Summary
Resident peritoneal macrophages (resMØs) are crucial for repairing peritoneal injuries and controlling infections by forming mesothelium-bound resMØ-aggregates in the peritoneal wall and omentum. Here we present a protocol to analyze these structures in mouse models of peritoneal inflammation. We describe the dissection, fixation, immunofluorescent staining, and mounting of whole peritoneal wall and omentum samples and subsequent confocal microscopy imaging of resMØ-aggregates. We also detail the steps to isolate resMØ-aggregates for additional studies, including flow cytometry and electron-microscopy-based analysis.
For complete details on the use and execution of this protocol, please refer to Vega-Pérez et al. (2021).1
Subject areas: Cell Biology, Cell Isolation, Flow Cytometry/Mass Cytometry, Immunology, Microscopy, Model Organisms
Graphical abstract
Highlights
-
•
Imaging of mouse peritoneal wall and omentum by whole-mount confocal microscopy
-
•
Isolation of mesothelium-bound resMØ-aggregates formed during peritoneal inflammation
-
•
Processing of resMØ-aggregates for flow cytometry and electron microscopy
-
•
Can be adapted to imaging of the thoracic cavity
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Resident peritoneal macrophages (resMØs) are crucial for repairing peritoneal injuries and controlling infections by forming mesothelium-bound resMØ-aggregates in the peritoneal wall and omentum. Here we present a protocol to analyze these structures in mouse models of peritoneal inflammation. We describe the dissection, fixation, immunofluorescent staining, and mounting of whole peritoneal wall and omentum samples and subsequent confocal microscopy imaging of resMØ-aggregates. We also detail the steps to isolate resMØ-aggregates for additional studies, including flow cytometry and electron-microscopy-based analysis.
Before you begin
The protocol below describes the specific steps for imaging mesothelium-bound immune cell aggregates, hereafter resMØ-aggregates (for resident peritoneal macrophage aggregates), formed in the peritoneal wall or omentum during peritoneal inflammation1,2,3,4 by whole mount immunofluorescence (WMI), i.e., in whole peritoneal walls and omenta, combined with confocal microscopy, and to isolate these aggregates to perform additional analyses. Examples of expected outcomes included in the present protocol refer to the bacterial infection model described by our group,1 based on the intraperitoneal injection of the Escherichia coli strain M6L4. This protocol can also be used for imaging the surface of other peritoneal organs, such as the mesentery, spleen, gut or ovary, as well as the inner wall of the thoracic cavity.
Institutional permissions
All the experiments were approved by the Animal Care and Use Committee of the Centro Nacional de Biotecnología-CSIC, Madrid. Ethical approvals from the relevant institutions are required prior to starting this procedure.
Establishing a mouse model of peritoneal inflammation
The experimental model used to analyze the innate immune system response to peritoneal inflammation relied on a single intraperitoneal inoculum of the E. coli M6L4 strain in C57BL/6 mice, at a 1 × 107 cfu sublethal dose, in 0.2 mL of PBS. Analyses were performed during the early phase of the response, i.e., at 0–18 h post infection.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ∗Anti-CD11b, PE-Cy7-conjugate, clone M1/70 (1:3000) | eBioscience | Cat # 25-0112-81 RRID: AB_469587 |
| ∗Anti-CD11c, APC-Cy7-conjugate, clone HL3 (1:100) | BD Biosciences | Cat # 561241 RRID: AB_10611727 |
| ∗Anti-CD16/CD32, purified, clone 2.4G2 (1:100) | BD Biosciences | Cat # 553142 RRID: AB_394657 |
| ∗Anti-CD19, PE-conjugate, clone 1D3 (1:400) | BD Biosciences | Cat # 553786 RRID: AB_395050 |
| ∗Anti-CD45, FITC-conjugate, clone 30-F11 (1:200) | BioLegend | Cat # 103108 RRID: AB_312973 |
| ∗Anti-CD45, Pacific Blue-conjugate, clone 30-F11 (1:800) | BioLegend | Cat # 103126 RRID: AB_493535 |
| ∗Anti-CD45R/B220, Pacific Blue-conjugate, clone RA3-6B2 (1:100) | BioLegend | Cat # 103227 RRID: AB_492876 |
| ∗Anti-CD90.2, PE-conjugate, clone 53-2.1 (1:500) | BD Biosciences | Cat # 553006 RRID: AB_394545 |
| ∗Anti-F4/80, APC-Cy7-conjugate, clone BM8 (1:100) | BioLegend | Cat # 123118 RRID: AB_893477 |
| ∗Anti-F4/80, FITC-conjugate, clone BM8 (1:100) | eBioscience | Cat # 11-4801-82 RRID: AB_2637191 |
| ∗Anti-MHC II (I-A/I-E), APC-conjugate, clone M5/114.15.2 (1:1000) | eBioscience | Cat # 17-5321-81 RRID: AB_469454 |
| ∗Anti-I-A/I-E, FITC-conjugate, clone 2G9 (1:500) | BD Biosciences | Cat # 553623 RRID: AB_394958 |
| ∗Anti-ICAM-1, FITC-conjugate, clone 3E2 (1:400) | BD Biosciences | Cat # 553252 RRID: AB_394734 |
| ∗Anti-Ly6C, PerCP-Cy5.5-conjugate, clone AL-21 (1:100) | BD Biosciences | Cat # 560525 RRID: AB_1727558 |
| ∗Anti-Ly6G, PE-conjugate, clone 1A8 (1:200) | BD Biosciences | Cat # 551461 RRID: AB_394208 |
| ∗Anti-Tim4, BV421-conjugate, clone 21H12 (1:1000) | BD Biosciences | Cat # 742773 RRID: AB_2741037 |
| ∗∗Anti-CD19, Alexa Fluor 647-conjugate, clone 6D5 (1:500) | BioLegend | Cat # 115522 RRID: AB_389329 |
| ∗∗Anti-F4/80, Alexa Fluor 488-conjugate, clone BM8 (1:500) | BioLegend | Cat # 123120 RRID: AB_893479 |
| ∗∗Anti-F4/80, Alexa Fluor 594-conjugate, clone BM8 (1:500) | BioLegend | Cat # 123140 RRID: AB_2563241 |
| ∗∗Anti-F4/80, Alexa Fluor 647-conjugate, clone BM8 (1:1000) | BioLegend | Cat # 123122 RRID: AB_893480 |
| ∗∗Anti-Fibrinogen, rabbit polyclonal (1:1000) | Agilent Dako | Cat# A008002, RRID: AB_578481 |
| ∗∗Anti-Ly6G, Alexa Fluor 647-conjugate, clone 1A8 (1:500) | BioLegend | Cat # 127609, RRID: AB_1134162 |
| ∗∗Anti-Podoplanin, Alexa Fluor 488-conjugate, clone 8.1.1 (1:1000) | BioLegend | Cat # 127406 RRID: AB_2161930 |
