Abstract
The term “alkaliphile” is used for microorganisms that grow optimally or very well at pH values above 9 but cannot grow or grow only slowly at the near-neutral pH value of 6.5. Alkaliphiles include prokaryotes, eukaryotes, and archaea. Many different taxa are represented among the alkaliphiles, and some of these have been proposed as new taxa. Alkaliphiles can be isolated from normal environments such as garden soil, although viable counts of alkaliphiles are higher in samples from alkaline environments. The cell surface may play a key role in keeping the intracellular pH value in the range between 7 and 8.5, allowing alkaliphiles to thrive in alkaline environments, although adaptation mechanisms have not yet been clarified. Alkaliphiles have made a great impact in industrial applications. Biological detergents contain alkaline enzymes, such as alkaline cellulases and/or alkaline proteases, that have been produced from alkaliphiles. The current proportion of total world enzyme production destined for the laundry detergent market exceeds 60%. Another important application is the industrial production of cyclodextrin by alkaline cyclomaltodextrin glucanotransferase. This enzyme has reduced the production cost and paved the way for cyclodextrin use in large quantities in foodstuffs, chemicals, and pharmaceuticals. It has also been reported that alkali-treated wood pulp could be biologically bleached by xylanases produced by alkaliphiles. Other applications of various aspects of alkaliphiles are also discussed.
DEFINITION OF ALKALIPHILES
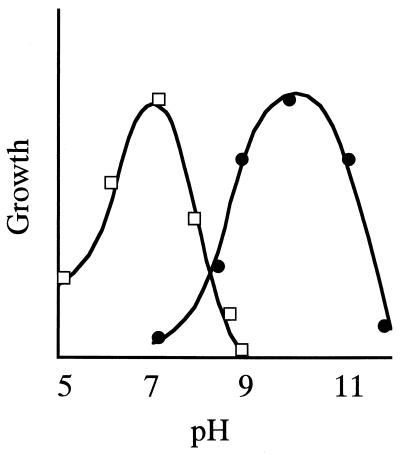
There are no precise definitions of what characterizes an alkaliphilic or alkalitolerant organism. Several microorganisms exhibit more than one pH optimum for growth depending on the growth conditions, particularly nutrients, metal ions, and temperature. In this review, therefore, the term “alkaliphile” is used for microorganisms that grow optimally or very well at pH values above 9, often between 10 and 12, but cannot grow or grow only slowly at the near-neutral pH value of 6.5 (Fig. 1).
FIG. 1.
pH dependency of alkaliphilic microorganisms. The typical pH dependency of the growth of neutrophilic and alkaliphilic bacteria is shown by open squares and solid circles, respectively.
HISTORY OF ALKALIPHILES
The discovery of alkaliphiles was fairly recent. Only 16 scientific papers on the topic could be found when I started experiments on alkaliphilic bacteria in 1968. The use of alkaliphilic microorganisms has a long history in Japan, since from ancient times indigo has been naturally reduced under alkaline conditions in the presence of sodium carbonate. Indigo from indigo leaves is reduced by particular bacteria that grow under these highly alkaline conditions in a traditional process called indigo fermentation. The most important factor in this process is the control of the pH value. Formerly, indigo reduction was controlled only by the skill of the craftsman. Microbiological studies of the process, however, were not conducted until the rediscovery of these alkaliphiles by me (69). Alkaliphiles remained little more than interesting biological curiosities, and at that time no further industrial application was attempted or even contemplated.
Since then, my colleagues and I have isolated a large number of alkaliphilic microorganisms and purified many alkaline enzymes. The first paper concerning an alkaline protease was published in 1971 (67). Over the past two decades, our studies have focused on the enzymology, physiology, ecology, taxonomy, molecular biology, and genetics of these isolates to establish a new microbiology of alkaliphilic microorganisms. Industrial applications of these microorganisms have also been investigated extensively, and some enzymes, such as alkaline proteases, alkaline amylases, and alkaline cellulases, have been put to use on an industrial scale. Alkaliphiles have clearly gained large amounts of genetic information by evolutionary processes and exhibit an ability in their genes to cope with particular environments; therefore their genes are a potentially valuable source of information waiting to be explored and exploited by the biotechnologists. Genes responsible for the alkaliphily of Bacillus halodurans C-125 and Bacillus firmus OF4 have been analyzed (49, 181). Recently, the complete genome of B. halodurans C-125 has been sequenced to obtain more information on industrial applications and on the molecular basis of alkaliphily. However, in this review, structural, physiological, and genetic aspects of alkaliphiles are not discussed; instead, the review focuses on studies of extracellular alkaline enzymes.
DISTRIBUTION AND ISOLATION OF ALKALIPHILES
Alkaliphiles consist of two main physiological groups of microorganisms; alkaliphiles and haloalkaliphiles. Alkaliphiles require an alkaline pH of 9 or more for their growth and have an optimal growth pH of around 10, whereas haloalkaliphiles require both an alkaline pH (>pH 9) and high salinity (up to 33% [wt/vol] NaCl). Alkaliphiles have been isolated mainly from neutral environments, sometimes even from acidic soil samples and feces. Haloalkaliphiles have been mainly found in extremely alkaline saline environments, such as the Rift Valley lakes of East Africa and the western soda lakes of the United States.
Alkaliphiles
Aerobic alkaliphiles.
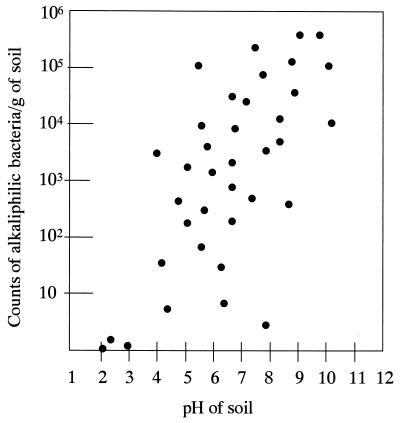
Alkaliphilic microorganisms coexist with neutrophilic microorganisms, as well as occupying specific extreme environments in nature. Figure 2 illustrates the relationship between the occurrence of alkaliphilic microorganisms and the pH of the sample origin. To isolate alkaliphiles, alkaline media must be used. Sodium carbonate is generally used to adjust the pH to around 10, because alkaliphiles usually require at least some sodium ions. Table 1 shows the makeup of an alkaline medium suitable for their isolation. The frequency of alkaliphilic microorganisms in neutral “ordinary” soil samples is 102 to 105/g of soil, which corresponds to 1/10 to 1/100 of the population of the neutrophilic microorganisms (65). Recent studies show that alkaliphilic bacteria have also been found in deep-sea sediments collected from depths up to the 10,898 m of the Mariana Trench (180). Many different kinds of alkaliphilic microorganisms, including bacteria belonging to the genera Bacillus, Micrococcus, Pseudomonas, and Streptomyces and eukaryotes such as yeasts and filamentous fungi, have been isolated from a variety of environments (30, 50, 65).
FIG. 2.
Distribution of alkaliphilic microorganisms in environments at various pHs.
TABLE 1.
Basal media for alkaliphilic microorganisms
| Ingredient | Amt (g/liter) in:
|
|
|---|---|---|
| Horikoshi-I | Horikoshi-II | |
| Glucose | 10 | 0 |
| Soluble starch | 0 | 10 |
| Yeast extract | 5 | 5 |
| Polypeptone | 5 | 5 |
| K2HPO4 | 1 | 1 |
| Mg2SO4 · 7H2O | 0.2 | 0.2 |
| Na2CO3 | 10 | 10 |
| Agar | 20 | 20 |
Anaerobic alkaliphiles.
The first brief report on anaerobic alkaliphiles was communicated by Niimura et al. (144), although no taxonomic details were reported. Subsequently, many anaerobic spore-forming alkaliphiles were isolated by conventional procedures, and a few applications have been studied. Podkovyrov and Zeikus recorded the isolation and purification of a cyclomaltodextrin glucanotransferase from Clostridium thermohydrosulfuricum 39E (155). Wiegel and colleagues (32, 117) have isolated and characterized a range of thermophilic anaerobic alkaliphiles. Nine moderately alkalitotolerant thermophilic bacteria with similar properties were isolated from water and soil samples obtained from Yellowstone National Park. Strain JW/YL-138T and eight similar strains represent a new genus and species, Anaerobranca horikoshii. At 60°C the pH range for growth is 6.9 to 10.3 and the optimum pH is 8.5. Cook et al. reported that Clostridium paradoxum, a novel alkali-thermophile, could maintain pH homeostasis inside the cell and thrive well in culture media in the pH range between 7.6 and 9.8 (23).
Kodama and Koyama (113) studied unique characteristics of anaerobic alkaliphiles belonging to Amphibacillus xylanus. The transport of leucine and glucose was remarkably stimulated by NH4+. An (NH4+ + Na+)-activated ATPase of A. xylanus was also reported by Takeuchi et al. (183). Primary amines also activated the enzyme, but secondary and tertiary amines were ineffective (183).
Until recently, few microbiologists thought that alkaliphiles thrived in hydrothermal areas. Stetter’s group (184) isolated Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeon growing on polysulfide at alkaline pH and at temperatures between 56 and 90°C, from a shallow marine hydrothermal system at Vulcano Island, Italy. The pH range for growth was 6.5 to 10.5, with an optimum around 9.0. Polysulfide and elemental sulfur were reduced to H2S. Then, from a shallow beach (depth 1.0 m) situated at the base of the reef close to Porto di Levante at Vulcano Island, Italy, Dirmeier et al. isolated a new hyperthermophilic member of the archaea (29). The cells are coccoid and have up to five flagella. They grow between 56 and 93°C (optimum, 85°C) and at pH 5.0 to 9.5 (optimum, 9.0). The organism is strictly anaerobic and grows heterotrophically on defined amino acids and complex organic substrates such as Casamino Acids, yeast extract, peptone, meat extract, tryptone, and casein. DNA-DNA hybridization and 16S rRNA partial sequences indicated that the new isolate belongs to the genus Thermococcus and represents a new species, Thermococcus acidaminovorans. An outstanding review on anaerobic alkalithermophiles has recently been published by Wiegel (198).
Haloalkaliphiles
The most remarkable examples of naturally occurring alkaline environments are soda deserts and soda lakes. Extremely alkaline lakes, for example, Lake Magadi in Kenya and the Wadi Natrun in Egypt, are probably the most stable highly alkaline environments on Earth, with a consistent pH of 10.5 to 12.0 depending on the site. Many organisms isolated from alkaline and highly saline environments such as soda lakes also require high salinity, which is achieved by adding NaCl to the isolation medium. In particular, hypersaline soda lakes are populated by alkaliphilic representatives of halophilic archaea. These red-pigmented microbes reach numbers of 107 to 108/ml in soda lake brines. A novel haloalkaliphilic archaeon was isolated from Lake Magadi (125). Cells of this organism contain large gas vacuoles in the stationary phase of growth, and colonies produced by these archaea are bright pink. The G+C content of the DNA is 62.7 mol%. The name Natronobacterium vacuolata sp. nov. is proposed. The type strain is designated NCIMB 13189. Lodwick et al. (121) determined the nucleotide sequence of the 23S and 5S rRNA genes from the haloalkaliphilic arachaeon Natronobacterium magadii. A phylogenetic tree for the halobacteria was produced by alignment of the 23S rRNA sequence with those of other archaea.
Subsequently, Kamekura et al. (81) studied the diversity of alkaliphilic halobacteria on the basis of phylogenetic tree reconstructions, signature bases specific for individual genera, and sequences of spacer regions between 16S and 23S rRNA genes. They proposed the following changes: Natronobacterium pharaonis to be transferred to Natronomonas gen. nov. as Natronomonas pharaonis gen. nov. comb. nov.; Natronobacterium vacaolatum to be transferred to the genus Halorubrum as Halorubrum vacuolatum comb. nov.; and Natronobacterium magadii to be transferred to the genus Natrialba as Natrialba magadii.
Recently, Xu et al. (200) isolated two haloalkaliphilic archaea from a soda lake in Tibet. The two strains were gram negative, pleomorphic, flat, nonmotile, and strictly aerobic. Growth required at least 12% NaCl and occurred between pH 8.0 and 11 with an optimum at pH 9.0 to 9.5. On 16S rRNA phylogenetic trees, the two strains formed a monophyletic cluster. They differed from their closest neighbors, Halobacterium trapanicum and Natrialba asiatica, in polar lipid composition, as well as physiological and phenotypic characteristics. DNA-DNA hybridization indicated that the two strains belonged to different species of the same genus. The results indicated that the strains should be classified in a new genus, Natronorubrum gen. nov.
Recently, Kevbrin et al. (89) isolated an alkaliphilic, obligately anaerobic, fermentative, asporogenous bacterium with a gram-positive cell wall structure from soda deposits in Lake Magadi, Kenya. 16S rDNA sequence analysis of this bacterium showed that it belongs phylogenetically to cluster XI of the low-G+C gram-positive bacteria. On the basis of its distinct phylogenetic position and unique physiological properties, they proposed a new genus and new species, Tindallia magadii, for this strain.
A novel osmolyte, 2-sulfotrehalose, was discovered in several Natronobacterium species of haloalkaliphilic archaea (26). The concentration of this novel disaccharide increased with increasing concentrations of external NaCl, behavior consistent with its identity as an osmolyte. Other common osmolytes (glycine, betaine, glutamate, and proline) were not accumulated or used for osmotic balance in place of the sulfotrehalose by these halophilic archaea. The ecology, physiology, and taxonomy of haloalkaliphiles have been recently reviewed by Jones et al. (80).
Methanogens
Boone et al. (16) had originally isolated halophilic methanogens from various neutral salinas and natural hypersaline environments. These strains grew fastest at temperatures near 40°C and, in medium containing 0.5 to 2.5 M NaCl, at pH values near 7. Zhilina and Zavarzin (206) then described bacterial communities in alkaline lakes, and in particular the diversity of anaerobic bacteria developing at pH 10 was discussed. A new obligate alkaliphilic, methylotrophic methanogen was isolated from Lake Magadi. According to its phenotypic and genotypic properties, the isolate belonged to Methanosalsus (Methanohalophilus) zhilinaeae (88). It exhibited 91% homology to the type strain of this formerly monotypic species. The strain did not require Cl− but was obligately dependent on Na+ and HCO3−. It was an obligate alkaliphile and grew within a pH range of 8 to 10 (92).
Cyanobacteria
Gerasimenko et al. (48) reported the discovery a wide diversity of alkaliphilic cyanobacteria (16 genera and 34 species were found). So far, no industrial application has been reported (11, 31, 128, 156, 199). Buck and Smith (19) reported an Na+/H+ electrogenic antiporter in alkaliphilic Synechocystis sp. Singh (165) partially purified a urease from an alkaliphilic diazotrophic cyanobacterium, Nostoc calcicola. The purified enzyme showed optimum activity at pH 7.5 and 40°C. The enzyme was found to be sensitive to metal cations, particularly Hg2+, Ag+, and Cu2+. 4-Hydroxymercuribenzoate (a mercapto-group inhibitor) and acetohydroxamic acid (a chelating agent of nickel) inhibited the enzyme activity completely. These results suggest the involvement of an SH group and Ni2+ in the activity of urease from N. calcicola.
