Highlights
-
•
Replication of diffuse WM injury.
-
•
Replication of diffuse WM injury correlations with clinical outcome.
-
•
Nature of injury not replicated (high Cho and Cr vs originally reported low NAA).
-
•
High sensitivity of MRS approach may be more important than choice of WM region.
Keywords: Replicability, Mild traumatic brain injury (mTBI), MR spectroscopic imaging (MRSI), Quantitative MR, Linear regression, Glasgow Outcome Scale Extended (GOSE)
Abbreviations: CC, Corpus callosum; CDE, Common Data Element; CENTER, Collaborative European NeuroTrauma Effectiveness Research; Cho, Choline; Cr, Creatine; CRLB, Cramér-Rao lower bound; DAI, Diffuse axonal injury; DTI, Diffusion tensor imaging; EPSI, Echo-planar spectroscopic imaging; FLAIR, Fluid-Attenuated Inversion Recovery; FOV, Field of view; Glx, Glutamate plus glutamine; GM, Gray matter; GCS, Glasgow Coma Scale; GOSE, Glasgow Outcome Scale – Extended; 1H MRS, Proton magnetic resonance spectroscopy; 1H MRSI, Proton magnetic resonance spectroscopic imaging; ISMRM, International Society for Magnetic Resonance in Medicine; LOC, Loss of consciousness; mI, myo-inositol; MIDAS, Metabolite Imaging and Data Analysis Software; MINT, Map Integrated Spectrum; MPRAGE, Magnetization Prepared Rapid Gradient Echo; mTBI, Mild traumatic brain injury; MWU, Mann-Whitney U test; NAA, N-acetyl-aspartate; NINDS, National Institute of Neurological Disorders and Stroke; PCS, Post-concussive symptoms; PRANA, Project Review and Analysis; PTA, Post-traumatic amnesia; ROI, Region-of-interest; SNR, Signal-to-noise ratio; SWI, Susceptibility-weighted imaging; TBI, Traumatic brain injury; TRACK, Transforming Research and Clinical Knowledge; TE, Echo time; TI, Inversion time; TR, Repetition time; VOI, Volume-of-interest; WM, White matter
Abstract
Purpose
Proton magnetic resonance spectroscopy (1H MRS) offers biomarkers of metabolic damage after mild traumatic brain injury (mTBI), but a lack of replicability studies hampers clinical translation. In a conceptual replication study design, the results reported in four previous publications were used as the hypotheses (H1-H7), specifically: abnormalities in patients are diffuse (H1), confined to white matter (WM) (H2), comprise low N-acetyl-aspartate (NAA) levels and normal choline (Cho), creatine (Cr) and myo-inositol (mI) (H3), and correlate with clinical outcome (H4); additionally, a lack of findings in regional subcortical WM (H5) and deep gray matter (GM) structures (H6), except for higher mI in patients’ putamen (H7).
Methods
26 mTBI patients (20 female, age 36.5 ± 12.5 [mean ± standard deviation] years), within two months from injury and 21 age-, sex-, and education-matched healthy controls were scanned at 3 Tesla with 3D echo-planar spectroscopic imaging. To test H1-H3, global analysis using linear regression was used to obtain metabolite levels of GM and WM in each brain lobe. For H4, patients were stratified into non-recovered and recovered subgroups using the Glasgow Outcome Scale Extended. To test H5-H7, regional analysis using spectral averaging estimated metabolite levels in four GM and six WM structures segmented from T1-weighted MRI. The Mann-Whitney U test and weighted least squares analysis of covariance were used to examine mean group differences in metabolite levels between all patients and all controls (H1-H3, H5-H7), and between recovered and non-recovered patients and their respectively matched controls (H4). Replicability was defined as the support or failure to support the null hypotheses in accordance with the content of H1-H7, and was further evaluated using percent differences, coefficients of variation, and effect size (Cohen’s d).
Results
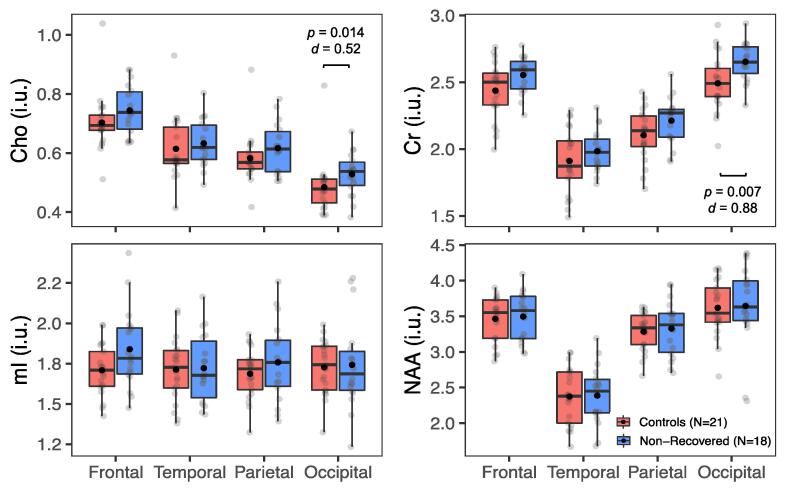
Patients’ occipital lobe WM Cho and Cr levels were 6.0% and 4.6% higher than controls’, respectively (Cho, d = 0.37, p = 0.04; Cr, d = 0.63, p = 0.03). The same findings, i.e., higher patients’ occipital lobe WM Cho and Cr (both p = 0.01), but with larger percent differences (Cho, 8.6%; Cr, 6.3%) and effect sizes (Cho, d = 0.52; Cr, d = 0.88) were found in the comparison of non-recovered patients to their matched controls. For the lobar WM Cho and Cr comparisons without statistical significance (frontal, parietal, temporal), unidirectional effect sizes were observed (Cho, d = 0.07 – 0.37; Cr, d = 0.27 – 0.63). No differences were found in any metabolite in any lobe in the comparison between recovered patients and their matched controls. In the regional analyses, no differences in metabolite levels were found in any GM or WM region, but all WM regions (posterior, frontal, corona radiata, and the genu, body, and splenium of the corpus callosum) exhibited unidirectional effect sizes for Cho and Cr (Cho, d = 0.03 – 0.34; Cr, d = 0.16 – 0.51).
Conclusions
We replicated findings of diffuse WM injury, which correlated with clinical outcome (supporting H1-H2, H4). These findings, however, were among the glial markers Cho and Cr, not the neuronal marker NAA (not supporting H3). No differences were found in regional GM and WM metabolite levels (supporting H5-H6), nor in putaminal mI (not supporting H7). Unidirectional effect sizes of higher patients’ Cho and Cr within all WM analyses suggest widespread injury, and are in line with the conclusion from the previous publications, i.e., that detection of WM injury may be more dependent upon sensitivity of the 1H MRS technique than on the selection of specific regions. The findings lend further support to the corollary that clinic-ready 1H MRS biomarkers for mTBI may best be achieved by using high signal-to-noise-ratio single-voxels placed anywhere within WM. The biochemical signature of the injury, however, may differ and therefore absolute levels, rather than ratios may be preferred. Future replication efforts should further test the generalizability of these findings.
1. Introduction
Traumatic brain injury (TBI) is a global health concern, with an estimated annual incidence of 69 million worldwide (Dewan et al., 2018). Mild TBI (mTBI), which accounts for 75–90% of cases, is diagnosed based on a Glasgow Coma Scale (GCS) score of 13–15, loss of consciousness (LOC) lasting <30 min, and post-traumatic amnesia (PTA) lasting <24 h (Prince and Bruhns, 2017, Kay and Adams, 1993). These criteria, however, lack prognostic utility for identifying the concerning subset of patients who suffer from ongoing cognitive, behavioral, and/or physical impairments beyond the post-acute recovery period (Permenter and Fernandez-de Thomas, 2021). Predictive biomarkers of post-concussive symptoms (PCS) following mTBI are therefore needed to improve management and patient outcomes (Dadas et al., 2018).
In mTBI, diffuse axonal injury (DAI), the main pathophysiological substrate of PCS (Bigler and Maxwell, 2012, McKee and Daneshvar, 2015), is rarely detected using conventional (T1- and T2-weighted) MR imaging (MRI). While hemorrhagic DAI is revealed by susceptibility-weighted imaging (SWI), most exams yield no findings. DAI, however, is known to occur in normal-appearing tissue and changes related to its presence can be detected by quantitative MR (qMR) methods, such as proton magnetic resonance spectroscopy (1H MRS) (Marino et al., 2011, Cecil et al., 1998, Ross et al., 1998). Unfortunately, despite decades of research, neither 1H MRS, nor other potentially quantitative techniques such as diffusion tensor imaging (DTI), feature prominently in routine clinical protocols. The challenges to adoption largely encompass the lack of fulfilling specific standards (Manikis et al., 2017, deSouza et al., 2019, Abramson et al., 2015, Weingärtner et al., 2021) established by the wider qMR community, including: (1) validation, (2) clinical qualification, and (3) utilization/dissemination. While single- and multi-voxel 1H MRS packages have been validated and are available from the major MR vendors, unsatisfactory progress in (2) has hampered their use in mTBI. One reason is the inability to extract generalizable information from past studies, due to their large heterogeneity in 1H MRS technique, patient population and study design. Harmonization of past data and advice for standardization of future studies (Bartnik-Olson et al., 2021) can help, but require wide collaborations and are time-intensive. An essential part of the solution are replication studies, which in recent years have been advocated widely in scientific research (Replicating scientific results is tough, 2021, Baker, 2016) and in the qMR field (Poldrack et al., 2017, Raunig et al., 2015).
Our purpose, therefore, was to investigate whether the 1H MR spectroscopic imaging (MRSI) findings in mTBI from our laboratory over the last decade and published across four publications (Kirov et al., 2013a, Kirov et al., 2013b, Kierans et al., 2014, Davitz et al., 2019), can be replicated. As a conceptual replication (Crandall and Sherman, 2016, Nosek and Errington, 2017) study design, key variables were kept constant, among them: acquisition type (MRSI), post-processing (regional and global analysis), patient source (metropolitan Emergency Department and concussion clinic), proportion of complicated mTBI (mostly MR-negative), and time from injury (∼20 days post-TBI). Our hypotheses (H1-H7) were our previously reported findings: Utilizing global analysis, patients would show diffuse abnormalities (H1), confined to white matter (WM) (H2), comprising low N-acetyl-aspartate (NAA) (H3) (Kirov et al., 2013a), and only in non-recovered patients (H4) (Kirov et al., 2013b). Utilizing regional analysis, patients would show a lack of findings in six subcortical WM regions (H5) (Davitz et al., 2019) and four deep gray matter (GM) structures (H6), except for higher patients’ myo-inositol (mI) in the putamen (H7) (Kierans et al., 2014) (Table 1). Support for the above hypotheses, i.e., replicating our previous results, would strengthen the argument that these findings can be generalized to other cohorts of similar characteristics. Specifically, they would support our previous conclusion (Davitz et al., 2019) that 1H MRSI changes are of a generally diffuse nature, and therefore their detection depends more on the sensitivity of the 1H MRS technique than on selection of specific regions; hence, single-voxels placed anywhere in WM may provide the most pragmatic and clinic-ready implementation of 1H MRS for mTBI.
Table 1.
Summary of hypotheses and replication status.
| Approach | Hypothesis on metabolic abnormalities | Source | Status | |
|---|---|---|---|---|
|
H1 |
Global Analysis |
Diffuse distribution |
(Kirov et al., 2013a) |
Replicated |
|
H2 |
Localized to white matter |
Replicated |
||
|
H3 |
Neuronal etiology |
Not Replicated |
||
| H4 | Present in functionally non-recovered individuals, absent in functionally recovered individuals* | (Kirov et al., 2013b) | Replicated | |
| H5 | Regional Analysis – White Matter | Absent in the body, genu, and splenium of the corpus callosum, the corona radiata, and the frontal and occipital white matter structures | (Davitz et al., 2019) | Replicated |
|
H6 |
Regional Analysis – Gray Matter | Absent in the caudate, putamen (NAA, Cr, Cho, Glx), globus pallidus, and thalamus |
(Kierans et al., 2014) | Replicated |
| H7 | Elevated mI in the putamen | Not Replicated | ||
*Original study dichotomized individuals according to presence or absence of PCS.
