To the Editor:
Two new proposals for the classification of myeloid malignancies have been presented: the 5th edition of the WHO Classification (WHO 2022 [1]) and the International Consensus Classification (ICC [2]). Here we address differences in entity defining criteria within myelodysplastic neoplasms (MDS), in particular in MDS with low blasts, and discuss accompanying hurdles.
MDS is a very heterogeneous disease representing clonal disorders of hematopoietic cells characterized by morphologic dysplasia, peripheral cytopenias, ineffective hematopoiesis and increased risk of leukemic transformation [3]. Somatic mutations in splicing pathway genes are detected in about half of MDS patients with SF3B1 as the most commonly mutated one, typically found in MDS with ring sideroblasts (RS) [4, 5]. SF3B1 mutations further define a distinct MDS subtype showing favorable prognosis and indolent disease course [6, 7]. Thus, MDS with low blasts and SF3B1 mutation (MDS-SF3B1) is considered a separate MDS entity in both WHO 2022 [1] and ICC [2] with slightly deviating defining entity criteria (Suppl. Table S1). Both WHO 2022 and ICC classifications require the presence of an SF3B1 mutation (WHO: VAF ≥ 5%, ICC: VAF ≥ 10%), a bone marrow (BM) blast count <5% and the absence of certain cytogenetic abnormalities and biallelic TP53 inactivations. The ICC further requires the absence of RUNX1 mutations. In contrast to ICC, in WHO 2022 the term “MDS with low blasts and ring sideroblasts” (MDS-LB-RS) is retained as an acceptable alternative to be used for cases with wild-type SF3B1 and ≥15% ring sideroblasts allowing the inclusion of driver mutations in other splicing components.
Here, we address the differences between WHO and ICC regarding MDS-SF3B1 and in particular if the WHO term “MDS with low blasts and ring sideroblasts” is meaningful as an alternative for SF3B1 wild-type cases. Therefore, we analyzed this “alternative group” referred to as MDS-LB-RS in an MDS cohort with respect to incidence, presence of other splicing gene mutations and clinical outcome.
We selected 704 de novo MDS patients with sample material available to perform WGS sent to our laboratory between 09/2005 and 12/2019 (male/female: 407/297; median age: 73 [23–93]; 409 with <5% BM blasts). Diagnoses were made based on cytomorphology, cytogenetics and molecular genetics as previously published [8]. All samples were subjected to amplification-free WGS (median coverage >100x) as reported previously [9, 10]. The validation cohort comprised 1804 de novo MDS patients (male/female: 1160/644; median age: 76 [24–96]; 1015 with <5% BM blasts) whose samples were subjected to targeted panel sequencing during routine diagnostics between 07/2017 and 07/2022 as previously described [11]. Details on statistics see supplement. All patients had given written informed consent to the use of genetic and clinical data according to the Declaration of Helsinki and the study was approved by the internal review board.
In 660/704 (94%) MDS cases data on the presence of RS were available and the basis for further analyses. Of these, 40% (262/660) showed RS ≥ 15% (Fig. 1A). 299/660 patients had low blasts (LB; BM blasts <5%) and also did not fulfill the criteria for the WHO 2022 entities MDS with low blasts and isolated 5q deletion (MDS-5q; n = 98) or MDS with biallelic TP53 inactivation (MDS-biTP53; n = 41). RS < 15% were detected in 115/299 (38%) while 184/299 (62%) patients showed RS ≥ 15%.
Fig. 1. MDS cohort overview according.
