Abstract
Floral color plays a major role in pollinator specificity, and changes in color may result in pollinator shifts and pollinator-mediated speciation. In the purple flowers of Platycodon grandiflorus, anthocyanins are the major pigment metabolites, whereas white flowers result due to the absence of anthocyanins. The lack of anthocyanins may be due to the inhibition of the anthocyanin biosynthesis pathway. However, the molecular mechanism of anthocyanin biosynthesis in P. grandiflorus is not fully understood. Hence, we identified R2R3-MYB transcription factor, PlgMYBR1, as a negative regulator for anthocyanin biosynthesis using sequence homology and tissue-specific expression pattern analyses. A heterologous co-expression assay suggested that PlgMYBR1 inhibited the function of AtPAP1 (Arabidopsis thaliana production of anthocyanin pigment 1), indicating that PlgMYBR1 plays as a repressor of anthocyanin biosynthesis in P. grandiflorus. Our results provide a foundation for future efforts to understand the anthocyanin biosynthesis in P. grandiflorus and, thereby, to improve flower color through genetic engineering.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03490-6.
Keywords: Anthocyanin, Flower color, R2R3-MYB transcription factor, Platycodon grandiflorus
Introduction
Platycodon grandiflorus is a monotypic species belonging to the Campanulaceae family, which is a widely used ornamental, edible, and traditional Chinese medicinal herb in East Asia (Zhang et al. 2015). In traditional Chinese medicine, rhizomes of P. grandiflorus are administered to treat cough, phlegm, chest tightness, sore throats, and other disorders (Ji et al. 2020). Although 75 triterpenoid glycosides have been identified from P. grandiflorus, platycodin D is considered to be the main active compound, which possesses multiple biological effects, such as anticancer, anti-inflammatory, antiobesity, and antidiabetic properties (Jeon et al. 2019; Ji et al. 2020). In addition, flavonoids (e.g., luteolin-7-O-glucoside, flavoplatycoside, and apigenin-7-O-glucoside), phenolic acids (e.g., caffeic acid, ferulic acid, and iobetyolin), polyacetylene (e.g., lobetyolinin and lobetyolin), and sterols (e.g., β-sitosterol, spinasterol, and betulin) have been isolated from this plant (Ji et al. 2020). Although considerable attention has been focused on the pharmacologic properties of the roots and rhizomes of P. grandiflorus, EtOH extract of P. grandiflorus flowers have also been shown to exhibit antioxidant, cholesterol adsorption inhibitory, and α-glucosidase inhibitory activities (Kang et al. 2019), indicating the potential of flowers as a source of dietary health supplements.
In Korea, two varieties with purple flowers (PFs) or white flowers (WFs) are more common, and there is a difference in pharmaceutical properties between these varieties (Park et al. 2007; Han et al. 2014). Therefore, SSR (sequence-characterized amplified region) and SNP (single nucleotide polymorphism) markers were developed for predicting flower color in P. grandiflorus (Park et al. 2007; Yu et al. 2021). Although these markers facilitate the analysis of genetic diversity and marker-assisted selection breeding, the molecular mechanisms of flower color variation in P. grandiflorus are still not fully understood.
Flower color is mainly due to the accumulation of pigments such as flavonoids/anthocyanins, carotenoids, and chlorophylls (Grotewold et al. 2006). Among them, anthocyanins are major pigmentation compounds, which primarily cause the formation of red, orange, blue, and purple flowers (Grotewold et al. 2006). In higher plants, anthocyanin accumulation is strongly correlated with the expression of anthocyanin structural genes controlled by MYB-bHLH-WD40 (MBW) activation complex, whereas R2R3-MYB with R2 and R3 repeats and R3-MYB with only R3 repeat are verified to be anthocyanin repressors via interfering with MBW complex formation (Mekapogu et al. 2020). This indicates that MYB transcription factors are the most specific and conspicuous regulators of anthocyanin biosynthesis among the MBW complex (Zimmermann et al. 2004; Mekapogu et al. 2020). Although the regulatory mechanisms by which MYBs affect plant color have been widely investigated in eudicot plants such as Petunia, Arabidopsis, Mimulus, grapevine, apple, and peach (Yuan et al. 2014; Liu et al. 2015; Hu et al. 2016; Tohge et al. 2017; Ma et al. 2018; Ding et al. 2020), the functional analysis of MYBs regulating anthocyanin biosynthesis are still relatively unknown in P. grandiflorus.
