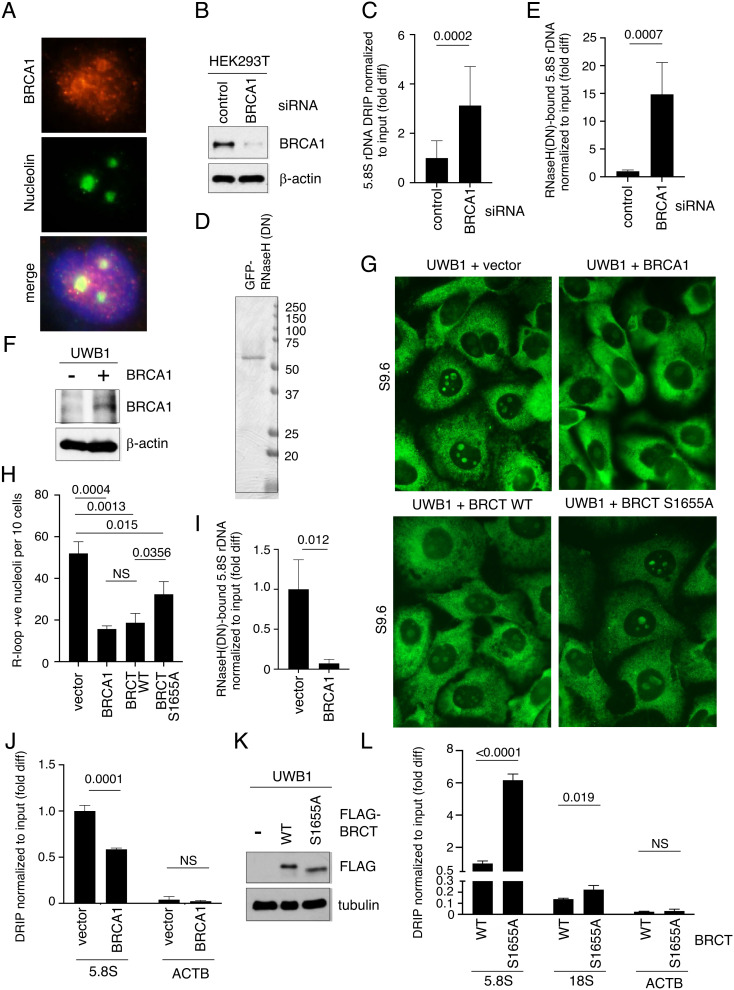
BRCA1 suppresses RNAPII-dependent R-loops via its interactions with transcription factors to regulate RNAPII pausing and termination (6, 8, 22). Because BRCA1 has been shown to localize to the nucleolus and promote RNAPI-dependent rRNA transcription (24, 26), we asked whether BRCA1 also contributes to R-loop regulation at rDNA loci. Using an antibody against BRCA1, we confirmed by immunofluorescence staining that BRCA1 was enriched in the nucleoli in unperturbed human osteosarcoma U2OS cells (Fig. 1A and SI Appendix, Fig. S1). We next analyzed the effect of BRCA1 on ribosomal R-loops in HEK293T cells treated with control or BRCA1 siRNA (Fig. 1B). The isolated DNA was subjected to DNA:RNA immunoprecipitation (DRIP) using an R-loop-specific antibody (S9.6), followed by quantitative PCR (qPCR) using primers flanking the 5.8S rDNA locus and normalized to the input DNA. Any RNase H-resistant nucleic acid binding to S9.6 was subtracted as background. We found that BRCA1 knockdown (KD) cells contained more R-loops at the rDNA loci compared to those of control KD cells (Fig. 1C). We also isolated R-loop-containing DNA from BRCA1 KD cells using purified recombinant RNase H1 D210N (DN) mutant proteins (Fig. 1D), which are catalytically inactive but show strong binding specificity toward R-loops (27). After subtracting nonspecific DNA binding to the beads, we confirmed that R-loops at rDNA loci were elevated in the BRCA1 KD cells compared to those of control cells (Fig. 1E).
Fig. 1.
BRCA1 BRCT suppresses R-loops at rDNA. (A) Representative images showing immunofluorescent staining of BRCA1 (red, Top) and nucleolin (green, Middle) in U2OS cells. DAPI (blue) in the merged image (Bottom) was used to detect nuclei. (B) Western blot analysis of BRCA1 using an anti-BRCA1 antibody in whole cell extracts prepared from HEK293T cells transfected with control siRNA or BRCA1-specific siRNA. β-actin is used as a loading control. (C) DRIP analysis using S9.6 antibody to measure RNase H-sensitive, R-loop levels normalized to input DNA at 5.8S rDNA loci isolated from HEK293T cells transfected with control or BRCA1 siRNA. Y-axis represents fold difference (fold diff) relative to control siRNA. (D) Recombinant GFP-His6-RNaseH1 (DN) proteins purified from E. coli, separated by SDS-PAGE and stained with Coomassie blue. (E) Quantification of R-loops copurified with GFP-His6-RNaseH1 (DN) proteins normalized to input DNA at 5.8S rDNA loci in HEK293T cells transfected with control or BRCA siRNAs. Y-axis represents fold difference relative to control siRNA. (F) Western blot analysis of BRCA1 using an anti-BRCA1 antibody in whole cell extracts prepared from brca1-deficient UWB1 cells with or without exogenously expressed full-length BRCA1 proteins. β-actin is used as a loading control. (G) Representative images showing immunofluorescent staining of R-loops (green) using S9.6 antibody in UWB1 cells expressing indicated BRCA1, BRCA1 BRCT WT, or S1566A mutant fragment. Empty vector was used as transfection control. (H) Quantification of the number of nucleoli per ten cells showing positive nucleolar signals from samples shown in G. At least 150 cells were analyzed per sample. (I) Quantification of R-loops copurified with GFP-His6-RNaseH1 (DN) proteins normalized to input DNA at 5.8S rDNA loci in UWB1 cells with or without exogenously expressed BRCA1 protein. Y-axis represents fold difference relative to the UWB1 cells transfected with the vector alone. (J) DRIP analysis using S9.6 antibody to measure RNase H-sensitive, R-loop levels normalized to input DNA at 5.8S rDNA loci (Left) and ACTB EX3 (Right) isolated from UWB1 cells with or without exogenously expressed BRCA1. Y-axis represents fold difference relative to the 5.8S rDNA DRIP signal in the UWB1 cells transfected with the vector alone. (K) Western blot analysis of FLAG-BRCT using an anti-FLAG antibody in whole cell extracts prepared from brca1-deficient UWB1 cells with or without an exogenously expressed BRCA1 BRCT WT or S1655A fragment. Tubulin is used as a loading control. (L) RNase H-sensitive, R-loop levels measured by DRIP at 5.8S rDNA locus (Left), 18S rDNA locus (Middle), and ACTB EX3 region (Right) normalized to input DNA in UWB1 cells expressing WT or S1655A BRCT mutant fragment. Y-axis represents fold difference relative to the 5.8S rDNA DRIP signal in UWB1 cells transfected with WT BRCT.
To further correlate BRCA1 expression with ribosomal R-loops, we ectopically expressed a FLAG-tagged full-length (FL) BRCA1 in BRCA1-null UWB1.289 (UWB1) ovarian cancer cells (Fig. 1F). Similar to the endogenous BRCA1 (SI Appendix, Fig. S1), ectopically expressed FLAG-BRCA1 proteins were enriched in the nucleoli of the UWB1 cells (SI Appendix, Fig. S2, 2nd column). Under the same exposure, UWB1 cells contained more intense nucleolar R-loop staining compared to that of BRCA1-proficient HEK293T cells (SI Appendix, Fig. S3A). Importantly, FLAG-BRCA1 expression reduced nucleolar R-loop signals in UWB1 cells (Fig. 1G, compare two Top panels; Fig. 1H, compare two Left columns). Similar R-loop reduction was also observed in BRCA1-null HCC1937 breast cancer cells transfected with FLAG-BRCA1 (SI Appendix, Fig. S3B). Dot blot analysis supported the conclusion that R-loop signals, which were eliminated by RNase H treatment, were reduced in both UWB1 (SI Appendix, Fig. S3 C and D) and HCC1937 cells (SI Appendix, Fig. S3 E and F) complemented with BRCA1 compared to cells transfected with empty vector alone. We further performed R-loop pulldown using RNase H (DN) mutant proteins (Fig. 1I) and S9.6 DRIP (Fig. 1J) to demonstrate that ectopic expression of BRCA1 decreased R-loop levels at rDNA loci in UWB1 cells. In contrast, the level of R-loops detected at the ACTB EX3 locus, which has been shown to form R-loops in cells (28), did not change significantly after BRCA1 complementation (Fig. 1J).
