Significance
Cytoskeletal proteins are tightly controlled to ensure their proper dynamics and function. Here, we study the regulation of the bacterial tubulin homolog FtsZ, a core component of the bacterial cell division apparatus, by its alphaproteobacterial regulator MipZ. FtsZ forms polarized, treadmilling filaments at the division site, passing through different conformational states during its polymerization cycle. Using a range of different approaches, including hydrogen–deuterium exchange and fluorescence correlation spectroscopy, we show that MipZ can sequester FtsZ monomers, affect the conformation of FtsZ, and block the polymerization interface at the (+)-end of FtsZ filaments, thereby disrupting their treadmilling dynamics and inducing filament disassembly. This combination of mechanisms may help to fine-tune the regulatory effect of MipZ and ensure robust division site placement.
Keywords: bacterial cell division, divisome, division site placement, FtsZ inhibitor, cytoskeleton
Abstract
The spatiotemporal regulation of cell division is a fundamental issue in cell biology. Bacteria have evolved a variety of different systems to achieve proper division site placement. In many cases, the underlying molecular mechanisms are still incompletely understood. In this study, we investigate the function of the cell division regulator MipZ from Caulobacter crescentus, a P-loop ATPase that inhibits the polymerization of the treadmilling tubulin homolog FtsZ near the cell poles, thereby limiting the assembly of the cytokinetic Z ring to the midcell region. We show that MipZ interacts with FtsZ in both its monomeric and polymeric forms and induces the disassembly of FtsZ polymers in a manner that is not dependent but enhanced by the FtsZ GTPase activity. Using a combination of biochemical and genetic approaches, we then map the MipZ–FtsZ interaction interface. Our results reveal that MipZ employs a patch of surface-exposed hydrophobic residues to interact with the C-terminal region of the FtsZ core domain. In doing so, it sequesters FtsZ monomers and caps the (+)-end of FtsZ polymers, thereby promoting their rapid disassembly. We further show that MipZ influences the conformational dynamics of interacting FtsZ molecules, which could potentially contribute to modulating their assembly kinetics. Together, our findings show that MipZ uses a combination of mechanisms to control FtsZ polymerization, which may be required to robustly regulate the spatiotemporal dynamics of Z ring assembly within the cell.
The Z ring was the first prokaryotic cytoskeletal structure to be identified (1, 2). It is formed by the tubulin homolog FtsZ and serves to recruit other divisome components to midcell prior to cell division (3–5). Like tubulin, FtsZ shows the ability to bind and hydrolyze guanosine-5'-triphosphate (GTP) (6–8). It has a conserved domain architecture that includes i) a variable N-terminal region; ii) a GTPase domain, composed of an N-terminal subdomain (NTD) and a C-terminal subdomain (CTD) that are separated by a long central helix (helix 7); iii) a long flexible linker; and iv) a conserved C-terminal peptide followed by a short variable region (9–14). FtsZ polymerization is a highly dynamic process that involves the association of molecules in the GTP-bound state and their dissociation upon nucleotide hydrolysis (15). Upon polymerization, FtsZ is thought to undergo a conformational change (16, 17), similar to eukaryotic cytoskeletal proteins such as actin and tubulin (18–20). In support of this notion, recent structural analyses of the polymeric (open-cleft) form of FtsZ revealed a downward shift of helix H7 and a rotation of the CTD compared to the monomeric (closed-cleft) form (2, 21–23). As also suggested for actin (19), this conformational change may promote intersubunit interactions, which in turn favor nucleotide hydrolysis (22–24), thereby destabilizing the subunit interface and promoting filament disassembly. Recent in vitro (25, 26) and in vivo (27, 28) work has shown that FtsZ polymers treadmill, whereby new subunits are preferentially added at the (+)-end and released from the (−)-end of the filament. This feature turned out to be essential for proper remodeling of the peptidoglycan layer during septum formation (27–29). The intrinsic polarity of FtsZ filaments is thought to result from differential interactions between the two terminal (open-cleft) subunits of the polymer and free (closed-cleft) monomers, which translate into distinct kinetics of subunit addition at the two ends (30). Notably, it has been suggested that new FtsZ subunits attach to the filament through their nucleotide-binding interface, yielding a directionality opposite to that of tubulin (31).
FtsZ polymerization needs to be tightly regulated to ensure correct cell division. To position their divisome at midcell, bacteria use both positive and negative regulators that stabilize the Z ring at the division plane (32–36) or inhibit its assembly at eccentric positions (37–40). Moreover, various regulatory factors are in place to modulate Z ring assembly in response to the growth conditions (41–43). The molecular mechanisms that have evolved to negatively regulate FtsZ assembly are diverse and include, for instance, the sequestration of monomers (44, 45), the capping of the C-terminal polymerization interface (plus-end) (46), and the fragmentation of polymers (47). Similar principles also apply to the control of tubulin dynamics in eukaryotic cells (48). However, some of the regulatory mechanisms found for tubulins, such as severing (48) or active disassembly (49), have not been observed for FtsZ to date. Yet, it is unclear whether these mechanisms are truly specific for the eukaryotic domain.
Many alphaproteobacteria lack common negative regulators of FtsZ such as the MinCD or nucleoid occlusion systems. Instead, they possess the MipZ protein, a gradient-forming P-loop ATPase of the ParA/MinD family, that uses the dynamics of chromosome segregation to control division site placement (39, 50). Like its family relatives, MipZ relies on its ATPase cycle for proper activation and localization (51, 52). MipZ monomers interact with complexes of the chromosome partitioning protein ParB at the cell poles, which stimulates their adenosine-5’-triphosphate (ATP)-dependent dimerization (52). The resulting dimers are released from ParB and interact nonspecifically with chromosomal DNA (51, 53), giving rise to a dynamic bipolar concentration gradient with minimum in the midcell region. Dimerization enables MipZ to interact with FtsZ and thus inhibit Z ring formation along the MipZ gradient, thereby limiting cell division to the cell center (39). The intrinsic ATPase activity of MipZ eventually promotes dissociation of the dimeric complex, generating monomers that reassociate with ParB to start the next localization cycle (39, 51).
In this study, we use a multipronged approach to investigate the molecular mechanism underlying the inhibition of FtsZ polymerization by MipZ. Our results indicate that the regulatory activity of MipZ is based on a combination of different effects. At the cell poles, where its concentration is high, it may be able to sequester FtsZ monomers. In addition, MipZ can cap the (+)-end of FtsZ polymers, thereby disrupting their treadmilling dynamics and stimulating their disassembly. We further show that MipZ binding affects the conformational dynamics of FtsZ molecules, which could provide an additional means to modulate their polymerization kinetics. Thus, MipZ emerges as a versatile cell division regulator that employs various mechanisms to efficiently control division site placement.
Results
MipZ Directly Interacts with FtsZ to Inhibit Its Polymerization.
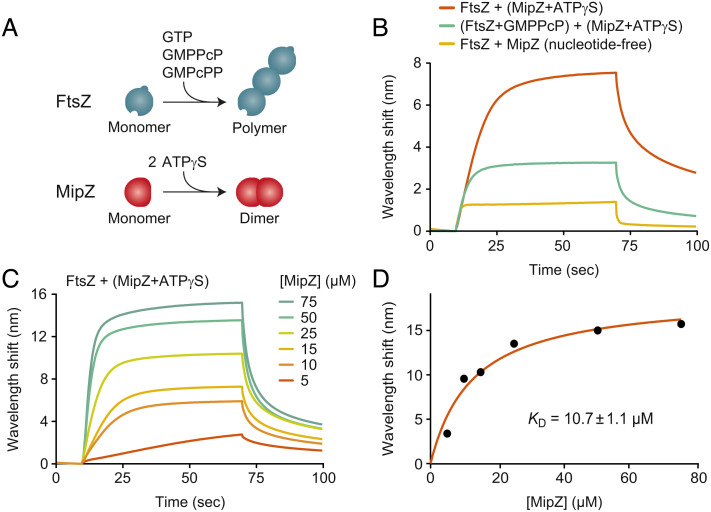
Previous work has revealed that, in the presence of MipZ, FtsZ has a moderately increased GTPase activity and forms shorter polymers that display a reduced tendency to sediment during ultracentrifugation (39). However, so far, no direct interaction between MipZ and FtsZ has been detected in vitro. To study the nature of the inhibitory effect of MipZ in more detail, we purified the two proteins and verified their functionality in vitro (SI Appendix, Fig. S1 A–D). Subsequently, we investigated the binding of MipZ to FtsZ by biolayer interferometry (54), using biotinylated FtsZ monomers coupled to a streptavidin-coated sensor as a bait (Fig. 1). While monomeric MipZ bound FtsZ with only low affinity, dimeric MipZ obtained by incubation of the protein with the slowly hydrolysable ATP analog adenosine 5'-[γ-thio]-triphosphate (ATPγS) showed an interaction with an apparent equilibrium dissociation constant (KD) of ~11 µM (Fig. 1 B–D). Notably, MipZ also interacted with biotinylated FtsZ that had been preincubated with guanosine-5'-[(β,γ)-methyleno]triphosphate (GMPPcP), a slowly hydrolysable GTP analog inducing the formation of stable short, curved polymers that can be readily immobilized on the sensor chip. However, in this case, the number of interacting MipZ molecules was significantly lower, while the binding affinity remained largely constant (Fig. 1B and SI Appendix, Fig. S2A), suggesting that polymerization may reduce the number of sites available for MipZ binding. To confirm that MipZ can interact with FtsZ polymers, we tested the effect of MipZ dimers on FtsZ sedimentation in the presence of GTP, GMPPcP, or guanosine-5'-[(α,β)-methyleno]triphosphate (GMPcPP), another slowly hydrolyzable GTP analog that produces very long, rigid FtsZ polymers. In all conditions, the amount of FtsZ recovered from the pellet after ultracentrifugation decreased when MipZ was included in the reaction (SI Appendix, Fig. S2B). At the same time, MipZ cosedimented with the remaining FtsZ polymers, supporting the notion that it interacts with the polymeric form of FtsZ.
