Abstract
Background
The limited variation observed among severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) consensus sequences makes it difficult to reconstruct transmission linkages in outbreak settings. Previous studies have recovered variation within individual SARS-CoV-2 infections but have not yet measured the informativeness of within-host variation for transmission inference.
Methods
We performed tiled amplicon sequencing on 307 SARS-CoV-2 samples, including 130 samples from 32 individuals in 14 households and 47 longitudinally sampled individuals, from 4 prospective studies with household membership data, a proxy for transmission linkage.
Results
Consensus sequences from households had limited diversity (mean pairwise distance, 3.06 single-nucleotide polymorphisms [SNPs]; range, 0–40). Most (83.1%, 255 of 307) samples harbored at least 1 intrahost single-nucleotide variant ([iSNV] median, 117; interquartile range [IQR], 17–208), above a minor allele frequency threshold of 0.2%. Pairs in the same household shared significantly more iSNVs (mean, 1.20 iSNVs; 95% confidence interval [CI], 1.02–1.39) than did pairs in different households infected with the same viral clade (mean, 0.31 iSNVs; 95% CI, .28–.34), a signal that decreases with increasingly stringent minor allele frequency thresholds. The number of shared iSNVs was significantly associated with an increased odds of household membership (adjusted odds ratio, 1.35; 95% CI, 1.23–1.49). However, the poor concordance of iSNVs detected across sequencing replicates (24.8% and 35.0% above a 0.2% and 1% threshold) confirms technical concerns that current sequencing and bioinformatic workflows do not consistently recover low-frequency within-host variants.
Conclusions
Shared within-host variation may augment the information in consensus sequences for predicting transmission linkages. Improving sensitivity and specificity of within-host variant identification will improve the informativeness of within-host variation.
Keywords: genomics, SARS-CoV-2, transmission, viral evolution, within-host diversity
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic sequencing has been powerfully used to reconstruct the virus’ evolutionary dynamics at broad temporal and spatial scales [1–3]. However, the virus’ relatively slow substitution rate compared with its short serial interval limits the viral diversity observed in many outbreaks, and viral consensus sequences—which represent the most common allele along the viral genome—are often identical or nearly so [4, 5].
Closely related SARS-CoV-2 consensus sequences have provided important evidence of recent shared transmission. For example, 4 individuals on the same international flight were infected with identical SARS-CoV-2 consensus genomes, evidence that the virus could be transmitted during air travel [6]. The majority, 75%, of SARS-CoV-2 consensus sequences from a fishing boat outbreak were identical to at least 1 other sequence, and the remaining sequences were closely related, suggesting rapid transmission from a single viral introduction [7]. Similarly, genomic surveillance in Boston during 2020 reported that 59 of 83 (71%) genomes sequenced from a skilled nursing facility were identical, implicating transmission within the facility [8].
Although consensus sequences have been harnessed to implicate or exclude the possibility of a shared recent transmission history, the utility of consensus sequences to infer specific transmission events—who infected whom—depends largely on linked epidemiological data, such as contact tracing information, household data, date of symptom onset, and timing between coronavirus disease 2019 testing and SARS-CoV-2 sequencing. Contextualizing outbreak sequence data within a broader sample of population-based sequence diversity is also important. For example, in an outbreak in Heilongjiang Province, China including more than 70 individuals, the majority of sequenced isolates (18 of 21) had identical consensus genomes and the remaining were closely related (within 2 single-nucleotide polymorphisms [SNPs]) [9]. In a large outbreak of the Delta variant in Provincetown, Massachusetts [10], 158 identical consensus genomes were found in the dominant genomic cluster in the outbreak, representing 41% of genomes in the cluster [10]. In these previous studies, consensus sequences alone were insufficient for reconstructing transmission lineages; instead, detailed epidemiological data were linked to genomic data to infer transmission events and to identify a likely single introduction from an asymptomatic traveler for the Heilongjiang Province outbreak [9] and multiple likely introductions for the Provincetown outbreak [10].
The SARS-CoV-2 genomic surveillance has created a rich, expanding source of epidemiological information. In the absence of detailed accompanying epidemiological data, it is not yet known whether routine, population-based sequencing data can be used to reconstruct transmission linkages of who infected whom or identify locations or individuals that may drive transmission.
Genomic studies of human immunodeficiency virus and other viral and bacterial pathogens have begun to harness the pathogen variation within individual infections, or within-host diversity, to reconstruct transmission linkages [11–13]. Previous studies have reported low levels of SARS-CoV-2 diversity within individual hosts and have estimated the size of a narrow transmission bottleneck that limits the viral diversity shared across hosts [7, 14–16]. Some previous studies have begun to harness within-host variation to support transmission linkages [10, 17]. However, previous research has not yet directly quantified the informativeness of within-host SARS-CoV-2 variation nor evaluated the effects of variant identification approaches on transmission inferences [16].
To investigate the potential for within-host SARS-CoV-2 diversity to be harnessed for studies of transmission, we deep sequenced SARS-CoV-2 samples collected from household members, allowing us to directly compare shared within-host variants among epidemiologically linked individuals and those with no known linkage, providing a test case for the transmission information contained within individual infections. We additionally sequenced artificial mixtures of SARS-CoV-2 variants to examine tradeoffs between sensitivity and specificity in within-host variation identification.
