Abstract
Genetic evidence showed two non-Mendelian genetic elements of Saccharomyces cerevisiae, called [URE3] and [PSI], to be prions of Ure2p and Sup35p, respectively. [URE3] makes cells derepressed for nitrogen catabolism, while [PSI] elevates the efficiency of weak suppressor tRNAs. The same approach led to identification of the non-Mendelian element [Het-s] of the filamentous fungus Podospora anserina, as a prion of the het-s protein. The prion form of the het-s protein is required for heterokaryon incompatibility, a normal fungal function, suggesting that other normal cellular functions may be controlled by prions. [URE3] and [PSI] involve a self-propagating aggregation of Ure2p and Sup35p, respectively. In vitro, Ure2p and Sup35p form amyloid, a filamentous protein structure, high in β-sheet with a characteristic green birefringent staining by the dye Congo Red. Amyloid deposits are a cardinal feature of Alzheimer’s disease, non-insulin-dependent diabetes mellitus, the transmissible spongiform encephalopathies, and many other diseases. The prion domain of Ure2p consists of Asn-rich residues 1 to 80, but two nonoverlapping fragments of the molecule can, when overproduced, induce the de nova appearance of [URE3]. The prion domain of Sup35 consists of residues 1 to 114, also rich in Asn and Gln residues. While runs of Asn and Gln are important for [URE3] and [PSI], no such structures are found in PrP or the Het-s protein. Either elevated or depressed levels of the chaperone Hsp104 interfere with propagation of [PSI]. Both [URE3] and [PSI] are cured by growth of cells in millimolar guanidine HCl. [URE3] is also cured by overexpression of fragments of Ure2p or fusion proteins including parts of Ure2p.
INTRODUCTION
Based on their genetic properties, we proposed that two non-Mendelian genetic elements of Saccharomyces cerevisiae, called [URE3] and [PSI], are prions (infectious proteins) of Ure2p and Sup35p, respectively (102). If a genetic element is a prion, it is a gene composed of protein instead of nucleic acid. Here we examine the basis for these proposals and what is known of the mechanism by which these prion genes propagate. The idea of an “infectious protein” arose in 1967 from studies of the mammalian transmissible spongiform encephalopathies (TSEs) (2, 48), and considerable evidence has accumulated that scrapie is caused by such an agent (83, 99). However, the difficulty of studying these systems has so far precluded obtaining proof of the “protein-only” model for the mammalian diseases.
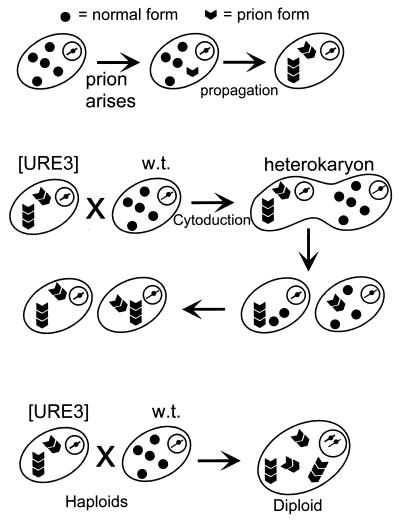
The known infectious proteins are altered forms of a cellular protein which have lost their normal functions but have acquired the ability to convert the normal form of the protein into the same abnormal form (Fig. 1). [URE3] makes cells derepressed for nitrogen catabolism (1, 59), while [PSI] elevates the efficiency of weak suppressor tRNAs (23, 24). The same genetic approach led to identification of the non-Mendelian element [Het-s] of the filamentous fungus Podospora anserina as a prion of the het-s protein (21). The prion form of the het-s protein is required for heterokaryon incompatibility, a normal fungal function, suggesting that other normal cellular functions may be controlled by prions.
FIG. 1.
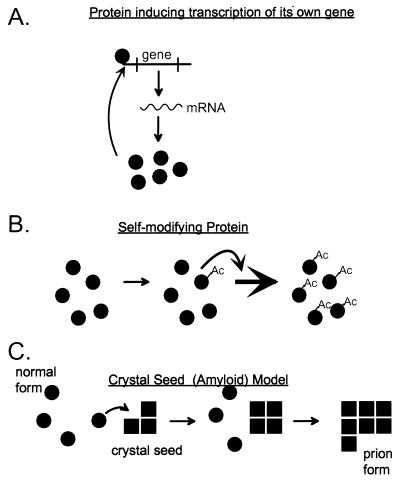
Definition of a prion. “Prion” means an infectious protein. Many mechanisms can be imagined for such an entity (48), including a protein that induces its own gene’s transcription (A); a self-propagating covalent protein modification, such as acetylation, in which the modified form of the protein is much better at self-acetylation than the unacetylated form (B); and a self-propagating change in conformation, such as amyloid formation—the likely mechanism for the known prions (C).
[URE3] and [PSI] involve a self-propagating aggregation of Ure2p (37, 68) and Sup35p (77, 79, 81), respectively. In vitro, Ure2p (90) and Sup35p (46, 55) form amyloid. Amyloid is a filamentous protein structure, high in β-sheet structures and with a characteristic green birefringent staining by the dye Congo red. Amyloid deposits are a cardinal feature of Alzheimer’s disease, non-insulin-dependent diabetes mellitus, the TSEs, and many other diseases.
Chaperones, particularly Hsp104, are critical for [PSI] propagation (14, 15, 71). Either elevated or depressed levels of Hsp104 interfere with the propagation of [PSI]. Both [URE3] and [PSI] are cured by growth of cells in millimolar guanidine HCl (M. Aigle, cited in reference 24; 96, 102). [URE3] is cured by overexpression of fragments of Ure2p or fusion proteins including parts of Ure2p (37). The prion domain of Ure2p consists of Asn-rich residues 1 to 80 (65, 68), but either of two nonoverlapping fragments of the molecule can, when overproduced, induce the de novo appearance of [URE3] (65). The prion domain of Sup35p consists of residues 1 to 114, also rich in Asn and Gln residues (92). While runs of Asn and Gln are important for [URE3] (65) and [PSI] (31), no such structures are found in PrP or the Het-s protein.
Many excellent reviews of this subject have appeared, and they should be consulted for different views and areas of emphasis (28, 58, 61, 63, 95, 103–105, 107, 108).
BACKGROUND AND HISTORY
Before Avery, McCloud, and McCarty showed that DNA could be the genetic material, many believed that all genes were made of proteins. The reemergence of the notion that proteins can mediate inheritance begins with even earlier events, with the recognition of scrapie, a uniformly fatal disease of sheep, in several countries in Europe in the early 18th century. The name of the disease derives from the apparent itching which leads the affected animals to rub their fur against trees or other structures, scraping off much of their coats. The human form of the disease was first described in the 1920s by German and Austrian physicians, whose names are immortalized in the various clinical forms of the conditions, Creutzfeldt-Jakob disease (CJD) and Gerstmann-Straussler-Scheinker disease (GSS) (25, 41, 42). At the time, there was no suspicion that these conditions were related to scrapie.
In 1936, the infectivity of scrapie by intraocular injection of sheep was demonstrated by Cuille and Chelle in France (27). Transmission from sheep to goats was demonstrated, the first of many interspecies transmissions that were to be achieved in later years (26). These results were extensively reproduced and extended in the United Kingdom by groups at Compton and Edinburgh (47, 109). It soon became clear that the scrapie agent was extraordinarily resistant to treatments, such as heat or fixation with formaldehyde, which affect most known bacteria and viruses (78). However, since the assays involved inoculation of sheep or goats and waiting for over 1 year, progress in purification of the agent was slow.
In 1957, Zigas and Gajdusek discovered kuru, an epidemic fatal neurological disease among the Fore, a Stone Age tribe in the New Guinea highlands (112). Infection was considered, among many other possible etiologies, but the animals inoculated with pathological material were discarded after they failed to develop disease in the first few months.
Hadlow noted the similarity of the pathology of kuru to that of scrapie and suggested that the diseases might be related (50). He suggested that an infectious etiology of kuru again be examined but that the animals be kept much longer in view of the long incubation times for scrapie transmission. Gibbs and Gajdusek inoculated monkeys and found that they indeed succumbed to a disease with all of the signs and pathologic findings typical of kuru or scrapie (43, 44). Klatzo et al. noted the similarity of kuru to CJD and GSS (56), and autopsy material from patients with CJD and GSS was likewise found to be infectious (43, 44).
These diseases became known as the transmissible spongiform encephalopathies (TSEs), spongiform because the large vesicles and extensive neuronal loss in the affected tissue gave the brain a spongy appearance in pathologic sections. Deposits of amyloid, a homogeneously staining material composed of protein (in spite of its name), was also typical of kuru, although this feature is actually unusual in other forms of the human TSEs. As discussed below in some detail, amyloid is a special filamentous protein structure composed of β-sheets, which may prove to be the unifying feature of a large group of diseases, including those due to the yeast prions [URE3] and [PSI].
Early Findings That Suggest an Infectious-Protein Etiology of the TSEs
The human TSEs could appear in an epidemic form (kuru), a familial form (including GSS), and, most commonly, an apparently spontaneous form (generally called CJD). Gajdusek and coworkers found that all three forms were infectious for monkeys (43, 44). This result is difficult to explain by classical disease patterns, but, as appreciated only much later, this is a pattern expected for an infectious protein (prion).
In 1966, Alper et al. showed that the scrapie agent was remarkably resistant to UV irradiation, far more resistant than DNA or RNA viruses (3). This led to speculation that the scrapie agent might replicate without an essential nucleic acid (2). These findings also led Griffith to suggest several means by which a protein could be infectious without an essential nucleic acid and not violate the essential tenets of molecular biology (48). One suggestion was essentially the prion model as understood today and was summed up by the author as follows: “There is an obvious analogy between the idea presented here and the idea that a gas can only condense on nuclei which are already present: many of the more general schemes could be summed up by saying that the subunits can only polymerize by utilizing ’condensation nuclei’ of polymer which are already there.” As apt as this formulation may appear today, it seems to have had little impact at the time. One has difficulty finding mention of this notion through the 1970s, and even Prusiner’s 1982 review coining the term “prion” does not mention this concept as a possible explanation of the “protein-only” model.
Amyloid plaques were well known in scrapie and kuru, and studies in the late 1960s defined amyloid as a protein structure rich in β-sheet (reviewed in reference 45), but just as amyloid is often secondary to chronic infections, it was not considered the central feature of scrapie at the time.
