Abstract
Engineered mesenchymal stem cells (MSCs) have been investigated extensively for gene delivery and, more recently, for targeted small molecule delivery. While preclinical studies demonstrate the potential of MSCs for targeted delivery, clinical studies suggest that tumor homing of native MSCs may be inefficient. We report here a surprising finding that loading MSCs with the anticancer drug paclitaxel (PTX) by nanoengineering results in significantly improved tumor homing compared to naïve MSCs. Loading PTX in MSCs results in increased levels of mitochondrial reactive oxygen species (ROS). In response to this oxidative stress, MSCs upregulate two important set of proteins. First were critical antioxidant proteins, most importantly nuclear factor erythroid 2-like 2 (Nrf2), the master regulator of antioxidant responses; upregulation of antioxidant proteins may explain how MSCs protect themselves from drug-induced oxidative stress. The second was CXCR4, a direct target of Nrf2 and a key mediator of tumor homing; upregulation of CXCR4 suggested a mechanism that may underlie the improved tumor homing of nanoengineered MSCs. In addition to demonstrating the potential mechanism of improved tumor targeting of nanoengineered MSCs, our studies reveal that MSCs utilize a novel mechanism of resistance against drug-induced oxidative stress and cell death, explaining how MSCs can deliver therapeutic concentrations of cytotoxic payload while maintaining their viability.
Keywords: Mesenchymal stem cells, Tumor homing, Nanoengineering, Targeted drug delivery, Drug resistance, Oxidative stress
Graphical abstract
1. Introduction
Cell-based delivery systems offer the possibility of precise and truly active targeted delivery of therapeutic agents to specific tissues in the body. Cellular carriers investigated for targeted delivery include macrophages, natural killer cells, erythrocytes, and mesenchymal stem (or stromal) cells (MSCs). In addition to improving drug bioavailability at the target, all these carriers provide other advantages including prolonged delivery times and biocompatibility. MSCs in particular, possess several unique properties [1], which make them highly attractive for tumor targeted drug delivery. Unlike embryonic stem cells, MSCs do not form teratomas or malignancies [2]. They can be easily isolated from many adult tissues including adipose tissue and bone marrow [[3], [4], [5], [6]]. MSCs can be expanded, engineered in vitro, and subsequently re-grafted [7]. Clinical trials investigating MSCs have successfully utilized both autologous and allogeneic sources [8]. Further, MSCs do not express, or express at very low levels, HLA class I and II molecules and thus do not suffer from significant immunogenicity concerns [9,10]. In addition, absence of co-stimulatory molecules CD40, 80 and 86, which play a key role in the initiation of immune response, allows for the use of allogeneic MSCs [[11], [12], [13], [14], [15], [16]]. Because of their ability to migrate through tissue barriers, MSCs can infiltrate the tumor matrix effectively and enhance the intra-tumoral distribution of the payload they carry [17]. Genetically modified MSCs expressing various cytokines have been shown to inhibit the growth of solid tumors [7,18]. We have previously demonstrated that MSCs can be functionalized with nano drug delivery systems (nanoengineering), extending their application to small molecule drug delivery [[19], [20], [21], [22]]. While these advances are highly exciting and demonstrate the promise for anticancer delivery, MSC-based anti-cancer therapies have not yet progressed in clinical trials. A recent phase I clinical study in prostate cancer patients observed the lack of a detectable number of MSCs in primary tumors [23].
Although data from the above clinical trial suggests that native MSCs do not home effectively to tumors, several pre-clinical studies show that MSCs engineered to carry cytotoxic drugs such as paclitaxel (PTX) actively home to lung tumors and facilitate sustained delivery of the payload at the target site and effective inhibition of tumor growth in multiple mouse tumor models [[24], [25], [26]]. We posited that this apparent inconsistency might be because nanoengineering of MSCs impacts their biology, resulting in a favorable alteration of their in vivo disposition. To test this idea and to identify targetable mechanisms that might underlie the efficient homing of PTX loaded MSCs, we performed global label free unbiased proteomics and follow-up characterization studies on nanoengineered MSCs. Our studies show that loading PTX into MSCs upregulated two important set of proteins. First were critical antioxidant proteins, most importantly nuclear factor erythroid 2-like 2 (Nrf2), the master regulator of antioxidant responses; upregulation of antioxidant proteins may explain how loaded MSCs protect themselves from drug-induced oxidative stress. The second was CXCR4, a direct target of Nrf2 and a key mediator of tumor homing; upregulation of CXCR4 suggested a mechanism that may underlie the improved tumor targeting of nanoengineered MSCs.
For successful drug delivery, the carrier cells must be resistant to the pharmacological effects of the payload. This is especially relevant for cytotoxic payloads such as chemotherapy drugs. In addition to demonstrating the potential mechanism of improved tumor targeting of nanoengineered MSCs, our studies reveal that MSCs utilize a novel mechanism of resistance against drug-induced oxidative stress and cell death.
2. Materials and methods
2.1. Materials
PTX and polyvinyl alcohol (PVA) were purchased from Sigma (St. Louis, MO). Polymer poly (dl-lactide-co-glycolide) (50:50 lactide to glycolide ratio; inherent viscosity: 0.55–0.75 dL/g) (PLGA) was obtained from Lactel Absorbable Polymers (Birmingham, AL). Methanol and acetonitrile were purchased from Fisher Scientific (Hampton, NH). Cell culture supplies (Fetal bovine serum (FBS) and penicillin/streptomycin, Dulbecco's phosphate-buffered saline (DPBS), Dulbecco's modified eagle medium (DMEM), and trypsin-EDTA were procured from Invitrogen Corporation (Carlsbad, CA). Allophycocyanin (APC) labeled anti-human CXCR4 and phycoerythrin (PE) labeled anti-human CXCR7 antibodies were purchased from BioLegend Antibodies (San Diego, CA, USA). N-azidoacetylmannosamine-tetraacylated (Ac4ManNAz) was purchased from Click Chemistry Tools (Scottsdale, AZ). The Seahorse XF Cell Mito Stress Test Kit containing oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) and rotenone/antimycin A was purchased from Agilent Technologies, (Santa Clara, CA). The human Nrf2 ELISA kit was purchased from Ray Biotech (Peachtree Corners, GA). TaqMan Gene Expression Assays were obtained from Applied Biosystems (Waltham, MA). MitoSOX™ reagent, Calcein AM, Hoechst 33,324, N-acetylcysteine (NAC) and tert-butyl hydroperoxide (TBHP) were purchased from Life Technologies, Invitrogen Corporation (CA, USA).
2.2. Methods
2.2.1. Cell culture
Human bone marrow derived MSCs (ScienCell Research Laboratories, Carlsbad, CA, USA) were cultured in human MSC media (ScienCell Research Laboratories, Carlsbad, CA). Lewis lung carcinoma (LL/2-luc, PerkinElmer Health Sciences, Shelton, CT, USA) were grown in DMEM supplemented with 10% v/v FBS and 1% v/v penicillin and streptomycin. All cell lines were maintained and monitored for morphology and growth characteristics at 5% carbon dioxide, 37 °C controlled humidity incubator. Cells were passaged two to three times prior to using them for in vivo or in vitro experiments.
2.2.2. Fabrication of PTX loaded nanoparticles
Two different PLGA nanoparticle formulations, non-surface functionalized and surface functionalized with PEG-dibenzocyclooctyne (DBCO), were fabricated as described below.
Non-surface functionalized, PLGA nanoparticles loaded with PTX (PTX NP) were formulated using an emulsion-solvent evaporation technique as described earlier [19,20,22]. In brief, PLGA (32 mg) and PTX (8 mg) were dissolved in chloroform and emulsified with aqueous PVA solution (2.5% w/v) to generate an oil-in-water emulsion using probe sonication (18–21 W, 5 min). The emulsion was stirred overnight under ambient conditions (∼18 h, 25 °C) and then for I hr under vacuum to remove chloroform. Nanoparticles were separated using ultracentrifugation (35,000 rpm; 35 min; 4 °C, Optima XPN-80 Ultracentrifuge, 70 Ti rotor, Beckman Coulter, Brea, CA) and washed three times to remove unencapsulated drug and residual PVA. Resulting suspension was centrifuged at 1000 rpm for 5 min (Allegra X-30 R, SX 4400 rotor, Beckman Coulter) to remove any large aggregates. Final nanoparticles were obtained by lyophilization (Labconco, FreeZone 4.5, Kansas City, MO). Nanoparticles without PTX (Blank NP) were manufactured similarly.
DBCO-functionalized PLGA nanoparticles (DBCO-PTX NP) loaded with PTX were formulated using an established surface functionalization technique described previously [19]. The oil-in-water emulsion comprising of PLGA and PTX in the chloroform phase and PVA aqueous phase was prepared as described above. Block co-polymer PLA-PEG-DBCO was synthesized as described earlier [27]. The block co-polymer was dissolved in chloroform (8 mg in 200 μL) and added slowly to the oil-in-water emulsion under continuous stirring. Chloroform was evaporated and nanoparticles collected by repeated ultracentrifugation steps and lyophilized as described above. Blank DBCO-functionalized drug-free nanoparticles were manufactured similarly.
