Abstract
Integral membrane proteins are found in all cellular membranes and carry out many of the functions that are essential to life. The membrane-embedded domains of integral membrane proteins are structurally quite simple, allowing the use of various prediction methods and biochemical methods to obtain structural information about membrane proteins. A critical step in the biosynthetic pathway leading to the folded protein in the membrane is its insertion into the lipid bilayer. Understanding of the fundamentals of the insertion and folding processes will significantly improve the methods used to predict the three-dimensional membrane protein structure from the amino acid sequence. In the first part of this review, biochemical approaches to elucidate membrane protein topology are reviewed and evaluated, and in the second part, the use of similar techniques to study membrane protein insertion is discussed. The latter studies search for signals in the polypeptide chain that direct the insertion process. Knowledge of the topogenic signals in the nascent chain of a membrane protein is essential for the evaluation of membrane topology studies.
Integral membrane proteins represent an important class of proteins that are involved in a wide variety of cellular functions. Knowledge of the structure of proteins is crucial to understanding their function. Unfortunately, there are no general and reliable methods for forming three-dimensional crystals of membrane proteins suitable for crystallographic analysis, and to date, only a handful of high-resolution membrane protein structures have been solved whereas several thousands of three-dimensional structures of globular proteins are known. Because of this, biochemical and prediction methods were needed to obtain structural information about membrane proteins.
Although integral membrane proteins come in a variety of sizes and shapes, they have common basic architectural principles, most probably due to the lipid environment in which they are embedded. The membrane-spanning portions of the so-called α-helix bundle proteins, which are the subjects of this review, contain one or more transmembrane α-helices, each of which is a stretch of approximately 20 amino acids with largely hydrophobic side chains. The α-helices are oriented more or less perpendicular to the plane of the membrane. In bitopic membrane proteins, a single helix connects two domains of the protein on either side of the membrane. In polytopic membrane proteins, the membrane-spanning portion of the protein consists of multiple α-helices that are connected by extramembranous domains, i.e., the loops. Three-dimensional structures show that the helices of polytopic membrane proteins are packed intimately in the membrane. Analysis of the hydrophobic moments of transmembrane α-helices in polytopic membrane proteins of known structures indicates that the most polar face of each helix is buried in the interior of the molecule while the least polar face is exposed to the lipids (148, 149).
A fundamental aspect of the structure of polytopic membrane proteins is the membrane topology, i.e. the number of transmembrane segments and their orientation in the membrane. Fortunately, despite the difficulties encountered in obtaining high-resolution structures, the physicochemical constraints imposed by the lipid environment provide a simple method to predict the topology of a membrane protein. The predicted topology can be verified by a variety of molecular and biochemical techniques. Membrane protein topology predictions are based on the observations that (i) the transmembrane α-helices have a high overall hydrophobicity and (ii) the charge distribution of the hydrophilic loops that connect the transmembrane segments follows the “positive inside” rule, which states that nontranslocated loops are enriched in positively charged residues compared to translocated loops (191). The first observation is used to identify the transmembrane segments in the amino acid sequence by analyzing the hydropathic properties of the amino acid sequence (39, 103, 175, 191), and the second observation is used to predict the overall orientation of the protein in the membrane. The biochemical techniques used to verify the predicted membrane topology are, without exception, based on modifications of the proteins by engineering the structural genes coding for the proteins. These techniques are reviewed and evaluated in the first part of this review.
The success of biochemical approaches to determining membrane protein topologies will increase dramatically with the understanding of the dynamics of the biosynthetic pathway leading to the folded protein in the membrane. Thus, as well as knowledge of membrane protein synthesis and targeting to the appropriate membrane, understanding of the process of insertion into the membrane and formation of the final three-dimensional structure is necessary to determine and understand the topology of membrane proteins (Fig. 1). Many important aspects of membrane protein biosynthesis seem to rely on rather well-defined signals encoded in the polypeptide chain, such as targeting signals and topogenic signals. The membrane topology is formed in a process in which the topogenic signals in the nascent polypeptide chain are recognized and translated by the insertion machinery. Topology studies and prediction methods will become much more reliable when all the topogenic signals present in the amino acid sequence of a membrane protein are recognized and understood and when it is known how the insertion machinery deals with them. Although several targeting and topological signals have been investigated extensively, a lot of topogenic information is still hidden in the polypeptide chains of membrane proteins as unknown signals. The second part of this review focuses on studies that specifically search for topogenic signals by using similar techniques to those used in the studies of membrane topology.
FIG. 1.
Membrane protein biogenesis. In many membrane topology studies, the membrane protein is modified by engineering of the structural gene, while the results are analyzed at the level of the folded protein. The structure of a membrane protein can be deduced from its amino acid sequence only if the different steps leading to the structure are understood.
MEMBRANE TOPOLOGY
Principles
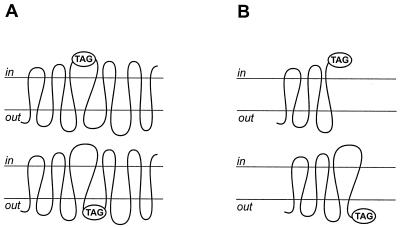
Plasma membrane proteins cross the membrane in a zigzag fashion and expose their hydrophilic loops alternately in the two compartments that are separated by the membrane. To verify predicted membrane protein topology models, the existence of all the putative transmembrane domains must be verified and the hydrophilic loops must be localized to one of the compartments. The simple structure of membrane proteins, the nature of the membrane, and the possibility of genetically modifying proteins allowed the development of a variety of biochemical and molecular techniques for the elucidation of the membrane topology of integral membrane proteins. In vivo, the membrane represents a barrier that separates different intracellular compartments from each other or separates the interior of the cell from the external environment. Due to the impermeability of the membrane to hydrophilic molecules, parts of a membrane protein that lie on opposite sides of the membrane are differently accessible to various agents. Easily identified target sites are inserted in the polypeptide, and membrane-impenetrable reagents are used to determine their accessibility at one side of the membrane (Fig. 2A). Examples of target sites include N-glycosylation sites, Cys residues, iodinatable sites, antibody epitopes, and proteolytic sites that are introduced at specific positions in the protein by site directed mutagenesis. By inserting the tag at different positions in the protein, the complete topology can be determined.
FIG. 2.
Biochemical approaches to determination of membrane protein topology. (A) Insertions. (B) Fusions.
A second approach to generate topological information makes use of gene fusions. A reporter molecule is attached to a hydrophilic domain of a membrane protein, and the cellular location of the reporter is determined by the topogenic information in the membrane protein (Fig. 2B). The reporters are typically molecules whose properties (for example, enzymatic activity) depend on their subcellular location. The gene encoding the reporter protein is fused at different sites in the gene encoding the membrane protein, and the properties of the resulting fusion proteins reveal at which side of the membrane the fusion sites reside. In the resulting fusion proteins, the C-terminal part of the membrane protein is deleted and replaced by the reporter moiety. Therefore, the fusions are termed C-terminal deletion fusions.
Enzyme Tags
The enzymes: alkaline phosphatase, β-galactosidase, and β-lactamase.
The most commonly used reporter protein for gene fusion studies in prokaryotes is alkaline phosphatase, encoded by the Escherichia coli phoA gene (24, 118, 175). Alkaline phosphatase (PhoA) is a metalloenzyme that consists of two identical subunits and resides in the bacterial periplasm (30). Each of the subunits contains two intramolecular disulfide bridges. PhoA is initially synthesized as a precursor with an N-terminal signal sequence that is removed during translocation across the membrane. In the periplasm, the mature part of PhoA is oxidized. The cysteine residues form disulfide bridges, triggering the correct folding of PhoA, a partially protease resistant conformation. The folding process is completed by dimer formation, yielding the active enzyme complex (5). The process of folding and assembly of PhoA occurs only after export to the periplasmic space, because various factors prevent formation of the disulfide bonds in the cytoplasm (48). This location-specific character of PhoA allowed its wide use as a reporter molecule in membrane protein topology studies in prokaryotic cells (47) and is applicable to heterologously expressed eukaryotic membrane proteins as well (21, 67, 108). In these studies, the mature part of PhoA, lacking its signal sequence, is fused behind various C-terminal truncated parts of a membrane protein. The truncated protein takes over the function of the signal sequence. If PhoA is fused to a domain of a membrane protein that normally is located in the cytoplasm, the PhoA molecule also remains in the cytoplasm and is inactive. In contrast, when PhoA is fused to a domain that normally is exported across the membrane, the PhoA moiety of the fusion protein is also exported across the membrane and is folded and assembled into the active state. Thus, the enzymatic activity of the fusion protein reveals the cellular location of the fusion site. The alkaline phosphatase activity of the fusion proteins is most easily determined by a plate assay using a chromogenic substrate such as XP (5-bromo-4-chloro-3-indolylphosphate) or, more quantitatively, by spectroscopic enzyme assays (124). Commercially available antibodies directed against PhoA enable the detection and quantitation of the fusion proteins by immunoblotting.
Fusions of the E. coli enzyme β-galactosidase (LacZ), a large tetrameric cytoplasmic enzyme, are used in a complementary fashion to the PhoA fusions. In contrast to PhoA, LacZ exhibits enzymatic activity in the cytoplasm. When attached downstream of an export signal, the enzyme is trapped in the membrane, which prevents proper folding; therefore, the enzyme is inactive (164). Thus, fusions of LacZ to a periplasmic fusion site of a membrane protein are inactive, while fusions to a cytoplasmic domain are active. Similar to PhoA fusions, β-galactosidase is visualized by a plate assay using the chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) or is measured quantitatively using the spectroscopic enzyme assay (63). Antibodies directed against LacZ to quantify the expression level are commercially available as well.
The third commonly used reporter molecule, which has been used as an alternative to PhoA, is the mature form of the monomeric periplasmic enzyme β-lactamase, encoded by the bla gene (28). Periplasmic β-lactamase protects cells against lysis by β-lactam antibiotics such as ampicillin, which otherwise inactivate enzymes essential for cell wall biosynthesis that are anchored to the outer surface of the cytoplasmic membrane. Cells expressing cytoplasmic β-lactamase are sensitive to the antibiotics because cytoplasmic β-lactamase cannot intervene between the incoming antibiotic and their periplasmic targets. Thus, cells expressing fusion proteins in which the mature form of β-lactamase is fused to a cytoplasmic domain of a membrane protein fail to grow on plates with a high ampicillin concentration while cells expressing fusions to periplasmic domains are able to grow on these plates. Plating the cells at a high density identifies in-frame cytoplasmic fusions. Under these conditions, lysis of a fraction of the cell population liberates enough of the cytoplasmic β-lactamase to permit growth of the cells. Plates with different ampicillin concentrations are used to determine the localization of the β-lactamase moiety (27).
Complementarity.
Various topology models have been tested using a combination of PhoA and LacZ fusions (45, 57, 76, 91, 92, 106, 170, 202, 206). Random fusions to both reporter molecules are often constructed, and this is followed by screening for positive clones on indicator plates. Positive LacZ fusions represent fusions to cytoplasmic domains, while positive PhoA fusions represent fusions to periplasmic domains. Genetic tricks have been developed which make it possible to exchange the LacZ reporter molecules in positive fusions for the PhoA molecule and vice versa (117, 200), resulting in fusions with complementary enzymatic properties (76, 170). The use of PhoA as a reporter molecule in these studies is advantageous in principle because a positive result (i.e., a high-activity fusion) requires the active export of the mature reporter enzyme moiety into the periplasm. In contrast, the use of LacZ as a complementary cytoplasmic reporter may give a positive phenotype as a result of many artifacts. Saturation of the export machinery, especially when using overexpression systems, and the liberation of an initially membrane-embedded, inactive portion of β-galactosidase may explain the unexpectedly high β-galactosidase activity. Conflicting results, i.e., high LacZ and PhoA activity at the same fusion site, have been reported in a number of membrane topology studies (45, 63, 68, 70, 77, 184). Furthermore, in a few cases, the truncated membrane proteins adopt alternate conformations depending on which reporter protein is present. For unknown reasons, the correct folding in the membrane is more often achieved with PhoA as the reporter (57, 106). In recent years, most topology studies have been done with just PhoA or β-lactamase.
Expression levels and specific activity.
It is important to demonstrate that the low activity of an alkaline phosphatase fusion protein is due to a cytoplasmic location of the PhoA moiety and not to a lower level of expression of the fusion protein. Thus, the level of expression is usually determined by Western blot analysis using antibodies raised against PhoA to quantitatively relate the enzymatic activity to the protein concentration. If all full-length fusion proteins in a set show more or less the same level of expression, irrespective of their localization and activity, the activities of the fusion proteins can easily be corrected for the level of expression or may be taken to indicate topology without correction (152, 169). On the other hand, if different amounts of fusion protein are detected within a set, this may indicate differential stability or differential synthesis, which can be resolved by pulse-chase immunoblot experiments (156). Instability of the membrane protein moiety results in the liberation of alkaline phosphatase from the fusion protein. The fate of the alkaline phosphatase then depends on the side of the membrane at which it is released. Release into the periplasm results in stable and active PhoA. Immunoblots show mixtures of full-length fusion proteins and free PhoA in ratios that depend on the stability of the membrane-embedded part of the proteins. In contrast, due to improper folding of the PhoA moiety in the cytoplasm, in cytoplasmic fusion proteins the free PhoA moiety is rapidly degraded and is not or poorly detected by Western blotting. As a consequence, fusions with a periplasmic PhoA moiety show larger amounts of immunoreactive products on a Western blot than do cytoplasmic fusions (45, 106, 143, 184). This systematic behavior can be used as an additional method to discriminate between fusions with a periplasmic or cytoplasmic PhoA moiety, but it is important that it is not used to normalize the activities. To demonstrate the synthesis of the fusion proteins, pulse and/or pulse-chase experiments are generally used (26, 45, 151, 152, 168, 170, 206, 208).
Sandwich fusions.
In the PhoA sandwich approach (55), the alkaline phosphatase sequence, lacking the signal sequence, is inserted into the membrane protein rather than replacing different C-terminal membrane protein sequences. Thus, the entire membrane protein is present and the approach does not suffer from the drawbacks pertinent to the C-terminal deletion fusion approach. This approach may give a more accurate picture of the topology if the final topology of a protein is affected by interactions between amino- and carboxyl-terminal sequences. Similar to the fusion approach, high alkaline phosphatase activity of a sandwich construct reveals a periplasmic location of the domain bearing the insertion while low activity corresponds to a cytoplasmic location.
The sandwich approach was used solely or in addition to PhoA fusions to determine the topology of a number of bacterial proteins including the maltose transporter subunit MalF (55), the aromatic amino acid permease AroP (42), the phenylalanine-specific permease PheP (137), the tryptophan transporter Mtr (157), and the eukaryotic multidrug resistance protein MDR (21). To minimize the disruption of known topological signals, such as positively charged residues in cytoplasmic loops, the fusions were positioned in the C-terminal portion of each hydrophilic loop (see also the next section). While in principle sandwich fusions may be the better approach, in practice many insertion constructs are inactive (like C-terminal deletion fusions). Only 3 of 25 AroP-PhoA sandwich fusions (42) and 5 of 25 PheP-PhoA sandwich-fusions (137) retained some transport activity. As was also observed with the Mtr permease (157), the active constructs involved insertions into periplasmic domains. Since it is possible that insertion of the large PhoA domain alters the topology of the protein, inactive sandwich proteins are risky for use in topology studies and do not provide an advantage relative to C-terminal deletion fusions.