Other Alkaliphiles
Sorokin et al. have extensively studied sulfur-oxidizing alkaliphiles. The strains oxidize sulfide, elemental sulfur, and thiosulfate to tetrathionate. Recently, sulfide dehydrogenase was isolated and purified from these alkaliphilic chemolithoautotrophic sulfur-oxidizing bacteria (166–169).
Zhilina et al. isolated and characterized Desulfonatronovibrio hydyogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium, from Lake Magadi. Strain Z-7935(T) is an obligatory sodium-dependent alkaliphile, which grows in sodium carbonate medium and does not grow at pH 7; the maximum pH for growth is more than pH 10, and the optimum pH is 9.5 to 9.7. The optimum NaCl concentration for growth is only 3% (wt/vol) (206–208).
Several alkaliphilic spirochetes were also isolated from Lake Magadi and from Lake Khatyn, Central Asia, by Zhilina et al. Analysis of the genes encoding 16S rRNA indicated a possible fanning out of the phylogenetic tree of spirochetes (207).
Khmelenina et al. (91) isolated two strains of the halotolerant alkaliphilic obligate methanotrophic bacterium Methylobacter alcaliphilus sp. nov. from moderately saline soda lakes in Tuva (Central Asia). The strains grow fastest at pH 9.0 to 9.5 and much more slowly at pH 7.0. No growth occurred at pH less than or equal to 6.8. They require NaHCO3 or NaCl for growth in alkaline medium.
SPECIAL PHYSIOLOGICAL FEATURES OF ALKALIPHILES
Internal pH
Most alkaliphiles have an optimal growth pH at around 10, which is the most significant difference from well-investigated neutrophilic microorganisms. Therefore, the question arises how these alkaliphilic microorganisms can grow in such an extreme environment. Is there any difference in physiological and structural aspects between alkaliphilic and neutrophilic microorganisms? Internal cytoplasmic pH can be estimated from the optimal pH of intracellular enzymes. For example, α-galactosidase from an alkaliphile, Micrococcus sp. strain 31-2, had its optimal catalytic pH at 7.5, suggesting that the internal pH is around neutral. Furthermore, the cell-free protein synthesis systems from alkaliphiles optimally incorporate amino acids into protein at pH 8.2 to 8.5, only 0.5 pH unit higher than that of the neutrophilic Bacillus subtilis (69).
Another method to estimate internal pH is to measure in cells the inside and outside distribution of weak bases, which are not actively transported by cells. The internal pH was maintained at around 8, despite a high external pH of 8 to 11, as shown in Table 2 (5, 51, 65, 171). Therefore, one of the key features in alkaliphily is associated with the cell surface, which discriminates and maintains the intracellular neutral environment separate from the extracellular alkaline environment.
TABLE 2.
Intracellular pH values in alkaliphilic Bacillus strains at different external pH values
| Microorganism | External pH | Internal pH |
|---|---|---|
| B. alcalophilus | 8.0 | 8.0 |
| 9.0 | 7.6 | |
| 10.0 | 8.6 | |
| 11.0 | 9.2 | |
| B. firmus | 7.0 | 7.7 |
| 9.0 | 8.0 | |
| 10.8 | 8.3 | |
| 11.2 | 8.9 | |
| 11.4 | 9.6 | |
| Bacillus strain YN-2000 | 7.5 | 8.5 |
| 8.5 | 7.9 | |
| 9.5 | 8.1 | |
| 10.2 | 8.4 | |
| B. halodurans C-125 | 7.0 | 7.3 |
| Intact cells | 7.5 | 7.4 |
| 8.0 | 7.6 | |
| 8.5 | 7.8 | |
| 9.0 | 7.9 | |
| 9.5 | 8.1 | |
| 10.0 | 8.2 | |
| 10.5 | 8.4 | |
| Protoplasts | 7.0 | 7.5 |
| 8.0 | 7.9 | |
| 8.5 | 8.2 | |
| 9.0 | 8.4 | |
| 9.3 | 8.6 |
Cell Walls
Acidic polymers.
Since the protoplasts of alkaliphilic Bacillus strains lose their stability in alkaline environments, it has been suggested that the cell wall may play a key role in protecting the cell from alkaline environments. Components of the cell walls of several alkaliphilic Bacillus spp. have been investigated in comparison with those of the neutrophilic B. subtilis. In addition to peptidoglycan, alkaliphilic Bacillus spp. contain certain acidic polymers, such as galacturonic acid, gluconic acid, glutamic acid, aspartic acid, and phosphoric acid (4). The negative charges on the acidic nonpeptidoglycan components may give the cell surface its ability to adsorb sodium and hydronium ions and repulse hydroxide ions and, as a consequence, may assist cells to grow in alkaline environments.
Peptidoglycan.
The peptidoglycans of alkaliphilic Bacillus spp. appear to be similar to that of B. subtilis. However, their composition was characterized by an excess of hexosamines and amino acids in the cell walls compared to that of the neutrophilic B. subtilis. Glucosamine, muramic acid, d- and l-alanine, d-glutamic acid, meso-diaminopimelic acid, and acetic acid were found in hydrolysates. Although some variation in the amide content among the peptidoglycans from alkaliphilic Bacillus strains was found, the variation in pattern was similar to that known in neutrophilic Bacillus species.
Na+ Ions and Membrane Transport
Alkaliphilic microorganisms grow vigorously at pH 9 to 11 and require Na+ for growth (116). The presence of sodium ions in the surrounding environment has proved to be essential for effective solute transport through the membranes of alkaliphilic Bacillus spp. According to the chemiosmotic theory, the proton motive force in the cells is generated by the electron transport chain or by excreted H+ derived from ATP metabolism by ATPase. H+ is then reincorporated into the cells with cotransport of various substrates. In Na+-dependent transport systems, the H+ is exchanged with Na+ by Na+/H+ antiporter systems, thus generating a sodium motive force, which drives substrates accompanied by Na+ into the cells. The incorporation of a test substrate, α-aminoisobutyrate, increased twofold as the external pH shifted from 7 to 9, and the presence of sodium ions significantly enhanced the incorporation; 0.2 N NaCl produced an optimum incorporation rate that was 20 times the rate observed in the absence of NaCl. Other cations, including K+, Li+, NH4+, Cs+, and Rb+, showed no effect, nor did their counteranions (108).
Mechanisms of Cytoplasmic pH Regulation
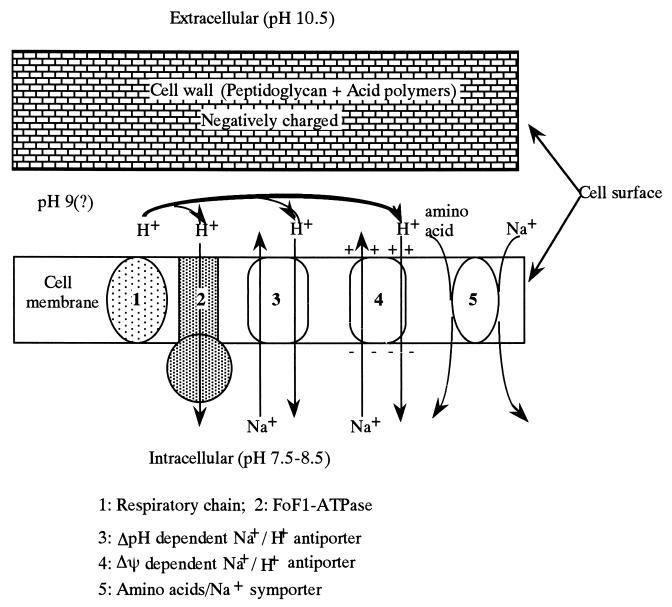
The cells have two barriers to reduce pH values from 10.5 to 8 (Fig. 3).
FIG. 3.
Schematic representation of cytoplasmic pH regulation.
Cell walls containing acidic polymers function as a negatively charged matrix and may reduce the pH value at the cell surface. The surface of the plasma membrane must presumably be kept below pH 9, because the plasma membrane is very unstable at alkaline pH values (pH 8.5 to 9.0) much below the pH optimum for growth.
Plasma membranes may also maintain pH homeostasis by using the Na+/H+ antiporter system (Δψ dependent and ΔpH dependent), the K+/H+ antiporter, and ATPase-driven H+ expulsion. Recent studies on the critical antiporters in several laboratories have begun to clarify the number and characteristics of the porters that support active mechanisms of pH homeostasis (53, 76, 109, 114, 130).
My group isolated a nonalkaliphilic mutant strain (mutant 38154) from B. halodurans C-125 as the host for cloning genes related to alkaliphily. A 3.7-kb parental DNA fragment (pALK fragment) from the parental strain restored the growth of mutant 38154 at alkaline pH. The transformant was able to maintain an intracellular pH that was lower than the external pH and contained an electrogenic Na+/H+ antiporter driven only by Δψ (membrane potential, interior negative). Membrane vesicles prepared from mutant 38154 did not show membrane potential Δψ-driven Na+/H+ antiporter activity. These results indicate that mutant 38154 affects, either directly or indirectly, electrogenic Na+/H+ antiporter activity. This was the first report of a DNA fragment responsible for a Na+/H+ antiporter system in the mechanism of alkaliphily (53, 56, 107, 115, 162).
Krulwich et al. (49, 51, 76, 114, 171, 172) have focused their studies on the facultative alkaliphile Bacillus firmus OF4, which is routinely grown on malate-containing medium at either pH 7.5 or 10.5. Current work is directed toward clarification of the characteristics and energetics of membrane-associated proteins that must catalyze inward proton movements. One such protein is the Na+/H+ antiporter, which enables cells to adapt to a sudden upward shift in pH and to maintain a cytoplasmic pH that is 2 to 2.3 units below the external pH in the most alkaline range of pH for growth. Another is the proton-translocating ATP synthase that catalyzes production of ATP under conditions in which the external proton concentration and the bulk chemiosmotic driving force are low. Three gene loci that are candidates for Na+/H+ antiporter, carrying genes with roles in Na+-dependent pH homeostasis, have been identified. All of them have homologs in B. subtilis, in which pH homeostasis can be carried out with either K+ or Na+. The physiological importance of one of the B. firmus OF4 loci, nhaC, has been studied by targeted gene disruption, and the same approach is being extended to the others. The atp genes, which encode the F1F0-ATP synthase of the alkaliphile have interesting motifs in areas of putative importance for proton translocation. As an initial step in studies that will probe the importance and possible role of these motifs, the entire atp operon from B. firmus OF4 has been cloned and functionally expressed in an Escherichia coli mutant with its atp genes completely deleted. The transformant does not exhibit growth on succinate but shows reproducible, modest increases in the aerobic growth yields on glucose as well as membrane ATPase activity that exhibits characteristics of the alkaliphile enzyme (114). As a result of these mechanisms, the membrane proteins play a key role in keeping the intracellular pH values in the range between 7.0 and 8.5.
PHYSICAL MAPS OF CHROMOSOMAL DNAS OF ALKALIPHILIC BACILLUS STRAINS
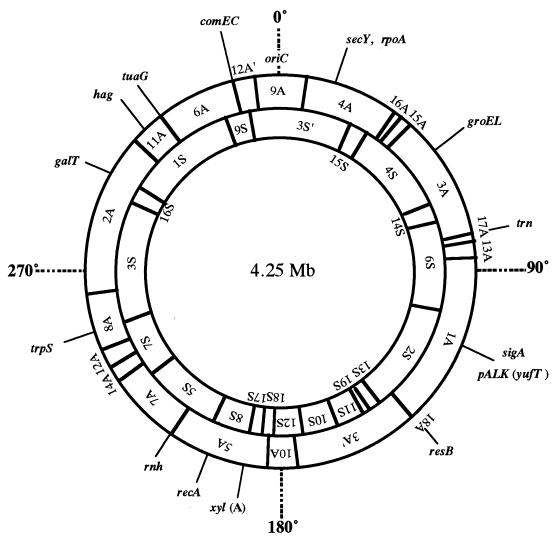
How alkaliphiles adapt to their alkaline environments is one of the most interesting and challenging topics that might be clarified by genome analysis. Two physical maps for alkaliphilic Bacillus strains have been published: those for B. firmus OF4 and B. halodurans C-125. A physical map of B. firmus OF4 is consistent with a circular chromosome of approximately 4 Mb, with an extrachromosomal element of 110 kb (49, 172). Although analysis is still in progress, several open reading frames for Na+/H+ antiporters that may play roles in pH homeostasis have been detected. A physical map of the chromosome of B. halodurans C-125 has been constructed to facilitate whole-genome analysis (181, 182). In this strain, the genome size is 4.25 Mb. Many open reading frames showing significant similarities to those of B. subtilis have been positioned on the physical map. As shown in Fig. 4, the oriC regions of the C-125 chromosome have been identified by Southern blot analysis with a DNA probe containing the gyrB region (181, 182).
FIG. 4.
Physical and genetic map of the chromosome of B. halodurans C-125. The outer and inner circles shows the AscI and Sse83871 physical maps, respectively. The locations of several genes are indicated on the maps. Dashed lines indicate the approximate positions of the genes.
Whole-genome analysis will inevitably give us useful information about adaptation to alkaline environments.
ALKALINE ENZYMES
Studies of alkaliphiles have led to the discovery of many types of enzymes that exhibit interesting properties. The first report concerning an alkaline enzyme, published in 1971, described an alkaline protease produced by Bacillus sp. strain 221. More than 35 new enzymes have been isolated and purified in my laboratory. Some of these have been produced on an industrial scale.
Alkaline Proteases
In 1971, Horikoshi (66) reported the production of an extracellular alkaline serine protease from alkaliphilic Bacillus sp. strain 221. This strain, isolated from soil, produced large amounts of alkaline protease that differed from the subtilisin group. The optimum pH of the purified enzyme was 11.5, and 75% of the activity was maintained at pH 13.0. The enzyme was completely inhibited by diisopropylfluorophosphate or 6 M urea but not by EDTA or p-chloromercuribenzoate. The molecular weight of the enzyme was 30,000, which is slightly higher than those of other alkaline proteases. The addition of a 5 mM solution of calcium ions was reflected in a 70% increase in activity at the optimum temperature (60°C). Subsequently, two Bacillus strains, AB42 and PB12, were reported which also produced an alkaline protease (9). These strains exhibited a broad pH range (pH 9.0 to 12.0), with a temperature optimum of 60°C for AB42 and 50°C for PB12. Since these reports, many alkaline proteases have been isolated from alkaliphilic microorganisms (35, 146, 187–190). Fujiwara et al. (34) purified a thermostable alkaline protease from a thermophilic alkaliphile, Bacillus sp. strain B18. The optimum pH and temperature for the hydrolysis of casein were pH 12 to 13 and 85°C, both of which are higher than those for alkaline proteases. Han and Damodaran (55) reported the purification and characterization of an extracellular endopeptidase from a strain of Bacillus pumilus displaying high stability in 10% (wt/vol) sodium dodecyl sulfate and 8 M urea. Some of the enzymes are now commercially available as detergent additives.
