Abstract
A plant breeding program involves hundreds of experiments, each having number of entries, genealogy information, linked experimental design, lists of treatments, observed traits, and data analysis. The traditional method of arranging breeding program information and data recording and maintenance is not centralized and is always scattered in different file systems which is inconvenient for retrieving breeding information resulting in poor data management and the loss of crucial data. Data administration requires a significant amount of manpower and resources to maintain nurseries, trials, germplasm lines, and pedigree records. Further, data transcription in scattered spreadsheets and files leads to nomenclature and typing mistakes, which affects data analysis and selection decisions in breeding programs. The accurate data recording and management tools could improve the efficiency of breeding programs. Recent interventions in data management using computer-based breeding databases and informatics applications and tools have made the breeder's life easier. Because of its digital nature, the data obtained is improved even further, allowing for the acquisition of images, voice recording and other specific data kinds. Public breeding programs are far behind the industry in the use of data management tools and softwares. In this article, we have compiled the information on available data recording tools and breeding data management softwares with major emphasis on potato breeding data management.
Keywords: Breeding data management, Data digitalization, Phenotyping, Precision breeding
1. Introduction
Large amounts of phenotypic data are generated by the thousands of experiments conducted in the greenhouse/field conditions. The management of such a huge amount of data is crucial to breeding research [1]. In many situations, these tasks are performed by several people at various times, in different places, and different ways, resulting in varying levels of complexity and inaccuracy in data collection. Due to inconsistencies in data that may overlap across silos, data quality often suffers [2]. For breeding operations to scale up in the 21st century, the digitalization of breeding data is critical. The adoption of digital breeding platforms helps in maintaining track of breeding materials, store experimental results, genotype data, record phenotype data, and apply algorithms for analysis, prediction, and selection [3]. Digitalization also allows the breeder to create crosses, nurseries, design field layouts, track pedigree records and analyze data for meaningful information in a fraction of time to develop superior varieties and clones.
The concept of digitalization for phenotypic data collection in plant breeding was proposed in the 1990s but the programs were limited in scope due to non-availability of high-performance computing systems and applications [4]. No matter which phenotyping strategy breeders choose, a breeding program's success depends on its ability to manage data effectively [5]. This is a crucial factor to consider when using high throughput large volume data. Data access by collaborators, simple analytics, good line selection timing, and secure mid to long-term data storage are all dependent on the proper design and implementation of a data streaming pipeline. The development of a software that allows for the digital collection of phenotypic data in the field has been aided by the introduction of high-performance computers. Although advanced breeding informatics softwares for breeding data management and analytics have been developed on a global scale, its implementation rate in public sector breeding programs still remains low. The majority of public sector breeding organizations still do manual data collection and administration processes using paper and pen to record data which is then transcribed in the excel sheets [2,6]. As a result, many breeding programs lack access to historical data sets while others don't have the resources to build the long-term reliable data sets essential to informed breeding decisions. It is crucial to move toward digitalization of acquired data considering the vast volume of information gathered through genotyping, greenhouse/field experiments, as well as the expense and labor involved in gathering that information. In this review article, we discuss various data management tools and their applications, allowing multiple users to collaborate on the same project at the same time to facilitate the breeders to make informed decisions.
2. Digital data recording tools
-
1.
Integrated Breeding (IB) FieldBook: IB FieldBook is an open-source tool that assists breeders in creating field books, collecting phenotypic data, and analyzing collected data for single-environment and multi-environments. This tool has been used for various crops i.e., chickpeas, groundnuts, cassava, beans, cowpea, wheat, rice, sorghum, and maize [7]. IB Fieldbook accepts both the Microsoft Excel Spreadsheet (XLS) and Comma Separated Values (CSV) format. The imported data can be edited in a user-friendly interface and saved to the database, where it can then be exported to a specific R programming format for statistical analysis.
-
2.PhenoApp: PhenoApp is an Android-based open-source tool that helps breeding programs to digitalize data collection. The phenoApps project aims to boost the overall efficiency of breeding programs by integrating and improving existing image processing, modeling, and data collection programs through user-friendly mobile applications [8]. It includes Field Book for data collection and Intercross for managing crosses.
-
a)Field Book: Field Book, the user-friendly field-based phenoApp, has been used in breeding programs all around the world with great success [2]. Field Book eliminates transcription mistakes, captures data in real-time, and enables quick data analysis. It gives breeders an easy way to collect data from field plots. It also allows developers to further customize it to match their individual data collection needs. Field Book generates numerous folders such as field export, field import, plot data, resources, database, and traits. Both XLS and CSV files can be loaded into Field Book. In Field Book, data is gathered by creating traits. The traits panel allows the user to manage the existing traits as well as create new traits or alter existing ones. There are twelve different trait formats available in Field Book viz., numeric, percent, categorical, date, text, boolean, counter, photo, disease rating, location, multicat, and audio. The Import and Export option on the toolbar can be used to create and transfer lists of traits between different devices. The application can be downloaded from https://excellenceinbreeding.org/toolbox/tools/field-book. Field Book is compatible with the ‘Breeding Management System’ (BMS) software and the files created in BMS can be transferred to Field Book and vice versa. The Field Book application can be synchronized with BMS for direct data transfer in BMS.
-
b)Intercross: Intercross, designed and developed by Ref. [9]; is an open-source Android application for planning, tracking, and managing crosses. Breeders can use two distinct interfaces to track the progress of targeted crosses: a basic list with the different combinations and a grid with specific parental combinations. Intercross determines whether the cross is open-pollinated, self-pollinated, polycross, or biparental on the basis of the parental combination. Intercross can also capture the entire new seed lot or family generation process. Intercross also features built-in barcode scanning, allowing users to quickly browse individual cross pages by searching for specific cross IDs via barcode. When using complex sample identification, barcode integration dramatically reduces error. Intercross allows users to group persons, establish a unique identification, and print a barcode label to make tracking crosses easier in programs that create polycrosses. Intercross can be accessed from https://play.google.com/store/apps/details?id=org.phenoapps.intercross&hl=en&gl=US.
-
a)
-
3.
Phenobook: Phenobook is another easy and user-friendly tool for organizing, collecting, and saving experimental data for subsequent analysis [10]. Phenobook was created to collect phenotypic observations in a simple and cost-effective manner. It includes web-based software for experiment design, data input and visualization, and export, as well as a mobile app for collecting data remotely. Phenobook is expected to improve the related procedures by reducing input mistakes, resulting in improved research quality and dependability.
-
4.
PhenoTyper: Another simple and inexpensive tool, the PhenoTyper is open-source application for data management that employs mobiles or personal digital assistants (PDA) [11]. Schemes are created from published ontologies. The PDA's interface stores data in Extensible Markup Language (XML) and CSV format. The Plant Ontology (PO) Consortium was used to create the controlled vocabulary in PhenoTyper [12].
-
5.
FieldLab: It is an Android based application for collection of data and storing images of the plots and is used by scientists at International Rice Research Institute (IRRI), Los Banos, Philippines for promoting the digitalization. It allows you to import and export observed data to Independent Commodity Intelligence Services (ICIS) format. FieldLab is also integrated with barcode reader to retrieve data. It can be downloaded from the website http://bbi.irri.org/products/fieldlab.