METHODS
Collection of Residual Severe Acute Respiratory Syndrome Coronavirus 2 Samples for Deep Sequencing
We assembled a collection of samples from 4 prospective SARS-CoV-2 research studies. The first was a prospective household transmission study, in which index cases with at least 1 reverse-transcription quantitative polymerase chain reaction (RT-qPCR)-confirmed SARS-CoV-2 test were enrolled along with household members. Participants were tested daily for SARS-CoV-2 ribonucleic acid (RNA) via RT-qPCR, using self-collected lower nasal swabs, and households were followed until all members tested negative for 7 consecutive days [18]. The second was a randomized, single-blind, placebo-controlled trial of Peginterferon Lambda-1a (Lambda) for reducing the duration of viral shedding or symptoms [19] in which oropharyngeal swabs were collected for 28 days after enrollment. The third was a phase 2, double-blind, randomized controlled outpatient trial of the antiviral favipiravir for reducing the duration of viral shedding in which participants self-collected daily anterior nasal swabs for 28 days after enrollment [20]. Neither Lambda nor favipiravir was found to shorten the duration of SARS-CoV-2 viral shedding [19, 20]. The fourth was a study of a noninvasive mask sampling method to quantify SARS-CoV-2 shedding in exhaled breath [21].
All study participants provided written consent and all studies were approved by the Institutional Review Board of Stanford University (Numbers 55479, 57686, 56032, and 55619). We identified members of the same household through either (1) participation in the household transmission study, which enrolled all household members simultaneously, or (2) address matching of participants in the 2 clinical trials and mask sampling study. We excluded household pairs with sampling dates greater than 14 days apart, which we predicted to reflect infection from outside the household. After filtering, the maximal time between samples collected from the same household was 7 days, within the range of expected SARS-CoV-2 generation times [22]. However, we were not able to confirm transmission pathways with epidemiological metadata, and we use household membership as a proxy for epidemiological linkage.
Sample Reverse-Transcription Quantitative Polymerase Chain Reaction Testing
We collected nasal swabs in 500 µL Primestore MTM (Longhorn Vaccines & Diagnostics) RNA-stabilizing media. For exhaled breath samples, we extracted RNA from gelatin membrane filters processed in 1 mL PrimeStore MTM media. The RNA was extracted using the MagMAX Viral/Pathogen Ultra Nucleic Acid Isolation Kit (catalog number A42356; Applied Biosystems) and eluted in 50 µL elution buffer [14] (Supplementary Methods).
Library Preparation and Sequencing
We followed the ARTIC v3 Illumina library preparation and sequencing protocols [23] and sequenced amplicons on an Illumina MiSeq platform (Supplementary Methods). Sequence data are available at Sequence Read Archive ([SRA] (BioProject ID: PRJNA842503).
Variant Identification
We used the nf-core/viralrecon v.2.4 bioinformatic pipeline to perform variant calling and generate consensus sequences from raw sequencing reads [24]. In brief, we removed primer sequences with iVar [26], removed reads mapping to the host genome with Kraken2 [27]; mapped reads to the MN908947.3 reference genome with Bowtie 2 [25]; called variants with respect to the reference genome with iVar [26]; generated consensus sequences with bcftools [28], which applies variants identified with an allele frequency greater than or equal to 75%; and assigned Nextclade lineages [29]. We used default pipeline parameters, except for modifying the pipeline to include variants with an alternate allele frequency ≥0.2%.
We included samples with a median coverage of 100X and with >70% of the genome covered by a depth of >10X. We focused our analysis on SNP variants and excluded SNPs occurring at previously reported problematic sites [30].
To test whether commonly applied filters would improve overall accuracy, we applied 5 variant filters: (1) a filter for intrahost single-nucleotide variant (iSNV) quality from iVar [26] (PASS = TRUE), (2) a variant quality score filter (Phred score >40), (3) a depth filter (of both major and minor alleles >5X), (4) a filter of false-positive iSNVs repeated in more than 1 sample in the artificial strain mixture experiment (below), and (5) all filters. We additionally excluded iSNVs occurring in primer binding sites (except for the unfiltered variant set).
To identify shared within-host diversity across samples, we compared each unique pair of samples meeting our quality criteria. We identified shared iSNVs as shared variant positions in which a variant was not fixed and with the same alternate allele call. We additionally determined the geometric mean of the sum of minor allele frequencies at shared iSNVs for each sample in a pair as a measure of shared viral population diversity. To exclude potential shared iSNVs attributable to sequencing batch, we excluded samples sequenced on the same Illumina sequencing lane in pairwise comparisons. We sequenced 12 samples in duplicate, including them in 2 different sequencing batches, to explore the replicability of iSNV detection.
Statistical Analysis
We fit a Poisson regression model for the number of iSNVs identified within a single sample including sequencing batch and participant as random effects. We additionally fit a Poisson regression model for the number of pairwise shared iSNVs as a function of pair type and distance between consensus sequences, including pair as a random effect. Finally, we fit a binomial regression model for predicting household membership as a function of the number of shared pairwise iSNVs and an indicator variable for close consensus sequences (pairwise distance ≤1 SNP), including the earliest samples collected from each pair to exclude multiple pairwise comparisons. We fit all models with the R package lme4 [31], and we included the set of variants after applying all filters, including iSNVs with a minor allele frequency of ≥0.2%. We excluded samples sequenced in the same sequencing batch.
Replicating Analysis in an Independent Deep Sequencing Dataset From Wisconsin
We additionally investigated patterns of shared within-host variation in a previously published dataset from a household transmission study in Wisconsin [14]. Specifically, we reanalyzed variants called by the previous study and filtered to include iSNVs with a minor allele frequency ≥1% and to exclude variants occurring at primer binding sites [14].
Samples in the previous study were sequenced in duplicate. To generate a set of variant calls that were comparable to those from our California dataset, we took the union of iSNVs identified in each replicate sample; for iSNVs detected in both samples, we included the mean minor allele frequency at that position. As in the previous study, we excluded iSNVs called in genomic positions <54 or >29 837 or at position 6669, which was identified as a problematic site. As described above, we excluded positions previously reported as problematic sites [30] from variant calls.
