SUMMARY
Human induced pluripotent stem cell (iPSC) technologies offer a unique resource for modeling neurological diseases. However, iPSC models are fraught with technical limitations including abnormal aggregation and inefficient maturation of differentiated neurons. These problems are in part due to the absence of synergistic cues of the native extracellular matrix (ECM). We report on the use of three artificial ECMs based on peptide amphiphile (PA) supramolecular nanofibers. All nanofibers display the laminin-derived IKVAV signal on their surface but differ in the nature of their non-bioactive domains. We find that nanofibers with greater intensity of internal supramolecular motion have enhanced bioactivity toward iPSC-derived motor and cortical neurons. Proteomic, biochemical, and functional assays reveal that highly mobile PA scaffolds caused enhanced β1-integrin pathway activation, reduced aggregation, increased arborization, and matured electrophysiological activity of neurons. Our work highlights the importance of designing biomimetic ECMs to study the development, function, and dysfunction of human neurons.
Keywords: iPSC-derived neurons, supramolecular nanofibers, peptide amphiphiles, IKVAV, supramolecular molecular motion, dynamics, neuronal maturation, extracellular matrix, laminin
eTOC Blurb
An eTOC blurb should also be included that is no longer than 50 words describing the context and significance of the findings for the broader journal readership. When writing this paragraph, please target it to non-specialists by highlighting the major conceptual point of the paper in plain language, without extensive experimental detail. The blurb must be written in the third person and refer to “Corresponding Author Last Name and colleagues.”
The utilization of iPSC technologies to model neurological diseases in vitro is challenging due to the inherent tendency of neurons to aggregate and their immature profile. Kiskinis and colleagues, developed artificial extracellular matrix biomimetic molecules exhibiting unprecedented molecular motion that promote advanced functional neuronal maturation and facilitate modeling of neurodegeneration.
Graphical Abstract
INTRODUCTION
The groundbreaking technology of induced pluripotent stem cells (iPSCs) and their differentiation into neural cells has provided unprecedented access to the human central nervous system (CNS). It has enabled the assembly of models for the investigation of neurodevelopment and neurological diseases, which have led to significant advancements in our understanding of these processes1–3. However, culturing stem cell-derived neurons in vitro remains challenging. In particular, neurons grown in cell autonomous systems exhibit insufficient levels of maturation and reduced long-term viability. To develop more physiological culture conditions, it is important to recapitulate the resident microenvironment of the nervous system. A critical and often overlooked component of this microenvironment is the extracellular matrix (ECM), which plays a pivotal role in neuronal maturation, signaling and ageing4–6.
The ECM in the CNS is an intercellular scaffold composed of glycosaminoglycans, proteoglycans, collagens, and proteins including laminin and fibronectin7–10. These components are organized in a highly dynamic fashion and vary in different regions and developmental stages of the CNS9,10. The ECM serves multiple purposes such as providing structural support and integrity, acting as a reservoir of soluble proteins including cytokines and growth factors, and mediating cellular signaling11–14. It modulates the transduction of cell surface receptors that internalize signals controlling cellular functions including migration, survival, neurite sprouting, synaptic plasticity and ageing15–20.
Current ECM approaches for culturing iPSC-derived neurons in vitro include purified or recombinant proteins such as laminin on glass or plastic-coated surfaces. Although these platforms allow initial attachment, neurons tend to form tight clusters of aggregated cells rendering single-cell resolution analyses by microscopy or patch clamp physiology unattainable, and ultimately fall short of promoting advanced maturation. Matrigel lacks CNS-specific ECM components, and contains variable quantities of cellular growth factors21,22. Primary mammalian glial cells are very effective in promoting neuronal maturation but they are derived from postnatal animals and are costly, heterogenous and exhibit high batch-to-batch variability.
A promising alternative is the use of synthetic biomaterials that can generate scaffolds with the capacity to mimic the topographical, biochemical, and mechanical properties of the ECM18,23–25. Chemically defined platforms with adaptable mechanical properties can modulate a series of cellular properties such as proliferation, differentiation, maturation, and cytoarchitecture in 2D and 3D CNS in vitro models20,26,27. Synthetic systems not only need to emulate the filamentous architecture and mechanics of natural matrices but additional features, including the hierarchical structure across scales, and the dynamics of their constituent molecules. The importance of ECM dynamics requires special consideration given the continued remodeling of neural microenvironments28, and the mobility of cell surface receptors engaged in intracellular signaling.
Supramolecular polymers in which monomeric components combine through secondary interactions rather than covalent bonds have emerged as new materials that can have both ordered structures and tunable dynamics, in contrast to ordinary hydrogels that are typically based on crosslinked covalent polymers29–33. These systems have a unique combination of order and dynamics and can integrate biological signals that trigger cellular responses such as cell adhesion, migration, proliferation and differentiation34–40. Thus, they are great candidates for designing biomimetic artificial matrices to interface with and signal to cells. The earliest example of a supramolecular biomaterial designed for bioactivity involved the self-assembly of a peptide amphiphile (PA) monomer into nanofibers and engrafted with bioactive sequences derived from ECM proteins41,42.
Here, we investigated how supramolecular PA nanofibers, displaying on their surfaces the same laminin α−1 derived sequence IKVAV43 but harboring different degrees of mobility, impact the functionality of human iPSC-derived neurons. We observed that supramolecular scaffolds with similar nanofiber architecture and chemical composition displayed remarkable enhancement of bioactivity when they exhibited more intense supramolecular motion. Neurons cultured on matrices with highly mobile PA molecules exhibited several features consistent with increased functional maturation.
RESULTS
Characterization of PA Supramolecular Nanofibers with a Laminin-Derived Peptide Sequence
In order to develop a neuronal culture platform that can recapitulate the properties of the ECM found in the adult CNS, we functionalized synthetic peptide amphiphile (PA) nanofibers with the bioactive pentapeptide sequence IKVAV that is derived from the laminin α−1 chain43. We selected laminin α−1 because we found it to be developmentally upregulated, exhibiting significantly higher levels in the adult relative to the neonatal spinal cord ECM (Figures 1A and 1B). The IKVAV sequence is known to interact with murine neural cells and promote attachment, growth, and maturation20,44–47, and is therefore a formidable candidate molecule for facilitating the maturation of iPSC-derived neurons. We utilized PAs to functionalize IKVAV on account of their ability to self-assemble into nanofiber scaffolds. We found that the IKVAV-PA showed an architectural topography similar to that of the adult spinal cord ECM after decellularization (Figure 1C).
Figure 1. Characterization of IKVAV-PA Supramolecular Nanofibers with Different Degrees of Supramolecular Motion.
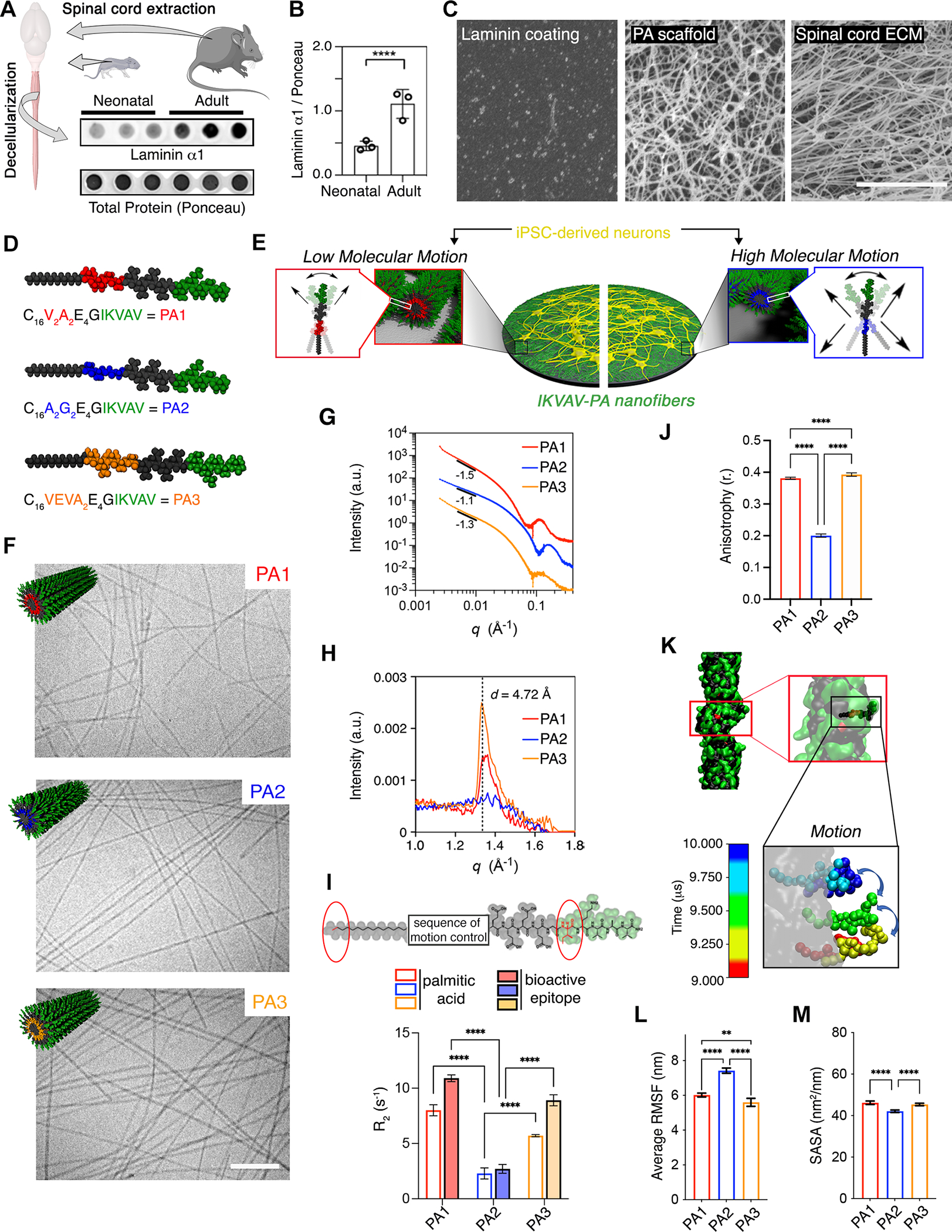
(A) Dot blot showing the protein level of laminin α1 in decellularized neonatal and adult mice spinal cords. Ponceau was used to assess total protein levels.
(B) Bar plot representing the normalized protein levels of laminin α1 in neonatal and adult mice spinal cords. Each dot represents values from independent biological replicates.
(C) SEM micrographs of laminin coating (left), PA scaffold (middle), and adult mouse decellularized spinal cord (right).
(D) Schematic representation of the chemical structure of each IKVAV-PA (V2A2= PA1, A2G2= PA2, and VEVA2= PA3).
(E) Schematic representation of hiPSC-derived neuron cultures on ECM-mimetic PA substrates with different degrees of molecular motion.
(F) Cryogenic TEM micrographs of PA1, PA2, and PA3 along with the molecular graphic representation (top, left) of each of the supramolecular fibers displaying IKVAV in green.
(G) Line graph showing the SAXS patterns of PA1, PA2, and PA3 nanofibers.
(H) Line graph showing the WAXS profiles of PA1, PA2, and PA3 nanofibers.
(I) Top: Chemical structure of IKVAV-PA sequence highlighting the first C in the aliphatic tail and I in the bioactive sequence analyzed by T2-NMR. Bottom: Bar graph showing the relaxation time for PA1, PA2 and PA3 molecules.
(J) Bar graph showing the fluorescence anisotropy (λex = 336 nm, λem = 450 nm) of PA1, PA2, and PA3 embedded 1–6-phenyl-1,3,5-hexatriene p-toluenesulfonate (DPH).
(K) Self-assembled structure of a single IKVAV-PA fiber after 10 μs of coarse-grained molecular dynamic simulation. Inset: Motion of a single peptide within the fiber during the last 1 F06Ds after equilibrating the structure. The illustrations include partial periodic images through the fiber formation axis. Water and ions are omitted for clarity.
(L) Bar graph showing the average of root-mean-square fluctuation (RMSF) values for the different IKVAV-PAs during the last 5 μs after equilibrating the fiber structures.
(M) Bar graph showing the SASA values of the IKVAV epitope displayed on the different IKVAV-PAs fibers in the equilibrated structure.
Scale bars: C=5 μm, F=250 nm. All the values are presented as the mean ± SD.
Based on our recent findings on the substantial effects of supramolecular motion in vivo48, we synthesized a small library of IKVAV-PAs where the hydrogen bonding segment was mutated within the peptide sequences: V2A2 (red), A2G2 (blue) and VEVA2 (orange), referred to as PA1, PA2, and PA3, respectively (Figure 1D). The IKVAV-PAs are composed of four main segments: (1) a hydrophobic fatty acid alkyl tail to drive aggregation of in water, (2) a peptide segment that forms intermolecular hydrogen bonds and controls the physical properties of supramolecular nanofibers, (3) a tetra-peptide of glutamic acid residues to enhance the solubility of the fibers in water, and (4) a terminal bioactive IKVAV sequence separated by a G residue to facilitate its display on the surface of nanofibers (Figure 1D). The chemical structure of the peptide segment next to the alkyl tail is a key factor affecting multiple properties of the PA nanofibers including their rigidity, the molecular dynamics within the fibers, and even their ability to enhance growth factor signaling32,48–52. The three IKVAV-PA nanofibers we designed provide us with the opportunity to interrogate the effects of supramolecular motion on neuronal responses (Figure 1E).
Using cryogenic transmission electron and scanning electron microscopy (cryo-TEM and SEM, respectively) we confirmed that the three IKVAV-PAs self-assemble into supramolecular nanofibers (Figures 1F and S1A). Synchrotron solution small-angle-x-ray scattering (SAXS) analysis confirmed the formation of high-aspect-ratio nanofibers with a slope between −1.1 and −1.5 in the Guinier region (Figure 1G). The three IKVAV-PAs form hydrogels with similar porosity and mechanical properties ranging from 1–7 kPa, which is within the range of spinal cord tissue (Figures S1B–C)53. We next characterized the secondary structure of each IKVAV-PA by solution phase synchrotron radiation wide-angle X-ray scattering (WAXS) (Figure 1H). We observed that PA1 and PA3 displayed a Bragg peak with a d-spacing of 4.72 Å, indicative of crystalline packing of the molecules in a β-sheet arrangement54. Importantly, this peak was not observed in PA2, indicating the lack of crystalline order (Figure 1H). This result was validated by circular dichroism (CD) and Fourier-transform infrared (FTIR) spectroscopy. Both revealed the presence of β-sheet secondary structure for PA1 and PA3 and random coil for PA2 (Figures S1D–E). We next measured the elastic modulus of the PA nanofibers by AFM (Figure S1F). PA1 and PA3 showed a higher Youngś modulus of 13 MPA and 20 MPa respectively, which correlates with the denser packing observed by WAXS. PA2 fibers showed a lower elastic modulus of 0.28 MPa, indicative of a more fluid nature (Figure S1F). Collectively, these results demonstrate that the A2G2 tetrapeptide segment of PA2 exists as a random coil within fibers, which may confer greater supramolecular dynamics in this matrix.
To probe the differences in supramolecular dynamics between the three IKVAV-PAs, we used transverse relaxation nuclear magnetic resonance (T2-NMR) spectroscopy (Figure 1I). These experiments analyzed the relaxation rate of the methyl protons of the palmitic acid tail and the delta carbon of the I residue in the IKVAV sequence (observed at 0.93 and 0.86 parts per million ppm, respectively) (Figure 1I, top). IKVAV-PA1 showed the highest relaxation rate consistent with a low degree of motion, while PA2 showed the lowest relaxation rate consistent with a greater degree of motion (Figure 1I, bottom). We further characterized the molecular dynamics using fluorescence depolarization (FD) by exciting encapsulated fluorophores with polarized light and then measuring the anisotropy of the emitted light (Figure 1J)55. PAs were mixed with 1–6-phenyl-1,3,5-hexatriene p-toluenesulfonate (DPH), which is known to be anchored at the hydrophilic-hydrophobic interface of amphiphilic assemblies56,57. Consistent with the T2-NMR results, PA1 and PA3 nanofibers displayed an anisotropy value of 0.39 and 0.38, while PA2 displayed a significantly lower value of 0.24 (Figure 1J), confirming the higher mobility within the PA2 nanofibers.
We next used coarse-grained molecular dynamic simulations to analyze the internal molecular dynamics of the three IKVAV-PA nanofibers58–64. The extent of molecular mobility, which is defined as the spatial fluctuations within the equilibrated filaments, was found to be significantly greater in PA2 relative to PA1 and PA3 filaments (Figures 1K–L and S1G–H). Furthermore, PA2 assemblies had a less ordered spatial localization of the amphiphilic molecules into a core-shell structure, increasing the extent of water contact within the core of the fiber, and reducing the presentation of IKVAV on the surface (solvent accessible surface area -SASA- values in PA2: 42.2% ± 0.8 vs 45.7% ± 0.4 and 45.6% ± 0.2 in PA1 and PA3, respectively) (Figures 1M, S1I–J). However, the bending of the fibers was similar in the three IKVAV-PAs (PA1: 5.3%±3.2, PA2: 3.9%± 2.6, and PA3: 8.3%± 3.7) (Figures S1K–M). Collectively, the experiments and simulations suggest that PA2 molecules within the supramolecular filaments are significantly more fluid and hydrated, and consequently more mobile than those in PA1 and PA3.
The Highly Mobile IKVAV-PA2 Activates β1-Integrin Associated Intracellular Pathways with High Efficiency
We next investigated the efficiency of the three laminin-derived IKVAV-PA scaffolds to transduce laminin-related responses onto human iPSC (hiPSC)-derived motor neurons (MNs), which are the main neuronal subtypes of the spinal cord. On day 14 of the differentiation protocol65, MN-enriched cultures were re-plated on the three different IKVAV-PA coatings (Figures 2A and S2A–D). Since IKVAV is known to bind to the transmembrane receptor β1-INTEGRIN (ITGB1)20,66–69, we first investigated the ability of the three IKVAV-PA nanofibers to recruit ITGB1. As a control, we used a standard coating of commercial laminin 111 (technical details in Methods section). Structured illumination microscopy (SIM) showed that MNs cultured for 3 days on the highly mobile PA2 exhibited significantly higher levels of ITGB1 on their processes, relative to the ones cultured on PA1, PA3, or natural laminin (Figures 2B–C). To ask whether adhesion to the PA coatings was exclusively mediated by an IKVAV-ITGB1 interaction, we pre-treated MN cultures with an ITGB1 or an ITGB4 antibody (the other β integrin highly expressed in MNs). We found that treatment with ITGB1 completely blocked cell attachment, while ITGB4 had no effect (Figures 2D–F and S2E). To assess the specificity of the IKVAV pentapeptide displayed on the PA nanofibers, we seeded MNs on PAs with a scrambled VVIAK sequence (scr-PA1, scr-PA2 and scr-PA3) and found very low levels of MN attachment on these coatings relative to the IKVAV-PAs or laminin (Figures S2F–G). To assess the significance of the PA nanofibers for IKVAV bioactivity, we immobilized the IKVAV pentapeptide sequence on glass alone (using APTES as a control) and found that while it outperformed the scrambled PA conditions, it caused significantly lower levels of cell attachment relative to the IKVAV-PAs or laminin (Figures S2F–G).
Figure 2. Highly Mobile IKVAV-PA Supramolecular Scaffold Enhances Integrin-Dependent Effects on hiPSC-Derived MNs.
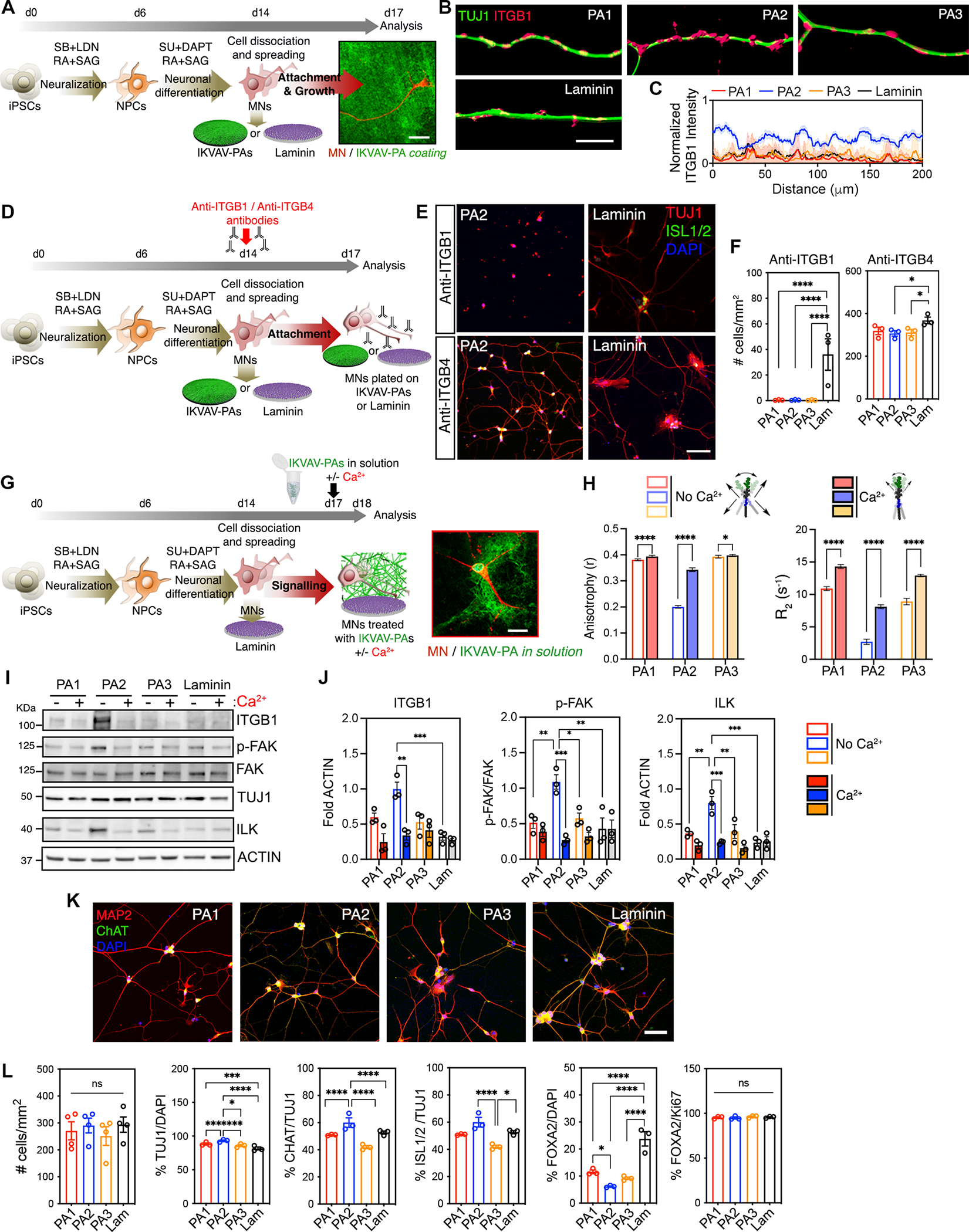
(A) Schematic representation of the experimental workflow for short-term culture experiments with hiPSC-derived MNs on IKVAV-PA scaffolds or laminin. Right: structured illumination microscopy micrograph of an hiPSC-derived MN labeled with TUJ1 (red) after 72h cultured on PA2 covalently linked to alexa-488 dye (green).