-
6.
Phenom App: Phenom App is an open-source Android application that improves the efficiency of collecting phenotypic data in the open-field/greenhouse/nursery etc. Phenom App is developed by Phenome Networks Ltd, Israel. It replaces the old pen-and-paper method, saving time, minimizing typing errors, connecting photos to traits data, and allowing users to browse past data for better decision-making. It is being used by over 70 private organizations across the globe (https://phenome-networks.com/en/our-customers/).
-
7.
GridScore: GridScore is plant phenotyping tool for collection, visualization and sharing of data. Features like GPS referencing, image tagging, and speech recognition are available in GridScore [13]. GridScore makes use of the Web Speech API, IndexedDB, and High Dots Per Inch (HiDPI) displays. It integrates visual representation of the field plan view with barcode system. The data of each stage of the breeding program can be seen, retrieved, and printed in the form of barcodes.
-
8.
Digital Voice Recording (DVR) for Data Collection: Voice recorders have been used since advent of Dictaphone by Nuance Communications, Inc. To mini recorders built into portable electronic devices. In voice recorders, sound files can be electronically produced and transmitted with usual data files. Voice-activated digital recorders with microphone are used for data recording. As computers now have built-in microphones and sound cards, so voice recording requires no additional equipment. The voice recognition software converts data into formats required for further studies including numerical format. Inclusion of a speech interface enables hands-free field data collection, and many more instructions can be used without the use of additional buttons. The speech interface might also be made available to non-English speaking users as speech-based software has been created for various languages. Smatrix (https://smatrix.systems/en/) is Windows and Android based data collection software that offers voice commands for data entry and navigation through its “Command & Control” feature. Smatrix offers a wide range of languages and one can easily record data in their native language. Moreover, Smatrix can manage accents or dialects since it adapts to your pronunciation. Smatrix with a suitable headset enables a noise cancelling system to reduce unwanted background noise. Further, if values are spoken beyond the defined range of values, an error message is generated. Depending on the experiment, users have reported data collection speeds up to four times faster using Smatrix.
Another pipeline for voice recording and dictation to gather, enter, and verify field data has been developed by Ref. [14]. This pipeline fully automates the data entry step while doing away with the necessity for a partner throughout the data collection and proofing steps. This means that one person can collect data just as effectively as a team of two or more persons. This pipeline makes use of a digital audio recording device and free software. The steps involved in the pipeline are as follows.
-
a)
Data is collected by an individual, who then records it as a digital audio file.
-
b)
The data are automatically entered into a spreadsheet while the audio recording is playing on a computer using dictation software.
-
c)
A single person reads the written data and listens to the original audio file, correcting any transcription errors.
In addition, Field Book also supports voice data recording. The “Plot Data folder” in Field Book contains information related to plots in the form of audio and images. Similarly, FieldLab and Seida also features voice data recording. Once the Seida software is installed, various plant characteristics, including numbers (plant height), dates (flowering), lists (disease rankings), images (flowering period), along with text can be entered by keyboard or converted by voice [15]. Another platform, GridScore also has speech recognition feature [13]. GridScore makes use of the Web Speech Application Programming Interface (API), which converts user's speech to text and stores it together with the recorded data. Moreover, applying speech analytics may enable the automation of various monitoring tasks, such as detection of files. The use of voice recording tools in plant breeding will further reduce the burden of typing data for thousands of entries in each season.
3. Handheld devices for data recording
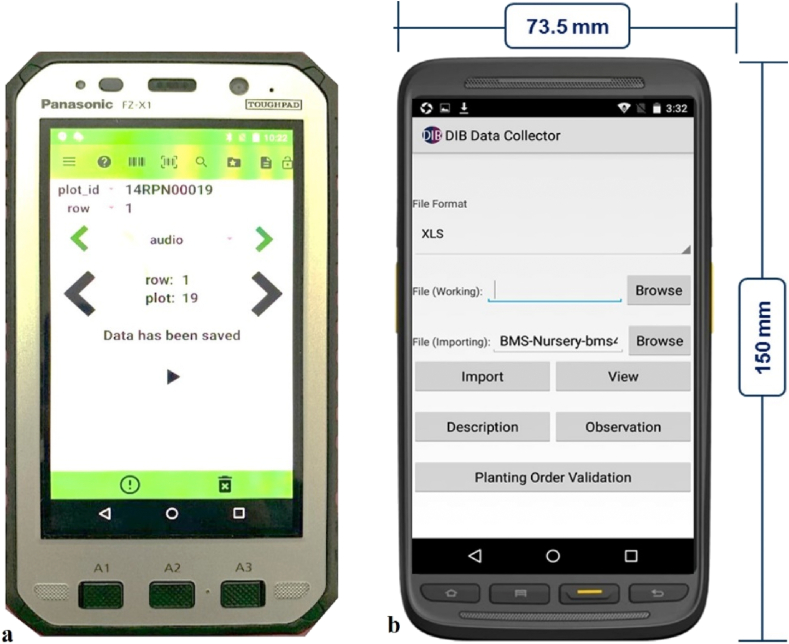
There are number of devices available for digital data recording of plant breeding programs and even smartphones can be used for loading data recording applications. However, rugged devices like toughpads are specially designed for these applications (Fig. 1).
-
1.
Toughpads: Breeders can have access to the recorded data through toughpads. Toughpads developed and designed by Panasonic, provides access to breeding software apps such as Field Book. Toughpad tablets have a fully robust design to endure drop, shock, spills, vibration, humidity, altitude, extreme temperature conditions, and rough handling. Moreover, these devices offer long battery life and pressure resistance, allowing them to meet the demands of breeders in field conditions. Data can also be readily transmitted with BMS for further storage and analysis (Fig. 1a).
-
2.
Digital Data Collector (DIB): DIB is a handheld device, created with an intuitive user interface to assist users in their breeding activities for recording data in the field. It offers a variety of features for field trial data collection, may perform on-site data recording, modifying, viewing, searching, and barcode reading. The handheld gadget data can be easily transferred with other programs, particularly BMS 4.0 and higher. (http://www.biosci.global/softwar-en/dib/) (Fig. 1b).
Fig. 1.
Handheld devices for data recording a) Panasonic toughpad b) Digital data collector.
4. Plant breeding data management platforms
-
1.
AGROBASE/4: AGROBASE/4 version 1.0 is the most widely used and commercially accessible breeding software for the management of germplasm files, pedigree data, genetic analysis, and multi-trait selection. AGROBASE/4 is used in breeding programs for maize, legume, small grains, and hops [16]. Now the revised version AGROBASE Generation II has been released.
AGROBASE Generation II®: It is the most advanced relational database that helps in designing and managing nurseries, managing germplasm, customizing pedigree nomenclature, and genotyping (https://www.agronomix.com/AGROBASE.aspx). This module also supports breeding methods in open-pollinated crops, cross-pollinating crops, clonal crops, polycrosses, and crops with cytoplasmic male sterility. Plethoras of optional analysis modules are available to meet a wide range of requirements. Data can be exported into a variety of formats, including XLS and ASCII plain text. It can be accessed via the Cloud, Citrix/ZenApp®, local area networks (LANs), or virtual private network (VPN) connections and runs on SQL-Server or SQL-Express or SQL-Server Standard Edition from Microsoft®. Many academic centers, government institutions, and seed companies have implemented AGROBASE Generation II. Genovix®: Genovix® (https://www.agronomix.com/genovix/) is a plant breeding and variety testing tool which is built with the most up-to-date application technology and runs on Microsoft® SQL-Server. Genovix offers a more advanced relational database design, which gives the user additional options. Many algorithms have been improved, allowing for more powerful and faster database processing.