2. Materials and methods
2.1. Study population
2.1.1. Recruitment
This study was approved by the Institutional Review Board (IRB) and written informed consent was obtained according to the Declaration of Helsinki. Patient recruitment took place from four Emergency Departments [NYULH Tisch Hospital, NYULH Brooklyn (Level I Trauma Center), NYULH Cobble Hill, and Bellevue Hospital Center (Level I Trauma Center)], and NYULH’s Concussion Center. Out of ∼700 mild TBI patients screened from these multi-site referrals, 566 were “potentially eligible”, but ∼67% could not be contacted and ∼28% were deemed ineligible based on an in-depth phone screen. The rest, 31 closed-head mTBI patients (23 female; age 37 ± 13 [average ± standard deviation (SD)] years, range 18 – 60 years; education 16 ± 2 years) were prospectively enrolled between November 2018 and December 2019. The average time between the accident and testing was 22 ± 10 days (range 5 – 53 days). Primary inclusion criteria were: (i) 18 – 65 years of age, (ii) less than two months from injury date, (iii) mTBI diagnosis according to the American Congress of Rehabilitation Medicine (ACRM, 1993); (iv) no previous mTBI within the past two years, (v) no history of moderate or severe TBI or other neurological disease, (vi) no history of disqualifying neurological or psychiatric conditions, or substance abuse, and (vii) no MRI contraindications.
Twenty-one age-, sex-, and education-matched healthy volunteers (14 female; age 34 ± 12 years, range 22 – 61 years; education 16 ± 3 years) were recruited as controls. Their inclusion criteria were the same as those for the patients, but with the absence of any TBI history.
2.1.2. Clinical outcome assessments
All patients were administered the Glasgow Outcome Scale – Extended (GOSE), a core measure (Wilson et al., 1998, Madhok et al., 2020) of global outcome after TBI, listed as part of the National Institute of Neurological Disorders and Stroke (NINDS) Common Data Elements (CDEs) (Wilde et al., 2010, Hicks et al., 2013). The GOSE is an eight-point scale that evaluates the overall impact of TBI on independence, work capacity, social relationships, and cognition. A score of 8 indicates full recovery and resumption of normal life, and descending scores from 7 to 3 reflect increasing degrees of mental and/or physical disability. Patients were dichotomized into “recovered” (GOSE = 8) and “non-recovered” (GOSE ≤ 7) groups, as commonly done (Madhok et al., 2020, Yuh et al., 2021) and recommended in a validation study (Nelson et al., 2017) using data from the multicenter Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) consortium.
Additionally, patients were evaluated on the presence of five post-concussion symptoms reported by the World Health Organization (WHO) Collaborating Center Task Force on mTBI (Borg et al., 2004) in 2004 as being the most common acute symptoms after TBI (headache, dizziness, sleep disturbance, memory problems, and blurred vision). Patients who endorsed at least one of these symptoms were considered “PCS-positive”, otherwise they were classified as “PCS-negative”, as done in our past work (Kirov et al., 2013b).
2.2. MRI and 1H MRSI data acquisition and post-processing
2.2.1. Data acquisition
All experiments were performed in a 3 T MRI scanner (MAGNETOM Prisma [syngo MR E11], Siemens Healthcare, Erlangen, Germany) using a 20-channel transmit-receive head coil (Siemens Healthcare, Erlangen, Germany). The protocol included the following clinically-used sequences: (i) 3D T1-weighted magnetization prepared rapid gradient echo (MPRAGE): repetition time (TR) = 2400 ms, echo time (TE) = 2.2 ms, inversion time (TI) = 1060 ms, voxel size = 0.8 × 0.8 × 0.8 mm3, field-of-view (FOV) = 256 × 240 × 208 mm3, 6:38 min; (ii) 2D T2-weighted fluid-attenuated inversion recovery (FLAIR): TR = 9000 ms, TE = 81 ms, TI = 2500 ms, voxel size = 0.7 × 0.7 × 5 mm3, FOV = 220 × 220 × 30 mm3, 2:44 min; (iii) SWI: TR = 28 ms, TE = 20 ms, voxel size = 0.7 × 0.7 × 3 mm3, FOV = 220 × 192 × 56 mm3, 3:46 min. To guide 1H MRSI placement, the MPRAGE images were re-sliced into an axial orientation parallel to the anterior commissure – posterior commissure (AC-PC) axis with 1 mm slice resolution. The FLAIR and SWI images were used for identification of any mTBI-related abnormalities.
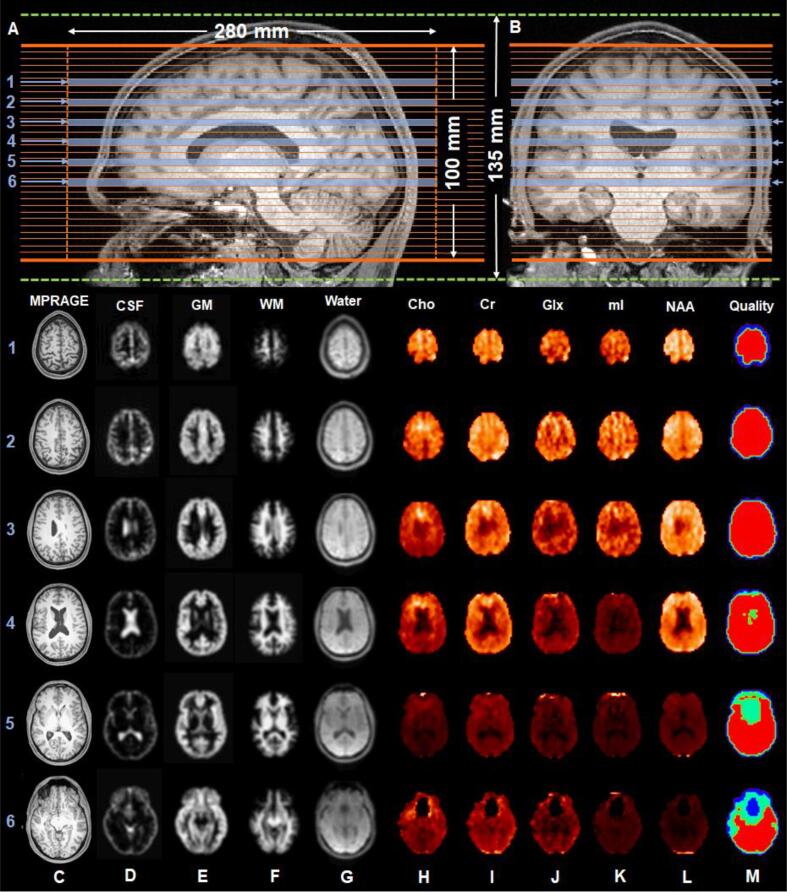
Three-dimensional 1H MRSI data were acquired using a validated (Ding et al., 2015) and widely used (e.g., Lecocq et al., 2015, Sabati et al., 2015, Ding et al., 2016, Zhang et al., 2018, Maghsudi et al., 2020a, Maghsudi et al., 2020b, Zhang et al., 2020) echo-planar spectroscopic imaging (EPSI) prototype sequence with lipid inversion nulling: TR = 1710 ms, TE = 17.6 ms, flip angle = 73°, TI = 198 ms, FOV = 280 × 280 × 135 mm3, matrix = 50 × 50 × 18, nominal voxel volume = 0.235 cm3, 16:49 min. A 280 × 280 × 100 mm3 excitation slab covered the base of the occipital lobe through the top the brain, parallel to the AC-PC axis, as shown in Fig. 1A-B. An outer volume saturation band was placed over the sinuses and eyeballs to reduce their magnetic field inhomogeneity effects. Automatic shimming, followed by a manual adjustment of first-order shims to a water frequency linewidth of <27 Hz for the whole head was performed for all subjects. The EPSI sequence included an interleaved, gradient-echo acquisition with the same spatial resolution as that of the metabolite acquisition for unsuppressed water, termed the “internal water” dataset (TR = 591 ms, TE = 6.3 ms, flip angle = 20°) (Barker et al., 1993).
Fig. 1.
1H MRSI EPSI sequence positioning and processing output. (A-B): Reconstructed sagittal and coronal T1-weighted MRI images of a control subject, superimposed with the 280 × 280 × 135 mm3 field-of-view (dotted green lines) and the 280 × 280 × 100 mm3 excitation slab (solid orange lines) partitioned into 32 slices, corresponding to the MRSI-resolution of 0.08 cm3 after interpolation. (C): Six reconstructed axial T1-weighted MRI images in accordance with the numbered slices (blue arrows and bars) designated in (A) and (B). (D-F): Tissue segmentation maps of CSF, GM, and WM, derived from the T1-weighted MRI in (C) and down-sampled to the MRSI-resolution. (G): The water reference image, used to support image registration, spatial transformations, and signal normalization procedures. (H-L): Metabolite maps and an accompanying quality map (M), in which red indicates voxels with metabolite linewidth <12 Hz and signal <3 standard deviations from the mean. Voxels within regions that are colored dark blue, cyan, and green on the quality map were excluded from the analysis. Abbreviations: MPRAGE, Magnetization Prepared Rapid Gradient Echo; CSF, cerebrospinal fluid; GM, gray matter; WM, white matter; Cho, choline; Cr, creatine; Glx, glutamate plus glutamine; mI, myo-inositol; NAA, N-acetyl-aspartate. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2.2. 1H MRSI initial data processing
Datasets were processed using the standard workflow of the Metabolite Imaging and Data Analyses System (MIDAS) software package (Maudsley et al., 2009, Maudsley et al., 2006). Briefly, spectra were corrected for frequency shifts due to B0 field inhomogeneities and fitted with a prior knowledge basis set, specific to the EPSI parameters and scanner’s field strength (Soher et al., 1998). A quality control procedure excluded (i) spectra whose metabolite linewidths were >12 Hz, and (ii) “outliers” defined as voxels with signal ≥3 times the SD from the mean of all voxels (Kreis, 2004, Maudsley et al., 2010) (Fig. 1M). The internal water dataset provided the time-domain correction functions (i.e., B0 shifts, phase, eddy-current correction, lineshape distortions) for spectral analysis, and was used in the metabolite signal intensity normalization procedure. In this pipeline, cerebrospinal fluid (CSF), gray matter (GM), and white matter (WM) maps were first obtained from FSL/FAST tissue segmentation of the high-resolution MPRAGE and subsequent convolution with the 1H MRSI point-spread function, to yield images interpolated to 64 × 64 × 32 points, with a voxel volume of ∼0.08 cm3 (Fig. 1D-F). Next, scaling was accomplished by taking these down-sampled CSF, GM, and WM maps and applying the following literature (Neeb et al., 2006) tissue water fractions: CSF = 0.98; GM = 0.726; WM = 0.634. For bias field correction and metabolite quantification, the following literature (Wansapura et al., 1999, Lu et al., 2005) tissue water T1 relaxation values (measured at 3 T) were used: CSF T1 = 4300 ms; GM T1 = 1350 ms; WM T1 = 840 ms. Finally, metabolite signals were normalized to the internal water signal, yielding relative metabolite levels in institutional units (i.u.). Following this normalization step, the internal water dataset (Fig. 1G) provided spatial transformation parameters to allow registration between the reconstructed axial MPRAGE (Fig. 1C) and the 1H MRSI-resolution metabolite data (Fig. 1H-L).
After the above steps, the data were processed within two separate pipelines, one for global and one for regional analysis.
