Abstract
Vα24-invariant natural killer T cells (NKTs) possess innate antitumor properties that can be exploited for cancer immunotherapy. We have shown previously that the CD62L+ central memory-like subset of these cells drives the in vivo antitumor activity of NKTs, but molecular mediators of NKT central memory differentiation remain unknown. Here, we demonstrate that relative to CD62L− cells, CD62L+ NKTs express a higher level of the gene encoding the Wnt/β-catenin transcription factor lymphoid enhancer binding factor 1 (LEF1) and maintain active Wnt/β-catenin signaling. CRISPR/Cas9-mediated LEF1 knockout reduced CD62L+ frequency after antigenic stimulation, while Wnt/β-catenin activator Wnt3a ligand increased CD62L+ frequency. LEF1 overexpression promoted NKT expansion and limited exhaustion following serial tumor challenge and was sufficient to induce a central memory–like transcriptional program in NKTs. In mice, NKTs expressing a GD2-specific chimeric-antigen receptor (CAR) with LEF1 demonstrated superior control of neuroblastoma xenograft tumors compared to control CAR-NKTs. These results identify LEF1 as a transcriptional activator of the NKT central memory program and advance development of NKT cell–based immunotherapy.
Synopsis
NKT cells employ an undefined mechanism to undergo central memory–like differentiation, which is associated with increased antitumor potential. The authors demonstrate that LEF1 drives central memory programming in human NKTs, informing rational design of NKT-based cancer immunotherapy.
Introduction
Vα24-invariant natural killer T cells (NKTs) are an evolutionarily conserved subset of innate lymphocytes that express an invariant TCR α-chain Vα24–Jα18 and recognize glycolipids presented by monomorphic MHC class I–like molecule CD1d (1). NKTs are characterized by innate antitumor properties including an ability to traffic to tumor sites in response to tumor-derived chemokines (2,3), to kill tumor-associated macrophages in the tumor microenvironment (3,4), and to trans-activate antitumor responses mediated by T and NK cells (1). In patients, infiltration of NKTs into primary tumors correlates with favorable outcome in several cancer types (2,5). These observations prompted the development of NKT-based immunotherapies for cancer patients. Early-stage clinical trials of adoptively transferred NKTs or chimeric-antigen receptor (CAR)-redirected NKTs showed that therapeutic NKTs are well tolerated and can produce objective responses in patients with various types of cancer (6,7). However, NKT-cell immunobiology remains poorly understood, limiting the development of NKT-based therapeutics.
NKTs constitute only 0.01% – 1% of human PBMCs (8), meaning that robust ex vivo expansion methods are required to generate cell numbers sufficient for clinical use. However, extensive expansion of T lymphocytes ex vivo is often coupled with terminal differentiation, exhaustion, and impaired therapeutic efficacy in preclinical tumor models (9) and in patients (10). Conversely, preservation of T cells that have stem-like properties such as T central memory or memory stem cells is associated with increased therapeutic efficacy of adoptive immunotherapy products (11). Although primary human NKTs in adult peripheral blood mostly exhibit an effector memory–like phenotype, we recently demonstrated that antigen-induced ex vivo expansion leads to accumulation of CD62L+ central memory–like NKTs, which are largely responsible for in vivo persistence and antitumor activity of CAR-redirected NKTs (12). Key mediators of this central memory-like program in NKTs remain to be identified.
T-cell factor 1 (TCF1) and the related transcription factor lymphoid enhancer-binding factor 1 (LEF1) are key regulators of Wnt/β-catenin signaling (13). In mature T cells, TCF1 and Wnt/β-catenin signaling play crucial roles in supporting the formation of central memory and stem cell memory cells (14-16), while LEF1 has a redundant and less critical role (17). In contrast, murine studies of NKT-cell development demonstrated that LEF1 is critical for the early stages of NKT-cell differentiation in the thymus (18), and we previously reported that LEF1 was among the top overexpressed immune-related genes in human CD62L+ NKTs compared to CD62L− cells (12). However, the causal role of LEF1 in NKT cell central memory differentiation has not been described. Here, we have demonstrated that LEF1 drives a central memory–like transcriptional program in human NKTs. We found that ectopic overexpression of LEF1 alleviated exhaustion and preserved stemness in NKTs after multiple rounds of T-cell receptor (TCR) stimulation. Further, NKTs that co-express LEF1 with a GD2-CAR demonstrated superior long-term antitumor activity in an in vivo neuroblastoma model. These findings identify LEF1 as a key regulator of the central memory–like transcriptional program in human NKTs and pave the way for the development of next-generation NKT-based cancer immunotherapy products.
Materials and Methods
Cell lines and culture conditions
K562 and 293T cells were purchased from the American Type Culture Collection. The artificial antigen-presenting cell, the B-8-2 clone of the K562 cell line was derived previously in our laboratory (12). CHLA-255 and Jurkat J32 cell lines were established and maintained as previously described (19,20). CHLA-255 with luciferase expression was generated by transducing parental CHLA-255 cells with the GFP.FFLuc retroviral plasmid (See Retro- and lentiviral constructs and retrovirus production). CHLA-255 and 293T were cultured in IMDM (HyClone, SH30259.01) and all other cell lines were maintained in RPMI 1640 (HyClone, SH30096.01). All cell culture medium was supplemented with 2 mM GlutaMAX-I (Life Technologies, 35050061) and 10% FBS (HyClone, SH30071.01) (complete RPMI) with the exception of CHLA-255 medium, which was supplemented with 20% FBS (complete IMDM). Cell lines with low passage number were thawed and used within 10 passages. Cell line authentication was performed using STR fingerprinting at MD Anderson Cancer Center within one year of use, and cell lines were checked for mycoplasma contamination (Lonza MycoAlert, LT07-318) every two months.
NKT-cell isolation, transduction, expansion, and sorting
Peripheral blood of healthy donors was purchased from Gulf Coast Regional Blood Center. PBMCs were isolated from buffy coats by Ficoll density gradient centrifugation (GE Healthcare, 17-5442-03). NKTs were purified using anti-iNKT microbeads according to the manufacturer’s instructions (Miltenyi Biotec, 130-094-842), and the negative PBMC fraction was irradiated (40 Gy) and aliquoted at 5 x 106 cells per ml for each donor. Approximately 1 x 106 NKTs were stimulated with an aliquot of 5 x 106 autologous PBMCs pulsed with α-galactosylceramide (αGalCer); 100 ng/mL; Avanti Polar Lipids, 86700P-1mg) per well in a 24-well plate. The culture was supplemented every other day with recombinant IL2 (200 U/mL; National Cancer Institute/TECIN™, 23-6019) in complete RPMI. NKTs were expanded for 10 to 12 days and then restimulated at a 2:1 or 4:1 NKT to B-8-2 ratio using 1 x 106 B-8-2 cells (irradiated with 100 Gy) per well in a 24-well plate. Twenty four–well, non-tissue culture treated plates were coated with RetroNectin (Takara Bio, T100B) overnight. On day 7 after primary stimulation or day 2 after restimulation, RetroNectin-coated plates were washed, inoculated with 1 ml of retroviral supernatant (See Retro- and lentiviral constructs and retrovirus production), and spun for 60 minutes at 4000 g. Viral supernatant was then removed, and 5 x 105 stimulated NKTs were added per well to complete media and 200 U/ml IL2. Cells were removed from the plate after 48 hours, washed, resuspended at 1 x 106 cells/ml in complete RPMI with IL2, and plated for continued expansion. NKT number was determined by trypan blue (Life Technologies, 15250061) counting. When indicated, NKTs were labeled with CD62L-PE mAb (DREG-56; BD Biosciences) and anti-PE microbeads (Miltenyi Biotec, 130-048-801) followed by magnetic sorting into CD62L+ and CD62L− subsets according to the manufacturer’s instructions. Sorted cell phenotype was determined by flow cytometry (see Flow cytometry).
To activate the Wnt/β-catenin signaling pathway, freshly isolated NKTs (approximately 1 x 106) were treated with 5 μM TWS119 (Sigma-Aldrich, 361554) on day 7 after primary stimulation or with 500 ng/mL recombinant Wnt3a (R&D Systems, 5036-WN-010) on days 0, 1, and 2 after primary stimulation as indicated. To inhibit Wnt/β-catenin signaling, NKTs were treated with 3 μM ICG-001 (Cayman Chemical, 16257) on days 0, 1, and 2 after primary stimulation as indicated.
Where indicated, 10 ng/ml of human recombinant IL21 (PeproTech, 200-21) was added to NKT cells in culture every other day to preserve the CD62L+ subset.
Retro- and lentiviral constructs and retrovirus production
CAR.GD2 constructs were generated as previously described (21) and included the following components: a single-chain variable fragment (scFv) from the GD2-specific antibody 14G2a connected via a short spacer derived from the IgG1 hinge region to the transmembrane domain of CD8α, followed by the signaling endodomain of 4-1BB fused with the CD3ζ signaling chain. As indicated, CAR.GD2 constructs also included a sequence encoding LEF1 downstream of CD3ζ separated by a p2A sequence. The LEF1 sequence was obtained from Origene and was inserted into the same retroviral vector backbone provided by Dr. Gianpietro Dotti (University of North Carolina) used for the CAR.GD2 constructs. Non-CAR retroviral constructs used in this study included GFP.FireflyLuciferase (GFP.FFLuc; plasmid pSFG-GFP-effluc from Dr. Brian A. Rabinovich) and GFP.LEF1, which consisted of GFP as a transduction marker separated from the over-expressed target protein by a p2A sequence. Retroviral supernatants were produced by transfecting 293T cells using GeneJuice (EMD Milliopore, 70967) according to the manufacturer’s protocol with a combination of the relevant retroviral plasmid, the RDF plasmid (gift from Dr. Mary Collins, Baylor College of Medicine (BCM)) encoding the RD114 envelope protein, and the PeqPam3 plasmid (gift from Dr. Elio Vanin, BCM) encoding the Moloney murine leukemia virus gag-pol fusion. The lentiviral construct encoding the 7-TGC Wnt reporter construct (22) was a generous gift from Drs. J. Rosen and K. Roarty (BCM), and the envelope plasmid pMD2.G and packaging plasmid δ8.2 were generous gifts from Drs. K. Scott and Y.H. Tsang (BCM). Lentiviral supernatants were similarly generated from 293T cells using GeneJuice and transfected with the relevant lentiviral construct, pMD2.G, and δ8.2. To enhance transduction efficiency, lentiviral supernatants were concentrated using Retro-X Concentrator (Takara Bio, 631456) according to the manufacturer’s protocol.