Interestingly, BRCA1 BRCT, when expressed as a FLAG-tagged protein fragment in UWB1 cells (Fig. 1K), also showed concentrated nucleolar localization (SI Appendix, Fig. S2, 3rd column) and was sufficient to reduce nucleolar R-loop signals to a level comparable to the expression of the FL BRCA1 protein (Fig. 1 G and H). On the contrary, a BRCT fragment containing a point mutation at residue S1655, which is important for protein–protein interaction and tumor suppression (29, 30), only partially suppressed nucleolar R-loops compared to the WT BRCT-expressing cells (Fig. 1 G and H). DRIP analysis further confirmed that in UWB1 cells expressing the S1655A BRCT mutant, the R-loop levels were elevated within rDNA loci, such as 5.8S and 18S, but not at ACTB EX3, compared to WT BRCT-expressing cells (Fig. 1L). These observations indicate that BRCA1 BRCT alone is sufficient to compensate for the loss of the endogenous BRCA1 and suppress ribosomal R-loops, but this function is partially impaired by the pathogenic S1655 mutation.
BRCA1 BRCT Enhances Double-Stranded RNA (dsRNA) Formation between rRNA and as-rRNA.
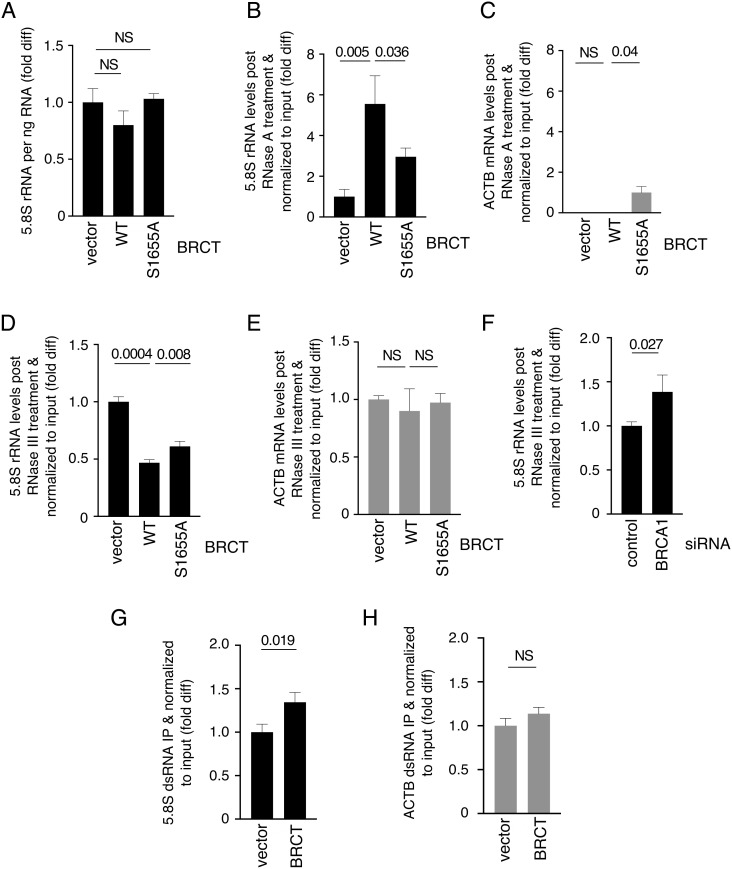
Next, we investigated the mechanism by which BRCA1 BRCT suppresses ribosomal R-loops. We found that the S1655A mutation did not affect BRCT nucleolar localization (SI Appendix, Fig. S2, right column), suggesting that the ribosomal R-loop accumulation observed in the BRCT S1655A mutant cells was not due to mislocalization of the fragment. BRCT expression in UWB1 cells also did not affect rRNA levels (Fig. 2A). Because RNase A cleaves single-stranded RNA (ssRNA) but not RNA as part of an R-loop, it was expected that expressing BRCA1 BRCT in UWB1 cells would enhance rRNA sensitivity to RNase A due to ribosomal R-loop reduction. However, we found that in the presence of BRCA1 BRCT, more rRNA molecules were resistant to RNase A (Fig. 2B), and this phenomenon was not observed with ACTB mRNA (Fig. 2C). S1655A mutation reduced BRCT-mediated resistance of 5.8S rRNA to RNase A (Fig. 2B), even though cells expressing the mutant proteins contained higher levels of ribosomal R-loops than the WT BRCT-expressing cells (Fig. 1H). In addition to R-loop, dsRNA is also more resistant to RNase A cleavage than ssRNA. Formation of dsRNA between rRNA and as-rRNA has been reported in various organisms (31–37). Therefore, we treated RNA with RNase III, which specifically cleaves dsRNA. We found that BRCT expression sensitized rRNA (Fig. 2D), but not ACTB mRNA (Fig. 2E), to RNase III cleavage compared to UWB1 cells transfected with vector alone. S1655A mutation reduced the BRCT-induced rRNA sensitivity to RNase III (Fig. 2D). Consistent with this, depleting BRCA1 in HEK293T also decreased rRNA sensitivity to RNase III treatment (Fig. 2F). Using J2 monoclonal antibody specific to dsRNA, we immunoprecipitated dsRNA and observed that BRCA1 BRCT increased dsRNA formation within rRNA (Fig. 2G) but not ACTB mRNA (Fig. 2H), further supporting a role of BRCA1 BRCT in promoting ds-rRNA.
Fig. 2.
BRCA1 BRCT promotes ds-rRNA. (A) 5.8S rRNA per ng RNA from UWB1 cells with or without ectopic expression of WT or S1655A BRCA1 BRCT was quantified using RT-qPCR. Y-axis represents fold difference relative to the UWB1 cells transfected with vector alone. (B and C) Intact 5.8S rRNA (B) and ACTB (C) post RNase A treatment normalized to total 5.8S rRNA from UWB1 cells with or without ectopic WT or S1655A BRCA1 BRCT expression were quantified using RT-qPCR. Y-axis represents fold difference relative to the UWB1 cells transfected with vector alone. (D and E) Intact 5.8S rRNA (D) and ACTB (E) post RNase III treatment normalized to total 5.8S rRNA from UWB1 cells with or without ectopic expression of WT or S1655A BRCA1 BRCT were quantified using RT-qPCR. Y-axis represents fold difference relative to UWB1 cells transfected with vector alone. (F) Intact 5.8S rRNA post RNase III treatment normalized to total 5.8S rRNA from HEK293T cells transfected with control or BRCA1 siRNA was quantified using RT-qPCR. Y-axis represents fold difference relative to control siRNA. (G and H) The amounts of ds-5.8SRNA (G) and ds-ACTB mRNA (H) immunopurified using J2 dsRNA antibody normalized to input RNA from UWB1 cells with or without exogenously expressed BRCA1 BRCT fragment were quantified using RT-qPCR. Y-axis represents fold difference relative to the UWB1 cells transfected with vector alone.
BRCA1 BRCT Exhibits Annealing Activities.
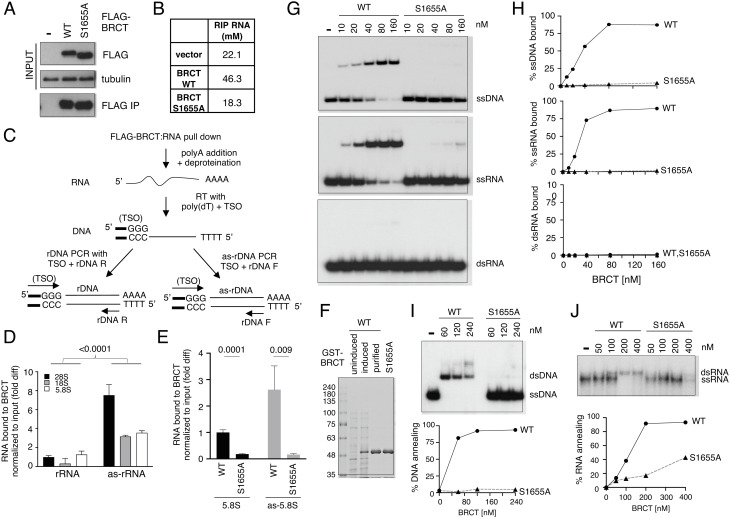
Because BRCA1 BRCT exhibits affinity toward DNA (18), we asked whether BRCA1 BRCT binds to RNA and promotes annealing between complementary RNA strands to generate dsRNA. To test this, we first performed RNA immunoprecipitation (RIP) using FLAG-BRCA1 BRCT expressed in HEK293T cells (Fig. 3A) and found RNA copurified with BRCA1 BRCT from cell extracts (Fig. 3B). BRCA1 BRCT–RNA interaction was reduced to the background level (i.e., vector) by the S1655A mutation (Fig. 3B). We next tested whether both rRNA and as-rRNA were present in cells and bound by BRCA1 BRCT. For this, we reverse transcribed and PCR amplified BRCA1 BRCT-bound RNA using oligo (dT) and TSO, followed by TA cloning. We found that in cells, BRCA1 BRCT preferentially bound to rRNA and as-rRNA sequences (SI Appendix, Fig. S4A). Using BRCT RIP followed by primer-specific RT coupled with qPCR (RT-qPCR) to distinguish between rRNA and as-rRNA (Fig. 3C), we detected the presence of 5.8S, 18S, and 28S sequences both in the form of rRNA (Fig. 3 D, Left) and as-rRNA (Fig. 3 D, Right) among the RNA species copurified with BRCA1 BRCT. Interestingly, BRCA1 BRCT exhibited a preference toward as-rRNA compared to rRNA (Fig. 3D), and the associations of BRCA1 BRCT with both rRNA and as-rRNA were impaired by the S1655A mutation (Fig. 3E).