| ∗∗Anti-Tim4, Alexa Fluor 647-conjugate, clone 21H12 (1:500) | BD Biosciences | Cat # 564177 RRID: AB_2647750 |
| ∗∗Anti-ZO-1, Alexa Fluor 594-conjugate, clone ZO1-1A12 (1:500) | Invitrogen | Cat # 339194 RRID: AB_2532188 |
| ∗∗Goat anti-rabbit Ig, Alexa Fluor 546-conjugate (1:1000) | Thermo Fisher | Cat # A11035 RRID: AB_143051 |
| ∗∗Goat-anti-rabbit Ig, Alexa Fluor 647-conjugate (1:1000) | Thermo Fisher | Cat # A21245 RRID: AB_2535813 |
| Bacterial and virus strains | ||
| Escherichia coli M6L4 strain | Dr. G. Núñez (University of Michigan Medical School) Zeng et al.5 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1,4-Diazabicyclo[2.2.2]octane | Sigma-Aldrich | Cat # D27802 |
| Acetone | VWR | Cat # 83683.230 |
| BDMA (benzyl dimethylamine) | TAAB | Cat # B008 |
| Bovine serum albumin | Sigma-Aldrich | Cat # A7030 |
| Bovine serum albumin - low fatty acid | Sigma-Aldrich | Cat # A8806 |
| DDSA (dodecenyl succinic anhydride) EM grade | TAAB | Cat # D027 |
| Dulbecco’s Modified Eagle’s Medium (DMEM)-low glucose | Sigma-Aldrich | Cat # D6046 |
| EDTA | Merck | Cat # 108418 |
| Epon 812 | TAAB | Cat # T026 |
| DAPI | Sigma-Aldrich | Cat # 28718-90-3 |
| DNase I | Roche | Cat # 10104159001 |
| FBS (fetal bovine serum) | Sigma-Aldrich | Cat # F7524 |
| Glutaraldehyde 25% | TAAB | Cat # G002 |
| Glycerol 85% | Sigma-Aldrich | Cat # 1040941000 |
| HEPES | Merck | Cat # H3375 |
| HEPES buffer 1 M | Biowest | Cat # LO180-500 |
| Liberase TL | Roche | Cat # 05401020001 |
| MNA (methyl nadic anhydride) | TAAB | Cat # M012 |
| Mowiol 4-88 | Merck | Cat # 91381 |
| NaH2PO4 | Merck | Cat # 106346 |
| Na2HPO4 | Merck | Cat # 106586 |
| NaCl | Merck | Cat # 116224 |
| Osmium tetroxide 2% | TAAB | Cat # O018/1 |
| Paraformaldehyde 4% aqueous solution, EM grade | Electron Microscopy Sciences | Cat # 157-4 |
| Paraformaldehyde 16% aqueous solution, EM grade | Electron Microscopy Sciences | Cat # 15700 |
| Sodium cacodylate trihydrate | Sigma-Aldrich | Cat # C0250 |
| Toluidine blue | Sigma-Aldrich | Cat # T-3260 |
| Tris base | Roche | Cat # 03573826001 |
| Triton X-100 | Sigma-Aldrich | Cat # X100 |
| Uranyl acetate | Electron Microscopy Sciences | Cat # 22400 |
| Critical commercial assays | ||
| BD Cytofix/cytoperm kit | BD Biosciences | Cat # 554714 |
| eBioscience Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher | Cat # 00-5523-00 |
| FITC Annexin V Apoptosis Detection Kit with 7-AAD | BioLegend | Cat # 640922 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J (8–10-week-old females) | Charles River | Strain code: 632 |
| Software and algorithms | ||
| Fiji | ImageJ Schindelin et al.6 | https://imagej.net/Fiji |
| LAS X | Leica | Htttps://www.leica-microsystems.com/products/ microscope-software |
| FlowJo X | FlowJo | https://www.flowjo.com/ solutions/flowjo |
| Prism 8 | GraphPad Software | http://www.graphpad.com/ scientific-software/prism/ |
| Other | ||
| 1.5 mL eppendorf tubes | Eppendorf | Cat # 0030121023 |
| 15 mL Falcon conical polypropylene tubes | Falcon | Cat # 352096 |
| 12-well plates, not treated | Falcon | Cat # 351143 |
| 5 mL polypropylene tubes | Corning | Cat # 352002 |
| 50 mL Falcon conical polypropylene tubes | Falcon | Cat # 352070 |
| 60-mm Petri dishes | Falcon | Cat # 353004 |
| 96-well plates, flat-bottom, not treated | Fisher Scientific | Cat # 267578 |
| 96-well plates, V-bottom, not treated | Fisher Scientific | Cat # 249570 |
| CO2 euthanasia chamber | Plexx | Cat # TT-8200 |
| Dissection board | Fisher Scientific | Cat # 36-119 |
| Glass funnel 55 mm | Fisher Scientific | Cat # 11572423 |
| Forceps, 45° angled serrated, 16 cm | FST | Cat # 11080-02 |
| Forceps, standard straight serrated, 13 cm | FST | Cat # 11000-13 |
| Forceps, fine straight serrated, 13 cm | FST | Cat # 11008-13 |
| Magnetic stirrer | IKA | Cat # 3622000 |
| μ-Dish 35 mm dishes with No. 1.5 ibidi Polymer Coverslip | ibidi | Cat # 81156 |
| Mouse lab animal cages (Eurostandard type II L) | Tecniplast | Cat # 1284L |
| Needles 25 G | BD Biosciences | Cat # 300600 |
| 200 μL P200 micropipette | Gilson | Cat # FA10005M |
| Scissors, straight blunt, 11 cm | FST | Cat #14074-11 |
| Scissors, straight sharp, 8.5 cm | FST | Cat #14084-08 |
| Solid aluminum cylindrical weights | This paper | supplied on demand |
| Syringes 10 mL | BD Biosciences | Cat # 305959 |
| BD LSR II Flow Cytometer | BD Biosciences | N/A |
| Roker 2D Digital rocking shaker | IKA-Werke GmbH | Cat # 4003000 |
| Jeol 1011 Transmission Electron microscope | Jeol Ltd. | N/A |
| Leica DM IL LED phase contrast inverted microscope | Leica microsystems | N/A |
| Leica EM UC6 Ultramicrotome | Leica microsystems | N/A |
| Leica DM 2500 Microscope | Leica microsystems | N/A |
| Leica TCS SP8 Confocal Microscope | Leica microsystems | N/A |
| Leica Stellaris 5 Confocal Microscope | Leica microsystems | N/A |
Note: ∗Antibodies for Flow Cytometry; ∗∗Antibodies for Confocal Microscopy.
Materials and equipment
Aluminum weights
In-house designed solid aluminum weights are used for pressing peritoneal wall or omentum samples against the bottom of μ-Dish 35 mm dishes, for optimal confocal microscopy imaging (see Figures 1A and 1B). We can provide these weights on-demand at cost price; alternatively these weights can be ordered from the machining company Erosimar (Madrid, Spain. https://www.erosimar.com).
PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| NaH2PO4 | 1.8 mM | 0.25 g |
| Na2HPO4 | 8.3 mM | 1.2 g |
| NaCl | 150 mM | 8.77 g |
| double distilled water (ddH20) | N/A | 1,000 mL |
| Total | N/A | 1,000 mL |
Note: Adjust pH to 7.4, sterilize by autoclaving, and store at room temperature (20°C–22°C) for up to 3 months.
