Abstract
DNA integration is a unique enzymatic process shared by all retroviruses and retrotransposons. During integration, double-stranded linear viral DNA is inserted into the host genome in a process catalyzed by the virus-encoded integrase (IN). The mechanism involves a series of nucleophillic attacks, the first of which removes the terminal 2 bases from the 3′ ends of the long terminal repeats and of the second which inserts the viral DNA into the host genome. IN specifically recognizes the DNA sequences at the termini of the viral DNA, juxtaposing both ends in an enzyme complex that inserts the viral DNA into a single site in a concerted manner. Small duplications of the host DNA, characteristic of the viral IN, are found at the sites of insertion. At least two host proteins, HMG-I(Y) and BAF, have been shown to increase the efficiency of the integration reaction.
Integration is an obligatory replication step for all retroviruses. The process begins after the virus enters the cell and the RNA genome is reverse transcribed into a double-stranded DNA. The direct precursor to integration is the linear form of viral DNA (4, 64). Circular DNA forms are detected in cells but are considered to be dead-end reverse transcription products (49) or products resulting from autointegration events (63). Linear viral DNA contains at its termini long terminal repeats (LTR) sequences. The ends of these LTRs are specifically recognized by the viral integrase (IN). Preintegration complexes, capable of catalyzing integration in vitro, can be isolated from infected cells (30). These complexes contain linear viral DNA, several viral proteins including matrix (7), reverse transcriptase (63), nucleocapsid (59), and IN (30) and at least two cellular proteins, high-mobility-group [HMG-I(Y)] (29) and barrier to autointegration factor (BAF) (62). Depending upon the retrovirus, preintegration complexes either enter the nuclei of nondividing cells through the nuclear pore (e.g., human immunodeficiency virus [HIV]) or wait until the nuclear membrane dissolves during cell division (e.g., Moloney murine leukemia virus [MoMuLV]) (75). In some retroviruses, nuclear localization signals are associated with various viral proteins, including IN, and facilitate migration to the nucleus (34, 55, 84). Once the preintegration complex associates with the host chromosome, viral IN catalyzes the insertion of the viral sequences into the host DNA. The two LTR ends of the linear viral DNA are brought together into a ternary complex with IN and host DNA, where the insertion occurs in a coordinated or concerted reaction. A 2-bp sequence is lost from each end of the viral DNA, and a short duplication of 4 to 7 bp from the host, depending upon the viral IN, is introduced into the host sequence flanking the viral DNA. Repair of damage to the host DNA by integration is presumably mediated by cellular enzymes. Integrated viral DNA is termed the provirus.
IN was first detected in avian myeloblastosis virus as a nonspecific endonuclease of 32 kDa. Sequences from peptide fragments of the protein showed sequence homology to the β chain of avian sarcoma virus (ASV) reverse transcriptase (77). An active enzyme consists of a multimeric structure of at least a dimer (47). The primary evidence establishing that the endonuclease was encoded in the viral genome came from analysis of an IN mutant ASV, LA335, which was temperature sensitive for replication and possessed a temperature-sensitive DNA endonuclease (36). These observations were confirmed by experiments in which mutations were introduced into conserved residues of IN and blocked replication of the virus (73). The first biochemical evidence linking the DNA endonuclease to integration was the demonstration that the enzyme could specifically cleave viral LTR ends (19). It was subsequently shown that IN could also preferentially bind to the LTR termini of viral DNA (54, 69). However, it was not until DNA oligodeoxynucleotides representing the ends of the viral LTRs were used as substrates for integration in vitro that IN was shown to be both necessary and sufficient to mediate integration (51) and that an energy source such as ATP was not required for the reaction.
STRUCTURE OF INTEGRASE
IN, encoded in the pol gene of the virus, is translated as part of a large Gag-Pol polyprotein and is processed into its mature form by the virus-encoded protease (PR). Based upon evidence from limited protease digestion studies (24) and alignment of the primary sequences of several IN proteins (46), which identified clusters of conserved residues, IN is thought to possess three structural domains. The domains consist of an N-terminal domain of 50 amino acids with a putative zinc binding motif resembling a zinc finger (HHCC), a central domain of 160 amino acids with a D,D(35)E motif, and a less highly conserved C-terminal domain of 80 amino acids. The crystal structures of the central ASV and HIV-1 IN core domains have been solved (6, 20). Nuclear magnetic resonance spectroscopy (NMR) solution structural data is available for the HIV-1 IN N and C-terminal domains (11, 12, 21, 22, 65). Figure 1 displays each of the known fragment structures of HIV-1 IN. Unfortunately, there is no structural data for an intact protein with or without a substrate mimic, so that we do not know how these fragments fit together in the holoenzyme.