Fig. 3.
BRCA1 BRCT binds to rRNA and as-rRNA to promote dsRNA formation. (A) Western blot analysis of WT and S1655A FLAG-BRCT using an anti-FLAG antibody before (Top) and after FLAG pulldown using M2 agarose beads (Bottom). Tubulin was used as input loading control (Middle). (B) Concentrations of RNA copurified with WT and S1655A BRCT from A measured by nanodrop. (C) Schematic of BRCT RIP coupled with RT-qPCR procedure. TSO: template switch oligo. rDNA F: rDNA forward primer. rDNA R: rDNA reverse primer. (D) Quantification of rRNA and as-rRNA copurified with WT FLAG-BRCA1 BRCT normalized to input RNA measured by RT-qPCR. Y-axis represents fold difference relative to 28S rRNA bound to BRCT. (E) 5.8S rRNA (Left) and 5.8S as-rRNA (Right) copurified with either WT or S1655A FLAG-BRCA1 BRCT normalized to input RNA were measured by RT-qPCR. Y-axis represents fold difference relative to 5.8S rRNA bound to WT BRCT. (F) Recombinant WT and S1655A GST-tagged BRCA1 BRCT protein fragments overexpressed and purified from E. coli, separated by SDS-PAGE and stained with Coomassie blue. (G) Representative images of the electrophoretic mobility shift assays of WT and S1655A GST-BRCT protein fragments binding to 32P-end labeled ssDNA (Top), ssRNA (Middle), or dsRNA (Bottom) substrate. (H) Quantification of the percentage of ssDNA, ssRNA, or ssRNA bound by WT and S1655A GST-BRCT protein fragments from G by Image J. (I, Top) Representative image of DNA:DNA annealing activity of purified recombinant BRCT WT and S1655A mutant protein fragments measured using 32P-end-labeled ssDNA with complementary ssDNA oligonucleotides. Quantification of the percentage of DNA annealing by Image J was shown (Bottom). (I, Top) Representative image of RNA:RNA annealing activity of purified recombinant BRCT WT and S1655A mutant protein fragments measured using 32P-end-labeled ssRNA with complementary ssRNA oligonucleotides. Quantification of the percentage of RNA annealing by Image J was shown (Bottom).
To further test if BRCA1 BRCT directly binds to RNA, we carried out in vitro binding analysis using recombinant GST-tagged BRCT fragments derived from BRCA1 amino acids 1582–1863 and purified to homogeneity from E. coli (Fig. 3F). Electrophoretic mobility shift assays showed that BRCA1 BRCT bound to both ssDNA and ssRNA, but not dsRNA (Fig. 3 G and H). However, in the presence of equal concentrations of the oligo substrates, BRCA1 BRCT did not show a preference toward ribosomal sequence in vitro (SI Appendix, Fig. S4 B and C). Likely, the preferential association of BRCA1 BRCT to rRNA and as-rRNA in cells is due to BRCA1 BRCT nucleolar localization (SI Appendix, Fig. S3). This observation also indicates that BRCA1 nucleolar localization is independent of BRCT binding to either DNA or RNA, because S1655A mutation impaired BRCA1 BRCT interaction with DNA/RNA without affecting its nucleolar localization.
Next, we tested whether BRCA1 BRCT exhibits annealing activity by incubating complementary ssDNA or ssRNA with purified recombinant BRCT protein fragments. Indeed, we observed increasing dsDNA (Fig. 3I) and dsRNA (Fig. 3J) formation in the presence of increasing amounts of WT BRCT, and S1655A mutation diminished this activity. These results together support a model that BRCA1 BRCT binds and promotes the annealing of complementary RNA strands to form dsRNA, and this activity depends on the S1655 residue.
BRCA1 BRCT Facilitates as-rRNA Production to Promote Ribosomal dsRNA.
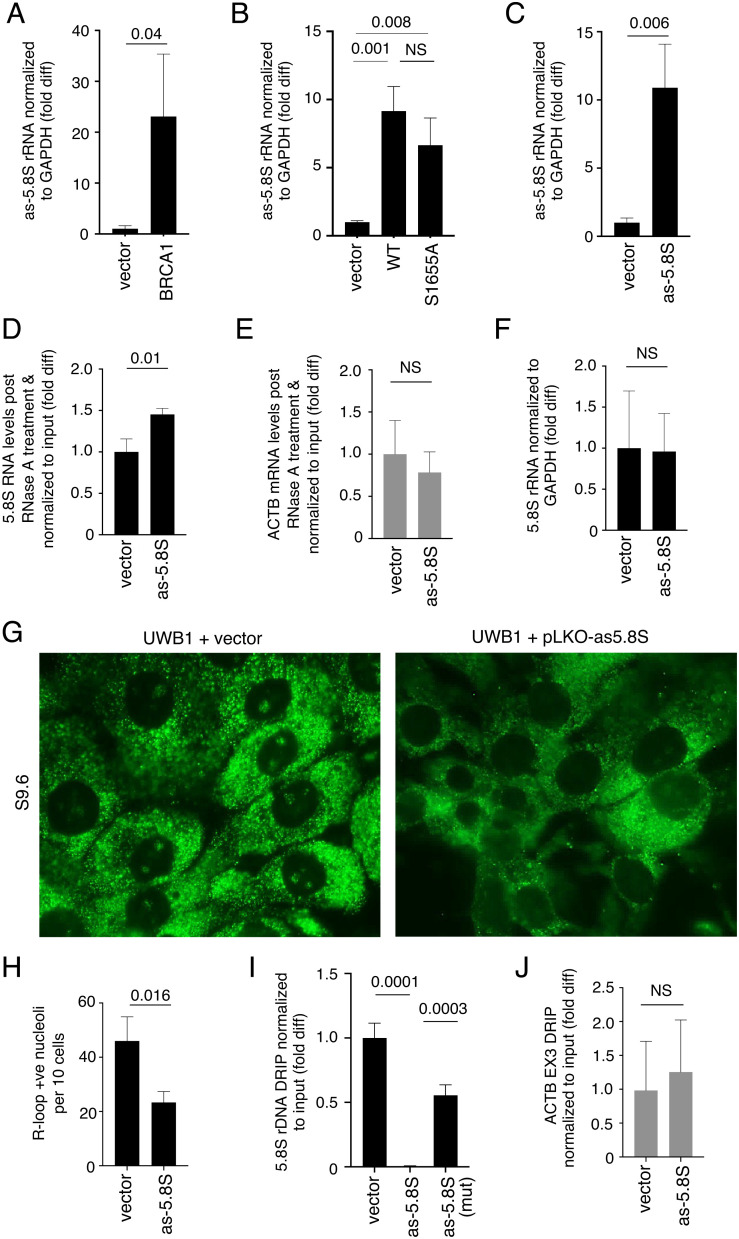
BRCA1 BRCT S1655A mutant expression was able to partially sensitize rRNA in UWB1 cells to RNase III treatment compared to cells transfected with BRCT WT fragment (Fig. 2D). However, S1655A mutation impaired BRCA1 BRCT binding to RNA for RNA annealing, suggesting that BRCA1 BRCT may possess additional activity independent of S1655 to promote dsRNA formation within rRNA. In quiescent cells, dsRNA formation within rRNA is driven by as-rRNA production (31). Because BRCA1 has been implicated in rRNA transcription (24), we asked whether BRCA1 is also involved in as-rRNA production. Indeed, expressing FLAG-BRCA1 in UWB1 cells elevated as-rRNA levels (Fig. 4A). Using both RT-qPCR (Fig. 4B) and northern blot analysis (SI Appendix, Fig. S5A), we further showed that this increase was also achieved by the expression of WT BRCA1 BRCT domain alone. Importantly, this phenomenon was not affected by S1655A mutation (Fig. 4B).
Fig. 4.
BRCA1 BRCT enhances as-rRNA production to promote ds-rRNA formation. (A and B) Quantification of as-5.8S rRNA levels normalized to GAPDH mRNA in UWB1 cells with or without ectopic BRCA1 FL expression (A) and with or without ectopic WT or S1655A BRCT expression (B) by RT-qPCR. Y-axis represents fold difference relative to UWB1 cells transfected with vector alone. (C) Quantification of as-5.8S rRNA levels normalized to GAPDH mRNA in UWB1 cells with or without ectopic as-5.8S rRNA expression by RT-qPCR. Y-axis represents fold difference relative to UWB1 cells transfected with vector alone. (D and E) Intact 5.8S rRNA (D) and ACTB (E) post RNase A treatment and normalized to input RNA in UWB1 cells with or without ectopic as-5.8S rRNA expression were quantified by RT-qPCR. Y-axis represents fold difference relative to UWB1 cells transfected with vector alone. (F) Quantification of 5.8S rRNA levels normalized to GAPDH mRNA in UWB1 cells with or without ectopic as-5.8S rRNA expression by RT-qPCR. Y-axis represents fold difference relative to UWB1 cells transfected with vector alone. (G) Representative images showing immunofluorescent staining of R-loops (green) using S9.6 antibody in UWB1 cells with (Right) or without (Left) exogenously expressed as-5.8S rRNA. (H) Quantification of the number of nucleoli per ten cells showing positive nucleolar signals from samples shown in G. At least 150 cells were analyzed per sample. (I) DRIP analysis using S9.6 antibody to measure RNase H-sensitive R-loop levels at 5.8S rDNA loci normalized to the input DNA in UWB1 cells with or without exogenously expressed as-5.8S rRNA or as-5.8S mutant (mut) rRNA. Y-axis represents fold difference relative to the vector-transfected UWB1 cells. (J) DRIP analysis using S9.6 antibody to measure RNase H-sensitive R-loop levels at ACTB EX3 normalized to the input DNA in UWB1 cells with or without exogenously expressed as-5.8S rRNA. Y-axis represents fold difference relative to the vector-transfected UWB1 cells.