PrP and Scrapie
PrP, the protein central to scrapie, was first identified as a mouse gene controlling the scrapie incubation period and therefore was named Sinc by Dickinson et al. (35). However, it was not suspected that the product of this gene was itself the infectious element. Merz et al. identified filamentous structures specifically in infectious materials, the “scrapie-associated filaments” (69). The purification of the infectious material by Prusiner and coworkers identified PrP, a 27- to 30-kDa species which was relatively resistant to protease digestion (8). This protease-resistant species was not found in uninfected brain. With the cloning of the PrP gene (17, 73), it was clear that PrP was a host-encoded protein whose form but not expression was tied to the disease. The identification of Dickinson’s Sinc gene as the PrP gene (12) and the finding that patients with familial CJD had mutations in their PrP gene (52, 74) showed that PrP was central to the disease process.
The demonstration that making mice transgenic for hamster PrP (84) made them susceptible to hamster scrapie agent suggested that PrP was actually essential for this disease. This was proven when it was shown that PrP knockout mice were immune to scrapie (10).
Is PrPSc the scrapie agent?
Since PrP is necessary for the propagation of scrapie and since an altered form of the protein (PrPSc) is found in the purified infectious material, it is believed that PrPSc is identical to the infectious material. This conclusion has been quite controversial (16, 39), however, because efforts to demonstrate that PrPSc is sufficient for scrapie were unsuccessful. Overproduction of PrP in certain transgenic lines leads to death, but the dead animals contain no infectious material (100). Expression in mice of PrP with mutations that lead to CJD in man result in death with classical scrapie pathology, but again no infectivity for normal mice (53). Attempts to create infectious material from PrP produced in bacteria or yeast have failed (see, e.g., reference 98). However, this type of negative evidence does not prove that PrP is not the infectious agent, and no other candidates have been found.
GENETIC CRITERIA FOR A PRION
In yeast and other fungi, viruses are infectious via the cytoplasmic mixing that occurs on mating or heterokaryon formation (reviewed in references 101 and 106), not by going out of one cell and into another. They are, in effect, sexually transmitted diseases. Some mammalian viruses, particularly those of the herpesvirus group and human immunodeficiency virus, are known to be transmitted in this way as well. The sigma virus of Drosophila was first found as a non-Mendelian genetic element conferring sensitivity to CO2 (91). This implies that an infectious protein in a yeast or fungus should appear as a non-Mendelian (nonchromosomal) genetic element.
We started with the concept of an infectious protein (prion) as an altered form of a cellular protein which has lost its normal function but which can convert the normal form of the protein into the same abnormal form, and we proposed three genetic criteria by which one could recognize prions in microorganisms such as yeast (Fig. 2) (102). The main purpose of these criteria was to distinguish prions from nucleic acid replicons such as viruses and plasmids.
FIG. 2.
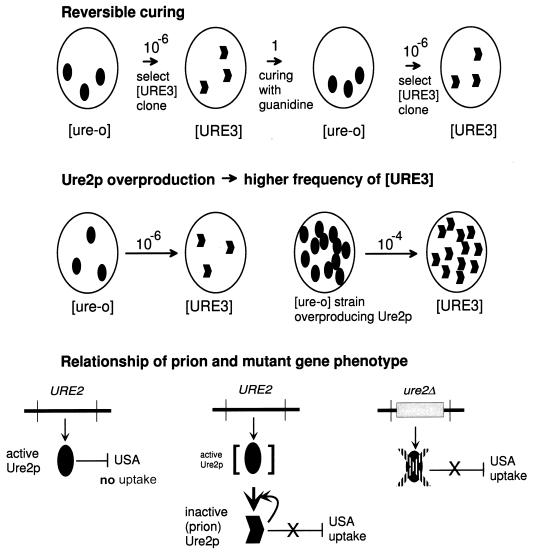
Genetic criteria for a prion illustrated by [URE3]. These genetic properties are expected of a prion but not of a nucleic acid replicon (102). Reversible curability means that a cured strain can again develop the prion de novo. Overproduction of the normal form of the protein increases the probability that the prion will arise. The presence of the prion causes the same phenotype as deletion of the chromosomal gene for the protein, because both lack the normal form, and the chromosomal gene for the protein is necessary for propagation of the prion. The requirement for a chromosomal gene for propagation of a nonchromosomal genetic element or infectious entity is frequently seen and is not an indication that the genetic element is a prion.
Reversible Curability
If a prion can be cured, it should be possible for the prion to arise again spontaneously in the cured strain. Whatever the spontaneous change that gives rise to the prion originally can occur again in the cured strain. This is generally not the case for viruses and plasmids, which, once cured, will not arise again at measurable frequencies without being reintroduced from outside.
Appearance of the Prion Induced by Overproduction of the Protein
Overproduction of the protein produces more molecules that have the potential to undergo the spontaneous prion change. This should increase the frequency with which the prion arises spontaneously. In contrast, overproduction of a chromosomally encoded polymerase or other factor involved in propagation of a plasmid or virus would not be expected to induce the appearance of the nucleic acid replicon.
Phenotypes of the Presence of the Prion and Mutation of the Gene for the Protein
The chromosomal gene for the protein that has the potential to become a prion is necessary for the propagation of the prion because it propagates by converting the normal form into the prion form. Both mutation of the gene and the presence of the prion form result in absence of an active protein. In the first case, the protein is not made; in the second, it is made but is quickly inactivated by being converted to the prion form. Thus, the phenotypes are the same.
The presence of a nucleic acid replicon generally confers the opposite phenotype on cells to that conferred by mutation in a chromosomal gene needed for its propagation. For example, mutation of a gene for a DNA polymerase needed for propagation of mitochondrial DNA results in loss of the mitochondrial DNA and inability of the cells to grow on glycerol or ethanol. In contrast, the presence of the mitochondrial DNA makes cells able to grow on glycerol (Table 1).
TABLE 1.
Relationship of phenotypes of a prion and mutations in the gene encoding the proteina
| Non-Mendelian element | Phenotype due to:
|
Relation of the two phenotypes | Does replacing the gene restore the phenotype? | |
|---|---|---|---|---|
| Presence of non-Mendelian element | Chromosomal mutant that loses the element | |||
| M dsRNA | Killer + | Killer − | Opposite | No |
| mitDNA | Glycerol + | Glycerol − | Opposite | No |
| Suppressive petite mitDNA | Glycerol − | Glycerol − | Same | No |
| Prion | Defective | Defective | Same | Yes |
| [URE3] | USA uptake + | USA uptake + | Same | Yes |
| [PSI] | Suppressor ⇑ | Suppressor ⇑ | Same | Yes |
Usually the presence of a nucleic acid replicon produces the opposite phenotype to that produced by a mutation in a gene needed for its propagation. However, there is a circumstance in which the phenotypes for a nucleic acid replicon and mutation in one of its maintenance genes could be the same. Certain deletion mutants of mitochondrial DNA (mitDNA) (called suppressive petites) make their presence known by efficiently eliminating the normal mitochondrial DNA and thus producing the same defective phenotype as the absence of the mitochondria DNA. Such a mutant mitochondrial DNA depends for its replication on the same chromosomal genes as does the wild-type mitochondrial DNA. The difference is that these mutant mitochondrial DNAs are dominant to the wild type, so that they appear as the presence of a non-Mendelian genetic element. However, this type of mutant may be distinguished from a prion by the result of replacing the chromosomal maintenance gene. Deletion of the potential prion protein produces the defective phenotype, and replacement of the gene restores the normal phenotype. Deletion of the gene needed for propagation of the defective mitDNA gives the defective phenotype, but replacement of the gene does not repair the defect, since mitochondrial DNA is still missing.
As described below, we used these genetic properties to identify two prion systems in Saccharomyces (102), and Coustou et al. have identified a third prion in Podospora (21). It is remarkable that none of these are known to be properties of scrapie, so that this approach has contributed evidence for the existence of prions not yet available in the mammalian system.
[URE3], A PRION FORM OF URE2P AFFECTING NITROGEN CATABOLISM
Discovery of [URE3]
The first step in uracil biosynthesis is catalyzed by aspartate transcarbamylase, and its product is ureidosuccinate (Fig. 3). Ureidosuccinate (USA) is normally not taken up from the medium in the presence of ammonium. Francois Lacroute isolated mutants that could take up ureidosuccinate under these conditions (USA+) and named them ure for ureidosuccinate. Most mutants had recessive chromosomal defects, including ure2 (Fig. 3). One mutant was dominant and showed irregular segregation in meiosis (59). The USA+ trait from this strain was also transmissible by transfer of cytoplasm from cell to cell by cytoduction, indicating that it was due to a nonchromosomal genetic element (1).
FIG. 3.
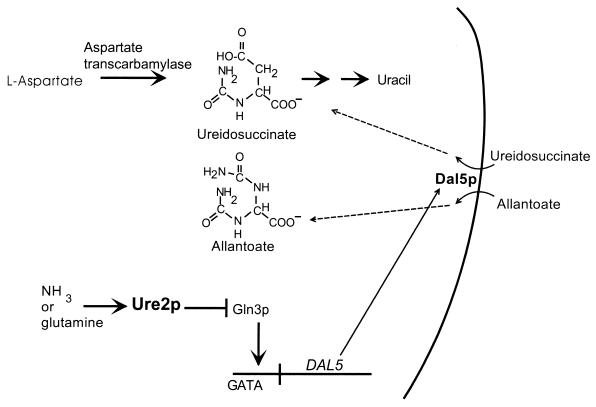
How ureidosuccinate uptake is controlled by the nitrogen source. Yeast cells growing on a rich source of nitrogen (such as ammonia or glutamine) repress the expression of transporters and metabolic enzymes needed for utilization of poor nitrogen sources. This is called nitrogen catabolite repression (19, 66). Ureidosuccinate (USA) is the product of aspartate transcarbamylase (ura2), the first step in uracil biosynthesis. The chance chemical resemblance of USA to the poor nitrogen source allantoate results in the ability of the allantoate transporter, Dal5p, to import USA. The presence of a rich nitrogen source, such as ammonia, is transmitted to Ure2p, which then blocks the action of the positive transcription factor, Gln3p. Thus, ammonia represses USA uptake, preventing ura2 cells from using USA in place of uracil. Modified from reference 105 with permission of the publisher.
Prion Genetic Criteria Satisfied by [URE3]
Aigle and Lacroute showed that although the phenotypes of [URE3] strains were the same as those of ure2 mutants, the ure2 mutants could not propagate [URE3] (1). In his thesis, Aigle recognized that this is a paradoxical finding and suggested that perhaps “the [URE3] factor renders inactive the product of URE2.” We confirmed this result (102) and, as discussed above, suggested that this is one of the properties expected if [URE3] is a prion of Ure2p (Fig. 2). Since ure2 deletion mutants can be complemented by a plasmid carrying the URE2 gene (20), [URE3] is not a defective interfering derivative of some putative wild-type nucleic acid replicon depending on URE2 (see above and the footnote to Table 1).
