Summary
Knowledge about the spatial organization of RNAs in eukaryotic cells is crucial for understanding their functions. Here, we present a detailed MERR APEX-seq protocol to achieve spatiotemporally resolved mapping of the subcellular transcriptome in cultured mammalian cells. This protocol provides detailed description of constructing cell lines stably expressing APEX2, immunofluorescence characterization, MERR APEX labeling, enrichment of biotinylated RNA, library construction and high-throughput sequencing, and MERR APEX-seq data analysis.
For complete details on the use and execution of this protocol, please refer to Li et al. (2022).1
Subject areas: Sequence Analysis, Sequencing, RNAseq, Molecular Biology, Molecular/Chemical Probes, Biotechnology and Bioengineering, Chemistry
Graphical abstract

Highlights
-
•
Protocol for proximity-dependent RNA labeling with APEX2
-
•
Protocol for metabolic labeling of newly synthesized RNA
-
•
Efficient enrichment of biotinylated RNA and reverse transcription
-
•
Detailed description of MERR APEX-seq data analysis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Knowledge about the spatial organization of RNAs in eukaryotic cells is crucial for understanding their functions. Here, we present a detailed MERR APEX-seq protocol to achieve spatiotemporally resolved mapping of the subcellular transcriptome in cultured mammalian cells. This protocol provides detailed description of constructing cell lines stably expressing APEX2, immunofluorescence characterization, MERR APEX labeling, enrichment of biotinylated RNA, library construction and high-throughput sequencing, and MERR APEX-seq data analysis.
Before you begin
The protocol below describes the specific steps in human embryonic kidney 293T (HEK293T) cells stably expressing APEX2 in the mitochondrial matrix. However, we have also applied this protocol in HEK293T cells stably expressing APEX2 in the endoplasmic reticulum and nuclear lamina.
Prepare cell culture
Timing: 2–4 days
-
1.
Prepare the complete cell culture medium consisting of Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, C11995500BT) supplemented with 10% fetal bovine serum (Gibco, 10099141).
-
2.Mammalian cell culture.
-
a.Thaw frozen HEK293T cells (∼ 1 × 107 cells per vial) at 37°C for 1 min.
-
b.Transfer cell suspension into a 15 mL centrifuge tube. Add 9 mL of complete medium to dilute the cell freezing medium.
-
c.Centrifuge the tube containing cell suspension at 300 × g for 3 min. Discard the supernatant containing cell freezing medium.
-
d.Resuspend the cell pellet with the complete medium and transfer cells into a 10-cm cell culture dish. Incubate cells at 37°C with 5% CO2 for 2 days.
-
e.Passage cells at 90% confluency. After washing cells twice with PBS, added 1 mL of 0.25% trypsin and incubate cells at 37°C for 1 min. Thereafter, quench the tryptic digestion with 4 mL complete medium and split cells at a 1:5 ratio.
-
f.Following two rounds of passages, cells can be used for APEX labeling.
-
g.Cells should be discarded after 20 passages.
-
a.
CRITICAL: All steps should be performed in a sterile environment. Cells should be handled in a biological safety cabinet.
Note: Following centrifugation, carefully discard supernatant to remove residual DMSO from the medium as much as possible.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-V5 (1:1000 dilution) | Biodragon | Cat# B1005 |
| Goat anti-mouse-Alexa Fluor 488 (1:1000 dilution) | Thermo Fisher | Cat# A-11029; RRID: AB_2534088 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Gibco | Cat# C11995500BT |
| Trypsin | Life | Cat# 25200056 |
| Fetal bovine serum | Gibco | Cat# 10099141 |
| LIPO-2000 | Invitrogen | Cat# 11668019 |
| Opti-MEM | Gibco | Cat# 31985062 |
| Blasticidin | Selleck | Cat# S7419 |
| 4-Thiouridine (s4U) | Sigma | Cat# T4509 |
| 6-Thioguanosine (s6G) | Sigma | Cat# 858412 |
| Hydrogen peroxide aqueous solution | Xilong | Cat# S6364 |
| Formaldehyde solution | Sigma | Cat# 252549 |
| TRIzol reagent | Invitrogen | Cat# 15596018 |
| Sodium ascorbate | Aladdin | Cat# S105024 |
| 6-Hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) | Sigma | Cat# 238813 |
| Dimethyl sulfoxide (DMSO) | Sigma | Cat# D5879 |
| 1 M Tris-HCl buffer, pH 7.5 | Invitrogen | Cat# 15567027 |
| 5 M NaCl, RNase free | Ambion | Cat# AM9759 |
| Tween-20 | Solarbio | Cat# T8200 |
| Ultrapure water | Beyotime | Cat# ST872 |
| Matrigel matrix | Corning | Cat# 356234 |
| PBS | Solarbio | Cat# P1020 |
| Paraformaldehyde | Sinopharm | Cat# 80096692 |
| Triton X-100 | Sigma | Cat# T8787 |
| BSA | Sangon | Cat# A500023-0100 |
