Abstract
Replication of the 30-kilobase genome of SARS-CoV-2, responsible for COVID-19, is a key step in the coronavirus life cycle that requires a set of virally encoded nonstructural proteins such as the highly conserved Nsp13 helicase. However, the features that contribute to catalytic properties of Nsp13 are not well established. Here, we biochemically characterized the purified recombinant SARS-CoV-2 Nsp13 helicase protein, focusing on its catalytic functions, nucleic acid substrate specificity, nucleotide/metal cofactor requirements, and displacement of proteins from RNA molecules proposed to be important for its proofreading role during coronavirus replication. We determined that Nsp13 preferentially interacts with single-stranded DNA compared with single-stranded RNA to unwind a partial duplex helicase substrate. We present evidence for functional cooperativity as a function of Nsp13 concentration, which suggests that oligomerization is important for optimal activity. In addition, under single-turnover conditions, Nsp13 unwound partial duplex RNA substrates of increasing double-stranded regions (16–30 base pairs) with similar efficiency, suggesting the enzyme unwinds processively in this range. We also show Nsp13-catalyzed RNA unwinding is abolished by a site-specific neutralizing linkage in the sugar–phosphate backbone, demonstrating continuity in the helicase-translocating strand is essential for unwinding the partial duplex substrate. Taken together, we demonstrate for the first time that coronavirus helicase Nsp13 disrupts a high-affinity RNA–protein interaction in a unidirectional and ATP-dependent manner. Furthermore, sensitivity of Nsp13 catalytic functions to Mg2+ concentration suggests a regulatory mechanism for ATP hydrolysis, duplex unwinding, and RNA protein remodeling, processes implicated in SARS-CoV-2 replication and proofreading.
Keywords: COVID-19, coronavirus, SARS-CoV-2, helicase, replication
Abbreviations: AMP-PNP, adenyl-imidodiphosphate; ATPγS, adenosine 5′-O-(3-thio)triphosphate; bp, base pair; BME, mercaptoethanol; BSA, bovine serum albumin; CoV, coronavirus; ds, double strand; eIF4A, eukaryotic initiation factor 4A; HCV, hepatitis C virus; MERS, Middle East respiratory syndrome; Nsp, nonstructural; PEG, poly(hexa)ethylene glycol; NTP, nucleoside triphosphate; PNK, T4 polynucleotide kinase; RT, room temperature; RTC, replication–transcription complex; SARS, severe acute respiratory syndrome; ss, single strand; TBK1, Tank-binding kinase 1
The COVID-19 pandemic caused by the novel severe acute respiratory syndrome (SARS)-coronavirus (CoV)-2 has necessitated prompt action by biomedical researchers to study essential steps in the coronavirus life cycle and characterize the molecular interactions and biochemical functions of the structural and nonstructural protein (Nsp)s encoded by the SARS-CoV-2 genome. Among the key players in SARS-CoV-2 replication is the virally encoded RNA helicase Nsp13. Nsp13, in complex with the Nsp12 RNA-dependent RNA polymerase, processivity factors (Nsp7, Nsp8), and proofreading exonuclease (Nsp14), presumably coordinates with host cell factors in a still not well understood mechanism to replicate the coronavirus RNA genome (1, 2, 3). Structural studies of the SARS-CoV-2 replication complex suggest the dimeric form of Nsp13 helicase exists at the leading edge of a multiprotein complex (Nsp7-8-12-14) with template-primer RNA poised to conduct nascent RNA synthesis (1). Multiple studies have addressed structural properties of the Nsp13 helicase in the context of the SARS-CoV-2 replication–transcription complex (RTC) (1, 2, 3, 4, 5) or in isolation (6). However, mechanistic details of Nsp13’s catalytic functions in SARS-CoV-2 coronavirus replication, or its proofreading activity in which Nsp13 is implicated, are lacking.
Recombinant Nsp13 helicases encoded by the sequence-related MERS-CoV (7), SARS-CoV-1 (8, 9, 10, 11, 12, 13, 14), and SARS-CoV-2 (6, 15, 16) genomes, all expressed in bacterial systems, have been biochemically characterized. Nsp13 helicases from SARS-CoV and SARS-CoV-2 differ by just one amino acid at residue 570 (isoleucine in SARS-CoV and valine in SARS-CoV-2) (17). An alignment of Nsp13 from SARS BJ01 and Bat SARS-like coronavirus shows over 99% similarity as well (18). Nsp13 has a low mutation rate with only 23 mutations observed in a sample of 439 SARS-CoV-2 viral strains (18). A BLASTp alignment of the MERS (QKF93417.1) and SARS-CoV-2 (YP_009725308.1) Nsp13 amino acid sequences revealed 85% positives and 72% identities over 590 amino acids indicating significant sequence conservation.
Purified recombinant SARS-CoV-1 Nsp13 was shown to be active as a DNA helicase (13) or RNA helicase (9) on a partial duplex substrate with a 5ʹ single-strand (ss) overhang, but not a 3ʹ ss overhang. These results suggest a 5ʹ to 3ʹ directionality of unwinding via its unidirectional translocation on the partial duplex with the 5ʹ ss overhang, where the helicase loads. Recombinant SARS-CoV-1 Nsp13 helicase was reported to unwind double-stranded (ds) DNA and dsRNA substrates with similar efficiency (8, 9), whereas a more recent study found that a recombinant SARS-CoV-1 Nsp13 helicase domain fragment displayed strong preferential unwinding of a 5ʹ ssDNA-tailed DNA duplex substrate compared with a 5ʹ ssRNA-tailed RNA duplex substrate (10). Consistent with the latter findings, Mickolajczyk et al. (15) provided kinetic evidence from bulk measurements using a recombinant SARS-CoV-2 Nsp13 protein that a 5ʹ ss-tailed duplex (18-bp) consisting entirely of DNA in both strands was unwound with an ∼2- to 3-fold greater rate than the same sequence substrate consisting entirely of RNA in both strands.
In the current work, we have undertaken a biochemical analysis of a purified recombinant SARS-CoV-2 Nsp13 helicase expressed using a baculovirus–insect cell system, which provides fitting surroundings for posttranslational modifications of eukaryotic expressed proteins (19). During viral infection, SARS-CoV-2 proteins are posttranslationally modified, suggesting regulatory mechanisms (20). Nsp13 directly binds to Tank-binding kinase 1 (TBK1) and regulates its activation (21), but whether Nsp13 is phosphorylated by TBK1 has not been examined. Nsp13 also interacts with the deubiquitinating enzyme USP13, which is proposed to stabilize Nsp13 via its deubiquitylation (22). Such reports suggest that Nsp13 posttranslational modifications in the eukaryotic host are relevant to its stability and function.
Here, we focus on Nsp13’s catalytic activity and nucleic acid substrate specificity. These studies provide new insights into the molecular functions of SARS-CoV-2 Nsp13 by clarifying the critical substrate element dictating nucleic acid preference and the requirements governing the delicate balance of nucleotide:metal cofactor ratio for RNA helicase activity. Our studies demonstrate a novel RNA–protein remodeling function of Nsp13 not previously reported for coronavirus helicases that is relevant to the proposed role of Nsp13 in the SARS-CoV-2 proofreading mechanism.
Results
Nsp13 RNA helicase activity characterization under multiple-turnover conditions
To generate recombinant SARS-CoV-2 Nsp13 protein (hereafter referred to as Nsp13), the Nsp13 coding sequence was synthesized with an additional sequence and inserted into pFastBac HTB vector creating pFastBac-HTB-Nsp13-FLAG containing a sequence coding for an N-terminal cleavable 6xHis tag and a C-terminal FLAG tag. This allowed for purification of full-length product and allowed for the removal of the 6xHis tag. pFastBac-HTB-Nsp13-FLAG was transformed into DH10Bac competent cells to generate baculovirus DNA, which was transfected and amplified in Sf9 insect cells to generate high-titer baculovirus. We infected Hi5 insect cells with the high-titer Nsp13 baculovirus and harvested cells. Cell pellets were stored at −80 °C, thawed on ice and lysed, centrifuged and lysate filtered before loading onto a 1-ml HisTrap HP column. The column was washed, and the protein was eluted off the HisTrap HP column with 400 mM imidazole. The eluted protein was then dialyzed to remove imidazole, and TEV protease was added to remove the N-terminal 6xHis tag. Following removal of 6xHis tag, the sample was bound to anti-FLAG M2 affinity beads. The beads were washed, and protein was eluted twice using 3× FLAG peptide. Both eluted fractions were collected by gravity flow, pooled and dialyzed against Storage Buffer, and stored at −80 °C. See Experimental procedures for details.
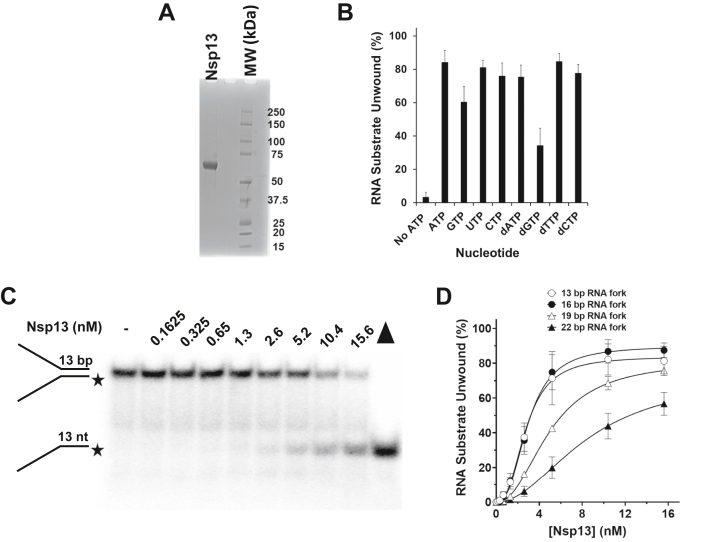
To begin characterizing the purified recombinant Nsp13 (Fig. 1A), we first tested its nucleoside triphosphate (NTP) dependence for unwinding a short 13-base pair (bp) RNA forked duplex substrate resembling the structure of a partial duplex RNA molecule during unwinding. Reaction conditions were similar to those used for the SARS-CoV-1 Nsp13 helicase (13). At an Nsp13 concentration of 5.2 nM, unwinding of greater than 75% of the RNA forked duplex substrate (0.25 nM) was achieved during the 15-min incubation in the presence of all ribo- and deoxyribonucleotides except GTP and dGTP, which displayed 60% ± 9 and 34% ± 10, respectively (Fig. 1B). The omission of NTP showed little to no Nsp13 helicase activity on the 13-bp RNA forked duplex substrate.
Figure 1.
Biochemical analysis of RNA helicase activity by purified recombinant Nsp13 protein as a function of nucleotide and duplex length.A, purified recombinant Nsp13 protein (5000 ng) after final dialysis (see Experimental procedures), resolved by SDS-PAGE and detected by Coomassie staining. B, nucleoside triphosphate requirements for Nsp13 helicase activity. Nsp13 (2.6 nM) was incubated with a partial duplex (13-bp) RNA substrate (0.25 nM) and the indicated nucleotide (2 mM) and 5 mM Mg2+ for 15 min at 37 °C. Reaction products were analyzed by native polyacrylamide gel electrophoresis and quantified as described under Experimental procedures. C and D, Nsp13 helicase activity as a function of protein concentration under multiple-turnover conditions. C, representative native polyacrylamide gel analysis of products from Nsp13 reaction mixtures with a 13-bp RNA partial duplex substrate conducted in the presence of 2 mM ATP and 5 mM Mg2+ for 15 min at 37 °C. Filled triangle represents heat-denatured RNA substrate control. D, quantitative assessment of Nsp13 helicase activity as a function of protein concentration on increasing-length duplex RNA substrates, as indicated. Data represent the average of at least three independent experiments with standard deviations (SD) indicated by error bars.
To address if the observed catalytic RNA helicase activity was intrinsic to the purified recombinant Nsp13 protein, we expressed and purified an engineered Walker A box (motif I) Nsp13-K288R, in which the invariant lysine of the ATPase domain was replaced with an arginine, in the same way as the nonmutated recombinant Nsp13 protein. Nsp13-K288R was devoid of any detectable helicase activity and displayed no measurable ATPase activity (Fig. S1). These results indicate that the observed catalytic activities of nonmutated Nsp13 are intrinsic to the recombinant helicase enzyme overexpressed and purified from insect cells.