Assumptions and rules.
Experimental support for a topological model requires a minimum of one fusion in each extra membranous domain. If the protein contains stretches of residues of intermediate hydrophobicity that cannot unambiguously be identified as membrane spanning, fusions should be made approximately every 30 residues (26).
The major assumption in the fusion approach to membrane protein topology is that truncation of a membrane protein does not affect its native topology. The fusion technique, especially using the reporter molecule alkaline phosphatase, has been used in a large number of topology studies (for a list of proteins until 1994, see reference 24), and several of the models that were obtained have been confirmed by other biochemical data (29, 143, 180) or even by crystallographic data (36, 85, 86, 177, 208), indicating that the assumption is valid for at least a number of proteins. On the other hand, different types of anomalous behavior of fusion proteins have been reported, which has led to the formulation of rules to optimize the analysis and to avoid pitfalls (24, 108, 175). One type of anomalous behavior is the unexpectedly high activity that is observed when alkaline phosphatase is fused near the N-terminal end of a cytoplasmic loop. An extensive study of MalF in which PhoA was fused at different sites in a cytoplasmic loop suggested that the high activity of fusions near the N-terminal end may be due to loss of C-terminal hydrophilic sequences containing positively charged residues that would lock the loop in the cytoplasm (25). It was proposed that fusions should be constructed to the C-terminal end of cytoplasmic loops or at least downstream of basic amino acid residues (26). A comparative study using β-lactamase as the reporter domain demonstrated that the equivalent MalF–β-lactamase fusions did not confer anomalous high resistance to ampicillin, indicating that the β-lactamase domain was correctly retained in the cytoplasm. The difference in behavior of PhoA and β-lactamase as the reporter domain may be the result of differences in the ability of the reporter proteins to fold into export-incompetent states in the cytoplasm; β-lactamase would fold more rapidly than PhoA (144). Similar to the β-lactamase fusions, the equivalent fusions of MalF and the biotin acceptor domain of the Propionibacterium shermanii transcarboxylase (see “The BAD tag” below) as the reporter did not result in anomalous behavior (87). The data obtained with truncated MalF fusions suggested that a truncated membrane protein may adopt different topologies depending on which reporter protein is present. However, the membrane protein also plays a role. β-Lactamase fusions in the first cytoplasmic domain of the rhamnose/H+ symporter of Salmonella enterica serovar Typhimurium (174) conferred an anomalous high resistance to ampicillin, indicating that with this protein β-lactamase is not retained in the cytoplasm. Also, PhoA fusions in two different positively charged cytoplasmic loops of the melibiose permease MelB resulted, independent of the positions of the fusion sites relative to the positive charges, in low-activity fusions, demonstrating that the positive charges were not essential for locking the reporter in the cytoplasm (143). Apparently, the behavior of fusion proteins is dependent on the combination of membrane protein and reporter protein.
A second type of mislocation of the reporter molecule was observed when PhoA was fused in periplasmic loops following a transmembrane segment containing one or more charged residues. The low activity of the fusions indicated that the membrane segment is retained in the cytoplasm (6, 29). The export ability of the segment may be reduced because of an overall lower hydrophobicity or because C-terminal sequences of the membrane protein are required to stabilize the segment in the membrane through the formation of interhelical salt bridges. Replacing the PhoA moiety in these fusions with β-lactamase resulted in the same anomalous behavior (144).
An artefact occurs with membrane proteins whose N termini face the periplasm, such as the E. coli inner membrane protein ProW (76), which is a component of the ProU osmoregulatory system (71). Fusions of the reporter molecule to the N-terminal domain lack transmembrane sequences and are bound to remain in the cytoplasm (76, 109, 144, 198).
An extensive PhoA fusion analysis study with the lactose permease, LacY, and the melibiose permease, MelB, of E. coli led to remarkable new insights. Fusions constructed at intervals of 3 to 5 residues in each predicted transmembrane segment showed a sharp change from high to low PhoA activity. The hypothesis was proposed that the cytoplasmic half of an outgoing segment is enough to promote translocation of the alkaline phosphatase moiety to the periplasmic space while the periplasmic half of an ingoing segment is enough to prevent translocation (29, 143, 180). This rule was used to determine the approximate middle of transmembrane segments VII and XI in LacY by constructing PhoA fusions at alternating residues. The fusion point of the chimera that showed a sharp increase in alkaline phosphatase activity was set at the middle of the transmembrane segment (180). A similar sharp transition in low and high alkaline phosphatase activity for fusions that were 6 to 10 residues apart was observed with the glutamate transporter of Bacillus stearothermophilus, GltT (169). Clearly, these observations and explanations are in marked contrast to those discussed above.
Proper assembly of the truncated membrane protein part of a fusion protein in the membrane seems to depend on the membrane protein, the reporter protein, and the location of the fusion site. Therefore, the fusion approach is successful for certain membrane proteins but not for others. The folding of the fusion proteins depends on features of the nascent chain such as charges and hydrophobicity, but it may also depend on the rate of folding of extramembranous domains and, clearly, on other still unknown features. It should be noted that the fusions that behave anomalously are only a minor fraction of all the fusions. They may be especially helpful in understanding additional features that give a particular stretch of residues a transmembrane disposition (see “Membrane insertion” below). When we understand all these features, we may be able to deduce the complete membrane topology from the results of the reporter fusion technique.
Glycosylation Tags
Glycosylation of membrane proteins.
In eukaryotic cells, glycosylation activity is found in the lumen of the endoplasmic reticulum (ER) and is accomplished by the enzyme oligosaccharyl transferase (OST), which adds oligosaccharides to the amino group of asparagine (Asn) residues of the consensus sequence Asn-X-Thr/Ser (196). N glycosylation is a common feature of eukaryotic membrane proteins, and the consensus sequence is usually found in the largest luminal exposed loop of the protein (104). The presence of endogenous sites in multiple loops is possible as well (171). Since modification of the glycosylation site occurs in a compartment-specific manner, the presence of glycosylation provides topological information. The loop(s) carrying the oligosaccharide is identified by successive elimination of potential glycosylation sites by site-directed mutagenesis (83). The state of glycosylation of native or mutant proteins is assayed in an in vivo expression system or in vitro in cell-free systems consisting of reticulocyte lysate or wheat germ lysate supplemented with microsomes, membranes derived from the ER. Addition of the oligosaccharide chain to a single site results in an increase in the apparent molecular mass of approximately 2.5 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Glycosylation can be manipulated by treatment with glycosidases (for example, Endo H [134, 187] and N-glycosidase F [132, 189]) or by expression of the protein in the presence of glycosylation inhibitors (for example, tunicamycin [18]) or competitive acceptor peptides (for example, Ac-Asn-Tyr-Thr-NH2 [140] and N-benzoyl–Asn-Leu-Thr–N-methylamide [130, 187]).
Glycosylation scanning mutagenesis and glycosylation fusions make use of the glycosylation properties in the ER lumen to study the complete topology of membrane proteins in the ER membrane. They have become the counterpart of the PhoA fusion and sandwich techniques to analyze eukaryotic membrane proteins. In the glycosylation scanning mutagenesis approach, consensus glycosylation sites or domains bearing a glycosylation site are introduced into membrane proteins devoid of glycosylation sites. Localization of the insertion site on the luminal or cytoplasmic side of the membrane is inferred from the presence or absence of glycosylation of the engineered protein, respectively. The technique has proven very useful in defining the folding pattern of various membrane proteins (18, 32, 34, 84, 131, 132, 140, 167, 178). In the glycosylation fusion approach, a domain bearing one or more glycosylation sites is fused behind different C-terminal deletion mutants of a membrane protein. The fused glycosylation site functions as a reporter molecule in which glycosylation of the domain indicates a luminal position of the fusion point while no glycosylation indicates a cytoplasmic location (13, 132, 187).
Distance between OST and the membrane.
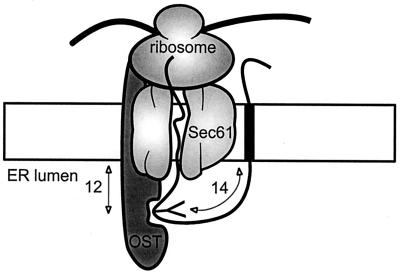
The OST enzyme, which is responsible for the glycosylation of eukaryotic membrane proteins, is embedded in the ER membrane and has its catalytic site facing the luminal compartment (196). Successful N glycosylation by the active site of OST requires adequate spacing of the acceptor site in the luminal loop of the membrane protein from the membrane surface (131, 197). The exact distance has been determined using E. coli leader peptidase as a model protein (130). It was found that there is a precise distance constraint that allows glycosylation of acceptor sites only when the Asn residue is located a minimum of 12 residues upstream or 14 residues downstream of a transmembrane segment (Fig. 3). A survey of polytopic membrane proteins revealed that luminal loops containing N-linked oligosaccharides have a minimum size of about 30 residues, with the N-glycosylation acceptor site at least 10 residues away from the predicted transmembrane segment (104). N-glycosylation scanning mutagenesis of extracytosolic loops in the anion exchanger of the erythrocyte membrane Band 3 demonstrated that the distance requirements inferred from Leader peptidase applied to this protein (140). Introduced acceptor sites were glycosylated only in loops larger than 25 residues, and a sharp cutoff was observed in glycosylation of sites positioned too close to the membrane. Furthermore, insertion of a small glycosylation tag in short loops did not result in N glycosylation, whereas these loops were efficiently glycosylated when a large domain bearing a glycosylation site was inserted. The results account for the lack of glycosylation of endogenous (104) or introduced (140, 178) sites that are close to the membrane. The distance constraint between OST and the glycosylation site was used to map the ends of the transmembrane segments of Band 3, resulting in a refined topology model (140).
FIG. 3.
ER translocase complex. The luminal OST is responsible for the glycosylation of membrane proteins in the ER. The active site of OST is located ∼14 residues away from the end of a downstream transmembrane segment and ∼12 residues away from an upstream hydrophobic segment.
Glycosylation scanning.
To study the topology of membrane proteins by using glycosylation scanning mutagenesis, the endogenous glycosylation sites in the protein must be removed and novel sites must be introduced at various positions. The mutations necessary to remove the endogenous sites and the resulting absence of glycosylation are not expected to have profound effects on the topology of membrane proteins. Indeed, the activities of several deglycosylated proteins, such as Band 3 (74), the erythrocyte glucose transporter GLUT1 (84), the glycine transporter GLYT1 (132), and the human Na+/glucose cotransporter SGLT1 (178), were comparable to those of the wild-type proteins, implying that glycosylation is not necessary for function and that the mutations to remove the glycosylation sites do not affect activity. The deglycosylated Na+/Cl−-coupled γ-aminobutyric acid (GABA) transporter from rat brain (GAT-1) exhibited only 38% of wild-type activity, but it was shown that the reduction in activity was due at least in part to reduced trafficking to the plasma membrane (18).
Insertion of novel glycosylation sites in the hydrophilic domains of membrane proteins has a dramatic effect on activity. A 4-amino-acid insertion (NNSS) in the human Na+/glucose cotransporter (SGLT1) (178) at 13 different positions resulted in 10 insertion mutants with abolished transport activity. Transport was completely abolished upon insertion of one or two glycosylation sites in 6 of the 11 hydrophilic loops of the GABA transporter GAT-1, and activity in the remaining loops was strictly dependent on the position in the loop (18). In contrast, all four cystic fibrosis transmembrane conductance regulator (CFTR) glycosylation insertion mutants were fully functional (32) and 10 of 15 mutants of the erythrocyte glucose transporter Glut1 carrying a 41-amino-acid insert bearing a glycosylation site retained 2.5 to 30% of the deoxyglucose transport activity in oocytes (84). Since it cannot be ruled out that the inserted sequences scramble the topology of a protein, the use of inactive mutants for topology mapping is risky. On the other hand, it is generally believed that misfolded proteins are retained in biosynthetic compartments and rapidly degraded, implying that a construct which reaches the plasma membrane has a good chance of having the correct topology (84). In N-glycosylation scanning mutagenesis, internal loops score negative, but the accessibility of an external loop to the glycosylation enzymes may also be limited because of steric hindrance. Consequently, absence of glycosylation does not always disprove an extracellular localization. For example, insertion of several glycosylation sites at different positions in the most C-terminal loop of the glycine transporter (GLYT1) did not result in glycosylated proteins even though data obtained with other biochemical methods supported an extracellular location of this loop (132). Creation of a glycosylation site by replacement of two amino acids in the third extracellular loop of the GABA transporter resulted in a glycosylated protein, while creation of a double glycosylation site by insertion of 4 amino acids (NNSS) at the same position did not result in glycosylation (18). The results demonstrate that the ability to undergo glycosylation can be critically dependent on the position of the site in the loop and on the nature of the insert.
Steric factors, such as membrane proximity, that prevent glycosylation can be overcome by using longer insertion epitopes. The native glycosylation domain of SGLT1 or the entire first glycosylated luminal domain of GLUT4, consisting of 48 and 41 amino acids, respectively, resulted in significantly higher levels of glycosylation when inserted in SGLT1 (178) and GLUT1 (84) than was observed with smaller tags.
Glycosylation fusions.
Domains bearing a glycosylation site have been used as reporter molecules in topology studies in very much the same way as alkaline phosphatase in PhoA fusions: the reporter molecule containing one or more glycosylation sites is fused behind different C-terminal deletions of a membrane protein. The domain will be glycosylated only when it is translocated to the luminal side of the ER membrane. Often, the technique is used in combination with in vitro translation and insertion in the presence or absence of insertion-competent microsomes derived from the ER membrane. Translation in the absence of microsomes results in a protein with the apparent molecular mass of the full-length fusion protein, while translation in the presence of microsomes results in the same or a higher apparent molecular mass depending on the cytoplasmic or luminal localization of the domain, respectively (see “Glycosylation of membrane proteins” above). The β-subunit of the gastric H+,K+-ATPase contains five N-linked glycosylation sites and has been used as a reporter molecule for the topological analysis of a number of membrane proteins, including the gastric H+,K+-ATPase (13), two rat Ca2+ ATPases (16) the rabbit gastric cholecystokinin A (CCK-A) receptor (17), and the glycine transporter GLYT1 (132). The membrane topology of the glutamate carrier GLT1 was studied by fusing a glycosylation consensus sequence behind a series of C-terminally truncated proteins (194). The in vitro translation-transcription system was also used to study the topology of the P-type ATPase from Helicobacter pylori (123) and the sodium ion-dependent citrate carrier of Klebsiella pneumoniae CitS (187), demonstrating that bacterial proteins can be expressed and analyzed in the ER system as well.
Chemical Modification
Cysteine residues in membrane proteins.
The cysteine residue is a relatively hydrophobic, nonbulky residue, and its introduction at most positions in a membrane protein is likely to be tolerated. This feature and the ease of specific chemical modification with sulfhydryl reagents are the basis of several methods aiming at topological and structure-function-related information on membrane proteins. In cysteine scanning mutagenesis, a series of single cysteine mutants is created and the reactivity of the single cysteine mutant to various sulfhydryl reagents is assessed under different conditions. To obtain single Cys mutants, the native cysteine residues present in the membrane protein are removed and novel cysteine residues are introduced at specific positions in the Cys-less protein. The native cysteine residues are generally removed by replacement by serine residues, which usually renders an active protein. Substitution of amino acids with cysteine residues appears to be tolerated at most positions as well. The eight native cysteine residues of the lactose permease (LacY) of E. coli have been replaced simultaneously to yield a Cys-less permease that retained at least 50% of its wild-type activity (188). Furthermore, it was demonstrated that except for four amino acids, all the amino acids of LacY could be replaced by a cysteine residue without disrupting the function of the transporter (for reviews, see references 62 and 93). Thus, cysteine-scanning mutagenesis permits topology assessment and other structure-function studies under conditions in which the proteins are active and therefore, presumably, the protein structure is not seriously altered.