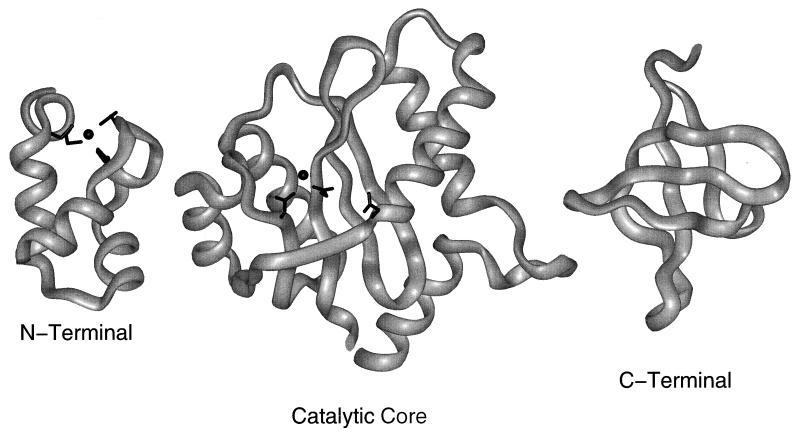
FIG. 1.
Ribbon diagrams of the structures of HIV-1 IN fragments. The N-terminal domain of HIV IN is shown on the left. It is based upon PDB file 1WJA (11), coordinating a single zinc cation (black sphere) by using the HHCC motif. This motif consists of residues His12, His16, Cys40, and Cys43, depicted as black sticks. The core domain, center, is based upon 1B14 (68), showing the active form of the HIV IN catalytic domain. The active site has a D,D(35)E motif. Residues Asp116, Asp64, and Glu152 are depicted as black sticks, in order from left to right. Side chains of the two Asp residues bind a single magnesium cation, shown here as a black sphere. The C-terminal DNA binding domain is based upon coordinates in PDB file 1IHV (65). The N- and C-terminal domains were solved by NMR; the catalytic core domain was solved by X-ray diffraction.
N-Terminal Domain
Within the N-terminal domain of IN is a putative zinc finger of the HHCC type (Fig. 1). Amino acid substitutions in any or all of the HHCC residues in HIV-1 IN (24, 25), nearly or completely abolish end-processing and joining reactions in vitro and in vivo, respectively. These results imply that the HHCC motif is important for IN catalytic function, but they do not demonstrate whether these residues are involved in coordinating zinc. Using a zinc binding assay, Bushman et al. (8) reported that wild-type HIV IN binds zinc. Moreover, a substitution at any of the HHCC residues reduced the level of zinc binding. Recently a solution structure of the N-terminal domain was determined and revealed a dimeric structure having an HHCC zinc binding motif that coordinates zinc. The folds of the N termini are similar to those of other DNA binding proteins in having a helix-turn-helix structural motif (12). N-terminal deletions of the first 49 residues also adversely affected the Mg2+-dependent catalytic activities of IN (24). However, a truncated Rous sarcoma virus IN containing residues 54 to 286 was still capable of end-processing and limited joining reactions in the presence of Mn2+ (9). Mn2+ ions had previously been shown to enhance the rate of 3′-end processing but in a nonspecific manner (51). N-terminal truncations of IN are also partly capable of reversing the integration process in a reaction referred to as disintegration (8). Taken together, these results suggest that the N terminus influences the catalytic activity of IN but does not contain its catalytic core. Moreover, Andrake and Skalka (2) using chemical cross-linking and size exclusion chromatography, determined that the N terminus of ASV IN was not involved in multimerization, since an N-terminally truncated fragment containing only residues 39 to 286 formed multimeric structures.
Central Core Domain
The central core domain comprises residues 50 to 235 and has been shown to coordinate divalent cations. The crystal structures of the catalytic core domains for HIV-1 (20) and ASV (6) have been solved. Crystallization of the HIV-1 core was dependent upon a F185K substitution, which increased its solubility (20, 44, 45); this is shown in Fig. 1. While an intact IN protein is required for complete activity, an ASV fragment is capable of end processing but not joining. The HIV-1 core fragment is not capable of either reaction (6, 40). Both cores, however, catalyze the disintegration reaction (6, 40, 45). The overall folded structure of the catalytic core is similar to that of nucleases such as Escherichia coli RNase H, the HIV-1 RNase H domain of reverse transcriptase and E. coli RuvC (6, 74, 88).
