Significance
Mycovirus-mediated hypovirulence is a prime example of multitrophic interactions and has attracted attention for its biocontrol potential. However, direct release of mycovirus-infected fungi is risky due to potential loss of virus overtime, overshadowing its application. Besides, little is known about how hypovirulence is conferred in virus-infected fungi at the molecular level. Here, we have discovered a mycovirus SlAV1 from plant pathogen S. lycopersici, which confers hypovirulence and pigmentation loss by inhibiting fungal biosynthesis of a phytotoxin Altersolanol A. Genomic integration and expression of a key SlAV1 gene in fungal host converts the pathogen into a biocontrol agent and provides enhanced plant resistance against virulent strains. This opens a promising and low-risk path to contain plant diseases via biocontrol.
Keywords: mycovirus, hyprovirulence, phytotoxin, biocontrol
Abstract
Mycovirus-infected fungi can suffer from poor growth, attenuated pigmentation, and virulence. However, the molecular mechanisms of how mycoviruses confer these symptoms remain poorly understood. Here, we report a mycovirus Stemphylium lycopersici alternavirus 1 (SlAV1) isolated from a necrotrophic plant pathogen Stemphylium lycopersici that causes altered colony pigmentation and hypovirulence by specifically interfering host biosynthesis of Altersolanol A, a polyketide phytotoxin. SlAV1 significantly down-regulates a fungal polyketide synthase (PKS1), the core enzyme of Altersolanol A biosynthesis. PKS1 deletion mutants do not accumulate Altersolanol A and lose pathogenicity to tomato and lettuce. Transgenic expression of SlAV1 open-reading frame 3 (ORF3) in S. lycopersici inhibits fungal PKS1 expression and Altersolanol A accumulation, leading to symptoms like SlAV1-infected fungal strains. Multiple plant species sprayed with mycelial suspension of S. lycopersici or S. vesicarium strains integrating and expressing ORF3 display enhanced resistance against virulent strains, converting the pathogenic fungi into biocontrol agents. Hence, our study not only proves inhibiting a key enzyme of host phytotoxin biosynthesis as a molecular mechanism underlying SlAV1-mediated hypovirulence of Stemphylium spp., but also demonstrates the potential of mycovirus-gene integrated fungi as a potential biocontrol agent to protect plants from fungal diseases.
Fungal pathogens threaten global agriculture and food safety by causing devastating plant diseases, a problem worsened by the surging climate change and ecosystem deterioration. Finding an effective and environment-friendly disease management strategy is crucial to sustainable agriculture that feeds the hungry, improves human health, and protects the Earth’s ecosystem. Hypovirulence (mycovirus-mediated virulence attenuation) is a phenomenon extensively studied in plant pathogenic fungi and exploited to control fungal diseases in agriculture and forestry (1). Mycoviruses or fungal viruses are ubiquitous in all major taxonomic fungal groups including many plant pathogens (2). Unlike most bacterial viruses, mycoviruses do not lyse fungal cells and rarely produce symptoms to their fungal hosts. However, some mycoviruses such as hypoviruses do cause symptoms such as reduced virulence on plant hosts and reduced sporulation, altered colony morphology, and pigmentation. Therefore, mycoviruses conferring hypovirulence are considered as potential biological agents to control fungal diseases (3). For instance, “Cryphonectria hypovirus 1 (CHV1)” has been successfully used to control chestnut blight caused by Cryphonectria parasitica in Europe (4). In addition, Rosellinia necatrix megabirnavirus 1 and Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 also have the potential to prevent diseases caused by their hosts (5–7). Furthermore, it is shown recently that some mycoviruses could convert pathogenic fungi into beneficial endophytes and activate plant immunity (8, 9). Despite showing promise in disease control, direct release of mycovirus-infected fungal cells in field is risky as the hypovirulent fungi can be converted into virulent strains by potentially losing the viruses overtime. Therefore, integrating the responsible viral gene(s) into fungal genomes could allow a durable hypovirulence, an ideal scenario for biocontrol. This approach so far has not been used to control plant diseases due to a poor understanding of the mechanisms of how the mycoviruses confer symptoms to their hosts.
The genus Stemphylium (phylum Ascomycota, teleomorph Pleospora) includes plant pathogenic, endophytic, and saprophytic fungal species distributed worldwide. Until now, more than 200 Stemphylium species are known (10), many of which are plant pathogens causing Stemphylium blight, one of the most devastating diseases in many commercial crops, such as pear (11), tomato (12), onion (13), cotton (14), sugar beet (15), garlic (16), and so on (17–19), yielding substantial economic losses. Despite being environment-unfriendly, fungicide application remains the mostly used method to control Stemphylium blight. However, long-term and excessive use of fungicides has led to increased antifungal resistance (16, 18, 20–24) and threatened environmental safety and human health. Therefore, it is critically urgent to develop effective and environment-friendly disease control strategies.
Fungal secondary metabolites (SMs) are small molecules with potent biological activities, many of which are phytotoxic and key pathogenicity factors (25, 26). Fungal SMs consist of four main chemical classes: polyketides, nonribosomal peptides, terpenes, and indole alkaloids (27). Phytopathogenic fungi especially necrotrophs often secrete phytotoxins such as HC-toxin, T-toxin, and alternariol to kill host cells during infections (25, 28). Altersolanol A is a polyketide and non-host-specific phytotoxin from Stemphylium spp., Alternaria spp., and Phomopsis spp. (29–35), producing necrotic lesions on tomato (Solanum lycopersicum), potato (S. tuberosum), pea (Pisum sativum), and garlic (Allium sativum). Interestingly, mycoviruses can modulate SMs of their hosts, most notably affecting fungal colony pigmentation. For instance, CHV1-EP713 causes reduction in the orange pigment in C. parasitica colony (36). We also observe that Stemphylium hypovirulent strains produce little or no yellow–brown pigment compared with virus-free strains. Although some mycoviruses can alter host colony pigmentation and reduce virulence, which secondary metabolite(s) contributes to the pigment and how exactly the virus affects its accumulation and fungal pathogenicity at the molecular level are yet to be determined.
Here, we identify an alternavirus Stemphylium lycopersici alternavirus 1 (SlAV1) in a hypovirulent S. lycopersici strain. Through a careful molecular, genetic, and metabolic dissection, we show that a polyketide phytotoxin Altersolanol A is largely responsible for pigmentation and essential for pathogenicity of S. lycopersici and its sister species S. vesicarium. Polyketide synthases (PKSs) are core biosynthetic enzymes of polyketide biosynthesis (37) and several fungal PKS genes are involved in melanin biosynthesis and virulence (38, 39). We show that SlAV1 confers hypovirulence by suppressing the biosynthesis of Altersolanol A via down-regulating the core biosynthetic enzyme PKS1. We further demonstrate that open reading frame 3 (ORF3) is the critical SlAV1 ORF for conferring Altersolanol A downregulation. This finding permitted the integration and transgenic expression of ORF3 in S. lycopersici and S. vesicarium genomes via bioengineering, which converts the pathogens into biocontrol agents and enhances plant resistance to pathogen infection. Our study thus provides an avenue of protecting plants from fungal pathogen infections: through integration of responsible mycovirus gene into fungal pathogen genome, working as a potential biocontrol agent in disease control.
Results
A Mycovirus Discovered in S. lycopersici.