Fig. 1.
MipZ binds directly to FtsZ. (A) Schematics depicting FtsZ polymerization in the presence of GTP or its nonhydrolyzable analogs GMPPcP or GMPcPP (Top) and MipZ dimerization in the presence of ATP or the nonhydrolyzable analog ATPγS (Bottom). (B) Biolayer interferometry analysis of the interaction between FtsZ and MipZ. Biotinylated FtsZ immobilized on a streptavidin-coated biosensor was probed with MipZ (15 µM) in the absence or presence of 1 mM ATPγS or 2 mM GMPPcP. At the end of association phase, the sensor was washed with protein-free buffer to follow MipZ dissociation. Components in parentheses were preincubated together before the start of the binding reaction. Note that 77% of the FtsZ molecules were associated with guanosine-5'-diphosphate (GDP) after purification. (C) Titration series analyzing the interaction of immobilized FtsZ with increasing concentrations (5 to 75 µM) of MipZ in the presence of ATPγS. (D) Binding affinity of MipZ•ATPγS dimers for FtsZ monomers. The wavelength shifts measured at the end of the association phase (Bmax) from panel c were plotted against the protein concentration. The apparent equilibrium dissociation constant (KD) of the FtsZ·MipZ complex was obtained by fitting the data to a one-site saturation ligand binding model. The KD value represents the average of three independent experiments (±SD). The graph shows the results of a representative experiment.
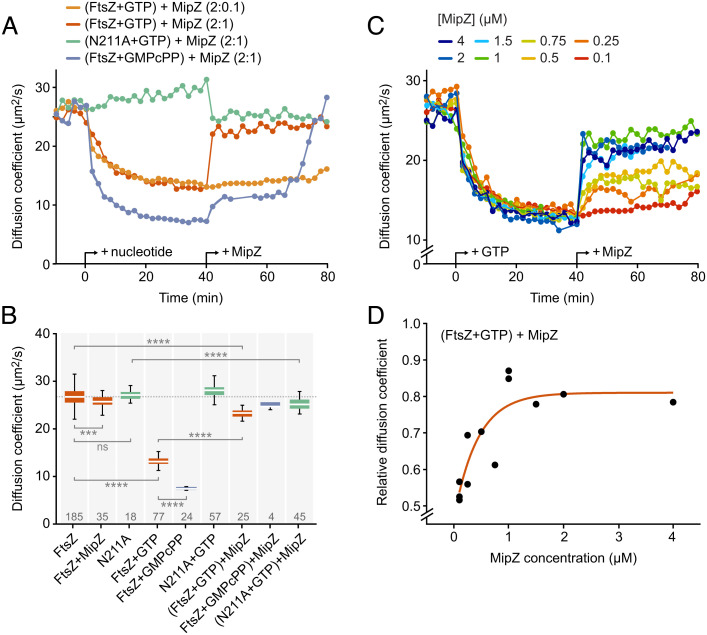
Next, we monitored the effect of MipZ on FtsZ polymerization over time using fluorescence correlation spectroscopy (FCS). This technique determines the diffusional mobility of molecules in solution and thus provides a measure of their average particle sizes (55). Upon the addition of GTP, the diffusion coefficient of FtsZ gradually increased to a value much lower than the one observed for the monomeric form, reflecting the formation of slowly diffusing polymers (Fig. 2 and SI Appendix, Fig. S3). By contrast, no change was observed for an FtsZ variant (FtsZN211A) carrying a mutation in the C-terminal polymerization interface that prevents its assembly into higher-order structures (56) (SI Appendix, Figs. S1B and S4A). Importantly, FtsZ•GTP polymers disassembled rapidly when mixed with MipZ dimers at a 2:1 (FtsZ:MipZ) ratio (Fig. 2 A and B). Titration experiments showed that the inhibitory effect of MipZ was concentration dependent (Fig. 2 A, C, and D). Notably, its maximum was already reached at substoichiometric MipZ concentrations, suggesting that MipZ may not, or at least not exclusively, act by sequestering FtsZ monomers (Fig. 2 C and D). However, even at high MipZ concentrations, the diffusion coefficient of FtsZ•GTP remained slightly (19%) lower than the one observed for the monomeric form. This effect may be, at least partially, explained by an interaction of MipZ with the FtsZ monomers released in the depolymerization reaction, because a similar increase in the diffusion coefficient was observed when MipZ was added to constitutively monomeric FtsZN211A•GTP (Fig. 2B). Moreover, it is conceivable that the rapid assembly kinetics of FtsZ•GTP make it difficult for MipZ to convert FtsZ to a purely monomeric state. Together, these results show that MipZ is a potent FtsZ inhibitor, whose action relies on a highly dynamic interaction with its target.
Fig. 2.
Effect of MipZ on the dynamics of FtsZ. (A) Time evolution of the diffusion coefficient of FtsZ measured by FCS. FtsZ or its monomeric variant FtsZN211A (2 µM), each containing 10% AlexaFluor488-labeled protein, was analyzed at 2 min intervals prior to and after the addition of the indicated nucleotide (2 mM for GTP, 0.2 mM for GMPcPP) and MipZ•ATPγS (at the indicated ratios), generated by preincubation of MipZ with 1 mM ATPγS. Data represent the average of 2 to 3 experiments. See SI Appendix, Fig. S3B for individual measurements. (B) Diffusion coefficients of FtsZ or FtsZN211A (2 µM) after the addition of MipZ•ATPγS (1 µM) in different conditions. The data shown represent averages of 2 to 3 consecutive FCS measurements made after signal stabilization. They are represented as box plots, with the white horizontal line indicating the median, the box the interquartile range, and the whiskers extending to the lowest and highest values within 1.5 times the interquartile range from the hinges. The number of data points obtained per condition is indicated at the bottom. The significance of differences to the respective MipZ-free condition was tested using the Student’s t test (ns: not significant, ***P < 0.001, ***P < 0.0001). (C) Time evolution of the diffusion coefficient of FtsZ after the addition of different concentrations of MipZ•ATPγS. FtsZ (2 µM) was analyzed at 2 min intervals prior and after the addition of GTP (2 mM) and, later, MipZ•ATPγS (at the indicated ratios). Data represent the (averaged) results of one to three experiments. (D) Change in the steady-state diffusion coefficient of FtsZ measured in the presence of GTP and MipZ•ATPγS relative to the diffusion coefficient of FtsZ-GDP alone as a function of the MipZ concentration. The values shown represent the average of consecutive measurements, obtained as described in (B). The data were fitted to the equation D(FtsZ + GTP + MipZ)/D(FtsZ − GTP − MipZ) = −0.34 * exp([MipZ]/−0.43) + 0.81.
The FtsZ GTPase Activity is Not Essential But Conducive for the Inhibitory Effect of MipZ.