The central core is thought to be the catalytic domain of the enzyme. Using chimeric IN proteins, Katzman and Sudol proposed that the DNA binding domain of the conserved CA dinucleotide in the viral LTR resides in the catalytic core, implying that the core contains the active site (52). The core domain contains a catalytic triad of three highly conserved residues, D,D(35)E. The triad for ASV is D64, D121, and E157. Substitutions of any of these residues generally abolish end-processing and/or joining reactions (57). The comparable triad for HIV-1 IN is D64, D116, and E152 (Fig. 1). Here again, most substitutions inactivate the enzyme, with two exceptions: a D116N substitution results in a more active HIV-1 IN than wild type in vivo (25), and an enzyme with D116E and E152D substitutions, while incapable of end processing and joining, catalyzes detectable disintegration reactions (24). Crystal structures of the catalytic core, coordinating a divalent cation, have been determined for ASV by using Mg2+ and Mn2+ (5) and for HIV by using Mg2+ (35, 68). The divalent cations were found to be coordinated by the two conserved aspartic acid residues of the catalytic triad. A comparison of HIV-1 and ASV cores indicated that the two aspartic acid residues are similarly positioned within the respective HIV-1 and ASV structures (5, 6, 20, 35, 68). A relatively extensive mutational analysis of conserved residues in the catalytic core of the intact IN has been performed (18, 20, 24, 44, 45, 53, 57, 58, 60). While some of the substitutions affect end-processing, joining, and disintegration reactions to various extents, others are less disruptive and in some cases allow end processing but prevent joining. These results imply that not all conserved residues in the core are essential to catalytic activity.
C-Terminal Domain
The C terminus of IN is the least highly conserved of the three domains (13, 67). An HIV-1 fragment representing residues 235 to 288 binds nonspecifically to DNA (26, 53, 66, 71, 82, 87). The structure of the C-terminal domain of HIV-1 IN, residues 220 to 270, has been determined by NMR (21, 65) and contains an SH3 fold motif (Fig. 1). Such structures have been found in a number of proteins but only in one other protein involved in DNA binding, Sso7d (3, 21, 65). In addition, multimerization of IN appears to be defective, suggesting that the C terminus of the protein is needed for dimerization or multimerization of the enzyme. Nuclear localization signals, which facilitate the entry of preintegration complexes into the nucleus, have been mapped to the C-terminal IN domain of several retroviruses and transposons. In ASV IN, the nuclear localization signal contains both basic and proline residues (55).
MULTIMERIZATION OF INTEGRASE
Several groups have examined the parameters of multimerization both in vitro and in vivo (23, 32, 80). While truncated N- or C-terminal HIV-1 IN fragments are incapable of supporting end-processing or joining reactions, a mixture of N-terminally truncated IN and C-terminally truncated IN has detectable activity (23, 80). These results imply that HIV-1 integrase is active as a dimer or higher-order multimer and that only a single N- or C-terminal domain is required for activity. In other mixing experiments, IN proteins containing single substitutions in the catalytic triad of the core domain could be complemented by IN proteins containing truncations to either the N- or C-terminal domains (23, 80). By using IN monomers having truncations in the N or C terminus, cis to the catalytic core and complemented with monomers having the missing truncated region, it was shown that the C-terminal domains cis to the catalytic site were active in end-processing and joining reactions in vivo whereas N-terminal truncations were not active in cis (23, 80).