[URE3] is curable by growth in the presence of low concentrations of guanidine HCl, but cells which have again acquired [URE3] can be isolated from the cured strains (102). This is another property expected of a prion but not a nucleic acid replicon. [URE3] arises in the cured strains with about the same frequency as it arose in the original parent strain (102). This shows that the isolation of [URE3] strains does not generally select a chromosomal mutant which is predisposed to give rise to the prion. The analogous problem in mammals has long been an unresolved question. The inherited forms of CJD are due to a dominant mutation in the gene for PrP, which results in the patient developing the disease with almost 100% certainty instead of the 1 in 106 per year chance of spontaneous CJD. It has often been speculated that the “spontaneous” form of CJD may be due to somatic mutation producing one of the changes in the PrP gene identical to those seen in the inherited form. However, the cells in which the disease starts would be among the first neurons killed, and so this speculation can never be verified. In yeast, the [URE3] disease is not lethal and the curability of [URE3] permits this question to be answered. It should also be emphasized that the fact that [URE3] is curable by the protein denaturant guanidine does not imply that it is a prion. The concentrations used in the curing are only 1 to 5 mM, not enough to denature any known protein. Moreover, RNA replication of poliovirus has long been known to be inhibited by similar low concentrations of guanidine, and mutants resistant to this “curing” are altered in the poliovirus RNA-dependent RNA polymerase (89).
The third property expected of a prion is that overproduction of the protein should increase the frequency with which the prion form arises. Overproduction of Ure2p results in a 20- to 200-fold increase in the frequency with which [URE3] appears de novo (102). A more detailed examination of this important experiment is given below.
Protease Resistance of Ure2p in [URE3] Strains
Extracts of isogenic [URE3] and [ure-o] strains were treated with proteinase K as a test of whether Ure2p was altered in [URE3] strains, a strong prediction of the prion hypothesis for [URE3]. It was found that Ure2p was more protease resistant in [URE3] strains, with 30- to 32-kDa fragments of the 40-kDa Ure2p showing persistence for longer times in the extracts of prion-carrying strains. One [URE3] strain showed a very stable 10-kDa fragment (68). Curing [URE3] resulted in the pattern returning to that of the wild-type extract. Mixing an extract of [URE3] cells with that of [ure-o] cells resulted in an additive result, indicating that the protease resistance of Ure2p was not due to the absence of a protease or the presence of a protease inhibitor in the [URE3] cells (68). The state of nitrogen regulation does not affect the protease resistance of the Ure2p. [ure-o] cells grown on proline or ammonia as the nitrogen source have the same protease sensitive form of Ure2p (67). This indicates that [URE3] has an abnormal form of Ure2p and this causes the derepression of nitrogen catabolism.
Prion Domain(s) of Ure2p
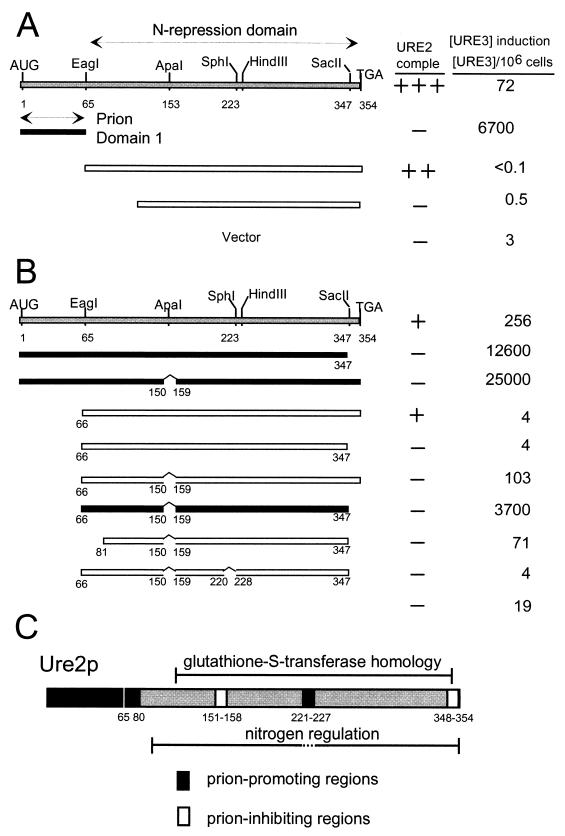
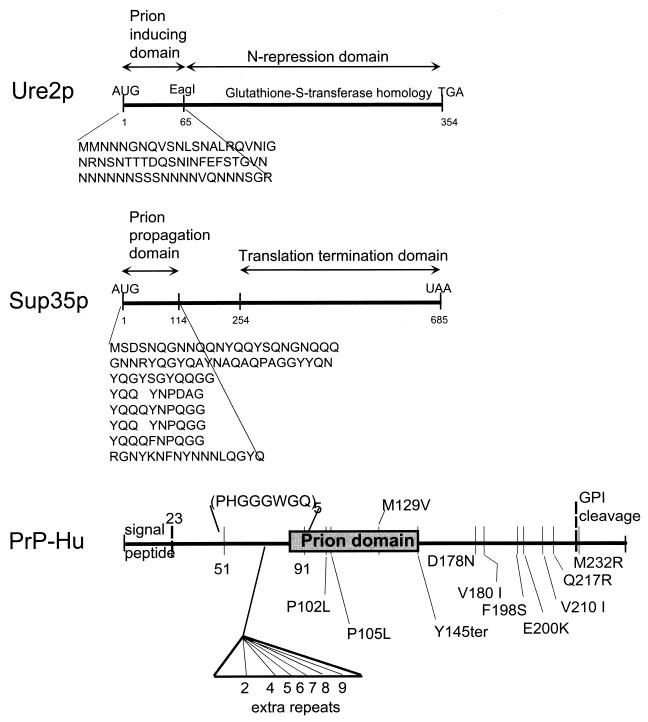
Deletion analysis showed that the N-terminal 65 amino acid residues are sufficient, when overexpressed in a strain with an intact chromosomal copy of URE2, to induce the appearance of [URE3] at high frequency. While overexpression of the intact Ure2p increases the frequency of [URE3] arising to 20- to 200-fold above the background rate, overexpression of this 65-residue fragment results in a 2,000- to 10,000-fold increase in the rate (68). In contrast, the C-terminal part of Ure2p, lacking precisely this N-terminal 65 residues, is capable of carrying out nitrogen regulation (20, 68). This N-terminal domain was therefore named the prion-inducing domain, and the C-terminal domain from residues 66 to 354 was dubbed the nitrogen regulation domain (Fig. 4A). The fact that a domain sufficient to induce [URE3] at high efficiency and the nitrogen regulation domain can be separated completely argues that [URE3] is not a stable transcriptional state. Transcriptional regulation and prion induction are apparently separate functions of a single molecule.
FIG. 4.
Prion domain(s) of Ure2p. Deletion mutants overexpressing fragments of Ure2p from a plasmid were assayed for the ability to induce the de novo formation of [URE3] in a strain expressing the full-length Ure2p from the chromosomal gene, and, separately, for the ability to complement the nitrogen regulation defect of a ure2Δ mutation. (A) The region from positions 1 to 65 is sufficient for prion induction. (B) A fragment of Ure2p lacking the region from positions 1 to 65 can also induce [URE3]. (C) Regions that promote prion formation (black) and block prion formation (white) are shown. Modified from reference 65 with permission of the publisher.
More detailed studies showed that the prion-inducing activity is not restricted to residues 1 to 65. The prion domain consists of 40% asparagine residues with several runs and includes 20% serine plus threonine. In fact, the asparagine-rich character of Ure2p extends to residue 80, and it was found that residues 1 to 80 induce the appearance of [URE3] significantly more efficiently than do residues 1 to 65 (65, 67). Deletion of any of the asparagine-rich regions (including, as one case, residues 66 to 80) from the full-length molecule reduces the prion-inducing ability of the remaining part, indicating that the asparagine runs are important (65). One deletion in the C-terminal part of the molecule, residues 221 to 227, abolishes the prion-inducing activity of Ure2p without affecting the nitrogen regulation function (65). This region has no asparagine or glutamine residues.
Further studies of Ure2p have shown two nonoverlapping regions of Ure2p capable of inducing the de novo appearance of [URE3] when overexpressed (65). Deletion of either residues 151 to 158 or the C-terminal 7 residues results in a marked increase in prion-inducing activity by the remaining molecule. These regions were interpreted as prion-inhibiting domains. Starting with a molecule lacking the original prion domain (residues 1 to 65), deletion of these two prion-inhibiting domains results in a second fragment of Ure2p that can induce the appearance of [URE3] (65). Thus, two parts of Ure2p, not overlapping except for the N-terminal Met-Met sequence, can induce the appearance of [URE3].
Further Properties of [URE3] Support Its Being a Prion
The genetic evidence described in the preceding sections indicates that [URE3] is not a nucleic acid replicon, such as a virus or plasmid. However, other “epigenetic” phenomena that are not prions have been described. A stable transcription regulation state can appear as a non-Mendelian genetic element. Escherichia coli may be unable to grow on low levels of lactose because the lac operon begins in an uninduced state and the levels of lactose in the medium are insufficient to induce it. However, if the same cells have been induced by a high level of lactose and then transferred to medium with the same low level, they could grow because the lac permease is already induced and can concentrate the lactose in the medium. Thus, once induced, the cells pass on this induced state to their progeny while growing on this medium (72). Since Ure2p is a transcription regulator, it was particularly important to determine whether [URE3] was a stable transcription state of this type. We have obtained several lines of evidence to show that this is not the case.
Selection of [URE3] strains from a [ure-o] parent was done with cells grown on a repressing nitrogen source (ammonia) or a poor (derepressing) nitrogen source (proline). There was no difference in the frequency of [URE3] colonies (67), suggesting that [URE3] is not a stable transcription state based on nitrogen regulation. This point is discussed in more detail below.
We have mentioned above that the possibility of separating the prion functions and nitrogen regulation functions of Ure2p argue against the “stable-transcription-state” model.
Interactions of Prion and Nitrogen Regulation Domains
Introduction of [URE3] into a cell expressing only the C-terminal nitrogen regulation domain has no effect. Nitrogen regulation is not interrupted. This shows that the fragment of Ure2p lacking the prion domain is not inactivated by [URE3]. Furthermore, it has been shown that [URE3] is not propagated by such cells (67). In cells expressing only the prion domain, Ure2p1–65, nitrogen regulation is completely defective. [URE3] can be introduced by cytoplasmic mixing (cytoduction) and is stably propagated, as shown by the fact that it can be passed by cytoduction to a wild-type strain (67).
When both the prion domain and the nitrogen regulation domain are expressed as separate molecules in the same cell, nitrogen regulation is normal and is unaffected by the introduction of [URE3]. The prion is also propagated stably, even though the cells remain in the nitrogen-repressed state (67). Thus, the [URE3] prion can be stably propagated in cells that are either repressed or derepressed for nitrogen degradation systems, indicating again that [URE3] is not a stable regulatory state.