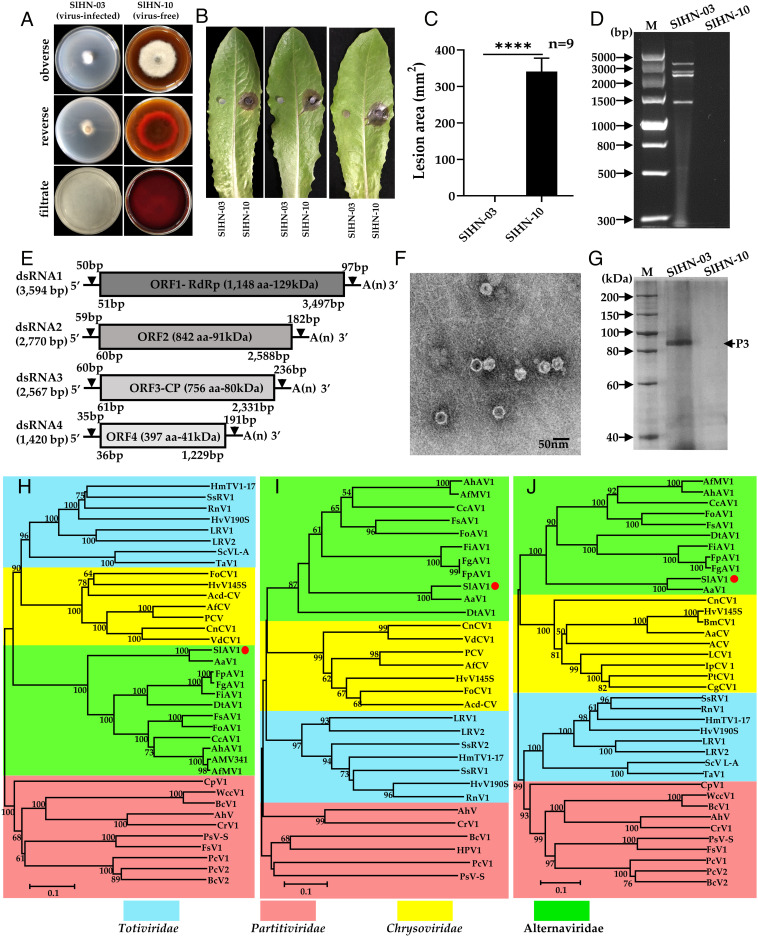
During a prior identification of lettuce leaf spot pathogens (40), a particular S. lycopersici strain Stemphylium lycopersici Hu Nan-03 (SlHN-03) drew our attention given its abnormal phenotype (Fig. 1A). Compared with other strains such as Stemphylium lycopersici Hu Nan-10 (SlHN-10), SlHN-03 showed slower growth and reduced pigmentation (Fig. 1A) and lack of pathogenicity when inoculated on detached lettuce and tomato leaves (Fig. 1 B and C and SI Appendix, Fig. S1). We suspected that such hypovirulence traits of SlHN-03 may be a sign of mycoviral infection. Since majority of known mycoviruses are double-strand RNA (dsRNA) viruses, we extracted dsRNA segments from SlHN-03, obtaining four dsRNA segments ranging from 1.4 to 3.6 kb (Fig. 1D). The full sequences of four dsRNAs were obtained by RNA sequencing (RNA-seq), and each dsRNA represents a single ORF. ORF1 and ORF3 encode putative RNA-dependent RNA polymerase (RdRp) and coat protein (CP), respectively, while ORF2 and ORF4 encode two proteins of unknown function (Fig. 1E). Basic Local Alignment Search Tool (BLAST) search suggests ORF1, ORF2, and ORF3 have 80.35%, 80.92%, and 81.23% amino acid sequence identities with those of Alternaria alternata virus 1 (AaV1) and other alternavirus, respectively, while ORF4 shares only 35.53% identities with AaV1 ORF4 (SI Appendix, Table S1). We then purified SIHN-03 viral particles via ultracentrifugation and observed them using a transmission electron microscope. Isometric viral particles with an average diameter of 34 nm were observed (Fig. 1F). SDS-PAGE of purified viral particles identified a single major protein with a molecular mass of ~ 91 kDa (Fig. 1G) corresponding to ORF3 based on mass spectrometry (SI Appendix, Fig. S2 and Table S2). Phylogenetic analysis showed that this virus is clustered with the AaV1 and other mycoviruses that belong to the proposed family “Alternaviridae” (41) (Fig. 1 H−J). In addition, multiple sequence alignment of the 5′- and 3′-terminal regions of the four dsRNA are highly conserved (SI Appendix, Fig. S3). This characteristic was similar to that described for the proposed family “Alternaviridae”. Hence, this virus represents a strain of a species within the proposed family Alternaviridae, and thus we named it SlAV1. To date, no virus of the family “Alternaviridae” has been shown to confer hypovirulence, except for Fusarium oxysporum alternavirus 1 (FoAV1), which exhibited biological control prospect against Fusarium wilt (42), although molecular mechanisms of FoAV1 hypovirulence remain unknown.
Fig. 1.
Hypovirulence-associated traits of strain SlHN-03 of S. lycopersici and characterization of Stemphylium lycopersici alternavirus 1 (SlAV1). (A) Abnormal colony morphology of virus-free strain SlHN-10 vs virus-infected strain SlHN-03 grown on a PDA plate at 28°C for 7 d. (B) and (C) Virulence assay on detached lettuce leaves (SlHN-10 on the Right and SlHN-03 on the Left). Data are represented as mean ± SD from three replicates with ****P < 0.0001 indicating the statistically significant difference by two-tailed Student’s t test. (D) dsRNA extracted from mycelia of strains SlHN-03 and SlHN-10, lane M: DNA marker (DL5000 bp, Vazyme). (E) Schematic genome organization of SlAV1, gray boxed indicates the putative ORFs encoded by dsRNA1-4, and UTRs as black lines. (F) Electron micrograph of viral particles. (G) Protein components of viral particles were analyzed by SDS-PAGE (8% agar gel); Lane M, protein marker (200 kDa ladder); Neighbor-joining phylogenetic trees constructed based on the amino acid sequences of viral RdRp (H), ORF2 (I) and CP (J) of SlAV1 and selected mycoviruses in the Totiviridae, Chrysoviridae, Partitiviridae, and Alternaviridae families. RdRp, RNA-dependent RNA polymerase. CP, coat protein. The numbers at nodes represent bootstrap values as percentages estimated by 1,000 replicates. The scale bar represents a genetic distance of 0.1 amino acid substitutions per site.
SlAV1 Is a Mycovirus Conferring Hypovirulence and Transfers Horizontally.
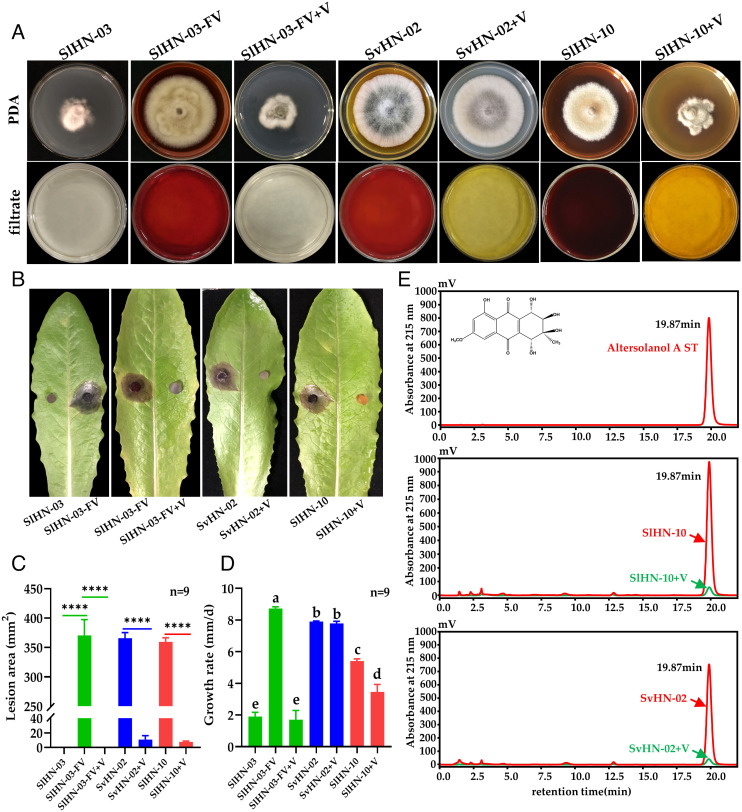
To investigate the effects of SlAV1 on its fungal host, we eliminated SlAV1 from SlHN-03 by hyphal tip isolation and protoplast regeneration, obtaining 32 regenerated strains (SI Appendix, Fig. S4A). Among them, eight strains were dsRNA-free with the colony morphology similar to that of SlHN-10 (SI Appendix, Fig. S4); three of those similar to SlHN-10 were used for virulence assays. As expected, the virus-cured strains restored growth rate, pigmentation, and virulence to lettuce, and hereinafter defined as SlHN-03-FV strains (Fig. 2 A–D). To determine whether SlAV1 is transmissible among Stemphylium species or even different fungal genera, each of virus-free strains S. vesicarium Hu Nan-02 (SvHN-02), A. alternata Hu Nan-06 (AaHN-06), and Fusarium sporotrichioides Hu Nan-11 (FsHN-11) were cocultured with SlHN-03 for 5 d in darkness at 28°C, followed by SlAV1 detection in recipient isolates. The results showed that SlAV1 successfully transmitted from SlHN-03 to SvHN-02 but not to AaHN-06 or FsHN-11, suggesting the role of vegetative incompatibility in preventing the spread of virus (SI Appendix, Fig. S5). However, AaHN-06 wounded hyphae were infected when cultured in filtrate containing SlAV1 (SI Appendix, Fig. S6). The virus-infected strain SvHN-02+V showed decreased pigmentation compared with SvHN-02 (Fig. 2A), and hardly caused lesion on detached lettuce leaves, resembling SlHN-03 (Fig. 2 B and C). Intriguingly, mycelium growth rates of SvHN-02 and SvHN-02+V are comparable (Fig. 2D), suggesting that hypovirulence caused by SlAV1 infection is linked to the suppressed fungal pigmentation by SlAV1, instead of abnormal mycelial growth. To test this hypothesis, we reinfected SlHN-03-FV and another S. lycopersici strain SlHN-10 showing strong pigmentation using SlAV1 to generate two strains SlHN-03-FV+V and SlHN-10+V, respectively (Fig. 2A and SI Appendix, Figs. S5B and S7). The phenotypes of SlHN-03-FV+V and SlHN-10+V match those of SlHN-03 in terms of colony pigmentation and hypovirulence (Fig. 2). In addition, the SlHN-10+V exhibited shrunk cell membranes, excessive shorter hyphal branching with few normal mitochondria (SI Appendix, Fig. S8). Therefore, we conclude that the SlAV1-mediated hypovirulence is strongly associated with reduced pigmentation.