MECHANISM OF RETROVIRAL INTEGRATION
Integration occurs in two well-characterized catalytic steps, referred to as end processing and joining, respectively. End processing involves removal of a dinucleotide, adjacent to a highly conserved CA dinucleotide, from the 3′ strand of the U3 and U5 viral DNA LTRs in a reaction involving a water molecule or other nucleophile (27, 83) (Fig. 2). This exposes a 3′ hydroxyl group, whose oxygen is used as an attacking nucleophile on the target DNA during the joining reaction, in which the viral DNA is inserted into the cellular DNA (27, 83). Engleman et al. identified three different forms of the cleaved dinucleotide resulting from end processing, depending upon the nucleophile used in the reaction (27). The most abundant form was a dinucleotide with a 5′ phosphate and a 3′ hydroxyl, which would arise when a water molecule acted as the nucleophile. It is believed that a Mg2+ atom coordinated [through the conserved D,D(35)E residues] in the active site of IN facilitates the deprotonation of the water to activate it as a nucleophile. This mechanism is analogous to the polymerization reaction catalyzed by DNA polymerases, including reverse transcriptase, or to activity catalyzed by adenosine cyclase (48, 79, 89), although in this case IN is believed to coordinate one rather than two Mg2+ atoms. A second dinucleotide product was detected when glycerol, introduced together with IN, acted as the nucleophile. This species was poorly phosphorylated and was resistant to phosphatase activity, implying the absence of a 5′ phosphate (27, 83). The third dinucleotide was identified as a 3′-5′ cyclic pGpTOH. This form arises when the hydroxyl of the terminal T acts as the attacking nucleophile to break a phosphodiester bond on the same strand (27).
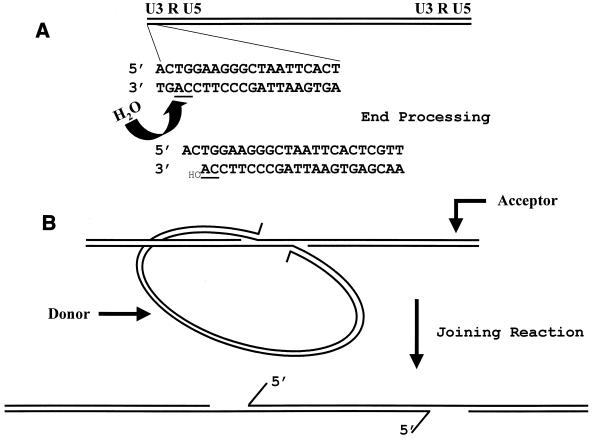
FIG. 2.
Diagrammatic representation of the mechanism of integration. (A) End-processing reaction. A viral DNA is depicted with the U3 LTR sequence on the left. Magnified below is the 20-bp sequence of the terminal HIV-1 U3 LTR. In an initial reaction, there is a loss of 2 bases from the 3′ strand, adjacent to a highly conserved CA dinucleotide (underlined), via a nucleophilic attack by a water molecule. (B) Joining reaction. The processed LTR DNA ends are brought together into a complex with the target DNA. Insertion of the donor into the target DNA involves a nucleophilic attack by using the 3′ hydroxyl groups on the exposed 3′ strand. The result is a gapped DNA with 5′ 2-base overhangs. Overhang removal, gap, and nick repair would complete the integration reaction.
The actual mechanism of viral DNA insertion was shown, by use of a phosphothioate-substituted target, to invert the chirality of the phosphate at the site of insertion (27). This indicates that a single-step transesterification reaction occurs and eliminates the possibility that IN forms a covalent intermediate with DNA. Such enzyme-DNA intermediates are found for topisomerases (14) and for IN of lambda bacteriophage (70, 81), where the chirality of the electrophile phosphate is maintained in joining the DNA.
The IN recognition sequence within the LTRs is relatively short. MuLV IN recognizes 11 to 12 bp (76), ASV IN recognizes 15 bp (51), and HIV-1 IN recognizes 20 bp (78). Cross-linking and substitution of bases in the LTR of HIV-1 have demonstrated that specific interactions between IN and the terminal LTR sequences are required for end-processing and joining reactions (28, 38, 39). The sequences at the U5 and U3 LTR ends are both derived from nearly perfect inverted repeats. Addition or deletion of sequences at or near the conserved CA dinucleotide at the termini of the LTRs alters the efficiency of DNA integration (17). For example, addition of sequence 3′ to the conserved CA dinucleotide in MuLV LTRs resulted in a delay in growth of the virus by several days. However, subsequent passages of the mutant virus selected for viruses with wild-type growth rates and concomitant nucleotide changes in the LTR sequences. One revertant contained a C-to-A transition 4 bases from the 3′ termini; another contained an 11-base deletion such that integration utilized a new internal CA dinucleotide. Replacement of 2 bases in the ASV U5 LTR adjacent to the CA dinucleotide or 4 bases adjacent to and including the C of the CA dinucleotide also decreased the efficiency of integration and resulted in a delayed-growth phenotype (16).