2.2.3. Global analysis
Using the Project Review and Analysis (PRANA) module in MIDAS, spectra and tissue fractions of individual voxels were integrated into a linear regression model, as described previously (Maudsley et al., 2009, Maudsley et al., 2006), to obtain average metabolite values corresponding to 100% GM and 100% WM within eight regions specified by PRANA’s default lobar atlas: bilateral frontal, parietal, temporal, and occipital lobes (Fig. 2A). Quality criteria excluded (i) lobes with <1000 voxels, (ii) edge voxels with <100% contribution for each atlas-defined region, and (iii) voxels with a CSF fraction >30%. Additionally, a correction for signal loss due to CSF partial volume was applied to voxels with a CSF fraction <0.3, expressed as
| (1) |
where is the uncorrected metabolite signal and is the fraction of CSF in the 1H MRSI voxel (Sabati et al., 2015). Bilateral metabolite levels were averaged to obtain a single GM and WM value per brain lobe.
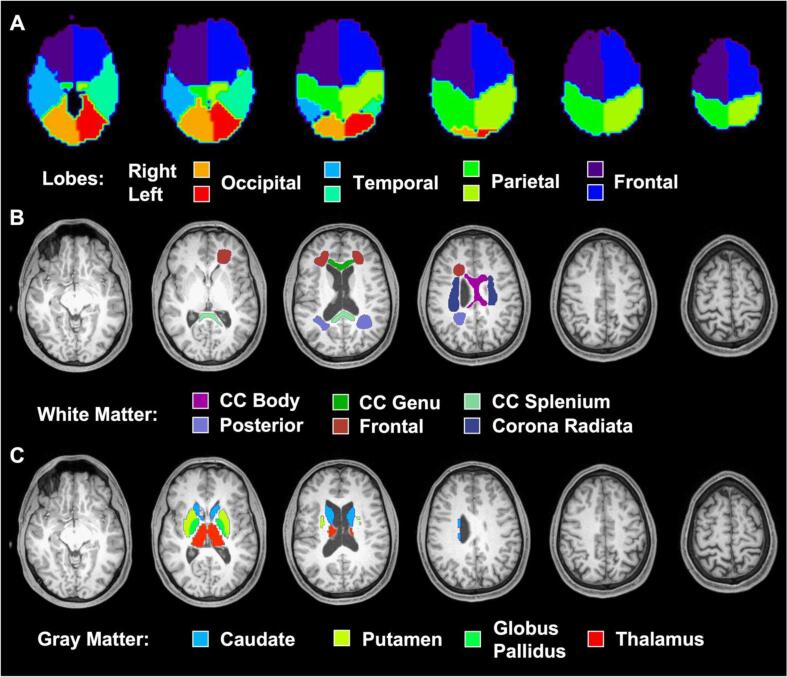
Fig. 2.
Segmented regions used in global and regional analyses. Axial T1-weighted MPRAGE slices from a control subject (as shown in Fig. 1C) overlaid with the (A) atlas-defined lobar regions from MIDAS (Maudsley et al., 2009, Maudsley et al., 2006), (B) manually traced white matter regions from FireVoxel (Rusinek et al., 2013), and (C) auto-segmented gray matter regions from FreeSurfer (Fischl et al., 2012). Abbreviations: CC, corpus callosum.
2.2.4. Regional analysis
Six WM regions-of-interest (ROIs) were manually drawn on each subject’s reconstructed axial MPRAGE using FireVoxel (Rusinek et al., 2013): bilateral frontal white matter (FWM), posterior white matter (PWM), corona radiata (CorRad), and the genu, body, and splenium of the corpus callosum (GCC, BCC, and SCC, respectively) (Fig. 2B). Previously, we had chosen these ROIs based on findings from 1H MRS (Lin et al., 2012), DTI (Hulkower et al., 2013), functional MRI (Sharp et al., 2014), and histopathology (Cecil et al., 1998, Browne et al., 2011, Mouzon et al., 2012) literature, while considering the inherent spatial resolution limitations of 1H MRS, which prohibited finer parcellation.
Four deep GM ROIs were extracted from each subject’s axial MPRAGE using FreeSurfer v6.0.0′s (Fischl, 2012) automatic segmentation pipeline: bilateral caudate, putamen, globus pallidus, and thalamus (Fig. 2C). Our previous decision to examine deep GM structures was motivated by prior neuroimaging literature, which implicated impaired glucose metabolism (Garcia-Panach et al., 2011) and blood perfusion (Ge et al., 2009) in thalamic and basal ganglia nuclei after TBI. FreeSurfer has been shown to delineate these ROIs reliably (Iscan et al., 2015, Hedges et al., 2022), particularly yielding thalamic volume estimates consistent with those obtained from gold-standard manual stereology (Keller et al., 2012). However, since the thalamus’ dorsal surface forms the floor of the lateral ventricles, and its medial surface extends entirely along the lateral wall of the third ventricle, it is susceptible to partial volume errors from neighboring CSF. As a result, the thalamus was eroded by one voxel (1 mm3) in all subjects, to reduce CSF signal contamination.
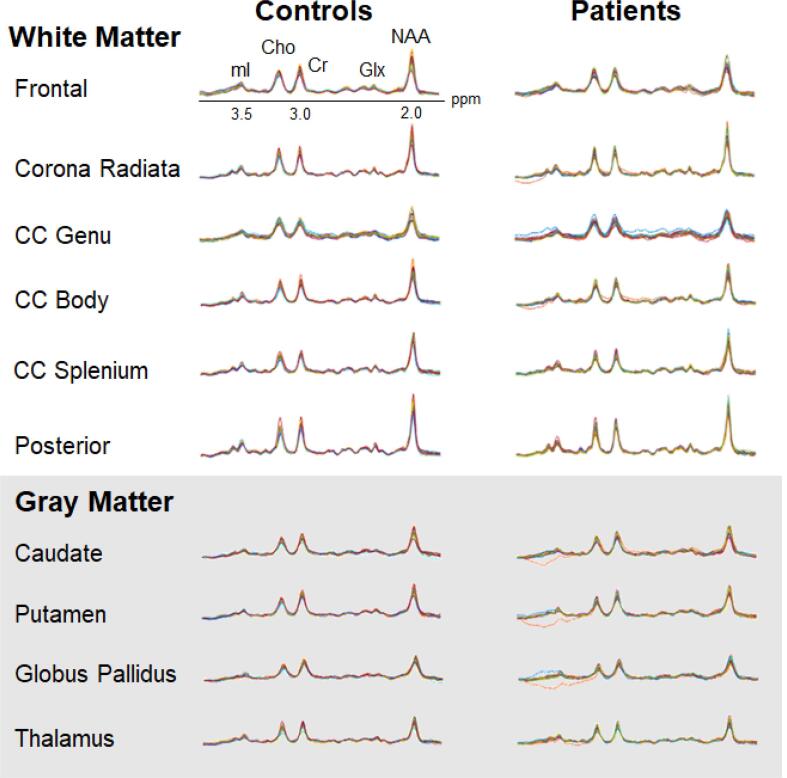
All ten ROIs were mapped to the 1H MRSI matrices using the Map Integrated Spectrum (MINT) module in MIDAS, which performed spectral averaging, i.e., all spectra within an ROI were averaged before fitting a single spectrum per ROI, per subject (Zhang et al., 2018, Goryawala et al., 2016) (Fig. 3). Quality criteria excluded (i) ROIs with <10 voxels per ROI, (ii) edge voxels with <90% contribution for each ROI, and (iii) voxels with a CSF fraction >30%. After the fitting procedure, a CSF partial volume correction (Eq. (1)) was applied to all metabolites within each ROI, and these corrected metabolite signals were then normalized by water. The ROIs in the left and right hemispheres were combined within a single mask, and therefore spectral averaging yielded a single metabolite level for each bilateral structure.
Fig. 3.
Regional 1H MRSI spectra. Averaged spectra from each of the 21 controls (left) and 26 patients (right) overlaid for each white matter (top) and gray matter (bottom) region, except for the genu, where only 19 controls and 23 patients contributed data which passed the quality control. The real part of the 1H spectra is shown, on common frequency (1.5 to 4.0 ppm) and intensity scales. Abbreviations: CC, corpus callosum; Cho, choline; Cr, creatine; Glx, glutamine plus glutamate; mI, myo-inositol; NAA, N-acetyl-aspartate.
2.3. Study design
Our study is classified as a conceptual replication study, in which the goal is to test the same hypothesis from an original study, but the approach (i.e., experimental technique) can vary (Nosek and Errington, 2017). In contrast, in a direct replication study, the goal is again to test the same hypothesis, but the approach is strictly constrained to that of the original study. Conceptual replication does not require matching every single parameter, and therefore allows controlling of variables not previously considered, and importantly, broader generalizability of findings (Crandall and Sherman, 2016). For example, compared to the original publications, the current study uses a different type of 1H MRSI acquisition and post-processing package, one that has become the most widely used 1H MRSI platform for studying the brain and its associated disorders (e.g., Klietz et al., 2019, Verma et al., 2019, Kahl et al., 2020), including TBI (e.g., Govindaraju et al., 2004, Gasparovic et al., 2009, Govind et al., 2010). This can be considered an improvement (as standards of research change over time) and an opportunity to see if the results replicate, not just with unique, lab-specific tools as used previously, but with more current methodology and assessments. Indeed, a recent article on reproducible MRS data analysis states that the use of locally developed tools across different studies comes at the expense of reproducibility (Soher et al., 2022).
Supplementary Tables 1 and 2 compare the human subjects’ characteristics in the original studies to those in this replication study. Supplementary Table 3 compares the respective MRI and 1H MRSI methods. A discussion on the similarities and differences of these methodological (i.e., cohort composition and technical) aspects across studies is included in the Supplementary Material (sections S1.1 – S1.3).
2.3.1. Hypotheses generation and assessment of replicability
The findings from our previous papers were used as the hypotheses of the current study. Importantly, negative findings were also subject to replication, i.e., we considered lack of findings an informative result, since they are important for sensitivity analyses, meta-analyses and guiding future research. Only in one instance replication of a negative finding was not incorporated in a hypothesis. Given the importance of diffuse injury as a concept in TBI, we formulated H1 to include both WM and GM injury, even though the latter was not found in the original study.
One additional change was made to a hypothesis in comparison with the original findings. While we had found that metabolic changes were present only in PCS-positive individuals, in the current study we stratify not based on PCS absence or presence, but on recovery and lack thereof in global functioning (assessed with the GOSE). This hypothesis change was made after data collection, due to the lack of PCS-negative subjects (see section 3.1 below and Supplementary Material, sections S1.1 – S1.3), which precluded testing the previous finding of lack of metabolic abnormalities in such a group. In consideration of the original findings, however, a comparison between PCS-positive patients and controls was still made. It is also noteworthy that while GOSE is a measure of global functional outcome, there is a well-known relationship between the presence of PCS, and higher PCS frequency, with a lower (worse) GOSE score (Voormolen et al., 2018).
The results from the previous publications were considered replicated if the null hypotheses were either supported or not supported in accordance with the content of H1-H7. Specifically, replication would entail the following: In the assessment of global changes, statistically significant differences between patients and controls in any WM or GM metabolite from the lobar linear regression analyses (H1). From the same analyses, findings only in WM (and not in GM) metabolites (H2); lower NAA in patients, and normal Cho, Cr, and mI in WM and GM (H3). Finally, that the above findings are present in functionally non-recovered individuals, and absent in functionally recovered individuals (H4). In the assessment of regional changes, lack of statistically significant differences between patients and controls for all metabolites, for all six WM (H5) and four GM (H6) regions, except for higher putaminal mI in patients (H7). Statistical significance, however, depends not only on biological differences, but also on the variability associated with every technical aspect of the experiment, type of statistical test, decisions of whether or not to account for covariates, etc. Indeed, the Committee on Reproducibility and Replicability in Science (National Academies of Sciences, Engineering, and Medicine, 2019) gives a definition which does not include a statistical clause, i.e., “replicability is obtaining consistent results across studies aimed at answering the same scientific question, each of which has obtained its own data. Two studies may be considered to have replicated if they obtain consistent results given the level of uncertainty inherent in the system under study.” We therefore also report within-group coefficients of variation (CVs), and take into consideration the percent difference between patients and controls, which minimizes confounders. Finally, effect sizes are also reported to compare them with those obtained previously and qualify the size of the effect (Nosek and Errington, 2017), based on Cohen’s d cut-off values.