RESULTS
Assembling a Collection of Longitudinally Sampled Individuals and Transmission Pairs
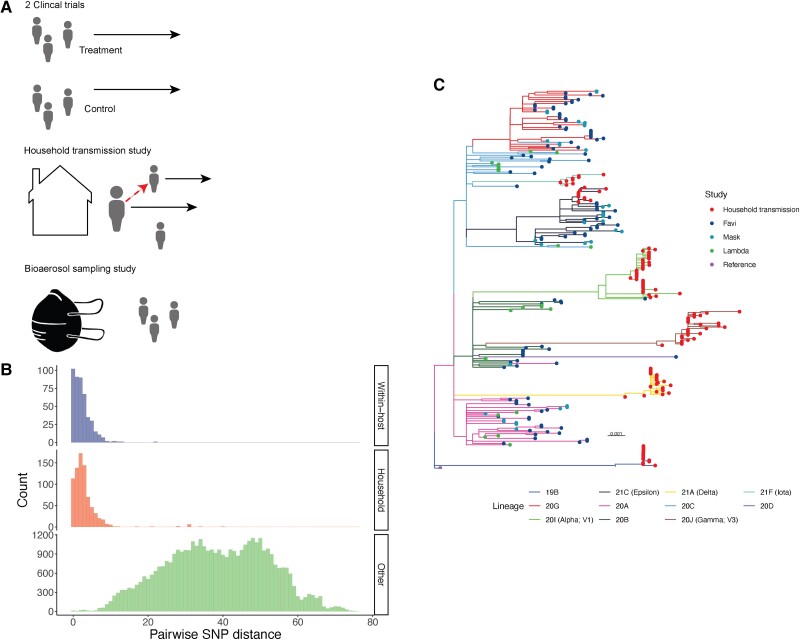
We aggregated residual nasal swabs from 4 studies collected from March 2020 through May 2021 and deep sequenced SARS-CoV-2 genomes using the ARTIC v3 tiled amplicon sequencing protocol (Figure 1A). Three hundred seven SARS-CoV-2 sequences from 286 unique biological samples from 135 participants met our quality and coverage filters, including 130 samples from 32 individuals in 14 households and 57 longitudinally sampled individuals.
Figure 1.
Genetic diversity of sampled severe acute respiratory syndrome coronavirus 2. (A) We identified household transmission pairs and longitudinal samples by address matching from 4 participants enrolled in studies including a household transmission study, clinical trials of Favipiravir and Lambda, and a mask shedding study. (B) Histogram of pairwise single nucleotide polymorphism distances between consensus sequences from samples longitudinally sampled from the same individual, from different individuals within the same household, and between individuals from different households. (C) A maximum likelihood phylogeny inferred from consensus sequences with IQ-Tree with branches colored by clade. Clade assignments are made with Nextclade [46] through the nf-core viralrecon pipeline [24]. Branch lengths are in distances of substitutions per site. FP, false positive; TP, true positive.
Samples had a median coverage depth of 1714 reads with a median of 99.0% of the SARS-CoV-2 genome covered by at least 10 reads. As expected, coverage depth was inversely correlated with RT-qPCR cycle threshold (Pearson's r = −0.15, P = .012), reflecting a positive correlation with SARS-CoV-2 RNA burden.
Samples were distributed across many of the major SARS-CoV-2 lineages circulating at the time of collection (Figure 1B). Overall, consensus sequences had a mean pairwise distance of 37.4 fixed SNPs between consensus sequences (see Methods) (range, 0–76; n = 42 325) (Figure 1B). A total of 42.3% (193 of 456) pairs of consensus sequences sampled longitudinally from the same individual differed by 0–1 SNP (mean pairwise distance, 2.37; range, 0–22). The single individual with consensus sequences that differed by 22 SNPs had been a participant in a SARS-CoV-2 clinical trial and had received the antiviral drug favipiravir [20]; samples were taken 10 days apart. A total of 32.3% (251 of 778) of pairs of consensus sequences sampled from different individuals in the same household differed by 0–1 SNP (mean pairwise distance, 3.06; range, 0–40), consistent with the relatively slow SARS-CoV-2 substitution rate [4]. In contrast, only a small minority, 0.49% (20 of 41 053), of pairs of consensus sequences sampled from different households were within 0–1 SNP (mean pairwise distance, 38.52; range, 0–76).
A Subset of Within-Host Diversity Is Consistently Recovered Over Time
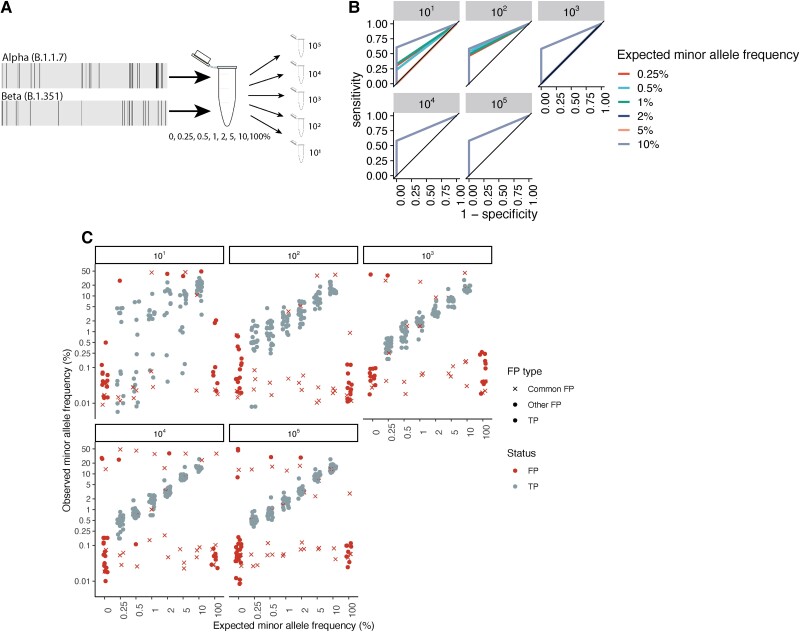
A major challenge in studies of within-host pathogen diversity is in distinguishing true, low-frequency intrahost nucleotide variants (iSNVs) from sequencing or bioinformatic errors [32]. By sequencing artificial strain mixtures of the Alpha and Beta variants, we established that we could reliably recover minority variants to minor allele frequencies as low as 0.25% with 103 viral copies/mL (Figure 2, Supplementary Methods, Supplementary Text), with a minimal cost of false-positive iSNVs and with accurate recovery of input minor allele frequencies (Supplementary Figure 1, Supplementary Figure 2).