EARLY IN VITRO-RECONSTITUTED INTEGRATION SYSTEMS
Fujiwara and Craigie (33) described the first cell-free integration system that used extracts of MoMuLV, a large linear donor DNA containing modified LTR termini with 5′ overhangs, and lambda DNA as the target. The donor DNA was constructed by placing an NdeI cleavage site between tandemly linked U5 and U3 LTR termini inserted in a plasmid. After restriction digestion, a linear donor was produced with LTR termini that differed from wild-type LTR termini by 1 bp and contained 5′ 2-base AT overhangs rather than blunt ends. Katz et al. (50) used purified ASV IN to reconstitute integration with a similar NdeI-constructed ASV donor DNA. Integrants in both systems were analyzed by packaging the lambda acceptor DNA, introducing the phage into cells, plaque purifying, and sequencing. Integrants displayed properties characteristic of in vivo retroviral integration, including the loss of 2 bp from the ends of both LTR termini, short duplications of the lambda DNA at the site of integration, and random distribution of integration sites.
CONCERTED DNA INTEGRATION
Murphy and Goff (72) demonstrated that when deletions were placed in the U3 LTR, 5′ to the conserved CA dinucleotide, end processing in both U3 and U5 LTRs was adversely affected. This result implies that integration in vivo occurs by a concerted mechanism in which the two LTR ends of the viral DNA are inserted into a single target site, such that a mutation in one LTR can influence the processing of the other. Unfortunately, oligodeoxynucleotide model substrates do not exhibit the concerted properties characteristic of in vivo integration. One exception was described by Kukolj and Skalka (56), who designed short duplex substrates whose sequences matched those of U3 and U5 ends of ASV and HIV-1 DNA but were covalently synapsed across the termini by short single-stranded linkers. These substrates were used more efficiently than were unlinked oligodeoxynucleotides duplexes. Moreover, substrates with a paired wild-type and mutated terminus were cleaved poorly at both ends, indicating that when termini were juxtaposed, the processing of both ends displayed concerted behavior. By using tethered donor molecules, the optimum spacing for the ASV system was shown to be 2 nucleotides. This placed the two conserved CA dinucleotide-processing sites 6 nucleotides apart, a separation equal to the staggered break introduced into the target DNA. The optimum separation of the HIV-1 conserved CA dinucleotides was 5 nucleotides, again matching the staggered break introduced into the target DNA in vivo. If both strands were tethered, the efficiency of the reaction was considerably decreased, due in part to a loss of torsional flexibility imparted by the gap in one strand. These results provided biochemical evidence that molecular communication must take place between IN bound to both viral DNA ends.
RECONSTITUTION OF AVIAN SARCOMA VIRUS CONCERTED DNA INTEGRATION
A concerted integration system using purified IN was reported by Fitzgerald et al. (31). The system included purified AMV IN; lambda as the linear acceptor DNA; a donor DNA, 3.4 kb in length, containing 30 bp from the ends of the LTRs with preprocessed 5′ AT overhangs; and a supF suppressor gene as a selectable marker. The presence of the supF tRNA gene in the mini-donor DNA provides for genetic selection of individual integrants in bacterial cells containing an expression vector with antibiotic resistance genes with amber mutations in the coding sequences. The linear donor was again constructed by using the NdeI restriction enzyme site (31). A small percentage of the integrants isolated from the reaction exhibited concerted integration and displayed properties expected of ASV DNA integration in vivo. However, the remainder arose through nonconcerted integration events, which produced deletions in the acceptor DNA. Vora et al. (86) established a similar system by using a 487-bp donor DNA that was also with precleaved NdeI ends. Here again, products arose from both concerted and nonconcerted reactions. Further analysis of the concerted integration products showed that they resulted from two one-ended integration events by different donor DNAs into the same acceptor rather than from both ends being provided by the same donor. As a consequence, U3-U5, U3-U3, or U5-U5 donor combinations were detected, complicating the analysis of the integration product (31, 86). In subsequent reports, changing of the buffer conditions (31, 85, 86) improved the overall efficiency of integration but with different percentages of concerted products detected.
The ASV reconstituted system that appears to most closely approximate the concerted integration in vivo uses purified recombinant ASV IN, a 3.4-kb supercoiled or linear target DNA, and a “mini” linear donor DNA substrate of only 294 bp with blunt authentic unprocessed viral termini (1, 42). This small size was chosen to maximize the probability that ends from the same donor would come in contact to facilitate concerted integration. The donor contains a supF suppressor tRNA gene flanked by only 15 bp from the ASV LTR termini. To increase the integration efficiency of this system, a host cell protein was also added to the reaction mixture by Aiyar et al. (1). The host cell protein was from the HMG family and has the ability to bend DNA that could assist in juxtaposing the U3 and U5 LTR termini in an integration complex. This bending of the donor DNA could favor concerted DNA integration. While only a small percentage of the donor DNA was integrated into the acceptor in this system, more than 90% of the integrants detected used a concerted mechanism with a single donor molecule (1). Removal of either the U3 or the U5 LTR sequences from the donor substrate resulted in a substantial reduction in the total number of integration products detected.