2.2.3. Characterization of nanoparticles
Nanoparticles were characterized for hydrodynamic diameter and morphology using photon correlation spectroscopy and transmission electron microscopy (TEM), respectively. In brief, nanoparticle dispersion in deionized water (0.1 mg/mL) was used to measure particle size using a Nanobrook Omni (Brookhaven Instruments, Holtsville, NY) operating at a 165° constant scattering angle. For TEM, a dispersion of nanoparticles was added onto a copper grid covered by a continuous thin carbon film (400 mesh, Ted Pella Inc., Redding, CA, USA) and air-dried overnight. Images were acquired on a FEI Tecnai G2 F30 S-TWIN instrument at 300 kV with a Gatan UltraScan 4000 CCD camera (Gatan, Pleasanton, CA, USA) and using Gatan Digital Micrograph 3.9.5 software (Gatan, Pleasanton, CA, USA). A 3–6 μm of under-focus was used to improve the phase contrast. Drug loading in nanoparticles was determined following extraction in methanol. Nanoparticle dispersion in methanol (1 mg/mL) was agitated overnight using a tube rotator (VWR International, Radnor, PA). The following day, dispersion was centrifuged at 13,000 rpm for 30 min to remove nanoparticles. The supernatant was quantified for PTX amount using reversed-phase high-performance liquid chromatography (HPLC) as described previously (Agilent Technologies, Santa Clara, CA) [22].
2.2.4. Nanoengineering of MSCs
Two different nanoengineering strategies were used to incorporate PTX in MSCs. First strategy consisted of incubation of MSCs with PTX loaded polymeric nanoparticles (PTX-NP) for endocytic uptake [19,22]. MSCs were incubated with PLGA-PTX NP (100 μg/mL) in Opti-MEM reduced serum media for 4 h at 37 °C, washed with PBS, and used for further studies. The second strategy consisted of covalent conjugation of PTX loaded, DBCO-functionalized polymeric nanoparticles (DBCO-PTX NP) to the surface of MSCs with azide functional groups (MSC-Az) [27]. MSC-Az were prepared by incubating MSCs with Ac4ManNAz sugars in growth medium (20 μM) for 3 days [19]. MSC-Az were incubated with PTX DBCO NP (100 μg/mL) in Opti-MEM reduced serum media for 4 h at 37 °C. The cells were washed repeatedly with PBS to remove unconjugated/unencapsulated nanoparticles prior to their use in further studies.
2.2.5. Effect of nanoengineering on CXCR4/7 expression in MSCs
Effect of nanoengineering on CXCR4 and CXCR7 expression was determined using flow cytometry. MSCs were nanoengineered, washed with PBS, and incubated with complete or serum free media for 16 h. Cells were then stained with APC labeled anti-human CXCR4 and PE labeled anti-human CXCR7 at 4 °C for 1 h. Cells were washed with DBPS thrice and, APC and PE associated fluorophore intensity were monitored by flow cytometry (LSRII, BD Bioscience, Franklin Lakes, NJ). Naïve MSCs stained similarly were used as controls.
2.2.6. Effect of nanoengineering on MSC proteome
Label-free proteomics using a modified in-stage tip (iST) was performed as previously described [28]. Briefly, following cell lysis, proteins were extracted with 6 M guanidium hydrochloride buffer, digested with Lys-C and followed by a second digestion with trypsin. The solution was then acidified and centrifuged. The supernatants were fractionated using activated in-house-made cation stage tips [29,30] and the peptides were eluted using elution buffers as previously described [31]. Mass spec analyses were performed on these fractions using the Q Exactive mass spectrometer. Mass spectra processing was performed with Proteome Discoverer v2.5. The generated de-isotoped peak list was submitted to an in-house Mascot server 2.2.07, Sequest HT and MS Amanda 2.0 for searching against the Swiss-Prot database. The search parameters were set as follows: species, homo sapiens; enzyme, trypsin with maximal two missed cleavage; fixed modification, cysteine carboxymethylation; 10 ppm mass tolerance for precursor peptide ions; 0.02 Da tolerance for MS/MS fragment ions.
Ingenuity Pathway Analysis (IPA): To identify the biological networks of differentially expressed proteins, we performed pathway analysis using Ingenuity Pathways Analysis software as previously described [29,[32], [33], [34], [35], [36], [37], [38]]. Ingenuity Pathways Analysis was optimized to include up to 35 proteins in a network. Proteins were considered upregulated when the levels were >1.3-fold compared to control and down regulated when the levels were <0.7-fold compared to control.
2.2.7. Effect of nanoengineering on ROS production
Effect of nanoengineering on intracellular ROS generation was studied using High-Content Analysis with CellROX® Oxidative Stress Reagents (Life Technologies, Invitrogen, USA). Cells were plated at a density of 1.6 × 104 cells/well in collagen coated PhenoPlate™ 384-well microplate (PerkinElmer, USA). The cells were nanoengineered as described previously. Negative control samples were pre-treated with 1000 μM of antioxidant, N-acetylcysteine (NAC) for 1 h before adding the inducer tert-butyl hydroperoxide (TBHP), while positive control samples were incubated with 200 μM of TBHP for 60 min. Following nanoengineering, cells in some of the wells were further incubated in either serum-free or serum containing MSC medium for 16 h. Following incubation with different treatment groups, cells were washed twice with 1X PBS and stained with 5 μM of MitoSOX™ reagent (510/580 nm) for 10–15 min. Cell viability was determined using 2 μM of Calcein AM (496/516 nm). The cells were then imaged real-time using the Operetta® CLSTM High Content Analysis System (PerkinElmer, USA). ROS production was determined using average fluorescence intensity of ∼2000 cells in 24 different fields from six replicates. Untreated cells were used as controls and stained simultaneously. Unstained cells were used to correct for any background autofluorescence. Single-cell intensity of ROS staining was quantified using Harmony 4.8 software from PerkinElmer.
2.2.8. Effect of nanoengineering on mitochondrial function
Effect of nanoengineering on mitochondrial function was determined using JC-1 mitochondrial potential assay kit (Cayman Chemicals). MSCs were nanoengineered, washed with PBS and stained immediately or incubated with complete or serum free media for 16 h. Cells were then incubated with JC-1 staining solution for 15–20 min and then washed at 400×g for 5 min. After washing, J-aggregates were monitored via fluorescence measurements at excitation and emission wavelengths of 535 nm and 595 nm, respectively. J-monomers were monitored via fluorescence measurements at excitation and emission wavelengths of 485 nm and 535 nm, respectively. The ratio of the fluorescence intensity of J-aggregates to fluorescence intensity of monomers was used as an indicator of cell health.
2.2.9. Effect of nanoengineering on cellular metabolic function
Oxygen consumption rate (OCR), a key indicator of mitochondrial respiration and glycolysis as well as ATP production rate, of nanoengineered and control MSCs was measured using Agilent Seahorse XFe96 analyzer (Agilent, Santa Clara, CA). MSCs were cultured at the cell density of 10,000 cells/well in 96-well microplates for 24 h. On the day of the assay, cells were incubated with nanoparticles (100 μg/mL) for 4 h, washed once, and then incubated in XF Seahorse Base Medium supplemented with 10 mM glucose, 1 mM sodium pyruvate, and 2 mM l-glutamine. Seahorse XF96 Analyzer was calibrated using the sensor cartridge that was pre-hydrated in Seahorse XF Calibrant. OCR readings over time were recorded under basal conditions and after the addition of mitochondrial inhibitors oligomycin (25 μM), carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP, 7.5 μM) and rotenone & antimycin A (5 μM).
2.2.10. Effect of nanoengineering on Nfe2l2 (Nrf2) expression
Nfe2l2 protein levels were analyzed using ELISA while the mRNA expression was quantified using real-time polymerase chain reaction (RT-PCR). MSCs were nanoengineered as described above. The cells were then washed with PBS and harvested immediately or incubated with complete or serum free media for 16 h and then harvested. Cells were then lysed, and Nfe2l2 levels were quantified by ELISA as per manufacturer's (RayBiotech) protocol. For mRNA expression, total RNA was extracted from the cell lysate using RNeasy mini kit (Qiagen, Hilden, Germany). RNA concentrations were determined with a spectrophotometer (NanoDrop; Thermo Fisher Scientific). RNA was reverse transcribed (RT) using Moloney murine leukemia virus reverse transcriptase (Ambion) and a mixture of anchored oligo-dT and random decamers (Integrated DNA Technologies). Two reverse-transcription reactions were performed for each sample using either 50 or 12.5 ng of input RNA in a final volume of 50 μL. SYBR Green assays were used in combination with PowerUp SYBR Green Master mix (Applied Biosystems/ThermoFisher Scientific) and run on a QuantStudio 6 detection system (Applied Biosystems/ThermoFisher). Two μl of cDNA were used per PCR reaction (10 μl final volume). Cycling conditions were 95 °C, 15 min, followed by 40 (two-step) cycles (95 °C, 15 s; 60 °C, 60 s). The Ct (cycle threshold) values for each sample were averaged from two independent PCRs. RPLP0 was used as the normalizer. The sequences of the primers are RPLP0-F: CCCATTCTATCATCAACGGGTACAA, RPLP0-R and CAGCAAGTGGGAAGGTGTAATCC, NFEL2-F: ATCCATTCCTGAGTTACAGTGTC and NFEL2-R: ACTTCTGTCAGTTTGGCTTCT. Relative expression was determined using Ct values (QuantStudio™ 3 Real-Time PCR System, Applied Biosystems), and fold change was calculated using normalized Ct values of RPLPO, housekeeping gene, as per 2^-(ΔΔCt) method [39].
