Abstract
While an increasing number of studies have evaluated tobacco microbiomes, comparative microbiome analyses across diverse tobacco products are non-existent. Moreover, to our knowledge, no previous studies have characterized the metabolically-active (live) fraction of tobacco bacterial communities and compared them across products. To address these knowledge gaps, we compared bacterial communities across four commercial products (cigarettes, little cigars, cigarillos and hookah) and one research cigarette product. After total DNA extraction (n=414) from all samples, the V3V4 region of the 16S rRNA gene was sequenced on the Illumina HiSeq platform. To identify metabolically-active bacterial communities within these products, we applied a coupled 5-bromo-2'-deoxyuridine labeling and sequencing approach to a subset of samples (n=56). Each tobacco product was characterized by its signature microbiome, along with a shared microbiome across all tobacco products consisting of Pseudomonas aeruginosa, P. putida, P. alcaligenes, Bacillus subtilis, and Klebsiella pneumoniae. Comparing across products (using Linear discriminant analysis Effect Size (LEfSe)), a significantly higher (p < 0.05) relative abundance of Klebsiella and Acinetobacter was observed in commercial cigarettes, while a higher relative abundance of Pseudomonas and Pantoea was observed in research cigarettes. Methylorubrum and Paenibacillus were higher in hookah, and Brevibacillus, Lactobacillus, Bacillus, Lysinibacillus, and Staphylococcus were higher in little cigars and cigarillos. Across all products, the majority of the metabolically-active bacterial communities belonged to the genus Pseudomonas, followed by several genera within the Firmicutes phylum (Bacillus, Terribacillus, and Oceanobacillus). Identification of some metabolically-active pathogens such as Bacillus cereus and Haemophilus parainfluenzae in commercial products is of concern because of the potential for these microorganisms to be transferred to users’ respiratory tracts via mainstream smoke. Future work is warranted to evaluate the potential impact of these tobacco bacterial communities on users’ oral and lung microbiomes, which play such an important role on the spectrum from health to disease.
Keywords: Tobacco, bacteria, microbiome, additive, menthol, metabolically-active
1. INTRODUCTION
Since the implementation of the 2009 Family Smoking Prevention and Tobacco Control Act, tobacco manufacturers in the United States (U.S.) have been required to test and disclose the ingredients in tobacco products under section 904(a)(3) of the Food, Drug, and Cosmetic Act (FD&C Act) (FDA 2020). To facilitate this process, the Food and Drug Administration (FDA) published a list of 93 harmful and potentially harmful constituents (HPHCs) in tobacco products and selected 20 of these HPHCs to be tested, with the findings reported back to FDA (FDA 2021). While these 20 HPHCs include a comprehensive list of chemicals that are potentially present in commercial tobacco products, microbiological constituents that could be harmful to tobacco users’ health are not addressed.
Nevertheless, over the past 50 years, multiple research groups have detected and characterized a plethora of microorganisms (bacteria and fungi) in commercial tobacco products using both culture-dependent and -independent techniques (Kurup et al. 1983; Eaton et al. 1995; Larsson et al. 2008; Sapkota et al. 2010; Tyx et al. 2016; Chopyk et al. 2017b; Smyth et al. 2017; Hani et al. 2018; Malayil et al. 2020a). For example using culture-based techniques, Eaton et al. (1995) cultured Mycobacterium spp. from cigarettes, and Kurup et al. (1983) isolated Aspergillus spp. from cigarette tobacco (Kurup et al. 1983; Eaton et al. 1995). Subsequently, multiple bacterial genera (e.g., Bacillus, Pseudomonas aeruginosa, Klebsiella, and Methylobacterium) were identified in cigarettes via 16S rRNA-based microarrays, cloning and sequencing (Sapkota et al. 2010), while Halomonas, Staphylococcus, and Pseudomonas were detected in hookah tobacco (Hani et al. 2018), and Bacillus, Staphylococcus, and Lactobacillus were identified in smokeless tobacco products (Tyx et al. 2016; Al-hebshi et al. 2017; Smyth et al. 2017) via either qPCR or 16S rRNA V1-V4 sequencing. More recently, our group and others have extensively characterized tobacco microbiomes using 16S rRNA gene sequencing approaches, demonstrating differences in bacterial diversity and abundance across brands, lots, and additives (menthol/flavors) of traditional cigarettes, little cigars, cigarillos, hookah, and smokeless tobacco (Tyx et al. 2016; Chopyk et al. 2017a; Chopyk et al. 2017b; Hani et al. 2018; Chattopadhyay et al. 2019; Smyth et al. 2019; Malayil et al. 2020a; Malayil et al. 2022b).
The high-throughput sequencing approaches used in these studies have enabled us to catalog the vast array of bacteria (including low abundant members) associated with tobacco products; however, DNA sequencing approaches cannot differentiate between live and dead bacteria within these products. To overcome this challenge, DNA labeling methods have been coupled with sequencing approaches to tease out the metabolically-active or live fraction of microbial communities present in complex environmental samples (Gast et al. 2018; Ni et al. 2020; Fareed Mohamed Wahdan et al. 2020; Malayil et al. 2020b; Hirohara et al. 2021; Millette et al. 2021). Specifically, the DNA labeling dye 5-bromo-2’-deoxyuridine (BrdU), a synthetic thymidine analog capable of incorporating into replicating DNA or dividing cells, has enabled researchers to identify live, replicating bacterial community members within complex communities. Previous studies have coupled BrdU labeling with next-generation sequencing, quantitative PCR, or immunocytochemistry assays to identify metabolically-active bacterial communities in soil and water, as well as to evaluate the response of live fungal species to plant litter substrates (Hamasaki 2006; Allison et al. 2007; McGuire et al. 2010; Robbins et al. 2011; Galand et al. 2013b; Tada and Grossart 2014; Treseder et al. 2014; Malayil et al. 2020b; Ambaye et al. 2020; Wahdan et al. 2021; Pold et al. 2021). However, to our knowledge, there are no published studies that have applied coupled BrdU labeling and 16S rRNA gene sequencing approaches to evaluate the metabolically-active fraction of tobacco bacterial communities. Moreover, there are no data comparing overall tobacco microbiomes across diverse products such as cigarettes, little cigars and hookah.
To address these knowledge gaps, we performed a detailed comparative analysis of the bacterial microbiomes present across different types of tobacco products. Additionally, we utilized a coupled BrdU labeling and 16S rRNA gene sequencing approach to identify the metabolically-active fraction of the total heterogeneous bacterial communities present across these products.
2. METHODS
2.1. Selection of tobacco products
Commercially-available cigarettes, little cigars, cigarillos, and hookah, as well as custom made research cigarettes were included (Table 1). The commercially-available tobacco products were purchased from stores in the College Park, MD area, as well as online. Tobacco products were selected based on their market availability, accessibility, and popularity among U.S. tobacco users. Each type of tobacco product included brands with or without additives (menthol or flavors). Commercially-available cigarettes included Marlboro menthol (MMB), Marlboro red (MRB), Newport non-menthol (NNM), Newport menthol (NMB2) and Newport menthol gold (NMG). The research cigarettes were custom-made SPECTRUM cigarettes (obtained from the National Institute of Drug Abuse, MD (NIDA)) and included six pairs of mentholated and non-mentholated varieties with varying concentrations of nicotine. The little cigars included Swisher Sweets Cherry (SSC), Cheyenne menthol (CMB), and Cheyenne Full Flavor (CFF); and the cigarillo product was Swisher Sweets Original cigarillos (SSO). Finally, the hookah products included Al Fakher mint (AFMF), Al Fakher watermelon (AFWF), Al Fakher Two Apple (AFTA), Fumari Ambrosia (FAMB), Fumari Mint Chocolate Chill (FMCC), and Fumari White Gummy Bear (FWGB).