Cysteine-scanning mutagenesis.
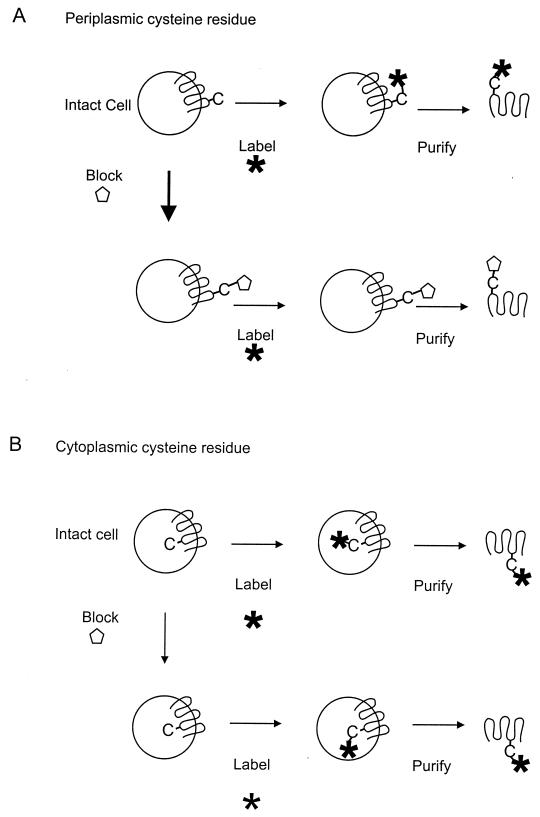
In topology studies, amino acids that are thought to reside in the putative extracellular or intracellular loops of a membrane protein are replaced by cysteine residues and the orientation with respect to the membrane is evaluated using membrane-permeable and -impermeable sulfhydryl reagents. Sulfhydryl reagents are available that contain either a biotin group (115), a fluorescent group (212), or a radiolabel (97), allowing detection of labelled proteins by streptavidin binding, fluorescence, or autoradiography, respectively. Labelling of a cysteine residue with a membrane-permeable reagent containing a detectable group after pretreatment of whole cells, spheroplasts, or right-side-out-oriented membrane vesicles with a membrane-impermeable reagent is indicative of a cytoplasmic cysteine residue, while the absence of labelling reveals a periplasmic location (Fig. 4). In addition to subcellular localization, labelling of a cysteine residue may depend on reactivity, possible steric constraints due to local structure, and, in cells and spheroplasts, scavenging of the label by thiol groups on cytoplasmic proteins. Therefore, it is essential to analyze labelling from both sides of the membrane either by performing the complementary experiment using inside-out membrane vesicles or by showing that the protein can be labelled in a noncompartmentalized system, i.e., in detergent solution. The sulfhydryl reagent reacts with cysteine residues present in all other proteins in the membrane, and therefore immunoprecipitation of the membrane protein or a purification step is necessary to show specific labelling.
FIG. 4.
Cysteine accessibility assay. Labeling of a periplasmic (A) and a cytoplasmic (B) cysteine residue in intact cells. The cells are treated with a detectable and membrane-permeant cysteine reagent (Label; from left to right) with or without pretreatment with a membrane-impermeable cysteine reagent (Block). Following the treatment, the protein is purified from the cells and assayed for labeling.
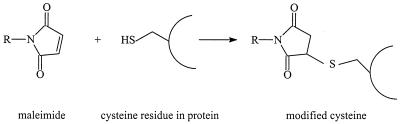
Maleimide-based sulfhydryl reagents, which form a thioether bond with cysteine residues (Fig. 5), have been used in several topology studies including those of the MotA protein (212), P-glycoprotein (115), transposon 10-encoded metal-tetracycline/H+ antiporter (97), Na+/proline transporter (92), and subunit a of the E. coli F0F1-ATP synthase (114, 183). The permeable maleimide compounds biotinmaleimide (92, 114, 115), N-[14C]ethylmaleimide (97), or fluoresceinmaleimide (183, 212) were used in combination with the membrane-impermeable maleimide compound acetomaleimide disulfonate.
FIG. 5.
Reaction of a thiol with a maleimide.
A limitation of the cysteine-scanning technique is that not all residues are accessible to the sulfhydryl compounds. In particular, cysteine residues close to or within a putative transmembrane domain were found to be less reactive with biotin-containing compounds than those located closer to the middle of a hydrophilic loop (115). For this reason, several reagents should be tried. On the other hand, the use of small impermeant reagents allows the detection of residues in small loops that may not be accessible to macromolecular reagents such as antibodies, proteases, or the glycosylation machinery (35, 212). Native cysteine residues present in membrane proteins have been reported to be inaccessible to the sulfhydryl reagents as well. In those cases, it may not be necessary to construct a Cys-less protein prior to the introduction of new cysteine residues (188).
Additional advantages.
An attractive feature of the use of cysteine scanning mutagenesis is that the library of functional single-cysteine mutants can be used for a more detailed structural analysis of the membrane protein. The E. coli LacY protein is the most complete example of cysteine-scanning mutagenesis to date, and its library of cysteine mutants (single and double Cys mutants) has been analyzed by a number of biophysical techniques aiming at obtaining both structural and dynamic information (62, 93). Helix packing, helix tilt, and ligand-induced conformational changes have been investigated using sulfhydryl-specific cross-linkers, spin labels, and fluorescent probes as well as site-directed peptide bond cleavage. Furthermore, cysteine-scanning mutagenesis in putative transmembrane segments has been used to explore the secondary structure of the segment in detail and to map channel-lining residues. Similar studies have been done for several other membrane proteins including the nicotinic acetylcholine receptor (1, 2, 4), the GABA receptor (203, 204), the cystic fibrosis transmembrane conductance regulator (CFTR) (3), the UhpT transporter (205), potassium channels (136), the tetracycline antiporter TetB (98), the erythrocyte anion exchanger AE1 (172), the GABA transporter GAT-1 (207), the oxalate/formate antiporter OxlT (64), and the serotonin transporter SERT (35). In these studies, sulfhydryl reagents that differ in charge, size, and hydrophilicity were used (88). These approaches are not discussed any further here.
Conclusions.
Cysteine-scanning mutagenesis is the most useful technique developed for topology studies so far, at least for bacterial systems. The advantages are as follows: (i) the analysis is done on the complete and active protein; (ii) the modifications of the protein are minimal and restricted to a single amino acid residue change, and so they are not likely to disturb the membrane topology; (iii) the point mutations are well tolerated, usually leaving the proteins active; (iv) the actual analysis is done by chemical modification after insertion of the protein into the membrane; (v) the chemical modification can be done using whole cells, thereby avoiding problems related to the conversion of cells into membrane vesicles with a uniform orientation; and (vi) the library of single Cys mutants forms the basis for many other types of investigations. Disadvantages of the technique are as follows: (i) a Cys-less mutant of the protein has to be constructed, and (ii) the Cys residues in the single Cys mutants may not be accessible to the thiol reagents. The Cys-scanning mutagenesis approach has also been used for eukaryotic membrane proteins (18, 75, 93, 160) but has not yet been used in combination with in vitro translation/insertion using microsomes derived from the ER membrane, which may have additional advantages. Besides cysteine residues, other amino acids may be useful. In a study to test the accessibility of predicted external loops of the serotonin neurotransmitter transporter, SERT, lysine residues were introduced as targets for the impermeant biotinylating reagent NHS-SS-biotin, which reacts specifically with lysine residues (35). A mutant SERT protein in which four native lysine residues in loop regions were replaced by arginine or glutamine residues showed normal transport activity. Similarly, a series of active mutant proteins containing novel single external lysine residues could be constructed, indicating that the replacement of lysine residues is tolerated, at least in external loops. A disadvantage of using lysine residues is that most proteins contain many lysine residues, making it less easy to construct a functional protein devoid of lysine residues.
The BAD Tag
BAD.
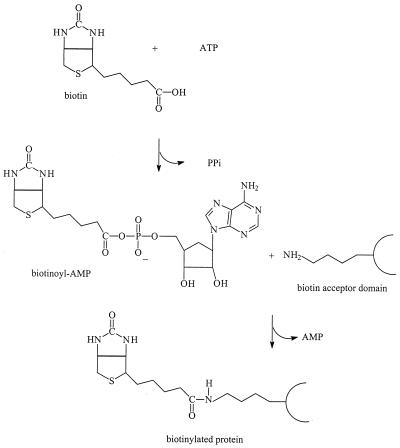
Biotin carboxylases are a family of enzymes that use a covalently attached biotin moiety as a mobile carboxy carrier to catalyze the transfer of CO2 between metabolites (155). The biotin group is attached posttranslationally to a specific lysine residue of newly synthesized biotin carboxylases by the cytoplasmic enzyme biotin ligase, which uses ATP to activate the carboxy group of biotin during the reaction (Fig. 6) (163). Biotinylated proteins are rare in nature, varying from one to five in different organisms. The biotin carboxyl carrier protein of acetyl coenzyme A carboxylase is the only biotinylated protein found in E. coli (59). A high degree of sequence homology is observed around the biotin attachment site of all biotinylated proteins, and it was demonstrated that the biotin ligase enzyme from different sources is able to biotinylate acceptor proteins from various species both in vivo (43) and in vitro (121). The minimum size of the biotin domain necessary for biotinylation is about 75 to 80 residues (43, 110), and this domain, the biotin acceptor domain (BAD), exists as a stable entity. The crystal structure of the BAD of the E. coli biotin carboxyl carrier protein has been solved (11). Expression of gene fusions of the BAD of various carboxylases to the C terminus of different soluble proteins resulted in in vivo biotinylated proteins. Biotin binds to avidin with unusually high affinity (the dissociation constants are in the order of 10−15 M), yielding a complex that is extremely stable over a wide range of temperatures and pH values (72). The biotin-avidin system is used to detect biotinylated proteins with high sensitivity and to purify biotinylated proteins in one step by affinity chromatography using immobilized avidin (33, 43). Similar to soluble proteins, membrane proteins fused to BAD result in biotinylated proteins (41, 141, 184). Lac permease constructs with the BAD of the oxaloacetate decarboxylase of K. pneumoniae either inserted in the middle cytoplasmic loop or fused at the C terminus catalyze active transport and become biotinylated in vivo (180). Due to incomplete in vivo biotinylation, only a fraction of the transport protein was recovered upon purification using immobilized avidin. Recently, a method for in vitro biotinylation of the LacY-BAD fusion protein, resulting in complete biotinylation, has been described (142). Similarly, fusions of the same BAD to the C terminus of the citrate transporter CitS resulted in active and in vivo biotinylated protein and could be purified in a functional state (141). Membrane topology studies based on the BAD make use of the compartment-specific in vivo biotinylation of the domain (87, 209) or the high sensitivity to detect the biotinylated proteins in combination with proteolysis (184).
FIG. 6.
In vivo biotinylation. Shown is the condensation reaction of biotin and a lysine residue, yielding a biotinylated protein. The reaction is catalyzed by the enzyme biotin ligase.
BAD in topology studies.
The membrane topology of the lactose transporter of E. coli LacY has been determined by a variety of biochemical techniques. The BAD of the oxaloacetate decarboxylase from K. pneumoniae was inserted into various loops to investigate the suitability of the BAD for topology studies. The rationale was that BAD inserted in a cytoplasmic domain would be biotinylated because it is accessible to the cytoplasmic biotin ligase enzyme and, consequently, BAD inserted in a periplasmic domain would not be biotinylated. The following observations led to the conclusion that the approach was hindered by unexpected effects. Insertion of the BAD in two out of four cytoplasmic loops of LacY did not result in biotinylation of the domain (209). Lack of biotinylation was also observed when the BAD was inserted at one specific position in the most C-terminal cytoplasmic domain of the citrate transporter CitS protein, while efficient biotinylation was observed when the domain was inserted at a different position in the same loop (M. van Geest and J. S. Lolkema, submitted for publication). Possibly, the biotinylation site is buried in the structure of the folded membrane protein, rendering the domain inaccessible to the biotin ligase enzyme. Alternatively, interactions of the domain with other regions of the membrane protein may prevent the proper folding of the BAD necessary for recognition by the biotin ligase enzyme. Insertions of the BAD into periplasmic domains of LacY resulted in biotinylated proteins that were all inactive in lactose transport (209). It could be shown that the domain was not translocated to the periplasm. Insertion of the BAD into a periplasmic loop blocks the translocation of the loop and, in all likelihood, the insertion of the two adjacent helices, resulting in a misfolded, inactive protein (209). In contrast, fusion of the BAD to the periplasmic C terminus of CitS resulted in an active fusion protein, and it was shown that the domain was translocated to the periplasm (184). Apparently, translocation of the domain is blocked only when sandwiched in a hydrophilic domain and not when fused to a hydrophilic domain, suggesting that the BAD is useful as a topological marker when using C-terminal deletion fusions. Biotinylation of the BAD of the 1.3S subunit of Propionibacterium shermanii transcarboxylase fused behind a series of C-terminal deletion mutants of the inner membrane protein MalF was indeed in agreement with the known topology of MalF (87). In fact, MalF-BAD fusions did not suffer from the problems encountered with MalF-PhoA fusions, in which the fusion site was at the beginning of a cytoplasmic loop, which resulted erroneously in the export of the PhoA moiety (see also “Complementarity” above) (26). In the corresponding MalF-BAD fusion, the BAD was securely locked in the cytoplasm, suggesting that the domain requires a less strong signal for cytoplasmic localization than does PhoA (87). This feature may be related to the fact that PhoA is a periplasmic protein while the BAD is part of a cytoplasmic protein.
In conclusion, C-terminal deletion fusions of the BAD may be a good alternative to LacZ fusions as positive indicators of cytoplasmic localization, with PhoA as the complementary positive indicator of periplasmic localization. Insertions of the BAD in putative cytoplasmic and periplasmic loops seem to result in many artifacts that make the domain less suitable as a topological marker. Only insertions that leave the protein active and that result in biotinylation report a trustworthy localization of the loop in the cytoplasm. Insertions of the BAD into putative cytoplasmic loops of CitS that did not compromise transport activity were used to verify a topology model of CitS that was based on C-terminal deletion PhoA fusions (van Geest and Lolkema, submitted) (see “Proteolysis” below).
Folding kinetics.