We next set out to test Nsp13 for unwinding partial duplex RNA substrates with increased lengths of ds regions as a function of protein concentration under multiple-turnover conditions. An example from a typical experiment with the 13-bp RNA forked duplex substrate is shown in Figure 1C. Both the 13-bp and 16-bp RNA substrates were unwound by Nsp13 to a very similar extent with 12% unwound at 1.3 nM and 36% unwound at 2.6 nM helicase concentration (Fig. 1D). For both the 13-bp and 16-bp substrates, a plateau of approximately 80 to 85% substrate unwound was achieved at 10.4 nM Nsp13. In contrast, there was a marked reduction in unwinding of the 19-bp and 22-bp RNA forked duplex substrates throughout the same Nsp13 protein titration range. At 1.3 nM Nsp13, only 3% of the 19-bp substrate was unwound, a 4-fold decrease compared with the 13-bp and 16-bp substrates. Although greater unwinding of the 19-bp partial duplex was observed with increasing Nsp13 concentrations, helicase activity was significantly reduced throughout the enzyme titration compared with the substrates only 3-bp or 6-bp shorter. The effect of duplex length on Nsp13 helicase activity was even more dramatic for the 22-bp RNA duplex substrate. For example, there was an approximately 8-fold reduction in substrate unwound for the 22-bp partial duplex compared with the 13-bp or 16-bp substrates at an Nsp13 concentration of 2.6 nM. Even at the highest Nsp13 concentration tested (15.6 nM), there was a significant reduction in the percentage of 22-bp partial duplex substrate unwound compared with the other RNA substrates tested.
Based on the results, we conclude that Nsp13 catalyzes only limited unwinding as a function of increasing RNA duplex length, even over a relatively narrow range of base pair tract lengths (13–22 bp), under the reaction conditions tested. Nsp13-catalyzed unwinding of the partial duplex RNA substrates displayed a sigmoidal character as a function of enzyme concentration (Fig. 1D), as indicated by the Hill coefficients (Table 1), suggesting functional cooperativity among Nsp13 molecules.
Table 1.
Hill coefficients of selected sigmoidal curves fit to Nsp13 unwinding of RNA and DNA substrates
| RNA forked duplex length (bp) | Hill coefficient (h) |
|
|---|---|---|
| h | R2 | |
| 13 | 2.6 | 0.97 |
| 16 | 2.6 | 0.99 |
| 19 | 2.2 | 1 |
| 22 | 2.0 | 0.98 |
Hill coefficient (h) and correlation coefficient (R2) values are shown for the best fit curves of the multi-turnover helicase assays described in Experimental procedures under 5 mM MgCl2 reaction conditions. Values were determined in GraphPad Prism using nonlinear regression and the “Specific binding with Hill slope” equation.
ATP and Mg2+ concentrations for Nsp13 helicase activity
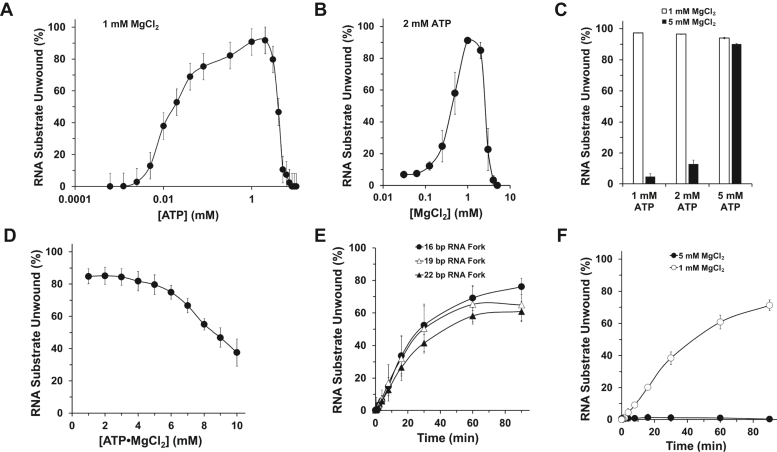
Previously, ATP concentration was determined to be an important factor for optimal helicase activity by recombinant SARS-CoV-1 Nsp13 (10). With the exception of a single study conducted using a fixed concentration of ATP (5 mM) (16), the importance of divalent cation concentration for Nsp13 helicase activity has not been examined. Given the essentiality of the divalent metal ion cofactor for ATP hydrolysis and helicase activity, we assessed if the concentration of ATP, as well as Mg2+, affected Nsp13 RNA helicase activity. Given that 1 mM Mg2+ represents a physiological concentration in the cytosol of mammalian cells (23, 24), we examined Nsp13 unwinding as a function of ATP concentration with 1 mM Mg2+ present in the reaction mixtures. As shown in Figure 2A, Nsp13 RNA helicase activity was dependent on ATP concentration with an optimum of 1 to 2 mM ATP. A steep decline of Nsp13 RNA helicase activity was observed at an ATP concentration of 3 mM and greater.
Figure 2.
Nsp13 helicase activity as a function of ATP and Mg2+ concentration.A, quantitative assessment of helicase activity by Nsp13 (0.325 nM) as a function of ATP concentration at 1 mM MgCl2. B, quantitative assessment of helicase activity by Nsp13 (0.325 nM) as a function of MgCl2 concentration at 2 mM ATP. C, quantitative assessment of helicase activity by Nsp13 (0.325 nM) at the indicated ATP concentration in the presence of 1 mM MgCl2 or 5 mM MgCl2. D, quantitative assessment of helicase activity by Nsp13 (0.325 nM) as a function of ATP-MgCl2, fixed at a 1:1 molar ratio. All experiments were performed using a partial duplex 13-bp RNA substrate (0.25 nM) incubated with Nsp13 for 15 min at 37 °C. E, single-turnover kinetic analysis of Nsp13 helicase (0.1625 nM) as a function of RNA duplex length using reaction conditions of 2 mM ATP and 1 mM MgCl2. Quantitative assessment of Nsp13 helicase activity is shown. F, single-turnover kinetic analysis of reaction mixtures containing Nsp13 (0.1625 nM) and the 30-bp partial duplex RNA substrate (0.25 nM) using 2 mM ATP and either 1 mM MgCl2 or 5 mM MgCl2 incubated at 37 °C for the indicated time points.
Based on these results, we performed a Mg2+ titration using a fixed 2 mM ATP concentration. Under these conditions, the optimal Mg2+ concentration was determined to be 1 to 2 mM, with a steep 4-fold decrease at 3 mM Mg2+ and a 27-fold decrease at 4 mM Mg2+ (Fig. 2B), consistent with a previous observation that a recombinant Nsp13 with a maltose-binding protein tag expressed and purified from Escherichia coli displayed much lower RNA helicase activity under conditions of 8 mM Mg2+ and 5 mM ATP (16). Importantly, our findings demonstrate that Nsp13 helicase is very sensitive to even a 3 mM Mg2+ concentration under the reaction conditions tested, establishing that optimal dsRNA unwinding activity is achieved at 1 to 2 mM Mg2+ paired with 2 mM ATP.
We reasoned that the observed strong inhibition of Nsp13 helicase activity by the higher Mg2+ concentrations might reflect an imbalance of the Mg2+:ATP ratio, favoring excess free Mg2+ that was inhibitory to Nsp13’s catalytic activity. As expected from our results in Figure 2, A and B, 5 mM Mg2+ greatly inhibited Nsp13 helicase activity when either 1 mM ATP or 2 mM ATP was present in the reaction mixture (Fig. 2C). However, at 5 mM ATP, inhibition of Nsp13 helicase activity by 5 mM Mg2+ was suppressed (Fig. 2C), supporting the conclusion that free Mg2+ impairs Nsp13 helicase activity. Furthermore, we tested increasing concentrations of ATP:Mg2+ at a fixed 1:1 ratio for their effect on Nsp13-catalyzed unwinding. As shown in Figure 2D, Nsp13 helicase activity was inhibited as the equimolar concentration of ATP:Mg2+ increased, especially above 5 mM, suggesting that excess ATP:Mg2+ complex was inhibitory to Nsp13 helicase activity.
Having determined optimal Mg2+ and ATP concentrations for SARS-CoV-2 Nsp13 RNA helicase activity using the 13-bp partial duplex RNA substrate, we set out to test if Nsp13 might be better able to unwind partial duplex RNA substrates of increasing lengths under more stringent single-turnover conditions that do not afford the opportunity for multiple binding events by Nsp13 molecules that either failed to stably bind during the preincubation phase or dissociated during the incubation with ATP. In control experiments, preincubation of increasing dT200 concentrations followed by the addition of radiolabeled 30-bp RNA partial duplex substrate allowed us to determine that 100 nM dT200 completely inhibited Nsp13 helicase activity (Fig. S2). The single-turnover conditions were achieved by the co-addition of 100 nM dT200 with ATP to serve as an Nsp13 protein trap. Using a substoichiometric ratio of Nsp13 (0.1625 nM) compared with the indicated RNA partial duplex substrate (0.25 nM) under single-turnover reaction conditions, an initial lag phase of 30 to 60 s was observed followed by a kinetic increase in RNA duplex unwinding for all substrate duplex lengths (16- to 22-bp) until an apparent plateau was approached by 60 to 90 min (Fig. 2E). Although there was a trend for an inverse relationship between RNA duplex length and amplitude, the ability of Nsp13 to unwind partial duplex RNA substrates of increasing ds regions with comparable efficiency suggests the enzyme displays similar processivity in this RNA duplex range. Consistent with this, we observed that the 30-bp RNA partial duplex substrate was unwound with similar efficiency as the other RNA partial duplex substrates (Fig. 2F). In contrast to the reaction conditions using 1 mM Mg2+, only a very small percent of the 30-bp RNA substrate was unwound by Nsp13 in reaction mixtures containing 5 mM Mg2+ throughout the 90-min time course. (Fig. 2F).
Strand-specific inhibition of Nsp13 helicase by an RNA backbone modification
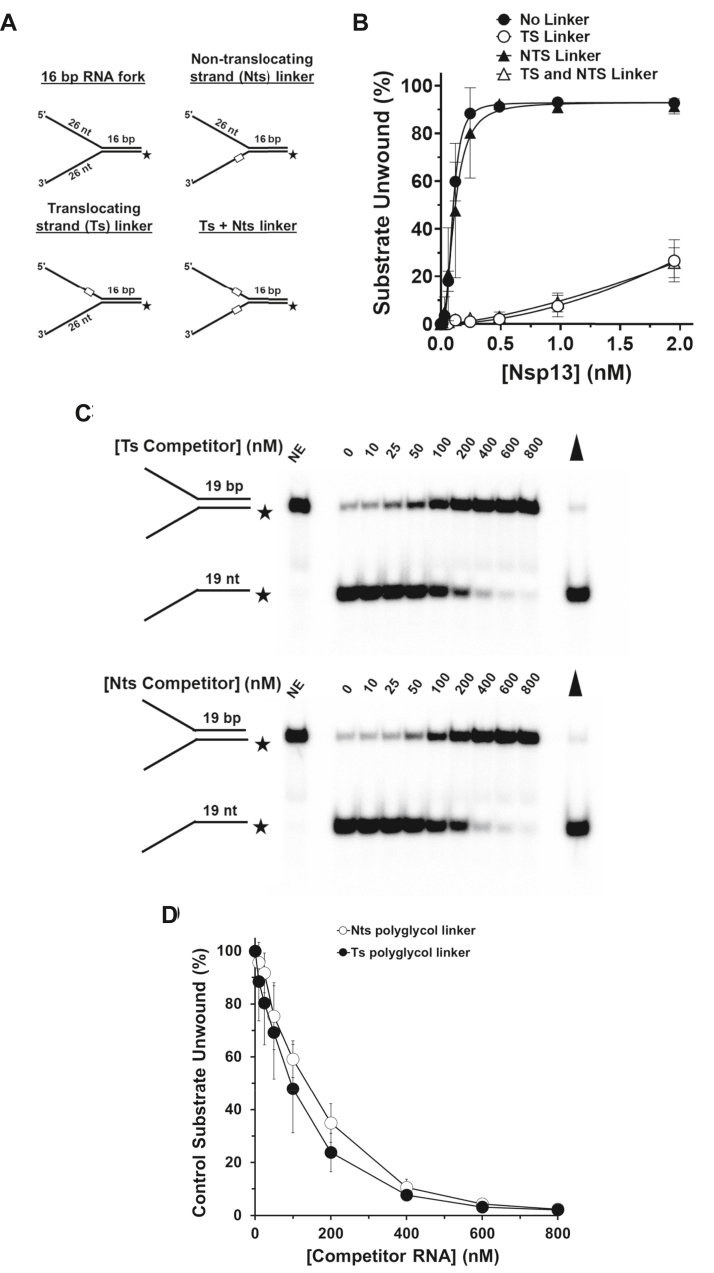
Given the lack of mechanistic information pertaining to the substrate preference of Nsp13, we sought to examine if its interactions with the ss tails of the forked duplex RNA substrate were important for its ability to unwind the adjacent ds region. Therefore, we tested Nsp13 on a series of forked duplex substrates in which a synthetic 18-atom poly(hexa)ethylene glycol (PEG) linkage of neutral charge that spans 3 RNA nucleotides (25) replaced the sugar–phosphate backbone and was positioned three nucleotides prior to the ss:ds junction in either the top or bottom RNA strands of the forked duplex substrate (Fig. 3A). Some helicases require sugar–phosphate backbone continuity in the ss tail to effectively load and unwind the duplex component of the substrate (25, 26), whereas E. coli Rep helicase tolerates backbone disruption (27), suggesting unique mechanisms of loading and initiation of duplex unwinding.