Figure 2.
Measuring the accuracy of within-host severe acute respiratory syndrome coronavirus 2 variant identification. (A) Diagram of artificial strain mixture experiment. We conducted a serial dilution experiment, mixing synthetic ribonucleic acid (RNA) controls (Twist Biosciences) of the Alpha (B.1.1.7) and Beta (B.1.351) variants so that the minor variant comprised 0%–10% of the source material and then serially diluted mixtures to a total of 101–105 total RNA copies. We conducted amplicon-based sequencing of artificial mixtures, identified intrahost single-nucleotide variants (iSNVs) with the viralrecon pipeline [24], and determined the sensitivity and specificity of our variant calling pipeline to recovering true variation within our synthetic mixtures. (B) Receiver operator characteristic curve showing 1 specificity versus sensitivity in the recovery of true minority variants, colored by total RNA dilution. Lines corresponding to each dilution include results from 5 artificial strain mixtures, including minority variants present at 0.5%–10% of the total viral pool. (C) Observed minor allele frequency versus expected minor allele frequency for artificial strain mixtures. Points indicate iSNV assignment (false-positive [FP] iSNV; true positive [TP] iSNV). For FP iSNVs, point shape indicates whether FPs were commonly repeated across samples (common FP, FP identified in 10 or more samples; other FP, any other FP iSNV). Points were jittered for visualization. Horizontal facets indicate the synthetic RNA copy number in units of genome copies per microliter.
Most (95.8%, 294 of 307) samples harbored at least 1 iSNV (median, 28; interquartile range [IQR], 8–56) above a minor allele frequency of 0.2% and applying all filters. As expected, the magnitude of recovered within-host diversity decreases to a median of 21 iSNVs (IQR, 5–45) and 7 (IQR, 2–22) with more conservative minor allele frequency thresholds of 0.5% and 1%, respectively (Supplementary Figure 2). The RT-PCR cycle threshold (Ct) value was positively associated with within-host diversity, as measured by iSNV richness above a 0.2% minor allele frequency (adjusted odds ratio [aOR], 1.12; 95% confidence interval [CI], 1.10–1.13), in a general linear model including batch and participant as random effects. Neither days after symptom onset nor study from which the sample was collected were associated with iSNV richness. In study-specific models, participant treatment and sampling method were significantly associated with within-host diversity (Supplementary Text), and, to exclude the influence of sampling method and treatment arm, we focused subsequent analyses on samples from the household transmission study only.
As previously reported [7, 14, 33], iSNVs are not consistently recovered within serial samples. Among individuals with recovered within-host diversity, a mean of 26.5% within-host iSNVs above a minor allele frequency of 0.2% were recovered between samples collected from the same host on 2 subsequent days; this proportion declined with time between samples (r = −0.26, P < .005). The variable recovery of within-host variation is consistent with previous reports that minor allele frequencies are poorly correlated within longitudinally sampled individuals [34], potentially reflecting both sampling bottlenecks, in which only a subset of within-host variation is collected, and sequencing bottlenecks, in which a subset of sampled variation is amplified and represented in sequencing data, as well as a dynamic within-host viral population (Box 1). Low frequency (<2% minor allele frequency) iSNVs were dynamic in longitudinally sampled participants (Supplementary Figure 3).
Box 1: Determinants of within-host SARS-CoV-2 diversity. Potential contributors to recovered SARS-CoV-2 diversity include biological determinants in addition to sampling methods.
-
Biological determinants:
True viral population diversity, including diversity present in the infecting inoculum and diversity generated through both neutral and selective within-host processes, which in turn may be driven by the host environment, host immune status, and immune history (including natural and vaccine-acquired immunity), treatment regimen, and viral genotype [40, 43, 44].
Viral population size within an individual, reflecting individual infection dynamics.
-
Study design:
Viral sampling technique and physical site of sampling may vary across studies.
Sequencing approach including amplicon-based, metagenomic sequencing, or other pathogen enrichment steps and sequencing platform often vary across studies.
Sequencing depth of coverage.
Prospective household transmission studies may enable infections to be identified and sampled earlier compared to samples collected through passive surveillance.
-
Bioinformatic choices:
Read filtering, mapping, and variant identification algorithms vary in sensitivity and specificity.
Some previous studies have required iSNVs to be identified in technical replicates [7, 14].
Minor allele frequency thresholds vary across studies, with previous studies applying filters ranging from 2% to 6% [14, 45].
In 12 samples sequenced in replicate, iSNV recovery was variable (Supplementary Figure 4, Supplementary Figure 5), with concordance, as measured by the Jaccard similarity coefficient, of 24.8% of identifying iSNVs above a minor allele frequency of 0.2% between the 2 sequencing replicates. Concordance increased with more strict minor allele frequency thresholds (Jaccard similarity coefficient: 35.0% and 52.6% with 1% and 2% minor allele frequency thresholds, respectively) (Supplementary Figure 5).