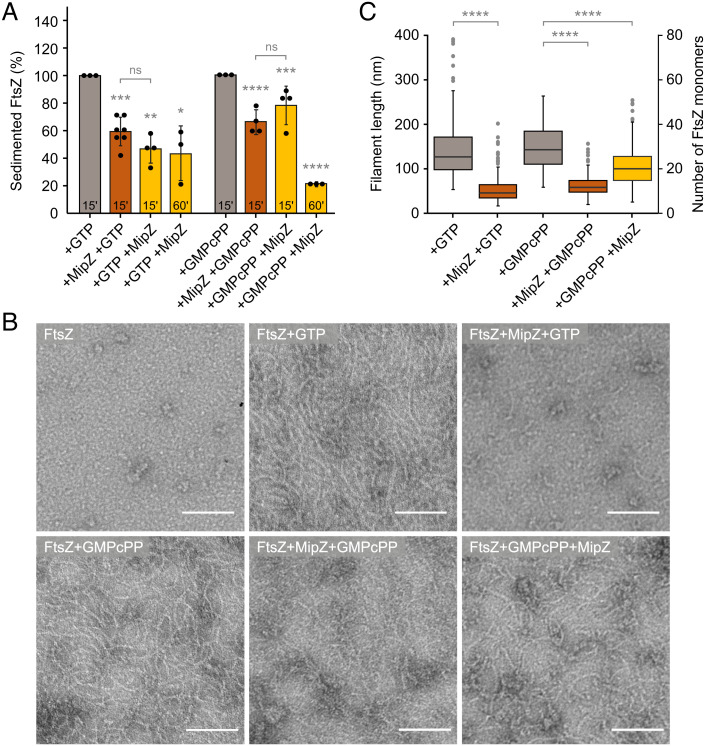
All FtsZ inhibitors known to date appear to require the GTPase activity of FtsZ to prevent FtsZ polymerization (31, 45, 57). Similarly, MipZ was reported to increase the GTPase activity of FtsZ by twofold (39). To assess the mechanistic significance of this stimulatory effect, we monitored the influence of MipZ on the mobility of FtsZ•GMPcPP polymers by FCS. In the presence of GMPcPP, FtsZ formed larger filaments that disassembled only slowly after the addition of MipZ (Fig. 2). Interestingly, the disassembly process was biphasic, with an initial slow phase that led to partial depolymerization of FtsZ, followed by a rapid second phase culminating in its complete disassembly (Fig. 2A). As a complementary approach, we compared the activity of MipZ on FtsZ•GTP or FtsZ•GMPcPP polymers by sedimentation assays and transmission electron microscopy. When FtsZ monomers were incubated with MipZ dimers prior to the start of polymerization, MipZ was able to efficiently prevent polymerization regardless of the nature of the nucleotide (Fig. 3). However, when preassembled in the absence of MipZ dimers, FtsZ•GMPcPP polymers showed less sensitivity to MipZ than FtsZ•GTP polymers, consistent with the results of the FCS analysis. A similar behavior was observed for FtsZ in the presence of the alternative GTP analog, GMPPcP, and for GTP-induced polymers of FtsZD213A (SI Appendix, Figs. S1E and S4A), a variant with strongly reduced GTPase activity (SI Appendix, Fig. S1B). Thus, MipZ appears to rely on the FtsZ GTPase activity for full inhibition, although it also acts on hydrolysis-impaired polymers, albeit with reduced efficiency.
Fig. 3.
Role of GTP hydrolysis in the MipZ-dependent regulation of FtsZ polymerization. (A) Sedimentation assay of FtsZ in the presence of MipZ and GTP or GMPcPP. FtsZ (3 µM) was incubated for 15 min with the indicated nucleotide (2 mM for GTP, 0.2 mM for GMPcPP), mixed with MipZ•ATPγS (6 µM) and incubated for another 15 to 60 min. Alternatively, FtsZ was first incubated with MipZ•ATPγS prior to the supplementation of nucleotide and additional 15 to 60 min of incubation. FtsZ polymers were collected by ultracentrifugation and quantified after SDS gel electrophoresis. The bar chart shows the average (±SD) of the percentage of pelleted FtsZ from three to seven independent experiments (dots). ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test). (B) Transmission electron micrographs of FtsZ incubated with MipZ and GTP or GMPcPP. FtsZ (5 µM) was incubated in the presence or absence of GTP or GMPcPP (2 mM) and/or MipZ•ATPγS (7.5 µM) in the indicated order, stained with uranyl acetate and visualized by transmission electron microscopy. (Scale bars, 100 nm). (C) Quantification of the polymer lengths in the images from (B). The data are represented as box plots, with the thick horizontal line indicating the median, the box the interquartile range, and the whiskers extending to the lowest and highest values within 1.5 times the interquartile range from the hinges. At least 100 filaments were counted for every condition. ****P < 0.0001 (Student’s t test). The difference in the results obtained for FtsZ with both GMPcPP and MipZ in different orders of addition was not statistically significant.
FtsZ Specifically Interacts with the S6-H7 Region of MipZ.
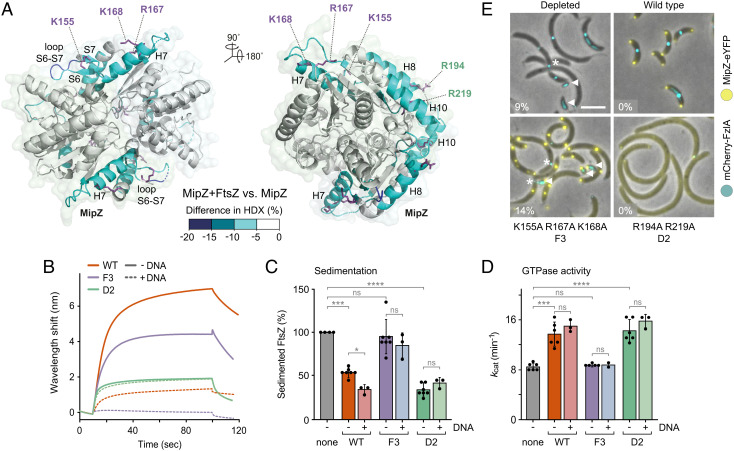
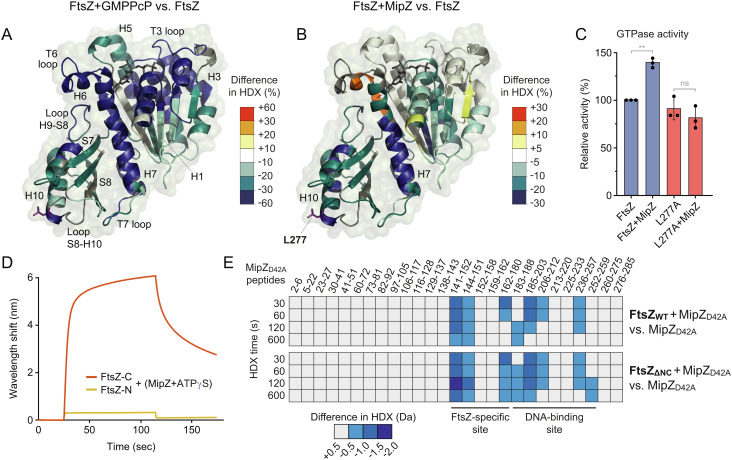
MipZ dimers interact with both chromosomal DNA and FtsZ, and they are thought to inhibit FtsZ polymerization while being dynamically associated with the nucleoid in vivo (39, 51). Whereas the DNA-binding site of MipZ has been recently narrowed down to a positively charged patch of amino acids at the interface between the two monomers (53), the determinants mediating its interaction with FtsZ are still unknown. To better understand the regulatory effect of MipZ, we aimed to map the binding interfaces between the two proteins. For this purpose, MipZ dimers were subjected to hydrogen–deuterium exchange (HDX) analysis (58) in the absence or presence of FtsZ to monitor local shifts in the accessibility of backbone amide hydrogens caused by FtsZ binding. Peptides of MipZ that showed a reduced rate of deuterium uptake in the presence of FtsZ clustered in two distinct regions of the protein (Fig. 4A and Dataset S1 and SI Appendix, Fig. S5A). One of these regions (residues 141 to 180) was specifically involved in FtsZ binding, encompassing helix H7 as well as a structural feature comprising sheet S6/S7 and loop S6-S7 that differentiates MipZ from homologous P-loop ATPases such as ParA and MinD (51). The second region (195 to 205, 238 to 259), by contrast, including helices H8 and H10, overlapped with the previously characterized, highly positively charged DNA-binding site (53). Notably, the DNA-binding site is located at the rim of the dimer interface and composed of residues from both subunits. The FtsZ-specific site, by contrast, only comprises residues from within each subunit, yielding two sites per MipZ dimer, located on the opposite sides of the complex (Fig. 4A).
Fig. 4.