Multiplex cytokine quantification assay
CAR.GD2-NKTs were stimulated in IMDM with 20% FBS for 24 hours with CHLA-255 neuroblastoma cells at a 1:1 effector to target (E:T) ratio. Supernatants were collected and analyzed using the MILLIPLEX MAP Human Cytokine/Chemokine Immunoassay panel (Millipore, HCYTOMAG-60K-07) for Luminex analysis according to the manufacturer’s protocol.
Flow cytometry
NKT-cell phenotype was assessed using fluorescently conjugacted mAbs for CD3 (UCHT1), Vα24-Jα18 (6B11), CD4 (RPA-T4), CD62L (DREG-56), PD-1 (EH12.1), TIM-3 (7D3), CCR7 (2-L1-A), CD27 (L128), CCR4 (L291H4), CD39 (A1), CD160 (BY55), LAG-3 (3DS223H), and Vβ11 (C21). All antibodies were purchased from BD Biosciences except for CCR4 (BioLegend), CD39 (BioLegend), CD160 (BioLegend), LAG-3 (Invitrogen), and Vβ11 (Beckman Coulter). CAR.GD2 expression by transduced NKTs was detected with the 14G2a anti-idiotype 1A7 mAb (purified from 1A7 mouse hybridoma (ATCC, HB-11786), which was custom-conjugated to Alexa Fluor 647 or Alexa Fluor 488 by BioLegend. Mitochondrial mass was measured using 25 nM MitoTracker Deep Red (ThermoFisher Scientific, M22426) or MitoTracker Green (ThermoFisher Scientific, M7514) according to the manufacturer’s protocol. Transcription factor staining was performed using the eBioscience Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher Scientific, 00-5523-00) with mAbs for long isoforms of LEF1 (C12A5) and TCF1 (C63D9), both from Cell Signaling Technology, and all isoforms of LEF1 (EP2030Y, Abcam).
Cytokine and degranulation staining were performed on expanded NKTs that were stimulated using PMA (50 ng/ml; Sigma-Aldrich, P1585) and ionomycin (500 ng/ml; Sigma-Aldrich, I9657) in the presence of GolgiPlug (BD Biosciences, 555029, GolgiStop (BD Biosciences, 554724), and CD107a antibody (H4A3; Biolegend). After a four-hour incubation, NKTs were washed then stained with eFluor 780 fixable viability dye (eBioscience, 65-0865-18) and for surface markers. Cells were then fixed and permeabilized using the Foxp3/Transcription factor buffer set following the manufacturer’s protocol. Permeabilized cells were stained overnight with LEF1, IL2 (MQ1-17H12, BD Biosciences), and IFNγ (B27, BD Biosciences) antibodies.
Fluorochrome- and isotype-matching mAbs for CD62L, PD-1, TIM-3, CCR7, CD27, CCR4, CD39, CD160, LAG-3, LEF1, TCF1, CD107a, IL-2, and IFNγ suggested by BD Biosciences, Cell Signaling Technology, or R&D Systems were used as negative controls. Analysis was performed on an LSR II or Symphony A5 five-laser flow cytometer (BD Biosciences) using BD FACSDiva software version 6.0 and FlowJo 10.8 (Tree Star).
In vitro cytotoxicity assay
CAR-mediated cytotoxicity of CAR.GD2-NKTs against CHLA-255 cells was evaluated using a 4-hour luciferase assay as previously described (21).
In vitro repeat killing assay
J32 leukemia cells were pulsed with αGalCer (100 ng/mL) overnight followed by CellTrace Violet (ThermoFisher Scientific, C34557) staining. Labeled J32 cells were co-cultured with NKTs transduced with GFP.LEF1 or GFP.FFLuc at the indicated E:T ratio in the presence of IL2 (50 U/mL). On day 3, tumor killing and NKT-cell expansion were measured by flow cytometry using CountBright Absolute Counting Beads (ThermoFisher Scientific, C36950). Fresh αGalCer-pulsed J32 cells were added to the culture according to NKT-cell number to re-establish the E:T ratio. This step was repeated every three days of co-culture. NKT-cell exhaustion/memory phenotype was assessed by flow cytometry after six days of resting from J32 stimulation using mAbs for PD-1, TIM-3 and CD62L.
LEF1 CRISPR-Cas9 gene editing
Two guide RNAs (gRNAs) specific for the LEF1 gene (CCCGGAATAACTCGAGTAGG and GTCACTGTAAGTGATGAGGG) were designed using CRISPRscan and COSMID algorithms (23,24). The LEF1 gene was genomically disrupted in NKTs according to the published protocol (25). Briefly, the 20-nucleotide sequence complementary to the specific gene locus was incorporated into an oligonucleotide primer and used to amplify the gRNA scaffold from the PX458 plasmid (a gift from Feng Zhang; Addgene 48138). gRNAs were generated through in vitro transcription using the High-Yield RNA Synthesis Kit (NEB Bio Labs, E2040S) from the DNA template following the manufacturer’s instructions and purified using the RNA Clean & Concentrator-25 kit (Zymo Research, R1017). The Neon Transfection System (ThermoFisher Scientific, MPK5000) was used to electroporate 5 μg of gRNA and 10 μg of Cas9 protein (PNA Bio, CP03) into 2 x 106 activated NKTs in 100 μL of buffer R using three 1600-V 10-milliseconds pulses. Following electroporation, NKTs were incubated in complete RPMI overnight. NKTs were then expanded in media supplemented with 200 U/mL IL2.
Seahorse assays
The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using the Agilent Seahorse XFe96 Analyzer (Agilent) per the manufacturer’s instructions. GFP.FFLuc-NKTs and GFP.LEF1-NKTs were assayed on day 12 post-secondary stimulation after normalization of transduction rate.
Quantitative real-time PCR (qRT-PCR)
Wnt signaling components and downstream gene expression were measured in NKTs by qPCR with specific primers. Briefly, RNA was extracted from unsorted or CD62L+/− sorted NKTs using the RNeasy Plus Mini Kit (QIAGEN, 74134). Twenty nanograms of isolated RNA was then subjected to reverse transcription and qPCR reaction using the Power SYBR Green RNA-to-CT 1-Step Kit (ThermoFisher Scientific, 4389986) on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, 1845096). Relative expression of Wnt target genes in CD62L+ versus CD62L− NKTs was determined using the ΔCt method with GAPDH as housekeeping gene. To detect expression of Wnt receptors, co-receptors, and ligands on expanded NKTs, Ct values of target gene expression were compared to Ct values of positive (GAPDH) and negative (CD19) controls. Ct values that fell within the range of GAPDH and CD19 Ct values were considered significantly expressed. For peripheral blood NKTs before expansion, NKTs were isolated from healthy donor buffy coats as previously described (see NKT-cell isolation, transduction, expansion, and sorting) and further enriched by FACS to improve purity. To increase the amount of available cDNA for qPCR, 10 ng of isolated RNA from each donor was subjected to whole transcriptome amplification using the QuantiTect Whole Transcriptome Kit (Qiagen, 207045) according to the manufacturer’s instructions. After amplification, cDNA was diluted 1:250 and used for qPCR reactions in triplicate for each target gene. Primers were used at the final concentration of 200 nM and are listed in Table 1.
Table 1.
List of primers used in real-time quantitative PCR
| Primer ID | Sequence |
|---|---|
| GAPDH F | 5’-GAAGGTGAAGGTCGGAGTC-3’ |
| GAPDH R | 5’-GAAGATGGTGATGGGATTTC-3’ |
| CD19 F | 5’-TGCCCCGTCTTATGGAAACC-3’ |
| CD19 R | 5’-CTCTTCTTCTGGGCCCACTC-3’ |
| AXIN 2 F | 5’-ACTGCCCACACGATAAGGAG-3’ |
| AXIN 2 R | 5’-CTGGCTATGTCTTTGCACCA-3’ |
| TCF1 F | 5’-TGCAGCTATACCCAGGCTGG-3’ |
| TCF1 R | 5’-CCTCGACCGCCTCTTCTTC-3’ |
| MYC F | 5’-TACCCTCTCAACGACAGCAG-3’ |
| MYC R | 5’-TCTTGACATTCTCCTCGGTG-3’ |
| WNT1 F | 5’-CGGCGTTTATCTTCGCTATC-3’ |
| WNT1 R | 5’-TTCGATGGAACCTTCTGAGC-3’ |
| WNT2 F | 5’-AAAGAAGATGGGAAGCGCCA-3’ |
| WNT2 R | 5’-TTCATCAGGGCTCTGGCATC-3’ |
| WNT3A F | 5’-TGTTGGGCCACAGTATTCCT-3’ |
| WNT3A R | 5’-GGGCATGATCTCCACGTAGT-3’ |
| WNT6 F | 5’-CAGCCCCTTGGTTATGGAC-3’ |
| WNT6 R | 5’-AACTGGAACTGGCACTCTCG-3’ |
| WNT8A F | 5’-AGGCTGAGAAGTGCTACCAGA-3’ |
| WNT8A R | 5’-CCATTGTTTGACCCATCACA-3’ |
| FZD1 F | 5’-CATCGTCATCGCCTGCTACT-3’ |
| FZD1 R | 5’-TAGCGTAGCTCTTGCAGCTC-3’ |
| FZD2 F | 5’-CTTCTCACAGGAGGAGACGC-3’ |
| FZD2 R | 5’-AAATGATAGGCCGCTCTGGG-3’ |
| FZD3 F | 5’-ACAGCAAAGTGAGCAGCTACC-3’ |
| FZD3 R | 5’-CTGTAACTGCAGGGCGTGTA-3’ |
| FZD4 F | 5’-TCAAGAGACGCTGTGAACCC-3’ |
| FZD4 R | 5’-GGTCGTTCTGTGGTGGGAAT-3’ |
| FZD5 F | 5’-GCACAACCACATCCACTACG-3’ |
| FZD5 R | 5’-GCACAACCACATCCACTACG-3’ |
| FZD6 F | 5’-AGGCTTGCACCGTTTTGTTC-3’ |
| FZD6 R | 5’-TGCTCGATGGCTTCACAACT-3’ |
| FZD7 F | 5’-CGCCTCTGTTCGTCTACCTC-3’ |
| FZD7 R | 5’-TCATGATGGTGCGGATACGG-3’ |
| FZD8 F | 5’-CGCCTCTGTTCGTCTACCTC-3’ |
| FZD8 R | 5’-TCATGATGGTGCGGATACGG-3’ |
| FZD9 F | 5’-TGCTCACCTTCTTGCTGGAG-3’ |
| FZD9 R | 5’-GCCAGCGAGTAGACGTTGTA-3’ |
| FZD10 F | 5’-AAGAAGAGCCGGAGAAAACC-3’ |
| FZD10 R | 5’-GACTGGGCAGGGATCTCATA-3’ |
| LRP5 F | 5’-CACCACCTTCTTGCTGTTCA-3’ |
| LRP5 R | 5’-GCTTTGACGTTCCTCAGTCC-3’ |
| LRP6 F | 5’-GACTGGGTTGCACGAAATCT-3’ |
| LRP6 R | 5’-CGGGGTTCCTCTAAGTCCTC-3’ |
In vivo experiments
Animal experiments were performed in accordance with and with the approval of the BCM Institutional Animal Care and Use Committee (IACUC) under protocol AN-5194. NSG mice were obtained from The Jackson Laboratory and maintained at the BCM animal care facility. To test the anti-neuroblastoma effect of CAR.GD2-NKTs, mice were injected i.v. with 1 x 106 luciferase-transduced CHLA-255 neuroblastoma cells to initiate tumor growth. On day 7, mice were injected i.v. with 4 x 106 CAR.GD2-NKTs followed by i.p. injection of IL2 (2000 U/mouse) every other day for 2 weeks. Tumor growth was assessed once per week by bioluminescent imaging (Perkin Elmer IVIS Lumina III in vivo imaging system, CLS136334, Small Animal Imaging Facility, Texas Children’s Hospital) with intraperitoneal injection of luciferin (Perkin Elmer, 122799) and five minute exposure.