2.4. Statistical analyses
Controls were matched to patients in age, sex, and education using a frequency matching approach where patients and controls were successfully matched if (i) they were of the same sex, and if (ii) their ages and education levels were within five years of each other, inclusive.
The Mann-Whitney U (MWU) test (performed with R Statistical Software, v4.1.0; R Core Team 2021) was used to compare mean metabolite levels for (1) all mTBI patients and their matched healthy controls, (2) GOSE-defined non-recovered mTBI patients and their matched healthy controls, and (3) WHO-defined PCS-positive mTBI patients and their matched healthy controls. Tests were run for every metabolite within (a) each tissue type (GM and WM) for the global analysis, and (b) each ROI for the regional analysis. A weighted least squares (wLS) analysis of covariance (ANCOVA; performed with SAS 9.3, SAS Institute, Cary, NC) was also used to test (a) and (b), but only for (4) GOSE-defined recovered mTBI patients and all controls. Nonparametric MWU tests, which do not require any assumptions about the underlying data distribution, were used for the fully matched cohorts in (1) to (3), whereas parametric ANCOVA tests, which do assume that the data are normally distributed with equal variance between groups, were used for (4). In this latter condition, three control subjects were not frequency matched in age with any patient, but their ages remained well within the age range of all patients. As a result, comparisons were adjusted for age and sex. Furthermore, a wLS ANCOVA accounted for inter-subject variation in terms of the numbers of voxels used in each brain region, by weighting each observation equal to the square root of the number of voxels used (i.e., according to the statistical precision with which it was measured). Note that Davitz et al. (2019) also performed stratified group analyses with the number of voxels per ROI as a weighting factor. All tests were conducted at the two-sided 5% significance level without correction for multiple comparisons.
CVs, defined as the SD divided by the mean metabolite level, and expressed as a percentage, were calculated as a measure of within-group (patients or controls) variability.
Percent differences, defined as the absolute difference between mean metabolite levels divided by the average of the two values, and expressed as a percentage (Cole and Altman, 2017), were calculated as a measure of magnitude change for the differences between patients and controls.
The Cohen’s d statistic, defined as the difference between mean metabolite levels divided by the pooled SD, was calculated as a measure of effect size for the differences between patients and controls:
| (2) |
where , , , , , and , denote the patients’ (“p”) and controls (“c”) sample sizes, metabolite level means and SDs, following Davitz et al. (2019). A “small” effect was defined as |d| ≤ 0.5, a “medium” effect was defined as 0.5 < |d| < 0.8, and a “large” effect was defined as |d| ≥ 0.8, as done in Gerhalter et al., 2021.
In post-hoc analyses, Spearman rank and Pearson correlations were performed to assess the association of time from injury to data acquisition with levels of lobar WM Cho and Cr. Both correlations were computed since the discrepancy between them can be informative (e.g., a substantially larger magnitude for the Spearman correlation may indicate that the association was monotonic, but nonlinear). These statistical tests were conducted at the two-sided 5% significance level using SAS version 9.4 software (SAS Institute, Cary, NC).
3. Results
3.1. Study population
Selection bias was assessed by comparing the sample of enrolled patients (n = 31) with the sample of potentially eligible patients (n = 566). The cohorts were comparable in terms of age and proportions of cause of injury; and in that most patients in both groups did not experience any period of loss of consciousness. The enrolled sample had a higher proportion of patients referred from the Concussion Clinic, and a higher proportion of female patients.
Of the 31 mTBI patients recruited, 26 (20 female; age 36.5 ± 12.5 years, range 18 – 60 years; education 15.9 ± 2.3 years, range 11 – 22 years) completed the full protocol and were included in the analyses. Reasons for exclusion were: missing 1H MRSI data (n = 1), claustrophobia (n = 2), presence of extensive non-specific white matter disease not believed to be consistent with traumatic aetiologies (n = 1), and inconsistent account of injury circumstances, which led to doubts about whether the definition of mTBI was met (n = 1). The time after injury for the remaining patients was 22.1 ± 10.3 days. All 21 age-, sex-, and education-matched healthy volunteers (14 female; age 34.2 ± 11.5 years, range 22 – 61 years; education 16.3 ± 3.5 years, range 10 – 26 years) successfully completed the full protocol and were included in the analyses. Patients’ race and ethnicity were categorized into the following mutually exclusive groups: non-Hispanic White (n = 13), non-Hispanic Black (n = 4), Hispanic of any race (n = 6), Asian (n = 1), mixed race (n = 1), and not reported (n = 1). Controls’ race and ethnicity yielded similar groupings: non-Hispanic White (n = 12), non-Hispanic Black (n = 2), Hispanic of any race (n = 5), and Asian (n = 2). All demographics and patients’ injury characteristics from the current study are presented in Supplementary Table 1, alongside those from the previous work (Kirov et al., 2013a, Kirov et al., 2013b, Kierans et al., 2014, Davitz et al., 2019). A breakdown of participant demographics and patient injury characteristics by classification instrument (i.e., either WHO or GOSE criteria) are provided in Supplementary Table 2, alongside those from the previous work (Kirov et al., 2013a, Kirov et al., 2013b, Kierans et al., 2014, Davitz et al., 2019). Out of the 26 mTBI patients who completed the current study, 24 were classified as PCS-positive based on the WHO criteria used in the previous study (Kirov et al., 2013b), and 18 were classified as non-recovered based on the GOSE (n = 1 had a score of 5 [Lower Moderate Disability]; n = 14 had a score of 6 [Upper Moderate Disability]; n = 3 had a score of 7 [Lower Good Recovery]). Correspondingly, two were PCS-negative and eight were GOSE-recovered (had a score of 8 [Upper Good Recovery]).
3.2. 1H MRSI
Shown in Table 1 is a determination of whether H1-H7 were supported or not supported, based on the results of the global and regional analyses (below) and the criteria outlined in section 2.3.1.
3.2.1. Global analysis
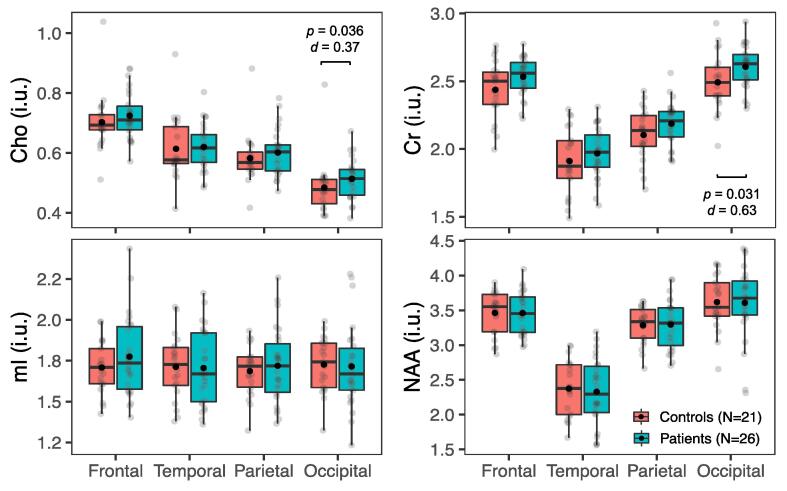
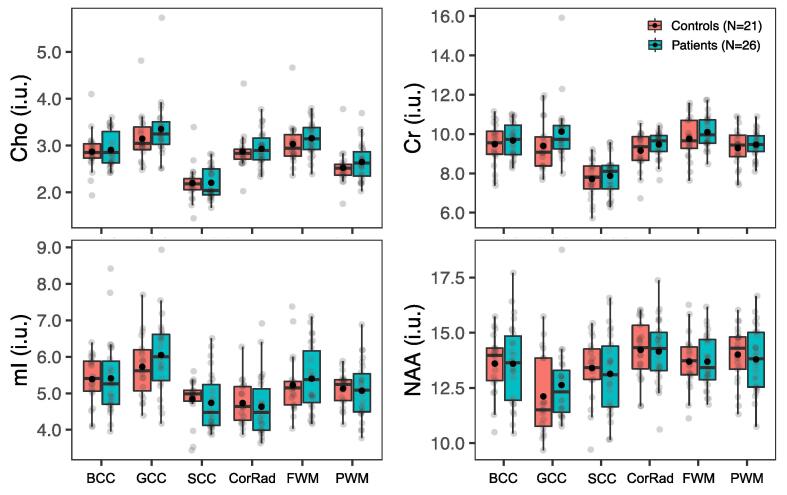
Boxplots of the global WM and GM metabolite distributions, respectively, in all patients (n = 26) and controls (n = 21) are shown in Fig. 4 and Supplementary Fig. 1. Statistically significant higher occipital WM Cho and Cr were found in patients (MWU: Cho p = 0.04; Cr p = 0.03), while no statistically significant group differences were found for mI and NAA in WM, nor for any metabolite in GM (MWU: p ≥ 0.05).
Fig. 4.
Metabolite levels in global white matter (WM) in all patientsvs. all controls. Boxplots displaying 1st, 2nd (median), and 3rd quartiles (box), ±95% (whiskers), and means (●) of Cho, Cr, mI, and NAA distributions within frontal, temporal, parietal, and occipital lobe WM between all patients and their matched controls. Note the statistically significant findings in the occipital lobe (Mann-Whitney U test, p < 0.05), with small-to-moderate effect sizes (Cohen’s d), and visually elevated Cho and Cr in all lobes (i.e., greater means and medians in patients compared to controls).
Comparisons of findings, percent differences, effect sizes (d), and CVs with those of the original study (Kirov et al., 2013a) are compiled in Supplementary Tables 4 and 5; and summarized as follows. The original publication (Kirov et al., 2013a) reported lower global WM NAA in patients (6.7% difference, d = -0.7). The change in occipital WM Cho was of similar absolute magnitude (5.9%) but had lower absolute effect size (d = 0.4). The change in occipital WM Cr was of lower absolute magnitude (4.5%), but of similar absolute effect size (d = 0.6). The average lobar CVs were comparable to the global CVs from the original study (maximum difference between patient groups = 3%), except for much lower Cho CV for the controls in the original study (7% vs. 16%).
3.2.2. Global analysis: Relationship to clinical outcome
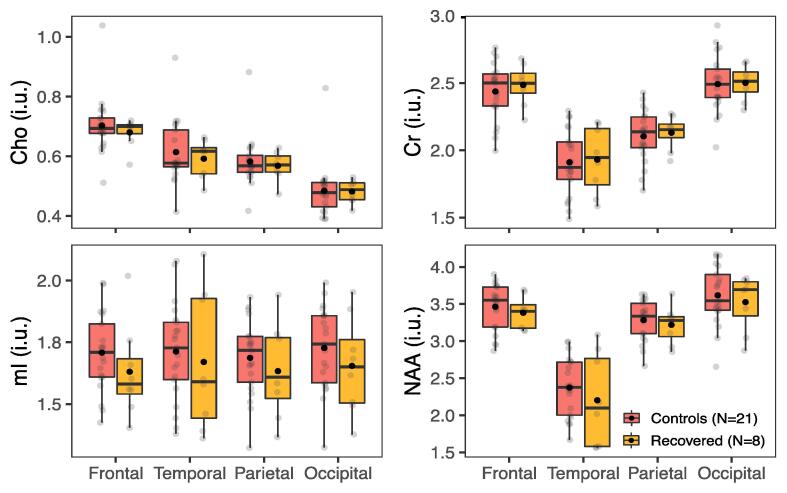
Boxplots of the global WM and GM metabolite distributions, respectively, in non-recovered patients (GOSE ≤ 7, n = 18) and their matched controls (n = 21) are shown in Fig. 5 and Supplementary Fig. 2. Similar to the unstratified whole-group comparison, there were statistically significant higher occipital WM Cho and Cr in patients than in controls (MWU: Cho p = 0.01; Cr p = 0.01). No statistically significant differences were found between recovered patients (GOSE = 8, n = 8) and controls (n = 21) (wLS ANCOVA: p ≥ 0.05) in either global WM or GM (Fig. 6 and Supplementary Fig. 3, respectively). Alterations in the same metabolites were observed when comparing PCS-positive patients (n = 24) and matched controls (n = 21), i.e., higher patients’ occipital WM Cho (MWU: p = 0.027, d = 0.4) and Cr (MWU: p = 0.015, d = 0.7).