Fig. 2.
Effects of SlAV1 on fungal morphology, growth, pathogenicity, and pigment synthesis. (A) Representative morphology of the SlAV1-infected S. lycopersici strain SlHN-03, virus-cured isolate (SlHN-03-FV), virus-reinfected isolate (SlHN-03-FV+V), S. vesicarium strain SvHN-02 and its virus-infected strain SvHN-02+V, S. lycopersici strain SlHN-10 and its virus-infected strain SlHN-10+V. Colony morphology of the obverse on potato dextrose agar (PDA) (Top panel) and the culture supernatants (Bottom panel). (B and C) Virulence assay of fungal strains on detached lettuce leaves. Data are represented as mean ± SD of nine replicates. **** indicates P < 0.0001 in two-tailed Student’s t test. (D) Growth rate measurement of fungal strains on PDA. Different letters indicate the statistically significant difference by a one-way ANOVA (P < 0.05). (E) Chromatographs of fungal strains for Altersolanol A detection using standard as reference (ST). Inlet of the chromatograph shows the chemical structure of Altersolanol A.
Yellow–Brown Pigmentation Suppressed by SlAV1 Is a Phytotoxin of Stemphylium spp.
Prior studies reported polyketide phytotoxin Altersolanol A gave the yellow–brown pigment to S. botryosum and S. solani and contributed to the fungal pathogenicity (29, 30, 43). To test whether the yellow–brown pigment produced by S. lycopersici and S. vesicarium contains Altersolanol A, the culture filtrate of SlAV1-free strains extracted using ethyl acetate were analyzed with a high-performance liquid chromatography coupled with a mass spectrometry (HPLC/MS) using Altersolanol A standard solution as reference. The results showed that the yellow–brown pigment produced by S. lycopersici (SlHN-10) and S. vesicarium (SvHN-02) had a retention time of 19.87 min and molecular mass of 336.09 (m/z = 337.09 [M + H]+) corresponding to Altersolanol A and confirmed by mass spectrometry (Fig. 2E and SI Appendix, Fig. S9). By contrast, Altersolanol A was barely detectable in SlAV1-infected strains SlHN-10+V and SvHN-02+V (Fig. 2E and SI Appendix, Table S3), suggesting Altersolanol A was a predominant component of the yellow–brown pigment. These results indicated that SlAV1 significantly suppressed the production of Altersolanol A in its host. (SI Appendix, Table S3).
SlAV1 Causes Hypovirulence through Suppressing the Expression of Altersolanol A Core Biosynthetic Enzyme PKS1.
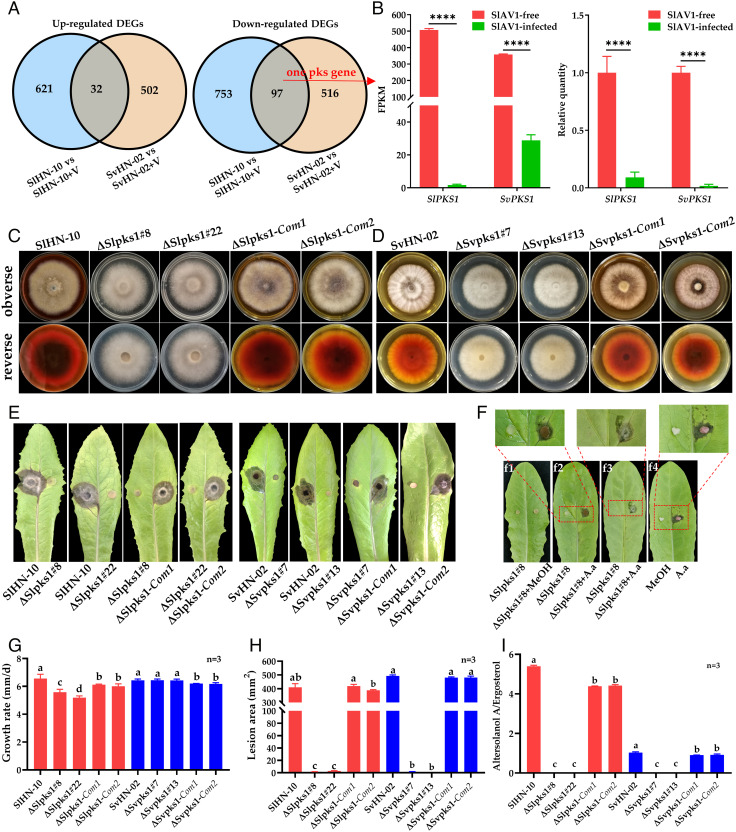
We further investigated the mechanisms underlying the SlAV1 suppression of Altersolanol A synthesis in S. lycopersici and S. vesicarium by identifying differentially expressed genes (DEGs) in virus-infected vs. virus-free strains using transcriptomic analysis. A total of 1,503 and 1,147 DEGs (fold change ≥ 2 and false discovery rate ≤ 0.05) were identified in SlHN-10 vs. SlHN-10+V and SvHN-02 vs. SvHN-02+V, respectively. For the two DEG sets, 216 DEGs are homologous and 129 DEGs showed conserved expression change between two species including 97 down-regulated and 32 up-regulated DEGs (Fig. 3A). Since Altersolanol A is a polyketide, we specifically looked for DEGs that encode key enzymes for polyketide biosynthesis and indeed found one down-regulated gene encoding a PKS. This was subsequently confirmed by real-time RT-PCR (RT-qPCR) (Fig. 3B), indicating that downregulation of PKS by SlAV1 may contribute to reduction in Altersolanol A biosynthesis in virus-infected strains. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment of SlAV1-induced DEGs indicated that pathways such as meiosis, cell cycle, ribosome biogenesis, spliceosome, RNA transport, and DNA replication were significantly enriched (SI Appendix, Fig. S10), reflecting the virus manipulation of host cell machinery due to SlAV1 infection (44).
Fig. 3.
SlPKS1 and SvPKS1 are required for biosynthesis of Altersolanol A, a key pathogenic factor of S. lycopersici. (A) A comparison of differentially expressed genes (DEGs) in virus-infected (+V) vs. virus-free SlHN-10 and SvHN-02 shown by Venn diagrams. (B) Expression levels of SlPKS1 and SvPKS1 based on RNA-seq and RT-qPCR. FPKM, fragments per kilobase per million mapped reads. (C) Colony morphology of the wild-type (SlHN-10), two independent SlPKS1 deletion mutants (ΔSlpks1#8 and ΔSlpks1#22), and two complemented strains (ΔSlpks1-Com1 and ΔSlpks1-Com2) grown on potato dextrose agar (PDA) for 7 d. (D) Colony morphology of the wild-type (SvHN-02), two independent SvPKS1 deletion mutants (ΔSvpks1#7 and ΔSvpks1#13), and two complemented strains (ΔSvpks1-Com1 and ΔSvpks1-Com2) grown on PDA for 7 d. (E) and (H) Virulence assay on detached lettuce leaves. (F) Symptoms on lettuce leaves inoculated with ΔSlpks1#8, ΔSlp ks1#8 added 5 μL methanol (f1) and ΔSlpks1#8 added 5 μL Altersolanol A (A.a) standards (10 μg/mL) (f2). (f3) shows the backside of f2 leaf. (f4), lettuce leaf inoculated with sterilized cotton balls soaked with 5 μL A.a standards (10 μg/mL), respectively. (G) Growth rate measurement of fungal strains on PDA. (I) Quantification of Altersolanol A production in S. lycopersici and S. vesicarium strains normalized by ergosterol levels proportional to fungal biomass. Error bars indicate SDs (n = 3). Different letters indicate the statistically significant difference by a one-way ANOVA (P < 0.05).