In this study, we aimed to investigate the molecular mechanism responsible for the different flower colors of P. grandiflorus and elucidate the role of MYB transcription factors by analyzing the expression pattern in different tissue and flower development stages. Using transient color assays, we performed functional characterization of P. grandiflorus MYB repressor 1 (PlgMYBR1). Taken together, these results suggest that the variation in PlgMYBR1 expression level between two varieties affects the accumulation of anthocyanins and flower color.
Materials and methods
Plant materials
Seeds of P. grandiflorus with WFs and PFs, obtained from the Danong, Co., Ltd, in South Korea, and of tobacco (Nicotiana tabacum cv. Petit Havana) were germinated and grown in a growth chamber with a long photoperiod (16-h light/8-h dark). To analyze the anthocyanin content and gene expression patterns, buds and flowers were harvested by stages according to bud diameter and open flower and immediately frozen in liquid nitrogen.
Determination of anthocyanin and chlorophyll contents
Total anthocyanin content was quantified as described by Shin et al. (2007). The absorbance was determined at 530 nm (A530) and 657 nm (A657), respectively, and the content was calculated using the following formula: (A530 − 0.33 × A657)/g of fresh weight.
Ten milligrams of samples were extracted in 95% ethanol at 80 °C for 30 min, and an aliquot of the ethanolic extracts was used for the determination of chlorophyll (Chl) contents. The contents of Chl a and b and total Chl (Chl a + b) were calculated according to Czyczyło-Mysza et al. (2013). Total anthocyanin and Chl contents in each sample were determined in triplicates.
Isolation and expression pattern of MYB repressors (MYBRs) in P. grandiflorus
To enlist the complete family of MYBR in P. grandiflorus, BLAST searches (p < 0.01) were performed on P. grandiflorus (variety with WFs) sequencing data (Kim et al. 2020). The amino acid sequences of candidate genes were analyzed to examine the presence of the characteristic conserved domains using Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/).
To analyze the tissue-specific expression pattern of P. grandiflorus, MYB repressors (PlgMYBRs), RNA-Seq data derived from eight different tissues were downloaded from NCBI GenBank (variety with WFs, SRR8712510–SRR8712517). As described by Kim et al. (2020), transcription levels of each gene were estimated in fragments per kilobase of transcript per million mapped read values.
RNA isolation and quantitative real-time PCR
Total RNA from P. grandiflorus and tobacco samples was extracted using the FavorPrep Plant Total RNA Mini Kit, and reverse-transcribed into cDNA using the cDNA synthesis kit (Toyobo, Co., Ltd., Osaka, Japan). Quantitative real-time PCR (qRT-PCR) was performed using the Toyobo SYBR‐Green Master Mix. Specific primer pairs for each gene (Table S1) were used, and the transcription levels of target genes were normalized to PlgActin or NtGAPDH. All analyses were performed in three biological replicates and two technical replicates.
Cloning and transient expression of PlgMYBR1 in tobacco
The full-length PlgMYBR1 gene amplified using gene-specific primers was cloned into the pENTR/D-TOPO vector and subcloned into a gateway binary vector pGWB505. For transient PlgMYBR1 expression, Agrobacterium tumefaciens GV3101 containing 35S:PlgMYBR1 or 35S:Arabidopsis thaliana production of anthocyanin pigment 1 (AtPAP1) construct was inoculated into 3 mL LB medium with antibiotics at 28 °C, and agro-infiltration was performed as described by Ji et al. (2021). Leaf color and anthocyanin content were monitored at 7 days after infiltration. Transient expression was assessed in three biological replicates.