2.2.11. Biodistribution of nanoengineered MSCs
Animal model: All the experiments involving animals were performed according to the guidelines of Temple University's IACUC. Temple University's laboratory animal care and use program has been accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC, A3594-01). For these studies, we used the orthotopic LL/2 murine lung tumor model. This model is syngeneic to immunocompetent C57/Bl6 mouse, and thus considers a fully functional immune system, which is important for cell-based systems. Also, since we are interested in developing MSC-based therapies for non-small cell lung cancer, this is an appropriate model to use in our studies. The LL/2-luc cells (1 × 106 cells in 200 μL of DPBS) were injected through tail vein in C57BL/6 to develop syngeneic lung tumors. Tumor growth was monitored using bioluminescence imaging. D-luciferin potassium salt solution (150 mg/kg; Gold Biotechnology, St. Louis, MO) was injected intraperitoneally and tumor associated bioluminescence was measured using an In Vivo Imaging System (IVIS, Caliper Life Sciences, Hopkinton, MA).
Tumor homing studies: Tumor homing of nanoengineered MSCs was determined by analyzing the MSCs levels in lung tumors. Once the tumor bioluminescence reached 5 × 106 photons/sec, either nanoengineered MSCs (1 × 106 cells in 200 μL of DPBS) or naïve MSCs were injected through the tail vein. A set of mice were euthanized 6 h and 7 days after injection and lung tissue was preserved in RNA-later. Preserved lung tissue was then homogenized using PowerGen 125 tissue homogenizer (Fisher Scientific, Waltham, MA) and total RNA was extracted using a RNeasy mini kit (Qiagen, Hilden, Germany). An equivalent amount of total RNA (2 μg) for each sample was reverse-transcribed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA). The resulting cDNA was used for qPCR using the TaqMan Gene Expression Master Mix (Applied Biosystems, Waltham, MA) and TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA) for THY1 (Hs06633377_s1) and GAPDH (Hs02786624_g1). Real-time PCR reactions were performed in quadruplets QuantStudio 12 K Flex detection system (Applied Biosystems, Waltham, MA) according to the recommended cycling program (2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C). Relative expression was quantified using Ct values and fold change in expression was calculated by normalization to the Ct of control MSC group, according to 2^-(ΔΔCt) method [39].
3. Results
3.1. Nanoengineering of MSCs
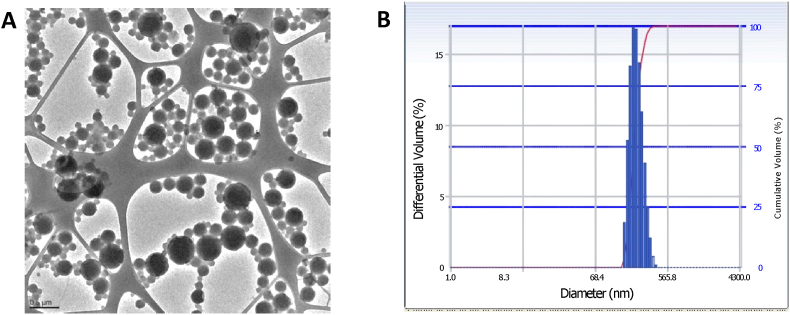
We utilized two different nanoengineering strategies to incorporate different amounts of PTX in MSCs. These included simple incubation with PTX loaded nanoparticles for endocytic uptake into MSCs and covalent conjugation of drug-loaded nanoparticles to the surface of MSCs. For endocytic uptake, nanoparticles were fabricated with a mean hydrodynamic diameter of 217 ± 2.9 nm and a polydispersity index of 0.21. Nanoparticles exhibited a net negative surface charge of −13.61 ± 8.2 mV. PTX loading in these nanoparticles was 18.5 ± 1.9% w/w. For surface conjugation, nanoparticles surface functionalized with DBCO functional groups were fabricated with a mean hydrodynamic diameter of 228.7 ± 44.2 nm, net negative surface charge of −11.64 ± 7.3 mV and a polydispersity index of 0.21. As can be seen from the TEM image in Fig. 1, PTX loaded, DBCO surface functionalized nanoparticles were spherical in shape and were in the 150–300 nm size range. As previous studies have shown, particle size obtained through dynamic light scattering technique is typically larger than that from microscopic techniques because of hydration and aggregation effects [40]. PTX loading in DBCO surface functionalized nanoparticles was 21.6 ± 10.9% w/w. Similar to that shown in our previous studies [22,27], surface conjugation of nanoparticles resulted in significantly higher PTX loading in MSCs (43.8 ± 2.5 pg/cell) relative to that with endocytic uptake (8.3 ± 0.6 pg/cell). We have previously shown that neither loading method adversely affects MSC viability, migratory, or differentiation properties [22,27].
Fig. 1.
Particle size distribution as measured by A) transmission electron microscopy and B) photon correlation spectroscopy of DBCO functionalized nanoparticles.
3.2. PTX loading triggers antioxidant response in MSCs
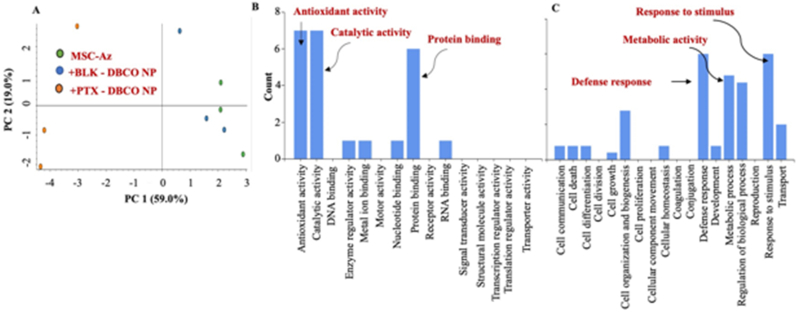
We performed global label-free, unbiased proteomics on nanoengineered MSCs using modified inStage technology as we reported before [29,[32], [33], [34], [35], [36], [37], [38]]. These studies show that MSCs nanoengineered with PTX underwent significant changes in the overall proteome (Fig. 2a), while the proteome of MSCs treated with blank nanoparticles was not significantly different from that of untreated MSCs. Analysis of molecular function profile revealed that loading PTX in MSCs significantly enhanced the expression of proteins involved in antioxidant and catalytic activity as well as in protein binding (Fig. 2b). Biological process profile also showed a similar increase in defense response as well as metabolic processes (Fig. 2c). Fig. 3a is a heat map depicting increases in proteins involved in antioxidant activity while Fig. 3b is a heat map showing increases in proteins involved in defense response.
Fig. 2.
Global label-free unbiased proteomics on MSCs nanoengineered using DBCO-functionalized nanoparticles. (A) PCA plot. (B) Molecular function profile (C) Biological process profile.
Fig. 3.
Global label-free unbiased proteomics on MSCs nanoengineered using DBCO-functionalized nanoparticles. (A) Grouped heat map showing the specific antioxidant proteins impacted by PTX loading (n = 3, p < 0.05). (B) Heat map showing the specific defense response proteins affected by PTX loading (n = 3, p < 0.05).
Further analysis of the various proteins that were upregulated showed that loading PTX results in the upregulation of antioxidant proteins including hemeoxygenase-1 (HO-1) and superoxide dismutase (Mn-SOD) (Fig. 4).
Fig. 4.
Hemeoxygenase-1 (left) and Mn-superoxide dismutase (right) levels in MSCs nanoengineered using DBCO-functionalized nanoparticles. Results significant at ∗p < 0.01, ∗∗p < 0.001.
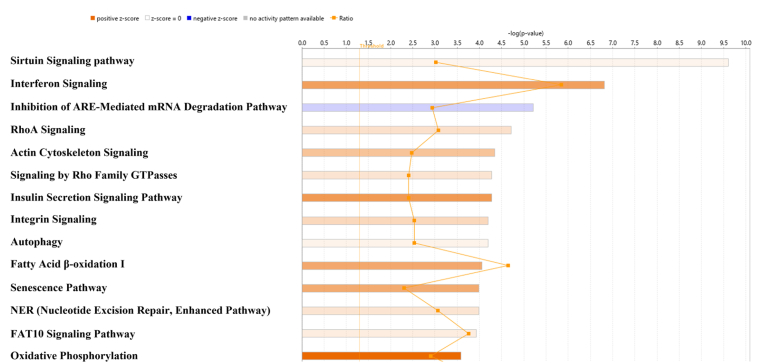
To determine physical and functional interactions, we performed canonical pathway analysis using IPA software comparing MSCs nanoengineered with PTX NP to MSCs nanoengineered with blank nanoparticles. The canonical pathways with largest differences between MSCs with and without PTX are shown in Fig. 5 and includes oxidative phosphorylation where complex I was downregulated whereas complex II, complex III and complex V, which are responsible for generating oxidative stress, were upregulated (Fig. 6).
