Abstract
Background
The BestCyte® Cell Sorter Imaging System (BestCyte) facilitates algorithmic discrimination of clinically relevant cells in Pap test cytopathology by classifying and projecting images of cells in galleries based on cytomorphology. Warranted is awareness of potential BestCyte advantages as measured through 3 cytologists’ interobserver diagnostic concordance, specificity and sensitivity differentials, and equivalency grading relative to manual microscopy (MM).
Objectives
Using 500 MM-reported ThinPrep thin-layers, analyze: (1) cytologists’ blinded BestCyte screening to raise Bethesda diagnoses; (2) correlate BestCyte and MM diagnoses (i.e., predicate) to establish Truth Reference Diagnoses (TRDx) from concordance between 4 possible diagnoses; (3) analyze cytologists’ and MM predicate diagnoses through 4 diagnostic thresholds defined by TRDx: NILM (Negative) for specificity, and ASCUS+, LSIL+, and ASCH+ (Positive) for graded sensitivity (with abnormal cells decreasing in size with increasing dysplasia); and, (4) statistically determine cytologists’ equivalency grading to MM using 95% Confidence Interval (CI) ranges.
Results
500 TRDx breakdown (n/%): NILM (241/48.2), ASCUS (79/15.8), ASCH (9/1.80), AGUS (2/0.40), LSIL (86/17.2), HSIL (68/13.6), CA (2/0.40), UNSAT (13/2.60). TRDx breakdown (n/%) per 4 of 4, 3 of 4, 2 of 4 diagnostic concordances: 264 (52.8%), 182 (36.4%), 54 (10.8%), respectively. No cases of discordant diagnoses were recorded. HSIL TRDx were established from 66.2% of 4 of 4 concordances, followed by NILM (59.3%), LSIL (46.5%), ASCUS (41.8%); antithetically, from 4.40% of 2 of 4 concordances. Specificity for MM predicate (NILM): 67.08%; for Cytologists 1, 2, and 3: 89.71%, 82.30%, 97.53%, respectively. For NILM threshold, cytologists revealed Significantly Superior equivalency to MM. Sensitivity for ASCUS+, LSIL+, and ASCH+ thresholds: MM (91.36%, 86.67%, 74.36%); Cytologist 1 (95.88%, 96.97%, 94.87%); Cytologist 2 (95.47%, 95.76%, 93.59%), Cytologist 3 (94.65%, 95.15%, 98.72%), respectively. Cytologists revealed Significantly Superior equivalency to MM for graded Positive thresholds; with Cytologist 3 for ASCUS+ being: Superior.
Conclusions
BestCyte detects and efficiently displays abnormal cells in strategic galleries standardizing objectivity by systematizing mosaics of cell-types for cytologists’ consideration. BestCyte fosters consistent, enhanced cytologists’ sensitivity values for the ASCUS+, LSIL+, and ASCH+ Positive thresholds relative to MM. Also, BestCyte facilitates improved specificity and superior equivalency grading to MM reflecting efficient screening, and reduced labor. Confident interpretations of small dysplastic epithelial cells characteristic of HSIL led to exceptional interobserver diagnostic concordance inferring BestCyte is primed for effective cervical cancer screening practice.
Keywords: BestCyte cell sorter imaging system, Artificial Intelligence-based cytology, Virtual Pap test thin-layer primary screening, Interobserver analysis, Sensitivity, Specificity, Truth reference diagnoses, Diagnostic thresholds, Equivalency grading
Background
Digital imaging systems exploiting algorithms and Artificial Intelligence (AI) are scalable and broadly redefining contemporary medicine by quantitating aspects of subjectively interpreted diagnostic screening practices as, for instance, detection of skin cancer in dermatopathology, or Covid-19 in pulmonary radiography.1 Equally, innovative systems as the BestCyte® Cell Sorter Imaging System (BestCyte) (CellSolutions LLC, Greensboro, NC, USA) may facilitate efficient discrimination of abnormal epithelial cells in Pap test gynecological cytopathology.2, 3, 4
As digital imaging systems in cytopathology benefit directly from advancements in whole slide image (WSI) scanning technology and the availability of cost-efficient processing power, memory capacity, and networking capabilities, they may be primed for deployment in Pap test cervical cancer screening practice.4,5 As BestCyte maps the cellular milieus on glass slides with precise conversion into digitized slides (i.e., WSI), it supports complex algorithmic computations on clinically relevant cells in fields of view (FOVs) to classify and strategically project images of cells in galleries based on cytomorphology.3,4 BestCyte may therefore standardize objectivity between cytologists and inversely decrease interpretive subjectivity, hence diagnostic variance; thus, may prove superior to conventional manual microscopy (MM).3,4,6 Raised awareness of the advantages potentially availed from BestCyte technology is warranted as measured through interobserver diagnostic concordance, specificity, and sensitivity differentials, and cytologists’ equivalency grading relative to MM.
Historically, in 1992 the PAPNET system (Neuromedical Systems Inc, Suffern, NY, USA) introduced a new era in cytopathology by transforming screening practice, whereby abnormal epithelial cells in Papanicolaou-stained Pap test smears were digitally differentiated from normal cells through neural networks.2,7, 8, 9, 10 PAPNET was automated and interactive; it displayed images of cells for human assessment to trigger MM in cases deemed normal or inadequate. PAPNET was thus integrated into quality control (QC) workflows aiming to minimize false-negative reporting.8, 9, 10 Practically, the system extended the diagnostic sensitivity intrinsic in MM to improve the detection rates of abnormal cells that possibly remained unseen due to various factors including but not limited to: Inconsistencies in smear cellularity, obscuration of cells, suboptimal technical quality or Papanicolaou staining, or to cytologists’ interpretive skill sets or bias.3,8,9 In turn, such limitations and the variable physical topographies of cellular milieus in Pap test smears inspired the development of liquid-based cytology (LBC) preparation platforms. LBC produces thin-layers of randomized cells from representative subpopulations of clinical specimens aiming to increase abnormal cell detection rates, and the efficacy of image analysis systems.2
Thereafter, the ThinPrep Imaging System (TIS) (Hologic, Bedford, MD, USA) and the FocalPoint GS Imaging System (Becton Dickinson, Burlington, NC, USA) were introduced for primary screening in 2004 and 2008, respectively, to complement proprietary (i.e., ThinPrep and SurePath) thin-layers and to facilitate final case reporting using Bethesda nomenclature.2 Comparative studies by Boon et al11 in 2010 reported significantly higher odds ratios for detecting abnormal cells in the ASCUS, CIN I-II, and CIN III+ diagnostic categories, and notably in asymptomatic routinely screened women, thus they concluded improved abnormal cell detection rates may be realized when applying TIS on ThinPrep thin-layers relative to PAPNET.
With the successive emergence of AI-driven digital cell image classification, Delga et al3 investigated novel BestCyte technology in 2014 for thin-layer screening and reporting based on its capacity to detect, classify, rank, and organize images of cells in dedicated galleries for targeted displays of cytomorphology. Through a double-blind study, Delga et al3 subjected ThinPrep and CellSolutions’ BestPrep® thin-layers to BestCyte image analysis and concluded it may supplant MM given its efficient image displays consuming shorter screening times to diagnosis; also, that BestCyte’s diagnostic specificity and sensitivity were equivalent to those of MM when using ASCUS as the cutoff diagnosis for the positive threshold (i.e., ASCUS+).3
In 2022, Chantziantoniou4 explored assertions raised by Delga et al3 by investigating BestCyte intraobserver primary screening outcomes as challenged by adjudicative WSI rescreening, whereby screening time formed a surrogate indicator for diagnostic confidence in overall system functionality. Chantziantoniou4 likewise concluded BestCyte may be an effective screening device relative to MM based on its innovative design fostering favorable user learning curves and substantially reduced screening review time expenditures meanwhile maintaining diagnostic confidence. Likewise, that small dysplastic cells, as those arising from HISIL, may be efficiently detected and effectively differentiated from other cells in galleries as demonstrated by exceptional reproducibility kappa coefficients.4
This investigation builds upon the conclusions reached by Delga et al3 and Chantziantoniou.4 The underlying objective was to investigate the contention that BestCyte may efficiently detect and display abnormal epithelial cells arising from cervical lesions of incremental pathobiological severity leading to optimal interobserver diagnostic concordance between screening cytologists as abnormal cells typically present with decreasing sizes with increasing intraepithelial dysplasia from LSIL through to HSIL. Using 500 randomized, MM-reported ThinPrep thin-layer cases this study has 4 salient steps: (1) Blinded, independent primary screening of 500 thin-layers by 3 expert cytologists using BestCyte to raise respective Bethesda diagnoses; (2) correlation of cytologists’ BestCyte diagnoses with MM diagnoses (i.e., predicate) to set Truth Reference Diagnoses (TRDx) based on diagnostic concordance between 4 possible diagnoses; (3) analysis of cytologists’ diagnoses relative to MM predicate diagnoses based on 4 diagnostic thresholds as defined by TRDx (NILM for negative, to calculate specificity; and ASCUS+, LSIL+, and ASCH+ for positive, to calculate graded sensitivity); and, (4) equivalency grading of the 3 cytologists’ BestCyte diagnoses relative to the predicate MM variable based on respective statistical calculations using 95% Confidence Interval (CI) ranges.