Bulk RNAseq and data analysis
To determine the effect of LEF1 overexpression on the NKT transcriptome, bulk RNAseq was conducted on six donor-paired samples of GFP+ cells sorted by FACs from GFP.LEF1- or GFP.FFLuc-transduced NKTs. To evaluate expression of Wnt components, bulk RNAseq was conducted on thirteen samples of NKTs that had been expanded for 17–20 days (primary and secondary expansion). RNA was isolated from all NKT cell samples using the RNeasy Plus Mini Kit (QIAGEN, 74134). Novogene performed library synthesis and paired-end sequencing on isolated RNA samples using Illumina PE150 technology.
Adaptor sequences were removed from raw RNAseq reads. Data quality was evaluated using FastQC (version 0.11.2) software to determine whether further data filtering criteria should be applied. Clean reads were mapped onto the human reference genome (GRCh38.p13 assembly) using HISAT2 software (v2.1.0). Following genome alignment, mapped reads were assembled into transcripts or genes using StringTie software (v1.3.5). Differential gene expression analysis was performed using the DESeq2 R package based on raw counts. Genes with greater than two-fold change difference (FC≥2 or FC≤0.5) between two groups with p-adj <0.05 were retained as significantly expressed genes.
Gene set enrichment analysis (GSEA) was conducted using GSEA desktop software on C7 immunological signature gene sets. Enrichment for central memory T-cell signature (GSE3782), naïve versus NK-like signature (GSE22886), effector memory T-cell signature (GSE11057), effector T-cell signature (26,27), and naïve T-cell signature (GSE15930) were highlighted.
Nanostring gene expression analysis
Total RNA was collected from CD62L+ and CD62L− cells that were sorted by MACS from NKTs expanded for 10 days, using the TRIzol reagent (Qiagen, 79306) according to the manufacturer's protocol. Gene expression analysis was performed using Immunology Panel version 2 (NanoString) at BCM Genomic and RNA Profiling Core with the nCounter Analysis System. Data were analyzed using nSolver 2.0 software (NanoString).
GSEA was performed similarly as in Bulk RNAseq and data analysis. Enrichment of central memory T-cell-like signature (GSE11057) was highlighted.
Statistics
The Shapiro-Wilk test was used to assess normality of continuous variables. Normality was rejected when the p value was less than 0.05. For non-normally distributed data, we used the Mann-Whitney U test to evaluate differences in continuous variables between two groups. To evaluate differences in continuous variables, we used two-sided, paired Student t tests to compare two groups, one-way ANOVA with post-test Bonferroni correction to compare more than two groups, and two-way ANOVA with the Sidak post hoc test to compare in a two-by-two setting. Survival was analyzed using the Kaplan-Meier method with the log-rank (Mantel-Cox) test to compare two groups. Statistics were computed using GraphPad Prism 7 (GraphPad Software). Differences were considered significant when the p value was less than 0.05.
Data Availability
All requests for raw and analyzed data and materials beyond the information provided in the main text and supplementary figures should be directed to the corresponding author. These requests will be promptly reviewed by the BCM Licensing Group to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement. All raw data for RNA sequencing has been deposited in the Gene Expression Omnibus (GEO) at accession number GSE217281.
Results
LEF1-expressing NKTs phenotypically and functionally resemble central memory T cells
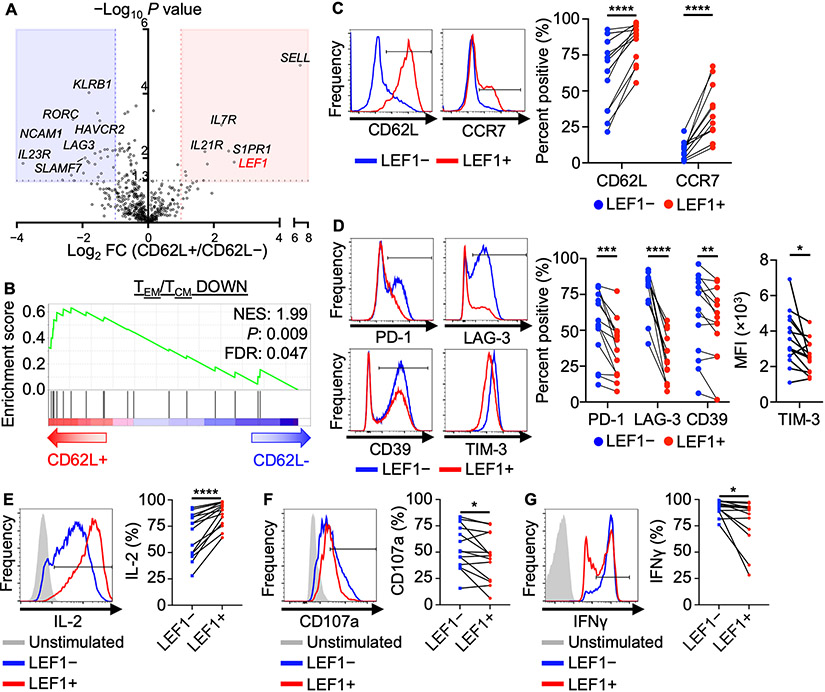
To identify genes that regulate the CD62L central memory–like program in NKTs, we performed immune-related gene expression analysis on CD62L+ and CD62L− sorted NKTs using the nCounter platform. Wnt transcription factor LEF1 was the top overexpressed gene in the CD62L+ subset except for SELL, which encodes CD62L (Fig. 1A). Next, we utilized gene-set enrichment analysis (GSEA) to determine whether CD62L+ and CD62L− NKTs follow a similar dichotomy in transcriptional programming to that observed in central memory (TCM) versus effector memory (TEM) T cells. We found that CD62L+ NKTs were significantly enriched for an established CD4+ TCM gene signature (P = 0.009), suggesting that they are similar to TCM at the mRNA level (Fig. 1B). Next, we confirmed by flow cytometry that LEF1+ NKTs express higher levels of central memory markers including CD62L and CCR7 (Fig. 1C). We found that only the long isoforms of LEF1 that contain the β-catenin–interacting domain were upregulated in CD62L+ NKTs, not total LEF1 including both short and long isoforms (Supplementary Fig. S1A-B). Given that TCF1 plays a more crucial role than LEF1 in the formation of TCM (17), we evaluated expression patterns of both proteins in primary human NKTs. While LEF1 expression was largely restricted to the CD62L+ subset, TCF1 was expressed at a similar level in both CD62L+ and CD62L− populations (Supplementary Fig. S1C), confirming the importance of LEF1 in distinguishing CD62L+ central memory–like NKTs.
Figure 1: CD62L+ NKTs express elevated levels of Wnt transcription factor LEF1 and are phenotypically central memory–like.
A, Twelve days after primary stimulation with αGalCer-pulsed autologous PBMCs, NKTs were magnetically sorted into CD62L+ and CD62L− subsets and processed for RNA isolation and Nanostring gene expression analysis. Volcano plot shows differential gene expression in CD62L+ versus CD62L− NKTs with P value cutoff at 0.05. B, Gene set enrichment analysis (GSEA) plot showing enrichment for a central memory CD4+ T-cell signature (GSE11057) in CD62L+ NKTs. C-D, Twelve days after stimulation, NKTs were gated into LEF1− (blue) and LEF1+ (red) populations, and surface expression of (C) central memory markers and (D) activation/exhaustion markers were measured by flow cytometry. Representative donor histograms and paired percent positive or MFI (n = 16 donors) are shown. E–G, Ten days after stimulation, NKTs were activated by 50 ng/ml PMA and 500 ng/ml ionomycin in the presence of GolgiPlug and GolgiStop and analyzed for (E) IL2, (F) CD107a, and (G) IFNγ expression by flow cytometry. Representative histograms and paired percent positive (n = 12 donors) are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns: not significant, paired Student’s t test for paired result from each donor.