Fig. 5.
Ingenuity Pathways Analysis for differentially regulated proteins. Orange squares connected with line represent ratio of the number of altered proteins to the total number of proteins in the pathway. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Schematic representation of effect of nanoengineering on mitochondrial protein expression. Red, up-regulated proteins; green, down-regulated proteins; white, proteins known to be in the network but were not identified in our analysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. MSCs are resistant to PTX induced oxidative stress
Since proteome data suggested that mitochondrial oxidative phosphorylation was impacted by PTX, which is known to induce ROS generation and oxidative stress [41,42], we next determined the effect of PTX loading on ROS levels in MSCs. These studies showed that incorporation of PTX in MSCs using either nanoengineering technique resulted in increased mitochondrial ROS generation at the end of 4 h incubation compared to that in untreated cells or in cells treated with blank nanoparticles (Fig. 7A and B). Blank nanoparticles induced slightly increased mitochondrial ROS generation; however, the ROS levels were lower than in cells treated with PTX nanoparticles. Interestingly, ROS levels returned to baseline levels once the nanoparticle treatments were removed and the cells were further incubated in serum-free or serum containing medium.
Fig. 7.
Effect of nanoengineering on intracellular ROS generation. A) The rate of ROS production is shown MSCs immediately after 4 h of nanoengineering, 16 h in serum-containing medium (SM) or 16 h in serum-free medium (SFM) after 4 h of nanoengineering. B) Representative live cell confocal images. Mitochondrial ROS was detected using MitoSox staining (orange) in cells (blue nuclei). (Scale bars: 50 μm). Data significant at ∗∗∗∗P < 0.00001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
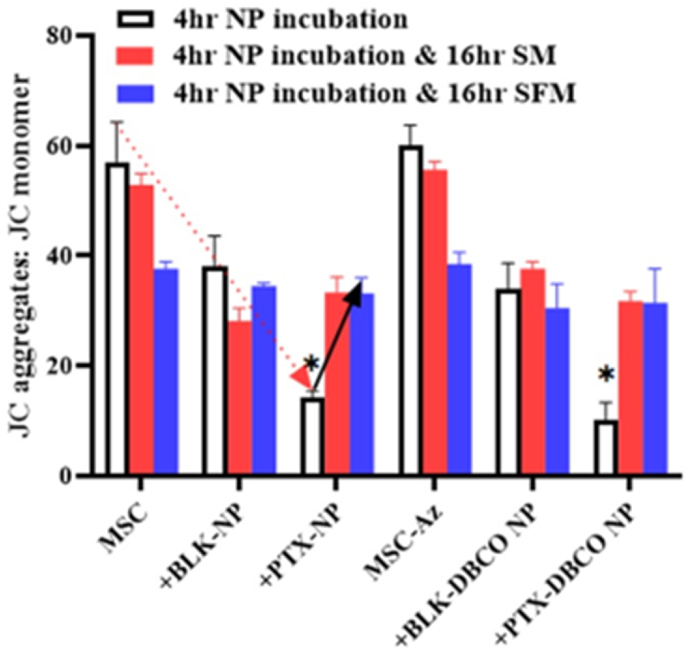
ROS generation in the mitochondria can result in oxidative damage to the organelle and can impact its ability to carry out a range of metabolic activities [43]. This mitochondrial disruption includes changes in the membrane potential and alterations to the redox potential of the mitochondria. In our studies, we evaluated the effect of nanoengineering on mitochondrial membrane potential using the JC-1 dye, which exhibits potential-dependent accumulation in mitochondria and a shift in fluorescence emission from ∼529 nm for the monomeric form to ∼590 nm for J-aggregates. Loss of mitochondrial potential is indicated by a decrease in the red/green fluorescence intensity (aggregate to monomer) ratio. Nanoengineering with PTX resulted in a significant depolarization of mitochondrial membrane potential immediately after nanoengineering (Fig. 8, red arrow). Cells appear to reverse this effect over the next 16 h (Fig. 8, black arrow), and this reversal was independent of whether cells were subjected to additional stress (serum-free conditions) or are provided exogenous antioxidant activity (albumin from serum). These studies suggests that MSCs quickly quench the increased ROS levels in response to PTX loading by generating antioxidant proteins, which likely scavenge any further ROS generated in serum-free medium.
Fig. 8.
Effect of nanoengineering on mitochondrial membrane potential. Mitochondrial dysfunction (JC-1 aggregate/monomer ratio) is shown for MSCs immediately after 4 h of nanoengineering, 16 h in serum-containing medium (SM) or 16 h in serum-free medium (SFM) after 4 h of nanoengineering. Results significant at ∗p < 0.001.
Loss of mitochondrial membrane potential is often associated with the opening of the mitochondrial permeability transition pore (MPTP), which allows the equilibration of ions and decoupling of the respiratory chain [44]. The effect of loading PTX on MSC cellular bioenergetics was assessed using the Seahorse extracellular flux analyzer as previously reported [44]. This technique uses selected inhibitors, which enables the estimation of main parameters that describe mitochondrial function including basal oxygen consumption rate (OCR), ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity, and non-mitochondrial OCR. These studies show no effect of nanoengineering on MSC bioenergetics at 4 h (Suppl Fig. S1). When nanoengineered MSCs were further incubated under serum-free conditions, we noted a decline in non-mitochondrial OCR and spare respiratory capacity (Fig. 9). The former was somewhat unexpected because non-mitochondrial OCR has been shown to increase in the presence of stressors such as ROS that affect mitochondrial integrity [45]. Cell exposure to nanoparticles (both blank and drug-loaded) likely affected oxygen consuming cellular processes since non-mitochondrial OCR declined for both blank and PTX nanoparticle-treated MSCs. Previous studies show that oxidative stress can reduce the reserve capacity, and if the reserve capacity drops below the basal respiration rate, then cell death is triggered [45]. In our studies, loading PTX did not affect basal respiration or overall ATP production. This suggests that MSCs quickly balance the increased oxidative stress by generating an antioxidant response and preventing significant mitochondrial damage .
Fig. 9.
Kinetic OCR (A) and maximal respiration (B) response of control MSCs (MSC-Az) as well as MSCs surface conjugated with blank nanoparticles (MSC-Az + BLK DBCO NP) or PTX loaded nanoparticles (MSC-Az + PTX DBCO NP). AP: ATP Production; BR: Basal Respiration; NOC; Non-mitochondrial oxygen consumption. Results significant at ∗∗p < 0.0001.
3.4. Upregulation of Nrf2 in response to PTX loading
Redox balance in cells is maintained by Nrf2, a major regulator of antioxidant responses. Nrf2 expression is upregulated and stabilized by oxidative stress, which in turn induces the expression of antioxidants and cytoprotective genes [46]. In agreement with our data demonstrating upregulation of antioxidant proteins, nanoengineering resulted in a significant increase in Nrf2 protein and mRNA levels 16 h after nanoengineering (Fig. 10). There was a good correlation between PTX loading and Nrf2 expression. MSCs loaded via simple endocytosis resulted in a ∼95% increase in Nrf2 expression while higher loading achieved via surface conjugation resulted in a ∼135% increase (in serum-free conditions).
Fig. 10.
Upregulation of Nrf2 in response to PTX loading. Levels of Nrf2 protein (A) and Nfe2l2 mRNA transcripts (B) in nanoengineered MSCs relative to untreated MSCs or those treated with blank nanoparticles analyzed after incubation in either serum containing (SM) or serum free media (SFM) for 16 h. Nanoengineering was achieved by simple incubation (MSC + PTX NP) or via surface conjugation (MSC-Az + PTX DBCO NP). Results significant at ∗p < 0.001.