| Streptavidin-Alexa Fluor 637 | Thermo Fisher | Cat# S21374 |
| DAPI | Thermo Fisher | Cat# D1306 |
| DNase I | NEB | Cat# M0303 |
| 0.5 M EDTA, pH 8.0 | Amresco | Cat# E522 |
| 1 N NaOH, BioReagent, suitable for cell culture | Sigma | Cat# S2770 |
| Yeast tRNA | Gibco | Cat# 15401011 |
| Glycogen, RNA grade | Fermentas | Cat# R0551 |
| RNase-free PBS | Life | Cat# AM9624 |
| Formamide | Sigma | Cat# F9037 |
| D-biotin | Invitrogen | Cat# B20656 |
| Chloroform | Tongguang | Cat# 112049 |
| Isopropanol | Tongguang | Cat# 106030 |
| SYBR Green Master Mix | Life | Cat# A25742 |
| VAHTS DNA Clean Beads | Vazyme | Cat# N411-02 |
| Critical commercial assays | ||
| RNA Clean and Concentrator-25 | Zymo Research | Cat# R1018 |
| SuperScript III Reverse Transcriptase | Invitrogen | Cat# 18080044 |
| NEBNext Ultra II RNA Library Prep Kit for Illumina | NEB | Cat# E7770 |
| NEBNext® Poly(A) mRNA Magnetic Isolation Module | NEB | Cat# E7490 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE192739 |
| Experimental models: Cell lines | ||
| Human Embryonic Kidney (HEK293T) cells | Laboratory of Prof. Jing Yang, Peking University | N/A |
| Oligonucleotides | ||
| RT-qPCR primers for MTCO1 gene Fwd: AGCCCACTTCCACTATGTCC Rev: TGGCGTAGGTTTGGTCTAGG |
Wang et al.2 | N/A |
| RT-qPCR primers for MTCYB gene Fwd: TCGGAGGACAACCAGTAAGC Rev: GTTTTCAATTAGGGAGATAGTTGGT |
Wang et al.2 | N/A |
| RT-qPCR primers for MTND2 gene Fwd: ACACTACTCCCCATATCTAACAAC Rev: AGGTAGGAGTAGCGTGGTAA |
Wang et al.2 | N/A |
| RT-qPCR primers for GAPDH gene Fwd: TGTCAAGCTCATTTCCTGGTAT Rev: CTCTCTTCCTCTTGTGCTCTTG |
Wang et al.2 | N/A |
| Recombinant DNA | ||
| MITO-APEX2 | Zhou et al.3 | N/A |
| pVSVG | Laboratory of Prof. Alice Ting, Stanford University | N/A |
| dR8.9 | Laboratory of Prof. Alice Ting, Stanford University | N/A |
| Software and algorithms | ||
| MultiQC package | Ewels et al.4 | RRID: SCR_014982 |
| HISAT2 | Kim et al.5 | RRID: SCR_015530 |
| HTSeq | Anders et al.6 | RRID: SCR_005514 |
| Bioconductor | Gentleman et al.7 | RRID: SCR_006442 |
| DESeq2 | Love et al.8 | RRID: SCR_015687 |
| Other | ||
| NanoDrop™ One Spectrophotometers | Thermo Fisher | Model: ND-ONE-W |
| Fragment Analyzer | Agilent Technologies | Model: M5310AA |
| StepOne Plus | Thermo Fisher | Model: 4376599 |
Materials and equipment
Recipes of solution and buffer mentioned in this protocol are described and listed in the tables below.
s6G solution
Dissolve 12.0 mg s6G (Sigma, 858412) in 200 μL of DMSO (final conc. = 200 mM). The s6G solution can be stored at −20°C for up to one year. The s6G solution should be diluted at 1:1000 (final conc. = 200 μM) for MERR APEX-seq metabolic labeling, or at other concentrations as needed.
s4U solution
Dissolve 10.4 mg s4U (Sigma, T4509) in 200 μL of ddH2O (final conc. = 200 mM). The s4U solution can be stored at −20°C for up to one year. The s4U solution should be diluted at 1:1000 (final conc. = 200 μM) for MERR APEX-seq metabolic labeling, or at other concentrations as needed.
Biotin-phenol (BP) solution
Dissolve 36.3 mg Biotin-phenol in 200 μL of DMSO (final conc. = 500 mM) to obtain BP stock solution. The stock solution can be stored at −80°C for up to two years. The stock solution should be diluted at 1:1000 (final conc. = 0.5 mM) for MERR APEX labeling.
Note: BP solution should be aliquoted into small volumes (e.g., 50 μL) to avoid freeze-thaw cycles.
Quencher buffer
Dissolve 65.0 mg sodium azide in 1 mL of ddH2O (final conc. = 1 M) to obtain a sodium azide stock solution. The stock solution can be stored at 20°C–25°C for one year.
Dissolve 21.8 mg sodium ascorbate in 110 μL of ddH2O (final conc. = 1 M). This solution should be freshly prepared each time.
Dissolve 13.8 mg Trolox in 110 μL of DMSO (final conc. = 500 mM). This solution should be freshly prepared each time.
Mix the above solution in PBS as described in the following table.
| Reagent | Final concentration (mM) | Volume (mL) |
|---|---|---|
| Sodium ascorbate (1 M) | 10 | 0.1 |
| Trolox (500 mM) | 5 | 0.1 |
| Sodium azide (1 M) | 10 | 0.1 |
| PBS | N/A | 9.7 |
| Total | N/A | 10.0 |
Note: The quencher buffer should be temporarily stored at 4°C and used within half an hour after preparation.
CRITICAL: Sodium azide is on the Hazardous Substance List as it is sensitive to heat and shock and it can react with water to form hydrazoic acid. Inhalation of or skin contact with sodium azide should be avoided. Metallic containers and spatula should be avoided when handling sodium azide.
4% (w/v) formaldehyde buffer
Dissolve 0.16 g paraformaldehyde in 4 mL of PBS to obtain formaldehyde buffer. This buffer could be stored at −20°C for half a year.
