Abstract
Myelofibrosis (MF) is a myeloproliferative disorder that exhibits considerable biological and clinical heterogeneity. At the two ends of the disease spectrum are the myelodepletive or cytopenic phenotype and the myeloproliferative phenotype. The cytopenic phenotype has a high prevalence in primary MF (PMF) and is characterized by low blood counts. The myeloproliferative phenotype is typically associated with secondary MF (SMF), mild anemia, minimal need for transfusion support, and normal to mild thrombocytopenia. Differences in somatic driver mutations and allelic burden, as well as the acquisition of non-driver mutations further influences these phenotypic differences, prognosis, and response to therapies such as JAK2 inhibitors. The outcome of patients with the cytopenic phenotype are comparatively worse and frequently pose a challenge to treat given the inherent exacerbation of cytopenias. Recent data indicate that an innate immune deregulated state that hinges on the myddosome-IRAK-NFκB axis favors the cytopenic myelofibrosis phenotype and offers opportunity for novel treatment approaches. We will review the biological and clinical features of the MF disease spectrum and associated treatment considerations.
Subject terms: Myeloproliferative disease, Myeloproliferative disease
Introduction
The BCR-ABL1-negative myeloproliferative neoplasms (MPNs) are a group of chronic hematopoietic stem and progenitor cell (HSPC) derived hematologic malignancies, which include essential thrombocythemia (ET), polycythemia vera (PV) and primary myelofibrosis (PMF). ET and PV can both progress to a form of secondary myelofibrosis (sMF) that collectively with PMF are simply termed myelofibrosis (MF). Hyperactivity of the JAK-STAT signaling pathway is the central biologic hallmark of these diseases and somatic mutations involving the JAK2, CALR and MPL genes comprise 90% of driver mutations [1]. Additionally, non-driver mutations frequently occur in genes involving signal transduction, epigenetic modifiers, spliceosome and tumor suppressor pathways that further influence phenotype and prognosis [2]. MF is characterized at a biological level by expansion of a malignant HSPC with aberrant trafficking to extramedullary sites of hematopoiesis [3]. The histopathological consequences are typified by bone marrow hypercellularity, reticulin and collagen fibrosis, and a high frequency of circulating CD34 + cells. The clinical picture is heterogeneous but in general includes progressive cytopenias, organomegaly, debilitating systemic symptoms, and potential for evolution to acute myeloid leukemia. The incidence of MF is 0.44 per 100,000 person-years, with a median age at time of diagnosis of approximately 68 years and a median survival of 5.2–5.9 years [4–7]. Given the highly variable clinical presentation, prognostic scales have been developed to guide treatment decisions.
Prognostication in myelofibrosis
Prognostication for MF has evolved over the years and originated with the Lille classification which included leukocytosis or leukopenia and anemia to define three prognostic risk categories (Table 1) [8]. The International Prognostic Scoring System (IPSS) which is applied at the time of diagnosis includes advanced age, leukocytosis, anemia, systemic symptoms, and peripheral blood blasts to create four prognostic categories [6]. The Dynamic IPSS (DIPSS) utilizes the same risk variables but can be applied at any point in the disease course [9]. Subsequently, the DIPSS-plus incorporated thrombocytopenia (<100 ×109/L), adverse karyotype, and red blood cell transfusion dependence to predict overall survival (OS) and determined that adverse karyotype or thrombocytopenia also predict leukemia free survival (LFS) [10]. The modern Mutation-enhanced International Prognostic Scoring System (MIPSS) and Genetically Inspired Prognosis Scoring System (GIPSS) further refine prognostication by integrating cytogenetic and molecular data [11, 12]. The Myelofibrosis Secondary to PV and ET-Prognostic Model (MYSEC-PM) is a prognostic scale specific for patients with sMF that integrates both clinical and molecular features for prognostication [13]. The current benefit of utilizing any of these prognostic models is either in the context of determining clinical trial eligibility or risk-adapted treatment decision making and particularly when considering the role of transplantation [14].
Table 1.
Prognostication in myelofibrosis includes diverse disease-specific variables that contribute to clinical heterogeneity.
| Lille | IPSS | DIPSS | DIPSSplus | MIPSS70 | GIPSS | MYSEC-PM | |
|---|---|---|---|---|---|---|---|
| Age | X | X | X | X | |||
| Leukocytosis | Xa | X | X | X | X | ||
| Anemia | X | X | X | X | X | X | |
| Symptoms | X | X | X | X | X | ||
| Circulating blasts | X | X | X | X | X | ||
| Thrombocytopenia | X | X | X | ||||
| RBC transfusion dependent | X | ||||||
| Adverse Karyotypeb | X | X | X | ||||
| BMF | X | ||||||
| Non-CALR type 1 | X | X | X | ||||
| HMR = 1 | Xc | Xd | |||||
| HMR > 1 | Xc |
IPSS International Prognostic Scorcing System, DIPSS Dynamic International Prognsotic Scoring System, MIPSS Mutation-Enhanced International Prognostic Scoring System 70, GIPSS Genetically Inspired Prognostic Scoring System, MYSEC-PM Myelofibrosis Secondary to PV and ET-Prognostic Model, RBC red blood cell, BMF bone marrow fibrosis, HMR high molecular risk.
aLeukocytosis and leukopenia are variables in the Lille prognostic Model.
bComplex karyotype or abnormalities including +8, -7/7q-, i(17q), -5/5q-, 12p-, inv(3) or 11q23 rearrangement.
cHMR mutations in MIPSS70: ASXL1, EZH2, IDH1/2, SRSF2.
dHMR mutations in GIPSS: ASXL1, SRSF2, U2AF1Q157.
Mutational impact on disease phenotype and therapeutic response in myelofibrosis
Of the three driver genes recurrently mutated in MPN, the JAK2 V617F mutation is present in 60% of PMF cases, CALR mutation accounts for 20-30% of cases with predominance of Type 1/1-like mutation, and MPLW515L/K mutation is found in 5-10% of cases. In sMF, JAK2V617F is present in almost all cases of PPV-MF and in PET-MF accounts for 50% of them, while CALR (Type 1 is the most prevalent) and MPL mutation for 30% and 10%, respectively [15]. The JAK2V617F variant allele frequency (VAF) ranges from a very low percentage to 100% with a median of approximately 50%; and frequently increases with the transition from PV and ET to sMF, reflecting clonal dominance [16, 17]. Conversely, a low JAK2V617F VAF (<25%) is associated with certain features (lower leukocyte count and hemoglobin) of a cytopenic rather than myeloproliferative MF phenotype and represents an independent variable associated with shortened survival in patients with PMF [18]. The absence of any driver mutation is operationally defined as “triple negative” (TN), and accounts for roughly 10% of PMF patients [19]. So called non-canonical JAK2 and MPL mutations may be found in a minority of TN patients by sequencing all gene coding regions by next generation sequencing (NGS) [20]. Triple-negativity is an independent variable for shortened survival in PMF [21]; data in sMF are scant [22].
In addition to the driver mutations, 40–60% of patients with MF compared to <20% of PV and ET [23] harbor deleterious mutations in a variety of myeloid-neoplasm associated genes, including DNA methylation and epigenetic regulators, members of the spliceosome, oncogenes and transcription factors [11, 24–26]. Mutated ASXL1 is the most common additional genetic abnormality in MF (25–40% of patients) that harbors unfavorable prognostic significance [27–29], and is included among the High Molecular Risk (HMR) mutations [24] together with EZH2 (4–7%), IDH1 and IDH2 (1–3% each), SRSF2 (8–15%) and U2AF1Q157 (8–16%) [30]. The presence of any HMR mutation confers shorter OS and LFS to patients with pre-fibrotic and overt PMF, which is compounded by the presence of more than one HMR mutation [2]. Accordingly, HMR and the number of HMR mutations are embedded in the MIPSS70 scores (MIPSS70/plus [11] and v2.0 [31]. On the other hand, the role of ASXL1 and HMR mutations in general in sMF remains uncertain [26, 32], and the detrimental value of ASXL1 mutation when it is the only additional mutation has been recently questioned in two independent cohorts of patients [26]. ASXL1 stands, together with a non-CALR/MPL mutated genotype, as the only genetic abnormality that informs survival and non-relapse mortality after stem cell transplantation in MF [14]. Other mutated genes reported at <5% frequency with uncertain impact on survival include TET2, DNMT3A, NFE2, SH2B3, CUX1, CBL, RUNX1, NOTCH1, N/KRAS and TP53. Myeloid mutations are enriched in patients with cytopenic versus myeloproliferative MF phenotype [33], including HMR and U2AF1 gene mutations (AM Vannucchi et al., 2022, submitted), consistent with prior data indicating clustering with anemia and thrombocytopenia [34].
The frequency of TP53 mutations, and/or chromosome 7 deletions and/or amplifications of genes encoding negative regulators of p53, such as MDM2, is increased at the time of evolution to secondary acute myeloid leukemia (sAML) [35]. However, a single, stable, low VAF, TP53 mutation detected at chronic phase has not been unequivocally associated with shorter OS and LFS, suggesting that haploinsufficiency of TP53 per se may not be sufficient for evolution to sAML [36–38]. Also, the mechanisms by which loss of TP53 function in association with dysregulated JAK-STAT signaling leads to leukemia remains unclear, but may be related to genetic instability leading to numerous chromosome abnormalities [39]. Of note, some cases of sAML originate from the background of previously JAK2V617F –positive hematopoiesis while in other cases blasts are JAK2 wildtype suggesting de novo origin [40]. One important question is whether these latter leukemias originated from an antecedent clonal hematopoiesis of indeterminate potential (CHIP), different from the one that established the chronic phase MPN. Single cell studies reinforce the extreme complexity and heterogeneity of leukemic evolution in MF with multiple subclones branching from the originating leukemic clone [41, 42].