(B) Representative 3D SIM shadow reconstructions of neurites of MNs cultured on IKVAV-PAs or laminin, immunolabeled for TUJ1 (green) and ITGB1 (pink).
(C) Line graph showing intensity analysis of ITGB1 labeling on neurites of MNs cultured on IKVAV-PAs orlaminin for 72 h.
(D) Schematic representation of the experimental workflow for short-term experiments with hiPSC-derived MNs cultured on IKVAV-PA scaffolds or laminin and treated with anti-ITGB1 or anti-ITGB4 antibodies.
(E) Representative confocal micrographs of MNs cultured on different IKVAV-PA fibers or laminin and treated with anti-ITGB1- or anti-ITGB4-blocking antibodies for 72 h. Cells were stained with neuronal (TUJ1, red) and MN (ISL1/2, green) markers, and nuclei were stained with DAPI (blue).
(F) Bar graphs representing the number of cells per mm2 on hiPSC-derived MN cultures after 72 h on IKVAV-PAs or laminin and treated with anti-ITGB1 or anti-ITGB4 antibodies.
(G) Schematic representation of the experimental workflow of hiPSC-derived MNs cultured on laminin coatings and IKVAV-PAs in solution with or without calcium treatment. Right: Confocal microscopy micrograph of a MN labeled with MAP2 (red) after 24 h treatment with alexa-488-labeled PA2 (green) in solution.
(H) Left: Bar graph showing fluorescence anisotropy of PA1, PA2, and PA3 with DPH in the presence or absence of calcium. Error bars represent to 3 independent experiments. Right: Bar graph of Isoleucine (I) relaxation time of the IKVAV-PAs in the presence or absence of calcium. Error bars correspond to 3 independent runs per condition.
(I) Representative WB of ITGB1 and downstream kinases of the integrin-transduction pathway (ILK, p-FAK, FAK) in MNs treated with IKVAV-PAs in the presence or absence of calcium for 72 h. TUJ1 was used to assess the neuronal-specific protein levels across conditions and ACTIN as a loading control. Laminin condition refers to MN cultures on laminin coating and non-treated with PAs in solution.
(J) Bar graphs representing the normalized protein levels of ITGB1, p-FAK, and ILK in MN cultures with IKVAV-PAs in solution with or without calcium treatment for 72 h.
(K) Representative confocal micrographs of hiPSC-derived MN cultures on IKVAV-PAs and laminin coatings stained for neuronal (MAP2, red) and MN (ChAT, green) markers. Nuclei stained with DAPI (blue).
(L) Bar graphs representing the number of cells per mm2, the percentage of TUJ1/DAPI, ChAT/TUJ1, ISL1/2/TUJ1, FOXA2/DAPI and FOXA2/Ki67 labeled cells in hiPSC-derived MN cultures on IKVAV-PAs and laminin coatings at 72 h.
Data was obtained from at least 3 independent differentiations. All values are presented as the mean ± SEM except for (J) where the values are presented as the mean ± SD. Each dot in graphs represents average values of multiple fields from a specific differentiation. Scale bars: A= 20 μm, B= 10 μm, E and K=100 μm, G= 25 μm.
Using western blot (WB) analysis we confirmed that MNs exposed to IKVAV-PA2, either as a coating or in solution, expressed significantly higher levels of ITGB1 protein, as well as integrin-linked kinase (ILK) and focal adhesion kinase (FAK) phosphorylation, two key signaling scaffold proteins that act downstream of ITGB1 and intersect many intracellular pathways in response to integrin-dependent ECM stimuli (Figures S2H–I). To interrogate the correlation between supramolecular motion and ITGB1 activation, we used 5 mM CaCl2, which is known to electrostatically crosslink negatively charged PA fibers48,70,71 (Figure 2G). Addition of Ca2+ ions to PA solutions caused a substantial decrease in supramolecular motion as measured by FD and T2-NMR (Figure 2H). Consistent with these results, addition of CaCl2 to MN cultures grown with IKVAV-PA2 in solution caused a substantial decrease in the activation of ITGB1 and its downstream intracellular pathway (ILK, pFAK/FAK) (Figures 2I–J). These results show a strong positive correlation between dynamics and bioactivity.
To assess any potential effects of the PAs on the efficiency of terminal MN differentiation, we used immunocytochemistry (ICC) to quantify the percentage of neural markers (Figures 2K–L). Cultures plated for 72 h on the three IKVAV-PAs or laminin coatings showed similar levels of cell attachment and survival (Figures 2L and S2J). However, the three IKVAV-PA coatings showed a higher percentage of β-III tubulin positive (TUJ1+) neurons (PA1=88%±1.2, PA2=93%±1.4, PA3=86%±1.5), compared to ones plated on laminin-coated coverslips (81%±1.3) (Figure 2L). Notably, plating on the IKVAV-PA2 nanofibers, which exhibit high supramolecular motion, led to a significantly higher percentage of ChAT+ and ISL1/2+ postmitotic MNs (71%±1.6 and 60%±1.7) compared to IKVAV-PA1 (59%±2.1, 51%±1.6), IKVAV-PA3 (57%±1.4, 42%±1.3) or laminin (62%±1.8, 53%±1.3) (Figures 2K–L). Conversely, plating on the PAs resulted in a decreased percentage of proliferating Ki67+/FOXA2+ floor plate progenitor cells that as we have previously reported65, often contaminate this differentiation protocol (Figures 2K–L). Critically, the PAs did not affect the proliferation capacity of FOXA2+ cells or cause any preferential specification towards GABAergic interneurons (Figures 2L, S2C–D). These results show that IKVAV-PA2 nanofibers improve neuronal purity of differentiated hiPSC cultures by mediating selective MN attachment and specification. Lastly, we performed live imaging analysis to assess the effects of the different IKVAV-PA substrates on cell motility, given the well-described role of the laminin-ITGB1 pathway on neuronal migration43,72–75. We found that hiPSC-derived MNs plated on the highly mobile IKVAV-PA2 coating exhibited higher velocity and distance of migration compared to MNs cultured on the more static IKVAV-PA1 and PA3, or laminin coatings (Figures S2K–L, Movies S1–2).
Proteomic Profiling and Morphological Characterization of hiPSC-Derived MNs Reveals Substantial Effects of IKVAV-PA2 Nanofibers on Maturation
We next assessed the effects of the IKVAV-PAs by plating MNs on these coating platforms for a prolonged time of up to 61 days in vitro (Figure 3A). Profilometry analysis showed that while all four coatings had an initial surface thickness of 200nm, this was well preserved only for the three IKVAV-PAs and dramatically reduced in the case of natural laminin at 60 days in vitro (Figures 3B–C and S3A–B). Accordingly, the higher levels of ITGB1 and ILK in MN cultures grown on the highly mobile PA2 nanofiber coatings were sustained even after 45 days in culture (Figures 3D–E).
Figure 3. Unbiased Characterization of the Effects of IKVAV-PA Nanofibers with High Supramolecular Motion on Long-Term hiPSC-Derived MN Cultures.
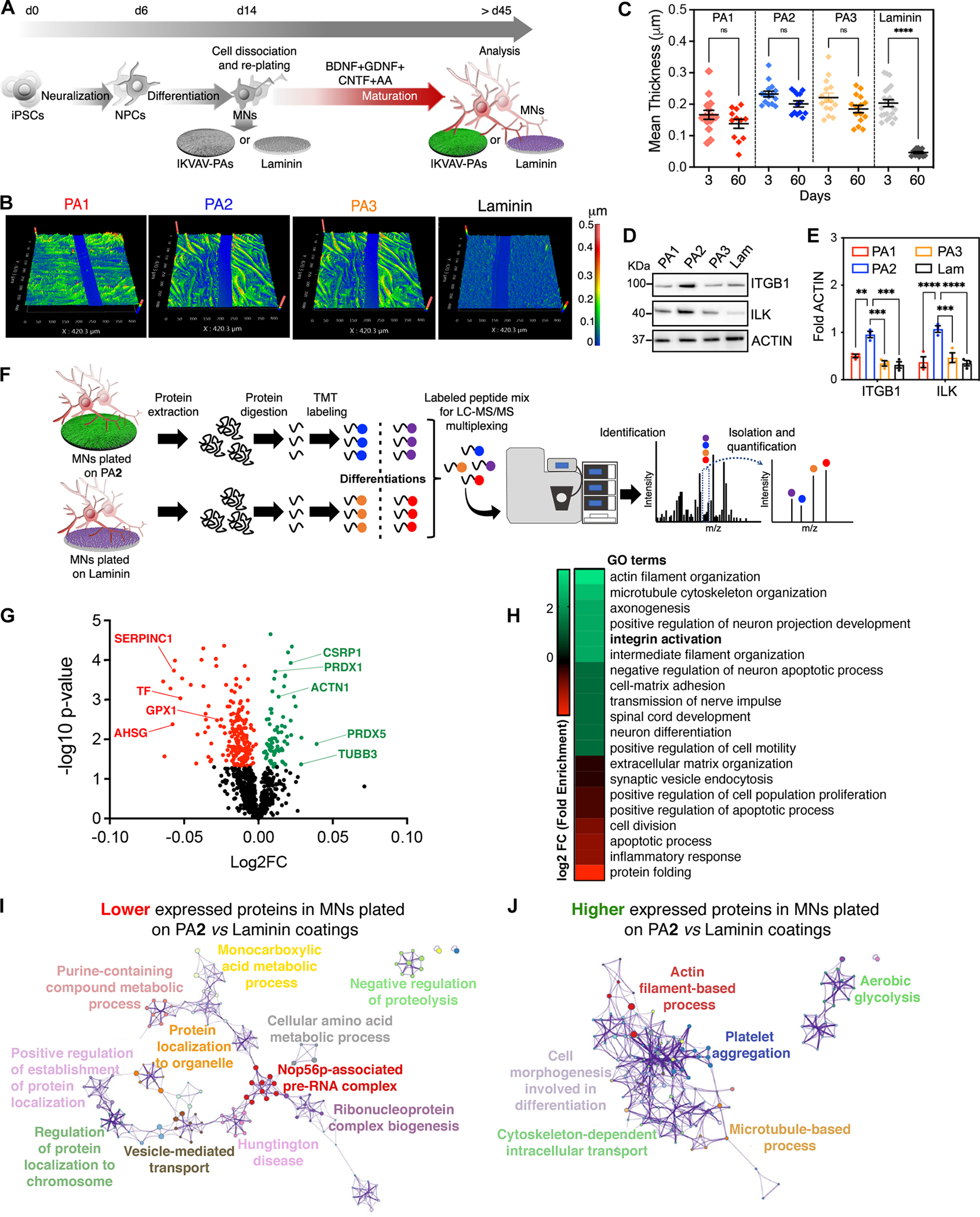
(A) Schematic representation of the experimental workflow for long-term culture experiments with hiPSC-derived MNs on IKVAV-PA scaffolds or laminin.
(B) Profilometry images of IKVAV-PAs and laminin coatings after 60 days in vitro.
(C) Dot plot representing the thickness of the different IKVAV-PA and laminin coatings after 3 and 60 days in vitro. Each dot represents values from independent coverslips.
(D) Representative WB of ITGB1 and ILK in MNs cultured on the different IKVAV-PAs or laminin for 45 days. ACTIN was used as a loading control.
(E) Bar graphs representing the normalized protein levels of ITGB and ILK in MNs cultured on different IKVAV-PAs or laminin for 45 days. Each dot represents the values from independent differentiations.
(F) Schematic representation of the quantitative proteomic analysis carried out in MNs cultured on PA2 or laminin for 45 days.
(G) Volcano plot displaying proteomic changes in hiPSC-derived MNs cultured on PA2 vs. laminin. Averages of log2 fold change (x-axis) vs. −log10 P-value (y-axis) values of each protein are represented by individual dots. Proteins up- and down-regulated by 2-fold and FDR < 0.05 are labeled with green and red dots, respectively.
(H) Subset of the most significant comparative GO terms enriched in the up-regulated (green) and down-regulated (red) group of proteins identified in hiPSC-derived MNs cultured on PA2 vs. laminin.
(I) Cytoscape pathway enrichment analysis of down-regulated proteins in hiPSC-derived MNs cultured on PA2 vs. laminin.
(J) Cytoscape pathway enrichment analysis of up-regulated proteins in hiPSC-derived MNs cultured on PA2 vs. laminin.
The internalization of the ITGB1 signaling cascade is known to have critical functional ramifications, although their exact nature in human neurons remains unclear. To uncover these effects, we used quantitative mass spectrometry (MS) and analyzed MN samples grown on PA2 or commercial laminin for 45 days in vitro (Figure 3F). We identified 892 proteins, with normalized intensity differences between the two conditions in 30.3% of them (Figures 3G and Table S1). To contextualize these changes, we performed comparative gene ontology (GO) and pathway enrichment analyses for the down-regulated (196) and up-regulated (76) groups of proteins (Figures 3H–J). The most significant term differences between the two groups were observed in biological processes associated with “cell-matrix adhesion”, “actin and microtubule organization”, “regulation of cell motility”, “neuron differentiation and projection”, “synapse organization”, and “apoptotic process” (Figures 3H–J and S3C). Interestingly, most of these processes can be modulated by the activation of “integrin pathways” (reviewed in76–80), a GO term that was strongly enriched in the group of upregulated proteins in MN cultured on IKVAV-PA2 coatings (Figure 3H). Among the most upregulated proteins in MNs cultured on PA2 were PRDX1 and PRDX5, which are cytoprotective81, the cytoskeletal protein ACTIN-1 that is involved in adhesion and motility82, and the cysteine and glycine-rich protein CSRP1 implicated in spinal cord regeneration83 (Figure 3G).
We next performed a systematic characterization of MNs cultured on the PAs to determine if the proteomic changes we identified translated to corresponding morphological and functional outcomes (Figure 4A). The interaction of MNs with all three IKVAV-PAs led to increased cell survival relative to laminin (Figure 4B). In accordance with the induction of cytoskeletal organization and axogenesis pathways, microfluidic-based axon sprouting assays and morphometric analysis showed that MNs cultured on the mobile PA2 nanofiber coatings displayed larger somas, a higher number of processes that were longer, and increased complexity of neuronal branching relative to other conditions (Figures 4C–J and S4A). This shift towards more elaborate morphologies was denoted by a distinctly more uniform spread and less clumping of neurons across the surfaces coated with the three IKVAV-PAs (Figure 4K). We quantitated this observation by confocal imaging of immunolabeled 60-day-old MNs as a readout of cell-cell and cell-substrate interactions (Figure 4L). We found that MNs plated on PA2 nanofiber coatings outperformed a range of commercial coatings including Matrigel, recombinant fibronectin and different types of commercial laminins, in terms of homogenous distribution as well as survival and ITGB1 protein expression levels (Figures S4B–F). This resolves a long-standing technical limitation of culturing hiPSC-derived neurons in vitro, which typically form large clusters of inaccessible cells.
Figure 4. The Effect of IKVAV-PA Nanofibers with High Supramolecular Motion on Long-Term Survival, Adhesion and Neuronal Morphogenesis on hiPSC-Derived MN Cultures.
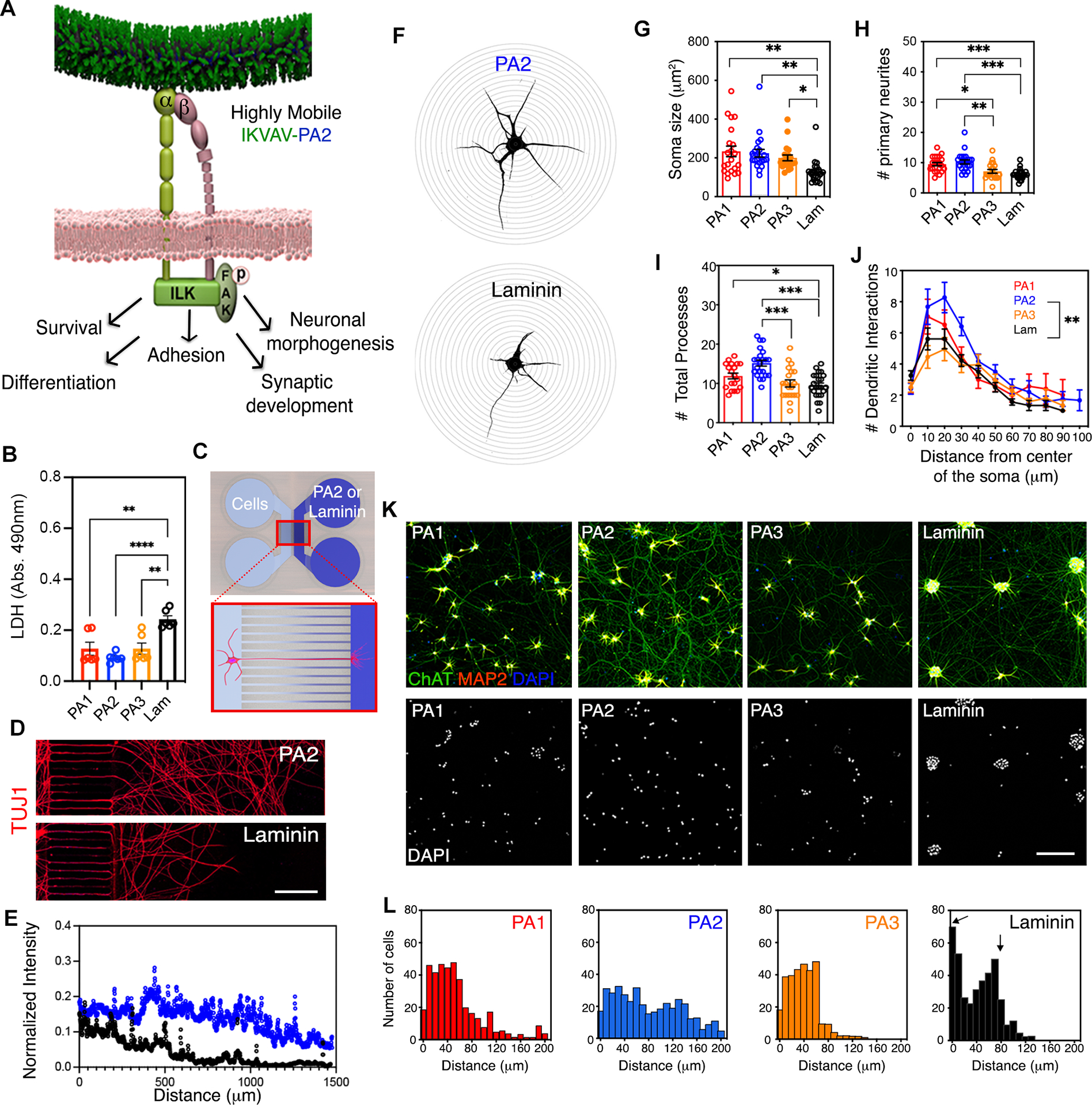
(A) Schematic representation of integrin signal transduction pathways that might mediate cellular behavior changes upon highly mobile presentation of IKVAV by the PA2.
(B) Bar graph representing LDH levels in hiPSC-derived MN cultures after 45 days on PA1, PA2, PA3 and laminin.
(C) Schematic representation of a microfluidic device system utilized to measure axonal growth on PA2 and laminin.
(D) Representative confocal micrographs of hiPSC-derived MN neurites along microchannels treated with PA2 or laminin after 30 days in culture. Neuronal processes were stained with TUJ1.
(E) Line graph displaying the fluorescence intensities of TUJ1+ neurite staining in hiPSC-derived MN cultures grown in microfluidic devices treated with PA2 or commercial laminin.
(F) Representative confocal micrographs of hiPSC-derived MNs on PA2 (top) or laminin (bottom) used for Sholl analysis.
(G) Bar graph depicting the soma size in hiPSC-derived MNs cultured on IKVAV-PAs or laminin for >45 days.
(H) Bar graph depicting the number of primary neurites in hiPSC-derived MNs cultured on IKVAV-PAs or laminin for >45 days.