Further, LEF1+ NKTs expressed lower levels of activation/exhaustion markers including PD-1, TIM-3, LAG-3, and CD39 than LEF1− NKTs, suggesting the former may be less prone to exhaustion (Fig. 1D). Functionally, LEF1+ NKTs produced more IL2, a cytokine associated with central memory and stem-like T cells (28), than LEF1− NKTs (Fig. 1E). Consistent with the more quiescent phenotype associated with the central memory state, LEF1+ NKTs produced lower levels of IFNγ and expressed lower levels of the degranulation marker CD107a than LEF1− NKTs following stimulation (Fig. 1F,G). Together, these findings suggest that LEF1 expression defines a subset of NKTs with a central memory–like phenotype.
LEF1 expression and Wnt/β-catenin signaling support formation of central memory–like NKTs
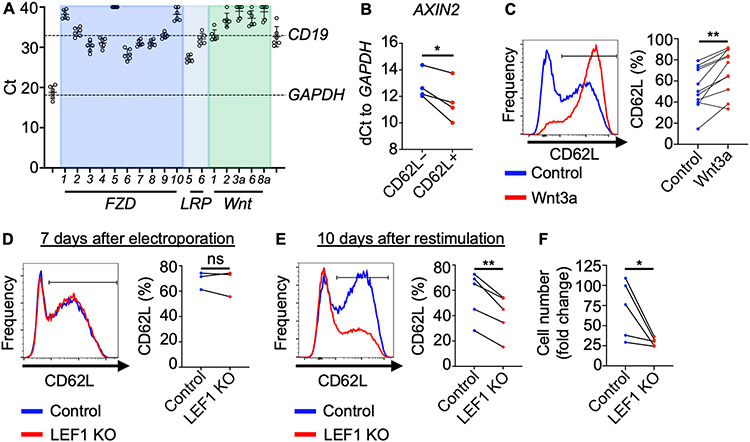
Since LEF1 is a known Wnt transcription factor, we used qRT-PCR to determine whether NKTs express receptors and ligands that can activate Wnt/β-catenin signaling. GAPDH and CD19 were used to set a range of Ct values from maximum to undetectable expression, respectively, between which expression levels of FZD receptors, LRP co-receptors, and Wnt ligands were evaluated. Primary-expanded NKTs expressed several FZD receptors including FZD3 and FZD6 as well as co-receptor LRP5, but none of the five selected Wnt ligands that have been reported to activate Wnt/β-catenin signaling (13) (Fig.2A). This suggests that NKTs receive signals from exogenous Wnt ligands. RNA isolated from NKTs was also subject to next-generation sequencing, and results for Wnt receptor expression were consistent with qRT-PCR findings (Supplementary Fig. S2A). To determine expression of Wnt receptors and ligands in freshly isolated primary NKTs, we isolated NKT cells from PBMCs using a combination of magnetic and FACS sorting techniques prior to placing the cells in culture. We found that freshly isolated NKTs expressed lower levels of Wnt receptors than NKTs that have been expanded in vitro (Supplementary Fig. S2B). To determine whether increased LEF1 expression in CD62L+ NKTs is associated with downstream Wnt/β-catenin signaling, we evaluated expression of Wnt target genes including AXIN2, MYC, and TCF7 (29) in CD62L+ and CD62L− NKTs by qRT-PCR. Expression levels of all three genes were elevated in CD62L+ relative to CD62L− cells, suggesting that Wnt/β-catenin signaling is active in the CD62L+ subset (Fig. 2B; Supplementary Fig. S2C). To detect Wnt signaling in NKTs, we employed lentiviral construct 7-TGC, which specifically reports a high level of β-catenin transcriptional activity (22) (Supplementary Fig. S2D). NKTs transduced with 7-TGC were detected as GFP+ after treatment with Wnt activator TWS119, which functions by inhibiting GSK3β, indicating active Wnt signaling (13) (Supplementary Fig. S2E). A significantly higher proportion of NKTs transduced with 7-TGC were CD62L+ in the GFP+ versus GFP− population; that is, there were more CD62L+ NKTs in the Wnt/β-catenin signaling-active population (Supplementary Fig. S2F).
Figure 2: CD62L+ NKTs undergo elevated Wnt/β-catenin signaling, favoring formation of central memory-like NKTs.
A, NKTs were expanded ex vivo for 10 days and qRT-PCR was performed on isolated RNA to determine expression levels of Wnt receptor, co-receptor, and ligand mRNA. GAPDH (positive control) and CD19 (negative control) expression were used to set a range of significant Ct values. Mean ± SD are from a total of six donors. B, On day 10 of ex vivo expansion, NKTs were magnetically sorted into CD62L+ and CD62L− subsets and expression of AXIN2 as Wnt target gene was determined from isolated RNA using qRT-PCR and the dCt method (n = 4 donors). C, NKTs were treated with 500 ng/mL human recombinant Wnt3a or PBS for three days following stimulation. CD62L expression was determined 10–12 days after initial TCR stimulation. Representative donor histogram and paired CD62L+ percentage (n = 11 donors) are shown. D–F, NKTs were electroporated with Cas9 only (control) or Cas9 with LEF1 gRNA. CD62L expression was analyzed by flow cytometry (D) seven days after electroporation, prior to secondary stimulation with αGalCer-pulsed B-8-2 cells (n = 3 donors), and (E) 10 days after secondary stimulation. Representative donor histograms and mean ± SEM CD62L+ percentage (n = 5 donors) are shown. (F) NKT number was measured 10 days after secondary stimulation, and expansion fold change was calculated from day 0. Mean fold change ± SEM (n = 5 donors) is shown. *P < 0.05, **P < 0.01, ns: not significant, paired Student’s t test for paired result from each donor.
Next, to determine whether CD62L expression depends on Wnt/β-catenin signaling, we stimulated NKTs with αGalCer and treated them with ICG-001, which inhibits downstream Wnt signaling by blocking interaction of β-catenin and transcriptional co-factor cAMP response element-binding protein (CREB) (30). NKTs treated with ICG-001 during ex vivo expansion had reduced CD62L expression (Supplementary Fig. S2G), suggesting that basal Wnt signaling plays a role in the generation and maintenance of CD62L+ NKTs after antigenic stimulation. To determine whether exogenous activation of Wnt/β-catenin signaling increases the frequency of CD62L+ NKTs, we treated NKTs with human recombinant Wnt3a ligand during ex vivo expansion and found that it significantly increased the rate of CD62L expression in NKTs after primary expansion (Fig. 2C). Wnt3a treatment did not impact expression of two other central memory markers, CCR7 and CD27, and moderately reduced expression of the activation/exhaustion marker LAG-3 (Supplementary Fig. S3A,B). Treatment with TWS119 during ex vivo expansion also increased the frequency of CD62L+ NKTs (Supplementary Fig. 3C).
We recently reported that IL21 specifically protects CD62L+ NKTs from apoptosis (31). To determine whether Wnt signaling and IL21 signaling additively increase the frequency of CD62L+ cells, we expanded NKTs with IL2 alone (default condition) or IL2 with IL21, Wnt3a, or both. We found that addition of either IL21 or Wnt3a promoted formation of CD62L+ NKTs, and the the combination of both increased the frequency of CD62L+ NKTs beyond addition of either agent alone (Supplementary Fig. 3D). Similar results were obtained when IL21 was combined with TWS119 (Supplementary Fig. 3E). To determine optimal timing for Wnt signaling to maximize formation of CD62L+ NKTs, we treated NKTs with Wnt3a ligand during early (days 0–2) or late (days 6–8) expansion, or throughout (days 0, 3, and 8). Wnt3a only promoted formation of CD62L+ NKTs when activated soon after TCR stimulation (Supplementary Fig. 3F), suggesting that Wnt signaling must occur in a limited time window after NKT stimulation to preserve or expand the central memory–like subset.
To understand the role of LEF1 in the central memory–like NKT program, we utilized CRISPR-Cas9 technology to knockout (KO) LEF1 in primary human NKTs (Supplementary Fig. S4A). Seven days after electroporation of Cas9 and gRNA specific to LEF1, CD62L expression (Fig. 2D) showed no change. In contrast, after re-stimulating NKTs with αGalCer-pulsed B-8-2 cells, CD62L+ frequency and NKT-cell expansion were dramatically reduced in LEF1 KO NKTs (Fig. 2E,F). Further, LEF1 KO NKTs exhibited a more activated/exhausted phenotype than control NKTs as indicated by higher TIM-3 expression levels in the KO cells (Supplementary Fig. S4B). These findings indicate that, consistent with the increased level of LEF1 expression, primary human CD62L+ NKTs undergo more Wnt/β-catenin signaling than CD62L− NKTs. Further, elevated LEF1 expression and associated downstream Wnt signaling are crucial for the preservation and expansion of central memory–like NKTs following antigenic stimulation.