Results and discussion
The expression levels of anthocyanin biosynthetic genes in two flower color varieties of P. grandiflorus
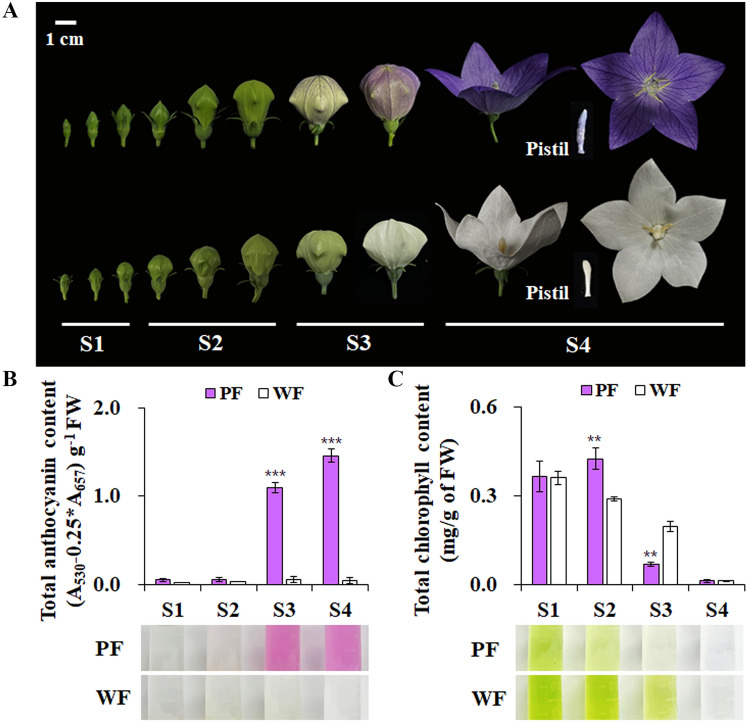
As described earlier, the chemical compounds determining the flower colors mainly include flavonoids/anthocyanins, carotenoids, and chlorophylls. In the PF of P. grandiflorus, delphinidin and its polyacylated derivatives including platyconin (delphinidin 3-dicaffeylrutinoside-5-glucoside) were identified as a major anthocyanin (Kondo et al. 2021; Lv et al. 2021). This suggests that anthocyanins are major pigmentation compounds, although a comparative analysis between WF and PF varieties of P. grandiflorus is still needed. Hence, we initially determined the contents of total anthocyanin and total Chl from petals of four different flowering stages of WF and PF. As shown in Fig. 1, the total anthocyanin contents of PF and WF were similar in S1 and S2. However, total anthocyanin content in PF increased at S3 and reached the maximum level at S4, whereas total anthocyanin content in WF remained unchanged in all stages. Total Chl content in both PF and WF increased to the highest at S1 and decreased as the flowers bloomed (Fig. 1).
Fig. 1.
Change in the color, anthocyanin content, and chlorophyll (Chl) content during flower development of P. grandiflorus. A The images of the flowers of P. grandiflorus varieties. S1: < 1 cm diameter, S2: 1–2 cm diameter, S3: 2–3 cm diameter, and S4: fully open. Analyses of anthocyanin accumulation (B) and total Chl contents (C) during the flower development in both varieties. The data are representative of three independent experiments (mean ± SE). *p < 0.05, **p < 0.01, and ***p < 0.001, as compared with those of purple petals
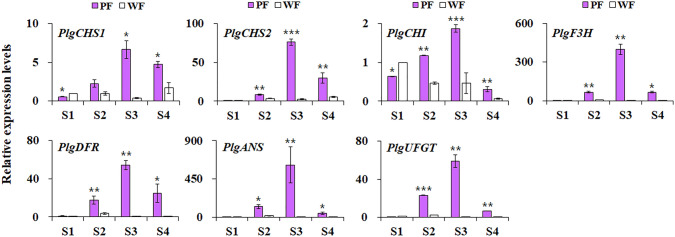
Differences in anthocyanin contents between two flower color varieties prompted us to investigate whether this phenomenon is mediated by the altering expression of genes involved in the anthocyanin biosynthetic pathway. As shown in Fig. 2, the expression levels of anthocyanin biosynthetic genes in PF were significantly higher than those of WF. In PF, the expression level of these genes increased initially, then decreased, and finally reached a peak at S3, whereas these genes were expressed at low levels and were almost unchanged or slightly decreased during the developmental process in WF (Fig. 2). Similarly, the differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation underlie color divergence of flowers, fruits, and leaves in various plants (Wei et al. 2011, 2015; Zhang et al. 2017; Naing et al. 2018; Le Maitre et al. 2019; Ye et al. 2021). Taken together, this indicates that anthocyanins are the main contributors to the coloration of P. grandiflorus petals. Thus, we focused on the differential expression levels of anthocyanin biosynthetic genes in the two contrasting flower color varieties of P. grandiflorus.
Fig. 2.