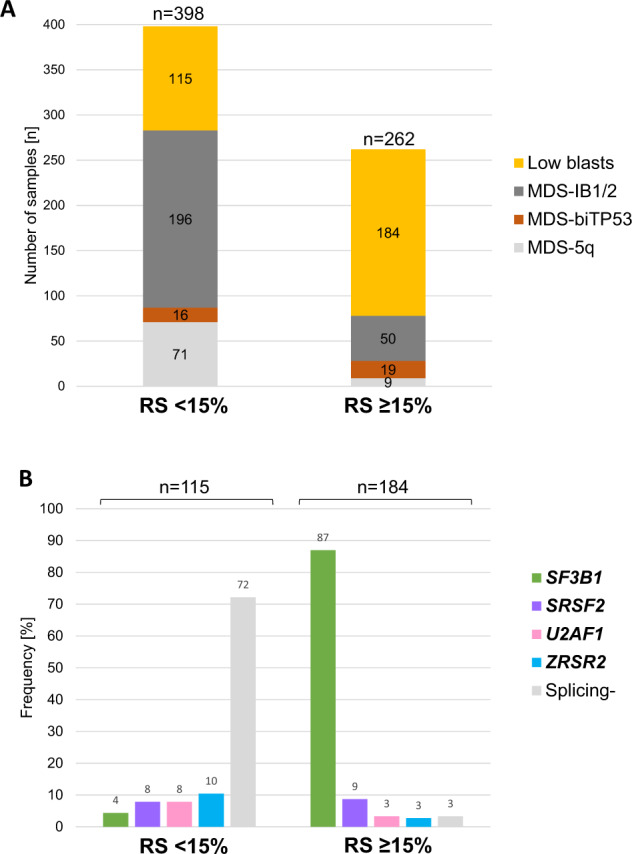
A Distribution of subsets within MDS patients according to WHO 2022 dependent on the presence of ring sideroblasts (RS). B Frequency of mutations in splicing genes SF3B1, SRSF2, U2AF1 or ZRSR2 within low blast MDS with RS < 15% or ≥15%. Splicing-: no mutation in splicing genes SF3B1, SRSF2, U2AF1 or ZRSR2.
In LB cases with RS ≥ 15% splicing mutations were found in 178/184 (97%) (SF3B1: 87%, SRSF2: 9%, U2AF1: 3%, ZRSR2: 3%), while only 28% (32/115) of patients with LB and RS < 15% harbored mutations in at least one of four analyzed splicing genes (SF3B1: 4%, SRSF2: 8%, U2AF1: 8%, ZRSR2: 10%; Suppl. Fig. S1A; Fig. 1B). Next, we analyzed the association of the distinct splicing gene mutations (SF3B1, SRSF2, U2AF1, ZRSR2) with the presence of RS (<15% vs. ≥15%) and BM blasts (<5% vs. ≥5%). SF3B1 mutations were significantly associated with BM blasts <5% (p < 0.001), while mutations in all other splicing genes showed association with BM blasts ≥5% (SRSF2: p < 0.001, U2AF1: p = 0.006, ZRSR2: p = 0.083; Supplementary Fig. S1B). These observations were mirrored in the large validation cohort (SF3B1: p < 0.001, SRSF2: p < 0.001, U2AF1: p = 0.003, ZRSR2: p = 0.037) where the trend in ZRSR2 reached statistical significance (Suppl. Fig S1B). We found that SF3B1 mutations were significantly associated with RS ≥ 15% (p < 0.001), while mutations in SRSF2, U2AF1 and ZRSR2 were not. For SRSF2 (p = 0.056) and ZRSR2 (p = 0.016) mutations we even observed a trend towards an association with RS < 15%. This was again confirmed in our validation cohort (SF3B1: p < 0.001, SRSF2: p < 0.001, ZRSR2: p < 0.001) where the trend in SRSF2 mutations became statistically significant. U2AF1 mutations were not found to be associated with RS in either cohort (p = 0.130 and p = 0.125, respectively; Supplementary Fig S1B). Together, these data clearly indicate that other splicing factor mutations cannot be used as a substitute for SF3B1 mutations and are not useful for the classification of MDS-LB-RS in the absence of SF3B1 mutations.
In this line, 87% (160/184) of cases with RS ≥ 15% showed SF3B1 mutations (Fig. 1B) indicating a high chance for detecting an SF3B1 mutation if RS ≥ 15%. In cases with RS < 15%, only 4% (5/115) harbored an SF3B1 mutation while mutations in SRSF2, U2AF1 and ZRSR2 were more frequent in this group (SRSF2 and U2AF1: each 9/115; 8%; ZRSR2: 12/115; 10%). Thus on the other hand, 13% (24/184) of cases with RS ≥ 15% were SF3B1 wild-type including two cases harboring complex karyotypes (Supplementary Fig. S2). Thus, 22 cases qualify to be assigned to MDS-LB-RS according to WHO 2022. In this group 17/22 (77%) cases harbored mutations in other spliceosome genes (U2AF1: n = 5, SRSF2: n = 12; VAF ≥ 10% in all cases; Suppl. Figure S2). Notably, in both RS groups (<15% vs. ≥15%) splicing gene mutations were not mutually exclusive, 13 cases harbored two splicing mutations (for details see Suppl. Results). Complex karyotypes and RUNX1 mutations were detected in 5 and 10 cases with mutated splicing genes, respectively (Supplementary Fig. S2).