LEF1 overexpression is sufficient to induce a central memory program in NKTs
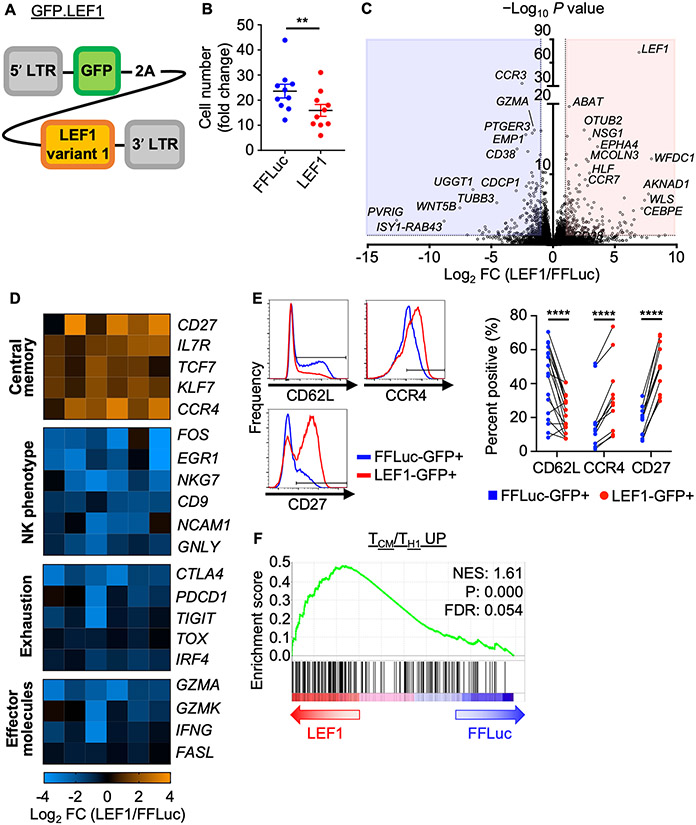
Next, we generated a retroviral construct to overexpress LEF1 (transcript variant 1, long isoform) in NKTs using GFP as a reporter (GFP.LEF1, Fig. 3A). We transduced NKTs with GFP.LEF1 or a control GFP/firefly luciferase (GFP.FFLuc) construct and confirmed LEF1 overexpression in the former and GFP expression in both (Supplementary Fig. S4C). LEF1 overexpression reduced NKT-cell expansion compared to the control (Fig. 3B). As Wnt signaling has been reported to support mitochondrial biogenesis (32) and enhanced mitochondrial mass promotes antitumor activity of adoptive T cells (33), we evaluated the metabolic effect of LEF1 overexpression on NKTs. LEF1-overexpressing NKTs had significantly increased mitochondrial mass compared with control cells (Supplementary Fig. S4D). This observation correlated with enhanced mitochondrial metabolic activity, as reflected by elevated basal, maximal, and spare respiratory capacity determined by Seahorse assay (Supplementary Fig. S4E,F).
Figure 3: LEF1 overexpression promotes a central memory–like program in human NKTs.
A, Design of the GFP.LEF1 gammaretroviral construct for overexpression of LEF1 long isoform. B–F, NKTs underwent 10 days of primary expansion followed by restimulation with αGalCer-pulsed B-8-2 cells. They were then transduced with the GFP.LEF1 construct or GFP.FFLuc two days after secondary stimulation and phenotypes were studied eight days later. (B) Cell number was determined and fold change was calculated relative to input cell number. Mean ± SEM (n = 10 donors) are shown. **P < 0.01, paired Student’s t test. (C) GFP+ cells were FACS sorted from GFP.FFluc and GFP.LEF1 NKTs. RNA isolated from the sorted cells was processed for bulk RNAseq analysis. Volcano plot shows differential gene expression (LEF1/FFLuc) with P value cutoff at 0.05 and fold change ≤ −2 / ≥ 2. (D) Differentially expressed genes of interest from (C) were grouped by shared phenotype/function. Heat maps show fold change in expression (LEF1/FFLuc). Each column represents a donor with a total of six donors tested; false discovery rate (FDR) < 0.05 (FDR < 0.20 for PDCD1) in LEF1-GFP+ and FFLuc-GFP+ cells. (E) Surface expression of central memory markers on GFP+ cells from FFLuc- or LEF1-transduced NKTs were measured by flow cytometry. Representative donor histograms and paired percent positive (n ≥ 11 donors) are shown. **P < 0.01, ****P < 0.0001, paired Student’s t test for paired result from each donor. (F) GSEA plot showing enrichment for a central memory T–cell signature in LEF1-overexpressing NKTs.
To determine whether LEF1 overexpression is sufficient to induce a central memory transcriptional program in ex vivo–expanded NKTs, we sorted GFP+ cells from GFP.LEF1 and GFP.FFLuc NKTs and subjected them to RNA sequencing analysis. We identified 436 differentially expressed genes in GFP.LEF1 versus GFP.FFLuc cells (Fig. 3C); LEF1 overexpression led to a significant enrichment for central memory–related gene expression (CD27, TCF7, IL7R) (34) and reduced expression of genes encoding effector molecules (GZMA, GZMK, FASL) (35) and genes associated with cell exhaustion (TOX, CTLA4, TIGIT) (36) (Fig. 3D). NKTs overexpressing LEF1 also showed a significant reduction in expression of NK-cell signature genes, which have been associated with CAR T–cell dysfunction in patients (37) (Fig. 3D; Supplementary Fig. S5A). LEF1 overexpression downregulated CD62L surface expression on NKTs, but consistent with the global shift towards a central memory gene expression program, upregulated other central memory markers including CD27 (38) and CCR4 (39) on the cell surface (Fig. 3D,E). Further, GSEA revealed that LEF1 overexpression in NKTs imparted a transcriptional program significantly enriched for a central memory signature (Fig. 3F; Supplementary Fig. S5B).
Conversely, the control group transcriptome was significantly enriched for effector, activated, and exhausted signatures compared with LEF1-overexpressing cells (Supplementary Fig. S5C,D). It has been reported that the CD39−CD69− subset of CD8+ T cells represents stem-like cells in adoptive cell therapy (11) and that CX3CR1− cells are precursors of the central memory subset (40). Consistent with these findings, LEF1 overexpression significantly reduced the transcript levels of these markers (Supplementary Fig. S6A,B). We also found that expression of HLF, which encodes a transcription factor that has been identified as a master regulator of stemness in hematopoietic stem cells (41), was elevated in NKTs overexpressing LEF1 (Supplementary Fig. S6C). Together, these results show that LEF1 promotes quiescent and central memory transcriptional signatures in NKTs.
Overexpression of LEF1 enhances NKT expansion following serial tumor challenge
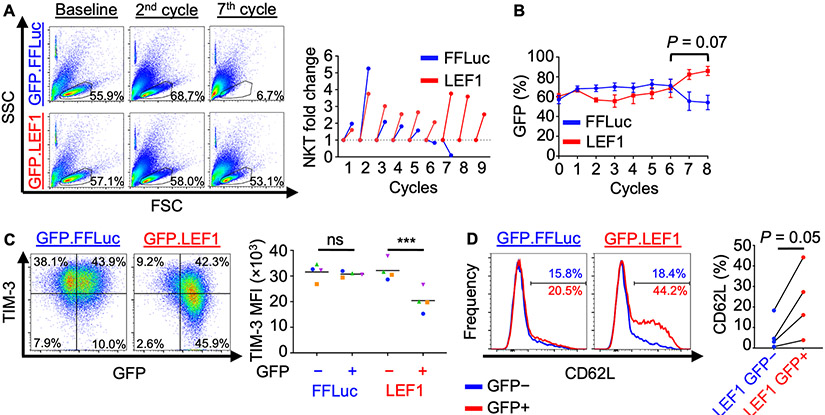
To determine whether LEF1 overexpression in NKTs confers the ability to maintain numeric expansion after multiple stimulations, we performed a repeat tumor challenge assay. In this experiment, GFP.LEF1 and GFP.FFLuc NKTs were challenged every three days for nine cycles with fresh CD1d+ J32 Jurkat cells to maintain an E:T ratio of 1:1. We found that while both groups eliminated tumor cells at this ratio for the first six cycles (Supplementary Fig. S7A), only GFP.LEF1-transduced NKTs continued to expand at late stages of the assay (Fig. 4A). This observation correlated with a sustained increase in the percentage of GFP+ cells in the GFP.LEF1 group but not the control group (Fig. 4B), indicating that LEF1 overexpression protects NKTs subjected to the pressure of repeated antigenic stimulation.
Figure 4: Overexpression of LEF1 improves NKT expansion and tumor control and reduces exhaustion following serial tumor challenge.
A–E, NKTs transduced with GFP.FFLuc or GFP.LEF1 were repeatedly challenged with CD1d+ J32 leukemia cells at a 1:1 ratio every three days. (A) NKT cell number was determined using counting beads and flow cytometry following each challenge cycle. FSC-SSC dot plots show approximate live cell gate representing NKTs at early (2nd) and late (7th) co-culture cycles. Results from a representative donor (n = 4 donors, two independent experiments) and NKT fold-change at each cycle are shown. (B) GFP expression was monitored over time using flow cytometry as a proxy for transduced cells. Mean GFP+ NKT percentage ± SEM is shown at each cycle (n = 4 donors). P = 0.07 for AUC analysis for cycles 6 to 8. C, D, Following cycle 5, NKTs were rested for six days following antigenic stimulation and (C) TIM-3 and (D) CD62L expression were assessed by flow cytometry. Representative results and mean TIM-3 MFI or CD62L percentage (n = 4 donors) are shown. ***P < 0.001, ns: not significant, one-way ANOVA with Sidak’s post-test.
To understand how LEF1 overexpression contributes to sustained NKT expansion, NKTs were rested for six days after the fifth challenge cycle (before the control group failed to expand), and exhaustion marker and CD62L expression were evaluated. LEF1-overexpressing NKTs included a large population of TIM-3lo cells that was not present in the control group, which was predominantly TIM-3hi after multiple rounds of antigenic stimulation (Fig. 4C). Additionally, we detected a population of CD62L+ cells in LEF1-overexpressing NKTs but not in control NKTs, which consisted mainly of CD62L− cells after multiple stimulations (Fig. 4D). Further, CD62L+ cells preserved by LEF1 overexpression were found in the TIM-3lo population but not in the control group (Supplementary Fig. S7B). LEF1 protein levels in LEF1-transduced NKTs were similar at the beginning and end of the seven stimulation cycles (Supplementary Fig. S7C). In summary, in the context of multiple rounds of antigenic stimulation, LEF1 overexpression in NKTs confers resistance to exhaustion and preserves the central memory–like subset and antitumor functionality.