Figure 3.
Strand-specific effect of backbone discontinuity on Nsp13 helicase activity.A, depiction of the partial duplex RNA substrates with a polyglycol linker (PEG) in the top or bottom strand. B, quantitative assessment of Nsp13 helicase activity from a 15-min incubation at 37 °C with 1 mM MgCl2 and 2 mM ATP as a function of protein concentration on the backbone-modified substrates (0.25 nM) shown in (A). Data represent the average of at least three independent experiments with SD indicated by error bars. C, representative native polyacrylamide gel analyses of reactions products from Nsp13 reaction mixtures designed to assess sequestration by RNA partial duplex molecules containing the PEG linker in the top or bottom strand, as indicated. Nsp13 was preincubated with the specified RNA partial duplex molecules in the presence of 2 mM ATP and 1 mM Mg2+ followed by the addition of a radiolabeled forked duplex tracker substrate as described in Experimental procedures. Filled triangle represents heat-denatured RNA substrate control. D, quantitative assessment of Nsp13 helicase activity as a function of the indicated concentration of the competitor RNA partial duplex with PEG in the top or bottom strands.
As shown in Figure 3B, we observed that Nsp13 helicase was profoundly sensitive to the PEG modification in the translocating strand (using the 1 mM Mg2+ condition), whereas little to no effect was observed by the nontranslocating strand linker. Only ∼25% of the RNA substrate with the translocating strand PEG was detected at the highest concentration of Nsp13 tested (1.95 nM), whereas approximately 90% of the control (unmodified) substrate was unwound at 0.5 nM Nsp13. Like the effect exerted by the top strand PEG, strong inhibition of Nsp13 helicase activity was observed for the substrate in which the PEG was placed in both the top and bottom strands of the forked duplex RNA substrate. Similarly, the translocating PEG in the forked duplex RNA substrate strongly inhibited Nsp13 helicase activity in which reaction mixtures contained 5 mM Mg2+ (Fig. S3). These results suggest that a sugar–phosphate backbone modification in the preexisting 5ʹ ssRNA arm impairs Nsp13’s ability to efficiently unwind the adjacent RNA duplex, providing insight into the apparent basis for the 5ʹ ss tail loading requirement of Nsp13 on forked duplex substrates with only 1 ss overhang (13).
It is plausible that the PEG modification may interfere with Nsp13 binding to the forked duplex RNA substrate. To address this issue, we performed helicase assays that enable us to assess sequestration of Nsp13 by the RNA forked duplex harboring the PEG in the top or bottom ss arm. Nsp13 was preincubated with increasing concentrations of the unlabeled version of forked duplex RNA in the presence of ATP, followed by the addition of a radiolabeled tracker substrate and further incubation. Reactions were then quenched and analyzed by native gel electrophoresis to score unwinding of the tracker substrate. As shown in Figure 3, C and D, Nsp13 helicase activity was comparably sensitive to inhibition of unwinding the tracker substrate by preincubation with the RNA forked duplex containing a PEG linker in the top strand or bottom strand. These results suggest that despite the strong inhibition of Nsp13 helicase activity by the presence of the PEG linker in the top strand, Nsp13 was not preferentially sequestered by the linker compared with the PEG in the bottom strand.
Strand-specific interaction of Nsp13 dictates its ability to load and unwind forked duplex substrates
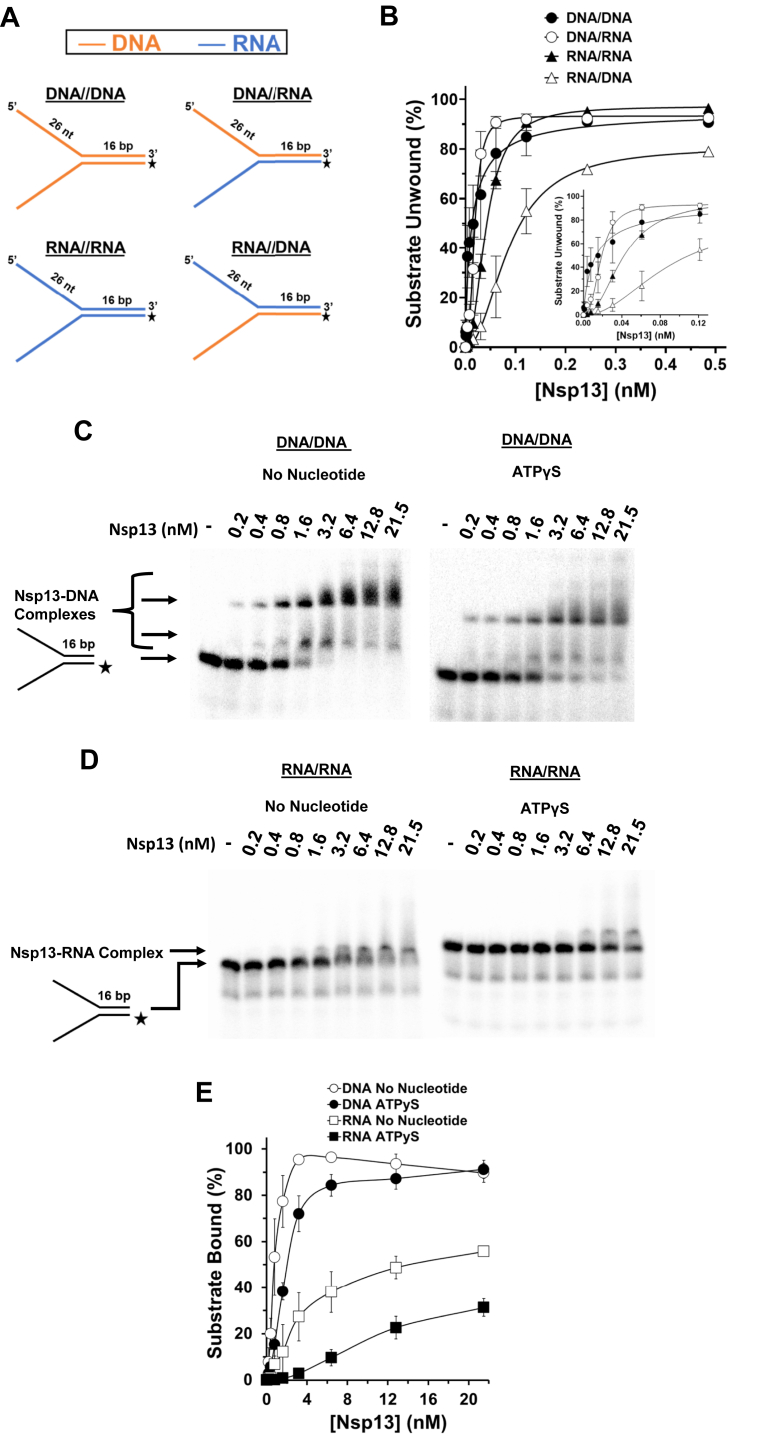
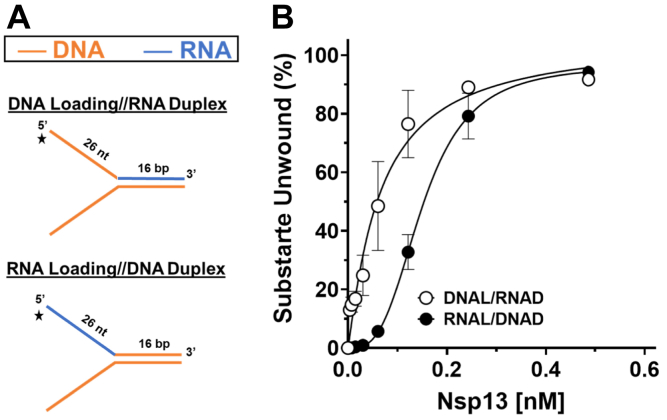
The importance of backbone continuity in the ssRNA segment of the forked duplex substrate that Nsp13 helicase is presumed to load prompted us to ask if the identity of the strand (RNA versus DNA) played a role in its substrate specificity. To address this, we tested Nsp13 for unwinding a series of forked duplex substrates very similar to those used for the PEG backbone-modified substrates, only they did not harbor a site-specific modification but rather consisted of a top and/or bottom strand that was either RNA or DNA (Fig. 4A). The forked duplex substrates containing a DNA top strand were unwound with significantly greater efficiency than those substrates consisting of an RNA top strand throughout the Nsp13 titration except at the highest helicase concentrations where unwinding reached a plateau of 90% substrate unwound (Fig. 4B). Nsp13 unwound a greater percentage of the DNA (top)/RNA (bottom) substrate (melting temperature (Tm) = 36.2 °C) compared with RNA (top)/DNA (bottom) substrate (Tm = 36.8 °C), suggesting that a difference in duplex stability is unlikely to contribute to the observed difference in duplex unwinding for the two substrates. At very low Nsp13 concentrations (15.2 pM, 7.59 pM, 3.79 pM), Nsp13 unwound a significantly greater percentage of the DNA–DNA substrate compared with the DNA (top)/RNA (bottom) hybrid, suggesting that Nsp13 interactions with the nontranslocating strand may contribute to unwinding efficiency. Unwinding of the forked duplex RNA, RNA/DNA, or DNA/RNA substrates by Nsp13 displayed a sigmoidal character as a function of enzyme concentration (Fig. 4B), indicated by the Hill coefficients (Table S1). These results, like those shown in Figure 1D and Table 1, suggest functional cooperativity among Nsp13 molecules. Beyond these observations, the results demonstrate that Nsp13 strongly prefers to unwind forked duplex substrates in which the top strand is composed of DNA. Results from experiments with the same substrates using reaction mixtures that contained 5 mM Mg2+ as opposed to 1 mM Mg2+ also demonstrate that forked duplex substrates with a 5ʹ ssDNA arm were preferentially unwound (Fig. S4). However, we did observe that, at the high Mg2+ concentration, the forked duplex with RNA in the top and bottom strands was unwound least efficiently.
Figure 4.
Chemical identity of nucleic acid translocating strand dictates Nsp13 helicase substrate preference.A, depiction of the partial duplex substrates with DNA or RNA in the top or bottom strand. B, quantitative assessment of Nsp13 helicase activity from a 15-min incubation at 37 °C with 1 mM Mg2+ and 2 mM ATP as a function of protein concentration on the nucleic acid substrates (0.25 nM) shown in (A). Inset shows the lower Nsp13 concentrations depicted in the larger graph. C and D, representative native polyacrylamide gel analyses of Nsp13 binding mixtures from Nsp13 reaction mixtures designed to assess binding of protein to DNA and RNA forked substrates conducted in the presence and absence of nucleotide. The indicated concentrations of Nsp13 were incubated with the specified forked duplex substrate (0.25 nM) for 15-min incubation at RT with no nucleotide or ATPγS (2 mM). E, quantitative assessment of Nsp13 binding to DNA (C) and RNA (D). Data represent the average of at least three independent experiments with SD indicated by error bars.
In order to determine if the more efficient unwinding of DNA versus RNA forked substrates by Nsp13 was due to having a higher binding affinity for DNA substrates, we performed electrophoretic mobility shift assays using Nsp13 with DNA and RNA forked duplex substrates (Fig. 4, C and D). At the same titration of enzyme concentrations, Nsp13 was able to bind with significantly greater efficiency on the DNA substrates than the RNA (Fig. 4E). At the highest concentrations, the percent substrate bound for DNA reached a plateau of approximately 90% for both the no nucleotide and the ATP analog adenosine 5'-O-(3-thio)triphosphate (ATPγS) conditions. However, at lower concentrations there was a difference between the DNA substrate nucleotide conditions. For example, at 3.2 nM, the no nucleotide condition had approximately 95% substrate bound compared with around 70% bound in the ATPγS condition. In comparison, the RNA substrate only approached but never achieved a plateau of substrate bound around 55% in the absence of nucleotide and 31% in the presence of ATPγS. The apparent dissociation constant (Kd) was determined for each reaction condition. In the absence of nucleotide, the Kd was 1.1 ± 0.2 and 13.2 ± 3 for DNA and RNA, respectively, and in the presence of ATPγS, 2.6 ± 0.6 and 38.9 ± 11 (Table 2). These results demonstrate that Nsp13 strongly prefers binding to DNA rather than RNA forked substrates.