Note: Paraformaldehyde is poorly soluble in PBS. The buffer could be heated at 60°C to speed up the dissolving procedure.
WARNING: Paraformaldehyde and the formaldehyde buffer are both toxic. Gloves and masks should be worn, and the buffer should be handled in a fume hood.
Alternatives: Formaldehyde solution of 37% (w/v) (Sigma, 252549) can also be used to prepare the 4% (w/v) formaldehyde buffer.
Permeabilization buffer
| Reagent | Final concentration (v/v) | Volume (mL) |
|---|---|---|
| Triton-X 100 | 0.1% | 0.004 |
| PBS | N/A | 4.0 |
| Total | N/A | 4.0 |
Note: Permeabilization buffer can be stored at 20°C–25°C for 6 months.
PBST buffer
| Reagent | Final concentration (v/v) | Volume (mL) |
|---|---|---|
| Tween-20 | 0.1% | 0.004 |
| PBS | N/A | 4.0 |
| Total | N/A | 4.0 |
Note: PBST buffer can be stored at 20°C–25°C for 6 months.
3% BSA (w/v) blocking buffer
Dissolve 300 mg BSA into PBST buffer to a total volume of 10 mL. 3% BSA (w/v) blocking buffer can be stored at −20°C for 6 months.
Bead wash buffer
| Reagent | Final concentration (mM) | Volume (mL) |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 5 | 0.25 |
| NaCl (5 M) | 1,000 | 10.00 |
| EDTA (500 mM) | 0.5 | 0.05 |
| Ultrapure H2O | N/A | 39.70 |
| Total | N/A | 50.00 |
Note: Bead wash buffer can be stored at 20°C–25°C for 6 months.
Solution A
| Reagent | Final concentration (M) | Volume (mL) |
|---|---|---|
| NaOH (1 M) | 0.1 | 0.10 |
| NaCl (5 M) | 0.05 | 0.01 |
| Ultrapure H2O | N/A | 0.89 |
| Total | N/A | 1.00 |
Note: Solution A should be prepared fresh each time. The solution of NaOH (1 M) should be sealed for storage to avoid reacting with atmospheric CO2.
Solution B
| Reagent | Final concentration (M) | Volume (mL) |
|---|---|---|
| NaCl (5 M) | 0.1 | 0.2 |
| Ultrapure H2O | N/A | 9.8 |
| Total | N/A | 10.0 |
Note: Solution B can be stored at 20°C–25°C for 6 months.
Bead blocking buffer
Dissolve 10 mg BSA in 1 mL of ultrapure H2O (final conc. = 10 mg/mL). The BSA solution can be stored at −20°C for one year after filtering through a 0.22 μm filter.
Dissolve 25 mg yeast tRNA in 2.5 mL of ultrapure H2O (final conc. = 10 mg/mL). The yeast tRNA solution can be stored at −20°C for one year.
| Reagent | Final concentration (mg/mL) | Volume (μL) |
|---|---|---|
| BSA (10 mg/mL) | 1 | 100.0 |
| Yeast tRNA (10 mg/mL) | 1 | 100.0 |
| Glycogen (20 mg/mL) | 0.05 | 2.5 |
| Ultrapure H2O | N/A | 797.5 |
| Total | N/A | 1,000.0 |
Note: Bead blocking buffer should be prepared on the day of use.
4 M NaCl wash buffer
| Reagent | Final concentration (M) | Volume (mL) |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 0.1 | 5.0 |
| NaCl (5 M) | 4 | 40.0 |
| EDTA (0.5 M) | 0.01 | 1.0 |
| Tween-20 | 0.2% (v/v) | 0.1 |
| Ultrapure H2O | N/A | 3.9 |
| Total | N/A | 50.0 |
Note: 4 M NaCl wash buffer can be stored at 20°C–25°C for 6 months.
Bead binding buffer
| Reagent | Final concentration (M) | Volume (mL) |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 0.1 | 1.00 |
| NaCl (5 M) | 1 | 2.00 |
| EDTA (0.5 M) | 0.01 | 0.20 |
| Tween-20 | 0.2% (v/v) | 0.02 |
| Ultrapure H2O | N/A | 6.78 |
| Total | N/A | 10.00 |
Note: Bead binding buffer can be stored at 20°C–25°C for 6 months.
2× bead binding buffer
| Reagent | Final concentration (M) | Volume (mL) |
|---|---|---|
| Tris-HCl (1 M, pH 7.5) | 0.2 | 2.00 |
| NaCl (5 M) | 2 | 4.00 |
| EDTA (0.5 M) | 0.02 | 0.40 |
| Tween-20 | 0.4% (v/v) | 0.04 |
| Ultrapure H2O | N/A | 3.56 |
| Total | N/A | 10.00 |
Note: 2×bead binding buffer can be stored at 20°C–25°C for 6 months.
Elution solution
| Reagent | Final concentration (mM) | Volume (mL) |
|---|---|---|
| Formamide | 95% (v/v) | 4.75 |
| D-biotin (50 mM) | 1.5 | 0.15 |
| EDTA (500 mM) | 10 | 0.10 |
| Total | N/A | 5.00 |
Note: Elution solution should be prepared on the day of use and temporarily stored at 20°C–25°C prior to use.
mRNA elution buffer
| Reagent | Volume (μL) |
|---|---|
| 5× First-Strand Buffer in SuperScript III kit | 8 |
| NEBNext Random Primers in NEBNext Ultra II RNA Library Prep Kit for Illumina kit | 2 |
| Nuclease-free water | 10 |
| Total | 20 |
Note: mRNA elution buffer should be prepared on the day of use and temporarily stored at 20°C–25°C prior to use.