A Signal of Transmission Linkage in Within-host Diversity
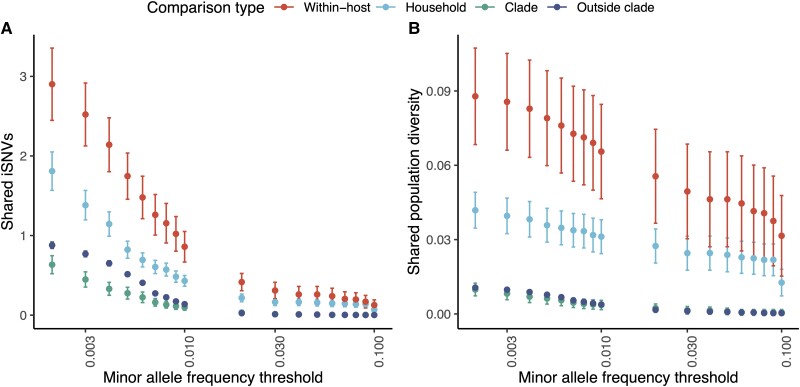
We tested whether within-host SARS-CoV-2 diversity could be used to identify transmission linkages using household membership as a proxy for probable epidemiological linkage (Supplementary Methods). After applying all variant filters, individual infections harbored a mean of 44 iSNVs (95% CI, 38–50) above a 0.2% minor allele frequency threshold. Pairs of individuals in the same household shared significantly more iSNVs (mean, 1.8 iSNVs; 95% CI, 1.6–1.1) than did pairs in different households infected with the same viral clade (mean, 0.63 iSNVs; 95% CI, .52–.74) or pairs in different households infected with a different viral clade (mean, 0.88; 95% CI, .84–.92) (Figure 3A). Overall, pairs of individuals in the same household were more likely to share 1 or more iSNVs (58% of pairs, 194 of 330) than were pairs in different households infected with the same viral clade (28%, 859 of 3055) or pairs in different households infected with a different viral clade (31% of pairs, 11 399 of 37 008) (Supplementary Figures 6–9).
Figure 3.
Shared within-host variants hold a signal of severe acute respiratory syndrome coronavirus 2 transmission. For our genomic collection from a household transmission study in California, (A) pairwise comparisons of the number of shared intrahost single-nucleotide variants (iSNVs), defined as a shared minor allele present at the same genomic position, and (B) shared population diversity, as measured by the geometric mean of the sum of within-host minor allele frequencies for shared iSNVs, identified across different minor allele frequency thresholds, after applying all variant filters (see Methods). Points and error bars indicate mean and 95% confidence intervals and are colored by comparison type. Each pair is assigned to a unique category. Within-host, pairs of samples from the same individual collected on different days; Household, pairs of individuals from the same household; Clade, pairs of individuals outside households infected with the same Nextclade clade; and Outside clade, pairs of individuals outside households infected with different Nextclade clades. Pairwise comparisons include only samples sequenced in different sequencing batches.
Applying an increasingly stringent minor allele frequency threshold up to an allele frequency threshold of 10% dramatically reduced the number of observed iSNVs within individual samples and shared between sample pairs (Figure 3A, Supplementary Figure 8, Supplementary Figure 10), but it did not eliminate the signal of greater levels of shared within-host diversity among household pairs than among epidemiologically unlinked pairs. For example, applying a more commonly used minor allele frequency threshold of 1%, a mean of 6.9 × 10−2 iSNVs (95% CI, 4.0 × 10−2 to 9.8 × 10−2) is shared between household pairs compared to 2.7 × 10−3 iSNVs (95% CI, −2.6 × 10−3 to 8.1 × 10−3) iSNVs shared between individuals in different households infected with the same viral clade.
We hypothesized that minor allele frequencies of shared variants would contribute additional information about transmission beyond the number of shared variant positions; shared variants at higher within-host frequency would be more likely to reflect true shared variation. We therefore measured shared population-level diversity as the geometric mean of the sum of within-host minor allele frequencies for shared iSNVs, which we refer to below as population diversity (see Methods). Across all minor allele frequency thresholds, pairs of individuals in the same household share significantly more population diversity (at a 0.2% minor allele frequency threshold, after filtering; mean, 3.8 × 10−2; 95% CI, 3.2 × 10−2 to 4.5 × 10−2) than do pairs in different households infected with the same viral clade (mean, 7.8 × 10−3; 95% CI, 7.0 × 10−3 to 8.6 × 10−3) and different viral clades (mean, 8.4 × 10−3; 95% CI, 8.1 × 10−3 to 8.6 × 10−3) (Figure 3B, Supplementary Figure 11).
In a generalized linear model for household membership, the number of shared iSNVs was significantly associated with an increased odds of household membership (aOR, 1.35; 95% CI, 1.23–1.49), and genetic distance between consensus sequences was significantly associated with reduced odds of household membership (aOR, 0.25; 95% CI, .20–.29). After excluding multiple comparisons between participants who had been sampled on multiple days, our sample size was small, including 23 unique household pairs, and the number of shared iSNVs did not remain significantly associated with household membership, when included in a model with distance between consensus sequences. Shared diversity, as measured as the standardized sum of shared minor allele frequencies between pairs, was similarly significantly associated with an increased odds of household membership (aOR, 1.32; 95% CI, 1.24–1.42) in a model also including genetic distance between consensus sequences, but it did not remain significant after excluding multiple comparisons between participants.