-
2.
Enterprise Breeding System (EBS): EBS is an open-source breeding application developed by the Consultative Group on International Agricultural Research (CGIAR) for crop breeding projects in Asia, Africa, and Latin America. EBS enables the user to manage germplasm, trials, nurseries and can connect data to advanced analytics and decision-making tools. EBS can be downloaded from the website https://excellenceinbreeding.org/toolbox/tools/enterprise-breeding-system-ebs. BMS is the main breeding data management software of EBS, which have been discussed in detail below:
Breeding Management System (BMS): BMS is a web-based program (https://bms.icar.gov.in/ibpworkbench/controller/auth/login) developed by Integrated Breeding Platform (IBP) that allows safe and effective data administration, including data storage, retrieval, and analysis, as well as real-time tracking of breeding status (https://excellenceinbreeding.org/toolbox/tools/breeding-management-system-bms). It has been specifically designed to meet the needs of modern plant breeders and has a suite of tools that support multiple breeding strategies from conventional to molecular breeding. BMS is accessible over the internet and compatible browsers such as Google Chrome, Mozilla Firefox, Internet Explorer, Safari, and Opera. BMS contains database-integrated tools that connect to the specific crop database. BMS minimizes the need to manually create, save and import files. Permissions in the system can be tailored to fit the needs of institutions with a variety of user specializations.
-
3.
Plant Research Information Sharing Manager (PRISM): PRISM (https://www.teamcssi.com/index.html) is a user-friendly web-based system that offers easy-to-use tools for keeping track of the breeding data. You can pre-configure inventory books, entry list books, and field books in PRISM. Moreover, additional book types can also be created in Enterprise Manager. PRISM can be customized to include specific plant breeding procedures and terminology to meet the specific requirements of a particular organization.
-
4.
Progeno: Progeno (https://www.progeno.net/software) is another user-friendly breeding program that allows plant breeders to integrate all the phenotypic and genotypic information in a single platform, thus helping in improving the selection efficiency of an experiment. To ensure that the Progeno software is suitable for use by practical breeders, a significant number of private breeding companies actively participated in its creation and assessment. Three key parts make up the Progeno software framework: a central database, the processing unit, and a web-based front end. Pedigree details, (statistically adjusted) phenotypic observations, and molecular marker scores, if available, are all stored in the database. The computational engine receives these typically enormous and imbalanced data sets and uses them to generate prediction models and breeding values for all relevant features.
-
5.
PHENOME: PHENOME is high-throughput phenotyping software developed by Ref. [17]. It allows researchers to collect and manage enormous volumes of plant phenotypic data in the lab/field using a Personal Digital Assistant (PDA) with an in-built barcode scanner. The PHENOME program can be customized to meet the needs of the user's experiments.
-
6.
CropSight: CropSight is Internet of Things (IoT)-powered platform designed for automated data collection, storage, and management [18]. CropSight supports RESTful API that syncs images of representative crop growth for visual crop assessment. Besides, CropSight mediates ongoing as well as historical crop performance.
-
7.
Breeding Information Management System (BIMS): BIMS is an online, free, and secure database system that allows individual breeders to have access to tools like data import/export, data analysis, and data archiving. A group of breeders can share data using BIMS as part of a collaborative endeavour. The Citrus Genome Database, Genome Database for Vaccinium, Pulse Crop Database, CottonGen, and Genome Database for Rosaceae have implemented BIMS [19]. Besides, BIMS allows users to browse publicly available breeding data in the above-mentioned community databases. BIMS was created as an extension module of Tripal which is an open-source toolkit for building online database web portals [20]. The BIMS handbook, training materials, presentations, webinars, and links to various community databases are available on the BIMS website https://www.breedwithbims.org.
-
8.
Database Management System (DBMS): DBMS enables quick and simple access, storage, and interrelation of potato data, as well as automatic reporting functions to support the research program's presentation and analytical needs. Information on potato plant genealogy and progeny performance in early selection, regional adaptation, and disease resistance trials is provided by DBMS tools. Database structures have been built to make it easier to collect and verify new data, connect to a hand-held data collecting device, and export recorded data and charts to conventional word processing and presentation software [21].
-
9.
Breedbase: Breedbase is a web-based comprehensive breeding data management and analyses software program. The Breedbase system has evolved from the Sol Genomics Network (SGN), Cassavabase and related sites [22]. It is an online breeding management software that includes field layout planning, phenotypic data collection, genotypic data storage, and advanced trial analysis. Breedbase is available on the website https://breedbase.org/and supports Breeding Application Programming Interface (BrAPI). Breeders can learn about ontologies, designing trials, collecting and analysing data, and genomic selection from the Breedbase workshops [23]. have integrated ‘waves’ into Breedbase improving its usability for spectral data analysis during plant breeding. Waves, an open-source R package, allow simplified analysis of spectral data along with several cross-validation techniques to evaluate prediction accuracy. Breedbase databases have been built for diverse crops, including cassava, rice, wheat, banana, yams, other Roots, Tuber and Banana (RTB) crops, sweetpotato, oat, kelp, maize, tomato, potato, other Solanaceae, milkweed, Erysimum, and fern [24].
-
10.
Doriane: Doriane is another plant breeding software that offers a variety of ready-to-use friendly interfaces as well as highly customised tools to meet your research and development needs. The client-server architecture makes it easier for researchers and laboratories to share information. Doriane improves the research workflow by integrating agronomical expertise, adaptability, collaboration, IT expertise, accompaniment and scalability. It boosts decision making and reach faster and more often to varieties that work in the market. The data management platform is being used by more than 30 organizations across the globe. For more details, visit https://www.doriane.com/blog/plant-breeding-software.
-
11.
PhenomeOne: The PhenomeOne app was created by Phenome Networks, a private company founded by Prof. Dani Zamir and Dr. Yaniv Semel (2008). PhenomeOne (https://phenome-networks.com/en/) manages all breeding and variety testing data for a wide range of crops, including vegetables, ornamentals, field crops, cannabis, herbs, berries, trees, and many more. This software platform assists in breeding field design, planning, managing selections, and pedigree tracking. The main system module serves as the foundation for all research data management. The remote data collection is done by the PhenoTop module. The inventory module digitizes the seed and plant lot tracking processes. The analysis module is a dashboard that includes a variety of functional and statistical analysis. The genomics module analyses Quantitative Trait Locus (QTL) and Genome Wide Association Study (GWAS) data. The PhenomeBI module generates a list of parent lines to cross to locate the best hybrids. Breeders can choose any above-mentioned modules to include in their PhenomeOne package and complementary modules can be purchased as needed.