The contribution of mutational profile to the response or resistance to the JAK 1/2 inhibitor ruxolitinib has been thoroughly investigated. In one report, a JAK2V617F VAF > 50% was associated with better spleen response to ruxolitinib [43]. Presence of HMR mutations does not significantly impact short-term response to ruxolitinib [44], however it may be associated with a shorter duration of response [45], and acquisition of new mutations configuring clonal progression that contributes to therapy resistance [45–47]. Involvement of the RAS/CBL pathway, that per se predicts shorter OS and LFS [48, 49], may be associated with reduced symptom and spleen response to JAK inhibitors, highlighting the opportunity for dual targeting of JAK-STAT and RAS/MAPK signaling.
Spectrum of clinical phenotype in myelofibrosis
The heterogeneous nature of MPNs was recognized by Dameshek across the entire spectrum of these diseases, but arguably it is most apparent for patients with MF; in the next sections we explore this theme of clinical heterogeneity within MF particularly focusing upon advanced MF [50].
The mutational landscape of MF has been better delineated in recent years and most non-driver mutations in MF are also prevalent in MDS and AML. Some authors have suggested that certain cases of MF appear not to be a pure MPN, but instead also have features of MDS – including dysplastic morphology and cytopenias etc. Indeed, the presence of non-driver mutations is reportedly correlated with the likelihood of myelodysplastic features and the severity of MF [51]. This highlights the MF disease spectrum ranging from myelodysplastic with consequent cytopenias to myeloproliferative MF. Other authors have focused upon a different terminology and describe advanced MF using the term “myelodepletive MF”. Here the degree of cytopenia associated with the myelodepletive MF phenotype is characterized by severe pancytopenia with low leukocyte count, platelet count, anemia, and frequently requiring transfusions. In contrast, the myeloproliferative MF phenotype is associated with leukocytosis, variable platelet counts, anemia, more frequently massive splenomegaly and a symptom profile associated with abdominal pain and night sweats [33].
These nosological models of myelodysplastic (mutational/morphological profile) and myelodepletive (thrombocytopenia/anemia) MF are likely not mutually exclusive. Whether the preferred term is cytopenic or myelodepletive, which recognizes a distinction from MDS and the WHO entity MDS with fibrosis, we will explore features of this phenotype in more detail below. These phenotypes are not always perfectly represented in individual studies due to the complex pathophysiology driving the clinical heterogeneity of MF, but instead simply represent a framework in which a spectrum of clinical phenotype can be better appreciated. Additionally, the paradox of atypical megakaryocyte hyperplasia which is a pathognomonic bone marrow feature of MF still underlies the thrombocytopenia overrepresented but not exclusive to the cytopenic MF profile. This is in contrast to the myelodepletive concept that may be best suited to convey a pathobiology that more closely resembles myelodysplastic syndrome.
Thrombocytopenia, (platelet count <100 × 109/) a key feature of cytopenic MF, is present in approximately 20% of MF patients at diagnosis with ∼11% presenting with a platelet count of <50 × 109/L and 30% at one year [33]. Thrombocytopenia is a recognized marker of poor prognosis. In the IPSS model, platelet counts <100 × 109/L were associated with decreased survival; however, due to its correlation with anemia, it was discarded as a separate factor [6]. In contrast, the DIPSS-plus [10] and subsequent prognostic scales also retain thrombocytopenia [12, 14]. The MYSEC includes platelets <150 × 109/L and hemoglobin <11 g/dL as poor prognostic markers [13], suggesting that in both PMF and sMF, the degree of thrombocytopenia has independent prognostic impact and serves as a marker of advanced disease.
It is also well established that thrombocytopenic MF patients have less frequent response and a shorter overall duration of response to ruxolitinib [52]. Barosi and colleagues reported that spleen response rates to ruxolitinib were reduced for those with lower rather than a higher JAK2V617F VAF. Here ≥ 50% JAK2V617F VAF was associated with a 5.5-fold greater probability of a spleen volume response compared with patients with <50% JAK2V617F VAF or another mutation [43]. The authors suggest that biology of the disease explains the higher response rate and that targeting JAK2 downstream signaling effectors with ruxolitinib would be more effective in persons with a high JAK2V617F VAF.
The previous findings may not however be generalizable across JAK2 inhibitors. Tremblay and colleagues assessed the efficacy of pacritinib, a JAK2/IRAK1 inhibitor, in MF patients with low JAK2V617F VAF in a post hoc analysis of the PERSIST-1 and −2 trials [53]. In that study, patients with lower JAK2V617F VAF had smaller baseline spleen size and lower hemoglobin and platelet counts as compared to patients with a higher VAF or JAK2 wildtype MF. Here, pacritinib treatment led to superior spleen and symptom burden reduction compared with BAT in patients with absent/low JAK2V617F allele burden, thus, suggesting that pacritinib may be uniquely suited for patients with cytopenic MF.
However, allelic burden is not universally found to correlate with advanced MF and cytopenias, suggesting that the underlying causality is likely more complex. In a cohort of 594 WHO-defined MF patients from Florence, a cytopenic phenotype, defined by ≥1 cytopenia without accompanying cytosis (leukocytes > 15 × 109/L, hemoglobin >16.5 g/dL for male and >16 g/dL for female, platelets >450 × 109/L) was identified in 166 patients [54]. Differences between PMF and sMF were also explored. Cytopenic PMF was associated with male gender (p = 0.0468), older age (p = 0.0002), lower peripheral blast count (p = 0.0006), higher prevalence of splenomegaly (p = 0.0142), constitutional symptoms (p < 0.0001), and BM fibrosis grade ≥2 (p < 0.0001). Also cytopenic MF patients were more likely to have very high-risk karyotypes (p = 0.0002), lack a driver mutation (TN; p < 0.0001), and also harbor a mutation involving ASXL1 (p = 0.0074), IDH1/2 (p = 0.064), N/KRAS (p = 0.0014), U2AF1 (p < 0.0001), or CUX1 (p = 0.0002). Karyotypic abnormalities (p = 0.0084), very high-risk cytogenetics (p = 0.0343), CBL (p = 0.0.171) and U2AF1 (p = 0.0148) mutations were significantly enriched in cytopenic patients with ≥2 cytopenias. In this study, OS was much lower in cytopenic PMF (median, 55 vs 103 months, respectively; P < .0001). Phenotypic differences for cytopenic MF were less evident in sMF, except for older age (p = 0.0207), and a molecular landscape which was enriched for mutations in TP53 (p = 0.0024), U2AF1 (P < 0.0001), and SETBP1 (p = 0.0125). Again, cytopenic sMF patients had shorter OS (median, 44 vs 105 months; p < 0.0001). Median OS was significantly inferior in those with ≥2 cytopenias compared with one cytopenia (median, 27 vs 58 months, respectively; p < 0.0001) [55].
Two large studies identified MF patients with platelets <50 ×109/L in particular as advanced disease. In a multivariable analysis of 1100 MF patients, thrombocytopenia was an independent negative prognostic variable for OS. Patients with platelets <50 ×109/L had other myelodepletive features (lower hemoglobin level, leukocytes, transfusion dependence, circulating blasts, older age, and abnormal/unfavorable karyotype). In this cohort, myelodepletive sMF did not appear to have substantially different clinical characteristics than myelodepletive PMF [56].
A study of the Spanish Registry data compared 57 such patients with 834 patients with a platelet count of ≥50 ×109/L [57]. This severely thrombocytopenic group was more likely to experience additional cytopenias, circulating blasts, MF-3 bone marrow fibrosis, and hemorrhage. Leukemic transformation was more common in the severely thrombocytopenic group (7.0 vs. 2.6 per 100 patient-years; p = 0.02), with a median projected survival of 2.2 years. No specific cytogenetic profile was associated with severe thrombocytopenia; however, there was a trend towards a lower frequency of JAK2V617F, and higher risk IPSS/DIPSS-plus score. Given the short OS, management of MF patients with severe thrombocytopenia constitutes a major unmet clinical need [57].
Additional studies have also demonstrated inferior outcome in PMF patients with low JAK2V617F allele burden, however, the relationship to severe thrombocytopenia was not established; Guglielmelli and colleagues stratified 186 PMF patients into: 1% to 25%, 25% to 50%, 50% to 75% and >75% JAK2V617F VAF [18]. The lowest quartile developed anemia and leukopenia more rapidly, but not particularly thrombocytopenia. In a prognostic model based on mutation status using an MD Anderson cohort of 344 PMF patients, a 50% cut-off dichotomized JAK2V617F patients into those with high JAK2V617F VAF and favorable survival and those with low JAK2V617F VAF and unfavorable survival [58].
Role of inflammatory signaling in myelofibrosis
In healthy individuals, inflammation is driven by a delicate interplay between cellular responses and stimulatory factors. Dysregulation of this inflammatory cascade is a hallmark of MPNs and the chronic inflammatory state of MF in particular, that is implicated in the debilitating constitutional symptoms and cytopenias characteristic of the disease [59, 60]. In Philadelphia chromosome-negative MPNs, driver mutations converge upon the JAK2/STAT3/STAT5 pathway [51], leading to its constitutive activation that can drive cytokine hypersensitivity, myeloid and megakaryocyte proliferation and differentiation [61]. NFκ-B remains the central transcriptional regulator of a wide array of inflammatory cytokines aberrantly expressed in MF that include interleukin (IL)−6, IL-1β, IL-8, tumor necrosis factor (TNF)-α, transforming growth factor beta (TGF)β and many others [62, 63]. NFκ-B is activated downstream of Toll-like receptors (TLR) whose signaling is upregulated in both malignant and non-malignant stromal cells of MF [64]. A constitutively activated JAK/STAT pathway can also drive sustained NFκ-B activation. The JAK2V617F gain of function mutation promotes p53 degradation through the accumulation of the E3 ubiquitin ligase HDM2 (Human Double Minute 2) via the La translational promoter in MPNs [65]. As a result, HDM2 increases NFκB activity by directly binding the Sp1 promoter site of NFκB p65 to activate its transcription [66]. Importantly, aberrant JAK2 signaling in MF and other MPNs leads to epigenetic changes that can also enhance NF-kB signaling [67].