We then characterized the function of this PKS in S. lycopersici and S. vesicarium. Full genomic sequences of the two PKSs were cloned from SlHN-10 and SvHN-02, named SlPKS1 and SvPKS1. The ΔSlpks1 and ΔSvpks1 mutant strains were created by replacing the SlPKS1 and SvPKS1 with a hygromycin resistance cassette, respectively (SI Appendix, Fig. S11 A and B). Single colony of two independent deletion mutant strains per species, ΔSlpks1#8 and ΔSlpks1#22, ΔSvpks1#7 and ΔSvpks1#13 were obtained and gene deletion was confirmed by Southern blot (SI Appendix, Fig. S11 C and D). The ΔSlpks1 and ΔSvpks1 mutants barely synthesized yellow–brown pigment compared with wild type (Fig. 3 C and D). The pigment was rescued in complemented strains containing a functional copy of PKS1 (Fig. 3 C and D). HPLC–MS analysis showed that Altersolanol A production in PKS1 deletion mutants was nondetectable compared with wild-type and their complemented strains (Fig. 3I). S. lycopersici strains overexpressing PKS1 gene had significantly stronger ability of Altersolanol A accumulation (SI Appendix, Fig. S12C and Table S3). Taken together, PKS1 is essential for the biosynthesis of Altersolanol A, which gives the yellow–brown pigment in S. lycopersici and S. vesicarium.
The pathogenicity of wild-type, PKS1 mutants and complemented strains were tested by inoculating detached lettuce (Fig. 3 E and H) and tomato leaves (SI Appendix, Fig S13). The results showed that PKS1 mutants lost pathogenicity, whereas wild-type and complemented strains had normal virulence. Furthermore, PKS1-overexpressing strains showed enhanced virulence on leaves of lettuce and tomato (SI Appendix, Fig. S12 D and E), indicating the essential role of PKS1 in fungal pathogenicity. Therefore, we reasoned that Altersolanol A, biosynthesized by PKS1 and other unidentified enzymes, is a fungal pathogenicity factor. To further confirm this, detached lettuce leaves were inoculated with SlPKS1 deletion strain ΔSlpks1#8, ΔSlpks1#8 plus methanol and ΔSlpks1#8 plus Altersolanol A (10 μg/mL dissolved in methanol), respectively. Neither ΔSlpks1#8 nor ΔSlpks1#8 plus methanol caused any lesion on lettuce leaves (Fig. 3F). By contrast, inoculation of ΔSlpks1#8 plus Altersolanol A produced obvious lesions on lettuce leaves with its mycelia penetrating and expanding to the backside of leaves at 3 d post inoculation (dpi). However, the lesion failed to expand after 3dpi, suggesting that the invasion and mycelial colonization of S. lycopersici require a continuous biosynthesis of Altersolanol A. In addition, lesions were induced by a direct application of Altersolanol A (10 μg/mL) on leaves (Fig. 3F) showing that Altersolanol A is sufficient to produce the leaf lesions without the presence of the pathogen. Since Altersolanol A has been reported in several plant pathogenic fungi (29–35) including Alternaria solani, we wonder whether the function of PKS1 is conserved among these fungi. Therefore, we conducted RNA interference of AsPKS1, a homolog of SlPKS1, and found AsPKS1 was also vital for Altersolanol A biosynthesis and pathogenicity of A. solani (SI Appendix, Fig. S14). Collectively, these results confirm that Altersolanol A is essential for pathogenicity and plant colonization of S. lycopersici, and the hypovirulence of SlAV1-infected strain is caused by its reduced Altersolanol A production due to downregulation of SlPKS1. We report that a mycovirus can inhibit the core biosynthetic enzyme of a phytotoxin as a mechanism underlying the host pigmentation reduction and hypovirulence.
ORF3 of SlAV1 Attenuates the Biosynthesis of Altersolanol A.
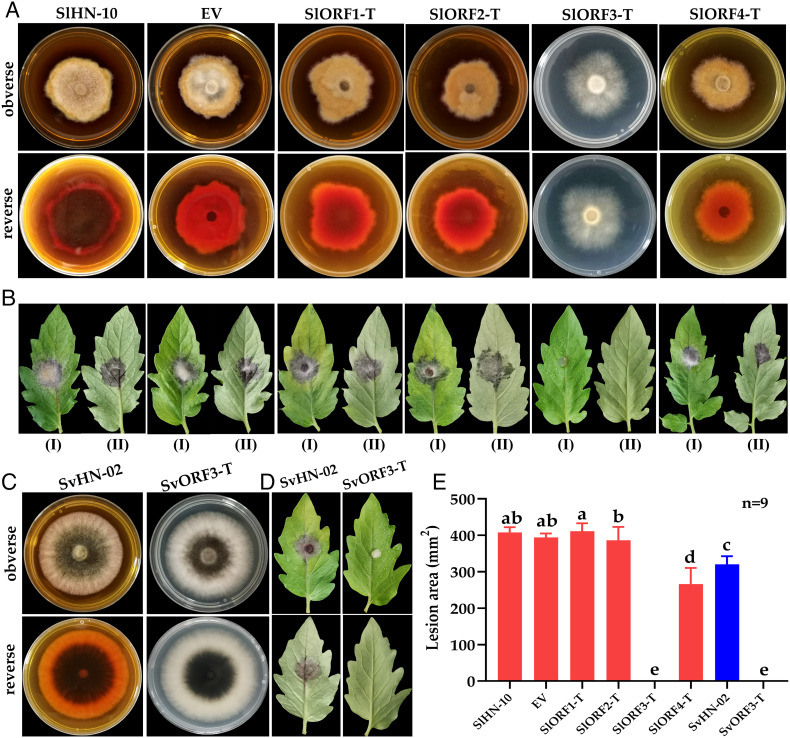
To determine which SlAV1 ORFs are responsible for the hypovirulence and attenuated biosynthesis of Altersolanol A in virus-infected strains, we expressed ORF1, ORF2, ORF3, and ORF4 of SlAV1 individually in the S. lycopersici virus-free strain SlHN-10 to obtain SlORF1-T, SlORF2-T, SlORF3-T, and SlORF4-T, respectively. Similar to wild-type strain SlHN-10 (Fig. 4A), SlORF1-T, SlORF2-T, and SlORF4-T had normal colony morphology, pigmentation and induced significantly necrosis lesion on tomato leaves (Fig. 4 B and E). However, SlORF3-T displayed decreased production of pigmentation and barely caused leaf lesions, similar to SlAV1-infected strains. Furthermore, we have quantified the ORF3 expression using RT-qPCR, and the results shows that the virus-infected strains have more ORF3 expression than ORF3 transgenic strains do (SI Appendix, Fig. S15). In addition, RNA-seq data and RT-qPCR results showed that PKS1 expression was suppressed in SIORF3-T (SI Appendix, Fig. S16). HPLC–MS analysis showed that the biosynthesis of Altersolanol A was significantly reduced in SlORF3-T (SI Appendix, Table S3). In addition, transgenic expression of SlAV1 ORF3 in S. vesicarium virus-free strain SvHN-02 yielded almost the same phenotypes as SlAV1-infected strains (Fig. 4 C and D and SI Appendix, Table S3). Although the exact regulatory mechanism is unclear, the inhibition of fungal PKS1 by SlAV1-ORF3 is likely indirect, since the ORF3 encodes a virus CP lacking any DNA-binding domain typically associated with transcription factors. Overall, these results demonstrate that ORF3 is responsible for the SlAV1-inflicted phenotypes in S. lycopersici and S. vesicarium.
Fig. 4.