PBS 2% BSA
| Reagent | Final concentration | Amount |
|---|---|---|
| Bovine Serum Albumin (BSA) | 2% w/v | 2 g |
| PBS | N/A | 100 mL |
| Total | N/A | 100 mL |
Note: Store at 4°C for up to 1 week.
PBS-EDTA-FBS
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA | 5 mM | 1.86 g |
| FBS | 3% v/v | 30 mL |
| PBS | N/A | 970 mL |
| Total | N/A | 1,000 mL |
Note: Store at 4°C for up to 1 week.
Antibody staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Bovine Serum Albumin (BSA) | 2% w/v | 0.2 g |
| DAPI | 0.005 mg/mL | 50 μL |
| PBS | N/A | 10 mL |
| Total | N/A | 10 mL |
Note: Prepare before use.
Mowiol-based WMI mounting solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Mowiol 4-88 | 10% w/v | 2.4 g |
| Glycerol 85% | 25% v/v | 6 mL |
| Tris 0.2 M stock | 100 mM | 12 mL |
| ddH2O | N/A | 6 mL |
| 1,4-Diazabicyclo[2.2.2] octane | 2.5% w/v | 0.6 g |
| Total | N/A | 25 mL |
Note: Mix 2.4 g Mowiol 4-88, 6 mL Glycerol 85%, 12 mL Tris 0.2 M and 6 mL ddH2O in a 50 mL Falcon tube in a magnetic stirrer overnight (12–16 h). Heat at 70°C in a waterbath for 10 min to fully dissolve Mowiol. Clarify by centrifugation at 5,000 g for 15 min at 4°C. Transfer the supernatant to a new 50 mL Falcon tube, add 0.6 g 1,4-Diazabicyclo[2.2.2] octane and mix gently by inversion of the tube. Store at −20°C in 2 mL single-use aliquots for up to 6 months.
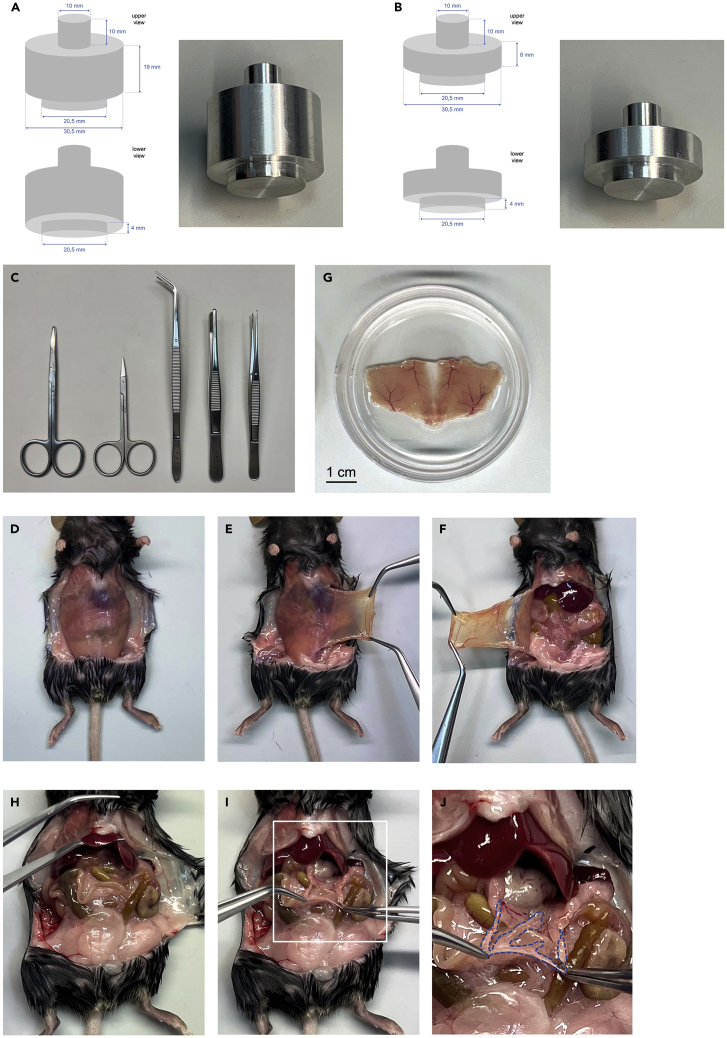
Figure 1.
Preparation of peritoneal wall and omentum samples for WMI and confocal microscopy studies
(A) Description of the aluminum weight used for WMI and confocal microscopy imaging of peritoneal wall samples.
(B) Description of the aluminum weight used for WMI and confocal microscopy imaging of omentum samples.
(C) Surgical tool set used for this protocol including (from left to right) straight blunt 11 cm scissors, straight sharp 8.5 cm scissors, 45° angled serrated 16 cm forceps, standard straight serrated 13 cm forceps and fine straight serrated 13 cm forceps.
(D) Incision in the skin in the abdominal area.
(E and F) Dissection of the peritoneal wall.
(G) Peritoneal wall flattened and stretched on the bottom of the 60-mm Petri dish.
(H–J) Dissection of the omentum. (I) Magnification of the area marked by a white lined square in (H).
Tris 0.2 M stock: Dissolve 2.42 g Tris Base in 100 mL ddH2O and adjust pH to 8.5. Store at room temperature (20°C–22°C) up to 3 months.
Enzymatic digestion solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Liberase TL 2.5 mg/mL stock | 0.05 mg/mL | 10 μL |
| DNase I 2.5 mg/mL stock | 0.25 mg/mL | 50 μL |
| HEPES 1 M | 50 mM | 25 μL |
| PBS BSA-low fatty acid (10×) stock | 0.01 mg/mL | 50 μL |
| DMEM-low glucose | N/A | 365 μL |
| Total | N/A | 500 μL |
Note: Prepare before use.
Liberase TL 2.5 mg/mL stock: Dissolve 5 mg Liberase TL in 2 mL ddH2O. Store at −20°C in 50 μL single-use aliquots for up to 6 months.
DNase I 2.5 mg/mL stock: Dissolve 2.5 mg DNase I in 1 mL ddH2O. Store at 4°C for up to 1 week.
PBS BSA-low fatty acid (10×) stock: Dissolve 1 g BSA-low fatty acid in 10 mL PBS. Store at 4°C for up to 1 week.
Fixation solution for electron microscopy
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde 16% | 1.5% | 4.7 mL |
| Glutaraldehyde 25% | 1.5% | 3 mL |
| HEPES 0.4 M stock | 0.15 M | 18.8 mL |
| ddH2O | N/A | 23.5 mL |
| Total | N/A | 50 mL |
Note: Store at 4°C for up to 24 h.
HEPES 0.4 M stock: Dissolve 9.53 g HEPES in 100 mL ddH2O. Adjust pH to 7.2 and store at 4°C for up to 3 months.
CRITICAL: Paraformaldehyde and glutaraldehyde are toxic; wear a lab coat, gloves and safety googles. Work in a chemical safety hood and dispose waste in accordance with local regulations.