The expression levels of genes involved in anthocyanin biosynthesis in the petal of two P. grandiflorus varieties. The expression levels of genes from the white petals were compared with those of the purple petals. The data are representative of three independent experiments (mean ± SE). *p < 0.05, **p < 0.01, and ***p < 0.001 as compared with those of the purple petals. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone hydroxylase; DFR, dihydroflavanol-4-reductase; ANS, anthocyanin synthase; UFGT, flavonoid-3-O-glucosyltransferase
Identification of MYBR genes in P. grandiflorus
In Arabidopsis, R2R3-MYB genes belonging to subgroup 6 (e.g., Arabidopsis thaliana MYB75, 90, and 114) are known as MYB activators that positively regulate the anthocyanin biosynthetic pathway, whereas the subgroup 4 R2R3-MYB genes (e.g., Arabidopsis thaliana MYB3, 4, 7, and 32) play as MYBRs that inhibit the anthocyanin biosynthesis (Ma and Constabel 2019; Deng et al. 2020). Similarly, three apple MYBs (Malus domestica MYB1, 10, and MYBA) regulate anthocyanin accumulation by directly activating the expression of anthocyanin structural genes, whereas Malus domestica MYB16, 111, and 308 negatively regulate pigmentation in apple (Lin-Wang et al. 2010; Ma and Constabel 2019; Wang et al. 2022). This indicates that the biosynthesis of anthocyanin is coregulated by MYB activators and MYBRs, and the identification of both regulators is important to understand flower or fruit color variation. To identify MYB activators and MYBRs in P. grandiflorus genome, amino acid sequences of MYB activators and MYBRs from various plants were used as queries, and the redundant sequences were removed, resulting in a total of 10 candidate R2R3-MYBR genes. However, we could not find MYB activators in the draft genomic data of P. grandiflorus, probably due to sequencing errors, the missing sequence that included exons or parts of exons, and misassemblies. As shown in Fig. 3, the sequence alignment of these candidate PlgMYBR proteins indicated that all had the conserved R2 and R3 domains in the N-terminal, together with C1/GIDP motif. The subgroup 4 R2R3 type MYBRs are characterized by two conserved motifs: C1/GIDP motif and C2/EAR (pdLNLD/EL), indicating that PGJG195470 belongs to the subgroup 4 R2R3 type MYB family. However, it is uncertain whether these C1/GIDP and C2/EAR motifs are the sole determinant of the repressive activity. In addition, Arabidopsis thaliana MYB3, Petunia hybrid MYB27, and Fragaria × ananasa MYB1 do not contain C2/EAR motif, although they are well characterized as R2R3-type MYBRs (Yan et al. 2021). R2R3-type MYBRs are divided into two clades: Arabidopsis thaliana MYB4-like clade (inhibition by direct binding to the promoter of anthocyanin biosynthetic genes) and Fragaria × ananasa MYB1-like clade (acting on bHLH and interfering with the proper assembly of the MBW complex) (Yan et al. 2021). MYBRs belonging to Fragaria × ananasa MYB1-like clade usually possess TLLLFR motif, which seems to be their suppression role (Matsui et al. 2008; Yan et al. 2021). Another difference between Arabidopsis thaliana MYB4-like and Fragaria × ananasa MYB1-like clades is the DNEI and DNEV sequences in the R3 domain, respectively (Chen et al. 2019). As shown in Fig. 3 except PGJG319100, all putative PlgMYBR proteins have a DNEI sequence, indicating that they are more closely related to the Arabidopsis thaliana MYB4-like clade and may play as MYBR by directly binding to the promoters of the target genes.
Fig. 3.