Phenotypes of S. lycopersici and S. vesicarium transgenic lines expressing SIAV1 ORFs. (A) The colony morphology of S. lycopersici strains SlHN-10, EV (empty vector), SlORF1-T, SlORF2-T, SlORF3-T, SlORF4-T grown for 5 d on potato dextrose agar (PDA). (B) Symptoms of detached tomato leaves inoculated with S. lycopersici strains pictured at the fifth day post inoculation. (C) The colony morphology of S. vesicarium strains SvHN-02 and SvORF3-T grown for 5 d on PDA. (D) Symptoms of detached tomato leaves inoculated with SvHN-02 and SvORF3-T pictured at the fifth day post inoculation. (E) Measurement of lesion area induced by each strain on detached tomato leaves. Error bars indicate SDs (n = 9). Different letters indicate the statistically significant difference by a one-way ANOVA (P < 0.05).
Spraying Mycelia of ORF3 Transgenic Strains Confers Plant Resistance to Stemphylium spp.
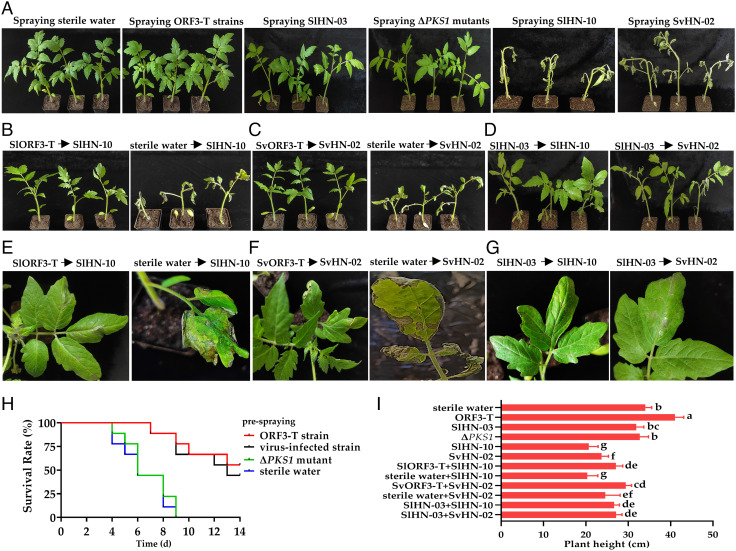
Previous studies reported the direct application of mycovirus-infected fungi could confer plant resistance against pathogenic fungi in field (8). Considering the potential risk of virus-infected fungal cells that may regain pathogenicity in field if they become cured of the virus, we sought an alternative approach by integrating the viral sequence in host genome before applying them to plants. To test whether integrating SlAV1 sequences into host genome could be used to control fungal pathogens, hyphal fragment suspension prepared from transgenic Stemphylium strains expressing SlAV1-ORF3 (ORF3-T) was sprayed on various host plants including tomato, pepper, eggplant, and cucumber, followed by a second spray of hyphal fragment suspensions of their virulent strains after 48 h. Almost no leaf spot was observed on plants presprayed with ORF3-T or SlAV1-infected strains before being sprayed with virulent strains (Fig. 5A). Critically, prespraying plants with ORF3-T- or SlAV1-infected strains showed resistance to virulent strain SlHN-10 and SvHN-02, which caused higher mortality on plants presprayed with only sterile water or ΔPKS1 mutants (Fig. 5 B–H and SI Appendix, Fig. S17). Compared with plants presprayed with sterile water, tomato plants presprayed with ORF3-T grew significantly taller (Fig. 5I). In addition, prespraying hyphal fragment suspensions of SlHN-10-ORF3-T or SvHN-02-ORF3-T also conferred resistance on pepper, eggplant, and cucumber (SI Appendix, Figs. S18 and S19). The results suggest that spraying hyphal fragment suspensions of ORF3-T strains on host plants could enhance their resistance to Stemphylium spp. infection.
Fig. 5.
Transgenic expression of SlAV1 ORF3 in S. lycopersici and S. vesicarium strains enhances disease resistance of tomato plants. (A) Tomato plants presprayed with hyphal fragment suspensions of ORF3-T strains, SlAV1-infected strains, ΔPKS1 mutants, virulent strains, or sterile water. (B–D) Tomato plants presprayed with hyphal fragment suspensions of SlHN-10-ORF3-T, SvHN-02-ORF3-T, or SlHN-03 but not sterile water show enhanced resistance against virulent S. lycopersici and S. vesicarium strain, respectively. The hyphal fragment suspension of virulent strains SlHN-10 or SvHN-02 is sprayed on leaves 48 h after spraying plants with their ORF3-transgenic expressed or SlHN-03 hyphal fragment suspensions, respectively. (E–G) The leaf closeup photos of (B–D), respectively. (H) Survival rates of infected tomato plants after spraying hyphal fragment suspensions of ORF3-T, SlHN-03, ΔPKS mutants, or sterile water. (I) Height measurement of plants with single or sequential spray of fungal strains or sterile water. Photos were taken 5 d post inoculation, plant survival rates of each day for fourteen consecutive days were calculated from nine tomato plants used for each treatment. Error bars indicate SDs (n = 6). Different letters indicate the statistically significant difference by a one-way ANOVA (P < 0.05).
To unveil the molecular mechanism underlying the enhanced resistance, we quantified the expression of the salicylic acid (SA) inducible pathogenesis-related protein 1 acidic (PR1a) gene and jasmonic acid (JA) inducible defensin gene (PDF1) (45) in tomato leaves sprayed with fungal mycelia. The PR1a gene was strongly induced in tomato plants sprayed with SlORF3-T compared with plants sprayed with SlHN-10, while PDF1 of plants sprayed with SlORF3-T or SlHN-10 was down-regulated (SI Appendix, Fig. S20), suggesting that SlORF3-T likely activated systemic acquired resistance (SAR). In addition, tomato leaves sprayed with virus-free or ORF3-transgenic SlHN-10 were stained with wheat germ agglutinin allowing observation for fungal hyphae colonization under a confocal microscope. Virus-free SlHN-10 strain colonized leaf tissue extensively, whereas ORF3-transgenic SlHN-10 exhibited minimal colonization as neither an endophyte nor a pathogen. Instead, ORF3-transgenic SlHN-10 grew epiphytically on leaf surface, and its growth was quite limited compared with virus-free SlHN-10 (SI Appendix, Fig. S21). It suggests that ORF3-transgenic SlHN-10 induces host resistance against pathogens, despite having an impaired ability to infect tomato leaves. Although integration of mycoviral genes into fungal genomes to study viral gene functions has been reported before, using the viral-gene integrated fungal cells as biocontrol agents to protect plants against pathogens has not been shown until this study.
Discussion
Fungal hypovirulence caused by mycovirus infections is an example of multitrophic interactions involving three or more life forms, having been reported in Ascomycete (Cryphonectria, Fusarium, Sclerotinia etc.) and Basidiomycete (Rhizoctonia). Fungi infected by a mycovirus are typically symptomless but occasionally show reduction in mycelial growth, sporulation, virulence, and altered colony pigmentation and secondary metabolic profiles. We have observed most of these symptoms on SlAV1-infected Stemphylium strains, suggesting that SlAV1 is a pathogenic mycovirus and the first isolated in this plant fungal pathogen. Although studies have similarly shown that pigmentation and SMs of host cells were modulated by some mycoviruses, the proof that a mycovirus actually targets the biosynthetic genes of the pigments has not been shown in previous research. In this work, we report the discovery of a fungal PKS1 being suppressed by the mycovirus SlAV1, specifically by one of its ORFs, leading to a reduction in biosynthetic product Altersolanol A and loss of fungal pathogenicity. We also present evidence that Altersolanol A is essential for fungal pathogenicity, and its continuous accumulation by Stemphylium during infection is required for disease progression. The exact molecular mechanism of viral interference of PKS1 is a matter for future investigation. Mycoviruses are known to modulate host cell signaling pathways and transcriptional regulation (8, 46). Belonging to serine/threonine (Ser/Thr) kinases, mitogen-activated protein kinases mediate intracellular signal transduction from cell surface to nucleus in response to various extracellular stimuli (47). RNA-seq data of this study revealed a total of seven (five up-regulated and two down-regulated) and eight (seven up-regulated and one down-regulated) differentially expressed Ser/Thr kinase genes in response to virus-infection of SlHN-10 vs SlHN-10+V and SvHN-02 vs. SvHN-02+V, respectively (SI Appendix, Fig. S22). Besides, three transcription factors were significantly down-regulated in virus-infected SlHN-10, while their homologs showed similar expression patterns in virus-infected SvHN-02 (SI Appendix, Fig. S23). Therefore, it is likely that SlAV1 specifically targets certain signaling proteins or transcription factors that regulate the expression of PKS1 and other unidentified biosynthetic genes for Altersolanol A. The most critical component of SlAV1 is its ORF3 in terms of inducing symptoms. The ORF3 gene encodes a structural protein instead of a regulatory protein (e.g., transcription factor or kinase) domain and therefore likely inhibits Altersolanol A biosynthetic gene PKS1 indirectly such as via perturbing unknown gene regulatory networks.