Biotin acceptor domain fusions have been used to study the rate of translocation of secretory proteins or periplasmic domains of membrane proteins (87, 153). The degree of biotinylation is taken as a measure of the time spent in the cytoplasm before translocation. The degree of biotinylation is determined by the rate of biotinylation of the BAD by the biotin ligase in the cytoplasm and the rate of translocation to the periplasm catalyzed by the export machinery. When the BAD was fused to the periplasmic proteins PhoA or maltose binding protein, little or no biotinylation of the domain was observed in E. coli. Biotinylation of the domain was observed when the protein secretion process was slowed by mutations in components of the export machinery or by azide treatment, which specifically inhibits the ATP driven translocation. Thus, the biotinylation of the BAD functions as an indicator of impaired protein export. The rates of translocation of several periplasmic domains of MalF were studied upon sodium azide treatment or depletion of SecE (a component of the export machinery), demonstrating the involvement of the export machinery in the insertion mechanism of the MalF protein (87). Similar studies have been performed with the BAD fused to the periplasmic C terminus of the citrate transporter CitS. The level of biotinylation was much lower than observed for the BAD fused to the cytoplasmic N terminus, consistent with the periplasmic location of the domain. Upon addition of sodium azide to the growth medium, the level of biotinylation of the C-terminal fusion increased significantly (M. van Geest and J. S. Lolkema, unpublished results).
Proteolysis
Membrane proteins, like all proteins, contain cleavage sites for various proteolytic enzymes. Externally added proteolytic enzymes cannot cross the membrane, and therefore cytoplasmic sites are protected against cleavage by the membrane upon exposure of bacterial right-side-out membranes or spheroplasts whereas these sites are not protected against cleavage in inside-out membranes. Similarly, in ER microsomes, the luminally exposed sites are protected against cleavage. Cleavage of the site(s) can be analyzed after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using antibodies directed against the protein. The technique has a clear advantage when it can be applied to native proteins without the need for modification. The limitations of the technique are that the naturally occurring cleavage sites are often not sensitive to the proteases and that more sites may be present, which complicates the analysis. To overcome the latter problem, proteolytic sites for specific proteases, for instance factor Xa, have been engineered into different loops of membrane proteins (154, 199). Similar to this are methods in which the accessibility of hydrophilic loops to nonspecific proteases, such as proteinase K, is assayed after the insertion of a detectable proteineous tag in the loop. Insertion into a cytoplasmic domain results in degradation of the tag upon exposure of inside-out membranes to proteinase K, and the protein or fragments thereof is no longer detectable. In contrast, treatment of spheroplasts or right-side-out membranes will result in the detection of a protected fragment. Insertion into a periplasmic loop gives the opposite result. Tags that have been used are the BAD of the oxaloacetate decarboxylase of K. pneumoniae (184, 209), a tag consisting of six histidine residues (His tag) (122, 185), and several antigenic epitopes, such as an epitope of the hemagglutinin of influenza virus (94–96), the 8-amino-acid FLAG epitope (135), the Myc epitope (31), and the secretory protein prolactin (38). Insertion of antigenic epitopes into membrane proteins which are expressed in eukaryotic cells allow the determination of the sidedness of the tag directly by immunofluorescence microscopy (94–96). The accessibility of an epitope tag or protease site to the antibody or protease has been improved by inserting multiple copies of the tag into the membrane protein (95, 96, 154). As with all these approaches, only functional insertion mutants are reliable for topology mapping, since their use guarantees that insertion of the tag did not alter the native membrane topology of the protein.
MEMBRANE INSERTION
Topology and Insertion
The previous part of this review presented an overview of biochemical approaches aimed at elucidating the membrane topology of polytopic membrane proteins. Some of the techniques work with intact and functional membrane proteins, but most techniques involve modified or truncated proteins. The changes to the proteins are made at the level of the coding sequences, while the results are interpreted in terms of the structure of the proteins as they sit in the membrane, thereby ignoring the steps in between that lead to the mature protein. Certain aspects of membrane protein structure may be properly understood only in the light of how these proteins are inserted into the lipid bilayer. Since membrane proteins contain domains that must be translocated across the bilayer, it is no surprise that the enzymatic machinery responsible for translocating proteins across the membrane also plays a role during the assembly of integral membrane proteins. An important notion is that the insertion machinery may not only insert the membrane protein but also determine its topology. Depending on the “communication” between the nascent polypeptide chain and the insertion machinery, the polypeptide is inserted into the membrane. Only when we are able to understand the “language” used by the polypeptide chain and the insertion machinery will it become possible to predict the complete structure of a membrane protein from the amino acid sequence. The success of the biochemical approaches that involve engineering of the proteins to determine membrane topology may improve significantly when we understand how the signals in the amino acid sequence interact with the insertion machinery. The next section briefly reviews the insertion machineries in the ER membrane and the bacterial cytoplasmic membrane and provides a discussion of several techniques used to detect topogenic signals in the polypeptide chain that trigger a certain response in the insertion machinery and thereby to determine the folding of the protein in the membrane.
Membrane Protein Biogenesis
Insertion machinery in the ER membrane.
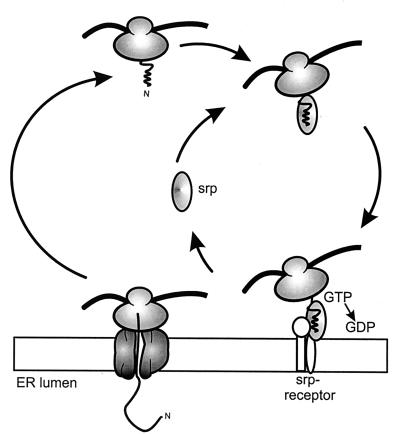
In the ER, insertion of membrane proteins into the membrane and preprotein translocation into the lumen proceed by similar mechanisms (for reviews, see references 147 and 195). The process of targeting of membrane and secretory proteins begins in the cytosol when a hydrophobic segment of a nascent polypeptide chain, either a signal sequence or the first transmembrane segment, emerges from the ribosome and is recognized by the signal recognition particle (SRP) (Fig. 7). Binding of the SRP to the ribosome results in translational arrest, and the whole complex is targeted to translocation sites in the membrane via interactions with the SRP receptor. Translation resumes after dissociation of the SRP from the ribosome, and the nascent chain is transferred cotranslationally into the translocon. The ordered binding and recognition steps along this pathway are controlled by GTP binding and hydrolysis and GDP-GTP exchange (Fig. 7) (12, 40, 125, 146). The cotranslational integration of the membrane proteins is mediated by the translocon, consisting of the heterotrimeric Sec61 complex (120, 138, 159) and a number of other membrane proteins, including the translocating chain-associated membrane protein (TRAM) in mammalian cells (81, 82) or the heterotetrameric Sec62/63p complex in yeast (138). Since the Sec61 complex both translocates secretory proteins and integrates membrane proteins into the lipid bilayer, it must coordinate a number of different functions. For membrane proteins, cytoplasmic domains have to be left in the cytoplasm, luminal domains have to be passed through the membrane, and transmembrane domains have to be properly oriented and inserted into the membrane.
FIG. 7.
Membrane protein insertion into the ER membrane. The SRP recognizes the first transmembrane segment emerging from the ribosome and targets the whole complex to the ER membrane. The ribosome-nascent chain complex is delivered at the translocon, where translation resumes and the protein is inserted into the membrane.
Insertion into the bacterial cytoplasmic membrane.
(i) Bacterial translocation machinery.
It is a matter of debate whether the same pathway is used in E. coli for export of preproteins and insertion of membrane proteins. In contrast to membrane protein insertion, translocation of preproteins across the inner membrane of E. coli has been studied extensively (50–52). Rather than occurring cotranslationally, preprotein translocation across the bacterial membrane occurs in a posttranslational manner and is carried out by a system involving chaperones like SecB and GroEL-GroES and the inner membrane components of the secretory (Sec) machinery, SecA, SecY, SecE, SecG, SecD, and SecF. Secretory proteins are synthesized as preproteins with an amino-terminal signal sequence. The signal sequence and/or mature domain of the preprotein is recognized in the cytoplasm by chaperones which ensure that the nascent secretory proteins do not fold or aggregate prematurely and which help to deliver the preproteins to specific receptors on the cytoplasmic membrane (for a review, see reference 60). The integral membrane components SecY, SecE, and SecG, which are homologous to subunits of the eukaryotic Sec61 complex, are believed to form a channel through which the nascent chain is extruded. SecA, which is peripherally bound to the SecYEG complex in the membrane and which does not have an eukaryotic counterpart, carries out an ATP-driven cycle of insertion and deinsertion into the translocase, during which 20 to 30 residues of the nascent chain are moved through the membrane (54, 179). In the bacterial system, SecA is the engine that drives translocation, while in the ER, translocation is driven by synthesis of the nascent chain on the ribosomes. The membrane-bound SecD and SecF components that are not essential for protein translocation per se stabilize SecA in its active conformation (53).
(ii) Sec-dependent and Sec-independent insertion of membrane proteins.
Recent evidence has changed our views about membrane protein insertion in bacteria. Previously, it was believed that insertion occurred by two different mechanisms, a Sec-dependent mechanism and a Sec-independent one. Proteins could use one mechanism exclusively, or domains within a protein could use one or the other mechanism. A Sec-dependent mechanism was demonstrated for inner membrane proteins containing large periplasmic domains, such as the maltose transporter subunit MalF and leader peptidase Lep (176, 201). In contrast, insertion of proteins without large periplasmic domains was proposed to be Sec independent (14, 201). Sec dependency would be related to the length of periplasmic loops (9) and to the number of charged residues in the translocated loops (10). Sec-dependent insertion was believed to occur in a sequential, N- to C-terminal fashion, similar to that in the ER membrane (see “Insertion mechanism” below). Sec-independent insertion was, for thermodynamic reasons, thought to occur via “helical hairpins” composed of interacting antiparallel α-helices formed from two consecutive hydrophobic segments and their short intervening polar loops (58). Formation of the helical hairpin would sufficiently increase the net hydrophobicity of the sequence to drive insertion into the lipid bilayer. In summary, the Sec machinery would assist only in situations when insertion could not follow a simple spontaneous mechanism based on the physicochemical properties of the polypeptide chain and the membrane.
More recently, evidence has been accumulating that the E. coli proteins Ffh and FtsY (19, 150) play an important role in the assembly of inner membrane proteins. The Ffh and FtsY proteins are homologous to subunits of the eukaryotic SRP and SRP receptor proteins, suggesting an insertion pathway similar to the one in the ER (46, 162, 181, 182). Moreover, structural similarities between the E. coli SecYEG complex and the eukaryotic Sec61 complex (see also reference 113) make it likely that in E. coli, after targeting, membrane insertion proceeds via the Sec machinery. It was realized that studies that had indicated Sec-independent insertion were based on negative results obtained in experiments with conditional SecA and SecY strains that were selected primarily for secretion defects rather than for defects in the assembly of membrane proteins. Sec-independent insertion had been observed after sodium azide treatment, which is known to inhibit the ATPase activity of the SecA subunit only partially. Furthermore, it cannot be assumed a priori that translocon mutations have equal effects on the export of preproteins and insertion of membrane proteins (176). Very recently, Sec-dependent insertion in E. coli was demonstrated for three small bitopic membrane proteins with and without large periplasmic domains by using an E. coli strain with the SecE subunit depleted. Also, these proteins required the E. coli SRP for correct assembly into the inner membrane (46). These observations make it less likely that insertion of more complicated polytopic proteins bypasses the translocon. Recent reports actually showed that the SecY protein plays a role in determining the topology and insertion of membrane proteins in the bacterial membrane (128, 145).
Membrane protein insertion into the bacterial cytoplasmic membrane may be very similar to the process in the ER membrane. This view is supported by functional, heterologous expression of bacterial proteins in eukaryotic membranes and vice versa (73, 80).
Insertion mechanism.
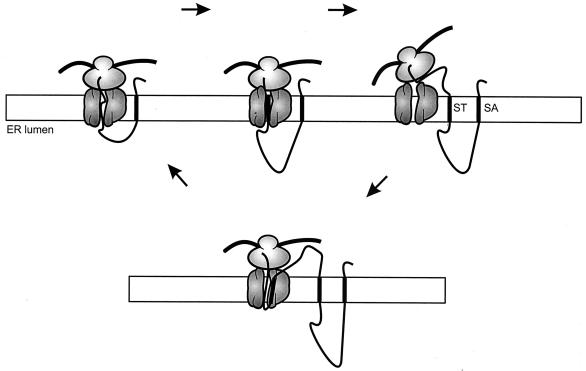
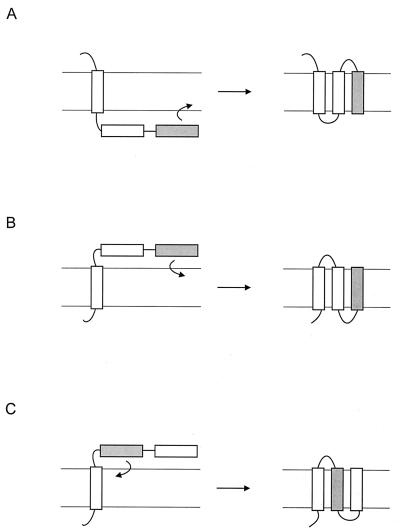
For reasons discussed above, most data on the actual insertion mechanism by the Sec machinery come from studies involving the eukaryotic ER system. The translocon forms an aqueous channel that permits the translocation of hydrophilic polypeptides (44, 69, 165) and opens laterally to allow the exit of transmembrane segments into the lipid bilayer (119, 127). Membrane integration is regulated by topogenic signals in the amino acid sequence of the nascent chain emerging from the ribosome (20). In a simple view, a series of alternating signal anchor (SA) and stop transfer (ST) sequences directs the integration of polytopic membrane proteins (22, 61, 112). A SA sequence is defined as a hydrophobic segment that has the ability to insert into the membrane with the N terminus in the cytoplasm and the C terminus in the lumen (Ncyt-Clum orientation). The sequence following the SA will be translocated to the lumen until translation of the next hydrophobic region, the ST sequence, which is retained in the membrane as an Nlum-Ccyt transmembrane segment. The following sequences will have a cytoplasmic location until the next SA sequence emerges from the ribosome and the cycle starts all over again (Fig. 8). Thus, according to this view, insertion of the polypeptide into the membrane occurs sequentially in an N- to C-terminal fashion, with the hydrophobic segments inserting into the membrane with alternating orientations. Consequently, the N-terminal segment determines the orientation of the protein in the membrane.
FIG. 8.
Sequential insertion of hydrophobic sequences. Hydrophobic segments insert into the membrane as they emerge from the ribosome. The orientation of the segment is opposite to the orientation of the previous segment. A segment with an Ncyt-Clumen orientation is termed signal anchor (SA), and a segment with an Nlumen-Ccyt orientation is termed stop transfer (ST).
Although the assembly of some membrane proteins in the ER appears to occur via this mechanism, observations such as the exclusion of hydrophobic segments from the membrane (184, 187), the insertion of less hydrophobic sequences (13, 16, 17, 133), and observations made with artificial proteins (65, 66) and with C-terminally truncated membrane proteins (37, 199) have suggested that the insertion process may be more complicated, depending on the coordinate action of two or more topogenic sequences in the nascent chain. Recently, it has been documented that the ribosome plays a role in the insertion process (49, 111, 127). The ribosome would detect a newly synthesized transmembrane segment and use this information to influence the activity of the translocon (111). Structural changes at the ribosome may allow the ribosome and/or cytosolic proteins to facilitate the folding and processing of the nascent chain and/or determine the orientation of the nascent chain in the membrane. The transmembrane segments of the multidrug transporter P-glycoprotein did not integrate into the lipid bilayer until termination of synthesis, suggesting that the transmembrane segments remain associated with the insertion machinery until the polypeptide is released from the ribosome (23). Such observations suggest that the nascent chain contains, in addition to SA and ST segments, novel topogenic sequences that signal to and control the activity of the insertion machinery.