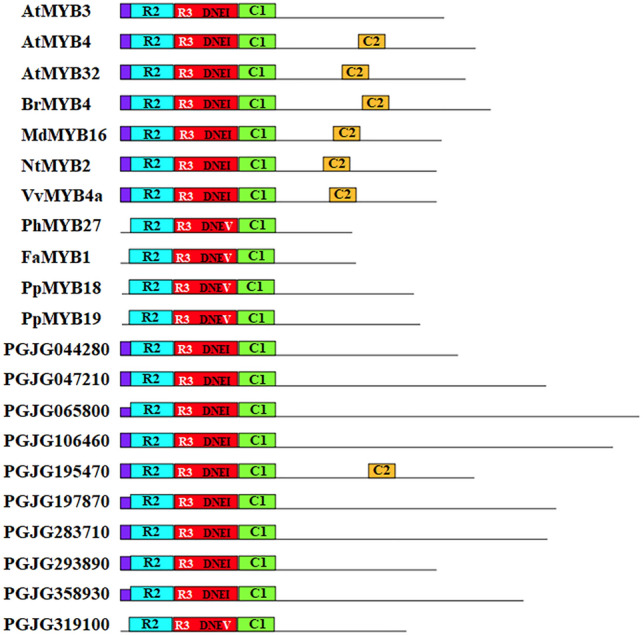
Comparison of putative PlgMYB repressors with other plant R2R3 type MYB repressors. Motif distributions were investigated using the MEME web server
Expression pattern of putative PlgMYBRs in various organs and flower developmental stages
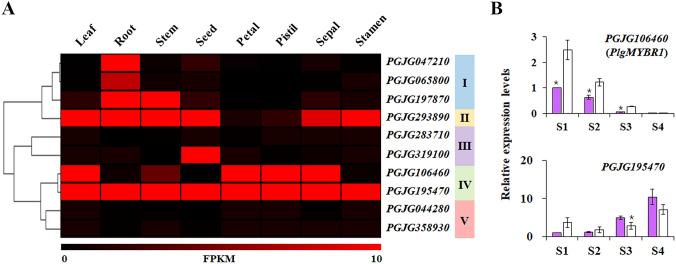
The analysis of organ-specific expression patterns should be helpful to determine whether target genes play a role in defining the function of given organs. A PF has purple petals with a purple pistil, while a WF has white petals with a white pistil (Fig. 1). Thus, we hypothesized that the difference in the anthocyanin accumulation between PF and WF is due to differentially expressed PlgMYBRs in petals and pistil. To test this, we analyzed the expression patterns of PlgMYBRs in different organs using RNA-seq data generated from P. grandiflorus variety with WFs (Kim et al. 2020), and expression patterns were organized by hierarchical clustering into 5 clusters (Fig. 4a). Cluster 1 contained genes that were expressed in root and stem, and cluster 3 genes were highly expressed in most organs, except petal and pistil. Interestingly, only cluster 4 genes were highly expressed in petal and pistil. Similar to our findings, MYBRs such as chrysanthemum MYB7 (CmMYB#7) and Freesia hybrida MYBx were markedly differentially expressed between colored and noncolored petals or tissues (Xiang et al. 2019; Li et al. 2020). This indicates the role of the two MYBRs, PGJG106460 (PlgMYBR1) and PGJG195470, in the biosynthesis of anthocyanins, considering their high transcript levels in white petals and pistil. Further, we compared the expression levels of PlgMYBR1 and PGJG195470 in PFs and WFs. As shown in Fig. 4b, the expression of PlgMYBR1 decreased during flower development, and PlgMYBR1 was highly expressed in WFs compared to that in PFs. The expression pattern of PGJG195470 was highly correlated with the accumulation pattern of anthocyanins in PFs. In chrysanthemum, the changes of Chrysanthemum morifolium MYB#7 transcript levels induced alterations in anthocyanin content, resulting in the production of red or white petals (Xiang et al. 2019). This suggests that PlgMYBR1 is an important factor in the two contrasting flower color varieties of P. grandiflorus too.
Fig. 4.
Expression pattern of putative PlgMYB repressors (PlgMYBRs). A Tissue-specific expression pattern of PlgMYBRs. Data represent FPKM values of RNA-Seq data generated from eight different tissues of P. grandiflorus with white flower. B The expression patterns of the selected PlgMYBRs were analyzed using qRT-PCR. The expression levels of genes from the white petals were compared with those of the purple petals. *p < 0.05. FPKM, fragments per kilobase of transcript per million mapped reads
Functional characterization of putative PlgMYBR1 in tobacco
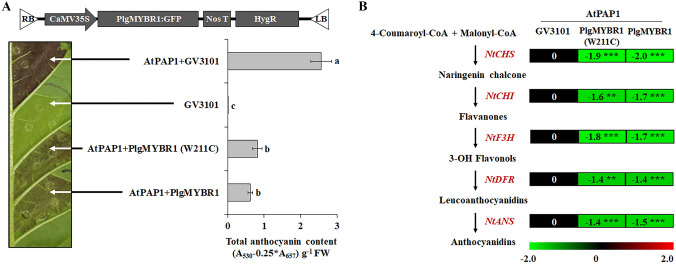
Transient color assays in tobacco leaves have been commonly used to characterize the function of MYB activators and MYBRs in various plants (Wang et al. 2017; Tirumalai et al. 2019; Xiang et al. 2019; Zhou et al. 2019). To characterize the function of PlgMYBR1, we co-infiltrated PlgMYBR1 with a well-known MYB activator, AtPAP1. As shown in Fig. 5a, transient expression of AtPAP1 resulted in the production of abundant anthocyanins after 7 days, whereas PlgMYBR1 significantly inhibited AtPAP1-induced anthocyanin accumulation. Interestingly, PlgMYBR1 sequence obtained from PFs exhibited a difference in single nucleotide sequence that changed the amino acid sequence, W211C, compared with P. grandiflorus genomic data (Fig. S1). Thus, we hypothesized that this inconsistent protein sequence seemed important for catalytic activity. To test this, we compared the inhibitory effect of PlgMYBR1 with that of PlgMYBR1(W211C). As shown in Fig. 5a, there was no difference in inhibitory effects between PlgMYBR1 and PlgMYBR1 (W211C) variants. In addition, the expression levels of AtPAP1-induced anthocyanin biosynthetic genes were significantly reduced by PlgMYBR1 and PlgMYBR1 (W211C). These indicated that PlgMYBR1 is a negative regulator of anthocyanin biosynthesis and floral color variation in P. grandiflorus is associated with the transcript level of PlgMYBR1 rather than the single amino acid difference between WFs and PFs.