The transmembrane TLRs contribute to this initial inflammatory signal, as monocytes from MPN patients are hyper-responsive to TLR ligands, which directs excess inflammatory cytokine production [68] (Fig. 1). TLRs, together with the IL-1 receptors, are part of a superfamily of pattern recognition receptors essential to the innate immune response that recognize pathogen-associated molecular patterns (PAMPs) from various microbes and self-derived molecules from damaged cells, known as damage-associated molecular patterns (DAMPs) that are over-expressed in both MPN patients and murine models [59, 64, 69]. Signaling downstream of TLRs is initiated by conformational changes induced by ligand-binding that leads to the recruitment of Toll/IL-1 receptor (TIR) domain-containing adaptor proteins that bind to the corresponding cytoplasmic TIR regions of TLRs [70, 71]. Five TLR adaptor proteins are known to interact, specifically Myeloid differentiation factor 88 (MyD88), TIR-domain containing adaptor molecule (TRIF), MyD88-adaptor-like (MAL, also known as TIR domain containing adaptor protein [TIRAP]), TRIF-related adaptor molecule (TRAM), and sterile α- and armadillo-motif-containing protein (SARM) [72]. MyD88-driven signaling primarily leads to the production of inflammatory cytokines such as TNF, IL-6, IL-1 and chemokines (CCL4), whereas TRIF induces the expression of type I and type II IFNs [71].
Fig. 1. Myddosome signaling.
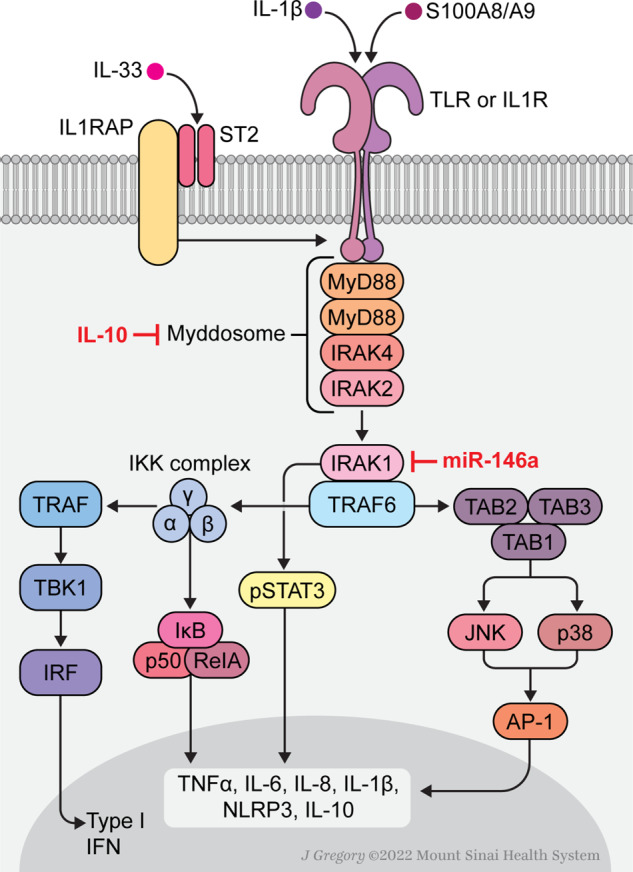
Ligand engagement of TIR -domain containing receptors triggers their dimerization and myddosome assembly through recruitment of the TIR-domain containing adaptor protein, MyD88. The N-terminal death domains (DD) of MyD88 proteins interact with the DD-containing, serine/threonine IRAK family kinases to create the active macromolecular protein signaling complex that converges upon IRAK1 transphosphorylation. Phosphorylated IRAK1 dissociates from the myddosome complex to activate the E3-ubiquitin ligase TRAF6 that is responsible for the activation of several transcription factors including NF-kB, AP-1, and interferon (IFN) regulatory factors (IRFs). Collectively, these proteins induce the expression of pro-inflammatory cytokines, IFNs and IFN-stimulated genes (ISGs), components of the NLRP3 inflammasome as well as the anti-inflammatory cytokine IL-10, which serves to quench myddosome signaling. The microRNA, miR-146a, also suppresses myddosome signaling through degradation of IRAK-1, TRAF-6 and TGF-β gene transcripts. IRAK1 also directly phosphorylates STAT3, triggering its nuclear translocation independently of the Janus kinases.
Myddosome deregulation
Two large signaling complexes referred to as supramolecular organizing centers or SMOCs direct innate immune response to TLR activation, namely the TLR-associated myddosome and the cytosolic, inflammasome complexes [73]. After TLR activation by PAMPs or DAMPs, the MyD88 adaptor rapidly initiates assembly of the myddosome complex via association with its death domain [70]. Key effectors include the serine/threonine kinase interleukin-1 receptor-associated kinase (IRAK)4, which phosphorylates its homolog IRAK1, triggering its auto-phosphorylation and dissociation from the myddosome, whereby it associates with the TNF receptor-associated factor (TRAF)-6. Poly-ubiquitination by TRAF6 activates IRAK1 signaling and the downstream induction of a large group of inflammatory cytokines through the activation of a variety of transcription factors including NF-κB, interferon regulatory factor (IRF)5, activator protein 1 (AP-1), and cAMP response element binding protein (CREB), as well as the activation of c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase (MAPK). IL-10 is the only anti-inflammatory cytokine generated by the IRAK1/NF-kB axis that serves as a negative feedback loop to extinguish myddosome-signaling [74]. IRAK1 also directly phosphorylates the transcription factor STAT3, triggering its nuclear translocation independent of the Janus kinases. Upon joint nuclear translocation with STAT3, IRAK1 phosphorylates Histone H3 thereby enhancing promoter binding by NF-kB to up-regulate inflammatory cytokines [75]. Cytokines under transcriptional control of NF-kB (e.g., TNFα, IL-6 & IL-8) are not extinguished by JAK1 or JAK2 inhibition in MF; however, they are suppressed by IRAK1 inhibition accompanied by a reduction in CD34 + colony formation [67, 76]. The activation of IRFs results in the transcription of a host of interferon-stimulated genes through the engagement of JAK1-associated interferon receptors [77].
A recent study by Muto et al. showed that TRAF6 can act as a tumor repressor and that loss of TRAF6 in pre-leukemic cells, which is associated with MYC-dependent signals, leads to overt myeloid leukemia [78]. Critically, the repression of TRAF6 has been observed in a subset of patients with myeloid malignancy, suggesting that dysregulation of TRAF6 can lead to acute leukemia. Moreover, miR-146a, which targets TRAF6 and IRAK1 mRNA for degradation, was shown to act as a tumor suppressor in the hematopoietic compartment and can control myeloproliferation in the spleen and BM through negative regulation of NF-kB [79], hence further linking chronic inflammation through MyD88 in MPNs. Importantly, somatic mutations of U2AF1 that occur in 10–15% of patients with PMF cause alternate splicing of IRAK4 gene transcripts to yield a longer isoform retaining exon 4, encoding a protein, IRAK4-Long (L) that is oncogenic in myeloid malignancies and can alone drive proliferation of the malignant clone through sustained myddosome activation [80].
Many studies suggest that inflammation supports MPN pathogenesis and development, yet it remains unclear whether inflammation is an event that initiates myeloproliferation and disease development, or simply a consequence or “byproduct” of the disease. For example, the hyper-responsiveness of monocytes and other hematopoietic progenitors to TLR ligands in MF relates in part to myddosome resistance to physiologic quenching by IL-10 [81]. Moreover, excess generation of TNFα paradoxically fosters the clonal expansion of JAK2V617F hematopoietic stem and progenitor cells (HSPC) while suppressing the growth of normal progenitors, indicating that the inflammatory bone marrow microenvironment in MF is conducive to propagate JAK2V617F-positive clones [82]. Interestingly, TLR4-directed myddosome signaling in CD34+ progenitors from patients with MF induces overexpression of the micro-RNA (miR)155, which degrades Jumonji And AT-Rich Interaction Domain Containing 2 (JARID2) gene transcripts giving rise to megakaryocytic hyperplasia that is characteristic of the disease [83]. More importantly, Rahman et. al. recently showed using gene silencing and cytokine neutralization approaches that IL-1 receptor/myddosome signaling is indispensable for clonal expansion, megakaryocyte proliferation and progression of MF in a JAK2V617F knock-in mouse model [84]. These findings were confirmed in a separate study by Rai et. al. which, in addition, demonstrated that serum IL-1β was derived from JAK2V617-mutant HSPC while HSPC IL-1 receptor expression and serum cytokine concentration directly correlated with JAK2V617F mutant allele fraction [85]. Moreover, antibody neutralization of IL-1β in a JAK2V617-mutant murine model reduced myelofibrosis and osteosclerosis, which was additive to the effects of ruxolitinib, suggesting that strategies that effectively mitigate IL-1 receptor signaling could be disease modifying.