Identification of the FtsZ-binding site(s) of MipZ. (A) Maximum differences in deuterium uptake between the MipZD42A•ATPγS-FtsZ complex and MipZD42A•ATPγS alone observed throughout the whole time course (0.5, 1, 2, and 10 min) mapped on the crystal structure of MipZ. Blue color indicates regions protected (lower HDX) upon FtsZ binding. (B) Biolayer interferometry analysis of the interaction between FtsZ and the F3 (K155A R167A K168A) and D2 (R194A R219A) variants of MipZ. Biotinylated FtsZ immobilized on a streptavidin-coated biosensor was probed with the indicated MipZ variants (15 µM) in the presence of 1 mM ATPγS. When indicated, a 14-bp dsDNA oligonucleotide (7.5 µM) was included in the reaction. (C) Effects of different MipZ variants on FtsZ polymerization. FtsZ (3 µM) was incubated with 2 mM GTP in the absence or presence of the indicated MipZ•ATPγS variants (6 µM). When indicated, a 14-bp dsDNA oligonucleotide (3 µM) was included in the reactions. After ultracentrifugation, the amount of sedimented FtsZ was quantified by SDS-PAGE. The bar chart shows the average (±SD) amount of sedimented FtsZ recovered from the pellet. Data represent the results of 3 to 7 independent experiments (dots). The significance of differences to the MipZ-free condition was tested using the Student’s t test (ns: not significant, *P < 0.05, ***P < 0.001, ****P < 0.0001). (D) GTPase activity of FtsZ in the presence of different MipZ variants. FtsZ (3 µM) was assayed in the absence (none) or presence of the indicated MipZ•ATPγS variants (4 µM). When indicated, a 14-bp dsDNA oligonucleotide (3 µM) was included in the reactions. The bar chart shows the average (±SD) of 2 to 6 independent experiments (dots). The significance of differences to the MipZ-free condition was tested using the Student’s t test (ns: not significant, ***P < 0.001, ****P < 0.0001). (E) Subcellular localization and functionality of different MipZ-eYFP variants. Shown are overlays of phase contrast and fluorescence images of MipZ-depleted cells (LC33, LC34, and LC36) producing the indicated MipZ-eYFP variants as well as mCherry-FzlA (false colored in cyan) as a proxy for the divisome. Minicells (white arrows) and mislocalized divisomes (asterisks) are indicated. The percentage of minicells produced by each strain is given in the lower left corner of the images. (Scale bar, 3 µm.)
To clarify the role of the two binding sites in the interaction with FtsZ, we purified two MipZ variants containing either three amino acid exchanges (K155A R167A K168A) in the FtsZ-specific site (MipZ-F3) or two exchanges (R194A R219A) in the previously characterized DNA-binding site (MipZ-D2). The resulting mutant proteins had normal ATPase activity, suggesting that protein folding, nucleotide binding, and dimerization were unaffected (SI Appendix, Fig. S1 C and D). As expected, both variants showed a marked decrease in their affinity for FtsZ in biolayer interferometry assays (Fig. 4B), confirming the contribution of the two sites to the MipZ–FtsZ interaction in vitro. Interestingly, MipZ-D2 exhibited a stronger reduction in the binding signal than MipZ-F3. However, only mutation of the FtsZ-specific site abolished the ability of MipZ to prevent the sedimentation of FtsZ polymers or stimulate the FtsZ GTPase activity, whereas exchanges in the DNA-binding site had hardly any influence on the regulatory activity of MipZ (Fig. 4 C and D). To corroborate this result, we tagged wild-type MipZ and its mutant derivatives with yellow fluorescent protein (eYFP) and analyzed their localization behavior and function in cells depleted of the native MipZ protein. In doing so, we colocalized the fusion proteins with a red fluorescent protein (mCherry)-tagged derivative of the cell division protein FzlA (32) as a proxy for FtsZ localization. As expected, exchanges in the FtsZ-specific site did not affect the ability of MipZ to interact with the nucleoid and condense into ParB-proximal patches. Nevertheless, the cells showed a phenotype similar to the one observed upon MipZ depletion, producing a mixture of minicells and short filaments with FzlA located at eccentric positions (Fig. 4E and SI Appendix, Fig. S6 A and B). These results suggest that the FtsZ-specific site is required for MipZ to productively interact with FtsZ and mediate the positioning of FtsZ at midcell. Defects in the DNA-binding site, by contrast, led to dispersion of MipZ within the cell and completely inhibited cell division (Fig. 4E), likely because MipZ retained its ability to interact with FtsZ but was no longer associated with the nucleoid, thereby inhibiting FtsZ polymerization throughout the cell (53).
To further investigate the role of the DNA-binding site in the regulatory activity of MipZ, we reinvestigated the association of MipZ with FtsZ in the presence of DNA. Notably, DNA reduced the FtsZ-binding activity of wild-type MipZ to a level slightly lower than that observed for MipZ-D2. Moreover, it completely abolished the binding of MipZ-F3, whereas it had no effect on the interaction with MipZ-D2 (Fig. 4B), indicating that FtsZ and DNA compete for interaction with the MipZ DNA-binding site. Importantly, however, the obstruction of the DNA-binding site by DNA had no adverse effects on the ability of MipZ to prevent the sedimentation of FtsZ or stimulate its GTPase activity (Fig. 4 C and D). Thus, only the FtsZ-specific site is required for MipZ to inhibit FtsZ polymerization, although its DNA-binding site contributes to the interaction with FtsZ, thereby potentially modulating MipZ activity in vivo.
The FtsZ-specific site and its surroundings contain a conspicuous array of exposed hydrophobic or charged residues (SI Appendix, Fig. S7A). To further corroborate the importance of this region for the regulatory effect of MipZ, we analyzed additional MipZ variants with exchanges in helix H7 (L161A, W164A, K168A, and L172A) and at the C-terminal end of helix H8 (K208A R209A). Cells producing these variants in place of the wild-type protein again displayed mild-to-severe cell division defects (SI Appendix, Fig. S6). Moreover, most of the variants also showed defects in GTPase stimulation and/or FtsZ binding (SI Appendix, Fig. S7 B and C). Together, these findings support the notion that the FtsZ-specific site is critical for the inhibitory activity of MipZ in vitro and in vivo.
MipZ Inhibits FtsZ Assembly by Interacting with the C-terminal Polymerization Interface.
Knowledge about the binding site of an FtsZ inhibitor can help to understand the underlying regulatory mechanism. We, therefore, probed the effect of MipZ on the accessibility of the FtsZ surface by HDX analysis (Dataset S2). As a reference, we first monitored shifts in the HDX pattern resulting from GMPPcP-induced polymerization of FtsZ in the absence of MipZ (Fig. 5A and SI Appendix, Fig. S5B). As expected, we observed drastic changes throughout the entire GTPase domain of FtsZ, which can be explained by the sum of three events: i) nucleotide binding and protection of the polymerization interfaces, which comprise loops H6-H7 and S3-H3 (T3) within the NTD as well as loop H7-H8 (T7) and helix H10 in the CTD (59); ii) protection of lateral interaction surfaces, including helices H3 and H5 and sheet S7 (60); and iii) polymerization-associated conformational changes, affecting helices H1 and H6, loop T6, helix H7, loop T7, and helices H9-S8 (30).
Fig. 5.
Identification of the MipZ-binding site of FtsZ. (A) Maximum differences in deuterium uptake between FtsZ in the presence or absence of GMPPcP (2 mM) observed throughout the whole time course (0.5, 1, 2, and 10 min) mapped on a structural model of the FtsZ GTPase domain. Blue color indicates regions protected (lower HDX) in the presence of GMPPcP. (B) Maximum differences in deuterium uptake between the FtsZ-MipZD42A complex and FtsZ alone in the presence of ATPγS. Blue and red color indicate regions of FtsZ that are protected (lower HDX) or deprotected (higher HDX), respectively, in the presence of MipZ. (C) Effect of MipZ on the GTPase activities of FtsZ and its L277A derivative. The indicated FtsZ proteins (2 µM) were assayed in the absence or presence of MipZ•ATPγS (4 µM). The apparent kcat values obtained were normalized to the value measured for wild-type FtsZ without MipZ. The bar chart shows the average (±SD) from three independent experiments (dots). Statistical significance was tested using a Student’s t test (ns: not significant, **P < 0.01). (D) Biolayer interferometry analysis of the interaction of MipZ with an N-terminal (FtsZ-N) fragment and a C-terminal (FtsZ-C) fragment of FtsZ, performed as described in Fig. 1B. (E) HDX analysis of the interaction of MipZ with full-length StrepII-FtsZ and a truncated variant comprising only helix H7 and the CTD (FtsZΔNC). The heat plots show the changes in the HDX pattern of MipZ induced by the addition of the indicated FtsZ proteins for a series of representative peptides (see Dataset S3 for the full list of peptides). Blue color indicates reduced deuterium uptake in the presence of the interactor.
On mixing FtsZ monomers with MipZ•ATPγS dimers (Fig. 5B and SI Appendix, Fig. S5B), we observed a strong protection of helix 7 and the CTD at the C-terminal polymerization interface of FtsZ, which likely reflects an interaction of MipZ with this region of the protein. Smaller changes in HDX, including an increased exposure of some segments, were also detected in the adjacent regions of the NTD, likely reflecting conformational changes induced by the binding event. Together, these results indicate that MipZ interacts with regions that are exposed at the (+)-end of FtsZ polymers.