3.5. PTX loading upregulates CXCR4 expression and signaling
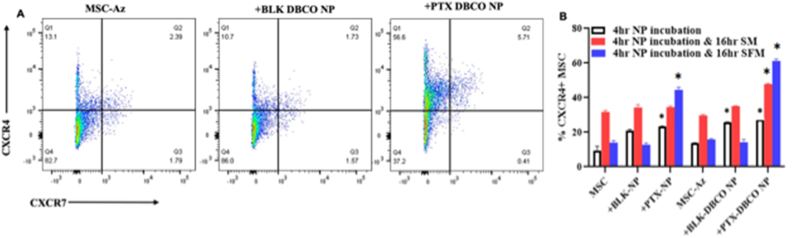
In addition to its role as a key regulator of the oxidative stress response, Nrf2 plays a critical role in stem cell migration and their retention in the niche [47]. It was previously shown that Nrf2 directly binds to CXCR4 promoter and activates its expression [48]. Nrf2−/− cells lack the homing ability and overexpression of CXCR4 in these cells restored the homing characteristics, suggesting that Nrf2 exerts at least some of its effects through regulation of CXCR4 signaling. The increase in Nrf2 transcript and protein levels following nanoengineering and the previous studies demonstrating direct activation of CXCR4 by Nrf2 directed us to investigate the effect of PTX loading on CXCR4 expression. As with the ROS studies, we investigated the impact of PTX loading extent (8.3 vs 43.8 pg/cell), time to recovery after 4 h of nanoengineering (0 vs 16 h) and the presence of serum on CXCR4 expression. The baseline expression of CXCR4 in MSCs was ∼15% (CXCR4+ and CXCR4+/7+; Fig. 11). Nanoengineering MSCs with PTX increased the number of MSCs expressing CXCR4 by 5-fold (∼65%, Fig. 11). The increase in CXCR4 expression was proportional to PTX loading in cells and the increase was lower when the cells were allowed to recover in the presence of serum for 16 h. There was not a significant effect of PTX loading on CXCR7 expression (Suppl Fig. S2).
Fig. 11.
Effect of nanoengineering on CXCR4 expression. (A) Representative dot plots of nonengineered MSCs after 16 h in serum free media (n = 3). (B) %CXCR4+ MSC just after 4 h NP incubation or followed by either 16 h serum containing (SM) or serum free media (SFM). Results significant at ∗p < 0.01.
We then turned to global proteomics to evaluate whether CXCR4 signaling is active in MSCs nanoengineered with PTX nanoparticles. As can be seen in Fig. 12, expression of several proteins in the Gα13-Rho signaling axis downstream to CXCR4 was upregulated in nanoengineered MSCs even in the absence of exogenous SDF-1 stimulation, suggesting that enhanced expression of CXCR4 following nanoengineering also results in increased downstream signaling events that could lead to increased tumor homing of MSCs.
Fig. 12.
Global proteomic analysis of proteins (highlighted red in the pathway) downstream to CXCR4 in MSCs nanoengineered using DBCO-functionalized nanoparticles. Example expression data are shown for some of the overexpressed proteins (n = 3). All results significant at ∗p < 0.01; ∗∗p < 0.001; ∗∗∗∗p < 0.00001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Effect of PTX loading on tumor homing of MSCs
We then determined the effect of loading PTX on tumor homing of MSCs in the syngeneic lung orthotopic LL/2 tumor model. We used RT-PCR to quantitate human MSC-specific mRNA transcripts of CD90 and GAPDH. This study showed that relative to controls (untreated MSCs and MSCs treated with blank nanoparticles), MSCs loaded with PTX demonstrated greatly enhanced tumor homing and retention in the tumor (for at least 7 days following the treatment) (Fig. 13). The extent of tumor homing and retention was higher for MSCs that were incubated in serum-free medium compared to those that were allowed to recover in the presence of serum, correlating with increased ROS production and greater CXCR4 expression observed in this group.
Fig. 13.
Biodistribution of MSCs nanoengineered using DBCO-functionalized nanoparticles. Fold increase of MSCs levels in lung was quantified at 6 h and 7 days by RT-PCR, as determined by CD90 (Thy-1) and GAPDH as human MSC marker genes. Extent of tumor homing and retention was two to six-fold higher with MSCs loaded with PTX and incubated in either serum or serum free media. (∗∗p < 0.0001, compared to control MSC group; #p < 0.001, compared to same group at different time points).
4. Discussion
Effectiveness of MSC-based therapies is reliant on successful migration of MSCs to the target tissue. Tumor homing of native MSCs, however, is likely not efficient. A recent phase I clinical study in prostate cancer patients failed to detect MSCs in the primary tumors [23]. This study concluded that although systemically infused allogeneic MSCs were safe, native MSCs do not home to primary tumors in sufficient levels. Yet, preclinical studies from our group [19,20,22,27] and that of others [1,18,49] show that MSCs carrying anticancer drugs accumulate in solid tumors and result in improved delivery of the payload and enhanced anticancer efficacy.
A key consideration in the use of MSCs and other cellular carriers is optimizing the drug payload in the cells. Loading high concentrations of a cytotoxic drug could directly kill the MSCs while loading too little may not result in therapeutic effect. In the current studies, we utilized two different nanoengineering strategies because they allowed for loading variable amounts of the drug in MSCs. The surface conjugation technique results in greater payload capacity. However, in certain instances (for example, to incorporate a fluorescence label on the membrane surface), it may be advantageous to load the drug via endocytosis rather than by surface conjugation. In our previous studies, priming MSCs with free PTX did not result in efficient drug incorporation in MSCs [50]. Our studies further showed that using nanoparticles results in stable drug loading in MSCs. Following endocytic uptake, MSCs internalize polymeric nanoparticles and a fraction of the internalized nanoparticles are exocytosed over a period of 4 h [51]. After 4 h, concentration of nanoparticles inside the cells remains steady for several hours [52]. For surface conjugation, our previous studies show that a fraction of nanoparticles are internalized while a significant fraction is attached to the cell membrane [52]. It is likely that even the membrane-bound nanoparticles are eventually internalized due to membrane turnover. Our studies show that loading PTX through either technique does not affect migration or other native properties of MSCs [52].
We have previously shown that nanoengineered MSCs result in enhanced tumor-targeted drug delivery and significantly less off-target deposition, as evidenced by significantly higher lung tumor-to-liver and lung tumor-to-spleen ratios for the drug compared to that following free or nanoparticle-encapsulated drug [19]. Also, plasma and tumor concentrations of PTX after IV injection of the nanoengineered MSCs was characterized by a slow, bi-exponential clearance of the drug over nearly 2 weeks [19]. Our studies also show that compared to PTX solution and PTX encapsulated in nanoparticles, which resulted in elevated alanine aminotransferase levels and a decrease in the alkaline phosphatase levels, nanoengineered MSCs did not alter any of the liver enzymes [19,53].
We have also previously established the anticancer efficacy of MSCs nanoengineered with PTX nanoparticles in different tumor models [19,20,22,27] including in the orthotopic A549 human lung adenocarcinoma model as well as in the syngeneic LL/2 murine orthotopic lung tumor model. Further, both nanoengineered mouse and human MSCs were effective in inhibiting tumor growth in the immunocompetent LL/2 mouse tumor model.
We posited that nanoengineering of MSCs impacts their biology, resulting in a favorable alteration of their in vivo disposition. Our studies suggest that MSCs counteract PTX-induced oxidative stress by upregulating antioxidant response mediated by Nrf2. CXCR4, a direct target of Nrf2 and a key mediator of tumor homing, is also upregulated, pointing to a potential mechanism of improved tumor targeting of nanoengineered MSCs.
MSC homing is a multistep process [54]. MSCs initially tether to the endothelial cells mediated by the binding of CD44 expressed on MSCs to selectins expressed by endothelial cells. This initial binding causes MSCs to roll along the vessel wall [55]. MSCs are then activated by their chemokine receptors (especially CXCR), in response to SDF-1 [56]. MSCs are then arrested, a step that is facilitated by MSC membrane integrins that bind to VCAM-1 on endothelial cells [[57], [58], [59]]. Next, MSCs travel via transmigration or diapedesis across the endothelial cell layer through the basement membrane. Our studies show that naïve MSCs have low CXCR4 expression, which could explain the low efficiency of tumor homing (% injected cells reaching the tumor) with native MSCs. Various strategies have been investigated for improving MSC homing, including targeted (local) administration, use of external magnetic fields to guide MSCs loaded with superparamagnetic nanoparticles, genetic modification to express chemokine receptors, cell surface engineering, among others [54,60]. Our studies suggest that tumor homing can be significantly improved by loading specific drugs that induce oxidative stress and/or upregulate CXCR4. The results of our studies agree with previous studies that demonstrate improved tumor homing of MSCs following genetic overexpression of CXCR4 [61,62].
Previous studies suggest that MSCs are resistant to chemotherapy drugs (for example, PTX) because they express transporters such as P-glycoprotein that rapidly efflux the drugs out [49,63] or because they adopt a non-proliferative fibroblastic state resistant to drugs that act on dividing cells [64]. While these mechanisms are potentially important, the upregulation of antioxidant response is also a key mechanism. Upregulation of Nrf2-Keap1 has been associated with the acquisition of resistance to traditional chemotherapy in NSCLC [65]. The EGFR pathway can activate Nrf2 in EGFR wild-type tumor cells after ligand-receptor binding. Also, activation of downstream signaling of the mutated EGFR pathway leads to constitutive expression of Nrf2 [66]. Our studies suggest that MSCs utilize a similar pathway to overcome PTX-induce oxidative stress and cell death.
While MSCs quickly quench the increased ROS levels induced by PTX loading, additional incubation of MSCs in serum-free medium, a known stressor, appears to further increase antioxidant response. Nrf2 is a critical transcription factor and an important regulator of the antioxidant response [67]. Under normal conditions, Nrf2 is present only in the cytoplasm as part of the Kelch-like ECH-associated protein 1 (Keap1)–Nrf2 complex. In the presence of oxidative stress, the Keap1–Nrf2 complex dissociates, allowing Nrf2 to translocate into the nucleus. Nrf2 binds with antioxidant response elements and regulates the transcription of downstream target genes, such as HO-1 and SOD [68,69]. Previous studies show that Nrf2 overexpression in MSCs protects them from oxidative stress-induced cell death by upregulating SOD and HO-1 [70], similar to the effect seen with PTX loading in our studies.