Recent investigations implicate deregulation of miR-146a in the constitutive activation of myddosome signaling and peripheral blood cytopenias of MF. miR-146a targets IRAK-1, TRAF-6 and TGFβ gene transcripts for degradation and its expression is down-regulated in peripheral blood granulocytes of MF patients [86, 87]. De-repression of these genes in miR-146a knock-out mice results in sustained STAT3 activity associated with development of extensive medullary fibrosis, megakaryocytic hyperplasia, anemia, thrombocytopenia and splenomegaly, features that closely phenocopy cytopenic MF [79]. In addition, upregulation of the macrophage colony-stimulating factor receptor (CSF1R) was found in the myeloid cell population. These findings were reversed by selective knock-down of NF-κB p50, confirming the critical role of constitutive myddosome signaling in the pathogenesis of cytopenic MF. The precise pathobiology underlying the impairment of hematopoiesis may relate in part to the downstream effects of interferon gamma (IFNγ) induction [88]. IFNγ receptor stimulation leads to release of the alarmin, high mobility group box-1 protein (HMGB1), which disrupts the bone marrow endothelial niche, while deletion of IFNγ prevents HMGB1 release and is sufficient to reverse the endothelial defect and restore myelopoiesis [89]. HMGB1 was also recently identified as a key mediator of the anemia of inflammation by physically displacing the binding of erythropoietin to its cognate receptor [90]. As a result, HMGB1 reduces the proliferation and increases cell death of erythroid precursors to exacerbate anemia. Additionally, sustained STAT3 activation upregulates transcription of the GLI1 gene in MF fibrocytes to activate pro-fibrotic pathways in fibrocyte progenitors [91]. Interestingly, a polymorphism in the miR-146a gene, rs2431697, was identified in MPN patients at higher risk for progression to sMF [86]. In a large cohort analysis, the rs2431697 TT genotype was found to be an independent co-variate for higher risk of progression to sMF, findings that were confirmed in a separate cohort analysis of PV and ET patients [92]. Importantly, patients with the rs2431697 TT genotype in both studies had significantly higher levels of plasma inflammatory cytokines.
The second key innate immune SMOC, i.e., the inflammasome, is emerging in importance in the pathobiology of MF. Inflammasome complexes initiate caspase-1 mediated maturation of IL-1β and IL-18 as well as an inflammatory, lytic cell death termed pyroptosis [93]. Among the cytosolic NOD-like receptors (NLRs) that serve as sensors for specific inflammasomes, the pyrin domain containing 3 (NLRP3) inflammasome is the most studied and is critical in both sterile and non-sterile inflammation. The NLRP3 inflammasome, which is abundant in myeloid cells, is composed of the intracellular sensor NLRP3, the adaptor apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC), pro-caspase-1 and finally its substrates pro-IL-1β, -IL-18 and the pyroptosis executioner, gasdermin-D [93, 94].
Upon NLRP3 inflammasome activation, pro-caspase-1 undergoes auto-catalytic cleavage to functional caspase-1, which in turn transforms pro-IL-1β and pro-IL-18 into their mature, active forms that are subsequently released from the cell [95]. NLRP3 activation is particularly interesting, as it requires two stimuli for activation. The first or “priming” signal is mediated by TLRs in response to PAMPs, stress-associated signals released from DAMPs or IL-1 receptor engagement, or alternatively, after stimulation by TNF-α or IL-6, which have been implicated in priming of the NLRP3 inflammasome with advancing age [68, 96, 97]. This results in transcriptional upregulation of each of the necessary inflammasome components via NF-κB. The functional activation of the NLRP3 inflammasome is mediated by “signal 2”, which can take the form of exogenous or endogenous PAMPs and DAMPs. DAMPs, which are released after cell damage, can include extracellular adenosine triphosphate (eATP), mitochondrial reactive oxygen species (ROS), HMGB1, calcium-modulated proteins including S100A9 and S100A8, uric acid crystals, and extracellular DNA and RNA fragments. This is particularly relevant as various DAMPs are up-regulated and have been implicated in the pathobiology of MPNs, such as S100A9 and IL-33 amongst others [64, 69].
Recent data from Zhou et al. showed that inflammasome-related genes, such as NLRP3, are highly expressed in the bone marrow of MPN patients and that increased expression was associated with JAK2V617F, leukocytosis, and splenomegaly [98]. A second inflammasome that is relevant in MPN is the Absence In Melanoma 2 (AIM2) inflammasome that recognizes double-stranded DNA, functioning to protect against pathogens and dsDNA released from apoptotic or dying cells, which in turn leads to the release of proinflammatory cytokines and sterile inflammation [99]. In vitro work using the D9 cell line showed that Aim2 inflammasome-related genes, such as AIM2, CASP1 and IL1β, are upregulated upon induction of JAK2V617F, further linking MPN and inflammasome activation [100]. Although the role of the NLRP3 inflammasome is well described in MDS, its role in MPN pathogenesis remains understudied [101].
DAMPS and the S100A8/S100A9 heterodimer
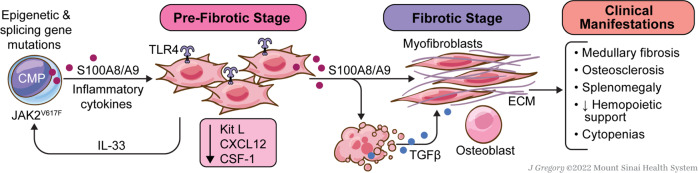
Recent studies have shown that the alarmin protein heterodimeric tetramer S100A8/S100A9 or calprotectin, which is primarily expressed in monocytes and granulocytes, is significantly upregulated in both murine models of MF and human stromal cells, i.e., cell populations that normally do not express S100A8/S100A9 at steady state [64] (Fig. 2). Critically, S100A8/S100A9 expression in mesenchymal stromal cells (MSCs) occurred with disease progression in both murine models and patient stroma, indicating that this may serve as an advanced disease biomarker. Indeed, calprotectin from MSCs stimulated the TLR4/Myddosome pathway in megakaryocytes to elaborate TGF-β and drive fibrosis progression and splenomegaly in MF patients. Most importantly, the S100A8/S100A9 heterodimer can be targeted in MPN murine models using the small molecule inhibitor tasquinimod, which significantly ameliorated the MF phenotype by reducing bone marrow fibrosis accompanied by spleen size reduction in JAK2V617F-driven models. As inhibition of alarmins in MDS can ameliorate the pathognomonic anemia through inhibition of an inflammatory cascade, it can be assumed that inhibition of alarmins in MF can also have a positive effect on debilitating cytopenias [101].
Fig. 2. TLR-directed pathogenesis of cytopenic myelofibrosis (MF).
JAK2V617F-mutant HSPC overexpress the heterodimeric alarmin, S100A8/S100A9, also termed calprotectin. Paracrine stimulation of TLR4 in bone marrow mesenchymal stromal cells (MSCs) instructs aberrant upregulation by MSCs. Calprotectin from MSCs stimulates the TLR4/Myddosome pathway in megakaryocytes to elaborate TGF-β and drive fibrosis progression, splenomegaly and exacerbation of cytopenias in MF.
Cytopenias, and anemia in particular, can be exacerbated by high circulating levels of calprotectin. Calprotectin sequesters cationic transition metals (e.g., calcium, iron, and zinc) that underlies its antimicrobial properties, while Fe+2 chelation restricts availability to the erythron, thereby contributing to iron-restricted anemia [102]. Moreover, the inflammatory proteins S100A9, TNFα and IL-1β each suppress transcription and cellular elaboration of erythropoietin, thereby reducing its availability to and stimulation of the erythron [103]. Molecular and proteomic analyses of CD34+ progenitors and granulocytes from patients with MPNs have shown that S100A8 and S100A9 are profoundly overexpressed in MF, with plasma levels that exert proliferative signals via the TLR4 and RAGE receptors, unrelated to JAK2V617F status [104, 105]. Of interest, plasma concentrations of S100A8 and S100A9 were highest in those patients with lower JAK2V617F VAF, a molecular feature more common in patients with cytopenic MF. Increased calprotectin production in cytopenic MF arises in part from the traditional non-driver, somatic gene mutations common to this disease subset. Although unrestrained JAK2/STAT activation induces S100A8/S100A9 in clonal hematopoietic progenitors and the surrounding stroma, both epigenetic regulatory and mRNA spicing gene mutations induce S100A8 and S100A9 overexpression to compound ineffective blood production in cytopenic MF [64, 106, 107]. Importantly, overexpression of S100A9 in a transgenic mouse model is alone sufficient to cause pancytopenia as a result of ineffective hematopoiesis that was ameliorated by NLRP3 inflammasome inhibition [108]. S100A9 also directs ineffective hematopoiesis by inducing the expression of the PD-1 death receptor on HSPC and its corresponding ligand, PD-L1, on myeloid-derived suppressor cells (MDSCs) [109].
Interleukin- 33
Interleukin-33 (IL-33), a member of the IL-1 cytokine family, signals via the myddosome analogous to its TLR family members. Full-length IL-33 is biologically active and is primarily expressed by epithelial and endothelial cells, however, IL-33 may exert a dual function in that it acts as an “alarmin” (DAMP) extracellularly, or as a nuclear factor modulating gene expression [110, 111]. Increased levels of IL-33 are demonstrable in the bone marrow and splenic vascular endothelium of MPN patients compared to controls, similar to expression of its accessory receptor ST2, also known as IL1RL1, which is necessary for IL-33 binding to the receptor complex and is upregulated in CD34 + HSPCs [69]. Binding of IL-33 to its cognate ST2 receptor initiates complex formation with the IL-1 receptor via the IL-1R accessory protein (IL-1RAcP) subunit. Signaling by the heterodimeric receptor is thereby mediated through the myddosome [112]. Mager et al. showed that IL-33 is an important contributor to the development of JAK2V617F-driven MPN in mice and exogenous IL-33 promotes colony formation of human primary CD34 + MPN HSPCs [69]. Interestingly, the IL-33/ST2 pathway can be activated in both the hematopoietic compartment and in non-hematopoietic MSC. IL-33 also stimulates the secretion of other DAMPs such as S100A8/S100A9 [113], which are strongly implicated in directing the MF phenotype [64].