We demonstrate that Altersolanol A produced by Stemphylium spp. contributes to the colony pigmentation and plays a key role in pathogenicity as a phytotoxin. Besides showing phytotoxicity, Altersolanol A is also an anticancer agent through the inhibition of NF-κB activity in cancer cells (48). Therefore, elucidation of its biosynthesis pathway will enable its mass production as a cancer drug through metabolic engineering. We identified the key PKS1 responsible for Altersolanol A biosynthesis by using transcriptomic analysis and functional characterization. Since production of fungal SMs is often controlled by biosynthesis gene clusters, we suspect that there may be an Altersolanol A BGC to which PKS1 belongs. In fact, our comparative transcriptomic data analysis revealed several genes coexpressed with PKS1 appeared to be clustered in genomes of S. lycopersici and S. vesicarium. However, biochemical functions of these genes related to Altersolanol A biosynthesis need further investigation. In addition, Altersolanol A is also produced by other plant pathogenic fungi, including Alternaria spp. (31–34) and Phomopsis spp. (35). It likely acts as a conserved fungal pathogenicity factor among different Altersolanol A producers, as shown by our disruption of PKS1 homologs in S. lycopersici, S. vesicarium, and A. solani. The evolutionary histories and mechanisms of Altersolanol A biosynthesis in fungi remain unknown. A full elucidation of the biosynthetic pathway of Altersolanol A in the future is vital not only to understanding how the pathway evolves among close-related fungi, but also to mitigating the accumulation of this phytotoxin by plant pathogens for disease control.
Hypovirulence has attracted broad interests owing to its application in plant disease control, where virus-infected fungal pathogens are applied to infected plants as biocontrol agents as reported previously (3–8). These applications directly use intact viruses that can suppress pathogenic strains via hyphal fusion. Although this approach indeed can reduce the disease severity upon application, the caveat is that the cytoplasmic mycoviruses are unstable in their host fungi. Some studies have proven that mycoviruses almost disappeared after successive culture of the fungal hosts carrying them (49, 50). We also found that the presence of SlAV1 within fungal host is not persistent. SlAV1-infected A. alternata strain AaHN-06+V did initially show phenotypic alterations including hypovirulence (SI Appendix, Fig S6), but SlAV1 became nondetectable after repeated subculture for 20 generations. Therefore, using fungal strains with intact mycoviruses for biocontrol can introduce risks via potentially converting back to pathogenic forms by potentially losing the viruses. In this study, transgenic Stemphylium strains that expressed ORF3 of SlAV1 could enhance plant resistance to virulent strains. We have shown that integrating and expressing ORF3 of SlAV1 in a pathogenic fungus is sufficient to confer hypovirulence. Applying virus-integrated fungal mycelia on tomato, lettuce, eggplant, and cucumber plants protected them from pathogenic strains, demonstrating its biocontrol effect. Furthermore, the ORF3-transgenic strain can enhance plant growth compared with water treatment, indicating the growth modulation of the hypovirulent strain. The enhanced disease resistance by prespraying virus-integrated strain is likely an elevated host immunity, since the SA-inducible PR1a gene was strongly activated after inoculation of SlORF3-T strain compared with SlHN-10. In traditional opinion, SA signaling pathway is associated with plant resistance to biotrophic and hemibiotrophic pathogens, while JA signaling pathway is involved in defense against necrotrophic pathogens (51). However, this binary view has been challenged by some recent reports. For instance, Zhao et al. (52) revealed the involvement of the SA pathway in the defense of chrysanthemum plants against the necrotrophic fungus Alternaria sp. In our study, the SA signaling pathway marker PR1a gene was significantly activated in plants sprayed with SlORF3-T compared with SlHN-10, indicating a plant SAR conferred by ORF3 transgenic strain. Different from traditional approaches, the genome-integrated viral gene and its transcript remain detectable in the fungal genome after many generations of subculture, suggesting its stable presence and expression in host (SI Appendix, Fig. S24). Whether transgenic fungal strains applied on plants will be selected against in fields as a risk of transgene loss remains a matter for further investigation. Previously, a few genes of other mycoviruses were associated with hypovirulence (53, 54) and offer potential biocontrol resources for the prevention of fungal diseases. Our proof-of-concept study presents a successful example of utilizing mycoviral gene integration as a potential biocontrol agent to enhance plant disease resistance, an effective but safer approach to control plant diseases.
Materials and Methods
Fungal strains, Plasmids, Primers, and Plant Materials.
S. lycopersici strains SlHN-03 (infected by SlAV1), SlHN-10, and S. vesicarium strain SvHN-02 were originally isolated from diseased lettuce leaves (40). A. alternata strain AaHN-06 and A. solani strain ZYAS-11 were isolated from leaves of tobacco brown spot and potato early blight, respectively. F. sporotrichioides strain FsHN-11 was kindly provided by Dr. Xiubin Liu of Hunan Agricultural University. Gene targeting vector pCX62 and expression vector KSTNP were kindly provided by Prof. Zhengguang Zhang of Nanjing Agricultural University and Dr. Yanling Li of Hunan Agricultural University, respectively. Four ORFs of SlAV1 were synthesized (Sangon Biotech, Shanghai) and inserted to overexpression vector pBC-Hygro, generating the recombinant plasmids pBC-Hygro-ORF1, pBC-Hygro-ORF2, pBC-Hygro-ORF3, and pBC-Hygro-ORF4, respectively. All primers used in this study are listed in SI Appendix, Tables S4 and S5. All plants were grown in a plant-growth room at 22°C ± 2°C with a photoperiod of 16 h/8 h (day/night).
Extracting dsRNAs, Sequencing and Analysis, Viral Particle Extraction, Observation, and SDS-PAGE.
Fungal dsRNAs were prepared as by previously described (55). The full sequences of dsRNAs were obtained by RNA-seq and a ligase-mediated terminal amplification as previously described (56). The viral particle extraction, observation, and SDS-PAGE were conducted as previously described (5, 55, 56). Protein bands were individually excised and subjected to peptide mass analysis using mass spectrometry as described previously (57). Sequence analyses including search of homologs, ORFs, and conserved domains were performed using databases and tools in the National Center for Biotechnology Information (NCBI). Sequence analysis and multiple alignments were conducted using ClustalX 2.0 (58). The neighbor-joining phylogenetic trees were constructed using MEGA 7.0 (59) with 1,000 bootstraps.
Curing of Virus from Strain SlHN-03.
Single hyphae tips were cut with a sterilized needle and inoculated onto fresh potato dextrose agar (PDA) with 1.0 μg/mL cycloheximide for additional five or six cycles. Then the hyphal tips were transferred to a potato dextrose broth (PDB) medium without cycloheximide and inoculated for 5 d for protoplast preparation. Protoplasts were prepared using 2% Kitalase (Wako cat #114-00373), following the method described previously (60). All regenerated isolates were cultured in PDA plates to observe colony morphology and then detected for mycovirus infections.
Virus Transmission Assay.
Virus-free S. vesicarium strain SvHN-02 (recipient), A. alternata strain AaHN-06 (recipient), and F. sporotrichioides strain FsHN-11 (recipient) were individually cultured 1 cm apart from virus-infected S. lycopersici strain SlHN-03 (donor) on the same PDA dish at 28°C in dark, respectively. After culturing for 5 d, mycelial agar plugs were taken from the edge of each colony of the recipient to obtain recipient derivative isolates. In addition, we also infected the strains mentioned above using the filtrate of SlHN-03 containing SlAV1 using the method described by Urayama et al. (61). All recipient derivative isolates were analyzed for the infection of virus via dsRNA extraction and RT-PCR.
Fungal Transformation for Construction of Targeted Gene Deletion, Complementation, and Transgenic Strains.