Differences in the defining entity criteria between WHO 2022 and ICC change the assignment into diagnostic categories of 30/299 MDS cases with low blasts (Fig. 2; Supplementary Fig. S3). The 22 cases classified as MDS-LB-RS according to WHO 2022 were classified as MDS, NOS based on ICC criteria. In addition, 8 cases assigned to the MDS-SF3B1 entity according to WHO belong to MDS, NOS according to ICC due to presence of RUNX1 mutations (n = 7) or SF3B1 VAF < 10% (n = 1). To evaluate whether or not MDS-LB-RS according to WHO 2022 is associated with a comparable favorable outcome as MDS-SF3B1 we performed survival analyses. Overall survival (OS) was significantly shorter in MDS-LB-RS (n = 22) compared to MDS-SF3B1 (n = 161; median: 5.3 vs. 7.9 years; p = 0.032; Supplementary Fig. S4A), mainly mediated by MDS-LB-RS cases harboring splicing mutations (p = 0.046; Supplementary Fig. S4B, C). In addition, MDS-LB-RS showed comparable outcome to other MDS-LB (n = 116) not fulfilling the criteria for MDS-SF3B1 or MDS-LB-RS (median: 5.3 vs. 6.2 years; p = 0.373; Supplementary Fig. S4D) and thus grouped together showed shorter OS compared to the MDS-SF3B1 entity (median: 5.8 vs. 7.9 years; p = 0.038; Supplementary Fig. S4E).
Fig. 2. Changes in sample categorization within low blast MDS.
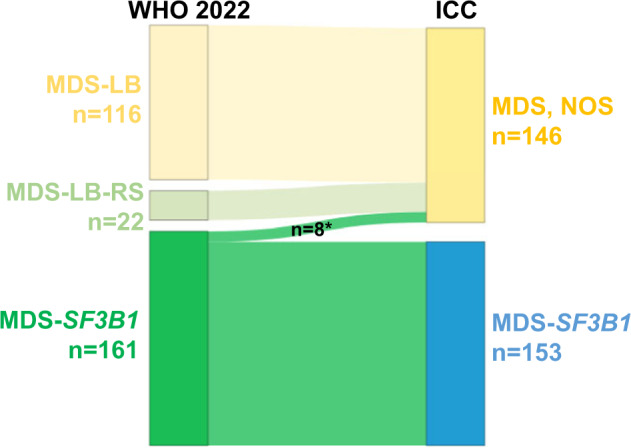
Cases (n = 299) were categorized according to WHO 2022 (left) and ICC (right) classifications. VAF: variant allelic frequency; * RUNX1mut (n = 7) and SF3B1mut VAF < 10% (n = 1); mut mutation.
Of note, within the WHO MDS-SF3B1 entity (n = 161) RUNX1 mutated cases (n = 7) were associated with shorter OS compared to RUNX1 wild-type cases (median: 2.1 vs. 8.3 years; p < 0.001; Supplementary Fig. S4F; for details on further co-mutations see Supplementary Results). Excluding RUNX1 mutated cases from WHO MDS-SF3B1 led to a more significant separation regarding OS between MDS-SF3B1 cases and non-MDS-SF3B1 cases with LB (p = 0.003 vs. p = 0.038; Supplementary Fig. S4G/E). Based on ICC, excluding RUNX1 mutations and SF3B1 VAFs <10% from the MDS-SF3B1 entity, a similar significant separation regarding OS was achieved between ICC MDS-SF3B1 cases and non-MDS-SF3B1 cases represented by MDS, NOS (p = 0.005; Supplementary Fig. S4H).
Following this, with regard to prognosis excluding RUNX1 mutations from the prognostically favorable MDS-SF3B1 entity is rational, concordant with the proposal of the IWG-PM [7]. In this line, several studies demonstrated the negative prognostic value of RUNX1 co-mutations in SF3B1 mutated MDS [6, 12–14], thereby highlighting the role as potential driver gene associated with worse OS and a higher leukemic transformation rate within SF3B1 mutated patients [15]. In addition, we previously showed that in SF3B1 mutated MDS del(5q) and/or RUNX1 mutations have a negative impact on outcome while a BM blast threshold of <5%, which is used by ICC and WHO 2022, has no independent impact on OS [15].