Non-driver mutations and association with the cytopenic MF profile
The nuclear factor erythroid-2 gene (NF-E2), a hematopoietic transcription factor, is critical for proper differentiation of erythroblasts and megakaryocytes [114]. A study of 2,035 MPN patients (PMF 184; PV 411; ET 577) showed that the cohort harboring NF-E2 mutations frequently was JAK2 V617F homozygous, and NF-E2 mutations were acquired significantly later in the disease course [25]. Another study also showed that mutations in NF-E2 were detected in MPN patients who harbored JAK2 V617F, provided a proliferative advantage to the doubly mutant clone; and in a murine model, NF-E2 mutations caused a myeloproliferative phenotype (erythrocytosis and thrombocytosis) while predisposing to leukemic transformation [115]. Guglielmelli et al. reported that NF-E2 mutations were twofold more frequent among 631 MPN patients who had JAK2 VAF > 50%, however, there were no clear prognostic or meaningful clinical/hematological correlates [116].
Conversely, in a larger recent study, multivariate analysis of the data from 707 patients with MPNs (113 PMF, 233 PV, 332 ET) and available NGS data demonstrated that NF-E2 mutations (VAF ≥ 5%) carried a hazard ratio (HR) of 10.29 for transformation to AML (independently from age and co-occurring HMR mutations) and an HR of 8.24 for OS [117]. The HR of NF-E2 mutations was about fivefold higher for leukemogenesis and fourfold higher for OS compared to HMR mutations, respectively, thereby associating NF-E2 gene mutations with an aggressive disease course [117]. Notably, the NF-E2 VAF decreased at the time of leukemic transformation compared to the chronic phase, indicating that NF-E2-mutated cells may be outcompeted by another new dominant clone. In this case, NF-E2 acts as a “sentinel” mutation, dramatically increasing the likelihood of acquiring other mutations and leukemogenesis via a paracrine effect [118].
Patients harboring NF-E2 mutations had a higher median hematocrit than non-mutated patients in line with its association with the myeloproliferative phenotype and higher incidence in PV (7.3%) vs. PMF (5.3%) and ET (3.6%) [117]. As previously noted, NF-E2 mutations were acquired later in the disease course [25], induced significantly lower rates of hematological responses leading to the necessity for more lines of treatment, and were detected in 40% of the patients who lost response to treatment [117]. On the basis of these findings, analysis of NF-E2 mutations can be performed at diagnosis and in follow-up or upon loss of response to treatment. In patients harboring NF-E2 mutations, histone deacetylase inhibitors may be a rational therapeutic given their downregulation of NF-E2 expression [119].
Spliceosome U2AF1 mutations are detected in 10–15% of PMF patients [51], and are associated with the cytopenic MF phenotype and with ≥2 cytopenias [54]. In addition, U2AF1Q157 was associated with inferior survival compared to wild type-U2AF1 [30]. For this reason, U2AF1Q157 is included with other HMR mutations in the MIPSS70-plus v.2.0 and GIPSS prognostic models for PMF [120, 121]. Tefferi et al. noted a phenotypic correlation of the spliceosome pathway mutations U2AF1 and SRSF2 with anemia [122]. In PMF, both U2AF1Q157 and U2AF1S34 mutations were strongly associated with severe anemia (Hg < 10 g/dL); and U2AF1Q157 specifically with thrombocytopenia (platelet count <100×109/L) [30, 34]. The strong association of mutant U2AF1 with anemia and thrombocytopenia was sustained when mutated-JAK2 and wild type JAK2 patients were analyzed independently; U2AF1 mutations directly associated with JAK2V617F [34].
Summary
MF is a hematopoietic stem and progenitor cell-derived malignancy with complex molecular underpinnings and an associated immune deregulated state. The phenotypic spectrum ranges from the cytopenic to proliferative MF clinical and hematological profile that relates in part to somatic JAK-STAT driver mutations, the presence of accompanying non-driver mutations, and aberrant cell-intrinsic and -extrinsic inflammatory pathways. The cytopenic MF patient population is enriched for the PMF subtype, clinically typified by cytopenias and less extensive splenomegaly, and molecularly characterized by wild-type JAK2 or low JAK2V617F VAF frequently accompanied by somatic mutations involving the spliceosome, epigenome, and apoptotic pathways. Due to inferior OS, higher risk of leukemic transformation and increased resistance to ruxolitinib therapy, cytopenic MF patients pose a therapeutic challenge and represent an unmet medical need. Recognition of the role of TLR signaling and downstream myddosome activation of NFκB mediated pro-inflammatory cytokine expression has provided novel therapeutic targets for MF, such as IRAK1. Tailoring treatment with JAK inhibitors to genotype and phenotype will extend the potential for clinical benefit across the disease spectrum.
Author contributions
JM and AFL conceived this manuscript and JM, HFEG, HTC, CH, SV, AV, RBK, and AFL drafted the first version. All the authors further revised, approved the final version and agree to be held accountable for the content.
Competing interests
JM receives research funding paid to his institution from Incyte, CTI Bio, Novartis, PharmaEssentia, Merck, Kartos, Geron, BMS, Roche, and consulting fees from Incyte, CTI Bio, Novartis, Roche, BMS, Celgene, Geron, PharmaEssentia, Morphosys, Sierra Oncology, GSK, Galecto, Karyopharm, Kartos, and Imago. HFEG has nothing to declare. HTC has nothing to declare. CNH received research funding paid to the institutional from Novartis, Celgene, and Constellation Pharmaceuticals, MorphoSys and consulting fees from Keros, Galecto, AOP and Roche, Novartis, Celgene, CTI BioPharma, AbbVie, Janssen, Geron, Promedior, AbbVie, AOP Pharma. SV receives research funding paid to the institution from Incyte, Roche, NS Pharma, Celgene, Gilead, Promedior, CTI BioPharma, Blueprint Medicines, Novartis, Sierra Oncology, PharmaEssentia, Protagonist Therapeutics, Kartos Galecto and consulting fees from Constellation, Sierra Oncology, Incyte, Novartis, BMS, Kartos, Galecto, Protagonist. AMV receives consulting fees from Novartis, BMS, Incyte, Blueprint, and Abbvie. RKR receives research funding paid to the institution from Research funding: Constellation, Incyte, Stemline, Zentalis and consulting feels from Constellation, Incyte, Celgene/BMS, Novartis, Promedior, CTI, Jazz Pharmaceuticals, Blueprint, Stemline, Galecto, Pharmaessentia, Abbvie, Sierra Oncology, Disc Medicines, Sunimoto Dainippon, Zentalis. JJK receives consulting fees from Novartis, Abbvie, BMS, Sierra Oncology, CTI Biopharma, PharmaEssentia, AOP Orphan. WV has nothing to disclose. RH received research grant funding to the institution from BMS, Citi Bio, Curis, Inc., Genentech, Kartos Therapeutics, Kymera Therapeutics, OncoMyx Therapeutics, Protagonist Therapeutics, Repare Therapeutics, Scholar Rock, Translational Drug Development (TD2), Turning Point Therapeutics and consulting fees from AbbVie, Ionis Pharmaceuticals, Novartis, Protagonist Therapeutics, Silence Therapeutics. RKS has nothing to declare. AFL receives consulting fees from Halia Therapeutic, Rigel Pharmaceuticals, CTI Biopharma, and Aileron Inc.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guglielmelli P, Lasho TL, Rotunno G, Score J, Mannarelli C, Pancrazzi A, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28:1804–10. doi: 10.1038/leu.2014.76. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Zhang W, Ishii T, Sozer S, Wang J, Xu M, et al. Correction of the abnormal trafficking of primary myelofibrosis CD34+ cells by treatment with chromatin-modifying agents. Cancer Res. 2009;69:7612–8. doi: 10.1158/0008-5472.CAN-09-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verstovsek S, Yu J, Scherber RM, Verma S, Dieyi C, Chen CC, et al. Changes in the incidence and overall survival of patients with myeloproliferative neoplasms between 2002 and 2016 in the United States. Leuk Lymphoma. 2022;63:694–702. doi: 10.1080/10428194.2021.1992756. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes F, Dupriez B, Passamonti F, Vannucchi AM, Morra E, Reilly JT, et al. Improving survival trends in primary myelofibrosis: an international study. J Clin Oncol. 2012;30:2981–7. doi: 10.1200/JCO.2012.42.0240. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes F, Dupriez B, Pereira A, Passamonti F, Reilly JT, Morra E, et al. New prognostic scoring system for primary myelofibrosis based on a study of the International Working Group for Myelofibrosis Research and Treatment. Blood. 2009;113:2895–901. doi: 10.1182/blood-2008-07-170449. [DOI] [PubMed] [Google Scholar]
- 7.Tefferi A, Guglielmelli P, Larson DR, Finke C, Wassie EA, Pieri L, et al. Long-term survival and blast transformation in molecularly annotated essential thrombocythemia, polycythemia vera, and myelofibrosis. Blood. 2014;124:2507–13. doi: 10.1182/blood-2014-05-579136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupriez B, Morel P, Demory JL, Lai JL, Simon M, Plantier I, et al. Prognostic factors in agnogenic myeloid metaplasia: a report on 195 cases with a new scoring system. Blood. 1996;88:1013–8. [PubMed] [Google Scholar]