Methods
BestCyte cell sorter imaging system
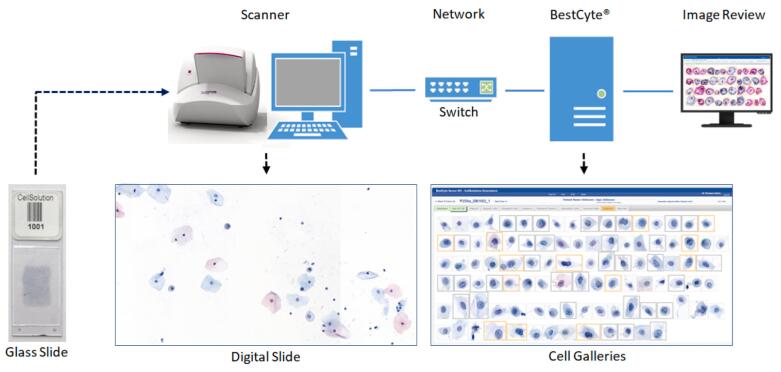
Fig. 1 illustrates the main components of the BestCyte® system: The glass slides with clinical samples containing cells (i.e., target objects for image analysis), a scanner (coupled to a controlling computer) connected to the BestCyte server accessible via image review workstation(s) through a network (or Internet). The image review workstations include standardized displays (i.e., monitors) to project images of cells in galleries with specified settings to ensure optimal contrast and color ranges. BestCyte may also be accessed through WiFi browser connectivity.
Fig. 1.
Main components of the BestCyte Cell Sorter Imaging System.
During scanning, digital images of overlapping FOVs are acquired and merged to generate a seamless high resolution digitized slide (i.e., WSI) mapping the cellular milieu in its entirety. The WSI transfers automatically to the BestCyte server for proprietary classification of the acquired objects into dedicated cell image galleries. BestCyte integrates 8 independent galleries identified through header tabs2, 3, 4 sequenced as: Overview, High N/C, Halos, Atypical, Elongated, Clusters, Endocerv(T-Zone), and InternalCtrl. All galleries are configured to compose images of cells (i.e., tiles) of specific cytomorphology as denoted by tab labeling. For instance, the leading Overview gallery incorporates highest-ranking tiles from the remainder 7 galleries to compose a case preview, forming glimpses of overall image capture, staining intensity, and degree of cell-type diversity and polymorphism. The InternalCtrl gallery displays images of reference cells in decreasing size, from mature superficial squamous epithelial cells through to basal or inflammatory cells appearing predominantly singly in square tiles. The InternalCtrl gallery also functions as an indicator for prescribed classifier performance.
Any given gallery may compose a maximum of 100 tiles based on ranking specifications, if as many tiles are ranked for that specific gallery. The actual number of images collated in any given gallery is indicated in the tab label (except for the Overview gallery). Therefore, as many as 800 tiles may be viewed for any given case from the vast number of high-power fields acquired through scanning and subsequently computed. Cell images in galleries and in corresponding WSIs are exhibited on the networked review stations supporting local or remote access for cytologists to render primary, secondary, or final diagnoses using an interactive Bethesda nomenclature menu. The BestCyte system also accommodates multiple users, therefore tracks respective diagnostic outcomes in accordance with QC metrics as stipulated through Clinical Laboratory Improvement Amendments of 1988.
BestCyte scanning involves perimeter demarcation of the glass slide area of interest prior to scanning (i.e., clinical sample), generation of the initial focus map, and selection of the focusing algorithm (i.e., single, or multi-plane focusing). BestCyte WSIs are optimized through specific optical, illumination, sensor, pixel resolution, and color mode specifications.
BestCyte supports digital diagnostic annotation of cells appearing in either sorted tiles in galleries, or in FOVs through WSI viewing. Therefore, marking of cells, as in conventional microscopy, is emulated whereby images annotated through 40x magnification are digitally tagged to converge in the reporting tab comprising the array of cytomorphology spotted to defend the diagnosis raised for follow-on secondary review and reporting. Selection of any tile in galleries automatically launches a browser tab redirecting the cytologist to a full-screen display of that precise 40x high-power field centrally located, allowing for dynamic, wider assessment thus selective inspection of the adjacent cellular milieu through virtual panning. Reselection of the BestCyte browser gallery tab closes the WSI display reverting the cytologist back to the gallery displays. BestCyte enables WSIs or galleries to be viewed exclusively, not simultaneously juxtaposed within a single full-screen browser display.
Omnidirectional panning is enabled by the computer mouse as is selection between 5x, 10x, 20x, or 40x magnification using either a dedicated on-screen menu, or the mouse scroll-wheel for progressive or regressive zooming through the 4 magnification options. Slide coverage and screening completeness are assured using an on-screen viewfinder with green color tracking that adaptively corrects coverage perimeters depending on selected magnification.
The BestCyte configuration for this study included single plane scanning using a Pannoramic® P250 Flash III RX scanner fitted with a CIS VCC-FC60FR19CL sensor (3DHistech Ltd. H-1141 Budapest, Öv utca 3., Hungary). Acquired WSI files had a pixel resolution of approximately 0.25 μm in a proprietary format using a 20x Zeiss Plan-Apochromat objective with 0.8 numerical aperture. Simulated digital magnification is enabled by the high-resolution camera so that cells exhibited in galleries may appear comparable to those as would be seen in high-power fields through conventional 40x light microscope objectives. The BestCyte WSI reflects the cellular sample deposited within a 20 mm diameter circular glass slide area as is standard for ThinPrep thin-layers (∼314 mm2).
With single plane focusing, and due to its high-resolution optics and camera with flash illumination technology, the 3DHistech Pannoramic® P250 scanner produced a digitized slide reflecting the entire cell deposition area from each ThinPrep thin-layer, on average, within 1 min and 20 sec (∼1.33 min). Approximately, 1 gigabyte digital storage capacity was required for the acquired WSI. During operation, the scanner follows the detailed focus map set prior to scanning. Coupled with AI-driven computational algorithms developed after comprehensive training, BestCyte distinguishes between out-of-focus, slightly defocused, or well-in-focus objects. Therefore, representative cell galleries may be constructed with a predominance of well-in-focus images from the computed FOVs based on ranking. This innovation is suited for slide preparations as ThinPrep or BestPrep® thin-layers even if cellular milieus are topographically varied. Nevertheless, by also enabling an ‘extended focus’ mode, the 3DHistech Pannoramic® P250 scanner allows for the construction of digitized slides composed of merged well-in-focus FOVs acquired through multiple focal planes. Therefore, BestCyte may accommodate and compensate for potential 3-dimensionality or uneven cellular topographies in other preparations as SurePath thin-layers.
Study slide case set
Courtesy of Marlboro-Chesterfield Pathology (Pinehurst, NC, USA), 500 ThinPrep Pap test thin-layer slides (‘Pinehurst Case Set’ (PCS)) were made available to support research and development (R&D) studies of BestCyte technology. These cases were screened commercially by MM and formally reported in 2016. The PCS consisted of non-sequentially accessioned, deidentified, and diagnostically randomized cases reflecting the Bethesda reporting spectrum. The PCS was however enriched with abnormal cases including LSIL and HSIL to support studies on dysplastic epithelial cells arising from significant cervical pathobiology, and for statistical confidence. All PCS thin-layers were cleared of markings prior to onsite scanning at Marlboro-Chesterfield Pathology.
BestCyte cell sorter imaging system primary screening
Three expert cytologists acquainted with BestCyte functionality were authorized to access the archived PCS WSI files. As all patient clinical information and Marlboro-Chesterfield Pathology-issued diagnoses were withheld, all screening exercises were blinded. The PCS thin-layer glass slides were likewise withheld for exclusively virtual exercises. The 500 cases were accessed and viewed independently on workstations with 32” 4K flat-panel HP pavilion monitors set at 100% zoom and 9:16 ratio full-screen display.
All 500 PCS thin-layers were primary screened by the 3 cytologists confidentially by inspecting the images of cells downloading after case selection, sorting adaptively into the 8 BestCyte galleries. Inspection of cells in WSI FOVs or wider WSI panning prior to commitment to diagnosis was optional and permitted. Tiles depicting cells of interest were annotated by the cytologists as were cells in FOVs using the interactive diagnosis menu.