-
12.KDDart: The KDDart (https://www.kddart.org/) infrastructure is built for a wide range of scientific research and experimentation. The KDDart application is developed by Diversity Array Technology (DArT) Pty Ltd, Canberra, Australia. The KDDart data platform has following components:
-
a)KDSmart application allows the collection of phenotypic data in the field. The breeder can create and manage traits, trials, and tags. KDSmart can import/export trial data using CSV files. It also allows users to scan traits and plot barcodes.
-
b)KDSens that allows users to collect, manage, store and integrate environmental data.
-
c)KDXplore displays the main trial navigator window which shows trials, selected trial details, and status with action buttons for activities that can be completed for that particular trial. KDXplore assists users in dealing with data in either a graphical or tabular format.
-
d)KDManage allows users to perform administrative and day-to-day management tasks.
-
e)KDCompute allows efficient data analysis within a secure framework. KDCompute manages both basic breeding applications as well as advanced analytics.
-
a)
-
13.
Easy Breed: Easy Breed (https://www.easybreed.com/en/) is a plant breeding data management system from WINTERSTEIGER. Easy Breed uses the open-source R software. It manages field trials, phenotypic data, genotypic data, and pedigree data. Easy Breed can be used for all the crops. The software allows the breeder to create crosses, and design randomized/non-randomized field experiments. Easy Breed can integrate data from different machines and from various suppliers, including seeders, plot combines, and laboratory equipment. Even data from drones and appraisal apps are effortlessly integrated and can be incorporated into the analysis. Many major organizations like Bayer, BASF, Australian Grain Technologies Pty Ltd, Cargill, CIMMYT, Corteva, FAO, ICARDA, KWS, Limagrain, Pioneer, Syngenta are using Easy Breed in their breeding programs (https://www.wintersteiger.com/en/Plant-Breeding-and-Research/References).
-
14.
GENEFLOW: GENEFLOW (http://www.geneflowinc.com/) is breeding software that integrates phenotype, genotype, and pedigree data stored in separated databases for studying phenotypic and genetic diversity. GENEFLOW permits the breeder to keep track of the genetic fingerprints of a particular variety and identify the ancestor responsible for a particular trait.
-
15.
Seida: A breeding application designed for assisting breeders in the collection of data quickly and accurately [15]. The phenotypic data can be connected with a variety of databases for subsequent decision-making processes by providing an enhanced option for conventional breeding, QTL localisation, marker assisted selection, and genomic studies.
-
16.
E-Brida:E-Brida is a user-friendly interface for plant breeders that provide information on the number of crosses, selections, and propagations performed over time. The trials can be labelled with a barcode that is generated automatically. It also enables breeders to navigate various attributes of a pedigree tree using a barcode label/RFID chip (https://www.e-brida.com/plant-breeding-software-e-brida/).
-
17.
Banana Breeding Tracker (BBT): BBT is a web-based breeding tracker that enables onsite digital data recording of banana [3]. It offers the relevant data in the form of a Quick Response (QR) code, reducing labelling errors. A fresh QR code can be generated for every development made in the breeding stages to avoid transcription errors. The QR code must be updated with phenotypic and genotypic data and made publicly available in order to improve banana breeding program. BBT is compatible with Google Chrome, Opera, Safari, Mozilla Firefox, Internet Explorer, and Microsoft Edge and provides access to multiple users from anywhere at any time through the link http://nrcbbioinfo.byethost33.com/bbtbase/index.php.
Most of these software frameworks are typically hosted on a dedicated server inside the breeding organizations walls and is only available to breeders with the necessary privileges. The number of android/iOS applications for field data recording and databases for plant breeding data management are increasing day by day due to more emphasis on precise phenotyping and data management. Therefore, it is difficult to list all available applications in an article. However, we believe that we have reported all major applications being used till date (Table 1).
Table 1.
Different breeding data handling databases implemented in various crops.
| Breeding database | Crops/Crop family | Reference |
|---|---|---|
| AGROBASE/4 | Corn, food legumes, hop, and canola | [16] |
| BMS | Maize, Sorghum, Pearl millet, Chickpea, Pigeonpea | https://excellenceinbreeding.org/toolbox/tools/breeding-management-system-bms |
| PRISM | Maize, soybeans, sorghum, cotton, rice, pumpkin, sunflower, wheat and popcorn | http://www.sytseed.com/product_prismen.html |
| Progeno | Wheat, Maize | https://www.progeno.net/ |
| PHENOME | Tomato | [17] |
| CropSight | Wheat | [18] |
| Field Book and BIMS | Genome Database for Rosaceae, CottonGEN, Pulse Database, Citrus and Vaccinium Genome Database | [19] |
| DBMS | Potato | [21] |
| Breedbase | Cassava, rice, wheat, banana, yams, other Roots, Tuber and Banana (RTB) crops, sweet potato, oat, kelp, maize, tomato, potato, other Solanaceae, milkweed, Erysimum, and fern | [24] |
| Doriane | Cucurbitaceae | [25] |
| PhenomeOne | Vegetables, ornamentals, field crops, cannabis, herbs, berries, trees etc. | https://phenome-networks.com/en/ |
| KDDart | Wheat, Mungbean | [26]; https://www.aciar.gov.au/sites/default/files/2022-03/CIM-2014-079-final-report_0.pdf |
| Easy Breed | All major crops | https://www.wintersteiger.com/en/Plant-Breeding-and-Research/References |
| E-Brida | Black currant, Vegetable and flowers | [27]; https://www.e-brida.com/plant-breeding-software-e-brida/ |
| BBT | Banana | [3] |
| Phenobook | Wheat | [28] |
5. Breeding interface/ontology tool
-
1.
Breeding Application Programming Interface (BrAPI): BrAPI (https://brapi.org/brapps) is a useful tool for data access, exchange, and integration between systems and applications [29]. BrAPI enables breeders to collect and transfer data, visualize geographic information, and search different parameter combinations.
-
2.
Crop Ontology (CO): Use of field books in the collection of phenotypic data improves its consistency and quality which requires the usage of ontologies approved by the end-users [30]. Adding high-quality and widely used ontologies to data annotations improves its discoverability, interoperability, and reusability. Ontologies must be created if they are not available which is a time-consuming process. CO (https://cropontology.org/) is a free, online curation and annotation tool developed by CGIAR that allows users to browse a wide database of crop terminology organized by phenotypic, breeding, germplasm, and trait categories. Breeders can have access to these COs through two web-based online services: the ‘CO Lookup Service’ and the ‘Crops Terminizer’. Ontologies and trait dictionaries are available online for banana, common bean, cowpea, chickpea, soybean, maize, potato, cassava, groundnut, wheat, rice, sorghum, and Solanaceous crops [31]. This collaborative crop ontology project is funded by the Generation Challenge Programme (GCP).
-
3.
Protege: Protege is the most widely used tool for building and maintaining ontologies. Simple and complex ontology-based applications can be created in a single workspace using Protege's fully customizable user interface [32]. RDF and OWL 2 Web Ontology Language specifications are fully supported by Protege.