- 9.Passamonti F, Cervantes F, Vannucchi AM, Morra E, Rumi E, Pereira A, et al. A dynamic prognostic model to predict survival in primary myelofibrosis: a study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment) Blood. 2010;115:1703–8. doi: 10.1182/blood-2009-09-245837. [DOI] [PubMed] [Google Scholar]
- 10.Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S, et al. DIPSS plus: A refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29:392–7. doi: 10.1200/JCO.2010.32.2446. [DOI] [PubMed] [Google Scholar]
- 11.Guglielmelli P, Lasho TL, Rotunno G, Mudireddy M, Mannarelli C, Nicolosi M, et al. MIPSS70: mutation-enhanced international prognostic score system for transplantation-age patients with primary myelofibrosis. J Clin Oncol. 2018;36:310–8. doi: 10.1200/JCO.2017.76.4886. [DOI] [PubMed] [Google Scholar]
- 12.Tefferi A, Guglielmelli P, Nicolosi M, Mannelli F, Mudireddy M, Bartalucci N, et al. GIPSS: genetically inspired prognostic scoring system for primary myelofibrosis. Leukemia. 2018;32:1631–42. doi: 10.1038/s41375-018-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passamonti F, Giorgino T, Mora B, Guglielmelli P, Rumi E, Maffioli M, et al. A clinical-molecular prognostic model to predict survival in patients with post polycythemia vera and post essential thrombocythemia myelofibrosis. Leukemia. 2017;31:2726–31. doi: 10.1038/leu.2017.169. [DOI] [PubMed] [Google Scholar]
- 14.Gagelmann N, Ditschkowski M, Bogdanov R, Bredin S, Robin M, Cassinat B, et al. Comprehensive clinical-molecular transplant scoring system for myelofibrosis undergoing stem cell transplantation. Blood. 2019;133:2233–42. doi: 10.1182/blood-2018-12-890889. [DOI] [PubMed] [Google Scholar]
- 15.Rotunno G, Pacilli A, Artusi V, Rumi E, Maffioli M, Delaini F, et al. Epidemiology and clinical relevance of mutations in postpolycythemia vera and postessential thrombocythemia myelofibrosis: A study on 359 patients of the AGIMM group. Am J Hematol. 2016;91:681–6. doi: 10.1002/ajh.24377. [DOI] [PubMed] [Google Scholar]
- 16.Passamonti F, Rumi E, Pietra D, Elena C, Boveri E, Arcaini L, et al. A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia. 2010;24::1574–9. doi: 10.1038/leu.2010.148. [DOI] [PubMed] [Google Scholar]
- 17.Loscocco GG, Guglielmelli P, Gangat N, Rossi E, Mannarelli C, Betti S, et al. Clinical and molecular predictors of fibrotic progression in essential thrombocythemia: A multicenter study involving 1607 patients. Am J Hematol. 2021;96:1472–80. doi: 10.1002/ajh.26332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guglielmelli P, Barosi G, Specchia G, Rambaldi A, Lo Coco F, Antonioli E, et al. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood. 2009;114:1477–83. doi: 10.1182/blood-2009-04-216044. [DOI] [PubMed] [Google Scholar]
- 19.Rumi E, Pietra D, Pascutto C, Guglielmelli P, Martínez-Trillos A, Casetti I, et al. Clinical effect of driver mutations of JAK2, CALR or MPL in primary myelofibrosis. Blood. 2014;124:1062–9. doi: 10.1182/blood-2014-05-578435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milosevic Feenstra JD, Nivarthi H, Gisslinger H, Leroy E, Rumi E, Chachoua I, et al. Whole-exome sequencing identifies novel MPL and JAK2 mutations in triple-negative myeloproliferative neoplasms. Blood. 2016;127:325–32. doi: 10.1182/blood-2015-07-661835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guglielmelli P, Pacilli A, Rotunno G, Rumi E, Rosti V, Delaini F, et al. Presentation and outcome of patients with 2016 WHO diagnosis of prefibrotic and overt primary myelofibrosis. Blood. 2017;129:3227–36. doi: 10.1182/blood-2017-01-761999. [DOI] [PubMed] [Google Scholar]
- 22.Passamonti F, Mora B, Giorgino T, Guglielmelli P, Cazzola M, Maffioli M, et al. Driver mutations’ effect in secondary myelofibrosis: an international multicenter study based on 781 patients. Leukemia. 2017;31:970–3. doi: 10.1038/leu.2016.351. [DOI] [PubMed] [Google Scholar]
- 23.Tefferi A, Guglielmelli P, Lasho TL, Coltro G, Finke CM, Loscocco GG, et al. Mutation-enhanced international prognostic systems for essential thrombocythaemia and polycythaemia vera. Br J Haematol. 2020;189:291–302. doi: 10.1111/bjh.16380. [DOI] [PubMed] [Google Scholar]
- 24.Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–9. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- 25.Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379:1416–30. doi: 10.1056/NEJMoa1716614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luque Paz D, Riou J, Verger E, Cassinat B, Chauveau A, Ianotto JC, et al. Genomic analysis of primary and secondary myelofibrosis redefines the prognostic impact of ASXL1 mutations: a FIM study. Blood Adv. 2021;5:1442–51. doi: 10.1182/bloodadvances.2020003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotunno G, Mannarelli C, Brogi G, Pacilli A, Gesullo F, Mannelli F, et al. Spectrum of ASXL1 mutations in primary myelofibrosis: prognostic impact of the ASXL1 p.G646Wfs*12 mutation. Blood. 2019;133:2802–8. doi: 10.1182/blood.2018879536. [DOI] [PubMed] [Google Scholar]
- 28.Tefferi A, Guglielmelli P, Lasho TL, Rotunno G, Finke C, Mannarelli C, et al. CALR and ASXL1 mutations-based molecular prognostication in primary myelofibrosis: an international study of 570 patients. Leukemia. 2014;28:1494–1500. doi: 10.1038/leu.2014.57. [DOI] [PubMed] [Google Scholar]
- 29.Tefferi A, Lasho TL, Finke C, Gangat N, Hanson CA, Ketterling RP, et al. Prognostic significance of ASXL1 mutation types and allele burden in myelofibrosis. Leukemia. 2018;32::2274–8. doi: 10.1038/leu.2017.318. [DOI] [PubMed] [Google Scholar]
- 30.Tefferi A, Finke CM, Lasho TL, Hanson CA, Ketterling RP, Gangat N, et al. U2AF1 mutation types in primary myelofibrosis: phenotypic and prognostic distinctions. Leukemia. 2018;32:2274–8. doi: 10.1038/s41375-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tefferi A, Guglielmelli P, Lasho TL, Gangat N, Ketterling RP, Pardanani A, et al. MIPSS70+ version 2.0: mutation and karyotype-enhanced international prognostic scoring system for primary myelofibrosis. J Clin Oncol. 2018;36:1769–70. doi: 10.1200/JCO.2018.78.9867. [DOI] [PubMed] [Google Scholar]
- 32.Guglielmelli P, Coltro G, Mannelli F, Rotunno G, Loscocco GG, Mannarelli C, et al. ASXL1 mutations are prognostically significant in PMF, but not MF following essential thrombocythemia or polycythemia vera. Blood Adv. 2022;6:2927–31. doi: 10.1182/bloodadvances.2021006350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcellino BK, Verstovsek S, Mascarenhas J. The myelodepletive phenotype in myelofibrosis: clinical relevance and therapeutic implication. Clin Lymphoma Myeloma Leuk. 2020;20:415–21. doi: 10.1016/j.clml.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Tefferi A, Finke CM, Lasho TL, Wassie EA, Knudson R, Ketterling RP, et al. U2AF1 mutations in primary myelofibrosis are strongly associated with anemia and thrombocytopenia despite clustering with JAK2V617F and normal karyotype. Leukemia. 2014;28:431–3. doi: 10.1038/leu.2013.286. [DOI] [PubMed] [Google Scholar]
- 35.Dunbar AJ, Rampal RK, Levine R. Leukemia secondary to myeloproliferative neoplasms. Blood. 2020;136:61–70. doi: 10.1182/blood.2019000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubesova B, Pavlova S, Malcikova J, Kabathova J, Radova L, Tom N, et al. Low-burden TP53 mutations in chronic phase of myeloproliferative neoplasms: association with age, hydroxyurea administration, disease type and JAK2 mutational status. Leukemia. 2018;32:450–61. doi: 10.1038/leu.2017.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–8. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 38.Arranz L, Sanchez-Aguilera A, Martin-Perez D, Isern J, Langa X, Tzankov A, et al. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature. 2014;512:78–81. doi: 10.1038/nature13383. [DOI] [PubMed] [Google Scholar]
- 39.Goyal H, Chachoua I, Pecquet C, Vainchenker W, Constantinescu SN. A p53-JAK-STAT connection involved in myeloproliferative neoplasm pathogenesis and progression to secondary acute myeloid leukemia. Blood Rev. 2020;42:100712. doi: 10.1016/j.blre.2020.100712. [DOI] [PubMed] [Google Scholar]
- 40.Beer PA, Delhommeau F, LeCouedic JP, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115:2891–2900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 41.Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587:477–82. doi: 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parenti S, Rontauroli S, Carretta C, Mallia S, Genovese E, Chiereghin C, et al. Mutated clones driving leukemic transformation are already detectable at the single-cell level in CD34-positive cells in the chronic phase of primary myelofibrosis. NPJ Precis Oncol. 2021;5:4. doi: 10.1038/s41698-021-00144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barosi G, Klersy C, Villani L, Bonetti E, Catarsi P, Poletto V, et al. JAK2(V617F) allele burden 50% is associated with response to ruxolitinib in persons with MPN-associated myelofibrosis and splenomegaly requiring therapy. Leukemia. 2016;30:1772–5. doi: 10.1038/leu.2016.45. [DOI] [PubMed] [Google Scholar]