To probe the putative MipZ binding site suggested by HDX analysis, we purified an FtsZ variant (L277A) bearing a single amino acid exchange in a region of helix H10 that was highly protected upon the addition of MipZ (Fig. 5B and SI Appendix, Fig. S1A). The mutant protein showed close-to-normal GTPase activity (Fig. 5C) and formed polymers similar to those of the wild-type protein (SI Appendix, Fig. S4D), indicating that the exchange did not have any significant effect on the nucleotide-dependent assembly of FtsZ, even though it was located in the vicinity of the polymerization interface. We then analyzed the ability of MipZ to interact productively with the mutant FtsZ protein. Importantly, MipZ was no longer able to stimulate the GTPase activity of FtsZL277A (Fig. 5C) nor could it induce the depolymerization of FtsZL277A filaments (SI Appendix, Fig. S4D), supporting the notion that helix H10 and the surrounding regions in the CTD of FtsZ are central components of the MipZ-binding interface. To further verify the MipZ binding site, we purified two FtsZ fragments, comprising either the N-terminal region of the protein up to the beginning of helix H7 (FtsZ-N; amino acids 1 to 182) or the adjacent C-terminal region (FtsZ-C; amino acids 183 to 508), and studied their interaction with MipZ by biolayer interferometry (Fig. 5D). While MipZ was unable to bind FtsZ-N, its interaction with FtsZ-C was similar to that observed with the full-length protein (compare Fig. 1B), consistent with the hypothesis that the MipZ-binding site is located in the CTD.
The conserved C-terminal peptide following the long variable linker of FtsZ is targeted by many different regulators (34, 44, 61). However, nonstructured protein regions show very high HDX rates, which makes it difficult to detect their interaction with other factors based on HDX analysis. To explore whether the flexible C-terminal region of FtsZ could contribute to MipZ binding, we therefore purified two FtsZ variants lacking the C-terminal linker (Ctl) or the conserved C-terminal peptide (Ctp) (SI Appendix, Fig. S1A), and analyzed their interaction with MipZ using the effect of MipZ on their GTPase activity and polymerization (as observed by TEM) as a proxy. We found that both variants were still sensitive to MipZ (SI Appendix, Figs. S1B and S4A), suggesting that the regulatory function of MipZ relies on interactions with the FtsZ core domain. In a complementary approach, we used HDX analysis to study the interaction of truncated FtsZ variants with MipZ. Notably, a fragment comprising only helix H7 and the CTD (FtsZΔNC, amino acids 183 to 320) changed the HDX pattern of MipZ in the same way as the full-length FtsZ protein (Fig. 5E and Dataset S3). These results show that the C-terminal region of the FtsZ GTPase domain contains all determinants mediating the interaction of FtsZ with the FtsZ-specific site and the DNA-binding site of MipZ.
To obtain insights into the structural basis of the MipZ–FtsZ interaction, we performed computational modeling studies with AlphaFold-Multimer (62), using a MipZ dimer and an FtsZ fragment (amino acids 183 to 325) comprising helix H7 and the CTD as a starting point. Strikingly, the predicted structure was perfectly in line with the experimental data and suggested an interaction between the FtsZ-specific site of MipZ and the CTD of FtsZ as identified by HDX analysis (SI Appendix, Fig. S8 A–C). The interaction determinants in MipZ include all residues in strand S7, helix H7, and helix H8 shown to be important for the regulatory effect of MipZ in this study (compare Fig. 4C and SI Appendix, Fig. S6A). Their side chains form hydrogen bonds and a dense network of hydrophobic interactions with residues in helix H10, loop S8-H10, and loop T7 of the CTD, including the verified interaction determinant L277. A similar interface was also obtained in predictions using full-length FtsZ (SI Appendix, Fig. S8D) or an MipZ monomer (SI Appendix, Fig. S8E). These findings further corroborate the central role of the FtsZ-specific site in MipZ function.
MipZ Binding Stabilizes FtsZ in a Distinct Conformation.
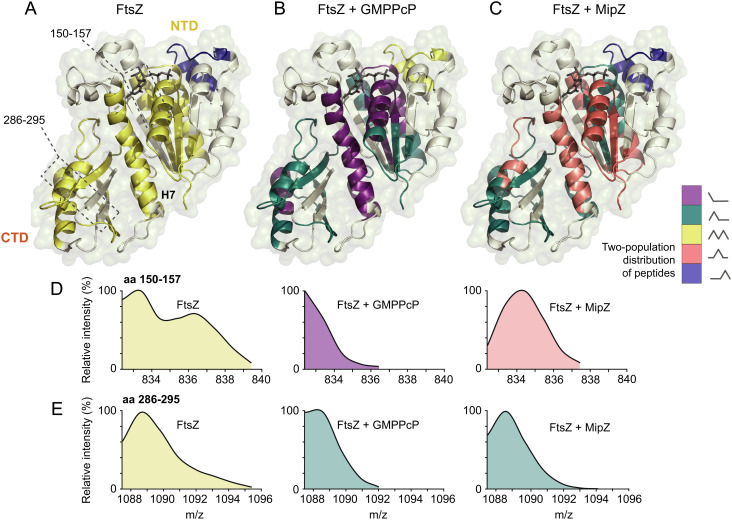
Similar to tubulin and other eukaryotic cytoskeletal elements, FtsZ was shown to exist in two distinct conformations (the closed- and open-cleft states) that differ in the arrangement of the NTD relative to helix H7 and the CTD. Monomers are typically found in the closed-cleft state, lacking a complete polymerization interface, and transition to the open-cleft state upon integration into a polymer (30). Interestingly, when further analyzing the HDX profiles of FtsZ monomers, we observed that the mass/charge profiles of multiple peptides showed a bimodal distribution (Fig. 6 A, D, and E and Dataset S2 and SI Appendix, Fig. S9A). This was particularly true for peptides located in helix H7 or the CTD, which both move significantly relative to the NTD during the closed-to-open-cleft transition (30) as well as in regions surrounding H7 (including H1, which is in direct contact with H7). This finding indicates structural flexibility in certain regions of the protein, possibly due to spontaneous transitions between the closed- and open-cleft states. A similar modality profile, albeit with slight differences in the proportions of the different populations, was obtained for a constitutively monomeric FtsZ variant carrying mutations in both polymerization interfaces (F182E L276E) (31) (SI Appendix, Figs. S1 A and B and S9 B and C and Dataset S4), excluding the possibility that the observed bimodal behavior reflects a dynamic equilibrium between monomers and small oligomers. Based on this finding, we next analyzed the modality profile of polymeric FtsZ, generated by incubation with GMPPcP. Under this condition, essentially all peptides switched to unimodal distributions (Fig. 6B and SI Appendix, Fig. S9A) that either resembled the distributions observed for the slow-exchanging subpopulation of FtsZ monomers or were shifted to even lower mass/charge ratios, indicating further increased protection. These results support the notion that polymerization stabilizes FtsZ in the open-cleft conformation (30) and causes a global transition of the protein to a more protected state. The only exception was a peptide (amino acids 68 to 84) in the T3 loop region of the NTD GTPase subdomain, which switched from a unimodal (unprotected) to a bimodal distribution in the presence of GMPPcP, consistent with the flexibility of this region observed in crystallographic studies (30).
Fig. 6.
Conformational dynamics of FtsZ upon polymerization or MipZ binding. The modality of the distributions of the peptide mass/charge ratios obtained in HDX analyses of (A) FtsZ alone, (B) FtsZ in the presence of GMPPcP, and (C) FtsZ in the presence of MipZ•ATPγS was plotted onto a structural model of C. crescentus FtsZ. The graphs in (D) and (E) show the distribution of the mass/charge ratios for two representative peptides (amino acids 150 to 157 and amino acids 286 to 295) after 60 s of deuteration in the three conditions described in panels A–C.
Importantly, incubation of FtsZ monomers with MipZ•ATPγS also induced a low-flexibility state, in which all peptides adopted unimodal mass/charge distributions (Fig. 6C and SI Appendix, Fig. S9A). Peptides close to the T7 loop and helix H10 (213 to 243, 254 to 265, and 278 to 307) were strongly protected, likely due to direct interaction with MipZ. By contrast, those in other regions of the protein, including helices H7 and H1 (amino acids 34 to 43, 150 to 157, 168 to 172, 183 to 203, and 243 to 253), mostly adopted intermediate mass/charge distributions, with maxima different from the ones identified for monomeric FtsZ alone. These results suggest that MipZ binding stabilizes FtsZ in a defined conformation that differs from the typical open- and closed-cleft states.