We next tested if increasing as-rRNA levels may bypass the loss of BRCA1 in UWB1 cells and enhance ds-rRNA formation. For this, we exogenously expressed as-rRNA complementary to 5.8S rRNA sequence in UWB1 cells (Fig. 4C). We found that ectopic expression of as-5.8S rRNA enhanced 5.8S rRNA resistance to RNase A cleavage (Fig. 4D) without affecting control ACTB mRNA (Fig. 4E). These results suggest that BRCA1 BRCT enhances ds-rRNA formation not only by promoting RNA annealing but also by increasing as-rRNA production. We suggest that even though S1655A mutation impaired the annealing activity of BRCA1 BRCT, the mutation did not affect as-rRNA production, allowing partial increases in RNase A resistance and RNase III sensitivity within rRNA compared to the increases induced by WT BRCA1 BRCT expression.
as-rRNA Production Suppresses Ribosomal R-Loops.
Previous studies in yeast and C. elegans showed that as-rRNA is transcribed in response to cellular stress to form ds-rRNA for targeted degradation (34–37). However, BRCT expression enhanced as-5.8S rRNA production in UWB1 cells (Fig. 4B) without significantly decreasing 5.8S rRNA levels (Fig. 2A). Similarly, ectopic expression of as-5.8S rRNA in UWB1 cells did not lead to a reduction in total 5.8S rRNA signals detected by RT-qPCR (Fig. 4F). Because ds-rRNA formation may prevent rRNA from hybridizing to rDNA, thereby suppressing ribosomal R-loops, we examined the effect of ectopic expression of as-5.8S rRNA on ribosomal R-loops. Indeed, overexpressing as-5.8S rRNA, but not vector alone, is sufficient to reduce nucleolar R-loop signals in UWB1 cells (Fig. 4 G and H). DRIP-qPCR analysis confirmed R-loop reduction within or near 5.8S rDNA in UWB1 cells overexpressing as-5.8S rRNA (Fig. 4I). This R-loop suppression was significantly weakened by mutations within as-5.8S rRNA that decreased complementarity with 5.8S rRNA (Fig. 4I). As expected, as-5.8S rRNA production did not affect R-loop levels at ACTB gene loci (Fig. 4J).
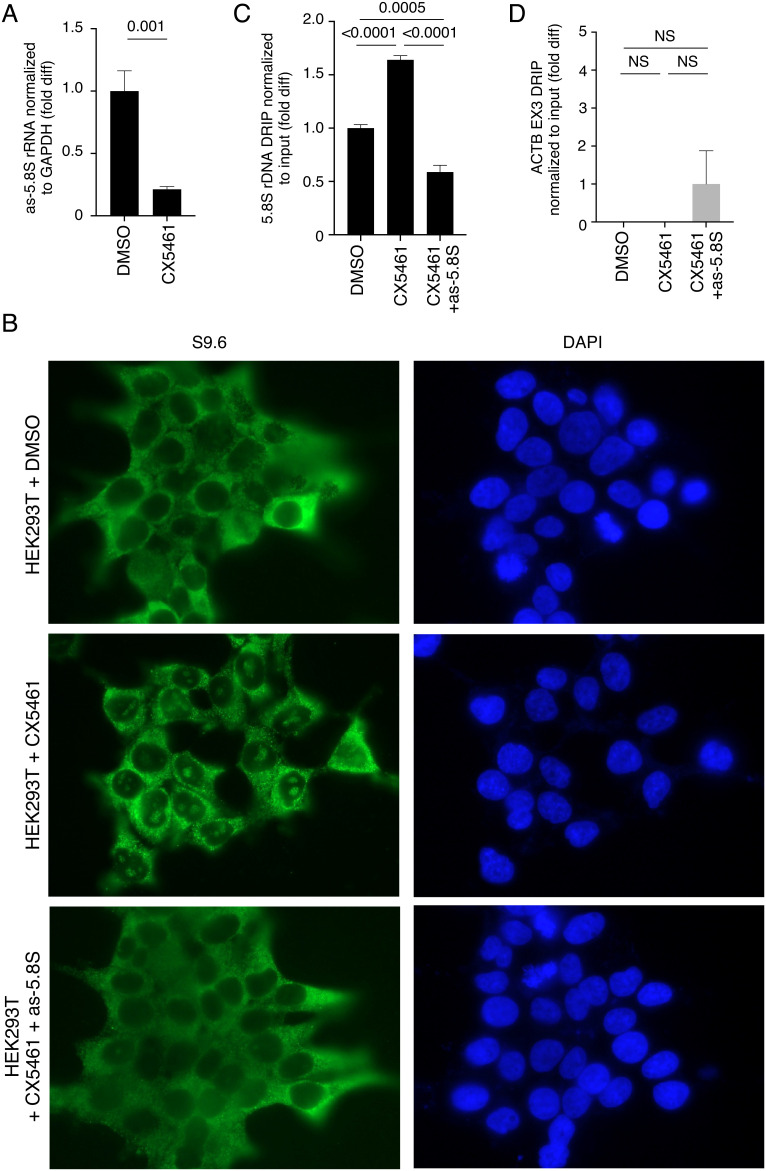
RNAPI inhibitor CX5461 has been shown to stabilize ribosomal R-loops (38). Because we found that CX5461 also inhibited as-rRNA production in HEK293T cells (Fig. 5A), we asked whether as-rRNA reduction by CX5461 contributes to the increase in ribosomal R-loops. Consistent with the previous study, we found that HEK293T cells treated with CX5461 increased nucleolar R-loops staining compared to cells without treatment (Fig. 5B, compare Fig. 5 B, Upper and Middle). Importantly, ectopic expression of as-5.8S rRNA in HEK293T cells decreased nucleolar R-loop staining induced by CX5461 (Fig. 5B, compare Fig. 5 B, Middle and Bottom). DRIP-qPCR analysis confirmed that as-5.8S rRNA expression suppressed CX5461-induced R-loops near 5.8S rDNA region (Fig. 5C). As expected, CX5461 treatment with or without as-5.8S rRNA expression had no significant effect on R-loop levels near the ACTB EX3 locus (Fig. 5D).
Fig. 5.
as-rRNA expression suppresses ribosomal R-loops induced by CX5461. (A) Quantification of as-5.8S rRNA levels normalized to GAPDH mRNA by RT-qPCR in HEK293T cells with or without treatment with RNAPI inhibitor CX5461. Y-axis represents fold difference relative to HEK293T cells without CX5461. (B) Representative images showing immunofluorescent staining of R-loops (green) using S9.6 antibody in HEK293T cells with or without treatment with CX5461 and ectopic expression of as-5.8S rRNA. DAPI is used to show nuclei. (C and D) DRIP analysis using S9.6 antibody to measure RNase H-sensitive R-loop levels at 5.8S rDNA loci (C) and ACTB EX3 region (D) normalized to the input DNA in HEK293T cells with or without CX5461 treatment and as-5.8S rRNA overexpression. Y-axis represents fold difference relative to HEK293T cells without CX5461 or as-5.8S rRNA overexpression.
UWB1 cells are defective in DNA damage repair due to BRCA1 deficiency. Therefore, we treated UWB1 cells with ultraviolet light (UV) to examine the effect of DNA damage on ribosomal R-loops. We found that UV treatment reduced R-loops at 5.8S rDNA (SI Appendix, Fig. S5B), likely due to the inhibition of rRNA synthesis by UV irradiation (39), but not at ACTB EX3 region (SI Appendix, Fig. S5C). as-5.8S expression further reduced R-loops in the UV-irradiated cells (SI Appendix, Fig. S5B). R-loop is known to induce DNA breakage and genomic instability (4). Consistent with the UV-induced R-loop reduction, we found that γH2AX levels at 5.8S rDNA loci were proportionally reduced after UV irradiation compared to those of nontreated UWB1 cells (SI Appendix, Fig. S5D). As expected, as-5.8S rRNA expression further decreased γH2AX molecules at 5.8S rDNA loci (SI Appendix, Fig. S5D). This observation highlights the significance of R-loops in contributing to rDNA damage. Together, these results indicate that BRCA1 promotes RNAPI-mediated as-rRNA expression to reduce ribosomal R-loop formation.
BRCA1-Mediated as-rRNA Production Negatively Regulates Pre-rRNA Processing.