HEPES 0.15 M buffer
Dissolve 3.57 g HEPES in 100 mL ddH2O. Adjust pH to 7.2 and store at 4°C for up to 3 months.
Cacodylate buffer 0.1 M
Dissolve 0.64 g sodium cacodylate trihydrate in 15 mL ddH2O. Adjust pH to 7.4 and store at 4°C for up to 3 months.
Cacodylate buffer 0.2 M
Dissolve 1.28 g sodium cacodylate trihydrate in 15 mL ddH2O. Adjust pH to 7.4 and store at 4°C for up to 3 months.
Osmium tetroxide solution
Mix 2 mL osmium tetroxide 2% and 2 mL cacodylate buffer 0.2 M. Prepare before use.
CRITICAL: Osmium tetroxide is acutely toxic; wear a lab coat, gloves and safety googles. Work in a chemical safety hood and dispose waste in accordance with local regulations.
Uranyl acetate solution
Dissolve 0.2 g uranyl acetate in 10 mL ddH2O. Store at 4°C for up to 3 months.
CRITICAL: Uranyl acetate is radioactive and highly toxic; wear a lab coat, gloves and safety googles. Work in a chemical safety hood behind a protective screen and dispose waste in accordance with local regulations.
Epoxy resin
| Reagent | Final concentration | Amount |
|---|---|---|
| Epon 812 | 48% | 2.4 g |
| DDSA EM grade | 19% | 0.95 g |
| MNA | 33% | 1.65 g |
| BDMA | 3% | 0.15 g |
| Total | N/A | 5 g |
Note: Using a precision balance, add sequentially with a Pasteur pipette 2.4 g 812 Epon resin, 0.95 g DDSA EM grade, 1.65 g MNA and 0.15 g BDMA in a 50 mL Falcon tube. Mix gently in a magnetic stirrer at room temperature (20°C–22°C) until the solution is completely homogenized. Centrifuge at 15,000 g for 5 min to remove air bubbles. Prepare before use.
Step-by-step method details
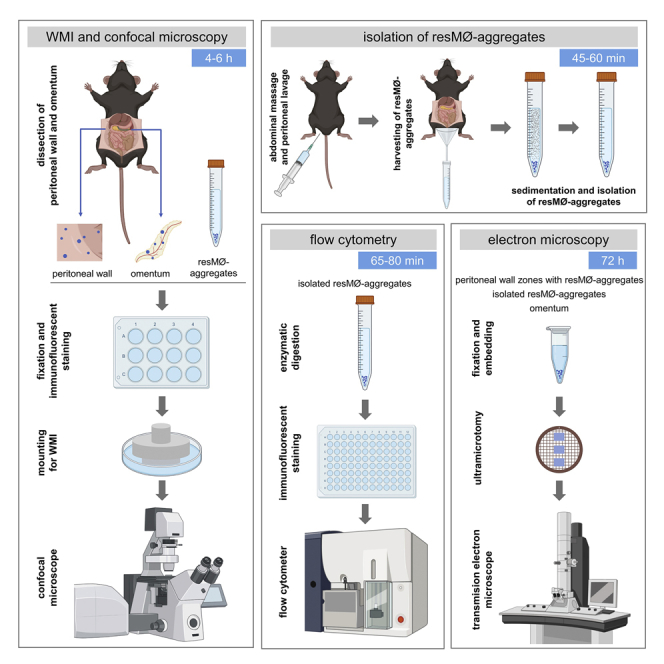
Whole mount immunofluorescence and confocal microscopy imaging of resMØ-aggregates
Timing: 4–6 h/mouse
This step describes how to carry out the dissection, fixation, immunofluorescent staining and mounting of whole peritoneal wall and omentum samples to perform WMI/confocal microscopy studies.
-
1.Dissection and fixation of the peritoneal wall.
-
a.Euthanize mice by CO2 inhalation.
-
b.Place the mouse on its back on a dissection board.
-
c.Spray the abdomen with 70% ethanol.
-
d.Using straight blunt scissors (see Figure 1C, showing the surgical tool set used) make an incision in the skin in the abdominal area, starting in the lower abdominal midline, without penetrating the abdominal wall, and extend the incision until the ribcage.
-
e.Extend the incision to the hind and forelegs (Figure 1D).
-
f.Dissect the whole peritoneal wall using straight sharp scissors and 45° angled serrated forceps (Figures 1E and 1F).
-
g.Transfer the peritoneal wall to a 60-mm Petri dish.
 CRITICAL: Make sure that the side of the peritoneal wall facing the peritoneal cavity is placed upwards during immunofluorescence staining.
CRITICAL: Make sure that the side of the peritoneal wall facing the peritoneal cavity is placed upwards during immunofluorescence staining. -
h.Flatten and stretch out the peritoneal wall on the bottom of the 60-mm Petri dish using standard straight serrated forceps (See Figure 1G).
-
i.Add 5 mL of 4% paraformaldehyde solution and incubate for 5 min at 4°C.
-
j.Wash twice in PBS for 5 min.
-
k.Cut the peritoneal wall along the midline in two halves and trim the edges so each can fit in a well of a 12-well plate.
-
a.
-
2.Dissection and fixation of the omentum.
-
a.Follow the steps 1a to 1e described above.
-
b.Using 45° angled serrated and fine serrated forceps, dissect the omentum by lifting the liver up, and isolating the omentum from the adipose tissue associated to pancreas, stomach and spleen (Figures 1H–1J; Methods video S1).Methods video S1. Dissection of the omentum, related to step 2Download video file (15.3MB, mp4)
-
c.Transfer the omentum to into a well of a 12-well plate containing 1 mL of 4% paraformaldehyde solution and incubate for 5 min at 4°C.
-
d.Wash twice in PBS for 5 min.Note: Fixed peritoneal wall or omentum samples can be kept in PBS overnight (12–16 h), at 4°C, before proceeding with immunofluorescence staining, although this can compromise the correct detection of some antigens as well as tissue integrity.
 CRITICAL: Paraformaldehyde is hazardous; wear gloves, work in chemical safety hood and dispose waste in accordance with local regulations.
CRITICAL: Paraformaldehyde is hazardous; wear gloves, work in chemical safety hood and dispose waste in accordance with local regulations.
-
a.
-
3.Immunofluorescence staining of whole peritoneal wall and omentum samples.Note: All staining steps are performed in 12-well plates (Figure 2A) under continuous agitation using a rocking shaker, in the dark.
-
a.Incubate each sample in 1 mL PBS 2% BSA for 30 min at 4°C for blocking unspecific binding of primary antibodies.
-
b.Incubate each sample in 1 mL of fluorophore-conjugated antibodies, diluted as indicated in the key resources table, in antibody staining solution, for 2–4 h at 4°C.
-
c.Wash twice in PBS for 5 min.Note: When using primary unconjugated antibodies, incubate the samples with unconjugated antibodies, diluted as indicated in the key resources table, in PBS 2% BSA for 2 h at 4°C, wash twice in PBS for 5 min and incubate with secondary antibodies in antibody staining solution, for 2 h at 4°C. For staining of intracellular molecules incubate the samples with 0.1% Triton X-100 for 15 min at 4°C prior to incubation with antibodies.Note: A complete list of the antibodies used in our WMI studies, and the corresponding working dilutions, is included in the key resources table. If using antibodies distinct from those listed in this protocol, the working dilution and time of incubation should be defined on the basis of titration experiments.