Studies aiming at unravelling the complicated insertion mechanism follow two lines of research: (i) mechanistic studies of the insertion machinery and (ii) identification of insertion and topogenic signals in the polypeptide sequence. The remainder of this review discusses the latter studies. Two steps are discriminated. The first is identification of segments in the amino acid sequence that have the potential to insert into the membrane in one or both orientations in the absence of other putative transmembrane segments or other topogenic signals. The second step is reevaluation of the insertion potencies of the putative transmembrane segments when combined with adjacent regions on the polypeptide chain that may or may not contain known insertion signals like charged residues. The second step reveals the relative strength of the signals and the presence of unknown signals. A putative transmembrane segment may be excluded from the membrane because its insertion is overruled by another stronger signal; alternatively, a less hydrophobic segment may be inserted because of strong orientation signals in adjacent transmembrane segments. Unexpected behavior may be helpful in decoding unknown signals in the amino acid sequence.
Insertion of Isolated Hydrophobic Segments
Principles.
Sequences throughout the amino acid sequence of a polytopic membrane protein are identified that are able to insert independently of each other into the ER membrane, with a preferred orientation. Different algorithms (56, 99) that predict putative membrane-spanning sequences are used to guide the initial selection of potential candidates. The glycosylation fusion approach (see “Glycosylation fusions” above) has been used mainly to assay the sequences for their membrane insertion properties. Briefly, a reporter molecule containing one or more glycosylation sites is fused behind the sequence encoding the putative transmembrane segment and in vitro transcription-translation is carried out in the presence of microsomes. Glycosylation of the reporter corresponds to insertion of the segment into the ER membrane, indicating that the segment exhibits SA activity, whereas a lack of glycosylation corresponds to a lack of SA activity. Fusion of a sequence encoding a transmembrane segment in front of the putative segment indicates insertion of the segment in the opposite orientation, i.e., ST activity. Different types of systems have been developed for these studies. Insertion vehicles based on the H+/K+-ATPase (M0 and M1 vectors), the E. coli inner membrane protein leader peptidase (Lep), and the prolactin molecule have been used to analyze both eukaryotic (13, 133) and bacterial (123, 187) membrane proteins in the ER. Possible methods to perform similar studies in the bacterial membrane are discussed below (see “Similar studies in E. coli”).
Insertion vehicles.
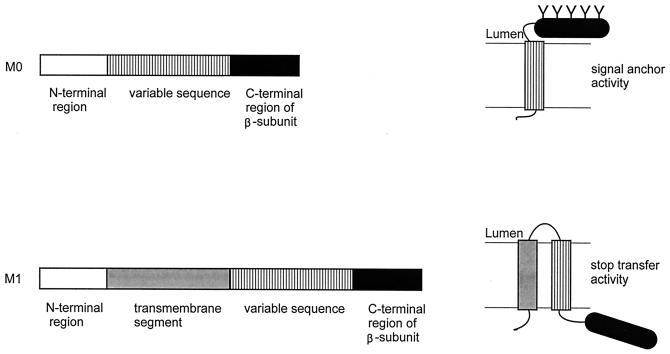
(i) M0 and M1 system.
The H+/K+-ATPase is a P-type ATPase that pumps H+ and K+ in opposite directions across the gastric membrane, driven by ATP hydrolysis. The enzyme consists of two subunits, α and β, that both are integral membrane proteins. The C-terminal region of the β-subunit contains five N-linked glycosylation sites. A vector was developed containing the DNA sequences encoding the cytoplasmic N-terminal region of the α-subunit excluding (M0 vector) or including (M1 vector) the first transmembrane segment, followed by a variable region representing the amino acid sequence under investigation, which is followed by the C-terminal region of the β-subunit containing the glycosylation sites (Fig. 9). The M0 translation product, without an insertion in the variable region, was not glycosylated when synthesized in the presence of microsomes, demonstrating that the β-subunit fragment does not insert into the membrane (13). The M0 vector assays for the SA capacity of the inserted sequences, while the M1 vector identifies segments with ST activity. Membrane insertion sequences of the H+/K+-ATPase (13), the ER and sarcoplasmic Ca2+-ATPases (16), a P-type ATPase of Heliobacter pylori (123), and the rabbit gastric cholecystokinin A receptor (17) have been identified using the M0 and M1 system (15). Topogenic properties of putative transmembrane segments of the glycine transporter GLYT1 were analyzed in a similar system (132).
FIG. 9.
M0 and M1 system. The variable sequence is preceded by the N-terminal region excluding (M0) or including (M1) the first transmembrane segment of the α-subunit of the gastric H+,K+ P-type ATPase and followed by the C-terminal region of the β-subunit of the same enzyme to assay for ST and SA activity, respectively (left). Insertion of the variable sequence into the membrane results in glycosylation of the C-terminal region of the β-subunit in the M0 vector and in the lack of glycosylation in the M1 vector (right).
A drawback of the M0 fusion vector is the lack of an ER-targeting sequence preceding the variable region. As a result, the vector assays for both targeting and translocation and the intrinsic signal anchor capacity of a single segment cannot be measured. Insertion of a segment into the M0 vector, which results in a nonglycosylated product, does not necessarily reflect a segment incapable of insertion, as was concluded by various authors (13, 16, 17, 123).
(ii) Lep system.
The system based on the E. coli membrane protein leader peptidase (Lep) does not suffer from the above problem. Leader peptidase is involved in the removal of targeting sequences from exported proteins. Lep is anchored to the cytoplasmic membrane by two N-terminal transmembrane segments, H1 and H2, connected by a cytoplasmic P1 domain, and it also contains the periplasmic catalytic C-terminal P2 domain. The insertion of Lep into the microsomal membrane has been studied extensively (65, 89, 192) using in vitro transcription-translation. In the presence of microsomes, the Lep molecule inserts into the membrane with both the N and C termini facing the lumen and with the P1 and P2 domains in the cytoplasm and lumen, respectively. The H1 domain has an intrinsic ability to target to the ER membrane and to insert with the N terminus in the lumen. The orientation of the H1 segment is determined largely by the highly positively charged P1 loop. An engineered glycosylation site C-terminal of H2 is efficiently glycosylated upon correct insertion into the microsomal membrane (89, 130). In the insertion vehicle, the H2 domain is replaced by segments encoding individual putative transmembrane segments of membrane proteins and glycosylation of the P2 domain is used as an indicator of the SA activity of the segment (Fig. 10). Since all constructs contain the H1 domain and the P1 loop, targeting of the molecules to the membrane is mediated by H1 and the N-terminal end of the variable sequence is locked in the cytoplasm. Thus, the construct measures the “intrinsic” SA function of various fragments that have already been targeted to the microsomal membrane. A similar Lep construct, in which the variable sequence was inserted in between the H2 and P2 domain, was used to measure the ST efficiency of a segment. The constructs have been used to study ST activities of artificial peptides with different hydrophobicities (153) and SA and ST activities of the putative transmembrane segments of the bacterial citrate transporter CitS (187).
FIG. 10.
Lep system. Vectors based on Leader peptidase of E. coli. The variable sequence replaces the second transmembrane segment, H2, of Lep (top) to measure SA activity or is inserted in the P2 domain to assay for ST activity (bottom). Membrane insertion of the variable sequence results in glycosylation and lack of glycosylation, respectively (right).
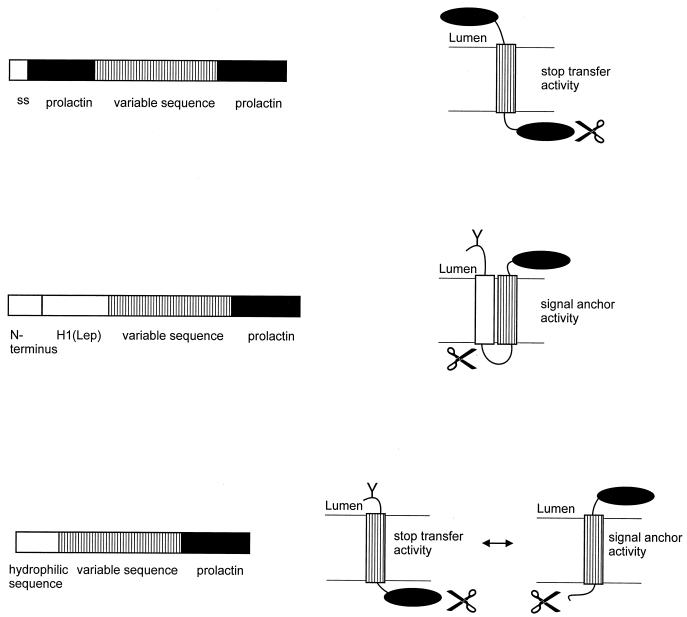
(iii) Prolactin system.
Three types of insertion vehicles for isolated putative segments have been developed based on prolactin (Fig. 11). Prolactin is a secreted protein, and preprolactin consists of a signal sequence followed by the mature prolactin domain. A vector described by Kuroiwa et al. (100) measures ST activity. The signal peptide of prolactin, mature prolactin, an inserted transmembrane segment, and a second mature prolactin molecule are arranged sequentially. Upon in vitro transcription-translation in the presence of microsomes, the whole construct is targeted to the membrane by the signal sequence and the first prolactin moiety is exported to the lumen. ST activity of the inserted transmembrane segment results in a cytoplasmic location of the second prolactin moiety, where it is sensitive to externally added proteinase K. Lack of ST activity results in translocation of the molecule to the lumen, where it is fully resistant to the protease.
FIG. 11.
Prolactin system. Shown are vectors based on the secretory protein prolactin. The variable sequence is inserted between preprolactin and prolactin (top), the H1 domain of leader peptidase and prolactin (middle), and a hydrophilic domain and prolactin (bottom). The assay of the membrane insertion activity of the variable sequence is based on protection of the prolactin domain against proteolytic degradation (right). The scissors represents the protease.
An insertion vehicle designed to measure SA activity consists of the H1 domain of leader peptidase followed by the variable region, followed by the prolactin domain. An engineered glycosylation site in front of the H1 domain was used in an independent assay to ensure that the H1 domain inserted correctly. SA activity of the inserted transmembrane segment results in translocation of the prolactin domain to the lumen, where it is resistant to externally added proteinase K. When the inserted segment does not function as an SA, the molecule remains outside the membrane and is degraded by the protease (133).
A third vector was designed to examine the preferred orientation of the segment under investigation (133). The segment is preceded by a hydrophilic domain containing a glycosylation site and followed by mature prolactin. If the segment prefers to insert in an ST orientation, a glycosylated molecule is obtained in which the prolactin moiety is sensitive to proteases, while a preferred SA orientation results in the absence of glycosylation and a prolactin moiety that is protected against proteolytic attack.
The prolactin vectors have been used to study the topogenic functions of transmembrane segments of the human anion exchanger Band 3 (133), the multidrug transporter P-glycoprotein (78, 126, 210), the γ-aminobutyrate transporter GAT-1 (37), and the cystic fibrosis transmembrane conductance regulator, CFTR (116); they have also been used to study SA and ST functions of artificial sequences (101, 102). In some studies, the insertion vehicles contained different reporter molecules, such as interleukin-2 (101).
SA and ST activities of isolated transmembrane segments.
Many hydrophobic segments of membrane proteins function equally efficient as SA and ST sequences when assayed in the M0 and M1 system. These include the four N-terminal segments of the H+/K+-ATPase (13) and the Ca2+-ATPase (16) and most of the transmembrane segments of the CCK-A receptor (17). For the P-type ATPase of H. pylori (123), the ability to insert in both orientations seemed to correlate with the orientation of the segments in the native protein. All four segments expected to have an SA orientation in the native protein showed both SA and ST activity when assayed as individual segments. In contrast, three of the four segments expected to have an ST orientation in the native protein were not able to act as SA sequences but did show ST activity. Similar observations were made with ST segments of the H+/K+-ATPase (13), the Ca2+-ATPase (16), and the CCK-A receptor (17), suggesting that SA activity is more critically dependent on a particular segment than is ST activity. It should be noted, though, that the M0 and M1 system does not discriminate between SA activity and targeting (see above). Since the average hydrophobicity of the ST segments was not lower than the average hydrophobicity of the SA segments, factors other than hydrophobicity must play a role in membrane insertion or targeting.
The 12 hydrophobic segments of the citrate transporter CitS all demonstrated SA activity when assayed in the Lep insertion vehicle. Thus, in contrast to what was observed above, all ST segments in the native protein inserted equally well as SA sequences in the membrane as isolated segments. No correlation between the hydrophobicity of the segment and insertion efficiency was seen. Interestingly, one hydrophobic segment of CitS that is located in the periplasm in the native protein (184, 187) was fully functional as SA and exhibited significant but incomplete ST activity. Apparently, signals present in the remaining part of the CitS polypeptide mediate the exclusion of this hydrophobic segment from the membrane during the insertion of CitS (see below).
Various sequences in the region of the fifth and sixth transmembrane domains of the H+/K+-ATPase (13) and the Ca2+-ATPase (16) did not show any membrane-inserting properties when assayed as individual segments in the M0 and M1 system. The average hydrophobicity of these segments was lower than that of the other segments. Similarly, one transmembrane segment of the CCK-A receptor did not show any membrane insertion properties and 4 out of 14 segments of the anion exchanger Band 3 that were supposed to be SA sequences showed insufficient SA activity. Since these segments are found to be transmembrane segments in the native proteins, their insertion in the membrane must be dependent on other signals in the polypeptide chain.
Similar studies in E. coli.
The topogenic functions of isolated transmembrane segments in the E. coli membrane have not been studied as extensively as those in the ER membrane, and approaches that combine insertion vehicles and in vitro translation-insertion have not been explored to their full potential. The efficiency of ST activity of a set of pseudo-random amino acid segments was measured both in the ER membrane and in E. coli (153). The E. coli study was done in vivo using a modified Lep molecule as the insertion vehicle in which the P2 domain was replaced by the mature part of alkaline phosphatase (PhoA). Most sequences behaved similarly in both systems, indicating that the criteria for functional ST sequences are largely conserved between prokaryotes and eukaryotes, but subtle differences were detected.
The insertion properties of various isolated sequences of the ArsB protein, a subunit of the bacterial arsenical resistance pump, were assayed in vivo in E. coli by fusing the segments in front of the β-lactamase reporter protein (202). Similarly, the first transmembrane segment of the E. coli serine chemoreceptor Tsr was fused in front of both β-galactosidase and the mature part of alkaline phosphatase (see “The enzymes: alkaline phosphatase, β-galactosidase, and β-lactamase” above) to assay the tolerance of its signal anchor function to mutations. It was concluded that a threshold hydrophobicity of the segment is required for efficient insertion into the membrane (107). The C-terminal transmembrane segment of the citrate transporter CitS, which has an Ncyt-Cper orientation, was fused with its C terminus to the mature part of PhoA. Expression of the construct resulted in efficient secretion of the PhoA molecule to the periplasm, demonstrating independent insertion of the segment (van Geest and Lolkema, unpublished). Extension of these methods to other segments and other membrane proteins would allow a detailed comparison between interactions of the eukaryotic and bacterial insertion systems with nascent polypeptide chains and may provide important information about the insertion mechanisms in both systems.
Interaction between Multiple Insertion Signals
Charge distribution and hydrophobic segments.