Eight Bethesda diagnoses were applicable for this investigation: NILM, ASCUS, ASCH, AGUS, LSIL, HSIL, CA (Carcinoma), and UNSAT. [Notation: The Bethesda nomenclature used for reporting the 500 ThinPrep PCS in 2016 included the diagnosis of AGUS to communicate the presence of ‘atypical glandular cells’. As such, AGUS was utilized in this study instead of Atypical Glandular Cells (AGC) to maintain diagnostic uniformity].
Study protocol
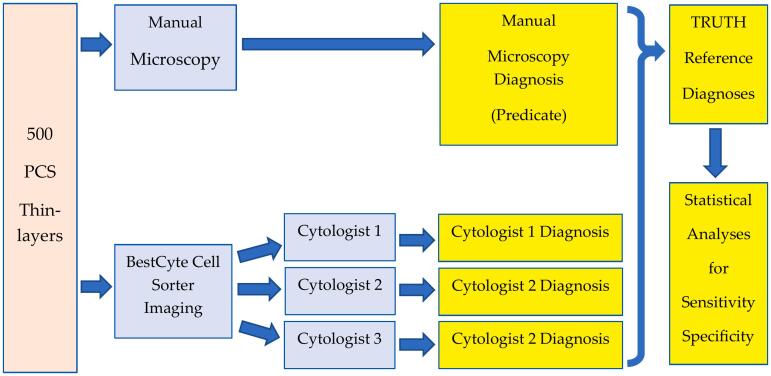
Fig. 2 summarizes the study protocol. All data were recorded in a master Excel spreadsheet. A running list for the 500 PCS thin-layers served to organize the PCS MM predicate diagnoses against the respective diagnoses committed by the 3 cytologists using BestCyte, designated arbitrarily as Cytologist 1, 2, and 3.
Fig. 2.
Summarized study protocol.
Based on this study design, and the 8 applicable Bethesda diagnoses, any combination of 4 independent diagnoses was possible for each of the 500 PCS thin-layers. However, 1 Bethesda TRDx was set for each case based on diagnostic concordance or reconciliation. TRDx were set automatically when 4 of 4, or 3 of 4, independent diagnoses were concordant, whereby the concurred Bethesda diagnosis established the TRDx. In cases with 2 concordant diagnoses of 4, or 4 discordant diagnoses, the WSIs were reviewed to reconcile a TRDx following completion of blinded screening.
Once all 500 TRDx were established, cytologists’ BestCyte diagnoses were correlated against the MM predicate diagnoses and analyzed independently through 4 dedicated Excel pivot tables reflecting 4 diagnostic thresholds as defined by TRDx: NILM for negative; and ASCUS+, LSIL+, and ASCH+, for graded positive reflecting increasing cervical lesion severity. For instance, the ASCUS+ threshold was inclusive of all abnormal diagnoses from ASCUS through to CA. In comparison, the ASCH+ threshold included exclusively ASCH, HSIL, and CA diagnoses.
Diagnostic data & statistical analyses
The pivot tables reflecting the 4 thresholds allowed for the numbers of negative and non-negative, and positive and non-positive, diagnoses to be determined for each cytologist and the MM predicate variable. From these data, respective specificity and sensitivity values were calculated using traditional equations:
-
•
Specificity = True negatives/True negatives + False positives
-
•
Sensitivity = True positives/True positives + False negatives
Cytologists’ equivalency gradings relative to MM predicate diagnoses were calculated through Normal Approximation statistical analyses12 equating the respective specificity and sensitivity values with their 95% CI ranges (i.e., lower and upper limits), and the differences in values, against those of MM predicate diagnoses. Individual cytologists’ equivalency gradings for the NILM, ASCUS+, LSIL+, and ASCH+ thresholds were interpreted according to the following schema drawing from CI ranges:
-
•
Significantly Inferior (CI lower limit is greater than zero)
-
•
Inferior (CI lower limit is less than or equal to zero; upper limit is greater than 0.05)
-
•
Non-inferior (CI upper limit is equal to or less than 0.05)
-
•
Equivalent (CI lower limit is equal to or greater than -0.05; upper limit equal to or less than 0.05)
-
•
Superior (CI lower limit is less than 0.05; upper limit is equal to or greater than zero)
-
•
Significantly Superior (CI upper limit is less than zero).
Data converging into 3 tables are illustrated through 4 bar plots (with superimposed linear trend lines where practical).
Results
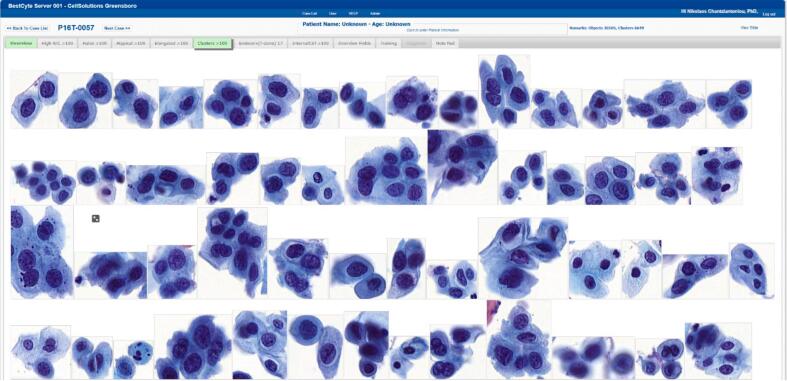
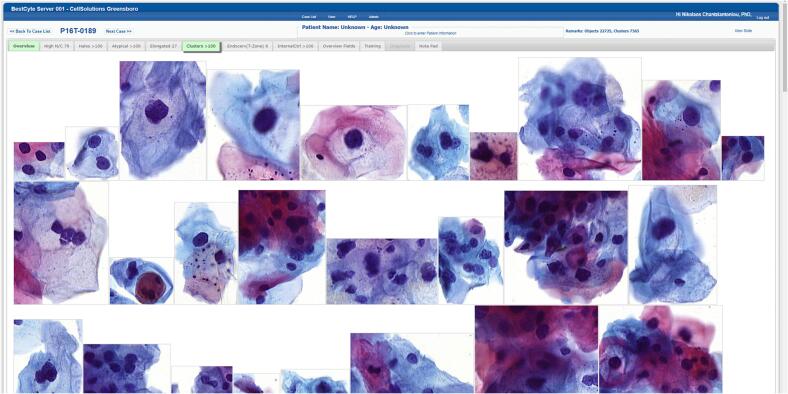
Tiles downloaded, sorted, and displayed adaptively into the 8 BestCyte galleries following ranking sequences within 6 sec of case selection without transfer interruption. The dimensions and orientation, thus the spatial placement, of sorted tiles in galleries was dependent upon the nature of digitized cells or clusters thereof in 40x magnification. For instance, small cells either isolated or in small clusters (i.e., inflammatory cells, histiocytes, basal cells, endocervical cells, or severely dysplastic cells) appeared positioned centrally in smaller square or rectangular tiles (Fig. 3). In contrast, larger cells (i.e., koilocytotic squamous cells, sheets of endocervical, metaplastic or squamous epithelial cells, or elongated cells) appeared positioned randomly in proportionately larger square or rectangular tiles organized in either portrait or landscape layouts (Fig. 4).
Fig. 3.
BestCyte Clusters gallery displaying 54 of the highest ranked of 100 sorted images of cells (40x) in 100% zoom and 9:16 ratio full-screen display (Study case 057: HSIL).
Fig. 4.
BestCyte Clusters gallery displaying 26 of the highest ranked of 100 sorted images of cells (40x) in 100% zoom and 9:16 ratio full-screen display (Study case 189: LSIL).