-
a.
-
4.Mounting samples for confocal microscopy.
-
a.Transfer peritoneal wall or omentum samples to μ-Dish 35 mm dishes with No. 1.5 ibidi Polymer Coverslip, containing 300 μL of mowiol-based WMI mounting solution (Figure 2B).Note: The surface of the peritoneal wall or omentum to be observed under the microscope should face the bottom of the dish. Place the samples carefully to avoid the formation of bubbles.
-
b.Place an aluminum weight, for either peritoneal wall or omentum samples, on top of each sample, to flatten and press the samples against the bottom of the dish (Figure 2C).Note: The weight has to be kept on top of the samples during imaging (Figure 2D). These samples are ready to be analyzed by confocal microscopy.Note: An example of a standard staining combination allowing the detection of resMØ-aggregates in the peritoneal wall would include Alexa Fluor 488-conjugated anti-podoplanin (mesothelium), Alexa Fluor 594-conjugated anti-F4/80 (resMØs), and unconjugated anti-fibrinogen (fibrin network) followed by Alexa Fluor 647-conjugated secondary antibody and DAPI (Figures 2E and 2F). Low magnification confocal microscopy imaging allows to explore the distribution of resMØ-aggregates, as shown in Figure 2G. For additional information related to the analysis of resMØ-aggregates by WMI, please refer to Vega-Pérez et al.1
 CRITICAL: The use of aluminum weights significantly improves the quality of WMI/confocal microscopy imaging.
CRITICAL: The use of aluminum weights significantly improves the quality of WMI/confocal microscopy imaging.
-
a.
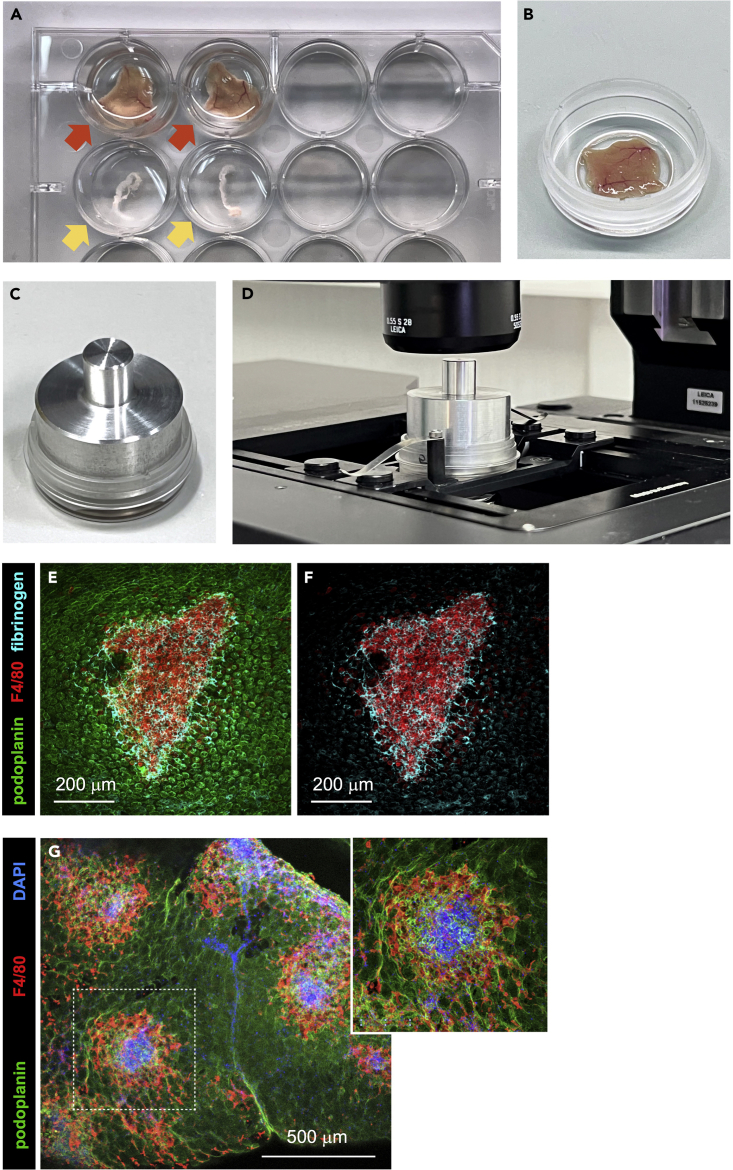
Figure 2.
Immunofluorescence staining of whole peritoneal wall and omentum samples
(A) Peritoneal wall (red arrows) and omentum (yellow arrows) samples transferred to wells of a 12-well plate to carry out the immunofluorescence staining process.
(B) Peritoneal wall sample transferred to a μ-Dish 35 mm dish with No. 1.5 ibidi Polymer Coverslip containing 300 μL of mowiol-based WMI mounting solution.
(C) Peritoneal wall sample mounted in a μ-Dish 35 mm dish with No. 1.5 ibidi Polymer Coverslip, with 300 μL of mowiol-based WMI mounting solution, and a peritoneal wall aluminum weight on top.
(D) Peritoneal wall sample mounted in a μ-Dish 35 mm dish with No. 1.5 ibidi Polymer Coverslip with aluminum weight on top, placed on the stage of a confocal microscope.
(E and F) WMI/confocal microscopy imaging of a resMØ-aggregate in the peritoneal wall, at 4 h after E. coli infection, after immunofluorescent staining with Alexa Fluor 488-conjugated anti-podoplanin (mesothelium), Alexa Fluor 594-conjugated anti-F4/80 (resMØs), and unconjugated anti-fibrinogen (fibrin network) followed by Alexa Fluor 647-conjugated secondary antibody. (E) podoplanin, F4/80 and fibrinogen staining; (F) F4/80 and fibrinogen staining. Images were acquired on a multispectral Leica TCS SP8 confocal microscope and analyzed using ImageJ software.
(G) Low magnificaction confocal microscopy image of the omentum, at 4 h after E. coli infection, showing the distribution of resMØ-aggregates after immunofluorescent staining with Alexa Fluor 488-conjugated anti-podoplanin (mesothelium), Alexa Fluor 594-conjugated anti-F4/80 (resMØs), and DAPI (nuclei). Insert: enlargement of the area marked in G with a white dotted line. Images were acquired on a multispectral Leica SP8 confocal microscope and analyzed using ImageJ software. Figures F and G reprinted with permission from Vega-Pérez at al.1
Isolation of resMØ-aggregates
Timing: 45–60 min/mouse
This step describes how to isolate mesothelium-bound resMØ-aggregates formed during inflammatory processes in different organs of the peritoneal cavity, particularly in the peritoneal wall and omentum. resMØ-aggregates induced by E coli infection can be easily isolated during the first 6 h post infection while, from 18 h post infection, resMØ-aggregates start to disaggregate and are more difficult to isolate. Isolated resMØ-aggregates can be subsequently analyzed by flow cytometry or electron microscopy, as detailed below, or be subjected to other studies.