Replication in a Wisconsin Study
We tested the replicability of our findings in an independent study conducted in Wisconsin where SARS-CoV-2 was deep sequenced from 133 acutely infected individuals, including members of 19 households [14]. At a frequency threshold of 0.5%, we found a similar signal that pairs of individuals in the same household shared significantly more iSNVs (mean, 9.63 iSNVs; 95% CI, 8.08–11.15) than did pairs in different households infected with the same viral clade (mean, 4.12 iSNVs; 95% CI, 4.02–4.21) or pairs in different households infected with a different viral clade (mean, 1.36; 95% CI, 1.31–1.40), in variants in filtered VCF files made publicly available from this earlier study [14] (Supplementary Figure 12a; Methods). Our findings were consistent across minor allele frequency thresholds, although, as in the California data, a signal of household membership was strongest when using minor allele frequency thresholds of ≤1% (Supplementary Figure 12a). We found a similar signal when measuring shared population diversity as the sum of shared minor allele frequencies (Supplementary Figure 12b). However, household pairs did not share significantly more diversity than epidemiologically unrelated pairs when applying a minor allele frequency threshold ≥3% (Supplementary Figure 12).
DISCUSSION
Although most SARS-CoV-2 genomic studies focus on consensus sequences, consensus sequences may not provide the resolution needed to reconstruct transmission linkages and identify potential sources of transmission in outbreak settings, where many cases may be closely genetically related. In this study, we explore the potential for harnessing within-host SARS-CoV-2 genomic variation as a source of information on transmission. Although previous studies of SARS-CoV-2 within-host diversity have often applied relatively conservative minor allele frequency thresholds, we tested whether a more liberal variant identification approach may reveal a signal of transmission, and we explored tradeoffs between sensitivity and specificity in within-host variant identification. We found that epidemiologically linked individuals share more iSNVs than unlinked individuals, yet, because of the current challenges in accurately recovering low frequency iSNVs, shared within-host variation may not consistently augment the information contained in viral consensus sequences.
Transmission is never directly observed and transmission inferences are strengthened by approaches that integrate multiple sources of data, such as consensus genomes, infection time, epidemiological priors on infection time distribution, and, spatial information [35]. Our findings are consistent with studies of other pathogens which have reported that within-host variation provides an additional source of transmission information, although it may not be sufficient as an independent source of transmission information [36]. We envision that SARS-CoV-2 within-host variation could provide epidemiological information in population-wide genomic surveillance and for high-resolution outbreak investigation. For example, the finding of shared within-host variation in population genomic surveillance data could provide additional evidence that a cluster of identical or closely related consensus SARS-CoV-2 genomes indicates an outbreak and could motivate public health investigation. In an outbreak investigation, patterns of shared within-host diversity could similarly be used as evidence supporting highly likely transmission linkages and, therefore, to identify common sources or locations of transmission, again to guide public health control [10]. Future improvements in sequencing and variant identification that improve detection of low-frequency within-host variation may open new possibilities in this regard.
As others have reported [7, 14, 26, 33], excluding sources of noise from within-host pathogen genomic data remains a major challenge, and future studies harnessing within-host variation need to adequately control for uncertainty in within-host variant identification. We sequenced artificial strain mixtures of 2 SARS-CoV-2 variants of concern and found significant tradeoffs between sensitivity and specificity in recovery of true within-host variants as increasingly strict variant filters were applied. Applying strict minor allele frequency thresholds therefore excludes much potential within-host variation that may contain epidemiological information. Our sequencing and bioinformatic pipeline was highly accurate in recovery of within-host variants, and we observed that minor allele frequencies were closely correlated with actual minor allele frequencies at medium to high RNA viral loads. We identified several filters and controls to minimize potential shared false-positive iSNVs (Box 2). Implementing controls such as the filtering of iSNVs that are repeated across samples or that consistently occur in low-coverage genomic regions and controlling for an effect of sequencing batch will be critical to recover a signal of true shared within-host variation.
Box 2: Potential explanations for shared iSNVs. As with the SARS-CoV-2 diversity present within individuals, observed shared within-host diversity could be attributable to a biological signal or the observation process.
-
False positive:
Convergent or homoplastic iSNVs reflecting highly mutable sites along the genome or sites under selection.
Sequencing batch effects due to contamination or adapter switching during a sequencing run.
Artefacts of common sampling approach reflecting contamination due to similar sampling or processing environment.
Bioinformatic errors falling in consistent genomic regions that are difficult to map and/or identify variants.
Some false-positive iSNVs likely persist when applying a liberal variant identification threshold and may contribute to the observed signal of shared within-host variation. Within-host variation shared between epidemiologically unlinked pairs within the same or a different viral clade likely indicate homoplastic shared iSNVs or false-positive iSNVs. We found that within-sample iSNV richness increases with sample Ct, suggesting that some low-frequency iSNVs may be false positive or, alternatively, may reflect an increase in within-host diversity over the duration of infection, as viral population size diminishes. Furthermore, the low concordance of iSNV detection across replicate samples underscores the challenges in low-frequency variant identification and low accuracy in low-frequency variant recovery in our samples. Concordance increased with increasingly strict minor allele frequency thresholds, possibly indicating either poor specificity (ie, false-positive iSNVs detected in only a single replicate) or poor sensitivity (ie, the false absence of iSNVs in one of the replicates) in iSNV detection. We also observed low sensitivity in the serial dilution experiment, which had a maximal sensitivity of less than 60% for detecting variants present at a frequency of 0.2%, in addition to low specificity, and the presence of false-positive iSNVs that were only detected in a single sample. However, our finding that the distribution of shared iSNVs among likely epidemiological pairs exceeds that of epidemiological unlinked individuals, a signal that persists after excluding samples sequenced in the same batch, suggests that the majority of these low-frequency iSNVs may reflect variation shared through transmission.