It should be noted that although PLGA nanoparticles result in sustained drug release, they are also characterized by a typical burst release over 8–12 h [50,51,71]. It is possible that the observed increases in ROS production initially is due to the burst release, while after 16 h, the drug is released slowly and at low concentrations, which could explain the decrease in ROS levels. This would also explain why the ∼10-fold higher PTX loading achieved with surface conjugation only resulted in slightly higher ROS production than with loading PTX using simple endocytic technique.
Pharmacological modulation of Nrf2 has been shown to alter MSC biology in response to different stresses. For example, ginger oleoresin was shown to protect MSCs from ionizing radiation induced DNA damage by activating nuclear translocation of Nrf2 and upregulating the expression of cytoprotective genes such as HO-1 [72]. Similarly, cyclic helix B peptide protected MSCs from starvation-induced apoptosis by activating the Nrf2/sirtuin (SIRT)3/FoxO3a pathway [73]. Another study showed that 17 b-estradiol conferred protection to human umbilical cord blood MSCs against high glucose-induced mitochondrial ROS production and cell death by activating nuclear translocation of Nrf2, followed by SIRT3 upregulation and Mn-SOD activation [74]. On the other hand, inhibition of Nrf2 by triclosan resulted in increased oxidative stress and impaired MSC proliferation [75]. Also, a previous study showed a 2-fold reduction in CXCR4 expression in hematopoietic stem cells from Nrf2−/− knockout mice [47]. These studies point to the important role of Nrf2 in MSC biology. In addition to PTX, other chemotherapy drugs are known to induce high levels of oxidative stress (for example, crizotinib, gefitinib, erlotinib and cisplatin) [76,77]. It will be interesting to determine whether these drugs also induce CXCR4 overexpression and result in improved tumor homing of MSCs. Similarly, it will also be important to determine whether MSCs can be used as carriers for drugs that do not induce oxidative stress.
In addition to the antioxidant mechanism described here, there are other potential explanations for how MSCs can deliver therapeutic concentrations of cytotoxic payload while maintaining their viability. First, it is possible that concentration of PTX in individual MSC is low enough to not kill the individual cell but sufficient number of MSCs accumulate in the tumor to cumulatively deliver therapeutic levels of PTX to kill the tumor cells. For example, nanoengineering by surface conjugation results in a loading of ∼40 pg of PTX/cell while simple endocytic loading results in a PTX loading of ∼4 pg of paclitaxel/cell. Intravenous injection of nanoengineered MSCs results in a lung tumor concentration of >1000 ng/g of PTX (Cmax), suggesting >250,000 MSCs (for 4 pg/cell loading) accumulate in the lungs during the initial time points. This translates to about 25% of the injected dose (typically 1 million MSCs/animal) reaching the target. Second, within MSCs, the drug is present inside nanoparticles and released slowly over several days. Thus, at any given time, the amount of free drug that MSCs are exposed to intracellularly is low but because of sustained and efficient accumulation of MSCs in the tumor and slow release of the drug, it may be possible to achieve therapeutic levels of the drug in the tumor.
5. Conclusions
Our studies show that MSCs counteract PTX-induced oxidative stress by upregulating Nrf2-mediated antioxidant response and CXCR4 expression. Thus, our studies reveal a novel mechanism of resistance that MSCs utilize against drug-induced oxidative stress and cell death. Importantly, these studies show that incorporating PTX induces upregulation of CXCR4 expression and improves tumor homing. Further, our studies suggests that modulation of the redox pathway may be a potential approach to enhancing CXCR4 levels in MSCs and their tumor homing.
Credit author statement
Swayam Prabha, Jayanth Panyam: Conceptualization, Methodology, Writing- Reviewing and Editing, Writing- Original draft preparation. Salim Merali and Carlos Barrero: Methodology, Writing- Reviewing and Editing, supervision; Carmen Merali, Drishti Sehgal, Nitu Bhaskar, Emmanuelle Nicolas, Magda Flores: Methodology and Data curation, Writing- Reviewing and Editing. Shubhmita Bhatnagar/Susheel Nethi - Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Vishnu Revuri for his assistance with initial studies. We acknowledge the help of Jignesh Nagar in animal studies. RT-PCR studies were performed at Fox Chase Cancer Center (FCCC) genomics facility and Penn Molecular Profiling Facility at University of Pennsylvania. SP received funding support from the National Institutes of Health (EB220558), OVPR (Temple University), Basser Cancer Center and American Cancer Society (RSG-22-123-01-ET). JP, SM are partially supported by OVPR catalytic award, Temple University. Funding support for JP (EB019893) from National Institutes of Health is acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100567.
Contributor Information
Swayam Prabha, Email: swayam.prabha0001@temple.edu.
Jayanth Panyam, Email: jayanth.panyam@temple.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Hamada H., Kobune M., Nakamura K., Kawano Y., Kato K., Honmou O., Houkin K., Matsunaga T., Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96(3):149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei X., Yang X., Han Z.P., Qu F.F., Shao L., Shi Y.F. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol. Sin. 2013;34(6):747–754. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianco P., Robey P.G., Simmons P.J. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P.S. In 't Anker. Scherjon S.A., Kleijburg-van der Keur C., de Groot-Swings G.M., Claas F.H., Fibbe W.E., Kanhai H.H. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cell. 2004;22(7):1338–1345. doi: 10.1634/stemcells.2004-0058. [DOI] [PubMed] [Google Scholar]
- 5.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 6.Zuk P.A., Zhu M., Mizuno H., Huang J., Futrell J.W., Katz A.J., Benhaim P., Lorenz H.P., Hedrick M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7(2):211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 7.Shah K. Mesenchymal stem cells engineered for cancer therapy. Adv. Drug Deliv. Rev. 2012;64(8):739–748. doi: 10.1016/j.addr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squillaro T., Peluso G., Galderisi U. Clinical trials with mesenchymal stem cells: an update. Cell Transplant. 2016;25(5):829–848. doi: 10.3727/096368915X689622. [DOI] [PubMed] [Google Scholar]
- 9.Klyushnenkova E., Mosca J.D., Zernetkina V., Majumdar M.K., Beggs K.J., Simonetti D.W., Deans R.J., McIntosh K.R. T cell responses to allogeneic human mesenchymal stem cells: immunogenicity, tolerance, and suppression. J. Biomed. Sci. 2005;12(1):47–57. doi: 10.1007/s11373-004-8183-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Huang J., Gong L., Yu D., An C., Bunpetch V., Dai J., Huang H., Zou X., Ouyang H., Liu H. The plasticity of mesenchymal stem cells in regulating surface HLA-I. iScience. 2019;15:66–78. doi: 10.1016/j.isci.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atoui R., Asenjo J.F., Duong M., Chen G., Chiu R.C., Shum-Tim D. Marrow stromal cells as universal donor cells for myocardial regenerative therapy: their unique immune tolerance. Ann. Thorac. Surg. 2008;85(2):571–579. doi: 10.1016/j.athoracsur.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Chen L., Tredget E.E., Liu C., Wu Y. Analysis of allogenicity of mesenchymal stem cells in engraftment and wound healing in mice. PLoS One. 2009;4(9) doi: 10.1371/journal.pone.0007119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kebriaei P., Isola L., Bahceci E., Holland K., Rowley S., McGuirk J., Devetten M., Jansen J., Herzig R., Schuster M., Monroy R., Uberti J. Adult human mesenchymal stem cells added to corticosteroid therapy for the treatment of acute graft-versus-host disease, Biology of blood and marrow transplantation. journal of the American Society for Blood and Marrow Transplantation. 2009;15(7):804–811. doi: 10.1016/j.bbmt.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Le Blanc K., Rasmusson I., Sundberg B., Gotherstrom C., Hassan M., Uzunel M., Ringden O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363(9419):1439–1441. doi: 10.1016/S0140-6736(04)16104-7. [DOI] [PubMed] [Google Scholar]