-
5.
Follow steps 1a to 1c described above.
-
6.
Make an incision in the skin in the abdominal area, starting in the lower abdominal midline, without penetrating the abdominal wall, and extend the incision until the neck.
-
7.
Extend the incision to the hind and forelegs and remove all the abdominal and thoracic skin.
-
8.
Inject 6–9 mL of cold PBS in the peritoneal cavity at the lower left quadrant of the abdomen, using a 10 mL syringe with a 25 G needle.
Note: The volume of PBS to be injected has to be adjusted to the age/size of the mice used. For 8–10-week-old C57BL/6 females 8 mL were injected.
-
9.
Massage the abdomen vigorously to detach resMØ-aggregates from the mesothelium (Methods video S2).
-
10.
Make an incision in the peritoneal wall starting in the lower abdominal midline and extend the incision until the ribcage (Figure 3A).
-
11.
Collect the peritoneal lavage containing the resMØ-aggregates directly into a 15 mL Falcon tube using a glass funnel (Figure 3A).
-
12.
Place the tube on ice and leave resMØ-aggregates to sediment for 5 min (Figure 3B; Methods video S3).
-
13.
Remove the supernatant carefully, leaving 0.5 mL with resMØ-aggregates at the bottom of the tube.
-
14.
Add carefully 5 mL of cold PBS.
-
15.
Repeat steps 12 and 13 as needed in order to obtain a resMØ-aggregate preparation as clean as possible of free cells.
Note: Check the quality of the preparation under an inverted phase contrast microscope after transferring the resMØ-aggregates into a well of a flat-bottom 96-well plate (Figure 3C).
CRITICAL: Use a 200 μL micropipette with tips in which the front end has been cut off to transfer isolated resMØ-aggregates, in order to avoid their disruption.
-
16.
Keep isolated resMØ-aggregates in 1 mL PBS in a 15 mL Falcon tube.
CRITICAL: The intensity/strength of the abdominal massage is critical to harvest resMØ-aggregates properly. An insufficient massage will not detach resMØ-aggregates while a too vigorous massage may disintegrate resMØ-aggregates.
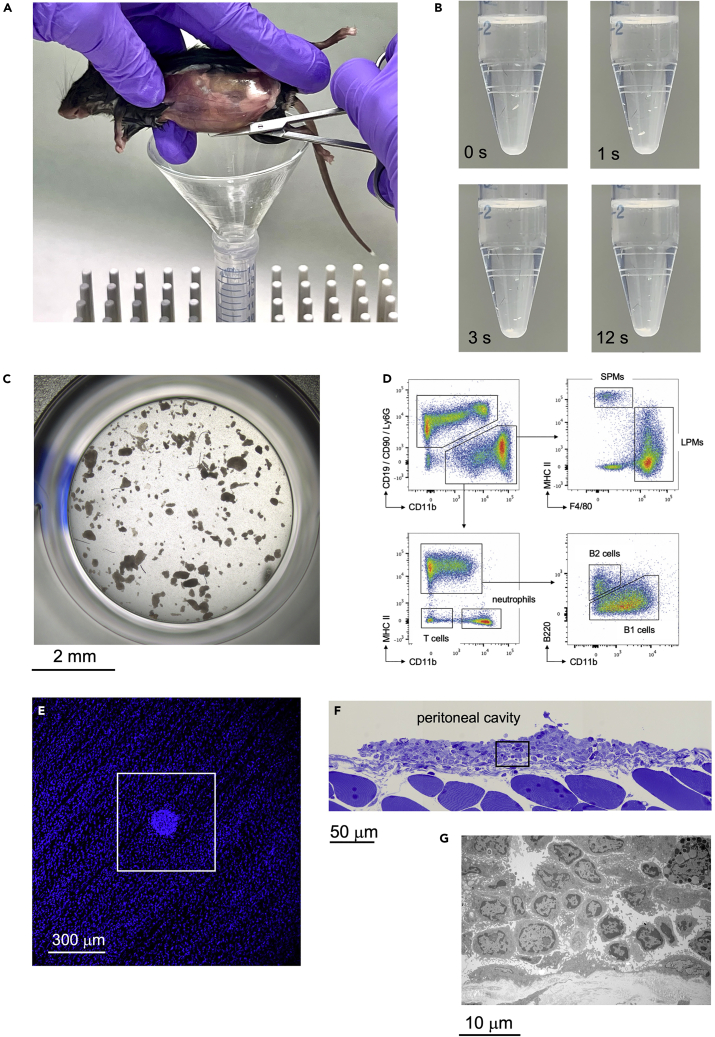
Figure 3.
Flow cytometry and electron microscopy analysis of isolated resMØ-aggregates
(A) Harvesting of resMØ-aggregates by making an incision in the peritoneal wall, after injecting 8 mL PBS intraperitoneally and performing a peritoneal massage.
(B) Sedimentation of resMØ-aggregates harvested at 4 h after E. coli infection; the time after starting of the sedimentation process is indicated.
(C) Phase contrast microscopy image of resMØ-aggregates isolated at 4 h after E. coli infection, and transferred into a well of a flat-bottom 96-well plate.
(D) Flow cytometry analysis of resMØ-aggregates isolated at 4 h after E. coli infection, after immunofluorescent staining with PE-Cy7-conjugated anti-CD11b, APC-Cy7-conjugated anti-F4/80, APC-conjugated anti-MHC II, PE-conjugated anti-Ly6G, PE-conjugated anti-CD19, Pacific Blue-conjugated anti-B220 and PE-conjugated anti-CD90. Data were acquired on a Becton Dickinson LSR II flow cytometer and analyzed using FlowJo X software.
(E) WMI/confocal microscopy image of the peritoneal wall after DAPI staining, allowing the detection of resMØ-aggregates at 4 h after E. coli infection.
(F) Semithin section of the peritoneal wall at 4 h after E. coli infection, showing a resMØ-aggregate. Toluidine blue staining.
(G) Electron microscopy image of the area marked by a black lined square in (F). Figures F and G reprinted with permission from Vega-Pérez at al.1
Processing of isolated resMØ-aggregates for flow cytometry
Timing: 20 min/mouse
This step describes how to disaggregate isolated resMØ-aggregates by enzymatic digestion to obtain a cell suspension containing the cells forming these aggregates, which can be subsequently analyzed by flow cytometry.
-
17.
Centrifuge the isolated resMØ-aggregates preparation obtained in step 16 at 500 g for 5 min at 4°C in a 15 mL Falcon tube.
-
18.
Remove the supernatant carefully.
-
19.
Add 500 μL of the enzymatic digestion solution and incubate in a waterbath at 37°C for 5 min.
-
20.
Disrupt resMØ-aggregates mechanically by gentle pipetting using a 200 μL micropipette.
Note: Check resMØ-aggregate disruption under an inverted phase contrast microscope, after transferring the cell suspension into a well of a flat-bottom 96-well plate.
-
21.
Add 10 mL PBS-EDTA-FBS to stop the enzymatic digestion.
-
22.