In conclusion, we again confirmed the association of SF3B1 mutations with low BM blasts and increased RS. We also showed that mutations in other splicing genes (SRSF2 and ZRSR2) were significantly associated with high BM blasts and low RS. Thus, we suggest the alternative term “MDS with low blasts and ring sideroblasts” (MDS-LB-RS) as a second best classification only for cases when SF3B1 mutation analysis is not available as in this setting RS ≥ 15% represent a good but not perfect surrogate for SF3B1 mutations. Conversely, MDS-LB-RS with wild-type SF3B1 are suggested to be classified as MDS-LB and separated from MDS-SF3B1, as biology seems to be different as indicated by a less favorable prognosis. We further demonstrated the negative prognostic impact of RUNX1 mutations in SF3B1 mutated patients and therefore suggest excluding these cases from the MDS-SF3B1 entity.
Supplementary information
Acknowledgements
We would like to thank all co-workers at MLL Munich Leukemia Laboratory for their dedicated work. The authors would also like to thank all physicians for providing samples and caring for patients as well as collecting data.
Author contributions
SH and CH designed the study, SH and GH interpreted the data, SH wrote the manuscript. CH was responsible for chromosome banding and FISH analyses, MM, CB and StH for molecular and bioinformatic analyses, IS and IF for cohort assembling, WK for immunophenotyping and TH for cytomorphologic analyses. All authors read and contributed to the final version of the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
CH, WK and TH declare part ownership of MLL Munich Leukemia Laboratory. CB, SH, StH, MM, IS, IF and GH are employed by MLL Munich Leukemia Laboratory.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01783-y.
References
- 1.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka H-M, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 6.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–41. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood. 2020;136:157–70. doi: 10.1182/blood.2020004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haferlach T, Kern W, Schoch C, Hiddemann W, Sauerland MC. Morphologic dysplasia in acute myeloid leukemia: importance of granulocytic dysplasia. J Clin Oncol. 2003;21:3004–5. doi: 10.1200/JCO.2003.99.091. [DOI] [PubMed] [Google Scholar]
- 9.Stengel A, Baer C, Walter W, Meggendorfer M, Kern W, Haferlach T, et al. Mutational patterns and their correlation to CHIP-related mutations and age in hematological malignancies. Blood Adv. 2021;5:4426–34. doi: 10.1182/bloodadvances.2021004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höllein A, Twardziok SO, Walter W, Hutter S, Baer C, Hernandez-Sanchez JM, et al. The combination of WGS and RNA-Seq is superior to conventional diagnostic tests in multiple myeloma: Ready for prime time? Cancer Genet. 2020;242:15–24. doi: 10.1016/j.cancergen.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrmann I, Lenk M, Haferlach T, Stengel A, Hutter S, Baer C, et al. AML, NOS and AML-MRC as defined by multilineage dysplasia share a common mutation pattern which is distinct from AML-MRC as defined by MDS-related cytogenetics. Leukemia. 2022;36:1939–42. doi: 10.1038/s41375-022-01631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komrokji R, Volpe V, Chan O, Al Ali N, Swoboda D, Kuykendall A, et al. Validation of the international working group proposal for SF3B1 mutant myelodysplastic syndromes. Blood. 2021;138:989–92. doi: 10.1182/blood.2021010831. [DOI] [PubMed] [Google Scholar]
- 13.Venable ER, Chen D, Chen CP, Bessonen KR, Nguyen PL, Oliveira JL, et al. Pathologic Spectrum and Molecular Landscape of Myeloid Disorders Harboring SF3B1 Mutations. Am J Clin Pathol. 2021;156:679–90. doi: 10.1093/ajcp/aqab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard E, Tuechler H, Greenberg Peter L, Hasserjian Robert P, Arango Ossa Juan E, Nannya Y, et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022;1:EVIDoa2200008. doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 15.Huber S, Hutter S, Meggendorfer M, Hoermann G, Walter W, Baer C, et al. The role of SF3B1 mutations in myelodysplastic syndromes. EHA Libr. 2022;357597:P735. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