HMG-1 was the initial HMG protein family member added to this system. It stimulated the integration reaction about fourfold compared with the activity observed in the presence of IN alone (1). Subsequently, Farnet and Bushman (29) reported that an HMG protein family member, HMG-I(Y), could be detected in HIV-1 preintegration complexes isolated from infected-cells and that integration was dependent upon the continued presence of HMG-I(Y). The addition of HMG-I(Y) to the ASV mini-donor DNA reconstituted system stimulated integration by more than 10-fold, with the mini-donor DNA being integrated via a concerted mechanism during the course of the reaction (42). Individual integrants, isolated from reactions reconstituted in the presence of HMG-1 or HMG-I(Y), showed end processing, concerted insertion with base pair duplication of acceptor DNA flanking the integrated donor DNA, and non-sequence-specific integration of the donor into the target, all characteristic of in vivo integration (1, 42).
MUTATIONS IN THE U5 OR U3 AVIAN SARCOMA VIRUS LONG TERMINAL REPEATS INFLUENCE INTEGRATION
The value of reconstituted systems lies in the ability to rapidly analyze mutations that influence integration. In ASV IN, the percentage of nonconcerted integration events can be increased by introducing base changes into the LTR sequences (1, 41, 85), which presumably alter the binding affinity of IN for the LTR recognition sequences. By changing the reaction conditions to favor IN-DNA contacts, the concerted nature of the integration can be rescued and individual integrants can be sequenced (41). When a 4-bp substitution was placed in the ASV U5 LTR, changing CTTCATT to GAAGATT, it resulted in a slight decrease in the efficiency of integration activity compared to that for a donor with a wild-type U5 LTR (1). However, one of every seven integrants sequenced contained deletions in the LTRs. In one case, 10 bases were removed from the U5 LTR, so that IN used the first internal CA dinucleotide for the nucleophilic attack. In a second integrant, IN left the mutation in the U5 LTR but deleted sequences in the wild-type U3 LTR, utilizing the first internal GA dinucleotide to drive the integration reaction (1). This latter result reproduced genetic changes observed in vivo (72) when a mutation placed at one LTR altered the processing of the other. Note also that mutations placed in U3 and U5 have similar effects on integration in terms of specificity. However, mutations placed in U3 have a threefold more deleterious effect on the efficiency of integration in vitro than do the same mutations introduced into the U5 LTR (41, 51, 85).
RECONSTITUTION OF HUMAN IMMUNODEFICIENCY VIRUS CONCERTED DNA INTEGRATION
Goodarzi et al. (37) have reported a reconstituted concerted HIV-1 IN-dependent integration system involving a 469-bp donor DNA constructed with the NdeI preprocessed ends. This system still exhibits insertion of two donor DNA molecules into a target, and only about half of the integrants resulted from a concerted DNA mechanism. An HIV-1 mini-donor DNA integration system comparable to the ASV system described by Aiyar et al. (1) has also been developed by using recombinant HIV-1 IN (42). This system appears to approximate integration in vivo more closely. The HIV-1 donor DNA contains only 20 bp of the HIV-1 LTR termini flanking the supF suppressor gene and uses the same target DNA and HMG protein family members as does the ASV system. In contrast to the ASV reconstituted system, where 60% of the sequenced integrants had 6-bp acceptor DNA duplications characteristic of ASV integration in vivo, the duplication of the acceptor DNA at the site of HIV-1 donor integration was almost exclusively 5 bp, characteristic of HIV-1 integration in vivo (42). The differences in base pair duplications in the ASV and HIV-1 reconstituted systems may reflect differences in the stability or conformational characteristics of protein-protein interactions among the respective ASV and HIV-1 IN dimers or tetramers, which form complexes with the ends of the donor and the acceptor DNA. Such differences could influence the spacing of staggered breaks introduced into the acceptor DNA, thereby altering the size of the duplications. The HIV-1 IN-dependent reactions also differ from those of ASV in that less than half or approximately 75% of the HMG-2 and HMG-I(Y) integrants examined, respectively, resulted from a concerted mechanism. The remainder resulted from multiple independent one-ended donor integration events that produce deletions in the target DNA (42). While all of the HMG proteins tested stimulated integration in vitro, HMG-I(Y) yielded the most concerted DNA integration products, consistent with the finding of this protein in HIV-1 preintegration complexes (29).