As others have highlighted, our findings underscore the need to control for other potential explanations for shared iSNVs while still prioritizing sensitivity (Box 2). We found that the signal of shared within-host variation across transmission pairs is strongest when including iSNVs at low minor allele frequency thresholds. Our findings suggest that for transmission inference, privileging sensitivity in variant identification may greatly improve sensitivity for recovering within-host variation, at a small cost of false-positive variant calls. Previous studies have reported relatively small SARS-CoV-2 bottlenecks when applying relatively strict thresholds; however, estimated bottleneck size is dependent on the variant calling approach and, specifically, the minor allele frequency threshold [14]. Previous work has also reported that SARS-CoV-2 transmission bottlenecks [14], such as SARS-CoV-2 viral loads [37], are highly dispersed, with the majority of transmission events including a small number of founding virions and a minority involving much larger founding populations [14]. The pattern we describe in which the majority of epidemiologically linked pairs do not share within-host variation and a minority share low-frequency variants is consistent with the previous evidence of overdispersion of the transmission bottleneck size; the transmission pairs that share low-frequency iSNVs may represent cases of a wide transmission bottleneck.
The optimal variant identification approach likely differs across applications—for example, measurements of transmission bottleneck are highly sensitive to allele frequency threshold [14, 38] and may prioritize specificity, whereas studies of transmission might prioritize sensitivity to identify potential transmission linkages. Previous studies of within-host SARS-CoV-2 variation have frequently applied minor allele frequency thresholds of 1%–3% [7, 14, 38], and/or they have excluded minority variants that are not identified in sequencing replicates [14], to maximize specificity of minor allele identification. Variant identification approach also depends on sequencing capacity. Although mean coverage of our samples was high (2508X), most population-based sequencing studies do not use deep sequence samples and thus need to use more conservative frequency and/or depth filters.
Our measures of within-host SARS-CoV-2 diversity are consistent with those measured in previous studies when applying similar thresholds: a mean of 3 (range, 0–5) iSNVs at a minor allele frequency ≥2% were identified in an outbreak on a fishing boat [7], and a mean of 3 iSNVs was reported in individuals sampled in a household study in Wisconsin above a 3% minor allele frequency threshold and consistent across sequencing replicates [14]. Furthermore, our finding that iSNVs can be shared between epidemiologically linked individuals is consistent with previous reports (1) that household membership is the most significant predictor of shared within-host variation [14], (2) that have identified the transmission of minor alleles [10], and (3) that have identified fixation of alleles over transmission chains [17]. Overall, SARS-CoV-2 within-host diversity is lower than that identified in other viral pathogens, and, as previously reported, we find that within-host viral diversity is frequently lost during transmission [14].
Our study has several limitations. First, we focused on a convenience sample of residual samples with accompanying household information collected in California from March 2020 through May 2021. Replicating these findings in other settings and with more recently emerged SARS-CoV-2 lineages is critical to understand the generalizability of our findings. Second, we used household membership as a proxy for epidemiological linkage and were not able to confirm links with contact tracing. It is possible that household members may have been misclassified as epidemiologically linked if they were infected outside of the home or, in a household with 3 or more people, 2 individuals may not have been directly linked through transmission. This misclassification would result in an underestimation of the effect of household membership on shared within-host variation. Third, our study focused on the potential epidemiological value of within-host viral variation. Our focus was on transmission linkage rather than in viral evolutionary dynamics or transmission bottlenecks, which might have different optimal variant identification approaches. Forth, many groups have hypothesized that evolution within immune-compromised or immune-suppressed populations may be an important driver of the emergence of new variants of concern or interest [39–43]. Our sample collection did not enable us to test these hypotheses. Fifth, the epidemiological utility of within-host variation depends on SARS-CoV-2 sampling and sequencing. Routine sequencing may not always generate sufficient depth to accurately recover within-host variation. Finally, the accuracy of minority variant identification we measured in the serial dilution experiment may represent an upper bound in accuracy, because artificial strain mixtures were constructed from RNA synthetic controls and did not include human or microbial nucleic acids and other sequencing contaminants. To maximize the number of household pairs for comparison, we included all possible individuals with a household member also in our collection and did not exclude samples based on viral load. Mean Ct for samples in the household transmission study, with available information on viral load, was 22.8, corresponding to a viral load far exceeding 103 input copies.
CONCLUSIONS
In conclusion, we find that SARS-CoV-2 variation within individual hosts may be shared across transmission pairs and could be used to confirm transmission linkage on a backdrop of limited diversity among consensus sequences. However, our results confirm technical concerns that current sequencing and bioinformatic workflows do not consistently recover low-frequency within-host variants. More broadly, pathogen diversity within individual infections holds largely untapped information that may enhance the resolution of transmission inferences.
Supplementary Material
Acknowledgments
Financial support. KSW received support from a Thrasher Research Foundation Early Career Award. We acknowledgment financial support from School of Medicine, Stanford University, Innovative Medicines Accelerator and operational support from Stanford ChEM-H.
Presented in part: California Department of Public Health COVIDNet Expert Panel, COVIDNet, California Department of Public Health, Richmond, CA, September 21, 2021.
Potential conflicts of interest. All authors: No reported conflicts of interest.
Contributor Information
Katharine S Walter, Division of Epidemiology, University of Utah, Salt Lake City, Utah, USA.
Eugene Kim, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Renu Verma, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Jonathan Altamirano, Department of Epidemiology and Population Health, Stanford University School of Medicine, Stanford, California, USA.
Sean Leary, Department of Pediatrics, Stanford University School of Medicine, Stanford, California, USA.
Yuan J Carrington, Department of Pediatrics, Stanford University School of Medicine, Stanford, California, USA.
Prasanna Jagannathan, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA; Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, California, USA.
Upinder Singh, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA; Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, California, USA.
Marisa Holubar, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Aruna Subramanian, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Chaitan Khosla, Stanford ChEM-H, Stanford University, Stanford, California, USA; Department of Chemistry and Chemical Engineering, Stanford University, Stanford, California, USA.
Yvonne Maldonado, Department of Epidemiology and Population Health, Stanford University School of Medicine, Stanford, California, USA; Department of Pediatrics, Stanford University School of Medicine, Stanford, California, USA.