Takami et al. (179) isolated a new alkaline protease from alkaliphilic Bacillus sp. strain AH-101. The enzyme was most active toward casein at pH 12 to 13 and stable for 10 min at 60°C and pH 5 to 13. The optimum temperature was about 80°C in the presence of 5 mM calcium ions. The alkaline protease showed a higher hydrolyzing activity against insoluble fibrous natural proteins such as elastin and keratin than did subtilisins and proteinase K (177, 178). Cheng et al. (21) reported a keratinase of a feather-degrading Bacillus licheniformis strain, PWD-1. This enzyme was stable from pH 5 to 12. The optimal reaction pHs for feather powder and casein were 8.5 and 10.5 to 11.5, respectively. Zaghloul et al. also reported the isolation, identification, and keratinolytic activity of several feather-degrading bacteria isolated from Egyptian soil. These isolates could degrade chicken feathers (205).
Structural analysis.
Schmidt et al. analyzed a series of serine protease genes of alkaliphilic Bacillus sp. strain LG12. Four tandem subtilisin-like protease genes were found on a 6,854-bp DNA fragment. The two downstream genes (sprC and sprD) appear to be transcribed independently, while the two upstream genes (sprA and sprB) seem to be part of the same transcript (161). Yeh et al. improved the translational efficiency of an alkaline protease YaB gene with different initiation codons in B. subtilis and alkaliphilic Bacillus YaB (201). Martin et al. found that the solution structure of serine protease PB92 from B. alcalophilus presents a well-defined global fold with a flexible structure-binding site. The well-defined global fold was rigid with the exception of a restricted number of sites (123).
Kobayashi et al. isolated and purified an alkaline protease from alkaliphilic Bacillus sp. strain KSM-K16 that was suitable for use in detergents (110). Shirai et al. studied the crystal structure of the M protease of alkaliphilic Bacillus sp. strain KSM-K16 by X-ray analysis to understand the alkaline adaptation mechanism of the enzyme (164). This analysis revealed a decrease in the number of negatively charged amino acids (aspartic acid and glutamic acid) and lysine residues, and an increase in arginine and neutral hydrophilic amino acids (histidine, asparagine, and glutamine) residues during the course of adaptation.
Other proteases.
Several alkaline proteases produced by alkaliphilic actinomycetes have been reported. Tsuchiya et al. (191–193) isolated thermostable alkaline protease from alkaliphilic Thermoactinomyces sp. strain HS682. The strain grew in alkaline media between pH 7.5 and 11.5. Maltose gave the highest productivity of protease at a concentration of 10 g/liter. The protease had its maximum proteolytic activity around pH 11.0 and at 70°C. In the presence of Ca2+ ions, maximum activity was observed at 80°C. An intracellular alkaline serine protease gene of alkaliphilic Thermoactinomyces sp. strain HS682 was cloned and expressed in E. coli. Yum et al. (204) purified an extracellular alkaline serine protease produced by Streptomyces sp. YSA-130. The optimum temperature and pH for the enzyme activity were 60°C and 11.5, respectively. The enzyme was stable at 50°C and between pH 4 and 12.
A serine protease from the keratin-degrading Streptomyces pactum DSM 40530 was purified by casein agarose affinity chromatography by Bockle et al. (15). The proteinase was optimally active in the pH range from 7 to 10 and at temperatures from 40 to 75°C. After incubation with the purified proteinase, rapid disintegration of whole feathers was observed.
Extracellular proteolytic activity was also detected in the haloalkaliphilic archaeon Natronococcus occultus as the culture reached the stationary growth phase (170). Proteolytic activity was precipitated with ethanol and subjected to preliminary characterization. Optimal conditions for activity were attained at 60°C and 1 to 2 M NaCl or KCl. Gelatin zymography in the presence of 4 M betaine revealed a complex pattern of active species with apparent molecular masses ranging from 50 to 120 kDa.
Industrial applications.
(i) Detergent additives.
The main industrial application of alkaliphilic enzymes is in the detergent industry, and detergent enzymes account for approximately 30% of total worldwide enzyme production. Not all of these are produced by alkaliphilic bacteria. However, many alkaline proteases have been produced by alkaliphilic Bacillus strains and are commercially available.
(ii) Dehairing.
Alkaline enzymes have been used in the hide-dehairing process, where dehairing is carried out at pH values between 8 and 10. These enzymes are commercially available.
(iii) Other applications.
An interesting application of alkaline protease was developed by Fujiwara and coworkers (35, 36, 75). They reported the use of an alkaline protease to decompose the gelatinous coating of X-ray films, from which silver was recovered. Protease B18′ had a higher optimum pH and temperature, around 13.0 and 85°C. The enzyme was most active toward gelatin on film at pH 10.
Starch-Degrading Enzymes
The first alkaline amylase was produced in Horikoshi-II medium by cultivating alkaliphilic Bacillus sp. strain A-40-2 (67). Several types of alkaline starch-degrading enzymes were observed. No alkaline amylases produced by neutrophilic microorganisms have so far been reported. Recent studies revealed that the starch-degrading enzymes α-amylase and cyclomaltodextrin glycosyltransferase (CGTase) are functionally and structurally closely related. α-Amylases have three molecular domains (called A, B, and C), and CGTases contain two additional domains (called D and E).
α-Amylases of alkaliphilic Bacillus strains.
The alkaline amylase was first produced in alkaliphilic Bacillus sp. strain A-40-2 (ATCC 21592), which was selected from about 300 colonies of bacteria grown in Horikoshi-II medium (67). The enzyme is most active at pH 10.0 to 10.5 and retains 50% of its activity between pH 9.0 and 11.5. The enzyme is not inhibited by 10 mM EDTA at 30°C but is completely inactivated by 8 M urea. The enzyme can hydrolyze 70% of starch to yield glucose, maltose, and maltotriose, and it is a type of saccharifying α-amylase. Boyer and Ingle reported an alkaline amylase in strain NRRL B-3881, which was the second report of an alkaline amylase (17, 18). The B-3881 amylase has its optimum pH for enzyme action at 9.2. It yields maltose, maltotriose, and a small amount of glucose and maltotetraose, all of which have a β-configuration. Considerable diversity of α-amylases has been reported. Kim et al. reported that the alkaliphilic Bacillus sp. strain GM8901 produced five alkaline amylases in a culture broth (97). McTigue et al. studied the alkaline amylases of three alkaliphilic Bacillus strains (126, 127). Bacillus halodurans A-59 (ATCC 21591), Bacillus sp. strain NCIB 11203, and Bacillus sp. strain IMD370 produced alkaline α-amylases with maxima for activity at pH 10.0. Kelly et al. (86) found that the alkaline amylase of Bacillus sp. strain IMD370 could hydrolyze raw starch. The enzyme digested raw cornstarch to glucose, maltose, maltotriose, and maltotetraose. The maximum pH for raw starch hydrolysis was pH 8.0, compared to pH 10.0 for soluble starch hydrolysis. It is of interest that degradation of raw starch was stimulated sixfold in the presence of β-cyclodextrin. Lin et al. have recently reported the production and properties of a raw-starch-degrading amylase from the thermophilic and alkaliphilic Bacillus sp. strain TS-23 (119). Activity staining revealed that two amylases with molecular masses of 150 and 42 kDa were produced. The 42-kDa amylase hydrolyzed only raw starch. Igarashi et al. isolated a novel liquefying α-amylase (LAMY) from cultures of an alkaliphilic Bacillus isolate, strain KSM-1378 (73). The enzyme had a pH optimum of 8.0 to 8.5 and displayed maximum activity at 55°C. The structural gene for the amylase contained a single open reading frame 1,548 bp in length, encoding to 516 amino acids that included a signal peptide of 31 amino acids. The four conserved regions were found in the deduced amino acid sequence. Essentially, the sequence of LAMY was consistent with the tertiary structures of reported amylolytic enzymes, which are composed of domains A, B, and C. Furthermore, Igarashi et al. improved the thermostability of the amylase by deleting an arginine-glycine residue in that molecule (74).
Other α-amylases.
Kimura and Horikoshi isolated a number of starch-degrading psychrotrophic microorganisms from the environment (102, 104, 106). Recently, a gene coding for a new amylolytic enzyme from Pseudomonas sp. strain KFCC 10818 was cloned, and its nucleotide sequence was determined (131). A deduced amino acid sequence contained four highly conserved regions of α-amylases. The haloalkaliphilic Natronococcus sp. strain Ah-36 produced an extracellular maltotriose-forming amylase (112). The amylase exhibited maximal activity at pH 8.7 and 55°C in the presence of 2.5 M NaCl. Kobayashi et al. have cloned this α-amylase and expressed it in Haloferax volcanii (111).
Cyclomaltodextrin glucanotransferases.
Nakamura and Horikoshi discovered many alkaliphilic Bacillus strains producing CGTases. The crude enzyme of Bacillus sp. strain 38-2 was a mixture of three enzymes: acidic CGTase, having an optimum pH for enzyme action at 4.6, neutral CGTase, having an optimal pH at 7.0, and alkaline CGTase, having an optimal pH at 8.5 (135–138). In 1975, Matsuzawa et al. established the industrial production of cyclodextrin (CD) by using crude CGTase of Bacillus sp. strain 38-2 (124). Since then, many alkaliphilic microorganisms producing CGTases have been reported (145, 147). Georganta et al. isolated CGTase-producing psychrophilic alkaliphilic bacteria from samples of deep-sea bottom mud (47). Isolate 3-22 grew at 4°C and showed activity at broad temperature ranges and from pH 5 to 9. The CGTase produced predominantly β-CD, with minor amounts of α- and γ-CDs. The formation of various CDs was observed after accumulation of maltooligosaccharides with degrees of polymerization more than seven (1). Salva et al. isolated 68 CGTase-producing alkaliphilic bacteria from among 400 soil bacteria (157). The enzyme of isolate 76 was partially purified by starch adsorption, and some of its properties were investigated. These properties seem to be quite similar to those of the Bacillus sp. strain 38-2 enzyme (135). Chung et al. isolated a thermostable CGTase from Bacillus stearothermophilus ET1 (22). The optimum pH for the enzyme-catalyzed reaction was pH 6.0, and the optimum temperature was observed at 80°C. A 13% (wt/vol) cornstarch solution was liquefied and converted to CDs solely by using this enzyme. The cornstarch conversion rate was 44%, and α-, β-, and γ-CDs were produced in the ratio of 4.2:5.9:1. Terada et al. studied the initial reaction of CGTase from the alkaliphilic Bacillus sp. strain A2-5a on amylose (186). Cyclic α-1,4-glucans with degrees of polymerization ranging from 9 to more than 60, in addition to α-, β-, and γ-CD, were detected at early stages of enzymatic reactions. Subsequently, large cyclic α-1,4-glucans were converted into smaller cyclic α-1,4-glucans. The final major product was β-CD. From the industrial point of view, γ-CD is of practical interest because it is rare and can encapsulate larger compound molecules. Recently, Parsiegla et al. reported that mutation of Bacillus circulans 8 was effective in increasing γ-CD production (154). Yamane’s group has extensively investigated the molecular structure of the CGTase of alkaliphilic Bacillus sp. strain 1011 (91–101, 113, 133, 134). Kimura et al. isolated the enzyme, purified and cloned its gene (99, 100). The enzyme, consisting of 686 amino acid residues, was crystallized and subjected to X-ray analysis. The molecule consists of five domains, designated A to E, and its backbone structure is similar to the structure of other bacterial CGTases. The molecule has two calcium-binding sites where calcium ions are coordinated by seven ligands, forming a distorted pentagonal bipyramid. Three histidine residues in the active center of CGTase participate in the stabilization of the transition state. His-327 is especially important for catalysis over the alkaline pH range (133). Analysis of the three-dimensional structures of CGTases has revealed that four aromatic residues, which are highly conserved among CGTases but not in α-amylases, are located in the active center (134).
Industrial production of cyclodextrins.
In 1969, Corn Products International Co. in the United States began producing β-CD by using B. macerans CGTase. Teijin Ltd. in Japan also produced β-CD by using the B. macerans enzyme in a pilot plant. However, there were two serious problems in both production processes: (i) the yield of CD from starch was not high, and (ii) Toxic organic solvents such as trichloroethylene, bromobenzene, and toluene were used to precipitate CD owing to the low conversion rate. The use of CGTase of alkaliphilic Bacillus sp. strain 38-2 overcame all these weak points and led to the mass production of crystalline α-, β-, γ-CD at low cost without using any organic solvents. The yield of CD was 85 to 90% from amylose and 70 to 80% from potato starch on a laboratory scale. Owing to the high conversion rate, CDs could be directly crystallized from the hydrolysate of starch without the addition of organic solvents (124). These simple methods reduced the cost of β-CD from $1,000/kg to $5/kg and that of a α-CD to within $15/kg. This has paved the way for its use in large quantities in foodstuffs, chemicals, and pharmaceuticals. Several other processes to produce CDs have been developed since the original report by Matsuzawa et al. (124). Kim et al. reported an enzymatic production of CDs from milled corn starch in an ultrafiltration membrane bioreactor to reduce product inhibition (96). In a batch operation with ultrafiltration, the conversion yield was increased 57% compared with that without ultrafiltration. Okada et al. also developed a bioreactor system with the enzyme immobilized on a capillary membrane (148). The percentage of CDs to total sugar obtained was slightly more than 60% under most operating conditions. Abraham immobilized CGTase for the continuous production of CDs (2). The immobilized enzyme had a pH optimum shifted to the alkaline side (from 6.5 to 7.5) and had a reduced temperature optimum (from 60°C to 50–55°C). Lima et al. developed a unique CD production process (118). A 10% (wt/vol) solution of cassava starch, liquefied with α-amylase, was incubated with CGTase by using only the enzyme and added ethanol (from 1 to 5%), followed by the addition of the yeast Saccharomyces cerevisiae (12% [wt/vol]) plus nutrients. However, the production costs were not reported.
Pullulanases.
In 1976, Nakamura et al. discovered an alkaline pullulanase of Bacillus sp. strain 202-1 (139). The enzyme has a pH optimum for enzyme action at pH 8.5 to 9.0 and is stable for 24 h at pH 6.5 to 11.0 at 4°C. The enzyme is most active at 55°C and is stable up to 50°C for 15 min in the absence of substrate. Kelly et al. found that alkaliphilic B. halodurans A-59 (ATCC 21591) produced three enzymes, α-amylase, pullulanase, and α-glucosidase, in culture broth (87). These three enzymes were separately produced, and the levels of α-glucosidase and pullulanase reached their maxima after 24 h of cultivation at the initial pH 9.7. Although this pullulanase was not purified, the indicated pH optimum was at 7.0.
Two highly alkaliphilic pullulanase-producing bacteria were isolated from Korean soil (94, 95). The two isolates were extremely alkaliphilic, since bacterial growth and enzyme production occurred at pH values ranging from pH 6.0 to 12.0 for Micrococcus sp. strain Y-1 and pH 6.0 to 10.0 for Bacillus sp. strain S-1. The enzyme displayed a temperature optimum of around 60°C and a pH optimum of around pH 9.0. The extracellular enzymes of both bacteria were alkaliphilic and moderately thermoactive; optimal activity was detected at pH 8.0 to 10.0 and between 50 and 60°C.