- 15.Prasad V.K., Lucas K.G., Kleiner G.I., Talano J.A., Jacobsohn D., Broadwater G., Monroy R., Kurtzberg J. Efficacy and safety of ex vivo cultured adult human mesenchymal stem cells (Prochymal) in pediatric patients with severe refractory acute graft-versus-host disease in a compassionate use study, Biology of blood and marrow transplantation. journal of the American Society for Blood and Marrow Transplantation. 2011;17(4):534–541. doi: 10.1016/j.bbmt.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Sundin M., Ringden O., Sundberg B., Nava S., Gotherstrom C., Le Blanc K. No alloantibodies against mesenchymal stromal cells, but presence of anti-fetal calf serum antibodies, after transplantation in allogeneic hematopoietic stem cell recipients. Haematologica. 2007;92(9):1208–1215. doi: 10.3324/haematol.11446. [DOI] [PubMed] [Google Scholar]
- 17.Bexell D., Gunnarsson S., Svensson A., Tormin A., Henriques-Oliveira C., Siesjo P., Paul G., Salford L.G., Scheding S., Bengzon J. Rat multipotent mesenchymal stromal cells lack long-distance tropism to 3 different rat glioma models. Neurosurgery. 2012;70(3):731–739. doi: 10.1227/NEU.0b013e318232dedd. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Lin X., Zhao J., Shi W., Zhang H., Wang Y., Kan B., Du L., Wang B., Wei Y., Liu Y., Zhao X. A tumor-selective biotherapy with prolonged impact on established metastases based on cytokine gene-engineered MSCs. Mol. Ther. 2008;16(4):749–756. doi: 10.1038/mt.2008.3. [DOI] [PubMed] [Google Scholar]
- 19.Layek B., Sadhukha T., Panyam J., Prabha S. Nano-engineered mesenchymal stem cells increase therapeutic efficacy of anticancer drug through true active tumor targeting. Mol. Cancer Therapeut. 2018;17(6):1196–1206. doi: 10.1158/1535-7163.MCT-17-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layek B., Shetty M., Nethi S.K., Sehgal D., Starr T.K., Prabha S. Mesenchymal stem cells as guideposts for nanoparticle-mediated targeted drug delivery in ovarian cancer. Cancers. 2020;12(4) doi: 10.3390/cancers12040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moku G., Layek B., Trautman L., Putnam S., Panyam J., Prabha S. Improving payload capacity and anti-tumor efficacy of mesenchymal stem cells using TAT peptide functionalized polymeric nanoparticles. Cancers. 2019;11(4) doi: 10.3390/cancers11040491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadhukha T., O'Brien T.D., Prabha S. Nano-engineered mesenchymal stem cells as targeted therapeutic carriers. J. Contr. Release. 2014;196:243–251. doi: 10.1016/j.jconrel.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Schweizer M.T., Wang H., Bivalacqua T.J., Partin A.W., Lim S.J., Chapman C., Abdallah R., Levy O., Bhowmick N.A., Karp J.M., De Marzo A., Isaacs J.T., Brennen W.N., Denmeade S.R. A phase I study to assess the safety and cancer-homing ability of allogeneic bone marrow-derived mesenchymal stem cells in men with localized prostate cancer. STEM CELLS Translational Medicine. 2019;8(5):441–449. doi: 10.1002/sctm.18-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Gao J., Ouyang X., Wang J., Sun X., Lv Y. Mesenchymal stem cells loaded with paclitaxel-poly(lactic-co-glycolic acid) nanoparticles for glioma-targeting therapy. Int. J. Nanomed. 2018;13:5231–5248. doi: 10.2147/IJN.S167142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Yao S., Liu C., Jiang Y. Tumor tropic delivery of doxorubicin-polymer conjugates using mesenchymal stem cells for glioma therapy. Biomaterials. 2015;39:269–281. doi: 10.1016/j.biomaterials.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Layek B., Sehgal D., Argenta P.A., Panyam J., Prabha S. Nanoengineering of mesenchymal stem cells via surface modification for efficient cancer therapy. Advanced Therapeutics. 2019;2 [Google Scholar]
- 28.Quinn C., Rico M.C., Merali C., Merali S. Dysregulation of S-adenosylmethionine metabolism in nonalcoholic steatohepatitis leads to polyamine flux and oxidative stress. Int. J. Mol. Sci. 2022;23(4) doi: 10.3390/ijms23041986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Pandey S., Travers M., Sun H., Morton G., Madzo J., Chung W., Khowsathit J., Perez-Leal O., Barrero C.A., Merali C., Okamoto Y., Sato T., Pan J., Garriga J., Bhanu N.V., Simithy J., Patel B., Huang J., Raynal N.J., Garcia B.A., Jacobson M.A., Kadoch C., Merali S., Zhang Y., Childers W., Abou-Gharbia M., Karanicolas J., Baylin S.B., Zahnow C.A., Jelinek J., Grana X., Issa J.J. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell. 2018;175(5):1244–1258 e26. doi: 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molina-Franky J., Plaza D.F., Merali C., Merali S., Barrero C., Arevalo-Pinzon G., Patarroyo M.E., Patarroyo M.A. A novel platform for peptide-mediated affinity capture and LC-MS/MS identification of host receptors involved in Plasmodium invasion. J. Proteonomics. 2021;231 doi: 10.1016/j.jprot.2020.104002. [DOI] [PubMed] [Google Scholar]
- 31.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11(3):319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 32.Patel A., Kostyak J., Dangelmaier C., Badolia R., Bhavanasi D., Aslan J.E., Merali S., Kim S., Eble J.A., Goldfinger L., Kunapuli S. ELMO1 deficiency enhances platelet function. Blood advances. 2019;3(4):575–587. doi: 10.1182/bloodadvances.2018016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanmughapriya S., Tomar D., Dong Z., Slovik K.J., Nemani N., Natarajaseenivasan K., Carvalho E., Lu C., Corrigan K., Garikipati V.N.S., Ibetti J., Rajan S., Barrero C., Chuprun K., Kishore R., Merali S., Tian Y., Yang W., Madesh M. FOXD1-dependent MICU1 expression regulates mitochondrial activity and cell differentiation. Nat. Commun. 2018;9(1):3449. doi: 10.1038/s41467-018-05856-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrero C.A., Perez-Leal O., Aksoy M., Moncada C., Ji R., Lopez Y., Mallilankaraman K., Madesh M., Criner G.J., Kelsen S.G., Merali S. Histone 3.3 participates in a self-sustaining cascade of apoptosis that contributes to the progression of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2013;188(6):673–683. doi: 10.1164/rccm.201302-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boden G., Cheung P., Kresge K., Homko C., Powers B., Ferrer L. Insulin resistance is associated with diminished endoplasmic reticulum stress responses in adipose tissue of healthy and diabetic subjects. Diabetes. 2014;63(9):2977–2983. doi: 10.2337/db14-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boden G., Duan X., Homko C., Molina E.J., Song W., Perez O., Cheung P., Merali S. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes. 2008;57(9):2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boden G., Homko C., Barrero C.A., Stein T.P., Chen X., Cheung P., Fecchio C., Koller S., Merali S. Excessive caloric intake acutely causes oxidative stress, GLUT4 carbonylation, and insulin resistance in healthy men. Sci. Transl. Med. 2015;7(304):304re7. doi: 10.1126/scitranslmed.aac4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S.J., Hoffman N.E., Shanmughapriya S., Bao L., Keefer K., Conrad K., Merali S., Takahashi Y., Abraham T., Hirschler-Laszkiewicz I., Wang J., Zhang X.Q., Song J., Barrero C., Shi Y., Kawasawa Y.I., Bayerl M., Sun T., Barbour M., Wang H.G., Madesh M., Cheung J.Y., Miller B.A. A splice variant of the human ion channel TRPM2 modulates neuroblastoma tumor growth through hypoxia-inducible factor (HIF)-1/2alpha. J. Biol. Chem. 2014;289(52):36284–36302. doi: 10.1074/jbc.M114.620922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Panyam J., Sahoo S.K., Prabha S., Bargar T., Labhasetwar V. Fluorescence and electron microscopy probes for cellular and tissue uptake of poly(d,l-lactide-co-glycolide) nanoparticles. Int. J. Pharm. 2003;262(1):1–11. doi: 10.1016/s0378-5173(03)00295-3. [DOI] [PubMed] [Google Scholar]
- 41.André N., Carré M., Brasseur G., Pourroy B., Kovacic H., Briand C., Braguer D. Paclitaxel targets mitochondria upstream of caspase activation in intact human neuroblastoma cells. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2002;532(1–2):256–260. doi: 10.1016/s0014-5793(02)03691-8. [DOI] [PubMed] [Google Scholar]
- 42.Jiang H., Zhang X.-W., Liao Q.-L., Wu W.-T., Liu Y.-L., Huang W.-H. Electrochemical monitoring of paclitaxel-induced ROS release from mitochondria inside single cells. Small. 2019;15(48) doi: 10.1002/smll.201901787. [DOI] [PubMed] [Google Scholar]