LEF1 mitigates CAR-NKT exhaustion and suppresses immediate effector function
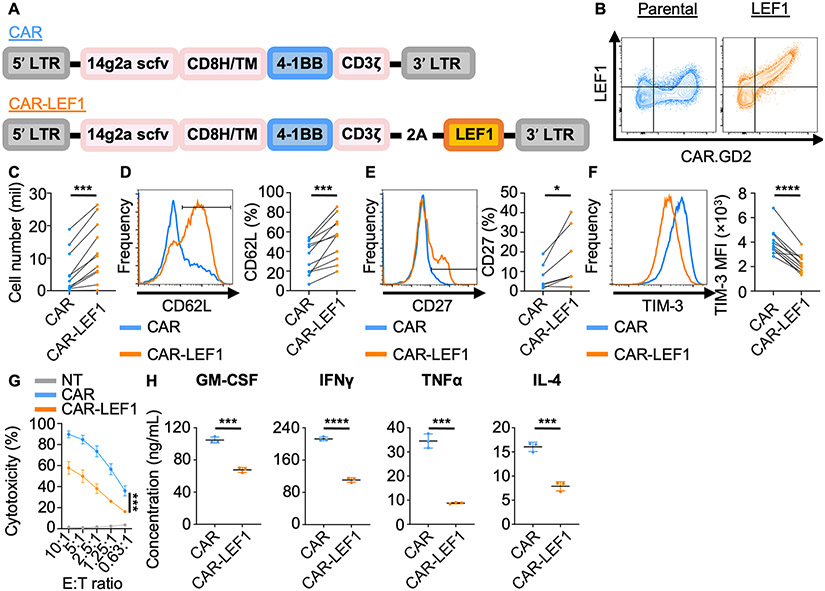
Given that LEF1 overexpression improves persistence and prevents exhaustion of NKTs challenged through the native NKT TCR, we asked whether we could exploit these functional advantages in the context of a CAR. We modified a CAR construct specific for the neuroblastoma antigen GD2 (21) (CAR.GD2) to include the LEF1 coding sequence downstream of the CAR modules and validated co-expression of CAR.GD2 and LEF1 in transduced NKTs (Fig. 5A,B). Incorporation of LEF1 into the CAR enhanced numeric expansion as well as CD62L and CD27 expression in CAR.GD2-NKTs (Fig. 5C-E). The CAR.GD2 construct is known to induce some degree of exhaustion in T cells due to tonic signaling (42). Consistent with the observation that LEF1 limited the susceptibility of NKT cells to exhaustion (Fig. 1D,3D), co-expression of LEF1 with CAR.GD2 reduced expression of all six tested activation/exhaustion markers in NKTs (36,43) (Fig. 5F; Supplementary Fig. S8). To evaluate the short-term antitumor activity of CAR.GD2-LEF1 NKTs against GD2+ tumor cells, we co-cultured CAR.GD2 and CAR.GD2-LEF1 NKTs with GD2+ CHLA-255 neuroblastoma cells for four hours. Both groups of NKTs killed the target cells, with slightly lower levels of cytotoxicity observed for CAR.GD2-LEF1 NKTs (Fig. 5G). Additionally, following CAR stimulation, we found that CAR.GD2-LEF1 NKTs produced lower levels of immediate effector cytokines including TH1 (GM-CSF, IFNγ, TNFα) and TH2 (IL4) mediators compared to CAR.GD2-NKTs (Fig. 5H), consistent with the reduction in effector molecule gene expression levels observed in LEF1-overexpressing NKTs (Fig. 3D). Taken together, NKTs transduced with a CAR.GD2 construct co-expressing LEF1 showed increased capacity for expansion, lower levels of exhaustion, and reduced immediate effector function against GD2+ target cells, observations in line with the notion that LEF1 restricts terminal differentiation of NKTs.
Figure 5: Incorporation of LEF1 into CAR.GD2 construct mitigates exhaustion and suppresses immediate effector function of transduced NKTs.
A, CAR constructs containing the anti-GD2 14g2a scFv, CD8 hinge and transmembrane domains, 4-1BB co-stimulatory domain, and CD3 zeta domain, with LEF1 following a 2A sequence (CAR-LEF1) or without (CAR). B–G, Two days after secondary stimulation with αGalCer-pulsed aAPCs, NKTs were transduced with parental or LEF1-containing CAR.GD2 constructs and phenotypes were studied eight days after. (B) Surface CAR and intracellular LEF1 expression were determined by flow cytometry. Representative dot plots show LEF1 expression relative to CAR expression from one of two donors. (C) NKT cell number was determined by trypan blue exclusion assay. Mean cell count ± SEM (n = 11 donors) is shown. (D) CD62L, (E) CD27, and (F) TIM-3 expression were assessed by flow cytometry in CAR+-gated NKTs. Representative histograms and mean ± SEM of CD62L, CD27 percentage or TIM-3 MFI (n = 11 donors) are shown. *P < 0.05, ***P < 0.001, ****P < 0.0001, paired Student’s t test for paired result from each donor. (G) Luciferase-transduced GD2+ CHLA-255 cells were co-cultured with CAR or CAR-LEF1 NKTs for four hours. Cytotoxicity was calculated from luminescence intensity using non-transduced (NT) NKTs as control. Results are from five donors tested in two independent experiments. Mean ± SEM is shown. ***P < 0.001 for AUC analysis between CAR and CAR-LEF1 groups. H, CAR and CAR-LEF1 NKTs were stimulated with GD2+ CHLA-255 cells, and supernatants were collected at 24 hours. GM-CSF, IFNγ, TNFα, and IL4 levels were measured by Luminex assay. Results are from one of two donors tested with similar results. ***P < 0.001, ****P < 0.0001, unpaired Student’s t test.
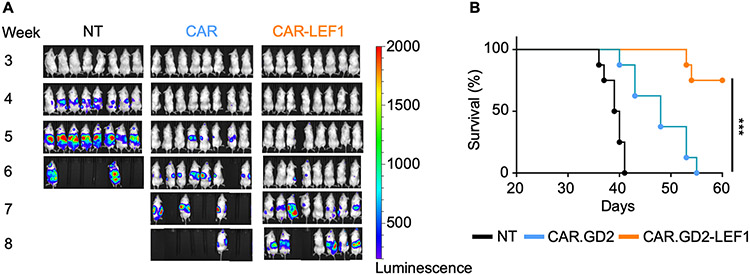
CAR.GD2-LEF1 NKTs have superior in vivo therapeutic activity versus parental CAR.GD2 NKTs
Next, we sought to determine whether co-expression of LEF1 with CAR.GD2 improves the antitumor function of transduced NKTs or durability of response in vivo. NSG mice were i.v. injected with luciferase-transduced GD2+ CHLA-255 neuroblastoma cells and seven days later were divided into three groups to receive non-transduced NKTs, CAR.GD2 NKTs, or CAR.GD2-LEF1 NKTs. Both CAR.GD2 NKTs and CAR.GD2-LEF1 NKTs prolonged the survival of treated animals compared with non-transduced control NKTs (P < 0.0001). However, CAR.GD2-LEF1 NKTs mediated more durable antitumor responses and improved the survival of treated mice compared with CAR.GD2 NKTs (Fig. 6). These results show that LEF1 mediates a transcriptional program in NKTs that, by restricting terminal effector differentiation and reducing exhaustion, ultimately improves the in vivo therapeutic activity of NKT cells. These findings may inform the development of effective NKT cell–based adoptive therapy in the clinic.
Figure 6: CAR-LEF1 NKTs have superior in vivo therapeutic activity versus parental CAR.GD2 NKTs.
A, NKTs were expanded with IL2 and transduced with either CAR.GD2 (CAR) or CAR.GD2-LEF1 (CAR-LEF1), with a non-transduced (NT) control group. NSG mice (n = 8 mice per group) were i.v. injected with 1 x 106 luciferase-transduced CHLA-255 cells on day 0. On day 7, mice received an i.v. injection of NT, CAR, or CAR-LEF1 NKT preparations (4 x 106 CAR+ cells per mouse). IL2 was injected i.p. three times a week for two weeks after NKT injection. Tumor growth was monitored using bioluminescence imaging once per week. One experiment was performed. B, A survival plot was generated using the Kaplan–Meier method. Differences in survival probability were compared using the log-rank test. ***P < 0.001.
Discussion
In this study, we report for the first time that Wnt transcription factor LEF1 drives the central memory program in human NKTs. After finding that CD62L+ central memory–like NKTs express elevated levels of LEF1 using immune-related gene expression analysis, we determined that LEF1+ cells expressed higher levels of central memory markers and lower levels of activation/exhaustion markers than NKTs that do not express LEF1. Next, we found that CD62L+ cells demonstrated more Wnt/β-catenin signaling activity than CD62L− cells, and that NKTs express several required receptors and a co-receptor for Wnt ligands. Moreover, LEF1 and Wnt/β-catenin signaling were found to play a crucial role in mediating the NKT central memory–like transcriptional program, as both genetic knockout of LEF1 and pharmacological inhibition of Wnt/β-catenin signaling blocked formation of CD62L+ NKTs. Consistent with these observations, activation of Wnt/β-catenin signaling by recombinant Wnt3a ligand significantly increased the frequency of CD62L+ NKTs. Further, ectopic overexpression of LEF1 was sufficient to induce a central memory–like program in NKTs. Functionally, LEF1 overexpression preserved central memory–like cells when NKTs underwent repeat antigenic stimulation. Finally, co-expression of LEF1 with a CAR.GD2 construct in NKTs reduced immediate effector function but promoted central memory differentiation. In vivo, CAR.GD2-LEF1 NKTs mediated more durable antitumor responses and increased the survival of treated mice compared with CAR.GD2-NKTs in an aggressive neuroblastoma xenograft mouse model.
Unlike conventional T cells that have a distinct naïve-to-memory differentiation hierarchy, human NKTs bypass the naïve stage and acquire a predominantly activated-memory phenotype during thymic differentiation (44). A prevalent theory in the NKT biology field is that NKT cells display a fast-acting, tissue-trafficking effector memory phenotype characterized by minimal CD62L expression and largely influenced by the transcriptional master regulator PLZF that is induced after positive selection in the thymus (45). Our group, however, has recently reported the presence of a novel CD62L-expressing NKT subset that mediates robust in vivo cell expansion, persistence, and durable tumor control (12). These findings prompted a more thorough study of the CD62L+ subset to maximize the therapeutic efficacy of NKTs.