- 44.Guglielmelli P, Biamonte F, Rotunno G, Artusi V, Artuso L, Bernardis I, et al. Impact of mutational status on outcomes in myelofibrosis patients treated with ruxolitinib in the COMFORT-II Study. Blood. 2014;123:2157–60. doi: 10.1182/blood-2013-11-536557. [DOI] [PubMed] [Google Scholar]
- 45.Patel KP, Newberry KJ, Luthra R, Jabbour E, Pierce S, Cortes J, et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood. 2015;126:790–7. doi: 10.1182/blood-2015-03-633404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pacilli A, Rotunno G, Mannarelli C, Fanelli T, Pancrazzi A, Contini E, et al. Mutation landscape in patients with myelofibrosis receiving ruxolitinib or hydroxyurea. Blood Cancer J. 2018;8:122. doi: 10.1038/s41408-018-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spiegel JY, McNamara C, Kennedy JA, Panzarella T, Arruda A, Stockley T, et al. Impact of genomic alterations on outcomes in myelofibrosis patients undergoing JAK1/2 inhibitor therapy. Blood Adv. 2017;1:1729–38. doi: 10.1182/bloodadvances.2017009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coltro G, Rotunno G, Mannelli L, Mannarelli C, Fiaccabrino S, Romagnoli S, et al. RAS/CBL mutations predict resistance to JAK inhibitors in myelofibrosis and are associated with poor prognostic features. Blood Adv. 2020;4:3677–87. doi: 10.1182/bloodadvances.2020002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos FPS, Getta B, Masarova L, Famulare C, Schulman J, Datoguia TS, et al. Prognostic impact of RAS-pathway mutations in patients with myelofibrosis. Leukemia. 2019;34:799–810. doi: 10.1038/s41375-019-0603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dameshek W. Some speculations on the myeloproliferative syndromes. Blood. 1951;6:372–5. [PubMed] [Google Scholar]
- 51.Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–79. doi: 10.1182/blood-2016-10-695940. [DOI] [PubMed] [Google Scholar]
- 52.Harrison CN, Schaap N, Mesa RA. Management of myelofibrosis after ruxolitinib failure. Ann Hematol. 2020;99:1177–91. doi: 10.1007/s00277-020-04002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tremblay D, Mesa R, Scott B, Buckley S, Roman-Torres K, Verstovsek S, et al. Pacritinib demonstrates spleen volume reduction in patients with myelofibrosis independent of JAK2V617F allele burden. Blood Adv. 2020;4:5929–35. doi: 10.1182/bloodadvances.2020002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coltro G, Mannelli F, Loscocco GG, Mannarelli C, Rotunno G, Maccari C, et al. A myelodepletive phenotype is associated with distinctive molecular features and adverse outcomes in patients with myelofibrosis. Blood. 2021;138:1498. [Google Scholar]
- 55.Coltro G, Mannelli F, Loscocco GG, Mannarelli C, Rotunno G, Maccari C, et al. Differential prognostic impact of cytopenic phenotype in prefibrotic vs overt primary myelofibrosis. Blood Cancer J. 2022;12:116. doi: 10.1038/s41408-022-00713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masarova L, Daver N, Pemmaraju N, Bose P, Pierce S, Manshouri T, et al. Do patients with post-essential thrombocythemia and post-polycythemia vera differ from patients with primary myelofibrosis? Blood. 2015;126:4069. doi: 10.1016/j.leukres.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernandez-Boluda JC, Correa JG, Alvarez-Larran A, Ferrer-Marin F, Raya JM, Martinez-Lopez J, et al. Clinical characteristics, prognosis and treatment of myelofibrosis patients with severe thrombocytopenia. Br J Haematol. 2018;181:397–400. doi: 10.1111/bjh.14601. [DOI] [PubMed] [Google Scholar]
- 58.Rozovski U, Verstovsek S, Manshouri T, Dembitz V, Bozinovic K, Newberry K, et al. An accurate, simple prognostic model consisting of age, JAK2, CALR, and MPL mutation status for patients with primary myelofibrosis. Haematologica. 2017;102:79–84. doi: 10.3324/haematol.2016.149765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol. 2011;29:1356–63. doi: 10.1200/JCO.2010.32.9490. [DOI] [PubMed] [Google Scholar]
- 60.Geyer HL, Dueck AC, Scherber RM, Mesa RA. Impact of inflammation on myeloproliferative neoplasm symptom development. Mediators Inflamm. 2015;2015:284706. doi: 10.1155/2015/284706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hasselbalch HC. Perspectives on chronic inflammation in essential thrombocythemia, polycythemia vera, and myelofibrosis: is chronic inflammation a trigger and driver of clonal evolution and development of accelerated atherosclerosis and second cancer? Blood. 2012;119:3219–25. doi: 10.1182/blood-2011-11-394775. [DOI] [PubMed] [Google Scholar]
- 62.Vaidya A, Kale VP. TGF-beta signaling and its role in the regulation of hematopoietic stem cells. Syst Synth Biol. 2015;9:1–10. doi: 10.1007/s11693-015-9161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher DAC, Miner CA, Engle EK, Hu HR, Collins TB, Zhou A, et al. Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NF kappa B signaling. Leukemia. 2019;33:1978–95. doi: 10.1038/s41375-019-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leimkuhler NB, Gleitz HFE, Ronghui L, Snoeren IAM, Fuchs SNR, Nagai JS, et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. 2021;28:637–52. doi: 10.1016/j.stem.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakatake M, Monte-Mor B, Debili N, Casadevall N, Ribrag V, Solary E, et al. JAK2(V617F) negatively regulates p53 stabilization by enhancing MDM2 via La expression in myeloproliferative neoplasms. Oncogene. 2012;31:1323–33. doi: 10.1038/onc.2011.313. [DOI] [PubMed] [Google Scholar]
- 66.Gu LB, Findley HW, Zhou MX. MDM2 induces NF-kappa B/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood. 2002;99:3367–75. doi: 10.1182/blood.v99.9.3367. [DOI] [PubMed] [Google Scholar]
- 67.Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, et al. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33:785–7. doi: 10.1016/j.ccell.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleischman AG, Luty SB, Garbati MR, Edmiston JB, Royer LR, LaTocha DH, et al. Exaggerated response to toll-like receptor agonist contributes to excessive TNF production in myeloproliferative neoplasm. Blood. 2013;122:4097–4097. [Google Scholar]
- 69.Mager LF, Riether C, Schurch CM, Banz Y, Wasmer MH, Stuber R, et al. IL-33 signaling contributes to the pathogenesis of myeloproliferative neoplasms. J Clin Invest. 2015;125:2579–91. doi: 10.1172/JCI77347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balka KR, De Nardo D. Understanding early TLR signaling through the Myddosome. J Leukoc Biol. 2019;105:339–51. doi: 10.1002/JLB.MR0318-096R. [DOI] [PubMed] [Google Scholar]
- 71.De Nardo D. Toll-like receptors: Activation, signalling and transcriptional modulation. Cytokine. 2015;74:181–9. doi: 10.1016/j.cyto.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 72.O’Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 73.Tan Y, Kagan JC. Innate immune signaling organelles display natural and programmable signaling flexibility. Cell. 2019;177:384–98. doi: 10.1016/j.cell.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang YS, Li T, Sane DC, Li LW. IRAK1 serves as a novel regulator essential for lipopolysaccharide-induced interleukin-10 gene expression. J Biol Chem. 2004;279:51697–703. doi: 10.1074/jbc.M410369200. [DOI] [PubMed] [Google Scholar]
- 75.Liu G, Park YJ, Abraham E. Interleukin-1 receptor-associated kinase (IRAK)-1-mediated NF-kappa B activation requires cytosolic and nuclear activity. Faseb J. 2008;22:2285–96. doi: 10.1096/fj.07-101816. [DOI] [PubMed] [Google Scholar]
- 76.Vaidya R, Gangat N, Jimma T, Finke CM, Lasho TL, Pardanani A, et al. Plasma cytokines in polycythemia vera: phenotypic correlates, prognostic relevance, and comparison with myelofibrosis. Am J Hematol. 2012;87:1003–5. doi: 10.1002/ajh.23295. [DOI] [PubMed] [Google Scholar]
- 77.Vainchenker W, Leroy E, Gilles L, Marty C, Plo I, Constantinescu SN. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Res. 2018;7:82. doi: 10.12688/f1000research.13167.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muto T, Guillamot M, Yeung J, Fang J, Bennett J, Nadorp B, et al. TRAF6 functions as a tumor suppressor in myeloid malignancies by directly targeting MYC oncogenic activity. Cell Stem Cell. 2022;29:298–314. doi: 10.1016/j.stem.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao JL, Rao DS, Boldin MP, Taganov KD, O’Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci USA. 2011;108:9184–9. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith MA, Choudhary GS, Pellagatti A, Choi K, Bolanos LC, Bhagat TD, et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat Cell Biol. 2019;21:640–50. doi: 10.1038/s41556-019-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai HY, Brooks SA, Craver BM, Morse SJ, Nguyen TK, Haghighi N, et al. Defective negative regulation of Toll-like receptor signaling leads to excessive TNF-alpha in myeloproliferative neoplasm. Blood Adv. 2019;3:122–31. doi: 10.1182/bloodadvances.2018026450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fleischman AG, Aichberger KJ, Luty SB, Bumm TG, Petersen CL, Doratotaj S, et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 2011;118:6392–8. doi: 10.1182/blood-2011-04-348144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124:e21–32. doi: 10.1182/blood-2013-12-544197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahman MF, Yang Y, Le BT, Dutta A, Posyniak J, Faughnan P, et al. Interleukin-1 contributes to clonal expansion and progression of bone marrow fibrosis in JAK2V617F-induced myeloproliferative neoplasm. Nat Commun. 2022;13:5347. doi: 10.1038/s41467-022-32928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rai S, Grockowiak E, Hansen N, Luque Paz D, Stoll CB, Hao-Shen H, et al. Inhibition of interleukin-1beta reduces myelofibrosis and osteosclerosis in mice with JAK2-V617F driven myeloproliferative neoplasm. Nat Commun. 2022;13:5346. doi: 10.1038/s41467-022-32927-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ferrer-Marin F, Arroyo AB, Bellosillo B, Cuenca EJ, Zamora L, Hernandez-Rivas JM, et al. miR-146a rs2431697 identifies myeloproliferative neoplasm patients with higher secondary myelofibrosis progression risk. Leukemia. 2020;34:2648–59. doi: 10.1038/s41375-020-0767-3. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Zhu H, Wei X, Li H, Yu Z, Zhang H, et al. LPS induces HUVEC angiogenesis in vitro through miR-146a-mediated TGF-beta1 inhibition. Am J Transl Res. 2017;9:591–600. [PMC free article] [PubMed] [Google Scholar]