Table 1:
Tobacco products included in this study.
| Additives | |||
|---|---|---|---|
| Product | Brand | Mentholation | Flavoring |
| Commercial cigarettes (n=90) | Marlboro red (MRB) | Non menthol | |
| Marlboro menthol (MMB) | Menthol | ||
| Newport non-menthol (NNM) | Non menthol | ||
| Newport menthol (NMB2) | Menthol | ||
| Newport menthol gold (NMG) | Menthol | ||
| Hookah (n=108) | Al Fakher two apple (AFTA) | Two apple | |
| Al Fakher mint (AFMF) | Mint | ||
| Al Fakher watermelon (AFWF) | Watermelon | ||
| Fumari ambrosia (FAMB) | Ambrosia | ||
| Fumari mint chocolate chill (FMCC) | Mint chocolate | ||
| Fumari white gummi bear (FQGB) | White gummi bear | ||
| Little cigars (n=108) | Cheyenne menthol (CMB) | Menthol | |
| Cheyenne full flavor (CFF) | Non menthol | ||
| Swisher sweets cherry (SSC) | Cherry | ||
| Cigarillos (n=36) | Swisher sweets original (SSO) | Non flavored | |
| Research cigarettes (n=72) | NRC 102 (0.44 mg/g nicotine) | Non menthol | |
| NRC 200 (1.33 mg/g nicotine) | Non menthol | ||
| NRC 300 (2.38 mg/g nicotine) | Non menthol | ||
| NRC 400 (5.02 mg/g nicotine) | Non menthol | ||
| NRC 404 (7.74 mg/g nicotine) | Non menthol | ||
| NRC 600 (15.54 mg/g nicotine) | Non menthol | ||
| NRC 103 (0.62 mg/g nicotine) | Menthol | ||
| NRC 201 (1.22 mg/g nicotine) | Menthol | ||
| NRC 301 (2.38 mg/g nicotine) | Menthol | ||
| NRC 401 (5.18 mg/g nicotine) | Menthol | ||
| NRC 501 (12.44 mg/g nicotine) | Menthol | ||
| NRC 601 (15.42 mg/g nicotine) | Menthol | ||
2.2. Extraction of total DNA and sequencing
Total DNA was extracted from 414 samples (90 cigarettes, 108 hookahs, 108 little cigars, 36 cigarillos, and 72 research cigarettes), which included six replicates of each type of product, using a previously published protocol (Fadrosh et al. 2014; Chopyk et al. 2017b; Holm et al. 2019). Briefly, after dissecting the tobacco product aseptically, 0.2 g of tobacco was measured and placed into Lysing Matrix B tubes, and 1 mL of ice-cold 1X Phosphate buffer solution (PBS; Gibco-Life Technologies, NY) was added. Negative control samples contained 1X PBS and no tobacco. Samples were then incubated twice at 37°C and 55°C with enzyme cocktails (Cocktail A: lysozyme from chicken egg white (Sigma-Aldrich, St. Louis, MO), lysostaphin from Staphylococcus staphylolyticus (Sigma-Aldrich, St. Louis, MO), and mutanolysin from Streptomyces globisporus ATCC 21553 (Sigma-Aldrich, St. Louis, MO); and Cocktail B: Proteinase K (Invitrogen-Life Technologies, Grand Island, NY, USA) and 10% (w/v) sodium dodecyl sulfate (SDS) (BioRad, Hercules, CA)), before mechanical lysis of cells using the MP Biomedical Fastprep 24 (Santa Ana, CA). Extracted DNA was purified using the Qiagen DSP DNA Mini Kit according to the manufacturer’s protocol, and then PCR-amplified targeting the V3V4 hypervariable region of the 16S rRNA gene using the universal primers 319F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT). Amplicons were visualized on a 1% agarose gel and then sequenced on an Illumina HiSeq2500 (Illumina, San Diego, CA) using a previously published protocol (Fadrosh et al. 2014; Holm et al. 2019).
2.3. BrdU treatment and immunocapture of BrdU
For a subset of tobacco samples (n=56), each tobacco product was aseptically dissected, and 0.2 g of each product was weighed out into Lysing Matrix B tubes (MP Biomedicals, OH). The samples were then incubated at room temperature in the dark for 48 h with 26 μL of 7.69 mM BrdU for incorporation of BrdU into replicating DNA. Concurrently, control samples without the addition of BrdU (no treatment (NOTRT)) were incubated similarly. After incubation, total DNA was extracted and purified following the above-mentioned procedures. After DNA extraction, immunocapture and isolation of BrdU-labeled DNA were performed according to a previously published protocol (Urbach et al. 1999; Malayil et al. 2020b) and sequenced as mentioned above.
2.4. Sequencing reads analysis
16S rRNA reads were screened for low quality and short length and assembled using PANDAseq, demultiplexed, and chimera trimmed using UCHIME. Taxonomic assignments and relative abundance of bacterial taxa (for each sample) used for comparative analyses were determined with the Kraken 2 classification tool (Wood and Salzberg 2014) and Bracken abundance estimator (Lu et al. 2017) from quality trimmed pair-end reads. Quality reads for BrdU and NO TRT samples were incorporated into QIIME v1.9, clustered de-novo using VSEARCH and taxonomies were assigned using the Greengenes database, using a 0.97 confidence threshold. Downstream statistical analysis and visualization of data were performed in R Statistical computing software (v. 0.99.473), Microbiome Analyst (Dhariwal et al. 2017; Chong et al. 2020), Power BI (v 3.0.3.0), and Cytoscape (v 3.7.2). Core and shared microbiomes were analyzed using network analyses of the relative abundance of bacterial species within products (visualized at > 5% relative abundance) and brands (visualized >1% relative abundance). A chord plot was used to visualize bacterial species between commercial and research cigarettes above 5% relative abundance, and a Sankey plot was used to visualize bacterial taxa between BrdU treated and non-treated samples above 10% relative abundance.
2.5. Statistical analysis
After quality trimming and removing samples that had a Goods coverage value of ≤ 0.95, 365 samples (out of 414) were included in downstream analyses. Alpha diversity was computed using the Vegan R package and the Dunn’s test was performed as a post-hoc test for the Kruskal-Wallis test to evaluate differences between products: p-values were adjusted using the Holm method, and p < 0.05 was considered statistically significant for all statistical tests performed in this study. Beta diversity analysis was performed using non-metric multidimensional scaling (NMDS) on the Bray-Curtis distance matrix (using Vegan R package) and statistical significance was computed using permutational multivariate analysis of variance (ADONIS) with 999 permutations after Wisconsin double standardization. A pairwise ADONIS test (on 999 permutations) was performed to compute adjusted p-values between pairs of tobacco products. Bacterial abundance profiles were compared with Linear Discriminant Analysis (LDA) Effective Size analysis (LEfSe) using Kruskal-Wallis sum-rank test (Segata et al. 2011) to detect differentially abundant taxa and their effect size. The False Discovery Rate (FDR)-adjusted p-value was set to < 0.05 and the LDA was set to >2.0.
2.6. Data availability
Data concerning the samples included in this study are deposited under the following NCBI BioProject accession numbers: PRJNA635703, PRJNA473598, PRJNA601146, and PRJNA641233.
3. RESULTS
3.1. Bacterial microbiome across products
Comparing alpha diversity (measured with the Shannon diversity index) across products, hookah had the lowest diversity, while cigarillos had the highest diversity. Statistically significant differences (p < 0.05) in bacterial genera diversity were found between all pairs of compared products (Figure 1a). Our beta diversity comparison showed that 19% (ADONIS R2 = 0.19; p < 0.001) of the variation in bacterial community composition was explained by product type (Figure 1b). The greatest variation in bacterial community composition structure was found among the little cigar samples, while the least was among the commercial cigarettes. A pairwise ADONIS test of significance also demonstrated significant variability in bacterial community composition between all pairs of tobacco products (Table S1). While the least effect size was between little cigars and cigarillos (ADONIS R2 = 0.02), the greatest effect size was found between cigarettes and research cigarettes (ADONIS R2 = 0.24).
Figure 1:
Diversity and abundance of bacterial communities across all tobacco products. (a) Alpha diversity across the five tobacco products measured using the Shannon diversity index. Statistical significance between products was determined using the Kruskal-Wallis test followed by a post hoc Dunn’s test with Holm’s adjusted p-values reported. P < 0.001 (***). (b) Principal coordinate analysis plot of Bray-Curtis computed distances across products. Ellipses are drawn at 95% confidence intervals. Differences in beta diversity between products were measured using ADONIS. (c) Heatmap showing the average relative abundance of the top bacterial genera present across tobacco products. Significantly higher differentially abundant genera were determined using a LEfSe analysis. P < 0.05 (*). Res. cigarette = Research cigarette.