Discussion
The control of FtsZ polymerization has been studied intensively, but for many of the known regulatory factors the mechanisms of action are still incompletely understood. The well-characterized examples mainly act by sequestering FtsZ monomers or capping polymers (44–46). This study now reports a bacterial cell division inhibitor that induces a conformational change in FtsZ which could potentially also destabilize filaments at the (+)-end, thus resembling eukaryotic cytoskeletal regulators.
We show that MipZ binds monomeric FtsZ with moderate affinity (Fig. 1A) and is thus, in principle, able to act as a sequesterer. However, this is unlikely to be its only role, since sequesterers do not increase GTP turnover but, on the contrary, reduce it by limiting the incorporation of new FtsZ•GTP subunits. Moreover, to efficiently compete with homotypic interactions between FtsZ subunits, sequesterers usually need to bind FtsZ with affinities that are comparable to or higher than those between FtsZ subunits within the polymer, as for instance shown for Escherichia coli SulA and SlmA (38, 44, 45, 63). While the KD value of the interaction between MipZ and FtsZ monomers is in the range of 10 to 20 µM, the critical concentration of FtsZ is only 0.3 µM (SI Appendix, Fig. S1F). Thus, sequestration may only be effective at relatively high MipZ concentrations. MipZ indeed strongly impaired FtsZ assembly when added at a concentration (6 µM) close to the KD value, in particular when preincubated with FtsZ before the start of the polymerization reaction (Fig. 3). However, marked effects on FtsZ polymerization were already observed at concentrations in the nanomolar range, with half of the maximum inhibitory effect obtained at an FtsZ:MipZ dimer ratio of 1:0.12 (Fig. 2C). These results suggest that the inhibitory activity of MipZ is based on a mechanism that goes beyond sequestration.
Consistent with a more complex inhibitory mechanism, we found that MipZ also binds to FtsZ polymers (Fig. 1 and SI Appendix, Fig. S1), which is plausible because the MipZ binding site is located in the helix H10 region of FtsZ (Fig. 4A), whose local conformation is unaffected by the closed-to-open-cleft transition during polymerization (30). However, since this region is an integral part of the FtsZ polymerization interface, the only region accessible to MipZ in an FtsZ polymer is the first subunit at the (+)-end. This hypothesis is consistent with the slow activity of MipZ on stabilized GMPcPP-bound polymers (Fig. 2 A and B). In this condition, depolymerization may largely depend on random fragmentation, since the absence of nucleotide hydrolysis prevents the release of individual subunits at the (−)-end. Thus, right after the addition of MipZ to FtsZ polymers, only a low number of binding sites are available to MipZ. However, over time, spontaneous fragmentation may gradually increase the number of free (+)-ends, which are again capped by MipZ and thus prevented from rejoining, leading to a progressive, superlinear decay in polymer length (compare Fig. 2A).
Notably, the ability of MipZ to block the polymerization interface of FtsZ by interacting with the helix H10 region of the CTD is reminiscent of previously characterized FtsZ inhibitors, such as E. coli MinC (47) or Bacillus subtilis MciZ (46, 64). Our experimental results indicate that hydrophobic residues in the FtsZ-specific region are critical for the regulatory effect of MipZ, and computational modeling suggests that these residues interact with the helix H10/loop S8-H10/loop T7 region in the CTD of FtsZ. Notably, this region forms a deep hydrophobic pocket that can accommodate the exposed W164 residue of MipZ (SI Appendix, Fig. S8B), which emerged to be a key determinant of the MipZ–FtsZ interaction in this study. A binding mode based on hydrophobic interactions has also been reported for B. subtilis MciZ, although the structural details may differ (46, 64). Thus, while bacterial FtsZ regulators are highly diverse in sequence and evolution, their activities are based on common mechanistic principles. Interestingly, while MipZ, MinC, and MciZ target the same region of FtsZ, they show highly different binding affinities, which may be explained by their distinct modes of action. MipZ and MinC are part of dynamic gradient-forming systems that specifically inhibit FtsZ assembly in the pole-proximal regions of the cell, requiring transient, low-affinity interactions (KD~ 10 µM) (51, 65). MciZ, by contrast, serves to completely block cell division in terminally differentiated cells (66) and thus forms a stable complex with FtsZ (KD ~ 100 nM).
Although dimerization is required for MipZ to inhibit FtsZ polymerization efficiently (39, 51), we found that MipZ monomers were also able to interact with FtsZ, albeit with lower affinity (Fig. 1B). This finding is consistent with the fact that the FtsZ-specific binding site of MipZ is located distal from the dimerization interface and thus, in principle, a constitutive feature of MipZ exists independently of its oligomerization state. In line with this notion, permanently monomeric MipZ variants are still able to inhibit cell division when overproduced from a replicating plasmid (SI Appendix, Fig. S10). However, unlike the wild-type protein, they are not able to disassemble the Z ring but rather appear to compromise its function, possibly by altering its architecture or dynamics. The differences in affinities between MipZ monomers and dimers may be due to slight differences in their conformations (51, 53) that may translate into distinct modes of interaction with FtsZ (SI Appendix, Fig. S8E).
Using HDX analysis, we were able to obtain more detailed mechanistic insights into the regulatory effects of MipZ. Interestingly, the mass/charge ratios of many peptides obtained from FtsZ monomers produced two distinct Gaussian distributions, indicative of a dynamic equilibrium between the closed- and open-cleft conformations. Importantly, after the addition of MipZ, several peptides, in particular from helix H7 and adjacent regions in the NTD, which are not directly contacted by MipZ, exhibited distributions that could not be attributed to either of these states (Fig. 6). These findings suggest that MipZ binding locks FtsZ in a distinct, inhibited conformation, thereby preventing sequestered monomers from reattaching to the existing polymers or inducing the release of terminal subunits from capped (+)-ends. The ability to actively disassemble FtsZ polymers could explain why MipZ can already act at concentrations that are far below the KD of the MipZ–FtsZ interaction and therefore insufficient to saturate FtsZ to an appreciable level. MipZ could thus potentially act in analogy to microtubule-depolymerizing kinesins (families 8 and 13) in eukaryotes. This group of ATPases binds to the (+)-ends of microtubules and locks the terminal subunits in a conformation that mimics the GDP-bound state, thereby reducing their affinity for the adjacent subunit and promoting their release from the polymer (49, 67). Similar to MipZ, the kinesin-8 motor KIF19A is able to reach its half-maximal inhibitory capacity at a 1:0.14 (tubulin:KIF19A) ratio (68). Thus, similar regulatory principles may apply to the spatiotemporal regulation of the tubulin cytoskeleton in bacterial and eukaryotic cells, although the structures of the proteins involved are clearly different. Notably, active FtsZ depolymerization could also explain the observed stimulation of GTP turnover in the presence of MipZ (Fig. 4D). However, a similar effect could also be exerted by capping, if it shifted the equilibrium toward shorter but more abundant FtsZ polymers and thus decreased the residence time of FtsZ molecules in polymeric assemblies without affecting the total fraction of polymerized protein.
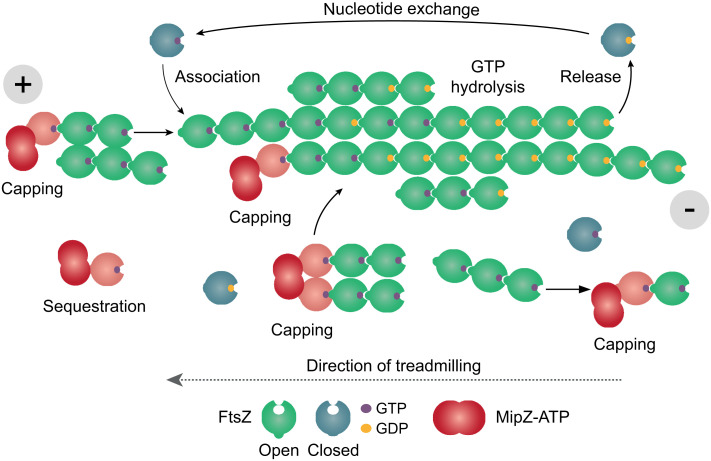
Taken together, our results suggest that MipZ can use multiple ways to control the dynamics of FtsZ polymerization (Fig. 7). On the one hand, it is able to sequester monomers or cap the (+)-end of FtsZ polymers – pathways that may be particularly effective at elevated MipZ concentrations. In addition, it induces a conformational change in interacting FtsZ molecules that could potentially favor their release from the (+)-end and thus actively promote filament disassembly. MipZ has a copy number of ~1,000 molecules per cell (39). However, its local concentration varies considerably between the poles and midcell, suggesting that different mechanisms may apply at these locations. At the poles, MipZ is highly concentrated, so that sequestration could make significant contributions to its regulatory function. However, closer to midcell, the MipZ concentration may not be sufficiently high for this process to work in an efficient manner, so that the control of FtsZ polymerization may largely rely on capping, possibly supported by active depolymerization.
Fig. 7.