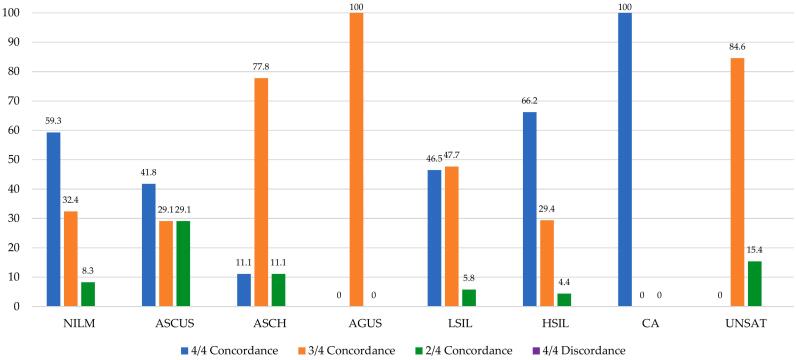
Table 1 outlines the diagnostic data (n/%) arising from the 500 PCS thin-layers analyzed for each of the 8 Bethesda TRDx: NILM (241/48.2), ASCUS (79/15.8), ASCH (9/1.80), AGUS (2/0.40), LSIL (86/17.2), HSIL (68/13.6), CA (2/0.40), and UNSAT (13/2.60). Table 1 also outlines TRDx data (n/%) depending on whether TRDx were established from diagnostic concordance or discordance. Of the 500 TRDx, 264 (52.8%), 182 (36.4%), and 54 (10.8%) were set from 4 of 4, 3 of 4, or 2 of 4, diagnostic concordances, respectively. No cases with 4 discordant independent diagnoses were recorded. These data are illustrated through one bar plot (Fig. 5). Notably, apart from CA, the HSIL TRDx were established from the highest percentage of 4 of 4 diagnostic concordances (66.2%) followed by NILM (59.3%), LSIL (46.5%), and ASCUS (41.8%). Antithetically, the HSIL TRDx were associated with the lowest percentage of cases with 2 of 4 diagnostic concordances (4.40%).
Table 1.
Number (n) and percent (%) of Truth Reference Diagnoses set from 4 of 4, 3 of 4, or 2 of 4 Diagnostic Concordances, or from 4 of 4 Diagnostic Discordances, per Bethesda Diagnosis, from the 500 PCS thin-layers.
| 4 of 4 diagnostic concordance |
3 of 4 diagnostic concordance |
2 of 4 diagnostic concordance |
4 of 4 diagnostic discordance |
TRDx |
TRDx |
||||
|---|---|---|---|---|---|---|---|---|---|
| TRDx | n= | % | n= | % | n= | % | n= | n= | % |
| NILM | 143 | 59.3 | 78 | 32.4 | 20 | 8.30 | 0 | 241 | 48.2 |
| ASCUS | 33 | 41.8 | 23 | 29.1 | 23 | 29.1 | 0 | 79 | 15.8 |
| ASCH | 1 | 11.1 | 7 | 77.8 | 1 | 11.1 | 0 | 9 | 1.80 |
| AGUS | 0 | 0.00 | 2 | 100 | 0 | 0.00 | 0 | 2 | 0.40 |
| LSIL | 40 | 46.5 | 41 | 47.7 | 5 | 5.80 | 0 | 86 | 17.2 |
| HSIL | 45 | 66.2 | 20 | 29.4 | 3 | 4.40 | 0 | 68 | 13.6 |
| CA | 2 | 100 | 0 | 0.00 | 0 | 0.00 | 0 | 2 | 0.40 |
| UNSAT | 0 | 0.00 | 11 | 84.6 | 2 | 15.4 | 0 | 13 | 2.60 |
| Values | 264 | 52.8 | 182 | 36.4 | 54 | 10.8 | – | 500 | – |
Fig. 5.
Bar plot illustrating Percent Truth Reference Diagnoses set from 4 of 4, 3 of 4, or 2 of 4 Diagnostic Concordances, per Bethesda Diagnosis, with inserted value labels. This figure does not reveal data for 4 of 4 Diagnostic Discordances as none were recorded in this study.
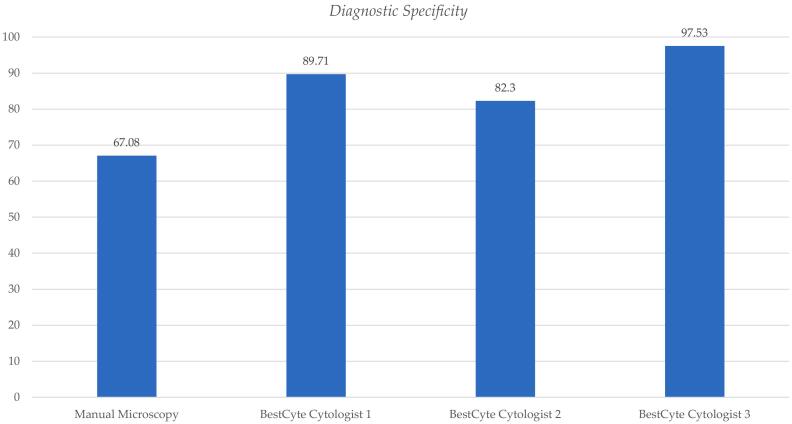
Table 2 outlines the diagnostic specificity differentials for the 3 cytologists’ diagnoses committed through BestCyte for the normal threshold (NILM) relative to MM. Table 2 includes respective differences in specificity values, 95% CI lower and upper limits, and equivalency grading relative to MM predicate diagnoses. The diagnostic specificity value calculated for the MM predicate PCS NILM diagnoses is 67.08% (0.6708). The BestCyte diagnostic specificity values for Cytologist 1, 2, and 3 are: 89.71% (0.8971), 82.30% (0.8230), and 97.53% (0.9753), respectively (mean = 89.85%). The corresponding 95% CI lower/upper limit values for Cytologist 1, 2, and 3 are: –0.2859/–0.1668, –0.2250/–0.0795, and –0.3667/–0.2423, respectively. Drawn from Normal Approximation calculations, the equivalency grading for all 3 cytologists relative to the predicate MM diagnoses for the NILM (negative) threshold is consistently interpreted as being: Significantly Superior (S. Superior). These data are illustrated through one bar plot (Fig. 6).
Table 2.
Diagnostic specificity differentials for the participating cytologists’ diagnoses committed through BestCyte Cell Sorter Imaging for the normal threshold: NILM; as defined by Truth Reference Diagnoses, including differences in specificity values, 95% Confidence Interval lower and upper limits, and equivalency grading relative to manual microscopy predicate diagnoses.
| Diagnostic specificity |
95% Confidence interval |
Equivalency |
|||||
|---|---|---|---|---|---|---|---|
| Diagnosis threshold | Cytologist | Manual microscopy | Cytologist | Difference | Lower limit | Upper limit | Grading |
| NILM | 1 2 3 |
0.6708 0.6708 0.6708 |
0.8971 0.8230 0.9753 |
–0.2263 –0.1523 –0.3045 |
–0.2859 –0.2250 –0.3667 |
–0.1668 –0.0795 –0.2423 |
S. Superior S. Superior S. Superior |
Fig. 6.
Bar plot illustrating differentials of diagnostic specificity for the participating Cytologists 1, 2, and 3 diagnoses committed through BestCyte Cell Sorter Imaging for the normal threshold: NILM; as defined by Truth Reference Diagnoses, relative to manual microscopy predicate diagnoses.
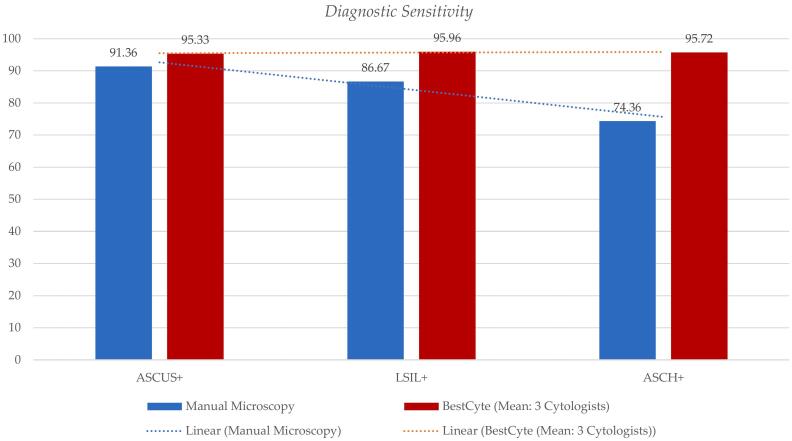
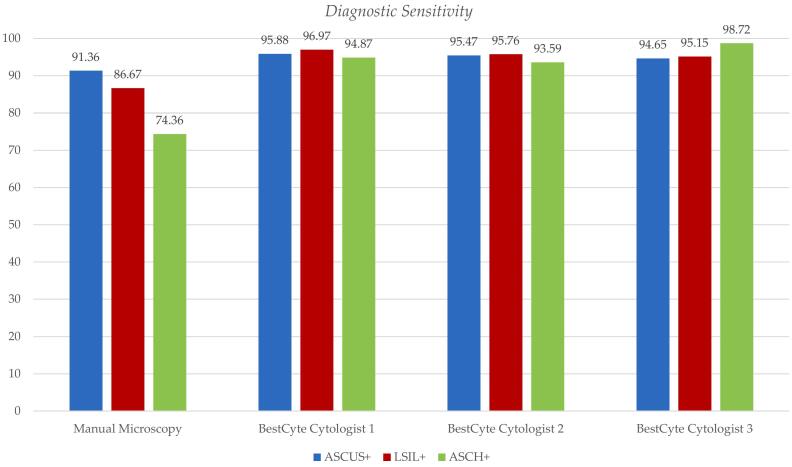
Table 3 outlines the diagnostic sensitivity differentials for the 3 cytologists’ diagnoses committed through BestCyte for the incremental positive thresholds (ASCUS+, LSIL+, and ASCH+) relative to MM. Table 3 includes differences in sensitivity values, 95% CI lower and upper limits, and equivalency grading relative to MM predicate diagnoses. The diagnostic sensitivity values for MM, per positive threshold ASCUS+, LSIL+, and ASCH+ are: 91.36% (0.9136), 86.67% (0.8667), and 74.36% (0.7436), respectively. The corresponding BestCyte diagnostic sensitivity values for the 3 cytologists for thresholds ASCUS+, LSIL+, and ASCH+, respectively are: Cytologist 1 (95.88%, 96.97%, 94.87%); Cytologist 2 (95.47%, 95.76%, 93.59%), and Cytologist 3 (94.65%, 95.15%, 98.72%). Drawn from Normal Approximation calculations, the corresponding equivalency grading for the 3 cytologists relative to the predicate MM diagnoses for thresholds ASCUS+, LSIL+, and ASCH+, is consistently interpreted as being: Significantly Superior (S. Superior), except for Cytologist 3 in the ASCUS+ threshold being: Superior. These data are illustrated through two bar plots (Fig. 7, Fig. 8).