Since both alpha and beta diversity metrics are unable to differentiate between the specific bacterial taxa that may be significantly different across tobacco products, the average relative abundance of bacterial genera identified across products was compared (Figure 1c). While the average relative abundance of Acinetobacter and Klebsiella was higher (FDR adjusted p < 0.05) in commercial cigarettes, relative abundance of Brucella, Enterobacter, Micrococcus, Pantoea and Pseudomonas was higher in research cigarettes (FDR adjusted p < 0.05). Little cigars had a significantly higher relative abundance of Moorella, Lactobacillus, Clostridium and Brevibacillus, and cigarillos had a higher relative abundance of Staphylococcus, Lysinibacillus, Corynebacterium, Blautia, Bacillus and Aerococcus (Figure 1c).
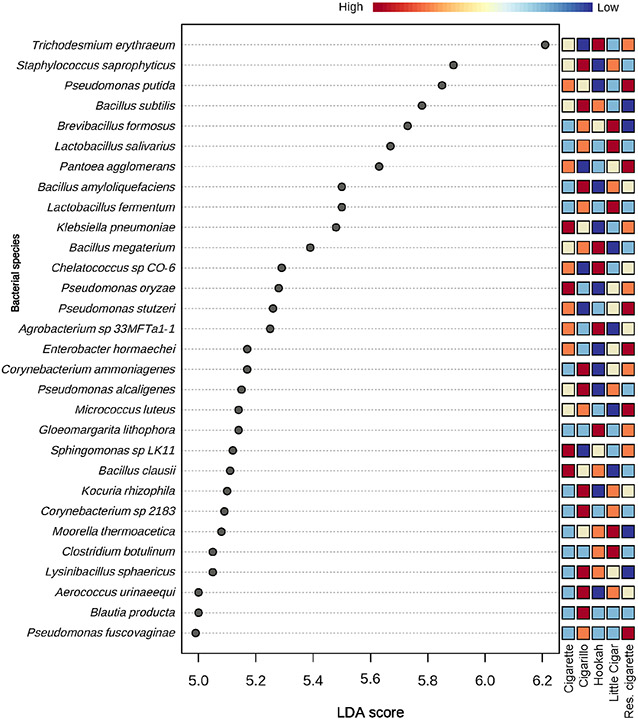
At the species level, both commercial cigarettes and research cigarettes had a higher relative abundance of Pseudomonas putida, Pseudomonas oryzae, Pseudomonas stutzeri, Pantoea agglomerans, Klebsiella pneumoniae, Enterobacter hormaechei, and Sphingomonas sp. LK11 compared to that in the little cigar, cigarillo, and hookah products (Figure 2). Bacillus subtilis, Brevibacillus formosus, Clostridium botulinum, Moorella thermoacetica, and Lysinibacillus sphaericus had a lower relative abundance in cigarettes (commercial and research) compared to hookah, cigarillos, and little cigars. Lactobacillus fermentum, Lactobacillus salivarius, Staphylococcus saprophyticus, B. formosus, C. botulinum, M. thermoacetica, and Aerococcus urinaeequi were at a higher relative abundance in little cigars.
Figure 2:
LEfSe analysis on the differential abundance of bacterial species across tobacco products. The color gradient on top shows the differential abundance of the taxa from low (blue) to high (red). Res. cigarette= Research cigarette
Comparing the cigarette products (commercial and research), B. subtilis, K. pneumoniae, Pseudomonas alcaligenes and P. oryzae were at a lower relative abundance in research cigarettes compared to commercial cigarettes and P. stutzeri, P. putida, P. aeruginosa, P. agglomerans, E. hormaechei, and Micrococcus luteus were at a higher relative abundance in research cigarettes compared to commercial cigarettes (Figure 3). Unique (absent in other products above 5% relative abundance) bacterial communities in commercial cigarettes comprised Ramlibacter tataouinensis, Bordetella genomosp. 13, and Acinetobacter baumannii, and that in research cigarettes included Corynebacterium ammoniagenes, A. urinaeequi, Enterococcus gilvus, and Bacillus amyloliquefaciens (Figure S1). The rest of the 11 bacterial species that had a relative abundance of greater than 5% were shared between the products.
Figure 3:
Chord plot showing the average relative abundance (above 5%) of bacterial species between commercial (yellow) and research (purple) cigarettes.
3.2. Core and shared microbiomes across products
Comparing across products, B. subtilis, K. pneumoniae, P. aeruginosa, P. putida, and P. alcaligenes were present in all five tobacco products forming the core microbiome across all products (Figure 4). M. thermoacetica, Bacillus amyloliquefaciens and A. urinaeequi was shared between cigarillo and little cigar products along with M. thermoacetica in hookah and B. amyloliquefaciens and A. urinaeequi in research cigarettes. Commercial and research cigarettes, along with cigarillos, shared S. saprophyticus and Micrococcus luteus. P. stutzeri, P. oryzae, and C. ammoniagenes was shared between little cigars and research cigarettes, while P. stutzeri and C. ammoniagenes was shared with hookah products and P. oryzae was shared with commercial cigarettes. Commercial cigarettes and little cigars shared B. genomosp. 13, A. baumannii, and L. fermentum.
Figure 4:
Network analysis showing shared and unique bacterial species across all tobacco products. The circles represent each bacterial species, and the hexagons represent the tobacco products. The nodes joining the shapes indicate the identification of the taxa within the product.
Meanwhile, some bacterial species were shared between two to four products, and each of the tobacco products harbored two to three unique bacterial species. Bacillus megaterium, Dickeya fangzhongdai, B. genomosp. 13, A. baumannii, L. fermentum, B. formosus, and C. botulinum were each shared by two products. While B. megaterium was shared between cigarillo products and hookah products, B. formosus and C. botulinum were shared between cigarillos and little cigars. While P. agglomerans and Bacillus clausii were absent in cigarillos, Enterobacter hormaechei was absent in hookah products. Cigarillos had three unique bacterial species while hookah and little cigars had two unique species each. The cigarette products each had a single unique bacterial species (Figure 4).
3.3. Effect of additives on the bacterial microbiome
To evaluate changes in the bacterial microbiome in response to additives (menthol and flavors) in the tobacco products, we compared the mentholated/flavored varieties to their non-mentholated/non-flavored counterparts (Figure S2). Bacterial genera with the highest relative abundance were Staphylococcus, Pseudomonas and Bacillus across all products. While the addition of menthol in commercial cigarettes had a significant effect on bacterial community composition (NMG vs. NNM pairwise ADONIS, p = 0.001; NMB2 vs. NNM pairwise ADONIS, p = 0.001; MRB vs. MMB pairwise ADONIS, p = 0.037), the addition of menthol in research cigarettes had a significant effect only between two pairs (NRC200 vs. NRC201 pairwise ADONIS, p = 0.01; NRC404 vs. NRC501 pairwise ADONIS, p = 0.045) (Table S2). Significant mentholation effects were also observed in little cigar products (CMB vs. CFF pairwise ADONIS, p = 0.002).
Comparing products with or without additives (menthol/flavors), a Random Forest algorithm was applied to construct decision trees to predict biomarker taxa that were best associated with the additives. Looking across bacterial genera, Corynebacterium, Brachybacterium, Methylorubrum, Propionibacterium, Xanthomonas, Brucella, Spingomonas and Bordetella were associated with mentholated cigarette products, and Pantoea, Aerococcus, Klebsiella, Enterococcus, Chrysobacterium and Agrobacterium were associated with non-mentholated cigarette products (Figure 5). Across all brands, the highest alpha diversity was observed among the non-menthol Marlboro cigarettes and non-flavored cigarillos, while the lowest alpha diversity was observed among the flavored products (little cigar SSC and all hookah products) (Figure S3). Comparing within brands, menthol Newport cigarettes (Shannon index average ± SD for NMB2, 2.23 ± 0.34 and NMG, 2.50 ± 0.13), mint flavors of Al Fakher (AFMF 1.81 ± 0.29) and Fumari (FMCC 1.94 ± 0.23) hookah, and menthol research cigarettes (2.24 ± 0.34) had higher alpha diversity compared to their non-menthol/mint counterparts (Shannon index average ± SD: 2.21 ± 0.17 (NNM); 1.74 ± 0.27 (AFWF); 1.63 ± 0.25 (AFTA); 2.0 ± 0.33 (FWGB); 1.66 ± 0.91 (FAMB), and 2.16 ± 0.39 for non-menthol research cigarettes); however, these differences were not statistically significant (Holm’s adjusted Dunn’s test p > 0.05). In contrast, lower alpha diversity was observed among mentholated little cigars and Marlboro cigarettes (CMB 2.35 ± 0.40; MMB 2.64 ± 0.28) when compared to non-mentholated (CFF 2.42 ± 0.44; MRB 2.74 ± 0.22) varieties. Comparing the little cigars, there were no statistically significant differences in the Shannon diversity index between mentholated and flavored varieties, although flavored little cigars had lower average values. All flavored hookah products had lower alpha diversity compared to all other brands, with the highest alpha diversity values observed for the mint flavors (Al Fakher Mint and Fumari Mint Chocolate Chill).