(I) Bar graph depicting the number of total neurite processes in hiPSC-derived MNs cultured on IKVAV-PAs or laminin for >45 days.
(J) Line graph depicting Sholl analysis of dendritic arborization of MNs cultured on the various coatings for >45 days in vitro.
(K) Representative confocal micrographs of hiPSC-derived MN cultures plated on IKVAV-PAs or laminin for >45 days. Cells were stained with neuronal (MAP2, red) and MN (ChAT, green) markers (top row). Nuclei were stained with DAPI (bottom row).
(L) Histogram analysis of cell distribution on the different coatings referred on K.
Data was obtained from at least 3 independent differentiations. All values are presented as the mean ± SEM; Each dot in graphs represents average values of multiple fields from a specific differentiation in B and individual cells in G-I. Scale bars: D=25 μm and E=100 μm.
The Highly Mobile PA2 Facilitates Advanced Functional Maturation of hiPSC-Derived MNs
The proteome and morphology of MNs suggest that PA2 nanofibers enhance their functional maturation. To substantiate these findings we next used biochemical, imaging, and electrophysiological approaches to characterize day 30–60 MNs. First, we quantified the amount of post-synaptic (PSD95) and pre-synaptic (Synapsin, SYN1, and Synaptophysin, SYP) markers. MNs grown on the dynamic PA2 coating expressed significantly higher levels of PSD95, SYN1 and SYP relative to MNs grown on PA1, PA3, and laminin (Figures 5A–B). SIM of pre- and post-synaptic terminals showed higher fluorescence intensity levels of PSD95 and SYN1 for MNs plated on PA2, demonstrating that that these proteins were properly distributed along neuronal processes (Figures 5C–D and S5A). To determine whether the increased expression of synaptic markers corresponded to the functional enhancement of synaptic connections, we next used whole cell manual patch clamp to record spontaneous excitatory postsynaptic currents (sEPSC) in MNs grown on PA2 and laminin coatings. We first categorized neurons into three groups defined by the maximum sEPSC amplitude in each case. We found that while 96% of MNs grown on PA2 displayed medium and large sEPSC events (categorized as >100pA), this percentage was reduced to 63% for MNs cultured on laminin, despite cells being held at the same voltage in all conditions (Figure 5E–F). Additionally, while there were no significant differences in mean values (Figure S5B), cumulative distribution analysis revealed that the sEPSC events recorded in MNs grown on PA2 were significantly larger in amplitude (pA) and occurred with significantly shorter inter-event intervals (IEI; ms) compared to MNs cultured on laminin (Figures 5G–I). These results may reflect synaptic enhancement through both pre-synaptic and post-synaptic mechanisms.
Figure 5. The Effect of Highly Mobile IKVAV-PA2 on Functional MN Maturation.
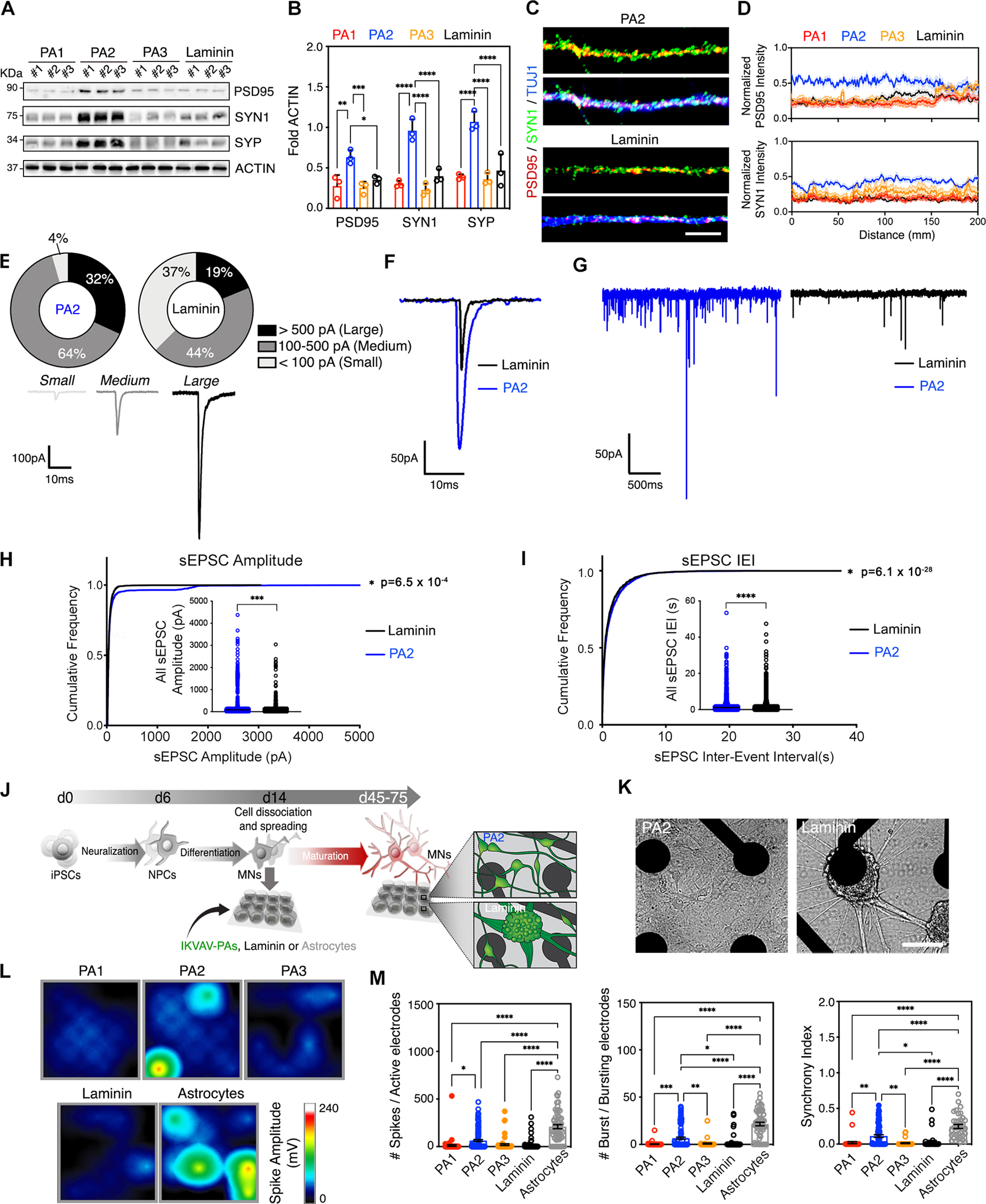
(A) Representative WB of pre-synaptic (SYN1 and SYP) and post-synaptic (PSD95) markers in hiPSC-derived MNs cultured on IKVAV-PAs or laminin at day 60. ACTIN was used as a loading control.
(B) Bar graphs representing normalized protein levels of PSD95, SYN1 and SYP in hiPSC-derived MNs cultured on supramolecular IKVAV-PAs or on laminin at day 60.
(C) Representative SIM micrographs of pre- and post-synaptic terminals (labeled with SYN1 and PS95, respectively) distributed along the neurites of MNs cultured on IKVAV-PA2 and laminin for > 45 days.
(D) Line graph displaying the analysis of fluorescence intensity of PSD95 and SYN1 along neuronal processes of MNs cultured on IKVAV-PAs and laminin.
(E) Top: Pie chart indicating the percentage of hiPSC-derived MNs cultured on PA2 or laminin for >30 days that generated large (>500 pA), medium (100–500 pA) or small (<100 pA) sEPC. n(PA2)=22 cells; n(laminin)=16 cells. Bottom: Representative traces of small, medium, and large sEPC. Analysis performed from contingency table showed statistical significance (p=0.014) between the two experimental conditions based on Chi-square (and Fisher’s exact) test.
(F) Representative sEPC traces from human MNs cultured on PA2 or laminin for 30 days.
(G) Expanded time scale magnification of sEPC traces from hiPSC-derived MNs cultured on PA2 or laminin for 30 days.
(H) Line graph displaying the cumulative frequency of sEPSC amplitude in hiPSC-derived MNs cultured on PA2 or laminin. Inside: Dot plot showing all recorded sEPSC amplitudes from multiple hiPSC-derived MNs cultured on PA2 or laminin.
(I) Line graph displaying the cumulative frequency of inter-event interval (IEI) of sEPSCs in human MNs cultured on PA2 or laminin. Inside: Dot plot showing all recorded IEI values from multiple hiPSC-derived MNs cultured on PA2 or laminin.
(J) Schematic representation of the experimental paradigm for continuous electrical activity recordings through a microelectrode array (MEA) system. hiPSC-derived MNs were cultured on MEA plates coated with IKVAV-PAs or laminin, or co-cultured with mouse astrocytes. On the right, drawings depicting the differential cell aggregation observed between cultures grown on IKVAV-PA2 or laminin.
(K) Bright field images of MNs cultured on an MEA plate coated with IKVAV-PA2 or laminin at day 40.
(L) Activity map of single MEA wells with hiPSC-derived MNs grown for >20 days on IKVAV-PAs, laminin or astrocytic monolayers.
(M) Bar graphs showing the differences in number of spikes per active electrode, number of bursts per active electrode, and the synchrony index of hiPSC-derived MNs cultured on IKVAV-PAs, laminin or astrocytic monolayers.
Data was obtained from at least 3 independent differentiations. All values are presented as the mean ± SEM. Each dot in graphs represents values from independent differentiation in B, of individual cells in H, I, and of individual wells in M. Scale bars: C= 20 μm; K=100 μm.
Using patch clamp, we tracked the physiological development of MNs, marked by the progressive ability to fire repetitive action potentials and the production of larger amplitude spikes84,85. After >30 days on PA2 coatings, 100% of MNs were capable of firing repetitively, while only 61% of the MNs did so when cultured on laminin (Figures S5C–D). Moreover, the action potentials of MNs on PA2 were larger in amplitude with faster rates of rise and fall (Figure S5E, Table S2), partially resembling the physiological properties of MNs co-cultured with glial cells65.
Next, we evaluated if the inherent electrophysiological changes at single cell level affected neuronal network activity by using multi-electrode arrays (MEAs), which allow for the recording of spontaneous electrical activity of cellular populations (Figures 5J–L and S5F). MNs cultured on PA2 exhibited an increased number of active electrodes compared to PA1, PA3 or laminin cultures (Figure S5G), in line with a more homogenous distribution of cells. Moreover, MN cultures grown on PA2 exhibited a higher number of action potential spikes and bursts per active electrode, as well as higher synchronized activity compared to MNs grown on the other IKVAV-PAs or laminin (Figures 5L–M). To put these effects into better context, we compared the activity of MNs plated on PA2 with ones plated on primary cortical glial cells, that are known to promote the maturation of human neurons in vitro86–88. We found that while PA2 outperformed laminin and in most cases PA1 and PA3, MNs on PA2 lagged in many metrics relative to ones on glial cells, suggesting that IKVAV-PA2 alone is not sufficient in replacing astrocytic monolayers (Figures 5M, S5G). Collectively, these experiments demonstrate that the highly mobile IKVAV-PA2 nanofibers induce more advanced synaptic and electrophysiological maturation of MNs relative to other non-cell-based ECM platforms.
The Response of hiPSC-Derived MNs to the Mobile IKVAV-PA2 is Dependent on ITGB1
To directly interrogate the contribution of IKVAV-ITGB1 signaling on PA2-dependent neuronal behavior changes, we next manipulated the expression of ITGB1 in MNs cultured on different coatings (Figure 6A). First, we knocked-down ITGB1 using small interfering RNA (siRNA-ITGB1) in MNs grown on IKVAV-PA2 scaffold coatings and observed clusters of aggregated neurons like the aggregation of MNs grown on laminin, while ones treated with a scramble siRNA control (siRNA-scr) were unaffected (Figures 6B–C). Knock-down of ITGB1 dramatically reduced the levels of intracellular kinases p-FAK and ILK, as well as the synaptic proteins PSD95 and SYP (Figures 6D–E). In contrast, lentiviral overexpression of ITGB1 in MNs cultured on laminin or on the less mobile IKVAV-PA1 scaffold coating reduced neuronal aggregation and promoted homogenous cellular distribution throughout the plate (Figures 6F–G and S6A–D). Moreover, it caused higher expression levels of synaptic proteins (Figures 6H–J and S6A–D). Lastly, we tested whether the beneficial effects of IKVAV-PA2 were consistent across different hiPSC-lines or other neuronal differentiation protocols. Indeed, we found that MNs derived from the hiPSC-11a line as well as cortical excitatory neurons, differentiated through a modified NGN2 overexpression protocol89,90, expressed higher levels of the ITGB1 receptor, exhibited reduced aggregation, increased survival, and elevated synaptic protein expression, when grown on IKVAV-PA2 scaffolds, relative to neurons cultured on PA1, PA3, and commercial laminin coatings (11a-MNs, Figures S6E–K; Cortical neurons, Figures S7A–E).
Figure 6. The modulation of ITGB1 expression by the highly mobile IKVAV-PA2 triggers changes in cellular behavior.
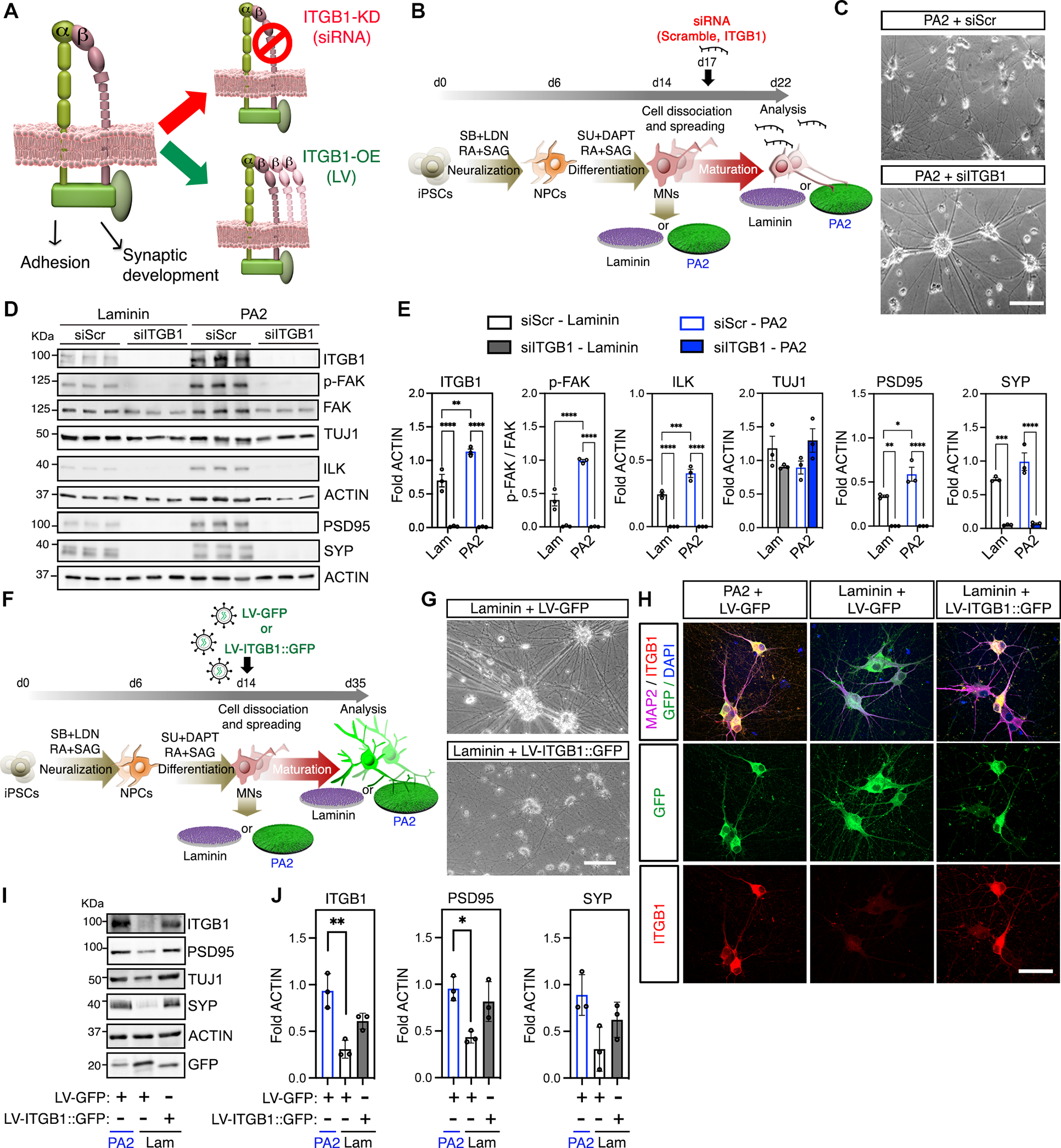
(A) Schematic representation of ITGB1 knock down (KD) and overexpression (LV) strategies to assess the importance of integrin-dependent effects on hiPSC-derived MNs cultured on IKVAV-PA2 or laminin.
(B) Schematic representation of the experimental workflow with hiPSC-derived MNs cultured on IKVAV-PA2 or laminin and transfected with scrambled (siScr) or ITGB1 (siITGB1) short interfering RNAs (siRNAs).
(C) Representative bright field images of hiPSC-derived MNs cultured on IKVAV-PA2 and transfected with scrambled (siScr) or ITGB1 (siITGB1) siRNAs.
(D) Representative WB of ITGB1, key downstream kinases of the integrin-transduction pathways (ILK, p-FAK, FAK) and synaptic proteins (PSD95, SYP) from MNs cultured on IKVAV-PA2 or laminin and transfected with scrambled (siScr) or ITGB1 (siITGB1) siRNAs. TUJ1 was used as neuronal marker and ACTIN as a loading control.
(E) Bar graphs representing the normalized protein levels of ITGB1, p-FAK, ILK, TUJ1, PSD95 and SYP in human MNs cultured on IKVAV-PA2 or laminin and transfected with scrambled (siScr) or ITGB1 (siITGB1) siRNAs.
(F) Schematic representation of the experimental workflow with hiPSC-derived MNs cultured on IKVAV-PA2 or laminin and transduced with GFP alone (LV-GFP) or ITGB1-GFP (LV-ITGB1::GFP) lentiviruses.
(G) Representative bright field images of MN cultures grown on commercial laminin and transduced with LV-GFP or LV-ITGB1-GFP.
(H) Representative confocal images of hiPSC-derived MNs cultured on IKVAV-PA2 or commercial laminin and transduced with LV-GFP or LV-ITGB1-GFP. Cells were labeled with GFP (green) and immunostained for ITGB1 (red) and the neuronal marker TUJ1 (magenta). Nuclei were labeled with DAPI (blue).
(I) Representative WB of ITGB1, TUJ1 and synaptic proteins (PSD95, SYP) from MN cultures on IKVAV-PA2 or laminin and transduced with LV-GFP or LV-ITGB1-GFP. GFP was used to determine transduction efficiencies and ACTIN as a loading control.
(J) Bar graphs representing normalized protein levels of ITGB1, PSD95 and SYP in human MNs cultured on IKVAV-PA2 or laminin and transduced with LV-GFP or LV-ITGB1-GFP.
Data was obtained from at least 3 independent differentiations. All values are presented as the mean ± SEM. Each dot in graphs represents values from independent differentiations. Scale bars: C=100μm; G= 100μm; H=50 μm.
Culturing Mutant ALS-SOD1 hiPSC-Derived MNs on IKVAV-PA2 Facilitates the Formation of Pathological SOD1 Aggregations
The advanced morphological and functional maturation of hiPSC-derived MNs grown on IKVAV-PA2 scaffolds, coupled with the ability to maintain isolated MNs for an extended timeline in vitro without the formation of cellular clusters, prompted us to investigate whether this ECM platform can facilitate the modeling of neurodegenerative pathology. We focused on the adult-onset neurodegenerative disease amyotrophic lateral sclerosis (ALS), which is characterized by the selective degeneration of spinal MNs, which often accumulate protein aggregates91. We specifically used a well-characterized model of an ALS patient cell line harboring a heterozygous dominant missense alanine-to-valine mutation at codon 4 (A4V) in the Cu/Zn superoxide dismutase (SOD1) gene and an isogenic-corrected control cell line92–94. We asked whether growing mutant SOD1 MNs on IKVAV-PA2, which enhances MN maturation without the trophic support of glial feeder layers, might reveal neuropathological aggregation (Figure 7A). We found that 2-month-old mutant (SOD1+/4AV) but not isogenic control (SOD1+/+) MNs, exhibited microscopically visible cytoplasmic SOD1 aggregation, as well as ubiquitin protein inclusions reminiscent of pathology in postmortem ALS-SOD1 tissue (Figures 7B–E). Critically, these protein aggregates were found in a small percent of MNs in both the IKVAV-PA2 and laminin coating conditions, although there was a significant increase in the proportion of MNs with inclusions in the presence of IKVAV-PA2 (PA2: 2.11%± 0.24 Laminin: 0.86%± 0.14 for SOD1 and PA2: 4.31%± 0.32 Laminin: 1.86% ± 0.18 for UB).
Figure 7. Highly Mobile IKVAV-PA2 Supramolecular Scaffold Enhances Protein Inclusion Phenotypes in ALS-SOD1 patient-derived MNs.