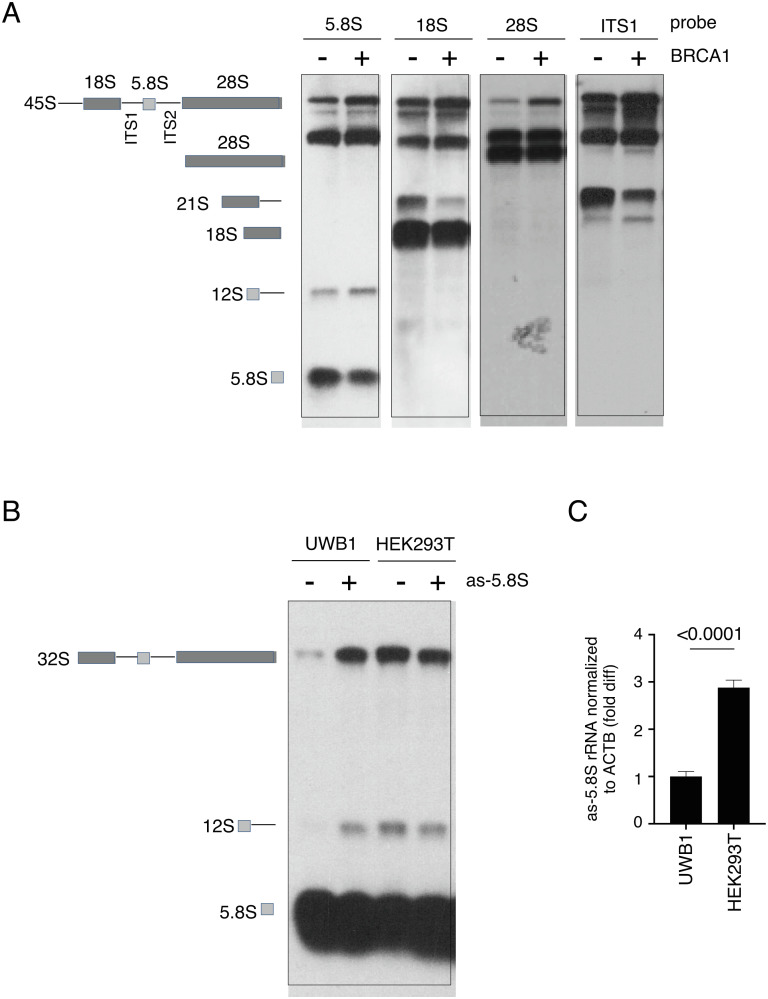
In mammalian cells, elevated as-rRNA levels perturb rRNA biogenesis (32). Therefore, we asked if complementing BRCA1 to increase as-rRNA production also affects rRNA processing in UWB1 cells. Using northern blot analysis, we analyzed rRNA maturation in UWB1 cells with or without ectopic expression of FL BRCA1. We observed a decrease in the mature 5.8S rRNA by BRCA1 expression and this was accompanied by an accumulation of the 12S rRNA precursor containing 5.8S when compared to UWB1 cells without BRCA1 expression (Fig. 6 A, 1st panel from the Left). Using probes against 18S (Fig. 6 A, 2nd panel from the Left) and ITS1 (Fig. 6 A, Right), we also observed a decrease in the ratio of 18S and its 21S precursor to its 45S rRNA precursor, which also showed accumulation in the blot using 28S as a probe (Fig. 6 A, 3rd panel from the Left). Similar accumulation of pre-rRNA intermediates was also observed in BRCA1-deficient HCC1937 and MDA436 cells transfected with FL BRCA1 (SI Appendix, Fig. S6A). Consequently, restoring BRCA1 in UWB1 cells also reduced protein synthesis rate (SI Appendix, Fig. S6B), supporting a role of BRCA1 in negatively regulating ribosomal biogenesis. On the contrary, BRCA2 KD in HEK293T cells using BRCA2-specific siRNA (SI Appendix, Fig. S6C) did not alter rRNA intermediates (SI Appendix, Fig. S6D). Pulse label with puromycin also showed comparable protein synthesis rates between control and BRCA2 KD cells (SI Appendix, Fig. S6E).
Fig. 6.
BRCA1-mediated as-rRNA expression negatively regulates rRNA processing. (A, Left) Schematic diagram of pre-rRNA and its processing into 18S, 5.8S, and 28S rRNA. (Right) Representative images showing northern blot analysis of the RNA samples prepared from UWB1 cells with or without ectopic BRCA1 expression using a fluorescent oligo probe complementary to either 5.8S, 18S, 28S, or ITS1 sequence. (B) Schematic diagram of 5.8S rRNA processing (Left). (Right) Representative images showing northern blot analysis of RNA samples prepared from either UWB1 cells (lanes 1 and 2) or HEK293T cells (lanes 3 and 4) with or without ectopic as-5.8S expression using an oligo probe complementary to 5.8S rRNA. (C) Quantification of as-5.8S rRNA levels normalized to ACTB mRNA by RT-qPCR in UWB1 and HEK293T cells. Y-axis represents fold difference relative to UWB1 cells.
To further confirm that the reduced efficiency in processing rRNA precursors by the BRCA1 expression in UWB1 cells is due to the elevated as-rRNA levels, we exogenously expressed as-5.8S rRNA in UWB1 and HEK293T cells. By northern blot analysis using an oligo complementary to 5.8S, we found that ectopic expression of as-5.8S rRNA enhanced 32S and 12S rRNA precursors in UWB1 cells (Fig. 6B, compare left 2 lanes). On the contrary, overexpressing as-5.8S rRNA did not change the levels of the rRNA precursors in HEK293T cells (Fig. 6B, compare right 2 lanes). RT-qPCR revealed higher endogenous as-5.8S rRNA levels in HEK293T cells compared to those of UWB1 cells (Fig. 6C), explaining for why additional expression of as-5.8S rRNA exerted little effect on rRNA processing in HEK293T cells. Based on these results, we conclude that BRCA1-dependent expression of as-rRNA contributes to the negative regulation of rRNA processing.
Discussion
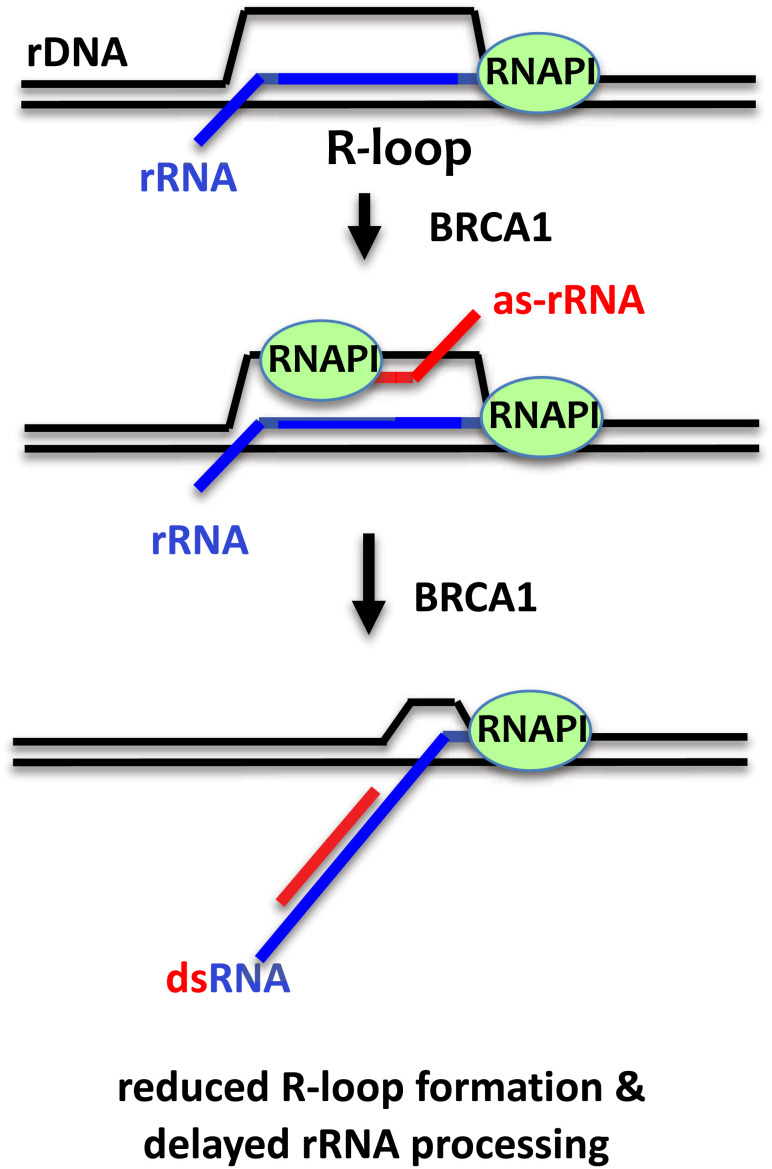
Our data suggest a model in which BRCA1 negatively regulates rRNA processing and maturation, and this function aids to suppress ribosomal R-loops. R-loop is known to promote as-RNA transcription across the mammalian genome (40). We propose that BRCA1 suppresses nucleolar R-loops via its BRCT in response to the transient formation of R-loops in rDNA loci by increasing as-rRNA production to enhance the formation of ds-rRNA and reduce the availability of nascent rRNA for hybridizing the rDNA template (Fig. 7). BRCA1 BRCT also facilitates as-rRNA annealing to rRNA via its RNA binding and annealing activity. The formation of ds-rRNA reduces the efficiency for RNA endo- and exonucleases to cleave rRNA precursors, leading to a slowdown in rRNA maturation. As we detected approximately tenfold higher levels of R-loops at 5.8S rDNA region compared to 18S rDNA (Fig. 1L), it is possible that R-loops are enriched within or near 5.8S. Hence, as-5.8S rRNA expression is sufficient to reduce nucleolar R-loop signals. Furthermore, because 28S, 18S, and 5.8S rRNAs are transcribed and processed from a single pre-rRNA molecule and 5.8S rRNA is located at the center of the pre-rRNA, it is also possible that dsRNA formation at the center of the pre-rRNA is sufficient to destabilize R-loops across the rDNA loci. In addition to binding to DNA and RNA, BRCA1 BRCT is also involved in protein–protein interactions (9, 15–17). It remains to be determined whether, in addition to RNA binding, altered protein–protein interaction(s) or posttranslational modification(s) also contribute to reduced ds-rRNA formation by the S1655A BRCT mutant. It would be of great interest in future studies to determine whether BRCA1-mediated as-RNA production also takes place in non-rDNA loci.