6. Data analytical support of breeding software
It should be obvious that, compared to the larger difficulty of integrating breeding data from different sources to guide selection, digitizing data collection is a rather simple problem to address. It is necessary to create platforms that integrate phenotype, genotype, pedigree, and ultimately meteorological and climate data in order to analyze multi-location, multi-year, and multivariate data on creations of linked lines. It is necessary to develop and integrate sophisticated breeding informatics systems into breeding workflows to improve predictions regarding the performance of diverse genotypes in diverse environments. In order to be effective, they must be computationally powerful, accessible in remote locations, and user-friendly [33]. give a thorough assessment of some of the top contenders, however, no single system is in general usage across the crops [6]. The data analyses facilities are available in most of the breeding data management software, which allow data analysis to support breeding decision in a more efficient and faster way. For example, in BMS the data from single and multiple sites can be analysed to draw a valid conclusion. There is no need to change the format of data as the format in which data is entered in software can be directly used for analysis. It also supports both genotypic and phenotypic data which is otherwise difficult to handle in excel sheets. At present, most of these pipelines does not have inbuilt data analytics capabilities but the data files exported from these pipelines can be used directly in advanced data analysis software's. These breeding pipelines support a) Data cleaning, b) Basic statistics, c) Data plotting in form of box plots and histograms, and d) Multilocation stability analysis to identify better wide adaptable genotypes and location specific genotypes. A fully operational and interconnected system should operate without interruption for a) permit simplified trial design, management, and phenotyping processes that help breeders and breeding teams to implement reliable statistical designs fast and simply b) utilize yield trial analytics that include phenotypic spatial correction, evaluations of the quality of the location data, multi-location analytical capabilities, and use of pedigree and genomic estimated breeding values (GEBVs) for selection of superior parental lines c) To make better predictions, summaries, and interpretations of variance components, such as genotype, location, year, agronomic management regime, and their interactions d) making the breeding program simple, intuitive, and powerful through progression of germplasm historically e) efficiently monitor breeding operations, spending, and activities over various timeframes [6].
7. Digitalization of potato breeding program in India
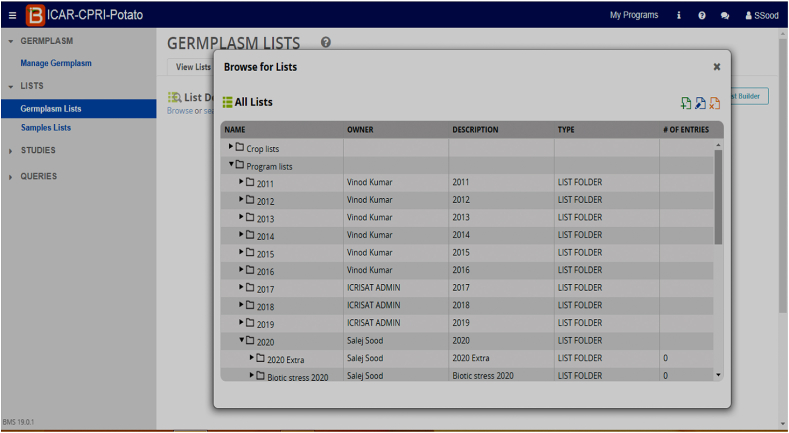
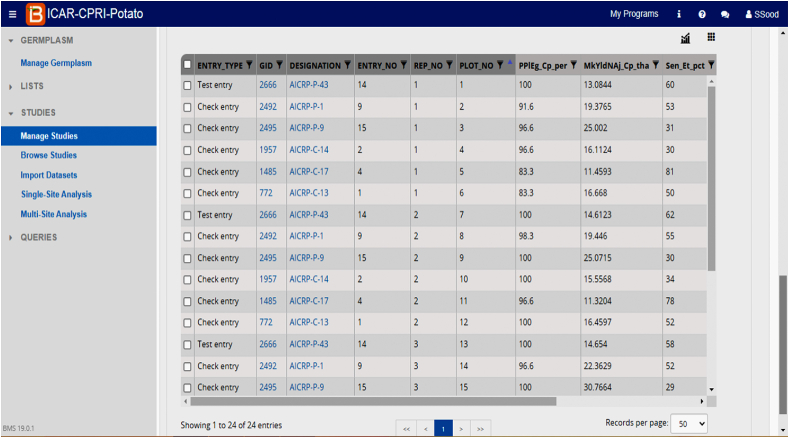
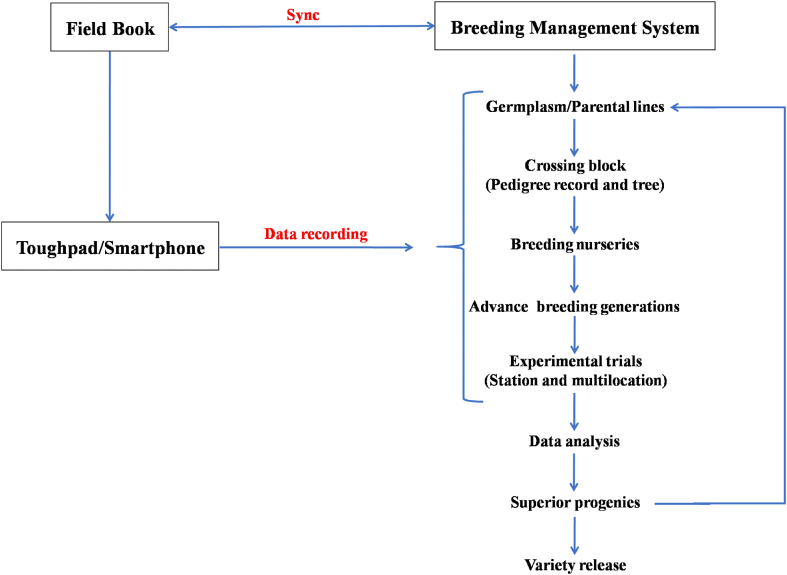
At ICAR-Central Potato Research Institute (CPRI), Shimla, we have started using BMS software (Fig. 2) under the collaborative project between ICAR and Bill Melinda Gates Foundation (BMGF) entitled “Application of Next-Generation Breeding, Genotyping and Digitalization Approaches for improving the Genetic Gain in Indian Staple Crops”. In the year 2019, we started migrating our parental lines in BMS and at present 4918 germplasm lines have been added which includes recurrent parental lines, advanced breeding clones, and varieties (both Indian as well as Exotic). Out of eight breeding programs ongoing at ICAR-CPRI, Shimla, we have started making crossing blocks and breeding nurseries for two breeding programs and efforts are being made to replicate the work in other programs too. Under the All India Coordinated Research Project (AICRP) on potato, all major multilocational trials were designed in BMS from the year 2020 onwards and data was imported directly as excel sheets received from various AICRP centers in BMS format. Besides, all AICRP potato trials for the past 10 years have been redesigned in BMS and uploaded as an online repository for future use (Figs. 3–6). The unified single platform from the selection of parental lines to the final AICRP multilocational testing will reduce the complexities and errors in breeding programs.
Fig. 4.
BMS interface displaying potato germplasm list from 2011 to 2020.