Fig. 5.
Transient activation of PlgMYBR1 in tobacco leaves. A Transient expression of PlgMYBR1-expressing constructs in tobacco introduced through agro-infiltration. AtPAP1 infiltration led to high accumulation of anthocyanin, whereas PlgMYBR1 inhibited the accumulation of AtPAP1-induced anthocyanin. B Effect of PlgMYBR1 on the expression of AtPAP1-induced genes involved in the anthocyanin biosynthetic pathway. The level of expression is represented as log2 ratio. Data are means (± SE) of three biological replicates per construct. Different letters indicate statistically significant differences between the samples by Duncan multiples: **p < 0.01, ***p < 0.001
Transposon insertions and deletions in promoter regions of MYBs lead to changes in the transcript levels of MYBs, resulting in flower or skin color variation between cultivars of apple, grape, and dahlia (Poudel et al. 2008; Espley et al. 2009; Ohno et al. 2011). Therefore, one possible explanation should be that transposable element influences on the expression of PlgMYBR1. In addition, the transcript levels of Malus domestica MYB1 change by DNA methylation in its promoter region, resulting in determining anthocyanin content in the fruit peel of two apple cultivars (Ma et al. 2018). Similarly, histone H3K9 demethylase JMJ25 directly affects the expression of poplar MYB182 (anthocyanin repressor) to modulate the biosynthesis of anthocyanins in poplar (Fan et al. 2018). Based on these findings, we hypothesized that altering the expression levels of PlgMYBR1 by epigenetic modification modulate anthocyanin biosynthesis in flowers of two P. grandiflorus varieties, as other possible explanations.
Conclusion
In this study, we identified that PlgMYBR1 as a negative regulator in anthocyanin biosynthesis by analyzing sequence homology and expression patterns. In addition, the heterologous co-expression assay suggested that PlgMYBR1 inhibited the function of AtPAP1, indicating that the transcript level of PlgMYBR1 determines anthocyanin contents in the petal of two P. grandiflorus varieties. Although the function of PlgMYBR1 has been inferred in heterologous systems and further experiments are required to understand the variation of its expression levels, our results have provided insights into the function of PlgMYBR1, which is predicted to regulate flower color formation through the regulation of anthocyanin biosynthesis.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
EK and TKH conceived and designed the experiments. EK performed the experiments. EK and TKH wrote the manuscript.