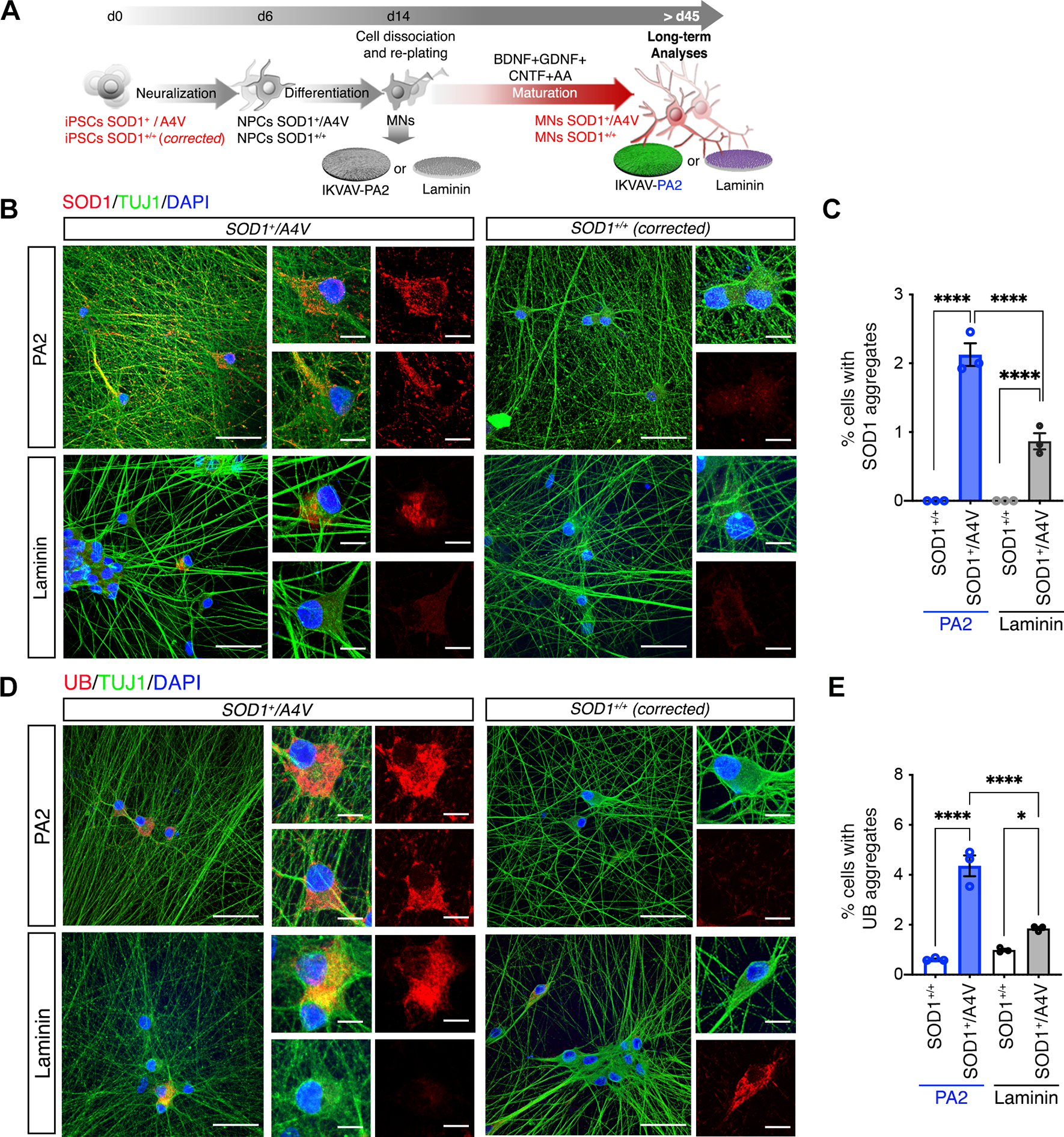
(A) Schematic representation of the experimental workflow with ALS-SOD1 MNs, differentiated from SOD1A4V/+ and isogenic control (SOD1+/+) iPSC lines cultured on IKVAV-PA2 or laminin.
(B) Representative confocal images of mutant SOD1 and control MNs cultured on IKVAV-PA2 or laminin, and immunolabeled for SOD1 (red) and TUJ1 (green). Nuclei were labeled with DAPI (blue).
(C) Bar graph representing the percentage of mutant SOD1 and control MNs exhibiting SOD1+ aggregates cultured on IKVAV-PAs or laminin for > 40 days.
(D) Representative confocal images of mutant SOD1 and control MNs cultured on IKVAV-PA2 or laminin, and immunolabeled for Ubiquitin (UB, red) and TUJ1 (green). Nuclei were labeled with DAPI (blue).
(E) Bar graph representing the percentage of mutant SOD1 and control MNs exhibiting Ubiquitin (UB)+ aggregates when cultured on IKVAV-PAs or laminin for > 40 days.
Data was obtained from at least 3 independent differentiations. All values are presented as the mean ± SEM. Each dot in graphs represents values from independent differentiations. Scale bars: B, D=50 μm; B, D insets=10 μm.
DISCUSSION
Pluripotent stem cells are widely used to generate the multitude of neural cells affected by neurological diseases. While differentiation protocols for specialized cells of the CNS are continuously being refined, most hiPSC-derived neurons are functionally immature and challenging to maintain long-term1–3. We developed an ECM mimetic platform based on scaffolds of supramolecular nanoscale fibrils formed by PA molecules to address these limitations. We designed supramolecular polymers, which displayed on their surface the bioactive laminin-derived IKVAV epitope with high molecular motion. The mobile PA2 scaffolds were efficient at activating cell-matrix-associated pathways and had profound effects on cultured neurons compared to low-mobility PA1 and PA3 scaffolds with the same IKVAV epitope (Movies S3 and S4). Our work demonstrates the importance of incorporating dynamically controllable features into synthetic ECM scaffolds that can provide significant improvements to stem cell-based neuronal models.
The enhancement of supramolecular motion within fibrils of IKVAV-PA2 scaffolds and its remarkable impact on hiPSC-neurons represents a breakthrough in the molecular engineering of artificial ECMs. It’s an opportunity that has emerged from the development of supramolecular biomaterials initiated with bioactive, artificial PA matrices31,33,41,42, and rooted in the non-covalent connection among monomeric components in these systems, a property that is in stark contrast to conventional macromolecules. Even in extensive recent reviews on artificial matrices, this feature has not been recognized as an opportunity to mimic the dynamic nature of natural ECMs95. The type of dynamic behavior demonstrated here is related to the varying degrees of motion that ECM molecules may have at the nanoscale level, and its impact on the way they interact with cell receptors potentially affecting multiple cellular functions15–19.
Our experimental data illustrates that the main difference between the most bioactive and less bioactive PAs is the internal order of the molecules within the nanofiber. PA2 molecules have less cohesive interactions and more fluid-like behavior within the nanofiber assembly similar to amphiphilic lipids in cell membranes96,97. Our results suggest an important role for motion in cell signaling beyond simple conformational changes of individual molecules. This dynamism is also present in lipids and proteins within cell membranes, where lipid raft mobility49 and receptor clustering98,99 can be directly influenced by the mobility of the ligands in the extracellular space. Our success in implementing this feature into a synthetic ECM-mimetic platform provides direct evidence for the critical importance of signal motion in mediating cellular behavior.
The PA-based ECM mimetic technology we describe here offers biological and technical advantages relative to current approaches for culturing stem cell-derived neurons in vitro. Firstly, the elevated engagement of the ITGB1 receptor mediates the inherently-driven, increased level of functional maturation. Secondly, the bioactivity of the mimetic epitope is steadily preserved over time, in stark contrast to commercial substrates that are rapidly biodegraded and require continuous supplementation. Thirdly, neuronal aggregation is drastically inhibited, essentially recapitulating the plating conditions that were previously attainable only by co-culturing on a glial layer. Lastly, the flexibility in the design of these synthetic matrices can be used to create tailorable platforms with a combination of multiple bioactive ECM epitopes that could further reproduce the various spatiotemporal aspects of the neuronal microenvironment31. Implementing cellular and acellular microenvironment cues in a spatiotemporal manner is crucial for the biofidelic reproduction of developmental and disease processes in vitro 21.
We anticipate that the PA platform will be of great interest to the stem cell community focused on developing hiPSC-based models of neurodevelopmental, neurological, and neurodegenerative diseases. The adaptive nature and inherent flexibility in the design of the PA supramolecular materials can facilitate the development of additional ECM mimetic platforms in the future.
Limitations of the study
While the IKVAV-PA platform we developed had dramatic effects on the functional maturation of hiPSC-derived neurons, a major challenge that remains to be resolved is the induction of neuronal ageing in vitro1,100. This is critical as it would facilitate better hiPSC-based models of adult and late-onset neurodegenerative diseases. Although we found that more mutant ALS-SOD1 MNs exhibited cytosolic protein aggregations, this is likely related to the ability of IKVAV-PA2 to sustain neurons long-term in vitro without cellular clustering or trophic glial support. We anticipate that this phenotype could be further exacerbated by exposure to an aged ECM platform or other ageing factors. The composition of the ECM in the CNS is known to change with age, becoming richer in collagens, proteoglycans, and inflammatory cytokines9,101,102, and could be mediating aspects of age-associated neurodegeneration. Future efforts guided towards creating artificial coatings that mimic the ageing ECM could prove useful. Similarly, there is a need to design ECM platforms that can facilitate the development and reproducibility of 3D organoid and assembloid-based CNS models that allow for the investigation of cell-cell interactions, circuits, and other neurodevelopmental processes.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Evangelos Kiskinis (evangelos.kiskinis@northwestern.edu).
Materials Availability
PA materials in this study were synthesized at the Peptide Synthesis Core at Northwestern University. Since additional scale up studies and characterization is ongoing, and the patent protecting the intellectual property related to these materials is still under review, Northwestern University requests that these materials only be distributed via a formal Materials Transfer Agreement (MTA) with the University.
Data and Code Availability
All the data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal protocols
All animal housing and procedures were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. All procedures were approved by the Northwestern University Institutional Animal Care and Use Committee.
Astrocyte cultures
Glial cells were derived from the cerebral cortex of newborn mice (P0, Charles River) as previously described 103. Passage 1 cells were cultured at a density of 2 × 105 cells/cm2 for 3 days in Neurobasal containing 3% normal human serum (NHS, Thermofisher), 1% penicillin–streptomycin (pen–strep, ThermoFisher), and 2 mml-glutamine on MEA plates with poly-D-lysine (Sigma Aldrich) and laminin (Gibco) coating. iPSC-derived MNs were culture in the presence of astrocytes for 40 days.
Human iPSC culture conditions
iPSC lines were derived by retroviral transduction of skin fibroblasts from healthy control individuals (11a: male, 36 years old; 18a: female, 48 years old) 104. Most of the experiments were performed in the 18a line, except for the results shown in Figures S6E–K, were we tested the effect of the distinct matrices on a distinct cell line (11a). For testing the effect of our highly mobile IKVAV-PA2 nanofibers in an ALS disease model, we have utilized a line carrying a disease-associated mutation in the SOD1 gene (A4V) and its respective isogenic control 92 (Figure 7). All iPSC lines were karyotyped and tested for pluripotency by ICC and the Scorecard assay 92,104. Human iPSCs were maintained on Matrigel (BD Biosciences) with mTeSR1 media (Stem Cell Technologies) and passaged on a weekly basis using 1mM EDTA or Accutase (Sigma). All cell cultures were maintained at 37 °C and 5% CO2 and tested for mycoplasma monthly.
Cortical neuron differentiation and long-term culture conditions
Human iPSCs were differentiated into cortical neurons using a modified version of a protocol based on NGN2 overexpression 89. Single-cell hiPSCs dissociated with Accutase were transduced in suspension for 5–10 min with lentiviruses (FUW-M2rtTA, TetO-Ngn2-Puro, TetO-FUW-EGFP) before plating (95000 cells/cm2). After 24h (day 1), medium with lentivirus was replaced with KOSR medium containing KnockOut DMEM supplemented with knockout replacement serum (KSR), NEAA, Glutamax, 55 μM β-mercaptoethanol (Gibco), 10 μM SB431542, 100 nM LDN-193189, 2 μM XAV939 (DNSK International) and 2 μg/ml of doxycycline (Sigma). On day 2, media was replaced with a 1:1 ratio of KOSR to neural induction media (NIM) which contain DMEM: F12 supplemented with NEAA, Glutamax, N2, 0.16 % D-glucose (Sigma) and 2 μg/ml heparin sulfate (Sigma). To the final 1:1 medium doxycycline and puromycin were added (2μg/ml; Sigma). On day 3, the media was replaced with NIM containing doxycycline and puromycin (2 μg/ml). Cultures were dissociated with Accutase on day 4, resuspended in NBM medium supplemented with doxycycline, BDNF and Rock inhibitor, and plated at a density of 20000–25000/cm2. Cortical neurons were feed 3 times a week with NBM supplemented with BDNF, 1 % FBS and doxycycline.
Motor neurons differentiation and long-term culture conditions
At 70 % confluency, hiPSC cultures were dissociated using Accutase and plated at a density of 105 cells/cm2 with 10 μM ROCK inhibitor (Y-27632, DNSK International) in mTeSR1. Next day (day 0), media was replaced with N2B27 medium (50 % DMEM: F12, 50% Neurobasal, supplemented with non-essential amino acids (NEAA), Glutamax, N2 and B27; Gibco, Life Technologies) containing 10 μM SB431542 (DNSK International), 100nM LDN-193189 (DNSK International), 1 μM Retinoic Acid (RA, Sigma) and 1 μM of Smoothened-Agonist (SAG, DNSK International). The culture medium was changed daily until day 6, then switched to N2B27 medium supplemented with 1 μM RA, 1 μM SAG, 5 μM DAPT (DNSK International) and 4 μM SU5402 (DNSK International). Cells were fed daily until day 14, when MNs were dissociated using TrypLE Express (Gibco, Life Technologies) supplemented with DNase I (Worthington) and plated at a density of 50,00 cells, 450,000–500,000 or 800,000 cells/well in 24 wells, 12 wells or 6 wells respectively, onto pre-coated poly-D-lysine (Sigma-Aldrich) with the various IKVAV-PAs or with commercial laminin (see “PA and laminin coating preparation” section below). For most of the analyses we utilized natural laminin 111 isolated from Engelbreth-Holm-Swarm (EHS) sarcoma (ThermoFisher Scientific, and Sigma-Aldrich). For comparative studies among different laminins (Figure S4), we used Laminins 211 (BioLamina), 521 (BioLamina) and Fibronectin (Thermofisher). hiPSC-derived MNs were feed 3 times a week with NBM medium (Neurobasal, NEAA, Glutamax, N2 and B27) supplemented with 1 % FBS, Ascorbic acid (0.2 μg/ml; Sigma-Aldrich), BDNF, CNTF and GDNF (10 ng/mL, R&D systems).
METHOD DETAILS
Material synthesis, purity characterization and PA preparation
PA synthesis and preparation: IKVAV-PA (C16VVAAEEEEGIKVAV, C16AAGGEEEEGIKVAV, C16VEVAAGIKVAV) and VVIAK-PA molecules (C16VVAAEEEEGVVIAK, C16AAGGEEEEGVVIAK, C16VEVAAGVVIAK) were synthesized by standard fluorenylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis using a CEM model Liberty Blue Microwave Assisted Peptide Synthesizer on rink amide MBHA resin. Automated coupling reactions were performed using 4 equiv. Fmoc-protected amino acid, 4 equiv. of N,N’-diisopropylcarbodiimide (DIC), and 8 equiv. ethyl(hydroxyimino)cyanoacetate (Oxyma pure). Removal of the Fmoc groups was achieved with 20 % 4-methylpiperidine in DMF. Peptides were cleaved from the resin using standard solutions of 95 % TFA, 2.5 % water, 2.5 % triisopropylsilane (TIS) and precipitated with cold ether. Basic purification using reverse-phase high-performance liquid chromatography (HPLC) was then performed using a Phenomenex Gemini NX-C18 column, (C18 stationary phase, 5um, 110 Å pore size, 150 ´ 30 mm) on a Shimadzu model Prominence modular HPLC system, two LC-20AP solvent delivery units, SPD-M20A diode array detector and a FRC-10A fraction collector, using H2O/CH3CN gradient containing 0.1 % NH4OH (v/v) as an eluent at a flow rate of 25.0 mL/min. The purity of lyophilized PAs was analyzed by liquid chromatography-mass spectrometry (LC-MS) using a Phenomenex Jupiter 4 μm Proteo 90 Å column (C12 stationary phase, 4 μm, 90 Å pore size, 1 × 150 mm) or Phenomenex Gemini C18, (C18 stationary phase, 5 μm, 110 Å pore size, 150 ´ 1 mm) on an Agilent model 1200 Infinity Series binary LC gradient system, using H2O/CH3CN gradient containing 0.1 % formic acid or NH4OH (v/v) as eluents, respectively, with a flow rate of 50 μL/min. Electrospray ionization mass (ESI-mass) spectrometry was performed in positive scan mode on an Agilent model 6510 Quadrupole Time-of-Flight LC-MS. For covalently linked dye PAs, the IKVAV-PA molecules mentioned previously were synthesized on Rink amide MBHA resin using the CEM Liberty microwave-assisted peptide synthesizer and protocols described above. The purified PAs were dissolved in tris(2-carboxyethyl) phosphine (TCEP) hydrochloride (5 eq with respect to the PA) in pH 8 Tris buffer and reacted with maleimide functionalized Alexa Fluor® 488. The final product was purified by HPLC and stored until use.
PA and laminin coating preparation
After lyophilization, the PA powder was reconstituted in 150 mM NaCl and 3 mM KCl solution and adjusted to a pH of 7.4 using 1 μL additions of 1N NaOH to ensure cell compatibility and material consistency. PA solutions were annealed at 80 °C for 30 min and then slowly cooled down at 1 °C per minute to reach a final temperature of 27 °C using a thermocycler (Eppendorf Mastercycler) for even and controlled heating and cooling of all samples. To prepare a PA coated substrate, 24-, 12-, or 6-well polystyrene cell culture plate or 12 mm and 18 mm glass coverslips (German Glass, Fisher Scientific, NC0319857) were coated with poly-D-Lysine (PDL, 0.01 mg/mL in water, Sigma-Aldrich) or poly-L-Ornithine (Ornithine, 0.01 mg/ml in borate buffer pH=8.4, Sigma Aldrich) for at least 3 h at 37 °C. The plates were then rinsed with Milli Q water three times and allowed to dry for 4 h. PAs were painted on the coverslips or tissue culture plates by dragging a pipette (8–30 μL of annealed PAs (1 wt%)) to extrude a thin, even coating of material across the surface without scratching the PDL or Ornithine coating. PA coatings were incubated for 2–3 h inside the hood. For dye-labeled PAs experiments, Alexa488-labeled-IKVAV-PAs were co-assembled at 1 mol% with their corresponding non-labeled PA counterparts. The plates were gently rinsed with media and the media was left for 1 h before further use. For the laminin control, after washing the PDL or Ornithine coatings three times with Milli Q water, Laminin (10 μg/ml, Thermofisher) was incubated for at least 1 h at 37 °C. Cells were seeded after aspirating the laminin solution.
Immobilization of IKVAV peptide on glass surface
Borosilicate glass coverslips (12 mm in diameter; Corning) were modified with synthetic IKVAV peptide following a technique described previously 105. Borosilicate glass coverslips were cleaned with 2 % (v/v) micro-90 detergent (Sigma Aldrich, Z281565) for 30 min at 60 °C, rinsed six times with distilled water, rinsed with ethanol and then dried. Coverslips were plasma-etched (Harrick Plasma PDC-001-HP) with O2 for 30 s, then immediately incubated in a 2 % (v/v) solution of (3-aminopropyl) triethoxysilane (Sigma Aldrich) in ethanol for 15 min. Coverslips were then rinsed twice with ethanol and twice with water and then dried in the oven. IKVAV peptide was then prepared at 50 nmol/mL in a 1.25 mg/mL solution of 1-ethyl-3-(dimethyl-aminopropyl) carbodiimide (Arcos Organics) with 2 % DMF (Dimethylformamide, Sigma Aldrich). Coverslips were incubated with this solution for 3.5 hrs at 40 °C. After incubation, coverslips were washed successively with 100 % acetic anhydride (Fisher Chemical), 2 M hydrochloric acid (Fisher Chemical), and 0.2 M sodium bicarbonate. After rinsing with an excess amount of water, samples were sonicated in 4 M urea for 10 min followed by 1 M NaCl for 10 min and then rinsed with an excess amount of water and dried at 100 °C for 1 h.
Conventional transmission electron microscopy (TEM)
An aliquot of 5 μL of sample solution ([PA] = 0.01 wt% in H2O) was deposited on a copper TEM grid with carbon support film (Electron Microscopy Science), and held in place with tweezers for 5 min. The sample solution was removed by capillary action using filter paper, and the grid was dried for 10 min. The sample was then stained with 10 μL of aqueous uranyl acetate (2 wt%, Sigma Aldrich) for 3 min and the solution was removed by capillary action using filter paper. The grid was dried for at least 2 h before imaging. TEM images were obtained using a Hitachi model HT-7700 electron microscope operating at 120 kV, equipped with an Orius SC 1000A camera.
Cryogenic transmission electron microscope (cryo-TEM)
Plunge-freezing for cryo-TEM samples was carried out using a FEI model Vitrobot Mark III. 6.5 μL of sample solution ([PA] = 0.01 wt% in H2O) was placed on a plasma-cleaned copper TEM grid with holey carbon support film (Electron Microscopy Science) and held with tweezers mounted on the Vitrobot. The specimen was blotted in an environment with 100 % humidity at room temperature (blot offset: 0.5 mm, blot total: 1, wait time: 0 s, blot time: 5 s, drain time: 0 s), and plunged into a liquid ethane reservoir cooled by liquid nitrogen. The vitrified samples were stored in liquid nitrogen and then transferred to a Gatan cryo-TEM holder. Cryo-TEM images were obtained using a Hitachi model HT-7700 or JEOL1230 electron microscope operating with an accelerating voltage of 120 kV, equipped with an Orius SC 1000A camera or a Gatan 831 CCD camera.