These results give the impression of the two domains having no interaction with each other. However, the fact that deletions of the C-terminal domain dramatically increase the frequency with which [URE3] is induced suggests that the C-terminal domain stabilizes the N-terminal prion domain (68). Two-hybrid studies show that C-terminal and N-terminal parts of Ure2p can interact, supporting this view (40).
[URE3] Induction De Novo Due to Ure2 Protein Overproduction
One of the central pieces of evidence for the prion explanation of scrapie is that the purified infectious material is highly enriched for the protease-resistant form of PrP, called PrPres or PrPSc. Critics have repeatedly pointed out that the highly aggregated and heterogeneous nature of this material makes complete purification impossible, making it possible that a conventional virus is the basis of infectivity. None of the yeast or fungal prions has been purified as infectious material.
Expression of high levels of Ure2p increases the frequency with which [URE3] arises by 20- to 200-fold. This was done by introducing URE2 on a high-copy-number plasmid. It was important to show that it was the overproduction of Ure2p, and not the overproduction of URE2 mRNA or the gene itself in high copy number, that produced this effect. Since introducing URE2 under the control of the GAL1,10 promoter induced [URE3] to arise only when cells were grown on galactose, it was not the gene in high copy number that had this effect (102). To show that it was the Ure2 protein, and not the mRNA, we introduced frameshift mutations in the gene, altering a large part of the prion domain (67). A single −1 frameshift mutation at codon 44 changed the amino acid sequence of most of the prion domain and completely eliminated prion-inducing activity. In contrast, a single +1 frameshift at codon 80 gave very high prion-inducing activity. Adding the frameshift at codon 44 to that at 80 completely inactivated the prion-inducing activity, but because the frame was restored at codon 80, a full-length protein was made with full activity in nitrogen regulation (67). The level and size of the mRNA of this double mutant were the same as those of the wild-type mRNA. To determine whether the single base eliminated at codon 44 was in itself of special importance for prion induction, we deleted the entire codon 44 and found that this did not significantly alter prion induction by either the full-length Ure2p or the prion domain fragment (65). Thus, it is not the mRNA or the gene in high copy number but the overproduced Ure2p itself that induces the high-frequency appearance of [URE3]. This experiment is the molecular biological equivalent of transmission of [URE3] by protein alone. The Ure2 protein is sufficient to initiate the appearance of the infecitous [URE3] prion.
Ure2p Aggregation in [URE3] Strains
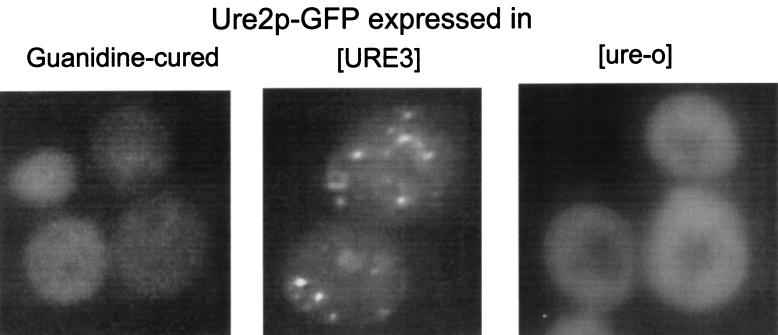
Fusions of full-length Ure2p or the prion domain, Ure2p1–65, or the C-terminal nitrogen regulation domain, Ure2p66–354, with the green fluorescent protein (GFP) were expressed in strains carrying [URE3], and the location of the fusion protein was monitored by fluorescence microscopy (37). In [URE3] cells, the fusion proteins that included the prion domain were seen in clumps, distributed throughout the cells (Fig. 5). Each cell had several clumps. When [URE3] was cured by growth in the presence of low concentrations of guanidine, the distribution of Ure2p returned to the same even distribution seen in wild-type strains. The C-terminal nitrogen regulation domain of Ure2p, when fused to GFP, did not appear clumpy, whether in a [URE3] strain or in a wild-type strain. These results suggest that the transcriptional regulator Ure2p is normally distributed throughout the cytoplasm and that [URE3] involves an aggregation of Ure2p (37).
FIG. 5.
Ure2p is aggregated in [URE3] cells. A Ure2p-GFP fusion was expressed from a single-copy plasmid with the URE2 promoter, and the distribution of fluorescence was examined in [ure-o] (wild-type), [URE3], and guanidine-cured strains. Aggregation was detected specifically in [URE3] strains (37).
Amyloid Formation In Vitro by Native Ure2p Initiated by the Prion Domain
Because the 65-residue amino-terminal fragment of Ure2p is capable of inducing the de novo formation of [URE3] in vivo, the properties of the synthetic peptide with this sequence were examined. Ure2p1–65 spontaneously and rapidly formed filaments 40 to 45 Å in diameter with essentially pure β-sheet structure (Fig. 6) (90). These filaments stained with Congo red to give the green birefringence diagnostic of amyloid (Fig. 7) and were resistant to digestion with proteinase K (90). Thus, the filaments formed by Ure2p1–65 have all of the characteristics of amyloid. A peptide from the C-terminal nitrogen regulation domain of Ure2p does not form such filaments, and amyloid formation is well known to be a reaction specific for a limited number of peptides and proteins.
FIG. 6.
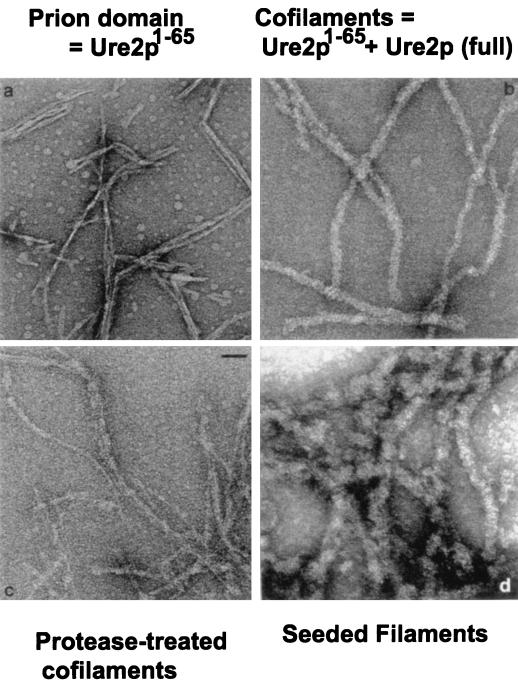
Amyloid is formed in vitro by Ure2p. (a) Filaments (45 Å) formed by Ure2p1–65. (b) Ure2p1–65/Ure2p 1:1 cofilaments 200 Å in diameter. (c) Ure2p1–65/Ure2p cofilaments digested with proteinase K, leaving the prion domain protease resistant in the form of narrow filaments. (d) Filaments (280 to 400 Å) of native Ure2p formed by seeding with Ure2p1–65/Ure2p cofilaments. Modified from reference 90 with permission of the publisher.
FIG. 7.
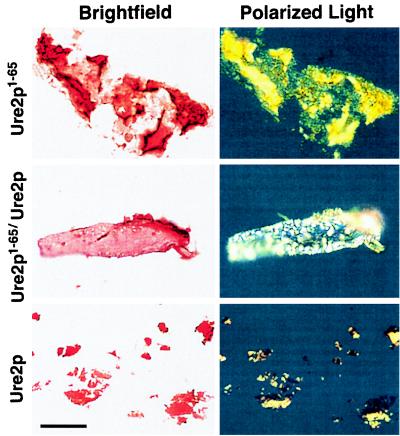
Ure2p filaments show birefringence on staining with Congo red. Filaments composed of Ure2p1–65, cofilaments of equimolar Ure2p1–65 and full-length Ure2p, and full-length Ure2p seeded with cofilaments were stained with Congo red and observed under bright-field conditions (left) or in a polarizing microscope (right). The green-yellow birefringence seen here is typical of amyloid. Reprinted from reference 90 with permission of the publisher.
Mixing Ure2p1–65 with native full-length Ure2p produces filaments consisting of a 1:1 mixture of the two proteins (Fig. 6b) (90). Under the same conditions, intact Ure2p itself does not form filaments or any other specific structure. Ure2p1–65 specifically forms filaments only with Ure2p and not with a number of other proteins tested. In addition, amyloid formation by the Alzheimer’s disease peptide Aβ1–42 does not result in cofilament formation with Ure2p. These Ure2p1–65/Ure2p cofilaments are also high in β-sheet structure and show the green birefringence on staining with Congo red (Fig. 7) (90). The Ure2p1–65/Ure2p cofilaments are 180 to 220 Å in diameter, wider than those formed of Ure2p1–65 alone (Fig. 7). Protease digestion of the cofilaments leaves Ure2p fragments of 7 to 10 kDa that include the N-terminal part of the molecule, the prion domain. In showing the partial protease resistance of Ure2p in [URE3] strains, it was found that one strain of [URE3] resulted in precisely this pattern (68). Examination of the protease-digested cofilaments by electron microscopy shows that they have become thinner, almost as thin as the filaments formed from Ure2p1–65 alone (Fig. 6c). These results indicate that, as with the prion induction and prion propagation process in vivo, this in vitro amyloid formation involves the interaction of the N-terminal parts of the prion-inducing Ure2p1–65 and the native Ure2p.
Addition of a small amount of the Ure2p1–65/Ure2p cofilaments to a large amount of native Ure2p results in the formation of filaments which are 290 to 400 Å in diameter, thicker, and more irregular than the cofilaments (Fig. 6d) (90). This shows that the filament formation is self-propagating.
Several lines of evidence indicate that these reactions reflect the events in the generation and propagation of [URE3]: (i) filament formation is initiated by the part of Ure2p that initiates [URE3] formation; (ii) cofilaments are formed with native Ure2p but not with other proteins tested; (iii) Aβ protein does not form cofilaments with Ure2p; (iv) the part of native Ure2p that becomes protease resistant in the cofilaments is the same part that is needed for participation in the [URE3] prion, namely, the N-terminal domain; (v) the cofilaments, once formed, can promote filament formation by native Ure2p without added Ure2p1–65; and (vi) Ure2p-GFP fusion protein is aggregated in vivo in [URE3] strains but evenly distributed in [ure-o] cells.
Thus, we have proposed that [URE3] involves amyloid formation and propagation (Fig. 8). Identifying a genetic element as a prion (propagating by whatever mechanism) implies that this element is a protein acting as a gene, as shown in Fig. 9. However, final proof that [URE3] is an amyloid form of Ure2p requires isolation and further characterization of the [URE3] form of Ure2p or demonstration that the amyloid formed in vitro can be introduced into [ure-o] cells and can make them convert to [URE3] cells at a frequency higher than does the native protein.