Table 2.
Kd values for Nsp13 binding to RNA and DNA
| Nucleotide condition | Kd (DNA) | Kd (RNA) |
|---|---|---|
| No nucleotide | 1.1 ± 0.2 | 13.2 ± 3 |
| ATPγS | 2.6 ± 0.6 | 38.9 ± 11 |
Kd values were calculated from at least three independent electrophoretic mobility shift assay experiments in which Nsp13 (0.2–21.5 nM) was incubated with the 16-bp DNA or RNA forked substrates (0.25 nM) for 15 min at RT as described in Experimental procedures. All experiments were performed using 1 mM MgCl2 in the presence or absence of 2 mM ATPγS.
RNA/DNA chemistry of the 5ʹ tail is a critical substrate feature for Nsp13 helicase activity
While our findings demonstrated Nsp13’s preferential unwinding of forked duplex substrates harboring a DNA translocating strand, they did not specify whether it is the DNA in the 5ʹ ss tail or the duplex region of the top strand that conferred preferential Nsp13 helicase activity. To address this question, two chimeric DNA/RNA oligonucleotides were prepared and used for designated helicase substrates: i) a 5ʹ ssDNA tail followed by a ssRNA segment annealed to a bottom (complementary) sequence within the duplex region; ii) a 5ʹ ssRNA tail followed by a ssDNA segment annealed to a bottom (complementary) sequence with the duplex region. In both cases, these chimeric oligonucleotides were annealed to a nonchimeric bottom (complementary) ssDNA to generate two forked duplexes (theoretical Tm values of 34.8 °C and 36.8 °C, respectively) from which Nsp13 helicase activity could be accessed. The results from these experiments are shown in Figure 5. The forked duplex containing a 5ʹ ssDNA tail followed by ssRNA was unwound with significantly greater efficiency than its counterpart, consisting of a 5ʹ ssRNA tail followed by ssDNA, throughout the Nsp13 protein titration. At an Nsp13 concentration of 0.03 nM, there was little to no detectable unwinding of the chimeric substrate with a ssRNA 5′ tail, whereas nearly 25% of the chimeric substrate with a ssDNA 5′ tail was unwound. At 0.06 nM Nsp13, only 5% of the substrate with a ssRNA 5′ tail was unwound, whereas nearly 50% of the chimeric substrate with a ssDNA 5′ tail was unwound.
Figure 5.
RNA/DNA chimeric substrates reveal that the chemistry of the preexisting 5ʹ single-stranded tail of the top strand is the key determinant for Nsp13 helicase activity on forked duplex substrates.A, depiction of the partial duplex substrates harboring chimeric RNA/DNA nucleotides in the 5ʹ tail of the top strand. B, quantitative assessment of Nsp13 helicase activity from a 15-min incubation at 37 °C with 1 mM Mg2+ and 2 mM ATP as a function of protein concentration on the nucleic acid substrates (0.25 nM) shown in (A). Data represent the average of at least three independent experiments with SD indicated by error bars.
These results demonstrate that Nsp13 preferentially unwinds a forked duplex substrate with a 5ʹ ssDNA tail as opposed to a 5ʹ ssRNA tail, irrespective of the RNA/DNA chemistry of the top strand’s duplex region. Thus, the specific RNA/DNA identity of the top strand 5ʹ ss tail is the critical feature of a forked duplex substrate that determines Nsp13’s efficiency of unwinding forked duplex substrates of differing constitution.
The demonstrated preference of Nsp13 to unwind forked duplex substrates flanked by a translocating single strand of DNA composition suggested to us that the enzyme preferentially interacts with ssDNA compared with ssRNA to allow the enzyme’s more efficient loading. To address this further, we determined the concentration of ss nucleic acid effector to achieve one-half maximal velocity for ATPase activity (Keff) (Table 3). The Keff values for ssRNA and ssDNA were 117 ± 64 nM and 12.8 ± 6.4 nM, respectively, with a p value < 0.05. The 9-fold greater Keff value for ssRNA compared with ssDNA suggests that Nsp13 more favorably interacts with ssDNA than with ssRNA. However, the Vmax is 1.4-fold greater for ssRNA compared with ssDNA (p value < 0.05), suggesting that at a saturating concentration of polynucleotide effector Nsp13 efficiently hydrolyzes ATP in the presence of ssRNA or ssDNA.
Table 3.
Keff and Vmax parameters for Nsp13-catalyzed ATP hydrolysis
| Parameter | ssRNA | ssDNA |
|---|---|---|
| Keff (nM) | 117 ± 64 | 12.8 ± 6.4 |
| Vmax (nM ATP∗sec−1) | 134 ± 20 | 97.6 ± 14 |
Keff and Vmax values were calculated from at least three independent experiments with a range of 60-nt-length effector concentrations (4–1000 nM). All experiments were performed using 1 mM ATP, 1 mM MgCl2, 0.05 nM Nsp13, for a 12.5-min time course at 37 °C.
Nsp13’s significantly reduced Keff for ssDNA compared with ssRNA prompted us to determine the kcat for ATPase activity using a subsaturating concentration (31 nM) of ssRNA or ssDNA (Table 4). The 3.7-fold greater kcat for Nsp13 ATP hydrolysis in the presence of ssDNA versus ssRNA (p value < 0.05) upheld the results from the Keff experiments. The lower Keff for ssDNA and greater kcat for ATP hydrolysis at a subsaturating effector concentration of ssDNA compared with ssRNA suggests that Nsp13 preferentially interacts with ssDNA compared with ssRNA. However, the difference in kcat values for ATP hydrolysis using the ssDNA versus ssRNA effector could be overcome by a saturating polynucleotide effector concentration of 1000 nM, consistent with the Vmax determinations.
Table 4.
Turnover rate constants (kcat) for Nsp13-catalyzed ATP hydrolysis as a function of polynucleotide effector
| [Effector] (nM) | ssRNA kcat (sec−1) | ssDNA kcat (sec−1) |
|---|---|---|
| 31 | 311 ± 88 | 1145 ± 207 |
| 1000 | 1526 ± 234 | 1245 ± 33.3 |
kcat values are based on average of three independent experiments as a function of stated 60-nt-length effector concentrations. All experiments were performed using 1 mM ATP, 1 mM MgCl2, 0.05 nM Nsp13, for a 10-min time course at 37 °C.
Nsp13 disrupts the high-affinity interaction of streptavidin bound to a biotinylated RNA molecule in a unidirectional manner
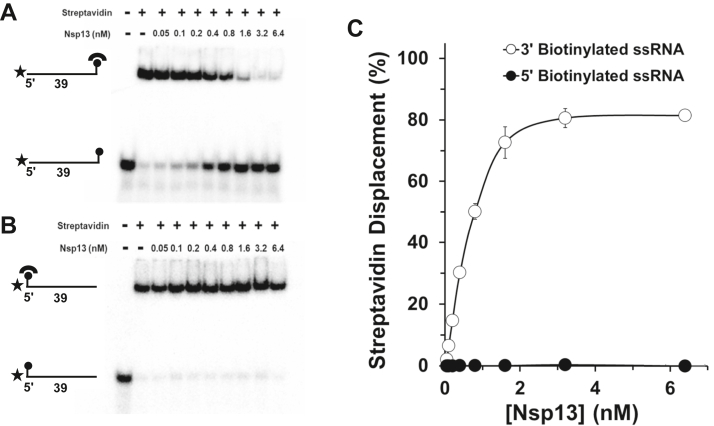
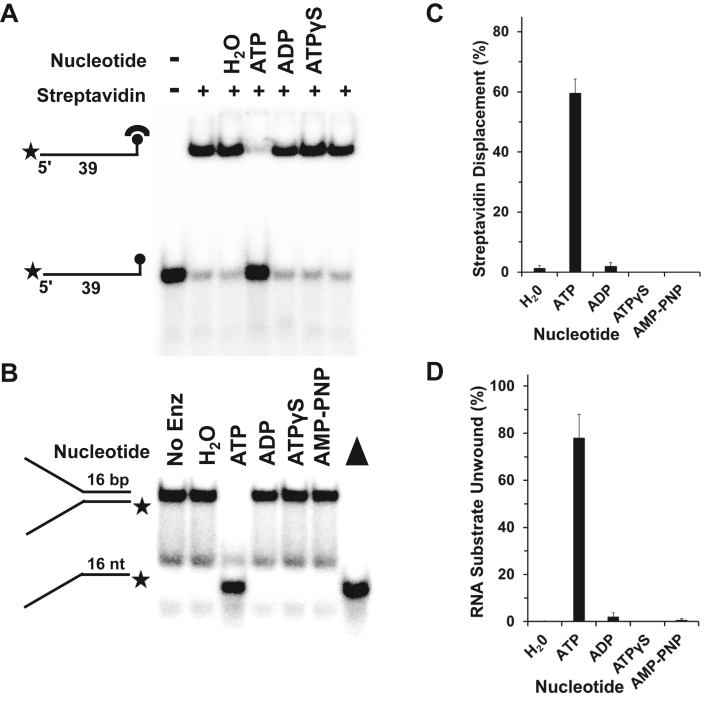
To further characterize the catalytic functions of Nsp13, we examined the enzyme’s ability to use its motor ATPase function to translocate and displace streptavidin from a biotin moiety covalently linked to either the 5ʹ end or 3ʹ end of a 39-mer ribonucleotide tract (Table S3) (0.25 nM final concentration in reaction mixture). The biotin–streptavidin complex with an apparent dissociation constant (Kd) of 10−14 M (28) and diameter of a streptavidin tetramer, ∼45 Å (29), represents a surrogate for a very-high-affinity protein–RNA interaction positioned at a defined location on the substrate. As a function of Nsp13 concentration, an increased percentage of the streptavidin was displaced from biotin covalently linked to the 3ʹ end of the 39-mer (Fig. 6A), whereas Nsp13 failed to displace streptavidin from biotin attached to the 5ʹ end of the same RNA oligonucleotide throughout the entire helicase protein titration, up to 6.4 nM Nsp13 (Fig. 6B). Even at a relatively low Nsp13 concentration of 0.3 nM, just exceeding the substrate concentration of 0.25 nM, 30% streptavidin displacement from the 3ʹ end biotin was achieved (Fig. 6C). The ability of Nsp13 to displace streptavidin bound to biotin conjugated to the 3ʹ end but not the 5ʹ end of the ssRNA suggests that Nsp13 does not disrupt the streptavidin–biotin interaction nonspecifically but rather in a manner that is consistent with 5ʹ to 3ʹ translocation on the ssRNA molecule, consistent with its blockage by a PEG linker in a strand-specific manner (this study).
Figure 6.
Nsp13 disrupts the high-affinity interaction of streptavidin bound to biotinylated RNA in a unidirectional manner.A, streptavidin displacement as a function of Nsp13 concentration under multi-turnover conditions. Representative polyacrylamide gel analysis of Nsp13 enzyme titration in streptavidin displacement reactions incubated for 15 min at 37 °C in the presence of 1 mM MgCl2 and 2 mM ATP using a 39-mer RNA substrate (0.25 nM) with a biotin moiety at the 3ʹ end (A) or 5ʹ end (B). Biotinylated oligonucleotide was preincubated with 3 nM streptavidin monomer for 10 min, and reactions were initiated by the addition of 1 μM biotin immediately followed by the indicated Nsp13 concentration. C, quantitative assessment of Nsp13-catalyzed streptavidin displacement. Data represent the average of three independent experiments with SD indicated by error bars.
We further tested Nsp13’s preferential displacement of streptavidin bound to 3ʹ biotinylated substrates by conducting multi-turnover kinetic assays on the same 39-mer ribonucleotide tract and its deoxyribonucleotide version (0.25 nM) (Table S3). The reactions contained a single concentration of Nsp13 (1.6 nM) and streptavidin (0.75 nM), and were quenched at time points from 1 to 16 min. The experimental data, shown in Figure S5, demonstrated that Nsp13 preferentially displaces streptavidin bound to 3′ biotinylated ssDNA compared with ssRNA. Displacement of streptavidin from the ssDNA substrate was significantly greater than from ssRNA at all time points. Moreover, the extent of streptavidin displacement reached ∼90% for ssDNA compared with ∼50% for ssRNA at the 16 min time point (Fig. S5). These data complement our results from helicase assays in which Nsp13 preferentially unwound partial duplex DNA substrates compared with RNA substrates.