Model of the molecular mechanism underlying the regulation of FtsZ polymerization by MipZ. FtsZ polymerizes by the addition of new subunits at the (+)-end and the release of subunits at the (−)-end, leading to treadmilling. The asymmetry of FtsZ filaments is based on a switch of FtsZ from the closed to the open conformation during polymerization. Since hydrolysis occurs in a time-dependent manner, subunits at the younger (+)-end of the filament are mostly bound to GTP. Those at the older (−)-end, by contrast, are bound to GDP, which induces instability and thus promotes the release of the terminal subunits. Filament growth occurs by the addition of individual subunits or oligomers to the (+)-end, and bundling will occur through lateral interactions. MipZ induces a conformational change in interacting FtsZ molecules (indicated in salmon). When bound to FtsZ monomers (sequestration), it can transiently arrest them in a polymerization-incompetent state, thereby reducing the rate of subunit addition and shifting the equilibrium toward depolymerization. MipZ can also bind to the (+)-ends of oligomers or polymers (capping), again preventing the growth of FtsZ polymers and favoring their disassembly. In addition, MipZ may potentially also induce a conformational change in the terminal FtsZ subunit and thus promote its release from the polymer.
Our work describes a bacterial cytoskeletal regulator that induces a conformational change in its target protein. It will be interesting to obtain more detailed structural insights into the conformation that FtsZ adopts in the presence of MipZ and to clarify whether this structural change affects the interactions between FtsZ subunits and, thus, their polymerization capacity.
Methods
Plasmids and Strains.
The bacterial strains, plasmids, and oligonucleotides used in this work are listed in SI Appendix, Tables S1–S3. Their construction and the growth conditions used are detailed in the SI Appendix. All plasmids were verified by DNA sequencing.
Fluorescence Microscopy.
Cells were grown to exponential phase in vanillate-containing medium, washed, and cultivated for 6 h in medium containing 0.3% (w/v) xylose to deplete the native MipZ protein and induce the production of the different MipZ-eYFP variants. After immobilization of the cells on agarose pads, images were acquired as detailed in the SI Appendix. The data were analyzed with Fiji 1.49 (69). Minicells were defined as cells whose length was smaller than the minimum length of the wild-type strain.
Transmission Electron Microscopy.
FtsZ (5 µM) was incubated with the indicated nucleotides in the absence or presence of MipZ (7.5 µM) at 25 °C in buffer P (50 mM HEPES/NaOH pH 7.2, 50 mM KCl, and 5 mM MgCl2). Aliquots of the reactions were transferred to glow-discharged carbon-coated grids. Protein was then stained with uranyl acetate and visualized by transmission electron microscopy. Details are given in the SI Appendix. Fiji 1.49 (69) was used for data analysis.
Immunoblot Analysis.
Immunoblot analysis was performed as described (39) using a polyclonal rabbit anti-MipZ antibody (39) or a monoclonal anti-GFP antibody (Sigma-Aldrich; Cat. #: G1544; RRID: AB_439690).
Protein Overproduction and Purification.
Proteins were overproduced in E. coli Rosetta(DE3)pLysS transformed with suitable plasmids. MipZ-His6 and its mutant derivatives were purified by Ni-NTA affinity chromatography, followed by cation exchange chromatography when indicated. FtsZ derivatives were overproduced as His6-SUMO fusions. After purification by Ni-NTA affinity chromatography, the His6-SUMO tag was removed by treatment with Ulp1-His6 protease (70) and a second round of Ni-NTA affinity chromatography. StrepII-tagged derivatives were further purified by gel filtration. Protein concentrations were determined before every experiment with the Bradford assay, using the Roti-Nanoquant reagent (Carl Roth) with bovine serum albumin (Carl Roth). A detailed description of the purification procedures is given in the SI Appendix.
Biolayer Interferometry.
Biolayer interferometry analysis was performed with a BLItz System (Sartorius) using Dip and Read High-Precision Streptavidin Biosensors. Biotinylated FtsZ (equivalent to a wavelength shift of 2 nm) was immobilized on the sensor surface and probed with MipZ or its mutant derivatives under the indicated conditions in buffer PG (50 mM HEPES/NaOH pH 7.2, 50 mM KCl, 5 mM MgCl2, and 10% (v/v) glycerol) containing 0.01% (v/v) Triton X-100 and 0.01 mM BSA. To determine equilibrium dissociation constants, the maximal wavelength shifts measured at the end of the association phases were plotted against the corresponding protein concentrations. Details are provided in the SI Appendix.
FCS.
Fluorescently labeled FtsZ was prepared by derivatization with a thiol-reactive dye, taking advantage of the fact that Caulobacter crescentus FtsZ contains a single cysteine residue (C123), which is exposed on the protein surface outside the polymerization interface. To this end, FtsZ or its mutant derivatives were incubated with dithiothreitol (DTT) for 10 min at room temperature. DTT was removed by passage of the protein through a PD SpinTrap G-25 desalting column (GE Healthcare) equilibrated with buffer PG (50 mM HEPES/NaOH pH 7.2, 50 mM KCl, 5 mM MgCl2, and 10% glycerol). The proteins (75 µM) were then incubated for 16 h at 4 °C with 10 mg ml−1 of the thiol-reactive fluorescent dye Alexa Fluor 488 C5-maleimide (Alexa488; Life Technologies; diluted in DMSO). Unreacted reagent was removed by extensive dialysis against buffer PG at 4 °C. All experiments were performed with a mixture of 90% nonlabeled protein and 10% labeled protein to avoid adverse effects of the label on the dynamics of FtsZ polymerization. MipZ was preincubated for 15 min with 1 mM ATPγS prior to its addition to the reaction. All FCS measurements were performed in a self-made chamber, using an LSM780 microscope equipped with a ConfoCor3 unit (Zeiss) in pseudo-crosscorrelation mode, as described previously (71). Details of the procedure are provided in the SI Appendix. To assess the time evolution of FtsZ polymerization after the addition of MipZ, FtsZ was added at the respective concentration. The chamber was then mixed thoroughly by pipetting, and 10 consecutive 2-min FCS measurements were performed. Subsequently, 2 mM GTP was added and 20 additional 2-min measurements were conducted before MipZ was added at the indicated concentrations. The diffusion coefficient was determined for each of the 2-min intervals.
Nucleotide Hydrolysis Assays.
GTPase and ATPase activities were measured using a continuous, regenerative coupled-enzyme assay (72), as detailed in the SI Appendix.
Sedimentation Assays.
FtsZ (3 µM) was preincubated with nucleotides in the absence or presence of MipZ (6 µM) and sedimented by ultracentrifugation. Pelleted protein was then applied to an SDS-polyacrylamide gel, followed by staining and quantification with an imaging system. Details are given in the SI Appendix.
HDX-Mass-Spectrometry (HDX-MS).
To elucidate the conformational dynamics of MipZ and FtsZ as well as the interaction interfaces of the two proteins, four separate sets of HDX-MS experiments were conducted that differed in the composition of the samples investigated. To determine the FtsZ-binding interface of dimeric MipZ (Dataset S1), 50 µM MipZD42A was incubated for 15 min at room temperature in buffer B6 supplemented with 1 mM ATPγS in the absence or presence of 100 µM FtsZ. The subsequent HDX reactions were conducted in D2O-containing buffer B6 supplemented with 1 mM ATPγS. To determine the MipZ-binding interface of FtsZ and compare the conformational changes induced by GMPPcP (Dataset S2), 50 µM FtsZ was incubated for 15 min at room temperature in buffer B6 containing 1 mM ATPγS in the absence of either interaction partner, in the presence of 100 µM MipZ, or in the presence of 2 mM GMPPcP. The HDX reactions were conducted in D2O-containing buffer B6 either supplemented with 1 mM ATPγS (determination of the MipZ-binding interface) or with both 1 mM ATPγS and 2 mM GMPPcP (comparison of conformational changes). To compare the HDX profiles of MipZ in the presence of wild-type FtsZ or truncated FtsZ variants (Dataset S3), 50 µM MipZD42A was incubated either alone or in the presence of 100 µM of FtsZ, FtsZΔN, or FtsZΔNC for 15 min at room temperature in buffer B6 supplemented with 1 mM ATPγS. The subsequent HDX reactions were conducted in D2O-containing buffer B6 supplemented with 1 mM ATPγS. To compare the conformational dynamics of FtsZ and its monomeric variant FtsZF182E/L276E (Dataset S4), the two proteins were incubated for approximately 15 min at room temperature in buffer B6, and HDX reactions were carried out in D2O-containing buffer B6. The mass spectrometric analysis of the HDX patterns was carried out essentially as described previously (53). Details on the procedure are given in the SI Appendix. The raw data and an in-depth analysis of the HDX profiles obtained are provided in Datasets S1–S4.