Fig. 7.
Model of the role of BRCA1 in suppressing ribosomal R-loop formation and rRNA processing. Schematic of the proposed mechanism by which BRCA1 promotes as-5.8S synthesis and delays rRNA processing to suppress rDNA R-loop synthesis. DNA is shown in black. Sense RNA is shown in blue and as-rRNA is shown in red. See text for details.
In quiescent cells, RNAPII drives as-rRNA transcription to silence rRNA transcription (31). Here, we showed that as-rRNA production in proliferating cells is dependent on RNAPI, as RNAPI inhibition by CX5461 suppresses as-rRNA synthesis. Likely, as-rRNA production depends on different RNAPs in response to various nutrient and growth conditions. In our study, we also observed as-rRNA complementary to 28S, 18S, and 5.8S rRNA in our BRCA1 BRCT RIP analysis. This study reports the detection of as-5.8S rRNA in human cells. It is possible that similar to RNAPII-mediated as-rRNA transcription (41, 42), as-rRNAs are also transcribed by RNAPI from the same precursor as a single RNA molecule containing antisense sequences complementary to all the three rRNA. Because BRCA1 interacts with RNAPI to facilitate rDNA transcription (24), it will be important for future studies to determine whether BRCA1-mediated as-rRNA production occurs via protein–protein interaction(s) with RNAPI or RNAPI-associated transcription factors. Alternatively, in C. elegans, production of stress-induced antisense ribosomal small interfering RNA (risiRNA) to degrade rRNA utilizes RNA-dependent RNA polymerases (RdRNPs) to synthesize risiRNAs using rRNA as a template (34). Even though BRCA1-induced as-rRNA expression did not play a role in rRNA degradation, it remains to be determined whether BRCA1-induced as-rRNA is also synthesized by RdRNP, and whether RNAPI is required to generate rRNA template for RdRNP-dependent RNA synthesis.
BRCT is a signature motif primarily identified in proteins involved in DNA replication and DNA damage repair (29). Interestingly, PES1, which is a component of the PeBoW complex that is important for rRNA maturation, contains a BRCT motif (43). The PES1 BRCT motif has been shown to promote PES1 nucleolar localization and PeBoW complex stability for processing rRNA precursors (44). Recently, the PeBoW complex was shown to copurify with the FA protein FANCI independent of its DNA damage-induced monoubiquitinated state, providing evidence for a non-DNA repair role of FANCI in rRNA processing and ribosomal biogenesis (45). In addition to FANCI, a recent study unveiled a role for FANCA in ribosomal biogenesis also independent of its function as a component of the FA core complex in DNA repair (46). FA is characterized by inherited bone marrow failure, which has been linked to defects in ribosomal maturation (47), and BRCA1 mutations have been linked to the FA phenotype in patients and mouse models (12, 48). Our discovery of a role of BRCA1 in rRNA processing suggests a potential connection between rRNA processing deregulation and BRCA1-associated FA. Furthermore, in cancer cells, rRNA processing is accelerated to meet the increasing demand for protein production for cell growth, and the expression of the alternate reading frame tumor suppressor blocks cancer cell growth by delaying rRNA maturation (49). Our studies open the possibility of targeting ribosomal biogenesis in BRCA1-associated cancer pathogenesis and progression.
Materials and Methods
Plasmids.
A pCDH-BRCA1 plasmid for the mammalian expression of FL BRCA1 was generated by subcloning FL BRCA1 cDNA from pMH-SFB-BRCA1 (Addgene). Plasmids expressing FLAG-tagged WT or S1655A mutant BRCA1 BRCT domains were generated by replacing FL BRCA1 with the corresponding cDNAs along with a nuclear localization signal in pMH-SFB-BRCA1. To generate a GST-tagged BRCT fragment, the corresponding coding sequence was PCR amplified from pMH-SFB-BRCA1 and cloned into pGEX-4T1 (GE Healthcare) between the EcoRI and SalI sites. The mutagenesis primer 5′-GAA TGT CCA TGG TGG TGG CTG GCC TGA CCC CAG AA -3′ was used to generate pGEX-4T1-S1655A. To induce ectopic expression of as-5.8S rRNA, the corresponding as-5.8S rRNA sequence was amplified by PCR using a 5′-AAG CGA CGC TCA GAC AGG CG-3′ forward primer and a 5′-CGA CTC TTA GCG GTG GAT CA-3′ reverse primer, and the PCR product was cloned into an inducible lentiviral pLKO vector (Addgene). as-5.8S mutant rRNA sequence was generated using pLKO-as-5.8S as the template with the following mutagenesis primers: 5′- AGA CAG GCG TAG CCC CGG GCC GCA AGT GCG TT-3′, 5′-CGG GCC GCA AGT GCG TTG TGT GTC CTG CAA TTC ACA TT-3′, 5′-TCC TGC AAT TCA CAT TGG GCC GCA AGT GCG TT -3′, 5′-TGG GCC GCA AGT GCG TTC GAG TGA TCC ACC GCT A-3′. Plasmid 1GFP/RNase H1 D210N for expressing GFP-His6-RNase H1 D210N in E. coli was purchased from Addgene.
Cell Culture and Transfection.
Human embryonic kidney HEK293T cells and human osteosarcoma U2OS cells were cultured in DMEM supplemented with 10% v/v FBS (Sigma-Aldrich) and streptomycin/penicillin (100 U/ml) and maintained at 37 °C and 5% CO2. UWB1 ovarian cancer cells (ATCC) were cultured in a 1:1 mixture of Mammary Epithelial Cell Growth Medium and RPMI supplemented with 10% v/v FBS and streptomycin/penicillin (100 U/ml). HCC1937 and MDA-436 breast cancer cells were cultured in RPMI with 10% v/v FBS and streptomycin/penicillin (100 U/ml). DNA transfection was carried out using Lipofectamine Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. To inhibit RNAPI, 1 µM CX5461 (Selectchem) was added to HEK293T cells for 4 h before harvesting. BRCA2 siRNA (sc-29825) was purchased from Santa Cruz. BRCA1 siRNA (#12519S) was purchased from Cell Signaling. siRNA was transfected using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s protocol.
Antibodies.
Mouse anti-BRCA1 (MS110; OP92), mouse anti-R-loop (S9.6; MABE1095), mouse anti-dsDNA (AE-2; MAB1293), and mouse anti-dsRNA (rJ2; MABE1134) were purchased from EMD Millipore. Mouse anti-BRCA2 mAb (OP95) was purchased from Calbiochem. Rabbit anti-nucleolin (D4C70; 14574), rabbit anti-β-actin (13E5; 4970), and rabbit anti-β-tubulin (9F3; 2128) were purchased from Cell Signaling Technology. Mouse anti-GAPDH (AC002) was purchased from ABClonal. Rabbit anti-FLAG (F7425) was purchased from Sigma-Aldrich.
Protein Expression and Purification.