Table 3.
Differentials of diagnostic sensitivity for the participating Cytologists 1, 2, and 3, diagnoses committed through BestCyte Cell Sorter Imaging for the incremental positive thresholds: ASCUS+, LSIL+, and ASCH+, as defined by Truth Reference Diagnoses, including differences in sensitivity values, 95% Confidence Interval lower and upper limits, and equivalency grading relative to manual microscopy predicate diagnoses.
| Diagnostic sensitivity |
95% Confidence interval |
Equivalency |
|||||
|---|---|---|---|---|---|---|---|
| Diagnosis threshold | Cytologist | Manual microscopy | Cytologist | Difference | Lower limit | Upper limit | Grading |
| ASCUS+ | 1 | 0.9136 | 0.9588 | –0.0453 | –0.0852 | –0.0053 | S. Superior |
| 2 | 0.9136 | 0.9547 | –0.0412 | –0.0709 | –0.0114 | S. Superior | |
| 3 | 0.9136 | 0.9465 | –0.0329 | –0.0784 | 0.0125 | Superior | |
| LSIL+ | 1 | 0.8667 | 0.9697 | –0.1030 | –0.1524 | 0.0537 | S. Superior |
| 2 | 0.8667 | 0.9576 | –0.0909 | –0.1487 | 0.0332 | S. Superior | |
| 3 | 0.8667 | 0.9515 | –0.0848 | –0.1464 | 0.0233 | S. Superior | |
| ASCH+ | 1 | 0.7436 | 0.9487 | –0.2051 | –0.3015 | 0.1087 | S. Superior |
| 2 | 0.7436 | 0.9359 | –0.1923 | –0.3105 | 0.0741 | S. Superior | |
| 3 | 0.7436 | 0.9872 | –0.2436 | –0.3453 | 0.1419 | S. Superior | |
Fig. 7.
Bar plot illustrating differentials of mean diagnostic sensitivity values for the participating Cytologists 1, 2, and 3 diagnoses committed through BestCyte Cell Sorter Imaging per incremental positive thresholds: ASCUS+, LSIL+, and ASCH+ (magenta) as defined by Truth Reference Diagnoses, relative to respective sensitivity values for manual microscopy predicate diagnoses (blue) with respective linear trend lines.
Fig. 8.
Bar plot illustrating diagnostic sensitivity values per incremental positive thresholds: ASCUS+ (blue), LSIL+ (magenta), and ASCH+ (green) as defined by Truth Reference Diagnoses, for manual microscopy predicate diagnoses, relative to Cytologist 1, 2, and 3 diagnoses committed through BestCyte Cell Sorter Imaging.
Fig. 7 denotes the mean diagnostic sensitivity values from the participating cytologists’ diagnoses committed through BestCyte per incremental positive thresholds: ASCUS+, LSIL+, and ASCH+ (magenta), relative to the corresponding actual sensitivity values for MM predicate diagnoses (blue) with linear trend lines. Fig. 7 reveals decreasing diagnostic sensitivity values for MM with thresholds sequenced ASCUS+, LSIL+, and ASCH+. In contrast, the corresponding mean diagnostic sensitivity values between the participating cytologists’ diagnoses committed through BestCyte per incremental positive thresholds ASCUS+, LSIL+, and ASCH+ (magenta) revealed a plateau trend due to essentially identical mean sensitivity values: ASCUS+ (95.33%), LSIL+ (95.96%), and ASCH+ (95.72%). These data are alternatively illustrated in Fig. 8 by plotting the actual diagnostic sensitivity values for MM predicate diagnoses against the 3 cytologists’ BestCyte diagnoses per positive thresholds: ASCUS+ (blue), LSIL+ (magenta), and ASCH+ (green).
Conclusions
Assimilation of AI-driven computational systems in contemporary diagnostic medicine depends upon their defined scope of application and effectiveness; however, of paramount significance would be their established relevance in clinical practice.1,2,13, 14, 15, 16 Equally, deployment of dedicated imaging systems in gynecological cytopathology would follow evidence of tangible advantages over conventional MM, including reduced labor and associated costs, overall method simplification, and improved diagnostic certainty between cytologists; collectively translating into enhanced diagnostic concurrence.2, 3, 4 Within this broad context, robust AI-driven BestCyte technology may prelude a formative paradigm in Pap test screening for improved abnormal cell detection for the prevention of invasive cervical cancer. To assure relevance nonetheless, digital imaging systems ought to recreate the fundamentals of diagnostic cytopathology albeit unconventionally; they ought to facilitate and reenable the interpretive conceptualizations applied in clinical practice5 through adequate digital detection of abnormal cells in clinical cases, and their effective display.
Basically, diagnostic interpretations in Pap test practice involve subjective assessments of cytomorphology depending on interpretive constructs, clinical context, and cytologists’ expertise; they thus remain a complex skill set.2,5,6 The raising of diagnoses is based upon detection of recognized diagnostic criteria through screening and after prudent consideration for likely cytomorphologic overlap and diagnostic pitfalls.5 Since the introduction of Papanicolaou’s vaginal fluid smear method in 194317 false-negative reporting remains an inherent limitation and foremost indicator of overall practice.2,5,8,10,18, 19, 20, 21, 22, 23, 24 Digital technology may be primed to lessen such limitations. The data reported herein support the contention that BestCyte technology may pose a robust alternate to MM by enhancing interobserver diagnostic concordance by standardizing objectivity; thus, may also lead to improved specificity and sensitivity values.3,4
According to the literature, false-negative reporting in the conventional Pap smear experience ranges 10%–50%18,20, 21, 22, 23 primarily because of 2 probable factors: (i) Sampling error, and (ii) laboratory error.18,23 In the former, suboptimal clinical sampling may result in poorly sampled cervical lesions, thus in specimens that may contain few abnormal cells. In the latter, suboptimal laboratory processing or screening practice may prove singularly or collectively deficient to spot and interpret few salvaged abnormal cells with precision.18,20,23,24 A relevant object-perception phenomenon described by DeCresce and Lifshitz19 is “Psychological Habituation”. In the cytopathology setting, either through primary screening or rescreening for reporting, cytologists are searching for rare events when the “vast majority of objects examined are not of concern”.19 That is, cytologists may become unintentionally desensitized to subtle cellular aberrations when normal cells exceedingly outnumber abnormal cells. This phenomenon may become increasingly adverse if few diagnostically significant cells in samples are randomized without spatial patterns as in smears. Mukherjee et al25 investigated aspects of this phenomenon by measuring patterns of students’ eye fixation upon clinically significant cells relative to non-significant cells. Nevertheless, due to the physical and mental processes involved in Pap test screening practice and reporting, and particularly for conventional smears, the overall practice is labor-intensive and exacerbated by tediousness, monotony, and fatigue,18,23 all which may potentially contribute to reporting error.