Nsp13’s ability to displace streptavidin from the 3ʹ-Biotinylated RNA substrate and catalyze ATP hydrolysis is regulated by Mg2+ concentration
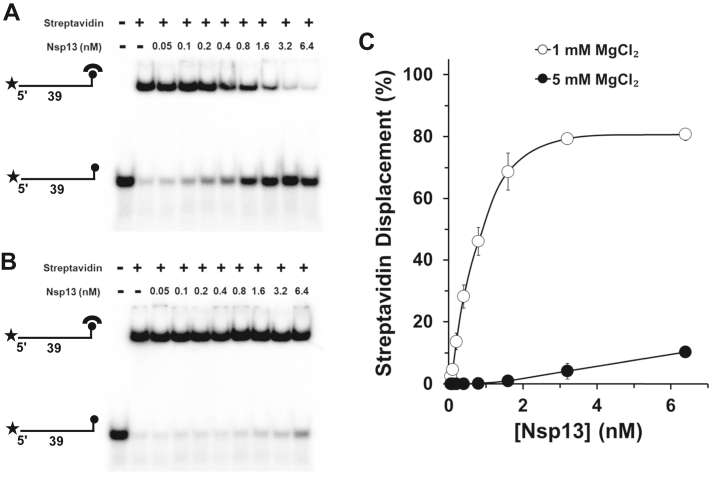
The ability of Nsp13 to translocate unidirectionally in a manner dependent on nucleoside triphosphate hydrolysis to displace the streptavidin bound to the RNA substrate prompted us to ask if Mg2+ concentration modulates Nsp13 protein displacement activity, as it did for duplex RNA unwinding. As shown in Figure 7, A–C, Nsp13-catalyzed streptavidin displacement was dramatically greater at 1 mM Mg2+ compared with 5 mM Mg2+ throughout the Nsp13 protein titration. For example, at 1 mM Mg2+ nearly 70% of the streptavidin-bound biotinylated RNA was disrupted by 1.6 nM Nsp13, whereas only 1 to 2% streptavidin displacement was observed in reaction mixtures containing 5 mM Mg2+. These results suggest that Mg2+ regulates Nsp13’s ability to displace streptavidin bound to the RNA molecule.
Figure 7.
Nsp13-catalyzed streptavidin displacement as a function of MgCl2 concentration. Representative polyacrylamide gel analysis of Nsp13 titration in streptavidin displacement reactions incubated for 15 min at 37 °C using 39-mer RNA substrate (0.25 nM) with a 3ʹ biotin moiety in the presence of 2 mM ATP and 1 mM MgCl2 (A) or 5 mM MgCl2 (B). Biotinylated oligonucleotide was preincubated with 3 nM streptavidin monomer for 10 min and reactions were initiated by the addition of 1 μM biotin immediately followed by the indicated Nsp13 concentration. C, quantitative assessment of Nsp13-catalyzed streptavidin displacement. Data represent the average of three independent experiments with SD indicated by error bars.
The increased Nsp13-catalyzed streptavidin displacement and RNA helicase activity in the presence of 1 mM Mg2+ compared with 5 mM Mg2+ prompted us to determine the effect of Mg2+ on the enzyme’s ATPase activity. Using an ATP concentration of 1 mM, we determined that the kcat values for Nsp13 ATPase activity in the presence of ssRNA using 1 mM Mg2+ or 5 mM Mg2+ were 1282 s−1 and 337 s−1, respectively (Table 5). The 4-fold reduction in Nsp13 ATPase activity in the presence of 5 mM Mg2+ compared with 1 mM Mg2+ suggests that the compromised streptavidin displacement and helicase activity at the elevated divalent cation concentration reflects a reduced ability to hydrolyze ATP.
Table 5.
Turnover rate constants (kcat) for Nsp13-catalyzed ATP hydrolysis as a function of MgCl2 concentration
| MgCl2 concentration | kcat (sec−1) |
|---|---|
| 1 mM | 1382 ± 189 |
| 5 mM | 337 ± 69 |
kcat values are based on average of three independent experiments as a function of stated MgCl2 concentrations. All experiments were performed using 1 mM ATP, 31 nM ssRNA 60-nt effector, 0.05 nM Nsp13, for a 10-min time course at 37 °C.
The inhibition of Nsp13 ATPase activity by free Mg2+ may be mediated by an allosteric effect on the enzyme, or a function strength in the reaction mixture. To address the specificity of Mg2+ inhibition, we tested other divalent cations (Mn2+, Ca2+, Co2+) for their ability to substitute for Mg2+ in the Nsp13 ATPase reaction with the 60-nt RNA effector and 1 mM ATP present. We observed very low to no Nsp13 ATPase activity in the presence of CaCl2 or CoCl2 at 1, 2, or 5 mM metal ion concentrations, whereas MnCl2 partially fulfilled the metal requirement but still a 2-fold or greater reduction in Nsp13 ATP hydrolysis, depending on the MnCl2 concentration (Fig. S6A). Next, we tested mixtures of Mg2+ (1 mM) with Mn2+, Ca2+, or Co2+ at concentrations of 1 mM or 4 mM with RNA effector and 1 mM ATP present, as before. Here we observed a significant reduction in Nsp13 ATPase activity by all three metals (Mn2+, Ca2+, or Co2+) at 1 mM concentration, and an even greater reduction at 4 mM (Fig. S6B). These results suggest that, while excess free Mg2+ exerts inhibition of Nsp13 ATPase activity, the effect is much greater for the other three metals, in a quantitative manner that was dependent on the identity of the divalent cation. Thus, the inhibition of Nsp13 ATPase activity was not specific to Mg2+. Lastly, we sought to address if the Mg2+ inhibition of Nsp13 ATPase activity would persist even when the RNA effector was absent from the reaction mixture. We observed that there was no inhibition of Nsp13 ATPase activity, even at 10 mM MgCl2, suggesting that the mechanism of inhibition was mediated in an RNA-dependent manner (Fig. S6C). It is plausible that excess free Mg2+ interferes with Nsp13’s favorable interaction with the RNA effector, disallowing the conformational change in the enzyme most favorable for ATP hydrolysis.
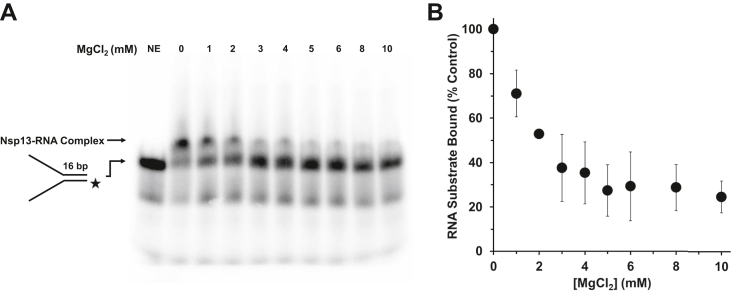
The reduction of Nsp13 ATPase activity by excess Mg2+ prompted us to determine the effect of the metal ion on the enzyme’s ability to bind RNA. As shown in Figure 8, increasing concentrations of Mg2+ inhibited Nsp13 binding to the RNA forked duplex; at the highest Mg2+ concentration tested (10 mM), there was a ∼70% reduction in binding compared to the omission of Mg2+. We also tested the effect of Mg2+ on Nsp13 binding to DNA (Fig. S7). Here, we only observed modest reduction of Nsp13 binding, even at the highest concentration of 10 mM Mg2+.
Figure 8.
Effect of MgCl2 on Nsp13’s binding to the RNA substrate. Nsp13 (12.8 nM) was incubated with the 16-bp RNA forked duplex substrate (0.25 nM) for 15 min at RT. A, binding mixtures were assessed by electrophoretic mobility shift assay, as described in Experimental procedures. B, quantitative analysis is shown. Data represent the average of three independent experiments with SD indicated by error bars.
Nsp13-catalyzed streptavidin displacement is ATP dependent
It was reported that the RNA helicase eukaryotic initiation factor 4A (eIF4A) (30) and the replicative CMG DNA helicase (31) hydrolyze the ATP analogue ATPγS, which in turn can fuel unwinding of duplex RNA and DNA, respectively. This is a rather unusual property of helicases, as a number of previously characterized helicase proteins, including UvrD (32) and Rep (33), are devoid of detectable unwinding activity in the presence of ATPγS. To address the nucleotide requirements for Nsp13-catalyzed streptavidin displacement and RNA helicase activity, we tested Nsp13 to perform the two catalytic activities in the presence of ATP, the poorly hydrolyzable ATP analogue ATPγS, the nonhydrolyzable ATP analogue adenyl-imidodiphosphate (AMP-PNP), ADP, or the omission of nucleotide altogether. The results from the streptavidin displacement experiments with the 3ʹ-biotinylated RNA substrate demonstrate that only ATP, but not ATPγS, AMP-PNP, or ADP, suffice to support Nsp13-catalyzed disruption of the RNA–protein complex (Fig. 9, A and B). Similarly, only ATP, but not ATPγS, AMP-PNP, or ADP, fueled Nsp13 helicase activity on the 16-bp RNA forked duplex substrate (Fig. 9, C and D). These results suggest that a threshold of chemical energy provided by an efficiently hydrolyzable nucleoside triphosphate (e.g., ATP) is required to disrupt the high-affinity streptavidin–ssRNA complex or unwind duplex RNA.
Figure 9.
Chemical energy requirements for Nsp13 to disrupt a high-affinity RNA–protein interaction compared with RNA helicase activity. Representative polyacrylamide gel analysis of Nsp13 enzyme titration using the indicated nucleotide (2 mM) in the presence of 1 mM Mg2+ for streptavidin displacement reactions using 3ʹ-biotinylated RNA substrate preincubated with streptavidin (3 nM monomer) for 10 min and initiated by addition of biotin (1 μM) and Nsp13 (A) or helicase reactions using 16-bp RNA partial duplex substrate (0.25 nM) (C). Note in (B), filled triangle represents heat-denatured RNA substrate control. In both helicase and streptavidin displacement experiments, reaction mixtures were incubated for 15 min at 37 °C. Data for streptavidin displacement (C) or RNA helicase activity (D) represent the average of three independent experiments with SD indicated by error bars.
Discussion
In this work, we found that recombinant SARS-CoV-2 Nsp13, purified from eukaryotic cells, unwound a partial duplex substrate with a 5ʹ ssDNA tail with the greatest efficiency, irrespective if the opposite strand is DNA or RNA. Thus, the critical element in substrate preference of SARS-CoV-2 Nsp13 is the chemical nature of the translocating single strand, i.e., DNA versus RNA. Nsp13’s preference for ssDNA compared with ssRNA for helicase loading was verified by our experiments with chimeric RNA–DNA or DNA–RNA oligonucleotides. We also confirmed Nsp13 preferentially displaces streptavidin bound to biotinylated ssDNA than ssRNA through our multi-turnover streptavidin kinetics assays. Moreover, the ATPase measurements to determine Keff or kcat also demonstrated a preferential interaction of Nsp13 with ssDNA compared with ssRNA at subsaturating concentrations of polynucleotide effector, which was upheld by Nsp13 electrophoretic mobility shift assay using DNA versus RNA forked duplex substrates.
For comparison of Nsp13 catalytic activities expressed in and purified from bacterial systems (7, 10, 34) with insect cells (current study), data for SARS-CoV-1, -2, and MERS Nsp13 proteins are listed in Table S2. It is difficult to conclude differences in specific catalytic activities given that the substrates and reaction conditions, as well as enzyme to substrate ratios, were distinct.
Altogether, the combined results demonstrating preferential interaction of Nsp13 with ssDNA compared with ssRNA combined with the demonstrated Nsp13 translocation directionality 5ʹ to 3ʹ help to explain the apparent substrate specificity for unwinding. In the absence of binding of Nsp13 specifically to the fork structure, the more efficient unwinding of a forked duplex substrate composed of two RNA strands compared with a substrate in which the top strand is RNA and the bottom strand is DNA can be rationalized because Nsp13 would preferentially bind to the DNA bottom strand and engage in nonproductive translocation away from the junction of the partial duplex substrate. In contrast, Nsp13 would not bind preferentially to the bottom strand of the forked duplex substrate consisting entirely of two RNA strands annealed to each other; consequently, the substrate would not be predisposed to nonproductive translocation by Nsp13. This work helps to clarify previous conflicting reports on the nucleic acid substrate specificity of Nsp13 (as described previously) and further characterizes the enzyme’s preferential interaction with a DNA 5ʹ tail on the top strand in the helicase substrate. Like Nsp13, the well-characterized hepatitis C virus NS3 helicase protein also preferentially unwinds duplex DNA compared with duplex RNA substrates (35).