Data availability
The data presented in this study are available on request from the corresponding author.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Chen L, Hu B, Qin Y, Hu G, Zhao J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol Bioch. 2019;136:178–187. doi: 10.1016/j.plaphy.2019.01.024. [DOI] [PubMed] [Google Scholar]
- Czyczyło-Mysza I, Tyrka M, Marcińska I, Skrzypek E, Karbarz M, Dziurka M, Hura T, Dziurka K, Quarrie SA. Quantitative trait loci for leaf chlorophyll fluorescence parameters, chlorophyll and carotenoid contents in relation to biomass and yield in bread wheat and their chromosome deletion bin assignments. Mol Breed. 2013;32:189–210. doi: 10.1007/s11032-013-9862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Wu D, Shi J, Balfour K, Wang H, Zhu G, Liu Y, Wang J, Zhu Z. Multiple MYB activators and repressors collaboratively regulate the juvenile red fading in leaves of Sweet potato. Front Plant Sci. 2020;11:941. doi: 10.3389/fpls.2020.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Patterson EL, Holalu SV, Li J, Johnson GA, Stanley LE, Greenlee AB, Peng F, Bradshaw HD, Jr, Blinov ML, Blackman BK, Yuan YW. Two MYB proteins in a self-organizing activator-inhibitor system produce spotted pigmentation patterns. Curr Biol. 2020;30:802–814. doi: 10.1016/j.cub.2019.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Wang X, Tang X, Ye X, Ren S, Wang D, Luo K. Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. Plant J. 2018;96:1121–1136. doi: 10.1111/tpj.14092. [DOI] [PubMed] [Google Scholar]
- Grotewold E. The genetics and biochemistry of floral pigments. Annu Rev Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Han E-H, Son Y-W, Kim M-B, Shin Y-W, Cho Y-S, Lee S-W. Establishment of tissue culture and acclimation of white balloon flower (Platycodon grandiflorum DC. cv. Jangback) for the raising of in vitro propagated seedlings. J Plant Biotechnol. 2014;41:134–139. doi: 10.5010/JPB.2014.41.3.134. [DOI] [Google Scholar]
- Hu DG, Sun CH, Ma QJ, You CX, Cheng L, Hao YJ. MdMYB1 Regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 2016;170:1315–1330. doi: 10.1104/pp.15.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon D, Kim SW, Kim HS. Platycodin D, a bioactive component of Platycodon grandiflorum, induces cancer cell death associated with extreme vacuolation. Anim Cells Syst. 2019;23:118–127. doi: 10.1080/19768354.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji MY, Bo A, Yang M, Xu JF, Jiang LL, Zhou BC, Li MH. The Pharmacological effects and health benefits of Platycodon grandiflorus-a medicine food homology species. Foods. 2020;9:142. doi: 10.3390/foods9020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji HS, Bang SG, Ahn MA, Kim G, Kim E, Eom SH, Hyun TK. Molecular cloning and functional characterization of heat stress-responsive superoxide dismutases in garlic (Allium sativum L.) Antioxidants. 2021;10:815. doi: 10.3390/antiox10050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JR, Kang MJ, Cho AR, Shin JH (2019) Physiochemical characteristics of Platycodon grandiflorum flower ethanolic extracts. Korean J Food Preserv 26:785–795. 10.11002/kjfp.2019.26.7.785
- Kim J, Kang SH, Park SG, Yang TJ, Lee Y, Kim OT, Chung O, Lee J, Choi JP, Kwon SJ, Lee K, Ahn BO, Lee DJ, Yoo SI, Shin IG, Um Y, Lee DY, Kim GS, Hong CP, Bhak J, Kim CK. Whole-genome, transcriptome, and methylome analyses provide insights into the evolution of platycoside biosynthesis in Platycodon grandiflorus, a medicinal plant. Hortic Res. 2020;7:112. doi: 10.1038/s41438-020-0329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Hagihara S, Takaya Y, Yoshida K. Polyacylated anthocyanins in bluish-purple petals of Chinese bellflower Platycodon grandiflorum. Int J Mol Sci. 2021;22:4044. doi: 10.3390/ijms22084044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Maitre NC, Pirie MD, Bellstedt DU. Floral color, anthocyanin synthesis gene expression and control in Cape erica species. Front Plant Sci. 2019;10:1565. doi: 10.3389/fpls.2019.01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shan X, Gao R, Han T, Zhang J, Wang Y, Kimani S, Wang L, Gao X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun Biol. 2020;3:396. doi: 10.1038/s42003-020-01134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P. MYB Transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8:689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Lv Y, Tong X, Zhang P, Yu N, Gui S, Han R, Ge D (2021) Comparative transcriptomic analysis on white and blue flowers of Platycodon grandiflorus to elucidate genes involved in the biosynthesis of anthocyanins. Iran J Biotechnol 19: e2811. 10.30498/ijb.2021.239899.2811 [DOI] [PMC free article] [PubMed]
- Ma D, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019;24:275–289. doi: 10.1016/j.tplants. [DOI] [PubMed] [Google Scholar]