Centrifuge at 500 g for 5 min at 4°C.
-
23.
Remove the supernatant and resuspend in 500 μL PBS-EDTA-FBS.
Note: resMØ-aggregates isolated during the first 60 min post-infection can be disaggregated without enzymatic digestion. In this case avoid steps 17–19 and 21.
Note: From 1.5 × 105 to 2.5 × 105 cells are usually obtained from resMØ-aggregates isolated at 4 h after E. coli infection of a 8–10-week-old C57BL/6 female.
Immunofluorescent staining of resMØ-aggregate cell suspensions for flow cytometry
Timing: 45–60 min
This step describes how to process resMØ-aggregate cell suspensions to be analyzed by flow cytometry. Immunofluorescent staining is performed in 96-well V-bottom plates in the dark.
-
24.
Transfer the cells obtained in step 23 into a 96-well V-bottom plate at ≥105 cells/well.
-
25.
Centrifuge the 96-well plate at 500 g for 5 min at 4°C.
-
26.
Discard the supernatant, resuspend the cells in 30 μL of anti-CD16/32 antibody to block Fc receptors, and incubate for 15 min at 4°C.
-
27.
Wash with 100 μL PBS-EDTA-FBS per well and centrifuge at 500 g for 5 min at 4°C.
-
28.
Discard the supernatant, resuspend the cells in 30 μL of fluorophore-conjugated antibodies, diluted as indicated in the key resources table in PBS-EDTA-FBS, and incubate for 15 min at 4°C.
-
29.
Wash with 100 μL PBS-EDTA-FBS per well and centrifuge at 500 g for 5 min at 4°C.
-
30.
Discard the supernatant, resuspend in 200 μL PBS-EDTA-FBS and transfer the cells to 5 mL polypropylene tubes. These samples are ready to be analyzed in a flow cytometer.
Note: A complete list of the antibodies used in our flow cytometry studies of resMØ-aggregate cells, and the corresponding working dilutions, is included in the key resources table. An example of a standard staining combination allowing the detection by flow cytometry of the main cell subsets composing resMØ-aggregates at 4 h after E. coli infection, i.e., LPMs (large resident peritoneal macrophages), SPMs (small peritoneal macrophages), B1 cells, B2 cells, T cells and neutrophils would include PE-Cy7-conjugated anti-CD11b, APC-Cy7-conjugated anti-F4/80, APC-conjugated anti-MHC II, PE-conjugated anti-Ly6G, PE-conjugated anti-CD19, Pacific Blue-conjugated anti-B220 and PE-conjugated anti-CD90 (Figure 3D). For additional information related to the analysis of resMØ-aggregates by flow cytometry, please refer to Vega-Pérez et al.1
Processing of resMØ-aggregates for electron microscopy imaging
Timing: 72 h
This step describes how to process isolated resMØ-aggregates, or peritoneal wall zones harboring resMØ-aggregates, for electron microscopy studies.
-
31.Processing isolated resMØ-aggregates for electron microscopy.Note: Removal of the different reagents described below has to be performed using a Pasteur pipette and avoiding centrifugation. All the steps should be carried out in a chemical safety hood; process carefully to avoid damaging resMØ-aggregates.
-
a.Transfer isolated resMØ-aggregates obtained in step 16 into a 1.5 mL eppendorf tube.
-
b.Add carefully 0.5 mL of the Fixation Solution for Electron Microscopy, and incubate overnight (12–16 h) at 4°C.
-
c.Remove the Fixation Solution and wash (x2) with 1 mL HEPES 0.15 M buffer for 5 min at 4°C.
-
d.Remove HEPES 0.15 M buffer and wash (x4) with 1 mL cacodylate buffer 0.1 M for 5 min at 4°C.
-
e.Remove cacodylate buffer 0.1 M and incubate with 0.5 mL osmium tetroxide solution for 1 h at 4°C in the dark.
-
f.Remove osmium tetroxide solution and wash (x4) with 1 mL cacodylate buffer 0.1 M for 5 min at 4°C.
-
g.Remove cacodylate buffer 0.1 M and wash (x2) with 1 mL ddH2O for 5 min at 4°C.
-
h.Remove ddH2O and incubate with 0.5 mL uranyl acetate solution for 60 min at 4°C in the dark.
-
i.Remove uranyl acetate solution and wash (x2) with 1 mL ddH2O for 5 min at 4°C.
-
j.Remove ddH2O and incubate with 1 mL 70% acetone for 10 min at 4°C.
-
k.Remove 70% acetone and incubate with 1 mL 90% acetone for 10 min at 4°C.
-
l.Remove 90% acetone and incubate with 1 mL 100% acetone for 10 min at 4°C.
-
m.Exchange 100% acetone and incubate for 10 min at room temperature (20°C–22°C).
-
n.Incubate in 1 mL 1:1 epoxy resin/100% acetone for 30 min at room temperature (20°C–22°C).Note: Epoxy resin should be prewarmed at 40°C.
-
o.Incubate in 1 mL epoxy resin for 2 h at room temperature (20°C–22°C).
-
p.Exchange epoxy resin and incubate for 3 h at room temperature (20°C–22°C).
-
q.Transfer to shape molds, add epoxy resin and incubate for 48 h at 60°C. These samples are ready to be processed by ultramicrotomy.
-
a.
-
32.Isolation of zones of the peritoneal wall harboring resMØ-aggregates.
-
a.Follow steps 1a to 1f.
-
b.Transfer the peritoneal wall to a 60-mm Petri dish containing 5 mL of the Fixation Solution for Electron Microscopy, and incubate overnight (12–16 h) at 4°C.
-
c.Remove the Fixation Solution and wash twice with 5 mL PBS for 5 min at 4°C.
-
d.Cut the peritoneal wall in two halves and trim the edges so each can fit in a well of a 12-well plate.
-
e.Remove the PBS and incubate with 5 mL DAPI at 1:200 in PBS for 15 min at 4°C in the dark.
-
f.Remove DAPI and wash with PBS for 5 min at 4°C.
-
g.Transfer the peritoneal wall samples to μ-Dish 35 mm dishes with No. 1.5 ibidi Polymer Coverslip containing 300 μL of PBS.
 CRITICAL: Do not use mowiol-based WMI mounting solution in this step.
CRITICAL: Do not use mowiol-based WMI mounting solution in this step. -
h.Place an aluminum weight for peritoneal wall on top of each sample.
-
i.Analyze the samples by confocal microscopy to locate resMØ-aggregates, which can be identified as spots with a high density DAPI staining (Figure 3E), and choose zones harboring one or several noticeable resMØ-aggregates.
-
j.Once spotted under the microscope by the DAPI staining, remove the aluminum weight carefully; the selected area can be then located by the laser beam. Next determine the position of the piece of the peritoneal wall to be dissected using as reference the position and morphology of the vasculature of this area.
-
k.Dissect carefully small pieces (∼2 × 2 mm) of peritoneal wall containing the zones defined in the previous step.
-
a.
-
33.Processing of peritoneal wall zones harboring resMØ-aggregates for electron microscopy.
-
a.Transfer the peritoneal wall pieces obtained in step 32j into a 1.5 mL eppendorf tube.