The substrate preference of Nsp13 to unwind duplex nucleic acid substrates with a loading/translocating ssDNA tract may bear relevance in vivo. The cytosolic accumulation of polynucleic acids extruded from the nucleus triggers the cGAS-STING pathway, which is implicated in chronic inflammation (36); presumably, Nsp13’s preferential interaction with the cytosolic DNA molecules may modulate the inflammatory response. Experimental evidence suggests a role of Nsp13 to modulate type 1 interferon production by targeting TBK1 for degradation (37); however, whether the nucleic acid interaction properties of Nsp13 are implicated remains to be determined.
Nsp13’s substrate specificity dictated by the chemical nature of the 5ʹ ss overhang suggests that the loading/translocation properties of Nsp13 strongly influence the enzymatic efficiency for unwinding the adjacent duplex. We further examined this issue for Nsp13-catalyzed unwinding of the dsRNA forked duplexes by assessing Nsp13 helicase activity on substrates that contained a backbone disruption (PEG) in either the 5ʹ ssRNA or 3ʹ ssRNA arm. Here, we observed strong inhibition of Nsp13 helicase when the PEG linker was positioned in the 5ʹ ssRNA overhang, i.e., the strand the helicase is presumed to translocate, immediately adjacent to the duplex. These results suggest that Nsp13 unwinds duplex RNA by a mechanism in which it is sensitive to the top strand sugar–phosphate backbone integrity.
Very recently, several conformational states of Nsp13 in the SARS-CoV-2 RTC with ss template RNA were solved by cryo-EM, providing evidence that the nsp13 “thumbs”-engaged protomer makes extensive hydrogen bonds and ionic interactions with the RNA phosphate backbone (4). These findings are consistent with ours, which show for the first time in a functional manner that Nsp13 helicase activity is strongly sensitive to a local neutralizing disruption of the sugar–phosphate backbone in the helicase translocating strand.
Previously, optical tweezers single-molecule measurements showed that an external constant force applied to a 180-bp RNA hairpin substrate activated Nsp13 helicase activity (15), suggesting that Nsp13 may operate (to some degree) via a passive unwinding mechanism in which thermal fraying of the duplex at the helicase entry point, favored by the force application, enables Nsp13 to capture the exposed ss nucleotides. The ability of the 5ʹ ss-positioned PEG linker to impede Nsp13 translocation and helicase activity would interfere with either passive or active mechanisms of unwinding because in either model Nsp13 would be unable to destabilize nucleic acid duplex ahead of the translocating enzyme. Reaction conditions with low Mg2+ allowed some detectable unwinding of the RNA forked duplex substrate with the 5ʹ PEG linker at higher Nsp13 concentrations; however, the potency of inhibition by the 5ʹ PEG was still quite strong at lower Nsp13 concentrations, suggesting that reduced Mg2+ did not dramatically alter Nsp13’s loading mechanism. The unique properties of Nsp13’s substrate loading may be important for its role during coronavirus replication or proofreading (see below).
Unwinding processivity is an important parameter of a helicase, leading us to investigate the ability of SARS-CoV-2 Nsp13 to unwind dsRNA substrates with increasing duplex lengths under multiple-turnover or single-turnover conditions. Initially, we performed helicase assays with partial duplex RNA substrates characterized by ds regions increasing by 3-bp increments from 13- to 22-bp under reaction mixture conditions (2 mM ATP, 5 mM Mg2+) similar to those used for the characterization of SARS-CoV-1 Nsp13 (13). Results from multiple-turnover experiments with increasing Nsp13 concentrations showed a marked decrease in the amount of substrate unwound as the duplex length was increased from 16-bp to 19-bp, and an even further reduction in percent substrate unwound by increasing the duplex length to 22-bp, suggesting that Nsp13 helicase is very sensitive to duplex RNA length under the reaction conditions tested.
Given that the replicated SARS-CoV-2 genome of 29,800-bp is the longest in the coronavirus family (38) and replication is essential in the coronavirus life cycle, we were struck by Nsp13’s sensitivity to duplex length dependence for RNA helicase activity, albeit under in vitro reaction conditions. Nsp13 interacts with other proteins of the coronavirus RTC (1, 3), and the rate of SARS-CoV-1 Nsp13 helicase activity is enhanced modestly 1.5-fold (11) or 2-fold (8) by the RNA-dependent RNA polymerase Nsp12. Therefore, we reasoned that the reaction conditions used for Nsp13 helicase assays could be optimized. Given the importance of nucleoside triphosphate as the chemical energy source and its essential cofactor Mg2+, we focused on these two components. Previously, bulk biochemical studies of SARS-CoV-2 Nsp13 had used 2 mM ATP and 5 mM Mg2+ (15). Because the estimated in vivo concentration of Mg2+ (1 mM) is much lower, we examined this issue carefully. From a series of biochemical experiments, we determined that optimal Nsp13 helicase activity was detected at a Mg2+ concentration of 1 mM and ATP concentration of 2 mM. The optimized ATP and Mg2+ concentrations exerted a substantial impact on Nsp13’s ability to unwind conventional partial duplex RNA substrates. This was demonstrated when we examined Nsp13’s activity on a 30-bp partial duplex substrate, which was very poorly unwound by Nsp13 using the original reaction conditions of 5 mM Mg2+. In stark contrast, in the presence of 1 mM Mg2+ 70% of the RNA partial duplex substrate was unwound by Nsp13 under single-turnover conditions. Further studies will be required to determine how Nsp13 helicase processivity relates to its role(s) in SARS-CoV-2 genome transactions. It is tempting to speculate that the RNA secondary structure of the coronavirus genome would be resolved by Nsp13 helicase to allow smooth polynucleotide synthesis by the SARS-CoV-2 RTC during replication/transcription. The calculated median base-pairing distance in a recently proposed SARS-CoV-2 consensus model is 25 nt (39), suggesting that the apparent processivity of Nsp13 helicase acting alone may be sufficient to resolve at least some of the intramolecular stem–loop structures.
The role of protein oligomerization in unwinding efficiency has been a topic of interest, as some RNA helicases are known to multimerize, which increases their specific activity (40). For example, Putnam et al. (41) observed that Ded1p showed a sigmoidal functional RNA helicase activity under pre-steady-state and steady-state conditions, providing evidence for the importance of oligomerization for optimal unwinding. We also observed that Nsp13 displayed sigmoidal behavior for helicase activity, suggesting functional cooperativity. In previously published work, chemical cross-linking of a bacterially expressed and purified recombinant SARS-CoV Nsp13 protein followed by SDS-PAGE analysis demonstrated that a small fraction could oligomerize to the dimeric and trimeric forms (12). Functional analysis of the higher-order oligomers was not performed; however, it was observed that longer ssDNA tracts in the partial duplex DNA substrate allowed enhanced Nsp13 helicase activity, leaving the authors to suggest functional cooperativity between monomers. More recently, cryo-EM was used to determine the structure of a SARS-CoV-2 mini-RTC (3). It was determined that two Nsp13 protomers reside within the complex and interact with each other via the conserved 1B domain in the helicase core. Site-directed mutagenesis of the Nsp13-2 protomer resulting in a T216A replacement significantly impaired the interaction of Nsp13 protomers with each other and the mini RTC helicase activity on a partial duplex DNA substrate, suggesting that the physical interaction of both Nsp13 protomers within the mini RTC is important for optimal unwinding. Our findings that purified recombinant Nsp13 displayed cooperativity for helicase activity as a function of enzyme concentration, as determined by Hill plot analysis, prompts further studies to determine the importance of these observations in the SARS-CoV-2 life cycle and nucleic acid processing in the infected host cell.
Having determined that Nsp13 helicase activity was enhanced under reaction conditions of low Mg2+, we sought to address if this preference held for Nsp13’s other catalytic functions. First, we examined streptavidin displacement from the 3ʹ-biotin position on the RNA substrate, which provides an indication of Nsp13 ssRNA translocation because the enzyme completely fails to displace streptavidin from the 5ʹ end. Moreover, RNA translocation is a prerequisite for Nsp13 helicase activity. The dramatic activation of Nsp13 streptavidin displacement at 1 mM Mg2+ compared with 5 mM Mg2+ suggests that the divalent cation concentration regulates Nsp13 RNA helicase activity by improving its RNA translocation activity. Second, we examined Nsp13 ATPase activity because the chemical energy derived from ATP hydrolysis fuels RNA translocation as well as helicase unwinding. We found that the rate of ATP hydrolysis by Nsp13 is significantly greater (4-fold) under the optimal conditions for unwinding, i.e., 1 mM Mg2+. Thus, Nsp13’s ATP hydrolysis, which drives translocation and duplex RNA unwinding activity, is regulated by Mg2+ concentration in the reaction mixture. The reduced ability of Nsp13 to bind the RNA substrate (or the DNA substrate) at greater Mg2+ concentrations is a probable contributing factor to its reduced catalytic functions.
While it is tempting to speculate that Nsp13 follows a general principle in which free Mg2+ inhibits helicase-catalyzed unwinding of ds nucleic acid substrates, as is the case for hepatitis C virus (HCV) NS3 RNA helicase (42) or E. coli RecQ DNA helicase (43), inhibition of helicase-catalyzed unwinding by excess Mg2+ relative to ATP is not always observed (44), suggesting unique helicase-specific effects. In the case of E. coli RecQ, it was suggested there is a requirement for Mg2+ and ATP to be bound to the enzyme as a complex (43), but this concept was not experimentally addressed. For HCV NS3, analyses of steady-state rates of DNA unwinding at various ATP and Mg2+ concentrations, combined with a scheme for the analysis of dissociation constants, suggested a model in which either free Mg2+ or free ATP binds to HCV helicase and competes with the binding of the ATP–Mg2+ complex, the entity that actually fuels enzymatic duplex strand separation by the helicase (42). Following this model, we determined experimentally from ATP and MgCl2 titrations that nearly equimolar concentrations of ATP and Mg2+ are required for optimal Nsp13 RNA helicase activity. A contributing factor for the sensitivity of Nsp13 helicase to excess Mg2+ may be the increased ionic strength of the reaction mixture, which would be predicted to alter the electrostatic interaction of the helicase to the RNA substrate (40).
Structural and biochemical studies indicate that ligand-induced changes in the conformation of a given helicase may be caused in part by Mg2+ coordination, which is linked to DNA activation of the ATPase cycle (45, 46). Recently, X-ray crystal structures of SARS-CoV-2 Nsp13 in multiple nucleotide-bound and conformational states were determined (6). It was revealed that the Mg2+ exists in an octahedral coordination, contacting both the β and γ phosphates of the bound AMP-PNP, as well as the side chain of Ser289 residing immediately adjacent to the invariant Lys288 of the Walker A box (motif I) that interacts with oxygen atoms of the α and γ phosphates. The Nsp13-bound Mg2+ interacts as well with three nearby water molecules that are also contacted by the conserved Asp374 and Glu375 of the Walker B box (motif II) implicated in ATP hydrolysis via facilitation of hydroxyl nucleophilic attack. Presumably, free Mg2+ cripples the coordination of nucleic acid binding to Nsp13-catalyzed ATPase activity, which in turn negatively affects duplex strand separation. Consistent with this, we observe that the rate of ATP hydrolysis by Nsp13 is compromised by the excess Mg2+ condition. Given the heightened interest in small molecule inhibitors of Nsp13 for the development of antiviral drugs (47, 48, 49, 50), it is plausible that incorporation of the Mg2+-binding site into the ATP-binding and hydrolysis site might improve drug specificity. Knowledge-based pharmacophore and solvent mapping strategies suggest a ranking in which a site proximal to the ATP-binding site of Nsp13 and within 3.5 Å of the catalytic Mg2+-binding site is optimal for virtual screening by molecular docking of small molecules that directly interfere with Nsp13 ATP hydrolysis (47). Nonetheless, specificity for inhibition of Nsp13’s ATPase activity by small molecules without interference in the catalytic functions of host ATPases and helicases could pose a challenge.