CELLULAR PROTEINS
HMG proteins represent a large family of nonhistone DNA binding proteins which are localized primarily in the nucleus of eukaryotic cells and which can be extracted in the presence of acid and salts. The proteins are classified into several families including HMG-1/-2, HMG-14/-17, and HMG-I(Y). The HMG proteins are relatively small, ranging from approximately 11 kDa for HMG-I(Y) and HMG-14/-17 to 25 kDa for HMG-1/-2, and are known to modulate chromatin structure and function (10). HMG proteins have common functional features, including (i) binding to the minor groove of double-stranded DNA; (ii) recognizing DNA structure rather than sequence; (iii) preferentially interacting with bent, supercoiled, or distorted DNA structures; (iv) binding to non-B-form DNA structures such as four-way-junctions and cisplatin adducts; (v) unwinding, bending, and supercoiling DNA substrates in the absence of ATP hydrolysis; and (vi) selectively interacting with other sequence-specific transcription factors as part of gene transcription regulatory complexes.
HMG protein family members increase the efficiency of integration in vitro by acting on the donor DNA (1) without forming stable complexes with IN or the LTR IN recognition sequences, as judged from gel shift and coprecipitation experiments (42). A truncated HMG-I(Y) protein (Δ50–90), which preserves the region of HMG-I(Y) that binds most tightly to substrate DNAs (43), is capable of stimulating integration as well as wild-type HMG-I(Y) does. In contrast, another mutant of HMG-I(Y) (II, III) which has several point mutations preventing protein-DNA interactions while retaining protein-protein interactions, does not stimulate integration in vitro. Taken together, these results imply that HMG-I(Y) needs to associate with the DNA to stimulate integration.
In addition to HMG proteins, another host protein may be essential for in vivo integration. BAF functions as a required factor for efficient integration by preventing autointegration. Lee and Coffin first noted that MoMuLV was resistant to autointegration and hypothesized that the lack of autointegration was due to incomplete reverse transcription (63). Lee and Cragie found that a cellular protein, BAF, prevented autointegration, and Chen and Engleman demonstrated that BAF could stimulate integration, implying that BAF maintains a competent integration complex by binding the viral DNA into a “open-mesh” complex (15, 61, 62).
PROSPECTIVE
With the availability of reconstituted concerted DNA integration assays that closely mimic the genetic complexities of integration in vivo, our understanding of the basic mechanism of integration should dramatically increase. Moreover, these systems will provide the means to screen for drugs targeted at HIV-1 IN, which will potentially open a third avenue of therapy to attack HIV-1-induced AIDS.
ACKNOWLEDGMENTS
We thank Mark Andrake, Fox Chase Cancer Center, Philadelphia, Pa., for helpful comments in revising this manuscript and Jerry Alexondratos, Macromolecular Structure Laboratory, NCI-FCRDC, Frederick, Md., for creating the HIV-1 IN structural image.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 3.Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 5.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 6.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman F D, Wang B. Rous sarcoma virus integrase protein: mapping functions for catalysis and substrate binding. J Virol. 1994;68:2215–2223. doi: 10.1128/jvi.68.4.2215-2223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 11.Cai M, Huang Y, Caffrey M, Zheng R, Craigie R, Clore G M, Gronenborn A M. Solution structure of the His12→Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium. Protein Sci. 1998;7:2669–2674. doi: 10.1002/pro.5560071221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 13.Cannon P M, Byles E D, Kingsman S M, Kingsman A J. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J Virol. 1996;70:651–657. doi: 10.1128/jvi.70.1.651-657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champoux J J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci USA. 1977;74:3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colicelli J, Goff S P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 18.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 19.Duyk G, Leis J, Longiaru M, Skalka A M. Selective cleavage in the avian retroviral long terminal repeat sequence by the endonuclease associated with the alpha beta form of avian reverse transcriptase. Proc Natl Acad Sci USA. 1983;80:6745–6749. doi: 10.1073/pnas.80.22.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 21.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 22.Eijkelenboom A P, van den Ent F M, Vos A, Doreleijers J F, Hard K, Tullius T D, Plasterk R H, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 23.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 28.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 30.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fletcher T M, III, Soares M A, McPhearson S, Hui H, Wiskerchen M, Muesing M A, Shaw G M, Leavitt A D, Boeke J D, Hahn B H. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16:5123–5138. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara T, Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci USA. 1989;86:3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 35.Goldgur Y, Dyda F, Hickman A B, Jenkins T M, Craigie R, Davies D R. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Natl Acad Sci USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golomb M, Grandgenett D P, Mason W. Virus-coded DNA endonuclease from avian retrovirus. J Virol. 1981;38:548–555. doi: 10.1128/jvi.38.2.548-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodarzi G, Im G J, Brackmann K, Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heuer T S, Brown P O. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 39.Heuer T S, Brown P O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 40.Hickman A B, Palmer I, Engelman A, Craigie R, Wingfield P. Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J Biol Chem. 1994;269:29279–29287. [PubMed] [Google Scholar]
- 41.Hindmarsh, P., and J. Leis. 1998. Unpublished observations.