Besides hydrophobic segments, the distribution of positively charged amino acids appears to be an important determinant in membrane topology, in both eukaryotic and bacterial membranes. Statistical studies of bacterial cytoplasmic membrane proteins have demonstrated that cytoplasmic domains contain more positive charged amino acids than do periplasmic domains, which resulted in the so-called positive inside rule (190, 191). Statistical analyses indicated that a similar positive inside rule is applicable to eukaryotic membrane proteins (79, 90, 105, 166, 193), although the tendency for positive charges to be absent from extracellular domains is somewhat weaker (65). The influence of positive charges on the topology of single and double membrane spanning proteins (7, 8, 129, 190, 193) as well as of polytopic membrane proteins (65, 66) has been experimentally demonstrated. The positive charges prevent the translocation of N- and C-terminal segments as well as the connecting loops of polytopic membrane proteins and therefore determine to some extent the orientation of a transmembrane segment. Thus, hydrophobic segments and positive charges in the connecting loops represent two types of topogenic signals.
In marked contrast to the above results, introduction of a positively charged lysine residue into the cytoplasmic domain just downstream of the second transmembrane segment of the serine chemoreceptor protein Tsr of E. coli resulted in enhanced export of the domain (161). The lysine residue was introduced into a stretch of 11 residues that is strongly amphipatic when folded as an α-helix. A total of 10 different mutations in the amphipatic stretch resulted in a correlation between enhanced export of the cytoplasmic domain and the amphipathicity of the stretch. It was suggested that an amphipathic sequence located immediately downstream of a transmembrane sequence represents a determinant of membrane topology.
Model proteins, consisting of four transmembrane segments and a distribution of positively charged residues in the loops that does not conform the positive inside rule in either of the two orientations in the membrane (“frustrated proteins”), have been characterized in the E. coli (66) and the ER (65) membrane. In both membrane types, some proteins adopted a “leave-one-out” topology, in which only three segments inserted into the membrane but in which the highly positively charged loops were in the cytoplasm. Leave-one-out topologies were also observed for P-glycoprotein mutants (211) and GLUT1 glucose transporter mutants (158) as a result of a change in the distribution of positively charged residues in domains flanking transmembrane segments. The leave-one-out topologies demonstrate that the topogenic signal imposed by the distribution of positively charged residues may overrule the topogenic signal imposed by the hydrophobic segment; the distribution of positively charged residues is a stronger signal than a hydrophobic segment. These studies demonstrate that mechanisms exist by which a hydrophobic segment can be excluded from the membrane.
Interaction between different hydrophobic segments.
(i) Insertion depending on upstream segments.
In the topology model of the anion exchanger Band 3, which is based on various experimental approaches (for a review, see reference 173), the first transmembrane segment, an SA segment, is connected by a short loop to the second transmembrane segment, an ST segment. A C-terminal deletion mutant containing the first two segments folded as in the complete molecule. However, when the second segment was separated from the first transmembrane segment by a loop containing 200 residues, the second segment exhibited insufficient ST activity (133). ST activity of the second segment improved when it was located closer to the first segment, as in the original Band 3 molecule. Thus, the distance between the two segments was critical for the topogenic process. The authors proposed that with the short connecting loop the preceding sequence might still be in the translocon, allowing a direct interaction between the two hydrophobic segments which would make it easier for the second segment to stop in the translocon. In this way, a segment with a lower hydrophobicity may become integrated into the membrane. With the long connecting loop, the first transmembrane segment may leave the translocation channel prematurely and cannot “help” with the insertion of the second segment (133).
The citrate transporter CitS contains a hydrophobic segment that in the complete molecule is translocated to the periplasm (187). The segment exhibited both SA and ST activities when assayed as an individual segment. It appeared that the presence of the preceding transmembrane segment in CitS was largely responsible for the exclusion of the segment from the membrane. Possibly, interactions between the two segments in the translocon occur which direct the second hydrophobic segment from the translocon to the periplasm. The nature of these interactions is unknown.
(ii) Insertion depending on downstream segments.
Some transmembrane segments of Band 3 which are inserted in the membrane in an SA orientation in the complete protein exhibited either insufficient or no insertion activity when assayed as individual segments (133). The segments are followed by highly hydrophobic segments with a strong preference for the ST orientation over the SA orientation. The authors suggested that segments with weak insertion signals might be integrated into the membrane by the immediate downstream segments (134) (Fig. 12A).
FIG. 12.
Hypothetical folding intermediates. In the folding intermediates (left), two successive transmembrane segments are not inserted in the membrane. The hydrophobic segment, indicated as a gray box, drives the insertion of the preceding (A and B) or following (C) segment to yield the membrane topologies indicated on the right. The intermediates are based on observations made with the anion exchanger Band 3 (A), the protein translocation complex subunit Sec61 (A and B), and the citrate transporter CitS (C).
Recently, it was demonstrated that instead of inserting into the membrane in an Nper-Ccyt orientation, transmembrane segment VIII of the citrate transporter CitS is translocated into the periplasm in the absence of C-terminal sequences of the protein. The presence of downstream transmembrane segment IX appeared to be sufficient to mediate the insertion of both segments (186). In contrast to the Band 3 case above, segment VIII is a highly hydrophobic segment, which makes it difficult to understand why it would require the presence of the less hydrophobic segment IX for insertion. Instead, it could be segment IX that requires the presence of segment VIII for insertion. First, segment VIII would be translocated to the periplasm. Only when segment IX arrives in the periplasm would segment VIII drive the insertion of the helical hairpin consisting of segments VIII and IX (Fig. 12C). This sequence of events requires a mechanism that prevents segment VIII from inserting before segment IX is translated and translocated to the periplasm. Surprisingly, the same phenomenon was not observed upon insertion of CitS in the ER membrane (187).
A requirement for downstream sequences was demonstrated for two transmembrane segments, with limited hydrophobicity, of the Sec61 protein from yeast (199). Segment TM2 required the presence of its downstream segment to stop translocation, while segment TM5 required C-terminal sequences to function as an SA sequence. Interestingly, in a construct consisting of the N-terminal five segments, TM5 was a transmembrane segment when the C-terminal sequences were coexpressed, suggesting that the insertion took place after the protein had left the translocon (Fig. 12B).
A requirement for downstream sequences was also observed for some of the transmembrane segments of the K+,H+-ATPase (13) and for the amino-terminal segment TM1 of the CFTR (116).
(iii) Folding intermediates.
The apparent interdependence between transmembrane segments for insertion may be a manifestation of the presence of folding intermediates during the insertion process. The intermediates may serve to insert segments that are relevant for catalysis and hence less hydrophobic. Figure 12 shows a number of hypothetical folding intermediates based on the examples above. Each intermediate requires very specific properties of the involved segments, resulting in specific actions of the insertion machinery to prevent a sequential insertion mechanism of the hydrophobic segments. A moderately hydrophobic segment at the cytoplasmic side of the membrane may be inserted into the membrane by the following hydrophobic segment (the “driving” segment) when the latter has a strong ST activity (Fig. 12A [based on Band 3 and Sec61 topologies]). In the folding intermediate, the hydrophobic driving segment does not enter the translocon as an SA sequence but is retained in the cytoplasm. Only when both segments are completed do the two insert as a helical hairpin. The latter action may take place in the translocon or after lateral movement into the lipid bilayer, either spontaneous or assisted by chaperone-like proteins. Similarly, a moderately hydrophobic periplasmic or luminal segment may be inserted by a downstream hydrophobic segment (Fig. 12B [based upon the Sec61 topology]). In the folding intermediate, the driving segment is exported to the periplasm. The loop between the two segments is initially translocated across the membrane to end up in the cytoplasm after the coordinate insertion of the two segments. Finally, the driving segment may also precede the less hydrophobic segment (Fig. 12C [based upon the CitS topology]). The insertion machinery translocates the driving segment across the membrane, where it waits for the less hydrophobic segment to be exported before both insert in concert.
The presence of folding intermediates as described above would imply that the folding of the polypeptide into the transmembrane segments cannot be understood solely based upon the energetics of transmembrane helix partitioning between the aqueous and membrane phases (139). The folding would be determined by the insertion machinery, and not all transmembrane domains may form autonomous folding domains. Importantly, such domains are likely to play a pivotal role in the function of the protein. For such membrane proteins, it is unlikely that they will refold from the denatured state into the native structure in vitro. The catalytically active structure of the protein is trapped in a local free energy minimum that is higher than the overall free energy minimum of the polypeptide and that cannot be reached in the absence of the biosynthetic machinery.
PROSPECTS
High-resolution three-dimensional structures are indispensable to our understanding of protein function. Given the enormous number of sequences that are produced in genome-sequencing projects, it is not realistic to assume that the structures of all the encoded proteins will be generated by crystallographic approaches, especially for membrane proteins. The structures will have to be generated by computational techniques. This review emphasizes that biochemical approaches provide, besides topological information, important information about the insertion process. Only when the molecular mechanism behind the insertion of a membrane protein is understood will it become possible to predict the three-dimensional structures from the amino acid sequence by a simple successive decoding of all the topogenic signals present in the amino acid sequence. Knowledge about new topogenic sequences present in the nascent chain of membrane proteins and about the interactions between different topogenic signals will help to develop better prediction algorithms and biochemical techniques to predict and determine membrane protein structure.
REFERENCES
- 1.Akabas M H, Karlin A. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the alpha-subunit. Biochemistry. 1995;34:12496–12500. doi: 10.1021/bi00039a002. [DOI] [PubMed] [Google Scholar]
- 2.Akabas M H, Kaufmann C, Archdeacon P, Karlin A. Identification of acetylcholine receptor channel-lining residues in the entire M2 segment of the alpha subunit. Neuron. 1994;13:919–927. doi: 10.1016/0896-6273(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 3.Akabas M H, Kaufmann C, Cook T A, Archdeacon P. Amino acid residues lining the chloride channel of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:14865–14868. [PubMed] [Google Scholar]
- 4.Akabas M H, Stauffer D A, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 5.Akiyama Y, Ito K. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J Biol Chem. 1993;268:8146–8150. [PubMed] [Google Scholar]
- 6.Allard J D, Bertrand K P. Membrane topology of the pBR322 tetracycline resistance protein. TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J Biol Chem. 1992;267:17809–17819. [PubMed] [Google Scholar]
- 7.Andersson H, Bakker E, von Heijne G. Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem. 1992;267:1491–1495. [PubMed] [Google Scholar]
- 8.Andersson H, von Heijne G. Position-specific Asp-Lys pairing can affect signal sequence function and membrane protein topology. J Biol Chem. 1993;268:21389–21393. [PubMed] [Google Scholar]
- 9.Andersson H, von Heijne G. Sec dependent and sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J. 1993;12:683–691. doi: 10.1002/j.1460-2075.1993.tb05702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson H, von Heijne G. Positively charged residues influence the degree of SecA dependence in protein translocation across the E. coli inner membrane. FEBS Lett. 1994;347:169–172. doi: 10.1016/0014-5793(94)00530-3. [DOI] [PubMed] [Google Scholar]
- 11.Athappilly F K, Hendrickson W A. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 12.Bacher G, Lutcke H, Jungnickel B, Rapoport T A, Dobberstein B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- 13.Bamberg K, Sachs G. Topological analysis of H+,K+-ATPase using in vitro translation. J Biol Chem. 1994;269:16909–16919. [PubMed] [Google Scholar]
- 14.Bassilana M, Gwizdek C. In vivo membrane assembly of the E. coli polytopic protein, melibiose permease, occurs via a Sec-independent process which requires the protonmotive force. EMBO J. 1996;15:5202–5208. [PMC free article] [PubMed] [Google Scholar]
- 15.Bayle D, Weeks D, Hallen S, Melchers K, Bamberg K, Sachs G. In vitro translation analysis of integral membrane proteins. J Recept Signal Transduct Res. 1997;17:29–56. doi: 10.3109/10799899709036593. [DOI] [PubMed] [Google Scholar]
- 16.Bayle D, Weeks D, Sachs G. The membrane topology of the rat sarcoplasmic and endoplasmic reticulum calcium ATPases by in vitro translation scanning. J Biol Chem. 1995;270:25678–25684. doi: 10.1074/jbc.270.43.25678. [DOI] [PubMed] [Google Scholar]
- 17.Bayle D, Weeks D, Sachs G. Identification of membrane insertion sequences of the rabbit gastric cholecystokinin-A receptor by in vitro translation. J Biol Chem. 1997;272:19697–19707. doi: 10.1074/jbc.272.32.19697. [DOI] [PubMed] [Google Scholar]
- 18.Bennett E R, Kanner B I. The membrane topology of GAT-1, a (Na+ + Cl−)-coupled gamma-aminobutyric acid transporter from rat brain. J Biol Chem. 1997;272:1203–1210. doi: 10.1074/jbc.272.2.1203. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 20.Bibi E. The role of the ribosome-translocon complex in translation and assembly of polytopic membrane proteins. Trends Biochem Sci. 1998;23:51–55. doi: 10.1016/s0968-0004(97)01134-1. [DOI] [PubMed] [Google Scholar]
- 21.Bibi E, Beja O. Membrane topology of multidrug resistance protein expressed in Escherichia coli N-terminal domain. J Biol Chem. 1994;269:19910–19915. [PubMed] [Google Scholar]
- 22.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borel A C, Simon S M. Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell. 1996;85:379–389. doi: 10.1016/s0092-8674(00)81116-2. [DOI] [PubMed] [Google Scholar]
- 24.Boyd D. Use of gene fusions to determine membrane protein topology. In: White S H, editor. Membrane protein structure. Experimental approaches. Oxford, United Kingdom: Oxford University Press; 1994. pp. 144–163. [Google Scholar]