One of the advances in this study is the demonstration that Nsp13 can efficiently disrupt RNA–protein interactions in a unidirectional manner. ATPγS, often viewed as a poorly hydrolyzable analogue of the naturally occurring ATP, failed to support Nsp13-catalyzed streptavidin displacement. The inability of ATPγS, ADP, or AMP-PNP to support Nsp13 streptavidin displacement suggests that nucleotide binding by Nsp13 is not sufficient for protein–RNA disruption, despite evidence from structural studies of a conformational change in Nsp13 induced by AMP-PNP binding (6, 11). Like protein displacement from RNA, ATPγS did not support Nsp13 helicase activity on the 16-bp RNA partial duplex, indicating that the chemical energy of nucleoside hydrolysis is required to fuel duplex unwinding as well as protein remodeling on an RNA molecule.
Based largely on structural studies (1, 2, 5), it is proposed that Nsp13 may use its ATPase-driven RNA translocase activity to push the coronavirus RNA-dependent RNA polymerase (Nsp12) complex backward on its template RNA when a 3ʹ terminal mismatch is inadvertently incorporated during coronavirus replication so that the proofreading nuclease (Nsp14) can remove the nascently synthesized nucleotide to ensure polymerase fidelity. In this scenario, Nsp13 with its 5ʹ to 3ʹ directionality of translocation would move on the same RNA strand that serves as a template for the Nsp12 RNA-dependent RNA polymerase, only in the opposite direction of the advancing polymerase to push it backward. In our current study, we provide biochemical evidence that Nsp13 requires backbone continuity on the 5ʹ tail of the top strand for unwinding of the adjacent RNA duplex and can efficiently disrupt a very strong protein–RNA complex in an ATP hydrolysis–dependent manner. We note that the recent characterization of Nsp13 helicase conformational structures in the replication–transcription complex suggests that a swivel of the Nsp13 1B domain by an allosteric mechanism transitions the Nsp13 “thumbs” protomer from helicase active to helicase inactive, which may be relevant to a switch between RNA synthesis and backtracking (4); however, how this regulation takes place is still poorly understood. RNA structural elements (e.g., 3ʹ -terminal mismatch and sugar–phosphate backbone integrity) and interactions with other protein factors are likely to influence Nsp13 enzymology and steric access to push backward the RNA-dependent RNA polymerase complex to enable proofreading or template switching during subgenomic transcription.
Experimental procedures
Plasmid construction
The SARS-CoV-2 coding sequence displays some codon bias (51), which potentially reduces recombinant protein expression in eukaryotic systems. Therefore, to generate recombinant SARS-CoV-2 Nsp13 protein (hereafter referred to as Nsp13), the Nsp13 coding sequence was synthesized as a fragment with codon optimization (Fig. S8) for insect cell expression. pFastBac-HTB-Nsp13-FLAG was created by inserting a DNA fragment containing the Nsp13 open reading frame with a 5ʹ-flanking BamHI restriction site and 3ʹ-flanking FLAG tag and XhoI restriction site (Genewiz) into the pFastBac-HTB plasmid (Thermo Fisher). This allowed for purification of full-length product and the removal of the 6xHis tag, which was shown to significantly reduce SARS-CoV-1 Nsp13 helicase activity (8).
The codon AAG coding for the invariant lysine residue K288 in the Walker A box (Motif I) of the Nsp13 helicase was mutated to AGG (arginine) through site-directed mutagenesis of the pFastBac HTB-Nsp13-FLAG plasmid to generate the recombinant Nsp13-K288R protein. Baculovirus expressing Nsp13-K288R was created, and the protein was purified and stored using the same conditions as the nonmutated Nsp13.
Recombinant Nsp13 protein expression and purification
pFastBac-HTB-Nsp13-FLAG was transformed into DH10Bac competent cells (Thermo Fisher) to generate baculovirus DNA containing the Nsp13 coding sequence. Baculovirus DNA was transfected into 2 x 106 Spodoptera frugiperda Sf9 insect cells, grown in Grace’s Insect Cell Medium supplemented with 10% fetal bovine serum and penn/strep, using six-well dishes to generate baculovirus. This baculovirus in turn was used to infect 2 × 107 Sf9 insect cells to generate high-titer baculovirus.
Hi5 insect cells (Thermo Fisher), grown in ExpressFive SFM Medium supplemented with L-glutamine and penn/strep, were infected with the high-titer baculovirus derived from pFastBac-HTB-Nsp13-FLAG plasmid. Forty-eight hours post infection, the baculovirus-infected Hi5 insect cells were harvested by centrifugation at 500g for 8 min in 250-ml conical-bottom centrifuge tubes (Nalgene). Cells were washed with 35 ml 1× PBS, transferred to 50-ml conical tubes (Falcon), and centrifuged again at 500g for 5 min. Cells were washed again with 1× PBS containing protease inhibitor (Roche) and centrifuged once more at 500g for 5 min. Cell pellets were stored at −80 °C until used for extraction and recombinant protein purification by sequential affinity chromatography (Fig. S9).
Briefly, cell pellets representing 2 × 108 infected Hi5 insect cells were thawed on ice and resuspended in 20 ml Lysis Buffer (50 mM Tris-HCl [pH 7.4], 500 mM NaCl, 0.5% NP40, 10% glycerol, 5 mM β-mercaptoethanol (BME), and protease inhibitor without EDTA). The cells were lysed for 45 min at 4 °C with gentle rotation. The cell lysate was centrifuged at 48,380g for 10 min at 4 °C, and the supernatant was filtered through a 0.45-micron PVDF filter (Millipore). The clarified supernatant was applied to a 1-ml HisTrap HP column pre-equilibrated in Buffer TN (50 mM Tris-HCl [pH 7.4], 500 mM NaCl, 10% glycerol, and 5 mM BME) with 10 mM imidazole and protease inhibitor without EDTA. The column was washed with 5 ml each of Buffer TN with 10 mM, 20 mM, and 40 mM imidazole and protease inhibitor without EDTA. The protein was eluted off the HisTrap HP column with 5 ml of Buffer TN with 400 mM imidazole. The eluted protein was then dialyzed into Buffer TN with no imidazole using an Amicon 30K centrifugal filter. Protein concentration was estimated with Pierce 660 nm protein assay kit (Thermo Fisher). TEV protease, 1 μg, was added for every 25 μg of protein, and the protein was cleaved overnight at 4 °C with gentle rotation. Following cleavage of the protein with TEV protease, the sample was dialyzed into NETN Buffer (50 mM Tris-HCl [pH 7.4], 500 mM NaCl, 0.5% NP40, 1 mM EDTA) added to 500 μl pre-equilibrated anti-FLAG M2 affinity beads and rotated for 2 h at 4 °C. The beads were washed twice with NETN Buffer. Protein was eluted from the beads by adding 500 μl Storage Buffer (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 5 mM BME) with 3× FLAG peptide (250 μg), incubating for 1 h with rotation at 4 °C and pouring the beads into a small column. Gravity was used to collect the elution. This step was repeated, and the fractions from both 3× FLAG elutions were pooled and dialyzed against Storage Buffer using another Amicon 30K centrifugal filter, followed by storage at −80 °C. Protein concentration was estimated, and samples from each step of the purification and the final protein sample were analyzed by electrophoresis using a Bis-Tris 4 to 12% gradient polyacrylamide gel followed by Coomassie staining. Purity of the recombinant Nsp13 protein was determined to be at least 90% homogeneous (Fig. 1A).
Preparation of nucleic acid substrates
The oligonucleotides used to prepare the partial duplex substrates used in this study are shown (Table S3). To prepare RNA, DNA, DNA–RNA hybrid, and poly(hexa)ethylene glycol (PEG) forked substrates, 10 pmol of ss RNA/DNA was incubated at 37 °C for 40 min in T4 Polynucleotide Kinase (PNK) buffer (70 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 5 mM dithiothreitol [DTT], 30 μCi γ-32P-ATP, and PNK) (New England Biolabs) in a 20-μl reaction mixture. PNK was heat-inactivated for 20 min at 75 °C. Excess γ-32P-ATP was removed by centrifugation of the sample in a G25 spin column (Cytiva). Top (complementary) strand, 25 pmol, was added to the radiolabeled ss RNA/DNA, and the final volume was brought up to 50 μl with ddH2O. Substrates were annealed by heating at 95 °C for 5 min followed by cooling to room temperature (RT) overnight. Substrates were stored at 4 °C.
Theoretical Tm values of selected partial duplex DNA, RNA, or DNA/RNA hybrid substrates were determined using the calculator https://penchovsky.atwebpages.com/TMres.php (52). These Tm values take into account duplex DNA concentration and monovalent cation concentration.
Multiple-turnover helicase assays
Nsp13 (indicated in monomer concentration throughout) was incubated with RNA partial duplex substrates ranging in duplex length from 13-bp to 22-bp. The 20-μl reaction mixtures contained 0.25 nM partial RNA substrate, 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 μg/ml bovine serum albumin (BSA), 5 nM unlabeled oligonucleotide (same sequence as radiolabeled oligonucleotide), and the indicated concentration of ATP (unless specified otherwise) and MgCl2, as noted in the figure legends. Reactions were initiated by the addition of Nsp13 and incubated at 37 °C for 15 min unless specified otherwise. Reaction mixtures were quenched with the addition of 20 μl 2× Stop buffer (18 mM EDTA, 0.6% SDS, 25% glycerol, 0.04% xylene cyanol, 0.04% bromophenol blue). Reaction products were resolved on native 12% polyacrylamide gels electrophoresed at 200 V for 2 h at RT. Gels were exposed to a phosphorimager screen and scanned with a Typhoon FLA 9500 imager. ImageQuantTL software was utilized for quantification of gel images. All experiments with DNA, DNA–RNA hybrid, and PEG-modified substrates followed this general procedure used for the RNA forked duplex substrates.
For Hill plot analysis to assess the sigmoidal nature of the multiple-turnover helicase data as a function of Nsp13 concentration, the Hill equation (53) was applied to helicase activity rather than ligand binding. Hill slopes (Hill coefficients) and R2 values (correlation coefficients) were determined by entering mean and standard deviation data values into GraphPad Prism software and utilizing the nonlinear regression option “Specific binding (unwinding) with Hill slope” described by the equation:
where
Bmax is the maximum specific binding (unwinding) achievable;
Kd is the concentration of enzyme that achieves ½ maximal binding (unwinding);
X is the concentration of enzyme;
h is the Hill slope (coefficient).
PEG sequestration helicase assays
For helicase sequestration experiments, Nsp13 (0.078 nM) was preincubated for 8 min at 37 °C in 20-uL reaction mixtures containing 2 mM ATP, 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 μg/ml BSA, 5 nM unlabeled oligonucleotide (same sequence as radiolabeled oligonucleotide), 1 mM MgCl2, and the indicated amounts of unlabeled partial duplex competitor RNA substrates containing the PEG linker in either the top or bottom strand. Subsequently, 5 fmol of radiolabeled 19-bp forked duplex tracker substrate was added to the mixture and incubated for an additional 10 min at 37 °C. The helicase reactions were quenched and resolved on 12% polyacrylamide gels and visualized as described under “Multiple-turnover helicase assays.”
Single-turnover kinetic helicase assays
The specified concentration of Nsp13 (see figure legends) was preincubated with 0.25 nM RNA partial duplex containing the specified number of base pairs (13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30) for 5 min at RT in 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 μg/ml BSA, and the indicated concentration of MgCl2, as noted in the figure legends. Reactions were initiated by the addition of 2 mM ATP, 5 nM unlabeled oligonucleotide (same as radiolabeled oligonucleotide), and 100 nM dT200 protein trap. The reaction mixture was incubated at 37 °C with 20-μl aliquots removed every 30 s for 2.5 min. A separate cocktail was made for no enzyme control and heat-denatured substrate control. Each aliquot was quenched by the addition of 20 μl 2× Stop buffer, as described above. Reaction products were resolved by native gel electrophoresis and analyzed as described in the multi-turnover helicase assay section. The intermediate band is a secondary structure formed by the radiolabeled strand not considered in quantitative helicase assessment.
Streptavidin displacement assays
The streptavidin displacement reaction mixtures (20 μl) contained the indicated radiolabeled RNA (and/or DNA, for multi-turnover kinetic assays) substrate (0.25 nM) with a strategically positioned biotin covalently attached to the oligonucleotide (see Table S3; 5ʹ end: biotin linked to thymidine base; 3ʹ end: biotin linked to 3′ hydroxyl), 2 mM ATP, 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 mg/ml BSA, and 1 ml RNase inhibitor (20 U of Invitrogen SUPERase-In or 40 U of Promega RNasin Plus) and the indicated concentration of MgCl2, as noted in the figure legends. RNA substrate was preincubated with 3 nM streptavidin monomer (Invitrogen) for 10 min at 37 °C. Reactions were then initiated by adding 1 mM biotin (Pierce) followed by Nsp13 and incubated at 37 °C for 15 min. Reactions were quenched with the addition of 20 ml of 2× Strep Displacement Stop buffer (18 mM EDTA, 25% glycerol, 0.04% xylene cyanol, 0.04% bromophenol blue). For multi-turnover kinetic assays, the streptavidin and Nsp13 concentrations were 1.6 nM and 0.75 nM, respectively. Reactions were quenched at 1, 2, 4, 8, 12, and 16 min time points. Products were resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) polyacrylamide gels electrophoresed at 200 V for 2 h at RT. A PhosphorImager (Typhoon FLA 9500 imager) was used for detection, and as above, the ImageQuantTL software (GE Healthcare) was used for quantitation of the reaction products.