Step-by-step method details
Stably expressing APEX2 construct in HEK293T cells
Timing: 2 weeks
The following steps describe the stable expression of APEX2 fusion constructs in HEK293T cells, including lentivirus preparation (step 1) and lentivirus infection (step 2) of APEX2 fusion constructs.
-
1.Lentivirus preparation.
-
a.Seed 4 × 105 HEK293T cells (passages < 10) to each well of a 6-well plate. Prepare 2 wells of HEK293T cells for each lentivirus package.
-
b.At approximately 60% confluency, replace cell culture medium with 2 mL fresh DMEM per well.
-
i.Cells are transfected with endotoxin-free plasmids of APEX2 fusion construct (2 μg), pVSVG (1.4 μg), and dR8.9 (2 μg) for each well, using Lipofectamine 2000.
-
ii.Mix the plasmids in 80 μL opti-MEM in tube #1.
-
iii.Add 10.8 μL Lipofectamine 2000 reagent in 80 μL Opti-MEM in tube #2.
-
iv.After 5 min, slowly add contents of tube #1 to tube #2 and let stand at 20°C–25°C for 15 min. Add the plasmid-Lipofectamine 2000 mixture to the two wells gently. Incubate the plate at 37°C with 5% CO2.
-
i.
-
c.Following incubation at 37°C for 4–6 h, change to complete cell culture medium.
-
d.After 36–48 h, collect the medium containing lentivirus and filter it through a 0.45 μm filter to remove particulates and cell debris.
-
e.The medium containing lentivirus should be aliquoted and flash-frozen in liquid nitrogen. Aliquots could be stored at −80°C for 6 months.
-
a.
-
2.Lentivirus infection.
-
a.Seed 4 × 105 HEK293T cells (passages < 10) to each well of a 6-well plate.
-
b.Grow HEK293T cells in 2 mL of complete cell culture medium in each well and incubate at 37°C with 5% CO2 until cells reach 50% confluency.
-
c.Infect HEK293T cells in each well by replacing 2 mL of complete cell culture medium with 1 mL of medium containing lentivirus.
-
d.Culture cells for 48 h at 37°C with 5% CO2.
-
e.The infected cells should be selected by 5 μg/mL blasticidin in complete cell culture medium for ∼7 days before further analysis.
-
a.
Note: APEX2 expression in HEK293T cells can be verified by V5-tag immunofluorescence in the following experiments.
Immunofluorescence characterization of MERR APEX2 labeling
Timing: 3 days
The following steps describe the protocols of immunofluorescence characterization of MERR APEX2 labeling in HEK293T cells. Cells are seeded on glass coverslips (step 3) for MERR APEX labeling (step 4) and subsequent immunofluorescence imaging analysis (steps 5–10).
-
3.Cell preparation.
-
a.Prepare one 24-well plate for immunofluorescence experiments. Place 12 mm glass coverslips in two wells of one 24-well plate.
-
b.Put 80 μL matrigel matrix (50× diluted with DMEM) on each glass coverslip. Incubate at 37°C for 6–10 h.
-
c.Wash glass coverslips twice with PBS for 2 min each time.
-
d.Seed 1 × 105 HEK293T cells to each well containing pre-coated glass coverslips. Incubate at 37°C with 5% CO2.
-
e.Incubate cells in complete cell culture medium at 37°C with 5% CO2 until cells reach 60% confluency.
-
a.
-
4.MERR APEX labeling.
-
a.Metabolic labeling.
-
i.Prepare medium containing 100 μM s6G (or s4U) by adding 0.5 μL of 200 mM s6G (or s4U) stock solution into 1 mL of the fresh complete cell culture medium.
-
ii.For the MERR APEX sample, replace the old medium with the fresh medium containing 100 μM s6G (or s4U).
-
iii.For the negative control sample, replace the old medium with 1 mL fresh medium without non-canonical nucleosides.
-
i.
-
b.Incubate cells at 37°C with 5% CO2 for 4 h.
-
c.BP probe incubation.
-
i.For the MERR APEX sample, transfer 0.5 mL of the medium from the well containing 100 μM s6G (or s4U) to a 1.5 mL tube.
-
ii.Add 0.5 μL BP solution, then mix thoroughly by the vortex.
-
iii.Replace the remaining medium with the medium containing BP.
-
iv.Incubate cells at 37°C with 5% CO2 for 30 min.
-
i.
-
d.Quencher buffer preparation.
-
i.Prepare quencher buffer consisting of 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox in 3 mL of PBS buffer in a sterile tube and mix well (See materials and equipment section).
-
ii.Place the tube containing the quencher buffer at 4°C before labeling.Note: The quencher buffer should be used within 30 min after preparation.
-
i.
-
e.APEX2 labeling.
-
i.Dilute 10 μL 10 M H2O2 in 990 μL ddH2O (final conc. = 100 mM) and mix by inversion.
-
ii.Add 5 μL 100 mM H=O2 solution to each well containing 500 μL medium.
-
iii.Gently shake the plate for 1 min to ensure that H2O2 will be spread evenly.
-
i.
-
f.Quenching APEX2 reaction.
-
i.Discard the medium of each well completely.