- 25.Boyd D, Beckwith J. The role of charged amino acids in the localization of secreted and membrane proteins. Cell. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- 26.Boyd D, Traxler B, Beckwith J. Analysis of the topology of a membrane protein by using a minimum number of alkaline phosphatase fusions. J Bacteriol. 1993;175:553–556. doi: 10.1128/jb.175.2.553-556.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 28.Broome-Smith J K, Tadayyon M, Zhang Y. Beta-lactamase as a probe of membrane protein assembly and protein export. Mol Microbiol. 1990;4:1637–1644. doi: 10.1111/j.1365-2958.1990.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 29.Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc Natl Acad Sci USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C N, Kuang W J, Chen E Y. Nucleotide sequence of the alkaline phosphatase gene of Escherichia coli. Gene. 1986;44:121–125. doi: 10.1016/0378-1119(86)90050-8. [DOI] [PubMed] [Google Scholar]
- 31.Chang H C, Bush D R. Topology of NAT2, a prototypical example of a new family of amino acid transporters. J Biol Chem. 1997;272:30552–30557. doi: 10.1074/jbc.272.48.30552. [DOI] [PubMed] [Google Scholar]
- 32.Chang X B, Hou Y X, Jensen T J, Riordan J R. Mapping of cystic fibrosis transmembrane conductance regulator membrane topology by glycosylation site insertion. J Biol Chem. 1994;269:18572–18575. [PubMed] [Google Scholar]
- 33.Chapman-Smith A, Turner D L, Cronan J E, Jr, Morris T W, Wallace J C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem J. 1994;302:881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavez R A, Hall Z W. The transmembrane topology of the amino terminus of the alpha subunit of the nicotinic acetylcholine receptor. J Biol Chem. 1991;266:15532–15538. [PubMed] [Google Scholar]
- 35.Chen J G, Liu-Chen S, Rudnick G. Determination of external loop topology in the serotonin transporter by site-directed chemical labeling. J Biol Chem. 1998;273:12675–12681. doi: 10.1074/jbc.273.20.12675. [DOI] [PubMed] [Google Scholar]
- 36.Chepuri V, Gennis R B. The use of gene fusions to determine the topology of all of the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. J Biol Chem. 1990;265:12978–12986. [PubMed] [Google Scholar]
- 37.Clark J A. Analysis of the transmembrane topology and membrane assembly of the GAT-1 gamma-aminobutyric acid transporter. J Biol Chem. 1997;272:14695–14704. doi: 10.1074/jbc.272.23.14695. [DOI] [PubMed] [Google Scholar]
- 38.Clark J A. Analysis of transporter topology using deletion and epitope tagging. Methods Enzymol. 1998;296:293–307. doi: 10.1016/s0076-6879(98)96022-0. [DOI] [PubMed] [Google Scholar]
- 39.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 40.Connolly T, Gilmore R. GTP hydrolysis by complexes of the signal recognition particle and the signal recognition particle receptor. J Cell Biol. 1993;123:799–807. doi: 10.1083/jcb.123.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consler T G, Persson B L, Jung H, Zen K H, Jung K, Prive G G, Verner G E, Kaback H R. Properties and purification of an active biotinylated lactose permease from Escherichia coli. Proc Natl Acad Sci USA. 1993;90:6934–6938. doi: 10.1073/pnas.90.15.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosgriff A J, Pittard A J. A topological model for the general aromatic amino acid permease, AroP, of Escherichia coli. J Bacteriol. 1997;179:3317–3323. doi: 10.1128/jb.179.10.3317-3323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cronan J E., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 44.Crowley K S, Liao S, Worrell V E, Reinhart G D, Johnson A E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 45.Danielsen S, Boyd D, Neuhard J. Membrane topology analysis of the Escherichia coli cytosine permease. Microbiology. 1995;141:2905–2913. doi: 10.1099/13500872-141-11-2905. [DOI] [PubMed] [Google Scholar]
- 46.De Gier J W, Scotti P A, Saaf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derman A I, Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derman A I, Prinz W A, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 49.Do H, Falcone D, Lin J, Andrews D W, Johnson A E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 50.Driessen A J, Fekkes P, van der Wolk J P. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 51.Driessen A J M. Translocation of proteins across the bacterial cytoplasmic membrane. In: Konings W N, Kaback H R, Lolkema J S, editors. Transport processes in membranes. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1996. pp. 759–790. [Google Scholar]
- 52.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 53.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 55.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 57.Eitinger T, Friedrich B. A topological model for the high-affinity nickel transporter of Alcaligenes eutrophus. Mol Microbiol. 1994;12:1025–1032. doi: 10.1111/j.1365-2958.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 58.Engelman D M, Steitz T A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 59.Fall R R. Analysis of microbial biotin proteins. Methods Enzymol. 1979;62:390–398. doi: 10.1016/0076-6879(79)62246-2. [DOI] [PubMed] [Google Scholar]
- 60.Fekkes P, Driessen A J. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedlander M, Blobel G. Bovine opsin has more than one signal sequence. Nature. 1985;318:338–343. doi: 10.1038/318338a0. [DOI] [PubMed] [Google Scholar]
- 62.Frillingos S, Sahin-toth M, Wu J, Kaback H R. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 63.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 64.Fu D, Maloney P C. Structure-function relationships in OxlT, the oxalate/formate transporter of Oxalobacter formigenes. Topological features of transmembrane helix 11 as visualized by site-directed fluorescent labeling. J Biol Chem. 1998;273:17962–17967. doi: 10.1074/jbc.273.28.17962. [DOI] [PubMed] [Google Scholar]
- 65.Gafvelin G, Sakaguchi M, Andersson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- 66.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 67.Geller D, Taglicht D, Edgar R, Tam A, Pines O, Michaelis S, Bibi E. Comparative topology studies in Saccharomyces cerevisiae and in Escherichia coli. The N-terminal half of the yeast ABC protein Ste6. J Biol Chem. 1996;271:13746–13753. doi: 10.1074/jbc.271.23.13746. [DOI] [PubMed] [Google Scholar]
- 68.Georgiou C D, Dueweke T J, Gennis R B. Beta-galactosidase gene fusions as probes for the cytoplasmic regions of subunits I and II of the membrane-bound cytochrome d terminal oxidase from Escherichia coli. J Biol Chem. 1988;263:13130–13137. [PubMed] [Google Scholar]
- 69.Gilmore R, Blobel G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell. 1985;42:497–505. doi: 10.1016/0092-8674(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 70.Gott P, Boos W. The transmembrane topology of the sn-glycerol-3-phosphate permease of Escherichia coli analysed by phoA and lacZ protein fusions. Mol Microbiol. 1988;2:655–663. doi: 10.1111/j.1365-2958.1988.tb00074.x. [DOI] [PubMed] [Google Scholar]
- 71.Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989;171:1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Green N M. Avidin. Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 73.Grisshammer R, Tate C G. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- 74.Groves J D, Tanner M J. Role of N-glycosylation in the expression of human band 3-mediated anion transport. Mol Membr Biol. 1994;11:31–38. doi: 10.3109/09687689409161027. [DOI] [PubMed] [Google Scholar]
- 75.Grunewald M, Bendahan A, Kanner B I. Biotinylation of single cysteine mutants of the glutamate transporter GLT-1 from rat brain reveals its unusual topology. Neuron. 1998;21:623–632. doi: 10.1016/s0896-6273(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 76.Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hagting A, van der Velde J, Poolman B, Konings W N. Membrane topology of the di- and tripeptide transport protein of Lactococcus lactis. Biochemistry. 1997;36:6777–6785. doi: 10.1021/bi963068t. [DOI] [PubMed] [Google Scholar]
- 78.Han E S, Zhang J T. Mechanism involved in generating the carboxyl-terminal half topology of P-glycoprotein. Biochemistry. 1998;37:11996–12004. doi: 10.1021/bi980702p. [DOI] [PubMed] [Google Scholar]
- 79.Hartmann E, Rapoport T A, Lodish H F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hennessey E S, Hashemzadeh-Bonehi L, Hunt L A, Broome-Smith J K. Assembly of eukaryotic class III (N-out, C-in) membrane proteins into the Escherichia coli cytoplasmic membrane. FEBS Lett. 1993;331:159–161. doi: 10.1016/0014-5793(93)80317-n. [DOI] [PubMed] [Google Scholar]
- 81.High S, Andersen S S, Gorlich D, Hartmann E, Prehn S, Rapoport T A, Dobberstein B. Sec61p is adjacent to nascent type I and type II signal-anchor proteins during their membrane insertion. J Cell Biol. 1993;121:743–750. doi: 10.1083/jcb.121.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.High S, Martoglio B, Gorlich D, Andersen S S, Ashford A J, Giner A, Hartmann E, Prehn S, Rapoport T A, Dobberstein B. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751. [PubMed] [Google Scholar]
- 83.Hipfner D R, Almquist K C, Leslie E M, Gerlach J H, Grant C E, Deeley R G, Cole S P. Membrane topology of the multidrug resistance protein (MRP). A study of glycosylation-site mutants reveals an extracytosolic NH2 terminus. J Biol Chem. 1997;272:23623–23630. doi: 10.1074/jbc.272.38.23623. [DOI] [PubMed] [Google Scholar]
- 84.Hresko R C, Kruse M, Strube M, Mueckler M. Topology of the Glut 1 glucose transporter deduced from glycosylation scanning mutagenesis. J Biol Chem. 1994;269:20482–20488. [PubMed] [Google Scholar]
- 85.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 86.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 87.Jander G, Cronan J E, Jr, Beckwith J. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J Bacteriol. 1996;178:3049–3058. doi: 10.1128/jb.178.11.3049-3058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Javitch J A. Probing structure of neurotransmitter transporters by substituted-cysteine accessibility method. Methods Enzymol. 1998;296:331–346. doi: 10.1016/s0076-6879(98)96025-6. [DOI] [PubMed] [Google Scholar]
- 89.Johansson M, Nilsson I, von Heijne G. Positively charged amino acids placed next to a signal sequence block protein translocation more efficiently in Escherichia coli than in mammalian microsomes. Mol Gen Genet. 1993;239:251–256. doi: 10.1007/BF00281625. [DOI] [PubMed] [Google Scholar]
- 90.Jones D T, Taylor W R, Thornton J M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 91.Jung H. Topology and function of the Na+/proline transporter of Escherichia coli, a member of the Na+/solute cotransporter family. Biochim Biophys Acta. 1998;1365:60–64. doi: 10.1016/s0005-2728(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 92.Jung H, Rubenhagen R, Tebbe S, Leifker K, Tholema N, Quick M, Schmid R. Topology of the Na+/proline transporter of Escherichia coli. J Biol Chem. 1998;273:26400–26407. doi: 10.1074/jbc.273.41.26400. [DOI] [PubMed] [Google Scholar]
- 93.Kaback H R. The lactose permease of Escherichia coli: past, present and future. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Transport processes in eukaryotic and prokaryotic organisms. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 203–227. [Google Scholar]
- 94.Kast C, Canfield V, Levenson R, Gros P. Membrane topology of P-glycoprotein as determined by epitope insertion: transmembrane organization of the N-terminal domain of mdr3. Biochemistry. 1995;34:4402–4411. doi: 10.1021/bi00013a032. [DOI] [PubMed] [Google Scholar]
- 95.Kast C, Canfield V, Levenson R, Gros P. Transmembrane organization of mouse P-glycoprotein determined by epitope insertion and immunofluorescence. J Biol Chem. 1996;271:9240–9248. doi: 10.1074/jbc.271.16.9240. [DOI] [PubMed] [Google Scholar]
- 96.Kast C, Gros P. Epitope insertion favors a six transmembrane domain model for the carboxy-terminal portion of the multidrug resistance-associated protein. Biochemistry. 1998;37:2305–2313. doi: 10.1021/bi972332v. [DOI] [PubMed] [Google Scholar]
- 97.Kimura T, Ohnuma M, Sawai T, Yamaguchi A. Membrane topology of the transposon 10-encoded metal-tetracycline/H+ antiporter as studied by site-directed chemical labeling. J Biol Chem. 1997;272:580–585. doi: 10.1074/jbc.272.1.580. [DOI] [PubMed] [Google Scholar]
- 98.Kimura-Someya T, Iwaki S, Yamaguchi A. Site-directed chemical modification of cysteine-scanning mutants as to transmembrane segment II and its flanking regions of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a transmembrane water-filled channel. J Biol Chem. 1998;273:32806–32811. doi: 10.1074/jbc.273.49.32806. [DOI] [PubMed] [Google Scholar]
- 99.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 100.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. Structural requirements for interruption of protein translocation across rough endoplasmic reticulum membrane. J Biochem. 1990;108:829–834. doi: 10.1093/oxfordjournals.jbchem.a123288. [DOI] [PubMed] [Google Scholar]
- 101.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. Systematic analysis of stop-transfer sequence for microsomal membrane. J Biol Chem. 1991;266:9251–9255. [PubMed] [Google Scholar]
- 102.Kuroiwa T, Sakaguchi M, Omura T, Mihara K. Reinitiation of protein translocation across the endoplasmic reticulum membrane for the topogenesis of multispanning membrane proteins. J Biol Chem. 1996;271:6423–6428. doi: 10.1074/jbc.271.11.6423. [DOI] [PubMed] [Google Scholar]
- 103.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 104.Landolt-Marticorena C, Reithmeier R A. Asparagine-linked oligosaccharides are localized to single extracytosolic segments in multi-span membrane glycoproteins. Biochem J. 1994;302:253–260. doi: 10.1042/bj3020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Landolt-Marticorena C, Williams K A, Deber C M, Reithmeier R A. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 106.LeBlanc H N, Beatty J T. Topological analysis of the Rhodobacter capsulatus PucC protein and effects of C-terminal deletions on light-harvesting complex II. J Bacteriol. 1996;178:4801–4806. doi: 10.1128/jb.178.16.4801-4806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee E, Manoil C. Mutations eliminating the protein export function of a membrane-spanning sequence. J Biol Chem. 1994;269:28822–28828. [PubMed] [Google Scholar]
- 108.Lee M, Manoil C. Molecular genetic analysis of membrane protein topology. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Transport processes of eukaryotic and prokaryotic organisms. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 189–201. [Google Scholar]
- 109.Lewis M J, Chang J A, Simoni R D. A topological analysis of subunit alpha from Escherichia coli F1F0-ATP synthase predicts eight transmembrane segments. J Biol Chem. 1990;265:10541–10550. [PubMed] [Google Scholar]
- 110.Li S J, Cronan J E J. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:855–863. [PubMed] [Google Scholar]
- 111.Liao S, Lin J, Do H, Johnson A E. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 112.Lingappa V R, Lingappa J R, Blobel G. Chicken ovalbumin contains an internal signal sequence. Nature. 1979;281:117–121. doi: 10.1038/281117a0. [DOI] [PubMed] [Google Scholar]
- 113.Lolkema J S, Slotboom D J. Estimation of structural similarity of membrane proteins by hydropathy profile alignment. Mol Membr Biol. 1998;15:33–42. doi: 10.3109/09687689809027516. [DOI] [PubMed] [Google Scholar]
- 114.Long J C, Wang S, Vik S B. Membrane topology of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J Biol Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 115.Loo T W, Clarke D M. Membrane topology of a cysteine-less mutant of human P-glycoprotein. J Biol Chem. 1995;270:843–848. doi: 10.1074/jbc.270.2.843. [DOI] [PubMed] [Google Scholar]
- 116.Lu Y, Xiong X, Helm A, Kimani K, Bragin A, Skach W R. Co- and posttranslational translocation mechanisms direct cystic fibrosis transmembrane conductance regulator N terminus transmembrane assembly. J Biol Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- 117.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 119.Martoglio B, Hofmann M W, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 120.Matlack K E, Mothes W, Rapoport T A. Protein translocation: tunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- 121.McAllister H C, Coon M J. Further studies on the properties of liver propionyl coenzyme A holocarboxylase synthetase and the specificity of holocarboxylase formation. J Biol Chem. 1966;241:2855–2861. [PubMed] [Google Scholar]
- 122.McKenna E, Hardy D, Kaback H R. Insertional mutagenesis of hydrophilic domains in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:11954–11958. doi: 10.1073/pnas.89.24.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Melchers K, Weitzenegger T, Buhmann A, Steinhilber W, Sachs G, Schafer K P. Cloning and membrane topology of a P type ATPase from Helicobacter pylori. J Biol Chem. 1996;271:446–457. doi: 10.1074/jbc.271.1.446. [DOI] [PubMed] [Google Scholar]