Scanning electron microscopy (SEM)
PA samples were fixed in a mixture of paraformaldehyde (2.0 %, Electron Microscopy Sciences), glutaraldehyde (2.5 %, Electron Microscopy Sciences) in phosphate buffered saline (1 X, Gibco) for 20 min. The fixative was removed, and the water was exchanged with ethanol by incubating the samples in a gradation of ethanol solutions with increasing concentration (30–100 %) of 200 proof ethanol (Decon Laboratories, Inc). Critical point drying (Tousimis Samdri-795) was used to remove the excess water. A purge cycle of 15 min was used. The resulting dehydrated sample coverslips were mounted on stubs using 12 mm carbon adhesive tape (Electron Microscopy Sciences) and coated with approximately 6 nm of osmium (Filgen, OPC-60A) in order to make the sample surface conductive for imaging. All images were taken with an accelerating voltage of 2 kV with a Hitachi SU8030 SEM instrument.
Porosity
Nitrogen physisorption isotherms were collected using a Micromeritics 3Flex instrument. To generate dry gels of the PA scaffolds, PA solutions at 1 wt% were gelled with 50 mM CaCl2aq (1:2 v/v) and let sit for 2 h. After aspirating the excess CaCl2 solution, the gels were rinsed with water in a cell strainer (Fisherbrand) and flash frozen in an aluminum dish floated on liquid nitrogen, then lyophilized. The samples were degassed using the Smart VacPrep instrument (Micromeritics) at 50 °C for roughly 72 h. Data points below 0.01 relative pressure were collected using fixed volume dosing (3 cm3/g STP) and an equilibration interval of 20 s. Data points above 0.01 relative pressure were collected at targeted pressures using a minimum equilibration interval of 10 s. Brunauer-Emmett-Teller (BET) fitting ranges and model results are presented below. The total surface area for each sample was approximately 1 m2, which results in ~17 % instrument uncertainty in the BET specific surface area values (included in the reported ± range reported in Figure S1B) according to the Micromeritics application notes (https://www.micromeritics.com/Repository/Files/micro_tech_tip_14-surface-area-analyses.pdf).
| Sample | BET Range (p/p0) | BET Surface Area (m2/g) | C-value | Sample Mass (mg) | Surface Area (m2) |
|---|---|---|---|---|---|
| PA1 | 0.05–0.35 | 51.65 | 8.62 | 17.2 | 0.89 |
| PA2 | 0.05–0.35 | 47.54 | 8.36 | 18.1 | 0.82 |
| PA3 | 0.05–0.35 | 53.65 | 8.60 | 19.6 | 1.05 |
Profilometry analysis
PA and laminin coated coverslips were prepared as described above and a Zygo Nexview 3D Optical Profilometer was used to image their surfaces. PA1, PA2, PA3, laminin-coated coverslips, and blank coverslips were tested at 72 h and 60 days in vitro. A 10X objective lens with a 2 X Zoom and 10-μm-scan length was used to obtain images. Samples were allowed to dry for ten min before imaging. Each coverslip was scratched using a surgical blade and at least 9 images from 9 coverslips were taken per condition. The surface thickness was analyzed using the Region tool in Zygo’s Mx software. The scratch surface was set as the reference plane, and the measured coating thickness was calculated as the average depth of the coating with respect to the reference plane. For visual comparison, all images were normalized to a standardized scale with a −0.5 μm minimum and 1.2 μm maximum.
Synchroton X-ray scattering measurements
PA solutions at 1 wt% were loaded in home-built sample cells comprised of two 30 μm thick AS32eco ultrathin glass (Schott AG) as window materials, attached to both sides of a 2.0 mm thick acrylic plate using 9474LE double-stick sheets (3M) as adhesives. The acrylic plates and the double-stick sheets were laser-cut to form sample cells with 6 mm height and 3 mm width, and around 38 μL of the solution was used for each cell. The sealed samples were then mounted on a translational stage at the beamline. Background samples containing buffer solutions were also collected to perform background subtraction. Experiments were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) 5-ID-D station at the Advanced Photon Source (APS), Argonne National Laboratory 106. Data was collected on a triple area detector system with an X-ray energy at 17 keV. The wavevector q is defined as = (4π/λ) sin(θ/2), where θ is the total scattering angle. Three different spots on each well were measured for one frame at 5 s exposure time, and the data was averaged based on these 3 frames. The acquired 2D scattering data were then reduced to 1D intensity vs. wavevector plots via azimuthal integration around the beam center in GSAS-II software 107 and were subtracted against buffer scattering profile before analysis on small angle power law and Bragg peaks at wide angles were performed. The software SasView 5.0.4 (https://www.sasview.org), accessed on 12/10/21 was used for linear fitting to obtain the slopes or fitting with the core shell parallelepiped model for other detailed structural features.
Fourier-transform Infrared (FTIR) spectroscopy
FTIR spectra of PA samples were recorded on a Bruker model Tensor 37 spectrometer. 100 μL of sample solution ([PA] = 1 wt% in D2O in aqueous NaCl and KCl ([NaCl] = 150 mM and [KCl] = 3 mM) were placed between two CaF2 windows with 50 μm separation. The spectra obtained are the result of 25 scans with 1 cm−1 resolution. Additional IR on dried samples used lyophilized powder from 100 μL of sample ([PA] = 1 wt% in aqueous NaCl and KCl ([NaCl] = 150 mM and [KCl] = 3 mM) placed on an attenuated total reflectance sample stage equipped with a germanium crystal. IR spectra were scanned 32 times using 1 cm−1 resolution and then averaged.
Circular dichroism (CD) Spectroscopy
Each IKVAV-PA sample was diluted to concentrations between 0.01–0.04 wt% in either H2O (no salt samples) or buffer containing 150 mM NaCl and 3 mM KCl (high salt). CD spectra were recorded on a JASCO model J-815 spectropolarimeter using a quartz cell of 0.5 mm optical path length. A continuous scanning mode was used with a speed of 100 nm per minute with the sensitivity set to standard mode. High Tension (HT) voltage was recorded for each sample to ensure that the measurement was not saturated. An accumulation of three measurements was used and a buffer sample was background-subtracted to obtain final spectra. The final spectra were normalized to the final concentration of each sample.
Transverse relaxation nuclear magnetic resonance spectroscopy (T2-NMR)
NMR spectra were acquired at 600 MHz on a Brucker Neo system with QCI-F cryoprobe. NMR spectra for IKVAV-PAs were recorded at 25 °C using H2O/D2O in 9/1 ratio (D2O contains 0.05 wt% 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid, sodium salt, Merck) as solvents. Chemical shifts are reported in part per million (ppm). For Ca2+ experiments, IKVAV-PA solution was mixed with 5uL of 20mM CaCl2 and 5 min after mixing the solution samples were run. The spin-spin relaxation rates were measured using the Carr–Purcell–Gill–Meiboom pulse sequence with a delay time of 0.2 ms in a variable loop. The peak intensity data was fit to and exponential in the form:
Where τ is the length of the delay time, R2 is the spin-spin relaxation rate and b are the baseline108.
DPH-embedded samples for fluorescence studies
An aqueous solution of 100 μL of annealed PA ([PA] = 6 mM, [KCl] = 3 mM, [NaCl] = 150 mM) was added a THF solution (2 μL, Sigma Aldrich) of 1,6-diphenyl-1,3,5-hexatriene (DPH; 1.4 mM, Sigma-Aldrich, D208000), and the mixture was incubated for 30 min at 25°C. Then, the mixture was diluted with aqueous KCl and NaCl (1900 μL, [KCl] = 3 mM, [NaCl] = 150 mM), incubated for 10–30 min at 25 °C to afford a solution of DPH (1.4 μM)-embedded PA (300 μM). To obtain IKVAV-PA2 (A2G2) in the presence of CaCl2, 5 mM CaCl2 was then added to the DPH-embedded PA aqueous solution to afford PA: CaCl2 at a molar ratio of 6:1. DPH was exited at 336 nm and emission was recorded at 450 nm on an ISS model PC1 spectrofluorometer with a 300 W xenon arc lamp with power of 18 A. Excitation slit and emission slit widths were set as 1 mm (8 nm bandwidth). Anisotropy was calculated using the following equation:
Where 𝐹∥ represents the parallel intensity to the excitation plane, 𝐹⊥ is the perpendicular intensity to the excitation plane, g is grating factor (G-factor) that represents the intensity ratio of the sensitivity of the detection system for vertically and horizontally polarized light. G-factors were determined individually in each measurement. Results were averaged based on 34 iterations from two measurements.
Atomic force microscopy (AFM)
AFM imaging and force measurements were performed at room temperature on a Bioscope Resolve BioAFM/Nanoscope V system (Bruker, Santa Barbara), integrated onto an Axio Oberver.D1m inverted optical microscope (Carl Zeiss, Inc.). Silicon nitride triangular probes (ScanAsyst Fluid, Bruker) having nominal tip radius ~20 nm and spring constant of ~0.7 N/m were used for imaging and indentations of thin fibers. SiO2 beads with a diameter of 1 μm attached to silicon nitride triangular cantilevers with 30 nm gold coating (Novascan Technologies, Inc.) were used for indentation experiments on thick gels. The deflection sensitivity of each probe was calibrated by repeated indentation on a clean glass slide in MilliQ water and the spring constant of the cantilever was estimated by thermal noise method. The effective tip radius of conical (sharp) probes was estimated before each measurement as a function of the indentation depth, using polycrystalline titanium tip characterizer sample (RS-15M, Bruker) with a tip estimation function (NanoScope Analysis software, Bruker).
Indentations were performed in MilliQ water by bringing the AFM probe in contact with the sample surface at a controlled load force and recording force-displacement curves during the loading-unloading cycles. During indentations a maximum load of <10 nN was applied at each data point to avoid plastic deformation and to keep the indentations within the elastic range. Measurements were performed by acquiring ~ 250 force curves per sample. To fit the force curves, we used the Protein Unfolding and Nano-indentation Analysis Software (PUNIAS). The elastic modulus of the thick gels (several μm-thick) was obtained by fitting the loading force curves through the Hertz model:
Where F is the force, δ the sample deformation, E the Young’s modulus, ν the Poisson ratio, and R the radius of the indenting probe. Fitting of the force curves was performed by controlling the indentation depth to 200 nm, i.e. within 10 % of the total film thickness, in order to minimize the rigid substrate effect. Force curves performed with sharp probes on thin fibers were analyzed through the DMT (Derjaguin, Muller and Toporov) model. The DMT model, which is based on the Hertz model yet includes a description of adhesion, is the standard model employed in mechanical studies on fibrils 109. Force curves are described in the DMT model by the following equation:
With F0 the adhesion force. The fitting of the force curves was performed by controlling the indentation depth to 4–5 nm, i.e., within 10 % of the total film thickness in order to minimize the rigid substrate effect. All samples were considered incompressible, with a Poisson ratio of 0.5.
Rheology
PA materials were prepared using methods described above. An MCR302 Rheometer (Anton Paar) was used for all rheological studies. PA liquid was placed on the sample stage (150 μL) and 150 mM CaCl2 solution (30 μL, for a final concentration of 25 mM CaCl2) was pipetted onto the underside of a 25 mm cone plate above the material. The instrument stage was set to 37 °C to simulate in vitro culture conditions. The plate was slowly lowered to the measuring position and a humidity collar was used to enclose the sample plunger and prevent sample evaporation during each 45 min experimental run. During the first interval of each experiment, the sample was equilibrated for 30 min with a constant angular frequency of 10 [rad/s] and 0.1 % strain. The storage and loss modulus (G’ and G”) were recorded at the end of the interval, after a plateau occurred. The angular frequency was incremented from 100 rad/s to 1 rad/s over 21 points. G’ and G” were recorded for all frequencies. Lastly, the % strain was increased incrementally from 0.1 to 100 % over 31 points and G’ and G” were recorded.
Simulation procedures
The PAs for the simulations were created in Avogadro 110 and transformed to MARTINI force field 62–64,111 CG representation using a modified version of martinize.py (martinize.py, 2.0; <http://cgmartini.nl/images/tools/martinize/martinize-2.6/martinize.py>) to include the aliphatic tail 58 and using coiled coil as choice for secondary structure. The last two E’s (furthest from aliphatic tail) and the K are charged while the two first E’s are treated as protonated as this was found to be ideal for fiber formation in preliminary simulations. This difference in protonation state between assembled and free peptides has been previously reported 112. Therefore, the final charge is (−2+1=) −1. Initial structures consist on 300 molecules arranged randomly and spaced a minimum of 3 Å, solvated with CG water and enough ions were added to neutralize the system in a cubic box 21.5 × 21.5 × 21.5 nm3. This corresponds to a concentration of 50 mM (7.8, 7.4 and 8.3 wt% for PA1, PA2, PA3, respectively). This is within the range of concentrations commonly used to speed up self-assembly simulations, which can be up to 10 times higher than the experimental systems59,113,114. All visualizations were rendered using Visual molecular dynamics software (VMD)115. Additionally, bending simulations were set up by placing equilibrated fibers from the previous simulations in a cubic box big enough, 30.0 × 30.0 × 30.0 nm3, to ensure that fibers remain as discrete, non-periodic, by not being able to reach opposite walls within the box. This is because periodicity would add unrealistic constrains for the bending of the fibers and this would not be studied reliably.
Coarse grained Molecular Dynamic (CG-MD) simulations were performed in GROMACS 5.0.4 software116 which was also used for the analysis of the simulations. A cut-off of 1.1 nm was used for intermolecular interactions using reaction field with a relative dielectric constant of 15 for electrostatics and potential-shift for Lennard-Jones interactions 117. All systems were minimized for 5000 steps or until the forces in atoms converged below 2000 pN. Self-assembly simulations were run using a 25 fs time step in an NPT ensemble using V-rescale algorithm for the temperature (303 K, τT = 1 ps) 118 and Berendsen for the pressure (1 bar, τP = 3 ps) 119. Simulations were run for 100,000,000 steps corresponding to 10 μs effective time 63,111. The water contacts of the PAs were calculated using the integration of the radial distribution function for the first solvation sphere of each backbone and aliphatic tail bead (Figure S1J) and applying the 4x factor to convert CG water to water molecules. Motion is measured using the root mean square fluctuations (RMSF) through the last 5 μs of simulation (given that fibers are equilibrated after 5 μs as shown by the root mean square fluctuations, RMSD, plots in Figures S1G–H). The solvent accessible surface area (SASA) is computed to measure the exposure of the IKVAV epitope through the last 5 μs of the simulations, and divided by the fiber length to obtain the normalized value of nm2 per nm of fiber. Fiber bending is the only feature analyzed from discrete fibers in 30 nm side boxes. Bending is computed as the variation in the maximum distance between atoms in the same fiber, divided by the initial fiber length. Bending value is given as the average of the last 5 μs of simulations.
Spinal cord dissection
Spinal cords were harvested under aseptic conditions from postnatal (postnatal day 0 (P0), Charles River) and adult (12 weeks, Charles River) CD1 mice and immediately placed in phosphate-buffered saline (PBS, Gibco) at 4 °C.
Spinal cord decellularization
Decellularized spinal cords were prepared by following previously reported protocol 120. Briefly, the spinal cords were rinsed twice (1 h each) in PBS. After 2 h, the PBS was replaced with 1 % Triton X-100 (Sigma Aldrich) solution and agitated for 3 h. The tissue was then rinsed with washing solution 0.01 % PBS three times (1 h per rinse). Next, the washing solution was replaced by 1% sodium deoxycholate solution (Sigma Aldrich) for 3 h. This process was repeated twice. Finally, tissue segments were washed three times (1 h each) in a 0.01 % PBS solution and stored in the same solution at 4 °C until use. All washing steps were carried out at 25 °C with agitation.
Cell seeding on PA coatings
Parameters such as cells density, cell dissociation or cell plating are crucial to get a homogenous culture (even when we culture on laminin coatings). For this study, MN cells harvested from Matrigel were dissociated carefully making sure that no aggregates of cells were seeded initially on PA and laminin coatings. We have optimized the number of cells cultured on coated glass coverslips or tissue culture plate to avoid unnecessary clustering. The density of cells plated varies from 50–75.000 cells in 24 well plates and between 350–500.000 cells in 12 well plates.
PA treatments with and without Calcium
For IKVAV-PA treatments, MNs were cultured on poly-DL-lysine (0.01 %, Sigma Aldrich) and Laminin (10 μg/ml) coatings in 12 well and 24 well plates at a density of 500,000 cells/well and 50,000 cells/well respectively. MNs were treated with 20 mM of IKVAV-PAs or commercial laminin protein (30 μg/mL) in solution. IKVAV-PAs mixed with 5 mM CaCl2 at a ratio PA:CaCl2 6:1 was used to treat MNs. For 2D coating experiments, MNs were cultured on the different IKVAV-PAs ([PA] = 6 mM, [KCl] = 3 mM, [NaCl] = 150 mM) or laminin (10 μg/ml) coatings in 6 well and 24 well plates at densities mentioned above.
Cell viability assay
To assess cell viability, we utilized CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega), a colorimetric assay that quantitatively measures lactate dehydrogenase (LDH), a stable cytosolic enzyme released upon cell lysis. Cell media from cells cultured in the various coated surfaces was collected at different cultured time points and the extracellular levels of LDH enzyme measured by quantifying the conversion of a tetrazolium salt into a red formazan product, read by 490–492 nm absorbance. The analysis was done in at least 3 independent differentiations with a minimum of 3 technical replicates per condition.
Western blot and dot blot analysis
Cells and decellularized spinal cords were harvested in RIPA buffer (10 mM Tris-HCl pH 8.0, 140mM NaCl, 1mM EDTA, 0.1 % SDS), supplemented with 1 % Triton X-100 (Triton, Sigma Aldrich), 1mM phenylmethylsulphonyl fluoride (DOT Scientific), and protease inhibitors (Millipore). For WB analysis lysates were sonicated and protein extracts were separated by SDS-PAGE followed by electrotransfer to a nitrocellulose membrane (Bio-Rad). For dot-blot analysis, 10 μg of protein was loaded through a 96 well Acrylic Dot-Blot System (Whatman, GE Healthcare Life Sciences) directly onto a nitrocellulose membrane. The membranes were blocked in Tris-buffered saline (TBS, Bio-Rad) + 5 % non-fat dry milk (Bio-rad) and then incubated overnight at 4 °C with primary antibodies. Primary antibodies were diluted in TBS + 0.1 % Tween + 5 % BSA (Sigma Aldrich). After several washes in TBS + 0.1 % Tween, membranes were incubated with their corresponding secondary HRP-conjugated antibodies (ThermoFisher). A radiance bioluminescent ECL substrate kits (Radiance ECL, Azure Biosystems or Pierce ECL Western Blotting Substrate, Thermofisher) was used to detect protein signals through Azure c300 Gel Imager (Azure) or Image Quant LAS4000 mini-imager (General Electric) on the automatic setting. Densitometry analysis for the bands was performed using Fiji software (ImageJ, NIH Image).
Immunocytochemistry
For immunofluorescence, fixed human hiPSCs-derived neurons were incubated with primary antibodies overnight at 4 °C, and then with their appropriate rabbit, mouse and goat anti- Alexa 488, Alexa 555 or Alexa 647 secondary antibodies (Thermofisher, A21202, A11055, A21206, A31572, A-31570, A21432, A31571, A78952 at 1:500–1:1000). DAPI (Molecular Probes D3571 at 1:1000) was used to stain nuclei. Finally, the preparations were cover-slipped with Immu-Mount (Thermofisher) for imaging.
Primary Antibodies
Primary antibodies used for WB and Immunocytochemistry: ISL1/2 (DSHB, 39.4D5-c), CHAT (Millipore, AB144P), ITGB1 (Millipore, clone HUTS-4, MAB2079Z), ITGB4 (Millipore, MAB1964), ILK (Cell Signaling, 3862), p-FAK (Cell Signaling, 3281S), FAK (Cell Signaling, 3285S), ACTIN (Sigma Aldrich, A2066), KI67 (Abcam, ab66155), MAP2 (Biolegend, 840601), TUJ1 (Biolegend, 802001), PSD95 (NeuroMab, 75–028), SYNAPTOPHYSIN (Abcam, ab32127), SYNAPSIN-1 (Millipore, AB1543), FOXA-2 (Santa Cruz Biotechnology, sc-101060), NeuN (Biolegend, 834502), Laminin alpha-1 (Santa Cruz, sc-74417), GFP (Abcam, ab6673), SOD1 (Proteintech, 10269–1-AP), Ubiquitin (Santa Cruz Biotechnology, sc-8017) and GABA (Sigma Aldrich, A2052).
Image acquisition and analysis
For protein expression studies, images were acquired by Nikon A1R confocal laser-scanning microscope with GaAsP detectors, Nikon structural illumination microcopy (N-SIM) using a ×100 oil immersion objective (NA=1.4) as z-series of 3–8 images, taken at 0.125 μm intervals, 1024×1024-pixel resolution or Zeiss LSM 800 confocal laser scanning microscope. The acquisition parameters were kept the same for all scans. Two-dimensional average projection reconstructions of images, fluorescence analysis, and quantification were done using Fiji software (National Institute of Health, USA) and Imaris. The fluorescence intensity of images taken for Figure 7 was adjusted in order to avoid overexposed signal in aggregate-positive neurons. This adjustment might underrepresent endogenous levels of SOD1 and Ubiquitin (UB). For cell distribution, images were acquired by Nikon A1R confocal laser-scanning microscope with GaAsP detectors using a ×20 objective at 1024×1024-pixel resolution. Cell position was analyzed by NIS-Elements software and plotted using Matlab Knnsearch plugin.