Figure 5:
Predicted association between the top 30 bacterial genera and specific tobacco products with/without additives. The color gradient shows the degree of association from low (blue) to high (red). C: cigarette, LC: little cigar, CG: cigarillo, H: hookah, and RC: research cigarette.
Comparing the menthol and non-menthol commercial cigarettes, there were 24 non-menthol cigarette samples containing unique bacterial genera, with nine samples resembling the menthol samples. Among the research cigarettes, 11 menthol samples had unique genera, while 18 samples were associated with genera similar to that of the menthol research cigarettes. Twenty-seven non-menthol research cigarettes samples had unique genera and seven had similar taxa found in their menthol counterparts. Comparing the little cigars samples, 24 flavored little cigar samples had unique genera and 16 non-flavored little cigars samples had unique genera. All 99 flavored hookah samples had unique bacterial genera not found in any other tobacco product.
3.4. Core and shared microbiomes across brands and flavors
Across the little cigar products, L. salivarius, L. fermentum, B. formosus, and S. saprophyticus formed the core microbiome of all products (Figure 6a). The shared bacterial species among the two Cheyenne products were B. subtilis, C. botulinum, P. agglomerans and E. hormaechei, which were absent in the Swisher Sweets. Meanwhile, bacterial species that were unique to the Cheyenne products were P. alcaligenes, P. oryzae, A. urinaeequi and M. thermoacetica, and the only species that was unique to the Swisher Sweets products was C. ammoniagenes.
Figure 6:
Network plots showing the shared and unique bacterial species within a) hookah, b) little cigars, c) cigarettes, and d) research cigarettes. The circles represent each bacterial species, and the hexagons represent the tobacco products. The nodes joining the shapes depict the identified taxa within the products.
Comparing the different hookah products, only B. subtilis formed the core microbiome across the four flavors, while B. megaterium, B. clausii and P. aeruginosa were shared between three flavors (Figure 6b). The Al Fakher flavor, two apple, did not share any bacterial species with any of the other products. Fumari ambrosia and white gummy bear harbored three and one unique bacterial species respectively. Two other hookah flavors, Fumari mint chocolate chill, and the Al Fakher mint flavor, did not have any bacterial species identified at a relative abundance higher than 1%, and hence, are not included in the figure.
Among the research cigarettes, K. pneumoniae, P. agglomerans, M. luteus, P. putida, P. aeruginosa and S. saprophyticus comprised the core microbiome of both menthol and nonmenthol research cigarettes (Figure 6c). While five unique bacterial species were found among the menthol research cigarettes, only one species was unique to the non-menthol varieties.
Among the commercial cigarettes, there were no core bacterial species (>1% relative abundance) that were identified (Figure 6d). B. subtilis and P. putida were shared between 4 brands of commercial cigarettes except for Newport non-menthols. Three of the five cigarette brands (Newport menthol gold, Newport non-menthol and Marlboro red) had unique bacterial species, while the other two (Newport menthol 2 and Marlboro menthol) did not have any unique bacterial species.
3.5. Metabolically-active bacterial communities across products
Comparing our subset of samples tested for total (NOTRT) and metabolically-active (BRDU) bacterial communities, alpha diversity was significantly (p < 0.001) lower in BrdU-treated samples with regard to both the observed number of species and Shannon diversity index metrics (Figure S4a). Beta diversity was also significantly different (PERMANOVA R2 = 0.5005; p < 0.001) with more than 50% of the variation explained by BrdU treatment (Figure S4b). Metabolically-active bacterial genera among the tobacco brands included Pseudomonas (highest relative abundance), followed by Terribacillus, Lactobacillus, and Gardnerella (Figure 7). Oceanobacillus was only identified among the BrdU-treated samples. Other taxa that were only identified in the BrdU-treated samples included Uncl. Clostridiales and Uncl. Aeromonadaceae. While the above-mentioned bacterial genera were present at a relative abundance of >10%, there were several bacterial taxa identified at the species level with a relative abundance of <10% including Atopobium vaginae, Alcaligenes faecalis, Bacillus cereus, Haemophilus parainfluenzae, Rothia mucilaginosa, Streptococcus agalactiae, and Propionibacterium acnes (Table S3).
Figure 7:
Sankey plot showing the metabolically-active (BrdU) bacterial species identified within the total bacterial community (NOTRT) across all tobacco brands and flavors. AFMF: Al Fakher mint, AFWF: Al Fakher watermelon, FMCC: Fumari mint chocolate chill, FWGB: Fumari white gummy bear, AFTA: Al Fakher two apple, NMG: Newport menthol gold, MMB: Marlboro menthol box, FAMB: Fumari Ambrosia, NNM: Newport non-menthol, MRB: Marlboro red, SSC: Swisher sweets cherry, SSO: Swisher sweets original, CFF: Cheyenne full flavor, CMB: Cheyenne menthol box.
4. DISCUSSION
In this study, we characterized total and metabolically-active bacterial communities across diverse tobacco products (cigarettes, hookah, little cigars and cigarillos) that are widely used in the U.S. While cigarillos were characterized by the highest diversity in bacterial communities (measured by the Shannon diversity index), hookah products had the lowest diversity compared to all other products. Bacterial community composition present within the tobacco products was significantly different across product types, with each product type harboring a unique microbiome signature. In addition, the majority of the metabolically-active bacteria across all products belonged to the genus Pseudomonas, followed by several genera within the Firmicutes phylum (Bacillus, Terribacillus, and Oceanobacillus).
While the most abundant genera among all tobacco products were Pseudomonas, Staphylococcus, and Bacillus, all five tobacco products harbored a core microbiome that included the following bacterial species: Pseudomonas aeruginosa, P. putida, P. alcaligenes, Bacillus subtilis and Klebsiella pneumoniae. Previous work from our group and others has demonstrated the presence of the above-mentioned species as members of the “core microbiome” of cigarettes, little cigars, and cigarillo products with noted differences in diversity and abundance across brands, lots, and flavors (Rooney et al. 2005; Sapkota et al. 2010; Chopyk et al. 2017a; Chopyk et al. 2017b; Smyth et al. 2017; Hani et al. 2018; Chattopadhyay et al. 2019; Smyth et al. 2019; Malayil et al. 2020a). These organisms being a part of the core microbiome of all tested tobacco products is of concern since several of them are pathogenic and have been associated with adverse health effects. For example, P. aeruginosa can cause chronic infections, particularly in people with chronic lung diseases like cystic fibrosis, bronchiectasis, and chronic obstructive pulmonary disease and is known to cause irreversible airway decline, poorer quality of life, and early mortality in these sensitive sub-populations (Erb-Downward et al. 2011; Fodor et al. 2012; Gellatly and Hancock 2013; Moradali et al. 2017; Jacobs et al. 2020). In addition, although considered a non-pathogenic probiotic, B. subtilis strain G7 isolates can be characterized by unique genetic features which are potentially lethal to vertebrates (Gu et al. 2019). Lastly, one of the most common nosocomial pathogens, K. pneumoniae, is known to express antibiotic resistance and virulence factors that aid the bacterium in spreading through the respiratory system, causing pneumonia, bloodstream infections and meningitis (Martin and Bachman 2018; Gonzalez-Ferrer et al. 2021).
In addition to the shared microbiome across all products, we also observed that each type of product harbored a unique microbiome signature. For instance, a significantly higher relative abundance of Staphylococcus and Bacillus was observed in cigarillos and a higher relative abundance of Lactobacillus and Brevibacillus was observed in little cigars. Previous studies from our group also had similar findings (Chattopadhyay et al. 2019; Smyth et al. 2019). Interestingly, we also observed the lowest alpha diversity (measured by the Shannon diversity index) in hookah products compared to all other tested products. This could be due to the higher levels of glycerol and honey that constitute about 70% of hookah tobacco (Khater et al. 2008; Uebelacker et al. 2019). Humectants, such as glycerol and propylene glycol, are usually added to tobacco filler to maintain moisture, dilute nicotine to a viscous consistency, and act as a carrier solvent. Glycerol (> 5% solutions) also has bactericidal activity, inhibiting bacterial growth and cellulolytic activity (Roger et al. 1992; Nalawade et al. 2015). Interestingly, bacterial species within a few genera (Lactobacillus, Klebsiella, Enterobacter, Citrobacter, and Clostridium) can metabolize glycerol to produce the antimicrobial reuterin, which also possesses antimicrobial properties against commensal bacterial populations (Cleusix et al. 2008; De Weirdt et al. 2010). Similarly, honey—which is one of the oldest known antimicrobials—also exhibits bactericidal activity against a variety of bacteria (Morroni et al. 2018; Almasaudi 2021).