In screening alkaline cellulases from detergent additives, Ara and coworkers isolated a novel alkaline pullulanase from alkaliphilic Bacillus sp. strain KSM-1876, which was identified as a relative of Bacillus circulans (8, 72). The enzyme had an optimum pH for enzyme action of around 10.0 to 10.5. This enzyme is a good candidate for use as an additive to dishwashing detergents. Ito’s group then found another alkaline amylopullulanase from alkaliphilic Bacillus sp. strain KSM-1378 (7). This enzyme efficiently hydrolyzed the α-1,4 and α-1,6 linkages of amylose, amylopectin, and glycogen at alkaline pH values. The kinetic studies revealed two independent active sites for the α-1,4 and α-1,6 hydrolytic reactions. Incubation of the enzyme at 40°C and pH 9.0 caused complete inactivation of the amylase activity within 4 days, but the pullulanase activity remained at the original level under the same conditions (7). This alkaline amylopullulanase can therefore be considered to be a “two-headed” enzyme molecule. Limited proteolysis with papain also revealed that the α-1,6- and α1,4 hydrolytic activities were associated with two different active sites (6). Furthermore, analysis of the amino acid sequence and molecular structure of the enzyme showed two different active sites (57). The enzyme was observed by transmission electron microscopy; it appeared to be a bent dumbbell-like molecule with a diameter of approximately 25 nm.
Takagi et al. reported diversity in the size and alkaliphily of thermostable α-amylase-pullulanases produced by recombinant Escherichia coli, B. subtilis and the wild-type Bacillus sp. strain XAL601 (176). It was revealed that the noncatalytic C-terminal region may be responsible for the high optimum pH of the enzyme activity. These observations were also reported for CGTase (82, 83) and alkaline carboxymethyl cellulase (CMCase) (132).
Lin et al. purified a thermostable pullulanase from the thermophilic alkaliphilic Bacillus sp. strain TS-23 (120). This purified enzyme had both pullulanase and amylase activities. The temperature and pH optima for both pullulanase and amylase activities were 70°C and pH 8 to 9, respectively. The enzyme remained more than 96% active at temperatures below 65°C, and both activities were retained at temperatures up to 90°C in the presence of 5% sodium dodecyl sulfate (120).
In some food industries, enzymes showing activity at lower temperatures have been requested for use in food processing. Psychrotrophic bacteria are thought to be potential producers of these enzymes. Kimura and Horikoshi (103, 105) reported that an alkalipsychrotrophic strain, Micrococcus sp. strain 207, extracellularly produced amylase and pullulanase. The pullulanase was purified to an electrophoretically homogeneous state by conventional methods. The purified enzyme was free of α-amylase activity. It had a pH optimum at 7.5 to 8.0 and was relatively thermostable (up to 45°C). The enzyme could hydrolyze the α-1,6-linkages of amylopectins, glycogens, and pullulan. Although many alkaline pullulanases have been reported as described above, no industrial application has been developed yet.
Cellulases
Alkaline cellulases of alkaliphilic Bacillus strains.
Commercially available cellulases display optimum activity over a pH range from 4 to 6. No enzyme with an alkaline optimum pH for activity (pH 10 or higher) had been reported before the rediscovery of alkaliphiles. Horikoshi and coworkers found bacterial isolates (Bacillus sp. strains N4 and 1139) producing extracellular alkaline CMCases (37, 71). One of these, alkaliphilic Bacillus sp. strain N-4 (ATCC 21833), produced multiple CMCases that were active over a broad pH range (pH 5 to 10). Sashihara et al. cloned the cellulase genes of Bacillus sp. strain N-4 in E. coli HB101. Several cellulase-producing clones with different DNA sequences were obtained (159). Another bacterium, Bacillus sp. strain 1139, produced one CMCase, which was purified and shown to have optimum pH for activity at pH 9.0. The enzyme was stable over the pH range from 6 to 11 (for 24 h at 4°C and 10 min at up to 40°C). The CMCase gene of Bacillus sp. strain 1139 was also cloned in E. coli (38–42). Beppu’s group (132, 153) constructed many chimeric cellulases from B. subtilis and Bacillus sp. strain N-4 enzyme genes in an effort to understand the alkaliphily of N-4 enzymes. Although the genes have high homology, the pH activity profiles of the two enzymes are quite different; the B. subtilis enzyme (BSC) has its optimum pH at 6 to 6.5, whereas the Bacillus sp. strain N-4 enzyme (NK1) is active over a broad pH range from 6 to 10.5. The chimeric cellulases showed various chromatographic behaviors, reflecting the origins of their C-terminal regions. The pH activity profiles of the chimeric enzymes in the alkaline range could be classified into either the BSC or NK1 type, depending mainly on the origins of the fifth C-terminal regions.
Cellulases as laundry detergent additives.
The discovery of alkaline cellulases created a new industrial application of cellulase as a laundry detergent additive. Ito (79a) mixed alkaline cellulases with laundry detergents and studied their effect by washing cotton underwear. The best results were obtained by one of the alkaline cellulases produced by an alkaliphilic Bacillus strain. However, the yield of enzyme was not sufficient for industrial purposes. Ito and coworkers (79, 203) then isolated the alkaliphilic Bacillus sp. strain KSM-635 from the soil and succeeded in producing an alkaline cellulase as a laundry detergent additive on an industrial scale.
Besides the KSM-635 enzyme, Shikata et al. isolated three strains, alkaliphilic Bacillus strains KSM-19, KSM-64, and KSM-520, producing alkaline cellulases for laundry detergents (163). The activities of these enzymes (pH optima; 8.5 to 9.5) were not inhibited at all by metal ions or various components of laundry products, such as surfactants, chelating agents, and proteinases. With a view to increasing industrial production, Sumitomo et al. (173, 174) overexpressed the alkaline cellulase of alkaliphilic Bacillus sp. strain KSM-64 by using B. subtilis harboring the vector pHSP64 carrying the cellulase gene. By this process, they produced 30 g of alkaline cellulase in 1 liter. After the discovery of the industrial application of alkaline cellulase as a detergent additive, many microbiologists have extensively studied alkaline cellulases (52, 58, 77, 93, 129, 158). Further details are reviewed by Ito (78).
Alkaline cellulases from other alkaliphiles.
Park et al. (152) and Damude et al. (24) studied a semialkaline cellulase produced by alkaliphilic Streptomyces strain KSM-9. Dasilva et al. (25) reported two alkaliphilic microorganisms, Bacillus sp. strain B38-2 and Streptomyces sp. strain S36-2. The optimum pH and temperature of the crude enzyme activities ranged from 6.0 to 7.0 at 55°C for the Streptomyces strain and 7.0 to 8.0 at 60°C for the Bacillus strain. However, the results indicated that the properties of these enzymes were not appropriate for industrial purposes.
Lipases
Although the initial motivation for studying alkaline lipase was its application to detergents, many alkaline lipases were significantly inhibited in the presence of either alkylbenzene sulfate or dodecyl benzene sulfonate.
Watanabe et al. (197) conducted an extensive screening for alkaline lipase-producing microorganisms from soil and water samples. Two bacterial strains were selected as potent producers of alkaline lipase. These were identified as Pseudomonas nitroreducens nov. subsp. thermotolerans and P. fragi. The optimum pH of the two lipases was 9.5. Both enzymes were inhibited by bile salts such as sodium cholate, sodium deoxycholate, and sodium taurocholate at 0.25%. However, further work has not been reported.
A thermophilic lipase-producing bacterium was isolated from a hot-spring area of Yellowstone National Park (196). The organism, characterized as a Bacillus sp., grew optimally at 60 to 65°C and in the pH range from 6 to 9. The partially purified lipase preparation had an optimum temperature of 60°C and an optimum pH of 9.5. It retained 100% of the original activity after being heated at 75°C for 30 min. It was active on triglycerides containing fatty acids having a carbon chain length of C16:0 to C22:0, as well as on natural fats and oils.
Bushan et al. (12) found a lipase produced from an alkaliphilic Candida species in a solid-state fermentation. The lipase from this microorganism had temperature and pH optima of 40°C and 8.5, respectively, and was stable at 45°C for 4 h. The enzyme activity was stimulated by Ni2+ and Ca2+ ions but inhibited by Fe2+ and Fe3+ ions.
Xylanases
The first paper describing the isolation of a xylanase from alkaliphilic bacteria was published in 1973 by Horikoshi and Atsukawa (70). The purified enzyme of Bacillus sp. strain C-59-2 exhibited a broad optimal pH ranging from 6.0 to 8.0. In the culture broth of Bacillus halodurans C-125, two xylanases were found (64). Xylanase A had a molecular weight of 43,000, and xylanase N had a molecular weight of 16,000. Xylanase N was most active at pH 6 to 7, and xylanase A was most active pH 6 to 10 and had some activity at pH 12. The xylanase A gene was cloned, sequenced, and expressed in E. coli (59–63). Four thermophilic alkaliphilic Bacillus strains (W1 [JCM2888], W2 [JCM2889], W3, and W4) produced xylanases (149, 150). The pH optima for enzyme action of strains W1 and W3 was 6.0, and that for strains W2 and W4 was between 6 and 7. The enzymes were stable between pH 4.5 and 10.5 at 45°C for 1 h. The optimum temperatures of xylanases of W1 and W3 were 65°C, and those of W2 and W4 were 70°C. The degree of hydrolysis of xylan was about 70% after 24 h of incubation.
Since the demonstration that alkali-treated wood pulp could be biologically bleached by xylanases instead of by the usual environmentally damaging chemical process involving chlorine, the search for thermostable alkaline xylanases has been extensive. Dey et al. isolated an alkaliphilic thermophilic Bacillus strain (NCIM 59) that produced two types of cellulase-free xylanase at pH 10 and 50°C (27). Khasin et al. reported that alkaliphilic B. stearothermophilus T-6 produced an extracellular xylanase that optimally bleached pulp at pH 9 and 65°C (90). Nakamura et al. also reported that an alkaliphilic Bacillus strain, 41M-1, isolated from soil, produced multiple xylanases extracellularly (140, 142, 143). One of the enzymes, xylanase J, was most active at pH 9.0. The optimum temperature for the activity at pH 9.0 was around 50°C. Then, an alkaliphilic and thermophilic Bacillus strain, TAR-1, was isolated from soil (141). The xylanase was most active over a pH range of 5.0 to 9.5 at 50°C. Optimum temperatures of the crude xylanase preparation were 75°C at pH 7.0 and 70°C at pH 9.0. These xylanases did not act on cellulose, indicating a possible application of the enzyme in biological debleaching processes.
Blanco et al. reported that an enzyme from Bacillus sp. strain BP-23 facilitated the chemical bleaching of pulp, generating savings of 38% in terms of chlorine dioxide consumption (14). Recently, Garg et al. reported a biobleaching effect of Streptomyces thermoviolaceus xylanase on birchwood kraft pulp. S. thermoviolaceus xylanase had the advantage of activity and stability at 65°C (43, 44).
Subsequently, many thermostable alkaline xylanases have been produced from various alkaliphiles isolated from geothermal areas (28, 122, 175).
Pectinases
The first paper on alkaline endopolygalacturonase produced by alkaliphilic Bacillus sp. strain P-4-N was published in 1972 (68). The optimum pH for enzyme action was 10.0 for pectic acid. Fogarty and coworkers (33, 85) then reported that Bacillus sp. strain RK9 produced endopolygalacturonate lyase. The optimum pH for the enzyme activity toward acid-soluble pectic acid was 10.0. Subsequently, several papers on potential applications of alkaline pectinase have been published. The first application of alkaline pectinase-producing bacteria in the retting of Mitsumata bast was reported by Yoshihara and Kobayashi (202). The pectic lyase (pH optimum 9.5) produced by the alkaliphilic Bacillus sp. strain GIR 277 has been used in improving the production of a type of Japanese paper. A new retting process produced a high-quality, nonwoody paper that was stronger than the paper produced by the conventional method. Tanabe et al. tried to develop a new waste treatment by using the alkaliphilic Bacillus sp. strain GIR 621-7 (184, 185). Cao et al. isolated four alkaliphilic bacteria, NT-2, NT-6, NT-33, and NT-82, producing pectinase and xylanase (20). One strain, NT-33, had an excellent capacity for degumming ramie fibers.
Chitinases
Tsujibo et al. isolated chitinases from the alkaliphilic Nocardiopsis albus subsp. prasina OPC-131 (195). The isolate produced two types of chitinases. The optimum pH of chitinase A was pH 5.0, and that of chitinase B was pH 7.0. Recently, Bhushan and Hoondal isolated the alkaliphilic, chitinase-producing Bacillus sp. strain BG-11 (13). The purified chitinase exhibited broad pH and temperature optima of 7.5 to 9.0 and 45 to 55°C, respectively. The chitinase was stable between pH 6.0 and 9.0 at 50°C for more than 2 h. Ag+, Hg2+, dithiothreitol, β-mercaptoethanol, glutathione, iodoacetic acid, and iodoacetamide inhibited the activity up to 50%. No further studies have been reported.
METABOLITES PRODUCED BY ALKALIPHILES
2-Phenylamine
Hamasaki et al. found that a large amount of 2-phenylethylamine was synthesized by cells of the alkaliphilic Bacillus sp. strain YN-2000. This amine was secreted in the medium during cell growth (54).
Carotenoids of Alkaliphilic Bacillus Strains
Aono and Horikoshi reported that alkaliphilic Bacillus sp. strains A-40-2, 2B-2, 8-1, and 57-1 produce yellow pigments in the cells (3) and that these are triterpenoid carotenoids. However, A-59, M-29, and Y-25, white strains when grown in Horikoshi-II medium, did not produce carotenoids. The physiological role of the yellow pigments was not reported.
Siderophores
Gascoyne et al. isolated a siderophore-producing alkaliphilic bacterium that accumulated iron, gallium, and aluminum (45, 46). Enrichment cultures initiated with samples from a number of alkaline environmental sources yielded 10 isolates. From this group, selections were made on the basis of growth at high pH and the gallium-binding capacity of the siderophores. It was found that some isolates grew well and high concentrations of siderophore were detected whereas others grew well in the presence of much lower concentrations of siderophore. The effect of iron, gallium, and aluminum on growth and siderophore production in batch culture was investigated for six isolates. The presence of iron greatly decreased the siderophore concentration in these cultures, whereas the response to added gallium or aluminum was dependent upon the isolate.