- 124.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miller J D, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 126.Moss K, Helm A, Lu Y, Bragin A, Skach W R. Coupled translocation events generate topological heterogeneity at the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2681–2697. doi: 10.1091/mbc.9.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mothes W, Heinrich S U, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport T A. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 128.Newitt J A, Bernstein H D. A mutation in the Escherichia coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
- 129.Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- 130.Nilsson I M, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 131.Olender E H, Simon R D. The intracellular targeting and membrane topology of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1992;267:4223–4235. [PubMed] [Google Scholar]
- 132.Olivares L, Aragon C, Gimenez C, Zafra F. Analysis of the transmembrane topology of the glycine transporter GLYT1. J Biol Chem. 1997;272:1211–1217. doi: 10.1074/jbc.272.2.1211. [DOI] [PubMed] [Google Scholar]
- 133.Ota K, Sakaguchi M, Hamasaki N, Mihara K. Assessment of topogenic functions of anticipated transmembrane segments of human band 3. J Biol Chem. 1998;273:28286–28291. doi: 10.1074/jbc.273.43.28286. [DOI] [PubMed] [Google Scholar]
- 134.Ota K, Sakaguchi M, von Heijne G, Hamasaki N, Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell. 1998;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- 135.Pan C J, Lei K J, Annabi B, Hemrika W, Chou J Y. Transmembrane topology of glucose-6-phosphatase. J Biol Chem. 1998;273:6144–6148. doi: 10.1074/jbc.273.11.6144. [DOI] [PubMed] [Google Scholar]
- 136.Pascual J M, Shieh C C, Kirsch G E, Brown A M. Multiple residues specify external tetraethylammonium blockade in voltage-gated potassium channels. Biophys J. 1995;69:428–434. doi: 10.1016/S0006-3495(95)79915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pi J, Pittard A J. Topology of the phenylalanine-specific permease of Escherichia coli. J Bacteriol. 1996;178:2650–2655. doi: 10.1128/jb.178.9.2650-2655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pohlschroder M, Prinz W A, Hartmann E, Beckwith J. Protein translocation in the three domains of life: variations on a theme. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 139.Popot J L, Engelman D M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 140.Popov M, Tam L Y, Li J, Reithmeier R A. Mapping the ends of transmembrane segments in a polytopic membrane protein. Scanning N-glycosylation mutagenesis of extracytosolic loops in the anion exchanger, band 3. J Biol Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 141.Pos K M, Bott M, Dimroth P. Purification of two active fusion proteins of the Na(+)-dependent citrate carrier of Klebsiella pneumoniae. FEBS Lett. 1994;347:37–41. doi: 10.1016/0014-5793(94)00502-8. [DOI] [PubMed] [Google Scholar]
- 142.Pouny Y, Weitzman C, Kaback H R. In vitro biotinylation provides quantitative recovery of highly purified active lactose permease in a single step. Biochemistry. 1998;37:15713–15719. doi: 10.1021/bi981519z. [DOI] [PubMed] [Google Scholar]
- 143.Pourcher T, Bibi E, Kaback H R, Leblanc G. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusion analysis. Biochemistry. 1996;35:4161–4168. doi: 10.1021/bi9527496. [DOI] [PubMed] [Google Scholar]
- 144.Prinz W A, Beckwith J. Gene fusion analysis of membrane protein topology: a direct comparison of alkaline phosphatase and beta-lactamase fusions. J Bacteriol. 1994;176:6410–6413. doi: 10.1128/jb.176.20.6410-6413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Prinz W A, Boyd D H, Ehrmann M, Beckwith J. The protein translocation apparatus contributes to determining the topology of an integral membrane protein in Escherichia coli. J Biol Chem. 1998;273:8419–8424. doi: 10.1074/jbc.273.14.8419. [DOI] [PubMed] [Google Scholar]
- 146.Rapiejko P J, Gilmore R. Signal sequence recognition and targeting of ribosomes to the endoplasmic reticulum by the signal recognition particle do not require GTP. Mol Biol Cell. 1994;5:887–897. doi: 10.1091/mbc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rapoport T A, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 148.Rees D C, DeAntonio L, Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 149.Rees D C, Komiya H, Yeates T O, Allen J P, Feher G. The bacterial photosynthetic reaction center as a model for membrane proteins. Annu Rev Biochem. 1989;58:607–633. doi: 10.1146/annurev.bi.58.070189.003135. [DOI] [PubMed] [Google Scholar]
- 150.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 151.Rothman A, Padan E, Schuldiner S. Topological analysis of NhaA, a Na+/H+ antiporter from Escherichia coli. J Biol Chem. 1996;271:32288–32292. doi: 10.1074/jbc.271.50.32288. [DOI] [PubMed] [Google Scholar]
- 152.Saaf A, Monne M, De Gier J W, von Heijne G. Membrane topology of the 60-kDa oxa1p homologue from Escherichia coli. J Biol Chem. 1998;273:30415–30418. doi: 10.1074/jbc.273.46.30415. [DOI] [PubMed] [Google Scholar]
- 153.Saaf A, Wallin E, von Heijne G. Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur J Biochem. 1998;251:821–829. doi: 10.1046/j.1432-1327.1998.2510821.x. [DOI] [PubMed] [Google Scholar]
- 154.Sahin-Toth M, Dunten R L, Kaback H R. Design of a membrane protein for site-specific proteolysis: properties of engineered factor Xa protease sites in the lactose permease of Escherichia coli. Biochemistry. 1995;34:1107–1112. doi: 10.1021/bi00004a001. [DOI] [PubMed] [Google Scholar]
- 155.Samols D, Thornton C G, Murtif V L, Kumar G K, Haase F C, Wood H G. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- 156.San Millan J L, Boyd D, Dalbey R, Wickner W, Beckwith J. Use of phoA fusions to study the topology of the Escherichia coli inner membrane protein leader peptidase. J Bacteriol. 1989;171:5536–5541. doi: 10.1128/jb.171.10.5536-5541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sarsero J P, Pittard A J. Membrane topology analysis of Escherichia coli K-12 Mtr permease by alkaline phosphatase and beta-galactosidase fusions. J Bacteriol. 1995;177:297–306. doi: 10.1128/jb.177.2.297-306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sato M, Hresko R, Mueckler M. Testing the charge difference hypothesis for the assembly of a eucaryotic multispanning membrane protein. J Biol Chem. 1998;273:25203–25208. doi: 10.1074/jbc.273.39.25203. [DOI] [PubMed] [Google Scholar]
- 159.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 160.Seal R P, Amara S G. A reentrant loop domain in the glutamate carrier EAAT1 participates in substrate binding and translocation. Neuron. 1998;21:1487–1498. doi: 10.1016/s0896-6273(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 161.Seligman L, Manoil C. An amphipathic sequence determinant of membrane protein topology. J Biol Chem. 1994;269:19888–19896. [PubMed] [Google Scholar]
- 162.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 163.Shenoy B C, Wood H G. Purification and properties of the synthetase catalyzing the biotination of the aposubunit of transcarboxylase from Propionibacterium shermanii. FASEB J. 1988;2:2396–2401. doi: 10.1096/fasebj.2.8.3360240. [DOI] [PubMed] [Google Scholar]
- 164.Silhavy T J, Shuman H A, Beckwith J, Schwartz M. Use of gene fusions to study outer membrane protein localization in Escherichia coli. Proc Natl Acad Sci USA. 1977;74:5411–5415. doi: 10.1073/pnas.74.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Simon S M, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 166.Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 167.Skach W R, Shi L B, Calayag M C, Frigeri A, Lingappa V R, Verkman A S. Biogenesis and transmembrane topology of the CHIP28 water channel at the endoplasmic reticulum. J Cell Biol. 1994;125:803–815. doi: 10.1083/jcb.125.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Skvirsky R C, Reginald S, Shen X. Topology analysis of the colicin V export protein CvaA in Escherichia coli. J Bacteriol. 1995;177:6153–6159. doi: 10.1128/jb.177.21.6153-6159.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Slotboom D J, Lolkema J S, Konings W N. Membrane topology of the C-terminal half of the neuronal, glial, and bacterial glutamate transporter family. J Biol Chem. 1996;271:31317–31321. doi: 10.1074/jbc.271.49.31317. [DOI] [PubMed] [Google Scholar]
- 170.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tam L Y, Landolt-Marticorena C, Reithmeier R A. Glycosylation of multiple extracytosolic loops in Band 3, a model polytopic membrane protein. Biochem J. 1996;318:645–648. doi: 10.1042/bj3180645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang X B, Fujinaga J, Kopito R, Casey J R. Topology of the region surrounding Glu681 of human AE1 protein, the erythrocyte anion exchanger. J Biol Chem. 1998;273:22545–22553. doi: 10.1074/jbc.273.35.22545. [DOI] [PubMed] [Google Scholar]
- 173.Tanner M J. The structure and function of band 3 (AE1): recent developments. Mol Membr Biol. 1997;14:155–165. doi: 10.3109/09687689709048178. [DOI] [PubMed] [Google Scholar]
- 174.Tate C G, Henderson P J. Membrane topology of the l-rhamnose-H+ transport protein (RhaT) from enterobacteria. J Biol Chem. 1993;268:26850–26857. [PubMed] [Google Scholar]
- 175.Traxler B, Boyd D, Beckwith J. The topological analysis of integral cytoplasmic membrane proteins. J Membr Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 176.Traxler B, Murphy C. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J Biol Chem. 1996;271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]
- 177.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 178.Turk E, Kerner C J, Lostao M P, Wright E M. Membrane topology of the human Na+/glucose cotransporter SGLT1. J Biol Chem. 1996;271:1925–1934. doi: 10.1074/jbc.271.4.1925. [DOI] [PubMed] [Google Scholar]
- 179.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:30862–30868. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 180.Ujwal M L, Jung H, Bibi E, Manoil C, Altenbach C, Hubbell W L, Kaback H R. Membrane topology of helices VII and XI in the lactose permease of Escherichia coli studied by lacY-phoA fusion analysis and site-directed spectroscopy. Biochemistry. 1995;34:14909–14917. doi: 10.1021/bi00045a036. [DOI] [PubMed] [Google Scholar]
- 181.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 182.Valent Q A, Scotti P A, High S, De Gier J W, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Valiyaveetil F I, Fillingame R H. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J Biol Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 184.van Geest M, Lolkema J S. Membrane topology of the sodium ion-dependent citrate carrier of Klebsiella pneumoniae. Evidence for a new structural class of secondary transporters. J Biol Chem. 1996;271:25582–25589. doi: 10.1074/jbc.271.41.25582. [DOI] [PubMed] [Google Scholar]
- 185.Reference deleted.
- 186.van Geest M, Lolkema J S. Transmembrane segment VIII of the Na+/citrate transporter CitS requires downstream TMS IX for insertion in the Escherichia coli membrane. J Biol Chem. 1999;274:29705–29711. doi: 10.1074/jbc.274.42.29705. [DOI] [PubMed] [Google Scholar]
- 187.van Geest M, Nilsson I, von Heijne G, Lolkema J S. Insertion of a bacterial secondary transport protein in the endoplasmic reticulum membrane. J Biol Chem. 1999;274:2816–2823. doi: 10.1074/jbc.274.5.2816. [DOI] [PubMed] [Google Scholar]
- 188.van Iwaarden P R, Pastore J C, Konings W N, Kaback H R. Construction of a functional lactose permease devoid of cysteine residues. Biochemistry. 1991;30:9595–9600. doi: 10.1021/bi00104a005. [DOI] [PubMed] [Google Scholar]
- 189.Vannier B, Zhu X, Brown D, Birnbaumer L. The membrane topology of human transient receptor potential 3 as inferred from glycosylation-scanning mutagenesis and epitope immunocytochemistry. J Biol Chem. 1998;273:8675–8679. doi: 10.1074/jbc.273.15.8675. [DOI] [PubMed] [Google Scholar]
- 190.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 191.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 192.von Heijne G. Membrane proteins: from sequence to structure. Annu Rev Biophys Biomol Struct. 1994;23:167–192. doi: 10.1146/annurev.bb.23.060194.001123. [DOI] [PubMed] [Google Scholar]
- 193.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 194.Wahle S, Stoffel W. Membrane topology of the high-affinity l-glutamate transporter (GLAST-1) of the central nervous system. J Cell Biol. 1996;135:1867–1877. doi: 10.1083/jcb.135.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 196.Welply J K, Shenbagamurthi P, Lennarz W J, Naider F. Substrate recognition by oligosaccharyltransferase. Studies on glycosylation of modified Asn-X-Thr/Ser tripeptides. J Biol Chem. 1983;258:11856–11863. [PubMed] [Google Scholar]
- 197.Wessels H P, Spiess M. Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell. 1988;55:61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- 198.Whitley P, Zander T, Ehrmann M, Haardt M, Bremer E, von Heijne G. Sec-independent translocation of a 100-residue periplasmic N-terminal tail in the E. coli inner membrane protein proW. EMBO J. 1994;13:4653–4661. doi: 10.1002/j.1460-2075.1994.tb06788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wilkinson B M, Critchley A J, Stirling C J. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J Biol Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- 200.Wilmes-Riesenberg M R, Wanner B L. TnphoA and TnphoA′ elements for making and switching fusions for study of transcription, translation, and cell surface localization. J Bacteriol. 1992;174:4558–4575. doi: 10.1128/jb.174.14.4558-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wolfe P B, Rice M, Wickner W. Effects of two sec genes on protein assembly into the plasma membrane of Escherichia coli. J Biol Chem. 1985;260:1836–1841. [PubMed] [Google Scholar]
- 202.Wu J, Tisa L S, Rosen B P. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J Biol Chem. 1992;267:12570–12576. [PubMed] [Google Scholar]
- 203.Xu M, Akabas M H. Amino acids lining the channel of the gamma-aminobutyric acid type A receptor identified by cysteine substitution. J Biol Chem. 1993;268:21505–21508. [PubMed] [Google Scholar]
- 204.Xu M, Covey D F, Akabas M H. Interaction of picrotoxin with GABAA receptor channel-lining residues probed in cysteine mutants. Biophys J. 1995;69:1858–1867. doi: 10.1016/S0006-3495(95)80056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yan R T, Maloney P C. Residues in the pathway through a membrane transporter. Proc Natl Acad Sci USA. 1995;92:5973–5976. doi: 10.1073/pnas.92.13.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Young C S, Beatty J T. Topological model of the Rhodobacter capsulatus light-harvesting complex I assembly protein LhaA (previously known as ORF1696) J Bacteriol. 1998;180:4742–4745. doi: 10.1128/jb.180.17.4742-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yu N, Cao Y, Mager S, Lester H A. Topological localization of cysteine 74 in the GABA transporter, GAT1, and its importance in ion binding and permeation. FEBS Lett. 1998;426:174–178. doi: 10.1016/s0014-5793(98)00333-0. [DOI] [PubMed] [Google Scholar]
- 208.Yun C H, Van Doren S R, Crofts A R, Gennis R B. The use of gene fusions to examine the membrane topology of the L subunit of the photosynthetic reaction center and of the cytochrome b subunit of the bc1 complex from Rhodobacter sphaeroides. J Biol Chem. 1991;266:10967–10973. [PubMed] [Google Scholar]
- 209.Zen K H, Consler T G, Kaback H R. Insertion of the polytopic membrane protein lactose permease occurs by multiple mechanisms. Biochemistry. 1995;34:3430–3437. doi: 10.1021/bi00010a035. [DOI] [PubMed] [Google Scholar]
- 210.Zhang J T, Chen M, Han E, Wang C. Dissection of de novo membrane insertion activities of internal transmembrane segments of ATP-binding-cassette transporters: toward understanding topological rules for membrane assembly of polytopic membrane proteins. Mol Biol Cell. 1998;9:853–863. doi: 10.1091/mbc.9.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhang J T, Lee C H, Duthie M, Ling V. Topological determinants of internal transmembrane segments in P-glycoprotein sequences. J Biol Chem. 1995;270:1742–1746. doi: 10.1074/jbc.270.4.1742. [DOI] [PubMed] [Google Scholar]
- 212.Zhou J, Fazzio R T, Blair D F. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]