-
ii.Wash cells with 500 μL pre-cold quencher buffer twice with 2 min each time.
-
iii.Remove the quencher buffer completely.
 CRITICAL: Proceed to the next steps without delay to avoid drying out cells.
CRITICAL: Proceed to the next steps without delay to avoid drying out cells.
-
i.
-
a.
-
5.
Fix cells with 500 μL 4% (w/v) formaldehyde buffer (See materials and equipment section) at 20°C–25°C for 15 min. Rinse cells twice with PBS.
-
6.
Permeabilize cells with 500 μL pre-chilled permeabilization buffer (See materials and equipment section) at 4°C for 15 min. Wash 3 times with PBS for 2 min each time.
-
7.
Block cells with 3% BSA (w/v) blocking buffer at 20°C–25°C for 30 min with a gentle shake.
-
8.Primary antibodies incubation.
-
a.Incubate cells with 300 μL primary antibody mixture (1:1000 dilution of mouse anti-V5 antibody in 3% BSA (w/v) blocking buffer) at 20°C–25°C for 1 h.
-
b.Wash 3 times with PBST at 20°C–25°C with shaking for 15 min each time.
-
a.
Note: The primary antibody mixture can be stored at −20°C.
-
9.Fluorophore-conjugated mixture incubation.
-
a.Incubate cells with 300 μL fluorophore-conjugated mixture (1:1000 dilution of Alexa Fluor goat anti-mouse-488 and 1:1000 dilution of Streptavidin-Alexa Fluor 647 in 3% BSA (w/v) blocking buffer) at 20°C–25°C for 1 h.
-
b.Wash 3 times with PBST at 20°C–25°C with shaking for 5 min each time.
-
a.
-
10.DAPI incubation for imaging.
-
a.Incubate cells with 300 μL DAPI mixture (1 μg/mL DAPI in PBS) at 20°C–25°C for 15 min.
-
b.Wash 3 times with PBS at 20°C–25°C with shaking for 5 min each time.
-
c.Samples are prepared for confocal imaging.
-
a.
MERR APEX labeling for RNA enrichment
Timing: 2 days
The following steps describe the protocols of MERR APEX2 labeling in living HEK293T cells for further RNA enrichment (steps 11 and 12).
-
11.Cell sample preparation.
-
a.Cell seeding.
-
i.Seed 4 × 105 HEK293T cells stably expressing APEX2 to one well of a 6-well plate.
-
ii.Prepare cells of one well for MERR APEX labeling and another well of cells for negative control.
-
i.
-
b.Incubate cells in complete cell culture medium at 37°C with 5% CO2 until cells reach 60% confluency.
-
a.
-
12.MERR APEX labeling.
-
a.Metabolic labeling.
-
i.For the MERR APEX labeling sample, replace medium with 100 μM s6G (1 μL s6G solution in 2 mL of the fresh complete cell culture medium) or 100 μM s4U (1 μL s6G solution in 2 mL of the fresh complete cell culture medium) to cells.
-
ii.For the negative control sample, replace the medium with 2 mL of the fresh complete cell culture medium to keep consistent.
-
iii.Incubate at 37°C with 5% CO2 for 4 h.
-
i.
-
b.BP probe incubation.
-
i.For the MERR APEX sample, transfer 1 mL of the cell culture medium containing 100 μM noncanonical nucleosides to a 1.5 mL tube.
-
ii.Add 1 μL BP solution and mix thoroughly with a vortex mixer.
-
iii.Replace the cell culture medium with 1 mL of medium containing BP.
-
iv.Incubate the plate at 37°C for 30 min.
-
i.
-
c.Quencher buffer preparation.
-
i.Prepare quencher buffer consisting of 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox in 5 mL PBS buffer (See materials and equipment section).
-
ii.Place the quencher buffer at 4°C after mixing well.Note: The quencher buffer should be used within 30 min after preparation.
-
i.
-
d.APEX2 labeling.
-
i.Dilute 10 μL 10 M H2O2 in 990 μL ddH2O (final conc. = 100 mM) and mix by inversion.
-
ii.Add 10 μL of 100 mM H2O2 solution to each well containing 1 mL medium. Shake the plate gently for 1 min.
-
i.
-
e.Quenching APEX2 labeling.
-
i.Discard the medium of each well completely.
-
ii.Wash cells with 1 mL pre-chilled quencher buffer twice for 2 min.
-
iii.Discard the quencher buffer. Follow the next step immediately.
 CRITICAL: Proceed to the next steps without delay to avoid drying out cells.
CRITICAL: Proceed to the next steps without delay to avoid drying out cells.
-
i.
-
a.
Enrichment of biotinylated RNA
Timing: 2 days
The following steps describe the protocols of biotinylated RNA enrichment after MERR APEX2 labeling in living HEK293T cells. Total RNA is extracted (steps 13–16) and enriched with streptavidin-coated magnetic beads (steps 17–19).
-
13.Total RNA extraction.
-
a.After MERR APEX labeling for RNA enrichment, cells in each 6-well plate should be lysed with 1 mL TRIzol reagent according to the manufacturer’s instructions.
-
b.Dissolve total RNA extracted from each well with 50 μL Ultrapure H2O. Heat RNA samples at 60°C for 5–10 min to promote dissolution.
-
c.Measure the concentration of total RNA using NanoDrop™ One Spectrophotometers. Approximately 50 μg of total RNA should be recovered for each sample.
-
a.
Note: An A260/A280 ratio of ∼2.0 indicates high purity for RNA.
-
14.