Video time-lapse analysis
For video time-lapse analysis of cell migration, cells were plated on the various coatings and placed in a Nikon BioStation IM-Q time-lapse imaging system with a 1.3-megapixel cooled monochrome camera at 37 °C with 5 % CO2. Cells were imaged in phase contrast, and acquired every 5–16 min for 24–96 h in vitro. Cell displacement, speed and trajectory were calculated with the aid of the ‘Manual Tracking’ plug-in of the ImageJ software (National Institutes of Health, USA).
Sholl analysis
Neuronal morphology analysis was performed in Fiji software from confocal images of neurons immunolabeled with anti-MAP2 antibody. Maximum-reconstructions were calibrated and adjusted for brightness and contrast for a subsequent semi-automatic tracing process using Simple Neurite Tracer (SNT). Resultant traced-processes images were used to analyze the branching complexity of the neurons by the Fiji plugin Sholl analysis. This tool creates a series of concentric circles around the neuron cell body and calculates the number of neuronal processes crossing the different circles. The graphical representation of these data will indicate the branching complexity along the neuronal arbor of one specific neuron. For data obtained from a minimum of 3 independent differentiations with a normal distribution, two-way ANOVA followed by a Bonferroni post hoc test for comparisons of the four experimental groups.
Tandem Mass Tag Mass Spectrometry (TMT-MS)
MNs cultured for 60 days on PA2, and laminin coatings were digested using lysis buffer (0.5 % SDS (sigma Aldrich), 50 mM AmBic (fisher), 50 mM NaCl (Sigma-Aldrich) and HALT Protease and Phosphatase Inhibitor cocktail (ThermoFisher Scientific), and vortexed briefly. Samples were sonicated during 30 s three times. A BCA assay (ThermoFisher Scientific) was performed to determine the protein concentration in processed samples, and 100 μg of protein/condition were used for the analysis. Peptides were analyzed by LC-MS/MS using a Dionex UltiMate 3000 Rapid Separation nanoLC and a Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (ThermoFisher Scientific). The peptide was separated on a 180-min analytical gradient from 5 % ACN/0.1 % FA to 40 % ACN/0.1 % FA. The top 15 most abundant precursor ions in each MS1 scan were selected for fragmentation. Precursors were selected with an isolation width of 2 Da and fragmented by higher-energy collisional dissociation (HCD) at 30 % normalized collision energy in the HCD cell.
All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.5.1). Mascot was set up to search the SwissProt_2017_11 database (selected for Homo sapiens, unknown version, 20244 entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.050 Da and a parent ion tolerance of 10.0 PPM. Carbamidomethyl of cysteine was specified in Mascot as a fixed modification. Deamidated of asparagine and glutamine, oxidation of methionine, acetyl of the n-terminus and TMT6plex of lysine and the n-terminus were specified in Mascot as variable modifications. Scaffold (version Scaffold_4.8.4, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95 % probability by the Peptide Prophet algorithm 121 with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.9 % probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm 122. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Gene ontology enrichment of differentially expressed proteins was performed with DAVID and subsequent comparative integration analysis with the online tool https://www.comparativego.com. Metascape (Cytoscape) was utilized to obtained pathway analyses of up- and down-regulated proteins in MNs grown on PA2 vs. laminin.
RNA isolation and quantitative PCR (qPCR) analysis
RNA was isolated from harvested day 0 (hiPSC) and day 14 (differentiated MN cultures) cell cultures with TRIzol Reagent (Invitrogen) in Phasemaker Tubes. Precipitation, wash and solubilization of RNA was carried out following manufacturer’s guidelines. Next, 1 μg of RNA was used to generate cDNA using Superscript IV Reverse Transcriptase (Invitrogen). Real-time qPCR reactions were performed utilizing iTaq Universal SYBR Green Supermix (Bio-Rad) protocol on a CFX system (Bio-Rad). Results were obtained from 2 independent differentiations and 3 technical replicates. The average cycle of threshold (Ct) value of two housekeeping genes (GPI and RPL0) was subtracted from the Ct value of the gene of interest to obtain the Delta Ct. Relative gene expression was determined as 2–DeltaCt (Delta-Delta Ct) and expressed relative to the day 0 hiPSC sample. We used the following primers for qPCR:
TUBB3: GGCCAAGGGTCACTACACG (Forward); GCAGTCGCAGTTTTCACACTC (Reverse).
ISL1: AGATCAGCCTGCTTTTCAGC (Forward); TCATGCCTCAATAGGACTGG (Reverse).
MNX1: GAACACCAGTTCAAGCTCAACA (Forward); GCTGCGTTTCCATTTCATTCG (Reverse).
LHX3: GGAGAGCGTTTACTGCAAGGA (Forward); TTGGCGGTTTCGTAGTCCG (Reverse).
FOXA2: GGAGCAGCTACTATGCAGAGC (Forward); CGTGTTCATGCCGTTCATCC (Reverse).
GPI: GTGTACCTTCTAGTCCCGCC (Forward); GGTCAAGCTGAAGTGGTTGAAGC (Reverse).
RPL0: TCTACAACCCTGAAGTGCTTGAT (Forward); CAATCTGCAGACAGACACTGG (Reverse).
ITGB1 Knockdown and Overexpression Experiments
ITGB1 knockdown experiments were performed by transfecting siRNAs (Ambion Silencer Select) with Lipofectamine RNAiMAX (Invitrogen) following manufacturer guidelines. Briefly, for 1 well of a 12-well plate with 450,000 cells/well, 10 pmol of siRNA was mixed with 3 μL of Lipofectamine RNAiMAX reagent in 100 μL Opti-MEM medium and incubated for 15 min at RT. Afterwards, we mixed it with 400 μL of cell culture medium and added to cells. Analyses of siRNA transfection experiments were performed 5 days after transfection. For ITGB1 overexpression, we designed a polycistronic lentiviral plasmid with ITGB1 under the promoter of EF1A and GFP under the promoter of CMV (VectorBuilder). Virus production from lentiviral constructs was performed using HEK293-FT cells by transfecting packaging pMD2.G and psPAX2 vectors with HilyMax (Dojindo Molecular Technologies). Viruses were collected from cell media 48 to 96 h after transfection. Lentiviral concentrated stocks were obtained after filtering collecting media and centrifugation at 25,000 g for 2 h at 4 °C. Viral titering was performed in 293FT cells. hiPSC-derived MNs were seeded at a density of 50,000 cells/well or 450,000 cells/well in 24 well or 12 well plates respectively. Cells were infected with a previously tittered viral burden for 24 h, and transduced cells were analyzed 21 days after infection.
Whole Cell Patch Clamp
Whole cell patch clamp was performed using 2–4MΩ glass electrodes pulled from glass capillary tubes (Item #TW150F-4, World Precision Instruments, Sarasota, FL, USA) with a Flaming-Brown P-97 (Sutter Instrument Company, Novato, CA, USA). Electrodes were positioned using a Sutter Instrument MP-285 motorized micromanipulator (Sutter Instrument Company). Whole-cell patch clamp measurements were performed at room temperature using the Multiclamp700B amplifier (Molecular Devices, Burlingame, CA, USA) and Winfluor software (University of Strathclyde, Glasgow, Scotland). Briefly, coverslips were perfused with a modified Ringer’s solution containing (in mM): 111 NaCl, 3.09 KCl, 25.0 NaHCO3, 1.10 KH2PO4, 1.26 MgSO4, 2.52 CaCl2, and 11.1 glucose. The solution was oxygenated with 95 % O2 and 5 % CO2 and the perfusion rate were 1.5–2.0 ml/min. Patch electrodes contained (in mM) 138 K-gluconate, 10 HEPES, 5ATP-Mg, 0.3 GTP-Li and Texas Red dextran (75 μM, 3000 MW, from Invitrogen, Life Technologies, Grand Island, NY, USA). In voltage-clamp mode, fast and slow capacitance transients, as well as whole-cell capacitance was compensated using the automatic capacitance compensation on the Multiclamp. In current clamp, neurons were subjected to depolarizing current ramps for testing I-on (the current level at firing onset), I-off (the current level at cessation of firing), and the frequency–current relationship. Hyperpolarizing current was used to hold neurons near −80 mV in between stimuli. Neurons selected were large (input resistance <1000 MΩ) and had a resting membrane potential −35 mV or less. The first action potential evoked by a depolarizing current ramp was used to measure all parameters, including I-ON (current at firing onset). Threshold voltage was defined as the voltage at which the slope exceeded 10 V/s. Action potential size was measured using overshoot (past 0 mV) minus threshold voltage. Duration of the action potential is measured at half of action potential height. Rates of rise and fall are defined as the peak and the trough of the first derivative of the action potential profile.
Spontaneous Excitatory Postsynaptic Currents (sEPSC) recordings and analysis:
Individual coverslips with neurons (DIV 49–53) were transferred to a recording chamber continuously perfused with 32 °C oxygenated aCSF bath solution (in mM): 125 NaCl, 26 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 1 MgSO4, 22 glucose, 2 CaCl2, pH 7.35 at 32–35 °C; osmolality 310–315 mOsm/K. Neurons were visualized using an inverted Olympus IX51 microscope equipped with a 40X objective. Recording pipettes were made of glass capillaries using a horizontal Sutter P-1000 puller yielding a 2–4 MΩ resistance pipette when filled with standard K-methyl sulfate intracellular solution containing (in mM): 120 K-MeSO4, 10 KCl, 10 HEPES, 10 Na2-phosphocreatine, 4 Mg-ATP, 0.4 Na3-GTP, pH 7.35 adjusted with KOH; osmolality 285–290 mOsm/Kg. In order to identify spontaneous excitatory post-synaptic potentials, neurons were held at −70 mV and whole-cell voltage clamp recordings were acquired in gap-free acquisition using a Multiclamp 700B amplifier and pCLAMP 11.1 software (Molecular Devices, USA). Signals were digitized with a sampling rate of 20 kHz (interval 50 μs) and lowpass filter frequency of 10 kHz. Data were analyzed using Easy Electrophysiology software (www.easyelectrophysiology.com). Synaptic events were identified as negative current events and included in analysis if peak amplitude was greater than a threshold value determined by baseline noise. For each neuron, sEPSC frequency was calculated from the number of sEPSC that occurred within the first 4 min of recording. Inter-event interval was calculated as the time between consecutive events and sEPSC amplitude was measured as the delta pA between baseline and peak current. Each cell produced a sEPSC frequency, mean sEPSC amplitude, and mean inter-event interval. Means from each cell were combined for each condition and tested with student’s t-test. To compare the distribution of sEPSC amplitude and IEI, the first 100 events from each cell were combined and tested with Kolmogorov-Smirnov.
Microelectrode array (MEA) analysis
For MEA electrophysiology studies, 12 and 24 well MEA plates were coated with polyethylenimine (PEI, Sigma Aldrich) and laminin according to Axion Biosystems protocol or PEI and IKVAV-PAs. Human iPSC-derived MNs were seeded at a density of 300,000 cells/well (12-well plates) and 50,000–70,000 cells/well (24-well plates) on the center of the well and cultured until day 40. As a control, primary mouse cortical astrocytes were seeded 2 days before culturing the MNs. Spontaneous network and synchronized activity were recorded using Axion Biosystems Maestro 768 channel amplifier and Axion Integrated Studios (AxIS) v2.4 software. The amplifier recorded from all channels simultaneously using a gain of 1200× and a sampling rate of 12.5 kHz/channel. After passing the signal through a Butterworth band-pass filter (300–5000 Hz) on-line spike detection (threshold = 6× the root-mean-square of noise on each channel) was done with the AxIS adaptive spike detector. All recordings were conducted at 37 °C with appropriate 5 % CO2 / 95 % O2. Spontaneous network activity was recorded for 5 min 3 times a week starting at day 20. Active electrodes, defined as having >5 spikes/min during the baseline-recording period, were used in the analysis. Synchronized activity was defined as spike and burst activity that occurred on 25 % of the electrodes or more in a well within 100 ms of each other. The mean firing rate (Hz), network burst duration (s), and number of spikes per network burst were used as a measure of neuronal activity as this demonstrates maturity of neuronal functional properties. All data reflects well-wide averages, where the reported value of n represents the number of wells per condition.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical tests and parameters including the definitions and exact values of n’s are reported in the corresponding Figure Legends. Results were considered as significant if p < 0.05. All data are reported as mean ± SEM unless otherwise stated.
We classified an independent biological replicate as an independent hiPSC differentiation. The average of individual values of each differentiation was shown as a dot in the graphs. For each statistical analysis, we first evaluated if datasets fit into Gaussian distribution using the D’Agostino-Pearson omnibus normality test. Accordingly, we utilized a student’s t test (parametric) or Kolmogórov–Smirnov test (non-parametric) to compare two experimental conditions. To compare ≥ 3 experimental conditions, one-way analysis of variance (ANOVA) followed by a Tukey post hoc test (parametric), or Kruskal-Wallis one-way analysis of variance (non-parametric) was performed. We also used repeated-measure two-way ANOVA followed by Tukey’s multiple comparison test for multipoint comparisons.
We tested each biological replicate within each differentiation for differences in variance by the Brown-Forsythe test. In cases where biological replicates tested negative, data from independent biological replicates were pooled per experimental condition and tested for significance. In cases where biological replicates tested positive, each independent differentiation was tested for significant differences separately. In such cases, we deemed a biological change as significant only if all independent biological replicates tested were significant, and we display in the corresponding figure the least significant, i.e., the highest p value from all comparisons.
Contingency table analysis was used to compare distinct electrophysiological categories between hiPSC-derived MNs cultured on PA2 or Laminin, and cumulative distribution analysis was used to determine sEPSC amplitude differences between both conditions. The distinct statistical methods applied, and p values from all main figures are:
(B) t test, ****P<0.0001.
(I) ANOVA followed by Tukey’s multiple comparison test; ****P<0.0001.
(L) ANOVA followed by Tukey’s multiple comparison test; **P<0.01, ****P<0.0001.
(M) ANOVA followed by Tukey’s multiple comparison test; ****P<0.0001.
(F) ANOVA followed by Tukey’s multiple comparison test (anti-ITGB1) or Kruskal-Wallis followed by Dunn’s multiple comparison test (anti-ITGB4); *P <0.05 and ****P<0.0001.
(H) ANOVA followed by Tukey’s multiple comparison test; ****P<0.0001.
(J) ANOVA followed by Tukey’s multiple comparison test; *P <0.05, **P<0.01, and ***P<0.001.
(L) ANOVA followed by Tukey’s multiple comparison test (ChAT/TUJ1, ISL1/2/TUJ1, FOXA2/DAPI and the number of cells per mm2) or Kruskal-Wallis followed by Dunn’s multiple comparison test (TUJ1/DAPI and FOXA2/Ki67); *P <0.05, ***P<0.001, and ****P<0.0001; ns=not significant.
(C) t test; ****P<0.0001; ns: no significant differences.
(E) ANOVA followed by Tukey’s multiple comparison test; **P <0.01, ***P<0.001, and ****P<0.0001.
(B) ANOVA followed by Tukey’s multiple comparison test; **P<0.01, ****P<0.0001.
(G) ANOVA followed by Tukey’s multiple comparison test; *P<0.05, **P<0.01.
(H) ANOVA followed by Tukey’s multiple comparison test; *P<0.05, **P<0.01, ***P<0.001.
(I) ANOVA followed by Tukey’s multiple comparison test; *P<0.05, ***P<0.001.
(J) Two-way ANOVA followed by Tukey’s multiple comparison test; **P<0.01.
(B) ANOVA followed by Tukey’s multiple comparison test; *P <0.05, **P <0.01, ***P<0.001, and ****P<0.0001.
(H) Kolmogórov-Smirnov t test; *P <0.05 (outside histogram); t test; ***P<0.001 (inside dot plot).
(I) Kolmogórov-Smirnov t test; *P <0.05 (outside histogram); t test; ****P<0.0001 (inside dot plot).
(M) ANOVA followed by Tukey’s multiple comparison test; *P <0.05, **P <0.01, ***P<0.001, ****P<0.0001.
(E) ANOVA followed by Tukey’s multiple comparison test; *P <0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
(J) ANOVA followed by Tukey’s multiple comparison test; *P <0.05 and **P<0.01.
(C) ANOVA followed by Tukey’s multiple comparison test; ****P<0.0001.
(E) ANOVA followed by Tukey’s multiple comparison test; *P<0.05; ****P<0.0001.
All the statistical analyses were performed using GraphPad Prism 7 software.
Supplementary Material
Supplementary Movie 1, related to Figure 2. hiPSC-derived MNs on IKVAV-PA2 coatings. Live imaging of human MNs on IKVAV-PA2 coatings during the first 63 h in culture. Images were captured once every 16 min using a Nikon BioStation IM-Q imaging system with a 1.3-megapixel cooled monochrome camera (40 × len) and processed using the Image J program. Total time = 63 h 44 min. Scale bar: 10 μm.
Supplementary Movie 2, related to Figure 2. hiPSC-derived MNs on commercial laminin coating. Live imaging of human MNs on commercial laminin coating during the first 63 h in culture. Images were captured once every 16 min using a Nikon BioStation IM-Q imaging system with a 1.3-megapixel cooled monochrome camera (40 × len) and processed using the Image J program. Total time = 63 h 44 min. Scale bar: 10 μm.
Supplementary Movie 3, related to Figure 2. Schematic video of the high mobile IKVAV-PA2 nanofibers interacting with ITGB1 on the surface of a human MNs.
Supplementary Movie 4, related to Figure 2. Schematic video of the slow mobile IKVAV-PA1 nanofibers interacting with ITGB1 on the surface of a human MNs.
Supplementary Movie 4, related to Figure 2. Schematic video of the slow mobile IKVAV-PA1 nanofibers interacting with ITGB1 on the surface of a human MNs.
Table S1, related to Figure 3. Normalized TMT-labeling data acquired by LC-MS/MS of hiPSC-derived MNs cultured on commercial laminin and IKVAV-PA2 for 45 days.