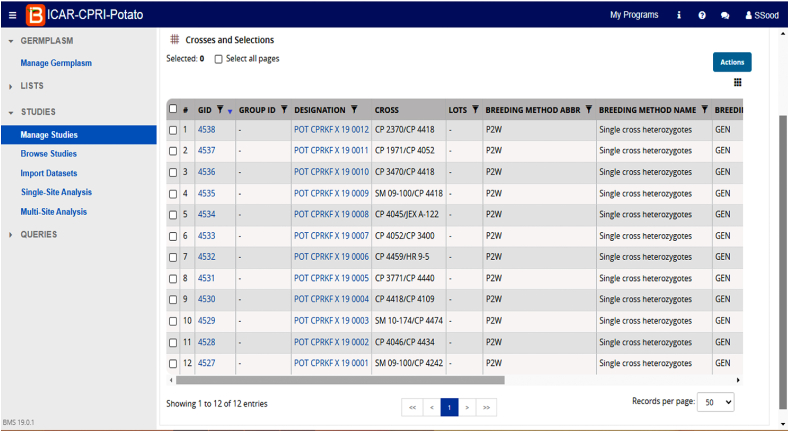
Fig. 5.
BMS interface displaying trials studies.
Fig. 2.
Flow chart of potato breeding digitalization scheme using BMS and Field Book.
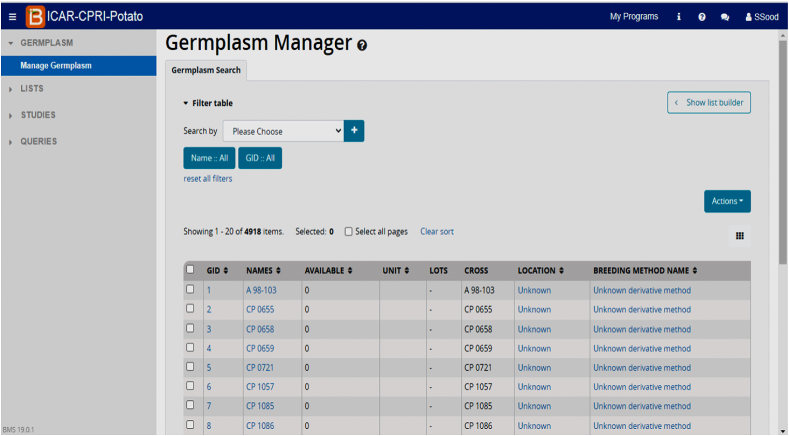
Fig. 3.
BMS interface displaying germplasm manager.
Fig. 6.
BMS interface displaying nurseries studies.
8. Work done at international potato center (CIP)
The International Potato Center (CIP), Lima, Peru, is a CGIAR research center founded in 1971. It provides access to affordable, nutritious food, and promote equitable, long-term business and employment growth. The methods for generating and processing data used by plant breeders have changed dramatically over the past few decades. The use of high-throughput technologies has resulted in an increase in both the quantity and complexity of data but deciphering the significance of the information hidden in the data requires linking with previous data or data collected by others. A robust and practical data management solution is therefore necessary in order to conduct meaningful data analysis. The CIP has created a global trial data management system that is accessible online (https://research.cip.cgiar.org/gtdms/). This system consists of three key components i.e., CIPCROSS, HIDAP, and Field Book Registry [34]. Through CIPCROSS, breeding materials can be tracked from crossing blocks to botanical seed inventories and eventually to seed distribution. It has two main tools: (a) Crossing Management System, a barcode-based tool that labels parents and crosses, stores the information in a database, and generates automated reports; (b) The Botanical Seed Inventory System tool that facilitates information about crosses and name of breeder's, location of seed in the storage, documentation of germination tests, as well as updates on seed stocks.
CIP has developed a highly integrated data analysis platform for potato and sweet potato breeders, called HIDAP in order to unify data collection, quality checking, and analysis. More than 100 R packages are included in this package, which has been built using the statistical platform R that includes the R shiny tools, the knitr package, and the Agricolae package. This software is available for download at https://research.cip.cgiar.org/gtdms/hidap/. It is possible to track clones and families that have been generated by breeding using HIDAP. Through this connectivity, it is possible to verify and maintain the identity of clones across different selection stages and to trace the pedigree of individuals during selection and breeding. Researchers can share field books with coworkers, regional breeding programs, and/or collaborating organizations through the HIDAP network. Another application “Field Book Registry” (https://research.cip.cgiar.org/cipfieldbookregistry/) which allows field books to be updated in real-time and their status to be viewed in the database. Data generated by the potato breeding program at CIP is stored in “Global Roots & Tubers Base” utilizing the free BioMart software (https://research.cip.cgiar.org/gtdms/biomart/). The database is structured to allow the storage of genotypic, phenotypic, pedigree, environmental, and geographical data, as well as the analysis of phenotypic and genotypic data through the use of software tools. A key objective is to identify effective models that are capable of predicting phenotypic traits and outcomes as well as providing valuable insights into the genetics of traits [34].
9. Benefits of data management using digitalization
The information and data stored can be promptly transferred at the touch of a button on the device of choice to the breeders on embracing digitalization (Fig. 7) [33].
-
1.
Data upload: Uploading data from the field is a major challenge with hundreds of spreadsheets. The applications allow you to collect data in the field and then upload it to the central database in seconds.
-
2.
Data accuracy: Data can be typed or copied incorrectly. Digitalization tools use very precise and well-defined import algorithms to ensure that historical data is imported accurately.
-
3.
Access to information: Digitalized documents are accessible and shareable from nearly anywhere within your team. Digitalization permits everyone in the team to have access to the recorded data, thus improving coordination between breeders.
-
4.
Data privacy and security: Digitalization necessitates a high level of security and permits permission-based sharing. User and password protection are available on both the web server and the mobile application. With the appropriate breeding digitalization tool, a breeder can set up particular authorization settings and alerts to allow information to flow between the members of a breeding program. The information is accessible to only specific breeders with valid credentials.
-
5.
Data analysis: It is a very time-consuming process to go through each spreadsheet for data analysis. Breeding software allows the breeder to analyze hundreds of experiments and many other attributes in a fraction of time. This is a big-time saver when a new season is approaching and breeders have to make selections for the next planting.
-
6.
Provide timely decisions: Each year, hundreds of crosses are made in breeding programs. Some crosses may be repeated over time simply because they were made many years ago and forgotten. A breeder can easily tell if a cross has been done before or not using breeding applications. Digitalization helps to keep track of crosses, saving both time and manpower hours. Thus, an up-to-date breeding record allows you to refer to your records quickly for future research planning by providing meaningful information in time.
-
7.
Provides a thorough understanding of the breeding program: You can maintain ongoing, up-to-date information using digital record management. Any new breeder will easily understand the breeding program from the available information in the database.
-
8.
Selection efficiency: Digitalization reduces data transcription errors and maintains proper record keeping of parental lines, crosses, nurseries, and trials that help in selection decisions. Thus, elite lines are selected which increases the breeding program efficiency.
-
9.
Genetic gain: Digitalized data helps in keeping proper records of the pedigree of progenies in different generations and breeding cycles, resulting in the selection of superior parents for recycling in the recurrent selection breeding scheme. Over generations better lines will be produced, resulting in high genetic gain per unit of time.
-
10.
Register hybrids: Hybrid registrations require a detailed history of their development, including data, parents, testing locations, and so on. Retrieving data from a large number of spreadsheets can take a long time. Occasionally, all of the data is missing or there are inaccuracies. This can be accomplished with digitalized tools in a matter of seconds, saving you a significant amount of time.
-
11.