Fig. 5.
Metabolite levels in global white matter (WM) in non-recovered patients vs. all controls. Boxplots displaying 1st, 2nd (median), and 3rd quartiles (box), ±95% (whiskers), and means (●) of Cho, Cr, mI, and NAA distributions within frontal, temporal, parietal, and occipital lobe WM between GOSE-defined, functionally non-recovered mTBI patients and their matched controls. Note higher Cho and Cr in the occipital lobe of patients (Mann-Whitney U test, p < 0.05), with moderate-to-large effect sizes (Cohen’s d), as well as visually elevated Cho and Cr in all lobes (i.e., greater means and medians in non-recovered patients compared to controls). Abbreviations: GOSE, Glasgow Outcome Scale – Extended.
Fig. 6.
Metabolite levels in global white matter (WM) in recovered patients vs. all controls. Boxplots displaying 1st, 2nd (median), and 3rd quartiles (box), ±95% (whiskers), and means (●) of Cho, Cr, mI, and NAA distributions within frontal, temporal, parietal, and occipital lobe WM between GOSE-defined, functionally recovered mTBI patients and all controls. Note an absence of statistically significant group differences (wLS ANCOVA, p ≥ 0.05) across all metabolites and all lobes. Abbreviations: wLS ANCOVA, weighted least squares analysis of covariance; GOSE, Glasgow Outcome Scale – Extended.
Comparisons of findings, percent differences, effect sizes, and CVs with those of the original study (Kirov et al., 2013b) are compiled in Supplementary Table 6, and summarized as follows. Kirov et al. (2013b) reported lower global WM NAA in PCS-positive patients, but normal metabolism in PCS-negative patients. The global WM NAA reduction in PCS-positive patients represented a 140% increase in effect size magnitude compared to that from the original whole-group analysis (Kirov et al., 2013a), which included both PCS-positive and PCS-negative patients. This large effect (d = -1.6) corresponded to a larger concentration difference between groups (12.1%). The current study’s comparison between non-recovered patients and controls yielded a 41% increase in effect size for occipital WM Cho (8.6% difference, d = 0.5) and a 40% increase in effect size for occipital WM Cr (6.3% difference, d = 0.9) compared to the whole-group analysis. The original study only reported global NAA CVs, which were lower than the average lobar NAA CVs from the current study (maximum difference across patient and control groups = 5%).
3.2.3. Global analysis: Relationship to elapsed time from injury
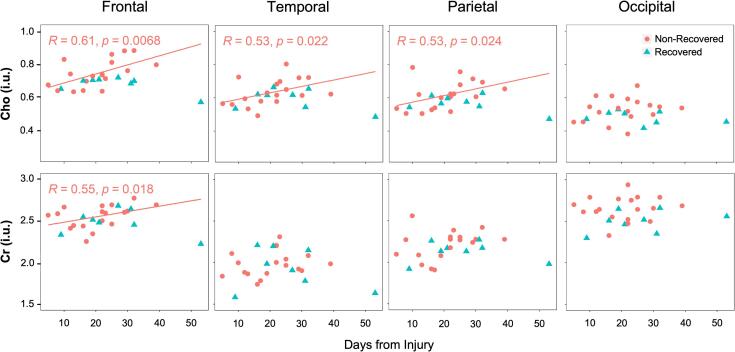
In the entire patient group, Spearman rank and Pearson correlations yielded no statistically significant associations of time from injury with levels of lobar WM Cho and Cr (p > 0.05). Among non-recovered patients, there were statistically significant associations with frontal (Spearman: r = 0.61, p = 0.007; Pearson: r = 0.58, p = 0.011), parietal (Spearman: r = 0.53, p = 0.024), and temporal (Spearman: r = 0.53, p = 0.022) WM Cho, and with frontal (Spearman: r = 0.55, p = 0.018) WM Cr. Among recovered patients, both Spearman rank and Pearson correlations yielded no statistically significant associations of time from injury with levels of lobar WM Cho and Cr (p > 0.05). These post-hoc analyses (not part of H1-H7) are presented in Fig. 7, Supplementary Fig. 4, and Supplementary Table 7, and are discussed in section 4.6.
Fig. 7.
Scatterplots of the time from accrual to image acquisition versus the levels of Cho (top row) and Cr (bottom row) in the frontal, temporal, parietal, and occipital white matter (WM) among GOSE-stratified patients. Results are in pink (●) for the non-recovered patients, and in blue (▲) for the recovered patients. Spearman rank correlation (r), corresponding p value, and the least squares regression line to predict the metabolite level as a function of time from accrual to image acquisition are presented for the statistically significant associations of Cho in the frontal, temporal, and parietal WM, and of Cr in the frontal WM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.4. Regional analysis
Boxplots of the regional WM and GM metabolite distributions, respectively, in all patients (n = 26) and controls (n = 21) are shown in Fig. 8, Fig. 9. All subjects contributed data for all structures, except for three patients and two controls, who did not meet the quality control threshold of ≥10 voxels in the GCC, and thus were excluded from that analysis. No statistically significant group differences were found for any metabolite in any of the six WM and the four deep GM ROIs (MWU: p ≥ 0.05). Comparisons between PCS-positive patients (n = 24) and matched controls (n = 21) also yielded an absence of findings.
Fig. 8.
Metabolite levels in regional white matter (WM) in all patients vs. all controls. Boxplots displaying 1st, 2nd (median), and 3rd quartiles (box), ±95% (whiskers), and means (●) of Cho, Cr, mI, and NAA distributions within segmented WM regions between all mTBI patients and their matched controls. Note an absence of statistically significant group differences (Mann-Whitney U test, p ≥ 0.05) across all metabolites and all regions, yet visually elevated Cho and Cr in all WM regions (i.e., greater means in patients compared to controls). Abbreviations: BCC, body of the corpus callosum; GCC, genu of the corpus callosum; SCC, splenium of the corpus callosum; CorRad, corona radiata; FWM, frontal white matter; PWM, posterior white matter.
Fig. 9.
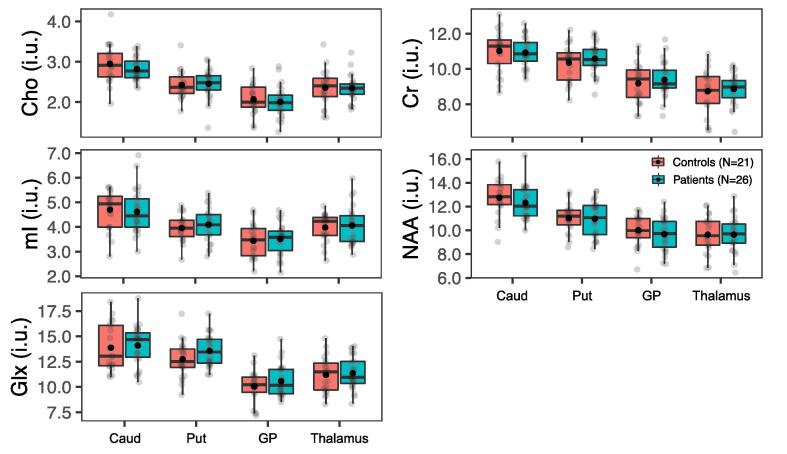
Metabolite levels in deep gray matter (GM) in all patients vs. all controls. Boxplots displaying 1st, 2nd (median), and 3rd quartiles (box), ±95% (whiskers), and means (●) of Cho, Cr, Glx, mI, and NAA distributions within segmented deep GM structures between all mTBI patients and their matched controls. Note an absence of statistically significant group differences (Mann-Whitney U test, p ≥ 0.05) across all metabolites in all regions. Abbreviations: Caud, caudate; Put, putamen; GP, globus pallidus; Thal, thalamus.
Comparisons of findings, percent differences, effect sizes, and CVs with those of the original regional WM (Davitz et al., 2019) and GM (Kierans et al., 2014) studies are compiled in Supplementary Tables 8 and 9, respectively, and summarized as follows. The original regional WM publication (Davitz et al., 2019) reported normal metabolism in PCS-positive patients, which mirrored our current findings. The original regional GM publication (Kierans et al., 2014) reported higher mI in the putamen (21% difference, d = 1.07). The statistically non-significant difference in putaminal mI in the current study was much lower, in both absolute magnitude and effect size (3.5% difference, d = 0.2). The current study more frequently yielded lower CVs compared to the original regional WM study (maximum difference across patients = 12%; controls = 27%) and to the original regional GM study (maximum difference across patients = 24%; controls = 25%).
4. Discussion
The clinical and scientific dissemination of a validated biomarker requires aggregation of disparate studies into a unified prescription for clinical use. The principal challenge, however, is that human studies incorporate many experimental variables, all of which can influence the results. In mTBI, qMR sequences, post-processing methods, sample sizes, types of trauma, patient populations, time from injury, clinical outcome measures, statistical approaches and others vary among studies, and each can influence the apparent relationship between the qMR measurement and the clinical endpoint. An additional confounder is the concern for type I and II errors, the latter of which is exacerbated by the well-known journal bias against negative results (Munafò et al., 2017). Therefore, replication studies are key for addressing the above issues and are essential for either advancing a candidate qMR technique or confirming its shortcomings.
With its high signal-to-noise ratio (SNR) and short acquisition time (e.g., ∼5 min), single-voxel 1H MRS is well-placed for clinical use, but it requires an a priori decision of voxel placement and usually only one region is sampled. Moreover, voxels placed in GM incur WM partial volume effects, which can diminish the sensitivity of the technique to detect GM changes. 1H MRSI can cover multiple brain regions, but has longer acquisition times (e.g., ∼20 min), more complex post-processing, and regional metabolism is observed with worse reproducibility than with single-voxel MRS, because of the smaller voxels’ lower SNR (Zhang et al., 2018). An advantage of 1H MRSI is its ability to assess global metabolism via linear regression (Tal et al., 2012, Hoch et al., 2017) or spectral decomposition (Goryawala et al., 2018) over many voxels. Such application can deliver reproducibility on par with that of single-voxel 1H MRS (Zhang et al., 2018), with the added benefit of negligible partial volume effects (Tal et al., 2012). Given these different strengths and weaknesses, the choice of acquisition and post-processing approaches for the evaluation of mTBI depends on how injury is distributed within the brain, at what magnitude, and how it relates to clinical metrics. Our previous 1H MRSI work, published in four publications from 2012 to 2019, has provided answers to these questions, but their generalizability remains unknown. Therefore, the goal of the current project was to test whether the results replicate in a new cohort.
Each original study is described in its own section below (4.1 to 4.4) under a heading which denotes the type of post-processing (global or regional) and target tissue (GM, WM) analysis done in the respective work. Each of these sections is structured identically as follows: the first paragraph states the scientific question posed in the previous published work and its findings; the second paragraph compares them to those of the replication study; the third paragraph contains a short discussion of the wider MRS literature on the topic.