Cholic Acid Derivatives
Kimura et al. isolated an alkaliphilic Bacillus strain from soil that grew well in media containing cholic acid (CA) at 5% (wt/vol) or higher concentrations (98). The 7α- and 12α-hydroxyl groups of CA were efficiently converted to keto groups, with the conversion rate for both hydroxyl groups reaching 100% by 72 h of cultivation. The strain also converted a 3α-hydroxyl group to a keto group, but the conversion rate was about 5% at 72 h. The strain neither affected any other part of the CA molecule nor oxidized 7β- or 12β-hydroxyl groups. By NTG mutagenesis, they isolated five mutants that selectively produced the following compounds from CA at high yield (close to 100%): strains M-4 and M-5 produced 7,12-diketolithocholic acid, strain 250 produced 7-ketodeoxycholic acid, and strains 124 and 336 yielded ketochenodeoxycholic acid. Furthermore, strains M-4, M-5, and 250 produced only 7-ketolithocholic acid from chenodeoxycholic acid.
Organic Acids
During the cultivation of alkaliphiles, the pH values of the culture media often decrease sharply due to the production of organic acids, which are produced by growth on carbohydrates. Paavilainen et al. reported comparative studies of organic acids produced by alkaliphilic bacilli (151). Four bacilli, Bacillus sp. strain 38-2 (ATCC 21783), B. alkalophilus subsp. halodurans (ATCC 27557), B. alcalophilus (ATCC 27648), and Bacillus sp. strain 17-1 (ATCC 31007), were cultured in the presence of various 1% (wt/vol) sugars and related compounds such as sugar alcohols. All these alkaliphiles produced acetic acid (4.5 to 5 g/liter at the maximum), while formic acid was produced by only one of the strains. In contrast to neutrophilic bacilli, acetoin, butanediol, and ethanol were not detected. Moderate amounts of isobutyric, isovaleric, α-oxoisovaleric, α-oxo-β-methylvaleric, α-oxoisocaproic, and phenylacetic acids were generated by three of the alkaliphiles.
Antibiotics and Enzyme Inhibitors
Since alkaliphiles were rediscovered, many Japanese pharmaceutical companies have tried using alkaline media to isolate new microorganisms producing new antibiotics. Although several have been found and reported, none are as yet commercially available.
The first report was published in 1980 by Sato et al. (160), where they recorded the isolation of Paecilomyces lilacinus 1907 from soil by using an alkaline medium (pH 10.5). New antibiotics were produced only under alkaline conditions (pH 9 to 10.5). P. lilacinus 1907 produced a major product, 1907-II, and a minor product, 1907-VIII, which had antibacterial and antifungal activities.
Subsequently, an alkaliphilic Nocardiopsis dassonvillei strain, OPC-15, that produced phenazine antibiotics under alkaline culture conditions was isolated (194). Strain OPC-15 grew well at pH 10 and accumulated two phenazine antibiotics in the mycelia, 1,6-dihydroxyphenazine (I) and 1,6-dihydroxyphenazine 3,5-dioxide (III), at different growth temperatures. Antibiotic I was formed when the organism was cultured at 27°C for 6 to 8 days, whereas the antibiotic III was formed only when the organism was grown at 27°C for 6 days and incubated further at 4°C for 2 days.
Bahn et al. isolated a novel aldose reductase inhibitor, YUA001, from the alkaliphilic Corynebacterium sp. strain YUA25-1. Compound YUA001 was purified from the supernatant of culture broth by successive silica gel column chromatography steps. The molecular formula of YUA001 was determined to be C13H19NO2. The compound had no antimicrobial activity against some gram-positive and gram-negative bacteria, fungi, and yeasts (10).
Besides these antibiotics, many compounds have been found and screened by pharmaceutical companies. However, there is one weak point in the direct production of antibiotics by alkaliphilic microorganisms. Many antibiotics are very unstable under alkaline condition, and it is highly possible that antibiotics produced are being destroyed during cultivation. However, some alkaliphiles, especially actinomycetes, can grow in neutral media under specified conditions, offering a strategy for the recovery of bioactive compounds.
FUTURE OF ALKALIPHILES
Since the rediscovery of alkaliphilic bacteria, more than 1,000 papers on many aspects of alkaliphiles and alkaliphily have been published. The alkaliphiles are unique microorganisms, with great potential for microbiology and biotechnological exploitation. The aspects that have received the most attention in recent years include (i) extracellular enzymes and their genetic analysis, (ii) mechanisms of membrane transport and pH regulation, and (iii) the taxonomy of alkaliphilic microorganisms. What will be the next line of development? It is unclear, but it may be the wider application of enzymes. Alkaline enzymes should find additional uses in various fields of industry, such as chiral-molecule synthesis, biological wood pulping, and more production of sophisticated enzyme detergents. Furthermore, alkaliphiles may be very good general genetic resources for such applications as production of signal peptides for secretion and promoters for hyperproduction of enzymes.
REFERENCES
- 1.Abelyan V A, Yamamoto T, Afrikyan E G. Isolation and characterization of cyclomaltodextrin glucanotransferases using cyclodextrin polymers and their derivatives. Biochemistry (Engl Transl Biokhimiya) 1994;59:573–579. [Google Scholar]
- 2.Abraham T E. Immobilized cyclodextrin glycosyl transferase for the continuous production of cyclodextrins. Biocatal Biotransform. 1995;12:137–146. [Google Scholar]
- 3.Aono R, Horikoshi K. Carotenes produced by alkaliphilic yellow pigmented strains of Bacillus. Agric Biol Chem. 1991;55:2643–2645. [Google Scholar]
- 4.Aono R, Horikoshi K. Chemical composition of cell walls of alkalophilic strains of Bacillus. J Gen Microbiol. 1983;129:1083–1087. [Google Scholar]
- 5.Aono R, Ito M, Horikoshi K. Measurement of cytoplasmic pH of the alkaliphile Bacillus lentus C-125 with a fluorescent pH probe. Microbiology. 1997;143:2531–2536. doi: 10.1099/00221287-143-8-2531. [DOI] [PubMed] [Google Scholar]
- 6.Ara K, Igarashi K, Hagihara H, Sawada K, Kobayashi T, Ito S. Separation of functional domains for the α-1,4 and α-1,6 hydrolytic activities of a Bacillus amylopullulanase by limited proteolysis with papain. Biosci Biotechnol Biochem. 1996;60:634–639. doi: 10.1271/bbb.60.634. [DOI] [PubMed] [Google Scholar]
- 7.Ara K, Igarashi K, Saeki K, Ito S. An alkaline amylopullulanase from alkalophilic Bacillus sp KSM-1378: kinetic evidence for two independent active sites for the α-1,4 and α-1,6 hydrolytic reactions. Biosci Biotechnol Biochem. 1995;59:662–666. [Google Scholar]
- 8.Ara K, Igarashi K, Saeki K, Kawai S, Ito S. Purification and some properties of an alkaline pullulanase from alkaliphilic Bacillus sp. KSM-1876. Biosci Biotechnol Biochem. 1992;56:62–65. [Google Scholar]
- 9.Aunstrup K, Ottrup H, Andresen O, Dambmann C. Proceedings of the 4th International Fermentation Symposium. Kyoto, Japan: Society of Fermentation Technology; 1972. Proteases from alkalophilic Bacillus species; pp. 299–305. [Google Scholar]
- 10.Bahn Y S, Park J M, Bai D H, Takase S, Yu J H. YUA001, a novel aldose reductase inhibitor isolated from alkalophilic Corynebacterium sp. YUA25. I. Taxonomy, fermentation, isolation and characterization. J Antibiot. 1998;51:902–907. doi: 10.7164/antibiotics.51.902. [DOI] [PubMed] [Google Scholar]
- 11.Bakels R H A, Vanwalraven H S, Krab K, Scholts M J C, Kraayenhof R. On the activation mechanism of the H+-ATP synthase and unusual thermodynamic properties in the alkalophilic Cyanobacterium Spirulina-Platensis. Eur J Biochem. 1993;213:957–964. doi: 10.1111/j.1432-1033.1993.tb17840.x. [DOI] [PubMed] [Google Scholar]
- 12.Bhushan B, Dosanjh N S, Kumar K, Hoondal G S. Lipase production from an alkalophilic yeast sp. by solid state fermentation. Biotechnol Lett. 1994;16:841–842. [Google Scholar]
- 13.Bhushan B, Hoondal G S. Isolation, purification and properties of a thermostable chitinase from an alkalophilic Bacillus sp. BG-11. Biotechnol Lett. 1998;20:157–159. [Google Scholar]
- 14.Blanco A, Vidal T, Colom J F, Pastor F I J. Purification and properties of xylanase A from alkali-tolerant Bacillus sp. strain BP-23. Appl Environ Microbiol. 1995;61:4468–4470. doi: 10.1128/aem.61.12.4468-4470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bockle B, Galunsky B, Muller R. Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl Environ Microbiol. 1995;61:3705–3710. doi: 10.1128/aem.61.10.3705-3710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boone D R, Mathrani I M, Liu Y, Menaia J A G F, Mah R A, Boone J E. Isolation and Characterization of Methanohalophilus portucalensis sp. nov. and DNA reassociation study of the genus Methanohalophilus. Int J Syst Bacteriol. 1993;43:430–437. [Google Scholar]
- 17.Boyer E W, Ingle M B. Extracellular alkaline amylase from a Bacillus species. J Bacteriol. 1972;110:992–1000. doi: 10.1128/jb.110.3.992-1000.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer E W, Ingle M B, Mercer G D. Bacillus alcalophilus subsp. halodurans subsp. nov.: an alkaline-amylase-producing alkalophilic organisms. Int J Syst Bacteriol. 1973;23:238–242. [Google Scholar]
- 19.Buck D P, Smith G D. Evidence for a Na+/H+ electrogenic antiporter in an alkaliphilic cyanobacterium Synechocystis. FEMS Microbiol Lett. 1995;128:315–320. [Google Scholar]
- 20.Cao J, Zheng L, Chen S. Screening of pectinase producer from alkalophilic bacteria and study on its potential application in degumming of ramie. Enzyme Microb Technol. 1992;14:1013–1016. [Google Scholar]
- 21.Cheng S W, Hu H M, Shen S W, Takagi H, Asano M, Tsai Y C. Production and characterization of keratinase of a feather-degrading Bacillus licheniformis PWD-1. Biosci Biotechnol Biochem. 1995;59:2239–2243. doi: 10.1271/bbb.59.2239. [DOI] [PubMed] [Google Scholar]
- 22.Chung H J, Yoon S H, Lee M J, Kim M J, Kweon K S, Lee I W, Kim J W, Oh B H, Lee H S, Spiridonova V A, Park K H. Characterization of a thermostable cyclodextrin glucanotransferase isolated from Bacillus stearothermophilus ET1. J Agric Food Chem. 1998;46:952–959. [Google Scholar]
- 23.Cook G M, Russell J B, Reichert A, Wiegel J. The intracellular pH of Clostridium paradoxum, an anaerobic, alkaliphilic, and thermophilic bacterium. Appl Environ Microbiol. 1996;62:4576–4579. doi: 10.1128/aem.62.12.4576-4579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damude H G, Gilkes N R, Kilburn D G, Miller R C, Antony R, Warren J. Endoglucanase CasA from alkalophilic Streptomyces strain KSM-9 is a typical member of family B of β-1,4-glucanases. Gene. 1993;123:105–107. doi: 10.1016/0378-1119(93)90547-g. [DOI] [PubMed] [Google Scholar]
- 25.Dasilva R, Yim D K, Asquieri E R, Park Y K. Production of microbial alkaline cellulase and studies of their characteristics. Rev Microbiol. 1993;24:269–274. [Google Scholar]
- 26.Desmarais D, Jablonski P E, Fedarko N S, Roberts M F. 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J Bacteriol. 1997;179:3146–3153. doi: 10.1128/jb.179.10.3146-3153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey D, Hinge J, Shendye A, Rao M. Purification and properties of extracellular endoxylanases from alkalophilic thermophilic Bacillus sp. Can J Microbiol. 1992;38:436–442. [Google Scholar]
- 28.Dimitrov P L, Kambourova M S, Mandeva R D, Emanuilova E I. Isolation and characterization of xylan-degrading alkali-tolerant thermophiles. FEMS Microbiol Lett. 1997;157:27–30. [Google Scholar]
- 29.Dirmeier R, Keller M, Hafenbradl D, Braun F J, Rachel R, Burggraf S, Stetter K O. Thermococcus acidaminovorans sp. nov., a new hyperthermophilic alkalophilic archaeon growing on amino acids. Extremophiles. 1998;2:109–114. doi: 10.1007/s007920050049. [DOI] [PubMed] [Google Scholar]
- 30.Duckworth A W, Grant W D, Jones B E, Vansteenbergen R. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol. 1996;19:181–191. [Google Scholar]
- 31.Dwivedi A, Srinivas U K, Singh H N, Kumar H D. Regulatory effect of external pH on the intracellular pH in alkalophilic cyanobacteria Microcystis aeruginosa and Hapalosiphon welwitschii. J Gen Appl Microbiol (Tokyo) 1994;40:261–263. [Google Scholar]
- 32.Engle M, Li Y H, Woese C, Wiegel J. Isolation and characterization of a novel alkalitolerant thermophile, Anaerobranca horikoshii gen. nov., sp. nov. Int J Syst Bacteriol. 1995;45:454–461. doi: 10.1099/00207713-45-3-454. [DOI] [PubMed] [Google Scholar]
- 33.Fogarty W M, Ward P O. Pectinases and pectic polysaccharides. Vol. 13. Edinburgh, United Kingdom: Churchill Livingstone; 1977. [Google Scholar]
- 34.Fujiwara N, Masui A, Imanaka T. Purification and properties of the highly thermostable alkaline protease from an alkaliphilic and thermophilic Bacillus sp. J Biotechnol. 1993;30:245–256. doi: 10.1016/0168-1656(93)90117-6. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara N, Yamamoto K. Decomposition of gelatin layers on X-ray film by the alkaline protease from Bacillus sp. B21. J Ferment Technol. 1987;65:531–534. [Google Scholar]