Such challenges inspired the development of LBC. Yet, whereas LBC thin-layers yield randomized cells from subpopulations of clinical specimens, they may also yield fewer abnormal cells relative to smears.19 Following commercial PAPNET rollout in 1992, investigative work to assess its impact in detecting abnormal cells by prompting QC MM produced insights pertaining to the problem of unseen diagnostic cells in smears. While varied, reported insights raised needed awareness of the parameters impacting screening performance. In 1996, Farnsworth et al23 reported a 7% increase in the detection rates for missed abnormalities when MM was triggered by PAPNET. Also in 1996, after reviewing cases (preceding the emergence of HSIL cervical lesions), Mango21 reported that most false-negative cases had few undetected diagnostic cells in smears and typically of smaller size. Furthermore, Boon and Kok24 reported that from 10 false-negative conventional smears discovered through PAPNET-triggered MM, 8 cases revealed cervical cancer cells entirely in epithelial fragments; and, less than 5 abnormal cells were identified in another 2 cases studied. Consequently, in 1997, Wilbur26 stressed that in cases reassessed as being HSIL after focused MM rescreening, the detection of few, isolated, small ‘immature metaplastic’ dysplastic cells manifested a common observation.
Of importance, therefore, are innovative tools that may attract cytologists’ attention alerting them to the likely presence of abnormal cells, and preferably to diagnostic criteria upon which to raise confident interpretations. The balance between digital detection of potentially few abnormal cells in cases and cytologists’ distraction from multitudes of normal cells in sample milieus were core objectives in the overall design, and R&D of BestCyte technology.4
Table 1 and Fig. 5 illustrate the conceivable association between cells of typically decreasing sizes appearing in ThinPrep thin-layers and cytologists’ interpretive concurrence using BestCyte galleries. Apart from 2 cases of CA in this study, HSIL TRDx were established from the highest number of 4 of 4 concordant diagnoses (66.2%) reflecting 45 of the 68 HSIL cases raised between the MM predicate variable and the 3 independent cytologists using BestCyte. Similarly, apart from 11 of the 13 UNSAT cases, ASCH TRDx were set from 77.8% of 3 of 4 concordant diagnoses reflecting 7 of the 9 ASCH diagnoses considered based on few, immature abnormal cells. Antithetically, HSIL TRDx were set from the lowest number (4.4%) of 2 of 4 concordant diagnoses reflecting 3 of the 68 HSIL diagnoses raised. These findings infer that the HSIL TRDx were associated with the least degree of diagnostic uncertainty compared to all other Bethesda TRDx established in this study; most of which HSIL cases produced images of small dysplastic cells appearing either singly in tiles or in small clusters as noted in Fig. 3. Notably, no cases with 4 discordant independent diagnoses were recorded in this study. Overall, these findings are taken to reflect BestCyte’s capacity to detect and display images of cells in dedicated galleries effectively; however, also due to the standardized projections of diagnostic criteria conducive to diagnostic concurrence between cytologists.
These assertions are furthermore supported by the NILM TRDx in this study. Normal cases may include greater varieties of small cell-types of potentially equivalent size to dysplastic cells characteristic of HSIL depending on degree of fixation, cytopreparation, and Papanicolaou staining, as: Basal cells, parabasal cells, immature metaplastic cells, histiocytes, lymphocytes, and isolated endometrial cells. Such cells and their potentially similar cytomorphology constitute differential considerations for HSIL diagnoses. Yet, the NILM TRDx are associated with the second highest number (59.3%) of 4 of 4 concordant diagnoses raised in this study reflecting 143 of the 241 NILM diagnoses, and inversely with 8.3% of 2 of 4 diagnostic concordances.
The lower reproducibility values typically experienced between cytologists with ASCUS diagnoses in routine practice due to interpretive and diagnostic gray zones2,4 are also reflected in the data herein reported as 41.8%, 29.1%, and 29.1% of ASCUS TRDx were set from 4 of 4, 3 of 4, and 2 of 4 diagnostic concordances respectively (Fig. 5). These findings suggest the participating cytologists experienced similar interpretive challenges throughout most cases reported as ASCUS. In contrast, however, given the defined diagnostic criteria for LSIL, 81 of the 86 (94.2%) LSIL TRDx were set from 4 of 4 and 3 of 4 diagnostic concordances collectively, and 5.8% from 2 of 4 concordances (Table 1, Fig. 4, Fig. 5). These findings, overall, reveal favorable associations between cytologists’ interpretive concordance rates (i.e., diagnostic interobserver precision) and BestCyte performance4; thus, ultimately, the potential impact of standardized objectivity in clinical screening practice particularly for precancerous HSIL cases.
Another contributing factor to optimal diagnostic concordance is arguably cell image organization and presentation, to also minimize screening labor.4 Depending on configuration, PAPNET displayed 64 or 128 images of cells as latticed square tiles.18,19,24 As missed cervical cancer cells in smears may appear as epithelial fragments following QC MM prompted by PAPNET,20,24 the Hologic TIS introduced location-guided MM screening whereby cytologists navigate through 22 pre-determined FOVs in ThinPrep thin-layers using automated microscopes for primary or secondary screening and case reporting. Subsequently, Hologic developed a digital imaging system projecting a lattice of 24 equidistant thumbnails (composed of 4 rows of 6 tiles) depicting diagnostically significant cells in 10x magnification. The lattice of images is positioned to the left of a half-screen demarcation, and on the right would appear the WSI ‘cell spot’ displaying the FOV containing the cells depicted in any given thumbnail.16 This system may display an additional 24 thumbnails depending on viewer’s preference. In comparison, BestCyte was engineered to facilitate intuitive classification of images based on arrayed projections of cytomorphologic features in galleries to optimize screening reproducibility, thus time to diagnosis.2, 3, 4 Moreover, whereas PAPNET proved effective for QC monitoring of likely missed dysplastic cells in smears, its projection of 128 latticed images may have influenced cytologists’ screening bias or sense of tediousness not unlike smears.5,18,19,23 As such, the Hologic TIS attempted to alleviate such drawbacks by prompting the cytologist to manual focusing through guided screening to improve discrimination of cells that may appear in larger groups or in 3-dimensional epithelial fragments as those experienced by Boon and Kok.24
It may be equally argued that a direct association exists between the magnitude of psychological habituation in any given case and respective screening labor; and if so, then the greater the degree of cytologists’ distraction the greater the likely degree of attention redirected towards subtle cellular alterations, hence reduced screening labor and time to diagnosis. This probable relationship seems upheld by the strategic assortment of variably sized tiles in the BestCyte galleries maximizing cytomorphologic contrasts.4 Moreover, reduced labor may in turn foster improved diagnostic precision.4
Variations in epithelial cell staining intensity due to shifts in cervical physiology is evidently another variable for consideration in digital system performance assessment. Comparative studies between MM and TIS by Tanaka et al in 202027 explored cases deemed ‘unreadable’ by the TIS. Their study concluded potential cytoplasmic over-staining, as may occur due to cervicitis from bacterial vaginosis, interfered with the system’s capacity to differentiate between nucleus and cytoplasm. Unreadable cases using TIS would dictate MM screening. Such observations underscore the importance for digital systems to adequately compute and discriminate between unlike cell-types that may appear inconsistently stained although present within a given high-power field or FOV. Irrespective of nuclear or cytoplasmic staining intensity, the BestCyte system adequately classifies arrays of tiles between 8 galleries to accentuate cytomorphologic diversity. By such grouping of cytomorphologic elements through galleries, this system facilitates the mental abstractions involved in subjective cytologic interpretations intuitively. Elements include cells with high nucleus/cytoplasmic (N/C) ratios, nuclear or cytoplasmic hyper-chromasia, spindle shapes, subtle cytoplasmic thinning, or overt clearing, and, cells that may appear spatially singly, isolated or in clusters, or in larger 3-dimensional epithelial fragments. For instance, the BestCyte High N/C gallery displays predominantly single, small cell-types with reduced cytoplasmic volume relative to nuclear volume, such as: Epithelial basal cells, parabasal cells, naked squamous cell nuclei typical of squamous atrophy or bacterial cytolysis, lymphocytes, histiocytes, and dysplastic cells characteristic of HSIL.4 Chantziantoniou4 reported optimal intraobserver kappa reproducibility coefficients for HSIL cases diagnosed through BestCyte technology as the characteristic diagnostic criteria were displayed unequivocally through 40x magnification in tiles populating the Overview gallery. BestCyte design facilitated confident commitment to HSIL diagnoses within 1.52 min on average.4
Such achievements are tantamount to the magnitude of data inputted to enable cell-type classification through algorithmic computations. The images programmed into the BestCyte algorithms through which to detect cytomorphologic features ensured their follow-on reidentification through screening. For instance, the High N/C gallery projects images of various cell-types with high N/C ratios discussed above. These advancements in digital detection and display of cytomorphology facilitated rapid discrimination between unlike cell-types minimizing screening times.3,4 Equally, such innovations simplify the fundamental constructs involved in cytologic interpretation,5 and particularly in cases with pronounced cytomorphologic overlap as in those involving differential considerations for HSIL.