4.1. Global GM and WM
The objective of our first publication was “to test if DAI is quantifiable with 1H MRSI” (Kirov et al., 2013a). To specifically capture the diffuse component of DAI, we applied a recently developed (at the time) linear regression approach for assessment of global injury with 1H MRSI. It yielded tissue-specific concentrations of Cho, Cr, mI, and NAA over the entire 360 cm3 1H MRSI volume-of-interest (VOI). We found lower NAA in mTBI patients in WM, but we did not detect any differences between mTBI patients and controls for any metabolite in GM. The findings were described as diffuse injury (H1) only in WM (H2), and affecting axons, not glia (i.e., lower NAA, but normal Cho, Cr, and mI; H3).
Each linear regression result from the current study is derived from a single bilateral lobe, i.e., it represents the average metabolite level within a large brain volume (lobes range from ∼100 cm3 to ∼400 cm3) (Brain Development Cooperative Group, 2012) which is on the scale of the volume interrogated in the original study (∼340 cm3, after accounting for ∼6% CSF content in the VOI (Kirov et al., 2009). Therefore, the Cho and Cr elevations in the occipital lobe WM of patients constitute a replication of H1 and H2 and a failure to support H3. The large brain coverage of the replication study enabled an examination of the scale of this injury. Given that only the occipital lobe showed a statistically significant finding, it can be concluded that diffuse injury does not extend elsewhere. As seen in Fig. 4 and Supplementary Table 4, however, the mean WM Cr and Cho values of patients were above those of the controls in every lobe. For Cr, the magnitude of the statistically non-significant differences in the parietal and frontal lobes were only ∼0.6 % smaller than the statistically significant result in the occipital lobe. It could therefore be conjectured that diffuse injury extends to other lobes, but its slightly lower magnitude and/or worse CVs there rendered it undetectable.
Prior 1H MRS literature (Gasparovic et al., 2009, Govind et al., 2010, Yeo et al., 2011) also support the notion of diffuse injury distribution after mTBI. Its manifestations, however, may differ across cohorts: while there were no cross-sectional differences, Mayer et al. (2015) observed a longitudinal decrease of global WM NAA in athletes at high risk for repetitive mTBI. There is also evidence for diffuse metabolic changes in moderate-to-severe TBIs (Signoretti et al., 2008, Lin et al., 2022), including in children (Babikian et al., 2018, Holshouser et al., 2019).
4.2. Global GM and WM: Relationship to clinical outcome
Linear regression for obtaining global GM and WM has not only technical advantages (minimal partial volume effects, high sensitivity), but is also practical for clinical applications because it obviates the need to (subjectively) choose a sampling region, as all voxels within an 1H MRSI acquisition can be used for the global concentrations. Therefore, to probe its potential as a biomarker, the second study’s objective was “to test whether the previously identified WM NAA decline correlated with patients’ PCS” (Kirov et al., 2013b). For that purpose, the patient cohort was stratified into two groups, PCS-positive and PCS-negative, and global metabolite levels in each were compared to those in matched controls. While there were no differences between PCS-negative patients and controls, WM NAA was lower in the PCS-positive patients. The findings were described as global WM NAA correlating with PCS (H4).
The current study used the GOSE to test whether findings from the whole-group analysis correlated with patients’ functional outcome. Thus, global metabolite levels in each subgroup were compared to those in matched controls, and the results mirrored those from the previous study in two notable ways. First, stratification showed that the whole-group findings were only reflected in the impaired cohort (i.e., only non-recovered patients vs. controls, as shown in Fig. 5, exhibited a statistically significant group difference). Second, we observed larger effect sizes for the elevated occipital WM Cho and Cr levels in non-recovered patients vs. controls (Supplementary Table 6), compared to those from the whole-group analysis (Supplementary Table 4). While we could not perform a comparison for the two PCS-negative patients, we did examine PCS-positive patients vs. controls, and as expected, found elevated occipital WM Cho (d = 0.4) and Cr (d = 0.7) in PCS-positive patients. Taken together, this replication study supported H4 with regards to global WM abnormalities correlating with worse outcome.
While relatively few 1H MRS studies in adult mTBI have examined the associations between metabolite levels and clinical outcomes, those that have done so often report statistically significant correlations between reduced NAA or NAA/Cr and greater disability (Ross et al., 1998, Garnett et al., 2000), worse global function (Garnett et al., 2000, Friedman et al., 1999), and worse neuropsychological performance (Govind et al., 2010, Friedman et al., 1999). Of note, George et al. (2014) found a direct correlation between Cr concentration and cognitive ability in the centrum semiovale, in mTBI patients examined at the early subacute stage of injury. Lack of further reports on Cho and Cr as mediators of clinical outcome may be attributed to the relative absence of Cho and Cr alterations in mTBI (Vagnozzi et al., 2010), which itself may be due in part to the widespread use of Cr levels as a reference for metabolite quantification.
4.3. Regional WM
The two original publications described above were followed by a regional analysis to determine if “1) WM damage is homogeneously diffuse, or if specific regions are more affected; and 2) partial-volume-corrected, structure-specific 1H MRSI voxel averaging is sensitive to regional WM metabolic abnormalities” (Davitz et al., 2019). These questions were to be answered by comparing the results and effect sizes across six WM regions to those obtained with global linear regression. Surprisingly, the regional WM analysis yielded no statistically significant differences (H5). Patients’ mean and median NAA, however, were lower than controls’ in every WM region, with effect sizes of similar magnitude. Given the statistically significant result of lower global WM NAA from the linear regression (Kirov et al., 2013a), we concluded that WM damage was homogenously distributed, but 1H MRSI regional averaging lacked the sensitivity to detect it on a regional basis.
The current replication study supported H5 (no statistically significant differences for any metabolite in any region). It also supported the assertion of insufficient sensitivity of regional 1H MRSI, given the lack of finding within the PWM vs. the statistically significant effect for the whole occipital lobe. Intriguingly, patients’ mean Cho and Cr were higher than controls’ in every WM region, mirroring the previous NAA observations. In contrast, however, their effect sizes and percent differences were not of similar magnitude across the different regions. Such observation may point to inhomogeneously distributed injury, i.e., higher Cho and Cr within all WM, but to a different extent in different regions.
There is a paucity of data on multiple WM ROIs examined in the same session, likely due to the lower number of 1H MRSI studies in mTBI compared to single-voxel. One rare study found unidirectionally lower NAA/Cho in patients compared to controls in all of the 12 studied regions, but only in 5 the difference was statistically significant (Govindaraju et al., 2004). In more severe TBI, both in adults (Signoretti et al., 2008) and children (Holshouser et al., 2019); 1H MRSI has yielded statistically significant metabolic alterations in much larger percentage (or even all, depending on severity) of regionally defined ROIs. It is therefore plausible that the nature of 1H MRS-detectable WM injury in TBI is truly diffuse, but its small magnitude in mTBI can often render it undetectable.
4.4. Regional GM
The global analysis grouped together cortical and deep GM, and therefore averaged out any differences between and within these two GM types. The last original study focused on the latter, with the purpose “to obtain quantitative neurometabolite measurements, specifically mI and Glx […] and compare these measurements against normal healthy controls” (Kierans et al., 2014). There were no statistically significant differences in any metabolite for the caudate, thalamus and globus pallidus and for Cho, Cr, Glx, and NAA in the putamen (H6). In the latter structure, however, mTBI patients showed higher mI (H7).
In the replication dataset, the negative results were replicated (supporting H6), while the positive one was not, as no differences were found for any metabolite in any structure (not supporting H7). We note that the original study also used ratios to Cr as a proxy for changes in the level of the metabolite in the numerator, assuming that Cr levels are stable across mTBI patients and controls. We opted against repeating this strategy, considering the accumulated evidence of altered Cr levels in mTBI in animal models (Signoretti et al., 2010) and human subjects (Gasparovic et al., 2009, Yeo et al., 2011, Vagnozzi et al., 2010). We note that the current replication study provides more support against the concept of stable Cr in TBI.
To our knowledge, there has been only one other study of the putamen in mTBI, and it also yielded negative findings (George et al., 2014). There has been more interest in the thalamus, with some groups reporting changes (George et al., 2014, Sours et al., 2015), and others not (Widerström-Noga et al., 2016). In early work from our laboratory using long-TE 1H MRSI in mostly chronic mTBI, we did not find thalamic findings (Kirov et al., 2007). We note more evidence of thalamic involvement in pediatric TBI (Holshouser et al., 2019) and severe TBI (Uzan et al., 2003, Tollard et al., 2009).
4.5. Key findings and clinical implications
The main conclusions from the four original publications and the current replication study are that, in mTBI cohorts with similar characteristics, 1H MRSI-identified injury: (i) has a diffuse WM distribution, which (ii) can differ in the type of altered metabolism; and which (iii) is present only in patients with unfavorable clinical outcome. Because the magnitude of this injury is approximately at or below 6% (6.7% from the previous study, ≤5.9% from the current study), we highlight the importance of high sensitivity 1H MRS approaches. Quantification using metabolite ratios should be made with caution, given the different metabolic injury profiles in each patient cohort, including higher Cr.
Guidance on acquisition strategies can also be extrapolated. Insights original to this study suggest that, in generalizing about the entire cerebrum, this type of injury can be described as inhomogeneously diffuse: unidirectional metabolic changes in all or most of WM, but with varying local profiles. The evidence suggests that this may be true on both regional (few cm3) and global (hundreds cm3) scales, but with larger heterogeneity on the smaller scale. This is in line with the expectation that focal injury will vary in accordance with the unique biomechanical consequences of each mTBI event. While such injury may not be generalizable, over large brain regions, focal differences would average out, decreasing individual heterogeneity. In such scenario, large single voxels, or linear regression from 1H MRSI over large brain areas would be well-suited to detect mTBI injury. Our previous findings of homogeneous diffuse injury dictated a similar recommendation, highlighting the use of single voxel anywhere in the WM as the most straightforward approach. Taken together, all our studies can be used to motivate a slightly modified version, emphasizing on the large voxel volume and a suggestion for placement in another WM area to account for possible differences in injury magnitude.
4.6. Limitations
An important aspect of a clinical recommendation is the timing of the 1H MRS exam, given that metabolite levels have been reported to change as a function of elapsed time from injury (Joyce et al., 2022). A cross-sectional study is unable to meaningfully contribute to this topic, but given the need to augment the above guidance with a timing recommendation, we conducted an exploratory post-hoc analysis (not part of H1-H7) which can guide future serial studies. The results suggest that the lack of a statistically significant correlation between time from injury and lobar WM Cr and Cho levels in all patients was at least partially due to the fact that the correlations were exclusively positive among non-recovered patients, but were primarily negative among recovered patients (Supplementary Table 7). Hence, the data suggest that time from injury had opposite effects on the metabolite levels within the two groups, with longer time associated with higher lobar WM Cr and Cho levels among non-recovered patients, and with lower levels of these metabolites among recovered patients (Fig. 7). This can be interpreted as ongoing, increasing metabolic abnormalities only in the impaired cohort. Two of our previous publications also explored this topic, but did not find a relationship between lower global WM NAA and time from injury, either in the whole (Kirov et al., 2013a), or in the PCS-stratified (Kirov et al., 2013b) cohorts. Interestingly, however, longer time from injury was associated with higher global WM Cr and Cho, but in that study these metabolites were not altered in patients (Kirov et al., 2013a).
By replicating the negative findings in global and deep GM, and the lack of replication of the sole previous GM finding, we did not find any evidence of GM injury. Though in line with DAI being the predominant pathological mechanism, we note that our findings do not negate the presence of focal cortical GM injury in either of the two cohorts. Cortical GM was only studied with global linear regression, an analysis which is not sensitive to changes in a small number of voxels (Tal et al., 2012).
The relatively small sample sizes of both studies dictate that the hypotheses should be further evaluated in larger samples and at different sites. Such studies will benefit from employing 1H MRSI and multiple WM single voxels within the same session in order to draw more robust conclusions about clinical applicability. These future studies should also incorporate a more diverse set of clinical outcome measures.