FIG. 8.
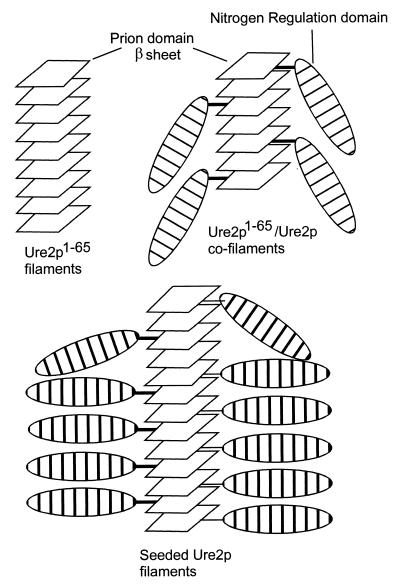
Model of [URE3] amyloid formation. The protease-resistant core of amyloid formed by Ure2p1–65 and full-length Ure2p is the N-terminal prion domain. The prion domain alone can form amyloid (left) and promotes amyloid formation by the full-length molecule (right and below).
FIG. 9.
A prion in yeast or fungi is a protein acting as a gene. Prions pass information from cell to cell by the transfer of the altered form of the protein. They are, in that sense, proteins acting as genes. w.t., wild type.
Curing of [URE3] by Fragments of Ure2p and Fusion Proteins
In the course of studying the distribution of Ure2p-GFP fusion proteins in [URE3] strains, it was noted that some such fusions resulted in the efficient curing of [URE3] (37). In fact, if sufficiently strongly overproduced, any of the Ure2p-GFP fusion proteins examined could cure [URE3]. Overexpression of the full-length Ure2p only rarely cured [URE3], but similarly overexpressed fragments, such as Ure2p1–65 or Ure2p66–354, cured most of the cells in which they were expressed (37). These results were explained by supposing that these fragments acted as terminators of the growing “crystal” of Ure2p. The nitrogen regulation domain, when present in excess, may interact with the C-terminal domain of full-length molecules, preventing their incorporation into the growing filaments.
[URE3] is also cured by millimolar concentrations of guanidine. Meiosis of stable [URE3] diploids often gives rise to meiotic segregants, few of which carry [URE3], although all are capable of propagating [URE3] if it is introduced by cytoduction. The reasons for the instability of [URE3] under these conditions remain to be elucidated.
[PSI], A PRION FORM OF SUP35P, A TRANSLATION RELEASE FACTOR
Discovery of [PSI]
In 1965, Cox discovered a nonchromosomal genetic element that made the weak ochre suppressor, SUQ5, become more efficient (23, 24). This genetic element, which he named [PSI], made strong suppressors into lethal suppressors (23a). Although [PSI] seemed at first to be specific for ochre (UAA) suppressors, later studies showed that suppression of all termination codons was affected (62, 75). In fact, [PSI] could result in a low level of nonsense suppression in strains without a recognizable suppressor tRNA mutation (62).
Not long before the discovery of [PSI], Inge-Vechtomov and Andrianova (54) and Hawthorne and Mortimer (51) had described sup35 mutations which led to the “omnipotent suppressor” phenotype. The sup35 mutants suppressed all types of nonsense mutations, much like [PSI]. There was, however, no apparent connection between SUP35 and [PSI].
Recently, Sup35p and Sup45p were shown to be the subunits of the translation termination factor, responsible for recognizing termination codons and cleaving the completed peptide from the final tRNA (88, 111). In the presence of a suppressor tRNA, there is competition between (i) the recognition of a termination codon by this suppressor tRNA, leading to insertion of an amino acid and continuation of the peptide chain, and (ii) recognition of the termination codon by the termination factor (Sup35p-Sup45p), and the resultant termination of the peptide chain. Any sup35 or sup45 mutation that impairs the function of these proteins will have the effect of increasing the apparent efficiency of suppression of the nonsense mutation.
Efforts to locate a nucleic acid corresponding to [PSI] were unsuccessful (reviewed in references 22 and 24). The mitochondrial DNA, 2μm DNA, and the L-A and M double-stranded RNA (dsRNA) viruses were easily ruled out, but the L-BC dsRNA virus and 20S and 23S single-stranded RNA (ssRNA) replicons were not tested. Early evidence that the rDNA plasmid called 3μm DNA was involved in [PSI] has not been confirmed.
Prion Genetic Criteria Satisfied by [PSI]
The precise parallel of the genetic properties of [PSI] to those of [URE3], outlined above, led to the proposal that [PSI], like [URE3], is a prion (102).
Reversible curing of [PSI].
Growth of cells carrying [PSI] in high-osmotic-strength medium results in the curing of [PSI] (87). Even without substantial cell growth, these conditions result in the conversion of [PSI+] cells to [psi−]. Further investigation of this phenomenon showed that the [psi−] cells generated were capable of becoming [PSI+] again at some low frequency (64). Growth in the presence of 1 to 5 mM guanidine HCl was found to also cure [PSI] (96), and it appeared that this curing was irreversible, but later studies showed that even the most thoroughly guanidine-cured strains could again become [PSI+] without its introduction from another cell (13).
[PSI] induction by overproduction of Sup35.
Overproduction of Sup35p, by introducing the gene on a high-copy-number plasmid, results in a suppressor effect in all cells. However, after the plasmid has been lost from the cells, many of the colonies are found to have become [PSI+], and the rate of conversion to [PSI+] was estimated to be about 100-fold higher than the spontaneous rate (13). That it is overproduction of Sup35p and not the SUP35 mRNA or the gene in high copy number that induced the de novo appearance of [PSI] was shown by introducing a frameshift mutation in SUP35 which abrogated the induction of [PSI] formation but had little effect on SUP35 mRNA levels (34).
Phenotypes of sup35 mutants and [PSI] strains.
As described above, either the presence of [PSI] or a sup35 mutation gives an “omnipotent-suppression” phenotype, with increased readthrough of any translation termination codon. However, certain sup35 mutants are unable to propagate [PSI] (92), and a mutant selected for its inability to propagate [PSI] is altered in SUP35 (36). As discussed above, this is the relationship expected if [PSI] is a prion of Sup35p, not if [PSI] is a nucleic acid replicon dependent on Sup35p for its replication (102).
Dominance of [psi−] to [PSI+] in vitro
Extracts of yeast programmed with model mRNAs and suppressor tRNAs read through the termination codons only if the extracts are made from cells carrying [PSI] (93). Although the [PSI] non-Mendelian genetic element, conferring increased readthrough, is dominant to the wild type in vivo, the result in the in vitro system is, surprisingly, the opposite. Mixing extracts of [PSI+] and [psi−] strains gives a mixture that behaves like the [psi−] extract alone, with very little readthrough (94). If [PSI] were a nucleic acid replicon whose phenotype resulted from its encoding a protein promoting readthrough, the in vitro result should be like the in vivo result, with [PSI+] dominant to [psi−]. We interpret this result as support for the prion model for [PSI] (108). If [PSI] is a prion, it produces a phenotype by inactivation of the normal protein. The wild-type extract should have the normal active protein and so should be dominant in vitro. However, in vivo the altered prion form appears dominant because it inactivates the normal form before the assay is done. The prion form could be dominant in vitro only if conversion of normal to prion forms occurred more rapidly than the reaction being measured. This apparently was not the case.
Purification of the component from [psi−] extracts that promoted chain termination resulted in isolation of a ribosome-associated factor (94) that fractionated like Sup35p (38).
N-Terminal Prion Domain of Sup35p
The C-terminal part of Sup35p is essential for growth and has homology to elongation factor 1α (EF-1α), indicating that it is this part of the molecule that is responsible for the translation termination function. In contrast, the N-terminal 253 residues are not essential for growth, and it is mutations in the N-terminal 123 residues that result in failure to propagate [PSI] (31, 36, 92) (Fig. 10). By expressing fragments of Sup35p from a plasmid in an initially [psi−] host, it was shown that overexpression of the N-terminal domain is sufficient to induce [PSI] (13, 34) and that deletions of the C-terminal part increase the efficiency of prion induction by the remaining N-terminal fragment (57). This result parallels that found in [URE3], where deletions of the C-terminal part of Ure2p dramatically increase the efficiency with which the N-terminal prion domain induces the appearance of [URE3] (68).
FIG. 10.
Comparison of prion proteins and prion domains. The prion domains of Ure2p and Sup35p are rich in Asn and Gln residues, and these residues are important in prion generation and propagation. PrP and the het-s protein are not Asn or Gln rich. The prion domain of the het-s protein has not been determined. For Ure2p, Sup35p, and PrP, mutations outside the prion domain may have dramatic effects on prion generation or propagation (see the text). Modified from reference 105 with permission of the publisher.
The N-terminal prion domain of Sup35p, like that of Ure2p, is quite rich in Asn and Gln residues, and Perutz et al. have shown that polyglutamine, such as is found in some of the triplet repeat diseases, can promote the aggregation of proteins (82). In fact, the Asn and Gln residues of Sup35p are critical for propagation of [PSI] (31).
Sup35p Aggregation in [PSI] Strains
Extracts of [PSI+] strains have most Sup35p in a rapidly sedimenting form (77, 81). Moreover, Sup35pN-GFP fusion proteins are aggregated in vivo specifically in strains with [PSI] (77). In each case, the aggregation depends on the N-terminal prion domain. This indicates that [PSI] concerns an aggregated state of Sup35p.
Hsp104 and Other Chaperones and [PSI]
In a screen for plasmids whose overproduction altered the efficiency of a weak suppressor in a [PSI+] strain, a clone of Hsp104 which eliminated the [PSI] effect was isolated (15). Further investigation showed that overexpression of Hsp104 cured [PSI] while deletion of Hsp104 also cured [PSI] (14). Hsp104 is one of the few true heat shock proteins in that it is specifically involved in protecting the cell from heat shock and is not otherwise necessary for growth (86). Hsp104 promotes the disaggregation of proteins aggregated as a result of the heat treatment (76).
The fact that Hsp104 levels are critical for propagation of [PSI] both supports the prion model for [PSI] and suggests a new avenue for treatment of prion diseases. While there have been many suggestions that chaperones might be involved in scrapie, the work of Chernoff et al. (14, 15) was the first proof for involvement of a chaperone in any prion.