Bioinformatic Tools.
The structural model of C. crescentus FtsZ used to illustrate the HDX data was generated with I-TASSER (73) using Staphylococcus aureus FtsZ (PDB ID: 4DXD) as a template. Protein structures were visualized with the PyMOL Molecular Graphics System (Schrödinger). Boxplots were generated using R version 3.5.1 (http://www.r-project.org). Protein modeling was performed with AlphaFold-Multimer v2.1.0 (62), as implemented in Google Colab, without the use of homologous structures as templates.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
We thank Till Ringel, Jaspara Knopp, and Leonie Tomm for their support during the early phases of this work and Maria Billini for constructing strains. This work was supported by the German Research Foundation (DFG) (TRR 174 – project 269423233 to G.B., P.S., and M.T. and DFG Core Facility for Interactions, Dynamics and Macromolecular Assembly – project 324652314 to G.B.) and the Max Planck Society (Max Planck Fellowship; to M.T.). L.C.-G. was supported by the Horizon 2020 Research and Innovation Program of the European Commission (Marie Sklodowska-Curie grant, agreement no. 659174 to L.C-G.).
Author contributions
L.C.-G. and M.T. designed research; L.C.-G., W.S., B.R., J.M., J.R., Y.R., and T.H. performed research; L.C.-G., W.S., B.R., J.M., J.R., Y.R., T.H., G.B., P.S., and M.T. analyzed data; L.C.-G., G.B., P.S., and M.T. acquired funding; G.B., P.S., and M.T. supervised the study; and L.C.-G. and M.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Laura Corrales-Guerrero, Email: laucorge@us.es.
Martin Thanbichler, Email: thanbichler@uni-marburg.de.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Du S., Lutkenhaus J., At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 27, 781–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagstaff J., Löwe J., Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol. 16, 187–201 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Du S., Lutkenhaus J., Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Errington J., Wu L. J., Cell cycle machinery in Bacillus subtilis. Subcell. Biochem. 84, 67–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goley E. D., et al. , Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80, 1680–1698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer P., Crossley R., Rothfield L., The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254–256 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee A., Dai K., Lutkenhaus J., Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. U.S.A. 90, 1053–1057 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.RayChaudhuri D., Park J. T., Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Buske P. J., Levin P. A., Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrales-Guerrero L., et al. , FtsZ of filamentous, heterocyst-forming cyanobacteria has a conserved N-terminal peptide required for normal FtsZ polymerization and cell division. Front. Microbiol. 9, 2260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X., et al. , Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol. 179, 6788–6797 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundararajan K., et al. , The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 6, 7281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buske P. J., Levin P. A., A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohan M. C., Eddelbuettel A. M. P., Levin P. A., Pappu R. V., Dissecting the functional contributions of the intrinsically disordered C-terminal tail of Bacillus subtilis FtsZ. J. Mol. Biol. 432, 3205–3221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C., Reedy M., Erickson H. P., Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182, 164–170 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y., Erickson H. P., Conformational changes of FtsZ reported by tryptophan mutants. Biochemistry 50, 4675–4684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz J. F., et al. , Activation of cell division protein FtsZ. Control of switch loop T3 conformation by the nucleotide gamma-phosphate. J. Biol. Chem. 276, 17307–17315 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Alushin G. M., et al. , High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157, 1117–1129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino F., et al. , Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 25, 528–537 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fujita J., et al. , Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus. J. Struct. Biol. 198, 65–73 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Matsui T., Han X., Yu J., Yao M., Tanaka I., Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 289, 3501–3509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Aportela E., Lopez-Blanco J. R., Andreu J. M., Chacon P., Understanding nucleotide-regulated FtsZ filament dynamics and the monomer assembly switch with large-scale atomistic simulations. Biophys. J. 107, 2164–2176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natarajan K., Senapati S., Probing the conformational flexibility of monomeric FtsZ in GTP-bound, GDP-bound, and nucleotide-free states. Biochemistry 52, 3543–3551 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Loose M., Mitchison T. J., The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez-Diaz D. A., et al. , Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 16, e2004845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisson-Filho A. W., et al. , Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., et al. , GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McQuillen R., Xiao J., Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 49, 309–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagstaff J. M., et al. , A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio 8, e00254-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du S., Pichoff S., Kruse K., Lutkenhaus J., FtsZ filaments have the opposite kinetic polarity of microtubules. Proc. Natl. Acad. Sci. U.S.A. 115, 10768–10773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goley E. D., Dye N. A., Werner J. N., Gitai Z., Shapiro L., Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gueiros-Filho F. J., Losick R., A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher D., et al. , The PomXYZ proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev. Cell 41, 299–314 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Gündoğdu M. E., et al. , Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 30, 617–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos-Leon F., et al. , A conserved cell division protein directly regulates FtsZ dynamics in filamentous and unicellular actinobacteria. Elife 10, e63387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z., Mukherjee A., Pichoff S., Lutkenhaus J., The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. U.S.A. 96, 14819–14824 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonthat N. K., et al. , Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 30, 154–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanbichler M., Shapiro L., MipZ, A spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126, 147–162 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Adams D. W., Wu L. J., Errington J., Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 34, 491–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill N. S., Buske P. J., Shi Y., Levin P. A., A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 9, e1003663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trusca D., Scott S., Thompson C., Bramhill D., Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 180, 3946–3953 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weart R. B., et al. , A metabolic sensor governing cell size in bacteria. Cell 130, 335–347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dajkovic A., Mukherjee A., Lutkenhaus J., Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J. Bacteriol. 190, 2513–2526 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Milam S. L., Erickson H. P., SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51, 3100–3109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bisson-Filho A. W., et al. , FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc. Natl. Acad. Sci. U.S.A. 112, E2130–2138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park K. T., Dajkovic A., Wissel M., Du S., Lutkenhaus J., MinC and FtsZ mutant analysis provides insight into MinC/MinD-mediated Z ring disassembly. J. Biol. Chem. 293, 5834–5846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akhmanova A., Steinmetz M. O., Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Walczak C. E., Gayek S., Ohi R., Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 29, 417–441 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Toro-Nahuelpan M., et al. , A gradient-forming MipZ protein mediating the control of cell division in the magnetotactic bacterium Magnetospirillum gryphiswaldense. Mol. Microbiol. 112, 1423–1439 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Kiekebusch D., Michie K. A., Essen L. O., Löwe J., Thanbichler M., Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol. Cell 46, 245–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiekebusch D., Thanbichler M., Spatiotemporal organization of microbial cells by protein concentration gradients. Trends Microbiol. 22, 65–73 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Corrales-Guerrero L., et al. , Molecular architecture of the DNA-binding sites of the P-loop ATPases MipZ and ParA from Caulobacter crescentus. Nucleic Acids Res. 48, 4769–4779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdiche Y., Malashock D., Pinkerton A., Pons J., Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 377, 209–217 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Rigler R., Mets Ü., Widengren J., Kask P., Fluorescence correlation spectroscopy with high count rate and low background: Analysis of translational diffusion. Eur. Biophys. J. Biophys. Lett. 22, 169–175 (1993). [Google Scholar]
- 56.Scheffers D. J., de Wit J. G., den Blaauwen T., Driessen A. J., GTP hydrolysis of cell division protein FtsZ: Evidence that the active site is formed by the association of monomers. Biochemistry 41, 521–529 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Arumugam S., Petrasek Z., Schwille P., MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl. Acad. Sci. U.S.A. 111, E1192–1200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konermann L., Pan J., Liu Y. H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Lu C., Stricker J., Erickson H. P., Site-specific mutations of FtsZ – effects on GTPase and in vitro assembly. BMC Microbiol. 1, 7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan F., et al. , Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. Elife 7, e35578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du S., Lutkenhaus J., SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet. 10, e1004460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2022). 10.1101/2021.10.04.463034 (Accessed 20 November 2022). [DOI]
- 63.Monterroso B., et al. , Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 20, e45946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Araujo-Bazan L., Huecas S., Valle J., Andreu D., Andreu J. M., Synthetic developmental regulator MciZ targets FtsZ across Bacillus species and inhibits bacterial division. Mol. Microbiol. 111, 965–980 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Raskin D. M., de Boer P. A., Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 4971–4976 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Handler A. A., Lim J. E., Losick R., Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol. 68, 588–599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W., et al. , Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat. Commun. 8, 70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang D., et al. , Motility and microtubule depolymerization mechanisms of the kinesin-8 motor, KIF19A. Elife 5, e18101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marblestone J. G., et al. , Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 15, 182–189 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramm B., Glock P., Schwille P., In vitro reconstitution of self-organizing protein patterns on supported lipid bilayers. J. Vis. Exp. 137, e58139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingerman E., Nunnari J., A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 404, 611–619 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.