- 36.Fujiwara N, Yamamoto K, Masui A. Utilization of thermostable alkaline protease from an alkalophilic thermophile for the recovery of silver from used X-ray film. J Ferment Bioeng. 1991;72:306–308. [Google Scholar]
- 37.Fukumori F, Kudo T, Horikoshi K. Purification and properties of a cellulase from alkalophilic Bacillus sp. no. 1139. J Gen Microbiol. 1985;131:3339–3345. [Google Scholar]
- 38.Fukumori F, Kudo T, Horikoshi K. Truncation analysis of an alkaline cellulase from an alkalophilic Bacillus species. FEMS Microbiol Lett. 1987;40:311–314. [Google Scholar]
- 39.Fukumori F, Kudo T, Narahashi Y, Horikoshi K. Molecular cloning and nucleotide sequence of the alkaline cellulase gene from the alkalophilic Bacillus sp. strain 1139. J Gen Microbiol. 1986;132:2329–2335. doi: 10.1099/00221287-132-8-2329. [DOI] [PubMed] [Google Scholar]
- 40.Fukumori F, Kudo T, Sashihara N, Nagata Y, Ito K, Horikoshi K. The third cellulase of alkalophilic Bacillus sp. strain N-4: evolutionary relationships within the cel gene family. Gene. 1989;76:289–298. doi: 10.1016/0378-1119(89)90169-8. [DOI] [PubMed] [Google Scholar]
- 41.Fukumori F, Ohishi K, Kudo T, Horikoshi K. Tandem location of the cellulase genes on the chromosome of Bacillus sp. strain N-4. FEMS Microbiol Lett. 1987;48:65–68. [Google Scholar]
- 42.Fukumori F, Sashihara N, Kudo T, Horikoshi K. Nucleotide sequences of two cellulase genes from alkalophilic Bacillus sp. strain N-4 and their strong homology. J Bacteriol. 1986;168:479–485. doi: 10.1128/jb.168.2.479-485.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garg A P, McCarthy A J, Roberts J C. Biobleaching effect of Streptomyces thermoviolaceus xylanase preparations on birchwood kraft pulp. Enzyme Microb Technol. 1996;18:261–267. [Google Scholar]
- 44.Garg A P, Roberts J C, McCarthy A J. Bleach boosting effect of cellulase-free xylanase of Streptomyces thermoviolaceus and its comparison with two commercial enzyme preparations on birchwood kraft pulp. Enzyme Microb Technol. 1998;22:594–598. [Google Scholar]
- 45.Gascoyne D J, Connor J A, Bull A T. Capacity of siderophore-producing alkalophilic bacteria to accumulate iron, gallium and aluminum. Appl Microbiol Biotechnol. 1991;36:136–141. [Google Scholar]
- 46.Gascoyne D J, Connor J A, Bull A T. Isolation of bacteria producing siderophores under alkaline conditions. Appl Microbiol Biotechnol. 1991;36:130–135. [Google Scholar]
- 47.Georganta G, Kaneko T, Nakamura N, Kudo T, Horikoshi K. Isolation and partial properties of cyclomaltodextrin glucanotransferase-producing alkaliphilic Bacillus spp. from a deep-sea mud sample. Starch. 1993;45:95–99. [Google Scholar]
- 48.Gerasimenko L M, Dubinin A V, Zavarzin G A. Alkaliphilic cyanobacteria from soda lakes of Tuva and their ecophysiology. Microbiology. 1996;65:736–740. [Google Scholar]
- 49.Grønstad A, Jaroszewicz E, Ito M, Sturr M G, Krulwich T A, Kolstø A-B. Physical map of alkaliphilic Bacillus firmus OF4 and detection of a large endogenous plasmid. Extremophiles. 1998;2:447–453. doi: 10.1007/s007920050091. [DOI] [PubMed] [Google Scholar]
- 50.Groth I, Schumann P, Rainey F A, Martin K, Schuetze B, Augsten K. Bogoriella caseilytica gen. nov., sp. nov., a new alkaliphilic actinomycete from a soda lake in Africa. Int J Syst Bacteriol. 1997;47:788–794. doi: 10.1099/00207713-47-3-788. [DOI] [PubMed] [Google Scholar]
- 51.Guffanti A A, Hicks D B. Molar growth yields and bioenergetic parameters of extremely alkaliphilic Bacillus species in batch cultures, and growth in a chemostat at pH 10.5. J Gen Microbiol. 1991;137:2375–2379. doi: 10.1099/00221287-137-10-2375. [DOI] [PubMed] [Google Scholar]
- 52.Hakamada Y, Koike K, Yoshimatsu T, Mori H, Kobayashi T, Ito S. Thermostable alkaline cellulase from an alkaliphilic isolate, Bacillus sp. KSM-S237. Extremophiles. 1997;1:151–156. doi: 10.1007/s007920050028. [DOI] [PubMed] [Google Scholar]
- 53.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 54.Hamasaki N, Shirai S, Niitsu M, Kakinuma K, Oshima T. An alkalophilic Bacillus sp. produces 2-phenylethylamine. Appl Environ Microbiol. 1993;59:2720–2722. doi: 10.1128/aem.59.8.2720-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han X Q, Damodaran S. Purification and characterization of protease Q: A detergent- and urea-stable serine endopeptidase from Bacillus pumilus. J Agric Food Chem. 1998;46:3596–3603. [Google Scholar]
- 56.Hashimoto M, Hamamoto T, Kitada M, Hino M, Kudo T, Horikoshi K. Characteristics of alkali-sensitive mutants of alkaliphilic Bacillus sp. strain C-125 that show cellular morphological abnormalities. Biosci Biotechnol Biochem. 1994;58:2090–2092. [Google Scholar]
- 57.Hatada Y, Igarashi K, Ozaki K, Ara K, Hitomi J, Kobayashi T, Kawai S, Watabe T, Ito S. Amino acid sequence and molecular structure of an alkaline amylopullulanase from Bacillus that hydrolyzes α-1,4 and α-1,6 linkages in polysaccharides at different active sites. J Biol Chem. 1996;271:24075–24083. doi: 10.1074/jbc.271.39.24075. [DOI] [PubMed] [Google Scholar]
- 58.Hayashi K, Nimura Y, Ohara N, Uchimura T, Suzuki H, Komacata K, Kozaki M. Low-temperature-active cellulase produced by Acremonium alcalophilum JCM 7366. Seibutsu-Kogaku Kaishi. 1996;74:A7–A10. [Google Scholar]
- 59.Honda H, Kudo T, Horikoshi K. Extracellular production of alkaline xylanase of alkalophilic Bacillus sp. by Escherichia coli carrying pCX311. J Ferment Technol. 1986;64:373–377. [Google Scholar]
- 60.Honda H, Kudo T, Horikoshi K. Molecular cloning and expression of the xylanase gene of alkalophilic Bacillus sp. strain C-125 in Escherichia coli. J Bacteriol. 1985;161:784–785. doi: 10.1128/jb.161.2.784-785.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Honda H, Kudo T, Horikoshi K. Production of extracellular alkaline xylanase of alkalophilic Bacillus sp. C-125 by Escherichia coli carrying pCX311. Syst Appl Microbiol. 1986;8:152–157. [Google Scholar]
- 62.Honda H, Kudo T, Horikoshi K. Purification and partial characterization of alkaline xylanase from Escherichia coli carrying pCX311. Agric Biol Chem. 1985;49:3165–3169. [Google Scholar]
- 63.Honda H, Kudo T, Horikoshi K. Selective excretion of alkaline xylanase by Escherichia coli carrying pCX311. Agric Biol Chem. 1985;49:3011–3015. [Google Scholar]
- 64.Honda H, Kudo T, Ikura Y, Horikoshi K. Two types of xylanases of alkalophilic Bacillus sp. no. C-125. Can J Microbiol. 1985;31:538–542. [Google Scholar]
- 65.Horikoshi K. Microorganisms in alkaline environments. Tokyo, Japan: Kodansha-VCH; 1991. [Google Scholar]
- 66.Horikoshi K. Production of alkaline enzymes by alkalophilic microorganisms. I. Alkaline protease produced by Bacillus no. 221. Agric Biol Chem. 1971;35:1407–1414. [Google Scholar]
- 67.Horikoshi K. Production of alkaline enzymes by alkalophilic microorganisms. II. Alkaline amylase produced by Bacillus No. A-40-2. Agric Biol Chem. 1971;35:1783–1791. [Google Scholar]
- 68.Horikoshi K. Production of alkaline enzymes by alkalophilic microorganisms. III. Alkaline pectinase of Bacillus no. P-4-N. Agric Biol Chem. 1972;36:285–293. [Google Scholar]
- 69.Horikoshi K, Akiba T. Alkalophilic microorganisms: a new microbial world. Heidelberg, Germany: Springer-Verlag KG; 1982. [Google Scholar]
- 70.Horikoshi K, Atsukawa Y. Xylanase produced by alkalophilic Bacillus no. C-59-2. Agric Biol Chem. 1973;37:2097–2103. [Google Scholar]
- 71.Horikoshi K, Nakao M, Kurono Y, Sashihara N. Cellulases of an alkalophilic Bacillus strain isolated from soil. Can J Microbiol. 1984;30:774–779. [Google Scholar]
- 72.Igarashi K, Ara K, Saeki K, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene that encodes an neopullulanase from an alkalophilic Bacillus. Biosci Biotechnol Biochem. 1992;56:514–516. doi: 10.1271/bbb.56.514. [DOI] [PubMed] [Google Scholar]
- 73.Igarashi K, Hatada Y, Hagihara H, Saeki K, Takaiwa M, Uemura T, Ara K, Ozaki K, Kawai S, Kobayashi T, Ito S. Enzymatic properties of a novel liquefying α-amylase from an alkaliphilic Bacillus isolate and entire nucleotide and amino acid sequences. Appl Environ Microbiol. 1998;64:3282–3289. doi: 10.1128/aem.64.9.3282-3289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Igarashi K, Hatada Y, Ikawa K, Araki H, Ozawa T, Kobayashi T, Ozaki K, Ito S. Improved thermostability of a Bacillus α-amylase by deletion of an arginine-glycine residue is caused by enhanced calcium binding. Biochem Biophys Res Commun. 1998;248:372–377. doi: 10.1006/bbrc.1998.8970. [DOI] [PubMed] [Google Scholar]
- 75.Ishikawa H, Ishimi K, Sugiura M, Sowa A, Fujiwara N. Kinetics and mechanism of enzymatic hydrolysis of gelatin layers of X-ray film and release of silver particles. J Ferment Bioeng. 1993;76:300–305. [Google Scholar]
- 76.Ito M, Guffanti A A, Zemsky J, Ivey D M, Krulwich T A. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J Bacteriol. 1997;179:3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ito S. Alkaline cellulases from alkaliphilic Bacillus: enzymatic properties, genetics, and application to detergents. Extremophiles. 1997;1:61–66. doi: 10.1007/s007920050015. [DOI] [PubMed] [Google Scholar]
- 78.Ito S, Kobayashi T, Ara K, Ozaki K, Kawai S, Hatada Y. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles. 1998;2:185–190. doi: 10.1007/s007920050059. [DOI] [PubMed] [Google Scholar]
- 79.Ito S, Shikata S, Ozaki K, Kawai S, Okamoto K I, Takei A, Ohta Y, Satoh T. Alkaline cellulase for laundry detergents: production by Bacillus sp. KSM-635 and enzymatic properties. Agric Biol Chem. 1989;53:1275–1281. [Google Scholar]
- 79a.Ito, S. Personal communication.
- 80.Jones B E, Grant W D, Duckworth A W, Owenson G G. Microbial diversity of soda lakes. Extremophiles. 1998;2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- 81.Kamekura M, DyallSmith M L, Upasani V, Ventosa A, Kates M. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int J Syst Bacteriol. 1997;47:853–857. doi: 10.1099/00207713-47-3-853. [DOI] [PubMed] [Google Scholar]
- 82.Kaneko K, Kudo T, Georganta G, Horikoshi K. The Fifth International Symposium of Cyclodextrins. 1990. Construction of chimeric series of Bacillus cyclomaltodextrin glucanotransferase and analysis of the thermal stabilities and pH optima of the enzymes and yield of cyclodextrins; p. 19. [Google Scholar]
- 83.Kaneko T, Song K B, Hamamoto T, Kudo T, Horikoshi K. Construction of a chimeric series of Bacillus cyclomaltodextrin glucanotransferases and analysis of the thermal stabilities and pH optima of the enzymes. J Gen Microbiol. 1989;135:3447–3457. doi: 10.1099/00221287-135-12-3447. [DOI] [PubMed] [Google Scholar]
- 84.Keller M, Braun F J, Dirmeier R, Hafenbradl D, Burggraf S, Rachel R, Stetter K O. Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeum growing on polysulfide at alkaline pH. Arch Microbiol. 1995;164:390–395. [PubMed] [Google Scholar]
- 85.Kelly C T, Fogarty W M. Production and properties of polygalacturonate lyase by an alkalophilic microorganisms, Bacillus sp. RK9. Can J Microbiol. 1978;24:1164–1172. doi: 10.1139/m78-190. [DOI] [PubMed] [Google Scholar]
- 86.Kelly C T, Mctigue M A, Doyle E M, Fogarty W M. The raw starch-degrading alkaline amylase of Bacillus sp. IMD 370. J Ind Microbiol. 1995;15:446–448. [Google Scholar]
- 87.Kelly C T, O’Reilly F, Fogarty W M. Extracellular α-glucosidase of an alkalophilic microorganism, Bacillus sp. ATCC 21591. FEMS Microbiol Lett. 1983;20:55–59. [Google Scholar]
- 88.Kevbrin V V, Lysenko A M, Zhilina T N. Physiology of the alkaliphilic methanogen Z-7936, a new strain of Methanosalsus zhilinaeae isolated from Lake Magadi. Microbiology. 1997;66:261–266. [Google Scholar]
- 89.Kevbrin V V, Zhilina T N, Rainey F A, Zavarzin G A. Tindallia magadii gen. nov., sp. nov.: an alkaliphilic anaerobic ammonifier from soda lake deposits. Curr Microbiol. 1998;37:94–100. doi: 10.1007/s002849900345. [DOI] [PubMed] [Google Scholar]
- 90.Khasin A, Alchanati I, Shoham Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol. 1993;59:1725–1730. doi: 10.1128/aem.59.6.1725-1730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khmelenina V N, Kalyuzhnaya M G, Starostina N G, Suzina N E, Trotsenko Y A. Isolation and characterization of halotolerant alkaliphilic methanotrophic bacteria from Tuva soda lakes. Curr Microbiol. 1997;35:257–261. [Google Scholar]
- 92.Khmelenina V N, Kalyuzhnaya M G, Trotsenko Y A. Physiological and biochemical properties of a haloalkalitolerant methanotroph. Microbiology. 1997;66:365–370. [Google Scholar]
- 93.Khyamihorani H. Partial purification and some properties of an alkaline cellulase from an alkalophilic Bacillus sp. World J Microbiol Biotechnol. 1996;12:525–529. doi: 10.1007/BF00419467. [DOI] [PubMed] [Google Scholar]
- 94.Kim C H, Choi H I, Lee D S. Pullulanases of alkaline and broad pH range from a newly isolated alkalophilic Bacillus sp. S-1 and a Micrococcus sp. Y-1. J Ind Microbiol. 1993;12:48–57. [Google Scholar]