5. Conclusions
A single-site conceptual replication study replicated evidence of injury which is diffuse, confined to WM, and which correlates with clinical outcome. Unlike the original studies, however, this injury was found to be of glial (Cr, Cho), and not of neuronal (NAA) origin. These results, and the low magnitude of injury found in all studies, suggest that the choice of WM region is secondary to the need for high sensitivity 1H MRS approaches; and that caution should be exercised when performing quantification using metabolite ratios, especially to Cr. It can be further extrapolated that clinic-friendly and high signal-to-noise ratio single-voxels placed anywhere in WM may provide the simplest 1H MRS biomarker for mTBI. Such conclusion has implications for clinical applications and should be further tested in larger sample sizes and in cohorts with different characteristics.
6. Additional Information
We thank all participants for taking part in this study.
Funding: This work was supported by the United States’ National Institutes of Health (NIH) Grant No R01NS097494. GM acknowledges support from grant number R01EB026456, and all authors acknowledge the support of the Center for Advanced Imaging Innovation and Research (CAI2R) at NYU Langone Health under grant number P41EB017183. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, which had no role in study design; in the collection, analysis and interpretation; the writing of the report; and decision to submit the article for publication.
Informed consent: All subjects provided written informed consent prior to their examination.
CRediT authorship contribution statement
Anna M. Chen: Investigation, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. Teresa Gerhalter: Investigation, Data curation, Writing – review & editing. Seena Dehkharghani: Writing – review & editing. Rosemary Peralta: Project administration, Writing – review & editing. Mia Gajdošík: Project administration, Writing – review & editing. Martin Gajdošík: Investigation, Methodology, Software, Writing – review & editing. Mickael Tordjman: Investigation, Writing – review & editing. Julia Zabludovsky: Investigation, Writing – review & editing. Sulaiman Sheriff: Software, Resources, Writing – review & editing. Sinyeob Ahn: Software, Resources, Writing – review & editing. James S. Babb: Formal analysis, Methodology, Writing – review & editing. Tamara Bushnik: Writing – review & editing. Alejandro Zarate: Writing – review & editing. Jonathan M. Silver: Writing – review & editing. Brian S. Im: Writing – review & editing. Stephen P. Wall: Writing – review & editing. Guillaume Madelin: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing. Ivan I. Kirov: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2023.103325.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abramson R.G., et al. Methods and challenges in quantitative imaging biomarker development. Acad. Radiol.. 2015;22:25–32. doi: 10.1016/j.acra.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Congress of Rehabilitation Medicine, 1993. Definition of mild traumatic brain injury. Journal of Head Trauma Rehabilitation 8, 86-87. https://www.acrm.org/wp-content/uploads/pdf/TBIDef_English_10-10.pdf.
- Babikian T., et al. Whole Brain Magnetic Resonance Spectroscopic Determinants of Functional Outcomes in Pediatric Moderate/Severe Traumatic Brain Injury. J. Neurotrauma. 2018;35:1637–1645. doi: 10.1089/neu.2017.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. 1,500 scientists lift the lid on reproducibility. Nature. 2016;533:452–454. doi: 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- Barker P.B., et al. Quantitation of proton NMR spectra of the human brain using tissue water as an internal concentration reference. NMR Biomed.. 1993;6:89–94. doi: 10.1002/nbm.1940060114. [DOI] [PubMed] [Google Scholar]
- Bartnik-Olson B.L., et al. The clinical utility of proton magnetic resonance spectroscopy in traumatic brain injury: recommendations from the ENIGMA MRS working group. Brain Imaging Behav.. 2021;15:504–525. doi: 10.1007/s11682-020-00330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigler E.D., Maxwell W.L. Neuropathology of mild traumatic brain injury: relationship to neuroimaging findings. Brain Imaging Behav.. 2012;6:108–136. doi: 10.1007/s11682-011-9145-0. [DOI] [PubMed] [Google Scholar]
- Borg J., et al. Diagnostic procedures in mild traumatic brain injury: results of the WHO Collaborating Centre Task Force on Mild Traumatic Brain Injury. J. Rehabil. Med. 2004;61–75 doi: 10.1080/16501960410023822. [DOI] [PubMed] [Google Scholar]
- Brain Development Cooperative, G. Total and regional brain volumes in a population-based normative sample from 4 to 18 years: the NIH MRI Study of Normal Brain Development. Cereb Cortex 22, 1-12 (2012). doi:10.1093/cercor/bhr018. [DOI] [PMC free article] [PubMed]
- Browne K.D., Chen X.H., Meaney D.F., Smith D.H. Mild traumatic brain injury and diffuse axonal injury in swine. J. Neurotrauma. 2011;28:1747–1755. doi: 10.1089/neu.2011.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil K.M., et al. Proton magnetic resonance spectroscopy for detection of axonal injury in the splenium of the corpus callosum of brain-injured patients. J. Neurosurg. 1998;88:795–801. doi: 10.3171/jns.1998.88.5.0795. [DOI] [PubMed] [Google Scholar]
- Cecil K.M., Lenkinski R.E., Meaney D.F., McIntosh T.K., Smith D.H. High-field proton magnetic resonance spectroscopy of a swine model for axonal injury. J. Neurochem. 1998;70:2038–2044. doi: 10.1046/j.1471-4159.1998.70052038.x. [DOI] [PubMed] [Google Scholar]
- Cole T.J., Altman D.G. Statistics Notes: What is a percentage difference? BMJ. 2017;358 doi: 10.1136/bmj.j3663. [DOI] [PubMed] [Google Scholar]
- Crandall C.S., Sherman J.W. On the scientific superiority of conceptual replications for scientific progress. J. Exp. Soc. Psychol. 2016;66:93–99. doi: 10.1016/j.jesp.2015.10.002. [DOI] [Google Scholar]
- Dadas A., Washington J., Diaz-Arrastia R., Janigro D. Biomarkers in traumatic brain injury (TBI): a review. Neuropsychiatr. Dis. Treat. 2018;14:2989–3000. doi: 10.2147/NDT.S125620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davitz M.S., et al. Quantitative multivoxel proton MR spectroscopy for the identification of white matter abnormalities in mild traumatic brain injury: Comparison between regional and global analysis. J. Magn. Reson. Imaging. 2019;50:1424–1432. doi: 10.1002/jmri.26718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deSouza N.M., et al. Validated imaging biomarkers as decision-making tools in clinical trials and routine practice: current status and recommendations from the EIBALL* subcommittee of the European Society of Radiology (ESR) Insights Imaging. 2019;10:1–16. doi: 10.1186/s13244-019-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan M.C., et al. Estimating the global incidence of traumatic brain injury. J. Neurosurg. 2018;1–18 doi: 10.3171/2017.10.JNS17352. [DOI] [PubMed] [Google Scholar]
- Ding X.Q., et al. Reproducibility and reliability of short-TE whole-brain MR spectroscopic imaging of human brain at 3T. Magn. Reson. Med. 2015;73:921–928. doi: 10.1002/mrm.25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.Q., et al. Physiological neuronal decline in healthy aging human brain - An in vivo study with MRI and short echo-time whole-brain (1)H MR spectroscopic imaging. Neuroimage. 2016;137:45–51. doi: 10.1016/j.neuroimage.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.D., et al. Quantitative proton MRS predicts outcome after traumatic brain injury. Neurology. 1999;52 doi: 10.1212/wnl.52.7.1384. [DOI] [PubMed] [Google Scholar]
- Garcia-Panach J., et al. A voxel-based analysis of FDG-PET in traumatic brain injury: regional metabolism and relationship between the thalamus and cortical areas. J. Neurotrauma. 2011;28:1707–1717. doi: 10.1089/neu.2011.1851. [DOI] [PubMed] [Google Scholar]
- Garnett M.R., et al. Early proton magnetic resonance spectroscopy in normal-appearing brain correlates with outcome in patients following traumatic brain injury. Brain. 2000;123(Pt 10):2046–2054. doi: 10.1093/brain/123.10.2046. [DOI] [PubMed] [Google Scholar]
- Gasparovic C., et al. Neurometabolite concentrations in gray and white matter in mild traumatic brain injury: an 1H-magnetic resonance spectroscopy study. J. Neurotrauma. 2009;26:1635–1643. doi: 10.1089/neu.2009-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., et al. Assessment of thalamic perfusion in patients with mild traumatic brain injury by true FISP arterial spin labelling MR imaging at 3T. Brain Inj. 2009;23:666–674. doi: 10.1080/02699050903014899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E.O., et al. Longitudinal and Prognostic Evaluation of Mild Traumatic Brain Injury: A 1H-Magnetic Resonance Spectroscopy Study. J. Neurotrauma. 2014;31:1018–1028. doi: 10.1089/neu.2013.3224. [DOI] [PubMed] [Google Scholar]
- Gerhalter T., et al. Global decrease in brain sodium concentration after mild traumatic brain injury. Brain Commun. 2021;3 doi: 10.1093/braincomms/fcab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryawala M.Z., Sheriff S., Maudsley A.A. Regional distributions of brain glutamate and glutamine in normal subjects. NMR Biomed. 2016;29:1108–1116. doi: 10.1002/nbm.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryawala M.Z., Sheriff S., Stoyanova R., Maudsley A.A. Spectral decomposition for resolving partial volume effects in MRSI. Magn. Reson. Med. 2018;79:2886–2895. doi: 10.1002/mrm.26991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind V., et al. Whole-brain proton MR spectroscopic imaging of mild-to-moderate traumatic brain injury and correlation with neuropsychological deficits. J. Neurotrauma. 2010;27:483–496. doi: 10.1089/neu.2009.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraju V., et al. Volumetric proton spectroscopic imaging of mild traumatic brain injury. AJNR Am. J. Neuroradiol. 2004;25:730–737. [PMC free article] [PubMed] [Google Scholar]
- Hedges E.P., et al. Reliability of structural MRI measurements: The effects of scan session, head tilt, inter-scan interval, acquisition sequence, FreeSurfer version and processing stream. Neuroimage. 2022;246 doi: 10.1016/j.neuroimage.2021.118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks R., et al. Progress in developing common data elements for traumatic brain injury research: version two–the end of the beginning. J. Neurotrauma. 2013;30:1852–1861. doi: 10.1089/neu.2013.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S.E., Kirov I.I., Tal A. When are metabolic ratios superior to absolute quantification? A statistical analysis. NMR Biomed. 2017;30 doi: 10.1002/nbm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshouser B., et al. Longitudinal Metabolite Changes after Traumatic Brain Injury: A Prospective Pediatric Magnetic Resonance Spectroscopic Imaging Study. J. Neurotrauma. 2019;36:1352–1360. doi: 10.1089/neu.2018.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulkower M.B., Poliak D.B., Rosenbaum S.B., Zimmerman M.E., Lipton M.L. A decade of DTI in traumatic brain injury: 10 years and 100 articles later. AJNR Am. J. Neuroradiol. 2013;34:2064–2074. doi: 10.3174/ajnr.A3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscan Z., et al. Test-retest reliability of freesurfer measurements within and between sites: Effects of visual approval process. Hum. Brain Mapp. 2015;36:3472–3485. doi: 10.1002/hbm.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce J.M., La P.L., Walker R., Harris A. Magnetic resonance spectroscopy of traumatic brain injury and subconcussive hits: A systematic review and meta-analysis. J. Neurotrauma. 2022 doi: 10.1089/neu.2022.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl K.G., et al. Altered neurometabolism in major depressive disorder: A whole brain (1)H-magnetic resonance spectroscopic imaging study at 3T. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2020;101 doi: 10.1016/j.pnpbp.2020.109916. [DOI] [PubMed] [Google Scholar]
- Kay, T. H., D. E.; Adams, R., 1993. Definition of mild traumatic brain injury. J Head Trauma Rehabil 8, 86-87.