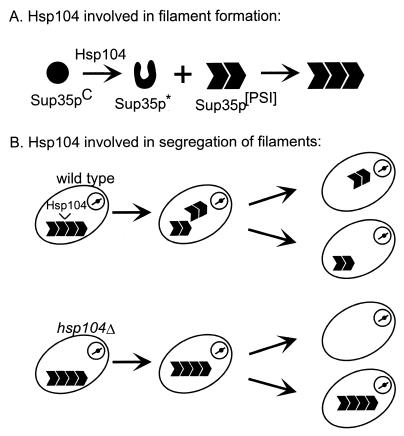
The mechanism of Hsp104 involvement in [PSI] is not completely resolved (Fig. 11). Overproduction of Hsp104 probably eliminates [PSI] by disaggregation of the Sup35p complexes. However, the positive role of Hsp104 in [PSI] propagation could be either a direct role in catalysis of the formation of the [PSI] form of Sup35p (14) or by facilitating the segregation of the aggregated form to the daughter cells by breaking up a single large aggregate into many smaller aggregates (81).
FIG. 11.
Possible roles of Hsp104 chaperone in [PSI] propagation. Hsp104 may be directly involved in the prion propagation reaction (top) or act to ensure the segregation to all cells of some of the altered form by breaking a few large aggregates into many smaller aggregates (bottom).
Although overexpression of Hsp104 cures [PSI] in up to 90% of cells (14), heat shock and other treatments that induce the high-level synthesis of Hsp104 do not detectably eliminate [PSI] (87, 96). One possible explanation for this paradox is that heat shock denatures many proteins, so that Hsp104 cannot concentrate its efforts on Sup35PSI. Another explanation is that other chaperones induced by heat shock prevent Hsp104 from curing [PSI] (71). Specifically, overproduction of Ssa1p, a member of the Hsp70 family, partially blocks curing of [PSI] by overproduction of Hsp104 (71).
In Vitro Propagation of [PSI]: Filament Formation by Sup35p
Using lysates of a [psi−] strain as a source of normal Sup35p, Paushkin et al. found that addition of small amounts of an extract of a [PSI+] strain would induce the aggregation of the normal Sup35p (79). The aggregation-inducing material was purified and corresponded to the aggregated Sup35p in the [PSI+] extract (79). The aggregated product of the conversion reaction could be used to prime a new reaction, so that the reaction could be continued apparently indefinitely.
In another approach, it was shown that the N-terminal prion domain peptide of Sup35p, made in E. coli, could form filaments in vitro with the properties of amyloid (55). Treating the peptide at pH 2 with acetonitrile led to the formation of filaments which were slightly protease resistant, had β-sheet structure, and showed green birefringence with Congo red. Although no details were reported, it was said that similar results were obtained with the Ure2p prion domain (55).
A third approach used Sup35p made in E. coli as the substrate. This material, on dilution from denaturant into buffer, spontaneously formed filaments after prolonged incubation (46). It was shown that only fragments of Sup35p that contained the prion domain could form filaments. The filament formation was accelerated by addition of an extract from a [PSI+] strain but not by addition of an extract from a [psi−] strain. Both of these facts argue that the filament formation observed was related to the [PSI] phenomenon.
The filaments formed by Sup35p1–123 are 87 Å in diameter, while those of Sup35p1–253 and the full-length protein are 115 and 170 Å in diameter, respectively, although the value for the latter species varies depending on the ionic conditions (46). Circular dichroism spectra indicate that the fibers have a structure rich in β-sheet. The Sup351–253 fibers bind Congo red and show the yellow-green birefringence typical of amyloid fibers (63a).
Inhibition of [PSI] Induction by Sup45p Overexpression
Sup35p is one of two subunits of the translation termination factor, the other being Sup45p. Sup35p has two binding sites for Sup45p, one in the C-terminal essential domain and the other requiring the N-terminal (N) prion domain and the middle (M) joining region (80). There is disagreement about whether Sup45p is present in the Sup35p aggregates formed in strains containing [PSI]. One group found Sup45p in such aggregates, but only if the aggregates include Sup35p containing binding sites for Sup45p (80). The authors suggested that a component of the suppression produced by [PSI] is the sequestration of Sup45p. Another group reported no difference in sedimentation properties of Sup45p in [PSI] strains (77). A third study showed that Upf1, a component of the nonsense-mediated mRNA decay pathway, is also present in these [PSI] aggregates (29).
Overexpression of Sup45p blocks the de novo formation of [PSI] induced by the overexpression of Sup35p (32). This effect is not due to lethality of [PSI+] strains with Sup45p overexpression, and [PSI] is not impaired in its ability to promote suppression. On the contrary, overexpression of Sup35p slows cell growth, particularly in [PSI+] strains, and this slowing is reversed by overexpression of Sup45p. Furthermore, overexpression of Sup45p actually increases the efficiency of suppression in strains already carrying [PSI+], so that it is not blocking detection of [PSI] or curing [PSI]. Finally, overexpression of Sup45p does not affect the [PIN] status of the cell (see below) (32).
Because Sup35p is normally found as a heterodimer with Sup45p, the blocking of [PSI] generation (but not propagation) by overexpression of Sup45p suggests that it is free Sup35p that is more likely to initiate the conversion to the prion form (32).
[ETA] and Strains of [PSI]
Scrapie strains, which show reproducible differences in incubation time, distribution of brain lesions, and, in some cases, physical properties of the PrPSc molecule, have been described by several groups (see, e.g., references 7 and 9).
It has now been shown that there are different strains of [PSI] (34). Different isolates of [PSI] from the same strain can vary in their efficiencies of suppression, stability as judged by frequency of spontaneous loss, and sensitivity to curing. [PSI] isolates are classified as strong, moderate, or weak, depending on their level of suppression. Strong isolates are more stable and resist curing better than weak isolates do. Elimination of [PSI] from strong and weak isolates and reisolation of new [PSI] derivatives yields the same spectrum of strong and weak isolates in each cured strain as in the original parent. This shows that the differences among strains of [PSI] is not due to a chromosomal mutation but is a difference of the [PSI] element itself (34).
Liebman and All-Robyn have previously described a non-Mendelian genetic element [ETA] as a factor that produced lethality in combination with certain sup35 mutations (60). [ETA] is, like [PSI], readily curable by guanidine HCl, but the phenotypic properties of [ETA] are clearly distinguished from those of [PSI]. It is now clear that [ETA] is a strain of [PSI], since it depends on the N-terminal part of Sup35p for its propagation and is lost on overproduction or elimination of Hsp104 (61, 110).
[PIN+], a Non-Mendelian Genetic Element Controlling Inducibility of [PSI]
Although it is stated above that curing of [PSI] is reversible, there is an apparent exception. Curing [PSI] by growth in the presence of guanidine results in two types of cured colonies: those that will again become [PSI+] when Sup35p is overexpressed (called Pin+ for [PSI] induction), and those that will not (Pin−) (33). The Pin+ phenotype is dominant to Pin− and segregates 4Pin+:0Pin− in seven tetrads of a cross of a Pin+ strain with a Pin− strain. Growth of Pin+ cells on 5 mM guanidine HCl results in conversion of over 90% of colonies to Pin−. These results indicate that Pin+ is determined by a nonchromosomal genetic element that the authors named [PIN+] (33).
The curability by guanidine suggests that [PSI+] is a prion, but it should be noted that growth in guanidine also frequently results in mutations of mitochondrial DNA and that guanidine blocks poliovirus replication (89). The prion model for [PIN+] is supported by the finding that deletion of HSP104 leads to loss of [PIN+] (33). Unlike [PSI], [PIN+] does not depend on the Sup35p N-terminal prion domain, leading to the suggestion that it is a prion form of the C-terminal domain (33). This possibility is difficult to test because the C-terminal domain is essential for growth.
[HET-S] PRION CONTROL OF VEGETATIVE INCOMPATIBILITY IN PODOSPORA
The [Het-s] non-Mendelian genetic element has been known since the 1940s, but a recent reinvestigation of this phenomenon has led to the proposal that it is in fact a prion necessary for normal functioning of the fungal cell (21).
Discovery of [Het-s]
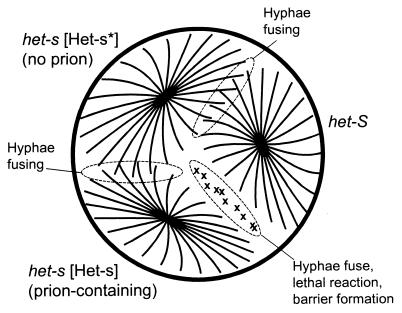
Filamentous fungi are capable of two forms of mating. Sexual mating is designed to generate diversity through meiosis and so occurs only between strains of different genotypes, ensured by a requirement for differences at one or more mating type loci. Heterokaryon formation (or vegetative anastomosis or hyphal anastomosis [anastomosis = fusion]) is a process designed for the purpose of cooperation between genetically identical individuals. When two fungal colonies grow toward each other, their hyphae (cellular processes) fuse to allow exchange of cytoplasm (and even nuclei, hence “heterokaryon formation”) between the two colonies. This heterokaryon formation allows sharing of nutrients, but it also allows fungal viruses to pass from one colony to the other, a potentially harmful event. Perhaps to prevent this infection, hyphal anastomosis is genetically controlled so that only colonies identical at many different loci (nine het loci in Podospora) are able to join. These close relatives would presumably already be carrying the same viruses. When hyphal anastomosis occurs between two individuals with differences at one or more het loci, the first fused hyphae rapidly undergo degeneration and death and form a barrier to further hyphal fusions. This is called vegetative incompatibility and is a normal function of most filamentous fungi (reviewed in reference 4).
Most of the het loci controlling vegetative incompatibility are unremarkable in that they show no unusual genetic behavior, but one such locus, het-s, is different. There are two alleles of het-s, called het-s and het-S, whose products differ by 14 amino acids in a protein of 289 residues (97). Remarkably, a single difference at residue 33 is sufficient to impart the incompatibility (30). Rizet discovered that het-s strains could have either of two phenotypes (85). Those called [Het-s] showed the incompatibility when paired with a het-S strain but could fuse with any other het-s strain. In contrast, those designated [Het-s*] were “neutral,” forming anastomoses equally with other het-s strains or with het-S strains (Fig. 12).
FIG. 12.
The [Het-s] prion of Podospora and heterokaryon incompatibility. Only cells carrying the [Het-s] prion can carry out heterokaryon incompatibility (21). Modified from reference 103 with permission of the publisher.
[Het-s] acts like the presence of a non-Mendelian genetic element, and [Het-s*] acts like its absence. When a het-s [Het-s] strain undergoes hyphal anastomosis with a het-s [Het-s*] strain, the [Het-s] trait quickly spreads through what was the het-s [Het-s*] colony.
Two Prion Genetic Criteria Satisfied by [Het-s]
[Het-s] can be, in effect, cured by mating [Het-s] males with [Het-s*] females. Because the male gametes are microspores with essentially no cytoplasm, the nonchromosomal genetic element [Het-s] is largely excluded and lost. However, from these cured meiotic segregants, cells which have again acquired the [Het-s] trait, which arises spontaneously with a frequency of about 10−7, can again be isolated (6). This is equivalent to the reversible-curing criterion for prions.