RNA digestion with 2.5 μL of DNase I in 60 μL of the solution. Incubate at 37°C for 30 min to remove residual DNA.
Note: Incubate for no more than 30 min in this step to avoid excessive RNA degradation.
-
15.Purification of INPUT RNA.
-
a.Purify both samples using RNA Clean & Concentrator kit according to the manufacturer’s instructions.
-
b.Elute total RNA with 200 μL Ultrapure H2O to obtain MERR APEX labeling INPUT and negative control INPUT. The INPUT samples will be used for biotin enrichment.
-
c.Measure the concentration of total RNA using NanoDrop™ One Spectrophotometers. INPUT samples can be stored at −80°C for 6 months.
-
a.
-
16.
Check RNA integrity with Fragment Analyzer following the manufacturer’s instructions.
CRITICAL: Only INPUT samples of RQN > 8.0 can be used for downstream enrichment.
-
17.Preparation of C1 beads.
-
a.Buffer removal for C1 beads.
-
i.Pipette 10–15 μL well-mixed Dynabeads MyOne Streptavidin C1 (Invitrogen, 65002, short as C1 beads) for each sample to one 1.5 mL DNA LoBind tube.
-
ii.Place the tube on a magnetic stand (Mich, Magpow-8) for 2 min to collect the beads. Discard the supernatant carefully.
-
i.
-
b.Bead wash buffer wash for C1 beads.
-
i.Resuspend C1 beads with 200 μL bead wash buffer (See materials and equipment section) and collect the beads with a magnetic stand.
-
ii.Discard the supernatant.
-
iii.Repeat the washing steps twice more.
-
i.
-
c.Solution A wash for C1 beads.
-
i.Resuspend C1 beads with 200 μL solution A (See materials and equipment section) with 2 min standing for RNase removal.
-
ii.Collect the beads with a magnetic stand and discard the supernatant.
-
iii.Repeat this step once more.
-
i.
-
d.Solution B wash for C1 beads.
-
i.Wash C1 beads with 200 μL solution B (See materials and equipment section).
-
ii.Collect the beads with a magnetic stand and discard the supernatant.
-
i.
-
e.Resuspend the well-washed C1 beads with 200 μL bead blocking buffer (See materials and equipment section) for every 10 μL of original C1 beads.
-
f.Block C1 beads at 20°C–25°C for 2 h.
-
a.
Note: Up to 50 μL C1 beads can be prepared in one 1.5 mL tube. Let the 1.5 mL tube stand in the magnetic rack for 2 min to achieve good separation of supernatant and beads.
CRITICAL: Do not prepare more than 50 μL of C1 beads in a 1.5 mL tube to avoid excessive non-specific adsorption.
-
18.Binding of biotinylated RNA to C1 beads.
-
a.Collect the beads with a magnetic stand and discard the supernatant.
-
b.Wash C1 beads 3 times with 200 μL 4 M NaCl wash buffer (See materials and equipment section). Collect the beads with a magnetic stand and discard the supernatant.
-
c.Wash C1 beads with 200 μL bead binding buffer (See materials and equipment section). Collect the beads with a magnetic stand and discard the supernatant.
-
d.Resuspend C1 beads with 200 μL 2× bead binding buffer (See materials and equipment section). Divide the C1 beads into two aliquots of 100 μL for C1 beads binding.
-
e.C1 beads binding for INPUT samples.
-
i.Dilute INPUT samples to 250 ng/μL with Ultrapure H2O.
-
ii.Mix C1 beads in 2× bead binding buffer and 100 μL of the two INPUT samples (25 μg for each sample).
-
iii.Bind at 20°C–25°C for 45 min with 1,200 rpm rotation on a rotating mixer.
-
i.
-
a.
-
19.Elution of biotinylated RNA.
-
a.Collect the beads with a magnetic stand and discard the supernatant.
-
b.Wash C1 beads with 200 μL 4 M NaCl wash buffer (See materials and equipment section) 3 times at 20°C–25°C and twice at 50°C for 3 min each to strip away nonspecific adsorption. Collect the beads with a magnetic stand and discard the supernatant.
-
c.Wash C1 beads with 200 μL 1× RNase-free PBS twice at 20°C–25°C Collect the beads with a magnetic stand and discard the supernatant.
-
d.RNA elution.
-
i.Add 50 μL elution solution and mix thoroughly (See materials and equipment section).
-
ii.Heat the slurry at 65°C for 5 min and 90°C for another 5 min while rotating at 1,200 rpm to achieve efficient elution.
-
i.
-
e.Collect the beads with a magnetic stand and transfer the supernatant into a 1.5 mL DNA LoBind tube.
-
f.Biotinylated RNA extraction with TRIzol reagent treatment.
-
i.Purify biotinylated RNA with 1 mL TRIzol reagent for each sample.
-
ii.Add 200 μL chloroform into the TRIzol-RNA mixture.
-
iii.Shake thoroughly to mix well.
-
iv.Centrifuge at 12,000 × g, 4°C for 10 min and the RNA should be distributed in the aqueous phase.
-
i.
-
g.Biotinylated RNA purification from the aqueous phase.
-
i.Pipette the aqueous phase into a clean 1.5 mL DNA LoBind tube.
-
ii.Add 20 μg glycogen to promote precipitation.
-
iii.Add an equal volume of isopropanol to precipitate RNA from the aqueous phase and store at −20°C for 12–24 h.
-
iv.Extract RNA following the manufacturer’s instructions from this step.
-
i.