- 95.Kim C H, Choi H I, Lee D S. Purification and biochemical properties of an alkaline pullulanase from alkalophilic Bacillus sp. S-1. Biosci Biotechnol Biochem. 1993;57:1632–1637. doi: 10.1271/bbb.57.1632. [DOI] [PubMed] [Google Scholar]
- 96.Kim T J, Lee Y D, Kim H S. Enzymatic production of cyclodextrins from milled corn starch in an ultrafiltration membrane bioreactor. Biotechnol Bioeng. 1993;41:88–94. doi: 10.1002/bit.260410112. [DOI] [PubMed] [Google Scholar]
- 97.Kim T U, Gu B G, Jeong J Y, Byun S M, Shin Y C. Purification and characterization of a maltotetraose-forming alkaline α-amylase from an alkalophilic Bacillus strain, GM8901. Appl Environ Microbiol. 1995;61:3105–3112. doi: 10.1128/aem.61.8.3105-3112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimura H, Okamura A, Kawaide H. Oxidation of 3-, 7-, and 12-hydroxyl groups of cholic acid by an alkalophilic Bacillus sp. Biosci Biotechnol Biochem. 1994;58:1002–1006. [Google Scholar]
- 99.Kimura K, Kataoka S, Ishii Y, Takano T, Yamane K. Nucleotide sequence of the β-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. strain 1011 and similarity of its amino acid sequence to those of α-amylases. J Bacteriol. 1987;169:4399–4402. doi: 10.1128/jb.169.9.4399-4402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kimura K, Takano T, Yamane K. Molecular cloning of the β-cyclodextrin synthetase gene from an alkaliphilic Bacillus and its expression in Escherichia coli and Bacillus subtilis. Appl Microbiol Biotechnol. 1987;26:147–153. [Google Scholar]
- 101.Kimura K, Tsukamoto A, Ishii Y, Takano T, Yamane K. Cloning of a gene for maltohexaose producing amylase of an alkalophilic Bacillus and hyperproduction of the enzyme in Bacillus subtilis cells. Appl Microbiol Biotechnol. 1988;27:372–377. [Google Scholar]
- 102.Kimura T, Horikoshi K. Carbohydrate use by alkalopsychrotrophic bacteria. Agric Biol Chem. 1989;53:2019–2020. [Google Scholar]
- 103.Kimura T, Horikoshi K. Characterization of pullulan-hydrolyzing enzyme from an alkalopsychrotrophic Micrococcus sp. Appl Microbiol Biotechnol. 1990;34:52–56. [Google Scholar]
- 104.Kimura T, Horikoshi K. Effects of temperature and pH on the production of maltotetraose by α-amylases of an alkalopsychrotrophic Micrococcus. J Ferment Bioeng. 1990;70:134–135. [Google Scholar]
- 105.Kimura T, Horikoshi K. Production of amylase and pullulanase by an alkalopsychrotrophic Micrococcus sp. Agric Biol Chem. 1989;53:2963–2968. [Google Scholar]
- 106.Kimura T, Horikoshi K. Purification and characterization of α-amylases of an alkalopsychrotrophic Micrococcus sp. Starch. 1990;42:403–407. [Google Scholar]
- 107.Kitada M, Hashimoto M, Kudo T, Horikoshi K. Properties of two different Na+/H+ antiport systems in alkaliphilic Bacillus sp. strain C-125. J Bacteriol. 1994;176:6464–6469. doi: 10.1128/jb.176.21.6464-6469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kitada M, Horikoshi K. Sodium ion-stimulated α-[1-14C]aminoisobutyric acid uptake in alkalophilic Bacillus species. J Bacteriol. 1977;131:784–788. doi: 10.1128/jb.131.3.784-788.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitada M, Morotomi S, Horikoshi K, Kudo T. K+/H+ antiporter in alkaliphilic Bacillus sp. no. 66 (JCM 9763) Extremophiles. 1997;1:135–141. doi: 10.1007/s007920050026. [DOI] [PubMed] [Google Scholar]
- 110.Kobayashi T, Hakamada Y, Adachi S, Hitomi J, Yoshimatsu T, Koike K, Kawai S, Ito S. Purification and properties of an alkaline protease from alkalophilic Bacillus sp. KSM-K16. Appl Microbiol Biotechnol. 1995;43:473–481. doi: 10.1007/BF00218452. [DOI] [PubMed] [Google Scholar]
- 111.Kobayashi T, Kanai H, Aono R, Horikoshi K, Kudo T. Cloning, expression, and nucleotide sequence of the α-amylase gene from the haloalkaliphilic archaeon Natronococcus sp. strain Ah-36. J Bacteriol. 1994;176:5131–5134. doi: 10.1128/jb.176.16.5131-5134.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kobayashi T, Kanai H, Hayashi T, Akiba T, Akaboshi R, Horikoshi K. Haloalkaliphilic maltotriose-forming α-amylase from the archaebacterium Natronococcus sp. strain Ah-36. J Bacteriol. 1992;174:3439–3444. doi: 10.1128/jb.174.11.3439-3444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kodama H, Koyama N. Unique characteristics of anaerobic alkalophiles belonging to Amphibacillus xylanus. Microbios. 1997;89:7–14. [Google Scholar]
- 114.Krulwich T A, Ito M, Hicks D B, Gilmour R, Guffanti A A. pH Homeostasis and ATP synthesis: studies of two processes that necessitate inward proton translocation in extremely alkaliphilic Bacillus species. Extremophiles. 1998;2:217–222. doi: 10.1007/s007920050063. [DOI] [PubMed] [Google Scholar]
- 115.Kudo T, Hino M, Kitada M, Horikoshi K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J Bacteriol. 1990;172:7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kurono Y, Horikoshi K. Alkaline catalase produced by Bacillus no. Ku-1. Agric Biol Chem. 1973;37:2565–2570. [Google Scholar]
- 117.Li Y H, Engle M, Weiss N, Mandelco L, Wiegel J. Clostridium thermoalcaliphilum sp. nov., an anaerobic and thermotolerant facultative alkaliphile. Int J Syst Bacteriol. 1994;44:111–118. doi: 10.1099/00207713-44-1-111. [DOI] [PubMed] [Google Scholar]
- 118.Lima H O S, DeMoraes F F, Zanin M. β-Cyclodextrin production by simultaneous fermentation and cyclization. Appl Biochem Biotechnol. 1998;70:789–804. doi: 10.1007/BF02920189. [DOI] [PubMed] [Google Scholar]
- 119.Lin L L, Chyau C C, Hsu W H. Production and properties of a raw-starch-degrading amylase from the thermophilic and alkaliphilic Bacillus sp. TS-23. Biotechnol Appl Biochem. 1998;28:61–68. [PubMed] [Google Scholar]
- 120.Lin L L, Tsau M R, Chu W S. Purification and properties of a 140-kDa amylopullulanase from thermophilic and alkaliphilic Bacillus sp. strain TS-23. Biotechnol Appl Biochem. 1996;24:101–107. [Google Scholar]
- 121.Lodwick D, McGenity T J, Grant W D. The phylogenetic position of the haloalkaliphilic archaeon Natronobacterium magadii, determined from its 23S ribosomal sequence. Syst Appl Microbiol. 1994;17:402–404. [Google Scholar]
- 122.Lopez C, Blanco A, Pastor F I J. Xylanase production by a new alkali-tolerant isolate of Bacillus. Biotechnol Lett. 1998;20:243–246. [Google Scholar]
- 123.Martin J R, Mulder F A A, Karimi Nejad Y, van der Zwan J, Mariani M, Schipper D, Boelens R. The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site. Structure. 1997;5:521–532. doi: 10.1016/s0969-2126(97)00208-6. [DOI] [PubMed] [Google Scholar]
- 124.Matsuzawa M, Kawano M, Nakamura N, Horikoshi K. An improved method for the production of Schardinger β-dextrin on an industrial scale by cyclodextrin glycosyltransferase of an alkalophilic Bacillus sp. Starch. 1975;27:410–413. [Google Scholar]
- 125.McGenity T J, Grant W D. The haloalkaliphilic archaeon (archaebacterium) Natronococcus occultus represents a distinct lineage within the Halobacteriales, most closely related to the other haloalkaliphilic lineage (Natronobacterium) Syst Appl Microbiol. 1993;16:239–243. [Google Scholar]
- 126.McTigue M A, Kelly C T, Doyle E M, Fogarty W M. The alkaline amylase of the alkalophilic Bacillus sp IMD 370. Enzyme Microb Technol. 1995;17:570–573. [Google Scholar]
- 127.McTigue M A, Kelly C T, Fogarty W M, Doyle E M. Production studies on the alkaline amylases of three alkalophilic Bacillus spp. Biotechnol Lett. 1994;16:569–574. [Google Scholar]
- 128.Miller A G, Tukrpin D H, Canvin D T. Na+ requirement for growth, photosynthesis, and pH regulation in the alkalotolerant cyanobacterium Synechococcus leopoliensis. J Bacteriol. 1984;159:100–106. doi: 10.1128/jb.159.1.100-106.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miyatake M, Imada K. A gene encoding endo-1,4-β-glucanase from Bacillus sp. 22–28. Biosci Biotechnol Biochem. 1997;61:362–364. doi: 10.1271/bbb.61.362. [DOI] [PubMed] [Google Scholar]
- 130.Morotomi S, Kitada M, Usami R, Horikoshi K, Kudo T. Physiological properties of a neutral-sensitive mutant derived from facultative alkaliphilic Bacillus sp. C-125. Biosci Biotechnol Biochem. 1998;62:788–791. doi: 10.1271/bbb.62.788. [DOI] [PubMed] [Google Scholar]
- 131.Na H K, Kim E S, Lee H B, Yoo O J, Jhon D Y. Cloning and nucleotide sequence of the α-amylase gene from alkalophilic Pseudomonas sp. KFCC 10818. Mol Cells. 1996;6:203–208. [Google Scholar]
- 132.Nakamura A, Fukumori F, Horinouchi S, Masaki H, Kudo T, Uozumi T, Horikoshi K, Beppu T. Construction and characterization of the chimeric enzymes between the Bacillus subtilis cellulase and an alkalophilic Bacillus cellulase. J Biol Chem. 1991;266:1579–1583. [PubMed] [Google Scholar]
- 133.Nakamura A, Haga K, Yamane K. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry. 1993;32:6624–6631. doi: 10.1021/bi00077a015. [DOI] [PubMed] [Google Scholar]
- 134.Nakamura A, Haga K, Yamane K. Four aromatic residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of replacements on substrate binding and cyclization characteristics. Biochemistry. 1994;33:9929–9936. doi: 10.1021/bi00199a015. [DOI] [PubMed] [Google Scholar]
- 135.Nakamura N, Horikoshi K. Characterization and some culture conditions of a cyclodextrin glycosyltransferase-producing alkalophilic Bacillus sp. Agric Biol Chem. 1976;40:753–757. [Google Scholar]
- 136.Nakamura N, Horikoshi K. Characterization of acid-cyclodextrin glycosyl-transferase of an alkalophilic Bacillus sp. Agric Biol Chem. 1976;40:1647–1648. [Google Scholar]
- 137.Nakamura N, Horikoshi K. Purification and properties of cyclodextrin glycosyltransferase of an alkalophilic Bacillus sp. Agric Biol Chem. 1976;40:935–941. [Google Scholar]
- 138.Nakamura N, Horikoshi K. Purification and properties of neutral-cyclodextrin glycosyl-transferase of an alkalophilic Bacillus sp. Agric Biol Chem. 1976;40:1785–1791. [Google Scholar]
- 139.Nakamura N, Watanabe K, Horikoshi K. Purification and some properties of alkaline pullulanase from a strain of Bacillus no. 202-1, an alkalophilic microorganism. Biochim Biophys Acta. 1975;397:188–193. doi: 10.1016/0005-2744(75)90192-8. [DOI] [PubMed] [Google Scholar]
- 140.Nakamura S, Nakai R, Kubo T, Wakabayashi K, Aono R, Horikoshi K. Structure-function relationship of the xylanase from alkaliphilic Bacillus sp. strain 41M-1. Nucleic Acids Symp Ser. 1995;34:99–100. [PubMed] [Google Scholar]
- 141.Nakamura S, Nakai R, Wakabayashi K, Ishiguro Y, Aono R, Horikoshi K. Thermophilic alkaline xylanase from newly isolated alkaliphilic and thermophilic Bacillus sp. strain TAR-1. Biosci Biotechnol Biochem. 1994;58:78–81. doi: 10.1271/bbb.58.78. [DOI] [PubMed] [Google Scholar]
- 142.Nakamura S, Wakabayashi K, Nakai R, Aono R, Horikoshi K. Production of alkaline xylanase by a newly isolated alkaliphilic Bacillus sp. strain 41M-1. World J Microbiol Biotechnol. 1993;9:221–224. doi: 10.1007/BF00327842. [DOI] [PubMed] [Google Scholar]
- 143.Nakamura S, Wakabayashi K, Nakai R, Aono R, Horikoshi K. Purification and some properties of an alkaline xylanase from alkaliphilic Bacillus sp. strain 41M-1. Appl Environ Microbiol. 1993;59:2311–2316. doi: 10.1128/aem.59.7.2311-2316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Niimura Y, Yanagida F, Uchimura T, Ohara N, Suzuki K, Kozaki M. A new facultative anaerobic xylan-using alkalophile lacking cytochrome, quinone and catalase. Agric Biol Chem. 1987;51:2271–2275. [Google Scholar]
- 145.Nomoto M, Chen C-C, Shen D-C. Purification and characterization of cyclodextrin glucanotransferase from an alkalophilic bacterium of Taiwan. Agric Biol Chem. 1986;50:2701–2707. [Google Scholar]
- 146.Nomoto M, Lee T-C, Su C-S, Liao C-W, Yen T-M, Yang C-P. Alkaline proteinases from alkalophilic bacteria of Taiwan. Agric Biol Chem. 1984;48:1627–1628. [Google Scholar]
- 147.Nomoto M, Shew D-C, Chen S-J, Yen T-M, Liao C-W, Yang C-P. Cyclodextrin glucanotransferase from alkalophilic bacterium of Taiwan. Agric Biol Chem. 1984;48:1337–1338. [Google Scholar]
- 148.Okada T, Ito M, Hibino K. Immobilization of cyclodextrin glucanotransferase on capillary membrane. J Ferment Bioeng. 1994;77:259–263. [Google Scholar]
- 149.Okazaki W, Akiba T, Horikoshi K, Akahoshi R. Production and properties of two types of xylanases from alkalophilic thermophilic Bacillus sp. Appl Microbiol Biotechnol. 1984;19:335–340. [Google Scholar]
- 150.Okazaki W, Akiba T, Horikoshi K, Akahoshi R. Purification and characterization of xylanases from alkalophilic thermophilic Bacillus spp. Agric Biol Chem. 1985;49:2033–2039. [Google Scholar]
- 151.Paavilainen S, Helisto P, Korpela T. Conversion of carbohydrates to organic acids by alkaliphilic bacilli. J Ferment Bioeng. 1994;78:217–222. [Google Scholar]
- 152.Park J-S, Horinouchi S, Beppu T. Characterization of leader peptide of an endo-type cellulase produced by an alkalophilic Streptomyces strain. Agric Biol Chem. 1991;55:1745–1750. [PubMed] [Google Scholar]
- 153.Park J S, Hitomi J, Horinouchi S, Beppu T. Identification of two amino acids contributing the high enzyme activity in the alkaline pH range of an alkaline endoglucanase from a Bacillus sp. Protein Eng. 1993;6:921–926. doi: 10.1093/protein/6.8.921. [DOI] [PubMed] [Google Scholar]