Overproduction of the het-s protein leads to an increase in the frequency with which [Het-s] arises (21). Indeed, deletion of het-s leads to loss of [Het-s] and inability to propagate this genetic element (21). This is the second of the expected properties of a prion.
Since deletion of the het-s gene does not result in the same phenotype as the presence of the putative prion, [Het-s], the third genetic criterion is not available in this system. This does not, however, argue against [Het-s] being a prion of the het-s protein. It means that [Het-s], like PrPSc, produces a phenotype by a positive action, not simply by preventing the het-s protein from carrying out its function.
Immunoblotting of protease-treated extracts of [Het-s*] and [Het-s] extracts showed that the het-s protein in the latter was substantially more resistant. This further supports the prion model for [Het-s].
Requirement of [Het-s] Prion for Normal Cell Function
The unique importance of the [Het-s] system is that the prion form of the protein is required for a normal function of the fungal cell. While scrapie, [URE3], and [PSI] are diseases, [Het-s] makes cells able to carry out a function that is necessary for their health. This suggests that other normal cellular functions may involve a prion-like mechanism.
STRUCTURE-BASED INHERITANCE: CORTICAL INHERITANCE IN PARAMECIUM AND OTHER CILIATES
Paramecium and other single-celled ciliates have an outer cellular layer called the cortex, on which is arrayed a specific pattern of cilia. These organelles function in motility and acquisition of food. Beisson and Sonneborn showed that accidental disruptions in the normal pattern of cilia that occurred in the process of cell division or mating were generally passed on to the progeny of the cell which had the original altering event (5). In an extension of this work, it was found that surgical alterations of the pattern of cilia could be carried out and would likewise be inherited. Similar results were later obtained with other ciliates (49).
This constitutes inheritance of a cellular structure, and is quite analogous to the inheritance of protein structures seen in [URE3], [PSI], and [Het-s]. It is tempting to speculate that other cellular structures that similarly template their own duplication will be found.
COMPARISON OF PRION SYSTEMS
Disease versus Normal Function
Scrapie is clearly a disease, and [URE3] causes slow growth of yeast on media with a good nitrogen source. [PSI] does not slow the growth of cells on the usual media, but it seems likely that inappropriate readthrough of translation termination codons is not adaptive except in rather exceptional circumstances. Indeed, Chernoff, Newnam, and Kumar (cited in reference 105) have found that [PSI+] cells are significantly less viable in deep stationary phase (that is, in expired medium) than are isogenic [psi−] cells. [Het-s] is clearly a case in which a prion is helping to carry out a normal cellular function.
Source of Prion Phenotype
[URE3] and [PSI] were identified as prions in part because the phenotype of the presence of the prion corresponded to that resulting from the absence of or insufficiency of the normal protein. In contrast, deletion of the Prnp gene encoding PrP results in little or no change in phenotype (11). Likewise, unlike [Het-s] strains, those with the het-s gene deleted have a “neutral” phenotype, in which the fungus is able to form heterokaryons with either het-s or het-S strains. This implies that while the phenotypes of [URE3] and [PSI] are produced by simple inactivation of Ure2p and Sup35p, respectively, PrPSc and the Het-s protein are positively doing something that the normal protein does not do, resulting in the observed phenotype or disease. All these prions are functionally dominant, in a genetic sense, but in the case of [URE3] and [PSI], they are dominant because of the propagation of the abnormal protein form, while for scrapie and [Het-s] the prion form does something directly that produces the phenotype, much like the usual genetic meaning of dominance.
Prion Domain Sequences
For Ure2p and Sup35p, the prion domains have been well delimited by extensive genetic studies, and in each case asparagine and glutamine residues are important for prion generation and propagation (Fig. 10). However, neither PrP nor the Het-s protein has asparagine-rich or glutamine-rich regions. The sequences responsible for amyloid formation by several proteins have been studied extensively, but it is not yet possible to predict which sequences will form this special structure. The availability of yeast prion systems promises to allow the isolation of prion-forming sequences from a library of genomic sequences, and this may lead to an understanding of what the requirements are for amyloid structure.
Identification of Prions
Efficient, self-propagating in vitro amyloid formation has been demonstrated for the Alzheimer’s disease Aβ peptide, for amylin (non-insulin-dependent diabetes mellitus), for fragments of immunoglobulin light chains such as cause amyloid accumulation in multiple myeloma, for transthyretin, and for others. In spite of intensive study, there is no evidence that any of these diseases is infectious. Recently, it has been suggested that many (or even most) proteins can form amyloid under some in vitro conditions (18). Thus, this type of in vitro criterion does not seem useful in identifying prions, although it is certainly critical in studying the mechanisms of prion propagation, since all prion proteins seem to form amyloid.
If one can purify and characterize the infectious material, the biochemical route is available to demonstrate the presence of a prion. This was the main approach taken initially for the TSEs, but the experimental difficulties of this system left considerable doubt.
The genetic approach to identifying prions has so far been reliable. No known phenomenon satisfying the genetic requirements has been found to operate by another mechanism. While both [URE3] and [PSI] appear to propagate by filament formation, the genetic approach is independent of the precise mechanism of propagation and may turn up prions based on other mechanisms, such as covalent self-modification.
The basic mechanism that appears to underlie the scrapie, [URE3], and [PSI] prions is that of formation of a linear crystal that appears as a filament. The free energy released in joining the crystal drives each of PrP, Ure2p, and Sup35p to change their conformation substantially. What other cellular structures would be determined in this way? Microtubules are capable of assembling from tubulins in a number of different forms in vitro, with the structure formed patterned after a seed. Any regularly repeating structure might display this type of feature.
Conclusions and Future Directions
It is evident that the study of yeast and fungal prions has had a dramatic impact on the prion field. While doubts remain about whether the infectious agent of scrapie is “protein only,” this issue seems to have been settled for the yeast systems. A role in heredity for prions was first detected in the yeast systems, largely because infection and heredity are the same thing in yeast. Thus, there are genes composed of altered proteins rather than nucleic acids. The involvement of chaperones in scrapie has often been proposed, but the first clear evidence for a role for a chaperone in a prion was that shown for Hsp104 in the [PSI] system. That both yeast prions appear to cause amyloidoses is striking. Although mammalian prions are intriguing and important, they are a relatively rare cause of human disease. Amyloidoses are thousands of times more frequent than are prion diseases, and these yeast systems are now a useful model for these common disorders. The [Het-s] system shows that normal functions can be determined by prions and raises the possibility that such a mechanism is widely used.
Many problems are posed by these interesting systems. Is Ure2p in [URE3] cells or Sup35p in [PSI] cells present in the form of amyloid? Must all prions be amyloids or filaments? Are other amyloid diseases due to prions? Can all amyloids be prions? How does guanidine cure [URE3] and [PSI]? How does high osmolarity cure [PSI]? What other prions are there? Where are they likely to be found? What is the structure of the Het-s protein in [Het-s] cells? Because of the difficulty of carrying out structural studies on filaments, the detailed structure of amyloid (in all systems) remains a subject of conjecture. It is expected that the yeast and fungal systems will continue to contribute to the broader prion and amyloid fields.
REFERENCES
- 1.Aigle M, Lacroute F. Genetical aspects of [URE3], a non-Mendelian, cytoplasmically inherited mutation in yeast. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 2.Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 3.Alper T, Haig D A, Clarke M C. The exceptionally small size of the scrapie agent. Biochem Biophys Res Commun. 1966;22:278–284. doi: 10.1016/0006-291x(66)90478-5. [DOI] [PubMed] [Google Scholar]
- 4.Begueret J, Turq B, Clave C. Vegetative incompatibility in filamentous fungi: het genes begin to talk. Trends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 5.Beisson J, Sonneborn T M. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc Natl Acad Sci USA. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beisson-Schecroun J. Incompatibilte cellulaire et interactions nucleo-cytoplasmiques dans les phenomenes de barrage chez Podospora anserina. Ann Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- 7.Bessen R A, Marsh R F. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton D C, McKinley M P, Prusiner S B. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 9.Bruce M E, Fraser H. Scrapie strain variation and its implications. Cur Top Microbiol Immunol. 1991;172:125–138. doi: 10.1007/978-3-642-76540-7_8. [DOI] [PubMed] [Google Scholar]
- 10.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 11.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp H P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 12.Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Linkage of prion protein and scrapie incubation time genes. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 13.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 14.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 15.Chernoff Y O, Ono B-I. Dosage-dependent modifiers of PSI-dependent omnipotent suppression in yeast. In: Brown A J P, Tuite M F, McCarthy J E G, editors. Protein synthesis and targeting in yeast. Berlin, Germany: Springer-Verlag KG; 1992. pp. 101–107. [Google Scholar]
- 16.Chesebro B. BSE and prions: uncertainties about the agent. Science. 1998;279:42–43. doi: 10.1126/science.279.5347.42. [DOI] [PubMed] [Google Scholar]
- 17.Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith J M, Garon C, Hasse A. Identification of scrapie prion protein-specific mRNA in scrapie-infected brain. Nature. 1985;315:331–333. doi: 10.1038/315331a0. [DOI] [PubMed] [Google Scholar]
- 18.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- 20.Coschigano P W, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox B S. Psi phenomena in yeast. In: Hall M N, Linder P, editors. The early days of yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 219–239. [Google Scholar]
- 23.Cox B S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 23a.Cox B S. A recessive lethal super-suppressor mutation in yeast and other PSI phenomena. Heredity. 1971;26:211–232. doi: 10.1038/hdy.1971.28. [DOI] [PubMed] [Google Scholar]
- 24.Cox B S, Tuite M F, McLaughlin C S. The Psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–179. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 25.Creutzfeldt H G. Uber eine eigenartige herdformige Erkrankung des Zentralnervensystems. Neurol Psychiatry. 1920;57:1–18. [Google Scholar]