Wnt/β-catenin signaling regulates development of both NKTs and T cells in that β-catenin expression is temporarily upregulated upon TCR engagement and subsequently downregulated to ensure proper NKT- and T-cell development (46,47). Consistent with the kinetics of β-catenin upregulation in developing NKTs and T cells, we observed that Wnt ligands boosted the frequency of central memory–like NKTs only when administered early after TCR stimulation; these results suggest that there is an optimal window for activating Wnt/β-catenin signaling in mature NKTs as well. Wnt/β-catenin signaling also plays a crucial role in supporting central memory formation in mature T cells (48). Specifically, T cells with higher Wnt activity yield central memory cells that proliferate and survive better than T cells with lower Wnt signaling, which instead progress to terminal differentiation (16). In line with this, we found that CD62L+ central memory–like NKTs undergo more Wnt signaling activity than CD62L− cells. While the Wnt reporter employed has mainly been used in the context of hematopoietic and cancer stem cells studies (49,50) and likely detects NKTs with the highest level of Wnt signaling only, we also showed that CD62L+ NKTs have higher expression of several specific Wnt-downstream target genes. This indicates that initiation of the central memory program via increased Wnt/β-catenin signaling may be conserved in both T and NKT cells. Although less is known about the importance of Wnt signaling in mature NKTs, it has been reported that activation of such signaling soon after stimulation of the NKT TCR in mice curbs IFNγ and enhances IL4 production in these cells (51), consistent with the role of LEF1 in NKT thymic development (18). Moreover, murine NKTs also express Wnt receptors and co-receptors and respond to Wnt ligands derived from myeloid cells (51). We show that human primary NKTs also express Wnt receptors and co-receptors, though at lower levels than in vitro–expanded NKTs. These results are consistent with the fact that peripheral blood NKTs largely exhibit an effector-memory phenotype while central memory–like cells accumulate during ex vivo expansion (12). Further work will determine whether human NKTs receive Wnt signals from the antigen-presenting cells upon antigen recognition that may support preferential expansion or survival of NKTs with the higher levels of Wnt receptor expression.
The results of this study identify LEF1 as a critical mediator of the central memory–like program in human NKTs. In conventional T cells, TCF1 (14,15), which belongs to the same protein family of proteins as LEF1, and Wnt/β-catenin signaling (48) are crucial to stemness and formation of the central memory subset. While LEF1 plays a supportive role in T cells in the absence of TCF1, it does not drive the central memory status of T cells (17). In contrast, the results of our knockout and overexpression studies show that the central memory program in NKT cells depends on LEF1, while TCF1 is expressed at high levels in both CD62L+ and CD62L− subsets. Relative dominance of LEF1 over TCF1 has also been observed in murine NKT-cell development (18). Future studies will shed light on the mechanisms that govern dominance of LEF1 in NKTs versus TCF1 in T cells. Since CD62L+ NKTs may be either LEF1+ or LEF1−, it will be important to identify additional or alternative phenotypic markers that exclusively define the LEF1+ NKT-cell subset.
LEF1-expressing NKTs have a TIM-3lo phenotype like antigen-experienced T cells that express TCF1 and exhibit properties of stemness (52), as opposed to terminally differentiated cells that are TIM-3hi. Functionally, LEF1-overexpressing NKTs are metabolically fitter than wild-type NKTs, have higher proliferative potential, and give rise to both CD62L+ and CD62L− cells after multiple rounds of stimulation. Our transcriptomic studies also revealed that LEF1 overexpression significantly reduces effector function and exhaustion level of NKTs, while supporting gene expression associated with the central memory program. Consistent with the function of LEF1 in mitigating exhaustion, LEF1 overexpression drastically reduced the expression of PVRIG, a checkpoint inhibitor recently reported in NK cells (53) and CD8+ T cells (54). Future studies will help determine whether PVRIG plays a role in NKT exhaustion. LEF1 overexpression also led to downregulation of an NK-like signature, which was recently reported to be associated with CAR T–cell dysfunction in patients (37). Our findings identify LEF1 as a key factor that governs stemness in central memory–like NKTs and support the parallel in central memory programming between LEF1+ NKTs and TCF1hi T cells.
LEF1 has been reported to support proliferation and survival of murine NKTs during thymic development, likely through induction of CD127 and c-myc expression (18). While we evaluated mature peripheral human NKTs, we also found that CD62L+ NKTs, which express elevated levels of LEF1, upregulate CD127 (12) and MYC. Knocking out LEF1 in mature human NKTs did not reduce viable cell number but inhibited NKT expansion and formation of CD62L+ cells after antigenic re-stimulation. Therefore, LEF1 appears to be crucial for NKT proliferation/survival and central memory differentiation after antigenic challenge and less important for homeostatic survival of mature NKTs. Similarly, TCF1 knockout in human CD8+ T cells reduces cell expansion after TCR stimulation (55). LEF1 also drives GATA3 expression, favoring differentiation of TH2 NKTs during thymic development (18); while we have observed co-expression of LEF1 and GATA3 in human peripheral NKTs (12), LEF1-overexpressing NKTs did not show elevated expression of TH2-cytokines such as IL4 and IL13, but rather a global reduction of effector cytokines following CAR stimulation.
We also reported that ectopic overexpression of LEF1 in primary lymphocytes reduces short-term effector function but alleviates NKT exhaustion and increases expansion potential after serial antigen challenge. These observations phenocopy what occurs in T cells when TCF1 is overexpressed during chronic infection (55) or in the tumor setting (56). LEF1 overexpression reduced NKT numeric expansion and CD62L expression during basal NKT culture but preserved NKT proliferation and CD62L expression over the course of multiple TCR stimulations. The decreased expansion of LEF1-transduced NKTs in basal culture is consistent with the transition to a central memory state, which is associated with induction of quiescence in the absence of antigenic stimulation. The same memory properties allow LEF1-transduced NKTs to undergo robust proliferation in response to repeated stimulation. The higher level of CD62L expression in LEF1-transduced compared to control NKTs following multiple rounds of stimulation is more likely due to faster loss of CD62L during terminal effector differentiation than LEF1-mediated CD62L downregulation. Finally, LEF1 overexpression also enhances mitobiogenesis and by extension metabolic fitness in NKTs, likely through Wnt signaling activity as reported in muscle cells (32). This effect is crucial because the persistence and antitumor activity of tumor-specific T cells can be improved by increasing mitochondrial mass and metabolic fitness (33). Future work will aim to elucidate the metabolic contribution of LEF1 and Wnt signaling to the long-term antitumor efficacy of NKTs.
The effects of LEF1 overexpression highlight the promise of CAR-LEF1 NKTs as a potential clinical therapy. Despite reducing immediate effector functionality, co-expression of LEF1 with CAR.GD2 in NKTs improved long-term tumor control in vivo. These results suggest that LEF1 overexpression increases the pool of NKT central memory cells that can maintain themselves, resist exhaustion, and give rise to NKT or CAR-NKT effector memory or effector cells following repeated antigenic challenge resulting in enhanced tumor control in vivo. While LEF1 expression has been described in various cancers (57,58), it has also been reported as a tumor suppressor gene (59,60). We have not observed any evidence that LEF1 overexpression induces transformation in NKT cells. Future studies are necessary to ensure the safety of CAR-LEF1 NKTs, including the use of clinically tested safety switches such as an inducible caspase-9 (61).
In summary, we have identified LEF1 as a key transcription factor that governs central memory and stemness in human NKTs. Promoting LEF1 expression or downstream Wnt signaling activity preserves central memory-like NKTs, renders NKTs resistant to exhaustion, and improves NKT TCR- and CAR-mediated long-term antitumor efficacy. These results pave the way for pharmacological and genetic enhancement of NKT cell–based cancer immunotherapy.
Supplementary Material
Acknowledgements
The authors are grateful to the staff of the Flow Cytometry Core Laboratory of the Texas Children’s Cancer and Hematology Center and Small Animal Imaging core facility at Texas Children’s Hospital. This work was supported by grants from the National Institutes of Health (CA262250 and CA126752 to L.S.M. and CA016303 to J.M.R.), and the Cancer Prevention and Research Institute of Texas (BCM Comprehensive Cancer Center Training Program, RP160283 to H.N.).
Footnotes
Competing Interests Statement
H.N., G.T., A.N.C., and L.S.M. are co-inventors on patent applications that relate to the use of NKTs in cancer immunotherapy and have been licensed by Baylor College of Medicine to Athenex, Inc. J.C.B., G.A.B., C.Z., L.G., X.X., M.S.W., J.M.D, T.D., C.M.S., C.N.C., E.J.D.P., and J.M.R. declare no competing financial interests.