ATPase assays
For experiments to determine kcat, ATPase reactions (45 μl) contained 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 μg/ml BSA, 60-nucleotide ssDNA or ssRNA effector (1 μM), a trace amount of γ-32P-ATP (∼2 nM) mixed with 1 mM cold ATP, and either 1 mM or 5 mM MgCl2 as specified. The reactions were initiated with the addition of Nsp13 protein and incubated at 37 °C for a total of 10 min with 5 μl samples removed every 2.5 min, including one sample removed immediately after the addition of Nsp13 before the incubation period. Samples were quenched in ATPase STOP solution (33.3 mM EDTA, 6.7 mM ATP, 6.7 mM ADP, the last two to visualize ATP and ADP on the thin layer chromatography [TLC]) and spotted on PEI cellulose TLC sheets (10 cm x 20 cm). The solvent (1 M formic acid, 0.8 mM LiCl) was allowed to migrate up to 1 cm from the top of the sheet, and the sheet was exposed to a phosphoimager screen for 0.5 to 1 h and scanned. ImageQuant TL software was used to quantitate hydrolyzed ATP. Less than 10% of the ATP substrate was consumed in the reaction over the entire time course of the experiment. The kcat values were expressed as the mean of at least three independent determinations with standard deviation. ATPase assays to determine Keff were similar to those for assays to determine kcat with a few exceptions. ATPase assay reactions (15 μl) contained the same buffer components, including 2 mM ATP and 1 mM MgCl2, but with increasing effector concentrations (4–1000 nM) of ssRNA or ssDNA effector. Reactions were again initiated by the addition of Nsp13 (0.05 nM) and incubated for 12 min at 37 °C. Samples (5 μl) were removed at 0 and 12 min and quenched by the addition of STOP solution and resolved as described above. The Keff values were expressed as the mean of at least three independent determinations with standard deviation.
Electrophoretic mobility shift assay
Nsp13 (indicated in monomer concentration throughout) was incubated with DNA or RNA 16-bp forked duplex substrate. The 20-μl reaction mixtures contained 0.25 nM DNA or RNA 16-bp forked substrate, 20 mM Hepes-KOH (pH 7.5), 20 mM NaCl, 1 mM DTT, 100 μg/ml BSA, 1 mM MgCl2 or the indicated concentration, and 2 mM ATPγS when indicated in the figure legend. Reactions were initiated by the addition of Nsp13 and incubated at RT for 15 min. Reaction mixtures were quenched with the addition of 4 μl Stop Buffer (74% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue). Reaction products were resolved on native 5% polyacrylamide gels electrophoresed at 200 V for 2 h 15 min at 4 °C. Gels were exposed to a phosphorimager screen and scanned with a Typhoon FLA 9500 imager. ImageQuantTL software was utilized for quantification of gel images.
Data availability
All the data described herein are located within the article or the supplementary information. Therefore, all data are readily available to be shared from the article. There are no exceptions to the sharing of data, materials, and software.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Intramural Training Program, NIA, National Institutes of Health, and a Special COVID-19 Grant from the Office of the Scientific Director, NIA, National Institutes of Health. We thank Dr Tomasz Kulikowicz (NIA, National Institutes of Health) who supplied the purified recombinant TEV protease and provided technical assistance on some kinetic helicase assays. We wish to thank members of the Helicases and Genomic Integrity Section, Translational Gerontology Branch, NIA, National Institutes of Health, for helpful discussion and input. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
J. A. S., L. N. L., M. P. J., R. A. L., C. E. H., A. J. D., R. M. B. conceptualization; J. A. S., L. N. L., M. P. J., R. A. L., C. E. H., A. J. D., R. M. B. methodology; J. A. S., L. N. L., M. P. J., R. A. L., C. E. H., A. J. D., R. M. B. formal analysis; J. A. S., L. N. L., M. P. J., R. A. L., C. E. H., A. J. D., R. M. B. writing – original draft; J. A. S., L. N. L., M. P. J., R. A. L., C. E. H., A. J. D., R. M. B. writing – review & editing.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Supporting information
References
- 1.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Wang Q., Malone B., Llewellyn E., Pechersky Y., Maruthi K., et al. Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex. Nat. Struct. Mol. Biol. 2022;29:250–260. doi: 10.1038/s41594-022-00734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., Olinares P.D.B., et al. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman J.A., Douangamath A., Yadzani S., Yosaatmadja Y., Aimon A., Brandão-Neto J., et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adedeji A.O., Lazarus H. Biochemical characterization of Middle East respiratory syndrome coronavirus helicase. mSphere. 2016;1 doi: 10.1128/mSphere.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PloS One. 2012;7 doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang K.J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee N.R., Kwon H.M., Park K., Oh S., Jeong Y.J., Kim D.E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee W.T.C., Yin Y., Morten M.J., Tonzi P., Gwo P.P., Odermatt D.C., et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021;12:2525. doi: 10.1038/s41467-021-22830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mickolajczyk K.J., Shelton P.M.M., Grasso M., Cao X., Warrington S.E., Aher A., et al. Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase. Biophys. J. 2021;120:1020–1030. doi: 10.1016/j.bpj.2020.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu T., Huang M., Wu D., Ren Y., Zhang X., Han Y., et al. SARS-Coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can Be inhibited by Bismuth salts. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samdani M.N., Morshed N., Reza R., Asaduzzaman M., Islam A. Targeting SARS-CoV-2 non-structural protein 13 via helicase-inhibitor-repurposing and non-structural protein 16 through pharmacophore-based screening. Mol. Divers. 2022;12:1–19. doi: 10.1007/s11030-022-10468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller L.K. Insect baculoviruses: powerful gene expression vectors. Bioessays. 1989;11:91–95. doi: 10.1002/bies.950110404. [DOI] [PubMed] [Google Scholar]
- 20.Adams C., Boonen K., Laukens K., Bittremieux W. Open modification searching of SARS-CoV-2-human protein interaction data reveals novel viral modification sites. Mol. Cell Proteomics. 2022;21:100425. doi: 10.1016/j.mcpro.2022.100425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chathuranga K., Weerawardhana A., Dodantenna N., Lee J.S. Regulation of antiviral innate immune signaling and viral evasion following viral genome sensing. Exp. Mol. Med. 2021;53:1647–1668. doi: 10.1038/s12276-021-00691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo G., Gao M., Gao X., Zhu B., Huang J., Luo K., et al. SARS-CoV-2 non-structural protein 13 (nsp13) hijacks host deubiquitinase USP13 and counteracts host antiviral immune response. Signal Transduct Target Ther. 2021;6:119. doi: 10.1038/s41392-021-00509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy E. Mysteries of magnesium homeostasis. Circ. Res. 2000;86:245–248. doi: 10.1161/01.res.86.3.245. [DOI] [PubMed] [Google Scholar]
- 24.Murphy E., Steenbergen C., Levy L.A., Raju B., London R.E. Cytosolic free magnesium levels in ischemic rat heart. J. Biol. Chem. 1989;264:5622–5627. [PubMed] [Google Scholar]
- 25.Beran R.K., Bruno M.M., Bowers H.A., Jankowsky E., Pyle A.M. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 26.Kawaoka J., Jankowsky E., Pyle A.M. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 27.Amaratunga M., Lohman T.M. Escherichia coli rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 28.Green N.M. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickson W.A., Pähler A., Smith J.L., Satow Y., Merritt E.A., Phizackerley R.P. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peck M.L., Herschlag D. Adenosine 5'-O-(3-thio)triphosphate (ATPgammaS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA. 2003;9:1180–1187. doi: 10.1261/rna.2103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao N.Y., Zhang D., Yurieva O., O'Donnell M.E. CMG helicase can use ATPγS to unwind DNA: implications for the rate-limiting step in the reaction mechanism. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2119580119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matson S.W., George J.W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J. Biol. Chem. 1987;262:2066–2076. [PubMed] [Google Scholar]
- 33.Wong I., Lohman T.M. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 34.Yue K., Yao B., Shi Y., Yang Y., Qian Z., Ci Y., et al. The stalk domain of SARS-CoV-2 NSP13 is essential for its helicase activity. Biochem. Biophys. Res. Commun. 2022;601:129–136. doi: 10.1016/j.bbrc.2022.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang P.S., Jankowsky E., Planet P.J., Pyle A.M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbari M., Shanley D.P., Bohr V.A., Rasmussen L.J. Cytosolic self-DNA-A potential source of chronic inflammation in aging. Cells. 2021;10:3544. doi: 10.3390/cells10123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui C., Xiao T., Zhang S., Zeng H., Zheng Y., Liu B., et al. SARS-CoV-2 NSP13 inhibits type I IFN production by degradation of TBK1 via p62-dependent selective autophagy. J. Immunol. 2022;208:753–761. doi: 10.4049/jimmunol.2100684. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 39.Huston N.C., Wan H., Strine M.S., de Cesaris Araujo Tavares R., Wilen C.B., Pyle A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021;81:584–598.e5. doi: 10.1016/j.molcel.2020.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venus S., Jankowsky E. Measuring the impact of cofactors on RNA helicase activities. Methods. 2022;204:376–385. doi: 10.1016/j.ymeth.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putnam A.A., Gao Z., Liu F., Jia H., Yang Q., Jankowsky E. Division of labor in an oligomer of the DEAD-box RNA helicase Ded1p. Mol. Cell. 2015;59:541–552. doi: 10.1016/j.molcel.2015.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frick D.N., Banik S., Rypma R.S. Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: magnesium provides a bridge for ATP to fuel unwinding. J. Mol. Biol. 2007;365:1017–1032. doi: 10.1016/j.jmb.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harmon F.G., Kowalczykowski S.C. Biochemical characterization of the DNA helicase activity of the escherichia coli RecQ helicase. J. Biol. Chem. 2001;276:232–243. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 44.Choudhary S., Sommers J.A., Brosh R.M., Jr. Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J. Biol. Chem. 2004;279:34603–34613. doi: 10.1074/jbc.M401901200. [DOI] [PubMed] [Google Scholar]
- 45.Soultanas P., Dillingham M.S., Velankar S.S., Wigley D.B. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 1999;290:137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- 46.Toseland C.P., Webb M.R. ATPase mechanism of the 5'-3' DNA helicase, RecD2: evidence for a pre-hydrolysis conformation change. J. Biol. Chem. 2013;288:25183–25193. doi: 10.1074/jbc.M113.484667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freidel M.R., Armen R.S. Mapping major SARS-CoV-2 drug targets and assessment of druggability using computational fragment screening: identification of an allosteric small-molecule binding site on the Nsp13 helicase. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Lemus G.R., Menéndez C.A., Alvarado W., Byléhn F., de Pablo J.J. Toward wide-spectrum antivirals against coronaviruses: molecular characterization of SARS-CoV-2 NSP13 helicase inhibitors. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abj4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt A.N., Gallazzi F., Quinn T.P., Lorson C.L., Sönnerborg A., Singh K. Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opin. Ther. Pat. 2021;31:339–350. doi: 10.1080/13543776.2021.1884224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng J., Weissmann F., Bertolin A.P., Posse V., Canal B., Ulferts R., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 2021;478:2405–2423. doi: 10.1042/BCJ20210201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kames J., Holcomb D.D., Kimchi O., DiCuccio M., Hamasaki-Katagiri N., Wang T., et al. Sequence analysis of SARS-CoV-2 genome reveals features important for vaccine design. Sci. Rep. 2020;10:15643. doi: 10.1038/s41598-020-72533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaloudas D., Pavlova N., Penchovsky R. EBWS: essential Bioinformatics web services for sequence analyses. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018;16:342–953. doi: 10.1109/TCBB.2018.2816645. [DOI] [PubMed] [Google Scholar]
- 53.Hill A.V. The combinations of haemoglobin with oxygen and with carbon monoxide. I. Biochem. J. 1913;7:471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described herein are located within the article or the supplementary information. Therefore, all data are readily available to be shared from the article. There are no exceptions to the sharing of data, materials, and software.