- Keller S.S., et al. Volume estimation of the thalamus using freesurfer and stereology: consistency between methods. Neuroinformatics. 2012;10:341–350. doi: 10.1007/s12021-012-9147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierans A.S., et al. Myoinositol and glutamate complex neurometabolite abnormality after mild traumatic brain injury. Neurology. 2014;82:521–528. doi: 10.1212/WNL.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov I., et al. Characterizing 'mild' in traumatic brain injury with proton MR spectroscopy in the thalamus: Initial findings. Brain Inj. 2007;21:1147–1154. doi: 10.1080/02699050701630383. [DOI] [PubMed] [Google Scholar]
- Kirov I.I., et al. MR spectroscopy indicates diffuse multiple sclerosis activity during remission. J. Neurol. Neurosurg. Psychiatry. 2009;80:1330–1336. doi: 10.1136/jnnp.2009.176263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov I., et al. Diffuse axonal injury in mild traumatic brain injury: a 3D multivoxel proton MR spectroscopy study. J. Neurol. 2013;260:242–252. doi: 10.1007/s00415-012-6626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov I., et al. Proton MR spectroscopy correlates diffuse axonal abnormalities with post-concussive symptoms in mild traumatic brain injury. J. Neurotrauma. 2013;30:1200–1204. doi: 10.1089/neu.2012.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klietz M., et al. Altered Neurometabolic Profile in Early Parkinson's Disease: A Study With Short Echo-Time Whole Brain MR Spectroscopic Imaging. Front. Neurol. 2019;10:777. doi: 10.3389/fneur.2019.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis R. Issues of spectral quality in clinical 1H-magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17:361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- Lecocq A., et al. Whole-brain quantitative mapping of metabolites using short echo three-dimensional proton MRSI. J. Magn. Reson. Imaging. 2015;42:280–289. doi: 10.1002/jmri.24809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.P., et al. Metabolic imaging of mild traumatic brain injury. Brain Imaging Behav. 2012;6:208–223. doi: 10.1007/s11682-012-9181-4. [DOI] [PubMed] [Google Scholar]
- Lin J.C., et al. Investigating whole-brain metabolite abnormalities in the chronic stages of moderate or severe traumatic brain injury. PM R. 2022;14:472–485. doi: 10.1002/pmrj.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., et al. Routine clinical brain MRI sequences for use at 3.0 Tesla. J. Magn. Reson. Imaging. 2005;22:13–22. doi: 10.1002/jmri.20356. [DOI] [PubMed] [Google Scholar]
- Madhok D.Y., et al. Clinical Predictors of 3- and 6-Month Outcome for Mild Traumatic Brain Injury Patients with a Negative Head CT Scan in the Emergency Department: A TRACK-TBI Pilot Study. Brain Sci. 2020;10 doi: 10.3390/brainsci10050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghsudi H., et al. Regional Metabolite Concentrations in Aging Human Brain: Comparison of Short-TE Whole Brain MR Spectroscopic Imaging and Single Voxel Spectroscopy at 3T. Clin. Neuroradiol. 2020;30:251–261. doi: 10.1007/s00062-018-00757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghsudi H., et al. Age-related Brain Metabolic Changes up to Seventh Decade in Healthy Humans : Whole-brain Magnetic Resonance Spectroscopic Imaging Study. Clin. Neuroradiol. 2020;30:581–589. doi: 10.1007/s00062-019-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikis G.C., Papanikolaou N., Matos C. Springer; 2017. Imaging Biomarkers 115–122. [DOI] [Google Scholar]
- Marino S., Ciurleo R., Bramanti P., Federico A., De Stefano N. 1H-MR spectroscopy in traumatic brain injury. Neurocrit. Care. 2011;14:127–133. doi: 10.1007/s12028-010-9406-6. [DOI] [PubMed] [Google Scholar]
- Maudsley A.A., et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley A.A., et al. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn. Reson. Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley A.A., Domenig C., Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR Biomed. 2010;23:251–256. doi: 10.1002/nbm.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A.R., et al. A Longitudinal Assessment of Structural and Chemical Alterations in Mixed Martial Arts Fighters. J. Neurotrauma. 2015;32:1759–1767. doi: 10.1089/neu.2014.3833. [DOI] [PubMed] [Google Scholar]
- McKee A.C., Daneshvar D.H. The neuropathology of traumatic brain injury. Handb. Clin. Neurol. 2015;127:45–66. doi: 10.1016/B978-0-444-52892-6.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzon B., et al. Repetitive mild traumatic brain injury in a mouse model produces learning and memory deficits accompanied by histological changes. J. Neurotrauma. 2012;29:2761–2773. doi: 10.1089/neu.2012.2498. [DOI] [PubMed] [Google Scholar]
- Munafò M.R., et al. A manifesto for reproducible science. Nat. Hum. Behav. 2017;1:0021. doi: 10.1038/s41562-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering & Medicine., 2019. Reproducibility and replicability in science. National Academies Press. [PubMed]
- Neeb H., Zilles K., Shah N.J. Fully-automated detection of cerebral water content changes: study of age- and gender-related H2O patterns with quantitative MRI. Neuroimage. 2006;29:910–922. doi: 10.1016/j.neuroimage.2005.08.062. [DOI] [PubMed] [Google Scholar]
- Nelson L.D., et al. Validating Multidimensional Outcome Assessment Using the TBI Common Data Elements: An Analysis of the TRACK-TBI Pilot Sample. J. Neurotrauma. 2017 doi: 10.1089/neu.2017.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek B.A., Errington T.M. Making sense of replications. Elife. 2017;6 doi: 10.7554/eLife.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permenter, C. M., Fernandez-de Thomas, R. J. & Sherman, A., 2021. in: Postconcussive Syndrome. [PubMed]
- Poldrack R.A., et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18:115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince C., Bruhns M.E. Evaluation and Treatment of Mild Traumatic Brain Injury: The Role of Neuropsychology. Brain Sci. 2017;7 doi: 10.3390/brainsci7080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunig D.L., et al. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat. Methods Med. Res. 2015;24:27–67. doi: 10.1177/0962280214537344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replicating scientific results is tough - but essential. Nature 600, 359-360 (2021). doi:10.1038/d41586-021-03736-4. [DOI] [PubMed]
- Ross B.D., et al. 1H MRS in acute traumatic brain injury. J. Magn. Reson. Imaging. 1998;8:829–840. doi: 10.1002/jmri.1880080412. [DOI] [PubMed] [Google Scholar]
- Rusinek H., et al. Fully automatic segmentation of white matter lesions: error analysis and validation of a new tool. Int. J. Comput. Assist. Radiol. Surg. 2013;8:289–291. [Google Scholar]
- Sabati M., et al. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn. Reson. Med. 2015;74:1209–1220. doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Scott G., Leech R. Network dysfunction after traumatic brain injury. Nat. Rev. Neurol. 2014;10:156–166. doi: 10.1038/nrneurol.2014.15. [DOI] [PubMed] [Google Scholar]
- Signoretti S., et al. Assessment of mitochondrial impairment in traumatic brain injury using high-resolution proton magnetic resonance spectroscopy. J. Neurosurg. 2008;108:42–52. doi: 10.3171/JNS/2008/108/01/0042. [DOI] [PubMed] [Google Scholar]
- Signoretti S., et al. Transient alterations of creatine, creatine phosphate, N-acetylaspartate and high-energy phosphates after mild traumatic brain injury in the rat. Mol. Cell. Biochem. 2010;333:269–277. doi: 10.1007/s11010-009-0228-9. [DOI] [PubMed] [Google Scholar]
- Soher B.J., Young K., Govindaraju V., Maudsley A.A. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn. Reson. Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- Soher B.J., Clarke W.T., Wilson M., Near J., Oeltzschner G. Community-Organized Resources for Reproducible MRS Data Analysis. Magn. Reson. Med. 2022 doi: 10.1002/mrm.29387. [DOI] [PubMed] [Google Scholar]
- Sours C., George E.O., Zhuo J., Roys S., Gullapalli R.P. Hyper-connectivity of the thalamus during early stages following mild traumatic brain injury. Brain Imaging Behav. 2015;9:550–563. doi: 10.1007/s11682-015-9424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal A., Kirov I.I., Grossman R.I., Gonen O. The role of gray and white matter segmentation in quantitative proton MR spectroscopic imaging. NMR Biomed. 2012;25:1392–1400. doi: 10.1002/nbm.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollard E., et al. Experience of diffusion tensor imaging and 1H spectroscopy for outcome prediction in severe traumatic brain injury: Preliminary results*. Crit. Care Med. 2009;37:1448–1455. doi: 10.1097/CCM.0b013e31819cf050. [DOI] [PubMed] [Google Scholar]
- Uzan M., et al. Thalamic proton magnetic resonance spectroscopy in vegetative state induced by traumatic brain injury. J. Neurol. Neurosurg. Psychiatry. 2003;74:33–38. doi: 10.1136/jnnp.74.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi R., et al. Assessment of metabolic brain damage and recovery following mild traumatic brain injury: a multicentre, proton magnetic resonance spectroscopic study in concussed patients. Brain. 2010;133:3232–3242. doi: 10.1093/brain/awq200. [DOI] [PubMed] [Google Scholar]
- Verma G., et al. Three-dimensional echo planar spectroscopic imaging for differentiation of true progression from pseudoprogression in patients with glioblastoma. NMR Biomed. 2019;32:e4042. doi: 10.1002/nbm.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voormolen D.C., et al. Divergent Classification Methods of Post-Concussion Syndrome after Mild Traumatic Brain Injury: Prevalence Rates, Risk Factors, and Functional Outcome. J. Neurotrauma. 2018;35:1233–1241. doi: 10.1089/neu.2017.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansapura J.P., Holland S.K., Dunn R.S., Ball W.S., Jr. NMR relaxation times in the human brain at 3.0 tesla. J. Magn. Reson. Imaging. 1999;9:531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Weingärtner S., et al. Development, validation, qualification, and dissemination of quantitative MR methods: Overview and recommendations by the ISMRM quantitative MR study group. Magn. Reson. Med. 2021 doi: 10.1002/mrm.29084. [DOI] [PubMed] [Google Scholar]
- Widerström-Noga E., Govind V., Adcock J.P., Levin B.E., Maudsley A.A. Subacute Pain after Traumatic Brain Injury Is Associated with Lower Insular N-Acetylaspartate Concentrations. J. Neurotrauma. 2016;33:1380–1389. doi: 10.1089/neu.2015.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde E.A., et al. Recommendations for the use of common outcome measures in traumatic brain injury research. Arch. Phys. Med. Rehabil. 2010;91:1650–1660 e1617. doi: 10.1016/j.apmr.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Wilson J.T., Pettigrew L.E., Teasdale G.M. Structured interviews for the Glasgow Outcome Scale and the extended Glasgow Outcome Scale: guidelines for their use. J. Neurotrauma. 1998;15:573–585. doi: 10.1089/neu.1998.15.573. [DOI] [PubMed] [Google Scholar]
- Yeo R.A., et al. A longitudinal proton magnetic resonance spectroscopy study of mild traumatic brain injury. J. Neurotrauma. 2011;28:1–11. doi: 10.1089/neu.2010.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh E.L., et al. Pathological Computed Tomography Features Associated With Adverse Outcomes After Mild Traumatic Brain Injury: A TRACK-TBI Study With External Validation in CENTER-TBI. JAMA Neurol. 2021;78:1137–1148. doi: 10.1001/jamaneurol.2021.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., et al. Comparison of reproducibility of single voxel spectroscopy and whole-brain magnetic resonance spectroscopy imaging at 3T. NMR Biomed. 2018;31:e3898. doi: 10.1002/nbm.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., et al. Reproducibility of whole-brain temperature mapping and metabolite quantification using proton magnetic resonance spectroscopy. NMR Biomed. 2020;33:e4313. doi: 10.1002/nbm.4313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.