References
- 1.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol 2004;4(3):231–7 doi 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 2.Metelitsa LS, Wu HW, Wang H, Yang Y, Warsi Z, Asgharzadeh S, et al. Natural killer T cells infiltrate neuroblastomas expressing the chemokine CCL2. J Exp Med 2004;199(9):1213–21 doi 10.1084/jem.20031462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song L, Asgharzadeh S, Salo J, Engell K, Wu HW, Sposto R, et al. Valpha24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest 2009;119(6):1524–36 doi 10.1172/jci37869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortesi F, Delfanti G, Grilli A, Calcinotto A, Gorini F, Pucci F, et al. Bimodal CD40/Fas-Dependent Crosstalk between iNKT Cells and Tumor-Associated Macrophages Impairs Prostate Cancer Progression. Cell Rep 2018;22(11):3006–20 doi 10.1016/j.celrep.2018.02.058. [DOI] [PubMed] [Google Scholar]
- 5.Tachibana T, Onodera H, Tsuruyama T, Mori A, Nagayama S, Hiai H, et al. Increased intratumor Valpha24-positive natural killer T cells: a prognostic factor for primary colorectal carcinomas. Clin Cancer Res 2005;11(20):7322–7 doi 10.1158/1078-0432.Ccr-05-0877. [DOI] [PubMed] [Google Scholar]
- 6.Exley MA, Friedlander P, Alatrakchi N, Vriend L, Yue S, Sasada T, et al. Adoptive Transfer of Invariant NKT Cells as Immunotherapy for Advanced Melanoma: A Phase I Clinical Trial. Clin Cancer Res 2017;23(14):3510–9 doi 10.1158/1078-0432.Ccr-16-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heczey A, Courtney AN, Montalbano A, Robinson S, Liu K, Li M, et al. Anti-GD2 CAR-NKT cells in patients with relapsed or refractory neuroblastoma: an interim analysis. Nat Med 2020;26(11):1686–90 doi 10.1038/s41591-020-1074-2. [DOI] [PubMed] [Google Scholar]
- 8.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med 2002;195(5):637–41 doi 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klebanoff CA, Crompton JG, Leonardi AJ, Yamamoto TN, Chandran SS, Eil RL, et al. Inhibition of AKT signaling uncouples T cell differentiation from expansion for receptor-engineered adoptive immunotherapy. JCI Insight 2017;2(23) doi 10.1172/jci.insight.95103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med 2018;24(5):563–71 doi 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishna S, Lowery FJ, Copeland AR, Bahadiroglu E, Mukherjee R, Jia L, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 2020;370(6522):1328–34 doi 10.1126/science.abb9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian G, Courtney AN, Jena B, Heczey A, Liu D, Marinova E, et al. CD62L+ NKT cells have prolonged persistence and antitumor activity in vivo. J Clin Invest 2016;126(6):2341–55 doi 10.1172/jci83476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17(1):9–26 doi 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 2010;33(2):229–40 doi 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pais Ferreira D, Silva JG, Wyss T, Fuertes Marraco SA, Scarpellino L, Charmoy M, et al. Central memory CD8(+) T cells derive from stem-like Tcf7(hi) effector cells in the absence of cytotoxic differentiation. Immunity 2020;53(5):985–1000.e11 doi 10.1016/j.immuni.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Boudousquié C, Danilo M, Pousse L, Jeevan-Raj B, Angelov GS, Chennupati V, et al. Differences in the transduction of canonical Wnt signals demarcate effector and memory CD8 T cells with distinct recall proliferation capacity. J Immunol 2014;193(6):2784–91 doi 10.4049/jimmunol.1400465. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Xue HH. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol 2012;189(6):2722–6 doi 10.4049/jimmunol.1201150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr T, Krishnamoorthy V, Yu S, Xue HH, Kee BL, Verykokakis M. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. J Exp Med 2015;212(5):793–807 doi 10.1084/jem.20141849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeger RC, Rayner SA, Banerjee A, Chung H, Laug WE, Neustein HB, et al. Morphology, growth, chromosomal pattern and fibrinolytic activity of two new human neuroblastoma cell lines. Cancer Res 1977;37(5):1364–71. [PubMed] [Google Scholar]
- 20.Makni H, Malter JS, Reed JC, Nobuhiko S, Lang G, Kioussis D, et al. Reconstitution of an active surface CD2 by DNA transfer in CD2-CD3+ Jurkat cells facilitates CD3-T cell receptor-mediated IL-2 production. J Immunol 1991;146(8):2522–9. [PubMed] [Google Scholar]
- 21.Xu X, Huang W, Heczey A, Liu D, Guo L, Wood M, et al. NKT Cells Coexpressing a GD2-Specific Chimeric Antigen Receptor and IL15 Show Enhanced In Vivo Persistence and Antitumor Activity against Neuroblastoma. Clin Cancer Res 2019;25(23):7126–38 doi 10.1158/1078-0432.Ccr-19-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 2010;5(2):e9370 doi 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 2015;12(10):982–8 doi 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 2014;3(12):e214 doi 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep 2016;17(5):1453–61 doi 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 2002;111(6):837–51 doi 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 27.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A 2006;103(9):3304–9 doi 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20(3):326–36 doi 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Göke B, et al. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics 2014;15:74 doi 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc Natl Acad Sci U S A 2004;101(34):12682–7 doi 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngai H, Tian G, Courtney AN, Ravari SB, Guo L, Liu B, et al. IL-21 Selectively Protects CD62L(+) NKT Cells and Enhances Their Effector Functions for Adoptive Immunotherapy. J Immunol 2018;201(7):2141–53 doi 10.4049/jimmunol.1800429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev 2010;24(14):1507–18 doi 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 2016;45(2):374–88 doi 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gattinoni L, Klebanoff CA, Restifo NP. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer 2012;12(10):671–84 doi 10.1038/nrc3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu Z, et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest 2005;115(6):1616–26 doi 10.1172/jci24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15(8):486–99 doi 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Good CR, Aznar MA, Kuramitsu S, Samareh P, Agarwal S, Donahue G, et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 2021;184(25):6081–100.e26 doi 10.1016/j.cell.2021.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochsenbein AF, Riddell SR, Brown M, Corey L, Baerlocher GM, Lansdorp PM, et al. CD27 expression promotes long-term survival of functional effector-memory CD8+ cytotoxic T lymphocytes in HIV-infected patients. J Exp Med 2004;200(11):1407–17 doi 10.1084/jem.20040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casciano F, Diani M, Altomare A, Granucci F, Secchiero P, Banfi G, et al. CCR4(+) Skin-Tropic Phenotype as a Feature of Central Memory CD8(+) T Cells in Healthy Subjects and Psoriasis Patients. Front Immunol 2020;11:529 doi 10.3389/fimmu.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, et al. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 2016;45(6):1270–84 doi 10.1016/j.immuni.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehnertz B, Chagraoui J, MacRae T, Tomellini E, Corneau S, Mayotte N, et al. HLF expression defines the human hematopoietic stem cell state. Blood 2021;138(25):2642–54 doi 10.1182/blood.2021010745. [DOI] [PubMed] [Google Scholar]
- 42.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med 2015;21(6):581–90 doi 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TJ, Park G, Kim J, Lim SA, Kim J, Im K, et al. CD160 serves as a negative regulator of NKT cells in acute hepatic injury. Nat Commun 2019;10(1):3258 doi 10.1038/s41467-019-10320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Der Vliet HJ, Nishi N, de Gruijl TD, von Blomberg BM, van den Eertwegh AJ, Pinedo HM, et al. Human natural killer T cells acquire a memory-activated phenotype before birth. Blood 2000;95(7):2440–2. [PubMed] [Google Scholar]
- 45.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 2008;29(3):391–403 doi 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyaram K, Sen JM, Chang CH. Temporal regulation of Wnt/β-catenin signaling is important for invariant NKT cell development and terminal maturation. Mol Immunol 2017;85:47–56 doi 10.1016/j.molimm.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Wetering M, de Lau W, Clevers H. WNT signaling and lymphocyte development. Cell 2002;109 Suppl:S13–9 doi 10.1016/s0092-8674(02)00709-2. [DOI] [PubMed] [Google Scholar]
- 48.Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 2009;15(7):808–13 doi 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter J, Stanley EG, Ng ES, Elefanty AG, Traver D, Willert K. WNT9A Is a Conserved Regulator of Hematopoietic Stem and Progenitor Cell Development. Genes (Basel) 2018;9(2) doi 10.3390/genes9020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giambra V, Jenkins CE, Lam SH, Hoofd C, Belmonte M, Wang X, et al. Leukemia stem cells in T-ALL require active Hif1α and Wnt signaling. Blood 2015;125(25):3917–27 doi 10.1182/blood-2014-10-609370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kling JC, Jordan MA, Pitt LA, Meiners J, Thanh-Tran T, Tran LS, et al. Temporal Regulation of Natural Killer T Cell Interferon Gamma Responses by β-Catenin-Dependent and -Independent Wnt Signaling. Front Immunol 2018;9:483 doi 10.3389/fimmu.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Im SJ, Konieczny BT, Hudson WH, Masopust D, Ahmed R. PD-1+ stemlike CD8 T cells are resident in lymphoid tissues during persistent LCMV infection. Proc Natl Acad Sci U S A 2020;117(8):4292–9 doi 10.1073/pnas.1917298117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Whelan S, Kotturi MF, Meyran D, D'Souza C, Hansen K, et al. PVRIG is a novel natural killer cell immune checkpoint receptor in acute myeloid leukemia. Haematologica 2021;106(12):3115–24 doi 10.3324/haematol.2020.258574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan S, Ophir E, Kotturi MF, Levy O, Ganguly S, Leung L, et al. PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8(+) T-cell Function. Cancer Immunol Res 2019;7(2):257–68 doi 10.1158/2326-6066.Cir-18-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rutishauser RL, Deguit CDT, Hiatt J, Blaeschke F, Roth TL, Wang L, et al. TCF-1 regulates HIV-specific CD8+ T cell expansion capacity. JCI Insight 2021;6(3) doi 10.1172/jci.insight.136648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shan Q, Hu S, Chen X, Danahy DB, Badovinac VP, Zang C, et al. Ectopic Tcf1 expression instills a stem-like program in exhausted CD8(+) T cells to enhance viral and tumor immunity. Cell Mol Immunol 2020. doi 10.1038/s41423-020-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santiago L, Daniels G, Wang D, Deng FM, Lee P. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am J Cancer Res 2017;7(6):1389–406. [PMC free article] [PubMed] [Google Scholar]
- 58.Petropoulos K, Arseni N, Schessl C, Stadler CR, Rawat VP, Deshpande AJ, et al. A novel role for Lef-1, a central transcription mediator of Wnt signaling, in leukemogenesis. J Exp Med 2008;205(3):515–22 doi 10.1084/jem.20071875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez A, Sanda T, Ma W, Zhang J, Grebliunaite R, Dahlberg S, et al. Inactivation of LEF1 in T-cell acute lymphoblastic leukemia. Blood 2010;115(14):2845–51 doi 10.1182/blood-2009-07-234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dräger J, Simon-Keller K, Pukrop T, Klemm F, Wilting J, Sticht C, et al. LEF1 reduces tumor progression and induces myodifferentiation in a subset of rhabdomyosarcoma. Oncotarget 2017;8(2):3259–73 doi 10.18632/oncotarget.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med 2011;365(18):1673–83 doi 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All requests for raw and analyzed data and materials beyond the information provided in the main text and supplementary figures should be directed to the corresponding author. These requests will be promptly reviewed by the BCM Licensing Group to verify if the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement. All raw data for RNA sequencing has been deposited in the Gene Expression Omnibus (GEO) at accession number GSE217281.