- Ma C, Jing C, Chang B, Yan J, Liang B, Liu L, Yang Y, Zhao Z. The effect of promoter methylation on MdMYB1 expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol. 2018;18:108. doi: 10.1186/s12870-018-1320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- Mekapogu M, Vasamsetti BMK, Kwon OK, Ahn MS, Lim SH, Jung JA. Anthocyanins in floral colors: biosynthesis and regulation in chrysanthemum flowers. Int J Mol Sci. 2020;21:6537. doi: 10.3390/ijms21186537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naing AH, Park DY, Park KI, Kim CK. Differential expression of anthocyanin structural genes and transcription factors determines coloration patterns in gerbera flowers. 3 Biotech. 2018;8:393. doi: 10.1007/s13205-018-1408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, Nakashima A, Deguchi A, Tatsuzawa F, Doi M, Iida S, Yazawa S. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis) J Exp Bot. 2011;62:5105–5116. doi: 10.1093/jxb/err216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CG, Bang KH, Kim OT, Jin DC, Kim DH, Sung JS, Seong NS, Park HW, Lee SC. Development of SCAR marker for discriminating between violet flowered lines and white flowered lines in Chinese bellflower (Platycodon grandiflorum A.) Korean J Med Crop Sci. 2007;15:1–5. [Google Scholar]
- Poudel PR, Goto-Yamamoto N, Mochioka R, Kataoka I, Beppu K. Expression analysis of UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) gene in an interspecific hybrid grape between Vitis ficifolia var. ganebu and Vitis vinifera cv. Muscat of Alexandria. Plant Biotechnol Rep. 2008;2:233. doi: 10.1007/s11816-008-0069-0. [DOI] [Google Scholar]
- Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- Tirumalai V, Swetha C, Nair A, Pandit A, Shivaprasad PV. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J Exp Bot. 2019;70:4775–4792. doi: 10.1093/jxb/erz264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, de Souza LP, Fernie AR. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot. 2017;68:4013–4028. doi: 10.1093/jxb/erx177. [DOI] [PubMed] [Google Scholar]
- Wang S, Chu Z, Ren M, Jia R, Zhao C, Fei D, Su H, Fan X, Zhang X, Li Y, Wang Y, Ding X. Identification of anthocyanin composition and functional analysis of an anthocyanin activator in Solanum nigrum fruits. Molecules. 2017;22:876. doi: 10.3390/molecules22060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang Z, Li LX, Wang HB, Zhou H, Chen XS, Feng SQ. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 2022;110:1021–1034. doi: 10.1111/tpj.15720. [DOI] [PubMed] [Google Scholar]
- Wei YZ, Hu FC, Hu GB, Li XJ, Huang XM, Wang HC. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of Litchi chinensis Sonn. PLoS One. 2011;6:e19455. doi: 10.1371/journal.pone.0019455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Chen X, Zong X, Shu H, Gao D, Liu Q. Comparative transcriptome analysis of genes involved in anthocyanin biosynthesis in the red and yellow fruits of sweet cherry (Prunus avium L.) PLoS One. 2015;10:e0121164. doi: 10.1371/journal.pone.0121164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, Chen K. CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019;70:3111–3123. doi: 10.1093/jxb/erz121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Pei X, Zhang H, Li X, Zhang X, Zhao M, Chiang VL, Sederoff RR, Zhao X. MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci. 2021;22:3103. doi: 10.3390/ijms22063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye LJ, Mӧller M, Luo YH, Zou JY, Zheng W, Wang YH, Liu J, Zhu AD, Hu JY, Li DZ, Gao LM. Differential expressions of anthocyanin synthesis genes underlie flower color divergence in a sympatric Rhododendron sanguineum complex. BMC Plant Biol. 2021;21:204. doi: 10.1186/s12870-021-02977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GE, Shin Y, Subramaniyam S, Kang SH, Lee SM, Cho C, Lee SS, Kim CK. Machine learning, transcriptome, and genotyping chip analyses provide insights into SNP markers identifying flower color in Platycodon grandiflorus. Sci Rep. 2021;11:8019. doi: 10.1038/s41598-021-87281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YW, Sagawa JM, Frost L, Vela JP, Bradshaw HD., Jr Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus) New Phytol. 2014;204:1013–1027. doi: 10.1111/nph.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Yang D, Zhang C, Zhang N, Li M, Liu Y. Platycodon grandiflorus—an ethnopharmacological, phytochemical and pharmacological review. J Ethnopharmacol. 2015;164:147–161. doi: 10.1016/j.jep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu B, Wu T, Yang Y, Fan L, Wen M, Sui J. Transcriptomic profiling of two Pak Choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in anthocyanin biosynthetic pathway. BMC Genom. 2017;18:288. doi: 10.1186/s12864-017-3677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang F, Espley RV, Ren F, Zhao J, Ogutu C, He H, Jiang Q, Allan AC, Han Y. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019;221:1919–1934. doi: 10.1111/nph.15486. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.