Precision breeding: The use of barcode labels using handheld devices/smartphones will remove the errors in genotype and phenotype data. The true-to-type plant selection for genotype and its association with phenotype across the locations will improve the breeding decisions.
Fig. 7.
Benefits of data management in crop breeding programs.
10. Future perspective
The management of thousands of experimental breeding lines established over various sites and years demands an appropriate breeding data management and analytical systems. Building parallel computing and data management infrastructure that is compatible with big data is necessary to usher in the era of optimum breeding, and new algorithms must also be developed to extract insights from the vast amounts of data. Global projects for community integration become more significant since one organization cannot independently produce all the data necessary for the optimal decision across a variety of locations and crops. Adopting appropriate data management rules and practises that assure data harmonisation, integration, and reusability is essential for the success of such community integration programs. Artificial intelligence (AI) powered data processing and mining is crucial for attaining breeding goals to promote data interoperability, globality, and shareability. AI facilitates the integration of multiple sensors into a single portable device and inbuilt data analysis software to acquire and manage various phenotypic aspects of large populations of plants. With the incorporation of multiple data sources, predictive phenomics would also be feasible, enabling the investigation of the relationship between genotype, environment, and management (G × E × M). Recent developments in data analytics, image processing, and machine learning could be prospective approach to enable automated rapid phenotyping and subsequent decision-making in plant research. We anticipate that these digital data management tools will aid in accelerating the pace of genetic gain to further enable real-time, automated stress trait detection and quantification for plant breeding.
11. Conclusion
A successful breeding process depends on reliable and effective data administration, including data storage, retrieval, and analysis. The traditional methods of data recording and collection are typically time-consuming, costly, and usually involve a lot of human intervention which increases the possibilities of making transcription mistakes. Moreover, breeding requires broad and precise phenotyping of plant populations for the successful application of genomic strategies to introduce desired traits. The data volume is increasing day by day making it difficult to interpret manually and make decisions. Recent intervention of digitalization tools and breeding data management softwares demonstrate real-time tracking and monitoring of the breeding program and securing data storage for future applications. Several breeding data management applications are available and being used in various crops including potatoes. The recent technological advancements in speech analytics have also made speech recognition software accessible to researchers for an audio recording of the data. The adoption of data management tools in breeding programs will improve the data quality, speed and efficiency in terms of disease resistant, high-yielding and better nutritional quality cultivars.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This work was supported by ICAR [1011165], BMGF [1011165].
Data availability statement
No data was used for the research described in the article.
Declaration of interest's statement
The authors declare the following conflict of interests: The corresponding author is an advisory board member of the Agriculture section in Heliyon Journal.
References
- 1.Pieruschka R., Schurr U. Plant phenotyping: past, present, and future. Plant Phenomics. 2019;2019:1–6. doi: 10.34133/2019/7507131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rife T.W., Poland J.A. Field book: an open-source application for field data collection on android. Crop Sci. 2014;54:1624–1627. [Google Scholar]
- 3.Kumar B.V., Backiyarani S., Chandrasekar A., Saranya S., Ramajayam D., Saraswathi M.S., Durai P., Kalpana S., Uma S. Strengthening of banana breeding through data digitalization. Database. 2020;2020:1–6. doi: 10.1093/database/baz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berke T.G., Baenziger P.S. Portable and desktop computer integrated field book and data collection system for agronomists. Agron. J. 1992;84:119–121. doi: 10.2134/agronj1992.00021962008400010023x. [DOI] [Google Scholar]
- 5.Reynolds M., Chapman S., Crespo-Herrera L., Molero G., Mondal S., Pequeno D.N.L., Pinto F., Pinera-Chavez F.J., Poland J., Rivera-Amado C., Pierre C.S., Sukumaran S. Breeder friendly phenotyping. Plant Sci. 2020;295 doi: 10.1016/j.plantsci.2019.110396. [DOI] [PubMed] [Google Scholar]
- 6.Cobb J.N., Juma R.U., Biswas P.S., et al. Enhancing the rate of genetic gain in public-sector plant breeding programs: lessons from the breeder's equation. Theor. Appl. Genet. 2019;132:627–645. doi: 10.1007/s00122-019-03317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lugo-Espinosa O., Sanchez-Gutierrez T.M., Camarena-Sagredo J.G., Vargas M., Alvarado G., Jarquin D., Burgueno J., Crossa J., Sanchez-Villeda H. IBFieldbook, an integrated breeding field book for plant breeding. Rev. Fitotec. Mex. 2013;36:201–208. [Google Scholar]
- 8.Rockel F., Schreiber T., Schuler D., Braun U., Krukenberg I., Schwander F., Peil A., Brandt C., Willner E., Gransow D., Scholz U., Kecke S., Maul E., Lange M., Topfer R. PhenoApp: a mobile tool for plant phenotyping to record field and greenhouse observations. F1000Research. 2022;11:15. doi: 10.12688/f1000research.74239.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rife T.W., Courtney C., Bauchet G., Neilsen M., Poland J.A. Intercross: an Android app for plant breeding and genetics cross management. Crop Sci. 2022;62:820–824. doi: 10.1002/csc2.20702. [DOI] [Google Scholar]
- 10.Crescente J.M., Guidobaldi F., Demichelis M., Formica M.B., Helguera M., Vanzetti L.S. Phenobook: an open-source software for phenotypic data collection. GigaScience. 2017;6:1–5. doi: 10.1093/gigascience/giw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohl K., Gremmels J. A software tool for the input and management of phenotypic data using personal digital assistants and other mobile devices. Plant Methods. 2015;11:25. doi: 10.1186/s13007-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal P., Avraham S., Ilic K., Kellogg E.A., McCouch S., Pujar A., et al. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genom. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raubach S., Schreiber M., Shaw P.D. GridScore: a tool for accurate, cross-platform phenotypic data collection and visualization. BMC Bioinf. 2022;23:214. doi: 10.1186/s12859-022-04755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toczydlowski R.H. An efficient workflow for collecting, entering, and proofing field data: harnessing voice recording and dictation software. Bull. Ecol. Soc. Am. 2017;98(4):291–297. [Google Scholar]