The statistical analyses to calculate specificity (Table 2, Fig. 6) through the NILM threshold for negative produced a value of 67.08% for the MM predicate variable. In comparison, specificity values for Cytologists 1, 2, and 3, are: 89.71%, 82.30%, and 97.53%, respectively. Based on 95% CI ranges of specificity values, all 3 cytologists have a Significantly Superior equivalency grading relative to MM (Table 2). Similar studies by Tanaka et al27 to investigate correlative specificity between MM and TIS analyses on 4011 ThinPrep thin-layers reported a specificity value of 88.87% for MM, and 89.55% for TIS; concluding that TIS was equivalent albeit non-inferior to MM. Data herein reported suggest improved specificity values are possible through BestCyte technology.
Table 3 and Fig. 7, Fig. 8 reveal the sensitivity values herein calculated for the MM predicate diagnosis variable relative to the 3 cytologists’ diagnoses for the 3 thresholds for graded positive as established by TRDx: ASCUS+, LSIL+, and ASCH+. The sensitivity values for MM for these thresholds are: 91.36%, 86.67%, and 74.36%, respectively. The gradually decreasing sensitivity values for MM across these thresholds is evident in Fig. 7 (with linear trend line) and in Fig. 8. In contrast, the sensitivity values for the 3 cytologists across these thresholds are relatively equivalent. With thresholds sequenced ASCUS+, LSIL+, and ASCH+, the mean sensitivity values between the 3 cytologists are: 95.33%, 95.96%, and 95.72%, respectively (Fig. 7, Fig. 8). All 3 cytologists have a Significantly Superior equivalency grading relative to MM baring 1 cytologist being Superior to MM in the ASCUS+ threshold (Table 3). When using thresholds defined by biopsy diagnoses, Tanaka et al27 investigated diagnostic sensitivity between MM and TIS. They reported equivalent MM/TIS sensitivity values of 91.8%/92.5% for CIN1+, 92.5%/94.0% for CIN2+, and 92.7%/94.0% for CIN3+, respectively, suggesting equivalent values across these thresholds. Therefore, despite using a broader, more inclusive threshold for positive (i.e., ASCUS+), data herein reported suggest improved specificity and sensitivity values may be achieved between independent cytologists, and with greater precision using BestCyte technology relative to MM, leading to superior equivalency gradings across the spectrum of Bethesda diagnoses.
Digital detection of rare cells, or events, through AI-driven algorithms is a promising advancement that may produce favorable balancing between diagnostic accuracy and reduced labor for improved cervical cancer screening outcomes.4 Study case 057 in this investigation is a case in point. Fig. 3 depicts the BestCyte Clusters gallery from HSIL study case 057 illustrating 54 of the 100 tiles ranked for this gallery through 9:16 ratio full-screen display. Nonetheless, the total of 717 tiles in simulated 40x magnification sorted and viewed through 8 galleries for this case were compiled following computations assessing 30105 relative objects from the 1150 FOVs acquired through 20x scanning typical for ThinPrep thin-layers based on cell deposition surface area (unpublished data). Therefore, BestCyte conducts considerable magnitudes of computations before displaying intuitive projections of cells to facilitate the greatest likelihood that diagnostically relevant cytomorphology and diagnostic criteria are noted through enhanced cytologists’ attention to detail. BestCyte galleries are not unlike the compositions of colorful cells illustrated in full-page Plates in Papanicolaou’s earlier publications to display cytomorphologic templates.17,28 Ultimately, BestCyte aligns cytologists’ attention to crucial cytomorphologic baselines4,29 to facilitate validation or rejection of interpretive suspicions. Such advantages availed through digital image analysis may prove substantial in the academic or clinical settings.30,31
Statistical analyses for equivalency grading as Normal Approximation12 produce a numerical value by which a cytologist’s equivalency to MM may be interpreted as being either superior or inferior depending on specificity and sensitivity values and their 95% CI ranges. However, for a screening cytologist or reporting pathologist practicing in a subjective diagnostic discipline as cytopathology, digital platform inferiority or superiority to MM may also attest to a sense of collective nuances inclusive of confidence and comfortability relative to career observations from conventional light microscope practice (emulated by digital imaging systems as BestCyte). The corresponding author investigated the BestCyte digital imaging system in a R&D setting to assess intra-4 and interobserver reproducibility arising from primary screening relative to known diagnoses established through MM. These studies honored various Recommendations and Good Practice Statements proposed for digital system assessment and validation.32, 33, 34 Accordingly, concurrent with formal clinical trials and system deployment in the clinical setting, BestCyte may be subjected to a rigorous suite of exercises relative to known cases and diagnoses involving primary screening and senior reviewer cytologists (i.e., cytotechnologists) and pathologists,35,36 and the quality control and assurance workflows applicable in diagnostic practice. All abnormal cases in the 500 ThinPrep PCS were formally reported by a pathologist in 2016. In this study, as the entire PCS was examined by 3 experienced cytologists exclusively within the primary screening realm, diagnostic concordance was taken to reflect comparable screening expertise and ensuing interpretive judgments incorporating established diagnostic criteria as in formal clinical practice.
Summary: The data reported herein substantiate speculations raised previously by Delga et al3 and Chantziantoniou.4 By strategically organizing digital images depicting diagnostically relevant cytomorphology in dedicated galleries, BestCyte technology standardizes objectivity between screening cytologists by systematizing the mosaics of cells (i.e., objects) projected for diagnostic consideration. In turn, BestCyte minimizes potential cytologists’ screening and interpretive effort, thus also object-perception biases; therefore, it may minimize interpretive subjectivity leading to enhanced interobserver diagnostic concurrence with superior equivalency relative to conventional MM. Due to its efficient detection, classification, and display of abnormal cells, BestCyte is primed to foster confident interpretations of small dysplastic epithelial cells characteristic of HSIL relative to other potentially mimicking cell-types. Moreover, BestCyte is versatile; it may accommodate ThinPrep, SurePath, BestPrep®, or thin-layer slides from other modalities.37 Such robust cell-type discrimination through organization of images in galleries using AI-driven computational innovation is fundamentally unique compared to navigational location-guided screening systems as TIS. BestCyte may thus pose a novel dimension in gynecological cytopathology. Ultimately, BestCyte may be deployed for primary and review screening, and final case sign-out; and therefore, primed for the identification of cervical precancerous lesions with improved diagnostic specificity and sensitivity for effective Pap test practice through digital image-based technology. BestCyte’s web-based connectivity adds yet another noteworthy advantage in the cervical cancer screening setting by facilitating robust synergy between expertise and knowledge transfer, thus efficient use of human resources and currently exploitable innovations in digital technology.
Statement of Ethics
All 500 PCS ThinPrep thin-layers analyzed for this research were deidentified prior to digital imaging, and glass slides and respective Marlboro-Chesterfield Pathology-issued diagnoses with patient-related clinical information were withheld for blinded studies.
Funding Sources
The research work conducted for this article was supported through Consultation service to CellSolutions LLC, NC, USA. No other funding was received for this work.
Author Contributions
The corresponding author, Dr. Nikolaos Chantziantoniou, PhD, CFIAC, conceptualized this study, acquired, analyzed, and interpretated the data presented, and completed the manuscript in its entirety.
Data Availability Statement
Data generated and analyzed are herein presented completely through Tables and Figures. Further inquiries may be directed to the corresponding author.
Conflict of Interest Statement
The corresponding author is contracted Consultant to CellSolutions LLC, Greensboro, NC, USA.
Acknowledgments
The author gratefully acknowledges support from Dr. Dell Dembosky, MD, and cytology staff at Marlboro-Chesterfield Pathology (Pinehurst, NC, USA) for availability of the 500 ThinPrep thin-layers to support this research. Likewise acknowledged is support from Dr. Thomas Gahm, PhD, President of Select Laboratory Software development, and optical engineer at CellSolutions, for providing the technical overview of the BestCyte system; and from Dr. Robert P. Hirsch, PhD, President of Stat-Aid Consulting, for his insights and for performing the statistical calculations for specificity, sensitivity, and equivalency grading.