- 154.Parsiegla G, Schmidt A K, Schulz G E. Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the γ-cyclodextrin production. Eur J Biochem. 1998;255:710–717. doi: 10.1046/j.1432-1327.1998.2550710.x. [DOI] [PubMed] [Google Scholar]
- 155.Podkovyrov S M, Zeikus J G. Structure of the gene encoding cyclomaltodextrinase from Clostridium thermohydrosulfuricum-39E and characterization of the enzyme purified from Escherichia coli. J Bacteriol. 1992;174:5400–5405. doi: 10.1128/jb.174.16.5400-5405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prasad P, Kashyap A K. Ammonium transport in the alkalophilic diazotropic cyanobacterium Nostoc calcicola—influence of phosphate limitation and metabolic inhibitors. J Plant Physiol. 1991;138:244–247. [Google Scholar]
- 157.Salva T D G, deLima V B, Pagan A P. Screening of alkalophilic bacteria for cyclodextrin glycosyltransferase production. Rev Microbiol. 1997;28:157–164. [Google Scholar]
- 158.Sancheztorres J, Perez P, Santamaria R I. A cellulase gene from a new alkalophilic Bacillus sp. (strain N186-1). Its cloning, nucleotide sequence and expression in Escherichia coli. Appl Microbiol Biotechnol. 1996;46:149–155. doi: 10.1007/s002530050797. [DOI] [PubMed] [Google Scholar]
- 159.Sashihara N, Kudo T, Horikoshi K. Molecular cloning and expression of cellulase genes of alkalophilic Bacillus sp. strain N-4 in Escherichia coli. J Bacteriol. 1984;158:503–506. doi: 10.1128/jb.158.2.503-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sato M, Beppu T, Arima K. Properties and structure of a novel peptide antibiotic no. 1970. Agric Biol Chem. 1980;44:3037–3040. [Google Scholar]
- 161.Schmidt B F, Woodhouse L, Adams R M, Ward T, Mainzer S E, Lad P J. Alkalophilic Bacillus sp. strain LG12 has a series of serine protease genes. Appl Environ Microbiol. 1995;61:4490–4493. doi: 10.1128/aem.61.12.4490-4493.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seto Y, Hashimoto M, Usami R, Hamamoto T, Kudo T, Horikoshi K. Characterization of a mutation responsible for an alkali-sensitive mutant, 18224, of alkaliphilic Bacillus sp. strain C-125. Biosci Biotechnol Biochem. 1995;59:1364–1366. doi: 10.1271/bbb.59.1364. [DOI] [PubMed] [Google Scholar]
- 163.Shikata S, Saeki K, Okoshi H, Yoshimatsu T, Ozaki K, Kawai S, Ito S. Alkaline cellulase for laundry detergents: production by alkalophilic strains of Bacillus and some properties of the crude enzymes. Agric Biol Chem. 1990;54:91–96. [Google Scholar]
- 164.Shirai T, Suzuki A, Yamane T, Ashida T, Kobayashi T, Hitomi J, Ito S. High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism. Protein Eng. 1997;10:627–634. doi: 10.1093/protein/10.6.627. [DOI] [PubMed] [Google Scholar]
- 165.Singh S. Partial purification and some properties of urease from the alkaliphilic cyanobacterium Nostoc calcicola. Folia Microbiol Prague. 1995;40:529–533. [Google Scholar]
- 166.Sorokin D Y, deJong G A H, Robertson L A, Kuenen G J. Purification and characterization of sulfide dehydrogenase from alkaliphilic chemolithoautotrophic sulfur-oxidizing bacteria. FEBS Lett. 1998;427:11–14. doi: 10.1016/s0014-5793(98)00379-2. [DOI] [PubMed] [Google Scholar]
- 167.Sorokin D Y, Lysenko A M, Mityushina L L. Isolation and characterization of alkaliphilic chemoorganoheterotrophic bacteria oxidizing reduced inorganic sulfur compounds to tetrathionate. Microbiology. 1996;65:326–338. [Google Scholar]
- 168.Sorokin D Y, Mityushina L L. Ultrastructure of alkaliphilic heterotrophic bacteria oxidizing sulfur compounds to tetrathionate. Microbiology. 1998;67:78–85. [Google Scholar]
- 169.Sorokin D Y, Muyzer G, Brinkhoff T, Kuenen J G, Jetten M S M. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Arch Microbiol. 1998;170:345–352. doi: 10.1007/s002030050652. [DOI] [PubMed] [Google Scholar]
- 170.Studdert C A, DeCastro R E, Seitz K H, Sanchez J J. Detection and preliminary characterization of extracellular proteolytic activities of the haloalkaliphilic archaeon Natronococcus occultus. Arch Microbiol. 1997;168:532–535. doi: 10.1007/s002030050532. [DOI] [PubMed] [Google Scholar]
- 171.Sturr M G, Guffanti A A, Krulwich T A. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J Bacteriol. 1994;176:3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sturr M G, Krulwich T A, Kolsto A B. Physical map of alkaliphilic Bacillus firmus OF4 and detection of a large endogenous plasmid. Extremophiles. 1998;2:447–453. doi: 10.1007/s007920050091. [DOI] [PubMed] [Google Scholar]
- 173.Sumitomo N, Ozaki K, Hitomi J, Kawaminami S, Kobayashi T, Kawai S, Ito S. Application of the upstream region of a Bacillus endoglucanase gene to high-level expression of foreign genes in Bacillus subtilis. Biosci Biotechnol Biochem. 1995;59:2172–2175. doi: 10.1271/bbb.59.2172. [DOI] [PubMed] [Google Scholar]
- 174.Sumitomo N, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene for an alkaline endoglucanase from an alkalophilic Bacillus and its expression in Escherichia coli and Bacillus subtilis. Biosci Biotechnol Biochem. 1992;56:872–877. doi: 10.1271/bbb.56.872. [DOI] [PubMed] [Google Scholar]
- 175.Sunna A, Prowe S G, Stoffregen T, Antranikian G. Characterization of the xylanases from the new isolated thermophilic xylan-degrading Bacillus thermoleovorans strain K-3d and Bacillus flavothermus strain LB3A. FEMS Microbiol Lett. 1997;148:209–216. doi: 10.1111/j.1574-6968.1997.tb10290.x. [DOI] [PubMed] [Google Scholar]
- 176.Takagi M, Lee S P, Imanaka T. Diversity in size and alkaliphily of thermostable α-amylase-pullulanases (AapT) produced by recombinant Escherichia coli, Bacillus subtilis and the wild-type Bacillus sp. J Ferment Bioeng. 1996;81:557–559. [Google Scholar]
- 177.Takami H, Akiba T, Horikoshi K. Substrate specificity of thermostable alkaline protease from Bacillus sp. no. AH-101. Biosci Biotechnol Biochem. 1992;56:333–334. doi: 10.1271/bbb.56.333. [DOI] [PubMed] [Google Scholar]
- 178.Takami H, Akiba T, Horikoshi K. Characterization of an alkaline protease from Bacillus sp. no. AH-101. Appl Microbiol Biotechnol. 1990;33:519–523. doi: 10.1007/BF00172544. [DOI] [PubMed] [Google Scholar]
- 179.Takami H, Akiba T, Horikoshi K. Production of extremely thermostable alkaline protease from Bacillus sp. no. AH-101. Appl Microbiol Biotechnol. 1989;30:120–124. [Google Scholar]
- 180.Takami H, Inoue A, Fuji F, Horikoshi K. Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett. 1997;152:279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x. [DOI] [PubMed] [Google Scholar]
- 181.Takami H, Nakasone K, Hirama C, Takai Y, Masui N, Fuji F, Nakamura Y, Inoue A. An improved physical and genetic map of the genome of alkaliphilic Bacillus sp. C-125. Extremophiles. 1999;3:21–28. doi: 10.1007/s007920050095. [DOI] [PubMed] [Google Scholar]
- 182.Takami H, Takaki Y, Nakasone K, Hirama C, Inoue A, Horikoshi K. Sequence analysis of a 32-kb region including the major ribosomal protein gene clusters from alkaliphilic Bacillus sp. strain C-125. Biosci Biotechnol Biochem. 1999;63:452–455. doi: 10.1271/bbb.63.452. [DOI] [PubMed] [Google Scholar]
- 183.Takeuchi M, Kaieda N, Koyama N. Effect of amines on the (NH4+ + Na+)-activated ATPase of an anaerobic alkaliphile, Amphibacillus xylanus. Microbios. 1997;90:201–208. [Google Scholar]
- 184.Tanabe H, Kobayashi Y, Akamatsu I. Pretreatment of pectic wastewater with pectate lyase from an alkalophilic Bacillus sp. Agric Biol Chem. 1988;52:1855–1856. [Google Scholar]
- 185.Tanabe H, Yoshihara K, Tamura K, Kobayashi Y, Akamatsu I, Niyomwan N, Footrakul P. Pretreatment of pectic wastewater from orange canning process by an alkalophilic Bacillus sp. J Ferment Technol. 1987;65:243–246. [Google Scholar]
- 186.Terada Y, Yanase M, Takata H, Takaha T, Okada S. Cyclodextrins are not the major cyclic α-1,4-glucans produced by the initial action of cyclodextrin glucanotransferase on amylose. J Biol Chem. 1997;272:15729–15733. doi: 10.1074/jbc.272.25.15729. [DOI] [PubMed] [Google Scholar]
- 187.Tsai Y, Lin S, Li Y, Yamasaki M, Tamura G. Characterization of an alkaline elastase from alkalophilic Bacillus YA-B. Biochim Biophys Acta. 1986;883:439–447. [Google Scholar]
- 188.Tsai Y, Yamasaki M, Tamura G. Substrate specificity of a new alkaline elastase from an alkalophilic Bacillus. Biochem Int. 1984;8:283–288. [PubMed] [Google Scholar]
- 189.Tsai Y, Yamasaki M, Yamamoto-Suzuki Y, Tamura G. A new alkaline elastase of an alkalophilic Bacillus. Biochem Int. 1983;7:577–583. [PubMed] [Google Scholar]
- 190.Tsuchida O, Yamagata Y, Ishizuka T, Arai T, Yamada J, Takeuchi M, Ichishima E. An alkaline proteinase of an alkalophilic Bacillus sp. Curr Microbiol. 1986;14:7–12. [Google Scholar]
- 191.Tsuchiya K, Ikeda I, Tsuchiya T, Kimura T. Cloning and expression of an intracellular alkaline protease gene from alkalophilic Thermoactinomyces sp. HS682. Biosci Biotechnol Biochem. 1997;61:298–303. doi: 10.1271/bbb.61.298. [DOI] [PubMed] [Google Scholar]
- 192.Tsuchiya K, Nakamura Y, Sakashita H, Kimura T. Purification and characterization of a thermostable alkaline protease from alkalophilic Thermoactinomyces sp. HS682. Biosci Biotechnol Biochem. 1992;56:246–250. doi: 10.1271/bbb.56.246. [DOI] [PubMed] [Google Scholar]
- 193.Tsuchiya K, Sakashita H, Nakamura Y, Kimura T. Production of thermostable alkaline protease by alkalophilic Thermoactinomyces sp. HS682. Agric Biol Chem. 1991;55:3125–3127. [Google Scholar]
- 194.Tsujibo H, Sato T, Inui M, Yamamoto H, Inamori Y. Intracellular accumulation of phenazine antibiotics production by an alkalophilic actinomycete. Agric Biol Chem. 1988;52:301–306. [Google Scholar]
- 195.Tsujibo H, Yoshida Y, Miyamoto K, Hasegawa T, Inamori Y. Purification and properties of two types of chitinases produced by an alkalophilic actinomycete. Biosci Biotechnol Biochem. 1992;56:1304–1305. [Google Scholar]
- 196.Wang Y X, Srivastava K C, Shen G J, Wang H Y. Thermostable alkaline lipase from a newly isolated thermophilic Bacillus, strain A30-1 (ATCC 53841) J Ferment Bioeng. 1995;79:433–438. [Google Scholar]
- 197.Watanabe N, Ota Y, Minoda Y, Yamada K. Isolation and identification of alkaline lipase-producing microorganisms, culture conditions and some properties of crude enzymes. Agric Biol Chem. 1977;41:1353–1358. [Google Scholar]
- 198.Wiegel J. Anaerobic alkalithermophiles, a novel group of extremophiles. Extremophiles. 1998;2:257–267. doi: 10.1007/s007920050068. [DOI] [PubMed] [Google Scholar]
- 199.Xu C H, Nejidat A, Belkin S, Boussiba S. Isolation and characterization of the plasma membrane by two-phase partitioning from the alkalophilic cyanobacterium Spirulina platensis. Plant Cell Physiol. 1994;35:737–741. [Google Scholar]
- 200.Xu Y, Zhou P J, Tian X Y. Characterization of two novel haloalkaliphilic archaea Natronorubrum bangense gen. nov., sp. nov., and Natronorubrum tibetense gen. nov., sp. nov. Int J Syst Bacteriol. 1999;49:261–266. doi: 10.1099/00207713-49-1-261. [DOI] [PubMed] [Google Scholar]
- 201.Yeh C M, Chang H K, Hsieh H M, Yoda K, Yamasaki M, Tsai Y C. Improved translational efficiency of subtilisin YaB gene with different initiation codons in Bacillus subtilis and alkalophilic Bacillus YaB. J Appl Microbiol. 1997;83:758–763. doi: 10.1046/j.1365-2672.1997.00306.x. [DOI] [PubMed] [Google Scholar]
- 202.Yoshihara K, Kobayashi Y. Retting of Mitsumata bast by alkalophilic Bacillus in paper making. Agric Biol Chem. 1982;46:109–117. [Google Scholar]
- 203.Yoshimatsu T, Ozaki K, Shikata S, Ohta Y, Koike K, Kawai S, Ito S. Purification and characterization of alkaline endo-1,4-β-glucanases from alkalophilic Bacillus sp. KSM-635. J Gen Microbiol. 1990;136:1973–1979. [Google Scholar]
- 204.Yum D Y, Chung H C, Bai D H, Oh D H, Yu J H. Purification and characterization of alkaline serine protease from an alkalophilic Streptomyces sp. Biosci Biotechnol Biochem. 1994;58:470–474. doi: 10.1271/bbb.58.470. [DOI] [PubMed] [Google Scholar]
- 205.Zaghloul T I, AlBahra M, AlAzmeh H. Isolation, identification, and keratinolytic activity of several feather-degrading bacterial isolates. Appl Biochem Biotechnol. 1998;70:207–213. doi: 10.1007/BF02920137. [DOI] [PubMed] [Google Scholar]
- 206.Zhilina T N, Zavarzin G A. Alkaliphilic anaerobic community at pH 10. Curr Microbiol. 1994;29:109–112. [Google Scholar]
- 207.Zhilina T N, Zavarzin G A, Rainey F, Kevbrin V V, Kostrikina N A, Lysenko A M. Spirochaeta alkalica sp. nov., Spirochaeta africana sp. nov., and Spirochaeta asiatica sp. nov., alkaliphilic anaerobes from the Continental soda lakes in Central Asia and the East African Rift. Int J Syst Bacteriol. 1996;46:305–312. doi: 10.1099/00207713-46-1-305. [DOI] [PubMed] [Google Scholar]
- 208.Zhilina T N, Zavarzin G A, Rainey F A, Pikuta E N, Osipov G A, Kostrikina N A. Desulfonatronovibrio hydyogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int J Syst Bacteriol. 1997;47:144–149. doi: 10.1099/00207713-47-1-144. [DOI] [PubMed] [Google Scholar]