-
h.Dissolve biotinylated RNA with 20 μL Ultrapure H2O as an enriched sample (ENRICH). ENRICH samples can be stored at −80°C for 6 months.
-
a.
CRITICAL: All experiments for RNA should be performed in an RNase-free environment, for example in the AirClean 600 PCR WorkStation.
Real time-qPCR (RT-qPCR)
Timing: 3 h
The following steps describe the protocols for reverse transcription and RT-qPCR of the INPUT and ENRICH samples (steps 20 and 21).
-
20.
Reverse transcription.
Take 1 μL INPUT (250 ng) and 5 μL ENRICH samples to perform reverse transcription using SuperScript III following the manufacturer’s instructions with random primers in 20 μL of the reaction solution.
-
21.RT-PCR.
-
a.Load 0.75 μL cDNA products to each well of a 96-well qPCR plate for RT-qPCR.
-
b.Perform RT-qPCR with PowerUp SYBR Green Master Mix in 10-μL of reaction solution on an ABI StepOne Plus instrument. For each detected gene, set 3–4 replicates for each targeted gene.
-
a.
Note: Before the experiments, the specificity of RT-qPCR primers should be evaluated by melting curve detection in the ABI StepOne Plus system.
cDNA library preparation
Timing: 2 days
The following steps describe the protocols of cDNA library construction for INPUT and ENRICH samples (steps 22 and 23). Analyze the cDNA length distribution of cDNA libraries before sequencing and purify cDNA with desired size if necessary (step 24).
-
22.Polyadenylation.
-
a.Take ∼300 ng INPUT and 5–10 μL ENRICH samples for mRNA isolation using Poly(A) mRNA Magnetic Isolation Module following the manufacturer’s instructions.
-
b.Elute mRNA from the magnetic beads with 11.5 μL mRNA elution buffer (See materials and equipment section).
-
a.
CRITICAL: All experiments for RNA should be performed in RNase-free apparatus.
-
23.cDNA libraries construction.
-
a.Construct cDNA libraries using NEBNext Ultra II RNA Library Prep Kit for Illumina following the manufacturer’s instructions with some modifications.
-
b.Adjust 2.5 μL diluted adapter to 0.55 μL for ENRICH samples.
-
c.Change 10 μL primer mixtures into 2 μL for each sample. Set 11 PCR cycles for INPUT samples and 15–20 PCR cycles for ENRICH samples.
-
d.Replace DNA purification beads with VAHTS DNA Clean Beads.
-
a.
Note: Over 100 ng of cDNA should be obtained after DNA purification for further quality control and sequencing.
-
24.
Detect DNA length distribution of all cDNA libraries using Fragment Analyzer following the manufacturer’s instructions.
CRITICAL: If there are fragments less than 200 nt, perform another round of 0.9× DNA beads purification and repeat DNA length analysis.
Note: cDNA must be QC detected using Fragment Analyzer before NGS.
Expected outcomes
RT-qPCR results of MERR APEX labeling
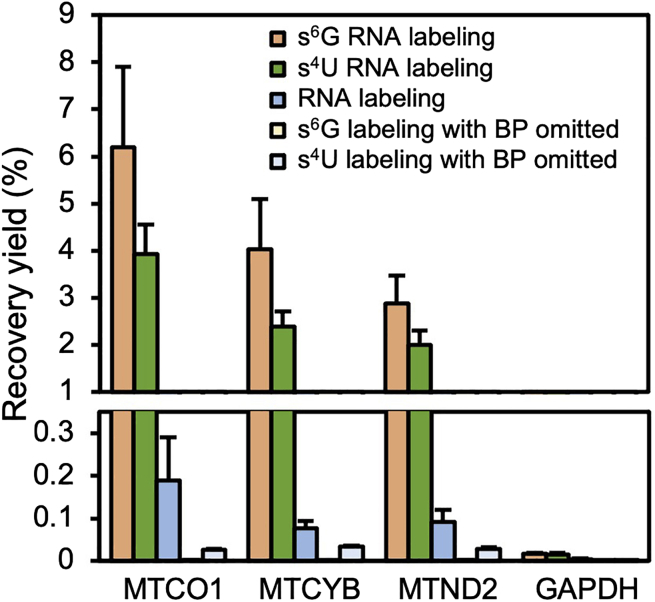
After the RT-qPCR step is completed, Ct values of each sample and each gene are obtained. Following the quantification of RT-qPCR results, enrichment fold changes can be calculated. The expected bar plot of RT-qPCR results has been shown in Figure 1. MTCO1, MTCYB, and MTND2 are encoded by the mitochondrial genome and transcribed within the mitochondrial matrix, whereas GAPDH is encoded by the nuclear genome and transcribed within the cytoplasm. The mitochondrial mRNAs are expected to be highly enriched with recovery yield ranging between 1%–10% with s6G/s4U MERR APEX-seq. Note that the recovery yield of MERR APEX-seq is signigicantly higher than APEX-seq (RNA labeling). The negative controls omitting biotin-phenol (BP) probe should have recovery yields much lower than 0.1%.
Figure 1.
RT-qPCR analysis of the enrichment yields for MT-mRNAs
The recovery rate is calculated from the Ct values of ENRICH and INPUT samples across four technical replicates. Figure from Li et al.1 Error bars represent standard deviations.