Furthermore, in our comparison between products, we observed significantly different microbiome signatures between mentholated versus non-mentholated products. Comparing within products, mentholated brands had lower alpha diversity compared to their non-mentholated varieties. Previous research has demonstrated that bacterial diversity is different between menthol and non-mentholated cigarettes and little cigars (Chattopadhyay et al. 2019; Chattopadhyay et al. 2019; Smyth et al. 2019; Malayil et al. 2020a), and Chopyk et al. (2017) found that mentholated products harbor taxa that are known to survive harsh conditions such as Anoxybacillus and Schlegella (Chopyk et al. 2017a).
In addition to evaluating the effects of mentholation, we found that all flavored varieties of tested tobacco products (Swisher Sweets Cherry and all hookah products) had the lowest alpha diversity compared to all other products. Even though we could not do a direct comparison between a flavored product with a non-flavored counterpart for the same type of tobacco product, our data potentially indicate that additives such as fruit flavorings may have a greater impact on bacterial diversity compared to other additives such as menthol or mint. Among the flavored little cigar and hookah products, we also found differences in the relative abundance of bacterial genera across products; however, we were unable to make comparisons against non-flavored counterparts. The observed differences in the relative abundance of bacterial taxa may be due to different curing techniques during tobacco processing and manufacturing. Given the limited data on bacterial community differences between mentholated/flavored tobacco products and non-flavored products, more in-depth studies are needed, particularly with regard to cigarettes, little cigars, and hookah products.
While we observed significant differences in the relative abundance of different bacterial species across the five tobacco products, we also identified known or emerging pathogens that could be important in terms of users’ health. For example, commercial and research cigarettes harbored P. putida, and P. agglomerans, both opportunistic human pathogens (Tian et al. 2016; Zhao et al. 2018). Two more pathogenic species identified among the research cigarettes were Aerococcus urinaeequi and Enterococcus gilvus, which have previously been found to be sensitive to antimicrobials (Vrabec et al. 2015; Rasmussen 2016). Our data also identified the bacterial pathogens M. luteus and E. hormaechei in four of the five tobacco products. Even though it is a part of the normal oral microflora, M. luteus can be an opportunistic pathogen (Khan et al. 2019) and E. hormaechei is a well-known causative agent of nosocomial infections, exhibiting resistance to multiple antibiotics (Monahan et al. 2019; Wang et al. 2020; Gou et al. 2020).
Furthermore, we found that both cigarillos and little cigars harbored the spore-forming bacteria C. botulinum, and cigarillos alone harbored L. sphaericus. While C. botulinum is a well-known anaerobic species producing the botulinum neurotoxin (Abdel-Moein and Hamza 2016), L. sphaericus produces insecticidal proteins leading to cytopathological effects in insect larvae (de Fátima Gomes Cavados et al. 2017). A recent case report also described L. sphaericus as a causative agent of severe sepsis (Wenzler et al. 2015). Another unique species identified in cigarillos was Saccharomonospora viridis. This thermophilic species is usually found in hay and compost, and prolonged exposure to its spores is known to cause farmer’s lung disease, bagassosis (a type of interstitial lung disease), and humidifier fever (Pati et al. 2009).
Given the plethora of bacterial species that 1) were present in the tested tobacco products and 2) have the potential to be transmitted via aerosols to users’ upper respiratory tracts, it was important to evaluate their viability. By coupling BrdU-labeling and sequencing, we were able to identify the metabolically-active fraction of the total bacterial communities identified across the tested tobacco products. Alpha diversity in BrdU-treated samples was lower than in non-BrdU-treated samples, which was consistent with previous studies utilizing this coupled approach to detect metabolically-active bacterial communities in water samples (Malayil et al. 2020b). Interestingly, we identified Pseudomonas—the bacterial genera detected as one of the most abundant overall in the tobacco products—to be metabolically-active. Other metabolically-active bacterial genera identified across the majority of tested products included Bacillus and Paenibacillus, while metabolically-active bacterial species included Atopobium vaginae, Alcaligenes faecalis, Bacillus cereus, Haemophilus parainfluenzae, Rothia mucilaginosa, Streptococcus agalactiae, and Cutibacterium acnes. These data are supported by previous findings from our group where we identified viable Bacillus and Paenibacillus in mainstream cigarette smoke extract using culture-dependent techniques (Malayil et al. 2022a). While previous studies have described a few of the identified species in our study as pathogenic (A. faecalis, B. cereus, Pseudomonas viridiflava, Rothia aeria, R. mucilaginosa, S. agalactiae) (Bottone 2010; Michon et al. 2010; Geng et al. 2012; Sarris et al. 2012; Maraki and Papadakis 2015; Huang 2020; Greve et al. 2021), some species (Methylobacterium adhaesivum, Acinetobacter johnsonii, A. lwoffii) (Kozińska et al. 2014; Szwetkowski and Falkinham 2020) are emerging animal/human pathogens, while others (P. acnes, H. parainfluenzae) are known to be associated with multiple diseases (e.g., inflammatory infections, bone and joint infections) (Portillo et al. 2013; O’Neil et al. 2016). It is important to note that two of the genera, Pseudomonas and Bacillus, that incorporated BrdU, are dominant members of the tobacco microbiome, and a few of the above-mentioned pathogenic species within these genera were also viable/metabolically-active. Hence, these organisms have the potential to be transferred to users during tobacco use, colonize the upper respiratory tract and potentially cause adverse health effects.
Strengths of this study include the sample size, the inclusion of multiple tobacco products, and the incorporation of a coupled BrdU-labeling/sequencing technique. However, like other sequencing-based studies, there were multiple limitations, including challenges with regard to species-level taxonomic assignments. Here, we used Kraken to perform sequence classification and estimated species abundance using Bracken (Wood and Salzberg 2014; Valenzuela-González et al. 2016; Lu et al. 2017). The kmer matching technique used by Kraken has been shown to supersede those pipelines using gene markers in terms of sensitivity and specificity (Miossec et al. 2020; Lu and Salzberg 2020), reducing runtime and memory usage (Wood et al. 2019) and reducing sensitivity to structural variations (e.g., inversions) within reads (Pearman et al. 2020). However, as with other classifiers, Kraken 2 also misclassifies reads at the species level, specifically with regard to genera or species sharing a high genomic identity (Wood et al. 2019).
In addition, there are only a few studies that have utilized the coupled BrdU-labeling/sequencing technique used here. This technique and other widely used approaches such as the incorporation of radiolabeled precursors (like thymidine and leucine) can have methodological limitations, including contrasting uptake of these compounds by different bacterial phyla (Pérez et al. 2010; Hellman et al. 2011; Galand et al. 2013a). Finally, since this labeling technique is limited to bacterial taxa that are capable of incorporating exogenous nucleotide precursors into replicating DNA (Urbach et al. 1999), additional culture-dependent and independent studies are required to better characterize the capacity of BrdU incorporation across diverse bacterial communities in environmental samples.
In closing, smoking tobacco products is known to impact the user’s oral microbiome, shifting the normal microflora towards a pathogen prevalent community. To better understand the association between members of the tobacco microbiome and the oral microbiome, we need in-depth comparative characterizations of the microbiome of diverse tobacco products. Here, we not only characterized the unique/signature bacterial taxa in each tested tobacco product but also identified the core bacterial communities and metabolically-active bacteria that were present across all products. Future work should evaluate the role of the identified tobacco bacterial species in the potential shifting of smokers’ oral and lung bacterial community compositions, which can ultimately lead to disease development in the oral cavity, overall respiratory tract and other body systems.
Supplementary Material
Figure S1: Network plot showing the unique and shared bacterial species between commercial and research cigarettes.
Figure S2: Average relative abundance of bacterial genera across all tobacco products and brands.
Figure S3: Bacterial alpha diversity by tobacco product, brand and flavor.