Jason R Andrews, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Turakhia Y, Thornlow B, Hinrichs AS, et al. Ultrafast sample placement on existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat Genet 2021; 53:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lemey P, Hong SL, Hill V, et al. Accommodating individual travel history and unsampled diversity in Bayesian phylogeographic inference of SARS-CoV-2. Nat Commun 2020; 11:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182:812–27.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duchene S, Featherstone L, Haritopoulou-Sinanidou M, Rambaut A, Lemey P, Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol 2020; 6:veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borges V, Isidro J, Macedo F, et al. Nosocomial outbreak of SARS-CoV-2 in a “non-COVID-19” hospital ward: virus genome sequencing as a key tool to understand cryptic transmission. Viruses 2021; 13:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi EM, Chu DKW, Cheng PKC, et al. In-flight transmission of SARS-CoV-2. Emerg Infect Dis 2020; 26:2713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hannon WW, Roychoudhury P, Xie H, et al. Narrow transmission bottlenecks and limited within-host viral diversity during a SARS-CoV-2 outbreak on a fishing boat. Virus Evol. 2022; 8(2):veac052. Available from: 10.1093/ve/veac052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lemieux JE, Siddle KJ, Shaw BM, et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science 2021; 371:eabe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J, Huang J, Xiang D. Large SARS-CoV-2 outbreak caused by asymptomatic traveler, China. Emerg Infect Dis 2020; 26:2260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddle KJ, Krasilnikova LA, Moreno GK, et al. Transmission from vaccinated individuals in a large SARS-CoV-2 Delta variant outbreak. Cell 2022; 185:485–92.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tonkin-Hill G, Ling C, Chaguza C, et al. Pneumococcal within-host diversity during colonisation, transmission and treatment. Nat Microbiol 2022; 7:1791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wymant C, Hall M, Ratmann O, et al. PHYLOSCANNER: inferring transmission from within- and between-host pathogen genetic diversity. Mol Biol Evol 2018; 35:719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leitner T. Phylogenetics in HIV transmission: taking within-host diversity into account. Curr Opin HIV AIDS 2019; 14:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Braun KM, Moreno GK, Wagner C, et al. Acute SARS-CoV-2 infections harbor limited within-host diversity and transmit via tight transmission bottlenecks. PLoS Pathog 2021; 17:e1009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin MA, Koelle K. Comment on “Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2”. Sci Transl Med 2021; 13:1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. San JE, Ngcapu S, Kanzi AM, et al. Transmission dynamics of SARS-CoV-2 within-host diversity in two major hospital outbreaks in South Africa. Virus Evol 2021; 7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li B, Deng A, Li K, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat Commun 2022; 13:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altamirano J, Govindarajan P, Blomkalns A, et al. 401. Natural history of shedding and household transmission of severe acute respiratory syndrome coronavirus 2 using intensive high-resolution sampling. Open Forum Infect Dis 2021; 8:S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jagannathan P, Andrews JR, Bonilla H, et al. Peginterferon lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat Commun 2021; 12:1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holubar M, Subramanian A, Purington N, et al. Favipiravir for treatment of outpatients with asymptomatic or uncomplicated COVID-19: a double-blind randomized, placebo-controlled, phase 2 trial. Clin Infect Dis 2022; 75:1883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verma R, Kim E, Degner N, Walter KS, Singh U, Andrews JR. Variation in SARS-CoV-2 bioaerosol production in exhaled breath. Open Forum Infect Dis 2021; 9:ofab600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hart WS, Miller E, Andrews NJ, et al. Generation time of the alpha and delta SARS-CoV-2 variants: an epidemiological analysis. Lancet Infect Dis 2022; 22:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benjamin F, Diana R, Betteridge E, et al. COVID-19 ARTIC v3 Illumina library construction and sequencing protocol V.5. Available at: 10.17504/protocols.io.bibtkann. Accessed May 10, 2020. [DOI]
- 24. Ewels PA, Peltzer A, Fillinger S, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020; 38:276–8. [DOI] [PubMed] [Google Scholar]
- 25. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grubaugh ND, Gangavarapu K, Quick J, et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 2019; 20:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 2014; 15:R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011; 27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Maio N, Walker C, Borges R, Weilguny L, Slodkowicz G, Goldman N. Masking strategies for SARS-CoV-2 alignments. 2020. Available from: https://virological.org/t/masking-strategies-for-sars-cov-2-alignments/480. Accessed July 20, 2020.
- 31. Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67:1–48. [Google Scholar]
- 32. Mccrone JT, Lauring S. Measurements of intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J Virol 2016; 90:6884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valesano AL, Rumfelt KE, Dimcheff DE, et al. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog 2021; 17:e1009499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lythgoe KA, Hall M, Ferretti L, et al. SARS-CoV-2 within-host diversity and transmission. Science 2021; 372:eabg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Didelot X, Fraser C, Gardy J, Colijn C, Malik H. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol Biol Evol 2017; 34:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Worby CJ, Lipsitch M, Hanage WP. Shared genomic variants: identification of transmission routes using pathogen deep-sequence data. Am J Epidemiol 2017; 186:1209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science 2021; 373:eabi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin MA, Koelle K. Comment on “Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2.”. Sci Transl Med 2021; 13:1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Available at: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563. Accessed December 20, 2020.
- 40. Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021; 592:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weigang S, Fuchs J, Zimmer G, et al. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat Commun 2021; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bessière P, Volmer R. From one to many: the within-host rise of viral variants. PLoS Pathog 2021; 17:e1009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021; 371:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. De Maio N, Worby CJ, Wilson DJ, Stoesser N. Bayesian reconstruction of transmission within outbreaks using genomic variants. PLoS Comput Biol 2018; 14:e1006117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.