- 42.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javaherian K, Sadeghi M, Liu L F. Nonhistone proteins HMG1 and HMG2 unwind DNA double helix. Nucleic Acids Res. 1979;6:3569–3580. doi: 10.1093/nar/6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins T M, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 45.Jenkins T M, Hickman A B, Dyda F, Ghirlando R, Davies D R, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones K S, Coleman J, Merkel G W, Laue T M, Skalka A M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 48.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 49.Junghans R P, Boone L R, Skalka A M. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J Virol. 1982;43:544–554. doi: 10.1128/jvi.43.2.544-554.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 51.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katzman M, Sudol M. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J Virol. 1998;72:1744–1753. doi: 10.1128/jvi.72.3.1744-1753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knaus R J, Hippenmeyer P J, Misra T K, Grandgenett D P, Muller U R, Fitch W M. Avian retrovirus pp32 DNA binding protein. Preferential binding to the promoter region of long terminal repeat DNA. Biochemistry. 1984;23:350–359. doi: 10.1021/bi00297a026. [DOI] [PubMed] [Google Scholar]
- 55.Kukolj G, Katz R A, Skalka A M. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene. 1998;223:157–163. doi: 10.1016/s0378-1119(98)00169-3. [DOI] [PubMed] [Google Scholar]
- 56.Kukolj G, Skalka A M. Enhanced and coordinated processing of synapsed viral DNA ends by retroviral integrases in vitro. Genes Dev. 1995;9:2556–2567. doi: 10.1101/gad.9.20.2556. [DOI] [PubMed] [Google Scholar]
- 57.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 61.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee M S, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee Y M, Coffin J M. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobel L I, Murphy J E, Goff S P. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J Virol. 1989;63:2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 66.Lutzke R A, Plasterk R H. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lutzke R A, Vink C, Plasterk R H. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maignan S, Guilloteau J P, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 69.Misra T K, Grandgenett D P, Parsons J T. Avian retrovirus pp32 DNA-binding protein. I. Recognition of specific sequences on retrovirus DNA terminal repeats. J Virol. 1982;44:330–343. doi: 10.1128/jvi.44.1.330-343.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 71.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinn T P, Grandgenett D P. Genetic evidence that the avian retrovirus DNA endonuclease domain of pol is necessary for viral integration. J Virol. 1988;62:2307–2312. doi: 10.1128/jvi.62.7.2307-2312.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice P, Craigie R, Davies D R. Retroviral integrases and their cousins. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 75.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 77.Schiff R D, Grandgenett D P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978;28:279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steitz T A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 80.van Gent D C, Vink C, Groeneger A A, Plasterk R H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Mansfeld A D, van Teeffelen H A, Baas P D, Jansz H S. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986;14:4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vink C, Oude Groeneger A M, Plasterk R H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vink C, Yeheskiely E, van der Marel G A, van Boom J H, Plasterk R H. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991;19:6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vora A C, Chiu R, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats. Role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 86.Vora A C, McCord M, Fitzgerald M L, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Woerner A M, Klutch M, Levin J G, Marcus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 88.Yang W, Steitz T A. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 89.Zimmermann G, Zhou D, Taussig R. Mutations uncover a role for two magnesium ions in the catalytic mechanism of adenylyl cyclase. J Biol Chem. 1998;273:19650–19655. doi: 10.1074/jbc.273.31.19650. [DOI] [PubMed] [Google Scholar]