- 26.Cuille J, Chelle P L. Experimental transmission of trembling to the goat. C R Seances Acad Sci. 1939;208:1058–1060. [Google Scholar]
- 27.Cuille J, Chelle P L. Pathologie animale. La maladie dite tremblant du mouton est-elle inoculable? C R Acad Sci (Paris) 1936;203:1552–1554. [Google Scholar]
- 28.Cullin C. Les prions, un mecanisme genetique conserve de l’homme a la levure. Med Sci. 1999;15:97–101. [Google Scholar]
- 29.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deleu C, Clave C, Begueret J. A single amino acid difference is sufficient to elicit vegetative incompatibility in the fungus Podospora anserina. Genetics. 1993;135:45–52. doi: 10.1093/genetics/135.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePace A H, Santoso A, Hillner P, Weissman J S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 32.Derkatch I L, Bradley M E, Liebman S W. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc Natl Acad Sci USA. 1998;95:2400–2405. doi: 10.1073/pnas.95.5.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derkatch I L, Bradley M E, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickinson A G, Meikle V M H, Fraser H. Identification of a gene which controls the incubation period of some strains of scrapie in mice. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 36.Doel S M, McCready S J, Nierras C R, Cox B S. The dominant PNM2− mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edskes H K, Gray V T, Wickner R B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eustice D C, Wakem L P, Wilhelm J M, Sherman F. Altered 40S ribosomal subunits in omnipotent suppressors of yeast. J Mol Biol. 1986;188:207–214. doi: 10.1016/0022-2836(86)90305-0. [DOI] [PubMed] [Google Scholar]
- 39.Farquhar C F, Somerville R A, Bruce M E. Straining the prion hypothesis. Nature. 1998;391:345–346. doi: 10.1038/34818. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Bellot E, Guillemet E, Baudin-Baillieu A, Gaumer S, Komar A A, Cullin C. Characterization of the interaction domains of Ure2p, a prion-like protein of yeast. Biochem J. 1999;338:403–407. [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstmann J. Uber ein noch nicht beschriebenes Reglexphanomen bei einer Erkrankung des zerebellum systems. Wien Med Wochenschr. 1928;78:906–908. [Google Scholar]
- 42.Gerstmann J, Straussler E, Scheinker I. Uber eine eigenartige hereditar-familiare erkrankung des zerebellaren systems. Neurology. 1936;154:736–762. [Google Scholar]
- 43.Gibbs C J, Gajdusek D C. Infection as the etiology of spongiform encephalopathy (Creutzfeldt-Jakob disease) Science. 1969;165:1023–1025. doi: 10.1126/science.165.3897.1023. [DOI] [PubMed] [Google Scholar]
- 44.Gibbs C J, Gajdusek D C, Asher D M, Alpers M P, Beck E, Daniel P M, Matthews W B. Creutzfeldt-Jakob disease (spongiform encephalopathy): transmission to chimpanzees. Science. 1968;161:388–389. doi: 10.1126/science.161.3839.388. [DOI] [PubMed] [Google Scholar]
- 45.Glenner G G. The bases of the staining of amyloid fibers: their physicochemical nature and the mechanism of their dye-substrate interaction. Prog Histochem Cytochem. 1981;13:1–37. doi: 10.1016/s0079-6336(81)80003-4. [DOI] [PubMed] [Google Scholar]
- 46.Glover J R, Kowal A S, Shirmer E C, Patino M M, Liu J-J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 47.Gordon W S. Advances in veterinary science. Vet Rec. 1946;58:516–520. [PubMed] [Google Scholar]
- 48.Griffith J S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 49.Grimes G W, Aufderheide K J. Cellular aspects of pattern formation: the problem of assembly. S. Basel, Switzerland: Karger; 1991. [PubMed] [Google Scholar]
- 50.Hadlow W J. Scrapie and kuru. Lancet. 1959;ii:289–290. [Google Scholar]
- 51.Hawthorne D C, Mortimer R K. Genetic mapping of nonsense suppressors in yeast. Genetics. 1968;60:735–742. doi: 10.1093/genetics/60.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsiao K, Baker H F, Crow T J, Poulter M, Owen F, Terwilliger J D, Westaway D, Ott J, Prusiner S B. Linkage of a prion protein missense variant to Gerstmann-Straussler syndrome. Nature. 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 53.Hsiao K K, Scott M, Foster D, Groth D F, DeArmond S J, Prusiner S B. Spontaneous neurodegeneration in transgenic mice with mutant prion protein of Gerstmann-Straussler syndrome. Science. 1990;250:1587–1590. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- 54.Inge-Vechtomov S G, Andrianova V M. Recessive supersuppressors in yeast. Genetika. 1970;6:103–115. [Google Scholar]
- 55.King C-Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klatzo I, Gajdusek D C, Zigas V. Pathology of Kuru. Lab Investig. 1959;8:799–847. [PubMed] [Google Scholar]
- 57.Kochneva-Pervukhova N V, Poznyakovski A I, Smirnov V N, Ter-Avanesyan M D. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae. Curr Genet. 1998;34:146–151. doi: 10.1007/s002940050379. [DOI] [PubMed] [Google Scholar]
- 58.Kushnirov V V, Ter-Avanesyan M D. Structure and replication of yeast prions. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 59.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liebman S W, All-Robyn J A. A non-Mendelian factor, [eta+], causes lethality of yeast omnipotent suppressor strains. Curr Genet. 1984;8:567–573. doi: 10.1007/BF00395701. [DOI] [PubMed] [Google Scholar]
- 61.Liebman S W, Derkatch I L. The yeast [PSI+] prion: making sense out of nonsense. J Biol Chem. 1999;274:181–184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 62.Liebman S W, Sherman F. Extrachromosomal [PSI+] determinant suppresses nonsense mutations in yeast. J Bacteriol. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindquist S. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 63a.Lindquist, S. Personal communication.
- 64.Lund P M, Cox B S. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 65.Maddelein M-L, Wickner R B. Two prion-inducing regions of Ure2p are nonoverlapping. Mol Cell Biol. 1999;19:4516–4524. doi: 10.1128/mcb.19.6.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 283–317. [Google Scholar]
- 67.Masison D C, Maddelein M-L, Wickner R B. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masison D C, Wickner R B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 69.Merz P A, Somerville R A, Wisniewski H M, Iqbal K. Abnormal fibrils from scrapie infected brain. Acta Neuropathol. 1981;54:63–74. doi: 10.1007/BF00691333. [DOI] [PubMed] [Google Scholar]
- 70.Reference deleted.
- 71.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B, Aebersold R, Barry R A, Tempst P, Templow D B, Hood L E, Prusiner S B, Weissmann C. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 74.Owen F, Poulter M, Lofthouse R, Collinge J, Crow T J, Risby D, Baker H F, Ridley R M, Hsiao K, Prusiner S B. Insertion in prion protein gene in familial Creutzfeldt-Jakob disease. Lancet. 1989;i:51–52. doi: 10.1016/s0140-6736(89)91713-3. [DOI] [PubMed] [Google Scholar]
- 75.Palmer E, Wilhelm J, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 76.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 77.Patino M M, Liu J-J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 78.Pattison I H. Resistance of the scrapie agent to formalin. J Comp Pathol. 1965;75:159–164. doi: 10.1016/0021-9975(65)90006-x. [DOI] [PubMed] [Google Scholar]
- 79.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 80.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol Cell Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 82.Perutz M F, Johnson T, Suzuki M, Finch J T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prusiner S B. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 85.Rizet G. Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev Cytol Biol Veg. 1952;13:51–92. [Google Scholar]
- 86.Sanchez V, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 87.Singh A C, Helms C, Sherman F. Mutation of the non-Mendelian suppressor [PSI] in yeast by hypertonic media. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamm I, Eggers H J. Specific inhibition of replication of animal viruses. Science. 1963;142:24–33. doi: 10.1126/science.142.3588.24. [DOI] [PubMed] [Google Scholar]
- 90.Taylor K L, Cheng N, Williams R W, Steven A C, Wickner R B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 91.Teninges D, Bras F, Dezelee S. Genome organization of the sigma rhabdovirus: six genes and a gene overlap. Virology. 1993;193:1018–1023. doi: 10.1006/viro.1993.1219. [DOI] [PubMed] [Google Scholar]
- 92.TerAvanesyan A, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tuite M F, Cox B S, McLaughlin C S. In vitro nonsense suppression in [psi+] and [psi−] cell-free lysates of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:2824–2828. doi: 10.1073/pnas.80.10.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tuite M F, Cox B S, McLaughlin C S. A ribosome-associated inhibitor of in vitro nonsense suppression in [psi−] strains of yeast. FEBS Lett. 1987;225:205–208. doi: 10.1016/0014-5793(87)81158-4. [DOI] [PubMed] [Google Scholar]
- 95.Tuite M F, Lindquist S L. Maintenance and inheritance of yeast prions. Trends Genet. 1996;12:467–471. doi: 10.1016/0168-9525(96)10045-7. [DOI] [PubMed] [Google Scholar]
- 96.Tuite M F, Mundy C R, Cox B S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turcq B, Deleu C, Denayrolles M, Begueret J. Two allelic genes responsible for vegetative incompatibility in the fungus Podospora anserina are not essential for cell viability. Mol Gen Genet. 1991;288:265–269. doi: 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 98.Weiss S, Famulok M, Edenhofer F, Wang Y-H, Jones I M, Groschup M, Winnacker E-L. Overexpression of active Syrian golden hamster prion protein PrPC as a glutathione S-transferase fusion in heterologous systems. J Virol. 1995;69:4776–4783. doi: 10.1128/jvi.69.8.4776-4783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weissmann C. Molecular genetics of transmissible spongiform encephalopathies. J Biol Chem. 1999;274:3–6. doi: 10.1074/jbc.274.1.3. [DOI] [PubMed] [Google Scholar]
- 100.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S-L, Torchia M, Carlson G A, Prusiner S B. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 101.Wickner R B. Double-stranded RNA viruses of yeast. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wickner R B. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 103.Wickner R B. A new prion controls fungal cell fusion incompatibility. Proc Natl Acad Sci USA. 1997;94:10012–10014. doi: 10.1073/pnas.94.19.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wickner R B. Prion diseases of mammals and yeast: molecular mechanisms and genetic features. R. G. Austin, Tex: Landes Co.; 1997. [Google Scholar]
- 105.Wickner R B. Prions and RNA viruses of Saccharomyces cerevisiae. Annu Rev Genet. 1996;30:109–135. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 106.Wickner R B. Viruses of yeasts, fungi and parasitic microorganisms. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. New York, N.Y: Raven Press; 1996. pp. 557–585. [Google Scholar]
- 107.Wickner R B, Chernoff Y. Prions of yeast and fungi: [URE3], [PSI] and [Het-s] discovered as heritable traits. In: Prusiner S B, editor. Prions, 0. 229–272. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 108.Wickner R B, Masison D C, Edskes H K. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 109.Wilson D R, Anderson R D, Smith W. Studies in scrapie. Comp Pathol. 1950;60:267–282. doi: 10.1016/s0368-1742(50)80025-5. [DOI] [PubMed] [Google Scholar]
- 110.Zhou P, Derkatch I L, Uptain S M, Patino M M, Lindquist S, Liebman S W. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhouravleva G, Frolova L, LeGoff X, LeGuellec R, Inge-Vectomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zigas V, Gajdusek D C. Kuru: clinical study of a new syndrome resembling paralysis agitans in natives of the Eastern Highlands of Australian New Guinea. Med J Aust. 1957;2:745–754. doi: 10.5694/j.1326-5377.1957.tb60287.x. [DOI] [PubMed] [Google Scholar]