The SlPKS1/SvPKS1 gene disruption vector was constructed as described by Chen et al. (62). The final disruption plasmid of SlPKS1/SvPKS1 was transformed into protoplasts of SlHN-10 or SvHN-02, respectively (63). The transformants were first screened on PDA containing hygromycin (300 μg/mL) and then checked by PCR and Southern blotting. For complementation, the SlPKS1 and SvPKS1 genes with their native promoters were amplified and inserted into the KSTNP expression vector, generating KSTNP-SlPKS1-Com and KSTNP-SvPKS1-Com, respectively. The two vectors were transformed into the ΔSlpks1#8 and ΔSvpks1#7, respectively. Transformants were screened on PDA with neomycin (200 μg/mL) and then confirmed by semi-quantitative RT-PCR. The SlAV1 ORF transgenic expression vectors were transformed into SlHN-10 protoplasts. Positive transformants selected on PDA plates containing 300 μg/mL hygromycin B and further confirmed by RT-PCR.
Southern Blotting and RT-qPCR Analysis.
The standard Southern blot protocol was used according to Sambrook and Russell (64). Probe labeling, hybridization, and detection were performed using the DIG High Prime DNA Labeling and Detection Starter Kit (Roche Applied Science, Penzberg, Germany). RT-qPCR was performed using the ABI 7300 real-time PCR system (Applied Biosystems, USA), and the transcription levels were analyzed using 7300 system SDS software. Relative gene expression was calculated according to the 2−ΔΔCt method. RT-qPCR analysis was conducted with three independent experiments, and including three technical replicates.
Growth Rate and Pathogenicity Assay.
Mycelial growth rate and colony morphology were evaluated according to the procedures described previously (60). Pathogenicity assay was conducted on detached lettuce and tomato leaves, incubated at 28°C with 60% humidity. Leaves of 30-day-old lettuce cultivar Sijijianye and tomato cultivar Lingyue were used for inoculation. Pathogenicity of the isolates were assessed by the presence or absence of lesion, and the lesion areas were measured with ImageJ software. All biological characterization experiments were conducted at least three times.
Altersolanol A Extraction, Qualitative and Quantitative Analyses.
Altersolanol A extraction procedure followed Zheng et al.’s (30). Five-ml culture filtrate of each sample was extracted three times with ethyl acetate, the upper fraction was collected, evaporated with an N2 stream, and dissolved in 1 ml methanol. Ergosterol concentrations were used to normalize Altersolanol A content per unit fungal mass as previously described (65). The samples were analyzed using HPLC/MS. Detailed information is given in Sl Appendix, Materials and Methods.
Biocontrol Ability Assessment.
To simulate the biocontrol ability of ORF3 transgenic strains in the field, we sprayed chamber-grown plants with hyphal fragment suspensions of SlORF3-T and SvORF3-T and evaluated their resistance to Stemphylium spp. Forty-eight hours after the spray of test strains, the hyphal fragment suspensions of virulent strains SlHN-10 and SvHN-02 were sprayed, respectively. All strains were grown in a 40 mL PDB medium at 28°C and shaken under 180 rpm for 5 d. The mycelia were collected and then homogenized in a blender, and the hyphal fragment suspension was diluted with sterile water to 2.0 OD600 units as described by Zhang et al. (8). Plants sprayed with equal sterile water were used as controls. The inoculated plants were placed in an incubator at 28°C and 100% relative humidity. Photographs were taken at 5 dpi. Experiments were repeated at least three times, and at least six samples were used for each individual test.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to thank Dr. Zhengguang Zhang, Dr. Xiubin Liu, and Dr. Yanlin Li for kindly providing strains and vectors for this study and reviewers for their critical and constructive comments and suggestions to improve this manuscript. This work was financially supported by the Scientific Research Fund of Hunan Provincial Education Department (19K040), the Excellent Doctoral Dissertation Cultivating Fund of Hunan Agricultural University (YB2019005) and the Hunan Provincial Natural Science Fund (2018JJ3220). L.G. is supported by the Taishan Scholars Program.
Author contributions
H.L., X.L.L, B.G., L.G., X.W.D., and Q.Z. designed research; H.L., H.W., X.L., D.S., W.G., J.Z., H.Z., and X.P. performed research; H.L., H.W., L.G., X.W.D., and Q.Z. analyzed data; and H.L., L.G., X.W.D., and Q.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: T.M., The Pennsylvania State University; J.X., Huazhong Agriculture University; and J.-R.X., Purdue University.
Contributor Information
Li Guo, Email: li.guo@pku-iaas.edu.cn.
Xing Wang Deng, Email: deng@pku.edu.cn.
Qian Zhou, Email: zhouqian2617@hunau.edu.cn.
Data, Materials, and Software Availability
Statistical analysis was conducted using the IBM SPSS Statistics (version 22.0) program with a one-way analysis of variance (ANOVA) and Tukey post hoc test. For two-group comparisons, two-tailed Student’s t test was conducted with a P value smaller than 0.05 being considered statistically significant. The graphs were produced using the GraphPad Prism 8 software. The four cDNA sequences of SlAV1 dsRNA have been deposited in GenBank under accession numbers MH040331-MH040334. The SlPKS1 and SvPKS1 DNA sequences have been deposited in the GenBank under accession numbers OM249968 and OM249969. The RNA-seq raw data have been deposited in NCBI under BioProject accession numbers PRJNA622902 and PRJNA623067.
Supporting Information
References
- 1.Nuss L. D., Hypovirulence: Mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3, 632–642 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Kotta-Loizou I., Mycoviruses and their role in fungal pathogenesis. Curr. Opin. Microbiol. 63, 10–18 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Xie J., Jiang D., New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Anagnostakis S. L., Biological control of chestnut blight. Science 215, 466–471 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Chiba S., et al. , A novel bipartite double-stranded RNA Mycovirus from the white root rot Fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83, 12801–12812 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X., et al. , A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 8387–8392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X., et al. , Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. U.S.A. 110, 1452–1457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., et al. , A 2-kb Mycovirus converts a pathogenic fungus into a beneficial endophyte for Brassica protection and yield enhancement. Mol. Plant 13, 1420–1433 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Zhou L., et al. , A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 15, 1893–1906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laura G., Gil-Serna J., Marta G., Iglesias C., Palmero D., Stemphylium leaf blight of garlic (Allium sativum) in Spain: Taxonomy and In Vitro fungicide response. Plant Pathol. J. 32, 388–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llorente I., Moragrega C., Ruz L., Montesinos E., An update on control of brown spot of pear. Trees 26, 239–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H., et al. , Mapping and screening of the tomato Stemphylium lycopersici resistance gene, Sm, based on bulked segregant analysis in combination with genome resequencing. BMC Plant Biol. 17, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay F., et al. , Stemphylium leaf blight: A re-emerging threat to onion production in eastern North America. Plant Dis. 105, 3780–3794 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Fracovig P. C., Mehta Y. R., Fonseca N. S., Reis E. M., Sources of resistance to Stemphylium solani in cotton cultivars. Summa Phytopathol. 25, 217–222 (1999). [Google Scholar]
- 15.Hanse B., Raaijmakers E. E. M., Schoone A. H. L., van Oorschot P. M. S., Stemphylium sp., the cause of yellow leaf spot disease in sugar beet (Beta vulgaris L.) in the Netherlands. Eur. J. Plant Pathol. 142, 319–330 (2015). [Google Scholar]
- 16.Laura G., Gil-Serna J., Marta G., Iglesias C., Palmero D., Stemphylium leaf blight of garlic (Allium sativum) in Spain: Taxonomy and in vitro fungicide response. Plant Pathol. J. 32, 388–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falloon P. G., Etiology and epidemiology of Stemphylium leaf spot and purple spot of asparagus in California. Phytopathology 77, 407–413 (1987). [Google Scholar]
- 18.Rahman T., Ahmed A. U., Islam M. R., Hosen M. I., Physiological study and both in vitro and in vivo antifungal activities against Stemphylium botryosum causing Stemphylium blight disease in Lentil (Lens culinaris). Plant Pathol. J. 9, 179–187 (2010). [Google Scholar]
- 19.Kim B. S., Yu S. H., Cho H. J., Hwang H. S., Gray leaf spot in peppers caused by Stemphylium solani and S. lycopersici. Plant Pathol. J. 20, 85–91 (2004). [Google Scholar]
- 20.Alberoni G., Collina M., Pancaldi D., Brunelli A., Resistance to dicarboximide fungicides in Stemphylium vesicarium of Italian pear orchards. Eur. J. Plant Pathol. 113, 211–219 (2005). [Google Scholar]