Table S2, related to Figure 4. Electrophysiological properties of hiPSC-derived neurons cultured for >25 days in IKVAV-PA2 and laminin coatings, and comparison with previously published data of MNs cocultured with mouse astrocytes.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACTIN | Sigma-Aldrich | Cat# A2066 RRID:AB_476693 |
| Alexa Donkey anti-Chicken IgG (H+L) 647 | Thermo Fisher Scientific | Cat# A78952 RRID:AB_2921074 |
| Alexa Donkey anti-Goat IgG (H+L) 488 | Thermo Fisher Scientific | Cat# A11055 RRID:AB_2534102 |
| Alexa Donkey anti-Goat IgG (H+L) 555 | Thermo Fisher Scientific | Cat# A21432 RRID:AB_2535853 |
| Alexa Donkey anti-Mouse IgG (H+L) 488 | Thermo Fisher Scientific | Cat# A210202 RRID:AB_141607 |
| Alexa Donkey anti-Mouse IgG (H+L) 555 | Thermo Fisher Scientific | Cat# A31570 RRID:AB_2536180 |
| Alexa Donkey anti-Mouse IgG (H+L) 647 | Thermo Fisher Scientific | Cat# A31571 RRID:AB_162542 |
| Alexa Donkey anti-Rabbit IgG (H+L) 488 | Thermo Fisher Scientific | Cat# A21206 RRID:AB_2535792 |
| Alexa Donkey anti-Rabbit IgG (H+L) 555 | Thermo Fisher Scientific | Cat# A31572 RRID:AB_162543 |
| ChAT | Millipore | Cat#AB144P RRID:AB_2079751 |
| HRP Donkey anti-Goat IgG (H+L) Secondary Antibody | Thermo Fisher Scientific | Cat# PA1-28664 RRID:AB_10990162 |
| HRP Goat anti-Mouse IgG (H+L) Secondary Antibody | Thermo Fisher Scientific | Cat# 31430 RRID:AB_228307 |
| HRP Goat anti-Rabbit IgG (H+L) Secondary Antibody | Thermo Fisher Scientific | Cat# 31460 RRID:AB_228341 |
| ITGB1 | Millipore | Cat#MAB2079Z RRID:AB_2233964 |
| ITGB4 | Millipore | Cat#MAB1946 RRID:AB_2924963 |
| FAK | Cell Signaling | Cat#3285S RRID:AB_2269034 |
| FOXA2 | Santa Cruz Biotechnology | Cat#sc-101060 RRID:AB_1124660 |
| GABA | Sigma-Aldrich | Cat#A2052 |
| GFP | Abcam | Cat#ab6673 |
| ILK | Cell Signaling | Cat#3862 |
| ISL1/2 | DSHB | Cat# 39.4D5, RRID:AB_2314683 |
| Ki67 | Abcam | Cat#ab66155 RRID:AB_1140752 |
| Laminin alpha-1 | Santa Cruz Biotechnology | Cat#sc-74417 RRID:AB_1125087 |
| MAP2 | Biolegend | Cat#840601 RRID:AB_2565455 |
| NEUN | Biolegend | Cat#834502 RRID:AB_2734601 |
| p-FAK | Cell Signaling | Cat#3281S RRID:AB_331079 |
| PSD95 | NeuroMab | Cat#75028 RRID:AB_2292909 |
| TUJ1 | Biolegened | Cat#802001 RRID:AB_2564645 |
| SOD1 | Proteintech | Cat#10269-1-AP RRID:AB_2193750 |
| SYN1 | Millipore | Cat#AB1543 RRID:AB_2200400 |
| SYP | Abcam | Cat#ab32127 RRID:AB_2286949 |
| Ubiquitin (UB) | Santa Cruz Biotechnology | Cat#sc-8017 RRID:AB_628423 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C737303 |
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C404010 |
| HIV-1-based lentivirus | This paper | Zufferey et al., 1998 [123] |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | Innovative Cell Technologies | Cat# AT 104-500 |
| (3-Aminopropyl)triethoxysilane | Sigma-Aldrich | Cat# 440140 |
| Ascorbic Acid | Sigma-Aldrich | Cat# A4403 |
| B27 supplement | Thermo Fisher Scientific | Cat# 17504001 |
| BDNF | R&D Systems | Cat# 248-BD |
| CNTF | R&D Systems | Cat# 257NT/CF |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat# D1306 |
| DAPT | DNSK International | Cat# 208255-80-5 |
| Dimethylformamide | Sigma-Aldrich | Cat# RTC000098 |
| 1,6-Diphenyl-1,3,5-hexatriene, | Sigma-Aldrich | Cat# D208000 |
| DMEM/F12 | Thermo Fisher Scientific | Cat# 11320-082 |
| DNase (D2) | Worthington | Cat# LK003172 |
| Doxycycline | Sigma-Aldrich | Cat# D9891-5G |
| ECL (PierceTM ECL) substrate | Thermo Fisher Scientific | Cat# 32106 |
| ECL (Radiance ECL) substrate | Azure Biosystems | Cat# AC2204 |
| Epredia Immu-Mount | Thermo Fisher Scientific | Cat# 9990402 |
| FBS - HyClone | VWR | Cat# 16777-006 |
| Fibronectin | Thermo Fisher Scientific | Cat# PHE0023 |
| GDNF | R&D Systems | Cat# 212-GD |
| 45% glucose solution | Sigma-Aldrich | Cat# G8769-100ML |
| Glutamax | Thermo Fisher Scientific | Cat# 35050-061 |
| Glutaraldehyde | Fisher Scientific | Cat# 50-262-18 |
| Heparin Sulfate | Sigma Aldrich | Cat# H3149 |
| Halt protease and phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat# 78447 |
| IKVAV-PA1: C16VVAAEEEEGIKVAV | Alvarez et al, 2021 [48]. | N/A |
| IKVAV-PA2: C16AAGGEEEEGIKVAV | Alvarez et al, 2021 [48]. | N/A |
| IKVAV-PA3: C16VEVAAEEEEGIKVAV | This paper | N/A |
| IKVAV peptide | Olbrich et al, 1996 [48]. | N/A |
| Knockout DMEM | Thermo Fisher Scientific | Cat# 10829-018 |
| Knockout Replacement Serum | Thermo Fisher Scientific | Cat# 10828-010 |
| Laminin | Life Technologies | Cat# 23017-015 |
| Laminin 211 | BioLamina | Cat# LN211-0501 |
| Laminin 521 | BioLamina | Cat# LN521-05 |
| LDN-193189 | DNSK International | Cat# 1062368-24-4 |
| Matrigel | Corning | Cat# BD354277 |
| 2-Mercaptoethanol | Thermo Fisher Scientific | Cat# 21985-023 |
| Micro-90 detergent | Sigma-Aldrich | Cat# Z281565 |
| mTeSR1 media | STEMCELL | Cat# 85850 |
| N2 supplement | Thermo Fisher Scientific | Cat# 17502001 |
| NEAA | Fisher Scientific | Cat# 11140050 |
| Neurobasal | Thermo Fisher Scientific | Cat# 21103-049 |
| Normal Donkey Serum | Jackson Immuno Research | Cat# 017-000-001 |
| Paraformaldehyde 16% | Fisher Scientific | Cat# 50-980-487 |
| Polyethylenimine hydrochloride | Sigma-Aldrich | Cat# 764965 |
| 1X PBS (TC) | Fisher Scientific | Cat# MT21040CV |
| Poly-D-Lysine hydrobromide | Sigma-Aldrich | Cat# P6407 |
| Poly-DL-ornithine hydrobromide | Sigma-Aldrich | Cat# P0671 |
| Pen/Strep | Fisher Scientific | Cat# 45000-650 |
| Puromycin | Sigma-Aldrich | Cat# P8833 |
| Retinoic Acid | Sigma-Aldrich | Cat# R2625 |
| ROCK inhibitor (Y-27632) | DNSK International | Cat# 129830-38-2 |
| SB431542 | DNSK International | Cat# 301836-41-9 |
| Scr-PA1: C16VVAAEEEEGVVIAK | Alvarez et al, 2021 [48]. | N/A |
| Scr-PA2: C16AAGGEEEEGVVIAK | Alvarez et al, 2021 [48]. | N/A |
| Scr-PA3: C16VEVAAEEEEGVVIAK | This paper | N/A |
| Smoothened-Agonist (SAG) | DNSK International | Cat# 364590-63-6 |
| SU5402 | DNSK International | Cat# 215542-92-3 |
| 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid, sodium salt | Sigma-Aldrich | Cat# 269913 |
| Tetrahydrofuran (THF) | Sigma-Aldrich | Cat# 78445 |
| TrypLE Express | Thermo Fisher Scientific | Cat# 12604-013 |
| XAV939 | Stemgent | Cat# 04-00046 |
| Critical commercial assays | ||
| BCA protein assay | Thermo Fisher Scientific | Cat# 23225 |
| CytoTox 96 Non-Radioactive Cytotoxicity Assay | Promega | Cat# G1780 |
| HilyMax | Dojindo Molecular Technologies | Cat# H357-10 |
| Lipofectamine RNAiMAX | Invitrogen | Cat# 13778030 |
| Superscript IV Reverse Transcriptase | Invitrogen | Cat# 18090010 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725124 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| 10plex Tandem Mass Tag System | Thermo Fisher Scientific | Cat# 90110 |
| Deposited data | ||
| TMT LC-MS/MS data | This paper | Table S1 |
| Experimental models: Cell lines | ||
| Human iPSC line 18a | Boulting et al, 2011 [104]. Harvard University | RRID:CVCL_8993 |
| Human iPSC line 11a | Boulting et al, 2011 [104]. Harvard University. | RRID:CVCL_8987 |
| Human iPSC SOD1+/4AV (39b) | Kiskinis et al., 2014 [92]. Harvard University | RRID:CVCL_X725 |
| Human iPSC SOD1+/+ (39b-cor) | Kiskinis et al., 2014 [92]. Harvard University | N/A |
| HEK293-FT | ATCC | RRID:CVCL_6911 |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| TUBB3: GGCCAAGGGTCACTACACG (Forward); GCAGTCGCAGTTTTCACACTC (Reverse). | This paper | N/A |
| ISL1: AGATCAGCCTGCTTTTCAGC (Forward); TCATGCCTCAATAGGACTGG (Reverse) | This paper | N/A |
| MNX1: GAACACCAGTTCAAGCTCAACA (Forward); GCTGCGTTTCCATTTCATTCG (Reverse). | This paper | N/A |
| LHX3: GGAGAGCGTTTACTGCAAGGA (Forward); TTGGCGGTTTCGTAGTCCG (Reverse). | This paper | N/A |
| FOXA2: GGAGCAGCTACTATGCAGAGC (Forward); CGTGTTCATGCCGTTCATCC (Reverse). | This paper | N/A |
| GPI: GTGTACCTTCTAGTCCCGCC (Forward); GGTCAAGCTGAAGTGGTTGAAGC (Reverse). | This paper | N/A |
| RPL0: TCTACAACCCTGAAGTGCTTGAT (Forward); CAATCTGCAGACAGACACTGG (Reverse). | This paper | N/A |
| siITB1 (siRNA ID:S7576) | Thermo Fisher Scientific | Cat# 4390824 |
| siScramble | Thermo Fisher Scientific | Cat# 4390843 |
| Recombinant DNA | ||
| FUW-M2rtTA | Addgene | RRID:Addgene_20342 |
| Lentiviral GFP plasmid | This paper / VectorBuilder | VB181102-1195sys |
| Lentiviral ITGB1::GFP plasmid | This paper / VectorBuilder | VB211104-1470afj |
| pMD2.G | Addgene | RRID:Addgene_12259 |
| psPAX2 | Addgene | RRID:Addgene_12260 |
| pTet-O-Ngn2-Puro | Addgene | RRID:Addgene_52047 |
| Tet-O-FUW-EGFP | Addgene | RRID:Addgene_30130 |
| Software and algorithms | ||
| Adobe Photoshop 7.0 | Adobe Systems Incorporated | RRID:SCR_014199 |
| Avogadro | Open Molecules | RRID:SCR_015983 |
| Axion’s Integrated Studio (AxIS) software | Axion Biosystems | RRID:SCR_016308 |
| BioRender | BioRender | RRID:SCR_018361 |
| pCLAMP 11.1 (Clampex 11 software) | Molecular devices | RRID:SCR_011323 |
| Crystallography Data Analysis Software | Toby and Von Dreele, 2013 [124]. | https://subversion.xray.aps.anl.gov/trac/pyGSAS |
| Cytoscape / Metascape | Zhou et al., 2019 [125]. | RRID:SCR_016620 |
| Comparative GO | Fruzangohar et al., 2017 [126]. | https://www.comparativego.com |
| Database for Annotation, Visualization and Integrated Discovery (DAVID) | Huang et al., 2009 [127]. | RRID:SCR_001881 |
| Easy Electrophysiology | Easy Electrophysiology Ltd | RRID:SCR_021190 |
| Fiji | Schindelin et al., 2012 [128]. | RRID:SCR_002285 |
| GROMACS 5.0.4 Software | GROMACS development team | https://doi.org/10.5281/zenodo.7323393. |
| Imaris | Oxford Instruments Group | RRID:SCR_007370 |
| Martinize.py, 2.0 | Martini - General Purpose Coarse-Grained Force Field | http://cgmartini.nl/images/tools/martinize/martinize-2.6/martinize.py |
| MATLAB | The MathWorks, Inc. | RRID:SCR_001622 |
| Mx™ Software | Zygo Corporation | https://www.zygo.com |
| NanoScope analysis software | Bruker | http://nanoscaleworld.bruker-axs.com/nanoscaleworld/media/p/775.aspx |
| NIS-Elements Advanced Research version 5 | Nikon Instruments Inc. | RRID:SCR_002776 |
| Prism7 | GraphPad Software | RRID:SCR_002798 |
| Protein unfolding nano-indentation analysis software | Punias | punias@free.fr |
| Scaffold v5 | Proteome Software Inc. | RRID:SCR_014345 |
| Small Angle Scattering Analysis Software | SasView | https://www.sasview.org |
| VMD-Visual Molecular Dynamics | University of Illinois at Urbana-Champaign | https://www.ks.uiuc.edu/Research/vmd/ |
| WinFluor Software | University of Strathclyde | http://spider.science.strath.ac.uk/sipbs/software_winfluor.htm |
| Cinema 4D version R20 | Maxon Computer GMBH | https://www.maxon.net/es/downloads/cinema-4d-past-versions |
| Other | ||
| CD-1 IGS (Crl:CD1(ICR)) mice for primary glia cultures and spinal cord tissue | Charles River | Cat#22; RRID:IMSR_CRL:022 |
Highlights.
ECM-mimetic PAs with distinct non-bioactive domains show different molecular motion.
Culturing iPSC-neurons on highly mobile IKVAV-PA2 nanofibers enhances ITGB1 activation.
IKVAV-PA2 coatings reduce neuronal aggregation, increase functional maturity.
IKVAV-PA2 coatings facilitate modeling neurodegenerative pathology in vitro.
ACKNOWLEDGMENTS
We are grateful to the following funding sources: US National Institutes of Health (NIH) National Institute on Neurological Disorders and Stroke and NIH National Institute on Aging R01NS104219 (E.K), the Les Turner ALS Foundation (E.K), the New York Stem Cell Foundation (E.K), the NIH National Institute of Biomedical Imaging and Bioengineering 5R01EB003806 (S.I.S), NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health R01AR0727 (S.I.S), the Center for Regenerative Nanomedicine at the Simpson Querrey Institute (S.I.S), U.S. Department of Energy DE-SC0001329 (S.I.S), Grant PID2021–124839OA-I00 and PID2020–114407RA-I00 funded by MCIN/ AEI/10.13039/501100011033 (Z.A. and J.A.O), Mike Lane fellowship from Castellers de Vila de Gracia 2021 (Z.A), AFM-Telethon Trampoline Grant #23648 (J.A.O), Beatriu de Pinos 2019 BP 00058 (Z.A), Ramon y Cajal fellowship RYC2020–028732-I (Z.A) and RYC2019–026980-I (J.A.O), Gipuzkoa Foru Aldundia 2019-FELL-000017–01 and Maria de Maeztu Units of Excellence MDM-2017–0720 (I.R.S), and NINDS R01 NS104436 (K.A.Q). We thank the Peptide Synthesis Core Facility and the Analytical BioNanoTechnology Core Facility, supported by the SHyNE Resource (NSF ECCS-2025633), the Center for Advanced Molecular Imaging (NCI CCSG P30 CA060553), the Proteomics Core Facility (NCI CCSG P30 CA060553), the National Resource for Translational and Developmental Proteomics (P41 GM108569), the Materials Research Science and Engineering Center (NSF DMR-1720139), the REACT Facility of the Northwestern University Center for Catalysis and Surface Science, Prof. N. Schweitzer for his assistance with the physisorption measurements, the SasView application developed under NSF award DMR-0520547 and under the SINE2020 project (#654000), the Keck Biophysics Facility (Grant #P30 CA060553); Northwestern University Center for Advanced Microscopy (NCI CCSG P30 CA060553), the Scanned Probe Imaging and Development, BioCryo, and Micro/Nano Fabrication facilities of NU’s NUANCE Center (NSF ECCS-2025633), the IIN and Northwestern’s MRSEC program (NSF DMR-1720139), the IBEC Core Facility and Microscopy Characterization Facility, Unit 7 of ICTS “NANBIOSIS” from CIBER-BBN both at Institute for Bioengineering of Catalonia. We thank M. Ziller for assisting with gene expression analysis and M. Seniw for the preparation of graphic illustrations. E.K is a New York Stem Cell Foundation – Robertson Investigator.
Footnotes
DECLARATION OF INTEREST
A patent pertaining to this work has been filed and is pending: “Dynamics Within Supramolecular IKVAV Matrices Enhance Functional Maturation of Human hiPSCs-Derived Neurons and Regeneration” (inventors: Z.A., J.A.O., K.S., E.K. and S.I.S.). E.K., N.A.S. and S.I.S. are co-founders of NeuronGrow.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ichida JK, and Kiskinis E (2015). Probing disorders of the nervous system using reprogramming approaches. Embo j 34, 1456–1477. 10.15252/embj.201591267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu YY, Chiu FL, Yeh CS, and Kuo HC (2019). Opportunities and challenges for the use of induced pluripotent stem cells in modelling neurodegenerative disease. Open Biol 9, 180177. 10.1098/rsob.180177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soliman MA, Aboharb F, Zeltner N, and Studer L (2017). Pluripotent stem cells in neuropsychiatric disorders. Mol Psychiatry 22, 1241–1249. 10.1038/mp.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagers AJ (2012). The stem cell niche in regenerative medicine. Cell Stem Cell 10, 362–369. 10.1016/j.stem.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Sprenger CC, Plymate SR, and Reed MJ (2010). Aging-related alterations in the extracellular matrix modulate the microenvironment and influence tumor progression. International Journal of Cancer. Journal International du Cancer 127, 2739–2748. 10.1002/ijc.25615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry BJ, Smith AST, Young JE, and Mack DL (2018). Advances and Current Challenges Associated with the Use of Human Induced Pluripotent Stem Cells in Modeling Neurodegenerative Disease. Cells Tissues Organs 205, 331–349. 10.1159/000493018. [DOI] [PubMed] [Google Scholar]
- 7.Barros CS, Franco SJ, and Müller U (2011). Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol 3, a005108. 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long KR, and Huttner WB (2019). How the extracellular matrix shapes neural development. Open Biol 9, 180216. 10.1098/rsob.180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sood D, Cairns DM, Dabbi JM, Ramakrishnan C, Deisseroth K, Black LD 3rd, Santaniello S, and Kaplan DL (2019). Functional maturation of human neural stem cells in a 3D bioengineered brain model enriched with fetal brain-derived matrix. Sci Rep 9, 17874. 10.1038/s41598-019-54248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauth S, Grevesse T, Pantazopoulos H, Campbell PH, Maoz BM, Berretta S, and Parker KK (2016). Extracellular matrix protein expression is brain region dependent. J Comp Neurol 524, 1309–1336. 10.1002/cne.23965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau LW, Cua R, Keough MB, Haylock-Jacobs S, and Yong VW (2013). Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat Rev Neurosci 14, 722–729. 10.1038/nrn3550. [DOI] [PubMed] [Google Scholar]
- 12.Benarroch EE (2015). Extracellular matrix in the CNS: Dynamic structure and clinical correlations. Neurology 85, 1417–1427. 10.1212/WNL.0000000000002044. [DOI] [PubMed] [Google Scholar]
- 13.Yue B (2014). Biology of the extracellular matrix: an overview. J Glaucoma 23, S20–23. 10.1097/IJG.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boekhoven J, and Stupp SI (2014). 25th anniversary article: supramolecular materials for regenerative medicine. Adv Mater 26, 1642–1659. 10.1002/adma.201304606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gattazzo F, Urciuolo A, and Bonaldo P (2014). Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta 1840, 2506–2519. 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daley WP, Peters SB, and Larsen M (2008). Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 121, 255–264. 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 17.Nirwane A, and Yao Y (2018). Laminins and their receptors in the CNS. Biol Rev Camb Philos Soc. 10.1111/brv.12454. [DOI] [PubMed] [Google Scholar]
- 18.Frantz C, Stewart KM, and Weaver VM (2010). The extracellular matrix at a glance. J Cell Sci 123, 4195–4200. 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorne JT, Segal TR, Chang S, Jorge S, Segars JH, and Leppert PC (2015). Dynamic reciprocity between cells and their microenvironment in reproduction. Biol Reprod 92, 25. 10.1095/biolreprod.114.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrukh A, Ortega F, Fan W, Marichal N, Paez JI, Berninger B, Campo AD, and Salierno MJ (2017). Bifunctional Hydrogels Containing the Laminin Motif IKVAV Promote Neurogenesis. Stem Cell Reports 9, 1432–1440. 10.1016/j.stemcr.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth JG, Huang MS, Li TL, Feig VR, Jiang Y, Cui B, Greely HT, Bao Z, Paşca SP, and Heilshorn SC (2021). Advancing models of neural development with biomaterials. Nature Reviews Neuroscience 22, 593–615. 10.1038/s41583-021-00496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aisenbrey EA, and Murphy WL (2020). Synthetic alternatives to Matrigel. Nature Reviews Materials 5, 539–551. 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong L, Cao L, Shen Z, Shao L, Gao S, Zhang C, Lu J, and Li W (2018). Materials for Neural Differentiation, Trans-Differentiation, and Modeling of Neurological Disease. Adv Mater 30, e1705684. 10.1002/adma.201705684. [DOI] [PubMed] [Google Scholar]
- 24.Lutolf MP, Gilbert PM, and Blau HM (2009). Designing materials to direct stem-cell fate. Nature 462, 433–441. 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engler AJ, Sen S, Sweeney HL, and Discher DE (2006). Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 126, 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 26.Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, and Lutolf MP (2016). Neural tube morphogenesis in synthetic 3D microenvironments. Proc Natl Acad Sci U S A 113, E6831–e6839. 10.1073/pnas.1603529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long KR, Newland B, Florio M, Kalebic N, Langen B, Kolterer A, Wimberger P, and Huttner WB (2018). Extracellular Matrix Components HAPLN1, Lumican, and Collagen I Cause Hyaluronic Acid-Dependent Folding of the Developing Human Neocortex. Neuron 99, 702–719 e706. 10.1016/j.neuron.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Simão D, Silva MM, Terrasso AP, Arez F, Sousa MFQ, Mehrjardi NZ, Šarić T, Gomes-Alves P, Raimundo N, Alves PM, and Brito C (2018). Recapitulation of Human Neural Microenvironment Signatures in iPSC-Derived NPC 3D Differentiation. Stem Cell Reports 11, 552–564. 10.1016/j.stemcr.2018.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stupp SI, LeBonheur V, Walker K, Li LS, Huggins KE, Keser M, and Amstutz A (1997). Supramolecular Materials: Self-Organized Nanostructures. Science 276, 384–389. 10.1126/science.276.5311.384. [DOI] [PubMed] [Google Scholar]