Differential gene expression results of MERR APEX-seq
After next-generation sequencing, raw data can be obtained. After data processing as described in the analysis of sequencing results, differential expression levels of each gene can be analyzed. The expected volcano plot of MERR APEX-seq shows in Figure 2. MT-RNAs are expected to be highly enriched, with log2Fold Change exceeding 2.0 and adjusted p value less than 0.05.
Figure 2.
Volcano plot of DESeq2 analysis of transcripts enriched by s6G MERR APEX-seq in the mitochondrial matrix
The cut-off of FDR is chosen as 0.05 (horizontal dotted line), and the cut-off of log2 fold change of ENRICH versus INPUT is set at 3 (vertical dotted line).
Quantification and statistical analysis
Quantification of RT-qPCR results
To characterize the enrichment of fold change and the recovery of representative genes in each sample, collect the cycle threshold (Ct) value of each RT-qPCR well. Ct value should be averaged by all replicates. The enrichment fold change (FC) can be calculated: FC = .
Note: The recovery rate depends on both enrichment fold change and the sample volume.
Analysis of sequencing results
The following steps describe the analysis of the cDNA libraries sequenced on the Illumina HiSeq X Ten platform. Please refer to Li et al.'s experimental model and subject details for suggestions on analyzing libraries.1
-
1.
The processing of NGS data is done in the Linux system. Perform quality control of the RNA-seq reads by FastQC (v0.11.8) and summarize by MultiQC (v1.8).4 Trim adapters for high-quality genome mapping.2 Map data with hisat2 (v2.1.0).5 Count the mapped reads by htseq-count (v0.7.2)6 with the option ‘–stranded no’.
-
2.
The differential expressed transcripts relative to negative control can be identified by DESeq2 with at least 2 replications using R language. The cutoff for enrichment transcripts can be FDR < 0.05 and log2 fold change > 0, as enriched by MERR APEX-seq.
Limitations
A low expression level of APEX2 in HEK293T cells would decrease the efficiency of MERR APEX labeling.
Extending the metabolic time of noncanonical nucleosides can increase the recovery yield of RNA enrichment, but it would have certain cytotoxicity.
Troubleshooting
Problem 1
When constructing HEK293T cells stably expressing APEX2, no cells survive blasticidin treatment.
Potential solution
The lentivirus preparation step may fail, or the lentivirus titer may be too low to infect cells successfully. Measure the titer of the lentivirus, then infect cells with a suitable volume. If that still does not work, repackage the lentivirus.
Problem 2
All cells survive blasticidin treatment.
Potential solution
Ensure the cell line for lentivirus infection is not resistant to blasticidin. Perform kill curve experiments with blasticidin before use.
Problem 3
APEX2 expression level is lower than 50%.
Potential solution
To avoid this problem, we recommend a fluorescent protein (e.g., GFP) can be fused to either the N-terminal or the C-terminal of APEX2. Then following the lentivirus package and infection, positive cells can be sorted by flow cytometry.
Problem 4
Magnetic beads have different degrees of residue on the tube wall of different samples at the C1 beads preparation step.
Potential solution
At the C1 beads preparation step, solution A, solution B, and bead blocking buffer do not have detergent, so some beads can remain on the tube wall. After 4 M NaCl wash buffer addition at the C1 beads binding step, beads remaining on the tube wall should be treated by sucking and blowing many times by tips until all the beads are resuspended.
Problem 5
RNAs are degraded during the experiment.
Potential solution
Ensure all treatment on RNA should be performed in RNase-free apparatus at AirClean 600 PCR WorkStation.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Peng Zou, zoupeng@pku.edu.cn.
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Acknowledgments
We thank G. Wang and X. Zhou for their helpful discussions. This work was supported by the Ministry of Science and Technology (2017YFA0503600, 2018YFA0507600, 2022YFA1304700) and the National Natural Science Foundation of China (32088101). P.Z. is sponsored by Bayer Investigator Award. We thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with Fragment Analyzer 12.
Author contributions
P.Z. conceived the project. R.L. and P.Z. designed experiments. R.L. performed all experiments in this paper. R.L. and P.Z. analyzed data and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ran Li, Email: li.ran@pku.edu.cn.
Peng Zou, Email: zoupeng@pku.edu.cn.
Data and code availability
All data presented are available in the main text. This paper’s accession number for the raw sequencing data is Gene Expression Omnibus (GEO): GSE192739. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Li R., Zou Z., Wang W., Zou P. Metabolic incorporation of electron-rich ribonucleosides enhances apex-seq for profiling spatially restricted nascent transcriptome. Cell Chem. Biol. 2022;29:1218–1231.e8. doi: 10.1016/j.chembiol.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Wang P., Tang W., Li Z., Zou Z., Zhou Y., Li R., Xiong T., Wang J., Zou P. Mapping spatial transcriptome with light-activated proximity-dependent rna labeling. Nat. Chem. Biol. 2019;15:1110–1119. doi: 10.1038/s41589-019-0368-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Wang G., Wang P., Li Z., Yue T., Wang J., Zou P. Expanding apex2 substrates for proximity-dependent labeling of nucleic acids and proteins in living cells. Angew. Chem. Int. Ed. Engl. 2019;58:11763–11767. doi: 10.1002/anie.201905949. [DOI] [PubMed] [Google Scholar]
- 4.Ewels P., Magnusson M., Lundin S., Käller M. Multiqc: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D., Langmead B., Salzberg S.L. Hisat: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anders S., Pyl P.T., Huber W. Htseq--a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented are available in the main text. This paper’s accession number for the raw sequencing data is Gene Expression Omnibus (GEO): GSE192739. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.