- 43.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J., Landar A., Darley-Usmar V.M. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 2011;51(9):1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill B.G., Dranka B.P., Zou L., Chatham J.C., Darley-Usmar V.M. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J. 2009;424(1):99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vomund S., Schäfer A., Parnham M.J., Brüne B., von Knethen A. Nrf2, the master regulator of anti-oxidative responses. Int. J. Mol. Sci. 2017;18(12) doi: 10.3390/ijms18122772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai J.J., Dudakov J.A., Takahashi K., Shieh J.-H., Velardi E., Holland A.M., Singer N.V., West M.L., Smith O.M., Young L.F., Shono Y., Ghosh A., Hanash A.M., Tran H.T., Moore M.A.S., van den Brink M.R.M. Nrf2 regulates haematopoietic stem cell function. Nat. Cell Biol. 2013;15(3):309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai X., Yan X., Wintergerst K.A., Cai L., Keller B.B., Tan Y. Nrf2: redox and metabolic regulator of stem cell state and function. Trends Mol. Med. 2020;26(2):185–200. doi: 10.1016/j.molmed.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Pessina A., Bonomi A., Coccè V., Invernici G., Navone S., Cavicchini L., Sisto F., Ferrari M., Viganò L., Locatelli A., Ciusani E., Cappelletti G., Cartelli D., Arnaldo C., Parati E., Marfia G., Pallini R., Falchetti M.L., Alessandri G. Mesenchymal stromal cells primed with paclitaxel provide a new approach for cancer therapy. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadhukha T., O'Brien T.D., Prabha S. Nano-engineered mesenchymal stem cells as targeted therapeutic carriers. J. Contr. Release. 2014;196:243–251. doi: 10.1016/j.jconrel.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 51.Layek B., Sadhukha T., Prabha S. Glycoengineered mesenchymal stem cells as an enabling platform for two-step targeting of solid tumors. Biomaterials. 2016;88:97–109. doi: 10.1016/j.biomaterials.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Layek B., Sehgal D., Argenta P.A., Panyam J., Prabha S. Nanoengineering of mesenchymal stem cells via surface modification for efficient cancer therapy. Advanced Therapeutics. 2019;2(9) [Google Scholar]
- 53.Layek B.S., D. Argenta P.A., Panyam J., Prabha S. Nanoengineering of mesenchymal stem cells via surface modification for efficient cancer therapy. Advamced therapeutics. 2019;2 [Google Scholar]
- 54.Ullah M., Liu D.D., Thakor A.S. Mesenchymal stromal cell homing: mechanisms and strategies for improvement. iScience. 2019;15:421–438. doi: 10.1016/j.isci.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sackstein R. The lymphocyte homing receptors: gatekeepers of the multistep paradigm. Curr. Opin. Hematol. 2005;12(6):444–450. doi: 10.1097/01.moh.0000177827.78280.79. [DOI] [PubMed] [Google Scholar]
- 56.Lau T.T., Wang D.A. Stromal cell-derived factor-1 (SDF-1): homing factor for engineered regenerative medicine. Expet Opin. Biol. Ther. 2011;11(2):189–197. doi: 10.1517/14712598.2011.546338. [DOI] [PubMed] [Google Scholar]
- 57.Rüster B., Göttig S., Ludwig R.J., Bistrian R., Müller S., Seifried E., Gille J., Henschler R. Mesenchymal stem cells display coordinated rolling and adhesion behavior on endothelial cells. Blood. 2006;108(12):3938–3944. doi: 10.1182/blood-2006-05-025098. [DOI] [PubMed] [Google Scholar]
- 58.Segers V.F., Van Riet I., Andries L.J., Lemmens K., Demolder M.J., De Becker A.J., Kockx M.M., De Keulenaer G.W. Mesenchymal stem cell adhesion to cardiac microvascular endothelium: activators and mechanisms. Am. J. Physiol. Heart Circ. Physiol. 2006;290(4):H1370–H1377. doi: 10.1152/ajpheart.00523.2005. [DOI] [PubMed] [Google Scholar]
- 59.Steingen C., Brenig F., Baumgartner L., Schmidt J., Schmidt A., Bloch W. Characterization of key mechanisms in transmigration and invasion of mesenchymal stem cells. J. Mol. Cell. Cardiol. 2008;44(6):1072–1084. doi: 10.1016/j.yjmcc.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 60.De Becker A., Riet I.V. Homing and migration of mesenchymal stromal cells: how to improve the efficacy of cell therapy? World J. Stem Cell. 2016;8(3):73–87. doi: 10.4252/wjsc.v8.i3.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W., Li M., Cheng H., Yan Z., Cao J., Pan B., Sang W., Wu Q., Zeng L., Li Z., Xu K. Overexpression of the mesenchymal stem cell Cxcr4 gene in irradiated mice increases the homing capacity of these cells. Cell Biochem. Biophys. 2013;67(3):1181–1191. doi: 10.1007/s12013-013-9632-6. [DOI] [PubMed] [Google Scholar]
- 62.Kalimuthu S., Oh J.M., Gangadaran P., Zhu L., Lee H.W., Rajendran R.L., Baek S.H., Jeon Y.H., Jeong S.Y., Lee S.W., Lee J., Ahn B.C. Vivo tracking of chemokine receptor CXCR4-engineered mesenchymal stem cell migration by optical molecular imaging. Stem Cell. Int. 2017;2017 doi: 10.1155/2017/8085637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonomi A., Silini A., Vertua E., Signoroni P.B., Coccè V., Cavicchini L., Sisto F., Alessandri G., Pessina A., Parolini O. Human amniotic mesenchymal stromal cells (hAMSCs) as potential vehicles for drug delivery in cancer therapy: an in vitro study. Stem Cell Res. Ther. 2015;6(1):155. doi: 10.1186/s13287-015-0140-z. 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosco D.B., Kenworthy R., Zorio D.A., Sang Q.X. Human mesenchymal stem cells are resistant to Paclitaxel by adopting a non-proliferative fibroblastic state. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0128511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013;27(20):2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamadori T., Ishii Y., Homma S., Morishima Y., Kurishima K., Itoh K., Yamamoto M., Minami Y., Noguchi M., Hizawa N. Molecular mechanisms for the regulation of Nrf2-mediated cell proliferation in non-small-cell lung cancers. Oncogene. 2012;31(45):4768–4777. doi: 10.1038/onc.2011.628. [DOI] [PubMed] [Google Scholar]
- 67.Silva-Islas C.A., Maldonado P.D. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol. Res. 2018;134:92–99. doi: 10.1016/j.phrs.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 68.Raghunath A., Sundarraj K., Nagarajan R., Arfuso F., Bian J., Kumar A.P., Sethi G., Perumal E. Antioxidant response elements: discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297–314. doi: 10.1016/j.redox.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao G.J., Hou N., Cai S.A., Liu X.W., Li A.Q., Cheng C.F., Huang Y., Li L.R., Mai Y.P., Liu S.M., Ou C.W., Xiong Z.Y., Chen X.H., Chen M.S., Luo C.F. Contributions of Nrf2 to puerarin prevention of cardiac hypertrophy and its metabolic enzymes expression in rats. J. Pharmacol. Exp. Therapeut. 2018;366(3):458–469. doi: 10.1124/jpet.118.248369. [DOI] [PubMed] [Google Scholar]
- 70.Mohammadzadeh M., Halabian R., Gharehbaghian A., Amirizadeh N., Jahanian-Najafabadi A., Roushandeh A.M., Roudkenar M.H. Nrf-2 overexpression in mesenchymal stem cells reduces oxidative stress-induced apoptosis and cytotoxicity. Cell Stress Chaperones. 2012;17(5):553–565. doi: 10.1007/s12192-012-0331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Layek B., Sadhukha T., Panyam J., Prabha S. Nano-engineered mesenchymal stem cells increase therapeutic efficacy of anticancer drug through true active tumor targeting. Mol. Cancer Therapeut. 2018;17(6):1196–1206. doi: 10.1158/1535-7163.MCT-17-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji K., Fang L., Zhao H., Li Q., Shi Y., Xu C., Wang Y., Du L., Wang J., Liu Q. Ginger oleoresin alleviated γ-ray irradiation-induced reactive oxygen species via the Nrf2 protective response in human mesenchymal stem cells. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/1480294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S., Zhang C., Niyazi S., Zheng L., Li J., Zhang W., Xu M., Rong R., Yang C., Zhu T. A novel cytoprotective peptide protects mesenchymal stem cells against mitochondrial dysfunction and apoptosis induced by starvation via Nrf2/Sirt3/FoxO3a pathway. J. Transl. Med. 2017;15(1):33. doi: 10.1186/s12967-017-1144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oh J.Y., Choi G.E., Lee H.J., Jung Y.H., Chae C.W., Kim J.S., Lee C.K., Han H.J. 17β-Estradiol protects mesenchymal stem cells against high glucose-induced mitochondrial oxidants production via Nrf2/Sirt3/MnSOD signaling. Free Radic. Biol. Med. 2019;130:328–342. doi: 10.1016/j.freeradbiomed.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 75.Yoon D.S., Choi Y., Cha D.S., Zhang P., Choi S.M., Alfhili M.A., Polli J.R., Pendergrass D., Taki F.A., Kapalavavi B., Pan X., Zhang B., Blackwell T.K., Lee J.W., Lee M.H. Triclosan disrupts SKN-1/Nrf2-Mediated oxidative stress response in C. elegans and human mesenchymal stem cells. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-12719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conklin K.A. Chemotherapy-associated oxidative stress: impact on chemotherapeutic effectiveness. Integr. Cancer Ther. 2004;3(4):294–300. doi: 10.1177/1534735404270335. [DOI] [PubMed] [Google Scholar]
- 77.Teppo H.-R., Soini Y., Karihtala P. Reactive oxygen species-mediated mechanisms of action of targeted cancer therapy. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/1485283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.