- 30.Cui H, Webber MJ, and Stupp SI (2010). Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers 94, 1–18. 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aida T, Meijer EW, and Stupp SI (2012). Functional supramolecular polymers. Science 335, 813–817. 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortony JH, Newcomb CJ, Matson JB, Palmer LC, Doan PE, Hoffman BM, and Stupp SI (2014). Internal dynamics of a supramolecular nanofibre. Nat Mater 13, 812–816. 10.1038/nmat3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freeman R, Han M, Alvarez Z, Lewis JA, Wester JR, Stephanopoulos N, McClendon MT, Lynsky C, Godbe JM, Sangji H, et al. (2018). Reversible self-assembly of superstructured networks. Science 362, 808–813. 10.1126/science.aat6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berns EJ, Sur S, Pan L, Goldberger JE, Suresh S, Zhang S, Kessler JA, and Stupp SI (2014). Aligned neurite outgrowth and directed cell migration in self-assembled monodomain gels. Biomaterials 35, 185–195. 10.1016/j.biomaterials.2013.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edelbrock AN, Àlvarez Z, Simkin D, Fyrner T, Chin SM, Sato K, Kiskinis E, and Stupp SI (2018). Supramolecular Nanostructure Activates TrkB Receptor Signaling of Neuronal Cells by Mimicking Brain-Derived Neurotrophic Factor. Nano Lett 18, 6237–6247. 10.1021/acs.nanolett.8b02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SS, Fyrner T, Chen F, Alvarez Z, Sleep E, Chun DS, Weiner JA, Cook RW, Freshman RD, Schallmo MS, et al. (2017). Sulfated glycopeptide nanostructures for multipotent protein activation. Nat Nanotechnol 12, 821–829. 10.1038/nnano.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuoka AJ, Morrissey ZD, Zhang C, Homma K, Belmadani A, Miller CA, Chadly DM, Kobayashi S, Edelbrock AN, Tanaka-Matakatsu M, et al. (2017). Directed Differentiation of Human Embryonic Stem Cells Toward Placode-Derived Spiral Ganglion-Like Sensory Neurons. Stem Cells Transl Med 6, 923–936. 10.1002/sctm.16-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sleep E, Cosgrove BD, McClendon MT, Preslar AT, Chen CH, Sangji MH, Perez CMR, Haynes RD, Meade TJ, Blau HM, and Stupp SI (2017). Injectable biomimetic liquid crystalline scaffolds enhance muscle stem cell transplantation. Proc Natl Acad Sci U S A 114, E7919–E7928. 10.1073/pnas.1708142114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webber MJ, Tongers J, Newcomb CJ, Marquardt KT, Bauersachs J, Losordo DW, and Stupp SI (2011). Supramolecular nanostructures that mimic VEGF as a strategy for ischemic tissue repair. Proc Natl Acad Sci U S A 108, 13438–13443. 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubert Perez CM, Alvarez Z, Chen F, Aytun T, and Stupp SI (2017). Mimicking the Bioactivity of Fibroblast Growth Factor-2 Using Supramolecular Nanoribbons. ACS Biomater Sci Eng 3, 2166–2175. 10.1021/acsbiomaterials.7b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartgerink JD, Beniash E, and Stupp SI (2001). Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 294, 1684–1688. 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 42.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, and Stupp SI (2004). Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science 303, 1352–1355. 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 43.Tashiro K, Sephel GC, Weeks B, Sasaki M, Martin GR, Kleinman HK, and Yamada Y (1989). A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. J Biol Chem 264, 16174–16182. [PubMed] [Google Scholar]
- 44.Goldberger JE, Berns EJ, Bitton R, Newcomb CJ, and Stupp SI (2011). Electrostatic control of bioactivity. Angew Chem Int Ed Engl 50, 6292–6295. 10.1002/anie.201100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, and Launey T (2012). A hybrid nanofiber matrix to control the survival and maturation of brain neurons. Biomaterials 33, 545–555. 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tysseling VM, Sahni V, Pashuck ET, Birch D, Hebert A, Czeisler C, Stupp SI, and Kessler JA (2010). Self-assembling peptide amphiphile promotes plasticity of serotonergic fibers following spinal cord injury. J Neurosci Res 88, 3161–3170. 10.1002/jnr.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, and Kessler JA (2008). Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J Neurosci 28, 3814–3823. 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Álvarez Z, Kolberg-Edelbrock AN, Sasselli IR, Ortega JA, Qiu R, Syrgiannis Z, Mirau PA, Chen F, Chin SM, Weigand S, et al. (2021). Bioactive scaffolds with enhanced supramolecular motion promote recovery from spinal cord injury. Science 374, 848–856. doi: 10.1126/science.abh3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newcomb CJ, Sur S, Lee SS, Yu JM, Zhou Y, Snead ML, and Stupp SI (2016). Supramolecular Nanofibers Enhance Growth Factor Signaling by Increasing Lipid Raft Mobility. Nano Lett 16, 3042–3050. 10.1021/acs.nanolett.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pashuck ET, Cui H, and Stupp SI (2010). Tuning Supramolecular Rigidity of Peptide Fibers through Molecular Structure. Journal of the American Chemical Society 132, 6041–6046. 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.da Silva RM, van der Zwaag D, Albertazzi L, Lee SS, Meijer EW, and Stupp SI (2016). Super-resolution microscopy reveals structural diversity in molecular exchange among peptide amphiphile nanofibres. Nat Commun 7, 11561. 10.1038/ncomms11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boekhoven J, Rubert Pérez CM, Sur S, Worthy A, and Stupp SI (2013). Dynamic Display of Bioactivity through Host–Guest Chemistry. Angewandte Chemie International Edition 52, 12077–12080. 10.1002/anie.201306278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper JG, Sicard D, Sharma S, Van Gulden S, McGuire TL, Cajiao MP, Tschumperlin DJ, and Kessler JA (2020). Spinal Cord Injury Results in Chronic Mechanical Stiffening. J Neurotrauma 37, 494–506. 10.1089/neu.2019.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng PN, Pham JD, and Nowick JS (2013). The supramolecular chemistry of beta-sheets. J Am Chem Soc 135, 5477–5492. 10.1021/ja3088407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossi AM, and Taylor CW (2011). Analysis of protein-ligand interactions by fluorescence polarization. Nature Protocols 6, 365–387. 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser RD, and London E (1998). Location of Diphenylhexatriene (DPH) and Its Derivatives within Membranes: Comparison of Different Fluorescence Quenching Analyses of Membrane Depth. Biochemistry 37, 8180–8190. 10.1021/bi980064a. [DOI] [PubMed] [Google Scholar]
- 57.Lakowicz JR (2006). Principles of Fluorescence Spectroscopy. 3 ed. Springer US. [Google Scholar]
- 58.Lee O-S, Cho V, and Schatz GC (2012). Modeling the Self-Assembly of Peptide Amphiphiles into Fibers Using Coarse-Grained Molecular Dynamics. Nano Letters 12, 4907–4913. 10.1021/nl302487m. [DOI] [PubMed] [Google Scholar]
- 59.Frederix PW, Scott GG, Abul-Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, and Tuttle T (2015). Exploring the sequence space for (tri-)peptide self-assembly to design and discover new hydrogels. Nat Chem 7, 30–37. 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- 60.Guo C, Arnon ZA, Qi R, Zhang Q, Adler-Abramovich L, Gazit E, and Wei G (2016). Expanding the Nanoarchitectural Diversity Through Aromatic Di-and Tri-Peptide Coassembly: Nanostructures and Molecular Mechanisms. ACS Nano 10, 8316–8324. [DOI] [PubMed] [Google Scholar]
- 61.Mazza M, Notman R, Anwar J, Rodger A, Hicks M, Parkinson G, McCarthy D, Daviter T, Moger J, and Garrett N (2013). Nanofiber-based delivery of therapeutic peptides to the brain. ACS Nano 7, 1016–1026. [DOI] [PubMed] [Google Scholar]
- 62.de Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schäfer LV, Periole X, Tieleman DP, and Marrink SJ (2013). Improved Parameters for the Martini Coarse-Grained Protein Force Field. Journal of Chemical Theory and Computation 9, 687–697. 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 63.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, and de Vries AH (2007). The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. The Journal of Physical Chemistry B 111, 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 64.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, and Marrink S-J (2008). The MARTINI Coarse-Grained Force Field: Extension to Proteins. Journal of Chemical Theory and Computation 4, 819–834. 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 65.Ziller MJ, Ortega JA, Quinlan KA, Santos DP, Gu H, Martin EJ, Galonska C, Pop R, Maidl S, Di Pardo A, et al. (2018). Dissecting the Functional Consequences of De Novo DNA Methylation Dynamics in Human Motor Neuron Differentiation and Physiology. Cell Stem Cell 22, 559–574 e559. 10.1016/j.stem.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freitas VM, Vilas-Boas VF, Pimenta DC, Loureiro V, Juliano MA, Carvalho MR, Pinheiro JJ, Camargo AC, Moriscot AS, Hoffman MP, and Jaeger RG (2007). SIKVAV, a laminin alpha1-derived peptide, interacts with integrins and increases protease activity of a human salivary gland adenoid cystic carcinoma cell line through the ERK 1/2 signaling pathway. Am J Pathol 171, 124–138. 10.2353/ajpath.2007.051264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caniggia I, Liu J, Han R, Wang J, Tanswell AK, Laurie G, and Post M (1996). Identification of receptors binding fibronectin and laminin on fetal rat lung cells. Am J Physiol 270, L459–468. 10.1152/ajplung.1996.270.3.L459. [DOI] [PubMed] [Google Scholar]
- 68.Pan L, North HA, Sahni V, Jeong SJ, McGuire TL, Berns EJ, Stupp SI, and Kessler JA (2014). beta1-Integrin and integrin linked kinase regulate astrocytic differentiation of neural stem cells. PLoS One 9, e104335. 10.1371/journal.pone.0104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Liu X, Josey B, Chou CJ, Tan Y, Zhang N, and Wen X (2014). Short laminin peptide for improved neural stem cell growth. Stem Cells Transl Med 3, 662–670. 10.5966/sctm.2013-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stendahl JC, Rao MS, Guler MO, and Stupp SI (2006). Intermolecular Forces in the Self-Assembly of Peptide Amphiphile Nanofibers. Advanced Functional Materials 16, 499–508. 10.1002/adfm.200500161. [DOI] [Google Scholar]
- 71.Greenfield MA, Hoffman JR, de la Cruz MO, and Stupp SI (2010). Tunable mechanics of peptide nanofiber gels. Langmuir 26, 3641–3647. 10.1021/la9030969. [DOI] [PubMed] [Google Scholar]
- 72.Franco SJ, and Müller U (2011). Extracellular matrix functions during neuronal migration and lamination in the mammalian central nervous system. Developmental neurobiology 71, 889–900. 10.1002/dneu.20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmid RS, Shelton S, Stanco A, Yokota Y, Kreidberg JA, and Anton ES (2004). alpha3beta1 integrin modulates neuronal migration and placement during early stages of cerebral cortical development. Development 131, 6023–6031. 10.1242/dev.01532. [DOI] [PubMed] [Google Scholar]
- 74.Fujioka T, Kaneko N, Ajioka I, Nakaguchi K, Omata T, Ohba H, Fassler R, Garcia-Verdugo JM, Sekiguchi K, Matsukawa N, and Sawamoto K (2017). beta1 integrin signaling promotes neuronal migration along vascular scaffolds in the post-stroke brain. EBioMedicine 16, 195–203. 10.1016/j.ebiom.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marchetti G, Escuin S, van der Flier A, De Arcangelis A, Hynes RO, and Georges-Labouesse E (2010). Integrin alpha5beta1 is necessary for regulation of radial migration of cortical neurons during mouse brain development. Eur J Neurosci 31, 399–409. 10.1111/j.1460-9568.2009.07072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colognato H, and Tzvetanova ID (2011). Glia unglued: how signals from the extracellular matrix regulate the development of myelinating glia. Dev Neurobiol 71, 924–955. 10.1002/dneu.20966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gardiner NJ (2011). Integrins and the extracellular matrix: key mediators of development and regeneration of the sensory nervous system. Dev Neurobiol 71, 1054–1072. 10.1002/dneu.20950. [DOI] [PubMed] [Google Scholar]
- 78.Kazanis I, and ffrench-Constant C (2011). Extracellular matrix and the neural stem cell niche. Dev Neurobiol 71, 1006–1017. 10.1002/dneu.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Myers JP, Santiago-Medina M, and Gomez TM (2011). Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Dev Neurobiol 71, 901–923. 10.1002/dneu.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park YK, and Goda Y (2016). Integrins in synapse regulation. Nat Rev Neurosci 17, 745–756. 10.1038/nrn.2016.138. [DOI] [PubMed] [Google Scholar]
- 81.Novitch BG, and Butler SJ (2009). Reducing the mystery of neuronal differentiation. Cell 138, 1062–1064. 10.1016/j.cell.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Kurklinsky S, Chen J, and McNiven MA (2011). Growth cone morphology and spreading are regulated by a dynamin-cortactin complex at point contacts in hippocampal neurons. J Neurochem 117, 48–60. 10.1111/j.1471-4159.2011.07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma L, Yu Y-M, Guo Y, Hart RP, and Schachner M (2012). Cysteine- and glycine-rich protein 1a is involved in spinal cord regeneration in adult zebrafish. Eur J Neurosci 35, 353–365. 10.1111/j.1460-9568.2011.07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carrascal L, Nieto-Gonzalez JL, Cameron WE, Torres B, and Nunez-Abades PA (2005). Changes during the postnatal development in physiological and anatomical characteristics of rat motoneurons studied in vitro. Brain Res Brain Res Rev 49, 377–387. 10.1016/j.brainresrev.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 85.Tadros MA, Lim R, Hughes DI, Brichta AM, and Callister RJ (2015). Electrical maturation of spinal neurons in the human fetus: comparison of ventral and dorsal horn. J Neurophysiol 114, 2661–2671. 10.1152/jn.00682.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muratore CR, Srikanth P, Callahan DG, and Young-Pearse TL (2014). Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS One 9, e105807. 10.1371/journal.pone.0105807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR, and Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. 10.1038/nature24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krencik R, Seo K, van Asperen JV, Basu N, Cvetkovic C, Barlas S, Chen R, Ludwig C, Wang C, Ward ME, et al. (2017). Systematic Three-Dimensional Coculture Rapidly Recapitulates Interactions between Human Neurons and Astrocytes. Stem Cell Reports 9, 1745–1753. 10.1016/j.stemcr.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, Barrett LE, Limone F, Worringer KA, Kommineni S, et al. (2018). Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Rep 23, 2509–2523. 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor JP, Brown RH Jr., and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, et al. (2014). Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 14, 781–795. 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, et al. (2014). Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep 7, 1–11. 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiskinis E, Kralj JM, Zou P, Weinstein EN, Zhang H, Tsioras K, Wiskow O, Ortega JA, Eggan K, and Cohen AE (2018). All-Optical Electrophysiology for High-Throughput Functional Characterization of a Human iPSC-Derived Motor Neuron Model of ALS. Stem cell reports 10, 1991–2004. 10.1016/j.stemcr.2018.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nicolas J, Magli S, Rabbachin L, Sampaolesi S, Nicotra F, and Russo L (2020). 3D Extracellular Matrix Mimics: Fundamental Concepts and Role of Materials Chemistry to Influence Stem Cell Fate. Biomacromolecules. 10.1021/acs.biomac.0c00045. [DOI] [PubMed] [Google Scholar]
- 96.Ramadurai S, Holt A, Krasnikov V, van den Bogaart G, Killian JA, and Poolman B (2009). Lateral Diffusion of Membrane Proteins. Journal of the American Chemical Society 131, 12650–12656. 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 97.Weiß K, Neef A, Van Q, Kramer S, Gregor I, and Enderlein J (2013). Quantifying the Diffusion of Membrane Proteins and Peptides in Black Lipid Membranes with 2-Focus Fluorescence Correlation Spectroscopy. Biophysical Journal 105, 455–462. 10.1016/j.bpj.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Changede R, and Sheetz M (2017). Integrin and cadherin clusters: A robust way to organize adhesions for cell mechanics. Bioessays 39, 1–12. 10.1002/bies.201600123. [DOI] [PubMed] [Google Scholar]
- 99.Sun Z, Costell M, and Fassler R (2019). Integrin activation by talin, kindlin and mechanical forces. Nat Cell Biol 21, 25–31. 10.1038/s41556-018-0234-9. [DOI] [PubMed] [Google Scholar]
- 100.Vera E, and Studer L (2015). When rejuvenation is a problem: challenges of modeling late-onset neurodegenerative disease. Development 142, 3085–3089. 10.1242/dev.120667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morawski M, Filippov M, Tzinia A, Tsilibary E, and Vargova L (2014). Chapter 10 - ECM in brain aging and dementia. In Progress in Brain Research, Dityatev A, Wehrle-Haller B, and Pitkänen A, eds. (Elsevier; ), pp. 207–227. 10.1016/B978-0-444-63486-3.00010-4. [DOI] [PubMed] [Google Scholar]
- 102.Birch HL (2018). Extracellular Matrix and Ageing. Subcell Biochem 90, 169–190. 10.1007/978-981-13-2835-0_7. [DOI] [PubMed] [Google Scholar]
- 103.Ortega JA, and Alcantara S (2010). BDNF/MAPK/ERK-induced BMP7 expression in the developing cerebral cortex induces premature radial glia differentiation and impairs neuronal migration. Cereb Cortex 20, 2132–2144. 10.1093/cercor/bhp275. [DOI] [PubMed] [Google Scholar]
- 104.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. (2011). A functionally characterized test set of human induced pluripotent stem cells. Nature biotechnology 29, 279–286. 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Olbrich KC, Andersen TT, Blumenstock FA, and Bizios R (1996). Surfaces modified with covalently-immobilized adhesive peptides affect fibroblast population motility. Biomaterials 17, 759–764. 10.1016/0142-9612(96)81412-8. [DOI] [PubMed] [Google Scholar]
- 106.Weigand SJ, and Keane DT (2011). DND-CAT’s new triple area detector system for simultaneous data collection at multiple length scales. Nucl Instrum Meth A 649, 61–63. 10.1016/j.nima.2010.12.045. [DOI] [Google Scholar]
- 107.Toby BH, and Von Dreele RB (2013). GSAS-II: the genesis of a modern open-source all purpose crystallography software package. J Appl Crystallogr 46, 544–549. 10.1107/S0021889813003531. [DOI] [Google Scholar]
- 108.Palmer AG 3rd (1993). Dynamic properties of proteins from NMR spectroscopy. Curr Opin Biotechnol 4, 385–391. 10.1016/0958-1669(93)90002-e. [DOI] [PubMed] [Google Scholar]
- 109.Neugirg BR, Koebley SR, Schniepp HC, and Fery A (2016). AFM-based mechanical characterization of single nanofibres. Nanoscale 8, 8414. 10.1039/c6nr00863a. [DOI] [PubMed] [Google Scholar]
- 110.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, and Hutchison GR (2012). Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4, 17. 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marrink SJ, de Vries AH, and Mark AE (2004). Coarse Grained Model for Semiquantitative Lipid Simulations. The Journal of Physical Chemistry B 108, 750–760. 10.1021/jp036508g. [DOI] [Google Scholar]
- 112.Tang C, Smith AM, Collins RF, Ulijn RV, and Saiani A (2009). Fmoc-Diphenylalanine Self-Assembly Mechanism Induces Apparent pKa Shifts. Langmuir 25, 9447–9453. 10.1021/la900653q. [DOI] [PubMed] [Google Scholar]
- 113.Guo C, Luo Y, Zhou R, and Wei G (2012). Probing the self-assembly mechanism of diphenylalanine-based peptide nanovesicles and nanotubes. ACS Nano 6, 3907–3918. 10.1021/nn300015g. [DOI] [PubMed] [Google Scholar]
- 114.Mayans E, Ballano G, Casanovas J, Diaz A, Perez-Madrigal MM, Estrany F, Puiggali J, Cativiela C, and Aleman C (2015). Self-Assembly of Tetraphenylalanine Peptides. Chemistry 21, 16895–16905. 10.1002/chem.201501793. [DOI] [PubMed] [Google Scholar]
- 115.Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 116.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, and Lindahl E (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. [Google Scholar]
- 117.De Jong DH, Baoukina S, Ingólfsson HI, and Marrink SJ (2016). Martini straight: boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7. [Google Scholar]
- 118.Bussi G, Donadio D, and Parrinello M (2007). Canonical sampling through velocity rescaling. J Chem Phys 126, 014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 119.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, and Haak JR (1984). Molecular-Dynamics with Coupling to an External Bath. Journal of Chemical Physics 81, 3684–3690. Doi 10.1063/1.448118. [DOI] [Google Scholar]
- 120.Guo SZ, Ren XJ, Wu B, and Jiang T (2010). Preparation of the acellular scaffold of the spinal cord and the study of biocompatibility. Spinal Cord 48, 576–581. 10.1038/sc.2009.170. [DOI] [PubMed] [Google Scholar]
- 121.Keller A, Nesvizhskii AI, Kolker E, and Aebersold R (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- 122.Nesvizhskii AI, Keller A, Kolker E, and Aebersold R (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- 123.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, and Trono D (1998). Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72, 9873–9880. 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Toby BH, and Von Dreele RB (2013). GSAS-II: the genesis of a modern open-source all purpose crystallography software package. Journal of Applied Crystallography 46, 544–549. doi: 10.1107/S0021889813003531. [DOI] [Google Scholar]
- 125.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fruzangohar M, Ebrahimie E, and Adelson DL (2017). A novel hypothesis-unbiased method for Gene Ontology enrichment based on transcriptome data. PLOS ONE 12, e0170486. 10.1371/journal.pone.0170486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 128.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1, related to Figure 2. hiPSC-derived MNs on IKVAV-PA2 coatings. Live imaging of human MNs on IKVAV-PA2 coatings during the first 63 h in culture. Images were captured once every 16 min using a Nikon BioStation IM-Q imaging system with a 1.3-megapixel cooled monochrome camera (40 × len) and processed using the Image J program. Total time = 63 h 44 min. Scale bar: 10 μm.
Supplementary Movie 2, related to Figure 2. hiPSC-derived MNs on commercial laminin coating. Live imaging of human MNs on commercial laminin coating during the first 63 h in culture. Images were captured once every 16 min using a Nikon BioStation IM-Q imaging system with a 1.3-megapixel cooled monochrome camera (40 × len) and processed using the Image J program. Total time = 63 h 44 min. Scale bar: 10 μm.
Supplementary Movie 3, related to Figure 2. Schematic video of the high mobile IKVAV-PA2 nanofibers interacting with ITGB1 on the surface of a human MNs.
Supplementary Movie 4, related to Figure 2. Schematic video of the slow mobile IKVAV-PA1 nanofibers interacting with ITGB1 on the surface of a human MNs.
Supplementary Movie 4, related to Figure 2. Schematic video of the slow mobile IKVAV-PA1 nanofibers interacting with ITGB1 on the surface of a human MNs.
Table S1, related to Figure 3. Normalized TMT-labeling data acquired by LC-MS/MS of hiPSC-derived MNs cultured on commercial laminin and IKVAV-PA2 for 45 days.
Table S2, related to Figure 4. Electrophysiological properties of hiPSC-derived neurons cultured for >25 days in IKVAV-PA2 and laminin coatings, and comparison with previously published data of MNs cocultured with mouse astrocytes.
Data Availability Statement
All the data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