- 88.Wong WJ, Baltay M, Getz A, Fuhrman K, Aster JC, Hasserjian RP, et al. Gene expression profiling distinguishes prefibrotic from overtly fibrotic myeloproliferative neoplasms and identifies disease subsets with distinct inflammatory signatures. Plos One. 2019;14:e0216810. doi: 10.1371/journal.pone.0216810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gopal A, Ibrahim R, Fuller M, Umlandt P, Parker J, Tran J, et al. TIRAP drives myelosuppression through an Ifngamma-Hmgb1 axis that disrupts the endothelial niche in mice. J Exp Med. 2022;219:e20200731. [DOI] [PMC free article] [PubMed]
- 90.Dulmovits BM, Tang Y, Papoin J, He M, Li J, Yang H, et al. HMGB1-mediated restriction of EPO signaling contributes to anemia of inflammation. Blood. 2022;139:3181–93. doi: 10.1182/blood.2021012048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manshouri T, Veletic I, Li P, Yin CC, Post SM, Verstovsek S, et al. GLI1 activates pro-fibrotic pathways in myelofibrosis fibrocytes. Cell Death Dis. 2022;13:481. doi: 10.1038/s41419-022-04932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aref S, Atia D, Al Tantawy A, Al Boghdady M, Gouda E. Predictive Value of miR-146a rs2431697 Polymorphism to Myelofibrosis Progression in Patients with Myeloproliferative Neoplasm. Asian Pac J Cancer Prev. 2021;22:3585–9. doi: 10.31557/APJCP.2021.22.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tourneur L, Witko-Sarsat V. Inflammasome activation: Neutrophils go their own way. J Leukoc Biol. 2019;105:433–6. doi: 10.1002/JLB.3CE1118-433R. [DOI] [PubMed] [Google Scholar]
- 94.Di Battista V, Bochicchio MT, Giordano G, Napolitano M, Lucchesi A. Genetics and Pathogenetic Role of Inflammasomes in Philadelphia Negative Chronic Myeloproliferative Neoplasms: A Narrative Review. Int J Mol Sci. 2021;22:1–16. doi: 10.3390/ijms22020561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–73. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, et al. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem. 2012;287:34474–83. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14:576–90. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 98.Zhou Y, Yan S, Liu N, He N, Zhang A, Meng S, et al. Genetic polymorphisms and expression of NLRP3 inflammasome-related genes are associated with Philadelphia chromosome-negative myeloproliferative neoplasms. Hum Immunol. 2020;81:606–13. doi: 10.1016/j.humimm.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 99.Sharma BR, Karki R, Kanneganti TD. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur J Immunol. 2019;49:1998–2011. doi: 10.1002/eji.201848070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liew EL, Araki M, Hironaka Y, Mori S, Tan TZ, Morishita S, et al. Identification of AIM2 as a downstream target of JAK2V617F. Exp Hematol Oncol. 2015;5:2. doi: 10.1186/s40164-016-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Basiorka AA, McGraw KL, Eksioglu EA, Chen XH, Johnson J, Zhang L, et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 2016;128:2960–75. doi: 10.1182/blood-2016-07-730556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakashige TG, Zhang B, Krebs C, Nolan EM. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol. 2015;11:765–71. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cluzeau T, McGraw KL, Irvine B, Masala E, Ades L, Basiorka AA, et al. Pro-inflammatory proteins S100A9 and tumor necrosis factor-alpha suppress erythropoietin elaboration in myelodysplastic syndromes. Haematologica. 2017;102:2015–20. doi: 10.3324/haematol.2016.158857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cokic VP, Mitrovic-Ajtic O, Beleslin-Cokic BB, Markovic D, Buac M, Diklic M, et al. Proinflammatory cytokine IL-6 and JAK-STAT signaling pathway in myeloproliferative neoplasms. Mediators Inflamm. 2015;2015:453020. doi: 10.1155/2015/453020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kovacic M, Mitrovic-Ajtic O, Beleslin-Cokic B, Djikic D, Suboticki T, Diklic M, et al. TLR4 and RAGE conversely mediate pro-inflammatory S100A8/9-mediated inhibition of proliferation-linked signaling in myeloproliferative neoplasms. Cell Oncol. 2018;41:541–53. doi: 10.1007/s13402-018-0392-6. [DOI] [PubMed] [Google Scholar]
- 106.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22:672–8. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McLemore AF, Hou HA, Meyer BS, Lam NB, Ward GA, Aldrich AL, et al. Somatic gene mutations expose cytoplasmic DNA to co-opt the cGAS/STING/NLRP3 axis in myelodysplastic syndromes. JCI Insight. 2022;7:e159430. doi: 10.1172/jci.insight.159430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen X, Eksioglu EA, Zhou J, Zhang L, Djeu J, Fortenbery N, et al. Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest. 2013;123:4595–611. doi: 10.1172/JCI67580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheng P, Eksioglu EA, Chen X, Kandell W, Le Trinh T, Cen L, et al. S100A9-induced overexpression of PD-1/PD-L1 contributes to ineffective hematopoiesis in myelodysplastic syndromes. Leukemia. 2019;33:2034–46. doi: 10.1038/s41375-019-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, et al. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol. 2011;187:1609–16. doi: 10.4049/jimmunol.1003080. [DOI] [PubMed] [Google Scholar]
- 111.Wasmer MH, Krebs P. The role of IL-33-dependent inflammation in the tumor microenvironment. Front Immunol. 2016;7:682. doi: 10.3389/fimmu.2016.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pinto SM, Subbannayya Y, Rex DAB, Raju R, Chatterjee O, Advani J, et al. A network map of IL-33 signaling pathway. J Cell Commun Signal. 2018;12:615–24. doi: 10.1007/s12079-018-0464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, Davis C, Shah S, Hughes D, Ryan JC, Altomare D, et al. IL-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol Carcinog. 2017;56:272–87. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levin J, Peng JP, Baker GR, Villeval JL, Lecine P, Burstein SA, et al. Pathophysiology of thrombocytopenia and anemia in mice lacking transcription factor NF-E2. Blood. 1999;94:3037–47. [PubMed] [Google Scholar]
- 115.Jutzi JS, Bogeska R, Nikoloski G, Schmid CA, Seeger TS, Stegelmann F, et al. MPN patients harbor recurrent truncating mutations in transcription factor NF-E2. J Exp Med. 2013;210:1003–19. doi: 10.1084/jem.20120521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guglielmelli P, Pacilli A, Coltro G, Mannelli F, Mannelli L, Contini E, et al. Characteristics and clinical correlates of NFE2 mutations in chronic Myeloproliferative neoplasms. Am J Hematol. 2020;95:E23–E26. doi: 10.1002/ajh.25668. [DOI] [PubMed] [Google Scholar]
- 117.Marcault C, Zhao LP, Maslah N, Verger E, Daltro de Oliveira R, Soret-Dulphy J, et al. Impact of NFE2 mutations on AML transformation and overall survival in patients with myeloproliferative neoplasms. Blood. 2021;138:2142–8. doi: 10.1182/blood.2020010402. [DOI] [PubMed] [Google Scholar]
- 118.Pahl HL. Sentinel mutations: the roses in the vineyard. Blood. 2021;138:2019–20. doi: 10.1182/blood.2021012345. [DOI] [PubMed] [Google Scholar]
- 119.Chifotides HT, Bose P, Verstovsek S. Givinostat: an emerging treatment for polycythemia vera. Expert Opin Inv Drug. 2020;29:525–36. doi: 10.1080/13543784.2020.1761323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rumi E, Trotti C, Vanni D, Casetti IC, Pietra D, Sant’Antonio E. The genetic basis of primary myelofibrosis and its clinical relevance. Int J Mol Sci. 2020;21:8885. doi: 10.3390/ijms21238885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Loscocco GG, Guglielmelli P, Vannucchi AM. Impact of mutational profile on the management of myeloproliferative neoplasms: a short review of the emerging data. OncoTargets Ther. 2020;13:12367–82. doi: 10.2147/OTT.S287944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tefferi A, Lasho TL, Finke CM, Elala Y, Hanson CA, Ketterling RP, et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1:105–11. doi: 10.1182/bloodadvances.2016000208. [DOI] [PMC free article] [PubMed] [Google Scholar]