For purifying GST-BRCT WT or S1655A recombinant proteins, proteins were expressed and harvested as described (50). Briefly, pGEX-4T-BRCA1 BRCT plasmid-transformed E. coli BL21 cells were cultured in Luria broth medium containing ampicillin (100 µg/ml) until the cell density (OD600) was 0.4. Isopropyl-β-D-thio-galactoside was then added to a final concentration of 0.1 mM, and cultures were incubated at 16 °C for 16 h. The bacteria were harvested and lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40, 1× protease inhibitor (Roche), 1 mM PMSF, and 0.2 mg/ml lysozyme), incubated on ice for 30 min, and sonicated at 4 °C. After centrifugation at 16,000 × g, the flow-through was applied to a GST column and washed sequentially with wash buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.05% NP-40), and the GST-tagged proteins were eluted with elution buffer (50 mM Tris pH 8.0, 150 mM NaCl, and 10 mM GSH). The concentrations of the purified proteins were measured using Pierce BCA Protein Assay Kit. To purify recombinant GFP-His6-RNase H1 D210N for R-loop isolation from cells, plasmid 1GFP/RNase H1 was transformed into BL21(DE3) and cultured in LB media supplemented with kanamycin (50 µg/ml) and induced by IPTG (final concentration of 0.3 mM) at 16 °C for 18 h. Cells were collected by centrifugation at 10,000 × g for 10 min, resuspended in 5 ml Ni binding buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 10 mM imidazole, 1 mM β-mercaptoethanol, 1 mM PMSF), and EDTA-free protease inhibitor cocktail tablet, followed by sonication. Supernatant containing soluble proteins was collected after centrifugation at 20,000 × g for 30 min and incubated with Ni–nitrilotriacetic acid Superflow beads. After binding, the beads were washed five times with 10 ml Ni wash buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 20 mM imidazole, 1 mM β-mercaptoethanol, 1 mM PMSF). The bound proteins were eluted with Ni elution buffer (50 mM NaH2PO4, 5 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.3% NP-40, 10% glycerol, 300 mM imidazole, 1 mM β-mercaptoethanol, plus protease inhibitors), followed by dialysis into the storage buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.3% NP-40, 10% glycerol, 1 mM β-mercaptoethanol, 1 mM PMSF). The dialyzed samples were flash frozen and stored at −80 °C.
DNA/RNA Annealing and DNA/RNA Binding Assays.
32P-radiolabeled ssDNA and ssRNA substrates were prepared by labeling non-rDNA bottom oligonucleotides (5′-ATC CCA GCA CCA GAT TCA GCA ATT AAG CTC TAA GCC ATG AAT TCA AAT GAC CTC TTA TCA-3′), rDNA oligonucleotides (5′-CGA CGC TCA GAC AGG CGTAGC CCC GGG AGG AAC CCG GGG CCG CAA GTG CGT TCG AAG TGT CGA TGA TCA ATG TGT CCT GCA ATT CAC ATT AAT TCT CGC A-3′), and non-rRNA bottom oligonucleotides (5′-AUCCCAGC ACCAGAUUCAGCAAUUAAGCUCUAAGCCAUGAAUUCAAAUGACCUCUUAUCA-3′) with [γ-32P] ATP. For dsRNA substrate, 32P-labeled RNA bottom oligonucleotides were annealed to unlabeled RNA top oligonucleotides (5′-UGA UAA GAG GUC AUU UGA AUU CAU GGC UUA GAG CUU AAU UGC UGA AUC UGG UGC UGG GAU-3′) (51). The 32P-labeled ssDNA, ssRNA, and annealed products were gel purified prior to analysis. DNA binding reactions were carried out by incubating recombinant BRCA1 BRCT WT or S1655A proteins with 4 pg of 32P-labeled ssDNA, ssRNA, or dsRNA in DNA binding buffer (30 mM Tris pH 7.5, 1 mM DTT, 100 µg/ml BSA) on ice for 15 min, and then the protein–DNA/RNA complexes were analyzed on 5% native polyacrylamide gels. Annealing assays were performed by preheating 32P-labeled substrates at 90 °C for 2 min in annealing buffer (30 mM Tris pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol), followed by snap cooling on ice for 10 min before adding either BRCA1 BRCT WT or S1655A mutant into the reaction. The reactions were incubated at room temperature for 15 min, followed by incubation with stop buffer to remove proteins by proteinase K for 10 min. RNase inhibitor (NEB, #M0314L) was added to all buffers containing RNA oligos. The reactions were deproteinized and analyzed on 8% polyacrylamide gels.
Immunofluorescence Assay.
To detect BRCA1, FLAG, and R-loops in UWB, HCC1937, HEK293T, and U2OS cells, cells cultured on coverslips were fixed using 3% paraformaldehyde at 4 °C for 10 min, then permeabilized using 0.1% Triton X-100 at room temperature for 5 min. The coverslips were incubated in 8% goat serum for 1 h to block nonspecific protein–antibody interactions, then incubated overnight with primary antibodies (S9.6, MS110, and nucleolin) at 4 °C. The coverslips were incubated with Alexa Fluor Plus 488 (Invitrogen) at 37 °C for 1 h, followed by DAPI staining for 5 min at room temperature, and the cells were covered with mounting medium (SlowFade Gold Antifade Mountant, Invitrogen). The coverslips were mounted onto glass slides and visualized with OLYMPUS IX71 inverted fluorescence microscope. All the images were acquired with cellSens standard (Version 1.3) software under OLYMPUS IX71 inverted fluorescence microscope equipped with a UPlanSApo 60X/1.35 oil immersion objective at room temperature. Identical contrast and brightness adjustments were used on images for all experiments.
RNA Isolation, RIP, and RT-qPCR.
HEK293T cells with or without expression of FLAG-BRCA1-BRCT were collected and lysed in RIP buffer (10 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1mM DTT, 2 mM Ribonucleoside Vanadyl Complex, 100 U/ml RNase inhibitor) with sonication (0.5 s sonication with 1 s break) and centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was collected and incubated overnight with FLAG M2 beads (Sigma-Aldrich) at 4 °C with rotation. The beads were sequentially washed with cold RIP buffer. An aliquot of the bound proteins was analyzed by western blotting. For the remaining bound BRCT-RNA complexes, poly(A) was then added to the end of the RNA by E. coli poly(A) polymerase (New England BioLabs) in the presence of RNase inhibitor. After poly(A) addition, bound BRCT-RNA complexes were washed and deproteinized in RIP buffer containing 30 µg proteinase K and 0.1% SDS for 20 min at 37 °C. RNA was purified using Trizol (Invitrogen) according to manufacturer’s manual, and their concentrations were determined by OD260 vs OD280 measurement using a NanoDrop ND1000 spectrophotometer. RT-qPCR was performed by converting RNA to DNA by superscript III (Invitrogen) reverse transcriptase in the presence of oligo-dT primer and template switching oligo (TSO). RT was followed by qPCR in triplicate using SYBR Green Master Mix (Applied Biosystems) on a Light Cycler 7500 Fast (Applied Biosystems). Primers used for qPCR were: 5′-ACTCTACGCGATCCGGG-3′ as TSO; 5′-ACTCGGCTCGTGCGTC-3′ as 5.8S Forward (F); 5′-GCGACGCTCAGACAGG-3′ as 5.8S Reverse (R); 5′-CTCAACACGGGAAACCTCA C-3′ as 18S F; 5′-CGCTCCACCAACTAAGAACG-3′ as 18S R; 5′-AGAGGTAAACGGGTGGGGTC-3′ as 28S F; and 5′-GGGGTCGGGAGGAACGG-3′ as 28S R.
DRIP, dsRNA IP, RNase A, and RNase III Treatments.
dsRNA IP protocol was adapted from ref. 52. Briefly, UWB1 cells were washed and lysed in NET-2 buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40). After centrifuge at 16,000 × g 10 min, 5 ug rJ2 antibody was added and incubated at 4 °C for 2 h, followed by incubation with protein G beads for 1 h. The beads were washed 4 times with NET-2 buffer. J2-bound dsRNA was extracted with Trizol reagent and quantified by RT-qPCR. Total RNA was harvested from 10% input lysate using a Quick-RNA Miniprep kit (Zymo). For RNase A treatment to detect dsRNA formation, 1 µg RNA was treated with or without 0.2 ng/ul RNase A at room temperature for 10 min. For RNase III treatment, 0.5 µg RNA was treated with or without 1 U RNase III at 37 °C for 1 h, followed by the addition of RNase inhibitor (Thermo Scientific), prior to cDNA synthesis using a High-Capacity RNA-to-cDNA Kit (Applied Biosystems). DRIP-qPCR for ACTB exon 3 (EX3) was performed using the primers as described (28). For R-loop isolation using purified recombinant GFP-His6-RNase H1 D210N, human cells were collected and lysed with 0.5 ml of SDS/Proteinase K buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS and 300 µg ml−1 proteinase K) at 37 °C overnight, followed by phenol/chloroform extraction and ethanol precipitation to purify nucleic acids. Nucleic acids were then sheared by sonication to generate DNA fragments of <1 kb, followed by incubation with 2 µg of GFP-RNASE H D210N in binding buffer (2XSSC, 0.25% Triton X100, 5% BSA plus protease inhibitors) overnight for every 10 µg of fragmented nucleic acids. The protein–DNA complexes were then incubated with precoated Ni-NTA beads for 4 h at 4 ˚C and washed extensively with the binding buffer. The bound nucleic acids were eluted with SDS/Proteinase K buffer at 55 °C for 2 h and purified by ChIP DNA Clean and Concentrator kit (Zymo Research, catalog 11-379c).
Northern Blotting and Protein Synthesis Rate.