- b.
-
a.
Expected outcomes
As pointed out above, during peritoneal inflammatory reactions caused by peritoneal damage or infection, resMØ-aggregates, which are crucial for repairing peritoneal injuries and controlling infections, are formed predominantly in the peritoneal wall and omentum. However, they can also develop in the mesothelial surface of other peritoneal organs, i.e., the mesentery, gonadal fat and intestine and, thus, the number of resMØ-aggregates detectable in the peritoneal wall or omentum is variable. At 4 h after E. coli infection, the number of resMØ-aggregates detectable in the peritoneal wall or the omentum is variable and usually ranges from a few (2–3) until many (10–15) in both locations.
On the other hand, the size of resMØ-aggregates formed in the peritoneal wall during the first hours after E. coli infection usually ranges from 100 to >800 μm. The structure and composition resMØ-aggregates formed in response to other inflammatory reactions may differ and therefore needs to be investigated.
As indicated above, from 1.5 × 105 to 2.5 × 105 cells are usually obtained after disaggregation of resMØ-aggregates isolated at 4 h after E. coli infection of an 8–10-week-old C57BL/6 female. However, the number of cells obtainable from resMØ-aggregates can differ depending on the age, sex, time of analysis and, particularly, on the experimental model of peritoneal aggression being analyzed.
Limitations
This protocol describes how to proceed for imaging resMØ-aggregates, formed in the peritoneal wall or omentum during peritoneal inflammation, by WMI combined with confocal microscopy. Although, as pointed out above, this protocol can be used for imaging other peritoneal organs in which resMØ-aggregates can form, such as mesentery, spleen, gut or ovary, as well as the inner wall of the thoracic cavity, it might be necessary in order to optimize the imaging in these organs to set up some steps of our protocol, and to modify the weight of the aluminum weights.
Examples of expected outcomes regarding the composition, kinetics and estimation of the number of resMØ-aggregates, described in the present protocol, refer to our experiments of peritoneal E. coli infection, but these parameters might differ depending on the peritoneal inflammation model used.
Similarly, as described in our protocol, resMØ-aggregates can be isolated to perform additional studies. However the possibility of isolating resMØ-aggregates is limited, in our E. coli infection model, to a time frame between around 1 h and 18 h after infection, determined by the composition, adhesion to the mesothelium and onset of the resMØ-aggregate disruption process. Time restrictions for isolating resMØ-aggregates may differ depending on the experimental model used.
As pointed out above, in our peritoneal E.coli infection model, resMØ-aggregates are preferentially formed in the peritoneal wall and omentum, but can also form in other peritoneal organs. Consequently, resMØ-aggregates isolated as described in this protocol might potentially contain resMØ-aggregates formed in all these locations and therefore, when analyzing them by flow cytometry, electron microscopy or other techniques, it is important that to take into account that the composition resMØ-aggregates may differ depending on the organ in which they are formed.
Troubleshooting
Problem 1
Mounting samples for confocal microscopy (step 4).
Potential solution
Commercial mounting media exist as an alternative to the homemade mowiol-based WMI mounting solution described in this protocol, but we recommend using of the homemade mowiol-based WMI mounting solution due to its superior optical quality. As pointed out above the use of aluminum weights to flatten peritoneal wall and omentum samples significantly improves the quality of imaging by WMI and confocal microscopy, and therefore their use is strongly recommended. On the other hand, special care should be taken during the processing of peritoneal wall and omentum samples to prevent them from excessive bending and twisting, respectively.
Problem 2
Isolating resMØ-aggregates (steps 9–15).
Potential solution
resMØ-aggregates are particularly fragile and therefore special care has to be taken when performing the abdominal massage, and when manipulating them, to preserve their integrity.
Problem 3
Processing isolated resMØ-aggregates for electron microscopy (step 31).
Potential solution
As pointed out above, resMØ-aggregates are particularly fragile and, therefore, special care has to be taken when manipulating them along all the steps of processing for electron microscopy to preserve their integrity.
Problem 4
Isolation of zones of the peritoneal wall harboring resMØ-aggregates (step 32).
Potential solution
As indicated above, the number of resMØ-aggregates detectable in the peritoneal wall or omentum is variable, ranging from a few (2–3) until many (10–15) at 4 h after E. coli infection. Consequently, in some cases it may be difficult to spot zones of the peritoneal wall containing resMØ-aggregates to be dissected and processed for electron microscopy, and thus it may be necessary to analyze a number of samples to find a convenient zone.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Carlos Ardavín (ardavin@cnb.csic.es).
Materials availability
Aluminum weights used WMI and confocal microscopy imaging can be provided on-demand at cost price. Alternatively these weights can be ordered from the machining company Erosimar (Madrid, Spain. https://www.erosimar.com).
Acknowledgments
This work was supported by the Ministerio de Ciencia, Innovación y Universidades of Spain (grant PGC 2018-101899-B-100 to C.A.), Ministerio de Ciencia e Innovación of Spain (grant PID2021-122748OB-I00 to C.A.), and Comunidad de Madrid (grant B2017/BMD-3731 to C.A.). We thank the Advanced Light Microscopy and the Electron Microscopy facilities of the CNB for technical advice and sample processing.
Author contributions
C.A. designed research, designed aluminum weights for WMI, analyzed data, and wrote the manuscript. M.F., A.V.-P., and A.G.-G. performed experiments, analyzed data, and contributed to review and editing of the manuscript. N.A.-L. performed experiments and contributed to review and editing of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102079.
Data and code availability
This study did not generate new unique datasets or code.
References
- 1.Vega-Pérez A., Villarrubia L.H., Godio C., Gutiérrez-González A., Feo-Lucas L., Ferriz M., Martínez-Puente N., Alcaín J., Mora A., Sabio G., et al. Resident macrophage-dependent immune cell scaffolds drive anti-bacterial defense in the peritoneal cavity. Immunity. 2021;54:2578–2594.e5. doi: 10.1016/j.immuni.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Shintani Y., Fields L., Shiraishi M., Podaru M.N., Kainuma S., Yamashita K., Kobayashi K., Perretti M., Lewis-McDougall F., Suzuki K. Cell barrier function of resident peritoneal macrophages in post-operative adhesions. Nat. Commun. 2021;12:2232. doi: 10.1038/s41467-021-22536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N., Czepielewski R.S., Jarjour N.N., Erlich E.C., Esaulova E., Saunders B.T., Grover S.P., Cleuren A.C., Broze G.J., Edelson B.T., et al. Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J. Exp. Med. 2019;216:1291–1300. doi: 10.1084/jem.20182024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zindel J., Peiseler M., Hossain M., Deppermann C., Lee W.Y., Haenni B., Zuber B., Deniset J.F., Surewaard B.G.J., Candinas D., Kubes P. Primordial GATA6 macrophages function as extravascular platelets in sterile injury. Science. 2021;371:eabe0595. doi: 10.1126/science.abe0595. [DOI] [PubMed] [Google Scholar]
- 5.Zeng M.Y., Cisalpino D., Varadarajan S., Hellman J., Warren H.S., Cascalho M., Inohara N., Núñez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new unique datasets or code.