Figure S4: Bacterial a) alpha and b) beta diversity between BrdU-treated (BRDU) and non-treated (NOTRT) tobacco samples.
Highlights:
We compared total and live bacterial communities across diverse tobacco products.
Cigarillos had the highest bacterial alpha diversity among all tested products.
Each tested tobacco product was characterized by a unique microbiome signature.
A core microbiome was also present among all tested tobacco products.
Metabolically-active (live) bacteria and pathogens were present in all products.
Acknowledgments:
We would like to thank Prachi Kulkarni, Emma Claye, Kelsey Babik, and Molly Reid from the University of Maryland School of Public Health (College Park, MD) for their help with the laboratory processing of samples. The authors are thankful to Dr. Lauren Hittle from the Institute of Genome Sciences, University of Maryland, Baltimore (Baltimore, MD) for her help with performing the sequencing of all samples.
FUNDING:
This work was supported by the University of Maryland Tobacco Center of Regulatory Science (UMD TCORS) “Rapid Response Characterization of New and Manipulated Tobacco Products” awarded by the National Institute of Health (NIH) and the Food and Drug Administration (FDA)—Award # P50-CA-180523-01. SC and ARS were supported by NRT-INFEWS: UMD Global STEWARDS (STEM Training at the Nexus of Energy, WAter Reuse and FooD Systems) that was awarded to the University of Maryland School of Public Health by the National Science Foundation National Research Traineeship Program, Grant number 1828910.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Competing interests: The authors declare that they have no competing interests.
References
- Abdel-Moein KA, Hamza DA (2016) Occurrence of human pathogenic Clostridium botulinum among healthy dairy animals: an emerging public health hazard. Pathog Glob Health 110:25–29. 10.1080/20477724.2015.1133107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-hebshi NN, Alharbi FA, Mahri M, Chen T (2017) Differences in the Bacteriome of Smokeless Tobacco Products with Different Oral Carcinogenicity: Compositional and Predicted Functional Analysis. Genes (Basel) 8. 10.3390/genes8040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SD, Hanson CA, Treseder KK (2007) Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biology and Biochemistry 39:1878–1887. 10.1016/j.soilbio.2007.02.001 [DOI] [Google Scholar]
- Almasaudi S (2021) The antibacterial activities of honey. Saudi Journal of Biological Sciences 28:2188–2196. 10.1016/j.sjbs.2020.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambaye TG, Vaccari M, van Hullebusch ED, Amrane A, Rtimi S (2020) Mechanisms and adsorption capacities of biochar for the removal of organic and inorganic pollutants from industrial wastewater. Int J Environ Sci Technol. 10.1007/s13762-020-03060-w [DOI] [Google Scholar]
- Bottone EJ (2010) Bacillus cereus, a Volatile Human Pathogen. Clin Microbiol Rev 23:382–398. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Smyth EM, Kulkarni P, Babik KR, Reid M, Hittle LE, Clark PI, Mongodin EF, Sapkota AR (2019) Little cigars and cigarillos harbor diverse bacterial communities that differ between the tobacco and the wrapper. PLOS ONE 14:e0211705. 10.1371/journal.pone.0211705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Liu P, Zhou G, Xia J (2020) Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 15:799–821. 10.1038/s41596-019-0264-1 [DOI] [PubMed] [Google Scholar]
- Chopyk J, Chattopadhyay S, Kulkarni P, Claye E, Babik KR, Reid MC, Smyth EM, Hittle LE, Paulson JN, Cruz-Cano R, Pop M, Buehler SS, Clark PI, Sapkota AR, Mongodin EF (2017a) Mentholation affects the cigarette microbiota by selecting for bacteria resistant to harsh environmental conditions and selecting against potential bacterial pathogens. Microbiome 5:22. 10.1186/s40168-017-0235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopyk J, Chattopadhyay S, Kulkarni P, Smyth EM, Hittle LE, Paulson JN, Pop M, Buehler SS, Clark PI, Mongodin EF, Sapkota AR (2017b) Temporal Variations in Cigarette Tobacco Bacterial Community Composition and Tobacco-Specific Nitrosamine Content Are Influenced by Brand and Storage Conditions. Front Microbiol 8:358. 10.3389/fmicb.2017.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleusix V, Lacroix C, Vollenweider S, Le Blay G (2008) Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiology Ecology 63:56–64. 10.1111/j.1574-6941.2007.00412.x [DOI] [PubMed] [Google Scholar]
- de Fátima Gomes Cavados C, Pires ES, Chaves JQ, Alvarez DN, Benites Gil H, Braz Ribeiro de Oliveira I, de Barros Pinto Viviani Cunha A, Pereira da Cunha de Araújo-Coutinho CJ (2017) Isolation and genetic characterization of Lysinibacillus sphaericus strains found in mosquito larvae (Diptera: Culicidae). Res Rep Trop Med 8:17–20. 10.2147/RRTM.S124066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weirdt R, Possemiers S, Vermeulen G, Moerdijk-Poortvliet TCW, Boschker HTS, Verstraete W, Van de Wiele T (2010) Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiology Ecology 74:601–611. 10.1111/j.1574-6941.2010.00974.x [DOI] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research 45:W180–W188. 10.1093/nar/gkx295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton T, Falkinham JO, von Reyn CF (1995) Recovery of Mycobacterium avium from cigarettes. J Clin Microbiol 33:2757–2758. 10.1128/jcm.33.10.2757-2758.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, Martinez FJ, Huffnagle GB (2011) Analysis of the Lung Microbiome in the “Healthy” Smoker and in COPD. PLOS ONE 6:e16384. 10.1371/journal.pone.0016384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. 10.1186/2049-2618-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareed Mohamed Wahdan S, Hossen S, Tanunchai B, Schädler M, Buscot F, Purahong W (2020) Future Climate Significantly Alters Fungal Plant Pathogen Dynamics during the Early Phase of Wheat Litter Decomposition. Microorganisms 8:908. 10.3390/microorganisms8060908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2020) Listing of Ingredients in Tobacco Products. In: U.S. Food and Drug Administration. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/listing-ingredients-tobacco-products. Accessed 29 Oct 2021 [Google Scholar]
- FDA (2021) Harmful and Potentially Harmful Constituents (HPHCs). FDA [Google Scholar]
- Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC (2012) The Adult Cystic Fibrosis Airway Microbiota Is Stable over Time and Infection Type, and Highly Resilient to Antibiotic Treatment of Exacerbations. PLOS ONE 7:e45001. 10.1371/journal.pone.0045001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand P, Alonso-Sáez L, Bertilsson S, Lovejoy C, Casamayor E (2013a) Contrasting activity patterns determined by BrdU incorporation in bacterial ribotypes from the Arctic Ocean in winter. Frontiers in Microbiology 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Alonso-Sáez L, Bertilsson S, Lovejoy C, Casamayor EO (2013b) Contrasting activity patterns determined by BrdU incorporation in bacterial ribotypes from the Arctic Ocean in winter. Front Microbiol 4. 10.3389/fmicb.2013.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast RJ, Fay SA, Sanders RW (2018) Mixotrophic Activity and Diversity of Antarctic Marine Protists in Austral Summer. Frontiers in Marine Science 5 [Google Scholar]
- Gellatly SL, Hancock REW (2013) Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathogens and Disease 67:159–173. 10.1111/2049-632X.12033 [DOI] [PubMed] [Google Scholar]