UWB1 or HEK293T cells were first lysed in NETN100 followed by centrifugation at 12,000 × g for 10 min to remove all cytosolic contents. The pellet obtained after centrifugation was washed thrice using NETN100, followed by the addition of Trizol reagent for phase separation-based RNA isolation. 1 µg RNA for detecting rRNA or 6 µg RNA for detecting as-rRNA was separated on a 1% denaturing agarose gel containing formaldehyde in MOPS buffer at low voltage (60 V) for 6 h. Once separated, the RNA was transferred overnight to a positively charged nylon membrane under capillary action-based transfer in SSC buffer. RNA was crosslinked onto the nylon membrane, and the membrane was incubated in ULTRAhyb™ Ultrasensitive Hybridization Buffer (Invitrogen) containing the indicated biotinylated probe overnight at 42 °C. For as-rRNA detection, the membrane was incubated with a mixture of biotinylated probes: 5.8S (5′–GTC GAT GAA CGC AGC TAG CTG CGA G-3′), 18S (5′–GCT TGT CTC AAA GAT TAA GCC ATG CAT GTC–3′), and 28S (5′–TAA GCA TAT TAG TCA GCG GAG GAA AAG–3′). The hybridized probes were detected using the Chemiluminescent Nucleic Acid Detection Module system (Thermo Fisher Scientific). To measure protein translation rate (53), cells were treated with 2 or 10 µM puromycin for 10 min, harvested, and lysed in RIPA buffer. After sonication and centrifugation at 12,000 × g for 15 min, protein containing supernatant was analyzed by western blot with anti-puromycin antibody.
Quantifications, Statistics, and Reproducibility.
For each figure, three or more independent biological experiments were performed. For graphs, each data point represents the mean of three technical replicates ± SD. At least two different sets of purified recombinant proteins were used to perform all in vitro analyses. DRIP was quantified as the fold difference (relative to the control sample) in the fluorescent signal obtained from the qPCR of the S9.6 IP signal normalized to the input DNA minus the S9.6 IP signal using DNA pretreated with RNase H normalized to RNase H-treated input DNA. RNase H (DN) pull down was quantified as the fold difference (relative to the control sample) in the fluorescent signal of the DNA bound to the RNase H (DN) protein beads normalized to the input DNA minus the amount of DNA being pulled down by the beads alone. Two-tailed Student’s t tests were used to calculate p values for statistically significant differences. For all graphs, P values were shown over the horizontal lines, and NS stands for not significant. Quantifications of R-loop-positive nucleoli in Figs. 1H and 4H were performed manually by blind observers.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Sarah Wilkinson for her comments and expert editing of this manuscript. Y.L. was supported by grants from NIH (R01GM127602, R01CA225842, and CA130899). We thank Dr. Xiaochun Yu (Westlake University) for supervising A.K.S. execution of the northern blot experiment as shown in Fig. 6A. Research reported in this publication included work performed in the Light Microscopy Digital Imaging Core, Integrative Genomics Core, and Drug Discovery & Structural Biology Core supported by the NCI of NIH under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions
C.-W.C., A.K.S., M.L., L.G., and Y.L. designed research; C.-W.C., A.K.S., M.L., L.G., N.L., K.O., and F.L. performed research; C.-W.C., A.K.S., and M.L. contributed new reagents/analytic tools; C.-W.C., A.K.S., M.L., L.G., and Y.L. analyzed data; and Y.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Chiang H. C., et al. , BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic. Acids Res. 47, 5086–5099 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison D. F., Wang G. G., R-loops: Formation, function, and relevance to cell stress. Cell Stress. 3, 38–46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuduri S., et al. , Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera A., García-Muse T., R loops: From transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Li X., Manley J. L., Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Hatchi E., et al. , BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill S. J., et al. , Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes. Dev. 28, 1957–1975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X., et al. , Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 8, 15908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C. C., Feng W., Lim P. X., Kass E. M., Jasin M., Homology-directed repair and the Role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2, 313–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anantha R. W., et al. , Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife 6, e21350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nyberg T., et al. , Prostate cancer risks for male BRCA1 and BRCA2 mutation carriers: A prospective cohort study. Eur. Urol. 77, 24–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawyer S. L., et al. , Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 5, 135–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Densham R. M., Morris J. R., The BRCA1 Ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus 8, 116–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarsounas M., Sung P., The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 21, 284–299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semmler L., Reiter-Brennan C., Klein A., BRCA1 and breast cancer: A review of the underlying mechanisms resulting in the tissue-specific tumorigenesis in mutation carriers. J. Breast. Cancer 22, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams R. S., Lee M. S., Hau D. D., Glover J. N., Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 11, 519–525 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Clapperton J. A., et al. , Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 11, 512–518 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Yamane K., Katayama E., Tsuruo T., The BRCT regions of tumor suppressor BRCA1 and of XRCC1 show DNA end binding activity with a multimerizing feature. Biochem. Biophys. Res. Commun. 279, 678–684 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Simons A. M., et al. , BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 66, 2012–2018 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Anderson S. F., Schlegel B. P., Nakajima T., Wolpin E. S., Parvin J. D., BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 19, 254–256 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Fan S., et al. , BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science 284, 1354–1356 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Nair S. J., et al. , Genetic suppression reveals DNA repair-independent antagonism between BRCA1 and COBRA1 in mammary gland development. Nat. Commun. 7, 10913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berthel E., et al. , Uncovering the translational regulatory activity of the tumor suppressor BRCA1. Cells 9, 941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston R., et al. , The identification of a novel role for BRCA1 in regulating RNA polymerase I transcription. Oncotarget 7, 68097–68110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veras I., Rosen E. M., Schramm L., Inhibition of RNA polymerase III transcription by BRCA1. J. Mol. Biol. 387, 523–531 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Tulchin N., et al. , BRCA1 protein and nucleolin colocalize in breast carcinoma tissue and cancer cell lines. Am. J. Pathol. 176, 1203–1214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crossley M. P., et al. , Catalytically inactive, purified RNase H1: A specific and sensitive probe for RNA-DNA hybrid imaging. J. Cell Biol. 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Pokharel S., Wang J. T., Xu X., Liu Y., RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat. Commun. 6, 6720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung C. C., Glover J. N., BRCT domains: Easy as one, two, three. Cell Cycle. 10, 2461–2470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B., BRCA1 tumor suppressor network: Focusing on its tail. Cell Biosci. 2, 6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bierhoff H., Schmitz K., Maass F., Ye J., Grummt I., Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold. Spring Harb. Symp. Quant. Biol. 75, 357–364 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Hwang C. J., Fields J. R., Shiao Y. H., Non-coding rRNA-mediated preferential killing in cancer cells is enhanced by suppression of autophagy in non-transformed counterpart. Cell Death Dis. 2, e239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu C., et al. , Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc. Natl. Acad. Sci. U.S.A. 115, 10082–10087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X., et al. , RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol. 24, 258–269 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Yan Q., Zhu C., Guang S., Feng X., The functions of non-coding RNAs in rRNA regulation. Front. Genet. 10, 290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buhler M., Spies N., Bartel D. P., Moazed D., TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15, 1015–1023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Z., Senturk N., Song C., Grummt I., lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes. Dev. 32, 836–848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanij E., et al. , CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 11, 2641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nocentini S., Inhibition and recovery of ribosomal RNA synthesis in ultraviolet-irradiation mammalian cells. Biochim. Biophys. Acta. 454, 114–128 (1976). [DOI] [PubMed] [Google Scholar]
- 40.Tan-Wong S. M., Dhir S., Proudfoot N. J., R-loops promote antisense transcription across the mammalian genome. Mol. Cell 76, 600–616 e606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham K. J., et al. , Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585, 298–302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaliatsi E. G., Giarimoglou N., Stathopoulos C., Stamatopoulou V., Non-coding RNA-driven regulation of rRNA biogenesis. Int. J. Mol. Sci. 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holzel M., et al. , Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170, 367–378 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holzel M., et al. , The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic. Acids Res. 35, 789–800 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sondalle S. B., Longerich S., Ogawa L. M., Sung P., Baserga S. J., Fanconi anemia protein FANCI functions in ribosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 2561–2570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gueiderikh A., et al. , Fanconi anemia A protein participates in nucleolar homeostasis maintenance and ribosome biogenesis. Sci. Adv. 7, eabb5414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J. M., Ellis S. R., Ribosomes and marrow failure: Coincidental association or molecular paradigm? Blood 107, 4583–4588 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Park D., et al. , Ablation of the Brca1-Palb2 interaction phenocopies fanconi anemia in mice. Cancer Res. 80, 4172–4184 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Gaviraghi M., Vivori C., Tonon G., How cancer exploits ribosomal rna biogenesis: A journey beyond the boundaries of rRNA transcription. Cells 8, 1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang C. W., et al. , Pathogenic mutations reveal a role of RECQ4 in mitochondrial RNA:DNA hybrid formation and resolution. Sci. Rep. 10, 17033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Z., et al. , Binding of FANCI-FANCD2 complex to RNA and R-loops stimulates robust FANCD2 monoubiquitination. Cell reports 26, 564–572.e565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dhir A., et al. , Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aviner R., The science of puromycin: From studies of ribosome function to applications in biotechnology. Comput. Struct. Biotechnol. J. 18, 1074–1083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.