- 15.Liu M. ASA, CSSA and SSSA International Annual Meetings, VIRTUAL, Phoenix, AZ. 2020. Seida APP: phenotypic DATA collection for plant breeding (Abstract)https://scisoc.confex.com/scisoc/2020am/meetingapp.cgi/Paper/127512 [Google Scholar]
- 16.Mulitze D.K. AGROBASE/4: a microcomputer database management and analysis system for plant breeding and agronomy. Agron. J. 1990;82:1016–1021. doi: 10.2134/agronj1990.00021962008200050035x. [DOI] [Google Scholar]
- 17.Vankadavath R.N., Hussain A.J., Bodanapu R., Kharshiing E., Basha P.O., Gupta S., Sreelakshmi Y., Sharma R. Computer aided data acquisition tool for high-throughput phenotyping of plant populations. Plant Methods. 2009;5:18. doi: 10.1186/1746-4811-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds D., Ball J., Bauer A., Davey R., Griffiths S., Zhou J. CropSight: a scalable and open-source information management system for distributed plant phenotyping and IoT-based crop management. GigaScience. 2019;8:1–11. doi: 10.1093/gigascience/giz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung S., Lee T., Gasic K., Campbell B.T., Yu J., Humann J., Ru S., Edge-Garza D., Hough H., Main D. The Breeding Information Management System (BIMS): an online resource for crop breeding. Database. 2021;2021:1–15. doi: 10.1093/database/baab054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spoor S., Cheng C.H., Sanderson L.A., et al. Tripal v3: an ontology-based toolkit for construction of FAIR biological community databases. Database. 2019:77. doi: 10.1093/database/baz077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozub J.G., Lynch D.R., Kozub G.C., Kawchuk L.M., Fujimoto D.K. A relational database system for potato breeding programs. Am. J. Potato Res. 2000;77:95–101. doi: 10.1007/BF02853736. [DOI] [Google Scholar]
- 22.Mueller L.A., Menda N., Strickler S.R., Saha S., Morales N., Flores M., Tecle I.Y., Fernandez-Pozo N., Bauchet G., Ogbonna A., York T., Foerster H., Ellerbrock B., Hosmani P. Boyce Thompson Institute for Plant Research, Cornell University; 2019. Breedbase.https://data.nal.usda.gov/dataset/breedbase Accessed 2022-07-25. [Google Scholar]
- 23.Hershberger J., Morales N., Simoes C.C., Ellerbrock B., Bauchet G., Mueller L.A., Gore M.A. Making waves in Breedbase: an integrated spectral data storage and analysis pipeline for plant breeding programs. Plant Phenome J. 2021;4 doi: 10.1002/ppj2.20012. [DOI] [Google Scholar]
- 24.Morales N., Ogbonna A.C., Ellerbrock B.J., Bauchet G.J., et al. Breedbase: a digital ecosystem for modern plant breeding. 2022. G3. [DOI] [PMC free article] [PubMed]
- 25.Sillon J.F., Royer F., Royer F.I. In: Cucurbitaceae 2008, Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Avignon (France), May 21-24th, 2008. Pitrat M., editor. 2008. Flexibility in Cucurbitaceae breeding management with an integrated research software; pp. 477–482.http://hdl.handle.net/2174/253 [Google Scholar]
- 26.Fazel-Najafabadi M., Peng J., Peairs F.B., Simkova H., Kilian A., Lapitan N.L.V. Genetic mapping of resistance to Diuraphis noxia (Kurdjumov) biotype 2 in wheat (Triticum aestivum L.) accession CI2401. Euphytica. 2015;203:607–614. doi: 10.1007/s10681-014-1284-0. [DOI] [Google Scholar]
- 27.Currie A.J., Alspach P.A. Estimating breeding parameters in a New Zealand blackcurrant (Ribes nigrum L.) breeding programme. Acta Hortic. 2016;1127:185–190. doi: 10.17660/ActaHortic.2016.1127.29. [DOI] [Google Scholar]
- 28.Basile S.M.L., Ramirez I.A., Crescente J.M., Conde M.B., Demichelis M., Abbate P., Rogers W.J., Pontaroli A.C., Helguera M., Vanzetti L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019;19:553. doi: 10.1186/s12870-019-2015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selby P., Abbeloos R., Backlund J.E., Salido M.B., et al. BrAPI-an application programming interface for plant breeding applications. Bioinformatics. 2019;35:4147–4155. doi: 10.1093/bioinformatics/btz190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnaud E., Laporte M.A., Kim S., Aubert C., Leonelli S., Miro B., Cooper L., et al. The ontologies community of practice: a CGIAR initiative for big data in agrifood systems. Patterns. 2020;1 doi: 10.1016/j.patter.2020.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrestha R., Matteis L., Skofic M., Portugal A., McLaren G., Hyman G., Arnaud E. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 2012;3:326. doi: 10.3389/fphys.2012.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musen M.A. The Protege Project: a look back and a look forward. AI Matters. 2015;1:4–12. doi: 10.1145/2757001.2757003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rathore A., Singh V.K., Pandey S.K., Rao C.R., Thakur V., Pandey M.K., Kumar V.A., Das R.R. Current status and future prospects of next-generation data management and analytical decision support tools for enhancing genetic gains in crops. Adv. Biochem. Eng. Biotechnol. 2018 doi: 10.1007/10_2017_56. [DOI] [PubMed] [Google Scholar]
- 34.Bonierbale M.W., Amoros W.R., Salas E., de Jong W. In: The Potato Crop. Campos H., Ortiz O., editors. Springer; Cham: 2020. Potato breeding. [DOI] [Google Scholar]
Web sources
- 35.Field Book. https://excellenceinbreeding.org/toolbox/tools/field-book. Accessed on 28/June/2022.
- 36.Intercross.https://play.google.com/store/apps/details?id=org.phenoapps.intercross&hl=en&gl=US. Accessed on 28/June/2022.
- 37.FieldLab. http://bbi.irri.org/products/fieldlab. Accessed on 28/June/2022.
- 38.PhenomApp.https://play.google.com/store/apps/details?id=com.phn.phenomapp&hl=en&gl=US. Accessed on 28/June/2022.
- 39.AGROBASE Generation II https://www.agronomix.com/AGROBASE.aspx. Accessed on 28/06/2022
- 40.Genovix. https://www.agronomix.com/genovix/. Accessed on 28/June/2022.
- 41.BMS. https://bms.icar.gov.in/ibpworkbench/controller/auth/login. Accessed on 28/June/2022.
- [42.PRISM. https://www.teamcssi.com/index.html. Accessed on 30/June/2022.
- [43.Progeno. https://www.progeno.net/software. Accessed on 30/June/2022.
- [44.BIMS. https://www.breedwithbims.org. Accessed on 30/June/2022.
- [4.Breedbase.https://breedbase.org/. Accessed on 30/June/2022.
- [46.Doriane. https://www.doriane.com/blog/plant-breeding-software. Accessed on 30/June/2022.
- [47.PhenomeOne. https://phenome-networks.com/en/. Accessed on 30/June/2022.
- [48.KDDart.https://www.kddart.org/. Accessed on Accessed on 30/June/2022.
- [49.Easy Breed. https://www.easybreed.com/en/. Accessed on Accessed on 30/June/2022.
- [50.GENEFLOW. http://www.geneflowinc.com/. Accessed on Accessed on 30/June/2022.
- [51.EBS. https://excellenceinbreeding.org/toolbox/tools/enterprise-breeding-system-ebs. Accessed on 30/June/2022.
- [52.E-Brida. https://www.e-brida.com/plant-breeding-software-e-brida/. Accessed on 30/June/2022.
- [53.Smatrix. https://smatrix.systems/en/. Accessed on 30/June/2022.
- [54.ImageBreed. https://imagebreed.org. Accessed on 30/June/2022.
- [55.Global Trial Data Management System. https://research.cip.cgiar.org/gtdms/. Accessed on 30/June/2022.
- [56.HIDAP. https://research.cip.cgiar.org/gtdms/hidap/. Accessed on 30/June/2022.
- [57.Field Book Registry. https://research.cip.cgiar.org/cipfieldbookregistry/. Accessed on 30/June/2022.
- 58.BioMart.https://research.cip.cgiar.org/gtdms/biomart/. Accessed on 30/June/2022. https://www.aciar.gov.au/sites/default/files/2022-03/CIM-2014-079-final-report_0.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.