- Geng Y, Wang KY, Huang XL, Chen DF, Li CW, Ren SY, Liao YT, Zhou ZY, Liu QF, Du ZJ, Lai WM (2012) Streptococcus agalactiae, an Emerging Pathogen for Cultured Ya-Fish, Schizothorax prenanti, in China. Transboundary and Emerging Diseases 59:369–375. 10.1111/j.1865-1682.2011.01280.x [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ferrer S, Peñaloza HF, Budnick JA, Bain WG, Nordstrom HR, Lee JS, Van Tyne D (2021) Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis. Infection and Immunity 89:e00693–20. 10.1128/IAI.00693-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J-J, Liu N, Guo L-H, Xu H, Lv T, Yu X, Chen Y-B, Guo X-B, Rao Y-T, Zheng B-W (2020) Carbapenem-Resistant Enterobacter hormaechei ST1103 with IMP-26 Carbapenemase and ESBL Gene blaSHV-178. Infect Drug Resist 13:597–605. 10.2147/IDR.S232514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve D, Moter A, Kleinschmidt MC, Pfäfflin F, Stegemann MS, Kursawe L, Grubitzsch H, Falk V, Kikhney J (2021) Rothia aeria and Rothia dentocariosa as biofilm builders in infective endocarditis. International Journal of Medical Microbiology 311:151478. 10.1016/j.ijmm.2021.151478 [DOI] [PubMed] [Google Scholar]
- Gu H-J, Sun Q-L, Luo J-C, Zhang J, Sun L (2019) A First Study of the Virulence Potential of a Bacillus subtilis Isolate From Deep-Sea Hydrothermal Vent. Front Cell Infect Microbiol 9:183. 10.3389/fcimb.2019.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki K (2006) Comparison of bromodeoxyuridine immunoassay with tritiated thymidine radioassay for measuring bacterial productivity in oceanic waters. J Oceanogr 62:793–799. 10.1007/s10872-006-0098-7 [DOI] [Google Scholar]
- Hani J, Abdel Nour G, Matta J, Jazzar B, Pfaffl MW, Hanna-Wakim L, Abdel Nour AM (2018) Shisha microbiota: the good, the bad and the not so ugly. BMC Research Notes 11:446. 10.1186/s13104-018-3553-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman M, Berg J, Brandt KK, Hallin S (2011) Survey of bromodeoxyuridine uptake among environmental bacteria and variation in uptake rates in a taxonomically diverse set of bacterial isolates. Journal of Microbiological Methods 86:376–378. 10.1016/j.mimet.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Hirohara T, Tsuri K, Miyagawa K, Paine RTR, Yamanaka H (2021) The Application of PMA (Propidium Monoazide) to Different Target Sequence Lengths of Zebrafish eDNA: A New Approach Aimed Toward Improving Environmental DNA Ecology and Biological Surveillance. Frontiers in Ecology and Evolution 9 [Google Scholar]
- Holm JB, Humphrys MS, Robinson CK, Settles ML, Ott S, Fu L, Yang H, Gajer P, He X, McComb E, Gravitt PE, Ghanem KG, Brotman RM, Ravel J (2019) Ultrahigh-Throughput Multiplexing and Sequencing of >500-Base-Pair Amplicon Regions on the Illumina HiSeq 2500 Platform. mSystems 4:e00029–19. 10.1128/mSystems.00029-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C (2020) Extensively drug-resistant Alcaligenes faecalis infection. BMC Infectious Diseases 20:833. 10.1186/s12879-020-05557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DM, Ochs-Balcom HM, Noyes K, Zhao J, Leung WY, Pu CY, Murphy TF, Sethi S (2020) Impact of Pseudomonas aeruginosa Isolation on Mortality and Outcomes in an Outpatient Chronic Obstructive Pulmonary Disease Cohort. Open Forum Infect Dis 7:ofz546. 10.1093/ofid/ofz546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Aung TT, Chaudhuri D (2019) The First Case of Native Mitral Valve Endocarditis due to Micrococcus luteus and Review of the Literature. Case Rep Cardiol 2019:5907319. 10.1155/2019/5907319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khater AEM, Abd El-Aziz NS, Al-Sewaidan HA, Chaouachi K (2008) Radiological hazards of Narghile (hookah, shisha, goza) smoking: activity concentrations and dose assessment. Journal of Environmental Radioactivity 99:1808–1814. 10.1016/j.jenvrad.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Kozińska A, Paździor E, Pękala A, Niemczuk W (2014) Acinetobacter johnsonii and Acinetobacter lwoffii - the emerging fish pathogens. Journal of Veterinary Research 58:193–199. 10.2478/bvip-2014-0029 [DOI] [Google Scholar]
- Kurup VP, Resnick A, Kagen SL, Cohen SH, Fink JN (1983) Allergenic fungi and actinomycetes in smoking materials and their health implications. Mycopathologia 82:61–64. 10.1007/BF00436948 [DOI] [PubMed] [Google Scholar]
- Larsson L, Szponar B, Ridha B, Pehrson C, Dutkiewicz J, Krysińska-Traczyk E, Sitkowska J (2008) Identification of bacterial and fungal components in tobacco and tobacco smoke. Tob Induc Dis 4:4. 10.1186/1617-9625-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser FP, Thielen P, Salzberg SL (2017) Bracken: Estimating species abundance in metagenomics data. PeerJ Computer Science 2017:e104. 10.7717/peerjcs.104 [DOI] [Google Scholar]
- Lu J, Salzberg SL (2020) Ultrafast and accurate 16S rRNA microbial community analysis using Kraken 2. Microbiome 8:124. 10.1186/s40168-020-00900-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayil L, Chattopadhyay S, Bui A, Panse M, Cagle R, Mongodin EF, Sapkota AR (2022a) Viable bacteria abundant in cigarettes are aerosolized in mainstream smoke. Environmental Research 113462. 10.1016/j.envres.2022.113462 [DOI] [PubMed] [Google Scholar]
- Malayil L, Chattopadhyay S, Kulkarni P, Hittle L, Clark PI, Mongodin EF, Sapkota AR (2020a) Mentholation triggers brand-specific shifts in the bacterial microbiota of commercial cigarette products. Appl Microbiol Biotechnol 104:6287–6297. 10.1007/s00253-020-10681-1 [DOI] [PubMed] [Google Scholar]
- Malayil L, Chattopadhyay S, Mongodin EF, Sapkota AR (2022b) Bacterial communities of hookah tobacco products are diverse and differ across brands and flavors. Appl Microbiol Biotechnol 106:5785–5795. 10.1007/s00253-022-12079-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malayil L, Ramachandran P, Chattopadhyay S, Cagle R, Hittle L, Ottesen A, Mongodin EF, Sapkota AR (2020b) Metabolically-active bacteria in reclaimed water and ponds revealed using bromodeoxyuridine DNA labeling coupled with 16S rRNA and shotgun sequencing. Water Research 184:116185. 10.1016/j.watres.2020.116185 [DOI] [PubMed] [Google Scholar]
- Maraki S, Papadakis IS (2015) Rothia mucilaginosa pneumonia: a literature review. Infectious Diseases 47:125–129. 10.3109/00365548.2014.980843 [DOI] [PubMed] [Google Scholar]
- Martin RM, Bachman MA (2018) Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Frontiers in Cellular and Infection Microbiology 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire KL, Bent E, Borneman J, Majumder A, Allison SD, Treseder KK (2010) Functional diversity in resource use by fungi. Ecology 91:2324–2332. 10.1890/09-0654.1 [DOI] [PubMed] [Google Scholar]
- Michon J, Jeulin D, Lang J-M, Cattoir V (2010) Rothia aeria acute bronchitis: the first reported case. Infection 38:335–337. 10.1007/s15010-010-0012-5 [DOI] [PubMed] [Google Scholar]
- Millette NC, da Costa M, Mora JW, Gast RJ (2021) Temporal and spatial variability of phytoplankton and mixotrophs in a temperate estuary. Marine Ecology Progress Series 677:17–31. 10.3354/meps13850 [DOI] [Google Scholar]
- Miossec MJ, Valenzuela SL, Pérez-Losada M, Johnson WE, Crandall KA, Castro-Nallar E (2020) Evaluation of computational methods for human microbiome analysis using simulated data. PeerJ 8:e9688. 10.7717/peerj.9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan LG, DeMaere MZ, Cummins ML, Djordjevic SP, Roy Chowdhury P, Darling AE (2019) High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathogens 11:3. 10.1186/s13099-019-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali MF, Ghods S, Rehm BHA (2017) Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol 7:39. 10.3389/fcimb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morroni G, Alvarez-Suarez JM, Brenciani A, Simoni S, Fioriti S, Pugnaloni A, Giampieri F, Mazzoni L, Gasparrini M, Marini E, Mingoia M, Battino M, Giovanetti E (2018) Comparison of the Antimicrobial Activities of Four Honeys From Three Countries (New Zealand, Cuba, and Kenya). Frontiers in Microbiology 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalawade TM, Bhat K, Sogi SHP (2015) Bactericidal activity of propylene glycol, glycerine, polyethylene glycol 400, and polyethylene glycol 1000 against selected microorganisms. J Int Soc Prev Community Dent 5:114–119. 10.4103/2231-0762.155736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Hatori S, Wang Y, Li Y-Y, Kubota K (2020) Uncovering Viable Microbiome in Anaerobic Sludge Digesters by Propidium Monoazide (PMA)-PCR. Microb Ecol 79:925–932. 10.1007/s00248-019-01449-w [DOI] [PubMed] [Google Scholar]