- 21.Alberoni G., Cavallini D., Collina M., Brunelli A., Characterisation of the first Stemphylium vesicarium isolates resistant to strobilurins in Italian pear orchards. Eur. J. Plant Pathol. 126, 453–457 (2010). [Google Scholar]
- 22.Alberoni G., Collina M., Lanen C., Leroux P., Brunelli A., Field strains of Stemphylium vesicarium with a resistance to dicarboximide fungicides correlated with changes in a two-component histidine kinase. Eur. J. Plant Pathol. 128, 171–184 (2010). [Google Scholar]
- 23.Wu D., et al. , Resistance risk assessment for fludioxonil in Stemphylium solani. Ann. Appl. Biol. 167, 277–284 (2015). [Google Scholar]
- 24.Hay F., et al. , Emergence of Stemphylium leaf blight of onion in New York associated with fungicide resistance. Plant Dis. 103, 3083–3092 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Pusztahelyi T., Holb I. J., Pócsi I., Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6, 573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina R., et al. , Secondary metabolites synthesized by Stemphylium lycopersici and Fulvia fulva, necrotrophic and biotrophic fungi pathogen of tomato plants. Curr. Plant Biol. 20, 100122 (2019). [Google Scholar]
- 27.Keller N. P., Turner G., Bennett J. W., Fungal secondary metabolism from bio-chemistry to genomics. Nat. Rev. Microbiol. 3, 937–947 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Howlett B. J., Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr. Opin. Plant Biol. 9, 371–375 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Assante G., Nasini G., Identity of the phytotoxin stemphylin from Stemphylium botryosum with Altersolanol A. Phytochemistry 26, 703–705 (1987). [Google Scholar]
- 30.Zheng L., Lv R., Huang J., Jiang D., Hsiang T., Isolation, purification, and biological activity of a phytotoxin produced by Stemphylium solani. Plant Dis. 94, 1231–1237 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Stoessl A., Unwin C. H., Stothers J. B., Metabolites of Alternaria solani part V. Biosynthesis of Altersolanol A and incorporation of Altersolanol A-13Cx into Altersolanol B and macrosporin. Tetrahedron Lett. 20, 2481–2484 (1979). [Google Scholar]
- 32.Suemitsu R., Nakamura A., Isolation and identification of Altersolanol A from the culture liquid of Alternaria porri (Ellis) Ciferri. Agric. Biol. Chem. 45, 2363–2364 (1981). [Google Scholar]
- 33.Stoessl A., Unwin C. H., Stothers J. B., On the biosynthesis of some polyketide metabolites in Alternaria solani: 13C and 2HMR studies. Can. J. Chem. 61, 372–377 (2011). [Google Scholar]
- 34.Holenstein J. E., Stoessl A., Metabolites of Alternaria solani, part IX: Phytotoxicity of Altersolanol A. J. Phytopathol. 108, 143–147 (1983). [Google Scholar]
- 35.Wheeler M. M., Wheeler D., Peterson G. W., Anthraquinone pigments from the phytopathogen Phomopsis juniperovora hahn. Phytochemistry 14, 288–289 (1975). [Google Scholar]
- 36.Sun L. Y., Nuss D. L., Suzuki N., Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87, 3703–3714 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Medina R., et al. , Secondary metabolite gene clusters arrangement and conservation within the genome of Stemphylium lycopersici codes the pathways for the synthesis of specific and non-specific toxins. Australas. Plant Pathol. 50, 51–72 (2021). [Google Scholar]
- 38.Alhawatema M. S., Gebril S., Cook D., Creamer R., RNAi-mediated down-regulation of a melanin polyketide synthase (pks1) gene in the fungus Slafractonia leguminicola. World J. Microbiol. Biotechnol. 33, 1–9 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Shimizu T., Ito T., Kanematsu S., Functional analysis of a melanin biosynthetic gene using RNAi-mediated gene silencing in Rosellinia necatrix. Fungal Biol. 118, 413–421 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Liu H., et al. , First report of Stemphylium lycopersici and Stemphylium vesicarium causing leaf spot on lettuce (Lactuca sativa) in China. Plant Dis. 103, 2957 (2019). [Google Scholar]
- 41.Kozlakidis Z., et al. , Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch. Virol. 158, 267–272 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Wen C., et al. , Molecular characterization of the first alternavirus identified in Fusarium oxysporum. Viruses 13, 2026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L., Liu W., Chen C. S., Jiang D. H., Huang J. B., Structural identification of SS-toxin produced by Stemphylium solani causing garlic leaf blight [in Chinese]. Acta Phytopathol. Sin. 44, 478–485, (2014). [Google Scholar]
- 44.López-Lastra M., et al. , Translation initiation of viral mRNAs. Rev. Med. Virol. 20, 177–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jogaiah S., Abdelrahman M., Tran L. S. P., Ito S. I., Different mechanisms of Trichoderma virens-mediated resistance in tomato against Fusarium wilt involve the jasmonic and salicylic acid pathways. Mol. Plant Pathol. 19, 870–882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S., et al. , Virus-mediated suppression of host non-self recognition facilitates horizontal transmission of heterologous viruses. PLoS Pathog. 13, e1006234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dickman M. B., Yarden O., Serine/threonine protein kinases and phosphatases in filamentious fungi. Fungal Genet. Biol. 26, 99–117 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Teiten M. H., et al. , Anticancer effect of Altersolanol A, a metabolite produced by the endophytic fungus Stemphylium globuliferum, mediated by its pro-apoptotic and anti-invasive potential via the inhibition of NF-κB activity. Bioorg. Med. Chem. 21, 3850–3858 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Kanematsu S., et al. , A Reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology 94, 561–568 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Darissa O., Adam G., Schäfer W., A dsRNA mycovirus causes hypovirulence of Fusarium graminearum to wheat and maize. Eur. J. Plant Pathol. 134, 181–189 (2012). [Google Scholar]
- 51.Glazebrook J., Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Zhao X., et al. , The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus Alternaria sp. Hortic. Res. 7, 80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bormann J., et al. , Expression of a structural protein of the mycovirus FgV-ch9 negatively affects the transcript level of a novel symptom alleviation factor and causes virus infection-like symptoms in Fusarium graminearum. J. Virol. 92, e00326-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao Z., et al. , ORF Ι of mycovirus SsNSRV-1 is associated with debilitating symptoms of Sclerotinia sclerotiorum. Viruses 12, 456 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H., et al. , A victorivirus and two novel mitoviruses co-infected the plant pathogen Nigrospora oryzae. Viruses 11, 83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong J., et al. , Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch. Virol. 159, 3063–3070 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Lutz T., et al. , Characterization of a novel Alternavirus infecting the fungal pathogen Fusarium solani. Virus Res. 317, 198817 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Larkin M. A., et al. , Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong J., Chen D., Zhu H. J., Gao B. D., Zhou Q., Hypovirulence of Sclerotium rolfsii caused by associated RNA mycovirus. Front. Microbiol. 7, 1798 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urayama S., et al. , Mycoviruses related to chrysovirus affect vegetative growth in the rice blast fungus Magnaporthe oryzae. J. Gen. Virol. 91, 3085–3094 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Chen Y., et al. , MoTup1 is required for growth, conidiogenesis and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 16, 799–810 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turgeon B. G., Condon B., Liu J., Zhang N., “Protoplast transformation of filamentous fungi” in Molecular and Cell Biology Methods for Fungi (Humana Press, 2010) pp. 3–19. [DOI] [PubMed] [Google Scholar]
- 64.Davis L. G.,Dibner M. D.,Battey J. F.,“Southern blot” in Basic Methods in Molecular Biology. (Elsevier, 1986) pp. 62–65. [Google Scholar]
- 65.Jedličková L., Gadas D., Havlová P., Havel J., Determination of ergosterol levels in barley and malt varieties in the Czech Republic via HPLC. J. Agric. Food Chem. 56, 4092–4095 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Statistical analysis was conducted using the IBM SPSS Statistics (version 22.0) program with a one-way analysis of variance (ANOVA) and Tukey post hoc test. For two-group comparisons, two-tailed Student’s t test was conducted with a P value smaller than 0.05 being considered statistically significant. The graphs were produced using the GraphPad Prism 8 software. The four cDNA sequences of SlAV1 dsRNA have been deposited in GenBank under accession numbers MH040331-MH040334. The SlPKS1 and SvPKS1 DNA sequences have been deposited in the GenBank under accession numbers OM249968 and OM249969. The RNA-seq raw data have been deposited in NCBI under BioProject accession numbers PRJNA622902 and PRJNA623067.