References
- 1.Cadario R., Longoni C., Morewedge C.K. Understanding, explaining, and utilizing medical artificial intelligence. Nat Human Behav. 2021;5:1636–1642. doi: 10.1038/s41562-021-01146-0. [DOI] [PubMed] [Google Scholar]
- 2.Lew M., Wilbur D.C., Pantanowitz L. Computational cytology: lessons learned from Pap test computer-assisted screening. Acta Cytol. 2021;65:286–300. doi: 10.1159/000508629. [DOI] [PubMed] [Google Scholar]
- 3.Delga A., Goffin F., Kridelka F., Maree R., Lambert C., Delvenne P. Evaluation of CellSolutions BestPrep® automated thin-layer liquid-based cytology Papanicolaou slide preparation and BestCyte® cell sorter imaging system. Acta Cytol. 2014;58(5):469–477. doi: 10.1159/000367837. [DOI] [PubMed] [Google Scholar]
- 4.Chantziantoniou N. BestCyte® cell sorter imaging system: primary and adjudicative whole slide image rescreening review times of 500 ThinPrep Pap test thin-layers - an intra-observer, time-surrogate analysis of diagnostic confidence potentialities. J Pathol Inform. 2022;13 doi: 10.1016/j.jpi.2022.100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chantziantoniou N., Mukherjee M., Donnelly A.D., Pantanowitz L., Austin R.M. Digital applications in cytopathology: problems, rationalizations, and alternative approaches. Acta Cytol. 2018;62:68–76. doi: 10.1159/000484434. [DOI] [PubMed] [Google Scholar]
- 6.Clary K.M., Davey D.D., Naryshkin S., et al. The role of monitoring interpretive rates, concordance between cytotechnologist and pathologist interpretations before sign-out, and turnaround time in gynecologic cytology quality assurance. Arch Pathol Lab Med. 2013;137:164–174. doi: 10.5858/arpa.2012-0120-CC. [DOI] [PubMed] [Google Scholar]
- 7.Pouliakis A., Karakitsou E., Margari N., et al. Artificial neural networks as decision support tools in cytopathology: past, present, and future. Biomed Eng Computat Biol. 2016;7:1–18. doi: 10.4137/BECB.S31601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koss L.G., Lin E., Schreiber K., Elgert P., Mango L. Evaluation of the PAPNET cytologic screening system for quality control of cervical smears. Am J Clin Pathol. 1994;101(2):220–229. doi: 10.1093/ajcp/101.2.220. [DOI] [PubMed] [Google Scholar]
- 9.Denaro T.J., Herriman J.M., Shapira O. PAPNET testing system – technical update. Acta Cytol. 1997;41:65–73. doi: 10.1159/000332307. [DOI] [PubMed] [Google Scholar]
- 10.Wilbur D.C. False negatives in focused rescreening of Papanicolaou smears: how frequently are ‘abnormal’ cells detected in retrospective review of smears preceding cancer or high-grade intraepithelial neoplasia? Arch Pathol Lab Med. 1997;121(3):273–276. [PubMed] [Google Scholar]
- 11.Boon M.E., Ouwerkerk-Noordam E., Maijer-Marres E.M., Bontekoe T.R. Switching from neural networks (PAPNET) to the Imager (HOLOGIC) for computer-assisted screening. Acta Cytol. 2011;55:163–166. doi: 10.1159/000323310. [DOI] [PubMed] [Google Scholar]
- 12.Agresti A. 2nd Edition. Wiley; 2007. An Introduction to Categorical Data Analysis; pp. 246–247. [Google Scholar]
- 13.Reyna M.A., Nsoesie E.O., Clifford G.D. Rethinking algorithm performance metrics for artificial intelligence in diagnostic medicine. JAMA. 2022 doi: 10.1001/jama.2022.10561. Published online July 8, 2022. [DOI] [PubMed] [Google Scholar]
- 14.Sornapudi S., Hagerty J., Stanley R.J., et al. EpithNet: deep regression for epithelium segmentation in cervical histology images. J Pathol Inform. 2020;11:10. doi: 10.4103/jpi.jpi_53_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui M., Zhang D.Y. Artificial intelligence and computational pathology. Lab Investig. 2021;101:412–422. doi: 10.1038/s41374-020-00514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantanowitz L., Harrington S. Experience reviewing digital Pap tests using a gallery of images. J Pathol Inform. 2021;12:7. doi: 10.4103/jpi.jpi_96_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papanicolaou G.N., Traut H.F. The Commonwealth Fund; New York, USA: 1943. Diagnosis of Uterine Cancer by the Vaginal Smear. [Google Scholar]
- 18.Koss L.G. Cervical (Pap) smear – new directions. Cancer. 1993;71:1406–1412. doi: 10.1002/cncr.2820710405. [DOI] [PubMed] [Google Scholar]
- 19.DeCresce R.P., Lifshitz M.S. PAPNET cytological screening system. Lab Med. 1991;22(24):276–280. [Google Scholar]
- 20.Faraker C.A., Boxer M.E. Rapid review (partial screening) of cervical cytology. Four years experience and quality assurance implications. J Clin Pathol. 1996;49:587–591. doi: 10.1136/jcp.49.7.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mango L.J. Reducing false negatives in clinical practice: the role of neural network technology. Am J Obstet Gynecol. 1996;175(4 Pt2):1114–1119. doi: 10.1016/s0002-9378(96)70014-5. [DOI] [PubMed] [Google Scholar]
- 22.O’Leary T.J., Telledo M., Buckner S.B., Ali I.S., Stevens A., Ollayos C.W. PAPNET-assisted rescreening of cervical smears – cost and accuracy compared with a 100% manual rescreening strategy. JAMA. 1998;279(3):235–237. doi: 10.1001/jama.279.3.235. [DOI] [PubMed] [Google Scholar]
- 23.Farnsworth A., Chambers F.M., Goldschmidt C.S. Evaluation of the PAPNET system in a general pathology service. Med J Aust. 1996;165(8):429–431. [PubMed] [Google Scholar]
- 24.Boon M.E., Kok L.P. Neural network processing can provide means to catch errors that slip through human screening of pap smears. Diagn Cytopathol. 1993;9(4):411–416. doi: 10.1002/dc.2840090408. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee M., Donnelly A., Rose B., et al. Eye tracking in cytotechnology: “visualizing” students becoming experts. JASC. 2020;9:76–83. doi: 10.1016/j.jasc.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Wilbur D.C. False negatives in focused rescreening of Papanicolaou smears: how frequently are ‘abnormal’ cells detected in retrospective review of smears preceding cancer or high-grade intraepithelial neoplasia? Arch Pathol Lab Med. 1997;121(3):273–276. [PubMed] [Google Scholar]
- 27.Tanaka K., Aoki D., Tozawa-Ono A., et al. Comparison of ThinPrep integrated imager-assisted screening versus manual screening of ThinPrep liquid-based cytology specimens. Acta Cytol. 2020;64:486–491. doi: 10.1159/000507910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chantziantoniou N. The Pap test – Celebrating 100 years in the making and beyond. JASC. 2014;3:143–150. doi: 10.1016/j.jasc.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Evered A., Dudding N. Accuracy and perceptions of virtual microscopy compared with glass slide microscopy in cervical cytology. Cytopathology. 2011;22(2):82–87. doi: 10.1111/j.1365-2303.2010.00758.x. [DOI] [PubMed] [Google Scholar]
- 30.Sorbye S.W., Suhrke P., Reva B.W., Berland J., Maurseth R.J., Al-Shibli K. Accuracy of cervical cytology: comparison of diagnoses of 100 Pap smears read by four pathologists at three hospitals in Norway. BMC Clin Pathol. 2017;17:18. doi: 10.1186/s12907-017-0058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girolami I., Pantanowitz L., Marletta S., et al. Diagnostic concordance between whole slide imaging and conventional light microscopy in cytopathology: a systematic review. Cancer Cytopathol. 2020;128:17–28. doi: 10.1002/cncy.22195. [DOI] [PubMed] [Google Scholar]
- 32.Pantanowitz L., Sinard J.H., Henricks W.H., et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137(12):1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans A.J., Brown R.W., Bui M.M., et al. Validating whole slide imaging systems for diagnostic purposes in pathology: guideline update from the College of American Pathologists in collaboration with the American Society for Clinical Pathology and the Association for Pathology Informatics. Arch Pathol Lab Med. 2022;146:440–450. doi: 10.5858/arpa.2020-0723-CP. [DOI] [PubMed] [Google Scholar]
- 34.Antonini P., Santonicco N., Pantanowitz L., et al. Relevance of the College of American Pathologists guideline for validating whole slide imaging for diagnostic purposes to cytopathology. Cytopathology. 2022;00:1–10. doi: 10.1111/cyt.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Society for Clinical Pathology . 2021. ASCP Board of certification BOG adopts “Cytologist” as new certification moniker. Published online November 8. [Google Scholar]
- 36.McAlpine E.D., Michelow P. The cytopathologist’s role in developing and evaluating artificial intelligence in cytopathology practice. Cytopathology. 2020;31(5) doi: 10.1111/cyt.12799. [DOI] [PubMed] [Google Scholar]
- 37.Gelwan E., Zhang M.L., Allison D.B., et al. Variability among observers utilizing the CellSolutions BestCyte Cell Sorter imaging system for the assessment of urinary tract cytology specimens. J Am Soc Cytopathol. 2019;8(1):18–26. doi: 10.1016/j.jasc.2018.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated and analyzed are herein presented completely through Tables and Figures. Further inquiries may be directed to the corresponding author.