- O’Neil CR, Wilson E, Missaghi B (2016) Bone and Joint Infections due to Haemophilus parainfluenzae: Case Report and Review of the Literature. Can J Infect Dis Med Microbiol 2016:4503025. 10.1155/2016/4503025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati A, Sikorski J, Nolan M, Lapidus A, Copeland A, Glavina Del Rio T, Lucas S, Chen F, Tice H, Pitluck S, Cheng J-F, Chertkov O, Brettin T, Han C, Detter JC, Kuske C, Bruce D, Goodwin L, Chain P, D’haeseleer P, Chen A, Palaniappan K, Ivanova N, Mavromatis K, Mikhailova N, Rohde M, Tindall BJ, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk H-P (2009) Complete genome sequence of Saccharomonospora viridis type strain (P101T). Stand Genomic Sci 1:141–149. 10.4056/sigs.20263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearman WS, Freed NE, Silander OK (2020) Testing the advantages and disadvantages of short- and long- read eukaryotic metagenomics using simulated reads. BMC Bioinformatics 21:220. 10.1186/s12859-020-3528-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez MT, Hörtnagl P, Sommaruga R (2010) Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ Microbiol 12:74–82. 10.1111/j.1462-2920.2009.02043.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pold G, Schimel JP, Sistla SA (2021) Soil bacterial communities vary more by season than with over two decades of experimental warming in Arctic tussock tundra. Elementa: Science of the Anthropocene 9:00116. 10.1525/elementa.2021.00116 [DOI] [Google Scholar]
- Portillo ME, Corvec S, Borens O, Trampuz A (2013) Propionibacterium acnes: An Underestimated Pathogen in Implant-Associated Infections. BioMed Research International 2013:e804391. 10.1155/2013/804391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M (2016) Aerococcus: an increasingly acknowledged human pathogen. Clinical Microbiology and Infection 22:22–27. 10.1016/j.cmi.2015.09.026 [DOI] [PubMed] [Google Scholar]
- Robbins S, Jacob J, Lu X, Moran MA, Mou X (2011) Bromodeoxyuridine (BrdU) Labeling and Subsequent Fluorescence Activated Cell Sorting for Culture-independent Identification of Dissolved Organic Carbon-degrading Bacterioplankton. J Vis Exp. 10.3791/2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger V, Fonty G, Andre C, Gouet P (1992) Effects of glycerol on the growth, adhesion, and cellulolytic activity of rumen cellulolytic bacteria and anaerobic fungi. Curr Microbiol 25:197–201. 10.1007/BF01570719 [DOI] [PubMed] [Google Scholar]
- Rooney AP, Swezey JL, Wicklow DT, McAtee MJ (2005) Bacterial species diversity in cigarettes linked to an investigation of severe pneumonitis in U.S. Military personnel deployed in operation iraqi freedom. Curr Microbiol 51:46–52. 10.1007/s00284-005-4491-z [DOI] [PubMed] [Google Scholar]
- Sapkota AR, Berger S, Vogel TM (2010) Human Pathogens Abundant in the Bacterial Metagenome of Cigarettes. Environ Health Perspect 118:351–356. 10.1289/ehp.0901201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Trantas EA, Mpalantinaki E, Ververidis F, Goumas DE (2012) Pseudomonas viridiflava, a Multi Host Plant Pathogen with Significant Genetic Variation at the Molecular Level. PLOS ONE 7:e36090. 10.1371/journal.pone.0036090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth EM, Chattopadhyay S, Babik K, Reid M, Chopyk J, Malayil L, Kulkarni P, Hittle LE, Clark PI, Sapkota AR, Mongodin EF (2019) The Bacterial Communities of Little Cigars and Cigarillos Are Dynamic Over Time and Varying Storage Conditions. Front Microbiol 10:2371. 10.3389/fmicb.2019.02371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth EM, Kulkarni P, Claye E, Stanfill S, Tyx R, Maddox C, Mongodin EF, Sapkota AR (2017) Smokeless tobacco products harbor diverse bacterial microbiota that differ across products and brands. Appl Microbiol Biotechnol 101:5391–5403. 10.1007/s00253-017-8282-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwetkowski KJ, Falkinham JO (2020) Methylobacterium spp. as Emerging Opportunistic Premise Plumbing Pathogens. Pathogens 9:149. 10.3390/pathogens9020149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Grossart H-P (2014) Community shifts of actively growing lake bacteria after N -acetyl-glucosamine addition: improving the BrdU-FACS method. ISME J 8:441–454. 10.1038/ismej.2013.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Zhao Y, Yuan X, Yi J, Fan J, Xu Z, Hu B, De Boer SH, Li X (2016) Dickeyafangzhongdai sp. nov., a plant-pathogenic bacterium isolated from pear trees (Pyrus pyrifolia). Int J Syst Evol Microbiol 66:2831–2835. 10.1099/ijsem.0.001060 [DOI] [PubMed] [Google Scholar]
- Treseder KK, Bent E, Borneman J, McGuire KL (2014) Shifts in fungal communities during decomposition of boreal forest litter. Fungal Ecology 10:58–69. 10.1016/j.funeco.2013.02.002 [DOI] [Google Scholar]
- Tyx RE, Stanfill SB, Keong LM, Rivera AJ, Satten GA, Watson CH (2016) Characterization of Bacterial Communities in Selected Smokeless Tobacco Products Using 16S rDNA Analysis. PLOS ONE 11:e0146939. 10.1371/journal.pone.0146939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebelacker L, Bunk S, Hochstöger J, Hackenberg S, Poier N, Ickrath P, Kleinsasser N (2019) [In vitro exposure of the shisha tobacco ingredient glycerol to human mucosa cells and lymphocytes]. Laryngorhinootologie 98:398–407. 10.1055/a-0885-1826 [DOI] [PubMed] [Google Scholar]
- Urbach E, Vergin KL, Giovannoni SJ (1999) Immunochemical Detection and Isolation of DNA from Metabolically Active Bacteria. Appl Environ Microbiol 65:1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-González F, Martínez-Porchas M, Villalpando-Canchola E, Vargas-Albores F (2016) Studying long 16S rDNA sequences with ultrafast-metagenomic sequence classification using exact alignments (Kraken). Journal of Microbiological Methods 122:38–42. 10.1016/j.mimet.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Vrabec M, Lovayová V, Dudriková K, Gallo J, Dudriková E (2015) Antibiotic Resistance and Prevalence of Enterococcus Spp. and Escherichia Coli Isolated from Bryndza Cheese. Italian Journal of Animal Science 14:3968. 10.4081/ijas.2015.3968 [DOI] [Google Scholar]
- Wahdan SFM, Heintz-Buschärt A, Sansupa C, Tanunchai B, Wu Y-T, Schädler M, Noll M, Purahong W, Buscot F (2021) Targeting the Active Rhizosphere Microbiome of Trifolium pratense in Grassland Evidences a Stronger-Than-Expected Belowground Biodiversity-Ecosystem Functioning Link. Frontiers in Microbiology 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Duan L, Liu F, Hu Y, Leng C, Kan Y, Yao L, Shi H (2020) First report of Enterobacter hormaechei with respiratory disease in calves. BMC Veterinary Research 16:1. 10.1186/s12917-019-2207-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzler E, Kamboj K, Balada-Llasat J-M (2015) Severe Sepsis Secondary to Persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus Bacteremia. International Journal of Infectious Diseases 35:93–95. 10.1016/j.ijid.2015.04.016 [DOI] [PubMed] [Google Scholar]
- Wood DE, Lu J, Langmead B (2019) Improved metagenomic analysis with Kraken 2. Genome Biology 20:257. 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tian Y, Li X, Hu B (2018) Complete Genome Sequence of a Dickeya fangzhongdai Type Strain Causing Bleeding Canker of Pear Tree Trunks. Genome Announcements 6:e00177–18. 10.1128/genomeA.00177-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Network plot showing the unique and shared bacterial species between commercial and research cigarettes.
Figure S2: Average relative abundance of bacterial genera across all tobacco products and brands.
Figure S3: Bacterial alpha diversity by tobacco product, brand and flavor.
Figure S4: Bacterial a) alpha and b) beta diversity between BrdU-treated (BRDU) and non-treated (NOTRT) tobacco samples.
Data Availability Statement
Data concerning the samples included in this study are deposited under the following NCBI BioProject accession numbers: PRJNA635703, PRJNA473598, PRJNA601146, and PRJNA641233.